Abstract
Synthesis of glutamate, the cell's major donor of nitrogen groups and principal anion, occupies a significant fraction of bacterial metabolism. In Bacillus subtilis, the gltAB operon, encoding glutamate synthase, requires a specific positive regulator, GltC, for its expression. In addition, the gltAB operon was shown to be repressed by TnrA, a regulator of several other genes of nitrogen metabolism and active under conditions of ammonium (nitrogen) limitation. TnrA was found to bind directly to a site immediately downstream of the gltAB promoter. As is true for other genes, the activity of TnrA at the gltAB promoter was antagonized by glutamine synthetase under certain growth conditions.
Nitrogen-containing groups of almost all intracellular compounds in bacteria are derived from glutamate or glutamine. The amino and amido groups of glutamate and glutamine are derived ultimately from ammonium. The two enzymes responsible for the unique pathway of ammonium assimilation and concomitant biosynthesis of glutamine and glutamate in Bacillus subtilis are glutamine synthetase, encoded by the glnA gene (16, 34), and glutamate synthase, whose two subunits are encoded by the gltAB operon (9, 34).
Expression of the gltAB operon is subject to nitrogen source-dependent regulation. Transcription is high when cells are grown in glucose-ammonium medium and low when cells are grown in glucose-glutamate medium; an intermediate level of transcription is seen in cells grown in glucose-glutamine or glucose-ammonium-glutamate (2, 9, 10). The best-characterized regulator of the gltAB operon is GltC, a member of the LysR family of bacterial transcription regulatory proteins (33). The gltC gene is located upstream of and is transcribed divergently from the gltAB operon (2, 10). GltC is also a negative autoregulator (2, 10). A gain-of-function mutant form of GltR, a protein related to GltC, also activates transcription from the gltAB promoter (3). Whether this activation reflects a normal activity of GltR under some undetermined physiological condition remains to be seen.
In our previous work, we have assumed that GltC is the principal nitrogen source-responsive regulator of gltAB. However, inactivation of GltC reduces gltAB expression so drastically that it is difficult to measure any expression, let alone regulation, in its absence. Nonetheless, certain results led us to ask whether another protein might also contribute to nitrogen source-dependent regulation of gltAB. First, the very low residual expression of gltAB seen in a gltC null mutant still seems to be subject to nitrogen source-dependent regulation (2). Second, cells producing the mutant form of GltR described above not only express gltAB in the total absence of GltC but do so in a nitrogen source-dependent manner (3).
In the work described below, we demonstrate that glutamate-dependent repression of gltAB is mediated principally by the TnrA protein. TnrA, an important regulator of many genes involved in nitrogen metabolism, activates its own gene (15, 30), the nasABCDEF genes (nitrate and nitrite utilization [27]), the nrgAB operon (ammonium transport [41]), the ureABC operon (urea utilization [39]), the gabP gene (γ-aminobutyrate permease [41]), the genes of purine utilization (H. Saxild, personal communication), and some other targets (30, 37) and represses the glnRA operon (40). TnrA is active only when cells are grown with a slowly metabolizable nitrogen source, such as glutamate (15, 40). TnrA and its homolog, GlnR (the principal regulator of the glnRA operon), are members of the MerR family of transcriptional regulators (35, 40). TnrA binds to a dyad symmetry element with the consensus sequence 5′-TGTNAN7TNACA-3′ (40, 42). The positive regulatory activity of TnrA is modulated by glutamine synthetase through a mechanism that is not fully understood (15, 40). As shown below, glutamine synthetase is also implicated in TnrA-dependent repression of the gltAB operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The B. subtilis strains used in this study, listed in Table 1, were grown at 37°C in TSS minimal medium (19) with 0.5% glucose and 0.2% nitrogen source or in DS nutrient broth medium (19). The same media with the addition of agar were used for growth of bacteria on plates. L broth or L agar (26) was used for growth of Escherichia coli strains. The following antibiotics were used when appropriate: chloramphenicol (2.5 μg/ml), spectinomycin (50 μg/ml), or a combination of erythromycin (0.5 μg/ml) and lincomycin (12.5 μg/ml) for B. subtilis strains, and ampicillin (50 μg/ml), kanamycin (25 μg/ml), or chloramphenicol (10 μg/ml) for E. coli strains.
TABLE 1.
B. subtilis strains used
| Strain | Genotype | Source or reference |
|---|---|---|
| SMY | Wild type | P. Schaeffer |
| SMY-S | ΔgltC::spc | 2 |
| PY150 | gltC150::(Tn917 erm) | P. Youngman |
| SX150103 | gltC150::(Tn917 erm) gltAp3 | This work |
| SF62 | tnrA62::(Tn917 erm) trpC2 | 40 |
| SF73 | glnA73 | 14 |
| BB278 | tnrA62::(Tn917 erm) | SMY × SF62 DNA |
| SF73-T | glnA73 tnrA62::(Tn917 erm) | SF73 × BB278 DNA |
| BB614 | ΔgltC::spc gltR24 ΔamyE::[cat Φ(gltC′p+-gusA) Φ(gltA′p+-lacZ)] | 3 |
| BB873 | ΔgltC::spc gltR24 tnrA62::(Tn917 erm) ΔamyE::[cat Φ(gltC′p+-gusA) Φ(gltA′p+-lacZ)] | BB614 × BB278 DNA |
| LG200 | ΔamyE::[cat Φ(gltC′p+-gusA) Φ(gltA′p+-lacZ)] | SMY × pLG200 (2) |
| LG200-S | ΔgltC::spc ΔamyE::[cat Φ(gltC′p+-gusA) Φ(gltA′p+-lacZ)] | 2 |
| LG203 | ΔamyE::[cat Φ(gltC′p3-gusA) Φ(gltA′p3-lacZ)] | SMY × pLG203 |
| LG203-S | ΔgltC::spc ΔamyE::[cat Φ(gltC′p3-gusA) Φ(gltA′p3-lacZ)] | SMY-S × LG203 DNA |
| LG203-T | tnrA62::(Tn917 erm) ΔamyE::[cat Φ(gltC′p3-gusA) Φ(gltA′p3-lacZ)] | BB278 × LG203 DNA |
| LG203-ST | ΔgltC::spc tnrA62::(Tn917 erm) ΔamyE::[cat Φ(gltC′p3-gusA) Φ(gltA′p3-lacZ)] | LG203-S × BB278 DNA |
| LG219 | ΔamyE::[cat Φ(gltC′p19-gusA) Φ(gltA′p19-lacZ)] | SMY × pLG219 |
| LG219-S | ΔgltC::spc ΔamyE::[cat Φ(gltC′p19-gusA) Φ(gltA′p19-lacZ)] | LG219 × SMY-S DNA |
