Abstract
The gram-negative coccobacillus, Actinobacillus actinomycetemcomitans, is the putative agent for localized juvenile periodontitis, a particularly destructive form of periodontal disease in adolescents. This bacterium has also been isolated from a variety of other infections, notably endocarditis. Fresh clinical isolates of A. actinomycetemcomitans form tenacious biofilms, a property likely to be critical for colonization of teeth and other surfaces. Here we report the identification of a locus of seven genes required for nonspecific adherence of A. actinomycetemcomitans to surfaces. The recently developed transposon IS903φkan was used to isolate mutants of the rough clinical isolate CU1000 that are defective in tight adherence to surfaces (Tad−). Unlike wild-type cells, Tad− mutant cells adhere poorly to surfaces, fail to form large autoaggregates, and lack long, bundled fibrils. Nucleotide sequencing and genetic complementation analysis revealed a 6.7-kb region of the genome with seven adjacent genes (tadABCDEFG) required for tight adherence. The predicted TadA polypeptide is similar to VirB11, an ATPase involved in macromolecular transport. The predicted amino acid sequences of the other Tad polypeptides indicate membrane localization but no obvious functions. We suggest that the tad genes are involved in secretion of factors required for tight adherence of A. actinomycetemcomitans. Remarkably, complete and highly conserved tad gene clusters are present in the genomes of the bubonic plague bacillus Yersinia pestis and the human and animal pathogen Pasteurella multocida. Partial tad loci also occur in strikingly diverse Bacteria and Archaea. Our results show that the tad genes are required for tight adherence of A. actinomycetemcomitans to surfaces and are therefore likely to be essential for colonization and pathogenesis. The occurrence of similar genes in a wide array of microorganisms indicates that they have important functions. We propose that tad-like genes have a significant role in microbial colonization.
Actinobacillus actinomycetemcomitans is a gram-negative bacterium associated with several human diseases (8, 36, 47). The most predominant of these is known as localized juvenile periodontitis, a severe disease of adolescents that is characterized by bone and tissue destruction and ultimately loss of teeth if untreated. A. actinomycetemcomitans is also a member of a clinically important group of bacteria implicated in infective endocarditis (39). These bacteria are referred to as the HACEK group (Haemophilus aphrophilus, A. actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, and Kingella kingae) (9). Vegetative growths and inflammation of the heart valves caused by the HACEK bacteria result in serious complications owing to the formation of bacterial masses on the valves.
A. actinomycetemcomitans expresses several potential virulence factors (8), but only the RTX-type leukotoxin has been studied in detail at the molecular and genetic levels (14, 24). Other possible factors include a cytolethal distending toxin, iron and hemin binding proteins, a trypsin-like protease, an OmpA family member, capsular polysaccharide biosynthetic proteins, catalase, and a GroEL-like protein (5, 12, 13, 23, 26, 28, 35, 41, 43, 46). However, their potential roles in pathogenesis are unknown. A. actinomycetemcomitans is also able to invade epithelial cells, and the bacteria can be transferred between cells (8, 27).
A striking and characteristic property of fresh clinical isolates of A. actinomycetemcomitans is their ability to form tenacious biofilms on solid surfaces, including glass, plastics, and hydroxyapatite (6, 21, 30). This property is very likely required for pathogenesis by allowing for colonization of teeth in an environment of continuous salivary flow. Clinical isolates form rough-appearing colonies, autoaggregate, and express bundles of fimbria-like structures that may be important for adherence and colonization (18, 30, 34).
Genetic analysis of rough, adherent strains has proven to be difficult. The distinctive adherence property of clinical strains is easily lost, as nonadherent, smooth-colony variants readily emerge during subculture (7, 45). In addition, while DNA can be introduced into A. actinomycetemcomitans by transformation (37) or conjugation (11), the efficiency of DNA transfer into rough, adherent strains is too low for standard transposon mutagenesis protocols involving suicide vectors. Recently we reported the development of transposon IS903φkan, which carries a cryptic kanamycin resistance gene that can be activated upon insertion of the transposon into an expressed gene, and we have demonstrated its utility in the direct selection of random insertions in genetically recalcitrant bacteria, such as A. actinomycetemcomitans (38). Here we report the use of IS903φkan to obtain adherence-defective mutants of A. actinomycetemcomitans CU1000, a well-characterized rough clinical isolate (7, 20, 30). Genetic and nucleotide sequence analyses of these mutants have allowed us to identify a locus of seven novel genes that are required for tenacious adherence, autoaggregation, and the production of bundled fibers. Examination of genome sequences of Bacteria and Archaea revealed that surprisingly diverse microorganisms are predicted to carry tad-related genes. We discuss the likely function of these genes and the significance of their widespread occurrence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. actinomycetemcomitans CU1000N is a spontaneous nalidixic acid-resistant (Nalr) derivative of the rough clinical isolate CU1000 (6). CU1060N is a Nalr derivative of CU1060 (6), a spontaneous smooth-colony variant of CU1000. A. actinomycetemcomitans strains were stored and cultured as previously described (6). Bacteria were streaked from −70°C frozen stocks onto agar plates containing A. actinomycetemcomitans growth medium (AAGM) (11) and incubated in a sealed chamber enriched with 5% CO2 at 37°C for several days. Individual colonies were inoculated into AAGM broth (11) in sealed, screw-cap tubes and incubated at 37°C. For light microscopy, bacteria were grown in broth for 1 day (CU1060N) or 2 days (CU1000N). For CU1000N, the cells were scraped from the walls and resuspended before preparation of wet mounts. To assay adherence of cells onto the surface of culture tubes, 2-day cultures were subjected to vortex mixing and the tube contents were discarded. An equal volume of 5-μg/ml ethidium bromide solution was added to the tubes, left for 10 min at room temperature, and then poured off, and the tubes were washed with water. The tubes were photographed in UV light.
