Abstract
We describe a genetic system that allows in vivo screening or selection of site-specific proteases and of their cognate-specific inhibitors in Escherichia coli. This genetic test is based on the specific proteolysis of a signaling enzyme, the adenylate cyclase (AC) of Bordetella pertussis. As a model system we used the human immunodeficiency virus (HIV) protease. When an HIV protease processing site, p5, was inserted in frame into the AC polypeptide, the resulting ACp5 protein retained enzymatic activity and, when expressed in an E. coli cya strain, restored the Cya+ phenotype. The HIV protease coexpressed in the same cells resulted in cleavage and inactivation of ACp5; the cells became Cya−. When the entire HIV protease, including its adjacent processing sites, was inserted into the AC polypeptide, the resulting AC-HIV-Pr fusion protein, expressed in E. coli cya, was autoproteolysed and inactivated: the cells displayed Cya− phenotype. In the presence of the protease inhibitor indinavir or saquinavir, AC-HIV-Pr autoproteolysis was inhibited and the AC activity of the fusion protein was preserved; the cells were Cya+. Protease variants resistant to particular inhibitors could be easily distinguished from the wild type, as the cells displayed a Cya− phenotype in the presence of these inhibitors. This genetic test could represent a powerful approach to screen for new proteolytic activities and for novel protease inhibitors. It could also be used to detect in patients undergoing highly active antiretroviral therapy the emergence of HIV variants harboring antiprotease-resistant proteases.
Site-specific proteases play a key role in many biological processes such as signal transduction, apoptosis, and development (for a review, see reference 36 and references therein). In addition, proteases are involved in the processing of polyprotein precursors of several viruses, such as picornaviruses and retroviruses (17, 18). Efficient tools for characterizing specific proteases and identifying specific inhibitors could provide new insights into the physiological role of proteases as well as new possibilities for therapy of infectious and noninfectious human diseases.
Several genetic assays that can detect proteolytic activity in vivo in bacteria as well as in yeasts have been described. They are based on different reporter enzymes or regulatory circuitries, but they have limited application (2, 6, 19, 32, 34) mainly because their sensitivity is insufficient to allow straightforward in vivo screening or selection procedures.
We describe here a novel bacterial genetic system in Escherichia coli that allows an easy functional characterization of proteases. This system is based on the specific proteolytically induced inactivation of a signaling enzyme, the adenylate cyclase (AC) of Bordetella pertussis (8, 21). When expressed in an E. coli strain deficient in its endogenous AC, encoded by cya, B. pertussis AC synthesizes a regulatory molecule, cyclic AMP (cAMP), that triggers the transcriptional activation of numerous genes, including genes involved in the catabolism of carbohydrates (35), and gives rise to a selectable Cya+ phenotype. The protease-mediated inactivation of AC can therefore be easily detected as the cells turn to a Cya− phenotype.
Here, we tested the human immunodeficiency virus (HIV) protease for use in a model system. This protease is required for the proteolytic processing of the polyprotein precursor Gag/Pol into mature viral proteins (28, 37). HIV protease is essential to generate infectious viral particles (17), and, as a consequence, protease inhibitors are widely used in therapeutic treatment of AIDS (14, 33, 39). As shown here, AC is an exquisitely sensitive reporter for testing the proteolytic activity of the HIV protease and its inhibition by known inhibitors. Furthermore, we show that this genetic test is able to distinguish between wild-type protease and inhibitor-resistant variants (5, 11, 25) that were isolated from patients undergoing highly active antiretroviral therapy (HAART). This approach could be used in phenotyping resistance tests to detect in AIDS patients preexisting or emerging minor subpopulations of viruses carrying antiprotease-resistant proteases (9, 27, 29, 30).
MATERIALS AND METHODS
Strain and growth media.
DHT1 [F− glnV44(AS) recA1 endA1 gyrA96 (Nalr) thi-1 hsdR17 spoT1 rfbD1 cya-854 ilv-691∷Tn10] is an AC-deficient (cya) derivative of DH1 that was constructed by cotransduction (24) of the cya-854 mutation (4) and the ilv-691∷Tn10 mutation (38). Transformation of DHT1 was performed by standard techniques (CaCl2 treatment or electroporation) (31). The growth medium used was the rich Luria-Bertani (LB) medium. Antibiotic concentrations were 100 μg of ampicillin, 50 μg of kanamycin, and 25 μg of tetracycline per ml. Screening for the ability to ferment sugars was performed on MacConkey agar plates containing 1% maltose (24). Indinavir sulfate (Crixivan [Merck], dissolved in water at a concentration of 20 mM) and saquinavir mesylate (Invirase [Roche], dissolved in ethanol at a concentration of 10 mM) were directly diluted into bacterial growth media at the indicated concentrations.
Plasmids.