| LG219-T | tnrA62::(Tn917 erm) ΔamyE::[cat Φ(gltC′p19-gusA) Φ(gltA′p19-lacZ)] | LG219 × SF62 DNA |
| LG219-ST | ΔgltC::spc tnrA62::(Tn917 erm) ΔamyE::[cat Φ(gltC′p19-gusA) Φ(gltA′p19-lacZ)] | LG219-S × BB278 DNA |
| LG219/24 | ΔamyE::[cat Φ(gltC′p19/24-gusA) Φ(gltA′p19/24-lacZ)] | SMY × pLG219/24 |
| LG219/24-T | tnrA62::(Tn917 erm) ΔamyE::[cat Φ(gltC′p19/24-gusA) Φ(gltA′p19/24-lacZ)] | LG219/24 × BB278 DNA |
| BB1045 | gltC150::(Tn917 erm) gltAp3 ΔamyE::[cat Φ(gltC′p19-gusA) Φ(gltA′p19-lacZ)] | SX150103 × LG219 DNA |
| BB1068 | tnrA62::(Tn917 erm::spc) | BB278 × p917::spc (36a) |
| BB1082 | gltC150::(Tn917 erm) gltAp3 ΔamyE::[cat Φ(gltC′p3-gusA) Φ(gltA′p3-lacZ)] | SX150103 × LG203 DNA |
| BB1083 | gltC150::(Tn917 erm) tnrA62::(Tn917 erm)::spc ΔamyE::[cat Φ(gltC′p3-gusA) Φ(gltA′p3-lacZ)] | BB1082 × BB1068 DNA |
| BB1194 | glnA73 ΔamyE::[cat Φ(gltC′p19-gusA) Φ(gltA′p19-lacZ)] | SF73 × LG219 DNA |
| BB1195 | glnA73 tnrA62::(Tn917 erm) ΔamyE::[cat Φ(gltC′p19-gusA) Φ(gltA′p19-lacZ)] | SF73-T × LG219 DNA |
| BB1199 | glnA73 ΔamyE::[cat Φ(gltC′p19/24-gusA) Φ(gltA′p19/24-lacZ)] | SF73 × LG219/24 DNA |
| LG700 | ΔamyE::[cat Φ(gltC′p700-gusA) Φ(gltA′p700-lacZ)] | 2 |
| LG700-S | ΔgltC::spc ΔamyE::[cat Φ(gltC′p700-gusA) Φ(gltA′p700-lacZ)] | SMY-S × LG700 DNA |
| LG700-ST | ΔgltC::spc tnrA62::(Tn917 erm) ΔamyE::[cat Δ(gltC′p700-gusA) Φ(gltA′p700-lacZ)] | LG700-S × BB278 DNA |
| LG702 | ΔamyE::[cat Φ(gltC′p702-gusA) Φ(gltA′p702-lacZ)] | 2 |
| LG702-S | ΔgltC::spc ΔamyE::[cat Φ(gltC′p702-gusA) Φ(gltA′p702-lacZ)] | SMY-S × LG702 DNA |
| LG702-ST | ΔgltC::spc tnrA62::(Tn917 erm) ΔamyE::[cat Φ(gltC′p702-gusA) Φ(gltA′p702-lacZ)] | LG702-S × BB278 DNA |
| LG708 | ΔamyE::[cat Φ(gltC′p708-lacZ) Φ(gltA′p708-gusA)] | SMY × pLG708 |
| LG711 | ΔamyE::[cat Φ(gltC′p711-lacZ) Φ(gltA′p711-gusA)] | SMY × pLG711 |
The plasmids used in this work are described below. E. coli strain JM107 (43) was used for the propagation of most plasmids. Strain BU1255 (dam-3 dcm-6 gal lac ara thr leu/F+) was used for experiments requiring unmethylated DNA. Plasmids of the pLG102 series were integrated at the amyE locus of B. subtilis as described previously (2).
DNA manipulations and transformation.
The methods for plasmid isolation, agarose and polyacrylamide gel electrophoresis, use of restriction and DNA modification enzymes, DNA ligation, PCR, transformation of electroporation-competent E. coli cells, and transformation of competent B. subtilis cells were described previously (6).
Isolation of Glt+ pseudorevertants.
Strain PY150 (gltC::Tn917 Glt− Eryr) was treated with 30 to 300 μg of N-methyl-N-nitro-N-nitrosoguanidine/ml (10 to 0.1% survival frequencies), and Glt+ pseudorevertants were isolated on TSS glucose-ammonium plates containing erythromycin and lincomycin but lacking glutamate. Chromosomal DNA from Glt+ pseudorevertants was used to transform the wild-type strain SMY to erythromycin resistance. Most of the transformants were Glt+, indicating that the Glt+ suppressor mutations were closely linked to the Tn917 insertion in gltC. A particular Glt+ derivative of a gltC::Tn917 mutant of strain SMY was named SX150103, and the suppressor mutation, which proved to be in the gltA promoter region, was named gltAp3.
Cloning of the gltAp3 mutation.
Plasmid pBB559 (′proJ′ neo) (5) was integrated by a single-crossover event into the chromosome of strain SX150103 (gltC::Tn917 gltAp3) at the proJ (previously called unk [5]) locus, 1.0 kb downstream from gltC. Chromosomal DNA of the transformant was isolated, digested with PstI, self-ligated, and introduced into E. coli cells. The resultant plasmid, pBB748, carries a 3′ part of the proJ gene, the entire gltC gene with the inserted Tn917, the gltCA intergenic region (within which was found the gltAp3 mutation), and a 5′ part of the gltA gene.
Construction of gltA-lacZ fusions.
To construct a gltA-lacZ fusion containing the gltAp3 mutation, we first used PCR to generate the mutant gltCA regulatory region from the chromosomal DNA of strain SX150103. The oligonucleotides oLS13 (5′-GGGCCACATGCAAATGATCA), corresponding to positions −175 to −156 with respect to the gltA transcriptional start point (2) (Fig. 1) and overlapping the BclI site (underlined), and oBB39 (5′-CTAGGGGGATCCGTCGACCTGCATCATGTTCAAATTCAG), containing an engineered BamHI site (underlined) and nucleotides corresponding to positions +99 to +119, served as 5′ and 3′ primers, respectively. The 0.29-kb BclI-BamHI fragment of the PCR product was cloned in the BglII site of a bidirectional fusion vector, pLG102 (2), creating plasmid pLG203, which contains gltAp3-lacZ and gltCp3-gusA fusions. The structure of pLG203 was confirmed by DNA sequencing. (This plasmid differed from pLG200 [2], containing a wild-type gltCA regulatory region, only at the site of the gltAp3 mutation.)
FIG. 1.
The gltCA intergenic region. Likely transcription start sites of the gltC and gltA genes, their −10 and −35 regions, and the gltA initiation codon are in boldface. Directions of transcription and translation are indicated by horizontal arrows. The mutations in the region are shown above the sequence with their allele numbers. The apparent GltC-binding Box I and Box II sequences (2) are underlined and double underlined, respectively. A TnrA binding sequence is underlined with dashes. Above this site the nucleotides that appear to interact with TnrA in the DNase I protection assay are shown by a horizontal line, and the TnrA consensus sequence (15) is presented. The fusions in pLG200, pLG203, and pLG219 contain fragments with coordinates from −160 to +119. The vertical arrows indicate upstream or downstream junctions for the truncated fusions in plasmids pLG700 (positions −56 to +119), pLG702 (positions −45 to +119), pLG708 (positions −160 to +24), and pLG711 (positions −160 to +3). The numbering of the sequence is with respect to the gltA transcription start point (2).
