Significance
Cyanobacteria have the simplest known circadian clock, but the mechanisms that connect the oscillator to cellular physiology are incompletely understood. We identify a previously unknown component of the circadian system, which we name KidA. We show that PAS domains in KidA bind to the core oscillator protein KaiB and shorten the clock period. Through both experiments and simulations, we show that the speed of the oscillator can be tuned by modulating alternative KaiB folds. The discovery of KidA suggests a general role for PAS domains in prokaryotic timekeeping and may be a link between circadian rhythms and bis-(3'-5')-cyclic dimeric guanosine monophosphate, a molecule implicated in the regulation of biofilm and the motile-sessile transition.
Keywords: circadian rhythms, cyanobacteria, diguanylate cyclase
Abstract
The cyanobacterial clock presents a unique opportunity to understand the biochemical basis of circadian rhythms. The core oscillator, composed of the KaiA, KaiB, and KaiC proteins, has been extensively studied, but a complete picture of its connection to the physiology of the cell is lacking. To identify previously unknown components of the clock, we used KaiB locked in its active fold as bait in an immunoprecipitation/mass spectrometry approach. We found that the most abundant interactor, other than KaiC, was a putative diguanylate cyclase protein predicted to contain multiple Per-Arnt-Sim (PAS) domains, which we propose to name KidA. Here we show that KidA directly binds to the fold-switched active form of KaiB through its N-terminal PAS domains. We found that KidA shortens the period of the circadian clock both in vivo and in vitro and alters the ability of the clock to entrain to light-dark cycles. The dose-dependent effect of KidA on the clock period could be quantitatively recapitulated by a mathematical model in which KidA stabilizes the fold-switched form of KaiB, favoring rebinding to KaiC. Put together, our results show that the period and amplitude of the clock can be modulated by regulating the access of KaiB to the fold-switched form.
Circadian rhythms are internally generated oscillations that allow organisms throughout the tree of life to anticipate the daily cycle in their environments. These systems typically include a core pacemaker along with a suite of additional factors responsible for transducing signals into and out of the oscillator. While much is now understood about the basic mechanisms that produce rhythms, a full picture of the extended clock network in any organism is missing. Cyanobacteria have emerged as a model system for studying circadian rhythms where genetic analysis is straightforward and biochemical mechanism is uniquely accessible. The core circadian pacemaker can be reconstituted with a mixture of three proteins, KaiA, KaiB, and KaiC, and phosphorylation rhythms in this purified system show many of the properties of the intact clock (1–3). The ability to add precisely defined components to this test-tube clock allows us to characterize how the core oscillator interacts with other components, making it a powerful tool to map out the circadian organization of the entire cell. This approach has been successfully used to extend the KaiABC oscillator to study the role of two signaling kinases, SasA and CikA, that regulate the phosphorylation status of a key output transcription factor RpaA (4). However, our understanding of the full circadian regulatory network and its relationship to the purified oscillator is far from complete.
Biochemical analysis of the Kai oscillator has made it clear that some interactions are regulated to occur only at specific clock times. This presents a challenge for protein-protein interaction studies because some protein complexes may not form unless the oscillator can be trapped in a physiologically relevant state. A key difference between the daytime and nighttime forms of the oscillator complex is that KaiB fold-switches into an alternative secondary structure to interact with KaiC during the subjective night. KaiB mutants that are locked into this fold (fsKaiB) have been used to successfully capture protein complexes for structural analysis (5, 6).
In this study, we sought to identify previously unknown components that interact with the Kai oscillator using an fsKaiB mutant as bait for coimmunoprecipitation from native cell lysate. This strategy has the potential to detect interactors that may have escaped previous screens, including hypothetical nighttime signaling pathways, a phase of the cycle where clock physiology is poorly understood. This approach led us to identify a previously uncharacterized protein, which we named KidA, as an interactor of the Kai oscillator. We show that deleting kidA or increasing the expression level of KidA leads to changes in the properties of the clock, including a dose-dependent shortening of the period. This shortening of the period is due to direct interaction of KidA with the Kai proteins and can be reconstituted in vitro. We trace this interaction to the N-terminal PAS domains of KidA, which bind to fold-switched KaiB. The trends in period and amplitude emerge with little parameter tuning when we introduce terms representing KidA binding to and stabilizing fold-switched KaiB to a molecularly detailed mathematical model of the Kai oscillator. This suggests that this mechanism is sufficient to account for the observed behavior. Put together, our results both qualitatively and quantitatively show how regulating the switching of a protein fold can tune the oscillator and how the clock may couple to other systems within the cell.
Results
Identification of KidA via co-IP/MS of Fold-Switched KaiB.