All in vitro DNA manipulations were performed according to standard protocols (31) using E. coli XL1-Blue (Stratagene) as the recipient. The plasmid pUCHIV is an expression vector for the HIV protease. The gene encoding this protein was amplified by PCR from plasmid pNH1, which harbors the Gag/Pol HIV DNA sequence (a kind gift from N. Heveker, ICGM, Paris, France) by using primers P1 (GCGGTCGACTCATATGGGACTGTATCCTTTAAC) and P2 (CGCGGATCCAGTTTCAATAGGAC). The amplified sequence was cleaved with SalI and BamHI and cloned into the SalI and BamHI sites of pUC19 (40).
Plasmids pUCB1, pUCB3, pUCV1, and pUCV2 express, respectively, HIV protease variants B1, B3, V1, and V2 (Table 1) under the control of the lac promoter. Viral DNAs encoding these variants were isolated from patients' blood by F. Clavel (Hospital Bichat-Claude Bernard, Paris, France) and amplified by PCR using primers P1 and P2 (see above). The amplified DNAs were then cleaved with SalI and BamHI and cloned into pUC19.
TABLE 1.
Phenotypic assay of HIV protease activity on liquid culturesa
| Plasmids | No inhibitor
|
Indinavir (200 μM)
|
Saquinavir (40 μM)
|
|||
|---|---|---|---|---|---|---|
| cAMP level | β-Gal activity | cAMP level | β-Gal activity | cAMP level | β-Gal activity | |
| pKT25 + pUC19 | <20 | 100 | <20 | 100 | <20 | 100 |
| pKACp5 + pUC19 | 3,000 | 6,200 | 3,000 | 6,700 | 3,000 | 6,500 |
| pKACp5 + pUCHIV | 75 | 300 | 800 | 2,700 | 400 | 3,000 |
DHT1 bacteria cotransformed with the indicated plasmids were grown overnight at 30°C in LB medium plus ampicillin and kanamycin and in the presence of the indicated HIV protease inhibitors. cAMP levels (in picomoles per milligram [dry weight]) and β-galactosidase (β-Gal) activities (in units per milligram [dry weight]) were measured as described in Materials and Methods.
Plasmid pKT25 is a derivative of the low-copy-number vector pSU40 (harboring a kanamycin resistance selectable marker) (1) that expresses the N-terminal (T25) fragment (codons 1 to 224) of cyaA under the transcriptional and translational controls of the lacZ gene. It was constructed by subcloning a 1,044-bp HindIII-EcoRI fragment from pT25 (13) into pSU40 linearized with HindIII and EcoRI and then by deleting a 236-bp NheI-HindIII fragment.
Plasmid pKAC is a pKT25 derivative that expresses the full catalytic domain of AC (i.e., the first 384 codons of cyaA). It was generated by subcloning the 0.9-kb AatII-EcoRI fragment of pCmAHL1 (13) into pKT25.
Plasmid pKACPr expresses an AC-HIV protease chimeric protein in which the 99 residues of the mature HIV protease and its two flanking regions (26 residues on each side) encompassing the processing sites p5 and p6 (18) were inserted between residues 224 and 225 of AC. First, a 450- bp-long DNA fragment encoding the mature HIV protease and its two flanking processing sites (p5 and p6) was amplified by PCR using plasmid NH1 as the target and oligonucleotides P3 (GGGGCTAGCGGTAGAGACAACAACTCC) and P4 (CCCGGTACCTTCTTCTGTCAATGGCC) as primers. The amplified DNA was digested with NheI and KpnI and subcloned into the corresponding sites of plasmid pACM224p815A (12). Then a 1,544-bp AatII-EcoRI DNA fragment from the resulting plasmid was subcloned into the same sites of pKT25. DNAs encoding protease variants B1, B3, V1, and V2 were similarly amplified with primers P3 and P4 and then subcloned into the NheI/KpnI sites of pKACPr to generate plasmids pKACB1, pKACB3, pKACV1, and pKACV2, respectively.
Plasmid pKACp5 expresses a recombinant AC in which the p5 HIV protease cleavage site was inserted between codons 224 and 225 of the AC gene. Two complementary oligonucleotides, GTACCCCAAAGAGTGATCTGAGGGAAGTTAAAGGATACAGTG and CTAGCACTGTATCCTTTAACTTCCCTCAGTCACTCTTTGGG, encoding the amino acid sequence TVSFNFPQITLW (p5 site), were hybridized and ligated into pKACPr cleaved at the NheI and KpnI sites.
Analytical methods.
β-Galactosidase assays were performed on toluenized bacterial suspensions, as described by Pardee et al. (26). One unit of activity corresponds to 1 nmol of o-nitrophenyl-β-d-galactoside hydrolyzed per min at 28°C. cAMP measurements were done by an enzyme-linked immunosorbent assay as described previously (13).
