Abstract
The genetic profile that is needed to identify talents has been studied extensively in recent years. The main objective of this investigation was to approach, for the first time, the study of genetic variants in several polygenic profiles and their role in elite endurance and professional football performance by comparing the allelic and genotypic frequencies to the non-athlete population. In this study, genotypic and allelic frequencies were determined in 452 subjects: 292 professional athletes (160 elite endurance athletes and 132 professional football players) and 160 non-athlete subjects. Genotyping of polymorphisms in liver metabolisers (CYP2D6, GSTM1, GSTP and GSTT), iron metabolism and energy efficiency (HFE, AMPD1 and PGC1a), cardiorespiratory fitness (ACE, NOS3, ADRA2A, ADRB2 and BDKRB2) and muscle injuries (ACE, ACTN3, AMPD1, CKM and MLCK) was performed by Polymerase Chain Reaction-Single Nucleotide Primer Extension (PCR-SNPE). The combination of the polymorphisms for the “optimal” polygenic profile was quantified using the genotype score (GS) and total genotype score (TGS). Statistical differences were found in the genetic distributions between professional athletes and the non-athlete population in liver metabolism, iron metabolism and energy efficiency, and muscle injuries (p<0.001). The binary logistic regression model showed a favourable OR (odds ratio) of being a professional athlete against a non-athlete in liver metabolism (OR: 1.96; 95% CI: 1.28–3.01; p = 0.002), iron metabolism and energy efficiency (OR: 2.21; 95% CI: 1.42–3.43; p < 0.001), and muscle injuries (OR: 2.70; 95% CI: 1.75–4.16; p < 0.001) in the polymorphisms studied. Genetic distribution in professional athletes as regards endurance (professional cyclists and elite runners) and professional football players shows genetic selection in these sports disciplines.
Introduction
The role of genetics in athletic performance has been shown in numerous studies to be important in defining the status of both an endurance athlete and a strength athlete [1–4]. Sports performance is a sum of various factors, both extrinsic and intrinsic, that can predict sports performance [5]. At the end of 2020, the total number of deoxyribonucleic acid (DNA) polymorphisms that are significant to athlete status was 97 (35 endurance-related, 24 power-related, and 38 strength-related) [6]. It should be borne in mind however, that hundreds and even thousands of polymorphisms are needed for the prediction of sports performance [7].
Even though numerous studies show an association of genetics with sports performance, there is still a very limited understanding of the role of genetics [8]. Despite this, recent years have witnessed the rise of an emerging market of direct-to-consumer marketing (DTC) tests that claim to be able to identify children’s athletic talents [9, 10]. It is vitally important that sport and exercise medicine practitioners are fully aware of the state of the evidence concerning genetic testing and the limitations of current knowledge [9, 11]. The genetic variants tested most frequently by the companies providing DTC genetic tests related to sport and exercise since 2015 have been those in the α-actinin 3 (ACTN3) and angiotensin-converting enzyme (ACE) genes, which presumably reflects the fact that more research has been conducted on these polymorphisms than any others in the context of sport and exercise [12–15]. Although the true role of the ACTN3 c.1729C>T (rs1815739) and ACE I/D (rs4340) polymorphisms in skeletal muscle performance and strength traits remains controversial [15–19], in a systematic review the ACE II genotype was associated with physical performance, especially endurance performance, while the ACTN3 CC genotype was associated with speed and power performance [12, 20]. Genetic information may represent a potentially useful tool for current procedures to identify future talent, enhance training or prevent sport-related injuries [7].
For many years, genes with allelic variants have been identified as predisposing individuals to elite performance, including ACTN3 [19], ACE [21], Homeostatic Iron Regulator (HFE) [22], Adenosine Monophosphate deaminase 1 (AMPD1) [23], among others, but to date, there is limited information on association analysing a larger set of genetic markers regarding sports performance.
Metabolism is key in sports performance because it is ultimately responsible for the ability to perform the necessary skills. Genetics play a relevant role in sports metabolism and several studies have presented the relevance of genes involved in liver metabolism [24, 25], energy efficiency and iron absorption [5] using polygenic profiles as determinants of athletic performance in endurance sports. Previous investigations have found that carrying the T allele in the c.34C>T polymorphism in the AMPD1 gene (rs17602729) might reduce VO2max trainability and lower response to endurance training [23, 26]. The peroxisome proliferator-activated receptor ɤ coactivator 1-α (PGC1α) regulates the expression of several genes associated with substrate oxidation, mitochondrial biogenesis, and muscle fibre conversion [27], suggesting that the GG genotype of the c.1444 G>A polymorphism (rs8192678) might facilitate endurance performance [28–30]. Also, genetics play a significant role in interindividual differences in serum iron parameters. The HFE gene regulates iron reabsorption [31–33]. Individuals with CG or GG genotypes in the c.187C>G polymorphism (rs1799945) possessed higher circulating iron concentrations inducing a higher haemoglobin concentration [34], associated with greater VO2max in professional athletes [35].
The measure of an individual’s peak capacity to perform dynamic aerobic exercise is dependent on the synergistic action of pulmonary, cardiovascular and muscle tissue via a suite of physiological actions that effectively transport and deliver oxygen from the atmosphere to the mitochondria in working muscles [36, 37].
A previous study shows that 125 single nucleotide polymorphisms (SNPs) were analysed regarding cardiorespiratory fitness, but an association with VO2max was only found in ACE (rs4340), Angiotensin II Receptor Type 1 (AGTR1) (rs275652) and Myostatin (GDF8) (rs7570532). Ninety-seven genes have been identified as possible predictors of VO2max trainability [38]. To verify the strength of these findings and to identify if there are more genetic variants and/or mediators, further tightly controlled studies are required that measure a range of biomarkers across ethnicities [38, 39].
Liver metabolism has recently shown its implication in the performance of elite endurance athletes through several polymorphisms; cytochrome Member 6 of subfamily D of family 2 P450 (CYP2D6) c.506-1G>A (rs3892097), isoform 1 of glutathione-S transferase mu (GSTM1), glutathione S-transferase pi (GSTP) c.313A>G (rs1695) and glutathione S-transferase theta (GSTT) implied in detoxify capacity, oxidative stress clearance and recovery of systemic homeostasis in high performance in endurance sports [24].
Several polymorphisms have been associated with muscle injury risk [40]; the creatine kinase isoenzyme MM (CKM) gene encodes the cytosolic muscle isoform of creatine kinase responsible for the rapid regeneration of ATP during intensive muscle contraction. The c.*800A>G (rs8111989) polymorphism plays a vital role in the energy homeostasis of muscle cells [41]. The G allele reduce activity of the skeletal muscle in endurance athletes [42, 43]. Also, the c.34C>T polymorphism (rs17602729) of the AMPD1 gene plays a vital role in the energy metabolism of muscle cells [5]. Subjects with the T allele have diminished exercise capacity and cardiorespiratory responses to exercise in the sedentary state compared to C allele carriers [23, 44]. The TT genotype is associated to metabolic myopathy, with exercise-induced muscle symptoms such as early fatigue, cramps and/or myalgia which are related to the risk of muscle injury [40, 45], especially in football [46]. The ACTN3 gene has been associated with strength in successful sprinters or track cyclists. The CC genotype is nearly always present among elite power athletes, whereas TT homozygosity, associated with a premature stop codon that produces complete α-actinin-3 deficiency, is more prevalent in some populations of elite endurance athletes, such as marathon runners [47–49]. The TT genotype is associated with a higher incidence and severity of injuries in endurance runners [50, 51] and professional football players [52] with regard to the CC counterparts. Myosin light chain kinase (MLCK), a calcium-calmodulin-dependent multi-functional enzyme, plays a critical role in the regulation of smooth muscle contraction [53]. Polymorphisms in the gene that codifies MLCK (c.37885C>A (rs28497577) and c.49C>T (rs2700352)) may alter regulatory light chain (RLC) phosphorylation, thus decreasing the ability to produce force and resist tension during voluntary muscle, showing that the C allele of c.37885C>A (rs28497577) polymorphism [53] and T allele of c.49C>T (rs2700352) polymorphism [54] could predispose to higher values of muscle damage during endurance competitions. In the ACE gene (rs4340), the deletion (D) allele is habitually more associated with higher activity of the enzyme angiotensin-converting enzyme than the insertion (I) allele [55]. Also, the I allele is associated with the resistance phenotype and the D allele with speed and power phenotypes [19]. Genetic variation in the ACE gene might be associated with many heritable traits, including physical, physiological and skill parameters and physical performance [56, 57]. II genotype is associated in the susceptibility to developing muscle injuries among professional football players [58].
The purpose of this study was to compare the differences in liver metabolism, iron metabolism and energy efficiency, cardiorespiratory fitness and muscle injuries polygenic profiles among elite endurance athletes and professional football players with a non-athlete population. In turn, it was intended to demonstrate, in a pioneering way, the genetic differences in these profiles between elite endurance athletes and professional football players.
Results
All the polymorphisms analysed met the Hardy-Weinberg equilibrium (HWE).
The athletes had a mean age of 24.9 years (±4.9 years): elite endurance athletes 26.1 years (±4.5 years) and professional soccer players 23.4 years (±5.1 years) and the non-athlete population 27.9 years (±4.5 years).
Polygenic profile of liver metabolism
When adding the genotype scores of CYP2D6, GSTM, GSTP and GSTT polymorphisms, the mean value of the TGS in the professional athletes had a value of 69.9 a.u. (±17.3 a.u.), statistical kurtosis: -0.40 (±0.28). The value for the group of elite endurance athletes was 69.7 a.u. (±18.9 a.u.), statistical kurtosis: -0.47 (±0.38) and in professional football players it was 70.2 a.u. (±15.4 a.u.), statistical kurtosis: -0.59 (±0.42). The mean value of the TGS in non-athletes was 63.2 a.u. (±17.8 a.u.) statistical kurtosis: -0.70 (±0.43). The TGS values of the professional athletes and non-athletes were statistically significant (p < 0.001). Similar between elite endurance athletes and professional football players with non-athletes (p = 0.004 and p < 0.001 respectively).
