Abstract
Sugarcane (Saccharum spp.) is a special crop plant that underwent anthropogenic evolution from a wild grass species to an important food, fodder, and energy crop. Unlike any other grass species which were selected for their kernels, sugarcane was selected for its high stem sucrose accumulation. Flowering in sugarcane is not favored since flowering diverts the stored sugar resources for the reproductive and developmental energy needs. Cultivars are vegetatively propagated and sugarcane breeding is still essentially focused on conventional methods, since the knowledge of sugarcane genetics has lagged that of other major crops. Cultivar improvement has been extremely challenging due to its polyploidy and aneuploidy nature derived from a few interspecific hybridizations between Saccharum officinarum and Saccharum spontaneum, revealing the coexistence of two distinct genome organization modes in the modern variety. Alongside implementation of modern agricultural techniques, generation of hybrid clones, transgenics and genome edited events will help to meet the ever-growing bioenergy needs. Additionally, there are two common biotechnological approaches to improve plant stress tolerance, which includes marker-assisted selection (MAS) and genetic transformation. During the past two decades, the use of molecular approaches has contributed greatly to a better understanding of the genetic and biochemical basis of plant stress-tolerance and in some cases, it led to the development of plants with enhanced tolerance to abiotic stress. Hence, this review mainly intends on the events that shaped the sugarcane as what it is now and what challenges ahead and measures taken to further improve its yield, production and maximize utilization to beat the growing demands.
Keywords: Sugarcane, Origin and domestication, Nobilization, Breeding, Genetic transformation, Transcriptomics, Genome sequencing, Marker-assisted selection
Introduction
Sugarcane (Saccharum spp.) is a robust and vigorous tropical plant, with superior growth over most other crop species partially because of its high photosynthetic efficiency as a C4 grass. Further, it contributes about 70% of the world sugar production and holds a great potential for the biomass as well as ethanol-based biofuel production. The global sugarcane production in 2018 has crossed 1,907,024,730 tons (190.7 million metric tons/MMT) with Brazil and India being as top producers (FAO 2020, 2021). Evolutionarily, the genus Saccharum has diverged from maize around 15–20 million years ago (mya) and split from sorghum as recently as 6–9 mya (Paterson et al. 2004). Molecular tracing techniques have suggested the origin of sugarcane in South-East Asia and New Guinea (Lebot 1999) and the domestication of sugarcane was happened about 10,000 years ago. Daniels and Roach (1987) has proposed S. robustum E. W. Brandes & Jeswiet ex Grassl to be the wild progenitor of S. officinarum L. (Thirugnanasambandam et al. 2018), while S. robustum itself is an introgressed hybrid of S. spontaneum L. along with species of other genera such as Erianthus and Miscanthus. Notable contributions from S. barberi, S. sinense Roxb. and species of Narenga are also involved (Daniels and Roach, 1987; Altpeter and Oraby 2010). The Dutch plant breeders in Java have also contributed for intensive sugarcane breeding to develop modern cultivars that subsequently enriched the sugarcane gene pool (D’Hont et al. 1996). Interspecific hybridization during domestication and by recent plant breeders who made the sugarcane genome much more complex than many other familiar crops (Dillon et al. 2007). Domesticated sugarcane is a vegetatively propagated, polyploid, aneuploid and an interspecific hybrid having an estimated genome size of > 10 Gb. Much of the genome contributions came from S. officinarum (80%) and S. spontaneum (10–15%), and with some recombinant chromosomes (5–10%) (Garsmeur et al. 2018; Zhang et al. 2018). S. robustum-derived S. officinarum that originated in New Guinea is a stout, juicy, less-fibrous with good amount of sugar and less disease resistant. Whereas S. spontaneum, which originated from India, has low sugar content but with higher stress and disease tolerance. The capability to accumulate more sucrose in the stem and the ability to tolerate variety of stress conditions are considered as the major desirable characteristics in sugarcane breeding and these two major genome contributors influence the sugarcane agronomic characters (Altpeter and Oraby 2010).
As cane sugar has been the sweetener of choice for over 2000 years, the ever-increasing demand for sugar and bioenergy raw materials for a rapidly growing population has pushed farmers to exploit the sugarcane gene pool to the maximum limits. In the modern age, conventional breeding and transgenic technologies are employed to generate sugarcane plants with higher yield and resilience against adverse abiotic and biotic factors. Intense sugarcane breeding practices by Dutch colonials in the Dutch East Indies have further expanded the gene pool of cultivated sugarcane and produced sugarcane varieties with better yield as well as more resilient characteristics (Stevenson 1965). The first transgenic sugarcane was reported in 1992 and following this, several research groups were came up with different transgenic events against various factors (Bower and Birch 1992; Nerkar et al. 2018). The arrival of high-throughput sequencing and accumulating transcriptomic data had shed light on specific genes with critical and agronomically important functions (Lovejot Kaur et al. 2017). As in any other crop, whole genome sequencing (WGS) (Garsmeur et al. 2018; Zhang et al. 2018) will further extend the knowledge about the crop and will lead the researchers to generate crops with better performance through functional genomics. Scientists further expose sugarcane with cutting edge genome editing technologies to tap the potential for greater extent (Jung and Altpeter 2016; Kannan et al. 2018). Unlike the model rice crop (Ramkumar et al. 2015), limited gene silencing studies were also done in sugarcane (Ingelbrecht et al. 1999). Due to the general complexity of abiotic stress tolerance and the difficulty in phenotypic selection for stress tolerance, Marker assisted selection (MAS) has been considered as an effective approach to improve plant stress tolerance (Foolad and Sharma 2005). However, the use of this approach requires identification of genetic markers that are associated with genes or QTL’s affecting whole plant stress tolerance or individual components contributing to it. Furthermore, applications of metabolomics and transcriptomics approaches will lead to a better understanding of the molecular basis of plant life, starting from signal perception to signal transduction and expression of genes and regulatory elements for combined or multiple abiotic stress as well as cross tolerance in sugarcane. In this review, we time-lined the journey of sugarcane from its wild progenitor to a modern crop plant equipped with a higher yield and well-armed against adverse factors like abiotic and biotic stress to feed the people and fuel the world.
Origin and domestication
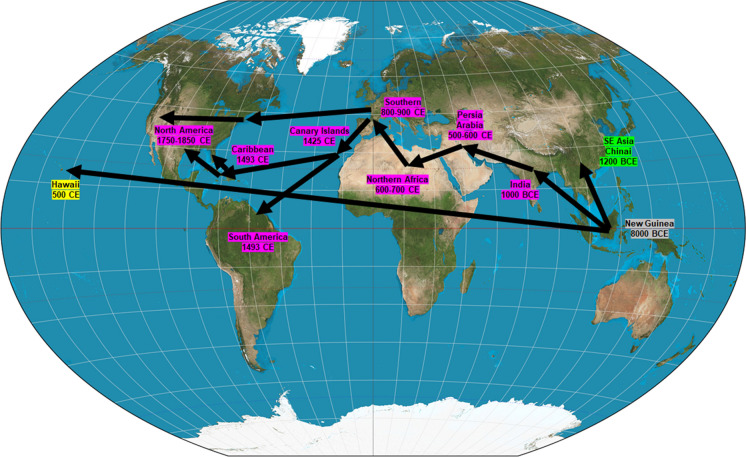
Sugarcane has enhanced its status from an unknown wild grass plant to the world’s largest cultivated cash crop with its incredible ability to synthesize and accumulate sucrose in its stem. Sugarcane domestication was started around 8000 BC in the region of New Guinea and later it reached Southeast Asia and India. During fifth century AD, a novel strategy for crystallization of sugar was invented by an Indian scientist who facilitated the storage and transportation of crystallized sugar substances easier. Following this invention, the process of sugarcane cultivation, refining sugarcane juice and production of granulated sugar was spread to China, Persia and the Mediterranean regions. Sugarcane has started spreading across the Asia, Europe and Arab nations where people started to cultivate sugarcane, especially after they conquered Egypt. Sugarcane has reached Spain around 715 AD and certain countries (Spain, Portugal, Italy, Cyprus and Azores) have tried to stabilize their economy using sugarcane production during sixteenth and seventeenth centuries. As the New World has offered better climatic conditions for the growth and better productivity, sugarcane varieties were also quickly introduced in the United States (Fig. 1).
Fig. 1.
Timeline of the anthropogenic intercontinental distribution of sugarcane. The major distribution routes were through India, then to the Middle East, Africa, Europe and finally to the Americas
Sugarcane belongs to the genus Saccharum, family Poaceae with the tribe Andropogoneae. Genera such as Miscanthus, Sclerostachya, Erianthus, Narenga, and Saccharum are closely related and forming the Saccharum complex. However, S. officinarum is the original sugarcane species and is supposed to have originated in the Indonesian Archipelago (Fig. 1). The species does not occur in the natural wild conditions but was grown and maintained for a long time by the island natives. The colonial Dutch workers have called S. officinarum as the ‘noble cane’ as it was used as the main source material for sugar production. The process of crossing and back-crossing was designed towards the development of hardy and disease resistance traits with sweeter noble canes is termed as ‘nobilization’ (Stevenson 1965). Present day sugarcane is a man-made hybrid clone produced from S. officinarum, and S. spontaneum with a few genes incorporated from S. barberi, and S. sinense and to a limited extent from S. robustum.
Saccharum spontaneum is a polymorphic wild grass distributed in the tropics and sub tropics (Panje and Babu 1960)
Saccharum robustum is a tall and thick canes spread in riverbanks of New Guinea and Indonesia (Mukherjee 1957)
Saccharum officinarum, the noble sugarcane, is present only under domesticated conditions in New Guinea and Indonesia (Parthasarathy 1948)
Saccharum barberi Jesw. and S. sinense Roxb. are North Indian and Chinese canes that were under cultivation for sugar production, and are believed to be produced from interspecific crosses between S. officinarum and local varieties of S. spontaneum (D’Hont et al. 2002). More recently, S. barberi and S. sinense are considered as single species with S, sinense as acccepted name and S. barberi as a synonym (World Flora online).
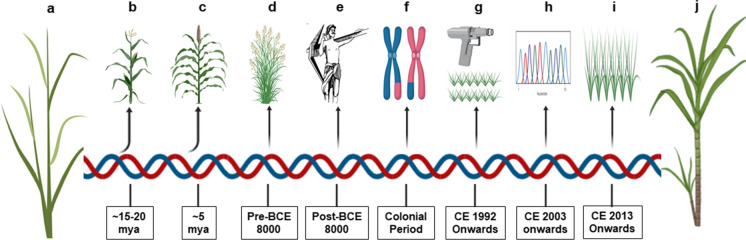
Saccharum edule Hassk. is a polymorphic species with aneuploidy and is cultivated in the Pacific islands for its aborted inflorescence, which is used as a vegetable. The cane of S. edule resembles that of S. robustum, and is a natural hybrid involving S. robustum, S. officinarum and Miscanthus spp (Roach 1972). However, in the recent years S. edule has been identified as S. spontaneum var. edulis (Hassk.) K.Schum. (World Flora online). S. officinarum is evolutionarily derived from the three groups of S. robustum, namely Red fleshed, Port Moresby and Teboe Salah, which are again evolved through interspecific introgression between S. spontaneum, S. arundinaceum Retz. and M. sinensis (Daniels J and Roach BT 1987; Daniels, J et al. 1989). It is hypothesized that S. officinarum is anthropogenically selected from S. robustum in New Guinea, as a sweeter chewing plant in riverbanks and lowlands or as a sweeter fencing plant in highlands or as a plant that was selectively damaged by rats and pigs for its sweetness, which drew human attention (Fig. 2).