RESULTS
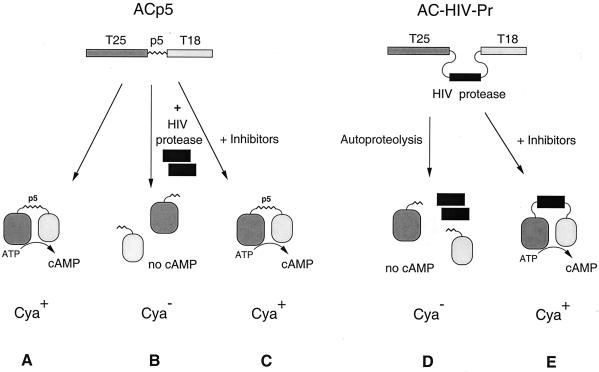
The catalytic domain of B. pertussis AC (400 amino acids) is composed of two subdomains, T25 and T18, that are both required for enzymatic activity (20, 21). It is a remarkably flexible molecule that can tolerate large in-frame polypeptide insertions between the two subdomains without loss of its enzymatic activity. However, when the fragments T25 and T18 are produced in E. coli as independent polypeptides, they are unable to reassociate to form an active enzyme (13). Hence, if a given proteolytic processing site is inserted into the intact AC between T25 and T18, the AC activity of the recombinant enzyme should be abolished upon specific cleavage by a site-specific protease. cAMP production, catalyzed by AC, can be easily monitored in vivo in E. coli as this molecule, through specific binding to the catabolic gene activator protein, controls the transcription of catabolic operons, such as lactose or maltose (35). As a consequence, both Cya+ and Cya− phenotypes can be easily scored on indicator plates or selected under appropriate conditions (Fig. 1): cells which produce cAMP will be able to use lactose or maltose as a unique carbon source; in contrast, cells which do not synthesize cAMP are unable to grow on minimal medium plus lactose (or maltose) and are resistant to the antibiotic mecillinam (10, 35).
FIG. 1.
Principle of an E. coli protease assay system based on AC. (Left) trans-Proteolysis. T25 and T18 are the two fragments of the catalytic domain of B. pertussis AC. In ACp5 (encoded by pKACp5), a 12-amino-acid linker corresponding to the p5 processing site of the HIV Gag/Pol polyprotein was inserted between the two fragments. ACp5 is active and, when expressed in a cya strain, restores a Cya+ phenotype (A). When coexpressed with HIV protease, ACp5 is cleaved at the p5 site and inactivated; the recipient cells are Cya− (B). The addition of protease inhibitor prevents proteolytic inactivation and restores a Cya+ phenotype (C). (Right) Autoproteolysis. In AC-HIV-Pr, the mature HIV protease and its flanking processing sites p5 and p6 were inserted between T25 and T18. This chimeric AC undergoes autoproteolysis, which results in inactivation of its cAMP-synthesizing activity (D); the recipient cells are Cya−. This processing is blocked in the presence of protease inhibitors (E).
First test: proteolysis of AC by HIV protease in trans.
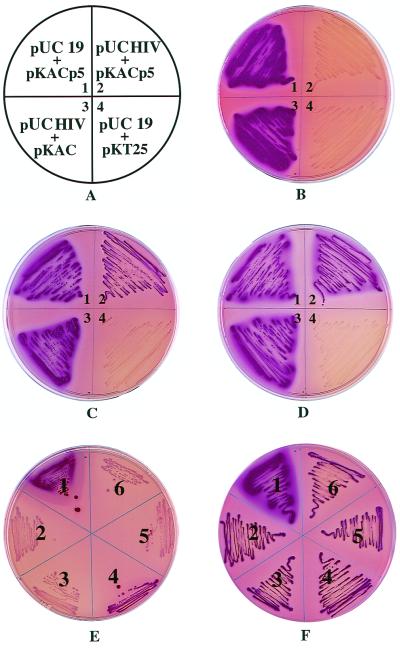
To test the principle described above, we constructed plasmid pKACp5 that codes for a chimeric AC, ACp5, in which an amino acid sequence, p5, corresponding to one of the processing sites of the Gag/Pol polyprotein precursor of HIV, was inserted in frame between the T25 and T18 fragments (Fig. 1). When transformed in DHT1, an E. coli cya strain, plasmid pKACp5 restored a Cya+ phenotype, as evidenced by the red color of the colonies on MacConkey-maltose medium (Fig. 2B, panel 1). cAMP and β-galactosidase assays on liquid cultures confirmed that these cells express an active chimeric AC (Table 1). When pKACp5 was cotransformed in DHT1 with a compatible plasmid that expresses the wild-type HIV protease, pUCHIV, the cells exhibited a Cya− phenotype (white colonies on MacConkey-maltose medium [Fig. 2B, panel 2]). This suggests that in vivo, ACp5 was split by the coexpressed HIV protease and inactivated. Indeed, only background levels of cAMP and β-galactosidase activity were measured in liquid cultures of these cells (Table 1). In the presence of an HIV protease inhibitor, saquinavir or indinavir, the Cya+ phenotype of the cells was restored, as shown on indicator plates (Fig. 2C, panel 2, and 2D, panel 2) and confirmed by cAMP and β-galactosidase measurements (Table 1). As expected, the inhibitors had no effect on control cells carrying pKACp5 and pUC19 and did not inhibit cell growth. These results indicate that the HIV protease is specifically involved in the ACp5 inactivation in vivo. In vitro experiments showed that the purified ACp5 polypeptide was specifically and rapidly cleaved when added to a cellular extract of DHT1 cells expressing the HIV protease with a concomitant loss of AC activity. Indinavir or saquinavir fully inhibited this degradation (data not shown).
