Abstract
The avrBs2 avirulence gene of the bacterial plant pathogen Xanthomonas campestris pv. vesicatoria triggers disease resistance in pepper plants containing the Bs2 resistance gene and contributes to bacterial virulence on susceptible host plants. We studied the effects of the pepper Bs2 gene on the evolution of avrBs2 by characterizing the molecular basis for virulence of 20 X. campestris pv. vesicatoria field strains that were isolated from disease spots on previously resistant Bs2 pepper plants. All field strains tested were complemented by a wild-type copy of avrBs2 in their ability to trigger disease resistance on Bs2 plants. DNA sequencing revealed four mutant alleles of avrBs2, two of which consisted of insertions or deletions of 5 nucleotides in a repetitive region of avrBs2. The other two avrBs2 alleles were characterized by point mutations with resulting single amino acid changes (R403P or A410D). We generated isogenic X. campestris pv. vesicatoria strains by chromosomal avrBs2 gene exchange to study the effects of these mutations on the dual functions of avrBs2 in enhancing bacterial virulence and inducing plant resistance by in planta bacterial growth experiments. The deletion of 5 nucleotides led to loss of avrBs2-induced resistance on Bs2 pepper plants and abolition of avrBs2-mediated enhancement of fitness on susceptible plants. Significantly, the point mutations led to minimal reduction in virulence function of avrBs2 on susceptible pepper plants, with either minimal (R403P allele) or an intermediate level of (A410D allele) triggering of resistance on Bs2 plants. Consistent with the divergent selection pressures on avrBs2 exerted by the Bs2 resistance gene, our results show that avrBs2 is evolving to decrease detection by the Bs2 gene while at the same time maintaining its virulence function.
Plant disease resistance genes provide effective protection against pathogens expressing cognate avirulence (avr) genes (9). In agricultural settings this protection is usually short-lived, as pathogen strains with altered or missing avr genes that are able to cause disease on previously resistant plants are selected (23, 27). This raises the question of what selective advantages avr genes provide for the pathogen (4, 10, 23, 37). Understanding the evolutionary emergence of virulence and the function of avr genes in pathogenicity is therefore of paramount agricultural significance.
A role in virulence for a bacterial avr gene was first directly demonstrated for avrBs2 from Xanthomonas campestris pv. vesicatoria (19), the causal agent of bacterial spot disease of pepper and tomato (15, 27). X. campestris pv. vesicatoria strains expressing avrBs2 induce a hypersensitive response (HR), a form of localized programmed cell death associated with plant defense (12), on pepper plants carrying the cognate Bs2 resistance gene (27). On susceptible pepper plants lacking Bs2, however, avrBs2 was shown to increase bacterial fitness (19). A role in virulence in the form of promoting pathogen aggressiveness or fitness was subsequently shown for several other avr genes, including avrE, avrA, and avrRpm1 from Pseudomonas syringae and pthA, avrb6, and avrXa7 from various Xanthomonas species (for reviews, see references 4 and 23).
Further evidence for a role in virulence for bacterial avr genes comes from the finding that expression of avr genes and hrp genes in P. syringae are coregulated (13, 17, 30). Although coregulation was not observed in X. campestris pv. vesicatoria, hrp genes in both species control the ability to cause disease on susceptible plants and to trigger a hypersensitive response (HR) on resistant plants (25). Furthermore, a subset of hrp genes show homology to genes encoding type III protein secretion systems of mammalian bacterial pathogens such as Escherichia coli, Salmonella enterica serovar Typhimurium, Shigella spp., and Yersinia spp. (8, 14). These observations led to the hypothesis that avr gene products, together with other bacterial effector proteins, are transferred from the bacterial cytoplasm to the plant cytosol via a type III protein secretion system. Consistent with this hypothesis, it was found that the site of action of several avr gene products for triggering an HR is within the plant cytosol (13, 24, 29, 33, 36). In addition, expression of the Xanthomonas citri pathogenicity gene pthA in host plant cells was sufficient to induce the disease symptoms associated with the pathogen (7). In the case of avr gene products, plant cells have then evolved mechanisms to recognize bacterial effector proteins via resistance gene products, directly or indirectly, to trigger defense responses.
The avrBs2 gene encodes a putative protein with homology to agrocinopine synthase (ACS) of Agrobacterium tumefaciens and the glycerophosphoryl diester phosphodiesterase UgpQ of E. coli (34). ACS and UgpQ function in synthesizing and hydrolyzing phosphodiester linkages, respectively (3, 31). Currently, it is not known whether this homology is of relevance for the fitness-enhancing function of avrBs2. The avrBs2 locus resides in the genome and is present in most X. campestris pathovars, representing a diverse host range (19). The plant Bs2 gene has been introduced by breeding it into cultivated pepper varieties (Capsicum annuum) from the wild relative Capsicum chacoense. Bs2 has recently been cloned and encodes a resistance protein of the NBS-LRR class (35). Bs2 has been deployed widely in the field and remarkably has provided long-lasting field resistance. It has been hypothesized that this durability is based on Bs2 targeting a gene present in all X. campestris pv. vesicatoria strains and necessary for X. campestris pv. vesicatoria to attain full virulence (19). However, Bs2 efficacy is jeopardized by the emergence of new X. campestris pv. vesicatoria strains that cause disease on previously resistant pepper plants (20, 21).
