Abstract
This study describes the occurrence and molecular identification of Monogenea from blue mackerel Scomber australasicus (Cuvier) (Perciformes: Scombridae), an edible fish, from Australian waters. Previous studies have provided either morphological or genetic results, whereas this study combines both methods of species identification. A total of 50 fish sourced from the waters off the south-eastern Australian coastline were examined and 71 Monogenea were recovered from the gills. The overall prevalence, mean intensity, and mean abundance were 64%, 2.22, and 1.42, respectively. Monogenea were initially classified morphologically as five species belonging to two families. Family Mazocraeidae was represented by Kuhnia scombri (Kuhn, 1829) Sproston, 1945, K. scombercolias Nasir & Fuentes Zambrano, 1983 and Pseudokuhnia minor (Goto, 1894) Rohde & Watson, 1985 and family Gastrocotylidae by Gastrocotyle kurra Unnithan, 1968 and Allogastrocotyle bivaginalis Nasir & Fuentes Zambrano, 1983. Molecular identification of Monogenea was conducted through sequencing of the mitochondrial cytochrome c oxidase subunit 1 (cox1) gene. The host S. australasicus was barcoded (cox1) to confirm the specific identity. There was no comparable sequence available in GenBank for K. scombercolias. Also, limited sequences were available in GenBank for the gastrocotylid Monogenea identified in this study. However, phylogenetic analyses of cox1 sequences of the Monogenea identified in this study clustered according to their familial groups. Gastrocotyle kurra and A. bivaginalis were identified for the first-time on S. australasicus in Australian waters. This study provides the first sequencing of cox1 gene for K. scombercolias. The outcomes of the study provide a basis for future Monogenea research in Australian waters, as well as for other Scomber spp.
Keywords: Fish, Monogenea, cox1, Mazocraeidae, Gastrocotylidae, Australia
Graphical abstract

Highlights
-
•
Five Monogenea were identified from the Australian blue mackerel Scomber australasicus.
-
•
The cox1 gene that has been sequenced in this study is novel for Kuhnia scombercolias.
-
•
A new host record was established for Gastrocotyle kurra and Allogastrocotyle bivaginalis.
-
•
The most prevalent monogenean was found to be the Gastrocotyle kurra at 50%.
-
•
Kuhnia scombri had the highest overall intensity at 2.64
1. Introduction
Blue mackerel Scomber australasicus (Cuvier) (Perciformes: Scombridae) is a small to medium-sized, schooling, teleost fish, which feed on plankton, copepods and crustaceans while young and, as adults, may predate small fish and squids (Froese and Pauly, 2018). The distribution of S. australasicus includes the western Pacific Ocean, waters of Taiwan and Japan and extends to Australia and New Zealand waters (Chen and Shih, 2015; Chou et al., 2011; Froese and Pauly, 2018; Quiazon et al., 2011). In Australia, S. australasicus is fished commercially and by recreational anglers (FRDC, 2020; Ward et al., 2009). The charter boat industry also uses S. australasicus as live bait for pelagic sports fishing and this species is also fed to maricultured tuna (Lowry et al., 2006; Neira and Keane, 2008).
Monogenea are ectoparasitic platyhelminthes that mainly parasitise the gills of fish (Whittington and Chisholm, 2008). Approximately 3500 Monogenea species have been described from marine fish (Rohde, 2005). In general, these parasites cause severe damage to the gills due to the invasiveness of their clamps and hooks at the site of attachment (Hutson et al., 2007; Whittington and Chisholm, 2008). Serious pathology and marked pathogenicity may lead to the death of the fish (Deveney et al., 2001).
To date, extensive surveys have been conducted to identify Monogenea infection in four Scomber (Linnaeus) species of mackerel from multiple locations in the Indo-Pacific and Atlantic Oceans (Rohde, 1986, 1989b; Rohde and Watson, 1985a, b). In studies by Rohde (1986, 1989b) and Rohde and Watson (1985a, 1985b), large numbers of Scomber hosts from several locations were examined. The authors concluded that geographical variation in morphology is common in the mazocraeid Monogenea. Therefore, populations of Monogenea from different geographical areas that differ only slightly in morphological features are not necessarily different species (Rohde, 1989b).
At the present time, five Monogenea species (Kuhnia scombri (Kuhn, 1829) Sproston, 1945, K. scombercolias Nasir & Fuentes Zambrano, 1983, K. sprostonae Price, 1961, Pseudokuhnia minor (Goto, 1894) Rohde & Watson, 1985 and Grubea australis Rohde, 1987), have been reported to infect S. australasicus in Australian waters (Table 1 and Rohde (1989b)). Previous studies on the parasites of Scomber species were based on morphological species identification and earlier morphological identification of Monogenea species from Scomber hosts showed ambiguity and many challenges according to Rohde (1989b) and Rohde and Watson (1985a, 1985b). These authors concluded that serious consideration should be given before naming species based on small morphometric and meristic variations. There have been no studies that have used combined morphological and molecular methods to identify and describe Monogenea species from S. australasicus. Therefore, specific identification for every species of Monogenea from S. australasicus using combined morphology and molecular tools is warranted.
Table 1.
Previous reports of Monogenea belonging to the family Mazocraeidae found on the gills of blue mackerel Scomber australasicus in Australian waters and other parts of the world. Abbreviations: NSW=New South Wales, VIC= Victoria, SA=South Australia, QLD = Queensland, WA=Western Australia.
| Parasite taxa | Localities | Reference |
|---|---|---|
| Kuhnia scombri | Cape Moreton, QLD; Gulf St. Vincent, SA; Fremantle, WA; and New Zealand | Schmarr et al. (2011) |
| K. scombercolias | ||
| Pseudokuhnia minor | ||
| Grubea australis | ||
| K. scombri | Location unspecified | Korotaeva (1974) |
| P. minor (syn. K. minor) | ||
| Kuhnia sp. | ||
| K. scombri | Jervis Bay and Sydney Fish Market, NSW; Golden Bay, WA; and New Zealand | Rohde, 1989a, Rohde, 1989b |
| K. sprostonae1 | Sydney Fish Market, Eden, NSW; Perth, WA | Rohde, 1989a, Rohde, 1989b |
| K. scombercolias2 | Coffs Harbour, NSW, Northern NSW; Tasmania; WA; Hawaii, USA | Rohde, 1989a, Rohde, 1989b |
| Grubea australis | Coast of NSW, Australia | Rohde (1986) |
| K. scombri | Jervis Bay and Sydney Fish Market, NSW, Australia | Rohde (1987) |
| K. sprostonae | ||
| P. minor | ||
| Grubea australis | ||
| P. minor | Jervis Bay and Sydney Fish Market, NSW, Australia; Philippines; Amoy, China | Rohde and Watson (1985a) |
| K. scombri | Eden, NSW, Australia | Hayward et al. (1998) |
| K. sprostonae | ||
| K. scombercolias | ||
| Grubea australis | ||
| P. minor | ||
| K. scombri | Eden, NSW, Australia | Perera (1993) |
| K. sprostonae | ||
| K. scombercolias | ||
| Grubea australis | ||
| P. minor | ||
| A microcotylid Monogenea3 | ||
| Mazocraeidae III, IV, V, VI (species name not mentioned in this publication) | South-eastern Australia | Rohde (1988) |
| K. scombri | Jervis Bay and Sydney Fish Market, NSW, Australia; Golden Bay, WA; New Zealand; Alumahan Bato, Philippines | Rohde and Watson (1985b) |
| K. sprostonae | NSW, Australia; Hawaii, USA | Rohde and Watson (1985b) |
Note:1Microhabitat of this Monogenea parasite is pseudobranchs. 2According to Rohde, 1989a, Rohde, 1989b, Kuhnia sprostonae species identified and described in the publications of Rohde and Watson (1985a, 1985b) would be considered as the combination of K. sprostonae and K. scombercolias. 3Family name of this parasite is Microcotylidae.
Hebert et al. (2003) and Ward et al. (2005) refer to the amplification of the mitochondrial cox1 gene as “the core of a global bio identification system for animals”. The molecular method has been extensively used for the accurate identification of many parasite species (McManus and Bowles, 1996). The molecular characterisation may also lead to the discovery of new or cryptic species (Ayadi et al., 2017; Bouguerche et al., 2019a; Oliva et al., 2014). For example, the cox1 gene has been used to differentiate among species of Monogenea (Catalano et al., 2010; Jovelin and Justine, 2001; Oliva et al., 2014). In previous studies, the characterisation of many Monogenea species was based on small sample numbers and amplification of a highly conserved region of the nuclear gene (28S rRNA, for example), which results in a lower degree of apparent genetic diversity. Therefore, the aim of the present study was to morphologically identify Monogenea species from S. australasicus and to characterise the species genetically, based on partial mitochondrial cox1 gene, in order to validate their taxonomic status.