FIG. 2.
Phenotypic assay of proteolysis by HIV protease in trans. (A through D) DHT1 bacteria transformed with the indicated plasmids (A) were plated on MacConkey-maltose plates containing ampicillin and kanamycin and supplemented with no protease inhibitors (B), 40 μM saquinavir (C), or 200 μM indinavir (D). Plates were incubated at 30°C for 36 h. (E and F) Phenotypic assay of mutant HIV protease activities. DHT1 bacteria were cotransformed with pKACp5 and pUC19 (1) pUCHIV (2), pUCB1 (3), pUCB3 (4), pUCV1 (5), or pUCV2 (6). Transformants were plated on MacConkey-maltose plates containing ampicillin and kanamycin and supplemented with either no protease inhibitor (E) or 40 μM saquinavir (F). Plates were incubated at 30°C for 36 h. Transformants plated on plates containing indinavir (200 μM) instead of saquinavir exhibited the same phenotype as in panel F.
To further demonstrate that HIV protease specifically cleaves ACp5 at the p5 site in vivo, DHT1 cells were cotransformed with pUCHIV and pKAC, which codes for the wild-type AC without the processing site p5. As shown in Fig. 2B, panel 3, these cells exhibited a Cya+ phenotype indicating that, in vivo, the wild-type AC was not significantly degraded by the HIV protease.
Altogether, these results demonstrate that this genetic test, based on the specific proteolysis and inactivation of a recombinant AC, offers a sensitive phenotypic assay for the HIV protease activity in vivo in E. coli.
We then tried to apply this test to probe for resistance towards inhibitors of different HIV protease variants isolated from patients undergoing HAART. Four different variants provided by F. Clavel (Hopital Bichat) were included in this study. Two of them, B1 and V1, exhibited normal sensitivity to saquinavir and indinavir, whereas variants B3 and V2 were resistant to indinavir and saquinavir, respectively, as determined in recombinant-virus assays (Table 2). The DNAs encoding these modified proteases were cloned in pUC19, and the resulting plasmids were cotransformed with pKACp5 into DHT1. As shown in Fig. 2E, DHT1 expressing protease B1, V1, or V2 exhibited a Cya− phenotype as expected, whereas the DHT1 cells that expressed protease B3 exhibited a Cya+ phenotype. This suggested that variant B3 is unable to completely inactivate ACp5, most likely because of a reduced catalytic efficacy. The decrease in catalytic activity of HIV protease as a consequence of mutations associated with resistance to inhibitors has been documented (9, 22, 23, 30, 42). As expected, when the transformants were plated on MacConkey-maltose medium supplemented with high concentrations of indinavir (Fig. 2F) or saquinavir (data not shown), they all acquired a Cya+ phenotype.
TABLE 2.
Characteristics of the HIV protease variants
| Modified protease | Mutation(s) | Phenotypic resistancea
|
|
|---|---|---|---|
| Saquinavir | Indinavir | ||
| B1 | V77I | 1 | 1 |
| B3 | M46I, V77I, V82T | 3 | 13 |
| V1 | L63P | 1 | 1 |
| V2 | L10I, L63P, L90M | 53 | 4 |
Fold increase in the concentration of inhibitors required for 50% inhibition of the viral replication in recombinant-virus assays (15), compared to wild-type HIV. These data were kindly provided by F. Clavel.
The inability of this test to detect the proteolytic activity of the in vivo-active variant B3 precluded its utilization for general screening of HIV proteases resistant to antiproteases and prompted us to design a more sensitive test.
Second test: autoproteolysis of a recombinant AC-HIV protease fusion protein.