Fig. 2.
Timeline of sugarcane. a. Progenitor grass species. b. Evolutionary split of maize. c. Evolutionary split of sorghum. d. Genus Saccharum pre-domestication. e. Selection and domestication. f. Breeding and improvement. g. Transgenic development. h. Omics research. i. market release of transgenic sugarcane. j. sugarcane with better traits
Sugarcane is a cross-pollinated crop with negligible selfing. Even though sugarcane flowers have reduced male fertility, they are rarely male sterile. Sugarcane pollens are very small with a half-life of just 12 min. They become non-viable beyond 35 min, when exposed to 26° C and 67% relative humidity (Moore 1976). Therefore, viable pollens are not expected to travel faraway in the field. However, they can be stored at a reduced temperature and increased relative humidity, where they retain some viability for up to 14 days. The flowering behavior of sugarcane is considerably significant for breeders to develop new varieties, whereas it is not a favorable character for farmers (Jeswiet J 1925).
Global economic importance
Sugarcane is a major industrial crop. Ever since 1961 to 2018, the global area under sugarcane cultivation has been increased from 8,911,979 hectares to 26,269,819 hectares, while the production has increased from 44.7 MMT to 190.7 MMT (FAO 2020). This increased production has significantly improved the livelihood of the sugarcane farmers worldwide. Furthermore, sugarcane stands as a major raw material for more than 75% of sugar production in the world. The worldwide demand for the cane sugar increases on each year and high demand for the sugar and sugar based products efficiently drives the sugarcane agriculture and sugar industries towards a profitable future. Sugarcane has been projected as a preferred cultivation crop for not only towards sugar production, but also for the production of bioenergy and bioethanol. Many of the European Union (EU) member states are already using some percentage of bioethanol or biodiesel in diesel or petrol fuels. Bioethanol is commonly mixed with petrol in various proportions starting from 10 to 85% along with case modification in the vehicles. In the case of Norway, nearly about 7% of bioethanol is used in standard octane petrol (Sundvor and Lopez-Aparicio 2014).
Compared to any other crop residues, the sugarcane residues are a rich source of lignocellulosic materials and most of the sugarcane residues are used for the fuel ethanol synthesis or as a source of energy. Bioethanol can be readily produced from various sugarcane residues, particularly from sugarcane juice and bagasse, as well as from the residues of the shoot system. Bioethanol production from sugarcane is a viable option for the use of huge deposits of sugarcane industry byproducts which are cost effective and can reduce the emission of greenhouse gases effectively. There are separate bioethanol producing mills that are maintained in Brazil and at given operating conditions, these ethanol distilleries can produce 82 to 85 L.t −1 (Liters per metric ton) of ethanol from freshly crushed sugarcane. At present, the transport sector accounts for about 23% of global energy consumption, which is a major cause of greenhouse gas emissions and eventually leads global warming (REN21 2019). The World Energy Council (WEC) has predicted a 200–300% rise in demand for transportation fuel, especially from China and India. Moreover, countries should achieve the mandated emission levels by 2050, which is possible only through a reduction in fossil fuel consumption. This increased demand for the transport fuel has been met with the production and utilization of bioethanol from the sugarcane by-products and this demand even increases the sugarcane production globally. Additionally, the sugarcane industries produce huge quantities of byproducts which are largely used as a major source of organic nutrients in the agricultural crop production. The direct application of these byproducts from sugarcane industries such as press mud or filter cake and bagasse in the field enhances the physical, chemical as well as biological properties of soil and increases the quality of cultivated crops and their yield. The use of these sugarcane byproducts efficiently cuts down the requirement for chemical fertilizers and reduces the recommended dose of fertilizers (Dotaniya et al. 2016). The major ten sugarcane producing countries around the globe are listed in Table 1.
Table 1.
Major sugarcane producing countries
| Countries | Production (Tonnes) |
|---|---|
| Brazil | 746,828,157 |
| India | 376,900,000 |
| China | 108,718,971 |
| Thailand | 104,360,867 |
| Pakistan | 67,173,975 |
| Mexico | 56,841,523 |
| Colombia | 36,276,860 |
| USA | 31,335,984 |
| Philippines | 24,730,820 |
| Indonesia | 21,744,000 |
| World | 1,907,024,730 |
The global production of sugarcane for 2019–20 is estimated to get reduced to 174 MMT, due to the reduced production in India, Brazil and US. Furthermore, the sugar consumption in India is anticipated to increase in parallel with a rising population of an anticipated 1.65 billion by 2050, demanding more sugar production. Unfortunately, the cropping area may not expand beyond 5 to 5.5 million ha in India and the increased demand must be met by improved sugarcane productivity and sugar recovery. This can be achieved by not only through practicing modern agricultural technologies and modernizing industrial sugar extraction, but also by integrating cutting edge research technologies to generate sugarcane with novel traits for better yield and resilience.
Economic importance in India
In India, Sugarcane is cultivated as a major cash crop in the tropical and sub-tropical regions. It is used as a main source of raw material for sugar production in the Indian subcontinent. With nearly about 5 million farmers involved, India ranks as the second largest producer of sugarcane (Table. 1). Sugarcane is grown in 2.57% of the total cultivated area, with a gross contribution of 1.1% to the national GDP. With respect to agricultural GDP, sugarcane contribution has jumped from a mere 5% in 1990–91 to 10% in 2010–11. Sugar industries are situated mostly in rural areas and contribute significantly to the rural economy by providing employment for nearly about 4% of the population. Alongside sugar, various by-products of the sugar industry (Jaggery, ethanol, electricity, paper and high value biomolecules) contribute to the economic growth as well. Various other byproducts of sugarcane industries such as bagasse, vinasse and molasses derived from the process of sugar production, are widely used for various purposes in rural India (Dotaniya et al. 2016; Santos et al. 2020). Bagasse is mainly utilized for heat and power production in boilers and cogeneration units during milling and distillation stages. Molasses have high energy value and are widely used as a food ingredient and as a raw material for ethanol production. Vinasse is mainly used in the irrigation process to increase the soil fertility (Steffi Formann et al. 2020). The sugar price in India is the lowest selling price in the world. The growth of sugarcane agriculture in the country has been increased from 1.17 mha in 1930–31, to 5.01 mha by 2013–14; a fourfold increase (Vision 2050, SBI, ICAR 2015). During this period, the productivity is also raised from 31 t/ha to 68 t/ha, the sugarcane production increased from 3.7 MMT to 35.5 MMT, and the sugar production increased from 0.12 MMT to 27.9 MMT. Sugar recovery has also showed an improvement from 9.05% to 10.27%. The number of sugar factories in operation has also gone up from 29 to over 527. This tremendous growth in cane and sugar production was contributed by two factors: a fourfold increase in crop area and an improvement in productivity by more than 100%. Both these became possible because of the development of new, well adapted varieties along with efficient crop production and crop protection technologies developed by the ICAR- SBI since 1912 (Vision 2050, SBI, ICAR 2015; Solomon 2016).
At present, India blends petrol with 5% of ethanol, and is targeting to reach 10% by 2022, a gross mismatch when compared with Brazil (27%). A policy shift towards higher ethanol utilization in the Indian energy sector is unavoidable. The molasses-mediated ethanol production is insufficient to meet this and it demands diverting of excess sugar production towards direct ethanol production from sugarcane juice. It is necessary to consider establishing energy plantations for ethanol and biomass production. Since expansion of cane area is not possible, such plantations must be established in marginal and underutilized lands with allied canes such as Erianthus, energy canes and recognized high biomass producing sugarcane clones. Hence to meet the increased demand for sugar, ethanol and power, the sugarcane production should be 55 MMT with 11% sugar recovery. Based on this prediction, sugarcane production has been increased by 5 million tons annually, along with increased sugar recovery. On the other hand, there are overriding concerns about the static productivity, high cost of cultivation, depletion of natural resources and climate change etc. Improving productivity against these constraints to achieve the projected production targets of 2050 is the real challenge to the sugarcane research and development establishments of the country. These challenges can be met with improved sugarcane hybrids and transgenics with agriculturally important traits.
Conventional breeding and trait improvement
The genetic bottleneck tightened by selective domestication has often reduced the gene pool of crop species. Despite a number of limitations such as narrow genetic pool, polyploid genome, poor fertility and very long breeding cycle (10–15 years), the conventional breeding approaches had significantly contributed to the crop improvement. Moreover, the recent sugarcane hybrids have a variable chromosome number (2n = 100–120) with an inhibited flowering nature. To sustain/improve sugarcane production and productivity with the limited available land, tolerance to abiotic and biotic stresses, integrated nutrient as well as pest management and increased sugar recovery are standing as real challenges. Both the conventional and molecular techniques need to be integrated. Although crop tolerance to biotic and abiotic stresses has been selected by conventional breeding programs, reducing the breeding cycles is the most crucial factor in developing improved varieties. The cultivated commercial sugarcane hybrids are developed by crossing Saccharum officinarum, (noble cane); with Saccharum barberi, Saccharum sinense, and clones of two other wild species such as Saccharum spontaneum and Saccharum robustum. Among these plants, the genes for sucrose accumulation are inherited from Saccharum officinarum, and Saccharum barberi and Saccharum sinense as well. Whereas Saccharum spontaneum and Saccharum robustum have genetically contributed for biotic and abiotic stress tolerance and higher biomass production.
During the initial period of sugarcane cultivation, the majority of sugarcane industries were totally dependent on the noble canes and a few were dependent on S. barberi and S. sinense. But these varieties were poor in yield as well as in adaptability. They are susceptible to diseases and pests. In order to overcome these constrains, the colonial Dutch plant breeders in the East Indies have played a crucial role in breeding sugarcane varieties to develop high yield varieties. Their backcross experiments (nobilization) have helped to produce resistant sugarcane varieties. These resistant interspecific sugarcane varieties are distributed throughout the world and are used as a foundation clones for many of the sugarcane breeding programs. Later, in 1912, the Imperial Sugarcane Breeding Station, Coimbatore, was established in India, where Co.205—the first generation of the commercial sugarcane variety was produced by the cross between S. officinarum and S. spontaneum. Later, clones of S. officinarum, S. spontaneum, and S. barberi were backcrossed to S. officinarum and the progenies of these species were utilized for the development of improved sugarcane varieties (Sreenivasan et al. 1987). In the Sugarcane breeding institute (SBI), more emphasis was given to the production of inter-specific hybrids (ISH) and a total of 486 ISH clones were produced using S. officinarum, S. spontaneum, S. barberi, S. sinense, S. robustum, and some indigenous as well as exotic ISH derivatives as parents. Today, over 90% of the Indian sugarcane cultivation areas are occupied by the varieties developed by the SBI as well as by the State Sugarcane Research Stations across the country.
