Summary
The GAI-RGA- and -SCR (GRAS) proteins regulate a myriad of biological functions in plants. The C-terminus of GRAS proteins is highly conserved, whereas the N-terminus is hypervariable. So far, GRAS proteins have been reported in more than 50 plant species. However, not many GRAS proteins are characterized, thus limiting the revelation of their many functions. This review provides a recent update on GRAS proteins, including their structural features, evolutionary gene family expansion/diversification, and interacting protein partners. Also, a mechanistic insight on GRAS protein-mediated plant growth and abiotic stress response is provided. For this, we assessed the transcriptional dynamics of GRAS genes in rice (monocot) and Arabidopsis (dicot) at different developmental stages and under several abiotic stresses. Lastly, the usage of genome-editing tools such as the CRISPR/Cas9 system to understand GRAS molecular functions is highlighted, with the ultimate goal of developing improved agronomic and climate-resilient traits in plants.
Subject areas: Biological sciences, Natural sciences, Plant biology, Plant physiology
Graphical abstract
Biological sciences; Natural sciences; Plant biology; Plant physiology
Introduction
The GAI-RGA- and -SCR (GRAS) transcription factors (TFs) are an important plant-specific TF family that derives the name from three members, GIBBERELLIN-ACID INSENSITIVE (GAI), REPRESSOR of GA1 (RGA), and SCARECROW (SCR). This TF is characterized by the presence of a conserved C- terminus GRAS domain consisting of five distinct motifs, including LHRI (Leucine Heptad Repeat I), VHIID, LHR II (Leucine Heptad Repeat II), PYRE, and SAW, and a highly variable N-terminus region containing homopolymeric stretches of specific amino acids, intrinsically disordered proteins (IDD) and conserved DELLA domain (Hakoshima, 2018; Sun et al., 2011). These structural features of GRAS proteins suggest that GRAS can potentially function as TFs in plants. Moreover, GRAS proteins contain nuclear localization sequences (NLSs), suggesting their site of action is the nucleus (Tian et al., 2004). The regulatory role of GRAS has been shown in various plant developmental and stress responses. For instance, GRAS proteins act as a key regulator of root development in Arabidopsis (Pauluzzi et al., 2012; Petricka et al., 2012). SHR, also known as a moving transcriptional regulator, moves from the stele into the adjacent layer. Further, the heterodimerization of SHR-SCR leads to the sequestration of SHR into the nucleus that blocks SHR movement in the endodermis. The formation of the SHR-SCR complex results in the upregulation of several TF genes, including zinc finger (ZF) of the BIRD/IDD family (Long et al., 2015; Welch et al., 2007). This regulatory network comprising BIRD/IDD and SCR with SHR dictates tissue specificity and developmental plasticity (Moreno-Risueno et al., 2015). Comparable homologs of AtSCR are found in maize (ZmSCR) and rice (OsSCR), which showed almost similar expression patterns with slight variability in OsSCR expression, suggesting regulatory activity during radial root patterning in dicots and monocots (Kamiya et al., 2003; Lim et al., 2005). Also, NSP1 and NSP2 can form homo and heterodimers to direct the nodulation process (Hirsch et al., 2009). Also, NSP2 can interact with RAM1 protein (Gobbato et al., 2012). Moreover, RAM1 also interacts with RAD1 for transcriptional regulation of several downstream genes (Park et al., 2015; Xue et al., 2015). Evidence that shows direct DNA-binding of GRAS proteins with promoters is not available, however, in vivo chromatin immunoprecipitation (ChIP) assays show association with several putative DELLA target genes (Zentella et al., 2007). Likewise, in vivo study showed that SHR binds to its own promoter and the promoters of other SHR targets (Cui et al., 2007). The GRAS protein isolated from lily activated the transcription in yeast and plant systems, suggesting GRAS acts as a transcriptional activator (Morohashi et al., 2003). The confirmatory evidence on the role of GRAS as classic TFs was shown in Medicago truncatula where in vivo and in vitro studies revealed that NSP1 binds directly to the promoter of a nodulation gene (Hirsch et al., 2009).
Being a plant-specific TF, the GRAS family has been reported in more than 50 species, including Arabidopsis (Tian et al., 2004), Oryza sativa (Tian et al., 2004), Glycine max (Wang et al., 2020a), Manihot esculenta (Shan et al., 2020), Hordeum vulgare (To et al., 2020), Brassica rapa (Song et al., 2014), Solanum lycopersicum (Huang et al., 2015), Nicotiana tobacum (Chen et al., 2015b), Camellia sinensis (Wang et al., 2018), and Rosa chinensis (Kumari et al., 2022). Based on conserved domains and functions, the GRAS family was initially classified into eight different subfamilies, including SCR, SCARECROW-LIKE3 (SCL3), SHORT-ROOT (SHR), DELLA, LATERAL SUPPRESSOR (LS), LIGHT SIGNALING through INTERACTIONS LIGHT-RESPONSIVE TRANSCRIPTION FACTOR PIFs (LISCL), HAIRY MERISTEM (HAM) and PHYTOCHROME A SIGNAL TRANSDUCTION1 (PAT1) (Fan et al., 2017; Kumari et al., 2022; Lee et al., 2008; Tian et al., 2004); however, in some plant species number of subfamilies reported the higher number of subfamilies (for details, see the later section). These TFs are reported to regulate multiple biological processes and molecular functions, including gibberellic acid (GA) signaling (Ikeda et al., 2001; Peng et al., 1997), brassinosteroid (BR) signaling (Tong et al., 2009), shoot and axillary meristem maintenance (Greb et al., 2003; Stuurman et al., 2002), radial organization of the root (Helariutta et al., 2000), phytochrome signal transduction (Bolle et al., 2000), nodulation and arbuscular mycorrhiza (AM) symbiosis (Gobbato et al., 2012; Heckmann et al., 2006; Kaló et al., 2005; Pimprikar et al., 2016; Rich et al., 2017; Shtark et al., 2016) and abiotic stress responses (Guo et al., 2019; Ma et al., 2010).
The green revolution in the 1960s also exploited the GRAS genes as mutants of Reduced height1 (Rht1) and Rht2 alleles encode DELLA proteins, which have reduced sensitivity for the GA (Hedden, 2003). These alleles result in the development of semi-dwarf plant stature with improved yield and lodging resistance. Further, SHR and SCR form a complex to regulate asymmetric cell division in roots (Cui et al., 2007). A GRAS protein, AtSCL14, interacts with TGACG motif-binding factor (TGA) TFs and regulates stress-response in Arabidopsis (Fode et al., 2008). Another GRAS gene, PeSCL7 provides tolerance to high salinity, osmotic, and drought stresses (Ma et al., 2010). These reports demonstrate the diversified functions of GRAS TFs, which made them interesting candidates for further studies to delineate their precise roles in plant growth, development, and stress response.
Genome-scale studies in different plant species have identified several GRAS TFs candidates that are not functionally characterized to date. Implementing functional genomics approaches coupled with overexpression and genome editing tools will enable the development of overexpression and mutant lines, which can further be studied to understand the involvement of GRAS TFs in regulating agronomic as well as stress-resilient traits. Thus, the functional characterization of GRAS TFs possesses excellent application potential.
Given this, the present review provides comprehensive information on the GRAS protein structure, GRAS-interacting proteins, and their involvement in various biological processes, such as growth and development, and phytochrome-mediated signaling, AM association, and response to biotic and abiotic stresses. Importantly, RNA sequencing data mining was performed to elucidate the expression dynamics of GRAS TFs in different tissues across the developmental scale and under abiotic stresses. In the end, we summarize genetic manipulation studies on GRAS by using genome editing approaches, and future perspectives on the usage of GRAS to improve agronomic traits and develop climate-resilient crops are discussed.
Structural features of the GRAS proteins
GRAS proteins are ∼400–770 amino acids long, and their carboxyl (C-) termini have a conserved GRAS domain, and amino (N-) termini have a variable region. The GRAS domain comprises ∼390 amino acids containing five distinct motifs in a specific order, i.e., leucine heptad repeat I (LHR I), VHIID, LHR II, PFYRE SAW motif (Hakoshima, 2018). Among these motifs, LHRI is conserved across all GRAS proteins and has nuclear localization signals (NLSs) (Sun et al., 2012).
The fourth motif, PFYRE, contains the following three distinct sequence signatures: (i) proline (P) residue, (ii) phenylalanine and tyrosine (FY) residues, and (iii) arginine and glutamic acid (RE) residues. The SAW motif contains the sequence signatures WX7G, LXW, and SAW and GRAS proteins are well conserved. The exact function of these two motifs PFYRE and SAW are not well defined in plants; however, their conserved nature may suggest that these two motifs are essential for the structural or functional integrity of the GRAS proteins. Mutations in these motifs result in phenotypic changes. For instance, mutations in PFYRE and SAW motifs of SLR1 (SLENDER RICE1), RGA, and NSP1 impact grain size, plant growth, and nodulation, respectively (Heckmann et al., 2006; Itoh et al., 2002). The N-terminus of GRAS proteins are highly variable (Pysh et al., 1999), comprised of variable length and unique sequences of intrinsically disordered regions (IDRs) (Sun et al., 2012). These features contribute to protein–protein interactions and gene-specific functions, therefore imparting functional specificity to GRAS proteins to act as activators (Figure 1). For instance, the DELLA subfamily of GRAS contains three motifs (DELLA, VHYNP, and LR/KXI motifs) with repeated hydrophobic or aromatic residues (Figure 1A). Similarly, in the case of other GRAS subfamilies, the repeated hydrophobic/aromatic residues (SHR; poly-Q, poly-T, Poly-S/H, and SCR; poly-S/P, poly-S, polyQ/P) play a similar role in the binding of GRAS proteins with their interacting partners (Sun et al., 2012) (Figures 1B and 1C).
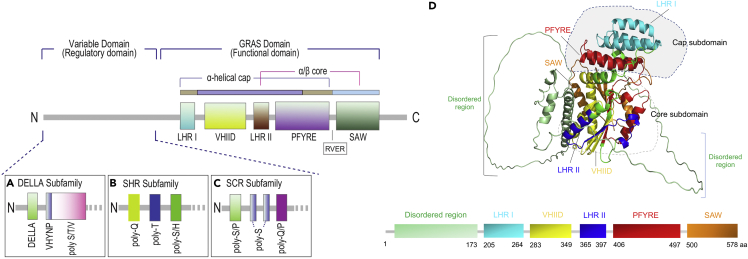
Figure 1.
Schematic depicting domains of the GRAS proteins
(A–C). The conserved C-terminus GRAS domain comprises five distinct motifs, LHRI, VHIID, LHRII, PYRE, and SAW. The LHRI, VHIID, LHRII, PFYRE motifs are part of the α-helical cap, whereas LHRII, PFYRE, and SAW are the components of the α/β core subdomain. The N-terminus comprises variable motifs and regions. (A) DELLA subfamily consists of two motifs DELLA and VHYNP and a poly-S/T/V region, (B) Short-root (SHR) subfamily comprises of three poly-Q, poly-T, and poly-S/H regions, and (C) SCARECROW (SCR) subfamily comprises of three poly-S/P, poly-S, and poly-Q/P regions on their N-terminus.
(D) Ribbon diagram of the GRAS protein of Os-SCL7. The conserved motifs and regions are shown in different colors: LHRI in cyan, VHIID in yellow, LHRII in blue, PFYRE in red, SAW in orange; disordered regions in green.
The structural characterization of GRAS was unavailable, which limited the understanding of GRAS protein function and its physiological roles. The GRAS proteins contain GAI N-terminus DELLA domain complex with GA3-bound GID1A (Murase et al., 2008). These motifs interact with the GA-bound receptor GA-INSENSITIVE DWARF 1 (GID1) and play a crucial role in GA signaling and plant development (Murase et al., 2008). The second study reported the crystal structure of the GRAS domain from rice (OsSCL7) and revealed the features of motifs to atomic resolution and pinpointed the dimerization mechanisms of GRAS proteins to act as dimers (Li et al., 2016). We also downloaded the predicted structure of OsSCL7 from Alpha-Fold database (Jumper et al., 2021) and visualized it on PhyMol (http://www.pymol.org/pymol.). The GRAS domain is located on C-terminal and has five different conserved motifs (LRI, VHIID, LRII, PFYRE, and SAW). The N-terminal has disordered regions (1–173 aa) (Figure 1D). The 3D structure of the GRAS domain has 12 α-helices, 8 β-sheets, and 5 310 helices (Figure 1D). The GRAS domain was comprised of two subdomains: cap and core. The cap subdomain at the top has a helical bundle formed of three α-helices from the LRI and a noncontiguous helical bundle insert of two α-helices from the PFYRE motif. The core subdomain is made up of α-β-α three-layer sandwiched Rossmann fold-like structure (Figure 1D). More structural studies are needed to shed light on the molecular functions of GRAS proteins, which is crucial for the characterization of other GRAS family members.
