Abstract
Purpose:
Recently, deep learning-based methods have been established to denoise the low-count PET images and predict their standard-count image counterparts, which could achieve reduction of injected dosage and scan time, and improve image quality for equivalent lesion detectability and clinical diagnosis. In clinical settings, the majority scans are still acquired using standard injection dose with standard scan time. In this work, we applied a 3D U-Net network to reduce the noise of standard-count PET images to obtain the virtual-high-count (VHC) PET images for identifying the potential benefits of the obtained VHC PET images.
Methods:
The training datasets, including down-sampled standard-count PET images as the network input and high-count images as the desired network output, were derived from 27 whole-body PET datasets, which were acquired using 90-minute dynamic scan. The down-sampled standard-count PET images were rebinned with matched noise level of 195 clinical static PET datasets, by matching the normalized standard derivation (NSTD) inside 3D liver region of interests (ROIs). Cross-validation was performed on 27 PET datasets. Normalized mean square error (NMSE), peak signal to noise ratio (PSNR), structural similarity index (SSIM), and standard uptake value (SUV) bias of lessons were used for evaluation on standard-count and VHC PET images, with real-high-count PET image of 90 minutes as the gold standard. In addition, the network trained with 27 dynamic PET datasets was applied to 195 clinical static datasets to obtain VHC PET images. The NSTD and mean/max SUV of hypermetabolic lesions in standard-count and virtual high-count PET images were evaluated. Three experienced nuclear medicine physicians evaluated the overall image quality of randomly selected 50 out of 195 patients’ standard-count and VHC images and conducted 5-score ranking. A Wilcoxon signed-rank test was used to compare differences in the grading of standard-count and VHC images.
Results:
The cross-validation results showed that VHC PET images had improved quantitative metrics scores than the standard-count PET images. The mean/max SUVs of 35 lesions in the standard-count and true-high-count PET images did not show significantly statistical difference. Similarly, the mean/max SUVs of VHC and true-high-count PET images did not show significantly statistical difference. For the 195 clinical data, the VHC PET images had a significantly lower NSTD than the standard-count images. The mean/max SUVs of 215 hypermetabolic lesions in the VHC and standard-count images showed no statistically significant difference. In the image quality evaluation by three experienced nuclear radiologists, standard-count images and VHC images received scores with mean and standard deviation of 3.34±0.80 and 4.26±0.72 from Physician 1, 3.02±0.87 and 3.96±0.73 from Physician 2, and 3.74±1.10 and 4.58±0.57 from Physician 3, respectively. The VHC images were consistently ranked higher than the standard-count images. The Wilcoxon signed-rank test also indicated that the image quality evaluation between standard-count and VHC images had significant difference.
Conclusions:
A deep learning method is proposed to convert the standard-count images to the VHC images. The VHC images have reduced noise level. No significant difference in mean/max SUV to the standard-count images is observed. VHC images improve image quality for better lesion detectability and clinical diagnosis.
I. Introduction
Positron emission tomography (PET) is a nuclear medicine imaging modality that produces 3D in-vivo observation of the functional process in the body. It has been widely applied in clinic for the diagnosis of various diseases in oncology1, neurology2,3 and cardiology4,5,6.
In spite of the immediate benefits of PET, the injection of radioactive tracer exposes patients and healthcare providers to ionizing radiation. Therefore, the dose needs to be kept as low as reasonably achievable (ALARA) in clinical practice7. However, reducing the injection dose could reduce the number of detected photons. This further compromises PET image quality with increased image noise and decreased signal-to-noise ratio (SNR), subsequently affecting diagnostic efficacy and quantitative accuracy, as noise could easily results in bias of mean and maximum standard uptake value (SUVmean and SUVmax)8. In oncological PET, higher image noise can cause substantial bias on SUVmean and overestimation of SUVmax, causing lesion quantification errors9,10. The same challenge of increased noise also applies to data of shortened scan time and obese patients.
To reduce PET image noise, numerous within-reconstruction and post-processing approaches have been developed. Many penalized likelihood reconstruction approaches have been proposed to incorporate a prior distribution of the image, such as total variation11, and entropy12. However, the optimization function and hyper-parameter tuning are difficult to choose in different applications. Among post-processing PET image denoising approaches, Gaussian filtering is the most commonly used denoising technique but with the cost of image blurring or loss of details. Many alternatives, such as non-local mean (NLM) filter with or without anatomical information13,14, block-matching 3D filters15, curvelet and wavelet16,17,18, have been proposed to provide a significant reduction of noise while preserving important structures.
Recently, artificial intelligence (AI) using deep learning (DL) have been established to convert the low-count PET images to their standard-count image counterparts19. These methods usually employed a deep convolutional neural network (CNN) with a supervised training scheme using low-count and standard-count PET image pairs as training data. Such image pairs can be generated by performing rebinning on the standard-count list-mode data with a specific down-sampling rate. A variety of network architectures have been utilized, such as deep auto-context CNN20, encoder-decoder residual deep network21, U-Net network22,23,24,25,26, generative adversarial network (GAN)27,28,29,30,31,32,33,34. Unsupervised PET denoising methods have also been explored in the scenario of obtaining clean PET images infeasible due to the injection dose limits, body motions, or short half-life of radio-tracer35,36,37,38,39.
Studies have demonstrated that the deep learning-based methods outperformed the than the state-of-the-art conventional denoisers and the AI-enhanced low-count PET images have reduced noise level and increased contrast-to-noise ratio when compared with their low-count counterparts. In40,41,42,43,44,45,46,47, they reported that the AI-enhanced low-count PET images have equivalent lesion detectability and clinical diagnosis as the standard-count PET images. For instance, Xu et al.46 used an FDA-cleared AI-based proprietary software package SubtlePET™ to enhance the low-dose noisy PET images of lymphoma patients and reported that the standard-count and AI-enhanced low-count PET images led to similar lymphoma staging outcomes based on the assessments by two nuclear radiologists. Katsari et al.47 also used the same software package to process 66%-count PET images. Their results showed that there was no significant difference between the AI-enhanced low-dose images and standard-count images in terms of lesion detectability and SUV-based lesion quantification.
While most prior studies denoised low-count PET images to the level of standard-count PET images, to our best knowledge, there is no study on noise reduction of standard-count PET images to obtain virtual-high-count (VHC) PET images. The primary obstacle is the challenge of obtaining high-count PET images as training label, as such high-count images can only be obtained by injecting more radiotracer into patient that is not ethical, scanning longer that is not clinically feasible, or scan using ultra-high-sensitivity scanner that is not widely available. In this paper, we took advantage of 90-minute dynamic PET data and utilized a deep learning-based method to obtain virtual-high-statistics PET images from the typical clinical standard-count PET images. The obtained VHC PET images were evaluated in terms of the change of noise and SUVs, to identify the benefits of the obtained VHC PET images.
II. Materials and Methods
II.A. Study Datasets
Two groups of human PET datasets were used in this study.