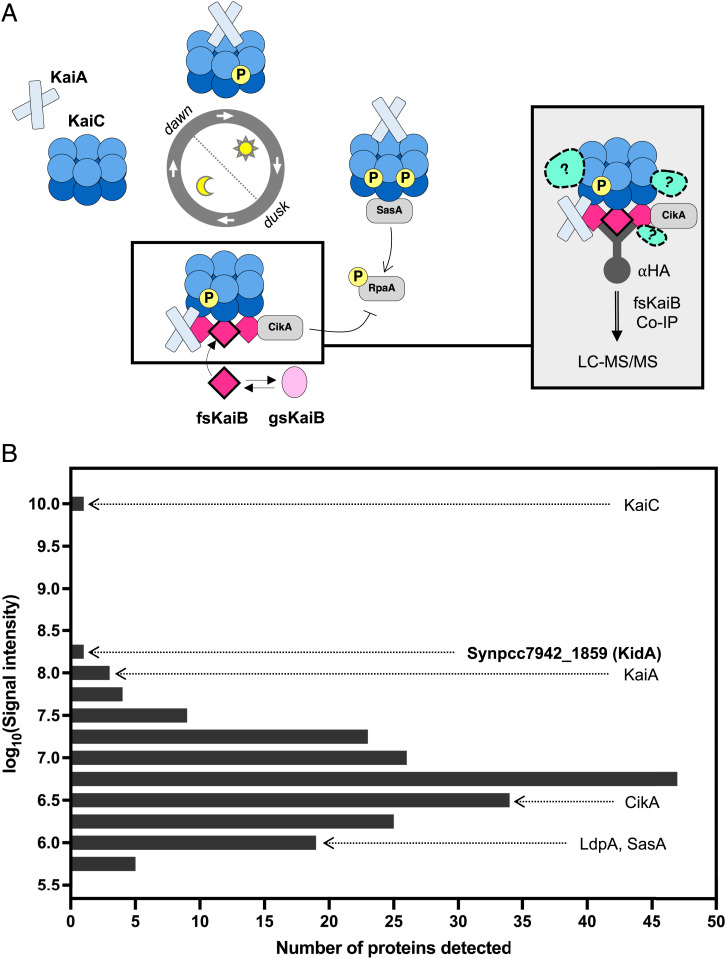
To search for previously unknown clock interactors, we utilized a co-immunoprecipitation/mass spectrometry (co-IP/MS) approach using a Synechococcus elongatus strain expressing an epitope-tagged KaiB mutant that constitutively adopts a fold-switched secondary structure found in KaiB•KaiC complexes (Fig. 1A). We generated a ranked list of candidate interactors based on the integrated signal intensity in liquid chromatography with tandem mass spectrometry (LC-MS/MS) for all peptides derived from a given protein, excluding KaiB itself and proteins found in a negative control wildtype sample. The strongest signal came from the core clock protein KaiC, known to interact directly with KaiB. Other oscillator components and known clock interactors, KaiA, CikA, LdpA, and SasA ranked #5, #118, #179, and #182, respectively. Unexpectedly, the protein with the second highest intensity was a previously uncharacterized protein that corresponds to Synpcc7942_1859 (Fig. 1B and SI Appendix, Tables 2 and 3). Domain prediction of this protein by Basic Local Alignment Search Tool (BLAST) identified C-terminal GGDEF and EAL domains, typically involved in bis-(3'-5')-cyclic dimeric guanosine monophosphate (c-di-GMP) production and hydrolysis. On this basis, we propose to name this gene kidA (kai-interacting diguanylate cyclase A). Alignment of the KidA sequence with enzymes with active GGDEF or EAL domains shows that KidA contains conserved residues needed for enzymatic activity (SI Appendix, Fig. S1).
Fig. 1.
KidA is identified as a potential clock interactor from fsKaiB co-IP/MS. (A) Experimental scheme to identify potential interactors of the nighttime state of the cyanobacterial oscillator. (Left) the circadian rhythm manifests as an oscillation of protein complexes. Near dusk, fold-switched KaiB molecules bind to KaiC. (Right) Unknown interactors may be detected by coprecipitation with a fold-switched KaiB mutant (G88A;D90R). (B) Histogram of proteins that were detected in the fsKaiB-HA co-IP/MS with 19x or higher signal intensity compared with the negative control (WT strain). Bins containing Kai proteins, known Kai protein interactors, and a previously undescribed candidate (KidA) are marked on the histogram.
The Period of the Circadian Rhythm Varies with KidA Expression Level.
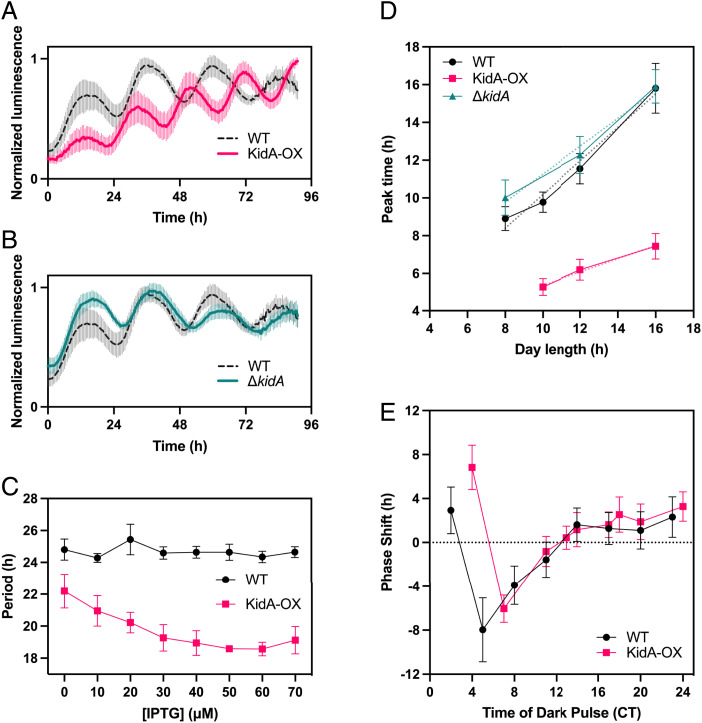
To determine the effect of KidA on the circadian rhythm, we monitored the in vivo clock rhythms of WT, ΔkidA, and inducible overexpression of KidA (KidA-OX) strains using a bioluminescent clock reporter assay (Fig. 2 A–C and SI Appendix, Fig. S2A). Induction of KidA led to a marked shortening of the free-running period compared with the wildtype; 10 μM (Isopropyl β-D-1 thiogalactopyranoside) IPTG induction shortened the free-running period by ∼3 h (Fig. 2A), while ΔkidA slightly lengthened the period by ∼2 h (Fig. 2B). Moreover, the period-shortening effect of KidA-OX depended on the concentration of the inducer, up to ∼6 h with maximal induction (Fig. 2C). Leaky expression in the absence of inducer showed a smaller but detectable effect. While point mutations in kaiC can cause large period changes, the period-shortening effect we observe here is large compared with the effect of known period mutants outside of the kai genes themselves (7), e.g., ldpA mutants (1 h) (8), cikA mutants (2.5 h) (9), and sasA mutants (3 h) (10). The short-period rhythms in the KidA-OX strain at low induction levels retain high amplitude. Above 40 μM IPTG, synchronized rhythms are not sustained after the first three cycles in constant light (SI Appendix, Fig. S2B).