In this paper, we report the molecular characterization of 20 X. campestris pv. vesicatoria field strains isolated from bacterial disease lesions on Bs2 pepper plants. These strains did not elicit an HR on Bs2 plants. In each of the 20 strains, we found a molecular lesion in the avrBs2 gene. Two mutant alleles of avrBs2 were found to lead to single amino acid changes in the predicted AvrBs2 protein. These novel avrBs2 alleles resulted in intermediate virulence and resistance phenotypes, representing a fine-tuning of AvrBs2 activity. This supports the hypothesis that avrBs2 is required for attaining full virulence. Our results reveal that avrBs2 is evolving to maintain its virulence function while avoiding Bs2-mediated recognition and activation of plant disease resistance.
MATERIALS AND METHODS
Bacterial strains and plasmids.
X. campestris pv. vesicatoria field strains described in this report were isolated from leaf spots of commercial F1 hybrid pepper plants containing the Bs2 gene grown in Florida. Spontaneous rifampin-resistant colonies were isolated by growing field strains on nutrient yeast growth medium (5) containing 100 μg of rifampin per ml. Plasmids were mobilized from E. coli strain DH5α into recipient X. campestris pv. vesicatoria strains by triparental mating using the helper plasmid pRK2013 (6). For conjugation of avrBs2+ into field strains, the vector p81546 (avrBs2 in the broad-host-range vector pRI40 [18, 34]) was used. For protein expression experiments, plasmid pDD62 was constructed from the vector pVSP61 (16) by inserting a linker containing start and stop codons in all three frames separated by a unique BamHI site downstream of the lacZ promoter. For chromosomal gene exchanges, the counterselectable suicide vector pSD800 was constructed by isolating the 6-kb band of pUCD800 (11) after complete digestion with BamHI and partial digestion with HindIII. This process eliminated the broad-host-range origin of DNA replication pSa. The 6-kb fragment was recircularized in the presence of the BamHI-HindIII fragment of the pBluescript polylinker (Stratagene, La Jolla, Calif.). The resulting vector, pSD800, contained a unique BamHI cloning site, the counterselectable sacB gene with its regulatory sequence sacR that leads to lethality on sucrose, a kanamycin resistance marker, and the 322 origin of DNA replication for propagation in E. coli.
Plant growth and inoculations.
For bacterial inoculations, the near-isogenic pepper cultivars ECW-0 (bs2/bs2) and ECW-20R (Bs2/Bs2) (15) were used. Two-month-old greenhouse-grown plants were shifted to a growth chamber with 16 h of light and with a temperature of 24°C 1 day before inoculations. For HR assays, leaves were infiltrated with a bacterial suspension with an optical density at 600 nm of 0.2 (≈2 × 108 CFU/ml) in 1 mM MgCl2 with a needleless syringe and scored after 24 h unless otherwise noted. For in planta bacterial growth experiments, leaves were syringe infiltrated with a bacterial suspension adjusted to ≈105 CFU/ml. At the time points indicated in Fig. 3 and 4, leaf tissue was harvested in triplicate and bacterial growth was determined as described previously (19).
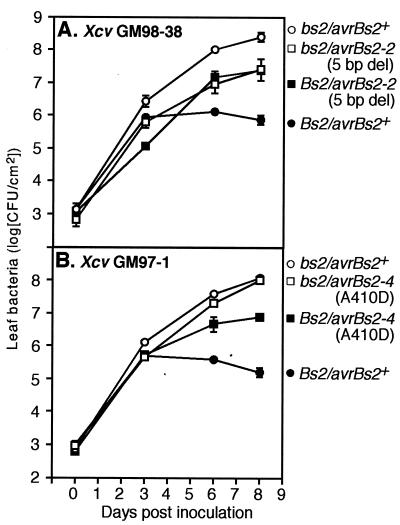
FIG. 3.
In planta growth of X. campestris pv. vesicatoria (Xcv) strains GM98-38 (A) and GM97-1 (B) in the pepper cultivars ECW-0 (bs2/bs2, open symbols) and ECW-20R (Bs2/Bs2, filled symbols). The growth of original field strains (squares) and isogenic strains expressing avrBs2+ (circles) is shown. Values are means of results with triplicate samples. Error bars denote standard deviation and are shown where values were larger than those represented by the symbols. Similar results were obtained in two independent experiments.
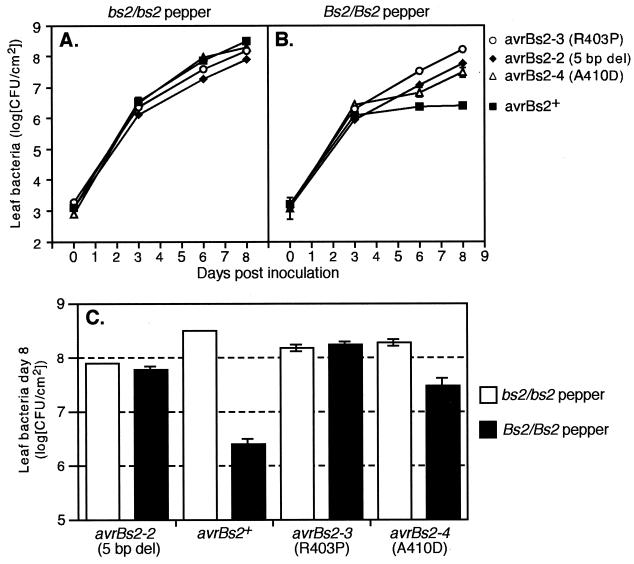
FIG. 4.