For network training, we included data of 27 subjects (female: 10, male: 17, age: 56±14 years) who underwent 18F-FDG PET/CT scan on a Siemens Biograph mCT scanner at Yale PET Center. The injected dose ranged from 255.7 MBq to 380.4 MBq, with mean and std 334.5±32.1 MBq. The patients’ body mass index (BMI) ranged from 21.7 kg· m2 to 37.2 kg· m2. Each subject underwent a continuous-bed-motion (CBM) 19 passes dynamic whole-body scan of 90 minutes immediately following tracer injection. Although the data were acquired for the dynamic study, we generated a high-count prompt sinogram by combining the list-mode data of all passes. The images were reconstructed with the Siemens E7 tool using the time-of-flight (TOF) ordered subset expectation maximization (OSEM) algorithm (2 iterations and 21 subsets) with corrections for random, attenuation, scatter, normalization, and decay. The voxel size was 4.0×4.0×3.0 mm3. Gaussian filter with a 5mm 3D filter kernel was applied. The reconstructed images were converted to SUV images based on the injected dose and patient weight.
For evaluation, we included 195 (male 122, female 73) standard-count clinical 18F-FDG datasets acquired on a Siemens Biograph mCT PET/CT scanner at Yale-New Haven Hospital. The data were acquired with single pass using continuous bed motion. The targeted post-injection times for clinical scans was 60 minutes. The injected dose ranges from 303.4 MBq to 436.6 MBq, with mean and standard deviation of 370.0±29.6 MBq. Table.1 outlines the clinical characteristics of the patients, including age, weight, height, and BMI. The images were reconstructed using the same reconstruction settings as above, using the Siemens E7 tool with the TOF OSEM algorithm (2 iterations and 21 subsets), voxel size 4.0×4.0×3.0 mm3 and a 5-mm 3D Gaussian post-filtering.
Table 1:
The age, height, weight, and BMI distribution of the 195 patients enrolled in clinical PET/CT scan.
| Age (year) | Weight (kg) | Height (m) | BMI (kg·m2) | |
|---|---|---|---|---|
| Range | 22.0–88.1 | 37.6–178.4 | 1.42–1.91 | 10.9–51.9 |
| Mean±Std | 63.9±14.2 | 77.3±22.9 | 1.67±0.1 | 27.8±6.9 |
II.B. Count Level Estimation
For network training, it is important to match the noise level of training input images with the noise level of the clinical PET data to achieve the best network performance48. We used the normalized standard derivation (NSTD) inside a spherical region of interest (ROI) with radii of 4×4×5 voxels in healthy liver parenchyma as the metric to measure the image noise. It is defined as
| (1) |
where Ij is the value at voxel j, is the mean value of the ROI, and n is the number of voxels in the ROI.
The NSTD of the 195 clinical PET/CT scans was calculated as a surrogate for the input count level of the standard-count PET image in the training. Then, we rebinned the total list-mode data of all passes in the 27 multi-pass dynamic PET datasets to get the down-sampled list-mode data, which matched the count level of the 195 clinical data based on the Poisson thinning process that discards coincidence events randomly by specific down-sampling factors. With the optimized down-sampling ratios, the standard-count PET images of the 27 multi-pass PET dynamic datasets were generated.
NSTD in the predefined liver ROI was calculated with the 195 clinical PET/CT scans to estimate the count level distribution of clinical standard-count PET images. Due to the variation in patients’ weight and injected dose, the noise level of clinical standard-count PET images usually varies largely among individuals. Based on the NSTD distribution, the noise level ranges from 0.052 to 0.380 with mean and standard deviation of 0.099±0.037. Fig.1(a) shows that the majority of NSTD values range from 0.06 to 0.12 (85%). Since there were only 27 multi-pass dynamic datasets, using a single down-sampling rate was not enough to match the noise level distribution of the clinical data. Therefore, we used the down-sampling rates of 20%, 30%, and 40% to rebin each 90-minute dynamic PET datasets. Therefore, each multi-pass dynamic dataset have three different down-sampled standard-count PET images. The obtained rebinned 90-minute dynamic datasets have a NSTD distribution which is close to that of 195 clinical data. Fig.1(a) shows that the histogram of NSTD for the down-sampled standard-count images derived from the multi-pass dynamic datasets matched that for the standard-count clinical data. Fig.1(b) displays a representative coronal slice for a standard-count clinical data, a down-sampled standard-count data and its corresponding true-high-count PET image.
Figure 1:
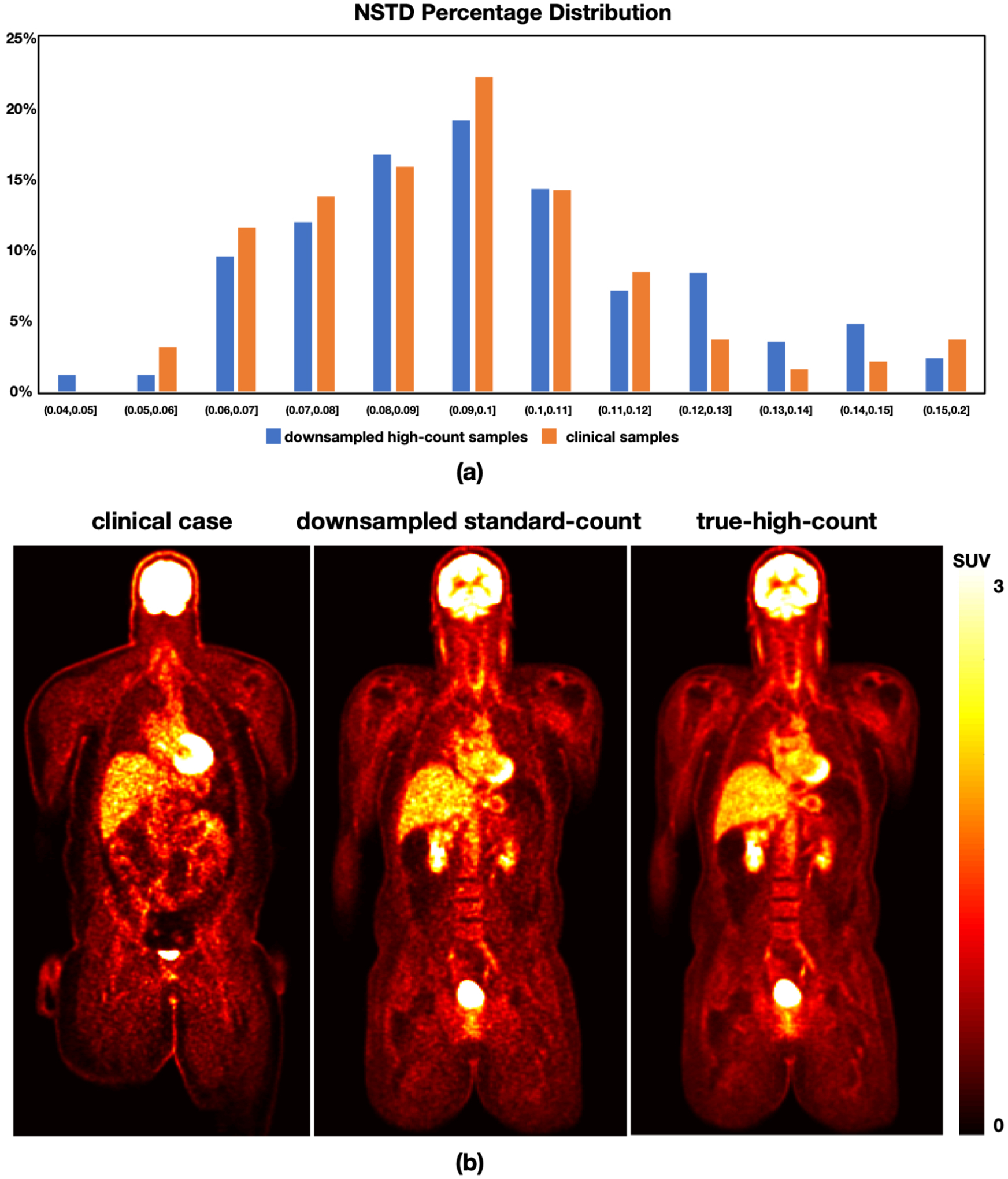
(a) The histogram of NSTD in the rebinned multi-pass dynamic data and 195 clinical data. (b) Visualization of a clinical standard-count image (left) from a sample patient (male, BMI 40.2 kg· m2). A down-sampled standard-count image (middle) and the corresponding true-high-count image (right) from a sample subject (male, BMI 30.8 kg· m2) from the multi-pass dynamic datasets are provided as well.
