Abstract
Motivation
Recombination is an essential driver of virus evolution and adaption, giving rise to new chimeric viruses, structural variants, sub-genomic RNAs and defective RNAs. Next-generation sequencing (NGS) of virus samples, either from experimental or clinical settings, has revealed a complex distribution of recombination events that contributes to intrahost diversity. We and others have previously developed alignment tools to discover and map these diverse recombination events in NGS data. However, there is no standard for data visualization to contextualize events of interest, and downstream analysis often requires bespoke coding.
Results
We present ViReMaShiny, a web-based application built using the R Shiny framework to allow interactive exploration and point-and-click visualization of viral recombination data provided in BED format generated by computational pipelines such as ViReMa (Viral-Recombination-Mapper).
Availability and implementation
The application is hosted at https://routhlab.shinyapps.io/ViReMaShiny/ with associated documentation at https://jayeung12.github.io/. Code is available at https://github.com/routhlab/ViReMaShiny.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Viruses exist as dynamic populations of diverse genomes (often referred to as intra-host diversity) which is maintained by the error-prone nature of viral replication (Lauring et al., 2013). In addition to single-nucleotide variations (SNVs), viral recombination contributes to intrahost diversity through the production of structural variants (SVs), sub-genomic RNAs (sgmRNAs), defective RNAs (D-RNAs) and chimeric viruses that seed the emergence of novel virus strains (Simon-Loriere and Holmes, 2011). Viral recombination has contributed to the generation of notable variants in SARS-CoV-2, such as conserved insertions and deletions in the Spike region of the Alpha, Delta and Omicron variants of concern (VOCs). In phages, recombination generates ‘virus mosaicism’ as co-infection of bacterial hosts and subsequent horizontal genetic exchange between phages is common (Hatfull, 2008). Consequently, recombination has an important influence on viral evolution both within single hosts and on ecological scales.
Improved high-throughput sequencing capabilities have enabled unprecedented characterization of the diversity of viral recombination events within virus populations. These technical advancements have led to a corresponding demand for bioinformatic software that can discover and map these events. ViReMa (Viral-Recombination-Mapper) (Routh and Johnson 2014) is a python package enabling viral read mapping from next-generation sequencing (NGS) data that was developed for this purpose. ViReMa has been used to characterize the full gamut of different recombination events in multiple studies, including sub-genomic mRNA production in coronaviruses (Gribble et al., 2021), drug-resistance development in HIV samples (Wang et al., 2022), the evolution of D-RNAs during serial virus passaging of Flock House virus in culture (Jaworski and Routh, 2017), demonstrated differences in D-RNA abundance between intra- and extra-cellular compartments of alphaviruses (Langsjoen et al., 2020), and compared recombination events from experimental and clinical isolates of SARS-CoV-2 (Jaworski et al., 2021).
Due to the combination of skills required, these studies are commonly collaborations between wet-lab experimentalists and bioinformaticians. The collaborative process can require multiple iterations of analyses to home in on data that are both valid and biologically interesting. Data exploration using easily accessible, GUI-based applications can improve turn-around times between iterations and allows experimentalists with limited coding experience to actively engage in analysis. R, a popular programming language for data visualization, includes the Shiny package which allows developers to create dashboards. The Shiny framework provides interactivity, extensibility and flexibility with local and web-hosted options available.
We present an R Shiny application, ViReMaShiny, that enables rapid visualization of viral recombination data from ViReMa or other applications that output recombination events using BED files. This application seeks to standardize the representation of key features in viral recombination events such as their frequency and position relative to important genomic elements.
2 Results
The ViReMaShiny application was created using the R Shiny framework and relies on the ggplot2 and circlize (Gu et al., 2014) packages for plotting. The initial input of user files requires BED files, an output of ViReMa (Sotcheff et al., 2022) and canonical splice-aware mappers such as HISAT2 (Kim et al., 2015) and STAR (Dobin et al., 2013), and are hosted locally. Either a single BED file or multiple BED files from biological or experimental replicates can be uploaded. The BED files follow the standardized format as depicted in Figure 1A. Briefly, the genome reference, strand and coordinates of the donor and acceptors sites of the recombination junction (the nucleotide index where the junction begins and ends, respectively) are provided in addition to the number of reads that map to each unique event. When reads are mapped using ViReMa, additional columns also provide the read coverage at each junction site, and the sequence of a fixed number of nucleotides both up- and down-stream of the donor and the acceptor sites. This latter information allows scrutiny of the sequence composition of each recombination junction. Once uploaded, users will be able to subset data visualized by file name and include reference sequences through drop-down menu selectors.
