Abstract
SMC protein is required for chromosome condensation and for the faithful segregation of daughter chromosomes in Bacillus subtilis. The visualization of specific sites on the chromosome showed that newly duplicated origin regions in growing cells of an smc mutant were able to segregate from each other but that the location of origin regions was frequently aberrant. In contrast, the segregation of replication termini was impaired in smc mutant cells. This analysis was extended to germinating spores of an smc mutant. The results showed that during germination, newly duplicated origins, but not termini, were able to separate from each other in the absence of SMC. Also, DAPI (4′,6′-diamidino-2-phenylindole) staining revealed that chromosomes in germinating spores were able to undergo partial or complete replication but that the daughter chromosomes were blocked at a late stage in the segregation process. These findings were confirmed by time-lapse microscopy, which showed that after duplication in growing cells the origin regions underwent rapid movement toward opposite poles of the cell in the absence of SMC. This indicates that SMC is not a required component of the mitotic motor that initially drives origins apart after their duplication. It is also concluded that SMC is needed to maintain the proper layout of the chromosome in the cell and that it functions in the cell cycle after origin separation but prior to complete segregation or replication of daughter chromosomes. It is proposed here that chromosome segregation takes place in at least two steps: an SMC-independent step in which origins move apart and a subsequent SMC-dependent step in which newly duplicated chromosomes condense and are thereby drawn apart.
Bacteria face the challenge of packaging a chromosome that is about 1 mm in contour length into a structure called the nucleoid that is only about 1 μm across its long axis. Yet the highly compacted chromosome in the nucleoid must be capable of undergoing duplication into daughter chromosomes that can be segregated to the progeny cells with high fidelity. Some insights into the arrangement of the chromosome in the cell and its segregation have come from recent advances in bacterial cytology, which have made it possible to visualize individual sites on chromosomes in living and fixed cells. These studies have shown that the replication origin regions of the chromosome localize toward opposite poles of the cell, while the region of the replication terminus generally remains located near the middle of the cell (5, 6, 17, 21, 30). Thus, the chromosome appears to be folded in an ordered arrangement, with the origin region located near one end of the nucleoid, the terminus near the other end, and the quarter points on the chromosome located in between (28). Time-lapse microscopy has revealed that the movement of the origin regions toward the poles occurs rapidly and abruptly during the cell cycle (5, 6, 25, 28, 29). Terminus regions of the chromosome also separate from each other but not as rapidly or as far apart as the origin regions (29). Thus, chromosome segregation is a dynamic process in bacteria.
A key challenge is to understand how these chromosome movements are choreographed. An important insight has come from the discovery in bacteria of homologs of the eukaryotic SMC (structural maintenance of chromosomes) proteins, a family of proteins that is now known to be present in bacteria and archaea, as well as in eukaryotes (9, 26). In eukaryotes, these large proteins (>135 kDa) play a key role in chromosome condensation and segregation during mitosis, as well as in DNA recombination and transcriptional silencing. Also, eukaryotes have multiple SMC proteins, multiple members of which assemble in complexes, such as the condensin complex, which mediates chromosome condensation, and the cohesion complex, which mediates the cohesion of sister chromatids. In contrast, most bacteria for which a complete sequence is known have a single smc gene.
In Bacillus subtilis, the smc gene is indispensable for the maintenance of chromosome compaction and for viability above 24°C (3, 7, 20). Escherichia coli does not have an SMC homolog. Instead, it produces a protein called MukB whose function is similar to that of SMC (22). Despite the lack of sequence similarity, MukB bears strong structural similarity to SMCs: both proteins are composed of an N-terminal domain containing an ATPase motif, two central coiled coil domains that are connected by a flexible linker sequence, and a globular C-terminal domain (14). Based on its structural similarity to kinesin, MukB was proposed to be a force-generating motor for chromosome segregation (18, 22). Also, MukB and SMC form antiparallel homodimers (19), such that the N-terminal domain of one molecule would interact with the C terminus of its partner and vice versa. For both types of proteins, it has been shown that the C-terminal domain, which contains a helix-turn-helix motif, is essential for binding to DNA (1, 24) and for the association of SMC with the chromosome (7). B. subtilis SMC was shown to have a higher affinity for single-stranded DNA than for double-stranded DNA (8), although in yeast the opposite is the case (1). A model for how SMC could cause chromosomes to compact by the introduction of supercoils has been proposed (12, 13).
I wished to investigate the role of SMC in the arrangement of the chromosome in the cell and the movement and segregation of the origin and terminus regions. For this purpose, I took advantage of a previously devised procedure for visualizing specific regions on the chromosome in living cells based on the use of green fluorescent protein (GFP)-LacI to decorate a cassette of the lactose operon operator that had been inserted into the chromosome. Using this system in combination with time-lapse microscopy, it was discovered that newly duplicated origin regions move rapidly apart toward opposite poles of the cell in the complete absence of gene for smc, although the position of origin regions in mutant cells was aberrant. On the other hand, segregation of the terminus region was impaired in an smc mutant strain. A two-step model for chromosome segregation is proposed in which origin proximal regions are separated in an SMC-independent manner but the final separation of daughter chromosomes is mediated by SMC.