2. Materials and methods
2.1. Fish collection
A total of 50 fish were purchased from fishermen from two retail fish markets in Australia. The fish had been caught from two separate locations off the coast of New South Wales (NSW; n = 20; July 23, 2018) and off the coast of Victoria (VIC; n = 30; September 29, 2018). Fish were transferred fresh to the Parasitology Laboratory of Charles Sturt University, Wagga Wagga Campus, in an insulated box filled with ice. All fish from each batch were examined on the day of arrival at the University. Fish were identified using morphological keys provided by Gommon et al. (2008).
2.2. Parasite collection
Each fish was dissected to remove the gills and the gills were separated and placed in an individual Petri dish containing salt water (1000 ml of water to 35 g of salt). The surfaces of all gills were thoroughly inspected under a dissecting microscope (Leica EZ4 Stereo Microscope, China) for the presence of Monogenea. Parasites were removed from gills using fine dissection needles. Collected parasites were counted and preserved in 70% ethanol for further morphological and molecular analyses.
2.3. Morphological examination
Mature Monogenea that were neither shattered, folded, or twisted were chosen for morphological identification. The specimens were rehydrated (70 percent ethanol to 50 percent ethanol to distilled water for 10–15 min each time), dyed with acetocarmine, then dehydrated in an increasing graduated ethanol series (50 percent, 70 percent, 90 percent, 95 percent, and 100 percent for 10–15 min each immersion). After that, the specimens were xylene-cleared and mounted in Canada balsam (Barton et al., 2009). Initial morphological analyses were conducted using a compound microscope (Upright Motorized Microscope ECLIPSE Ni-E, Nikon, Japan) fitted with a computer screen. Monogenea were initially grouped based on their key morphological traits including body shape and size; morphology and morphometry of the sucker, haptor and genital atrium; number and organisation of clamps; shape, size, number of anchors and marginals according to Nasir and Fuentes Zambrano (1983), Rohde (1986, 1989b), Rohde and Watson (1985a, 1985b), Sproston (1945), Unnithan (1968), and Bouguerche et al. (2019b). Characteristics were measured directly with an eyepiece micrometre (BX-43 Olympus Microscope, Olympus Corporation, Japan). All measurements are in micrometres and are given as the range followed by the mean in parentheses unless otherwise stated. A dash (−) indicates that measurements could not be made or were not available. A microscope (BX-43 Olympus Microscope, Olympus Corporation, Japan) with a drawing tube (Camera Lucida) was used for drawings. Voucher material (specimens) were deposited in the South Australian Museum under the accession numbers 36952 – 36966.
2.4. Molecular barcoding of host and parasite
A small piece of host muscle tissue and a small piece from each parasite were transferred into separate 1.5ml autoclaved Eppendorf tubes for molecular analyses. The remaining anterior and posterior regions of the parasites were processed for microscopy and morphological studies.
DNA was extracted using DNeasy Blood & Tissue Kits (Qiagen, Hilden, Germany), with some modifications to the manufacturer's instructions (Shamsi et al., 2019), and eluted in 40 μl of elution buffer. PCR amplification of the fragment of the mitochondrial cox1 gene of both mackerel and parasite was carried out using the following primer sets. For mackerel, FishF1 (forward: 5′- TCA ACC AAC CAC AAA GAC ATT GGC AC-3′) and FishR1 (reverse: 5′- TAG ACT TCT GGG TGG CCA AAG AAT CA -3′) were used (Ward et al., 2005). For Monogenea, COI-ASmit1 (forward: 5′-TTT TTT GGG CAT CCT GAG GTT TAT-3′) and COI-ASmit2 (reverse: 5′-TAA AGA AAG AAC ATA ATG AAA ATG-3′) were used (Littlewood et al., 1997). The cycling conditions to amplify the host's cox1 gene was an initial 95 °C for 2 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 54 °C for 30 s, and extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. The cox1 gene of Monogenea was amplified according to protocol provided in Hossen et al. (2020). An aliquot (3 μl) of each amplicon was examined on a 1.5% w/v agarose gel stained with GelRed® and photographed using a gel documentation system.
Representative samples from hosts and parasites were sent to the Australian Genome Research Facility (AGRF), Queensland, Australia, and were subjected to Sanger sequencing using the same primer sets as for PCR. Sequence data including chromatograms were observed initially through Sequence Scanner Software (Applied Biosystems Genetic Analysers). The host and parasite sequences were compared to the GenBank database content with BLAST and deposited in GenBank. A single alignment file was created for all the sequences and then aligned by the MUSCLE (in MEGA v.10) (Kumar et al., 2016). The evolutionary (pairwise) genetic distance was calculated using MEGA v.10 (Kumar et al., 2016).
2.5. Phylogenetic analysis
Almost all mazocreid and gastrocotylid species sequences available in GenBank were included in a preliminary phylogenetic analyses. The phylogenetic tree was constructed from the sequences generated in this study with sequences from GenBank which had both morphological and cox1 molecular data (Table 2). All sequences were then aligned with MUSCLE in MEGA v. 7 (Kumar et al., 2016). The phylogenetic relationships among species were determined using the Bayesian method using MrBayes v.3.2 (Ronquist and Huelsenbeck, 2003). The HKY+G model was applied for mitochondrial genes as suggested by jModelTest 2 (Darriba et al., 2012). Choricotyle australiensis Roubal, Armitage & Rohde, 1983 (Sequence ID: MT783686) identified from snapper Chrysophrys auratus (Förster) in Australian waters was used as the outgroup for phylogenetic analyses (Hossen et al., 2020). Sample frequency was set at 1000 and calculated for 1,500,000 generations for cox1 regions until the p-value < 0.01. After the ‘mcmc’ run, the first 25% samples were discarded, and the ‘sumt’ command was used to summarise the phylogenetic trees. Figtree v.1.4.3 was used to visualise the phylogenetic trees (Rambaut, 2014).
Table 2.
Details of the sequences used in the present study to construct the phylogenetic trees based on cox1 data. Abbreviations: NSW=New South Wales, VIC= Victoria.
| Monogenea species | Monogenea family | Host species | Host family | Geographical origin | Species morphology | GenBank ID cox1 | Reference |
|---|---|---|---|---|---|---|---|
| Gastrocotyle kurra Unnithan, 1968 | Gastrocotylidae | Scomber australasicus (Cuvier) | Scombridae | Australia: Off the coast of NSW | Yes | MZ273876–78 | Present study |
| Allogastrocotyle bivaginalis Nasir & Fuentes Zambrano, 1983 | Gastrocotylidae | S. australasicus | Scombridae | Australia: Off the coast of VIC | Yes | MZ273879–81 | Present study |
| Kuhnia scombri (Kuhn, 1829) Sproston, 1945 | Mazocraeidae | S. australasicus | Scombridae | Australia: Off the coast of VIC | Yes | MZ273888–91 | Present study |
| K. scombercolias Nasir & Fuentes Zambrano, 1983 | Mazocraeidae | S. australasicus | Scombridae | Australia: Off the coast of VIC | Yes | MZ273882–87 | Present study |
| Pseudokuhnia minor (Goto, 1894) Rohde & Watson, 1985 | Mazocraeidae | S. australasicus | Scombridae | Australia: Off the coast of VIC | Yes | MZ273892–93 | Present study |
| Allogastrocotyle bivaginalis | Gastrocotylidae | Trachurus picturatus (Bowdich) | Carangidae | Algeria | Yes | MN192391–92 | Bouguerche et al. (2019) |
| Pellonicola elongatusUnnithan, 1968 | Gastrocotylidae | Ilisha megaloptera (Swainson) | Pristigasteridae | India | No | KU872043* | Unpublished |
|
Engraulicola thrissocles (Tripathi, 1959) Lebedev, 1971 |
Gastrocotylidae | Thryssa hamiltonii (Gray) | Engraulidae | India | No | KU872046* | Unpublished |
| Pseudaxine trachuri Parona & Perugia, 1889 | Gastrocotylidae | Trachurus trachurus (Linnaeus) | Carangidae | France | No | AY009168 | Jovelin and Justine (2001) |
| Pseudaxine trachuri | Gastrocotylidae | Trachurus trachurus | Carangidae | Algeria | Yes | MN192393 | Bouguerche et al. (2019) |
| Gotocotyla sawara Ishii, 1936 | Gotocotylidae | Scomberomorus | Scombridae | China: Eight localities along the coast of China | No | KF739594 | Shi et al. (2014) |
| niphonius (Cuvier) | |||||||
| Kuhnia scombri | Mazocraeidae | Scomber japonicus | Scombridae | China: Ten localities along the coast of China | No | KU380080–82 | Yan et al. (2016) |
| Pseudokuhnia minor | Mazocraeidae | Scomber japonicus (Houttuyn) | Scombridae | China | No | KU379830–32 | Yan et al. (2016) |
| Mazocraeoides gonialosae Tripathi, 1959 | Mazocraeidae | Konosirus punctatus (Temminck & Schlegel) | China: Seven localities along the coast of China | No | JF773397 | Li et al. (2011) | |
| Gastrocotyle kurra | Gastrocotylidae | – | – | – | No | KF804042* | Unpublished |
| Gastrocotyle trachuri Van Beneden & Hesse, 1863 | Gastrocotylidae | Trachurus trachurus | Carangidae | France: Sete | No | AY009167** | Jovelin and Justine (2001) |
| Choricotyle australiensis Roubal, Armitage & Rohde, 1983 (Outgroup) | Diclidophoridae | Chrysophrys auratus (Forster) | Sparidae | Australia: New South Wales | Yes | MT783686 | Hossen et al. (2020) |
Note: *Sequences published in GenBank only. **The sequence deposited for Gastrocotyle trachuri in GenBank under the accession number of AY009167 has later been considered as Pseudaxine trachuri by Bouguerche et al. (2020).