IS903φkan mutagenesis of A. actinomycetemcomitans.
Plasmid pVJT128, which carries a chloramphenicol resistance (Cmr) gene and the cryptic IS903φkan transposon, was mobilized into rough strain CU1000N by conjugation with an Escherichia coli donor strain (SK338) that contains pVJT128 and an oriT-defective derivative of RK2, pRK21761 (38). CU1000N(pVJT128) transconjugants were selected on AAGM medium containing nalidixic acid (20 μg/ml) and chloramphenicol (4 μg/ml) and screened for sensitivity to kanamycin to confirm that pRK21761 had not transferred and that transposition of IS903φkan had not occurred. One such CU1000N(pVJT128) transconjugant was grown in AAGM broth containing chloramphenicol and then plated onto medium containing 4 μg of chloramphenicol per ml and 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (to induce transposition of IS903φkan), and colonies were grown for 3 days. Several pools of colonies were each plated onto solid medium containing 20 or 40 μg of kanamycin per ml. Because the kanamycin-resistant (Kmr) colonies were small on this medium, the colonies from each pool were mixed and replated onto nonselective medium, where colony morphology was easily distinguished. To ensure that the mutants were independent isolates, only one smooth-colony mutant was kept from each original pool. Mutants were grown in broth in the absence of chloramphenicol to allow loss of the resident pVJT128 plasmid, as described previously (38).
DNA procedures.
Preparation of plasmid DNA from E. coli was done by the alkaline lysis protocol (1). Agarose gel electrophoresis has been described previously (33). DNA manipulations with restriction endonucleases and T4 DNA ligase were done according to the manufacturers' recommendations. Amplification of DNA by PCR was done with Taq DNA polymerase (32). All cloned PCR products were confirmed by nucleotide sequencing. Transformation of E. coli was done by the method of Cohen et al. (4). Inverse PCR with IS903φkan insertion mutants was done as previously described (38). Approximately 10 to 20 μg of genomic DNA was digested with EcoRI (which does not cleave IS903φkan), followed by ethanol precipitation, dilution, and ligation to circularize genomic fragments. PCR amplification was carried out for 30 cycles using primers directed outward from the ends of IS903φkan. Amplified products were cloned into pCR2.1 (Invitrogen, Carlsbad, Calif.) or pT7blue-3 (Novagen, Madison, Wis.) for sequencing. Nucleotide sequence determination was done by the Columbia University DNA Sequencing Facility using a Perkin-Elmer Applied Biosystems Automated DNA sequencer 373A.
Genetic complementation analysis.
The desired open reading frame (ORF) was amplified by PCR, cloned into pCR2.1, sequenced, and then subcloned into the IncQ expression vector pJAK16 (a gift of J. Kornacki), a derivative of the tacp expression vector pMMB67 (10). The pJAK16 derivatives were mobilized by conjugation from E. coli donors to the appropriate A. actinomycetemcomitans strains (38). Because the tacp promoter is leaky even in the presence of functional LacI repressor, complementation did not require induction by IPTG.
Electron microscopy.
Colonies were resuspended in 0.1 M Tris-HCl (pH 7.5), and a drop of the suspension was placed on a Formvar- and carbon-coated copper grid (EM Sciences, Fort Washington, Pa.) and left for approximately 10 min. Grids were immersed in a drop of 1% uranyl acetate and removed immediately; the excess stain was wicked away. Grids were viewed in a JEOL 1200EX transmission electron microscope at 80.0 kV.
Sequence analysis and similarity searching.
Nucleotide sequences potentially coding for proteins similar to the tad gene products were identified using the BLAST alignment program to search both GenBank and the Unfinished Genomes Database available on the National Center for Biotechnology Information web page (http://www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html). Default settings of the basic BLAST program were used. All reported sequences significantly exceeded the default criteria and were among the best hits when compared to the corresponding Tad polypeptides. Contiguity and sequential order of tad-related ORFs were established using the ORF finder program to characterize the region around each tad-like sequence. The putative products of all neighboring ORFs were compared to sequences in GenBank using the pBlast function. Preliminary sequence data for Methanococcus jannaschii, Archeoglobus fulgidus, and Chlorobium tepidum were obtained from The Institute for Genomic Research website at http://www.tigr.org. Bordetella pertussis and Yersinia pestis sequences were produced by the respective Sanger Centre sequencing groups (http://www.sanger.ac.uk/), Pyrococcus horikoshii sequences were produced by NITE (http://www.nite.go.jp), the Pyrococcus abyssi sequence was produced by Genoscope (http://www.genoscope.cns.fr), the Pyrococcus furiosus sequence was produced by the Utah Genome Center (http://www.genome.utah.edu), the Pseudomonas aeruginosa sequence was produced by the Pseudomonas Genome Project (http://www.pseudomonas.com), the Methanobacterium thermoautotrophicum sequence was produced by the Genome Therapeutics Center (http://www.genomecorp.com), and the Pasteurella multocida sequence was produced by the University of Minnesota P. multocida Genome Project (http://www.cbc.umn.edu/ResearchProjects/AGAC/Pm/index.html). A compilation of Intein sequences is found in InBase, the New England BioLabs Intein Database (http://www.neb.com/inteins). Accession numbers for tadA- and tadB-related sequences, respectively, in other organisms are as follows: M. jannaschii, Q58191 and Q58189; P. abyssi, CAB50295 and CAB50294; P. horikoshii, BAA29741 and BAA29744; A. fulgidus, AAB90582 and AAB90583; and M. thermoautotrophicum, AAB86174 and AAB85476. The GenBank accession number for plasmid pCL1 from Chlorobium limicola is U77780, that for KlbA (TrbB) is C44020, and that for VirB11 is AAA88655.