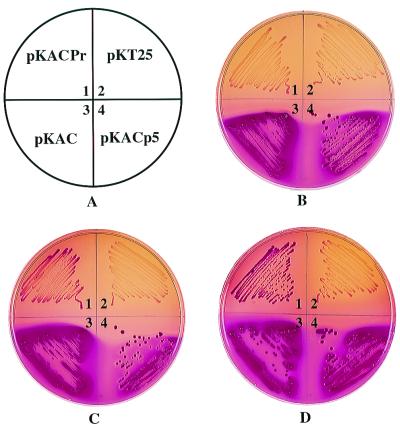
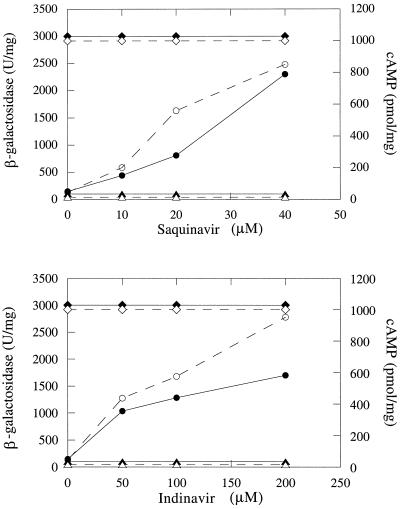
In order to increase the sensitivity of our test, we constructed a chimeric AC, AC–HIV-Pr, in which the entire HIV protease—a 150-residue fragment from the Gag/Pol polyprotein precursor, encompassing the mature HIV protease (99 residues) and its flanking regions (25 residues on each side), including the processing sites p5 and p6—was inserted in frame between T25 and T18 (Fig. 1). When expressed in DHT1 (Fig. 3B, panel 1), this fusion protein was rapidly split, due to autoproteolysis, and inactivated; no cAMP was produced, and the cells displayed a Cya− phenotype (white colonies on MacConkey-maltose plates). However, when plated on MacConkey-maltose medium supplemented with the protease inhibitor indinavir or saquinavir, the DHT1/pKACPr cells exhibited a red Cya+ phenotype (Fig. 3C, panel 1, and 3D, panel 1). This indicates that, in the presence of the protease inhibitors, the autoproteolysis of AC–HIV-Pr was inhibited and as a consequence its AC activity was preserved; cAMP measurements confirmed that this was indeed the case (Fig. 4). Quantification of the antiprotease effect was carried out in liquid cultures by determining β-galactosidase activities of DHT1 cells expressing either parental AC or AC–HIV-Pr. As shown in Fig. 4, there is a dose-dependent relation between β-galactosidase activities and cAMP levels of cells expressing AC–HIV-Pr and indinavir or saquinavir concentrations.
FIG. 3.
Phenotypic assay of HIV protease autoproteolysis. DHT1 bacteria transformed with the indicated plasmids (A) were plated on MacConkey-maltose plates containing kanamycin and either no protease inhibitors (B), 40 μM saquinavir (C), or 200 μM indinavir (D). Plates were incubated at 30°C for 36 h.
FIG. 4.
Quantitative analysis of HIV protease inhibition. DHT1 cells were transformed with pKACp5 (diamonds), pKACPr (circles), and pKT25 (triangles) and grown at 30°C overnight in LB broth containing kanamycin, in the presence of the indicated concentrations of HIV protease inhibitors. β-Galactosidase activities (closed symbols) and cAMP levels (open symbols) were determined as described in Materials and Methods.
As a control, we constructed a modified form of AC–HIV-Pr, in which the essential Asp residue at position 25 of the mature protease was replaced with an Asn residue by site- directed mutagenesis; this modification has been shown to abolish the proteolytic activity of the HIV protease (17). As expected, the resulting chimeric AC, AC–HIV-Pr-D25N, was not autoproteolytically processed, and when the mutant protein was expressed in DHT1, the cells displayed a Cya+ phenotype (data not shown).
Detection of antiprotease-resistant HIV proteases.
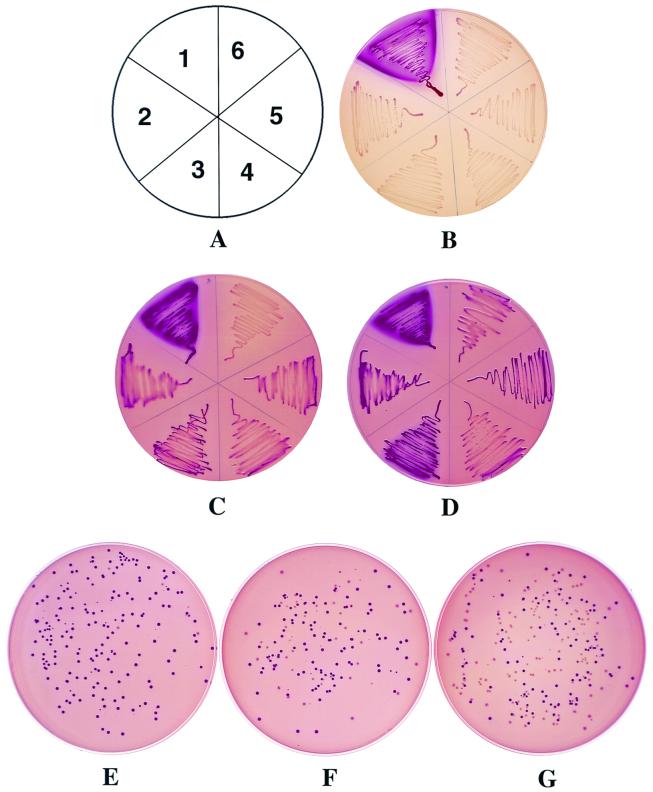
We then tested whether this new design could allow us to detect the protease activity of variants that are less active than the wild-type HIV protease, like the B3 variant described above. The DNAs containing the full protease coding region and the flanking processing sequences were amplified from the variants B1, B3, V1, and V2 and cloned in frame in place of the wild-type HIV protease sequence into pKAC-HIV-Pr. The resulting plasmids were transformed into DHT1, and cells were plated on MacConkey-maltose plates. As shown in Fig. 5B, all transformants expressing AC-protease fusions, including protease B3, now exhibited a Cya− phenotype. This indicates that all these variants were able to autoproteolyse efficiently in vivo with a simultaneous inactivation of the AC activity of the fusion proteins.
