Abstract
The receptor binding domain (RBD) of the SARS-CoV-2 spike protein is the primary target of neutralizing antibodies and is a component of almost all current vaccines. Here, RBD immunogens were created with stabilizing amino acid changes that improve the neutralizing antibody response, as well as characteristics for production, storage, and distribution. A computational design and in vitro screening platform identified three improved immunogens, each with approximately nine amino acid changes relative to the native RBD sequence, and four key changes conserved between immunogens. The changes are adaptable to all vaccine platforms and compatible with mutations in emerging variants of concern. The immunogens elicit higher levels of neutralizing antibodies than native RBD, focus the immune response to structured neutralizing epitopes, and have increased production yields and thermostability. Incorporating these variant-independent amino acid changes in next-generation COVID vaccines may enhance the neutralizing antibody response and lead to longer duration and broader protection.
A design pipeline identifies amino acid changes to the RBD of spike, improving biophysical characteristics and vaccine efficacy in mice.
INTRODUCTION
Coronavirus disease 2019 (COVID-19) remains a global health issue, killing nearly a million people and sickening millions more in early 2022 alone (1). Several severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines effectively prevent severe disease and temporarily prevent infection, but efficacy against infection wanes over time and is diminished against emerging variants. Furthermore, regular booster requirements will continue to strain global distribution efforts of existing vaccines. Therefore, SARS-CoV-2 vaccine development is still required to produce next-generation vaccines with improved efficacy, stability, manufacturability, and duration of protection.
Structure-based antigen design has proven successful for SARS-CoV-2, and extending these efforts could further improve these vaccines. The most effective SARS-CoV-2 vaccines contain an engineered spike protein antigen with stabilizing amino acid changes in the S2 domain (2–5) that improve the neutralizing antibody response (6–8). These changes eliminate a furin protease cleavage site and stabilize the prefusion conformation of the spike protein, increasing the structural and conformational stability of the antigen. Stabilizing mutations have also improved respiratory syncytial virus (RSV), HIV, foot and mouth disease virus (FMDV), and other vaccine candidates in preclinical animal studies, establishing antigen stabilization as an effective method of vaccine design (9). Similarly, undesired nonneutralizing immunodominant epitopes can be removed to focus the immune response toward protective epitopes and improve vaccine candidates (10). However, these designs are largely human-guided efforts limited to predictable noncombinatorial amino acid changes. Computational methods have improved markedly in the past several years, and the software suite ROSETTA can calculate the energetic effects of combinatorial amino acid changes. ROSETTA has been used to stabilize antigens in vitro, isolate neutralizing epitopes, target germline antibodies, and create novel nanoparticles (11–15). However, there are few examples of computationally stabilized or immunofocused antigens eliciting enhanced protection in animals. Here, we sought to create a computational and experimental antigen design pipeline that could enhance SARS-CoV-2 vaccine efficacy and be broadly applicable to other antigens and pathogens.
The receptor binding domain (RBD) of the SARS-CoV-2 spike protein is one of the most promising targets for structure-based antigen design. The RBD engages the Ace2 receptor to mediate viral entry and is consequently the target of the most potent SARS-CoV-2 neutralizing antibodies (Fig. 1, A and B) (16–20). However, potent neutralizing antibodies are rare in convalescent patients, relative to nonneutralizing antibodies that bind epitopes elsewhere on the spike protein (19–21). This phenomenon could be due to structural instability in neutralizing epitopes of the RBD and the ability of the RBD to adopt a “down” conformation that obscures neutralizing epitopes in the spike trimer (Fig. 1A). The RBD is thought to transiently sample an “up” conformation compatible with Ace2 binding, which exposes neutralizing epitopes including some that may cross-protect against other coronaviruses (22–26). Consistent with these observations, the native RBD sequence is an effective vaccine (27, 28). However, the RBD contacts other domains in the context of the full-length (FL)–spike protein, contains nonneutralizing epitopes, and may not stably present all neutralizing epitopes. Here, we have used our design pipeline to stabilize the RBD structure, focus the immune response to neutralizing epitopes and away from nonneutralizing epitopes, and expose the RBD either as a stand-alone vaccine or in the RBD-up conformation of the spike trimer. These designed RBD immunogens have improved characteristics in vitro, improve the neutralizing antibody response in mice, and would be expected to increase the efficacy of spike-based vaccines.
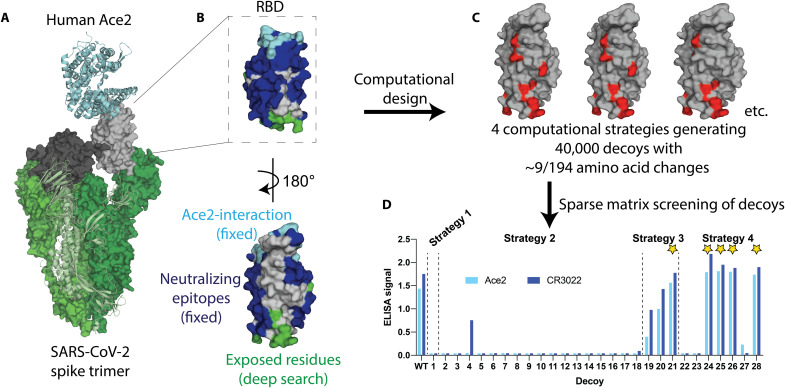
Fig. 1. Overview of SPEEDesign pipeline used to create RBD immunogens.
(A) The SARS-CoV-2 spike trimer (green) binds human Ace2 (cyan) to mediate viral entry. This interaction is mediated by the up conformation of the RBD (gray), which can also exist in a down conformation (black). (B) The RBD design process retained the Ace2-interaction surface (cyan) and all known SARS neutralizing epitopes (blue). Residues exposed upon isolation of the RBD (green) were heavily designed, while all other residues (gray) were designed more conservatively. (C) Four computational strategies were used to create 40,000 decoys, each of which has an average of nine amino acid changes (red) from the native SARS-CoV-2 sequence. (D) Twenty-eight sequences sampling the top scoring decoys were screened in vitro, identifying five lead candidates (stars).
RESULTS
SPEEDesign of novel RBD immunogens
We used a ROSETTA-based computational design and in vitro screening approach to develop improved RBD vaccine candidates. The objectives of this approach are to improve the protective efficacy of a given antigen by (i) focusing the immune response toward potently neutralizing antibody epitopes on the antigen; (ii) reducing or eliminating immune responses to poorly neutralizing and/or immunodominant epitopes within the antigen; (iii) optimizing the thermal stability of the antigen to increase its durability in vivo following immunization; and (iv) promoting conformational states of a protein antigen that may be hidden, for example, the up state of the RBD in the spike protein. These four objectives are achieved by careful definition of the role of individual amino acids within the antigen and, by extension, their mutability in ROSETTA design strategies, coupled to a clustering and in vitro screening approach. Given the objectives outlined above, we refer to this computational and in vitro screening pipeline as Stabilizer for Protein Expression and Epitope Design (SPEEDesign) in the subsequent sections.
The Ace2 interface and all known neutralizing epitopes within the RBD were retained by preventing amino acid changes, because the designed immunogen is expected to focus the immune response to these segments (Fig. 1B). Residues that are exposed in the RBD-up conformation, or that are exposed upon extraction of the RBD from the FL-spike trimer, were thoroughly searched during the design process to identify amino acid changes that would stabilize an accessible RBD. All remaining residues, which likely include surface residues that comprise poorly or nonneutralizing epitopes, were allowed to sample a limited sequence space defined by energetic and evolutionary restraints (29).
Four different computational ROSETTA design strategies, which differ in the depth of design for each class of residues (see Materials and Methods), were used to identify amino acid changes that would stabilize the RBD (Fig. 1C). Ten thousand decoys were produced for each computational strategy, each of which had approximately nine amino acid changes from the native RBD sequence. Twenty-eight decoys were selected using a clustering algorithm designed to broadly sample the 40,000 computational decoys (Fig. 1D). These decoys were distributed between strategies according to the sequence diversity produced by each strategy. For example, more decoys were selected from strategy 2 than strategy 1, because strategy 2 sampled a much larger sequence space.
The 28 representative sequences were screened in vitro for expression and presentation of neutralizing epitopes (Fig. 1D). We used Ace2 to probe the integrity of the most potent neutralizing epitope and the monoclonal antibody (mAb) CR3022, which recognizes an orthogonal surface conserved between coronaviruses. All successful lead immunogen candidates were derived from strategies 3 and 4, which sampled a moderate sequence space, rather than the highly restricted strategy 1 or highly divergent strategy 2. In addition, the immunogens with the best ROSETTA score from each strategy (decoys 1, 2, 19, and 22) were not successful, indicating that ROSETTA score alone is not a suitable predictor for a successful design. The clustering and screening strategies unique to the SPEEDesign pipeline are a critical advancement that allows identification of improved immunogens among the many decoys produced during ROSETTA design.