Fig. 2.
KidA overexpression (KidA-OX) mutants exhibit shorter in vivo clock rhythms and altered entrainment. (A and B) Bioluminescence (PpsbAI::luxAB, PpsbAI::luxCDE) time traces of WT, KidA-OX, and ΔkidA cultures induced with 10 µM IPTG. (A) WT vs. KidA-OX (B) WT vs. ΔkidA. Each trace was normalized by dividing by the maximum value. The solid and dotted lines show the mean normalized signal and the spread shows the SD. Average period obtained from fitting: WT = 23.5 ± 0.2 h, n = 8; KidA-OX = 20.1 ± 0.1 h, n = 7; ΔkidA = 25.3 ± 0.4 h, n = 8. Traces are aligned so that t = 0 occurs at a trough after three cycles in constant light. (C) The mean best-fit periods of the KidA-OX strain induced using different concentrations of IPTG and the WT strain characterized in the same conditions. Average period obtained from fitting at 50 µM IPTG: WT = 24.6 ± 0.6 h; KidA-OX = 18.6 ± 0.1 h. Error bars show the SD (n = 3–4). (D) Clock phases of WT, KidA-OX, and ΔkidA induced with 10 µM IPTG after entrainment to different photoperiods (10L:14D, 12L:12D, 16L:8D). The average time of the first peak of the bioluminescence signal after dawn following four LD cycles is plotted. Error bars show SD (n = 11–23 for each sample). Dotted lines show linear regression to the data: mWT = 0.89 (CI95% = [0.47, 1.31], R2 = 0.98), mKidA-OX = 0.35 (CI95% = [–0.08, 0.78], R2 = 0.99), mΔkidA = 0.74 (CI95% = [–0.54, 2.01], R2 = 0.98). (E) Phase response curves of WT and KidA-OX induced with 10 µM IPTG. Entrained cultures were allowed to free-run in constant light and then were subjected to a 5 h dark pulse at various times. The mean difference between the first peak of the reporter after the dark pulse and the closest peak time in the control (phase shift) is plotted. Error bars show the SD (n = 3–20 for each sample). CT, circadian time.
KidA OverexpressionAlters the Relationship of Entrained Phase to Photoperiod.
Conceptually, circadian clocks can be divided into oscillator mechanisms that maintain self-sustaining rhythms and input mechanisms that use signals from the environment to adjust the phase of the oscillation. To determine whether KidA plays an input-specific role in the clock, we first investigated whether KidA affects the entrainment of the clock to different photoperiods. Previous work has shown that a reporter of kaiBC gene expression has a linear dependence of peak phase on day length, with a slope of about 0.5 (midday tracking), when entrained with 24 h cycles of varied photoperiods (3). We performed a similar experiment using the KidA-OX strain, subjecting the cells to 8:16, 10:14, 12:12, or 16:8 light-dark entrainment cycles. We found that the phase of the psbAI reporter signal from the KidA-OX strain had a very different dependence on photoperiod (KidA-OX slope m = 0.35) compared with matched wildtype (WT) and ΔkidA strains (WT slope m = 0.89, ΔkidA slope m = 0.74) (Fig. 2D). A change in the slope of entrained phase vs. photoperiod is not expected solely from the shortened free-running period seen in KidA-OX and likely requires a change in the response of the clock to the day-night cycle (3).
A possible explanation for this effect could be that the KidA-OX strain is hypersensitive to dark pulses, and the phase is reset every night. We thus investigated whether the KidA-OX strain had an altered phase response curve for 5 h dark pulses, a condition that can produce strong resetting from out-of-phase dark pulses in WT cells (11). We were additionally motivated to test this possibility because a known interactor of fsKaiB, CikA, is implicated in the dark pulse-induced resetting of the clock (9). However, after correcting for the shortened period, the KidA-OX strain did not have a noticeable difference from the WT both in terms of the maximum phase advance and maximum phase delay, suggesting that the photoperiodic entrainment phenotype is not caused simply by increased sensitivity to a short dark pulse (Fig. 2E).
The N-Terminal Region of KidA Is Composed of Multiple PAS Domains, Which Are Responsible for the Period-Shortening Effect of KidA.
