ABSTRACT
The SARS-CoV-2 Omicron variants were first detected in November 2021, and several Omicron lineages (BA.1, BA.2, BA.3, BA.4, and BA.5) have since rapidly emerged. Studies characterizing the mechanisms of Omicron variant infection and sensitivity to neutralizing antibodies induced upon vaccination are ongoing by several groups. In the present study, we used pseudoviruses to show that the transmembrane serine protease 2 (TMPRSS2) enhances infection of BA.1, BA.1.1, BA.2, and BA.3 Omicron variants to a lesser extent than ancestral D614G. We further show that Omicron variants have higher sensitivity to inhibition by soluble angiotensin-converting enzyme 2 (ACE2) and the endosomal inhibitor chloroquine compared to D614G. The Omicron variants also more efficiently used ACE2 receptors from 9 out of 10 animal species tested, and unlike the D614G variant, used mouse ACE2 due to the Q493R and Q498R spike substitutions. Finally, neutralization of the Omicron variants by antibodies induced by three doses of Pfizer/BNT162b2 mRNA vaccine was 7- to 8-fold less potent than the D614G. These results provide insights into the transmissibility and immune evasion capacity of the emerging Omicron variants to curb their ongoing spread.
IMPORTANCE The ongoing emergence of SARS-CoV-2 Omicron variants with an extensive number of spike mutations poses a significant public health and zoonotic concern due to enhanced transmission fitness and escape from neutralizing antibodies. We studied three Omicron lineage variants (BA.1, BA.2, and BA.3) and found that transmembrane serine protease 2 has less influence on Omicron entry into cells than on D614G, and Omicron exhibits greater sensitivity to endosomal entry inhibition compared to D614G. In addition, Omicron displays more efficient usage of diverse animal species ACE2 receptors than D614G. Furthermore, due to Q493R/Q498R substitutions in spike, Omicron, but not D614G, can use the mouse ACE2 receptor. Finally, three doses of Pfizer/BNT162b2 mRNA vaccination elicit high neutralization titers against Omicron variants, although the neutralization titers are still 7- to 8-fold lower those that against D614G. These results may give insights into the transmissibility and immune evasion capacity of the emerging Omicron variants to curb their ongoing spread.
KEYWORDS: SARS-CoV-2 Omicron variants, virus entry pathways, transmembrane serine protease 2, animal ACE2 receptors, vaccine booster, breakthrough infections, neutralization resistance
INTRODUCTION
Since the origin of the COVID-19 pandemic, several SARS-CoV-2 variants of concern (VOCs) with enhanced transmissibility, ACE2 binding affinity, and immune-evasive properties have emerged, including the most recent Omicron VOCs. Currently, Omicron (B.1.1.529) is comprised of five main lineages designated BA.1 (and its sublineage BA.1.1), BA.2 (and its sublineages BA.2.12.1 and BA.2.75), BA.3, BA.4, and BA.5. BA.1 was first identified in November 2021 in Botswana, and it rapidly replaced the then dominant Delta (B.1.617.2) VOC to become globally prevalent due to its enhanced transmissibility and ability to evade antibody neutralization (1–6). By early 2022, an alarming rise of BA.2 was seen in several parts of the world, leading to the replacement of BA.1 and BA.1.1. In comparison to BA.1, BA.2 was demonstrated to have faster replication kinetics, enhanced fusogenicity, and greater pathogenicity in hamsters (7, 8). BA.4 and BA.5 are two new lineages that are presently emerging in South Africa.
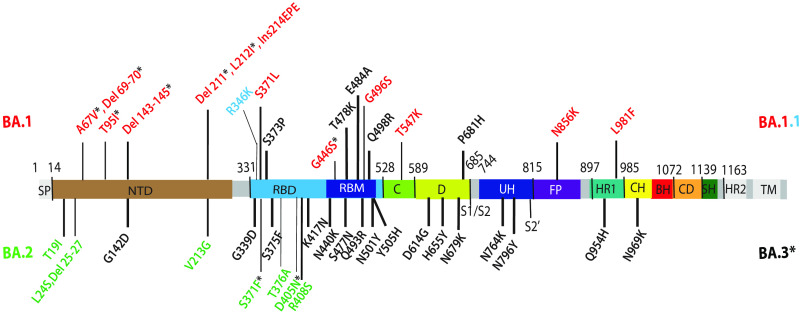
The spike protein of Omicron variants bears an unprecedented degree of antigenic divergence with the highest number of substitutions compared to ancestral B.1 (Wuhan-Hu-1 and D614G) variants and earlier VOCs. These include 21 spike substitutions shared by the three main lineages of Omicron in the N-terminal domain (NTD) (G142D), receptor binding domain (RBD) (G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H), and furin cleavage site proximity (D614G, H655Y, N679K, P681H) of the S1 subunit, as well as substitutions in fusion peptide proximity (N764K, D796Y) and heptad repeat region 1 (HR1) (Q954H, N969K) of the S2 subunit (Fig. 1). Additionally, 16 unique insertions/deletions/substitutions in BA.1 and 9 unique insertions/substitutions in BA.2 are present. BA.3 shares 10 unique substitutions/deletions with BA.1 and two unique substitutions with BA.2. BA.1.1 differs from BA.1 by one RBD substitution (R346K).
FIG 1.
Omicron lineage substitutions in spike. Substitutions in Omicron spikes are shown in a primary structure of the SARS-CoV-2 spike protein, with various domains and cleavage sites indicated. SP, signal peptide; NTD, N-terminal domain; RBD, receptor binding domain; RBM, receptor binding motif; C, domain C; D, domain D; S1/S2, furin cleavage junction of S1/S2 subunits; UH, upstream helix; FP, fusion peptide; HR1/2, heptad repeat 1/2; CH, central helix; BH, beta hairpin; CD, connector domain; SH, stem helix; TM, transmembrane domain. Substitutions common to Omicron (BA) are shown in black. Substitutions unique to BA.1 are shown in red. R346K substitution additionally present in BA.1.1 is shown in light blue. Substitutions unique to BA.2 are shown in green. Substitutions in BA.1 and BA.2 shared by BA.3 are shown as residues marked with an asterisk.
Several reports continue to demonstrate total loss of or severely dampened neutralizing activity of serum or plasma obtained from previously convalesced individuals, recipients of two doses of the Pfizer-BioNTech BNT162b2 or Moderna mRNA-1273 vaccine, as well as several therapeutic monoclonal antibodies against the BA.1 and BA.2 VOCs (1, 3, 9–14). A third booster dose of either Pfizer-BioNTech BNT162b2 or Moderna mRNA-1273 vaccine, however, recalls and expands preexisting antigen-specific memory B cell clones and generates novel B cell clones resulting in enhanced neutralizing antibody titers and breadth toward BA.1 and BA.2 VOCs (15, 16). Booster-elicited neutralizing antibody titers remain durable for at least 4 months (17, 18).
Apart from humans, SARS-CoV-2 was also found to naturally infect diverse domestic and wild animal species, including farm minks (19, 20), companion pets (e.g., cats, dogs, ferrets, Syrian hamsters) (21–23), zoo animals (e.g., lions, tigers, cougars, snow leopards, gorillas, otters, hippopotami) (24), and free-ranging white-tailed deer (25). Furthermore, experimental infections in livestock species have determined low-level replication of ancestral variants in cattle (26, 27), pigs (28), and sheep (27, 29). These observations signify the broad host range of SARS-CoV-2 and the risk of infection by heavily mutated variants to give rise to potential reservoirs that may pose a further risk for spillover back to humans.
The primary genetic determinant of SARS-CoV-2 host range is spike interaction with a species-specific receptor to allow subsequent viral entry. SARS-CoV-2 enters host cells by interacting with angiotensin-converting enzyme 2 (ACE2) in a species-specific manner. For instance, ancestral SARS-CoV-2 spike does not interact with murine ACE2, and therefore, human ACE2 (hACE2) transgenic murine models (30–32) or mouse-adapted SARS-CoV-2 (33) were developed to study SARS-CoV-2 pathogenesis. The trimeric spike is a class I fusion protein that is cleaved into the S1 and S2 subunits, which are noncovalently associated on the surface of virions. Following the interaction of the RBD (residues 331 to 528) of the S1 subunit with the N terminus of ACE2 and further proteolytic processing of the S2 subunit at the S2′ site by the host proteases (cathepsin L in endosomes, or TMPRSS2 on the plasma membrane), extensive and irreversible conformational changes occur in the S2 subunit to facilitate membrane fusion. The insertion of S2 fusion peptide in the target cell membrane and the interaction between HR1 and HR2 of the S2 subunit result in the formation of a stable six-helix bundle that brings the viral and cell membranes into proximity for fusion and subsequent viral entry.
Here, we report that compared to ancestral D614G, Omicron variants’ (BA.1, BA.1.1, BA.2, and BA.3) infection is less influenced by TMPRSS2, and Omicron variants are therefore relatively more susceptible to endosomal entry inhibition. Additionally, Omicron variants exhibit greater sensitivity to soluble ACE2 (sACE2) inhibition. Furthermore, Omicron variants’ spikes have more efficient usage of ACE2 orthologs from nine diverse animal species than D614G. In addition, we found that Q493R and Q498R substitutions in the Omicron variant spikes promote usage of mouse ACE2 for entry, whereas the Q493R substitution prevents usage of Chinese rufous horseshoe bat ACE2. Finally, sera obtained from fully vaccinated (three doses of the Pfizer/BNT162b2 vaccine) individuals and fully vaccinated individuals with a breakthrough infection of Omicron potently neutralize pseudoviruses bearing spike proteins of D614G and Omicron variants. However, neutralization titers against the Omicron variants are 7- to 8-fold lower than those of D614G. Our findings expand the current knowledge that Omicron variants BA.1, BA.1.1, BA.2, and BA.3 show less TMPRSS2 dependence and greater resistance to vaccine-induced antibody neutralization than D614G. However, sensitivity to sACE2 is 3-fold higher for BA.1, BA.1.1, and BA.2, whereas BA.3 is relatively less sensitive (1.75-fold) than D614G.
RESULTS
Infectivity and endosomal entry of Omicron variant pseudoviruses.
Pseudoviruses bearing spike proteins of BA.1, BA.1.1, BA.2, and BA.3 Omicron variants displayed similar infectivity as D614G in 293T-ACE2 and 293T-ACE2-TMPRSS2 cells (Fig. 2A). The presence of TMPRSS2 resulted in a 2.4- to 14.2-fold enhancement of infection for all the pseudoviruses in 293T-ACE2-TMPRSS2 cells compared to 293T-ACE2 cells. However, D614G pseudovirus infection was greatly enhanced (14.2-fold; P ≤ 0.0001) as observed previously (34), whereas infection with Omicron variants was comparatively less enhanced (2.4- to 4.9-fold; P ≤ 0.0001) in the presence of TMPRSS2 (Fig. 2A and B). These findings indicate that TMPRSS2 confers relatively less enhancement of Omicron entry into cells than D614G.
FIG 2.
