Abstract
Consumption of live microorganisms “Probiotics” for health benefits and well-being is increasing worldwide. Their use as a therapeutic approach to confer health benefits has fascinated humans for centuries; however, its conceptuality gradually evolved with methodological advancement, thereby improving our understanding of probiotics-host interaction. However, the emerging concern regarding safety aspects of live microbial is enhancing the interest in non-viable or microbial cell extracts, as they could reduce the risks of microbial translocation and infection. Due to technical limitations in the production and formulation of traditionally used probiotics, the scientific community has been focusing on discovering new microbes to be used as probiotics. In many scientific studies, probiotics have been shown as potential tools to treat metabolic disorders such as obesity, type-2 diabetes, non-alcoholic fatty liver disease, digestive disorders (e.g., acute and antibiotic-associated diarrhea), and allergic disorders (e.g., eczema) in infants. However, the mechanistic insight of strain-specific probiotic action is still unknown. In the present review, we analyzed the scientific state-of-the-art regarding the mechanisms of probiotic action, its physiological and immuno-modulation on the host, and new direction regarding the development of next-generation probiotics. We discuss the use of recently discovered genetic tools and their applications for engineering the probiotic bacteria for various applications including food, biomedical applications, and other health benefits. Finally, the review addresses the future development of biological techniques in combination with clinical and preclinical studies to explain the molecular mechanism of action, and discover an ideal multifunctional probiotic bacterium.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12602-022-09992-8.
Keywords: Probiotics, Hormones, Immunity, Metabolites, Metabolomics, Microbiota
Introduction
In 1908, Ellie Metchnikoff introduced probiotics as useful microbes to favorably improve the human gut microbiota, replacing harmful microbes, thereby boosting human health [1]. The term “probiotic” was first introduced by Lilly and Stillwell in 1965, as secreted microbial by-products that strengthen the growth of its neighbors [2]. Previous studies have illustrated probiotics as favorable therapeutics to treat gut microbiota imbalance [3, 4]. Probiotics are “live microorganisms” that confer beneficial health effects to the host when administered in adequate amounts [5]. Probiotics are inclusive of a broad range of microbes and their application, and they differentiate live microbes used as sources of useful compounds from those that are used for health benefits only [5]. Currently, the probiotics market is growing exponentially estimated to reach USD 61.1 billion in 2021, and is projected to reach USD 91.1 billion by 2026 (http://www.marketsandmarkets.com/PressReleases/probiotics.asp).
Probiotics are described as free of pathogens and antibiotic resistance and their activity, viability, and growth efficacy should be properly established [6, 7]. The most common species used in probiotics research and development belong to Lactobacillus spp. and Bifidobacterium spp. Moreover, other species are available in the market including the Bacillus spp., Enterococci, Escherichia coli, Weissella spp., and Saccharomyces [8]. Probiotics have been commercialized as lyophilized pills, but also as supplements to various food sources such as cheese, yogurt, and nutritional bars to enhance human health. Most of the probiotic strains belonging to the Lactobacilli and Bifidobacterium have been given the Generally Regarded as Safe (GRAS) status in the USA, and qualified for the safety status given by EFSA (European Food Safety Authority). The multifactorial selection criteria of probiotics used in various food sources are often divided into three categories: resistant to acidic gastric conditions, capable of attaching to the gastrointestinal mucosa, and immune system modulator [9, 10]. Diverse approaches have been undertaken by Asian countries mainly Indonesia, Malaysia, Philippines, Singapore, Thailand, and Vietnam to regulate probiotics marketing [11]. Among these countries, only Indonesia, Malaysia, Philippines, and Thailand have enacted specific regulations related to the legal definition of probiotics. Malaysia, Philippines, and Thailand have permitted to use of probiotics foods or health supplements; however, all six Southeast Asian countries have permitted for application of new microorganism to be used following different approaches and requirements [11].
Probiotics have previously been classified into mono- or multi-strain microbial products [12]. Microbial strains in multi-species probiotics belong to Bifidobacterium breve, Bifidobacterium infantis, Bifidobacterium longum, Eubacterium faecium, Lactobacillus acidophilus, Lactiplantibacillus plantarum, Lacticaseibacillus casei, and Streptococcus thermophilus [12]. Multi-strain/multi-species probiotic strains due to their symbiotic nature show a cumulative positive effect on health in terms of treating antibiotic-associated diarrhea, improving growth performance and mortality in broilers, and protecting against Salmonella typhimurium infection [12]. Similarly, Chapman et al. [13] showed that probiotic mixtures comprised of different strains produced a superior effect than that of single-strain probiotics, against pathogenic growth. Among the multi-species, Bifidobacterium lactis W51, L. acidophilus W22, Lactiplantibacillus plantarum W21, and Lactococcus lactis W19 have been proven to strengthen the gut barrier function and are also currently commercialized worldwide [14]. These probiotic mixtures were proven to be beneficial to patients with food intolerances by increasing the IL-10 levels and decreasing the cytokine Th2 [15]. Classical probiotics comprising the Lactobacillus and Bifidobacterium groups were shown as promising biological tools for the treatment of a wide variety of inflammatory and metabolic diseases [9].
The results of many clinical trials involving humans and mice studies have shown the efficacy of probiotics against multiple diseases including their ability to suppress hypertension [16], reduce irritable bowel symptoms [17], prevent inflammatory bowel disease [18], and post-operative complications [19]. Similarly, probiotics showed higher efficacy against allergic disorders [20], potent antimicrobial [21], and anti-colorectal cancer properties [22]. At the molecular level, probiotics initiate the gene activation to regulate the host cells’ immune response including the regulation of brain behaviour through bidirectionalneuronal signaling [23].
The human gut microbiome in the form of probiotics can act as potential therapeutics against various issues like metabolic disorders, obesity, bacterial infection, and immunity [24]. Ingested microbes in the form of probiotics act as a source of useful compounds such as vitamins, bacteriocins, fatty acids, oligosaccharides, and various other immune-modulatory compounds that control the host immune system [25]. The human gut contains different microbes such as bacteria, fungi, and viruses; however, the consumption of probiotic bacteria alters the gut microbial composition in a strategic way to combat the pathogenic microbes in the intestinal niche [26]. However, in-depth studies in humans are urgently required to assess the probiotics-induced changes in gut microflora and these changes are associated with clinical benefits in the host. The recent discovery of high-throughput sequencing techniques coupled with other experimental approaches such as improved culturing methodologies, cost-effective genome and metagenome sequencing, and more powerful tools to unravel the bacterial genomes allows focusing on probiotics’ relevant biological questions, facilitating patient-oriented therapeutics [27]. The development of new tools integrated with the analysis of the microbial community composition, transcriptome shotgun sequencing, and proteomics approaches allows the identification of potent probiotic strains [28]. Novel sampling systems provide information about the immune system, metabolism, and microbiome alteration, driving this field forward [29]. Ultimately, an integrated approach will support the establishment of mechanistic insight into effector molecules and will pave the way for researchers to developnovel, emerging concepts like postbiotics [30].
Accumulating evidence addresses the increasing importance of the identification/discovery of novel probiotic bacteria with robust functionalities that hold great potential to commercialize worldwide as functional products and therapeutics. In the present review, we summarize the use of bacterial strains as probiotics and provide an overview of microbial selection, screening trials, metabolic engineering, and molecular biology approach in combination with synthetic biology to address research developments in the rational design and engineering of probiotic bacteria.
Mechanistic and Molecular Insight of Probiotic Function
Previous studies have shown diverse probiotic effects through multifarious mechanisms like pathogenic protection, enhancement of immunomodulation, alteration of gut microbiota, improvement ofthe barrier function of gut epithelium, and short-chain fatty acids (SCFAs) production (Fig. 1a) [31, 32]. The efficacy of probiotics to treat various diseases has been summarized in Table 1. Probiotics activity requires either direct contact or proximity to host cells for induction of inflammatory responses. The role of probiotics at an immune marker levelcomprises the modulation of mitogen-activated protein kinase (MAPK) signaling pathway, interleukin (IL)-6 & IL-8, tumor necrosis factor (TNF)-α, phosphoinositide 3-kinase (Akt), Interferon (IFN)-γ, and other contact-dependent mechanisms [72]. The specific effect of probiotics includes Lactobacillus (effector: lipoteichoic acid) mediated secretion of TNF-α, through toll-like receptor 2 (TLR-2), B. longum (effector: cell surface pilli) stimulated IL-10 secretion, modulation of proinflammatory cytokine and helper-T cell response by B. longum 36,524 (effector: exopolysaccharide), and L. rhamnosus (effector: cell surface appendages) mediated modulation of TNF-α, IL-6/10/12 in the intestinal mucous. Additional examples include L. rhamnosus (effector: pilli) induced generation of reactive oxygen species (ROS) and inhibition of NF-κB activation, Ligilactobacillus salivarius Ls33 (effector: peptidoglycan) mediated protection from colitis in IL-10 dependent manner, L. acidophilus L-92 (effector: surface layer protein slpA) mediated immune modulation, and B. lactis mediated induced IgA secretion [73, 74].
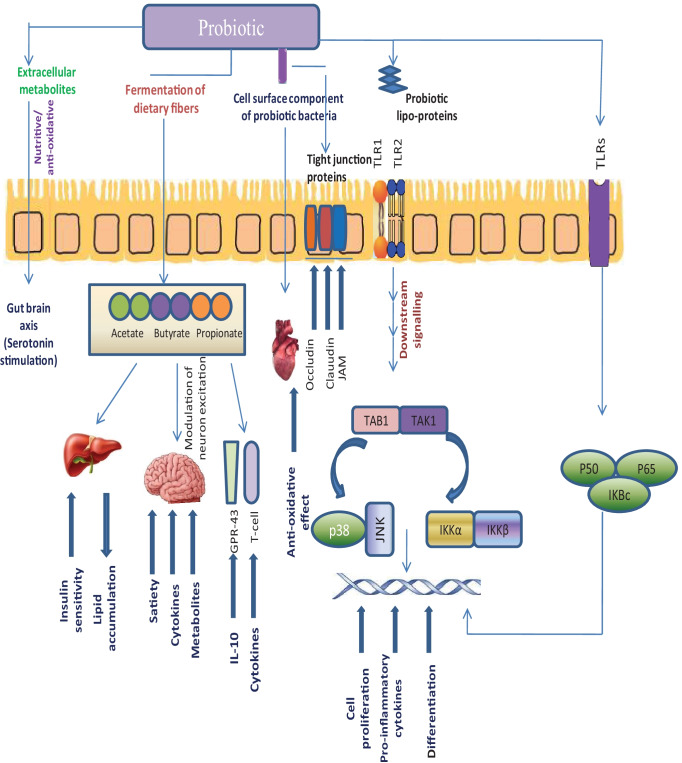
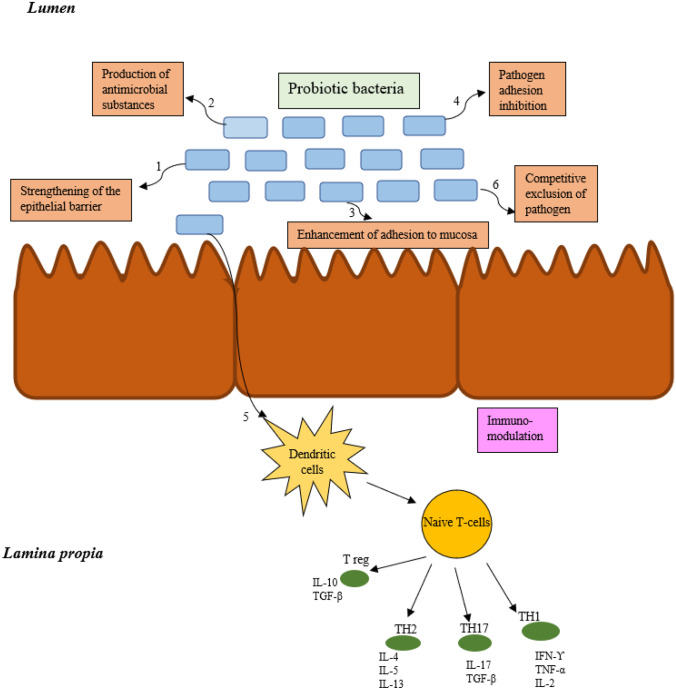
Fig. 1.
a Mechanism of action of probiotic bacteria; (i) probiotic bacteria can be used to cure depression by enhancing beneficial bacteria population which further improve mood through gut-brain axis; (ii) probiotic-produced SCFAs (acetate, butyrate, propionate) confers beneficial effects in the gut, brain, and also enhace the relese of IL-10 and cytokines; (iii) probiotic bacteria also alters the tight junction protein that forms a seal between adjacent epithelial cells near apical surfaces; (iv) probiotic bacteria interact with epithelial cells through TLR1/TLR2 and induces an increase in downstream signalling that are involved in immune response, and other processes like cell-proliferation, pro-inflmmatory cytokines productions, and differentiations. b Transport of probiotic and their active metabolites/factors from intestinal lumen to mucosa. Probiotic bacteria and their active metabolites confers several beneficial effects by (1) strengthening the epithelial barrier, (2) antimicrobial productions, (3) enhancement of adhesion protein to mucosa, (4) inhibiting the pathogen attachement, (5) activating the dendritic cells, and (6) competitive exlude the pathogens
Table 1.
Probiotic strains and their use in metabolic disorder and disease prevention
| Strain | Disease prevention | References |
|---|---|---|
| L. rhamnosusGG | Allergy and Immune Response | [33] |
| Enterococcus mundtii ST4SA | Antibiotic removal | [34] |
| Lactobacillus strains | [35] | |
| L. brevis KB290 | ||
| Bifidobacterium strains | ||
| L. plantarum 299v | Cardiovascular disease | [36] |
| E. coli 1917 | Colitis | [37] |
| E. faecalis | Colon cancer | [38] |
| L. plantarum | Diarrhea | [39] |
| L. caseiDN-114 001 | [40] | |
| B. bifidum | Eczema | [41] |
| B. lactis | [42] | |
| Escherichia coli | Food allergies | [43] |
| L. casei | Gastroenteritis | [44] |
| E. faecium M-74 | Hypercholesterolemia | [45] |
| Propionibacterium freudenreichii | [46] | |
| L. reuteri | Infant colic syndrome | [47] |
| Lactobacillus strains | Intestinal dysbiosis | [48] |
| L. johnsoniiN6.2 | ||
| B. infantis35624 | Irritable bowel syndrome | [49] |
| B. bifidum | [50] | |
| E. coliDSM17252 | [51] | |
| Lactobacillus strains | ||
| Saccharomyces cerevisiae CNCM I-3856 | [52] | |
| L. acidophulus | Lactose intolerance | [53] |
| S. cerevisiae | Pain relief | [54] |
| L.acidophulus | Peptic ulcer disease | [55] |
| L. rhamnosus GR-1 | Urinary tract infection | [56] |
| L. reuteri RC-14 | ||
| L. acidophilus | Ulcerative colitis | [57] |
| E. coli | ||
| Bifidobacterium | [58] | |
| L. rhamnosus GR-1 | Vaginal candidiasis | [59] |
| L. pentosus B281 | Inhibition of cancer cell proliferation | [60] |
| L. plantarum B282 | and cell cycle arrest | |
| L. casei ATCC 393 | Induction of apoptosis | [61] |
| Bacillus polyfermenticus KU3 | [62] | |
| Enterococcus faecium RM11 | [63] | |
| L. fermentum RM28 | ||
| L. acidophilus SNUL | Suppressed proliferation of tumor cells | [64] |
| L. casei YIT9029 | ||
| B. longum HY8001 | ||
| L. reuteri PTCC 1655 | Prevention of gastric cancer | [65] |
| L. kefiri P-IF | [66] | |
| Bacillus natto, | Prevention of colorectal carcinoma | [67] |
| L. acidophilus | ||
| Propionibacterium freudenreichii | Prevention of liver cancer | [68] |
| Streptococcus thermophilus | Decreased the risk of colorectal cancer | [69] |
| Lactobacillus delbruckii | ||
| L. acidophilus L1 | Decreased the risk of bladder cancer | [70] |
| L. casei Shirota (LcS) |
Minimized the human papilloma virus (HPV) associated infection |
[71] |
Most mice studies have demonstrated that probiotics inhibit pathogenic colonization either by attachment to epithelium cells through competition for mucosal binding sites, by producing antimicrobial compounds, or by blocking the ability of pathogens to adhere (Fig. 1b). Collectively, most of these responses were shown in different probiotic strains in mice models. Also, the response is strain-dependent (Table 2). Many lactic acid bacteria (LAB) produce compound such as bacteriocins which show antimicrobial activity [83]. Bacteriocins have been shown to disrupt the quorum sensing (QS) signaling pathway. Production of bacteriocin Abp118 by L. salivarius UCC118 inhibits the infection of L. monocytogenes in mice [84]. L. acidophilus La-5 has been shown to inhibit the virulence factor expression of pathogenic E. coli O157:H7 in an in-vitro mice model [85]. L. acidophilus A4 antagonized the adhesion of E. coli to the intestinal epithelium by upregulating the secretion of IL-8, TNF-α, and mucin-2 [86]. Among other lactic acid bacteria, L. acidophilus GP1B improved the survival of mice subjects, following infection with Clostridium difficile. Additionally, Limosilactobacillus reuteri RC-14 repressed the virulence of Staphylococcus aureus through toxic shock syndrome toxin-1 [87]. Among other functionalities, probiotic bacteria have been shown to maintain gut barrier function through the up-regulation of tight junction proteins like claudin-1 and occluding [88]. The hydroxy cis-12-octadecenoic acid (HYA) produced by L. plantarum has been demonstrated to modulate the tight junction proteins by inducing the secretion of INF-γ and TNF-α, via mitogen-activated protein kinase (MAP-K) and extracellular signal-regulated kinase (ER-K) pathway [89]. Additionally, many probiotics can secrete metabolites, improving colonic epithelial resistance, enhancing the upregulation of mucous secretion (MUC1, MUC2, MUC3) in colonic epithelial cells, and modulating the microbiome population.
Table 2.
Microorganism employed as probiotics
| Species | Response | References |
|---|---|---|
| A. muciniphila | Enhance the immunity and gut barrier function, production of Vitamin B12 | [75] |
| B. uniformis | Intestinal homeostasis and immune | [75] |
| B. coagulans, B. subtilis, B. laterosporu | Control of infections and increased survival of host | [76] |
| B. longum, B. catenulatum, B. breve, B. animalis, B. bifidum | Enhanced maturation of DCs and production of IL-10 & IL-12, upregulation of c-myc and il-6 genes | [77] |
| E. faecium | Prevention of diarrhea and minimizing the chronic sinusitis and bronchitis | [78] |
| L. plantarum, L. paracasei, L. acidophilus, L. casei, L. reuteri,L. rhamnosus, L. curvatus, L. crispatus | Induction of mucin secretion, induction of mucosal, humoral and cellular immune responses | [79] |
| L. lactis | Activate the innate immune response and protection against pathogen infection | [80] |
| P. productus | Improvement of nervous system functioning | [75] |
| P. jensenii, P. freudenreichii | [75] | |
| S. boulardii | Prevention of Clostridium difficile infection and antibiotic associated diarrhea | [81] |
| S. sanguis, S. oralis, S. mitis, S. thermophilus, S. salivarius | Inhibition of fungal infection, prevention of biofilm formation by pathogenic bacteria | [82] |
In mice, probiotic metabolite HYA ameliorated the pathogen-induced epithelial barrier disruption by preventing the degradation of E-cadherin and beta-catenin in a GPR 40-dependent pathway [90]. L. rhamnosus secreted proteins p40 and p75, which play a key role in maintaining intestinal epithelial stability by inhibiting cytokine-mediated apoptosis. In an in-vitro cell-based assay, Lactobacillus strains have been shown to secrete MUC-3 in HT29 cells and MUC-2 in Caco-2 cells; however, these effects require direct mucosal adherence. Various Bifidobacterium and Lactobacillus resist bile salts by producing the bile salt hydrolase (BSH), thus minimizing weight gain and also reducing the cholesterol, and triglyceride levels in mice [91]. Sarkar et al. [92] reported that probiotics can also regulate the host’s antidepressant and anxiolytic effects via regulating the signaling to the central nervous system. In L. rhamnosus JB-1 administrated mice, corticosterone response was attenuated through the expression of mRNA for γ-GABA-A and GABA-B brain receptors [93]. Mice fed with L. reuteri (strain ATCC PTA 6475) restored the oxytocin level, and improved their social behaviour when fed with the high-fat diet, which resulted in gut dysbiosis [94].