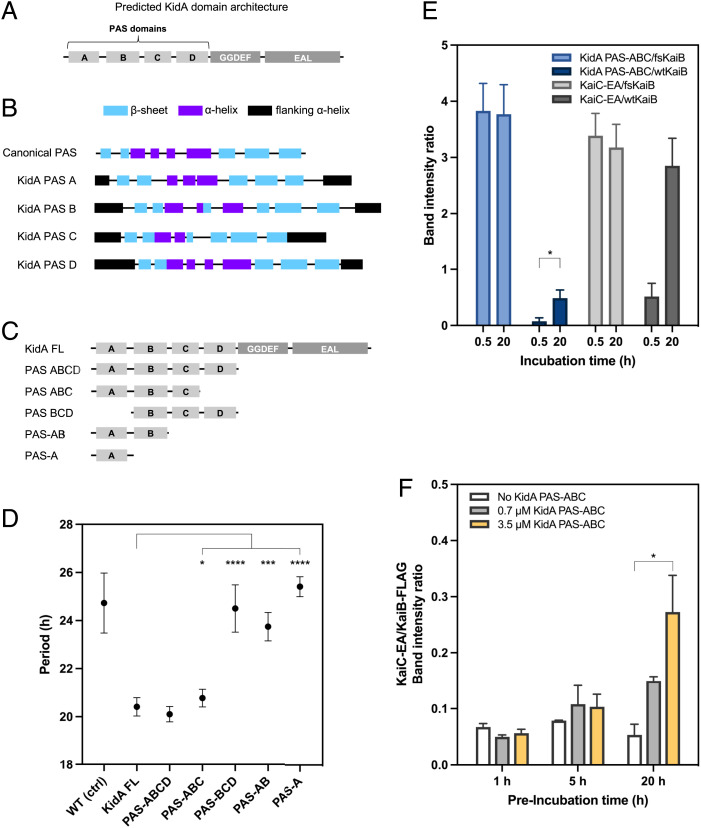
To understand the biochemical basis of the period-shortening phenotype of the KidA-OX strain, we first used structural prediction to further dissect KidA’s domain architecture beyond the GGDEF and EAL domains predicted by BLAST in the C-terminal region. We used the Phyre2 algorithm to generate local three-dimensional folding predictions for the N-terminal region of the protein. Four tandem local structures that matched the canonical Per-Arnt-Sim (PAS) domain topology were predicted to form in the N-terminal region (12) (Fig. 3B). Based on these results, we propose that the full-length KidA protein architecture is composed of four PAS-like domains followed by a GGDEF domain and an EAL domain (Fig. 3A). We denote these predicted domains PAS-A, -B, -C, and -D, respectively.
Fig. 3.
An N-terminal fragment of KidA is sufficient for the clock phenotype and binds to fold-switched KaiB. (A) Proposed KidA domain architecture derived from BLAST alignment and secondary structure prediction. (B) Phyre2 secondary structure prediction suggests four tandem domains with PAS-like folds. Canonical PAS architecture was modified from Möglich et al. (23). In the schematic, the first flanking alpha helix is the same as the last flanking alpha helix of the previous PAS domain. Predicted domain boundaries between the first and last β-sheets: PAS-A (22–114); PAS-B (145– 246); PAS-C (270–350); PAS-D (373–472). (C) Domain truncation mutants of KidA expressed in vivo. (D) Free-running clock periods of S. elongatus strains expressing KidA fragments described in (C). The average periods are plotted, and error bars show the SD (n = 4–16). The PAS-A construct expressed poorly compared with the other constructs. (E) In vitro co-IP of KidA PAS-ABC fragment (3.5 µM) using fsKaiB-FLAG or wtKaiB-FLAG (3.5 µM) as bait in a 30-min or 20-h coincubation; KaiC S431E;T432A (KaiC-EA) served as a positive control for KaiB interaction. Band intensities were determined by densitometry after background subtraction. Mean band intensity ratios are plotted, error bars show the SD (n = 3 for each condition). (F) In vitro co-IP of KaiC-EA using wtKaiB-FLAG (3.5 µM) as bait after preincubating wildtype KaiB-FLAG with varying amounts (0, 0.7, or 3.5 µM) of KidA PAS-ABC for varying lengths of time (1, 5, or 20 h). KaiC-EA was added 5 min prior to IP. Band intensities were determined by densitometry after background subtraction. Mean band intensity ratios are plotted, error bars show the SD (n = 2–3 for each condition). (D–F) All significance analyses were performed using Welch’s t test. * = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001.
Many PAS domains are known to bind to small-molecule ligands. We attempted to predict ligand-binding sites by aligning the KidA domain sequences with consensus sequences for PAS domain families whose structure and ligand-binding sites have been well characterized (13). This approach did not yield positive results for PAS-A, PAS-B, or PAS-C. However, the PAS-D sequence contains some of the conserved residues present in both the light-oxygen-voltage-sensing (LOV) domain’s flavin mononucleotide (FMN)-binding region and the MmoS/NifL domain’s flavin adenine dinucleotide (FAD)-binding region; this suggests that KidA PAS-D may interact with a flavin cofactor, though it lacks the cysteine involved in canonical LOV domain photochemistry (SI Appendix, Fig. S3).
We next sought to identify a fragment necessary and sufficient for the period-shortening effect of KidA-OX. We created a series of domain truncation mutants of KidA and characterized the circadian rhythms of strains overexpressing these fragments. This analysis revealed that the first three PAS domains (KidA PAS-ABC) are sufficient to phenocopy full-length KidA’s period-shortening effect. Constructs that lack either PAS-A or PAS-C did not shorten the period (Fig. 3D). Thus, we conclude that the GGDEF and EAL domains are not involved in the period phenotype, PAS-D is dispensable, and KidA engages the oscillator through the three N-terminal PAS domains.
KidA Directly Binds Fold-Switched KaiB through PAS Domains In Vitro.
Because we initially identified KidA via pulldown with KaiB, we investigated whether the two purified proteins directly interact by using an in vitro co-IP approach. We used the KidA fragment spanning the first three PAS domains (KidA PAS-ABC) shown to be sufficient for the in vivo period-shortening effect (Fig. 3 C and D). Coincubation of KidA PAS-ABC with wildtype KaiB (wtKaiB-FLAG) resulted in direct binding but with slow kinetics, reaching half-maximal binding after ∼12 h (Fig. 3E and SI Appendix, Fig. S4). This behavior is similar to the kinetics observed for the binding of KaiC with KaiB, which is slow partially because of the fold-switching of KaiB (Fig. 3E) (5). We thus hypothesized that KidA PAS-ABC might bind selectively to fold-switched KaiB. To test this, we performed the same co-IP experiment using a mutant KaiB that is locked in the fold-switched form (fsKaiB-FLAG) to pull down KidA PAS-ABC (14). Much more KidA PAS-ABC was pulled down with fsKaiB-FLAG than with wtKaiB-FLAG, and protein complexes had fully formed after only a 30 min incubation (Fig. 3E). A KaiC phosphomimetic for the nighttime state (KaiC-EA) also rapidly formed complexes with fsKaiB, serving as a control for a known fsKaiB interactor (Fig. 3E). Thus, the KidA PAS-ABC region directly interacts with fold-switched KaiB. We interpret the slow kinetics of KidA PAS-ABC binding to wtKaiB as reflecting the slow conversion of the KaiB ground-state fold to the fold-switched form (5), which is then stabilized in complex with KidA. Consistent with this interpretation, KaiB that is preincubated with KidA PAS-ABC becomes competent to bind rapidly to KaiC-EA, and the amount of rapid binding increases with the preincubation time and the amount of KidA PAS-ABC(Fig. 3G). Furthermore, KidA PAS-ABC does not inhibit the assembly of KaiABC complexes when all Kai proteins are present (SI Appendix, Fig. S5).