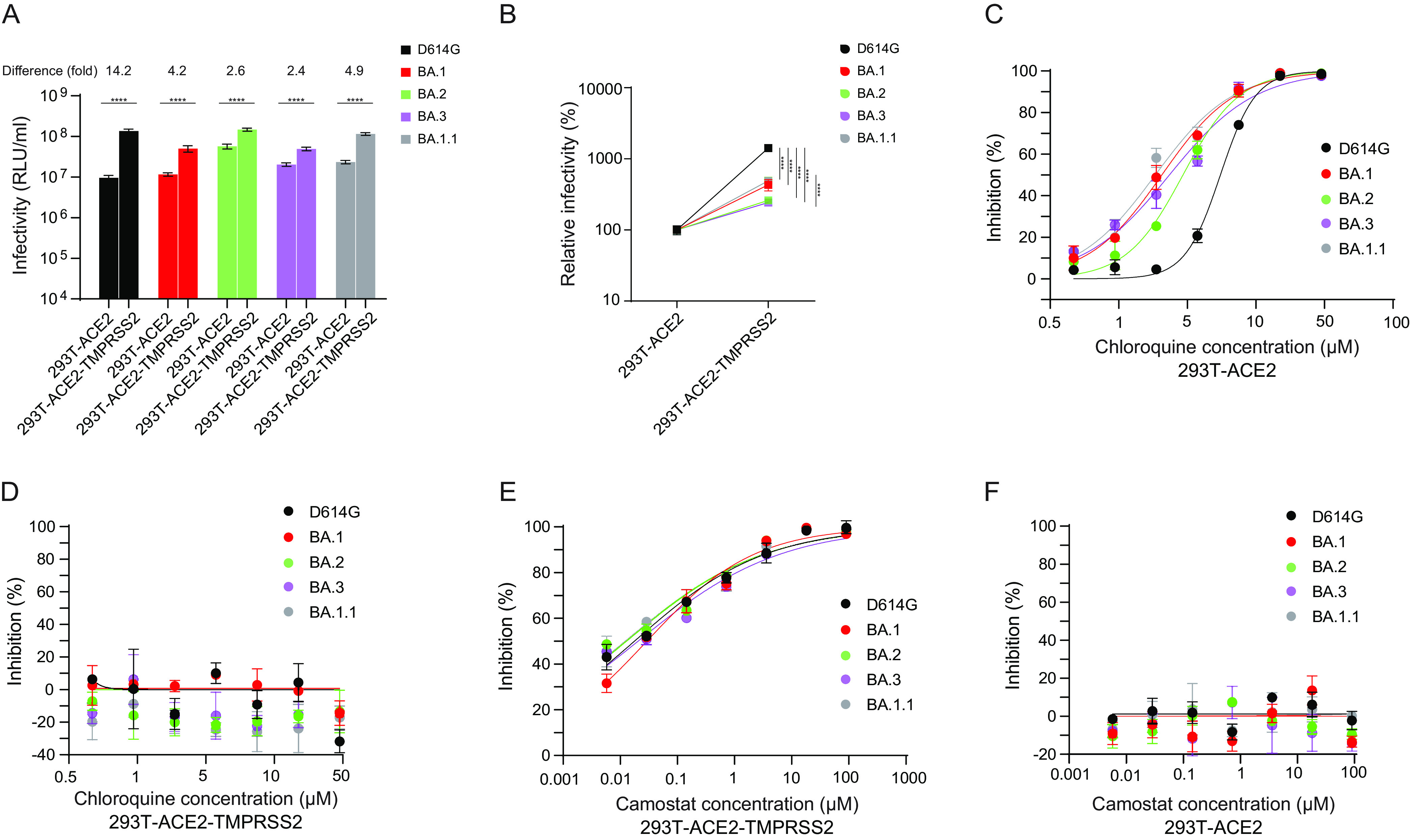
Infection and cell entry mechanisms of Omicron lineage pseudoviruses. Each cell type was infected with D614G and Omicron lineage pseudoviruses. (A) Infectivity of D614G and Omicron (BA) lineage pseudovirus stocks on 293T-ACE2 and 293T-ACE2-TMPRSS2 cells. (B) Relative infectivity of D614G and Omicron lineage pseudoviruses in stable 293T-ACE2 cells compared to stable 293T-ACE2-TMPRSS2 cells. (C and D) Chloroquine sensitivity of D614G and Omicron lineage pseudoviruses in stable 293T-ACE2 (C) and 293T-ACE2-TMPRSS2 cells (D). Cells were pretreated with the indicted concentration of chloroquine for 2 h prior to infections with pseudoviruses in medium containing the inhibitor. (E and F) Camostat sensitivity of D614G and Omicron lineage pseudoviruses in stable 293T-ACE2-TMPRSS2 (E) and 293T-ACE2 cells (F). Cells were pretreated with the indicted concentration of camostat mesylate for 2 h prior to infection with pseudoviruses in medium containing the inhibitor. The x axis indicates the concentration of inhibitor. The y axis indicates the percentage of inhibition compared to pseudovirus infection without inhibitor treatment. Results shown are the average of three independent experiments. Asterisks (*) denote significance: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Several recent studies showed delayed or attenuated replication of BA.1, BA.1.1, and BA.2 variants compared to B.1 and Delta VOCs in TMPRSS2-expressing cell lines (Calu-3, Caco-2, VeroE6/TMPRSS2) compared to VeroE6 cells, along with higher sensitivity to endosomal inhibitors (chloroquine, bafilomycin A) and less sensitivity to TMPRSS2 inhibitor (camostat mesylate) (6, 8, 35, 36). In cells expressing ACE2 but not TMPRSS2, spike-mediated entry follows the endosomal route, whereas in cells expressing both ACE2 and TMPRSS2, spike-mediated entry occurs mainly at the cell surface. We investigated ACE2 preference and TMPRSS2 usage by examining the sensitivity of D614G and the Omicron (BA.1, BA.1.1, BA.2, and BA.3) variant pseudoviruses to endosomal and TMPRSS2 inhibitors, chloroquine and camostat mesylate, respectively. In 293T-ACE2 cells, chloroquine potently inhibited viral entry of both D614G and Omicron variants, but Omicron pseudoviruses were more sensitive to chloroquine (BA.1 50% inhibitory concentration [IC50], 2.06 μM; P < 0.001; BA.2 IC50, 2.94 μM; P < 0.005; BA.3 IC50, 2.34 μM; P < 0.001; BA.1.1 IC50, 1.85 μM; P < 0.002) than D614G (IC50, 5.51 μM) (Fig. 2C). No chloroquine inhibition was observed in 293T-ACE2-TMPRSS2 cells (Fig. 2D), suggesting that the presence of TMPRSS2 facilitates entry via the cell surface for all variants. D614G (IC50, 0.02 μM) and Omicron (BA.1 IC50, 0.03 μM; BA.2 IC50, 0.01 μM; BA.3, 0.02 μM; BA.1.1 IC50, 0.01 μM) pseudoviruses displayed similar sensitivity to camostat mesylate in 293T-ACE2-TMPRSS2 cells (Fig. 2E). However, camostat mesylate had no effect on pseudovirus infections in 293T-ACE2 cells (Fig. 2F). These findings indicate increased sensitivity of Omicron pseudoviruses to endosomal entry inhibition compared to D614G.
Sensitivity of Omicron variants to soluble ACE2 neutralization.
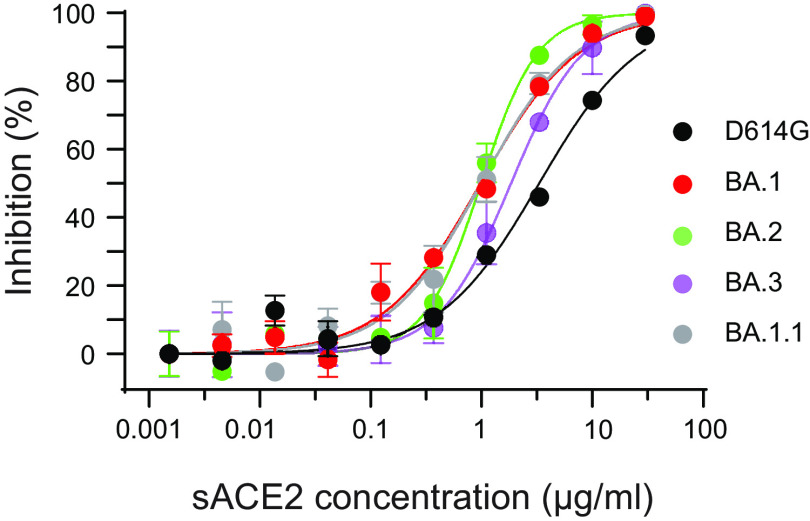
We further investigated ACE2 binding of Omicron variants by analyzing the sensitivity of D614G and Omicron pseudoviruses to soluble human ACE2 (sACE2) in a neutralization assay. Compared to D614G (IC50, 3.31 μg/mL), the BA.1 (IC50, 0.98 μg/mL; P < 0.0001), BA.2 (IC50, 0.99 μg/mL; P < 0.0004) and BA.1.1 (IC50, 0.99 μg/mL; P < 0.0005) variants demonstrated approximately 3-fold greater sensitivity to sACE2, whereas the BA.3 variant (IC50, 1.85 μg/mL; P < 0.0005) showed 1.75-fold greater sensitivity to sACE2 (Fig. 3). The sACE2 IC50 values reported here are comparable to previously reported values for D614G and BA.1 (37).
FIG 3.
The neutralization of soluble ACE2 to Omicron lineage pseudoviruses. The sensitivity of D614G and Omicron lineage pseudoviruses to soluble human ACE2 was evaluated on stable 293T-ACE2-TMPRSS2 cells. Results shown are representative of three independent experiments.
Thermal stability of Omicron variant pseudoviruses.
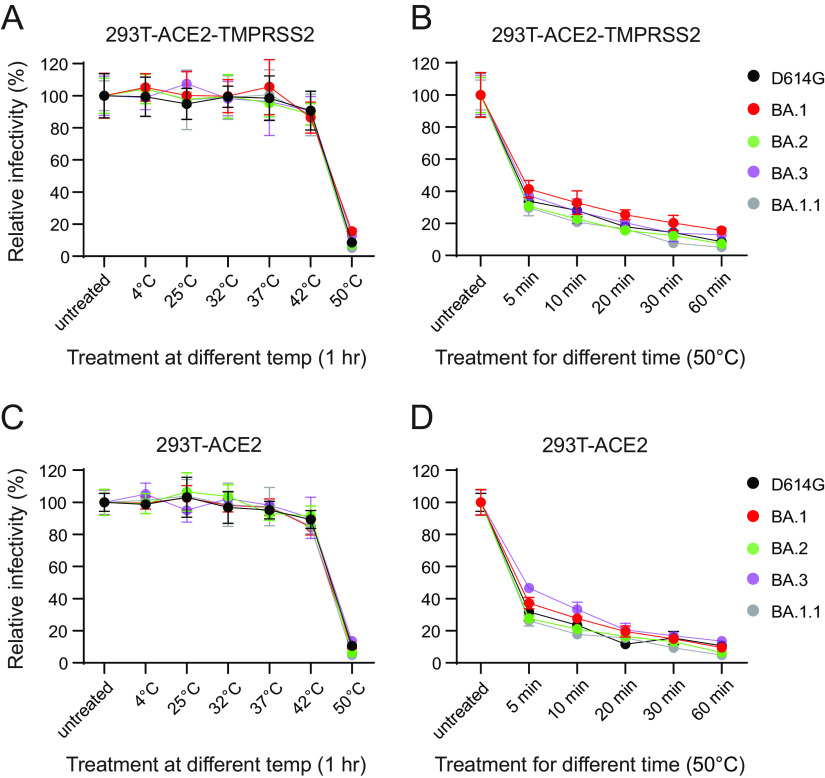
Preincubation of viruses at different temperatures can reveal differences in the stability of the viral glycoprotein trimers, including SARS-CoV-2 (37, 38). We evaluated the effect of temperature on spike glycoprotein stability, and thus infectivity, by incubating D614G and Omicron variant pseudoviruses at 4°C, 25°C (room temperature [RT]), 32°C, 37°C, 42°C, and 50°C for an hour or 50°C for various periods of time before measuring their infectivity on 293T-ACE2 and 293T-ACE2-TMPRSS2 cells. We observed no significant differences in the infectivity of D614G and Omicron variant pseudoviruses after incubation at 4°C, RT, 32°C, 37°C, and 42°C (Fig. 4). All viruses were relatively stable at 4°C, RT, 32°C, 37°C, and 42°C. Our findings are in agreement with a previous study where no differences in infectivity were observed between vesicular stomatitis virus (VSV)-pseudotyped D614G and BA.1 VOC pseudoviruses upon extended incubation (72 h) at 4°C, RT, and 37°C (37). However, infectivity of all pseudoviruses dramatically declined upon incubation at 50°C, with no significant difference between D614G and Omicron variant pseudoviruses (Fig. 4).
FIG 4.
Thermal stability of Omicron pseudoviruses. The temperature stability of D614G and Omicron lineage pseudoviruses on 293T-ACE2-TMPRSS2 and 293T-ACE2 cells was evaluated. (A and C) Pseudoviruses were untreated or subjected to various temperatures for an hour prior to infectivity on 293T-ACE2-TMPRSS2 (A) and 293T-ACE2 (C) cells. (B and D) Pseudoviruses were untreated or subjected to 50°C for different durations prior to infectivity on 293T-ACE2-TMPRSS2 (B) and 293T-ACE2 (D) cells. Results shown are the average of three independent experiments.
pH stability of Omicron variant pseudoviruses.