Up to now, a few in-vivo proteomics studies involving large-scale characterization of microbiota-mediated changes have been displayed especially in gastrointestinal tract (GIT) models [95]. Proteomics studies involving probiotics have provided insight into molecular mechanisms and metabolic pathways, required to overcome the challenges of acidic pH in GIT (pH 2.0) and bile salts in the small intestine. Most proteomic studies highlighted the over-expression of proteins regulating the carbohydrate metabolism, over-expression of Clp family proteins (GroES, GroEL, DnaK, DnaJ), and proton translocating ATPase to maintain cytoplasmic homeostasis. Additionally, the expression level of proteins involved in transcription, translation, DNA repair, nucleotide, and amino acid biosynthesis were highly affected [96, 97]. Furthermore, proteomic evidence showed enhanced exopolysaccharides (EPS) production, supporting the hypothesis that EPS may confer protection to bacterium against the adverse environment in the GIT. On the other hand, in L. rhamnosus the under-expression of EPS biosynthesis genes resulted in a less protective EPS layer, which was found to assist in gut adhesion [98]. Moreover, proteomic evidence supports that Lactobacillus exposure to bile salts triggers the expression of Nag A/Nag B proteins, maintaining the redox balance and preventing oxidative damages [99, 100].
An in-vitro proteomic study identified the major proteins associated with adhesion processes as well as the probiotic-host effect was interestingly shown by cell appendages/or envelopes [101]. Furthermore, proteins such as enolase (ENO), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and elongation factor Tu (EF-Tu) promote probiotic adhesion to GIT. However, these proteins are also used by pathogens to bind to the mucous layer of GIT, illustrating the key role of probiotics in preventing enteropathogenic bacteria to attach to the intestinal mucosa [102]. Moonlighting proteins such as GroEL and EF-Tu, present in the probiotic strain Lactobacillus johnsonii NCC533, enhance IL-8 secretion in macrophages, illustrating their role in immune modulation. Izquierdo and associates suggested that differences in the surface proteome of Lactobacillus strains were responsible for their diversity in adhesion ability to mucous of GIT [103]. A higher ratio of proteins like GroEL and DnaK responsible for mucin binding was present in highly adhesive L. plantarum WHE 92, when compared to less adhesive L. plantarum 299v and CECT 4185. The proteomic map of the L. plantarum 299v identified 29 proteins, highlighting their role in mucin binding [101]. Ashida et al. [104] reported the surface layer protein (SlpA) of L. acidophilus in adhesion to Caco-2 cells. The immuno-stimulatory activity and protective role of SlpA in L. helveticus against Salmonella typhimurium have been reported [105]. A mutational study featuring the SlpA of L. acidophilus confirmed the SlpA’s role in binding to dendritic cells for enhanced pro-inflammatory cytokine production [106]. Candela et al. [107] reported that exposure to bile acids upregulates Plg receptors such as ENO/DnaK and phosphoglycerate mutase in Bifidobacterium animalis BI07. Similar changes were observed in the surface layer proteome of B. longum under bile stress [108]. These findings illustrate the role of bile as signaling trigger molecule to enhance the probiotic-host cross-talk and further colonization processes.
Recently, Caenorhabditis elegans has attracted attention as an Ideal model for testing the efficacy of suitable probiotic candidates [109]. As compared to cell culture, C. elegans has a certain advantage of being an entire organism with different cell and tissue structures. Additionally, the absence of ethical issues and lower cost allows a safe orientation of future experiments on mammals and also decrease the number of animals to use/or sacrifice. A study showed that probiotic administration decreased the bacterial load in its intestine and also increased its lifespan. Some Lactobacilli and Bifidobacterial strains (B. infantis, B. longum) increased the survival of C. elegans from 17% to a maximum of 46% [110–112]. A recent study observed that probiotic bacterium L. paracasei D3-5 and L. plantarum SK-9 administration increased the function of C. elegans with an improvement in physical functions and high muscle mass [112]. Probiotic bacteria enhance the longevity of C. elegans via modulating the p38 mitogen-activated protein kinase (p38 MAPK) signaling pathway and through induction of the SKN-1 genes encoding antioxidant proteins. A study showed that probiotic intake increased the longevity of C. elegans through involving the pmk-1 and NHR (nuclear hormone receptor) family; however, the detail mechanism is still unclear [113]. Recently, it was observed that probiotic B. infantis administration significantly increased the lifespan of C. elegans through mutation of Toll-like receptor homolog TOL-1 [114]. These shreds of evidence clearly indicate that C. elegans model represents a cheap, easy-to-handle and rapid tool to screen the new probiotic microorganisms and also to explore the molecular aspects of interactions. Additionally, in the future C. elegans will emerge as powerful, bioethical and pertinent model host between in-vitro and mammalian models.
Probiotics: Important Source of Bioactive Molecules
Probiotics are important sources of various bioactive compounds (Table 3). Among these bioactive compounds, bacteriocins are proteinaceous toxins produced by a wide variety of bacteria [129]. Target-specific bacteriocins are non-toxic in nature and limit the progression of pathogens to neighboring cells. Many LAB-produced bacteriocins are emerging as antibiotics with proven potential in in-vitro and in-vivo systems [130, 131]. LAB-bacteriocins are very specific to their target species, displaying bacteriostatic and bactericidal activity and do not affect their neighboring bacteria. Therefore, simultaneous applications of bacteriocins and antibiotics are appearing as possible therapeutic agents [132–135]. Members of the LAB-group belong to the hetero-fermentative group and have the ability to produce various organic acids. The produced organic acids act on the outer surface of pathogenic bacteria and hamper the protein synthesis machinery resulting in increased fatalities [136]. The lactic acid produced by LAB completely inhibits the growth of pathogens, such as E. coli, Salmonella, and L. monocytogenes even at 0.5% (v/v) concentration. Additionally, the mucous layer of epithelial cells provides protection against damages caused by these acids. Similarly, diacetyl produced by many LAB-groups, interacts with arginine receptors of Gram-negative bacteria, leading to reduced arginine availability [137]. Some LAB-groups have the ability to produce H2O2, which inhibits pathogenic bacteria through a toxic oxidation mechanism [138]. The reuterin produced by L. reuteri inhibited DNA replication and was shown to compromise the survival of E. coli and Candida spp. [139].
Table 3.
Bioactive compounds of probiotic bacteria and its effect on health system
| Probiotic strain | Bioactive compounds | Health effects | References |
|---|---|---|---|
| B. coagulansRK-02 | Exopolysaccharides (EPS) | Amelioration of toxic oxidative free radicals | [115] |
| B.adolescentis DSM 18,350 | Folate vitamins | Biosynthesis of nucleic acid | [116] |
| B. pseudocatenulatum | |||
| Bifidobacterium spp. | Pyridoxine (Vitamin B6) | Amino acid metabolism | [117] |
| Clostridium spp. | Amino acids | Provides essential amino acid | [118] |
| E. casseli flavus MI001 | Enterocins | Antimicrobial activity | [119] |
| Enterobacteriaceae | Metabolite like amino acids | Nutrient support | [118] |
| Enterococcus spp. | Aromatic amino acids | Improve the male reproductive system | [118] |
| Fusobacterium varium | amino acids (arginine) | Improve the male/female reproductive system | [118] |
| F. prausnitzii, | Butyric acid | Energy source for colonocytes | [120] |
| Lactobacillus spp. | Lactic acid | Improve the metabolism | [121] |
| Lactobacillus spp. | Vitamin B1 | Synthesis of nucleic acid/fatty acids | [122] |
| Lactobacillus sp. G3 | Amylase | Starch hydrolysis | [123] |
| L. gasseri strains DSM 20,604 and 20,077 | Inumins/Levans | Minimize fat/cholesterol absorption | [124] |
| L. fermentum E-3 | Superoxide dismutase/Catalase | Antioxidant activity | [125] |
| L. fermentum E-18 | |||
| L. fermentum CECT 5716 | Vitamin B9 | Energy metabolism | [126] |
| L. reuteriJCM1112 | Vitamin B12 | Improves the RBC formation | [127] |
| P. freudenreichii | Propionic acid | Play a role in gluconeogenesis | [128] |
| P. freudenreichii | β-galactosidase | Hydrolysis of β-galactoside | [128] |
Probiotic bacteria have the ability to produce EPSs in bulk [140]. Microbial EPSs have caught attention for their cumulative health effect. These have been shown to promote an immunostimulatory response, antitumor and antioxidant activity, and also lower blood cholesterol [141]. Enzymes such as glycosyltransferase and glycantransferase can metabolize sugar nucleotides into EPSs. EPSs contain multiple sources of oligosaccharides, which are cross-linked polymers, composed of monomer units. Probiotics together with other gut residing bacteria ferment non-digestible oligosaccharides to monosaccharides, therefore, providing nutrients for the proliferation of gut microflora [142]. Dietary oligosaccharides boost the immunoglobulin-A (IgA) level, which plays a natural role in the host’s defense. In animal studies, fructooligosaccharide (FOS) induced the synthesis of IgA [143]. Similarly, FOS and galactooligosaccharides (GOS) have been used for nutritional therapy in case of constipation [142].
Probiotics are a source of various important enzymes like lactase, amylase, esterase, and lipases. Lactase hydrolyses lactose to glucose and galactose, which are further converted to SCFAs [144]. LAB reduces the amount of lactose in yoghurt through β-galactosidase, lowering lactose intolerance [145]. Similarly, amylase and peptidase are also probiotically produced enzymes that severely affect the biochemical reaction of the host, modulating its metabolism. Probiotic bacteria also play an important role in flavor development through the biochemical conversion of amino acids to aldehydes, alcohols, and acids. Small molecules like acyl-homoserine lactones produced by gut bacteria play an important role in quorum sensing and biofilm formation [146]. Many LAB produces small peptides and amino acids by proteolysis of casein [147].
Many probiotic bacteria produce water and fat-soluble B-group vitamins. Water-soluble vitamins are absorbed in the intestine, whereas fat-dissolvable vitamins are ingested as micelles in the intestinal tract [148]. They are required for various metabolic processes of carbohydrates, fats, amino acids, and nucleic acid synthesis. Many Bacillus and E. coli strains have been shown to produce riboflavin [149]. Similarly, Bifidobacteria strains produce B-complex vitamins, which help in the maintenance of the overall health of the gut [150]. Interestingly, few bacterial strains are sufficient sources of B12, which is not synthesized by plants and is important for blood formation and nervous system function. The LAB probiotic strain L. reuteri is able to produce the cobalamin. Propionibacterium shermani also produces B12, propionic acid, and other useful metabolites for industrial application. Another group of vitamins, thiamine, is produced by Bifidobacterium [150].
Metabolic and Functional Potential of Probiotics
Gut bacteria ferment the undigested carbohydrates to produce energy which can be used for multiple body functions. These probiotic bacteria maintain a healthy symbiotic relationship with the host’s gut flora. Under optimal intestinal conditions, they modulate their metabolic pathway machinery to metabolize undigested sugar into SCFAs, such as acetate, butyrate, and propionate [121]. It is estimated that most of the produced SCFAs (up to 95%) are utilized by the gut microbiota for their energy-dependent processes. However, their profile varies with different prebiotic sources. The produced SCFAs act in a beneficial way, specifically as an energy source and also counteracting pathogenic microorganisms [151]. In humans, they enhance signaling pathways like AMP kinase in muscle, and promote fatty acid oxidation, thereby decreasing lipid accumulation [152]. Among SCFAs, acetate crosses the blood–brain barrier and is associated with regulatory neuropeptides that favor appetite suppression [153]. Butyrate is used as an energy source for the propagation of colonocytes and enterocytes. Additionally, butyrate has shown therapeutic potential in diseases like diarrhea, colon cancer, cardiovascular disease, and other inflammatory responses [154, 155]. Propionate diffuses into the portal vein system to be utilized for hepatic gluconeogenesis processes [156]. Moreover, SCFAs reduce the secretion of cytokines/or chemokines by lowering the local infiltration of macrophages, and enhance adipogenesis in a G-protein coupled receptors such as the GPR43-dependent mechanism, by activating the peroxisome proliferator receptor [152]. To regulate carbohydrate and fat metabolism, probiotic bacteria produce many amino acids and their derived molecules (Supplementary Figure 1) [144]. The amino acids produced in the gut are further fermented by bacteria to produce phenol and indoles, maintain the energy balance, and inhibit pathogen infections. Moreover, the fermentation of amino acids generates several chains of esters, alcohols, and organic acids, which form the aroma and flavor of various food products [157].
Few probiotic bacterial strains contain exopolysaccharide-metabolization enzymes such as glycosyltransferases and glycantransferases, which convert sugars to EPSs. Besides EPS-producing, probiotic bacteria offer further advantages in antitumor and antioxidant activity, immune-stimulatory behavior, and also lower blood cholesterol [158]. The human body develops intolerance to lactose due to a deficiency in the enzyme lactase; however, LAB possesses the enzyme β-galactosidase which decreases the amount of lactose in food products, especially yoghurt [159]. Additionally, LAB also produces enzymes like amylase and peptidase that improve the metabolism of their host [160]. The biological effect of administrated probiotics is severely affected by the enzymatic activities in the gut, being estimated that Lactobacillus and Bifidobacteria exhibit over 20 different enzymatic functions. B. longum administration changes the intestinal microbiota, lowering the β-glucuronidase level [161]. In addition, B. longum increases ATP production through the phosphoketolase pathway leading to enhanced acetic acid production [162]. Similarly, B. animalis activates the pathway of intermediate metabolites like formate, resulting in enhanced oxalate consumption [99].
Furthermore, clinical trials using probiotics to treat non-alcoholic fatty liver disease (NAFLD), showed a reduction in liver aminotransferase activity [163]. A random clinical study, featuring 30 healthy individuals fed with yogurt containing two probiotic Lactobacillus strains, compared with healthy individuals fed with standard yogurt, detected nineteen enzymatic activities in the feces of probiotic-treated volunteers. The activity profile tested by both groups was constant; however, the naphthol-AS-BI-phosphohydrolase and leucine arylamidase activities were higher in the feces of the probiotic administrated group [164]. In another study, probiotic-treated group received L. gasseri CECT 5714 and L. coryniformis CECT 5711, showing higher fecal butyrate levels, when compared to the control group. Changes in fecal acid contents could be used as biomarkers for identifying individuals who benefited from probiotic treatment; however, the impact of the probiotic intake on microbiota and metabolites production is associated with the gut microbial ecosystem [165].
Another clinical study featuring 33 healthy individuals of different age groups (26 to 76), were given an oral dose of L. plantarum strain Lp-8, showed higher levels of acetate and propionate when compared to butyrate, illustrating that the production of fecal metabolites is bacteria-dependent [166]. A 4-week random clinical trial involving 20 human volunteers fed with Bifidobacterium reported higher levels of metabolites like acetate, butyrate, isobutyrate, and succinate when compared to control groups [167]. Probiotic bacteria either by themselves or in symbiosis upon administration to elderly people could be able to improve their metabolic functions [168]. In a trial of 96 days with probiotic yeast Saccharomyces boulardii, the strain was found to be helpful for decreasing the occurrence of diarrhea receiving the total enteral nutrition. The level of fecal butyrate was found to be lower in patients as compared to healthy individuals. A trial with S. boulardii boosted the SCFAs content in patients and increased fecal SCFAs, potentially contributing to protection against diarrhea [169]. Probiotic L. plantarum strain 299v enhanced the fecal SCFAs content especially butyrate in patients suffering from C. difficile infection, which contributed to minimizing the effects of antibiotics. The SCFAs level was found to be decreased in the group treated with the antibiotic metronidazole when compared to the probiotic-treated group. However, after the study, the total SCFAs level regressed to pre-antibiotic treatment levels [169].
A clinical study with L. casei featuring 77 subjects of 84 years of age showed a decrease in norovirus-gastroenteritis processes, while a significant amount of fecal acetic acid content was observed [170]. Considering the microbiome population, Bifidobacterium and Lactobacillus are the dominant genera, while the population of Enterobacteriaceae decreased in the fecal content of probiotic-treated individuals. Recently, Nagata et al. [171] documented those individuals treated with probiotic L. casei strain Shirota, recorded higher fecal acetate content with significantly lower fever incidence and enhanced bowel movement. Additionally, the C. difficile count was lower in the probiotic-supplemented group. Likewise, L. paracasei Lpc-37 and B. lactis HN019 consumption reduced the risk of diarrhea in children, and the concentration of fecal SCFAs and branched-chain fatty acids (BCFAs) were higher in probiotic-treated subjects. L. paracasei Lpc-37 intake led to increased Bifidobacterium population, isobutyrate, and isovalerate levels. B. lactis HN019 administration was found to be positively correlated with the microbiota population and negatively correlated with the propionate level [172]. Consumption of probiotic L. salivarius CRCT5713 increased the fecal Lactobacilli population and the fecal amino acid content, especially of butyric acid [173]. Oral application of probiotic B. lactis Bb12 lowered the fecal pH in newborn infants and is positively correlated with higher acetate and lactate content [174]. Among other probiotic strains, B. bifidum W23 and B. animalis W52 exhibited higher SCFA and lactate concentrations and lower succinate/lactose concentrations in children with eczema. The observed outcome emphasizes the role of SCFAs and other metabolites in the regulation of the immune system [161]. The use of probiotics provided beneficial effects in terms of improving carbohydrate metabolism and fasting blood glucose levels, reducing metabolic stress, and influencing microbial ecology and immunity [175]. A probiotic preparation containing Bifidobacterium, Lactobacillus, and S. salivarius attenuated NAFLD, primarily at the level of amino acid metabolism, nucleic acid degradation, and microbial-mediated amino acids metabolism. In addition to lowering liver lipids concentration, probiotics also play an important role in NAFLD and some of the fecal metabolites act as biomarkers to evaluate the response [176].