Reconstitution of the Period-Shortening Effect of KidA In Vitro.
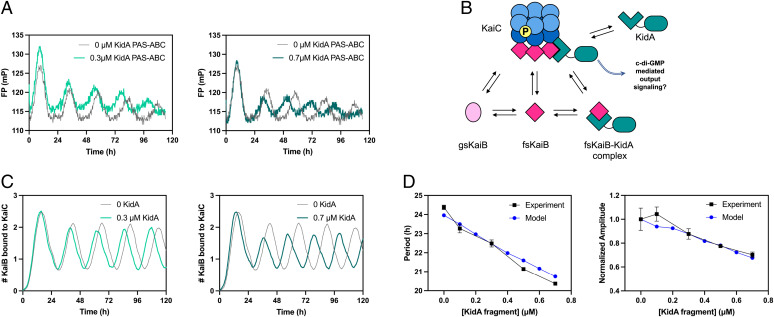
To investigate the mechanism of KidA’s period-shortening effect, we first asked whether the phenotype results from KidA’s direct interaction with the Kai proteins themselves rather than other factors in vivo. We added KidA PAS-ABC to the in vitro reconstituted Kai oscillator system composed of purified KaiA, KaiB, and KaiC, and monitored oscillations using a fluorescence polarization (FP) label on KaiB. The addition of KidA PAS-ABC shortened the period of the KaiABC oscillator (Fig. 4A), and the magnitude of period shortening was dependent on the concentration of KidA PAS-ABC (Fig. 4D, Left, black) in a manner quantitatively similar to the in vivo trend (Fig. 2A). Additionally, the amplitude of the FP rhythms decreased with increasing KidA PAS-ABC (Fig. 4D, Right, black).
Fig. 4.
In vitro and in silico reconstitution of the KidA period-shortening effect. (A) In vitro reconstitution of the period-shortening effect of KidA. The KidA PAS-ABC fragment was added to a standard mixture of KaiA, KaiB, and KaiC. Oscillations were measured by fluorescence polarization of labeled KaiB (averaged traces shown from duplicate experiments). (B) Cartoon overview of a mathematical model based on the hypothesis that KidA stabilizes the fold-switched form of KaiB by direct binding. The Paijmans model was modified to explicitly describe the interconversion between the ground-state and fold-switched state of KaiB.The protein complexes and reaction arrows shown were added to allow KidA to bind fsKaiB (SI Appendix). (C) Time course of oscillations of KaiC-bound KaiB molecules per hexamer in the mathematical model. Model parameters were chosen to have period-dependence close to the experimental value. (D) Plots of the period and amplitude of the fluorescence polarization rhythm (as shown in Fig. 4A) as a function of [KidA] and the predictions from the model with the same parameter set (as shown in Fig. 4C). (Left) Period and (Right) amplitude normalized by dividing by the amplitude value at 0 µM KidA. Mean values are plotted, and the error bars show the SD (Experimental n = 2, Model n = 20).
The enzymatic rates of KaiC are important for setting the pace of phosphorylation and dephosphorylation, and thus contribute to the overall period of oscillation (15,16). However, we could not detect any influence of KidA on phosphorylation or dephosphorylation kinetics in partial reactions that lacked KaiB (SI Appendix, Fig. S6). Thus, we turned to a mathematical model to ask whether binding interactions with KaiB alone are sufficient to change the oscillator period and amplitude.
KidA Binding to KaiB in the Fold-Switched Form Shortens the Period in a Mathematical Model.
Our data suggest a model in which KidA acts by binding and stabilizing the fsKaiB fold. To test whether this mechanism can account for our observations, we modified an existing mathematical model of the Kai oscillator to include a KidA-KaiB binding equilibrium. Specifically, we built on a model published by Paijmans et al. that accurately describes the phosphorylation dynamics and their dependence on the adenosine triphosphate:adenosine diphosphate ratio ([ATP]/[ADP]) (17). The original model assumes an inexhaustible pool of KaiB that binds slowly to KaiC. Since we hypothesized that KidA shortens the period by increasing the concentration of free fold-switched KaiB, we modified the model to track an explicit pool of KaiB monomers that can slowly interconvert between ground-state and fold-switched forms. The fold-switched molecules can bind rapidly to KaiC. KidA is introduced to the model as a species that binds specifically to fsKaiB. The model contains 10 additional parameters beyond the original model (SI Appendix, Table 4).
To characterize the KidA-dependent clock properties of the model, we used Markov chain Monte Carlo to explore parameters that supported circadian oscillations both with and without KidA present. The majority of sampled parameter sets (77%) showed shortening of the period with the addition of KidA, though the median slope of period vs. KidA concentration was less negative than that observed experimentally (–0.74 h/μM vs. –5.6 h/μM, SI Appendix, Fig. S7A). Most parameter sets (76%) also showed reduced amplitude of the KaiB•KaiC complex rhythm with increasing KidA (SI Appendix, Fig. S7B). In these analyses, only parameter sets that have close to a 24 h period without KidA were considered (SI Appendix, Fig. S7C). The predominance of parameter sets leading to period shortening and reduced amplitude without having selected for these features suggests that the qualitative effects on period and amplitude are generic features of mechanisms in which a binding partner, such as KidA, stabilizes the fold-switched KaiB state.
