Abstract
Intercellular cross talk between cancer cells and stromal and immune cells is essential for tumor progression and metastasis. Extracellular vesicles and particles (EVPs) are a heterogeneous class of secreted messengers that carry bioactive molecules and that have been shown to be crucial for this cell–cell communication. Here, we highlight the multifaceted roles of EVPs in cancer. Functionally, transfer of EVP cargo between cells influences tumor cell growth and invasion, alters immune cell composition and function, and contributes to stromal cell activation. These EVP‐mediated changes impact local tumor progression, foster cultivation of pre‐metastatic niches at distant organ‐specific sites, and mediate systemic effects of cancer. Furthermore, we discuss how exploiting the highly selective enrichment of molecules within EVPs has profound implications for advancing diagnostic and prognostic biomarker development and for improving therapy delivery in cancer patients. Altogether, these investigations into the role of EVPs in cancer have led to discoveries that hold great promise for improving cancer patient care and outcome.
Keywords: biomarkers, cancer, extracellular vesicles and particles, metastasis, therapeutic deliverables
Subject Categories: Cancer, Membranes & Trafficking
The heterogeneity of secreted extracellular vesicles, their biogenesis, purification, and multifaceted roles in malignancies are covered by this extensive review.

Introduction
Extracellular vesicles (EVs), which constitute a heterogenous group of vesicles carrying various biomolecular materials that are secreted by most cells, have gained increasing attention due to their complex cargo and their ability to mediate long‐distance communication in normal development and physiology, as well as in several pathophysiological conditions. The two major groups of EVs that have received intensive study include exosomes and microvesicles (MVs, also called ectosomes) (Cocucci & Meldolesi, 2015; Meldolesi, 2018; van Niel et al, 2018). MVs vary in size, ranging from 70 nm to almost 1 μm, and they are shed directly from the plasma membrane into the extracellular space. Exosomes form within the endosomal system prior to their secretion and are typically 50–150 nm in size. Further dissection of EVs has led to the recent discoveries of subcategories with different canonical EV markers and possibly of different cellular origins (Kowal et al, 2016), as well as of distinct subclasses with different sizes and cargo, named exosome small (Exo‐S) and exosome large (Exo‐L), and of a new non‐membranous nanoparticle, named exomere (Zhang et al, 2018b). Thus, we refer to this collective secreted heterogeneous mixture consisting of MVs/ectosomes, exosomes, and exomeres as extracellular vesicles and particles (EVPs). Throughout this review, we will use the term EVP, or we will use more specific nomenclature (e.g., MV, exosome, exomere) when subtype of EVPs are known for a particular study. For further discussion on appropriate use of EV terminology, we refer readers to a detailed description of this matter by Thery et al (2018).
While many physiological processes, including neurotransmission and immune signaling, are mediated by EVPs (Saliba et al, 2019; Zhou et al, 2020b), the role of EVPs in systemic aspects of human diseases, and in particular cancer, has attracted much attention. The inhibition of exosome production by cancer and stromal cells is invariably associated with reduced cancer growth and metastasis in a series of experimental studies (Bobrie et al, 2012; Peinado et al, 2012; Matsumoto et al, 2017; Richards et al, 2017), supporting the notion that exosome secretion is pivotal to cancer development. A considerable body of literature has shown the involvement of EVPs in all aspects of cancer progression, including host–microbiota interaction, carcinogenesis, metastasis establishment, and systemic effects of cancer on distant organs. EVPs are found in all bodily fluids, and their cargo signature can be used to predict cancer type at early stages and therapeutic responses (Hoshino et al, 2020; Shimada et al, 2021). The innate low toxicity and broad tissue distribution of EVPs also make them desirable and autologous carriers of chemotherapeutics, genetic material, or imaging agents.
In this review, we first present fundamental aspects of EVPs, particularly as they relate to cancer, including their heterogeneity, their mechanisms of biogenesis and uptake, and their diverse biomolecular cargoes. Next, we briefly cover key methods for the isolation and use of EVPs for experimental purposes. We will then discuss the multifaceted functional roles of EVPs during cancer (Figure 1), illustrating how EVPs from tumor, stroma, and immune cells in the tumor microenvironment organically orchestrate tumor growth and invasion, progression to metastatic disease, and systemic effects of cancer. Finally, we will examine the potential of EVPs as cancer biomarkers, therapeutic deliverables, and therapeutic/prognostic targets, highlighting their promises and limitations.
Figure 1. Multifaceted roles of EVPs in cancer.
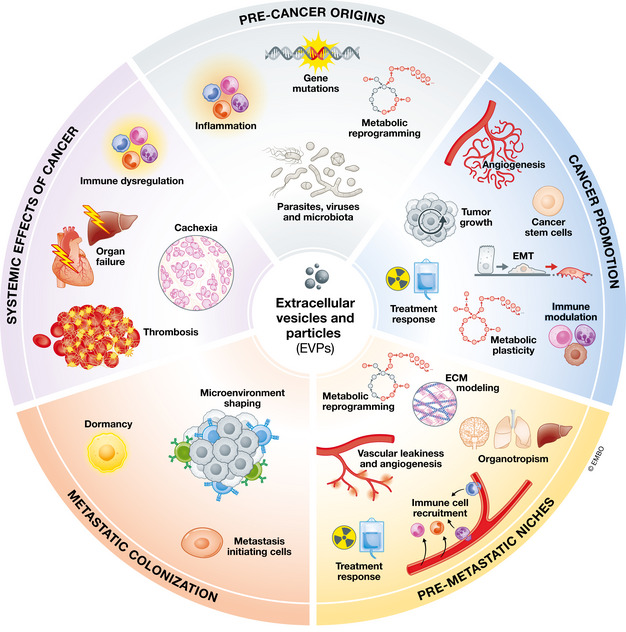
Diagram depicting the contribution of EVPs to different aspects of cancer initiation and progression, which is the subject of this review.
EVP heterogeneity and biogenesis
Complexity at the nanoscale level: EVP heterogeneity
EVPs represent a heterogeneous mixture of vesicles and particles that also vary in their biophysical properties, particularly with regard to size and density. Hence, characterization of the different subclasses is critical for understanding their contribution to cancer. By implementing asymmetric‐flow field‐flow fractionation (AF4) technology, Zhang et al (2018b) recently identified three distinct subpopulations of EVPs, named exosome small (Exo‐S, 60–80 nm) and exosome large (Exo‐L, 90–120 nm), alongside a newly discovered nanoparticle population, named exomere (<50 nm, with peak at ~ 35 nm), which lacks a membrane structure. In support, Zhang et al (2019e) also reported the isolation of exomeres from cultured cell lines using a modified ultracentrifugation strategy and demonstrated the transfer of functional exomere cargo to recipient cells. These novel nanoparticles were also found in human blood plasma by atomic force imaging (Bairamukov et al, 2020).
Exomeres exhibit a unique biomolecular composition compared to Exo‐S and Exo‐L. Specifically, they are more enriched in proteins involved in metabolic pathways, while Exo‐S and Exo‐L preferentially contain membrane proteins and signaling proteins. All three populations package DNA in a cell‐type dependent manner, whereas RNA is generally more enriched in Exo‐S and Exo‐L across cell types. Exomeres contain less lipids than Exo‐S and Exo‐L and display a distinct composition profile of different lipid classes. Besides exomeres, Zhang et al (2021) recently reported additional non‐membranous nanoparticles named supermeres, which were further isolated from EVs and exomere‐depleted cell culture conditioned medial via ultracentrifugation. The protein and RNA composition of supermeres differ from Exo‐S, Exo‐L, and exomeres. Remarkably, the majority of extracellular RNA was found associated with supermeres rather than exosomes and exomeres. The biogenesis, molecular and structural organization, and functional mechanisms of supermeres remain to be determined. Furthermore, a recent study has reported that cytotoxic T cells release perforin and granzymes in stable particles named supramolecular attack particles (SMAPs), which represent another type of non‐EV particle (Balint et al, 2020). The SMAPs are autonomously cytotoxic and ~ 120 nm in diameter, composed of a cytotoxic core and a shell of glycoproteins but lack a phospholipid membrane. More than 285 SMAP‐associated proteins have been identified, including perforin and granzymes. A C‐terminal fragment of thrombospondin‐1 has been found in the shell structure and may contribute to the targeting specificity of SMAPs. Whether SMAPs function only through the immunological synapse or via other modes of action requires further investigation.
Other secreted vesicles with potentially more specialized functions have also been described. Recently, D'Acunzo et al (2021) reported the identification of mitovesicles, a new population of brain‐derived double‐membraned EVs of mitochondrial origin. These mitovesicles overlap in size and cosediment with exosomes, but they can be further separated from exosomes via a high‐resolution density gradient step. They contain a specific subset of mitochondrial constituents whose levels and cargo change during pathophysiological processes involving mitochondrial dysfunction, such as in Down Syndrome, but their mechanism of release is unknown. In addition, several studies have identified various types of larger, micro‐sized vesicles. For example, adult neurons from C. elegans were found to extrude large vesicles called exophers (~ 4 μm), which contain protein aggregates and organelles (Melentijevic et al, 2017). In migrating cells, an additional class of large vesicles (~ 1 μm), named migrasomes, form at the tips and intersection of trailing edge retraction fibers and contain numerous smaller vesicles and cytosolic contents (Ma et al, 2015). Lastly, large oncosomes (0.5–10 μm) carrying oncoproteins such as AKT1 are shed from the plasma membrane of cancer cells (Minciacchi et al, 2017).
New beginnings: biogenesis of endosome‐ and plasma membrane‐derived EVPs
Exosome biogenesis begins with the formation of nano‐sized intralumenal vesicles (ILVs) that are contained within endocytic compartments known as multivesicular endosomes or multivesicular bodies (MVBs) (Simons & Raposo, 2009; Gruenberg, 2020). ILVs form by inward budding of the endosome limiting membrane and detachment of the bud as a vesicle into the endosome lumen. MVBs traffic to the plasma membrane where they fuse and release the ILVs extracellularly as exosomes. By contrast, plasma‐membrane‐derived MVs form by direct budding of plasma membrane into the extracellular space (Sedgwick & D'Souza‐Schorey, 2018; Clancy et al, 2021).
Intralumenal vesicle budding at multivesicular bodies
Pathways of ILV budding into MVBs during exosome biogenesis include those regulated by endosomal sorting complex required for transport (ESCRT) (Juan & Furthauer, 2018), by programmed cell death 6‐interacting protein (also known as ALG‐2‐interacting protein X (Alix)) (Bissig & Gruenberg, 2014), and by lipids (Skotland et al, 2017b) (Figure 2A). These pathways have been studied in cancer cells, as well as non‐cancer cell types that may be crucial microenvironmental regulators of tumor progression and metastasis.
Figure 2. EVP cargo, biogenesis, and uptake.
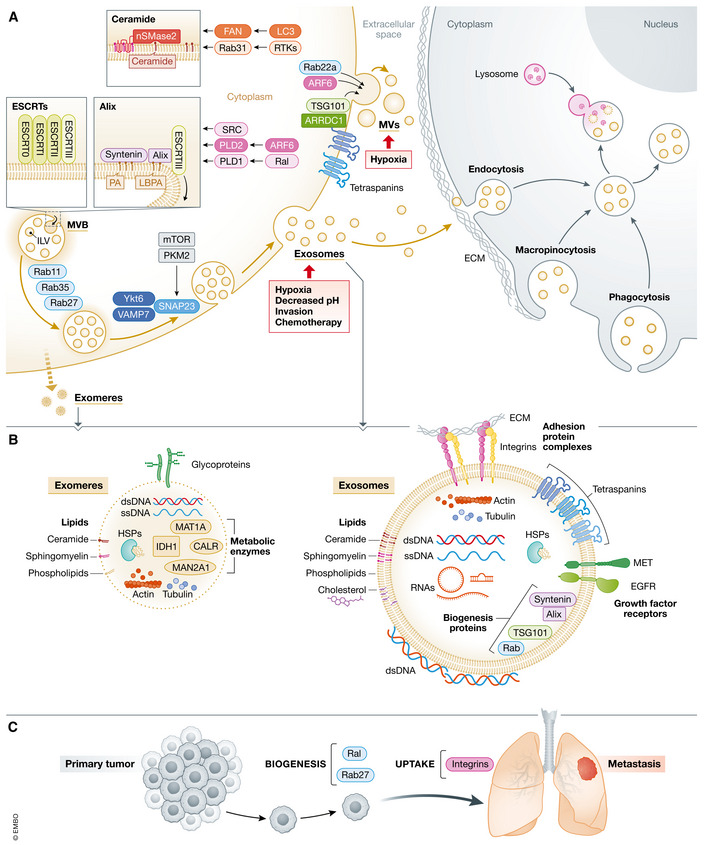
(A) EVP biogenesis occurs in MVB endosomes, giving rise to secreted exosomes, and at the plasma membrane, resulting in the generation of MVs, which are also termed ectosomes. Invagination of the endosome membrane leads to the formation of nanosized (50–150 nm) ILVs that are contained within the MVB lumen. ILV formation is regulated by various molecular processes at the MVB membrane that are each capable of capturing cargo and remodeling membranes for ILV generation and are also induced by upstream regulators. MVB trafficking is controlled by Rabs and SNARE‐complexes for secretion of exosomes at the cell surface. MVs/ectosomes range in size from 50 nm to almost 1 μm. Their budding occurs at plasma membrane microdomains enriched for ESCRT proteins, like TSG101, which is recruited by ARRDC1 for MV formation, and MV biogenesis is also stimulated by ARF6. In cancer, molecular pathways involving RTKs/Rab31, SRC, ARF6/PLD2, Ral/PLD1, and mTOR/PKM2 along with environmental and cellular factors related to hypoxia, pH, invasion, chemotherapy can all influence exosome biogenesis. Hypoxia and Rab22a promote MV formation in cancer cells. EVP uptake involves attachment of EVPs to extracellular matrix via adhesion molecules, such as integrins, on EVPs. Pathways of cellular uptake include endocytosis, macropinocytosis, and phagocytosis. Internalized EVPs traffic to the perinuclear area of recipient cells where they may fuse with lysosomes. (B) EVPs (including exomeres on the left and exosomes on the right) carry a variety of macromolecules, including proteins, nucleic acids, and lipids. Transmembrane proteins include adhesion molecules, like integrins, growth factor receptors, and tetraspanins, which are involved in biogenesis and which may also mediate adhesion. Cytosolic proteins such as actin, HSPs and other biogenesis factors are also commonly found in EVPs. Both dsDNA and ssDNA are found associated with EVPs. Double‐stranded DNA is present both inside and on the surface of EVPs. Various RNAs, such as miRNAs, mRNAs, and other short and long noncoding RNAs, are carried by EVPs. Lipids, particularly cholesterol, phospholipids, ceramides, and sphingomyelin are enriched in EVPs. (C) Biogenesis and uptake factors functionally regulate in vivo cancer metastasis. Inhibition of Ral and Rab GTPases involved in biogenesis impairs metastasis. Blockade of exosomal integrins reduces exosome uptake and metastasis. HSP, heat shock protein; ILV, intralumenal vesicle; MVB, multivesicular body; MV, macrovesicle; RTK, receptor tyrosine kinases; dsDNA, double‐stranded DNA; ssDNA, single‐stranded DNA.
The ESCRT pathway of ILV biogenesis involves a series of four main complexes, ESCRT‐0, ESCRT‐I, ESCRT‐II, and ESCRT‐III, which interact and assemble in an ordered, stepwise fashion on membranes (Hurley, 2015; Juan & Furthauer, 2018; Vietri et al, 2020). ESCRT‐0, ‐I, and ‐II subunits possess ubiquitin binding domains for capture of ubiquitinated cargo, while ESCRT‐I, ‐II, and ‐III promote membrane remodeling for ILV budding. The ATPase VPS4 interacts with ESCRT‐III to support completion of ILV formation by promoting membrane scission, resulting in ILVs pinching off into MVB lumens. Importantly, depletion of multiple ESCRT protein subunits or VPS4 affects exosome biogenesis by altering exosome number, size, and protein composition to varying extents (Tamai et al, 2010; Baietti et al, 2012; Colombo et al, 2013; Jackson et al, 2017; Banfer et al, 2018).
Intraluminal vesicle biogenesis mediated by ESCRT‐III is also induced by the ESCRT‐associated protein Alix (Bissig & Gruenberg, 2014). Targeting of Alix to endosomes for ESCRT‐III engagement occurs through multiple mechanisms that all have been shown to support exosome secretion in cancer cells. In MCF7 breast cancer cells, syntenin, a cytoplasmic adapter protein, recruits Alix to MVBs where interaction with ESCRT‐III induces ILV formation (Baietti et al, 2012; Roucourt et al, 2015). Syntenin may be targeted to endosomes through activation of phospholipase D (PLD)2 by the GTPase ADP‐ribosylation factor 6 (ARF6); PLD2, in turn, generates phosphatidic acid (PA) at the MVB limiting membrane to which syntenin can bind (Ghossoub et al, 2014). Generation of PA at endosomes can also occur via PLD1 activation by Ral GTPases to increase exosome biogenesis, and this function for Ral supports in vivo 4T1 mammary carcinoma metastasis (Ghoroghi et al, 2021). Localization of Alix to MVBs also occurs via association with the late endosome‐specific lipid lysobisphosphatidic acid (LBPA) to support ESCRT‐III‐dependent ILV formation and exosome production in HeLa cells (Matsuo et al, 2004; Larios et al, 2020).
The lipid ceramide has also been implicated in exosome biogenesis (Skotland et al, 2017b; van Niel et al, 2018). Neutral sphingomyelinase 2 (nSMase2), which is the enzyme that generates ceramide from sphingomyelin at endosomes, increases ILV and exosome biogenesis (Trajkovic et al, 2008). This function of ceramide at MVBs may be enabled by multiple, additional pathways to enhance exosome biogenesis. The autophagy‐related protein microtubule‐associated protein 1A/1B‐light chain 3 (LC3) may recruit FAN, an activator of nSMase (Adam‐Klages et al, 1996), to endosome membranes where FAN could stimulate ceramide‐mediated ILV formation (Leidal et al, 2020). Moreover, activated Rab31 can augment exosome production and packaging of epidermal growth factor receptor (EGFR) into cancer cell‐derived exosomes, and it was proposed that this occurs via the ceramide pathway of ILV production (Wei et al, 2021a), suggesting that it may be critical for cancer cell exosome biogenesis.
Trafficking and plasma membrane fusion of multivesicular bodies
The final stages of exosome biogenesis involve the trafficking of MVBs to the plasma membrane where they fuse and release ILVs as exosomes. Rab GTPase proteins, which are major regulators of intracellular membrane trafficking (Zhen & Stenmark, 2015), control the movement of MVBs toward the plasma membrane (Blanc & Vidal, 2018). Rab protein activity is regulated by GTPase‐activating proteins (GAPs) (Zhen & Stenmark, 2015), and Rab‐dependent pathways are further mediated by interaction with downstream effectors that are required for transport to and fusion of traveling vesicles with destination membranes (Fukuda, 2013). Alongside Rabs, these Rab GAPs and effectors have also been implicated in exosome release (Figure 2A).
Rab27 and Rab35 are among the most recognized Rabs that influence MVB to plasma membrane trafficking for exosome secretion, and they also have functional roles in cancer. Rab27a and Rab27b associate with MVBs and mediate efficient release of exosomes by promoting targeting and docking of MVBs to the cell surface in HeLa cells (Ostrowski et al, 2010). The Rab27 effectors Slp4 and Slac2b also support exosome release (Ostrowski et al, 2010). Rab35 and its GAPs TBC1D10A, TBC1D10B, and TBC1D10C were also shown to regulate transport and fusion of MVBs with the plasma membrane in oligodendroglial cells (Hsu et al, 2010). Importantly, Rab27a and Rab35 are necessary for the secretion of exosomes from tumors in vivo (Bobrie et al, 2012; Peinado et al, 2012; Pucci et al, 2016). Rab11 (Savina et al, 2002) and Rab7 (Baietti et al, 2012) may also function in this final stage of exosome biogenesis.
Additional factors residing on the MVB membrane and at the cell periphery with direct roles in promoting fusion of membranes also control exosome secretion. These molecules include vesicle‐ and target‐SNARES (v‐ and t‐SNARES), which localize to the vesicle membrane and plasma membrane, respectively (Jahn & Scheller, 2006). The t‐SNARE SNAP23 is phosphorylated and localizes to the intracellular face of the plasma membrane to promote exosome release in cancer cells (Wei et al, 2017b; Verweij et al, 2018; Yang et al, 2019b). Likewise, the v‐SNAREs VAMP7 and Ykt6 are also implicated in exosome secretion by facilitating MVB‐plasma membrane fusion in cancer cells (Fader et al, 2009; Gross et al, 2012; Sun et al, 2020a).
Biogenesis of endosome‐derived EVPs in cancer cells
Although these pathways of exosome biogenesis have been extensively characterized, it remains uncertain which are crucial in cancer cells and if certain pathways are preferentially upregulated in cancer cells compared with non‐transformed cells. As noted above, studies of some of these pathways have been conducted in cancer cell lines, and the functional roles of some biogenesis factors in mediating in vivo metastasis have been shown. However, more firmly establishing whether there are distinctions in mechanisms of exosome biogenesis pathways in cancer versus non‐cancer cells and further illuminating how such pathways are triggered will uncover possible routes for safe therapeutic targeting of exosomes for cancer treatment.
Insight into such specificity is beginning to emerge. For instance, the tyrosine kinase SRC can enhance exosome secretion by stimulating ILV budding through phosphorylating syndecans and syntenin (Imjeti et al, 2017) and interacting with Alix (Hikita et al, 2019). Because SRC is overexpressed or exhibits increased activation by growth factor and integrin signaling in multiple cancers (Kim et al, 2009), it may potentiate the syntenin‐Alix pathway of exosome biogenesis in cancer cells. Similarly, Rab31‐dependent upregulation of ceramide‐induced ILV formation may represent another cancer‐cell‐specific pathway of exosome biogenesis. Rab31 has been described to be overexpressed in cancer (Chua & Tang, 2015), and phosphorylation of Rab31 by various receptor tyrosine kinases often overactivated in cancer, such as EGFR, HER2, and MET, leads to abnormal activation of Rab31, which in turn could induce exosome biogenesis (Wei et al, 2021a). Overexpression or enhanced activation of other GTPases involved in exosome biogenesis, including ARF6 (Li et al, 2017d), Ral (Yan & Theodorescu, 2018), Rab27 (Li et al, 2018e), and Rab35 (Shaughnessy & Echard, 2018) in cancer cells has also been reported, indicating that pathways involving these factors may also be avenues by which exosome biogenesis is upregulated in cancer.
Microenvironmental and cellular stimuli frequently associated with cancer progression may also underlie tumor‐cell‐specific exosome production (Figure 2A). Notably, hypoxia, a common feature of the primary tumor microenvironment, has been shown to increase EVP production in various cancer types, including breast (King et al, 2012), lung (Hsu et al, 2017), prostate (Panigrahi et al, 2018), and ovarian cancer (Dorayappan et al, 2018) and melanoma (Park et al, 2019). Mechanistically, induction of HIF‐1α supports enhanced EVP release during hypoxia in breast cancer cells (King et al, 2012), while hypoxia appears to enhance MVB biogenesis and release of endosome‐derived exosomes in prostate cancer (Panigrahi et al, 2018) and ovarian cancer (Dorayappan et al, 2018). Additionally, alterations in pH associated with decreased extracellular and increased intracellular pH are prevalent in cancer cells (White et al, 2017), and augmented exosome biogenesis associated with changes in MVB biogenesis and transport has been observed when cancer cells are cultured in more acidic medium (Boussadia et al, 2018; Nakase et al, 2021). Impairment of lysosomal function can also promote exosome release and alter exosomal cargo by breast cancer cells (Latifkar et al, 2019). Progression of tumors to an invasive phenotype may similarly increase exosome biogenesis by supporting enhanced exosome secretion at sites of invadopodia, which are actin‐rich cellular protrusions that degrade extracellular matrix (ECM) (Eddy et al, 2017). MVBs were shown to dock at invadopodia in a Rab27‐dependent manner, and interfering with invadopodia reduced exosome secretion (Hoshino et al, 2013). MVB docking to the plasma membrane also relies on the actin‐binding protein cortactin, which promotes invadopodia formation (Artym et al, 2006; Sinha et al, 2016). Further work is needed to better understand the extent to which tumorigenesis influences exosome biogenesis and has the exciting potential to uncover roles for additional cancer‐associated phenotypes, such as altered metabolism, epithelial‐to‐mesenchymal transition (EMT), ECM stiffness, stromal activation, and immune cell infiltration. It is noteworthy that many of these phenotypes are interconnected; hence, their ability to mediate exosome production may converge on common molecular mediators that would be attractive therapeutic targets.
Biogenesis of plasma‐membrane‐derived microvesicles
Biogenesis of plasma‐membrane‐derived MVs (also known as ectosomes) involves budding of the plasma membrane out into the extracellular space and release of the bud as a shed vesicle (Clancy et al, 2021) (Figure 2A). Initiation of MV formation begins with the establishment of plasma membrane domains rich in lipids such as cholesterol and ceramide (Sedgwick & D'Souza‐Schorey, 2018). Additionally, discrete domains of the plasma membrane that are enriched with proteins involved in membrane reshaping, such as TSG101 and Vps4, have been associated with plasma membrane MV budding (Booth et al, 2006). Furthermore, targeting of TSG101 to the plasma membrane for MV biogenesis was demonstrated to occur via interaction with Arrestin Domain Containing 1 (ARRDC1) (Nabhan et al, 2012). Interestingly, ARRDC1 is distinctly localized to the plasma membrane and not the MVB, indicating that it may play a key role in dictating TSG101‐dependent exosome versus MV formation. TSG101 may also facilitate cargo recruitment through protein–protein interactions as has been proposed for endosome‐derived exosomes; indeed, TSG101 can regulate MV packaging of T cell receptors (Choudhuri et al, 2014). ARF6‐induced actomyosin contractility promotes the final shedding step of plasma membrane blebs (Muralidharan‐Chari et al, 2009).
In cancer cells, MV biogenesis is enhanced by hypoxia through an unclear mechanism involving upregulation of Rab22A (Wang et al, 2014). Protein targeting to MV in tumor cells has been linked to trafficking mediated by the SNARE protein vesicle‐associated membrane protein 3 (VAMP3) (Clancy et al, 2015), and miRNA cargo can be directed to tumor MVs via an interaction between ARF6, a regulator of MV biogenesis, and Exportin‐5, an RNA binding protein that mediates export of miRNA precursors out of the nucleus (Clancy et al, 2019). These studies have provided important insight into MV formation, but as with exosome biogenesis, further work is needed to understand mechanisms of MV biogenesis in cancer cells.
Biogenesis of exomeres
The biogenesis mechanisms of exomeres are still under investigation. While Exo‐S and Exo‐L are enriched in ESCRTs, Rabs, and SNARE‐related proteins, indicating that biogenesis may involve MVB trafficking or plasma membrane budding, proteins associated with exosome and MV biogenesis were shown to be lacking in exomeres, suggesting that exomere biogenesis may rely on different, yet uncharacterized mechanisms (Zhang et al, 2018b, 2019e). Subcellular localization analysis of exomere‐enriched proteins showed their specific association with endoplasmic reticulum and mitochondria, suggesting that their biogenesis may, at least partially, originate in these organelles. Enrichment in microtubule‐associated proteins in exomeres also implies the possibility of microtubule/cytoskeleton involvement in the secretion of exomeres. Furthermore, given the fact that exomere‐specific proteins are involved in metabolic processes, cell metabolic status might dictate exomere production and release. Lastly, future investigations into lipid species selectively enriched in exomeres, such as triglyceride, ceramide, and cholesteryl ester, might provide further information on their biogenesis (Zhang et al, 2018b, 2019e).
Taking it all in: mechanisms of EVP uptake
Intercellular communication involving EVP uptake by recipient cells is essential for EVP‐mediated cancer phenotypes. Therefore, understanding mechanisms of uptake may be key for identifying viable routes of therapeutic targeting of EVPs in cancer. In support, adjuvant treatment of mice with the drug reserpine, which was found to inhibit EVP uptake, appeared to eliminate lung metastasis of B16F10 melanoma cells (Ortiz et al, 2019), underscoring the potentially significant impact of targeting EVP uptake for cancer treatment.
The first step of EVP uptake involves attachment of vesicles to recipient cells. This binding can be mediated by surface molecules on EVPs. In particular, integrins and tetraspanins may regulate uptake either by directly promoting attachment to receptors on host cells or by supporting adhesion to cell‐adjacent ECM, which enables uptake. For instance, B‐cell‐derived EVPs carrying integrins β1 and β2 can bind activated fibroblasts and also fibronectin and collagen‐I (Clayton et al, 2004). Likewise, along with integrin α4 or β4, tetraspanin 8 regulates differential uptake by numerous cell types, including endothelial cells, lung fibroblasts, and bone marrow cells, and in multiple organs, such as the lung, liver, spleen, and pancreas (Nazarenko et al, 2010; Rana et al, 2012). In cancer, this function of EVP integrins is critical for target cell selection and uptake in pre‐metastatic niches; cancer cell EVPs can bind to laminin for uptake by fibroblasts and epithelial cells in the lungs or to fibronectin for uptake by Kupffer cells in the liver via integrin β4 or β5, respectively, and this ultimately determines metastatic organotropism (Hoshino et al, 2015).
Glycosylation of EVP surface proteins also influences EVP targeting and internalization. Increased glycosylation was shown to impede EVP uptake by ovarian cancer cells in vitro (Escrevente et al, 2011), and uptake of breast cancer cell EVPs by brain endothelial cells in vitro is also diminished by glycosylation (Nishida‐Aoki et al, 2020). Interestingly, changes in certain glycosylation patterns alter in vivo biodistribution. Specifically, while removal of N‐linked glycans did not appear to affect organ biodistribution of breast cancer cell EVPs, loss of O‐linked glycans enhanced uptake by the lungs and brain without affecting uptake by the spleen and liver (Nishida‐Aoki et al, 2020). Molecules on the surface of the receiving cell can also impact uptake of cancer cell EVPs. Cell surface 25‐hydroxycholesterol blocks EVP uptake (Ortiz et al, 2019), whereas heparan sulfate proteoglycans favor EVP uptake (Christianson et al, 2013). It would also be expected that additional integrin ligands or tetraspanin binding partners on host cells are required for uptake.
Methods employing fluorescent labeling of EVPs using lipophilic dyes and subsequent intracellular imaging of EVP fate in recipient cells have demonstrated that EVPs seem to be mainly internalized through regulated endocytosis, after which they enter the endocytic pathway and are trafficked to perinuclear late endosomes or lysosomes (Morelli et al, 2004; Tian et al, 2010, 2014a; Svensson et al, 2013; Costa Verdera et al, 2017). EVPs can also be taken up by macropinocytosis (Tian et al, 2014a; Nakase et al, 2015; Costa Verdera et al, 2017) and phagocytosis (Feng et al, 2010). Through these various modes of cellular uptake, EVPs would be expected to initially stay intact; hence, an outstanding question is how EVP cargoes are accessed by target cells. Backfusion of internalized EVPs with host endosomal membranes would facilitate liberation of intra‐EVP cargoes and allow membrane‐associated molecules to engage effectors by assuming the same orientation and topology relative to endosomal membranes as in donor cells. Direct monitoring and visualization of cargoes will be necessary to tease out these possibilities and more firmly corroborate direct and specific roles for EVP cargoes in eliciting changes in recipient cell phenotype.
Fully loaded: EVP cargo
EVP protein packaging
Packaging of particular proteins into EVPs functionally influences cancer progression and metastasis (Figure 2B). Most notably, EVP integrin profiles can distinguish cancers that metastasize to certain distant sites, and selective integrin packaging also plays a crucial role in dictating organ‐specific uptake of EVPs and consequent pre‐metastatic niche formation (Hoshino et al, 2015). EVPs derived from lung‐tropic breast cancer cells package more α6β4 and α6β1 integrins than brain metastatic breast cancer cells or liver metastatic pancreatic cancer cells, whereas αvβ5 is more highly represented in EVPs from liver‐tropic cells. Additionally, depletion of these integrins impaired organotropic EVP uptake and reduced metastasis. Other EVP molecules, such as cell migration‐inducing and hyaluronan‐binding protein (CEMIP), are also associated with metastasis to particular organs. Brain metastatic breast cancer cells preferentially package CEMIP into EVPs, and EVP CEMIP functionally supports brain metastasis (Rodrigues et al, 2019). Although levels of cellular CEMIP protein are equivalent between lung‐, bone‐, and brain‐tropic breast cancer cells, CEMIP was found markedly enriched in brain tropic cell‐derived EVPs. Collectively, these studies illustrate the critical role of specific EVP protein packaging in determining metastatic fate. As a result, monitoring cancer patient EVPs for selectively packaged proteins, such as specific integrins or CEMIP, may aid in selection of therapies most effective in treating future metastases at specific organs.
The oncoprotein EGFR has been identified in EVPs derived from glioma cells, squamous cell carcinoma cells, lung cancer cells, and gastric cancer cells. Oncogenic activation of EGFR and increased expression of wild‐type (WT) EGFR promote incorporation of EGFR into EVPs and allow for paracrine transfer of activated EGFR to less aggressive tumor cells and to endothelial cells to support tumor progression (Al‐Nedawi et al, 2008, 2009). EGFR could only be detected in serum EVPs from gastric cancer patients compared with healthy donors and its levels increased with cancer stage (Qu et al, 2017). Moreover, EGFR+ EVPs promoted gastric cancer liver metastasis. Interestingly, Rab31 was shown to promote packaging of EGFR into exosomes via the ceramide pathway of ILV biogenesis. Activated EGFR can phosphorylate Rab31, which stimulates Rab31‐dependent ILV formation and incorporation of EGFR into those ILVs (Wei et al, 2021a). Thus, although pathways of selective protein packaging into EVPs remain largely undefined, this study of EGFR packaging has begun to provide new insights into this process and may be blocked to inhibit EVP EGFR‐mediated phenotypes. Another oncoprotein, MET, has also been shown to be selectively packaged into cancer cell EVPs. Comparison of MET levels between EVPs from metastatic B16F10 mouse melanoma cells and from the less aggressive B16F1 variant showed that increased MET correlated with metastatic ability, and EVP MET was responsible for promoting melanoma lung metastasis by favoring premetastatic niche conditioning, corroborating the functional importance of selective protein packaging in metastasis (Peinado et al, 2012).
In addition to these functional studies of particular EVP proteins, proteomic analysis of EVPs has been instrumental in defining the broad repertoire of nuclear, cytoplasmic, and membrane proteins incorporated into cancer EVPs. These studies have identified common proteins that tend to include defined EVP markers, such as molecules associated with biogenesis. Importantly, these investigations have substantiated the importance and prevalence of distinct protein packaging for cancer‐associated EVPs, potentially making EVPs powerful tools for diagnosis and prognosis.
Analysis of EVPs from a panel of 60 cancer cell lines (NCI‐60) representative of nine tissue types identified greater than 6,000 unique proteins across EVPs from all cell lines (Hurwitz et al, 2016). 213 common proteins were identified that include proteins, such as Rabs, that are expected regulators of biogenesis. This study also demonstrated that proteomes of EVPs from different cell lines but of the same cancer type cluster together, and further analysis of individual cancers showed that samples also cluster based on stage or aggressiveness of disease. These exclusive proteins may therefore represent biomarkers for cancer type and prediction of disease state. These differences in proteins between EVPs generally reflected the varying levels of expression in the cells of origin, but this work also identified proteins that are preferentially enriched in certain EVPs even when similarly expressed between cell types, supportive of selective packaging.
A recent landmark study has reinforced the significance of EVP proteins as biomarkers through a large‐scale analysis of hundreds of human patient‐derived EVPs (Hoshino et al, 2020). This study aimed to identify EVP markers suitable for characterization of human patient EVPs, to establish whether cancer patient EVP proteomes are distinct from EVPs of healthy patients, and to determine if EVPs from patients with different types of cancer are distinct. Characterization of markers confirmed the presence of traditional EVP markers and also established new markers common to all samples. The conventional markers included HSP8 and Alix, which were also among the predominant ones found in the NCI‐60 study of cancer cell lines (Hurwitz et al, 2016), indicating a refined panel including these along with the newly established markers may be optimal for characterization of EVPs from patients. Hoshino and colleagues also mined for proteins that may be associated with cancer EVPs and discovered that EVPs from tumor tissue explants carry distinct proteins compared to EVPs from non‐tumor explants of the same tissue type. Furthermore, many of these proteins are significantly enriched in or exclusive to a particular cancer type, such as lung cancer or pancreatic cancer, but proteins common to different cancers were also identified in this tissue explant EVP analysis. Remarkably, these proteins were also observed to be enriched in EVPs from plasma of cancer patients compared to plasma‐derived EVPs from healthy individuals. Thus, select EVP proteins can potentially serve as clinically tractable liquid biopsy tools to identify and diagnose cancer. Hoshino and colleagues further identified additional cancer‐associated EVP proteins from other organ sources, such as immune organs, that are representative of systemic changes associated with cancer and therefore contribute to EVP proteome profiles which may help detect cancer via liquid biopsy.
Additional proteomic studies have more specifically focused on determining whether EVP proteomes may represent disease stage for types of cancer. For colon cancer, analysis of patient‐derived primary colorectal cancer cells and paired lymph node metastatic cells revealed that EVPs from both cell types carry approximately 800 proteins each but, less than half are similarly abundant between the samples, demonstrating that the majority are selectively enriched and could be used as predictors of disease stage (Choi et al, 2012). Likewise, a larger analysis of seven human melanoma cell lines uncovered that EVPs from more aggressive or metastatic melanoma carry distinct molecules compared to EVPs from cell lines representative of less advanced disease (Lazar et al, 2015). Additionally, proteomic characterization of glioblastoma EVPs also showed that the enrichment of certain proteins is associated with tumor grade and aggressiveness (Mallawaaratchy et al, 2017). Furthermore, multiple reports detailing the proteome of breast cancer EVPs established specific EVP protein signatures based on metastatic ability (Gangoda et al, 2017), primary tumor molecular subtype (Rontogianni et al, 2019), and treatment status and recurrence (Vinik et al, 2020). Altogether, these studies have identified distinct proteins selectively packaged into EVPs, which may have prognostic and predictive value for cancer detection, progression, and therapeutic response.
EVP DNA and RNA cargo
DNA present in EVPs may be a valuable source of circulating tumor DNA for liquid biopsy biomarker analysis. Cancer cell derived EVPs contain a variety of DNA molecules, including genomic DNA (Balaj et al, 2011;Kahlert et al, 2014; Lázaro‐Ibáñez et al, 2014; Thakur et al, 2014) and mitochondrial DNA (Guescini et al, 2010; Sansone et al, 2017). Genomic DNA represents the entire genome (Kahlert et al, 2014; Thakur et al, 2014) and may be single‐stranded (Balaj et al, 2011) or double‐stranded (Kahlert et al, 2014; Lázaro‐Ibáñez et al, 2014; Thakur et al, 2014) (Figure 2B). Moreover, imaging (Maire et al, 2021) and biochemical analysis (Thakur et al, 2014) of EVP DNA showed that it is present both on the surface of and within EVPs. DNA sequencing has identified the presence of oncogenes in various cancer‐derived EVPs, including amplified c‐Myc in medulloblastoma EVPs (Balaj et al, 2011), mutant BRAF in melanoma EVPs, mutant EGFR in non‐small‐cell lung cancer EVPs (Thakur et al, 2014), mutant KRAS and p53 in pancreatic cancer EVPs (Kahlert et al, 2014), and mutant PTEN in prostate cancer EVPs (Lázaro‐Ibáñez et al, 2014).
Mechanisms of EVP DNA incorporation remain understudied and obscure. The overall levels of DNA are higher in EVPs from cancer cells compared with normal fibroblasts (Balaj et al, 2011; Thakur et al, 2014), suggesting that tumorigenic phenotypes promote DNA packaging. Additionally, packaging of DNA into tumor‐derived EVPs may be induced by several stimuli, including oncogenic HRAS transformation (Lee et al, 2014), chemotherapy (Ke et al, 2017; Kitai et al, 2017; Yokoi et al, 2019), and radiation therapy (Diamond et al, 2018). Recent work also suggested that secretion of DNA via EVPs is cytoprotective by alleviating cellular stress associated with accumulation of harmful cytoplasmic DNA and micronuclei (Takahashi et al, 2017; Yokoi et al, 2019). Hence, cancer‐therapy‐induced DNA damage may promote EVP DNA packaging. Molecularly, tetraspanins, which are abundant in EVPs, may control EVP DNA loading through interaction with histones and DNA (Yokoi et al, 2019).
EVPs also transport various mRNA and noncoding RNA species, and many of these RNAs are significantly enriched in EVPs compared with the cell of origin, indicating that active mechanisms drive their packaging. For example, some mRNAs that are present in EVPs from mast cells could not be detected in parent cells, while some miRNAs were more abundant in EVPs (Valadi et al, 2007). Similarly, diverse RNAs are more highly represented in tumor‐derived EVPs compared to the tumor, including mRNAs in glioma cells (Skog et al, 2008), miRNAs in colorectal cancer cells (Cha et al, 2015), circular RNAs in liver cancer cells (Li et al, 2015c) and colon cancer cells (Dou et al, 2016), small nuclear RNAs in Lewis Lung Carcinoma (LCC) tumors (Liu et al, 2016d), and long noncoding RNAs in colorectal cancer cells (Hinger et al, 2018).
Whether or not EVPs transfer sufficient amounts of RNA to elicit phenotypic changes in recipient cells has been debated. However, RNAs are enriched in EVPs from cancer patients, and EVP RNAs impact disease progression by promoting pre‐metastatic niche formation and metastasis in mouse models (Xie et al, 2019; Möller & Lobb, 2020). Moreover, primary tumor cells may expose other cells in their immediate surroundings or at distant sites to a constant delivery of EVP‐encapsulated RNA that may indeed be critical for cancer progression. Accordingly, the mechanisms governing EVP RNA sorting have garnered considerable attention. In cancer cells, EVP packaging may be modulated by activation of oncogenes, such as mutant KRAS, which alters loading of various RNA molecules into EVPs (Cha et al, 2015; Dou et al, 2016; Hinger et al, 2018), in part by regulating association of the miRNA‐interacting protein Ago2 with MVBs (McKenzie et al, 2016). In addition, the nSMase2‐ceramide pathway was found to be dependent on 3′ UGGA and 3' UUU motifs, which agrees with prior work describing an increased prevalence of 3′‐end uridylated miRNA in EVPs compared with cells (Koppers‐Lalic et al, 2014). The RBP hnRNPA2B1 promotes selective sorting of miRNA into EVPs by binding specific motifs (EXOmotifs) contained within those miRNAs (Villarroya‐Beltri et al, 2013). Although it remains unclear how hnRNPA2B1 engages EVP biogenesis machinery, this study showed that hnRNPA2B1 associates with intracellular ceramide‐rich MVB structures, suggesting that the nSMase2‐ceramide pathway of exosome biogenesis may be involved. Moreover, this function of hnRNPA2B1 supports colorectal cancer liver metastasis (Zhao et al, 2020c) and bladder cancer lymphatic metastasis (Chen et al, 2019a) by regulating EVP sorting of tumor cell miRNA and long noncoding RNA, respectively. YBX1 is another RBP that mediates encapsulation of diverse small noncoding RNAs, including miRNA, tRNA, Y RNA, and Vault RNA, into EVPs (Shurtleff et al, 2016, 2017). Mechanistically, ubiquitination of YBX1, which supports interaction with TSG101 and consequent YBX1 secretion (Palicharla & Maddika, 2015), may further dictate loading of YBX1 and associated RNAs. Furthermore, secretion of EVP YBX1 was shown to be enhanced by EMT following HRAS‐mediated transformation of epithelial cells (Tauro et al, 2013b), suggesting that YBX1‐mediated packaging of miRNAs into EVPs may be augmented in cancer. Another recent study has identified an additional RBP‐mediated pathway of RNA sorting that is anchored by the autophagy‐related protein LC3 (Leidal et al, 2020). LC3 is targeted to MVB membranes and it interacts with the RBPs hnRNPK and SAFB. This interaction allows for capture and loading of small RNAs, namely snoRNAs and miRNAs, into exosomes. It remains unknown how this particular pathway may influence incorporation of RNA into cancer cell‐derived EVPs. Autophagy is well recognized for being upregulated as a vital coping mechanism in normal and cancer cells in response to environmental stressors (Galluzzi et al, 2015). Therefore, the contribution of LC3 to classical autophagy versus exosomal loading may be fine‐tuned to manage the homeostatic secretory and stress response needs of cells.
Overall, these studies of EVP RNA loading have provided considerable insight into selective packaging of bioactive molecules and may be key in guiding future interrogation of protein and DNA packaging. Such investigation could similarly be aimed at understanding how particular protein modifications influence packaging and how protein and DNA cargoes may interact with core biogenesis machineries.
EVP lipid content
Lipids represent an additional class of macromolecules that are packaged into EVPs (Figure 2B). They were first identified in EVPs from reticulocytes, which were reported to harbor cholesterol, sphingomyelin, and various phospholipids, including phosphatidylcholine, phosphatidylserine (PS), phosphatidylinositol, and phosphatidylethanolamine (Vidal et al, 1989). These lipids appeared mostly equivalently abundant in EVPs and cells, but subsequent studies of EVP lipid analysis have described an enrichment of lipids in EVP compared with cells, namely cholesterol, sphingomyelin, ceramide, and PS (Egea‐Jimenez & Zimmermann, 2019). B cell EVPs have an increased abundance of cholesterol, sphingomyelin, and ganglioside GM3 compared with levels in cells, whereas multiple phospholipid species were less enriched in EVPs compared with cells (Wubbolts et al, 2003). This lipid profile of B cell EVPs was also shown to be associated with detergent resistance properties similar to lipid raft microdomains found within cellular membranes. Likewise, sphingomyelin was found to be the main lipid enriched in EVPs from both mast cells and dendritic cells, whereas cholesterol was not found to be enriched, and the phospholipids phosphatidylcholine and phosphatidylethanolamine were decreased or increased, respectively, in EVPs compared with cells (Laulagnier et al, 2004).
In cancer, similar trends of EVP lipid content appear to exist. A large‐scale lipidomic analysis quantifying greater than 200 lipid species in EVPs from PC3 prostate cancer cells identified cholesterol, sphingomyelin, glycosphingolipids, such as ceramides, and PS as being more abundant in EVPs than cells, with other phospholipids generally lower in EVPs (Llorente et al, 2013). Furthermore, EVPs from urine of prostate cancer patients have higher levels of some lipids, namely ceramides, relative to urine‐derived EVPs from healthy patients, suggesting that these lipids could serve as fluid‐based biomarkers (Skotland et al, 2017a); however, in another study, ceramide levels were found to be decreased in urine EVPs from stage 2 benign prostate hyperplasia patients compared with urine EVPs from stage 3 prostate cancer patients (Clos‐Garcia et al, 2018), complicating the potential use of this lipid for biomarker purposes. Additionally, EVPs from colorectal cancer cells, glioblastoma cells, and hepatocellular carcinoma cells also display an enrichment of cholesterol, sphingomyelin, and PS compared with cells (Lydic et al, 2015; Haraszti et al, 2016). These studies unveil common themes in EVP lipid content, and further work establishing mechanisms of lipid packaging and functional roles for EVPs lipids may enhance their biomarker and therapeutic potential.
Exomere cargo
Following their recent discovery, exomeres have been thoroughly characterized for their molecular composition (Zhang et al, 2018b, 2019e). Proteomics analysis revealed unique protein profiles of exomeres that are quite distinct from that of EVs. As expected, membrane‐associated proteins are relatively low in exomeres, consistent with their lack of external membrane. Exomeres are instead enriched in metabolic enzymes and proteins involved in glycosylation, hypoxia, microtubule assembly, and coagulation. Gene Set Enrichment Analysis strikingly demonstrated that metabolic processes, including carbohydrate metabolism and protein synthesis, are selectively associated with exomere‐specific proteins. These bioinformatic analyses suggest potential roles for exomeres in modulating the metabolism in the recipient cells. Furthermore, the biological activity of exomere protein cargo has been demonstrated by the functional work carried out by Zhang and collaborators, where they showed that exomeres‐encapsulated β‐galactoside α2,6‐sialyltransferase 1 (ST6Gal‐I) and amphiregulin (AREG) mediate hypersialyation of membrane proteins and activation of EGFR signaling, respectively, in the recipient cells (Zhang et al, 2019e).
Posttranslational modifications of proteins are critical for cell signaling. Via lectin blotting and glycomic MS analysis, our group further evaluated the N‐ glycan profiles of exomere and exosome subsets (Zhang et al, 2018b). The extent of N‐glycosylation and the protein carriers present in exomeres were found different from that in Exo‐S and Exo‐L for the examined glycan species, including bisected and branched N‐glycans, structures related to fucosylation (fucose‐linked α ‐1,6) to GlcNAc or fucose‐linked (α ‐1,3) to GlcNAc‐related structures, and α ‐2,6‐sialylated glycans. Instead, complex N‐glycans with relatively high levels of sialylation are prevalent in all subsets. Glycomic studies further revealed differences in N‐glycan composition and structures among exomeres, Exo‐S, and Exo‐L, as evidenced by detection of unique ions in exomeres specifically. Notably, the N‐glycan profile of exomeres and exosomes is cell type‐specific.
Interestingly, exomeres contain lipids, though their total lipid content is three to fivefold lower than EVs, which is consistent with the lack of an external membrane in exomeres (Zhang et al, 2018b, 2019e). Additionally, lipidomic analysis showed distinct lipid composition among exomere and EVs. Major structural components of the plasma membrane lipid bilayer, such as phospholipids, sphingomyelin, and sterols, ranked top in both exomeres and EVs. Compared with other lipid classes, relatively higher levels of triglycerides and ceramides and a higher ratio of esterified to unesterified cholesterol were observed in exomeres compared with EVs, suggesting that exomeres may serve as a major carrier to transport these metabolites to recipient cells.
Similar to EVs, nucleic acids have also been found as part of exomere cargo. DNA content of exomeres is comparable with that of EVs and display cell type‐dependent patterns in their relative abundance (Zhang et al, 2018b, 2019e). As examined in a human pancreatic cancer cell line, DNA molecules carried by exomeres showed a slightly smaller size than those associated with EVs. In contrast to DNA, and regardless of cell type, exomeres contain less RNA and predominantly small RNAs (< 1,000 nucleotides). Interestingly, as examined in murine melanoma B16F10 cells, abundant small RNA peaks, likely composed of tRNAs, microRNAs, and other small RNAs, were detected in Exo‐S and Exo‐L, but not in exomeres (Zhang et al, 2018b).
Overall, the complex cargo of exomeres is starting to emerge, but questions remain regarding their packaging and regarding the biogenesis and biological functions of exomeres. Advanced, high‐resolution isolation platforms for single particle analysis and additional in vivo functional studies are desired to further investigate these aspects of exomeres biology.
Seeing is believing: isolation, labeling, and models for EVP studies
Methods for EVP isolation
Technology has advanced significantly in the field of EVP study, leading to the development of various methodologies for EVP isolation in the past decade. Based on the fundamental principles for separating EVPs from other types of entities in biofluids, these methods can be grouped into two main categories: one exploits the size, density, and charge of EVPs, while the other uses affinity capture techniques, such as immuno‐recognition of unique epitopes present on the EVP surface or specific ligand–receptor interaction.
The first category of EVP isolation and subtype separation methods includes differential ultracentrifugation (UC), density gradient, size exclusion chromatography (SEC), ultrafiltration (UF), anion exchange chromatography, and polymer precipitation (Thery et al, 2006; Merchant et al, 2010; Lasser et al, 2012; Tauro et al, 2012; Kim et al, 2016a). Additionally, AF4 has been successfully adapted to fractionate EVPs on the basis of hydrodynamic size. As we described (Zhang & Lyden, 2019), two perpendicular flows in a thin, flat, hollow channel with a semi‐permissive bottom wall membrane allow for separation and elution of EVP subtypes at different time points. Several key advantages offered by the AF4 technique include high separation resolution (down to a few nanometers), the ability to separate EVPs across a large size range of a few nanometers to micrometers, and being label‐free, gentle, rapid, and highly reproducible. However, due to the limited loading capacity, samples analyzed using AF4 usually need to be pre‐processed by other methods (such as UC) to first enrich and concentrate EVPs. By employing this technique, we have reported successful separation of distinct subsets of EVs and identification of exomeres from multiple cell lines (Zhang et al, 2018b). Several studies have described isolation and analysis of plasma and urine EVPs utilizing AF4 after initial isolation steps, such as UC, UF, SEC, and immunoaffinity capture (Yang et al, 2017a; Oeyen et al, 2018; Kim et al, 2020; Multia et al, 2020; Wu et al, 2020a), though the yield and purity of EVPs isolated from these samples need to be compared with other methods in parallel.
The application of UF‐based methods, such as dead‐end filtration and tangent flow filtration, for EVP isolation has increased greatly in the past few years (Liangsupree et al, 2021). EXODUS (exosome detection via the ultrafast‐isolation system) is a recently reported platform developed based on UF (Chen et al, 2021e). By enabling membrane vibration and generating transverse waves and acoustic streaming, EXODUS effectively limits the fouling effect and particle aggregation on the nanoporous membrane, thus increasing EVP isolation efficiency. Detailed characterization and comparison of EXODUS with other methods were conducted mainly on urine samples and showed superior performance in yield, purity, and speed. It can operate on a large range of sample volumes, from tens of microliters to hundreds of milliliters. Separating EVPs within different size ranges can be achieved by utilizing membranes with different pore sizes. In addition, the EXODUS workstation has the automatic operation feature, making it useful for high‐throughput study. However, more extensive analysis is needed to determine the performance of EXODUS for the isolation of EVPs from plasma. A general limitation for size‐based separation approaches, including EXODUS, is that it cannot separate EVPs from other types of molecular entities with similar sizes.
Wu et al (2017) described an acoustofluidic platform, which processes undiluted blood directly to isolate EVPs based on size. Two separation modules are integrated to first remove blood cells and platelets and subsequently separate EVPs from microparticles and other large bodies. The unique features of this approach include no requirement for blood pre‐processing, being label‐free and gentle, preservation of intact EVP morphology, flexibility to adjust the cutoff size for each separation module, and automation. However, as Wu and colleagues noted, the isolated samples may contain non‐EV particles (i.e., exomeres) and aggregates with sizes similar to that of EVs, such as lipoprotein particles. Refining the device configuration to separate EVPs from lipoproteins based on their different acoustic contrast factors has been proposed.
Due to the net negative charge carried by EVPs, charge‐based technologies, such as ion exchange and electrophoresis, have also been adapted to EVP isolation (Kim et al, 2016a; Kosanovic et al, 2017; Heath et al, 2018; Marczak et al, 2018; Chen et al, 2018a; Notarangelo et al, 2019; Kim & Shin, 2021). Ion exchange is a rapid and scalable approach, which can easily process samples in large volumes, an important application for large‐scale preparation of EVPs for therapeutic purposes. However, structural integrity and functionality of isolated EVPs have to be evaluated, especially in the case where buffers with extreme pH or high salt concentration have been used at the binding or elution steps. Analyzing complex samples, such as plasma, with charge‐based techniques will be challenging, and combination with other methods will be necessary to increase the purity of isolated EVPs.
