Abstract
Gut microbiota refers to those microorganisms in the human digestive tract that display activities fundamental in human life. With at least 4 million different bacterial types, the gut microbiota is composed of bacteria that are present at levels sixfold greater than the total number of cells in the entire human body. Among its multiple functions, the microbiota helps promote the bioavailability of some nutrients and the metabolization of food, and protects the intestinal mucosa from the aggression of pathogenic microorganisms. Moreover, by stimulating the production of intestinal mediators able to reach the central nervous system (gut/brain axis), the gut microbiota participates in the modulation of human moods and behaviors. Several endogenous and exogenous factors can cause dysbiosis with important consequences on the composition and functions of the microbiota. Recent research underlines the importance of appropriate physical activity (such as sports), nutrition, and a healthy lifestyle to ensure the presence of a functional physiological microbiota working to maintain the health of the whole human organism. Indeed, in addition to bowel disturbances, variations in the qualitative and quantitative microbial composition of the gastrointestinal tract might have systemic negative effects. Here, we review recent studies on the effects of physical activity on gut microbiota with the aim of identifying potential mechanisms by which exercise could affect gut microbiota composition and function. Whether physical exercise of variable work intensity might reflect changes in intestinal health is analyzed.
Key Points
| Nutrition and a healthy lifestyle ensure the maintenance of a functional physiological microbiota. |
| Interactions between physical activity and gut microbiota play a role in systemic and intestinal health. |
| Sports activities, diet composition, and probiotic intake may all influence the gut microbiota, which subsequently contributes to physical performance in endurance training. |
| Irregular, exhausting, or long-lasting training has a negative impact on intestinal microbiota, and the subsequent dysbiosis may contribute, at least in part, to impaired immune response and general health conditions in athletes. |
Introduction
Since ancient times, physical activity has been considered a powerful tool for preventing and improving disease onset and progression [1]. According to the World Health Organization (WHO), regular exercise may help to prevent cardiovascular risk and metabolic diseases (such as type 2 diabetes, insulin resistance, and obesity), some mental and cognitive disorders (such as anxiety and depression), and even certain cancers [2]. More recently, it has been suggested that a correlation between intestinal microbiota and exercise, including strong competitive sport activities, may help to explain the advantages of physical activity on overall body health. On the other hand, irregular or excessive physical activity as well as inappropriate endurance training may induce unfavorable changes in gut microbial composition with repercussions on athletic performance [3].
The human microbiota is defined as the set of living microorganisms in symbiosis with the human body and is estimated to include approximately 1014–1015 bacteria [4]. This microbial population spans the entire body (except for the brain and the circulatory system) and is mostly concentrated in the oral cavity, intestinal tract and skin [4, 5]. The human microbiota is represented by bacteria (more than 45,000 phyla of bacteria have been identified), archaea, fungi, viruses, bacteriophages, and protozoa [1, 6]. Microbes are found primarily in five regions: the skin, nose, oral cavity, gastrointestinal tract, and urogenital tract [4]. For all species of bacteria and archaea, nine hypervariable regions have been identified in the 16S gene, termed V1–V9, and contain 30–100 base pairs [5, 6]. The highly conserved regions can be used to design primers and sequence the gene. This information subsequently facilitates the classification of bacteria with the most conserved regions associated with the highest classification, whereas the least conserved regions are associated with the genus and species. Interestingly, these microbes are present in the human body from birth [7–9], suggesting that the proper functioning of the human organism depends not only on the expression of one's genes but also on the gene expression of the coexisting microorganisms. Based on this notion, the US National Institutes of Health (NIH) in 2008 launched the Human Microbiome Project [8], a scientific program with the main purpose of creating a reference database for sequences of microbial genetic material that exists in the human body. The goal was to detect the relationship between the microbiome and humans and to analyze the potential consequences of changes in the bacterial composition on human health and disease. Among the very large number of bacterial cells that make up the intestinal microbiota, approximately 2,000 species have been discovered [5, 9, 10], and more than 500 species have been classified into 12 different phyla: 93.5% belong to Pseudomonadota (e.g., Proteobacteria, 8%), Actinomycetota (e.g., Actinobacteria 3%), Bacteroidota (e.g., Bacteroidetes, 23%), and Bacillota (e.g., Firmicutes, 65%). Of the 12 genera found, three phyla contain only one species isolated from humans, as in the case of Akkermansia muciniphila (the sole representative of the Verrucomicrobia phylum) (Table 1) [10–12]. In addition, of the 386 obligatory anaerobic species identified in the human intestine, some have also been found in the mucosa of the oral cavity [7, 10, 12]. The stomach hosts the lowest number of bacterial cells (0.1–10%), which are mainly represented by Lactobacillus, Candida, Streptococcus, and Helicobacter pylori [10, 13]. Any modification in the amount of residing bacteria is associated with certain pathologies, as exemplified by the causative role of H. pylori in the pathogenesis of duodenal ulcers and, potentially, in gastric cancer [14, 15]. However, the acidic pH of the stomach limits the presence of bacteria, whereas the favorable pH in the colon promotes a more suitable habitat for bacteria, such as Bacteroides, Clostridium, Bifidobacterium, and Enterobacteriaceae [16, 17]. Most of these species are obligate anaerobic bacteria as the limited amount of oxygen is consumed by aerobic bacteria, such as Escherichia coli, which help to maintain the low oxygen environment of the colon [18].
Table 1.
Examples of “friendly” bacteria that colonize the human digestive system in bacterial eubiosis
| Species | Action |
|---|---|
| Akkermansia muciniphila | This bacterium represents 3–5% of typical intestinal bacterial members, and its presence is decreased in obese subjects. Its activity has been related to the thickness of the intestinal wall, resulting in reduced food absorption |
| Alistipes putredinis | Belonging to the Rikenellaceae family, phylum Bacteroidota, this species seems to be particularly represented in subjects with type 2 diabetes and obesity |
| Bacteroides vulgatus | Gram-negative bacillus, non-endospore-forming bacilli belonging to the common resident bacteria of the human microbiota. It is involved in numerous metabolic activities and can provide a certain level of protection from invasive pathogens |
| Bifidobacterium adolescentis | Gram-positive bacterium belonging to the Actinomycetota phylum. It is an organism normally present in healthy subjects. Its colonization occurs from birth. It tends to decrease in adulthood and in old age due to factors such as diet, stress, and antibiotic intake |
| Bifidobacterium longum | Gram-positive, anaerobic bacterium belonging to the phylum Actinomycetota. This bacterium is a human commensal and considered one of the first colonizers of the gastrointestinal tract of newborns. Several strains of this bacterium exhibit various protective functions and are often taken as a probiotic agent |
| Eubacterium rectale | Belonging to the Bacillota phylum, it is thought to play a beneficial role in the maintenance of the normal ecology of the large intestine based on the production of substances, such as butyric acid, which acts as a growth inhibitor for other bacteria |
| Faecalibacterium prausnitzii | Commensal microorganism belonging to the Ruminococcaceae family, phylum Bacillota. It plays a protective role in maintaining the correct intestinal ecosystem. It is scarcely present in subjects suffering from IBS and type 2 diabetes |
| Lactobacillus gasseri | This bacterium belongs to the Lactobacillaceae family, phylum Bacillota. Additionally, when used as a probiotic agent, it provides protection from pathogens. This bacterium is present in nonobese subjects |
| Lactobacillus rhamnosus | This bacterium belongs to the Lactobacillaceae family, phylum Bacillota. L. rhamnosus is regarded as a probiotic agent. It is mainly localized in the colon. Among its beneficial actions, it helps to defend against pathogens, such as Candida spp. |
| Streptococcus thermophilus | This bacterium belongs to the Streptococcaceae family, phylum Firmicutes. It is a thermophilic microorganism. Its optimal growth temperature is between 37 and 42 °C. It is a normal commensal, and S. thermophilus levels are increased in subjects with metabolic syndrome and IBS |
IBS irritable bowel syndrome
Interestingly, the recognition of cross-talk axes between the gut/lung, gut/brain, gut/skin, gut/muscle, gut/liver, and bladder/gut further underlines the potential role of gut bacteria in modulating the physiological function of multiple organs [9, 19].
