Abstract
Riboswitches are an outstanding example of genetic regulation mediated by RNA conformational switching. In these non-coding RNA elements, the occupancy status of a ligand-binding domain governs the mRNA’s decision to form one of two mutually exclusive structures in the downstream expression platform. Temporal constraints upon the function of many riboswitches, requiring folding of complex architectures and conformational switching in a limited co-transcriptional timeframe, make them ideal model systems for studying these processes. In this review, we focus on the mechanism of ligand-directed conformational changes in one of the most widely distributed riboswitches in bacteria: the cobalamin family. We describe the architectural features of cobalamin riboswitches whose structures have been determined by x-ray crystallography, which suggest a direct physical role of cobalamin in effecting the regulatory switch. Next, we discuss a series of experimental approaches applied to several model cobalamin riboswitches that interrogate these structural models. As folding is central to riboswitch function, we consider the differences in folding landscapes experienced by RNAs that are produced in vitro and those that are allowed to fold co-transcriptionally. Finally, we highlight a set of studies that reveal the difficulties of studying cobalamin riboswitches outside the context of transcription and that co-transcriptional approaches are essential for developing a more accurate picture of their structure-function relationships in these switches. This understanding will be essential for future advancements in the use of small-molecule guided RNA switches in a range of applications such as biosensors, RNA imaging tools, and nucleic acid-based therapies.
Keywords: riboswitch, gene regulation, cobalamin, co-transcriptional folding, RNA structure
Introduction
Discovered two decades ago, riboswitches are non-protein coding RNA elements primarily found in the 5’ leader of bacterial mRNAs that directly bind small molecule metabolites or ions to regulate expression of the message.1–4 The general architecture of a riboswitch consists of two functional domains: the ligand-binding aptamer domain and the downstream regulatory expression platform. The aptamer domain folds into an intricate structure often requiring multi-helix packing via long-range tertiary interactions5, 6, enabling the aptamer to host a ligand binding site capable of binding its effector molecule with high affinity and specificity.7 Occupancy of the aptamer is communicated to the downstream expression platform, which informs a conformational switch that dictates the expression of the message.8, 9 Central to riboswitches’ ability to regulate gene expression is the ability to direct their secondary structural conformational switch in an effector-dependent fashion.
Another feature of riboswitches that make them outstanding models of RNA folding processes, including conformational changes, is that they are generally temporally constrained.10, 11 For example, riboswitches regulating transcription via ligand-dependent formation of an intrinsic terminator—a hairpin followed by a polyuridine tract that induces RNAP to disengage from RNA synthesis12, 13—must be able to efficiently fold, productively bind the effector ligand, and promote alternative secondary structure formation within a limited temporal window before the polymerase escapes beyond the polyuridine tract of the terminator.8 Regulatory processes in which a decision must be made prior to the system being able to come to equilibrium are referred to as being under kinetic control (a good discussion of thermodynamic versus kinetic control of regulatory processes is given by Coppins et al.14). Riboswitches that control expression at the translational level are also subject to a limited temporal window of action because of processes such as the coupling of the pioneer round of translation to transcription15, 16 or by transcriptional termination by Rho.17–19 The kinetic control of riboswitches places these RNAs under significant pressure to fold and switch rapidly and with high fidelity to minimize inappropriate expression of the mRNA. Thus, most riboswitches evolved under significant pressure to evade various forms of misfolding, making them the “superfolders” of the RNA world.
Currently, over 55 different riboswitches have been validated20, indicating that they are a highly effective means of regulating gene expression in bacteria. While in aggregate these regulatory elements are found in all major groups of bacteria, only a few classes of riboswitches stand out as being broadly distributed across bacteria: thiamine pyrophosphate (TPP), cobalamin (also known as vitamin B12), and S-adenosylmethionine (SAM).21, 22 This indicates that these riboswitches are competitive or even superior to their protein counterparts such as the MetJ23 (SAM) and CarH24, 25 (B12) repressors in their ability to regulate gene expression. The observation that these riboswitches are extensively used to regulate gene expression across species representing a broad spectrum of environmental, physiological, and genetic contexts further reinforces that they are robust solutions to the challenge of maintaining cellular homeostasis of these essential metabolites.21, 22 Over the past two decades, these riboswitches have been extensively characterized using a spectrum of experimental approaches that span in vitro, in vivo, and in silico. Thus, each of these riboswitches serve as model systems for investigating RNA-mediated regulatory processes that operate through conformational switching. In this review, we focus on how cobalamin riboswitches—one of the few riboswitches for which there are crystal structures of the functional, ligand-bound regulatory sequence—regulate gene expression through the binding of cobalamin to drive a conformational change that dictates the mRNA’s expression fate.
Cobalamins are a large, complex metabolite essential for a set of biochemical reactions
Vitamin B12, the effector of cobalamin riboswitches, is the largest of the small molecule metabolites in biology and it plays essential roles in bacterial metabolism.26–29 All forms of cobalamin contain a porphyrin ring called a corrin that coordinates a central cobalt atom and is flanked by a set of methyl groups and propionamide groups (Fig. 1A). Unlike other porphyrin rings, the corrin ring contains two sites of unsaturation and thus has a pronounced pucker to its structure, as first revealed in the pioneering crystallographic work of Dorothy Hodgkin.30 One of the corrin flanking groups is a long flexible linker capped by 5,6-dimethylbenzimidzole, whose N3 nitrogen interacts with the fifth coordination site of cobalt, known as the alpha axial position (cyan, Fig. 1A). This nucleobase is not universal in biology, as other benzimidiazoles, purines, and imidazoles are also used in cobalamins.31 The sixth coordination site of cobalamins, known as the beta axial position (yellow, Fig. 1A), is the functional site of this coenzyme and bears different functional moieties depending on the reaction that the coenzyme helps to catalyze (Fig. 1B).26–29 5’-deoxyadenosylcobalamin (AdoCbl) contains an adenosyl group whose C5’ carbon forms a coordinate covalent bond with cobalt. This bond is extremely weak and easily homolytically dissociates to enable the adenosyl moiety to participate in free radical catalyzed carbon-carbon bond rearrangements that are particularly important in anaerobic metabolism. A second form is methylcobalamin (MeCbl) which is most notably used in the synthesis of methionine through donation of methyl group to homocysteine. Finally, the other two forms most relevant to this review are hydroxocobalamin (HyCbl) that contains a hydroxy group and is the form typically available in the diet, and cyanocobalamin (CNCbl), a synthetic compound whose cobalt-carbon bond is stable but readily metabolized to HyCbl and further converted into the biologically active forms AdoCbl and MeCbl. Importantly for cobalamin riboswitches, MeCbl, CNCbl, and HyCbl all have small beta axial moieties while AdoCbl has a sterically bulky beta axial moiety.
Figure 1.
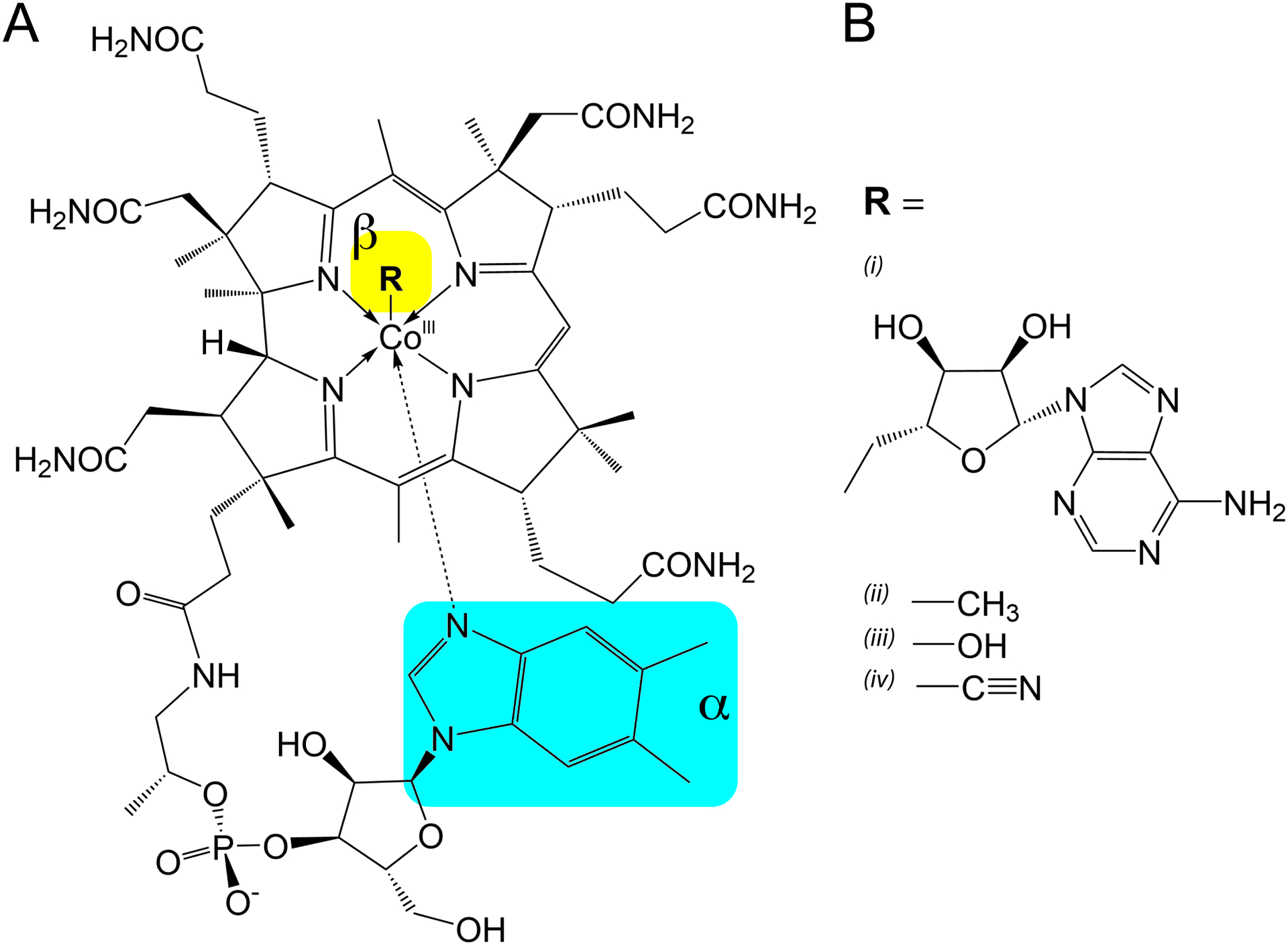
Structure of cobalamin with variable beta-axial moieties (R) which determine its form. (A) The common cobalamin corrin ring shared between all forms shown here, with the 5,6-dimethylbenzimidzole coordinated to the alpha-axial face. Arrows are used to depict coordinate bonds with the cobalt (III) ion as opposed to covalent bonding. The alpha- and beta-axial positions are highlighted in cyan and yellow, respectively. (B) Variable functional moieties that can be present in the beta-axial position of the corrin ring. (i) 5’-deoxyadenosylcobalamin (ii) methylcobalamin (iii) hydroxocobalamin (iv) cyanocobalamin.