Immunogen expression and stability
Five lead candidate immunogens (1 to 5, corresponding to decoys 28, 24, 25, 26, and 21, respectively) were expressed and purified to determine their biophysical characteristics. All immunogens were well-folded monomeric proteins that can be highly purified with yields much greater than native wild-type (WT) RBD (Fig. 2, A, B, and D). Three immunogens (1, 2, and 3) can be purified with final yields ≥200 mg/liter, or more than fourfold higher than WT RBD (Fig. 2D). Furthermore, these yields are approximately 6-fold higher than the yields reported for an optimized FL-spike ectodomain known as “hexapro” and 200-fold higher than the well-established 2P prefusion–stabilized FL-spike ectodomain (30). These yields were achieved without optimization of the expression and purification system, suggesting that process development could reach production levels sufficient for development of a highly cost-effective vaccine.
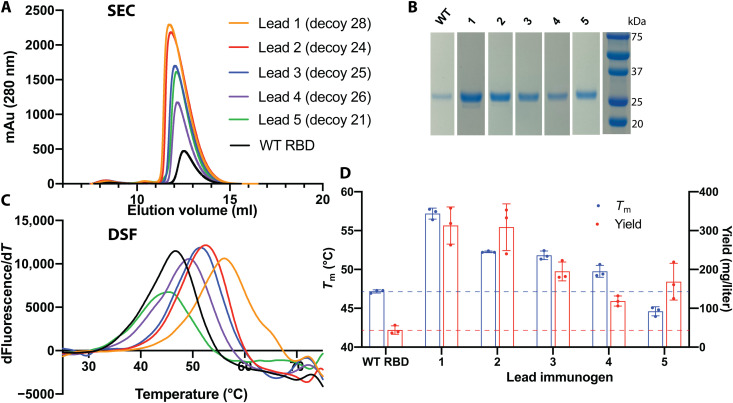
Fig. 2. Stability and yield are higher for immunogens than WT RBD.
(A) All five immunogens expressed at higher levels than WT RBD and eluted as monomers by size exclusion chromatography (SEC). (B) SDS–polyacrylamide gel electrophoresis confirmed the high purity and yield of RBD immunogens. (C) Differential scanning fluorimetry indicated that four immunogens have higher thermostability than WT RBD. (D) Tm and purification yield averages and SDs from three separate purifications.
The thermostability of four lead immunogens was also higher than WT RBD (Fig. 2, C and D). One immunogen (1) had a melting temperature (Tm) of 57° or 10°C higher than WT RBD, and two additional immunogens (2 and 3) had greater than 5°C increases in Tm. Expression yield roughly correlated with Tm, and immunogens 1, 2, and 3 had substantially higher yields and stability than WT RBD. Increased thermostability likely contributes to increased immunogen yield, will likely increase the half-life of the antigen in the body, and will likely improve stability during storage, transportation, and administration of a vaccine, alleviating cold-chain requirements.
Structural basis for immunogen stability
The five lead immunogens exhibited a distinct pattern of amino acid changes that provide a mechanistic basis for their enhanced biophysical characteristics. There are nine positions at which multiple lead immunogens had nonconservative amino acid substitutions relative to the native protein sequence (Fig. 3A and table S1). At four of these positions, the immunogens shared similar amino acid identities: (i) Alanine-363 was changed to a large hydrophobic residue in all five lead immunogens, (ii) I468T in all five lead immunogens, (iii) H519D in all five lead immunogens, and (iv) A522P in three of five lead immunogens.
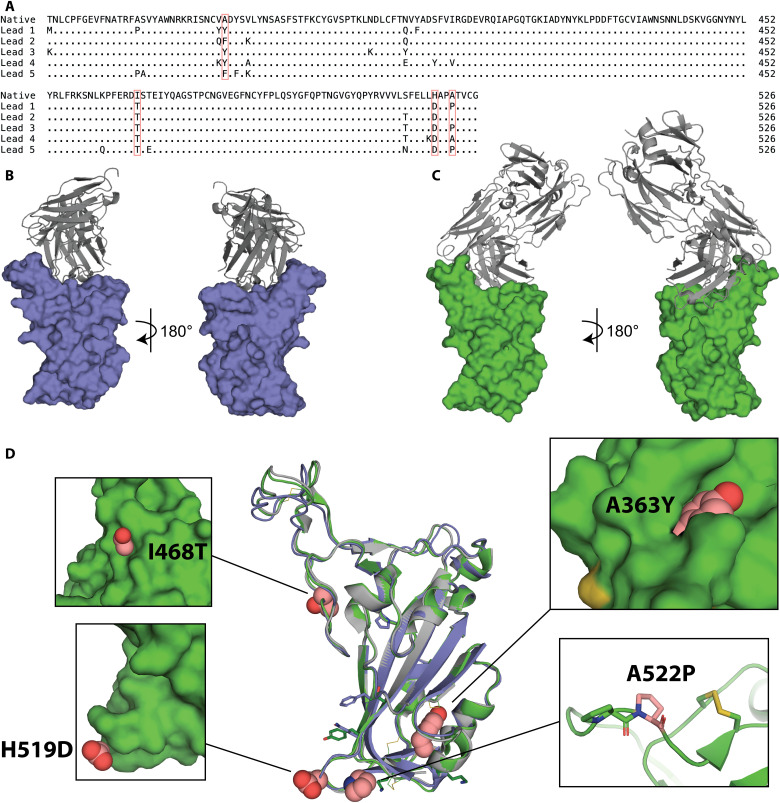
Fig. 3. Structural basis for immunogen stabilization.
(A) Sequence alignment of amino acid changes in lead immunogens relative to the native RBD sequence. Four recurring changes are highlighted in pink. (B) Crystal structure of lead 1 (slate) in complex with scFv C144 (gray). (C) Crystal structure of lead 3 (green) in complex with Fab P2B-2F6 (gray). (D) The crystal structures of lead immunogens 1 (slate) and 3 (green) are globally similar to WT RBD (gray; PDB:7BWJ) despite amino acid changes (recurring, spheres; unique, sticks). Insets illustrate key recurring substitutions (pink).
To determine how these amino acid changes improve the stability of the immunogens, we solved the crystal structures of lead immunogens 1 and 3 in complex with the neutralizing scFv C144 and Fab P2B-2F6, respectively (Fig. 3, B and C) (31, 32). The overall structures of the immunogens were very similar to the native RBD, with Cα root mean square deviations (RMSDs) of 0.913 and 0.476 Å for immunogens 1 and 3, respectively, indicating that the amino acid changes did not alter the overall shape, secondary structure, or tertiary structure of the RBD (Fig. 3D and table S2). The enhanced biophysical characteristics were driven by local structural changes around substituted side chains. For example, A363Y is a space-filling substitution that likely stabilizes the protein fold and proximal disulfide bonds. Ile468 and His519 are buried in the RBD-down conformation and become exposed in the RBD-up conformation (fig. S1). Therefore, the I468T and H519D substitutions increase the hydrophilicity of solvent-exposed residues, likely improving the solubility of RBD immunogens and potentially promoting the RBD-up conformation in the context of the spike trimer. Last, A522P creates a tandem proline sequence that likely promotes a sharp kink in the backbone adjacent to a disulfide bond. All amino acid changes in the immunogens, including the four key positions, are distinct from the changes found in naturally occurring variants of concern (e.g., Beta, Delta, and Omicron), suggesting that these immunogens are compatible with next-generation vaccines that derive from variant sequences (fig. S2). Immunogens 1 to 3 exhibited increased expression in the context of the Beta variant to an even greater extent than the original variant (fig. S2C). Thus, stabilizing mutations promoted the stability of the immunogens through diverse structural and energetic mechanisms that do not affect the global antigen structure.
Neutralizing epitopes on immunogens
We probed the Ace2 receptor–binding site and several key three-dimensional antibody epitopes to establish that the stabilizing mutations did not disrupt key interacting residues or neutralizing epitopes in the immunogens (fig. S3). Ace2 binds to the end of the RBD, as do potently neutralizing Ace2-blocking antibodies that include the mAbs REGN10933 and P2B-2F6 (17, 31, 33). Additional neutralizing mAbs CR3022 and S309 bind to opposing sides and do not overlap with each other or the Ace2 binding site (Fig. 3A) (34, 35). The Ace2 binding site and these four epitopes therefore report the structural integrity of almost all neutralizing surfaces retained during the design process. The designed immunogens bound Ace2, REGN10933, P2B-2F6, CR3022, and S309 at least as well as WT, with the exception of immunogen 5, which showed a slight decrease in binding to P2B-2F6, suggesting that the stabilizing changes made to the immunogens 1 to 4 did not perturb nearby surfaces and critical neutralizing epitopes (fig. S3B).
We further used biolayer interferometry (BLI) to measure the integrity of the Ace2 binding site and REGN10933 epitope in a more quantitative fashion (fig. S3C). Consistent with the enzyme-linked immunosorbent assay (ELISA) results, immunogens 1 to 4 bound both probes with biophysical parameters very similar to WT RBD (table S3). While these probes are only a small fraction of the antibodies reported to bind SARS-CoV-2, they represent all four classes of epitopes recognized by known RBD antibodies (36). Therefore, neutralizing epitopes were unperturbed on RBD immunogens 1 to 4, and the resulting immune response would be expected to recognize the native RBD protein and SARS-CoV-2.