MATERIALS AND METHODS
Media and bacterial strains.
Cells were grown in rich medium (Luria-Bertani) or in S750 defined minimal medium (10). Strain PG63 (lacO cassette at origin region, expression of GFP-LacI under control of pveg, smc::kan) was created by transformation of AT63 (pveg-gfp-lacI [Mlsr] pAT12 [lacO cassette, Cmr] integrated at the spo0J locus [359°]) (28, 29) with chromosomal DNA from strain PGΔ388 (smc::kan [7]), selecting for chloramphenicol (Cm, 5 μg/ml), Mls (erythromycin and lincomycin, 5 and 25 μg/ml, respectively), and kanamycin (Kan, 5 μg/ml) at 23°C. Similarly, strain PG6 (smc::kan, GFP-LacI, lacO cassette at terminus region) was created by transformation of AT62 (pveg-gfp-lacI [Mlsr] pAT12 [lacO cassette, Cmr] integrated at 180°) (28) with chromosomal DNA from PGΔ388, selecting for Cmr, Mlsr, and Kanr. Disruption of the smc gene was verified by PCR and Western blot.
Purification and germination of B. subtilis spores.
Spores of B. subtilis were purified and germinated by the procedure of Callister and Wake (4), with the following modifications. In order to avoid purification of spores that accumulated suppressors of smc (a frequent event), cells were streaked out on DSM plates and incubated at room temperature for 5 days. Cells with a deletion of smc create spores only at low frequency (3), 0.5% in contrast to 82% for wild-type cells. Suppressor colonies (smc colonies are small, smooth, and rounded, in contrast to more wrinkled and much larger colonies from the wild type) were excised from the plates, which were flooded with 1 M KCl–0.5 M NaCl. Cells were washed twice in distilled water (dH2O) and incubated in 10 mM Tris buffer (pH 7.5) containing 2 mg of lysozyme per ml for 60 min at 37°C, followed by successive washes in 1 M NaCl, H2O, 0.05% sodium dodecyl sulfate, and 50 mM Tris–10 mM EDTA (pH 7.5) and three times with distilled water. Purified spores were stored at 4°C or at −80°C in dH2O–5% glycerol.
For germination of spores, a culture with an optical density (OD) of 0.6 to 0.8 was washed in H2O and resuspended in 0.5% glucose containing S750 medium supplemented with 0.3% aspartate, 0.1% glutamate, 0.025% Casamino Acids, 0.05% yeast extract, and 0.025% sodium citrate. After a heat treatment at 70°C for 30 min, spores were centrifuged, resuspended in medium, and incubated at 30°C for 30 min, followed by incubation at 23°C. The 30-min incubation at 30°C had no effect on the germination of smc cells, but it increased the germination efficiency and synchronicity considerably. As a control for the possibility that the sporulation protocol selected for mutations that suppressed the smc mutant defect, germinated spores were grown to mid-exponential phase and resuspended in sporulation medium. After incubation for 5 days, spore-forming efficiency was tested by heat treatment (30-min incubation at 80°C and plating onto rich medium) and was found to be similar (0.4 to 2%) to that observed for the first round of sporulation.
Microscopy.
Microscopy was performed as described in Webb et al. (29). For the acquisition of single pictures, all strains were grown at 23°C to mid-exponential phase, and aliquots were observed on an Olympus BX60 microscope. Discrete fluorescent foci could be seen in 40 to 70% of the smc mutant cells (PG63 and PG6) compared to 80 to 90% of the cells in the case of the wild type. I attribute this difference in the efficiency with which foci could be detected to the observation that smc mutant cells are more difficult to flatten out on slides than are wild-type cells.
For time-lapse analysis, cells were harvested in early stationary phase and resuspended in fresh medium (at an OD of ca. 0.25 at 600 nm). Cultures were incubated at 23°C for 1.5 to 3 h, and aliquots were placed on a microscope slide with a pad of agarose containing growth medium. A coverslip was placed on the cells, excess agarose was cut away, and images were acquired every 4 min. Measurement of the position of origins in the cells were determined using Metamorph 3.0 software.
Immunofluorescence microscopy.
Antibodies against FtsZ protein were affinity purified according to standard methods (kind gift of J. Kemp, Harvard University). Then, 0.1-ml aliquots of mid-exponential-phase cells were diluted into 1.9 ml of methanol and fixed for 10 min at room temperature. After centrifugation, the cell pellet was dried. Immunofluorescence microscopy was performed according to the method of Pogliano et al. (23).
RESULTS
Abnormal chromosome arrangement in cells lacking SMC.