In the affinity‐based category of EVP isolation methods, the most commonly utilized approach is immunoaffinity capture (IAC) by antibodies recognizing either general EV markers (such as tetraspanins CD9, CD81, and CD63) or membrane proteins that are unique to EVPs derived from specific cell types (such as EpCAM) (Tauro et al, 2013a; Kowal et al, 2016; Wang et al, 2016a; Zhao et al, 2016b; Brett et al, 2017; Ko et al, 2018; Sharma et al, 2018; Katsu et al, 2019; Lo et al, 2020). Both conventional immunoprecipitation and fluorescence‐activated cell sorting have been adapted for IAC of EVP subsets. Microfluidics coupling IAC with different fluidics designs represents a popular approach for positive or negative selection of EVP subsets in biofluids (Contreras‐Naranjo et al, 2017; Wang et al, 2021b). The advantages of IAC include allowing isolation of select EVPs derived from a specific cell type and being a single‐step, rapid, and flexible procedure. However, IAC approaches cannot separate EVP subsets that share the same targeting epitopes, and eluting EVPs from binding antibodies can be challenging, making IAC incompatible with functional studies that require intact EVPs.
Recent innovation in aptamers has made them promising alternatives to antibody‐based probes for isolation of EVP subsets. Aptamers are chemically synthesized short RNA or single‐stranded DNA molecules with unique 3D structures that bind their cognate targets with high specificity and affinity, comparable with antibodies (Sun et al, 2016). Remarkably, profiling of serum EVP surface proteins utilizing a panel of seven fluorescently labeled aptamers along with thermophoretic enrichment and linear discriminant analysis can successfully detect early stage cancers and classify cancer types with high specificity and sensitivity (Liu et al, 2019a). Liu and colleagues showed that this assay was superior to PSA levels for discrimination of prostate cancer from benign prostate enlargement and for recurrence assessment post‐prostatectomy. Their study also indicated that the thermophoresis condition can be adjusted to further separate small EVPs from microparticles. A strategy for duplex detection of EpCAM and Her2 on a single EVP was further developed to improve the identification of breast cancer‐derived EVPs by integrating hybridization chain reaction with dual DNA aptamer‐mediated recognition of these two targets (Li et al, 2021c). Dong et al (2018) described a highly sensitive electrochemical method for detecting tumor‐derived EVPs based on aptamer recognition‐induced multi‐DNA release and cyclic enzymatic amplification. Aptamer capturing can also be used for isolation of EVP subsets, and the captured EVPs can be nondestructively released via disruption of the aptamer 3D structure by incubating with complementary sequences or by restriction enzyme cleavage, allowing for preservation of EVP bioactivity (Zhang et al, 2019c).
Commercial kits have been developed based on the reversible binding of Tim4 protein to PS on the surface of EVPs. This affinity‐based method is highly specific and calcium (Ca2+)‐dependent (Miyanishi et al, 2007), facilitating release of intact EVPs by adding Ca2+ chelators (Nakai et al, 2016). This technique has been applied to various sample types and utilized for isolation and for quantification by ELISA and flow cytometry. However, similar to other affinity‐based approaches, this method cannot distinguish EVPs of different sizes. Moreover, for lipid‐rich samples, such as plasma, it may be challenging to separate EVPs efficiently from other PS‐containing particles. Strategies based on other separation principles may have to be included to improve the purity of isolated EVPs. To a lesser extent, lectin probes have been used to separate EVPs carrying characteristic glycans on their surface (Shimoda et al, 2019; Yamamoto et al, 2019; Jankovic et al, 2020). Heparin and peptides that exhibit specific affinity for canonical heat shock proteins have also been tested for EVP isolation (Ghosh et al, 2014; Balaj et al, 2015; Mao et al, 2019a).
Although many approaches for EVP isolation have been developed, different methods may result in enrichment of specific subsets of the heterogenous EVP population due to their unique separation principles. Therefore, caution should be exercised when determining the molecular composition and functional role of EVPs isolated by the various methods. The choice of method for EVP isolation depends on the sample complexity and quantity, and the required yield, purity, and bioactivity for downstream use. Technological advancements are still urgently needed for complex sample processing, high‐throughput analysis, and large‐scale preparation of high‐quality EVPs for therapeutic applications. Advancing our understanding of EVP biogenesis and the physical and molecular features of distinct EVP subpopulations is necessary to guide further methodology development for their isolation.
Tracking EVP biodistribution and uptake in vivo
Tracking the in vivo fate of cancer cell EVPs in mice is essential for understanding their contribution to tumor progression and metastasis. Mapping EVP organ biodistribution and cellular uptake has primarily been accomplished by injecting mice with EVPs purified from in vitro cell lines. The administration of exogenous EVPs has the drawback of not fully recapitulating the endogenous release of tumor EVPs in mice. However, because tumors secrete additional factors, such as soluble proteins, it has the distinct and critical advantage of allowing for the study of EVP‐specific phenotypes in vivo. Visualizing these injected EVPs has relied mainly on labeling them prior to injection using fluorescent lipophilic dyes that can be detected ex vivo in whole organs or in tissue sections. These dyes have multiple advantages. They are available as different fluorochromes, providing flexibility for signal readout and combined immunofluorescence‐based analysis of cell type‐specific EVP uptake. Their use also does not require any prior knowledge of EVP biomolecule content, as they will label all lipid‐containing particles isolated by conventional EVP purification procedures. Finally, they can label EVPs isolated from samples, such as patient‐derived specimens, for which genetic‐mediated tagging of EVPs may not be feasible. However, limitations of the dyes include formation of aggregates that could lead to signal artifacts and fluorescent signal half‐lives that may not completely reflect the biological fate and turnover of circulating EVPs. Nevertheless, this approach combined with functional validation of EVP‐mediated phenotypes has been crucial for unraveling key aspects of cancer progression and metastasis (Peinado et al, 2012; Costa‐Silva et al, 2015; Hoshino et al, 2015; Rodrigues et al, 2019).
There remains a pressing need for imaging and tracking of EVPs secreted endogenously by tumors in vivo to functionally connect EVP biodistribution with EVP‐mediated phenotypes. Limited studies have made use of cancer cell lines stably expressing genetic reporters, allowing implanted cells to release tagged EVPs that can be traced. These reporters are typically fusion proteins that consist of a signal generating protein to visualize EVPs and an EVP targeting sequence to ensure EVP packaging of the fusion protein. In particular, expression of GFP or luciferase targeted to EVPs through fusion to CD63 or lipid anchoring domains has been used to track EVPs secreted by tumors in mice. Orthotopic mammary tumors were shown to secrete GFP‐CD63 EVPs into the surrounding microenvironment, where they are taken up by stromal cells (Suetsugu et al, 2013). Spontaneous metastasis of these cells was associated with EVP uptake in the lungs and the presence of GFP+ circulating EVPs in blood. Similarly, CD63‐GFP EVPs secreted by melanoma in vivo were observed to be taken up by macrophages in tumor‐draining lymph nodes (Pucci et al, 2016). Genetically stable expression of membrane‐bound, EVP‐targeted Gaussia luciferase was also used to show that melanoma tumors secrete EVPs that reach distant tissues by measuring luciferase activity in harvested organs. Fusion of GFP to a palmitoylation signal also targets GFP to EVPs and allows for tracing of EVPs within the tumor microenvironment of thymoma tumors in mice (Lai et al, 2015). Likewise, fusion of the high intensity luciferase NanoLuc (Nluc) to CD63 enabled in vivo detection of EVPs secreted by subcutaneous colon cancer xenograft tumors in the stomach and intestine (Hikita et al, 2018). Multiple other luciferase‐based fusion proteins have been developed, but their ability to mark EVPs in vivo has only been investigated in the context of exogenous administration of luciferase+ EVPs from cultured cells (Takahashi et al, 2013; Lai et al, 2014; Wang et al, 2020c). Overall, these studies using fluorescent and bioluminescent reporters demonstrate how tumor‐derived EVPs communicate with their local and distant environments, providing support for endogenous secretion of tumor EVPs in mediating metastasis.
In addition to tracing EVPs in vivo, tracking delivery of specific EVP cargo remains a considerable challenge. Gain‐ and loss‐of‐function approaches have been crucial in defining the importance of various cargoes in mediating EVP‐dependent phenotypes, but understanding whether cargoes are active in recipient cells in vivo will establish direct links between EVP molecules and the observed phenotypes. Studies exploiting the packaging of Cre mRNA into EVPs have made headway into addressing this question. In this approach, tumor cells expressing a Cre transgene package Cre mRNA into EVPs; in vivo injection of these tumor cells into Cre‐reporter mice allows for visualization of host cells that acquire EVP Cre mRNA. Intracranial injection of Cre+ glioma cells into Cre‐reporter mice showed that Cre mRNA is delivered mainly to CD45+ leukocytes and also to neurons, microglia, and endothelial cells (Ridder et al, 2015). Similarly, Lewis lung carcinoma cells also deliver exosomal Cre mRNA primarily to CD45+ leukocytes when injected intravenously or subcutaneously, and Cre mRNA can be detected in serum EVPs of tumor‐bearing mice (Ridder et al, 2015). This same approach has been used to demonstrate that B16 melanoma tumor EVPs can deliver Cre mRNA to the lymph nodes, lungs, and spleen and that aggressive breast cancer cells can deliver mRNA to less aggressive breast cancer cells in vivo (Zomer et al, 2015). These proof‐of‐principle studies have been valuable in tracking uptake and transfer of endogenous EVPs and EVP molecules, but more consistent implementation of similar approaches combined with functional analysis is needed.
EVP Functions
Under construction: pre‐cancer origins
Chronic inflammation
Prolonged or chronic inflammatory conditions associated with immune infiltration and cytokine release precede the development of various cancers, including colorectal and liver cancer (Greten & Grivennikov, 2019). Immune cells are a major source of circulating EVPs in this context. For instance, the concentration of monocyte‐derived and T‐cell‐derived EVPs is increased in the serum of patients with systemic lupus erythematosus and correlates with activation of monocytes, neutrophils, B cells, and CD4+ lymphocytes (Lopez et al, 2020). T‐cell‐derived EVPs were found significantly enriched in tRNA fragments in comparison to releasing cells (Chiou et al, 2018). This selective packaging was proposed to be a mechanism for disposing of tRNAs that inhibit T cell ability to home to lymph nodes, become activated, and produce cytokines. Myeloid‐derived suppressor cells (MDSCs), which expand during chronic infectious and inflammatory diseases (Gabrilovich & Nagaraj, 2009), are also major producers of EVPs. For example, MDSC‐derived EVPs from individuals with late chronic sepsis or human immunodeficiency virus (HIV) or hepatitis C virus (HCV) infections are involved in priming naïve myeloid cells for differentiation into immunosuppressive MDSCs and in inhibiting T cell activation via transfer of the long noncoding RNA transcript HOTAIRM1 (Wang et al, 2018b; Alkhateeb et al, 2020; Thakuri et al, 2020).
Among other inflammatory conditions, chronic pancreatitis is associated with release of circulating EVPs enriched in pro‐inflammatory miRNAs and proteins that may foster systemic disease. These EVPs home to distant organs, such as the liver, lungs, and intestines, and induce pyroptosis of alveolar macrophages and polarization macrophages to an inflammatory phenotype associated with release of cytokines such as IL‐1β, IL‐6, and CCL‐2, leading to vascular leakage and exacerbating lung injury (Bonjoch et al, 2016; Jimenez‐Alesanco et al, 2019; Wu et al, 2020f). EVPs were also found to be associated with the onset of inflammatory bowel diseases (IBDs), such as colitis and Crohn's disease, which predispose to the development of colorectal cancer (CRC) (Stidham & Higgins, 2018; Guan, 2019). In an experimental model of dextran sulfate sodium (DSS)‐induced colitis, circulating EVPs expressing a series of acute‐phase proteins and lncRNA NEAT1‐induced polarization of macrophages toward a pro‐inflammatory phenotype (Wong et al, 2016; Liu et al, 2018c). EVPs from the colon of mice with colitis were found to express proteins associated with cell proliferation (e.g., epithelial growth factor receptor, EGFR) and induce fibroblast proliferation via EGFR‐ERK signaling, suggesting that EVPs produced during IBD development may directly lead to CRC onset (Hasegawa et al, 2020).
Similar to a wound that does not heal, fibrotic diseases are associated with the chronic differentiation and accumulation of myofibroblasts and excessive deposition of ECM components such as collagen I and lead to a higher risk of organ failure, morbidity, and progression to malignancy (Distler et al, 2019). EVPs have a central role in the development of lung fibrosis. EVPs from macrophages promote the proliferation of pulmonary interstitial fibroblasts via miR‐328 transfer, aggravating fibrosis (Yao et al, 2019). Instead, EVPs from pulmonary fibroblasts suppress the differentiation of neighboring myofibroblasts by delivering anti‐fibrotic prostaglandin (PG)E2 (Lacy et al, 2019). EVPs are also involved in the etiology of liver fibrosis, where hepatic stellate cells (HSCs) proliferate and differentiate into pro‐tumorigenic myofibroblasts. EVPs derived from HSCs, hepatocytes, and inflammatory macrophages in fibrotic livers induce HSC proliferation, migration, and metabolic switch via their protein and miRNA cargo, promoting progression of liver fibrosis (Wang et al, 2015; Seo et al, 2016; Chen et al, 2018b, 2019b, 2020c; Wan et al, 2019; Gao et al, 2020a; Zhang et al, 2020f). In HSCs, the deregulation of autophagy pathways, such as the PDGF/SHP2/mTOR and TRIB3/SQSTM1 pathways, allows for an increased release of EVPs with fibrogenic properties (Gao et al, 2020a; Zhang et al, 2020f). Conversely, NK cell‐derived EVPs decrease TGF‐β1‐dependent HSC activation, proliferation, and autophagy (Wang et al, 2020g, 2020h). Finally, in diabetes or cardiac dysfunction, EVPs derived from macrophages, cardiomyocytes, CD4+ T cells, and endothelial progenitor cells promote a fibrogenic response in cardiac fibroblasts, leading to myocardial fibrosis (Ke et al, 2017; Nie et al, 2018; Cai et al, 2020; Govindappa et al, 2020).
Mutations in oncogenes and tumor suppressors
Oncogenes are drivers of cancer initiation, progression, and metastasis, and numerous studies have started to unravel how they promote cancer progression by regulating biogenesis and secretion of EVPs that contribute to the establishment of tumor‐supportive microenvironments.
Kras is one of the most frequently mutated oncogenes in many cancers, including pancreatic, colon, and lung cancers (Prior et al, 2012). Using isogenic CRC cell lines that differ only in Kras mutation status, Higginbotham and collaborators have pioneered studies aimed at understanding how Kras exerts non‐cell autonomous effects via EVPs and reported that activated mutant Kras controls the molecular composition and functions of EVPs. For example, elevated levels of amphiregulin (AREG), KRAS, EGFR, Src family kinases, and integrins were detected in Kras mutant EVPs (Higginbotham et al, 2011; Demory Beckler et al, 2013; Clark et al, 2016). In vitro functional studies consistently showed that EVPs derived from Kras mutant cells, but not from Kras‐WT cells, can enhance invasion and 3D growth of non‐transformed Kras‐WT cells (Higginbotham et al, 2011; Demory Beckler et al, 2013), implying that mutant Kras can alter the signals mediated via EVPs and confer a growth advantage for surrounding WT cells. Kras mutant cells also package functional GLUT1 in EVPs, which in turn regulates the balance between glycolysis and oxidative phosphorylation in recipient cells and within intestinal adenomas in vivo (Zhang et al, 2018c). Another report indicated that Rab13 is not only specifically recruited to EVPs but also required for the secretion of EVPs from Kras mutant cells, whereas Rab13 depletion has no effect on the EVP production in Kras‐WT cells, indicating that tumor cells with overactivated Kras employ distinct EVP biogenesis mechanisms (Hinger et al, 2020). An important unanswered question is how mutant Kras regulates EVP cargo sorting. RNA profiling analyses showed a Kras‐dependent selective exporting of miRNAs and long RNAs (mRNAs and ncRNAs) (Cha et al, 2015; Hinger et al, 2018), although the molecular mechanism is unknown. Together, these studies suggest that specific Kras mutant‐dependent EVP cargoes may serve as potential biomarkers for cancer detection and as therapeutic targets.
Oncogenic Hras also exerts paracrine activities by altering EVP production and cargo composition. For example, Hras‐transformed MDCK cells release EVPs enriched in proteases, integrins, VEGF‐associated proteins, and the master transcriptional regulator YBX1 (Tauro et al, 2013b). These EVPs induced angiogenesis, indicating that EVP‐mediated communication between tumor cells and endothelial cells commences during early stages in the metastatic cascade (Gopal et al, 2016). Fibroblasts expressing constitutively active Hras‐V12 undergo senescence and release EVPs with distinct lipid signatures enriched in hydroxylated sphingomyelin, lyso‐ and ether‐linked phospholipids, and sulfatides (Buratta et al, 2017). Lee and colleagues showed that in transformed rat intestinal epithelial cells oncogenic Hras stimulates release of EVPs containing chromatin‐associated double‐stranded DNA fragments covering the entire host genome, including full‐length Hras (Lee et al, 2014). EVPs containing oncogenic Hras DNA stimulated endothelial cell proliferation and migration and also increased p53 levels, phosphorylated γH2AX, and micronuclei formation, which are all reminiscent of a genotoxic stress response.
EGFR, which has a pivotal role in the pathogenesis of many human cancers, is also incorporated into EVPs (Al‐Nedawi et al, 2008, 2009; Skog et al, 2008) and is involved in regulating EVP biogenesis and EVP‐mediated signaling pathways. Constitutively active EGFR (EGFRvIII) is frequently detected in glioblastoma multiforme and reported to alter the expression of EVP‐regulating genes and EVP properties, including their protein composition (Choi et al, 2018). For instance, pro‐invasive proteins (CD44, basigin, and CD151) were shown to be associated with EVPs of EGFRvIII‐expressing glioma cells, whereas EVP markers (CD81 and CD82) were downregulated in EVPs of EGFRvIII‐negative cells. Increased EVP uptake by EGFRvIII‐positive glioma cells was also observed. EGFR and p53 mutations are common genetic alterations in NSCLC. Transformation of normal human bronchial epithelial cells by p53 knockdown and overexpression of EGFR L858R promotes secretion of EVPs enriched in proteins involved in E2F and Myc pathways, which may induce proliferative and migratory phenotypes in recipient cells (Lobb et al, 2017). In head and neck cancer cells, EGFR overexpression coupled with E‐cadherin blockade led to loss of EGFR and tissue factor (TF) from the plasma membrane, coinciding with a surge in emission of EVPs containing both receptors. These EVPs transferred TF to cultured endothelial cells, rendering them highly pro‐coagulant (Garnier et al, 2012). Thus, EVPs might have a role in connecting aberrant EGFR signaling in cancer cells with dysregulated coagulation, a key process in malignant cancer progression.
Specific p53 mutations have oncogenic functions and promote tumor progression and metastasis (Olive et al, 2004; Hingorani et al, 2005; Morton et al, 2010; Freed‐Pastor & Prives, 2012; Cooks et al, 2013; Zhu et al, 2015). Recent studies demonstrated that cells expressing such oncogenic p53 mutants (Mutp53) can utilize EVPs to reprogram recipient tumor cells, fibroblasts, and tumor‐associated macrophages. For instance, NSCLC expressing oncogenic p53R273H and p53R175H mutants produce EVPs that promote invasion and migration of other tumor cells (Novo et al, 2018). This process requires the ability of Mutp53 to control the levels of EVP podocalyxin, a sialomucin linked to cancer aggressiveness, and to increase Rab‐coupling protein (RCP)‐dependent integrin trafficking in target cells. EVPs from Mutp53‐expressing tumor cells promote integrin recycling to the plasma membrane of fibroblasts and influence their ECM deposition and remodeling, generating a supportive microenvironment for tumor initiation and cell invasion. In agreement, Ju and colleagues demonstrated that Mutp53s can activate stromal fibroblasts in colon cancer, a process dependent on transfer of specific EVP‐associated miRNAs (Ju et al, 2019). In addition, colon cancer cells expressing Mutp53 promote the differentiation of a distinctive, tumor‐supportive macrophage subpopulation via EVPs carrying miR‐1246. Co‐injection of tumor cells with these reprogrammed macrophages resulted in larger primary tumors and increased liver and lung metastatic burden (Cooks et al, 2018). Furthermore, p53 null and especially DNA contact Mutp53 (p53R273H) enhance Hsp90α secretion by cancer cells via RCP adaptor (Zhang et al, 2020c). Notably, administration of Hsp90α monoclonal antibody attenuated lung and liver metastases in mice carrying p53R270H (equivalent to R273H in humans) or p53‐null tumors. Taken together, these studies provide evidence that oncogenic p53 mutants influence the tumor microenvironment via an EVP‐mediated, paracrine fashion to promote malignancy, and that different Mutp53s utilize distinct mechanisms to regulate EVP‐transmitted oncogenic functions.
Besides oncogenes, Apc mutation increases EVP secretion via activation of Wnt pathway when introduced into WT small intestinal organoids (Szvicsek et al, 2019). Since Apc mutation is an early event in intestinal and colorectal tumorigenesis, this evidence implicates tumor‐derived EVPs at an early stage of tumor development. A newly discovered role of the tumor suppressor Lkb1 in EVP biogenesis and release was also described (Zhang et al, 2018a). Restoration of Lkb1 expression in lung cancer cells enhanced EVP secretion, and EVPs from Lkb1‐expressing cells promoted recipient cell migration by downregulating expression of migration‐suppressing miRNAs and EVP secretion. Lastly, in rhabdomyosarcoma, PAX3‐FOXO1 fusion drives the alteration of myoblast EVP content, particularly miR‐486‐5p, which mediates fibroblast migration and invasion (Ghamloush et al, 2019).
In conclusion, oncogenes take part in the biogenesis and secretion of EVPs, instruct selective packaging of EVP cargo molecules, act as active cargo themselves, and influence the uptake of EVPs and multiple signaling pathways in recipient cells. Crosstalk mediated by tumor‐derived EVPs, occurring at local or distant sites, contributes to key aspects of tumorigenesis and metastasis, such as immunosuppression, ECM organization, angiogenesis, and vasculature remodeling. Future studies incorporating multi‐omics characterization of EVP composition and genetic manipulation of key DNA components will be critical for further understanding the contribution of oncogenes to EVP biogenesis and cargo packaging. This knowledge will guide the development of novel therapeutic strategies that target EVPs for cancer intervention.
Metabolic reprogramming
Metabolic rewiring is one of the first steps of transformation that supports the higher nutrient demands of cancer cells (Fendt et al, 2020). It has been recognized that metabolic reprogramming during cancer initiation is both the cause and consequence of EVP excretion.
The metabolome of EVPs is relatively understudied compared with proteome and transcriptome, but recent studies identified an array of metabolites in EVPs, such as amino acids, organic acids, sugars and their conjugates, nucleotides and nucleosides, cyclic alcohols, carnitines, aromatic compounds, and vitamins (Altadill et al, 2016; Zhao et al, 2016a; Puhka et al, 2017; Clos‐Garcia et al, 2018; Luo et al, 2018; Zebrowska et al, 2019). Additionally, a diverse set of metabolic enzymes has been documented in EVPs derived from various sources. For example, glucose deprivation in cardiomyocytes increases the synthesis and release of EVPs loaded with functional glucose transporters and glycolytic enzymes, which in turn potentiate glucose uptake of recipient endothelial cells (Garcia et al, 2016). Similarly, human prostate‐derived EVPs carry functional glycolytic enzymes that produce ATP when supplied with substrates (Ronquist et al, 2013). Furthermore, neural stem/progenitor cell‐derived EVPs harbor the catalytically active asparaginase‐like protein 1 (Asrgl1) enzyme capable of increasing glutamate, GABA, and aspartate while decreasing asparagine in cell culture media (Iraci et al, 2017). Similarly, arginase‐1 activity is associated with hepatocyte‐derived EVPs and induces a significant change in arginine metabolites in serum (Royo et al, 2017), suggesting that EVPs are capable of modifying their metabolic environment before being internalized by target cells.
EVPs can also mediate the exchange of regulators of metabolic signaling pathways under a broad range of pathophysiological conditions, such as altered glucose metabolism and inflammation, which are predisposing factors for cancer initiation. Pancreatic β cells are a major source of EVPs with metabolic reprogramming potential, particularly in patients with type 2 diabetes (Li et al, 2020a). Increased glucose levels stimulate pancreatic β cell release of EVPs enriched in lncRNA‐p3134, which promotes insulin secretion and suppresses cell death from glucotoxicity (Ruan et al, 2018). Similarly, miR‐29 packaged in β cell EVPs stimulates chemotaxis and activation of pro‐inflammatory macrophages via induction of IL‐12, IL‐6, and IL‐1β. Consequently, systemic inflammation promotes insulin insensitivity, leading to predisposition to type 2 diabetes (Sun et al, 2021c). In addition, EVPs from serum of diabetic patients are enriched in miR‐20‐5b and are taken up by skeletal muscle cells, which in turn increase their glycogen synthesis via AKTIP/STAT3 regulation (Katayama et al, 2019).
A recent study by Goulielmaki and colleagues revealed a novel EVP‐based link between DNA damage, metabolic disorders, and inflammation (Goulielmaki et al, 2020). Using an engineered mouse model carrying an ERCC1‐XPF DNA repair defect (Er1F/‐ ), the authors showed that persistent DNA damage accumulation in Er1F/‐ tissue‐infiltrating macrophages triggers cytoplasmic stress and increases EVP biogenesis. These EVPs, which were also detected in Er1F/‐ animal sera, promoted glucose uptake in recipient pancreatic cells and hepatocytes by upregulating glucose transporters, such as GLUT1, and enhanced glucose tolerance in WT mice. Future studies are necessary to identify the specific EVP cargoes that mediate such metabolic reprogramming in recipient cells.
EVPs have also been implicated in the etiology of obesity‐induced insulin resistance, a known risk factor for cancer development (Kahn et al, 2006; Romeo et al, 2012; Johnson & Olefsky, 2013; Barazzoni et al, 2018). EVPs from adipocytes, especially those from obese mice, are enriched in enzymes and substrates of fatty acid oxidation and increase the motility of tumor cells (Lazar et al, 2016; Clement et al, 2020). In obesity and under lipolytic stimuli, adipocytes release EVPs enriched in aP2 (also called fatty acid binding protein 4), which can affect glucose and lipid metabolism in target cells and is involved in diabetes, fatty liver disease, and cancer (Ertunc et al, 2015). Furthermore, chronic inflammation and accumulation of proinflammatory macrophages, particularly in the adipose tissue and liver, are hallmarks of obesity and have been previously linked to obesity‐induced insulin resistance and cancer (Romeo et al, 2012; Li et al, 2015a, 2016; Lackey & Olefsky, 2016; Kita et al, 2019). Recent work by Ying and colleagues uncovered a new function for adipose tissue macrophages (ATMs), whereby they systemically modulate insulin action via EVPs (Ying et al, 2017). The authors reported that treating lean mice with EVPs derived from obese mice ATMs led to glucose intolerance and insulin resistance, whereas treating obese mice with EVPs derived from lean mice ATMs led to improved glucose tolerance and insulin sensitivity. MiR‐155 contained in ATM EVPs from obese mice emerged as a key factor regulating these processes in the liver, adipose tissue, and muscle, likely via downregulating GLUT4. In a similar fashion, EVPs derived from HSCs in fibrotic livers reprogram glucose metabolism in neighboring HSCs, Kupffer cells, and sinusoidal endothelial cells and induce a shift from oxidative phosphorylation to aerobic glycolysis, which has been associated with tumor development (Wan et al, 2019). Lastly, EVPs derived from adipocytes undergoing endoplasmic reticulum stress cause glucose and lipid metabolic changes in hepatocytes, leading to nonalcoholic hepatic steatosis, fibrosis, and inflammation (Gu et al, 2021).
Collectively, these studies demonstrate that EVPs are key players in metabolic reprogramming at local and systemic levels in different physiological and pathological conditions and may thus provide therapeutic targets to maintain metabolic homeostasis and prevent cancer initiation. In addition, metabolome studies on EVPs derived from different body fluids will identify biomarker candidates for disease detection and monitoring.
Parasites, viruses, and microbiota
EVPs have emerged as essential routes of bilateral communication between hosts and organisms (i.e., parasites, viruses, and bacteria) that govern cancer pathogenesis. Common parasites, such as Plasmodium species, Leishmania species, and Toxoplasmas gondii, produce EVs enriched in parasite antigens and nucleic acids that stimulate the host immune response and parasite survival and induce EVP release by host stromal cells (Wu et al, 2018; Liang et al, 2019). For example, macrophages stimulated with Plasmodium berghei, Leishmania, or T. gondii EVs overexpress CD40 ligand and release IL‐8, IL‐12, IFN‐γ, and TNF‐α (Couper et al, 2010; Silverman et al, 2010; Dlugonska & Gatkowska, 2016), inducing a protective immunity for infections and, potentially, cancer initiation. Similarly, EVs from Schistosoma mansoni are internalized by endothelial cells and induce a phenotype consistent with endothelial activation, thrombosis, and immune cell recruitment (Kifle et al, 2020).
EVPs have a central role in viral infections. Epstein‐Barr virus (EBV)‐infected B cells release EVPs containing the small viral RNA EBER1 and activate an anti‐viral immune response in recipient plasmacytoid DCs (Baglio et al, 2016). Interestingly, cells may also employ EVPs as a means to excrete viral DNA from cells (Takahashi et al, 2017). Human papilloma virus (HPV)‐infected cancer cells (e.g., HeLa cells) transfer long noncoding RNAs (lncRNAs) to uninfected cervical cells and affect their metabolism and viability (Hewson et al, 2016). EVPs from HVC‐infected hepatocytes activate TGF‐ β1 expression in HSCs via miR‐19a/miR‐192 shuttling, inducing their activation and expression of fibrogenic markers (Devhare et al, 2017; Kim et al, 2019b). HIV‐infected CD4+ T cells release EVPs enriched in pro‐hypoxic and pro‐inflammatory mediators (Duette et al, 2018). Furthermore, lymphatic endothelial cells infected with Kaposi's sarcoma‐associated herpesvirus (KSHV) release viral miRNA‐enriched EVPs that mediate metabolic reprogramming of non‐infected vascular and lymphatic cells, thereby increasing aerobic glycolysis, propensity to KSHV infection, angiogenesis, and migration, potentially promoting sarcoma development (Yogev et al, 2017). Finally, latent membrane protein 1 (LMP1) encoded by EBV‐infected cells is packaged into EVPs and induces activation of normal fibroblasts to cancer‐associated fibroblasts (CAFs) via regulation of the NF‐kB pathway and glucose metabolism (Wu et al, 2020d).
Bacteria, including gut microbiota, are an important source of EVs. As bacteria are deficient in canonical EV secretion systems, these vesicles originate from outer membrane budding and are thus indicated as outer membrane vesicles (OMVs) (Shen et al, 2012). Other mechanisms of EV release have been characterized but are less studied (Chronopoulos & Kalluri, 2020). Despite differences in secretion pathways of eukaryotes and prokaryotes, OMVs range from 20 to 100 nm in diameter, are similar to eukaryotic EVs and exomeres, and have the ability to communicate with the host immune system. Beneficial bacteria strains, such as Bacteroides fragilis, release OMVs enriched in surface bacterial capsular polysaccharides, which orchestrate an immune‐suppressive response involving Tregs, T cells, and DCs. Importantly, adoptive transfer of OMV‐stimulated DCs protects mice from DSS‐induced colitis (Shen et al, 2012). Similarly, OMVs from Bacteroides acidifaciens and Akkermansia muciniphila strains provide a protective effect against colitis‐associated weight loss, inflammatory cell infiltration, and cytokine release by colon epithelial cells, ultimately ameliorating the severity of IBD (Kang et al, 2013; Patten et al, 2017; Ashrafian et al, 2019), as well as reducing gut permeability in type 2 diabetes (Chelakkot et al, 2018). Conversely, strains of probiotic and commensal Escherichia coli produce LPS‐expressing OMVs that induce secretion of pro‐inflammatory and immunomodulatory cytokines, such as IL‐10, IL‐8, and TNF‐α, by peripheral blood mononuclear cells and intestinal epithelial cells (Ellis & Kuehn, 2010; Fabrega et al, 2016; Patten et al, 2017; Canas et al, 2018). As a result, OMV‐associated LPS was detected at significantly higher levels in the plasma of patients with IBD and chemotherapy‐induced intestinal mucositis in comparison to healthy controls and is a potential biomarker of intestinal barrier dysfunction (Tulkens et al, 2020). The composition of gut microbiome and corresponding OMVs dramatically changes upon colitis or IBD development in mice and humans, with a striking reduction in numbers of OMVs from less immune‐activating strains of Bacteroides acidifaciens and Akkermansia muciniphila strains (Kang et al, 2013) and a change of OMV content toward inducing oxidative stress (Zhang et al, 2018e). Hence, inflammatory gut syndromes are associated with the release of bacterial OMVs that might exacerbate advancement of malignant disease. More indirectly, myeloid DCs exposed to Helicobacter pylori release EVs that express bacterial components and elicit systemic immune reactions, such as CD4+ T cell activation, explaining skin eruptions in H. pylori‐infected patients (Ito et al, 2018).
Host and dietary EVPs can affect the gut microbiota. Common gut bacteria strains (e.g., Fusobacterium nucleatum and Escherichia coli) internalize EVPs from different host cells, including adipose and gut epithelial cells. These EVPs increase the proliferation rate and gene expression profile of gut bacteria, which may be involved in initiating the development of pre‐cancerous conditions, such as colitis (Liu et al, 2016b; Yu et al, 2019b). EVPs from dietary sources elicit different microbiota responses. To illustrate, EVPs from edible plants, such as ginger root, are preferentially taken up by Lactobacillaceae and promote their production of IL‐22, thereby providing protection against gut permeability and colitis (Teng et al, 2018). On the other hand, milk‐derived EVPs are preferentially internalized in Escherichia coli and Lactoplantibacillus plantarum and are enriched in miRNAs that influence bacterial expression of genes involved in adhesion and invasion (Yu et al, 2019b). Viable Fusobacterium nucleatum has been found in primary and distant metastatic sites of CRC patients and mouse xenografts, where it supports tumor growth and progression (Bullman et al, 2017). Hence, host–microbiota–diet interplay via EVPs may facilitate bacterial colonization of distant organs, possibly promoting cancer initiation and progression.
Build it up: cancer promotion
Cancer stem cells
A small fraction of pluripotent and mostly quiescent tumor cells named cancer stem cells (CSCs) have been shown to be responsible for tumor initiation. The resistance of CSCs to conventional chemotherapy and their ability to propagate largely account for the high rates of therapeutic failure and recurrence in primary and metastatic tumors (Batlle & Clevers, 2017). Cancer‐associated EVPs play a central role in maintaining CSC pluripotency, similarly to how embryonic stem cells maintain their pluripotency via EVP‐mediated intercellular communication (Hur et al, 2020) (Figure 3). Chen and colleagues were the first to demonstrate that culture media from several mouse tumor cell lines promoted the differentiation of mouse induced pluripotent cells into cancer initiating cells in vivo (Chen et al, 2012). Further, it was determined that cancer‐derived EVPs activate expression of core stemness drivers Nanog and Oct3/4 in mouse induced pluripotent cells, conferring on them properties of self‐renewal and plasticity (Yan et al, 2014b; Calle et al, 2016). Interestingly, EVPs from breast cancer and serous carcinoma contain the mRNA and protein of Nanog and other stemness drivers, and their levels correlate with poor overall survival (Rodriguez et al, 2015; Sherman‐Samis et al, 2019). These mRNAs, however, may not be responsible for promotion of pluripotency. Instead, fibronectin exposed on the surface of cancer‐associated EVPs may be responsible for CSC maintenance (Hur et al, 2020). CAFs are a major source of EVPs promoting CSC maintenance and chemoresistance by inducing the de‐differentiation of cancer cells and the activation of stemness expression pathways, such as Wnt/β‐catenin pathway, via EVP‐associated mRNAs, miRNA, and lncRNAs (Hu et al, 2015, 2019b; Ren et al, 2018; Rodrigues et al, 2018; Wang et al, 2019c; Liu et al, 2020a).
Figure 3. EVPs promote multiple aspects of cancer growth.
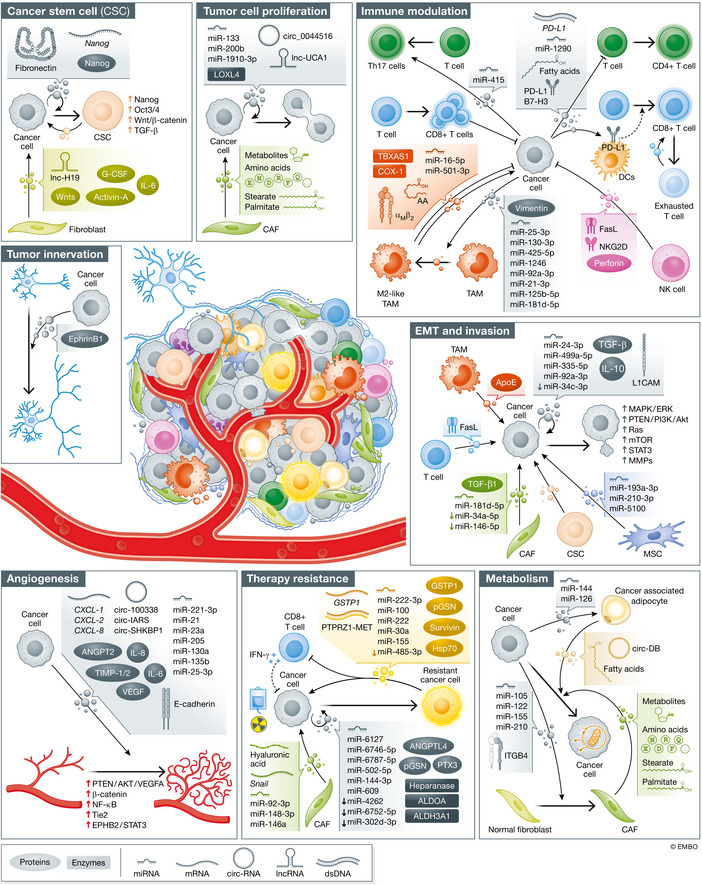
The growth of primary tumors is positively impacted by EVPs released by tumor cells and other cells in the tumor microenvironment, including stroma cells (fibroblasts, MSCs, and adipocytes) and immune cells (TAMs, DCs, T lymphocytes, NK cells, and neutrophils). These EVPs influence tumor cells directly by promoting tumor formation and progression via different means, including maintaining CSC pluripotency, promoting tumor cell proliferation, inducing tumor cell EMT and invasion, and altering tumor metabolic demands. Additionally, cancer cell‐derived EVPs generate a favorable microenvironment permissive for local tumor expansion by promoting angiogenesis and vascular remodeling, and modulating immune functions towards a pro‐tumorigenic and immunosuppressive phenotype. Finally, tumor‐ and stroma‐derived EVPs have a central role in inducing cancer resistance to chemotherapy and radiation therapy. Several EVP cargoes have been found responsible for these pro‐tumorigenic roles, including nucleic acids (mRNAs, miRNAs, lncRNAs, circRNAs, DNAs), proteins, enzymes, surface receptors and lipids. TAM, tumor‐associated macrophage; DC, dendritic cell; NK, Natural Killer; MSC, mesenchymal stem cell; CSC, cancer stem cell.
In turn, CSC‐derived EVPs support tumor progression through multiple pathways. For instance, pancreatic cancer CSCs induce a distinct transcriptomic change, including activation of EGF/VEGF and EMT pathways, in non‐CSCs cancer cells, rendering them apoptosis‐resistant, invasive, proliferative, and metastatic. Importantly, this reprogramming depends on the activation of EVP‐induced cellular signaling rather than on direct transfer of mRNAs/miRNAs (Wang et al, 2019d). Angiogenesis is also supported by CSCs EVPs, with lncRNA H19 and miR‐26a being central players (Conigliaro et al, 2015; Wang et al, 2019e). Finally, CSC EVPs promote the differentiation of normal fibroblasts into CAFs (Zhang et al, 2020a).
Thus, a growing body of evidence suggests that cancer‐associated EVPs, both cancer cell‐ and stroma‐derived, drive the dynamic balance between induction, maintenance, and differentiation of CSCs, which in turn promote tumor progression via EVP‐mediated communication.
Tumor growth
The importance of EVPs in sustaining tumor growth and tumor cell proliferation is demonstrated by the observation that treatment with GW4869, an inhibitor of ceramide‐mediated biogenesis, or knockout of Rab27a slows tumor growth in vivo (Bobrie et al, 2012; Matsumoto et al, 2017; Matsumoto et al, 2017; Richards et al, 2017). In contrast, exposure of cancer cell lines, such as pancreatic cancer, lung adenocarcinoma, and breast cancer cells, to endogenous EVPs or EVPs from more invasive cell lines promotes growth and cell cycle progression and inhibits apoptosis of cancer cells (Qu et al, 2009; Harada et al, 2017; Xie et al, 2020a; Shen et al, 2021). Pathway analysis has shown enrichment of proliferative pathways in cancer‐derived EVPs (Shi et al, 2020a). Several EVP cargos, including miRNAs, circRNA, and enzymes, such as lysyl oxidase‐like 4, have been found responsible for this proliferation‐promoting effect (Chen et al, 2014; Zhang et al, 2018f; Li et al, 2019d; Luan et al, 2020; Xie et al, 2020a; Wang et al, 2020d). Moreover, numerous growth factors are selectively packaged in cancer cell line‐ and patient‐derived EVPs (Hoshino et al, 2020) (Figure 3).
Host‐derived EVPs also play a prominent role in tumor growth, and CAF‐derived EVPs in particular enhance proliferation of cancer cells (Zhao et al, 2016a, 2020a; Zhou et al, 2021b). Notably, CAFs obtained from breast cancer biopsies, but not fibroblasts from adjacent tissue, release EVPs that induce breast cancer cell proliferation via transfer of LINC00355 and miR‐500a‐5p (Yan et al, 2020; Chen et al, 2021a). Metabolic reprogramming of cancer cells might be another major feature of CAF‐derived EVPs, as highlighted by the observation that CAF EVPs promote growth of pancreatic cancer cells by reducing mitochondrial respiration and enhancing glucose and glutamine metabolism via their cargo of amino acids, lipids, and other metabolic intermediates (Zhao et al, 2016a). Additionally, CAF EVPs also have a major role in driving chemoresistance, as reviewed below.
Recent evidence shows that innervation of the tumor mass, a process defined as axonogenesis, promotes growth and metastasis of different cancer types, such as head and neck and prostate cancer. Madeo and colleagues recently discovered that blood EVPs from patients with head and neck cancer and from several oropharyngeal squamous cancer cell lines induce neurite outgrowth in vitro and in vivo, an effect that depends on exosomal ephrin‐B and that is abrogated by Rab27a/b knockout (Madeo et al, 2018).
EMT, migration, and invasion
The phenotypic plasticity of cells is a physiological aspect of embryo development that has been adopted by tumor cells for enhanced motility and immune evasion that facilitates local and distant invasion (Brabletz et al, 2021). Higher motility and ability to undergo EMT can be transferred in an autocrine and paracrine manner via EVP cargo (Figure 3). For example, EVPs from highly metastatic cell lines can transfer EMT properties to lowly metastatic cell lines by regulating MAPK/ERK, PTEN/Akt/Snail, Ras and mTOR signaling, with miRNA shuttling playing a prominent role (Wang et al, 2016b; Chen et al, 2018c, 2020d; He et al, 2019b; Yang et al, 2020a; Sun et al, 2021a). Similarly, CAF‐derived EVPs induce EMT, migration, and ultimately metastasis of various cancer types, including castration‐resistant prostate cancer, ovarian cancer, and breast cancer (Li et al, 2017e; Novo et al, 2018; Wang et al, 2020a). Upregulation of E‐cadherin, vimentin, N‐cadherin, ZEB1, Snail, Slug, Twist1, and MMPs, and activation of PTEN/PI3K/AKT/β‐catenin and EGFR/ERK pathways in cells taking up CAF EVPs are all major drivers of tumor cell EMT (Li et al, 2017e; Novo et al, 2018; Wang et al, 2020a; Yang et al, 2020b; Zhang et al, 2020h). Further, intratumoral fibroblasts interacting with interferon‐stimulated gene responsive (ISG‐R) breast cancer cells undergo NOTCH1‐MYC activation and produce EVPs that, similar to viruses, are enriched in 5′‐triphosphate RNAs and induce an anti‐viral response in breast cancer cells, promoting pulmonary metastasis (Nabet et al, 2017). This signaling requires activation of the RIG‐I receptor in cancer cells by unshielded RN7SL1 RNA in stromal EVPs (Wang et al, 2010; Nabet et al, 2017). In hypoxic tumors, bone‐marrow‐derived mesenchymal stem cells (MSCs) promote EMT and invasion of lung cancer cells via transfer of different miRNAs and activation of STAT3 signaling (Zhang et al, 2019g). CSC‐derived EVPs can also promote EMT of neighboring differentiated cancer cells in clear cell renal cell carcinoma (CCRCC) patients by inducing activation of PTEN‐dependent EMT gene expression and, consequently, promoting pulmonary metastasis of CCRCC cells. EVPs from CSCs isolated from metastatic CCRCC patients were particularly potent drivers of tumor growth and lung metastasis (Wang et al, 2019a), suggesting the existence of functional changes in CSCs EVP cargoes in advanced disease.
EVP‐induced EMT program may also result in increased cell migration, although the two processes can be independent of one another (He et al, 2019c; Schelch et al, 2021). Direct autocrine and paracrine shuttling of EVP cargo, including tetraspanins, promotes in vitro migration of cancer cells (Pace et al, 2019; Matsumoto et al, 2020; Huang et al, 2020c). EVPs mediate in vivo communication between highly invasive MDA‐MB‐231 and less invasive T47D breast cancer cell lines to facilitate cancer cell motility, invasiveness, and metastatic potential (Zomer et al, 2015). Further, Luga and colleagues have shown that EVPs from fibroblast‐like L cells activate Wnt11‐dependent planar cell polarity signaling in cancer cells and promote the formation of protrusive invadopodia (Luga et al, 2012). Among other EVP factors, TGF‐β is a central regulator of EMT and cell migration and has been detected in EVPs from cancer cells and CAFs, but not in EVPs from other cancer‐associated cells, underscoring the importance of cancer cell and fibroblast communication in EMT and migration (Webber et al, 2010; Wang et al, 2016b; Li et al, 2017e; Ringuette Goulet et al, 2018; Batlle & Massague, 2019; Ferguson Bennit et al, 2021). In response, cells stimulated by TGF‐β release a second wave of EVPs that induce MMP‐2 expression in neighboring cancer cells, further expanding the migratory potential (Wu et al, 2018). Other evidence suggests that EVPs from tumor‐associated macrophages (TAMs) contribute to tumor cell invasion via shuttling ApoE and activation of the PI3K/Akt pathway in tumor cells, partially explaining the association between TAM density and poor prognosis (Zheng et al, 2018b; Lan et al, 2019). Interestingly, Fas ligand (FasL)+ EVPs from activated CD8+ T cells promote MMP‐9 expression and motility of tumor cells in vitro and lung invasion in vivo, while surprisingly lacking pro‐apoptotic ability via Fas‐FasL engagement (Cai et al, 2012). This suggests that in “hot tumors,” cancer cells exploit the EVP cargo of cytotoxic T cells to their advantage, hijacking the tumor‐suppressive role of T cell activation.
Angiogenesis
Cancer‐derived EVPs induce significant changes in the endothelial cell compartment associated with angiogenic switch (Figure 3). As vascular beds are the gateway for dissemination, the pro‐angiogenic properties of EVPs correlate with metastatic potential and poor prognosis (Zhou et al, 2014; Maji et al, 2017; Tang et al, 2018a).
Several EVP cargoes mediate a pro‐angiogenic effect. EVPs derived from glioblastoma cells support tube formation of brain endothelial cells via transfer of pro‐angiogenic IL‐6, IL‐8, VEGF, and TIMP‐1/2 selectively packaged in these EVPs (Skog et al, 2008). Likewise, pro‐angiogenic angiopoietin 2 (ANGPT2) was found expressed in HCC‐derived EVPs and was internalized and recycled by endothelial cells (Xie et al, 2020a). mRNAs of pro‐angiogenic cytokines, such as CXCL1, 2 and 8, are enriched in EVPs from melanoma cell lines but not normal melanocyte cultures (Bardi et al, 2019). Similarly, the cell adhesion molecule E‐cadherin is released by ovarian cancer cells into conditioned medium and patient‐derived ascites via EVPs and induces vascularization in vitro and in vivo (Tang et al, 2018a). EVPs from HCC cell lines promote growth, migration, and differentiation of HSCs into functional α‐SMA+ CAFs, via transfer of miR‐21 and activation PTEN/PDK1/AKT pathway, which in turn supports release of pro‐angiogenic factors and HCC tumor growth in vivo (Zhou et al, 2018). Interestingly, inflamed perivascular adipose tissue in obese mice releases EVPs enriched in miR‐221‐3p, which promote vascular remodeling by inducing proliferation and migration of vascular smooth cells (Mao et al, 2019b). The same miRNA was found to target thrombospondin‐2 (THBS2) in endothelial cells and increase endothelial cell migration, tube formation, and sprouting in cervical squamous cell carcinoma (Wu et al, 2019c). Several other EVP‐associated miRNAs and circRNAs increase angiogenesis and endothelial barrier permeability (Bao et al, 2018; Zeng et al, 2018; Yang et al, 2018a; Li et al, 2018b; He et al, 2019a; Du et al, 2020b; Huang et al, 2020d). These results are substantiated by preclinical models in which education with miRNA‐enriched cancer‐derived EVPs increased tumor microvessel density and intratumoral VEGF levels, promoting tumor growth (Zeng et al, 2018; Xie et al, 2020b). The release of EVPs with pro‐angiogenic properties is significantly affected by microenvironmental factors, such as nutrient availability and oxygen levels. EVPs from cancer cells under hypoxia or aerobic glycolysis have a more potent effect on tube formation and tumor vascularization in vivo than normoxic EVPs, due to the enrichment of pro‐angiogenic mRNA, miRNA, and VEGF cargo (Umezu et al, 2014; Mao et al, 2019b; Zhang et al, 2020d). Among others, the activation of PTEN/AKT/VEGFA, beta‐catenin, NF‐kB, Tie2, and EPHB2/STAT3 signaling pathways was observed in endothelial cells exposed to cancer‐derived EVPs (Tang et al, 2018a; Sato et al, 2019; He et al, 2019a; Xie et al, 2020a; Song et al, 2021).
It is interesting to note that also HUVEC‐derived EVPs reduce expression of tight junction proteins in neighboring endothelial cells via endoplasmic reticulum stress response and increase vascular permeability and metastasis in vivo (Lin et al, 2020), but the signaling controlling the release and cargo of EVPs from tumor‐associated endothelial cells needs to be further investigated.
Immune modulation
Immune cells are primary targets of cancer cell‐derived EVPs in mouse models, especially in the lungs and liver, with neutrophils and myeloid cells being the most avid takers (Hoshino et al, 2015; Ridder et al, 2015; Zomer et al, 2015; Wen et al, 2016). On the one hand, intratumoral release of EVPs correlates with immune cell recruitment. Infiltration of neutrophils in orthotopic 4T1 murine breast carcinoma tumors was dramatically reduced by Rab27a knockdown in tumor cells (Bobrie et al, 2012). MMP3/9 and chemoattractant G‐CSF were found enriched in these EVs, supporting neutrophil recruitment and tumor growth, but this effect might also be due to neutrophil differentiation from bone marrow precursor cells via other factors contained in EVs (Bobrie et al, 2012).
On the other hand, cancer‐derived EVPs induce modulation of immune functions, predominantly with pro‐tumorigenic consequences. While M1 and M2 macrophage phenotypes have been characterized in vitro and in mice, macrophage polarization is less dichotomous in humans. In general, tumor‐associated macrophages (TAMs) can be found in a classic pro‐inflammatory Th1/M1‐like and tumor suppressive phenotype, or an alternative anti‐inflammatory M2‐like phenotype that has been associated with increased cancer invasiveness, motility, and metastasis (Noy & Pollard, 2014; Laviron & Boissonnas, 2019). Several lines of evidence indicate that cancer‐cell‐derived EVPs induce polarization of TAMs toward an M2‐like tumor‐promoting phenotype. CRC cell lines exposed to CXCL12, a cytokine found in the CRC microenvironment, produce EVPs enriched in different miRNAs that induce the activation of PTEN/PI3K/Akt signaling pathway in TAMs, shifting them toward an M2 phenotype. Polarized macrophages then support tumor cell EMT, endothelial cell tube formation, and progression to liver metastatic CRC in vivo (Wang et al, 2020c). CRC tumor cell–derived EVPs reprogram macrophages to release MCP‐1 and TNF and to undergo cytoskeleton rearrangement and protrusion formation (Chen et al, 2016b). In the brain, glioma cells shed EVPs that induce activation of astrocytes, ultimately promoting glioma growth (Gao et al, 2020b). EVPs enriched in miR‐1246 from p53‐mutant CRC cells stimulate the enrichment of polarized TAMs in tumors that correlate with poor prognosis (Cooks et al, 2018). Other EVP miRNA cargos were found responsible for eliciting macrophage pro‐tumorigenic activation and causing intratumoral infiltration (Casadei et al, 2017; Hsieh et al, 2018; Chen et al, 2018d; Kwon et al, 2020; Zhao et al, 2020c). Similarly, T cells respond to miR‐415‐enriched EVPs from gastric cancer cells by undergoing mTOR activation and differentiation into Th17 cells, promoting tumor infiltration (Liu et al, 2018a). Circular RNA circPACRGL, expressed in CRC‐derived EVPs, serves as a sponge for miR‐142‐3p and miR‐506‐3p in cancer cells, resulting in upregulation and release of TGF‐β1, and induction of phenotypic switch from N1 to N2 neutrophils (Shang et al, 2020). N2 neutrophils have been found to promote tumor growth and progression elsewhere (Fridlender et al, 2009).