Genetic Engineering of Probiotics: a Promising Way Forward
In the human body, cellular metabolism and complex biological processes are regulated by various enzymes. Therefore, any enzyme deficiency leads to several metabolic disorders, where the accumulated toxic product produces abnormal responses, in some cases leading to death [177]. Thus, engineering the beneficial probiotic bacterial strains with specific enzymes or metabolic pathways resulted in protection against metabolic disorders [27, 178]. Additionally, the engineering of probiotics improves their performance in terms of their site-specificity and multi-host functionality [179]. In the present scenario, the interest is increasing in engineering the probiotics in combination with synthetic biology approaches to enhance the delivery of target molecules [178, 180], the sensing ability [181, 182], and anti-pathogenicity [183–186]. Additionally, various molecular biology tools targeting the multiple regulatory systems can be employed to engineer the beneficial probiotic strains (Fig. 2). Engineered probiotic strains have advantages over microbiota-mediated therapy by displaying specific functions that are not shown by gut microbiota. Individuals with diabetes may develop abnormal levels of blood glucose due to insufficient production of pancreatic insulin. Currently, researchers are designing recombinant probiotic strains for the treatment of diabetes. For example, Lactococcus lactis was engineered to secrete an immunoregulatory cytokine IL-10 and the oral delivery of this strain was found promising for the treatment of type 1 diabetes [187] (Fig. 3).
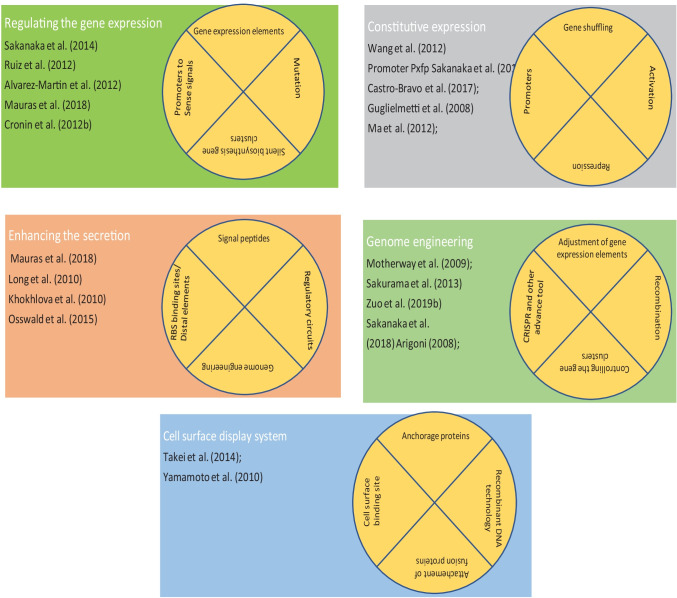
Fig. 2.
The use of molecular biology tools in combination with synthetic biology approach to engineer the probiotic bacteria to enhance their functionalities; the improvement can be done by a regulating the gene expression, b constitutive expression of gene under suitable promoters, c. enhancing the secretion of target enzyme by addition of signal peptides, d. edition of genes through genome engineering, and e. through development of cell surface display system
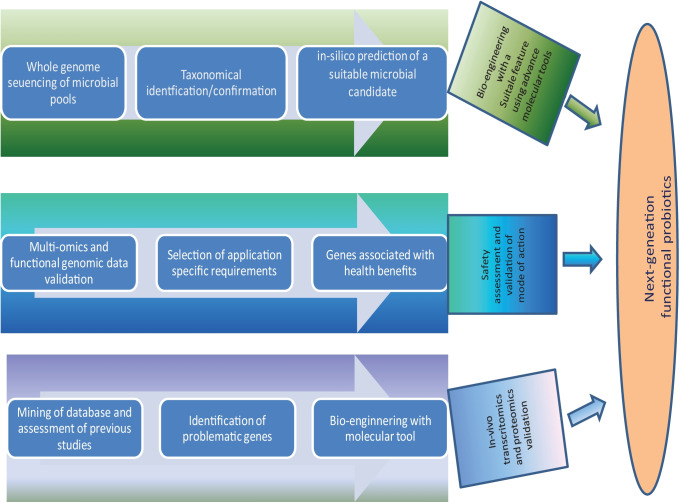
Fig. 3.
Schematic diagram summarizing the each route for development of next-generation probiotics; the probiotic bacteria can be engineered for enhancing their suitable feature using adavanced molecular biology tools, further its efficacy can be tested by in-vivo evaluation followed by evaluating the safety aspects, and validating the mode of action
In another study, a recombinant strain of L. gasseri was developed to include the glucagon-like peptide (GLP)-1 gene, which is known to induce insulin production both in vitro and in vivo. Oral delivery of the L. gasseri GLP-1 strain in diabetic rats resulted in enhanced insulin production from pancreatic beta cells as well as in the reduction of blood glucose (10 to 20%) [188]. Similarly, engineered L. lactis strains are able to secrete the autoantigen GAD65370–575, leading to a decrease in IL-10-induced pancreatic islet inflammation under hyperglycemic conditions [189]. Deficiency in phenylalanine hydroxylase (PAH) is the root cause of the metabolic disorder phenylketonuria, where the organism accumulates phenylalanine resulting in several health issues [190]. By recombinant development, phenylalanine ammonia-lyase (PAL) gene from Anabaena variabilis was introduced into L. reuteri, which reduced the blood phenylalanine level in an in-vivo mice study [191]. Similarly, the probiotic strain SYNB1618 was developed to secrete PAL and l-amino acid deaminase (LAAD), which greatly reduced the blood phenylalanine up to 38% [178]. Recently, strain SYNB1618 (thyA−, argR−) was developed by genomic insertion of argR215, and its oral administration greatly reduced the ammonia level, thereby enhancing the survival rate up to 50% [27].
Moreover, in recent years, various mutagenesis approaches like single-crossover-insertion [192], double deletion [193], homologus recombination [194], and inducible plasmid self-destruction (IPSD)-mediated genome engineering have been well established for probiotic Bifidobacterial strains. Many of these approaches possess several drawbacks like low-transformation efficiency, unstable mutations, loss of plasmid, and extensive screening of a large pool of colonies (Supplementary Figure 2) [195]. However, the CRISPR-Cas genome editing tool can be employed for rapid and efficient genome modifications [196, 197]. This newly discovered tool can be used for transcriptional regulation and control of the genetic circuits [196]. The CRISPR-Cas systems for genome engineering have been applied to various lactic acid bacterial groups like Lactobacillus, Lactococcus, and Streptococcus [196, 197]. The CRISPR-Cas in combination with IPSD will further improve the genome engineering in probiotic bacterial groups [198]. Therefore, the development of genetic tools including the plasmid expression systems, genome engineering approaches, etc. will promote the synthetic biology applications of probiotics. A number of broad-host range plasmids such as pSH71, pAMβ11, pBC1, and pTB6 have been characterized for gene expression in LAB [199, 200]. However, their functionality in other probiotic strains has not been well established.
Various approaches like transformation, conjugation, electroporation or phage transduction, etc. can be used to transfer plasmid DNA to other microbial strains. However, evolved bacterial host defence systems like CRISPR-Cas, and restriction-modification systems prevent the invasion of foreign DNA [201–203]. To overcome these issues, conjugation is an alternative tool to transfer plasmid DNA in a single-stranded form from donor to recipient bacteria [204]. Higher success was obtained by transferring the plasmid from E. coli to a variety of Bifidobacterial strains using a conjugative transfer system [205]. On the other hand, for efficient expression of the foreign gene products in a host, promoters, regulatory elements such as ribosome binding sites (RBS), cell-surface anchoring elements, and various secretion and localization signals need to be considered [206]. Promoters such as hup encoding a histone-like protein and gap encoding glyceraldehyde-3-phosphate dehydrogenase, induce the expression of genes in Bifidobacterial strains at high levels [207]. The promoter Pgap has been used for efficient homo and/or heterologous expression and for the production of various fluorescent proteins and therapeutic factors in Bifidobacteria [208, 209]. In response to certain signals, genetic circuit elements reprogramme the gene expression in a tightly controlled manner [210]. Bacterial one/two-component system, and quorum sensing are used as a tool to construct the genetic circuit to diagnose disease [181, 182], bacteriocidal activity [185, 186, 211], and to deliver therapeutic molecules [180]. Additionally, the gene expression system sensing the GI-signals for pH, bile salts, inflammation, etc. can be used to construct a genetic circuit for various diagnostic, and therapeutic purposes [182].
In recent years, engineered probiotics to treat bacterial infections and inhibit pathogen proliferation in the gastrointestinal tract have received great attention. These “programmed” probiotics are a safer therapy to efficiently target multidrug-resistant bacteria, through the secretion of antimicrobials, immunomodulation, inhibition of adhesion, and toxic protein production from pathogens [184, 212]. Among pathogens, biofilm-forming bacteria cause severe chronic infections leading to immune system imparity. Researchers across the globe have focused on engineering probiotics to inhibit biofilm formation [213, 214]. Attempts were made to develop an engineered E. coli strain to inhibit the Pseudomonas aeruginosa, through the modulation of quorum sensing. The engineered E. coli was developed with the addition of E7 lysis protein and pyocin S5 which minimized the P. aeruginosa growth by 99%, also reducing biofilm formation by 90% [214]. During an in-vivo study, dispersin B (anti-biofilm protein) was introduced to the constructed strain and the engineered strain showed higher efficacy in C. elegans and mice infection studies [184]. Furthermore, microcin H47 was introduced in E. coli N 1917 to counteract the Salmonella infections [215]. To counteract the overgrowth of Salmonella, atetrathionate sensing system was incorporated, which induced the production of H47 and was found to be effective against Salmonella [216]. Similarly, strain N 1917 was engineered to secrete the antimicrobial peptide Mccj25 and the administration of this strain was effective to decrease the population of Salmonella by 97%, while the homeostasis of the native microbiome was maintained.
On the other hand, intestinal infection with Vibrio cholera causes acute diarrheal disease and if it remains untreated, it can lead to shock and even death [217, 218]. Infections by this pathogen are regulated by the quorum-sensing system in a density proportional manner. Therefore, attempts were made to disrupt the sensing system of V. cholerae, using an engineered probiotic E. coli strain. Administration of the strain was found to be quite effective to decrease the toxin formation in a mouse model [219]. Additionally, oral delivery of L. lactis could prevent the V. cholerae infection through lactic acid secretion. Pursuing engineered probiotics, researchers focused on developing a strong sensing system to identify molecules or produce strong signals, making them an effective diagnostic device. In that sense, a diagnostic circuit was introduced in L. lactis through the fusion of CqsS (V. cholera) and NisK (L. lactis) domains. The constructed L. lactis strain was orally delivered to cholera-infected mice, enhancing their β-lactamase secretion, which was recorded by colorimetric shift [185]. Similarly, Sedlmayer et al. [220] coupled the formyl peptide sensor with quorum sensors to eliminate pathogenic infections. Expression cassettes encoding luciferase (lux CDABE) and β-galactosidase (lac Z) were introduced into E. coli to develop the strain PROP-Z, to detect liver metastasis by luminescent analysis [221, 222]. High levels of nitric oxide (NO) serve as a signaling device, as well as an inflammatory indicator in inflammatory bowel disease [223, 224]. Probiotics were engineered to detect NO levels which serve as markers for inflammation.
Therefore, the development of metabolic engineering in parallel with synthetic biology enabled the creation of novel probiotic strains with valuable features. The use of engineered probiotic strains should precede rigorous clinical trials, required to minimize the exposition of patients to any potential risks [225]. Gene encoding as a novel effector function must be evaluated in vitro and in vivo in terms of genetic stability during the production processes. Nonetheless, further research is needed to provide the adequate colonization of anecological niche in the gut by delivered strains.
Probiotics and COVID-19
Coronaviruses are a large family of viruses that have crown-like appendages on their surface. Human coronaviruses (CoV) were identified in the mid-1960s, and currently, the following seven types of CoV have been identified (https://www.cdc.gov/coronavirus/types.htm). Viral infections in the respiratory tract lead to disturbance in the gut microbiota [226]. COVID-19 infection resulted in an increase in disorders of the human stomach and intestine, lymphopenia, acute respiratory distress, and multi-organ failure [227]. Additionally, the overproduction of proinflammatory cytokines termed as “cytokine storm” was observed in COVID-19 patients [228]. The increase in cytokine can damage the lung, brain, liver, cardiovascular system, etc. [229].
A recent study demonstrated that COVID-19 infection reduced the count of Bifidobacterium spp. and Lactobacillus spp. [230]. Initially, the treatment option applied for coronavirus disease 2019 (COVID-19) included artificial ventilation and antiviral agents. In addition to the development of vaccines, the use of biological agents to target viral infections using immunomodulation has received great attention [231]. Baud et al. [232] listed the bacteria such as B. bifidum, B. breve, L. casei, L. gasseri, L. plantarum, Pediococcus pentosaceus, etc. as potent probiotics with the ability to decrease the burden of COVID-19 pandemic. Similarly, patients suffering from other complications like antibiotic-related diarrhea showed a higher proportion of COVID-19 infection. These recent findings further indicate that there is a requirement for microbiota balance in patients suffering from COVID-19 infection using some probiotics [230]. COVID-19 infection also causes severe hypoxia, whereas a reduction in probiotic species leads to an increase in the number of pathogens like Actinobacteria spp., Corynebacterium spp., and Ruthenibacterium spp. [233]. Available theoretical evidence suggests the biological role of probiotics in fighting the cytokine storm related to COVID-19 infection; however, there is an urgent need for clinical and laboratory evidence [232, 234].
Experimental evidence suggests that L. paracasei and L. plantarum have the capability to reduce the immune-inflammatory response [235]. The biological activity of probiotic bacteria belonging to Lactobacillus and Bifidobacterium controls the gastrointestinal dysbiosis associated with COVID infection [236]. Similarly, biofilm-forming probiotic bacteria like L. Reuteri and a few Lactobacillus spp. produces biologically active molecules that showed anti-inflammatory properties [237, 238]. Bottari et al. [239] illustrated the immune benefits of probiotics in COVID infection by priming mucosal immunity through stimulation of IgA secretion, improvement of biological functions of macrophages, and stimulation of regulatory cells. Experimental evidence suggests that probiotics also shape the T cell subsets [240], enhance the antimicrobial peptide production [241], and also direct the Th17 cells’ differentiations in the small intestine [242]. Probiotics belonging to Lactobacilli have been reported to produce peptides that interact with the ACE2 receptor, the host entry receptor of SARS-CoV-2 viruses [243]. A recent study by Minato et al. [244] find that Paenibacillus bacteria naturally produce carboxypeptidase homologous to the ACE2 receptor.
Recently, it was observed that acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike proteins binds to human angiotensin-converting enzyme 2 (ACE2). To counteract this problem, biongineered probiotics expressing ACE2 were developed to neutralize SARS-CoV-2 [245]. Probiotic L. paracasei (LP) expressing secretory human ACE2 (sACE2) controlled the viral spread by sequestering the virus or blocking the spike protein interaction with host cell-associated receptors. The sACE2 produced by probiotic bacteria confers systemic effects to control the viral entry at multiple organs including the lungs. Additionally, the bioengineered bacteria enhance the innate immunity and confer beneficial effects to control dysbiosis in the SARS-CoV-2 infected patients [245].
Probiotics and Cancer
Besides treating various metabolic diseases and disorders, probiotics also modulate cancer signaling [246], through induction of apotosis [247], and autophagy [248], inhibition of mutagenic and kinase activity [249], activation of tumor suppressors [250], and prevention of metastasis [251]. The structural components of probiotic bacteria, their secreted metabolites, and various other signaling molecules can alter the metabolic and regulatory reactions associated with the host [252].
The structural components constitute the microbe-associated molecular pattern and include the S-layer proteins, flagella, pili, capsular polysaccharides, lipoteichoic acid, and lipopolysaccharides [253]. The metabolites include the secreted proteins, extracellular vesicles, short-chain fatty acids, hydrogen peroxides, and bacteriocins [254]. Previous reports showed that probiotic bacteria induce apoptosis to inhibit tumorigenesis and progression via altering the tumor necrosis factor, inhibitors of apoptosis proteins, B cell lymphoma (Bcl)-2, caspases, and p53 gene [255, 256]. The bacteriocin produced by probiotic bacteria shows anticancer activity through the formation of minute pores on the plasma membrane which induce apoptosis and cell cycle arrest at the G1 phase [257]. The probiotic-derived ferrochrome act as tumor-suppressive molecule to inhibits the colon cancer progression via c-jun N-terminal Kinase (JNK)-mediated apoptosis [258]. Similarly, linoleic acid produced by probiotic L. plantarum initiate the apoptosis in breast cancer cells via downregulation of the NFκB pathway [259]. On the other hand, L. acidophilus and B. bifidum increased the cytotoxic activity against breast and colon cancer cells by upregulating IFN-γ and TNF-α expression, and downregulating Bcl2 expression [260]. Probiotic bacterium L. acidophilus induces apoptosis via increasing the mRNA expression of surviving [261]. Another study revealed that Propionibacterium enhanced apoptosis in colorectal carcinoma cells through the action of SCFAs on mitochondria [262].
The surface protein from L. acidophilus induced HCT116 cell death via altering the level of autophagy-linked protein (microtubule-associated protein 1) [263]. The exopolysaccharides of probiotic bacteria activate the autophagy in colon cancer cells via stimulating the Beclin1/GRP78 and apoptotic pathways Bcl-2 and Bak proteins [248]. A previous study demonstrated that probiotic-produced SCFAs target the tumor cell through epigenetic regulation of the tumor suppressor and oncogenes [264]. The probiotic bacterium L. acidophilus, L. rhamnosus MD14, and L. rhamnosus GG enhanced the expression of tumor suppressor genes in an experimental colon carcinogenesis model [265]. Similarly, probiotic bacterium B. longum induced the expression of tumor suppressor miR-15 and miR-145 in an experimental murine colorectal cancer [266]. Some studies demonstrated the probiotic-mediated tumor-suppressive effect by downregulation of oncogenes [267, 268]. The probiotic L. crispatus and L. rhamnosus alter the cancer progression by affecting the expression of mTOR-related genes and altering the proto-oncogenes (Wnt/β-catenin) pathways [269]. Similarly, probiotic bacteria also downregulate the KRAS proto-oncogene, thereby decreasing the progression of colon cancer [265].
Probiotic bacteria also modulate the kinase and phosphatase enzymes that play a major role in tumor biology such as cell propagation and metastasis etc. [270]. Previous studies showed that probiotic bacteria and their produced metabolites act as kinase inhibitors to treat diarrhea after cancer therapy. A study showed that probiotic secretory proteins modulate the protein kinase C (PKC) to protect the intestinal epithelial tight junctions from H2O2-induced damage [271]. Probiotic bacterium L. plantarum induces apoptosis via downregulating the MAP-kinases and upregulating the phosphatases [272]. These shreds of evidence clearly show the multifarious effects of probiotics against cancer. Although, researchers have discovered the apoptotic potential of probiotics on cancer, their molecular mechanisms still need to be discovered. Further, in-depth investigation and clinical trials w.r.t probiotic-mediated autophagy and its role in cancer elimination are urgently required.