In previous work, fusions of GFP to either LacI or Spo0J have been used to visualize the location of the replication origin in living cells. However, as noted by Britton et al. (3), the presence of an smc mutation interferes with the visualization of fluorescent foci from Spo0J-GFP. Likewise, Moriya et al. (20) have shown that the position of Spo0J foci is perturbed in smc mutant cells using immunofluorescence. An attractive explanation for this, as suggested by Britton et al. (3), is that Spo0J binds to multiple, widely separated sites in the origin region (16) and that the binding of the partition protein to the origin region involves the formation of a higher-order structure in a manner that is dependent on SMC. In the case of GFP-LacI, however, visualization of the origin region is achieved by use of a lacO cassette, which consists of multiple copies of the binding site for the lactose operon repressor arranged in tandem and inserted at a single site in the chromosome. The binding of GFP-LacI that is expressed in the cell to lacO sequences results in a fluorescent focus that can be visualized in living cells. Hence, fluorescent foci from GFP-LacI are not expected to be dependent upon a higher-order chromosome structure. I therefore reasoned that it should be possible to visualize GFP-LacI foci in cells lacking SMC and hence that the fluorescent fusion protein could be used to investigate the effect of an smc null mutation on the number, location, and movement of replication origin regions. To do this, an smc insertion-deletion mutation (smcΔ388) was introduced by transformation (see Materials and Methods) into strain AT63 (29), which harbors a GFP-LacI-producing construct and tandem copies of lacO near the origin of replication (359°). The presence of the smcΔ388 mutation in one such transformant (strain PG63) was confirmed both from its phenotype (it was temperature sensitive and defective in chromosome condensation and segregation [3, 7, 20]) and directly by means of the PCR.
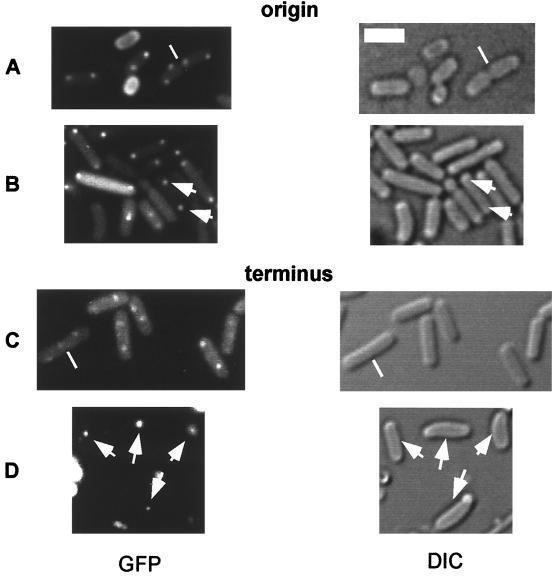
The numbers and positions of the origin regions of the chromosomes in cells of strains AT63 and PG63 growing at 23°C (doubling times of 50 and 185 min, respectively) were analyzed using fluorescence microscopy (Fig. 1). Septa were visualized using Normarski differential interference contrast, which visualizes septa as subtle demarcations between cells (Fig. 1A, B, and C). A high proportion of PG63 cells (ranging from 15 to 35% in three experiments) showed a higher-than-normal number of origin regions. Unlike the parent strain (AT63), in which only one, two, or four foci were generally visible (Fig. 1A), some smc mutant cells exhibited three (Fig. 1B) and as many as six (data not shown) foci. Cells mutant for smc are known to produce anucleate cells at high frequency (3, 7, 20). Yet, the overall DNA to protein ratio for mutant cells is similar to that of wild-type cells (20). The results indicate that the presence of anucleate cells (15 to 19% under these growth conditions) might be compensated for to some extent by the presence of other cells that have a higher than normal number of chromosomal origin regions and thus DNA.
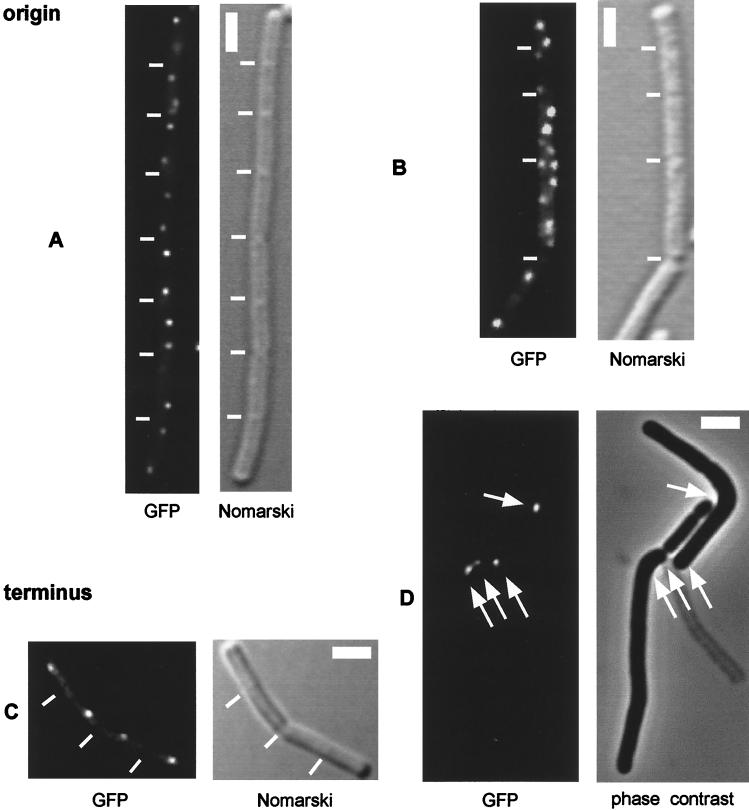
FIG. 1.
