Abstract
Silver nanoparticles (NPs) were synthesized using an efficient bioreducing agent from Fagonia cretica extract having the advantage of eco-friendliness over chemical and physical methods. The sharp color change and appearance of representative absorption peaks in the UV-visible spectra confirm the quick reduction of the Ag salt and evolution of Ag NPs. Morphological and structural aspects showed that the resulting Ag NPs are highly crystalline with an average size of 16 nm. Furthermore, compositional analysis of the extract confirmed the existence of active bioreducing and stabilizing agents in the Fagonia cretica extract. Furthermore, various concentrations of AgNO3 and the Fagonia cretica extract were employed to obtain a higher yield with better stability of Ag NPs. The resulting Ag NPs showed effective antibacterial activity against Proteus vulgaris, Escherichia coli, and Klebsiella pneumoniae. It is found that the Ag NPs induce maximum production of reactive oxygen species (ROS) in Proteus vulgaris as compared to Escherichia coli and Klebsiella pneumoniae which induce cell toxicity, while ROS production in the presence of Ag NPs is 30% higher than that in the presence of only the plant extract and control in all three bacterial strains. Thus, present findings show that plant extracts can be a useful natural resource to prepare functional nanomaterials for targeted applications especially in the field of biotechnology.
Silver nanoparticles (NPs) were synthesized using an efficient bioreducing agent from Fagonia cretica extract having the advantage of eco-friendliness over chemical and physical methods.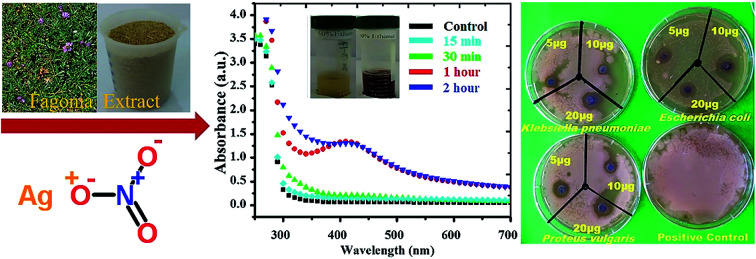
1. Introduction
Synthesis of functional nanomaterials is getting immense attention from researchers due to their vast applications like in biomedicine, drug delivery, cancer treatment, bio-imaging, molecular based detection etc. But synthesis of nanomaterials via physical and chemical methods has severe impacts on the environment and human health as these methods excrete tons of hazardous chemicals and toxic by-products.1–5
Therefore, there is a growing concern to develop clean, efficient, non-toxic, non-lethal, environment-friendly, cost-effective biological methods for the synthesis of nanoparticles (NPs) which have no threat to life. Recently, there have been many reports on synthesis of Ag NPs by using root, leaf and stem extracts of plants such as Dracaena cochinchinensis, Eucommia ulmoides, Gloriosa superba, Raphanus sativus and Sargassum polycystum.6–11 These biological entities have bioactive compounds in their extracts which have the ability to reduce metal ions into atoms, which results in nuclei that consequently provide a base for a variety of nanomaterials. For example, Hu et al. investigated that conversion of Ag ions to Ag NPs and their size distribution is totally dependent on the concentration of lignin which serves as the reducing and capping agent.12 Similarly, dragon's blood resin extract containing loureirin A & B also acts as strong reducing and capping agents to produce Ag NPs.13 Recent reports have shown that the presence of active functional groups such as sugar and phenolic acids is responsible for the reduction of metal ions into NPs.14 Keeping in mind the above studies, it is highly desirable to find alternative cheap and easily available bioreducing sources for the synthesis of various functional nanomaterials to compete with physical and chemical methods both in terms of cost and yield. It is also interesting to investigate the nature of biomaterials involved in the mechanism of the recognition–reduction process and synthesis of NPs. Moreover, there is a need to know about the biomolecules in the plant extract which lead to the formation of metal complexes, ultimately producing corresponding nanomaterials. Furthermore, this area is getting more attention as these biologically synthesized nanomaterials are in high demand for various biomedical and catalytic applications like drug delivery, early disease detection, antimicrobial, antifungal, and sensing applications.15–19
Considering the advantages of green synthesis over other methods, here, we have explored a new plant extract named Fagonia cretica as the reducing and stabilizing agent for the synthesis of Ag NPs due to its abundance and low cost as well as no involvement of any bacterial or fungal species like other biological synthesis methods which eliminates all associated clinical concerns which eliminates all associated clinical concerns, thus, making the Fagonia cretica extract a unique bioreducing source that is worth exploring for the mentioned purpose. The as-synthesized Ag NPs are highly crystalline and exhibited significant anti-microbial activities against various strains of Gram negative bacteria, confirming that biological reactors are a better option for the synthesis of nanomaterials for wide application in future not only in medical applications but also in environmental and energy applications.