Nucleotide sequence accession number. The nucleotide sequence of the 6,704-bp tad region has been submitted to GenBank (accession no. AF152598).
RESULTS AND DISCUSSION
Isolation of adherence-defective mutants by transposon mutagenesis.
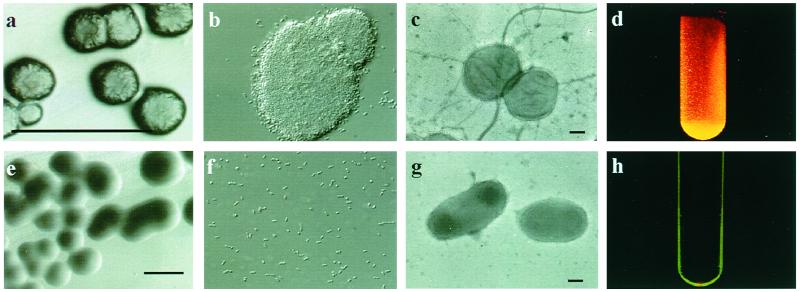
A. actinomycetemcomitans strain CU1000 was originally isolated from a 13-year-old, African-American female with localized juvenile periodontitis (7). To facilitate the study and genetic manipulation of CU1000, we isolated a spontaneous nalidixic acid-resistant variant, CU1000N. Like its parent, CU1000N exhibits the characteristic rough, opaque colony morphology typical of fresh clinical isolates (Fig. 1a). In broth, CU1000N adheres tightly to the walls of the culture vessels (Fig. 1d). The biofilm is resistant to vortex mixing or vigorous agitation, and the bacteria must be physically scraped from the walls. The rough colony phenotype can be maintained indefinitely during subcultures on solid medium. Rough A. actinomycetemcomitans strains give rise to nonadherent, planktonic variants, whose colonies on solid medium are smooth, translucent, and larger than those of rough strains (7, 20, 30, 45) (Fig. 1e and h). One such derivative of CU1000 is CU1060 (7), and its Nalr derivative is CU1060N. Cells of the smooth, nonadherent variants, including CU1060N, do not display the striking bundles of parallel, fimbria-like fibers normally present in abundance on cells of rough strains (Fig. 1c and 2). It has been suggested that these fibrils mediate adherence (30, 34). Cells of the rough strains and smooth variants also differ dramatically in their ability to autoaggregate (Fig. 1b and f).
FIG. 1.
Phenotypes of isogenic rough- and smooth-colony strains of A. actinomycetemcomitans. Top panels, rough-colony strain CU1000N; bottom panels: smooth-colony variant CU1060N. (a and e) Images from stereomicroscopy of 3-day-old colonies. Bars, 1.0 mm. (b and f) Images from light microscopy of bacterial cells using differential interference contrast optics. (c and g) Images from transmission electron microscopy of negatively stained cells. Bars, 0.2 μm. (d and h) Adherence of cells to the culture tube surface, as visualized by fluorescence of ethidium bromide-stained cells in UV light.
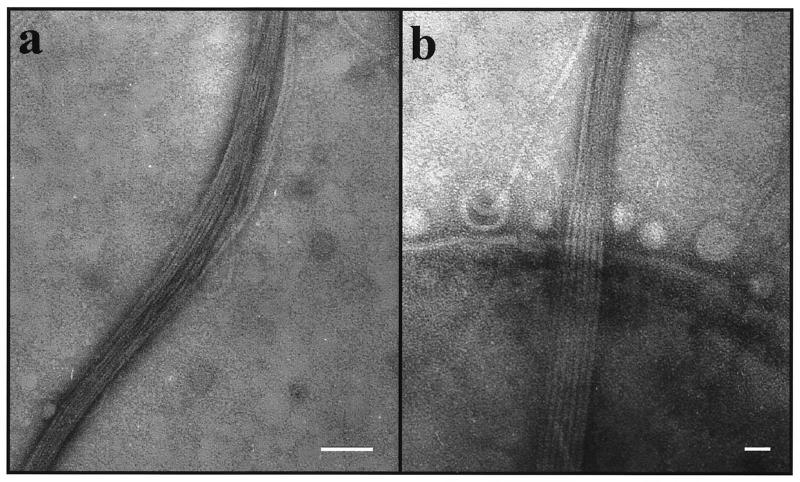
FIG. 2.
Electron micrograph of fibril bundles produced by the rough strain CU1000N. (a) Fibril bundle showing individual strands separating from the bundle at several places. Bar, 50 nm. (b) Parallel array of six or seven fibrils passing over the edge of a bacterial cell. The spherical structures at the bacterial cell surface are likely to be membranous vesicles, which are commonly seen in preparations of rough strains of A. actinomycetemcomitans (30). Bar, 20 nm.
To investigate the molecular mechanism of tight, nonspecific adherence of A. actinomycetemcomitans to surfaces, we first set out to identify the genetic determinants involved. We exploited the apparent correlation of smooth colony morphology with nonadherence to isolate transposon insertion mutants that are defective in adherence. We induced transposition of IS903φkan in the rough strain CU1000N, and insertion mutants were selected by plating on medium containing kanamycin. Several independent Kmr mutants exhibiting a smooth colony phenotype were chosen for study. Southern blot analysis revealed a single IS903φkan insertion in the chromosome of each smooth mutant (data not shown).