II.C. Network Architecture and Training
A 3D U-Net network architecture was used. The network was trained using the pairs of standard-count PET images (input) and high-count PET images (label) from the 27 dynamic datasets. Note that the whole-body PET images often show high inter-patient and intra-patient uptake variation, and the tracer concentration is much higher in the bladder than anywhere else. This could decrease the relative contrast among other organs and introduce difficulties in deep network training. Therefore, we used ITK’s ConnectedThresholdImageFilter49 to segment the bladder with seeds (SUV>20) and a lower threshold value of 3 and replace the segmented voxels with the value of 3, which is applied in inference as well.
During training, 64×64×64 image patches were randomly cropped by a data generator in real time to feed to network training. Random flipping along three axes was applied for data augmentation. Each epoch contained 100 steps with a mini-batch size of 16, resulting in 1600 patches used in each epoch. L2 loss was used, and Adam optimizer was applied with an initial learning rate of 0.0001. The learning rate exponentially decayed every 100 steps with a decay rate of 0.996. In inference, the trained network model takes the full image to get the VHC images. The code was implemented with TensorFlow 2.2.
II.D. Evaluation
3-fold cross-validation on the 27 dynamic PET datasets was applied with the standard-count and true-high-count PET images from 18 subjects used for network training and the remaining 9 datasets used for testing.
For quantitative evaluation, we used normalized root mean square error (NRMSE), peak signal to noise ratio (PSNR), and structural similarity index (SSIM) with the true-high-count PET image as the reference. NRMSE is widely used to quantify the differences between the predicted and the reference, which is defined as
| (2) |
where is the approximation estimation, x is the reference or ground-truth, N is the number of voxels in x, and μx is the average of x.
PSNR is the ratio between the maximum possible power of a signal and the power of corrupting noise that affects the fidelity of its representation.
| (3) |
where MAX is the maximum value of x.
SSIM is a combined measure obtained from three complementary components (luminance similarity, contrast similarity, and structural similarity) that better reflects the visual similarity of a representation to the reference.
| (4) |
where , ux is the average of and x respectively, and σx is the variance of and x respectively, is the covariance of and x, c1 and c2 are two variables to stabilize the division with weak denominator.
In the 27 dynamic PET datasets, 35 lesions were delineated by one experienced physician. The mean/max SUVs of the lesion ROIs were calculated in the standard-count, VHC, and true-high-count PET images.
In addition, another network was trained using all the 27 dynamic PET datasets as training data. The trained network was applied on the 195 clinical PET datasets to obtain their VHC images. In the 195 standard-count clinical images, 215 hypermetabolic lesions were manually segmented using the ITK-SNAP50. NSTD of the liver ROIs were calculated in the standard-count and VHC images for measuring the noise level changes. In addition, mean/max SUVs of the hypermetabolic lesions were obtained for quantitative evaluation. An in-house software was developed with the feature of sagittal, coronal and axial image visualization for image evaluation study. Three experienced nuclear medicine physicians evaluated the image quality of the standard-count and VHC images according to overall quality, image sharpness and noise. The images were ranked as 1 - very poor/non-diagnostic, 2 - poor, 3 - moderate, 4 - good, and 5 - excellent. 50 of the 195 patients were randomly chosen for evaluation and the selected 50 standard-count and 50 VHC images were assessed at a random order. The physicians were instructed to score the images based on the overall diagnostic quality. Because the data were not normally distributed, the Wilcoxon signed-rank test was performed to compare image quality grading of standard-count and VHC images for each physician.
III. Results
III.A. Cross-validation using 90-min Data
Table.2 lists the PSNR, NRMSE, and SSIM of the standard-count and VHC PET images with respect to the true-high-count images using cross-validation. The results show that the VHC images achieve superior NRMSE, PSNR and SSIM compared to the standard-count images.
Table 2:
Quantitative evaluation on the standard-count and virtual-high-count images with respect to the true-high-count images in terms of PSNR, NRMSE, and SSIM.
| NRMSE (%) | PSNR (dB) | SSIM (0–1) | |
|---|---|---|---|
| Standard-count | 7.84±2.77 | 35.24±2.88 | 0.990±0.005 |
| Virtual-high-count | 6.64±2.12 | 36.56±2.51 | 0.994±0.002 |
Fig.2 shows the standard-count, VHC, and true-high-count images for two sample datasets. The VHC images are less noisy compared to the standard-count counterparts and the fine structures are well-preserved. Fig.3 plots the line profiles in the standard-count, VHC, and high-count images. For Subject 1, the three profiles are close. For Subject 2, the line profile associated with the VHC image is closer to that of the true-high-count image, compared to the profile associated with the standard count image.
