Abstract
The emergence of a novel coronavirus, COVID-19, in December 2019 led to a global pandemic with more than 170 million confirmed infections and more than 6 million deaths (by July 2022). Studies have shown that infection with SARS-CoV-2 in cancer patients has a higher mortality rate than in people without cancer. Here, we have reviewed the evidence showing that gut microbiota plays an important role in health and is linked to colorectal cancer development. Studies have shown that SARS-CoV-2 infection leads to a change in gut microbiota, which modify intestinal inflammation and barrier permeability and affects tumor-suppressor or oncogene genes, proposing SARS-CoV-2 as a potential contributor to CRC pathogenesis
Abbreviations: SARS-CoV-2, Severe acute respiratory syndrome coronavirus-2; ACE, Angiotensin-converting enzyme; TMPRS, Transmembrane protease, serine; RSV, Respiratory syncytial virus; IBD, Inflammatory bowel syndrome; CRC, Colorectal cancer; HMP, Human microbiome project; SCFAs, Short-chain fatty acids; MCFAs, Medium-chain fatty acids; TH1, T-helper cell-1; ITIM, Immunoreceptor tyrosine-based inhibitory motif; mTOR, Mammalian target of rapamycin; ARDS, Acute respiratory distress syndrome; GI, Gastrointestinal
Keywords: COVID-19, Gut microbiome, Colorectal cancer
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) appeared late in 2019 in Wuhan, China, led to a worldwide pandemic with more than 500 million confirmed infected cases and over 6 million deaths related to this infection as of July 2022 [1]. SARS-CoV-2 is a positive-sense single-stranded RNA virus transmitted mainly through respiratory aerosol droplets and direct contact, which is the main source of human coronavirus disease 2019 (COVID-19) [2], [3], [4]. Spike glycoprotein (S1 domain) of the SARS-CoV2 recognizes and attaches to the angiotensin-converting enzyme-2 (ACE2) receptors expressed on target cells to facilitate the attachment of the virus to the host cells [5]. Afterward, host protease TMPRSS2 cleaves S, manifesting the S2 binding domain and facilitating host cell infection [6].
It has been demonstrated that for binding the virus to the host cells and subsequent entry, the ACE2 receptor and TMPRSS2 are needed. However, since other tissues express distinctive receptors and proteases, they may interact with the spike protein, revealing the multi-tissue affinity of SARS-CoV-2 [7], [8], [9], [10]. SARS-CoV-2 affects several organs, such as the brain, gut, and lungs. The symptoms vary from mildly symptomatic such as fatigue, diarrhea, vomiting, cough, and dyspnea, to severe conditions, such as acute respiratory distress syndrome and cytokine storm resulting in multiple organ failures requiring mechanical ventilation and eventually death. Immune system dysregulation, especially cytokine storm and up-regulation of inflammatory cytokines, is the main characteristic of COVID-19 [11]. The severity of the disease depends on co-morbidities, sex, and age [12]. In severely ill cases, the virus triggers tremendous inflammatory responses [13].
Cancer increases the risk of viral infections, so cancer patients are considered at high-risk for viral infections targeting the respiratory system [14]. In cancer patients, respiratory infections have a significant lethal burden due to dysregulated immune response that leads to a reduced ability to cope with disorders [15]. It has been demonstrated that respiratory syncytial virus (RSV) infection, along with the Coronaviridae family, inhibits and breaks down p53, a tumor suppressor that can hinder carcinogenesis [16], [17], [18]; this brings up the question, are cancer patients more feasible to develop deadly side effects after SARS-CoV-2? Recent studies have shown the relationship between SARS-CoV-2 and cancer, highlighting a high mortality rate among cancer patients infected by SARS-CoV-2 [19]. A great attempt has been accomplished over the last few years to gather data regarding the correlation between COVID-19 and cancer. These studies have revealed that some groups are at a higher risk of developing severe COVID-19, especially patients under treatment for hematologic, lung, or metastatic cancers [19], [20].
The pandemic is vague, and the increase in mortality rates is due to late diagnosis and poor cancer management [20]. Immune dysregulation due to SARS-CoV-2 can cause even more difficulties for subpopulations at risk. From a mechanical point of view, the interaction between the host immune environment and cancer or SARS-CoV-2 infections all apply almost identical pathways such as hypercoagulability, an aberrant immune response, such as altered expression of ACE-2 and TMPRSS2, altered cytokine levels, and prothrombotic status, make the human body highly unstable and may exacerbate the effects of SARS-CoV-2 in some cancer patients [20]. Limited information about the pathogenesis of COVID-19, especially the distinct immune response, has made it difficult to accurately and definitively treat the disease [21].
A pan-cancer analysis realized that ACE2 and TMPRSS2 are down-regulated in tumors while upregulated in digestive organs (both cancerous and normal) [22], [23]. Also, it has been demonstrated that colon epithelial cells contain high expression levels of ACE2 and can trigger gut barrier dysfunction due to SARS-CoV-2 viral replication [24]. Moreover, there are subsidiary host proteases for the processing of spikes and exposure of the S2 domain, including TMPRSS2 and TMPRSS4 [7]. The outcome is the local inflammation in the gut epithelia, and the systemic inflammation caused by the respiratory infection due to SARS-CoV-2 forcefully affects the composition of the intestinal microbiota [25]. Formerly, it has been shown that the disease caused by Influenza can remarkably modify the gut microbiota composition [26].
Interferons produced in the lungs can lead to dysbiosis by reducing obligate anaerobic bacteria and enriching proteobacteria in the gut. These interferons were shown to play an important role in the stomach by inhibiting antimicrobial and inflammatory responses during Salmonella-induced colitis, a risk factor for colon cancer due to increased intestinal colonization and dissemination of Salmonella [27], [28]. Moreover, patients suffering from inflammatory bowel syndrome (IBD) had an elevated risk of Influenza and IBD-influenced flu-like symptoms [28]. The fact that IBD and IBD-induced dysbiosis is a prominent risk factors for CRC is noteworthy and has reached high attention recently. It has also been identified that intestinal dysbiosis can affect the carcinogenesis and growth of CRC [29], [30], [31].