On solid medium, the mutants form smooth-appearing colonies (Table 1; Fig. 3a), although the colony size was routinely smaller than that observed for the spontaneous smooth strain CU1060N. The mutants showed obvious defects in surface adherence in broth culture (Table 1). All mutants grew in loose association with the walls of the culture tubes, and cells were easily dispersed into suspension upon vortex mixing. This phenotype differs markedly from the tight adherence (Tad+) phenotype of the parental rough strain CU1000N, and it is also distinct from the complete nonadherence phenotype of the spontaneous smooth isolate CU1060N, in which the bacteria show no association with the walls of the culture tube. Light microscopy of the mutants revealed that the ability of the cells to autoaggregate was greatly reduced, but not abolished, relative to that of the rough strain (data not shown). The diminished autoaggregation parallels the behavior of the resuspended mutants in broth. Cells of the parental rough strain (CU1000N) settle in clumps within minutes after being scraped from the walls and resuspended, whereas Tad− mutant cells formed clumps and settled to the bottom of the tube only after several hours. Cells of the spontaneous smooth-colony variant (CU1060N) grow in suspension and do not settle.
TABLE 1.
Phenotypes and genetic complementation of tad mutants
| Strain | Genotype of:
|
Phenotype
|
|||
|---|---|---|---|---|---|
| Strain | Test plasmida | Colonyb | Adherence | Fibrilsc | |
| CU1000N | Wild type rough | None | R++ | + | + |
| CU1060N | Smooth | None | S | − | − |
| tadA-G | S | − | − | ||
| flp-tadG | R+ | + | + | ||
| Aa1360 | tadA | Vector | S | − | − |
| tadA+ | R+ | + | + | ||
| Aa1332 | tadB | Vector | S | − | − |
| tadB+ | R+ | + | + | ||
| Aa1359 | tadC | Vector | S | − | − |
| tadC+ | R+ | + | + | ||
| Aa1577 | tadD | Vector | S | − | − |
| tadC+D+ | R+ | + | + | ||
| Aa1347 | tadE | Vector | S | − | − |
| tadE+ | R+ | + | + | ||
| Aa1512 | tadF | Vector | S | − | − |
| tadF+ | R+ | + | + | ||
| Aa1561 | tadG | Vector | S | − | − |
| tadG+ | R+ | + | + | ||
The vector is tacp promoter plasmid pJAK16 (see Materials and Methods). Complementation did not require induction by IPTG.
R, rough; S, smooth.
Determined by electron microscopy as described for Fig. 1.
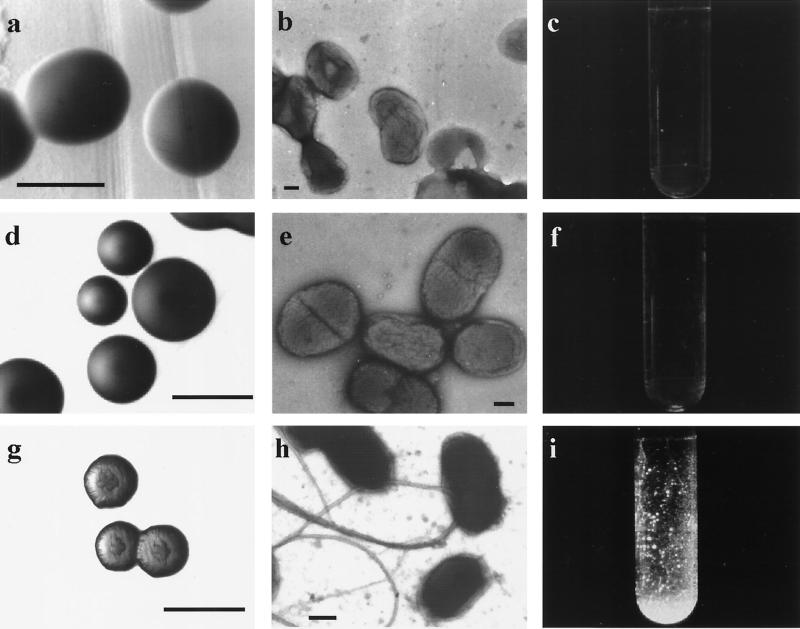
FIG. 3.
Phenotypes and genetic complementation of a tadC mutant. Top panels, mutant strain Aa1359 (tadC::IS903φkan). Middle panels, strain Aa1359 containing the pJAK16 vector plasmid. Bottom panels, Aa1359 containing pJAK16 with the cloned tadC ORF from CU1000N. The methods used are identical to those described for Fig. 1. Results for complementation of the other mutants were essentially the same (Table 1). Bars, 1.0 mm (a, d, and g), 0.2 μm (b and e), and 0.5 μm (h).
Identification of seven tad genes.
To map the sites of IS903φkan insertion for several Tad− mutants, we did inverse PCR on genomic DNA with primers directed outward from the ends of the transposon (38). Using these sequences and partial nucleotide sequences from the ongoing A. actinomycetemcomitans Genome Sequencing Project to design additional PCR primers, we found that all of the insertions mapped to a continuous 7-kb region of the A. actinomycetemcomitans genome. We determined the sequence for both strands of this region for the rough strain CU1000N. The sequence revealed a cluster of seven closely spaced ORFs oriented in the same direction (Fig. 4). At least one IS903φkan insertion mutant was isolated for each ORF, and all mutants were defective in adherence (Table 1) and autoaggregation (data not shown). Genetic complementation analysis with the appropriate ORFs on plasmids showed that the IS903φkan insertions are not polar on expression of downstream genes (Table 1). The complemented Tad− mutants produced rough-appearing colonies on solid medium, although not to the same degree as those of the wild-type rough strain (Fig. 3a, d, and g), presumably due to nonstoichiometric levels of expression from the plasmid-borne genes. However, cells of the complemented mutants clearly regained the tight adherence phenotype of the rough parental strain (Fig. 3c, f, and i) and the ability to autoaggregate (data not shown). All mutants with insertions in different ORFs were examined by electron microscopy, and all were found to lack fibrils (Fig. 3b), unless the complementing plasmid was present (Table 1, Fig. 3h). Controls with the vector (Fig. 3d, e, and f) or plasmids with noncognate ORFs (Table 1) showed no evidence of complementation for any of the phenotypes. These results confirmed that all seven ORFs are required for tight adherence, and we therefore designated them genes tadA, tadB, tadC, tadD, tadE, tadF, and tadG.
