Abstract
Background
Evidence describing the impact of diabetes mellitus (DM) on the recurrence and mutation rate of Mycobacterium tuberculosis (Mtb) is limited.
Methods
This study was nested in 3 cohort studies of tuberculosis (TB) patients with and without DM in India. Paired Mtb isolates recovered at baseline and treatment failure/recurrence underwent whole genome sequencing. We compared acquisition of single-nucleotide polymorphisms (SNPs), TB drug resistance mutations, and type of recurrence (endogenous reactivation [<8 SNPs] or exogenous reinfection [≥8 SNPs]) by DM status.
Results
Of 1633 enrolled in the 3 parent cohorts, 236 (14.5%) had microbiologically confirmed TB treatment failure/recurrence; 76 Mtb isolate pairs were available for sequencing (22 in TB-DM and 54 in TB-only). The SNP acquisition rate was overall was 0.43 (95% confidence interval [CI], .25–.64) per 1 person-year (PY); 0.77 (95% CI, .40–1.35) per 1 PY, and 0.44 (95% CI, .19–.86) per 1 PY at treatment failure and recurrence, respectively. Significant difference in SNP rates by DM status was seen at recurrence (0.21 [95% CI, .04–.61]) per 1 PY for TB-only vs 1.28 (95% CI, .41–2.98) per 1 PY for TB-DM; P = .02). No significant difference in SNP rates by DM status was observed at treatment failure. Acquired TB drug resistance was seen in 4 of 18 (22%) in TB-DM vs 4 of 45 (9%) in TB-only (P = .21). Thirteen (17%) participants had exogenous reinfection; the reinfection rate at recurrence was 25% (3/12) for TB-DM vs 17% (4/24) in TB-only (P = .66).
Conclusions
Considerable intrahost Mtb mutation rates were present at recurrence among patients with DM in India. One-fourth of patients with DM had exogenous reinfection at recurrence.
Keywords: diabetes mellitus, drug resistance and recurrence, India, tuberculosis, whole genome sequencing
Nested in 3 tuberculosis (TB) cohorts with and without diabetes mellitus (DM) in India, whole genome sequencing revealed considerable intrahost Mycobacterium tuberculosismutation rates among TB-DM patients at recurrence in India. Exogenous reinfection was identified in one-fourth of patients with DM.
Tuberculosis (TB) is one of the most lethal infectious disease globally. Despite the monomorphic population structure of the Mycobacterium tuberculosis (Mtb) pathogen [1, 2], TB disease presents with considerable clinical heterogeneity (eg, disease severity, treatment outcomes). Variable and adverse TB treatment outcomes are likely the result of Mtb microevolution—the acquisition of mutations over time [3–5]. Previous studies estimate that the Mtb genome accumulates an average of 0.3–0.6 single-nucleotide polymorphisms (SNPs) per year [6–9], which are in part due to stochastic mutations arising during bacterial replication. Oxidative DNA damage is a key driver of Mtb mutations within the host, and different environments and immunologic stresses, such as human immunodeficiency virus (HIV) and diabetes mellitus (DM), may contribute to Mtb replication [2, 10–12]. DM increases the risk of TB and may adversely impact TB treatment outcomes [13–15], but evidence describing the impact of DM on Mtb microevolution remains scarce.
DM is a known immune-altered state that is increasingly recognized as a comorbidity in the setting of TB and could have a significant impact on Mtb microevolution, including the acquisition of mutations conferring anti-TB drug resistance (similar to HIV), in high-burden settings [16–18]. To date, limited data suggest that TB recurrence in the context of DM may be due to TB reactivation [19], but this evidence is limited by several factors, including small sample size, that the recurrence rates were not compared to TB patients without DM, and that the results remain unconfirmed by other studies in high-TB-prevalence settings.
To address this knowledge gap, we conducted a study nested within 3 large cohort studies among patients with pulmonary TB with and without DM in India. Using whole genome sequencing (WGS) on Mtb strains among participants who had treatment failure or recurrence, we aimed to assess the impact of DM on the rate of mutation acquisition, including TB drug resistance, and type of recurrence-endogenous Mtb reactivation or exogenous Mtb reinfection, in India, a TB-DM epicenter [20, 21] where high TB prevalence persists due to high TB transmission rates.
MATERIALS AND METHODS
Study Design and Study Sites
The present study was nested within 3 large cohort studies among patients with new pulmonary TB with known DM status in India: Cohort for TB Research by the Indo-US Medical Partnership (C-TRIUMPh); Effects of Diabetes on TB Severity (EDOTS); and Impact of Diabetes Mellitus on TB Treatment Outcomes (TB-DM study) [20–23]. C-TRIUMPh and TB-DM recruited at Byramjee-Jeejeebhoy Government Medical College (BJGMC) in Pune and the National Institute for Research in Tuberculosis (NIRT) in Chennai; and EDOTS recruited at Professor M. Vishwanathan Diabetes Research Center (MVDRC) in Chennai. NIRT primarily enrolled participants from semirural and rural populations, the BJGMC site recruited participants from semiurban and urban populations, and MVDRC recruited from 10 urban TB units. At entry, the parent studies obtained consent from all participants for the parent studies as well as to store Mtb isolates and conduct future procedures. Separate ethics committee approval was obtained from all 3 institutions and the Johns Hopkins University Institutional Review Board for this study.