2. Experimental section
Materials & plant extract
A Fagonia cretica plant was obtained from the Agriculture Department of the Islamia University of Bahawalpur, Pakistan, the Herberium certificate of Fagonia cretica is given in the ESI for plant collection and habitat (Certificate 1 ESI †). After drying the pieces of the Fagonia cretica plant, they were ground to get a fine powder. Afterwards, 1.5 g of the Fagonia cretica powder was mixed with 50 mL of solutions containing different ethanol to water ratios (30, 50, 70 & 90% ethanol, as shown in Fig. S1†) in a 250 mL conical flask and shaken overnight at 37 °C. Finally, the extract was filtered using filter paper, and the filtrate was kept at 4 °C for further use.
Synthesis of Ag NPs & physical characterization
Initially 0.1 M aqueous solution of AgNO3 was prepared and mixed with the plant extract at 5 : 1, respectively. The mixture was stirred with a magnetic stirrer (150 rpm) at 50 °C for a time period of 15–120 min. The colour change in the reaction mixture was recorded through visual observation. The bioreduction of Ag ions in the aqueous solution was monitored by periodic sampling of aliquots (150 μL), and UV-vis spectra of the solution were measured. On completion of the reaction, the product was collected using a centrifuge (20 min at 6000 rpm). The supernatant was saved in separate falcon tubes and kept at 4 °C for high performance liquid chromatography (HPLC). Note: various concentrations of the AgNO3 aqueous solution (0.5 mM, 1 mM and 2 mM) and plant extract (10 mL, 20 mL and 40 mL) were used to determine their impact on the growth of Ag NPs. The physical characterization and antimicrobial activity of Ag NPs are given in the ESI.†
3. Results and discussion
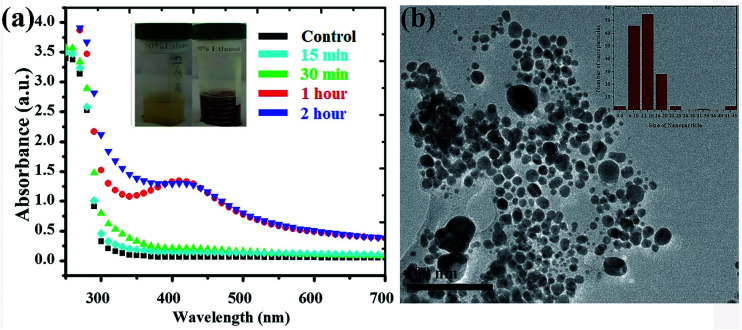
Silver NPs synthesized using the bioreducing agents extracted from Fagonia cretica are of great interest because of their unique characteristics. The reduction of AgNO3 by the Fagonia cretica extract is first observed by a change in color from light orange to dark red indicating the strong absorption of visible light due to the excitation of the NP surface plasmons (the inset of Fig. 1a), which depends on the size, shape and concentration of the Ag NPs.20 Furthermore, to confirm the reduction of Ag ions to metallic Ag NPs, UV-vis spectra were recorded at various time intervals of 15, 30, 60 & 120 min where the maximum absorbance was observed at 440 nm (Fig. 1a). The control sample does not show any peak due to the absence of the reducing agent (plant extract) resulting in no formation of Ag NPs, while the sample having the plant extract shows maximum intensity for 60 & 120 min assuring that the Fagonia cretica extract plays a main role in reducing Ag ions to form Ag NPs. The size of the NPs is a matter of great concern specially for their biological applications as the size of NPs strongly affects the rate of diffusion through biological membranes. Previously, it has been observed that small size NPs have more penetration power, but too small a size brings the issue of enhanced toxicity compared to larger size NPs; thus, an appropriate size is highly desirable for specific biological applications.6,21 Therefore, transmission electron microscopy (TEM) was used to examine the morphology and size of NPs prior to their biological application. Fig. 1b clearly shows that the as-synthesized NPs are spherical in shape and have a very narrow size distribution where 70% of the NPs have a size in the range of 11–15 nm, while 22% have a size between 16 and 45 nm and only 1% have a size smaller than 5 nm (the inset of Fig. 1b). The abundant NPs with a size range of 11–15 nm are highly suitable for biomedical applications as the size of the synthesized NPs is within the tolerable range for inducing toxicity within cells.