Cancer‐derived EVPs are also a significant source of secreted PD‐L1 in the tumor microenvironment, making them major regulators of immune checkpoints. NSCLC, glioblastoma, prostate, and CRC cell lines all release PD‐L1+ EVPs that block T cell activation and expansion in vitro and in lymph nodes in vivo (Ricklefs et al, 2018; Poggio et al, 2019; Kim et al, 2019a). The subsequent reduction of intratumoral CD8/CD4 ration and T cell exhaustion promote tumor growth. This evidence is further supported by the observation that Rab27a‐ or nSMase‐KO cell lines grow slower than their WT counterparts in immunocompetent mice, but to a similar extent in T cell deficient mice (Poggio et al, 2019). PD‐L1 exposure on EVPs also prevents the activation of an immune memory response against tumor cells (Poggio et al, 2019). EVPs might also transfer expression of PD‐L1 to other cells (Ricklefs et al, 2018; Yin et al, 2020; Liang et al, 2020b). For example, cancer cell‐derived EVPs induce PD‐L1 expression in DCs and decreases their antigen presenting and CD8+ T cell priming activity, supporting the generation of a tumor‐permissive microenvironment and resistance to immunotherapy. DC cell dysfunction is induced by lipid accumulation and fatty acid oxidation in DCs as a result of EVP fatty acid transfer (Yin et al, 2020). Of note, PD‐L1 expression in plasma EVPs associates with disease progression in different types of cancer (Theodoraki et al, 2018; Li et al, 2019a). Other inhibitory immune checkpoints, such as B7‐H3, were also found expressed on cancer‐derived EVPs, although less well studied compared with PD‐L1 (Purvis et al, 2020).
Conversely, EVPs derived from immune cells themselves have both tumor‐supportive and tumor‐suppressive properties, depending on the cell source. NK cell‐derived EVPs directly caused tumor cell cytotoxicity via shuttling of FasL, perforin, and NKG2D, and reduced tumor growth and metastasis in murine models of glioblastoma and melanoma (Lugini et al, 2012; Shoae‐Hassani et al, 2017; Zhu et al, 2017, 2018b), suggesting that NK EVP‐driven cytotoxicity might be a source of tumor control. The microenvironmental cues and factors leading to release of NK cell EVPs remain unclear. Most TAM‐derived EVPs do not share the immunosuppressive phenotype of the parental cells and, instead, are endowed with an immunomodulatory phenotype that induce T cell activation and expansion, ultimately promoting tumor cell cytotoxicity. In addition, lipids and several proteins involved in lipid metabolism were represented in TAM‐derived EVPs and induced the production of thromboxane (TXA2), but not other pro‐inflammatory eicosanoids such as PGE2, by tumor cells (Cianciaruso et al, 2019). PDAC‐associated TAMs release EVPs enriched in miR‐501‐3p that instead induce migration and apoptosis resistance of pancreatic cancer cells via TGF‐β pathway, angiogenesis, as well as primary tumor growth and metastasis in vivo (Yin et al, 2019). Furthermore, EVPs from HCC‐associated CD206+ M2‐like TAMs are induced a migratory phenotype in HCC cells, partially explaining the association between TAM infiltration and risk of metastasis in HCC patient. This effect relies on different mechanisms, including the transfer of exosomal αMβ2 to HCC cells leading to increased endothelial cell adhesion, as well as increased MMP‐9 activity in HCC cells that promotes their invasion of distant sites (Wu et al, 2020b). Finally, EVPs from exhausted CD8+ T cells in human HCC further promote the exhaustion of naive CD8+ T cells, supporting immune evasion (Wang et al, 2019b).
In conclusion, cancer‐associated immune cells are both the source and recipient of EVPs, with different immune modulating properties. Cancer‐derived EVPs preferentially induce polarization of immune cells toward a tumor‐promoting phenotype and prevent infiltration and activation of anti‐tumor lymphocytes, generating an immunosuppressive and permissive microenvironment. In turn, immune‐derived EVPs have shown both anti‐ and pro‐tumorigenic functions, highlighting the different contribution of immune cell‐derived EVPs in the progression of a range of tumor types.
Cancer cell metabolic plasticity
Reprogrammed energy metabolism is a hallmark of cancer (Hanahan & Weinberg, 2011). Cancer‐derived EVPs from culture or patient biofluids are particularly rich in mediators of metabolic reprogramming, including purine metabolites, glycolytic, and gluconeogenic enzymes, which confer higher invasive phenotype to recipient cells (Ronquist et al, 2016; Zhang et al, 2017c; Ludwig et al, 2020) (Figure 3). In NSCLC, EVP‐associated lncRNAs and circRNAs potentiate glucose uptake and lactate production, which are the main forms of energy sustaining cancer growth (Ding et al, 2020; Chen et al, 2021b). Our lab has determined that the distribution of metabolic mediators in different EVP subgroups is not equal. Instead, metabolic enzymes involved in glycolysis and mTOR signaling are specifically enriched in exomeres derived from different types of cancer cells (Zhang et al, 2018b, 2019e). These exomeres primarily target the liver in animal models, supporting the hypothesis that tumor‐derived exomeres can systemically influence metabolism of cancer patients (Zhang et al, 2018b).
Besides intrinsic regulation of tumor cell metabolism, several studies have highlighted the importance of EVP‐mediated metabolic crosstalk between cells in the tumor microenvironment as another tier of cancer metabolism regulation. Mast cell‐derived EVPs are particularly rich in regulators of eicosanoid metabolism, which is involved in DC maturation, inflammation, cell growth, angiogenesis, and thrombosis (Subra et al, 2010; Cianciaruso et al, 2019; Mizuno et al, 2019). Similarly, cancer‐associated adipocytes (CAAs) promote tumor progression via storage of energy molecules, cytokines, and growth factors and are associated with poor prognosis (Park et al, 2014). In order to reprogram adipocytes into CAAs, breast cancer cells shuttle EVP miRNAs involved in alteration of adipocyte homeostasis (Wu et al, 2019b). In turn, CAAs promote extensive metabolic remodeling in tumor cells, including increased glucose and fatty acid uptake and support an aggressive phenotype (Wu et al, 2019b). In HCC, adipocyte‐derived EVPs reduce DNA damage and promote cell cycle progression via USP7/Cyclin A2 (Zhang et al, 2019a). Adipocyte EVPs can also transfer fatty acids and stimulate fatty acid oxidation in melanoma cells, a process increased by obesity (Clement et al, 2020). These transferred fatty acids fuel fatty acid oxidation, which subsequently redistributes mitochondria to membrane protrusions of migrating cells and increases their migration capability.
The conversion of normal fibroblasts into tumor‐promoting CAFs relies on metabolic reprogramming, in part induced by cancer cell‐derived EVPs and their miRNAs. Breast‐cancer‐secreted, EVP‐encapsulated miR‐105 activates Myc signaling in CAFs, which reprograms their metabolism in favor of glycolysis and glutaminolysis and allows secretion of glucose‐ and glutamine‐derived metabolites to fuel adjacent tumor cells. In addition, by consuming metabolic byproducts and promoting extracellular acidification, EVP‐reprogrammed CAFs generate a nutrient‐rich and permissive microenvironment that promotes tumor cell growth and migration (Yan et al, 2018b). Breast‐cancer‐derived EVPs carry high levels of miR‐122, which suppresses glucose uptake by lung fibroblasts and brain astrocytes in the pre‐metastatic niche, thereby increasing the nutrient availability for tumor cells and facilitating metastasis (Fong et al, 2015). Additionally, in CAFs, glycolysis is favored to oxidative phosphorylation in response to EVP‐associated integrin β4 + and miRNAs, potentially providing lactate and pyruvate as means of energy for breast and melanoma tumor cells (Pavlides et al, 2009; Shu et al, 2018; Sung et al, 2020). Instead, EVPs from colorectal cancer cells lead to metabolic reprogramming of fibroblast by upregulating proteins required for glycogen metabolism, amino acid biosynthesis, and transporters for glucose, lactate, and amino acids (Rai et al, 2019). The functional contribution of such metabolic transformation to tumor progression needs to be evaluated. In return, EVPs released by CAFs increase glucose uptake and glycolysis and inhibit mitochondrial oxidative phosphorylation in prostate cancer cells (Zhao et al, 2016a). Metabolomic characterization of CAF EVPs revealed high levels of different amino acids, fatty acids, and TCA‐cycle intermediates, which can be readily utilized by host cells. Importantly, the authors provided evidence that CAF‐derived EVPs can supply metabolites to cancer cells and rescue their proliferation and growth under nutrient deprivation.
These studies collectively suggest that distinct types of metabolic interactions between tumor and stroma exist to facilitate tumor progression, and this may depend on tumor type, stage, and the metabolic conditions provided by the tumor microenvironment.
Intervention, radiation, and chemotherapy
A growing body of evidence suggests that standard of care treatment, such as surgery, radiation therapy, and chemotherapy, is associated with a surge in circulating EVPs that may come from tumor or stromal cells that survive treatment (Figure 3). Both therapy‐induced metabolic changes and selection of cells with altered EVP release might be potential underlying mechanisms. Surgery is associated with a change in EVPs levels and cargo in the bodily fluids of cancer patients (Campanella et al, 2015; Butz et al, 2016; Rodriguez Zorrilla et al, 2019). The levels of CD63+ EVPs were found to return to normal days after tumor resection, but high EVPs count immediately after surgery was predictive of poor overall survival in oral squamous cell carcinoma patients (Rodriguez Zorrilla et al, 2019).
Treatment of different cancer cells with sublethal doses of common chemotherapeutics, such as rapamycin, doxorubicin, cisplatin, panabinostat, bortezomib, carfilzomib, or melphalan, increases the release of EVPs, often called chemoexosomes, and changes their protein and miRNA cargo (Bandari et al, 2018; Samuel et al, 2018; Tubita et al, 2019; Wills et al, 2021; Li et al, 2021d). Different protein cargo was also identified in EVPs from irradiated cancer cells (Mutschelknaus et al, 2017; Abramowicz et al, 2019; Mo et al, 2020). EVPs released upon radiation therapy can affect multiple components of the tumor microenvironment. EVPs from irradiated lung cancer, head and neck cancer, and ovarian cancer cells stimulate tumor cell motility, migration, and proliferation (Mutschelknaus et al, 2017; Samuel et al, 2018; Mo et al, 2020; Wang et al, 2020b). Several mechanisms have been characterized, including packaging of angiopoietin‐like 4 in EVPs (Mo et al, 2020), EVP enrichment of metabolic enzymes, such as ALDOA and ALDH3A1 (Wang et al, 2020b), induction of the AKT and p38/JNK signaling pathway (Mutschelknaus et al, 2017; Samuel et al, 2018), and transfer of EVP ECM‐degrading heparanase in recipient cells (Bandari et al, 2018). Additionally, these EVPs increase angiogenesis via VEGF‐B upregulation in endothelial cells (Mo et al, 2020), as well as stimulate macrophage migration and secretion of TNF‐α, ultimately promoting tumor growth (Bandari et al, 2018).
Drug resistance can also be transmitted via EVPs, and thus EVP‐mediated communication might represent an additional mechanism of expansion of therapy‐resistant subclones and cell competition, as described in detail by Parker and colleagues in this Cancer Reviews series (Parker et al, 2021). Gemcitabine‐resistant NSCLC cells transfer resistance to parental cells via shuttling of EVP miR‐222‐3p, which targets suppressor of cytokine signaling 3 (SOCS3) followed by activation of JAK/STAT signaling. As expected, levels of miR‐222‐3p in serum EVPs from NSCLC patients negatively correlated with response to gemcitabine treatment and higher levels identified patients with no response and progressive disease (Wei et al, 2017a). EVPs from adriamycin‐resistant breast cancer cells transmit resistance to sensitive cells via transfer of GSTP1 and Hsp70, the latter of which reprograms the energy metabolism of recipient cells toward reduced mitochondria respiration and increased glycolysis (Yang et al, 2017c; Hu et al, 2021). Levels of GSTP1 mRNA were higher in serum EVPs from chemo‐resistant breast cancer patients in comparison to patients who achieve complete response (Yang et al, 2017c). EVPs from hypoxic ovarian cancer cells propagate cisplatin resistance by decreasing dsDNA damage and increasing survival (Dorayappan et al, 2018). MDA‐MB‐231 cells treated with different microtubule stabilizers release survivin‐enriched EVPs that promote survival of adjacent tumor cells and fibroblasts (Kreger et al, 2016). EVP‐encapsulated miR‐155 from chemoresistant cells confers resistance to recipient sensitive cells in breast cancer, oral squamous carcinoma, and PDAC (Mikamori et al, 2017; Santos et al, 2018; Kirave et al, 2020). Finally, EVPs from glioblastoma cells harboring the PTPRZ1‐MET fusion mutation mediate the horizontal transfer of chemoresistance to temozolomide in vitro and in patients (Zeng et al, 2017).
Multiple reports point to CAFs, which are innately chemoresistant (Richards et al, 2017), as a main source of EVPs promoting chemoresistance. EVPs from normal skin fibroblasts exposed to radiation therapy were found enriched in hyaluronic acid, which promotes different aspects of cancer progression (Zare et al, 2020). Additionally, EVPs from CAFs derived from CRC tissue, but not normal fibroblasts from control colorectal mucosa, induce resistance of cancer cells to 5‐FU/L‐OHP chemotherapy, and promote metastasis via transfer of miR‐92‐3p, which activates Wnt/β‐catenin pathway and inhibits mitochondrial apoptosis in recipient cells (Bandari et al, 2018; Hu et al, 2019a). Similarly, cultured CAFs induce resistance of bladder cancer cells to paclitaxel and doxorubicin via EVP‐mediated miR‐148‐3p and downregulation of PTEN (Shan et al, 2021). Lastly, EVPs from CAFs exposed to gemcitabine promote proliferation and chemoresistance of PDAC cells and orthotopic tumors via transfer of miR‐146a and Snail mRNA (Richards et al, 2017).
Our current knowledge on the mechanisms of EVP release relies on experiments performed on untreated cancer cells. The evidence summarized here suggests that the amount and cargo of EVPs are affected by chemo‐ and radiotherapy on both tumor, immune and stromal cell components, but further research is needed to understand the effect of clinical intervention on EVP biogenesis and release.
Creating a favorable soil: pre‐metastatic niches
EVPs from different cell and tissue sources have the “innate” tendency to distribute to pre‐metastatic sites, such as lungs, liver, bone marrow, and brain, that reflect the organotropism of the releasing cells (Peinado et al, 2012;Hoshino et al, 2015; Yoshida et al, 2019). Peinado and colleagues have shown that the protein content of EVPs correlates with their metastatic potential (Peinado et al, 2012). Building on this, integrin expression was found to be a major pattern of EVPs orchestrating the formation of pre‐metastatic niches at future sites of organotropic metastasis (Hoshino et al, 2015; Yoshida et al, 2019). EVP organotropism has important functional consequences, such as promoting metastatic seeding of the distant site by allowing formation of pre‐metastatic niches (Figure 3). This is demonstrated by the observation that inhibition of exosome exocytosis via Rab27a‐KO is sufficient to reduce the likelihood of distant metastasis in a plethora of models, including mammary carcinoma cells (Bobrie et al, 2012; Zhang et al, 2015b) and melanoma cells (Peinado et al, 2012; Guo et al, 2019a). It remains to be determined if inhibition of exomere release affects metastasis similarly. Due to their ability to selectively deliver their cargo at specific distant sites, EVPs might as well be the first and foremost messengers preparing a “congenial soil” for the “seed.”
Gateways to colonization: vascular leakiness and angiogenesis
The continuity of the endothelial lining of the blood and lymphatic system represents an important barrier to improper extravasation of immune cells and tumor cells and to potentially harmful therapeutics at distant sites. Peinado and colleagues were the first to show that murine melanoma B16F10‐derived EVPs, but not EVPs from non‐metastatic Melan‐A cell line, increased lung vascular permeability, an initial step in pre‐metastatic niche formation. This promoted the rate and extent of spontaneous lung and bone metastasis, while minimally affecting tumor growth (Peinado et al, 2012). Further, EVPs from highly brain‐invasive breast cancer cells increased permeability of the blood–brain barrier (BBB) and promoted brain metastasis of less invasive cells (Tominaga et al, 2015). Rodrigues and colleagues more recently confirmed this finding showing that brain‐tropic EVPs are taken up by CD31+Glut1+ brain endothelial cells and increased leakiness in brain capillaries (Rodrigues et al, 2019).
Endothelial cell junctions are a major target of cancer‐derived EVPs. miR‐105 and miR‐181 are restricted to EVPs from highly metastatic breast cancer cell lines, suppress expression of tight junction proteins, such as zona occludens (ZO)‐1, and dysregulate localization of N‐cadherin and actin in microvascular endothelial cells, leading to endothelial barrier disruption, cancer cell transendothelial migration, and lung and brain metastasis (Zhou et al, 2014; Tominaga et al, 2015). Functional miR‐105+ EVPs were also detected in the serum of patients with stage II and III breast cancer and correlate with risk of metastasis (Zhou et al, 2014). Similarly, highly invasive variants of hepatocellular carcinoma cell lines, but not non‐metastatic variants, release EVPs that reduce expression of VE‐cadherin and ZO‐1 in endothelial cells, leading to endothelial permeability and tumor cell transendothelial migration (Fang et al, 2018; Yokota et al, 2021). These EVPs were found enriched in different miRNAs, some of which associate with lower disease‐free survival and higher frequency of distant metastasis in HCC patients (Fang et al, 2018; Yokota et al, 2021). Other miRNAs were found highly expressed in EVPs from breast cancer, CRC, hepatoma, and ovarian cancer cells and patient plasma, and were shown to compromise the integrity of endothelial cell junctions via downregulation of VE‐cadherin, ZO‐1, and Claudin‐5, and silencing of KLF2/4, factors controlling transcription of VEGF and junction proteins, ultimately promoting metastasis to both livers and lungs (Di Modica et al, 2017; Fang et al, 2018; Zeng et al, 2018; Lin et al, 2020). More recent evidence has shown that tubulin tyrosine ligase like 4 (TTLL4) expressed in EVPs from breast cancer cell lines MDA‐MB‐231 and MDA‐MB‐468 contributes to EVP biogenesis, endothelial cell permeability, and tumor cell adhesion to endothelial cells (Arnold et al, 2020). Along the same lines, MDA‐MB‐231‐derived EVPs are enriched in nucleoside diphosphate kinase (NDPK) A and B in comparison to their non‐tumorigenic counterpart (HME1) and activate P2Y1 receptor signaling in lung microvascular cells, inducing cell migration and decreased junctional β‐catenin. This resulted in increased vascular leakiness and pulmonary metastasis, an effect that could be prevented by P2Y1 inhibitors (Duan et al, 2021). Finally, Yoshida and colleagues showed that EVPs isolated from high‐grade bladder cancer cells are carriers of tyrosine kinases, such as ErbB2 and CRK, and, in addition to promoting proliferation of tumor cells locally, stimulate FAK/AKT signaling, migration, and proliferation of endothelial cells, and promote lung metastasis in EVP‐educated mice (Yoshida et al, 2019).
A fertile soil for metastasis might also be “prepared” by EVPs at local or distant lymph nodes. Melanoma‐derived EVPs promote angiogenesis and chemotaxis in sentinel lymph nodes, facilitating tumor cell infiltration at later stages of disease (Hood et al, 2011). These findings were corroborated recently by other groups showing that EVP‐associated lncRNAs, miRNA, and CXCL4 promote lymph node remodeling and lymphatic metastasis in cancer types prone to this route of dissemination, such as breast cancer, cervical squamous carcinoma, and CRC (Li et al, 2018c; Zhou et al, 2019a; Chen et al, 2020a). A direct effect of EVPs on tube formation of lymphatic endothelial cells, MMP upregulation, and reprogramming of macrophages to produce lymphangiogenic VEGF‐C have also been described (Li et al, 2018c; Zhou et al, 2019a; Sun et al, 2019b; Chen et al, 2020a).
Immune cell and bone marrow cell recruitment
Tissue‐resident immunity is typically refractory to tumor cell colonization, but the accumulation of bone‐derived immune cells or the activation of resident immune cells at distant sites generates an immune suppressive environment that allows for metastatic dissemination and outgrowth (Kaplan et al, 2005; Murgai et al, 2017; Kaczanowska et al, 2021). In the liver niche, Pan02 EVPs harboring migration inhibitory factor (MIF) educate hepatic Kupffer cells to release TGF‐β, which induces differentiation of HSCs and deposition of fibronectin. the recruitment of bone marrow‐derived cells (BMDCs), including macrophages and neutrophils, to fibronectin‐rich microenvironments ultimately supports metastasis formation (Costa‐Silva et al, 2015). These findings were recently corroborated by the observation that EVPs from KPC murine pancreatic cancer cells promote enrichment of macrophages in the liver niche. However, these EVPs could not rescue the reduction of myeloid cell infiltration in livers of mice engrafted with Rab27a‐deficient KPC cells, suggesting that Rab27a has autologous functions in pre‐metastatic niche formation in addition to EVP‐mediated mechanisms (Kren et al, 2020). Several PDAC EVP cargo proteins, such as MET, ADAM9, S100A4, LGALS3, and integrins β4 and β5, are involved in immune modulation and cell recruitment, while EGFR, CLDN1, CAV1, and SDC1 are associated with angiogenesis, innate immune response, and cell migration in the pre‐metastatic liver (Emmanouilidi et al, 2019). Gastric cancer‐derived EVPs overexpress epithelial growth factor receptor (EGFR) in both murine models and patients, especially at advanced stage of disease. By upregulating hepatocyte growth factor (HGF) in the liver microenvironment, these EGFR+ EVPs act as a chemoattractant to tumor cells expressing c‐MET, the HGF receptor (Zhang et al, 2017b). Interestingly, EBV infection in the liver has been correlated with release of EVPs enriched in LMP1, which promotes expression of pre‐metastatic markers S100A8, fibronectin, and VEGFR1 in liver and lungs (Wu et al, 2020d).
Comparable changes to the immune profile of pre‐metastatic lungs have been observed in response to EVPs from lung‐tropic cancer types such as breast cancer and melanoma. Bobrie and collaborators showed that murine breast carcinoma tumors promote infiltration of neutrophils into pre‐metastatic lungs (Bobrie et al, 2012). Similarly, murine breast cancer E0771 and 4T1‐derived EVPs induce an immunosuppressive pre‐metastatic niche in lungs and liver of naïve mice, with increased granulocytic and myeloid MDSCs and, as a consequence, reduced infiltration of CD8+ T and NK cells (Wen et al, 2016). The tumor‐suppressive activity of NK cells and CD4+/CD8+ T cells was also directly suppressed by EVPs (Wen et al, 2016). EVPs from invasive breast cancer cell lines are enriched for Annexin II and prime macrophage activation at distant sites, including the brain and lungs, via activation of MAPK, NF‐kB, and STAT3 signaling, leading to the release of pro‐inflammatory IL‐6 and TNFα (Maji et al, 2017). Enrichment in insulin‐like growth factor 2 mRNA binding protein 1 (IGF2BP1) has been observed in EVPs from melanoma cell lines and increased the deposition of fibronectin as well as recruitment of CD45+ cells in lungs, further promoting lung cancer metastasis (Ghoshal et al, 2019). Recruitment of immune cells to lungs can also happen via education of other stroma cells, especially fibroblasts. Hoshino and colleagues have shown that lung‐tropic EVPs expressing integrin α6β4 upregulated pro‐inflammatory S100 proteins in lung fibroblasts (Hoshino et al, 2015), which function as damage‐associated molecular pattern (DAMP) molecules for the migration and activation of macrophages, neutrophils, and DCs (Xia et al, 2017). Similarly, EVPs from Lewis lung carcinoma cells (LLC) are mainly engulfed by pulmonary fibroblasts and activate their NF‐kB signaling via miR‐3473b transfer. In response, fibroblasts release pro‐inflammatory cytokines, such as IL‐6, CCL‐2, and CCL‐5, and drive recruitment of B cells, promoting pulmonary colonization (Du et al, 2020a). Moreover, EVPs produced upon direct interaction between breast cancer cells and fibroblasts in the primary tumor induce activation of distant myeloid and DC immune cells in the spleen (Nabet et al, 2017) and might thus be involved in the generation of pre‐metastatic niches.
While our knowledge of pre‐metastatic niches in the lungs and liver has exponentially grown in the last decade, we know very little about other pre‐metastatic sites. EVPs from the metastatic melanoma cell line B16F10 educate BMDCs and increase the production of c‐Kit+Tie2+ precursor cells. This effect is due to the horizontal transfer of oncogenic MET, which induces S6 and ERK phosphorylation in BMDCs and promotes the mobilization of vasculogenic and hematopoietic BMDC precursor cells from the bone marrow to future metastatic sites. When transplanted into lethally irradiated naïve mice, MET+ EVP‐educated BMDCs promote primary tumor growth as well as metastasis to multiple organ sites, including distant lymph nodes and brain (Peinado et al, 2012).
The brain has been known for a long time to be a sanctuary site for cancer metastasis, where tumor cells are protected from chemotherapy and immune surveillance. The mechanisms leading to preparation of a favorable soil for tumor dissemination are not yet fully understood. By employing an organotypic brain slice culture system, Rodrigues and colleagues have shown that brain tissue preconditioned with EVPs from brain tropic breast cancer cell, but not from their parental counterpart, is more receptive to tumor cell colonization (Rodrigues et al, 2019). CEMIP, which was found restricted to brain‐tropic EVPs, was the culprit of brain preconditioning by inducing transcriptional changes in brain endothelial cells consistent with altered morphogenesis, junction formation, and vascular permeability. The clinical implication of these findings is illustrated by the observation that CEMIP levels were much higher in brain metastasis tissue and their EVPs than adjacent and distant tissues and correlated with shorter survival (Rodrigues et al, 2019).
Finally, the contribution of neutrophil extracellular traps (NETs), which have been detected at primary and distant sites in a series of cancers, to pre‐metastatic niche formation is still lacking (Yang et al, 2015a; Tohme et al, 2016). In a seminal paper, Park and colleagues have shown that tumor‐driven NET formation in the lung vasculature is an essential step for breast cancer metastasis (Park et al, 2016). Recently, Leal and colleagues have determined that breast cancer cell–derived EVPs induce NET formation in vitro and in mice (Leal et al, 2017), but the underlying mechanisms are not yet known. Further research might unveil the role of EVP‐induced NETs in the establishment of pre‐metastatic niches at different sites.
Metabolic reprogramming
Similar to their role within the tumor microenvironment, cancer‐derived EVPs promote the metabolic reprogramming of stromal and immune cells at distant sites prior to tumor cell colonization. For example, prostate‐cancer‐derived EVPs transfer functional pyruvate kinase M2 (PKM2) to bone marrow fibroblasts, where it drives HIF‐1α‐dependent production of CXCL12 and stimulates tumor cell proliferation via the CXCL12:CXCR4 axis (Dai et al, 2019). Furthermore, melanoma‐derived EVPs and their miR‐155 and miR‐210 cargo promote glycolysis and inhibit oxidative phosphorylation in fibroblasts, inducing extracellular acidification that has been previously associated with pre‐metastatic niche formation (Shu et al, 2018).
Looking at immune cells, CD11b+ cells in the bone marrow take up EVPs from Pan02 murine pancreatic cancer cells and shift toward a tumor permissive phenotype. In particular, Pan02 EVPs induced significant transcriptional changes in bone marrow monocytes/macrophages, consistent decreased differentiation, increased polarization towards a tumor‐suppressive M1‐like phenotype, and upregulation of immunoglobulins (Ig) genes, which have been implicated in inflammation, immune cell recruitment, and activation (Maia et al, 2020). The role of EVPs in inducing macrophage polarization is reverted in hypoxic conditions, where tumor‐derived EVPs instead steer macrophage differentiation toward an M2‐like pro‐tumorigenic phenotype, with increased oxidative phosphorylation and suppressed mTOR pathways (Park et al, 2019). Finally, education with prostate‐cancer‐derived EVPs induces NF‐kB signaling and versican (VCAN) expression in myeloid cells and drives osteoclast proliferation and differentiation to a bone resorption phenotype. These effects lead to increased metastatic colonization of bones via tumor cell adhesion to VCAN‐rich bone marrow niche (Henrich et al, 2020). Interestingly, prostate cancer‐derived EVPs do not affect tumor cells directly, further supporting the idea that long‐distance changes in the pre‐metastatic stromal and immune compartments are essential for tumor cell seeding and metastasis (Dai et al, 2019).
ECM remodeling
A prerequisite for pre‐metastatic niche establishment is the formation of a remodeled ECM backbone that allows for improved tumor cell and immune infiltration and provides sufficient tissue stiffness for tumor cell invasion (Kai et al, 2019). Considering the distinct integrin expression profile of tumor cells and their EVPs with different organ tropism, ECM alteration at the distant sites might also direct tissue homing. Thus, by altering ECM of distant organs, tumor‐derived EVPs might achieve both increased and directional colonization.
Fibroblasts in the lungs and HSCs in the liver are major orchestrators of ECM composition. Fibroblasts educated with EVPs from p53 mutant lung and pancreatic cancer cells produce an ECM mesh that is dramatically different in structure, binding, and composition to fibroblasts educated with EVPs from p53 competent cells. In particular, fibrillar collagen in educated lungs appeared to have a more punctate and less organized structure, reminiscent of the unstructured vasculature of tumors. EVP sialomucin podocalyxin was found responsible for these changes (Novo et al, 2018). Similar fibroblast activation was observed in correlation with enhanced pulmonary metastasis in animal models, suggesting that Mutp53s potentially drive this phenomenon via EVPs. In support of this observation, Capaci and colleagues recently showed in a breast cancer model that Mutp53s induce Golgi tubule vesiculation and alter the secretome of tumor cells, ultimately enhancing tumor growth and metastatic colonization via ECM remodeling (Capaci et al, 2020). Though the contribution of EVPs in this process was not investigated, it is reasonable to speculate that EVPs as a major component of the secretome play an important role here.
Education of the pre‐metastatic liver with pancreatic cancer cell–derived EVPs induces activation of HSCs via Kupffer cell‐derived TGF‐β, allowing for a 10‐fold increase in fibronectin and collagen I deposition and repressing the deposition of vitronectin and tenascin C (Costa‐Silva et al, 2015; Xie et al, 2021). Activation of IGF‐1/PI3K/AKT pathways has been reported in PDAC EVP‐educated HSCs (Xie et al, 2021). HSC activation was also observed in LLC tumor‐bearing mice, where EVPs from myeloid BMDCs drive their production of collagen I, promoting recruitment of granulocytic MDSCs and cancer cell adhesion and extravasation. Enrichment of miR‐92a in BMDC‐derived EVPs, which decreases expression and phosphorylation of SMAD7 in HSCs, was responsible for their metastatic niche‐promoting activity. miR‐92a‐enriched EVPs were also found in the serum of lung cancer patients and induce a similar reprogramming of HSCs (Hsu et al, 2020). The mechanism through which primary lung cancer cells influence BMDCs to release pro‐metastatic EVPs is still unknown. Intratumor hypoxia might increase the ECM remodeling properties of EVPs, as shown by evidence that EVPs from hypoxic PC‐3 cells caused MMP‐dependent deposition of fibronectin and collagen IV at various pre‐metastatic sites (Deep et al, 2020).
In the bone niche, bone formation and resorption are the main ECM remodeling events. Certain cancers, such as prostate cancer, take advantage of osteoblast proliferation or activation and ECM mineralization to metastasize to bones, while other cancers, such as breast, lung, and kidney cancer, favor increased osteoclast activity. The EVP miRNA profiles of cancer cell lines inducing either phenotype are dramatically different. In particular, miR‐940 and ‐1260a were found enriched in EVPs from cells promoting osteoblastic lesions and were associated with osteoblast differentiation and osteogenesis (Hashimoto et al, 2018). In the pre‐metastatic bone, miR‐375+ prostate‐cancer‐derived EVPs directly promote formation of calcium nodules in osteoblasts, leading to ECM mineralization (Li et al, 2019e).
EVPs can be themselves carriers of ECM proteins and remodeling enzymes. Laminin, collagens, and cathepsin hydrolases were detected (Hood et al, 2011; Latifkar et al, 2019). In clinical settings, EVPs from pancreatic duct fluid, plasma, and tumor tissue of PDAC patients were enriched in tenascin C, laminin subunits, THBS1/2 and versican, consistent with the alteration of ECM composition in PDAC and pre‐metastatic liver (Zheng et al, 2018a; Hoshino et al, 2020).
Induction of pre‐metastatic niche by chemotherapy
Chemotherapy has been found associated with a higher risk of metastatic disease in preclinical models and in patients that do not achieve complete response (Liedtke et al, 2008; Volk‐Draper et al, 2014; Karagiannis et al, 2017; Keklikoglou et al, 2019; D'Alterio et al, 2020). It is believed that therapy‐induced tissue damage mimics early events associated with the establishment of pre‐metastatic niches, including release of cytokines, chemokines, and EVPs (Ratajczak et al, 2013). Doxorubicin treatment induces the overexpression of the pro‐inflammatory glycoprotein PTX3 in MDA‐MB‐231‐derived EVPs, which then establishes a favorable pulmonary pre‐metastatic niche for both highly metastatic and poorly metastatic TNBC cell lines (Wills et al, 2021). Similarly, mouse education with EVPs from paclitaxel‐treated breast cancer cells or from tumors of paclitaxel‐ or doxorubicin‐treated MMTV‐PyMT mice increases lung colonization. These EVPs were found to be enriched in annexin A6 (ANXA6), which induces release of CCL2 by endothelial cells, promoting the recruitment and expansion of Ly6C+CCR2+ monocytes in the pre‐metastatic lungs. ANXA6‐positive EVPs were also found in the plasma of breast cancer patients undergoing neoadjuvant chemotherapy (Keklikoglou et al, 2019). Furthermore, EVPs derived from rapamycin‐treated HCT116 cells are enriched in miRNAs that can functionally decrease expression of histone genes in lung fibroblasts, reprogramming them toward decreased DNA packaging and chromatin assembly. This epigenetic reprogramming might reduce the ability of fibroblasts to differentiate into myofibroblasts in pre‐metastatic sites (Tubita et al, 2019).
Not only chemotherapy itself, but also resistance to chemotherapy alters the amount and cargo of circulating EVPs, shifting it to a pre‐metastatic niche promoting one. To illustrate, ovarian cancer patients have higher serum concentration of EVPs in comparison to cisplatin sensitive patients, potentially due to intra‐tumor hypoxic conditions (Dorayappan et al, 2018). Moreover, temozolomide‐resistant glioblastoma cells selectively package lncRNA HOTAIR into their EVPs, which propagate chemoresistance and, potentially, metastatic ability of tumor cells (Yuan et al, 2020). Conversely, EVPs from doxorubicin‐ and panabinostat‐resistant cells are enriched in Bcl2‐associated athanogene 6 (BAG6), which induces transcriptomic changes in pre‐metastatic lungs consistent with reduced recruitment and activation of pro‐metastatic neutrophils and increased accumulation of Ly6Clow anti‐tumor patrolling monocytes. As a result, education with BAG6‐expressing EVPs reduced lung metastasis (Schuldner et al, 2019).
Determining the right destination: organotropic metastasis
Over the years, different theories have been proposed to explain the selective metastatic distribution of different cancer types. By proposing the “anatomical and mechanical” theory in 1858, Virchow and others speculated that metastatic tropism relied on the physical arrest of tumor cells in the vasculature of distant organs, and that circulatory patterns drive organ distribution (Ewing, 1928; Virchow, 1989). This theory could not explain the selective colonization of organs with similar blood supply and was then challenged by the famous “seed and soil” theory by the British surgeon Stephen Paget (Paget, 1989). Many experimental evidences have supported this theory by showing that the genetic makeup of tumor cells, the structural and molecular properties of the distant niches and the interaction of tumor cells with the metastatic microenvironment all drive metastasis organotropism (Gao et al, 2019). Recent evidence has added to this paradigm showing that distant sites are not always intrinsically receptive to tumor cells, but are rather remotely educated by primary tumor‐derived soluble factors and EVPs. In a seminal paper, Hoshino and colleagues have shown that EVPs share the same organ distribution pattern of the secreting cells and that EVP integrins are major determinants of this selective homing (Hoshino et al, 2015). In more details, integrins α6β4 and α6β1 were abundant in lung tropic EVPs, while integrin αvβ5 was found enriched in liver‐tropic EVPs. Similarly, Rodrigues and colleagues identified exosomal CEMIP as driver of brain metastatic colonization (Rodrigues et al, 2019). Other evidence has shown that in the complex ecosystem of the tumor microenvironment specific subpopulations of EVPs have different organotropism. For example, among all circulating EVPs in CCRCC patients, CSC‐derived CD103+ EVPs specifically home to primary tumors and pre‐metastatic lungs, while CD103− EVPs lack this selectivity (Wang et al, 2019a).
As reviewed in the previous sections, organotropic EVPs set the stage for tumor cell colonization by inducing vascular remodeling and inflammation in the pre‐metastatic niche (Peinado et al, 2012;Hoshino et al, 2015; Rodrigues et al, 2019). For example, lung‐tropic EVPs drive the colonization of the lungs by bone‐tropic tumor cells, showing for the first time that tumor‐derived EVPs can redirect the organotropic pattern of tumor cells and allow dissemination of tumor cells with poor intrinsic metastatic potential (Hoshino et al, 2015). At the same time, pre‐education of mice with B16F10‐derived EVPs allows their seeding not only to lungs, but also to contralateral lymph nodes, brain, and mesentery (Peinado et al, 2012). This evidence adds a new layer of complexity to the seed and soil theory, suggesting that the “congenial soil” is actively prepared by EVPs with selective organotropism. EVPs can define the pattern of organ distribution regardless of the innate organotropism of tumor cells, suggesting that intrinsic tumor features might not be sufficient prognostic markers of metastasis site. Instead, the levels of integrin β4, integrin αv, and CEMIP were found significantly higher in plasma EVPs of patients with lung, liver, and brain metastasis, respectively (Hoshino et al, 2015; Rodrigues et al, 2019). Together, these reports indicate that EVP markers are both drivers and bona fide predictors of organotropic metastasis.
A new life: metastatic colonization
The survival and outgrowth of tumor cells that have infiltrated a distant organ are critical bottlenecks in the metastatic cascade. Once in the parenchyma of a distant organ, tumor cells have to face a dramatic reduction in nutrient availability, as well as withstand an inhospitable and immune‐reactive microenvironment, resulting in only a few tumor cells being able to expand to a macrometastatic status. The establishment of a pre‐metastatic niche partially overcomes this limitation by manipulating distant tissues and allowing for better survival of colonizing immune cells. Our knowledge of early stages of metastatic outgrowth is still limited, but it appears that tumor cell‐ or stroma‐derived signaling is essential to clinically manifest metastasis. Similar to EVPs derived from primary tumor cells, those derived from metastatic cells themselves may shape the metastatic niche to allow survival and overt growth or, alternatively, maintain a temporary dormancy status leading to metastatic latency.
Metastasis‐initiating cells
Of the millions of tumor cells shed by a primary tumor per day, only a few metastasis‐initiating cells (MICs) will proceed to form macrometastasis. Although the features, emergence, and selection of these metastatic progenitor cells are not fully understood, some MIC traits are starting to emerge more clearly. Ganesh and colleagues have shown that L1CAM expression in CRC epithelial cells triggers the tissue regenerative, chemoresistant, and disseminating abilities of MICs (Ganesh et al, 2020). They showed that L1CAM mediates the interaction of MICs with laminin‐rich ECMs, such as in EVP‐educated pre‐metastatic lungs (Hoshino et al, 2015; Ganesh et al, 2020). L1CAM was detected in B16F10 Exo‐Ls (Zhang et al, 2018b), supporting the notion that EVPs from primary tumors may directly influence the emergence and metastatic spreading of MICs. Likewise, thrombospondin and collagen‐interacting CD36, a bona fide marker and driver of the MIC phenotype (Pascual et al, 2017), are ubiquitous markers of human EVPs, both from non‐tumor and tumor tissue sources (Hoshino et al, 2020).
It is also possible that tumor‐ or stroma‐derived EVPs may alter the expression profile of a proportion of CTCs, either before or after intravasation, transforming them into MICs. In breast cancer patients, circulating tumor cells (CTCs) with bone‐metastasis‐initiating potential were found to express CD44, CD47, and MET (Baccelli et al, 2013). Fibroblasts are a major source of CD47+ EVPs that can horizontally transfer CD47 to tumor cells, where it helps evade immune surveillance in the blood circulation and at distant organs (Kamerkar et al, 2017). In addition, EVPs from highly metastatic melanoma cell lines increase the expression of CD44 and MET in bone marrow progenitor cells, suggesting a similar transfer from breast cancer cells (Peinado et al, 2012).
Interestingly, the cargo of EVPs from metastatic sites can differ dramatically from the EVPs from primary sites. In a murine model of CRC, EVPs from metastasis‐bearing livers were found enriched in tumor‐suppressive miRNAs (e.g., miR‐19 and miR‐193a) and depleted of oncogenic miRNAs (e.g., miR‐21) in comparison to EVPs from primary colon tumor tissue. Teng and colleagues found that this difference relies on differential packaging of miRNA, whereby tumor suppressive miRNAs are actively shed by MICs and their progenitor cells via overexpression of miR‐binding major vault protein (MVP). As a consequence, human CRCs, which are poor in tumor suppressive miRNAs and rich in MVP, have a higher risk of metastasis (Teng et al, 2017). Understanding the mechanisms that lead to selection or adaptation of MICs with a different EVPs packaging profile will help identify how metastases initiate and progress.
Metastatic microenvironment
In the secondary site, co‐option of the metastatic microenvironment allows MICs to either maintain dormancy or sustain growth. Metastasis‐associated fibroblasts (MAFs) play a central role in promoting MIC outgrowth. Pein and colleagues have shown that, early in dissemination, factors such as IL‐1α and IL‐1β derived from micrometastatic lesions trigger the transition of lung fibroblasts into activated and pro‐inflammatory MAFs, which promote progression to macrometastasis via CXCL9/10 secretion (Pein et al, 2020). Similar cancer cell‐derived factors were detected in EVPs from ovarian cancer patients (Bretz et al, 2013) and lung‐tropic EVPs were found to induce activation of fibroblasts (Hoshino et al, 2015). Although direct evidence is still missing, a growing body of work has revealed a direct effect of cancer cell‐derived EVPs in the metabolic reprogramming of fibroblasts at primary and distant sites and suggests that EVP‐associated factors may be involved in inducing MAF differentiation.
In the bone niche, osteolytic activity of osteoclasts promotes release of growth factors and nutrients that support the initial division of tumor cells (Esposito et al, 2018). EVPs from prostate cancer MICs are rich in RANK ligand (RANKL) and EVPs from NSCLC cells promote osteocyte expression of RANKL (Taverna et al, 2017), a potent inducer of macrophage differentiation into osteoclasts (Shiao et al, 2016). Prostate cancer cell–derived EVPs induce osteoblast differentiation directly (Itoh et al, 2012; Inder et al, 2014; Ye et al, 2017; Hashimoto et al, 2018; Li et al, 2019e; Borel et al, 2020), while both breast cancer cell– and lung cancer cell–derived EVPs promote osteoclast differentiation (Taverna et al, 2017; Xu et al, 2018b; Tiedemann et al, 2019; Guo et al, 2019b; Loftus et al, 2020), reflecting the osteoblastic and osteoclastic metastatic niches induced by these different types of cancer.
EVPs also contribute to metastasis initiation and sustained growth in the brain. Conditioned medium of brain metastasis cells containing factors, such as EGF, TGF‐α, and MIF, the latter of which has been previously detected in EVPs from breast and pancreatic cancer cells (Costa‐Silva et al, 2015), stimulates STAT3 activation in astrocytes to support the initial steps of MIC growth. The hypothesis that cancer‐derived EVPs may be major drivers of astrocyte activation, even at pre‐metastatic stages, is supported by the observation that EVPs from brain‐tropic MDA‐MB‐231 cells are actively taken up by astrocytes, albeit in lower amounts than by endothelial cells and microglia (Rodrigues et al, 2019). In response, astrocyte‐derived EVPs induce PTEN loss in cancer cells locally by shuttling miR‐19a to MICs. As a result, NF‐κB phosphorylation and CCL2 secretion by cancer cells promote infiltration of CCR2+ myeloid cells in the brain metastases, further supporting metastasis growth and reducing survival (Zhang et al, 2015b). In addition, reactive astrocytes promote outgrowth of brain metastatic nodules via recruitment of Iba1+ microglia and reduction of CD8+ T cell anti‐tumor response (Priego et al, 2018). In a similar manner, VEGF‐A, TIMP‐1, and the ECM proteins collagen and tenascin‐C, all of which are found in the conditioned medium of reactive astrocytes and of patient brain metastasis explants, dampen CD8+ T cell activation (Priego et al, 2018; Hoshino et al, 2020).
The angiogenic switch is an essential event to sustain metastatic outgrowth and exit from dormancy and relies on local release of growth factors and recruitment of endothelial progenitor cells from the bone marrow (Gao et al, 2008). In primary tumors, EVPs are prime carriers of pro‐angiogenic factors, such as miRNAs, VEGF‐A, and IL‐6 (Skog et al, 2008; Umezu et al, 2014; Mao et al, 2019b; Zhang et al, 2020d), or induce their synthesis by endothelial cells (Tang et al, 2018a; Sato et al, 2019; He et al, 2019a; Xie et al, 2020a; Song et al, 2021). Moreover, EVPs from primary and potentially secondary melanoma tumors influence the expression of MET oncoproteins in vasculogenic c‐Kit+Tie2+ bone marrow precursors, inducing the activation of a signaling pathway involved in cell motility. Indeed, the numbers of CD45−c‐Kit+/TIE2+/low progenitor cells with increased MET activation were the highest in the blood of patients with Stage IV metastatic melanoma, suggesting that MIC‐derived EVPs may be involved in sustained angiogenesis in the metastatic niche (Peinado et al, 2012).
In conclusion, EVP cargo has potential roles in the bilateral tumor–microenvironment interplay at metastatic sites, with the lethal consequence of metastatic progression. Functional experiments will need to be conducted to understand the change in EVP release between cancer cells in the primary tumor and MICs as well as to fully puzzle out their role in metastatic growth.
Tumor cell dormancy
As discussed above, the metastasis microenvironment is a key determinant of MIC fate. MICs undergo metastatic dormancy via entry into a proliferative quiescence or failure to sustain proliferation due to tissue‐resident immunity, lack of angiogenesis, or nutrient deficiency (Goddard et al, 2018). Both direct and indirect evidences suggest that EVPs contribute to maintaining or awakening dormant tumor cells, particularly in breast cancer, where bone metastasis is a main cause of minimal residual disease and relapse. Ono and colleagues have shown that EVPs from human bone marrow MSCs induce dormancy and cell cycle arrest and impair tumor growth in vivo by targeting cell‐cycle‐related genes via exosomal miR‐23b (Ono et al, 2014). Noticeably, breast cancer cells with high miR‐23b levels and MSCs were found in close contact in the bone marrow of patients with breast cancer. More recently, Bliss and colleagues confirmed that miR‐222/‐223 encapsulated in EVPs from cancer‐educated MSCs, and to a lesser extent naïve MSCs, promotes cell cycle arrest and decreases chemosensitivity of breast cancer cells (Bliss et al, 2016). EVPs from bone marrow stromal cells, in particular macrophages polarized toward an M2‐like phenotype, maintain quiescence of breast cancer cells (Lim et al, 2011; Walker et al, 2019), while pro‐inflammatory macrophages tend to awaken dormant cells by activating NF‐ κB pathway and cell cycle progression (Walker et al, 2019). Sansone and colleagues have shown that, in hormonal therapy‐resistant breast cancer, mitochondrial genes packaged into CAF‐derived EVPs may be responsible for inducing oxidative phosphorylation in breast cancer CSCs, provoking the tumor cells to exit from dormancy and consequently fostering the recurrence of metastatic cancer (Sansone et al, 2017). The perivascular niche is a sanctuary for tumor cell quiescence at the metastatic site (Ghajar et al, 2013; Ghajar, 2015), with endothelial‐derived thrombospondin‐1 as a main inducer of breast cancer dormancy and TGF‐β1 and periostin inducing exit from senescence. All of these factors have been recently characterized in human cancer cell–derived EVPs (Hoshino et al, 2020). Similarly, a seminal paper by Lawson and colleagues reported that breast cancer cells from early‐stage metastasis harbor a dormant‐like expression signature with a high expression of quiescence‐associated genes, such as CDKN1B, CHEK1, TGFBR3, and TGFB2, while progression to macrometastatic disease is accompanied by expression of genes involved in dormancy escape, such as MYC, CDK2, and MMP‐1 (Lawson et al, 2015). It is tempting to speculate that EVPs from the primary tumor may be involved in the exit from dormancy, as suggested by the evidence that human cancer‐associated EVPs are enriched in MYC targets, CDK2, and MMPs (Hoshino et al, 2015; Rodrigues et al, 2019). Similarly, TGF‐β has cytostatic effects and has been directly linked to induction of dormancy in tumor cells (Massague & Ganesh, 2021). Both TGF‐β and proteins involved in TGF‐β signaling have also been found in various cancer cell‐derived EVPs (Webber et al, 2010; Wang et al, 2016b; Li et al, 2017e; Ringuette Goulet et al, 2018; Batlle & Massague, 2019; Ferguson Bennit et al, 2021; Hoshino et al, 2020). Anti‐mitogenic DKK is packaged into cancer cell‐derived EVPs, particularly those that display organotropisms to the brain, bone, and lung, major sites of tumor cell dormancy (Lim et al, 2012; Faict et al, 2018; Gan et al, 2020). Finally, NETs have been associated with the tumor cell exit from dormancy via proteolysis of extracellular laminin in the metastatic lung niche, which activates intracellular integrin α3β1 signaling in cancer cells (Albrengues et al, 2018). By inducing NET formation (Leal et al, 2017), cancer cell‐derived EVPs may prepare an environment that is permissive for dormancy evasion and metastatic progression.
Whether inhibiting or promoting exit from dormancy is better for targeting dormant cells is still under debate (Ghajar, 2015), our current knowledge indicates that EVPs may drive tumor cell dormancy in both directions, but further experiments are needed to address their contribution in different types of cancer and metastatic niches.
Within circulating distances: EVPs as mediators of the systemic effects of cancer
Due to their ability to signal at a long‐range distance, cancer‐associated EVPs mediate the most systemic and deadly aspects of cancer. It has been estimated that tumor‐derived EVPs represent up to 10% of the total EVPs found in plasma of cancer patients (Fraser et al, 2019). Moreover, Hoshino and colleagues have shown that among cancer‐associated EVPs in plasma, approximately 50% are from the primary tumor and its tumor microenvironment, whereas other cancer‐associated EVPs are produced by distant organ sites, such as the liver and immune organs (Hoshino et al, 2020). This evidence indicates that EVP‐mediated signaling in cancer is rarely the product of a single site, but rather is an orchestrated, multi‐organ process.
Thrombosis
Cancer patients have a higher risk of being affected by a hypercoagulable state, with deep vein thrombosis and pulmonary embolism being the most frequent and lethal complications (Caine et al, 2002). Preliminary evidence suggests that tumor‐derived EVPs may induce systemic thrombosis. Breast cancer cell–derived EVPs directly interact with platelets and induce their activation and subsequent aggregation (Gomes et al, 2017). Interestingly, the pro‐thrombotic effect of EVPs correlates with the metastatic potential of the releasing cells (Gomes et al, 2017). Moreover, induction of thrombosis in mice by EVPs from murine 4T1 breast cancer cells is associated with the formation of NETs (Leal et al, 2017), which have been previously implicated in cancer‐associated thrombosis (Thalin et al, 2019). Despite this evidence, our knowledge of the factors leading to EVP‐induced thrombosis is still lacking. EVPs from a variety of cancer cell lines and tumor tissues express coagulation factors, such as factor X, thrombospondin, and collagens (Zhang et al, 2018b; Hoshino et al, 2020). PS, a membrane lipid that supports the assembly of coagulation factor complexes during the coagulation cascade, is preferentially exposed on the outer leaflet of the tumor cell membrane and found in a range of EVPs, including tumor and immune cell‐derived EVPs (Utsugi et al, 1991; Tripisciano et al, 2017; Zhang et al, 2018b; Skotland et al, 2019). Furthermore, several breast cancer cell lines release EVPs carrying TF, a major initiator of the coagulation cascade (Garnier et al, 2012; Gomes et al, 2017; Leal et al, 2017; Tawil et al, 2021). TF seems to be preferentially associated with Exo‐L and EVPs from highly invasive cancer cell lines (Gomes et al, 2017; Zhang et al, 2018b). Neoadjuvant and adjuvant therapies can increase the ratio of TF/TF pathway inhibitor in plasma EVPs of patients with breast cancer, partially explaining the link between chemotherapy and thrombosis (Aharon et al, 2017). TF‐expressing EVPs directly induce platelet activation and blood clotting ex vivo and can transfer TF to endothelial cells, increasing their pro‐coagulant activity (Garnier et al, 2012; Gomes et al, 2017; Iyer et al, 2021; Tawil et al, 2021). However, this effect was not observed in patients as demonstrated by the evidence that TF is not present in EVPs from a wide range of cancers and tissues, not even in cancer types associated with the highest risk of thrombosis, such as pancreatic cancer and lung cancer (Hoshino et al, 2020). These observations suggest the existence of TF‐independent pathways of EVP‐induced thrombosis. Further research is needed to address this important but understudied field of cancer research.
Cancer cells are likely not the only source of pro‐thrombotic EVPs. For example, TAM‐derived EVPs are enriched in enzymes and lipid substrates of TXA2 synthesis pathway, which is a major activator of platelet aggregation (Cianciaruso et al, 2019), suggesting that they contribute to immuno‐thrombosis similar to their parent cells. Furthermore, activated platelets are a major source of blood EVPs, which can be taken up by several cell types, including other platelets, vascular smooth muscle cells, and endothelial cells (Heijnen et al, 1999; Srikanthan et al, 2014; Tan et al, 2016; Li et al, 2017c). Platelet‐derived EVPs are enriched in TF and PS and lead to the generation of thrombin in vesicle‐free plasma. This effect was inhibited by incubation with PS‐blocking Annexin V, but not with anti‐TF antibody (Tripisciano et al, 2017), further supporting the notion that TF might not be the main pro‐coagulant factor in EVPs. The release of EVPs by cancer‐educated platelets, the protein expression profile of platelet‐derived EVPs, and the role of platelet‐derived EVPs in cancer still need to be determined.
Immune dysregulation
DAMPs include a broad range of factors that are released by damaged or activated cells and that interact with pattern recognition receptors (PRRs) on immune cells to achieve their activation and defense response. Just like a wound that does not heal, primary and metastatic tumors release large amounts of EVPs enriched in different DAMPs. Among other DAMPs, dsDNA detected on the surface of cancer cell‐derived EVPs is a potent inducer of immune responses, including inflammatory responses and type‐I interferon signaling (Thakur et al, 2014; Lou & Pickering, 2018; Wang et al, 2018c; Maire et al, 2021). EVP DNA may induce systemic activation of STING in DCs, eliciting an anti‐tumor response via DC activation and CD8+ T‐cell infiltration, but also may induce the release of pro‐inflammatory cytokines by innate immune cells (Hernandez et al, 2016; Sharma & Johnson, 2020). The release of EVP‐associated DNA by CRC tumors during the course of irinotecan therapy and its uptake by intestinal macrophages and DCs, followed by activation of the AIM2 inflammasome, may partially explain the intestinal damage associated with chemotherapy (Lian et al, 2017). Similar responses may be achieved by extracellular DNA produced during the process of NETosis in response to cancer‐derived EVPs (Leal et al, 2017). DAMP proteins, such as versican and galectin 9, have also been found to be expressed in EVPs from pancreatic and lung cancer tissue, but not in EVPs from adjacent or distant tissues (Hoshino et al, 2020). These molecules can trigger secretion of pro‐ and anti‐inflammatory cytokines by distant immune cells and promote systemic inflammation and evasion from T cell anti‐tumor activity (Wight et al, 2020; Yang et al, 2021d).