Study Procedures
The study designs and procedures of the parent studies have been described elsewhere [21–23]. In brief, all 3 parent studies collected sputum at baseline for acid-fast bacilli (AFB) and mycobacteria growth indicator tube (MGIT) culture. All participants received standard thrice-weekly anti-TB treatment via directly observed therapy. The regimen consisted of 450 mg (600 mg for those ≥60 kg body weight) rifampin, 600 mg isoniazid, 1200 mg ethambutol, and 1500 mg pyrazinamide during the intensive phase, followed by rifampin and isoniazid at the same doses during the continuation phase. Sputum AFB and MGIT culture were repeated at month 5/6 and if recurrent TB was suspected after completion of TB treatment. For the purposes of this study, TB treatment failure was defined as the presence of symptoms consistent with TB and positive culture at month 5 or 6. Recurrence was defined as the presence of new symptoms consistent with TB and positive culture after completion of 6 months of TB treatment and cure. DM was defined as hemoglobin A1c (HbA1c) ≥6.5%, fasting blood glucose level of ≥126 mg/dL, random blood glucose >200 mg/dL, diabetes diagnosis by self-report, or current use of DM medication.
Whole Genome Sequencing
WGS was performed on Mtb strains isolated at baseline (screening or entry) and at treatment failure or recurrence. DNA was extracted from MGIT cultures using standardized protocols. The paired sputum samples obtained from subculturing in Lowenstein-Jensen media were subjected to DNA extraction using a standardized protocol. In brief, Ultra-Deep Microbiome Prep kit (Molzym, Bremen, Germany) version 2.1 was used for removal of other DNA, lysis of pathogen, DNA purification, and DNA elution. We collected extracted DNA in elution tubes and preserved it at –20°C for shipment and sequencing. Isolated genomic DNA of individual Mtb strains was sequenced using the Illumina MiSeq sequencer as per manufacturer instructions (Illumina, San Diego, California) with 2 × 150-bp pair-end reads and a minimum coverage of approximately 100-fold [24]. WGS was performed at Medgenome laboratories based in Bangalore, India. The WGS data analysis was performed using approaches similar to the methods described previously with some modifications [6, 7, 9]. In brief, the quality control of Illumina raw reads was examined by FastQC version 0.1.1.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and reads were trimmed to remove low-quality bases (average quality per base <20) using Trimmomatic version 0.39 [24]. The filtered reads were then aligned to the H37Rv Reference Genome (GenBank accession number NC_000962) with the BWA-mem algorithm (version 0.7.17) [25]. The resulting SAM files were then sorted, converted to BAM format, and processed for duplicate removal with Picard (http://broadinstitute.github.io/picard/) (version 2.24.0). The processed BAM files were then indexed with Samtools version 1.11 [26], followed by SNP and InDel calling using freebayes version 1.3.0 (github.com/freebayes). As an additional quality check, the coverage of the genome was evaluated using Samtools and all genomes had a coverage >98% with a depth of at least 10 using the H37Rv genome as the reference.
To remove low-quality base calls, we filtered any base calls using the thresholds similar to a previous study [27]: variant depth on the forward and reverse strands >1 (SAF >1 and SAR >1), variant depth at each side of the site >1 (RPR >1 & RPL >1), mapping quality (MOM) >30, Phred-scaled base quality score (QUAL) >100, high-quality read depth (DP) >10, and QUAL/alternate allele observation count (AO) >10. Fixed SNPs were classified as positions where >90% of reads were the alternative allele (ALT), while heterogeneous SNPs were classified as positions where >10% and <90% of reads were ALT [28]. For heterogeneous SNP calling, a minimal 10 reads mapping to the ALT is required (AO ≥10). For paired isolates, a previously suggested allele frequency change ≥70% (ΔAF ≥70%) was used as a cutoff to determine in-host evolution. The variation annotation was performed using snpEff version 4.3 [29]. Isolates with heterozygous SNPs at established lineage-specific sites were classified as mixed infections. The Mtb lineage was determined using a python script fast-lineage-caller version 1.0 developed from the Farhat laboratory (https://github.com/farhat-lab/fast-lineage-caller), which used the freebayes vcf output as the as the input file and returned the lineage/sublineage calls. The program included 5 SNP schemes for Mtb [30, 31-34].
SNPs located at the mobile genetic elements, PE, PPE, and PE-PGRS gene regions that could potentially cause incorrect read alignment, were excluded in the SNP and phylogenetic analysis. Absolute number of accumulated SNPs was calculated. Recombination free core SNP alignment was generated and a maximum likelihood phylogenetic tree was inferred from the resulting SNP alignment in RAxML version 8.2.4 using a general time-reversible model of nucleotide substitution and 4 discrete gamma categories of rate heterogeneity (GTRGAMMA option). High confident mutations conferring drug resistance to isoniazid, rifampin, ethambutol, pyrazinamide, streptomycin, levofloxacin, amikacin, and capreomycin were assessed using Mykrobe and TBProfiler using default settings [35, 36]. The Mykrobe and TBProfiler results were manually curated to check the consistency, and a combined resistance prediction was used.