In planta growth in ECW-0 (A) and ECW-20R (B) of X. campestris pv. vesicatoria strain GM98-38 expressing avrBs2+ (filled squares) and the mutant alleles avrBs2-2 (filled diamonds), avrBs2-3 (open circles), and avrBs2-4 (open triangles). Values are means of results with triplicate samples. Error bars denote standard deviation and are shown where values were larger than the values represented by the symbols. Similar results were obtained in three independent experiments. (C) Data from day 8 postinoculation in panels A and B in bar graph format for ease of comparison of levels of bacterial growth in ECW-0 (open bars) and ECW-20R (filled bars) pepper plants. Error bars denote standard deviation and are shown where values were larger than those represented by the bar outlines. del, deletion.
Molecular techniques and sequencing and subcloning of avrBs2 alleles.
Standard techniques were used for DNA manipulation and DNA gel blot analysis (32). X. campestris pv. vesicatoria genomic DNA was blotted onto Hybond-N+ nylon membranes (Amersham Pharmacia Biotech, Piscataway, N.J.) and hybridized according to the manufacturer's instructions. The avrBs2 alleles of field strains were sequenced by PCR using a PRISM Ready Reaction DyeDeoxy Terminator Cycle sequencing kit (PE Applied Biosystems, Foster City, Calif.) on an ABI 377 automatic sequencing unit.
Clones of avrBs2 alleles were constructed by amplifying the region surrounding the four classes of nucleotide changes with primers 5′-GCGGGCTGTTCGATAATC-3′ (forward, nucleotides 1002 to 1019) and 5′-AGTTGCGGTAAACGTAGTAGTC-3′ (reverse, nucleotides 1871 to 1850) using Pfu DNA polymerase (Stratagene). PCR products were subcloned into the vector pCR2.1-TOPO using a TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.) and sequenced to verify the absence of PCR errors. The region containing the nucleotide changes was excised with BspEI and NcoI and placed into p815-33B (avrBs2 in pUC118 as a 2.4-kb BamHI fragment) (34). All constructs were verified by sequencing the cloning junctions and the region containing the four classes of nucleotide changes. The resulting clones of avrBs2 alleles were subcloned into pDD62 and pSD800 as BamHI fragments.
Antibody production and protein gel blot analysis.
For antibody production, avrBs2 was cloned into the phage T5 expression vector pQE30 (Qiagen, Chatsworth, Calif.) with an in-frame N-terminal six-His epitope tag-encoding sequence (N-His6-avrBs2). N-His6-AvrBs2 protein was overexpressed in E. coli M15 cells (Qiagen) and purified using nickel-nitrilotriacetic acid agarose according to the instructions of the manufacturer (Qiagen). Polyclonal antiserum was raised against the purified N-His6-AvrBs2 fusion protein in rabbits (Covance, Richmond, Calif.).
Bacterial protein was extracted from saturated liquid cultures of X. campestris pv. vesicatoria by harvesting cells by centrifugation and resuspending cells in lysis buffer (8 M urea, 0.1 M NaH2PO4, 10 mM Tris-HCl [pH 8]). Soluble protein was collected following centrifugation at 14,000 × g for 10 min. The concentration of protein in the supernatant was quantified spectrophotometrically by measuring absorption at 280 nm and comparing values to those of a standard curve obtained with bovine serum albumin (2). Protein samples (30 μg) were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (22) and transferred to MSI nitrocellulose membranes (Micron Separations, Inc., Westborough, Mass.) by electroblotting the samples in transfer buffer containing 192 mM glycine, 3.5 mM SDS, 25 mM Tris base (pH 8.3), and 20% (vol/vol) methanol at 1 A for 1 h. AvrBs2 protein was detected with a rabbit polyclonal antibody (diluted 1:2,000) raised against the N-His6-AvrBs2 fusion protein using an ECL Western blotting kit (Amersham Pharmacia Biotech, Piscataway, N.J.).
Chromosomal gene exchanges in X. campestris pv. vesicatoria strains.
The avrBs2 alleles cloned into the counterselectable suicide vector pSD800 (see above) were introduced into recipient X. campestris pv. vesicatoria strains by conjugation. Colonies with single crossover events were identified after 6 days of growth on 25 μg of kanamycin per ml. Integration of pSD800 into the chromosome was verified by replica plating kanamycin-resistant (Kanr) colonies on kanamycin and 5% sucrose. Colonies that were Kanr and sucrose sensitive (Sucs) were inoculated onto ECW-20R plants to verify that integration of pSD800 did not cause a frameshift in avrBs2+. Positive colonies were pooled and grown overnight without selection. Cells were harvested and plated on medium containing 5% sucrose to identify colonies that had lost pSD800 because of a second crossover event. Candidate Sucr colonies were replica plated on medium containing kanamycin, and the phenotype of candidate Sucr Kans colonies was tested in HR assays with ECW-20R plants. The avrBs2 genes of strains with the desired phenotype were fully sequenced to verify the genotype.
Pulsed-field gel electrophoresis.