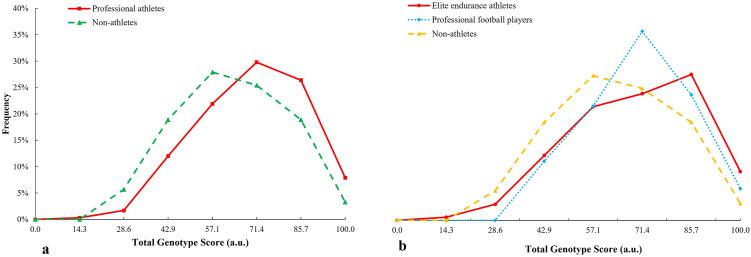
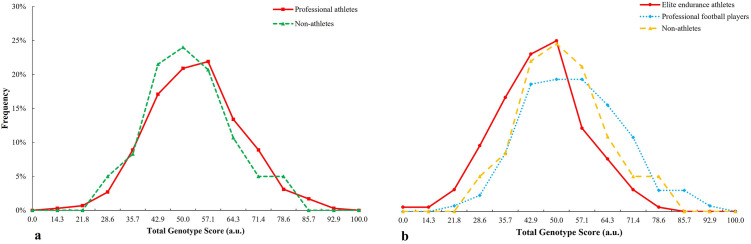
TGS distribution of liver-metabolising genes in the professional athletes is shifted to the right with respect to non-athletes (p = 0.022) (Fig 1a), similar between professional football players with non-athletes (p = 0.010) and shows statistical trends with respect to endurance athletes (p = 0.087) (Fig 1b).
Fig 1. TGS distribution of liver metabolism genes in a) professional athletes and non-athlete subjects and b) elite endurance athletes and professional football players with regard to non-athlete subjects.
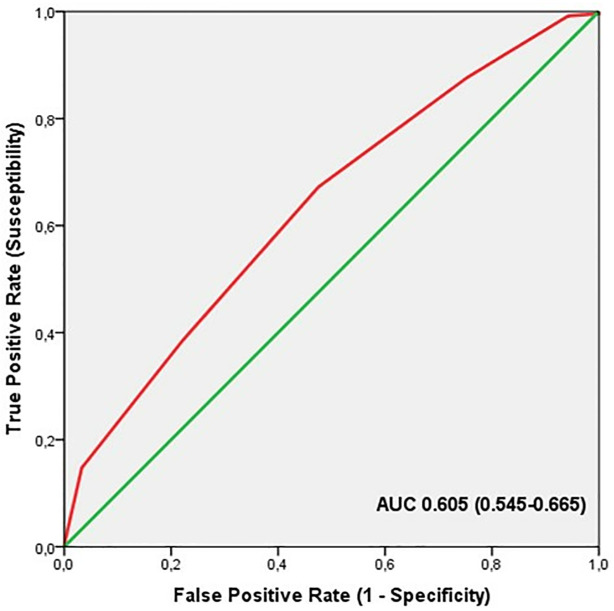
ROC analysis showed significant discriminatory accuracy of TGS in the identification of professional athletes (AUC = 0.605; 95% CI: 0.545–0.665; p = 0.001) (sensitivity = 0.640, specificity = 0.475) (Fig 2). The corresponding TGS value at this point was 64.2 a.u. Binary logistic regression analysis showed that subjects with a higher TGS of 64.2 a.u. had an odds ratio (OR) of 1.96 (95% CI: 1.28–3.01; p = 0.002) of being professional athletes, compared to those with a TGS below this value. The elite endurance athletes showed an OR at the cut-off point in comparison to the non-athlete population of 1.79 (95% CI: 1.11–2.88; p = 0.017) and the professional football players, in comparison to non-athlete subjects, had an OR of 2.20 (95% CI: 1.33–3.66; p = 0.001).
Fig 2. ROC curve summarising the ability of TGS of liver metabolism genes to distinguish potential professional athletes from non-athletes.
Genotype distribution of liver-metabolising genes in the professional athletes’ group, when compared with the non-athlete population, was statistically significant for CYP2D6 (p < 0.001), showing a higher frequency in the “optimal” genotype in athletes (GG 93.2%) than the non-athlete population (GG 61.1%) (Table 1). Between both groups of professional athletes (endurance and football players), statistically significant results were found in CYP2D6 (p = 0.002), which was more favourable in football players (GG 98.5%) than elite endurance athletes (88.80%), and the GSTP (p = 0.014) and GSTT genotypes (p<0.049), which presented a more favourable genetic score in elite endurance athletes than football players (56.9% vs. 42.4% and 45.0% vs. 38.6% respectively). Differences between endurance athletes and non-athletes were found only in the CYP2D6 polymorphism (p < 0.001), while in the professional football players and non-athlete population they were found in the CYPD2D6 (p < 0.001) and GSTT (p = 0.003) genes (Table 1).
Table 1. Genotype distribution in professional athletes, elite endurance athletes, professional football players and non-athletes of liver metabolism polymorphisms.
| Symbol | Gene | Polymorphism | dbSNP | Genotype Score | Professional athletes | Elite endurance athletes | Professional football players | Elite Endurance athletes vs. Professional football players p-value | non-athletes | Professional athletes vs. non-athletes p-value | Elite endurance athletes vs. non-athletes p-value | Professional football players vs. non-athletes p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP2D6 | cytochrome P450 family 2 subfamily D member 6 | c.506-1G>A | rs3892097 | 2 = GG | 272 (93.2%) | 142 (88.8%) | 130 (98.5%) | 0.002 | 98 (61.1%) | < 0.001 | < 0.001 | <0.001 |
| 1 = GA | 18 (6.2%) | 17 (10.6%) | 1 (0.8%) | 59 (37.0%) | ||||||||
| 0 = AA | 2 (0.7%) | 1 (0.6%) | 1 (0.8%) | 3 (1.9%) | ||||||||
| GSTM1 | glutathione-S transferase mu isoform 1 | "Null" polymorphism | 1 = + | 121 (41.4%) | 59 (36.9%) | 62 (47.0%) | 0.081 | 57 (35.7%) | 0.389 | 0.999 | 0.104 | |
| 0 = - | 171 (58.6%) | 101 (63.1%) | 70 (53.0%) | 103 (64.3%) | ||||||||
| GSTP | glutathione S-transferase pi | c.313A>G | rs1695 | 2 = AA | 147 (50.3%) | 91 (56.9%) | 56 (42.4%) | 0.014 | 80 (50.0%) | 0.975 | 0.306 | 0.423 |
| 1 = GA | 116 (39.7%) | 59 (36.9%) | 57 (43.2%) | 64 (39.9%) | ||||||||
| 0 = GG | 29 (10.0%) | 10 (6.3%) | 19 (14.4%) | 16 (10.1%) | ||||||||
| GSTT | glutathione S-transferase theta | +/- | 2 = +/+ | 123 (42.1%) | 72 (45.0%) | 51 (38.6%) | <0.001 | 64 (40.0%) | 0.205 | 0.779 | 0.003 | |
| 1 = +/- | 89 (30.5%) | 34 (21.3%) | 55 (41.7%) | 39 (24.4%) | ||||||||
| 0 = -/- | 80 (27.4%) | 54 (33.8%) | 26 (19.7%) | 57 (35.6%) |
Polygenic profile of iron metabolism and energy efficiency
When adding the genotype scores of HFE, AMPD1 and PGC1a polymorphisms, the mean value of the TGS in professional athletes was 49.7 a.u. (±12.0 a.u.), statistical kurtosis: 0.13 (±0.28). For the group of elite endurance athletes, it was 51.1 a.u. (±11.6 a.u.), statistical kurtosis: 0.38 (±0.38) and in the professional football players, it was 48.1 a.u. (±12.4 a.u.), statistical kurtosis: -0.11 (±0.41). The mean value of the TGS in the non-athletes was 43.3 a.u. (±11.9 a.u.) statistical kurtosis: -0.46 (±0.43). The TGS values of the professional athletes and non-athletes were statistically significant (p < 0.001). Similar between elite endurance athletes and professional football players with non-athletes (p = 0.004 and p = 0.002 respectively).
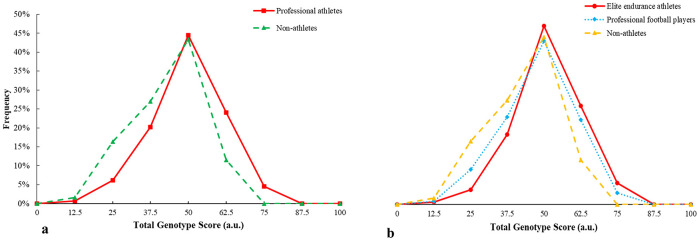
TGS distribution of iron metabolism and energy efficiency genes in the professional athletes is shifted to the right with respect to non-athletes (p < 0.001) (Fig 3a), similar between professional football players (p = 0.044) and endurance athletes (p < 0.001) with respect to non-athletes (Fig 3b).
Fig 3. TGS distribution of iron metabolism and energy efficiency in a) professional athletes and non-athlete subjects and b) elite endurance athletes and professional football players with regard to non-athlete subjects.
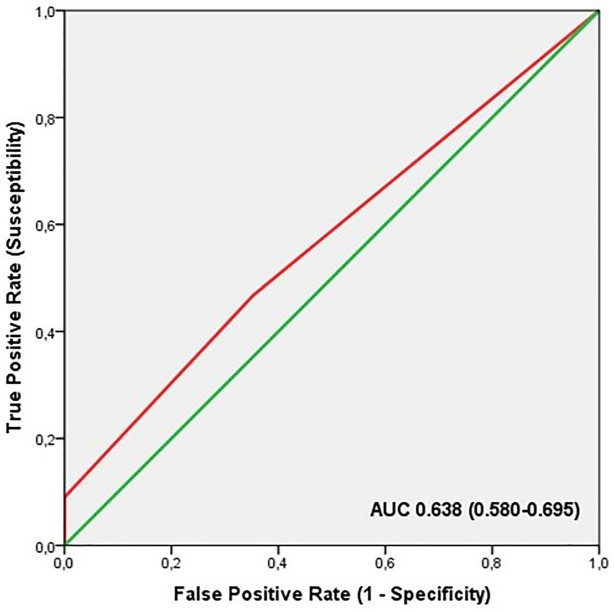
ROC analysis showed significant discriminatory accuracy of TGS in the identification of professional athletes (AUC = 0.638; 95% CI: 0.580–0.695; p < 0.001) (sensitivity = 0.729, specificity = 0.549) (Fig 4). The corresponding TGS value at this point was 43.7 a.u. Binary logistic regression analysis showed that subjects with a higher TGS of 43.7 a.u. had an OR of 2.21 (95% CI: 1.42–3.43; p < 0.001) of being professional athletes, compared to those with a TGS below this value. The elite endurance athletes showed an OR at the cut-off point in comparison to the non-athlete population of 2.82 (95% CI: 1.69–4.73; p < 0.001) and professional football players, in comparison to non-athlete subjects, had an OR of 1.69 (95% CI: 1.02–2.82; p = 0.041).
Fig 4. ROC curve summarising the ability of TGS of iron metabolism and energy efficiency genes to distinguish potential professional athletes from non-athletes.