The Evolution of the Human Gut Microbiota During Life
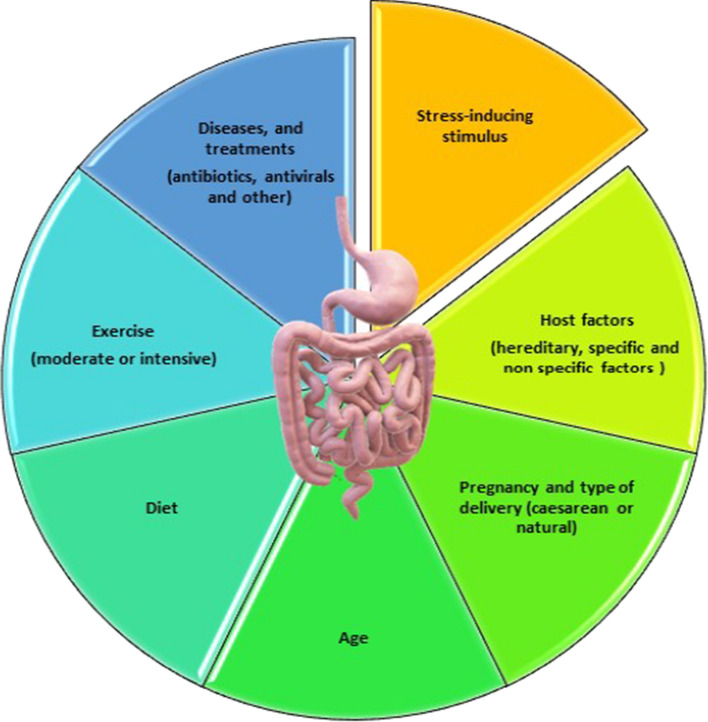
Gut microbial biodiversity evolves with aging and depends on several factors, starting with birth delivery procedures (Fig. 1) [20–22]. Even during pregnancy, maternal exposure to environmental factors, including microbes, might influence postnatal immune functioning and the subsequent development of allergic diseases [23, 24]. Newborns of mothers in contact with farm animals have shown a reduced predisposition to allergies and asthma. This finding might depend on the increased immune response associated with prenatal exposure to these agents and is potentially associated with a change in chordal blood regulatory T cells (Treg) and reduced Th2 cytokine secretion (increased Th2 cytokine secretion is a feature of an allergic response) [25–30]. The intestinal tract of the infant is rapidly colonized [8, 16]. The composition of microorganism communities in infants differs based on vaginal or cesarean birth. While Lactobacillus and Prevotella species prevail in the gut of infants born by natural delivery, Streptococcus, Propionibacterium, and Corynebacterium bacteria predominate in infants born by cesarean section [30, 31].
Fig. 1.
The main factors that influence the composition of the gut microbiota
At birth, the composition of the gut microbiota is mainly represented by E. coli, and a progressive increase in Lactobacillus and Bifidobacterium species is noted during the next few months of postnatal development. During the first weeks of life, the incomplete activity of Toll-like receptors (TLRs) allows the necessary formation of a stable bacterial community in the gut [9, 27, 32, 33].
Nutrition is one of the most important factors in colonization. With its high concentration of oligosaccharides, breast milk facilitates the growth of Lactobacillus and Bifidobacterium (bacteria able to produce short chain fatty acids (SCFAs) and promote the synthesis of IgG immunoglobulin) and, to a lesser extent, of Bacteroides spp. and Clostridia spp. [27, 33]. On the other hand, formula feeding mainly promotes the growth of Bifidobacteria, Clostridioides difficile, and Escherichia coli [33, 34].
With the introduction of solid foods, the diversity of the microbiota increases. The bacterial community profile reveals the onset of Bacteroides and a decrease in E. coli, whereas Lactobacillus levels remain constant [27, 31, 35].
In adulthood, the intestinal microbiota forms a relatively stable community (but variable between different individuals) that is mainly dominated by the Bacteroidota and Bacillota phyla as well as Escherichia and Lactobacillus to a lesser extent, whereas the presence of Bifidobacterium remains constant. In the elderly, Bifidobacterium species decrease in quantity, whereas Escherichia and Lactobacillus generally tend to increase [12, 35].
Dysbiosis, which is defined as the quantitative and qualitative imbalance in the microbiota composition and in the subsequent relevant changes in cytokine production (Table 2) [36–40], has been linked to several diseases.
Table 2.
Dysbiotic microbiota associated with inflammation and diseases, such as asthma, type 2 diabetes, obesity, irritable bowel syndrome (IBS), and inflammatory bowel disease (IBD)
| Species | Pathogenic action |
|---|---|
| Anaerotruncus colihominis | It is an anaerobic, Gram-negative, nonmotile and rod-shaped microbe. Produces indole from tryptophan and uses glucose and mannose as its main energy source. It can cause bacteremia under conditions of immune system deficiency |
| Bacteroides ovatus | Belonging to the Bacteroidota phylum, it was identified as the main cause of the systemic antibody response in IBS |
| Collinsella aerofaciens | Human commensal known for its ability to ferment a wide range of carbohydrates (including starch). This fermentation results in the formation of products, such as hydrogen and ethanol, which increase the presence of intestinal gas when present at high levels |
| Desulfovibrio piger | Gram-negative, sulfur-reducing bacterium belonging to the Desulfovibrionaceae family, phylum Pseudomonadota. Excessive presence of this microorganism is related to IBD |
| Dorea formicigenerans | This bacterium belongs to the Clostridiaceae family. It is particularly present in subjects with hepatic steatosis of nonalcoholic origin |
| Escherichia fergusonii | Opportunistic pathogen microorganism involved in IBS and found in obese subjects |
| Finegoldia magna | This bacterium belongs to the Peptostreptococcaceae family, phylum Bacillota. Overgrowth of this bacterium can lead to bacteremia, visceral and skin lesions. It has also been isolated in subjects suffering from joint prosthesis infections |
| Haemophilus influenzae | This bacterium belongs to the phylum Pseudomonadota and is responsible for potentially serious infections, especially in children, that are preferentially located in the respiratory tract and meninges |
| Parabacteroides distasonis | This bacterium belongs the Bacteroidota phylum. These opportunistic pathogens can cause severe infections when present in combination with other aerobic and anaerobic bacteria |
| Parabacteroides merdae | Microorganism belonging to the Bacteroidota phylum. This bacterium is mainly observed in subjects with type 2 diabetes and IBS |
| Peptostreptococcus anaerobius | This bacterium belongs to the Peptostreptococcaceae family, phylum Bacillota. In the context of immunosuppressive conditions, this bacterium can give rise to systemic infections by triggering infectious focuses in brain, neck, liver, breast, lungs, central nervous system, chest, abdomen, pelvis, skin, bones, joints, and soft tissues |
| Shigella boydii | Opportunistic pathogen involved in various inflammatory intestinal pathologies |
Intestinal dysbiosis may result from five main conditions: (a) deficiency: diet poor in soluble fibers and/or rich in packaged, refined, and sterilized foods or as a consequence of antibiotic treatments greatly impacting the microbiota species Bifidobacteria and Lactobacilli; (b) putrefaction: diet rich in animal fat and low in fibers promoting an increase in Bacterioides, Clostridia, Peptococci, and Eubacteria species; (c) fermentation: subsequent to a relative intolerance to carbohydrates or excessive consumption of simple sugars; (d) sensitization: resulting from an immune response to components of the intestinal microbiota and exemplified by a deficit in the immune barrier composed of secretory IgA; (e) fungal dysbiosis: diet rich in simple sugars, leavened foods, and refined carbohydrates and low in fibers, which favor excessive and unbalanced growth of Candida spp. and yeast microorganisms in the intestines [37, 38, 41–43].
In addition to diet, the microbiota is influenced by nonspecific and specific host factors, including lifestyle (urban or rural), geographic location, surgery, smoking habits, chronic alcoholism, xenobiotics (such as heavy metals), drugs, stress, mental conditions such as depression and, finally, exercise [44].
Nonspecific host factors include some molecules produced by intestinal epithelial cells to control the colon surface, and alterations in the structure of these factors may therefore influence microbial composition. Among them, those that define the mucus composition as well as antimicrobial peptides (AMPs) and IgA immunoglobulins may help the growth of some species of microorganisms and inhibit the growth of others [43, 45, 46]. In the large intestine, mucus plays a key role in blocking harmful interactions between microorganisms and intestinal epithelial cells [47, 48]. Mucin and mucin O-glucans play a fundamental role in the formation of the intestinal microbiota and in the selection of the most suitable microbial species for the health of the host. On the other hand, the amount of mucus is more limited in the small intestines; AMPs produced by Paneth cells through a mechanism involving pattern recognition receptors (PRRs) are involved in the formation of the microbiota [48, 49]. These PRRs are activated by various microbial components, such as lipopolysaccharides (LPS), via the microbe-associated molecular patterns (MAMPs) pathway [50]. The PRR-MAMP system sustains the efficient action of the barrier created by mucus by determining the production of the immunoglobulins IgA, mucin, and AMP, the highest concentrations of which are found inside intestinal crypts [51]. AMPs are the first line of defense against pathogenic microorganisms and carcinogenesis. Some species of the gut microbiota, such as the phylum Bacteroidetes, are resistant to high concentrations of AMP [51–53]. Their presence has been considered responsible for the secretion of several proteins, including those of the Regenerating (Reg) family. Moreover, plasma cells of the intestinal mucosa produce IgAs, and the ability of IgAs to camouflage bacteria, may help to control their numbers [45, 50, 54, 55].