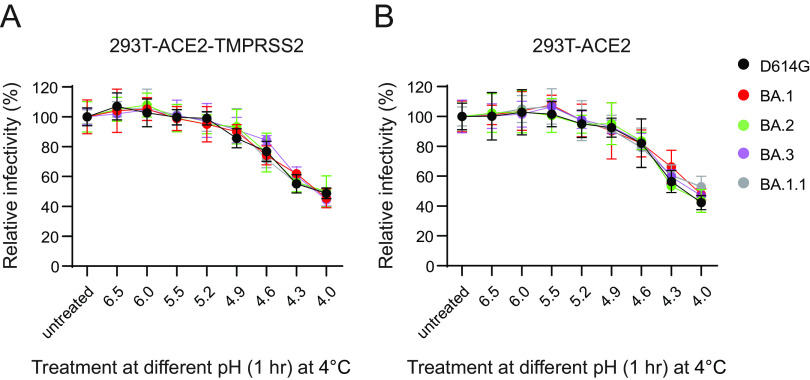
Since Omicron variants displayed greater sensitivity to endosomal inhibition than D614G, we hypothesized greater sensitivity or adaptation of Omicron variants to the lower-pH environment encountered in the endosomes. To address this possibility, we subjected D614G and Omicron pseudoviruses to different pH treatments prior to restoring pH to 7.0 and infecting 293T-ACE2 and 293-ACE2-TMPRSS2 cells. We noticed a comparable drop in infectivity between D614G and Omicron pseudoviruses at a lower pH starting at 4.9, and a much dramatic loss of infectivity was observed at pH 4.0 (Fig. 5A and B).
FIG 5.
pH stability of Omicron pseudoviruses. The pH stability of D614G and Omicron lineage pseudoviruses on 293T-ACE2-TMPRSS2 and 293T-ACE2 cells was evaluated. (A and B) Pseudoviruses were untreated or subjected to various pHs for an hour prior to adjusting the pH to 7.0 and infectivity on 293T-ACE2-TMPRSS2 (A) and 293T-ACE2 (B) cells. Results shown are the average of three independent experiments.
Omicron variants display distinct ACE2 receptor usage compared to D614G.
Substitutions in the RBD have been linked to SARS-CoV-2 adaptation to new hosts, such as ferret (39), mouse (33), mink (40), and white-tailed deer (41). The ongoing dominance and circulation of Omicron VOCs pose a significant risk for reverse zoonosis and spill-back into humans. Therefore, we investigated the ability of the Omicron (BA.1, BA.1.1, BA.2, and BA.3) variants to use ACE2 orthologs from 10 diverse host species, African green monkey, Chinese rufous horseshoe bat, ferret, mouse, Chinese hamster, Syrian golden hamster, white-tailed deer, swine, bovine, and Malayan pangolin. 293T cells transiently transfected with ACE2 receptors of each species were infected with pseudoviruses bearing spike proteins of D614G and the Omicron variants. We found that D614G pseudoviruses infected cells expressing ACE2 receptors of all the species well above the background except mouse, consistent with prior studies (3, 33, 42) (Fig. 6A). Conversely, ACE2 receptors of all species, except horseshoe bat, supported infection by Omicron pseudoviruses (Fig. 6B to E). 293T cells with stable expression of human ACE2 (293T-ACE2) were used as a positive control. We confirmed robust ACE2 expression for each species via Western blotting using the V5 tag at the C terminus of these ACE2 proteins (see Fig. S1 in the supplemental material). The Omicron pseudoviruses had significantly higher levels of infection than D614G pseudovirus in cells expressing African green monkey, ferret, mouse, Chinese hamster, Syrian golden hamster, white-tailed deer, swine, and bovine ACE2 receptors (Table 1). No significant difference between D614G and Omicron variants was observed with respect to Malayan pangolin ACE2 (Table 1). These findings indicate that the receptor binding motif (RBM) substitutions in the Omicron (BA.1, BA.1.1, BA.2, and BA.3) variants can modulate ACE2 receptor usage and, therefore, Omicron variants’ entry into cells.
FIG 6.
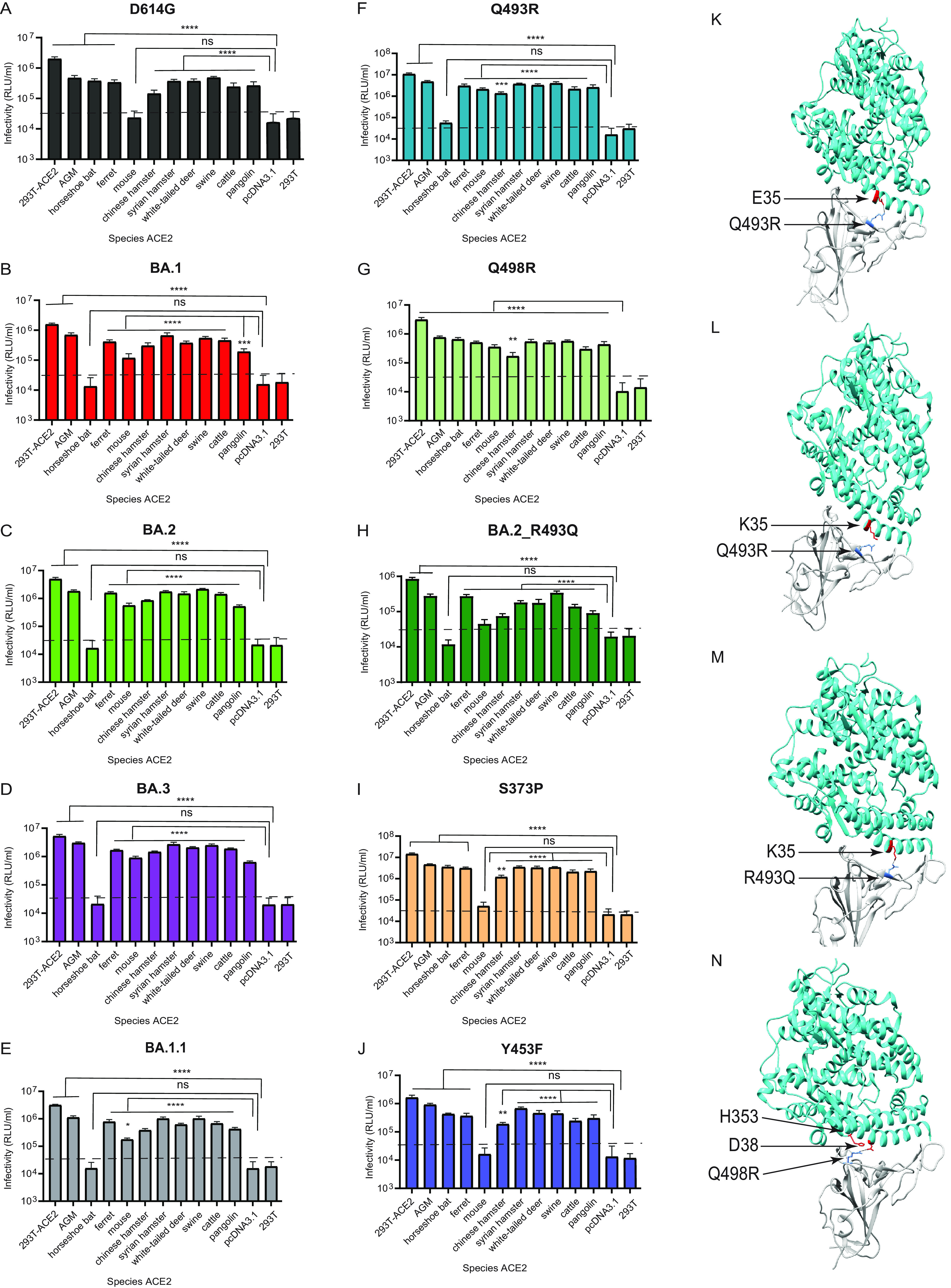
Omicron lineage pseudovirus entry into cells expressing ACE2 from different species. (A to J) Infectivity of D614G (A), BA.1 (B) BA.2 (C) BA.3 (D) BA.1.1 (E), Q493R (F), Q498R (G), BA.2_R493Q (H), S373P (I), and Y453F (J) pseudoviruses on 293T cells transiently transfected with ACE2 orthologs of the indicated species. The ACE2 of African green monkey is denoted as AGM. 293T cells expressing ACE2 of the indicated species, as well as control 293T-ACE2 cells stably expressing human ACE2, were simultaneously infected by the indicated pseudoviruses with titers of ~106 RLU/mL on 293T-ACE2 cells. Luciferase activities were determined 48 h postinfection. ns, not significant. Significant differences in infectivity between each species’ ACE2 compared to the pcDNA3.1 control for pseudoviruses are denoted by asterisks: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****,: P ≤ 0.0001. The dashed line indicates maximum background level infection on 293T cells. Results shown are the average of three independent experiments with eight intra-assay replicates. (K to N) The SARS-CoV-2 Omicron RBD-ACE2 interface (PDB: 7WBP) is shown with contacting residues as sticks at the RBD-ACE2 interface. The SARS-CoV-2 Omicron RBD and ACE2 are colored in gray and cyan, respectively. Positions in RBD (blue) that contact ACE2 (red) residues are highlighted. Residue positions are indicated by arrows. The SARS-CoV-2 RBD/ACE2 interactions between 493R/E35 (K), R493/K35 (L), Q493/K35 (M), and R498/D38-H353 (N) are shown.
TABLE 1.