Discovery that several 5’ leader sequences of bacterial mRNAs directly bind cobalamin
Cobalamin riboswitches were first characterized in the landmark study by the Breaker group describing the discovery of riboswitches.2 While the ability of RNA to bind small molecules was previously established through in vitro selection experiments that generated aptamers32 as well as binding of an exogenous guanosine nucleotide to group I introns33, this study laid the groundwork for the discovery of widespread use of small molecule-RNA interactions to drive regulatory conformation switches in biology. The ability of the 5’ leader regions of the mRNAs for Escherichia coli (Eco) btuB, a B12 transport gene, and Salmonella typhimurium cob, a B12 biosynthesis operon, to directly bind AdoCbl was demonstrated using “in-line” chemical probing2, an approach that exploits the ability of RNA to self-cleave in the presence of magnesium.34 Transcripts of the two leaders appeared to change their cleavage pattern in the presence AdoCbl, which was interpreted as a binding of AdoCbl and associated changes in RNA structure.2 Direct binding of AdoCbl to the btuB leader was confirmed using equilibrium dialysis, a classic technique to measure binding affinities of small molecule-protein interactions.35 Most convincingly, this study showed that the RNA was highly selective for AdoCbl over cobalamins including MeCbl and CNCbl but accepting of others such as 5’-deoxypurinylcobalamin (PurCbl) and 3-deaza-AdoCbl.2 The hypothesis that AdoCbl directly binds mRNA explained previous observations of AdoCbl repressed expression of btuB36 as well as a set of other genes related to Cbl biosynthesis and transport in S. typhimurium.37, 38
It is important to note, however, that the proposed model of the Eco btuB leader secondary structure in this study poorly correlates with later models and a compelling model for cobalamin-dependent regulatory switching by the RNA could not be provided. The inaccurate secondary structure could have been due, in part, to difficulties in homogeneously refolding the RNA, giving rise to in-line probing patterns that do not enable accurate secondary structure modeling. Instead, the subsequent use of comparative sequence analysis of this widely distributed element (see below) revealed patterns of base covariation that yielded significantly improved secondary structural models.2, 39, 40 Further, these analyses identified regions that might be involved in gene regulation via their proximity to transcriptional terminators or ribosome binding sites.
Cobalamin riboswitches are broadly distributed throughout bacteria and regulate a spectrum of B12 associated processes
The cobalamin riboswitch is the second most widely distributed riboswitch across bacteria, exceeded only by the TPP riboswitch.21, 22 A recent analysis of riboswitch distribution across bacteria found cobalamin riboswitches (called AdoCbl and AqCbl) in all major bacterial groups with the exception of Epsilonproteobacteria, Caldiserica, Dictyoglomi, and Elusimicrobia (4 out of 36 groups).22 In particular, obligate pathogenic bacteria that do not appear to have B12-dependent enzymes lack Cbl riboswitches.40 Comparative genomic analysis across over 100 bacterial species revealed that these riboswitches regulate a spectrum of cobalamin biosynthetic and transport genes, as well as proteins associated with cobalt metabolism (transporters, chelatases, and reductases) and salvage of corrinoids. Cobalamin riboswitches have been also observed in tandem regulatory systems that regulate methionine synthetase41 and as a small, trans-acting RNA that regulates ethanolamine utilization.42, 43
Bioinformatic analysis of the expression platforms of cobalamin riboswitches also enabled prediction of whether these riboswitches act at the level of transcription or translation.37, 39, 44 It was predicted that regulation mainly occurs at the transcriptional level for Gram-positive bacteria such as the Firmicutes (Bacilli and Clostridia), which generally contain rho-independent (intrinsic) terminator hairpins.39 In contrast, regulation was predicted to occur mainly at the translational level for Gram-negative bacteria such as the Proteobacteria and Cyanobacteria, which mostly has candidate regulatory hairpins in which the ribosome binding site (RBS) is embedded. Out of the two, cobalamin riboswitches more often act at the level of translation.21 Whether regulation of mRNA expression occurs at the transcriptional or translational levels, these analyses suggest that a conserved hairpin structure called P13 (see below) plays a central role in modulating a conformational change that manipulates the RBS or intrinsic terminator. As stated above, there is also an example of an AdoCbl riboswitch that is part of a small RNA that acts via an antisense mechanism to control multiple target messages in Enterococcus faecalis.42
Cobalamin riboswitches have a complex multi-domain architecture
The secondary structure of cobalamin riboswitches is divided into three distinct regions: the aptamer domain core, aptamer domain peripheral elements (subdomains 1 and 2), and the regulatory expression platform (Fig. 2).39, 45, 46 The core of the aptamer domain consists of a central four-way junction flanked by paired regions P3 – P6. Within both the helices and the joining regions of the junction (J3/4, J4/5, J5/6, and J6/3) are the majority of nucleotides with >97% conservation in cobalamin riboswitches, indicating that this region is critical for ligand binding or regulatory function.21, 22, 34, 39, 46 Included in this core region are the P4/L4 stem-loop, P6, and the internal loop IL6/7, which also contain conserved nucleotides.
Figure 2.
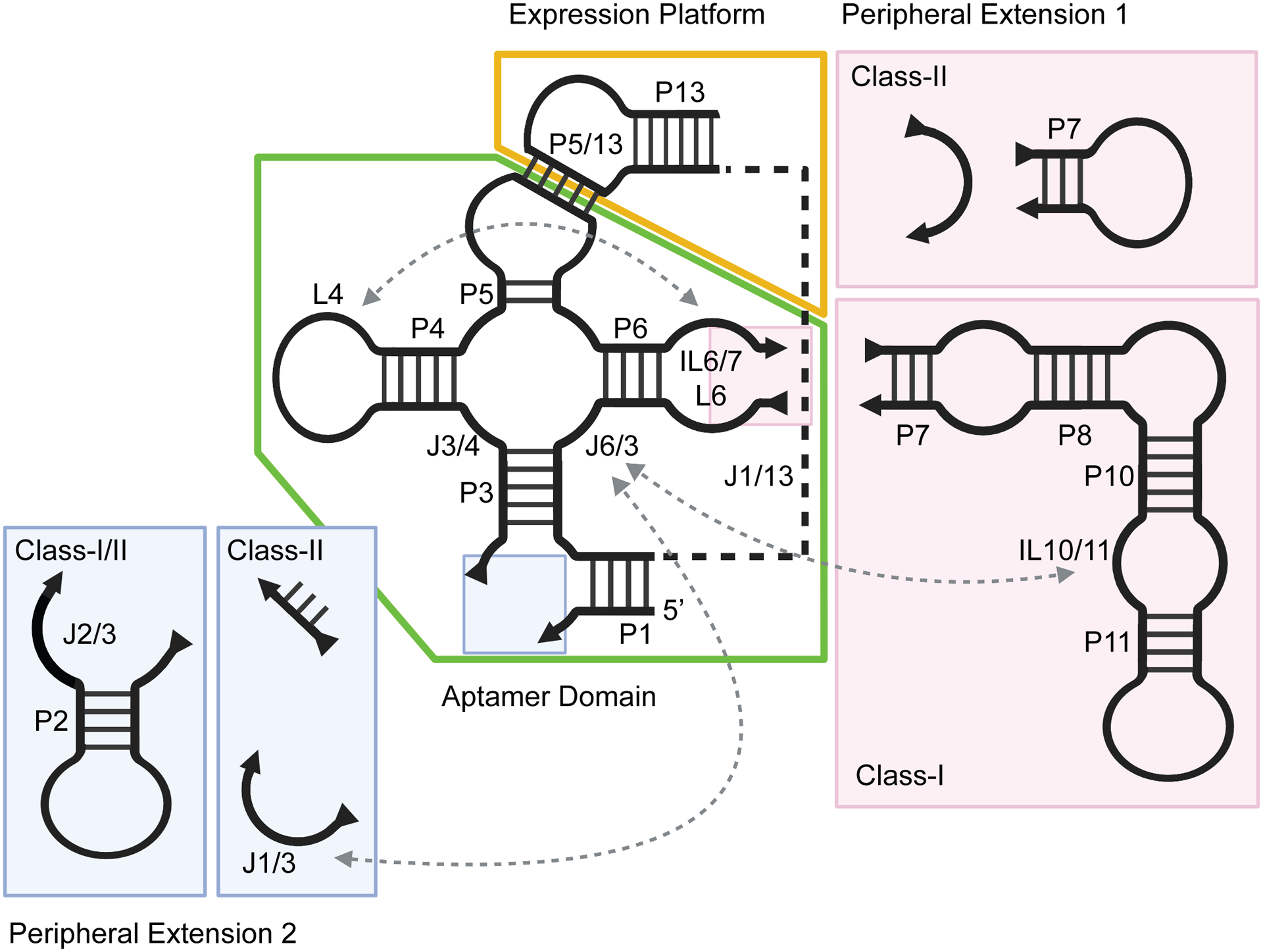
Secondary structure schematic of cobalamin riboswitches, delineating structural differences between class-I and class-II. The conserved aptamer domain is boxed in green and the expression platform in yellow. Peripheral extensions which vary between classes are colored in pink (subdomain 1) and blue (subdomain 2). Long-range tertiary contacts are indicated by dashed arrows. Created with Biorender.com.
Flanking the conserved aptamer core are two peripheral domains. The first, the P7-P12 peripheral extension, projects from the conserved IL6/7 of the core. This domain is quite variable in secondary structure across phylogeny, but invariably has a small internal loop motif, IL10/11, that contains a set of purine residues that are the only highly conserved nucleotides in this domain.39, 46 The second peripheral extension lies between P1 and P3. Again, this element is variable and is observed as a simple two-way junction (J1/3) or a more complex three-way junction motif (P2 and J2/3).
Finally, the expression platform of all cobalamin riboswitches comprises a joining region, J1/13, that separates the two domains and at least one stem-loop structure, P13/L13.39, 46 The joining region does not contain any nucleotide conservation patterns or secondary structure, indicating that this is a flexible linker spanning the two functional domains. Early phylogenetic analysis of secondary structure suggested that L5 of the aptamer domain interacts with L13 of the expression platform through a base pairing motif called a “kissing loop”.40, 46 Beyond the conserved P13/L13, expression platforms of various cobalamin riboswitches contain hairpins that act as RBS/anti-RBS sequester elements or intrinsic terminators that inform the expression machinery.
Currently, there are four crystal structures of cobalamin riboswitches that yield clear insights into the three-dimensional architecture of the RNA, ligand recognition, and the relationship between the aptamer domain and expression platform. Two of these structures are representative of “class-I” cobalamin riboswitches: the aptamer domain of the Cbl riboswitch upstream of Symbiobacterium thermophilum (Sth) cobQ, which encodes a cobyric acid synthase (PDB ID 4GXY)47, and the Cbl riboswitch that regulates the Thermoanearobacter tengcongensis (Tte) fecB gene, the periplasmic component of an ABC-type Fe3+-siderophore transport system (PDB ID 4GMA)45. Importantly, the Tte fecB riboswitch includes P13 of the expression platform and thus encompasses the complete regulatory element. Class-I cobalamin riboswitches are defined by their ability to specifically bind AdoCbl2, 46 and all possess the full P7-P12 extension with the conserved IL10/11 element.39 The global architecture of the Tte fecB riboswitch is shown in Figure 3A, showing the structural relationship between the different domains and bound cobalamin.45 Another set of structures represent a “class-II” cobalamin riboswitch found in a sequence derived from an oceanic metagenome (environmental sequence 8, or env8).48 One structure has the aptamer domain alone (PDB ID 4FRG) while the other has the full-length riboswitch (PDB ID 4FRN), both in complex with hydroxocobalamin.45 This class of riboswitches have variable selectivity for the different forms of cobalamin and lack the P7-P12 extension, although the env8 variant displays extremely high selectivity for cobalamins with a small beta axial moiety. The global architecture of the full-length env8 riboswitch is shown in Figure 3B. Finally, a recent structure of an “atypical” aptamer domain of the cobalamin riboswitch of Bacillus subtilis (Bsu) yvrC, an uncharacterized ABC transporter, in complex with AdoCbl (PDB ID 6VMY) was determined.49 This RNA is unusual in that while it lacks a P7-P12 peripheral extension, it does bind AdoCbl in addition to other forms of cobalamin.46 Thus, this RNA is more related to other class-II cobalamin riboswitches, some of which also bind multiple forms of cobalamin.50 Together, these structures provide a clear picture of common features of the architecture of cobalamin riboswitches, how peripheral domains facilitate ligand recognition, and the conformational switch that governs gene regulation, as we discuss below.