Two truncated gltA-lacZ fusions were constructed as follows. Plasmid pBB73 contains the 1.15-kb HindIII-BamHI fragment of the gltA-gltC region from pPJ1 (2) cloned in pBS(−) (Stratagene). pBB73 was cleaved with AgeI and BamHI (Fig. 1), blunt ended using the large (Klenow) fragment of DNA polymerase I, and self-ligated. The BamHI site was reconstituted within the resulting plasmid, pBB74. Incubation of pBB74 with BclI and BamHI liberated a 0.19-kb fragment, corresponding to positions −160 to +24, which was cloned in the BglII site of pLG102 (2). The resultant plasmid, containing truncated gltA-gusA and gltC-lacZ fusions, was called pLG708. A truncated gltA fusion without a putative TnrA binding site (see Results) was constructed as described above for pLG203 with oligonucleotide oBB42 (5′-CTAACAGATCTATAATCATTATAGGTTGTCAAAAC), containing an engineered BglII site (underlined) and nucleotides corresponding to positions −24 to +3, as a 3′ primer for PCR. oBB42 also contained nucleotides (boldface) corresponding to the gltAp3 mutation at position −10 (this mutation was also present in the chromosomal DNA used for PCR) and to the gltAp19 mutation at position −17 (Fig. 1). The gltAp19 mutation, described previously (2), does not affect expression or regulation from the gltA promoter but significantly increases expression from the gltC promoter and was used in some experiments to augment the sensitivity of gltC-gusA fusion assays. The plasmid containing truncated gltAp3/19-gusA and gltCp3/19-lacZ fusions was called pLG711. The structure of the insert within pLG711 was confirmed by sequencing.
Mutation in the TnrA binding site.
With plasmid pIPC119 (2), carrying the gltAp19 version (see above) of the gltA regulatory region, as a template and using the mutagenic oligonucleotide oBB37 (5′-GATAATACCGATCATAAAATCTAATAACTCTATAATC), corresponding to positions +31 to −7 of the gltAB transcription unit, we created, by the method of Kunkel et al. (2, 23), a triple mutant that has the original gltpA19 mutation and a double mutation, gltAp24 (shown in boldface in oBB37) (Fig. 1), in the putative TnrA binding site. The presence of the mutations in the resulting plasmid, pIPC119/24, was confirmed by DNA sequencing of the entire gltCA regulatory region. The 0.29-kb BclI-BamHI fragment of pIPC119/24 containing the gltCA regulatory region was cloned in pLG102 as described above. The plasmid containing the gltAp19/24-lacZ and gltCp19/24-gusA bidirectional fusions was called pLG219/24.
Labeling of DNA fragments.
The 291-bp BamHI-BclI fragment of pIPC10 (2) containing the gltC-gltA regulatory region with coordinates from −160 to +119 relative to the gltA transcription start was gel purified and labeled using the Klenow fragment of E. coli DNA polymerase I and [α-32P]ATP (31). Alternatively, the 277- or 346-bp fragment containing the gltC-gltA regulatory region from positions −175 to +102 or from −175 to +119 were synthesized by PCR using oligonucleotide oLS13 (5′-GGGCCACATGCAAATGATCA) (positions −175 to −156) as the 5′ primer and oBB13 (5′-CAGGACGGTAGAGACC) (positions 87 to 102) or oBB4 (5′-CTCATTCCCTGATCTCG) (corresponds to vector sequences) as the 3′ primer and plasmid pIPC119 or pIPC119/24 as a template. One of the primers for each PCR was labeled using T4 polynucleotide kinase and [γ-32P]ATP (31). The labeled PCR products were purified on an 8% nondenaturing polyacrylamide gel (31).
Gel shift experiments.
Purified TnrA was isolated as described previously (42). The conditions for incubation of TnrA with the gltA regulatory fragment were as described previously (42) or were modified by using 20 mM Tris-Cl (pH 8.0), 50 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 0.05% Nonidet P-40, and 5.0 to 7.5% glycerol as a binding buffer. In addition to 32P-labeled DNA and various amounts of TnrA, the 10-μl incubation mixture contained 1 μg of bovine serum albumin and 0.5 μg of poly(dI-dC) · poly(dI-dC). The samples were incubated for 10 to 15 min at room temperature or at 30°C and separated on 5 to 8% nondenaturing polyacrylamide gels in 0.5× Tris-borate buffer or in Tris-taurine buffer (U.S. Biochemical sequencing protocol).
To prepare B. subtilis crude extracts, 40 ml of cells were grown in TSS medium supplemented with ammonium chloride or proline as nitrogen source, centrifuged, and washed once with 50 mM Tris-Cl, pH 8.0. The cell pellets were kept at −70°C. The cells were resuspended in 2.5 to 3.0 ml of sonication buffer (20 mM Tris-Cl [pH 8.0], 50 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 5.0 to 7.5% glycerol) plus 1 to 2 mM phenylmethylsulfonyl fluoride and disrupted by sonication on ice. Cell debris was removed by centrifugation. The supernatant fluid was used as a source of protein in gel shift experiments.
DNase I protection experiments.
The 32P-labeled gltA promoter DNA fragment was incubated with purified TnrA in a total volume of 10 μl under the conditions described above. One microliter of binding buffer containing 0.125 U of RQ1 DNase I (Promega) and 10 mM CaCl2 was added, followed by the addition, after 1 min, of 4 μl of dye solution and subsequent heating of the samples at 80°C for 5 min. The samples were analyzed on a 5% urea-polyacrylamide DNA sequencing gel. A sequencing ladder obtained from pIPC119 using the primer identical to the labeled primer in each PCR served to precisely locate the protected region.
DNA sequencing.
DNA fragments containing the gltA region were sequenced on both strands by the dideoxy chain termination method of Sanger et al. (32) using vector- or gltA region-specific oligonucleotides as primers. Plasmid double-stranded DNA to be sequenced was purified with a QIAprep spin miniprep kit (Qiagen). A Sequenase reagent kit (U.S. Biochemicals) was used according to the protocol of the manufacturer. DNA and protein sequences were analyzed using the DNA Strider (24) and BLAST (1) programs.
Enzyme assays.
β-Galactosidase and β-glucuronidase activities were determined as described previously (2). All activities reported were the averages of at least two experiments, and the mean errors did not exceed 20%.
RESULTS
Expression of the gltAB operon is not abolished in the absence of GltC and is still nitrogen regulated.
A gltC mutant of B. subtilis expresses the gltAB operon at a very low level, indicating that GltC, a positive regulator, is almost absolutely required for gltAB expression (2, 10). Nevertheless, in the presence of ammonium plus glutamate, ammonium plus proline, or glutamine as a nitrogen source, residual levels of gltAB mRNA (10), gltA-lacZ (2), and gltA-gusA activities can be detected in gltC mutants, and the low activities of the fusions were further reduced when glutamate or proline served as the sole nitrogen source (Table 2, strain LG219-S, and data not shown). These results suggest the existence of a GltC-independent, nitrogen source-dependent mode of regulation of gltAB expression. Because they require a source of glutamate for growth, gltC mutants cannot be tested during growth in the presence of ammonium as the sole nitrogen source, i.e., the conditions under which the expression of glutamate synthase is highest in wild-type cells (9, 10).
TABLE 2.