Probiotics and Neurodegenerative Disease
Neurodegenerative diseases are serious disorders especially in the aging population and are characterized by degeneration of structure and function of the central nervous system (CNS) and peripheral nervous system (PNS). These diseases will impose an increasing socio-economic burden and incidences are also increasing year by year [273]. At present, the most common neurodegenerative diseases are Alzheimer’s disease (AD) and Parkinson’s disease (PD) and steadily progress due to the loss of neurons in the brain [274]. The etiology of these diseases is still unknown; however, inflammation may be involved in the progressive development of these diseases [275]. A recent study showed a decrease in the population of intestinal microbes and a higher abundance of pro-inflammatory bacteria in AD patients as compared to healthy ones [276]. Another study showed an increased population of gram-negative Bacilli in AD patients may lead to increased distribution of lipopolysaccharides from the gut to the systemic circulation and it enhances the pathology of AD through neuroinflammation [277]. Similarly, in PD patients, a higher population of hypothetical pathogens such as E. coli, Enterococcus, Proteus, and Streptococcus was observed [278]. The influence of the brain-gut axis has attracted attention especially in neurodegenerative diseases and gut microbiota affects immunity, inflammation, and neuroregulation through the brain-gut axis [279]. These evidences illustrate that maintaining a healthy microbiome benefits normal brain functioning, and boosts the immune response etc.
Probiotic intake helps the proliferation of intestinal microflora, ameliorating gastrointestinal function, reducing gut leakiness, and helping fight against pathogens by regulating the immune system [280]. A recent study showed that probiotic intake decreased the inflammatory cytokines IL-6 and TNF-α in serum and improved the anti-inflammatory cytokines IL-10 in serum and brain in PD mice [281]. Probiotic administration reduced the expression of pro-inflammatory IL-1, TNF-α, and increased the expression of anti-inflammatory TGF-β, and PPAR-γ in PD patients [282]. Additionally, the CBM (total bowel movement), spontaneous bowel movements (SBM), very low-density lipoproteins (VLDL), triglyceride, and Bristol Stool Scale improved in PD patients.
A recent in-vitro study with probiotic Lactobacillus and Bifidobacterium reduced oxidative stress, and pro-inflammatory cytokines in peripheral blood mononuclear cells isolated from PD patients [283]. A recent investigation showed that a mixture of probiotics conferred a neuroprotective response on dopaminergic neurons to counteract motor impairments in a PD mouse model [284]. Probiotic formulation VSL#3 controls the expression of various genes in the brain cortex, minimizes inflammation, and improves neuronal performance [285]. The innovative probiotic formulations SLAB51 (known as Sivomixx) effectively control the 6-hydroxydopamine-induced deleterious effects both in in-vivo and in-vitro PD models. The impairment involves the protection of dopaminergic neurons, restoration of pro-survival and neuroprotective pathways, induction of anti-inflammatory pathways, and amelioration of behavioral impairments [286]. These evidences clearly illustrates that probiotics intake modified the gut microbiome and influenced the protective response to CNS disease via the gut-brain axis, mediating different pathways involving neural, hormonal, inflammatory, antioxidant, and immune signaling [287, 288].
Next-Generation Probiotics
Probiotics are live microorganisms, usually isolated from gut bacteria; however, until their safety and health effects are fully characterized, they cannot be given the term “Probiotics.” Among various nomenclatures, probiotics are referred to as “functional food” or “beneficial bacteria” that are thought to prevent disturbances and balance the host’s system [289]. Previous research trials showed that the gut microbiota has limited therapeutical potential, therefore there is a high demand for novel and cost-effective formulations [290, 291]. Promising results have been observed in bacterial species Lactobacillus and Bifidobacterium in the prevention of diverse metabolic and inflammatory diseases. The information generated from recent clinical studies showed a positive outcome for inflammatory and metabolic disease, therefore making the path for selection of next-generation probiotics including the members of Akkermansia muciniphila, Bacteroides uniformis, Clostridium spp., and F. prausnitzii [292, 293]. A. muciniphila are well-known for mucin degradation and were linked to the maintenance of a healthier metabolic status. Under a high-fat diet, microbiota composition especially A. muciniphila decreased and was associated with several abnormalities like high-level blood glucose, insulin resistance, and increased plasma triglycerides [294]. In a clinical study with different diets, Dao et al. [295] found that an optimal or even increased population of A. muciniphila was associated with blood glucose maintenance as well as minimizing hypercholesterolemia. In another study, it was observed that the presence of A. muciniphila restored the gut barrier and also reduced the inflammation [296].
Among all microbes comprised in the gut, Bacteroides constitute approximately 25% of the total population. These commonly behave as commensal microorganisms and are transmitted from mother to child during delivery. Bacteroides are primarily responsible for the fermentation of complex sugars, producing volatile fatty acids in the gut. Among Bacteroides, B. fragilis and B. thetaiotamicron can efficiently metabolizes carbohydrates. Polysaccharides produced by B. fragilis activate the T cell-mediated immune response and therefore, strengthen the host immunity [297]. Round et al. [298] reported that B. fragilis-produced polysaccharide signals through TLR2 and activates Toll-like receptor (TLR) pathways. However, B. fragilis are benefitted from the presence of several virulence factors on their capsules and thereby avoiding the host immunogenic responses. Additionally, the presence of enterotoxins and proteases are responsible for the destruction of tight junctions in the intestinal epithelium and brush border enzymes, respectively. Capsular polysaccharides of B. fragiliss showed a positive effect on the host, reducing host inflammation and pathogenesis. B. uniformis isolated from the feces of infants were considered a potential probiotic strain. Administration of this strain improved the lipid profile, reduced glucose-insulin and leptins, and increased phagocytosis under a high-fat diet. Therefore, this strain ameliorated the diet-induced metabolic disorders and immunological dysfunctions in obese mice [299]. Butyrate-producing Eubacterium hallii are natural residents of the gut, demonstrated to lower mucosal inflammation, strengthening the epithelial barrier function, and providing the energy for colonocytes [300].
Moreover, various physiological processes related to energy metabolism and improved insulin sensitivity were observed following oral supply of this specific strain to obese and diabetic mice. Additionally, an increased dosage of E. hallii did not show any negative effect on physiological parameters and food intake, illustrating the strain as a safe and effective therapeutic agent. Various strains belonging to Clostridia clusters were effective in Treg cell differentiation. Atarashi et al. proposed that Clostridia spp. are strong metabolites producers including SCFAs [301]. The produced SCFAs regulate the Foxp3 gene, controlling the Treg-cell development. The author proposed that a cocktail of bacterial strains acted more effectively for treating disease as compared to a single strain. Faecalibacterium prausnitzii belonging to Clostridium cluster IV is one of the most predominant bacteria in human feces (3 to 5%). The absence of F. prausnitzii is commonly associated with gut-associated disorders. However, studies with F. prausnitzii in humans are still lacking; therefore, further investigations are needed to unravel the safety aspect of this bacterium in humans.
Probiotics and Its Safety Aspects
According to the Food and Drug Administration (FDA), probiotics are classified as dietary supplements, therefore having less stringent requirements in terms of their safety, dosage, and efficacy. Most of the probiotic strains fall into the GRAS category [302]. The use of probiotics has been growing exponentially and shown in a wide variety of applications such as the food industry and health care. Probiotics are live microorganisms gaining popularity because of their beneficial effects on human health; however, they may cause minor infections, especially in adults with secondary conditions (e.g., chronic infections, diabetes, immune deficiency) [303]. A total of 24 fungemia cases have been recorded associated with the probiotic S. boulardii [304]. Munoz et al. [305] reported three cases of S. cerevisiae infection, in an ICU dealing with S. boulardii therapy. However, in clinical trials, no reports of bacterial/fungal infection were observed following probiotic use. All cases of fungemia were observed in immuno-compromised, chronic or debilitated patients. Additionally, probiotic bacteria may be able to transfer antibiotic resistance genes to their neighbor pathogenic microorganisms. Several properties critical for probiotic survival should be properly monitored before commercializing a specific strain, as the evidence clearly shows variations in probiotic strains belonging to the same species. Additionally, various other properties like formulation, stability of the organism, colonization in the intestinal epithelium, anti-pathogenic response, immuneactivation, and various other important functionalities need to be carefully monitored. The industrial application requires the stability and survival of the used strains [306].
Limitation and Future Aspects
As probiotics are live organisms, their count in food items must be kept at an optimum level. According to the National Sanitary Surveillance Agency, approximately 108–109 CFU of probiotics in food are regarded as safe [307]. However, its level can be affected by bacteriophages that lead to cellular lysis [308]. Moreover, the accurate effect in the host depends on the ingested strain, its quantity, survival rate, and physiological condition of the host [309]. Moreover, methodologies to detect culturable bacteria require specific synthetic media under defined conditions. The selection of phage-resistant strains can be performed via plaque assay, qPCR, flow cytometry, and biosensors [310, 311]. Selective culture techniques are limited in providing the most holistic definition of viable probiotic bacteria for the dose available in the final product. Reliance on cell-culture techniques does not provide direct cues on microbe-microbe and/or microbe-host interactions, pertaining to the gut environment and is even not consistent in in-vivo trials. Many of the probiotic strains lose their viability during the processing of food products via acidification or in the presence of oxygen in the medium [312, 313]. Viability can be controlled by approaching classical microbiological methods such as growing in a specific cultivation medium [314]. Additionally, previous studies also showed poor colonization efficiency of exogenous probiotics compared to those of humans in the gut [315, 316]. Many quorum sensing disrupting probiotic bacteria have the potential to inhibit the pathogenic microbes; however, their signaling behaviour and complexity may differ in-vivo, when compared to in-vitro experiments. Additionally, manipulation of the quorum-sensing pathway may result in inhibition of commensal bacteria in vivo [317]. Although, the use of probiotics are more often related to the enhancement of beneficial gut microbiota or protecting the host against multiple diseases. However, the exact mechanisms by which probiotics alter the microbiota in in-vivo models are still not properly known.
Nonetheless, many debate that the effect of probiotics might not relate to their interaction with the host microbiome [5]. The systemic review of Kristensen et al. [23] reported the lack of evidence for probiotic-mediated modulation of microbiota in many of the analyzed studies. Similarly, previous studies summarized multiple trials associated with beneficial probiotics of which only 21% showed microbiome alterations [318]. Their establishment for broad function and replication in the host environment is not clear [319]. Further, human clinical studies are required to follow probiotic-mediated cell response to assess possible outcomes and observe the colonization potential of strain-specific probiotics. The identification and characterization of biomarkers linked to probiotic properties could lead to the evolution of novel strains with improved function and health effects [320]. Importantly, in-vivo probiotic effects in animal models do not guarantee effective translation to human subjects and therefore, future research should focus on accurately translating animal trial findings to human therapies. Despite having demonstrated their potential for various clinical and nutritional applications, further studies are required to strengthen the implementation of probiotics for human usage. Future research should be directed towards controlled human studies to determine the strain-specific role of probiotics and which dosages may guarantee effectiveness and safety.
Although the effects of probiotics in health care have been demonstrated in many studies, the molecular approach integrated with proteomics still requires further research efforts to unravel the probiotic-cross talk. Further in-depth studies are still needed to explore probiotics’ applications as therapeutic agents. More clinical information is required to provide the evidence to prevent primary and secondary Clostridium-derived infection (CDI), including the selection and use of bacterial strains that show better results in immunocompromised or critically ill patients [321]. Bacteroides spp. rapidly transfer antibiotic resistance gene among bacteria in the human colon, both within the Bacteroides genus and among Bacteroides species and Gram-positive bacteria, therefore this characteristic has to be taken into account while developing next-generation probiotics. Information regarding the safety profile of Faeccalibacterium and Akkermansia is not sufficient; therefore, further animal and human clinical studies are needed [322]. The development of more advanced technical tools will promote the production and formulation of next-generation probiotics, enabling their use as supplements. Using next-generation probiotics in food is limited to their safety and suitability to the target consumer groups when compared to traditional probiotics [320]. Moreover, recently developed marine probiotics showed their potential in the prevention of ecological damages. An integrated effort would help to develop better formulations that improve viability, safety, and higher efficacy. Moreover, the properties of a probiotic strain are important for the efficacy of the treatment, maintenance of their characteristics, and relative efficacy should be verified. The increasing evidences indicated that manufacturing and production procedures may influence the quality and safety of probiotics [323, 324]; thereby, attention should be given to choosing the specific formulation to be administered. Furthermore, before the use of probiotics for the prevention and treatment of disorders, greater investment in clinical trials is necessary.
Conclusions
Probiotics possess several promising attributes which fulfil our prompt nutritional and clinical necessities. Many probiotic microbes have been found to show positive responses against several diseases. The contribution of probiotics to the treatment of severe diseases such as cancer, obesity, diabetes, and infectious diseases is growing, representing a high demand in the research arena. Technological advancements linked to RNA sequencing, culturomics and metabolomics have propelled the field of probiotics, detailing the interaction with the indigenous microbiome. Presently, probiotic administration of food products presents a cost-effective alternative and fruitful source for the assessment of the effects of newly discovered probiotic strains. Moreover, further developments gathered in clinical trials highlighted the role of specific probiotic strains in terms of their safety (for clinical and food applications), antagonistic activity towards pathogens, and ability to tolerate acid and bile stress. The probiotics-based biotherapy shows enormous potential as a therapeutic agent against neurodegenerative disease; however, extensive clinical trials and characterization of the biochemical effects of probiotics are required to fully elucidate the scope of probiotics against specific diseases. There is still much to explore the connection between probiotics and gut microbiota with neurodegenerative disease, which will open a new possibility for researchers in the largely uncharted territory of neurodegenerative and gut microbiota. The studies on the relationship between gut microbiota and neurometabolites will provide a new concept for the prevention and intervention of neurodegenerative diseases. The development of probiotics demands close interaction between the pharmaceutical industries, research agencies, and regulatory bodies.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1 Probiotic bacteria induce several beneficial traits by production of vitamins, organic acids, short chain fatty acids (SCFAs), neurotransmitters, enzymes, amino acids, and antimicrobial peptides like bacterocins. (PDF 75 KB)
Supplementary Figure 2 Probiotic bacteria can be engineered for their functional improvement by employing genetic engineering approaches like CRISPR plasmid mediated gene insertion/deletion, LPSD plasmid mediated crossover, employing synthetic biological route, and through development of regulatory circuit. (PDF 47 KB)
Acknowledgements
We acknowledge the Department of Bioenginering and Biotechnology, BIT Mesra for providing necessary infrastructure.
Author Contribution
Conceptualization: Rajnish Prakash Singh. Original draft writing: Rajnish Prakash Singh. Editing: Afreen Shadan and Ying Ma. All authors read and approved the final manuscript.
Funding
The work was supported by the Department of Biotechnology, Government of India, Ramalingaswami Re-entry Fellowship (BT/RLF/2020–21) grant.
Declarations
Ethics Approval
Not required.
