Abstract
Background
Analysis of long noncoding RNA (lncRNA) localisation at both the tissue and subcellular levels can provide important insights into the cell types that are important for their function.
Methods
By applying new fluorescent in situ hybridisation technique called hybridisation chain reaction (HCR), we achieved a high-throughput lncRNA visualisation and evaluation of clinical samples.
Results
Assessing 1728 pairs of 16 lncRNAs and clear-cell renal-cell carcinoma (ccRCC) specimens, three lncRNAs (TUG1, HOTAIR and CDKN2B-AS1) were associated with ccRCC prognosis. Furthermore, we derived a new lncRNA risk group of ccRCC prognosis by combining the expression levels of these three lncRNAs. Examining genomic alterations underlying this classification revealed prominent features of tumours that could serve as potential biomarkers for targeting lncRNAs. We then derived combination of HCR with expansion microscopy and visualised nanoscale-resolution HCR signals in cell nuclei, uncovering intracellular colocalization of three lncRNA (TUG1, HOTAIR and CDKN2B-AS1) signals such as those located intra- or out of the nucleus or nucleolus in cancer cells.
Conclusion
LncRNAs are expected to be desirable noncoding targets for cancer diagnosis or treatments. HCR involves plural probes consisting of small DNA oligonucleotides, clinically enabling us to detect cancerous lncRNA signals simply and rapidly at a lower cost.
Subject terms: Cancer imaging, Renal cell carcinoma, Tumour biomarkers
Introduction
Over the past decade, the development of technology has revealed that over 80% of our genome is actively transcribed to RNAs, and only 2% of these RNAs are translated to proteins [1]. The remaining RNAs do not encode a protein, which is called a noncoding RNA. Noncoding RNAs with over 200 nucleotides are especially called long noncoding RNAs (lncRNAs). Many of them are uniquely expressed in differentiated tissues or specific cancer types [2]. Various interactions with chromatin, protein or RNA are reported as functions of lncRNAs [3]. Intracellular signalling networks caused or mediated by lncRNAs induce proliferation, growth suppression, motility, immortality, angiogenesis and viability in cancer cells [4]. Although investigations for unravelling the function of lncRNAs are rapidly advancing, the function and relation with cancer of most lncRNAs are still unknown.
Analysis of lncRNA localisation at both the tissue and subcellular levels by techniques such as fluorescent in situ hybridisation can provide important insights into the cell types that are important for their function [5]. Recently, further techniques for detecting lncRNAs called hybridisation chain reaction (HCR) were developed [6]. HCR involves only small DNA oligonucleotides [7], which self-assemble at the target lncRNA. Small nucleotides can penetrate deeper, and the self-assembled chain can amplify the signal ~200-fold [8]. In addition, HCR requires only two steps of tissue hybridisation and amplification, which enables us to detect HCR in a high-throughput manner at a lower cost [9].
In human clear-cell renal-cell carcinoma (ccRCC) tissues, it has been reported that a number of tumorigenic lncRNAs are upregulated and tumour suppressive lncRNAs are repressed [10]; rather, aberrant lncRNA expression is a marker for poor patient prognosis. Using a simple/rapid imaging technique of HCR, we herein investigated the expression of a variety of in situ lncRNAs in ccRCC. Uncovering the intracellular colocalization of lncRNAs facilitates functional analysis [11], and the understanding of the target lncRNAs will be deepened. Expansion microscopy is a recently developed technique that enables nanoscale-resolution imaging of preserved cells and tissues on conventional diffraction-limited microscopes via isotropic physical expansion of the specimens before imaging [12]. In this study, we revealed that the combination of HCR and expansion microscopy (HCR expansion fluorescence in situ hybridisation; HCR-ExFISH) [12] could visualise nanoscale-resolution HCR signals, achieving an unprecedented quantitative representation of lncRNA expression in clinical specimens for the first time.
Materials and methods
Human tumour samples
All human sample studies were reviewed and approved by the Institutional Review Board, Keio University Hospital. Surgical specimens from ccRCC patients who had been treated surgically with radical/partial/cytoreductive nephrectomy at our institution between 2000 and 2014 were used. Our cohort included relatively old cases before the targeted therapy/immune-oncology era; therefore, only three patients received immune checkpoint inhibitors. Radiation therapy was performed when treating brain metastasis or palliative lesions. All tumours were histologically confirmed to be ccRCC, an alcohol-based PAXgene (Qiagen) was used for fixation prior to paraffin embedding, and 4-mm cores of paraffin-embedded tumours were punched out from optimal cancerous areas, creating tissue microarray sections. The UICC TNM system was used for tumour staging, and nuclear grading was carried out according to the WHO/International Society of Urological Pathology grading system. All procedures were performed in compliance with the 1964 Helsinki Declaration and present ethical standards and the hospital’s ethical guidelines.
In situ hybridisation by a hybridisation chain reaction
In brief, the lncRNA signals from ccRCC samples were examined using the HCR RNA-FISH approach (Molecular Instruments) according to the manufacturer’s protocol. After deparaffinization, 10-μm-thick sections were further fixed with 4% paraformaldehyde for 20 min and treated with proteinase K. Sections were then hybridised with 2 nM probe solution at 37 °C overnight and washed at 37 °C using a decreasing gradient of probe wash buffers. Next, sections were amplified with 3 μM hairpin solution at room temperature overnight. Fluorescently labelled hairpins were pretreated by heating at 95 °C for 90 s and cooling to room temperature in a dark drawer for 30 min prior to use. After removing excess hairpins, coverslips were mounted on glass slides in Vectashield® mounting medium containing DAPI and visualised under a fluorescence microscope (IX8, Olympus, Tokyo, Japan) or a confocal microscope (FV3000, Olympus, Tokyo, Japan). All of the probes were designed and purchased from Molecular Instruments (Supplementary Table 1). In this study, all cut-off values for high or low lncRNA expression were based on the detection of three or more visible RNA signals identified under the ×10 magnification of fluorescence microscopy.
