Abstract
CRISPR-Cas9 gene editing has emerged as a powerful therapeutic technology, but the lack of safe and efficient in vivo delivery systems, especially for tissue-specific vectors, limits its broad clinical applications. Delivery of Cas9 ribonucleoprotein (RNP) owns competitive advantages over other options; however, the large size of RNPs exceeds the loading capacity of currently available delivery vectors. Here, we report a previously unidentified genome editing delivery system, named exosomeRNP, in which Cas9 RNPs were loaded into purified exosomes isolated from hepatic stellate cells through electroporation. ExosomeRNP facilitated effective cytosolic delivery of RNP in vitro while specifically accumulated in the liver tissue in vivo. ExosomeRNP showed vigorous therapeutic potential in acute liver injury, chronic liver fibrosis, and hepatocellular carcinoma mouse models via targeting p53 up-regulated modulator of apoptosis (PUMA), cyclin E1 (CcnE1), and K (lysine) acetyltransferase 5 (KAT5), respectively. The developed exosomeRNP provides a feasible platform for precise and tissue-specific gene therapies of liver diseases.
An exosome-based nanoplatform enables RNP-based–CRISPR-Cas9 genome-editing therapy for liver diseases.
INTRODUCTION
The RNA-guided clustered, regularly interspaced, short palindromic repeats (CRISPR)–associated nuclease protein 9 (Cas9)–based technologies have been developed and validated as powerful and precise tools for therapeutic genome editing (1). Because of its site-specific and multiplexing capability, CRISPR-Cas9–mediated somatic genome editing has shown great potential in treating various genetic disorders in animal models (2–4) and even in a recent phase 1 clinical trial for transthyretin amyloidosis (5). However, the efficient delivery of CRISPR-Cas9 systems remains challenging (6, 7). Major obstacles for CRISPR-Cas9–based genome editing include low delivery efficiency and lack of tissue specificity, which severely limit its clinical applications (8). For therapeutic genome editing, Cas9 nuclease and single guide RNA (sgRNA) have to be efficiently delivered into cells and ultimately into the nucleus. Three options are available for the delivery cargos, namely, plasmid DNA encoding Cas9 and sgRNA, Cas9 mRNA plus sgRNA, and Cas9 ribonucleoprotein (RNP; protein complexed with sgRNA) (1, 6, 9). RNP delivery avoids many pitfalls that DNA and mRNA encounter during transcription and translation processes, allowing for rapid onset of editing with low immune response and weak off-target activity (10, 11). However, therapeutic delivery of RNPs is currently bottlenecked by the large size of RNPs exceeding the loading capacity of both viral and nonviral vectors (7, 8). In addition, the degradation or denaturation during the formulation and bloodstream circulation further hinder the therapeutic application of RNP-based genome-editing systems (7). To date, investigation of in vivo RNP delivery mainly focuses on two classes of vehicles: polymer-based nanoparticles and liposomes (6, 12–14). Although these nonviral vectors can protect RNP from degradation in the bloodstream, tissue-specific genome editing remains elusive after the systemic administration (10, 15–17).
Exosomes are naturally released nanovesicles by cells with the size ranging from 40 to 160 nm (18). Their inherent biocompatibility, transportation capability, bloodstream stability, and engineerability have made exosomes potential delivery vehicles for therapeutic purposes (19). Compared with other nonviral vectors, exosomes provoke low immunogenicity and minimal toxicity (18, 20). Exosomes have homologous tissue-targeting ability, which depends on the phenotype of the source cells, composition, and tissue origin. Recently, endogenous exosomes obtained from the specific cells transfected with CRISPR-Cas9 plasmid have been successfully used to deliver Cas9 RNP to the target cells (21–23), indicating the promising potential of exosomes for RNP delivery. However, these methods are generally cumbersome and time-consuming. Several studies have indicated that through the electroporation, plasmid DNA or mRNA-based Cas9 editors can be loaded into the purified exosomes, representing a new generation of genome-editing carriers for safe and effective therapeutic delivery (24–26). Nevertheless, loading large proteins such as RNP into exosomes by electroporation has not been successfully reported up to now.
Liver is a popular target organ for therapeutic gene delivery, because of abundant hepatic blood flow and high uptake and accumulation of nanovectors (27). In addition, gene editing is a rational therapeutic approach for liver diseases because many liver-related disorders are highly associated with mutations with a single gene (28, 29). For instance, the U.S. Food and Drug Administration approved lipid nanoparticles for the delivery of small interfering RNA (siRNA) to the liver to treat transthyretin-induced amyloidosis in 2017 (30). However, many liver diseases, including acute liver injury, chronic liver fibrosis, and hepatocellular carcinoma (HCC), lack satisfactory treatment, and thus, alternative therapeutic options are urgently needed. In addition, liver-directed gene therapy for liver diseases is another problem that needs to be solved. For synthetic nonviral delivery systems, attaching ligands to their surface to enhance the targeting ability to the specific receptor can achieve liver-targeting ability, such as grafting galactose or glycoconjugates to target to the asialoglycoprotein receptor (31). However, these methods would provoke a certain degree of immunogenicity and liver toxicity. Compared with synthetic nonviral delivery vectors, naturally occurring endogenous vectors of therapeutic agents would avoid these problems. Hepatocyte-derived exosomes have liver-targeting ability due to the homologous tissue-targeting ability and is potential for the in vivo delivery of CRISPR-Cas9 RNP for therapeutic liver-tissue genome editing.
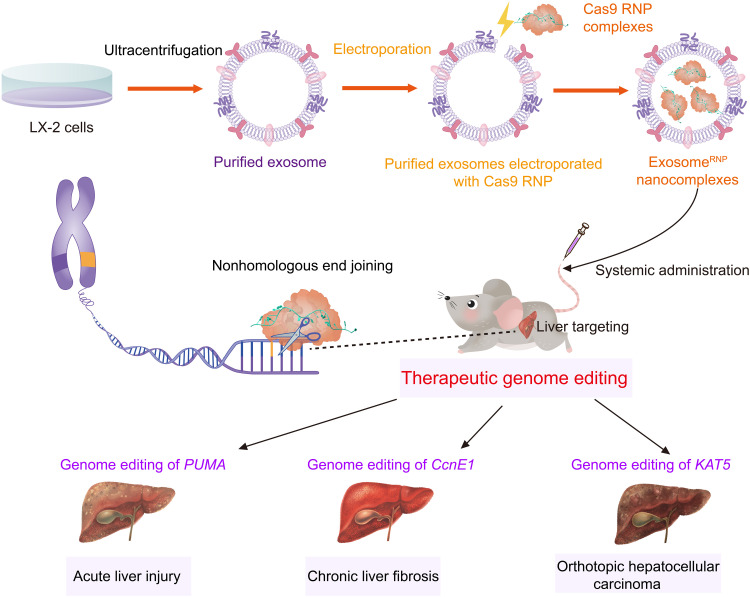
In this study, we developed a method where Cas9 RNP can be loaded into exosomes from hepatic stellate cells (HSCs; LX-2) for the treatment of different liver disorders. Compared to using primary cells, using immortalized cell lines has several advantages of homogeneity, repeatability, and operability in the production of exosomes. In addition, as nontransformed cells, LX-2 cells exhibit higher proliferation rate and superior exosome production capability than immortalized normal hepatocytes, such as THLE-2 (32). The genome-editing delivery system, termed as exosomeRNP, was obtained by loading Cas9 RNP into purified LX-2 exosomes through an optimized electroporation method (Fig. 1). As expected, exosomeRNP showed robust liver-specific genome-editing activity and potent therapeutic effects on liver injury, liver fibrosis, and orthotopic HCC mouse models. The exosomeRNP represent a new generation of endogenous vesicles for the efficient delivery of Cas9 RNP, which can generate liver-specific, robust genome editing and enable precision therapy of liver diseases.
Fig. 1. Schematic illustration of exosome for in vivo delivery of Cas9 RNP for the treatment of liver disorders.
ExosomeRNP shows vigorous therapeutic potential in acute liver injury, chronic liver fibrosis, and orthotopic HCC mouse models via targeting PUMA, CcnE1, and KAT5, respectively.