2.6. Epidemiological data analyses
The prevalence, mean intensity and mean abundance of the Monogenea were calculated as follows: Prevalence (%) = Number of infected fish/Total number of examined fish × 100; Mean intensity = Number of parasites/Number of infected hosts; and Mean abundance = Number of parasites/Total number of examined hosts (Bush et al., 1997).
3. Results
3.1. Molecular identification of fish
The preliminary identification of the host species using morphological characteristics was ascertained by the sequencing of cox1 gene. A search in GenBank for the representative sequence generated (Sequence ID: MZ274049–50) in this study showed 100% similarity with S. australasicus (Sequence ID: DQ107708) identified from Australian waters (Ward et al., 2005), thus confirming the host's taxonomic status.
3.2. Morphological identification of monogenea
Microscopic examination (which included morphological, morphometric and meristic data) revealed five species of Monogenea belonging to the families Mazocraeidae (Kuhnia scombri (Kuhn, 1829) Sproston, 1945, K. scombercolias Nasir & Fuentes Zambrano, 1983, and Pseudokuhnia minor (Goto, 1894) Rohde and Watson, 1985a, Rohde and Watson, 1985b) and Gastrocotylidae (Gastrocotyle kurra Unnithan, 1968 and Allogastrocotyle bivaginalis Nasir & Fuentes Zambrano, 1983).
A total of 71 Monogenea individuals were collected from 32 of the 50 fish examined. All Monogenea were recovered from the gills of S. australasicus. The overall prevalence, mean intensity, and mean abundance were 64%, 2.22 and 1.42, respectively. The intensity of infection of individual species of Monogenea was highest in the fish collected from off the coast of Victoria. Fish sourced from Victorian waters had a Monogenea prevalence of 73%, which was the highest overall. The diversity of Monogenea morphotypes was highest in Victorian samples with four species identified. Kuhnia scombri had the highest overall intensity at 2.64. The most prevalent Monogenea was found to be Gastrocotyle kurra at 50%. Allogastrocotyle bivaginalis was the least prevalent Monogenea species. The prevalence, mean intensity, and mean abundance of Monogenea from S. australasicus are presented in Table 3.
Table 3.
Prevalence and abundance of Monogenea in Scomber australasicus (Cuvier) examined in the present study. Abbreviations: NSW=New South Wales, VIC= Victoria.
| Source of fish (number examined) | Name of parasites | Fish infected | Range in infected fish | Prevalence (%) | Total number found | Mean intensity | Mean abundance |
|---|---|---|---|---|---|---|---|
| Off the coast of NSW, Australia (n = 20) | Gastrocotyle kurra | 10 | 1–4 | 50 | 22 | 2.2 | 1.1 |
| Date: 23-07-2018 | Total | 10 | 1–4 | 50 | 22 | 2.2 | 1.1 |
| Off the coast of VIC, Australia (n = 30) | Allogastrocotyle bivaginalis | 3 | 1–1 | 10 | 3 | 1 | 0.1 |
| Date: 29-09-2018 | Kuhnia scombri | 11 | 1–5 | 37 | 29 | 2.64 | 0.97 |
| K. scombercolias | 7 | 1–4 | 23 | 11 | 1.58 | 0.37 | |
| Pseudokuhnia minor | 5 | 1–2 | 17 | 6 | 1.2 | 0.2 | |
|
Total |
22 |
1–7 |
73 |
49 |
2.28 |
1.63 |
|
| Grand Total (n = 50) | 32 | 1–7 | 64 | 71 | 2.22 | 1.42 |
Parasite species collected in this study corresponded in morphology and measurements to previous descriptions as provided by Nasir and Fuentes Zambrano (1983), Rohde (1986, 1989b), Rohde and Watson (1985a, 1985b), and Sproston (1945), Unnithan (1968), and Bouguerche et al. (2019b).
3.2.1. Kuhnia scombri
Seventeen specimens were observed in Victorian waters (Fig. 1a–g and Table 4). Body lanceolate. Maximum width near mid-line, tapering to narrow anterior and posterior regions up until the haptor. Copulatory organ consists of a central pad with two rows of 5 genital hooks, and two smaller pads anterolaterally, each with one large hook (Fig. 1c). Haptor well separated from body proper, heart-shaped, contains eight clamps in two opposing rows of four clamps (Fig. 1a). Distance between clamp rows decreases towards anterior portion of haptor. The caeca posteriorly connected and nearly reach to the end of body. Anchors long, strong, stout, with a hook and ridged shaft (Fig. 1f and g). No vitellaria observed in haptor. Eggs elongated-ellipsoid, operculated, filaments at each pole (Fig. 1d).
Fig. 1.
Kuhnia scombri ex Scomber australasicus. a) Whole body; b) Oral suckers with pharynx; c) Copulatory organ; d) A typical egg; e) A typical clamp; f) Large anchors; g) Small anchors.
Table 4.
Comparative measurements of mazocreid Monogenea. Measurements are in micrometres unless otherwise stated and indicated as the range followed by the mean. Abbreviations: NSW=New South Wales, VIC= Victoria, WA=Western Australia.
| Source | Present study | Rohde and Watson (1985b) | Present study | Nasir and Fuentes Zambrano (1983) | Rohde, 1989a, Rohde, 1989b | Present study | Rohde and Watson (1985a) |
|---|---|---|---|---|---|---|---|
| Monogenea | Kuhnia scombri | K. scombri | K. scombercolias | K. scombercolias | K. scombercolias | Pseudokuhnia minor | P. minor |
| Hosts (Scientific name) | Scomber australasicus | S. australasicus | S. australasicus | S. colias | S. australasicus | S. australasicus | S. australasicus |
| Hosts (Common name) | Blue mackerel | Blue mackerel | Blue mackerel | Atlantic chub mackerel | Blue mackerel | Blue mackerel | Blue mackerel |
| Locality | Off the coast of VIC, Australia | Jervis Bay and Sydney Fish Market, NSW, Australia | Off the coast of VIC, Australia | Gulf of Cariaco, Venezuela |
Sydney fish market and Eden, NSW; Perth, WA | Off the coast of VIC, Australia | NSW, Australia |
| No. of specimens | Seventeen (n = 17) | Not specified | Eight (n = 8) | Three (n = 3) | Not specified | Six (n = 6) | Not specified |
| Total body length (mm) | 2.25–4.28 (3.30) | 1.49–4.33, 2.43 (13) | 1.85–2.65 (2.12) | 1120–1400 | 0.82–344, 1.55 (6) | 1.20–2.65 (1.86) | 0.70–1.45, 1.09 (8) |
| Maximum body width (mm) | 0.20–0.65 (0.48) | 0.16–0.73, 0.43 (13) | 0.40–0.65 (0.48) | 378–585 | 0.28–0.73, 0.42 (6) | 0.18–0.45 (0.32) | 0.17–0.35, 0.25 (8) |
| Haptor (opisthaptor) length | 275–400 (331.67) | 250–420, 310 (13) | 175–320 (242.50) | 120–130 | 160–450, 240 (6) | 220–300 (250) | 170–330, 240 (7) |
| Oral sucker length | 65–90 (75.53) | – | 75–100 (86.89) | 22–26 | – | 63–73 (67) | – |
| Oral sucker width | 40–60 (50.35) | – | 48–50 (49.11) | 21–24 | – | 30–38 (33.83) | – |
| Buccal suckers (diameter) | – | 36–52, 41 (10) | – | – | 38–65, 54 (5) | – | 21–44, 32 (7) |
| Pharynx length | 36–50 (44.31) | – | 40–50 (44.33) | 23–30 | – | 35–42 (39.33) | – |
| Pharynx width | 36–45 (40.31) | – | 40–46 (42.78) | 20–27 | – | 27–33 (29.17) | – |
| Pharynx (diameter) | – | 24–37, 31 (10) | – | – | 22–37, 31 (5) | – | 17–30, 25 (8) |
| Clamps number | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Clamp length (large) | 43–55 (48.29) | – | 43–60 (48.11) | 26–34 | – | 45–50 (47.17) | – |
| Clamp width (large) | 35–46 (42.14) | – | 40–50 (44.22) | 35–40 | – | 38–43 (40.17) | – |
| Largest clamp skeleton (diameter) | – | 35–59, 43 (13) | – | – | 35–66, 44 (6) | – | 30–39, 34 (8) |
| Genital atrium length | 55–75 (65.86) | – | 35–50 (45) | – | – | 45–80 (65) | – |
| Genital atrium width | 50–60 (53.21) | – | 35–50 (44.11) | – | – | 45–75 (62.17) | – |
| Large genital hooks length | 15–25 (19) | 20–27, 23.2 (21) | 14–16 (15) | – | 13–21, 16 (5) | 26–35 (30.33) | 28–37 |
| Small genital hooks length | 12–18 (15.81) | 14–18, 15.7 (21) | 8–12 (9.78) | – | 10–14, 11.2 (6) | 14–20 (16.50) | 16–17 |
| Number of genital atrial hooks | 8–12 (10.50) | 8–11, 9.9 (20) | 10–14 (11.71) | – | 10–11, 10.8 (4) | 7–8 | – |
| Distance genital atrium-anterior end | 200–250 (230) | – | 150–180 (165) | – | – | 140–220 (180) | – |
| Distance vitellaria-anterior end | 275.00 | – | 200–350 (251.88) | – | – | 370–450 (400) | – |
| Large hamulus length | 90–114 (103.53) | 106–110 | 40–50 (44.33) | 32–40 | 31–35 | 42–52 (46.17) | 26–36 |
| Small hamulus (marginal's) length | 15–27 (21.82) | 21–24, 22.5 (21) | 16–20 (17.11) | 07–09 | 17–21, 19 (2) | 13–18 (15) | 14–18 |
| Egg length (without filament) | 275–350 (300) | – | 250–320 (273.33) | 130.00 | – | 275 (n = 1) | – |
| Egg width | 80–90 (86.67) | – | 65–95 (75) | 30.00 | – | 82 (n = 1) | – |
3.2.2. Kuhnia scombercolias
Eight specimens were observed in Victorian waters (Fig. 2a–f and Table 4). Body elongated and looked dorsoventrally flattened but comparatively shorter than K. scombri. Copulatory organ similar to that of K. scombri, with an additional strong hook on central pad (Fig. 2b). Haptor not clearly separated from body proper. Haptor contains eight clamps and organised in two opposing separate rows of four clamps on each side and distance between each row of clamps decreases posteriorly (Fig. 2a). Anchors smaller than those observed in K. scombri (Fig. 2e and f).