Expansion-assisted in situ hybridisation
Herein, we have arranged the protocol presented by Asano et al., which they called ExFISH [12]. In the first step, we anchored the proteins and RNAs to the hydrogel matrix using small molecules called acryloyl-X SE, which binds to primary amine groups on proteins, and label-IT amine solutions, which bind to guanine in RNA and DNA. In the second step, HCR protocols are applied to the tissues in the swellable hydrogel polymer. Finally, the sample tissues were expanded and imaged in a low-salt buffer. As a procedure, we sectioned fresh-frozen ccRCC tissues to 10-μm thickness by using Cryostat (CM3050S, Leica, Tokyo. Japan). The tissue slices were mounted on slides and fixed with 4% paraformaldehyde for 5 min. Then, the samples were incubated with Label-IT amine (Mirus Bio, cat. no. MIR3900) solution, which enables RNA to be anchored to the polymer, and kept overnight at 37 °C. Next, the samples were incubated with Acryloyl-X SE (AcX; Invitrogen, cat. no. A20770)/DMSO solution, which enables proteins to be anchored to the hydrogel, and kept overnight at room temperature without shaking. After washing the samples with PBS, gelation with StockX, TEMED, 4HT and APS was demonstrated. The samples were taken into hydrogels with a thickness of 300 μm and removed from slides. The hydrogels were incubated with digestion buffer and kept overnight at room temperature in the dark. After digestion, we carried out the hybridisation and amplification steps described above. Then, the tissues were incubated with D523 DAPI solution and N511 Nucleolus Bright Green (DOJINDO, Kumamoto, Japan) to stain the nucleus and nucleolus. Finally, tissues were immersed in 0.05x SSCT and expanded. The visualisation was performed under confocal microscopy (FV3000, Olympus, Tokyo, Japan) and the dotted signals were counted manually.
DNA extraction and sequencing
Genomic DNA was extracted from matched fresh-frozen tissue samples to tissue microarrays with a DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s protocol. The DNA integrity number was 4.0, which was calculated using the Agilent 2000 TapeStation (Agilent Technologies, Waldbronn, Germany). A genomic DNA library was constructed using the GeneRead DNAseq Targeted Panel V2 (Human Comprehensive Cancer Panel), which covers more than 95% of the total exon region in 160 cancer-related genes [13, 14]; thereafter, it was amplified and sequenced using a GeneRead DNA I Amp Kit (Qiagen) and MiSeq (Illumina). The FastQ files were analysed using an original bioinformatics pipeline called GenomeJack (Mitsubishi Space Software, Tokyo, Japan) [14].
Statistical analysis
Human samples were randomly collected, and no statistical method was used to predetermine sample group sizes. All data are presented as medians and interquartile ranges (IQRs) for continuous variables and frequencies with percentages for categorical variables. Variables between groups were compared using the two-tailed Student’s t test and Mann–Whitney U test as appropriate. Categorical variables were compared using Fisher’s exact test. Web-based dataset analysis using Kaplan–Meier Plotter, a web-based genomic/clinical database (http://kmplot.com/analysis/index.php?p=background) [15], was used for prognostic assessment of lncRNAs currently evaluated (access data: April 2021). Survival curves were estimated using the Kaplan–Meier method and compared using the log-rank test. The Kaplan–Meier Plotter analysis provides statistical differences based on a cut-off value with the best performing threshold selected automatically. Univariate and multivariate Cox regression models with stepwise selection were used to evaluate variables associated with recurrence-free and overall survival. Differences among groups were regarded as significant when P < 0.05. All analyses were performed using the SPSS Version 27.0 statistical software package (IBM, Armonk, NY) and JMP version 16.0 (SAS Institute Inc., Cary, NC).