Cachexia
Cancer‐associated cachexia is a systemic disorder associated with body weight loss and unbalanced energy expenditure, which cannot be reversed via nutritional intervention (Argiles et al, 2018; Baracos et al, 2018). Inflammation and metabolic alterations in the skeletal muscle, liver, gut, and adipose tissues together contribute to the pathology of this paraneoplastic disorder. EVPs may be important mediators in the communication between different organs in cachexia, as suggested by evidence that EVPs from CRC, gastric cancer, and pancreatic cancer cells induce weight loss and muscle atrophy in mice (Argiles et al, 2018; Zhang et al, 2019b; Di et al, 2021). The role of EVPs in cancer‐induced cachexia is further supported by the finding that mice harboring tumors and treated with GW4869 or mice injected with Rab27 knockdown tumors did not develop muscle wasting (Zhang et al, 2017a; Qiu et al, 2020a).
The metabolic changes occurring in both adipose tissue and muscle are affected by cancer‐derived EVPs. To illustrate, white adipose tissue (WAT) browning is induced by EVPs from colon cancer and gastric cancer cell lines both in vitro and in mice, and it can be reversed by inhibiting exosome release via GW4869 (Zhang et al, 2019b; Di et al, 2021). These functions were driven by EVP cargo of miR‐146‐5p and ciRS‐133, which regulate genes involved in oxygen and glucose consumption, differentiation, and transcriptomic changes consistent with WAT browning (Zhang et al, 2019b; Di et al, 2021). In gastric cancer patients, EVP ciRS‐133 was found enriched more than 120 folds in comparison to healthy controls and was an independent biomarker of brown adipose tissue mass and body fat percentage (Zhang et al, 2019b). Lipolysis and adipogenesis by adipose tissue‐derived MSCs (hAD‐MSCs) are also perturbed in patients with cachexia. Pancreatic cancer EVPs, including those from patient blood, activate lipolysis of subcutaneous adipocytes via transfer of the hormone adrenomedullin early on during cancer progression (Sagar et al, 2016). Lipolysis and cachexia induced by LLC‐derived exosomes were suppressed by GW4869 in vitro and in vivo (Hu et al, 2018), while A549 lung cancer cell–derived EVPs activate TGF‐β signaling in hAD‐MSCs, reducing adipogenic differentiation and thus contributing to adipose tissue loss (Wang et al, 2017c).
Another aspect of cancer‐associated cachexia known to be affected by EVPs is muscle wasting, associated with degradation of myofibrillar proteins and changes in muscle metabolism (Fearon et al, 2012). The transfer of EVP miRNAs from oral squamous carcinoma cells induces endoplasmic reticulum stress in recipient muscle cells, resulting in myotube atrophy and apoptosis (Qiu et al, 2020a). Moreover, EVP‐associated Hsp70/90 released by a range of cachexic tumor types induces myotube catabolism and muscle wasting via TLR4/p38 MAPK signaling in muscle cells (Zhang et al, 2017a). Muscle‐derived stem cells are also affected by cancer‐derived EVPs, such as in the case of osteosarcoma‐derived EVPs that induce Notch signaling in MDSCs, leading to decreased myogenesis and muscle atrophy (Mu et al, 2016).
Organ failure
Secondary organ failure is a major cause of death in cancer patients and is linked to systemic tumor‐derived factors and to tissue injury upon surgery, chemotherapy, or radiotherapy. Acute liver failure is a common condition in patients at advanced stages of cancer and can develop in the absence of malignant invasion of the liver (Smith & James, 1998), suggesting the involvement of distant signaling via cytokines and EVPs. Hepatic failure can develop following microemboli, which can be directly induced by tumor‐derived EVPs (Gomes et al, 2017; Leal et al, 2017), or might be induced by the infiltration of immune cells in the liver in response to EVP inflammatory and chemoattractant mediators, such as DAMPs (Hoshino et al, 2020; Wu et al, 2010). Cardiovascular failure is another major aspect of cancer at late stage. By inducing thromboembolism, EVPs may contribute to the most fatal aspects of cardiovascular failure. Evidence also shows that, in patients under immune checkpoint blockade therapy, EVPs from PD‐1 inhibitor‐treated macrophages induce cardiac senescence in cardiomyocytes and may partially explain the cardiovascular adverse events of immunotherapy (Xia et al, 2020). The expression profile of serum EVPs from heart failure patients after transplant differs from patients with no organ rejection, in particular in terms of proteins involved in inflammation and immunity (Kennel et al, 2018), further pointing to a role for EVPs in the etiology of immune‐driven cardiac complications.
EVPs as promising nanotools in oncology
EVPs hold tremendous potential as prognostic and deliverable tools for the clinical management of cancer. Three major lines of research are actively pursued and will be the subject of this section. First of all, the involvement of EVPs in cancer progression and in response to clinical intervention is reflected in altered EVP levels or EVP cargoes in blood and tissues of cancer patients. These EVPs collectively form a reservoir of biomarkers to be mined for early cancer detection, treatment monitoring, and prognosis of disease development. Secondly, endogenous and engineered EVPs have been explored as delivery vehicles for various types of therapeutics and imaging reagents in animal models. They hold greater potential than conventional nanoparticles such as liposomes due to their low immunogenicity and toxicity, unique targeting specificity, which can be further engineered by expressing specific surface features, and capability to deliver a variety of therapeutics ranging from drugs, nucleic acids, to immune adjuvants and imaging molecules. Lastly, we will review the research effort on targeting the EVP production and uptake for cancer management. Given the fact that EVPs mediate the intercellular communication in many fundamental physiological processes, understanding how to target the production and uptake of EVPs specifically involved in cancer progression will be critical in this line of research.
Cancer diagnostic and prognostic biomarkers
Tissue‐derived biomarkers
The use of tissue biopsies to analyze the production and content of EVPs is an important tool in the discovery of cancer biomarkers. Tumor tissue‐derived EVPs in fact have protein expression profiles that are distinct from adjacent and distant tissues and thus can be utilized for cancer diagnosis (Figure 4). In the case of pancreatic cancer, pro‐inflammatory mediators, such as S100A13 and periostin, were exclusively found in tumor‐tissue‐derived EVPs (Hoshino et al, 2020). Several ubiquitous tissue‐derived EVP proteins (e.g., thrombospondin and versican) could be used to discriminate tumor tissue versus non‐tumor tissues with 90% sensitivity and 94% specificity, while other EVP proteins could discriminate between different tumor types (Hoshino et al, 2020). Among them, EVPs from pancreatic cancer tissues were particularly enriched in factors involved in coagulation and EMT, while lung cancer tissues produced EVPs enriched in RNA processing proteins, which suggests selective packaging or different microenvironmental sources of EVPs in these two cancer types.
Figure 4. EVPs contribute to organotropism and pmn formation.
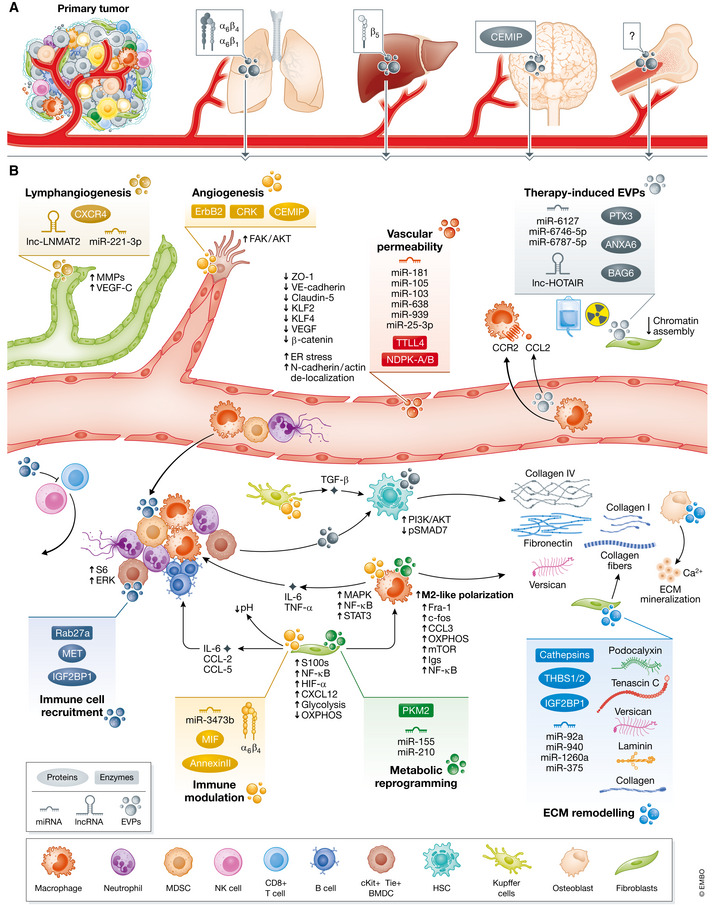
(A) Several EVP surface receptors drive organotropism to different metastatic sites. EVP integrins α6β4 and α6β1 drive lung tropism, integrin β5 drive liver tropism, and CEMIP drives brain tropism. The determinants of EVP bone tropism are yet not known. The tissue‐specific uptake of EVPs promotes pre‐metastatic niche formation and defines organotropism of disseminating tumor cells. (B) EVP uptake by stroma and immune cells at distant sites induces the generation of a favorable pre‐metastatic niches, highly receptive for tumor cell seeding and outgrowth. Endothelial and lymphatic cell proliferation and vascular permeability, recruitment of BMDCs, activation of resident immune cells, remodeling of ECM, and metabolic reprogramming of fibroblasts and immune cells are the major phenotypical changes associated with pre‐metastatic niche formation. EVPs released by the primary tumor after chemotherapy and radiotherapy are also involved in pre‐metastatic niche formation. CEMIP, Cell migration‐inducing and hyaluronan‐binding protein; ECM, extracellular matrix.
The profile of tissue‐derived EVPs is directly related to the composition of circulating EVPs. By analyzing the proteome of tissue‐ and plasma‐derived EVPs of pancreatic and lung cancer patients, Hoshino and collaborators demonstrated that several protein markers were present in both tissue‐derived and plasma‐derived EVPs and that they were selective for the cancer cell of origin (Hoshino et al, 2020). In contrast, other proteins were only present in plasma EVPs. In pancreatic cancer, many proteins restricted to immune cell lineages were found exclusively expressed in plasma EVPs, suggesting systemic immune dysregulation. EVP size distribution can also potentially distinguish EVPs derived from tumor cells and other cells or organs. In NSCLC, pulmonary vein EVPs were smaller than EVPs from peripheral veins and were associated with a higher risk of relapse and shorter overall survival (Navarro et al, 2019). It could be speculated that, during lung cancer progression, tumor‐derived EVPs become enriched in smaller particles, such as exomeres, with metabolism‐reprogramming and tumor‐promoting properties. These findings further underscore the heterogeneity of cell and organ sources that contribute to the pool of circulating EVPs.
Liquid biopsies
The use of EVPs from bodily fluids as predictive markers of cancer, cancer type, and stage of disease is of particular importance in the absence of tissue biopsies and for large‐scale screening of the general population, providing that the latest Minimal Information for Studies of Extracellular Vesicles (MISEV) guidelines are met for EVP preparation, analysis, and reporting (Thery et al, 2018). The use of EVPs as biomarkers might overcome some limitations associated with CTCs, including their very low concentration (Pantel & Alix‐Panabieres, 2019). To illustrate, 1 milliliter of plasma from glioblastoma patients contains more than 10 billion circulating EVPs and it has been estimated that at least 1 billion of them are cancer cell‐derived EVPs (Fraser et al, 2019). The high persistence of EVPs in blood is probably due to the fact that they may evade clearance by patrolling NK cells and phagocytes in the circulation and thus may represent a more stable picture of homeostasis. Moreover, EVP cargo reflects the contents of the cell of origin, including surface markers and oncogenes, thus allowing to capture the phenotypic heterogeneity and invasive potential of the primary tumor and aiding in the estimation of metastatic risk. In contrast, no more than 17% of CTCs in peripheral blood have a phenotype consistent with MICs (Lawson et al, 2015), suggesting that liquid biopsies based on CTCs may largely miss patients at high risk of metastasis. On a technical note, EVPs are stable and retain functional activity if stored frozen for several months and can thus be used in retrospective studies (Mendt et al, 2018). Finally, prognostic EVP markers may be more potent than cancer‐ and CTC‐derived biomarkers. Melo and colleagues showed that EVP glypican‐1 could be used to distinguish samples from healthy controls and from patients with pancreatic pre‐cancerous lesions, allowing for early detection, which is not achievable with the current CA19‐9 PDAC biomarker (Melo et al, 2015). Thus, EVPs can be utilized as bona fide liquid biopsies for cancer diagnosis and prognosis.
Many studies have highlighted the association between EVP‐based liquid biopsies and disease. Various EVP cargos, including RNAs (miRNAs, mRNAs, lncRNAs, circRNAs, and tsRNAs), DNA, proteins, enzymes, glycoproteins (Ko et al, 2018; Chen et al, 2020g), and lipids (Skotland et al, 2017a), are dysregulated in bodily fluids of cancer patients, as summarized in Figure 4. Other parameters found to be associated with cancer diagnosis and poor prognosis include increased EVP mRNA or protein (Peinado et al, 2012; Dijkstra et al, 2014), increased absolute EVP numbers (Kharmate et al, 2016; Galbo et al, 2017; Navarro et al, 2019; Moloney et al, 2020), and EVP size (Navarro et al, 2019). Notably, RNA editing in originating cells was also reflected by EVP cargo. Nigita and colleagues have shown that the edited forms of three miRNAs, miR‐381‐3p, miR‐589‐3p, and miR‐411‐5p were dysregulated in EVPs from patients with lung cancer, despite no changes in their rate of editing (Nigita et al, 2018). Hoshino and colleagues have recently characterized the complete proteomic profile of EVPs from plasma samples of 16 different cancer types and identified predictive proteins, mainly immunoglobulins, overrepresented or downregulated in cancer‐associated EVPs that could discriminate cancer versus non‐cancer or different types of cancers with more than 95% sensitivity and 90% specificity (Hoshino et al, 2020). This large study has provided evidence of the feasibility and potential use of liquid biopsy markers for the early diagnosis of cancer of unknown origin, which might have applications in the large‐scale screening of the general population (Figure 5).
Figure 5. EVPs serve as biomarkers in cancer.

List of the most recent EVP cargoes found dysregulated in bodily fluids or tissue biopsies in different cancer types. Only EVP biomarkers significantly correlating with clinicopathological parameters (such as overall survival or relapse‐free survival) are shown. Parameters correlating with therapeutic response are indicated. Arrows denote upregulated or downregulated markers.
Sequential tumor stages are also associated with specific EVP markers. To illustrate, stage III and stage IV melanoma patient EVPs had increasing amounts of tumor cell markers TYRP2 and pro‐metastatic MET/pMET, and patients with stage IV disease had EVPs with high levels of VLA‐4 and HSP70, supporting the value of these markers for the diagnosis and prognosis of melanoma (Peinado et al, 2012). In PDAC patient samples, EVP markers, such as S100A13, periostin, and basigin, could differentiate between various PDAC stages and distinguish between PDAC and chronic pancreatitis with high specificity and selectivity (Jiao et al, 2019; Hoshino et al, 2020; Yu et al, 2020; Huang et al, 2020a). For example, MIF has been found to be present at significantly higher levels in EVPs from patients with PDAC and in mouse models of pancreatic intraepithelial neoplasia (PanIN) and PDAC relative to healthy controls (Costa‐Silva et al, 2015). Preliminary evidence suggests that metastatic progression could be predicted by EVP markers in osteosarcoma (Wang et al, 2020f), gastric cancer (Ohzawa et al, 2020), CRC (Teng et al, 2017), oral squamous cell carcinoma (Li et al, 2019c), and PDAC (Melo et al, 2015). Larger studies are needed to determine the ability of EVP markers to predict the onset of metastatic disease or diagnose occult metastasis.
Interestingly, mutated oncogenes have also been detected in plasma of cancer patients. EGFRvIII, the mutant form of EGFR, was detected in tumor samples and plasma EVPs from glioblastoma patients (Skog et al, 2008; Fraser et al, 2019). KRAS G12D and TP53 R273H mutated DNAs were specifically detected in EVPs from the plasma of patients with chronic pancreatitis and PDAC, suggesting early onset of these mutations in the pancreatic microenvironment (Yang et al, 2017b). Similarly, mutant KRAS mRNA (either the KRAS G12D or KRAS G12V variant) was found in serum EVPs of patients with PDAC, but interestingly they were restricted to cancer cell‐derived EVPs expressing glypican‐1. Using c‐Met expression in plasma EVPs, Lux and colleagues were able to diagnose PDAC with a sensitivity of 72.4% and specificity of 89.5% (Lux et al, 2019). Thus, EVP DNA has the potential to provide clinical information on the mutational status of the tumor cell of origin and help drive personalized therapy. Mutated genes or proteins, however, may not be good targets to predict cancer incidence, as they are not detected in EVPs from healthy individuals (Yang et al, 2017b).
Measures of therapeutic responses
Therapeutic intervention changes the profile of circulating EVPs (see Build it up: Cancer promotion). Overall, levels of EVPs markers are decreased significantly in gastric and breast cancer patients after surgery, suggesting that they are mostly cancer‐derived (Tang et al, 2019a; Zheng et al, 2020a). Similarly, Gumireddy and colleagues showed that levels of AKAP4+ EVPs decrease after NSCLC resection but undergo a surge later only in patients experiencing recurrence (Gumireddy et al, 2015). After chemotherapy, patients with progressive disease had higher levels of expression of EVP markers than patients with partial or complete responses in CRC (Yang et al, 2018b), breast cancer (Aharon et al, 2017; Tang et al, 2019a), lung cancer (Yuwen et al, 2017, 2019; Ma et al, 2019b; Zhao et al, 2020d), rectal cancer (Kral et al, 2018), prostate cancer (Khan et al, 2012), ovarian cancer (Yang et al, 2019a), and rhabdomyosarcoma (Ghamloush et al, 2019). Similarly, circulating EVP numbers and size increase in patients with CRC and breast cancer after chemotherapy (Aharon et al, 2017; Bar‐Sela et al, 2020). EVP miRNAs and proteins were also found to be dysregulated in relation to radiotherapy response in several cancer types, including glioma (Li et al, 2020f), esophageal squamous cell carcinoma (Luo et al, 2019; Chen et al, 2021c), NSCLC (Dinh et al, 2016), and brain metastasis (Chen et al, 2021d). Importantly, EVPs may be used as liquid biopsies to determine the feasibility and efficacy of immunotherapy. Not only does blood EVP PD‐L1 reflect PD‐L1 and CD8+ T cell infiltration in tumors, but the EVP PD‐L1 and miRNA signature in plasma is also informative with respect to PD‐L1 expression and efficacy of immunotherapy, especially in NSCLC, allowing for the selection of patients most likely to benefit from checkpoint blockade inhibitors (Katakura et al, 2020; Peng et al, 2020; Shimada et al, 2021).
In conclusion, there is emerging evidence that EVP profiles change in response to therapeutic interventions, with more pronounced alterations in protein and RNA/miRNA levels in response to chemotherapy and radiation therapy. We could speculate that this effect is due to either induction of transcriptomic changes in treated cells (tumor adaptation) or a Darwinian selection of resistant clones, and thus EVP cargo might provide information of tumor evolution over the course of therapy (Vendramin et al, 2021). Although the exact nature, cause, and applicability of this expression change need to be further determined, we could hypothesize that EVPs might be employed as liquid biopsies to measure therapeutic readouts that cannot currently be assessed by conventional imaging methods, such as presence of occult residual disease or minimal (< 10%) tumor shrinkage (Martens et al, 2014). Additionally, circulating EVPs might offer early response prediction within days from treatment onset, instead of at the end of the therapy cycle, allowing for faster determination of therapeutic strategies and treatment plan adjustments.
Therapeutic delivery strategies
Advances in nanotechnology have led to the engineering of a new generation of therapeutics‐loaded nanomaterials with improved stability, tissue penetration, and intracellular targeting compared to traditional agents (Mitchell et al, 2021). Despite the potential of engineered nanoparticles to become a new frontier of precision medicine, some limitations, including clearance, toxicity, and nonspecific distribution, still remain. As a natural vehicle for proteins, lipids, and nucleic acids, EVPs present a few advantages over other delivery nanotools. First, EVPs are endowed with very low immunogenicity and can deliver cargoes to various cell types at distant sites without prior clearance by innate immune cells (Lai et al, 2014). EVP size may matter in this regard, as EVPs are in an optimal size range to avoid rapid excretion by the kidneys, while being sufficiently small to avoid opsonization and immune cell recognition (Mitchell et al, 2021). The presence of the “self” marker CD47 and the exposure of negatively charged lipids (e.g., PS) and proteins on EVP surfaces may also explain the resistance to phagocytosis (Kamerkar et al, 2017; Zhang et al, 2018b).
Further, EVPs have a broad tissue biodistribution, including most hematopoietic organs (i.e., liver, spleen, bone marrow, lymph nodes), lungs, and kidneys (Lai et al, 2014; Hoshino et al, 2015; Zhang et al, 2018b). Exomeres preferentially localize to the liver (Zhang et al, 2018b), but their mechanism of tissue homing needs to be further investigated. High accumulation in the liver and spleen has also been observed with engineered nanoparticles (Mitchell et al, 2021). EVPs can cross the blood–brain barrier after intranasal inhalation (Zhuang et al, 2011; Haney et al, 2015), with some limitations, however, due to dosing variability (Mitchell et al, 2021). Importantly, brain homing can be achieved after intracardiac delivery of EVPs from brain‐tropic cells, which preferentially localize to endothelial cells and microglia (Hoshino et al, 2015; Rodrigues et al, 2019), indicating a permeability of the blood–brain barrier to EVPs that has not yet been observed with engineered nanoparticles (Mitchell et al, 2021). Thus, EVPs could be particularly useful as delivery systems for organs with low‐permeability physical barriers, such as the central nervous system and the gastrointestinal tract.
Finally, non‐engineered nanoparticle drug carriers become concentrated preferentially at certain sites, such as the spleen and liver, by virtue of an enhanced permeability and retention effect of the vasculature as well as phagocyte accumulation (Schroeder et al, 2011; Mitchell et al, 2021). In contrast, EVPs have an innate organ distribution, with integrins being major determinants of their tropism (Hoshino et al, 2015). Furthermore, these EVP surface integrins and receptors achieve activation of intracellular signaling cascades with potential therapeutic applications (Costa‐Silva et al, 2015; Hoshino et al, 2015; Rodrigues et al, 2019).
Altogether, this growing evidence suggests that endogenous or engineered EVPs represent promising delivery tools for precision medicine in oncology.
Endogenous EVPs as therapeutic tools
Despite their distinctive tissue homing, cancer cell‐derived EVPs are not desirable therapeutic or delivery tools due to their systemic effects on oncogenesis, pre‐metastatic niche establishment, and thrombosis. EVPs from other cell sources have been evaluated and have shown endogenous functions.
MSCs have often been used as a source of therapeutic EVPs due to their self‐renewal and multipotent properties (Gabrilovich & Nagaraj, 2009). Moreover, MSC‐derived EVPs can mimic the regenerative and immunosuppressive properties of MSCs, but with increased tissue permeability due to their smaller size. A growing body of literature shows promising therapeutic properties of endogenous cargo of adipose‐, bone marrow–, or umbilical cord–derived EVPs from MSCs in ischemic stroke (Xin et al, 2013; Chen et al, 2016a; Huang et al, 2020b; Li et al, 2020b; Zhao et al, 2020e), diabetic retinopathy (Safwat et al, 2018; Gu et al, 2020), Alzheimer's Disease (Wendeln et al, 2018; Feng et al, 2020), and arthritis (Wu et al, 2020e). EVPs derived from MSCs ameliorate the severity of IBD in mice by reducing infiltration of macrophages in colon tissue and decreasing their expression of pro‐inflammatory mediators, such as TNF‐ α, IL‐1β, and IL‐7 (Mao et al, 2017; Ma et al, 2019c). MSC‐derived EVPs prevent development of pulmonary complications, such as bronchopulmonary dysplasia (Willis et al, 2020). MSC‐derived EVPs can also prevent liver fibrosis by reactivating HSC autophagy (Qu et al, 2017) and suppressing HSC activation and collagen deposition (Lou et al, 2017). While a beneficial effect of MSC‐derived EVPs has been identified in these pre‐cancerous conditions, multiple findings have shown a deleterious effect of these EVPs on cancer progression. In particular, MSC‐derived EVPs from different sources, including cancer‐associated MSCs, bone marrow aspirates, and adipose tissue, were found to promote cancer cell proliferation (Roccaro et al, 2013), increase tumor growth in mice (Roccaro et al, 2013; Vallabhaneni et al, 2015), suppress CD4+ T cell proliferation (Cheng et al, 2020), promote EMT and migration (Lin et al, 2013; Gu et al, 2016), support ECM remodeling (Yang et al, 2015b), potentiate angiogenesis (Zhang et al, 2015a), and sustain chemoresistance (Ji et al, 2015). Conversely, studies on glioma have reported anti‐tumor effects of MSC‐derived EVPs, such as a reduction of tumor cell proliferation and migration (Lee et al, 2013; Xu et al, 2019a). The nature of this dichotomy is not known. Roccaro and colleagues proposed that pro‐tumorigenic properties could be restricted to tumor‐educated MSCs, while normal MSCs mainly have anti‐tumorigenic effects (Roccaro et al, 2013). Nevertheless, MSC‐derived EVPs, especially from autologous sources, may not be an ideal candidate for cancer treatment.
Another main issue with MSC‐derived EVP isolation is that MSCs have limited self‐renewal capacity and undergo senescence within a few passages in culture. EVPs from senescent MSCs might have deleterious properties, including cancer induction (Severino et al, 2013). Different approaches have been employed to avoid MSC senescence and improve EVP manufacturing, including MSC immortalization via lentiviral MYC transduction (Chen et al, 2011), exposure to hypoxia (Gonzalez‐King et al, 2017; Zhu et al, 2018a), and treatment with small‐molecule inhibitors (Wang et al, 2020e). Despite successfully increasing EVP release, some of these approaches were shown to alter the cargo and functional effect of MSC‐derived EVP and thus strategies to improve EVP manufacturing need to be further evaluated. Among them, the embryonic cell line HEK293T has been tested for safety and efficacy in preclinical experiments (Liang et al, 2020a), but the role of their endogenous cargo still needs to be elucidated. Similarly, reticulocytes have been used to produce EVPs with low immunogenicity as scaffolds for drug and magnetic particle loading (Blanc et al, 2005; Qi et al, 2016). Finally, Pan and colleagues have shown high purity and yield of urinary EVPs for autologous delivery, with more than 3 mg of EVPs per half liter of urine (Pan et al, 2020).
Immune cells may be another valuable source of EVPs for cancer management, mainly due to their endogenous anti‐inflammatory and anti‐tumor properties, but their applicability entirely depends on the immune cell source. EVPs from bone marrow‐derived macrophages reduce hematopoiesis in atherosclerotic mice (Cianciaruso et al, 2019) and promote an anti‐tumor T cell response (Bouchareychas et al, 2020) via their cargo of miRNAs and lipid biosynthesis enzymes. On the contrary, EVPs from M2‐polarized macrophages mediate resistance to cisplatin and apoptosis of gastric cancer cells via miR‐21 delivery and activation of the PI3K/AKT signaling pathway (Zheng et al, 2017) and thus may not be an ideal tool for biomolecule delivery. EVPs from Treg cells reduced DSS‐induced IBD in mice by decreased apoptosis of intestinal epithelial cells through miR‐195a‐3p transfer (Liao et al, 2020). EVPs from NK cells also have high potential as therapeutic tools, as they share the same tumor cytotoxic properties of the producing cells (Lugini et al, 2012; Shoae‐Hassani et al, 2017; Zhu et al, 2017, 2018b). Other advantages of NK cell‐derived EVPs also include their selective uptake by tumor cells, but not by other resting PBMCs, and their selective homing to tumors in vivo (Lugini et al, 2012; Zhu et al, 2018b). To overcome the low availability of NK cell EVPs, Zhu and colleagues designed a novel protocol with a higher efficiency of isolation of NK EVP mimetics. Finally, further information on the immune properties of EVPs has been gained by studies on DC‐derived exosomes (Dex). Zitvogel and colleagues were the first to show that Dex from immature DCs expresses MHC‐I and MHC‐II complexes and that, when mature DCs are exposed to tumor‐specific antigens, their EVPs can halt tumor growth in vivo by inducing CD8+ T cell activation and immunize the host after a single injection, alone or in conjunction with adjuvants (Zitvogel et al, 1998; Chaput et al, 2004). Remarkably, Dex immunization was more effective at controlling tumor progression than DC adoptive transfer (Zitvogel et al, 1998). Further work by Thery and colleagues and other groups elucidated that Dex can activate CD4+ and CD8+ T cells in vivo via autocrine and paracrine mechanisms, with potential innate anti‐tumor effects. First, Dex from mature DCs, and to a lesser extent immature DCs, are taken up by other DCs, especially the CD8α− subtype, and B cells to present EVPs markers on their MHCs and engage CD4+ T cell activation into effector T cells (Thery et al, 2002; Segura et al, 2005a). Further, MHCs on Dex taken up by DCs work directly as adjuvant for their antigen‐presenting activity (Thery et al, 2002). Moreover, Dex themselves present peptides to T cells to induce T cell activation and proliferation, a process also dependent on Dex expression of ICAM‐1 and B7.2 surface proteins (Hwang et al, 2003; Segura et al, 2005a,b). Lastly, Dex can also prime NK cells via their cargo of IL‐15α and NKG2D ligand, thus eliciting an anti‐tumor immune response (Viaud et al, 2009). Several clinical trials using DC‐derived EVPs as cancer vaccines have been designed, with increasing levels of T cell responses due to improved Dex manufacturing (Lamparski et al, 2002; Escudier et al, 2005; Viaud et al, 2011; Damo et al, 2015).
Finally, EVPs from dietary sources hold great promise as natural nanoparticles for treatment and delivery. EVPs containing RNA (miRNA, mRNA, and lncRNA) and proteins are found in the whey fraction of human and animal milk (Izumi et al, 2015; van Herwijnen et al, 2016; Zeng et al, 2019). Milk EVPs are resistant to gastric, bile, and pancreatic juices, thus contributing to a major portion of nucleic acids in blood cells and tissues (Baier et al, 2014; Zeng et al, 2019; Wu et al, 2019a), and are well tolerated in vivo upon oral administration (Arntz et al, 2015; Somiya et al, 2018; Stremmel et al, 2020). Cow milk may represent the most available and cost‐effective source of EVPs. A number of studies have provided evidence that bovine milk‐derived EVPs are taken up by cecal microbiota and intestinal cells, where they reduce oxidative stress, and by immune cells, including human PBMCs (Arntz et al, 2015; Somiya et al, 2018; Zhou et al, 2019b; Wang et al, 2021c). The endogenous cargo of milk‐derived EVPs was shown to have immunoregulatory properties by reducing systemic inflammation and preventing the onset of arthritis and IBD (Arntz et al, 2015; Wu et al, 2019a; Stremmel et al, 2020). Importantly, milk EVPs were found to induce activation of NK cells and CD8+ T cells directly (Komine‐Aizawa et al, 2020), suggesting that these EVPs may directly mediate anti‐tumor immune responses. The uptake of EVPs from bovine raw milk was also observed for several cancer cell lines, including leukemia (Izumi et al, 2015), colon carcinoma (Wolf et al, 2015), and ovarian cancer cells (Benmoussa et al, 2020), although further investigations are necessary to determine the effect of the endogenous cargo of milk EVPs on cancer progression.
Similar to all other eukaryotes, plant cells are endowed with MVBs and can release exosome‐like vesicles (An et al, 2007). EVPs have been successfully isolated from the juice of edible plants, such as carrots, ginger root, grapes, and citrus fruits, including grapefruit, clementine, and lemon, and contain a cargo of RNAs and proteins (Mu et al, 2014; Baldini et al, 2018; Stanly et al, 2019). Lemon juice‐derived EVPs have distinctive antioxidant properties on MSCs due to their naturally occurring cargo of citrate, vitamin C, small RNAs, and protein transporters (Baldini et al, 2018). Although surface proteins might be altered at low pH in the stomach and intestine, Mu and colleagues showed active uptake of plant‐derived EVPs in intestinal macrophages and stem cells (Mu et al, 2014). In particular, ginger‐derived EVPs induce the synthesis of immunomodulatory IL‐10 and IL‐6 and promote activation of macrophage Nrf2 and intestinal Wnt/TCF4 pathways, inducing an anti‐inflammatory response in the gut. Although the nature and function of protein and nucleic acid cargo in plant‐derived EVPs need further elucidation, they may represent the new frontier of inter‐species deliverables with beneficial innate properties.
Engineered EVPs
Exogenous EVP cargo allows for tissue distribution and cellular uptake that is not achievable by naturally occurring EVPs or by unshielded therapeutic agents, making engineered EVPs ideal carriers for cancer therapeutic and imaging agents. Different EVP engineering workflows have been devised for this objective. Autologous EVPs may represent the best approach to ensure increased targeting (Liu et al, 2019d), high biocompatibility, and low toxicity, but other types of EVPs also may be valid alternatives in terms of availability, cost‐effectiveness, and endogenous beneficial properties.
Genetic engineering of EVP‐releasing cells allows for enhanced targeting and bioactive properties of EVPs. Lysosome‐associated membrane glycoprotein 2 (Lamp2) is highly packaged into EV membranes and has been used as part of an expression construct for the introduction of targeting moieties on EVs. Alvarez‐Erviti and colleagues were the first group to report the generation of autologous Dexs engineered to express a fusion construct between the extracellular N‐terminus of Lamp2 and peptides binding specific acetylcholine receptors in the brain and muscle of mice. These EVs were found to selectively home to these tissues in vitro and in vivo (Alvarez‐Erviti et al, 2011). Along the same lines, EVPs expressing CD63‐Apo‐A1 were found to have increased uptake in liver cancer cells via interaction of Apo‐A1 with their scavenger class B type 1 receptor (Liang et al, 2018), while expression of lipotropic peptide‐TNF‐α vectors in EVP‐producing cells allows anchorage of TNF‐α to the EVP membrane (Zhuang et al, 2020). Through a similar approach, other groups have engineered EVPs to express several targeting molecules: RGD peptides specific for αVβ3 integrin on breast cancer and gastric cancer cells (Tian et al, 2014b; Xin et al, 2021; Gong et al, 2019); Her2‐binding antibody mimetics and designed ankyrin repeated proteins (DARPins) to target Her2+ colorectal and breast cancer cells, respectively (Gomari et al, 2018; Limoni et al, 2019; Liang et al, 2020a); aminoethylanisamide, a ligand for sigma receptors in NSCLC (Choi et al, 2018); folic acid that recognizes folate receptors expressed on tumor cells (Pi et al, 2018; Li et al, 2018f; Feng et al, 2021), and tumor MUC‐1‐interacting 5TR1 aptamer (Schindler et al, 2019; Bagheri et al, 2020). These EVPs showed better uptake by cancer cells in vitro and accumulation in tumors in mice, were well tolerated in vivo, and did not cause any organ toxicity or hematological or histopathological abnormalities (Liang et al, 2020a).
EVPs can also be directly decorated with molecules not incorporated into their membranes. For example, allowing cells to produce EVPs in the presence of azide‐choline drives the incorporation of azide groups on membrane lipids of the EVP surface, followed by conjugation with antibodies for targeted delivery (Nie et al, 2020). This approach results in the local release of antibodies at low pH conditions, such as in the acidic tumor microenvironment. EVPs can be conjugated with PEGylated antibodies for direct cancer antigen targeting, such as in the case of antibodies against somatostatin receptor 2 on neuroendocrine tumor cells (Si et al, 2020). Similarly, EVPs can be coated with Fe3O4 superparamagnetic nanoparticles coupled with antibodies against A33 antigen, expressed in more than 95% of CRC biopsies (Li et al, 2018d). Other engineered EVP products have been reported. Morishita and colleagues successfully achieved the synthesis of EV particles expressing a fusion product of exosome‐specific lactadherin bound to extracellular streptavidin (Morishita et al, 2016), which works as a platform for EV coating with biotinylated DNAs, RNAs, and proteins. Astutely, Pi and colleagues devised a workflow to decorate EVPs with nucleic acids, peptides, or naturally occurring vitamins. Their innovative method makes use of the arrow‐shaped motor packaging RNA (pRNA) of the bacteriophage phi29 (pRNA‐3WJ), engineered to display cancer‐selective ligands, such as folate, selective for folate receptor on cancer cells, and RNA aptamers of prostate‐specific membrane antigen (PSMA) and EGFR. Additionally, cholesterol conjugation to different extremities of the arrow‐shaped pRNA‐3WJ allows for different conformations of RNA loading on EVP membranes or partial internalization in the EVP lumen (Pi et al, 2018).
Most intravenously injected tumor cell‐derived EVPs are quickly cleared by macrophages, especially in the liver and spleen (Imai et al, 2015). To overcome this, genetic engineering of donor cells or direct EVP manipulation can be employed to improve their circulation half‐life. In this regard, the expression of the “do not eat me” signal CD47 on EVPs results in better tumor distribution and lower clearance by circulating myeloid cells (Kamerkar et al, 2017; Mendt et al, 2018; Lv et al, 2020a). Kim and colleagues incorporated PEG into EVPs as a method to decrease immunogenicity and blood clearance and to add vector moieties to EVPs altogether (Choi et al, 2018).
The targeting ability of EVPs has also been achieved by coupling them with magnetic nanoparticles, such as superparamagnetic iron oxide nanoparticles, and driving their tumor infiltration via application of a local magnetic field (Li et al, 2018d; Zhuang et al, 2020). Through a different approach, Qi and colleagues employed the presence of transferrin receptors on the surface of reticulocyte‐derived EVs, which are produced during maturation of reticulocytes into erythrocytes, to achieve EV coating with superparamagnetic magnetite colloidal nanocrystal clusters (SMCNCs). These SMCNC‐coupled EVs could be directed to hepatoma tumors in mice upon application of a mild magnetic field (Qi et al, 2016). Similarly, EVPs loaded with sinoporphyrin sodium could be induced to accumulate in tumors via guided ultrasound (Liu et al, 2019d).
EVPs as therapeutic carriers
EVPs are naturally occurring carriers of functional genetic information and are endowed with low toxicity and broad tissue distribution in vivo. EVPs show more than 30 times higher cell uptake than other vectors, such as nanoparticles or liposomes (Kim et al, 2016b; Pan et al, 2020). Moreover, EVPs are resistant to harsh environmental conditions, such as low pH in gastric juices and shear stress in the blood, and it is thus conceivable that they can be employed as delivery vehicles for drugs, nucleic acids, and imaging agents in cancer patients. EVPs hold particularly great promise as delivery vehicles for anti‐cancer drugs, especially for compounds with low solubility. Because of their targeted delivery and their ability to cross most physical barriers in the body, EVP‐encapsulated drugs have the potential to improve localized treatments while minimizing side effects from off‐target delivery, which is the main limitation of cancer chemotherapy (Hadla et al, 2016; Schindler et al, 2019). EVP‐loaded chemotherapeutics also have a stronger cytotoxic effect than free drugs (Saari et al, 2015), providing room for reduced clinical doses. Drug loading has been achieved by incubation and co‐centrifugation of EVPs with drugs, leading to passive surface binding, EVP permeabilization, or EVP sonication or electroporation in the presence of soluble drugs, which achieves drug loading inside the EVP membrane (Tian et al, 2014b; Saari et al, 2015; Kim et al, 2016b; Liang et al, 2020a). Based on these approaches, EVPs from various cancer or macrophage cell lines have been successfully loaded with drugs, including doxorubicin (Hadla et al, 2016; Schindler et al, 2019; Wei et al, 2019; Bagheri et al, 2020; Tian et al, 2014b; Qi et al, 2016; Kim et al, 2016b; Gomari et al, 2018, 2019; Gong et al, 2019), paclitaxel (Saari et al, 2015; Kim et al, 2016b; Choi et al, 2018; Schindler et al, 2019; Bagheri et al, 2020; Zhang et al, 2020c), 5‐fluorouracil (Liang et al, 2020a), erastin (Yu et al, 2019a), aspirin (Tran et al, 2019), cisplatin (Li et al, 2020c; Zhang et al, 2020e), romidepsin (Si et al, 2020), cabazitaxel (Qiu et al, 2020b), and atorvastatin (Nooshabadi et al, 2020). EVP‐bound drugs were shown to be released into the cell cytoplasm after EVP endocytosis and caused cancer cell death, with higher efficiency and lower doses than drugs alone (Saari et al, 2015; Kim et al, 2016b; Gomari et al, 2019; Yu et al, 2019a). Growth of primary tumors and lung metastasis in mice was found to be reduced in response to EVPs loaded with doxorubicin (Tian et al, 2014b; Qi et al, 2016; Gong et al, 2019; Schindler et al, 2019; Bagheri et al, 2020) and paclitaxel (Kim et al, 2016b; Choi et al, 2018), respectively. Importantly, EVPs could be loaded with drug payloads that would be too toxic in their free form but caused no side effects if selectively delivered to tumors (Si et al, 2020). More indirectly, EVPs can be loaded with nanoparticle‐carrying drugs or other therapeutic agents, which makes them functional carriers of membrane‐soluble agents. Zhao and colleagues have reported the generation of breast cancer cell–derived EVPs encapsulating cationic bovine serum albumin (CBSA) conjugated with anti‐S100A4 siRNA, with EVPs dictating organ distribution while CBSA served as a non‐antigenic and biodegradable deliverable. These EVPs successfully localized to the lungs of mice and prevented metastasis formation in a breast cancer mouse model (Zhao et al, 2020b).
EVPs can also be employed for the sensitization of tumor cells to other types of chemotherapy, such as sonodynamic and hypothermic therapy. Cancer cell‐derived EVPs loaded with sinoporphyrin sodium, an organic sonosensitizer, spontaneously distributed to homotypic primary and metastatic tumors and simultaneously triggered DVDMS intracellular distribution to mitochondria and induction of cytotoxic ROS in response to ultrasound. The application of therapeutic ultrasound further potentiated the cytotoxic effect of EVP‐associated DVDMS, reducing growth of primary and metastatic tumors (Liu et al, 2019d). Lv and colleagues showed the efficacy of EVP‐thermosensitive liposomal hybrids to localize to metastatic peritoneal carcinoma and release their cargo of GM‐CSF and docetaxel in response to localized hypothermia. In turn, the EVP content inhibited tumor cell proliferation directly (via doxorubicin), while activating the phagocytic activity of macrophages against tumor cells and promoting an anti‐tumor adaptive immune response (via GM‐CSF) (Lv et al, 2020a).
EVP‐encapsulated nucleic acids are more protected from degradation by blood nucleases, ensuring better tissue delivery and achieving selective organotropism and thus have the potential to serve as tools for gene therapy delivery. For example, injection of siRNA encapsulated in Lamp2‐ligand‐expressing exosomes achieves a much more restricted tissue distribution than naked siRNA in mice and effectively depletes targets in recipient cells (Alvarez‐Erviti et al, 2011). Survivin siRNA‐loaded EVPs with tumor tropism dictated by surface PSMA aptamer‐pRNA‐3WJ, EGFR aptamer‐pRNA‐3WJ, and folate‐ pRNA‐3WJ abrogated the growth of prostate, breast, and colorectal cancer, respectively (Pi et al, 2018). Importantly, EVP siRNA did not induce immune stimulation in vivo and achieved a level of target silencing comparable to fivefold higher amounts of naked siRNA. Kamarkar, Mendt and colleagues successfully hindered pancreatic cancer development and metastasis by silencing oncogenic KrasG12D via siRNA and shRNAs loaded into fibroblast and MSC‐derived EVPs and with much higher efficiency than engineered liposomes (Kamerkar et al, 2017; Mendt et al, 2018). Anti‐miRNAs work as traps for endogenous miRNAs with known pro‐cancerous effects. Liang and colleagues used 5‐fluorouracil and anti‐miR‐21 loaded HEK293T EVPs to induce expression of tumor‐suppressor PTEN, growth arrest, and apoptosis of colorectal cancer cells, and reduction of tumor growth in vivo, while anti‐miR‐21 alone failed to reach a sufficient therapeutic effect (Liang et al, 2020a). Loading of miRNA‐26 on Apo‐A1‐engineered EVPs allowed selective silencing of their targets Cyclin E2 and CDK6 and reduced migration and proliferation in liver cancer HepG2 cells (Liang et al, 2018). Several other miRNAs with anti‐tumor functions were overexpressed in donor cells or directly transfected in EVPs, leading to their enrichment in EVPs that further induced cancer cell apoptosis and chemo‐ and radio‐sensitivity, and reduced tumor growth in vivo (Gong et al, 2019; Pomatto et al, 2019; Liu et al, 2019b; Kobayashi et al, 2020; Konishi et al, 2020; Kulkarni et al, 2020; Yao et al, 2020; Sharif et al, 2021). Nucleic acids were also loaded into EVPs from dietary sources. Some studies have shown successful transfection of exogenous miRNA or pro‐apoptotic and oncogene‐directed siRNAs into raw bovine milk EVPs and delivery to cancer cells in vitro and in vivo (Aqil et al, 2019; Tao et al, 2020a; Del Pozo‐Acebo et al, 2021; Munagala et al, 2021). In a similar manner, ginger‐derived EVPs were conjugated with cholesterol‐pRNA‐3WJ constructs and folic acid to deliver survivin siRNA to CRC cells expressing folate receptors (Li et al, 2018f). Together, these reports provide proof‐of‐principle evidence that engineered dietary EVPs are a nontoxic and cost‐effective delivery tool for therapeutic nucleic acids.
Preclinical studies suggest that EVPs could also be engineered to induce phagocytosis of tumor cells. HEK293T EVPs overexpressing signal regulatory protein α (SIRPα) interact with the “don't eat me” CD47 signal on tumor cells, thus enhancing tumor cell phagocytosis in vitro and inhibiting tumor growth in immunocompetent mice. Both direct macrophage phagocytosis and infiltration of anti‐tumor T cells were observed in tumors from mice treated with SIRPα‐EVPs (Koh et al, 2017). Similarly, RAW264.7‐derived EVPs decorated with anti‐CD47 and anti‐SIRPα antibodies induced phagocytosis of 4T1 cells via pH‐responsive antibody release and reduced tumor growth in vivo (Nie et al, 2020).
Finally, some studies have demonstrated successful use of EVPs as immune adjuvants, with potential applications for cancer immunotherapy. Exosomes from DCs exposed to or expressing tumor antigens induce T cell activation and anti‐tumor immunization (Zitvogel et al, 1998; Thery et al, 2002; Segura et al, 2005a; Lu et al, 2017; Li et al, 2018a). Additionally, by loading murine melanoma‐derived EVPs with immunomodulatory cytosine–guanine dinucleotide (CpG) DNA, Morishita and colleagues achieved immunization of mice to tumor‐specific antigens, protecting them from tumor initiation, growth, and metastasis. Specifically, CpG‐loaded EVPs activated DCs and induced antigen presentation to T cells, eliciting an anti‐tumor Th1 response (Morishita et al, 2016). Although similar results were obtained in response to CpG‐conjugated liposomes and nanoparticles co‐injected with tumor‐associated antigens (de Jong et al, 2007; Yan et al, 2014a), CpG‐loaded cancer EVPs contain a complete cargo capable of inducing activation and tumor antigen presentation within the same antigen‐presenting cell for anti‐tumor immunization.
EVPs as imaging tools
Engineered EVPs can be used as imaging agents. As discussed previously, optical imaging of fluorescence‐ or bioluminescence‐labeled EVPs is an invaluable tool to study EVP functions in preclinical models, but it has the limitations of rapid photobleaching and low tissue penetration that render it impractical for clinical imaging. Instead, positron emission tomography (PET) imaging of EVP‐encapsulated radioactive isotopes, such as 64Cu and 68Ga, allows for higher sensitivity localization of EVPs in animals (Shi et al, 2019; Jung et al, 2020). Despite these advantages, nuclear imaging may have safety limitations due to radionuclide handling and radiation exposure. Among all the available imaging techniques, intravital visualization of EVPs encapsulating or coated with superparamagnetic nanoparticles via magnetic resonance imaging (MRI) and CT scanning may be the safest (Qi et al, 2016; Li et al, 2018d; Zhuang et al, 2020; Cohen et al, 2021). Engineering of MSC exosomes to express a fusion product of membrane lactadherin and ferritin, naturally occurring MRI reporters, allows for exosome tracing in vivo via MRI imaging (Liu et al, 2020b). Alternatively, near‐infrared (NIR) laser irradiation for cancer imaging has been tested. EVPs labeled with photoluminescent quantum dots engineered to target cell nuclei enabled concomitant intratumoral visualization and hyperthermia‐mediated necrosis of tumor cells, due to their photothermal conversion when irradiated with an NIR laser (Cao et al, 2019). Autologous urinary EVPs were successfully loaded with nanocomposites of gold nanoparticles and the photosensitizer chlorine e6 to produce passion fruit‐like EVPs with low immunogenicity and high tumor homing and retention. The release of nanoparticle‐chlorine e6 complexes in response to laser irradiation induced ROS generation and apoptosis of cancer cells (Pan et al, 2020). Through a different approach, re‐assembled EVPs from pancreatic cancer cells were deprived of internal cargo and loaded with chlorine e6, enabling the photoacoustic imaging of subcutaneous murine melanoma tumors, due to their innate and potent tumor tropism. Simultaneously, these EVPs achieved active control of tumor growth by inducing both production of ROS in tumor cells upon laser irradiation and systemic release of macrophage‐derived chemoattractants, potentially owing to the transfer of tumor antigens to antigen‐presenting cells (Jang et al, 2021b). Although these experiments were performed using human EVPs injected into mice, which may achieve a stronger immune response, they provide proof‐of‐principle evidence for the application of EVPs to in vivo imaging and immunostimulation. It is anticipated that, in the future, labeling EVPs with distinct tropism via these methods will allow visualization of different tumor types, pre‐metastatic niches, and tumor prognostic features, such as vascular leakiness and dormancy.
Another potential frontier of cancer imaging is the labeling of endogenous EVPs. Chen and colleagues described the generation of a hydrogel‐gold nanoparticle‐based biosensor coupled with tumor‐specific DNA aptamers, such as PSMA found in prostate cancer cells and EVPs, which could be visualized via surface plasmon resonance imaging. When incubated with prostate cancer patient serum or cell conditioned medium, the biosensor allowed the measurement of cancer cell‐derived EVP levels in serum. This biosensor could potentially be applied to the magnetic separation of EVPs from bodily fluids, with possible analytical and therapeutic applications (Chen et al, 2020e). Further research in EVP‐specific prognostic biomarkers and probes compatible with intravital EVP imaging will open the way to a new era of cancer imaging and management, where labeled EVPs might allow visualization of sites of vascular leakiness and PMN formation, currently not detectable via conventional clinical imaging.
Prevention of EVP uptake and production for cancer management
Overall, the available data on EVP‐mediated cancer progression highlight the potentially immense utility of targeting EVP secretion and uptake for cancer treatment. Indeed, multiple studies have provided in vivo proof‐of‐principle evidence that interfering with EVP secretion and uptake reduces tumor growth, impedes metastatic progression, and inhibits systemic effects of cancer (Figure 2C). Notably, tumor cell depletion of Rab27 or Ral GTPases attenuated spontaneous lung metastasis of melanoma and mammary carcinoma in mice (Bobrie et al, 2012; Peinado et al, 2012; Ghoroghi et al, 2021). Similarly, treatment with GW4869 decreased tumor growth (Matsumoto et al, 2017; Richards et al, 2017) and impaired systemic effects of cancer, namely cachexia, in tumor‐bearing mice (Hu et al, 2018; Qiu et al, 2020a). Conversely, enhancement of EVP uptake by decreasing 25‐hydroxycholesterol, which blocks EVP internalization, on target cells via genetic deletion of cholesterol 25‐hydroxylase was shown to potentiate spontaneous melanoma metastasis to the lungs (Ortiz et al, 2019), suggesting that inhibiting uptake is anti‐metastatic.
However, several challenges in effectively targeting EVP biogenesis and uptake exist. First, EVPs influence key developmental, physiological, and homeostatic functions. For example, exosomal secretion of the transferrin receptor is necessary for maturation of reticulocytes into erythrocytes (Harding et al, 1983; Pan & Johnstone, 1983). Transfer of exosomes plays an important role in optimizing communication between immune cells (Raposo et al, 1996; Zitvogel et al, 1998; Théry et al, 2002). EVPs also can contribute to cellular fitness by removing cytotoxic DNA (Takahashi et al, 2017; Yokoi et al, 2019). In addition, many of the molecular mediators of EVP biogenesis serve additional functions, which could lead to inappropriate inhibition of other pathways upon their blockade (Jahn & Scheller, 2006; Hurley, 2015; Zhen & Stenmark, 2015). Hence, the biological functions of EVPs and of molecular regulators of biogenesis raise important questions about systemic side effects associated with their inhibition. Furthermore, as detailed above, there are redundant pathways for seemingly all steps of biogenesis, as well as uptake, which could complicate effective target selection and pathway inhibition. Development of EVP‐targeted therapies may therefore require a comprehensive understanding of how such pathways are regulated specifically in cancer and further interrogation into cancer‐associated inducers of EVP production and uptake may reveal novel targets that can be safely inhibited.
Interestingly, there have been some investigations into repurposing already developed drugs for targeting EVP biogenesis or uptake. Screening of compound libraries has identified multiple candidates, including the microbial metabolite manumycin A, tipifarnib, neticonazole, climbazole, ketoconazole, and triademenol, which all reduce biogenesis (Datta et al, 2017, 2018). The mechanisms of action of some of these drugs appear to involve inhibition of Ras‐mediated signaling, which results in reduced levels of biogenesis mediators, such as Alix, nSMase2, and Rab27. They may therefore be primarily active in cancer cells compared with normal cells, making them attractive candidates for further investigation of preclinical efficacy in mouse models of metastasis. The statin simvastatin also was shown to impair EVP biogenesis, and this reduction was associated with decreased levels of Alix (Kulshreshtha et al, 2019). Finally, multiple drugs, including the anti‐hypertensive drug reserpine and the anti‐coagulant heparin, can block EVP uptake. Reserpine was shown to reduce uptake of cancer‐cell‐derived EVPs by non‐cancer cells, preventing melanoma metastasis in vivo (Ortiz et al, 2019). Heparin can impair uptake of EVPs from glioblastoma and oral squamous cell carcinoma cell lines by other cancer and non‐cancer cells, leading to diminished in vitro tumor cell proliferation, migration, and invasion and reduced oral squamous cell carcinoma xenograft tumor growth in vivo (Christianson et al, 2013; Sento et al, 2016).