Figure 3.
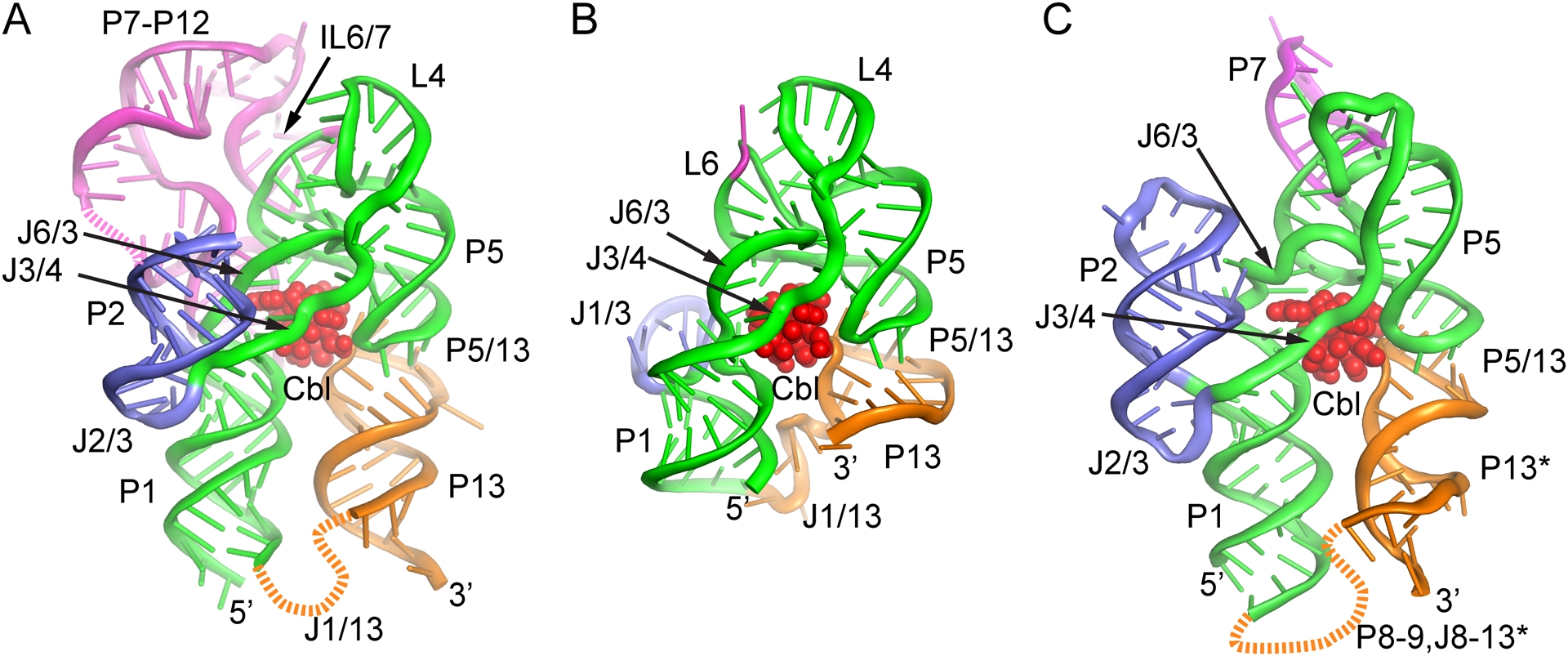
Global architecture of class-I and class-II cobalamin riboswitches. (A) Cartoon representation of the structure of the Tte class-I cobalamin riboswitch using the same color scheme in Fig. 2. Bound AdoCbl is shown as red spheres and strands not modeled are denoted as dashed lines. (PDB ID 4GMA). (B) Cartoon representation of the env8 class-II cobalamin riboswitch with bound HyCbl (red). The RNA is shown in the same orientation as in panel A to emphasize the similarity of structure of the core region, kissing loop interaction (P5/13) and cobalamin binding pocket. (PDB ID 4FRN). (C) Cartoon representation of the B. subtilis yvrC riboswitch bound to AdoCbl (red). The P13 helix (denoted by asterisk) shown in orange is the P9 helix of the crystallization construct that makes an intermolecular lattice contact (domain swap) that is proposed to mimic the native L5-L13 interaction. The 3’ region of the crystallized RNA (P13-P14) was not visible in the crystal structure. (PDB ID 6VMY). Figure created using PyMol (Schrödinger).
The central four-way junction mediates recognition of the variable beta-axial moiety of cobalamins
The central region of all cobalamin riboswitches contains critical structural features for cobalamin recognition. L4 has a long-range interaction with either IL6/7 (class-I and some class-II Cbl riboswitches) or L6 (only observed in class-II Cbl riboswitches). The terminal loop of P4 forms a classic T-loop with the structural features that define this motif51, 52, although the yvrC cobalamin riboswitch uses a slightly different “T-loop/tetraloop” hybrid structure.49 In all structures, L4 directly interacts with the IL6/7 or L6 loops, forming a functionally similar set of interactions. In-line and NMIA (“SHAPE”53, 54) probing of class-I and class-II Cbl riboswitches suggest that this long-range interaction forms independently of cobalamin binding and is likely an important structural element for organizing the Cbl binding pocket in the adjacent four-way junction by bringing P4 and P6 into a near parallel arrangement.45, 55
The four-way junction brings together the P3-P6 helices and organizes them through coaxial stacking of P3-P4 and P5-P6. This stacking is mediated by the joining strands J3/4 and J6/3, which also form the binding site for the beta-axial face of cobalamin.45, 47, 49 The J3/4 strand, which contains a highly conserved set of nucleotides (5’-GRAA), is observed to be in a nearly identical conformation in all four cobalamin riboswitch structures (green, Fig. 4). The first two purines contribute one half of a purine stacked “spine” (yellow, Fig. 4) that is the heart of the four-way junction. This purine spine has been observed in other RNAs as being critical for structural stability.56 A key ligand-RNA contact is mediated by this strand: a propionamide group of cobalamin is involved in hydrogen bonding contacts with the sugar edge of a guanine residue at the 3’-end of P3—one of the few such interactions in this region of the binding pocket.45, 47, 49 Conversion of this propionamide group to propionate is strongly deleterious to binding to the Eco btuB riboswitch.57 The two invariant adenosines at the 3’-end of J3/4 form A-minor triples58, 59 with the first base pair of P5 and a base pair between J4/5 and L6, which likely serves to anchor the P5/P6 coaxial stack and helps to orient L5 for recognition by L13.
Figure 4.
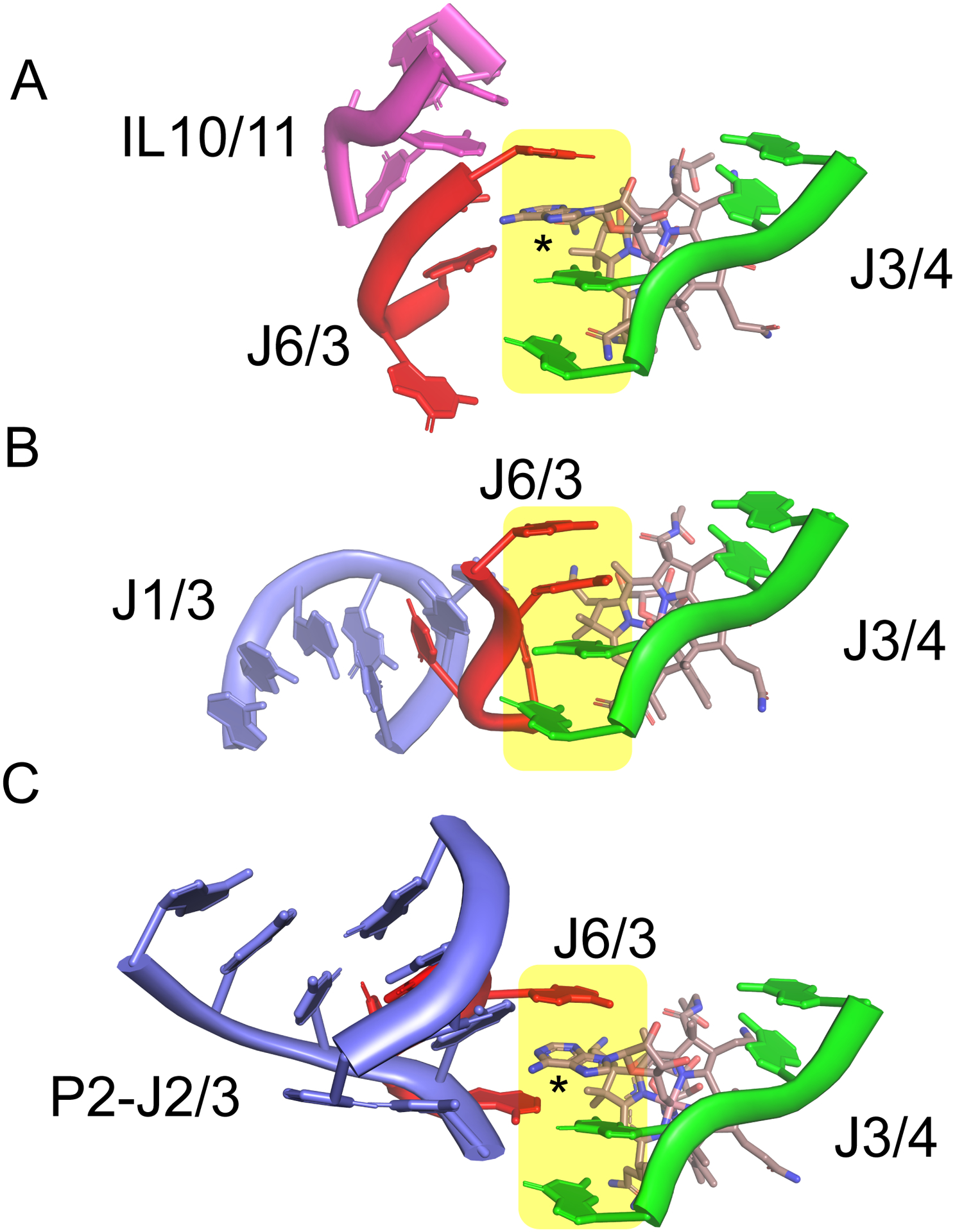
Organization of the cobalamin binding pocket in the aptamer domain. (A) Cartoon representation of the joining strands of the four-way junction that interact with AdoCbl in the Tte cobalamin riboswitch. The color scheme is the same as that used in Fig. 2, with cobalamin rendered as sticks and the central purine “spine” is highlighted in yellow. This purine spine is composed of two purines from J3/4 and one each from J6/3 and the 5’-deoxyadenosyl moiety of Cbl. An asterisk denotes the participation of a purine from the 5’-deoxyadenosyl moiety of Cbl in the central purine spine. (PDB ID 4GMA). (B) Cartoon representation of the two strands in the four-way junction, J3/4 and J6/3, that interact with HyCbl in the env8 cobalamin riboswitch. The orientation of the Cbl-RNA complex is that same as in panel A to emphasize the similar arrangement of J3/4 and cobalamin and the different conformation of J6/3 as enforced by interactions with J1/3 (magenta). (PDB ID 4FRN). (C) Cartoon representation of J3/4 and J6/3 of the Bsu yvrC cobalamin riboswitch in the same orientation as those in panels A and B; the asterisk denotes the 5’-deoxyadenosyl moiety of Cbl in the central purine spine. (PDB ID 6VMY). Figure created using PyMol (Schrödinger).