Results
Flow diagram of lncRNA selection on ccRCC outcome
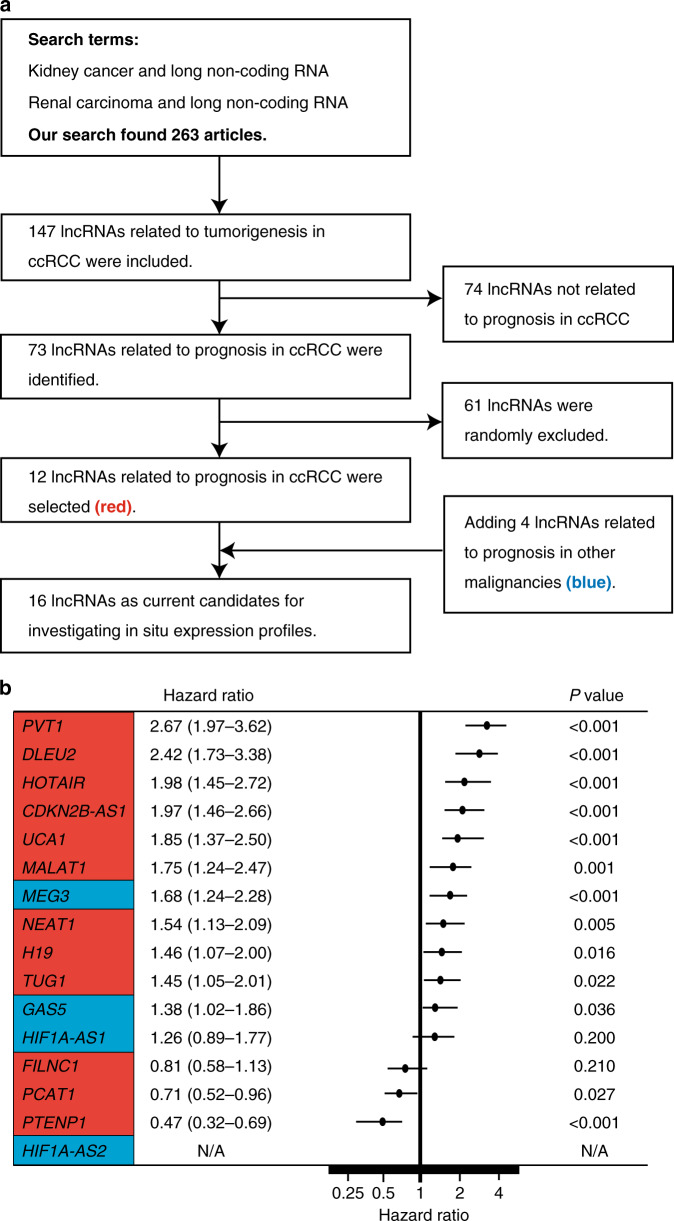
For extraction of potential lncRNAs in the study, we firstly searched articles on Pubmed in April 2019. We used the following search terms “long non-coding RNA and kidney cancer” or “long non-coding RNA renal carcinoma”; thereafter we selected lncRNAs considered to be candidates for predictors of ccRCC prognosis (Fig. 1a). Screening publications on the PubMed website, we initially found 263 articles and identified 147 lncRNAs involved in ccRCC tumorigenesis. Among them, we found 73 lncRNAs that were reported to affect the survival of ccRCC patients in the literature, and the TCGA ccRCC dataset revealed a significant relationship with survival for 12 lncRNAs (NEAT1, HOTAIR, TUG1, DLEU2, MALAT1, FILNC1, PVT1, H19, UCA1, CDKN2B-AS1, PCAT1 and PTENP1; see http://kmplot.com/analysis/index.php?p=background, Supplementary Table 2). Initiatively adding four lncRNAs (GAS5, MEG3, HIF1A-AS1 and HIF1A-AS2) that were major targets in other malignancies (Supplementary Table 2), we finally selected a total of 16 lncRNAs to clinically investigate in situ expression profiles by in-house ccRCC samples. Together, Fig. 1b shows the results of the log-rank test for ccRCC survival in the TCGA database. FILNC1 and HIF1A-AS1 had no relation with the survival of ccRCC, for HIF1A-AS2 no information is available in the TCGA database. The remaining 13 lncRNAs showed a significant relation between their expression and survival in ccRCC.
Fig. 1. The sixteen lncRNAs in this study.
a Study workflow for determining lncRNAs associated with the outcome of prognosis in ccRCC. b Forest plot of the hazard ratio for overall mortality of indicated lncRNAs via ccRCC patient data obtained from Kaplan–Meier plotter analysis, compared using the log-rank analysis.
Visualisation of lncRNAs by hybridisation chain reaction
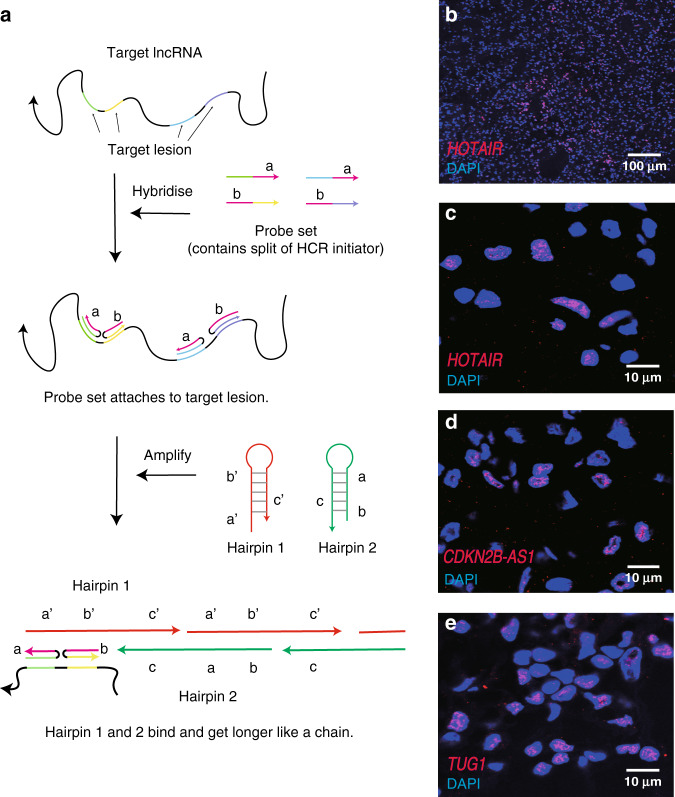
In this study, we applied the HCR system to image lncRNAs in clinical ccRCC specimens. HCR is a new technique that uses plural probes consisting of ~25–50 nucleotides [7]. In brief, short probes enable easier practice with low-cost accessibility. After binding to target lncRNAs, fluorophore-tagged oligonucleotides are trapped in a hairpin conformation and get longer like a chain (Fig. 2a), so that the amount of single mRNA signals can be detected in a high-throughput manner. All 16 lncRNAs were imaged in 108 individual ccRCCs and thereafter were robustly analysed. As shown in Fig. 2b–e, lncRNAs fluorescent by HCR were visualised as puncta.