Outcomes and Definitions
The outcomes were differences in SNPs and anti-TB drug resistance mutations among paired Mtb strains (isolated at baseline and treatment failure or recurrence) and type of recurrence by DM status. The type of recurrence was classified as endogenous reactivation (<8 new SNPs at treatment failure or recurrence) or exogenous reinfection (≥8 new SNPs at recurrence) [37]. Mtb lineage of isolated Mtb strains was also assigned.
Statistical Analysis
Participant characteristics and outcomes were summarized using descriptive statistics and compared by DM status using Fisher exact test. The rate of SNP acquisition per 1 person-year (PY), with 95% exact Poisson confidence intervals (CIs), was calculated as the number of new SNPs among paired Mtb strains divided by the time interval (days) between Mtb isolates. It was determined at treatment failure and recurrence and compared by DM status using Poisson regression. We excluded paired strains meeting definition of reinfection from SNP acquisition rate calculation.
RESULTS
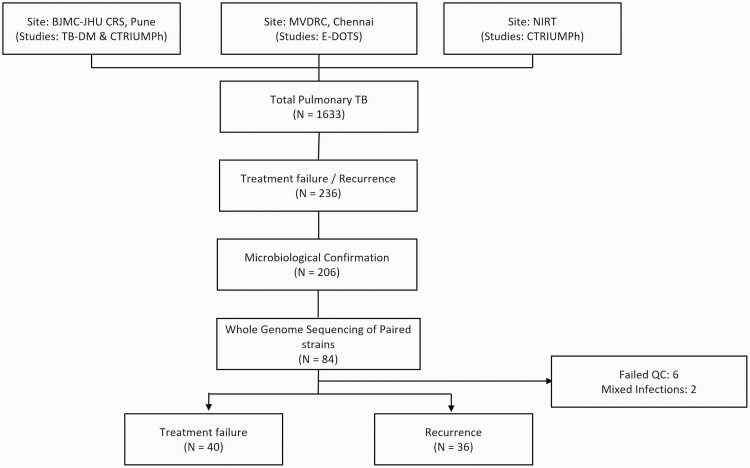
Among 1633 patients enrolled in the parent studies with new pulmonary TB, 236 (14.5%) had microbiologically confirmed TB treatment failure or recurrence (Figure 1). Of these, 84 of 236 (35.6%) had paired Mtb isolates available (on January 2017) at baseline and treatment failure/recurrence and underwent WGS. Paired Mtb isolates from 76 of 84 (91%) patients (40 with treatment failure and 36 with recurrence) were evaluable and are included in this analysis; those excluded (n = 8) had either the presence of mixed infection (n = 2) or had failed quality control (n = 6).
Figure 1.
Study flowchart. Abbreviations: BJGMC JHU CRS, Byramjee-Jeejeebhoy Government Medical College–Johns Hopkins University Clinical Research Site; C-TRIUMPh, Cohort for Tuberculosis Research by the Indo-US Medical Partnership; EDOTS, Effects of Diabetes on TB Severity; MVDRC, Professor M. Vishwanathan Diabetes Research Center; NIRT, National Institute for Research in Tuberculosis; QC, quality control; TB, tuberculosis; TB-DM, Impact of Diabetes Mellitus on TB Treatment Outcomes.
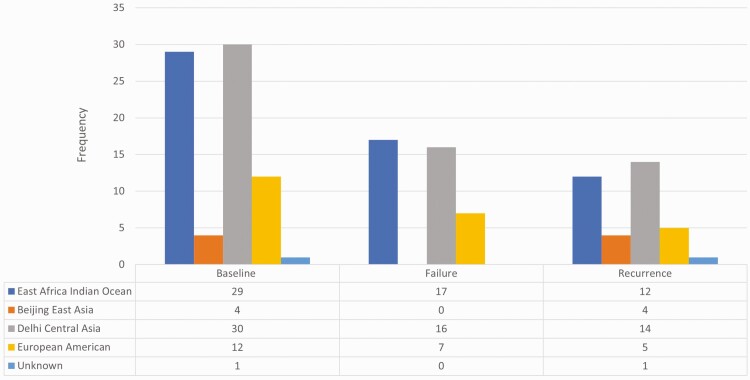
Baseline characteristics are shown in Table 1. TB-DM and TB-only comprised 29% (n = 22) and 71% (n = 54) of the study population, respectively. The median HbA1c for TB-DM was 9.0% (interquartile range [IQR], 7.0%–10.4%) vs 5.7% (IQR, 5.5%–6.0%) for TB-only (P < .001). As shown in Figure 2, baseline Mtb isolates had diverse lineages. The majority were Central Asia and East Africa Indian Ocean followed by European American and Beijing East Asia. Delhi Central Asia and East Africa Indian Ocean lineages predominated in Pune and Chennai, respectively.
Table 1.