Conflict of Interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Metchnikoff E. The prolongation of life: optimistic studies. New York: Putnam’s Sons; 1908. [Google Scholar]
- 2.Lilly DM, Stillwell RH. Probiotics: growth promoting factors produced by microorganisms. Science. 1965;147:747–748. doi: 10.1126/science.147.3659.747. [DOI] [PubMed] [Google Scholar]
- 3.Cani PD, Van Hul M. Novel opportunities for next-generation probiotics targeting metabolic syndrome. Curr Opin Biotechnol. 2015;32:21–27. doi: 10.1016/j.copbio.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Chang CS, Kao CY. Current understanding of the gut microbiota shaping mechanisms. J Biomed Sci. 2019;26:59. doi: 10.1186/s12929-019-0554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill C, et al. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 6.Plaza-Diaz J, Robles-Sanchez C, Abadia-Molina F, Saez-Lara MJ, Vilchez-Padial LM, Gil A, Gomez-Llorente C, Fontana L. Gene expression profiling in the intestinal mucosa of obese rats administered probiotic bacteria. Sci Data. 2017;4:170186. doi: 10.1038/sdata.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plaza-Diaz J, Ruiz-Ojeda FJ, Gil-Campos M, Gil A. Immune-mediated mechanisms of action of probiotics and synbiotics in treating pediatric intestinal diseases. Nutrients. 2018;10(1):42. doi: 10.3390/nu10010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Hage R, Hernandez-Sanabria E, Van de Wiele T. Emerging trends in “smart probiotics”: functional consideration for the development of novel health and industrial applications. Front Microbiol. 2017;8:1889. doi: 10.3389/fmicb.2017.01889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins JK. Thornton G, Sullivan GO: Selection of probiotic strains for human application. Int Dairy. 1998;J8:487–490. doi: 10.1016/S0958-6946(98)00073-9. [DOI] [Google Scholar]
- 10.Gueimonde M, Sánchez B. Enhancing probiotic stability in industrial processes. Microb Ecol Health Dis. 2012 doi: 10.3402/mehd.v23i0.18562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siong TE, Hardinsyah and Sook Sum CA, Status of probiotic regulations in Southeast Asia countries. Mal J Nutr. 2021;27(3):507–530. [Google Scholar]
- 12.Timmerman HM, Koning CJM, Mulder L, Rombouts FM, Beynen AC. Mono strain, multi strain and multispecies probiotics—a comparison of functionality and efficacy. Int J Food Microbiol. 2004;96:219–233. doi: 10.1016/j.ijfoodmicro.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Chapman C, Gibson G, Rowland I. Health benefits of probiotics: are mixtures more effective than single strains? Eur J Nutr. 2011;50:1–17. doi: 10.1007/s00394-010-0166-z. [DOI] [PubMed] [Google Scholar]
- 14.Asemi Z, Zare Z, Shakeri H, Sabihi SS, Esmaillzadeh A. Effect of multispecies probiotic supplements on metabolic profiles, Hs-CRP, and oxidative stress in patients with Type 2 diabetes. Ann Nutr Metab. 2013;6:1–9. doi: 10.1159/000349922. [DOI] [PubMed] [Google Scholar]
- 15.Besseling-van der Vaart I, Heath MD, Guagnini F, Kramer MF. In vitro evidence for efficacy in food intolerance for the multispecies probiotic formulation Ecologic R Tolerance (Syngut TM) Benef Microbes. 2016;7:111–118. doi: 10.3920/BM2015.0051. [DOI] [PubMed] [Google Scholar]
- 16.Lye HS, Kuan CY, Ewe JA, et al. The improvement of hypertension by probiotics: effects on cholesterol, diabetes, renin, and phytoestrogens. Int J Mol Sci. 2009;10:3755–3775. doi: 10.3390/ijms10093755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moayyedi P, Ford AC, Talley NJ, et al. The efficacy of probiotics in the treatment of irritable bowel syndrome: a systematic review. Gut. 2010;59:325–332. doi: 10.1136/gut.2008.167270. [DOI] [PubMed] [Google Scholar]
- 18.Golowczyc MA, Mobili P, Garrote GL, et al. Protective action of Lactobacillus kefir carryingS-layer protein against Salmonella enteric serovar enteritidis. Int J Food Microbiol. 2007;118:264–273. doi: 10.1016/j.ijfoodmicro.2007.07.042. [DOI] [PubMed] [Google Scholar]
- 19.Woodard GA, Encarnacion B, Downey JR, et al. Probiotics improve outcomes after Rouxen-Y gastric bypass surgery: a prospective randomized trial. J Gastrointest Surg. 2009;13:1198–1204. doi: 10.1007/s11605-009-0891-x. [DOI] [PubMed] [Google Scholar]
- 20.Rather IA, Bajpai VK, Kumar S, Lim J, Paek WK, Park YH. Probiotics and atopic dermatitis: an overview. Front Microbiol. 2016;7:507. doi: 10.3389/fmicb.2016.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karska-Wysocki B, Bazo M, Smoragiewicz W. Antibacterial activity of Lactobacillus acidophilus and Lactobacillus case against methicillin-resistant Staphylococcus aureus (MRSA) Microbiol Res. 2010;165:674–686. doi: 10.1016/j.micres.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Rafter J, Bennett M, Caderni G, et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am J Clin Nutr. 2007;85:488–496. doi: 10.1093/ajcn/85.2.488. [DOI] [PubMed] [Google Scholar]
- 23.Kristensen NB, Bryrup T, Allin KH, Nielsen T, Hansen TH, Pedersen O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 2016;8:1–11. doi: 10.1186/s13073-016-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patrice DC. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conlon M, Bird A. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2015;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langella P, Martín R. Emerging health concepts in the probiotics field: streamlining the definitions. Front Microbiol. 2019;10:1047. doi: 10.3389/fmicb.2019.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurtz CB, Millet YA, Puurunen MK, Perreault M, Charbonneau MR, Isabella VM, Kotula JW, Antipov E, Dagon Y, Denney WS (2019) An engineered E.coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci Transl Med 117975 [DOI] [PubMed]
- 28.Certain LK, Way JC, Pezone MJ, Collins JJ. Using engineered bacteria to characterize infection dynamics and antibiotic effects in vivo. Cell Host Microbe. 2017;22:263–268. doi: 10.1016/j.chom.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557(7705):434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santiago-Rodriguez TM, Hollister EB. Human virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses. 2019;11:7. doi: 10.3390/v11070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan FF, Liu JH, March JC. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes. 2015;641:794–803. doi: 10.2337/db14-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludwig IS, Broere F, Manurung S, Lambers TT, Vander Zee R, Eden WV. Lactobacillus rhamnosus GG-derived soluble mediators modulate adaptive immune cells. Front Immunol. 2018 doi: 10.3389/fimmu.2018.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Botes M, Loos B, Van Reenen CA, Dicks LM. Adhesion of the probiotic strains Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 to Caco-2 cells under conditions simulating the intestinal tract, and in the presence of antibiotics and anti-inflammatory medicaments. Arch Microbiol. 2008;190:573–584. doi: 10.1007/s00203-008-0408-0. [DOI] [PubMed] [Google Scholar]
- 35.Gharbi Y, FhoulaI R-M, Afef N, Boudabous A, Gueimonde M, Ouzari HI. In-vitro characterization of potentially probiotic Lactobacillus strains isolated from human microbiota: interaction with pathogenic bacteria and the enteric cell line HT29. Annals Microbiol. 2019;69:61–72. doi: 10.1007/s13213-018-1396-1. [DOI] [Google Scholar]
- 36.Benjamin C, Hofeld BC, Puppala VK, Tyagi S, Ahn KW, Anger A, Jia S, Salzman NH, Hessner MJ, Widlansky ME. Lactobacillus plantarum 299v probiotic supplementation in men with stable coronary artery disease suppresses systemic inflammation. Sci Rep. 2021;11:3972. doi: 10.1038/s41598-021-83252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, Fernández-Caballero JA, Garcia F, Rodríguez-Cabezas ME, Galvez J. The Administration of Escherichia coli Nissle 1917 ameliorates development of DSS-induced colitis in Mice. Front Pharmacol. 2018 doi: 10.3389/fphar.2018.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Balawi M, Morsy FM. Enterococcus faecalis is a better competitor than other lactic acid bacteria in the initial colonization of colon of healthy new-born babies at first week of their life. Front Microbiol. 2020 doi: 10.3389/fmicb.2020.02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hawrelak J. Probiotics: choosing the right one for your needs. J Aust Traditional-Med Soc. 2003;9(2):67–75. [Google Scholar]
- 40.Giralt J, Regadera JP, Verges R, Romero J, de la Fuente I, Biete A, Villoria J, Cobo JM, Guarner F. Effects of probiotic Lactobacillus casei DN-114 001 in prevention of radiation-induced diarrhea: results from multicentre, randomized, placebo-controlled nutritional trial. Int J Radiat Oncol Biol Phys. 2008;71:1213–1219. doi: 10.1016/j.ijrobp.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Marras L, Caputo M, Bisicchia S, Soato M, Bertolino G, Vaccaro S, Inturr R. The role of Bifidobacteria in predictive and preventive medicine: a focus on eczema and hypercholesterolemia. Microorganisms. 2021;9:836. doi: 10.3390/microorganisms9040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro-López V, Ramírez-Boscá A, Ramón-Vidal D, Ruzafa-Costas B, Genovés-Martínez S, Chenoll-Cuadros E, Carrión-Gutiérrez M, Parte JH, Prieto-Merino D, Codoñer-Cortés FM. Effect of oral administration of a mixture of probiotic strains on SCORAD index and use of topical steroids in young patients with moderate atopic dermatitis A randomized clinical trial. JAMA Dermatol. 2018;154(1):37–43. doi: 10.1001/jamadermatol.2017.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sestito S, D’Auria E, Baldassarre ME, Salvatore S, Tallarico V, Stefanelli E, Tarsitano F, Concolino D, Pensabene L. The role of prebiotics and probiotics in prevention of allergic diseases in infants. Front Pediatr. 2020;22(8):583946. doi: 10.3389/fped.2020.583946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hill D, Sugrue I, Tobin C, Hill C, Stanton C, Ross RP. The Lactobacillus casei group: history and health-related applications. Front Microbiol. 2018;9:2107. doi: 10.3389/fmicb.2018.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miremadi F, Sherkat F, Stojanovska L. Hypocholesterolaemic effect and anti-hypertensive properties of probiotics and prebiotics: A review. J Fun Foods. 2016;25:497–510. doi: 10.1016/j.jff.2016.06.016. [DOI] [Google Scholar]
- 46.An M, Park YH, Lim YH. Ant obesity and antidiabetic effects of the dairy bacterium Propionibacterium freudenreichii MJ2 in high-fat diet-induced obese mice by modulating lipid metabolism. Sci Rep. 2021;11:2481. doi: 10.1038/s41598-021-82282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schreck BA, Gregory PJ, Jalloh MA, Risoldi CZ, Hein DJ. Probiotics for the treatment infantile colic: a systematic review. J Pharm Pract. 2017;30:366–374. doi: 10.1177/0897190016634516. [DOI] [PubMed] [Google Scholar]
- 48.Teixeira LD, Kling DN, Lorca GL, Gonzalez CF. Lactobacillus johnsonii N6.2 diminishes caspase-1 maturation in the gastrointestinal system of diabetes prone rats. Benef Microbes. 2018;9:527–539. doi: 10.3920/BM2017.0120. [DOI] [PubMed] [Google Scholar]
- 49.Brenner DM, Chey WD. Bifidobacterium infantis 35624: a novel probiotic for the treatment of irritable bowel syndrome. Rev Gastroenterol Disord. 2009;9:7–15. [PubMed] [Google Scholar]
- 50.Guglielmetti S, Mora D, Gschwender M, PoppK, Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life–a double-blind, placebo-controlled study. Alim Pharmacol Therapeu. 2011;33:1123–1132. doi: 10.1111/j.1365-2036.2011.04633.x. [DOI] [PubMed] [Google Scholar]
- 51.Enck P, Zimmermann K, Menke G, Klosterhalfen S. Randomized controlled treatment trial of irritable bowel syndrome with a probiotic E. coli preparation (DSM17252) compared to placebo. J Gastroenterol. 2009;47:209–214. doi: 10.1055/s-2008-1027702. [DOI] [PubMed] [Google Scholar]
- 52.Cayzeele-Decherf A, Pelerin F, Leuillet S, et al. Saccharomyces cerevisiae CNCM I-3856 in irritable bowel syndrome: an individual subject meta-analysis. World J Gastroenterol. 2017;23:336–344. doi: 10.3748/wjg.v23.i2.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masoumi SJ, Mehrabani D, Saberifiroozi M, Fattahi MR, MoradiF NM. The effect of yogurt fortified with Lactobacillus acidophilus and Bifidobacterium sp. probiotic in patients with lactose intolerance. Food Sci Nutr. 2020;9:1704–1711. doi: 10.1002/fsn3.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pineton de Chambrun G, Neut C, Chau A, Cazaubiel M, Pelerin F, Justen P, Pierre Desreumaux P. A randomized clinical trial of Saccharomyces cerevisiae versus placebo in the irritable bowel syndrome. Dig Liver Dis. 2015;47(2):119–124. doi: 10.1016/j.dld.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Khoder G, Al-Menhali AA, Al-Yassir F, Karam SM. Potential role of probiotics in the management of gastric ulcer. Exp Ther Med. 2016;12(1):3–17. doi: 10.3892/etm.2016.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anukam KC, Hayes K, Summers K, Reid G. Probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14may help downregulate TNF-Alpha, IL-6, IL-8, IL-10 and IL-12(p70) in the neurogenic bladder of spinal cord injured patient with urinary tract infections: a two-case study. Adv Urol. 2009;2009:680363. doi: 10.1155/2009/680363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imaoka A, Shima T, Kato K, Mizuno S, Uehara T, Matsumoto S, Setoyama H, Hara T, Umesaki Y. Anti-inflammatory activity of probiotic Bifidobacterium: enhancement of IL-10 production in peripheral blood mononuclear cells from ulcerative colitis patients and inhibition ofIL-8 secretion in HT-29 cells. World J Gastroenterol. 2008;14:2511–2516. doi: 10.3748/wjg.14.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srutkova D, Schwarzer M, Hudcovic T, Zakostelska Z, Drab V, Spanova A, Rittich B, Kozakova H, Schabussova I. Bifidobacterium longum CCM 7952 promotes epithelial barrier function and prevents acute DSS-induced colitis in strictly strain-specific manner. PLoS ONE. 2015 doi: 10.1371/journal.pone.0134050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oerlemans EFM, Bellen G, Ingmar Claes I, Henkens T, Allonsius CN, Wittouck S, Van den Broek MFL, Wuyts S, Kiekens F, Gilbert GG, Donders GGG, Sarah Lebeer S. Impact of a Lactobacilli-containing gel on vulvovaginal candidiasis and the vaginal microbiome. Sci Rep. 2020;10:7976. doi: 10.1038/s41598-020-64705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saxami G, Karapetsas A, Lamprianidou E, Kotsianidis I, Chlichlia A, Tassou C, Zoumpourliset V, Galanis A. Two potential probiotic lactobacillus strains isolated from olive microbiota exhibit adhesion and anti-proliferative effects in cancer cell lines. J Funct Foods. 2016;24:461–471. doi: 10.1016/j.jff.2016.04.036. [DOI] [Google Scholar]
- 61.Tiptiri-Kourpeti A, Spyridopoulou K, Santarmaki V, Aindelis G, Tompoulidou E, Lampranidou E, Saxami G, Ypsilantis P, Lampri E, Simopoulos C, et al. Lactobacillus casei exerts anti-proliferative effects accompanied by apoptotic cell death and up-regulation of TRAIL in colon carcinoma cells. PLoS ONE. 2016;11:e0147960. doi: 10.1371/journal.pone.0147960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee NK, Son SH, Jeon EB, Jung GH, Lee JY, Paik HD. The prophylactic effect of probiotic Bacillus polyfermenticus KU3 against cancer cells. J Funct Foods. 2015;14:513–518. doi: 10.1016/j.jff.2015.02.019. [DOI] [Google Scholar]
- 63.Thirabunyanon M, Boonprasom P, Niamsup P. Probiotic potential of lactic acid bacteria isolated from fermented dairy milks on antiproliferation of colon cancer cells. Biotechnol Lett. 2009;31:571–576. doi: 10.1007/s10529-008-9902-3. [DOI] [PubMed] [Google Scholar]
- 64.Lee JW, Shin JG, Kim EH, Kang HE, Yim IB, Kim JY, Joo HG, Woo HJ. Immunomodulatory and antitumor effects in vivo by the cytoplasmic fraction of Lactobacillus casei and Bifidobacterium longum. J Vet Sci. 2004;5:41–48. doi: 10.4142/jvs.2004.5.1.41. [DOI] [PubMed] [Google Scholar]
- 65.Rasouli BS, Ghadimi-Darsajini A, Nekouian R, Iragian GR. In vitro activity of probiotic Lactobacillus reuteri against gastric cancer progression by downregulation of urokinase plasminogen activator/urokinase plasminogen activator receptor gene expression. J Cancer Res Ther. 2017;13:246–251. doi: 10.4103/0973-1482.204897. [DOI] [PubMed] [Google Scholar]
- 66.Ghoneum M, Felo N. Selective induction of apoptosis in human gastric cancer cells by Lactobacillus kefiri (PFT), a novel kefir product. Oncol Rep. 2015;34:1659–1666. doi: 10.3892/or.2015.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohigashi S, Hoshino Y, Ohde S, Onodera H. Functional outcome, quality of life, and efficacy of probiotics in postoperative patients with colorectal cancer. Surg Today. 2011;41:1200–1206. doi: 10.1007/s00595-010-4450-6. [DOI] [PubMed] [Google Scholar]
- 68.El-Nezami HS, Polychronaki NN, Ma J, Zhu H, Ling W, Salminen EK, Juvonen RO, Salminen SJ, Poussa T, Mykkänen HM. Probiotic supplementation reduces a biomarker for increased risk of liver cancer in young men from Southern China. Am J Clin Nutr. 2006;83:1199–1203. doi: 10.1093/ajcn/83.5.1199. [DOI] [PubMed] [Google Scholar]
- 69.Pala V, Sieri S, Berrino F, Vineis P, Sacerdote C, Palli D, Masala G, Panico S, Mattiello A, Tumino R. Yogurt consumption and risk of colorectal cancer in the Italian European prospective investigation into cancer and nutrition cohort. Int J Cancer. 2011;129:2712–2719. doi: 10.1002/ijc.26193. [DOI] [PubMed] [Google Scholar]
- 70.Ohashi Y, Nakai S, Tsukamoto T. Habitual intake of lactic acid bacteria and risk reduction of bladder cancer. Urol Int. 2002;68:273–280. doi: 10.1159/000058450. [DOI] [PubMed] [Google Scholar]
- 71.Ma EL, Choi YJ, Choi J, Pothoulakis C, Rhee SH, Imet E. The anticancer effect of probiotic Bacillus polyfermenticus on human colon cancer cells is mediated through ErbB2 and ErbB3 inhibition. Int J Cancer. 2010;127:780–790. doi: 10.1002/ijc.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas CM, Versalovic J. Probiotics–host communication: modulation of signalling pathways in the intestine. Gut Microbes. 2010;1:148–163. doi: 10.4161/gmic.1.3.11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fukushima Y, Kawata Y, Hara H, Terada A, Mitsuoka T. Effect of a probiotic formula on intestinal immunoglobulin A production in healthy children. Int J Food Microbiol. 1998;42:39–44. doi: 10.1016/S0168-1605(98)00056-7. [DOI] [PubMed] [Google Scholar]
- 74.Galdeano CM, Perdigón G. The probiotic bacterium Lactobacillus case induces activation of the gut mucosal immune system through innate immunity. Clin Vaccine Immunol. 2006;13:219–226. doi: 10.1128/CVI.13.2.219-226.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kobyliak N, Conte C, Cammarota G, Haley AP, Styriak I, Gaspar L, et al. Probiotics in prevention and treatment of obesity: a critical view. Nutr Metab. 2016;13:1–13. doi: 10.1186/s12986-016-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nguyen HT, Truong DH, Kouhounde S, Ly S, Raza findralambo H, Delvigne F, Biochemical engineering approaches for increasing viability and functionality of probiotic bacteria. Int J Mol Sci. 2016;17:1–18. doi: 10.3390/ijms17060867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Westermann C, Gleinser M, Corr SC, Riedel CU. A critical evaluation of Bifidobacterial adhesion to the host tissue. Front Microbiol. 2016;7:1–8. doi: 10.3389/fmicb.2016.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Onyenweaku F, Obeagu EI, Ifediora AC, Nwandikor UU. Health benefits of probiotics. Int J InnovAppl Res. 2016;4:21–30. [Google Scholar]