Among specific factors involved in the development and modification of the microbiota are miRNAs, which are small fragments of RNA that do not encode any genetic information. These miRNAs form in the nucleus, are transferred to the cytoplasm, are implicated in the regulation of distinct mRNAs, and may exit the cell and circulate in body fluids [56, 57]. Epithelial, intestinal, and Hopx-positive cells are the main sources of miRNA. Some miRNAs (such as miRNA515-5p for Fusobacterium nucleatum and miRNA-1226-5p for E. coli) have been demonstrated to be able to enter bacterial cells and induce gene expression, therefore facilitating bacterial growth [58].
As perhaps the most recognized factor that can perturb the composition of the microbiota, antibiotics have a profound effect on resident bacteria, and their misuse or overuse is widely acknowledged as one of the most important causes for the increase in antibiotic-resistant pathogens [59, 60].
The Main Functions of Gut Microbiota on Health
The intestinal microbiota is highly involved in strengthening the gastrointestinal barrier and participates in regular peristalsis and intestinal homeostasis. In fact, the recognition of commensal bacteria by toll-like receptors (TLRs) is necessary to stimulate the proliferation and physiological turnover of epithelial cells, protecting the epithelial surface from intestinal injury [60–63]. As mentioned above, in the epithelium of the small intestine, Paneth cells perceive enteric bacteria through the activation of TRLs and trigger the expression of various antimicrobial factors [61, 63, 64]. This process allows control and limits the penetration of the intestinal barrier by pathogenic bacteria. The microbiota participates in the development of the gut-associated lymphatic tissue (GALT) and the host immune system by stimulating the secretion of IgA and the production of antimicrobial molecules that inhibit the proliferation and colonization of pathogenic bacteria [64, 65]. Using ligands produced in commensal bacteria (such as LPS), the gut microbiota influences the development and function of the mucosal immune system [64]. The innate immune system can also recognize potentially pathogenic microbes by identifying the TLRs of molecules called pathogen-associated molecular patterns (PAMPs) and react by increasing the levels of cytokines and enhancing the activation of T cells against these pathogens [64, 65].
In addition, the microbiota participates in metabolic functions by processing nondigestible dietary residues that produce SCFAs (such as n-butyrate, acetate, and propionate), which subsequently contribute to the host energy balance by increasing the availability of nutrients [66]. SCFAs are secreted into the intestinal lumen, pass the epithelial barrier, are released into the bloodstream, and reach peripheral organs and tissues, where they will be used as substrates for energy metabolism. For example, hepatocytes use propionate for gluconeogenesis. SCFAs are mediators of the gut/brain axis and contribute to stimulating the release of peptide YY (PYY) and 5-hydroxytryptamine (5-HT) [9, 60]. SCFAs also act as signaling molecules to regulate immune and inflammatory responses. For instance, n-butyrate regulates the function and migration of neutrophils, increases the expression of tight junction proteins in the epithelial colon, reduces mucosal permeability, and inhibits the synthesis of inflammatory cytokines. In addition to the production of SCFAs, bacterial species of the intestinal microbiota synthesize glycans, amino acids, and vitamins (e.g., K, B12, biotin, folate, and thiamine), all essential components for host metabolism [9, 60].
Biomolecular Interactions Between Physical Exercise and Gut Microbiota
Physical activity protects against several chronic diseases, and the gut microbiota might be involved in many of these beneficial effects [67]. By playing a positive role in homeostasis and energy regulation, physical exercise induces changes in intestinal microbial composition. However, some specific differences should be considered based on various forms of exercise; exercise frequency, mode, or intensity; the peculiarities of aerobic training or resistance exercise; and the advantages and consequences in amateurs or athletes of competitive disciplines [67–69]. Salient differences between regular, noncompetitive physical activity and athletic exercise training are discussed below.
Regular Exercise Training and Active Lifestyle
Regular physical activity influences the gut/brain axis, resulting in an anti-inflammatory immunoregulatory state. By reducing the transient evacuation time and therefore the contact time between pathogens and the gastrointestinal mucus layer, low-intensity exercise may help to reduce the risk of colon cancer, diverticulosis, and IBD in individuals undergoing regular training sessions [9, 70]. Even in the presence of a high-fat diet, physical exercise is related to lower inflammatory infiltrates and better protection of the morphology and integrity of the intestine. In fact, especially when combined with sedentary behavior, a high-fat diet increases intestinal villi width due to plasmacytoid and lymphocyte infiltrates [71]. Regular exercise might prevent some of these changes by reducing the expression of cyclooxygenase 2 (Cox-2) in the proximal and distal intestine.
On the other hand, it has been observed that resistance exercise results in a transient decrease in splanchnic blood flow (up to 80% of baseline levels) with potential subsequent changes in the morphology and physiology of the intestinal tissues [67]. This reduction depends on the increased arterial resistance in the splanchnic vascular bed, secondary to enhanced activation of the sympathetic nervous system. Thus, when physical exercise is excessively prolonged, the increased intestinal permeability might favor bacterial translocation from the colon with a subsequent associated risk of gastrointestinal issues [67, 71]. In experimental studies on animals, voluntary running is associated with microbiota variation and concomitant increases in both the n-butyrate concentration and cecum diameter. Although this last condition might lead to exposure to gastrointestinal disturbances, n-butyrate-mediated control of NF-kB signaling pathways with subsequent protection against carcinogenesis might compensate for the overall risk of exercise-associated colonic diseases [72]. In this regard, it is important to recall that butyrate may inhibit the activity of histone deacetylases and therefore influence gene regulation, immune modulation, reduction of oxidative stress, suppression of carcinogenesis and cell differentiation, and, in terms of physiological activities, regulation of the intestinal barrier, visceral sensitivity, and modulation of intestinal motility [73]. Similarly, regular exercise prevents obesity development and produces changes in the percentage of major bacterial phyla in high-fat-fed obese mice. In this animal model, the total distance traveled by the animals was inversely correlated with the Bacteroidota-Bacillota phyla ratio [67, 69].
Increased production of immunoglobulin A (IgA) and a reduced number of B and T-CD4 cells were observed in the intestines of mice performing moderate long-term exercise compared to mice that did not undergo any physical training. These findings suggest that exercise in mice may enhance the strength of the commensal microbiota to counteract exogenous colonization and therefore help protect against infections by intestinal pathogens [72–74].
To add complexity, some other studies have observed a decrease in the genus Faecalibacterium prausnitzii, which is potentially responsible for pathologies in the fatty intestine, in exercising mice [74]. In this regard, it has been hypothesized that the association between an inadequate dietary restriction to the body's needs combined with exercise might be responsible for a decrease in “good” bacteria and an increase in harmful bacteria with possible alterations in the barrier function of the intestinal mucosa [67, 75].
These findings call attention to the relationship between nutritional status and exercise, especially during the juvenile period when the composition of the gut microbiota is modified with a relative increase in Bacteroidota and a concomitant decrease in the Bacillota phylum [72]. This shift is associated with appetite-related signaling, as serum leptin levels correlate positively with Bifidobacterium and Lactobacillus populations and negatively with the levels of Bacteroides spp. and Prevotella spp. [67, 69], whereas ghrelin serum levels exert opposite effects on these bacterial populations. Thus, early-life exercise may profoundly influence the composition of the gut microbiota by stimulating the development of bacteria capable of causing adaptive changes in host metabolism and contribute to optimizing the development of brain function [67, 76].
Regarding the influence of specific training activities, an inverse relationship has been noted between the quality and nature of physical activity and the amount of fecal bile acids, and this correspondence becomes stronger as physical activity intensifies. This is a specific example of how exercise frequency, mode, or intensity may affect the gut microbiota [12]. Given that the antimicrobial effects of various bile acids differ, the profile and the relative concentration of individual bile acids may play a role in favoring some species and reducing others. In rodents, integration of cholic acid in the diet changes the microbiota composition (both quantitatively and qualitatively) with an increase in the Bacillota (mainly Clostridia spp.) and a decrease in the Bacteroidota phylum. The microbiota may subsequently influence metabolic function through the synthesis of the so-called secondary bile acids that regulate the deposition of fat in the liver and muscles by activating hormone receptors, such as the farnesoid X receptor (FXR). Moreover, bile acids seem to be involved in increased energy expenditures in the muscles. Overall, these observations further reinforce the idea that gut bacteria actively participate in metabolic homeostasis and may therefore contribute to protection from obesity [77–80].