Little is known about the impact of SARS-CoV-2 on the host microbiome, especially in the niche environment, for example, the association between gut microbiota and CRC. According to the American Cancer Society, new cases of CRC are estimated to reach more than 140,000, and deaths will reach more than 50,000 by 2020 in the United States. The precise underlying relationship between SARS-CoV-2-induced gut microbial changes and CRC will be critical in this study area. [32].
2. The gut microbiome and CRC
One of the leading causes of cancer-related deaths in the United States is CRC. Meanwhile, the molecular mechanism of its development is not fully understood. According to studies conducted in the United States, the prevalence of CRC increased by 2 % from 2007 to 2016 among adults younger than 55 years, prompting recommendations to lower the age for colonoscopy screening for CRC [32], [33], [34]. In the past decade, the gut microbiota and its critical role in human health have been highlighted by the Human Microbiome Project (HMP). This vital role of gut microbiota is due to its protective, nutritional, and metabolic activities. Recently, microbiota-derived metabolites have been shown as a possible inducer of carcinogenesis, suggesting gut microbiota as a potential contributor to CRC pathogenesis [2], [35], [36]. Anaerobic microbes inhabiting the large intestine ferment food components to produce a wide range of metabolites that contain significant fermentation products, including gases and organic acids in healthy adults. They also have short-chain fatty acids (SCFAs), such as butyric acid, propionic acid, and acetic acid, as well as medium-chain fatty acids (MCFAs), such as oleic acid, lauric acid, and linoleic acid [37], [38]. These products are released into the lumen of the large intestine and act as signaling molecules between the host and bacteria [35]. Also, SCFAs have been shown to maintain the intestinal barrier and exert their anti-inflammatory effects by increasing the expression of anti-inflammatory cytokines and promoting differentiation of T lymphocytes to regulatory subtype (T-reg) [39]. Notably, these processes in the gut create a multi-organ axis, including the "gut-lung axis" and the "gut-brain axis," which maintain a healthy homeostatic state of these systems that depend on metabolic products of the gut [40], [41].
Intestinal dysbiosis introduces metabolites and products bacteria produce into the circulatory system, resulting in systemic inflammation [42]. Distinct dysbiosis in CRC patients from healthy groups has been detected by stool analysis [29]. Dysbiosis of gut microbiota in CRC patients can affect the immune system related to carcinogenesis and tumor progression. Studies on CRC have shown that frequent changes in the gut microbiota occur during tumor growth and account for tumor progression [43], [44], [45]. One study showed that colonization of the microbiota-derived from tumour-bearing mice significantly increased tumorigenesis in germ-free mice [46]. Common characteristics of CRC patients are a noticeable decrease in Bacteroides, Firmicutes, and Actinobacteria, along with increases in Porphyromonas and Fusobacterium populations [47]. Actinobacteria is one of the four main types of intestinal microbiota. However, they contain only a small percentage of the bacterial population. Actinobacteria are essential in maintaining gut homeostasis and immune tolerance [48]. An unbalanced population of Actinobacteria has been shown to lead to several pathological conditions [49], [50]. In particular, the reduction of CRC metabolites belonging to the phylum Actinobacteria, together with a proinflammatory cell-mediated immune response, cytotoxic T-helper cell-1 (Th1), induced by this group, leads to a poor prognosis of CRC [51]. Also, the reduction of bacteria that produce butyrate, especially Actinobacteria and Firmicutes phyla) leads to a reduction of the main energy source for colonocytes. It increases the pH of the colon and provides an unfavorable environment for colonocytes, and induces tumor formation [47]. Increased bile acid secretion has been shown to be associated with focal destruction of the intestinal epithelium. Subsequently, it stimulates repair mechanisms, including inflammatory reactions and hyperproliferation of undifferentiated cells [52].
It has been shown that microbes that convert primary bile acids into secondary acids play a role in developing progressive tumors [ 49,53]. Bile acids are molecules that play an important role in signaling and regulating digestive and physiological functions, immune homeostasis, and lipid and glucose metabolism. Cholesterol is a precursor for the synthesis of bile acids and forces them to be conjugated to glycine and taurine in liver cells [54]. Most of these bile acids are then released into the small intestine. Bile acids that reach the colon interact with gut microbes. Bile acids are considered toxic to most gut microbiota. Therefore, its presence in large amounts creates selective pressures that profoundly affect the gut microbiota, confirming a specific set of gut bacteria that can perform their enzymatic role on bile acids [55]. A prominent example is an excess taurine, excreted as taurocholic acid, a conjugated bile acid, and then converted to deoxycholic acid, which is genotoxic and tumorigenic [56].
Studies on mice have shown that probiotic supplements increase the ratio of Lactobacillus and Bifidobacterium in the intestinal microbiota, reduce the infiltration of inflammatory cells and CRC associated with colitis, and reduce chemokine expression [57]. Further research has shown that increased amounts of Proteobacteria, a small member of the intestinal microbiota, can be a possible microbial clue to epithelial dysfunction in CRC patients [58], [59]. Understanding the complex factors leading to dysbiosis remains a point of obscurity, and its consequences on CRC are unclear.
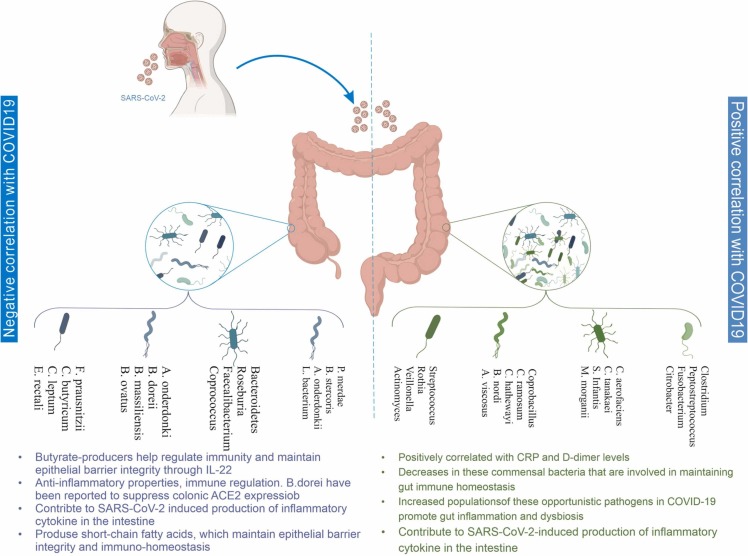
A positive and negative relationship between gut microbiome composition and COVID-19 disease has been summarized in Fig. 1.