Bacteria for pulsed-field gel electrophoresis were grown overnight in BBL nutrient broth (Becton Dickinson and Company, Cockeysville, Md.). Cells (109 CFU) were pelleted in microcentrifuge tubes, washed in 1 ml of sterile deionized water, and subsequently resuspended in 0.5 ml of TAE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8.0]). Cell suspensions were mixed with 0.5 ml of melted, cooled, low-melting-point agarose solution (10 mM Tris [pH 8.0], 10 mM MgCl2, 0.1 mM EDTA [pH 8.0], 2% [wt/vol] SeaPlaque GTG agarose [FMC Bioproducts, Rockland, Maine] in sterile deionized water) and pipetted into a Bio-Rad (Richmond, Calif.) block mold. Hardened agarose inserts were removed and transferred to lysing solution (0.5 mg of proteinase K per ml, 1% [wt/vol] N-lauroylsarcosine, 0.5% [wt/vol] SDS, 0.5 mM EDTA [pH 9.5]) in sterile disposable polypropylene tubes. Cells were lysed by placing tubes in a 50°C water bath for 16 h. After lysis, the inserts were removed from the lysis solution and washed in sterile TAE buffer. Subsequently, aliquots were placed into microcentrifuge tubes containing 200 μl of restriction enzyme buffer. After 30 min of incubation at room temperature, the restriction buffer was replaced with fresh buffer containing 10 U of the SpeI restriction enzyme (Promega, Madison, Wis.). Microcentrifuge tubes were incubated in a horizontal position for 16 h at 37°C. After incubation, the restriction buffer was removed and 0.5 ml of lysing solution (without proteinase K) was added. Samples were incubated at 50°C in a water bath twice for 2 h. Agarose sections were placed into the wells of a 1% gel (SeaKem GTG agarose; FMC Bioproducts). Wells were sealed with cooled 2% agarose, and the gel was placed in a Bio-Rad contour-clamped homogeneous electric field DR II unit and run at 200 V (16 V/cm) with linearly increased pulse times from 3 to 30 s over 22 h. Saccharomyces cerevisiae chromosome inserts (Bio-Rad) were used as molecular size markers. Gels were stained in ethidium bromide (0.5 mg/liter) and photographed on a UV transilluminator.
RESULTS
Complementation of HR-inducing phenotype by plasmid-borne avrBs2.
To study the molecular basis of altered virulence of X. campestris pv. vesicatoria in the field, we analyzed 20 X. campestris pv. vesicatoria field strains isolated in the years 1996 to 1998 from disease lesions on Bs2 pepper plants grown in Florida. These field strains did not elicit an HR, a marker for successful plant disease resistance (12), on Bs2 pepper plants. To determine if mutations in avrBs2 were responsible for disease on previously resistant Bs2 pepper plants, we conjugated a wild-type copy of avrBs2 on the broad-host-range vector pRI40 (18, 34) into a subset of 15 mutant strains. All 15 field strains elicited an HR on Bs2 plants when bearing avrBs2+, whereas empty vector controls did not elicit an HR (data not shown). The complementation experiments demonstrate that a mutation in avrBs2 was responsible for elimination of the HR phenotype on Bs2 plants. DNA gel blot hybridization experiments of chromosomal DNA isolated from field strains probed with full-length avrBs2 showed that none of the field strains had major rearrangements at the avrBs2 locus (data not shown).
Identification of molecular lesions in avrBs2.
We determined the nucleotide sequence of the avrBs2 gene in the 15 field strains assayed above and one additional field strain. Alterations within the coding sequence of avrBs2 were found in each case, revealing four mutant alleles of avrBs2 (Table 1). Three field isolates contained an avrBs2 allele (avrBs2-1) characterized by an insertion of five nucleotides, CGCGC, within a repetitive region of the gene containing 3.8 CGCGC repeats between nucleotide positions 1508 and 1526 of the avrBs2+ sequence (Fig. 1). One additional allele (avrBs2-2) had a CGCGC deletion in the same region (Fig. 1). Both the insertion and deletion cause a frameshift in the avrBs2 open reading frame, resulting in missense mutations after amino acid positions 449 and 448, respectively, and truncated proteins of 464 and 478 amino acids, respectively. The avrBs2-1 allele had been observed previously both in spontaneous mutants isolated from greenhouse-grown Bs2 pepper plants and in field strains from Australia (34). The other two avrBs2 mutant alleles were novel and consisted of point mutations leading to single amino acid changes, either at position 403 (R403P) for avrBs2-3 or at position 410 (A410D) for avrBs2-4 (Table 1). Both amino acids are conserved between AvrBs2 and ACS (Fig. 1), but not UgpQ. Eleven field strains contained the avrBs2-3 allele, whereas one contained avrBs2-4. The base change leading to the R403P mutation destroys an NaeI restriction site in avrBs2. This polymorphism was used to identify four additional field strains with the avrBs2-3 allele, verified by sequencing within this region. For the remainder of the study, we focused on the X. campestris pv. vesicatoria field strains GM98-38, GM98-16, and GM97-1, which contain the newly identified alleles avrBs2-2, avrBs2-3, and avrBs2-4, respectively (Table 1).
TABLE 1.
Mutations in avrBs2 identified in X. campestris pv. vesicatoria field strains
| avrBs2 allele | Mutation | Position(s)a | Effectb | No. of strains | Representative X. campestris pv. vesicatoria strain |
|---|---|---|---|---|---|
| avrBs2-1 | CGCGC insertion | 1508–1526 | Frameshift | 3 | GM97-157 |
| avrBs2-2 | CGCGC deletion | 1508–1526 | Frameshift | 1 | GM98-38 |
| avrBs2-3 | G to C | 1386 | R403P | 15 | GM98-16 |
| avrBs2-4 | C to A | 1407 | A410D | 1 | GM97-1 |
Numbering of nucleotides is according to the work of Swords et al. (34).