Fig. 1. (a) UV-vis spectra of Ag NPs with the Fagonia cretica extract at different time points (the inset shows the colour change before and after Ag ion reduction) and (b) TEM image of Ag NPs, while the inset shows the particle size distribution.
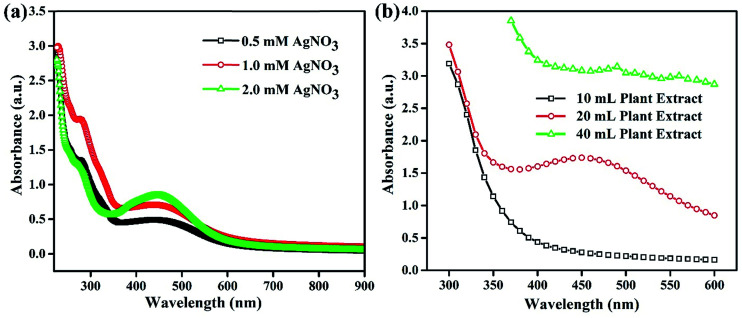
It is very common that the concentrations of the salt precursor (AgNO3) and Fagonia cretica extract will have a large impact on the nuclei formation, growth and yield of Ag NPs. To explore the effect of AgNO3 and the Fagonia cretica extract, various concentrations were employed while keeping the other one constant. The concentration of AgNO3 strongly influences the formation of Ag NPs as a sharp absorption peak for Ag NPs was noticed at the 2.0 mM concentration of AgNO3 as compared to 0.5 mM and 1.0 mM as shown in Fig. 2a, which confirms that an optimal concentration of Ag+ ions is required as a deficiency of ions at a lower concentration and excessive ions at a higher concentration result in the formation of improper and/or complex intermediates that ultimately bring about poor conversion efficiencies. Similarly, a change in the concentration of the Fagonia cretica extract also influenced the growth of Ag NPs; at a low concentration of the extract, no peak was observed in the UV-vis spectrum confirming that low concentrations are unable to provide enough bioreducing molecules for formation of stable Ag+ ion complexes and their conversion to metallic Ag NPs (Fig. 2b). However, increasing the concentration of the Fagonia cretica extract from 10 to 20 mL results in a sharp peak confirming the formation of appropriate metal–organic complexes that later on yield Ag NPs as shown in Fig. 2b. However, with further increase in the concentration of the Fagonia cretica extract up to 40 mL, we found that a reduction in the absorption peak corresponds to the poor yield of Ag NPs and overall high absorbance due to the large quantity of the plant extract (organic molecules). In addition to the reduction in the absorption peak, a change in the peak position is also observed which might be due to the formation of improper metal–organic complexes in the presence of a higher concentration of organic molecules from the Fagonia cretica extract (Fig. 2b). Thus, this confirms that to obtain Ag NPs with better material features, appropriate concentrations of AgNO3 and the Fagonia cretica extract are highly required; however, antimicrobial activity was assessed using Ag NPs prepared under the best conditions.
Fig. 2. UV-vis absorption spectra of Ag NPs synthesized at different concentrations of (a) AgNO3 and (b) the plant extract.