GRAS gene family: expansion and diversification in the plants
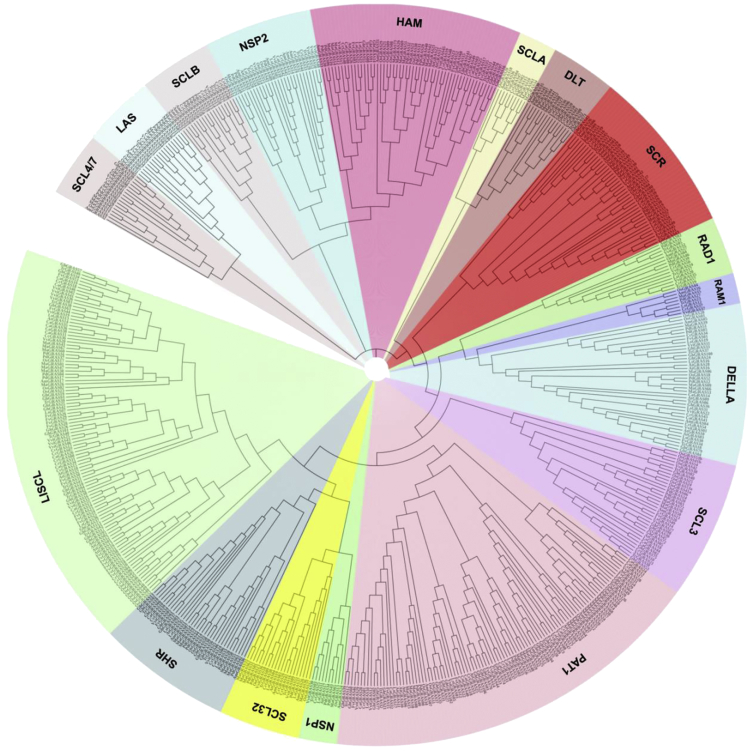
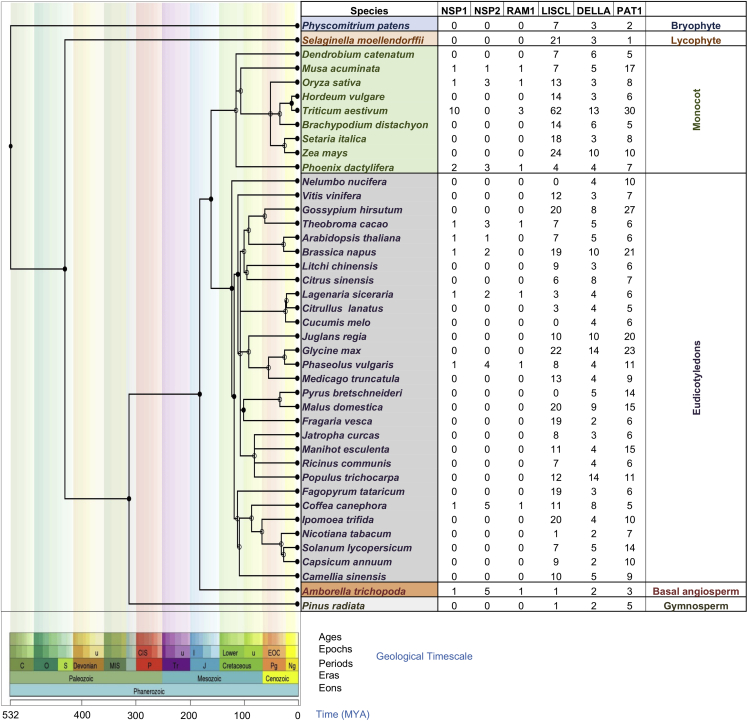
The GRAS genes have been reported in more than 50 plant species, and the number of genes ranged from 33 (Arabidopsis) to 188 (wheat) (Table 1). However, their classification into different subfamilies is still not standardized. Initially, the GRAS genes were clustered into eight subfamilies, including- (i) DELLA, (ii) HAM, (iii) LISCL, (iv) PAT1, (v) LS, (vi) SCR, (vii) SHR, and (viii) SCL9 in case of rice and Arabidopsis (Lee et al., 2008; Tian et al., 2004). Similar to Arabidopsis and rice, eight subfamilies of GRAS genes were also reported in apple (Fan et al., 2017), walnut (Quan et al., 2019), and barrel clover (Zhang et al., 2017). However, in other plant species, the number of subfamilies is higher; for instance, 13 subfamilies were identified in the case of populus (Liu and Widmer, 2014), tea (Wang et al., 2018), and castor bean (Xu et al., 2016) and 16 subfamilies were identified in bottle gourd (Sidhu et al., 2020). Whereas combined phylogenetic analysis using eight different angiosperm species, regroup GRAS TF into 17 subfamilies (Cenci and Rouard, 2017). To further understand the evolutionary relationship, we also performed the phylogenetic analysis using 581 GRAS proteins from 12 plant species and found that GRAS proteins were grouped into 17 subfamilies such as RAM1, NSP1, NSP2, etc. (Figures 2 and S1–S3). The studies mentioned above showed considerable divergence among the GRAS family genes in flowering plants. To understand the functional divergence of GRAS subfamilies in different species, we constructed an evolutionary Tree of Life for 42 plant species belonging to bryophyte, lycophyte, gymnosperm, basal angiosperm (Amborella), and angiosperms (monocot and dicot). The phylogenetic tree clearly shows the evolutionary emergence of NSP1, NSP2, and RAM1 in the higher plant species, suggesting the attainment of specialized functions such as nodulation or arbuscular mycorrhizal interactions (Figure 3). In the future, more studies on GRAS genes in the plant kingdom may identify more subfamilies that can help trace the evolutionary history of addition/deletion or duplication of subfamilies.
Table 1.
Details of GRAS genes reported in different plant species
| Species (common name) |
Family | Monocot/ Eudicot |
No. of GRAS | No. of subfamilies | References |
|---|---|---|---|---|---|
| A. thaliana (Arabidopsis) | Brassicaceae | Eudicot | 33 | 08 | Lee et al. (2008) |
| Brassica juncea (brown mustard) | Brassicaceae | Eudicot | 88 | 09 | Li et al. (2019) |
| Brachypodium distachyon (stiff brome) | Poaceae | Monocot | 48 | 10 | Niu et al. (2019) |
| Brassica napus (rapeseed) | Brassicaceae | Eudicot | 92 | 09 | Guo et al. (2019) |
| Brassica oleracea (cabbage) | Brassicaceae | Eudicot | 35 | 09 | Li et al. (2019) |
| Brassica rapa (mustard) | Brassicaceae | Eudicot | 48 | 08 | Song et al. (2014) |
| Camellia sinensis (tea plant) | Theaceae | Eudicot | 52 | 13 | Wang et al. (2018) |
| Capsicum annuum (pepper) | Solanaceae | Eudicot | 50 | 10 | Liu et al. (2018) |
| Chenopodium quinoa (quinoa) | Amaranthaceae | Eudicot | 52 | 14 | Zhu et al. (2021) |
| Cicer arietinum (chickpea) | Fabaceae | Eudicot | 46 | 9 | Yadav et al. (2022) |
| Citrullus lanatus (watermelon) | Cucurbitaceae | Eudicot | 37 | 10 | Lv et al. (2021) |
| Citrus sinensis (sweet orange) | Rutaceae | Eudicot | 50 | 11 | Zhang et al. (2019) |
| Cucumis melo (melon) | Cucurbitaceae | Eudicot | 37 | 9 | Bi et al. (2021) |
| Dendrobium catenatum (chained dendrobium) | Orchidaceae | Monocot | 47 | 11 | Zeng et al. (2019) |
| Fagopyrum tataricum (buckwheat) | Polygonaceae | Eudicot | 47 | 10 | Liu et al. (2019) |
| F. vesca (woodland strawberry) | Rosaceae | Eudicot | 54 | 14 | Chen et al. (2019a) |
| Glycine max (soybean) | Fabaceae | Eudicot | 117 | 09 | Wang et al. (2020a) |
| G. hirsutum (cotton) | Malvaceae | Eudicot | 150 | 14 | Zhang et al. (2018) |
| Hibiscus hamabo (hardy yellow hibiscus) | Malvaceae | Eudicot | 59 | 9 | Ni et al. (2022) |
| Hippophae rhamnoides (sea buckthorn) | Elaeagnaceae | Eudicot | 62 | 9 | Yu et al. (2021) |
| Hordeum vulgare (barley) | Poaceae | Monocot | 62 | 12 | To et al. (2020) |
| Ipomoea trifida (sweet potato) | Convolvulaceae | Eudicot | 70 | 11 | Chen et al. (2019b) |
| Jatropha curcas (physic nut) | Euphorbiaceae | Eudicot | 48 | 12 | Wu et al. (2015) |
| Juglans regia (english walnut) | Juglandaceae | Eudicot | 52 | 08 | Quan et al. (2019) |
| L. siceraria (bottle gourd) | Cucurbitaceae | Eudicot | 37 | 17 | Sidhu et al. (2020) |
| Litchi chinensis (litchi) | Sapindaceae | Eudicot | 48 | 14 | Chen et al. (2021) |
| Malus domestica (apple) | Rosaceae | Eudicot | 127 | 08 | Fan et al. (2017) |
| Manihot esculenta (casava) | Euphorbiaceae | Eudicot | 77 | 14 | Shan et al. (2020) |
| Medicago sativa (alfalfa) | Fabaceae | Eudicot | 51 | 9 | Zhang et al. (2021) |
| Medicago truncatula (barrel clover) | Fabaceae | Eudicot | 59 | 08 | Zhang et al. (2017) |
| Musa acuminata (banana) | Musaceae | Monocot | 73 | 16 | Cenci and Rouard (2017) |
| Nelumbo nucifera (sacred lotus) | Nelumbonaceae | Eudicot | 38 | 09 | Wang et al. (2016) |
| Nicotiana tabacum(tobacco) | Solanaceae | Eudicot | 21 | 08 | Chen et al. (2015b) |
| O. sativa (rice) | Poaceae | Monocot | 57 | 08 | Tian et al. (2004) |
| Panax ginseng (ginseng) | Araliaceae | Eudicot | 59 | 13 | Wang et al. (2020b) |
| Panicum virgatum (switchgrass) | Poaceae | Monocot | 144 | 8 | Wang et al. (2021) |
| Pennisetum glaucum (pearl millet) | Poaceae | Monocot | 57 | 8 | Jha et al. (2021) |
| Phaseolus vulgaris (common bean) | Fabaceae | Eudicot | 55 | 16 | Laskar et al. (2021) |
| Physcomitrella patens (moss) | Funariaceae | Monocot | 40 | 13 | Wu et al. (2014) |
| Populus tremula (populus) | Salicaceae | Eudicot | 106 | 13 | Liu and Widmer (2014) |
| Prunus mume (japanese apricot) | Rosaceae | Eudicot | 46 | 11 | Lu et al. (2015) |
| Prunus persica (peach) | Rosaceae | Eudicot | 48 | 13 | Jiang et al. (2022) |
| Pyrus bretschneideri (chinese white pear) | Rosaceae | Eudicot | 99 | 12 | Chang et al. (2021) |
| Ricinus communis (castor beans) | Euphorbiaceae | Eudicot | 48 | 13 | Xu et al. (2016) |
| Rosa chinensis (rose) | Rosaceae | Eudicot | 59 | 17 | Kumari et al. (2022) |
| Selaginella moellendorffii (lycophyte) | Selaginellaceae | Eudicot | 45 | 15 | Engstrom (2011) |
| Setaria italica (foxtail millet) | Poaceae | Monocot | 57 | 13 | Fan et al. (2021a) |
| Solanum lycopersicum (tomato) | Solanaceae | Eudicot | 54 | 10 | Niu et al. (2017) |
| Sorghum bicolour (Indian millet) | Poaceae | Monocot | 81 | 13 | Fan et al. (2021b) |
| Triticum aestivum (wheat) | Poaceae | Monocot | 188 | 12 | Liu and Wang (2021) |
| Vitis vinifera (grape vine) | Vitaceae | Eudicot | 52 | 13 | Grimplet et al. (2016) |
| Zea mays(maize) | Gramineae | Monocot | 86 | 08 | Guo et al. (2017) |
Figure 2.
Phylogenetic analysis of GRASs in plant species
Phylogenetic tree of GRAS proteins in Arabidopsis thaliana (Lee et al., 2008), Amborella trichopoda (Albert et al., 2013), Capsicum annum (Liu et al., 2018), Coffea canephora (Denoeud et al., 2014), Fragaria vesca (Chen et al., 2019a), Gossypium hirsutum (Zhang et al., 2018), Lagenaria siceraria (Sidhu et al., 2020), Musa acuminate (D’Hont et al., 2012), O. sativa (Tian et al., 2004), Phoenix dactylifera (Al-Mssallem et al., 2013), Theobroma cacao (Argout et al., 2011), and Vitis vinifera (Grimplet et al., 2016). GRAS protein sequences are aligned using MUSCLE package available in MEGA X software (Stecher et al., 2020). The phylogenetic tree was created using the neighbor-joining (NJ) method with the Poisson model, pairwise deletion, and 1,000 bootstraps values. The phylogenetic tree was visualized using the iTOL online tool (Letunic and Bork, 2021). The GRAS proteins are clustered into 17 subfamilies, marked by various colors.
Figure 3.
Phylogenetic tree of 42 species belonging to bryophyte, lycophyte, gymnosperm, basal angiosperm, and angiosperms (monocot and dicot) visualized as a species tree
The divergence times at different nodes are depicted in different geological time scales. The number of genes present/absent of NSP1, NSP2, RAM1, LISCL, DELLA, and PAT1 in different species are shown in columns. Dated phylogeny trees for 31 plant species were retrieved from TimeTree (Kumar et al., 2017).
GRAS proteins are not present in algae but are identified in bryophytes (Marchantia polymorpha) (Guo et al., 2019). GRAS protein sequences across different plant species, including lower plants and higher plant species, showed 12 subclades in a phylogenetic tree. In flowering plants, the GRAS family showed maximum divergence. There is enormous variability in the number of subfamilies of GRAS TFs among different plant species (Table 1), suggesting some species or lineage-specific retention of GRAS subfamilies during evolution. Further, the less conservation of the GRAS domains across different subfamilies, but more sequence similarities (up to 98%) within subfamilies, highlights similar functions of GRAS genes (Sun et al., 2012). For example, most of the genes characterized yet from the DELLA subfamily are involved in the GA signaling pathway. It has also been confirmed with GRAS protein functional characterization in different plants, which has indicated a conserved function/pattern among putative orthologues of each subfamily and/or subclade in different plants. Another good example is the NSP1 gene, conserved in leguminous and non-leguminous crops and associated with nodule formation (Heckmann et al., 2006; Smit et al., 2005).
In bryophytes, ESTs similar to GRAS gene sequences were found, suggesting that this protein family arose before the evolution of land plants (Nishiyama et al., 2003). The GRAS genes are mostly intronless in several plant species, including grapevine (88.46%), barley (88.2%), tomato (77.4%), capsicum (84%), mustard (83.3%), apricot (82.2%), Arabidopsis (67.6%), rice (55%), and populus (54.7%) (Grimplet et al., 2016; Huang et al., 2015; Liu et al., 2018; Liu and Widmer, 2014; Lu et al., 2015; Song et al., 2014; To et al., 2020). The probable causes for the intronless genes in eukaryotic genomes are duplication events and retroposition of intron-containing genes (Zou et al., 2011). Intronless genes are a typical prokaryote feature since GRAS-like proteins were initially reported in bacteria and involved in methylase activity (Zhang et al., 2012).
Intrinsically disordered proteins (IDPs) and functional versatility in GRAS
In eukaryotes, 25–30% of proteins are intrinsically disordered proteins (IDPs) and play a significant role in various molecular and cellular functions. More than 70% of signaling proteins and 82–94% of TFs have IDRs (Wang et al., 2007). IDR, present within IDP, enables proteins to undergo disorder-to-order transitions to recognize and bind at specific binding interfaces among different partners (Sun et al., 2011, 2012). Computational and bioinformatics analysis revealed that the GRAS proteins also fall under the IDP category (Sun et al., 2011). The GRAS protein’s N-terminus contains molecular recognition features (MoRFs), short interaction-prone sites within IDR that, upon interaction and recognition of partner’s proteins, allow specific disorder-to-order transitions to form functional complexes (Sun et al., 2011, 2012). The functional diversification of GRAS proteins is attributed to the intrinsically disordered N-domain, which is reflected by (a) its expansion into different subfamilies, (b) role in transcriptional regulation along with signaling pathways, and (c) some proteins can show cross-talk in different signal pathways forming homo- or heterodimers. For example, BdSLR1 and BdSLRL1 can form homodimers but cannot form heterodimers (Niu et al., 2019).
Intrinsic disorder characteristic in the GRAS proteins has provided them binding plasticity, which is directly correlated with the functional polymorphism of these proteins (Sun et al., 2011). In the DELLA subfamily, the N-terminus has been characterized as intrinsically disordered, whereas the C-terminus possesses only basic structural protein folding with a series of highly conserved motifs (Sun et al., 2011). This highly variable N-terminus domain with intrinsically disordered characteristics enables this gene family with multiple functions. The intrinsically disordered N-domain allows GRAS proteins to act as the key regulators in several signaling pathways.