Fig. 1.
Thoroughly shows the negative and positive correlation between gut microbiome composition with immune system dysregulation and GI-affected symptoms in COVID-19 patients.
3. SARS-CoV-2-induced gut microbiome dysbiosis and CRC
The most common feature of COVID-19 infection is respiratory symptoms. However, it can affect the digestive system and cause complications such as inflammation and intestinal blockage to diarrhea [60]. One theory is that SARS-CoV-2 disease downregulates ACE2, leading to decreased activation of the mammalian target of rapamycin (mTOR) and increased autophagy, leading to intestinal dysbiosis and diarrhea [60]. The ACE expression has reduced in mice infected with SARS-CoV-2 and mice injected with recombinant SARS protein. The downregulation, as mentioned above, may play a role in the pathogenesis of SARS and diseases developing into Acute respiratory distress syndrome (ARDS) [61], [62], [63]. Another theory is that the small intestine is likely to be a critical site for exacerbating the systemic inflammatory response, where ACE2 inhibition leads to increased levels of angiotensinogen and hyperactivation of the renin-angiotensin system, which in turn shuts down the amino acid transporter BA0T1 and subsequently leads to tryptophan deficiency in cells, which leads to decreased secretion of antimicrobial peptides and intestinal dysbiosis [64]. Recent studies have shown that patients with COVID-19 suffer from dysbiotic gut microbiota [65], [66], [67], [68], [69]. A study reported an alteration and correlation of the fecal microbiome of SARS-CoV-2 patients with the severity of the disease [30]. In patients with COVID-19, significant changes were observed in stool microbiomes with enrichment of opportunistic pathogens and reduction of beneficial compounds during hospitalization [70]. Surprisingly, following the elimination of SARS-CoV-2 and its respiratory symptoms, symbionts and intestinal dysbiosis reduction persisted during hospitalization [71]. The result of a study reported that Bacteroides dorei, Bacteroides thetaiotaomicron, Bacteroides massiliensis and Bacteroides ovatus stool-derived COVID-19 patients have a negative correlation with ACE2 expression and viral load in the body [72].
In another study, the changes in the frequency of ten predominant intestinal bacterial groups in COVID-19 patients with pneumonia were investigated to show the relationship between the mentioned bacterial groups and clinical indicators [66]. The results show that the changes in the intestinal microbial community and dysbiosis in COVID-19 patients are associated with the severity of the disease and blood parameters. Common opportunistic pathogens, such as Enterococcus (Ec) and Enterobacteriaceae (E), have been shown to be elevated in COVID-19 patients, mainly severe patients. The findings suggest that this group of bacteria can play the role of diagnostic biomarkers of COVID-19, and the Ec/E ratio can be applied in critically ill patients to predict death [73], [74].
Another study was conducted on COVID-19 patients and investigated the relationship between SARS-CoV-2 transcriptional activity and fecal microbiome changes [69]. Even without gastrointestinal (GI) symptoms, in some patients, signs of active viral infection become apparent up to 6 days after removal of SARS-CoV-2 from respiratory samples. High-level SARS-CoV-2 characters in stool samples showed a higher abundance of bacterial species Collinsella tanakaei, Collinsella aerofaciens, Morganella morganii, and Streptococcus infantis [75], [76] . In patients with active gastrointestinal problems infected with SARS-CoV-2, loss of beneficial bacteria increases functional capacity for biosynthesis of nucleotides and amino acids, and carbohydrate metabolism is characteristic of the gut microbiota [77]. Although patients with SARS-CoV-2 suffered from ARDS and recovered, they did not show any gastrointestinal symptoms during the illness; a long-term chronic gastrointestinal infection was observed among these patients. Altered gut bacterial composition with a higher relative abundance of opportunistic pathogens such as Rote, Streptococcus, Actinomyces, Veillonella and beneficial symbionts has been reported in patients with COVID-19 [65], [66], [67]. It has also been suggested that levels of Romboutsia, Erysipelatoclostridium, Fusicatenibacter, Actinomyces, and Intestinibacter have a correlation with COVID-19 diseases severity, suggesting the gut microbiota as a diagnostic biomarker and therapeutic target for COVID-19. Decreased bacterial diversity in the gut is associated with severe and long-term metabolic effects and disease pro-longevity [70], [78].
A significant decrease in the abundance of butyrate-producing bacteria such as Clostridium butyricum, Faecalibacterium prausnitzii, Eubacterium rectale, and Clostridium leptum was shown in patients with COVID-19, which differentiates patients with a critical condition from those with milder disease [66]. SCFA-producing bacteria such as Parabacteroides merdae, Bacteroides stercoris, Alistipes onderdonkii, and Lachnospiraceae bacteria showed higher abundance in stool samples collected from patients with mild infection caused by SARS-CoV-2 [69]. In the case of butyrate-producing bacteria, it is noteworthy that they are critical in maintaining the integrity of the gut barrier [79]. The role of SCFAs in maintaining gut-lung epithelial barrier integrity involves signaling for IL-22. Macrophages in the inflamed gut upregulate butyrate and other SCFAs and enhance the Warburg effect for neoplastic cell metabolism. Therefore, fatty acid oxidation is limited due to metabolic dependence on anaerobic glycolysis [80]. Also, in severe cases of COVID-19, bacteremia caused by Fusobacterium nucleatum has been observed and shown to induce colonic mucosa and associated mucosal inflammation [81].