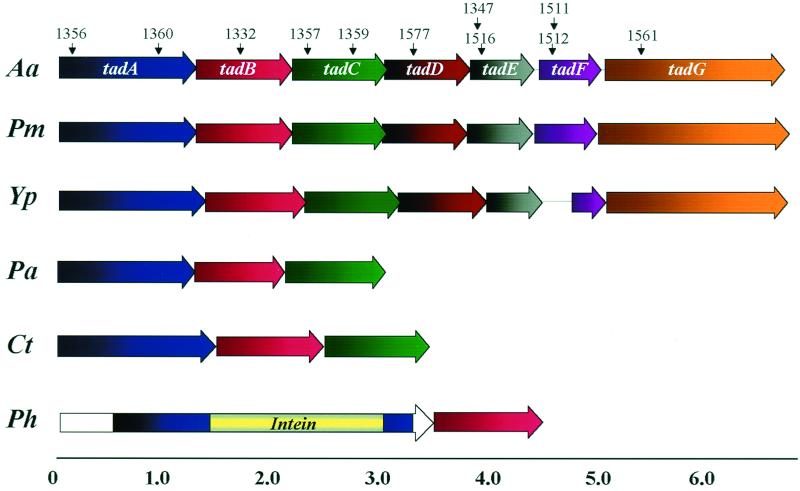
FIG. 4.
Arrangement of tad genes in A. actinomycetemcomitans and other microorganisms. The top line shows the tadABCDEFG genes of A. actinomycetemcomitans (Aa) strain CU1000N. The locations of the IS903φkan insertions in specific mutant strains are indicated by downward arrows. Below are tad-related genes from P. multocida (Pm), Y. pestis (Yp), P. aeruginosa (Pa), C. tepidum (Ct), and P. horikoshii (Ph). The tad sequences for organisms other than A. actinomycetemcomitans were obtained from individual genome sequencing projects as described in Materials and Methods. The scale bar at the bottom is in kilobase pairs.
Tad proteins may constitute a fibril secretion system.
We searched GenBank to identify other proteins that are related to the tad gene products. The predicted polypeptide products of the tadB, tadC, tadD, tadE, tadF, and tadG genes showed no significant similarities to any other proteins of known function in the database. However, PSORT (29) and TMPred (17) predicted the presence of six (TadB), five (TadC), and one (TadD, TadE, TadF, and TadG) membrane-spanning regions. Thus, these proteins are likely to be localized to the membrane.
The predicted amino acid sequence of the TadA polypeptide showed significant similarity (32% identity and 51% similarity) to VirB11, a protein involved in the export of macromolecules from bacterial cells (2). VirB11 is required for the transfer of T-DNA from the tumor-inducing bacterium Agrobacterium tumefaciens to plant cells (3). Other members of the VirB11 family include TrbB, which is required for the formation of bacterial mating pairs and conjugative transfer of the promiscuous antibiotic resistance plasmid RP4 (25), and PtlH of B. pertussis, which is needed for secretion of the pertussis toxin (42). These proteins are cytoplasmic but act at the inner membrane, possibly through interaction with a membrane protein (2). The VirB11 family of proteins all contain a Walker nucleotide-binding motif (40), as does TadA, and they are believed to provide energy for the export apparatus by hydrolysis of ATP (2). However, robust phylogenetic analyses have shown that tadA is the prototype of a distinct and novel gene subfamily that does not include trbB, virB11, or ptlH (P. J. Planet, S. C. Kachlany, R. DeSalle, and D. H. Figurski, unpublished results).
Genes for other proteins specifically expressed by rough A. actinomycetemcomitans strains map near the tad gene cluster. Haase et al. (15) recently identified two rough strain-specific outer membrane proteins expressed from adjacent genes, rcpA and rcpB. We found that these genes map upstream of the tad region, separated from tadA by an ORF whose function is unknown. At present the rcp gene products have not been implicated in adherence, but RcpA is similar to the GspD family of outer membrane proteins involved in secretion and pilus assembly (15, 31). Inoue et al. (19) have identified a 6.5-kDa protein (Flp) as a fibril subunit, and the flp gene was also found to map in the vicinity of the tad region, upstream of rcpA. It has been suggested by others that the fibrils displayed by rough strains of A. actinomycetemcomitans are responsible for surface adherence (18, 30, 34), which is consistent with our finding that all seven tad genes are required for adherence and fibril formation (Table 1). We found that a plasmid carrying the complete tadABCDEFG region from the rough strain is able to complement mutations in any of the tad genes (data not shown), but it is unable to complement the spontaneous smooth strain CU1060N (Table 1). However, a plasmid containing the upstream region, beginning from flp and continuing through tadG, is fully able to complement the spontaneous smooth strain to a rough, adherent phenotype (Table 1). Thus, it is likely that the entire 12-kb flp-tadG region is responsible for the tight adherence properties of A. actinomycetemcomitans. We also have preliminary evidence that flp is required for fibril formation and adherence (S. C. Kachlany, P. J. Planet, R. Aussenberg, D. H. Figurski, D. H. Fine, and J. B. Kaplan, unpublished data). Furthermore, new evidence implicates a similar locus in fibril formation in the distantly related α-proteobacteria. A region of the Caulobacter crescentus genome was found to contain highly similar genes in conserved order with the flp-tadA region of A. actinomycetemcomitans, and the genes were shown to be required for pili formation (35a). Because A. actinomycetemcomitans TadA is related to proteins involved in macromolecular transport and the other Tad proteins are potentially localized to the membrane, we suggest that the Tad proteins constitute a novel system involved in the export of factors (possibly RcpA, RcpB, or Flp) that are required for fibril formation and tight adherence of A. actinomycetemcomitans.
tad genes are widespread.