Moreover, phosphorylation and dephosphorylation have widely been observed for the functionality of GRAS proteins. Phosphorylation plays an essential role in adjusting disordered protein interactions and ultimately signal transduction events (Iakoucheva et al., 2004; Mittag et al., 2010). For instance, in rice, GID2 interacts specifically with phosphorylated SLR1 protein (a DELLA protein), not with unphosphorylated SLR1 protein; and strongly suggests that only the phosphorylated state of SLR1 protein can undergo GA-dependent degradation (Gomi et al., 2004). Moreover, a rice kinase may also directly phosphorylate SLR1 proteins (Dai and Xue, 2010). In SLR1 proteins, Ser residues present within DELLA/TVHYNP and polyS/T/V domain (at N-terminus domain) undergo phosphorylation (Itoh et al., 2005). The experimental demonstrations have confirmed that rice SLR1 and Brachypodium BdSLR1 show that the DELLA domain and TVHYNP motif are the key regulators for transcriptional activation activity and GID1 interaction (Hirano et al., 2012; Niu et al., 2019). Also, CCaMK (calcium- and calmodulin-dependent protein kinase) is required to facilitate nodulation signaling that requires the GRAS family transcriptional NSP1 and NSP2 (Gleason et al., 2006). The reversible phosphorylation also plays a significant role in regulating the stress-induced response of some GRAS proteins. For instance, NtGRAS1 in tobacco (Czikkel and Maxwell 2007), and CIGR1 (chitin-inducible gibberellin-responsive) and CIGR2 in the case of rice (Day et al., 2004) are good examples. Therefore, the phosphorylation and dephosphorylation states of DELLA proteins are critical to governing the stability or degradation of proteins and regulating intracellular signal transduction under different stress conditions (Czikkel and Maxwell, 2007; Day et al., 2004). Although the knowledge about GRAS proteins' phosphorylation and dephosphorylation is still elusive, disorder-assisted phosphorylation may provide a different dimension to study GRAS proteins and their recognition patterns during signaling mechanisms.
The IDPs also provided a functional advantage over structurally ordered proteins for molecular recognition via protein–protein interactions. To facilitate specific recognition, they undergo binding-induced conformational changes and display different binding sites to recognize different partners during protein interactions, giving binding plasticity to these proteins. The MoRFs are responsible for this disorder-to-order transition to recognize their interacting partners. The N-domain is highly variable between subfamilies, but it is highly conserved within the subfamily (Sun et al., 2011). For example, in the DELLA subfamily, repeated hydrophobic/aromatic residues are the restricted motifs in the N domain and are crucial for GID1 interaction, GA signaling, and ultimately plant growth (Murase et al., 2008). The GRAS TFs exhibit a higher level of divergence owing to MoRFs at their N-terminus region, which allows them to function as an activator in different signaling and molecular pathways. Whereas the C-terminus region is highly conserved in the GRAS family and interacts with downstream proteins (Bolle, 2004; Pysh et al., 1999). For instance, Leu heptad repeats in the conserved LHRI-VHIID-LHRII domain involved in protein–protein interactions, which has also been proved experimentally by following interacting pairs, where these interactions are conferred by either an individual motif or the entire region: DELLA–PIFs, DELLA–JAZ1, DELLA–GID2, SHR– SCR, NSP1–NSP2, SCL14-TGA, and SCL1–HDA19 (Cui et al., 2007; Fode et al., 2008; Gao et al., 2004; Hirsch et al., 2009; Hou et al., 2010). It has been observed that the GRAS proteins are mostly transcriptional coactivators, which may block or enhance the activity of interacting partners (Sun et al., 2012). Another conserved C-terminus, the VHIID domain is involved in protein–DNA interactions (Pysh et al., 1999). Therefore, this transition in protein structure for molecular recognition based upon intrinsic disorder-to-order transition owing to the presence of MoRFs provides a theoretical framework that can help explore more probable functioning of GRAS TFs in signaling and regulation pathways that can further be confirmed by experimental elucidations.
GRAS-interacting protein complexes
Generally, transcriptional regulation of a particular gene is regulated by the action of TFs that directly binds promoter cis-elements of target genes. However, transcriptional regulation is also mediated by regulatory complexes, such as TFs interact with other TFs or transcriptional regulators (TRs) (Gutjahr et al., 2015). GRAS proteins carry out their functions via forming homo- or heterodimers and/or interacting with other proteins. GRAS proteins primarily function via interaction with several protein partners such as TFs (e.g., IDD, TGA, MYB, SPL) and chromatin remodeling complexes (Pickle; PKL) (Zhang et al., 2012, 2014). In Arabidopsis, Indeterminate Domain (AtIDD) proteins regulate the expression of SCR by interacting with SHR and SCR, which are further involved in root tissue formation by accommodating their ZF motifs into the groove of the SHR GRAS domain (Aoyanagi et al., 2020; Hakoshima, 2018). Further, IDD proteins also act as transcriptional coactivators with DELLA proteins. The DELLA/IDD protein complex induces the expression of the SCL3 gene and regulates the GA signaling pathway in plants (Yoshida et al., 2014). The TGA TFs belong to group D of bZIP TFs and are found ubiquitously in eukaryotes (Johnson et al., 2003). TGA plays a vital role in Arabidopsis to recruit LlSCL/SCL9 branch member SCL14 (SCARECROW-LIKE 14) to its target promoter region. The TGA/SCL14 protein complex activates the detoxification mechanism in plants against xenobiotic chemicals such as isonicotinic acid 2,4,6-triiodobenzoic acid (Fode et al., 2008).
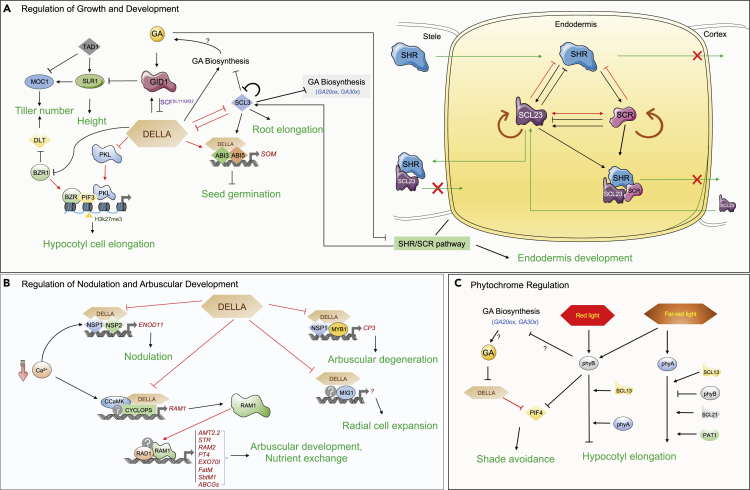
In eukaryotes, chromatin remodeling is essential for gene expression, and chromatin-remodeling factor PKL/Enhanced Photomorphogenic1(EPP1) suppresses phytochrome signaling in plants (Jing et al., 2013). PKL interacts with regulators of hypocotyl growth such as PIF3 and BZR1 to inhibit the histone methylation of genes involved in cell elongation (i.e., IAA19, PRE1, TCH4) (Zhang et al., 2014). Also, PKL interacts with DELLA proteins to negatively regulate its activity by constricting its binding ability. Therefore, the PIF-BZR1-PKL-DELLA module regulates H3K27me3 modification status and ensures plant seed etiolation (Figure 4A).
Figure 4.
Role of GRAS TFs in different biological processes in plants
(A–C). Regulatory circuits mediated by GRAS transcription factors (TFs) during plant growth and development (A), nodulation and arbuscular mycorrhizal development (B), and phytochrome regulation (C). Black arrows indicate transcriptional controls; protein–protein interaction is denoted by red arrows; green denoted the movement of proteins; bars denote negative regulation; and the cross represents the confined movement. BR, brassinosteroid; PKL, pickle; ABI, abscisic acid insensitive; SOM, SOMNUS; AM, arbuscular mycorrhizal; Ca2+, calcium; FR, far-red; R, red; PIF4, Phytochrome Interacting Factor 4.
Several GRAS TFs are involved in AM symbiotic association and regulation of plant developmental transitions. RAM1 is a central regulator of arbuscular development and is involved in hyphopodia formation in AM fungi, whose transcription is regulated explicitly by CYCLOPS and DELLA (Pimprikar et al., 2016). The calcium- and calmodulin-dependent kinase (CCaMK) interacts with CYCLOPS and directly binds to the cis-element (GGCGCC box/AM-CYC box) of the RAM1 promoter (Pimprikar et al., 2016). CYCLOPS also interacts with DELLA, which further boosts its capability to transactivate the RAM1 promoter along with the promoters of other nodulation genes such as NIN (NODULE INCEPTION) and ERN1(ERF REQUIRED FOR NODULATION1) (Jin et al., 2016; Pimprikar et al., 2016). These studies suggest that the complex formed by GRAS proteins is crucial for establishing arbuscular mycorrhiza symbiosis between plants and fungi. BOTRYTIS SUSCEPTIBLE1 INTERACTOR (BOI) is an Arabidopsis RING protein that interacts with DELLA and suppresses the expression of GA-responsive genes and eventually affects seed germination, juvenile-to-adult phase transition, and flowering (Park et al., 2013).
MicroRNA-based regulation of GRAS
MicroRNAs are 21–24 nucleotide long, highly conserved noncoding RNA sequences that regulate plant growth and developmental processes by directing cleavage of mRNA sequences or translational inhibition. The microRNAs regulate several developmental processes and environmental interactions in plants (Lanet et al., 2009; Mallory et al., 2004; Song et al., 2019). The predicted target of miR171 is the AtSCL6 gene (Llave et al., 2002). The three SCL6 transcripts targeted by miR171 are critical for differentiating axillary meristem and promoting shoot elongation (Wang et al., 2010). Moreover, miR171 target transcripts SCL6, SCL22, and SCL27 and control chlorophyll biosynthesis (Ma et al., 2014). Similarly, in rice and barley, miR171 functions in phase transitions and floral meristem determinacy, thereby highlighting the role of GRAS genes in regulating developmental processes in monocots and dicots (Curaba et al., 2013; Fan et al., 2015).
The miR171 targets NSP2 transcripts in legumes such as barrel clover and lotus, thus regulating nodule formation and mycorrhiza association (De Luis et al., 2012; Hofferek et al., 2014; Lauressergues et al., 2012). Likewise, in soybeans, the GmSCL-6 and GmNSP2 mRNAs are cleaved by miRNAo and miR171q to regulate the nodulation process (Hossain et al., 2019). In tomatoes, the functional genomic study of the miR171-SlGRAS24 module demonstrated that miR171 regulates various agronomical traits in tomatoes via gibberellin and auxin homeostasis (Huang et al., 2017). These studies emphasized the crucial role of miRNAs in regulating GRAS genes during different developmental stages of plants.
Biological functions regulated by GRAS
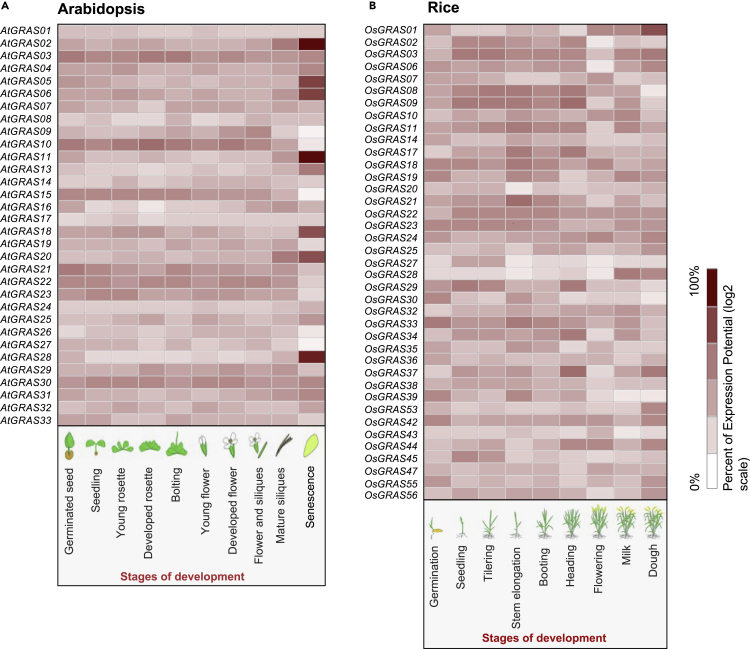
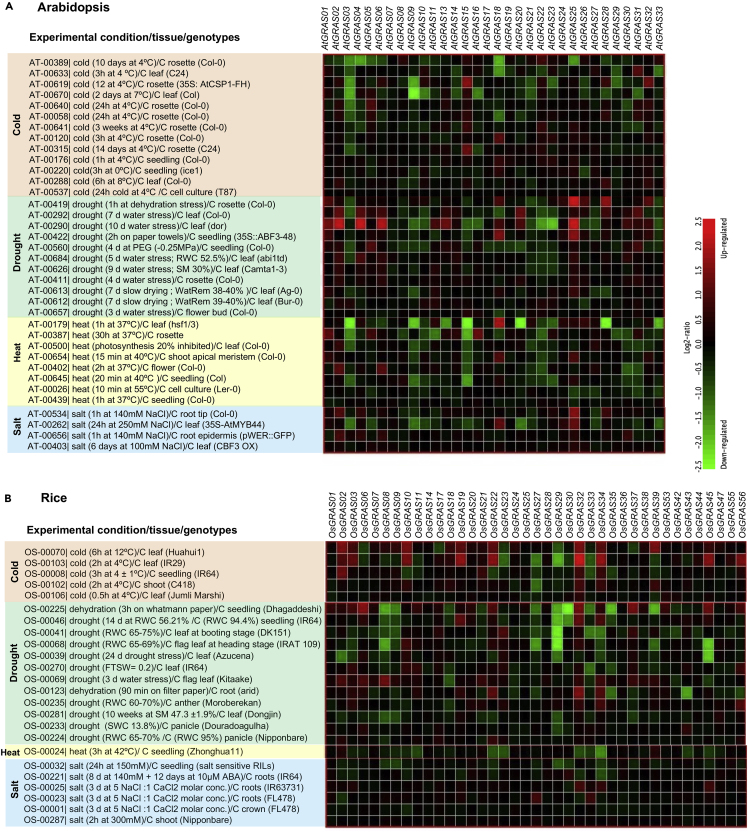
The GRAS TF family members play an essential role in regulating various biological processes, such as plant growth and development (GA signal transduction, shoot/root formation, male gametogenesis, phytochrome signaling, and symbiotic association) (Figure 4). Also, GRAS plays a crucial role in plant stress responses. The transcriptional reprogramming of GRAS genes occurs in different plant development stages (Figure 5) and abiotic stress responses (cold, drought, heat, and salt) (Figure 6) in Arabidopsis and rice. These diverse roles of GRAS TFs make them potential candidates for improving plants.
Figure 5.
Expression of GRAS genes at different developmental stages
(A and B) Heat maps representing GRAS gene expression patterns in (A) Arabidopsis and (B) rice during various developmental stages. The expression levels of GRAS genes in different developmental stages were analyzed using the Genevestigator tool (Hruz et al., 2008). Heatmap color represents the % of expression potential (log2 scale).
Figure 6.