TnrA effect on expression of gltA and gltC
| Strain | Genotype | gltA-gltC region of fusion | β-Galactosidase activity, Miller units (gltA-lacZ fusion)
|
β-Glucuronidase activity, Miller units (gltC-gusA fusion)
|
|||
|---|---|---|---|---|---|---|---|
| Ammonium | Ammonium + glutamate | Glutamate | Ammonium | Glutamate | |||
| LG200 | Wild type | Wild type | 75.8 | 26.1 | ≤2.1 | 0.7 | 1.1 |
| LG200-S | gltC | Wild type | NGb | 1.0 | 0.5 | NG | 8.0 |
| LG203 | Wild type | gltAp3 | 118.1 | 54.5 | 6.3 | 1.1 | 1.8 |
| LG203-S | gltC | gltAp3 | NG | 8.7 | 1.1 | NG | 11.9 |
| LG203-T | tnrA | gltAp3 | 120.1 | 52.7 | 56.7 | 1.2 | 2.8 |
| LG203-ST | gltC tnrA | gltAp3 | NG | 8.9 | 12.3 | NG | 23.4 |
| LG219 | Wild type | Wild typea | 74.2 | 30.8 | ≤2.0 | 14.6 | 16.9 |
| LG219-S | gltC | Wild type | NG | 0.9 | 0.3 | NG | 85.8 |
| LG219-T | tnrA | Wild type | 73.2 | 27.6 | 34.6 | 13.5 | 25.3 |
| LG219-ST | gltC tnrA | Wild type | NG | 0.9 | 1.0 | NG | 87.2 |
| LG219/24 | Wild type | gltAp24 | 74.4 | NDc | 32.6 | ND | 15.4 |
| LG219/24-T | tnrA | gltAp24 | 78.0 | ND | 31.2 | ND | ND |
Strains of the LG219 series contain a gltAp19 mutation (2) (Fig. 1) within their fusions. This mutation does not affect expression or regulation from the gltA promoter but strongly increases expression from the gltC promoter (compare LG200 and LC219) and was used to increase activity of the gltC-gusA fusion.
NG, no growth. gltC mutants cannot grow in ammonium medium because they require glutamate for growth.
ND, not determined.
Characterization of a cis-acting Glt+ mutation.
We sought to isolate mutations in the gltA promoter region that would allow a higher level of GltC-independent expression of the gltAB operon. To do so, we plated strain PY150 (gltC150::Tn917) on minimal medium without glutamate and isolated Glt+ pseudorevertants (see Materials and Methods). Chromosomal DNA from the gltCA regulatory region of one such pseudorevertant, strain SX150103, was cloned (see Materials and Methods). DNA sequencing revealed a single point mutation, gltAp3, in the −10 region of the gltA promoter that created a perfect match with the −10 region of the consensus B. subtilis ςA-type promoter (Fig. 1). Strain SX150103 (gltC150::Tn917 gltAp3) grew in the presence of ammonium as the sole nitrogen source with a generation time of 90 min compared to 60 min for the wild-type strain (the difference in the growth rates of these two strains disappeared when a source of glutamate was present in the medium). The ability of strain SX150103 to grow in the absence of glutamate indicates synthesis of a sufficient amount of glutamate synthase to generate glutamate intracellularly. An increased level of gltA mRNA in strain SX150103, in comparison to the parent strain (gltC150::Tn917), was demonstrated by S1 nuclease protection experiments (8) and was confirmed (see below) using gltA-lacZ fusions.
Expression of a gltAp3-lacZ fusion.
When a bidirectional gltA-lacZ gltC-gusA transcriptional fusion construct carrying the gltAp3 mutation was placed at the amyE locus of a gltC+ strain, LG203, expression of gltA-lacZ was 50% higher than that seen in a gltAp+ fusion strain (LG200) (Table 2), indicating that the gltAp3 promoter is stronger than the wild-type promoter even in the presence of GltC. In this strain, gltAB expression was still regulated by the nitrogen source in a manner similar to that seen with the wild-type promoter (Table 2). As expected from the Glt+ phenotype of strain SX150103, the gltAp3-lacZ fusion had significant activity in the absence of GltC (Table 2; compare strains LG203-S and LG200-S). This GltC-independent activity was also regulated by the nitrogen source in the medium in a manner similar to that described for GltC-dependent expression of the gltA-lacZ fusion containing a wild-type regulatory region (strain LG200) (Table 2) (2). Together with our results obtained with wild-type cells, these results strongly suggest that GltC is not the only factor involved in gltAB regulation and that at least one additional regulator responds to a nitrogen source-dependent signal.
Expression from the gltC promoter in the gltAp3 mutant fusion construct increased by 50% both in gltC+ cells and in gltC null mutant cells in which negative autoregulation of gltC was relieved (Table 2). This positive effect is probably due to the location of the gltAp3 mutation only 5 bp upstream of the −35 region of the gltC promoter (Fig. 1). Thus, a single gltAp3 mutation at the amyE locus increased the efficiencies of both the gltA and gltC promoters at the same locus and also made gltA-lacZ expression partially independent of GltC.
Nitrogen source-dependent regulation of gltA is impaired in a tnrA mutant.
Mutations in the tnrA gene have been shown to increase or reduce expression of several genes involved in nitrogen metabolism when glutamate or another poor nitrogen source is present as the sole nitrogen source (15, 40). A tnrA null mutation caused a 10-fold relief of glutamate-mediated repression of gltA-lacZ fusions containing either the wild-type regulatory region or the gltAp3 mutation (Table 2, strains LG219-T and LG203-T). No effect of a tnrA mutation on the gltA-lacZ fusions was observed when ammonium, glutamine, ammonium plus glutamate, or ammonium plus proline served as nitrogen sources (Table 2 and data not shown).
TnrA also appeared to be responsible for a 1.5- to 2-fold decrease in gltC-gusA expression in glutamate medium (Table 2). The mechanism of the small TnrA effect on gltC expression is unclear; it cannot contribute significantly to gltA repression because the same level of gltC expression in the presence of ammonium is sufficient for a high level of expression from the gltA promoter. The conclusion that TnrA-mediated repression of gltA is not due to an effect on gltC was confirmed by the loss of glutamate-dependent repression of gltAp+-lacZ and gltAp3-lacZ fusions in a gltC tnrA double mutant (Table 2, strains, LG219-ST and LG203-ST).
The ability of strain SX150103 (gltC gltAp3) to grow without a source of glutamate allowed us to extend our analysis of GltC-independent, nitrogen source-dependent regulation of gltA to the case of growth with ammonium as the sole nitrogen source, i.e., the condition under which glutamate synthase expression is maximal in wild-type cells (9, 10). Expression of the gltAp+-lacZ or gltAp3-lacZ fusion in the background of strain SX150103 (gltC gltAp3) was maximal under the same conditions as in wild-type cells despite the absence of GltC (Table 3). Introduction of a tnrA mutation into this strain significantly increased expression of the gltAp3-lacZ fusion in the presence of glutamate as the sole nitrogen source but did not completely abolish the difference in gltA-lacZ expression in the presence of various nitrogen sources (Table 3), as expected from the current notion that the TnrA protein is equally inactive in ammonium and ammonium-plus-glutamate media (15, 40). We conclude that, in the absence of GltC, nitrogen source-dependent regulation of gltA is mostly, but perhaps not exclusively, mediated by TnrA.