Fig. 2. Assessment of hybridisation chain reaction.
a Simplified flowchart of two HCR steps. First, probe sets containing a split of HCR initiator detect the target lncRNA (hybridisation step). Second, HCR hairpins bind to the initiator and get longer like a chain (amplification step). b Representatives of HCR images for lncRNA HOTAIR at ×10 objective. c–e Zoomed images of HCR for lncRNAs HOTAIR (c), TUG1 (d) and CDKN2B-AS1 (e) at ×100 oil immersion objective.
Screening for the effect of lncRNAs on ccRCC prognosis and genetics
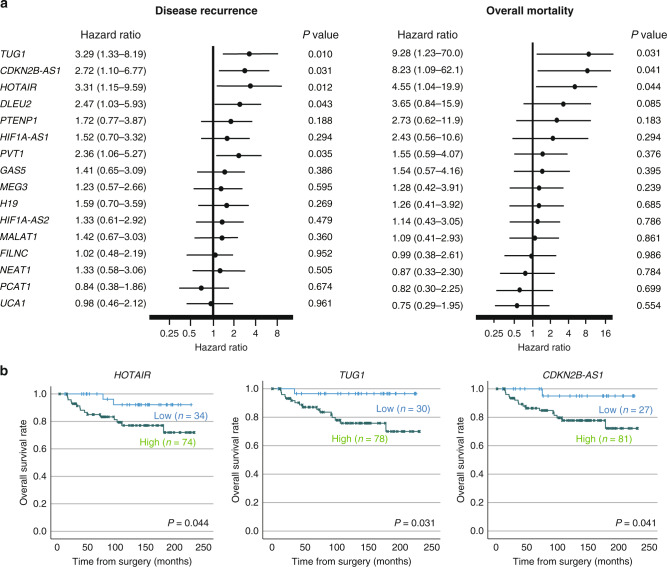
The clinicopathological characteristics of our in-house 108 ccRCC patients are shown in Table 1. The median follow-up period following surgery was 118 (IQR, 50–174) months. Disease recurrence and overall mortality were found in 27 (25.0%) and 17 (15.7%) patients, respectively. Five patients had synchronous metastatic lesions at their diagnosis; thereby these patients were excluded from the recurrence-free survival analysis. In this cohort of ccRCC, survival analyses revealed that high expression levels of 5 lncRNAs (HOTAIR, TUG1, PVT1, CDKN2B-AS1 and DLEU2, Fig. 3a) were significantly related to subsequent disease recurrence, and three (TUG1, HOTAIR and CDKN2B-AS1, Fig. 3a, b) were significantly related to overall mortality thereafter.
Table 1.
Clinicopathological characteristics associated with expressions of lncRNAs which showed a relation with survival.
| Characteristics | All patients (n = 108) | LncRNA risk group | |||
|---|---|---|---|---|---|
| low risk, n = 12 | intermediate risk, n = 37 | high risk, n = 59 | P value | ||
| Age, year, median, (IQR) | 59 (52–68) | 51 (40–61) | 60 (52–67) | 59 (52–69) | 0.144 |
| Sex, no (%) | 0.588 | ||||
| Male | 88 (81.5%) | 11 (91.7%) | 29 (78.4%) | 48 (81.4%) | |
| Female | 20 (18.5%) | 1 (8.3%) | 8 (21.6%) | 11 (18.6%) | |
| Nuclear grade, no (%) | 0.587 | ||||
| G1 + G2 | 93 (86.1%) | 11 (91.7%) | 33 (90.2%) | 49 (83.1%) | |
| G3 + G4 | 15 (13.9%) | 1 (8.3%) | 4 (10.8%) | 10 (16.9%) | |
| Pathological T stage, no (%) | 0.039 | ||||
| pT1 + pT2 | 94 (87.0%) | 12 (100.0%) | 35 (94.6%) | 47 (80.0%) | |
| pT3 + pT4 | 14 (13.0%) | 0 (0.0%) | 2 (5.4%) | 12 (20.0%) | |
| Tumour size, mm, median (IQR) | 40 (28–50) | 30 (18–43) | 33 (28–48) | 45 (30–59) | 0.042 |
| Venous Invasion | 0.431 | ||||
| Positive | 83 (76.9%) | 11 (91.7%) | 28 (75.7%) | 44 (74.6%) | |
| Negative | 25 (23.1%) | 1 (8.3%) | 9 (24.3%) | 15 (25.4%) | |
Fig. 3. Relationship between expression levels of 16 lncRNAs and outcome of prognosis in ccRCC patients.
a Forest plot of the hazard ratio for disease recurrence and overall mortality of ccRCC patients treated surgically, compared using the Cox regression analysis. b Kaplan–Meier curves of overall survival according to TUG1, CDKN2B-AS1 and HOTAIR expressions.