- 79.Dixit Y, Wagle A, Vakil B. Patents in the field of probiotics, prebiotics, synbiotics: a review. J Food Microbiol Saf Hygiene. 2016;1:1–13. [Google Scholar]
- 80.Chen CY, Tsen HY, Lin CL, Yu B, Chen CS. Oral administration of a combination of select lactic acid bacteria strains to reduce the Salmonella invasion and inflammation of broiler chicks. Poult Sci. 2012;91:2139–2147. doi: 10.3382/ps.2012-02237. [DOI] [PubMed] [Google Scholar]
- 81.Chen X, Yang G, Song JH, Xu H, Li D, Goldsmith J, et al. Probiotic yeast inhibits VEGFR signalling and angiogenesis in intestinal inflammation. PLoS ONE. 2013;8:1–7. doi: 10.1371/journal.pone.0064227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arora T, Singh S, Sharma RK. Probiotics: interaction with gut microbiome and anti obesity potential. Nutrition. 2013;29:591–596. doi: 10.1016/j.nut.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 83.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 84.Corr SC, Li Y, et al. Bacteriocin production as a mechanism for the anti-infective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Medellin-Peña MJ, Wang H, Johnson R, Anand S, Griffiths MW. Probiotics affect virulence-related gene expression in Escherichia coliO157:H7. Appl Environ Microbiol. 2007;73:4259–4267. doi: 10.1128/AEM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim Y, Kim SH, Whang KY, Kim YJ, Oh S. Inhibition of Escherichia coli O157:H7 attachment by interactions between lactic acid bacteria and intestinal epithelial cells. J Microbiol Biotechnol. 2008;18:1278–1285. [PubMed] [Google Scholar]
- 87.Li J, Wang W, Xu SX, Magarvey NA, McCormick JK. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc Natl Acad Sci USA. 2011;108:3360–3365. doi: 10.1073/pnas.1017431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohland CL, Macnaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol. 2010;298:G807–G819. doi: 10.1152/ajpgi.00243.2009. [DOI] [PubMed] [Google Scholar]
- 89.Miyamoto J, et al. A gut microbial metabolite of linoleic acid, 10-hydroxycis-12-octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40–MEK–ERK pathway. J Biol Chem. 2015;290:2902–2918. doi: 10.1074/jbc.M114.610733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamada M, et al. A bacterial metabolite ameliorates periodontal pathogen-induced gingival epithelial barrier disruption via GPR40 signaling. Sci Rep. 2018;8:9008. doi: 10.1038/s41598-018-27408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Joyce SA, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci USA. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sarkar A, et al. Psychobiotics and the manipulation of bacteria–gut–brain signals. Trends Neurosci. 2016;39:763–781. doi: 10.1016/j.tins.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bravo JA, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buffington SA, et al. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aires J, Butel MJ. Proteomics, human gut microbiota and probiotics. Expert Rev Prot. 2011;8:279–288. doi: 10.1586/epr.11.5. [DOI] [PubMed] [Google Scholar]
- 96.Whitehead K, Versalovic J, Roos S, Britton RA. Genomic and genetic characterization of the bile stress response of probiotic Lactobacillus reuteri ATCC 55730. Appl Environ Microbiol. 2008;74:1812–1819. doi: 10.1128/AEM.02259-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ruiz L, Ruas-Madiedo P, Gueimonde M, de Los Reyes-Gavila´n CG, Margolles A, Sa´ nchez B, How do bifidobacteriacounteract environmental challenges? Mechanisms involved and physiological consequences. Genes Nutr. 2011;6:307–318. doi: 10.1007/s12263-010-0207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koskenniemi K, Laakso K, Koponen J, Kankainen M, Greco D, Auvinen P, SavijokiK NTA, Surakka A, Salusjarvi T, et al. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol Cell Proteomics. 2011;10(M110):002741. doi: 10.1074/mcp.M110.002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sanchez B, Champomier-Verge MC, Stuer-Lauridsen B, Ruas-Madiedo P, Anglade P, Baraige F, de los Reyes-Gavila CG, Johansen E, Zagorec M, Margolles A, Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: a proteomic and physiological approach. Appl Environ Microbiol. 2007;73:6757–6767. doi: 10.1128/AEM.00637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hamon E, Horvatovich P, Izquierdo E, BringelF ME, Aoude-Werner D, Ennahar S. Comparative proteomic analysis of Lactobacillus plantarum for the identification of key proteins in bile tolerance. BMC Microbiol. 2011;11:63. doi: 10.1186/1471-2180-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beck HC, Feddersen S, Petersen J (2011) Application of probiotic proteomics in enteric cytoprotection. In: Probiotic Bacteriaand Enteric Infections. MalagoJF, Koniks JG, Marinsek-Logov R (Eds.) Springer Science + Business Media B.V Dodrecht The Netherlands pp 155–168
- 102.Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 103.Izquierdo E, Horvatovich P, Marchioni E, Aoude-Werner D, Sanz Y, Ennahar S. 2-DE and MS analysis of key proteins in the adhesion of Lactobacillus plantarum, a first step toward early selection of probiotics based on bacterial biomarkers. Electrophoresis. 2009;30:949–956. doi: 10.1002/elps.200800399. [DOI] [PubMed] [Google Scholar]
- 104.Ashida N, Yanagihara S, Shinoda T, Yamamoto N. Characterization of adhesive molecule with affinity to Caco-2cells in Lactobacillus acidophilus by proteome analysis. J Biosci Bioeng. 2011;112:333–337. doi: 10.1016/j.jbiosc.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 105.Beganovic J, Frece J, Kos B, Lebos Pavunc A, Habjanic K, Suskovic J. Functionality of the S-layer protein from the probiotic strain Lactobacillus helveticus M92. Antonie Van Leeuwenhoek. 2011;100:43–53. doi: 10.1007/s10482-011-9563-4. [DOI] [PubMed] [Google Scholar]
- 106.Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, VanKooyk Y. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA. 2008;105:19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Candela M, Centanni M, Fiori J, Biagi E, Turroni S, Orrico C, Bergmann S, Hammerschmidt S, Brigidi P. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology. 2010;156:1609–1618. doi: 10.1099/mic.0.038307-0. [DOI] [PubMed] [Google Scholar]
- 108.Ruiz L, CouteY SB, de los Reyes-Gavila CG, Sanchez JC, Margolles A, The cell-envelope proteome of bifidobacterium longum in an in vitro bile environment. Microbiology. 2009;155:957–967. doi: 10.1099/mic.0.024273-0. [DOI] [PubMed] [Google Scholar]
- 109.Lee J, Choe J, Kim J, Oh S, Park S, Kim S, et al. Heat-killed Lactobacillus spp. cells enhance survivals of Caenorhabditis elegans against Salmonella and Yersinia infections. Lett Appl Microbiol. 2015;61:523–530. doi: 10.1111/lam.12478. [DOI] [PubMed] [Google Scholar]
- 110.Sharma K, Pooranachithra M, Balamurugan K, Goel G. Probiotic mediated colonization resistance against E.coli infection in experimentally challenged Caenorhabditis elegans. Microb Pathog. 2019;127:39–47. doi: 10.1016/j.micpath.2018.11.041. [DOI] [PubMed] [Google Scholar]
- 111.Poupet C, Saraoui T, Veisseire P, Bonnet M, Dausset C, Gachinat M, et al. Lactobacillus rhamnosus Lcr35 as an effective treatment for preventing Candida albicans infection in the invertebrate model Caenorhabditis elegans: first mechanistic insights. PLoS ONE. 2019;14:e0216184. doi: 10.1371/journal.pone.0216184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang S, Ahmadi S, Nagpal R, Jain S, Mishra SP, Kavanagh K, et al. Lipoteichoic acid from the cell wall of a heat killed Lactobacillus paracasei D3–5 ameliorates aging-related leaky gut, inflammation and improves physical and cognitive functions: from C. elegans to mice. Geroscience. 2019;42:333–524. doi: 10.1007/s11357-019-00137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mooijaart SP, Brandt BW, Baldal EA, Pijpe J, Kuningas M, Beekman M, et al. C. elegans DAF-12, nuclear hormone receptors and human longevity and disease at old age. Ageing Res Rev. 2005;4:351–371. doi: 10.1016/j.arr.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 114.Sun S, Mizuno Y, Komura T, Nishikawa Y, Kage-Nakadai E. Toll-like receptor homolog TOL-1 regulates Bifidobacterium infantis-elicited longevity and behavior in Caenorhabditis elegans. Biosci Microbiota Food Heal. 2019;38:105–110. doi: 10.12938/bmfh.18-031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kodali VP, Lingala VK, Karlapudi AP, Indira M, Venkateswarulu TC, John Babu D. Biosynthesis and potential application of bacteriocins. J Pure Appl Microbiol. 2013;7:2933–2945. [Google Scholar]
- 116.Rossi M, Amaretti A, Raimondi S. Folate Production by probiotic bacteria. Nutrients. 2011;3:118–134. doi: 10.3390/nu3010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Patel A, Shah N, Prajapati JB. Biosynthesis of vitamins and enzymes in fermented foods by lactic acid bacteria and related genera: a promising approach. Croatian J Food Sci Technol. 2013;5:85–91. [Google Scholar]
- 118.Dai Z, Wu Z, Hang S, Zhu W, Wu G. Amino acid metabolism in intestinal bacteria and its potential implications for mammalian reproduction. MHR Basic Sci Reprod Med. 2015;21:389–409. doi: 10.1093/molehr/gav003. [DOI] [PubMed] [Google Scholar]
- 119.Indira M, Venkateswarulu TC, Peele KA, Prabhakar KV, Krupanidhi S. Characterization of bacteriocin producing probiotic properties of Enterococcus casseliflavus MI001 isolated from curd sample. Curr Trends Biotechnol Pharm. 2019;13:64–71. [Google Scholar]
- 120.Geirnaert A, Calatayud M, Grootaert C, Laukens D, Devriese S, Smagghe G, De Vos M, Boon N, Van de Wiele T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci Rep. 2017;7:11450. doi: 10.1038/s41598-017-11734-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensaland probiotic bacteria. Microb Cell Fact. 2017;16:79. doi: 10.1186/s12934-017-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gu Q, Li P. Biosynthesis of vitamins by probiotic bacteria, probioticsand prebiotics in human nutrition and health. London: In tech open; 2016. pp. 135–148. [Google Scholar]
- 123.Padmavathi T, Bhargavi R, Priyanka PR, Niranjan NR, Pavitra PV. Screening of potential probiotic lactic acid bacteria and production of amylase and its partial purification. J Genet Eng Biotechnol. 2018;16:357–362. doi: 10.1016/j.jgeb.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Anwar MA, Kralj S, Pique AV, Leemhuis H, van der Maarel MJ, Dijkhuizen L. Inulin and levan synthesis by probiotic Lactobacillus gasseri strains: characterization of three novel fructansucrase enzymes and their fructan products. Microbiology. 2010;156(4):1264–1274. doi: 10.1099/mic.0.036616-0. [DOI] [PubMed] [Google Scholar]
- 125.Wang Y, Wu Y, Wang Y, Xu H, Mei X, Yu D, Wang Y, Li W. Antioxidant properties of probiotic bacteria. Nutrients. 2017;9:521. doi: 10.3390/nu9050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cardenas N, Laino JE, Delgado S, Jimenez E, Del Valle MJ, De Giori GS, Sesma F, Mayo B, Fernandez L, LeBlanc JG, Rodriguez JM. Relationships between the genome and some phenotypical properties of Lactobacillus fermentum CECT 5716, a probiotics train isolated from human milk. Appl Microbiol Biotechnol. 2015;99:4343–4353. doi: 10.1007/s00253-015-6429-0. [DOI] [PubMed] [Google Scholar]
- 127.Santos F, Wegkamp A, de Vos WM, Smid EJ, Hugenholtz J. High-level folate production in fermented foods by the B12 producer Lactobacillus reuteri JCM1112. Appl Environ Microbiol. 2008;74:3291–3294. doi: 10.1128/AEM.02719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vorobjeva LI, Khodjaev EY, Vorobjeva NV. Propionic acid bacteria as probiotics. Microb Ecol Health Dis. 2008;20:109–112. [Google Scholar]
- 129.Indira M, Venkateswarulu TC, Prabhakar KV, Peele KA, Krupanidhi S. Isolation and characterization of bacteriocin producing Enterococcus casseliflavus and its antagonistic effect on Pseudomonas aeruginosa. Karbala Int J Mod Sci. 2018;4:361–368. doi: 10.1016/j.kijoms.2018.09.002. [DOI] [Google Scholar]
- 130.Caly DL, Chevalier M, Flahaut C, Cudennec B, Al Atya AK, Chataigné G, et al. The safe enterocin DD14 is a leaderless two-peptide bacteriocin with anti-Clostridium perfringens activity. Int J Antimicrob Agents. 2017;49:282–289. doi: 10.1016/j.ijantimicag.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 131.Seddik HA, Bendali F, Gancel F, Fliss I, Spano G, Drider D. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob Proteins. 2017;9:111–122. doi: 10.1007/s12602-017-9264-z. [DOI] [PubMed] [Google Scholar]
- 132.Naghmouchi K, Belguesmia Y, Baah J, Teather R, Drider D. Antibacterial activity of class I and IIa bacteriocins combined with polymyxin E against resistant variants of Listeria monocytogenes and Escherichia coli. Res Microbiol. 2011;162:99–107. doi: 10.1016/j.resmic.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 133.Hammami R, Fernandez B, Lacroix C, Fliss I. Anti-infective properties of bacteriocins: an update. Cell Mol Life Sci. 2013;70:2947–2967. doi: 10.1007/s00018-012-1202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Meade E, Slattery MA, Garvey M. Bacteriocins, potent antimicrobial peptides and the fight against multi-drug resistant species: resistance is futile? Antibiotics. 2020;9(1):32. doi: 10.3390/antibiotics9010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hammami R, Cotter PD, Rebuffat S, Said LS, Gaudreau H, Bédard F, Biron E, Drider D, Fliss I. Bacteriocins as a new generation of antimicrobials: toxicity aspects and regulations. FEMS Microbiol Rev. 2021;45:1–24. doi: 10.1093/femsre/fuaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Surendran Nair M, Amalaradjou MA, Venkitanarayanan K. Anti virulence properties of probiotics in combating microbial pathogenesis. New York, NY: Elsevier Ltd.; 2017. [DOI] [PubMed] [Google Scholar]
- 137.Singh VP. Recent approaches in food bio-preservation-a review. Open Vet J. 2018;8:104–111. doi: 10.4314/ovj.v8i1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mitchell A, Chang HYY, DaughertyL FM, Hunter S, Lopez R, et al. The Inter Pro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Helal MM, Hashem AM, Ghobashy MOI, Shalaby ASG. Some physiological and biological studies on reuterin production from Lactobacillus reuteri. J Probiotics Health. 2016;4:1000156. doi: 10.4172/2329-8901.1000156. [DOI] [Google Scholar]
- 140.Indira M, Venkateswarulu TC, Chakravarthy K, Reddy AR, Babu DJ, Kodali VP. Morphological and biochemical characterization of exopolysaccharide producing bacteria isolated from dairy effluent. J Pharm Sci Res. 2016;8:88–91. [Google Scholar]
- 141.Ates O. Systems biology of microbial exopolysaccharides production. Front Bioeng Biotech. 2015;3:200. doi: 10.3389/fbioe.2015.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Patel S, Goyal A. The current trends and future perspectives of prebiotics research a review. 3 Biotech. 2012;2:115–125. doi: 10.1007/s13205-012-0044-x. [DOI] [Google Scholar]
- 143.Nakamura Y, Nosaka S, Suzuki M, Nagafuchi S, Takahashi T, Yajima T, Takenouchi-ohkubo N, Iwase T, Moro I. Dietary fructooligosaccharides up-regulate immunoglobulin A response and polymeric immunoglobulin receptor expression in intestines of infant mice. Clin Exp Immunol. 2004;137:52–58. doi: 10.1111/j.1365-2249.2004.02487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hill D, Sugrue I, Tobin C, Hill C, Stanton C, Ross RP. The Lactobacillus casei group: history and health related applications. Front Microbiol. 2018;9:2107. doi: 10.3389/fmicb.2018.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Britton R (2017) In: Lactobacillus reuteri, In: The microbiota in gastrointestinal pathophysiology: implications for human health, prebiotics, probiotics, and dysbiosis. Academic Press, pp. 89–97
- 147.Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiel I, Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57:1–24. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ravisankar P, Reddy AA, Nagalakshmi B, Koushik OS, Kumar BV, Anvith PS. The comprehensive review on fat soluble vitamins. IOSR J Pharm. 2015;5(11):12–28. [Google Scholar]
- 149.Lin Z, Xu Z, Li Y, Wang Z, Chen T, Zhao X. Metabolic engineering of Escherichia coli for the production of riboflavin. Microb Cell Fact. 2014;13:104. doi: 10.1186/s12934-014-0104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Markowiak P, Slizewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients. 2017;9:1021. doi: 10.3390/nu9091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pylkas AM, Juneja LR, Slavin JL. Comparison of different fibres for in vitro production of short chain fatty acids by intestinal microflora. J Med Food. 2005;8(1):113–116. doi: 10.1089/jmf.2005.8.113. [DOI] [PubMed] [Google Scholar]
- 152.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11:577–591. doi: 10.1038/nrendo.2015.128. [DOI] [PubMed] [Google Scholar]
- 153.Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome–brain–cell axis to promote metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gill PA, VanZelm MC, Muir JG, Gibson PR. Short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Thera. 2018;48:15–34. doi: 10.1111/apt.14689. [DOI] [PubMed] [Google Scholar]
- 155.Moss JW, Williams JO, Ramji DP. Nutraceuticals as therapeutic agents for atherosclerosis. Biochimica et Biophysica Acta—Mol. Basis Dis. 2018;1864:1562–1572. doi: 10.1016/j.bbadis.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, B€ackhed F, Mithieux G, Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 157.Smit G, Smit BA, Engels WJ. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol Rev. 2005;29(3):591–610. doi: 10.1016/j.fmrre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 158.GhanKA EEA, Hamouda RA. Evaluation of antioxidant and antitumor activities of Lactobacillus acidophilus bacteria isolated from Egyptian infants. Int J Pharmacol. 2014;10:282–288. doi: 10.3923/ijp.2014.282.288. [DOI] [Google Scholar]
- 159.Sun Z, Harris HMB, McCann A, Guo C, Argimón S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea, MC, O’Sullivan O, Ritari J, Douillard FP, Paul Ross R, Yang R, Briner AE, Felis, GE, de Vos WM, Barrangou, R, Klaenhammer, TR, Caufield PW, Cui Y, Zhang H, O’Toole PW (2015) Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6:8322 [DOI] [PMC free article] [PubMed]
- 160.Savaiano DA. Lactose digestion from yogurt: mechanism and relevance. Am J Clin Nutr. 2014;99:1251S–1255S. doi: 10.3945/ajcn.113.073023. [DOI] [PubMed] [Google Scholar]
- 161.Kim DH, Jin YH. Intestinal bacterial beta-glucuronidase activity of patients with colon cancer. Arch Pharm Res. 2001;24:564–567. doi: 10.1007/BF02975166. [DOI] [PubMed] [Google Scholar]
- 162.Sanchez B, Champomier-Verge MC, Anglade P, Baraige F, deLos Reyes-Gavila CG, Margolles A, Zagorec M. Proteomic analysis of global changes in protein expression during bilesalt exposure of Bifidobacterium longum NCIMB 8809. J Bacteriol. 2005;187:5799–5808. doi: 10.1128/JB.187.16.5799-5808.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Buss C, Valle-Tovo C, Miozzo S, Alves de Mattos A. Probiotics andsynbiotics may improve liver aminotransferases levels in non-alcoholic fatty liver disease patients. Ann Hepatol. 2014;13:482–488. doi: 10.1016/S1665-2681(19)31246-3. [DOI] [PubMed] [Google Scholar]