Amino acids are numbered by assuming initiation of translation at the second Met (codon at nucleotide positions 179 to 181). See the text for details of frameshift mutations.
FIG. 1.
Schematic representation of AvrBs2 and positions of mutations. The region of homology with the phosphodiester synthase ACS is hatched. Nucleotide changes in avrBs2-1 and avrBs2-2 are shown above the scheme, with numbering denoting the nucleotide positions of avrBs2+ (34). Single amino acid (aa) changes generated by avrBs2-3 and avrBs2-4 are shown below, with numbering denoting amino acid positions and with the assumption of the start of transcription being at the codon of nucleotides 179 to 181 (34). The alignment between ACS and AvrBs2 (residues 173 to 714) was performed using the BLAST algorithm (1), and only the region surrounding the single amino acid changes is shown.
Expression of mutant AvrBs2 proteins in X. campestris pv. vesicatoria.
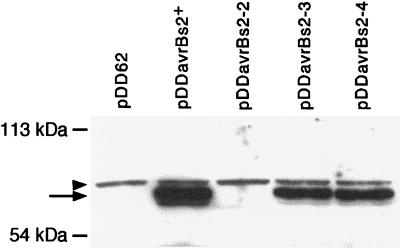
We tested the steady-state expression level of mutant AvrBs2 proteins in X. campestris pv. vesicatoria by protein blot analysis. Proteins isolated from crude lysate of X. campestris pv. vesicatoria expressing the avrBs2 alleles driven by the lacZ promoter were separated by SDS-PAGE and probed with a polyclonal antibody recognizing full-length AvrBs2 protein. Figure 2 shows that the AvrBs2-3 and AvrBs2-4 proteins accumulated to similar levels in X. campestris pv. vesicatoria, although the levels were slightly lower than for AvrBs2+. Conversely, the avrBs2-2 allele did not give rise to a full-length AvrBs2 protein. Conceptual translation of the avrBs2-2 allele predicts a truncated 52-kDa polypeptide, which was confirmed by overexposure of the protein blot, revealing low expression of a 52-kDa polypeptide (data not shown). These protein expression experiments indicate that AvrBs2-3 and AvrBs2-4 accumulate to steady-state levels comparable to those in wild-type X. campestris pv. vesicatoria but that AvrBs2-2 is both truncated and unstable. We therefore predict that the avrBs2-2 allele abolishes avrBs2+ function.
FIG. 2.
Protein gel blot analysis of X. campestris pv. vesicatoria crude lysate. The locations of the 113- and 54-kDa molecular mass markers are shown. The indicated avrBs2 alleles on the vector pDD62 were conjugated into strain GM98-38. Approximately 30 μg of total protein was loaded onto a 12% polyacrylamide gel. AvrBs2 was detected using polyclonal antibodies raised against full-length AvrBs2. The arrow shows the position of full-length AvrBs2 at approximately 80 kDa. The arrowhead indicates a nonspecific protein recognized by the AvrBs2 antiserum.
Engineering of stable chromosomal gene exchanges at the avrBs2 locus.
To test for complementation of the dual functions of AvrBs2+, we performed in planta bacterial growth experiments with vector-complemented GM98-38 strains. Preliminary results indicated that a plasmid-borne copy of avrBs2+ not only reestablished the induction of defense responses on resistant pepper plants as evidenced by suppression of bacterial growth but also enhanced the fitness of strains on susceptible pepper plants (data not shown). The results with other field strains, however, were difficult to interpret due to loss of plasmid pRI40 in a subset of individual colonies recovered from leaves several days postinoculation.
To circumvent uncertainties resulting from plasmid instability, variable plasmid copy numbers or gene expression levels, we constructed the counterselectable suicide vector pSD800 and by homologous recombination introduced avrBs2+ into the chromosomes of strains GM98-38 and GM97-1 (see Materials and Methods). This resulted in two sets of isogenic X. campestris pv. vesicatoria strains that differed only in the avrBs2 allele. Although we attempted several times to obtain strains with avrBs2+ using various field strains containing avrBs2-3, we were not able to do so because of the apparent failure of the vector pSD800, which contains avrBs2+, to integrate into the chromosome.
In planta growth of isogenic X. campestris pv. vesicatoria field strains.
Representative growth curves for GM98-38 and GM97-1 are shown in Fig. 3. With GM98-38, restoration of the avrBs2+ gene in the chromosome led to a 10-fold increase in growth by day 8 on susceptible plants, whereas growth was suppressed 30-fold relative to that of the original GM98-38 field strain due to the avrBs2-triggered resistance response on Bs2 plants (Fig. 3A). These experiments show that the fitness increase in avrBs2+-containing strains is not due to higher gene copy numbers or overexpression, since in the present study avrBs2+ was expressed from its native promoter in the chromosome. Results with GM97-1 and its isogenic avrBs2+ strain were remarkable in two respects. First, comparison of the growth behaviors of these two strains on susceptible pepper plants revealed that the avrBs2-4 allele is only minimally affected in its virulence function by day 8 (Fig. 3B). Second, comparison of growth of the original GM97-1 field strain on susceptible and resistant pepper plants indicated that avrBs2-4 has retained some of its function in triggering plant resistance. Growth of GM97-1 was suppressed 10-fold on Bs2 plants compared to growth on susceptible plants. Since growth of the corresponding reconstituted avrBs2+ strain was suppressed approximately 700-fold, GM97-1 was only partially recognized on Bs2 plants (Fig. 3B). Thus, the avrBs2-4 allele of GM97-1 represents a fine-tuning of avrBs2+ function, presumably as a consequence of selection pressure by the Bs2 resistance gene.