Genotype distribution of iron metabolism and energy efficiency genes in the professional athlete’s group, when compared with the non-athlete population, was statistically significant for HFE c.187C>G (p = 0.001), showing a higher frequency in the “optimal” genotype in athletes (GG 5.8%) than the non-athlete population (GG 0.0%) and AMPD1 CC genotype (94.2% vs. 62.1% respectively; p = 0.006) (Table 2). Between both groups of professional athletes (endurance and football players), statistically significant results were found in HFE c.187C>G showing genotypes more favourable in iron absorption in endurance athletes than in professional football players (p = 0.001). Differences between endurance athletes and the non-athlete population was found in the HFE c.187C>G polymorphism (p < 0.001), similar in professional football players and the non-athlete population (p = 0.013), presenting similar results in the AMPD1 polymorphism between endurance athletes and non-athletes (p = 0.010) and professional football players and the non-athlete population (p = 0.014) (Table 2).
Table 2. Genotype distribution in professional athletes, elite endurance athletes, professional football players and non-athletes of iron metabolism and energy efficiency polymorphisms.
| Symbol | Gene | Polymorphism | dbSNP | Genotype Score | Professional athletes | Elite endurance athletes | Professional football players | Elite endurance athletes vs. Professional football players p-value | non-athletes | Professional athletes vs. non-athletes p-value | Elite endurance athletes vs. non-athletes p-value | Professional football players vs. non-athletes p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HFE | Homeostatic Iron Regulator | c.187C>G | rs1799945 | 2 = GG | 16 (5.8%) | 8 (5.0%) | 8 (6.1%) | 0.001 | 0 (0.0%) | 0.001 | <0.001 | 0.013 |
| 1 = GC | 115 (39.0%) | 78 (48.8%) | 37 (28.0%) | 43 (26.8%) | ||||||||
| 0 = CC | 161 (55.2%) | 74 (46.2%) | 87 (65.9%) | 117 (73.2%) | ||||||||
| HFE | Homeostatic Iron Regulator | c.845G>A | rs1800562 | 2 = AA | 1 (0.3%) | 0 (0.0%) | 1 (0.8%) | 0.449 | 0 (0.0%) | 0.621 | 0.998 | 0.404 |
| 1 = GA | 16 (5.5%) | 10 (6.3%) | 6 (4.5%) | 10 (6.3%) | ||||||||
| 0 = GG | 275 (94.2%) | 150 (93.7%) | 125 (94.7%) | 150 (93.7%) | ||||||||
| AMPD1 | Adenosine monophosphate deaminase 1 | c.34C>T | rs17602729 | 2 = CC | 233 (79.8%) | 128 (80.0%) | 105 (79.6%) | 0.291 | 99 (62.1%) | 0.006 | 0.010 | 0.014 |
| 1 = CT | 57 (19.5%) | 32 (20.0%) | 25 (18.9%) | 60 (37.4%) | ||||||||
| 0 = TT | 2 (0.7%) | 0 (0.0%) | 2 (1.5%) | 1 (0.5%) | ||||||||
| PGC1a | Peroxisome proliferator activated receptor coactivator | c.1444 G>A | rs8192678 | 2 = GG | 192 (65.7%) | 105 (65.6%) | 87 (65.9%) | 0.347 | 88 (54.8%) | 0.232 | 0.523 | 0.119 |
| 1 = GA | 91 (31.1%) | 53 (33.1%) | 38 (28.8%) | 63 (39.7%) | ||||||||
| 0 = AA | 9 (3.2%) | 2 (1.3%) | 7 (5.3%) | 9 (5.5%) |
Polygenic profile of cardiorespiratory fitness
When adding the genotype scores of ACE, NOS3, ADRA2A, ADRB2 and BDKRB2 polymorphisms, the mean value of the TGS in professional athletes had a value of 53.6 a.u. (±13.0 a.u.), statistical kurtosis: 0.06 (±0.28). That of the group of elite endurance athletes was 51.1 a.u. (±11.6 a.u.), statistical kurtosis: 0.14 (±0.38) and in professional football players it was 54.8 a.u. (±13.6 a.u.), statistical kurtosis: -0.13 (±0.42). The mean value of the TGS in non-athletes was 51.7 a.u. (±12.0 a.u.) statistical kurtosis: -0.18 (±0.43). The TGS values of non-athletes and professional athletes were not statistically significant but there were differences between professional soccer players and the non-athlete population (p = 0.041).
TGS distribution of cardiorespiratory fitness genes in professional athletes was similar with respect to non-athletes (p = 0.590) (Fig 5a), similar between professional football players and endurance athletes (p = 0.282) with respect to non-athletes (p = 0.830) (Fig 5b).
Fig 5. TGS distribution of cardiorespiratory fitness in a) professional athletes and non-athlete subjects and b) endurance athletes and football players with regard to non-athlete subjects.
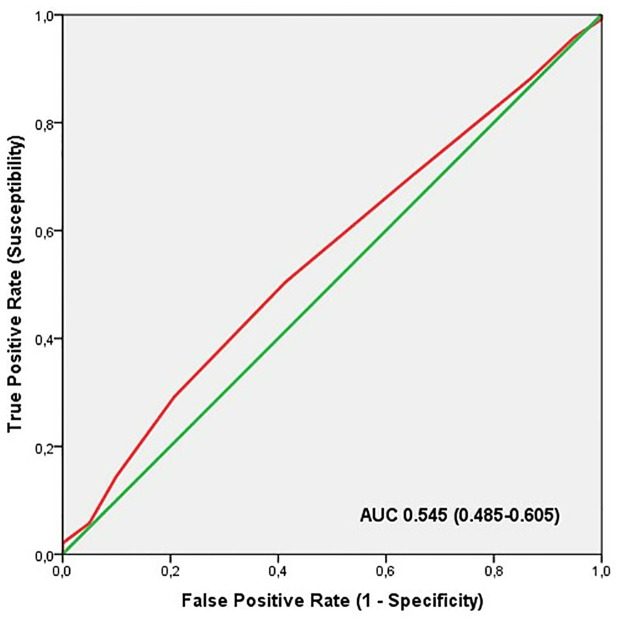
ROC analysis in this profile did not show significant discriminatory accuracy of TGS in the identification of professional athletes (AUC = 0.545; 95% CI: 0.485–0.605; p = 0.152) (sensitivity = 0.493, specificity = 0.413) (Fig 6). The corresponding TGS value at this point was 53.5 a.u. Binary logistic regression analysis showed that subjects with a higher TGS of 53.5 a.u. had an OR of 1.38 (95% CI: 0.90–2.12; p = 0.129) of being professional athletes, compared to those with a TGS below this value. The elite endurance athletes showed an OR at the cut-off point in comparison to the non-athlete population of 1.28 (95% CI: 0.79–2.06; p = 0.303) and professional football players, in comparison to non-athlete subjects, had an OR of 1.50 (95% CI: 0.91–2.48; p = 0.105).
Fig 6. ROC curve summarising the ability of TGS in cardiorespiratory fitness genes to distinguish potential professional athletes from non-athletes.
Genotype distribution of cardiorespiratory fitness genes in the professional athletes’ group when compared with the non-athlete population was statistically significant for ACE (p = 0.006), showing a higher frequency in the “non-optimal” genotype in professional athletes (DD 47.9%) than the non-athlete population (DD 38.7%), and similar in the ADRA2A c.-1291C>G GG genotype (11.5% vs. 3.3% respectively; p = 0.010). However, in the ADRB2 c.79C>G polymorphism, the professional athletes showed a higher frequency in the “optimal” genotype (CC 31.5%) than the non-athlete population (16.00%) (p < 0.001) (Table 5). Between both groups of professional athletes (endurance and football players), statistically significant results were found in NOS3 c.-786T>C showing a genotype more favourable in endurance athletes than in professional football players (p = 0.037), However, in the polymorphisms ADRB2 c.46A>G and BDKRB2–9/+9 more favourable genotypes were found in professional football players than in endurance athletes (p = 0.034 and p < 0.001 respectively). Differences between endurance athletes and non-athletes were found in the ACE I/D polymorphism (p = 0.011), NOS3 c.-786T>C (p = 0.005), BDKRB2–9/+9 (p = 0.003). Statistical differences were found in the ADRA2A c.-1291C>G and ADRB2 c.79C>G polymorphisms in professional football players regarding the non-athlete population (Table 3).
Table 3. Genotype distribution in professional athletes, elite endurance athletes, professional football players and non-athletes of cardiorespiratory fitness polymorphisms.
