Abstract
The carboxy-terminus of Hsp70-interacting protein (CHIP) is a ubiquitin ligase and co-chaperone belonging to Ubox family that plays a crucial role in the maintenance of cellular homeostasis by switching the equilibrium of the folding-refolding mechanism towards the proteasomal or lysosomal degradation pathway. It links molecular chaperones viz. HSC70, HSP70 and HSP90 with ubiquitin proteasome system (UPS), acting as a quality control system. CHIP contains charged domain in between N-terminal tetratricopeptide repeat (TPR) and C-terminal Ubox domain. TPR domain interacts with the aberrant client proteins via chaperones while Ubox domain facilitates the ubiquitin transfer to the client proteins for ubiquitination. Thus, CHIP is a classic molecule that executes ubiquitination for degradation of client proteins. Further, CHIP has been found to be indulged in cellular differentiation, proliferation, metastasis and tumorigenesis. Additionally, CHIP can play its dual role as a tumor suppressor as well as an oncogene in numerous malignancies, thus acting as a double agent. Here, in this review, we have reported almost all substrates of CHIP established till date and classified them according to the hallmarks of cancer. In addition, we discussed about its architectural alignment, tissue specific expression, sub-cellular localization, folding-refolding mechanisms of client proteins, E4 ligase activity, normal physiological roles, as well as involvement in various diseases and tumor biology. Further, we aim to discuss its importance in HSP90 inhibitors mediated cancer therapy. Thus, this report concludes that CHIP may be a promising and worthy drug target towards pharmaceutical industry for drug development.
Keywords: Chaperones (HSC70/HSP70 & HSP90), CHIP, Oncogene, Therapy, Tumor suppressor, Ubiquitin proteasome system (UPS)
Introduction
The functional state of proteins changes into malfunctioned state that results in serious ailments related to neurological, immune, cardiovascular systems and so on. As a prevention to these ailments, proteome will try to maintain the cellular homeostasis by two ways: (a) via proteasomal abasement of malfunctioned polypeptides and (b) via molecular chaperones mediated refolding. Cellular homeostasis is well maintained by proper balance between molecular chaperones and degradation machinery. Proteasome mediated abasement of malfunctioned polypeptides occur through ubiquitin proteasome system (UPS) or autophagy, which may works in an independent as well as synergistic manner.1,2 On the other hand, molecular chaperone (like HSP70, HSP90 etc.) behaves like an inspector that inspects and guides the folding machinery of proteome to recycle the malfunctioned polypeptides into their native form. Furthermore, molecular chaperones also assist the proteasome mediated degradation machinery along with their role in refolding machinery.3,4
UPS is a multi-component system that regulates cellular threshold of malfunctioned and misfolded proteins or polypeptide through degradation in some bacteria,5 all eukaryotic cells,6, 7, 8, 9, 10 and archaea.11 Proteasome mediated protein degradation is catalyzed by series of enzymatic reactions. UPS involves three well known enzymes viz ubiquitin activating enzymes (E1s), ubiquitin conjugating enzymes (E2s) and ubiquitin ligase enzymes (E3s) (Fig. 1).12 The fourth one is known as ubiquitin chain elongating enzymes (E4s) that also found in UPS mediated degradation machinery.13 Ubiquitin (∼8.5 kDa protein)14, 15, 16 is activated and conjugated to the E2 enzyme in the presence of ATP and E1 enzyme.17 Next, E2-ubiquitin complex18 is catalyzed in the presence of E3 ubiquitin ligases and transferred the ubiquitin molecule to the client protein. Peptide bonds are formed between specific lysine residues of client proteins and glycine residue in the carboxy terminus of ubiquitin molecule.19,20 E4 type of enzymes are also involved in the degradation machinery of UPS, responsible for polyubiquitination of client proteins. Seven types of ubiquitin lysine residues (K6, K11, K27, K29, K33, K48 and K63) are responsible for polyubiquitination. Polyubiquitinated client proteins are transferred and degraded by 26s proteasome.21 Secondly, autophagy is another system of maintaining homeostasis of proteome by lysosome mediated proteolysis. Cellular abnormal proteins are automatically engulfed by phagosome called autophagosome. Autophagosome is further engulfed by a lysosome complex, called auto-phagolysosome and is ultimately degraded through lysosomal hydrolysis.22 Furthermore, chaperones are also involved in lysosome mediated proteolysis viz, chaperone assisted selective autophagy (CASA) and chaperone-mediated autophagy (CMA).22, 23, 24 Over 500 E3 ligases are identified till date,25 here in this report, we are presenting an overview of up-to-date information of CHIP with special attention to its dual role in cancer and possible strategy for combinatorial use with HSP90 inhibitors.