Despite all the possibilities of improving sugarcane through introgression breeding, it is very difficult to anticipate its impact or success (Stalker 1980; Berding and Roach 1987). Hence, the introgression process in sugarcane breeding is therefore traditionally termed as a long-term and risky investment by Wang et al. 2010. Present day sugarcane cultivars are highly heterozygous, with several different alleles at each locus known as genomic redundancy, which may possess evolutionary advantage or the duplicated genes might have novel functions. However, marker-assisted selection (MAS) has arisen as a powerful tool to facilitate genetic manipulation through the identification of candidate genes for various traits of sugarcane and to develop saturated genetic maps. Molecular markers such as RAPD (al-Janabi et al. 1993), RFLP (Lu et al. 1994; da Silva et al. 1995), AFLP (Barbosa et al. 2003) and SSR (Selvi et al. 2003) have already been used in sugarcane for genotyping as well as genetic mapping and efforts have been made to develop molecular markers using these techniques (Lu et al. 1994; Glaszmann et al. 1997; Alix et al. 1998; Jannoo et al. 2007).
Genetic transformations and crop resilience
Unlike any other model or major crop species from 1983 (Ramkumar et al. 2020), a complete sugarcane genetic transformation and plant regeneration system was established a decade later (Bower and Birch 1992), even though the transformation of protoplast and cell suspension culture were reported earlier. This delay is a reflection of the lack of a plant regeneration system, which was established later. This laid down the foundation for a surge in developing sugarcane transgenics with novel traits. Though various techniques are followed for the transformation and genetic improvement of sugarcane, the Agrobacterium mediated gene transformation (Provides 85–100% of calli transformation with assured transgene integration) and Biolistic particle bombardment techniques (Shows 39% of transformation efficiency in broad range of sugarcane varieties) are considered as novel methods of gene delivery in many sugarcane varieties (Santosa et al. 2004; Ramasamy et al. 2018). Both of these methods are widely used to introduce different types of genes for the genetic improvement of sugarcane varieties. Details of various genes transferred to sugarcane, their sources and importance were listed in Table 2.
Table 2.
List of specific genes transformed to Sugarcane
| Name of the genes | Functional role | References |
|---|---|---|
| nptII | Kanamycin resistant gene | Bower and Birch 1992 |
| EPSPS | Glyphosate tolerant gene | Wang et al. 2017 |
| mutated ALS | In-vitro Selectable marker gene used to identify transformed callus | van der Vyver et al. 2013 |
| bar gene encoding a bacterial PPT acetyltransferase | Confers resistance against herbicide glufosinate ammonium and can be used as an effective selectable marker gene in sugarcane transformation | Gallo-Meagher and Irvine 1996; Falco et al. 2000; Budeguer et al. 2021 |
| Cry1a | Insecticidal endotoxin from Bacillus thuringiensis | Arencibia et al. 1997 |
| Cry1ac | Insecticidal endotoxin from Bacillus thuringiensis used against sugarcane stem borers | Weng et al. 2011; Zhou et al. 2018; Dessoky et al. 2021 |
| Cry1ab | Insecticidal endotoxin from Bacillus thuringiensis used to control Sugarcane borer Diatraea saccharalis | Cristofoletti et al. 2018 |
| Cry1aa3 | Insecticidal endotoxin from Bacillus thuringiensis used to induce resistance against Sugarcane borer insects Chilo sacchariphagus (Internode borer) and Scripophaga excerptalis (Early shoot borer) | Kalunke et al. 2009 |
| Cry2A | Insecticidal endotoxin from Bacillus thuringiensis used against Lepidopteran insects | Qamar et al. 2021 |
| Cry2ab | Insecticidal endotoxin from Bacillus thuringiensis used to control Sugarcane borer Diatraea saccharalis | Kumar and Udayasuriyan 2004; Cristofoletti et al. 2018 |
|
Snowdrop lectin (Galanthus nivalis agglutinin) |
Lectin used against Woolly aphid Ceratovacuna lanigera | Zhangsun et al. 2007 |
|
Amaranthus viridis agglutinin gene and soybean Kunitz tripsin inhibitor, AVAc-SKTI |
Fusion insect resistant gene construct used to induce resistant against Diatraea saccharalis and Ceratovacuna lanigera | Deng et al. 2008 |
| Potato proteinase inhibitor II | Used to induce resistance against Antitrogus consanguineus | Nutt et al. 2001 |
| albD gene | Albicidin detoxification gene used against against leaf scald disease causing bacterium Xanthomonas albilineans | Zhang and Birch 1997 |
| truncated endochitinase gene | Used against red rot fungal disease causing fungus Colletotrichum falcatum | Viswanathan et al. 2006 |
| β-1,3-glucanase gene | Used against red rot fungal disease causing fungus Colletotrichum falcatum | Nayyar et al. 2017 |
| Coat protein gene of sugarcane mosaic virus | Used to induce resistance against Sugarcane mosaic virus | Joyce et al. 1998; Yao et al. 2017 |
| Coat protein of Sorghum mosaic potyvirus | Used to induce resistance against Sorghum mosaic Potyvirus | Ingelbrecht et al. 1999 |
| Bacterial betA gene (Choline dehydrogenase) | Used to produce drought tolerant Sugarcane. betA gene increases Glycine betaine which acts as an osmoprotectant and help sugarcane to acclimate in water deficit conditions | Sugiharto 2017 |
| Trehalose synthase from Grifola frondosa | Used for the induction of drought tolerance in sugarcane | Zhang et al. 2006 |
| AVP1 | Kumar et al. 2014 | |
| DREB2 | Reis et al. 2014 | |
| HSP70 | Augustine et al. 2015b | |
| Pea DNA helicase 45 | Augustine et al. 2015a | |
| Scdr2 | Potential drought responsive gene in sugarcane varieties | Begcy et al. 2019 |
| P5CS | Used for the induction of drought and salinity tolerance in sugarcane | Molinari et al. 2007 |
| Isopentenyl transferase | Used for the induction of cold tolerance in sugarcane | Belintani et al. 2012 |
| Sucrose Phosphate synthase (SoSPS) | Used to increase the sucrose content by overexpression | Groenewald and Botha 2008; Anur et al. 2020 |
| pyrophosphate: fructose 6-phosphate 1-phosphotransferase (PFP) | Used to increase the sucrose content by downregulation | |
| Sucrose isomerase | Used in doubling of sucrose content by targeting to vacuoles | Wu and Birch 2007 |
| Caffeoyl-CoA O-methyltransferase (CCoAOMT) | Down regulated via RNAi based methods for the possible reduction of lignin content in sugarcane | Jung et al. 2013; Bewg et al. 2016 |
| ferulate 5-hydroxylase (F5H) | ||
| caffeic acid O-methyltransferase (COMT) | ||
| BAHD acyltransferase | Silencing of a BAHD acyltransferase in sugarcane increases biomass digestibility | Souza et al 2019 |
Generally, sugarcane lacks herbicide resistance and is susceptible to commercial herbicides. Once after the transformation of sugarcane with nptII gene by Bower and Birch (1992), several other genes such as bar, EPSPS and mutated ALS genes were also introduced against selected herbicides (reviewed in Nerkar et al. 2018) (Table 2). The Initial research with transgenic sugarcane plants resistant to a specific active molecule called Phosphinothricine (PPT) of the widely used herbicide BASTA was developed by Agrobacterium mediated transformation (Enríquez-Obregón et al., 1998). These transgenic sugarcane plants showed integration of the bar gene having PPT acetyltransferase and conferred resistance against BASTA. Recent experiments with sugarcane varieties transgenic for EPSPS (The gene responsible for glyphosate tolerance) have also shown a healthy growth even though they were exposed to 0.5% of Roundup. Whereas the wild type sugarcane plants die off when treated with 0.1% of Roundup (Wang et al. 2017).
For the development of insect resistance, different genes were introduced in sugarcane. Arencibia et al. 1997 were the first to introduce Cry1A gene from Bacillus thuringiensis (Bt) against Diatraea saccharalis, which was then followed by various Bt gene versions such as Cry1Ab, Cry1Ac and Cry1Aa3 against insects such as Chilo infuscatellus, Diatraea saccharalis, Helicoverpa armigera, Proceras venosatus, Scirpophaga excerptalis (reviewed in Srikanth et al. 2011; Wang et al. 2017; Cristofoletti et al. 2018; Zhou et al. 2018) (Table 2). Several other genes encoding proteinase inhibitors and lectins were also used for resistance against insects, including Antitrogus consanguineous, Dermolepida albohirtum, Eoreuma loftini, Diatraea saccharalis, Ceratovacuna lanigera and Scirpophaga excerptalis (Srikanth et al. 2011). Apart from Cry1A and Cry2A, there are other Bt genes such as Cry2Aa and Cry2Ab that are found to be promising proteins towards the management of resistance development in insects. These proteins differ from the Cry1A proteins in structure and insecticidal mechanism (Kumar and Udayasuriyan 2004). However, Cry2A gene containing transgenic sugarcane plants have shown increased resistance against stem borer and comparable sugar yield (Gao et al. 2018).
Sugarcane transgenics with albicidin detoxification (albD) gene have produced resistance against the leaf scald disease causing bacterium Xanthomonas albilineans (Zhang and Birch 1997). Similarly, expression of truncated cDNA of the endochitinase gene from Trichoderma harzianum (Viswanathan et al. 2006) and β-1,3-glucanase gene from Trichoderma sp (Nayyar et al., 2017) has yielded significant resistance against red rot fungal disease caused by Colletotrichum falcatum. In the case of viral diseases, Joyce et al. (1998) have demonstrated viral resistance against sugarcane mosaic virus (SCMV) by transgenic expression of SCMV coat protein (CP) gene. Yao et al. 2017 have studied the level of disease resistance and field performance of SCMV-CP expressing transgenic sugarcane lines such as B2, B36, B38, B48 and B51A. Their results showed that the transgenic sugarcanes developed from sugarcane variety ca. Badila was found to have superior disease resistant characteristics and the transgenic line B48 was found to be highly resistant to SCMV. They also confirmed the stable expression of the transgene over several generations with vegetative propagation. Similar type of result was obtained when CP gene of Sorghum mosaic potyvirus was expressed (Ingelbrecht et al. 1999) and further clarified that the resistance is post-transcriptional gene silencing (PTGS) mediated. Recent studies on the RNA interference mediated resistance to Sorghum mosaic virus (SrMV) in sugarcane was done by (Guo et al. 2015) (Table 2). They have developed a SrMV resistant transgenic sugarcane line from the disease resistant sugarcane cultivar ROC22. This transgenic ROC22 lines express a hairpin construct targeting 423 bp long conserved region of SrMV coat protein and subsequent SrMV challenge experiments by artificial inoculation showed superior level of resistance against SrMV.