Fig. 2.
Kuhnia scombercolias ex Scomber australasicus. a) Whole body; b) Copulatory organ; c) A typical egg; d) A typical clamp; e) Large anchors; f) Small anchors.
3.2.3. Pseudokuhnia minor
Six specimens were observed in Victorian waters (Fig. 3a–f and Table 4). Body proper lanceolate and dorsoventrally flattened (Fig. 3a). Haptor ventral and well separated from body proper. Oral suckers separated, and pharynx suckers smaller than the buccal suckers (Fig. 3b). Copulatory organ consists of a central muscular pad with 10 (may vary) median smaller genital hooks and two muscular pads anterolaterally, each with a single curved large hook (Fig. 3c). Terminal lappet contains one pair of anchors and one pair of marginals. Large shape and structure of anchors also differ from other Kuhnia species being distinctly hook-like at posterior end and forked anteriorly (Fig. 3f). Haptor containing two approximately parallel rows of clamps of equal size with four clamps in each row. Caeca terminating at the level of the posterior part of the haptor. Species contains two dorsolateral vaginas, or two vaginal cavities (posterior to gonopore) supported by numerous transverse ridges.
Fig. 3.
Pseudokuhnia minor ex Scomber australasicus. a) Whole body; b) Oral suckers with pharynx; c) Copulatory organ; d) A typical egg; e) A typical clamp; f) Large anchors.
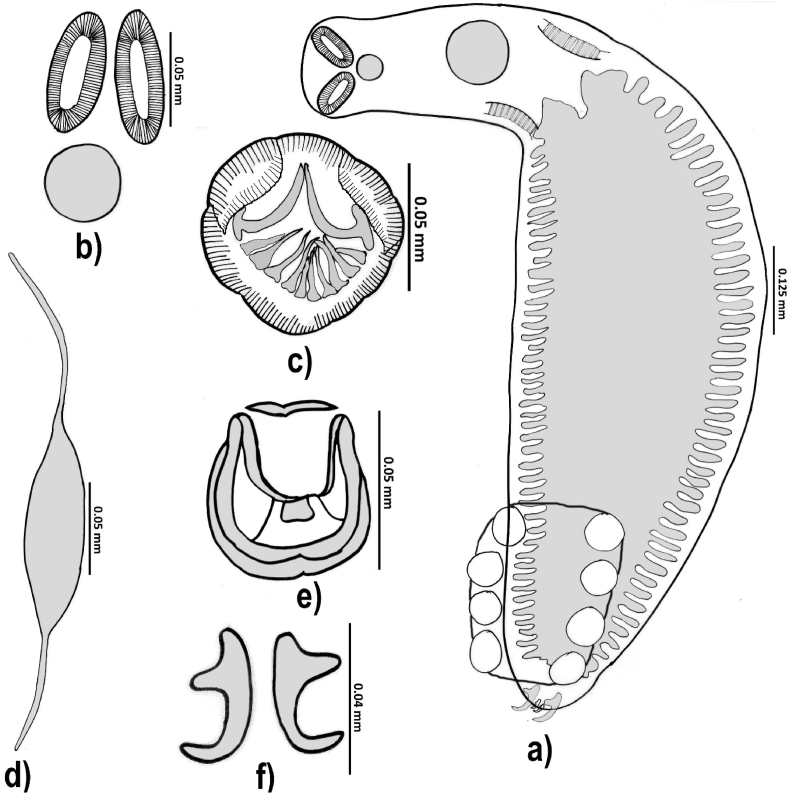
3.2.4. Gastrocotyle kurra
Ten specimens were observed in NSW waters (Fig. 4a–f and Table 5). Body typically gastrocotylid and body proper divided into two regions. Anterior (neck) region narrow and contains pharyngeal or post-pharyngeal area while main part of body proper (comparatively wider/leaf-like) contains a haptoral frill extending forwards from base of the terminal lappet to the centre and almost straight (Fig. 4a). Division between neck and body proper always clearly visible even in immature specimens. Maximum body width at level of the haptoral frill. Number of clamps 18–27 and arranged in a single row along the margin of the unilateral frill (Fig. 4a). Terminal lappet (Fig. 4f) in the form of a terminal knob containing three symmetrical pairs of oblique anchors: proximal anchors (largest), middle anchors (smallest), and distal anchors (intermediate between largest and smallest). Spindle-shaped eggs with a filament at each pole observed in a few specimens (Fig. 4d). Anterior filament slightly longer than the posterior filament.
Fig. 4.
Gastrocotyle kurra ex Scomber australasicus. a) Whole body; b) Oral suckers with pharynx; c) Copulatory organ; d) A typical egg; e) A typical clamp; f) Terminal lappet with anchors.
Table 5.
Comparative measurements of gastrocotylid Monogenea. Measurements are in micrometres unless otherwise stated and indicated as the range followed by the mean. Abbreviations: NSW=New South Wales.