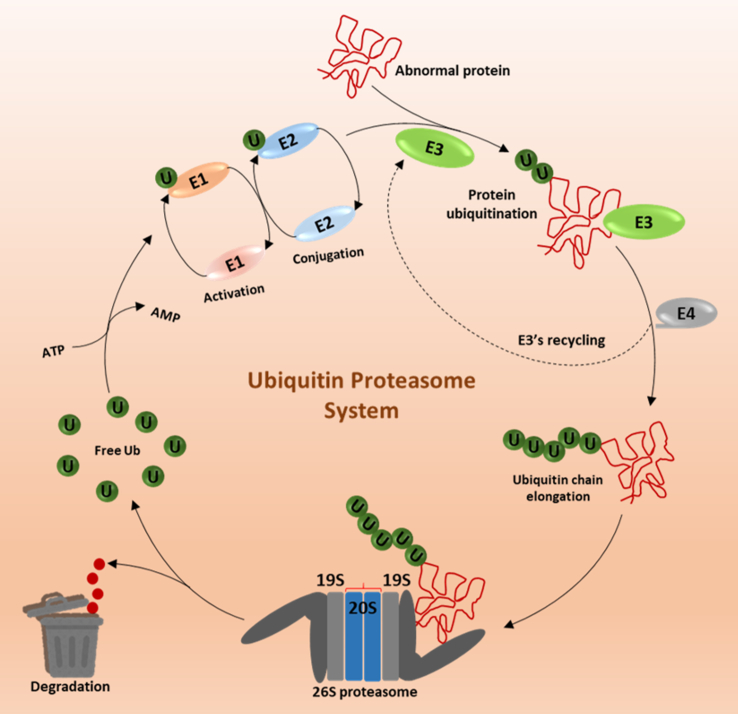
Figure 1.
Ubiquitin proteasome system (UPS). ATP mediated activation of free ubiquitin molecules occurs via binding of free ubiquitin to the ubiquitin binding enzyme (E1's) through a thioester linkage, subsequently the activated ubiquitin molecule gets transferred to ubiquitin conjugating enzymes (E2's). This Ub-E2's subsequently forms a complex with abnormal protein and with a specific E3 ubiquitin ligase enzyme which is responsible for transferring the ubiquitin moiety to the amino acid residue lysine present within the abnormal client protein that leads to poly-ubiquitination and 26S proteasome mediated degradation. Extensive role of E3 ubiquitin ligases as an E4 enzyme discovered recently as a new component of UPS is responsible for the elongation of ubiquitination chain. Finally, the ubiquitin molecules are recycled and again gets involved in next cycles of UPS.
CHIP: discovery, gene location, mutations, architectural alignment, tissues specific expression and sub-cellular localization
CHIP is playing important and selective role in the proteolysis of its client proteins through UPS. It is responsible for its dual action as a co-chaperone and an E3 ligase. It plays a major role in maintaining tissue homeostasis through maintenance of cellular threshold of client proteins including oncogenes and tumor suppressors by UPS/lysosomal degradation.26,27 In this section, we are providing a brief overview into its discovery, gene location, mutations, architectural alignment, tissues specific expression and its sub-cellular localization.