Similarly, changes produced by exercise in the profile of SCFAs add support to the relationship that ties physical activity to the muscle/microbiota axis. SCFAs produced by the microbiota can activate AMP-dependent kinase (AMPK), a master regulator of energy metabolism, in muscle cells [81, 82]. The activation of this kinase by SCFAs can occur directly by increasing the AMP/ATP ratio and/or indirectly through the leptin FFar2 pathway, thus controlling the activity of various factors involved in lipid metabolism, cholesterol, and glucose levels in the muscle. In addition, SCFAs produced in the colon compartment stimulate the FFar2/3 receptors and increase the plasma concentrations of peptide YY (PYY), a satiety hormone that strengthens the insulin-mediated disposal of glucose in muscles and adipose tissue [82–84].
Muscles concomitantly express TLR4 and TLR5 receptors, which could be activated by circulating LPS, the levels of which may vary according to the composition of the gut microbiota. Activation of TLRs by LPS from the membrane of some bacterial types leads to the production of inflammatory cytokines in muscles through activation of NF-kB [79–81], and muscle atrophy in mice injected with LPS is related to activation of TLR4 receptors. Interestingly, in rats fed a high-fat diet, both acute and chronic exercise may induce a significant decrease in the TLR4-mediated signaling pathway in liver, muscle, and adipose tissue accompanied by the concomitant reduction in serum LPS levels and improved insulin signaling and sensitivity in metabolic target tissues [82, 83].
During regular physical activity, myokines (cytokines and other peptides) released from muscle fibers exert paracrine and endocrine effects. Exercise stimulates muscle cells to produce IL-6, thereby increasing the total circulating levels of this cytokine and contributing to its metabolic and anti-inflammatory effects [84]. IL-6 enhances fat oxidation and glucose uptake through AMPK phosphorylation and activates the secretion of the anti-inflammatory cytokines IL-10, IL-1ra, and TNF-R, protecting against chronic diseases associated with low-grade inflammation. Thus, physical exercise may indirectly protect the microbiota from changes induced by inflammatory conditions (such as IBD and type 2 diabetes) [85, 86].
As briefly mentioned above, weight loss could cause changes in the composition of the gut microbiota, and exercise may induce weight loss. This aspect is of particular interest given that the composition of the microbiota differs in obese and nonobese individuals. Although the nature of these changes and how they are produced remains unknown, it is worth emphasizing that commensal bacteria are able to activate hormones and neurotransmitters (epinephrine, acetylcholine, histamine, serotonin, gamma aminobutyric acid) acting on the brain, and their receptors are sensitive to the same mediators released by the host brain [4, 87, 88].
The activity of the hypothalamic pituitary adrenal (HPA) axis has important consequences in terms of reciprocal modulation between the gut and brain (the gut/brain axis). This two-way communication may induce changes in certain populations of bacteria, and the specific hormones released may subsequently modify host behavior [6]. It is well known that under physical and psychological stress, activation of the HPA axis with subsequent release of various hormones (corticotropin, cortisol, noradrenaline, adrenaline, dopamine) may play a role in dysbiosis of the intestinal microbiota. The release of corticotropin-releasing factor (CRF) alters gastric acid secretion, gastrointestinal motility, and mucus production, all of which affect intestinal resident bacteria. Similarly, elevated plasma levels of noradrenaline under stress conditions impact the intestinal microbiota and increase the virulence of enteric pathogens, such as Salmonella enterica serotype typhimurium and E. coli [6].
During intense physical training, physical stress and homeostasis alterations occur when the body exceeds 60% of the maximal oxygen consumption (VO2max) or if the duration of the exercise exceeds 90 min (even when the intensity does not exceed 40% VO2max), leading to activation of the HPA axis [89, 90]. Similarly, in the precompetition periods, athletes face high levels of psychological stress that also trigger the HPA axis with similar consequences on the microbiota profile. Hence, the intensity of exercise performed is an important factor that may alter the gut microbiota (Fig. 2) [78, 91–95].
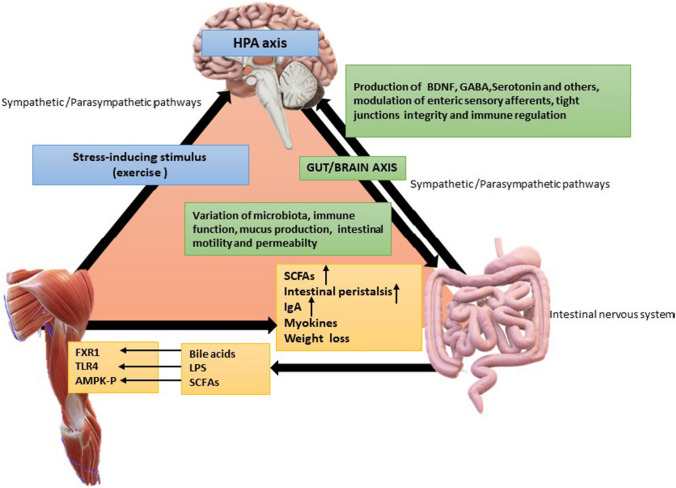
Fig. 2.
Main biomolecular interactions during regular physical exercise and training. The tight interplay between the gut microbiota and the gut/brain axis, HPA axis, or muscle/gut axis may help to explain the renowned beneficial effects of exercise on several organs and functions. Depending on the nature and intensity of physical training, the composition and activities of intestinal bacteria may vary. This process subsequently contributes to modulating immune function (by improving the sensitivity of Toll-like receptors that recognize bacterial DNA and through the production of butyrate), reducing intestinal inflammation (mediated by various myokines, such as IL-6, TNF-α, and IL-10) to lower salivary cortisol via the gut/brain axis, and improving the psychophysiological conditions of patients with inflammatory bowel disease or suffering from anxiety, stress-induced depression, obesity, mobility, musculoskeletal disorders and respiratory diseases. HPA hypothalamic pituitary adrenal axis, BDNF brain-derived neurotrophic factor, GABA γ-aminobutyric acid, SCFAs short-chain fatty acids, LPS lipopolysaccharides, FXR farnesoid X receptor, TLR4 toll-like receptor 4, AMPK AMP-activated protein kinase. Credits: Original figure by I. A. Charitos
As easily predictable, both the microbial profile and the fecal composition commonly observed in professional athletes differ significantly compared to individuals with a more sedentary lifestyle. As expected, intestinal metabolic activity is more intense in athletes, whose microbiota is mostly composed of good bacteria, such as F. prausnitzii, and is characterized by higher levels of butyrate, propionate, and acetate production [78].
Thus, regular athletic exercise training and an active lifestyle along with adequate specific dietary recommendations have indubitable advantages in athletes, and the appropriate gut microbial diversity may importantly contribute to strengthening their ability to cope with physical and mental stress [94, 95].
Interestingly, specific disciplines may have a different impact on gut microbiota. In a cross-sectional observational study, the effects of very intense exercise training, the plasma levels of creatine kinase (as a marker of extreme exercise), and the circulating levels of inflammatory cytokines (IL-6, TNF-α, IL-1) were compared between male professional rugby athletes and controls matched for physical size, age, and sex [96]. Not surprisingly, professional athletes and controls differed significantly with respect to inflammatory and metabolic markers with rugby athletes showing lower levels of proinflammatory cytokines. More importantly, rugby athletes exhibited a greater diversity of gut microorganisms (22 phyla) with respect to controls. Among these microorganisms, the Bacillota phylum and F. prausnitzii spp. were particularly represented in athletes, both of which are positively associated with favorable factors, such as longevity and health state. Akkermansia muciniphila prevents metabolic disorders and obesity, and Akkermansia spp. bacteria were more numerous in subjects with a low body mass index (BMI; < 25 kg/m2) compared with those with a high BMI (> 28 kg/m2) [96]. Similarly, in another study on rugby players and matched controls, BMI values were inversely correlated with fecal SCFAs and the microbial metabolite trimethylamine N-oxide (TMAO), suggesting that the microbiota composition of rugby athletes was characterized by an increased presence of gut bacteria with high biosynthetic activity [97].
In a 4-month prospective observational study (composed of 33 days of training and subsequent 90 days of follow-up), the differences in the gut microbiota were evaluated between endurance athletes and healthy controls. Considering their respective diet regimens, stool samples were collected from 14 marathon runners and 11 cross-country skiers and compared with 46 healthy sedentary subjects. Once more, endurance athletes showed a more diverse gut microbiota with respect to sedentary controls with a parallel enhanced production of butyrate (modulator of proper immune function in the host). These microbiota changes favor species with more efficient metabolic activities, and the corresponding increase in butyrate levels persisted over the 3 months of follow-up [98].