Baseline Characteristics and Acquisition of Single-Nucleotide Polymorphisms Among Patients With Pulmonary Tuberculosis With Treatment Failure or Recurrence in India
| Characteristic | Overall (N = 76) | Treatment Failure (n = 40) | TB Recurrence (n = 36) |
|---|---|---|---|
| Sex | |||
| Female | 14 (18) | 10 (25) | 4 (11) |
| Male | 62 (82) | 30 (75) | 32 (89) |
| Age, y, median (IQR) | 38 (26–48) | 38 (26–48) | 38 (27–47) |
| BMI, kg/m2, median (IQR) | 16 (15–19) | 17 (15–19) | 16 (14–19) |
| HIV | 1 (1) | 1 (3) | 0 |
| Smoker | 29 (38) | 15 (38) | 14 (39) |
| Uses alcohol | 44 (58) | 21 (53) | 23 (64) |
| Hemoglobin, g/dL, median (IQR) | 12.0 (10.7–13.5) | 11.5 (10.4–13.3) | 12.5 (11.2–13.7) |
| HbA1c, %, median (IQR) | 5.9 (5.6–6.7) | 5.8 (5.6–6.3) | 6.0 (5.6–7.3) |
| Diabetes mellitus | |||
| Yes | 22 (29) | 10 (25) | 12 (33) |
| No | 54 (71) | 30 (75) | 24 (67) |
| SNP acquisition excluding exogenous reinfection | |||
| <1 SNP | 58 (92) | 32 (94) | 26 (90) |
| >2 SNP | 5 (8) | 2 (6) | 3 (10) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: BMI, body mass index; DM, diabetes mellitus; HbA1c, hemoglobin A1c; HIV, human immunodeficiency virus; IQR, interquartile range; SNP, single-nucleotide polymorphism; TB, tuberculosis.
Figure 2.
Lineage of Mycobacterium tuberculosis strains isolated at baseline and tuberculosis treatment failure or recurrence in India.
Mutation Acquisition
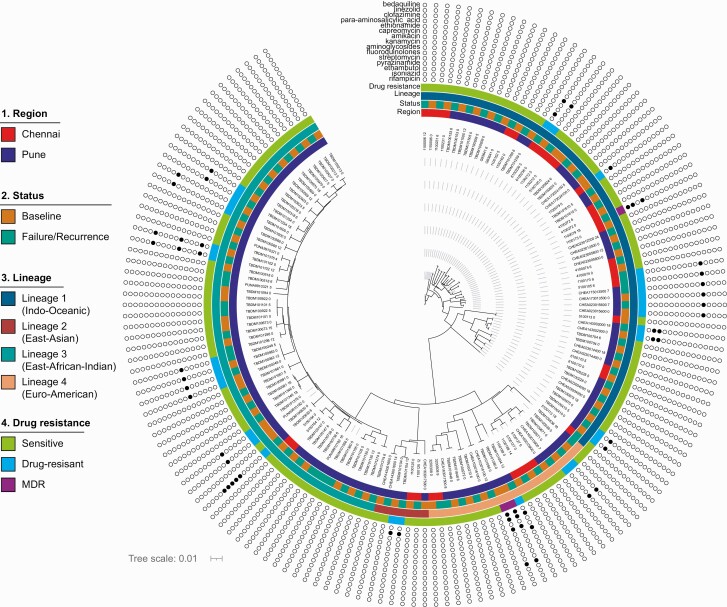
Phylogenetic reconstruction of all 76 paired Mtb strains shows genetically distinct isolates by participant origin (Pune or Chennai) (Figure 3). Overall, 57 (84%) pairs acquired at least 1 SNP at treatment failure or recurrence. After excluding 13 pairs with high SNP accumulation ranging from 21 to 1099 SNPs, the overall SNP acquisition rate was 0.43 (95% CI, .25–.64) per 1 PY; 0.77 (95% CI, .40–1.35) per 1 PY at treatment failure, and 0.44 (95% CI, .19–.86) per 1 PY at recurrence. The rate of SNP acquisition did not significantly differ by DM status at either treatment failure (0.61 [95% CI, .25–1.26] per 1 PY for TB-only vs 1.2 [95% CI, .39–2.80] per 1 PY for TB-DM; P = .27), but differed significantly for recurrence (0.21 [95% CI, .04–.61] per 1 PY for TB-only vs 1.27 [95% CI, .41–2.98] per 1 PY for TB-DM; P = .02).
Figure 3.
Phylogenetic reconstruction of 76 paired Mycobacterium tuberculosis strains showing clustering of isolates, with participant origin in Chennai or Pune, India, as well as presence of drug resistance mutations. Color schemes depict lineages. Abbreviation: MDR, multidrug-resistant.
Acquisition of Drug Resistance
Twenty-one (28%) had resistance to ≥1 TB drug wither at baseline or at treatment failure or recurrence (Table 2). Baseline Mtb isolates from 15 participants (2 with treatment failure and 6 with recurrence) had resistance to ≥1 TB drug. Despite SNP accumulation among paired Mtb strains, TB drug resistance patterns remained unchanged for 13 (89%) paired isolates at treatment failure or recurrence. Of the remaining 8 pairs, acquired TB drug resistance was more commonly seen among TB-DM participants than TB-only participants (4/18 [22%] in TB-DM vs 4/45 [9%] in TB-only; P = .21). At treatment failure, acquired drug resistance to isoniazid (katG), and ethionamide (ethA) was observed in 2 isolate pairs of TB-DM participants: Acquired resistance to fluoroquinolones (gyrA), capreomycin (rrs) in one pair and ethionamide (ethA) was observed in a TB-only participant (Table 2; Supplementary Materials). At recurrence, a TB-DM participant acquired new TB drug resistance mutations to multiple TB drugs (katG [isoniazid], rpoB [rifampicin], and pncA [pyrazinamide]), and 1 TB-DM participant acquired resistance to ethionamide. In TB-only participants, acquired resistance to isoniazid and streptomycin was observed in 1 pair each at recurrence (Table 2, Supplementary Materials). Due to the small number of TB-DM participants, the effect of DM on acquisition of drug resistance could not be assessed.