In addition to establishing how best to target EVP biogenesis and uptake, it will also be critical to define when to provide treatments blocking these pathways. Other common treatments such as chemotherapy can increase secretion of pro‐metastatic EVPs that enhance pre‐metastatic niche formation in the lungs and consequent breast cancer lung metastasis (Keklikoglou et al, 2019). Thus, targeting EVP biogenesis or uptake in combination with standard therapies against the primary tumor may potentially mitigate pro‐metastatic side effects that could result from therapy‐induced EVP biogenesis. Also, inhibiting EVP uptake using reserpine as an adjuvant/neoadjuvant therapy in combination with tumor resection was found to markedly reduce lung metastasis and improve survival of mice with melanoma tumors, whereas reserpine treatment alone only had marginal effects on survival (Ortiz et al, 2019). These results further indicate that targeting EVP‐dependent pathways for metastasis prevention may be most effective in the context of other standard treatments.
Conclusions and future perspectives
This review has summarized evidence that highlights the multifaceted effect of EVPs in cancer initiation, progression, and metastasis. One of the major revolutions in the field is the considerable technological advancement in the separation, analysis, and in vivo tracking of EVPs. This has led to the discovery of a previously unappreciated EVP heterogeneity and facilitated the molecular and functional characterization of distinct EVP subsets. As a result, EVP nomenclature is constantly evolving to fairly encompass and distinguish among the various EVP subclasses (Thery et al, 2018). The improvement of isolation strategies with higher resolution to remove contaminants and further separate specific EVP subsets is warranted to enhance the reproducibility and quality of studies involving EVPs and will further address the relevance of EVP subsets in cancer development. New visualization methods including super‐resolution microscopy and real‐time imaging will further advance our understanding of cargo packaging and biogenesis mechanisms, opening new avenues for therapeutic intervention, such as the discovery of more cell‐specific and safer druggable targets of EVP biogenesis. Our knowledge on EVPs from different sources has also rapidly expanded. EVPs from most tumor cells, stroma cells, immune cells, microorganisms, and dietary sources play pivotal roles in cell–cell communication, even between life domains, during cancer development. In addition to promoting tumor growth, a major property of EVPs is to travel long distances via the hematogenous and lymphatic routes in order to shape the physiology of distant organs with the formation of PMNs and the induction of systemic effects of cancer. As shown by multiple preclinical studies, tumor‐derived EVPs are determinants of organotropic metastasis, making them potential predictive/prognosis biomarkers and targets for metastasis‐preventive therapies. Research into the organotropic distribution of EVPs has also opened the way to engineering “designer EVPs,” which can be loaded with therapeutic or imaging molecules and endowed with the ability to image pre‐metastatic sites, to tailor treatment to a selective organ site, and to reduce systemic toxicity associated with free drugs in preclinical models. The introduction of EVPs as therapeutic deliverable tools has not reached the clinic, with a few exceptions (Lamparski et al, 2002; Escudier et al, 2005; Viaud et al, 2011; Damo et al, 2015). Further strategy development on the cellular source and standardization of manufacturing pipelines for EVPs with high clinical quality are prerequisites for their further therapeutic application. Finally, by representing the phenotypic heterogeneity and invasive potential of cells of origin and the effect of therapeutic intervention in cancer patients, EVPs are among the most promising sources of liquid biomarkers for cancer detection and therapeutic response assessment. Standardization of EVP isolation and analysis pipelines, developing assays with acceptable specificity and sensitivity, and addressing the reproducibility and rigor of assays in large patient cohort studies will be necessary to introduce EVP biomarkers as diagnostic and prognostic tools in clinical settings.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Acknowledgements
The authors gratefully acknowledge support from the National Cancer Institute (CA232093, CA163117, CA210240 and CA207983 to DL, and CA218513 to DL and HZ), the Thompson Family Foundation, the Tortolani Foundation, the Pediatric Oncology Experimental Therapeutics Investigator's Consortium, the Malcolm Hewitt Weiner Foundation, the Manning Foundation, the Sohn Foundation, the Theodore A. Rapp Foundation, the Hartwell Foundation, the Children's Cancer and Blood Foundation (all to DL), the AHEPA Vth District Cancer Research Foundation (to DL and SL), and the United States Department of Defense (W81XWH‐20‐1‐0263 to SL).
The EMBO Journal (2022) 41: e109288.
This review is part of the Cancer Review Series.
Contributor Information
Haiying Zhang, Email: haz2005@med.cornell.edu.
David Lyden, Email: dcl2001@med.cornell.edu.
References
- Abbastabar M, Sarfi M, Golestani A, Karimi A, Pourmand G, Khalili E (2020) Tumor‐derived urinary exosomal long non‐coding RNAs as diagnostic biomarkers for bladder cancer. EXCLI J 19: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramowicz A, Wojakowska A, Marczak L, Lysek‐Gladysinska M, Smolarz M, Story MD, Polanska J, Widlak P, Pietrowska M (2019) Ionizing radiation affects the composition of the proteome of extracellular vesicles released by head‐and‐neck cancer cells in vitro . J Radiat Res 60: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam‐Klages S, Adam D, Wiegmann K, Struve S, Kolanus W, Schneider‐Mergener J, Kronke M (1996) FAN, a novel WD‐repeat protein, couples the p55 TNF‐receptor to neutral sphingomyelinase. Cell 86: 937–947 [DOI] [PubMed] [Google Scholar]
- Aharon A, Sabbah A, Ben‐Shaul S, Berkovich H, Loven D, Brenner B, Bar‐Sela G (2017) Chemotherapy administration to breast cancer patients affects extracellular vesicles thrombogenicity and function. Oncotarget 8: 63265–63280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J (2008) Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 10: 619–624 [DOI] [PubMed] [Google Scholar]
- Al‐Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J (2009) Endothelial expression of autocrine VEGF upon the uptake of tumor‐derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci USA 106: 3794–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, Upadhyay P, Uyeminami DL, Pommier A, Kuttner V et al (2018) Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 361: eaao4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhateeb T, Bah I, Kumbhare A, Youssef D, Yao ZQ, McCall CE, Gazzar ME (2020) Long non‐coding RNA Hotairm1 promotes S100A9 support of MDSC expansion during sepsis. J Clin Cell Immunol 11: 600 [PMC free article] [PubMed] [Google Scholar]
- Altadill T, Campoy I, Lanau L, Gill K, Rigau M, Gil‐Moreno A, Reventos J, Byers S, Colas E, Cheema AK (2016) Enabling metabolomics based biomarker discovery studies using molecular phenotyping of exosome‐like vesicles. PLoS One 11: e0151339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29: 341–345 [DOI] [PubMed] [Google Scholar]
- An Q, van Bel AJ, Huckelhoven R (2007) Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal Behav 2: 4–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando W, Kikuchi K, Uematsu T, Yokomori H, Takaki T, Sogabe M, Kohgo Y, Otori K, Ishikawa S, Okazaki I (2019) Novel breast cancer screening: combined expression of miR‐21 and MMP‐1 in urinary exosomes detects 95% of breast cancer without metastasis. Sci Rep 9: 13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqil F, Munagala R, Jeyabalan J, Agrawal AK, Kyakulaga AH, Wilcher SA, Gupta RC (2019) Milk exosomes – natural nanoparticles for siRNA delivery. Cancer Lett 449: 186–195 [DOI] [PubMed] [Google Scholar]
- Argiles JM, Stemmler B, Lopez‐Soriano FJ, Busquets S (2018) Inter‐tissue communication in cancer cachexia. Nat Rev Endocrinol 15: 9–20 [DOI] [PubMed] [Google Scholar]
- Arnold J, Schattschneider J, Blechner C, Krisp C, Schluter H, Schweizer M, Nalaskowski M, Oliveira‐Ferrer L, Windhorst S (2020) Tubulin Tyrosine Ligase Like 4 (TTLL4) overexpression in breast cancer cells is associated with brain metastasis and alters exosome biogenesis. J Exp Clin Cancer Res 39: 205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arntz OJ, Pieters BC, Oliveira MC, Broeren MG, Bennink MB, de Vries M, van Lent PL, Koenders MI, van den Berg WB, van der Kraan PM et al (2015) Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol Nutr Food Res 59: 1701–1712 [DOI] [PubMed] [Google Scholar]
- Artym VV, Zhang Y, Seillier‐Moiseiwitsch F, Yamada KM, Mueller SC (2006) Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res 66: 3034–3043 [DOI] [PubMed] [Google Scholar]
- Ashrafian F, Behrouzi A, Shahriary A, Ahmadi Badi S, Davari M, Khatami S, Rahimi Jamnani F, Fateh A, Vaziri F, Siadat SD (2019) Comparative study of effect of Akkermansia muciniphila and its extracellular vesicles on toll‐like receptors and tight junction. Gastroenterol Hepatol Bed Bench 12: 163–168 [PMC free article] [PubMed] [Google Scholar]
- Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bauerle T, Wallwiener M et al (2013) Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 31: 539–544 [DOI] [PubMed] [Google Scholar]
- Baek DW, Kim G, Kang BW, Kim HJ, Park SY, Park JS, Choi GS, Kang MK, Hur K, Kim JG (2020) High expression of microRNA‐199a‐5p is associated with superior clinical outcomes in patients with locally advanced rectal cancer. J Cancer Res Clin Oncol 146: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri E, Abnous K, Farzad SA, Taghdisi SM, Ramezani M, Alibolandi M (2020) Targeted doxorubicin‐loaded mesenchymal stem cells‐derived exosomes as a versatile platform for fighting against colorectal cancer. Life Sci 261: 118369 [DOI] [PubMed] [Google Scholar]
- Baglio SR, van Eijndhoven MA, Koppers‐Lalic D, Berenguer J, Lougheed SM, Gibbs S, Leveille N, Rinkel RN, Hopmans ES, Swaminathan S et al (2016) Sensing of latent EBV infection through exosomal transfer of 5'pppRNA. Proc Natl Acad Sci USA 113: E587–E596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J (2014) MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK‐293 kidney cell cultures, and mouse livers. J Nutr 144: 1495–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E et al (2012) Syndecan‐syntenin‐ALIX regulates the biogenesis of exosomes. Nat Cell Biol 14: 677–685 [DOI] [PubMed] [Google Scholar]
- Bairamukov V, Bukatin A, Landa S, Burdakov V, Shtam T, Chelnokova I, Fedorova N, Filatov M, Starodubtseva M (2020) Biomechanical properties of blood plasma extracellular vesicles revealed by atomic force microscopy. Biology (Basel) 10: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L, Lessard R, Dai L, Cho Y‐J, Pomeroy SL, Breakefield XO, Skog J (2011) Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2: 180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L, Atai NA, Chen W, Mu D, Tannous BA, Breakefield XO, Skog J, Maguire CA (2015) Heparin affinity purification of extracellular vesicles. Sci Rep 5: 10266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini N, Torreggiani E, Roncuzzi L, Perut F, Zini N, Avnet S (2018) Exosome‐like nanovesicles isolated from Citrus limon L. exert antioxidative effect. Curr Pharm Biotechnol 19: 877–885 [DOI] [PubMed] [Google Scholar]
- Balint S, Muller S, Fischer R, Kessler BM, Harkiolaki M, Valitutti S, Dustin ML (2020) Supramolecular attack particles are autonomous killing entities released from cytotoxic T cells. Science 368: 897–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandari SK, Purushothaman A, Ramani VC, Brinkley GJ, Chandrashekar DS, Varambally S, Mobley JA, Zhang Y, Brown EE, Vlodavsky I et al (2018) Chemotherapy induces secretion of exosomes loaded with heparanase that degrades extracellular matrix and impacts tumor and host cell behavior. Matrix Biol 65: 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfer S, Schneider D, Dewes J, Strauss MT, Freibert SA, Heimerl T, Maier UG, Elsasser HP, Jungmann R, Jacob R (2018) Molecular mechanism to recruit galectin‐3 into multivesicular bodies for polarized exosomal secretion. Proc Natl Acad Sci USA 115: E4396–E4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, You B, Shi S, Shan Y, Zhang Q, Yue H, Zhang J, Zhang W, Shi Y, Liu Y et al (2018) Metastasis‐associated miR‐23a from nasopharyngeal carcinoma‐derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 37: 2873–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar‐Sela G, Cohen I, Avisar A, Loven D, Aharon A (2020) Circulating blood extracellular vesicles as a tool to assess endothelial injury and chemotherapy toxicity in adjuvant cancer patients. PLoS One 15: e0240994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracos VE, Martin L, Korc M, Guttridge DC, Fearon KCH (2018) Cancer‐associated cachexia. Nat Rev Dis Primers 4: 17105 [DOI] [PubMed] [Google Scholar]
- Barazzoni R, Gortan Cappellari G, Ragni M, Nisoli E (2018) Insulin resistance in obesity: an overview of fundamental alterations. Eat Weight Disord 23: 149–157 [DOI] [PubMed] [Google Scholar]
- Bardi GT, Al‐Rayan N, Richie JL, Yaddanapudi K, Hood JL (2019) Detection of inflammation‐related melanoma small extracellular vesicle (sEV) mRNA content using primary melanocyte sEVs as a reference. Int J Mol Sci 20: 1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E, Clevers H (2017) Cancer stem cells revisited. Nat Med 23: 1124–1134 [DOI] [PubMed] [Google Scholar]
- Batlle E, Massague J (2019) Transforming growth factor‐beta signaling in immunity and cancer. Immunity 50: 924–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmoussa A, Laugier J, Beauparlant CJ, Lambert M, Droit A, Provost P (2020) Complexity of the microRNA transcriptome of cow milk and milk‐derived extracellular vesicles isolated via differential ultracentrifugation. J Dairy Sci 103: 16–29 [DOI] [PubMed] [Google Scholar]
- Bijnsdorp IV, Geldof AA, Lavaei M, Piersma SR, van Moorselaar RJ, Jimenez CR (2013) Exosomal ITGA3 interferes with non‐cancerous prostate cell functions and is increased in urine exosomes of metastatic prostate cancer patients. J Extracell Vesicles 10.3402/jev.v2i0.22097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig C, Gruenberg J (2014) ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 24: 19–25 [DOI] [PubMed] [Google Scholar]
- Bjornetro T, Redalen KR, Meltzer S, Thusyanthan NS, Samiappan R, Jegerschold C, Handeland KR, Ree AH (2019) An experimental strategy unveiling exosomal microRNAs 486‐5p, 181a‐5p and 30d‐5p from hypoxic tumour cells as circulating indicators of high‐risk rectal cancer. J Extracell Vesicles 8: 1567219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc L, De Gassart A, Geminard C, Bette‐Bobillo P, Vidal M (2005) Exosome release by reticulocytes–an integral part of the red blood cell differentiation system. Blood Cells Mol Dis 35: 21–26 [DOI] [PubMed] [Google Scholar]
- Blanc L, Vidal M (2018) New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 9: 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss SA, Sinha G, Sandiford OA, Williams LM, Engelberth DJ, Guiro K, Isenalumhe LL, Greco SJ, Ayer S, Bryan M et al (2016) Mesenchymal stem cell‐derived exosomes stimulate cycling quiescence and early breast cancer dormancy in bone marrow. Cancer Res 76: 5832–5844 [DOI] [PubMed] [Google Scholar]
- Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, Ostrowski M, Thery C (2012) Rab27a supports exosome‐dependent and ‐independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res 72: 4920–4930 [DOI] [PubMed] [Google Scholar]
- Bonjoch L, Casas V, Carrascal M, Closa D (2016) Involvement of exosomes in lung inflammation associated with experimental acute pancreatitis. J Pathol 240: 235–245 [DOI] [PubMed] [Google Scholar]
- Booth AM, Fang Y, Fallon JK, Yang J‐M, Hildreth JEK, Gould SJ (2006) Exosomes and HIV Gag bud from endosome‐like domains of the T cell plasma membrane. J Cell Biol 172: 923–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel M, Lollo G, Magne D, Buchet R, Brizuela L, Mebarek S (2020) Prostate cancer‐derived exosomes promote osteoblast differentiation and activity through phospholipase D2. Biochim Biophys Acta Mol Basis Dis 1866: 165919 [DOI] [PubMed] [Google Scholar]
- Bouchareychas L, Duong P, Covarrubias S, Alsop E, Phu TA, Chung A, Gomes M, Wong D, Meechoovet B, Capili A et al (2020) Macrophage exosomes resolve atherosclerosis by regulating hematopoiesis and inflammation via MicroRNA cargo. Cell Rep 32: 107881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boufraqech M, Zhang L, Jain M, Patel D, Ellis R, Xiong Y, He M, Nilubol N, Merino MJ, Kebebew E (2014) miR‐145 suppresses thyroid cancer growth and metastasis and targets AKT3. Endocr Relat Cancer 21: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussadia Z, Lamberti J, Mattei F, Pizzi E, Puglisi R, Zanetti C, Pasquini L, Fratini F, Fantozzi L, Felicetti F et al (2018) Acidic microenvironment plays a key role in human melanoma progression through a sustained exosome mediated transfer of clinically relevant metastatic molecules. J Exp Clin Cancer Res 37: 245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz S, Schuhwerk H, Brabletz T, Stemmler MP (2021) Dynamic EMT: a multi‐tool for tumor progression. EMBO J 40: e108647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett SI, Lucien F, Guo C, Williams KC, Kim Y, Durfee PN, Brinker CJ, Chin JI, Yang J, Leong HS (2017) Immunoaffinity based methods are superior to kits for purification of prostate derived extracellular vesicles from plasma samples. Prostate 77: 1335–1343 [DOI] [PubMed] [Google Scholar]
- Bretz NP, Ridinger J, Rupp AK, Rimbach K, Keller S, Rupp C, Marme F, Umansky L, Umansky V, Eigenbrod T et al (2013) Body fluid exosomes promote secretion of inflammatory cytokines in monocytic cells via Toll‐like receptor signaling. J Biol Chem 288: 36691–36702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T et al (2017) Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358: 1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratta S, Urbanelli L, Sagini K, Giovagnoli S, Caponi S, Fioretto D, Mitro N, Caruso D, Emiliani C (2017) Extracellular vesicles released by fibroblasts undergoing H‐Ras induced senescence show changes in lipid profile. PLoS One 12: e0188840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz H, Nofech‐Mozes R, Ding Q, Khella HWZ, Szabo PM, Jewett M, Finelli A, Lee J, Ordon M, Stewart R et al (2016) Exosomal MicroRNAs are diagnostic biomarkers and can mediate cell‐cell communication in renal cell carcinoma. Eur Urol Focus 2: 210–218 [DOI] [PubMed] [Google Scholar]
- Cai Z, Yang F, Yu L, Yu Z, Jiang L, Wang Q, Yang Y, Wang L, Cao X, Wang J (2012) Activated T cell exosomes promote tumor invasion via Fas signaling pathway. J Immunol 188: 5954–5961 [DOI] [PubMed] [Google Scholar]
- Cai C, Zhang H, Zhu Y, Zheng P, Xu Y, Sun J, Zhang M, Lan T, Gu B, Li S et al (2019) Serum exosomal long noncoding RNA pcsk2‐2:1 as a potential novel diagnostic biomarker for gastric cancer. Onco Targets Ther 12: 10035–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Chao G, Li W, Zhu J, Li F, Qi B, Wei Y, Chen S, Zhou G, Lu X et al (2020) Activated CD4(+) T cells‐derived exosomal miR‐142‐3p boosts post‐ischemic ventricular remodeling by activating myofibroblast. Aging (Albany NY) 12: 7380–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine GJ, Stonelake PS, Lip GY, Kehoe ST (2002) The hypercoagulable state of malignancy: pathogenesis and current debate. Neoplasia 4: 465–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle AS, Nair N, Oo AK, Prieto‐Vila M, Koga M, Khayrani AC, Hussein M, Hurley L, Vaidyanath A, Seno A et al (2016) A new PDAC mouse model originated from iPSCs‐converted pancreatic cancer stem cells (CSCcm). Am J Cancer Res 6: 2799–2815 [PMC free article] [PubMed] [Google Scholar]
- Campanella C, Rappa F, Sciume C, Marino Gammazza A, Barone R, Bucchieri F, David S, Curcuru G, Caruso Bavisotto C, Pitruzzella A et al (2015) Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer 121: 3230–3239 [DOI] [PubMed] [Google Scholar]
- Canas MA, Fabrega MJ, Gimenez R, Badia J, Baldoma L (2018) Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1‐mediated immune responses in intestinal epithelial cells. Front Microbiol 9: 498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Wu T, Zhang K, Meng X, Dai W, Wang D, Dong H, Zhang X (2019) Engineered exosome‐mediated near‐infrared‐II region V2C quantum dot delivery for nucleus‐target low‐temperature photothermal therapy. ACS Nano 13: 1499–1510 [DOI] [PubMed] [Google Scholar]
- Capaci V, Bascetta L, Fantuz M, Beznoussenko GV, Sommaggio R, Cancila V, Bisso A, Campaner E, Mironov AA, Wisniewski JR et al (2020) Mutant p53 induces Golgi tubulo‐vesiculation driving a prometastatic secretome. Nat Commun 11: 3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadei L, Calore F, Creighton CJ, Guescini M, Batte K, Iwenofu OH, Zewdu A, Braggio DA, Bill KL, Fadda P et al (2017) Exosome‐derived miR‐25‐3p and miR‐92a‐3p stimulate liposarcoma progression. Cancer Res 77: 3846–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S et al (2015) KRAS‐dependent sorting of miRNA to exosomes. Elife 4: e07197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput N, Schartz NE, Andre F, Taieb J, Novault S, Bonnaventure P, Aubert N, Bernard J, Lemonnier F, Merad M et al (2004) Exosomes as potent cell‐free peptide‐based vaccine. II. Exosomes in CpG adjuvants efficiently prime naive Tc1 lymphocytes leading to tumor rejection. J Immunol 172: 2137–2146 [DOI] [PubMed] [Google Scholar]
- Chelakkot C, Choi Y, Kim DK, Park HT, Ghim J, Kwon Y, Jeon J, Kim MS, Jee YK, Gho YS et al (2018) Akkermansia muciniphila‐derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med 50: e450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TS, Arslan F, Yin Y, Tan SS, Lai RC, Choo AB, Padmanabhan J, Lee CN, de Kleijn DP, Lim SK (2011) Enabling a robust scalable manufacturing process for therapeutic exosomes through oncogenic immortalization of human ESC‐derived MSCs. J Transl Med 9: 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kasai T, Li Y, Sugii Y, Jin G, Okada M, Vaidyanath A, Mizutani A, Satoh A, Kudoh T et al (2012) A model of cancer stem cells derived from mouse induced pluripotent stem cells. PLoS One 7: e33544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ et al (2014) Exosomes from drug‐resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One 9: e95240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Chen CH, Wallace CG, Yuen CM, Kao GS, Chen YL, Shao PL, Chen YL, Chai HT, Lin KC et al (2016a) Intravenous administration of xenogenic adipose‐derived mesenchymal stem cells (ADMSC) and ADMSC‐derived exosomes markedly reduced brain infarct volume and preserved neurological function in rat after acute ischemic stroke. Oncotarget 7: 74537–74556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yang L, Cui Y, Zhou Y, Yin X, Guo J, Zhang G, Wang T, He QY (2016b) Cytoskeleton‐centric protein transportation by exosomes transforms tumor‐favorable macrophages. Oncotarget 7: 67387–67402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xu Y, Lu Y, Xing W (2018a) Isolation and visible detection of tumor‐derived exosomes from plasma. Anal Chem 90: 14207–14215 [DOI] [PubMed] [Google Scholar]
- Chen L, Chen R, Kemper S, Cong M, You H, Brigstock DR (2018b) Therapeutic effects of serum extracellular vesicles in liver fibrosis. J Extracell Vesicles 7: 1461505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Guo P, He Y, Chen Z, Chen L, Luo Y, Qi L, Liu Y, Wu Q, Cui Y et al (2018c) HCC‐derived exosomes elicit HCC progression and recurrence by epithelial‐mesenchymal transition through MAPK/ERK signalling pathway. Cell Death Dis 9: 513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhou J, Li X, Wang X, Lin Y, Wang X (2018d) Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor‐promoted phenotype. Cancer Lett 435: 80–91 [DOI] [PubMed] [Google Scholar]
- Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, Zhong G, Li Y, Li J, Huang J et al (2019a) Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Investig 130: 404–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Brenner DA, Kisseleva T (2019b) Combatting fibrosis: exosome‐based therapies in the regression of liver fibrosis. Hepatol Commun 3: 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, Zhong G, Li Y, Li J, Huang J et al (2020a) Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest 130: 404–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Cao P, Huang C, Wu Q, Chen S, Chen F (2020b) Serum exosomal miR‐7977 as a novel biomarker for lung adenocarcinoma. J Cell Biochem 121: 3382–3391 [DOI] [PubMed] [Google Scholar]
- Chen L, Yao X, Yao H, Ji Q, Ding G, Liu X (2020c) Exosomal miR‐103‐3p from LPS‐activated THP‐1 macrophage contributes to the activation of hepatic stellate cells. FASEB J 34: 5178–5192 [DOI] [PubMed] [Google Scholar]
- Chen QL, Xie CF, Feng KL, Cui DY, Sun SL, Zhang JC, Xiong CM, Huang JH, Chong Z (2020d) microRNAs carried by exosomes promote epithelial‐mesenchymal transition and metastasis of liver cancer cells. Am J Transl Res 12: 6811–6826 [PMC free article] [PubMed] [Google Scholar]
- Chen W, Li J, Wei X, Fan Y, Qian H, Li S, Xiang Y, Ding S (2020e) Surface plasmon resonance biosensor using hydrogel‐AuNP supramolecular spheres for determination of prostate cancer‐derived exosomes. Mikrochim Acta 187: 590 [DOI] [PubMed] [Google Scholar]
- Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P, Ye Y (2020f) Exosome‐transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett 475: 119–128 [DOI] [PubMed] [Google Scholar]
- Chen Z, Liang Q, Zeng H, Zhao Q, Guo Z, Zhong R, Xie M, Cai X, Su J, He Z et al (2020g) Exosomal CA125 as a promising biomarker for ovarian cancer diagnosis. J Cancer 11: 6445–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Sang Y, Song X, Zhang D, Wang L, Zhao W, Liang Y, Zhang N, Yang Q (2021a) Exosomal miR‐500a‐5p derived from cancer‐associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. Theranostics 11: 3932–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Li Y, Wang Y, Xu J (2021b) LncRNA HOTAIRM1 knockdown inhibits cell glycolysis metabolism and tumor progression by miR‐498/ABCE1 axis in non‐small cell lung cancer. Genes Genomics 43: 183–194 [DOI] [PubMed] [Google Scholar]
- Chen F, Xu B, Li J, Yang X, Gu J, Yao X, Sun X (2021c) Hypoxic tumour cell‐derived exosomal miR‐340‐5p promotes radioresistance of oesophageal squamous cell carcinoma via KLF10. J Exp Clin Cancer Res 40: 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Cheng JC, Chen YF, Yang JC, Hsu FM (2021d) Circulating exosomal integrin beta3 is associated with intracranial failure and survival in lung cancer patients receiving cranial irradiation for brain metastases: a prospective observational study. Cancers (Basel) 13: 380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhu Q, Cheng L, Wang Y, Li M, Yang Q, Hu L, Lou D, Li J, Dong X et al (2021e) Exosome detection via the ultrafast‐isolation system: EXODUS. Nat Methods 18: 212–218 [DOI] [PubMed] [Google Scholar]
- Cheng WC, Liao TT, Lin CC, Yuan LE, Lan HY, Lin HH, Teng HW, Chang HC, Lin CH, Yang CY et al (2019) RAB27B‐activated secretion of stem‐like tumor exosomes delivers the biomarker microRNA‐146a‐5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int J Cancer 145: 2209–2224 [DOI] [PubMed] [Google Scholar]
- Cheng A, Choi D, Lora M, Shum‐Tim D, Rak J, Colmegna I (2020) Human multipotent mesenchymal stromal cells cytokine priming promotes RAB27B‐regulated secretion of small extracellular vesicles with immunomodulatory cargo. Stem Cell Res Ther 11: 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou NT, Kageyama R, Ansel KM (2018) Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep 25: 3356–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR, Kim SS, Cho SW, Cheong JY (2020) Serum exosomal microRNA, miR‐10b‐5p, as a potential diagnostic biomarker for early‐stage hepatocellular carcinoma. J Clin Med 9: 281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D‐S, Choi D‐Y, Hong B, Jang S, Kim D‐K, Lee J, Kim Y‐K, Pyo Kim K, Gho Y (2012) Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J Extracell Vesicles 1: 18704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Montermini L, Kim DK, Meehan B, Roth FP, Rak J (2018) The impact of oncogenic EGFRvIII on the proteome of extracellular vesicles released from glioblastoma cells. Mol Cell Proteomics 17: 1948–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K, Llodrá J, Roth EW, Tsai J, Gordo S, Wucherpfennig KW, Kam LC, Stokes DL, Dustin ML (2014) Polarized release of T‐cell‐receptor‐enriched microvesicles at the immunological synapse. Nature 507: 118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson HC, Svensson KJ, Van Kuppevelt TH, Li JP, Belting M (2013) Cancer cell exosomes depend on cell‐surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci USA 110: 17380–17385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronopoulos A, Kalluri R (2020) Emerging role of bacterial extracellular vesicles in cancer. Oncogene 39: 6951–6960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua CE, Tang BL (2015) The role of the small GTPase Rab31 in cancer. J Cell Mol Med 19: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciaruso C, Beltraminelli T, Duval F, Nassiri S, Hamelin R, Mozes A, Gallart‐Ayala H, Ceada Torres G, Torchia B, Ries CH et al (2019) Molecular profiling and functional analysis of macrophage‐derived tumor extracellular vesicles. Cell Rep 27: 3062–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy JW, Sedgwick A, Rosse C, Muralidharan‐Chari V, Raposo G, Method M, Chavrier P, D'Souza‐Schorey C (2015) Regulated delivery of molecular cargo to invasive tumour‐derived microvesicles. Nat Commun 6: 6919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy JW, Zhang Y, Sheehan C, D'Souza‐Schorey C (2019) An ARF6–Exportin‐5 axis delivers pre‐miRNA cargo to tumour microvesicles. Nat Cell Biol 21: 856–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy JW, Schmidtmann M, D'Souza‐Schorey C (2021) The ins and outs of microvesicles. FASEB Bioadv 3: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DJ, Fondrie WE, Yang A, Mao L (2016) Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer‐derived exosomes. J Proteomics 133: 161–169 [DOI] [PubMed] [Google Scholar]
- Clayton A, Turkes A, Dewitt S, Steadman R, Mason MD, Hallett MB (2004) Adhesion and signaling by B cell‐derived exosomes: the role of integrins. FASEB J 18: 977–979 [DOI] [PubMed] [Google Scholar]
- Clement E, Lazar I, Attane C, Carrie L, Dauvillier S, Ducoux‐Petit M, Esteve D, Menneteau T, Moutahir M, Le Gonidec S et al (2020) Adipocyte extracellular vesicles carry enzymes and fatty acids that stimulate mitochondrial metabolism and remodeling in tumor cells. EMBO J 39: e102525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos‐Garcia M, Loizaga‐Iriarte A, Zuniga‐Garcia P, Sanchez‐Mosquera P, Rosa Cortazar A, Gonzalez E, Torrano V, Alonso C, Perez‐Cormenzana M, Ugalde‐Olano A et al (2018) Metabolic alterations in urine extracellular vesicles are associated to prostate cancer pathogenesis and progression. J Extracell Vesicles 7: 1470442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E, Meldolesi J (2015) Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol 25: 364–372 [DOI] [PubMed] [Google Scholar]
- Cohen O, Betzer O, Elmaliach‐Pnini N, Motiei M, Sadan T, Cohen‐Berkman M, Dagan O, Popovtzer A, Yosepovich A, Barhom H et al (2021) 'Golden' exosomes as delivery vehicles to target tumors and overcome intratumoral barriers: in vivo tracking in a model for head and neck cancer. Biomater Sci 9: 2103–2114 [DOI] [PubMed] [Google Scholar]
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G (2013) Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 126: 5553–5565 [DOI] [PubMed] [Google Scholar]
- Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M et al (2015) CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer 14: 155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras‐Naranjo JC, Wu HJ, Ugaz VM (2017) Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip 17: 3558–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks T, Pateras IS, Tarcic O, Solomon H, Schetter AJ, Wilder S, Lozano G, Pikarsky E, Forshew T, Rosenfeld N et al (2013) Mutant p53 prolongs NF‐kappaB activation and promotes chronic inflammation and inflammation‐associated colorectal cancer. Cancer Cell 23: 634–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG, Harris CC (2018) Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR‐1246. Nat Commun 9: 771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Verdera H, Gitz‐Francois JJ, Schiffelers RM, Vader P (2017) Cellular uptake of extracellular vesicles is mediated by clathrin‐independent endocytosis and macropinocytosis. J Control Release 266: 100–108 [DOI] [PubMed] [Google Scholar]
- Costa‐Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H et al (2015) Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nat Cell Biol 17: 816–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couper KN, Barnes T, Hafalla JC, Combes V, Ryffel B, Secher T, Grau GE, Riley EM, de Souza JB (2010) Parasite‐derived plasma microparticles contribute significantly to malaria infection‐induced inflammation through potent macrophage stimulation. PLoS Pathog 6: e1000744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, Cang SD (2019) Mechanism of exosomal microRNA‐224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol 25: 1890–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Lv Z, Ding H, Xing C, Yuan Y (2020a) MiR‐1539 and its potential role as a novel biomarker for colorectal cancer. Front Oncol 10: 531244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Chen Y, Hu M, Lin Y, Zhang S, Kong L, Chen Y (2020b) Diagnostic and prognostic value of the cancer‐testis antigen lactate dehydrogenase C4 in breast cancer. Clin Chim Acta 503: 203–209 [DOI] [PubMed] [Google Scholar]
- D'Acunzo P, Perez‐Gonzalez R, Kim Y, Hargash T, Miller C, Alldred MJ, Erdjument‐Bromage H, Penikalapati SC, Pawlik M, Saito M et al (2021) Mitovesicles are a novel population of extracellular vesicles of mitochondrial origin altered in Down syndrome. Sci Adv 7: eabe5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alterio C, Scala S, Sozzi G, Roz L, Bertolini G (2020) Paradoxical effects of chemotherapy on tumor relapse and metastasis promotion. Semin Cancer Biol 60: 351–361 [DOI] [PubMed] [Google Scholar]
- Dai J, Escara‐Wilke J, Keller JM, Jung Y, Taichman RS, Pienta KJ, Keller ET (2019) Primary prostate cancer educates bone stroma through exosomal pyruvate kinase M2 to promote bone metastasis. J Exp Med 216: 2883–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Zhou J, Wang H, Zhang M, Yang X, Song W (2020) miR‐424‐5p promotes the proliferation and metastasis of colorectal cancer by directly targeting SCN4B. Pathol Res Pract 216: 152731 [DOI] [PubMed] [Google Scholar]
- Damo M, Wilson DS, Simeoni E, Hubbell JA (2015) TLR‐3 stimulation improves anti‐tumor immunity elicited by dendritic cell exosome‐based vaccines in a murine model of melanoma. Sci Rep 5: 17622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Kim H, Lal M, McGee L, Johnson A, Moustafa AA, Jones JC, Mondal D, Ferrer M, Abdel‐Mageed AB (2017) Manumycin A suppresses exosome biogenesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration‐resistant prostate cancer cells. Cancer Lett 408: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Kim H, McGee L, Johnson AE, Talwar S, Marugan J, Southall N, Hu X, Lal M, Mondal D et al (2018) High‐throughput screening identified selective inhibitors of exosome biogenesis and secretion: a drug repurposing strategy for advanced cancer. Sci Rep 8: 8161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong S, Chikh G, Sekirov L, Raney S, Semple S, Klimuk S, Yuan N, Hope M, Cullis P, Tam Y (2007) Encapsulation in liposomal nanoparticles enhances the immunostimulatory, adjuvant and anti‐tumor activity of subcutaneously administered CpG ODN. Cancer Immunol Immunother 56: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deep G, Jain A, Kumar A, Agarwal C, Kim S, Leevy WM, Agarwal R (2020) Exosomes secreted by prostate cancer cells under hypoxia promote matrix metalloproteinases activity at pre‐metastatic niches. Mol Carcinog 59: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura M (2017) Exosomal microRNA in plasma as a non‐invasive biomarker for the recurrence of non‐small cell lung cancer. Oncol Lett 13: 1256–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozo‐Acebo L, Hazas MLL, Tome‐Carneiro J, Gil‐Cabrerizo P, San‐Cristobal R, Busto R, Garcia‐Ruiz A, Davalos A (2021) Bovine milk‐derived exosomes as a drug delivery vehicle for miRNA‐based therapy. Int J Mol Sci 22: 1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demory Beckler M, Higginbotham JN, Franklin JL, Ham AJ, Halvey PJ, Imasuen IE, Whitwell C, Li M, Liebler DC, Coffey RJ (2013) Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics 12: 343–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB (2017) Exosome‐mediated intercellular communication between hepatitis C virus‐infected hepatocytes and hepatic stellate cells. J Virol 91: e02225‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Modica M, Regondi V, Sandri M, Iorio MV, Zanetti A, Tagliabue E, Casalini P, Triulzi T (2017) Breast cancer‐secreted miR‐939 downregulates VE‐cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett 384: 94–100 [DOI] [PubMed] [Google Scholar]
- Di W, Zhang W, Zhu B, Li X, Tang Q, Zhou Y (2021) Colorectal cancer prompted adipose tissue browning and cancer cachexia through transferring exosomal miR‐146b‐5p. J Cell Physiol 236: 5399–5410 [DOI] [PubMed] [Google Scholar]
- Diamond JM, Vanpouille‐Box C, Spada S, Rudqvist N‐P, Chapman JR, Ueberheide BM, Pilones KA, Sarfraz Y, Formenti SC, Demaria S (2018) Exosomes shuttle TREX1‐sensitive IFN‐stimulatory dsDNA from irradiated cancer cells to DCs. Cancer Immunol Res 6: 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra S, Birker IL, Smit FP, Leyten GH, de Reijke TM, van Oort IM, Mulders PF, Jannink SA, Schalken JA (2014) Prostate cancer biomarker profiles in urinary sediments and exosomes. J Urol 191: 1132–1138 [DOI] [PubMed] [Google Scholar]
- Ding C, Xi G, Wang G, Cui D, Zhang B, Wang H, Jiang G, Song J, Xu G, Wang J (2020) Exosomal circ‐MEMO1 promotes the progression and aerobic glycolysis of non‐small cell lung cancer through targeting MiR‐101‐3p/KRAS axis. Front Genet 11: 962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TK, Fendler W, Chalubinska‐Fendler J, Acharya SS, O'Leary C, Deraska PV, D'Andrea AD, Chowdhury D, Kozono D (2016) Circulating miR‐29a and miR‐150 correlate with delivered dose during thoracic radiation therapy for non‐small cell lung cancer. Radiat Oncol 11: 61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler JHW, Gyorfi AH, Ramanujam M, Whitfield ML, Konigshoff M, Lafyatis R (2019) Shared and distinct mechanisms of fibrosis. Nat Rev Rheumatol 15: 705–730 [DOI] [PubMed] [Google Scholar]
- Dlugonska H, Gatkowska J (2016) Exosomes in the context of Toxoplasma gondii – host communication. Ann Parasitol 62: 169–174 [DOI] [PubMed] [Google Scholar]
- Dong H, Chen H, Jiang J, Zhang H, Cai C, Shen Q (2018) Highly sensitive electrochemical detection of tumor exosomes based on aptamer recognition‐induced Multi‐DNA release and cyclic enzymatic amplification. Anal Chem 90: 4507–4513 [DOI] [PubMed] [Google Scholar]
- Dorayappan KDP, Wanner R, Wallbillich JJ, Saini U, Zingarelli R, Suarez AA, Cohn DE, Selvendiran K (2018) Hypoxia‐induced exosomes contribute to a more aggressive and chemoresistant ovarian cancer phenotype: a novel mechanism linking STAT3/Rab proteins. Oncogene 37: 3806–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG et al (2016) Circular RNAs are down‐regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep 6: 37982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Duan X, Yao X, Wan J, Cheng Y, Wang Y, Yan Y, Zhang L, Zhu L, Ni C et al (2020a) Tumour‐derived exosomal miR‐3473b promotes lung tumour cell intrapulmonary colonization by activating the nuclear factor‐kappaB of local fibroblasts. J Cell Mol Med 24: 7802–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Liang Y, Li J, Zhao JM, Wang ZN, Lin XY (2020b) Gastric cancer cell‐derived exosomal microRNA‐23a promotes angiogenesis by targeting PTEN. Front Oncol 10: 326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Nordmeier S, Byrnes AE, Buxton ILO (2021) Extracellular vesicle‐mediated purinergic signaling contributes to host microenvironment plasticity and metastasis in triple negative breast cancer. Int J Mol Sci 22: 597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duette G, Pereyra Gerber P, Rubione J, Perez PS, Landay AL, Crowe SM, Liao Z, Witwer KW, Holgado MP, Salido J et al (2018) Induction of HIF‐1alpha by HIV‐1 infection in CD4(+) T cells promotes viral replication and drives extracellular vesicle‐mediated inflammation. MBio 9: e00757‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easom NJW, Marks M, Jobe D, Gillmore R, Meyer T, Maini MK, Njie R (2020) ULBP1 is elevated in human hepatocellular carcinoma and predicts outcome. Front Oncol 10: 971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy RJ, Weidmann MD, Sharma VP, Condeelis JS (2017) Tumor cell invadopodia: invasive protrusions that orchestrate metastasis. Trends Cell Biol 27: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea‐Jimenez AL, Zimmermann P (2019) Lipids in exosome biology. In Handbook of Experimental Pharmacology, Barrett JE (ed), pp 309–336. Berlin, Heidelberg: Springer International Publishing; [DOI] [PubMed] [Google Scholar]
- Ellis TN, Kuehn MJ (2010) Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanouilidi A, Paladin D, Greening DW, Falasca M (2019) Oncogenic and non‐malignant pancreatic exosome cargo reveal distinct expression of oncogenic and prognostic factors involved in tumor invasion and metastasis. Proteomics 19: e1800158 [DOI] [PubMed] [Google Scholar]
- Ertunc ME, Sikkeland J, Fenaroli F, Griffiths G, Daniels MP, Cao H, Saatcioglu F, Hotamisligil GS (2015) Secretion of fatty acid binding protein aP2 from adipocytes through a nonclassical pathway in response to adipocyte lipase activity. J Lipid Res 56: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escrevente C, Keller S, Altevogt P, Costa J (2011) Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 11: 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudier B, Dorval T, Chaput N, Andre F, Caby MP, Novault S, Flament C, Leboulaire C, Borg C, Amigorena S et al (2005) Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived‐exosomes: results of thefirst phase I clinical trial. J Transl Med 3: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M, Guise T, Kang Y (2018) The biology of bone metastasis. Cold Spring Harb Perspect Med 8: a031252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing J (1928) Neoplastic diseases; A treatise on tumors, 3d ed rev. and enl., with 546 illustrations. edn. Philadelphia, London: W.B. Saunders; [Google Scholar]
- Fabrega MJ, Aguilera L, Gimenez R, Varela E, Alexandra Canas M, Antolin M, Badia J, Baldoma L (2016) Activation of immune and defense responses in the intestinal mucosa by outer membrane vesicles of commensal and probiotic Escherichia coli strains. Front Microbiol 7: 705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader CM, Sánchez DG, Mestre MB, Colombo MI (2009) TI‐VAMP/VAMP7 and VAMP3/cellubrevin: two v‐SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta 1793: 1901–1916 [DOI] [PubMed] [Google Scholar]
- Faict S, Muller J, De Veirman K, De Bruyne E, Maes K, Vrancken L, Heusschen R, De Raeve H, Schots R, Vanderkerken K et al (2018) Exosomes play a role in multiple myeloma bone disease and tumor development by targeting osteoclasts and osteoblasts. Blood Cancer J 8: 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, Zhuang SM (2018) Hepatoma cell‐secreted exosomal microRNA‐103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology 68: 1459–1475 [DOI] [PubMed] [Google Scholar]
- Fang Z, Wang X, Wu J, Xiao R, Liu J (2020) High serum extracellular vesicle miR‐10b expression predicts poor prognosis in patients with acute myeloid leukemia. Cancer Biomark 27: 1–9 [DOI] [PubMed] [Google Scholar]
- Fearon KC, Glass DJ, Guttridge DC (2012) Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metab 16: 153–166 [DOI] [PubMed] [Google Scholar]
- Fendt SM, Frezza C, Erez A (2020) Targeting metabolic plasticity and flexibility dynamics for cancer therapy. Cancer Discov 10: 1797–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Zhao W‐L, Ye Y‐Y, Bai X‐C, Liu R‐Q, Chang L‐F, Zhou Q, Sui S‐F (2010) Cellular internalization of exosomes occurs through phagocytosis. Traffic 11: 675–687 [DOI] [PubMed] [Google Scholar]
- Feng Y, Guo M, Zhao H, Han S, Dong Q, Cui M (2020) Mesenchymal‐stem‐cell‐derived extracellular vesicles mitigate trained immunity in the brain. Front Bioeng Biotechnol 8: 599058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Xiong Z, Wang C, Xiao W, Xiao H, Xie K, Chen K, Liang H, Zhang X, Yang H (2021) Folic acid‐modified Exosome‐PH20 enhances the efficiency of therapy via modulation of the tumor microenvironment and directly inhibits tumor cell metastasis. Bioact Mater 6: 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson Bennit HR, Gonda A, Kabagwira J, Oppegard L, Chi D, Licero Campbell J, De Leon M, Wall NR (2021) Natural killer cell phenotype and functionality affected by exposure to extracellular survivin and lymphoma‐derived exosomes. Int J Mol Sci 22: 1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, Chow A, O'Connor ST, Li S, Chin AR et al (2015) Breast‐cancer‐secreted miR‐122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol 17: 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton AE, Prado MM, Lopez‐Jimenez E, Fajardo‐Puerta AB, Jawad ZAR, Lawton P, Giovannetti E, Habib NA, Castellano L, Stebbing J et al (2018) Glypican‐1 is enriched in circulating‐exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget 9: 19006–19013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser K, Jo A, Giedt J, Vinegoni C, Yang KS, Peruzzi P, Chiocca EA, Breakefield XO, Lee H, Weissleder R (2019) Characterization of single microvesicles in plasma from glioblastoma patients. Neuro Oncol 21: 606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredsoe J, Rasmussen AKI, Mouritzen P, Borre M, Orntoft T, Sorensen KD (2019) A five‐microRNA model (pCaP) for predicting prostate cancer aggressiveness using cell‐free urine. Int J Cancer 145: 2558–2567 [DOI] [PubMed] [Google Scholar]
- Freed‐Pastor WA, Prives C (2012) Mutant p53: one name, many proteins. Genes Dev 26: 1268–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, Worthen GS, Albelda SM (2009) Polarization of tumor‐associated neutrophil phenotype by TGF‐beta: “N1” versus “N2” TAN. Cancer Cell 16: 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F, Jiang W, Zhou L, Chen Z (2018) Circulating exosomal miR‐17‐5p and miR‐92a‐3p predict pathologic stage and grade of colorectal cancer. Transl Oncol 11: 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii N, Hirata H, Ueno K, Mori J, Oka S, Shimizu K, Kawai Y, Inoue R, Yamamoto Y, Matsumoto H et al (2017) Extracellular miR‐224 as a prognostic marker for clear cell renal cell carcinoma. Oncotarget 8: 109877–109888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M (2013) Rab27 effectors, pleiotropic regulators in secretory pathways. Traffic 14: 949–963 [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, Nagaraj S (2009) Myeloid‐derived suppressor cells as regulators of the immune system. Nat Rev Immunol 9: 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbo PM Jr, Ciesielski MJ, Figel S, Maguire O, Qiu J, Wiltsie L, Minderman H, Fenstermaker RA (2017) Circulating CD9+/GFAP+/survivin+ exosomes in malignant glioma patients following survivin vaccination. Oncotarget 8: 114722–114735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Bravo‐San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V et al (2015) Autophagy in malignant transformation and cancer progression. EMBO J 34: 856–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan DX, Wang YB, He MY, Chen ZY, Qin XX, Miao ZW, Chen YH, Li B (2020) Lung cancer cells‐controlled Dkk‐1 production in brain metastatic cascade drive microglia to acquire a pro‐tumorigenic phenotype. Front Cell Dev Biol 8: 591405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh K, Basnet H, Kaygusuz Y, Laughney AM, He L, Sharma R, O'Rourke KP, Reuter VP, Huang YH, Turkekul M et al (2020) L1CAM defines the regenerative origin of metastasis‐initiating cells in colorectal cancer. Nat Cancer 1: 28–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangoda L, Liem M, Ang C‐S, Keerthikumar S, Adda CG, Parker BS, Mathivanan S (2017) Proteomic profiling of exosomes secreted by breast cancer cells with varying metastatic potential. Proteomics 17: 1600370 [DOI] [PubMed] [Google Scholar]
- Gao D, Nolan DJ, Mellick AS, Bambino K, McDonnell K, Mittal V (2008) Endothelial progenitor cells control the angiogenic switch in mouse lung metastasis. Science 319: 195–198 [DOI] [PubMed] [Google Scholar]
- Gao Y, Bado I, Wang H, Zhang W, Rosen JM, Zhang XH (2019) Metastasis organotropism: redefining the congenial soil. Dev Cell 49: 375–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wei B, de Assuncao TM, Liu Z, Hu X, Ibrahim S, Cooper SA, Cao S, Shah VH, Kostallari E (2020a) Hepatic stellate cell autophagy inhibits extracellular vesicle release to attenuate liver fibrosis. J Hepatol 73: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang Z, Mashimo T, Shen B, Nyagilo J, Wang H, Wang Y, Liu Z, Mulgaonkar A, Hu XL et al (2020b) Gliomas interact with non‐glioma brain cells via extracellular vesicles. Cell Rep 30: 2489–2500 [DOI] [PubMed] [Google Scholar]
- Garcia NA, Moncayo‐Arlandi J, Sepulveda P, Diez‐Juan A (2016) Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc Res 109: 397–408 [DOI] [PubMed] [Google Scholar]
- Garnier D, Magnus N, Lee TH, Bentley V, Meehan B, Milsom C, Montermini L, Kislinger T, Rak J (2012) Cancer cells induced to express mesenchymal phenotype release exosome‐like extracellular vesicles carrying tissue factor. J Biol Chem 287: 43565–43572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, Almeida D, Koller A, Hajjar KA, Stainier DY et al (2013) The perivascular niche regulates breast tumour dormancy. Nat Cell Biol 15: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghajar CM (2015) Metastasis prevention by targeting the dormant niche. Nat Rev Cancer 15: 238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghamloush F, Ghayad SE, Rammal G, Fahs A, Ayoub AJ, Merabi Z, Harajly M, Zalzali H, Saab R (2019) The PAX3‐FOXO1 oncogene alters exosome miRNA content and leads to paracrine effects mediated by exosomal miR‐486. Sci Rep 9: 14242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoroghi S, Mary B, Larnicol A, Asokan N, Klein A, Osmani N, Busnelli I, Delalande F, Paul N, Halary S et al (2021) Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes. Elife 10: e61539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Davey M, Chute IC, Griffiths SG, Lewis S, Chacko S, Barnett D, Crapoulet N, Fournier S, Joy A et al (2014) Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS One 9: e110443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal A, Rodrigues LC, Gowda CP, Elcheva IA, Liu Z, Abraham T, Spiegelman VS (2019) Extracellular vesicle‐dependent effect of RNA‐binding protein IGF2BP1 on melanoma metastasis. Oncogene 38: 4182–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghossoub R, Lembo F, Rubio A, Gaillard CB, Bouchet J, Vitale N, Slavik J, Machala M, Zimmermann P (2014) Syntenin‐ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat Commun 5: 3477 [DOI] [PubMed] [Google Scholar]
- Goddard ET, Bozic I, Riddell SR, Ghajar CM (2018) Dormant tumour cells, their niches and the influence of immunity. Nat Cell Biol 20: 1240–1249 [DOI] [PubMed] [Google Scholar]
- Gomari H, Forouzandeh Moghadam M, Soleimani M (2018) Targeted cancer therapy using engineered exosome as a natural drug delivery vehicle. Onco Targets Ther 11: 5753–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomari H, Forouzandeh Moghadam M, Soleimani M, Ghavami M, Khodashenas S (2019) Targeted delivery of doxorubicin to HER2 positive tumor models. Int J Nanomedicine 14: 5679–5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes FG, Sandim V, Almeida VH, Rondon AMR, Succar BB, Hottz ED, Leal AC, Vercoza BRF, Rodrigues JCF, Bozza PT et al (2017) Breast‐cancer extracellular vesicles induce platelet activation and aggregation by tissue factor‐independent and ‐dependent mechanisms. Thromb Res 159: 24–32 [DOI] [PubMed] [Google Scholar]
- Gong C, Tian J, Wang Z, Gao Y, Wu X, Ding X, Qiang L, Li G, Han Z, Yuan Y et al (2019) Functional exosome‐mediated co‐delivery of doxorubicin and hydrophobically modified microRNA 159 for triple‐negative breast cancer therapy. J Nanobiotechnol 17: 93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐King H, Garcia NA, Ontoria‐Oviedo I, Ciria M, Montero JA, Sepulveda P (2017) Hypoxia inducible factor‐1alpha potentiates jagged 1‐mediated angiogenesis by mesenchymal stem cell‐derived exosomes. Stem Cells 35: 1747–1759 [DOI] [PubMed] [Google Scholar]
- Gopal SK, Greening DW, Hanssen EG, Zhu HJ, Simpson RJ, Mathias RA (2016) Oncogenic epithelial cell‐derived exosomes containing Rac1 and PAK2 induce angiogenesis in recipient endothelial cells. Oncotarget 7: 19709–19722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulielmaki E, Ioannidou A, Tsekrekou M, Stratigi K, Poutakidou IK, Gkirtzimanaki K, Aivaliotis M, Evangelou K, Topalis P, Altmuller J et al (2020) Tissue‐infiltrating macrophages mediate an exosome‐based metabolic reprogramming upon DNA damage. Nat Commun 11: 42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindappa PK, Patil M, Garikipati VNS, Verma SK, Saheera S, Narasimhan G, Zhu W, Kishore R, Zhang J, Krishnamurthy P (2020) Targeting exosome‐associated human antigen R attenuates fibrosis and inflammation in diabetic heart. FASEB J 34: 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greten FR, Grivennikov SI (2019) Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51: 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JC, Chaudhary V, Bartscherer K, Boutros M (2012) Active Wnt proteins are secreted on exosomes. Nat Cell Biol 14: 1036–1045 [DOI] [PubMed] [Google Scholar]