In contrast to the relatively fixed positioning of J3/4, the orientation of the J6/3 strand is significantly different between the four structures and appears to serve the role of the selectivity filter for different forms of cobalamin (red strand, Fig. 4). In the Sth and Tte class-I riboswitches bound to AdoCbl, the 5’-deoxyadenosyl moiety displaces one of the residues contributed by J6/3 of the central purine spine to participate directly in organizing the junctional RNA (denoted by asterisk in Fig. 4A).45, 47 This positioning of the Cbl 5’-deoxyadenosyl moiety is reinforced through a non-canonical A-A pair with an adenosine in the conserved IL10/11 of the P7-P12 peripheral extension. An extensive set of further RNA-RNA interactions between the peripheral extension around IL10/11 and the J6/3 strand further enforce a structure that creates a pocket for recognition of the sterically bulky 5’-deoxyadenosyl moiety. In contrast, J6/3 of the env8 class-II riboswitch, which does not contain the P7-P12 peripheral extension, instead interacts with a T-loop structure in J1/3, the other peripheral extension (Fig. 4B).45 This interaction forces J6/3 towards J3/4 and closes the pocket used for AdoCbl recognition in class-I riboswitches. Thus, this pocket sterically occludes the bulky 5’-deoxyadenosyl group but permits the smaller beta-axial groups of MeCbl, HyCbl, and CNCbl, accounting for this riboswitch’s strong discrimination against AdoCbl. Yet another set of interactions between J6/3 and the P2-J2/3 peripheral extension is observed in yvrC, showing an example of how RNAs without the P7-P12 extension can still recognize AdoCbl (Fig. 4C).49 However, this RNA is also capable of binding cobalamins with small beta-axial moieties, suggesting a malleable J6/3 element that can adjust to small versus large groups. Determining these alternative conformations of J6/3 awaits determining the yvrC RNA structure in the presence of MeCbl or CNCbl.
The promiscuous Cbl binding observed in the yvrC riboswitch is not exclusive to this RNA. A survey of AdoCbl and MeCbl binding by isothermal titration calorimetry (ITC)60 across a set of class-II riboswitches revealed that these structurally related RNAs have a range of selectivities, from >300-fold preference for AdoCbl to over 105-fold preference for MeCbl.50 Like yvrC, there were also RNAs that bind both cobalamin forms with high affinity. For example, env62 has KDs of 110 and 60 nM for AdoCbl and MeCbl, respectively. In an attempt to determine the specific sequence elements responsible for AdoCbl versus MeCbl selectivity, an AdoCbl-selective RNA was systematically mutagenized to convert it to MeCbl-selective. While a specific set of nucleotides whose identity dictated selectivity was not found, it was discovered that many Cbl riboswitches of class-IIa (a subclassification of class-II that share a consensus 5’-RGY (R, purine; Y, pyrimidine) sequence in J6/3) are capable of productively binding either form of Cbl with large or small beta-axial moieties. This analysis strongly suggested that ligand selectivity is determined by the interaction between J1/3 and J6/3, consistent with the crystal structure of env8. However, this finding is likely not generalizable to all class-II riboswitches as many members have very different J1/3 peripheral extensions from env8. It would appear that class-II riboswitches have multiple means to confer cobalamin selectivity or promiscuity and a simple examination of sequence cannot currently determine the binding properties of a class-II member. Thus, for now, while class-I Cbl riboswitches are likely all highly specific for AdoCbl due to the presence of the P7-P12 extension, class-II Cbl riboswitches exhibit an incompletely understood range of Cbl-binding properties.
The aptamer domain and expression platform interact through a conserved kissing loop interaction
The crystal structure of a cobalamin riboswitch containing both the aptamer domain and the native P13 allows direct insights into the coupling of ligand binding to the aptamer domain and the regulatory conformational switch. Unique amongst all current riboswitch structures, the sequence used to determine the structure of the full-length env8 class-II structure was experimentally demonstrated to contain all of the information necessary and sufficient for ligand-dependent regulation.45 Importantly, the RBS is embedded within L13 and positioned just upstream of the start codon of an ORF that encodes a protein homologous to other bacterial BtuB proteins. Comparison of this structure with the env8 aptamer domain-HyCbl complex revealed that L13 of the expression platform forms a kissing loop interaction with L5 of the aptamer domain to form a new helix, P5/13 (Fig. 5A). Formation of the kissing loop interaction between hairpins in the aptamer domain (L5) and expression platform (L13) occludes the RBS from interaction with the 30S ribosomal subunit (cyan, Fig. 5A). The kissing loop motif is a common tertiary structure motif in RNA in which the bases of the two interacting loops form canonical base pairs to form a new helix that is coaxially stacked between the stems of the hairpins.6 This interaction serves to organize global architecture as found in the lysine riboswitch61, 62 and the VS ribozyme63–66 and mediates RNA-RNA interactions as found in regulation of plasmid replication67, 68 and retroviral dimerization.69–71
Figure 5.
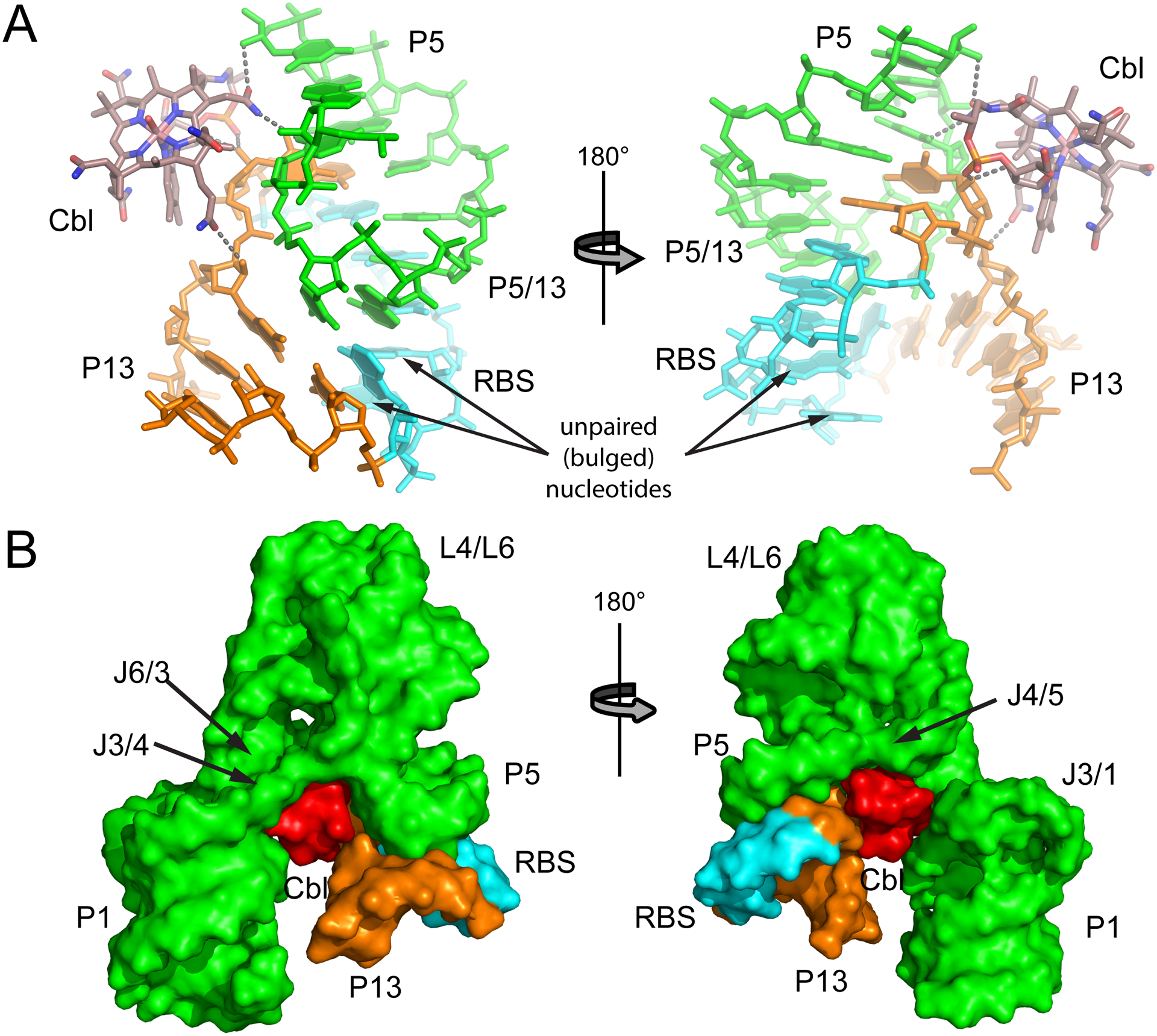
Structural relationship of bound cobalamin to the kissing loop interaction in the env8 class-II riboswitch. (A) Stick representation of the P5-P5/13-P13 kissing loop interaction using the same coloring scheme as in Fig. 2, except that the ribosome binding site (RBS) is highlighted in cyan. Hydrogen bonding interactions between HyCbl and RNA are denoted as grey dashed lines. (B) Surface representation of the env8 HyCbl-RNA complex emphasizing the extensive van der Waals contacts between bound cobalamin and the regulatory switch. Recruitment of the P13/L13 stem-loop to the aptamer domain-cobalamin complex uses a composite small-molecule/RNA surface. Figure created using PyMol (Schrödinger).
Although the env8 HyCbl structure provides the clearest example of the native kissing loop formation in cobalamin riboswitches, the other Cbl riboswitch crystal structures also yield insights into this important RNA-RNA interaction.45, 47, 49 An analogous kissing loop interaction is observed in the Tte class I riboswitch, although the structural details of the loop-loop interaction are different. This riboswitch regulates the formation of a transcriptional terminator, such that stable formation of P13 and the kissing loop between L5 and L13 fates the mRNA to form a downstream transcriptional terminator. In the absence of Cbl the mRNA forms the competitive antiterminator that promotes synthesis of the full message and expression of FecB, a Fe3+-citrate transporter. Less straightforward is the interaction in the yvrC riboswitch.49 While the RNA used for crystallization does contain P13 which regulates transcription via cobalamin-dependent formation of a transcriptional terminator, the riboswitch-AdoCbl complex instead crystallized with an intermolecular interaction between L5 and L9, part of a small hairpin immediately downstream of P1. This interaction is proposed to mimic the intramolecular L5-L13 interaction of the native riboswitch. Finally, while the RNA used to determine the Ste aptamer-AdoCbl crystal structure only contained the aptamer domain, an intermolecular lattice contact between L5 and the IL8/10 loop of a neighboring molecule was proposed to resemble the native L5-L13 interaction.47 Thus, in all four Cbl riboswitch structures, L5 forms an RNA-RNA interaction, suggesting that this loop is primed for formation of loop-loop interactions that serve as the conformational switch in these regulatory RNAs.