Vaccination with immunogens enhances neutralization
Mice were immunized with the five lead immunogens to determine whether amino acid changes improved the neutralizing antibody response (Fig. 4). CD-1 outbred mice were used to mimic the genetic diversity found in the human population more closely than inbred mice. A group of mice was also immunized with a trimeric FL-spike ectodomain antigen containing the stabilizing 2P and polybasic site mutations. This antigen is similar to the antigen used in most approved vaccines, providing an approximate performance benchmark. All immunogens generated antibodies that recognize trimeric FL-spike ectodomain, and immunogens 1, 2, and 3 generated geometric mean titers (GMTs) greater than WT RBD, consistent with their enhanced biophysical characteristics (Fig. 4B). The GMTs elicited by all RBD immunogens were lower than FL-spike, consistent with the removal of the N-terminal and S2 domains in the RBD antigens. We measured inhibition of RBD/Ace2 binding to evaluate the titers of functional antibodies elicited by each antigen (Fig. 4C). Again, immunogens 1, 2, and 3 generated Ace2-blocking GMTs up to 10-fold higher than WT RBD and comparable to FL-spike. At this point, immunogens were down-selected, and 4 and 5 were omitted from further analysis because they consistently performed worse than 1 to 3.
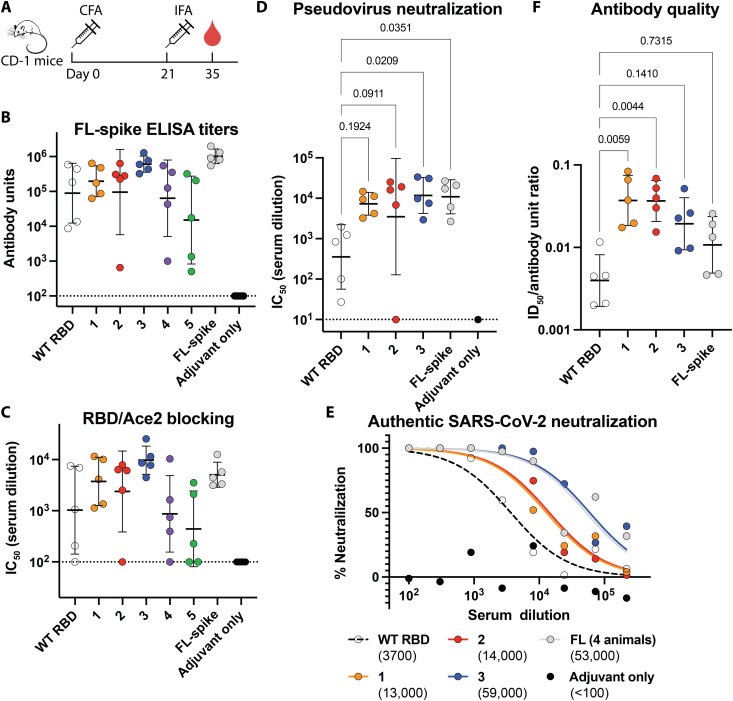
Fig. 4. Neutralizing antibody titers are higher in mice immunized with immunogens than WT RBD.
(A) Immunization and blood draw schedule for CD-1 mice. (B) Serum ELISA titers against trimeric FL-spike ectodomain. Dashed line indicates detection limit of assay, bars represent GMT, and error bars indicate geometric SD. (C) Titers of antibodies blocking Ace2/RBD interaction depicted as described in (B). (D) Pseudovirus neutralization titers depicted as described in (B). Statistical comparisons were made using a Kruskal-Wallis analysis of variance (ANOVA) followed by Dunn’s test corrected for multiple comparisons of immunogens and FL-spike with WT RBD. (E) Neutralization of WA-1 SARS-CoV-2 in a plaque assay. ID50 values are indicated in parentheses. Serum from each animal in a group was pooled, except for the animal with the lowest pseudovirus neutralization titers in the FL-group, which had insufficient volume. (F) Antibody quality measured by ratio of pseudovirus neutralization titers (D) and FL-spike ELISA titers (B), depicted as described in (B). Statistical test is identical to (D).
We measured neutralizing titers in a pseudoviral neutralization assay and again found that immunogens 1, 2, and 3 had GMTs greater than WT RBD and comparable to FL-spike. (Fig. 4D). Immunogen 3 elicited neutralizing titers significantly greater than WT RBD with a GMT more than 30-fold higher than WT, an even greater enhancement than the ~2-fold increase provided by the 2P and polybasic stabilizing mutations to FL-spike (37). Last, we measured the neutralizing activity of pooled serum against authentic SARS-CoV-2 (Fig. 4E). Again, all three immunogens have improved neutralizing titers. Immunogen 3 elicits approximately 10-fold higher titers than WT RBD and reaches levels comparable to FL-spike.
DISCUSSION
These results demonstrate that a handful of amino acid changes to the RBD can improve the protective antibody response. A 30-fold increase in neutralizing antibody titers equates to lengthening the lifetime of neutralizing antibodies by approximately five half-lives of decay, suggesting that immunogens may confer a much longer duration of antibody-mediated protection than WT RBD. In addition, this 30-fold enhancement is comparable to the reduction in neutralizing potential against new SARS-CoV-2 variants, such as Omicron, suggesting that these immunogens may increase the breadth of protection, in addition to duration (38–43).
The three lead immunogens appear to have achieved the objectives set out for SPEEDesign. While it is difficult to pinpoint the cause of improved vaccine performance, the three immunogens appear to have elicited a higher quality of antibody response than WT RBD. For example, immunogen 3 elicited 30-fold greater neutralizing antibody titers but only 7-fold greater spike ELISA titers, meaning that the increase in immunoglobulin G (IgG) levels alone is insufficient to explain the large improvement in neutralizing antibody titers. The antibody quality can be quantified by calculating the ratio of neutralizing antibodies to total antibodies in each animal (Fig. 4F). All three immunogens elicit a higher quality antibody response than WT RBD, and immunogens 1 and 2 have significantly higher quality antibodies than WT RBD. These results indicate that SPEEDesign achieved the major objectives of focusing the immune response to potently neutralizing epitopes and away from poorly neutralizing epitopes. We expect that SPEEDesign will achieve similar success with other antigens that have structurally characterized neutralizing epitopes and will be useful for designing vaccines against many other pathogens.
These monomeric designed RBD immunogens are expected to improve existing vaccines. The native RBD sequence is used in two approved vaccines (Corbevax and Soberana 2), and the immunogens reported here elicit higher neutralizing antibody titers than the native sequence. In addition to the enhanced neutralization, improvements in stability and yield suggest that recombinant RBD immunogens would be easily manufactured and distributed to meet the global need for a SARS-CoV-2 vaccine. These manufacturing and distribution benefits would be even more meaningful in the event that seasonal updates and mass vaccination campaigns are required. Last, these immunogens can be easily improved by established methods of foldon trimerization, multimerization, and/or nanoparticle display (44–48).
The designed immunogens can also be easily adapted to all other vaccine platforms by making amino acid changes to the RBD sequence within the FL-spike or within RBD-only vaccines. This study is limited to the investigation of a recombinant RBD antigen in a model adjuvant, but we expect that enhancing the antigen would benefit a vaccine regardless of delivery platform and adjuvant. Additional studies of specific vaccine candidates are required to determine the precise magnitude of benefit for each unique platform. The changes identified here do not exclude the existing improvements in spike, they are fully compatible with changes in the S2 domain, and they are compatible with all known changes in emerging SARS-CoV-2 variants of concern. The SPEEDesign changes stabilize the RBD in isolation and may provide additional benefit to an FL-spike vaccine by restricting the down conformation of the RBD. Regardless of the platform, these amino acid changes would be expected to increase RBD stability, enhance the protective immune response, and lengthen the lifetime and breadth of protection conferred by a SARS-CoV-2 vaccine.
MATERIALS AND METHODS
SPEEDesign residue definition
Each amino acid in the target antigen was categorized as fixed, intermediate, or deep search, defining the depth of the computational search at that position. RBD residues that form the interface with Ace2 or neutralizing antibodies were defined as fixed (33, 34, 49–55). These epitopes accurately predicted and covered the residues targeted by all known SARS-CoV-2 antibodies that target the RBD including those recently identified after the design process was initiated and therefore retained the SARS-CoV-2 neutralizing epitopes as fixed. The residues that comprise these epitopes were defined as those that have a >1-Å change in solvent-accessible surface area upon complex formation, and calculations were performed in PyMOL (56).
Those residues that are exposed in the RBD-up conformation or upon extraction of the RBD from the FL-spike protein were defined as deep search. Residues exposed upon domain extraction from a larger protein require unique handling during the design processes. These residues are buried or interacting with other residues in the larger protein, and they become fully solvent exposed once the domain is extracted. This marked change in chemical environment is accommodated by allowing deep search residues to vary greatly during the design process. Since these residues are not exposed in homologous proteins, conservation or evolutionary-based design principles may not prove sufficient to redesign these new nonnatural surfaces. We therefore classified these residues for deep search during design where all amino acids except cysteine are allowed.