Table 2.
Differences in Antituberculosis Drug Resistance Mutations Among Paired Mycobacterium tuberculosis Strains, According to Tuberculosis Treatment Outcome (n = 21)
| Patient ID | Origin | Outcome Timing | TB Drug Resistance Pattern, Mutation (TB Drug) | |
|---|---|---|---|---|
| Baseline | Follow-up | |||
| Treatment failure | ||||
| 1 | Pune | Month 5 | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) |
| 2 | Pune | Month 6 | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) |
| 3 | Pune | Month 6 | None | Rrs (capreomycin, kanamycin, aminoglycosides, amikacin) gyrA (fluoroquinolones, levofloxacin, ofloxacin, ciprofloxacin) |
| 4 | Pune | Month 5 |
rpoB (rifampicin) fabG1 (isoniazid, isoniazid) pncA (pyrazinamide) |
None |
| 5 | Pune | Month 6 | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) | None |
| 6 | Pune | Month 5 |
rpoB (rifampicin) fabG1 (isoniazid, ethionamide) pncA (pyrazinamide) |
None |
| 7a | Chennai | Month 5 | None | katG (isoniazid) |
| 8 | Pune | Month 5 | ahpC (isoniazid) | ahpC (isoniazid) |
| 9a | Pune | Month 5 | ethA (ethionamide) | ethA (ethionamide) |
| 10a | Chennai | Month 6 | None | ethA (ethionamide) |
| Recurrence | ||||
| 11 | Pune | Month 12 | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) |
| 12 | Pune | Month 12 |
katG (isoniazid) gyrB (fluoroquinolones, levofloxacin, moxifloxacin, ofloxacin, ciprofloxacin) ethA (ethionamide) |
katG (isoniazid) gyrB (fluoroquinolones, levofloxacin, moxifloxacin, ofloxacin, ciprofloxacin) ethA (ethionamide) |
| 13 | Pune | Month 12 | fabG1 (isoniazid) | fabG1 (isoniazid) |
| 14 | Chennai | Month 18 |
katG (isoniazid) embB (ethambutol) |
katG (isoniazid) embB (ethambutol) |
| 15a | Chennai | Month 18 | None |
katG (isoniazid) pncA (pyrazinamide) rpoB (rifampin) |
| 16a | Pune | Month 18 | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) |
| 17 | Pune | Month 12 | None |
gid (streptomycin) KatG (isoniazid) |
| 18 | Pune | Month 18 | gyrA (ciprofloxacin/moxifloxacin/ofloxacin, fluoroquinolones, levofloxacin) | None |
| 19a | Chennai | Month 9 | None | ethA (ethionamide) |
| 20a | Chennai | Month 7 | ethA (ethionamide) | ethA (ethionamide) |
| 21 | Chennai | Month 7 | ethA (ethionamide) | ethA (ethionamide) |
Abbreviations: ID, identification; TB, tuberculosis.
Represents patients with TB and diabetes mellitus.
Exogenous Reinfection
Overall, 63 (83%) participants had paired Mtb strains with <8 SNP differences, indicating endogenous TB reactivation of TB at treatment failure or recurrence (Table 1, Figure 3). The remaining 13 participants were categorized as having exogenous TB reinfection (6 at treatment failure and 7 at recurrence) with SNP differences among paired Mtb strains ranging from 21 to 1099 (Table 3). Overall, exogenous reinfections did not differ significantly among TB-DM when compared to TB-only (18% [4/22] vs 17% [9/54], respectively [n = 4/47]; P = .56). Exogenous reinfection rate at recurrence was 25% (3/12) for TB-DM vs 17% (4/24) for TB-only (P = .66).
Table 3.
Differences in Single-Nucleotide Polymorphisms and Lineage Among Paired Mycobacterium tuberculosis Strains Isolated From Participants With Exogenous Tuberculosis Reinfectiona, According to Diabetes Mellitus Status (n = 13)
| Patient ID | Participant Origin | Outcome | SNP Difference, No. | Lineage | |
|---|---|---|---|---|---|
| Baseline | Outcome | ||||
| TB-only | |||||
| 1 | Chennai | Recurrence | 522 | East Africa Indian Ocean | East Africa Indian Ocean |
| 2 | Chennai | Failure | 1092 | Beijing East Africa | European American |
| 3 | Pune | Recurrence | 186 | Delhi Central Asia | Delhi Central Asia |
| 4 | Pune | Failure | 267 | Delhi Central Asia | Delhi Central Asia |
| 5 | Pune | Failure | 1026 | European American | Delhi Central Asia |
| 6 | Pune | Recurrence | 1099 | Delhi Central Asia | European American |
| 7 | Pune | Failure | 24 | Unknown | Unknown |
| 8 | Pune | Failure | 155 | Delhi Central Asia | Delhi Central Asia |
| 9 | Pune | Failure | 1062 | European American | Delhi Central Asia |
| TB-DM | |||||
| 10 | Chennai | Recurrence | 21 | East Africa Indian Ocean | East Africa Indian Ocean |
| 11 | Chennai | Recurrence | 21 | East Africa Indian Ocean | East Africa Indian Ocean |
| 12 | Chennai | Recurrence | 532 | East Africa Indian Ocean | East Africa Indian Ocean |
| 13 | Chennai | Failure | 503 | East Africa Indian Ocean | East Africa Indian Ocean |
Abbreviations: DM, diabetes mellitus; ID, identification; SNP, single-nucleotide polymorphism; TB, tuberculosis.