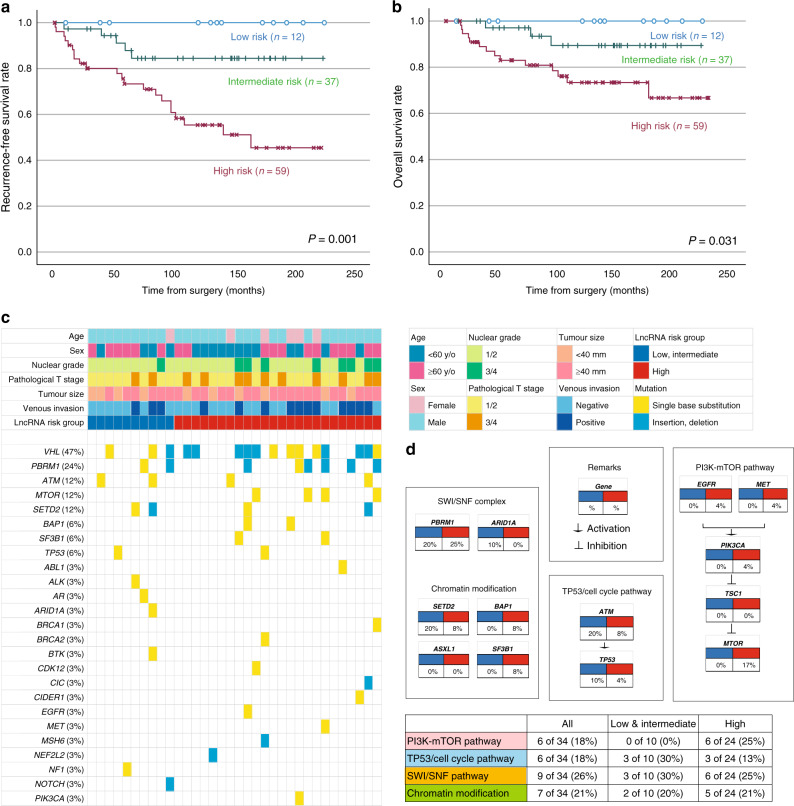
Standing on these results, we propose new risk groups of ccRCC. We divided 108 patients into three groups based on the lncRNA high expression status (low risk, 0; intermediate risk, 1–2; high risk, 3), which enabled the sequential stratification of patient prognosis. According to TUG1, HOTAIR and CDKN2B-AS1 lncRNA expression, 12 (11.1%) patients were classified as low risk, 37 (34.3%) were classified as intermediate risk, and 59 (54.6%) were classified as high risk. The Kaplan–Meier analysis showed recurrence-free (Fig. 4a) and overall survival rates (Fig. 4b) according to new risk groups. Notably, this scoring system demonstrated good discrimination for predicting ccRCC prognosis. Clinicopathological features among this classification are shown in Table 1. Pathological T stage and tumour size were significantly different among these risk groups. Importantly, multivariate Cox regression analyses identified pathological T stage (hazard ratio: HR 5.03, P < 0.001), tumour size (HR 2.67, P = 0.021), and high lncRNA risk group (HR 3.97, P = 0.007) as independent risk factors for tumour recurrence (Table 2). In addition, for overall survival, pathological T stage (HR 3.68, P = 0.029), tumour size (HR 6.67, P = 0.012), venous invasion (HR 5.86, P = 0.003) and high lncRNA risk group (HR 4.32, P = 0.026) were identified as independent risk factors (Table 2).
Fig. 4. Relationship between the lncRNA risk classification, outcome of prognosis and genetic alterations in ccRCC patients.
a, b Kaplan–Meier curves of disease-free (a) and overall survival (b) according to the lncRNA risk groups comprising three lncRNA (TUG1, CDKN2B-AS1 and HOTAIR) expression, compared using the Cox regression analysis. c Alteration landscape of 34 primary ccRCC tumour samples. Upper heatmap: age, sex, nuclear grade, pathological tumour stage, tumour size, venous invasion and the lncRNA risk groups. d Genomic alterations in tumorigenic signalling pathways related to ccRCC development on the lncRNA risk groups. The table shows the percentage of samples with alterations in each of the selected signalling pathways.
Table 2.
Parameters associated with postoperative disease recurrence and overall mortality in ccRCC patients after adjusting univariate and multivariate Cox regression analyses.
| Reccurence-free survival | Overall survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | Univariate | Multivariate | Univariate | Multivariate | ||||
| P value | HR | 95% CI | P value | P value | HR | 95% CI | P value | |
| Age (≥60 years vs. <60 years) | 0.620 | 1.32 | 0.54–4.26 | 0.516 | 0.140 | 1.59 | 0.57–4.51 | 0.361 |
| Sex (male vs. female) | 0.831 | 1.07 | 0.35–3.32 | 0.979 | 0.441 | 1.17 | 0.31–4.40 | 0.890 |
| Nuclear grade (G3/4 vs. G1/2) | 0.018 | 1.92 | 0.58–6.33 | 0.209 | <0.001 | 3.08 | 0.92–10.28 | 0.060 |
| Pathological T stage (pT3/4 vs. pT1/2) | <0.001 | 5.03 | 2.06–12.24 | <0.001 | <0.001 | 3.68 | 1.14–11.84 | 0.029 |
| Tumour size (≥40 mm vs. <40 mm) | 0.007 | 2.67 | 1.16–6.19 | 0.021 | 0.005 | 6.67 | 1.51–29.40 | 0.012 |
| Venous invasion (positive vs. negative) | <0.001 | 2.15 | 0.73–6.39 | 0.109 | <0.001 | 5.86 | 1.80–19.22 | 0.003 |
| LncRNA risk group (high vs. low & intermediate) | 0.001 | 3.97 | 1.45–10.89 | 0.007 | 0.022 | 4.32 | 1.24–15.08 | 0.026 |
An analysis of genomic alterations underlying these risk groups revealed prominent features of tumours that could serve as potential biomarkers for targeting lncRNAs. Herein, we analysed 34 ccRCC tumour samples comprising low/intermediate (n = 10) and high lncRNA risk (n = 24) groups for alterations in 160 cancer-associated genes (Fig. 4c). The most frequently altered genes in this cohort (>10%) were VHL, PBRM1, ATM, MTOR and SETD2. We compared changes in typical cancer-related pathways, and each of the lncRNA risk groups had unique genetic features. Alterations in the PI3K-mTOR pathway were highly prevalent in the high lncRNA risk group, occurring in 0% of the low/intermediate lncRNA risk and 25% of the high lncRNA risk group. On the other hand, alterations in the TP53/cell cycle pathway were highly prevalent in the low/intermediate lncRNA risk group (30%, as compared to 13% in the high lncRNA risk group). Collectively, these findings indicate that the genetic background associated with a poor prognosis in ccRCC may be related to the expression of lncRNAs [16, 17].