Expression of GRAS genes under different abiotic stresses
(A and B). Heat maps representing GRAS gene expression patterns in (A) Arabidopsis and (B) rice under abiotic stresses (cold, drought, heat, and salt). The expression levels of GRAS genes in response to different abiotic stresses were analyzed using the Genevestigator tool (Hruzet al. 2008) (red = upregulation and green = downregulation of genes).
Shoot meristem and plant tillering
As plants differ from animals, there is a continuous growth to form new organs during post-embryonic shoot development, which depends on shoot apical meristem (SAM). In tomatoes, the Lateral suppressor (LAS), a GRAS gene, was involved in axillary meristem initiation and lateral shoot formation through negative regulation of GA signal transduction (Schumacher et al., 1999). Similarly, in Arabidopsis, an AtLAS (ortholog of LeLs) gene was involved in axillary shoot formation (Greb et al., 2003), thus signifying a conserved mechanism for lateral shoot formation in tomato and Arabidopsis. The higher expression of AtLAS in leaf axile can be considered as a marker of meristem formation, and AtLAS along with STM gene switched on the expression of REVOLUTA (Rev) gene that is a potential regulator of shoot branching (Greb et al., 2003) However, the ortholog in rice, i.e., MOC1, regulates tiller number, signifying the attainment of functional differences between monocotyledonous and dicotyledonous. A GRAS gene, Hairy Meristem (HAM), was identified in Petunia involved in lateral organogenesis and meristem maintenance. The ham mutants of Petunia showed non-maintenance of the meristem. Further, HAM and WUSCHEL (WUS) work in parallel during the specification and maintenance of meristem, and HAM is essential for prolonged response to TERMINATOR and SHOOTMERISTEMLESS (PhSTM) in Petunia (Stuurman et al., 2002). Recently, the SCL28 gene was reported to regulate the mitotic cell cycle via MYB3R TFs and modulates the meristematic cell development in Arabidopsis (Goldy et al., 2021). It suggests that this pathway is implicated in meristem cell differentiation and maintenance.
MONOCULM 1 (MOC1) encodes a GRAS protein in rice, expressed in the axillary buds, and regulates axillary bud initiation, thus executing the tiller formation in rice. The moc1 null mutants exhibited only single culm (without tillering), whereas overexpression lines showed enhanced tillering (Lin et al., 2012). TILLER AND DWARF 1 (TAD1) recruit the MOC1 to ANAPHASE-PROMOTING COMPLEX (APC/C), and APC/C degrades the MOC1 owing to E3 ubiquitin ligase activity. In tad mutants, TAD1 gene is disrupted and thus could not recognize the MOC1, thus tad mutants show enhanced tillering (Xu et al., 2012). These studies suggested the MOC1 involved in downstream of TAD1 and control tillering in a positive manner, although the molecular mechanism behind it is still unilluminated. Gibberellin (GA) has long been known to inhibit tillering in plants (Li et al., 2003). Another gene, dwarf, and low-tillering (DLT), which encodes a GRAS protein, regulate plant morphology via BR signaling in rice. The mutant of the DLT gene showed a dwarf phenotype and low-tillering in rice suggested the involvement of the DLT gene in tillering, although the molecular mechanism is not known (Tong et al., 2009).
Microsporogenesis, fruit ripening, and seed germination
The GRAS proteins regulate anther development and fruit ripening. LiSCL is a nuclear-localized microsporocytes gene expressed explicitly during premeiotic anther developmental in Lilium longiflorum anthers. As shown by the transient expression assay, it is involved in meiosis and activates the pollen mother cell (Morohashi et al., 2003). The Lels mutants in tomatoes showed the blocked axillary meristem initiation (Schumacher et al., 1999). Tomato GRAS1 is also differentially expressed in breaker and mature fruits emphasizing its role in fruit development (Huang et al., 2015). Recently, it has been demonstrated that a GRAS TF, SlGRAS4, was involved in tomato ripening by regulating ethylene biosynthesis genes and MADS TFs (Liu et al., 2021). SlGRAS4 binds to the promoter sequence of the crucial ethylene biosynthesis genes (like SlACO1 and SlACO3), activates their expression, and enhances the fruit ripening. Further, SlGRAS4 also represses the SlMADS1 (negative regulator of fruit ripening) expression and accelerates the ripening (Liu et al., 2021).
Seeds germination is influenced by external (temperature, moisture, and light) and internal (phytohormones such as ABA and GA) cues. ABA regulates seed dormancy via DELLA protein that belongs to the GRAS family (Koornneef et al., 2002). The DELLA protein makes a complex with ABSCISIC ACID INSENSITIVE 3 (ABI3) and ABI5 and binds to the promoter of SOMNUS (SOM), which is involved in the negative regulation of the seed germination process (Figure 4A) (Lim et al., 2013). During unfavorable situations (heat stress), the levels of ABA increase, whereas GA decreases, which leads to the binding of the DELLA/ABI3/ABI5 protein complex on the SOM promoter. It causes transcription activation of SOM genes and inhibits seed germination during unfavorable environmental conditions (Lim et al., 2013).
Radial patterning of root and shoot
The asymmetric cell divisions pave the way for plant organ development and the formation of patterns. Almost 1.5 decades ago, AtSCR, a GRAS TF was identified to express in the cortex and endodermis (Di Laurenzio et al., 1996), shoot apical meristem (Fukaki et al., 1998), bundle sheath (Wysocka-Diller et al., 2000), and quiescent center (Sabatini et al., 2003). The scr mutant in Arabidopsis showed aberrant plant growth with defective roots, hypocotyl stem, and inflorescence. This suggests that AtSCR is essential in root and shoot formation radial patterning. SHORT-ROOT (SHR) is another GRAS TF required for quiescent center identity and its role in radial root patterning (Sabatini et al., 2003). AtSHR gene is exclusively expressed in provascular tissue (Nakajima et al., 2001), and shr mutants did not possess one layer of cortex, unlike in scr mutants (Helariutta et al., 2000). Presence of SHR and SCR is critical for periclinal division and endodermis formation (Cruz-Ramírez et al., 2012; Cui et al., 2007; Gallagher et al., 2004). In shr mutants, SCR expression levels are significantly low, suggesting that transcription of SCR is directly or indirectly controlled by the SHR gene (Nakajima et al., 2001). The SHR gene transcript is initially present in the stele, from where it moves to the adjacent cell layers and interacts with its target SCR. SHR functions downstream to SCR and is primarily responsible for asymmetric cell division in Arabidopsis (Di Laurenzio et al., 1996; Heidstra et al., 2004). However, SCR restricts SHR movement to endodermis (Cui et al., 2007). The SCL23 is another GRAS TF, which is operative in the same pathway. No alterations in the root pattern were observed in the single mutant under normal growth conditions, which suggests that it functions redundantly. However, the double mutant scrscl23 mimicked the shr phenotypes and resulted in smaller roots and plants. Therefore, SCL23, SCR, and SHR genes play a significant role in endodermis formation in the root meristem (Kim et al., 2017) (Figure 4A). Another GRAS gene SCL3 was found to be involved in the GA pathway and helps develop root endodermis (Heo et al., 2011; Yoshida et al., 2014). The scl3 null mutant showed decreased GA responses and enhanced GA biosynthesis gene expression specifying that SCL3 functions as a positive regulator of the GA signaling pathway (Heo et al., 2011; Zhang et al., 2011). However, the tissue-specific maintenance of the GA pathway in endodermis is regulated by the joint action of SCL3 and SHR/SCR genes, highlighting a cross-regulatory network of the GRAS TFs in the root endodermis. In Zea mays, SCARECROW (ZmSCR), an ortholog of AtSCR, was found to be localized to the endodermal cell layer. Despite variation in the size and configuration of the quiescent center in the two species, it suggests that conserved pathways are involved in radial patterning (Lim et al., 2000).
An ortholog of Arabidopsis SCR is OsSCR found in rice. OsSCR was upregulated in the endodermis, whereas it is downregulated in the daughter cortex. OsSCR transcripts were found in leaf primordium at the P3 stage during stomatal development and in guard mother cells (GMCs). Also, a higher transcript level of OsSCR was found in the ligule initiation tissues. It suggests that OsSCR has multiple functions, including cortex/endodermis, stomata, and ligule development (Kamiya et al., 2003).
Two orthologs of Arabidopsis SHR have been identified in rice, namely OsSHR1 and OsSCR. The OsSHR1 is expressed in leaves and roots, stomatal development, and specifically in P3 leaf primordia at stage 1. The OsSCR is predominately expressed in stomata, whereas the OsSHR1 transcript was almost ubiquitously found in the L1-layer cells and not restricted in the stomatal row. The co-expression of OsSHR1 and OsSCR during different developmental tissues suggests that their co-operative networking is essential for forming root endodermis, stomata, and subsidiary cells. SHR acts as a key player that activates SCL23 and SCR genes in the endodermis. SHR TF enters into endodermis and forms protein complexes of SHR-SCR, SHR-SCL23, or SHR-SCR- SCL23 to regulate the SCR and SCL23, respectively, and a negative feedback loop functions to control each other levels to maintain cell specificity in both roots and shoots (Yoon et al., 2016).
Stress responses
GRAS genes play a significant role in providing tolerance against different abiotic and biotic stresses. For instance, the functional characterization of the PeSCL7 gene of populus emphasizes its involvement in salinity, osmotic and water stresses (Ma et al., 2010). Further, overexpression of Brassica napus gene BnLAS in Arabidopsis showed an increase in chlorophyll content, stomatal density, and enhanced tolerance to drought and salt stress (Yang et al., 2011). Also, the enhanced epidermal wax deposition was observed in the leaves of transgenic plants, together with increased expression of wax biosynthesis and regulatory genes like CER1, CER2, KCS1, and KCS2. OsGRAS23 is a nuclear-localized stress-responsive GRAS TF, which belongs to the LISCL subfamily of GRAS proteins in rice. Overexpression of OsGRAS23 in rice led to enhanced tolerance against drought and oxidative stresses owing to decreased H2O2 accumulation (Xu et al., 2015). The OsGRAS23 positively regulates drought tolerance through binding to the promoters of several stress-related genes (anti-oxidants, protein kinases, proteinase inhibitors, and enzymes related to metabolism).
Moreover, under drought stress, the expression of several GRAS genes is modulated in castor beans (Xu et al., 2016). For instance, the expression of 16 RcGRAS members has been induced up to 5-fold and the expression of four RcGRAS genes has downregulated during drought stress.
The PAT1 proteins implicated in phytochrome signaling are also involved in regulating abiotic stress responses. Overexpression lines of the VaPAT1 gene showed increased tolerance against multiple abiotic stresses (i.e., cold, drought, and salt stresses). The VaPAT1 gene positively regulates several stress-related genes (SIZ, CBF1, MYB34, MYC2, COR15A, RD29A) and leads to enhanced concentration of proline and soluble sugar contents in plant cells and provides abiotic stress tolerance (Yuan et al., 2016).
In wheat, TaSCL14 GRAS gene showed enhanced expression under high-light stress. TaSCL14 acts as a multifunctional regulator as the silencing of TaSCL14 produces various defects, including (i) lowered tolerance to photooxidative stress, (ii) increased injury to biological membranes, (iii) increased rate of leaf senescence, (iv) decreased photosynthetic capacity, and (v) inhibited plant growth (Chen et al., 2015a).
The DELLA genes regulate GA signals and the JA pathway, both of which play distinct roles in plant disease resistance (Hou et al., 2010). Furthermore, DELLA also involved in signal transduction during bacteria infection in Arabidopsis, and provides resistance to such bacteria infection (Wild et al., 2012). In cassava, four MeDELLA proteins have been identified. The overexpression of these genes in Nicotiana benthamiana showed lesser bacterial growth, whereas the silencing of genes decreased resistance against bacteria, highlighting their role in bacterial blight resistance (Li et al., 2018). Likewise, ecotypic expression of SCL14 (LlSCL/SCL9 subfamily GRAS protein) in Arabidopsis showed enhanced tolerance to toxins chemicals (isonicotinic acid and 2,4,6- triiodobenzoic acid, 2,4-D, p-chlorophenoxyisobutyric acid), whereas the mutants were highly susceptible. These results showed that SCL14 is essential for imparting tolerance against xenobiotic stress (Fode et al., 2008). SCL28 is a nuclear-localized transcriptional activator found in the root meristem zone in Arabidopsis. SCL28 orchestrates cell division and elongation patterns of root growth during stress conditions (Choe et al., 2017). Also, GRAS homologs found in tobaco are induced by stress conditions such as antimycin A, H2O2, salicylic acid, and L-cysteine, which suggests their role under stress conditions (Czikkel and Maxwell, 2007). Collectively, GRAS proteins are also involved in managing various biotic and abiotic stresses in plants along with their role in growth and development. However, there are still gaps in knowledge of how GRASs function in many plants in response to stresses, and further functional study is required.
Symbiosis
The plant forms mutualistic partnerships with mycorrhizal fungi and with rhizobial bacteria to exchange nutrients (Gutjahr and Paszkowski, 2013; Pimprikar and Gutjahr, 2018). Plants recognize the Nod factor from rhizobial bacteria and Myc factor from mycorrhizal fungus, resulting in these interactions (Genre et al., 2013; Khan et al., 2022; Maillet et al., 2011; Venkateshwaran et al., 2013). The GRAS genes are actively involved in nodulation and the formation of fungal hyphae during the symbiotic associations (Gobbato et al., 2012; Khan et al., 2022). A conserved signaling route is involved in both Nod and Myc factor signaling cascade, and this signaling pathway divides into two branches, both of which contain GRAS TFs, enabling targeted activation of nodulation or mycorrhizal-associated responses (Gobbato et al., 2012). In this highly conserved signaling cascade (symbiosis-specific association), the GRAS TF has come into the picture after the induction of calcium signaling induced by CCaMK (Pimprikar et al., 2016; Singh et al., 2014).
The two GRAS genes, NSP1 and NSP2, are well characterized in M. truncatula and are involved Nod-factor signaling and nodule formation during rhizobial symbiotic association (Hirsch et al., 2009; Kaló et al., 2005; Smit et al., 2005). Further, LjNSP1 and LjNSP2 function as the Nod factor-activated transcription regulators in the lotus (Heckmann et al., 2006). NSP1 and NSP2 can form a heterodimer, which binds to the promoter of the Early Nodulin 11 (ENOD11) gene, and a single mutation in the LHR I domain of NSP2 affects its binding ability that causes nodule formation and nitrogen fixation (Hirsch et al., 2009). Other GRAS proteins also form homo- or heteropolymers, such as RAM1-NSP2 (Gobbato et al., 2012) and RAD1-LjNSP2 (Xue et al., 2015), RAM1-RAD1 (Park et al., 2015; Xue et al., 2015), and DIP1-RAM1 and regulates the nodulation process in plants.