| Symbol | Gene | Polymorphism | dbSNP | Genotype Score | Professional athletes | Elite endurance athletes | Professional football players | Elite endurance athletes vs. Professional football players p-value | non-athletes | Professional athletes vs. non-athletes p-value | Elite endurance athletes vs. non-athletes p-value | Professional football players vs. non-athletes p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACE | Angiotensin I-converting enzyme | Alu 287bp (I/D) | rs4340 | 2 = II | 38 (13.0%) | 25 (15.6%) | 13 (9.8%) | 0.175 | 16 (10.1%) | 0.034 | 0.011 | 0.276 |
| 1 = ID | 114 (39.1%) | 56 (35.0%) | 58 (43.9%) | 82 (51.2%) | ||||||||
| 0 = DD | 140 (47.9%) | 79 (49.4%) | 61 (46.3%) | 62 (38.7%) | ||||||||
| NOS3 | Nitric Oxide Synthase 3 | c.-786T>C | rs2070744 | 2 = TT | 130 (44.6%) | 82 (51.2%) | 48 (36.4%) | 0.037 | 48 (30.3%) | 0.044 | 0.005 | 0.506 |
| 1 = TC | 114 (39.0%) | 54 (33.8%) | 60 (45.5%) | 67 (41.8%) | ||||||||
| 0 = CC | 48 (16.4%) | 24 (15.0%) | 24 (18.1%) | 45 (27.9%) | ||||||||
| NOS3 | Nitric Oxide Synthase 3 | c.894G>T | rs1799983 | 2 = GG | 137 (46.9%) | 71 (44.4%) | 66 (50.0%) | 0.406 | 63 (39.1%) | 0.227 | 0.329 | 0.201 |
| 1 = GT | 136 (46.6%) | 80 (50.0%) | 56 (42.4%) | 81 (50.8%) | ||||||||
| 0 = TT | 19 (6.5%) | 9 (5.6%) | 10 (7.6%) | 16 (10.1%) | ||||||||
| ADRA2A | Adrenoceptor α-2a | c.-1291C>G | rs1800544 | 2 = CC | 135 (46.2%) | 71 (44.2%) | 64 (48.5%) | 0.759 | 90 (56.4%) | 0.010 | 0.008 | 0.033 |
| 1 = GC | 123 (42.3%) | 69 (43.5%) | 54 (40.9%) | 66 (41.3%) | ||||||||
| 0 = GG | 34 (11.5%) | 20 (12.3%) | 14 (10.6%) | 4 (2.3%) | ||||||||
| ADRB2 | Adrenergic receptor β-2 | c.46A>G | rs1042713 | 2 = AA | 33 (11.3%) | 15 (9.4%) | 18 (13.6%) | 0.034 | 14 (9.1%) | 0.607 | 0.751 | 0.119 |
| 1 = GA | 146 (50.0%) | 91 (56.9%) | 55 (41.7%) | 87 (54.4%) | ||||||||
| 0 = GG | 113 (38.7%) | 54 (33.7%) | 59 (44.7%) | 59 (36.5%) | ||||||||
| ADRB2 | Adrenergic receptor β-2 | c.79C>G | rs1042714 | 2 = CC | 92 (31.5%) | 43 (26.9%) | 49 (37.1%) | 0.056 | 26 (16.0%) | <0.001 | <0.001 | <0.001 |
| 1 = GC | 164 (56.2%) | 100 (62.5%) | 64 (48.5%) | 77 (48.3%) | ||||||||
| 0 = GG | 36 (12.3%) | 17 (10.6%) | 19 (14.4%) | 57 (35.7%) | ||||||||
| BDKRB2 | Bradykinin Receptor β2 | +9 pb/-9 pb | rs5810761 | 2 = -9/-9 | 74 (25.3%) | 33 (20.6%) | 41 (31.1%) | <0.001 | 38 (23.8%) | 0.153 | 0.003 | 0.373 |
| 1 = -9/+9 | 129 (44.2%) | 61 (38.1%) | 68 (51.5%) | 84 (52.4%) | ||||||||
| 0 = +9/+9 | 89 (30.5%) | 66 (41.3%) | 23 (17.4%) | 38 (23.8%) |
Polygenic profile for muscle injuries
When adding the genotype scores of ACE, ACTN3, AMPD1, MLCK and CKM polymorphisms, the mean value of the TGS in professional athletes had a value of 58.3 a.u. (±11.5 a.u.), statistical kurtosis: -0.20 (±0.28). That of the group of elite endurance athletes was 57.9 a.u. (±11.8 a.u.), statistical kurtosis: -0.29 (±0.38) and in professional football players was 58.7 a.u. (±11.1 a.u.), statistical kurtosis: -0.05 (±0.42). The mean value of the TGS in non-athletes was 51.2 a.u. (±10.8 a.u.) statistical kurtosis: 0.11 (±0.43). The TGS values of the professional athletes and non-athletes were statistically significant (p < 0.001). Similar between elite endurance athletes and professional football players with non-athletes (all p < 0.001).
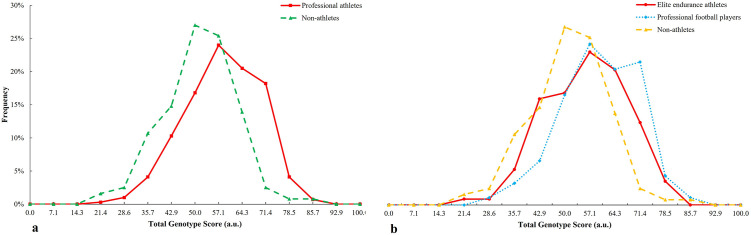
TGS distribution for muscle injuries genes in professional athletes is shifted to the right with respect to non-athletes (p < 0.001) (Fig 7a), showing similar results between elite endurance athletes and professional football players with non-athletes (p < 0.001) (Fig 7b).
Fig 7. TGS distribution for muscle injuries genes in a) professional athletes and non-athlete subjects and b) endurance athletes and football players with regard to non-athlete subjects.
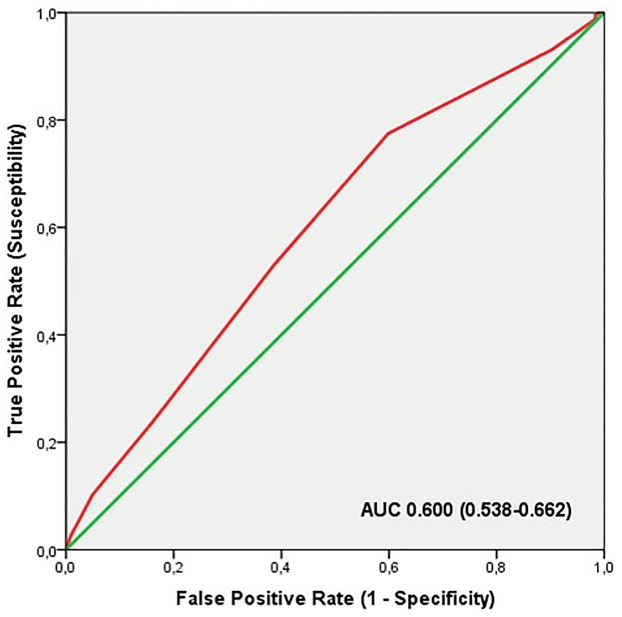
ROC analysis in this profile did not show significant discriminatory accuracy of the TGS in the identification of professional athletes (AUC = 0.600; 95% CI: 0.538–0.662; p = 0.002) (sensitivity = 0.775, specificity = 0.598) (Fig 8). The corresponding TGS value at this point was 53.5 a.u. Binary logistic regression analysis showed that subjects with a lov TGS of 53.5 a.u. had an OR of 2.70 (95% CI: 1.75–4.16; p < 0.001) of being professional athletes, compared to those with a TGS below this value. The elite endurance athletes showed an OR at the cut-off point in comparison to the non-athlete population of 2.48 (95% CI: 1.53–4.03; p < 0.001) and professional football players, in comparison to non-athlete subjects, had an OR of 2.99 (95% CI: 1.78–5.01; p < 0.001).
Fig 8. ROC curve summarising the ability of TGS for muscle injuries genes to distinguish potential professional athletes from non-athletes.
Genotype distribution for muscle injuries polymorphisms in the professional athletes’ group when compared with the non-athlete population was statistically significant for ACE I/D (p = 0.034), showing a higher frequency in the “protective” genotype in athletes (DD 47.9%) than the non-athlete population (DD 38.7%), AMPD1 CC genotype (79.8% vs. 62.1% respectively; p = 0.006) and “protective” c.37885C>A and c.49C>T MLCK polymorphisms (p < 0.001) (Table 4). Between both groups of professional athletes (endurance and football players), statistically significant results were found in MLCK polymorphisms showing genotypes more protective in professional football players than in endurance athletes (c.37885C>A; p = 0.002 and c.49C>T; p = 0.001). Differences between endurance athletes and non-athletes were found in the ACE polymorphism (p = 0.001), presenting similar results in the AMPD1 polymorphism between endurance athletes, and the non-athlete population (p = 0.013) and professional football players and non-athletes (p = 0.014). In MLCK c.37885C>A and c.49C>T polymorphisms statistically significant differences were found between both groups of endurance athletes and professional football players and the non-athlete population (p < 0.001) (Table 4).
Table 4. Genotype distribution in professional athletes, elite endurance athletes, professional football players and non-athletes for muscle injuries polymorphisms.
| Symbol | Gene | Polymorphism | dbSNP | Genotype Score | Professional athletes | Elite endurance athletes | Professional football players | Elite endurance athletes vs. Professional football players p-value | non-athletes | Professional athletes vs. non-athletes p-value | Elite endurance athletes vs. non-athletes p-value | Professional football players vs. non-athletes p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACE | Angiotensin I-converting enzyme | Alu 287bp (I/D) | rs4340 | 2 = DD | 140 (47.9%) | 79 (49.4%) | 61 (46.2%) | 0.175 | 62 (38.7%) | 0.034 | 0.011 | 0.216 |
| 1 = ID | 114 (39.0%) | 56 (35.0%) | 58 (43.9%) | 82 (51.2%) | ||||||||
| 0 = II | 38 (13.1%) | 25 (15.6%) | 13 (9.9%) | 16 (10.1%) | ||||||||
| ACTN3 | α-actinin 3 | c.1729C>T | rs1815739 | 2 = CC | 91 (31.2%) | 55 (34.4%) | 36 (27.3%) | 0.211 | 48 (30.0%) | 0.681 | 0.678 | 0.410 |
| 1 = TC | 150 (51.4%) | 82 (51.2%) | 68 (51.5%) | 91 (56.6%) | ||||||||
| 0 = TT | 51 (17.4%) | 23 (14.4%) | 28 (21.2%) | 21 (13.4%) | ||||||||
| AMPD1 | Adenosine Monophosphate Deaminase 1 | c.34C>T | rs17602729 | 2 = CC | 233 (79.8%) | 128 (80.0%) | 105 (79.6%) | 0.291 | 99 (62.1%) | 0.006 | 0.010 | 0.014 |
| 1 = CT | 57 (19.5%) | 32 (20.0%) | 25 (18.9%) | 60 (37.4%) | ||||||||
| 0 = TT | 2 (0.7%) | 0 (0.0%) | 2 (1.5%) | 1 (0.5%) | ||||||||
| CKM | Muscle-specific creatine kinase | c.*800A>G | rs8111989 | 2 = GG | 26 (8.9%) | 16 (10.0%) | 10 (7.6%) | 0.418 | 10 (5.9%) | 0.718 | 0.326 | 0.384 |
| 1 = GA | 142 (48.6%) | 88 (55.0%) | 54 (40.9%) | 72 (45.3%) | ||||||||
| 0 = AA | 124 (42.5%) | 56 (35.0%) | 68 (51.5%) | 78 (48.8%) | ||||||||
| MLCK | Myosin-light chain kinase | c.37885C>A | rs28497577 | 2 = AA | 1 (0.4%) | 0 (0.0%) | 1 (0.8%) | 0.002 | 1 (0.6%) | <0.001 | <0.001 | <0.001 |
| 1 = CA | 217 (74.3%) | 107 (66.9%) | 110 (83.3%) | 56 (35.3%) | ||||||||
| 0 = CC | 74 (25.3%) | 53 (33.1%) | 21 (15.9%) | 103 (64.1%) | ||||||||
| MLCK | Myosin-light chain kinase | c.49C>T | rs2700352 | 2 = CC | 167 (57.2%) | 79 (49.4%) | 88 (66.7%) | 0.001 | 31 (19.6%) | <0.001 | <0.001 | <0.001 |
| 1 = CT | 99 (33.9%) | 59 (36.8%) | 40 (30.3%) | 109 (68.1%) | ||||||||
| 0 = TT | 26 (8.9%) | 22 (13.8%) | 4 (3.0%) | 20 (12.3%) |
All genetic profiles, through binary logistic regression showed different prediction values of being professional athletes, both endurance athletes and professional football players, in the genes presented, with reference to non-athletes, as shown in Table 5.