Uncovering cellular colocalization of lncRNAs by an expansion-assisted hybridisation chain reaction
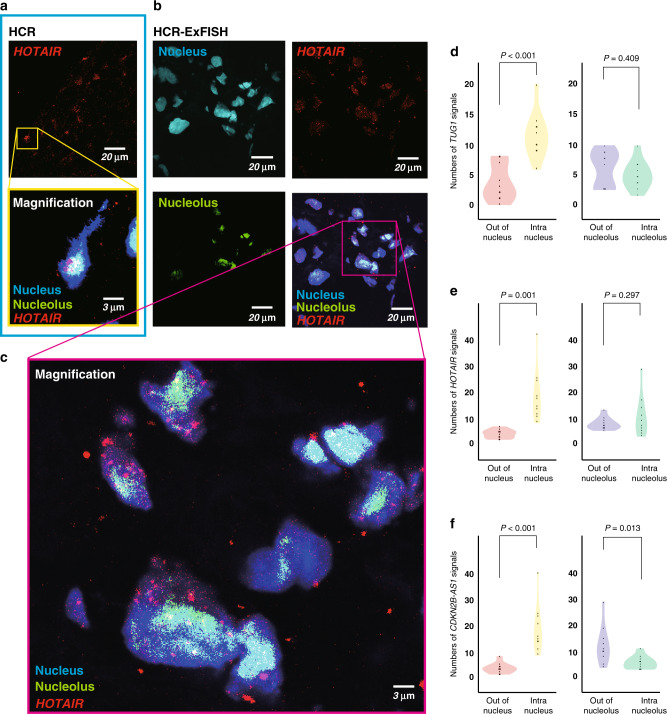
Detecting the HCR signals by conventional microscopy could not distinguish their colocalization in the cellular organelle, for example, intra- or out-of-nuclear or nucleolar (Fig. 5a); therefore, the information obtained by these images was inadequate for functional analysis. Herein, we applied a new imaging method combining HCR and expansion microscopy, so-called HCR-ExFISH, in clinical tumour samples, revealing that human ccRCC sections expanded 2–3-fold in one direction. In this analysis, we identified solo signals at the nanoresolution level (Fig. 5b, c). As a result, we counted the HCR signals of three lncRNAs (TUG1, HOTAIR and CDKN2B-AS1) to uncover colocalization of lncRNA signals, such as those located intra- or out of the nucleus or nucleolus.
Fig. 5. The HCR-ExFISH reveals intracellular colocalization of lncRNAs.
a, b LncRNA HOTAIR signals by conventional HCR (a) and nanoresolution HCR-ExFISH (b). Images are acquired using confocal microscopy. c High-magnification image of the boxed region in (b). d–f Violin plot shows the spatial heterogeneity of lncRNA TUG1 (d), HOTAIR (e) and CDKN2B-AS1 (f) signals in subcellular localisation, compared using the two-tailed Student’s t test.
In summary, HCR signals were more intranuclear than out of nuclear in all three lncRNAs. While the HCR signals of HOTAIR and TUG1 showed no significant difference in the intra- or out-of-nucleolus distribution (Fig. 5d, e), CDKN2B-AS1 was more out of the nucleolus than within the nucleolus (P = 0.013, Fig. 5f). Together, for the first time, an expansion-assisted hybridisation chain reaction approach could image cellular colocalization of lncRNAs in ccRCC, which may provide an effective tool to functionally analyse lncRNAs in the research community.
Discussion
Mounting evidence has revealed that lncRNAs may play an important role in carcinogenesis and tumour progression. In ccRCC, up- or downregulation of several kinds of lncRNAs was associated with tumorigenesis, e.g., Fuhrman grade, TNM stage, and lymph node or distant metastasis, resulting in disease recurrence and poor mortality [18]. Furthermore, since lncRNAs are expected to be desirable targets for cancer diagnosis or treatment [19], recent studies have examined the application of lncRNAs for the treatment or diagnosis of cancers. Antisense oligonucleotide technologies and nanoparticle-mediated RNA interference can be used to knock down oncogenic lncRNAs that are overexpressed in cancers, and Polovic et al. reported that this technique induced cell apoptosis in RCC cells [20]. Shi et al. revealed that blocking lncTASR gene transcription led to sensitivity of RCC cells to sunitinib therapy in vitro and suppression of tumour development in mouse RCC models [21]. Although technological challenges remain in diagnosing cancers, measuring the altered expression levels of lncRNAs in human body fluids (circulating lncRNAs) could contribute to early cancer detection [22, 23]. Taken together, the close relationships between cancers and lncRNAs are obvious. In this study, for the first time, we revealed two remarks.