All other residues were defined as intermediate. These residues were allowed to vary to a limited extent that is driven by conservation and evolutionary analysis of similar protein sequences to identify potential amino acid changes.
SPEEDesign ROSETTA strategies
All ROSETTA strategies left fixed residues unchanged to preserve neutralizing epitopes, and each strategy differs in the amino acid changes allowed for the intermediate and deep search categories of residues. In strategy 1, intermediate residues were unchanged and all amino acids except cysteine were allowed at deep search positions. In strategy 2, all amino acids were allowed at deep search positions, and intermediate positions were allowed to sample amino acids found in proteins with similar sequences (evolutionary constraints). Evolutionary constraints in strategy 2 were defined by creating a position-specific scoring matrix (PSSM) using PSI-BLAST and including variant amino acids in the ROSETTA design process if they had scores >0 in the PSSM (57). As a result, strategy 2 sampled a very large sequence space. This sequence space was constrained in strategy 3 by disallowing amino acid changes that are energetically unfavorable when made individually (energetic constraints), an approach adapted from the PROSS protocol (29). The energetic constraints in strategy 3 were imposed by using the ROSETTA FilterScan module, which estimates the energetic effect of individual amino acid changes (58). Amino acid changes from the PSSM were disallowed if they had energetic penalties (delta_filters) > 0.5 Rosetta energy units (R.e.u.). Strategy 4 placed the same evolutionary and energetic constraints on the intermediate residues as strategy 3 but with a more stringent energetic cutoff of −0.45 R.e.u. In addition, strategy 4 ignored all energetic and evolutionary restraints for deep search residues, allowing them to sample all amino acids. These amino acid constraints were used to create a ResFile for each strategy that was used by the FastDesign module in ROSETTA.
SPEEDesign clustering
For each computational strategy, decoys with scores in the 95th percentile were clustered by sequence similarity using CD-HIT and the top scoring decoy from each cluster was selected as a representative sequence (59). The number of clusters was selected on the basis of the sequence diversity produced in each computational strategy. For example, strategy 1 sampled a limited sequence space, while strategy 2 sampled a very large sequence space. Therefore, more clusters were created to sample strategy 2 than strategy 1.
SPEEDesign in vitro screening
Synthetic DNA coding for secreted RBD immunogens was cloned (GenScript) into a customized pHL-sec expression plasmid. pHL-sec was a gift from E. Y. Jones (Addgene plasmid no. 99845; http://n2t.net/addgene:99845; RRID:Addgene_99845) (60). Plasmid was transfected into human expi293F cells and grown in a 96-well plate according to the manufacturer’s instructions (Thermo Fisher Scientific). Cell-free supernatant was harvested after 5 days of expression.
Cell-free supernatant was diluted in phosphate-buffered saline–Tween 20 (PBST) + 2% bovine serum albumin (BSA) and added to Ni-NTA HisSorb Plates (Qiagen) to capture His-tagged immunogens. After incubation for 1 hour at room temperature, plates were washed three times with PBST. Neutralizing epitopes were probed using an Ace2-Fc(IgG1) fusion (0.2 μg per well) or a human IgG1 antibody containing the CR3022 variable domain (0.05 μg per well). After incubation for 1 hour at room temperature, plates were washed three times with PBST and 200 μl of 1:5000 peroxidase-conjugated anti-human IgG was added (Jackson ImmunoResearch Laboratories Inc., catalog no. 109-035-098). Plates were incubated for 30 min at room temperature and washed three times with PBST. Last, 70 μl of tetramethylbenzidine (TMB) (MilliporeSigma) was added and incubated for 5 min at room temperature before quenching with 70 μl of 2 M H2SO4. Absorbance at 450 nm (Abs450) was measured using a Biotek Synergy H1 plate reader.
Immunogen expression, purification, and calculation of yields
Recombinant RBD immunogens were expressed in expi293F cells, as described for SPEEDesign in vitro screening above. Trimeric FL-spike ectodomain was also expressed in expi293F cells using a modified pHL-sec vector. The construct contains amino acids 16 to 1208 of the spike protein followed by a foldon trimerization domain and a 6-His tag. This construct also contains the “2P” stabilizing mutations at K986P and V987P and mutation of the furin cleavage site (682-685 GSAS to RRAR) (25).
Cell-free supernatant was harvested after 4 days, and His-tagged immunogens were purified by gravity chromatography using Ni Sepharose excel resin according to the manufacturer’s instructions (Cytiva). Immunogens were further purified by size exclusion chromatography using a Superdex 75 Increase 10/300 GL column (RBD immunogens) or Superose 6 Increase 10/300 GL column (trimeric FL-spike) equilibrated in 1× PBS. Fractions corresponding to trimeric FL-spike or monomeric RBD were pooled, snap-frozen in liquid nitrogen, and stored at −80°C.
Transfection, expression, and purification were performed in triplicate on three separate days to calculate RBD immunogen purification yields. Each replicate consisted of a 30-ml culture, and yields were calculated by integrating the area under the monomeric peak on the Abs280 chromatogram during size exclusion chromatography. These yields closely matched yields calculated from pooled fractions. Extinction coefficients were calculated using the ExPASy ProtParam tool (61).
Antibody expression and purification
Antibodies for ELISAs were created by fusing the variable regions for the indicated antibody to the human IGHG*01, IGKC*01, or IGLC2*02 constant regions and cloning into the pHL-sec plasmid (GenScript). The Ace2-Fc fusion was expressed from pcDNA3: pcDNA3-sACE2(WT)-Fc(IgG1) was a gift from E. Procko (Addgene plasmid no. 145163; http://n2t.net/addgene:145163; RRID:Addgene_145163) (62). Heavy and light chain plasmids were mixed in equal amounts and transfected into expi293F cells according to the manufacturer’s instructions, and cell-free supernatant was harvested after 4 days of expression (Thermo Fisher Scientific).
Cell-free supernatant was batch-incubated with protein A agarose resin (GoldBio) for 1 hour at room temperature. Resin was collected and washed with 10 column volumes (CVs) of protein A IgG-binding buffer (Thermo Fisher Scientific). Protein was eluted with 10 CVs of IgG elution buffer (Thermo Fisher Scientific) and neutralized with 1 CV of 1 M tris (pH 9.0). Antibodies were concentrated and buffer-exchanged into PBS using an Amicon centrifugal filter (MilliporeSigma). Ace2-Fc was further purified by size exclusion chromatography using a Superdex 200 Increase 10/300 GL column (Cytiva) equilibrated in 1× PBS.
Differential scanning fluorimetry
Differential scanning fluorimetry (DSF) was performed using the Protein Thermal Shift Dye Kit according to the manufacturer’s instructions (Thermo Fisher Scientific). Final reactions contained purified immunogen (0.125 mg/ml), 1× Protein Thermal Shift buffer, 1× Thermal Shift Dye, and 0.63× PBS. Fluorescence was monitored using a 7500 Fast Real-Time polymerase chain reaction system (Thermo Fisher Scientific) as the temperature was increased from 25° to 95°C at a ramp rate of 1%. Tm was calculated as the peak of the derivative of the melt curve. DSF reactions were performed in technical quadruplicate on each plate and in biological triplicate using three different protein preps on three separate days. Technical replicates were averaged to calculate the Tm for a biological replicate, and the three biological replicates were averaged to calculate the reported Tm.
Crystallization
The antigen-binding fragment (Fab) of P2B-2F6 was used to promote crystallization of lead immunogen 3. P2B-2F6 Fab was produced by fusing the P2B-2F6 variable region to a His-tagged human IGHG*01 CH1 domain and cloning into pHL-sec (GenScript). Heavy chain Fab plasmid was cotransfected 1:1 with light chain in expi293F cells according to the manufacturer’s instructions, and cell-free supernatant was harvested after 4 days of expression (Thermo Fisher Scientific). His-tagged Fab was purified from cell-free supernatant by gravity chromatography using Ni Sepharose excel resin according to the manufacturer’s instructions (Cytiva). Fab was further purified by size exclusion chromatography using a Superdex 75 increase 10/300 GL column (Cytiva) equilibrated in 1× PBS.
Purified Fab was mixed with purified lead immunogen 3 in a 1.5:1 ratio and incubated for 30 min on ice. Complex was purified by size exclusion chromatography on a Superdex 200 Increase 10/300 GL column equilibrated in 10 mM Na-Hepes (pH 7.4) and 100 mM NaCl. Purified complex was concentrated to 18 mg/ml using an Amicon centrifugal filter (MilliporeSigma), and crystal trays were set up using a mosquito crystal robot (STP Labtech). Drops contained 0.2 μl of complex and 0.2 μl of reservoir solution [0.2 M sodium fluoride, 20% (w/v) polyethylene glycol (PEG) 3350]. Crystals were grown by hanging-drop vapor diffusion at 18°C for 13 days. Crystals were cryoprotected in a solution that contained 7 μl of well solution and 3 μl of 100% glycerol and flash-frozen in liquid nitrogen.