Discovery
Tetratricopeptide repeat (TPR) domain of CHIP is a key element responsible for protein–protein interactions. Based upon this fact Ballinger and his colleagues tried to find out the novel interacting proteins of TPR domain. Therefore, his team had started and successfully screened the human heart cDNA library with a fraction of cyclophilin-40 containing three TPR repeats. Through this screening, CHIP (a 34.8 kDa encoded protein) containing three TPR repeats at its N-terminus with Ubox domain at C-terminus was identified.28, 29, 30 Furthermore, amino acid arrangements of human CHIP have similarity with ∼98% of mouse CHIP and ∼60% of fruit fly CHIP. Surprisingly, Ubox domain regions of all three species have 87% amino acid similarity which indicates that 94 amino acid residues of Ubox domain of these species are similar and found to be highly conserved.31
Gene location and mutations
Protein CHIP is encoded by gene STUB1 (STIP1 homology and U-box containing gene-1) which is located at p13.3 in chromosome number 16. In humans, it consists of 7 exons and 6 introns and translated into 303 amino acid containing protein.28 Till date, two isoforms of human CHIP containing 303 & 231 amino acids are reported with Uniprot IDs Q9UNE7-1 and Q9UNE7-2 respectively; but no experimental proofs are available for second isoform.
Mutations are the changes in DNA sequence that can occur during copying of DNA or may be induced by some environmental factors (viz., UV light, smoking, chemical reagents etc.) that can produce serious illness.32,33 Recent reports state that STUB1 gene mutations are associated with various major diseases like Gorden Holmes syndrome (GHS),34,35 Machado-Joseph disease (MJD)36,37 and intracranial aneurysm (IA).38 GHS is characterized by hereditary cerebellar ataxia along with hypogonadotropic hypogonadism,39 whereas MJD is characterized by progressive cerebellar ataxia that results in loss of muscle co-ordination.40,41 Mutations in RNF216 and OTUD1 genes are also associated with GHS syndrome. Patients with these mutations have similar neurological characteristics to the patients with STUB1 gene mutations.35,42 Furthermore, STUB1 gene mutation also attenuates E3 ligase activity and results into inhibition of proteasome mediated degradation of client protein hypoxia inducible factor-1 alpha (HIF1α).43 Till date, the limited number of studies has been performed over the STUB1 gene mutations and its involvement in the disease prognosis. Thus, more attention and focused research is needed in the area of STUB1 gene mutations borne diseases.
Architectural alignment
CHIP contains three TPR repeats at amino terminus, Ubox domain at its carboxyl terminus and both the structural units are integrated through a bridge called helical hairpin (HH) (Fig. 2A).28 Three sets of anti-parallel alpha helices are present in all three TPR repeats. The structural linkage between 1st and 2nd TPR repeats or 2nd and 3rd TPR repeats look like a ‘Knob and Hole’ structure. Hydrophobicity of this structure plays a key role in protein–protein interaction through each TPR unit.44,45 Association of client proteins with TPR domain of CHIP mostly occurs through molecular chaperones like HSC70/HSP70 and HSP90 29,46,47. Additionally, some reports revealed that TPR domain may also interact directly with client proteins in a chaperone independent manner.48 The extended seventh helix present at the end of third set of TPR repeat is connected to helical hairpin. Two extended long alpha helices containing ‘Helical Hairpin’ (HH) further associates with Ubox domain. Ubox domain is organized as “Two beta hairpin - 1st alpha helix - 3rd beta hairpin - 2nd alpha helix”.49 Previously, E3 ligases are belonged to HECT and RING finger family. Later it was established that the E3 ligases cause poly-ubiquitination and found that it belongs to Ubox family.50,51 Ubox family is quiet similar to RING finger domain, only difference is that RING finger domain containing family is stabilized by Zn binding whereas Ubox family is stabilized by hydrogen bonding, and rest remains same.52,53
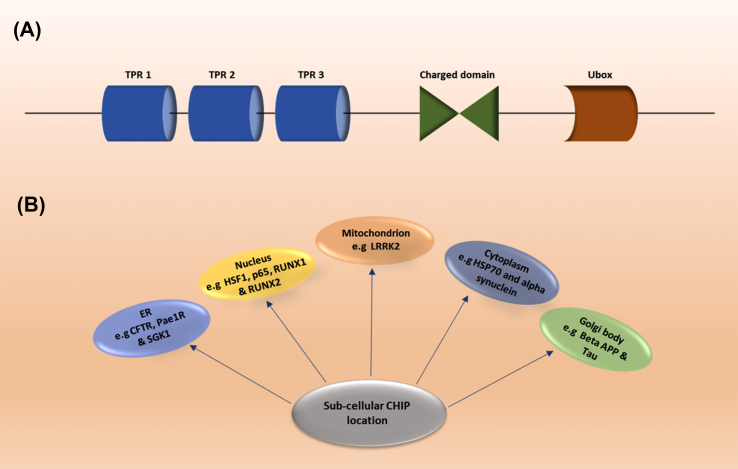
Figure 2.