RESULTS
Characterization of exosome and exosomeRNP
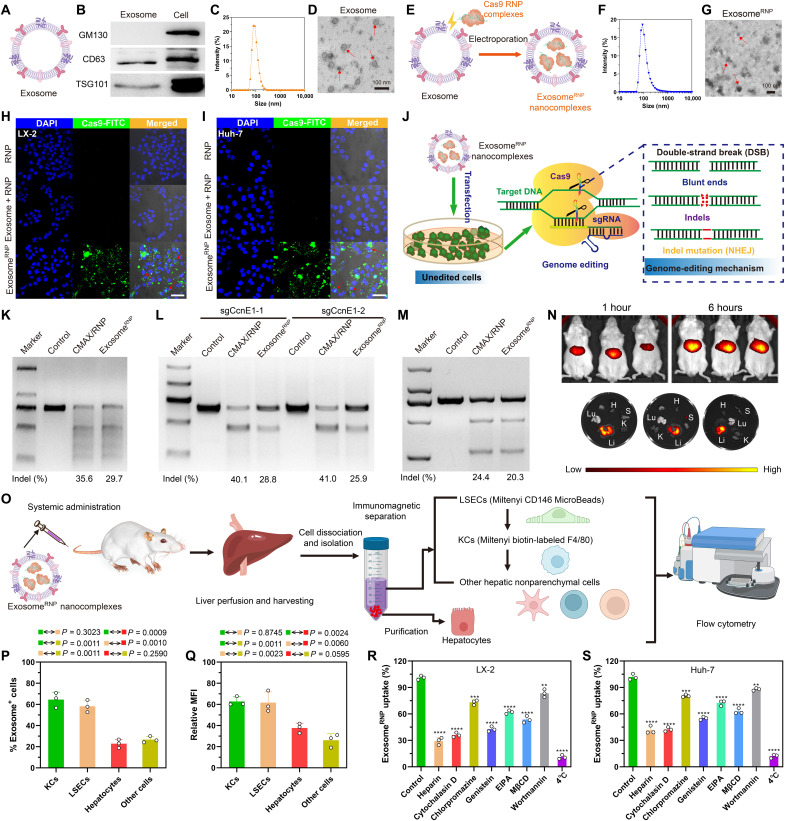
To verify the successful isolation of exosomes from LX-2 cells, we detected several exosome or cytoplasm markers by Western blotting. As expected, the lysate from LX-2–derived exosomes expressed exosome-specific markers CD63 and TSG101 but not the Golgi apparatus–associated GM130 protein (Fig. 2, A and B). LX-2–derived exosomes were next characterized by dynamic light scattering (DLS) with the size ranging from 50 to 200 nm (Fig. 2C). Furthermore, as shown in Fig. 2D and fig. S1, LX-2–derived exosomes showed typical saucer-shaped nanovesicles characterized by transmission electron microscopy (TEM). In the preliminary experiment, we tried to use electroporation, freeze-thawing, and sonication methods to load Cas9 RNP into purified exosomes, respectively. Cas9 RNP has been successfully loaded into the purified exosomes through an electroporation-based method as evidenced by Western blotting (fig. S2). However, for the other two methods, the low-intensity band of Cas9 indicated the inefficient loading of Cas9 RNP into the exosomes. Compared with other methods, electroporation would have higher loading efficiency with relative low cost (33), which is beneficial for loading large molecules, such as nucleic acid and protein drugs (fig. S3). No obvious changes on the size and morphology of exosomes can be detected (Fig. 2, F and G, and fig. S4) after loading of RNP by electroporation (Fig. 2E). Next, we used DLS analysis to evaluate the stability of exosomeRNP nanocomplexes. As shown in fig. S5, the prolonged incubation time in serum-free or serum-containing medium did not result in an increase of the particle size. Furthermore, 4 weeks of storage at −80°C and 7 days of storage at 4°C had no obvious effect on the main particle size of exosomes. In addition, the Cas9 protein entrapment efficiency of exosomeRNP complexes was about 20% calculated by Western blot analysis (fig. S6).
Fig. 2. Characterization, genome-editing activity, biodistribution and cellular uptake mechanism of exosomeRNP.
Characterization of purified exosome (A) and exosomeRNP complexes (E). (B) Biomarkers of exosome by western blotting. (C and D) DLS and TEM the image of purified exosome. The arrows show the typical exosome nanoparticles. (F and G) DLS and TEM image of exosomeRNP complexes. The arrows show the typical exosomeRNP nanoparticles. (H and I) Cytosolic delivery of Cas9-FITC into LX-2 (H) and Huh-7 (I) cells by exosomes for 4 hours. The red arrows point at the efficient translocation of RNP into the nuclei. Scale bars, 25 μm. DAPI, 4′,6-diamidino-2-phenylindole. (J) Exosome-mediated Cas9 RNP delivery for genome editing. (K) Frequency of PUMA indel mutation detected by T7E1 assay from AML-12 cells after the specified treatments. (L) Frequency of CcnE1 indel mutation detected by T7E1 assay from AML-12 cells after the specified treatments. (M) Frequency of KAT5 indel mutation detected by T7E1 assay from LX-2 cells after the specified treatments. (N) In vivo distribution of DiR-labeled exosomes in the whole mice (top) or in the dislodged organs from mice (bottom). H, heart; Lu, lung; Li, liver; K, kidney; S, spleen. (O) Schematic illustrating the procedure to isolate different hepatic cell types and determine exosomeRNP biodistribution. (P) Percentage of each hepatic cell type that is DiI-labeled exosomeRNP-positive. (Q) Relative MFI of each hepatic cell type. (R and S) Mechanism of cellular uptake of exosomeRNP nanocomplexes in LX-2 (R) and Huh-7 (S) cells by the addition of different inhibitors. The values are normalized to the control. Statistical significance was calculated by Students’ t test (mean ± SD, n = 3). **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To investigate the cellular uptake process, exosomes were labeled with red fluorescence CF640R wheat germ agglutinin (WGA). It was found that the red fluorescence mainly distributed in the perinuclear region, indicating successful cytosolic distribution of WGA-labeled exosome into LX-2 and Huh-7 cells (fig. S7). As shown in Fig. 2 (H and I) and figs. S8 and S9, exosomeRNP showed efficient cytosolic and nuclear delivery of Cas9-FITC into LX-2 and Huh-7 cells. In contrast, neither RNP nor the physical mixture of exosome and RNP (exosome + RNP) could effectively enter into LX-2 and Huh-7 cells. Next, we investigated the potential of exosome-mediated RNP delivery for the disruption of the gene in vitro (Fig. 2J). As shown in Fig. 2 (K to M), exosomeRNP delivery achieved an obvious indel frequency at different loci, suggesting that the delivery of exosomeRNP resulted in efficient genome editing in these different loci, which is comparable to the commercial RNP delivery reagent CRISPRMAX (CMAX). Last, the distribution of exosome after systemic administration was evaluated. DiR-labeled LX-2–derived exosomes mainly distributed in the liver tissue of mice (Fig. 2N), suggesting the potential benefit for delivery of therapies to treat liver-related disorders.
Biodistribution of exosomeRNP nanocomplexes within different hepatic cell types
We investigated the biodistribution of DiI-labeled exosomeRNP nanocomplexes in different hepatic cell populations including parenchymal cells (hepatocytes), nonparenchymal Kupffer cells (KCs), liver sinusoidal endothelial cells (LSECs), and other nonparenchymal cells after systemic administration in healthy mice. The isolation of hepatocytes was performed using density gradient–based separation. Then, magnetic beads were used for the isolation of KCs and LSECs (Fig. 2O), respectively, and the rest of cells are considered as other nonparenchymal cells. As shown in Fig. 2 (P and Q) , the exosome positive rate of KCs, LSECs, hepatocytes, and other nonparenchymal cells were 64.5, 58.2, 22.9, and 26.7%, respectively. In addition, the relative mean fluorescence intensity (MFI) of KCs (62.8) and LSECs (61.7) was also higher than those of hepatocytes (37.7) and other nonparenchymal cells (26.1). KCs and LSECs are the main composition of the hepatic reticuloendothelial system, and these two cell types are largely involved in the pathological process of liver diseases, including liver fibrosis, hepatic ischemia reperfusion injury, drug-induced injury, HCC, etc. (34–36). Hepatocytes are the predominant liver cells, comprising about 57% of total liver cells, followed by LSECs (23.2%), KCs (15%), and other liver cells (4.8%) (37). In considering both cell population and exosome uptake (relative MFI), most of the injected exosomeRNP nanocomplexes were taken up by hepatocytes, which are related to metabolic disorders and HCC.
Cellular uptake mechanisms of exosomeRNP
As reported in previous papers, the uptake of exosomes by target cells is predominantly mediated by endocytosis (38, 39). To address the detailed mechanisms, a panel of endocytosis inhibitors was used to investigate internalization pathways of exosomeRNP (38). As shown in Fig. 2 (R and S), the internalization of exosomeRNP nanocomplexes was markedly reduced when cells were incubated at 4°C, which indicated that the uptake is an active and energy dependent. Next, the cells were preincubated with different endocytosis inhibitors at the suggested concentrations, including heparin (10 μg/ml), cytochalasin D (1 μM), chlorpromazine (10 μM), 5-(N-ethyl-nisopropyl) amiloride (EIPA) (100 μM), wortmannin (50 nM), methyl-β-cyclodextrin (M-β-CD) (10 mM), and genistein (150 μM), respectively. As expected, the addition of two well-established endocytosis inhibitors (heparin and cytochalasin D) significantly inhibited the exosomeRNP internalization (Fig. 2, R and S). Both chlorpromazine (inhibitor of clathrindependent pathway) and genistein (inhibitor of clathrinindependent pathway) inhibited the cellular uptake of exosomeRNP nanocomplexes, but genistein exhibited superior blockage capability in comparison with chlorpromazine. In addition, M-β-CD, which is a well-established inhibitor of lipid raft-mediated (caveolin) and clathrin-independent endocytosis, showed stronger inhibition capability than chlorpromazine. Furthermore, both EIPA and wortmannin treatment reduced the uptake of exosomeRNP to some extent, indicating that the macropinocytosis and phagocytosis pathways may also contribute to the internalization of exosomeRNP. Similar results were observed in LX-2 cells and Huh-7 cells. Collectively, we found that exosomeRNP could be uptaken via multiple pathways, including clathrindependent endocytosis, caveolin-mediated endocytosis (relative dominance), macropinocytosis, and phagocytosis, which is consistent with other studies (40, 41).