His-tagged C144 scFv was expressed and purified as described above for other proteins. Immunogen 1 was mixed with C144 scFv in a 1:1 molar ratio. The admixture was purified by size exclusion chromatography. Purified complex (15 mg/ml) was mixed with 0.04 M potassium dihydrogen phosphate, 16% PEG 8000, and 20% glycerol in 1:1 ratio for crystallization. Crystals were grown using hanging-drop vapor diffusion at 18°C. The crystal was cryoprotected in well solution supplemented with 30% glycerol before flash-freezing in liquid nitrogen.
Data collection and structure determination
Crystal diffraction data were collected at 1.0 Å at 100°K on the GM/CA 23-ID-D beamline at the Advanced Photon Source. Data were processed using autoPROC or XDS (63, 64). Reflections were indexed and integrated using XDS (64). Data were scaled and merged using AIMLESS (65, 66). The P2B-2F6/WT RBD structure (Protein Data Bank: 7BWJ) was used as a starting model for rigid body refinement in PHENIX Refine (67). This model was then edited in COOT to incorporate the amino acid changes, and subsequent rounds of refinement and model building were performed with COOT and PHENIX Refine (68). The final model was evaluated with MolProbity, which showed good geometry, with 96.0% of the residues as Ramachandran favored and 0% outlier residues (table S2) (69). Software used in this project was curated by SBGrid (70).
ELISA analysis of immunogen epitopes
Nunc MaxiSorp plates (Thermo Fisher Scientific) were coated with 100 μl of purified immunogen (0.01 mg/ml) diluted in 50 mM Na-carbonate (pH 9.5). Plates were coated overnight at 4°C and then washed three times with PBST. Plates were blocked for 1 hour at room temperature with 2% BSA in PBST and then washed three times with PBST. Primary antibody (100 μl) was added to each well at the indicated concentration: Ace2, 3.1 ng/ml; REGN10933, CR3022, and S309, 1.5 ng/ml; and P2B-2F6, 7.5 ng/ml. Primary antibody was incubated for 1 hour at room temperature, then plates were washed three times with PBST, and 200 μl of 1:5000 peroxidase-conjugated anti-human IgG was added (Jackson ImmunoResearch Laboratories Inc., catalog no. 109-035-098). Plates were incubated for 30 min at room temperature and washed three times with PBST. Last, 70 μl of TMB (MilliporeSigma) was added and incubated for 10 min at room temperature before quenching with 70 μl of 2 M H2SO4. Abs450 was measured using a Biotek Synergy H1 plate reader.
Biolayer interferometry
The binding affinity of purified immunogens to REGN10933 and Ace2-Fc was measured using a kinetic BLI assay using an Octet-Red96e (Sartorius). REGN10933 IgG or Ace2-Fc was buffer-exchanged into HBS-EP buffer [10 mM Na-Hepes (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% (v/v) P20 surfactant] using Zeba spin desalting columns (Thermo Fisher Scientific). REGN10933 or Ace2-Fc was loaded onto Anti-hIgG Fc Capture (AHC) biosensors (Sartorius) over the course of 300 s until reaching a signal of ~0.6 nm. BLI pins were then immersed in immunogens twofold serially diluted in HBS-EP buffer (150 to 2.34 nM). After 300 s, pins were immersed in HBS-EP buffer to measure dissociation. Association rate (ka), dissociation rate (kdis), and dissociation constant (KD) were globally fit using a 1:1 binding model in Data Analysis HT 12.0 (Sartorius). Three independent protein preps (biological replicates) were each measured in technical triplicate. Values reported are the average and SD between biological replicates.
Mouse immunizations
Mouse immunogenicity studies were performed in an American Association for Accreditation of Laboratory Animal Care-accredited facility under the guidelines and approval of the Institutional Animal Care and Use Committee at the National Institutes of Health. Five 5- to 6-week-old female CD-1 mice (Charles River Laboratories) per group were immunized with 10 μg of antigen each. Antigen was formulated as a 1:1 ratio in Complete Freund’s Adjuvant (MilliporeSigma) on day 0 and Incomplete Freund’s Adjuvant (MilliporeSigma) on day 21, and 100 μl of formulated antigen was delivered by intraperitoneal injection. Blood was collected on day 35, and serum was separated and stored at −80°C.
Serum antibody titer ELISA
Nunc MaxiSorp plates (Thermo Fisher Scientific) were coated with 100 μl of purified trimeric FL-spike ectodomain (0.01 mg/ml) diluted in 50 mM Na-carbonate (pH 9.5). Plates were incubated overnight at 4°C and then washed three times with PBST. Plates were blocked for 1 hour at room temperature with 2% BSA in PBST and then washed three times with PBST. Serum was diluted in 2% BSA in PBST, and 100 μl was added to each well. After 1 hour of incubation at room temperature, plates were washed three times with PBST and 200 μl of 1:5000 peroxidase-conjugated anti-mouse IgG was added (Jackson ImmunoResearch Laboratories Inc., catalog no. 115-035-164). Plates were incubated for 30 min at room temperature and washed three times with PBST. Last, 70 μl of TMB (MilliporeSigma) was added and incubated for 20 min at room temperature before quenching with 70 μl of 2 M H2SO4. Abs450 was measured using a Biotek Synergy H1 plate reader.
Pooled serum from mice immunized with WT RBD was used as a standard curve on each plate to calculate the antibody titers of individual animals in all groups. One antibody unit (AU) was defined as the dilution of the standard serum required to achieve an Abs450 value of 1. Each plate included triplicate twofold serial dilutions of the standard serum from 20 to 0.01 AU. Serum from each animal was diluted such that the Abs450 fell in the informative portion of the standard curve between 0.1 and 2.0. The Abs450 values for the standard curve were fit to a four-parameter logistic curve, which was used to convert Abs450 values to AU for each individual animal. AU values for each individual animal were measured in triplicate on separate plates, and the average was reported.
RBD/Ace2-blocking assay
Nunc MaxiSorp plates (Thermo Fisher Scientific) were coated with 100 μl of purified WT RBD (0.32 μg/ml) diluted in 50 mM Na-carbonate (pH 9.5). Plates were incubated overnight at 4°C and then washed three times with PBST. Plates were blocked for 1 hour at room temperature with 2% BSA in PBST and then washed three times with PBST. Serum was diluted in 2% BSA in PBST in a threefold dilution series from 1:100 to 1:218,700. Serum (50 μl) was mixed with 10 μl of 30 nM Ace2-Fc or 10 μl of buffer as a background control. Serum mixture (50 μl) was added to RBD-coated plates and incubated for 1 hour at room temperature. Plates were washed three times with PBST, and 200 μl of 1:30,000 peroxidase-conjugated anti-human IgG was added (Jackson ImmunoResearch Laboratories Inc., catalog no. 109-035-098). Plates were incubated for 30 min at room temperature and washed three times with PBST. Last, 70 μl of TMB (MilliporeSigma) was added and incubated for 20 min at room temperature before quenching with 70 μl of 2 M H2SO4. Abs450 was measured using a Biotek Synergy H1 plate reader.
Ace2/RBD binding inhibition was calculated by first subtracting the Abs450 values of the background controls lacking Ace2-Fc. Eight wells without serum were used to calculate the maximum signal. Inhibition was calculated using the following formula
where X is the Abs450 of a well after background subtraction and max is the average of the eight samples without serum after background subtraction.
Percent inhibition values were measured for each serum dilution in triplicate, and average values were plotted in GraphPad Prism 8. Data were fit using a normalized dose response curve with a variable slope
where X is the serum dilution, Y is the % inhibition, and HillSlope and ID50 are calculated parameters corresponding to the slope of the curve and the dilution at which 50% inhibition occurs, respectively. ID50 values for each animal were log-transformed and plotted, along with the geometric mean value for each group.
Pseudoviral neutralization
Mouse serum was blinded and diluted in a duplicate fourfold series from 1:5 to 1:81,920. Serum was mixed 1:1 with pseudovirus containing the SARS-CoV-2 spike protein and a luciferase reporter in a 96-well plate (GenScript). After 1 hour of incubation at room temperature, 20,000 human embryonic kidney (HEK) 293 cells overexpressing Ace2 were added to each well. Cells and pseudovirus were incubated at 37°C, 5% CO2 for 48 hours, after which culture medium was removed and Bio-Glo luciferase reagent (Promega) was added to the wells. Luciferase signal was measured using an EnVision plate reader, and % inhibition was calculated using the following formula
where X is the luciferase signal, min is the average signal from duplicate wells without pseudovirus, and max is the average signal from duplicate wells without serum. Log ID50 values were calculated in the same manner described for the RBD/Ace2-blocking assay.
Plaque reduction neutralizing test against authentic SARS-CoV-2
Vero E6 cells stably expressing TMPRSS2 were plated in a 24-well plate 1 day before infection (71). Equal volumes of serum from each animal in a group were combined and heat-inactivated at 56°C for 30 min. The FL-spike group only contains serum from the four animals with the highest pseudovirus neutralizing titers because we did not have sufficient serum from the fifth animal for this assay. Serum was diluted 1:100 in infection medium [Dulbecco’s modified Eagle’s medium, 2% heat-inactivated fetal bovine serum, 1× GlutaMAX (Thermo Fisher Scientific), and hygromycin (0.25 mg/ml)], followed by threefold serial dilutions. Diluted serum was mixed 1:1 with WA-1 strain SARS-CoV-2 (GenBank, MN985325.1) and incubated for 30 min at 37°C. Approximately 30 plaque-forming units of virus were added to each well of Vero-TMPRSS2 cells and incubated for 30 min at 37°C before overlaying with 0.65% methyl cellulose 400 centipoise. Cells were incubated for 72 hours at 37°C before staining with 0.2% crystal violet in 10% neutral-buffered formalin.