TABLE 3.
Residual gltA expression in the absence of GltC and TnrAa
| Strain | Genotype | gltA-gltC region of fusion | β-Galactosidase activity, Miller units (gltA-lacZ fusion)
|
||
|---|---|---|---|---|---|
| Ammonium | Ammonium + glutamate | Glutamate | |||
| BB1045 | gltC gltAp3 | Wild type | 1.9 | 0.9 | 0.3 |
| BB1082 | gltC gltAp3 | gltAp3 | 18.6 | 10.5 | 2.5 |
| BB1083 | gltC gltAp3 tnrA | gltAp3 | 20.2 | 12.7 | 10.7 |
The various strains, derivatives of SX150103, were grown to mid-exponential phase in the indicated glucose minimal medium and assayed for β-galactosidase activity.
Role of TnrA in GltR24-dependent expression of gltAB.
GltR is a homolog of GltC of unknown physiological function; GltR24 is a gain-of-function mutant form of GltR (3). We previously demonstrated that expression of a gltAp+-lacZ fusion in a gltC gltR24 mutant was subject to repression in the presence of glutamate or proline despite the absence of GltC (3). This regulation could be due to a dependence of GltR24 activity on nitrogen source-dependent signals or to the activity of another protein. When a tnrA mutation was introduced into a gltC gltR24 double mutant, repression of the gltAp+-lacZ fusion by proline or glutamate was reduced more than 10-fold (Table 4 and data not shown). This result shows that TnrA, and not GltR24, is the protein primarily responsible for the nitrogen source-dependent regulation of gltA in a gltC gltR24 mutant.
TABLE 4.
Effect of a tnrA mutation on GltR24-dependent gltA expressiona
| Strain | Genotype | β-Galactosidase activity, Miller units (gltA-lacZ fusion)
|
||
|---|---|---|---|---|
| Ammonium | Ammonium + proline | Proline | ||
| LG200 | Wild type | 75.8 | 50.4 | <0.6 |
| LG200-S | gltC | NGb | 0.9 | 0.2 |
| BB614 | gltR24 gltC | 34.2 | 12.5 | 1.6 |
| BB873 | gltR24 gltC tnrA | 32.0 | NDc | 24.3 |
The various strains, all carrying a gltAp+-lacZ fusion, were grown to mid-exponential phase in the indicated glucose minimal medium and assayed for β-galactosidase activity.
NG, no growth. gltC mutants are glutamate auxotrophs and therefore cannot grow with ammonium as the sole nitrogen source.
ND, not determined.
Wild-type GltR protein is also not responsible for the residual regulation of gltA in a gltC tnrA double mutant, since a null mutation in gltR did not affect expression from the gltAp+ or gltAp3 promoter in such a strain (data not shown). Furthermore, expression from the gltAp3 promoter in a gltC mutant is not affected by a gltR null mutation (data not shown).
Role of TnrA in regulation of mutant gltA promoter regions defective in sites of GltC interaction.
Previously we showed that expression from the gltA promoter is dependent on the integrity of two upstream dyad symmetry sequences, Box I and Box II (2) (Fig. 1). These sequences were suggested to be GltC binding sites (2). When one or both of these boxes were deleted, there was still some residual expression of a gltA-lacZ fusion, and this expression was still reduced by the presence of proline or glutamate (Table 5 and data not shown). Expression of these fusions, as expected, was not affected by a gltC mutation under any growth conditions, but glutamate- or proline-mediated repression was relieved by a tnrA mutation (Table 5 and data not shown).
TABLE 5.
Effect of a tnrA mutation on Box I- and Box II-independent gltA expressiona
| Strain | Genotype | gltA regulatory regionb fused to lacZ | β-Galactosidase activity, Miller units (gltA-lacZ fusion)
|
||
|---|---|---|---|---|---|
| Ammonium | Ammonium + proline | Proline | |||
| LG700 | Wild type | gltAp700 | 2.5 | 1.7 | ≤0.2 |
| LG700-S | gltC | gltAp700 | NGc | 1.6 | ≤0.2 |
| LG700-ST | gltC tnrA | gltAp700 | NG | 1.9 | 1.9 |
| LG702 | Wild type | gltAp702 | 1.9 | 1.6 | ≤0.2 |
| LG702-S | gltC | gltAp702 | NG | 2.1 | ≤0.2 |
| LG702-ST | gltC tnrA | gltAp702 | NG | 2.1 | 2.2 |
The various strains were grown to mid-exponential phase in the indicated glucose minimal medium and assayed for β-galactosidase activity.
The gltAp700 promoter has Box I deleted and contains nucleotides from positions −56 to +119 with respect to the gltA transcription start point; the gltAp702 promoter had both Box I and Box II deleted and contains nucleotides from positions −45 to +119 (Fig. 1) (2).
NG, no growth. gltC mutants are glutamate auxotrophs and therefore cannot grow with ammonium as the sole nitrogen source.
Role of glutamine synthetase in gltAB expression.
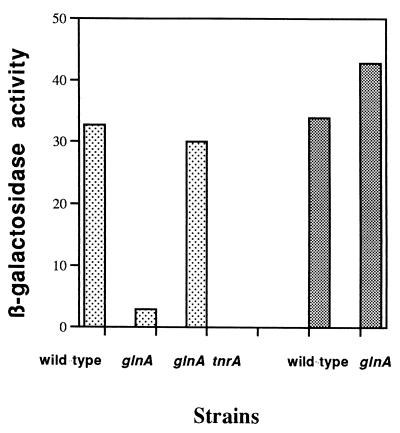
Expression of a gltAp+-lacZ fusion (Fig. 2) and accumulation of glutamate synthase activity (14, 17) are both reduced by mutations in the glnA gene, encoding glutamine synthetase. The effect of glutamine synthetase on the expression of genes involved in nitrogen metabolism has been attributed to inactivation of TnrA by glutamine synthetase in the presence of rapidly utilized nitrogen sources (15). In accord with this model, the reduction in gltAB expression caused by the glnA73 mutation was completely relieved by a tnrA mutation (Fig. 2).
FIG. 2.
Effects of glnA and tnrA mutations on gltA-lacZ expression. Strains LG219 (wild type), BB1194 (glnA73), and BB1195 (glnA73 tnrA) containing a gltAp19-lacZ fusion (lightly shaded bars) and strains LG219/24 (wild type) and BB1199 (glnA73) containing a gltAp19/24-lacZ fusion with the gltAp24 mutation in the TnrA binding site (darkly shaded bars) were grown to mid-exponential phase in glucose-glutamine minimal medium and assayed for β-galactosidase activity. Strains of the LG219 series contain a gltAp19 mutation (Fig. 1) within their fusions. This mutation does not effect expression or regulation from the gltA promoter (Table 2).
Roles of other nitrogen metabolism genes in glutamate synthase regulation.
Mutations in several genes regulated by TnrA, namely, nasA, nasB, nasD, nasE, ureC, nrgAB, nrgB, and glnR (15), had no effect on the expression of a gltA-lacZ fusion (data not shown). Moreover, mutations in the citA, citZ, citB, odhA, gltA, and proAB genes were unable to restore repression to a tnrA mutant strain (data not shown). Thus, no known target of TnrA (other than glutamine synthetase) and no enzyme involved in glutamate or proline synthesis substituted for or mitigated the effect of TnrA. Therefore, it seems likely that the effect of TnrA on gltAB is direct rather than through one of its known regulated genes.