On the other hand, F. nucleatum has a surface protein called Fap2 that binds to galactose and interacts with the T cell ITIM domain inhibitory receptor, leading to suppression and dysfunction of the immune system associated with COVID-19. In addition, F.nucleatum has been shown to correlate with Toll-like receptors 2 and 4, leading to increased expression of microRNA-21 (miRNA21), enhancing NF-κB induction and inflammation and increasing cell proliferation [82], [83], [84], [85]. It is also shown that IL-18, the intestinal inflammatory cytokine, is increased in the COVID-19 patients' serum [67]. These findings indicate that alteration in gut microbiota composition leads to SARS-CoV-2-induced inflammatory cytokines in the intestine.
The change of stool fungal microbiome (mycobiome) in Covid-19 patients has been investigated. In this regard, patients with COVID-19 had significant changes in their stool microbiome, characterized by increased proportions of opportunistic fungal pathogens such as Aspergillus flavus, Candida auris, and Candida albicans. It was shown that even after the removal of SARS-CoV-2 from the body, two respiratory-related fungal pathogens, A. Niger and A. Flavus, were detected in the stool samples of a group of COVID-19 patients [86], [87].
There is a condition with persistent and prolonged COVID-19-induced symptoms among survivors, termed "long haulers", characterized by prolonged symptoms last for more than 4–6 weeks from the onset of symptoms [88], [89]. This condition can result from post-acute COVID-19 infection, where immune system disturbances and gastrointestinal symptoms persist, and the patient's microbiome status is affected despite the elimination of SARS-CoV-2. Since the COVID-19 pandemic is a new disease, all pathological aspects have not yet been determined; in this regard, dysbiosis's timeline and long-term effects have not yet been elucidated, so further research is needed [90], [91]. Intestinal barrier dysfunction is a distinct phenomenon related to respiratory infection associated with a more severe clinical course of the disease. From a clinical point of view, it has been shown that regulation of the gut microbiota slows down the replication of the primary influenza virus in the lung epithelium, reducing enteritis and ventilator-associated pneumonia [92]. In this respect, it has been shown that Bifidobacterium species' development can hinder inflammation induced by dysbiotic events during and after influenza infection [93], [94]. Currently, there is no definitive clinical evidence for a therapeutic role of gut microbiota regulation in the treatment of COVID-19; however, clinical trials are ongoing [95], [96], [97]. By reducing proinflammatory signaling and maintaining intestinal barrier integrity, probiotics can support patients' immune homeostasis in the gut and prevent overactivation of the immune response. The human immune system is developed to deal with microorganisms inside and outside our body [98]. This subject indicates that self-limiting is the main feature of most infectious diseases caused by viruses or bacteria [99], [100]. Accordingly, one of the possible reasons for the asymptomatic and moderate nature of most cases of COVID-19 can be extracted from this issue [101]. Therefore, the mentioned microorganisms can play a role and benefit as a source of metabolites such as essential amino acids and fatty acids [102].
In conditions of intestinal dysbiosis, microorganisms and damaged tissue in the body can degrade and become a source of nutrition, leading to a temporary oversupply of nutrition. This event increases inflammation during acute infection [103] and can trigger chronic diseases, such as malignancies [104]. For example, some excess nutrients from damaged tissue and a dysbiotic gut microbiome can be converted into lipid mediators that lead to lipotoxicity and other tissue damage, emphasizing the importance of nutritional status [105]. These obtained observations provide a basis for more significant research and help to find the relationship between changes in the gut microbiota due to COVID-19 infection and its association with an increased risk for the development or progression of CRC [106], [107]. Given the global prevalence of this virus and the diagnosis of CRC as the third most common cancer in both sexes in the United States, research into the related effects of these two diseases has become a great necessity. As soon as these questions are clarified and obscure questions are answered, it becomes possible to develop efficient probiotic therapies, which can maintain a more homeostatic environment for the gut during the disease, thereby increasing the survival rate in CRC. Fig. 2, Fig. 3 reveal the relationship between the gut microbiome and COVID-19 infection leading to CRC.
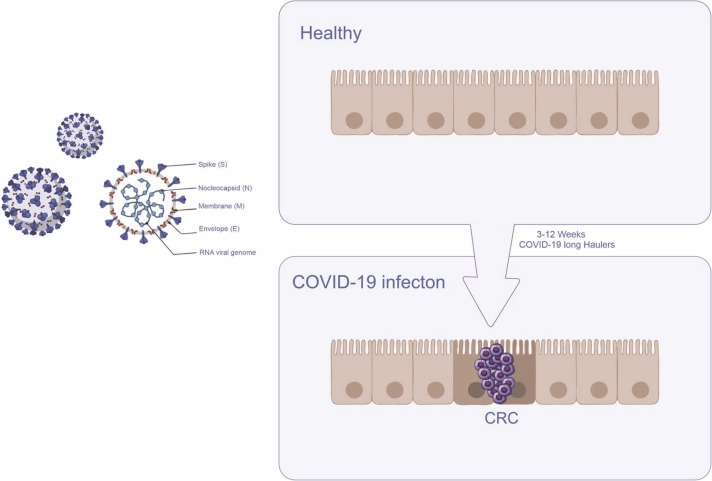
Fig. 2.
Shows the shape of the Covid-19 virus and its constituent parts and how healthy colon cells become cancerous after Covid-19 infection.
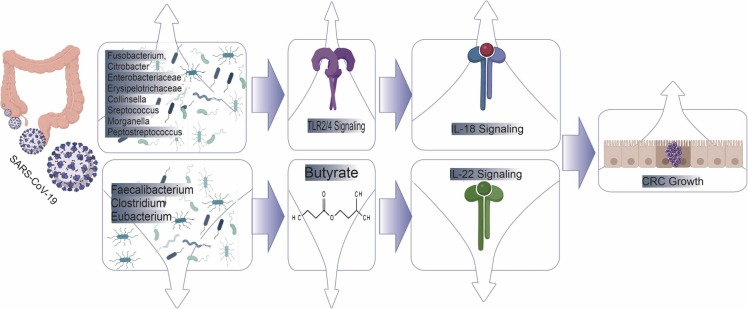
Fig. 3.