A GRAS gene called Reduced Arbuscular Mycorrhization 1 (RAM1) acts as a major gene in the transcriptional activation of the late arbuscule-related genes that are essential during the nutrient exchange by targeting carbohydrate and lipid metabolism genes (Bravo et al., 2017; Jiang et al., 2018; Luginbuehl and Oldroyd, 2017; Park et al., 2015). Moreover, in Petunia hybrida, the genes involved in the early stages are not affected by the ram1/ata mutation. However, the genes involved in late arbuscule functioning, for example, PT4 and STR, are activated explicitly by RAM1 (Rich et al., 2017). In M. truncatula, RAM1 and NSP2 are required to form cutin monomers during fungal entry (Murray et al., 2013). RAM1 from M. truncatula has also been reported to be involved in arbuscular development (Gobbato et al., 2012). In rice, DELLA INTERACTING PROTEIN (DIP1) is a crucial gene for mycorrhizal colonization. Mutations in DIP1, RAM1, and DELLA (SLENDER RICE1) showed a substantial reduction in AM formation, although the exact mechanism of arbuscular branching has not been investigated yet (Yu et al., 2014). Further, the expression of RAM1 is regulated by CCaMK/CYCLOPS and DELLA (Pimprikar et al., 2016). CCaMK interacts with CYCLOPS to activate RAM1 by directly binding at a conserved palindromic region (GGCGCC box) of the RAM1 promoter (Pimprikar et al., 2016). DIP1 interacts with mycorrhizal GRAS protein RAM1 to regulate mycorrhizal-associated gene expression (Yu et al., 2014). Other GRAS proteins are also involved in AM degeneration regulation, which involves the interaction of MYB1 with DELLA and NSP1. This interaction regulates the Cysteine protease 3 (CP3) gene, a well-known player involved in AM degeneration (Floss et al., 2017). MYCORRHIZA-INDUCED GRAS (MIG1), a GRAS-domain TF found in M. truncatula, expresses in arbuscule-containing cells, suggesting a specific function during fungal association. It was also reported that MIG1 also interacts with DELLA, and this complex regulates radial cell expansion in roots to adjust fungal infection structures during AM symbiosis (Heck et al., 2016) (Figure 4B). Overall, these studies suggest that nodulation and arbuscule formation involves calcium and hormone signaling. The RAM1 promoter act as a central integration node of this calcium (CCaMK/CYCLOPS) and hormonal (DELLA/GA) signaling (Figure 4B), which plays a vital role in the regulation of nodulation and AM development in plants. However, the function of GRAS proteins in nodulation and AM symbiosis remains largely unknown, and much more study is required to understand the details.
Phytohormones and growth regulation
Phytohormones are the master regulators of various biological processes in plants and play a central role in various growth and stress signaling pathways. For instance, GAs are involved in plant developmental processes, including leaf expansion, stem elongation, hypocotyl elongation, seed germination, pollen maturation, trichome development, and flowering. Most of the GA signaling pathway components have been identified in plants. The DELLA (a major GRAS TFs subfamily) acts as a central component of growth inhibition (Achard and Genschik, 2009; Davière and Achard, 2013; Fukazawa et al., 2014, 2017). In earlier reports, it has been shown that the GAI and RGA (DELLA proteins) degradation occurs via the E3 ligase components. A conformational change occurs by the interaction between GA-GID1-DELLA complex in the presence of GA, which is subsequently recognized by SCFSLY1/GID2 and causes the degradation of DELLA protein via the E3 ubiquitin ligase complex. The degradation of DELLA proteins subsequently activates the downstream genes (Dill and Sun, 2001; Fu et al., 2002). DELLA gain-of-function mutants showed dwarf phenotype and insensitivity against GA; however, loss-of-function mutants showed enhanced GA sensitivity resulting in slender or tall phenotypes (Cheng et al., 2004). Degradation of RGA proteins is delayed by Xanthomonas effector protein, which helps plants provide tolerance against different stress conditions. The ERF-associated amphiphilic repression (EAR) motif of Xanthomonas effector protein interacts with the DELLA motif of RGA proteins and regulates the GA-induced binding of GID1 (Tan et al., 2014). In contrast to Arabidopsis, only one highly conserved DELLA was found in tomatoes, grapevines, and cereals. In tomatoes, a DELLA gene called PROCERA (PRO) has been characterized by a missense mutation in the GRAS domain region (Bassel et al., 2008; Carrera et al., 2012). This mutant showed partial responsiveness to GA, whereas an enhanced GA responsive mutant suggested the retention of its partial activity (Van Tuinen et al., 1999), which confers that tomatoes have both DELLA-independent and DELLA-dependent pathways for GA-signaling (Livne et al., 2015).
During the “green revolution,” dwarf cultivars (RHT-B1 and RHT-D1, an ortholog of GAI) have been selected from wheat, which lacks GA response. These dwarf mutants (Rht-B1 and Rht-D1) have reduced GA response because of truncation near the DELLA domain (Peng et al., 1999). Similar truncation has been found in the D8 genes of maize. Maize contains two duplicated DELLA genes, DWARF PLANT 8 (D8) and DWARF PLANT 9 (D9), found on different chromosomes. Both D8 and D9 genes are gibberellin-insensitive and produce dwarfing phenotypes. The Arabidopsis transgenics carrying the dominant maize D8 and D9 alleles also showed a reduced length of rosette leaves, siliques, inflorescence, stems, root structures, shortened filaments, and produced dwarf floral structures with a few seeds (Lawit et al., 2010; Winkler and Freeling, 1994). In maize, D8 and D9 mutants also exhibited male sterility (Winkler and Freeling 1994). The dwarf plant 8 (d8) is a homolog of Reduced height (Rht1d) from wheat, Slender1 from barley, and Slender rice1 from rice (Lawit et al., 2010; Winkler and Freeling, 1994). The Arabidopsis transgenic carrying the grapevine Gibberellin Insensitive1 (VvGAI1) mutant alleles with three different promoters of wheat (Rht-B1b), barley (Sln1d), and Brassica (Brrga1-d) (Chandler et al., 2002; Muangprom et al., 2005) showed very short internodes and dwarf phenotype regardless of which promoter was used (Zhong and Yang, 2012). Two DELLA genes in Brachypodium distachyon (BdSLR1 and BdSLRL1) have been characterized in Arabidopsis and found to be involved in GA-mediated signaling and plant development. A similar function was also observed for Arabidopsis, rice, maize, and wheat orthologs (Niu et al., 2019). Single base deletion leading to frameshift mutation was observed in rice slender mutant, resulting in mutants exhibiting constitutive GA response phenotype (Ikeda et al., 2001) and reduced JA sensitivity (Yang et al., 2012), along with enhanced disease susceptibility (Yang et al., 2013). Unlike rice, in Arabidopsis, DELLA proteins are also found to promote susceptibility and resistance to the virulent biotrophs and necrotrophs, respectively. This differential function of DELLA proteins in Arabidopsis and rice might result from JA and SA signaling (Navarro et al., 2008). It also throws light on the diverged functions of DELLA in rice and Arabidopsis, which might be owing to the evolutionary or signaling divergence between monocots and dicots. In barley, the SLN (for Slender, rice SLR1 ortholog) gene has been identified (Chandler et al., 2002) and found to be degraded in response to GA (Fu et al., 2002).
Brassinosteroids (BRs) are steroidal phytohormones that regulate various biological processes, including agronomic traits in crop plants. OsDLT (DWARF AND LOW-TILLERING), a GRAS TF, was involved in BR signaling in rice, and dlt mutants showed phenotypic deformities including the reduced height, less tillering, and late-flowering, similar to that observed in BR-mutants (Tong et al., 2009, 2012). Gene OsDLT was down-regulated in the presence of BR; OsBZR1 can bind to its promoter to repress its expression, as experimentally proved by in vitro assays, which suggests that OsDLT and OsBZR1 regulate each other in opposite ways. In rice, upregulation of DWF4 (BR biosynthesis gene) enhances the tillering number, indicating a positive role of BR in tillering (Wu et al., 2008). In contrast, the BR negatively controls the OsDLT resulting in a reduction in tiller number and grain size in the mutants. Therefore, phenotypes of growth and tiller number are controlled by the expression of DLT in rice.
GRAS family members are also found to be involved in auxin signaling. An ortholog of AtSCL15 (Pysh et al., 1999) from Arabidopsis and BnSCL1 from B. napus are highly expressed in root tissue, and their expression is influenced by auxin (Gao et al., 2004). Under the influence of auxin, BnSCL1 expression is upregulated, and BnSCL1 interacts with histone deacetylase 19 (HDA19) to regulate downstream genes involved in the radial patterning of root. SCL1 genes in Pinus radiate (PrSCL1) and Castanea sativa (CsSCL1) are the two GRAS genes predominately expressed in roots under the influence of exogenous auxin (Sánchez et al., 2007).
Phytochrome signaling and growth regulation
In addition, to regulate various developmental pathways, GRAS proteins also play a crucial role in light signaling. For instance, SCL21, SCL5, and PAT1 have been involved in phytochrome A (PhyA) signal transduction. Proteins encoded by these three genes contain conserve EAISRRDL motif, which is absent in other family members, and the mutants of these three genes showed common Phy-A related deformities (Bolle et al., 2000; Torres-Galea et al., 2006, 2013). Further, SCL13 (Scarecrow-like 13), another member of the PAT1 subfamily is involved in the phytochrome B (PhyB) pathway. It is mainly involved in red light signaling and is also known to modulate phyA responses in a phyB-independent manner. The SCL3 antisense lines showed lower sensitivity toward red light and executed their role in hypocotyl elongation in plants (Torres-Galea et al., 2006). Two more cytoplasm and nuclear-localized proteins belonging to the PAT1 subfamily, i.e., AtPAT1 and AtSCL21, have also been characterized as positive regulators of phyA signal transduction in Arabidopsis. Moreover, the biochemical evidence showed that AtPAT1 and AtSCL21 interact physically. The double mutant showed an elongated hypocotyl under FR light, suggesting that SCL21 and PAT1 are essential for FR light-mediated signaling. These results suggested that the heterodimeric complex of SCL21 and PAT1 acts as a positive regulator of the phyA signaling pathway (Bolle et al., 2000; Torres-Galea et al., 2013).
GRAS proteins (especially DELLA) are also involved in regulation shade avoidance responses via phytochrome signaling in plants. In Arabidopsis, low red/far-red light ratio sense by the phyB photoreceptor and the phyB inactivates the PHYTOCHROME INTERACTING FACTOR 4 (PIF4) TFs through phosphorylation. PIF4 TFs are also a target by the GA signaling pathway, phyB promotes GA biosynthesis through regulating the expression of these two genes GA20ox and GA3ox. This leads to the degradation of DELLA proteins that otherwise prevent PIF4 function (Colebrook et al., 2014; Hisamatsu et al., 2005) (Figure 4C). Overall, these studies suggest that GRAS forms the core functional module of light signaling involving phytochrome.
Genome editing of gras genes for plant improvement
With recent advances in genome-editing technologies [i.e., zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), RNA-guided CRISPR-Cas nuclease system], several avenues are open for improving plant traits. These genome-editing technologies can precisely target any gene of interest. Here, a summary of recent studies utilizing CRISPR-Cas9 based genome editing to manipulate GRAS genes is provided. CRISPR-Cas9 technology has been implemented to target the GRAS gene in plants (Feng et al., 2013). The DELLA domain in GAI locus was targeted in Arabidopsis, and one amino acid was deleted. This resulted in DELLA proteins becoming insensitive to GA-induced degradation, causing repressed growth and a dwarf phenotype (Feng et al., 2013). Similarly, in tomatoes, the mutant generated by CRISPR, where one amino acid was deleted from DELLA protein, resulted in a dwarf phenotype (Tomlinson et al., 2019). In rice, SLR genes (which encode DELLA) were edited by targeting the DELLA domain, and 16 lines were developed involving six types of mutation in SLR genes (Jung et al., 2020). Out of these, two mutants were insensitive toward GA and were dwarf with shrunken leaf and short internode.
The SHR subfamily (involved in root radial patterning) has also been edited through the CRISPR-Cas system. In G. max, mutants were developed with anomalous root radial patterning by editing SHR genes (Cai et al., 2015). GRAS genes have also been edited in polyploid plant species where multiple paralogous genes are present. For instance, in B. napus (allotetraploid), four genes (BnaA9.RGA, BnaC9.RGA, BnaA6.RGA, and BnaC7.RGA) belonging to the RGA subfamily of GRAS proteins were edited through CRISPR-Cas (Yang et al., 2017). The quadruple mutant obtained was significantly taller than the wild type, implying that GRAS genes can be edited precisely in diploid and polyploid plant species. These studies indicated that the CRISPR-Cas technology has significant potential for crop improvement and extensive usage in agriculture. Till now only a few GRAS genes (DELLAs, SHRs and RGAs) have been targeted that mainly regulate plant height. As GRAS genes were also involved in various biological processes, we suggested that the CRISPR-Cas system should be used to target potential GRAS genes for crop improvement against grain yield, abiotic and biotic stress enhancement.
Conclusions and future outlook
GRAS proteins play a plethora of biological functions, including regulation of growth and development, phytohormones and phytochrome signaling, symbiosis, biotic and abiotic stress regulation. For instance, DELLA acts as a master regulator of the GA gibberellin signaling pathway and is involved in several biological events of plant development, stress regulation, phytochrome signaling, and symbiotic associations. The SCL subfamily is also reported to regulate endodermis development, seed germination, root elongation, and multiple abiotic stress regulations. Similarly, RAM1 act as a central regulator of arbuscular development and is involved in hyphopodia formation in AM fungi, whose transcription is regulated explicitly by CYCLOPS and DELLA (Pimprikar et al., 2016). However, predominant studies are limited to genome-scale identification and characterization using in silico tools, followed by expression profiling in different tissues, developmental stages, and stress treatments. Further functional studies are necessary to gain mechanistic insights into the precise role of GRAS TFs in regulating different aspects of plant life. Open access databases such as Plant Transcription Factor Database (http://planttfdb.gao-lab.org/family.php?fam=GRAS) contain the information of GRAS proteins in all the sequenced plant genomes and provide a base for functional studies. Also, efforts need to be directed toward identifying their upstream regulators and downstream target genes. This comprehensive information will help to delineate the cascade of molecular events involving GRAS proteins during the growth, development, and stress responses.
Importantly, crystal structure elucidation and in vivo studies are needed to reveal the mode of action of GRAS proteins with their interacting partners and substantiate that GRAS acts as TFs. The genome sequencing data of various lower and higher plant species are available now, which provides an opportunity to explore the evolution and diversification of GRAS genes on an evolutionary timescale. The conserved functional mechanisms of the GRAS family genes among different species suggest its conservation among plant species. For instance, LS, LAS, and MOC1 from tomatoes, Arabidopsis, and rice, respectively, share only about a 50% sequence similarity and show a conserved function in promoting shoot branching or tillering. LAS genes in Arabidopsis and tomato are functionally similar, and a conserved control mechanism exists between two distantly related species. However, the differences in the mutant phenotypes in both species may be attributed to redundant gene activities during flower development in Arabidopsis, leading to variability in phenotypes (Greb et al., 2003). The expression of many GRAS TFs is triggered upon stress exposure, suggesting their potential for plant genetic engineering and genome editing (ZFNs, TALENs, CRISPR/Cas9) approaches to improve agronomic traits that can lead to the development of higher-yielding crop varieties.
Acknowledgments
The authors acknowledge the Council of Scientific and Industrial Research (CSIR) for providing funds (MLP-201). V.J. and V.G. thank the Department of Science and Technology for the INSPIRE faculty award. V.J. also thanks the Science and Engineering Research Board (SERB) for the Early Career Research Award. P.K. also thanks CSIR for Junior Research Fellowship. This manuscript represents CSIR-IHBT communication number: 4793.