FIG. 5.
Phenotypic assay of mutant HIV protease activities. DHT1 bacteria were transformed with pKACp5 (1), pKACPr (2), pKACB1 (3), pKACB3 (4), pKACV1 (5), or pKACV2 (6). Transformants were plated on MacConkey-maltose plates containing kanamycin and either no protease inhibitor (B), 20 μM saquinavir (C), or 40 μM saquinavir (D). Plates were incubated at 30°C for 36 h. (E through G) Model screening of an HIV protease mutant resistant to saquinavir. DHT1 bacteria were transformed with mixtures of pKACV2 and pKACPr at ratios of 1/100 (E), 1/10 (F), and 1/1 (G). Transformants were plated on MacConkey-maltose plates containing kanamycin and 20 μM saquinavir. Plates were incubated at 30°C for 40 h.
When the transformants were plated on MacConkey-maltose medium supplemented with a high concentration of saquinavir (Fig. 5D), they acquired a Cya+ phenotype as expected. Importantly, with a lower concentration of this inhibitor, the cells expressing AC-V2 exhibited a Cya− phenotype whereas those expressing the AC fused to wild-type protease or other variants exhibited a Cya+ phenotype (Fig. 5C). These data indicate that under these conditions, protease V2 was still able to autoproteolyse and inactivate the corresponding fusion protein, as expected for an antiprotease-resistant variant.
In order to determine whether this system could be used to identify the minor fraction of protease variants that are resistant to a given inhibitor among an excess of sensitive proteases, we performed a model screening. Plasmid pKACV2 (encoding the AC-V2 fusion protein, which is resistant to saquinavir) was mixed with plasmid pKACPr at various ratios (1/1, 1/10, and 1/100) and the different mixtures were transformed into DHT1. Transformants were plated on MacConkey-maltose medium supplemented with 20 μM saquinavir or left unsupplemented. In the absence of the inhibitor, all transformants were Cya− (results not shown). In the presence of the drug, a mixture of Cya− and Cya+ colonies was evidenced; as shown in Fig. 5E to G, the ratios of Cya− cells to Cya+ cells paralleled the ratios of pKACV2 to pKACPr plasmids, suggesting that the Cya− colonies expressed the V2 variant resistant to saquinavir. Plasmid DNA analysis confirmed that all the Cya− colonies harbored pKACV2 whereas all the Cya+ colonies tested harbored pKACPr (data not shown). These experiments demonstrate that this genetic test permits an easy distinction, at the phenotypic level, between antiprotease-sensitive and antiprotease-resistant variants of the HIV protease. This test could be employed to detect a minor fraction of HIVs expressing resistance to a given protease inhibitor among a vast majority of sensitive ones.
DISCUSSION
We have designed a powerful genetic test that permits an easy in vivo characterization of specific proteases in E. coli. This system relies on coupling of a protease activity to the degradation of a signaling enzyme, the AC from B. pertussis. We took advantage of both the modular structure of the catalytic domain of AC and the well-known cAMP signaling cascade in E. coli (13, 21). AC can tolerate large in-frame polypeptide insertions between its two subdomains (T25 and T18) without alteration of its enzymatic activity; therefore, an exogenous polypeptide fragment encompassing the proteolytic site(s) of a given protease can be easily inserted between the two subdomains of AC to yield an active chimeric enzyme, which will be inactivated upon selective cleavage. In this study, either an HIV protease specific cleavage site or the HIV protease itself with its flanking processing sites was inserted in frame in AC. Proteolysis or autoproteolysis of the chimeric AC was efficient enough to abolish cAMP synthesis; as a consequence, the transformed E. coli cells displayed a Cya− phenotype, which could be easily monitored. Addition of protease inhibitors restored the Cya+ phenotype.
Previous attempts to design a genetic test for HIV protease in E. coli had limited success mainly because of the low catalytic efficiency of this protease. Most of these genetic assays were based on the proteolytic inactivation of reporter enzymes or regulatory proteins such as transcriptional activators or repressors (2, 6, 19, 32, 34). One key feature that determines the sensitivity of these assays is the extent of inactivation of the target protein under physiological conditions. In most cases, the residual activity exhibited by the fraction of uncleaved target did not allow a phenotypic distinction between cells that express the specific protease and those that do not.
The sensitivity of the AC inactivation assay is remarkable, as evidenced by its ability to detect proteolytic activity of modified proteases that are resistant to therapeutic inhibitors and, as a consequence, exhibit lower catalytic activity than wild-type HIV protease. This sensitivity can be accounted for, most likely, by two characteristics of AC (8, 21).
(i) AC exhibits a very low catalytic activity when expressed in E. coli cya (in the absence of its natural activator, the eukaryotic calmodulin protein). Therefore, the synthesis of cAMP required to confer a Cya+ phenotype to the E. coli cya cells needs a fairly large number of intact AC molecules. Hence, upon proteolytic inactivation, the fraction of uncleaved AC is probably not sufficient to maintain a Cya+ phenotype.