Figure 2:
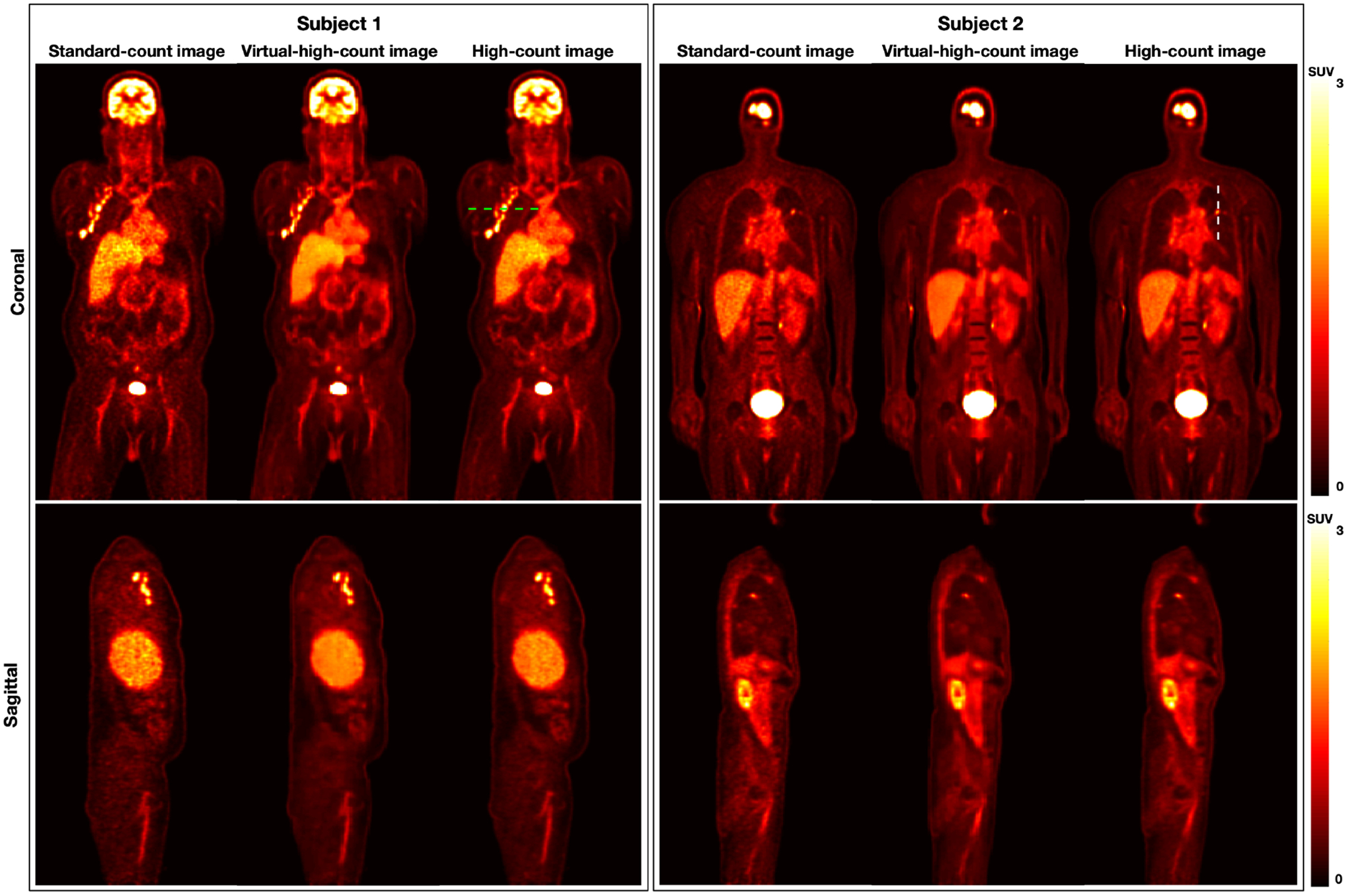
Visualization of standard-count, virtual-high-count, and true-high-count images from two sample subjects from the 90-min multi-pass dynamic datasets. The standard-count images are nosier than the true-high-count and predicted virtual-high-count images. Subject1 is male with a BMI of 32.3 kg· m2, while Subject2 is male with a BMI of 22.4 kg· m2.
Figure 3:
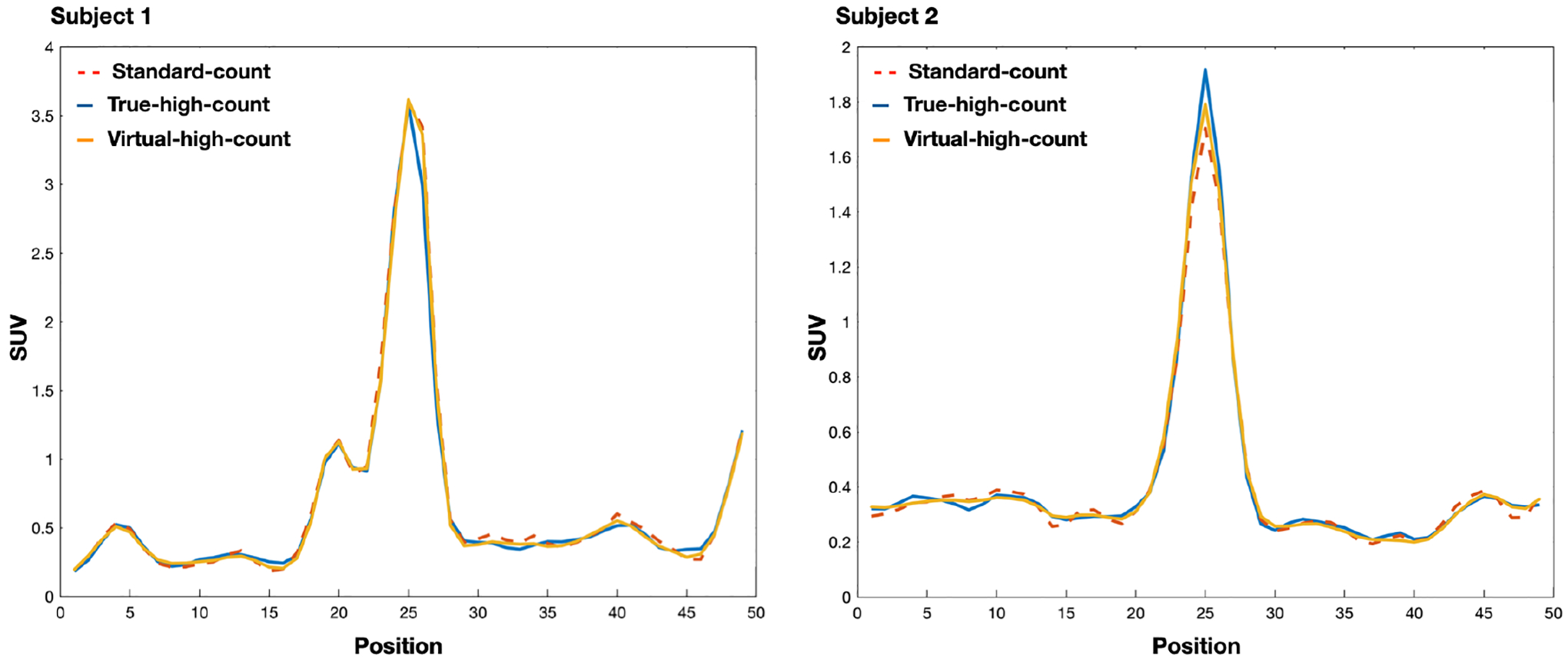
Comparison of the image profiles in Fig.2. The image profiles show that the standard-count, virtual-high-count, and true-high-count images are close.
Fig.4 plots the correlation between the true-high-count images and standard-count/VHC images in terms of the mean/max SUVs within ROIs. In the correlation plot of mean SUV between the true-high-count images and standard-count/VHC images, the distribution closely aligns with the line of identity, resulting in high correlation coefficients and R2 values. It is observed that in the correlation plot of max SUV between the true-high-count images and standard-count images, the distribution is slightly off the line of identity.