Fig. 1.
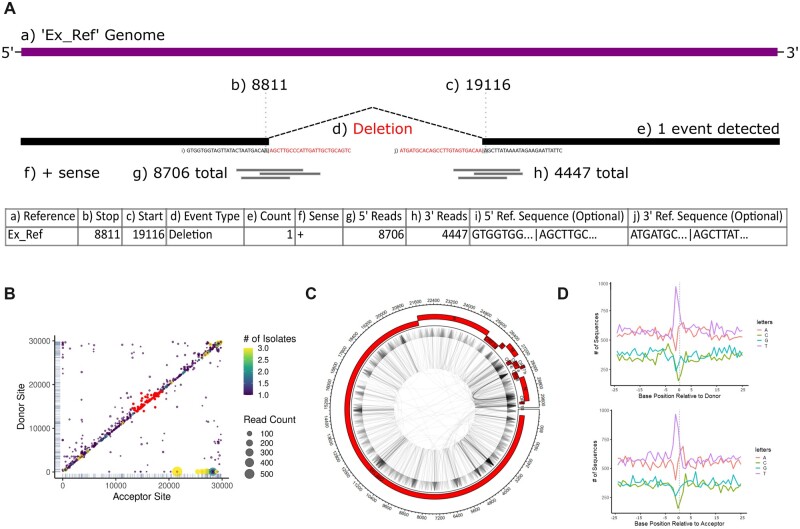
Example input data and plot outputs from ViReMaShiny using sample SARS-CoV-2 data from a previous study (Jaworski et al., 2021). (A) A table with an example recombination event in ViReMa output BED format. Subsection (a) refers to the reference sequence of the event; (b, c) indicate the nucleotide base positions spanning the event; (d) is the type of recombination event; (e) is the number of reads detected with this event; (f) is the sense or strandedness of the nucleic acid the event was detected on; (g, h) correspond to the number of reads spanning (b) and (c), respectively; (i, j) are reference-derived nucleotide sequences at the recombination junction, 25 bp upstream and downstream of both (b) and (c). (B) A scatterplot depicting recombination events by donor and acceptor site indexes. Read counts correspond to dot size while color encodes the number of isolates the event appears in. A subset of interest is highlighted in red. (C) An example Circos plot depicting all recombination events. The plot includes annotations for the NCBI RefSeq NC_045512.2 SARS-CoV-2 genome. (D) Nucleotide bias at donor (top) and acceptor sites (bottom) for all recombination events. Donor and acceptor sites correspond to (b) and (c) in (A), respectively (A color version of this figure appears in the online version of this article.)
Interactivity is centered around two plots depicting recombination events by the donor (y-axis) and acceptor (x-axis) sites of each unique recombination junction. If multiple BED files representing multiple unique samples are uploaded, then a color bar indicates the frequency with which specific recombination junctions are seen in multiple samples. We present an example of a recombination heatmap from samples of SARS-CoV-2 RNA obtained from three nasopharyngeal swabs from a previous study (Jaworski et al., 2021) (Figure 1B, Supplementary Data S1). This visualization quickly summarizes notable features of recombination events including favored acceptor or donor sites, evidence of recombination hotspots and the frequency or abundance of similar events within a dataset. For example, the conserved sgmRNAs of SARS-CoV-2 are visible as yellow spots in the lower right portion of the heatmap and abundant insertions and deletions are observed in the upper right end of the heatmap, representing diverse RNA recombination near the 3′UTR of SARS-CoV-2. Frequent small InDels are represented by the numerous points close to the x = y-axis. This approach has been extensively used to visualize recombination events in a range of viruses including Nodaviruses (Routh et al., 2012), alphaviruses (Langsjoen et al., 2020) and coronaviruses (Gribble et al., 2021). Scrubbing the scatterplots generates a filterable table, allowing users to identify events with specific features. A text box above this table alternatively allows for filter expressions in R syntax to be applied to data based on each of the parameters in the BED file(s). This allows users to sample specific events with desired features, such as (for example) only small InDels or only highly abundant events. Events in the table can be highlighted on the scatterplot using a toggle-able button or by clicking on a row in the sequence table.