Determination of the half-life of exosomeRNP in the liver
Lipophilic fluorescent dyes including DiR, DiI, DiD, and PKH are commonly used to label exosomes. However, these lipophilic dyes are highly stable and have a much longer half-life in vivo compared to the exosome itself, which could not provide the specific information to reflect the degradation of exosome (42). In contrast, bioluminescence tagged transmembrane proteins could provide sensitive and accurate signals to determine the half-life of exosome (43). Thus, we used the CD63-NanoLuc fusion protein to label exosomes according to a recent reported method (44). First, we constructed a plasmid DNA vector expressing CAG-CD63-NanoLuc (fig. S10), and CD63-NanoLuc-labeled exosomes can be isolated after transfection of cells. To evaluate the pharmacokinetics of exosomes in the liver, labeled exosomeRNP was intravenously injected into mice. At different time points, mice were euthanized, and liver tissues were lysed and analyzed by the Nano-Glo Luciferase Assay System (N1110, Beijing, Promega) according to the manufacturer’s protocol. The NanoLuc activities were determined by the GloMax Discover Microplate Reader (Beijing, Promega). The time-course data were analyzed using a two-compartmental model, and the half-life of the exosomeRNP in the liver was calculated according to a previously reported procedure (45). In the liver, as shown in fig. S11, intravenously injected CAG-CD63-NanoLuc exosomes went through a distribution phase with a half-life of 10.52 min (t1/2α) followed by an elimination phase with a longer half-life of 154.5 min (t1/2β), which is in good agreement with the results of other previous studies (42, 44).
Biocompatibility and immunogenicity evaluation of exosomeRNP
Immunogenicity and biocompatibility of this extracellular vesicle formulation were performed both in vitro and in vivo. We first evaluated cell toxicity [by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and lactate dehydrogenase (LDH) release] of exosomeRNP nanocomplexes (fig. S12). Figure S12 showed that the cytotoxicity of exosomeRNP nanocomplexes was almost negligible, when the total exosomal protein was in the range of 5 to 80 μg/ml. Subsequently, the hemolysis assay was performed. As expected, exosomeRNP nanocomplexes showed negligible hemolytic activity in vitro.
Next, to further assess exosomeRNP-induced immune response and toxicity, we dosed healthy BALB/c mice with exosomeRNP intravenously every other day for three times. As shown in fig. S13, no obvious pathological change was observed in liver sections after systemic administration of exosomeRNP compared with the control group. Blood chemistry measurements, including alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), and LDH also indicated the biosafety of exosomeRNP. To determine whether exosomeRNP would induce immune responses, key cytokines in blood were measured by enzyme-linked immunosorbent assay (ELISA) in BALB/c mice after intravenous injection for three doses. No significant changes in major immune cytokines [interleukin-6 (IL-6), interferon-γ-inducible protein-10 (IP-10), monocyte chemoattractant protein-1 (MCP-1), tumor necrosis factor–α (TNF-α), and interferon-γ (IFN-γ)] were observed in mice dosed with exosomeRNP. Collectively, these results demonstrated that exosomeRNP had excellent biocompatibility and low immunogenicity.
ExosomeRNP targeting PUMA ameliorates acute liver injury
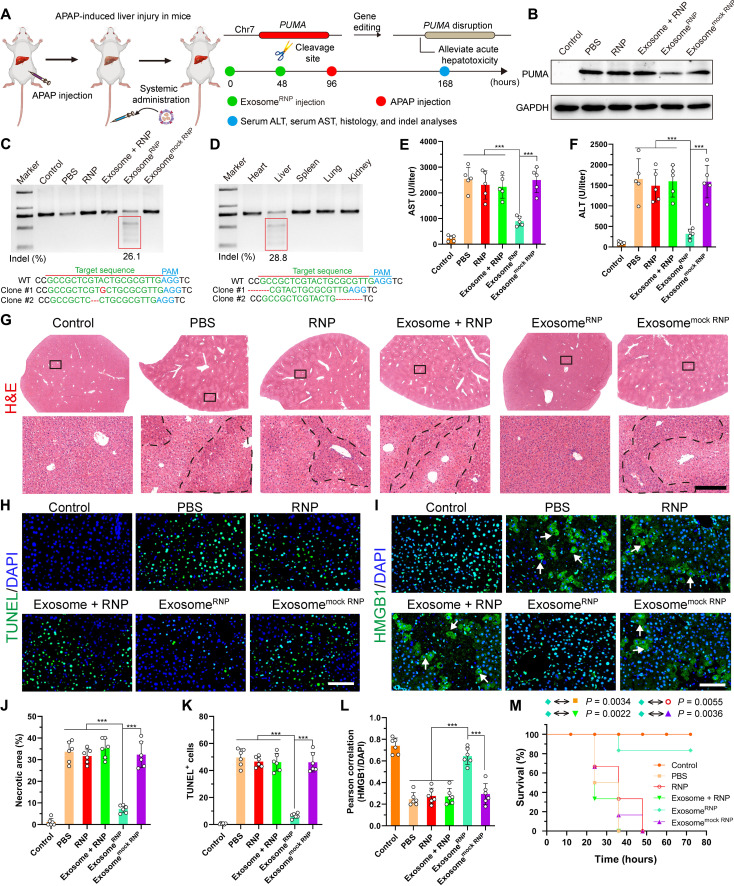
A single overdose of acetaminophen (APAP) or therapeutic misadventure is the leading cause of drug-induced acute liver failure (46, 47). Recently, Zhang and co-authors (48) have found that p53 up-regulated modulator of apoptosis (PUMA) plays a critical role in APAP-induced liver injury and is markedly induced after APAP treatment. Thus, for the therapy of acute liver injury, we designed sgRNA-targeting PUMA and delivered exosomeRNP to investigate its therapeutic efficacy for the acute liver injury (Fig. 3A). By screening different sequences of sgPUMA (fig. S14), a frequency of indels up to 31.3% was observed after the commercial transfection agent (CAMX) treatment. Next, we investigated the therapeutic potential of exosomeRNP in vivo. The delivery of exosomeRNP resulted in the indel frequency of 26.1% (Fig. 3C). The targeting property of exosomeRNP was further proved by T7 endonuclease I (T7E1) assay. As shown in Fig. 3D, among various major organs, obvious indel frequency was observed only in the liver but not in the heart, spleen, lung, and kidney. In addition, PUMA protein was markedly induced in the liver of APAP-treated mice (Fig. 3B and fig. S15), which correlated with increased serum AST and ALT levels (Fig. 3, E and F). However, AST and ALT levels and PUMA protein expression were significantly decreased after the exosomeRNP treatment (Fig. 3, E and F). Other groups, including phosphate-buffered saline (PBS), RNP, exosome + RNP, and exosomemock RNP, did not suppress APAP-induced serum ALT and AST levels.
Fig. 3. ExosomeRNP-mediated genome editing suppressed APAP-induced acute liver injury and lethality.
(A) Schematic illustration of exosome for in vivo delivery of Cas9 RNP for the treatment of APAP-induced liver injury. APAP was administrated through intraperitoneal injection, and exosome/RNP complexes were administered through tail vein. (B) Western blotting of PUMA in livers of mice after the specified treatments. (C) Frequency of indel mutation detected by T7E1 assay from liver tissue after the specified treatments. (D) Frequency of indel mutation detected by T7E1 assay from different organs after exosomeRNP treatment at day 7. (E and F) Serum AST (E) and ALT (F) levels in mice after the specified treatments. (G) Hematoxylin and eosin (H&E) staining of liver sections from mice after the specified treatments. Representative images with dotted line indicating example necrotic centrilobular areas. Scale bar, 200 μm. The regions within the dotted lines denote the accumulation of blood cells. (H) TUNEL (green) staining of liver sections after the specified treatments. Scale bar, 80 μm. (I) HMGB1 (green) staining of liver sections after the specified treatments. Representative images with arrows indicating example cells with cytoplasmic HMGB1 staining and hollow nuclei. Scale bar, 80 μm. (J) Quantification of necrotic areas by ImageJ software. (K) Quantification of TUNEL signals by ImageJ software. (L) Quantification of colocalization of HMGB1 and nuclei by ImageJ software. Means ± SD; n = 6 [one-way analysis of variance (ANOVA) with a Tukey’s post hoc test, ***P < 0.001]. (M) Survival rates of mice after the specified treatments. Statistical significance was calculated by log-rank test (means ± SD, n = 6).