- 164.Olivares M, Diaz-Ropero MA, Gomez N, Lara-Villoslada F, SierraS MJ, A, Martin R, Lopez-Huertas E, Rodriguez JM, Xaus J, Oral administration of two probiotic strains, Lactobacillus gasseri CECT5714 and Lactobacillus coryniformisCECT5711, enhances the intestinal function of healthy adults. Int J Food Microbiol. 2006;107:104–111. doi: 10.1016/j.ijfoodmicro.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 165.Ferrario C, Taverniti V, Milani C, Fiore W, Laureati M, NoniI De, Stuknyte M, Chouaia B, Riso P, Guglielmetti S. Modulation of Clostridiales bacteria and butyrate by probiotic intervention with Lactobacillus paracasei DG varies among healthy adults. J Nutr. 2014;144:1787–1796. doi: 10.3945/jn.114.197723. [DOI] [PubMed] [Google Scholar]
- 166.Wang L, Zhang J, Guo Z, Kwok L, MaC ZW, Lv Q, Huang W, Zhang H. Effect of oral consumption of probiotic Lactobacillus plantarum P-8 on fecal microbiota, SIgA, SCFAs, and TBAs of adults of different ages. Nutrition. 2014;30:776–783. doi: 10.1016/j.nut.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 167.Worthley DL, Le Leu RK, Whitehall VL, Conlon M, ChristophersenC BD, Mallitt KA, Hu Y, Irahara N, Ogino S, et al. A human, double-blind, placebo-controlled, crossover trial of prebiotic, probiotic, and synbiotic supplementation: effects on luminal, inflammatory, epigenetic, and epithelial biomarkers of colorectal cancer. Am J Clin Nutr. 2009;90:578–586. doi: 10.3945/ajcn.2009.28106. [DOI] [PubMed] [Google Scholar]
- 168.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 169.Schneider SM, Girard-Pipau F, Filippi J, Hebuterne X, MoyseD HGC, PompeiA RP. Effects of Saccharomyces boulardii on fecal short-chain fatty acids and microflora in patients on long-term total enteral nutrition. World J Gastroenterol. 2005;11:6165–6169. doi: 10.3748/wjg.v11.i39.6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Stadlbauer V, Leber B, Lemesch S, TrajanoskS,Bashir M, Horvath A,Tawdrous,M, Stojakovic T, Fauler G, Fickert P, Högenauer C, Klymiuk I, Stiegler P, Lamprecht M, Pieber TR, Tripolt NJ, Sourij H (2015) Lactobacillus casei shirota supplementation does not restore gut microbiota composition and gut barrier in metabolic syndrome: a randomized pilot study. Plos One 28:10(10):e0141399 [DOI] [PMC free article] [PubMed]
- 171.Nagata S, Asahara T, Wang C, Suyama Y, Chonan O, Takano K, Daibou M, Takahashi T, Nomoto K, Yamashiro Y. The effectiveness of Lactobacillus beverages in controlling infections among the residents of an aged care facility: a randomized placebo-controlled double-blind trial. Ann Nutr Metab. 2016;68:51–59. doi: 10.1159/000442305. [DOI] [PubMed] [Google Scholar]
- 172.Hemalatha R, Ouwehand AC, Saarinen MT, Prasad UV, Swetha K, Bhaskar V. Effect of probiotic supplementation on total lactobacilli, bifidobacteria and short chain fatty acids in 2–5-year-old children. Microb Ecol Health Dis. 2017;28:1298340. doi: 10.1080/16512235.2017.1298340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Maldonado J, Lara-Villoslada F, Sierra S, Sempere L, Gomez M, Rodriguez JM, Boza J, Xaus J, Olivares M. Safety and tolerance of the human milk probiotic strain Lactobacillus salivarius CECT5713 in 6-month-old children. Nutrition. 2010;26:1082–1087. doi: 10.1016/j.nut.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 174.Mohan R, Koebnick C, Schildt J, Mueller M, Radke M, Blaut M. Effects of Bifidobacterium lactis Bb12 supplementation on body weight, fecal pH, acetate, lactate, calprotectin, and IgA in preterm infants. Pediatr Res. 2008;64:418–422. doi: 10.1203/PDR.0b013e318181b7fa. [DOI] [PubMed] [Google Scholar]
- 175.Saez-Lara MJ, Robles-Sanchez C, Ruiz-Ojeda FJ, Plaza-Diaz J, Gil A. Effects of probiotics and synbiotics on obesity, insulin resistance syndrome, type 2 diabetes and non-alcoholic fatty liver disease: a review of human clinical trials. Int J Mol Sci. 2016;17:928. doi: 10.3390/ijms17060928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Miccheli A, Capuani G, Marini F, Tomassini A, PraticoG CS, Gnani D, BavieraG AA, Putignani L, et al. Urinary (1)H-NMR based metabolic profiling of children with NAFLD undergoing VSL#3treatment. Int J Obes (Lond) 2015;39:1118–1125. doi: 10.1038/ijo.2015.40. [DOI] [PubMed] [Google Scholar]
- 177.Picca S, Dionisi-Vici C (2019) Hyperammonaemia and metabolic diseases. Critical care pediatric nephrology and dialysis: a practical handbook. Sethi, S.K., Raina R, McCulloch M, Bunchman TE (Eds.) Springer: Singapore pp. 311–323
- 178.Isabella VM, Ha BN, Castillo M, Lubkowicz DJ, Rowe SE, Millet YA, Anderson CL, Li N, Fisher AB, West KA, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat Biotechnol. 2018;36:857–864. doi: 10.1038/nbt.4222. [DOI] [PubMed] [Google Scholar]
- 179.Mathipa MG, Thantsha MS. Probiotic engineering: towards development of robust probiotic strains with enhanced functional properties and for targeted control of enteric pathogens. Gut Pathog. 2017;9:28. doi: 10.1186/s13099-017-0178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gurbatri CR, Lia I, Vincent R, Coker C, Castro S, Treuting PM, Hinchliffe TE, Arpaia N, Danino T. Engineered probiotics for local tumour delivery of checkpoint blockade nanobodies. Sci Transl Med. 2020;12:530. doi: 10.1126/scitranslmed.aax0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Daeffler KNM, Galley JD, Sheth RU, Ortiz-Velez LC, Bibb CO, Shroyer NF, Britton RA, Tabor JJ. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol Syst Biol. 2017;13(4):923. doi: 10.15252/msb.20167416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Riglar DT, Giessen TW, Baym M, Kerns SJ, Niederhuber MJ, Bronson RT, Kotula JW, Gerber GK, Way JC, Silver PA. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat Biotechnol. 2017;35(7):653–658. doi: 10.1038/nbt.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Geldart K, Borrero J, Kaznessis YN. Chloride-inducible expression vector for delivery of antimicrobial peptides targeting antibiotic-resistant Enterococcus faecium. Appl Environ Microbiol. 2015;81(11):3889–3897. doi: 10.1128/AEM.00227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hwang IY, Koh E, Wong A, March JC, Bentley WE, Lee YS, Chang MW. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat Commun. 2017;8:15028. doi: 10.1038/ncomms15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mao N, Cubillos-Ruiz A, Cameron DE, Collins JJ (2018) Probiotic strains detect and suppress cholera in mice. Sci Transl Med 10:eaao02586 [DOI] [PMC free article] [PubMed]
- 186.Palmer JD, Piattelli E, McCormick BA, Silby MW, Brigham CJ, Bucci V. Engineered probiotic for the inhibition of Salmonella via tetrathionate-induced production of microcin H47. ACS Infect Dis. 2018;4(1):39–45. doi: 10.1021/acsinfecdis.7b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Takiishi T, Korf H, Van Belle TL, Robert S, Grieco FA, Caluwaerts S, Galleri L, Spagnuolo I, Steidler L, Van Huynegem K. Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice. J Clin Investig. 2012;122:1717–1725. doi: 10.1172/JCI60530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Agarwal P, Khatri P, Billack B, Low WK, Shao J. Oral delivery of glucagon like peptide-1 by a recombinant Lactococcus lactis. Pharm Res. 2014;31:3404–3414. doi: 10.1007/s11095-014-1430-3. [DOI] [PubMed] [Google Scholar]
- 189.Hvidberg A, Nielsen MT, Hilsted J, Ørskov C, Holst JJ. Effect of glucagonlikepeptide-1 (proglucagon 78–107amide) on hepatic glucose production in healthy man. Metabolism. 1994;43:104–108. doi: 10.1016/0026-0495(94)90164-3. [DOI] [PubMed] [Google Scholar]
- 190.Mitchell JJ, Trakadis YJ, Scriver CR. Phenylalanine hydroxylase deficiency. Genetics Med. 2011;13:697–707. doi: 10.1097/GIM.0b013e3182141b48. [DOI] [PubMed] [Google Scholar]
- 191.Durrer KE, Allen MS, Von Herbing IH. Genetically engineered probiotic for the treatment of phenylketonuria (PKU); assessment of a novel treatment in vitro and in the PAHenu2 mouse model of PKU. PLoS ONE. 2017;12:e0176286. doi: 10.1371/journal.pone.0176286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.O’Connell Motherway M, O’driscoll J, Fitzgerald GF, Van Sinderen D, Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol. 2009;2(3):321–332. doi: 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hirayama Y, Sakanaka M, Fukuma H, Murayama H, Kano Y, Fukiya S, Yokota A. Development of a double-crossover marker less gene deletion system in Bifidobacterium longum: functional analysis of the α-galactosidase gene for raffinose assimilation. Appl Environ Microbiol. 2012;78(14):4984–4994. doi: 10.1128/AEM.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sakaguchi K, He J, Tani S, Kano Y, Suzuki T. A targeted gene knockout method using a newly constructed temperature-sensitive plasmid mediated homologous recombination in Bifidobacterium longum. Appl Microbiol Biotechnol. 2012;95(2):499–509. doi: 10.1007/s00253-012-4090-4. [DOI] [PubMed] [Google Scholar]
- 195.O’Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hidalgo-Cantabrana C, O’Flaherty S, Barrangou R. CRISPR-based engineering of next-generation lactic acid bacteria. Curr Opin Microbiol. 2017;37:79–87. doi: 10.1016/j.mib.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 197.Roberts A, Barrangou R (2020) Applications of CRISPR-Cas systems in lactic acid bacteria. FEMS Microbiol Rev 1–15:fuaa016 [DOI] [PubMed]
- 198.Briner AE, Lugli GA, Milani C, Duranti S, Turroni F, Gueimonde M, Margolles A, Van Sinderen D, Ventura M, Barrangou R. Occurrence and diversity of CRISPR-Cas systems in the genus Bifidobacterium. PLoS ONE. 2015;10(7):e0133661. doi: 10.1371/journal.pone.0133661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Landete JM, Peirot´en ´A, Rodríguez E, Margolles A, Medina M, Arqu´es JL, Anaerobic green fluorescent protein as a marker of Bifidobacterium strains. Int J Food Microbiol. 2014;175:6–13. doi: 10.1016/j.ijfoodmicro.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 200.Watson D, Sleator RD, Hill C, Gahan CG. Enhancing bile tolerance improves survival and persistence of Bifidobacterium and Lactococcus in the murine gastrointestinal tract. BMC Microbiol. 2008;8(1):176. doi: 10.1186/1471-2180-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bottacini F, Morrissey R, Roberts RJ, James K, Van Breen J, Egan M, Lambert J, Van Limpt K, Knol J, O’Connell-Motherway M. Comparative genome and methylome analysis reveals restriction/modification system diversity in the gut commensal Bifidobacterium breve. Nucleic Acids Res. 2018;46(4):1860–1877. doi: 10.1093/nar/gkx1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.O’Callaghan A, Bottacini F, Motherway MC, Van Sinderen D. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genomics. 2015;16(1):832. doi: 10.1186/s12864-015-1968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lee JH, O’Sullivan DJ. Genomic insights into bifidobacteria. Microbiol Mol Biol Rev. 2010;74(3):378–416. doi: 10.1128/MMBR.00004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bron PA, Marcelli B, Mulder J, van der Els S, Morawska LP, Kuipers OP, Kok J, Kleerebezem M. Renaissance of traditional DNA transfer strategies for improvement of industrial lactic acid bacteria. Curr Opin Biotechnol. 2019;56:61–68. doi: 10.1016/j.copbio.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 205.Dominguez W, O’Sullivan DJ. Developing an efficient and reproducible conjugation-based gene transfer system for bifidobacteria. Microbiology. 2013;159(2):328–338. doi: 10.1099/mic.0.061408-0. [DOI] [PubMed] [Google Scholar]
- 206.Sun Z, Westermann C, Yuan J, Riedel CU. Experimental determination and characterization of the gap promoter of Bifidobacterium bifidum S17. Bioengineered. 2014;5(6):371–377. doi: 10.4161/bioe.34423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Klijn A, Moine D, Delley M, MercenierA AF, Pridmore R. Construction of a reporter vector for the analysis of Bifidobacterium longum promoters. Appl Environ Microbiol. 2006;72(11):7401–7405. doi: 10.1128/AEM.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sun Z, Baur A, Zhurina D, Yuan J, Riedel CU. Accessing the inaccessible: molecular tools for bifidobacteria. Appl Environ Microbiol. 2012;78(15):5035–5042. doi: 10.1128/AEM.00551-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zuo F, Yu R, Feng X, Khaskheli GB, Chen L, Ma H, Chen S. Combination of heterogeneous catalase and superoxide dismutase protects Bifidobacterium longum strain NCC2705 from oxidative stress. Appl Microbiol Biotechnol. 2014;98(17):7523–7534. doi: 10.1007/s00253-014-5851-z. [DOI] [PubMed] [Google Scholar]
- 210.Ruegg TL, Pereira JH, Chen JC, DeGiovanni A, Novichkov P, Mutalik VK, Tomaleri GP, Singer SW, Hillson NJ, Simmons BA, Adams PD, Thelen MP. Jungle Express is a versatile repressor system for tight transcriptional control. Nat Comm. 2018;9:3617–3617. doi: 10.1038/s41467-018-05857-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lubkowicz D, Ho CL, Hwang IY, Yew WS, Lee YS, Chang MW. Reprogramming probiotic Lactobacillus reuterias a biosensor for Staphylococcus aureus derived AIP-I detection. ACS Synth Biol. 2018;7(5):1229–1237. doi: 10.1021/acssynbio.8b00063. [DOI] [PubMed] [Google Scholar]
- 212.Renwick MJ, Brogan DM, Mossialos E. A systematic review and critical assessment of incentive strategies for discovery and development of novel antibiotics. J Antibiot. 2016;69:73–88. doi: 10.1038/ja.2015.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Saeidi N, Wong CK, Lo TM, Nguyen HX, Ling H, Leong SSJ, Poh CL, Chang MW. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol. 2011;7:521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gupta S, Bram EE, Weiss R. Genetically programmable pathogen sense and destroy. ACS Synthetic Biol. 2013;2:715–723. doi: 10.1021/sb4000417. [DOI] [PubMed] [Google Scholar]
- 215.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ali M, Nelson AR, Lopez AL, Sack DA. Updated global burden of cholera in endemic countries. PLoS Neglected Trop Dis. 2015;9:e0003832. doi: 10.1371/journal.pntd.0003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bennish ML (1994) Cholera: pathophysiology, clinical features, and treatment. In: Vibrio cholerae and cholera. molecular to global perspectives. Wachsmuth IK Blake PA OlsvikO (Eds.) Washington D.C. ASM Press pp 229–255
- 219.Duan F, March JC. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc Natl Acad Sci. 2010;107:11260–11264. doi: 10.1073/pnas.1001294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sedlmayer F, Hell D, Müller M, Ausländer D, Fussenegger M. Designer cells programming quorum-sensing interference with microbes. NatCommun. 2018;9:1822. doi: 10.1038/s41467-018-04223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Meighen EA. Molecular biology of bacterial bioluminescence. Microbiol Mol Biol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Riedel CU, Casey PG, Mulcahy H, O’Gara F, Gahan CG, Hill C. Construction of p16Slux, a novel vector for improved bioluminescent labelling of gram-negative bacteria. Appl Environ Microbiol. 2007;73:7092–7095. doi: 10.1128/AEM.01394-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Leonard N, Bishop A, Polak J, Talbot I. Expression of nitric oxide synthase in inflammatory bowel disease is not affected by corticosteroid treatment. J Clin Pathol. 1998;51:750–753. doi: 10.1136/jcp.51.10.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kimura H, Miura S, Shigematsu T, Ohkubo N, Tsuzuki Y, Kurose I, Higuchi H, Akiba Y, Hokari R, Hirokawa M. Increased nitric oxide production and inducible nitric oxide synthase activity in colonic mucosa of patients with active ulcerative colitis and Crohn’s disease. Dig Dis Sci. 1997;42:1047–1054. doi: 10.1023/A:1018849405922. [DOI] [PubMed] [Google Scholar]
- 225.Neef A, Sanz Y. Future for probiotic science in functional food and dietary supplement development. Curr Opin Clin Nutr Metab Care. 2013;16:679–687. doi: 10.1097/MCO.0b013e328365c258. [DOI] [PubMed] [Google Scholar]
- 226.Dhar D, Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285:198018. doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ceccarelli G, Scagnolari C, Pugliese F, Mastroianni CM, d’Ettorre G. Probiotics and COVID-19. Lancet Gastroenterol Hepatol. 2020;5(8):721–722. doi: 10.1016/S2468-1253(20)30196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu YI, Zhang LI, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo LI, XieJ., Wang G, Jiang R, Gao Z, Jin QI, Wang J, Cao B, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bhaskar S, Sinha A, Banach M, Mittoo S, Weissert R, Kass JS, Rajagopal S, PaiAR KS. Cytokine storm in COVID-19—Immunopathological mechanisms, clinical considerations, and therapeutic approaches: The REPROGRAM Consortium Position Paper. Front Immunol. 2020;11:1648. doi: 10.3389/fimmu.2020.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Xu K, Cai H, Shen Y, Ni Q, Chen Y, Hu S, Li J, Wang H, Yu L, Huang H (2020) Management of corona virus disease-19 (COVID-19): the Zhejiang experience. J Zhejiang Univ (medical science) 49 [DOI] [PMC free article] [PubMed]
- 231.Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Baud D, Agri VD, Gibson GR, Reid G, Giannoni E (2020) Using probiotics to flatten the curve of coronavirus disease COVID-2019 pandemic. Front Public Health 8:186. 10.3389/fpubh.2020.00186 [DOI] [PMC free article] [PubMed]
- 233.Yu L, Tong Y, Shen G, Fu A, Lai Y, Zhou X, Yuan Y, Wang Y, Pan Y, Yu Z. Immunodepleting with hypoxemia: a potential high risk subtype of coronavirus disease MedRxiv. 2020 doi: 10.1101/2020.03.03.20030650. [DOI] [Google Scholar]
- 234.Mak JWY, Chan FKL, Ng SC. Probiotics and COVID-19: one size does not fit all. Lancet Gastroenterol Hepatol. 2020;5(7):644–645. doi: 10.1016/S2468-1253(20)30122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Schmitter T, Fiebich BL, Fischer JT, GajfulinM LN, Rose T, Goetz MR. Ex vivo anti-inflammatory effects of probiotics for periodontal health. J Oral Microbiol. 2018;10(1):1502027. doi: 10.1080/20002297.2018.1502027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sundararaman A, Ray M, Ravindra PV, Halami PM. Role of probiotics to combat viral infections with emphasis on COVID-19. Appl Microbiol Biotechnol. 2020;104(19):8089–8104. doi: 10.1007/s00253-020-10832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ayyanna R, Ankaiah D, Arul V. Anti-inflammatory and antioxidant properties of probiotic bacterium Lactobacillus mucosae AN1 and Lactobacillus fermentum SNR1 in Wistar albino rats. Front Microbiol. 2018;9:3063. doi: 10.3389/fmicb.2018.03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Jones SE, Versalovic J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 2009;9(1):35. doi: 10.1186/1471-2180-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Bottari B, Castellone V, Neviani E. Probiotics and Covid-19. Int J Food Sci Nutr. 2020;12:1–7. doi: 10.1080/09637486.2020.1807475. [DOI] [PubMed] [Google Scholar]
- 240.Lee N, Kim WU. Microbiota in T-cell homeostasis and inflammatory diseases. Exp Mol Med. 2017;49(5):e340. doi: 10.1038/emm.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9(5):356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 242.Ivanov II, FrutosRdeL MN, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Li JQ, Zhao JJ, Wang XD, Qayum A, Hussain MA, Liang GZ et al (2019) Novel angiotensin-converting enzyme-inhibitory peptides from fermented bovine milk started by lactobacillus helveticus KLDS.31 and lactobacillus casei KLDS.105:Purification, identification, and interaction mechanisms. Front Microbiol 10:2643 [DOI] [PMC free article] [PubMed]