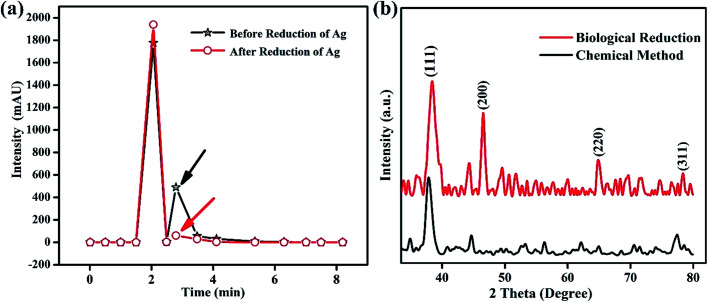
To identify the role of the Fagonia cretica extract in the reduction of Ag ions, HPLC studies of the Fagonia cretica extract were conducted before using it as the reducing agent and after it reduced Ag ions to Ag NPs to confirm its role in reduction, and the results are shown in Fig. 3a. A clear difference in absorbance on the basis of retention time was observed before and after the reduction reaction (full spectra are shown in Fig. S2†). A change in the peak height of about 30% and disappearance of the shoulder peak at 2.793 min (pointed by black and red arrows in Fig. 3a) confirm that the Fagonia cretica extract is responsible for the reduction of Ag ions to Ag NPs. Hence, it is proved that the Fagonia cretica extract contains the respective chemical entities that are involved in reduction of Ag NPs and act as the capping agent for their stability and suitability for biomedical applications.
Fig. 3. (a) Chromatographic profiling of the Fagonia cretica extract before and after reduction and (b) XRD spectra of Ag NPs synthesized using the Fagonia cretica extract and chemical method.
Fourier-transform infrared spectroscopy (FTIR) analysis was carried out to further identify the prominent bioactive molecules responsible for the stability and capping of the NPs. The FTIR spectrum of the crude Fagonia cretica extract was compared with that of the as-synthesised Ag NPs to investigate the biomolecules involved in bio-reducing and stabilizing NPs. The FTIR spectrum of the Fagonia cretica extract shows peaks at 3864.44, 3729.37, 3626.26, 3467.80, 2916.00, 1636.56, 1061.73, 800.64, 715.39 and 627.47 cm−1 (Fig. S3a†) while in the FTIR spectrum of Ag NPs, peaks at 3739.15, 2358.74, 1472.89, 1401.80, 1114.29, 1050.90, 869.08, 643.89, and 600.15 cm−1 are observed (Fig. S3b†). Therefore, the main functional groups involved in bioreducing Ag NPs from the Fagonia cretica extract are hydroxyl and amino groups. Normally, they are groups with highly electronegative elements and have lone pairs that can induce the reduction of metallic ions and create surface functionalized NPs. A shift in the absorbance peak from 3729.37 to 3739.15 cm−1 was observed, with an increase in band intensity due to the presence of hydroxyl groups on the NP surface. The peak at 1061.73 cm−1 in the spectrum of the crude plant extract representing the C H bending vibrations of ether linkages is shifted to 1050.89 cm−1 which may help in attaching carboxyl groups on the surface of Ag NPs. Another differential peak in the spectra of the plant extract of Fagonia cretica and Ag NPs was compared and it was found that the peak absorbance for C–H plane bending at 627.47 cm−1 (plant extract) is shifted to 643.89 cm−1 (Ag NPs). The existence of these peaks indicate that Ag NP growth is based on the reducing molecules of Fagonia cretica which not only reduce the Ag ions but also attach several functional groups e.g. hydroxyl and secondary amines on the surface of the as-synthesized Ag NPs that stabilize the particles and assist in providing better biological activity.13,22 These findings are well supported by the literature where ketone and hydroxyl groups are reported to be responsible for the reduction of silver ions to Ag NPs, which have better stability due to naturally existing capping agents.23–27
The crystallinity of Ag NPs was characterized by X-ray diffraction (XRD) that delineates their face centered cubic (fcc) structure (Fig. 3b). The XRD spectrum of the biologically synthesized Ag NPs showed distinct peaks at 2θ of 38°, 45°, 65 and 77° corresponding to lattice planes of (111), (200), (220) and (311), respectively, in good agreement with standard PDF no. 04-0783 while that of the chemically synthesized Ag NPs showed diffraction peaks at 38°, 45° and 77°. However, the presence of more and sharp diffraction peaks confirms the better crystallinity of the biologically synthesized Ag NPs, showing that biological synthesis is an effective way not only for biological applications but also for many other fields as it helps to produce large amounts in easy ways with possibilities to create various functionalities. The Scherrer equation (D = kλ/β cos θ) is applied to determine the crystallite size of Ag particles which is 1.66 nm (biologically) and 1.19 nm (chemically); the smaller crystallite size might be due to the presence of capping agents in the Fagonia cretica extract that control the growth to a smaller size and provide high stability. Furthermore, Fig. S4† shows the stability of the colloidal solution of Ag NPs at pH 4, where it is clear that the suspension showed high stability even after one month. A slight change in color after one month shows little sedimentation of NPs; however, no obvious aggregation of Ag NPs was observed in the one-month old solution which demonstrates their high stability and suitability for biomedical applications.