Architectural alignment and subcellular localization of CHIP. (A) The schematic diagram represents the architectural alignment of all the three domains (TPR, Ubox and Charged) of CHIP. TPR (tetratricopeptide repeats) domain is present at the N-terminus of CHIP, while Ubox domain is at C-terminus. The charged domain is localized in between TPR and Ubox domains of CHIP. (B) The diagram represents the sub-cellular localization of CHIP in endoplasmic reticulum (ER), mitochondria, cytoplasm, nucleus and golgi bodies.
Tissues specific expression and subcellular localization
In general, protein CHIP is expressed in almost all type of tissues, but the expression levels differ in different tissues. Tissues having high expression of CHIP are skeletal muscle, heart, and brain whereas pancreas, lung, liver, placenta, and kidneys express CHIP at low levels. Generally, tissues with more metabolic activity and protein turnover have high CHIP expression level.46
At the time of discovery, CHIP was studied as a cytoplasmic protein, but as the research proceeds in last few decades various studies revealed that CHIP is widely distributed throughout the cell28 (Fig. 2B). According to the literature CHIP is localized at various cellular regions and compartments but its expression is not fixed at any specific region or compartment of the cell. Client proteins of CHIP like CFTR,54 Pae1R55 and SGK156 co-localizes at endoplasmic reticulum (ER) for ubiquitination mediated proteasomal degradation. Furthermore, CHIP interacts with HSF1 and is localized within the nucleus.57 Some reports also revealed that co-localization of CHIP and its association with client proteins specifically occurs in nucleus rather than cytoplasm viz., p65 58, RUNX159 and RUNX2.60 Another study indicates that CHIP also maintain homeostasis of mitochondrial proteins along with cytoplasmic proteins e.g., leucine rich repeat kinase 2 (LRRK2).61 CHIP further maintains the balance of those proteins which are present in golgi body viz., β-amyloid precursor protein (β-APP)62 and Tau.63 Additionally, proteins such as HSP7064 and α-synuclein65 interact with CHIP at cytoplasm. Thus, the literature review revealed that CHIP is mostly present in cytoplasm and partly distributed in the nucleus. TPR domain of CHIP is associated with client proteins via chaperones (viz., HSP70, HSP90, etc.) for maintenance of cellular threshold. So, in the next section we are interested to discuss the protein folding-refolding mechanism and CHIP mediated disposal mechanism of abnormal proteins.
Molecular chaperones and CHIP
Molecular chaperone (viz., HSC70/HSP70 and HSP90) contains ATPase domain and carboxyl terminus domain (CTD) at its N-terminus and C-terminus respectively, plays important role in proper folding of the newly synthesized polypeptides as well as refolding of the abnormal/misfolded proteins.66 CHIP through its N-terminus TPR domain associates with the CTD of molecular chaperones (HSC70/HSP90). Refolding, proteasomal degradation and lysosomal degradation of abnormal or malfunctioned proteins depends on the interaction of molecular chaperones with the CHIP.28,67 Here, we classified the mechanism of association of E3 ligase CHIP with the molecular chaperones into three types: (a) HSC70/HSP70 dependent, (b) HSP90 dependent and (c) Direct mechanisms for degradation of client proteins (Fig. 3).