Table 5. Prediction values of polygenic profiles of being professional athletes, elite endurance athletes and professional football players.
| Polygenic Profiles | Professional athletes | Elite endurance Athletes | Professional football players | |||
|---|---|---|---|---|---|---|
| ORa (95% CI)b | p-value | ORa (95% CI)b | p-value | ORa (95% CI)b | p-value | |
| Liver metabolism | 1.96 (1.28–3.01) | 0.002 | 1.79 (1.11–2.88) | 0.017 | 2.20 (1.32–3.66) | 0.001 |
| Iron metabolism and energy efficiency | 2.21 (1.42–3.43) | <0.001 | 2.82 (1.69–4.73) | <0.001 | 1.67 (1.02–2.82) | 0.041 |
| Cardiorespiratory fitness | 1.38 (0.90–2.12) | 0.129 | 1.28 (0.79–2.07) | 0.303 | 1.51 (0.91–2.48) | 0.105 |
| Muscle Injuries | 2.70 (1.75–4.16) | <0.001 | 2.48 (1.53–4.03) | <0.001 | 2.99 (1.78–5.01) | <0.001 |
aOR: Odds Ratio
b95% CI: 95% Confidence interval
Discussion
Previous research has been satisfactory in finding links between potential genetic markers associated with enhanced physiological functioning and professional sports performance [8, 59, 60]. Although polygenic profiles of athletes have previously been identified, this is the first study to investigate the polygenic profiles of several distinct physiological systems in elite endurance athletes, professional football players and the non-athlete population in a sample of homogeneously selected individuals.
It is known that in competitions like cycling and elite running the accumulated efforts over several weeks affect performance, which is also the case in professional football, due to the alteration in the redox-system of the systemic homeostasis and withdrawal of toxic products generated by the high oxidative stress produced by these sports disciplines [61, 62]. This oxidative stress in professional athletes is a determinant of performance. In this respect, the comparisons presented in the polygenic profile of liver metabolism, previously defined by Varillas et al. [24], in which it showed that endurance athletes had a higher systemic recovery capacity than the non-athlete population, presenting a metabolism that scavenges free radicals and oxygen peroxides produced by high-performance sports, are further demonstrated in this research. We propose that professional football players also present optimal hepatic metabolism genetics with respect to the non-athlete population, even with an OR higher than elite endurance athletes, thus indicating that the contact and strength exercise adapts this cohort of football players to more effective systemic cleaning (Table 5).
By using a polygenic model, it has been shown for the first time that polymorphic variations in iron metabolism and energy efficiency genes had a joint effect on the probability of becoming a professional athlete, as previously shown in a recent article [5], adding this cohort of professional football players. The significant “favourability” in the genetic profile studied in professional athletes, elite endurance athletes and professional football players versus non-athletes, presenting in elite endurance athletes a favourable adaptative factor, especially conditioned by the c.187C>G polymorphism of the HFE gene, as well as the c.34C>T polymorphism of the AMPD1 gene in both groups of athletes (Table 4), has been shown in a previous study by Ruiz et al. [33]. in genes predictive of endurance with 7 different markers associated with sports performance.
Previous studies have shown the association of the c.187C>G polymorphism of the HFE gene with professional athletes [22, 35, 63], as well as the involvement of the AMPD1 polymorphism in early muscle fatigue [64–66], revealed for the first time in professional football players, presenting allelic frequencies in these genes similar to elite endurance athletes, indicating that professional football players could present genetic factor conditioning that makes them resemble elite endurance athletes in these polygenic profiles, both in iron metabolism, energy efficiency and liver metabolism.
Cardiorespiratory fitness refers to the capacity of the circulatory and respiratory systems to supply oxygen to skeletal muscle mitochondria for energy production needed during physical activity [67, 68]. Cardiorespiratory fitness is positively associated with power and endurance exercise performance [62] and is a strong prognostic factor of morbidity and mortality from all causes and, particularly, from cardiovascular disease (CVD) [69]. The measure of an individual’s peak capacity to perform dynamic aerobic exercise is dependent on the synergistic action of pulmonary, cardiovascular and muscle tissue via a suite of physiological actions that effectively transport and deliver oxygen from the atmosphere to the mitochondria in working muscles [36, 37].
Although these interindividual variations have been previously described, the polygenic profile in polymorphisms of ACE, NOS3, ADRA2A, ADRB2 and BDKRB2 genes was similar in elite endurance athletes and in the non-athlete population, suggesting that the combination of these genes does not determine endurance performance. However, differences in TGS in cardiovascular aptitude were only shown between professional football players and the non-athlete population, suggesting that these genetic markers could be selective for successful professional football practice due to more extreme cardiovascular work than that which occurs in endurance sports, showing for the first time an “optimal” genetic profile in cardiorespiratory fitness which differentiates professional football players, from endurance athletes and the non-athlete population.
The most important finding of this study was the “favourability” of the polygenic profile in muscle injuries to professional athlete status in elite endurance athletes and professional football players.
Although excellent muscle performance in endurance sports is facilitated by an optimal polygenic profile in several key polymorphisms by muscle fibres, elasticity and metabolism [5, 24, 70, 71], this analysis indicates that the influence of these five genes is strong enough to discriminate this profile and the risk of muscle injuries [40]. The most protectives polymorphisms for muscle injuries in professional athletes compared to the non-athlete population were those in the ACE, AMPD1 and MLCK (Table 4). The c.1729C>T polymorphism of ACTN3 gene widely studied and correlated with sports performance [12, 47, 72] in the largest cohort of professional athletes presented in the literature to date, which shows no association and relevance in the identification of sports talents due to its lack of relationship to muscle injuries and not affecting sports performance [20, 59], an aspect that should be expanded in the relationship of the polymorphism of ACTN3 c.1729C> T with sports injuries in subsequent studies.
In turn, there were differences between both groups of professional athletes and the non-athlete population in the AMPD1 and MLCK genes, these being the genes that most predict “optimal” metabolism, strength and possible prevention of sports injuries (Table 4). Between the two groups of professional athletes, the polymorphisms c.37885C>A and c.49C>T of the MLCK gene present more “optimal” genetic values in professional football players than in elite endurance athletes, being protective predictors of loss of muscle strength after exercise that is produced more in football matches compared to endurance competitions [53, 54], favouring greater muscular force in football players in comparison to elite endurance athletes, presenting for the first time this genetic adaptation between these sport modalities in the largest and most homogeneous cohort analysed, being "protective" in the risk of suffering muscular injuries, an aspect that increases the capacity for sports performance and thus favours the status of elite athlete (Table 4).
Whilst genetic testing has the potential to assist in the identification of future talented performers, genetic tests should be combined with other tools, such as physiological values, and injury data to obtain an accurate identification of those athletes predisposed to succeed in the sport. Previous studies show the importance of the use of total genotype scores, composed of a high number of performance-enhancing polymorphisms, which will likely be one of the best strategies in the utilisation of genetic information to identify talent in sport [11], as presented in this research. The genetic information may represent a potentially useful adjunct to existing talent identification procedures, enhancing the process of selection. Such information should not be used as a standalone, but as an adjunct to current talent identification processes, thereby allowing the training process to become more personalised, and enabling athletes to get ever closer to their maximum potential.
Currently, we cannot use genetic information for the identification of talent because few genes have been discovered related to sports performance and many of the results shown in the literature are still contradictory [9, 10]. These contradictory effects demonstrate that there is not a singular genetic profile that confers sporting success, but that the required genetic profiles are likely specific for the characteristics of each sport [9, 11].
Accordingly, at present, only a few of the genetic markers are known that likely associate with elite athlete status, making predictions of future sporting performance based on such information not only difficult but also probably inaccurate [9, 10, 73]. In addition, testing the utility and efficacy of genetic testing in the talent identification process should be conducted to ascertain whether the information provided by genetic testing and not obtained through other traditional non-genetic tests such as physical testing, is of relevance to increase the specificity of overall talent selection [73].
For the first time, to the best of our knowledge, the relationship between these polymorphisms involved in liver metabolism, iron metabolism and energy efficiency, cardiorespiratory fitness and muscle injuries target genes is shown, leading to defining the capacity to become a professional athlete. This is a new type of genetic study, showing a definitive model of the polygenic profiles that help the capacity of physical effort in this group of subjects contributing to understanding the multiple and complex mechanisms that define it.
Subsequent studies should be carried out that amplify these polygenic profiles in elite endurance athletes, professional football players and elite athletes in sprint events to determine their ability to reach a high sports performance to corroborate the results shown in this study and to be able to conclude that these genetic markers are predisposed to support optimal talent identification in high sports performance, in order to discriminate the good from the best athletes.
Our findings demonstrates that the genetic distribution in professional athletes as regards endurance (professional cyclist and elite runners) and professional football players is different to the non-athlete population, there being a favourable polygenic profile in terms of liver metabolism, iron metabolism and energy efficiency and muscle injuries.
These genetic data presented among the analysed professional athletes show a concordance between elite endurance athletes and professional football players, supporting for the first time that players of this sports modality present genetic characteristics similar to endurance sports athletes.
These results open up a new path of research into these gene groups to complete knowledge on talent identification for high sports performance in professional athletes.
Materials and methods
Study design
A transversal prospective study.
Subjects
Four hundred and fifty-two subjects were recruited: 160 elite male endurance athletes (112 professional cyclists from Union Cycliste Internationale (UCI) World Tour teams, competing in grand tours, being UCI world champions and Grand Tour winners, with victories in the UCI World Tour and Continental Pro Tour races; and 48 elite long-distance runners: 5,000 m to marathon athletes who have competed in top-level races at the Olympic Games, World Championships, and European Championships in the marathon, half-marathon, and cross country); 132 male professional football players (from the Spanish Liga Santander and Liga Smartbank with several of them competing in the Union of European Football Associations (UEFA) Champions League and UEFA Europa League competitions representing their respective countries in international events), and 160 non-athlete male subjects. The non-athletes were matched by age with the elite athletes, the inclusion criteria being that they be non-smokers, and not suffering from chronic or acute diseases, or obesity, at the time of sampling.
Participation and acceptance of inclusion of participants were obtained by signing the informed consent document. The study protocol was approved by the Ethics Committee of the Universidad Francisco de Vitoria (32/2020) and was in accordance with the Declaration of Helsinki for Research in Humans of 1964 (last modified in 2013).