Figure 4:
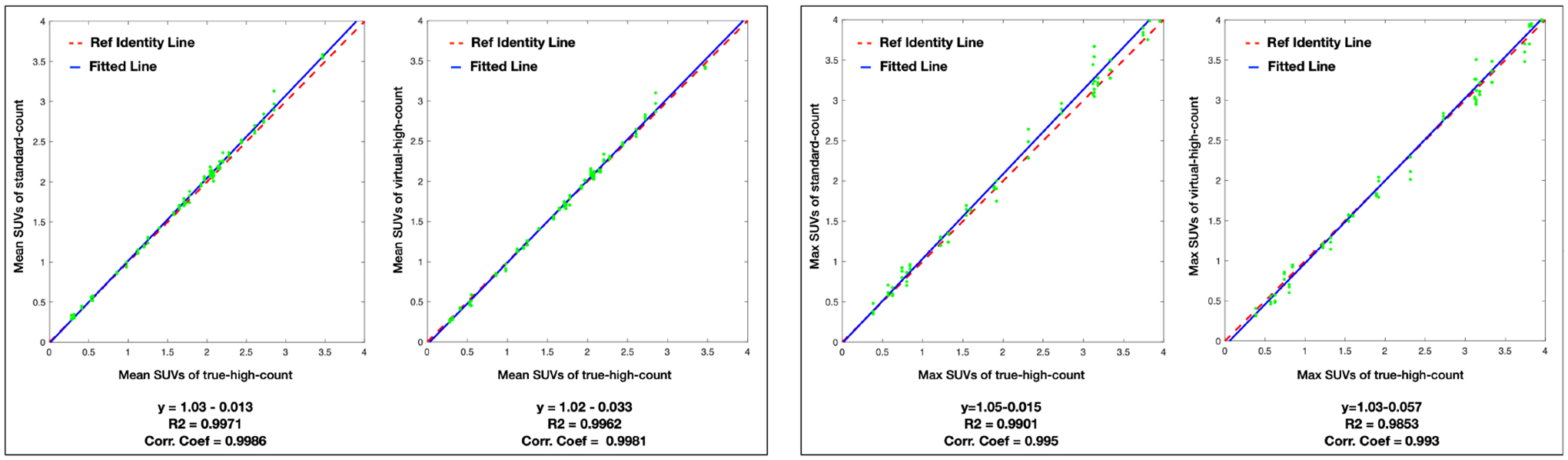
The correlation and linear fitting of ROI mean/max SUVs between the true-high-count and the standard-count/predicted virtual-high-count images. The fitted function, coefficient of determination (R2), and correlation coefficients (Corr. Coef.) are listed below. The red dash line is the line of identity, and the blue line is the fitted line.
The mean and standard deviation of the mean/max SUVs in the standard-count, VHC, and true-high-count images are 1.71±0.79 / 3.26±1.80, 1.68±0.79 / 3.15±1.77, and 1.68±0.77 / 3.11±1.70, respectively. A paired two-sample t-test is conducted to determine whether the mean/max SUVs in the standard-count, VHC, and true-high-count images are statistically different. Between the standard-count and true-high-count PET images, the p values are 0.75 and 0.56 for the mean and max SUVs, respectively. Between the VHC and true-high-count PET images, the p values are 0.96 and 0.90 for the mean and max SUVs, respectively. The results show that the mean/max SUVs in the standard-count, VHC, and true-high-count PET images are not significantly different.
III.B. Evaluation on Clinical Data
Fig.5 displays the standard-count and VHC PET images for three sample clinical datasets. The virtual high-count image achieves a lower noise level as compared to the standard-count image for all three patients. The virtual high-count images for patient 1 show substantial reduction of noise when the standard-count images have high noise level. For patient 2 and 3 with visually lower noise level than patient 1, the virtual high-count images preserve the fine structures.
Figure 5:
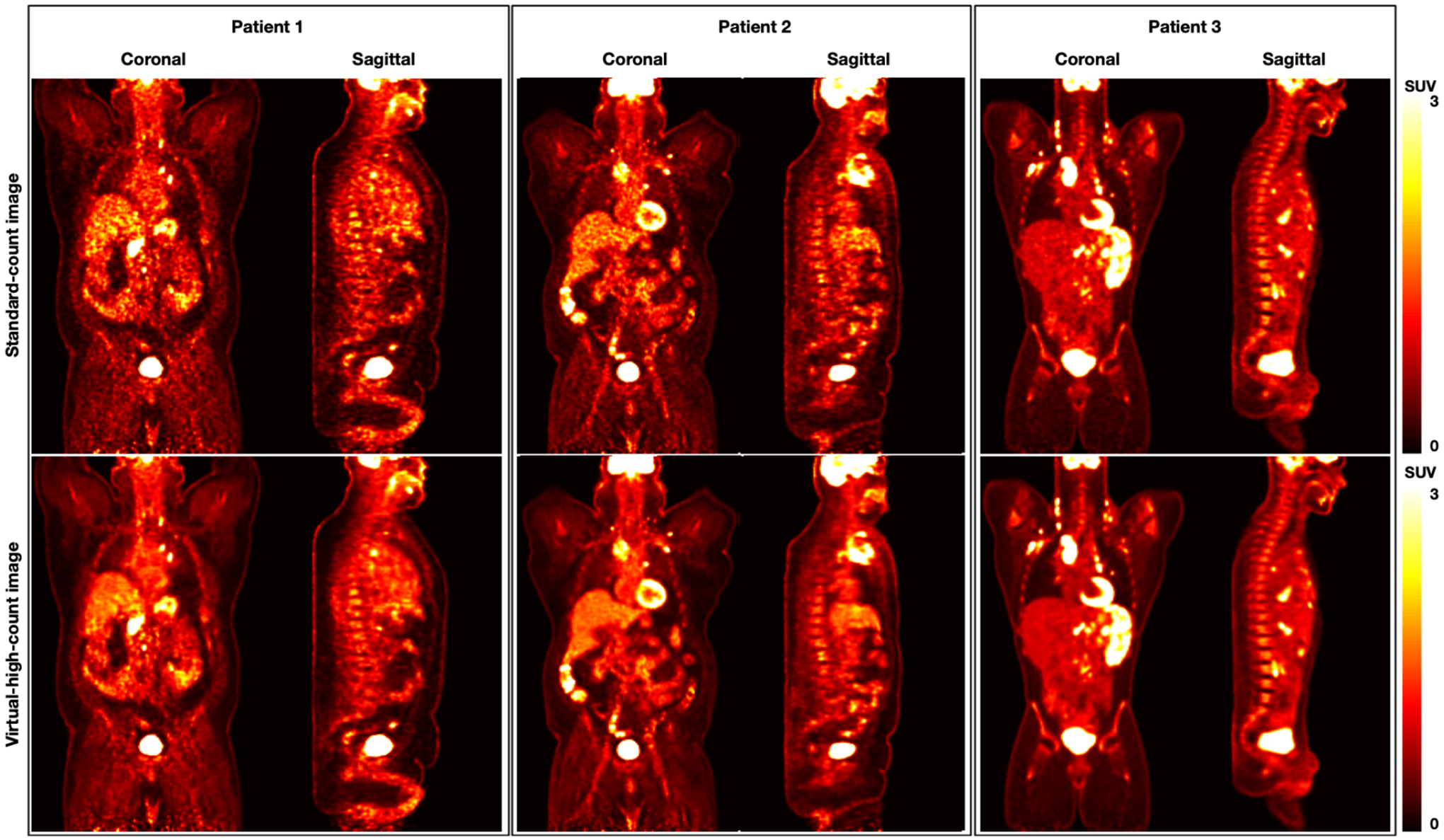
Visualization of the standard-count and virtual-high-count images from three sample patients (Patient 1: male, BMI 48.1 kg· m2; Patient 2: male, BMI 37.9 kg· m2; Patient 3: male, BMI 26.7 kg· m2).