We also compared each Tad protein individually to the Unfinished Microbial Genomes Database, and we were surprised to find significant similarities for each of the Tad proteins with predicted proteins of unknown function in Y. pestis, the causative agent of bubonic plague, and P. multocida, a pathogen that infects humans and animals. The coding regions for the Tad-like proteins in both organisms show all seven ORFs in an arrangement identical to that of the tadABCDEFG region of A. actinomycetemcomitans. Not only do the predicted P. multocida and Y. pestis proteins show significant percent identities with the predicted Tad proteins, but the sizes and isoelectric points (pIs) of the corresponding proteins are strikingly similar (Table 2). Recently, a complete tad region coding for putative polypeptides with high similarities to the seven A. actinomycetemcomitans tad gene products has been identified in the genome of Haemophilus ducreyi, the causative agent of chancroid (E. J. Hansen, personal communication).
TABLE 2.
Similarities among the A. actinomycetemcomitans, P. multocida, and Y. pestis Tad proteins
| Protein | Source | Molecular mass (kDa) | Theoretical pI | Identity to A. actinomycetemcomitans Tad protein (%) |
|---|---|---|---|---|
| TadA | A. actinomycetemcomitans | 47.1 | 5.51 | |
| P. multocida | 47.1 | 5.22 | 85 | |
| Y. pestis | 48.1 | 5.84 | 60 | |
| TadB | A. actinomycetemcomitans | 33.4 | 9.30 | |
| P. multocida | 34.4 | 9.86 | 70 | |
| Y. pestis | 33.7 | 9.70 | 33 | |
| TadC | A. actinomycetemcomitans | 32.2 | 9.13 | |
| P. multocida | 31.9 | 9.50 | 73 | |
| Y. pestis | 31.6 | 9.55 | 29 | |
| TadD | A. actinomycetemcomitans | 28.4 | 9.27 | |
| P. multocida | 28.7 | 9.45 | 59 | |
| Y. pestis | 28.7 | 7.82 | 35 | |
| TadE | A. actinomycetemcomitans | 21.7 | 9.42 | |
| P. multocida | 22.4 | 8.87 | 54 | |
| Y. pestis | 17.9 | 5.91 | 27 | |
| TadF | A. actinomycetemcomitans | 22.9 | 9.52 | |
| P. multocida | 21.7 | 9.47 | 47 | |
| TadG | A. actinomycetemcomitans | 60.4 | 9.32 | |
| P. multocida | 66.6 | 8.66 | 42 | |
| Y. pestis | 58.5 | 5.72 | 21 |
BLAST searches also identified partial tad loci in a remarkable variety of organisms among the domains Bacteria and Archaea (44). Genomic regions containing tad-like ORFs in the same order and predicted to code for polypeptides with significant similarities to TadA (>63%), TadB (>41%), and TadC (>46%) were found in several bacterial genomes (Fig. 4), including P. aeruginosa, C. tepidum, C. crescentus, Thiobacillus ferroxidans, B. pertussis, and an endogenous plasmid (pCL1) of C. limicola. In addition, Sinorhizobium meliloti and Bordetella bronchiseptica also contain ORFs whose predicted products show strong similarity to TadA, TadB, and TadC, but the contiguous order of the ORFs cannot yet be established because they appear on separate sequence contigs. We have also obtained evidence by PCR and nucleotide sequence analysis for the presence of tad-related genes in a strain of H. aphrophilus originally isolated from a patient with bacterial endocarditis, Y. enterocolitica, and Yersinia pseudotuberculosis (data not shown). However, we found no evidence of tad-like genes in any of several nonpathogenic Yersinia species. Remarkably, adjacent ORFs whose predicted polypeptide products have significant similarity to TadA (>58%) and TadB (>38%) exist in nearly all Archaea whose genomes have been partly or completely sequenced. These include P. furiosus, P. horikoshii (Fig. 4), P. abyssi, M. thermoautotrophicum, M. jannaschii, and A. fulgidus. Several of the archaeal TadA sequences contain all or part of the KlbA Intein (Fig. 4). The KlbA Intein was first identified in a polypeptide with similarity to the KlbA (TrbB) protein of promiscuous plasmid RK2 (New England BioLabs Intein Database). We found that this polypeptide has even greater similarity to A. actinomycetemcomitans TadA (40% identity over 413 amino acids) than to KlbA (35% identity over 297 amino acids) or VirB11 (32% identity over 259 amino acids).
The presence of a complete tad region in A. actinomycetemcomitans, Y. pestis, P. multocida, and H. ducreyi raises the question of whether these genes are conserved by descent or acquired through horizontal gene transfer. We found that the G+C contents of the complete tad regions are similar to each other (35 to 36%) but significantly different from those of the resident genomes for which sequence data exist (48% for A. actinomycetemcomitans [22] and 47% for Y. pestis [Sanger Centre]). These results are consistent with the hypothesis that the tad regions were inserted into these genomes following horizontal gene transfer from an as-yet-unidentified source.