These structures further revealed that the regulatory kissing loop as part of the P5-P5/13-P13 coaxial stack forms the other half of the cobalamin ligand binding pocket. Importantly, P5 and P5/13 directly contact the alpha-axial face of the bound cobalamin (Fig. 5A).45 These contacts include hydrogen bonding interactions between amide groups around the corrin ring and the minor groove of P5, the loop-proximal base pair of L13, and the last adenosine of J3/4 which forms an A-minor triple interaction with a base pair in L5. In addition, the 5,6-dimethylimidzole ring and other functional groups in the cobalamin make van der Waals contacts with nucleotides in the kissing loop interactions (Fig. 5B). A similar set of contacts are observed in the native P5-P5/13-P13 kissing loop structure in the Tte-AdoCbl complex structure, suggesting that these interactions are conserved throughout the cobalamin family.45 Presumably, these direct cobalamin-kissing loop interactions serve to promote the formation of the regulatory conformational change involving a long-range interaction between the two domains.
The conformational switch can form in the presence of elevated magnesium
As with all biological RNAs, including riboswitches, divalent magnesium cations are essential for both structure and function of the cobalamin riboswitch.72–76 In-line probing showed that the Eco btuB cobalamin riboswitch undergoes a series of Mg2+-dependent folding events in the absence of effector. This was interpreted as evidence of a stepwise hierarchical folding pathway as the divalent cation concentration is increased from 0 to 1.0 mM.55 Interestingly, besides Mg2+-induced protection of helical elements, significant protections of loops involved in tertiary architecture formation such as L4 and IL6/7 indicate that divalent ions serve to organize the RNA on a tertiary level as well, making it competent to bind AdoCbl. Subsequent Tb3+ probing of the Eco btuB riboswitch reinforced the model that the tertiary architecture of this RNA is substantially pre-organized by magnesium prior to effector binding.77 Similarly, the L4 and L6 loops in the env8 riboswitch shows significant protection with 1 mM Mg2+ without ligand present in comparison to no Mg2+ by SHAPE chemical probing, indicative of this aspect of tertiary structure forming in the absence of ligand.45 These data further support pre-organization of the aptamer domain by cations.
In contrast to the L4-IL6/7 tertiary interaction that serves to organize the conserved core of the aptamer domain, the interdomain kissing loop interaction is likely not formed under physiological magnesium ion concentrations. NMIA probing of the env8 and the E. coli btuB riboswitches indicate that protection of the L5 and L13 loops only occurs at elevated (>10 mM) magnesium ion concentrations in the absence of Cbl, indicating that this structural element does not form under physiological conditions.45 Thus, unlike many other kissing loop interactions that are stable under physiological divalent conditions, this motif in cobalamin riboswitches is significantly destabilized. This observation was corroborated with smFRET studies of the env8 riboswitch, which showed that without Mg2+, even saturating levels of HyCbl were unable to induce the L5-L13 interaction.78 Measurement of FRET that reports on L5-L13 docking as an ensemble and in single molecules indicates that the formation of the kissing loop has a [Mg2+]1/2 of ~2.2 mM. smFRET measurements also reveal rapid docking/undocking rates at 1 mM Mg2+, indicating that this interaction is unstable under physiological conditions. This is compelling evidence that while the regulatory kissing loop can form in the absence of effector, it is a weak and transitory interaction. These data are further supported by analysis of the Eco btuB riboswitch, which implicates the kissing loop interaction in ligand binding and regulatory control.79 However, it should be noted that a study of the magnesium-induced architecture of the Eco btuB riboswitch using Tb3+ suggests that its kissing loop does form under physiological conditions in the absence of AdoCbl.77
Cobalamin stabilizes the kissing loop interaction at physiological magnesium concentrations
Chemical probing of the Eco btuB and env4 class-II riboswitches provides strong evidence that the presence of effector bound to the aptamer domain stabilizes the L5-L13 interaction.45 SHAPE probing of these RNAs in the presence of ligand as a function of magnesium concentration reveals that the divalent cation concentration needed to stabilize the kissing loop drops to physiological concentrations (0.5 – 1.0 mM). In support of this finding, smFRET studies of env8 reveal that the presence of HyCbl drops the [Mg2+]1/2 of formation to 0.3 mM and dramatically decreases the dissociation (undocking) rate of the L5-L13 interaction.78 Coupled with the crystal structures showing direct interactions between the alpha axial face of the cobalamins and the kissing loop (Fig. 5), the role of Cbl in directly stabilizing the formation of one of the two regulatory conformations is clear.
Non-helical features of the kissing loop confer cobalamin dependent formation
To understand how the kissing loop interaction of cobalamin riboswitches is harnessed as an effector-dependent conformational switch, a structure-guided analysis of this interaction was performed.80 Electrophoretic mobility shift assays (EMSA)81 were used to investigate the ability of a series of L5 mutants to form the kissing loop in the presence of HyCbl. This analysis revealed that two bulged nucleotides in the middle of the P5/P13 helix (Fig. 5A) were highly destabilizing to kissing loop formation in the absence of cobalamin, but that kissing loop formation could be stabilized in the presence of cobalamin. Specifically, the presence of HyCbl reduced the KD between the two halves of the kissing loop by over 6,800-fold. Phylogenetic analysis of other class-II cobalamin riboswitches reveals that they all contain such features in their P5/P13 interactions, suggesting that this is a general mechanism for generating a cobalamin-dependent switch.80 This is similar to previous findings that non-canonical base pairs in an aptamer-HIV TAR element interaction are strongly destabilizing82 as well as a kissing loop interaction in the HIV dimerization initiation site.83 The EMSA experiments were supported by a cell-based reporter assay in which the env8 riboswitch regulates expression of a fluorescent reporter gene in E. coli.80 These data reinforced the central role of unpaired nucleotides in P5/13 that destabilize the kissing loop interaction in the absence of Cbl. Notably, the cobalamin-bound aptamer domain provides a composite small molecule/RNA interface for interacting with the RBS-containing P13 to form the regulatory kissing loop interaction (Fig. 5B). However, the riboswitch’s ligand-dependent regulatory function is most likely a process too complex to be fully explained simply by the change in interdomain affinity. Many factors within the intercellular environment, including folding kinetics, are expected to be at play.
Moving towards co-transcriptional models of conformational switching
The above studies have yielded key insights into the structures and mechanism for ligand-dependent conformational changes within the cobalamin riboswitch family. However, these studies have almost entirely relied upon classic in vitro methods that use full-length RNAs transcribed using T7 RNA polymerase and purified by denaturing methods.84–86 Following purification, these transcripts are generally heat-cooled to further reset any residual structure and promote formation of uniform secondary structure in a buffer containing low concentrations of monovalent salt. Folding is then initiated typically by addition of divalent magnesium cations, which promote formation of long-range tertiary interactions.87 These approaches are possible due to the high free energies of RNA folding and resulting structural stability which allows reformation after denaturation without the need for additional cofactors, something which is rarely the case for proteins.88 RNA synthesized, purified, and renatured using these approaches has been essential for development of much of the current understanding of RNA structure and function. However, it is well established that even simple RNAs may exhibit difficulties refolding to achieve complete biological activity under in vitro reconstitution conditions88, and cobalamin riboswitches are no exception. Across multiple studies there is strong evidence that model cobalamin riboswitches have difficulty refolding (see below). Given that they evolved to function in the context of transcription, it is increasingly clear that co-transcriptional approaches are essential for developing a more accurate picture of the structure-function relationship in these RNAs.8, 89–91
RNA folding in the context of transcription in the cellular environment experiences a fundamentally different landscape. In vivo, there are factors that favor proper RNA folding that do not exist with in vitro approaches. Co-transcriptional folding is the first level of these factors.92–94 That RNA folds as it is being transcribed means the final folded state is influenced by the 5’- to 3’-polarity of transcription, where upstream nucleotides can begin forming local secondary structures before those more downstream are even transcribed. This folding landscape cannot be accessed when the full-length RNA molecule is refolded simultaneously. The final folded state is also influenced by RNA polymerase (RNAP) speed11, 95, 96 and pausing97, 98, two other elements unaccounted for by in vitro refolding. These effects of co-transcriptional pausing can significantly constrain the folding landscape, enabling the RNA to avoid kinetic traps and stable misfolds. Finally, there is the surrounding factor of the cellular milieu. Molecular crowding and microenvironment concentrations of metal ions and other cellular solutes can have vast implications in RNA folding.99, 100 These conditions can be attempted to be replicated in vitro, but often they are not. Altogether, these factors combine to influence a final folded product of RNA that may be very different than RNA produced and purified in vitro.
Riboswitch folding and ligand affinity can be enhanced in the context of transcription
The importance of transcription on folding and binding is highlighted in the structural determination of the Bsu yvrC Cbl riboswitch bound to AdoCbl.49 Attempts to assemble a ligand-bound riboswitch using traditional methods of denaturing gel purification of an RNA transcript, refolding, and subsequent loading with ligand proved unsuccessful. Obtaining a bound complex for crystallization required that the riboswitch be transcribed in the presence of a cobalamin ligand and purified by non-denaturing polyacrylamide gel electrophoresis.49 Formation of AdoCbl, MeCbl, or HyCbl complexes with the yvrC riboswitch were assessed using electrophoretic mobility shift assays (EMSA) and by size exclusion chromatography and multiangle light scattering (SEC-MALS). Notably, these experiments also suggested that the conformation of the riboswitch differs between bound AdoCbl and MeCbl or HyCbl, i.e., the riboswitch folds differently depending on the ligand it binds. While it is highly likely that these changes involve different conformations of J6/3, as suggested by other cobalamin riboswitch structures (Fig. 4), this is currently speculative as only the yvrC crystal structure bound to AdoCbl was solved. It should be noted that other large, complex RNAs have also required synthesis and purification using native approaches from in vitro transcription reactions, such as the Oceanobacillus iheyensis group II intron101, 102 and the VS ribozyme103, and various methods have been developed to effect native purification of RNA.104 While these studies clearly indicate that the yvrC riboswitch cannot be refolded in a ligand-binding competent state49, what features of the RNA lead to misfolding were not determined. Approaches such as SHAPE chemical probing of the structure of these RNAs purified under different conditions may yield important insights into how co-transcriptional folding evades misfolding.105
Other cobalamin riboswitches also appear to have difficulty refolding. The wild type env8 riboswitch consistently displays multiphase binding curves in ITC measurements corresponding to a high affinity and a low affinity ligand binding interaction.50 This can be interpreted as a population that has a properly folded binding site capable of a high affinity interaction and another population with local misfolding that creates a lower affinity binding mode. Interestingly, mutants of the riboswitch in L5 that alter the kissing loop interaction alleviate this phenomenon80, suggesting a potential misfold involving the Cbl-dependent conformational switch. In turn, this indicates that conformational switching may be only fully achieved using RNAs that are co-transcriptionally folded and natively purified, although this has not been tested.