Plates were randomized and blinded, and plaques were counted manually. Percent neutralization was calculated relative to the average of 14 wells without serum. Percent neutralization values were measured for each serum dilution in duplicate, and average values were plotted in GraphPad Prism 8. Data were fit using a normalized dose response curve
where X is the serum dilution, Y is the % neutralization, and ID50 is calculated corresponding to the dilution at which 50% inhibition occurs.
Acknowledgments
We thank all the members of LMIV COVID group for discussions of results and N. Max and P. Patel for experimental assistance. R. Johnson and N. Lackemeyer at the NIAID SARS-CoV-2 virology core (SVC) provided training and resources for high-containment experiments that involved authentic SARS-CoV-2. This study used the Office of Cyber Infrastructure and Computational Biology (OCICB) High Performance Computing (HPC) cluster at the National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289. We thank J. P. Gorres for proofreading and editing the manuscript.
Funding: This work was funded by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health.
Author contributions: Conceptualization: N.H.T. and T.H.D. Data analysis: N.H.T. and T.H.D. Software: N.H.T. and T.H.D. Methodology: T.H.D. and N.H.T. Formal analysis: T.H.D. and N.H.T. Investigation: T.H.D., W.K.T., B.B., S.O.-G., and T.O. Visualization: T.H.D., W.K.T., and N.H.T. Funding acquisition: N.H.T. and T.H.D. Project administration: N.H.T., N.D.S., L.E.L., and T.H.D. Supervision: N.H.T. and L.E.L. Writing—original draft: T.H.D. and N.H.T. Writing—review and editing: T.H.D., N.H.T., W.K.T., and N.D.S.
Competing interests: N.H.T. and T.H.D. are inventors on a pending patent awaiting USPTO review related to this work filed by the United States of America, as represented by the Secretary, Dept. of Health and Human Services. U.S. Provisional Application no. 63/200,194, filed on 18 February 2021; PCT/US2022/070744, filed on 1 February 2022. The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession codes 8DCC and 8DCE. Plasmids can be provided by N.H.T. pending scientific review and a completed material transfer agreement. Requests should be submitted to N.H.T.
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
Tables S1 to S3
References
REFERENCES AND NOTES
- 1.Dong E., Du H., Gardner L., An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh E. E., Frenck R. W. Jr., Falsey A. R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M. J., Bailey R., Swanson K. A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P. Y., Türeci Ö., Tompkins K. R., Lyke K. E., Raabe V., Dormitzer P. R., Jansen K. U., Şahin U., Gruber W. C., Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 383, 2439–2450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J. S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M. B., Haupt R. E., Logue J., McGrath M., Weston S., Piedra P. A., Desai C., Callahan K., Lewis M., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J. D., Griffin P., Wilkinson B., Glenn G. M., Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 383, 2320–2332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson L. A., Anderson E. J., Rouphael N. G., Roberts P. C., Makhene M., Coler R. N., McCullough M., Chappell J. D., Denison M. R., Stevens L. J., Pruijssers A. J., McDermott A., Flach B., Doria-Rose N. A., Corbett K. S., Morabito K. M., O’Dell S., Schmidt S. D., Swanson P. A. II, Padilla M., Mascola J. R., Neuzil K. M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J. E., Graham B. S., Beigel J. H.; mRNA-1273 Study Group , An mRNA vaccine against SARS-CoV-2—Preliminary report. N. Engl. J. Med. 383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadoff J., Gars M. L., Shukarev G., Heerwegh D., Truyers C., de Groot A. M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., Berghmans P.-J., Kimmel M., Van Damme P., de Hoon J., Smith W., Stephenson K. E., De Rosa S. C., Cohen K. W., McElrath M. J., Cormier E., Scheper G., Barouch D. H., Hendriks J., Struyf F., Douoguih M., Van Hoof J., Schuitemaker H., Interim results of a phase 1–2a trial of Ad26.COV2.S Covid-19 vaccine. N. Engl. J. Med. 384, 1824–1835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbett K. S., Edwards D. K., Leist S. R., Abiona O. M., Boyoglu-Barnum S., Gillespie R. A., Himansu S., Schäfer A., Ziwawo C. T., DiPiazza A. T., Dinnon K. H., Elbashir S. M., Shaw C. A., Woods A., Fritch E. J., Martinez D. R., Bock K. W., Minai M., Nagata B. M., Hutchinson G. B., Wu K., Henry C., Bahl K., Garcia-Dominguez D., Ma L. Z., Renzi I., Kong W. P., Schmidt S. D., Wang L., Zhang Y., Phung E., Chang L. A., Loomis R. J., Altaras N. E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M. K., Shi W., Leung K., Yang E. S., West A., Gully K. L., Stevens L. J., Wang N., Wrapp D., Doria-Rose N. A., Stewart-Jones G., Bennett H., Alvarado G. S., Nason M. C., Ruckwardt T. J., McLellan J. S., Denison M. R., Chappell J. D., Moore I. N., Morabito K. M., Mascola J. R., Baric R. S., Carfi A., Graham B. S., SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pallesen J., Wang N., Corbett K. S., Wrapp D., Kirchdoerfer R. N., Turner H. L., Cottrell C. A., Becker M. M., Wang L., Shi W., Kong W. P., Andres E. L., Kettenbach A. N., Denison M. R., Chappell J. D., Graham B. S., Ward A. B., McLellan J. S., Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U.S.A. 114, E7348–E7357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bos R., Rutten L., van der Lubbe J. E. M., Bakkers M. J. G., Hardenberg G., Wegmann F., Zuijdgeest D., de Wilde A. H., Koornneef A., Verwilligen A., van Manen D., Kwaks T., Vogels R., Dalebout T. J., Myeni S. K., Kikkert M., Snijder E. J., Li Z., Barouch D. H., Vellinga J., Langedijk J. P. M., Zahn R. C., Custers J., Schuitemaker H., Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines 5, 91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheiblhofer S., Laimer J., Machado Y., Weiss R., Thalhamer J., Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev. Vaccines 16, 479–489 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ntumngia F. B., Pires C. V., Barnes S. J., George M. T., Thomson-Luque R., Kano F. S., Alves J. R. S., Urusova D., Pereira D. B., Tolia N. H., King C. L., Carvalho L. H., Adams J. H., An engineered vaccine of the Plasmodium vivax Duffy binding protein enhances induction of broadly neutralizing antibodies. Sci. Rep. 7, 13779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campeotto I., Goldenzweig A., Davey J., Barfod L., Marshall J. M., Silk S. E., Wright K. E., Draper S. J., Higgins M. K., Fleishman S. J., One-step design of a stable variant of the malaria invasion protein RH5 for use as a vaccine immunogen. Proc. Natl. Acad. Sci. U.S.A. 114, 998–1002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sesterhenn F., Yang C., Bonet J., Cramer J. T., Wen X., Wang Y., Chiang C.-I., Abriata L. A., Kucharska I., Castoro G., Vollers S. S., Galloux M., Dheilly E., Rosset S., Corthésy P., Georgeon S., Villard M., Richard C.-A., Descamps D., Delgado T., Oricchio E., Rameix-Welti M.-A., Más V., Ervin S., Eléouët J.-F., Riffault S., Bates J. T., Julien J.-P., Li Y., Jardetzky T., Krey T., Correia B. E., De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 368, eaay5051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Correia B. E., Bates J. T., Loomis R. J., Baneyx G., Carrico C., Jardine J. G., Rupert P., Correnti C., Kalyuzhniy O., Vittal V., Connell M. J., Stevens E., Schroeter A., Chen M., MacPherson S., Serra A. M., Adachi Y., Holmes M. A., Li Y., Klevit R. E., Graham B. S., Wyatt R. T., Baker D., Strong R. K., Crowe J. E., Johnson P. R., Schief W. R., Proof of principle for epitope-focused vaccine design. Nature 507, 201–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jardine J., Julien J.-P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P.-S., MacPherson S., Jones M., Nieusma T., Mathison J., Baker D., Ward A. B., Burton D. R., Stamatatos L., Nemazee D., Wilson I. A., Schief W. R., Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bale J. B., Gonen S., Liu Y., Sheffler W., Ellis D., Thomas C., Cascio D., Yeates T. O., Gonen T., King N. P., Baker D., Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L., Wang P., Nair M. S., Yu J., Rapp M., Wang Q., Luo Y., Chan J. F.-W., Sahi V., Figueroa A., Guo X. V., Cerutti G., Bimela J., Gorman J., Zhou T., Chen Z., Yuen K.