Fluorescence microscopy of B. subtilis cells producing GFP-LacI and carrying tandem copies of lacO near the origin (A and B) or terminus (C and D) of replication. (A) AT63 (wild type). (B) PG63 (smc::kan). (C) AT62 (wild type). (D) PG6 (smc::kan). The white lines indicate septa, which were visualized by differential interference contrast (Nomarski) microscopy. The scale bar (thick white line) represents 2 μm. Cells were grown in rich medium at 23°C, and images were collected during the mid-exponential phase of growth.
The positions of the fluorescent foci was determined by measuring the distance of each focus from the nearest pole of the cell using Metamorph software and were plotted as a function of cell length. Figure 2A and B present the results for wild-type cells; for simplicity, only the data for cells displaying one (Fig. 2A), two (Fig. 2A), or four (Fig. 2B) foci are shown. As can be seen in the figure, cells with two foci were generally smaller than cells with four foci. In the case of cells with two fluorescent foci, the signals tended to be located toward opposite poles of the cell (open and filled circles in Fig. 2A) (28, 30). Sometimes cells with two foci were observed in which the foci were close to the mid-cell position or in the same half of the cell (dotted line indicates mid-cell point; Fig. 2A). This was only observed for small cells and, on this basis and on the basis of previous observations made by time-lapse microscopy, I presume that these represent young cells in which the origin region had undergone duplication but that the resulting daughter origins have not yet migrated toward opposite poles of the cell. In the case of wild-type cells with four foci, the signals tended to be evenly spaced across the cell, with two foci located near opposite poles (filled circles and open squares) and the other two foci (open circles and filled squares in Fig. 2B) in between. These positions correspond to polar positions in the future daughter cells.
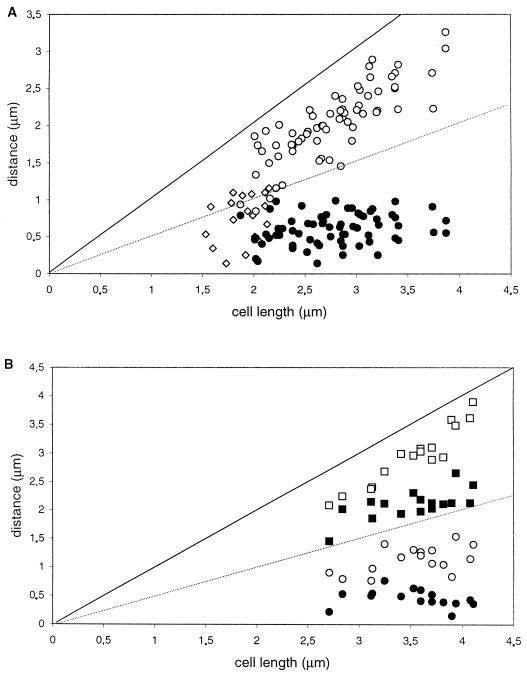
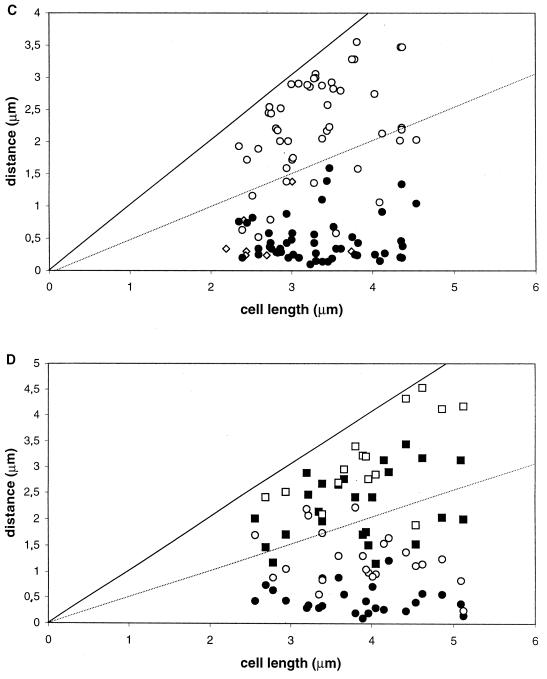
FIG. 2.