First, we achieved high-throughput lncRNA visualisation and evaluation of 1728 pairs using clinical specimens, revealing a subset of lncRNAs associated with ccRCC prognosis. In our study, three lncRNAs (TUG1, HOTAIR and CDKN2B-AS1) were related to poor overall survival. Furthermore, we derived a new risk group of ccRCC prognosis by combining the expression levels of these three lncRNAs. Previous studies reported that high expression of these three lncRNAs was associated with poor prognosis in ccRCC [24–26], thereby being consistent with TCGA database analysis and our current results. Of course, public database analysis is useful to comprehend outlines of selected molecules; however, it is important to re-evaluate real clinical specimens. In this sense, our study, which evaluated many clinical samples in a high-throughput manner, could provide new insight for future research.
Second, by fusing HCR and expansion microscopy (HCR-ExFISH), we identified the intracellular colocalization of lncRNAs. Prognostic TUG1, HOTAIR and CDKN2B-AS1 existed more in the nucleus than out of the nucleus. It was already reported that lncRNAs in the nucleus mainly serve as regulators that affect chromosomal spatial conformation, transcription factor activity, and alternative splicing; rather, lncRNAs in the cytoplasm predominantly affect mRNA stability and translation regulation [27]. Thus, the subcellular localisation of lncRNAs is an additional essential layer of complexity that must be taken into account to fully understand the roles of lncRNAs in any cellular function [11]. While MALAT1 and NEAT1 are known to exist predominantly in the nucleus, TUG1 and HOTAIR have both nuclear and cytoplasmic distributions [28]. To the best of our knowledge, this is the first report demonstrating that TUG1 and HOTAIR existed more intranuclei than out of the nucleus in ccRCC tissues by using nanoresolutional HCR-ExFISH. When colocalizing nucleolar signals, CDKN2B-AS1 was more out of the nucleolus than within the nucleolus. There are few reports about the function of lncRNAs in the nucleolus. RMRP is the noncoding RNA component of the RNA processing endoribonuclease that is essential for processing preribosomal RNA in the nucleolus [29]. However, by binding to Hu antigen R, RMRP is exported in the cytoplasm and targeted to the mitochondria and works to maintain mitochondrial structure and mediate oxidative phosphorylation and mitochondrial DNA replication [30]. For further functional analysis, colocalization assessment, which extends to other organelles, including nuclear speckles, paraspeckles, cytoplasmic ribosomes or mitochondria, is needed for lncRNAs.
However, some limitations remain to be addressed at this stage. First, our study was retrospective, and a limited number of patients were included in the analysis. Second, little data are currently available on the real impact of lncRNAs on the response to systemic therapy, e.g., anti-angiogenic treatments and immunotherapies. We punched out a 4-mm core of tumour centres to create tissue microarrays; therefore, we did not mention the effect of tumour heterogeneity on lncRNA expression studied here. Notably, two IncRNAs, i.e., HIF1A-AS1 and FILNC1, were not associated with the prognosis of ccRCC in the TCGA dataset; however, since the main aim of the present study was to demonstrate that conclusions such as this may be reached using our HCR method, we did not adopt strict criteria for lncRNA screening. For detailed detection of colocalization in the cellular organelle, fluorescent staining for cytoplasm should be required in HCR-ExFISH. Of 108 patients, our fresh-frozen tissue samples available for DNA extraction and sequencing limited to 34 tumours, potentially including a selection bias for genomic alteration analysis. Last, the cancerous effects of lncRNAs on patient outcome and tumorigenesis in malignancies need to be supported by biological evidence.
In conclusion, we revealed that a new lncRNA detection system, i.e., the HCR RNA-FISH approach, enabled us to evaluate lncRNA expression in a low-cost and high-throughput manner. Three lncRNAs (TUG1, HOTAIR and CDKN2B-AS1) could be an indicator for the prognosis of ccRCC. The combination of HCR and expansion microscopy uncovered colocalization of lncRNAs in cellular organelles at the nanoresolution level. This study is advanced and unique in using a combination of new techniques for the functional analysis of lncRNAs. We believe that our method will accelerate further functional analysis of various lncRNAs worldwide.
Supplementary information
Author contributions
RK, NT and MO designed the study. RK, KT and EA performed the experiments. YY, TT, KM, SM, TK, HN and RM provided conceptual advice. RK and NT wrote the manuscript.
Funding
This study was supported by Grants-in-Aid for Scientific Research (KAKENHI 19K18598 and 21K09356 to RK; 19H03792, 21K19414, and 22H03217 to NT; and 18H02939 to MO) and grants from the Kobayashi Foundation for Cancer Research (to NT), the SGH Foundation for Cancer Research (to NT), the JUA Research Grant (to NT), the Princess Takamatsu Cancer Research Fund (to NT), and the Keio Gijuku Academic Development Funds (to NT).