An increased presence of Veillonella spp. and a particular V. atypica strain was observed in stool samples from a group of marathon runners [99]. When V. atypica was grafted into the intestines of some guinea pigs, these animals demonstrated greater resistance on the wheel running test. V. atypica uses lactate as the only source of carbon for its metabolic processes, and the results from this study on marathon runners strongly suggest that the presence of this species improves the execution time of endurance exercise [99].
In addition, the intestinal microbiota composition was evaluated in professional and amateur-level cyclists. The results obtained suggest that the extent of exercising time during an average week correlates directly with the genus Prevotella, the abundance of which is accompanied by higher levels of branched chain amino acid metabolism. Compared to amateur cyclists, professional cyclists also show an increased abundance of Methanobrevibacter smithii, which is involved in the production of methane. Interestingly, when methane metabolism is upregulated, a similar upregulation occurs in other energy-signaling pathways, including carbohydrate metabolism pathways [100].
Improper, Irregular, and Exhausting Training Activity
When physical activity is too intense, all beneficial effects listed above may yield opposite results [57]. In addition, psycho-physical stress, which is a relatively common condition for competing athletes, exerts a major impact on the intestinal barrier, whose rapid cell turnover and high energy requirement make the structure particularly vulnerable [93, 94].
The risk of overtraining increases when intense workout days are not alternated with appropriate breaks to cool down or when the number of resting days in a week is not adequate for the athlete’s needs. This imbalance between the time/intensity of training and subsequent recovery is an important contributor to the onset of overtraining and associated symptoms [101]. Exhausting training can quantitatively and qualitatively change the composition of intestinal microbiota, promoting dysbiosis that favors inflammation and producing negative consequences in terms of metabolic balance. In mouse models, exhaustive exercise promotes intestinal inflammation and increases the growth of Ruminococcus gnavus, Butyrivibrio spp., Oscillospira spp., and Coprococcus spp., with a concomitant decrease in Turicibacter spp. [102]. When evaluated in a postexercise phase, immune function depression is more pronounced when the session training is continuous, prolonged for more than 90 min, and exhibits an intensity proximal to 65–75% of aerobic capacity; moreover, an inadequate diet may aggravate this status. Indeed, high endurance athletes and/or very long workout sessions are associated with an increased risk for viral and bacterial infections [103]. Neuroendocrine modifications have been regarded as potential mechanisms underlying this effect, in part after muscle microtrauma that triggers the release of cytokines and in part related to changes in the intestinal microbiota. This finding once again draws attention to the ability of intestinal bacteria to interact with several distant organs, including skeletal muscles [104]. In reciprocal regulation, the muscle-intestine axis promotes correct protein intake and participates in optimal protein deposition and muscle function, and the immune system is influenced and subsequently helps to shape microbial communities (Fig. 3) [105–107].
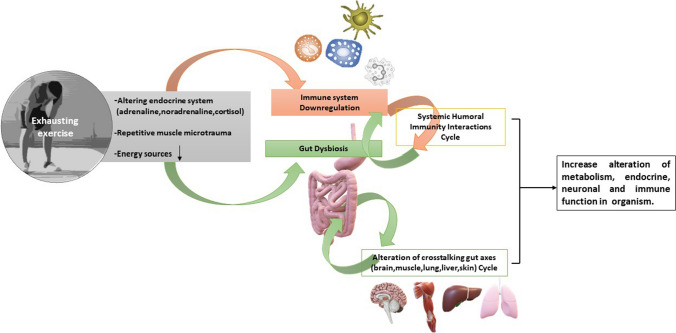
Fig. 3.
Each intense and prolonged training leads to physiological stress and transient but significant changes in immune defense, enhancing the release of stress hormones, pro- and anti-inflammatory cytokines and reactive oxygen species. Changes may affect a the activity of natural killer cells, b the number and the correct function of T and B cells, c the function of upper airway neutrophils, d the salivary concentration of IgA, and e the oxidative activities of granulocytes. MHC expression is suppressed for several hours during recovery from prolonged endurance exercise. Therefore, endocrinological alterations (such as an increase in cortisol secretion), repetitive muscle microtrauma, and a lack of energy can lead to both irregular immunomodulatory effects and intestinal dysbiosis [106, 107]. According to this hypothesis, altered function in two independently regulated pathways (the first concerning the influence of the immune system on the intestinal mucosa, the second related to the relationship between intestinal mucosa and several tissues) may contribute to creating a unifying vicious cycle responsible for both unhealthy status and poor performance. Credits: Original figure by I. A. Charitos
Competitive sport activity is undoubtedly associated with positive effects on cardiovascular conditioning, mitochondrial biogenesis, and increased sensitivity to insulin. Nevertheless, when inappropriately programmed or when the specific athlete’s needs are underestimated, intense sports activity may promote potential negative effects, such as increased oxidative stress, dehydration, immunosuppression, increased intestinal permeability or leaky gut syndrome (LGS), decreased intestinal barrier function, and increased production of inflammatory mediators. Indeed, athletes participating in high-intensity exercises suffer very often from gastrointestinal symptoms, including nausea, cramps, diarrhea or constipation, bloating, and even bleeding [95]. The severity of these clinical manifestations depends on several interconnected aspects, including the athletes’ physiologic conditions, the intensity and duration of the specific training activity and the adequate nutrition plan according to the sport disciplines. Therefore, intestinal eubiosis in professional athletes is crucial for achieving maximum athletic performance. In this respect, it is important to emphasize that dysbiosis-induced LGS may progressively exacerbate an endotoxemic condition determining susceptibility to infections and autoimmune diseases [95]. In addition, the production of SCFAs (such as butyrate) by microbiota is among the most effective methods by which the body increases its energy levels, counteracts the negative effects of inflammatory cytokines, regulates some neutrophil activities (such as the ability to migrate), improves the disposal of oxidative radicals, and regulates immunity [108]. High-intensity competitive training alters the microbiota profile in a variety of species, such as Dorea longicatena, B. vulgatus, F. prausnitzii, B. uniformis, Prevotella copri, and Eubacterium rectale, and modifies the proliferation of species producing butyrate, such as Roseburia hominis and members of the genus Subdoligranulum. These changes increase the metabolic potential of some genes with specific functions in well-trained athletes whose nutrition necessities differ from sedentary individuals. Simultaneously, energy, fiber, and macronutrient contents remain unchanged. In part, these effects may contribute to explaining how and to what extent the microbiota reacts to aerobic training in athletes participating in high-intensity competitions [109–112].
In a 32-year-old male ultramarathon runner, the effects of intense physical activity on the gut microbiota were observed during preparation and afterward in a 163-km race across the mountains. The ratio between the Bacteroidota/Bacillota phyla (now considered a reliable indicator of the microbiota composition) was relatively stable during the prerace training. However, 2 h after the conclusion of the race, an approximately 69% decrease in Bacteroides, Subdolingranulum, and Alloprevotella species with a concomitant increase in Pseudomonadota phylum, Haemophilus, Veillonella, and Streptococcus species was measured. As previously mentioned, Veillonella plays a key role in the lactic acid cycle, and the genus Haemophilus hosts various pathogenic species. Although no gastrointestinal infection or inflammation symptoms were reported in this case, either during or after the race conclusion, it is plausible that the proliferation of intestinal pathogens may contribute to the incidence of infections in athletes undergoing prolonged and intense physical exercise [113]. Indeed, the decreased activity of the immune system during the postexercise phase is well known and defined as the "open window" [114]. This condition is opposite to the activation of lymphocytes observed under physical exercise characterized by both moderate intensity/duration or an intense but short duration: only prolonged (greater than 1 h) and/or high-intensity (greater than 70% VO2 max) efforts can substantially decrease lymphocyte number and activities, thereby eliciting transient immunosuppression in the post-exercise phase [114]. Thus, in an otherwise unexplained performance deterioration in a professional athlete, the evaluation of his/her microbiota (eubiosis or dysbiosis) along with intestinal functions might provide some interesting hints to interpret the general conditions [112–114]. In this regard, the use of probiotics (Saccharomyces boulardii, Lactobacillus reuteri, and others) and prebiotics to maintain the eubiosis of the intestinal microbiota may represent an additional support for exercise performance capacity, training adaptations, and recovery from exercise [115, 116].