Genotypes
Target genes
In order to investigate the role of different genetic variants related to the status of elite athletes, the following polygenic profiles were selected with functional polymorphisms in the following target genes.
Liver metabolism
The polymorphisms studied were previously presented as CYP2D6 (rs3892097), GSTM1, GSTP (rs1695) and GSTT, and have been previously associated in relation to their influence on the detoxification capacity of elite athletes [24], having been presented previously as markers of drug metabolism in different pathologies [74–76].
Iron metabolism and energy efficiency
In this genetic profile, the polymorphisms of HFE c.187C>G (rs1799945) and c.845G>A (rs1800562), AMPD1 c.34C>T (rs17602729) and PGC1α c.1444 G>A (rs8192678), show a capacity for efficient iron absorption and muscle metabolism for optimal muscle performance in professional sports [5, 64, 65].
Cardiorespiratory fitness
The polymorphisms of ACE I/D (rs4340), nitric oxide synthase (NOS3) c.-786T>C (rs2070744) and c.894G>T (rs1799983), α2a adrenoceptor (ADRA2a) c.-1291C>G (rs1800544), β-2 adrenoceptor (ADRB2) c.46A>G (rs1042713) and c.79C>G (rs1042714) and bradykinin receptor B2 (BDKRB2 -9b/+9pb (rs5810761) have been shown to enhance cardiovascular training in sports, increasing VO2max, strength and sports performance [18, 39, 77–79].
Muscle injuries
Polymorphisms of the genes ACE I/D (rs4340), ACTN3 c.1729C>T (rs1815739), AMPD1 c.34C>T (rs17602729), CKM c.-800A>G (rs8111989) MLCK: c.37885C>A (rs28497577) and c.49C>T (rs2700352) have previously been linked to muscle injury incidence as well as muscle damage after competitive exercise [43, 54, 80].
Sample collection and genotyping
The extraction of DNA was carried out by oral smear with SARSTED swabs, and was kept cold until its extraction in the laboratory.
The extraction of genomic DNA from the oral mucosa samples was carried out by automatic extraction in QIACube equipment (QIAGEN, Venlo, Netherlands), with a DNA concentration yield of 25-40ng/ml, which was kept in solution in a volume of 100 microliters at -20ºC until genotyped.
All the selected genes were genotyped by multiplex analysis by Polymerase Chain Reaction-Single Nucleotide Primer Extension (PCR-SNPE), a multiplex-PCR for amplification of targeted sequences, followed by single-base extension assay of probe-primers using the commercial SNaPshot Kit (Applied Biosystems, Foster City, CA) in the Real Time-PCR instrument QuantStudio5 (QS5) (Thermofisher, CA).
Polygenic potential for detection of professional athlete status
The combined influence of each of the SNPs in the different profiles was calculated following the Williams and Folland procedure [81]. Typically, the candidate polymorphisms were bi-allelic, except for GSTM which only provided one allele. According to previous research in athletic performance, SNPs were scored as follows; a score of 2 points was given to an optimal genotype score (GS), 1 point was scored for heterozygotes and non-favourable homozygotes scored 0 points, except for the GSTM gene whose optimal GS had a value of 1.
For each polygenic profile, the scores obtained in each genetic polymorphism were added up for a perfect total genotype score that represents the optimal genotype, which was for the liver metabolic profile 7 arbitrary units (a.u.) (range 0–7 a.u.), profile energy and iron 8 a.u. (range 0–8 a.u.), cardiorespiratory fitness of 14 u.a. (range 0–14 a.u.) and muscle performance of 12 a.u. (range 0–12 a.u.). Finally, this value was transformed into a scale of 0–100 a.u. to facilitate interpretation, namely the Total Genotype Score (TGS), as follows.
Liver metabolism:
| (1) |
Iron metabolism and energy efficiency:
| (2) |
Cardiorespiratory fitness:
| (3) |
Muscle injuries:
| (4) |
As indicated above, a TGS of 100 a.u. represents a "perfect" profile and a TGS of 0 a.u. would be the "worst" profile possible when all GS have a score of 0 a.u. [81]. Finally, the distribution was evaluated of all TGS among the different profiles of professional athletes, elite endurance athletes, professional football players and non-athletes.
Statistical analysis
The statistical average and kurtosis were calculated using the Statistical Package for the Social Sciences (SPSS), v.21.0 for Windows (IBM Corp. Released 2012. IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY: IBM Corp., USA).
The HWE was tested for each SNP using χ2 tests. The probability of having an “optimal” soccer genotype for one to four genetic profiles between soccer players and non-athletes was calculated using the χ2 test with a fixed α 0.05.
The genotypic frequencies of the polymorphisms in liver-metabolising, energy and iron metabolising, cardiorespiratory fitness and muscle injuries profiles were compared between soccer players and non-athletes, using a χ2 test with fixed α 0.05. The ability of TGS to correctly distinguish potential professional athletes from non-athletes (0 = professional athlete, 1 = non-athlete) was assessed using receiver operating characteristic (ROC) curves [82]. With that purpose, the area under the ROC curve (AUC) was calculated with confidence intervals of 95% (95% CI). Finally, a binary logistic regression model was used to study the relationship between TGS and the elite endurance athlete and professional football player status.
Acknowledgments
The authors are grateful for the participation of all professional athletes (endurance and football players) and non-athlete population in this research.
Data Availability
All relevant data are within the paper. All manuscript files are available from the Research Square database (access number 10.21203/rs.3.rs-1296652/v1).
Funding Statement
This work was supported by grant from the Universidad Francisco de Vitoria (UFV) through the project “Genetic profile of elite and high-performance Caucasian athletes; comparison between endurance, power sports and non-athlete population” (UFV2020-18) (DV-D). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funder web site: UFV: https://www.ufv.es/.
References
- 1.Joyner MJ, Coyle EF. Endurance exercise performance: the physiology of champions. J Physiol. 2008;586(1):35–44. Epub 2007/09/29. doi: 10.1113/jphysiol.2007.143834 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmetov II, Egorova ES, Gabdrakhmanova LJ, Fedotovskaya ON. Genes and Athletic Performance: An Update. Med Sport Sci. 2016;61:41–54. Epub 2016/06/12. doi: 10.1159/000445240 . [DOI] [PubMed] [Google Scholar]
- 3.Tanisawa K, Wang G, Seto J, Verdouka I, Twycross-Lewis R, Karanikolou A, et al. Sport and exercise genomics: the FIMS 2019 consensus statement update. Br J Sports Med. 2020;54(16):969–75. Epub 2020/03/24. doi: 10.1136/bjsports-2019-101532 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maciejewska-Skrendo A, Leźnicka K, Leońska-Duniec A, Wilk M, Filip A, Cięszczyk P, et al. Genetics of Muscle Stiffness, Muscle Elasticity and Explosive Strength. J Hum Kinet. 2020;74:143–59. Epub 2020/12/15. doi: 10.2478/hukin-2020-0027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varillas Delgado D, Tellería Orriols JJ, Monge Martín D, Del Coso J. Genotype scores in energy and iron-metabolising genes are higher in elite endurance athletes than in nonathlete controls. Appl Physiol Nutr Metab. 2020;45(11):1225–31. Epub 2020/05/08. doi: 10.1139/apnm-2020-0174 . [DOI] [PubMed] [Google Scholar]
- 6.Ahmetov II, Hall ECR, Semenova EA, Pranckevičienė E, Ginevičienė V. Advances in sports genomics. Advances in Clinical Chemistry: Elsevier; 2021. doi: 10.1016/bs.acc.2021.07.004 [DOI] [PubMed] [Google Scholar]
- 7.Varillas-Delgado D, Del Coso J, Gutiérrez-Hellín J, Aguilar-Navarro M, Muñoz A, Maestro A, et al. Genetics and sports performance: the present and future in the identification of talent for sports based on DNA testing. Eur J Appl Physiol. 2022:1–20. Epub 20220416. doi: 10.1007/s00421-022-04945-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones N, Kiely J, Suraci B, Collins DJ, de Lorenzo D, Pickering C, et al. A genetic-based algorithm for personalized resistance training. Biol Sport. 2016;33(2):117–26. Epub 20160401. doi: 10.5604/20831862.1198210 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webborn N, Williams A, McNamee M, Bouchard C, Pitsiladis Y, Ahmetov I, et al. Direct-to-consumer genetic testing for predicting sports performance and talent identification: Consensus statement. Br J Sports Med. 2015;49(23):1486–91. Epub 2015/11/20. doi: 10.1136/bjsports-2015-095343 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickering C, Kiely J. Can Genetic Testing Predict Talent? A Case Study of 5 Elite Athletes. Int J Sports Physiol Perform. 2020;16(3):429–34. Epub 2020/12/04. doi: 10.1123/ijspp.2019-0543 . [DOI] [PubMed] [Google Scholar]
- 11.Pickering C, Kiely J, Grgic J, Lucia A, Del Coso J. Can Genetic Testing Identify Talent for Sport? Genes (Basel). 2019;10(12). Epub 2019/11/30. doi: 10.3390/genes10120972 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma F, Yang Y, Li X, Zhou F, Gao C, Li M, et al. The association of sport performance with ACE and ACTN3 genetic polymorphisms: a systematic review and meta-analysis. PLoS One. 2013;8(1):e54685. Epub 2013/01/30. doi: 10.1371/journal.pone.0054685 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmetov II, Fedotovskaya ON. Current Progress in Sports Genomics. Adv Clin Chem. 2015;70:247–314. Epub 2015/08/02. doi: 10.1016/bs.acc.2015.03.003 . [DOI] [PubMed] [Google Scholar]