Shows that the COVID-19 virus affects the microbial composition, leading to dysregulation of TLR signaling and butyrate production, ultimately leading to unbalanced interleukin secretion and CRC development.
4. Conclusions and future directions
The long-term effects of infection by SARS-CoV-2 remain an ambiguous point of this pandemic. This is critical for those with significant co-morbidities before the onset of COVID-19. Cancer is only one of the listed diseases. Understanding the role of COVID-19 concerning cancer is extremely important due to immunosuppressive therapies and the multifaceted nature of this disease. When birth date is taken into account, those born in 1990 have twice the risk of colon cancer compared to those born around 1950 [32]. The critical role of gut microbiota in the development and health of CRC has been proven. Significant changes in gut microbiota and microbiota have been demonstrated in early studies in patients with COVID-19. The SARS-CoV-2 infection leads to a change in the intestinal microbiota. The consequences of this disorder include the abundance of opportunistic pathogens and the reduction of beneficial compounds, the overall reduction of microbial diversity, the absence of butyrate-producing bacteria, and F.nucleatum bacteremia. Therefore, SARS-CoV-2-associated gut microbiome alteration could be a new contributor to colorectal cancer pathogenesis that can be considered for therapeutic goals.
CRediT authorship contribution statement
SAM, HN, ZV, AA, and GZ: Writing – original draft. SAM, AS, EM, BAZ, AEN, and FE: Writing – review & editing, Visualization. MA: Conceptualization, Supervision.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Funding
The authors received no financial support for this article's research, authorship, or publication.
Authors' agreement to publication
All authors and institutions have confirmed this manuscript for publication.
Competing interests
The authors declare that they have no competing interests.
Acknowledgment
None.
Data Availability Statement
Not applicable.
References
- 1.W. Coronavirus, Dashboard-Available from: 〈https://covid19.who.int〉, World Health Organization (WHO), Geneva, 2021.
- 2.Esmaeil Amini M., Shomali N., Bakhshi A., Rezaei S., Hemmatzadeh M., Hosseinzadeh R., et al. Gut microbiome and multiple sclerosis: new insights and perspective. Int. Immunopharmacol. 2020;88 doi: 10.1016/j.intimp.2020.107024. [DOI] [PubMed] [Google Scholar]
- 3.Mohammed R.N., Tamjidifar R., Rahman H.S., Adili A., Ghoreishizadeh S., Saeedi H., et al. A comprehensive review about immune responses and exhaustion during coronavirus disease (COVID-19) Cell Commun. Signal. 2022;20(1):1–10. doi: 10.1186/s12964-022-00856-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y., Yang C., Xu X.-f, Xu W., Liu S.-w. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharm. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zang R., Castro M.F.G., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc3582. eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helal M.A., Shouman S., Abdelwaly A., Elmehrath A.O., Essawy M., Sayed S.M., et al. Molecular basis of the potential interaction of SARS-CoV-2 spike protein to CD147 in COVID-19 associated-lymphopenia. J. Biomol. Struct. Dyn. 2022;40(3):1109–1119. doi: 10.1080/07391102.2020.1822208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akbari M., Shomali N., Faraji A., Shanehbandi D., Asadi M., Mokhtarzadeh A., et al. CD133: an emerging prognostic factor and therapeutic target in colorectal cancer. Cell Biol. Int. 2020;44(2):368–380. doi: 10.1002/cbin.11243. [DOI] [PubMed] [Google Scholar]
- 11.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B., Li R., Lu Z., Huang Y. Does comorbidity increase the risk of patients with COVID-19: evidence from meta-analysis. Aging. 2020;12(7):6049. doi: 10.18632/aging.103000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGill A.R., Kahlil R., Dutta R., Green R., Howell M., Mohapatra S., et al. SARS–CoV-2 immuno-pathogenesis and potential for diverse vaccines and therapies: opportunities and challenges. Infect. Dis. Rep. 2021;13(1):102–125. doi: 10.3390/idr13010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamilos G., Lionakis M.S., Kontoyiannis D.P. Are all patients with cancer at heightened risk for severe Coronavirus Disease 2019 (COVID-19)? Clin. Infect. Dis. 2021;72(2):351–356. doi: 10.1093/cid/ciaa1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultze J.L., Aschenbrenner A.C. COVID-19 and the human innate immune system. Cell. 2021;184(7):1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machado D., Pizzorno A., Hoffmann J., Traversier A., Endtz H., Lina B., et al. Role of p53/NF-κB functional balance in respiratory syncytial virus-induced inflammation response. J. Gen. Virol. 2018;99(4):489–500. doi: 10.1099/jgv.0.001040. [DOI] [PubMed] [Google Scholar]
- 17.Yuan L., Chen Z., Song S., Wang S., Tian C., Xing G., et al. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J. Biol. Chem. 2015;290(5):3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akbari M., Shanehbandi D., Asadi M., Shomali N., Faraji A., Khaze V., et al. Effects of CD133 silencing on survival and migration of HT-29 colorectal cancer cells. Iran. J. Immunol. 2019;16(3):246–257. doi: 10.22034/IJI.2019.80275. [DOI] [PubMed] [Google Scholar]