- 244.Minato T, Nirasawa S, Sato T, Yamaguchi T, Hoshizaki M, Inagaki T, et al. B38-CAP is a bacteria-derived ACE2-like enzyme that suppresses hypertension and cardiac dysfunction. Nat Commun. 2020;11(1):1058. doi: 10.1038/s41467-020-14867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Verma A, Xu K, Du T, Zhu P, Liang Z, Liao S, Zhang J, Raizada M, Grant M, Li Q. Expression of human ACE2 in Lactobacillus and beneficial effects in diabetic retinopathy in mice. Molecular Therapy - Methods & Clinical Development. 2019;14:161–170. doi: 10.1016/j.omtm.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Górska A, Przystupski D, Niemczura MJ, Kulbacka J. Probiotic Bacteria: a promising tool in cancer prevention and therapy. Curr Microbiol. 2019;76:939–949. doi: 10.1007/s00284-019-01679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Tiptiri-Kourpeti A, Spyridopoulou K, Santarmaki V, Aindelis G, Tompoulidou E, Lamprianidou EE, Saxami G, Ypsilantis P, Lampri ES, Simopoulos C, et al. Lactobacillus Casei exerts anti-proliferative effects accompanied by apoptotic cell death and up-regulation of TRAIL in colon carcinoma cells. PLoS ONE. 2016;11:e0147960. doi: 10.1371/journal.pone.0147960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kim Y, Oh S, Yun HS, Oh S, Kim SH. Cell-bound exopolysaccharide from probiotic bacteria induces autophagic cell death of tumour cells. Lett Appl Microbiol. 2010;51:123–130. doi: 10.1111/j.1472-765X.2010.02859.x. [DOI] [PubMed] [Google Scholar]
- 249.Uccello M, Malaguarnera G, Basile F, D’agata V, Malaguarnera M, Bertino G, Vacante M, Drago F, Biondi A. Potential role of probiotics on colorectal cancer prevention. BMC Surg. 2012;12:S35. doi: 10.1186/1471-2482-12-S1-S35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lamichhane P, Maiolini M, Alnafoosi O, Hussein S, Alnafoosi H, Umbela S, Richardson T, Alla N, Lamichhane N, Subhadra B, et al. Colorectal cancer and probiotics: Are bugs really drugs? Cancers. 2020;12:1162. doi: 10.3390/cancers12051162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Motevaseli E, Dianatpour A, Ghafouri-Fard S. The Role of probiotics in cancer treatment: emphasis on their in vivo and in vitro anti-metastatic effects. Int J Mol Cell Med. 2017;6:66–76. doi: 10.22088/acadpub.BUMS.6.2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shenderov BA. Metabiotics: novel idea or natural development of probiotic conception. Microb Ecol Health Dis. 2013 doi: 10.3402/mehd.v24i0.20399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lebeer S, Bron PA, Marco ML, Van Pijkeren JP, O’Connell Motherway M, Hill C, Pot B, Roos S, Klaenhammer T. Identification of probiotic effector molecules: present state and future perspectives. Curr Opin Biotechnol. 2018;49:217–223. doi: 10.1016/j.copbio.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 254.Kumar M, Nagpal R, Verma V, Kumar A, Kaur N, Hemalatha R, Gautam SK, Singh B. Probiotic metabolites as epigenetic targets in the prevention of colon cancer. Nutr Rev. 2013;71:23–34. doi: 10.1111/j.1753-4887.2012.00542.x. [DOI] [PubMed] [Google Scholar]
- 255.Sharma S, Singh RL, Kakkar P. Modulation of Bax/Bcl-2 and caspases by probiotics during acetaminophen induced apoptosis in primary hepatocytes. Food Chem Toxicol Int J Publ Br Ind Biol Res Assoc. 2011;49:770–779. doi: 10.1016/j.fct.2010.11.041. [DOI] [PubMed] [Google Scholar]
- 256.Karimi Ardestani S, Tafvizi F, Tajabadi Ebrahimi M. Heat-killed probiotic bacteria induce apoptosis of HT-29 human colon adenocarcinoma cell line via the regulation of Bax/Bcl2 and Caspases pathway. Human Exp Toxicol. 2019;38:1069–1081. doi: 10.1177/0960327119851255. [DOI] [PubMed] [Google Scholar]
- 257.Chumchalová J, Smarda J. Human tumor cells are selectively inhibited by colicins. Folia Microbiol. 2003;48:111–115. doi: 10.1007/BF02931286. [DOI] [PubMed] [Google Scholar]
- 258.Konishi H, Fujiya M, Tanaka H, Ueno N, Moriichi K, Sasajima J, Ikuta K, Akutsu H, Tanabe H, Kohgo Y. Probiotic derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nat Commun. 2016;7:12365. doi: 10.1038/ncomms12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Kadirareddy RH, Vemuri SG, Palempalli UMD. Probiotic conjugated linoleic acid mediated apoptosis in breast Cancer cells by downregulation of NFκB. Asian Pac J Cancer Prev APJCP. 2016;17:3395–3403. [PubMed] [Google Scholar]
- 260.Khosrovan Z, Haghighat S, Mahdavi M. The probiotic bacteria induce apoptosis in breast and colon cancer cells: an immunostimulatory effect. Immunoregulation. 2020;3:37–50. doi: 10.32598/IMMUNOREGULATION.3.1.5. [DOI] [Google Scholar]
- 261.Isazadeh A, Hajazimian S, Shadman B, Safaei S, Bedoustani AB, Chavoshi R, Shanehbandi D, Mashayekhi M, Nahaei M, Baradaran B. Anti-cancer effects of probiotic Lactobacillus acidophilus for colorectal cancer cell line caco-2 through apoptosis induction. Pharm Sci. 2020;27:262–267. doi: 10.34172/PS.2020.52. [DOI] [Google Scholar]
- 262.Jan G, Belzacq AS, Haouzi D, Rouault A, Métivier D, Kroemer G, Brenner C. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ. 2002;9:179–188. doi: 10.1038/sj.cdd.4400935. [DOI] [PubMed] [Google Scholar]
- 263.Wang H, Cheng X, Zhang L, Xu S, Zhang Q, Lu R. A surface-layer protein from Lactobacillus acidophilus NCFM induces autophagic death in HCT116 cells requiring ROS-mediated modulation of MTOR and JNK signaling pathways. Food Funct. 2019;10:4102–4112. doi: 10.1039/C9FO00109C. [DOI] [PubMed] [Google Scholar]
- 264.Sharma M, Shukla G. Metabiotics: one step ahead of probiotics; an insight into mechanisms involved in anticancerous effect in colorectal cancer. Front Microbiol. 2016;7:1940. doi: 10.3389/fmicb.2016.01940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Sharaf LK, Sharma M, Chandel D, Shukla G (2018) Prophylactic intervention of probiotics (L. acidophilus, L. rhamnosus GG) and celecoxib modulate bax-mediated apoptosis in 1,2-Dimethylhydrazine-induced experimental colon carcinogenesis. BMC Cancer 18:1111 [DOI] [PMC free article] [PubMed]
- 266.Fahmy CA, Gamal-Eldeen AM, El-Hussieny EA, Raafat BM, Mehanna NS, Talaat RM, Shaaban MT. Bifidobacterium Longum suppresses murine colorectal cancer through the modulation of oncomiRs and tumor uppressor MiRNAs. Nutr Cancer. 2019;71:688–700. doi: 10.1080/01635581.2019.1577984. [DOI] [PubMed] [Google Scholar]
- 267.Azam R, Ghafouri-Fard S, Tabrizi M, Modarressi MH, Ebrahimzadeh-Vesal R, Daneshvar M, Mobasheri MB, Motevaseli E. Lactobacillus Acidophilus and Lactobacillus Crispatus culture supernatants downregulate expression of cancer-testis genes in the MDA-MB-231 cell line. Asian Pac J Cancer Prev APJCP. 2014;15:4255–4259. doi: 10.7314/APJCP.2014.15.10.4255. [DOI] [PubMed] [Google Scholar]
- 268.Motevaseli E, Shirzad M, Akrami SM, Mousavi AS, Mirsalehian A, Modarressi MH. Normal and tumour cervical cells respond differently to vaginal Lactobacilli, independent of PH and lactate. J Med Microbiol. 2013;62:1065–1072. doi: 10.1099/jmm.0.057521-0. [DOI] [PubMed] [Google Scholar]
- 269.Taherian-Esfahani Z, Abedin-Do A, Nouri Z, Mirfakhraie R, Ghafouri-Fard S, Motevaseli E. Lactobacilli differentially modulate MTOR and Wnt/β-catenin pathways in different cancer lell Lines. Iran J Cancer Prev. 2016;9:e5369. doi: 10.17795/ijcp-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 271.Seth A, Yan F, Polk DB, Rao RK. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP Kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1060–G1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Asoudeh-Fard A, Barzegari A, Dehnad A, Bastani S, Golchin A, Omidi Y. Lactobacillus Plantarum induces apoptosis in oral cancer KB cells through upregulation of PTEN and downregulation of MAPK signalling pathways. Bioimpacts. 2017;7:193–198. doi: 10.15171/bi.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Heemels MT. Neurodegenerative diseases. Nature. 2016;539:179. doi: 10.1038/539179a. [DOI] [PubMed] [Google Scholar]
- 274.Fratiglioni L, Qiu C. Prevention of common neurodegenerative disorders in the elderly. Exp Gerontol. 2009;44:46–50. doi: 10.1016/j.exger.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 275.Dugger BN, Dickson DW. Pathology of neurodegenerative diseases. Cold Spring Harb Perspect Biol. 2017;9:a028035. doi: 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kesika P, Suganthy N, Sivamaruthi BS, Chaiyasut C. Role of gut-brain axis, gut microbial composition, and probiotic intervention in Alzheimer’s disease. Life Sci. 2021;264:118627. doi: 10.1016/j.lfs.2020.118627. [DOI] [PubMed] [Google Scholar]
- 277.Spielman LJ, Gibson DL, Klegerism A. Unhealthy gut, unhealthy brain: the role of the intestinal microbiota in neurodegenerative diseases. Neurochem Int. 2018;120:149–163. doi: 10.1016/j.neuint.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 278.Cirstea MS, Yu AC, Golz E, Sundvick K, Kliger D, Radisavljevic N, et al. Microbiota composition and metabolism are associated with gut function in Parkinson’s disease. Mov Disord. 2020;35:1208–1217. doi: 10.1002/mds.28052. [DOI] [PubMed] [Google Scholar]
- 279.Westfall S, Lomis N, Kahouli I, Dia SY, Singh SP, Prakash S. Microbiome, probiotics and neurodegenerative diseases: deciphering the gut brain axis. Cell Mol Life Sci. 2017;74:3769–3787. doi: 10.1007/s00018-017-2550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Pandey KR, Naik SR, Vakil BV. Probiotics, prebiotics and synbiotics- a review. J Food Sci Technol. 2015;52:7577–7587. doi: 10.1007/s13197-015-1921-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Perez Visñuk D, Savoy de Giori G, LeBlanc JG, de Moreno de LeBlanc A, Neuroprotective effects associated with immune modulation by selected lactic acid bacteria in a Parkinson’s disease model. Nutrition. 2020;79–80:110995. doi: 10.1016/j.nut.2020.110995. [DOI] [PubMed] [Google Scholar]
- 282.Borzabadi S, Oryan S, Eidi A, Aghadavod E, Daneshvar Kakhaki R, Tamtaji OR, et al. The effects of probiotic supplementation on gene expression related to inflammation, insulin and lipid in patients with Parkinson’s disease: a randomized, double-blind, placebocontrolled trial. Arch Iran Med. 2018;21:289–295. [PubMed] [Google Scholar]
- 283.Magistrelli L, Amoruso A, Mogna L, Graziano T, Cantello R, Pane M, Comi C. Probiotics may have beneficial effects in Parkinson’s disease: in vitro evidence. Front Immunol. 2019;10:969. doi: 10.3389/fimmu.2019.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Hsieh TH, Kuo CW, Hsieh KH, Shieh MJ, Peng CW, Chen YC, Chang YL, Huang YZ, Chen CC, Chang PK, Chen KY, Chen HY. Probiotics alleviate the progressive deterioration of motor functions in a mouse model of Parkinson’s disease. Brain Sci. 2020;10:206. doi: 10.3390/brainsci10040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Distrutti E, O’Reilly J-A, McDonald C, Cipriani S, Renga B, Lynch MA, Fiorucci S. Modulation of intestinal microbiota by the probiotic VSL#3 resets brain gene expression and ameliorates the age-related deficit in LTP. PLoS ONE. 2014;9:e106503. doi: 10.1371/journal.pone.0106503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Castelli V, d’Angelo M, Lombardi F, Alfonsetti M, Antonosante A, Catanesi M, Benedetti E, Palumbo P, Cifone MG, Giordano A, Desideri G, Cimini A. Effects of the probiotic formulation SLAB51 in in vitro and in vivo Parkinson’s disease models. Aging. 2020;12:4641–4659. doi: 10.18632/aging.102927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Boon Wong C, Kobayashi Y, Xiao J (2018) Probiotics for preventing cognitive impairment in Alzheimer’s disease. In: Gut Microbiota - Brain Axis (Evrensel A, Önen Ünsalver B, eds), pp85–104. London, UK: IntechOpen
- 288.Gazerani, Probiotics for Parkinson’s disease. Int J Mol Sci. 2019;20:4121. doi: 10.3390/ijms20174121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Bashiardes S, Godneva A, Elinav E, Segal E. Towards utilization of the human genome and microbiome for personalized nutrition. Curr Opin Biotechnol. 2018;51:57–63. doi: 10.1016/j.copbio.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 290.Suez J, Zmoraa N, Elina E. Probiotics in the next-generation sequencing era. Gut Microbes. 2020 doi: 10.1080/19490976.2019.1586039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Veiga P, Suez J, Derrien M, Elinav E. Moving from probiotics to precision probiotics. Nat Microbiol. 2020;5:878–880. doi: 10.1038/s41564-020-0721-1. [DOI] [PubMed] [Google Scholar]
- 292.Leshem A, Liwinski T, Elinav E. Immune-microbiota interplay and colonization resistance in infection. Mol Cell. 2020;21:597–613. doi: 10.1016/j.molcel.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 293.Foditsch C, Santos TM, Teixeira AG, Pereira RV, Dias JM, Gaeta N, et al. Isolation and characterization of Faecalibacterium prausnitzii from calves and piglets. PLoS ONE. 2014;9:e116465. doi: 10.1371/journal.pone.0116465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Schneeberger M, Everard A, Gómez-Valadés AG, Matamoros S, Ramírez S, Delzenne NM, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015;5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 296.Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in apoe-/-mice clinical. Circulation. 2016;133:2434–2446. doi: 10.1161/CIRCULATIONAHA.115.019645. [DOI] [PubMed] [Google Scholar]
- 297.Troy EB, Kasper DL. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci. 2010;15:25–34. doi: 10.2741/3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.RoundJL LSM, Li J, Tran G, Jabri B, Chatila TA, et al. The toll like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Yang JY, Lee YS, Kim Y, Lee SH, Ryu S, Fukuda S, et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. 2016;10:104–116. doi: 10.1038/mi.2016.42. [DOI] [PubMed] [Google Scholar]
- 300.Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. Potential beneficial effects of butyrate in intestinal and extra intestinal diseases. World J Gastroenterol. 2011;17:1519–1528. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 302.Liong MT. Safety of probiotics: translocation and infection. Nutr Rev. 2008;66:192–202. doi: 10.1111/j.1753-4887.2008.00024.x. [DOI] [PubMed] [Google Scholar]
- 303.Martin-Cabezas R, Davideau JL, Tenenbaum H, Huck O. Clinical efficacy of probiotics as an adjunctive therapy to non-surgical periodontal treatment of chronic periodontitis: a systematic review and meta-analysis. J Clin Periodontol. 2016;43(6):520–530. doi: 10.1111/jcpe.12545. [DOI] [PubMed] [Google Scholar]
- 304.Sulik-Tyszka B, Snarski E, Niedźwiedzka M, Augustyniak M, Myhre TN, Kacprzyk A, Kopec ES, Roszkowska M, Trojaczek JD, Jędrzejczak WW, Wróblewska M. Experience with Saccharomyces boulardii probiotic in onco-haematological patients. Probiotics and Antimicro Prot. 2018;10:350–355. doi: 10.1007/s12602-017-9332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Muñoz P, Bouza E, Cuenca-Estrella M, Eiros JM, Pérez MJ, Somolinos S, Rincón C, Hortal J, Peláez T. Saccharomyces cerevisiae Fungemia: An Emerging Infectious Disease. Clin Infect Dis. 2005;11:1625–1634. doi: 10.1086/429916. [DOI] [PubMed] [Google Scholar]
- 306.Sutton A. Product development of probiotics as biological drugs. Clin Infect Dis. 2008;46:S128–S132. doi: 10.1086/523325. [DOI] [PubMed] [Google Scholar]
- 307.Costa GN, Miglioranza LHS. Probiotics: the effects on human health and current prospects. In: Rigobelo EC, editor. Probiotics. London: Intech Open; 2012. pp. 367–384. [Google Scholar]
- 308.Leroy F, De Vuyst L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol. 2004;15:67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- 309.Sanders ME, Benson A, Lebeer S, Merenstein DJ, Klaenhammer TR. Shared mechanisms among probiotic taxa: implications for general probiotic claims. Curr Opin Biotechnol. 2018;49:207–216. doi: 10.1016/j.copbio.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 310.Garneau JE, Moineau S. Bacteriophages of lactic acid bacteria and their impacton milk fermentations. Microb Cell Factor. 2011;10:S20. doi: 10.1186/1475-2859-10-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Lucchini S, Sidoti J, BrussowH, Broad-range bacteriophage resistance inStreptococcus thermophilus by insertional mutagenesis. Virology. 2000;275:267–277. doi: 10.1006/viro.2000.0499. [DOI] [PubMed] [Google Scholar]
- 312.Antunes AEC, Cazetto TF, Abolini HM. Viability of probiotic micro-organisms during storage, post acidification and sensory analysis of fat-free yogurts with added whey protein concentrate. Int J Dairy Technol. 2005;58:169–173. doi: 10.1111/j.1471-0307.2005.00203.x. [DOI] [Google Scholar]
- 313.Pereira EPR, Cavalcanti RN, Esmerino EA, Silva R, Guerreiro LRM, Cunha RL, Bolini HMA, Meireles MA, Faria JAF, Cruz AG. Effect of incorporation of antioxidants on the chemical, rheological, and sensory properties of probiotic petit Suisse cheese. J Dairy Sci. 2016;99:1762–1772. doi: 10.3168/jds.2015-9701. [DOI] [PubMed] [Google Scholar]
- 314.Davis C. Enumeration of probiotic strains: review of culture-dependent and alternative techniques to quantify viable bacteria. J Microbiol Methods. 2014;103:9–17. doi: 10.1016/j.mimet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 315.Marcobal A, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME. 2013;J7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Zmora N, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–1405. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 317.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotictreatedgut microbiota. Cell Rep. 2015;10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 318.McFarland LV. Use of probiotics to correct dysbiosis of normal microbiota following disease or disruptive events: a systematic review. BMJ Open. 2014;4:e005047. doi: 10.1136/bmjopen-2014-005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Warda AK, Bettio PDH, Hueston CM, Di Benedetto G, Clooney AG, Hill C. Oral administration of heat-treated Lactobacilli modifies the murine microbiome and reduces Citrobacter induced colitis. Front Microbiol. 2020;11:69. doi: 10.3389/fmicb.2020.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Suez J, ZmoraN SE, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25:716–729. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- 321.Mills JP, Rao K, Young VB. Probiotics for prevention of Clostridium difficile infection. Curr Opin Gastroenterol. 2018;34(1):3–10. doi: 10.1097/MOG.0000000000000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kent AG, Vill AC, Shi Q, Satlin MJ, Brito IL. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat Commun. 2020;11:4379. doi: 10.1038/s41467-020-18164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Palumbo P, Lombardi F, Cifone MG, Cinque B. The epithelial barrier model shows that the properties of VSL#3 depend from where it is manufactured. Endocr Metab Immune Disord Drug Targets. 2019;19:199–206. doi: 10.2174/1871530318666181022164505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Plaza-Diaz J, Ruiz-Ojeda FJ, Gil-Campos M, Gil A. Mechanisms of action of probiotics. Adv Nutr. 2019;10:S49–S66. doi: 10.1093/advances/nmy063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Probiotic bacteria induce several beneficial traits by production of vitamins, organic acids, short chain fatty acids (SCFAs), neurotransmitters, enzymes, amino acids, and antimicrobial peptides like bacterocins. (PDF 75 KB)
Supplementary Figure 2 Probiotic bacteria can be engineered for their functional improvement by employing genetic engineering approaches like CRISPR plasmid mediated gene insertion/deletion, LPSD plasmid mediated crossover, employing synthetic biological route, and through development of regulatory circuit. (PDF 47 KB)