Several other genes involved in regulation of nitrogen metabolism in B. subtilis were checked for their possible involvement in gltA regulation. No effect of sigL, codY, or abrB mutations on gltA-lacZ expression was observed under any growth conditions tested (data not shown).
Localization of a DNA target of TnrA-mediated regulation.
The gltA fusions used in most of our experiments contain the gltA regulatory region in a DNA segment corresponding to positions −160 to +119 with respect to the gltA transcription start point (2). A truncated fusion (as in pLG708) containing the sequence from positions −160 to +24 (Fig. 1) had the same activity and was subject to the same regulation as the longer gltA fusion (data not shown). A fusion (as in pLG711) that contained nucleotides from positions −160 to +3 (Fig. 1) retained dependence on GltC but was not repressed in cells grown in glutamate (data not shown). These results suggest that a target of TnrA-mediated repression lies immediately downstream of the gltA transcriptional start point.
A sequence corresponding, with one mismatch, to the proposed TnrA binding consensus (28, 40, 41) is located at positions +6 to +22 with respect to the gltA transcriptional start point (Fig. 1). The double mutation gltAp24 (Fig. 1) (see Materials and Methods), in which two conserved nucleotides of the putative TnrA binding site were altered, abolished TnrA-dependent repression of a gltAp19-lacZ fusion (Table 2, strains LG219/24 and LG219/24-T). The glnA73 mutation had no effect on gltAp19/24-lacZ expression (Fig. 2), in accord with the inability of TnrA to repress this fusion.
Gel shift experiments.
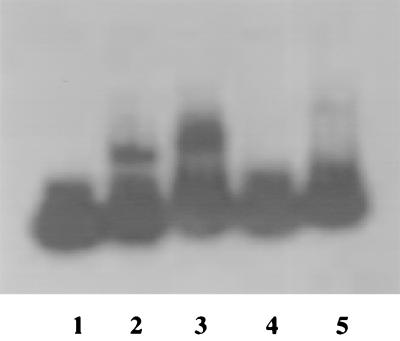
Crude extracts of wild-type B. subtilis cells grown in the presence of proline as the sole nitrogen source contained a factor that bound to the gltA regulatory region in gel mobility shift assays (Fig. 3). This binding was not affected by the absence of GltC (data not shown). The gltA binding activity was not detected in cell extracts from wild-type or gltC cells grown in the presence of ammonium (data not shown). Binding to the gltA promoter region was also not observed when extracts from tnrA mutant cells were used, irrespective of the nitrogen source used for their growth (Fig. 3).
FIG. 3.
Gel mobility shift assay with crude extracts. Cells of strains SMY (tnrA+) and BB278 (tnrA) were grown in glucose-proline medium to mid-exponential phase, harvested, and disrupted. The supernatant fluid from low-speed centrifugation was used as a crude extract for assays of binding to a radioactive 346-bp DNA fragment corresponding to positions −175 to +119 of the gltAB transcription unit. Lane 1, probe only; lanes 2 and 3, 1 and 4 μl of SMY extracts; lanes 4 and 5, 1 and 4 μl of BB278 extracts.
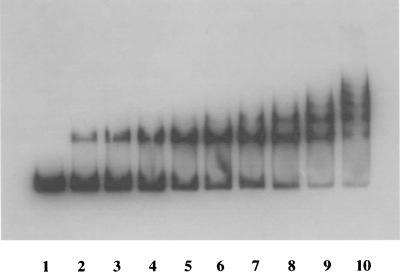
To test whether TnrA interacts directly with the gltA promoter region, a DNA fragment containing all sequences required for regulation was mixed with purified TnrA (Fig. 4). A complex of TnrA and gltA promoter region DNA was clearly seen; the equilibrium dissociation constant for this interaction, defined as the TnrA concentration required to shift 50% of the DNA molecules, was estimated to be 50 nM. The corresponding value for TnrA binding to the consensus binding site of the nrgAB promoter was about 8 nM (41).
FIG. 4.
Gel mobility shift analysis of the interaction between the gltA promoter and TnrA. A radioactively labeled 291-bp fragment containing the gltA promoter region with coordinates from −160 to +119 relative to the gltA transcription start point (lane 1) was incubated with increasing concentrations of purified TnrA (lanes 2 to 10). The TnrA concentrations were 5.5 (lane 2), 11 (lane 3), 22 (lane 4), 44 (lane 5), 88 (lane 6), 175 (lane 7), 350 (lane 8), 700 (lane 9), and 1,400 nm (lane 10).
At high concentrations of TnrA, additional complexes of lower mobility were seen (Fig. 4), suggesting that TnrA is able to interact with the gltC-gltAB regulatory region at several low-affinity binding sites not detected by footprinting (see below).
Footprinting of the TnrA binding site.
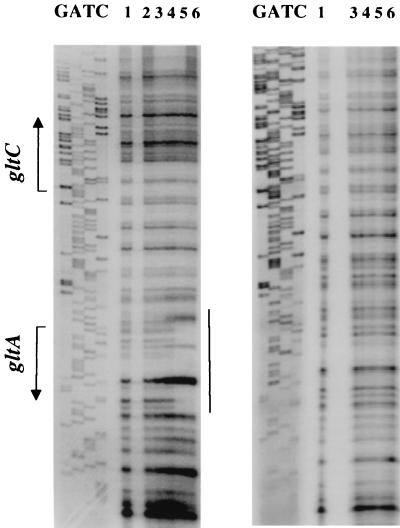
A DNase I protection experiment identified nucleotides from positions −2 to +28 with which TnrA appeared to interact (Fig. 5). Three hypersensitive sites in this region are separated from each other by approximately one helical turn of DNA (11 bp). No protection was observed for a similar DNA fragment containing the gltAp24 double mutation in the apparent TnrA binding site (Fig. 5).
FIG. 5.
DNase I footprinting analysis of TnrA binding to the gltA promoter. The 346-bp gltAp+ (left) and gltAp24 (right) promoter fragments, labeled on the nontemplate strand, were incubated with purified TnrA and then with DNase I. The sequence of the nontemplate strand of pIPC119 (2) determined using oBB13 as a primer is shown on the left. The apparent transcription start sites of gltA and gltC and the directions of transcription are shown by the bent arrows. The region of interaction with TnrA is indicated by a vertical line. Lanes 1, no TnrA; lane 2, 1.2 nM TnrA; lanes 3, 4.6 nM TnrA; lanes 4, 18.4 nM TnrA; lanes 5, 73.8 nM TnrA; lanes 6, 295 nM TnrA. The righthand panel has no lane corresponding to lane 2.
DISCUSSION
Many genes of nitrogen metabolism in B. subtilis are subject to regulation by multiple proteins. The glnRA operon is repressed by both GlnR and TnrA (35, 40), the gabP gene is activated by TnrA and repressed by CodY (13), the ureABC operon is activated by TnrA and repressed by GlnR and CodY (39), the roc regulon is activated by both RocR and AhrC (4, 20, 25), and, the bkd operon is activated by BkdR and repressed by CodY (12). Other genes of nitrogen metabolism, such as the dpp and hut operons, in addition to having nitrogen-specific regulators, are regulated by AbrB, a global regulator of many B. subtilis genes (18, 36). The present work reveals that the gltAB operon is both activated by GltC and repressed by TnrA.