Comparison of relative infectivity (versus infectivity on 293T-ACE2) of pseudoviruses for each species ACE2
| Species | Pseudovirus | Fold change compared to D614G | P valuea |
|---|---|---|---|
| African green monkey | D614G | 1 | |
| BA.1 | 1.86 | <0.0001**** | |
| BA.2 | 1.53 | 0.007** | |
| BA.3 | 2.41 | <0.0001**** | |
| BA.1.1 | 1.48 | 0.014* | |
| S373P | 1.34 | 0.08 | |
| Q493R | 1.85 | <0.0001**** | |
| Q498R | 1.04 | 0.8 | |
| BA.2_R493Q | 0.77 | 0.9 | |
| Y453F | 2.37 | <0.0001**** | |
| Chinese rufous horseshoe bat | D614G | 1 | |
| BA.1 | 0.01 | 0.018* | |
| BA.2 | 0.02 | 0.019* | |
| BA.3 | 0.02 | 0.019* | |
| BA.1.1 | 0.01 | 0.018* | |
| S373P | 1.27 | 0.509 | |
| Q493R | 0.09 | 0.03* | |
| Q498R | 1.12 | 0.8 | |
| BA.2_R493Q | 0.06 | 0.7 | |
| Y453F | 1.39 | 0.35 | |
| Ferret | D614G | 1 | |
| BA.1 | 1.55 | 0.0003*** | |
| BA.2 | 1.89 | <0.0001**** | |
| BA.3 | 1.87 | <0.0001**** | |
| BA.1.1 | 1.46 | 0.002** | |
| S373P | 1.27 | 0.064 | |
| Q493R | 1.65 | <0.0001**** | |
| Q498R | 0.98 | 0.875 | |
| BA.2_R493Q | 1.15 | 0.6 | |
| Y453F | 1.32 | 0.028* | |
| Mouse | D614G | 1 | |
| BA.1 | 6.55 | <0.0001**** | |
| BA.2 | 9.8 | <0.0001**** | |
| BA.3 | 14.96 | <0.0001**** | |
| BA.1.1 | 4.85 | <0.0001**** | |
| S373P | 0.61 | 0.611 | |
| Q493R | 16.95 | <0.0001**** | |
| Q498R | 9.88 | <0.0001**** | |
| BA.2_R493Q | 7.40 | 0.03* | |
| Y453F | 0.28 | 0.344 | |
| Chinese hamster | D614G | 1 | |
| BA.1 | 2.73 | <0.0001**** | |
| BA.2 | 2.42 | <0.0001**** | |
| BA.3 | 3.91 | <0.0001**** | |
| BA.1.1 | 1.72 | 0.004** | |
| S373P | 1.17 | 0.469 | |
| Q493R | 1.75 | 0.0025** | |
| Q498R | 0.78 | 0.367 | |
| BA.2_R493Q | 1.19 | 0.7 | |
| Y453F | 1.64 | 0.009** | |
| Syrian golden hamster | D614G | 1 | |
| BA.1 | 2.25 | <0.0001**** | |
| BA.2 | 1.89 | 0.0002*** | |
| BA.3 | 2.71 | <0.0001**** | |
| BA.1.1 | 1.7 | 0.003** | |
| S373P | 1.28 | 0.219 | |
| Q493R | 1.84 | 0.0004*** | |
| Q498R | 0.92 | 0.735 | |
| BA.2_R493Q | 0.89 | 0.5 | |
| Y453F | 2.22 | <0.0001**** | |
| White-tailed deer | D614G | 1 | |
| BA.1 | 1.28 | 0.072 | |
| BA.2 | 1.61 | 0.0002*** | |
| BA.3 | 2.13 | <0.0001**** | |
| BA.1.1 | 1.05 | 0.769 | |
| S373P | 1.2 | 0.193 | |
| Q493R | 1.62 | 0.0001*** | |
| Q498R | 0.88 | 0.426 | |
| BA.2_R493Q | 0.86 | 0. 83 | |
| Y453F | 1.52 | 0.001** | |
| Swine | D614G | 1 | |
| BA.1 | 1.43 | 0.0004*** | |
| BA.2 | 1.79 | <0.0001**** | |
| BA.3 | 1.96 | <0.0001**** | |
| BA.1.1 | 1.35 | 0.004** | |
| S373P | 0.97 | 0.821 | |
| Q493R | 1.51 | <0.0001**** | |
| Q498R | 0.75 | 0.134 | |
| BA.2_R493Q | 1.54 | 0.2 | |
| Y453F | 1.14 | 0.235 | |
| Bovine | D614G | 1 | |
| BA.1 | 2.36 | <0.0001**** | |
| BA.2 | 2.33 | <0.0001**** | |
| BA.3 | 2.89 | <0.0001**** | |
| BA.1.1 | 1.73 | 0.0011** | |
| S373P | 1.15 | 0.485 | |
| Q493R | 1.61 | 0.006** | |
| Q498R | 0.78 | 0.304 | |
| BA.2_R493Q | 1.17 | 0.45 | |
| Y453F | 1.24 | 0.266 | |
| Malayan pangolin | D614G | 1 | |
| BA.1 | 0.93 | 0.772 | |
| BA.2 | 0.81 | 0.423 | |
| BA.3 | 0.92 | 0.739 | |
| BA.1.1 | 1.02 | 0.917 | |
| S373P | 1.18 | 0.453 | |
| Q493R | 1.82 | 0.0007*** | |
| Q498R | 1.05 | 0.818 | |
| BA.2_R493Q | 1.06 | 0.56 | |
| Y453F | 1.39 | 0.095 |
P values were calculated by one-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison tests (Omicron [human ACE2 versus species ACE2] versus D614G [human ACE2 versus species ACE2]). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
We next investigated the residues in D614G and Omicron spikes that allow or prevent infection of cells expressing Chinese rufous horseshoe bat and mouse ACE2, respectively. Crystal structures of SARS-CoV-2 RBD-ACE2 complexes show the human ACE2 N-terminal helix cradled in the ridged concave surface formed by the spike RBM (43–46). A total of 17 RBM residues contact ACE2 residues in the N terminus (Table 2). Among wild-type SARS-CoV-2 RBM residues, Q498 interacts with D38, Y41, Q42, L45, and K353 residues of ACE2, while Q493 interacts with K31, H34, and E35 of ACE2, forming a network of hydrogen bonds (43–46). Structural and computational determination of Omicron RBD-ACE2 interactions revealed Q493R forming a new salt bridge with E35 while disrupting previous interactions of Q493 with K31 observed in the wild type. The Q498R substitution also forms a new hydrogen bond and a salt bridge with ACE2 D38 and Q42 (47–49).
TABLE 2.
ACE2 residues of species known to interact with SARS-CoV-2 spike receptor binding motifa
We assessed whether Q493R and Q498R substitutions in the RBM of Omicron spike facilitate entry of Omicron pseudoviruses in cells expressing mouse ACE2 or, conversely, confer resistance to entry into cells expressing Chinese rufous horseshoe bat ACE2. Pseudoviruses bearing Q493R and Q498R substitutions on the D614G background spike were therefore generated to infect 293T cells expressing ACE2 proteins of all the species described above. As expected, the Q493R substitution rescued pseudovirus infection in cells expressing mouse ACE2 but abolished pseudovirus infection in cells expressing horseshoe bat ACE2 (Table 1) (Fig. 6F). The Q498R substitution also rescued infection in cells expressing mouse ACE2 above the background but had no effect on cells expressing Chinese rufous horseshoe bat ACE2 (Table 1) (Fig. 6G).
We further examined the potential contribution of Q493R and Q498R in determining species tropism in the context of Omicron spike by reverting the R493 position of BA.2 spike to Q493 (BA.2_R493Q). The BA.2_R493Q pseudovirus efficiently used ACE2 receptors of all species, including mouse but not Chinese rufous horseshoe bat ACE2 (Table 1) (Fig. 6H). The usage of mouse ACE2 by BA.2_R493Q pseudovirus is anticipated due to the facilitation by the Q498R substitution. However, lack of Chinese rufous horseshoe bat ACE2 usage by BA.2_R493Q pseudovirus points to a role of other Omicron RBM mutations acting either independently or epistatically with Q493R to facilitate horseshoe bat ACE2 binding. However, in contrast to BA.2, BA.2_R493Q displayed reduced ACE2 utilization but similar efficiency as D614G and Q498R pseudoviruses for all the remaining species except Malayan pangolin (Table 1). These results suggest that Q493 plays an important role in BA.2 usage of ACE2 receptors belonging to different species. As controls, the Omicron non-RBM S373P and mink adapted Y453F substitutions had no effect on pseudovirus entry in cells expressing mouse ACE2 or horseshoe bat ACE2 (Table 1) (Fig. 6I and J).
It is likely that the Q493R basic substitution in Omicron VOCs forms a salt bridge with the acidic E35 residue of mouse ACE2, thus facilitating infection of cells expressing mouse ACE2. Conversely, the Q493R substitution on the D614G background likely disrupts interaction with the basic K35 residue of horseshoe bat ACE2, thus facilitating infection of cells expressing Chinese rufous horseshoe bat ACE2 (Table 2) (Fig. 6K and L). Alternatively, the K35 residue in Chinese rufous horseshoe bat ACE2 interacts with Q493 residue in D614G spike (Fig. 6M). On the other hand, substitutions similar to Q498R were previously observed in mouse-adapted SARS-CoV-2 (Q498H, Q498Y/P499T) to strengthen interaction with D38 of mouse ACE2, which otherwise forms an intramolecular salt bridge with H353 (50–52) (Table 2) (Fig. 6N). The interactions between R498 in Omicron spike and D38 and H353 in mouse ACE2 likely play an important role in infection.
For the remaining species, S373P, Y453F, Q493R, and Q498R substitutions were similar to those of D614G in ACE2 usage (Fig. 6I, J, F and G). However, the Q493R substitution significantly enhanced the usage of all the remainder species, highlighting the role played by this substitution in enhancing ACE2 usage of Omicron variants, as described above (Fig. 6G, Table 1). While S373P and Q498R substitutions had no significant effect compared to D614G, the mink-adapted Y453F substitution led to enhanced usage of African green monkey, ferret, Chinese hamster, Syrian golden hamster, and white-tailed deer ACE2 receptors (Table 1). A previous report has indicated that the Y453F substitution enhanced spike interaction with Mustela species ACE2 orthologs, including minks, ferrets, and stouts, while not compromising human ACE2 usage (Fig. 6J, Table 1) (40). Our findings extend this report and provide further insights into the effect of Y453F substitution for other species’ ACE2.
Enhanced neutralization of Omicron variants by vaccinee sera postbooster.
To assess potential antigenic differences among the Omicron variants, we next evaluated the serum neutralizing activity in serum from vaccinated individuals who received three immunizations (two-dose primary vaccine series and a third dose of booster) of the Pfizer/BNT162b2 vaccine. Receipt of a booster dose elicited significantly higher neutralization titers against D614G (geometric mean titers ([GMT], 7,527)) (Fig. 7A), compared to previously reported two-dose primary vaccine series (GMT, 1,310) (53). Using the same serum samples, we assessed neutralizing activity against the three Omicron variant pseudoviruses. Neutralization titers against BA.1 (GMT, 1,087), BA.2 (GMT, 961), BA.3 (GMT, 916), and BA.1.1 (GMT, 982) pseudoviruses were significantly reduced, 6.9-fold (P ≤ 0.05), 7.8-fold (P ≤ 0.001), 8.2-fold (P ≤ 0.0001), and 7.7-fold (P ≤ 0.001), respectively, compared to those of D614G (GMT, 7,527) (Fig. 7A).
FIG 7.
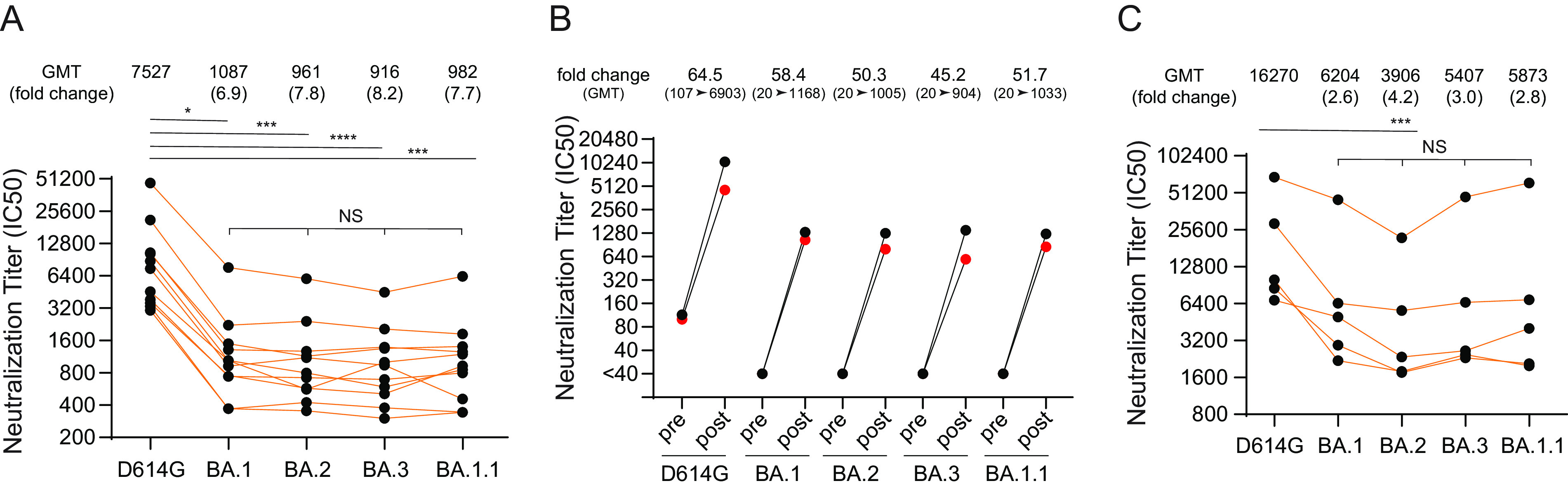
The sensitivity of Omicron lineage pseudoviruses to post-vaccine booster and vaccine breakthrough infection sera. (A) Neutralization of D614G and Omicron lineage pseudoviruses to vaccine booster-elicited sera obtained from individuals that received primary two-dose series and a third booster dose of Pfizer/BNT162b2 mRNA vaccine. (B) Sera neutralization titers pre- and post-booster receipt in individuals. (C) Titers of five vaccine breakthrough cases that experienced BA.1 or BA.1.1 infection. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. NS, not significant. Results shown are average two independent experiments.