Water deficit and drought are the major abiotic factor threatening sugarcane production worldwide. Recent advances in understanding drought stress biology and the use of biotechnological tools for the evaluation of specific stress related proteins against drought conditions have initiated a major development in engineering plants against drought stress (Ferreira et al. 2017). So far, specific genes such as Glycine Betaine and bacterial betA genes were successfully introduced in sugarcane for the enhancement of water stress tolerance (Sugiharto 2017). Transformation of Choline dehydrogenase gene into sugarcane is believed to be the first step for the commercial cultivation of transgenic sugarcane varieties with induced water stress tolerance. This Choline dehydrogenase gene is involved in the production of Glycine Betaine, which helps plant cells to maintain water potential by osmotic adjustment. Additionally, genes such as Trehalose synthase from the fungus Grifola frondosa, Arabidopsis vacuolar pyrophosphatase (AVP1), Sugarcane SoP5CS, Dehydration Responsive Element Binding 2 (DREB2) of Erianthus arundinaceus and Arabidopsis, Erianthus arundinaceus HSP70 (EaHSP70) and Pea DNA Helicase 45 (DH45) are introduced into sugarcane to increase the drought tolerant capacity (Ferreira et al. 2017).
Recent understanding on drought responsive genes of sugarcane has showed Scdr2 gene as a potential drought responsive gene in the sugarcane cultivars. Overexpressing Scdr2 gene in transgenic tobacco plants those subjected to drought and salinity stress has shown increased levels of photosynthesis, internal CO2 levels, reduced hydrogen peroxide accumulation, increased stomatal conductance and resistance to both drought and salinity based stress conditions. Whereas, wild type non-transgenic plants are affected by both type of stress conditions and showed increased respiration rates (Begcy et al. 2019). Similarly, sugarcane plants transgenic for the P5CS gene showed a positive correlation with high proline content, increased photochemical efficiency in photosystem II and an increase in biomass production under drought stress conditions. Experiments shows that P5CS gene (∆1-pyrroline-5-carboxylate synthetase) from Vigna aconitifolia has conferred salinity tolerance to sugarcane transgenics. Additionally, this stress induced accumulation of proline in the transgenic plants turns into a part of the anti-oxidant defense system and ensures protection against stressful drought conditions (Molinari et al. 2007). Similarly, increased cold tolerance in sugarcane was achieved by expressing isopentenyl transferase (ipt) under a cold inducible promoter (Belintani et al. 2012).
As sugarcane becomes more and more important for the future biofuel industry, efforts to increase the sucrose content by overexpressing S. officinarum sucrose phosphate synthase (SoSPS) and RNAi-mediated down regulation of pyrophosphate: fructose 6-phosphate 1-phosphotransferase (PFP) have become successful (Groenewald and Botha 2008; Anur et al. 2020). Notably, doubling of total sugar content was achieved earlier by vacuolar targeting of Sucrose isomerase (Wu and Birch 2007). In addition to sucrose, specific bacterial genes from Ralstonia eutropha, those involved in polyhydroxybutyrate synthesis (PHAA, PHAB and PHAC) are also utilized to accumulate biopolymers in sugarcane (Petrasovits et al. 2012).
Reduction of lignin content is another approach to increase the biofuel production through biomass conversion. Genes involved in lignin biosynthesis such as caffeoyl-CoA O-methyltransferase (CCoAOMT), ferulate 5-hydroxylase (F5H) and caffeic acid O-methyltransferase (COMT) are targeted via RNAi methods to reduce the lignin content (Jung et al. 2013; Bewg et al. 2016). Recent RNAi mediated gene silencing studies on BAHD acyltransferase in sugarcane have improved the overall biomass digestibility of sugarcane straw obtained from BAHD silenced transgenic plant lines (Souza et al. 2019). With the advent of genome editing technologies, successful editing of nearly 100 COMT gene copies using TALEN was reported (Jung and Altpeter 2016; Kannan et al. 2018). The CRISPR/Cas technology gets versatile, fine-tuned and improved day by day in model plant Arabidopsis (Arya et al. 2020). Recently, successful CRISPR/Cas-mediated gene targeted events for multi-copy ALS genes (Oz et al. 2021), and magnesium chelatase alleles (Eid et al. 2021) were reported.
Molecular approaches against drought and salinity
Under drought stress, gene expression alterations can induce cellular adaptation and the study of gene expression profiles suggests the complexity of defense mechanisms that are involved in abiotic stresses. Although, plant response to drought has been analyzed at all these levels in the past few years, focus has been shifted toward the molecular mechanisms underlying drought responses. Understanding the molecular mechanisms of drought response is important for the improvement of drought tolerance in plants using molecular techniques (Nakashima et al. 2014). Generally, water stress changes the expression of several genes that are supposed to show a vital role in stress response and tolerance. Among which, a number of stress responsive genes have also been recognized and characterized (Shinozaki and Yamaguchi-Shinozaki 2007; Todaka et al. 2015). Recent molecular studies on the responses of sugarcane to drought stress conditions have shown a drought inducible protein SoDip22 in the water stress tolerant phenotype of sugarcane. It is a small 22 Kda protein, detected in the bundle sheath cells of sugarcane leaves and plays a key role in the maintenance of water molecules during water deficit conditions (Santiago et al., 2018).
For the purpose of better understanding the molecular mechanism of the physiological retorts those that happen in sugarcane during stress environments, high throughput gene expression studies have been conducted by many researchers (Flavia Stal Papini-Terzi et al. 2005; Rocha et al. 2007; Flavia S. Papini-Terzi et al. 2009; F. A. Rodrigues et al. 2011). These studies are mainly focused on the signal transduction and the role of phytohormones. Upon analyses of these high throughput gene expression data, it becomes clear that subsequent water stress conditions stimulate wide spread signal transduction networks, including several transcription factors, protein kinases and phosphatases (Zhu 2002; Xiong et al. 2002; Singh et al. 2002; Rabbani et al. 2003). Additionally, a significant part of the plant’s responses to drought stress is ABA-dependent. Both water stress and exogenous ABA treatment share a number of differentially expressed genes (Himmelbach et al. 2003; Shinozaki and Yamaguchi-Shinozaki 2007). Rocha et al. 2007 reported about 93 differentially expressed genes in sugarcane plants exposed to drought using macro-array technology. This includes genes such as HSP70, catalases, expansines, tyrosine phosphatases, Fructose-bisphosphate aldolase and trehalose metabolism-related proteins.
Sequencing and real-time reverse transcription-PCR profiling of selected ESTs (Expressed sequence tags) clusters identified around 25 stress related clusters with more than twofold relative expression during water deficit stress in sugarcane (Gupta et al. 2010). Specific differentially expressed genes were identified under water stress conditions in tolerant as well as sensitive sugarcane plants using the macroarray technique using ESTs from leaf libraries generated by the SUCEST project (Rodrigues et al. 2009, 2011). A suppression subtractive hybridization (SSH) approach was also used to isolate ESTs from those up-regulated during water-deficit stress in sugarcane and elucidate their transcriptional regulatory mechanisms (Prabu et al. 2011). Iskandar et al. (2011) studied gene expression profiling of drought responsive genes using real time quantitative PCR during sucrose accumulation under water deficit conditions in sugarcane culms.
Differentially expressed drought responsive genes were identified using Supersage (Kido et al. 2012). Hybridization-based approaches using DNA arrays have been the most successful for large-scale gene expression profiling in sugarcane under water stress (Papini-Terzi et al. 2009; Lembke et al. 2012). M. D. da Silva et al. (2013) verified the expression profile of aquaporins in sugarcane roots under water stress and T. Kumar et al. (2014) studied drought tolerance in sugarcane through ectopic expression of AVP1 gene of Arabidopsis using an Agrobacterium-mediated transformation system. By using small RNA deep sequencing (Thiebaut et al. 2012, 2014; Ferreira et al. 2012; Gentile et al. 2013, 2015), miRNA sequences are identified in the sugarcane genome during drought stress (Selvi et al. 2021). Vantini et al. (2015) have identified drought responsive genes in water stress tolerant sugarcane roots using the cDNA-amplified fragment length polymorphism technique. Li et al. (2016) studied the gene expression profile of sugarcane plants which are subjected to different water stress conditions using cDNA arrays.
Recently, the drought transcriptome was studied by high throughput sequencing strategies and subsequent identification of genes and pathways that are involved in drought tolerance in sugarcane (Selvi et al. 2020) was identified. Differential expression for the genes like dehydrin (D1), late embryogenesis abundant protein (LEA protein), ethylene responsive transcription factor (ER1), ABA- responsive element binding factor, DREB 2, drought responsive proteins (DR1), SNF1 related protein kinase, sucrose phosphate synthase were observed in control and stressed plants. Up regulation of transcripts like ABA responsive element, SNF1 related protein kinase, DREB 2 and drought responsive proteins (DR1) were noticed in drought tolerant variety Co 06,022. The transcription factors viz., NAC, WRKY, AP2-EREBP showed higher expression on both leaves and shoots of Co 06,022 compared to control and susceptible genotype Co 8021 (Devi et al. 2019; Selvi et al. 2020). Identified potential candidate genes will be used further for developing functional markers for drought stress tolerance in sugarcane. Furthermore, a more relevant and practical approach could be a comparative analysis of the stress transcriptome between cultivated and wild plant species for the possible discovery of abiotic stress tolerant genes. This can become possible by studying Saccharum spontaneum and Erianthus arundinaceus, a wild relative of sugarcane which has mechanisms for tolerating extreme drought conditions and continues to grow even during severe long drought situations.
The other method for the identification of genes involved in abiotic stresses and widely adopted in the recent past is the comparative gene expression analysis between; (i) stressed and unstressed samples of the same species, (ii) contrasting genotypes of the same species and related species (Pandey 2015). These comparative techniques were highly successful for studying the salinity stress responsive genes through which a number of salinity responsive genes have been identified in many crop species (Kawasaki et al. 2001). Among the techniques available, SSH continues to be a highly proficient tool for species with limited genome sequence information next to RNA-Sequencing analysis.
To understand the molecular basis of salt-stress response in sugarcane, Pagariya and his coworkers have initiated physio-biochemical assays and cDNA-RAPD-based gene expression studies under high salt (2% NaCl) supply regimes. Among the 335 differentially expressed transcript-derived fragments, 156 up-regulated and 85 down-regulated transcript-derived fragments were functionally categorized as metabolism, DNA/RNA/cellular processes, signal transduction/cell rescue/defense, cell wall modifications, transcriptional regulation, transport/trafficking, and unknown/hypothetical proteins (Pagariya et al. 2012).