| Source | Present study | Unnithan (1968) | Present study | Bouguerche et al. (2019) | Bouguerche et al. (2019) | Nasir and Fuentes Zambrano (1983) |
|---|---|---|---|---|---|---|
| Monogenea | Gastrocotyle kurra | G. kurra | Allogastrocotyle bivaginalis | A. bivaginalis | A. bivaginalis | A. bivaginalis |
| Hosts (Scientific name) | Scomber australasicus | Caranx kurra (Syn. now as Decapterus kurroides) | Scomber australasicus | Trachurus picturatus | Trachurus lathami | Trachurus lathami |
| Hosts (Common name) | Blue mackerel | Redtail scad | Blue mackerel | Blue jack mackerel | Rough scad | Rough scad |
| Locality | Off the coast of NSW, Australia | Trivandrum and near places, Kerala, India | Off the coast of NSW, Australia | Off Algeria, Mediterranean Sea | Off Venezuela, Atlantic | Off Venezuela, Western Atlantic |
| No. of specimens | Ten (n = 10) | Seven (n = 7) | Three (n = 3) | Thirty-three (n = 33) | Holotype | Two specimens measured |
| Total body length (mm) | 1.60–2.10 (1.84) | 1.50–2.87 | 2.05–3.00 (2.54) | 1.18–2.68 (2.03; n = 26) | 1.76 | 1.75–2.21 |
| Body width (mm) | 0.32–0.60 (0.40) | 0.45–0.64 | 0.38–0.45 (0.43) | 0.31–1.93 (0.48; n = 26) | 0.42 | 0.46–0.50 |
| Anterior narrow neck (mm) | 0.70–1.00 (0.86) | – | – | – | – | – |
| Haptor length | 750–1200 (990) | 1880 | 730–925 (801.67) | 310–1870 (1143; n = 26) | 945.00 | 945.00 |
| Terminal lappet | 100–160 (122) x 75–100 (84) | 225 × 135 | 500 (n = 1) | – | – | 100–130 |
| Oral sucker length | 30–40 (34.50) | – | 38–40 (38.67) | 20–40 (29; n = 28) | 20 | 16–26 |
| Oral sucker width | 20–32 (25.40) | – | 35–40 (36.67) | 15–50 (29; n = 28) | 20 | 18–22 |
| Pharynx length | 42–50 (45.80) | 40–44 | 33–43 (37) | 30–50 (38; n = 24) | 30 | 54 |
| Pharynx width | 33–46 (38.50) | – | 32–40 (35.33) | 20–52 (36; n = 24) | 27 | 58 |
| Clamps number | 18–27 (23) | 16–29 | 26–28 (27) | 23–36 (32 ± 3; n = 29) | 32 | 32–33 |
| Clamp length (large) | 63–85 (77.50) | 60 × 76 | 55–58 (56) | 37–68 (55 ± 7; n = 48) | 55 | 50–56 |
| Clamp width (large) | 55–76 (63.60) | – | 45–48 (46.67) | 31–68 (42 ± 6; n = 46) | 33 | 41–44 |
| Genital atrium length | 40–48 (42.70) | – | 45–50 (46.67) | 25–50 (38 ± 6; n = 30) | 37 | – |
| Genital atrium width | 29–38 (32.40) | – | 40–45 (42.67) | 25–50 (39 ± 5; n = 30) | 30 | – |
| Number of atrial hooks | 17–19 (18) | – | 11–13 (12) | 11–13 (12; n = 20) | 12 | 12 |
| Atrial hooks length (large) | 14–17 (15.30) | – | 17–20 (18.50) | 13–25 (21 ± 3; n = 41) | 30 | 17–20 |
| Atrial hooks length (small) | 5–7 (5.70) | – | 10–14 (11.67) | – | – | – |
| Large hamuli (proximal anchor) length | 45–55 (50.30) | 40–44 | 33–35 (34) | 24–32 (29; n = 20) | 33 | 30–34 |
| Small hamuli (middle anchor) length | 17–20 (18) | 16 | – | – | – | – |
| Distal anchor (marginal's) length | 25–30 (26.50) | 20 | 10–15 (12.33) | 9–13 (11; n = 19) | 13 | – |
| Distance genital atrium-anterior end | 150–275 (233.57) | – | 160–200 (174.33) | 125–310 (206; n = 27) | 160 | 134–216 |
| Egg length | 300 (n = 1) | 225 × 75 to 255 × 90 | – | – | – | 80 |
| Egg width | 80 (n = 1) | – | – | – | – | 28 |
| Anterior filament | 250 (n = 1) | 300–375 | – | – | – | – |
| Posterior filament | 150 (n = 1) | 285–375 | – | – | – | – |
3.2.5. Allogastrocotyle bivaginalis
Three specimens were observed in Victorian waters (Fig. 5a–f and Table 5). Body typically gastrocotylid, elongated and anterior extremity constricted. Two ventrolateral vaginal openings present. Haptors long, unilateral, and asymmetrical and armed with 26–28 clamps and arranged in a single row (Fig. 5a). Single terminal lappet and armed with two pairs of anchors; large (Fig. 5e) and small (Fig. 5f). Large anchors sickle-shaped (Fig. 5e). Oesophagus bifurcates posterior to the genital atrium. Genital atrium (gonopore) armed with hooks and obliquely oriented (Fig. 5c).
Fig. 5.
Allogastrocotyle bivaginalis ex Scomber australasicus. a) Whole body; b) Oral suckers with pharynx; c) Copulatory organ; d) A typical clamp; e) Large anchors; f) Small anchors.
3.3. Molecular characterisation of monogenea
A total of 18 specimens (representatives from each species and each collection location) were sequenced for their cox1 regions.
3.3.1. Mazocraeid monogenea
Four specimens (voucher numbers 319-2, 319-3, 319-5 and 322-1) belonging to K. scombri from Victoria were subjected to sequencing. The length of the cox1 sequences generated for the specimens was 396 bp. However, the sequences were not identical. The pairwise genetic comparison between the sequences revealed 0.76–1.56% nucleotide variability. A search in GenBank for one of the four sequences (Sequence ID: MZ273888–91) generated in this study showed 99% similarity (349/350 with no nucleotide gaps) with K. scombri (Sequence ID: KU380242) identified from the chub mackerel S. japonicus (Houttuyn) in ten locations along the coast of China (Yan et al., 2016).
Two specimens (voucher numbers 317-1 and 321-1) belonging to P. minor from Victorian fish were sequenced. The length of the cox1 sequences generated for the specimens was 396 bp but were not identical. The pairwise genetic comparison between the sequences (Sequence ID: MZ273892 & MZ273899) revealed 0.25% nucleotide variability. A search in GenBank for the representative sequence showed 96% similarity (302/315 with no nucleotide gaps) with the P. minor (Sequence ID: KU379915) identified from the chub mackerel S. japonicus in ten locations along the coast of China (Yan et al., 2016).
Six specimens (voucher numbers 315-1, 315-2, 318, 319-4, 322-2 and 331-2) belonging to K. scombercolias from Victoria were subjected to sequencing. The length of the cox1 sequences generated for the specimens was 396 bp. However, the sequences were not identical. The pairwise genetic comparison between the sequences (Sequence ID: MZ273882–87) generated in the present study revealed 0.77% nucleotide variability. There is no comparable sequence data available in GenBank for this parasite. Therefore, no similarity index is provided for this species.
3.3.2. Gastrocotylid monogenea
Three specimens (voucher numbers 190, 194 and 199) belonging to G. kurra from NSW fish were subjected to sequencing. The length of the cox1 sequences generated for the specimens was 396 bp but not identical. The pairwise genetic comparison between the sequences (Sequence ID: MZ273876–78) revealed 1.54% nucleotide variation. A search in GenBank for one of the three sequences showed 90% similarity (255/282 with no nucleotide gaps) with the only sequence available for G. kurra (Sequence ID: KF804042).
Three specimens (voucher numbers 324-2, 331-1 and 335) belonging to A. bivaginalis from Victorian fish were subjected to sequencing. The length of the cox1 sequences generated for the specimens was 396 bp but not identical. The pairwise genetic comparison between the sequences (Sequence ID: MZ273879–81) revealed 0.25–0.51% nucleotide variation. A search in GenBank for one of the three sequences generated in this study showed 98% similarity (389/396 with no nucleotide gaps) with the A. bivaginalis (Sequence ID: MN192392) identified from the blue jack mackerel Trachurus picturatus (Bowdich) in Algeria, Mediterranean Sea (Bouguerche et al., 2019b).
3.4. Phylogenetic analyses
The Bayesian phylogenetic analyses grouped the Monogenea of Mazocraeidae and Gastrocotylidae independently. Kuhnia scombri clustered with K. scombri and P. minor grouped with P. minor. The sequences for K. scombercolias clustered with closely related Kuhnia species but with a clear distinction and a strong bootstrap value. The gastrocotylid Monogenea G. kurra clustered with G. kurra. Allogastrocotyle bivaginalis displayed a clear distinction from G. kurra and other gastrocotylid Monogenea species and the phylogenetic tree grouped with A. bivaginalis. The phylogenetic relationship of Monogenea species is shown in Fig. 6.
Fig. 6.
The Phylogenetic relationship of Monogenea identified from blue mackerel Scomber australasicus in this study compared with closely related species cox1 sequences available in GenBank (Table 2 for details). The tree has been constructed/inferred using the Bayesian method. *indicates the cox1 sequences generated in the present study. The Bayesian posterior probability values more than 95% were shown on the node.
4. Discussion
This study confirmed the presence and specific identification of three mazocraeid Monogenea species (K. scombri, K. scombercolias, and P. minor) and the first evidence of the occurrence and identification of two gastrocotylid Monogenea, G. kurra and A. bivaginalis, infecting S. australasicus from Australian waters. The studies conducted of S. australasicus by Rohde (1986, 1989b) and Rohde and Watson (1985a, 1985b) showed that five Monogenea species (K. scombri, K. scombercolias, K. sprostonae, P. minor, and Grubea australasicus) commonly occur in S. australasicus from Australian waters.