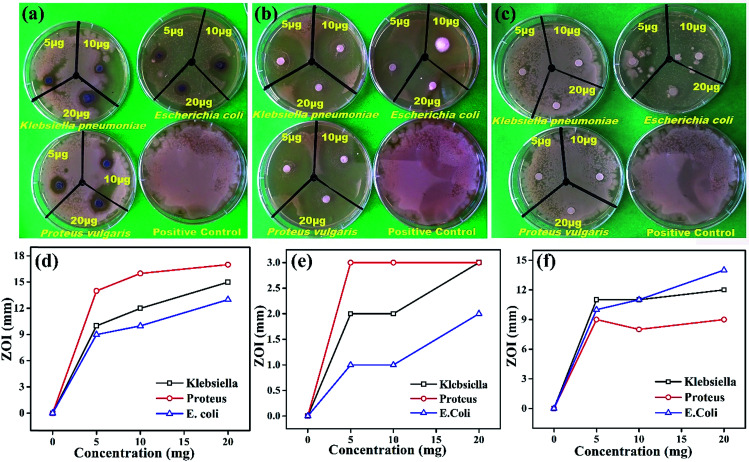
Comparative antimicrobial activities of the as-synthesized Ag NPs and standard ciprofloxacin against strains of Proteus vulgaris, Escherichia coli and Klebsiella pneumonia have been investigated. These microbes cause infections mainly in people with an impaired immunity system, and most of them may be a source of complicated urinary tract, wound and nosocomial infections. Our results showed that the antimicrobial activity of the synthesized Ag NPs is significant against the growth of Proteus vulgaris, Escherichia coli and Klebsiella pneumoniae strains with different concentrations while the Fagonia cretica extract (tested to eliminate its impact and validity of Ag NPs) with all different concentrations has not shown any significant antimicrobial activity (Fig. 4a–c). It is found that growth of all three bacterial strains (Proteus vulgaris, Escherichia coli, and Klebsiella pneumoniae) was significantly suppressed by treating with Ag NPs and ciprofloxacin compared to control experiments (Tables S1–S3†). Furthermore, Ag NPs have shown maximum antimicrobial activity for Proteus vulgaris while they show minimum antimicrobial activity in Escherichia coli for all concentrations (5, 10 & 20 μg), and a similar trend is observed for the Fagonia cretica extract but with much lower activity as shown in Fig. 4d and e. Ciprofloxacin showed better antimicrobial activity against Escherichia coli and Klebsiella pneumoniae at different concentrations and comparatively lower activity against Proteus vulgaris (Fig. 4e). However, Ag NPs show much better antimicrobial activity for all three strains as compared to commercial ciprofloxacin and the Fagonia cretica extract that shows that the as-synthesized Ag NPs are more effective than commercially available ciprofloxacin.
Fig. 4. Antibacterial activity of (a) the synthesized Ag NPs, (b) ciprofloxacin and (c) the Fagonia cretica extract via the disc method. A plot of the concentration of (d) Ag NPs, (e) ciprofloxacin and (f) the Fagonia cretica extract vs. the zone of bacterial inhibition.