- 14.Galeandro V, Notarnicola A, Bianco A, Tafuri S, Russo L, Pesce V, et al. ACTN3/ACE genotypes and mitochondrial genome in professional soccer players’ performance. J Biol Regul Homeost Agents. 2017;31(1):207–13. Epub 2017/03/25. . [PubMed] [Google Scholar]
- 15.Ginevičienė V, Pranculis A, Jakaitienė A, Milašius K, Kučinskas V. Genetic variation of the human ACE and ACTN3 genes and their association with functional muscle properties in Lithuanian elite athletes. Medicina (Kaunas). 2011;47(5):284–90. . [PubMed] [Google Scholar]
- 16.Pérusse L, Rankinen T, Hagberg JM, Loos RJ, Roth SM, Sarzynski MA, et al. Advances in exercise, fitness, and performance genomics in 2012. Med Sci Sports Exerc. 2013;45(5):824–31. doi: 10.1249/MSS.0b013e31828b28a3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagle JP, Carroll KM, Cunanan AJ, Wetmore A, Taber CB, DeWeese BH, et al. Preliminary Investigation Into the Effect of ACTN3 and ACE Polymorphisms on Muscle and Performance Characteristics. J Strength Cond Res. 2021;35(3):688–94. doi: 10.1519/JSC.0000000000002809 . [DOI] [PubMed] [Google Scholar]
- 18.Melián Ortiz A, Laguarta-Val S, Varillas-Delgado D. Muscle Work and Its Relationship with ACE and ACTN3 Polymorphisms Are Associated with the Improvement of Explosive Strength. Genes (Basel). 2021;12(8). Epub 20210729. doi: 10.3390/genes12081177 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eynon N, Hanson ED, Lucia A, Houweling PJ, Garton F, North KN, et al. Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med. 2013;43(9):803–17. Epub 2013/05/18. doi: 10.1007/s40279-013-0059-4 . [DOI] [PubMed] [Google Scholar]
- 20.Pickering C, Kiely J. ACTN3: More than Just a Gene for Speed. Front Physiol. 2017;8:1080. Epub 20171218. doi: 10.3389/fphys.2017.01080 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shahmoradi S, Ahmadalipour A, Salehi M. Evaluation of ACE gene I/D polymorphism in Iranian elite athletes. Adv Biomed Res. 2014;3:207. Epub 2014/11/06. doi: 10.4103/2277-9175.143242 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chicharro JL, Hoyos J, Gomez-Gallego F, Villa JG, Bandres F, Celaya P, et al. Mutations in the hereditary haemochromatosis gene HFE in professional endurance athletes. Br J Sports Med. 2004;38(4):418–21. Epub 2004/07/27. doi: 10.1136/bjsm.2002.003921 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucia A, Martin MA, Esteve-Lanao J, San Juan AF, Rubio JC, Olivan J, et al. C34T mutation of the AMPD1 gene in an elite white runner. BMJ Case Rep. 2009;2009. Epub 2009/01/01. doi: 10.1136/bcr.07.2008.0535 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varillas Delgado D, Telleria Orriols JJ, Martin Saborido C. Liver-Metabolizing Genes and Their Relationship to the Performance of Elite Spanish Male Endurance Athletes; a Prospective Transversal Study. Sports Med Open. 2019;5(1):50. Epub 2019/12/11. doi: 10.1186/s40798-019-0227-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarebska A, Jastrzebski Z, Ahmetov II, Zmijewski P, Cieszczyk P, Leonska-Duniec A, et al. GSTP1 c.313A>G polymorphism in Russian and Polish athletes. Physiol Genomics. 2017;49(3):127–31. Epub 2017/01/08. doi: 10.1152/physiolgenomics.00014.2016 . [DOI] [PubMed] [Google Scholar]
- 26.Thomaes T, Thomis M, Onkelinx S, Fagard R, Matthijs G, Buys R, et al. A genetic predisposition score for muscular endophenotypes predicts the increase in aerobic power after training: the CAREGENE study. BMC Genet. 2011;12:84. Epub 2011/10/05. doi: 10.1186/1471-2156-12-84 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peplonska B, Adamczyk JG, Siewierski M, Safranow K, Maruszak A, Sozanski H, et al. Genetic variants associated with physical and mental characteristics of the elite athletes in the Polish population. Scand J Med Sci Sports. 2017;27(8):788–800. Epub 2016/05/04. doi: 10.1111/sms.12687 . [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Wang D, Yan P, Yan S, Chang Q, Cheng Z. Meta-analyses of the association between the PPARGC1A Gly482Ser polymorphism and athletic performance. Biol Sport. 2019;36(4):301–9. Epub 2020/01/16. doi: 10.5114/biolsport.2019.88752 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petr M, Maciejewska-Skrendo A, Zajac A, Chycki J, Stastny P. Association of Elite Sports Status with Gene Variants of Peroxisome Proliferator Activated Receptors and Their Transcriptional Coactivator. Int J Mol Sci. 2019;21(1). Epub 2019/12/29. doi: 10.3390/ijms21010162 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tharabenjasin P, Pabalan N, Jarjanazi H. Association of PPARGC1A Gly428Ser (rs8192678) polymorphism with potential for athletic ability and sports performance: A meta-analysis. PLoS One. 2019;14(1):e0200967. Epub 2019/01/10. doi: 10.1371/journal.pone.0200967 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grealy R, Herruer J, Smith CL, Hiller D, Haseler LJ, Griffiths LR. Evaluation of a 7-Gene Genetic Profile for Athletic Endurance Phenotype in Ironman Championship Triathletes. PLoS One. 2015;10(12):e0145171. Epub 2015/12/31. doi: 10.1371/journal.pone.0145171 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janssen MC, Swinkels DW. Hereditary haemochromatosis. Best Pract Res Clin Gastroenterol. 2009;23(2):171–83. Epub 2009/05/06. doi: 10.1016/j.bpg.2009.02.004 . [DOI] [PubMed] [Google Scholar]
- 33.Ruiz JR, Gomez-Gallego F, Santiago C, Gonzalez-Freire M, Verde Z, Foster C, et al. Is there an optimum endurance polygenic profile? J Physiol. 2009;587(Pt 7):1527–34. Epub 2009/02/25. doi: 10.1113/jphysiol.2008.166645 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbara KH, Marcin L, Jedrzej A, Wieslaw Z, Elzbieta AD, Malgorzata M, et al. The impact of H63D HFE gene carriage on hemoglobin and iron status in children. Ann Hematol. 2016;95(12):2043–8. Epub 2016/11/04. doi: 10.1007/s00277-016-2792-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semenova EA, Miyamoto-Mikami E, Akimov EB, Al-Khelaifi F, Murakami H, Zempo H, et al. The association of HFE gene H63D polymorphism with endurance athlete status and aerobic capacity: novel findings and a meta-analysis. Eur J Appl Physiol. 2020. Epub 2020/01/24. doi: 10.1007/s00421-020-04306-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozemek C, Laddu DR, Lavie CJ, Claeys H, Kaminsky LA, Ross R, et al. An Update on the Role of Cardiorespiratory Fitness, Structured Exercise and Lifestyle Physical Activity in Preventing Cardiovascular Disease and Health Risk. Prog Cardiovasc Dis. 2018;61(5–6):484–90. Epub 2018/11/18. doi: 10.1016/j.pcad.2018.11.005 . [DOI] [PubMed] [Google Scholar]
- 37.Valenzuela PL, Maffiuletti NA, Joyner MJ, Lucia A, Lepers R. Lifelong Endurance Exercise as a Countermeasure Against Age-Related [Formula: see text] Decline: Physiological Overview and Insights from Masters Athletes. Sports Med. 2020;50(4):703–16. doi: 10.1007/s40279-019-01252-0 . [DOI] [PubMed] [Google Scholar]
- 38.Williams CJ, Williams MG, Eynon N, Ashton KJ, Little JP, Wisloff U, et al. Genes to predict VO(2max) trainability: a systematic review. BMC Genomics. 2017;18(Suppl 8):831. Epub 20171114. doi: 10.1186/s12864-017-4192-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varillas-Delgado D, Tellería Orriols JJ, Del Coso J. Genetic Profile in Genes Associated with Cardiorespiratory Fitness in Elite Spanish Male Endurance Athletes. Genes. 2021;12(8). doi: 10.3390/genes12081230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varillas D, Gutierrez-Hellín J, Maestro A. Genetic profile in genes associated with sports injuries in elite endurance athletes. Int J Sports Med. 2022. Epub 20220803. doi: 10.1055/a-1917-9212 . [DOI] [PubMed] [Google Scholar]
- 41.Echegaray M, Rivera MA. Role of creatine kinase isoenzymes on muscular and cardiorespiratory endurance: genetic and molecular evidence. Sports Med. 2001;31(13):919–34. doi: 10.2165/00007256-200131130-00003 . [DOI] [PubMed] [Google Scholar]
- 42.Zhou DQ, Hu Y, Liu G, Gong L, Xi Y, Wen L. Muscle-specific creatine kinase gene polymorphism and running economy responses to an 18-week 5000-m training programme. Br J Sports Med. 2006;40(12):988–91. Epub 2006/09/27. doi: 10.1136/bjsm.2006.029744 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fedotovskaia ON, Popov DV, Vinogradova OL, Akhmetov II. [Association of the muscle-specific creatine kinase (CKMM) gene polymorphism with physical performance of athletes]. Fiziol Cheloveka. 2012;38(1):105–9. . [PubMed] [Google Scholar]
- 44.Rico-Sanz J, Rankinen T, Rice T, Leon AS, Skinner JS, Wilmore JH, et al. Quantitative trait loci for maximal exercise capacity phenotypes and their responses to training in the HERITAGE Family Study. Physiol Genomics. 2004;16(2):256–60. Epub 2003/11/20. doi: 10.1152/physiolgenomics.00035.2003 . [DOI] [PubMed] [Google Scholar]
- 45.Gross M. Clinical heterogeneity and molecular mechanisms in inborn muscle AMP deaminase deficiency. J Inherit Metab Dis. 1997;20(2):186–92. doi: 10.1023/a:1005352605421 . [DOI] [PubMed] [Google Scholar]
- 46.McCabe K, Collins C. Can Genetics Predict Sports Injury? The Association of the Genes GDF5, AMPD1, COL5A1 and IGF2 on Soccer Player Injury Occurrence. Sports (Basel). 2018;6(1). Epub 20180305. doi: 10.3390/sports6010021 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, et al. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet. 2003;73(3):627–31. Epub 2003/07/25. doi: 10.1086/377590 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Druzhevskaya AM, Ahmetov II, Astratenkova IV, Rogozkin VA. Association of the ACTN3 R577X polymorphism with power athlete status in Russians. Eur J Appl Physiol. 2008;103(6):631–4. Epub 20080510. doi: 10.1007/s00421-008-0763-1 . [DOI] [PubMed] [Google Scholar]