For two of the vaccinated individuals described above, we obtained serum samples 1 week prior to a third booster dose, as well as 3 weeks after the third booster. We found 45.2- to 64.5-fold enhancement in postboost neutralization titer compared to preboost titer for D614G and all four Omicron variants (Fig. 7B).
We also evaluated convalescent-phase sera from five vaccinated individuals who experienced breakthrough infection postboost, when BA.1 or BA.1.1 variants were predominant (December 2021 to January 2022). Serum samples from all individuals had the highest neutralization activity against D614G pseudoviruses (GMT, 16,270) followed by lower neutralization against BA.1 (GMT, 6,204), BA.2 (GMT, 3,906), BA.3 (GMT, 5,407), and BA.1.1 (GMT, 5,873) variant pseudoviruses (Fig. 7C). Although the sample size is small, the results show similar resistance among these Omicron variants compared to D614G.
DISCUSSION
In the present study, we compared four Omicron lineage (BA.1, BA.1.1, BA.2, and BA.3) variant spikes for entry pathway, receptor usage, temperature and pH stability, and immune evasiveness properties. We found that TMPRSS2 enhances pseudovirus infection relatively for all four Omicron variants, but the enhancement is less than that observed for the D614G pseudovirus. In addition, compared to D614G, all Omicron lineage spikes more efficiently used ACE2 receptors from diverse animal species, including mouse ACE2 but not Chinese rufous horseshoe bat ACE2, for virus entry. Finally, all four Omicron variants had similar stability and immune-evasive properties in serum from individuals who received three doses of the Pfizer/BNT162b2 vaccine. The neutralization titers against Omicron were 7- to 8-fold lower than those against D614G.
Several recent studies demonstrated less enhancement of virus entry by TMPRSS2 for BA.1 pseudovirus infection compared to D614G and Delta pseudoviruses in ACE2-TMPRSS2-expressing cell lines (Calu3, Caco2, VeroE6-TMPRSS2), while no infection differences were observed in cell lines expressing ACE2 but not TMPRSS2 (VeroE6, H1299, HeLa-, HEK293T- and A549-ACE2) (6, 54, 55). Similar observations were reported in studies that performed authentic SARS-CoV-2 infections (6, 54, 55). Compared to D614G or the Delta variant, Omicron variants (BA.1, BA.1.1) had attenuated or inefficient replication in TMPRSS2-positive lower airway or gallbladder organoids, Calu3, and Caco2 lung cell lines but had no significant difference in cells with low or no TMPRSS2 expression (H1299, HeLa- and 293T-ACE2 cells) (6, 36, 55). Due to apparently less efficient use of TMPRSS2 compared to other variants, Omicron variants appear to favor the endocytic route of entry rather than TMPRSS2-mediated entry at the cell surface and consequently were more sensitive to endosomal inhibitors (chloroquine, bafilomycin A1 and E64d) (6, 35, 56). These findings and our findings together suggest that Omicron variants are better adapted to use the endosomal entry pathway in cells expressing only ACE2, though coexpression of TMPRSS2 could still enhance infection.
A possible mechanism that contributes to less efficient TMPRSS2 utilization by Omicron variants is the accumulation of multiple additional basic amino acid substitutions, leading to overall positively charged Omicron spike proteins compared to the D614G spike. These substitutions may confer greater sensitivity to low-pH-induced conformational changes in endosomes that could facilitate cleavage by cathepsin L or fusion-inducing conformational changes. Therefore, Omicron variants may be better adapted for entry in the low-pH environment encountered in the upper airway (6). While our temperature and pH treatment failed to demonstrate stability differences between Omicron variants and D614G, a recent preprint showed greater sensitivity of Omicron (BA.1, BA.2, and BA.4) pseudoviruses to endosomal entry inhibition (E64d) than pseudoviruses with D614G and Delta VOC spikes, but no inhibition by camostat in Caco-2 cells where both endosomal and TMPRSS2-mediated cell surface entry routes are active (57). This phenotype was attributed to the N969K S2 substitution in Omicron variants (56, 57). However, maximum likelihood fitting models comparing the entry routes of Omicron suggest that Omicron is also efficient in utilizing TMPRSS2 for entry into human nasal epithelial cells (57).
Alternatively, the BA.1 spike protein was shown to be less efficiently cleaved at the S1/S2 site compared to the wild type and Delta variant in authentic SARS-CoV-2-infected cells, as well as on virions (6, 7, 36, 55). Efficient cleavage at the S1/S2 site is known to be required for exposure of the S2′ site for TMPRSS2 processing after ACE2 binding. Substitutions proximal to the furin cleavage site (H655Y, N679K, and P681H), as well as substitutions in the S2 subunit, may reduce efficiency of TMPRSS2 cleavage of the Omicron spikes (36). Finally, higher affinities of BA.1 (6-fold) and BA.2 (11-fold) RBD for ACE2 coupled with transitions into the so called two- or three-RBD-up conformations may also contribute to the efficient use of ACE2 (48, 58, 59). In the present study, we report that BA.1, BA.2, and BA.1.1 were similarly inhibited by the soluble ACE2 monomer (3-fold versus D614G), while BA.3 and D614G were comparably less sensitive.
Our data further show that all four Omicron lineage spike proteins more efficiently use ACE2 receptors from diverse animal species for entry. Our findings showing Omicron lineage spikes’ use of mouse ACE2 for entry extend previous reports showing enhanced binding of BA.1 and BA.2 RBD to mouse ACE2 (3, 58). Similar (K417N, N501Y, Q493H/R) (33, 50, 60) or closely related (Q498H) (50, 61) mouse-adapted substitutions previously found in experimentally infected mice are observed in Omicron RBM that may contribute to binding to mouse ACE2. We show that Q493R and Q498R substitutions alone on the D614G background confer the ability to use mouse ACE2. On the other hand, the Omicron spike is unable to use Chinese rufous horseshoe bat (Rhinolophus sinicus) ACE2 for entry. While the Q493R substitution alone in the D614G background prevented Chinese rufous horseshoe bat ACE2 usage, a revertant R493Q substitution in the BA.2 spike background failed to rescue the infection of cells expressing Chinese rufous horseshoe bat ACE2, suggesting that other Omicron RBM substitutions play a role in determining horseshoe bat ACE2 use. These changes in species tropism are likely due to specific RBM-ACE2 interfacial residues that promote (R493-E35) or disrupt (R493-K35) interactions in mouse or horseshoe bat ACE2, respectively. In addition, the Q493R substitution also significantly enhanced the usage of all other species of ACE2, as demonstrated by Q493R and BA.2_R493Q substitution effects. Altogether, the spike substitutions in the BA.1, BA.1.1, BA.2, and BA.3 Omicron variants permit robust use of diverse ACE2 orthologues for entry and thus have the potential to broaden the risk of the Omicron variants to infect animal species and spill back to humans.
With the previous occurrences of SARS-CoV-2 interspecies transmission between humans and animals, including minks (62), pet hamsters (63), and several others (21–23), it has been hypothesized that the Omicron variants may have originated due to interspecies transmission between humans and rodents (3). While Omicron efficiently enters cells expressing mouse ACE2, a postentry block attenuates Omicron infection in laboratory mice, arguing against a mouse origin (3, 64, 65). Alternatively, prolonged and extensive replication in immunocompromised hosts may have given rise to Omicron as was seen for earlier variants (3).
Despite several amino acid differences in the spikes of the BA.1, BA.1.1, BA.2, and BA.3 variants, three immunizations with the Pfizer/BNT162b2 vaccine elicited high and comparable levels of neutralizing antibodies against BA.1, BA.1.1, BA.2, and BA.3 pseudoviruses, at least in the short term. Since three-dose mRNA vaccine-induced antibodies elicited robust neutralizing antibodies and were shown to protect against severe disease with BA.1 infection (1, 66), this protection likely extends to emerging BA.2 VOCs. These findings are consistent with several recent studies highlighting enhanced breadth and potency of three-dose mRNA vaccine-induced antibody response against BA.1, BA.1.1, and BA.2 variants (1, 3, 9–14, 66). Cross-neutralizing antibodies against Omicron were observed post-vaccine boost, but not post-2nd vaccination (1), suggesting recalled memory B cell or de novo induction of novel B cell clones that permit cross-neutralization (15, 16).
Breakthrough infections with BA.1 or BA.1.1 after three vaccinations induced high neutralization titers against all Omicron variant pseudoviruses, as was the case for after three doses of the Pfizer/BNT162b2 vaccine without breakthrough infection. Comparable marked enhancement of serum-neutralizing activity between three dose-vaccinated subjects, vaccine breakthrough, and infected/vaccinated cases was reported in an earlier study where the number of exposures and/or time period between exposures to SARS-CoV-2, either via vaccination or infection, correlated with the strength of neutralizing antibody responses, as well as resilience to variants (10). Furthermore, our findings extend a recent report where sera from vaccinated individuals with confirmed Omicron breakthrough infection showed higher neutralization titers than those of vaccinated individuals without breakthrough infection (67). Altogether, findings from us and others suggest that breakthrough infections can boost preexisting immunity induced by three doses of the Pfizer/BNT162b2 vaccine, thereby eliciting antibodies that neutralize not only Omicron and B.1 variants, but also Alpha, Beta, and Delta variants (67). Finally, a recent study used homologous hamster sera and antigenic cartography to visualize the antigenic evolution of SARS-CoV-2 variants, demonstrating distinct antigenicity of BA.1 and BA.2 variants, separate from ancestral and earlier SARS-CoV-2 variants (68).
Our study has several caveats, including the use of pseudoviruses instead of authentic SARS-CoV-2 for conducting experiments. However, our findings using pseudoviruses agree with those reported using authentic SARS-CoV-2. For instance, authentic BA.1 and BA.1.1 variants were shown to undergo attenuated replication in TMPRSS2-expressing cells compared to ancestral Wuhan-Hu-1 and Alpha, Beta, and Delta variants (6, 36). These reports also showed greater sensitivity of BA.1 pseudovirus entry to endosomal inhibitor E64d. While we used pseudovirus entry assays to determine Omicron variant usage of ACE2 receptors of various animal species, it remains unknown whether there may be intrinsic and/or innate host-specific factors that might act to inhibit live Omicron variants at an entry or postentry step. Furthermore, although we identified RBM substitutions in Omicron spike that conferred the ability to use mouse or horseshoe bat ACE2, we did not confirm ACE2 substitutions that permit or prevent Omicron spike binding. For instance, introducing K35E substitution in horseshoe bat ACE2 should permit its use by Omicron variants. Finally, analysis of a limited number of serum samples and short follow-up after the receipt of three doses of the Pfizer/BNT162b2 mRNA vaccine do not give us insights into the durability or maturation status of the antibody responses. While studies of antibody durability are ongoing, our findings indicate that three-dose immunizations with the Pfizer/BNT162b2 will likely contribute to protection from severe disease caused by the BA.2 variant.
MATERIALS AND METHODS
Ethics statement.
Sera were obtained from participants who received three doses of the Pfizer/BNT162b2 vaccine and had no serological evidence of SARS-CoV-2 infection prior to vaccination. The first two doses of the Pfizer/BNT162b2 vaccine were received before 1 March 2021, whereas the third dose of Pfizer/BNTech162b2 vaccine was received by 15 December 2021. Sera were also obtained from five individuals who experienced vaccine breakthrough infection between December 2021 and January 2022, when BA.1 and BA.1.1 were dominant. Sera were collected at the U.S. Food and Drug Administration with written consent under an approved institutional review board (IRB) protocol (FDA IRB study no. 2021-CBER-045).
Plasmids and cell lines.