(ii) AC is a modular protein that can tolerate large in-frame insertions between the subdomains T25 and T18. In addition, the isolated T25 and T18 fragments have no detectable affinity for each other, and, furthermore, the T18 fragment itself seems to be largely unstructured (in the absence of its activator calmodulin [D.L., unpublished observations]). Therefore, it is likely that the inserted polypeptide sequence, corresponding to a given proteolytic site, is subject to few structural constraints and as a consequence might be easily accessible to a coexpressed protease. This was probably not the case when protease-specific cleavage sites were inserted into permissive sites of enzymes such as β-galactosidase or thymidylate synthase (2, 19).
In addition, the great tolerance of AC to insertions was shown to be particularly advantageous when the full-length HIV protease was inserted into AC. Indeed, the autoproteolysis of the protease precursor was found to be more efficient than the design of proteolysis in trans, and it was therefore more sensitive in detecting weak protease activity, as shown here for variant B3.
We have shown here that this genetic test is able to identify protease variants that are resistant to a given protease inhibitor. When cloned into AC, these protease variants autoproteolysed even in the presence of the inhibitor; therefore, the transformants displayed a Cya− phenotype, whereas cells expressing the wild-type protease were Cya+. It should be possible to use this genetic test to identify, in HIV-infected patients undergoing HAART, viral clones that harbor protease variants resistant to a given inhibitor (3, 7, 16, 41). The ability to detect, at a very early stage or even before initiation of an antiprotease treatment, the emergence of HIV variants resistant to given drugs should have major clinical benefits (9, 27, 30).
Due to its simplicity, this genetic approach should be generally applicable to the cloning of new proteases, to the identification of target sites of known proteases whose physiological target proteins are unknown, or to large-scale screening of new protease inhibitors. Such screening requires a selection procedure for cells that express a protease that can inactivate the target AC. This could be easily set up, as it is known that Cya− bacteria are resistant to different antibiotics, including mecillinam (10), whereas Cya+ cells are sensitive. Alternative selection modes could be easily engineered by placing toxic genes under the control of a cAMP/catabolic gene activator protein-dependent promoter (15).
ACKNOWLEDGMENTS
We thank François Clavel for the kind gift of materials and stimulating discussions. We also thank Nicolaus Heveker for the gift of plasmid pNH1 and Marina Perrotte for kind gift of P1 lysate from a cya-deficient strain.
Financial support came from the Institut Pasteur and from the Centre National de la Recherche Scientifique (CNRS, URA 2185, Biologie Structurale et Agents Infectieux).
N. D. and G. K. contributed equally to this work.
REFERENCES
- 1.Bartolome B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 2.Baum E Z, Bebernitz G A, Gluzman Y. Beta-galactosidase containing a human immunodeficiency virus protease cleavage site is cleaved and inactivated by human immunodeficiency virus protease. Proc Natl Acad Sci USA. 1990;87:10023–10027. doi: 10.1073/pnas.87.24.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borman A M, Paulous S, Clavel F. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J Gen Virol. 1996;77:419–426. doi: 10.1099/0022-1317-77-3-419. [DOI] [PubMed] [Google Scholar]
- 4.Brickman E, Soll L, Beckwith J. Genetic characterization of mutations which affect catabolite-sensitive operons in Escherichia coli, including deletions of the gene for adenyl cyclase. J Bacteriol. 1973;116:582–587. doi: 10.1128/jb.116.2.582-587.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, et al. In vivo emergence of HIV-1 variants resistant to multiple protease. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 6.Dasmahapatra B, DiDomenico B, Dwyer S, Ma J, Sadowski I, Schwartz J. A genetic system for studying the activity of a proteolytic enzyme. Proc Natl Acad Sci USA. 1992;89:4159–4162. doi: 10.1073/pnas.89.9.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dulioust A, Paulous S, Guillemot L, Delavalle A M, Boue F, Clavel F. Constrained evolution of human immunodeficiency virus type 1 protease during sequential therapy with two distinct protease inhibitors. J Virol. 1999;73:850–854. doi: 10.1128/jvi.73.1.850-854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser P, Ladant D, Sezer O, Pichot F, Ullmann A, Danchin A. The calmodulin-sensitive adenylate cyclase of Bordetella pertussis: cloning and expression in Escherichia coli. Mol Microbiol. 1988;2:19–30. [PubMed] [Google Scholar]