Author contributions
Conceptualization, V.G.; writing – original draft, V.J., M.K., P.K., V.G., and G.Z.; writing – review and editing, V.J., V.G., G.Z., and S.K.; data curation, V.J., M.K., and P.K.; visualization, V.J., M.K., P.K., and V.G.; supervision, V.G., G.Z., and S.K.; project administration, V.G. and S.K.; funding acquisition, V.G., G.Z., and S.K.
Declaration of interests
The authors declare no conflict of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105026.
Contributor Information
Gaurav Zinta, Email: gzinta@gmail.com.
Vijay Gahlaut, Email: zone4vijay@gmail.com.
Supplemental information
References
- Achard P., Genschik P. Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 2009;60:1085–1092. doi: 10.1093/jxb/ern301. [DOI] [PubMed] [Google Scholar]
- Al-Mssallem I.S., Hu S., Zhang X., Lin Q., Liu W., Tan J., Yu X., Liu J., Pan L., Zhang T., et al. Genome sequence of the date palm Phoenix dactylifera L. Nat. Commun. 2013;4 doi: 10.1038/ncomms3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert V.A., Barbazuk W.B., De Pamphilis C.W., Der J., Leebens-Mack J., Ma Q.-H. The Amborella genome and the evolution of flowering plants. Science. 2013;342 doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- Aoyanagi T., Ikeya S., Kobayashi A., Kozaki A. Gene regulation via the combination of transcription factors in the INDETERMINATE DOMAIN and GRAS families. Genes. 2020;11:613. doi: 10.3390/genes11060613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argout X., Salse J., Aury J.-M., Guiltinan M.J., Droc G., Gouzy J., Allegre M., Chaparro C., Legavre T., Maximova S.N., et al. The genome of Theobroma cacao. Nat. Genet. 2011;43:101–108. doi: 10.1038/ng.736. [DOI] [PubMed] [Google Scholar]
- Bassel G.W., Mullen R.T., Bewley J.D. Procera is a putative DELLA mutant in tomato (Solanum lycopersicum): effects on the seed and vegetative plant. J. Exp. Bot. 2008;59:585–593. doi: 10.1093/jxb/erm354. [DOI] [PubMed] [Google Scholar]
- Bi Y., Wei B., Meng Y., Li Z., Tang Z., Yin F., Qian C. Genome-wide GRAS gene family analysis reveals the classification, expression profiles in Melon (Cucumis melo L.) Phyton. 2021;90:1161–1175. doi: 10.32604/phyton.2021.014396. [DOI] [Google Scholar]
- Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- Bolle C., Koncz C., Chua N.-H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 2000;14:1269–1278. doi: 10.1101/gad.14.10.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A., Brands M., Wewer V., Dörmann P., Harrison M.J. Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 2017;214:1631–1645. doi: 10.1111/nph.14533. [DOI] [PubMed] [Google Scholar]
- Cai Y., Chen L., Liu X., Sun S., Wu C., Jiang B., Han T., Hou W. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E., Ruiz-Rivero O., Peres L.E.P., Atares A., Garcia-Martinez J.L. Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiol. 2012;160:1581–1596. doi: 10.1104/pp.112.204552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci A., Rouard M. Evolutionary analyses of GRAS transcription factors in angiosperms. Front. Plant Sci. 2017;8:273. doi: 10.3389/fpls.2017.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler P.M., Marion-Poll A., Ellis M., Gubler F. Mutants at the Slender1 locus of barley cv himalaya. Molecular and physiological characterization. Plant Physiol. 2002;129:181–190. doi: 10.1104/pp.010917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Sun M., Zhang M., Tang Z., Sheng J., Liu Y., Song B., Li J., Zhao K., Wu J. Genome-wide comparison of the GRAS protein family in eight rosaceae species and GRAS gene expression analysis in Chinese white pear (Pyrus bretschneideri Rehder) N. Z. J. Crop Hortic. Sci. 2021 doi: 10.1080/01140671.2021.1936081. [DOI] [Google Scholar]
- Chen H., Li H., Lu X., Chen L., Liu J., Wu H. Identification and expression analysis of GRAS transcription factors to elucidate candidate genes related to stolons, fruit ripening and abiotic stresses in woodland strawberry (Fragaria vesca) Int. J. Mol. Sci. 2019;42:21–32. doi: 10.3390/ijms20184593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Yan Q., Li J., Feng L., Zhang Y., Xu J., Xia R., Zeng Z., Liu Y. The GRAS gene family and its roles in seed development in litchi (Litchi chinensis Sonn) BMC Plant Biol. 2021;21:1–15. doi: 10.1186/s12870-021-03193-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Li H., Chen Y., Zheng Q., Li B., Li Z. TaSCL14, a Novel Wheat (Triticum aestivum L.) GRAS gene, regulates plant growth, photosynthesis, tolerance to photooxidative stress, and senescence. J. Genet. Genomics. 2015;42:21–32. doi: 10.1016/j.jgg.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Chen Y.Q., Tai S.S., Wang D.W., Ding A.M., Sun T.T., Wang W.F., Sun Y.H. Homology-based analysis of the GRAS gene family in tobacco. Genet. Mol. Res.: GMR. 2015;14:15188–15200. doi: 10.4238/2015.November.25.7. [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhu P., Wu S., Lu Y., Sun J., Cao Q., Li Z., Xu T. Identification and expression analysis of GRAS transcription factors in the wild relative of sweet potato Ipomoea trifida. BMC Genom. 2019;20:911. doi: 10.1186/s12864-019-6316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Qin L., Lee S., Fu X., Richards D.E., Cao D., Luo D., Harberd N.P., Peng J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development. 2004;131:1055–1064. doi: 10.1242/dev.00992. [DOI] [PubMed] [Google Scholar]
- Choe J., Kim B., Yoon E.K., Jang S., Kim G., Dhar S., Lee S.A., Lim J. Characterization of the GRAS transcription factor SCARECROW-LIKE 28’s role in Arabidopsis root growth. J. Plant Biol. 2017;60:462–471. doi: 10.1007/s12374-017-0112-1. [DOI] [Google Scholar]
- Colebrook E.H., Thomas S.G., Phillips A.L., Hedden P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014;217:67–75. doi: 10.1242/jeb.089938. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A., Díaz-Triviño S., Blilou I., Grieneisen V.A., Sozzani R., Zamioudis C., Miskolczi P., Nieuwland J., Benjamins R., Dhonukshe P., et al. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell. 2012;150:1002–1015. doi: 10.1016/j.cell.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Curaba J., Talbot M., Li Z., Helliwell C. Over-expression of microRNA171 affects phase transitions and floral meristem determinacy in barley. BMC Plant Biol. 2013;13:6. doi: 10.1186/1471-2229-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czikkel B.E., Maxwell D.P. NtGRAS1, a novel stress-induced member of the GRAS family in tobacco, localizes to the nucleus. J. Plant Physiol. 2007;164:1220–1230. doi: 10.1016/j.jplph.2006.07.010. [DOI] [PubMed] [Google Scholar]
- D’Hont A., Denoeud F., Aury J.-M., Baurens F.-C., Carreel F., Garsmeur O., Noel B., Bocs S., Droc G., Rouard M., et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- Dai C., Xue H. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010;29:1916–1927. doi: 10.1038/emboj.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J.-M., Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- Day R.B., Tanabe S., Koshioka M., Mitsui T., Itoh H., Ueguchi-Tanaka M., Matsuoka M., Kaku H., Shibuya N., Minami E. Two rice GRAS family genes responsive to N-acetylchitooligosaccharide elicitor are induced by phytoactive gibberellins: evidence for cross-talk between elicitor and gibberellin signaling in rice cells. Plant Mol. Biol. 2004;54:261–272. doi: 10.1023/B:PLAN.0000028792.72343.ee. [DOI] [PubMed] [Google Scholar]
- De Luis A., Markmann K., Cognat V., Holt D.B., Charpentier M., Parniske M., Stougaard J., Voinnet O. Two MicroRNAs linked to nodule infection and nitrogen-fixing ability in the legume Lotus japonicus. Plant Physiol. 2012;160:2137–2154. doi: 10.1104/pp.112.204883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F., Carretero-Paulet L., Dereeper A., Droc G., Guyot R., Pietrella M., Zheng C., Alberti A., Anthony F., Aprea G., et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345:1181–1184. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/S0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dill A., Sun T.P. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159:777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom E.M. Phylogenetic analysis of GRAS proteins from moss, lycophyte and vascular plant lineages reveals that GRAS genes arose and underwent substantial diversification in the ancestral lineage common to bryophytes and vascular plants. Plant Signal. Behav. 2011;6:850–854. doi: 10.4161/psb.6.6.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Zhang D., Gao C., Zhao M., Wu H., Li Y., Shen Y., Han M. Identification, classification, and expression analysis of GRAS gene family in Malus domestica. Front. Physiol. 2017;8:253. doi: 10.3389/fphys.2017.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T., Li X., Yang W., Xia K., Ouyang J., Zhang M. Rice osa-miR171c mediates phase change from vegetative to reproductive development and shoot apical meristem maintenance by repressing four OsHAM transcription factors. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Wei X., Lai D., Yang H., Feng L., Li L., Niu K., Chen L., Xiang D., Ruan J., et al. Genome-wide investigation of the GRAS transcription factor family in foxtail millet (Setaria italica L.) BMC Plant Biol. 2021;21:1–19. doi: 10.1186/s12870-021-03277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Yan J., Lai D., Yang H., Xue G., He A., Guo T., Chen L., Cheng X.B., Xiang D.B., et al. Genome-wide identification, expression analysis, and functional study of the GRAS transcription factor family and its response to abiotic stress in sorghum [Sorghum bicolor (L.) Moench] BMC Genom. 2021;22:1–21. doi: 10.1186/s12864-021-07848-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D.-L., Wei P., Cao F., Zhu S., Zhang F., Mao Y., et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss D.S., Gomez S.K., Park H.-J., MacLean A.M., Müller L.M., Bhattarai K.K., Lévesque-Tremblay V., Maldonado-Mendoza I.E., Harrison M.J. A transcriptional program for arbuscule degeneration during AM Symbiosis is regulated by MYB1. Curr. Biol. 2017;27:1206–1212. doi: 10.1016/j.cub.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Fode B., Siemsen T., Thurow C., Weigel R., Gatz C. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell. 2008;20:3122–3135. doi: 10.1105/tpc.108.058974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Richards D.E., Ait-ali T., Hynes L.W., Ougham H., Peng J., Harberd N.P. Gibberellin-mediated proteasome-dependent degradation of the Barley DELLA protein SLN1 repressor. Plant Cell. 2002;14:3191–3200. doi: 10.1105/tpc.006197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Wysocka-Diller J., Kato T., Fujisawa H., Benfey P.N., Tasaka M. Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 1998;14:425–430. doi: 10.1046/j.1365-313X.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- Fukazawa J., Mori M., Watanabe S., Miyamoto C., Ito T., Takahashi Y. DELLA-GAF1 complex is a main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiol. 2017;175:1395–1406. doi: 10.1104/pp.17.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J., Teramura H., Murakoshi S., Nasuno K., Nishida N., Ito T., Yoshida M., Kamiya Y., Yamaguchi S., Takahashi Y. DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell. 2014;26:2920–2938. doi: 10.1105/tpc.114.125690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K.L., Paquette A.J., Nakajima K., Benfey P.N. Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 2004;14:1847–1851. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- Gao M., Parkin I., Lydiate D., Hannoufa A. An auxin-responsive SCARECROW-like transcriptional activator interacts with histone deacetylase. Plant Mol. Biol. 2004;55:417–431. doi: 10.1007/s11103-004-0892-9. [DOI] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Balzergue C., Puech-Pagès V., Novero M., Rey T., Fournier J., Rochange S., Bécard G., Bonfante P., et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B.W., Oldroyd G.E.D. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature. 2006;441:1149–1152. doi: 10.1038/nature04812. [DOI] [PubMed] [Google Scholar]
- Gobbato E., Marsh J.F., Vernié T., Wang E., Maillet F., Kim J., Miller J.B., Sun J., Bano S.A., Ratet P., et al. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 2012;22:2236–2241. doi: 10.1016/j.cub.2012.09.044. [DOI] [PubMed] [Google Scholar]
- Goldy C., Pedroza-Garcia J.-A., Breakfield N., Cools T., Vena R., Benfey P.N., De Veylder L., Palatnik J., Rodriguez R.E. The Arabidopsis GRAS-type SCL28 transcription factor controls the mitotic cell cycle and division plane orientation. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2005256118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K., Sasaki A., Itoh H., Ueguchi-Tanaka M., Ashikari M., Kitano H., Matsuoka M. GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 2004;37:626–634. doi: 10.1111/j.1365-313x.2003.01990.x. [DOI] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schäfer E., Müller D., Herrero R., Schmitz G., Theres K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003;17:1175–1187. doi: 10.1101/gad.260703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J., Agudelo-Romero P., Teixeira R.T., Martinez-Zapater J.M., Fortes A.M. Structural and functional analysis of the GRAS gene family in grapevine indicates a role of GRAS proteins in the control of development and stress responses. Front. Plant Sci. 2016;7:353. doi: 10.3389/fpls.2016.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Wen J., Yang J., Ke Y., Wang M., Liu M., Ran F., Wu Y., Li P., Li J., et al. Genome-wide survey and expression analyses of the GRAS gene family in Brassica napus reveals their roles in root development and stress response. Planta. 2019;250:1051–1072. doi: 10.1007/s00425-019-03199-y. [DOI] [PubMed] [Google Scholar]
- Guo Y., Wu H., Li X., Li Q., Zhao X., Duan X., An Y., Lv W., An H. Identification and expression of GRAS family genes in maize (Zea mays L.) PLoS One. 2017;12 doi: 10.1371/journal.pone.0185418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Paszkowski U. Multiple control levels of root system remodeling in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2013;4:1–8. doi: 10.3389/fpls.2013.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Sawers R.J., Marti G., Andrés-Hernández L., Yang S.Y., Casieri L., Angliker H., Oakeley E.J., Wolfender J.L., Abreu-Goodger C., et al. Transcriptome diversity among rice root types during asymbiosis and interaction with arbuscular mycorrhizal fungi. Proc. Natl. Acad. Sci. USA. 2015;112:6754–6759. doi: 10.1073/pnas.1504142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakoshima T. Structural basis of the specific interactions of GRAS family proteins. FEBS Lett. 2018;592:489–501. doi: 10.1002/1873-3468.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck C., Kuhn H., Heidt S., Walter S., Rieger N., Requena N. Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr. Biol. 2016;26:2770–2778. doi: 10.1016/j.cub.2016.07.059. [DOI] [PubMed] [Google Scholar]
- Heckmann A.B., Lombardo F., Miwa H., Perry J.A., Bunnewell S., Parniske M., Wang T.L., Downie J.A. Lotus japonicus Nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 2006;142:1739–1750. doi: 10.1104/pp.106.089508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. The genes of the green revolution. Trends Genet. 2003;19:5–9. doi: 10.1016/S0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Welch D., Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/S0092-8674(00)80865-X. [DOI] [PubMed] [Google Scholar]