Codon-optimized, full-length open reading frames of the spike genes of B.1 (D614G) and Omicron variants in the study synthesized into pVRC8400 (B.1, BA.1, BA.2, BA.2_R493Q, and BA.3) or pcDNA3.1(+) (BA.1.1) were obtained from the Vaccine Research Center (National Institutes of Health, Bethesda, MD, USA) and GenScript (Piscataway, NJ, USA). The codon optimization parameters for spike gene expression in human cells followed GenScript’s optimum gene algorithm as described previously (53). The spike substitutions present in the Omicron variants spikes are listed in Fig. 1. The HIV gag/pol packaging (pCMVΔR8.2) and firefly luciferase-encoding transfer vector (pHR′CMV-Luc) plasmids (69, 70) were obtained from the Vaccine Research Center (National Institutes of Health). ACE2 genes of various species (African green monkey, Chinese rufous horseshoe bat [Rhinolophus sinicus], ferret, mouse, Chinese hamster, Syrian golden hamster, white-tailed deer, swine, bovine, and pangolin) with a C-terminal V5 tag were synthesized by GenScript as described previously (42). 293T (ATCC, Manassas, VA, USA; catalog [cat.] no. CRL-11268), 293T-ACE2 (BEI Resources, Manassas, VA, USA; cat. no. NR-52511) (71), and 293T-ACE2-TMPRSS2 cells stably expressing human angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) (BEI Resources; cat. no. NR-55293) (34) were maintained at 37°C in Dulbecco’s modified Eagle medium (DMEM) supplemented with high glucose, l-glutamine, minimal essential medium (MEM) nonessential amino acids, penicillin/streptomycin, HEPES, and 10% fetal bovine serum (FBS).
SARS-CoV-2 pseudovirus production and neutralization assay.
HIV-based lentiviral pseudoviruses with the desired spike proteins (D614G, BA.1, BA.1.1, BA.2, and BA.3) were generated as previously described (34, 72). Pseudoviruses comprising the spike glycoprotein and a firefly luciferase (FLuc) reporter gene packaged within HIV capsid were produced in 293T cells by cotransfection of 5 μg of pCMVΔR8.2, 5 μg of pHR′CMVLuc, and 0.5 μg of pVRC8400 or 4 μg of pcDNA3.1(+) carrying a codon-optimized spike gene. Pseudovirus supernatants were collected approximately 48 h posttransfection, filtered through a 0.45-μm low-protein binding filter, and stored at −80°C. Pseudovirus titers were measured by infecting 293T-ACE2-TMPRSS2 cells for 48 h prior to measuring luciferase activity (luciferase assay reagent; Promega, Madison, WI, USA), as described previously (73). Pseudovirus titers were expressed as relative luminescence units per milliliter of pseudovirus supernatants (RLU/mL).
Neutralization assays were performed using 293T-ACE2-TMPRSS2 cells in 96-well plates as previously described (34, 72). Pseudoviruses with titers of approximately 106 RLU/mL of luciferase activity were incubated with serially diluted sera or inhibitors for 2 h at 37°C prior to inoculation onto the plates that had been preseeded 1 day earlier with 3.0 × 104 cells/well. Pseudovirus infectivity was determined 48 h postinoculation for luciferase activity by luciferase assay reagent (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The inhibitor concentration or inverse of the serum dilutions causing a 50% reduction of RLU compared to the control was reported as the neutralization titer. Titers were calculated using a nonlinear regression curve fit (GraphPad Prism Software, Inc., La Jolla, CA, USA). The mean titer from at least two independent experiments, each with intra-assay duplicates, was reported as the final titer. For experiments involving camostat mesylate (0.03 to 500 μM) and chloroquine (0.39 to 25 μM) inhibitors, each target cell type was pretreated with inhibitor for 2 h before pseudovirus infection in the presence of the respective inhibitor as described previously (34).
Thermal and pH stability of pseudoviruses.
For assessing thermal stability, pseudoviruses were adjusted to have similar levels of infectivity on ice and were then treated for a specific time period at 50°C or treated for an hour at different temperatures. All treatments were transferred to ice prior to infection experiments. For assessing pH stability, pseudoviruses were mixed with different pH 1 M citrate buffers to obtain a final concentration of 0.1 M citrate, and the mixtures were incubated at 4°C for 1 h. The samples were pH adjusted to physiological pH (7.0) using 1 M Tris (pH 8) prior to the infection experiments. The infection levels of untreated and treated pseudoviruses were compared 48 h postinfection.
Soluble ACE2 protein production.
His-tagged soluble human ACE2 was produced in FreeStyle 293-F cells by transfecting soluble human ACE2 (1 to 741 amino acids [aa]) expression vector plasmid DNA using 293fectin (Thermo Fisher) and purified using HiTrap chelating column charged with nickel (GE Healthcare) according to the manufacturer’s instructions. The eluate containing soluble ACE2 was concentrated to 1.0 mL. Protein-containing fractions were pooled and concentrated using an Amicon Ultra-15 ultracentrifugal unit. The purified proteins were analyzed on a 4 to 12% SDS-PAGE stained with Coomassie blue or membrane probed with mouse monoclonal 6×-His tag antibody (4A12E4) (Thermo Fisher, Waltham, MA, USA) (Fig. S2).
Soluble ACE2 neutralization using SARS-CoV-2 pseudoviruses.
Soluble human ACE2 neutralization assays were performed using 293T-ACE2-TMPRSS2 cells as previously described (53). Briefly, pseudoviruses were treated with 3-fold serial dilutions of soluble ACE2 for 1 h at 37°C. Pseudovirus and soluble ACE2 mixtures (100 μL) were then inoculated onto 96-well plates that had been preseeded with 3.0 × 104 cells per well 1 day prior to the assay. Pseudovirus firefly luciferase activity was determined 48 h postinoculation. The ACE2 concentration causing a 50% reduction of luciferase activity compared to untreated control was reported as the IC50 using a nonlinear regression curve fit (GraphPad Prism Software Inc., La Jolla, CA, USA).
Western blotting.
Cell lysates were resuspended in 1× Laemmli loading buffer containing 2-mercaptoethanol, heated at 70°C for 10 min, resolved by 4 to 20% SDS-PAGE, and transferred onto nitrocellulose membranes. Membranes were probed for the V5-tag and γ-actin using V5 epitope tag antibody (Novus Biologicals, Centennial, CO, USA) and mouse gamma actin polyclonal antibody (Thermo Fisher Scientific, Waltham, MA, USA), respectively.
Computational analysis.
Contact residues in Omicron RBD/human ACE2 complexes are shown as sticks on the Protein Data Bank entry (PDB) code 7WBP (47) using the UCSF Chimera program (http://www.cgl.ucsf.edu/chimera/). Substitutions in RBD/ACE2 complex were introduced by the rotamer function of the USCF Chimera program.
Statistical analyses.
One-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison tests (Omicron variants compared to D614G, and the comparison among Omicron variants) and geometric mean titers (GMT) with 95% confidence intervals were performed using GraphPad Prism software. P values of less than 0.05 were considered statistically significant. All neutralization titers were log2 transformed for analyses.
ACKNOWLEDGMENTS
We would like to thank Hongquan Wan (FDA) and Hailun Ma (FDA) for the critical review of the manuscript. We do not have commercial or other associations or competing interests that might pose a conflict of interest.
Financial support was provided by the U.S. Food and Drug Administration.
Footnotes
Supplemental material is available online only.
Contributor Information
Carol D. Weiss, Email: carol.weiss@fda.hhs.gov.
Wei Wang, Email: wei.wang@fda.hhs.gov.
Stacey Schultz-Cherry, St. Jude Children’s Research Hospital.
REFERENCES
- 1.Lusvarghi S, Pollett SD, Neerukonda SN, Wang W, Wang R, Vassell R, Epsi NJ, Fries AC, Agan BK, Lindholm DA, Colombo CJ, Mody R, Ewers EC, Lalani T, Ganesan A, Goguet E, Hollis-Perry M, Coggins SAA, Simons MP, Katzelnick LC, Wang G, Tribble DR, Bentley L, Eakin AE, Broder CC, Erlandson KJ, Laing ED, Burgess TH, Mitre E, Weiss CD. 2022. SARS-CoV-2 BA.1 variant is neutralized by vaccine booster-elicited serum but evades most convalescent serum and therapeutic antibodies. Sci Transl Med 14:eabn8543. 10.1126/scitranslmed.abn8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, San JE, Cromer D, Scheepers C, Amoako DG, Karim F, Bernstein M, Lustig G, Archary D, Smith M, Ganga Y, Jule Z, Reedoy K, Hwa S-H, Giandhari J, Blackburn JM, Gosnell BI, Abdool Karim SS, Hanekom W, Davies M-A, Hsiao M, Martin D, Mlisana K, Wibmer CK, Williamson C, York D, Harrichandparsad R, Herbst K, Jeena P, Khoza T, Kløverpris H, Leslie A, Madansein R, Magula N, Manickchund N, Marakalala M, Mazibuko M, Moshabela M, Mthabela N, Naidoo K, Ndhlovu Z, Ndung’u T, Ngcobo N, Nyamande K, Patel V, COMMIT-KZN Team , et al. 2022. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 602:654–656. 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, Pinto D, VanBlargan LA, De Marco A, di Iulio J, Zatta F, Kaiser H, Noack J, Farhat N, Czudnochowski N, Havenar-Daughton C, Sprouse KR, Dillen JR, Powell AE, Chen A, Maher C, Yin L, Sun D, Soriaga L, Bassi J, Silacci-Fregni C, Gustafsson C, Franko NM, Logue J, Iqbal NT, Mazzitelli I, Geffner J, Grifantini R, Chu H, Gori A, Riva A, Giannini O, Ceschi A, Ferrari P, Cippà PE, Franzetti-Pellanda A, Garzoni C, Halfmann PJ, Kawaoka Y, Hebner C, Purcell LA, Piccoli L, Pizzuto MS, Walls AC, Diamond MS, et al. 2022. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602:664–670. 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt F, Muecksch F, Weisblum Y, Da Silva J, Bednarski E, Cho A, Wang Z, Gaebler C, Caskey M, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2022. Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med 386:599–601. 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilhelm A, Widera M, Grikscheit K, Toptan T, Schenk B, Pallas C, Metzler M, Kohmer N, Hoehl S, Helfritz FA, Wolf T, Goetsch U, Ciesek S. 2021. Reduced neutralization of SARS-CoV-2 Omicron Variant by Vaccine Sera and monoclonal antibodies. medRxiv. 10.1101/2021.12.07.21267432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A, Kimura I, Yamasoba D, Gerber PP, Fatihi S, Rathore S, Zepeda SK, Papa G, Kemp SA, Ikeda T, Toyoda M, Tan TS, Kuramochi J, Mitsunaga S, Ueno T, Shirakawa K, Takaori-Kondo A, Brevini T, Mallery DL, Charles OJ, Bowen JE, Joshi A, Walls AC, Jackson L, Martin D, Smith KGC, Bradley J, Briggs JAG, Choi J, Madissoon E, Meyer KB, Mlcochova P, Ceron-Gutierrez L, Doffinger R, Teichmann SA, Fisher AJ, Pizzuto MS, de Marco A, Corti D, Hosmillo M, Lee JH, James LC, Thukral L, Veesler D, Sigal A, Sampaziotis F, Ecuador-COVID19 Consortium , et al. 2022. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603:706–714. 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, Ito J, Uriu K, Tsuda M, Zahradnik J, Shirakawa K, Suzuki R, Kishimoto M, Kosugi Y, Kobiyama K, Hara T, Toyoda M, Tanaka YL, Butlertanaka EP, Shimizu R, Ito H, Wang L, Oda Y, Orba Y, Sasaki M, Nagata K, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Kuramochi J, Seki M, Fujiki R, Kaneda A, Shimada T, Nakada T-a, Sakao S, Suzuki T, Ueno T, Takaori-Kondo A, Ishii KJ, Schreiber G, Sawa H, Saito A, Irie T, Tanaka S, Matsuno K, Fukuhara T, Ikeda T, The Genotype to Phenotype Japan (G2P-Japan) Consortium , et al. 2022. Virological characteristics of SARS-CoV-2 BA.2 variant. bioRxiv 10.1101/2022.02.14.480335. [DOI]