Data availability
All data supporting the findings of this study are included within the article and its Supplementary Information files (and Reporting summary). Also, the data will be shared upon reasonable request to the corresponding author from colleagues who want to analyse in deep our findings.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All procedures were performed in approval of the Research Ethics Committee of Keio University (Approval Nos.: 20180098 and 20190059) and in compliance with the 1964 Helsinki Declaration and present ethical standards. Both written informed consent and passive (opt-out) informed consent procedures have been applied to the experimental use of human samples. Opt-out informed consent from patients was obtained by exhibiting the research information on our department's website (Department of Urology, Keio University Hospital, Tokyo, Japan). The need to obtain written informed consent was waived if patients had finished their follow-up or had died, due to the study’s observational nature and the urgent need for cancer patient care. This was approved and reviewed by the Research Ethics Committee of Keio University, in accordance with the ethical guidelines for Medical and Health Research Involving Human Subjects (Public Notice of the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare as of July 2018; https://www.lifescience.mext.go.jp/files/pdf/n2181_01.pdf).
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01895-3.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- 4.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi HM, Chang JY, Trinh le A, Padilla JE, Fraser SE, Pierce NA. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol. 2010;28:1208–12. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 2018;145:dev165753. [DOI] [PMC free article] [PubMed]
- 8.Choi HM, Beck VA, Pierce NA. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano. 2014;8:4284–94. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, Deisseroth K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell. 2016;164:792–804. doi: 10.1016/j.cell.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen H, Luo G, Chen Q. Long noncoding RNAs as tumorigenic factors and therapeutic targets for renal cell carcinoma. Cancer Cell Int. 2021;21:110. doi: 10.1186/s12935-021-01805-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045. [DOI] [PMC free article] [PubMed]
- 12.Asano SM, Gao R, Wassie AT, Tillberg PW, Chen F, Boyden ES. Expansion microscopy: protocols for imaging proteins and RNA in cells and tissues. Curr Protoc Cell Biol. 2018;80:e56. doi: 10.1002/cpcb.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe K, Kosaka T, Aimono E, Hongo H, Mikami S, Nishihara H, et al. Japanese case of enzalutamide-resistant prostate cancer harboring a SPOP mutation with scattered allelic imbalance: response to platinum-based therapy. Clin Genitourin Cancer. 2019;17:e897–e902. doi: 10.1016/j.clgc.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Takamatsu K, Tanaka N, Hakozaki K, Takahashi R, Teranishi Y, Murakami T, et al. Profiling the inhibitory receptors LAG-3, TIM-3, and TIGIT in renal cell carcinoma reveals malignancy. Nat Commun. 2021;12:5547. doi: 10.1038/s41467-021-25865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Computational Struct Biotechnol J. 2021;19:4101–9. doi: 10.1016/j.csbj.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tumkur Sitaram R, Landström M, Roos G, Ljungberg B. Significance of PI3K signalling pathway in clear cell renal cell carcinoma in relation to VHL and HIF status. J Clin Pathol. 2021;74:216–22. doi: 10.1136/jclinpath-2020-206693. [DOI] [PubMed] [Google Scholar]
- 17.D’Avella C, Abbosh P, Pal SK, Geynisman DM. Mutations in renal cell carcinoma. Urologic Oncol. 2020;38:763–73. doi: 10.1016/j.urolonc.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Li Z, Li W, Zhou L, Jiang Y. Prognostic significance of long non-coding RNAs in clear cell renal cell carcinoma: a meta-analysis. Medicine. 2019;98:e17276. doi: 10.1097/MD.0000000000017276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polovic M, Dittmar S, Hennemeier I, Humpf HU, Seliger B, Fornara P, et al. Identification of a novel lncRNA induced by the nephrotoxin ochratoxin A and expressed in human renal tumor tissue. Cell Mol Life Sci: CMLS. 2018;75:2241–56. doi: 10.1007/s00018-017-2731-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H, Sun Y, He M, Yang X, Hamada M, Fukunaga T, et al. Targeting the TR4 nuclear receptor-mediated lncTASR/AXL signaling with tretinoin increases the sunitinib sensitivity to better suppress the RCC progression. Oncogene. 2020;39:530–45. doi: 10.1038/s41388-019-0962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miranda-Castro R, de-Los-Santos-Álvarez N, Lobo-Castañón MJ. Long noncoding RNAs: from genomic junk to rising stars in the early detection of cancer. Anal Bioanal Chem. 2019;411:4265–75. doi: 10.1007/s00216-019-01607-6. [DOI] [PubMed] [Google Scholar]
- 23.Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15:39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama H, Tamai K, Shibuya R, Nakamura M, Mochizuki M, Yamaguchi K, et al. Long non-coding RNA HOTAIR promotes cell migration by upregulating insulin growth factor-binding protein 2 in renal cell carcinoma. Sci Rep. 2017;7:12016. doi: 10.1038/s41598-017-12191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang PQ, Wu YX, Zhong XD, Liu B, Qiao G. Prognostic significance of overexpressed long non-coding RNA TUG1 in patients with clear cell renal cell carcinoma. Eur Rev Med Pharmacol Sci. 2017;21:82–6. [PubMed] [Google Scholar]
- 26.Xie X, Lin J, Fan X, Zhong Y, Chen Y, Liu K, et al. LncRNA CDKN2B-AS1 stabilized by IGF2BP3 drives the malignancy of renal clear cell carcinoma through epigenetically activating NUF2 transcription. Cell Death Dis. 2021;12:201. doi: 10.1038/s41419-021-03489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX, Hong W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547:1–9. doi: 10.1016/j.gene.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 28.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–77. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldfarb KC, Cech TR. Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev. 2017;31:59–71. doi: 10.1101/gad.286963.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016;30:1224–39. doi: 10.1101/gad.276022.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are included within the article and its Supplementary Information files (and Reporting summary). Also, the data will be shared upon reasonable request to the corresponding author from colleagues who want to analyse in deep our findings.