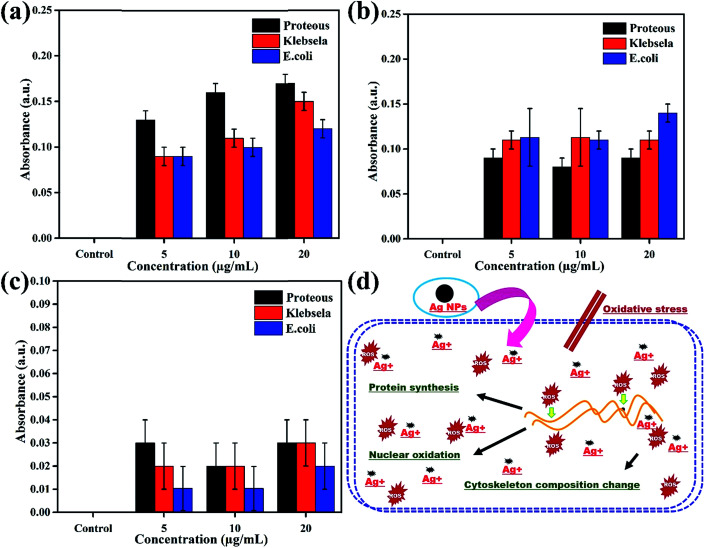
The oxidation of Ag NPs into Ag+ ions assist their diffusion into the biological system or put oxidative stress to produce reactive oxygen species (ROS) that change the permeability of membranes, damage DNA/RNA, proteins and lipids and finally induce cytotoxicity in prokaryotic cells.28,29 Therefore, investigation of the magnitude of ROS production in all three bacterial strains (Proteus vulgaris, Escherichia coli, and Klebsiella pneumoniae) in response to various concentrations (5, 10 & 20 μg) of Ag NPs, ciprofloxacin and the plant extract is very important to quantify the level of toxicity in comparison to control experiments, and the results are presented in Fig. 5a–c. The ROS generation was totally dependent on the dose of treatment as can be seen from Fig. 5a in which as the concentration of Ag NPs increased from 5 μg mL−1 to 20 μg mL−1 the production of ROS also increases to its maximum in all bacterial strains. However, ROS calculations have shown that Ag NPs are comparatively more effective towards Proteus vulgaris as compared to Escherichia coli and Klebsiella pneumoniae. On the other hand, a variation in the ciprofloxacin concentration did not influence the production of ROS much; however, ciprofloxacin shows maximum production of ROS in Escherichia coli as compared to Klebsiella pneumoniae and Proteus vulgaris as shown in Fig. 5b. The plant extract induces the production of a very small amount of ROS; therefore, its concentration did not induce much variation in ROS production and no specificity for any bacterial strain was found as shown in Fig. 5c.
Fig. 5. Production of ROS in various bacterial strains subject to different concentrations of (a) Ag NPs (b) ciprofloxacin and (c) the plant extract after 24 h exposure. (d) Schematic description of the possible mechanism of the action of the antibacterial activity of Ag NPs.
In short, the production of ROS in the presence of Ag NPs is 30% higher than for the positive control for all concentrations in the three bacterial strains. These results suggested that the interaction of Ag NPs with different bacterial cells causes ROS production, which induces toxicity in terms of oxidative stress in the cells, leading to cell death as shown schematically in Fig. 5d. The Ag NPs strongly affect the protein synthesis process in the cells which induces structural changes and ultimately death of the cells, a basic principle behind the antibacterial activity of the Ag NPs.30–34
4. Conclusions
In conclusion, we have successfully synthesized Ag NPs using Fagonia cretica extract in ethanol which is highly environmentally benign. The morphological and structural analysis shows that the as-synthesized Ag NPs are highly crystalline with an average particle size of 16 nm. Furthermore, when employed as an antimicrobial agent, the crystalline Ag NPs showed significant antibacterial activity against Proteus vulgaris, Escherichia coli, and Klebsiella pneumoniae by producing 30% more ROS than the control. The HPLC and FTIR analysis revealed that the Fagonia cretica extract has the ability to reduce Ag ions to Ag atoms which construct Ag NPs and consequently coat them with hydroxyl and amine groups for high stability. Hence, easy and fast production of functional Ag NPs makes the Fagonia cretica extract a promising candidate for the preparation of nanomaterials for various applications e.g. in biomedicine, pharmacokinetics, pharmaceutics and bio-nanotechnology.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the financial and technical support provided by the Islamia University of Bahawalpur, Pakistan, Peking University, China and RMIT University, Australia. The authors would also like to acknowledge the Vice-Chancellor Research Fellowship scheme at RMIT University for funding.