- 8.Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, Kosugi Y, Tsuda M, Orba Y, Sasaki M, Shimizu R, Kawabata R, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Suganami M, Oide A, Chiba M, Ito H, Tamura T, Tsushima K, Kubo H, Ferdous Z, Mouri H, Iida M, Kasahara K, Tabata K, Ishizuka M, Shigeno A, Tokunaga K, Ozono S, Yoshida I, Nakagawa S, Wu J, Takahashi M, Kaneda A, Seki M, Fujiki R, Nawai BR, Suzuki Y, Kashima Y, Abe K, Imamura K, Shirakawa K, Genotype to Phenotype Japan (G2P-Japan) Consortium ., et al. 2022. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603:700–705. 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheward DJ, Kim C, Ehling RA, Pankow A, Castro Dopico X, Dyrdak R, Martin DP, Reddy ST, Dillner J, Karlsson Hedestam GB, Albert J, Murrell B. 2022. Neutralisation sensitivity of the SARS-CoV-2 omicron (B.1.1.529) variant: a cross-sectional study. Lancet Infect Dis 22:813–820. 10.1016/S1473-3099(22)00129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls AC, Sprouse KR, Bowen JE, Joshi A, Franko N, Navarro MJ, Stewart C, Cameroni E, McCallum M, Goecker EA, Degli-Angeli EJ, Logue J, Greninger A, Corti D, Chu HY, Veesler D. 2022. SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell 185:872–880.e3. 10.1016/j.cell.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland W-H, Porrot F, Staropoli I, Lemoine F, Péré H, Veyer D, Puech J, Rodary J, Baele G, Dellicour S, Raymenants J, Gorissen S, Geenen C, Vanmechelen B, Wawina-Bokalanga T, Martí-Carreras J, Cuypers L, Sève A, Hocqueloux L, Prazuck T, Rey FA, Simon-Loriere E, Bruel T, Mouquet H, André E, Schwartz O. 2022. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602:671–675. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, Berrios C, Ofoman O, Chang CC, Hauser BM, Feldman J, Roederer AL, Gregory DJ, Poznansky MC, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. 2022. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 185:457–466.e4. 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, Vogels CBF, Malik AA, De la Cruz E, Jorge A, De los Santos M, Leon P, Breban MI, Billig K, Yildirim I, Pearson C, Downing R, Gagnon E, Muyombwe A, Razeq J, Campbell M, Ko AI, Omer SB, Grubaugh ND, Vermund SH, Iwasaki A. 2022. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat Med 28:481–485. 10.1038/s41591-022-01705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowen JE, Sprouse KR, Walls AC, Mazzitelli IG, Logue JK, Franko NM, Ahmed K, Shariq A, Cameroni E, Gori A, Bandera A, Posavad CM, Dan JM, Zhang Z, Weiskopf D, Sette A, Crotty S, Iqbal NT, Corti D, Geffner J, Grifantini R, Chu HY, Veesler D. 2022. Omicron BA.1 and BA.2 neutralizing activity elicited by a comprehensive panel of human vaccines. bioRxiv. 10.1101/2022.03.15.484542. [DOI] [PMC free article] [PubMed]
- 15.Goel RR, Painter MM, Lundgreen KA, Apostolidis SA, Baxter AE, Giles JR, Mathew D, Pattekar A, Reynaldi A, Khoury DS, Gouma S, Hicks P, Dysinger S, Hicks A, Sharma H, Herring S, Korte S, KC W, Oldridge DA, Erickson RI, Weirick ME, McAllister CM, Awofolaju M, Tanenbaum N, Dougherty J, Long S, D’Andrea K, Hamilton JT, McLaughlin M, Williams JC, Adamski S, Kuthuru O, Drapeau EM, Davenport MP, Hensley SE, Bates P, Greenplate AR, Wherry EJ. 2022. Efficient recall of Omicron-reactive B cell memory after a third dose of SARS-CoV-2 mRNA vaccine. bioRxiv. 10.1016/j.cell.2022.04.009. [DOI] [PMC free article] [PubMed]
- 16.Muecksch F, Wang Z, Cho A, Gaebler C, Tanfous TB, DaSilva J, Bednarski E, Ramos V, Zong S, Johnson B, Raspe R, Schaefer-Babajew D, Shimeliovich I, Daga M, Yao K-H, Schmidt F, Millard KG, Turroja M, Jankovic M, Oliveria TY, Gazumyan A, Caskey M, Hatziioannou T, Bieniasz PD, Nussenzweig MC. 2022. Increased potency and breadth of SARS-CoV-2 neutralizing antibodies after a third mRNA vaccine dose. bioRxiv. 10.1101/2022.02.14.480394. [DOI]
- 17.Xia H, Zou J, Kurhade C, Cai H, Yang Q, Cutler M, Cooper D, Muik A, Jansen KU, Xie X, Swanson KA, Shi P-Y. 2022. Neutralization and durability of 2 or 3 doses of the BNT162b2 vaccine against Omicron SARS-CoV-2. Cell Host Microbe 30:485–488.e3. 10.1016/j.chom.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanshylla K, Tober-Lau P, Gruell H, Münn F, Eggeling R, Pfeifer N, Le NH, Landgraf I, Kurth F, Sander LE, Klein F. 2022. Durability of Omicron-neutralising serum activity after mRNA booster immunisation in older adults. Lancet Infect Dis 22:445–446. 10.1016/S1473-3099(22)00135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude MB, Hakze-van der Honing RW, Gerhards N, Tolsma P, Bouwstra R, Sikkema RS, Tacken MG, de Rooij MM, Weesendorp E, Engelsma MY, Bruschke CJ, Smit LA, Koopmans M, van der Poel WH, Stegeman A. 2020. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 25:2001005. 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritter JM, Wilson TM, Gary JM, Seixas JN, Martines RB, Bhatnagar J, Bollweg BC, Lee E, Estetter L, Silva-Flannery L, Bullock HA, Towner JS, Cossaboom CM, Wendling NM, Amman BR, Harvey RR, Taylor D, Rettler H, Barton Behravesh C, Zaki SR. 2022. Histopathology and localization of SARS-CoV-2 and its host cell entry receptor ACE2 in tissues from naturally infected US-farmed mink (Neovison vison). Vet Pathol 59:681–695. 10.1177/03009858221079665. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Zhang H, Gao J, Huang K, Yang Y, Hui X, He X, Li C, Gong W, Zhang Y, Zhao Y, Peng C, Gao X, Chen H, Zou Z, Shi Z-L, Jin M. 2020. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg Microbes Infect 9:2013–2019. 10.1080/22221751.2020.1817796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson EI, Elia G, Grassi A, Giordano A, Desario C, Medardo M, Smith SL, Anderson ER, Prince T, Patterson GT, Lorusso E, Lucente MS, Lanave G, Lauzi S, Bonfanti U, Stranieri A, Martella V, Solari Basano F, Barrs VR, Radford AD, Agrimi U, Hughes GL, Paltrinieri S, Decaro N. 2020. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat Commun 11:6231. 10.1038/s41467-020-20097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Račnik J, Kočevar A, Slavec B, Korva M, Rus KR, Zakotnik S, Zorec TM, Poljak M, Matko M, Rojs OZ, Županc TA. 2021. Transmission of SARS-CoV-2 from human to domestic ferret. Emerg Infect Dis 27:2450–2453. 10.3201/eid2709.210774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koeppel KN, Mendes A, Strydom A, Rotherham L, Mulumba M, Venter M. 2022. SARS-CoV-2 reverse zoonoses to pumas and lions, South Africa. Viruses 14:120. 10.3390/v14010120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandler JC, Bevins SN, Ellis JW, Linder TJ, Tell RM, Jenkins-Moore M, Root JJ, Lenoch JB, Robbe-Austerman S, DeLiberto TJ, Gidlewski T, Kim Torchetti M, Shriner SA. 2021. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc Natl Acad Sci USA 118:e2114828118. 10.1073/pnas.2114828118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulrich L, Wernike K, Hoffmann D, Mettenleiter TC, Beer M. 2020. Experimental infection of cattle with SARS-CoV-2. Emerg Infect Dis 26:2979–2981. 10.3201/eid2612.203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosco-Lauth AM, Walker A, Guilbert L, Porter S, Hartwig A, McVicker E, Bielefeldt-Ohmann H, Bowen RA. 2021. Susceptibility of livestock to SARS-CoV-2 infection. Emerg Microbes Infect 10:2199–2201. 10.1080/22221751.2021.2003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickering BS, Smith G, Pinette MM, Embury-Hyatt C, Moffat E, Marszal P, Lewis CE. 2021. Susceptibility of domestic swine to experimental infection with severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 27:104–112. 10.3201/eid2701.203399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaudreault NN, Cool K, Trujillo JD, Morozov I, Meekins DA, McDowell C, Bold D, Carossino M, Balaraman V, Mitzel D, Kwon T, Madden DW, Artiaga BL, Pogranichniy RM, Roman-Sosa G, Wilson WC, Balasuriya UBR, García-Sastre A, Richt JA. 2022. Susceptibility of sheep to experimental co-infection with the ancestral lineage of SARS-CoV-2 and its alpha variant. Emerg Microbes Infect 11:662–675. 10.1080/22221751.2022.2037397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Wang W, Xue J, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao L, Liu P, Zhao L, Ye F, Wang H, Zhou W, Zhu N, Zhen W, Yu H, Zhang X, Guo L, Chen L, Wang C, Wang Y, Wang X, Xiao Y, Sun Q, Liu H, Zhu F, Ma C, Yan L, Yang M, Han J, Xu W, Tan W, Peng X, Jin Q, Wu G, Qin C. 2020. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583:830–833. 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 31.Jiang R-D, Liu M-Q, Chen Y, Shan C, Zhou Y-W, Shen X-R, Li Q, Zhang L, Zhu Y, Si H-R, Wang Q, Min J, Wang X, Zhang W, Li B, Zhang H-J, Baric RS, Zhou P, Yang X-L, Shi Z-L. 2020. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182:50–58.e8. 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun S-H, Chen Q, Gu H-J, Yang G, Wang Y-X, Huang X-Y, Liu S-S, Zhang N-N, Li X-F, Xiong R, Guo Y, Deng Y-Q, Huang W-J, Liu Q, Liu Q-M, Shen Y-L, Zhou Y, Yang X, Zhao T-Y, Fan C-F, Zhou Y-S, Qin C-F, Wang Y-C. 2020. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28:124–133.e4. 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leist SR, Dinnon KH, Schäfer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ, Gully KL, Scobey T, Brown AJ, Sheahan TP, Moorman NJ, Boucher RC, Gralinski LE, Montgomery SA, Baric RS. 2020. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183:1070–1085.e12. 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neerukonda SN, Vassell R, Herrup R, Liu S, Wang T, Takeda K, Yang Y, Lin T-L, Wang W, Weiss CD. 2021. Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLoS One 16:e0248348. 10.1371/journal.pone.0248348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H, Lu L, Peng Z, Chen L-L, Meng X, Zhang C, Ip JD, Chan W-M, Chu AW-H, Chan K-H, Jin D-Y, Chen H, Yuen K-Y, To KK-W. 2022. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect 11:277–283. 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuai H, Chan JF-W, Hu B, Chai Y, Yuen TT-T, Yin F, Huang X, Yoon C, Hu J-C, Liu H, Shi J, Liu Y, Zhu T, Zhang J, Hou Y, Wang Y, Lu L, Cai J-P, Zhang AJ, Zhou J, Yuan S, Brindley MA, Zhang B-Z, Huang J-D, To KK-W, Yuen K-Y, Chu H. 2022. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603:693–699. 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Anang S, Iketani S, Guo Y, Liu L, Ho DD, Sodroski JG. 2021. Functional properties of the spike glycoprotein of the emerging SARS-CoV-2 variant B.1.1.529. bioRxiv. 10.1016/j.celrep.2022.110924. [DOI] [PMC free article] [PubMed]