Fig.6 plots the correlation and linear fitting of NSTD and mean/max SUVs of 215 hypermetabolic lesions between the standard-count and VHC clinical images for the 195 clinical data. The NSTD values are scattered below the line of identity, meaning that the VHC images have decreased NSTD as compared with that of the stand-count images. The mean/max SUVs of the VHC images closely align with the line of identity, yielding high correlation coefficients. In the correlation plot of max SUV, the distribution is slightly below the line of identity, especially for high max SUVs (> 1.5).
Figure 6:
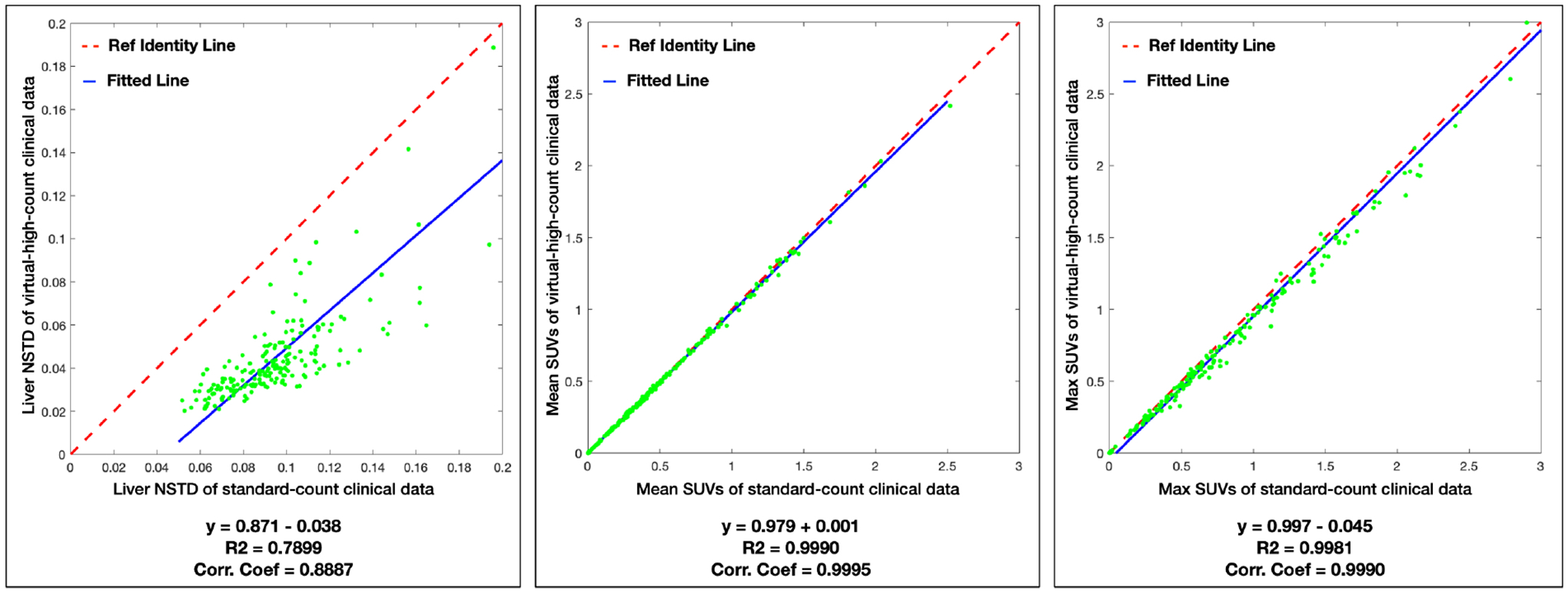
The correlation and linear fitting of NSTD and mean/max SUVs of 215 hypermetabolic lesions between the standard-count and predicted virtual-high-count images for the 195 clinical data. The fitted function, coefficient of determination (R2), and correlation coefficients (Corr. Coef.) are listed below. The red dash line is the line of identity, and the blue line is the fitted line.
The VHC images have reduced NSTD with mean and standard deviation of 0.10±0.04, compared with 0.05±0.04 for the stand-dose images. For the mean/max SUVs, the mean and standard deviation are 0.50±0.45 / 0.99±1.39 in the standard-count images and 0.49±0.44 / 0.94±1.39 in the VHC images. To determine whether the mean/max SUVs in the standard-count and VHC images are statistically different, a paired two-sample t test is conducted. The null hypothesis is that the distribution means are not statistically different when the significance value p > 0.05. The p values are 0.84 and 0.72 respectively for the mean and max SUVs. This means that there is no significant statistically difference in the mean and max SUVs between the standard-count and VHC PET images.
Three experienced nuclear medicine physicians completed the image evaluation task. The mean and standard deviation of scores of 50 standard-count images and 50 VHC images are are 3.34±0.80 and 4.26±0.72 for Physician 1, 3.02±0.87 and 3.96±0.73 for Physician 2, 3.74±1.10 and 4.58±0.57 for Physician 3. The VHC images were consistently ranked higher than the standard-count images across three physicians. The VHC images have significant improved ranking scores of 0.92, 0.92, and 0.84 for Physician 1, 2, 3 respectively. Fig.7 plots the evaluation ranking results, which clearly shows that the VHC images received higher ranking scores than the standard-count images. The Wilcoxon signed rank test led to p-values of 7e-8, 9.3e-8, and 4.9e-4 for Physician 1, 2, 3, respectively. This indicates that the image quality evaluation between standard-count and VHC images has substantial statistically difference.
Figure 7:
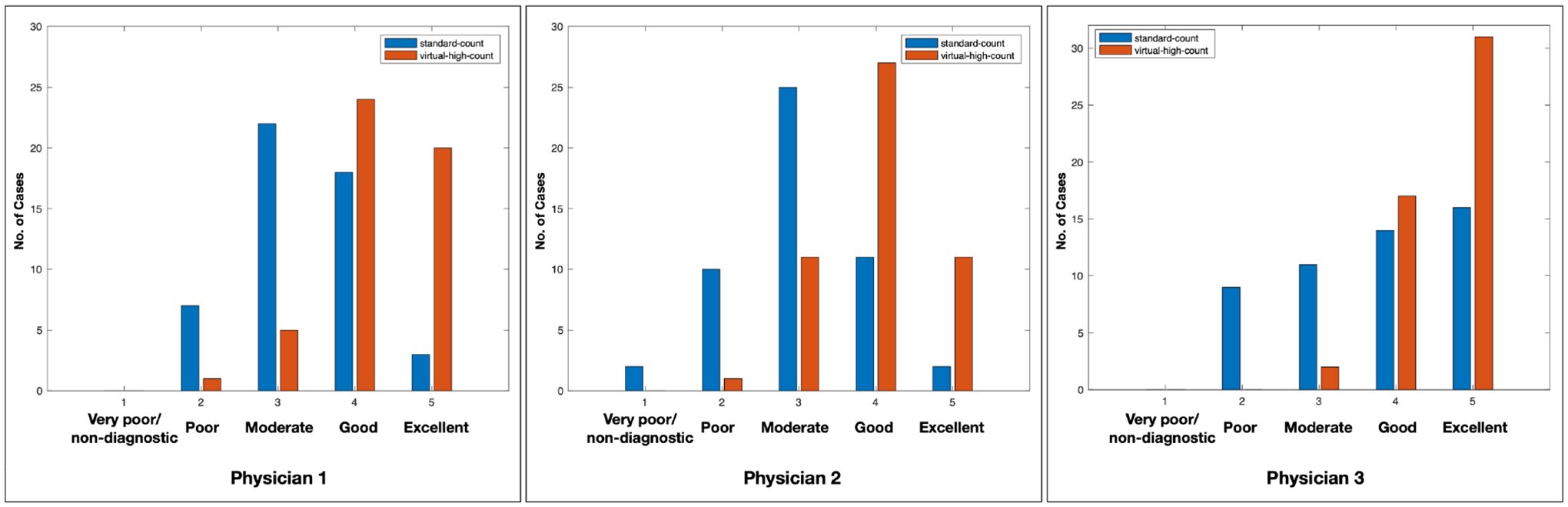
The evaluation scores of standard-count and virtual-high-count images from three nuclear medicine physicians. Three nuclear medicine physicians gave higher scores to the virtual-high-count images than standard-count images.