The striking adherence-defective phenotype of the Tad− mutants of A. actinomycetemcomitans and the conservation of apparently nonessential tad genes in many microorganisms raise the possibility that these genes are important for establishment of the organisms in their environments. Like A. actinomycetemcomitans, Y. pestis can form crinkled colonies and adhere to the walls of a culture vessel (9). In addition, for Y. pestis to be transmitted from the flea vector to the human host, the bacteria must form large clumps which obstruct the proventriculus of the flea (16). The starving flea eventually regurgitates the clumped bacteria into the bloodstream of the host, thereby transmitting plague. The Tad proteins, which we have shown are required for autoaggregation of A. actinomycetemcomitans, may likewise be involved in the clumping of Y. pestis in the flea. Much less is known about the genetic basis for virulence in P. multocida, which causes a variety of animal infections, such as fowl cholera, atrophic rhinitis in pigs, and hemorrhagic septicemia in cattle, resulting in the loss of several hundred million dollars annually in the animal industry (9). P. multocida is frequently found in the oral cavities of cats and dogs, and it is the most common organism isolated from human wounds inflicted by bites and scratches of cats and dogs. By analogy with A. actinomycetemcomitans, the tad genes of P. multocida may be important for adherence and colonization of the oral cavity. The elucidation of the specific roles of the Tad proteins in adherence and autoaggregation will be important to understanding A. actinomycetemcomitans colonization and pathogenesis. This knowledge may also provide significant clues about the functions of Tad proteins in other microorganisms.
ACKNOWLEDGMENTS
We thank A.-J. Silverman for her instruction and suggestions for the electron microscopy; H. Schreiner, J. Kaplan, D. Furgang, P. Goncharoff, R. Stevens, T. Rosche, J. Ferguson, and V. Thomson for their animated and helpful discussions; and K. Calame, H. Shuman, and S. Silverstein for their comments on the manuscript. We also thank E. Hansen, L. Shapiro, and J. Skerker for communication of unpublished results. We are grateful to Bruce Roe, Fares Z. Najar, Sandy Clifton, Tom Ducey, Lisa Lewis, and Dave Dyer for the use of unpublished nucleotide sequence data from the A. actinomycetemcomitans Genome Sequencing Project at the University of Oklahoma.
This work was supported in part by NIH research grants (to D. H. Figurski and D. H. Fine) and NIH traineeships (to S. C. Kachlany and P. J. Planet).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 2.Christie P J. Agrobacterium tumefaciensT-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christie P J, Ward J E, Gordon M P, Nester E W. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc Natl Acad Sci USA. 1989;86:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S N, Chang A C Y, Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coliby R-factor DNA. Proc Natl Acad Sci USA. 1972;69:2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dongari A I, Miyasaki K T. Sensitivity of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilusto oxidative killing. Oral Microbiol Immunol. 1991;6:363–372. doi: 10.1111/j.1399-302x.1991.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 6.Fine D H, Furgang D, Kaplan J, Charlesworth J, Figurski D H. Tenacious adhesion of Actinobacillus actinomycetemcomitansstrain CU1000 to salivary coated hydroxyapatite. Arch Oral Biol. 1999;44:1063–1076. doi: 10.1016/s0003-9969(99)00089-8. [DOI] [PubMed] [Google Scholar]
- 7.Fine D H, Furgang D, Schreiner H C, Goncharoff P, Charlesworth J, Ghazwan G, Fitzgerald-Bocarsly P, Figurski D H. Phenotypic variation in Actinobacillus actinomycetemcomitansduring laboratory growth: implications for virulence. Microbiology. 1999;145:1335–1347. doi: 10.1099/13500872-145-6-1335. [DOI] [PubMed] [Google Scholar]
- 8.Fives-Taylor P M, Meyer D H, Mintz K P, Brissette C. Virulence factors of Actinobacillus actinomycetemcomitans. Periodontology 2000. 1999;20:136–167. doi: 10.1111/j.1600-0757.1999.tb00161.x. [DOI] [PubMed] [Google Scholar]
- 9.Forbes B A, Sahm D F, Weissfeld A S. Bailey & Scott's diagnostic microbiology. St. Louis, Mo: Mosby, Inc.; 1998. [Google Scholar]
- 10.Fürste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacPexpression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 11.Goncharoff P, Yip J K K, Wang H, Schreiner H C, Pai J-A, Furgang D, Stevens R H, Figurski D H, Fine D H. Conjugal transfer of broad-host-range incompatibility group P and Q plasmids from Escherichia coli to Actinobacillus actinomycetemcomitans. Infect Immun. 1993;61:3544–3547. doi: 10.1128/iai.61.8.3544-3547.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulhen F, Hafezi A, Uitto V-J, Hinode D, Nakamura R, Grenier D, Mayrand D. Subcellular localization and cytotoxic activity of the GroEL-like protein isolated from Actinobacillus actinomycetemcomitans. Infect Immun. 1998;66:5307–5313. doi: 10.1128/iai.66.11.5307-5313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graber K R, Smoot L M, Actis L A. Expression of iron binding proteins and hemin binding activity in the dental pathogen Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1998;163:135–142. doi: 10.1111/j.1574-6968.1998.tb13037.x. [DOI] [PubMed] [Google Scholar]
- 14.Guthmiller J M, Kolodrubetz D, Kraig E. Mutational analysis of the putative leukotoxin transport genes in Actinobacillus actinomycetemcomitans. Microb Pathog. 1995;18:307–321. doi: 10.1006/mpat.1995.0028. [DOI] [PubMed] [Google Scholar]