The idea that the Cbl riboswitch may fold differently between Mg2+-induced refolding of a full-length denatured transcript and co-transcriptionally is further supported by studies of the Eco btuB riboswitch. In both oligonucleotide hybridization/RNase H digestion and SHAPE chemical footprinting, the Mg2+-refolded riboswitch has significantly decreased levels of protection in the aptamer region, the kissing loop, and the RBS hairpin in the presence of AdoCbl than its natively folded counterpart.79, 106 In addition, co-transcriptional folding of this Cbl riboswitch appears to affect its affinity for the Cbl ligand. In the oligonucleotide hybridization/RNase H digestion assays, the Kswitch value, which is the concentration of AdoCbl at which half of the riboswitch molecules are switched to the “off” state, is 55 nM. This is significantly lower than KD values determined using in vitro transcribed, purified, and refolded RNA (~250–300 nM).2, 45 This may be an artifact of other factors such as oligonucleotide hybridization efficiency, especially as DNA probes targeting other regions of the riboswitch gave other Kswitch values, but it nevertheless argues in favor of the importance of careful consideration of purification approaches for large RNAs. These examples suggest that misfolding may be a broader feature of cobalamin riboswitches prepared using traditional denaturing protocols.
Pausing is a critical feature of the cobalamin riboswitch folding
Programmed pausing in which sequences within the riboswitch induce RNAP to stall for a short period of time is a common feature of riboswitches.11, 107, 108 Studies of a flavin mononucleotide (FMN) riboswitch revealed that pause sites situated at key regions extend the timeframe in which the aptamer domain can fold and bind effector, allowing a conformational switch to form in the absence of a downstream competing sequence.11 Similarly, in vitro single turn-over transcription assays performed on the Eco btuB riboswitch as a function of time revealed three major RNAP pause sites that map to the expression platform (Fig. 6).106 The first pause site (PA) is proposed to allow the aptamer to fold without interference from downstream sequences in the expression platform that can induce alternate aptamer folding in the absence of ligand. Pause B (PB) appears to allow transitory formation of the L5-L13 kissing loop while the aptamer interrogates the cellular environment for AdoCbl. This is important as it gives the riboswitch time to bind ligand and stabilize that folded conformation before prematurely committing to the apo state. Depending on whether AdoCbl is bound, a third pause site (PC) can influence the fate of the anti-RBS sequence. Without bound effector, alternate aptamer folding is induced by helix formation between the anti-RBS and anti-anti-RBS sequence (situated within J6/3). Thus, pausing appears to facilitate two key steps in the pathway of folding in this riboswitch: binding and conformational switching. The critical nature of pausing was reinforced through assays that used mutant RNAPs with reduced transcriptional pausing. These RNAPs yielded transcripts with significantly impaired folding and conformational changes. Further analysis of folding rates of the riboswitch in the absence and presence of AdoCbl with wild type and mutant RNAP indicate that pausing primarily acts to facilitate tertiary folding of the aptamer domain. While the rates of folding of the aptamer domain and expression platform are balanced with use of the wild type RNAP, the rates diverge with use of the fast RNAP mutants, particularly in the presence of AdoCbl, such that the aptamer domain folds more slowly and the expression platform more rapidly.
Figure 6.
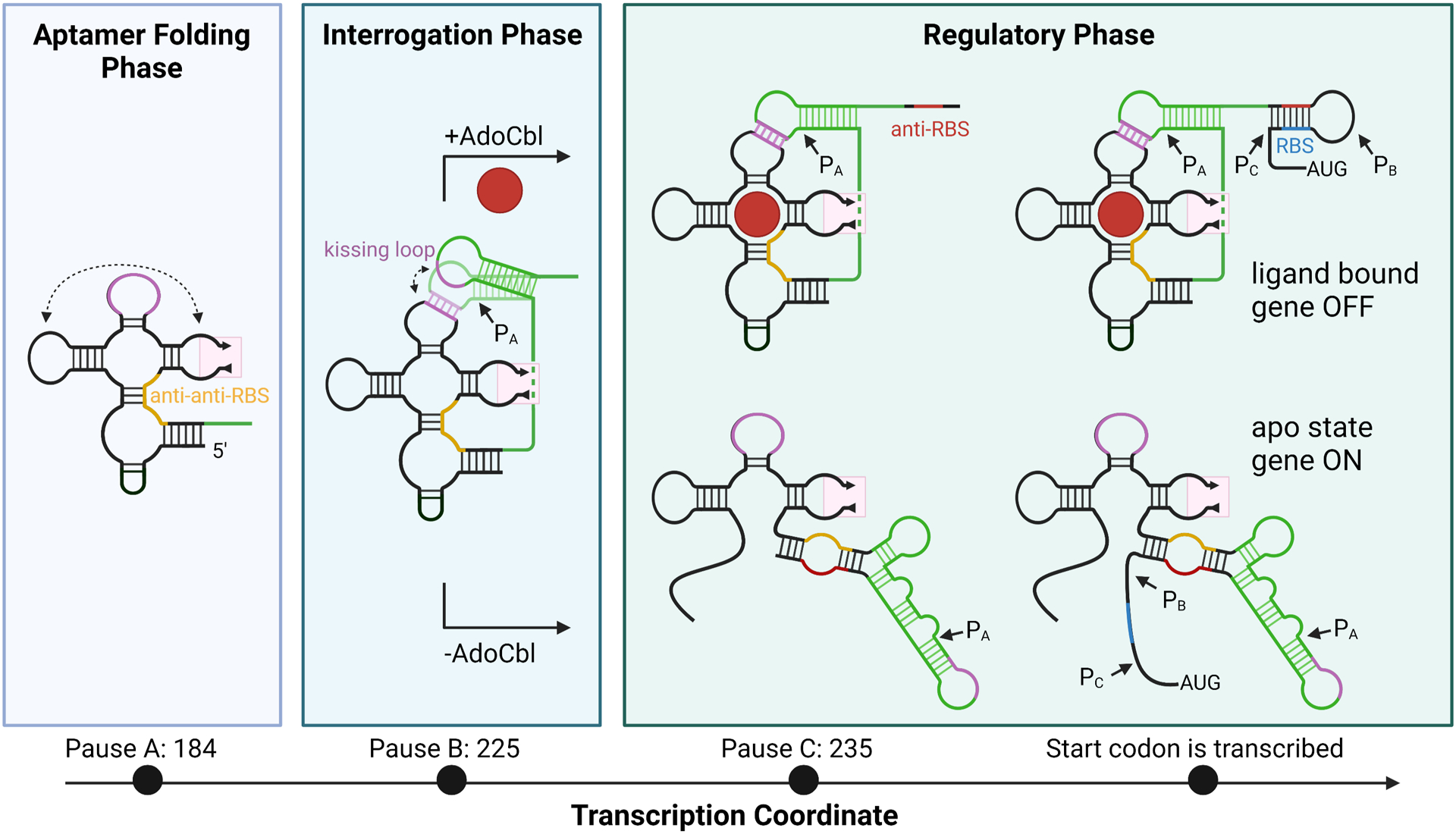
A model for E. coli btuB cobalamin riboswitch folding. Folding intermediaries for the transcript at each pause site are plotted by transcription coordinate. The regions that interact through a kissing loop are colored in purple. Peripheral extension 2 is omitted, indicated by the pink box. The anti-anti-RBS region is colored in yellow. P13 and surrounding regions which fold differentially depending on the presence of ligand are colored in green. The anti-RBS region is colored in red. The RBS is colored in blue. Each pause site is correlated with a folding phase. Pause sites are indicated when present on the transcript as PX with an arrow. Created with Biorender.com.
Cell-based studies of regulatory activity strongly implicate the kissing loop in driving regulatory activity
Given observed differences between Mg2+-induced folding and co-transcriptional folding of cobalamin riboswitches, it is important to assess the mechanism of the coupling of effector binding to gene regulation using approaches that examine the riboswitch in a native genetic and physiological context. To accomplish this, a series of mutant BtuB-LacZ fusions were incorporated into the E. coli genome and the ability of AdoCbl added to cell growth medium to repress expression of the reporter was assessed.79 These mutants were similar to those described to probe the kissing loop interaction in the env8 class II riboswitch using a cell-based assay.80 These experiments demonstrated that destabilization of the kissing loop by ablating canonical base pairing between L5 and L13 was highly deleterious to AdoCbl-dependent regulation. Interestingly, compensatory mutations in L5 and L13 that result in base pair transversions do not recapitulate the regulatory activity of the wild type riboswitch. Rather, compensatory mutants were significantly diminished in their ability to repress expression in the presence of AdoCbl. Further, it was shown that ablation of the kissing loop interaction significantly affected AdoCbl binding, consistent with prior calorimetric measurements of AdoCbl binding to the aptamer domain and full-length riboswitch.45 This contrasts with findings from the env8 riboswitch, in which the kissing loop has only a modest (~5-fold) impact on effector binding, suggesting that the kissing loop plays differing roles in promoting Cbl binding across the Cbl riboswitch family. These data also highlight the importance of pause site B in the Eco btuB riboswitch (Figure 6) since both the aptamer and P13 have been synthesized and are free to fold and form the kissing loop interaction.
Mutagenic analysis of the effect of kissing loop formation on the anti-RBS/RBS (P14) helix of Eco btuB was also examined. In this case, mutations in the stem of P13 were made that affect the structures in the conformational switch.79 These data indicate that P13 is critically important for regulation, but the details of its involvement are unresolved as the model does not allow for clean interpretation of the results. Further work needs to be done to examine the details of the conformational change that lead to the on, and especially the off, state. But together with the results of the kissing loop mutants discussed above, these data convincingly reveal the central role of the kissing loop on governing the regulatory conformational switch in response to the occupancy status of the aptamer domain. While these experiments largely confirmed prior structural and biochemical data using in vitro approaches, this work provided important nuances to our understanding of Cbl riboswitch function in the context of co-transcriptional folding and in its native environment.
Cell-based genetic screening to illuminate elements of ligand recognition
In addition to probing mechanism in the native environment, cell-based and in vivo approaches can incorporate powerful genetic selection and screening to yield insights not easily gained through in vitro methods. For example, in the env8 crystal an interaction between J1/3 and J6/3 (Fig. 4B) is important for ligand binding and regulatory activity.45 Using the cell-based reporter system established for investigating the kissing loop interaction (see above), a genetic screen was performed in which a set of nucleotides of J1/3 and J6/3 were fully randomized and the resultant library screened for functional variants.109 Functional variants identified in the screen defined the sequence space of this tertiary interaction that supports ligand binding and regulatory activity. This artificial phylogeny can be coupled to a fluorescence reporter assay to quantify the activity of each sequence variant, adding a functional component to the sequence alignment. This analysis revealed that highly functional sequences of the J1/3 element conform to a classic T-loop module in J1/3 and reinforce the functional significance of a C-G base pair between J1/3 and J6/3. Because of the sparse number of natural sequences for class II Cbl riboswitches48 and the high degree of sequence variation within their J1/3 elements, the artificial phylogeny assisted in establishing the structural and functional relevance of key tertiary interactions that could not be inferred from conservation patterns. In addition, these experiments can reveal alternative functional solutions not represented in biology. This approach applied to investigating the env8 L4-L6 interaction has revealed an expanded sequence space that supports the efficient co-transcriptional folding of the riboswitch (manuscript in preparation). Thus, exploiting experimental approaches that approximate the native functional environment of the riboswitch can yield new insights into folding and conformational switching.