Kuhnia scombri (Kuhn, 1829) Sproston, 1945 was first described by Kuhn in 1829 as Octostoma scombri. Goto (1894) followed by providing a detailed account of the internal anatomy of this species. To date, K. scombri has been identified from four species of Scomber fish: Scomber scombrus (Linnaeus) in the waters of North America, North Sea, Bay of Biscay, Mediterranean, east and west Atlantic and Guernsey; S. japonicus in the waters of Brazil, Mediterranean, Japan, Galapagos Islands, Ecuador, South Africa and Argentina; S. colias (Gmelin) from the Mediterranean; S. australasicus in NSW (south-east Australia), New Zealand and Philippines (where only a single specimen was observed) (Rohde and Watson, 1985b). Sproston (1945) also identified this parasite from S. scombrus from waters of Skagerrak, North Sea, English Channel, east and west Atlantic, and the Mediterranean; S. colias from the Mediterranean; and S. japonicus from Vladivostok and Misaki (Japan). Rohde and Watson (1985b) found extensive morphological variation of this species in different Scomber hosts from disparate geographical locations.
There are 164 cox1 sequences available in GenBank for K. scombri from a single study conducted in ten locations along the coast of China in 2016 (Yan et al., 2016). The parasite was identified from chub mackerel S. japonicus; however, no morphology data was provided in this publication and there was 0.1–5.6% (mean 2.4%) sequence divergence (Yan et al., 2016). The pairwise genetic distance between the sequences generated in the present study with representative GenBank sequences (KU380080–82 and KU380242) showed 0.57–3.88% nucleotide variability (Table 6). Due to the low genetic diversity between the present data and GenBank sequences, this variation was not considered as interspecific variation. Also, the morphometric and meristic data of the species showed a close or complete similarity with the earlier studies (Table 4). Three sequences of K. scombri for nuclear genes (two 28S and one 18S) were also available in GenBank; however, no morphological data were provided (Littlewood et al., 1999; Mollaret et al., 2000; Olson and Littlewood, 2002). This study explores, for the first time, the cox1 sequences of K. scombri from the S. australasicus in Australian waters in conjunction with the morphological examination.
Table 6.
Pairwise genetic distance (%) of Kuhnia scombri between the sequences generated in the present study with four representative GenBank sequences; *indicates the cox1 sequences generated in the present study.
| Sequence ID, Monogenea and host | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|
| 1 | *MZ273888 Kuhnia scombri ex Scomber australasicus | |||||||
| 2 | *MZ273889 K. scombri ex S. australasicus | 0.87 | ||||||
| 3 | *MZ273890 K. scombri ex S. australasicus | 1.15 | 0.86 | |||||
| 4 | *MZ273891 K. scombri ex S. australasicus | 1.16 | 0.86 | 0.57 | ||||
| 5 | KU380080 K. scombri ex S. japonicus | 1.79 | 0.87 | 1.75 | 1.76 | |||
| 6 | KU380081 K. scombri ex S. japonicus | 3.27 | 2.95 | 3.27 | 3.30 | 3.27 | ||
| 7 | KU380082 K. scombri ex S. japonicus | 1.77 | 1.46 | 1.74 | 1.75 | 1.16 | 3.88 | |
| 8 | KU380242 K. scombri ex S. japonicus | 0.58 | 0.29 | 0.57 | 0.57 | 1.17 | 3.27 | 1.16 |
Kuhnia scombercolias Nasir & Fuentes Zambrano, 1983, was first described by Nasir and Fuentes Zambrano (1983) from the Atlantic chub mackerel S. colias in Venezuelan waters. To date, the species K. scombercolias has been identified from S. australasicus in waters of Australia and Hawaii, USA; from S. colias in Venezuela; from S. japonicus in Ecuador, Brazil and the Arabian Sea (Table 1 and Rohde (1989b)). Morphologically, K. scombercolias is difficult to distinguish from other Kuhnia species, particularly K. sprostonae (Rohde, 1989b; Rohde and Watson, 1985b), as both species share several morphological characteristics (Rohde, 1989b) including: i) the haptor is not clearly distinguished from the rest of the body; ii) the copulatory organ has the same structure; iii) the number, organisation and shape/structure of clamps are similar; and iv) the anchor shape/structure is the same. These similarities created challenges and ambiguities during morphological species identification in the current study, making it difficult to differentiate these two species. Rohde and Watson (1985b) partially explain the greater morphological variations of K. sprostonae associated with hosts and geographical origins. However, in a subsequent publication, Rohde (1989b) concluded that some of the K. sprostonae species identified and described in Rohde and Watson (1985b) should be synonymised with K. scombercolias. Hence, morphological species identification of small mazocraeid Monogenea species is sometimes problematic. Rohde (1989a, 1989b) stated that “populations of Monogenea from the same host species or genus in different geographical areas are likely to be conspecific and should not be described as different species if they differ only slightly from each other”. In the current study, however, the similar genetic sequences resolved the issues. As a result, it is suggested that both species (K. sprostonae and K. scombercolias) be reviewed using genetic data to ensure that they are indeed two distinct species.
There is no comparable genetic sequence data available for either K. sprostonae or K. scombercolias and sequence data is available for only one Kuhnia species, K. scombri. A search in GenBank for one of the representative sequences generated for K. scombercolias showed 85% similarity (295/346 with no nucleotide gaps) with K. scombri (Sequence ID: KU380140) identified from the chub mackerel S. japonicus in ten locations along the coast of China (Yan et al., 2016). Our sequences are novel for K. scombercolias.
Pseudokuhnia minor (Goto, 1894) Rohde and Watson, 1985a, Rohde and Watson, 1985b, was first described by Goto (1894) from the gills of chub mackerel S. japonicus from Japan as Octocotyle minor (approximately 2 mm in length). Sproston (1945) synonymised Kuhnia spp. with Octocotyle spp. The species Kuhnia minor, since its original description by Goto (1894), has been included in this genus by other parasitologists. Rohde and Watson (1985a) described Kuhnia minor as having two vaginas. This important characteristic was overlooked by previous parasitologists. The absence of vagina (no vagina) is an important feature of the genus Kuhnia. Subsequently, a new genus, Pseudokuhnia, was established for Kuhnia minor under the family Mazocraeidae (Rohde and Watson, 1985a).
Pseudokuhnia minor has previously been identified from S. australasicus in NSW (south-east Australia), Philippines and China; S. japonicus in Atlantic South Africa (Capetown), Atlantic Spain, Haifa (Mediterranean Israel) and Japan; Indian Ocean (host not mentioned); S. scombrus in the west English Channel area (Table 1 and Rohde and Watson (1985a)). Rohde and Watson (1985a) found the extensive morphological variation of this species from the different hosts and geographical locations.
There are 250 cox1 sequences available in GenBank for P. minor from a single study conducted in ten locations along the coast of China in 2016 (Yan et al., 2016). The parasite was identified from chub mackerel S. japonicus however, no morphology data of the parasite was provided in this publication. No sequence data are available in GenBank for the nuclear genes of P. minor. In Yan et al. (2016), 0.1–4.8% (mean 1.6%) divergence was observed among the sequences. The pairwise genetic distance between the sequences generated in the present study with representative GenBank sequences (KU379830–32 and KU379915) showed 0.32–6.54% nucleotide variability (Table 7). Due to the low genetic diversity between the present data and GenBank sequences, the variation is not considered as interspecific variation. Although, the morphology, morphometric, and meristic data of the species showed close or complete similarity with earlier studies (Fig. 3a–f and Table 4) additional molecular work is required for definitive identification of this species. This study explores, for the first time, the cox1 sequences in combination with morphological characteristics of P. minor from S. australasicus in Australian waters. Rohde and Watson (1985a) observed greater morphological and geographical variations of P. minor from four Scomber fish species.
Table 7.
Pairwise genetic distance (%) of Pseudokuhnia minor between the sequences generated in the present study with four representative GenBank sequences; *indicates the cox1 sequences generated in the present study.
| Sequence ID, Monogenea and host | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| 1 | *MZ273892 Pseudokuhnia minor ex Scomber australasicus | |||||
| 2 | *MZ273893 P. minor ex S. australasicus | 0.32 | ||||
| 3 | KU379830 P. minor ex S. japonicus | 6.11 | 6.50 | |||
| 4 | KU379831 P. minor ex S. japonicus | 6.14 | 6.54 | 4.39 | ||
| 5 | KU379832 P. minor ex S. japonicus | 5.33 | 5.70 | 2.28 | 4.65 | |
| 6 | KU379915 P. minor ex S. japonicus | 4.32 | 4.68 | 1.95 | 3.65 | 1.63 |
Gastrocotyle kurra Unnithan, 1968 was first described by Unnithan (1968) from redtail scad Decapterus kurroides (Bleeker) (Syn. Caranx kurra) in Indian waters. No updated morphological description of G. kurra is available in the literature. The morphometric and meristic data for G. kurra in the present study closely or completely matched with the earlier descriptions (Fig. 4a–f and Table 5). There is a single cox1 sequence available in GenBank for G. kurra as a direct submission. However, the sequence obtained from GenBank did not have morphological data and information regarding the host and locality was not provided. One sequence for the nuclear 28S gene is available in GenBank; however, again, no morphological description is included (Tambireddy et al., 2016). As the pairwise genetic distance between the sequences generated in the present study and the GenBank sequence (Sequence ID: KF804042) showed 11.21% nucleotide variability, the GenBank sequence must be a misidentification. Without morphological description, the identity of this specimen will not be able to be determined. The present study describes, for the first time, the cox1 sequence and morphological identification of G. kurra from S. australasicus in Australian waters. This study also provides a new host record of this Monogenea species.