Molecular approaches against high temperature stress
The next generation sequencing method (RNA-Seq) was used to analyze the transcripts of sugarcane and characterized several candidate genes related to heat stress tolerance in the sugarcane variety Co 99,004, where several candidate genes were characterized during the formative stage of growth (Devi et al. 2019; Raju et al. 2020). A total of 1,42,859 transcripts were assembled and the longest sequence in the assembled transcripts was 21,435 bp. DGE analysis has revealed that a total of 1137 and 1171 transcripts were significantly up-regulated and down regulated, respectively. The functional annotation of these transcripts has revealed that they were involved in response to oxidative stress, heat shock protein and glutamate pathway. Nine transcripts were validated for their biological significance through qRT-PCR and RT-PCR for their expression in leaf tissues, and the results showed high consistency between qRT-PCR and RNA-Seq methods. The longest sequence in the assembled transcripts contains 21,435 bp which has an identity of 99% with Maturase K protein from Saccharum officinarum, the same has been submitted in NCBI and acquired an accession number (NCBI Accession no. SRR7252568—RAN-Seq of Control—Co 99,004; NCBI Accession no. SRR7252569—RAN-Seq of heat treated Co 99,004) (Raju et al. 2020).
Elucidating high temperature stress-responsive proteins as well as associated mechanisms is essential for improving heat tolerance in sugarcane. In a recent study, proteomic analysis was conducted in sugarcane varieties such as Co 99,004 (heat stress tolerant), Co 0315 (heat stress susceptible) and SES 150 (Saccharum spontaneum clone) that were subjected to high temperature stress. Two-dimensional gel electrophoresis (2-DE) and MALDI-TOF/MS based proteomics approaches have revealed a total of 25 differentially up-regulated protein spots, out of which 22 and 3 other proteins were expressed only in Co 99,004 and SES 150, respectively. Protein profiles clearly showed the variation in protein expression between the control and heat treated sugarcane varieties. Protein identification and annotation of differentially expressed abundant proteins has revealed the involvement of several mechanisms in high temperature stress tolerance, including protein folding, sorting and degradation, transcription, translation, cell growth and death, glycan biosynthesis and metabolism, DNA replication, repair and signal transduction. The high temperature stress responsive proteins were clearly identified in the sugarcane varieties viz., Poly [ADP-ribose] polymerase 2-B (32.11 kDa), REF/SRPP-like protein cpx (21.65 kDa), Putative F-box/kelch-repeat protein (31.56 kDa), Putative cyclin-dependent kinase F-2 (7.68 kDa), Probable F-box protein (10.51 kDa), Thioredoxin O2 (12.72 kDa), Dirigent protein (15.95 kDa), MYB family transcription factor (17.8 kDa), stress-response A/B barrel domain (11.87 kDa) and Protein TIFY 11d (9.11 kDa). These differentially expressed abundant proteins were further quantified by qRT-PCR and are relatively reliable. These proteins are identified in the study, which provides a strong basis to elucidate gene functions and to further explain the molecular mechanisms underlying the adaptation of sugarcane varieties to high temperature stress (Devi et al. 2019; Raju et al. 2020).
Research in omics era
The aneuploidy, polyploidy and complex nature of cultivated sugarcane with large genome size (> 10 Gb) makes WGS, assembly and annotation as one of the toughest tasks to accomplish, nevertheless a partial work has been done (Garsmeur et al. 2018). Zhang et al. (2012) estimated the monoploid genome sizes of S. officinarum (985 Mb) and S. spontaneum (843 Mb). However, genome sequencing has revealed the haploid genome size of S. officinarum (930 Mb) and S. spontaneum (750 Mb) (Garsmeur et al. 2018; Zhang et al. 2018), suggesting the monoploid genome size of cultivated sugarcane to be somewhere between 750 to 930 Mb. The release of WGS will kickstart gene family analysis, which is being done with conventional cDNA cloning (Li et al. 2017; Hua-ying et al. 2019) or with available EST data (Zhang et al. 2013; McIntyre et al. 2015; Li et al. 2017) from NCBI or with partially assembled genome sequences (Santiago et al. 2018; Li et al. 2020) from NCBI. A public database, GRASSIUS (https://www.grassius.org/index.php; (Yilmaz et al. 2009), with nucleotide and peptide sequences of transcription factors of selected cereal crop members, was also used to perform gene family analysis (Ramaswamy et al. 2017).
The transcriptomic analysis in sugarcane started long before the genome analysis. SUCEST (http://sucest-fun.org/; Vettore et al. 2003), a database by Brazilian consortium has made a greater contribution in pioneering large scale transcriptomic studies in sugarcane, reflecting the significance of sugarcane with Brazilian economy. Following this, several research groups have started analyzing sugarcane transcriptome data against various biotic, abiotic factors and developmental stages as well (Manners and Casu 2011; Lovejot Kaur et al. 2017; Ali et al. 2019). The NCBI SRA database has a total transcriptomic data of 973 runs, of which 750 are generated from sugarcane hybrid cultivars while 223 are generated from Saccharum officinarum (https://www.ncbi.nlm.nih.gov/sra). A list of databases other than NCBI is summarized in Table 3, of which a few databases were discontinued in due course. Being a crop meant for carbohydrate production, studies related to metabolomics are scarce. Studies on callus development (Mahmud et al. 2014), stem development against sucrose accumulation (Glassop et al. 2007) and to understand lignin biosynthesis (Bottcher et al. 2013) were done following a generalized metabolomic study (Bosch et al. 2003).
Table 3.
Sugarcane databases
| Database | Data type | Weblink |
|---|---|---|
| Sugarcane Genome Hub | Genome | https://sugarcane-genome.cirad.fr/ |
| GRASSIUS | Transcription factors | https://www.grassius.org/index.php |
| SUCEST | Transcriptome | http://sucest-fun.org/ |
| ISAAA | Commercial GM events | https://www.isaaa.org/default.asp |
| GrainGenes | Phenotypic and Molecular | Discontinued for sugarcane |
| SGDB | Germplasm | Discontinued |
| SymGRASS | Orthologous genes involved in arbuscular mycorrhiza and root nodule symbiosis | Discontinued |
Sugarcane—A platform for molecular farming
Plant-based production of pharmaceutical and nonpharmaceutical products is gaining momentum around the world. Many plant species have become promising alternative to the traditional expression systems for producing a variety of valuable or high-value molecules of pharmaceutical and nonpharmaceutical products. Plants are preferred as a platform for the production of recombinant proteins because of the low costs and greater scalability of plant production systems without incurring the high costs associated with downstream processing and purification. Of the plant systems, sugarcane represents an ideal candidate for biofactory applications due to its large biomass, rapid growth rate, efficient carbon fixation pathway, a well-developed storage tissue system, minimal transgene dispersal due to the vegetative method of propagation, high quantity of extractable juice with very low protein content (0.01–0.02%), and a well-established downstream processing technology. Sugarcane as a platform for molecular farming has been of interest for nearly two decades (Birch et al. 1995). In sugarcane, the culm contributes 70% of the dry weight, which is largely made of storage parenchyma cells where the vacuoles occupy most of the cell volume. The juice stored in the vacuoles contributes to 60% of the cane weight. Therefore, it is logical to target any recombinant protein to the vacuole, so as to attain the maximum protein yield. Therefore, sugarcane can be used as an efficient platform for molecular farming. The first attempt by Gnanasambandam and Birch (2004) to localize recombinant GFP to the vacuole using the N-terminal-targeting motif NPIR from a vacuolar protein, sporamin, of sweet potato has led to mis-targeting of r-GFP to the chloroplasts. Sugar-yielding plants such as sugarcane have attracted attention because of their high biomass production, especially the storage culm from which juice is extracted. The high quantity of extractable juice with very low protein content and a well-established juice processing technology have encouraged researchers to express proteins in the sugarcane culm. Many attempts have been made to express and purify recombinant proteins from sugarcane stem and leaves (Jackson et al. 2010).
Initial studies that aimed at expressing the protein in cytoplasm and purifying it from leaves have resulted in poor yields as leaves contain many proteins other than the introduced ones, which may interfere in downstream processing (Fischer et al. 2004). In stem parenchyma, as most of the cell volume is occupied by the vacuole, vacuolar targeting is supposed to lead to higher levels of r-proteins than when the cytoplasm is targeted. Palaniswamy et al. (2016) demonstrated that targeting of r-proteins to parenchymatous lytic vacuoles of sugarcane has resulted in high yield with relatively simple purification steps. Hence, there is a possibility of using sugarcane as an efficient platform for the production of large quantities of recombinant proteins by deploying newly identified vacuolar-targeting determinants (the 78-bp or 18-bp VT determinants) along with a strong constitutive promoter (Port ubi882). In addition, a downstream purification system could be easily developed, which offers a promising solution for the extraction and purification of recombinant proteins from the sugarcane juice. Although this is a promising biofarming technology, the presence of vacuolar proteolytic enzymes and the acidic nature of lytic vacuoles need to be given due consideration for the selection of the candidate recombinant proteins. Considering the practical situations, where the purity after the affinity chromatography stage is around 50%, we would be able to get an r-protein yield of 4.8–7.2 kg/acre (40,000 canes per acre) with purity comparable to that of the commercially equivalent r-proteins. The unique aspect of sugarcane is the extraction of a large juice volume (700 mL) by crushing 1 kg of cane. Therefore, sugarcane is possibly an efficient platform for molecular farming.
Conclusions and future perspectives
Human intervention with plants through yield-oriented agricultural selection has brought directional evolution of selected wild plants into domesticated crops. This can be termed anthropogenic coevolution. Sugarcane has been modified from a wild fibrous grass into a sweet, juicy crop. Trait improvement via conventional breeding practices and cutting-edge modern research activities has further polished sugarcane into a resilient high yielding crop. Omic approaches led the researchers to understand the sugarcane crop at a molecular level with higher resolution. Tolerance to water deficit stress involves a number of gene regulatory networks and is a very complex trait. Saccharum arundinaceum Retz., a wild relative of Saccharum, is well known for its high fibre, high biomass, tolerance to drought and water logging, pest and disease resistance with multi-ratooning ability. Functional genomic analysis on these genes involved in abiotic stress would enable dissection of the complex traits into component characters and would offer sources for various parameters in breeding for abiotic stress tolerance. The reports of tight linkages between the candidate genes and the QTLs conferring abiotic stress resistance are encouraging (Francia et al. 2005). For breeders, the knowledge of the precise locations of the QTLs that has large effects, have application for integration in MAS.
Sugarcane holds an important place as it is not only the source of sugar but also an important renewable energy source. Losses caused by drought can be reduced to a great extent by using genetic engineering and genome editing tools. In the future all these developments are expected to address the problem of abiotic stresses through the integration of breeding and molecular techniques. Apart from feeding the sugar needs, this C4 crop could potentially fuel the industry, which would boost the economy and energize the environment by cutting down on carbon emissions. As sugarcane stem vacuoles provide a huge storage factory for targeting r-proteins, its easy and cost efficient purification processing, and owing to its edible nature, sugarcane can be considered as a platform for production of edible vaccines and for several other r-proteins, biomolecules, industrial and pharmaceutical products through modern biotechnological approaches. Currently, the world is facing a critical time in the COVID-19 pandemic, leading to massive global human and economic losses. The high transmission rate and the emergence of diverse SARS-CoV-2 variants demand that several COVID-19 vaccines are being developed using different production systems. A plant-based production offers several advantages, such as low cost, rapidity, scalability, and safety. Hence, sugarcane stem vacuole based vaccine production may be one of the choices for shortening the global demand. If this system becomes successful in global vaccine production rather than as a sweet feeding crop and movement crop (biofuel production), our sugarcane will also become a lifesaving crop. As food, fuel and drugs in a single crop, sugarcane is climbing the slope of enlightenment and will soon reach its goal.