SNP difference >8 among paired Mycobacterium tuberculosis strains isolated at baseline and tuberculosis treatment outcome (treatment failure or recurrence).
DISCUSSION
The interaction between TB and DM has been recognized for decades. This nested cohort study of Mtb evolution among pulmonary TB patients (with and without DM) with treatment failure or recurrence in India found high overall rates of SNP acquisition at recurrence among people with TB-DM. Accumulation of mutations conferring TB drug resistance to 1 or more TB drugs during and after TB treatment were more commonly seen among TB-DM participants. Although recurrence events due to TB reactivation were common in our population, our analysis indicates that exogenous TB reinfection may be more common among patients with DM at recurrence compared to patients with TB alone. Overall, this study provides new evidence suggesting that comorbid DM may increase the intrapatient Mtb mutation rate as well as the likelihood of exogenous TB reinfection in high TB-DM settings where TB transmission is high.
The overall intrapatient Mtb mutation rate of 1.2 SNPs/genome/year among people with TB-DM at recurrence is much higher than rates estimated by prior human and nonhuman primate studies at 0.3–0.6 SNPs/genome/year [6, 7]. Furthermore, compared to TB alone, the SNP accumulation rate was several-fold higher among people with TB-DM. Furthermore, we observed that 84% of paired Mtb strains accumulated 1 or more SNPs spanning TB treatment through 12 months after treatment completion, which is relatively higher compared to prior studies. Although not statistically significant, acquisition of resistance conferring mutations to 1 or more drugs was >2-fold higher among people with TB-DM. Multiple factors, including host immunity and pathogen factors, may play a role [2, 10–12]. Although rates of SNP acquisition during TB treatment were similar among participants with and without DM, patients with DM had close to 2-fold higher SNP accumulation after treatment completion compared to patients with TB alone. We theorize that lower TB drug levels among patients with DM may have induced excessive oxidative DNA damage to Mtb strains, and could, in part, explain this finding [38, 39].
The majority of recurrence in our study population was due to endogenous TB reactivation. However, in contrast to a prior study from Mexico, we found that one-fourth of patients with DM had recurrence events due to exogenous Mtb reinfection at recurrence. Interestingly, although patients with DM in our study were more likely to have exogenous TB reinfection at recurrence, in contrast, exogenous reinfection causes the majority of recurrence among patients with HIV-TB coinfection [40, 41]. Both DM and HIV are immunosuppressed states and cause dysregulation of innate immune responses, which leads to high Mtb bacterial burden. However, our findings suggest that these immune-altered states may not be equal. Unlike HIV, DM adversely affects innate immune responses to TB followed by a hyperactive adaptive immune system, which likely provides some protection against exogenous reinfection after the first TB episode.
Acquired drug resistance was detected at both treatment failure and recurrence in our cohort. While 1 Mtb isolate pair acquired resistance to 2 key first-line anti-TB drugs, rifampin and pyrazinamide, 12 months after treatment completion, we observed that 7 more isolate pairs acquired resistance to both first- and second-line drugs—isoniazid, capreomycin, ethionamide, and fluoroquinolones—during and beyond TB treatment. While we did not observe primary drug resistance to anchor TB drugs, isoniazid and rifampin (a criterion for multidrug resistance), 4 baseline Mtb isolates were resistant to isoniazid and 5 were resistant to at least 1 first- or second-line TB drug. Notably, 4 isolate pairs had fluoroquinolone resistance, a finding with a major repercussion—this effective drug may not be used for multidrug-resistant infections in these individuals or among the general population in the region if the fluoroquinolone-resistant strains are widespread.
Key strengths of our study include the utilization of WGS, which is known to identify the full spectrum of mutations, including mutations conferring drug resistance to all first-and second-line TB drugs, as demonstrated in our study. Furthermore, we assessed and compared rates of SNP acquisition during and after TB treatment by DM status. A key limitation, however, is that our small sample size was not powered to detect a small difference in the rate of acquisition of SNPs or drug resistance mutations. Nonetheless, to our knowledge, our sample size is among the largest to investigate and compare the effect of DM on Mtb mutation acquisition and recurrence. The self-reported medication adherence was >95%, so we could not assess the impact of adherence on mutation rates. In addition, our analysis did not account for within-host diversity (ie, by looking at colonies or serial isolates), which potentially introduces misclassification bias in assigning exogenous or endogenous reinfection, but we attempted to look at heterogenous subpopulations informatically. Notably, the significant, diverse SNP differences (ranging between 21 and 1099) among paired Mtb strains implies that reinfection is the appropriate classification [42]. We did not analyze highly polymorphic regions of the genome, for example, PPE/PGRS. The difference in mutation rates between TB-DM and TB-only could be due to differential replication that we could not account for. Finally, distinguishing between within-host evolution and mixed infection may be difficult with closely related strains. Our phylogenetic tree analysis suggests that Mtb strains circulating in geographic regions are quite similar. Therefore, categorizing strains with small SNP differences as reactivation may underestimate the reinfection rate.