Allogastrocotyle bivaginalis Nasir & Fuentes Zambrano, 1983, was first described by Nasir and Fuentes Zambrano (1983) from rough scad Trachurus lathami (Nichols) from the waters of Venezuela, Western Atlantic. Allogastrocotyle bivaginalis found in the present study is recorded for the third time after its original description by Nasir and Fuentes Zambrano (1983). Previously, Bouguerche et al. (2019b) morphologically redescribed this species from blue jack mackerel T. picturatus from waters of the Algerian coast, and two cox1 sequences were deposited in GenBank to provide molecular validation for this species. The only two cox1 sequence data available in GenBank are from Bouguerche et al. (2019b). The pairwise genetic distance between the sequences of the present study and two GenBank sequences (Sequence ID: MN192391–92) showed 2.33% nucleotide variability, which is within the range of intraspecific variation (Bouguerche et al., 2019a). The specimens of A. bivaginalis that has been collected from S. australasicus in the present study morphologically slightly differ from those collected on T. lathami (by Nasir and Fuentes Zambrano, 1983) by body length, length of terminal lappet, dimensions of oral suckers and of pharynx; they differ from those collected on T. picturatus (by Bouguerche et al., 2019b) by haptor length, remarkably by clamp number and dimensions of clamps (Fig. 5a–f and Table 5). However, their genetic data supporting the identification A. bivaginalis, for the first time, in Australian waters from S. australasicus.
In the present study, the interspecific genetic variation for a species was found to be up to 10% for the cox1 sequences. According to Hebert et al. (2003), the cox1 sequence diversity greater than 2% indicates different species, and sequence diversity less than 2% indicates intraspecific variation. However, several studies have shown that the threshold values may not be applicable to all species and that this value is group-specific (Meyer and Paulay, 2005; Collins and Cruickshank, 2013). In a comparison of intraspecific variation for various representatives of the Polyopisthocotylea, Bouguerche et al. (2019a) found the range to be 0–5.6%, with the majority under 2%. Increasing the range of samples, from a wide range of Monogenea species, hosts and geographical distributions, as well as markers, are needed in future investigations to gain a deeper understanding of the evolutionary relationship between these Monogenea.
5. Conclusion
The overall prevalence of Monogenea in the study was 64% and the diversity and intensity of parasitic infection were highest in the fish sourced from Victorian waters. Five species of Monogenea from two families were identified and a new host record was established for G. kurra and A. bivaginalis. This study highlights that for species of Monogenea from S. australasicus of NSW and Victorian waters, geographical variation is common and should be considered when identifying other groups of Monogenea. This study further shows that populations of Monogenea from the same genera of hosts in different geographical areas are likely to be conspecific and should not be described as novel species unless genetic material from the original identification is also examined. Further morphological and molecular analyses of Monogenea parasites from different hosts and locations are required to explore the distribution of this understudied group.
Ethical approval
All applicable institutional, national, and international guidelines for the care and use of animals were followed.
Declaration of competing interest
The authors declare that they have no conflicts of interest.
Acknowledgements
Md. Shafaet Hossen is grateful to the Australian Research Training Program Scholarship for providing a PhD scholarship through Charles Sturt University.
References
- Ayadi Z.E.M., Gey D., Justine J.-L., Tazerouti F. A new species of microcotyle (monogenea: Microcotylidae) from Scorpaena notata (Teleostei: Scorpaenidae) in the Mediterranean Sea. Parasitol. Int. 2017;66:37–42. doi: 10.1016/j.parint.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Barton D., Beaufrère C., Justine J.L., Whittington I. Polyopisthocotylean monogeneans from carangid fishes off Queensland, Australia and New Caledonia, with a description of Heteromicrocotyloides megaspinosus sp. nov. Acta Parasitol. 2009;54:205–217. [Google Scholar]
- Bouguerche C., Gey D., Justine J.-L., Tazerouti F. Microcotyle visa n. sp. (monogenea: Microcotylidae), a gill parasite of Pagrus caeruleostictus (Valenciennes) (Teleostei: Sparidae) off the Algerian coast, western mediterranean. Syst. Parasitol. 2019;96:131–147. doi: 10.1007/s11230-019-09842-2. [DOI] [PubMed] [Google Scholar]
- Bouguerche C., Tazerouti F., Gey D., Justine J.-L. Redescription and molecular characterisation of Allogastrocotyle bivaginalis Nasir & Fuentes Zambrano, 1983 (monogenea: Gastrocotylidae) from Trachurus picturatus (Bowdich) (Perciformes: Carangidae) off the Algerian coast, Mediterranean Sea. Syst. Parasitol. 2019;96:681–694. doi: 10.1007/s11230-019-09883-7. [DOI] [PubMed] [Google Scholar]
- Bouguerche C., Tazerouti F., Gey D., Justine J.-L. No vagina, one vagina, or multiple vaginae? An integrative study of Pseudaxine trachuri (Monogenea, Gastrocotylidae) leads to a better understanding of the systematics of Pseudaxine and related genera. Parasite. 2020;27 doi: 10.1051/parasite/2020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush A.O., Lafferty K.D., Lotz J.M., Shostak A.W. Parasitology meets ecology on its own terms. J. Parasitol. 1997;83:575–583. [PubMed] [Google Scholar]
- Catalano S.R., Hutson K.S., Ratcliff R.M., Whittington I.D. Redescriptions of two species of microcotylid monogeneans from three arripid hosts in southern Australian waters. Syst. Parasitol. 2010;76:211–222. doi: 10.1007/s11230-010-9247-x. [DOI] [PubMed] [Google Scholar]
- Chen H.-Y., Shih H.-H. Occurrence and prevalence of fish-borne Anisakis larvae in the spotted mackerel Scomber australasicus from Taiwanese waters. Acta Trop. 2015;145:61–67. doi: 10.1016/j.actatropica.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Chou Y.-Y., Wang C.-S., Chen H.-G., Chen H.-Y., Chen S.-N., Shih H.-H. Parasitism between Anisakis simplex (Nematoda: Anisakidae) third-stage larvae and the spotted mackerel Scomber australasicus with regard to the application of stock identification. Vet. Parasitol. 2011;177:324–331. doi: 10.1016/j.vetpar.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Collins R.A., Cruickshank R.H. The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 2013;13:969–975. doi: 10.1111/1755-0998.12046. [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveney M., Chisholm L., Whittington I.D. First published record of the pathogenic monogenean parasite Neobenedenia melleni (Capsalidae) from Australia. Dis. Aquat. Org. 2001;46:79–82. doi: 10.3354/dao046079. [DOI] [PubMed] [Google Scholar]
- FRDC (Fisheries Research and Development Corporation) 2020. Blue Mackerel (2020)https://www.fish.gov.au/report/389-Blue-Mackerel-2020 [Google Scholar]
- Froese R., Pauly D. 2018. FishBase. version February 2018. [Google Scholar]
- Gommon M., Bray D., Kuiter R. New Holland Publisher; Australia: 2008. Fishes of Australia's Southern Coast. [Google Scholar]
- Goto S. Studies on the ectoparasitic trematodes of Japan. J. Coll. Sci. Imp. Univ. Tokyo, Japan. 1894;8:1–273. [Google Scholar]
- Hayward C.J., Perera K.M.L., Rohde K. Assemblages of ectoparasites of a pelagic fish, slimy mackerel (Scomber australasicus), from south-eastern Australia. Int. J. Parasitol. 1998;28:263–273. doi: 10.1016/s0020-7519(97)00186-0. [DOI] [PubMed] [Google Scholar]
- Hebert P.D.N., Cywinska A., Ball S.L., Dewaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B: Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossen M.S., Barton D.P., Zhu X., Wassens S., Shamsi S. Re-Description and molecular characterisation of Choricotyle australiensis Roubal, Armitage & Rohde, 1983 (monogenea: Diclidophoridae) infecting Chrysophrys auratus (Forster) (Perciformes: Sparidae) Syst. Parasitol. 2020;97:815–825. doi: 10.1007/s11230-020-09950-4. [DOI] [PubMed] [Google Scholar]