Distance of fluorescent foci from the cell pole as a function of cell size. (A and B) AT63 (GFP-LacI, lacO cassette at 359°). (C and D) PG63 (smc::kan, GFP-LacI, lacO cassette at 359°). Cells were grown in rich medium at 23°C, and images were obtained during the mid-exponential phase of growth. Panels A and C show cells with one (⧫) or two (● and ○) fluorescent foci; panels B and D show cells with four foci (●, ○, ■, and □, with “●” being the closest and “□” being the farthest focus from the pole chosen for measurements). The measurements in panels C and D do not include very large cells, in which origin signals tended to be even more random than in small cells.
In the case of smc mutant cells, a strikingly different pattern of origin positions was observed (Fig. 2C and D). First, many smc mutant cells had fluorescent foci located very close to one or both cell poles, significantly closer than was observed in wild-type cells. Second, the location of origins was more irregular than was observed for the wild type. Whereas in wild-type cells with two fluorescent foci the origins tended to localize toward opposite ends of the cell in a symmetric pattern (Fig. 2A), in mutant cells one origin was often very close to a pole but the other origin was located in a more or less random position (Fig. 2C). In contrast to wild-type cells with four foci, in which two origins were found in each half of the cell (Fig. 2B), in mutant cells three or four of the origins were often found in the same half of the cell (Fig. 2D).
Terminal regions of the chromosomes fail to separate in smc null cells.
In slow-growing cells, the terminus regions are generally located near the middle of the cell (28, 30) (Fig. 1C) and, following duplication, rapidly move apart (29). Thus, large cells usually have two fluorescent signals located near the quarter points of the cell just prior to cell division (data not shown). To analyze the position of termini in smc mutant cells, strain AT62 carrying the lacO cassette at the terminus region (181°) was transformed with chromosomal DNA from PGΔ388 to create strain PG6. Whereas termini in wild-type cells (AT62) were generally located at the mid-cell position (Fig. 1C), fluorescent signals from the terminus in PG6 cells were often found near the cell pole (Fig. 1D). Importantly, and contrary to chromosomal origins which were distributed throughout the filament in the mutant cells (Fig. 1B), terminus signals were often clustered at one site in the cell (Fig. 1D, arrows). This was observed in about 60% of the cells and especially in large cells or filaments. I interpret these observations to indicate that the absence of SMC causes a defect in replication through the terminus or a defect in the segregation of newly duplicated terminus regions.
Germinating spores of an smc mutant are defective in chromosome segregation.
A complication in studying the role of SMC in chromosome segregation during growth is the heterogeneity of the cells both with respect to the stage of the cell cycle and the number of replication origin regions (ranging up to six; see above). For this reason the effect of an smc mutation was examined during outgrowth of germinated spores, since only a single chromosome is packaged into the spore and spores can be germinated with partial synchrony by a regimen involving heat treatment followed by suspension in germination medium. Accordingly, spores from strains PY79, AT63, PGΔ388, PG63, and PG6 were purified. After heat treatment at 70°C for 30 min, the spores were suspended in germination medium and allowed to germinate at 30°C. After 30 min, outgrowth was allowed to proceed at room temperature (∼23°C).
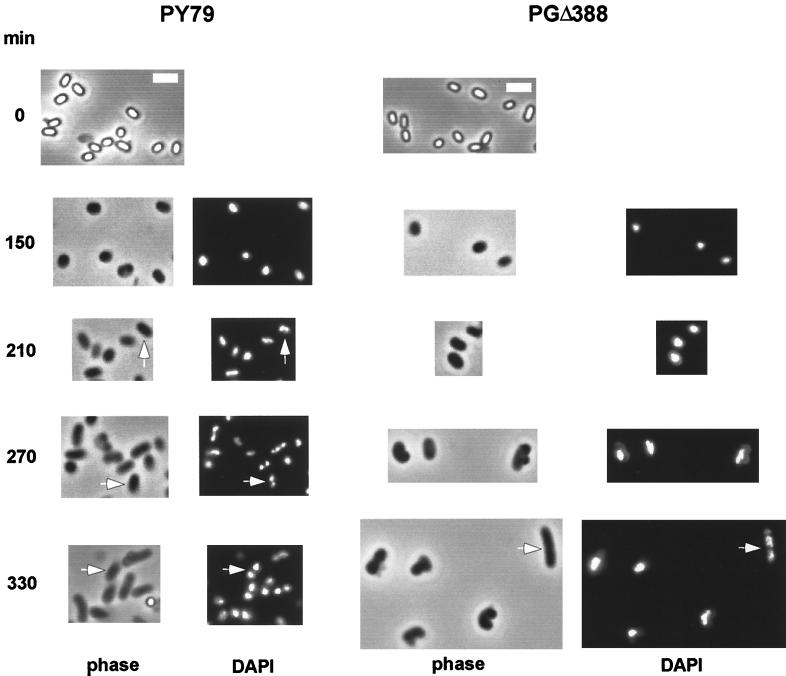
Spores from the smc mutant strains underwent germination and the initial stage of outgrowth (cell elongation) normally, as judged by the conversion of phase-bright spores to phase-dark cells during the 30-min period of germination and the ensuing elongation of cells emerging from the germinated spores during the first 150 min of the outgrowth period (Fig. 3). However, by 210 min, when most of the cells from the wild-type spores started to segregate their chromosomes, as seen by the bilobed appearance of nucleoids (Fig. 3, arrow), cells from the mutant spores still contained one nucleoid. In the wild type, clear separation of nucleoids commenced at about 270 min (arrow), and by 330 min all of the wild-type cells had two segregated nucleoids. In contrast, >95% of the smc mutant cells (PGΔ388 and PG63) had only one nucleoid mass at 330 min, although cell elongation was comparable to that of the wild type. Occasionally, some mutant cells were observed that had two nucleoid masses, but these nucleoids frequently had an abnormal morphology (330 min, arrow). After germination, the mutant cells continued to grow and entered exponential-phase growth but with a much longer doubling time (240 min) than for the wild type (70 min).