- 19.Dai M., Liu D., Liu M., Zhou F., Li G., Chen Z., et al. Patients with cancer appear more vulnerable to SARS-CoV-2: a multicenter study during the COVID-19 outbreak. Cancer Discov. 2020;10(6):783–791. doi: 10.1158/2159-8290.CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Dam P.A., Huizing M., Mestach G., Dierckxsens S., Tjalma W., Trinh X.B., et al. SARS-CoV-2 and cancer: are they really partners in crime? Cancer Treat. Rev. 2020;89 doi: 10.1016/j.ctrv.2020.102068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H., Li H.-B., Lyu J.-R., Lei X.-M., Li W., Wu G., et al. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect. Dis. 2020;96:19–24. doi: 10.1016/j.ijid.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao R., Hernandez K., Huang L., Luke J.J. ACE2 and TMPRSS2 expression by clinical, HLA, immune, and microbial correlates across 34 human cancers and matched normal tissues: implications for SARS-CoV-2 COVID-19. J. Immunother. Cancer. 2020;8:2. doi: 10.1136/jitc-2020-001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C., Wang K., Zhang M., Hu X., Hu T., Liu Y., et al. High expression of ACE2 and TMPRSS2 and clinical characteristics of COVID-19 in colorectal cancer patients. NPJ Precis. Oncol. 2021;5(1):1–7. doi: 10.1038/s41698-020-00139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamers M.M., Beumer J., Van Der Vaart J., Knoops K., Puschhof J., Breugem T.I., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizutani T., Ishizaka A., Koga M., Ikeuchi K., Saito M., Adachi E., et al. Correlation analysis between gut microbiota alterations and the cytokine response in patients with coronavirus disease during hospitalization. Microbiol. Spectr. 2022;10(2) doi: 10.1128/spectrum.01689-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deriu E., Boxx G.M., He X., Pan C., Benavidez S.D., Cen L., et al. Influenza virus affects intestinal microbiota and secondary salmonella infection in the gut through type I interferons. PLoS Pathog. 2016;12(5) doi: 10.1371/journal.ppat.1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zha L., Garrett S., Sun J. Salmonella infection in chronic inflammation and gastrointestinal cancer. Diseases. 2019;7(1):28. doi: 10.3390/diseases7010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tinsley A., Navabi S., Williams E.D., Liu G., Kong L., Coates M.D., et al. Increased risk of influenza and influenza-related complications among 140,480 patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2019;25(2):369–376. doi: 10.1093/ibd/izy243. [DOI] [PubMed] [Google Scholar]
- 29.Sánchez-Alcoholado L., Ramos-Molina B., Otero A., Laborda-Illanes A., Ordóñez R., Medina J.A., et al. The role of the gut microbiome in colorectal cancer development and therapy response. Cancers. 2020;12(6):1406. doi: 10.3390/cancers12061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuo T., Zhang F., Lui G.C., Yeoh Y.K., Li A.Y., Zhan H., et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159(3) doi: 10.1053/j.gastro.2020.05.048. (944-955. e8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akbari M., Adili A., Faraji A., Pakdel A., Aslaminabad R., Nasrabadi D., et al. Restoration of miR-124 serves as a promising therapeutic approach in CRC by affecting CDK6 which is itself a prognostic and diagnostic factor. Gene Rep. 2021;24 [Google Scholar]
- 32.Street W. Cancer facts & figures 2020. Am. Cancer Soc. 2020 [Google Scholar]
- 33.Azar M., Aghazadeh H., Mohammed H.N., Sara M.R.S., Hosseini A., Shomali N., et al. miR-193a-5p as a promising therapeutic candidate in colorectal cancer by reducing 5-FU and oxaliplatin chemoresistance by targeting CXCR4. Int. Immunopharmacol. 2021;92 doi: 10.1016/j.intimp.2020.107355. [DOI] [PubMed] [Google Scholar]
- 34.Tamjidifar R., Akbari M., Tarzi S., Sadeghzadeh M., Abolghasemi M., Poursaei E., et al. Prognostic and diagnostic values of miR-506 and SPON 1 in colorectal cancer with clinicopathological considerations. J. Gastrointest. Cancer. 2021;52(1):125–129. doi: 10.1007/s12029-019-00356-0. [DOI] [PubMed] [Google Scholar]
- 35.Louis P., Hold G.L., Flint H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 36.Guinane C.M., Cotter P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 2013;6(4):295–308. doi: 10.1177/1756283X13482996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Den Besten G., Van Eunen K., Groen A.K., Venema K., Reijngoud D.-J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva Y.P., Bernardi A., Frozza R.L. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 2020;11:25. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhar D., Mohanty A. Gut microbiota and Covid-19-possible link and implications. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang A.T., Marsland B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019;12(4):843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 41.Carabotti M., Scirocco A., Maselli M.A., Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol.: Q. Publ. Hell. Soc. Gastroenterol. 2015;28(2):203. [PMC free article] [PubMed] [Google Scholar]
- 42.Terruzzi I., Senesi P. Does intestinal dysbiosis contribute to an aberrant inflammatory response to severe acute respiratory syndrome coronavirus 2 in frail patients? Nutrition. 2020;79 doi: 10.1016/j.nut.2020.110996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scanlan P.D., Shanahan F., Clune Y., Collins J.K., O'Sullivan G.C., O'Riordan M., et al. Culture‐independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ. Microbiol. 2008;10(3):789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- 44.Gopalakrishnan V., Helmink B.A., Spencer C.N., Reuben A., Wargo J.A. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou S., Fang L., Lee M.H. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol. Rep. 2018;6(1):1–12. doi: 10.1093/gastro/gox031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zackular J.P., Baxter N.T., Iverson K.D., Sadler W.D., Petrosino J.F., Chen G.Y., et al. The gut microbiome modulates colon tumorigenesis. MBio. 2013;4(6):e00692–13. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raskov H., Burcharth J., Pommergaard H.-C. Linking gut microbiota to colorectal cancer. J. Cancer. 2017;8(17):3378. doi: 10.7150/jca.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vitetta L., Vitetta G., Hall S. Immunological tolerance and function: associations between intestinal bacteria, probiotics, prebiotics, and phages. Front. Immunol. 2018:2240. doi: 10.3389/fimmu.2018.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Binda C., Lopetuso L.R., Rizzatti G., Gibiino G., Cennamo V., Gasbarrini A. Actinobacteria: a relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 2018;50(5):421–428. doi: 10.1016/j.dld.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 50.Magne F., Gotteland M., Gauthier L., Zazueta A., Pesoa S., Navarrete P., et al. The firmicutes/bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients. 2020;12:5. doi: 10.3390/nu12051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwabuchi N., Takahashi N., Xiao J.z., Miyaji K., Iwatsuki K. In vitro Th1 cytokine‐independent Th2 suppressive effects of bifidobacteria. Microbiol. Immunol. 2007;51(7):649–660. doi: 10.1111/j.1348-0421.2007.tb03953.x. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen T.T., Ung T.T., Kim N.H., Jung Y.D. Role of bile acids in colon carcinogenesis. World J. Clin. Cases. 2018;6(13):577–588. doi: 10.12998/wjcc.v6.i13.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridlon J.M., Wolf P.G., Gaskins H.R. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes. 2016;7(3):201–215. doi: 10.1080/19490976.2016.1150414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiang J.Y. Bile acid metabolism and signaling. Compr. Physiol. 2013;3(3):1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molinero N., Ruiz L., Sánchez B., Margolles A., Delgado S. Intestinal bacteria interplay with bile and cholesterol metabolism: implications on host physiology. Front. Physiol. 2019:185. doi: 10.3389/fphys.2019.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernstein C., Holubec H., Bhattacharyya A.K., Nguyen H., Payne C.M., Zaitlin B., et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch. Toxicol. 2011;85(8):863–871. doi: 10.1007/s00204-011-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mendes M.C.S., Paulino D.S., Brambilla S.R., Camargo J.A., Persinoti G.F., Carvalheira J.B.C. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World J. Gastroenterol. 2018;24(18):1995. doi: 10.3748/wjg.v24.i18.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin N.-R., Whon T.W., Bae J.-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Rizzatti G., Lopetuso L., Gibiino G., Binda C., Gasbarrini A. Proteobacteria: a common factor in human diseases. BioMed Res. Int. 2017;2017 doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Oliveira A.P., Lopes A.L.F., Pacheco G., Nolêto I.R.d.S.G., Nicolau L.A.D., Medeiros J.V.R. Premises among SARS-CoV-2, dysbiosis and diarrhea: walking through the ACE2/mTOR/autophagy route. Med. Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.110243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Imai Y., Kuba K., Penninger J.M. The discovery of angiotensin‐converting enzyme 2 and its role in acute lung injury in mice. Exp. Physiol. 2008;93(5):543–548. doi: 10.1113/expphysiol.2007.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silhol F., Sarlon G., Deharo J.-C., Vaïsse B. Downregulation of ACE2 induces overstimulation of the renin–angiotensin system in COVID-19: should we block the renin–angiotensin system? Hypertens. Res. 2020;43(8):854–856. doi: 10.1038/s41440-020-0476-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mönkemüller K., Fry L.C., Rickes S. Systemic inflammatory response and thrombosis due to alterations in the gut microbiota in COVID-19. Rev. Esp. Enferm. Dig.: Organo Of. Soc. Esp. Patol. Dig. 2020;112(7):584–585. doi: 10.17235/reed.2020.7297/2020. [DOI] [PubMed] [Google Scholar]
- 65.Zheng B., Huang C., Guo F., Lu H., Gao H., Zhang H., et al. Alterations of the gut microbiota in patients with COVID-19 or H1N1 influenza. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang L., Gu S., Gong Y., Li B., Lu H., Li Q., et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering. 2020;6(10):1178–1184. doi: 10.1016/j.eng.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tao W., Zhang G., Wang X., Guo M., Zeng W., Xu Z., et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med. Microecol. 2020;5 doi: 10.1016/j.medmic.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H., Ai J.-W., Yang W., Zhou X., He F., Xie S., et al. Metatranscriptomic characterization of coronavirus disease 2019 identified a host transcriptional classifier associated with immune signaling. Clin. Infect. Dis. 2021;73(3):376–385. doi: 10.1093/cid/ciaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zuo T., Liu Q., Zhang F., Lui G.C.-Y., Tso E.Y., Yeoh Y.K., et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2021;70(2):276–284. doi: 10.1136/gutjnl-2020-322294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H., Wang H., Sun Y., Ren Z., Zhu W., Li A., et al. Potential associations between microbiome and COVID-19. Front. Med. 2021;8 doi: 10.3389/fmed.2021.785496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuo T., Zhang F., Lui G.C.Y., Yeoh Y.K., Li A.Y.L., Zhan H., et al. Alterations in gut microbiota of patients With COVID-19 during time of hospitalization. Gastroenterology. 2020;159(3):944–955. doi: 10.1053/j.gastro.2020.05.048. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patel P., Roper J. Gut microbiome composition is associated with COVID-19 disease severity. Gastroenterology. 2021;161(2):722–724. doi: 10.1053/j.gastro.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang L., Gu S., Gong Y., Li B., Lu H., Li Q., et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering. 2020;6(10):1178–1184. doi: 10.1016/j.eng.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venzon M., Bernard-Raichon L., Klein J., Axelrad J.E., Zhang C., Hussey G.A., et al. Gut microbiome dysbiosis during COVID-19 is associated with increased risk for bacteremia and microbial translocation. bioRxiv: Prepr. Serv. Biol. 2022 [Google Scholar]
- 75.Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veterini A.S., Andriyanto L., Hamzah H.A. Case report: respiratory manifestations of COVID-19 starting with a gastrointestinal complaint: a coincidence or a correlation? Afr. J. Infect. Dis. 2021;15(2 Suppl.):S31–S37. doi: 10.21010/ajidv15i2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Howell M.C., Green R., McGill A.R., Dutta R., Mohapatra S., Mohapatra S.S. SARS-CoV-2-induced gut microbiome dysbiosis: implications for colorectal cancer. Cancers. 2021;13(11) doi: 10.3390/cancers13112676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu T.F.D., Philippou E., Kolokotroni O., Siakallis G., Rahima K., Constantinou C. Gut and airway microbiota and their role in COVID-19 infection and pathogenesis: a scoping review. Infection. 2022;50(4):815–847. doi: 10.1007/s15010-021-01715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jayasimhan A., Mariño E. Dietary SCFAs, IL-22, and GFAP: the three musketeers in the gut–neuro–immune network in type 1 diabetes. Front. Immunol. 2019:2429. doi: 10.3389/fimmu.2019.02429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McNabney S.M., Henagan T.M. Short chain fatty acids in the colon and peripheral tissues: a focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients. 