- Hutson K.S., Ernst I., Whittington I.D. Risk assessment for metazoan parasites of yellowtail kingfish Seriola lalandi (Perciformes: Carangidae) in South Australian sea-cage aquaculture. Aquaculture. 2007;271:85–99. [Google Scholar]
- Jovelin R., Justine J.L. Phylogenetic relationships within the polyopisthocotylean monogeneans (Platyhelminthes) inferred from partial 28S rDNA sequences. Int. J. Parasitol. 2001;31:393–401. doi: 10.1016/s0020-7519(01)00114-x. [DOI] [PubMed] [Google Scholar]
- Korotaeva V.D. Helminths of some commercial marine fish of the sub-order Scomberoidei from the Australian region. Izv. Tikhookean. Nauchno-Issled. Inst. Rybn. Khoz. Okeanogr. 1974;88:61–66. [Google Scholar]
- Kuhn J. Kuhnia scombriEx gills of Scomber scombrus (Linnaeus) Mem. Mus. Hist. Nat. 1829;18:357–368. [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Shi S.-F., Brown C.L., Yang T.-B. Phylogeographical pattern of Mazocraeoides gonialosae (Monogenea, Mazocraeidae) on the dotted gizzard shad, Konosirus punctatus, along the coast of China. Int. J. Parasitol. 2011;41:1263–1272. doi: 10.1016/j.ijpara.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Littlewood D.T.J., Rohde K., Clough K.A. Parasite speciation within or between host species? –Phylogenetic evidence from site-specific polystome monogeneans. Int. J. Parasitol. 1997;27:1289–1297. doi: 10.1016/s0020-7519(97)00086-6. [DOI] [PubMed] [Google Scholar]
- Littlewood D.T.J., Rohde K., Clough K.A. The interrelationships of all major groups of Platyhelminthes: phylogenetic evidence from morphology and molecules. Biol. J. Linn. Soc. 1999;66:75–114. [Google Scholar]
- Lowry M., Steffe A., Williams D. Relationships between bait collection, bait type and catch: a comparison of the NSW trailer-boat and gamefish-tournament fisheries. Fish. Res. 2006;78:266–275. [Google Scholar]
- McManus D.P., Bowles J. Molecular genetic approaches to parasite identification: their value in diagnostic parasitology and systematics. Int. J. Parasitol. 1996;26:687–704. doi: 10.1016/0020-7519(96)82612-9. [DOI] [PubMed] [Google Scholar]
- Meyer C.P., Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 2005;3:2229–2238. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollaret I., Jamieson B.G.M., Justine J.L. Phylogeny of the Monopisthocotylea and Polyopisthocotylea (Platyhelminthes) inferred from 28S rDNA sequences. Int. J. Parasitol. 2000;30:171–185. doi: 10.1016/s0020-7519(99)00197-6. [DOI] [PubMed] [Google Scholar]
- Nasir P., Fuentes Zambrano J.L. Algunos tremátodos monogenéticos venezolanos. Riv. Parassitol. 1983;44:336–380. [Google Scholar]
- Neira F.J., Keane J.P. Ichthyoplankton‐based spawning dynamics of blue mackerel (Scomber australasicus) in south‐eastern Australia: links to the East Australian Current. Fish. Oceanogr. 2008;17:281–298. [Google Scholar]
- Oliva M.E., Sepulveda F.A., González M.T. Parapedocotyle prolatili gen. n. et sp. n., a representative of a new subfamily of the Diclidophoridae (Monogenea), a gill parasite of Prolatilus jugularis (Teleostei: Pinguipedidae) from Chile. Folia Parasitol. 2014;61:543. [PubMed] [Google Scholar]
- Olson P.D., Littlewood D.T.J. Phylogenetics of the Monogenea–evidence from a medley of molecules. Int. J. Parasitol. 2002;32:233–244. doi: 10.1016/s0020-7519(01)00328-9. [DOI] [PubMed] [Google Scholar]
- Perera K.M.L. No evidence for seasonality in the ectoparasitic fauna of slimy mackerel, Scomber australasicus. Mar. Freshw. Res. 1993;44:709–719. [Google Scholar]
- Quiazon K.M.A., Yoshinaga T., Ogawa K. Distribution of Anisakis species larvae from fishes of the Japanese waters. Parasitol. Int. 2011;60:223–226. doi: 10.1016/j.parint.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Rambaut A. 2014. FigTree v1.4.2, a Graphical Viewer of Phylogenetic Trees.http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- Rohde K. Grubea australis n. sp. (monogenea, Polyopisthocotylea) from Scomber australasicus in southeastern Australia, and Grubea cochlear Diesing, 1858 from S. scombrus and S. japonicus in the mediterranean and western Atlantic. Syst. Parasitol. 1986;9:29–38. [Google Scholar]
- Rohde K. Different populations of Scomber australasicus in New Zealand and south‐eastern Australia, demonstrated by a simple method using monogenean sclerites. J. Fish. Biol. 1987;30:651–657. [Google Scholar]
- Rohde K. Gill Monogenea of deepwater and surface fish in southeastern Australia. Hydrobiologia. 1988;160:271–283. [Google Scholar]
- Rohde K. Gill monogenea of Rastrelliger spp. (Scombridae) Syst. Parasitol. 1989;14:79–91. [Google Scholar]
- Rohde K. Kuhnia sprostonae Price, 1961 and K. scombercolias Nasir & Fuentes Zambrano, 1983 (monogenea: Mazocraeidae) and their microhabitats on the gills of Scomber australasicus (Teleostei: Scombridae), and the geographical distribution of seven species of gill monogenea of Scomber. spp. Syst. Parasitol. 1989;14:93–100. [Google Scholar]
- Rohde K. CSIRO publishing; Collingwood, Victoria, Australia: 2005. Marine Parasitology. [Google Scholar]
- Rohde K., Watson N. Morphology and geographical variation of Pseudokuhnia minor n.g., n. comb. (Monogenea: Polyopisthocotylea) Int. J. Parasitol. 1985;15:557–567. [Google Scholar]
- Rohde K., Watson N. Morphology, microhabitats and geographical variation of Kuhnia spp. (monogenea: Polyopisthocotylea) Int. J. Parasitol. 1985;15:569–586. [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schmarr D.W., Whittington I.D., Ovenden J.R., Ward T.M. American Fisheries Society; 2011. Discriminating Stocks of Scomber australasicus Using a Holistic Approach: a Pilot Study, American Fisheries Society Symposium; pp. 397–417. [Google Scholar]
- Shamsi S., Barton D.P., Zhu X. Description and characterisation of Terranova pectinolabiata n. sp. (Nematoda: Anisakidae) in great hammerhead shark, Sphyrna mokarran (Rüppell, 1837), in Australia. Parasitol. Res. 2019;118:2159–2168. doi: 10.1007/s00436-019-06360-4. [DOI] [PubMed] [Google Scholar]
- Shi S.-F., Li M., Yan S., Wang M., Yang C.-P., Lun Z.-R., Brown C.L., Yang T.-B. Phylogeography and demographic history of Gotocotyla sawara (Monogenea: Gotocotylidae) on Japanese Spanish mackerel (Scomberomorus niphonius) along the coast of China. J. Parasitol. 2014;100:85–92. doi: 10.1645/13-235.1. [DOI] [PubMed] [Google Scholar]
- Sproston N.G. The genus Kuhnia n.g. (Trematoda: monogenea) an examination of the value of some specific characters, including factors of relative growth. Parasitology. 1945;36:176–190. [Google Scholar]
- Tambireddy N., Gayatri T., Gireesh-Babu P., Pavan-Kumar A. Molecular characterization and phylogeny of some mazocraeidean monogeneans from carangid fish. Acta Parasitol. 2016;61:360–368. doi: 10.1515/ap-2016-0047. [DOI] [PubMed] [Google Scholar]
- Unnithan R.V. On six species of monogenetic trematodes, parasitic on the gills of marine fishes from the Indian seas. TREUBIA. 1968;27:141–164. [Google Scholar]
- Ward R.D., Zemlak T.S., Innes B.H., Last P.R., Hebert P.D. DNA barcoding Australia's fish species. Phil. Trans. Biol. Sci. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T., Rogers P., McLeay L., McGarvey R. Evaluating the use of the daily egg production method for stock assessment of blue mackerel, Scomber australasicus. Mar. Freshw. Res. 2009;60:112–128. [Google Scholar]
- Whittington I.D., Chisholm L. In: Fish Diseases. Eiras J.C., Segner H., Wahlii T., Kapoor B.G., editors. Science Publishers Ltd; Manchester, NH: 2008. Diseases caused by monogenea; pp. 683–816. [Google Scholar]
- Yan S., Wang M., Yang C.-P., Zhi T.-T., Brown C.L., Yang T.-B. Comparative phylogeography of two monogenean species (Mazocraeidae) on the host of chub mackerel, Scomber japonicus, along the coast of China. Parasitology. 2016;143:594–605. doi: 10.1017/S0031182016000160. [DOI] [PubMed] [Google Scholar]