Most genes in the TnrA regulon are positively regulated by this protein. The products of most of these genes are involved in obtaining ammonium directly from the medium or from less rapidly metabolized nitrogen sources. Only two operons, glnRA and gltAB, are known to be repressed by TnrA. It is interesting that TnrA stimulates the intracellular accumulation of ammonium while repressing the only enzymes that are involved in assimilation of ammonium. While the rates of expression of the key ammonium assimilation genes are determined by the extent to which GltC, TnrA, and GlnR are active as transcriptional regulators, the targets of these proteins are only partially overlapping. That is, TnrA regulates both glnRA (40) and gltAB (this paper), but GlnR has no detectable effect on gltAB expression and no effect of GltC on glnRA expression has been observed (unpublished results).
The nutritional environment determines the activities of these regulatory proteins. When cells in glucose-containing medium are provided with glutamine, the favored nitrogen source, GlnR is active, strongly repressing glnRA transcription (35). GltC is also active under these conditions, stimulating transcription of gltAB to a moderately high level (10), even though glutamine is an adequate source of glutamate. TnrA seems to be inactive in cells in glucose-glutamine medium and therefore plays no role in establishing the levels of gltAB or glnRA expression in such cells (15).
When cells are grown in glucose-ammonium medium, they need to have both glutamine synthetase and glutamate synthase activities and therefore express both the glnRA and gltAB operons at a higher level than in glutamine medium. While GltC stimulates gltAB transcription and partially active GlnR represses glnRA to a moderate extent in cells grown in glucose-ammonium medium, TnrA seems to be inactive in such cells.
When cells are grown in glucose-glutamate medium, glutamate synthase activity should be superfluous, because extracellular glutamate is apparently transported well. (A glutamate auxotroph grows at the wild-type rate in glucose-glutamate medium, indicating that glutamate uptake is not limiting. Moreover, derepression of gltAB expression due to a tnrA mutation does not improve the growth rate of cells in glucose-glutamate medium.) Under these conditions, gltAB expression is almost completely shut down by TnrA (this paper), overcoming high activity of GltC (unpublished results).
Despite the efficient transport of glutamate, it is a poor source of ammonium under the growth conditions used here, because the ammonium-liberating enzyme, glutamate dehydrogenase (encoded by rocG), is strongly repressed by glucose and requires ornithine for induction (6). Therefore, glutamine synthetase activity, which is needed for glutamine synthesis, is highly derepressed in cells grown in glucose-glutamate medium. High expression of glutamine synthetase presumably allows the enzyme to utilize the low level of available ammonium, which is limiting for growth. Under these conditions, GlnR is inactive but TnrA is active, serving to dampen the maximal level of glnRA expression by about fourfold (40), perhaps to avoid squandering ATP in the glutamine synthetase reaction. Thus, in cells grown in glucose-glutamate medium, TnrA acts at glnRA as a fine-tuning mechanism for modulating gene expression, while it acts as a major regulator of gltAB expression.
Repression of the gltAB operon by TnrA in cells grown in glucose-glutamate medium achieves two metabolic functions. First, it stops the unneeded synthesis of glutamate, thereby conserving the important metabolites 2-ketoglutarate, glutamine, and NADPH. This is likely to be a major contribution of TnrA to the cellular economy, since synthesis of glutamate, the cell's principal anion and principal donor of nitrogen groups, occupies a significant fraction of cellular metabolism. Second, the conservation of glutamine spares the requirement for ammonium, the limiting factor for growth in glucose-glutamate medium, and allows more glutamine to be available as a precursor for the synthesis of other compounds. The same physiological goals are achieved by TnrA-dependent repression of gltAB in the presence of proline, a good source of glutamate (Tables 4 and 5) (2).
If glutamate and ammonium or proline and ammonium are provided together, expression of the gltAB operon is not repressed by TnrA (Table 2, Fig. 2, and data not shown), even though there is no apparent need for glutamate synthase activity (as is the case in glucose-glutamine medium). It seems that TnrA does not respond to glutamate availability per se but rather to some other indicator of nitrogen availability. Perhaps the primary role of TnrA is to respond to ammonium or glutamine availability by activating genes whose products could provide ammonium and by repressing glutamate synthase in order to spare glutamine.
What, then, is the role of GltC in gltAB expression? GltC is clearly a positive regulator in whose absence so little expression of gltAB occurs under any growth conditions that the cells are auxotrophic for glutamate. But the existence of mutant forms of GltC that are partially constitutively active (5) suggests that its activity is subject to regulation. Although GltC is active in cells in glucose-glutamate medium, we have recently discovered (unpublished data) that GltC activity is greatly reduced when the medium contains arginine-related amino acids that are substrates of the RocR-dependent degradative pathway leading to glutamate and to its dissimilation to ammonium and 2-ketoglutarate (6, 20).
Our in vivo experiments indicate that GltC and TnrA interact with the gltCA regulatory region independently. TnrA is able to repress gltAB both in the presence and absence of GltC or its putative binding sites, and GltC is required for gltAB expression both in the presence and absence of TnrA. In addition, the apparent binding sites for the two proteins in the gltAB regulatory region are far apart. In the case of the gltAB promoter, the TnrA binding site overlaps with the transcription start site, consistent with simple mechanisms of repression, while GltC appears to bind upstream of the −35 region (2). A more complicated arrangement of TnrA and GlnR binding sites is found in the glnRA promoter region (11, 15, 21). One of these sites overlaps the −35 region of the promoter (11, 21). Genes activated by TnrA have TnrA binding sites located upstream of their promoters (15).
Interestingly, glutamate synthase expression is subject to various modes of regulation in different bacteria, reminiscent of different modes of regulation for the other enzyme of ammonium assimilation, glutamine synthetase (15, 29). In E. coli, expression of the glutamate synthase genes is positively regulated by the leucine responsive protein, LRP (38). In Klebsiella aerogenes, Nac, a distant relative of GltC, is a negative regulator of the glutamate synthase operon; expression of Nac itself is positively regulated by the NtrC system of nitrogen regulation (7). In addition, K. aerogenes LRP regulates glutamate synthase expression in the same way as in E. coli (22). This dual control by positive and negative regulators is formally analogous to the combined effects of GltC and TnrA on the B. subtilis gltAB operon, although the signals that control the activities of the regulators are different in gram-positive and gram-negative bacteria.
ACKNOWLEDGMENTS
We are grateful to K. Matsuno and M. Nakano for many helpful discussions, to M. Nakano for gifts of strains, and to M. Berne, Tufts University Protein and Nucleic Acid Analysis Facility, for synthesis of oligonucleotides.