Conclusions
Increasing research findings confirm the notion that regular physical activity and sport in general may influence both qualitative and quantitative changes in intestinal microbial composition with overall benefits for the host in terms of immune protection and metabolic advantages. Indeed, the diversity, stability, and enrichment of the microbial members of the microbiota is one of the fundamental aspects of intestinal tract homeostasis and physiology, but is also a key player in adequate signaling not only along the brain-gut axis but also in other gut crosstalk axes (such as the lung and liver). Exercise complements and reinforces the diversity of gut microflora by stimulating the proliferation of “friendly” bacteria that can modulate mucosal immunity and improve barrier functions, produce substances that protect against gastrointestinal disorders and colon cancer (such as SCFAs), and improve the Bacteroidota/Bacillota phyla ratio, which aid in controlling weight gain (fighting obesity). Therefore, regular physical activity should be regarded as a treatment to maintain eubiosis of the microbiota (or rebalance any dysbiosis), thus resulting in an improvement in the state of health. In this regard, further and more detailed studies on the specific modifications produced by physical activity on the microbiota composition could be useful to explore new approaches for the treatment of metabolic and inflammatory diseases in which the microbiota plays a fundamental role. Conversely, irregular and exhausting training (especially that experienced by professional athletes) may contribute to dysbiosis in the intestinal microbiota and trigger negative feedback that may also affect the intestinal-mediated modulation of other organs and tissues and contribute to impaired athletic performance. To prevent or restore this dysbiosis and promote the recovery of athletes, the integration of probiotics and prebiotics has been proposed in addition to other dietary interventions.
A deeper understanding of the mechanisms by which the healthy microbiota exerts protective effects will add useful information on some—still unclear—consequences of intense physical activity and help us to comprehend how the intensity, frequency, and duration of the training, cycles of rest and sleep, proper nutrition, and stress management may influence the gut microbiota and the extent to which microbiota activity may subsequently influence athlete performance.
Declarations
Funding
This research was funded by Accademia Italiana Medici Specializzandi (AIMS). The funder had no influence on the content and conclusion of the study. Open access funding provided by Università degli Studi di Bari Aldo Moro within the CRUI-CARE Agreement.
Conflict of interest
Angelika E. Wegierska, Ioannis A. Charitos, Skender Topi, Maria A. Potenza, Monica Montagnani, and Luigi Santacroce declare that they have no conflicts of interest relevant to the content of this review.
Availability of data and material
All useful data are available in the article.
Code availability
N/a.
Author contributions
Conceptualization: AEW, LS. Methodology: MAP, LS. Formal analysis and investigation: AEW, MM. Writing—original draft preparation: IAC. Writing—review and editing: IAC, MM, LS. Funding acquisition: LS; Resources: ST. Supervision: LS. All authors read and approved the final manuscript.
Ethics approval
N/a.
Consent to participate
N/a.
Consent for publication
N/a.
Footnotes
Monica Montagnani and Luigi Santacroce are equal co-last authors.
Change history
8/25/2022
Missing Open Access funding information has been added in the Funding Note.
References
- 1.Santacroce L, Charitos IA, Topi S, Bottalico L. The Alcmaeon's School of Croton: philosophy and science. Open Access Maced J Med Sci. 2019 doi: 10.3889/oamjms.2019.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (2020) WHO guidelines on physical activity and sedentary behaviour. World Health Organization. https://apps.who.int/iris/handle/10665/336656. License: CC BY-NC-SA 3.0 IGO Accessed 1 May 2021.
- 3.Cronin O, O'Sullivan O, Barton W, Cotter PD, Molloy MG, Shanahan F. Gut microbiota: implications for sports and exercise medicine. Br J Sports Med. 2017;51(9):700–701. doi: 10.1136/bjsports-2016-097225. [DOI] [PubMed] [Google Scholar]
- 4.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLOS Biol. 2016 doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24(1):4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- 6.Matijašić M, Meštrović T, Paljetak HČ, Perić M, Barešić A, Verbanac D. Gut microbiota beyond bacteria-mycobiome, virome, archaeome, and eukaryotic parasites in IBD. Int J Mol Sci. 2020;21(8):2668. doi: 10.3390/ijms21082668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sommer F, Bäckhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013 doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007 doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santacroce L, Man A, Charitos IA, Haxhirexha K, Topi S. Current knowledge about the connection between health status and gut microbiota from birth to elderly. A narrative review. Front Biosci (Landmark Ed). 2021;26. (in press). [DOI] [PubMed]
- 10.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017 doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang B, Wang Y, Qian PY. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinform. 2016 doi: 10.1186/s12859-016-0992-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, Mele MC. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottalico L, Castellaneta F, Charitos IA. From hydrotherapy to the discovery of the gut microbiota: the historical gastrointestinal health concept. Pharmacophore. 2020;11:82–90. [Google Scholar]
- 14.Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes. 2013 doi: 10.4161/gmic.26205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charitos IA, D’Agostino D, Topi S, Bottalico L. 40 years of Helicobacter pylori: a revolution in biomedical thought. Gastroenterol Insights. 2021;12(2):111–135. doi: 10.3390/gastroent12020011. [DOI] [Google Scholar]
- 16.Sung J, Kim N, Kim J, Jo HJ, Park JH, Nam RH, Seok YJ, Kim YR, Lee DH, Jung HC. Comparison of gastric microbiota between gastric juice and mucosa by next generation sequencing method. J Cancer Prev. 2016;21(1):60–65. doi: 10.15430/JCP.2016.21.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nardone G, Compare D. The human gastric microbiota: is it time to rethink the pathogenesis of stomach diseases? United Eur Gastroenterol J. 2015;3(3):255–260. doi: 10.1177/2050640614566846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Browne HP, Neville BA, Forster SC, Lawley TD. Transmission of the gut microbiota: spreading of health. Nat Rev Microbiol. 2017;15(9):531–543. doi: 10.1038/nrmicro.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meštrović T, Matijašić M, Perić M, Čipčić Paljetak H, Barešić A, Verbanac D. The role of gut, vaginal, and urinary microbiome in urinary tract infections: from bench to bedside. Diagnostics (Basel) 2020;11(1):7. doi: 10.3390/diagnostics11010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mändar R, Mikelsaar M. Transmission of mother's microflora to the newborn at birth. Biol Neonate. 1996;69(1):30–35. doi: 10.1159/000244275. [DOI] [PubMed] [Google Scholar]
- 21.Walker RW, Clemente JC, Peter I, Loos RJF. The prenatal gut microbiome: are we colonized with bacteria in utero? Pediatr Obes. 2017;12(Suppl 1):3–17. doi: 10.1111/ijpo.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;22(6):23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanabe H, Sakurai K, Nakanishi Y, Kato T, Kawasaki Y, Nakano T, Yamaide F, Taguchi-Atarashi N, Shiko Y, Takashima I, Watanabe M, Ochiai S, Ohno H, Fukuoka H, Shimojo N, Mori C. Association of the maternal gut microbiota/metabolome with cord blood CCL17. Nutrients. 2021;13(8):2837. doi: 10.3390/nu13082837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holt PG, Strickland DH. Soothing signals: transplacental transmission of resistance to asthma and allergy. J Exp Med. 2009;206(13):2861–2864. doi: 10.1084/jem.20092469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faa G, Gerosa C, Fanni D, Nemolato S, van Eyken P, Fanos V. Factors influencing the development of a personal tailored microbiota in the neonate, with particular emphasis on antibiotic therapy. J Matern Fetal Neonatal Med. 2013 doi: 10.3109/14767058.2013.829700. [DOI] [PubMed] [Google Scholar]
- 26.de Meer G, Janssen NAH, Brunekreef B. Early childhood environment related to microbial exposure and the occurrence of atopic disease at school age. Allergy. 2005;60(5):619–625. doi: 10.1111/j.1398-9995.2005.00746.x. [DOI] [PubMed] [Google Scholar]
- 27.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Björkstén B, Engstrand L, Andersson AF. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014 doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 28.Illi S, von Mutius E, Lau S, Nickel R, Niggemann B, Sommerfeld C, Wahn U, Multicenter Allergy Study Group The pattern of atopic sensitization is associated with the development of asthma in childhood. J Allergy Clin Immunol. 2001 doi: 10.1067/mai.2001.118786. [DOI] [PubMed] [Google Scholar]