- 49.Shang X, Huang C, Chang Q, Zhang L, Huang T. Association between the ACTN3 R577X polymorphism and female endurance athletes in China. Int J Sports Med. 2010;31(12):913–6. Epub 2010/10/12. doi: 10.1055/s-0030-1265176 . [DOI] [PubMed] [Google Scholar]
- 50.Gutiérrez-Hellín J, Baltazar-Martins G, Aguilar-Navarro M, Ruiz-Moreno C, Oliván J, Del Coso J. Effect of ACTN3 R577X Genotype on Injury Epidemiology in Elite Endurance Runners. Genes (Basel). 2021;12(1). Epub 2021/01/13. doi: 10.3390/genes12010076 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno V, Areces F, Ruiz-Vicente D, Ordovás JM, Del Coso J. Influence of the ACTN3 R577X genotype on the injury epidemiology of marathon runners. PLoS One. 2020;15(1):e0227548. Epub 2020/01/29. doi: 10.1371/journal.pone.0227548 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massidda M, Voisin S, Culigioni C, Piras F, Cugia P, Yan X, et al. ACTN3 R577X Polymorphism Is Associated With the Incidence and Severity of Injuries in Professional Football Players. Clin J Sport Med. 2019;29(1):57–61. Epub 2017/08/18. doi: 10.1097/JSM.0000000000000487 . [DOI] [PubMed] [Google Scholar]
- 53.Del Coso J, Valero M, Lara B, Salinero JJ, Gallo-Salazar C, Areces F. Myosin Light Chain Kinase (MLCK) Gene Influences Exercise Induced Muscle Damage during a Competitive Marathon. PLoS One. 2016;11(8):e0160053. Epub 2016/08/03. doi: 10.1371/journal.pone.0160053 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clarkson PM, Hoffman EP, Zambraski E, Gordish-Dressman H, Kearns A, Hubal M, et al. ACTN3 and MLCK genotype associations with exertional muscle damage. J Appl Physiol (1985). 2005;99(2):564–9. Epub 2005/04/09. doi: 10.1152/japplphysiol.00130.2005 . [DOI] [PubMed] [Google Scholar]
- 55.Yan X, Dvir N, Jacques M, Cavalcante L, Papadimitriou ID, Munson F, et al. ACE I/D gene variant predicts ACE enzyme content in blood but not the ACE, UCP2, and UCP3 protein content in human skeletal muscle in the Gene SMART study. J Appl Physiol (1985). 2018;125(3):923–30. Epub 2018/06/22. doi: 10.1152/japplphysiol.00344.2018 . [DOI] [PubMed] [Google Scholar]
- 56.Moran CN, Vassilopoulos C, Tsiokanos A, Jamurtas AZ, Bailey ME, Montgomery HE, et al. The associations of ACE polymorphisms with physical, physiological and skill parameters in adolescents. Eur J Hum Genet. 2006;14(3):332–9. Epub 2006/01/05. doi: 10.1038/sj.ejhg.5201550 . [DOI] [PubMed] [Google Scholar]
- 57.Scott RA, Moran C, Wilson RH, Onywera V, Boit MK, Goodwin WH, et al. No association between Angiotensin Converting Enzyme (ACE) gene variation and endurance athlete status in Kenyans. Comp Biochem Physiol A Mol Integr Physiol. 2005;141(2):169–75. Epub 2005/06/14. doi: 10.1016/j.cbpb.2005.05.001 . [DOI] [PubMed] [Google Scholar]
- 58.Massidda M, Miyamoto-Mikami E, Kumagai H, Ikeda H, Shimasaki Y, Yoshimura M, et al. Association between the ACE I/D polymorphism and muscle injuries in Italian and Japanese elite football players. J Sports Sci. 2020;38(21):2423–9. Epub 2020/07/03. doi: 10.1080/02640414.2020.1787683 . [DOI] [PubMed] [Google Scholar]
- 59.Guth LM, Roth SM. Genetic influence on athletic performance. Curr Opin Pediatr. 2013;25(6):653–8. doi: 10.1097/MOP.0b013e3283659087 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joyner MJ. Genetic Approaches for Sports Performance: How Far Away Are We? Sports Med. 2019. Epub 2019/11/07. doi: 10.1007/s40279-019-01164-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis NA, Howatson G, Morton K, Hill J, Pedlar CR. Alterations in redox homeostasis in the elite endurance athlete. Sports Med. 2015;45(3):379–409. Epub 2014/10/17. doi: 10.1007/s40279-014-0276-5 . [DOI] [PubMed] [Google Scholar]
- 62.Issurin VB. Biological Background of Block Periodized Endurance Training: A Review. Sports Med. 2019;49(1):31–9. Epub 2018/11/10. doi: 10.1007/s40279-018-1019-9 . [DOI] [PubMed] [Google Scholar]
- 63.Aranda N, Viteri FE, Montserrat C, Arija V. Effects of C282Y, H63D, and S65C HFE gene mutations, diet, and life-style factors on iron status in a general Mediterranean population from Tarragona, Spain. Ann Hematol. 2010;89(8):767–73. Epub 2010/01/29. doi: 10.1007/s00277-010-0901-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gineviciene V, Jakaitiene A, Pranculis A, Milasius K, Tubelis L, Utkus A. AMPD1 rs17602729 is associated with physical performance of sprint and power in elite Lithuanian athletes. BMC Genet. 2014;15:58. Epub 2014/06/03. doi: 10.1186/1471-2156-15-58 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fedotovskaya ON, Danilova AA, Akhmetov II. Effect of AMPD1 gene polymorphism on muscle activity in humans. Bull Exp Biol Med. 2013;154(4):489–91. Epub 2013/03/15. doi: 10.1007/s10517-013-1984-9 . [DOI] [PubMed] [Google Scholar]
- 66.Rubio JC, Martin MA, Rabadan M, Gomez-Gallego F, San Juan AF, Alonso JM, et al. Frequency of the C34T mutation of the AMPD1 gene in world-class endurance athletes: does this mutation impair performance? J Appl Physiol (1985). 2005;98(6):2108–12. Epub 2005/01/29. doi: 10.1152/japplphysiol.01371.2004 . [DOI] [PubMed] [Google Scholar]
- 67.Ross R, Blair SN, Arena R, Church TS, Després J-P, Franklin BA, et al. Importance of Assessing Cardiorespiratory Fitness in Clinical Practice: A Case for Fitness as a Clinical Vital Sign: A Scientific Statement From the American Heart Association. Circulation. 2016;134(24):e653–e99. doi: 10.1161/CIR.0000000000000461 [DOI] [PubMed] [Google Scholar]
- 68.Raghuveer G, Hartz J, Lubans DR, Takken T, Wiltz JL, Mietus-Snyder M, et al. Cardiorespiratory Fitness in Youth: An Important Marker of Health: A Scientific Statement From the American Heart Association. Circulation. 2020;142(7):e101–e18. doi: 10.1161/CIR.0000000000000866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaowa, Del Coso J, Gu Z, Gerile W, Yang R, Díaz-Peña R, et al. Interindividual Variation in Cardiorespiratory Fitness: A Candidate Gene Study in Han Chinese People. Genes (Basel). 2020;11(5). Epub 20200515. doi: 10.3390/genes11050555 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruiz JR, Arteta D, Buxens A, Artieda M, Gomez-Gallego F, Santiago C, et al. Can we identify a power-oriented polygenic profile? J Appl Physiol (1985). 2010;108(3):561–6. Epub 2010/01/02. doi: 10.1152/japplphysiol.01242.2009 . [DOI] [PubMed] [Google Scholar]
- 71.Muniesa CA, Gonzalez-Freire M, Santiago C, Lao JI, Buxens A, Rubio JC, et al. World-class performance in lightweight rowing: is it genetically influenced? A comparison with cyclists, runners and non-athletes. Br J Sports Med. 2010;44(12):898–901. Epub 2008/09/20. doi: 10.1136/bjsm.2008.051680 . [DOI] [PubMed] [Google Scholar]
- 72.Yang N, Garton F, North K. alpha-actinin-3 and performance. Med Sport Sci. 2009;54:88–101. Epub 20090817. doi: 10.1159/000235698 . [DOI] [PubMed] [Google Scholar]
- 73.Williams AG, Wackerhage H, Day SH. Genetic Testing for Sports Performance, Responses to Training and Injury Risk: Practical and Ethical Considerations. Med Sport Sci. 2016;61:105–19. Epub 20160610. doi: 10.1159/000445244 . [DOI] [PubMed] [Google Scholar]
- 74.Gronau S, Koenig-Greger D, Jerg M, Riechelmann H. Gene polymorphisms in detoxification enzymes as susceptibility factor for head and neck cancer? Otolaryngol Head Neck Surg. 2003;128(5):674–80. Epub 2003/05/16. doi: 10.1016/s0194-5998(03)00176-1 . [DOI] [PubMed] [Google Scholar]
- 75.Rana SV, Kamboj JK, Sharma SK, Ola RP, Sinha SK, Singh K. Antioxidant status and GST gene polymorphisms in antitubercular treatment-induced hepatotoxicity patients. Hepatol Int. 2013;7(3):876–82. Epub 2013/07/01. doi: 10.1007/s12072-013-9452-1 . [DOI] [PubMed] [Google Scholar]
- 76.Masoudi M, Saadat I, Omidvari S, Saadat M. Genetic polymorphisms of GSTO2, GSTM1, and GSTT1 and risk of gastric cancer. Mol Biol Rep. 2009;36(4):781–4. Epub 2008/04/10. doi: 10.1007/s11033-008-9245-0 . [DOI] [PubMed] [Google Scholar]
- 77.Sawczuk M, Maciejewska-Karlowska A, Cieszczyk P, Skotarczak B, Ficek K. Association of the ADRB2 Gly16Arg and Glu27Gln polymorphisms with athlete status. J Sports Sci. 2013;31(14):1535–44. Epub 2013/05/02. doi: 10.1080/02640414.2013.786184 . [DOI] [PubMed] [Google Scholar]
- 78.Saunders CJ, Xenophontos SL, Cariolou MA, Anastassiades LC, Noakes TD, Collins M. The bradykinin beta 2 receptor (BDKRB2) and endothelial nitric oxide synthase 3 (NOS3) genes and endurance performance during Ironman Triathlons. Hum Mol Genet. 2006;15(6):979–87. Epub 2006/02/08. doi: 10.1093/hmg/ddl014 . [DOI] [PubMed] [Google Scholar]
- 79.Gomez-Gallego F, Ruiz JR, Buxens A, Artieda M, Arteta D, Santiago C, et al. The -786 T/C polymorphism of the NOS3 gene is associated with elite performance in power sports. Eur J Appl Physiol. 2009;107(5):565–9. Epub 2009/08/25. doi: 10.1007/s00421-009-1166-7 . [DOI] [PubMed] [Google Scholar]
- 80.Lippi G, Longo UG, Maffulli N. Genetics and sports. Br Med Bull. 2010;93:27–47. Epub 2009/02/12. doi: 10.1093/bmb/ldp007 . [DOI] [PubMed] [Google Scholar]
- 81.Williams AG, Folland JP. Similarity of polygenic profiles limits the potential for elite human physical performance. J Physiol. 2008;586(1):113–21. Epub 2007/09/29. doi: 10.1113/jphysiol.2007.141887 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39(4):561–77. Epub 1993/04/01. . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper. All manuscript files are available from the Research Square database (access number 10.21203/rs.3.rs-1296652/v1).