This work was supported by grants from the U.S. Public Health Service to A.L.S. (GM36718) and to S.H.F. (GM51127) and from the Charlton Fund of Tufts University School of Medicine to B.R.B.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky B R, Janssen P J, Sonenshein A L. Sites required for GltC-dependent regulation of Bacillus subtilis glutamate synthase expression. J Bacteriol. 1995;177:5686–5695. doi: 10.1128/jb.177.19.5686-5695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky B R, Sonenshein A L. Altered transcription activation specificity of a mutant form of Bacillus subtilis GltR, a LysR family member. J Bacteriol. 1997;179:1035–1043. doi: 10.1128/jb.179.4.1035-1043.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belitsky B R, Sonenshein A L. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc Natl Acad Sci USA. 1999;96:10290–10295. doi: 10.1073/pnas.96.18.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belitsky B R, Sonenshein A L. Mutations in GltC that increase Bacillus subtilis gltA expression. J Bacteriol. 1995;177:5696–5700. doi: 10.1128/jb.177.19.5696-5700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belitsky B R, Sonenshein A L. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol. 1998;180:6298–6305. doi: 10.1128/jb.180.23.6298-6305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender R A. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol Microbiol. 1991;5:2575–2580. doi: 10.1111/j.1365-2958.1991.tb01965.x. [DOI] [PubMed] [Google Scholar]
- 8.Bohannon D E. Regulation of glutamate synthase expression. Ph.D. thesis. Boston, Mass: Tufts University; 1989. [Google Scholar]
- 9.Bohannon D E, Rosenkrantz M S, Sonenshein A L. Regulation of Bacillus subtilis glutamate synthase genes by the nitrogen source. J Bacteriol. 1985;163:957–964. doi: 10.1128/jb.163.3.957-964.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohannon D E, Sonenshein A L. Positive regulation of glutamate biosynthesis in Bacillus subtilis. J Bacteriol. 1989;171:4718–4727. doi: 10.1128/jb.171.9.4718-4727.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown S W, Sonenshein A L. Autogenous regulation of the Bacillus subtilis glnRA operon. J Bacteriol. 1996;178:2450–2454. doi: 10.1128/jb.178.8.2450-2454.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debarbouille M, Gardan R, Arnaud M, Rapoport G. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol. 1999;181:2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferson A E, Wray L V, Jr, Fisher S H. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol Microbiol. 1996;22:693–701. doi: 10.1046/j.1365-2958.1996.d01-1720.x. [DOI] [PubMed] [Google Scholar]
- 14.Fisher S H. Isolation and characterization of glutamine requiring mutants of Bacillus subtilis SMY. Ph.D. Thesis. Boston, Mass: Tufts University; 1978. [Google Scholar]
- 15.Fisher S H. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol Microbiol. 1999;32:223–232. doi: 10.1046/j.1365-2958.1999.01333.x. [DOI] [PubMed] [Google Scholar]
- 16.Fisher S H, Rosenkrantz M S, Sonenshein A L. Glutamine synthetase gene of Bacillus subtilis. Gene. 1984;32:427–438. doi: 10.1016/0378-1119(84)90018-0. [DOI] [PubMed] [Google Scholar]
- 17.Fisher S H, Sonenshein A L. Bacillus subtilis glutamine synthetase mutants pleiotropically altered in glucose catabolite repression. J Bacteriol. 1984;157:612–621. doi: 10.1128/jb.157.2.612-621.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher S H, Strauch M A, Atkinson M R, Wray L V., Jr Modulation of Bacillus subtilis catabolite repression by transition state regulatory protein AbrB. J Bacteriol. 1994;176:1903–1912. doi: 10.1128/jb.176.7.1903-1912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouet A, Sonenshein A L. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J Bacteriol. 1990;172:835–844. doi: 10.1128/jb.172.2.835-844.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardan R, Rapoport G, Debarbouille M. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol Microbiol. 1997;24:825–837. doi: 10.1046/j.1365-2958.1997.3881754.x. [DOI] [PubMed] [Google Scholar]
- 21.Gutowski J C, Schreier H J. Interaction of the Bacillus subtilis glnRA repressor with operator and promoter sequences in vivo. J Bacteriol. 1992;174:671–681. doi: 10.1128/jb.174.3.671-681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janes B K, Bender R A. Two roles for the leucine-responsive regulatory protein in expression of the alanine catabolic operon (dadAB) in Klebsiella aerogenes. J Bacteriol. 1999;181:1054–1058. doi: 10.1128/jb.181.3.1054-1058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 24.Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller C M, Baumberg S, Stockley P G. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol Microbiol. 1997;26:37–48. doi: 10.1046/j.1365-2958.1997.5441907.x. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 27.Nakano M M, Hoffmann T, Zhu Y, Jahn D. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J Bacteriol. 1998;180:5344–5350. doi: 10.1128/jb.180.20.5344-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano M M, Yang F, Hardin P, Zuber P. Nitrogen regulation of nasA and the nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. J Bacteriol. 1995;177:573–579. doi: 10.1128/jb.177.3.573-579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 391–407. [Google Scholar]
- 30.Robichon D, Arnaud M, Gardan R, Pragai Z, O'Reilly M, Rapoport G, Débarbouillé M. Expression of a new operon from Bacillus subtilis, ykzB-ykoL, under the control of the TnrA and PhoP-PhoR global regulators. J Bacteriol. 2000;182:1226–1231. doi: 10.1128/jb.182.5.1226-1231.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 34.Schreier H J. Biosynthesis of glutamine and glutamate and assimilation of ammonia. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 281–298. [Google Scholar]
- 35.Schreier H J, Brown S W, Hirschi K D, Nomellini J F, Sonenshein A L. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J Mol Biol. 1989;210:51–63. doi: 10.1016/0022-2836(89)90290-8. [DOI] [PubMed] [Google Scholar]
- 36.Slack F J, Mueller J P, Strauch M A, Mathiopoulos C, Sonenshein A L. Transcriptional regulation of a Bacillus subtilis dipeptide transport operon. Mol Microbiol. 1991;5:1915–1925. doi: 10.1111/j.1365-2958.1991.tb00815.x. [DOI] [PubMed] [Google Scholar]
- 36a.Steinmetz M, Richter R. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene. 1994;142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Grau R, Perego M, Hoch J A. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 1997;11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiese D E, II, Ernsting B R, Blumenthal R M, Matthews R G. A nucleoprotein activation complex between the leucine-responsive regulatory protein and DNA upstream of the gltBDF operon in Escherichia coli. J Mol Biol. 1997;270:152–168. doi: 10.1006/jmbi.1997.1057. [DOI] [PubMed] [Google Scholar]
- 39.Wray L V, Jr, Ferson A E, Fisher S H. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J Bacteriol. 1997;179:5494–5501. doi: 10.1128/jb.179.17.5494-5501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wray L V, Jr, Ferson A E, Rohrer K, Fisher S H. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:8841–8845. doi: 10.1073/pnas.93.17.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wray L V, Jr, Zalieckas J M, Ferson A E, Fisher S H. Mutational analysis of the TnrA-binding sites in the Bacillus subtilis nrgAB and gabP promoter regions. J Bacteriol. 1998;180:2943–2949. doi: 10.1128/jb.180.11.2943-2949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wray L V, Jr, Zalieckas J M, Fisher S H. Purification and in vitro activities of the Bacillus subtilis TnrA transcription factor. J Mol Biol. 2000;300:29–40. doi: 10.1006/jmbi.2000.3846. [DOI] [PubMed] [Google Scholar]
- 43.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]