- 29.Schaub B, Liu J, Höppler S, Schleich I, Huehn J, Olek S, Wieczorek G, Illi S, von Mutius E. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol. 2009 doi: 10.1016/j.jaci.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 30.Ege MJ, Bieli C, Frei R, van Strien RT, Riedler J, Ublagger E, Schram-Bijkerk D, Brunekreef B, van Hage M, Scheynius A, Pershagen G, Benz MR, Lauener R, von Mutius E, Braun-Fahrländer C, Parsifal Study team Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol. 2006 doi: 10.1016/j.jaci.2005.12.1307. [DOI] [PubMed] [Google Scholar]
- 31.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006 doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 32.Neu J, Rushing J. Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin Perinatol. 2011;38(2):321–331. doi: 10.1016/j.clp.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012 doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arboleya S, Watkins C, Stanton C, Ross RP. Gut bifidobacteria populations in human health and aging. Front Microbiol. 2016;19(7):1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fouhy F, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. Composition of the early intestinal microbiota: knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microbes. 2012;3(3):203–220. doi: 10.4161/gmic.20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;2(26):26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawrelak JA, Myers SP. The causes of intestinal dysbiosis: a review. Altern Med Rev. 2004;9(2):180–197. [PubMed] [Google Scholar]
- 38.Bull MJ, Plummer NT. Part 1: the human gut microbiome in health and disease. Integr Med (Encinitas) 2014;13(6):17–22. [PMC free article] [PubMed] [Google Scholar]
- 39.Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. A new genomic blueprint of the human gut microbiota. Nature. 2019;568(7753):499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut. 2004;53(1):1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dudek-Wicher RK, Junka A, Bartoszewicz M. The influence of antibiotics and dietary components on gut microbiota. Prz Gastroenterol. 2018;13(2):85–92. doi: 10.5114/pg.2018.76005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol. 2017;17(10):635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redondo-Useros N, Nova E, González-Zancada N, Díaz LE, Gómez-Martínez S, Marcos A. Microbiota and lifestyle: a special focus on diet. Nutrients. 2020;12(6):1776. doi: 10.3390/nu12061776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Artis D, Wang ML, Keilbaugh SA, et al. RELMbeta/FIZZ2 is a goblet cell-specific immuneeffector molecule in the gastrointestinal tract. Proc Natl Acad Sci USA. 2004 doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015 doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;16(7):e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hooper LV. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol. 2009 doi: 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]
- 50.Cockburn DW, Koropatkin NM. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 51.Carvalho FA, Aitken JD, Vijay-Kumar M, Gewirtz AT. Toll-like receptor-gut microbiota interactions: perturb at your own risk! Annu Rev Physiol. 2012 doi: 10.1146/annurev-physiol-020911-153330. [DOI] [PubMed] [Google Scholar]
- 52.Cullen TW, Schofield WB, Barry NA, et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015 doi: 10.1126/science.126058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ostaff MJ, Stange EF, Wehkamp J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol Med. 2013;5(10):1465–1483. doi: 10.1002/emmm.201201773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutzeit C, Magri G, Cerutti A. Intestinal IgA production and its role in host-microbe interaction. Immunol Rev. 2014;260(1):76–85. doi: 10.1111/imr.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polimeno L, Barone M, Mosca A, Viggiani MT, Joukar F, Mansour-Ghanaei F, Mavaddati S, Daniele A, Debellis L, Bilancia M, Santacroce L, Di Leo A. Soy metabolism by gut microbiota from patients with precancerous intestinal lesions. Microorganisms. 2020;8(4):469. doi: 10.3390/microorganisms8040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalla R, Ventham NT, Kennedy NA, et al. MicroRNAs: new players in IBD [published correction appears in Gut. 2015 Jun;64(6):1008] Gut. 2015 doi: 10.1136/gutjnl-2014-307891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012 doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, Comstock LE, Gandhi R, Weiner HL. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol. 2016;12(6):1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santacroce L, Charitos IA, Ballini A, Inchingolo F, Luperto P, De Nitto E, Topi S. The human respiratory system and its microbiome at a glimpse. Biology (Basel). 2020;9(10):318. doi: 10.3390/biology9100318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alam A, Neish A. Role of gut microbiota in intestinal wound healing and barrier function. Tissue Barriers. 2018;6(3):1539595. doi: 10.1080/21688370.2018.1539595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wegienka G, Havstad S, Zoratti EM, Woodcroft KJ, Bobbitt KR, Ownby DR, Johnson CC. Regulatory T cells in prenatal blood samples: variability with pet exposure and sensitization. J Reprod Immunol. 2009 doi: 10.1016/j.jri.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santacroce L, Sardaro N, Topi S, Pettini F, Bottalico L, Cantore S, Cascella G, Del Prete R, Dipalma G, Inchingolo F. The pivotal role of oral microbiota in health and disease. J Biol Regul Homeost Agents. 2020 doi: 10.23812/20-127-L-45. [DOI] [PubMed] [Google Scholar]
- 64.Kho ZY, Lal SK. The human gut microbiome—a potential controller of wellness and disease. Front Microbiol. 2018;14(9):1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar RD. Role of the normal gut microbiota. World J Gastroenterol. 2015 doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57(1):1–24. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mailing LJ, Allen JM, Buford TW, Fields CJ, Woods JA. Exercise and the gut microbiome: a review of the evidence, potential mechanisms, and implications for human health. Exerc Sport Sci Rev. 2019 doi: 10.1249/JES.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 68.Clauss M, Gérard P, Mosca A, Leclerc M. Interplay between exercise and gut microbiome in the context of human health and performance. Front Nutr. 2021;10(8):637010. doi: 10.3389/fnut.2021.637010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mohr AE, Jäger R, Carpenter KC, Kerksick CM, Purpura M, Townsend JR, West NP, Black K, Gleeson M, Pyne DB, Wells SD, Arent SM, Kreider RB, Campbell BI, Bannock L, Scheiman J, Wissent CJ, Pane M, Kalman DS, Pugh JN, Ortega-Santos CP, Ter Haar JA, Arciero PJ, Antonio J. The athletic gut microbiota. J Int Soc Sports Nutr. 2020 doi: 10.1186/s12970-020-00353-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manders RJ, Van Dijk JW, van Loon LJ. Low-intensity exercise reduces the prevalence of hyperglycemia in type 2 diabetes. Med Sci Sports Exerc. 2010;42(2):219–225. doi: 10.1249/MSS.0b013e3181b3b16d. [DOI] [PubMed] [Google Scholar]
- 71.Monda V, Villano I, Messina A, Valenzano A, Esposito T, Moscatelli F, Viggiano A, Cibelli G, Chieffi S, Monda M, Messina G. Exercise modifies the gut microbiota with positive health effects. Oxid Med Cell Longev. 2017 doi: 10.1155/2017/3831972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matsumoto M, Inoue R, Tsukahara T, Ushida K, Chiji H, Matsubara N, Hara H. Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci Biotechnol Biochem. 2008 doi: 10.1271/bbb.70474. [DOI] [PubMed] [Google Scholar]
- 73.Pant K, Peixoto E, Richard S, Gradilone SA. Role of histone deacetylases in carcinogenesis: potential role in cholangiocarcinoma. Cells. 2020;9(3):780. doi: 10.3390/cells9030780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Campbell SC. Faecalibacterium prausnitzii abundance in mouse and human gut can predict metabolism of oat avenanthramides. J Nutr. 2021 doi: 10.1093/jn/nxab086. [DOI] [PubMed] [Google Scholar]
- 75.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mika A, Fleshner M. Early-life exercise may promote lasting brain and metabolic health through gut bacterial metabolites. Immunol Cell Biol. 2016;94(2):151–157. doi: 10.1038/icb.2015.113. [DOI] [PubMed] [Google Scholar]