Next, we wanted to compare the effects of avrBs2-2 and avrBs2-4 directly and include avrBs2-3 in our analysis. Since significant in vitro growth differences were observed between individual field strains (data not shown), we generated an allelic series of avrBs2 in strain GM98-38. To generate chromosomal alleles of avrBs2-3 and avrBs2-4 in the GM98-38 background, we introduced these alleles on the counterselectable suicide vector pSD800 into the isogenic avrBs2+ strain shown in Fig. 3A and screened for loss of HR activity after two crossover events (see Materials and Methods). Figure 4 shows a direct comparison of the effects of avrBs2 alleles on the growth behavior of GM98-38 on susceptible (Fig. 4A) and resistant (Fig. 4B) pepper plants. The data for day 8 of Fig. 4A and B are replotted in Fig. 4C for ease of comparison. These experiments confirmed the results obtained with field isolate GM97-1, in that GM98-38 containing avrBs2-4 was partially recognized in Bs2 plants and attained a high level of growth on susceptible plants comparable to that of GM98-38 containing avrBs2+. Remarkably, GM98-38 expressing avrBs2-3 achieved the same level of virulence as the isogenic strain expressing avrBs2-4 on susceptible plants, without partially triggering a resistance response on Bs2 plants (Fig. 4C). The avrBs2-3 allele therefore appears to uncouple to a large extent the dual phenotype of AvrBs2 in enhancing bacterial virulence and inducing plant resistance.
HR phenotype of X. campestris pv. vesicatoria GM98-38 isogenic strains.
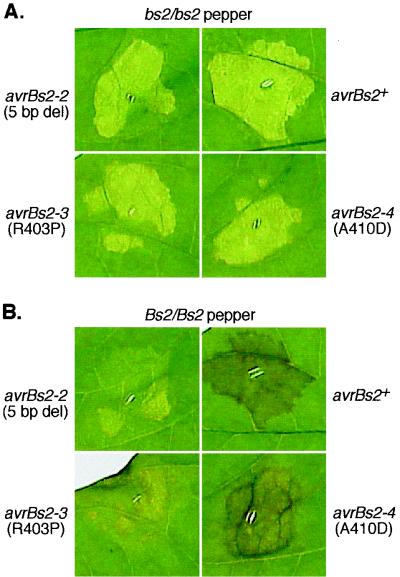
In initial HR assays at an inoculation density of 2 × 108 CFU/ml, no visible HR was observed on Bs2 plants infiltrated with either the field strain GM97-1 or strain GM98-38 expressing avrBs2-4. To verify the partial induction of plant resistance by avrBs2-4 observed in in planta growth experiments, we performed HR assays with higher inoculation densities. When inoculated at 109 CFU/ml, GM98-38 expressing avrBs2+ gave rise to a rapid HR response within 24 h, as evidenced by browning and desiccation of the infiltrated leaf area (data not shown). By 3 days postinoculation, isogenic strains expressing avrBs2-4, but not avrBs2-2 or avrBs2-3, induced a partial HR on Bs2 plants (Fig. 5). These results therefore confirm the findings from in planta growth experiments in that expression of the avrBs2-4 allele induced a partial plant resistance response dependent on the presence of the Bs2 gene.
FIG. 5.
Phenotype in leaves of ECW-0 (A) and ECW-20R (B) inoculated with 109 CFU of X. campestris pv. vesicatoria strain GM98-38 expressing avrBs2+ (upper right) and the mutant alleles avrBs2-2 (upper left), avrBs2-3 (lower left), and avrBs2-4 (lower right) per ml. Phenotypes were recorded 3 days postinoculation by placing leaves on a light box to distinguish clear water-soaked lesions (disease) from browning of the tissue associated with HR (resistance). del, deletion.
Genotypic polymorphism in X. campestris pv. vesicatoria field strains.
The field strains characterized in this study were isolated in pepper fields in Florida lying within a 50-mile radius. Strains harboring the avrBs2-3 allele were most highly represented in our sampling (15 out of 20). To determine whether this mutation arose several times independently, we explored the genotypes of the strains characterized here by pulsed-field gel electrophoresis. As shown in Fig. 6, all 15 field strains harboring avrBs2-3 and strain GM97-1 appear isogenic at this level of resolution, whereas a polymorphism can be detected in strain GM98-38. This result supports the hypothesis that the mutation generating the avrBs2-3 allele arose once, although it is likely that additional polymorphisms between the various field strains with avrBs2-3 could be detected at higher resolution.
FIG. 6.
Pulsed-field gel electrophoresis of genomic DNA isolated from the indicated X. campestris pv. vesicatoria (Xcv) field strains digested with SpeI. The locations of the 680-, 365-, and 225-kb markers are shown. Arrows indicate polymorphisms that distinguish strain GM98-38 from the other strains analyzed.