Electronic supplementary information (ESI) available: Part of the experimental section, FTIR and chromatography spectra, and three tables describing the details of zones of inhibition. See DOI: 10.1039/c8na00343b
References
- Ahmed S. Ahmad M. Swami B. L. Ikram S. J. Adv. Res. 2016;7:17–28. doi: 10.1016/j.jare.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M. Yang W. Ju Y. Chu X. Wang Y. Deng Y. Mahmood N. Hou Y. Nano Res. 2017;10:1912–1923. doi: 10.1007/s12274-016-1375-4. [DOI] [Google Scholar]
- Ji Z. X. Jin X. George S. Xia T. Meng H. Wang X. Suarez E. Zhang H. Y. Hoek E. M. V. Godwin H. Nel A. E. Zink J. I. Environ. Sci. Technol. 2010;44:7309–7314. doi: 10.1021/es100417s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M. Mustafa G. Iqbal J. Ashfaq M. Mahmood N. Toxicol. Res. 2018;7:84–92. doi: 10.1039/C7TX00248C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao R. Yu J. Ge Z. G. Zhao L. Y. Sheng F. G. Xu L. L. Li G. J. Hou Y. Nanoscale. 2013;5:11954–11963. doi: 10.1039/C3NR04157C. [DOI] [PubMed] [Google Scholar]
- Murtaza H. Zhongqiu T. Javed I. Umer A. Shiying M. Rongji D. Hong Q. Yulin D. Nanosci. Nanotechnol. Lett. 2013;5:780–784. [Google Scholar]
- Palanisamy S. Rajasekar P. Vijayaprasath G. Ravi G. Manikandan R. Prabhu N. M. Mater. Lett. 2017;189:196–200. doi: 10.1016/j.matlet.2016.12.005. [DOI] [Google Scholar]
- Devia M. S. Ashokkumar K. Annapoorani S. Mater. Lett. 2017;188:197–200. doi: 10.1016/j.matlet.2016.11.016. [DOI] [Google Scholar]
- Lü S. Wu Y. Liu H. Mater. Lett. 2017;196:217–220. doi: 10.1016/j.matlet.2017.03.068. [DOI] [Google Scholar]
- Hasan M. Ullah I. Zulfiqar H. Naeem K. Iqbal A. Gul H. Ashfaq M. Mahmood N. Mater. Today Chem. 2018;8:13–28. doi: 10.1016/j.mtchem.2018.02.003. [DOI] [Google Scholar]
- Sadia A. Syed H. K. Isfahan T. Wajid R. Nasir A. Murtaza H. Nanosci. Nanotechnol. Lett. 2017;9:2005–2012. doi: 10.1166/nnl.2017.2550. [DOI] [Google Scholar]
- Hu S. Hsieh Y. L. Int. J. Biol. Macromol. 2016;82:856–862. doi: 10.1016/j.ijbiomac.2015.09.066. [DOI] [PubMed] [Google Scholar]
- Murtaza H. Javed I. Umer A. Saeed Y. Ranran Y. Liang Y. Dai R. Deng Y. L. J. Nanosci. Nanotechnol. 2015;15:1320–1326. doi: 10.1166/jnn.2015.9090. [DOI] [PubMed] [Google Scholar]
- Paulkumar K. Gnanajobitha G. Vanaja M. Pavunraj M. Annadurai G. Adv. Nat. Sci.: Nanosci. Nanotechnol. 2017;8:035019. [Google Scholar]
- Dang H. Hasan M. Meng W. Zhao H. Iqbal J. Dai R. Lv F. J. Nanopart. Res. 2014;16:2338–2347. doi: 10.1007/s11051-014-2338-x. [DOI] [Google Scholar]
- Salomoni R. Léo P. Montemor A. F. Rinaldi B. G. Rodrigues M. F. A. Nanotechnol., Sci. Appl. 2017;10:115–121. doi: 10.2147/NSA.S133415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W. Jung J. H. Lamsa K. Kim Y. S. Min J. S. Lee Y. S. Mycobiology. 2012;40:53–58. doi: 10.5941/MYCO.2012.40.1.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza H. Javed I. Umer A. Nian X. Hao D. Baradi W. Yasmeen S. Kaleem U. Dai R. Yulin D. J. Nanosci. Nanotechnol. 2014;14:4066–4071. doi: 10.1166/jnn.2014.8201. [DOI] [PubMed] [Google Scholar]