Furthermore, typical features of hepatocyte necrosis were confirmed by hematoxylin and eosin (H&E) staining in the liver tissues (Fig. 3, G and J). As expected, reduced centrilobular cell necrosis and hyperemia was clearly observed in the mice treated with exosomeRNP complexes. Furthermore, terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL)–positive cells were markedly reduced of mice received with exosomeRNP treatment, in sharp contrast with other treatments (Fig. 3, H and K). High-mobility group box 1 (HMGB1) protein is a nuclear protein binding to chromatin, which can be released from nuclei undergoing necrosis. Typical cytoplasmic translocation in the liver was observed in the mice received with APAP treatment, which was only blocked in the mice received with exosomeRNP treatment (Fig. 3, I and L). In addition, exosomeRNP treatment not only significantly decreased AST and ALT levels but also effectively improved survival of mice at 72 hours (Fig. 3M). However, other treatments including PBS, RNP, exosome + RNP, and exosomemock RNP did not suppress APAP-induced acute liver injury and lethality.
ExosomeRNP targeting CcnE1 ameliorates chronic liver fibrosis
Chronic liver fibrosis represents a major global health problem for its high morbidity and mortality worldwide, especially no licensed antifibrotic therapies for its advanced stage (49–51). Thus, finding effective therapies for liver fibrosis is a tremendous medical challenge. Cyclin E1 (CcnE1), a member of the cyclin-dependent protein kinase family, facilitates the proliferation of HSCs and plays an essential role in liver fibrogenesis (52). Recently, Liedtke and co-authors (53) showed that therapeutic intervention with stabilized CcnE1-siRNA could attenuate the initiation and progression of fibrosis. In addition, inhibition of CcnE1 through stabilized CcnE1-siRNA displayed antifibrotic properties in primary human HSCs. Thus, genome editing of CcnE1 through exosomeRNP may also attenuate fibrosis initiation.
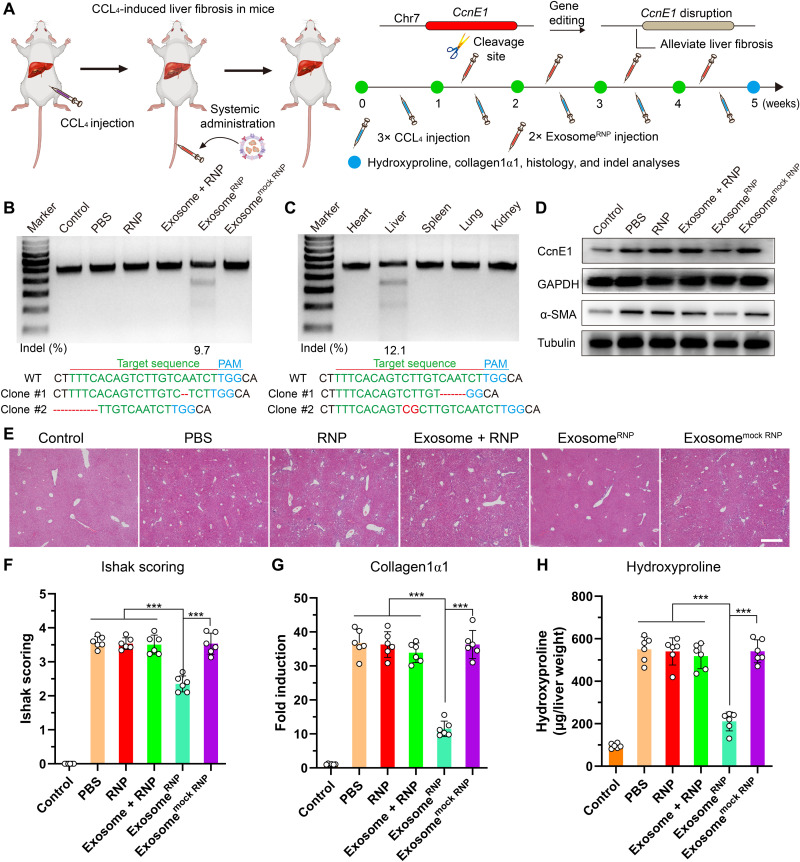
By screening different sequences of sgCcnE1 (fig. S16), a frequency of indels up to 39.8% was observed. Next, we investigated the therapeutic potential of exosomeRNP in vivo. To induce parenchymal liver fibrosis, three intraperitoneal injections of CCl4 per week for 5 weeks were performed. For therapy, mice received exosomeRNP nanocomplexes 7 days after the first CCl4 injection and twice a week thereafter (Fig. 4A). Treatment of exosomeRNP resulted in the indel frequency of 9.7% (Fig. 4B). In the meantime, the obvious digestion bands were only observed in the liver tissue among various organs (Fig. 4C), which indicated strong liver-targeting ability of exosomeRNP. Expression of CcnE1 and α–smooth muscle actin (α-SMA) is usually low in the normal liver but significantly increased after CCl4 treatment. As shown in Fig. 4D, exosomeRNP treatment resulted in significant reduced CcnE1 and α-SMA protein expression. In addition, total ASL, ALT, and bilirubin levels were not affected by various treatments (fig. S17), which was consistent with the previous report (53). As evidenced by H&E staining (Fig. 4E), Ishak scoring (Fig. 4F), collagen1α1 expression (Fig. 4G), and hydroxyproline content determination (Fig. 4H), therapeutic intervention with exosomeRNP targeting CcnE1 largely attenuated fibrosis initiation.
Fig. 4. ExosomeRNP-mediated genome editing suppressed CCL4-induced liver fibrosis.
(A) Schematic illustration of exosome for in vivo delivery of Cas9 RNP for the treatment of CCL4-induced liver fibrosis. CCL4 was administrated through intraperitoneal injection, and exosomeRNP complexes were administered through tail vein. (B) Frequency of indel mutation detected by T7E1 assay from liver tissues after the specified treatments at day 35. (C) Frequency of indel mutation detected by T7E1 assay from different organs after exosomeRNP treatment at day 35. (D) Western blotting of CcnE1 and α-SMA in livers of mice after the specified treatments. (E) H&E staining of the liver sections from the mice after the specified treatments. Scale bar, 200 μm. (F to H) Assessment of fibrosis progression by semiquantitative scoring (Ishak) (F), determination of morphometric collagen1α1 (G), and hydroxyproline content (H). Means ± SD; n = 6 (one-way ANOVA with a Tukey’s post hoc test, ***P < 0.001).
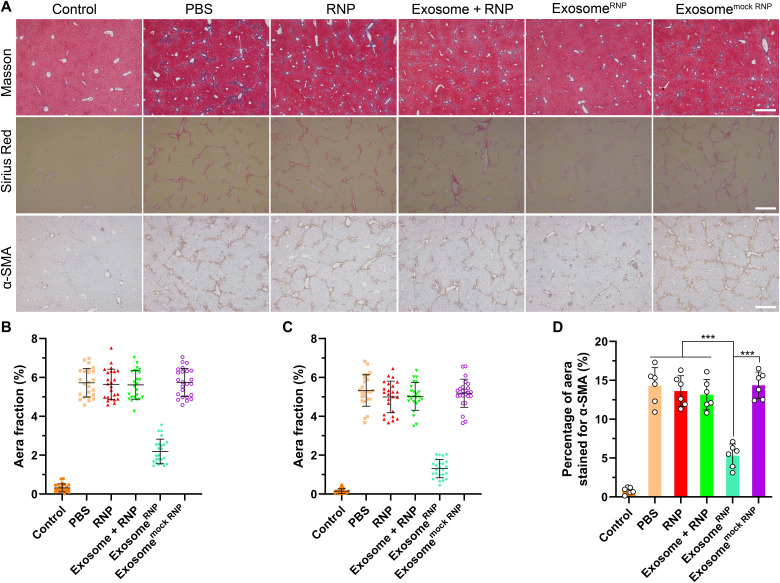
Next, we used Sirius Red and Masson staining to evaluate liver fibrosis. The positive areas occupied by liver collagen (Sirius Red or Masson) were significantly increased in the mice treated with CCL4 compared to the normal mice. In comparison with other treatments, genome editing of CcnE1 by exosomeRNP revealed significant antifibrotic effect demonstrated by lack of Sirius Red and Masson-positive area (Fig. 5, A to C). Furthermore, the percentage of α-SMA–positive area (activated HSCs in the liver) in random fields from each immunohistochemical section was quantified by image analysis (Fig. 5, A and D). As expected, genome editing of CcnE1 by exosomeRNP significantly reduced activated HSCs demonstrated by lack of α-SMA–positive areas. Collectively, these results demonstrated that exosomeRNP targeting CcnE1 strongly prevented CCL4-induced chronic liver fibrosis in mice.