- Gruenberg J (2020) Life in the lumen: the multivesicular endosome. Traffic 21: 76–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Zhang H, Gao Y (2020) Adipose mesenchymal stem cells‐secreted extracellular vesicles containing microRNA‐192 delays diabetic retinopathy by targeting ITGA1. J Cell Physiol 236: 5036–5051 [DOI] [PubMed] [Google Scholar]
- Gu H, Ji R, Zhang X, Wang M, Zhu W, Qian H, Chen Y, Jiang P, Xu W (2016) Exosomes derived from human mesenchymal stem cells promote gastric cancer cell growth and migration via the activation of the Akt pathway. Mol Med Rep 14: 3452–3458 [DOI] [PubMed] [Google Scholar]
- Gu H, Yang K, Shen Z, Jia K, Liu P, Pan M, Sun C (2021) ER stress‐induced adipocytes secrete‐aldo‐keto reductase 1B7‐containing exosomes that cause nonalcoholic steatohepatitis in mice. Free Radic Biol Med 163: 220–233 [DOI] [PubMed] [Google Scholar]
- Guan Q (2019) A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J Immunol Res 2019: 7247238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guescini M, Genedani S, Stocchi V, Agnati LF (2010) Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm 117: 1–4 [DOI] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Chang DH, Liu Q, Kossenkov AV, Yan J, Korst RJ, Nam BT, Xu H, Zhang L et al (2015) AKAP4 is a circulating biomarker for non‐small cell lung cancer. Oncotarget 6: 17637–17647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Lui GYL, Lai SL, Wilmott JS, Tikoo S, Jackett LA, Quek C, Brown DL, Sharp DM, Kwan RYQ et al (2019a) RAB27A promotes melanoma cell invasion and metastasis via regulation of pro‐invasive exosomes. Int J Cancer 144: 3070–3085 [DOI] [PubMed] [Google Scholar]
- Guo L, Zhu Y, Li L, Zhou S, Yin G, Yu G, Cui H (2019b) Breast cancer cell‐derived exosomal miR‐20a‐5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med 8: 5687–5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Wang Y, Jia J, Mao X, Stankiewicz E, Scandura G, Burke E, Xu L, Marzec J, Davies CR et al (2020a) The identification of plasma exosomal miR‐423‐3p as a potential predictive biomarker for prostate cancer castration‐resistance development by plasma exosomal miRNA sequencing. Front Cell Dev Biol 8: 602493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wang X, Wang K, He Y (2020b) Appraising the value of serum and serum‐derived exosomal LncRNA‐EXOC7 as a promising biomarker in cervical cancer. Clin Lab 10.7754/Clin.Lab.2019.191203 [DOI] [PubMed] [Google Scholar]
- Guo S, Hu C, Zhai X, Sun D (2021) Circular RNA 0006602 in plasma exosomes: a new potential diagnostic biomarker for hepatocellular carcinoma. Am J Transl Res 13: 6001–6015 [PMC free article] [PubMed] [Google Scholar]
- Hadla M, Palazzolo S, Corona G, Caligiuri I, Canzonieri V, Toffoli G, Rizzolio F (2016) Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine (Lond) 11: 2431–2441 [DOI] [PubMed] [Google Scholar]
- Han Z, Li Y, Zhang J, Guo C, Li Q, Zhang X, Lan Y, Gu W, Xing Z, Liang L et al (2020) Tumor‐derived circulating exosomal miR‐342‐5p and miR‐574‐5p as promising diagnostic biomarkers for early‐stage lung adenocarcinoma. Int J Med Sci 17: 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Shi WJ, Xie YB, Zhang ZG (2021) Diagnostic value of four serum exosome microRNAs panel for the detection of colorectal cancer. World J Gastrointest Oncol 13: 970–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, Patel T, Piroyan A, Sokolsky M, Kabanov AV et al (2015) Exosomes as drug delivery vehicles for Parkinson's disease therapy. J Control Release 207: 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Yamamoto H, Kishida S, Kishida M, Awada C, Takao T, Kikuchi A (2017) Wnt5b‐associated exosomes promote cancer cell migration and proliferation. Cancer Sci 108: 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraszti RA, Didiot M‐C, Sapp E, Leszyk J, Shaffer SA, Rockwell HE, Gao F, Narain NR, Difiglia M, Kiebish MA et al (2016) High‐resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J Extracell Vesicles 5: 32570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C, Heuser J, Stahl P (1983) Receptor‐mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol 97: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Kuwata K, Yoshitake J, Shimomura S, Uchida K, Shibata T (2020) Extracellular vesicles derived from inflamed murine colorectal tissue induce fibroblast proliferation via epidermal growth factor receptor. FEBS J 288: 1906–1917 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Ochi H, Sunamura S, Kosaka N, Mabuchi Y, Fukuda T, Yao K, Kanda H, Ae K, Okawa A et al (2018) Cancer‐secreted hsa‐miR‐940 induces an osteoblastic phenotype in the bone metastatic microenvironment via targeting ARHGAP1 and FAM134A. Proc Natl Acad Sci USA 115: 2204–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Zhu W, Chen Q, Yuan Y, Wang Y, Wang J, Wu X (2019a) Ovarian cancer cell‐secreted exosomal miR‐205 promotes metastasis by inducing angiogenesis. Theranostics 9: 8206–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Li Z, Yu Y, Zeng Q, Cheng Y, Ji W, Xia W, Lu S (2019b) Exosomal miR‐499a‐5p promotes cell proliferation, migration and EMT via mTOR signaling pathway in lung adenocarcinoma. Exp Cell Res 379: 203–213 [DOI] [PubMed] [Google Scholar]
- He W, Tang J, Li W, Li Y, Mei Y, He L, Zhong K, Xu R (2019c) Mutual regulation of JAG2 and PRAF2 promotes migration and invasion of colorectal cancer cells uncoupled from epithelial‐mesenchymal transition. Cancer Cell Int 19: 160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath N, Grant L, De Oliveira TM, Rowlinson R, Osteikoetxea X, Dekker N, Overman R (2018) Rapid isolation and enrichment of extracellular vesicle preparations using anion exchange chromatography. Sci Rep 8: 5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ (1999) Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha‐granules. Blood 94: 3791–3799 [PubMed] [Google Scholar]
- Henrich SE, McMahon KM, Plebanek MP, Calvert AE, Feliciano TJ, Parrish S, Tavora F, Mega A, De Souza A, Carneiro BA et al (2020) Prostate cancer extracellular vesicles mediate intercellular communication with bone marrow cells and promote metastasis in a cholesterol‐dependent manner. J Extracell Vesicles 10: e12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez C, Huebener P, Schwabe RF (2016) Damage‐associated molecular patterns in cancer: a double‐edged sword. Oncogene 35: 5931–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson C, Capraro D, Burdach J, Whitaker N, Morris KV (2016) Extracellular vesicle associated long non‐coding RNAs functionally enhance cell viability. Noncoding RNA Res 1: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham JN, Demory Beckler M, Gephart JD, Franklin JL, Bogatcheva G, Kremers GJ, Piston DW, Ayers GD, McConnell RE, Tyska MJ et al (2011) Amphiregulin exosomes increase cancer cell invasion. Curr Biol 21: 779–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita T, Miyata M, Watanabe R, Oneyama C (2018) Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci Rep 8: 14035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita T, Kuwahara A, Watanabe R, Miyata M, Oneyama C (2019) Src in endosomal membranes promotes exosome secretion and tumor progression. Sci Rep 9: 3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinger SA, Cha DJ, Franklin JL, Higginbotham JN, Dou Y, Ping J, Shu L, Prasad N, Levy S, Zhang B et al (2018) Diverse long RNAs are differentially sorted into extracellular vesicles secreted by colorectal cancer cells. Cell Rep 25: 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinger SA, Abner JJ, Franklin JL, Jeppesen DK, Coffey RJ, Patton JG (2020) Rab13 regulates sEV secretion in mutant KRAS colorectal cancer cells. Sci Rep 10: 15804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA (2005) Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7: 469–483 [DOI] [PubMed] [Google Scholar]
- Hofmann L, Ludwig S, Schuler PJ, Hoffmann TK, Brunner C, Theodoraki MN (2020) The potential of CD16 on plasma‐derived exosomes as a liquid biomarker in head and neck cancer. Int J Mol Sci 21: 3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JL, San RS, Wickline SA (2011) Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res 71: 3792–3801 [DOI] [PubMed] [Google Scholar]
- Horie K, Kawakami K, Fujita Y, Matsuda Y, Arai T, Suzui N, Miyazaki T, Koie T, Mizutani K, Ito M (2020) Serum exosomal gamma‐glutamyltransferase activity increased in patients with renal cell carcinoma with advanced clinicopathological features. Oncology 98: 734–742 [DOI] [PubMed] [Google Scholar]
- Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega‐Larson N, Tyska MJ, Weaver AM (2013) Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep 5: 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Costa‐Silva B, Shen T‐L, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S et al (2015) Tumour exosome integrins determine organotropic metastasis. Nature 527: 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H, Heissel S et al (2020) Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 182: 1044–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CH, Tai SK, Yang MH (2018) Snail‐overexpressing cancer cells promote M2‐like polarization of tumor‐associated macrophages by delivering MiR‐21‐abundant exosomes. Neoplasia 20: 775–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C, Morohashi Y, Yoshimura S, Manrique‐Hoyos N, Jung S, Lauterbach MA, Bakhti M, Gronborg M, Mobius W, Rhee J et al (2010) Regulation of exosome secretion by Rab35 and its GTPase‐activating proteins TBC1D10A‐C. J Cell Biol 189: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, Wu CY, Kuo PL (2017) Hypoxic lung cancer‐secreted exosomal miR‐23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO‐1. Oncogene 36: 4929–4942 [DOI] [PubMed] [Google Scholar]
- Hsu YL, Huang MS, Hung JY, Chang WA, Tsai YM, Pan YC, Lin YS, Tsai HP, Kuo PL (2020) Bone‐marrow‐derived cell‐released extracellular vesicle miR‐92a regulates hepatic pre‐metastatic niche in lung cancer. Oncogene 39: 739–753 [DOI] [PubMed] [Google Scholar]
- Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, Wu Y, Qin J (2015) Fibroblast‐derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS One 10: e0125625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Ru Z, Xiao W, Xiong Z, Wang C, Yuan C, Zhang X, Yang H (2018) Adipose tissue browning in cancer‐associated cachexia can be attenuated by inhibition of exosome generation. Biochem Biophys Res Commun 506: 122–129 [DOI] [PubMed] [Google Scholar]
- Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ et al (2019a) CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial‐mesenchymal transition in colorectal cancer. Mol Cancer 18: 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YB, Yan C, Mu L, Mi YL, Zhao H, Hu H, Li XL, Tao DD, Wu YQ, Gong JP et al (2019b) Exosomal Wnt‐induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene 38: 1951–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Xu Z, Zhu S, Sun W, Wang X, Tan C, Zhang Y, Zhang G, Xu Y, Tang J (2021) Small extracellular vesicle‐mediated Hsp70 intercellular delivery enhances breast cancer adriamycin resistance. Free Radic Biol Med 164: 85–95 [DOI] [PubMed] [Google Scholar]
- Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, Wang D, See W, Costello BA, Quevedo F et al (2015) Exosomal miR‐1290 and miR‐375 as prognostic markers in castration‐resistant prostate cancer. Eur Urol 67: 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Bockorny B, Paul I, Akshinthala D, Frappart PO, Gandarilla O, Bose A, Sanchez‐Gonzalez V, Rouse EE, Lehoux SD et al (2020a) PDX‐derived organoids model in vivo drug response and secrete biomarkers. JCI Insight 5: e135544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Hong Z, Xiao C, Li L, Chen L, Cheng S, Lei T, Zheng H (2020b) Effects of exosomes on neurological function recovery for ischemic stroke in pre‐clinical studies: a meta‐analysis. Front Cell Neurosci 14: 593130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Yan Y, Liu Y, Lin M, Ma J, Zhang W, Dai J, Li J, Guo Q, Chen H et al (2020c) Exosomes with low miR‐34c‐3p expression promote invasion and migration of non‐small cell lung cancer by upregulating integrin alpha2beta1. Signal Transduct Target Ther 5: 39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, Zhou J, Tang ZY (2020d) Exosomal circRNA‐100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res 39: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Tang S, Shen D, Li X, Liang L, Ding Y, Xu B (2021) Circulating plasma exosomal miRNA profiles serve as potential metastasis‐related biomarkers for hepatocellular carcinoma. Oncol Lett 21: 168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur YH, Feng S, Wilson KF, Cerione RA, Antonyak MA (2020) Embryonic stem cell‐derived extracellular vesicles maintain ESC stemness by activating FAK. Dev Cell 56: 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH (2015) ESCRTs are everywhere. EMBO J 34: 2398–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz SN, Rider MA, Bundy JL, Liu X, Singh RK, Meckes DG Jr (2016) Proteomic profiling of NCI‐60 extracellular vesicles uncovers common protein cargo and cancer type‐specific biomarkers. Oncotarget 7: 86999–87015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husna AA, Rahman MM, Lai YC, Chen HW, Hasan MN, Nakagawa T, Miura N (2021) Identification of melanoma‐specific exosomal miRNAs as the potential biomarker for canine oral melanoma. Pigment Cell Melanoma Res 34: 1062–1073 [DOI] [PubMed] [Google Scholar]
- Hwang I, Shen X, Sprent J (2003) Direct stimulation of naive T cells by membrane vesicles from antigen‐presenting cells: distinct roles for CD54 and B7 molecules. Proc Natl Acad Sci USA 100: 6670–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda C, Haga H, Makino N, Inuzuka T, Kurimoto A, Ueda T, Matsuda A, Kakizaki Y, Ishizawa T, Kobayashi T et al (2021) Utility of Claudin‐3 in extracellular vesicles from human bile as biomarkers of cholangiocarcinoma. Sci Rep 11: 1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Takahashi Y, Nishikawa M, Kato K, Morishita M, Yamashita T, Matsumoto A, Charoenviriyakul C, Takakura Y (2015) Macrophage‐dependent clearance of systemically administered B16BL6‐derived exosomes from the blood circulation in mice. J Extracell Vesicles 4: 26238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imjeti NS, Menck K, Egea‐Jimenez AL, Lecointre C, Lembo F, Bouguenina H, Badache A, Ghossoub R, David G, Roche S et al (2017) Syntenin mediates SRC function in exosomal cell‐to‐cell communication. Proc Natl Acad Sci USA 114: 12495–12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inder KL, Ruelcke JE, Petelin L, Moon H, Choi E, Rae J, Blumenthal A, Hutmacher D, Saunders NA, Stow JL et al (2014) Cavin‐1/PTRF alters prostate cancer cell‐derived extracellular vesicle content and internalization to attenuate extracellular vesicle‐mediated osteoclastogenesis and osteoblast proliferation. J Extracell Vesicles 10.3402/jev.v3.23784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraci N, Gaude E, Leonardi T, Costa ASH, Cossetti C, Peruzzotti‐Jametti L, Bernstock JD, Saini HK, Gelati M, Vescovi AL et al (2017) Extracellular vesicles are independent metabolic units with asparaginase activity. Nat Chem Biol 13: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Shiromizu T, Ohnishi S, Suzuki S, Mabe K, Hasegawa A, Ujiie H, Fujita Y, Sato Y, Terai S et al (2018) Potential role of extracellular vesicle‐mediated antigen presentation in Helicobacter pylori hypersensitivity during eradication therapy. J Allergy Clin Immunol 142: 672–676 [DOI] [PubMed] [Google Scholar]
- Itoh T, Ito Y, Ohtsuki Y, Ando M, Tsukamasa Y, Yamada N, Naoe T, Akao Y (2012) Microvesicles released from hormone‐refractory prostate cancer cells facilitate mouse pre‐osteoblast differentiation. J Mol Histol 43: 509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A, Humphries TLR, Owens EP, Zhao KN, Masci PP, Johnson DW, Nikolic‐Paterson D, Gobe GC, Fairlie DP, Vesey DA (2021) PAR2 activation on human kidney tubular epithelial cells induces tissue factor synthesis, that enhances blood clotting. Front Physiol 12: 615428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H, Tsuda M, Sato Y, Kosaka N, Ochiya T, Iwamoto H, Namba K, Takeda Y (2015) Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J Dairy Sci 98: 2920–2933 [DOI] [PubMed] [Google Scholar]
- Jackson CE, Scruggs BS, Schaffer JE, Hanson PI (2017) Effects of inhibiting VPS4 support a general role for ESCRTs in extracellular vesicle biogenesis. Biophys J 113: 1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH (2006) SNAREs — engines for membrane fusion. Nat Rev Mol Cell Biol 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Jang JY, Kim YS, Kang KN, Kim KH, Park YJ, Kim CW (2021a) Multiple microRNAs as biomarkers for early breast cancer diagnosis. Mol Clin Oncol 14: 31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y, Kim H, Yoon S, Lee H, Hwang J, Jung J, Chang JH, Choi J, Kim H (2021b) Exosome‐based photoacoustic imaging guided photodynamic and immunotherapy for the treatment of pancreatic cancer. J Control Release 330: 293–304 [DOI] [PubMed] [Google Scholar]
- Jankovic T, Goc S, Mitic N, Danilovic Lukovic J, Jankovic M (2020) Membrane‐associated gamma‐glutamyl transferase and alkaline phosphatase in the context of concanavalin A‐ and wheat germ agglutinin‐reactive glycans mark seminal prostasome populations from normozoospermic and oligozoospermic men. Ups J Med Sci 125: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janpipatkul K, Trachu N, Watcharenwong P, Panvongsa W, Worakitchanon W, Metheetrairut C, Oranratnachai S, Reungwetwattana T, Chairoungdua A (2021) Exosomal microRNAs as potential biomarkers for osimertinib resistance of non‐small cell lung cancer patients. Cancer Biomark 31: 281–294 [DOI] [PubMed] [Google Scholar]
- Ji R, Zhang B, Zhang X, Xue J, Yuan X, Yan Y, Wang M, Zhu W, Qian H, Xu W (2015) Exosomes derived from human mesenchymal stem cells confer drug resistance in gastric cancer. Cell Cycle 14: 2473–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji R, Zhang X, Gu H, Ma J, Wen X, Zhou J, Qian H, Xu W, Qian J, Lin J (2019) miR‐374a‐5p: a new target for diagnosis and drug resistance therapy in gastric cancer. Mol Ther Nucleic Acids 18: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YJ, Jin DD, Jiang F, Liu JX, Qu LS, Ni WK, Liu ZX, Lu CH, Ni RZ, Zhu J et al (2019) Characterization and proteomic profiling of pancreatic cancer‐derived serum exosomes. J Cell Biochem 120: 988–999 [DOI] [PubMed] [Google Scholar]
- Jimenez‐Alesanco A, Marcuello M, Pastor‐Jimenez M, Lopez‐Puerto L, Bonjoch L, Gironella M, Carrascal M, Abian J, de‐Madaria E, Closa D (2019) Acute pancreatitis promotes the generation of two different exosome populations. Sci Rep 9: 19887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Liu P, Wu Y, Meng X, Wu M, Han J, Tan X (2018) Exosomal zinc transporter ZIP4 promotes cancer growth and is a novel diagnostic biomarker for pancreatic cancer. Cancer Sci 109: 2946–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AM, Olefsky JM (2013) The origins and drivers of insulin resistance. Cell 152: 673–684 [DOI] [PubMed] [Google Scholar]
- Ju Q, Zhao L, Gao J, Zhou L, Xu Y, Sun Y, Zhao X (2019) Mutant p53 increases exosome‐mediated transfer of miR‐21‐3p and miR‐769‐3p to promote pulmonary metastasis. Chin J Cancer Res 31: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan T, Furthauer M (2018) Biogenesis and function of ESCRT‐dependent extracellular vesicles. Semin Cell Dev Biol 74: 66–77 [DOI] [PubMed] [Google Scholar]
- Jung KO, Kim YH, Chung SJ, Lee CH, Rhee S, Pratx G, Chung JK, Youn H (2020) Identification of lymphatic and hematogenous routes of rapidly labeled radioactive and fluorescent exosomes through highly sensitive multimodal imaging. Int J Mol Sci 21: 7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska S, Beury DW, Gopalan V, Tycko AK, Qin H, Clements ME, Drake J, Nwanze C, Murgai M, Rae Z et al (2021) Genetically engineered myeloid cells rebalance the core immune suppression program in metastasis. Cell 184: 2033–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, Zhang J, Weitz J, Chin L, Futreal A et al (2014) Identification of double‐stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 289: 3869–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn SE, Hull RL, Utzschneider KM (2006) Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444: 840–846 [DOI] [PubMed] [Google Scholar]
- Kai F, Drain AP, Weaver VM (2019) The extracellular matrix modulates the metastatic journey. Dev Cell 49: 332–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R (2017) Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 546: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka R, Iinuma H, Dejima H, Sakai T, Uehara H, Matsutani N, Kawamura M (2018) Usefulness of plasma exosomal microRNA‐451a as a noninvasive biomarker for early prediction of recurrence and prognosis of non‐small cell lung cancer. Oncology 94: 311–323 [DOI] [PubMed] [Google Scholar]
- Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ et al (2013) Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium‐induced colitis. PLoS One 8: e76520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan A, Wells RB, Sivakumar S, Komatsu S, Singh KP, Samten B, Philley JV, Sauter ER, Ikebe M, Idell S et al (2016) Mitochondrial reprogramming regulates breast cancer progression. Clin Cancer Res 22: 3348–3360 [DOI] [PubMed] [Google Scholar]
- Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA et al (2005) VEGFR1‐positive haematopoietic bone marrow progenitors initiate the pre‐metastatic niche. Nature 438: 820–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis GS, Pastoriza JM, Wang Y, Harney AS, Entenberg D, Pignatelli J, Sharma VP, Xue EA, Cheng E, D'Alfonso TM et al (2017) Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM‐mediated mechanism. Sci Transl Med 9: eaan0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katakura S, Kobayashi N, Hashimoto H, Kamimaki C, Tanaka K, Kubo S, Nakashima K, Teranishi S, Manabe S, Watanabe K et al (2020) MicroRNA‐200b is a potential biomarker of the expression of PD‐L1 in patients with lung cancer. Thorac Cancer 11: 2975–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama M, Wiklander OPB, Fritz T, Caidahl K, El‐Andaloussi S, Zierath JR, Krook A (2019) Circulating exosomal miR‐20b‐5p is elevated in type 2 diabetes and could impair insulin action in human skeletal muscle. Diabetes 68: 515–526 [DOI] [PubMed] [Google Scholar]
- Katsu M, Hama Y, Utsumi J, Takashina K, Yasumatsu H, Mori F, Wakabayashi K, Shoji M, Sasaki H (2019) MicroRNA expression profiles of neuron‐derived extracellular vesicles in plasma from patients with amyotrophic lateral sclerosis. Neurosci Lett 708: 134176 [DOI] [PubMed] [Google Scholar]
- Ke X, Yang D, Liang J, Wang X, Wu S, Wang X, Hu C (2017) Human endothelial progenitor cell‐derived exosomes increase proliferation and angiogenesis in cardiac fibroblasts by promoting the mesenchymal‐endothelial transition and reducing high mobility group box 1 protein B1 expression. DNA Cell Biol 36: 1018–1028 [DOI] [PubMed] [Google Scholar]
- Keklikoglou I, Cianciaruso C, Guc E, Squadrito ML, Spring LM, Tazzyman S, Lambein L, Poissonnier A, Ferraro GB, Baer C et al (2019) Chemotherapy elicits pro‐metastatic extracellular vesicles in breast cancer models. Nat Cell Biol 21: 190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennel PJ, Saha A, Maldonado DA, Givens R, Brunjes DL, Castillero E, Zhang X, Ji R, Yahi A, George I et al (2018) Serum exosomal protein profiling for the non‐invasive detection of cardiac allograft rejection. J Heart Lung Transplant 37: 409–417 [DOI] [PubMed] [Google Scholar]
- Khan S, Jutzy JM, Valenzuela MM, Turay D, Aspe JR, Ashok A, Mirshahidi S, Mercola D, Lilly MB, Wall NR (2012) Plasma‐derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One 7: e46737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharmate G, Hosseini‐Beheshti E, Caradec J, Chin MY, Tomlinson Guns ES (2016) Epidermal growth factor receptor in prostate cancer derived exosomes. PLoS One 11: e0154967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kifle DW, Chaiyadet S, Waardenberg AJ, Wise I, Cooper M, Becker L, Doolan DL, Laha T, Sotillo J, Pearson MS et al (2020) Uptake of Schistosoma mansoni extracellular vesicles by human endothelial and monocytic cell lines and impact on vascular endothelial cell gene expression. Int J Parasitol 50: 685–696 [DOI] [PubMed] [Google Scholar]
- Kim LC, Song L, Haura EB (2009) Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol 6: 587–595 [DOI] [PubMed] [Google Scholar]
- Kim DK, Nishida H, An SY, Shetty AK, Bartosh TJ, Prockop DJ (2016a) Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc Natl Acad Sci USA 113: 170–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O et al (2016b) Development of exosome‐encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 12: 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Kim H, Choi YJ, Kim SY, Lee JE, Sung KJ, Sung YH, Pack CG, Jung MK, Han B et al (2019a) Exosomal PD‐L1 promotes tumor growth through immune escape in non‐small cell lung cancer. Exp Mol Med 51: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee CH, Lee SW (2019b) Exosomal transmission of microRNA from HCV replicating cells stimulates transdifferentiation in hepatic stellate cells. Mol Ther Nucleic Acids 14: 483–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Choi MC, Jeong JY, Hwang S, Jung SG, Joo WD, Park H, Song SH, Lee C, Kim TH et al (2019c) Serum exosomal miRNA‐145 and miRNA‐200c as promising biomarkers for preoperative diagnosis of ovarian carcinomas. J Cancer 10: 1958–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Yang JS, Lee GB, Moon MH (2020) Evaluation of exosome separation from human serum by frit‐inlet asymmetrical flow field‐flow fractionation and multiangle light scattering. Anal Chim Acta 1124: 137–145 [DOI] [PubMed] [Google Scholar]
- Kim H, Shin S (2021) ExoCAS‐2: rapid and pure isolation of exosomes by anionic exchange using magnetic beads. Biomedicine 9: 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kim JS, Park NR, Nam H, Sung PS, Bae SH, Choi JY, Yoon SK, Hur W, Jang JW (2021) Exosomal miR‐125b exerts anti‐metastatic properties and predicts early metastasis of hepatocellular carcinoma. Front Oncol 11: 637247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HW, Michael MZ, Gleadle JM (2012) Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 12: 421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirave P, Gondaliya P, Kulkarni B, Rawal R, Garg R, Jain A, Kalia K (2020) Exosome mediated miR‐155 delivery confers cisplatin chemoresistance in oral cancer cells via epithelial‐mesenchymal transition. Oncotarget 11: 1157–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita S, Maeda N, Shimomura I (2019) Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J Clin Invest 129: 4041–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa T, Taniuchi K, Tsuboi M, Sakaguchi M, Kohsaki T, Okabayashi T, Saibara T (2019) Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol Oncol 13: 212–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitai Y, Kawasaki T, Sueyoshi T, Kobiyama K, Ishii KJ, Zou J, Akira S, Matsuda T, Kawai T (2017) DNA‐containing exosomes derived from cancer cells treated with topotecan activate a STING‐dependent pathway and reinforce antitumor immunity. J Immunol 198: 1649–1659 [DOI] [PubMed] [Google Scholar]
- Ko J, Bhagwat N, Black T, Yee SS, Na YJ, Fisher S, Kim J, Carpenter EL, Stanger BZ, Issadore D (2018) miRNA profiling of magnetic nanopore‐isolated extracellular vesicles for the diagnosis of pancreatic cancer. Cancer Res 78: 3688–3697 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Sawada K, Miyamoto M, Shimizu A, Yamamoto M, Kinose Y, Nakamura K, Kawano M, Kodama M, Hashimoto K et al (2020) Exploring the potential of engineered exosomes as delivery systems for tumor‐suppressor microRNA replacement therapy in ovarian cancer. Biochem Biophys Res Commun 527: 153–161 [DOI] [PubMed] [Google Scholar]
- Koh E, Lee EJ, Nam GH, Hong Y, Cho E, Yang Y, Kim IS (2017) Exosome‐SIRPalpha, a CD47 blockade increases cancer cell phagocytosis. Biomaterials 121: 121–129 [DOI] [PubMed] [Google Scholar]
- Kohaar I, Chen Y, Banerjee S, Borbiev T, Kuo HC, Ali A, Ravindranath L, Kagan J, Srivastava S, Dobi A et al (2021) A urine exosome gene expression panel distinguishes between indolent and aggressive prostate cancers at biopsy. J Urol 205: 420–425 [DOI] [PubMed] [Google Scholar]
- Komine‐Aizawa S, Ito S, Aizawa S, Namiki T, Hayakawa S (2020) Cow milk exosomes activate NK cells and gammadeltaT cells in human PBMCs in vitro . Immunol Med 43: 161–170 [DOI] [PubMed] [Google Scholar]
- Konishi H, Hayashi M, Taniguchi K, Nakamura M, Kuranaga Y, Ito Y, Kondo Y, Sasaki H, Terai Y, Akao Y et al (2020) The therapeutic potential of exosomal miR‐22 for cervical cancer radiotherapy. Cancer Biol Ther 21: 1128–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers‐Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX et al (2014) Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 8: 1649–1658 [DOI] [PubMed] [Google Scholar]
- Kosanovic M, Milutinovic B, Goc S, Mitic N, Jankovic M (2017) Ion‐exchange chromatography purification of extracellular vesicles. Biotechniques 63: 65–71 [DOI] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal‐Bengtson B, Dingli F, Loew D, Tkach M, Thery C (2016) Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA 113: E968–E977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kral J, Korenkova V, Novosadova V, Langerova L, Schneiderova M, Liska V, Levy M, Veskrnova V, Spicak J, Opattova A et al (2018) Expression profile of miR‐17/92 cluster is predictive of treatment response in rectal cancer. Carcinogenesis 39: 1359–1367 [DOI] [PubMed] [Google Scholar]
- Kreger BT, Johansen ER, Cerione RA, Antonyak MA (2016) The enrichment of survivin in exosomes from breast cancer cells treated with paclitaxel promotes cell survival and chemoresistance. Cancers (Basel) 8: 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren N, Michaud D, Bagchi S, Greene K, Pylayeva‐Gupta Y (2020) Rab27a plays a dual role in metastatic propensity of pancreatic cancer. Sci Rep 10: 7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni B, Gondaliya P, Kirave P, Rawal R, Jain A, Garg R, Kalia K (2020) Exosome‐mediated delivery of miR‐30a sensitize cisplatin‐resistant variant of oral squamous carcinoma cells via modulating Beclin1 and Bcl2. Oncotarget 11: 1832–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshreshtha A, Singh S, Ahmad M, Khanna K, Ahmad T, Agrawal A, Ghosh B (2019) Simvastatin mediates inhibition of exosome synthesis, localization and secretion via multicomponent interventions. Sci Rep 9: 16373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumata Y, Iinuma H, Suzuki Y, Tsukahara D, Midorikawa H, Igarashi Y, Soeda N, Kiyokawa T, Horikawa M, Fukushima R (2018) Exosomeencapsulated microRNA23b as a minimally invasive liquid biomarker for the prediction of recurrence and prognosis of gastric cancer patients in each tumor stage. Oncol Rep 40: 319–330 [DOI] [PubMed] [Google Scholar]
- Kwon Y, Kim M, Kim Y, Jung HS, Jeoung D (2020) Exosomal microRNAs as mediators of cellular interactions between cancer cells and macrophages. Front Immunol 11: 1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey DE, Olefsky JM (2016) Regulation of metabolism by the innate immune system. Nat Rev Endocrinol 12: 15–28 [DOI] [PubMed] [Google Scholar]
- Lacy SH, Woeller CF, Thatcher TH, Pollock SJ, Small EM, Sime PJ, Phipps RP (2019) Activated human lung fibroblasts produce extracellular vesicles with antifibrotic prostaglandins. Am J Respir Cell Mol Biol 60: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire C, Chen JW, Tannous BA, Breakefield XO (2014) Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 8: 483–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, Breakefield XO (2015) Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun 6: 7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamparski HG, Metha‐Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, Le Pecq JB (2002) Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods 270: 211–226 [DOI] [PubMed] [Google Scholar]
- Lan F, Qing Q, Pan Q, Hu M, Yu H, Yue X (2018) Serum exosomal miR‐301a as a potential diagnostic and prognostic biomarker for human glioma. Cell Oncol (Dordr) 41: 25–33 [DOI] [PubMed] [Google Scholar]
- Lan J, Sun L, Xu F, Liu L, Hu F, Song D, Hou Z, Wu W, Luo X, Wang J et al (2019) M2 macrophage‐derived exosomes promote cell migration and invasion in colon cancer. Cancer Res 79: 146–158 [DOI] [PubMed] [Google Scholar]
- Lan F, Yue X, Xia T (2020) Exosomal microRNA‐210 is a potentially non‐invasive biomarker for the diagnosis and prognosis of glioma. Oncol Lett 19: 1967–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larios J, Mercier V, Roux A, Gruenberg J (2020) ALIX‐ and ESCRT‐III‐dependent sorting of tetraspanins to exosomes. J Cell Biol 219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasser C, Eldh M, Lotvall J (2012) Isolation and characterization of RNA‐containing exosomes. J Vis Exp 59: e3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifkar A, Ling L, Hingorani A, Johansen E, Clement A, Zhang X, Hartman J, Fischbach C, Lin H, Cerione RA et al (2019) Loss of sirtuin 1 alters the secretome of breast cancer cells by impairing lysosomal integrity. Dev Cell 49: 393–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux J‐F, Kobayashi T, Salles J‐P, Perret B, Bonnerot C et al (2004) Mast cell‐ and dendritic cell‐derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem J 380: 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviron M, Boissonnas A (2019) Ontogeny of tumor‐associated macrophages. Front Immunol 10: 1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY et al (2015) Single‐cell analysis reveals a stem‐cell program in human metastatic breast cancer cells. Nature 526: 131–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar I, Clement E, Ducoux‐Petit M, Denat L, Soldan V, Dauvillier S, Balor S, Burlet‐Schiltz O, Larue L, Muller C et al (2015) Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment Cell Melanoma Res 28: 464–475 [DOI] [PubMed] [Google Scholar]
- Lazar I, Clement E, Dauvillier S, Milhas D, Ducoux‐Petit M, LeGonidec S, Moro C, Soldan V, Dalle S, Balor S et al (2016) Adipocyte exosomes promote melanoma aggressiveness through fatty acid oxidation: a novel mechanism linking obesity and cancer. Cancer Res 76: 4051–4057 [DOI] [PubMed] [Google Scholar]
- Lázaro‐Ibáñez E, Sanz‐Garcia A, Visakorpi T, Escobedo‐Lucea C, Siljander P, Ayuso‐Sacido Á, Yliperttula M (2014) Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: apoptotic bodies, microvesicles, and exosomes. Prostate 74: 1379–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal AC, Mizurini DM, Gomes T, Rochael NC, Saraiva EM, Dias MS, Werneck CC, Sielski MS, Vicente CP, Monteiro RQ (2017) Tumor‐derived exosomes induce the formation of neutrophil extracellular traps: implications for the establishment of cancer‐associated thrombosis. Sci Rep 7: 6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Finniss S, Cazacu S, Bucris E, Ziv‐Av A, Xiang C, Bobbitt K, Rempel SA, Hasselbach L, Mikkelsen T et al (2013) Mesenchymal stem cells deliver synthetic microRNA mimics to glioma cells and glioma stem cells and inhibit their cell migration and self‐renewal. Oncotarget 4: 346–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Chennakrishnaiah S, Audemard E, Montermini L, Meehan B, Rak J (2014) Oncogenic ras‐driven cancer cell vesiculation leads to emission of double‐stranded DNA capable of interacting with target cells. Biochem Biophys Res Commun 451: 295–301 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Lee J, Jung JH, Park HY, Moon PG, Chae YS, Baek MC (2021) Exosomal Del‐1 as a potent diagnostic marker for breast cancer: prospective cohort study. Clin Breast Cancer 21: e748–e756 [DOI] [PubMed] [Google Scholar]
- Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH et al (2020) The LC3‐conjugation machinery specifies the loading of RNA‐binding proteins into extracellular vesicles. Nat Cell Biol 22: 187–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Oh DY, Bandyopadhyay G, Lagakos WS, Talukdar S, Osborn O, Johnson A, Chung H, Maris M, Ofrecio JM et al (2015a) LTB4 promotes insulin resistance in obese mice by acting on macrophages, hepatocytes and myocytes. Nat Med 21: 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J (2015b) Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol 36: 2007–2012 [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015c) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25: 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Liu S, Lu M, Bandyopadhyay G, Oh D, Imamura T, Johnson AMF, Sears D, Shen Z, Cui B et al (2016) Hematopoietic‐derived galectin‐3 causes cellular and systemic insulin resistance. Cell 167: 973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Y, Guo X, Zhou L, Jia Z, Peng Z, Tang Y, Liu W, Zhu B, Wang L et al (2017a) GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med 21: 838–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li B, Ren C, Chen Y, Guo X, Zhou L, Peng Z, Tang Y, Chen Y, Liu W et al (2017b) The clinical significance of circulating GPC1 positive exosomes and its regulative miRNAs in colon cancer patients. Oncotarget 8: 101189–101202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tan M, Xiang Q, Zhou Z, Yan H (2017c) Thrombin‐activated platelet‐derived exosomes regulate endothelial cell expression of ICAM‐1 via microRNA‐223 during the thrombosis‐inflammation response. Thromb Res 154: 96–105 [DOI] [PubMed] [Google Scholar]
- Li R, Peng C, Zhang X, Wu Y, Pan S, Xiao Y (2017d) Roles of Arf6 in cancer cell invasion, metastasis and proliferation. Life Sci 182: 80–84 [DOI] [PubMed] [Google Scholar]
- Li W, Zhang X, Wang J, Li M, Cao C, Tan J, Ma D, Gao Q (2017e) TGFbeta1 in fibroblasts‐derived exosomes promotes epithelial‐mesenchymal transition of ovarian cancer cells. Oncotarget 8: 96035–96047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Huang S, Zhou Z, Lin W, Chen S, Chen M, Ye Y (2018a) Exosomes derived from rAAV/AFP‐transfected dendritic cells elicit specific T cell‐mediated immune responses against hepatocellular carcinoma. Cancer Manag Res 10: 4945–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X (2018b) Circular RNA IARS (circ‐IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res 37: 177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Lu Y, Xu Y, Wang J, Zhang C, Du Y, Wang L, Li L, Wang B, Shen J et al (2018c) Horizontal transfer of exosomal CXCR4 promotes murine hepatocarcinoma cell migration, invasion and lymphangiogenesis. Gene 676: 101–109 [DOI] [PubMed] [Google Scholar]
- Li Y, Gao Y, Gong C, Wang Z, Xia Q, Gu F, Hu C, Zhang L, Guo H, Gao S (2018d) A33 antibody‐functionalized exosomes for targeted delivery of doxorubicin against colorectal cancer. Nanomedicine 14: 1973–1985 [DOI] [PubMed] [Google Scholar]
- Li Z, Fang R, Fang J, He S, Liu T (2018e) Functional implications of Rab27 GTPases in cancer. Cell Commun Signal 16: 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang H, Yin H, Bennett C, Zhang HG, Guo P (2018f) Arrowtail RNA for ligand display on ginger exosome‐like nanovesicles to systemic deliver siRNA for cancer suppression. Sci Rep 8: 14644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li C, Zhi C, Liang W, Wang X, Chen X, Lv T, Shen Q, Song Y, Lin D et al (2019a) Clinical significance of PD‐L1 expression in serum‐derived exosomes in NSCLC patients. J Transl Med 17: 355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lv Y, Shao C, Chen C, Zhang T, Wei Y, Fan H, Lv T, Liu H, Song Y (2019b) Tumor‐derived exosomal lncRNA GAS5 as a biomarker for early‐stage non‐small‐cell lung cancer diagnosis. J Cell Physiol 234: 20721–20727 [DOI] [PubMed] [Google Scholar]
- Li C, Zhou Y, Liu J, Su X, Qin H, Huang S, Huang X, Zhou N (2019c) Potential markers from serum‐purified exosomes for detecting oral squamous cell carcinoma metastasis. Cancer Epidemiol Biomarkers Prev 28: 1668–1681 [DOI] [PubMed] [Google Scholar]
- Li R, Wang Y, Zhang X, Feng M, Ma J, Li J, Yang X, Fang F, Xia Q, Zhang Z et al (2019d) Exosome‐mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol Cancer 18: 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SL, An N, Liu B, Wang SY, Wang JJ, Ye Y (2019e) Exosomes from LNCaP cells promote osteoblast activity through miR‐375 transfer. Oncol Lett 17: 4463–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang W, Chen X, Ling H, Xie P, Chen Z, Adili A, Chen Z, Yang F, Zhang CY et al (2020a) Proteomic profiling of MIN6 cell‐derived exosomes. J Proteomics 224: 103841 [DOI] [PubMed] [Google Scholar]
- Li G, Xiao L, Qin H, Zhuang Q, Zhang W, Liu L, Di C, Zhang Y (2020b) Exosomes‐carried microRNA‐26b‐5p regulates microglia M1 polarization after cerebral ischemia/reperfusion. Cell Cycle 19: 1022–1035 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li J, Li N, Wang J (2020c) M1 macrophage‐derived exosome‐encapsulated cisplatin can enhance its anti‐lung cancer effect. Minerva Med 10.23736/S0026-4806.20.06564-7 [DOI] [PubMed] [Google Scholar]
- Li S, Zhang M, Zhang H, Hu K, Cai C, Wang J, Shi L, Ma P, Xu Y, Zheng P (2020d) Exosomal long noncoding RNA lnc‐GNAQ‐6:1 may serve as a diagnostic marker for gastric cancer. Clin Chim Acta 501: 252–257 [DOI] [PubMed] [Google Scholar]
- Li W, Ding X, Wang S, Xu L, Yin T, Han S, Geng J, Sun W (2020e) Downregulation of serum exosomal miR‐320d predicts poor prognosis in hepatocellular carcinoma. J Clin Lab Anal 34: e23239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ye L, Wang L, Quan R, Zhou Y, Li X (2020f) Identification of miRNA signatures in serum exosomes as a potential biomarker after radiotherapy treatment in glioma patients. Ann Diagn Pathol 44: 151436 [DOI] [PubMed] [Google Scholar]
- Li J, Wang N, Zhang F, Jin S, Dong Y, Dong X, Chen Y, Kong X, Tong Y, Mi Q et al (2021a) PIWI‐interacting RNAs are aberrantly expressed and may serve as novel biomarkers for diagnosis of lung adenocarcinoma. Thorac Cancer 12: 2468–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shan W, Hua Y, Chao F, Cui Y, Lv L, Dou X, Bian X, Zou J, Li H et al (2021b) Exosomal miR‐92b‐3p promotes chemoresistance of small cell lung cancer through the PTEN/AKT pathway. Front Cell Dev Biol 9: 661602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Deng J, Han Z, Liu C, Tian F, Xu R, Han D, Zhang S, Sun J (2021c) Molecular identification of tumor‐derived extracellular vesicles using thermophoresis‐mediated DNA computation. J Am Chem Soc 143: 1290–1295 [DOI] [PubMed] [Google Scholar]
- Li Z, Yang H, Ye L, Quan R, Chen M (2021d) Role of exosomal miRNAs in brain metastasis affected by radiotherapy. Transl Neurosci 12: 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Q, Xu J, Yan S, Huang M, Ding H, Sun X, Bi A, Ding J, Sun B, Geng M (2017) Chemotherapy‐induced intestinal inflammatory responses are mediated by exosome secretion of double‐strand DNA via AIM2 inflammasome activation. Cell Res 27: 784–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Kan S, Zhu Y, Feng S, Feng W, Gao S (2018) Engineered exosome‐mediated delivery of functionally active miR‐26a and its enhanced suppression effect in HepG2 cells. Int J Nanomedicine 13: 585–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Mao L, Zhang S, Guo X, Liu G, Wang L, Hou J, Zheng Y, Luo X (2019) Identification and molecular characterization of exosome‐like vesicles derived from the Taenia asiatica adult worm. Acta Trop 198: 105036 [DOI] [PubMed] [Google Scholar]
- Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si K, Sun B, Chen B, Xiao Z (2020a) Engineered exosomes for targeted co‐delivery of miR‐21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnol 18: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Liu Y, Zhang Q, Zhang H, Du J (2020b) Tumor‐derived extracellular vesicles containing microRNA‐1290 promote immune escape of cancer cells through the Grhl2/ZEB1/PD‐L1 axis in gastric cancer. Transl Res 231: 102–112 [DOI] [PubMed] [Google Scholar]
- Liangsupree T, Multia E, Riekkola ML (2021) Modern isolation and separation techniques for extracellular vesicles. J Chromatogr A 1636: 461773 [DOI] [PubMed] [Google Scholar]
- Liao J, Liu R, Shi YJ, Yin LH, Pu YP (2016) Exosome‐shuttling microRNA‐21 promotes cell migration and invasion‐targeting PDCD4 in esophageal cancer. Int J Oncol 48: 2567–2579 [DOI] [PubMed] [Google Scholar]
- Liao F, Lu X, Dong W (2020) Exosomes derived from T regulatory cells relieve inflammatory bowel disease by transferring miR‐195a‐3p. IUBMB Life 10.1002/iub.2385 [DOI] [PubMed] [Google Scholar]
- Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, Symmans WF, Gonzalez‐Angulo AM, Hennessy B, Green M et al (2008) Response to neoadjuvant therapy and long‐term survival in patients with triple‐negative breast cancer. J Clin Oncol 26: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA, Greco SJ, Bryan M, Patel PS, Rameshwar P (2011) Gap junction‐mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res 71: 1550–1560 [DOI] [PubMed] [Google Scholar]
- Lim JW, Mathias RA, Kapp EA, Layton MJ, Faux MC, Burgess AW, Ji H, Simpson RJ (2012) Restoration of full‐length APC protein in SW480 colon cancer cells induces exosome‐mediated secretion of DKK‐4. Electrophoresis 33: 1873–1880 [DOI] [PubMed] [Google Scholar]
- Limoni SK, Moghadam MF, Moazzeni SM, Gomari H, Salimi F (2019) Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl Biochem Biotechnol 187: 352–364 [DOI] [PubMed] [Google Scholar]
- Lin R, Wang S, Zhao RC (2013) Exosomes from human adipose‐derived mesenchymal stem cells promote migration through Wnt signaling pathway in a breast cancer cell model. Mol Cell Biochem 383: 13–20 [DOI] [PubMed] [Google Scholar]
- Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW et al (2018) Tumor‐originated exosomal lncUEGC1 as a circulating biomarker for early‐stage gastric cancer. Mol Cancer 17: 84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Zhang C, Xiang P, Shen J, Sun W, Yu H (2020) Exosomes derived from HeLa cells break down vascular integrity by triggering endoplasmic reticulum stress in endothelial cells. J Extracell Vesicles 9: 1722385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Eng C, Shen J, Lu Y, Takata Y, Mehdizadeh A, Chang GJ, Rodriguez‐Bigas MA, Li Y, Chang P et al (2016a) Serum exosomal miR‐4772‐3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 7: 76250–76260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, Comstock LE, Gandhi R, Weiner HL (2016b) The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 19: 32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L, Zheng G, Li P, Li C, Wang C (2016c) Exosomal long noncoding RNA CRNDE‐h as a novel serum‐based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 7: 85551–85563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, Huang B, Xu X, Zheng J, Cao X (2016d) Tumor exosomal RNAs promote lung pre‐metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell 30: 243–256 [DOI] [PubMed] [Google Scholar]
- Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, Xiang Y, Wu N, Wu L, Bai L et al (2017a) Circulating exosomal microRNAs as prognostic biomarkers for non‐small‐cell lung cancer. Oncotarget 8: 13048–13058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, Dai Z, Cao Y, Fan J, Zhou J (2017b) Serum exosomal miR‐125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther 10: 3843–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Bu Z, Zhao F, Xiao D (2018a) Increased T‐helper 17 cell differentiation mediated by exosome‐mediated microRNA‐451 redistribution in gastric cancer infiltrated T cells. Cancer Sci 109: 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MX, Liao J, Xie M, Gao ZK, Wang XH, Zhang Y, Shang MH, Yin LH, Pu YP, Liu R (2018b) miR‐93‐5p transferred by exosomes promotes the proliferation of esophageal cancer cells via intercellular communication by targeting PTEN. Biomed Environ Sci 31: 171–185 [DOI] [PubMed] [Google Scholar]
- Liu R, Tang A, Wang X, Chen X, Zhao L, Xiao Z, Shen S (2018c) Inhibition of lncRNA NEAT1 suppresses the inflammatory response in IBD by modulating the intestinal epithelial barrier and by exosome‐mediated polarization of macrophages. Int J Mol Med 42: 2903–2913 [DOI] [PubMed] [Google Scholar]
- Liu C, Zhao J, Tian F, Cai L, Zhang W, Feng Q, Chang J, Wan F, Yang Y, Dai B et al (2019a) Low‐cost thermophoretic profiling of extracellular‐vesicle surface proteins for the early detection and classification of cancers. Nat Biomed Eng 3: 183–193 [DOI] [PubMed] [Google Scholar]
- Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, Wang L, Li P, Zhao Y, Duan W et al (2019b) Exosome‐transmitted miR‐128‐3p increase chemosensitivity of oxaliplatin‐resistant colorectal cancer. Mol Cancer 18: 43 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu W, Li J, Zhang P, Hou Q, Feng S, Liu L, Cui D, Shi H, Fu Y, Luo Y (2019c) A novel pan‐cancer biomarker plasma heat shock protein 90alpha and its diagnosis determinants in clinic. Cancer Sci 110: 2941–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bai L, Guo K, Jia Y, Zhang K, Liu Q, Wang P, Wang X (2019d) Focused ultrasound‐augmented targeting delivery of nanosonosensitizers from homogenous exosomes for enhanced sonodynamic cancer therapy. Theranostics 9: 5261–5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang Z, Zhou L, Hu L, Yin C, Qing D, Huang S, Cai X, Chen Y (2020a) Cancer associated fibroblasts‐derived exosomes contribute to radioresistance through promoting colorectal cancer stem cells phenotype. Exp Cell Res 391: 111956 [DOI] [PubMed] [Google Scholar]
- Liu T, Zhu Y, Zhao R, Wei X, Xin X (2020b) Visualization of exosomes from mesenchymal stem cells in vivo by magnetic resonance imaging. Magn Reson Imaging 68: 75–82 [DOI] [PubMed] [Google Scholar]
- Liu W, Yang D, Chen L, Liu Q, Wang W, Yang Z, Shang A, Quan W, Li D (2020c) Plasma exosomal miRNA‐139‐3p is a novel biomarker of colorectal cancer. J Cancer 11: 4899–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Li B, Shi X, Zhang J, Chen AM, Xu J, Wang W, Huang K, Gao J, Zheng Z et al (2021a) Cross‐platform genomic identification and clinical validation of breast cancer diagnostic biomarkers. Aging (Albany NY) 13: 4258–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Peng X, Liu Y, Hao R, Zhao R, Zhang L, Zhao F, Liu Q, Liu Y, Qi Y (2021b) The diagnostic value of serum exosomal Has_circ_0000615 for breast cancer patients. Int J Gen Med 14: 4545–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Mo F, Song X, He Y, Yuan Y, Yan J, Yang Y, Huang J, Zhang S (2021c) Exosomal hsa‐miR‐21‐5p is a biomarker for breast cancer diagnosis. PeerJ 9: e12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente A, Skotland T, Sylvanne T, Kauhanen D, Rog T, Orlowski A, Vattulainen I, Ekroos K, Sandvig K (2013) Molecular lipidomics of exosomes released by PC‐3 prostate cancer cells. Biochim Biophys Acta 1831: 1302–1309 [DOI] [PubMed] [Google Scholar]
- Lo TW, Zhu Z, Purcell E, Watza D, Wang J, Kang YT, Jolly S, Nagrath D, Nagrath S (2020) Microfluidic device for high‐throughput affinity‐based isolation of extracellular vesicles. Lab Chip 20: 1762–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb RJ, Hastie ML, Norris EL, van Amerongen R, Gorman JJ, Moller A (2017) Oncogenic transformation of lung cells results in distinct exosome protein profile similar to the cell of origin. Proteomics 10.1002/pmic.201600432 [DOI] [PubMed] [Google Scholar]
- Loftus A, Cappariello A, George C, Ucci A, Shefferd K, Green A, Paone R, Ponzetti M, Delle Monache S, Muraca M et al (2020) Extracellular vesicles from osteotropic breast cancer cells affect bone resident cells. J Bone Miner Res 35: 396–412 [DOI] [PubMed] [Google Scholar]
- Lopez P, Rodriguez‐Carrio J, Caminal‐Montero L, Suarez A (2020) Relationship between T‐cell exosomes and cellular subsets in SLE according to type I IFN‐signaling. Front Med (Lausanne) 7: 604098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou G, Yang Y, Liu F, Ye B, Chen Z, Zheng M, Liu Y (2017) MiR‐122 modification enhances the therapeutic efficacy of adipose tissue‐derived mesenchymal stem cells against liver fibrosis. J Cell Mol Med 21: 2963–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Pickering MC (2018) Extracellular DNA and autoimmune diseases. Cell Mol Immunol 15: 746–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zuo B, Jing R, Gao X, Rao Q, Liu Z, Qi H, Guo H, Yin H (2017) Dendritic cell‐derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J Hepatol 67: 739–748 [DOI] [PubMed] [Google Scholar]
- Lu Y, Duan Y, Xu Q, Zhang L, Chen W, Qu Z, Wu B, Liu W, Shi L, Wu D et al (2020) Circulating exosome‐derived bona fide long non‐coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J Cell Mol Med 24: 1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y, Li X, Luan Y, Zhao R, Li Y, Liu L, Hao Y, Oleg Vladimir B, Jia L (2020) Circulating lncRNA UCA1 promotes malignancy of colorectal cancer via the miR‐143/MYO6 axis. Mol Ther Nucleic Acids 19: 790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig N, Gillespie DG, Reichert TE, Jackson EK, Whiteside TL (2020) Purine metabolites in tumor‐derived exosomes may facilitate immune escape of head and neck squamous cell carcinoma. Cancers (Basel) 12: 1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luga V, Zhang L, Viloria‐Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL (2012) Exosomes mediate stromal mobilization of autocrine Wnt‐PCP signaling in breast cancer cell migration. Cell 151: 1542–1556 [DOI] [PubMed] [Google Scholar]
- Lugini L, Cecchetti S, Huber V, Luciani F, Macchia G, Spadaro F, Paris L, Abalsamo L, Colone M, Molinari A et al (2012) Immune surveillance properties of human NK cell‐derived exosomes. J Immunol 189: 2833–2842 [DOI] [PubMed] [Google Scholar]