Analysis of the relationship between the parameters describing KidA and quantitative changes in period and amplitude revealed a notable trend (SI Appendix, Fig. S8A). The magnitude of the period and amplitude slopes are anticorrelated with both the dissociation constant (Kd) of the KidA-fsKaiB binding reaction and the off-rate (kKidAoff); tighter binding KidA exerts a stronger period-shortening effect (SI Appendix, Fig. S8 A–C). To ensure that our results were not idiosyncratic to the small number of parameter sets in the tails of the distributions in SI Appendix, Fig. S7 A–C, we harvested many additional parameter sets with period and amplitude slopes consistent with the experimentally observed dependence of the period on KidA concentration by including a restraint on the period slope during fitting (SI Appendix, Fig. S7 D–F). These parameter sets show similar correlations.
To understand this trend, we selected a parameter set that gave period and amplitude slopes close to the experimental values to study in detail (shown in Fig. 4 C and D). We found that KidA primarily affects rebinding of KaiB to KaiC after dissociation of a KaiB•KaiC complex (SI Appendix, Fig. S9A). Binding of KidA to fsKaiB increases the lifetime of fsKaiB•KidA complexes and in turn the likelihood that these complexes rebind to the N-terminal (CI) domain of KaiC before KaiB reverts to its ground-state fold. In this way, KidA increases the population of hexamers with a complete set of six KaiB monomers bound, which are the hexamers capable of sequestering KaiA on CI in the model (SI Appendix, Fig. S9C). Increased KaiA sequestration (SI Appendix, Fig. S9D) results in faster depletion of the KaiA pool available for catalyzing nucleotide exchange in the C-terminal (CII) domain and earlier dephosphorylation of KaiC hexamers, thus resulting in a shorter period. Moreover, the lower amount of free KaiA reduces the number of hexamers that enter the phosphorylation cycle and consequently the amplitude (SI Appendix, Fig. S9B). Consistent with this picture, the effect of KidA on the oscillator can be largely recapitulated by an effective model that approximates the role of KidA by adjusting the equilibrium constant between the ground-state and fold-switched forms of KaiB (SI Appendix, Fig. S10).
Discussion
In this study, we identified a previously unstudied protein that interacts with the Kai circadian oscillator using a co-IP/MS approach. KaiB, an essential component of the oscillator, is a metamorphic protein, and multiple interactions in the clock are gated by its transition between alternate folds. By using a mutant form of KaiB that is locked into its fold-switched state as bait, we attempted to stabilize complexes that might otherwise be difficult to detect. The top hit, which we named KidA, is predicted to contain multiple PAS-like domains and both GGDEF and EAL domains, involved in c-di-GMP synthesis and hydrolysis, respectively. Deletion or mild overexpression of KidA changes the free-running period of the clock; more severe overexpression causes arrhythmia. A fragment consisting of the first three PAS domains of KidA directly interacts with fold-switched KaiB and shortens the period of the KaiABC oscillator.
While mutations of KaiC are known to markedly change the period of the in vitro oscillator, typically causing a concomitant change in the KaiC ATPase cycle (18), most other manipulations, such as changing Kai protein or nucleotide concentrations produce only slight changes in the period (4). Here, we find that large period changes can also be caused by introducing a specific binder for the fold-switched state of KaiB, suggesting that regulating access to this fold plays an important role in timing.
KidA-OX in cells leads to an entrainment defect in which the clock does not appropriately set its phase relative to the photoperiod. It does not, however, show increased phase shifts in response to a brief pulse of darkness. This suggests that the regulation of KidA activity may be more complex in a LD cycle. The physiological functions of KidA are incompletely understood. One possibility is that KidA’s role is to act as a sensor for the fold-switched form of KaiB that appears in the subjective night. Information about the clock state might then be relayed to downstream processes, perhaps through diguanylate cyclase activity. Cyclic-di-GMP is a small molecule known to be involved in a variety of cellular processes in prokaryotic organisms, including regulation of biofilm formation and lifestyle transitions (19). Considering that many cyanobacteria engage in biofilm formation, the potential for KidA’s role in biofilm, and, by extension, clock regulation of biofilms is especially interesting (20).
Alternatively, a physiological role of KidA could be to adjust the period or entrainment properties of the oscillator, as occurs with mild overexpression. In this scenario, the activity of full-length KidA may be low under standard culture conditions, but with the potential for unknown signals to stimulate KidA to interact strongly with the oscillator. PAS domains can sense signals by binding a variety of small-molecule ligands as well as through protein-protein interactions (13). The presence of multiple PAS domains suggests the possibility that KidA could receive and integrate other signals besides the clock state (21), particularly since PAS-D, which is predicted by sequence to bind a flavin cofactor, is not required for the clock period phenotype. Notably, the kidA gene is adjacent to a heme oxygenase gene (Synpcc7942_1858), which suggests the possibility that KidA could bind heme, biliverdin (the product of the heme oxygenase reaction), or similar molecules.
Bacterial PAS domains frequently appear in tandem and are typically followed by enzymatic domains such as histidine kinase or diguanylate cyclase domains. The PAS domain was first described in eukaryotic proteins that associate with clock proteins (the P in PAS domain stands for the Drosophila clock protein PERIOD), and PAS domain-mediated clock protein interactions have been found throughout eukaryotes (22). Involvement of PAS domains in prokaryotic rhythms is only just becoming apparent (23). The identification of KidA provides a clear example of how a bacterial protein containing PAS domains can interact with a circadian oscillator.