- 9.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 10.Jaffe A, Chabbert Y A, Derlot E. Selection and characterization of beta-lactam-resistant Escherichia coli K-12 mutants. Antimicrob Agents Chemother. 1983;23:622–625. doi: 10.1128/aac.23.4.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan A H, Michael S F, Wehbie R S, Knigge M F, Paul D A, Everitt L, Kempf D J, Norbeck D W, Erickson J W, Swanstrom R. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc Natl Acad Sci USA. 1994;91:5597–5601. doi: 10.1073/pnas.91.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karimova G, Fayolle C, Gmira S, Ullmann A, Leclerc C, Ladant D. Charge-dependent translocation of Bordetella pertussis adenylate cyclase toxin into eukaryotic cells: implication for the in vivo delivery of CD8(+) T cell epitopes into antigen-presenting cells. Proc Natl Acad Sci USA. 1998;95:12532–12537. doi: 10.1073/pnas.95.21.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 15.Kellam P, Larder B A. Recombinant virus assay: a rapid phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1994;38:23–30. doi: 10.1128/aac.38.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klabe R M, Bacheler L T, Ala P J, Erickson V S, Meek J L. Resistance to HIV protease inhibitors: a comparison of enzyme inhibition and antiviral potency. Biochemistry. 1998;37:8735–8742. doi: 10.1021/bi972555l. [DOI] [PubMed] [Google Scholar]
- 17.Kohl N E, Emini E A, Schleif W A, Davis L J, Heimbach J C, Dixon R A, Scolnick E M, Sigal I S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo L C, Shafer J A. Retroviral proteases. Methods Enzymol. Vol. 241 1994. [Google Scholar]
- 19.Kupiec J J, Hazebrouck S, Leste L T, Sonigo P. Conversion of thymidylate synthase into an HIV protease substrate. J Biol Chem. 1996;271:18465–18470. doi: 10.1074/jbc.271.31.18465. [DOI] [PubMed] [Google Scholar]
- 20.Ladant D. Interaction of Bordetella pertussis adenylate cyclase with calmodulin: identification of two separated calmodulin-binding domains. J Biol Chem. 1988;263:2612–2618. [PubMed] [Google Scholar]
- 21.Ladant D, Ullmann A. Bordetella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol. 1999;7:172–176. doi: 10.1016/s0966-842x(99)01468-7. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y, Lin X, Hong L, Foundling S, Heinrikson R L, Thaisrivongs S, Leelamanit W, Raterman D, Shah M, Dunn B M, et al. Effect of point mutations on the kinetics and the inhibition of human immunodeficiency virus type 1 protease: relationship to drug resistance. Biochemistry. 1995;34:1143–1152. doi: 10.1021/bi00004a007. [DOI] [PubMed] [Google Scholar]
- 23.Loeb D D, Swanstrom R, Everitt L, Manchester M, Stamper S E, Hutchison C A., III Complete mutagenesis of the HIV-1 protease. Nature. 1989;340:397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 25.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 26.Pardee A B, Jacob F, Monod J. The genetic control and cytoplasmic expression of inducibility in the synthesis of β-galactosidase of Escherichia coli. J Mol Biol. 1959;1:165–168. [Google Scholar]
- 27.Perrin L, Telenti A. HIV treatment failure: testing for HIV resistance in clinical practice. Science. 1998;280:1871–1873. doi: 10.1126/science.280.5371.1871. [DOI] [PubMed] [Google Scholar]
- 28.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 29.Ridky T, Leis J. Development of drug resistance to HIV-1 protease inhibitors. J Biol Chem. 1995;270:29621–29623. doi: 10.1074/jbc.270.50.29621. [DOI] [PubMed] [Google Scholar]
- 30.Roberts N A, Craig J C, Sheldon J. Resistance and cross-resistance with saquinavir and other HIV protease inhibitors: theory and practice. AIDS. 1998;12:453–460. doi: 10.1097/00002030-199805000-00005. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sices H J, Kristie T M. A genetic screen for the isolation and characterization of site-specific proteases. Proc Natl Acad Sci USA. 1998;95:2828–2833. doi: 10.1073/pnas.95.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sommadossi J P. HIV protease inhibitors: pharmacologic and metabolic distinctions. AIDS. 1999;13(Suppl. 1):S29–S40. [PubMed] [Google Scholar]
- 34.Stebbins J, Deckman I C, Richardson S B, Debouck C. A heterologous substrate assay for the HIV-1 protease engineered in Escherichia coli. Anal Biochem. 1996;242:90–94. doi: 10.1006/abio.1996.0433. [DOI] [PubMed] [Google Scholar]
- 35.Ullmann A, Danchin A. Role of cyclic AMP in bacteria. Adv Cyclic Nucleotide Res. 1983;15:1–53. [Google Scholar]
- 36.Vanaman T C, Bradshaw R A. Proteases in cellular regulation minireview series. J Biol Chem. 1999;274:20047. doi: 10.1074/jbc.274.29.20047. [DOI] [PubMed] [Google Scholar]
- 37.Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- 38.Wanner B L. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1986;191:39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- 39.Wlodawer A, Erickson J W. Structure-based inhibitors of HIV-1 protease. Annu Rev Biochem. 1993;62:543–585. doi: 10.1146/annurev.bi.62.070193.002551. [DOI] [PubMed] [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 41.Yerly S, Kaiser L, Race E, Bru J P, Clavel F, Perrin L. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet. 1999;354:729–733. doi: 10.1016/S0140-6736(98)12262-6. [DOI] [PubMed] [Google Scholar]
- 42.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]