- Heo J.O., Chang K.S., Kim I.A., Lee M.H., Lee S.A., Song S.K., Lee M.M., Lim J. Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA. 2011;108:2166–2171. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Kouketu E., Katoh H., Aya K., Ueguchi-Tanaka M., Matsuoka M. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 2012;71:443–453. doi: 10.1111/j.1365-313X.2012.05000.x. [DOI] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E.D. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in medicago truncatulaw. Plant Cell. 2009;21:545–557. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu T., King R.W., Helliwell C.A., Koshioka M. The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol. 2005;138:1106–1116. doi: 10.1104/pp.104.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofferek V., Mendrinna A., Gaude N., Krajinski F., Devers E.A. MiR171h restricts root symbioses and shows like its target NSP2 a complex transcriptional regulation in Medicago truncatula. BMC Plant Biol. 2014;14:199. doi: 10.1186/s12870-014-0199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.S., Hoang N.T., Yan Z., Tóth K., Meyers B.C., Stacey G. Characterization of the spatial and temporal expression of two soybean miRNAs identifies SCL6 as a novel regulator of soybean nodulation. Front. Plant Sci. 2019;10:475. doi: 10.3389/fpls.2019.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y.C., Xia K., Yan Y., Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008 doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Peng S., Xian Z., Lin D., Hu G., Yang L., Ren M., Li Z. Overexpression of a tomato miR171 target gene SlGRAS24 impacts multiple agronomical traits via regulating gibberellin and auxin homeostasis. Plant Biotechnol. J. 2017;15:472–488. doi: 10.1111/pbi.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Xian Z., Kang X., Tang N., Li Z. Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant Biol. 2015;15:209. doi: 10.1186/s12870-015-0590-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva L.M., Radivojac P., Brown C.J., O’Connor T.R., Sikes J.G., Obradovic Z., Dunker A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Sasaki A., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Hasegawa Y., Minami E., Ashikari M., Matsuoka M. Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 2005;46:1392–1399. doi: 10.1093/pcp/pci152. [DOI] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha D.K., Chanwala J., Sandeep I.S., Dey N. Comprehensive identification and expression analysis of GRAS gene family under abiotic stress and phytohormone treatments in Pearl millet. Funct. Plant Biol. 2021;48:1039–1052. doi: 10.1071/FP21051. [DOI] [PubMed] [Google Scholar]
- Jiang C., Gao F., Li T., Chen T., Zheng X., Lian X., Wang X., Zhang H., Cheng J., Wang W., Ye X. Genome-wide analysis of the GRAS transcription factor gene family in peach (Prunus persica) and ectopic expression of PpeDELLA1 and PpeDELLA2 in Arabidopsis result in dwarf phenotypes. Sci. Hortic. 2022;298 doi: 10.1016/j.scienta.2022.111003. [DOI] [Google Scholar]
- Jiang Y., Xie Q., Wang W., Yang J., Zhang X., Yu N., Zhou Y., Wang E. Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol. Plant. 2018;11:1344–1359. doi: 10.1016/j.molp.2018.09.006. [DOI] [PubMed] [Google Scholar]
- Jin Y., Liu H., Luo D., Yu N., Dong W., Wang C., Zhang X., Dai H., Yang J., Wang E. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 2016;7:1–14. doi: 10.1038/ncomms12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Zhang D., Wang X., Tang W., Wang W., Huai J., Xu G., Chen D., Li Y., Lin R. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell. 2013;25:242–256. doi: 10.1105/tpc.112.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Boden E., Arias J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell. 2003;15:1846–1858. doi: 10.1105/tpc.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.J., Kim J.H., Lee H.J., Kim D.H., Yu J., Bae S., Cho Y.-G., Kang K.K. Generation and transcriptome profiling of Slr1-d7 and Slr1-d8 mutant lines with a new semi-dominant dwarf allele of SLR1 using the CRISPR/Cas9 system in Rice. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21155492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P., Gleason C., Edwards A., Marsh J., Mitra R.M., Hirsch S., Jakab J., Sims S., Long S.R., Rogers J., et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- Kamiya N., Itoh J.-I., Morikami A., Nagato Y., Matsuoka M. he SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J. 2003;36:45–54. doi: 10.1046/j.1365-313X.2003.01856.x. [DOI] [PubMed] [Google Scholar]
- Kim G., Dhar S., Lim J. The SHORT-ROOT regulatory network in the endodermis development of Arabidopsis roots and shoots. J. Plant Biol. 2017;60:306–313. doi: 10.1007/s12374-017-0134-8. [DOI] [Google Scholar]
- Khan Y., Xiong Z., Zhang H., Liu S., Yaseen T., Hui T. Expression and roles of GRAS gene family in plant growth, signal transduction, biotic and abiotic stress resistance and symbiosis formation—a review. Plant Biol. 2022;24:404–416. doi: 10.1111/plb.13364. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Bentsink L., Hilhorst H. Seed dormancy and germination. Curr. Opin. Plant Biol. 2002;5:33–36. doi: 10.1016/S1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Kumari P., Gahlaut V., Kaur E., Singh S., Kumar S., Jaiswal V. Genome-wide identification of GRAS transcription factors and their potential roles in growth and development of rose (Rosa chinensis) J. Plant Growth Regul. 2022 doi: 10.1007/s00344-022-10635-z. [DOI] [Google Scholar]
- Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crété P., Voinnet O., Robaglia C. Biochemical evidence for translational repression by Arabidopsis MicroRNAs. Plant Cell. 2009;21:1762–1768. doi: 10.1105/tpc.108.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D., Delaux P.-M., Formey D., Lelandais-Brière C., Fort S., Cottaz S., Bécard G., Niebel A., Roux C., Combier J.-P. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012;72:512–522. doi: 10.1111/j.1365-313X.2012.05099.x. [DOI] [PubMed] [Google Scholar]
- Laskar P., Bhattacharya S., Chaudhuri A., Kundu A. Exploring the GRAS gene family in common bean (Phaseolus vulgaris L.): characterization, evolutionary relationships, and expression analyses in response to abiotic stresses. Planta. 2021;254:1–21. doi: 10.1007/s00425-021-03725-x. [DOI] [PubMed] [Google Scholar]
- Lawit S.J., Wych H.M., Xu D., Kundu S., Tomes D.T. Maize DELLA proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant Cell Physiol. 2010;51:1854–1868. doi: 10.1093/pcp/pcq153. [DOI] [PubMed] [Google Scholar]
- Lee M.-H., Kim B., Song S.-K., Heo J.-O., Yu N.-I., Lee S.A., Kim M., Kim D.G., Sohn S.O., Lim C.E., et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008;67:659–670. doi: 10.1007/s11103-008-9345-1. [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Sun B., Xie F., Gong R., Luo Y., Zhang F., Yan Z., Tang H. Identification of the GRAS gene family in the Brassica juncea genome provides insight into its role in stem swelling in stem mustard. PeerJ. 2019;7 doi: 10.7717/peerj.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhao Y., Zhao Z., Wu X., Sun L., Liu Q., Wu Y. Crystal structure of the GRAS domain of SCARECROW-LIKE7 in Oryza sativa. Plant Cell. 2016;28:1025–1034. doi: 10.1105/tpc.16.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu W., Li B., Liu G., Wei Y., He C., Shi H. Identification and functional analysis of cassava DELLA proteins in plant disease resistance against cassava bacterial blight. Plant Physiol. Biochem. 2018;124:70–76. doi: 10.1016/j.plaphy.2017.12.022. [DOI] [PubMed] [Google Scholar]
- Li X., Qian Q., Fu Z., Wang Y., Xiong G., Zeng D., Wang X., Liu X., Teng S., Hiroshi F., et al. Control of tillering in rice. Nature. 2003;422:618–621. doi: 10.1038/nature01518. [DOI] [PubMed] [Google Scholar]
- Lim J., Helariutta Y., Specht C.D., Jung J., Sims L., Bruce W.B., Diehn S., Benfey P.N. Molecular analysis of the SCARECROW gene in maize reveals a common basis for radial patterning in diverse meristems. Plant Cell. 2000;12:1307–1318. doi: 10.1105/tpc.12.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Jung J.W., Lim C.E., Lee M.-H., Kim B.J., Kim M., Bruce W.B., Benfey P.N. Conservation and diversification of SCARECROW in maize. Plant Mol. Biol. 2005;59:619–630. doi: 10.1007/s11103-005-0578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Park J., Lee N., Jeong J., Toh S., Watanabe A., Kim J., Kang H., Kim D.H., Kawakami N., et al. ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell. 2013;25:4863–4878. doi: 10.1105/tpc.113.118604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Wang D., Dong H., Gu S., Cheng Z., Gong J., Qin R., Jiang L., Li G., Wang J.L., et al. Rice APC/CTE controls tillering by mediating the degradation of MONOCULM 1. Nat. Commun. 2012;3:752. doi: 10.1038/ncomms1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Sun Y., Xue J., Jia X., Li R. Genome-wide characterization and expression analysis of GRAS gene family in pepper (Capsicum annuum L.) PeerJ. 2018;6 doi: 10.7717/peerj.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Huang L., Ma Z., Sun W., Wu Q., Tang Z., Bu T., Li C., Chen H. Genome-wide identification, expression analysis and functional study of the GRAS gene family in Tartary buckwheat (Fagopyrum tataricum) BMC Plant Biol. 2019;19:342. doi: 10.1186/s12870-019-1951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Widmer A. Genome-wide comparative analysis of the GRAS gene family in populus, Arabidopsis and rice. Plant Mol. Biol. Report. 2014;32:1129–1145. doi: 10.1007/s11105-014-0721-5. [DOI] [Google Scholar]
- Liu Y., Shi Y., Su D., Lu W., Li Z. SlGRAS4 accelerates fruit ripening by regulating ethylene biosynthesis genes and SlMADS1 in tomato. Hortic. Res. 2021;8:3. doi: 10.1038/s41438-020-00431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang W. Characterization of the GRAS gene family reveals their contribution to the high adaptability of wheat. PeerJ. 2021;9 doi: 10.7717/peerj.10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livne S., Lor V.S., Nir I., Eliaz N., Aharoni A., Olszewski N.E., Eshed Y., Weiss D. Uncovering DELLA-independent gibberellin responses by characterizing new tomato procera mutants. Plant Cell. 2015;27:1579–1594. doi: 10.1105/tpc.114.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Long Y., Smet W., Cruz-Ramírez A., Castelijns B., de Jonge W., Mähönen A.P., Bouchet B.P., Perez G.S., Akhmanova A., Scheres B., et al. Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell. 2015;27:1185–1199. doi: 10.1105/tpc.114.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Wang T., Xu Z., Sun L., Zhang Q. Genome-wide analysis of the GRAS gene family in Prunus mume. Mol. Genet. Genomics. 2015;290:303–317. doi: 10.1007/s00438-014-0918-1. [DOI] [PubMed] [Google Scholar]
- Luginbuehl L.H., Oldroyd G.E.D. Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Curr. Biol. 2017;27:R952–R963. doi: 10.1016/j.cub.2017.06.042. [DOI] [PubMed] [Google Scholar]
- Lv G., Zheng X., Duan Y., Wen Y., Zeng B., Ai M., He B. The GRAS gene family in watermelons: identification, characterization and expression analysis of different tissues and root-knot nematode infestations. PeerJ. 2021;9 doi: 10.7717/peerj.11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H.-S., Liang D., Shuai P., Xia X.-L., Yin W.-L. The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 2010;61:4011–4019. doi: 10.1093/jxb/erq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Hu X., Cai W., Huang W., Zhou X., Luo Q., Yang H., Wang J., Huang J. Arabidopsis miR171-targeted scarecrow-like proteins bind to GT cis-elements and mediate gibberellin-regulated chlorophyll biosynthesis under light conditions. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet F., Poinsot V., André O., Puech-Pagès V., Haouy A., Gueunier M., Cromer L., Giraudet D., Formey D., Niebel A., et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T., Kay L.E., Forman-Kaya J.D. Protein dynamics and conformational disorder in molecular recognition. J. Mol. Recognit. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno M.A., Sozzani R., Yardımcı G.G., Petricka J.J., Vernoux T., Blilou I., Alonso J., Winter C.M., Ohler U., Scheres B., et al. Transcriptional control of tissue formation throughout root development. Science. 2015;350:426–430. doi: 10.1126/science.aad1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Minami M., Takase H., Hotta Y., Hiratsuka K. Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 2003;278:20865–20873. doi: 10.1074/jbc.M301712200. [DOI] [PubMed] [Google Scholar]
- Muangprom A., Thomas S.G., Sun T., Osborn T.C. A novel dwarfing mutation in a green revolution gene from Brassica rapa. Plant Physiol. 2005;137:931–938. doi: 10.1104/pp.104.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- Murray J.D., Cousins D.R., Jackson K.J., Liu C. Signaling at the root surface: the role of cutin monomers in mycorrhization. Mol. Plant. 2013;6:1381–1383. doi: 10.1093/mp/sst090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Navarro L., Bari R., Achard P., Lisón P., Nemri A., Harberd N.P., Jones J.D.G. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 2008;18:650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- Ni L., Wang Z., Liu X., Wu S., Hua J., Liu L., Yin Y., Li H., Gu C. Genome-wide study of the GRAS gene family in Hibiscus hamabo Sieb. et Zucc and analysis of HhGRAS14-induced drought and salt stress tolerance in Arabidopsis. Plant Sci. 2022;319 doi: 10.1016/j.plantsci.2022.111260. [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Fujita T., Shin-I T., Seki M., Nishide H., Uchiyama I., Kamiya A., Carninci P., Hayashizaki Y., Shinozaki K., et al. Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution. Proc. Natl. Acad. Sci. USA. 2003;100:8007–8012. doi: 10.1073/pnas.0932694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Chen S., Li J., Liu Y., Ji W., Li H. Genome-wide identification of GRAS genes in Brachypodium distachyon and functional characterization of BdSLR1 and BdSLRL1. BMC Genom. 2019;20:635. doi: 10.1186/s12864-019-5985-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Zhao T., Xu X., Li J. Genome-wide identification and characterization of GRAS transcription factors in tomato (Solanum lycopersicum) PeerJ. 2017;5 doi: 10.7717/peerj.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.-J.J., Floss D.S., Levesque-Tremblay V., Bravo A., Harrison M.J. Hyphal branching during arbuscule development requires reduced arbuscular mycorrhiza. Plant Physiol. 2015;169:2774–2788. doi: 10.1104/pp.15.01155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Nguyen K.T., Park E., Jeon J.-S., Choi G. DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell. 2013;25:927–943. doi: 10.1105/tpc.112.108951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauluzzi G., Divol F., Puig J., Guiderdoni E., Dievart A., Périn C. Surfing along the root ground tissue gene network. Dev. Biol. 2012;365:14–22. doi: 10.1016/j.ydbio.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Richards D.E., Hartley N.M., Murphy G.P., Devos K.M., Flintham J.E., Beales J., Fish L.J., Worland A.J., Pelica F., et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Petricka J.J., Winter C.M., Benfey P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimprikar P., Carbonnel S., Paries M., Katzer K., Klingl V., Bohmer M.J., Karl L., Floss D.S., Harrison M.J., Parniske M., et al. A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr. Biol. 2016;26:987–998. doi: 10.1016/j.cub.2016.01.069. [DOI] [PubMed] [Google Scholar]