IV. Discussion
In this work, we used a deep learning method for VHC PET image generation. To get the standard-count and true-high-count PET image pairs for network training, we used 27 datasets with 90min multi-pass dynamic scans. The true-high-count images were reconstructed from the prompt sinogram combining list-mode data of all passes. The NSTD of a predefined liver ROI was calculated and served as a surrogate for the count level. The multi-pass dynamic datasets were then rebinned based on the NSTD to generate down-sampled images with matched count level distribution to that of a selection of 195 clinical data. Comparing with previous deep learning methods that predicted the standard-count PET images from low-dose PET images, our method utilized the multi-pass dynamic datasets to translate the standard-count PET images to VHC PET images.
We found that the virtual high-count PET images have decreased noise level and increased image quality. The results showed that the mean/max SUVs derived from the virtual high-count PET images are not significantly different from those derived from the standard-count or true-high-count PET images. Based on visual assessment, the VHC images have better SNR and well-preserved fine details. From the overall image assessments by three experienced nuclear medicine physicians, the VHC images received higher evaluation scores than the standard-count images. In future studies, the lesion detectability observer study based on lesion detection and localization is needed to better understand the benefits of VHC images.
In this study, we focused on noise reduction of standard-count PET data. Existing deep learning-based PET denoising approaches, including SubtlePET™, can only denoise low-count images to the level of standard count PET images. In contrast, the biggest innovation and advantage of our proposed technology is that we are uniquely capable to denoise the standard-count images to the quality of high-count data, by utilizing various sources of high-count PET images as training that are acquired through long scans (90 minutes). 195 clinical PET/CT scans were used to get the count level distribution of standard-count clinical PET images. However, the count level distribution of the selected 195 clinical PET/CT data might not fully represent the count level distribution of all clinical PET/CT scans. In addition, since only 27 dynamic datasets were used for network training, there is a risk of low diversity in the training data which could in turn introduce overfitting, biased model evaluation, and generalization problems to the network. Therefore, the patch-based training and random flipping were used in the network training to eliminate the influence of limited training data. In addition, since the current network was trained using 18F-FDG dynamic whole-body PET/CT scan datasets, its generalization ability to different scanners, patient populations, radio-tracers was unknown and it is desirable to include a wide spectrum of training data to improve the model generalizability.
In the multi-pass dynamic data image reconstruction, the intra- and inter- pass motion effects were not corrected. This could introduce motion artifacts in the reconstructed PET images. However, since the Poisson thinning process discards coincidence events randomly, the true-high-count and down-sampled standard-count PET images have similar levels of motion corruptions. Therefore, the uncorrected motion should have minimal effects on the deep learning based VHC PET image generation presented in this study.
V. Conclusion
We have developed a deep convolutional neural network that is able to generate VHC PET images from clinical standard-count PET images. The U-Net was well-trained by the count-level-controlled long dynamic PET datasets, meaning that the count level of the input images for training matched that of the clinical standard-count PET images. The results showed that the VHC PET images can effectively reduce the noise, while introducing negligible biases to the mean/max SUVs. Such findings were confirmed by three experienced nuclear medicine physicians.
Supplementary Material
Acknowledgement
This work is supported by NIH grant R01EB025468.
Footnotes
Conflict of Interest
The authors have no conflicts to disclose.
References
- 1.Rohren EM, Turkington TG, and Coleman RE, Clinical applications of PET in oncology, Radiology 231, 305–332 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Politis M and Piccini P, Positron emission tomography imaging in neurological disorders, Journal of neurology 259, 1769–1780 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Schöll M, Damián A, and Engler H, Fluorodeoxyglucose PET in neurology and psychiatry, PET clinics 9, 371–390 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Takalkar A, Mavi A, Alavi A, and Araujo L, PET in cardiology, Radiologic Clinics 43, 107–119 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Gould KL, PET perfusion imaging and nuclear cardiology, Journal of Nuclear Medicine 32, 579–606 (1991). [PubMed] [Google Scholar]
- 6.Schindler TH, Schelbert HR, Quercioli A, and Dilsizian V, Cardiac PET imaging for the detection and monitoring of coronary artery disease and microvascular health, JACC: Cardiovascular Imaging 3, 623–640 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Strauss KJ and Kaste SC, The ALARA (as low as reasonably achievable) concept in pediatric interventional and fluoroscopic imaging: striving to keep radiation doses as low as possible during fluoroscopy of pediatric patients—a white paper executive summary, Radiology 240, 621–622 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Alessio A, Pierce L, Thielemans K, Wollenweber S, Ganin A, and Kinahan P, Quiescent period respiratory gating for PET/CT, Medical physics 37, 5037–5043 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinahan PE and Fletcher JW, Positron emission tomography-computed tomography standardized uptake values in clinical practice and assessing response to therapy, in Seminars in Ultrasound, CT and MRI, volume 31, pages 496–505, Elsevier, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riddell C, Carson RE, Carrasquillo JA, Libutti SK, Danforth DN, Whatley M, and Bacharach SL, Noise reduction in oncology FDG PET images by iterative reconstruction: a quantitative assessment, Journal of Nuclear Medicine 42, 1316–1323 (2001). [PubMed] [Google Scholar]
- 11.Rudin LI, Osher S, and Fatemi E, Nonlinear total variation based noise removal algorithms, Physica D: nonlinear phenomena 60, 259–268 (1992). [Google Scholar]
- 12.Somayajula S, Panagiotou C, Rangarajan A, Li Q, Arridge SR, and Leahy RM, PET image reconstruction using information theoretic anatomical priors, IEEE transactions on medical imaging 30, 537–549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buades A, Coll B, and Morel J-M, A non-local algorithm for image denoising, in 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), volume 2, pages 60–65, IEEE, 2005. [Google Scholar]
- 14.Dutta J, Leahy RM, and Li Q, Non-local means denoising of dynamic PET images, PloS one 8, e81390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabov K, Foi A, Katkovnik V, and Egiazarian K, Image denoising with block-matching and 3D filtering, in Image Processing: Algorithms and Systems, Neural Networks, and Machine Learning, volume 6064, page 606414, International Society for Optics and Photonics, 2006. [Google Scholar]