2017;9(12):1348. doi: 10.3390/nu9121348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolff L., Martiny D., Deyi V.Y.M., Maillart E., Clevenbergh P., Dauby N. COVID-19–associated Fusobacterium nucleatum bacteremia, Belgium. Emerg. Infect. Dis. 2021;27(3):975. doi: 10.3201/eid2703.202284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun C.-H., Li B.-B., Wang B., Zhao J., Zhang X.-Y., Li T.-T., et al. The role of Fusobacterium nucleatum in colorectal cancer: from carcinogenesis to clinical management. Chronic Dis. Transl. Med. 2019;5(03):178–187. doi: 10.1016/j.cdtm.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gur C., Ibrahim Y., Isaacson B., Yamin R., Abed J., Gamliel M., et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Y., Weng W., Peng J., Hong L., Yang L., Toiyama Y., et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κb, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152(4):851–866. doi: 10.1053/j.gastro.2016.11.018. (.e24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shomali N., Hatamnezhad L.S., Tarzi S., Tamjidifar R., Xu H., Shotorbani S.S. Heat shock proteins regulating toll-like receptors and the immune system could be a novel therapeutic target for melanoma. Curr. Mol. Med. 2021;21(1):15–24. doi: 10.2174/1566524020666200511091540. [DOI] [PubMed] [Google Scholar]
- 86.Zuo T., Zhan H., Zhang F., Liu Q., Tso E.Y., Lui G.C., et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159(4):1302–1310. doi: 10.1053/j.gastro.2020.06.048. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zuo T., Zhan H., Zhang F., Liu Q., Tso E.Y.K., Lui G.C.Y., et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159(4):1302–1310. doi: 10.1053/j.gastro.2020.06.048. (.e5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leviner S. Recognizing the clinical sequelae of COVID-19 in adults: COVID-19 Long-Haulers. J. Nurse Pract. 2021;17(8):946–949. doi: 10.1016/j.nurpra.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S., et al. Post-acute COVID-19 syndrome. Nat. Med. 2021;27(4):601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amenta E.M., Spallone A., Rodriguez-Barradas M.C., El Sahly H.M., Atmar R.L., Kulkarni P.A., editors. Postacute COVID-19: An Overview and Approach to Classification. Open Forum Infectious Diseases. Oxford University Press; US: 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carfì A., Bernabei R., Landi F. Persistent symptoms in patients after acute COVID-19. Jama. 2020;324(6):603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gao Q.Y., Chen Y.X., Fang J.Y. Novel coronavirus infection and gastrointestinal tract. J. Dig. Dis. 2019;21(3):125. doi: 10.1111/1751-2980.12851. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yasui H., Kiyoshima J., Hori T., Shida K. Protection against influenza virus infection of mice fed Bifidobacterium breve YIT4064. Clin. Diagn. Lab. Immunol. 1999;6(2):186–192. doi: 10.1128/cdli.6.2.186-192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Q., Hu J., Feng J.-W., Hu X.-T., Wang T., Gong W.-X., et al. Influenza infection elicits an expansion of gut population of endogenous Bifidobacterium animalis which protects mice against infection. Genome Biol. 2020;21(1):1–26. doi: 10.1186/s13059-020-02007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baindara P., Chakraborty R., Holliday Z., Mandal S., Schrum A. Elsevier; 2021. Oral Probiotics in Coronavirus Disease 2019: Connecting the Gut–lung Axis to Viral Pathogenesis, Inflammation, Secondary Infection and Clinical Trials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang L., Han H., Li X., Chen C., Xie X., Su G., et al. Probiotics use is associated with improved clinical outcomes among hospitalized patients with COVID-19. Ther. Adv. Gastroenterol. 2021;14 doi: 10.1177/17562848211035670. (17562848211035670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hung Y.P., Lee C.C., Lee J.C., Tsai P.J., Ko W.C. Gut dysbiosis during COVID-19 and potential effect of probiotics. Microorganisms. 2021;9:8. doi: 10.3390/microorganisms9081605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Broderick N.A. A common origin for immunity and digestion. Front. Immunol. 2015;6:72. doi: 10.3389/fimmu.2015.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Levin B.R., Antia R. Why we don't get sick: the within-host population dynamics of bacterial infections. Science. 2001;292(5519):1112–1115. doi: 10.1126/science.1058879. [DOI] [PubMed] [Google Scholar]
- 100.Levin B.R., Baquero F., Ankomah P.P., McCall I.C. Phagocytes, antibiotics, and self-limiting bacterial infections. Trends Microbiol. 2017;25(11):878–892. doi: 10.1016/j.tim.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 101.Azkur A.K., Akdis M., Azkur D., Sokolowska M., van de Veen W., Brüggen M.C., et al. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy. 2020;75(7):1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McFall-Ngai M., Hadfield M.G., Bosch T.C., Carey H.V., Domazet-Lošo T., Douglas A.E., et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Troisi J., Venutolo G., Tanyà M.P., Carri M.D., Landolfi A., Fasano A. COVID-19 and the gastrointestinal tract: source of infection or merely a target of the inflammatory process following SARS-CoV-2 infection? World J. Gastroenterol. 2021;27(14):1406. doi: 10.3748/wjg.v27.i14.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saltiel A.R., Olefsky J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Investig. 2017;127(1):1–4. doi: 10.1172/JCI92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garbarino J., Sturley S.L. Saturated with fat: new perspectives on lipotoxicity. Curr. Opin. Clin. Nutr. Metab. Care. 2009;12(2):110–116. doi: 10.1097/MCO.0b013e32832182ee. [DOI] [PubMed] [Google Scholar]
- 106.González O.A., Tobia C., Ebersole J.L., Novak M.J. Caloric restriction and chronic inflammatory diseases. Oral Dis. 2012;18(1):16–31. doi: 10.1111/j.1601-0825.2011.01830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Odun-Ayo F., Reddy L. Gastrointestinal microbiota dysbiosis associated with SARS-CoV-2 infection in colorectal cancer: the implication of probiotics. Gastroenterol. Insights. 2022;13(1):35–59. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.