There are numerous examples of PAS domain-containing genes appearing co-operonic with or adjacent to kai gene homologs in bacterial genomes outside of cyanobacteria that have kaiBC genes but no kaiA (24). This pattern of genomic organization suggests that the association of PAS domain-containing bacterial proteins with Kai proteins via PAS domains could be a generally conserved mechanism widespread in kai homologs. Further study of KidA promises to advance our understanding of how the core Kai oscillator might be linked to other cellular processes. Because it contains multiple PAS domains and enzymatic domains that can participate in c-di-GMP signaling, KidA may act as a bridge between environmental inputs, clock state, and second messenger signaling.
Materials and Methods
fsKaiB-HA co-IP/MS.
The S. elongatus PCC 7942 strain expressing HA-tagged fsKaiB (KaiBG88A;D90R) from an inducible promoter (MRC1109) was grown in BG-11M media (25) with shaking at 175 rpm in a Percival incubator with constant light (intensity of approximately ∼100 µmol photons m−2s−1 using cool white fluorescent bulbs), where each 250 mL flask contained 100 mL culture volume. When a high density (OD750 =1) was achieved, the culture was subjected to a 12:12 LD cycle. After the first 12 h dark pulse, expression was induced with 500 µM IPTG. After 12 h in light following the IPTG induction, the light sample (L) was harvested. After 12 h in dark following the light sample harvest, the dark sample (D) was harvested. A negative control sample was obtained by collecting WT cells under identical conditions in L and D and pooling them. The cell pellets were lysed and subjected to anti-HA co-IP using magnetic beads (see Anti-HA Co-IP from Cyanobacterial Lysate in SI Appendix, Supplementary Materials and Methods). The resulting eluate samples (fsKaiB L, fsKaiB D, and WT L+D) were analyzed at the Harvard Center for Mass Spectrometry (Cambridge, MA) (see LC-MS/MS Analysis of fsKaiB Co-IP Samples in SI Appendix, Supplementary Materials and Methods).
Monitoring In Vivo Clock Rhythms Using Bioluminescence Reporters.
Bioluminescence reporter strains expressed both luxAB luciferase genes derived from Vibrio harveyi under a rhythmic promoter (PpsbAI) and luxCDE genes allowing endogenous production of substrate. The sample preparation and data collection were performed as previously described (3) with slight modifications (see In Vivo Clock Rhythm Measurement in SI Appendix, Supplementary Materials and Methods). All experiments in Fig. 2 were performed with 10 µM IPTG unless otherwise noted, and the periods and peak times were estimated by fitting the data to the equation Y = Offset+Slope*X+Amplitude*sin((2*pi*((X-PhaseShift)/Period))) (see Circadian Rhythm Data Fitting in SI Appendix, Supplementary Materials and Methods for details). One 12L:12D cycle was used to entrain cells in the experiments shown in Fig. 2 A–C before release to constant light. Cells used in the photoperiod experiment (Fig. 2D) were subjected to four cycles of entrainment with the given photoperiod (8L:16D, 10L:14D, 12L:12D, or 16L:8D) followed by constant light. For the phase response experiment (Fig. 2E), cells were entrained by two 12L:12D cycles and were released to constant light, during which a 5 h dark pulse was given at the specified time. The earliest dark pulse start time was 48 h after the entrainment cycles. The dark pulse start time for each condition was converted into circadian time by multiplying the time by (24/average fitted period).
Protein Expression and Purification.
KaiA, KaiB, KaiB-FLAG, KaiC, and KaiC-EA proteins were purified as described previously (26, 27). The fsKaiB-FLAG protein (KaiB-1–99-Y7A-I87A-Y93A-FLAG) was a gift from Andy LiWang (14). For KidA PAS-ABC purification, the KidA coding sequence for residues 1–359, preceded by the (PreScission Protease) PSP cleavage site (LFQ/GP) sequence and followed by the HA tag sequence, was cloned into the pRSET-B vector. The expression plasmid was used to transform BL-21 (DE 3) Escherichia coli cells. We used 5 mL overnight culture to grow 1 L culture in LB to optical density (OD600) between 0.5 and 0.6 by shaking at 37 °C. Then 0.3 mM IPTG was added for 3 h induced expression at 30 °C. The cells were pelleted by centrifugation for 10 min at 5,000 g, 4 °C, snap-frozen in liquid nitrogen, and stored in –80 °C. For purification, the cell pellets were lysed using an Emulsiflex-C3 homogenizer (Avestin) and centrifugated for 30 min at 30,000 g, 4 °C. The supernatant containing the soluble protein was syringe-filtered using a 0.45 μm filter and was Ni-affinity purified by HisTrap HP column (GE) followed by cleavage by HRV3C protease (Pierce, Thermo Scientific). Next, the protein sample was subjected to anion exchange chromatography using a HiTrap Q column (GE) and eluted with a gradient from 20 mM Tris pH 8, 90 mM NaCl buffer to 20 mM Tris pH 8, 1 M NaCl. The protein eluted in two separate peaks in the chromatograph during anion exchange. The composition of each fraction was analyzed by SDS-PAGE. Each peak contained a doublet with approximately the expected molecular weight for KidA PAS-ABC. The fractions corresponding to each peak were pooled separately; the late-eluting peak was used for all experiments in this study. The material was then buffer-exchanged into Kai reaction buffer (10% glycerol, 20 mM Tris⋅HCl pH 8, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA pH 8, 1 mM ATP pH 8). The concentration was measured by Bradford assay.
In Vitro Co-IP and Fluorescence Polarization.