- 16.Turkheimer FE, Boussion N, Anderson AN, Pavese N, Piccini P, and Visvikis D, PET image denoising using a synergistic multiresolution analysis of structural (MRI/CT) and functional datasets, Journal of Nuclear Medicine 49, 657–666 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Mejia JM, Domínguez H. d. J. O., Villegas OOV, Máynez LO, and Mederos B, Noise reduction in small-animal PET images using a multiresolution transform, IEEE transactions on medical imaging 33, 2010–2019 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Le Pogam A, Hanzouli H, Hatt M, Le Rest CC, and Visvikis D, Denoising of PET images by combining wavelets and curvelets for improved preservation of resolution and quantitation, Medical image analysis 17, 877–891 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Malekzadeh M, Mirian N, Song T-A, Liu C, and Dutta J, Artificial intelligence-based image enhancement in pet imaging: Noise reduction and resolution enhancement, PET clinics 16, 553–576 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang L, Qiao Y, Nie D, An L, Lin W, Wang Q, and Shen D, Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI, Neurocomputing 267, 406–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen KT et al. , Ultra–low-dose 18F-florbetaben amyloid PET imaging using deep learning with multi-contrast MRI inputs, Radiology 290, 649–656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronneberger O, Fischer P, and Brox T, U-net: Convolutional networks for biomedical image segmentation, in International Conference on Medical image computing and computer-assisted intervention, pages 234–241, Springer, 2015. [Google Scholar]
- 23.Sano A, Nishio T, Masuda T, and Karasawa K, Denoising PET images for proton therapy using a residual U-net, Biomedical Physics & Engineering Express 7, 025014 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Schaefferkoetter J, Yan J, Ortega C, Sertic A, Lechtman E, Eshet Y, Metser U, and Veit-Haibach P, Convolutional neural networks for improving image quality with noisy PET data, EJNMMI research 10, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Wu J, Lu W, Onofrey JA, Liu Y-H, and Liu C, Noise reduction with cross-tracer and cross-protocol deep transfer learning for low-dose PET, Physics in Medicine & Biology 65, 185006 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Spuhler K, Serrano-Sosa M, Cattell R, DeLorenzo C, and Huang C, Full-count PET recovery from low-count image using a dilated convolutional neural network, Medical Physics 47, 4928–4938 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Zhao K, Zhou L, Gao S, Wang X, Wang Y, Zhao X, Wang H, Liu K, Zhu Y, and Ye H, Study of low-dose PET image recovery using supervised learning with CycleGAN, Plos one 15, e0238455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, Wu X, Zhou J, Shen D, and Zhou L, 3D conditional generative adversarial networks for high-quality PET image estimation at low dose, Neuroimage 174, 550–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L, Schaefferkoetter JD, Tham IW, Huang G, and Yan J, Supervised learning with cyclegan for low-dose FDG PET image denoising, Medical image analysis 65, 101770 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Ouyang J, Chen KT, Gong E, Pauly J, and Zaharchuk G, Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss, Medical physics 46, 3555–3564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong Y, Shan H, Teng Y, Tu N, Li M, Liang G, Wang G, and Wang S, Parameter-transferred wasserstein generative adversarial network (PT-WGAN) for low-dose pet image denoising, IEEE Transactions on Radiation and Plasma Medical Sciences 5, 213–223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, and Bengio Y, Generative adversarial nets, Advances in neural information processing systems 27 (2014). [Google Scholar]
- 33.Jeong YJ, Park HS, Jeong JE, Yoon HJ, Jeon K, Cho K, and Kang D-Y, Restoration of amyloid PET images obtained with short-time data using a generative adversarial networks framework, Scientific reports 11, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue H et al. , A 3D attention residual encoder–decoder least-square GAN for low-count PET denoising, Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 983, 164638 (2020). [Google Scholar]
- 35.Hashimoto F, Ohba H, Ote K, Teramoto A, and Tsukada H, Dynamic PET image denoising using deep convolutional neural networks without prior training datasets, IEEE Access 7, 96594–96603 (2019). [Google Scholar]
- 36.Hashimoto F, Ohba H, Ote K, Kakimoto A, Tsukada H, and Ouchi Y, 4D deep image prior: Dynamic PET image denoising using an unsupervised four-dimensional branch convolutional neural network, Physics in Medicine & Biology 66, 015006 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Cui J et al. , PET image denoising using unsupervised deep learning, European journal of nuclear medicine and molecular imaging 46, 2780–2789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song T-A, Yang F, and Dutta J, Noise2Void: unsupervised denoising of PET images, Physics in Medicine & Biology 66, 214002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yie SY, Kang SK, Hwang D, and Lee JS, Self-supervised PET denoising, Nuclear Medicine and Molecular Imaging 54, 299–304 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nai Y-H et al. , Validation of low-dose lung cancer PET-CT protocol and PET image improvement using machine learning, Physica Medica 81, 285–294 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Schaefferkoetter J, Sertic A, Lechtman E, and Veit-Haibach P, Clinical Evaluation of Deep Learning for Improving PET Image Quality, 2020. [DOI] [PMC free article] [PubMed]
- 42.Chen KT et al. , True ultra-low-dose amyloid PET/MRI enhanced with deep learning for clinical interpretation, European Journal of Nuclear Medicine and Molecular Imaging, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuchiya J, Yokoyama K, Yamagiwa K, Watanabe R, Kimura K, Kishino M, Chan C, Asma E, and Tateishi U, Deep learning-based image quality improvement of 18 F-fluorodeoxyglucose positron emission tomography: a retrospective observational study, EJNMMI physics 8, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ladefoged CN, Hasbak P, Hornnes C, Højgaard L, and Andersen FL, Low-dose PET image noise reduction using deep learning: application to cardiac viability FDG imaging in patients with ischemic heart disease, Physics in Medicine & Biology 66, 054003 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Sanaat A, Shiri I, Arabi H, Mainta I, Nkoulou R, and Zaidi H, Deep learning-assisted ultra-fast/low-dose whole-body PET/CT imaging, European Journal of Nuclear Medicine and Molecular Imaging, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu F et al. , Evaluation of Deep Learning Based PET Image Enhancement Method in Diagnosis of Lymphoma, 2020.
- 47.Katsari K, Penna D, Arena V, Polverari G, Ianniello A, Italiano D, Milani R, Roncacci A, Illing RO, and Pelosi E, Artificial intelligence for reduced dose 18F-FDG PET examinations: A real-world deployment through a standardized framework and business case assessment, EJNMMI physics 8, 1–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Q, Liu H, Niloufar M, Ren S, and Liu C, The impact of noise level mismatch between training and testing images for deep learning-based PET denoising, 2021.
- 49.Ibanez L, Schroeder W, Ng L, and Cates J, The ITK software guide, (2003).
- 50.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, and Gerig G, User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability, Neuroimage 31, 1116–1128 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