- 15.Haase E M, Zmuda J L, Scannapieco F A. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect Immun. 1999;67:2901–2908. doi: 10.1128/iai.67.6.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinnebusch B J, Perry R D, Schwan T G. Role of the Yersinia pestis hemin storage (hms) locus in the transition of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 18.Holt S C, Tanner A C, Socransky S S. Morphology and ultrastructure of oral strains of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. Infect Immun. 1980;30:588–600. doi: 10.1128/iai.30.2.588-600.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue T, Tanimoto I, Ohta H, Kato K, Murayama Y, Fukui K. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitansfimbriae. Microbiol Immunol. 1998;42:253–258. doi: 10.1111/j.1348-0421.1998.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 20.Inouye T, Ohta H, Kokeguchi S, Fukui K, Kato K. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1990;57:13–17. doi: 10.1016/0378-1097(90)90405-f. [DOI] [PubMed] [Google Scholar]
- 21.Kagermeier A S, London J. Actinobacillus actinomycetemcomitansstrains Y4 and N27 adhere to hydroxyapatite by distinctive mechanisms. Infect Immun. 1985;47:654–658. doi: 10.1128/iai.47.3.654-658.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan J B, Fine D H. Codon usage in Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1998;163:31–36. doi: 10.1111/j.1574-6968.1998.tb13022.x. [DOI] [PubMed] [Google Scholar]
- 23.Komatsuzawa H, Kawai T, Wilson M E, Taubman M A, Sugai M, Suginaka H. Cloning of the gene encoding the Actinobacillus actinomycetemcomitansserotype b OmpA-like outer membrane protein. Infect Immun. 1999;67:942–945. doi: 10.1128/iai.67.2.942-945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lally E T, Kieba I R. Molecular biology of Actinobacillus actinomycetemcomitans leukotoxin. In: Genco R, Hamada S, Lehner T, McGhee J, Mergenhagen S, editors. Molecular pathogenesis of periodontal disease. Washington, D.C.: American Society for Microbiology; 1994. pp. 69–82. [Google Scholar]
- 25.Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 26.Mayer M P A, Bueno L C, Hansen E J, DiRienzo J M. Identification of a cytolethal distending toxin gene locus and features of a virulence-associated region in Actinobacillus actinomycetemcomitans. Infect Immun. 1999;67:1227–1237. doi: 10.1128/iai.67.3.1227-1237.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer D H, Sreenivasan P K, Fives-Taylor P M. Evidence for invasion of a human oral cell line by Actinobacillus actinomycetemcomitans. Infect Immun. 1991;59:2719–2726. doi: 10.1128/iai.59.8.2719-2726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyasaki K T, Wilson M E, Reynolds H S, Genco R J. Resistance of Actinobacillus actinomycetemcomitans and differential susceptibility of oral Haemophilusspecies to the bactericidal effects of hydrogen peroxide. Infect Immun. 1984;46:644–648. doi: 10.1128/iai.46.3.644-648.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. Protein Struct Funct Genet. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 30.Rosan B, Slots J, Lamont R J, Listgarten M A, Nelson G M. Actinobacillus actinomycetemcomitansfimbriae. Oral Microbiol Immunol. 1988;3:58–63. doi: 10.1111/j.1399-302x.1988.tb00082.x. [DOI] [PubMed] [Google Scholar]
- 31.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 32.Saiki R K, Gelfand D H, Stoffel S, Scharf S I, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Scannapieco F A, Millar S J, Reynolds H S, Zambon J J, Levine M J. Effect of anaerobiosis on the surface ultrastructure and surface proteins of Actinobacillus actinomycetemcomitans (Haemophilus actinomycetemcomitans) Infect Immun. 1987;55:2320–2323. doi: 10.1128/iai.55.9.2320-2323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shenker B J, McKay T, Datar S, Miller M, Chowhan R, Demuth D. Actinobacillus actinomycetemcomitans immunosuppressive protein is a member of the family of cytolethal distending toxins capable of causing a G2arrest in human T cells. J Immunol. 1999;162:4773–4780. [PubMed] [Google Scholar]
- 35a.Skerker J M, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slots J, Ting M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalisin human periodontal disease: occurrence and treatment. Periodontology 2000. 1999;20:82–121. doi: 10.1111/j.1600-0757.1999.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 37.Sreenivasan P K, LeBlanc D J, Lee L N, Fives-Taylor P. Transformation of Actinobacillus actinomycetemcomitansby electroporation, utilizing constructed shuttle plasmids. Infect Immun. 1991;59:4621–4627. doi: 10.1128/iai.59.12.4621-4627.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson V J, Bhattacharjee M K, Fine D H, Derbyshire K M, Figurski D H. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J Bacteriol. 1999;181:7298–7307. doi: 10.1128/jb.181.23.7298-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Winkelhoff A J, Slots J. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalisin nonoral infections. Periodontology 2000. 1999;20:122–135. doi: 10.1111/j.1600-0757.1999.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 40.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P-L, Shirasu S, Shinohara M, Daito M, Fujii T, Kowashi Y, Ohura K. Purification and characterization of a trypsin-like protease from the culture supernatant of Actinobacillus actinomycetemcomitansY4. Eur J Oral Sci. 1999;107:147–153. doi: 10.1046/j.0909-8836.1999.eos107211.x. [DOI] [PubMed] [Google Scholar]
- 42.Weiss A A, Johnson F D, Burns D L. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci USA. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White P A, Nair S P, Kim M-J, Wilson M, Henderson B. Molecular characterization of an outer membrane protein of Actinobacillus actinomycetemcomitansbelonging to the OmpA family. Infect Immun. 1998;66:369–372. doi: 10.1128/iai.66.1.369-372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyss C. Selected low-cohesion variants of Actinobacillus actinomycetemcomitans and Haemophilus aphrophiluslack distinct antigens recognized by human antibodies. Arch Microbiol. 1989;151:133–136. doi: 10.1007/BF00414427. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida Y, Nakano Y, Yamashita Y, Koga T. Identification of a genetic locus essential for serotype b-specific antigen synthesis in Actinobacillus actinomycetemcomitans. Infect Immun. 1998;66:107–114. doi: 10.1128/iai.66.1.107-114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zambon J J. Actinobacillus actinomycetemcomitansin human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]