Declarations
Conflicts of interest
All the authors declare that they have no conflict of interest.
Human and animal rights
The authors declare that no research involving human participants and/or animals was conducted.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Raju Gomathi, Email: gomathi_sbi@yahoo.co.in.
Thakku R. Ramkumar, Email: ramkumar.thakkur@ufl.edu, Email: rrajaram@desu.edu
References
- Ali A, Khan M, Sharif R, et al. Sugarcane Omics: an update on the current status of research and crop improvement. Plants. 2019;8:344. doi: 10.3390/plants8090344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix K, Baurens FC, Paulet F, et al. Isolation and characterization of a satellite DNA family in the Saccharum complex. Genome. 1998;41:854–864. doi: 10.1139/g98-076. [DOI] [PubMed] [Google Scholar]
- Al-Janabi SM, Honeycutt RJ, McClelland M, Sobral BW. A genetic linkage map of Saccharum spontaneum L. SES 208. Genetics. 1993;134:1249–1260. doi: 10.1093/genetics/134.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altpeter F, Oraby H. Sugarcane. In: Kempken F, Jung C, editors. Genetic modification of plants: agriculture, horticulture and forestry. Berlin, Heidelberg: Springer; 2010. pp. 453–472. [Google Scholar]
- Anur RM, Mufithah N, Sawitri WD, et al. Overexpression of sucrose phosphate synthase enhanced sucrose content and biomass production in transgenic sugarcane. Plants. 2020;9:200. doi: 10.3390/plants9020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arencibia A, Vázquez RI, Prieto D, et al. Transgenic sugarcane plants resistant to stem borer attack. Mol Breeding. 1997;3:247–255. doi: 10.1023/A:1009616318854. [DOI] [Google Scholar]
- Arya SS, Mahto BK, Ramkumar TR, Lenka SK (2020) Sharpening gene editing toolbox in Arabidopsis for plants. J Plant Biochem Biotechnol 29:769–784
- Barbosa AMM, Geraldi IO, Benchimol LL, et al. Relationship of intra- and interpopulation tropical maize single cross hybrid performance and genetic distances computed from AFLP and SSR markers. Euphytica. 2003;130:87–99. doi: 10.1023/a:1022381008721. [DOI] [Google Scholar]
- Begcy K, Mariano ED, Lembke CG, et al. Overexpression of an evolutionarily conserved drought-responsive sugarcane gene enhances salinity and drought resilience. Ann Bot. 2019;124:691–700. doi: 10.1093/aob/mcz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belintani NG, Guerzoni JTS, Moreira RMP, Vieira LGE. Improving low-temperature tolerance in sugarcane by expressing the ipt gene under a cold inducible promoter. Biol Plant. 2012;56:71–77. doi: 10.1007/s10535-012-0018-1. [DOI] [Google Scholar]
- Berding N, Roach BT (1987) Germplasm Collection, Maintenance, and Use. In: Developments in Crop Science. Elsevier, pp 143–210
- Bewg WP, Poovaiah C, Lan W, et al. RNAi downregulation of three key lignin genes in sugarcane improves glucose release without reduction in sugar production. Biotechnol Biofuels. 2016;9:270. doi: 10.1186/s13068-016-0683-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch RG, Bower R, Elliott A, Potier B, Franks T, Cordeiro G (1995) Expression of foreign genes in sugarcane. InProceedings of the International Society of Sugarcane Technologists XXII Congress, Cartegena 1995 Sep (Vol. 2, pp. 368–373).
- Bosch S, Rohwer JM, Botha FC (2003) The Sugarcane Metabolome. In: Proceedings of South african sugar technological association. Proceedings of South african sugar technological association, pp 129–133
- Bottcher A, Cesarino I, Brombini dos Santos A, et al. Lignification in sugarcane: biochemical characterization, gene discovery, and expression analysis in two genotypes contrasting for lignin content. Plant Physiol. 2013;163:1539–1557. doi: 10.1104/pp.113.225250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Birch RG. Transgenic sugarcane plants via microprojectile bombardment. Plant J. 1992;2:409–416. doi: 10.1111/j.1365-313X.1992.00409.x. [DOI] [Google Scholar]
- Budeguer F, Enrique R, Perera MF et al (2021) Genetic transformation of sugarcane, current status and future prospects. Front Plant Sci 12:768609. 10.3389/fpls.2021.768609 [DOI] [PMC free article] [PubMed]
- Cristofoletti PT, Kemper EL, Capella AN, et al. Development of transgenic sugarcane resistant to sugarcane borer. Tropical Plant Biol. 2018;11:17–30. doi: 10.1007/s12042-018-9198-y. [DOI] [Google Scholar]
- D’Hont A, Grivet L, Feldmann P, et al. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Molec Gen Genet. 1996;250:405–413. doi: 10.1007/BF02174028. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Paulet F, Glaszmann JC. Oligoclonal interspecific origin of “North Indian” and “Chinese” sugarcanes. Chromosome Res. 2002;10:253–262. doi: 10.1023/a:1015204424287. [DOI] [PubMed] [Google Scholar]
- Deng Z-N, Wei Y-W, Lü W-L, Li Y-R (2008) Fusion insect-resistant gene mediated by matrix attachment region (MAR) sequence in transgenic sugarcane. Sugar Tech 10:87–90. 10.1007/s12355-008-0015-z
- Dessoky ES, Ismail RM, Elarabi NI et al (2021) Improvement of sugarcane for borer resistance using Agrobacterium mediated transformation of cry1Ac gene. GM Crops & Food 12:47–56. 10.1080/21645698.2020.1809318 [DOI] [PMC free article] [PubMed]
- da Silva J, Honeycutt RJ, Burnquist W, et al. Saccharum spontaneum L. ‘SES 208’ genetic linkage map combining RFLP- and PCR-based markers. Mol Breed. 1995;1:165–179. doi: 10.1007/BF01249701. [DOI] [Google Scholar]
- da Silva MD, de Silva RL, O, Costa Ferreira Neto JR, , et al. Expression analysis of sugarcane aquaporin genes under water deficit. J Nucl Acids. 2013;2013:e763945. doi: 10.1155/2013/763945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J, N. Paton, P. Smith, B.T. Roach (1989) Further studies on the origin of sugar canes Saccharum officinarum L., S. barberi Jesw. and S. sinense Roxb. using flavonoid chemotaxonomic markers. Sugar cane 7–15
- Daniels J, Roach BT. Sugarcane Improvement Through Breeding, Volume 11–1st Edition. 1. Amsterdam: Elsevier; 1987. Taxonomy and Evolution; pp. 7–84. [Google Scholar]
- de Souza WR, Pacheco TF, Duarte KE, et al. Silencing of a BAHD acyltransferase in sugarcane increases biomass digestibility. Biotechnol Biofuels. 2019 doi: 10.1186/s13068-019-1450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi K, Prathima PT, Gomathi R, et al. Gene expression profiling in sugarcane genotypes during drought stress and rehydration. Sugar Tech. 2019;21:717–733. doi: 10.1007/s12355-018-0687-y. [DOI] [Google Scholar]
- Dillon SL, Shapter FM, Henry RJ, et al. Domestication to crop improvement: genetic resources for sorghum and Saccharum (Andropogoneae) Ann Bot. 2007;100:975–989. doi: 10.1093/aob/mcm192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotaniya ML, Datta SC, Biswas DR, et al. Use of sugarcane industrial by-products for improving sugarcane productivity and soil health. Int J Recycl Org Waste Agricult. 2016;5:185–194. doi: 10.1007/s40093-016-0132-8. [DOI] [Google Scholar]
- Eid A, Mohan C, Sanchez S, Wang D, Altpeter F (2021) Multiallelic, targeted mutagenesis of magnesium chelatase with CRISPR/Cas9 provides a rapidly scorable phenotype in highly polyploid sugarcane. Front Genome Ed 3:654996. 10.3389/fgeed.2021.654996 [DOI] [PMC free article] [PubMed]
- Enríquez-Obregón GA, Vázquez-Padrón RI, Prieto-Samsonov DL, et al. Herbicide-resistant sugarcane (Saccharum officinarum L.) plants by Agrobacterium-mediated transformation. Planta. 1998;206:20–27. doi: 10.1007/s004250050369. [DOI] [Google Scholar]
- Falco MC, Tulmann Neto A, Ulian EC (2000) Transformation and expression of a gene for herbicide resistance in a Brazilian sugarcane. Plant Cell Reports 19:1188–1194. 10.1007/s002990000253 [DOI] [PubMed]
- FAO (2020) World Food and Agriculture - Statistical Yearbook 2020. pp 58
- FAO (2021) World Food and Agriculture – Statistical Yearbook 2021. pp 60
- Ferreira TH, Gentile A, Vilela RD, et al. microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp) PLoS ONE. 2012;7:e46703. doi: 10.1371/journal.pone.0046703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira THS, Tsunada MS, Bassi D, et al. Sugarcane water stress tolerance mechanisms and its implications on developing biotechnology solutions. Front Plant Sci. 2017 doi: 10.3389/fpls.2017.01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Stoger E, Schillberg S, Christou P, Twyman RM. Plant-based production of biopharmaceuticals. Curr Opin Plant Biol. 2004;7:152–158. doi: 10.1016/j.pbi.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Foolad MR, Sharma A (2005) Molecular markers as selection tools in tomato breeding. Acta Hortic 225–240. 10.17660/ActaHortic.2005.695.25
- Formann S, Hahn A, Janke L, Stinner W, Sträuber H, Logroño W, Nikolausz M. Beyond sugar and ethanol production: value generation opportunities through sugarcane residues. Front Energy Res. 2020;8:579577. doi: 10.3389/fenrg.2020.579577. [DOI] [Google Scholar]
- Francia E, Tacconi G, Crosatti C, et al. Marker assisted selection in crop plants. Plant Cell Tiss Organ Cult. 2005;82:317–342. doi: 10.1007/s11240-005-2387-z. [DOI] [Google Scholar]