- 77.Clarke SF, Murphy EF, Nilaweera K, Ross PR, Shanahan F, O'Toole PW, Cotter PD. The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes. 2012 doi: 10.4161/gmic.20168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cerdá B, Pérez M, Pérez-Santiago JD, Tornero-Aguilera JF, González-Soltero R, Larrosa M. Gut microbiota modification: another piece in the puzzle of the benefits of physical exercise in health? Front Physiol. 2016 doi: 10.3389/fphys.2016.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grigor'eva IN. Gallstone disease, obesity and the firmicutes/bacteroidetes ratio as a possible biomarker of gut dysbiosis. J Pers Med. 2020 doi: 10.3390/jpm11010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ali AH, Carey EJ, Lindor KD. Recent advances in the development of farnesoid X receptor agonists. Ann Transl Med. 2015 doi: 10.3978/j.issn.2305-5839.2014.12.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lahiri S, Kim H, Garcia-Perez I, Reza MM, Martin KA, Kundu P, Cox LM, Selkrig J, Posma JM, Zhang H, Padmanabhan P, Moret C, Gulyás B, Blaser MJ, Auwerx J, Holmes E, Nicholson J, Wahli W, Pettersson S. The gut microbiota influences skeletal muscle mass and function in mice. Sci Transl Med. 2019 doi: 10.1126/scitranslmed.aan5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lustgarten MS. The role of the gut microbiome on skeletal muscle mass and physical function: 2019 update. Front Physiol. 2019 doi: 10.3389/fphys.2019.01435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ortiz-Alvarez L, Xu H, Martinez-Tellez B. Influence of exercise on the human gut microbiota of healthy adults: a systematic review. Clin Transl Gastroenterol. 2020;11(2):e00126. doi: 10.14309/ctg.0000000000000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schnyder S, Handschin C. Skeletal muscle as an endocrine organ: PGC-1α, myokines and exercise. Bone. 2015;80:115–125. doi: 10.1016/j.bone.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gizard F, Fernandez A, De Vadder F. Interactions between gut microbiota and skeletal muscle. Nutr Metab Insights. 2020 doi: 10.1177/1178638820980490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yiu JH, Dorweiler B, Woo CW. Interaction between gut microbiota and toll-like receptor: from immunity to metabolism. J Mol Med (Berl) 2017 doi: 10.1007/s00109-016-1474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oliveira AG, Carvalho BM, Tobar N, Ropelle ER, Pauli JR, Bagarolli RA, Guadagnini D, Carvalheira JB, Saad MJ. Physical exercise reduces circulating lipopolysaccharide and TLR4 activation and improves insulin signaling in tissues of DIO rats. Diabetes. 2011 doi: 10.2337/db09-1907. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Nieman DC, Wentz LM. The compelling link between physical activity and the body's defense system. J Sport Health Sci. 2019 doi: 10.1016/j.jshs.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Radak Z, Zhao Z, Koltai E, Ohno H, Atalay M. Oxygen consumption and usage during physical exercise: the balance between oxidative stress and ROS-dependent adaptive signaling. Antioxid Redox Signal. 2013;18(10):1208–1246. doi: 10.1089/ars.2011.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Helgerud J, Høydal K, Wang E, Karlsen T, Berg P, Bjerkaas M, Simonsen T, Helgesen C, Hjorth N, Bach R, Hoff J. Aerobic high-intensity intervals improve VO2max more than moderate training. Med Sci Sports Exerc. 2007;39(4):665–671. doi: 10.1249/mss.0b013e3180304570. [DOI] [PubMed] [Google Scholar]
- 91.Dalton A, Mermier C, Zuhl M. Exercise influence on the microbiome-gut-brain axis. Gut Microbes. 2019;10(5):555–568. doi: 10.1080/19490976.2018.1562268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leal LG, Lopes MA, Batista ML., Jr Physical exercise-induced myokines and muscle-adipose tissue crosstalk: a review of current knowledge and the implications for health and metabolic diseases. Front Physiol. 2018 doi: 10.3389/fphys.2018.01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016 doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Booth FW, Roberts CK, Laye MJ. Lack of exercise is a major cause of chronic diseases. Compr Physiol. 2012 doi: 10.1002/cphy.c110025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut. 2019;68(8):1516–1526. doi: 10.1136/gutjnl-2019-318427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Clarke SF, Murphy EF, O'Sullivan O, Lucey AJ, Humphreys M, Hogan A, Hayes P, O'Reilly M, Jeffery IB, Wood-Martin R, Kerins DM, Quigley E, Ross RP, O'Toole PW, Molloy MG, Falvey E, Shanahan F, Cotter PD. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63(12):1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 97.Barton W, Penney NC, Cronin O, Garcia-Perez I, Molloy MG, Holmes E, Shanahan F, Cotter PD, O'Sullivan O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut. 2018;67(4):625–633. doi: 10.1136/gutjnl-2016-313627. [DOI] [PubMed] [Google Scholar]
- 98.Mohr AE, Jäger R, Carpenter KC, Kerksick CM, Purpura M, Townsend JR, West NP, Black K, Gleeson M, Pyne DB, Wells SD, Arent SM, Kreider RB, Campbell BI, Bannock L, Scheiman J, Wissent CJ, Pane M, Kalman DS, Pugh JN, Ortega-Santos CP, Ter Haar JA, Arciero PJ, Antonio J. The athletic gut microbiota. J Int Soc Sports Nutr. 2020;17(1):24. doi: 10.1186/s12970-020-00353-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, Wibowo MC, Wurth RC, Punthambaker S, Tierney BT, Yang Z, Hattab MW, Avila-Pacheco J, Clish CB, Lessard S, Church GM, Kostic AD. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med. 2019;25(7):1104–1109. doi: 10.1038/s41591-019-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Petersen LM, Bautista EJ, Nguyen H, Hanson BM, Chen L, Lek SH, et al. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome. 2017;5(1):98. doi: 10.1186/s40168-017-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mujika I. The influence of training characteristics and tapering on the adaptation in highly trained individuals: a review. Int J Sports Med. 1998;19(7):439–446. doi: 10.1055/s-2007-971942. [DOI] [PubMed] [Google Scholar]
- 102.Clark A, Mach N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: a systematic review for athletes. J Int Soc Sports Nutr. 2016;24(13):43. doi: 10.1186/s12970-016-0155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gleeson M, Bishop NC, Stensel DJ, Lindley MR, Mastana SS, Nimmo MA. The anti-inflammatory effects of exercise: mechanisms and implications for the prevention and treatment of disease. Nat Rev Immunol. 2011;11(9):607–615. doi: 10.1038/nri3041. [DOI] [PubMed] [Google Scholar]
- 104.Yoo JY, Groer M, Dutra SVO, Sarkar A, McSkimming DI. Gut microbiota and immune system interactions. Microorganisms. 2020;8(10):1587. doi: 10.3390/microorganisms8101587.Erratum.In:Microorganisms.2020Dec21;8(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ticinesi A, Lauretani F, Tana C, Nouvenne A, Ridolo E, Meschi T. Exercise and immune system as modulators of intestinal microbiome: implications for the gut-muscle axis hypothesis. Exerc Immunol Rev. 2019;25:84–95. [PubMed] [Google Scholar]
- 106.Sellami M, Gasmi M, Denham J, Hayes LD, Stratton D, Padulo J, Bragazzi N. Effects of acute and chronic exercise on immunological parameters in the elderly aged: can physical activity counteract the effects of aging? Front Immunol. 2018;10(9):2187. doi: 10.3389/fimmu.2018.02187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chiang LM, Chen YJ, Chiang J, Lai LY, Chen YY, Liao HF. Modulation of dendritic cells by endurance training. Int J Sports Med. 2007;28(9):798–803. doi: 10.1055/s-2007-964914. [DOI] [PubMed] [Google Scholar]
- 108.Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3(10):858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morita, Yokoyama, Imai, Takeda, Ota, Kawai, Okazaki. Aerobic exercise training with brisk walking increases intestinal bacteroides in healthy elderly women. Nutrients. 2019;11(4):868. [DOI] [PMC free article] [PubMed]
- 110.Taniguchi H, Tanisawa K, Sun X, Kubo T, Hoshino Y, Hosokawa M, Higuchi M. Effects of short-term endurance exercise on gut microbiota in elderlymen. Physiol Rep. 2018;6(23):e13935. doi: 10.14814/phy2.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Keohane DM, Woods T, O'Connor P, Underwood S, Cronin O, Whiston R, O'Sullivan O, Cotter P, Shanahan F, Molloy MGM. Four men in a boat: ultra-endurance exercise alters the gut microbiome. J Sci Med Sport. 2019;22(9):1059–1064. doi: 10.1016/j.jsams.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 112.Nieman DC, Henson DA, Austin MD, Sha W. Upper respiratory tract infection is reduced in physically fit and active adults. Br J Sports Med. 2011;45(12):987–992. doi: 10.1136/bjsm.2010.077875. [DOI] [PubMed] [Google Scholar]
- 113.Grosicki GJ, Durk RP, Bagley JR. Rapid gut microbiome changes in a world-class ultramarathon runner. Physiol Rep. 2019;7(24):e14313. doi: 10.14814/phy2.14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kakanis MW, Peake J, Brenu EW, Simmonds M, Gray B, Hooper SL, Marshall-Gradisnik SM. The open window of susceptibility to infection after acute exercise in healthy young male elite athletes. Exerc Immunol Rev. 2010;16:119–137. [PubMed] [Google Scholar]
- 115.Wosinska L, Cotter PD, O'Sullivan O, Guinane C. The potential impact of probiotics on the gut microbiome of athletes. Nutrients. 2019;11(10):2270. doi: 10.3390/nu11102270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Santacroce L, Charitos IA, Bottalico L. A successful history: probiotics and their potential as antimicrobials. Expert Rev Anti Infect Ther. 2019 doi: 10.1080/14787210.2019.1645597. [DOI] [PubMed] [Google Scholar]