-Y., Kwong P. D., Sodroski J. G., Yin M. T., Sheng Z., Huang Y., Shapiro L., Ho D. D., Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Hansen J., Baum A., Pascal K. E., Russo V., Giordano S., Wloga E., Fulton B. O., Yan Y., Koon K., Patel K., Chung K. M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W. Y., Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Romero Hernandez A., Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D. C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A. J., Stahl N., Yancopoulos G. D., Kyratsous C. A., Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., Vacca F., Cardamone D., de Santi C., Torres J. L., Ozorowski G., Benincasa L., Jang H., di Genova C., Depau L., Brunetti J., Agrati C., Capobianchi M. R., Castilletti C., Emiliozzi A., Fabbiani M., Montagnani F., Bracci L., Sautto G., Ross T. M., Montomoli E., Temperton N., Ward A. B., Sala C., Ippolito G., Rappuoli R., Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell 184, 1821–1835.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brouwer P. J. M., Caniels T. G., van der Straten K., Snitselaar J. L., Aldon Y., Bangaru S., Torres J. L., Okba N. M. A., Claireaux M., Kerster G., Bentlage A. E. H., van Haaren M. M., Guerra D., Burger J. A., Schermer E. E., Verheul K. D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M. J., Bijl T. P. L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N. A., Wiersinga W. J., Vidarsson G., Haagmans B. L., Ward A. B., de Bree G. J., Sanders R. W., van Gils M. J., Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dejnirattisai W., Zhou D., Ginn H. M., Duyvesteyn H. M. E., Supasa P., Case J. B., Zhao Y., Walter T. S., Mentzer A. J., Liu C., Wang B., Paesen G. C., Slon-Campos J., López-Camacho C., Kafai N. M., Bailey A. L., Chen R. E., Ying B., Thompson C., Bolton J., Fyfe A., Gupta S., Tan T. K., Gilbert-Jaramillo J., James W., Knight M., Carroll M. W., Skelly D., Dold C., Peng Y., Levin R., Dong T., Pollard A. J., Knight J. C., Klenerman P., Temperton N., Hall D. R., Williams M. A., Paterson N. G., Bertram F. K. R., Siebert C. A., Clare D. K., Howe A., Radecke J., Song Y., Townsend A. R., Huang K. Y. A., Fry E. E., Mongkolsapaya J., Diamond M. S., Ren J., Stuart D. I., Screaton G. R., The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 184, 2183–2200.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., Vacca F., Cardamone D., De Santi C., Torres J. L., Ozorowski G., Benincasa L., Jang H., Genova C. D., Depau L., Brunetti J., Agrati C., Capobianchi M. R., Castilletti C., Emiliozzi A., Fabbiani M., Montagnani F., Bracci L., Sautto G., Ross T. M., Montomoli E., Temperton N., Ward A. B., Sala C., Ippolito G., Rappuoli R., Extremely potent human monoclonal antibodies from Covid-19 convalescent patients. Cell 184, 1821–1835.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv Z., Deng Y.-Q., Ye Q., Cao L., Sun C.-Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., Chen Q., Wang N., Nie J., Cui Z., Zhu D., Shaw N., Li X.-F., Li Q., Xie L., Wang Y., Rao Z., Qin C.-F., Wang X., Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 369, eabc5881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H., Wu N. C., Yuan M., Bangaru S., Torres J. L., Caniels T. G., van Schooten J., Zhu X., Lee C.-C. D., Brouwer P. J. M., van Gils M. J., Sanders R. W., Ward A. B., Wilson I. A., Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity 53, 1272–1280.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rappazzo C. G., Tse L. V., Kaku C. I., Wrapp D., Sakharkar M., Huang D., Deveau L. M., Yockachonis T. J., Herbert A. S., Battles M. B., O’Brien C. M., Brown M. E., Geoghegan J. C., Belk J., Peng L., Yang L., Hou Y., Scobey T. D., Burton D. R., Nemazee D., Dye J. M., Voss J. E., Gunn B. M., McLellan J. S., Baric R. S., Gralinski L. E., Walker L. M., Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science 371, 823–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walls A. C., Park Y.-J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. E. Toledo-Romani, M. García-Carmenate, C. Valenzuela-Silva, W. Baldoquín-Rodríguez, M. Martínez-Pérez, M. Rodríguez-González, B. Paredes-Moreno, I. Mendoza-Hernández, R. González-Mujica, O. Samón-Tabio, P. Velazco-Villares, Juan Pablo Bacallao-Castillo, E. Licea-Martín, M. Rodríguez-Ortega, N. Herrera-Marrero, E. Caballero-González, L. Egües-Torres, R. Duartes-González, S. García-Blanco, S. Pérez-Cabrera, S. Huete-Ferreira, K. Idalmis-Cisnero, O. Fonte-Galindo, D. Meliá-Pérez, I. Rojas-Remedios, View ORCID Profile Delaram Doroud, M. M. Gouya, A. Biglari, P. Van der Stuyft, S. Fernández-Castillo, Y. Climent-Ruiz, Y. Valdes-Balbín, D. García-Rivera, V. Verez-Bencomo; the SOBERANA Phase team, Safety and efficacy of the two doses conjugated protein-based SOBERANA-02 COVID-19 vaccine and of a heterologous three-dose combination with SOBERANA-PLUS: Double-blind, randomised, placebo-controlled phase 3 clinical trial. Medrxiv 2021.10.31.21265703 (2021).
- 28.S. Thuluva, V. Paradkar, K. Turaga, S. R. Gunneri, V. Yerroju, R. Mogulla, P. V. Suneetha, M. Kyasani, S. K. Manoharan, S. Adabala, A. S. Javvadi, G. Medigeshi, J. Singh, H. Shaman, A. Binayke, A. Zaheer, A. Awasthi, C. Singh, A Venkateshwar Rao, I. Basu, K. A. A. Kumar, A. K. Pandey, Immunogenic superiority and safety of Biological E’s CORBEVAX™ vaccine compared to COVISHIELD™ (ChAdOx1 nCoV-19) vaccine studied in a phase III, single blind, multicenter, randomized clinical trial. Medrxiv 2022.03.20.22271891 (2022).
- 29.Goldenzweig A., Goldsmith M., Hill S. E., Gertman O., Laurino P., Ashani Y., Dym O., Unger T., Albeck S., Prilusky J., Lieberman R. L., Aharoni A., Silman I., Sussman J. L., Tawfik D. S., Fleishman S. J., Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol. Cell 63, 337–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh C. L., Goldsmith J. A., Schaub J. M., DiVenere A. M., Kuo H.-C., Javanmardi K., le K. C., Wrapp D., Lee A. G., Liu Y., Chou C. W., Byrne P. O., Hjorth C. K., Johnson N. V., Ludes-Meyers J., Nguyen A. W., Park J., Wang N., Amengor D., Lavinder J. J., Ippolito G. C., Maynard J. A., Finkelstein I. J., McLellan J. S., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L., Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H.-H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West A. P. Jr., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C., Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Yuan M., Wu N. C., Zhu X., Lee C.-C. D., So R. T. Y., Lv H., Mok C. K. P., Wilson I. A., A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto D., Park Y.-J., Beltramello M., Walls A. C., Tortorici M. A., Bianchi S., Jaconi S., Culap K., Zatta F., de Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J. B., Chen R. E., Havenar-Daughton C., Snell G., Telenti A., Virgin H. W., Lanzavecchia A., Diamond M. S., Fink K., Veesler D., Corti D., Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Barnes C. O., Jette C. A., Abernathy M. E., Dam K.-M. A., Esswein S. R., Gristick H. B., Malyutin A. G., Sharaf N. G., Huey-Tubman K. E., Lee Y. E., Robbiani D. F., Nussenzweig M. C., West A. P. Jr., Bjorkman P. J., SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amanat F., Strohmeier S., Rathnasinghe R., Schotsaert M., Coughlan L., García-Sastre A., Krammer F., Introduction of two prolines and removal of the polybasic cleavage site lead to higher efficacy of a recombinant spike-based SARS-CoV-2 vaccine in the mouse model. MBio 12, e02648-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C. O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J. A., Oliveira T. Y., Yang Z., Abernathy M. E., Huey-Tubman K. E., Hurley A., Turroja M., West K. A., Gordon K., Millard K. G., Ramos V., Silva J. D., Xu J., Colbert R. A., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P. J., Casellas R., Hatziioannou T., Bieniasz P. D., Nussenzweig M. C., mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 592, 616–622 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.-S., Winkler M. S., Lier M., Dopfer-Jablonka A., Jäck H.-M., Behrens G. M. N., Pöhlmann S., The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 185, 447–456.e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L., Iketani S., Guo Y., Chan J. F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M. S., Yu J., Chik K. K.-H., Yuen T. T.-T., Yoon C., To K. K.-W., Chen H., Yin M. T., Sobieszczyk M. E., Huang Y., Wang H. H., Sheng Z., Yuen K.