- Gao S, Yang Y, Xu L, et al. Particle bombardment of the cry2A gene cassette induces stem borer resistance in sugarcane. Int J Mol Sci. 2018;19:1692. doi: 10.3390/ijms19061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsmeur O, Droc G, Antonise R, et al. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat Commun. 2018;9:2638. doi: 10.1038/s41467-018-05051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile A, Ferreira TH, Mattos RS, et al. Effects of drought on the microtranscriptome of field-grown sugarcane plants. Planta. 2013;237:783–798. doi: 10.1007/s00425-012-1795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile A, Dias LI, Mattos RS, et al. MicroRNAs and drought responses in sugarcane. Front Plant Sci. 2015;6:58. doi: 10.3389/fpls.2015.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassop D, Roessner U, Bacic A, Bonnett GD. Changes in the sugarcane metabolome with stem development. Are they related to sucrose accumulation? Plant Cell Physiol. 2007;48:573–584. doi: 10.1093/pcp/pcm027. [DOI] [PubMed] [Google Scholar]
- Glaszmann JC, Dufour P, Grivet L, et al. Comparative genome analysis between several tropical grasses. Euphytica. 1997;96:13–21. doi: 10.1023/A:1002987620250. [DOI] [Google Scholar]
- Gnanasambandam A, Birch RG. Efficient developmental mis-targeting by the sporamin NTPP vacuolar signal to plastids in young leaves of sugarcane and Arabidopsis. Plant Cell Rep. 2004;23:435–447. doi: 10.1007/s00299-004-0860-5. [DOI] [PubMed] [Google Scholar]
- Groenewald J-H, Botha FC. Down-regulation of pyrophosphate: fructose 6-phosphate 1-phosphotransferase (PFP) activity in sugarcane enhances sucrose accumulation in immature internodes. Transgenic Res. 2008;17:85–92. doi: 10.1007/s11248-007-9079-x. [DOI] [PubMed] [Google Scholar]
- Guo J, Gao S, Lin Q, et al. Transgenic sugarcane resistant to sorghum mosaic virus based on coat protein gene silencing by RNA interference. Biomed Res Int. 2015;2015:e861907. doi: 10.1155/2015/861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Raghuvanshi S, Gupta A, et al. The water-deficit stress- and red-rot-related genes in sugarcane. Funct Integr Genomics. 2010;10:207–214. doi: 10.1007/s10142-009-0144-9. [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E. Relay and control of abscisic acid signaling. Curr Opin Plant Biol. 2003;6:470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Hua-ying M, Wen-ju W, Wei-hua S, et al. Genome-wide identification, phylogeny, and expression analysis of Sec14-like PITP gene family in sugarcane. Plant Cell Rep. 2019;38:637–655. doi: 10.1007/s00299-019-02394-1. [DOI] [PubMed] [Google Scholar]
- Ingelbrecht IL, Irvine JE, Mirkov TE. Posttranscriptional gene silencing in transgenic sugarcane. dissection of homology-dependent virus resistance in a monocot that has a complex polyploid genome1. Plant Physiol. 1999;119:1187–1198. doi: 10.1104/pp.119.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskandar HM, Casu RE, Fletcher AT, et al. Identification of drought-response genes and a study of their expression during sucrose accumulation and water deficit in sugarcane culms. BMC Plant Biol. 2011;11:12. doi: 10.1186/1471-2229-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MA, Nutt KA, Hassall R, Rae AL. Comparative efficiency of subcellular targeting signals for expression of a toxic protein in sugarcane. Funct Plant Biol. 2010;37:785–793. doi: 10.1071/FP09243. [DOI] [Google Scholar]
- Jannoo N, Grivet L, Chantret N, et al. Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J. 2007;50:574–585. doi: 10.1111/j.1365-313X.2007.03082.x. [DOI] [PubMed] [Google Scholar]
- Jeswiet J. Beschrijving der soorten van het suikerriet. Eerste Bijdrage. Morphlogie van het geslachet Saccharum. Arch V Suikerrindus. 1925;33:391–404. [Google Scholar]
- Joyce PA, McQualter RB, Bernard MJ, Smith GR. Engineering for resistance to SCMV in sugarcane. Acta Hortic. 1998;461:385–392. doi: 10.17660/ActaHortic.1998.461.44. [DOI] [Google Scholar]
- Jung JH, Altpeter F. TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol Biol. 2016;92:131–142. doi: 10.1007/s11103-016-0499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Vermerris W, Gallo M, et al. RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol J. 2013;11:709–716. doi: 10.1111/pbi.12061. [DOI] [PubMed] [Google Scholar]
- Kalunke RM, Kolge AM, Babu KH, Prasad DT (2009) Agrobacterium mediated transformation of sugarcane for borer resistance using Cry 1Aa3 gene and one-step regeneration of transgenic plants. Sugar Tech 11:355–359. 10.1007/s12355-009-0061-1
- Kannan B, Jung JH, Moxley GW, et al. TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol J. 2018;16:856–866. doi: 10.1111/pbi.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, et al. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13:889–905. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido EA, Ferreira Neto JRC, Silva RLO, Pandolfi V, Guimaraes ACR, Veiga DT, Benko-Iseppon AM. New insights in the sugarcane transcriptome responding to drought stress as revealed by super SAGE. Sci World J. 2012;2012:821062. doi: 10.1100/2012/821062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Udayasuriyan V. Cloning of cry2Aa and cry2Ab genes from new isolates of Bacillus thuringiensis and their expression in recombinant Bacillus thuringiensis and Escherichia coli strains. World J Microbiol Biotechnol. 2004;20:11–17. doi: 10.1023/B:WIBI.0000013285.15036.0d. [DOI] [Google Scholar]
- Kumar T, Uzma null, Khan MR, , et al. Genetic improvement of sugarcane for drought and salinity stress tolerance using Arabidopsis vacuolar pyrophosphatase (AVP1) gene. Mol Biotechnol. 2014;56:199–209. doi: 10.1007/s12033-013-9695-z. [DOI] [PubMed] [Google Scholar]
- Lebot V. Biomolecular evidence for plant domestication in Sahul. Genet Resour Crop Evol. 1999;46:619–628. doi: 10.1023/A:1008748504038. [DOI] [Google Scholar]
- Lembke CG, Nishiyama MY, Sato PM, et al. Identification of sense and antisense transcripts regulated by drought in sugarcane. Plant Mol Biol. 2012;79:461–477. doi: 10.1007/s11103-012-9922-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Nong Q, Solanki MK, et al. Differential expression profiles and pathways of genes in sugarcane leaf at elongation stage in response to drought stress. Sci Rep. 2016;6:25698. doi: 10.1038/srep25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Nong Q, Xie J, et al. Molecular characterization and co-expression analysis of the SnRK2 gene family in sugarcane (Saccharum officinarum L.) Sci Rep. 2017;7:17659. doi: 10.1038/s41598-017-16152-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hua X, Zhong W, et al. Genome-wide identification and expression profile analysis of WRKY family genes in the autopolyploid saccharum spontaneum. Plant Cell Physiol. 2020;61:616–630. doi: 10.1093/pcp/pcz227. [DOI] [PubMed] [Google Scholar]
- Lovejot Kaur, S. Dharshini, Bakshi Ram, C. Appunu (2017) Sugarcane Genomics and Transcriptomics. In: Sugarcane Biotechnology: Challenges and Prospects. Springer, pp 13–32
- Lu YH, D’Hont A, Paulet F, et al. Molecular diversity and genome structure in modern sugarcane varieties. Euphytica. 1994;78:217–226. doi: 10.1007/bf00027520. [DOI] [Google Scholar]
- Mahmud I, Thapaliya M, Boroujerdi A, Chowdhury K. NMR-based metabolomics study of the biochemical relationship between sugarcane callus tissues and their respective nutrient culture media. Anal Bioanal Chem. 2014;406:5997–6005. doi: 10.1007/s00216-014-8002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners JM, Casu RE. Transcriptome analysis and functional genomics of sugarcane. Tropical Plant Biol. 2011;4:9–21. doi: 10.1007/s12042-011-9066-5. [DOI] [Google Scholar]
- McIntyre CL, Goode ML, Cordeiro G, et al. Characterisation of alleles of the sucrose phosphate synthase gene family in sugarcane and their association with sugar-related traits. Mol Breeding. 2015;35:98. doi: 10.1007/s11032-015-0286-5. [DOI] [Google Scholar]
- Molinari HBC, Marur CJ, Daros E, et al. Evaluation of the stress-inducible production of proline in transgenic sugarcane (Saccharum spp.): osmotic adjustment, chlorophyll fluorescence and oxidative stress. Physiol Plant. 2007;130:218–229. doi: 10.1111/j.1399-3054.2007.00909.x. [DOI] [Google Scholar]
- Mukherjee SK. Origin and distribution of saccharum. Bot Gaz. 1957;119:55–61. doi: 10.1086/335962. [DOI] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci. 2014;5:170. doi: 10.3389/fpls.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayyar S, Sharma BK, Kaur A, et al. Red rot resistant transgenic sugarcane developed through expression of β-1,3-glucanase gene. PLoS ONE. 2017;12:e0179723. doi: 10.1371/journal.pone.0179723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerkar G, Thorat A, Sheelavantmath S, et al. Genetic transformation of sugarcane and field performance of transgenic sugarcane. In: Gosal SS, Wani SH, et al., editors. Biotechnologies of crop improvement transgenic approaches. Cham: Springer International Publishing; 2018. pp. 207–226. [Google Scholar]
- Nutt K, Allsopp P, Geijskes R, et al (2001) Canegrub resistant sugarcane. Australian Society of Sugar Cane Technologists, Mackay: International Society of Sugar Cane Technologists. Proceedings of the XXIV Congress, Brisbane, Australia, 17–21 September 2001. Volume 2.