An ancient ribozyme?
One interesting hypothesis regarding cofactor-binding riboswitches such as Cbl and SAM is that they may have once functioned as ribozymes using those cofactors in an RNA World.110 In support of this idea, several in vitro selections (SELEX) have recently raised methyltransferase ribozymes using m6G and SAM as cofactors.111–113 It was noted that the catalytic core of the MTR1 methyltransferase ribozyme has strong similarity to the guanine/hypoxanthine binding pocket of the guanine riboswitch.114 Even more strikingly, a preQ1 class I riboswitch has been shown to have self-methylation activity when bound to m6preQ.115 These results demonstrate that RNAs, and specifically riboswitches, have the capability to promote this chemical reaction.
Cobalamin riboswitches may also have catalytic capability, but this function has not been explored. As part of its role as an enzyme cofactor, specifically for methyltransferases such as methionine synthase, MeCbl participates in transmethylation reactions wherein the cobalamin is the methyl group donor.26–28 If the cobalamin riboswitch could promote a similar methylation reaction, it is expected to be self-methylating as is seen with the preQ1 riboswitch.115 This expectation is due to the positioning of the beta axial methyl group, which points towards the inside of the binding pocket in all of the solved structures of the cobalamin riboswitch. Although a Cbl riboswitch structure has not been solved bound to MeCbl, structural docking into the env8-HyCbl structure45 indicates that the methyl group could be in a position favorable for transfer to either the N3 or 2-’OH positions of A20. While previous SHAPE data does not seem to support methylation of the 2’-OH position, as this would result in a reverse transcription stop site which is not observed45, methylation of N3 remains possible. Studies to confirm or refute this potential functionality of the cobalamin riboswitch may be undertaken in the future and could deepen our understanding of this important RNA element in a new and exciting way.
Perspectives
Riboswitches serve as an important model system for exploring the nature of gene regulation driven by conformational changes in RNA. As we have presented, a broad spectrum of structural, biochemical, and cell-based approaches have revealed key aspects of the mechanism of coupling effector binding to the regulatory switch in cobalamin riboswitches. These studies have shown a direct physical relationship between cobalamin and stabilization of the conformational switch that governs mRNA expression. Unique amongst riboswitches, the structural studies of the cobalamin riboswitch have been able to access the full riboswitch, enabling detailed mechanistic studies of small molecule guided conformational changes. Critically, the use of co-transcriptional folding approaches has revealed mechanistic aspects such as pausing and the relationship between Cbl binding and kissing loop formation. We expect these co-transcriptional methods to become more widespread in the future to better simulate folding in the cellular context.
It is generally accepted that the free energy landscape of RNA folding is “rugged”.116, 117 While the native RNA folded state is very stable, other conformations are as well, allowing for RNA to become kinetically trapped in a misfolded state—at least while studying RNA folding outside the context of transcription. The purification and refolding process ubiquitous to in vitro studies most likely does not result in a homogenous population of RNA in one native state, but rather a heterogeneous population of several folded states. This problem of RNA misfolding in vitro is even more relevant today, where mass production of RNA for applications such as vaccines becoming increasingly common. Large-scale RNA production also relies on in vitro transcription and purification, which can leave a heterogeneous, misfolded product.118 Some alternative methods of purification to reduce heterogeneity have been suggested and employed88, but nonetheless the classic in vitro methods described here are most prevalent. A deeper understanding of the differences between whole-transcript and co-transcriptional folding pathways will greatly enrich our understanding of how to control the RNA folding process across many potential applications.
For the Cbl and other riboswitches, there are a number of applications that are emerging. As simple regulatory devices, riboswitches present themselves as powerful tools for synthetic biology to probe the metabolic environment and regulate gene expression. For example, the cobalamin riboswitch has been used as a sensor to monitor various aspects of cobalamin metabolism in E. coli.119, 120 Development of RNAs that sense novel small molecules requires not only raising aptamers against the desired effector, but also understanding how to couple it to a high fidelity regulatory switch. This latter process remains poorly understood and represents a major bottleneck in progress towards synthetic riboswitches. Cobalamin riboswitches also serve as the foundation for a novel class of tools for imaging RNAs in live cells.121 It was reasoned that riboswitches have evolved to efficiently fold in a variety of cellular environments, and thus may be superior to in vitro selected aptamers in this type of application. Understanding the co-transcriptional folding pathways of these RNAs will certainly facilitate the development of improved versions of these devices. Finally, as nucleic acid-based therapeutics become increasingly realized, riboswitches will play important roles as a means of regulating their expression via a bioavailable small molecule.122 Engineering practical devices that function in the context of human mRNAs123–127 will again require further analysis of how ligand-guided conformational changes drive gene expression across a spectrum of biological RNAs. While great strides have been made in developing mechanistic models of conformational changes in the Cbl family of riboswitches, this new understanding must now be applied to emerging RNA-based technologies.
Acknowledgements
This work was funded by grants from the National Institutes of Health to R.T.B. (R01 GM073850 and GM133184) and through support from the CU Boulder Molecular Biophysics Training program to S.R.L. (T32GM065103).
Footnotes
Conflict of interest
R.T.B. serves on the Scientific Advisory Board of Expansion Therapeutics, SomaLogic and MeiraGTx.
References
- 1.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–56. [DOI] [PubMed] [Google Scholar]
- 2.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. [DOI] [PubMed] [Google Scholar]
- 3.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–6. [DOI] [PubMed] [Google Scholar]
- 4.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A. 2002;99:15908–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batey RT, Rambo RP, Doudna JA. Tertiary Motifs in RNA Structure and Folding. Angew Chem Int Ed Engl. 1999;38:2326–43. [DOI] [PubMed] [Google Scholar]
- 6.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res. 2011;44:1302–11. [DOI] [PubMed] [Google Scholar]
- 7.Serganov A, Patel DJ. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys. 2012;41:343–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochim Biophys Acta. 2009;1789:584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. Chembiochem. 2003;4:1024–32. [DOI] [PubMed] [Google Scholar]
- 10.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–14. [DOI] [PubMed] [Google Scholar]
- 11.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. [DOI] [PubMed] [Google Scholar]
- 12.Ray-Soni A, Bellecourt MJ, Landick R. Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annu Rev Biochem. 2016;85:319–47. [DOI] [PubMed] [Google Scholar]
- 13.Roberts JW. Mechanisms of Bacterial Transcription Termination. J Mol Biol. 2019. [DOI] [PubMed] [Google Scholar]
- 14.Coppins RL, Hall KB, Groisman EA. The intricate world of riboswitches. Curr Opin Microbiol. 2007;10:176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan H, Conn AB, Williams PB, Diggs S, Hahm J, Gamper HB, et al. Transcription–translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucleic Acids Research. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatterjee S, Chauvier A, Dandpat SS, Artsimovitch I, Walter NG. A translational riboswitch coordinates nascent transcription-translation coupling. Proc Natl Acad Sci U S A. 2021;118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastet L, Chauvier A, Singh N, Lussier A, Lamontagne AM, Prevost K, et al. Translational control and Rho-dependent transcription termination are intimately linked in riboswitch regulation. Nucleic Acids Res. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, et al. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci U S A. 2012;109:5376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastet L, Turcotte P, Wade JT, Lafontaine DA. Maestro of regulation: Riboswitches orchestrate gene expression at the levels of translation, transcription and mRNA decay. RNA Biol. 2018;15:679–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breaker RR. The Biochemical Landscape of Riboswitch Ligands. Biochemistry. 2022;61:137–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. Riboswitch diversity and distribution. RNA. 2017;23:995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissbach H, Brot N. Regulation of methionine synthesis in Escherichia coli. Mol Microbiol. 1991;5:1593–7. [DOI] [PubMed] [Google Scholar]
- 24.Klug G Beyond catalysis: vitamin B12 as a cofactor in gene regulation. Mol Microbiol. 2014;91:635–40. [DOI] [PubMed] [Google Scholar]
- 25.Padmanabhan S, Jost M, Drennan CL, Elias-Arnanz M. A New Facet of Vitamin B12: Gene Regulation by Cobalamin-Based Photoreceptors. Annu Rev Biochem. 2017;86:485–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randaccio L, Geremia S, Demitri N, Wuerges J. Vitamin B12: unique metalorganic compounds and the most complex vitamins. Molecules. 2010;15:3228–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown KL. Chemistry and enzymology of vitamin B12. Chem Rev. 2005;105:2075–149. [DOI] [PubMed] [Google Scholar]
- 28.Giedyk M, Goliszewska K, Gryko D. Vitamin B12 catalysed reactions. Chem Soc Rev. 2015;44:3391–404. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee R, Ragsdale SW. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem. 2003;72:209–47. [DOI] [PubMed] [Google Scholar]
- 30.Hodgkin DC, Kamper J, Mackay M, Pickworth J, Trueblood KN, White JG. Structure of vitamin B12. Nature. 1956;178:64–6. [DOI] [PubMed] [Google Scholar]
- 31.Anderson PJ, Lango J, Carkeet C, Britten A, Krautler B, Hammock BD, et al. One pathway can incorporate either adenine or dimethylbenzimidazole as an alpha-axial ligand of B12 cofactors in Salmonella enterica. J Bacteriol. 2008;190:1160–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gold L, Janjic N, Jarvis T, Schneider D, Walker JJ, Wilcox SK, et al. Aptamers and the RNA world, past and present. Cold Spring Harb Perspect Biol. 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cech TR. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–68. [DOI] [PubMed] [Google Scholar]
- 34.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. [DOI] [PubMed] [Google Scholar]
- 35.Sebille B Methods of drug protein binding determinations. Fundam Clin Pharmacol. 1990;4 Suppl 2:151s–61s. [DOI] [PubMed] [Google Scholar]
- 36.Lundrigan MD, Koster W, Kadner RJ. Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12. Proc Natl Acad Sci U S A. 1991;88:1479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravnum S, Andersson DI. Vitamin B12 repression of the btuB gene in Salmonella typhimurium is mediated via a translational control which requires leader and coding sequences. Mol Microbiol. 1997;23:35–42. [DOI] [PubMed] [Google Scholar]
- 38.Richter-Dahlfors AA, Andersson DI. Cobalamin (vitamin B12) repression of the Cob operon in Salmonella typhimurium requires sequences within the leader and the first translated open reading frame. Mol Microbiol. 1992;6:743–9. [DOI] [PubMed] [Google Scholar]
- 39.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 2003;9:1084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278:41148–59. [DOI] [PubMed] [Google Scholar]
- 41.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, et al. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–4. [DOI] [PubMed] [Google Scholar]
- 42.DebRoy S, Gebbie M, Ramesh A, Goodson JR, Cruz MR, van Hoof A, et al. Riboswitches. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science. 2014;345:937–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, et al. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2009;106:4435–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci U S A. 2000;97:7190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson JE Jr., Reyes FE, Polaski JT, Batey RT. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492:133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peselis A, Serganov A. Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nat Struct Mol Biol. 2012;19:1182–4. [DOI] [PubMed] [Google Scholar]