FIG. 3.
Time course of germinating spores from strain PY79 (wild type) and PGΔ388 (smc::kan). Samples of cells at the indicated times after the start of germination were fixed and stained with DAPI prior to image acquisition. DAPI images of purified spores are not shown since spores show a strong blue autofluorescence. The scale bars (thick white lines) represent 2 μm. The arrows indicate partially (210-min time point) and completely (270- and 330-min time points) separated nucleoids.
One possibility for the failure of chromosome segregation to take place in germinated spores of the smc mutant could be a defect in the initiation of replication. The lacO cassette decorated with GFP-LacI was used to investigate this issue because it provided a means to visualize origin regions. Because maturation of GFP is a slow process, it was necessary to modify the procedure described above by suspending the germinating cells in a buffer (T-base) lacking nutrients for 1.5 h following 150 min in germination medium in order to obtain bright fluorescent foci. Following this procedure, two separated origin regions could be seen in all wild-type (Fig. 4A) and smc mutant cells (Fig. 4B, arrows, 81 cells counted). These findings are consistent with the idea that smc is not needed for the initiation of replication during germination or the partial separation of newly duplicated origin regions. Conversely, when spores of PG6, which carries the lacO cassette at the terminus, were germinated for 240 min and resuspended in T-base, only a single focus was detectable (Fig. 4D, arrows, 54 cells counted), in contrast to wild-type cells that had two well-separated foci. Further, many of the wild-type cells had undergone cell division (Fig. 4C, white bar). These results support a function for SMC in the complete separation or replication of daughter chromosomes.
FIG. 4.
Fluorescence and Nomarski DIC images of germinating spores of cells producing GFP-LacI and carrying tandem copies of lacO. Images were collected 1.5 h after resuspension in salt buffer. Panels A and C show wild-type cells (AT63 and AT62, respectively) and panels B and D show the smc mutant cells (PG63 and PG6, respectively) containing the lacO cassette near the origin (359°; A and B) or near the terminus (180°; C and D). The scale bars (thick white lines) represent 2 μm, the arrows indicate the position of fluorescent foci, and the white bars show the position of the septa.
SMC is not required for the rapid separation of chromosomal origins.
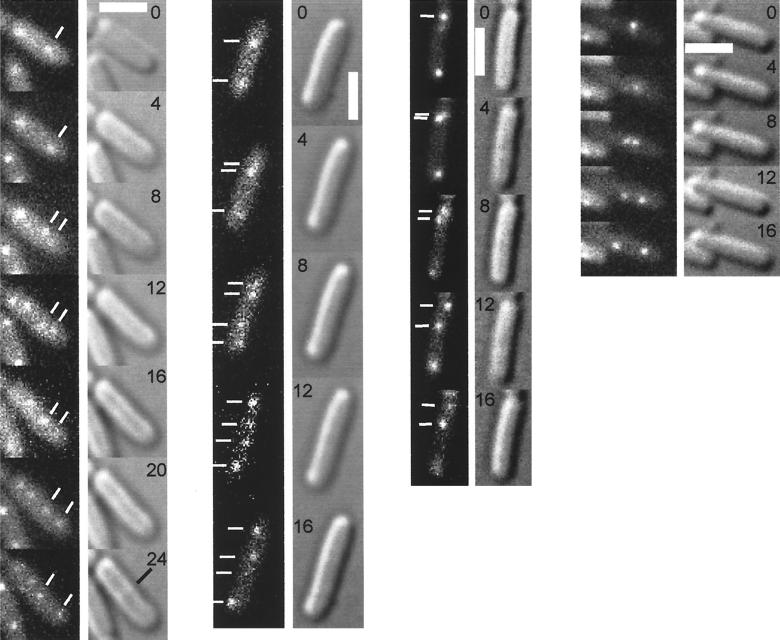
One of the striking findings of this investigation is that origin regions can and do localize toward the cell poles in the absence of SMC. This observation suggests that the “mitotic” motor that drives newly duplicated origins apart does not depend on SMC. To test this possibility directly, time-lapse microscopy was used to monitor origin movement in smc mutant cells. Cells of strain PG63 were grown until early stationary phase and resuspended in fresh minimal medium. Previous time-lapse microscopy experiments have shown that in wild-type cells the origin regions separate from each other abruptly during the cell cycle. Because smc mutant cells grow with a long doubling time (4 h at 25°C), it was necessary to observe a large number of cells in order to catch those that were at the stage of origin separation. Of 115 cells studied, 9 were observed in which origin separation was taking place. In all cases, the origin regions were observed to move apart in a dynamic fashion, similar to that observed in wild-type cells. The results of four such experiments are shown in Fig. 5, with the fluorescence signals shown in the left-hand column and the corresponding Nomarski DIC images in the right-hand column. In the first time-lapse sequence, a fluorescent focus (labeled with white lines) undergoes duplication into two foci between min 4 and 12. The two daughter foci then move apart during min 16 to 24. The black bar in the 24-min image indicates the position of a newly formed septum. A similar pattern of origin movement is seen in the second sequence. In the third sequence, a fluorescent focus located somewhat near the pole undergoes duplication with one of the daughter foci moving to the mid-cell position and the other remaining relatively stationary. Conversely, a focus located at the mid-cell position in the fourth sequence undergoes duplication near the cell middle, and the resulting daughter foci move apart toward opposite poles of the cell. Both the pattern of separation of newly duplicated foci from a polar position and from a mid-cell position have been observed previously in wild-type cells (29).
FIG. 5.