Purified proteins of interest were coincubated with KaiB-FLAG or fsKaiB-FLAG in 30 °C. After the incubation, the reactions were snap-frozen in liquid nitrogen and kept in –80 °C. The frozen reactions were thawed on ice and were promptly subjected to co-IP using anti-FLAG antibody-conjugated magnetic beads (Sigma-Aldrich, #M8823). The eluates from the co-IP were run in SDS-PAGE gel. The gel was stained using Sypro Ruby (Invitrogen), which was then imaged by ChemiDoc (Bio-Rad). The band intensity was used as a metric of the quantity of each protein that was pulled down by the anti-FLAG beads (see In Vitro Co-IP Using FLAG-Tagged Proteins as Bait in SI Appendix, Supplementary Materials and Methods for details). Fluorescence polarization measurements were performed by adding fluorescein-labeled KaiB to the purified KaiABC reaction as previously described (3, 28, 29) (see Fluorescence Polarization in SI Appendix, Supplementary Materials and Methods for details).
Mathematical Modeling.
We expanded the model of Paijmans et al. (17) to include the proposed KaiB and KidA dynamics. In brief, KaiB is treated as a monomer and allowed to convert between its ground-state and fold-switched forms. KidA binds and unbinds KaiB only when it is in the fold-switched form; KidA does not interfere with KaiB-CI or KaiB-KaiA binding. Detailed balance is enforced for all of the reactions added to the model. The model accounts for individual KaiC hexamers and their complexes, and its dynamics are simulated with the kinetic Monte Carlo procedure in (17). The parameters of the original model were set at their values in Paijmans et al. (17); parameters for the added reactions were manually set to reprise the KaiB populations in the original model and then sampled with a Markov chain Monte Carlo procedure similar to that in (30). Further model details are given in the SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Carrie Partch and Andy LiWang for useful discussions, and the LiWang lab for the gift of the fsKaiB mutant protein. We thank Rust lab members for their suggestions on the manuscript. We thank Bogdan Budnik and Renee Robinson at the Harvard Center for Mass Spectrometry for their analysis of samples. This work was supported by an HHMI Simons Faculty Scholar award and NIH Grant R01 GM107369 to M.J.R. and NSF Award MCB-1953402 to A.R.D.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202426119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Nakajima M., et al. , Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Rust M. J., Golden S. S., O’Shea E. K., Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science 331, 220–223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leypunskiy E., et al. , The cyanobacterial circadian clock follows midday in vivo and in vitro. eLife 6, e23539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chavan A. G., et al. , Reconstitution of an intact clock reveals mechanisms of circadian timekeeping. Science 374, eabd4453 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang Y. G., et al. , Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science 349, 324–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng R., et al. , Structural basis of the day-night transition in a bacterial circadian clock. Science 355, 1174–1180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shultzaberger R. K., Boyd J. S., Diamond S., Greenspan R. J., Golden S. S., Giving time purpose: The Synechococcus elongatus clock in a broader network context. Annu. Rev. Genet. 49, 485–505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama M., Kondo T., Xiong J., Golden S. S., ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. J. Bacteriol. 185, 1415–1422 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitz O., Katayama M., Williams S. B., Kondo T., Golden S. S., CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289, 765–768 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki H., et al. , A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101, 223–233 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Pattanayak G. K., Phong C., Rust M. J., Rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Curr. Biol. 24, 1934–1938 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry J. T., Crosson S., Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65, 261–286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heisler J., et al. , Structural mimicry confers robustness in the cyanobacterial circadian clock. BioRxiv [Preprint] (2020). 10.1101/2020.06.17.158394. Accessed 2 February 2022 [DOI]
- 15.Terauchi K., et al. , ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 16377–16381 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abe J., et al. , Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science 349, 312–316 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Paijmans J., Lubensky D. K., Ten Wolde P. R., A thermodynamically consistent model of the post-translational Kai circadian clock. PLOS Comput. Biol. 13, e1005415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito-Miwa K., Furuike Y., Akiyama S., Kondo T., Tuning the circadian period of cyanobacteria up to 6.6 days by the single amino acid substitutions in KaiC. Proc. Natl. Acad. Sci. U.S.A. 117, 20926–20931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenal U., Reinders A., Lori C., Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Parnasa R., et al. , Small secreted proteins enable biofilm development in the cyanobacterium Synechococcus elongatus. Sci. Rep. 6, 32209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann T. H., Shapiro L., Integration of cell cycle signals by multi-PAS domain kinases. Proc. Natl. Acad. Sci. U.S.A. 115, E7166–E7173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Möglich A., Ayers R. A., Moffat K., Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eelderink-Chen Z., et al. , A circadian clock in a nonphotosynthetic prokaryote. Sci. Adv. 7, eabe2086 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loza-Correa M., et al. , The Legionella pneumophila kai operon is implicated in stress response and confers fitness in competitive environments. Environ. Microbiol. 16, 359–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustos S. A., Golden S. S., Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J. Bacteriol. 173, 7525–7533 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phong C., Markson J. S., Wilhoite C. M., Rust M. J., Robust and tunable circadian rhythms from differentially sensitive catalytic domains. Proc. Natl. Acad. Sci. U.S.A. 110, 1124–1129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J., Chew J., Chockanathan U., Rust M. J., Mixtures of opposing phosphorylations within hexamers precisely time feedback in the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. U.S.A. 111, E3937–E3945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang Y. G., Tseng R., Kuo N. W., LiWang A., Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc. Natl. Acad. Sci. U.S.A. 109, 16847–16851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heisler J., Chavan A., Chang Y.-G., Liang A., Real-Time in Vitro Fluorescence Anisotropy of the Cyanobacterial Circadian Clock In: Brown S.A. (eds) Circadian Clocks. Methods in Molecular Biology vol 2130. (Springer, 2021) [DOI] [PubMed] [Google Scholar]
- 30.Hong L., et al. , Bayesian modeling reveals metabolite-dependent ultrasensitivity in the cyanobacterial circadian clock. Mol. Syst. Biol. 16, e9355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.