DISCUSSION
Here, we describe the molecular characterization of 20 X. campestris pv. vesicatoria field strains that evade host recognition mediated by the pepper resistance gene Bs2, which targets the X. campestris pv. vesicatoria gene avrBs2. Since loss of avrBs2 compromises fitness of the pathogen on susceptible host plants, evasion of Bs2-mediated resistance by inactivating avrBs2 comes at a cost in fitness to the pathogen. This loss of pathogen fitness was hypothesized to account for the effectiveness of Bs2-mediated resistance of pepper to bacterial spot disease in the field (19). In a previous study, Swords et al. (34) identified spontaneous mutants and field strains of X. campestris pv. vesicatoria from Australia and Barbados harboring avrBs2 alleles that gave rise to truncated AvrBs2 protein. These X. campestris pv. vesicatoria strains failed to elicit an HR on Bs2 pepper plants, and consistent with the above hypothesis, were compromised in pathogen fitness on susceptible plants. In our study, we found a molecular lesion in the avrBs2 gene in all 20 X. campestris pv. vesicatoria field strains examined, revealing four mutant alleles. The two mutant alleles comprising 5-bp deletions or insertions are likely to constitute null mutations of avrBs2 based on protein expression and in planta growth studies. Results with truly isogenic strains presented here, which obviate uncertainties regarding levels of expression of avrBs2, confirm that avrBs2+ functions in enhancing the fitness of X. campestris pv. vesicatoria on susceptible plants (19, 34).
Whereas deletion of full-length AvrBs2 led to attenuation of pathogen virulence, the point mutations in AvrBs2 characterized here had minimal effects on the virulence phenotype on susceptible pepper plants. The avrBs2-3 allele in addition abolished the resistance phenotype and therefore constitutes an almost complete uncoupling of the dual AvrBs2+ phenotype. Expression of the avrBs2-4 allele, in contrast, led to partial induction of resistance on Bs2 plants, as shown by in planta growth experiments and macroscopic HR assays. Since this finding was obtained with avrBs2-4 in two different strain backgrounds, it is unlikely that this phenotype is due to second-site mutations. Consequently, our results suggest that strains harboring avrBs2-3 have a significant fitness advantage for X. campestris pv. vesicatoria on Bs2 plants. The questions of whether avrBs2-3 confers a significant fitness advantage on Bs2 plants under field conditions and whether strains harboring avrBs2-3 are replacing other X. campestris pv. vesicatoria strains merit more careful studies.
Recently, Kousik and Ritchie (20) described X. campestris pv. vesicatoria field strains from pepper fields that overcome Bs2-mediated resistance and reported on the capacity of these strains to cause disease comparable to that caused by avrBs2+ strains. Although the molecular nature of these field strains is not known, these results suggest that avrBs2+ activity is dispensable in attaining full virulence (20). By using truly isogenic strains and quantitative in planta growth experiments, we were able to study separately the two aspects of avrBs2+ activity, namely, the capacity to elicit an HR on Bs2 plants and to increase pathogen fitness on susceptible plants. Significantly, we show that the avrBs2-3 allele, by uncoupling the dual avrBs2+ functions, promotes X. campestris pv. vesicatoria virulence on previously resistant Bs2 pepper plants to a level as high as that promoted by avrBs2+ on susceptible plants. The use of isogenic strains excludes effects of compensatory second-site mutations that increase the general fitness of bacteria. Our data therefore support the hypothesis that Bs2 targets an essential gene, while at the same time showing the result of divergent selection pressure on avrBs2.
A striking feature of the avrBs2 gene structure is the frequent occurrence of mutations within the CGCGC repeat region. Identification of mutant alleles with precise deletions or insertions of one repeat unit (this study and reference 34) is reminiscent of replication slippage at microsatellites (28). Interestingly, microsatellites in contingency genes encoding virulence factors of human pathogens such as Neisseria gonorrhoeae and Haemophilus influenzae cause expression of the corresponding proteins to be switched on and off at ≈10,000-fold higher rates than are expected from random mutations (26, 38). These contingency genes predominantly give rise to bacterial cell surface structures necessary for target host cell adhesion and/or invasion but which also trigger the host immune response. By rapidly modulating the expression of several of these surface structures, the likelihood of an advantageous combination of virulence factors in the changing environment within the host is increased for these pathogens (26, 38). Since the avrBs2 locus resides in the chromosome and cannot be abolished by plasmid loss, the microsatellite-like structure in avrBs2 may function in increasing the frequency of avrBs2 inactivation. Unlike missense mutations, inactivation by replication slippage is reversible, since the repeat structure in the gene is maintained (28). Further experiments are necessary to show whether the CGCGC repeat region in avrBs2 functions as a microsatellite in reversibly switching off avrBs2 expression under unfavorable conditions induced by the Bs2 resistance gene.
ACKNOWLEDGMENTS
We thank C. I. Kado for providing the vector pUCD800, K. L. Pernezny for supplying some of the X. campestris pv. vesicatoria field strains, Pauline Hsu for technical assistance, and Mary Beth Mudgett for advice on protein gel blot analysis and critical reading of the manuscript.
This work was supported by Novartis Agricultural Discovery Institute, Inc. (San Diego, Calif.). W.G. was supported in part by a fellowship from the Swiss National Science Foundation (823A-050350).