Users can view summary statistics and plots under the ‘Overview’ tab. A table summarizes the total and unique events for each sample. A Circos plot (Krzywinski et al., 2009) depicts directional recombination events relative to user-provided annotations (Figure 1C). Numerous visual options can be adjusted through sliders. Annotations can either be added manually through an editable table or included as a BED file. If certain columns are included in the inputted files, nucleotide plots demonstrate the abundance and any enrichment or depletion for nucleotides proximal and distal to donor and acceptor sites. For example, in SARS-CoV-2, recombination sites are most frequently flanked by U-rich tracks (Figure 1D). Manhattan plots show broad patterns in deleted and duplicated segments, as has previously been used to identify conserved functional motifs in the RNA genome of Flock House virus (Routh and Johnson, 2014). Export of all table data and plots are available in high-fidelity file formats such as TIFF and PDF.
Ultimately, scientific questions such as most common recombination event, nucleotide usage proximal and distal to recombination sites, and samples with the most unique events can be answered within minutes of file upload. Vignettes included in the documentation demonstrate how to use the application for these purposes. Associated documentation also includes tutorials for analysis of ViReMa output data in R.
3 Conclusion
ViReMaShiny standardizes outputs and improves the approachability of exploratory viral recombination analysis. This application is built on the outputs of the ViReMa python script, allowing for intuitive investigation of data with no coding requirement. We plan on expanding support for analysis in the R environment as well as providing options to visualize recombination between multiple genes of multi-partite viruses such as influenza virus (Alnaji et al., 2021).
Supplementary Material
Acknowledgements
We would like to thank Dr Fadi Alnaji (University of Illinois at Urbana-Champaign) for discussions and advice.
Funding
This work was supported by the National Institutes of Health (NIH) [R21AI151725 to A.L.R.] and CDC contract [200-2021-11195 to A.L.R.].
Conflict of Interest: none declared.
Contributor Information
Jason Yeung, John Sealy School of Medicine, The University of Texas Medical Branch, Galveston, TX 77550, USA.
Andrew L Routh, Department of Biochemistry and Molecular Biology, The University of Texas Medical Branch, Galveston, TX 77550, USA; Sealy Center for Structural Biology and Molecular Biophysics, The University of Texas Medical Branch, Galveston, TX 77550, USA; Institute for Human Infections and Immunity, University of Texas Medical Branch, Galveston, TX, USA.
References
- Alnaji F.G. et al. (2021) Influenza a virus defective viral genomes are inefficiently packaged into virions relative to wild-type genomic RNAs. mBio, 12, e0295921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A. et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble J. et al. (2021) The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathogens, 17, e1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z. et al. (2014) Circlize implements and enhances circular visualization in R. Bioinformatics, 30, 2811–2812. [DOI] [PubMed] [Google Scholar]
- Hatfull G.F. (2008) Bacteriophage genomics. Curr. Opin. Microbiol., 11, 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski E. et al. (2021) Tiled-ClickSeq for targeted sequencing of complete coronavirus genomes with simultaneous capture of RNA recombination and minority variants. Elife, 10, e68479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski E., Routh A. (2017) Parallel ClickSeq and nanopore sequencing elucidates the rapid evolution of defective-interfering RNAs in flock house virus. PLoS Pathogens, 13, e1006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods, 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. (2009) Circos: an information aesthetic for comparative genomics. Genome Res., 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langsjoen R.M. et al. (2020) Differential alphavirus defective RNA diversity between intracellular and extracellular compartments is driven by subgenomic recombination events. mBio, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S. et al. (2013) The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol., 11, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh A., Johnson J.E. (2014) Discovery of functional genomic motifs in viruses with ViReMa - a virus recombination mapper-for analysis of next-generation sequencing data. Nucleic Acids Res., 42, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh A. et al. (2012) Nucleotide-resolution profiling of RNA recombination in the encapsidated genome of a eukaryotic RNA virus by next-generation sequencing. J. Mol. Biol., 424, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E., Holmes E.C. (2011) Why do RNA viruses recombine? Nat. Rev. Microbiol., 9, 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotcheff S. et al. (2022) ViReMa: a Virus Recombination Mapper of Next-Generation Sequencing data characterizes diverse recombinant viral nucleic acids. bioRxiv 2022:2022.2003.2012.484090. 10.1101/2022.03.12.484090. [DOI] [PMC free article] [PubMed]
- Wang S. et al. (2022) Covariation of viral recombination with single nucleotide variants during virus evolution revealed by CoVaMa. Nucleic Acids Res., 50, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.