- Pimprikar P., Gutjahr C. Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol. 2018;59:678–695. doi: 10.1093/pcp/pcy024. [DOI] [PubMed] [Google Scholar]
- Pysh L.D., Wysocka-Diller J.W., Camilleri C., Bouchez D., Benfey P.N. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313X.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Quan S., Niu J., Zhou L., Xu H., Ma L., Qin Y. Genome-wide identification, classification, expression and duplication analysis of GRAS family genes in juglans regia L. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-48287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich M.K., Courty P.-E., Roux C., Reinhardt D. Role of the GRAS transcription factor ATA/RAM1 in the transcriptional reprogramming of arbuscular mycorrhiza in Petunia hybrida. BMC Genom. 2017;18:589. doi: 10.1186/s12864-017-3988-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C., Vielba J.M., Ferro E., Covelo G., Solé A., Abarca D., de Mier B.S., Díaz-Sala C. Two SCARECROW-LIKE genes are induced in response to exogenous auxin in rooting-competent cuttings of distantly related forest species. Tree Physiol. 2007;27:1459–1470. doi: 10.1093/treephys/27.10.1459. [DOI] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G., Theres K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA. 1999;96:290–295. doi: 10.1073/pnas.96.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Z., Luo X., Wu M., Wei L., Fan Z., Zhu Y. Genome-wide identification and expression of GRAS gene family members in cassava. BMC Plant Biol. 2020;20:46. doi: 10.1186/s12870-020-2242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtark O.Y., Sulima A.S., Zhernakov A.I., Kliukova M.S., Fedorina J.V., Pinaev A.G., Kryukov A.A., Akhtemova G.A., Tikhonovich I.A., Zhukov V.A. Arbuscular mycorrhiza development in pea (Pisum sativum L.) mutants impaired in five early nodulation genes including putative orthologs of NSP1 and NSP2. Symbiosis. 2016;68:129–144. doi: 10.1007/s13199-016-0382-2. [DOI] [Google Scholar]
- Sidhu N.S., Pruthi G., Singh S., Bishnoi R., Singla D. Genome-wide identification and analysis of GRAS transcription factors in the bottle gourd genome. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-71240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Katzer K., Lambert J., Cerri M., Parniske M. CYCLOPS, A DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe. 2014;15:139–152. doi: 10.1016/j.chom.2014.01.011. [DOI] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- Song X., Li Y., Cao X., Qi Y. MicroRNAs and their regulatory roles in plant–environment interactions. Annu. Rev. Plant Biol. 2019;70:489–525. doi: 10.1146/annurev-arplant-050718-100334. [DOI] [PubMed] [Google Scholar]
- Song X.M., Liu T.K., Duan W.K., Ma Q.H., Ren J., Wang Z., Li Y., Hou X.L. Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp. pekinensis) Genomics. 2014;103:135–146. doi: 10.1016/j.ygeno.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Stecher G., Tamura K., Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020;37:1237–1239. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman J., Jäggi F., Kuhlemeier C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002;16:2213–2218. doi: 10.1101/gad.230702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Jones W.T., Rikkerink E.H.A. GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 2012;442:1–12. doi: 10.1042/BJ20111766. [DOI] [PubMed] [Google Scholar]
- Sun X., Xue B., Jones W.T., Rikkerink E., Dunker A.K., Uversky V.N. A functionally required unfoldome from the plant kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 2011;77:205–223. doi: 10.1007/s11103-011-9803-z. [DOI] [PubMed] [Google Scholar]
- Tan L., Rong W., Luo H., Chen Y., He C. The Xanthomonas campestris effector protein XopDXcc8004 triggers plant disease tolerance by targeting DELLA proteins. New Phytol. 2014;204:595–608. doi: 10.1111/nph.12918. [DOI] [PubMed] [Google Scholar]
- Tian C., Wan P., Sun S., Li J., Chen M. Genome-Wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004;54:519–532. doi: 10.1023/B:PLAN.0000038256.89809.57. [DOI] [PubMed] [Google Scholar]
- To V.-T., Shi Q., Zhang Y., Shi J., Shen C., Zhang D., Cai W. Genome-Wide analysis of the GRAS gene family in barley (Hordeum vulgare L.) Genes. 2020;11:553. doi: 10.3390/genes11050553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson L., Yang Y., Emenecker R., Smoker M., Taylor J., Perkins S., Smith J., MacLean D., Olszewski N.E., Jones J.D.G. Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnol. J. 2019;17:132–140. doi: 10.1111/pbi.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Jin Y., Liu W., Li F., Fang J., Yin Y., Qian Q., Zhu L., Chu C. DWARF and LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009;58:803–816. doi: 10.1111/j.1365-313X.2009.03825.x. [DOI] [PubMed] [Google Scholar]
- Tong H., Liu L., Jin Y., Du L., Yin Y., Qian Q., Zhu L., Chu C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell. 2012;24:2562–2577. doi: 10.1105/tpc.112.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Galea P., Hirtreiter B., Bolle C. Two GRAS proteins, SCARECROW-LIKE21 and phytochrome A signal TRANSDUCTION1, function cooperatively in phytochrome A signal transduction. Plant Physiol. 2013;161:291–304. doi: 10.1104/pp.112.206607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Galea P., Huang L.-F., Chua N.-H., Bolle C. The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol. Genet. Genomics. 2006;276:13–30. doi: 10.1007/s00438-006-0123-y. [DOI] [PubMed] [Google Scholar]
- Van Tuinen A., Peters A.H.L.J., Kendrick R.E., Zeevaart J.A.D., Koornneef M. Characterisation of the procera mutant of tomato and the interaction of gibberellins with end-of-day far-red light treatments. Physiol. Plantarum. 1999;106:121–128. doi: 10.1034/j.1399-3054.1999.106117.x. [DOI] [Google Scholar]
- Venkateshwaran M., Volkening J.D., Sussman M.R., Ané J.-M. Symbiosis and the social network of higher plants. Curr. Opin. Plant Biol. 2013;6:118–127. doi: 10.1016/j.pbi.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Wang X.-S., Zhu H.-B., Jin G.-L., Liu H.-L., Wu W.-R., Zhu J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.) Plant Sci. 2007;172:414–420. doi: 10.1016/j.plantsci.2006.10.004. [DOI] [Google Scholar]
- Wang L., Ding X., Gao Y., Yang S. Genome-wide identification and characterization of GRAS genes in soybean (Glycine max) BMC Plant Biol. 2020;20:415. doi: 10.1186/s12870-020-02636-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Mai Y.-X., Zhang Y.-C., Luo Q., Yang H.-Q. MicroRNA171c-Targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol. Plant. 2010;3:794–806. doi: 10.1093/mp/ssq042. [DOI] [PubMed] [Google Scholar]
- Wang N., Wang K., Li S., Jiang Y., Li L., Zhao M., Jiang Y., Zhu L., Wang Y., Su Y., et al. Transcriptome-Wide identification, evolutionary analysis, and GA stress response of the GRAS gene family in Panax ginseng C. A. Meyer. Plants. 2020;9:190. doi: 10.3390/plants9020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li G., Sun Y., Qin Z., Feng P. Genome-wide analysis and characterization of GRAS family in switchgrass. Bioengineered. 2021;12:6096–6114. doi: 10.1080/21655979.2021.1972606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-X., Liu Z.-W., Wu Z.-J., Li H., Wang W.-L., Cui X., Zhuang J. Genome-wide identification and expression analysis of GRAS family transcription factors in tea plant (Camellia sinensis) Sci. Rep. 2018;8 doi: 10.1038/s41598-018-22275-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shi S., Zhou Y., Zhou Y., Yang J., Tang X. Genome-wide identification and characterization of GRAS transcription factors in sacred lotus (Nelumbo nucifera) PeerJ. 2016;4 doi: 10.7717/peerj.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch D., Hassan H., Blilou I., Immink R., Heidstra R., Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;21:2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M., Daviere J.M., Cheminant S., Regnault T., Baumberger N., Heintz D., Achard P. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell. 2012;24:3307–3319. doi: 10.1105/tpc.112.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler R.G., Freeling M. Physiological genetics of the dominant gibberellin-nonresponsive maize dwarfs, Dwarf8 and Dwarf9. Planta. 1994;193:341–348. doi: 10.1007/BF00201811. [DOI] [Google Scholar]
- Wu C.Y., Trieu A., Radhakrishnan P., Kwok S.F., Harris S., Zhang K., Wang J., Wan J., Zhai H., Takatsuto S., et al. Brassinosteroids regulate grain filling in rice. Plant Cell. 2008;20:2130–2145. doi: 10.1105/tpc.107.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Zhu Y., Song W., Li Y., Yan Y., Hu Y. Unusual tandem expansion and positive selection in subgroups of the plant GRAS transcription factor superfamily. BMC Plant Biol. 2014;14:373. doi: 10.1186/s12870-014-0373-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.Y., Wu P.Z., Chen Y.P., Li M.R., Wu G., Jiang H. Genome-wide analysis of the GRAS gene family in physic nut (Jatropha curcas L.) Genet. Mol. Res. 2015;14:19211–19224. doi: 10.4238/2015.December.29.31. [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller J.W., Helariutta Y., Fukaki H., Malamy J.E., Benfey P.N. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127:595–603. doi: 10.1242/dev.127.3.595. [DOI] [PubMed] [Google Scholar]
- Xu C., Wang Y., Yu Y., Duan J., Liao Z., Xiong G., Meng X., Liu G., Qian Q., Li J. Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat. Commun. 2012;3:750. doi: 10.1038/ncomms1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Chen S., Li T., Ma X., Liang X., Ding X., Liu H., Luo L. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 2015;15:141. doi: 10.1186/s12870-015-0532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Chen Z., Ahmed N., Han B., Cui Q., Liu A. Genome-Wide identification, evolutionary analysis, and stress responses of the GRAS gene family in Castor beans. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17071004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Cui H., Buer B., Vijayakumar V., Delaux P.-M., Junkermann S., Bucher M. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 2015;167:854–871. doi: 10.1104/pp.114.255430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Yadava Y.K., Kohli D., Meena S., Paul V., Jain P.K. Genome-wide identification and expression analysis of the GRAS gene family in response to drought stress in chickpea (Cicer arietinum L.) 3 Biotech. 2022;12:64. doi: 10.1007/s13205-021-03104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.-L., Yang Y., He Z. Roles of plant hormones and their interplay in rice immunity. Mol. Plant. 2013;6:675–685. doi: 10.1093/mp/sst056. [DOI] [PubMed] [Google Scholar]
- Yang D.-L., Yao J., Mei C.-S., Tong X.-H., Zeng L.-J., Li Q., Xiao L.-T., Sun T., Li J., Deng X.-W., et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA. 2012;109:1192–1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wu J.-J., Tang T., Liu K.-D., Dai C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-07871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Yang Q., Fu T., Zhou Y. Overexpression of the Brassica napus BnLAS gene in Arabidopsis affects plant development and increases drought tolerance. Plant Cell Rep. 2011;30:373–388. doi: 10.1007/s00299-010-0940-7. [DOI] [PubMed] [Google Scholar]
- Yoon E.K., Dhar S., Lee M.-H., Song J.H., Lee S.A., Kim G., Jang S., Choi J.W., Choe J.-E., Kim J.H., et al. Conservation and diversification of the SHR-SCR-SCL23 regulatory network in the development of the functional endodermis in Arabidopsis shoots. Mol. Plant. 2016;9:1197–1209. doi: 10.1016/j.molp.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Hirano K., Sato T., Mitsuda N., Nomoto M., Maeo K., Koketsu E., Mitani R., Kawamura M., Ishiguro S., et al. DELLA protein functions as a transcriptional activator through the DNA binding of the INDETERMINATE DOMAIN family proteins. Proc. Natl. Acad. Sci. USA. 2014;111:7861–7866. doi: 10.1073/pnas.1321669111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Zhang G., Lyu Z., He C., Zhang J. Genome-wide analysis of the GRAS gene family exhibited expansion model and functional differentiation in sea buckthorn (Hippophae rhamnoides L.) Plant Biotechnol. Rep. 2021;15:513–525. doi: 10.1007/s11816-021-00694-1. [DOI] [Google Scholar]
- Yu N., Luo D., Zhang X., Liu J., Wang W., Jin Y., Dong W., Liu J., Liu H., Yang W., et al. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 2014;24:130–133. doi: 10.1038/cr.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Fang L., Karungo S.K., Zhang L., Gao Y., Li S., Xin H. Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis. Plant Cell Rep. 2016;35:655–666. doi: 10.1007/s00299-015-1910-x. [DOI] [PubMed] [Google Scholar]
- Zeng X., Ling H., Chen X., Guo S. Genome-wide identification, phylogeny and function analysis of GRAS gene family in Dendrobium catenatum (Orchidaceae) Gene. 2019;705:5–15. doi: 10.1016/j.gene.2019.04.038. [DOI] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Liu J., Yang Z.E., Chen E.Y., Zhang C.J., Zhang X.Y., Li F.G. Genome-wide analysis of GRAS transcription factor gene family in Gossypium hirsutum L. BMC Genom. 2018;19:348. doi: 10.1186/s12864-018-4722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Iyer L.M., Aravind L. Bacterial GRAS domain proteins throw new light on gibberellic acid response mechanisms. Bioinformatics. 2012;28:2407–2411. doi: 10.1093/bioinformatics/bts464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Jing Y., Jiang Z., Lin R. The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell. 2014;26:2472–2485. doi: 10.1105/tpc.113.121848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Cao Y., Shang C., Li J., Wang J., Wu Z., Ma L., Qi T., Fu C., Bai Z., Hu B. Genome-wide characterization of GRAS family genes in Medicago truncatula reveals their evolutionary dynamics and functional diversification. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu X., Wang X., Sun M., Song R., Mao P., Jia S. Genome-wide identification of GRAS gene family and their responses to abiotic stress in Medicago sativa. Int. J. Mol. Sci. 2021;22:7729. doi: 10.3390/ijms22147729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Mi L., Xu L., Yu C., Li C., Chen C. Genome-wide identification, characterization, interaction network and expression profile of GRAS gene family in sweet orange (Citrus sinensis) Sci. Rep. 2019;9:2156. doi: 10.1038/s41598-018-38185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.L., Ogawa M., Fleet C.M., Zentella R., Hu J., Heo J.O., Lim J., Kamiya Y., Yamaguchi S., Sun T.P. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2011;108:2160–2165. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G.Y., Yang Y. Characterization of grape Gibberellin Insensitive1 mutant alleles in transgenic Arabidopsis. Transgenic Res. 2012;21:725–741. doi: 10.1007/s11248-011-9565-z. [DOI] [PubMed] [Google Scholar]
- Zhu X., Wang B., Wei X. Genome wide identification and expression pattern analysis of the GRAS family in quinoa. Funct. Plant Biol. 2021;48:948–962. doi: 10.1071/FP21017. [DOI] [PubMed] [Google Scholar]
- Zou M., Guo B., He S. The roles and evolutionary patterns of intronless genes in deuterostomes. Comp. Funct. Genomics. 2011 doi: 10.1155/2011/680673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.