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J G, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. [Google Scholar]
- 3.Brzoska P, Boos W. Characteristics of a ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the Ugp transport system of Escherichia coli. J Bacteriol. 1988;170:4125–4135. doi: 10.1128/jb.170.9.4125-4135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dangl J L. The enigmatic avirulence genes of phytopathogenic bacteria. In: Dangl J L, editor. Bacterial pathogenesis of plants and animals—molecular and cellular mechanisms. Berlin, Germany: Springer-Verlag; 1994. pp. 99–118. [DOI] [PubMed] [Google Scholar]
- 5.Daniels M J, Barber C E, Turner D C, Cleary W G, Sawczyc M K. Isolation of mutants of Xanthomonas campestris pv. campestris with altered pathogenicity. J Gen Microbiol. 1984;130:2447–2455. [Google Scholar]
- 6.Ditta G, Stanfield D, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan Y P, Castañeda A, Zhao G, Erdos G, Gabriel D W. Expression of a single, host-specific, bacterial pathogenicity gene in plant cells elicits division, enlargement, and cell death. Mol Plant-Microbe Interact. 1999;12:556–560. [Google Scholar]
- 8.Fenselau S, Balbo I, Bonas U. Determinants of pathogenicity in Xanthomonas campestris pv. vesicatoria are related to proteins involved in secretion in bacterial pathogens of animals. Mol Plant-Microbe Interact. 1992;5:390–396. doi: 10.1094/mpmi-5-390. [DOI] [PubMed] [Google Scholar]
- 9.Flor H. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 10.Gabriel D W. Why do pathogens carry avirulence genes? Physiol Mol Plant Pathol. 1999;55:205–214. [Google Scholar]
- 11.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman R N, Novacky A J. The hypersensitive reaction in plants to pathogens: a resistance phenomenon. St. Paul, Minn: APS Press; 1994. [Google Scholar]
- 13.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gough C L, Genin S, Zischek C, Boucher C A. hrp genes of Pseudomonas solanacearum are homologous to pathogenicity determinants of animal pathogenic bacteria and are conserved among plant pathogenic bacteria. Mol Plant-Microbe Interact. 1992;5:384–389. doi: 10.1094/mpmi-5-384. [DOI] [PubMed] [Google Scholar]
- 15.Hibberd A M, Bassett M J, Stall R E. Allelism tests of three dominant genes for hypersensitive resistance to bacterial spot of pepper. Phytopathology. 1987;77:1304–1307. [Google Scholar]
- 16.Hinsch M, Staskawicz B J. Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol Plant-Microbe Interact. 1996;9:55–61. doi: 10.1094/mpmi-9-0055. [DOI] [PubMed] [Google Scholar]
- 17.Huynh T V, Dahlbeck D, Staskawicz B J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 18.Innes R L, Bent A F, Kunkel B N, Bisgrove S R, Staskawicz B J. Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J Bacteriol. 1993;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kearney B, Staskawicz B J. Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2. Nature. 1990;346:385–386. doi: 10.1038/346385a0. [DOI] [PubMed] [Google Scholar]
- 20.Kousik C S, Ritchie D F. Disease potential of pepper bacterial spot pathogen races that overcome the Bs2 gene for resistance. Phytopathology. 1996;86:1336–1343. [Google Scholar]
- 21.Kousik C S, Ritchie D F. Response of bell pepper cultivars to bacterial spot pathogen races that individually overcome major resistance genes. Plant Dis. 1998;82:181–186. doi: 10.1094/PDIS.1998.82.2.181. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Leach J E, White F F. Bacterial avirulence genes. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 24.Leister R T, Ausubel F M, Katagiri F. Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindgren P B. The role of hrp genes during plant-bacterial interactions. Annu Rev Phytopathol. 1997;35:129–152. doi: 10.1146/annurev.phyto.35.1.129. [DOI] [PubMed] [Google Scholar]
- 26.Meyer T F. Pathogenic Neisseriae: complexity of pathogen-host cell interplay. Clin Infect Dis. 1999;28:433–441. doi: 10.1086/515160. [DOI] [PubMed] [Google Scholar]
- 27.Minsavage G V, Dahlbeck D, Whalen M C, Kearney B, Bonas U, Staskawicz B J, Stall R E. Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria-pepper interactions. Mol Plant-Microbe Interact. 1990;3:41–47. [Google Scholar]
- 28.Moxon E R, Wills C. DNA microsatellites: agents of evolution? Sci Am. 1999;280:94–99. doi: 10.1038/scientificamerican0199-94. [DOI] [PubMed] [Google Scholar]
- 29.Mudgett M B, Staskawicz B J. Protein signaling via type III secretion pathways in phytopathogenic bacteria. Curr Opinion Microbiol. 1998;1:109–114. doi: 10.1016/s1369-5274(98)80150-1. [DOI] [PubMed] [Google Scholar]
- 30.Pirhonen M U, Lidell M C, Rowley D L, Lee S W, Jin S, Liang Y, Silverstone S, Keen N T, Hutcheson S W. Phenotypic expression of Pseudomonas syringae avr genes in E. coli is linked to the activities of the hrp-encoded secretion system. Mol Plant-Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 31.Ryder M H, Tate M E, Jones G P. Agrocinopine A, a tumor-inducing plasmid-coded enzyme product, is a phosphodiester of sucrose and l-arabinose. J Biol Chem. 1984;259:9704–9710. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 34.Swords K M M, Dahlbeck D, Kearney B, Roy M, Staskawicz B J. Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2. J Bacteriol. 1996;178:4661–4669. doi: 10.1128/jb.178.15.4661-4669.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tai T H, Dahlbeck D, Clark E T, Gajiwala P, Pasion R, Whalen M C, Stall R E, Staskawicz B J. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 37.Vivian A, Gibbon M J. Avirulence genes in plant-pathogenic bacteria: signals or weapons? Microbiology. 1997;143:693–704. doi: 10.1099/00221287-143-3-693. [DOI] [PubMed] [Google Scholar]
- 38.Weiser J N, Pan N. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol Microbiol. 1998;30:767–775. doi: 10.1046/j.1365-2958.1998.01108.x. [DOI] [PubMed] [Google Scholar]