-Y., Ho D. D., Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602, 676–681 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Muik A., Lui B. G., Wallisch A. K., Bacher M., Mühl J., Reinholz J., Ozhelvaci O., Beckmann N., de la Caridad Güimil Garcia R., Poran A., Shpyro S., Finlayson A., Cai H., Yang Q., Swanson K. A., Türeci Ö., Şahin U., Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine–elicited human sera. Science 375, 678–680 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P., Nair M. S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P. D., Graham B. S., Mascola J. R., Chang J. Y., Yin M. T., Sobieszczyk M., Kyratsous C. A., Shapiro L., Sheng Z., Huang Y., Ho D. D., Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Pegu A., O’Connell S. E., Schmidt S. D., O’Dell S., Talana C. A., Lai L., Albert J., Anderson E., Bennett H., Corbett K. S., Flach B., Jackson L., Leav B., Ledgerwood J. E., Luke C. J., Makowski M., Nason M. C., Roberts P. C., Roederer M., Rebolledo P. A., Rostad C. A., Rouphael N. G., Shi W., Wang L., Widge A. T., Yang E. S.; The mRNA-1273 Study Group, Beigel J. H., Graham B. S., Mascola J. R., Suthar M. S., McDermott A. B., Doria-Rose N. A., Arega J., Beigel J. H., Buchanan W., Elsafy M., Hoang B., Lampley R., Kolhekar A., Koo H., Luke C., Makhene M., Nayak S., Pikaart-Tautges R., Roberts P. C., Russell J., Sindall E., Albert J., Kunwar P., Makowski M., Anderson E. J., Bechnak A., Bower M., Camacho-Gonzalez A. F., Collins M., Drobeniuc A., Edara V. V., Edupuganti S., Floyd K., Gibson T., Ackerley C. M. G., Johnson B., Kamidani S., Kao C., Kelley C., Lai L., Macenczak H., McCullough M. P., Peters E., Phadke V. K., Rebolledo P. A., Rostad C. A., Rouphael N., Scherer E., Sherman A., Stephens K., Suthar M. S., Teherani M., Traenkner J., Winston J., Yildirim I., Barr L., Benoit J., Carste B., Choe J., Dunstan M., Erolin R., ffitch J., Fields C., Jackson L. A., Kiniry E., Lasicka S., Lee S., Nguyen M., Pimienta S., Suyehira J., Witte M., Bennett H., Altaras N. E., Carfi A., Hurley M., Leav B., Pajon R., Sun W., Zaks T., Coler R. N., Larsen S. E., Neuzil K. M., Lindesmith L. C., Martinez D. R., Munt J., Mallory M., Edwards C., Baric R. S., Berkowitz N. M., Boritz E. A., Carlton K., Corbett K. S., Costner P., Creanga A., Doria-Rose N. A., Douek D. C., Flach B., Gaudinski M., Gordon I., Graham B. S., Holman L. S., Ledgerwood J. E., Leung K., Lin B. C., Louder M. K., Mascola J. R., McDermott A. B., Morabito K. M., Novik L., O’Connell S., O’Dell S., Padilla M., Pegu A., Schmidt S. D., Shi W., Swanson P. A. II, Talana C. A., Wang L., Widge A. T., Yang E. S., Zhang Y., Chappell J. D., Denison M. R., Hughes T., Lu X., Pruijssers A. J., Stevens L. J., Posavad C. M., Gale M. Jr., Menachery V., Shi P. Y., Durability of mRNA-1273 vaccine–induced antibodies against SARS-CoV-2 variants. Science 373, 1372–1377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan T. K., Rijal P., Rahikainen R., Keeble A. H., Schimanski L., Hussain S., Harvey R., Hayes J. W. P., Edwards J. C., McLean R. K., Martini V., Pedrera M., Thakur N., Conceicao C., Dietrich I., Shelton H., Ludi A., Wilsden G., Browning C., Zagrajek A. K., Bialy D., Bhat S., Stevenson-Leggett P., Hollinghurst P., Tully M., Moffat K., Chiu C., Waters R., Gray A., Azhar M., Mioulet V., Newman J., Asfor A. S., Burman A., Crossley S., Hammond J. A., Tchilian E., Charleston B., Bailey D., Tuthill T. J., Graham S. P., Duyvesteyn H. M. E., Malinauskas T., Huo J., Tree J. A., Buttigieg K. R., Owens R. J., Carroll M. W., Daniels R. S., McCauley J. W., Stuart D. I., Huang K. Y. A., Howarth M., Townsend A. R., A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 12, 542 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M., Yang M., Li Y., Cheng H., Yuan Y., Zhang W., Ke C., Wong G., Qi J., Qin C., Yan J., Gao G. F., A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell 182, 722–733.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walls A. C., Fiala B., Schäfer A., Wrenn S., Pham M. N., Murphy M., Tse L. V., Shehata L., O’Connor M. A., Chen C., Navarro M. J., Miranda M. C., Pettie D., Ravichandran R., Kraft J. C., Ogohara C., Palser A., Chalk S., Lee E.-C., Guerriero K., Kepl E., Chow C. M., Sydeman C., Hodge E. A., Brown B., Fuller J. T., Dinnon K. H. III, Gralinski L. E., Leist S. R., Gully K. L., Lewis T. B., Guttman M., Chu H. Y., Lee K. K., Fuller D. H., Baric R. S., Kellam P., Carter L., Pepper M., Sheahan T. P., Veesler D., King N. P., Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang Y. F., Sun C., Zhuang Z., Yuan R.-Y., Zheng Q., Li J.-P., Zhou P.-P., Chen X.-C., Liu Z., Zhang X., Yu X.-H., Kong X.-W., Zhu Q.-Y., Zhong Q., Xu M., Zhong N.-S., Zeng Y.-X., Feng G.-K., Ke C., Zhao J.-C., Zeng M.-S., Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano 15, 2738–2752 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., Qiao Y., Zhan Y., Liu J., Zhang J., Zhang X., Peng Z., Li Y., Lin Y., Liang L., Wang G., Chen Y., Chen Q., Pan T., He X., Zhang H., Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity 53, 1315–1330.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wrapp D., De Vlieger D., Corbett K. S., Torres G. M., Wang N., Van Breedam W., Roose K., van Schie L.; VIB-CMB COVID-19 Response Team, Hoffmann M., Pöhlmann S., Graham B. S., Callewaert N., Schepens B., Saelens X., McLellan J. S., Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 181, 1004–1015.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walls A. C., Xiong X., Park Y.-J., Alejandra Tortorici M., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F. A., Corti D., Veesler D., Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176, 1026–1039.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pak J. E., Sharon C., Satkunarajah M., Auperin T. C., Cameron C. M., Kelvin D. J., Seetharaman J., Cochrane A., Plummer F. A., Berry J. D., Rini J. M., Structural insights into immune recognition of the severe acute respiratory syndrome coronavirus S protein receptor binding domain. J. Mol. Biol. 388, 815–823 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwang W. C., Lin Y., Santelli E., Sui J., Jaroszewski L., Stec B., Farzan M., Marasco W. A., Liddington R. C., Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J. Biol. Chem. 281, 34610–34616 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D. S., Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 281, 15829–15836 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sui J., Li W., Murakami A., Tamin A., Matthews L. J., Wong S. K., Moore M. J., Tallarico A. S. C., Olurinde M., Choe H., Anderson L. J., Bellini W. J., Farzan M., Marasco W. A., Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 101, 2536–2541 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meulen J. T., van den Brink E. N., Poon L. L. M., Marissen W. E., Leung C. S. W., Cox F., Cheung C. Y., Bakker A. Q., Bogaards J. A., van Deventer E., Preiser W., Doerr H. W., Chow V. T., de Kruif J., Peiris J. S. M., Goudsmit J., Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLOS Med. 3, e237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LLC Schrodinger, The PyMOL Molecular Graphics System. Version 2.4 (2010).
- 57.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehead T. A., Chevalier A., Song Y., Dreyfus C., Fleishman S. J., de Mattos C., Myers C. A., Kamisetty H., Blair P., Wilson I. A., Baker D., Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat. Biotechnol. 30, 543–548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Aricescu A. R., Lu W., Jones E. Y., A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 62, 1243–1250 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A., ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan K. K., Dorosky D., Sharma P., Abbasi S. A., Dye J. M., Kranz D. M., Herbert A. S., Procko E., Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 369, 1261–1265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G., Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., Sliz P., Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X., Luongo C., Matsuoka Y., Park H.-S., Santos C., Yang L., Moore I. N., Afroz S., Johnson R. F., Lafont B. A. P., Martens C., Best S. M., Munster V. J., Hollý J., Yewdell J. W., le Nouën C., Munir S., Buchholz U. J., A single intranasal dose of a live-attenuated parainfluenza virus-vectored SARS-CoV-2 vaccine is protective in hamsters. Proc. Natl. Acad. Sci. U.S.A. 118, e2109744118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.-Y., Katsamba P. S., Sampson J. M., Schön A., Bimela J., Boyington J. C., Nazzari A., Olia A. S., Shi W., Sastry M., Stephens T., Stuckey J., Teng I.-T., Wang P., Wang S., Zhang B., Friesner R. A., Ho D. D., Mascola J. R., Shapiro L., Kwong P. D., Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe 28, 867–879.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Tables S1 to S3
References