- 48.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, et al. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan CW, Mondragon A. Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding. Nucleic Acids Res. 2020;48:7569–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polaski JT, Webster SM, Johnson JE, Jr., Batey RT. Cobalamin riboswitches exhibit a broad range of ability to discriminate between methylcobalamin and adenosylcobalamin. J Biol Chem. 2017;292:11650–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan CW, Chetnani B, Mondragon A. Structure and function of the T-loop structural motif in noncoding RNAs. Wiley Interdiscip Rev RNA. 2013;4:507–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagaswamy U, Fox GE. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA. 2002;8:1112–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc. 2005;127:4223–31. [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson KA, Merino EJ, Weeks KM. Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–6. [DOI] [PubMed] [Google Scholar]
- 55.Choudhary PK, Sigel RK. Mg(2+)-induced conformational changes in the btuB riboswitch from E. coli. RNA. 2014;20:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schroeder GM, Dutta D, Cavender CE, Jenkins JL, Pritchett EM, Baker CD, et al. Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation. Nucleic Acids Res. 2020;48:8146–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gallo S, Mundwiler S, Alberto R, Sigel RK. The change of corrin-amides to carboxylates leads to altered structures of the B12-responding btuB riboswitch. Chem Commun (Camb). 2011;47:403–5. [DOI] [PubMed] [Google Scholar]
- 58.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8:339–43. [DOI] [PubMed] [Google Scholar]
- 59.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilbert SD, Batey RT. Monitoring RNA-ligand interactions using isothermal titration calorimetry. Methods Mol Biol. 2009;540:97–114. [DOI] [PubMed] [Google Scholar]
- 61.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeAbreu DM, Olive JE, Collins RA. Additional roles of a peripheral loop-loop interaction in the Neurospora VS ribozyme. Nucleic Acids Res. 2011;39:6223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersen AA, Collins RA. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc Natl Acad Sci U S A. 2001;98:7730–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DasGupta S, Suslov NB, Piccirilli JA. Structural Basis for Substrate Helix Remodeling and Cleavage Loop Activation in the Varkud Satellite Ribozyme. J Am Chem Soc. 2017;139:9591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouchard P, Legault P. A remarkably stable kissing-loop interaction defines substrate recognition by the Neurospora Varkud Satellite ribozyme. RNA. 2014;20:1451–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takechi S, Yasueda H, Itoh T. Control of ColE2 plasmid replication: regulation of Rep expression by a plasmid-coded antisense RNA. Mol Gen Genet. 1994;244:49–56. [DOI] [PubMed] [Google Scholar]
- 68.Siemering KR, Praszkier J, Pittard AJ. Mechanism of binding of the antisense and target RNAs involved in the regulation of IncB plasmid replication. J Bacteriol. 1994;176:2677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci U S A. 1996;93:5572–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shetty S, Kim S, Shimakami T, Lemon SM, Mihailescu MR. Hepatitis C virus genomic RNA dimerization is mediated via a kissing complex intermediate. RNA. 2010;16:913–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Greatorex J The retroviral RNA dimer linkage: different structures may reflect different roles. Retrovirology. 2004;1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klein DJ, Ferre-D’Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–6. [DOI] [PubMed] [Google Scholar]
- 75.Lipfert J, Sim AY, Herschlag D, Doniach S. Dissecting electrostatic screening, specific ion binding, and ligand binding in an energetic model for glycine riboswitch folding. RNA. 2010;16:708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heppell B, Blouin S, Dussault AM, Mulhbacher J, Ennifar E, Penedo JC, et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol. 2011;7:384–92. [DOI] [PubMed] [Google Scholar]
- 77.Choudhary PK, Gallo S, Sigel RK. Tb(3+)-Cleavage Assays Reveal Specific Mg(2+) Binding Sites Necessary to Pre-fold the btuB Riboswitch for AdoCbl Binding. Front Chem. 2017;5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holmstrom ED, Polaski JT, Batey RT, Nesbitt DJ. Single-molecule conformational dynamics of a biologically functional hydroxocobalamin riboswitch. J Am Chem Soc. 2014;136:16832–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lussier A, Bastet L, Chauvier A, Lafontaine DA. A kissing loop is important for btuB riboswitch ligand sensing and regulatory control. J Biol Chem. 2015;290:26739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polaski JT, Holmstrom ED, Nesbitt DJ, Batey RT. Mechanistic Insights into Cofactor-Dependent Coupling of RNA Folding and mRNA Transcription/Translation by a Cobalamin Riboswitch. Cell Rep. 2016;15:1100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2:1849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duconge F, Toulme JJ. In vitro selection identifies key determinants for loop-loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA. 1999;5:1605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paillart JC, Westhof E, Ehresmann C, Ehresmann B, Marquet R. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J Mol Biol. 1997;270:36–49. [DOI] [PubMed] [Google Scholar]
- 84.Baronti L, Karlsson H, Marusic M, Petzold K. A guide to large-scale RNA sample preparation. Anal Bioanal Chem. 2018;410:3239–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edwards AL, Garst AD, Batey RT. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Methods Mol Biol. 2009;535:135–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Price SR, Ito N, Oubridge C, Avis JM, Nagai K. Crystallization of RNA-protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J Mol Biol. 1995;249:398–408. [DOI] [PubMed] [Google Scholar]
- 87.Yamagami R, Sieg JP, Bevilacqua PC. Functional Roles of Chelated Magnesium Ions in RNA Folding and Function. Biochemistry. 2021;60:2374–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uhlenbeck OC. Keeping RNA happy. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- 89.Lai D, Proctor JR, Meyer IM. On the importance of cotranscriptional RNA structure formation. RNA. 2013;19:1461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scharfen L, Neugebauer KM. Transcription Regulation Through Nascent RNA Folding. J Mol Biol. 2021;433:166975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gong S, Wang Y, Wang Z, Zhang W. Co-Transcriptional Folding and Regulation Mechanisms of Riboswitches. Molecules. 2017;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boyle J, Robillard GT, Kim SH. Sequential folding of transfer RNA. A nuclear magnetic resonance study of successively longer tRNA fragments with a common 5’ end. J Mol Biol. 1980;139:601–25. [DOI] [PubMed] [Google Scholar]
- 93.Kramer FR, Mills DR. Secondary structure formation during RNA synthesis. Nucleic Acids Res. 1981;9:5109–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–75. [DOI] [PubMed] [Google Scholar]
- 95.Saldi T, Fong N, Bentley DL. Transcription elongation rate affects nascent histone pre-mRNA folding and 3’ end processing. Genes Dev. 2018;32:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saldi T, Riemondy K, Erickson B, Bentley DL. Alternative RNA structures formed during transcription depend on elongation rate and modify RNA processing. Mol Cell. 2021;81:1789–801 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci U S A. 1999;96:9545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci U S A. 2007;104:17995–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leamy KA, Assmann SM, Mathews DH, Bevilacqua PC. Bridging the gap between in vitro and in vivo RNA folding. Q Rev Biophys. 2016;49:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ganser LR, Kelly ML, Herschlag D, Al-Hashimi HM. The roles of structural dynamics in the cellular functions of RNAs. Nat Rev Mol Cell Biol. 2019;20:474–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chillon I, Marcia M, Legiewicz M, Liu F, Somarowthu S, Pyle AM. Native Purification and Analysis of Long RNAs. Methods Enzymol. 2015;558:3–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suslov NB, DasGupta S, Huang H, Fuller JR, Lilley DM, Rice PA, et al. Crystal structure of the Varkud satellite ribozyme. Nat Chem Biol. 2015;11:840–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Batey RT. Advances in methods for native expression and purification of RNA for structural studies. Curr Opin Struct Biol. 2014;26:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dethoff EA, Weeks KM. Effects of Refolding on Large-Scale RNA Structure. Biochemistry. 2019;58:3069–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perdrizet GA 2nd, Artsimovitch I, Furman R, Sosnick TR, Pan T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc Natl Acad Sci U S A. 2012;109:3323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes Dev. 2009;23:2650–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wong TN, Pan T. RNA folding during transcription: protocols and studies. Methods Enzymol. 2009;468:167–93. [DOI] [PubMed] [Google Scholar]
- 109.Polaski JT, Kletzien OA, Drogalis LK, Batey RT. A functional genetic screen reveals sequence preferences within a key tertiary interaction in cobalamin riboswitches required for ligand selectivity. Nucleic Acids Res. 2018;46:9094–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Scheitl CPM, Ghaem Maghami M, Lenz AK, Hobartner C. Site-specific RNA methylation by a methyltransferase ribozyme. Nature. 2020;587:663–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Deng J, Wilson TJ, Wang J, Peng X, Li M, Lin X, et al. Structure and mechanism of a methyltransferase ribozyme. Nat Chem Biol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang HY, Gao YQ, Zhang L, Chen DR, Gan JH, Murchie AIH. The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences. Nat Catal. 2021;4:872-+. [Google Scholar]
- 114.Scheitl CPM, Mieczkowski M, Schindelin H, Hobartner C. Structure and mechanism of the methyltransferase ribozyme MTR1. Nat Chem Biol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Flemmich L, Heel S, Moreno S, Breuker K, Micura R. A natural riboswitch scaffold with self-methylation activity. Nat Commun. 2021;12:3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9:339–45. [DOI] [PubMed] [Google Scholar]
- 118.Gagnon P, Persic S, Goricar B, Cernigoj U, Strancar A. A new runway for purification of messenger RNA. BioProcess International. 2020;18:36–45. [Google Scholar]
- 119.Fowler CC, Brown ED, Li Y. Using a riboswitch sensor to examine coenzyme B(12) metabolism and transport in E. coli. Chem Biol. 2010;17:756–65. [DOI] [PubMed] [Google Scholar]
- 120.Fowler CC, Sugiman-Marangos S, Junop MS, Brown ED, Li Y. Exploring intermolecular interactions of a substrate binding protein using a riboswitch-based sensor. Chem Biol. 2013;20:1502–12. [DOI] [PubMed] [Google Scholar]
- 121.Braselmann E, Wierzba AJ, Polaski JT, Chrominski M, Holmes ZE, Hung ST, et al. A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat Chem Biol. 2018;14:964–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tickner ZJ, Farzan M. Riboswitches for Controlled Expression of Therapeutic Transgenes Delivered by Adeno-Associated Viral Vectors. Pharmaceuticals (Basel). 2021;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Finke M, Brecht D, Stifel J, Gense K, Gamerdinger M, Hartig JS. Efficient splicing-based RNA regulators for tetracycline-inducible gene expression in human cell culture and C. elegans. Nucleic Acids Res. 2021;49:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sporing M, Boneberg R, Hartig JS. Aptamer-Mediated Control of Polyadenylation for Gene Expression Regulation in Mammalian Cells. ACS Synth Biol. 2020;9:3008–18. [DOI] [PubMed] [Google Scholar]
- 125.Strobel B, Duchs MJ, Blazevic D, Rechtsteiner P, Braun C, Baum-Kroker KS, et al. A Small-Molecule-Responsive Riboswitch Enables Conditional Induction of Viral Vector-Mediated Gene Expression in Mice. ACS Synth Biol. 2020;9:1292–305. [DOI] [PubMed] [Google Scholar]
- 126.Mustafina K, Fukunaga K, Yokobayashi Y. Design of Mammalian ON-Riboswitches Based on Tandemly Fused Aptamer and Ribozyme. ACS Synth Biol. 2020;9:19–25. [DOI] [PubMed] [Google Scholar]
- 127.Vogel M, Weigand JE, Kluge B, Grez M, Suess B. A small, portable RNA device for the control of exon skipping in mammalian cells. Nucleic Acids Res. 2018;46:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]