Time-lapse microscopy. Cells from the smc mutant PG63, which produces GFP-LacI and contains the lacO cassette at 359°, were grown on agarose pads containing S750 medium at room temperature. The left-hand columns show fluorescence from GFP, and the right-hand columns show Nomarski DIC images. The numbers indicate the time in minutes at which the images were acquired. The scale bar (thick white line) represents 2 μm. The white lines indicate the position of fluorescent foci; the black lines indicate the emergence of a septum.
Upon quantifying the results for all nine time-lapse sequences, I find that fluorescent foci moved apart an average distance of 1.25 μm (standard deviation, 0.11 μm) over a period of 8 to 12 min in smc mutant cells compared to 1.4 μm in wild-type cells (29). The average velocity during the period of abrupt movement was calculated to be 0.125 μm/min in the mutant cells compared to the previously reported value of 0.17 μm/min in wild-type cells. I conclude that the separation of newly duplicated origins does not depend on SMC and that the extent and velocity of movement in cells lacking SMC is close to that observed in wild-type cells.
DISCUSSION
This report helps to clarify the role of SMC protein in the cell cycle of B. subtilis. The principal finding is that newly duplicated chromosomal origins undergo rapid and abrupt separation from each other in the absence of SMC, the dynamics of movement being similar to that observed in wild-type cells (29). I conclude from this that SMC is not needed in the initial stages of chromosome segregation. Lemon and Grossman have speculated that DNA replication is the driving force for chromosome separation in bacteria (15). Based on their observation that DNA polymerase localizes near the middle of the cell in B. subtilis, these workers propose a factory model in which the DNA polymerase remains stationary and the DNA template is threaded through the replication complex. The demonstration that SMC is not required for origin separation is consistent with this model in that the findings show that SMC is not the motor for moving origin regions toward the cell poles.
A second principal contribution of the present work was the demonstration of the aberrant placement of replication origins in cells lacking SMC. Compared to the bipolar placement of origins observed in wild-type cells, origins in mutant cells exhibited a somewhat random distribution. For example, in contrast to the symmetric distribution of origins found in wild-type cells, mutant cells were frequently observed in which origins were asymmetrically distributed to one half of the cell. Also, consistent with the idea that chromosomes are decondensed in the absence of SMC (3, 7, 20), origins were often found at a more extreme polar position in mutant cells than is observed in wild-type cells. Aberrant positioning of replication origins was unlikely to be due to an effect of the smc mutation on placement of the division septum since the use of immunofluorescence microscopy showed that Z-rings were generally not mispositioned in mutant cells, although their number was substantially reduced (data not shown). A similar role for SMC in the proper placement of the chromosome has recently been reported for Caulobacter crescentus. In wild-type cells of this aquatic bacterium, origins are generally found at one or both poles of the cell, but in cells lacking SMC origins are observed at intermediate positions (11). It is concluded that proper folding of the chromosome and its proper arrangement within the cell depends on SMC.
I have also discovered a role for SMC in the terminal stage of chromosomal replication or separation. This was seen most clearly in DAPI (4′,6′-diamidino-2-phenylindole)-stained images of germinating spores of a mutant lacking the smc gene. During germination such mutant spores underwent complete or partial chromosome duplication, but the two daughter chromosomes did not completely separate from each other. The results with GFP-LacI were consistent with this interpretation. As noted above, fluorescent foci corresponding to chromosomal origins in either growing cells or germinating spores of an smc mutant were found to resolve into two foci, which then moved apart from each other. In contrast, foci corresponding to the terminal region frequently did not separate or even resolve into two foci. This indicates either that replication did not proceed across the terminus or that the terminal regions did undergo replication but were blocked in subsequently moving apart from each other. In toto, the findings indicate that SMC functions after the initiation of replication but before chromosomal termini are segregated.