- Priya R. S. Geetha D. Ramesh P. S. Ecotoxicol. Environ. Saf. 2016;134:308–318. doi: 10.1016/j.ecoenv.2015.07.037. [DOI] [PubMed] [Google Scholar]
- Wang H. Joseph J. A. Free Radical Biol. Med. 1999;27:612–616. doi: 10.1016/S0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
- Bar H. Bhui D. K. Sahoo G. P. Sarkar P. De S. P. Misra A. Colloids Surf., A. 2009;339:134–139. doi: 10.1016/j.colsurfa.2009.02.008. [DOI] [Google Scholar]
- Li L. Sun J. Li X. Zhang Y. Wang Z. Wang C. Dai J. Wang Q. Biomaterials. 2012;33:1714–1721. doi: 10.1016/j.biomaterials.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Song J. Y. Jang H. K. Kim B. S. Process Biochem. 2009;44:1133–1138. doi: 10.1016/j.procbio.2009.06.005. [DOI] [Google Scholar]
- Kumara R. Roopanb S. M. Prabhakarnc A. Khannaa V. G. Chakroborty S. Spectrochim. Acta, Part A. 2012;90:173–176. doi: 10.1016/j.saa.2012.01.029. [DOI] [PubMed] [Google Scholar]
- Zahir A. A. Chauhan I. S. Bagavan A. Kamaraj C. Elango G. Shankar J. Arjaria N. Roopan S. M. Rahuman A. A. Neeloo S. Antimicrob. Agents Chemother. 2015;59:4782–4799. doi: 10.1128/AAC.00098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhumitha G. Elango G. Roopan S. M. J. Sol-Gel Sci. Technol. 2015;014:14-3591–14-3598. [Google Scholar]
- Anand K. Kaviyarasu K. Muniyasamy S. Roopan S. M. Gengan R. M. Chuturgoon A. A. J. Cluster Sci. 2017;28:2279–2291. doi: 10.1007/s10876-017-1212-2. [DOI] [Google Scholar]
- Thiyagarajan K. Bharti V. K. Tyagi S. Tayagi P. K. Ahuja A. Kumar K. Raj T. Kumar B. RSC Adv. 2018;8:23213–23229. doi: 10.1039/C8RA03649G. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Elbourne A. Coyle V. E. Troung V. K. Sabri Y. M. Kandjani A. E. Bhargava S. K. Ivanova E. P. Nanoscale Adv. 2018 doi: 10.1039/c8na00124c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivask A. ElBadway A. Kaweeteerawat C. Boren D. Fischer H. Ji Z. Chang C. H. Liu R. Tolaymat T. Telesca D. Zink J. I. Cohen Y. Holden P. A. Godwin H. A. ACS Nano. 2014;8:374–386. doi: 10.1021/nn4044047. [DOI] [PubMed] [Google Scholar]
- Tian X. Jiang X. Welch C. Croley T. R. Wong T. Chen C. Fan S. Chong Y. Li R. Ge C. Chen C. Yin J. ACS Appl. Mater. Interfaces. 2018;10:8443–8450. doi: 10.1021/acsami.7b17274. [DOI] [PubMed] [Google Scholar]
- Xiu Z. Zhang Q. Puppala H. L. Colvin V. L. Alvarez P. J. J. Nano Lett. 2012;12:4271–4275. doi: 10.1021/nl301934w. [DOI] [PubMed] [Google Scholar]
- Ouay B. L. Stellacci F. Nano Today. 2015;10:339–354. doi: 10.1016/j.nantod.2015.04.002. [DOI] [Google Scholar]
- Panáček A. Kvítek L. Smékalová M. Večeřová R. Kolář M. Röderová M. Dyčka F. Šebela M. Prucek R. Tomanec O. Zbořil R. Nat. Nanotechnol. 2018;13:65–71. doi: 10.1038/s41565-017-0013-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.