Fig. 5. Determination and quantification of liver fibrosis after the specified treatments.
(A) Fibrosis was visualized by Masson, Sirius Red, and α-SMA, respectively. Scale bars, 200 μm. Percentage of Masson (B), Sirius Red (C), and α-SMA-positive area (D) in randomly selected fields from each specimen, by computerized image analysis. Means ± SD (one-way ANOVA with a Tukey’s post hoc test, ***P < 0.001).
ExosomeRNP targeting KAT5 ameliorates orthotopic HCC
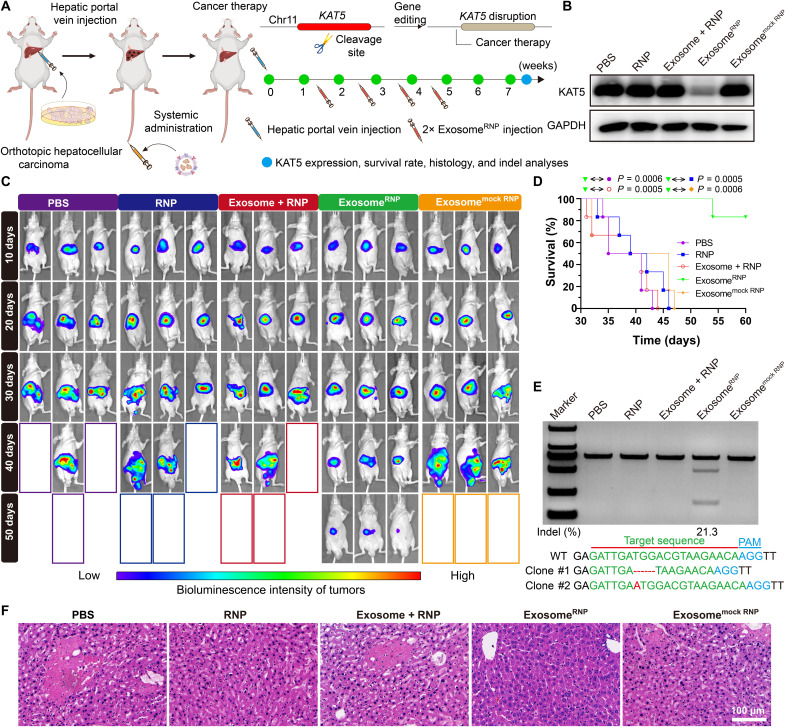
Because of the high recurrence rate and lack of effective therapy options, HCC is often associated with poor patient survival (54–56). New therapeutic options for HCC, including CRISPR-based technologies (57), have attracted wide attention in recent years. K (lysine) acetyltransferase 5 (KAT5) is required for HCC growth, and disruption of KAT5 inhibited tumor growth both in vitro and in vivo (58). Therefore, we designed sgRNA-targeting KAT5 and developed exosomeRNP to treat HCC (Fig. 6A). The therapeutic effect of exosomeRNP targeting KAT5 was tested in an orthotopic murine model of HCC. One week after the hepatic portal vein injection of luciferase-transfected Huh-7 (Huh-7-luci) cells, mice were intravenously injected with different treatments (PBS, RNP, exosome + RNP, exosomeRNP, and exosomemock RNP), and the tumor growth was evaluated using in vivo bioluminescence imaging. The mice treated with exosomeRNP-targeting KAT5 displayed the smallest tumor volume and the weakest intensity of bioluminescence, whereas other treatments had no obvious inhibitory (Fig. 6C). In addition, protein analysis indicated that the KAT5 expression level was remarkably reduced after the exosomeRNP treatment, while no changes were observed among other treatments (Fig. 6B). Furthermore, as compared with other groups, the survival of the mice treated with exosomeRNP was significantly prolonged as well (Fig. 6D). The in vivo genome editing was further validated by the T7E1 and Sanger sequencing, where significant genome disruption in the KAT5 site (21.3% indel mutation) was found in the liver tissue (Fig. 6E). Furthermore, a similar tendency was observed by the histological staining (Fig. 6F). As compared with other treatments, the liver slice of exosomeRNP group showed the fewest tumor cells. Collectively, these results demonstrated that exosomeRNP targeting KAT5 produced excellent anticancer activities in orthotopic HCC.
Fig. 6. ExosomeRNP-mediated genome editing for orthotopic HCC therapy.
(A) Schematic illustration of exosome for in vivo delivery of Cas9 RNP for the treatment of orthotopic HCC. Huh-7-luci cells were administrated through hepatic portal vein injection, and exosomeRNP complexes were administered through tail vein. (B) Western blotting analysis of KAT5 expression after the specified treatments. (C) In vivo luciferase expression of orthotopic HCC in the whole mice. (D) Survival rates after the specified treatments. Statistical significance was calculated by log-rank test (means ± SD, n = 6). (E) Frequency of indel mutation detected by T7E1 assay from liver tissues after the specified treatments. (F) H&E staining of the liver sections from the mice after the specified treatments. Scale bar, 100 μm.
Cytosolic delivery of various proteins
We hypothesized that the exosome-facilitated efficient cellular uptake of RNP could be extended to other proteins. To test this hypothesis, a series of bioactive proteins with different molecular weights and isoelectric points (pIs) were used in various assays. Cytosolic delivery of fluorescein isothiocyanate (FITC)–labeled bovine serum albumin (BSA) (FITC-BSA; 69.3 kDa, pI 4.7) by exosome was first investigated. Figures S18 and S19 showed efficient cytosolic uptake of FITC-BSA into LX-2 and Huh-7 cells after 1 hour of incubation with exosomeBSA-FITC. Similar results were observed for phycoerythrin (R-PE; 240 kDa, pI 4.3) with red-orange fluorescence. Furthermore, toxic proteins, including saporin (32.8 kDa, pI 9.3) and cytochrome C (Cyt C;12.4 kDa, pI 10.3), were also tested on Huh-7 cells (figs. S20 and S21). As shown in fig. S22, saporin (a ribosome inactivator) treatment showed minimal toxicity on Huh-7 cells, whereas the cytosolic delivery of saporin by exosome resulted in severe cytotoxicity, probably due to the efficient translocation of saporin into the cytosol to inactivate ribosome. A similar phenomenon was observed on Cyt C, which can bind with apoptosis related factor 1 to induce cell apoptosis after entering into cells (fig. S22). We also evaluated the exosome-mediated endocytosis-promoting effect on β-galactosidase (β-Gal; 430 kDa, pI 5.0), which is a native enzyme catalytically hydrolyzing β-Gal–containing substrate. We treated HeLa, LX-2, and Huh-7 cells with exosomeβ-Gal complexes and further added the substrate 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) for staining. β-Gal catalyzes the hydrolysis of X-Gal into an insoluble blue dye, revealing the intracellular amount of β-Gal in treated cells. Compared to β-Gal alone or the physical mixture of exosomes and β-Gal (exosome + β-Gal), exosomeβ-Gal showed significantly higher accumulation of blue-colored product (fig. S23) in cells, indicating the superior cellular uptake. Collectively, these results suggested that exosome may be a potent universal nonviral vector for efficient intracellular delivery of various proteins.
DISCUSSION
In this study, we developed an exosome-based nanoplatform enabling RNP-based–CRISPR-Cas9 genome-editing therapy for liver diseases. RNP can be effectively loaded into LX-2–derived exosomes through electroporation and specifically delivered to liver by exosomeRNP. ExosomeRNP showed vigorous therapeutic potential in acute liver injury, chronic liver fibrosis, and HCC mouse models via targeting PUMA, CcnE1, and KAT5, respectively. Our findings not only provide a practical strategy overcoming the delivery obstacles for RNP but also open a promising avenue for the precise and tissue-specific gene therapy of liver diseases.
MATERIALS AND METHODS
Materials
Dulbecco’s modified Eagle medium (DMEM), penicillin-streptomycin, 4′,6-diamidino-2-phenylindole, Lipofectamine CMAX, DiR, DiI, and PBS were purchased from Thermo Fisher Scientific (USA). Cas9 protein, T7E1 enzyme, and HiScribe T7 Quick High Yield RNA Synthesis Kit were obtained from New England Biolabs (Beijing, China). Fetal bovine serum (FBS) was purchased from Zhejiang Tianhang Biotech (Hangzhou, China). The MTT assay kit was purchased from MultiSciences Biotech (Hangzhou, China). The in situ X-Gal staining kit was purchased from Beyotime Biotech (Shanghai, China). The HOOK(TM) Dye Labeling Kit (FITC) was purchased from Sangon Biotech (Shanghai, China). APAP, WGA, and FITC were purchased from Sigma-Aldrich (St. Louis, MO). The In Situ Apoptosis Detection Kit was purchased from Millipore (MA, USA). The 2× Hieff PCR Master Mix reagent was purchased from Yeasen Biotechnology (Shanghai, China). Anti-PUMA (1:1000; ET1602-24) antibody was obtained from HUABIO (Hangzhou, China). Anti-CcnE1 (1:1000; ab211342) and anti–α-SMA (1:1000; ab5694) antibodies were obtained from Abcam (Cambridge, UK). Anti-KAT5 (1:1000; A01393) antibody was purchased from Boster Biological Technology (Shanghai, China). Anti–glyceraldehyde-3-phosphate dehydrogenase (1:1000; ab8245) and anti-tubulin antibodies (1:1000; ab179513) were obtained from Abcam (Cambridge, UK).