- Luo X, An M, Cuneo KC, Lubman DM, Li L (2018) High‐performance chemical isotope labeling liquid chromatography mass spectrometry for exosome metabolomics. Anal Chem 90: 8314–8319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo A, Zhou X, Shi X, Zhao Y, Men Y, Chang X, Chen H, Ding F, Li Y, Su D et al (2019) Exosome‐derived miR‐339‐5p mediates radiosensitivity by targeting Cdc25A in locally advanced esophageal squamous cell carcinoma. Oncogene 38: 4990–5006 [DOI] [PubMed] [Google Scholar]
- Lux A, Kahlert C, Grutzmann R, Pilarsky C (2019) c‐Met and PD‐L1 on circulating exosomes as diagnostic and prognostic markers for pancreatic cancer. Int J Mol Sci 20: 3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Cheng L, Lu Y, Zhang X, Wang Y, Deng J, Zhou J, Liu B, Liu J (2020a) Thermosensitive exosome‐liposome hybrid nanoparticle‐mediated chemoimmunotherapy for improved treatment of metastatic peritoneal cancer. Adv Sci (Weinh) 7: 2000515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Wang Y, Xu W, Dong X (2020b) Serum exosomal miR‐17‐5p as a promising biomarker diagnostic biomarker for breast cancer. Clin Lab 10.7754/Clin.Lab.2020.200127 [DOI] [PubMed] [Google Scholar]
- Lydic TA, Townsend S, Adda CG, Collins C, Mathivanan S, Reid GE (2015) Rapid and comprehensive ‘shotgun’ lipidome profiling of colorectal cancer cell derived exosomes. Methods 87: 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li Y, Peng J, Wu D, Zhao X, Cui Y, Chen L, Yan X, Du Y, Yu L (2015) Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res 25: 24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G, Song G, Zou X, Shan X, Liu Q, Xia T, Zhou X, Zhu W (2019a) Circulating plasma microRNA signature for the diagnosis of cervical cancer. Cancer Biomark 26: 491–500 [DOI] [PubMed] [Google Scholar]
- Ma Y, Yuwen D, Chen J, Zheng B, Gao J, Fan M, Xue W, Wang Y, Li W, Shu Y et al (2019b) Exosomal transfer of cisplatin‐induced miR‐425‐3p confers cisplatin resistance In NSCLC through activating autophagy. Int J Nanomedicine 14: 8121–8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZJ, Wang YH, Li ZG, Wang Y, Li BY, Kang HY, Wu XY (2019c) Immunosuppressive effect of exosomes from mesenchymal stromal cells in defined medium on experimental colitis. Int J Stem Cells 12: 440–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida T, Tomofuji T, Maruyama T, Yoneda T, Ekuni D, Azuma T, Miyai H, Mizuno H, Kato H, Tsutsumi K et al (2016) miR1246 and miR4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep 36: 2375–2381 [DOI] [PubMed] [Google Scholar]
- Madeo M, Colbert PL, Vermeer DW, Lucido CT, Cain JT, Vichaya EG, Grossberg AJ, Muirhead D, Rickel AP, Hong Z et al (2018) Cancer exosomes induce tumor innervation. Nat Commun 9: 4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Sasaki H, Ueda S, Miyamoto S, Terada S, Konishi H, Kogata Y, Ashihara K, Fujiwara S, Tanaka Y et al (2020) Serum exosomal microRNA‐34a as a potential biomarker in epithelial ovarian cancer. J Ovarian Res 13: 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia J, Otake AH, Pocas J, Carvalho AS, Beck HC, Magalhaes A, Matthiesen R, Strano Moraes MC, Costa‐Silva B (2020) Transcriptome reprogramming of CD11b(+) bone marrow cells by pancreatic cancer extracellular vesicles. Front Cell Dev Biol 8: 592518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire CL, Fuh MM, Kaulich K, Fita KD, Stevic I, Heiland DH, Welsh JA, Jones JC, Gorgens A, Ricklefs T et al (2021) Genome‐wide methylation profiling of glioblastoma cell‐derived extracellular vesicle DNA allows tumor classification. Neuro Oncol 23: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji S, Chaudhary P, Akopova I, Nguyen PM, Hare RJ, Gryczynski I, Vishwanatha JK (2017) Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol Cancer Res 15: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallawaaratchy DM, Hallal S, Russell B, Ly L, Ebrahimkhani S, Wei H, Christopherson RI, Buckland ME, Kaufman KL (2017) Comprehensive proteome profiling of glioblastoma‐derived extracellular vesicles identifies markers for more aggressive disease. J Neurooncol 131: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manda SV, Kataria Y, Tatireddy BR, Ramakrishnan B, Ratnam BG, Lath R, Ranjan A, Ray A (2018) Exosomes as a biomarker platform for detecting epidermal growth factor receptor‐positive high‐grade gliomas. J Neurosurg 128: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Mao F, Wu Y, Tang X, Kang J, Zhang B, Yan Y, Qian H, Zhang X, Xu W (2017) Exosomes derived from human umbilical cord mesenchymal stem cells relieve inflammatory bowel disease in mice. Biomed Res Int 2017: 5356760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W, Wen Y, Lei H, Lu R, Wang S, Wang Y, Chen R, Gu Y, Zhu L, Abhange KK et al (2019a) Isolation and retrieval of extracellular vesicles for liquid biopsy of malignant ground‐glass opacity. Anal Chem 91: 13729–13736 [DOI] [PubMed] [Google Scholar]
- Mao Y, Wang Y, Dong L, Zhang Y, Zhang Y, Wang C, Zhang Q, Yang S, Cao L, Zhang X et al (2019b) Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J Exp Clin Cancer Res 38: 389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczak S, Richards K, Ramshani Z, Smith E, Senapati S, Hill R, Go DB, Chang HC (2018) Simultaneous isolation and preconcentration of exosomes by ion concentration polarization. Electrophoresis 10.1002/elps.201700491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens MH, Lambregts DMJ, Kluza E, Beets‐Tan RGH (2014) Tumor response to treatment: prediction and assessment. Curr Radiol Rep 2: 62 [Google Scholar]
- Massague J, Ganesh K (2021) Metastasis‐initiating cells and ecosystems. Cancer Discov 11: 971–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Takahashi Y, Nishikawa M, Sano K, Morishita M, Charoenviriyakul C, Saji H, Takakura Y (2017) Accelerated growth of B16BL6 tumor in mice through efficient uptake of their own exosomes by B16BL6 cells. Cancer Sci 108: 1803–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Kano M, Murakami K, Toyozumi T, Suito H, Takahashi M, Sekino N, Shiraishi T, Kamata T, Ryuzaki T et al (2020) Tumor‐derived exosomes influence the cell cycle and cell migration of human esophageal cancer cell lines. Cancer Sci 111: 4348–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano Y et al (2015) Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer 113: 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R et al (2004) Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303: 531–534 [DOI] [PubMed] [Google Scholar]
- McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, Patton JG, Weaver AM (2016) KRAS‐MEK signaling controls Ago2 sorting into exosomes. Cell Rep 15: 978–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldolesi J (2018) Exosomes and ectosomes in intercellular communication. Curr Biol 28: R435–R444 [DOI] [PubMed] [Google Scholar]
- Melentijevic I, Toth ML, Arnold ML, Guasp RJ, Harinath G, Nguyen KC, Taub D, Parker JA, Neri C, Gabel CV et al (2017) C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature 542: 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N et al (2015) Glypican‐1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523: 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendt M, Kamerkar S, Sugimoto H, McAndrews KM, Wu CC, Gagea M, Yang S, Blanko EVR, Peng Q, Ma X et al (2018) Generation and testing of clinical‐grade exosomes for pancreatic cancer. JCI Insight 3: e99263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant ML, Powell DW, Wilkey DW, Cummins TD, Deegens JK, Rood IM, McAfee KJ, Fleischer C, Klein E, Klein JB (2010) Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin Appl 4: 84–96 [DOI] [PubMed] [Google Scholar]
- Mikamori M, Yamada D, Eguchi H, Hasegawa S, Kishimoto T, Tomimaru Y, Asaoka T, Noda T, Wada H, Kawamoto K et al (2017) MicroRNA‐155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci Rep 7: 42339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Chen L, Liu S, Yu Y, Guo Q, Li P, Zhu S (2019) Loss of circulating exosomal miR‐92b is a novel biomarker of colorectal cancer at early stage. Int J Med Sci 16: 1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minciacchi VR, Spinelli C, Reis‐Sobreiro M, Cavallini L, You S, Zandian M, Li X, Mishra R, Chiarugi P, Adam RM et al (2017) MYC mediates large oncosome‐induced fibroblast reprogramming in prostate cancer. Cancer Res 77: 2306–2317 [DOI] [PubMed] [Google Scholar]
- Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R (2021) Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov 20: 101–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S (2007) Identification of Tim4 as a phosphatidylserine receptor. Nature 450: 435–439 [DOI] [PubMed] [Google Scholar]
- Mizuno R, Kawada K, Sakai Y (2019) Prostaglandin E2/EP signaling in the tumor microenvironment of colorectal cancer. Int J Mol Sci 20: 6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo F, Xu Y, Zhang J, Zhu L, Wang C, Chu X, Pan Y, Bai Y, Shao C, Zhang J (2020) Effects of hypoxia and radiation‐induced exosomes on migration of lung cancer cells and angiogenesis of umbilical vein endothelial cells. Radiat Res 194: 71–80 [DOI] [PubMed] [Google Scholar]
- Möller A, Lobb RJ (2020) The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer 20: 697–709 [DOI] [PubMed] [Google Scholar]
- Moloney BM, Gilligan KE, Joyce DP, O'Neill CP, O'Brien KP, Khan S, Glynn CL, Waldron RM, Maguire CM, Holian E et al (2020) Investigating the potential and pitfalls of EV‐encapsulated microRNAs as circulating biomarkers of breast cancer. Cell 9: 141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli AE, Larregina AT, Shufesky WJ, Sullivan MLG, Stolz DB, Papworth GD, Zahorchak AF, Logar AJ, Wang Z, Watkins SC et al (2004) Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 104: 3257–3266 [DOI] [PubMed] [Google Scholar]
- Morishita M, Takahashi Y, Matsumoto A, Nishikawa M, Takakura Y (2016) Exosome‐based tumor antigens‐adjuvant co‐delivery utilizing genetically engineered tumor cell‐derived exosomes with immunostimulatory CpG DNA. Biomaterials 111: 55–65 [DOI] [PubMed] [Google Scholar]
- Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, Jamieson NB, Oien KA, Lowy AM, Brunton VG et al (2010) Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA 107: 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Zhuang X, Wang Q, Jiang H, Deng ZB, Wang B, Zhang L, Kakar S, Jun Y, Miller D et al (2014) Interspecies communication between plant and mouse gut host cells through edible plant derived exosome‐like nanoparticles. Mol Nutr Food Res 58: 1561–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Agarwal R, March D, Rothenberg A, Voigt C, Tebbets J, Huard J, Weiss K (2016) Notch signaling mediates skeletal muscle atrophy in cancer cachexia caused by osteosarcoma. Sarcoma 2016: 3758162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multia E, Liangsupree T, Jussila M, Ruiz‐Jimenez J, Kemell M, Riekkola ML (2020) automated on‐line isolation and fractionation system for nanosized biomacromolecules from human plasma. Anal Chem 92: 13058–13065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munagala R, Aqil F, Jeyabalan J, Kandimalla R, Wallen M, Tyagi N, Wilcher S, Yan J, Schultz DJ, Spencer W et al (2021) Exosome‐mediated delivery of RNA and DNA for gene therapy. Cancer Lett 505: 58–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Yamamoto CM, Akino T, Tanaka H, Fukuzawa N, Suzuki H, Osawa T, Tsuji T, Seki T, Harada H (2018) Bladder cancer detection by urinary extracellular vesicle mRNA analysis. Oncotarget 9: 32810–32821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan‐Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D'Souza‐Schorey C (2009) ARF6‐regulated shedding of tumor cell‐derived plasma membrane microvesicles. Curr Biol 19: 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgai M, Ju W, Eason M, Kline J, Beury DW, Kaczanowska S, Miettinen MM, Kruhlak M, Lei H, Shern JF et al (2017) KLF4‐dependent perivascular cell plasticity mediates pre‐metastatic niche formation and metastasis. Nat Med 23: 1176–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschelknaus L, Azimzadeh O, Heider T, Winkler K, Vetter M, Kell R, Tapio S, Merl‐Pham J, Huber SM, Edalat L et al (2017) Radiation alters the cargo of exosomes released from squamous head and neck cancer cells to promote migration of recipient cells. Sci Rep 7: 12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, Benci JL, DeMichele AM, Tchou J, Marcotrigiano J et al (2017) Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell 170: 352–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q (2012) Formation and release of arrestin domain‐containing protein 1‐mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA 109: 4146–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai W, Yoshida T, Diez D, Miyatake Y, Nishibu T, Imawaka N, Naruse K, Sadamura Y, Hanayama R (2016) A novel affinity‐based method for the isolation of highly purified extracellular vesicles. Sci Rep 6: 33935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Chen IH, Wang CC, Chen PJ, Tseng HP, Huang KT, Hu TH, Li LC, Goto S, Cheng YF et al (2019) Circulating exosomal miR‐92b: its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am J Transplant 19: 3250–3262 [DOI] [PubMed] [Google Scholar]
- Nakase I, Kobayashi NB, Takatani‐Nakase T, Yoshida T (2015) Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Sci Rep 5: 10300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase I, Ueno N, Matsuzawa M, Noguchi K, Hirano M, Omura M, Takenaka T, Sugiyama A, Bailey Kobayashi N, Hashimoto T et al (2021) Environmental pH stress influences cellular secretion and uptake of extracellular vesicles. FEBS Open Bio 11: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Molins L, Marrades RM, Moises J, Vinolas N, Morales S, Canals J, Castellano JJ, Ramirez J, Monzo M (2019) Exosome analysis in tumor‐draining pulmonary vein identifies NSCLC patients with higher risk of relapse after curative surgery. Cancers (Basel) 11: 249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, Lochnit G, Preissner KT, Zoller M (2010) Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome‐induced endothelial cell activation. Cancer Res 70: 1668–1678 [DOI] [PubMed] [Google Scholar]
- Nie X, Fan J, Li H, Yin Z, Zhao Y, Dai B, Dong N, Chen C, Wang DW (2018) miR‐217 promotes cardiac hypertrophy and dysfunction by targeting PTEN. Mol Ther Nucleic Acids 12: 254–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie W, Wu G, Zhang J, Huang LL, Ding J, Jiang A, Zhang Y, Liu Y, Li J, Pu K et al (2020) Responsive exosome nano‐bioconjugates for synergistic cancer therapy. Angew Chem Int Ed Engl 59: 2018–2022 [DOI] [PubMed] [Google Scholar]
- Nigita G, Distefano R, Veneziano D, Romano G, Rahman M, Wang K, Pass H, Croce CM, Acunzo M, Nana‐Sinkam P (2018) Tissue and exosomal miRNA editing in non‐small cell lung cancer. Sci Rep 8: 10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida‐Aoki N, Tominaga N, Kosaka N, Ochiya T (2020) Altered biodistribution of deglycosylated extracellular vesicles through enhanced cellular uptake. J Extracell Vesicles 9: 1713527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooshabadi VT, Khanmohammadi M, Shafei S, Banafshe HR, Malekshahi ZV, Ebrahimi‐Barough S, Ai J (2020) Impact of atorvastatin loaded exosome as an anti‐glioblastoma carrier to induce apoptosis of U87 cancer cells in 3D culture model. Biochem Biophys Rep 23: 100792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarangelo M, Zucal C, Modelska A, Pesce I, Scarduelli G, Potrich C, Lunelli L, Pederzolli C, Pavan P, la Marca G et al (2019) Ultrasensitive detection of cancer biomarkers by nickel‐based isolation of polydisperse extracellular vesicles from blood. EBioMedicine 43: 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo D, Heath N, Mitchell L, Caligiuri G, MacFarlane A, Reijmer D, Charlton L, Knight J, Calka M, McGhee E et al (2018) Mutant p53s generate pro‐invasive niches by influencing exosome podocalyxin levels. Nat Commun 9: 5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R, Pollard JW (2014) Tumor‐associated macrophages: from mechanisms to therapy. Immunity 41: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehme F, Krahl S, Gyorffy B, Muessle B, Rao V, Greif H, Ziegler N, Lin K, Thepkaysone ML, Polster H et al (2019) Low level of exosomal long non‐coding RNA HOTTIP is a prognostic biomarker in colorectal cancer. RNA Biol 16: 1339–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeyen E, Van Mol K, Baggerman G, Willems H, Boonen K, Rolfo C, Pauwels P, Jacobs A, Schildermans K, Cho WC et al (2018) Ultrafiltration and size exclusion chromatography combined with asymmetrical‐flow field‐flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J Extracell Vesicles 7: 1490143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzawa H, Kumagai Y, Yamaguchi H, Miyato H, Sakuma Y, Horie H, Hosoya Y, Kawarai Lefor A, Sata N, Kitayama J (2020) Exosomal microRNA in peritoneal fluid as a biomarker of peritoneal metastases from gastric cancer. Ann Gastroenterol Surg 4: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda Y, Shimura T, Iwasaki H, Katano T, Kitagawa M, Nishigaki R, Fukusada S, Natsume M, Tanaka M, Nishie H et al (2020) Serum exosomal dicer is a useful biomarker for early detection of differentiated gastric adenocarcinoma. Digestion 102: 640–649 [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T (2004) Mutant p53 gain of function in two mouse models of Li‐Fraumeni syndrome. Cell 119: 847–860 [DOI] [PubMed] [Google Scholar]
- Ono M, Kosaka N, Tominaga N, Yoshioka Y, Takeshita F, Takahashi RU, Yoshida M, Tsuda H, Tamura K, Ochiya T (2014) Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci Signal 7: ra63 [DOI] [PubMed] [Google Scholar]
- Ortiz A, Gui J, Zahedi F, Yu P, Cho C, Bhattacharya S, Carbone CJ, Yu Q, Katlinski KV, Katlinskaya YV et al (2019) An interferon‐driven oxysterol‐based defense against tumor‐derived extracellular vesicles. Cancer Cell 35: 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP et al (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12: 19–30 [DOI] [PubMed] [Google Scholar]
- Pace KR, Dutt R, Galileo DS (2019) Exosomal L1CAM stimulates glioblastoma cell motility, proliferation, and invasiveness. Int J Mol Sci 20: 3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget S (1989) The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev 8: 98–101 [PubMed] [Google Scholar]
- Palicharla VR, Maddika S (2015) HACE1 mediated K27 ubiquitin linkage leads to YB‐1 protein secretion. Cell Signal 27: 2355–2362 [DOI] [PubMed] [Google Scholar]
- Pan BT, Johnstone RM (1983) Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 33: 967–978 [DOI] [PubMed] [Google Scholar]
- Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, Zhang P, Qian H, Jiang PC, Xu WR et al (2017) Exosomes‐mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol 143: 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Qin J, Liu X, He B, Wang X, Pan Y, Sun H, Xu T, Xu M, Chen X et al (2019) Identification of serum exosomal hsa‐circ‐0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet 10: 1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Pei L, Zhang A, Zhang Y, Zhang C, Huang M, Huang Z, Liu B, Wang L, Ma L et al (2020) Passion fruit‐like exosome‐PMA/Au‐BSA@Ce6 nanovehicles for real‐time fluorescence imaging and enhanced targeted photodynamic therapy with deep penetration and superior retention behavior in tumor. Biomaterials 230: 119606 [DOI] [PubMed] [Google Scholar]
- Panigrahi GK, Praharaj PP, Peak TC, Long J, Singh R, Rhim JS, Abd Elmageed ZY, Deep G (2018) Hypoxia‐induced exosome secretion promotes survival of African‐American and Caucasian prostate cancer cells. Sci Rep 8: 3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K, Alix‐Panabieres C (2019) Liquid biopsy and minimal residual disease ‐ latest advances and implications for cure. Nat Rev Clin Oncol 16: 409–424 [DOI] [PubMed] [Google Scholar]
- Park J, Morley TS, Kim M, Clegg DJ, Scherer PE (2014) Obesity and cancer–mechanisms underlying tumour progression and recurrence. Nat Rev Endocrinol 10: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Wysocki RW, Amoozgar Z, Maiorino L, Fein MR, Jorns J, Schott AF, Kinugasa‐Katayama Y, Lee Y, Won NH et al (2016) Cancer cells induce metastasis‐supporting neutrophil extracellular DNA traps. Sci Transl Med 8: 361ra138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Dutta B, Tse SW, Gupta N, Tan CF, Low JK, Yeoh KW, Kon OL, Tam JP, Sze SK (2019) Hypoxia‐induced tumor exosomes promote M2‐like macrophage polarization of infiltrating myeloid cells and microRNA‐mediated metabolic shift. Oncogene 38: 5158–5173 [DOI] [PubMed] [Google Scholar]
- Parker TM, Gupta K, Palma AM, Yekelchyk M, Fisher PB, Grossman SR, Won KJ, Madan E, Moreno E, Gogna R (2021) Cell competition in intratumoral and tumor microenvironment interactions. EMBO J 40: e107271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, Berenguer A, Prats N, Toll A, Hueto JA et al (2017) Targeting metastasis‐initiating cells through the fatty acid receptor CD36. Nature 541: 41–45 [DOI] [PubMed] [Google Scholar]
- Patten DA, Hussein E, Davies SP, Humphreys PN, Collett A (2017) Commensal‐derived OMVs elicit a mild proinflammatory response in intestinal epithelial cells. Microbiology (Reading) 163: 702–711 [DOI] [PubMed] [Google Scholar]
- Pavlides S, Whitaker‐Menezes D, Castello‐Cros R, Flomenberg N, Witkiewicz AK, Frank PG, Casimiro MC, Wang C, Fortina P, Addya S et al (2009) The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 8: 3984–4001 [DOI] [PubMed] [Google Scholar]
- Pein M, Insua‐Rodriguez J, Hongu T, Riedel A, Meier J, Wiedmann L, Decker K, Essers MAG, Sinn HP, Spaich S et al (2020) Metastasis‐initiating cells induce and exploit a fibroblast niche to fuel malignant colonization of the lungs. Nat Commun 11: 1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa‐Silva B, Moreno‐Bueno G, Hergueta‐Redondo M, Williams C, Garcia‐Santos G, Ghajar C et al (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro‐metastatic phenotype through MET. Nat Med 18: 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng ZY, Gu RH, Yan B (2018) Downregulation of exosome‐encapsulated miR‐548c‐5p is associated with poor prognosis in colorectal cancer. J Cell Biochem 10.1002/jcb.27291 [DOI] [PubMed] [Google Scholar]
- Peng XX, Yu R, Wu X, Wu SY, Pi C, Chen ZH, Zhang XC, Gao CY, Shao YW, Liu L et al (2020) Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR /ALK wild‐type advanced non‐small cell lung cancer. J Immunother Cancer 8: e000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P, Li H, Haque F, Wang S, Croce CM et al (2018) Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol 13: 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao HY, Guo S, Wang Y, Zhang J (2020) Exosomal long non‐coding RNA CEBPA‐AS1 inhibits tumor apoptosis and functions as a non‐invasive biomarker for diagnosis of gastric cancer. Onco Targets Ther 13: 1365–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L et al (2019) Suppression of exosomal PD‐L1 induces systemic anti‐tumor immunity and memory. Cell 177: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomatto MAC, Bussolati B, D'Antico S, Ghiotto S, Tetta C, Brizzi MF, Camussi G (2019) Improved loading of plasma‐derived extracellular vesicles to encapsulate antitumor miRNAs. Mol Ther Methods Clin Dev 13: 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priego N, Zhu L, Monteiro C, Mulders M, Wasilewski D, Bindeman W, Doglio L, Martinez L, Martinez‐Saez E, Ramon YCS et al (2018) STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat Med 24: 1024–1035 [DOI] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C (2012) A comprehensive survey of Ras mutations in cancer. Cancer Res 72: 2457–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci F, Garris C, Lai CP, Newton A, Pfirschke C, Engblom C, Alvarez D, Sprachman M, Evavold C, Magnuson A et al (2016) SCS macrophages suppress melanoma by restricting tumor‐derived vesicle‐B cell interactions. Science 352: 242–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka M, Takatalo M, Nordberg ME, Valkonen S, Nandania J, Aatonen M, Yliperttula M, Laitinen S, Velagapudi V, Mirtti T et al (2017) Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer‐related changes. Theranostics 7: 3824–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis IJ, Velpula KK, Guda MR, Nguyen D, Tsung AJ, Asuthkar S (2020) B7‐H3 in medulloblastoma‐derived exosomes; a novel tumorigenic role. Int J Mol Sci 21: 7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Liu C, Long L, Ren Y, Zhang S, Chang X, Qian X, Jia H, Zhao J, Sun J et al (2016) Blood exosomes endowed with magnetic and targeting properties for cancer therapy. ACS Nano 10: 3323–3333 [DOI] [PubMed] [Google Scholar]
- Qiu JJ, Lin XJ, Tang XY, Zheng TT, Lin YY, Hua KQ (2018) Exosomal metastasis associated lung adenocarcinoma transcript 1 promotes angiogenesis and predicts poor prognosis in epithelial ovarian cancer. Int J Biol Sci 14: 1960–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Chen W, Wu C, Yuan Y, Li Y (2020a) Exosomes of oral squamous cell carcinoma cells containing miR‐181a‐3p induce muscle cell atrophy and apoptosis by transmissible endoplasmic reticulum stress signaling. Biochem Biophys Res Commun 533: 831–837 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Sun J, Qiu J, Chen G, Wang X, Mu Y, Li K, Wang W (2020b) Antitumor activity of cabazitaxel and MSC‐TRAIL derived extracellular vesicles in drug‐resistant oral squamous cell carcinoma. Cancer Manag Res 12: 10809–10820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu JL, Qu XJ, Zhao MF, Teng YE, Zhang Y, Hou KZ, Jiang YH, Yang XH, Liu YP (2009) Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis 41: 875–880 [DOI] [PubMed] [Google Scholar]
- Qu Y, Zhang Q, Cai X, Li F, Ma Z, Xu M, Lu L (2017) Exosomes derived from miR‐181‐5p‐modified adipose‐derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J Cell Mol Med 21: 2491–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbari M, Pecqueux M, Aust D, Stephan H, Tiebel O, Chatzigeorgiou A, Tonn T, Baenke F, Rao V, Ziegler N et al (2019) Expression of glypican 3 is an independent prognostic biomarker in primary gastro‐esophageal adenocarcinoma and corresponding serum exosomes. J Clin Med 8: 696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A, Greening DW, Chen M, Xu R, Ji H, Simpson RJ (2019) Exosomes derived from human primary and metastatic colorectal cancer cells contribute to functional heterogeneity of activated fibroblasts by reprogramming their proteome. Proteomics 19: e1800148 [DOI] [PubMed] [Google Scholar]
- Rana S, Yue S, Stadel D, Zöller M (2012) Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol 44: 1574–1584 [DOI] [PubMed] [Google Scholar]
- Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ (1996) B lymphocytes secrete antigen‐presenting vesicles. J Exp Med 183: 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak MZ, Jadczyk T, Schneider G, Kakar SS, Kucia M (2013) Induction of a tumor‐metastasis‐receptive microenvironment as an unwanted and underestimated side effect of treatment by chemotherapy or radiotherapy. J Ovarian Res 6: 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y (2018) Carcinoma‐associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 8: 3932–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R (2017) Cancer‐associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene 36: 1770–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs FL, Alayo Q, Krenzlin H, Mahmoud AB, Speranza MC, Nakashima H, Hayes JL, Lee K, Balaj L, Passaro C et al (2018) Immune evasion mediated by PD‐L1 on glioblastoma‐derived extracellular vesicles. Sci Adv 4: eaar2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder K, Sevko A, Heide J, Dams M, Rupp A‐K, Macas J, Starmann J, Tjwa M, Plate KH, Sültmann H et al (2015) Extracellular vesicle‐mediated transfer of functional RNA in the tumor microenvironment. OncoImmunology 4: e1008371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringuette Goulet C, Bernard G, Tremblay S, Chabaud S, Bolduc S, Pouliot F (2018) Exosomes induce fibroblast differentiation into cancer‐associated fibroblasts through TGFbeta signaling. Mol Cancer Res 16: 1196–1204 [DOI] [PubMed] [Google Scholar]
- Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, Azab F, Flores LM, Campigotto F, Weller E et al (2013) BM mesenchymal stromal cell‐derived exosomes facilitate multiple myeloma progression. J Clin Invest 123: 1542–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues CFD, Serrano E, Patricio MI, Val MM, Albuquerque P, Fonseca J, Gomes CMF, Abrunhosa AJ, Paiva A, Carvalho L et al (2018) Stroma‐derived IL‐6, G‐CSF and Activin‐A mediated dedifferentiation of lung carcinoma cells into cancer stem cells. Sci Rep 8: 11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues G, Hoshino A, Kenific CM, Matei IR, Steiner L, Freitas D, Kim HS, Oxley PR, Scandariato I, Casanova‐Salas I et al (2019) Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat Cell Biol 21: 1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Silva J, Herrera A, Herrera M, Pena C, Martin P, Gil‐Calderon B, Larriba MJ, Coronado MJ, Soldevilla B et al (2015) Exosomes enriched in stemness/metastatic‐related mRNAS promote oncogenic potential in breast cancer. Oncotarget 6: 40575–40587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Zorrilla S, Perez‐Sayans M, Fais S, Logozzi M, Gallas Torreira M, Garcia Garcia A (2019) A pilot clinical study on the prognostic relevance of plasmatic exosomes levels in oral squamous cell carcinoma patients. Cancers (Basel) 11: 429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo GR, Lee J, Shoelson SE (2012) Metabolic syndrome, insulin resistance, and roles of inflammation–mechanisms and therapeutic targets. Arterioscler Thromb Vasc Biol 32: 1771–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist KG, Ek B, Stavreus‐Evers A, Larsson A, Ronquist G (2013) Human prostasomes express glycolytic enzymes with capacity for ATP production. Am J Physiol Endocrinol Metab 304: E576–E582 [DOI] [PubMed] [Google Scholar]
- Ronquist KG, Sanchez C, Dubois L, Chioureas D, Fonseca P, Larsson A, Ullen A, Yachnin J, Ronquist G, Panaretakis T (2016) Energy‐requiring uptake of prostasomes and PC3 cell‐derived exosomes into non‐malignant and malignant cells. J Extracell Vesicles 5: 29877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontogianni S, Synadaki E, Li B, Liefaard MC, Lips EH, Wesseling J, Wu W, Altelaar M (2019) Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol 2: 325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roucourt B, Meeussen S, Bao J, Zimmermann P, David G (2015) Heparanase activates the syndecan‐syntenin‐ALIX exosome pathway. Cell Res 25: 412–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo F, Moreno L, Mleczko J, Palomo L, Gonzalez E, Cabrera D, Cogolludo A, Vizcaino FP, van‐Liempd S, Falcon‐Perez JM (2017) Hepatocyte‐secreted extracellular vesicles modify blood metabolome and endothelial function by an arginase‐dependent mechanism. Sci Rep 7: 42798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Lin N, Ma Q, Chen R, Zhang Z, Wen W, Chen H, Sun J (2018) Circulating LncRNAs analysis in patients with type 2 diabetes reveals novel genes influencing glucose metabolism and islet beta‐cell function. Cell Physiol Biochem 46: 335–350 [DOI] [PubMed] [Google Scholar]
- Saari H, Lazaro‐Ibanez E, Viitala T, Vuorimaa‐Laukkanen E, Siljander P, Yliperttula M (2015) Microvesicle‐ and exosome‐mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells. J Control Release 220: 727–737 [DOI] [PubMed] [Google Scholar]
- Safwat A, Sabry D, Ragiae A, Amer E, Mahmoud RH, Shamardan RM (2018) Adipose mesenchymal stem cells‐derived exosomes attenuate retina degeneration of streptozotocin‐induced diabetes in rabbits. J Circ Biomark 7: 1849454418807827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar G, Sah RP, Javeed N, Dutta SK, Smyrk TC, Lau JS, Giorgadze N, Tchkonia T, Kirkland JL, Chari ST et al (2016) Pathogenesis of pancreatic cancer exosome‐induced lipolysis in adipose tissue. Gut 65: 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba DG, Cespedes‐Donoso PF, Balint S, Compeer EB, Korobchevskaya K, Valvo S, Mayya V, Kvalvaag A, Peng Y, Dong T et al (2019) Composition and structure of synaptic ectosomes exporting antigen receptor linked to functional CD40 ligand from helper T cells. Elife 8: e47528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel P, Mulcahy LA, Furlong F, McCarthy HO, Brooks SA, Fabbri M, Pink RC, Carter DRF (2018) Cisplatin induces the release of extracellular vesicles from ovarian cancer cells that can induce invasiveness and drug resistance in bystander cells. Philos Trans R Soc Lond B Biol Sci 373: 20170065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandfeld‐Paulsen B, Aggerholm‐Pedersen N, Baek R, Jakobsen KR, Meldgaard P, Folkersen BH, Rasmussen TR, Varming K, Jorgensen MM, Sorensen BS (2016a) Exosomal proteins as prognostic biomarkers in non‐small cell lung cancer. Mol Oncol 10: 1595–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandfeld‐Paulsen B, Jakobsen KR, Baek R, Folkersen BH, Rasmussen TR, Meldgaard P, Varming K, Jorgensen MM, Sorensen BS (2016b) Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol 11: 1701–1710 [DOI] [PubMed] [Google Scholar]
- Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, Stepanova A, Iommarini L, Mastroleo C, Daly L et al (2017) Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy‐resistant breast cancer. Proc Natl Acad Sci USA 114: E9066–E9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JC, Lima NDS, Sarian LO, Matheu A, Ribeiro ML, Derchain SFM (2018) Exosome‐mediated breast cancer chemoresistance via miR‐155 transfer. Sci Rep 8: 829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Vasaikar S, Eskaros A, Kim Y, Lewis JS, Zhang B, Zijlstra A, Weaver AM (2019) EPHB2 carried on small extracellular vesicles induces tumor angiogenesis via activation of ephrin reverse signaling. JCI Insight 4: e132447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A, Vidal M, Colombo MI (2002) The exosome pathway in K562 cells is regulated by Rab11. J Cell Sci 115: 2505–2515 [DOI] [PubMed] [Google Scholar]
- Schelch K, Vogel L, Schneller A, Brankovic J, Mohr T, Mayer RL, Slany A, Gerner C, Grusch M (2021) EGF induces migration independent of EMT or invasion in A549 lung adenocarcinoma cells. Front Cell Dev Biol 9: 634371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Collinson A, Matthews C, Pointon A, Jenkinson L, Minter RR, Vaughan TJ, Tigue NJ (2019) Exosomal delivery of doxorubicin enables rapid cell entry and enhanced in vitro potency. PLoS One 14: e0214545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Heller DA, Winslow MM, Dahlman JE, Pratt GW, Langer R, Jacks T, Anderson DG (2011) Treating metastatic cancer with nanotechnology. Nat Rev Cancer 12: 39–50 [DOI] [PubMed] [Google Scholar]
- Schuldner M, Dorsam B, Shatnyeva O, Reiners KS, Kubarenko A, Hansen HP, Finkernagel F, Roth K, Theurich S, Nist A et al (2019) Exosome‐dependent immune surveillance at the metastatic niche requires BAG6 and CBP/p300‐dependent acetylation of p53. Theranostics 9: 6047–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick AE, D'Souza‐Schorey C (2018) The biology of extracellular microvesicles. Traffic 19: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E, Amigorena S, Thery C (2005a) Mature dendritic cells secrete exosomes with strong ability to induce antigen‐specific effector immune responses. Blood Cells Mol Dis 35: 89–93 [DOI] [PubMed] [Google Scholar]
- Segura E, Nicco C, Lombard B, Veron P, Raposo G, Batteux F, Amigorena S, Thery C (2005b) ICAM‐1 on exosomes from mature dendritic cells is critical for efficient naive T‐cell priming. Blood 106: 216–223 [DOI] [PubMed] [Google Scholar]
- Sento S, Sasabe E, Yamamoto T (2016) Application of a persistent heparin treatment inhibits the malignant potential of oral squamous carcinoma cells induced by tumor cell‐derived exosomes. PLoS One 11: e0148454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo W, Eun HS, Kim SY, Yi HS, Lee YS, Park SH, Jang MJ, Jo E, Kim SC, Han YM et al (2016) Exosome‐mediated activation of toll‐like receptor 3 in stellate cells stimulates interleukin‐17 production by gammadelta T cells in liver fibrosis. Hepatology 64: 616–631 [DOI] [PubMed] [Google Scholar]
- Severino V, Alessio N, Farina A, Sandomenico A, Cipollaro M, Peluso G, Galderisi U, Chambery A (2013) Insulin‐like growth factor binding proteins 4 and 7 released by senescent cells promote premature senescence in mesenchymal stem cells. Cell Death Dis 4: e911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan G, Zhou X, Gu J, Zhou D, Cheng W, Wu H, Wang Y, Tang T, Wang X (2021) Downregulated exosomal microRNA‐148b‐3p in cancer associated fibroblasts enhance chemosensitivity of bladder cancer cells by downregulating the Wnt/beta‐catenin pathway and upregulating PTEN. Cell Oncol (Dordr) 44: 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P et al (2020) Exosomal circPACRGL promotes progression of colorectal cancer via the miR‐142‐3p/miR‐506‐3p‐ TGF‐beta1 axis. Mol Cancer 19: 117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao B, Ye G, Guo J (2020) Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res 26: 1475–1482 [DOI] [PubMed] [Google Scholar]
- Shao H, Zhang Y, Yan J, Ban X, Fan X, Chang X, Lu Z, Wu Y, Zong L, Mo S et al (2021) Upregulated microRNA‐483‐3p is an early event in pancreatic ductal adenocarcinoma (PDAC) and as a powerful liquid biopsy biomarker in PDAC. Onco Targets Ther 14: 2163–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif S, Ghahremani MH, Soleimani M (2021) Differentiation induction and proliferation inhibition by a cell‐free approach for delivery of exogenous miRNAs to neuroblastoma cells using mesenchymal stem cells. Cell J 22: 556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Ludwig S, Muller L, Hong CS, Kirkwood JM, Ferrone S, Whiteside TL (2018) Immunoaffinity‐based isolation of melanoma cell‐derived exosomes from plasma of patients with melanoma. J Extracell Vesicles 7: 1435138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Johnson A (2020) Exosome DNA: critical regulator of tumor immunity and a diagnostic biomarker. J Cell Physiol 235: 1921–1932 [DOI] [PubMed] [Google Scholar]
- Shaughnessy R, Echard A (2018) Rab35 GTPase and cancer: linking membrane trafficking to tumorigenesis. Traffic 19: 247–252 [DOI] [PubMed] [Google Scholar]
- Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK (2012) Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Song Y, Zhao B, Xu Y, Ren X, Zhou Y, Sun Q (2021) Cancer‐derived exosomal miR‐7641 promotes breast cancer progression and metastasis. Cell Commun Signal 19: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman‐Samis M, Onallah H, Holth A, Reich R, Davidson B (2019) SOX2 and SOX9 are markers of clinically aggressive disease in metastatic high‐grade serous carcinoma. Gynecol Oncol 153: 651–660 [DOI] [PubMed] [Google Scholar]
- Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ (2018) Decreased levels of serum exosomal miR‐638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem 119: 4711–4716 [DOI] [PubMed] [Google Scholar]
- Shi S, Li T, Wen X, Wu SY, Xiong C, Zhao J, Lincha VR, Chow DS, Liu Y, Sood AK et al (2019) Copper‐64 labeled PEGylated exosomes for in vivo positron emission tomography and enhanced tumor retention. Bioconjug Chem 30: 2675–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Tan Q, Feng F, Huang H, Liang J, Cao D, Wang Z (2020a) Identification of core genes in the progression of endometrial cancer and cancer cell‐derived exosomes by an integrative analysis. Sci Rep 10: 9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang J, Yang X, Liu Z (2020b) Exosomal miR‐1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int J Clin Oncol 25: 89–99 [DOI] [PubMed] [Google Scholar]
- Shiao SL, Chu GC, Chung LW (2016) Regulation of prostate cancer progression by the tumor microenvironment. Cancer Lett 380: 340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda A, Sawada SI, Sasaki Y, Akiyoshi K (2019) Exosome surface glycans reflect osteogenic differentiation of mesenchymal stem cells: profiling by an evanescent field fluorescence‐assisted lectin array system. Sci Rep 9: 11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Matsubayashi J, Kudo Y, Maehara S, Takeuchi S, Hagiwara M, Kakihana M, Ohira T, Nagao T, Ikeda N (2021) Serum‐derived exosomal PD‐L1 expression to predict anti‐PD‐1 response and in patients with non‐small cell lung cancer. Sci Rep 11: 7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Park YH, Jung SH, Jang SH, Kim MY, Lee JY, Chung YJ (2021) Urinary exosome microRNA signatures as a noninvasive prognostic biomarker for prostate cancer. NPJ Genom Med 6: 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoae‐Hassani A, Hamidieh AA, Behfar M, Mohseni R, Mortazavi‐Tabatabaei SA, Asgharzadeh S (2017) NK cell‐derived exosomes from NK cells previously exposed to neuroblastoma cells augment the antitumor activity of cytokine‐activated NK cells. J Immunother 40: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S, Yang Y, Allen CL, Maguire O, Minderman H, Sen A, Ciesielski MJ, Collins KA, Bush PJ, Singh P et al (2018) Metabolic reprogramming of stromal fibroblasts by melanoma exosome microRNA favours a pre‐metastatic microenvironment. Sci Rep 8: 12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Temoche‐Diaz MM, Karfilis KV, Ri S, Schekman R (2016) Y‐box protein 1 is required to sort microRNAs into exosomes in cells and in a cell‐free reaction. Elife 5: e19276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Yao J, Qin Y, Nottingham RM, Temoche‐Diaz MM, Schekman R, Lambowitz AM (2017) Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci USA 114: E8987–E8995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y, Kim S, Zhang E, Tang Y, Jaskula‐Sztul R, Markert JM, Chen H, Zhou L, Liu XM (2020) Targeted exosomes for drug delivery: biomanufacturing, surface tagging, and validation. Biotechnol J 15: e1900163 [DOI] [PubMed] [Google Scholar]
- Silverman JM, Clos J, de'Oliveira CC, Shirvani O, Fang Y, Wang C, Foster LJ, Reiner NE (2010) An exosome‐based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J Cell Sci 123: 842–852 [DOI] [PubMed] [Google Scholar]
- Simons M, Raposo G (2009) Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol 21: 575–581 [DOI] [PubMed] [Google Scholar]
- Sinha S, Hoshino D, Hong NH, Kirkbride KC, Grega‐Larson NE, Seiki M, Tyska MJ, Weaver AM (2016) Cortactin promotes exosome secretion by controlling branched actin dynamics. J Cell Biol 214: 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Würdinger T, Van Rijn S, Meijer DH, Gainche L, Curry WT, Carter BS, Krichevsky AM, Breakefield XO (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10: 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotland T, Ekroos K, Kauhanen D, Simolin H, Seierstad T, Berge V, Sandvig K, Llorente A (2017a) Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur J Cancer 70: 122–132 [DOI] [PubMed] [Google Scholar]
- Skotland T, Sandvig K, Llorente A (2017b) Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res 66: 30–41 [DOI] [PubMed] [Google Scholar]
- Skotland T, Hessvik NP, Sandvig K, Llorente A (2019) Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res 60: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, James OF (1998) The failing malignant liver. Gut 42: 454–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda N, Iinuma H, Suzuki Y, Tsukahara D, Midorikawa H, Igarashi Y, Kumata Y, Horikawa M, Kiyokawa T, Fukagawa T et al (2019) Plasma exosome‐encapsulated microRNA‐21 and microRNA‐92a are promising biomarkers for the prediction of peritoneal recurrence in patients with gastric cancer. Oncol Lett 18: 4467–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somiya M, Yoshioka Y, Ochiya T (2018) Biocompatibility of highly purified bovine milk‐derived extracellular vesicles. J Extracell Vesicles 7: 1440132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Chen Y, Zhu G, Xie H, Yang Z, Li L (2020) Exosome‐mediated miR‐9‐5p promotes proliferation and migration of renal cancer cells both in vitro and in vivo by targeting SOCS4. Biochem Biophys Res Commun 529: 1216–1224 [DOI] [PubMed] [Google Scholar]
- Song Y, Wang M, Tong H, Tan Y, Hu X, Wang K, Wan X (2021) Plasma exosomes from endometrial cancer patients contain LGALS3BP to promote endometrial cancer progression. Oncogene 40: 633–646 [DOI] [PubMed] [Google Scholar]
- Srikanthan S, Li W, Silverstein RL, McIntyre TM (2014) Exosome poly‐ubiquitin inhibits platelet activation, downregulates CD36 and inhibits pro‐atherothombotic cellular functions. J Thromb Haemost 12: 1906–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanly C, Moubarak M, Fiume I, Turiak L, Pocsfalvi G (2019) Membrane transporters in citrus clementina fruit juice‐derived nanovesicles. Int J Mol Sci 20: 6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stidham RW, Higgins PDR (2018) Colorectal cancer in inflammatory bowel disease. Clin Colon Rectal Surg 31: 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremmel W, Weiskirchen R, Melnik BC (2020) Milk exosomes prevent intestinal inflammation in a genetic mouse model of ulcerative colitis: a pilot experiment. Inflamm Intest Dis 5: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YY, Sun L, Guo ZR, Li JC, Bai TT, Cai XX, Li WH, Zhu YF (2019) Upregulated expression of serum exosomal miR‐375 and miR‐1307 enhance the diagnostic power of CA125 for ovarian cancer. J Ovarian Res 12: 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, De Medina P, Monsarrat B, Perret B, Silvente‐Poirot S et al (2010) Exosomes account for vesicle‐mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res 51: 2105–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM (2013) Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude‐mouse models. Adv Drug Deliv Rev 65: 383–390 [DOI] [PubMed] [Google Scholar]
- Sun H, Tan W, Zu Y (2016) Aptamers: versatile molecular recognition probes for cancer detection. Analyst 141: 403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Li Y, Zhou Y, Ng TK, Zhao C, Gan Q, Gu X, Xiang J (2019a) Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol 234: 1416–1425 [DOI] [PubMed] [Google Scholar]
- Sun B, Zhou Y, Fang Y, Li Z, Gu X, Xiang J (2019b) Colorectal cancer exosomes induce lymphatic network remodeling in lymph nodes. Int J Cancer 145: 1648–1659 [DOI] [PubMed] [Google Scholar]
- Sun C, Wang P, Dong W, Liu H, Sun J, Zhao L (2020a) LncRNA PVT1 promotes exosome secretion through YKT6, RAB7, and VAMP3 in pancreatic cancer. Aging 12: 10427–10440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Liu X, Pan B, Hu X, Zhu Y, Su Y, Guo Z, Zhang G, Xu M, Xu X et al (2020b) Serum exosomal miR‐122 as a potential diagnostic and prognostic biomarker of colorectal cancer with liver metastasis. J Cancer 11: 630–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Lin F, Sun W, Zhu W, Fang D, Luo L, Li S, Zhang W, Jiang L (2021a) Exosome‐transmitted miRNA‐335‐5p promotes colorectal cancer invasion and metastasis by facilitating EMT via targeting RASA1. Mol Ther Nucleic Acids 24: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jin J, Jing H, Lu Y, Zhu Q, Shu C, Zhang Q, Jing D (2021b) ITIH4 is a novel serum biomarker for early gastric cancer diagnosis. Clin Chim Acta 523: 365–373 [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhou Y, Shi Y, Zhang Y, Liu K, Liang R, Sun P, Chang X, Tang W, Zhang Y et al (2021c) Expression of miRNA‐29 in pancreatic beta cells promotes inflammation and diabetes via TRAF3. Cell Rep 34: 108576 [DOI] [PubMed] [Google Scholar]
- Sung JS, Kang CW, Kang S, Jang Y, Chae YC, Kim BG, Cho NH (2020) ITGB4‐mediated metabolic reprogramming of cancer‐associated fibroblasts. Oncogene 39: 664–676 [DOI] [PubMed] [Google Scholar]
- Svensson KJ, Christianson HC, Wittrup A, Bourseau‐Guilmain E, Lindqvist E, Svensson LM, Mörgelin M, Belting M (2013) Exosome uptake depends on ERK1/2‐heat shock protein 27 signaling and lipid raft‐mediated endocytosis negatively regulated by caveolin‐1. J Biol Chem 288: 17713–17724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szvicsek Z, Oszvald A, Szabo L, Sandor GO, Kelemen A, Soos AA, Paloczi K, Harsanyi L, Tolgyes T, Dede K et al (2019) Extracellular vesicle release from intestinal organoids is modulated by Apc mutation and other colorectal cancer progression factors. Cell Mol Life Sci 76: 2463–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Nishikawa M, Shinotsuka H, Matsui Y, Ohara S, Imai T, Takakura Y (2013) Visualization and in vivo tracking of the exosomes of murine melanoma B16‐BL6 cells in mice after intravenous injection. J Biotechnol 165: 77–84 [DOI] [PubMed] [Google Scholar]
- Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, Takasugi M, Watanabe S, Kanemaki MT, Obuse C et al (2017) Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun 8: 15287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M, Ikeda Y, Shibuya M, Miura F, Sano K (2018) Usefulness of exosome‐encapsulated microRNA‐451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci 25: 155–161 [DOI] [PubMed] [Google Scholar]
- Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, Shiina M, Fukushima K, Hoshino T, Sano K et al (2010) Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT‐0 protein. Biochem Biophys Res Commun 399: 384–390 [DOI] [PubMed] [Google Scholar]
- Tan M, Yan HB, Li JN, Li WK, Fu YY, Chen W, Zhou Z (2016) Thrombin stimulated platelet‐derived exosomes inhibit platelet‐derived growth factor receptor‐beta expression in vascular smooth muscle cells. Cell Physiol Biochem 38: 2348–2365 [DOI] [PubMed] [Google Scholar]
- Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, Ayad NG (2018) Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer 17: 74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M, Baba H (2013) Clinical impact of serum exosomal microRNA‐21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 119: 1159–1167 [DOI] [PubMed] [Google Scholar]
- Tang MKS, Yue PYK, Ip PP, Huang RL, Lai HC, Cheung ANY, Tse KY, Ngan HYS, Wong AST (2018a) Soluble E‐cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat Commun 9: 2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Fu K, Sun H, Rong D, Wang H, Cao H (2018b) CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer 17: 137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Zheng K, Tang Y, Li Z, Zou T, Liu D (2019a) Overexpression of serum exosomal HOTAIR is correlated with poor survival and poor response to chemotherapy in breast cancer patients. J Biosci 44: 37 [PubMed] [Google Scholar]
- Tang X, Liu S, Liu Y, Lin X, Zheng T, Liu X, Qiu J, Hua K (2019b) Circulating serum exosomal aHIF is a novel prognostic predictor for epithelial ovarian cancer. Onco Targets Ther 12: 7699–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhao Y, Song X, Song X, Niu L, Xie L (2019c) Tumor‐derived exosomal miRNA‐320d as a biomarker for metastatic colorectal cancer. J Clin Lab Anal 33: e23004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Zhou J, Yuan C, Zhang L, Li D, Si D, Xiu D, Zhong L (2019) Metabolomics identifies serum and exosomes metabolite markers of pancreatic cancer. Metabolomics 15: 86 [DOI] [PubMed] [Google Scholar]
- Tao H, Xu H, Zuo L, Li C, Qiao G, Guo M, Zheng L, Leitgeb M, Lin X (2020a) Exosomes‐coated bcl‐2 siRNA inhibits the growth of digestive system tumors both in vitro and in vivo . Int J Biol Macromol 161: 470–480 [DOI] [PubMed] [Google Scholar]
- Tao Y, Tang Y, Yang Z, Wu F, Wang L, Yang L, Lei L, Jing Y, Jiang X, Jin H et al (2020b) Exploration of serum exosomal LncRNA TBILA and AGAP2‐AS1 as promising biomarkers for diagnosis of non‐small cell lung cancer. Int J Biol Sci 16: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ (2012) Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863‐derived exosomes. Methods 56: 293–304 [DOI] [PubMed] [Google Scholar]
- Tauro BJ, Greening DW, Mathias RA, Mathivanan S, Ji H, Simpson RJ (2013a) Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell‐derived organoids. Mol Cell Proteomics 12: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauro BJ, Mathias RA, Greening DW, Gopal SK, Ji H, Kapp EA, Coleman BM, Hill AF, Kusebauch U, Hallows JL et al (2013b) Oncogenic H‐ras reprograms Madin‐Darby canine kidney (MDCK) cell‐derived exosomal proteins following epithelial‐mesenchymal transition. Mol Cell Proteomics 12: 2148–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S, Pucci M, Giallombardo M, Di Bella MA, Santarpia M, Reclusa P, Gil‐Bazo I, Rolfo C, Alessandro R (2017) Amphiregulin contained in NSCLC‐exosomes induces osteoclast differentiation through the activation of EGFR pathway. Sci Rep 7: 3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawil N, Bassawon R, Meehan B, Nehme A, Montermini L, Gayden T, De Jay N, Spinelli C, Chennakrishnaiah S, Choi D et al (2021) Glioblastoma cell populations with distinct oncogenic programs release podoplanin as procoagulant extracellular vesicles. Blood Adv 5: 1682–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, Deng Z, Kumar A, Zhang L, Merchant ML et al (2017) MVP‐mediated exosomal sorting of miR‐193a promotes colon cancer progression. Nat Commun 8: 14448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, Hutchins E, Mu J, Deng Z, Luo C et al (2018) Plant‐derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe 24: 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Kang H, Chu Y (2019) Identification of an exosomal long noncoding RNA SOX2‐OT in plasma as a promising biomarker for lung squamous cell carcinoma. Genet Test Mol Biomarkers 23: 235–240 [DOI] [PubMed] [Google Scholar]
- Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa‐Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J et al (2014) Double‐stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 24: 766–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakuri BKC, Zhang J, Zhao J, Nguyen LN, Nguyen LNT, Khanal S, Cao D, Dang X, Schank M, Wu XY et al (2020) LncRNA HOTAIRM1 promotes MDSC expansion and suppressive functions through the HOXA1‐miR124 axis during HCV infection. Sci Rep 10: 22033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalin C, Hisada Y, Lundstrom S, Mackman N, Wallen H (2019) Neutrophil extracellular traps: villains and targets in arterial, venous, and cancer‐associated thrombosis. Arterioscler Thromb Vasc Biol 39: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL (2018) Clinical significance of PD‐L1(+) exosomes in plasma of head and neck cancer patients. Clin Cancer Res 24: 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoraki MN, Matsumoto A, Beccard I, Hoffmann TK, Whiteside TL (2020) CD44v3 protein‐carrying tumor‐derived exosomes in HNSCC patients' plasma as potential noninvasive biomarkers of disease activity. Oncoimmunology 9: 1747732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Duban L, Segura E, Veron P, Lantz O, Amigorena S (2002) Indirect activation of naive CD4+ T cells by dendritic cell‐derived exosomes. Nat Immunol 3: 1156–1162 [DOI] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, Clayton A (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3.22 [DOI] [PubMed] [Google Scholar]
- Théry C, Duban L, Segura E, Véron P, Lantz O, Amigorena S (2002) Indirect activation of naïve CD4+ T cells by dendritic cell–derived exosomes. Nat Immunol 3: 1156–1162 [DOI] [PubMed] [Google Scholar]
- Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin‐Smith GK et al (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7: 1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Wang Y, Wang H, Zhu Z, Xiao Z (2010) Visualizing of the cellular uptake and intracellular trafficking of exosomes by live‐cell microscopy. J Cell Biochem 111: 488–496 [DOI] [PubMed] [Google Scholar]
- Tian T, Zhu Y‐L, Zhou Y‐Y, Liang G‐F, Wang Y‐Y, Hu F‐H, Xiao Z‐D (2014a) Exosome uptake through clathrin‐mediated endocytosis and macropinocytosis and mediating miR‐21 delivery. J Biol Chem 289: 22258–22267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, Wei J, Nie G (2014b) A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 35: 2383–2390 [DOI] [PubMed] [Google Scholar]
- Tiedemann K, Sadvakassova G, Mikolajewicz N, Juhas M, Sabirova Z, Tabaries S, Gettemans J, Siegel PM, Komarova SV (2019) Exosomal release of L‐plastin by breast cancer cells facilitates metastatic bone osteolysis. Transl Oncol 12: 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohme S, Yazdani HO, Al‐Khafaji AB, Chidi AP, Loughran P, Mowen K, Wang Y, Simmons RL, Huang H, Tsung A (2016) Neutrophil extracellular traps promote the development and progression of liver metastases after surgical stress. Cancer Res 76: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga N, Kosaka N, Ono M, Katsuda T, Yoshioka Y, Tamura K, Lotvall J, Nakagama H, Ochiya T (2015) Brain metastatic cancer cells release microRNA‐181c‐containing extracellular vesicles capable of destructing blood‐brain barrier. Nat Commun 6: 6716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319: 1244–1247 [DOI] [PubMed] [Google Scholar]
- Tran PHL, Wang T, Yin W, Tran TTD, Nguyen TNG, Lee BJ, Duan W (2019) Aspirin‐loaded nanoexosomes as cancer therapeutics. Int J Pharm 572: 118786 [DOI] [PubMed] [Google Scholar]
- Tripisciano C, Weiss R, Eichhorn T, Spittler A, Heuser T, Fischer MB, Weber V (2017) Different potential of extracellular vesicles to support thrombin generation: contributions of phosphatidylserine, tissue factor, and cellular origin. Sci Rep 7: 6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y (2017) Circulating exosomal microRNA‐21 as a biomarker in each tumor stage of colorectal cancer. Oncology 92: 360–370 [DOI] [PubMed] [Google Scholar]
- Tubita V, Segui‐Barber J, Lozano JJ, Banon‐Maneus E, Rovira J, Cucchiari D, Moya‐Rull D, Oppenheimer F, Del Portillo H, Campistol JM et al (2019) Effect of immunosuppression in miRNAs from extracellular vesicles of colorectal cancer and their influence on the pre‐metastatic niche. Sci Rep 9: 11177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulkens J, Vergauwen G, Van Deun J, Geeurickx E, Dhondt B, Lippens L, De Scheerder MA, Miinalainen I, Rappu P, De Geest BG et al (2020) Increased levels of systemic LPS‐positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 69: 191–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH (2014) Exosomal miR‐135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor‐inhibiting HIF‐1. Blood 124: 3748–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsugi T, Schroit AJ, Connor J, Bucana CD, Fidler IJ (1991) Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res 51: 3062–3066 [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO (2007) Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659 [DOI] [PubMed] [Google Scholar]
- Vallabhaneni KC, Penfornis P, Dhule S, Guillonneau F, Adams KV, Mo YY, Xu R, Liu Y, Watabe K, Vemuri MC et al (2015) Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget 6: 4953–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Herwijnen MJ, Zonneveld MI, Goerdayal S, Nolte‐'t Hoen EN, Garssen J, Stahl B, Maarten Altelaar AF, Redegeld FA, Wauben MH (2016) Comprehensive proteomic analysis of human milk‐derived extracellular vesicles unveils a novel functional proteome distinct from other milk components. Mol Cell Proteomics 15: 3412–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, D'Angelo G, Raposo G (2018) Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19: 213–228 [DOI] [PubMed] [Google Scholar]
- Vendramin R, Litchfield K, Swanton C (2021) Cancer evolution: Darwin and beyond. EMBO J 40: e108389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij FJ, Bebelman MP, Jimenez CR, Garcia‐Vallejo JJ, Janssen H, Neefjes J, Knol JC, De Goeij‐De Haas R, Piersma SR, Baglio SR et al (2018) Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol 217: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S, Ploix S, Lapierre V, Thery C, Commere PH, Tramalloni D, Gorrichon K, Virault‐Rocroy P, Tursz T, Lantz O et al (2011) Updated technology to produce highly immunogenic dendritic cell‐derived exosomes of clinical grade: a critical role of interferon‐gamma. J Immunother 34: 65–75 [DOI] [PubMed] [Google Scholar]
- Viaud S, Terme M, Flament C, Taieb J, Andre F, Novault S, Escudier B, Robert C, Caillat‐Zucman S, Tursz T et al (2009) Dendritic cell‐derived exosomes promote natural killer cell activation and proliferation: a role for NKG2D ligands and IL‐15Ralpha. PLoS One 4: e4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Sainte‐Marie J, Philippot JR, Bienvenue A (1989) Asymmetric distribution of phospholipids in the membrane of vesicles released during in vitro maturation of guinea pig reticulocytes: evidence precluding a role for “aminophospholipid translocase”. J Cell Physiol 140: 455–462 [DOI] [PubMed] [Google Scholar]
- Vietri M, Radulovic M, Stenmark H (2020) The many functions of ESCRTs. Nat Rev Mol Cell Biol 21: 25–42 [DOI] [PubMed] [Google Scholar]
- Villarroya‐Beltri C, Gutiérrez‐Vázquez C, Sánchez‐Cabo F, Pérez‐Hernández D, Vázquez J, Martin‐Cofreces N, Martinez‐Herrera DJ, Pascual‐Montano A, Mittelbrunn M, Sánchez‐Madrid F (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4: 2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinik Y, Ortega FG, Mills GB, Lu Y, Jurkowicz M, Halperin S, Aharoni M, Gutman M, Lev S (2020) Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci Adv 6: eaba5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virchow R (1989) Cellular pathology. As based upon physiological and pathological histology. Lecture XVI‐‐Atheromatous affection of arteries. 1858. Nutr Rev 47: 23–25 [DOI] [PubMed] [Google Scholar]
- Volk‐Draper L, Hall K, Griggs C, Rajput S, Kohio P, DeNardo D, Ran S (2014) Paclitaxel therapy promotes breast cancer metastasis in a TLR4‐dependent manner. Cancer Res 74: 5421–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker ND, Elias M, Guiro K, Bhatia R, Greco SJ, Bryan M, Gergues M, Sandiford OA, Ponzio NM, Leibovich SJ et al (2019) Exosomes from differentially activated macrophages influence dormancy or resurgence of breast cancer cells within bone marrow stroma. Cell Death Dis 10: 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Xia T, Du Y, Liu J, Xie Y, Zhang Y, Guan F, Wu J, Wang X, Shi C (2019) Exosomes from activated hepatic stellate cells contain GLUT1 and PKM2: a role for exosomes in metabolic switch of liver nonparenchymal cells. FASEB J 33: 8530–8542 [DOI] [PubMed] [Google Scholar]
- Wan L, Chen X, Deng J, Zhang S, Tu F, Pei H, Hu R, Liu J, Yu H (2021) Plasma exosome‐derived B‐cell translation gene 1: a predictive marker for the prognosis in patients with non‐small cell lung cancer. J Cancer 12: 1538–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T et al (2010) Structural and functional insights into 5'‐ppp RNA pattern recognition by the innate immune receptor RIG‐I. Nat Struct Mol Biol 17: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Gilkes DM, Takano N, Xiang L, Luo W, Bishop CJ, Chaturvedi P, Green JJ, Semenza GL (2014) Hypoxia‐inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc Natl Acad Sci USA 111: E3234–E3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Ding Q, Yaqoob U, de Assuncao TM, Verma VK, Hirsova P, Cao S, Mukhopadhyay D, Huebert RC, Shah VH (2015) Exosome adherence and internalization by hepatic stellate cells triggers sphingosine 1‐phosphate‐dependent migration. J Biol Chem 290: 30684–30696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Guo R, Yang Y, Jacobs B, Chen S, Iwuchukwu I, Gaines KJ, Chen Y, Simman R, Lv G et al (2016a) The novel methods for analysis of exosomes released from endothelial cells and endothelial progenitor cells. Stem Cells Int 2016: 2639728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yi J, Chen X, Zhang Y, Xu M, Yang Z (2016b) The regulation of cancer cell migration by lung cancer cell‐derived exosomes through TGF‐beta and IL‐10. Oncol Lett 11: 1527–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, Zhang Y, Huang X (2017a) Circulating exosomal miR‐125a‐3p as a novel biomarker for early‐stage colon cancer. Sci Rep 7: 4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Wang L, Yang Y, Gong L, Xiao B, Liu X (2017b) A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun 493: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Wang S, Li X, Xu M, Wang J, Zhao RC (2017c) Reduced adipogenesis after lung tumor exosomes priming in human mesenchymal stem cells via TGFbeta signaling pathway. Mol Cell Biochem 435: 59–66 [DOI] [PubMed] [Google Scholar]
- Wang J, Yang K, Yuan W, Gao Z (2018a) Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Monit 24: 9307–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Cao D, Wang L, Zhao J, Nguyen LN, Dang X, Ji Y, Wu XY, Morrison ZD, Xie Q et al (2018b) HCV‐associated exosomes promote myeloid‐derived suppressor cell expansion via inhibiting miR‐124 to regulate T follicular cell differentiation and function. Cell Discov 4: 51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li Y, Guan X, Zhao J, Shen L, Liu J (2018c) Exosomal double‐stranded DNA as a biomarker for the diagnosis and preoperative assessment of pheochromocytoma and paraganglioma. Mol Cancer 17: 128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Song X, Liu L, Niu L, Wang X, Song X, Xie L (2018d) Circulating exosomes contain protein biomarkers of metastatic non‐small‐cell lung cancer. Cancer Sci 109: 1701–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang G, Zhao D, Wang J, Bai Y, Peng Q, Wang H, Fang R, Chen G, Wang Z et al (2019a) CD103‐positive CSC exosome promotes EMT of clear cell renal cell carcinoma: role of remote MiR‐19b‐3p. Mol Cancer 18: 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shen H, He Q, Tian W, Xia A, Lu XJ (2019b) Exosomes derived from exhausted CD8+ T cells impaired the anticancer function of normal CD8+ T cells. J Med Genet 56: 29–31 [DOI] [PubMed] [Google Scholar]
- Wang Y, Yin K, Tian J, Xia X, Ma J, Tang X, Xu H, Wang S (2019c) Granulocytic myeloid‐derived suppressor cells promote the stemness of colorectal cancer cells through exosomal S100A9. Adv Sci (Weinh) 6: 1901278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Sun H, Provaznik J, Hackert T, Zoller M (2019d) Pancreatic cancer‐initiating cell exosome message transfer into noncancer‐initiating cells: the importance of CD44v6 in reprogramming. J Exp Clin Cancer Res 38: 132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZF, Liao F, Wu H, Dai J (2019e) Glioma stem cells‐derived exosomal miR‐26a promotes angiogenesis of microvessel endothelial cells in glioma. J Exp Clin Cancer Res 38: 201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Mao JH, Wang BY, Wang LX, Wen HY, Xu LJ, Fu JX, Yang H (2020a) Exosomal miR‐1910‐3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF‐kappaB signaling pathway. Cancer Lett 489: 87–99 [DOI] [PubMed] [Google Scholar]
- Wang C, Xu J, Yuan D, Bai Y, Pan Y, Zhang J, Shao C (2020b) Exosomes carrying ALDOA and ALDH3A1 from irradiated lung cancer cells enhance migration and invasion of recipients by accelerating glycolysis. Mol Cell Biochem 469: 77–87 [DOI] [PubMed] [Google Scholar]
- Wang D, Wang X, Si M, Yang J, Sun S, Wu H, Cui S, Qu X, Yu X (2020c) Exosome‐encapsulated miRNAs contribute to CXCL12/CXCR4‐induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett 474: 36–52 [DOI] [PubMed] [Google Scholar]
- Wang H, Wei H, Wang J, Li L, Chen A, Li Z (2020d) MicroRNA‐181d‐5p‐containing exosomes derived from CAFs promote EMT by regulating CDX2/HOXA5 in breast cancer. Mol Ther Nucleic Acids 19: 654–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bonacquisti EE, Brown AD, Nguyen J (2020e) Boosting the biogenesis and secretion of mesenchymal stem cell‐derived exosomes. Cell 9: 660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang H, Sun X, Wang X, Ren T, Huang Y, Zhang R, Zheng B, Guo W (2020f) Exosomal PD‐L1 and N‐cadherin predict pulmonary metastasis progression for osteosarcoma patients. J Nanobiotechnol 18: 151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Y, Quan J (2020g) Exosomal miR‐223 derived from natural killer cells inhibits hepatic stellate cell activation by suppressing autophagy. Mol Med 26: 81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Y, Quan J (2020h) Exosomes derived from natural killer cells inhibit hepatic stellate cell activation and liver fibrosis. Hum Cell 33: 582–589 [DOI] [PubMed] [Google Scholar]
- Wang C, Wang J, Cui W, Liu Y, Zhou H, Wang Y, Chen X, Chen X, Wang Z (2021a) Serum exosomal miRNA‐1226 as potential biomarker of pancreatic ductal adenocarcinoma. Onco Targets Ther 14: 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma P, Kim DH, Liu BF, Demirci U (2021b) Towards microfluidic‐based exosome isolation and detection for tumor therapy. Nano Today 37: 101066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang X, Shi Z, Shen L, Zhang J, Zhang J (2021c) Bovine milk exosomes attenuate the alteration of purine metabolism and energy status in IEC‐6 cells induced by hydrogen peroxide. Food Chem 350: 129142 [DOI] [PubMed] [Google Scholar]
- Wang L, Wu J, Ye N, Li F, Zhan H, Chen S, Xu J (2021d) Plasma‐derived exosome MiR‐19b acts as a diagnostic marker for pancreatic cancer. Front Oncol 11: 739111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zhu H, Xiao M, Zhou S (2021e) Serum exosomal long noncoding RNA CRNDE as a prognostic biomarker for hepatocellular carcinoma. J Clin Lab Anal 35: e23959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Qian T, Bao S, Zhao H, Chen H, Xing Z, Li Y, Zhang M, Meng X, Wang C et al (2021f) Circulating exosomal miR‐363‐5p inhibits lymph node metastasis by downregulating PDGFB and serves as a potential noninvasive biomarker for breast cancer. Mol Oncol 15: 2466–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fang YX, Dong B, Du X, Wang J, Wang X, Gao WQ, Xue W (2021g) Discovery of extracellular vesicles derived miR‐181a‐5p in patient's serum as an indicator for bone‐metastatic prostate cancer. Theranostics 11: 878–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pei L, Yue Z, Jia M, Wang H, Cao LL (2021h) The potential of serum exosomal hsa_circ_0028861 as the novel diagnostic biomarker of HBV‐derived hepatocellular cancer. Front Genet 12: 703205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber J, Steadman R, Mason MD, Tabi Z, Clayton A (2010) Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res 70: 9621–9630 [DOI] [PubMed] [Google Scholar]
- Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng L, Ding L, Zhang Y, Zhang L, Li N et al (2017a) Exosomes derived from gemcitabine‐resistant cells transfer malignant phenotypic traits via delivery of miRNA‐222‐3p. Mol Cancer 16: 132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Wang D, Jin F, Bian Z, Li L, Liang H, Li M, Shi L, Pan C, Zhu D et al (2017b) Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome‐associated protein 23. Nat Commun 8: 14041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Chen J, Wang S, Fu F, Zhu X, Wu C, Liu Z, Zhong G, Lin J (2019) A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro . Int J Nanomedicine 14: 8603–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, Zhang R, Wu Y, Gao S, Kang T (2021a) RAB31 marks and controls an ESCRT‐independent exosome pathway. Cell Res 31: 157–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Li Z, Feng H, Ren L (2021b) Serum exosomal EphA2 is a prognostic biomarker in patients with pancreatic cancer. Cancer Manag Res 13: 3675–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen SW, Sceneay J, Lima LG, Wong CS, Becker M, Krumeich S, Lobb RJ, Castillo V, Wong KN, Ellis S et al (2016) The biodistribution and immune suppressive effects of breast cancer‐derived exosomes. Cancer Res 76: 6816–6827 [DOI] [PubMed] [Google Scholar]
- Wendeln AC, Degenhardt K, Kaurani L, Gertig M, Ulas T, Jain G, Wagner J, Hasler LM, Wild K, Skodras A et al (2018) Innate immune memory in the brain shapes neurological disease hallmarks. Nature 556: 332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Grillo‐Hill BK, Barber DL (2017) Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J Cell Sci 130: 663–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight TN, Kang I, Evanko SP, Harten IA, Chang MY, Pearce OMT, Allen CE, Frevert CW (2020) Versican‐a critical extracellular matrix regulator of immunity and inflammation. Front Immunol 11: 512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis GR, Fernandez‐Gonzalez A, Reis M, Yeung V, Liu X, Ericsson M, Andrews NA, Mitsialis SA, Kourembanas S (2020) Mesenchymal stromal cell‐derived small extracellular vesicles restore lung architecture and improve exercise capacity in a model of neonatal hyperoxia‐induced lung injury. J Extracell Vesicles 9: 1790874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills CA, Liu X, Chen L, Zhao Y, Dower CM, Sundstrom J, Wang HG (2021) Chemotherapy‐induced upregulation of small extracellular vesicle‐associated PTX3 accelerates breast cancer metastasis. Cancer Res 81: 452–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf T, Baier SR, Zempleni J (2015) The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma Caco‐2 cells and rat small intestinal IEC‐6 cells. J Nutr 145: 2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WY, Lee MM, Chan BD, Kam RK, Zhang G, Lu AP, Tai WC (2016) Proteomic profiling of dextran sulfate sodium induced acute ulcerative colitis mice serum exosomes and their immunomodulatory impact on macrophages. Proteomics 16: 1131–1145 [DOI] [PubMed] [Google Scholar]
- Woo J, Santasusagna S, Banks J, Pastor‐Lopez S, Yadav K, Carceles‐Cordon M, Dominguez‐Andres A, Den RB, Languino LR, Pippa R et al (2020) Urine extracellular vesicle GATA2 mRNA discriminates biopsy result in men with suspicion of prostate cancer. J Urol 204: 691–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worst TS, von Hardenberg J, Gross JC, Erben P, Schnolzer M, Hausser I, Bugert P, Michel MS, Boutros M (2017) Database‐augmented mass spectrometry analysis of exosomes identifies claudin 3 as a putative prostate cancer biomarker. Mol Cell Proteomics 16: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Han M, Chen T, Yan W, Ning Q (2010) Acute liver failure: mechanisms of immune‐mediated liver injury. Liver Int 30: 782–794 [DOI] [PubMed] [Google Scholar]
- Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, Li H, Li P, Quinn D, Dao M et al (2017) Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci USA 114: 10584–10589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DM, Deng SH, Liu T, Han R, Zhang T, Xu Y (2018) TGF‐beta‐mediated exosomal lnc‐MMP2‐2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Med 7: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Kittana H, Shu J, Kachman SD, Cui J, Ramer‐Tait AE, Zempleni J (2019a) Dietary depletion of milk exosomes and their microRNA cargos elicits a depletion of miR‐200a‐3p and elevated intestinal inflammation and chemokine (C‐X‐C Motif) ligand 9 expression in Mdr1a(‐/‐) mice. Curr Dev Nutr 3: nzz122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Li J, Li Z, Sun S, Zhu S, Wang L, Wu J, Yuan J, Zhang Y, Sun S et al (2019b) Exosomes from the tumour‐adipocyte interplay stimulate beige/brown differentiation and reprogram metabolism in stromal adipocytes to promote tumour progression. J Exp Clin Cancer Res 38: 223 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wu XG, Zhou CF, Zhang YM, Yan RM, Wei WF, Chen XJ, Yi HY, Liang LJ, Fan LS, Liang L et al (2019c) Cancer‐derived exosomal miR‐221‐3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis 22: 397–410 [DOI] [PubMed] [Google Scholar]
- Wu B, Chen X, Wang J, Qing X, Wang Z, Ding X, Xie Z, Niu L, Guo X, Cai T et al (2020a) Separation and characterization of extracellular vesicles from human plasma by asymmetrical flow field‐flow fractionation. Anal Chim Acta 1127: 234–245 [DOI] [PubMed] [Google Scholar]
- Wu J, Gao W, Tang Q, Yu Y, You W, Wu Z, Fan Y, Zhang L, Wu C, Han G et al (2020b) M2 macrophage‐derived exosomes facilitate hepatocarcinoma metastasis by transferring alphaM beta2 integrin to tumor cells. Hepatology 73: 1365–1380 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wu L, Zhou WB, Zhou J, Wei Y, Wang HM, Liu XD, Chen XC, Wang W, Ye L, Yao LC et al (2020c) Circulating exosomal microRNAs as novel potential detection biomarkers in pancreatic cancer. Oncol Lett 20: 1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou Z, Xu S, Liao C, Chen X, Li B, Peng J, Li D, Yang L (2020d) Extracellular vesicle packaged LMP1‐activated fibroblasts promote tumor progression via autophagy and stroma‐tumor metabolism coupling. Cancer Lett 478: 93–106 [DOI] [PubMed] [Google Scholar]
- Wu X, Zhu D, Tian J, Tang X, Guo H, Ma J, Xu H, Wang S (2020e) Granulocytic myeloid‐derived suppressor cell exosomal prostaglandin E2 ameliorates collagen‐induced arthritis by enhancing IL‐10(+) B cells. Front Immunol 11: 588500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XB, Sun HY, Luo ZL, Cheng L, Duan XM, Ren JD (2020f) Plasma‐derived exosomes contribute to pancreatitis‐associated lung injury by triggering NLRP3‐dependent pyroptosis in alveolar macrophages. Biochim Biophys Acta Mol Basis Dis 1866: 165685 [DOI] [PubMed] [Google Scholar]
- Wu Y, Wei J, Zhang W, Xie M, Wang X, Xu J (2020g) Serum exosomal miR‐1290 is a potential biomarker for lung adenocarcinoma. Onco Targets Ther 13: 7809–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubbolts R, Leckie RS, Veenhuizen PTM, Schwarzmann G, Möbius W, Hoernschemeyer J, Slot J‐W, Geuze HJ, Stoorvogel W (2003) Proteomic and biochemical analyses of human B cell‐derived exosomes. J Biol Chem 278: 10963–10972 [DOI] [PubMed] [Google Scholar]
- Xia C, Braunstein Z, Toomey AC, Zhong J, Rao X (2017) S100 proteins as an important regulator of macrophage inflammation. Front Immunol 8: 1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Chen H, Chen D, Ye Y, Xie C, Hou M (2020) PD‐1 inhibitor inducing exosomal miR‐34a‐5p expression mediates the cross talk between cardiomyocyte and macrophage in immune checkpoint inhibitor‐related cardiac dysfunction. J Immunother Cancer 8: e001293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao CT, Lai WJ, Zhu WA, Wang H (2020) MicroRNA derived from circulating exosomes as noninvasive biomarkers for diagnosing renal cell carcinoma. Onco Targets Ther 13: 10765–10774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Dong Z, Wang D, Liu M, Ding J, Chen W, Shang Z, Yue C, Zhang Y (2021) Clinical value of lncRNA CCAT1 in serum extracellular vesicles as a potential biomarker for gastric cancer. Oncol Lett 21: 447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie JY, Wei JX, Lv LH, Han QF, Yang WB, Li GL, Wang PX, Wu SB, Duan JX, Zhuo WF et al (2020a) Angiopoietin‐2 induces angiogenesis via exosomes in human hepatocellular carcinoma. Cell Commun Signal 18: 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y et al (2020b) Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR‐582‐3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer 19: 112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Dang W, Zhang S, Yue W, Yang L, Zhai X, Yan Q, Lu J (2019) The role of exosomal noncoding RNAs in cancer. Mol Cancer 18: 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Gao Y, Ho C, Li L, Jin C, Wang X, Zou C, Mao Y, Wang X, Li Q et al (2021) Exosome‐delivered CD44v6/C1QBP complex drives pancreatic cancer liver metastasis by promoting fibrotic liver microenvironment. Gut 71: 568–579 [DOI] [PubMed] [Google Scholar]
- Xin H, Li Y, Cui Y, Yang JJ, Zhang ZG, Chopp M (2013) Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J Cereb Blood Flow Metab 33: 1711–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Yuan YW, Liu C, Zhou LQ, Liu L, Zhou Q, Li SH (2021) Preparation of internalizing RGD‐modified recombinant methioninase exosome active targeting vector and antitumor effect evaluation. Dig Dis Sci 66: 1045–1053 [DOI] [PubMed] [Google Scholar]
- Xu H, Dong X, Chen Y, Wang X (2018a) Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med 56: 479–484 [DOI] [PubMed] [Google Scholar]
- Xu Z, Liu X, Wang H, Li J, Dai L, Li J, Dong C (2018b) Lung adenocarcinoma cell‐derived exosomal miR‐21 facilitates osteoclastogenesis. Gene 666: 116–122 [DOI] [PubMed] [Google Scholar]
- Xu H, Zhao G, Zhang Y, Jiang H, Wang W, Zhao D, Hong J, Yu H, Qi L (2019a) Mesenchymal stem cell‐derived exosomal microRNA‐133b suppresses glioma progression via Wnt/beta‐catenin signaling pathway by targeting EZH2. Stem Cell Res Ther 10: 381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Li J, Chen L, Guo L, Ye M, Wu Y, Ji Q (2019b) Plasma miR‐32 levels in non‐small cell lung cancer patients receiving platinum‐based chemotherapy can predict the effectiveness and prognosis of chemotherapy. Medicine (Baltimore) 98: e17335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhou J, Tang J, Min X, Yi T, Zhao J, Ren Y (2020) Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal 34: e23323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Zheng L, Kang L, Xu H, Gao L (2021) microRNA‐let‐7e in serum‐derived exosomes inhibits the metastasis of non‐small‐cell lung cancer in a SUV39H2/LSD1/CDH1‐dependent manner. Cancer Gene Ther 28: 250–264 [DOI] [PubMed] [Google Scholar]
- Xue X, Wang X, Zhao Y, Hu R, Qin L (2018) Exosomal miR‐93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys Res Commun 502: 515–521 [DOI] [PubMed] [Google Scholar]
- Yagi T, Iinuma H, Hayama T, Matsuda K, Nozawa K, Tsukamoto M, Shimada R, Akahane T, Tsuchiya T, Ozawa T et al (2019) Plasma exosomal microRNA‐125b as a monitoring biomarker of resistance to mFOLFOX6‐based chemotherapy in advanced and recurrent colorectal cancer patients. Mol Clin Oncol 11: 416–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Harada Y, Suzuki T, Fukushige T, Yamakuchi M, Kanekura T, Dohmae N, Hori K, Maruyama I (2019) Application of high‐mannose‐type glycan‐specific lectin from Oscillatoria agardhii for affinity isolation of tumor‐derived extracellular vesicles. Anal Biochem 580: 21–29 [DOI] [PubMed] [Google Scholar]
- Yan C, Theodorescu D (2018) RAL GTPases: biology and potential as therapeutic targets in cancer. Pharmacol Rev 70: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Rolfe BE, Zhang B, Mohammed YH, Gu W, Xu ZP (2014a) Polarized immune responses modulated by layered double hydroxides nanoparticle conjugated with CpG. Biomaterials 35: 9508–9516 [DOI] [PubMed] [Google Scholar]
- Yan T, Mizutani A, Chen L, Takaki M, Hiramoto Y, Matsuda S, Shigehiro T, Kasai T, Kudoh T, Murakami H et al (2014b) Characterization of cancer stem‐like cells derived from mouse induced pluripotent stem cells transformed by tumor‐derived extracellular vesicles. J Cancer 5: 572–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Dang G, Zhang X, Jin C, Qin L, Wang Y, Shi M, Huang H, Duan Q (2017) Downregulation of circulating exosomal miR‐638 predicts poor prognosis in colon cancer patients. Oncotarget 8: 72220–72226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Jiang Y, Liang C, Cheng M, Jin C, Duan Q, Xu D, Yang L, Zhang X, Ren B et al (2018a) Exosomal miR‐6803‐5p as potential diagnostic and prognostic marker in colorectal cancer. J Cell Biochem 119: 4113–4119 [DOI] [PubMed] [Google Scholar]
- Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J, Liu X, Chen CH, Fadare O, Pizzo DP et al (2018b) Cancer‐cell‐secreted exosomal miR‐105 promotes tumour growth through the MYC‐dependent metabolic reprogramming of stromal cells. Nat Cell Biol 20: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Wang P, Fang W, Liang C (2020) Cancer‐associated fibroblasts‐derived exosomes‐mediated transfer of LINC00355 regulates bladder cancer cell proliferation and invasion. Cell Biochem Funct 38: 257–265 [DOI] [PubMed] [Google Scholar]
- Yang C, Sun W, Cui W, Li X, Yao J, Jia X, Li C, Wu H, Hu Z, Zou X (2015a) Procoagulant role of neutrophil extracellular traps in patients with gastric cancer. Int J Clin Exp Pathol 8: 14075–14086 [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Bucan V, Baehre H, von der Ohe J, Otte A, Hass R (2015b) Acquisition of new tumor cell properties by MSC‐derived exosomes. Int J Oncol 47: 244–252 [DOI] [PubMed] [Google Scholar]
- Yang JS, Lee JC, Byeon SK, Rha KH, Moon MH (2017a) Size dependent lipidomic analysis of urinary exosomes from patients with prostate cancer by flow field‐flow fractionation and nanoflow liquid chromatography‐tandem mass spectrometry. Anal Chem 89: 2488–2496 [DOI] [PubMed] [Google Scholar]
- Yang S, Che SP, Kurywchak P, Tavormina JL, Gansmo LB, Correa de Sampaio P, Tachezy M, Bockhorn M, Gebauer F, Haltom AR et al (2017b) Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol Ther 18: 158–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Wang DD, Li J, Xu HZ, Shen HY, Chen X, Zhou SY, Zhong SL, Zhao JH, Tang JH (2017c) Predictive role of GSTP1‐containing exosomes in chemotherapy‐resistant breast cancer. Gene 623: 5–14 [DOI] [PubMed] [Google Scholar]
- Yang H, Zhang H, Ge S, Ning T, Bai M, Li J, Li S, Sun W, Deng T, Zhang L et al (2018a) Exosome‐derived miR‐130a activates angiogenesis in gastric cancer by targeting C‐MYB in vascular endothelial cells. Mol Ther 26: 2466–2475 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yang YN, Zhang R, Du JW, Yuan HH, Li YJ, Wei XL, Du XX, Jiang SL, Han Y (2018b) Predictive role of UCA1‐containing exosomes in cetuximab‐resistant colorectal cancer. Cancer Cell Int 18: 164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Kim HS, Park SJ, Lee EJ, Kim SI, Song G, Lim W (2019a) Inhibition of miR‐214‐3p aids in preventing epithelial ovarian cancer malignancy by increasing the expression of LHX6. Cancers (Basel) 11: 1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Peng X, Li Y, Zhang X, Ma Y, Wu C, Fan Q, Wei S, Li H, Liu J (2019b) Long non‐coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol Cancer 18: 78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Feng X, Liu H, Tong R, Wu J, Li C, Yu H, Chen Y, Cheng Q, Chen J et al (2020a) High‐metastatic cancer cells derived exosomal miR92a‐3p promotes epithelial‐mesenchymal transition and metastasis of low‐metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene 39: 6529–6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Yan Y, Yang Y, Hong X, Wang M, Yang Z, Liu B, Ye L (2020b) MiR‐210 in exosomes derived from CAFs promotes non‐small cell lung cancer migration and invasion through PTEN/PI3K/AKT pathway. Cell Signal 73: 109675 [DOI] [PubMed] [Google Scholar]
- Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, Ning T, Bai M, Li H, Zhu K et al (2020c) Hypoxia induced exosomal circRNA promotes metastasis of colorectal cancer via targeting GEF‐H1/RhoA axis. Theranostics 10: 8211–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Teng F, Chang L, Wang J, Liu DL, Cui YS, Li GH (2021a) Tumor‐derived exosomal circRNA_102481 contributes to EGFR‐TKIs resistance via the miR‐30a‐5p/ROR1 axis in non‐small cell lung cancer. Aging (Albany NY) 13: 13264–13286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wu H, Zhu Y, Chen X, Chen Y (2021b) Plasma exosomal caveolin‐1 predicts poor prognosis in ovarian cancer. J Cancer 12: 5005–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Chen M, Gu J, Niu K, Zhao X, Zheng L, Xu Z, Yu Y, Li F, Meng L et al (2021c) Novel biomarkers of dynamic blood PD‐L1 expression for immune checkpoint inhibitors in advanced non‐small‐cell lung cancer patients. Front Immunol 12: 665133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Sun L, Li CF, Wang YH, Yao J, Li H, Yan M, Chang WC, Hsu JM, Cha JH et al (2021d) Galectin‐9 interacts with PD‐1 and TIM‐3 to regulate T cell death and is a target for cancer immunotherapy. Nat Commun 12: 832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MY, Zhang WH, Ma WT, Liu QH, Xing LH, Zhao GF (2019) microRNA‐328 in exosomes derived from M2 macrophages exerts a promotive effect on the progression of pulmonary fibrosis via FAM13A in a rat model. Exp Mol Med 51: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Yin Y, Jin G, Li D, Li M, Hu Y, Feng Y, Liu Y, Bian Z, Wang X et al (2020) Exosome‐mediated delivery of miR‐204‐5p inhibits tumor growth and chemoresistance. Cancer Med 9: 5989–5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Li SL, Ma YY, Diao YJ, Yang L, Su MQ, Li Z, Ji Y, Wang J, Lei L et al (2017) Exosomal miR‐141‐3p regulates osteoblast activity to promote the osteoblastic metastasis of prostate cancer. Oncotarget 8: 94834–94849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Ma T, Huang B, Lin L, Zhou Y, Yan J, Zou Y, Chen S (2019) Macrophage‐derived exosomal microRNA‐501‐3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3‐mediated TGF‐beta signaling pathway. J Exp Clin Cancer Res 38: 310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Zeng W, Wu B, Wang L, Wang Z, Tian H, Wang L, Jiang Y, Clay R, Wei X et al (2020) PPARalpha inhibition overcomes tumor‐derived exosomal lipid‐induced dendritic cell dysfunction. Cell Rep 33: 108278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G, Kong W, Zheng S, Shan Y, Zhang J, Ying R, Wu H (2021) Exosomal miR‐130a‐3p promotes the progression of differentiated thyroid cancer by targeting insulin‐like growth factor 1. Oncol Lett 21: 283 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez‐Carretero A, Fu W et al (2017) Adipose tissue macrophage‐derived exosomal mirnas can modulate in vivo and in vitro insulin sensitivity. Cell 171: 372–384 [DOI] [PubMed] [Google Scholar]
- Yogev O, Henderson S, Hayes MJ, Marelli SS, Ofir‐Birin Y, Regev‐Rudzki N, Herrero J, Enver T (2017) Herpesviruses shape tumour microenvironment through exosomal transfer of viral microRNAs. PLoS Pathog 13: e1006524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi A, Villar‐Prados A, Oliphint PA, Zhang J, Song X, De Hoff P, Morey R, Liu J, Roszik J, Clise‐Dwyer K et al (2019) Mechanisms of nuclear content loading to exosomes. Sci Adv 5: eaax8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Noda T, Okumura Y, Kobayashi S, Iwagami Y, Yamada D, Tomimaru Y, Akita H, Gotoh K, Takeda Y et al (2021) Serum exosomal miR‐638 is a prognostic marker of HCC via downregulation of VE‐cadherin and ZO‐1 of endothelial cells. Cancer Sci 112: 1275–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama T, Gorry M, Sobo‐Vujanovic A, Lin Y, Vujanovic L, Gaither‐Davis A, Moss ML, Miller MA, Griffith LG, Lauffenburger DA et al (2018) ADAM10 sheddase activity is a potential lung‐cancer biomarker. J Cancer 9: 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Tsuda M, Matsumoto R, Semba S, Wang L, Sugino H, Tanino M, Kondo T, Tanabe K, Tanaka S (2019) Exosomes containing ErbB2/CRK induce vascular growth in premetastatic niches and promote metastasis of bladder cancer. Cancer Sci 110: 2119–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Iinuma H, Umemoto Y, Yanagisawa T, Matsumoto A, Jinno H (2018) Exosome‐encapsulated microRNA‐223‐3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol Lett 15: 9584–9592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Sawada K, Nakamura K, Kinose Y, Nakatsuka E, Kobayashi M, Miyamoto M, Ishida K, Matsumoto Y, Kodama M et al (2018) Exosomal miR‐99a‐5p is elevated in sera of ovarian cancer patients and promotes cancer cell invasion by increasing fibronectin and vitronectin expression in neighboring peritoneal mesothelial cells. BMC Cancer 18: 1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa N, Sugimoto K, Tameda M, Inagaki Y, Ikejiri M, Inoue H, Usui M, Ito M, Takei Y (2020) miR‐3940‐5p/miR‐8069 ratio in urine exosomes is a novel diagnostic biomarker for pancreatic ductal adenocarcinoma. Oncol Lett 19: 2677–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Gai C, Li Z, Ding D, Zheng J, Zhang W, Lv S, Li W (2019a) Targeted exosome‐encapsulated erastin induced ferroptosis in triple negative breast cancer cells. Cancer Sci 110: 3173–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Zhao Z, Xu X, Li M, Li P (2019b) Characterization of three different types of extracellular vesicles and their impact on bacterial growth. Food Chem 272: 372–378 [DOI] [PubMed] [Google Scholar]
- Yu S, Li Y, Liao Z, Wang Z, Wang Z, Li Y, Qian L, Zhao J, Zong H, Kang B et al (2020) Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut 69: 540–550 [DOI] [PubMed] [Google Scholar]
- Yuan Z, Yang Z, Li W, Wu A, Su Z, Jiang B (2020) Exosome‐mediated transfer of long noncoding RNA HOTAIR regulates temozolomide resistance by miR‐519a‐3p/RRM1 axis in glioblastoma. Cancer Biother Radiopharm 10.1089/cbr.2019.3499 [DOI] [PubMed] [Google Scholar]
- Yuwen DL, Sheng BB, Liu J, Wenyu W, Shu YQ (2017) MiR‐146a‐5p level in serum exosomes predicts therapeutic effect of cisplatin in non‐small cell lung cancer. Eur Rev Med Pharmacol Sci 21: 2650–2658 [PubMed] [Google Scholar]
- Yuwen D, Ma Y, Wang D, Gao J, Li X, Xue W, Fan M, Xu Q, Shen Y, Shu Y (2019) Prognostic role of circulating exosomal miR‐425‐3p for the response of NSCLC to platinum‐based chemotherapy. Cancer Epidemiol Biomarkers Prev 28: 163–173 [DOI] [PubMed] [Google Scholar]
- Zare N, Kefayat A, Javanmard SH (2020) Evaluation of radiation and ammonium lactate effects on hyaluronic acid expression as a pro‐cancerous factor in supernatant and exosome isolated from supernatant of primary mouse fibroblast cell culture. Int J Prev Med 11: 125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebrowska A, Skowronek A, Wojakowska A, Widlak P, Pietrowska M (2019) Metabolome of exosomes: focus on vesicles released by cancer cells and present in human body fluids. Int J Mol Sci 20: 3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng AL, Yan W, Liu YW, Wang Z, Hu Q, Nie E, Zhou X, Li R, Wang XF, Jiang T et al (2017) Tumour exosomes from cells harbouring PTPRZ1‐MET fusion contribute to a malignant phenotype and temozolomide chemoresistance in glioblastoma. Oncogene 36: 5369–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B, Chen T, Xie MY, Luo JY, He JJ, Xi QY, Sun JJ, Zhang YL (2019) Exploration of long noncoding RNA in bovine milk exosomes and their stability during digestion in vitro . J Dairy Sci 102: 6726–6737 [DOI] [PubMed] [Google Scholar]
- Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F et al (2018) Cancer‐derived exosomal miR‐25‐3p promotes pre‐metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun 9: 5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wu X, Zhang X, Sun Y, Yan Y, Shi H, Zhu Y, Wu L, Pan Z, Zhu W et al (2015a) Human umbilical cord mesenchymal stem cell exosomes enhance angiogenesis through the Wnt4/beta‐catenin pathway. Stem Cells Transl Med 4: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, Li P, Li M, Wang X, Zhang C et al (2015b) Microenvironment‐induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu Z, Ding H, Zhou Y, Doan HA, Sin KWT, Zhu ZJ, Flores R, Wen Y, Gong X et al (2017a) Tumor induces muscle wasting in mice through releasing extracellular Hsp70 and Hsp90. Nat Commun 8: 589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Deng T, Liu R, Bai M, Zhou L, Wang X, Li S, Wang X, Yang H, Li J et al (2017b) Exosome‐delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun 8: 15016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lu S, Zhou Y, Meng K, Chen Z, Cui Y, Shi Y, Wang T, He QY (2017c) Motile hepatocellular carcinoma cells preferentially secret sugar metabolism regulatory proteins via exosomes. Proteomics 10.1002/pmic.201700103 [DOI] [PubMed] [Google Scholar]
- Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y, Ming H (2017d) Serum long non coding RNA MALAT‐1 protected by exosomes is up‐regulated and promotes cell proliferation and migration in non‐small cell lung cancer. Biochem Biophys Res Commun 490: 406–414 [DOI] [PubMed] [Google Scholar]
- Zhang C, Xiao X, Chen M, Aldharee H, Chen Y, Long W (2018a) Liver kinase B1 restoration promotes exosome secretion and motility of lung cancer cells. Oncol Rep 39: 376–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L et al (2018b) Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field‐flow fractionation. Nat Cell Biol 20: 332–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Jeppesen DK, Higginbotham JN, Demory Beckler M, Poulin EJ, Walsh AJ, Skala MC, McKinley ET, Manning HC, Hight MR et al (2018c) Mutant KRAS exosomes alter the metabolic state of recipient colonic epithelial cells. Cell Mol Gastroenterol Hepatol 5: 627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ni M, Su Y, Wang H, Zhu S, Zhao A, Li G (2018d) MicroRNAs in serum exosomes as potential biomarkers in clear‐cell renal cell carcinoma. Eur Urol Focus 4: 412–419 [DOI] [PubMed] [Google Scholar]
- Zhang X, Deeke SA, Ning Z, Starr AE, Butcher J, Li J, Mayne J, Cheng K, Liao B, Li L et al (2018e) Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat Commun 9: 2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xing T, Chen Y, Xiao J (2018f) Exosome‐mediated miR‐200b promotes colorectal cancer proliferation upon TGF‐beta1 exposure. Biomed Pharmacother 106: 1135–1143 [DOI] [PubMed] [Google Scholar]
- Zhang H, Lyden D (2019) Asymmetric‐flow field‐flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat Protoc 14: 1027–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F et al (2019a) Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination‐related USP7. Oncogene 38: 2844–2859 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T, Yang H, Sun W, Wang X, Zhu K et al (2019b) Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR‐133/PRDM16 pathway. Int J Cancer 144: 2501–2515 [DOI] [PubMed] [Google Scholar]
- Zhang K, Yue Y, Wu S, Liu W, Shi J, Zhang Z (2019c) Rapid capture and nondestructive release of extracellular vesicles using aptamer‐based magnetic isolation. ACS Sens 4: 1245–1251 [DOI] [PubMed] [Google Scholar]
- Zhang P, Zhou X, Zeng Y (2019d) Multiplexed immunophenotyping of circulating exosomes on nano‐engineered ExoProfile chip towards early diagnosis of cancer. Chem Sci 10: 5495–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Higginbotham JN, Jeppesen DK, Yang YP, Li W, McKinley ET, Graves‐Deal R, Ping J, Britain CM, Dorsett KA et al (2019e) Transfer of functional cargo in exomeres. Cell Rep 27: 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J, Yan K, Duan W, Zhao Y, Wang L et al (2019f) Evaluation of serum exosomal LncRNA‐based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med 23: 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L, Li G, Tang J, Xiang J (2019g) Hypoxic BMSC‐derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3‐induced EMT. Mol Cancer 18: 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Li D, Shen L, Hu D, Tang B, Guo W, Wang Z, Zhang Z, Wei G, He D (2020a) Exosomes derived from Piwil2induced cancer stem cells transform fibroblasts into cancerassociated fibroblasts. Oncol Rep 43: 1125–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, Dai X, Zhou H, Zhu J, Zhang H et al (2020b) Circular RNA circSATB2 promotes progression of non‐small cell lung cancer cells. Mol Cancer 19: 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wang C, Ma B, Xu M, Xu S, Liu J, Tian Y, Fu Y, Luo Y (2020c) Mutant p53 drives cancer metastasis via RCP‐mediated Hsp90alpha secretion. Cell Rep 32: 107879 [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhang X, Huang S, Chen J, Ding P, Wang Q, Li L, Lv X, Li L, Zhang P et al (2020d) FOXM1D potentiates PKM2‐mediated tumor glycolysis and angiogenesis. Mol Oncol 15: 1466–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu L, Tang M, Li H, Guo X, Yang X (2020e) The effects of umbilical cord‐derived macrophage exosomes loaded with cisplatin on the growth and drug resistance of ovarian cancer cells. Drug Dev Ind Pharm 46: 1150–1162 [DOI] [PubMed] [Google Scholar]
- Zhang XW, Zhou JC, Peng D, Hua F, Li K, Yu JJ, Lv XX, Cui B, Liu SS, Yu JM et al (2020f) Disrupting the TRIB3‐SQSTM1 interaction reduces liver fibrosis by restoring autophagy and suppressing exosome‐mediated HSC activation. Autophagy 16: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Han T, Feng D, Li J, Wu M, Peng X, Wang B, Zhan X, Fu P (2020g) Screening of non‐invasive miRNA biomarker candidates for metastasis of gastric cancer by small RNA sequencing of plasma exosomes. Carcinogenesis 41: 582–590 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao J, Ding M, Su Y, Cui D, Jiang C, Zhao S, Jia G, Wang X, Ruan Y et al (2020h) Loss of exosomal miR‐146a‐5p from cancer‐associated fibroblasts after androgen deprivation therapy contributes to prostate cancer metastasis. J Exp Clin Cancer Res 39: 282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Jeppesen DK, Higginbotham JN, Graves‐Deal R, Trinh VQ, Ramirez MA, Sohn Y, Neininger AC, Taneja N, McKinley ET et al (2021) Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat Cell Biol 23: 1240–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, Marini JC, Tudawe T, Seviour EG, San Lucas FA et al (2016a) Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 5: e10250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Yang Y, Zeng Y, He M (2016b) A microfluidic ExoSearch chip for multiplexed exosome detection towards blood‐based ovarian cancer diagnosis. Lab Chip 16: 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Li H, Guo Q, Zhou A, Wang X, Li P, Zhang S (2020a) Exosomal Sonic Hedgehog derived from cancer‐associated fibroblasts promotes proliferation and migration of esophageal squamous cell carcinoma. Cancer Med 9: 2500–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Gu C, Gan Y, Shao L, Chen H, Zhu H (2020b) Exosome‐mediated siRNA delivery to suppress postoperative breast cancer metastasis. J Control Release 318: 1–15 [DOI] [PubMed] [Google Scholar]
- Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, Zhang Z, Cai S, Xu Y, Li X et al (2020c) Tumor‐derived exosomal miR‐934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol 13: 156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li M, Dai X, Yang Y, Peng Y, Xu C, Dai N, Wang D (2020d) Downregulation of exosomal miR1273a increases cisplatin resistance of nonsmall cell lung cancer by upregulating the expression of syndecan binding protein. Oncol Rep 44: 2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Gan Y, Xu G, Yin G, Liu D (2020e) MSCs‐Derived exosomes attenuate acute brain injury and inhibit microglial inflammation by reversing CysLT2R‐ERK1/2 mediated microglia M1 polarization. Neurochem Res 45: 1180–1190 [DOI] [PubMed] [Google Scholar]
- Zhen Y, Stenmark H (2015) Cellular functions of Rab GTPases at a glance. J Cell Sci 128: 3171–3176 [DOI] [PubMed] [Google Scholar]
- Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, Ma Y, Shen L (2017) Exosomal transfer of tumor‐associated macrophage‐derived miR‐21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res 36: 53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Hernandez JM, Doussot A, Bojmar L, Zambirinis CP, Costa‐Silva B, van Beek E, Mark MT, Molina H, Askan G et al (2018a) Extracellular matrix proteins and carcinoembryonic antigen‐related cell adhesion molecules characterize pancreatic duct fluid exosomes in patients with pancreatic cancer. HPB (Oxford) 20: 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Luo Q, Wang W, Li J, Wang T, Wang P, Chen L, Zhang P, Chen H, Liu Y et al (2018b) Tumor‐associated macrophages‐derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis 9: 434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, Lv Q, Qin C, Chu H, Wang M et al (2018c) Exosome‐transmitted long non‐coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer 17: 143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Zhang H, Gao H, Sun J, Li J, Zhang X, Gao L, Ma P, Li S (2020a) Plasma exosomal long noncoding RNA lnc‐SLC2A12‐10:1 as a novel diagnostic biomarker for gastric cancer. Onco Targets Ther 13: 4009–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Xu K, Zhou B, Chen T, Huang Y, Li Q, Wen F, Ge W, Wang J, Yu S et al (2020b) A circulating extracellular vesicles‐based novel screening tool for colorectal cancer revealed by shotgun and data‐independent acquisition mass spectrometry. J Extracell Vesicles 9: 1750202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GD, Xu ZY, Hu C, Lv H, Xie HX, Huang T, Zhang YQ, Chen GP, Fu YF, Cheng XD (2021) Exosomal miR‐590‐5p in serum as a biomarker for the diagnosis and prognosis of gastric cancer. Front Mol Biosci 8: 636566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Wang K, Li J, Xiao S, Wei W, Liu J (2020) Determination of serum exosomal H19 as a noninvasive biomarker for breast cancer diagnosis. Onco Targets Ther 13: 2563–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR et al (2014) Cancer‐secreted miR‐105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25: 501–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, Shi X (2018) Hepatocellular carcinoma‐derived exosomal miRNA‐21 contributes to tumor progression by converting hepatocyte stellate cells to cancer‐associated fibroblasts. J Exp Clin Cancer Res 37: 324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CF, Ma J, Huang L, Yi HY, Zhang YM, Wu XG, Yan RM, Liang L, Zhong M, Yu YH et al (2019a) Cervical squamous cell carcinoma‐secreted exosomal miR‐221‐3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1. Oncogene 38: 1256–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Paz HA, Sadri M, Cui J, Kachman SD, Fernando SC, Zempleni J (2019b) Dietary bovine milk exosomes elicit changes in bacterial communities in C57BL/6 mice. Am J Physiol Gastrointest Liver Physiol 317: G618–G624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Shen W, Zou H, Lv Q, Shao P (2020a) Circulating exosomal long non‐coding RNA H19 as a potential novel diagnostic and prognostic biomarker for gastric cancer. J Int Med Res 48: 300060520934297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Xie F, Wang L, Zhang L, Zhang S, Fang M, Zhou F (2020b) The function and clinical application of extracellular vesicles in innate immune regulation. Cell Mol Immunol 17: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Wei W, Ma J, Yang Y, Liang L, Zhang Y, Wang Z, Chen X, Huang L, Wang W et al (2021a) Cancer‐secreted exosomal miR‐1468‐5p promotes tumor immune escape via the immunosuppressive reprogramming of lymphatic vessels. Mol Ther 29: 1512–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Li J, Tang Y, Yang M (2021b) Exosomal LncRNA LINC00659 transferred from cancer‐associated fibroblasts promotes colorectal cancer cell progression via miR‐342‐3p/ANXA2 axis. J Transl Med 19: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, Barsyte‐Lovejoy D, Al‐awar R, Katona BW, Shilatifard A et al (2015) Gain‐of‐function p53 mutants co‐opt chromatin pathways to drive cancer growth. Nature 525: 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Kalimuthu S, Gangadaran P, Oh JM, Lee HW, Baek SH, Jeong SY, Lee SW, Lee J, Ahn BC (2017) Exosomes derived from natural killer cells exert therapeutic effect in melanoma. Theranostics 7: 2732–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lu K, Zhang N, Zhao Y, Ma Q, Shen J, Lin Y, Xiang P, Tang Y, Hu X et al (2018a) Myocardial reparative functions of exosomes from mesenchymal stem cells are enhanced by hypoxia treatment of the cells via transferring microRNA‐210 in an nSMase2‐dependent way. Artif Cells Nanomed Biotechnol 46: 1659–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Gangadaran P, Kalimuthu S, Oh JM, Baek SH, Jeong SY, Lee SW, Lee J, Ahn BC (2018b) Novel alternatives to extracellular vesicle‐based immunotherapy – exosome mimetics derived from natural killer cells. Artif Cells Nanomed Biotechnol 46: S166–S179 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Wang H, Pang Y, Hu H, Zhang H, Wang W (2020) Exosomal long non‐coding RNA UCA1 functions as growth inhibitor in esophageal cancer. Aging (Albany NY) 12: 20523–20539 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, Ju S, Mu J, Zhang L, Steinman L et al (2011) Treatment of brain inflammatory diseases by delivering exosome encapsulated anti‐inflammatory drugs from the nasal region to the brain. Mol Ther 19: 1769–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang M, Chen X, Du D, Shi J, Deng M, Long Q, Yin X, Wang Y, Rao L (2020) SPION decorated exosome delivery of TNF‐alpha to cancer cell membranes through magnetism. Nanoscale 12: 173–188 [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi‐Castagnoli P, Raposo G, Amigorena S (1998) Eradication of established murine tumors using a novel cell‐free vaccine: dendritic cell derived exosomes. Nat Med 4: 594–600 [DOI] [PubMed] [Google Scholar]
- Zomer A, Maynard C, Verweij FJ, Kamermans A, Schafer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SIJ et al (2015) In vivo imaging reveals extracellular vesicle‐mediated phenocopying of metastatic behavior. Cell 161: 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]