In conclusion, our study provides genomic insights on the heterologous evolution of Mtb strains during and after completion of TB treatment among immunocompetent (TB-only) and immune-altered hosts (TB-DM) in an epicenter of TB and DM. Importantly, TB drug resistance can emerge during TB treatment and beyond, and reactivation of TB is a major cause of TB recurrence, yet exogenous reinfection should be considered at recurrence among people with DM. Future work should confirm our study findings.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. V. M., N. G., A. G., B. M., L. C., U. D. R., S. K. S., J.E.G., and B. N. K. conceived the study, and V. M. obtained funding. V. M., D. K., V. V., R. L., A. K., N. N. P., S. B., M. S. P., S. D., and C. P. conducted the study and collected data. L. C., N. G., V. M., M. F., B. M., and B. N. K. performed data analyses and interpreted the data. V. M., L. C., and N. G. drafted the initial manuscript, and all authors assisted in manuscript preparation and approved the manuscript.
Acknowledgments. The authors thank the clinic and research staff of the National Institute for Research in Tuberculosis, B. J. Medical College-Sassoon General Hospital (SGH), and M. Vishwanathan Diabetes Research Center for their immense contributions. They acknowledge Katharine McIntire for copyediting.
Disclaimer. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Financial support. This work was supported by the US Civilian Research and Development Foundation (CRDF) (grant number OISE-17-63221 to V. M., U. D. R., and B. N. K.); the National Institutes of Health (NIH) (grant number R01A1I097494 to J. E. G.); the Regional Prospective Observational Research in Tuberculosis in India consortium funded by the Indo (Department of Biotechnology [DBT])–US (NIH) (grant number USB1-31147-XX-13 CRDF/NIH to A. G. and V. M.); and the NIH-funded Johns Hopkins Baltimore-Washington-India Clinical Trials Unit for the National Institute of Allergy and Infectious Diseases (NIAID) Networks (grant number UM1AI069465 to V. M., N. G., and A. G.).
Contributor Information
Vidya Mave, Byramjee-Jeejeebhoy Medical College–Johns Hopkins University Clinical Research Site, Pune, India; Johns Hopkins University School of Medicine, Baltimore, Maryland, USA; Johns Hopkins India, Pune, India.
Liang Chen, Hackensack Meridian Health, Center for Discovery and Innovation, Nutley, New Jersey, USA.
Uma Devi Ranganathan, ICMR-National Institute for Research in Tuberculosis, Chennai, India.
Dileep Kadam, Byramjee-Jeejeebhoy Government Medical College, Pune, India.
Vijay Vishwanathan, Professor M. Vishwanathan Diabetes Research Center, Chennai, India.
Rahul Lokhande, Byramjee-Jeejeebhoy Government Medical College, Pune, India.
Siva Kumar S, ICMR-National Institute for Research in Tuberculosis, Chennai, India.
Anju Kagal, Byramjee-Jeejeebhoy Government Medical College, Pune, India.
Neeta N Pradhan, Byramjee-Jeejeebhoy Medical College–Johns Hopkins University Clinical Research Site, Pune, India; Johns Hopkins India, Pune, India.
Shri Vijay Bala Yogendra Shivakumar, Johns Hopkins India, Pune, India.
Mandar S Paradkar, Byramjee-Jeejeebhoy Medical College–Johns Hopkins University Clinical Research Site, Pune, India; Johns Hopkins India, Pune, India.
Sona Deshmukh, Byramjee-Jeejeebhoy Medical College–Johns Hopkins University Clinical Research Site, Pune, India; Johns Hopkins India, Pune, India.
Jeffrey A Tornheim, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Hardy Kornfeld, University of Massachusetts, Boston, Massachusetts, USA.
Maha Farhat, Harvard Medical School, Boston, Massachusetts, USA.
Amita Gupta, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Chandrasekaran Padmapriyadarsini, ICMR-National Institute for Research in Tuberculosis, Chennai, India.
Nikhil Gupte, Byramjee-Jeejeebhoy Medical College–Johns Hopkins University Clinical Research Site, Pune, India; Johns Hopkins University School of Medicine, Baltimore, Maryland, USA; Johns Hopkins India, Pune, India.
Jonathan E Golub, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Barun Mathema, Columbia University, New York, New York, USA.
Barry N Kreiswirth, Hackensack Meridian Health, Center for Discovery and Innovation, Nutley, New Jersey, USA.
References
- 1. World Health Organization. Global tuberculosis report 2019. Available at: https://www.who.int/publications/i/item/9789241565714. Accessed 3 January 2022.