Cells and animals
Huh-7 and AML-12 cells were purchased from the American Type Culture Collection. Stable Huh-7 cell lines expressing luciferase (Huh-7-luci) and LX-2 cells were gifts from the First Affiliated Hospital of Zhejiang University. In general, Huh-7, Huh-7-luci, and LX-2 cells were cultured in DMEM containing 10% FBS at 37°C under 5% CO2. AML-12 mouse liver cells were cultured in DMEM/Ham’s F-12 nutrient medium supplemented with 10% FBS, dexamethasone (40 ng/ml; Sigma-Aldrich), and ITS premix (BD) at 37°C under 5% CO2.
Female C57BL/6 mice (6 to 8 weeks) and BALB/c mice (4 to 6 weeks) were fed in the Laboratory in Animals Centre (Zhejiang University). All animal treatments were approved by the Laboratory Animal Welfare and Ethics Committee of Zhejiang University.
Preparation of exosome and exosomeRNP
To prepare exosome-depleted FBS, FBS was ultracentrifuged at 100,000g, 4°C for 20 hours, and then filtered through 100-nm filters. For exosome isolation, LX-2 cells were cultured in DMEM containing exosome-depleted FBS. Then, the cultured medium was collected and centrifuged at 800g for 5 min, 3000g for 15 min, and 10,000g for 1 hour, respectively. The resulting supernatant was subjected to ultracentrifugation at 100,000g, 4°C for 2 hours in a SW32 Ti rotor (Beckman). Then, the purified exosome pellet was resuspended in cold PBS to be subjected to ultracentrifugation again. For electroporation, the pellet was resuspended in electroporation buffer [1.15 mM potassium phosphate (pH 7.2), 25 mM potassium chloride, and 21% OptiPrep working solution]. The suspended exosomes were then filtered through 0.22-μm filters. To prepare exosomeRNP complexes, Cas9 proteins were mixed well with sgRNA to from RNP complexes. Then, RNP complexes were added to exosomes at a weight ratio of 1:5, and then the mixture was electroporated by Gene Pulser Xcell (Bio-Rad) to form exosomeRNP complexes. After electroporation, exosomes were centrifuged at 100,000g, 4°C for 2 hours, and then the precipitation was resuspended in cold PBS solution.
Characterization of exosome and exosomeRNP
The size and morphology of exosome and exosomeRNP were characterized by DLS and TEM (HT7700, Hitachi, Japan). The total exosomal protein was quantified by bicinchoninic acid method, and the concentration of exosome was diluted to no more than 0.2 mg/ml. To detect exosomal markers, exosomes were treated with radioimmunoprecipitation assay (RIPA) lysis buffer. Then, exosome samples were incubated with primary anti-CD63 (1:1000; Abcam), anti-GM130 (1:1000; Abcam), or anti-TSG101 antibodies (1:1000; Abcam). The loading efficiency of Cas9 protein was quantified by Western blot. The gray levels of each band were quantified with ImageJ. Then, a standard curve was prepared and used to determine the bound Cas9 protein inside the exosome. The encapsulation efficiency (EE) could be calculated according to the equation EE (%) = C2/C1 × 100%, where C1 represents the original concentration of Cas9 protein in the solution and C2 represents the bound Cas9 protein inside the exosome. The stabilities of exosomeRNP nanocomplexes in the culture DMEM medium with or without serum, 4 weeks of storage at −80°C, and 7 days of storage at 4°C were determined by DLS.
Cytosolic delivery of RNP
Cytosolic delivery of RNP was tested on LX-2 and Huh-7 cells. The Cas9 protein was labeled with FITC according to the standard protocol. The cells were cultured in a 24-well plate overnight. Then, the exosomeRNP complexes were diluted with DMEM and added into the plate. After 1 hour of incubation, the cells were observed by laser scanning confocal microscopy (LSCM 880, Germany). To address the detailed uptake mechanisms, a panel of endocytosis inhibitors was used to investigate internalization pathways of exosomeRNP. The cells were preincubated with different endocytosis inhibitors, including heparin (10 μg/ml), cytochalasin D (1 μM), chlorpromazine (10 μM), EIPA (100 μM), wortmannin (50 nM), M-β-CD (10 mM), and genistein (150 μM), respectively. ExosomeRNP nanocomplexes were also incubated with cells at 4°C. The cells incubated at 37°C (no inhibitor) treated with exosomeRNP nanocomplexes only were set as the control group. The fluorescence intensity was determined by flow cytometry. The values are normalized to the control.
Biodistribution of exosomeRNP nanocomplexes within different hepatic cell types
Mice were euthanized, and liver tissues were collected after DiI-labeled exosomeRNP treatment. The liver was perfused with liver perfusion medium (Life Technologies) in a sterile culture dish and agitated for liver cell suspension preparation. The cell suspension was centrifuged at 50g for 5 min at 4°C to obtain hepatocytes. Then, cold isolation medium (DMEM, low glucose with penicillin-streptomycin and 15 mM Hepes buffer) was added to hepatocyte precipitation for purification. Then, we used commercially available magnetic beads (Miltenyi Biotec, USA) specific for KCs (biotin-labeled anti-mouse F4/80) and LSECs (mouse CD146) isolation according to a recent paper. The cell suspension was subsequently prepared for flow cytometry analysis.
Determination of the half-life of exosomeRNP in the liver
A plasmid DNA vector expressing CAG-CD63-NanoLuc was constructed according to a recent reported method (44). Then, CD63-NanoLuc–labeled exosomes in the culture supernatant were obtained after transfection with a plasmid DNA vector encoding CAG-CD63-NanoLuc. To evaluate the pharmacokinetics of exosomes in the liver, labeled exosomeRNP was intravenously injected into mice. At different time points, mice were euthanized, and liver tissues were lysed and analyzed by the Nano-Glo Luciferase Assay System (N1110, Beijing, Promega) according to the manufacturer’s protocol. The NanoLuc activities were determined by the GloMax Discover Microplate Reader (Beijing, Promega). The time-course data were analyzed using a two-compartmental model, and the half-life of the exosomeRNP in the liver was calculated as previously described (45).
Cytosolic delivery of fluorescent proteins, toxic proteins, and native enzyme
Cytosolic delivery of fluorescent proteins was tested on LX-2 and Huh-7 cells (BSA-FITC and R-PE). The BSA was labeled with FITC according to the protocol. To prepare exosomeBSA-FITC and exosomeR-PE complexes, BSA-FITC and R-PE were added to exosomes at a weight ratio of 1:1 and incubated on ice for 10 min, and then the mixture was electroporated by Gene Pulser Xcell (Bio-Rad). After electroporation, exosomes were washed and then centrifuged at 100,000g, 4°C for 2 hours. Last, then the precipitation was resuspended in cold PBS solution. The exosomeBSA-FITC and exosomeR-PE complexes were further diluted with DMEM containing FBS. Then, the exosomeBSA-FITC and exosomeR-PE complexes were added into the plate. After 1 hour of incubation, the cells were observed by LSCM.
Cytosolic delivery of toxic proteins (saporin and Cyt C) was tested on LX-2 and Huh-7 cells. Exosomesaporin and exosomeCyt C complexes were prepared as described above. The cytotoxicity of cells was evaluated by a commercial MTT assay kit.
Exosomeβ-Gal complexes were prepared with a similar method. Cytosolic delivery of β-Gal into HeLa, LX-2, and Huh-7 cells by exosome was determined by a commercial in situ X-Gal staining kit.
Immunogenicity and biocompatibility assays
Immunogenicity and biocompatibility of this formulation were performed both in vitro and in vivo. The cytotoxicity of cells treated with or without exosomeRNP was evaluated by commercial MTT and LDH assay. Hemolytic activity of the exosomeRNP nanocomplexes was evaluated according to a standard method. Next, to further assess exosomeRNP-induced immune response and toxicity, we dosed healthy BALB/c mice with exosomeRNP intravenously every other day for three times. Liver tissues were collected for further pathological analysis. Furthermore, blood was collected to separate the serum for determination of systemic toxicity and immunogenicity. Serum levels of ALT, AST, BUN, and LDH in healthy mice treated with or without exosomeRNP were determined using a biochemical autoanalyzer (TBA-40, Toshiba). Five major immune cytokines levels (IL-6, IP-10, MCP-1, TNF-α, and IFN-γ) in healthy mice treated with or without exosomeRNP were determined using ELISA kits following the manufacturer’s protocol (MultiSciences, China).
sgRNA design and synthesis
sgPUMA, sgCcnE1, and sgKAT5 were prepared by in vitro transcription reaction. Transcription templates of guide RNA were designed by online tools (http://crispr.mit.edu/ and http://chopchop.cbu.uib.no/). In addition, the in vitro transcription reaction to generate sgRNA was performed using the HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs, USA). The corresponding sgRNA sequences targeting PUMA, CcnE1, and KAT5 and DNA sequences for polymerase chain reaction (PCR) were listed in tables S1 and S2, respectively.