In previous work, immunolocalization images were obtained suggesting that SMC is not only associated with the chromosome but also with the cell poles, especially in young cells (7). Since SMC is not needed for newly duplicated origins to move apart, I speculate that SMC does not become associated with the chromosome until after the initiation of replication or after origins move apart and approach the cell poles. Like its homologs in yeast (2), SMC might bind preferentially to particular sites at scattered locations around chromosome and introduce supercoils in flanking DNA, thereby causing the chromosome to condense. This condensation could facilitate nucleoid segregation and the final separation of the terminal regions of the chromosome.
ACKNOWLEDGMENTS
The work was performed in the laboratory of Richard Losick, whom I thank for help in writing the manuscript. I also thank R. Britton for helpful comments.
P.L.G. was a postdoctoral fellow of the Deutsche Forschungsgemeinschaft. The work in Richard Losick's laboratory was supported by grant GM15868.
REFERENCES
- 1.Akhmedov A T, Frei C, Tsai-Pflugfelder M, Kemper B, Gasser S M, Jessberger R. Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998;273:24088–24094. doi: 10.1074/jbc.273.37.24088. [DOI] [PubMed] [Google Scholar]
- 2.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 3.Britton R A, Lin D C-H, Grossman A D. Characterization of a procaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callister H, Wake R G. Completion of the replication and division cycle in temperature-sensitive DNA initiation mutants of Bacillus subtilis 168 at the non-permissive temperature. J Mol Biol. 1977;117:71–84. doi: 10.1016/0022-2836(77)90023-7. [DOI] [PubMed] [Google Scholar]
- 5.Glaser P, Sharpe M E, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- 6.Gordon S, Sitnikov D, Webb C D, Teleman A, Losick R, Murray A W, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 7.Graumann P L, Losick R, Strunnikov A V. Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J Bacteriol. 1998;180:5749–5755. doi: 10.1128/jb.180.21.5749-5755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirano M, Hirano T. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 1998;17:7139–7148. doi: 10.1093/emboj/17.23.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- 10.Jaacks K J, Healy J, Losick R, Grossman A D. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen R B, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci USA. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 13.Kimura K, Rybenkov V V, Crisona N J, Hirano T, Cozzarelli N R. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- 14.Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 15.Lemon K P, Grossman A D. Localization of bacterial DNA polymerase evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 16.Lin D C-H, Grossman A D. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 17.Lin D C-H, Levin P A, Grossman A D. Bipolar localization of a chromosome partition protein to Bacillus subtilis. Proc Natl Acad Sci USA. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockhart A, Kendrick-Jones J. Nucleotide-dependent interaction of the N-terminal domain of MukB with microtubules. J Struct Biol. 1998;124:303–310. doi: 10.1006/jsbi.1998.4056. [DOI] [PubMed] [Google Scholar]
- 19.Melby T E, Ciampaglio C N, Briscoe G, Erickson H P. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriya S, Tsujikawa E, Hassan A K M, Asai K, Kodama T, Osagawara N. A Bacillus subtilis gene encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- 21.Niki H, Hiraga S. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 1998;12:1036–1045. doi: 10.1101/gad.12.7.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 1992;11:5101–5109. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pogliano K, Harry E, Losick R. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 24.Saleh A Z, Yamanaka K, Niki H, Ogura T, Yamazoe M, Hiraga S. Carboxyl terminal region of the MukB protein in Escherichia coli is essential for DNA binding activity. FEMS Microbiol Lett. 1996;143:211–216. doi: 10.1111/j.1574-6968.1996.tb08482.x. [DOI] [PubMed] [Google Scholar]
- 25.Sharpe M E, Errington J. A fixed distance for separation of newly duplicated copies of oriC in Bacillus subtilis: implications for co-ordination of chromosome segregation and cell division. Mol Microbiol. 1998;28:981–990. doi: 10.1046/j.1365-2958.1998.00857.x. [DOI] [PubMed] [Google Scholar]
- 26.Strunnikov A V. SMC proteins and chromosome structure. Trends Cell Biol. 1998;8:454–459. doi: 10.1016/s0962-8924(98)01370-1. [DOI] [PubMed] [Google Scholar]
- 27.Sun Q, Yu X C, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 28.Teleman A A, Graumann P L, Lin D C H, Grossman A D, Losick R. Chromosome arrangement within a bacterium. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 29.Webb C D, Graumann P L, Kahana J, Teleman A A, Silver P, Losick R. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol Microbiol. 1998;28:883–892. doi: 10.1046/j.1365-2958.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 30.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C-H, Grossman A D, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cels of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]