- 2. Cadena AM, Fortune SM, Flynn JL.. Heterogeneity in tuberculosis. Nat Rev Immunol 2017; 17:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGrath M, Gey van Pittius NC, van Helden PD, Warren RM, Warner DF.. Mutation rate and the emergence of drug resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 2014; 69:292–302. [DOI] [PubMed] [Google Scholar]
- 4. Ford CB, Shah RR, Maeda MK, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet 2013; 45:784–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Müller B, Borrell S, Rose G, Gagneux S.. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet 2013; 29:160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walker TM, Ip CLC, Harrell RH, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 2013; 13:137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bryant JM, Schürch AC, van Deutekom H, et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis 2013; 13:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hershberg R, Lipatov M, Small PM, et al. High functional diversity in mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol 2008; 6:e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ford CB, Lin PL, Chase MR, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet 2011; 43:482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Q, Wei J, Li Y, et al. Mycobacterium tuberculosis clinical isolates carry mutational signatures of host immune environments. Sci Adv 2020; 6:eaba4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ.. Within-host evolution of bacterial pathogens. Nat Rev Microbiol 2016; 14:150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Q, Via LE, Luo T, et al. Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci Rep 2015; 5:17507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baker MA, Harries AD, Jeon CY, et al. The impact of diabetes on tuberculosis treatment outcomes: a systematic review. BMC Med 2011; 9:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shewade HD, Jeyashree K, Mahajan P, et al. Effect of glycemic control and type of diabetes treatment on unsuccessful TB treatment outcomes among people with TB-diabetes: a systematic review. PLoS One 2017; 12:e0186697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jeon CY, Murray MB.. Diabetes mellitus increases the risk of active tuberculosis: a systematic review of 13 observational studies. PLoS Med 2008; 5:e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Allué-Guardia A, García JI, Torrelles JB.. Evolution of drug-resistant Mycobacterium tuberculosis strains and their adaptation to the human lung environment. Front Microbiol 2021; 12:612675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Narendran G, Menon PA, Venkatesan P, et al. Acquired rifampicin resistance in thrice-weekly antituberculosis therapy: impact of HIV and antiretroviral therapy. Clin Infect Dis 2014; 59:1798–804. [DOI] [PubMed] [Google Scholar]
- 18. Pérez-Navarro LM, Fuentes-Domínguez FJ, Zenteno-Cuevas R.. Type 2 diabetes mellitus and its influence in the development of multidrug resistance tuberculosis in patients from southeastern Mexico. J Diabetes Complications 2015; 29:77–82. [DOI] [PubMed] [Google Scholar]
- 19. Jimenez-Corona ME, Luis Pablo C-H, Garcia-Garcia L, et al. Association of diabetes and tuberculosis: impact on treatment and post-treatment outcomes. Thorax 2013; 68:214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mave V, Gaikwad S, Barthwal M, et al. Diabetes mellitus and tuberculosis treatment outcomes in Pune, India. Open Forum Infect Dis 2021; 8:ofab097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kornfeld H, Sahukar SB, Procter-Gray E, et al. Impact of diabetes and low body mass index on tuberculosis treatment outcomes. Clin Infect Dis 2020; 71:e392–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupte A, Padmapriyadarsini C, Mave V, et al. Cohort for tuberculosis research by the Indo-US medical partnership (CTRIUMPh): protocol for a multicentric prospective observational study. BMJ Open 2016; 6:e010542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mave V, Meshram S, Lokhande R, et al. Prevalence of dysglycemia and clinical presentation of pulmonary tuberculosis in western India. Int J Tuberc Lung Dis 2017; 21:1280–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin MA, Lee RS, Cowley LA, Gardy JL, Hanage WP.. Within-host Mycobacterium tuberculosis diversity and its utility for inferences of transmission. Microb Genom 2018; 4:e000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vargas R, Freschi L, Marin M, et al. In-host population dynamics of Mycobacterium tuberculosis complex during active disease. Elife 2021; 10:e61805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coll F, McNerney R, Guerra-Assunção JA, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 2014; 5:4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shitikov E, Kolchenko S, Mokrousov I, et al. Evolutionary pathway analysis and unified classification of East Asian lineage of Mycobacterium tuberculosis. Sci Rep 2017; 7:9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stucki D, Brites D, Jeljeli L, et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet 2016; 48:1535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lipworth S, Jajou R, de Neeling A, et al. SNP-IT tool for identifying subspecies and associated lineages of Mycobacterium tuberculosis complex. Emerg Infect Dis 2019; 25:482–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freschi L, Vargas R Jr, Husain A, et al. Population structure, biogeography and transmissibility of Mycobacterium tuberculosis. Nat Commun 2021; 12:6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bradley P, Gordon NC, Walker TM, et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 2015; 6:10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Phelan J, O’Sullivan DM, Machado D, et al. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med 2019; 11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Witney AA, Bateson AL, Jindani A, et al. ; RIFAQUIN Study Team. . Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial. BMC Med 2017; 15:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang MJ, Chae JW, Yun HY, et al. Effects of type 2 diabetes mellitus on the population pharmacokinetics of rifampin in tuberculosis patients. Tuberculosis (Edinb) 2015; 95:54–9. [DOI] [PubMed] [Google Scholar]
- 39. Alfarisi O, Mave V, Gaikwad S, et al. Effect of diabetes mellitus on the pharmacokinetics and pharmacodynamics of tuberculosis treatment. Antimicrob Agents Chemother 2018; 62:e01383-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sonnenberg P, Murray J, Glynn JR, Shearer S, Kambashi B, Godfrey-Faussett P.. HIV-1 and recurrence, relapse, and reinfection of tuberculosis after cure: a cohort study in South African mineworkers. Lancet 2001; 358:1687–93. [DOI] [PubMed] [Google Scholar]
- 41. Narayanan S, Swaminathan S, Supply P, et al. Impact of HIV infection on the recurrence of tuberculosis in south India. J Infect Dis 2010; 201:691–703. [DOI] [PubMed] [Google Scholar]
- 42. Guerra-Assunção J, Crampin AC, Houben R, et al. Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. eLife 2015; 3:e05166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.