Induction of liver injury and therapy
APAP was dissolved in normal saline, and an intraperitoneal injection at 500 mg/kg was performed in each mouse. For treatment, C57BL/6 mice were treated with exosomeRNP complexes 4 and 2 days before the administration of APAP, respectively. A dose of 50 μg of Cas9 protein was given in each mouse. Livers were collected for further pathological analysis. TUNEL staining was conducted according to the standard protocol. HMGB1 staining was conducted with anti-HMGB1 (ab18256, Abcam) at 1:1000 dilution.
Induction of liver fibrosis and therapy
For induction of chronic liver fibrosis, CCL4 was dissolved in corn oil. Briefly, mice were injected intraperitoneally with CCl4 at 0.6 ml/kg three times a week for 5 weeks. For treatment, C57BL/6 mice were treated with exosomeRNP complexes twice a week during the experiment. The mice were given a dose of 50 μg of Cas9 protein once. Collagen1α1 expression in whole-liver extracts was determined by the Mouse Collagen1α1 ELISA Kit (Sangon Biotech). Hydroxyproline content in whole-liver extracts was determined by the Hydroxyproline Content Assay Kit (Solarbio Life Sciences). For H&E, Masson, and Sirius Red staining, tissues were fixed in 4% paraformaldehyde. α-SMA staining was performed with anti–α-SMA (Abcam) at 1:1000 dilution.
Establishment of orthotopic HCC mouse model and therapy
BALB/c mice were intraperitoneally injected with pentobarbital solution for anesthesia induction. The thoracic region was sterilized, and a left median incision was made, followed by exteriorizing the whole liver tissue. A total of 1 × 107 Huh-7-luci cells were injected into the hepatic portal vein. After that, the liver was returned to the original location under anesthesia. One week after orthotopic inoculation, mice were treated with exosomeRNP complexes twice a week during the experiment. The mice were given a dose of 50 μg of protein once. The IVIS Spectrum machine was used to monitor the tumor growth (PerkinElmer, USA).
T7E1 assays and Sanger sequencing
The T7E1 experiment was used to evaluate the editing efficiency of target genomic loci. The cells or tissues were collected to extract the DNA through the FastPure Cell/Tissue DNA Isolation Mini Kit (Vazyme Biotech). Each specific target genomic locus was amplified by PCR using FastPure Gel DNA Extraction Mini Kit (Vazyme Biotech). Afterward, a standard T7E1 assay was performed. The disrupted lanes were imaged by a gel documentation system and analyzed by ImageJ software. Indel percentage analysis was calculated on the basis of the following formula: 100 × (1 − (1 − fraction cleaved)1/2), where fraction cleaved = band intensity of each digested band/(band intensity of each digested band + band intensity of undigested band). PCR products of the genomic region–flanking target sites of sgPUMA, sgCcnE1, and sgKAT5 were subcloned to the T-clone vector, and colonies were randomly picked for Sanger sequencing.
Western blotting
Liver samples were prepared, and proteins were extracted using RIPA buffer. Proteins were separated by SDS–polyacylamide gel electrophoresis gel according to the standard procedure. The polyvinylidene difluoride membrane was incubated with 5% BSA for 1.5 hours and then incubated with the antibody against PUMA, CcnE1, α-SMA, and KAT-5 at 4°C overnight.
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 8 software (GraphPad Software Inc.). All results were expressed as means ± SD. Biological replicates were used in all experiments unless otherwise stated. The statistical significance was analyzed using Students’ t test and analysis of variance (ANOVA). One-way ANOVA with a Tukey’s post hoc test was used when more than two groups were compared. Statistical significance of survival rates was calculated by log-rank test. P < 0.05 was considered significant. N.S., P > 0.05; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Acknowledgments
Funding: This work was supported by the National Key R&D Program of China (2021YFA0909900 and 2019YFA0802202), the National Natural Science Foundation of China (82073779, 21975218, and 51773176), the Natural Science Foundation of Zhejiang Province (LR21H300002 and LGF20E030004), the Fellowship of China Postdoctoral Science Foundation (2021 M702874), the Fellowship of China National Postdoctoral Program for Innovative Talents (BX2021264), and the Higher Education Discipline Innovation Project (111 Project, B13026).
Author contributions: X.L. and Y.P. conceived the project and designed the experiments. T.W., J.Z., and Q.P. performed all the experiments. T.W. analyzed the data. X.L., T.Z., and Y.P. supervised the project and wrote the manuscript.
Competing interests: Y.P., T.Z., X.L., T.W., J.Z., and Q.P. are inventors on a patent application related to this work filed by Zhejiang University (no. 202210644925.4, filed on 8 June 2022). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S23
Tables S1 and S2
REFERENCES AND NOTES
- 1.Doudna J. A., The promise and challenge of therapeutic genome editing. Nature 578, 229–236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Min Y.-L., Bassel-Duby R., Olson E. N., CRISPR correction of Duchenne muscular dystrophy. Annu. Rev. Med. 70, 239–255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodges C. A., Conlon R. A., Delivering on the promise of gene editing for cystic fibrosis. Genes Dis. 6, 97–108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan T., Ping Y., Delivery of genome-editing biomacromolecules for treatment of lung genetic disorders. Adv. Drug Deliv. Rev. 168, 196–216 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Gillmore J. D., Gane E., Taubel J., Kao J., Fontana M., Maitland M. L., Seitzer J., O’Connell D., Walsh K. R., Wood K., Phillips J., Xu Y., Amaral A., Boyd A. P., Cehelsky J. E., McKee M. D., Schiermeier A., Harari O., Murphy A., Kyratsous C. A., Zambrowicz B., Soltys R., Gutstein D. E., Leonard J., Sepp-Lorenzino L., Lebwohl D., CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021). [DOI] [PubMed] [Google Scholar]
- 6.van Haasteren J., Li J., Scheideler O. J., Murthy N., Schaffer D. V., The delivery challenge: Fulfilling the promise of therapeutic genome editing. Nat. Biotechnol. 38, 845–855 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Behr M., Zhou J., Xu B., Zhang H., In vivo delivery of CRISPR-Cas9 therapeutics: Progress and challenges. Acta Pharm. Sin. B. 11, 2150–2171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan L., Ouyang K., Xu X., Xu L., Wen C., Zhou X., Qin Z., Xu Z., Sun W., Liang Y., Nanoparticle delivery of CRISPR/Cas9 for genome editing. Front. Genet. 12, 673286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuo C., Zhang J., Lee J.-H., Jiao J., Cheng D., Liu L., Kim H.-W., Tao Y., Li M., Spatiotemporal control of CRISPR/Cas9 gene editing. Signal Transduct. Target. Ther. 6, 238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan T., Niu D., Wu C., Xu F.-J., Church G., Ping Y., Material solutions for delivery of CRISPR/Cas-based genome editing tools: Current status and future outlook. Mater. Today 26, 40–66 (2019). [Google Scholar]
- 11.Xu X., Wan T., Xin H., Li D., Pan H., Wu J., Ping Y., Delivery of CRISPR/Cas9 for therapeutic genome editing. J. Gene Med. 21, e3107 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Liu C., Zhang L., Liu H., Cheng K., Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release 266, 17–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong J., Wang H. X., Lao Y. H., Hu H., Vatan N., Guo J., Ho T. C., Huang D., Li M., Shao D., Leong K. W., A versatile nonviral delivery system for multiplex gene-editing in the liver. Adv. Mater. 32, 2003537 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong H., Ju E., Yi K., Xu W., Lao Y. H., Cheng D., Zhang Q., Tao Y., Li M., Ding J., Advanced Nanotheranostics of CRISPR/Cas for viral hepatitis and hepatocellular carcinoma. Adv. Sci. 8, 2102051 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Z., Ukidve A., Kim J., Mitragotri S., Targeting strategies for tissue-specific drug delivery. Cell 181, 151–167 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Mitchell M. J., Billingsley M. M., Haley R. M., Wechsler M. E., Peppas N. A., Langer R., Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20, 101–124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan X., Pan Q., Xin H., Chen Y., Ping Y., Genome-editing prodrug: Targeted delivery and conditional stabilization of CRISPR-Cas9 for precision therapy of inflammatory disease. Sci. Adv. 7, eabj0624 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalluri R., LeBleu V. S., The biology , function , and biomedical applications of exosomes. Science 367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horodecka K., Düchler M., CRISPR/Cas9: Principle, applications, and delivery through extracellular vesicles. Int. J. Mol. Sci. 22, 6072 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsharkasy O. M., Nordin J. Z., Hagey D. W., de Jong O. G., Schiffelers R. M., Andaloussi S. E. L., Vader P., Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 159, 332–343 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Chen R., Huang H., Liu H., Xi J., Ning J., Zeng W., Shen C., Zhang T., Yu G., Xu Q., Chen X., Wang J., Lu F., Friend or Foe? Evidence indicates endogenous exosomes can deliver functional gRNA and Cas9 protein. Small 15, 1902686 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Ye Y., Zhang X., Xie F., Xu B., Xie P., Yang T., Shi Q., Zhang C.