- 38.Wang Q, Nair MS, Anang S, Zhang S, Nguyen H, Huang Y, Liu L, Ho DD, Sodroski JG. 2021. Functional differences among the spike glycoproteins of multiple emerging severe acute respiratory syndrome coronavirus 2 variants of concern. iScience 24:103393. 10.1016/j.isci.2021.103393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J, Peacock TP, Brown JC, Goldhill DH, Elrefaey AME, Penrice-Randal R, Cowton VM, De Lorenzo G, Furnon W, Harvey WT, Kugathasan R, Frise R, Baillon L, Lassaunière R, Thakur N, Gallo G, Goldswain H, Donovan-Banfield I, Dong X, Randle NP, Sweeney F, Glynn MC, Quantrill JL, McKay PF, Patel AH, Palmarini M, Hiscox JA, Bailey D, Barclay WS. 2022. Mutations that adapt SARS-CoV-2 to mink or ferret do not increase fitness in the human airway. Cell Rep 38:110344. 10.1016/j.celrep.2022.110344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren W, Lan J, Ju X, Gong M, Long Q, Zhu Z, Yu Y, Wu J, Zhong J, Zhang R, Fan S, Zhong G, Huang A, Wang X, Ding Q. 2021. Mutation Y453F in the spike protein of SARS-CoV-2 enhances interaction with the mink ACE2 receptor for host adaption. PLoS Pathog 17:e1010053. 10.1371/journal.ppat.1010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marques AD, Sherrill-Mix S, Everett JK, Adhikari H, Reddy S, Ellis JC, Zeliff H, Greening SS, Cannuscio CC, Strelau KM, Collman RG, Kelly BJ, Rodino KG, Bushman FD, Gagne RB, Anis E. 2022. Evolutionary trajectories of SARS-CoV-2 Alpha and Delta variants in white-tailed deer in Pennsylvania. medRxiv. 10.1101/2022.02.17.22270679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S, Selvaraj P, Lien CZ, Nunez IA, Wu WW, Chou C-K, Wang TT, Subbarao K. 2021. The PRRA insert at the S1/S2 site modulates cellular tropism of SARS-CoV-2 and ACE2 usage by the closely related bat RaTG13. J Virol 95:e01751-20. 10.1128/JVI.01751-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y, Wang Q, Zhou H, Yan J, Qi J. 2020. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181:894–904.e9. 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 46.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han P, Li L, Liu S, Wang Q, Zhang D, Xu Z, Han P, Li X, Peng Q, Su C, Huang B, Li D, Zhang R, Tian M, Fu L, Gao Y, Zhao X, Liu K, Qi J, Gao GF, Wang P. 2022. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 185:630–640.e10. 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann M, Zhang L, Pöhlmann S. 2022. Omicron: master of immune evasion maintains robust ACE2 binding. Signal Transduct Target Ther 7:118. 10.1038/s41392-022-00965-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin W, Xu Y, Xu P, Cao X, Wu C, Gu C, He X, Wang X, Huang S, Yuan Q, Wu K, Hu W, Huang Z, Liu J, Wang Z, Jia F, Xia K, Liu P, Wang X, Song B, Zheng J, Jiang H, Cheng X, Jiang Y, Deng S-J, Xu HE. 2022. Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science 375:1048–1053. 10.1126/science.abn8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang K, Zhang Y, Hui X, Zhao Y, Gong W, Wang T, Zhang S, Yang Y, Deng F, Zhang Q, Chen X, Yang Y, Sun X, Chen H, Tao YJ, Zou Z, Jin M. 2021. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine 67:103381. 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinnon KH, III, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586:560–566. 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gawish R, Starkl P, Pimenov L, Hladik A, Lakovits K, Oberndorfer F, Cronin SJF, Ohradanova-Repic A, Wirnsberger G, Agerer B, Endler L, Capraz T, Perthold JW, Cikes D, Koglgruber R, Hagelkruys A, Montserrat N, Mirazimi A, Boon L, Stockinger H, Bergthaler A, Oostenbrink C, Penninger JM, Knapp S. 2022. ACE2 is the critical in vivo receptor for SARS-CoV-2 in a novel COVID-19 mouse model with TNF- and IFNγ-driven immunopathology. Elife 11:e74623. 10.7554/eLife.74623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neerukonda SN, Vassell R, Lusvarghi S, Wang R, Echegaray F, Bentley L, Eakin AE, Erlandson KJ, Katzelnick LC, Weiss CD, Wang W. 2021. SARS-CoV-2 Delta variant displays moderate resistance to neutralizing antibodies and spike protein properties of higher soluble ACE2 sensitivity, enhanced cleavage and fusogenic activity. Viruses 13:2485. 10.3390/v13122485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer A-S, Winkler MS, Lier M, Dopfer-Jablonka A, Jäck H-M, Behrens GMN, Pöhlmann S. 2022. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell 185:447–456.e11. 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng C, Evans JP, Qu P, Faraone J, Zheng Y-M, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, Saif LJ, Oltz EM, Mohler P, Xu K, Gumina RJ, Liu S-L. 2021. Neutralization and stability of SARS-CoV-2 Omicron variant. bioRxiv. 10.1101/2021.12.16.472934. [DOI]
- 56.Willett BJ, Grove J, MacLean OA, Wilkie C, De Lorenzo G, Furnon W, Cantoni D, Scott S, Logan N, Ashraf S, Manali M, Szemiel A, Cowton V, Vink E, Harvey WT, Davis C, Asamaphan P, Smollett K, Tong L, Orton R, Hughes J, Holland P, Silva V, Pascall DJ, Puxty K, da Silva Filipe A, Yebra G, Shaaban S, Holden MTG, Pinto RM, Gunson R, Templeton K, Murcia PR, Patel AH, Klenerman P, Dunachie S, Dunachie S, Klenerman P, Barnes E, Brown A, Adele S, Kronsteiner B, Murray SM, Abraham P, Deeks A, Ansari MA, de Silva T, Turtle L, Moore S, Austin J, PITCH Consortium , et al. 2022. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol 7:1161–1179. 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peacock TP, Brown JC, Zhou J, Thakur N, Sukhova K, Newman J, Kugathasan R, Yan AWC, Furnon W, De Lorenzo G, Cowton VM, Reuss D, Moshe M, Quantrill JL, Platt OK, Kaforou M, Patel AH, Palmarini M, Bailey D, Barclay WS. 2022. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. bioRxiv 10.1101/2021.12.31.474653. [DOI]
- 58.Xu Y, Wu C, Cao X, Gu C, Liu H, Jiang M, Wang X, Yuan Q, Wu K, Liu J, Wang D, He X, Wang X, Deng S-J, Xu HE, Yin W. 2022. Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.1 and BA.2 variants and their mouse origins. bioRxiv 10.1101/2022.04.12.488075. [DOI] [PMC free article] [PubMed]
- 59.Li L, Liao H, Meng Y, Li W, Han P, Liu K, Wang Q, Li D, Zhang Y, Wang L, Fan Z, Zhang Y, Wang Q, Zhao X, Sun Y, Huang N, Qi J, Gao GF. 2022. Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1. Cell 185: P2952– P2960. 10.1016/j.cell.2022.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun S, Gu H, Cao L, Chen Q, Ye Q, Yang G, Li R-T, Fan H, Deng Y-Q, Song X, Qi Y, Li M, Lan J, Feng R, Guo Y, Zhu N, Qin S, Wang L, Zhang Y-F, Zhou C, Zhao L, Chen Y, Shen M, Cui Y, Yang X, Wang X, Tan W, Wang H, Wang X, Qin C-F. 2021. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat Commun 12:5654. 10.1038/s41467-021-25903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Huang K, Wang T, Deng F, Gong W, Hui X, Zhao Y, He X, Li C, Zhang Q, Chen X, Lv C, Lin X, Yang Y, Sun X, Shi Z, Chen H, Zou Z, Jin M. 2021. SARS-CoV-2 rapidly adapts in aged BALB/c mice and induces typical pneumonia. J Virol 95:e02477-20. 10.1128/JVI.02477-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munnink BBO, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M, Bouwmeester-Vincken N, Harders F, Hakze-van der Honing R-v, Wegdam-Blans MCA, Bouwstra RJ, GeurtsvanKessel C, van der Eijk AA, Velkers FC, Smit LAM, Stegeman A, van der Poel WHM, Koopmans MPG. 2021. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371:172–177. 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yen H-L, Sit THC, Brackman CJ, Chuk SSY, Gu H, Tam KWS, Law PYT, Leung GM, Peiris M, Poon LLM, Cheng SMS, Chang LDJ, Krishnan P, Ng DYM, Liu GYZ, Hui MMY, Ho SY, Su W, Sia SF, Choy K-T, Cheuk SSY, Lau SPN, Tang AWY, Koo JCT, Yung L. 2022. Transmission of SARS-CoV-2 delta variant (AY.127) from pet hamsters to humans, leading to onward human-to-human transmission: a case study. Lancet 399:1070–1078. 10.1016/S0140-6736(22)00326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu S, Selvaraj P, Sangare K, Luan B, Wang TT. 2022. Spike protein-independent attenuation of SARS-CoV-2 Omicron variant in laboratory mice. bioRxiv. 10.1101/2022.02.08.479543. [DOI] [PMC free article] [PubMed]
- 65.Halfmann PJ, Iida S, Iwatsuki-Horimoto K, Maemura T, Kiso M, Scheaffer SM, Darling TL, Joshi A, Loeber S, Singh G, Foster SL, Ying B, Case JB, Chong Z, Whitener B, Moliva J, Floyd K, Ujie M, Nakajima N, Ito M, Wright R, Uraki R, Warang P, Gagne M, Li R, Sakai-Tagawa Y, Liu Y, Larson D, Osorio JE, Hernandez-Ortiz JP, Henry AR, Ciuoderis K, Florek KR, Patel M, Odle A, Wong L-YR, Bateman AC, Wang Z, Edara V-V, Chong Z, Franks J, Jeevan T, Fabrizio T, DeBeauchamp J, Kercher L, Seiler P, Gonzalez-Reiche AS, Sordillo EM, Chang LA, van Bakel H, Consortium Mount Sinai Pathogen Surveillance (PSP) study group , et al. 2022. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603:687–692. 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altarawneh HN, Chemaitelly H, Ayoub HH, Tang P, Hasan MR, Yassine HM, Al-Khatib HA, Smatti MK, Coyle P, Al-Kanaani Z, Al-Kuwari E, Jeremijenko A, Kaleeckal AH, Latif AN, Shaik RM, Abdul-Rahim HF, Nasrallah GK, Al-Kuwari MG, Butt AA, Al-Romaihi HE, Al-Thani MH, Al-Khal A, Bertollini R, Abu-Raddad LJ. 2022. Effects of previous infection and vaccination on symptomatic Omicron infections. N Engl J Med 387:21–34. 10.1056/NEJMoa2203965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suryawanshi RK, Chen IP, Ma T, Syed AM, Brazer N, Saldhi P, Simoneau CR, Ciling A, Khalid MM, Sreekumar B, Chen P-Y, Kumar GR, Montano M, Gascon R, Tsou C-L, Garcia-Knight MA, Sotomayor-Gonzalez A, Servellita V, Gliwa A, Nguyen J, Silva I, Milbes B, Kojima N, Hess V, Shacreaw M, Lopez L, Brobeck M, Turner F, Soveg FW, George AF, Fang X, Maishan M, Matthay M, Morris MK, Wadford D, Hanson C, Greene WC, Andino R, Spraggon L, Roan NR, Chiu CY, Doudna JA, Ott M. 2022. Limited cross-variant immunity from SARS-CoV-2 Omicron without vaccination. Nature 607:351–355. 10.1038/s41586-022-04865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mykytyn AZ, Rissmann M, Kok A, Rosu ME, Schipper D, Breugem TI, van den Doel PB, Chandler F, Bestebroer T, de Wit M, van Royen ME, Molenkamp R, Oude Munnink BB, de Vries RD, GeurtsvanKessel C, Smith DJ, Koopmans MPG, Rockx B, Lamers MM, Fouchier R, Haagmans BL. 2022. Omicron BA.1 and BA.2 are antigenically distinct SARS-CoV-2 variants. bioRxiv. 10.1101/2022.02.23.481644. [DOI] [PMC free article] [PubMed]
- 69.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 70.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 15:871–875. 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 71.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD. 2020. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12:513. 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neerukonda SN, Vassell R, Weiss CD, Wang W. 2022. Measuring neutralizing antibodies to SARS-CoV-2 using lentiviral spike-pseudoviruses. Methods Mol Biol 2452:305–314. 10.1007/978-1-0716-2111-0_18. [DOI] [PubMed] [Google Scholar]
- 73.Wang W, Butler EN, Veguilla V, Vassell R, Thomas JT, Moos M, Jr, Ye Z, Hancock K, Weiss CD. 2008. Establishment of retroviral pseudotypes with influenza hemagglutinins from H1, H3, and H5 subtypes for sensitive and specific detection of neutralizing antibodies. J Virol Methods 153:111–119. 10.1016/j.jviromet.2008.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download jvi.01140-22-s0001.pdf, PDF file, 1.6 MB (1.6MB, pdf)