- Oz MT, Altpeter A, Karan R et al (2021) CRISPR/Cas9-mediated multi-allelic gene targeting in sugarcane confers herbicide tolerance. Front Genome Editing 3:15 [DOI] [PMC free article] [PubMed]
- Pagariya MC, Devarumath RM, Kawar PG. Biochemical characterization and identification of differentially expressed candidate genes in salt stressed sugarcane. Plant Sci. 2012;184:1–13. doi: 10.1016/j.plantsci.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Palaniswamy H, Syamaladevi DP, Mohan C, Philip A, Petchiyappan A, Narayanan S. Vacuolar targeting of r-proteins in sugarcane leads to higher levels of purifiable commercially equivalent recombinant proteins in cane juice. Plant Biotechnol J. 2016;14:791–807. doi: 10.1111/pbi.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, editor. Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives. New York: Springer-Verlag; 2015. [Google Scholar]
- Panje RR, Babu CN. Studies in Saccharum spontaneum distribution and geographical association of chromosome numbers. Cytologia. 1960;25:152–172. doi: 10.1508/cytologia.25.152. [DOI] [Google Scholar]
- Papini-Terzi FS, Rocha FR, Nicoliello Vêncio RZ, et al. Transcription profiling of signal transduction-related genes in sugarcane tissues. DNA Res. 2005;12:27–38. doi: 10.1093/dnares/12.1.27. [DOI] [PubMed] [Google Scholar]
- Papini-Terzi FS, Rocha FR, Vêncio RZ, et al. Sugarcane genes associated with sucrose content. BMC Genomics. 2009;10:120. doi: 10.1186/1471-2164-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy N. Origin of Noble Sugar-Canes (Saccharum officinarum.) Nature. 1948;161:608–608. doi: 10.1038/161608a0. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. PNAS. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasovits LA, Zhao L, McQualter RB, et al. Enhanced polyhydroxybutyrate production in transgenic sugarcane: Improving PHB production in sugarcane. Plant Biotechnol J. 2012;10:569–578. doi: 10.1111/j.1467-7652.2012.00686.x. [DOI] [PubMed] [Google Scholar]
- Prabu G, Kawar PG, Pagariya MC, Prasad DT. Identification of water deficit stress upregulated genes in sugarcane. Plant Mol Biol Report. 2011;29:291–304. doi: 10.1007/s11105-010-0230-0. [DOI] [Google Scholar]
- Qamar Z, Nasir IA, Abouhaidar MG et al (2021) Novel approaches to circumvent the devastating effects of pests on sugarcane. Sci Rep 11:12428. 10.1038/s41598-021-91985-8 [DOI] [PMC free article] [PubMed]
- Rabbani MA, Maruyama K, Abe H, et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju G, Shanmugam K, Kasirajan L (2020) High-throughput sequencing reveals genes associated with high-temperature stress tolerance in sugarcane. 3 Biotech 10:198. 10.1007/s13205-020-02170-z [DOI] [PMC free article] [PubMed]
- Ramasamy M, Mora V, Damaj MB, et al. A biolistic-based genetic transformation system applicable to a broad-range of sugarcane and energycane varieties. GM Crops Food. 2018;9:211–227. doi: 10.1080/21645698.2018.1553836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy M, Narayanan J, Manickavachagam G, et al. Genome wide analysis of NAC gene family ‘sequences’ in sugarcane and its comparative phylogenetic relationship with rice, sorghum, maize and Arabidopsis for prediction of stress associated NAC genes. Agri Gene. 2017;3:1–11. doi: 10.1016/j.aggene.2016.10.003. [DOI] [Google Scholar]
- Ramkumar TR, Parameswari C, Sugapriya T, Veluthambi K. Effect of orientation of transcription of a gene in an inverted transferred DNA repeat on transcriptional gene silencing in rice transgenics—a case study. Physiol Mol Biol Plants. 2015;21:151–157. doi: 10.1007/s12298-014-0273-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkumar TR, Lenka SK, Arya SS, Bansal KC. A short history and perspectives on plant genetic transformation. In: Rustgi S, Luo H, editors. Biolistic DNA delivery in plants: methods and protocols. US, New York, NY: Springer; 2020. pp. 39–68. [DOI] [PubMed] [Google Scholar]
- Reis RR, Andrade Dias Brito da Cunha B, Martins PK, et al (2014) Induced over-expression of AtDREB2A CA improves drought tolerance in sugarcane. Plant Sci 221–222:59–68. 10.1016/j.plantsci.2014.02.003 [DOI] [PubMed]
- Roach BT. Chromosome numbers in Saccharum edule. Cytologia. 1972;37:155–161. doi: 10.1508/cytologia.37.155. [DOI] [Google Scholar]
- Rocha FR, Papini-Terzi FS, Nishiyama MY, et al. Signal transduction-related responses to phytohormones and environmental challenges in sugarcane. BMC Genomics. 2007;8:71. doi: 10.1186/1471-2164-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues FA, de Laia ML, Zingaretti SM. Analysis of gene expression profiles under water stress in tolerant and sensitive sugarcane plants. Plant Sci. 2009;176:286–302. doi: 10.1016/j.plantsci.2008.11.007. [DOI] [Google Scholar]
- Rodrigues FA, Graça JP, Laia ML, et al. Sugarcane genes differentially expressed during water deficit. Biol Plant. 2011;55:43–53. doi: 10.1007/s10535-011-0006-x. [DOI] [Google Scholar]
- Santiago TR, Pereira VM, de Souza WR, et al. Genome-wide identification, characterization and expression profile analysis of expansins gene family in sugarcane (Saccharum spp.) PLoS ONE. 2018;13:0191081. doi: 10.1371/journal.pone.0191081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Eichler P, Machado G et al (2020) By-products of the sugarcane industry. In: Sugarcane Biorefinery, Technology and Perspectives. Elsevier, pp 21–48. 10.1016/B978-0-12-814236-3.00002-0
- Santosa DA, Hendroko R, Farouk A, Greiner R. A rapid and highly efficient method for transformation of sugarcane callus. Mol Biotechnol. 2004;28:113–119. doi: 10.1385/MB:28:2:113. [DOI] [PubMed] [Google Scholar]
- Selvi A, Nair NV, Balasundaram N, Mohapatra T. Evaluation of maize microsatellite markers for genetic diversity analysis and fingerprinting in sugarcane. Genome. 2003;46:394–403. doi: 10.1139/g03-018. [DOI] [PubMed] [Google Scholar]
- Selvi A, Devi K, Manimekalai R, Prathima PT. Comparative analysis of drought-responsive transcriptomes of sugarcane genotypes with differential tolerance to drought. 3 Biotech. 2020;10:236. doi: 10.1007/s13205-020-02226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvi A, Devi K, Manimekalai R, Prathima PT, Valiyaparambth R, Lakshmi K. High-throughput miRNA deep sequencing in response to drought stress in sugarcane. 3 Biotech. 2021;11(7):1–8. doi: 10.1007/s13205-021-02857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5:430–436. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Solomon S. Sugarcane production and development of sugar industry in India. Sugar Tech. 2016;18:588–602. doi: 10.1007/s12355-016-0494-2. [DOI] [Google Scholar]
- Sreenivasan TV, Ahloowalia BS, Heinz DJ. Sugarcane improvement through breeding. Cytogenetics. 1987;1987(5):211–253. [Google Scholar]
- Srikanth J, Subramonian N, Premachandran MN. Advances in transgenic research for insect resistance in sugarcane. Tropical Plant Biol. 2011;4:52. doi: 10.1007/s12042-011-9077-2. [DOI] [Google Scholar]
- Stalker HT (1980) Utilization of Wild Species for Crop Improvement 11. Paper no. 6188 of the Journal series of the North Carolina Agricultural Research Service, Raleigh, North Carolina 27650. In: Brady NC (ed) Advances in Agronomy. Academic Press, 111–147
- Stevenson GC. Genetics and breeding of sugar cane. London: Longmans, Green & Co., Ltd.,; 1965. [Google Scholar]
- Sugiharto B. Biotechnology of drought-tolerant sugarcane. IntechOpen. 2017 doi: 10.5772/intechopen.72436. [DOI] [Google Scholar]
- Sundvor I, López-Aparicio S. Impact of bioethanol fuel implementation in transport based on modelled acetaldehyde concentration in the urban environment. Sci Total Environ. 2014;496:100–106. doi: 10.1016/j.scitotenv.2014.07.017. [DOI] [PubMed] [Google Scholar]
- Thiebaut F, Grativol C, Carnavale-Bottino M, et al. Computational identification and analysis of novel sugarcane microRNAs. BMC Genomics. 2012;13:290. doi: 10.1186/1471-2164-13-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut F, Rojas CA, Grativol C, et al. Genome-wide identification of microRNA and siRNA responsive to endophytic beneficial diazotrophic bacteria in maize. BMC Genomics. 2014;15:766. doi: 10.1186/1471-2164-15-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirugnanasambandam PP, Hoang NV, Henry RJ. The challenge of analyzing the sugarcane genome. Front Plant Sci. 2018;9:616. doi: 10.3389/fpls.2018.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Shinozaki K, Yamaguchi-Shinozaki K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front Plant Sci. 2015;6:84. doi: 10.3389/fpls.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vyver C, Conradie T, Kossmann J, Lloyd J (2013) In vitro selection of transgenic sugarcane callus utilizing a plant gene encoding a mutant form of acetolactate synthase. In Vitro CellDevBiol-Plant 49:198–206. 10.1007/s11627-013-9493-0 [DOI] [PMC free article] [PubMed]
- Vantini JS, Dedemo GC, Jovino Gimenez DF, et al. Differential gene expression in drought-tolerant sugarcane roots. Genet Mol Res. 2015;14:7196–7207. doi: 10.4238/2015.June.29.13. [DOI] [PubMed] [Google Scholar]
- Vettore AL, da Silva FR, Kemper EL, et al. Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res. 2003;13:2725–2735. doi: 10.1101/gr.1532103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision 2050—SBI ICAR. https://icar.org.in/files/ICAR-SBIVision2050-FinalDir-SBI.pdf
- Viswanathan R, Premkumari S, Amalraj R, Kathiresan T. Cloning partial endochitinase cDNA of Trichoderma harzianum antagonistic to Colletotrichum falcatum causing red rot of sugarcane. Curr Sci. 2006;91:951–956. [Google Scholar]
- Wang J, Roe B, Macmil S, et al. Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics. 2010;11:261. doi: 10.1186/1471-2164-11-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WZ, Yang BP, Feng XY, et al. Development and characterization of transgenic sugarcane with insect resistance and herbicide tolerance. Front Plant Sci. 2017;8:1535. doi: 10.3389/fpls.2017.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L-X, Deng H-H, Xu J-L et al (2011) Transgenic sugarcane plants expressing high levels of modified cry1Ac provide effective control against stem borers in field trials. Transgenic Res 20:759–772. 10.1007/s11248-010-9456-8 [DOI] [PubMed]
- Wu L, Birch RG. Doubled sugar content in sugarcane plants modified to produce a sucrose isomer. Plant Biotechnol J. 2007;5:109–117. doi: 10.1111/j.1467-7652.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu J-K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Ruan M, Qin L, et al. Field performance of transgenic sugarcane lines resistant to sugarcane mosaic virus. Front Plant Sci. 2017 doi: 10.3389/fpls.2017.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Nishiyama MY, Jr, Fuentes BG, et al. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol. 2009;149:171–180. doi: 10.1104/pp.108.128579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Birch RG. The gene for albicidin detoxification from Pantoea dispersa encodes an esterase and attenuates pathogenicity of Xanthomonas albilineans to sugarcane. PNAS. 1997;94:9984–9989. doi: 10.1073/pnas.94.18.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-Z, Yang B-P, Feng C-L et al (2006) Expression of the Grifola frondosa trehalose synthase gene and Improvement of drought-tolerance in sugarcane (Saccharum officinarum L.). J Int Plant Biol 48:453–459. 10.1111/j.1744-7909.2006.00246.x
- Zhang J, Nagai C, Yu Q, et al. Genome size variation in three Saccharum species. Euphytica. 2012;185:511–519. doi: 10.1007/s10681-012-0664-6. [DOI] [Google Scholar]
- Zhang J, Arro J, Chen Y, Ming R. Haplotype analysis of sucrose synthase gene family in three Saccharum species. BMC Genomics. 2013;14:314. doi: 10.1186/1471-2164-14-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang X, Tang H, et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet. 2018;50:1565–1573. doi: 10.1038/s41588-018-0237-2. [DOI] [PubMed] [Google Scholar]
- Zhangsun D, Luo S, Chen R, Tang K (2007) Improved Agrobacterium-mediated genetic transformation of GNA transgenic sugarcane. Biologia 62:386–393. 10.2478/s11756-007-0096-2
- Zhou D, Liu X, Gao S, et al. Foreign cry1Ac gene integration and endogenous borer stress-related genes synergistically improve insect resistance in sugarcane. BMC Plant Biol. 2018;18:342. doi: 10.1186/s12870-018-1536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]