-Y., Zhang Y., Chen J., Jiang X., Li J., An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater. Sci. 8, 2966–2976 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Gee P., Lung M. S. Y., Okuzaki Y., Sasakawa N., Iguchi T., Makita Y., Hozumi H., Miura Y., Yang L. F., Iwasaki M., Wang X. H., Waller M. A., Shirai N., Abe Y. O., Fujita Y., Watanabe K., Kagita A., Iwabuchi K. A., Yasuda M., Xu H., Noda T., Komano J., Sakurai H., Inukai N., Hotta A., Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun. 11, 1334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S. M., Yang Y., Oh S. J., Hong Y., Seo M., Jang M., Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control. Release 266, 8–16 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Gulei D., Berindan-Neagoe I., Activation of necroptosis by engineered self tumor-derived exosomes loaded with CRISPR/Cas9. Mol. Ther. - Nucleic Acids. 17, 448–451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z., Zhou X., Wei M., Gao X., Zhao L., Shi R., Sun W., Duan Y., Yang G., Yuan L., In vitro and in vivo RNA inhibition by CD9-HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 19, 19–28 (2019). [DOI] [PubMed] [Google Scholar]
- 27.LoDuca P. A., Hoffman B. E., Herzog R. W., Hepatic gene transfer as a means of tolerance induction to transgene products. Curr. Gene Ther. 9, 104–114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aravalli R. N., Steer C. J., Gene editing technology as an approach to the treatment of liver diseases. Expert Opin. Biol. Ther. 16, 595–608 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Aravalli R. N., Steer C. J., CRISPR/Cas9 therapeutics for liver diseases. J. Cell. Biochem. 119, 4265–4278 (2018). [DOI] [PubMed] [Google Scholar]
- 30.K. O. Paredes, J. Ruiz-Cabello, D. I. Alarcón, M. Filice, in Nucleic Acid Nanotheranostics (Elsevier, 2019), pp. 421–456. https://linkinghub.elsevier.com/retrieve/pii/B9780128144701000150
- 31.Ding B., Li T., Zhang J., Zhao L., Zhai G., Advances in liver-directed gene therapy for hepatocellular carcinoma by non-viral delivery systems. Curr. Gene Ther. 12, 92–102 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Xu L., Hui A. Y., Albanis E., Arthur M. J., O’Byrne S. M., Blaner W. S., Mukherjee P., Friedman S. L., Eng F. J., Human hepatic stellate cell lines, LX-1 and LX-2: New tools for analysis of hepatic fibrosis. Gut 54, 142–151 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrmann I. K., Wood M. J. A., Fuhrmann G., Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 16, 748–759 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Li P., He K., Li J., Liu Z., Gong J., The role of Kupffer cells in hepatic diseases. Mol. Immunol. 85, 222–229 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Poisson J., Lemoinne S., Boulanger C., Durand F., Moreau R., Valla D., Rautou P.-E., Liver sinusoidal endothelial cells: Physiology and role in liver diseases. J. Hepatol. 66, 212–227 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson A. L., Qurashi M., Shetty S., The role of sinusoidal endothelial cells in the axis of inflammation and cancer within the liver. Front. Physiol. 11, 990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J.-K., Utsumi T., Seo Y.-E., Deng Y., Satoh A., Saltzman W. M., Iwakiri Y., Cellular distribution of injected PLGA-nanoparticles in the liver. Nanomedicine 12, 1365–1374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulcahy L. A., Pink R. C., Carter D. R. F., Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 3, 24641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szabo G., Momen-Heravi F., Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 14, 455–466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian T., Zhu Y.-L., Hu F.-H., Wang Y.-Y., Huang N.-P., Xiao Z.-D., Dynamics of exosome internalization and trafficking. J. Cell. Physiol. 228, 1487–1495 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Costa Verdera H., Gitz-Francois J. J., Schiffelers R. M., Vader P., Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. J. Control. Release 266, 100–108 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto A., Takahashi Y., Chang H.-Y., Wu Y.-W., Yamamoto A., Ishihama Y., Takakura Y., Blood concentrations of small extracellular vesicles are determined by a balance between abundant secretion and rapid clearance. J. Extracell. Vesicles 9, 1696517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simonsen J. B., Pitfalls associated with lipophilic fluorophore staining of extracellular vesicles for uptake studies. J. Extracell. Vesicles 8, 1582237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta D., Liang X., Pavlova S., Wiklander O. P., Corso G., Zhao Y., Saher O., Bost J., Zickler A. M., Piffko A., Maire C. L., Ricklefs F. L., Gustafsson O., Llorente V. C., Gustafsson M. O., Bostancioglu R. B., Mamand D. R., Hagey D. W., Görgens A., Nordin J. Z., Andaloussi S. E. L., Quantification of extracellular vesicles in vitro and in vivo using sensitive bioluminescence imaging. J. Extracell. Vesicles 9, 1800222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morishita M., Takahashi Y., Nishikawa M., Sano K., Kato K., Yamashita T., Imai T., Saji H., Takakura Y., Quantitative analysis of tissue distribution of the B16BL6-derived exosomes using a streptavidin-lactadherin fusion protein and iodine-125-labeled biotin derivative After intravenous injection in mice. J. Pharm. Sci. 104, 705–713 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Bunchorntavakul C., Reddy K. R., Acetaminophen-related hepatotoxicity. Clin. Liver Dis. 17, 587–607 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Nakagawa H., Maeda S., Hikiba Y., Ohmae T., Shibata W., Yanai A., Sakamoto K., Ogura K., Noguchi T., Karin M., Ichijo H., Omata M., Deletion of apoptosis signal-regulating kinase 1 attenuates acetaminophen-induced liver injury by inhibiting c-Jun N-terminal kinase activation. Gastroenterology 135, 1311–1321 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Chen D., Ni H.-M., Wang L., Ma X., Yu J., Ding W.-X., Zhang L., p53 up-regulated modulator of apoptosis induction mediates acetaminophen-induced necrosis and liver injury in mice. Hepatology 69, 2164–2179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koyama Y., Brenner D. A., New therapies for hepatic fibrosis. Clin. Res. Hepatol. Gastroenterol. 39, S75–S79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiménez Calvente C., Sehgal A., Popov Y., Kim Y. O., Zevallos V., Sahin U., Diken M., Schuppan D., Specific hepatic delivery of procollagen α1(I) small interfering RNA in lipid-like nanoparticles resolves liver fibrosis. Hepatology 62, 1285–1297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fallowfield J. A., Ramachandran P., A relaxin-based nanotherapy for liver fibrosis. Nat. Nanotechnol. 16, 365–366 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Nevzorova Y. A., Bangen J.-M., Hu W., Haas U., Weiskirchen R., Gassler N., Huss S., Tacke F., Sicinski P., Trautwein C., Liedtke C., Cyclin E1 controls proliferation of hepatic stellate cells and is essential for liver fibrogenesis in mice. Hepatology 56, 1140–1149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bangen J.-M., Hammerich L., Sonntag R., Baues M., Haas U., Lambertz D., Longerich T., Lammers T., Tacke F., Trautwein C., Liedtke C., Targeting CCl 4 -induced liver fibrosis by RNA interference-mediated inhibition of cyclin E1 in mice. Hepatology 66, 1242–1257 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Marquardt J. U., Andersen J. B., Thorgeirsson S. S., Functional and genetic deconstruction of the cellular origin in liver cancer. Nat. Rev. Cancer 15, 653–667 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Zucman-Rossi J., Villanueva A., Nault J.-C., Llovet J. M., Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 149, 1226–1239.e4 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Lee T. K.-W., Guan X.-Y., Ma S., Cancer stem cells in hepatocellular carcinoma—From origin to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 19, 26–44 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Song X., Liu C., Wang N., Huang H., He S., Gong C., Wei Y., Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy. Adv. Drug Deliv. Rev. 168, 158–180 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Kwan S.-Y., Sheel A., Song C.-Q., Zhang X.-O., Jiang T., Dang H., Cao Y., Ozata D. M., Mou H., Yin H., Weng Z., Wang X. W., Xue W., Depletion of TRRAP induces p53-independent senescence in liver cancer by down-regulating mitotic genes. Hepatology 71, 275–290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S23
Tables S1 and S2