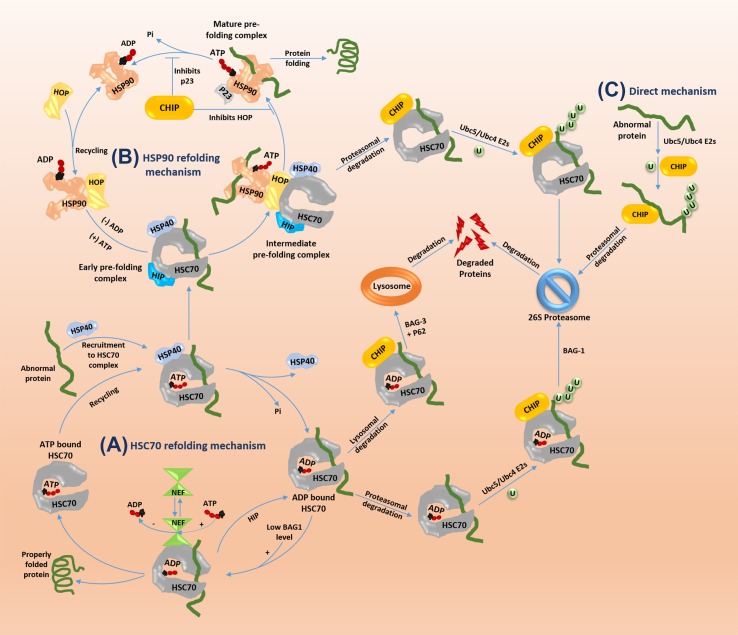
Figure 3.
Molecular chaperones (HSP70/HSC70 and HSP90) dependent, independent and direct mechanisms of CHIP mediated degradation of client proteins. CHIP mediated degradation of client proteins through inhibition of either (A) HSP70/HSC70 mediated refolding; (B) HSP90 mediated refolding or, (C) Chaperones independent route for degradation of client proteins. Arrows indicate either positive (solid) or negative (T shaped) regulation.
HSC70/HSP70 dependent mechanism
HSC70/HSP70 is a molecular chaperone; with the help of co-chaperones, it promotes either the proper refolding of the abnormal/unfolded/improperly folded proteins to their native states or CHIP mediated proteasomal or lysosomal degradation.28,67,68 HSC70 – HSP40 complex promotes protein folding while E3 ligase CHIP competes with HSC70-HSP40 complex for degradation.69, 70, 71, 72, 73, 74 Literature review suggests that chaperones viz., HSC70/HSP70 and HSP90 are actively competes with each other for re-folding of abnormal client proteins to their native form.75,76 Furthermore, in a normal physiological conditions ATP bound HSC70 is known for its low affinity for substrates and for its fast exchange rate while both the conditions are reverse for ADP bound HSC70/HSP70.28,46,77 Co-chaperones both HSC70 interacting protein (HIP) and HSP40 binds with the N-terminus ATPase domain of HSC70. HSP40 increases the ATPase activity of N-terminus domain of HSC70/HSP70 while HIP enhances the affinity of client proteins by stabilizing the ADP-bound form of HSC70/HSP70.46,78 ‘CHIP triage’ is a condition which decides chaperone bound client proteins either refolded back to its native state or degraded by proteasomal or lysosomal pathway.67 Additionally, recent reports revealed that chaperone and proteasome system compete, where chaperone system try to save proteins by proper folding while proteasome try to dispose-off the damaged proteins. Some reviews suggest that decision is also dependent on the state of misfolded protein or amount of abnormality in the protein. Chaperone system will try to refold the abnormal proteins, if not possible then proteasomal system will dispose-off the damaged proteins.46,75,79, 80, 81, 82 CHIP attenuates the HIP functionality and inhibits HSP40 ATPase activity, that results in the inhibition of HSP70 dependent refolding cycle.28 HSC70/HSP70 bound client proteins may be degraded either through proteasomal degradation (BAG1 mediated) or lysosomal degradation (BAG3 mediated). BAG1 is a HSP70 associated co-chaperone that in a low concentration promotes the folding of proteins while in a high concentration it promotes the degradation of abnormal proteins through the 26S proteasome.83, 84, 85, 86, 87, 88, 89 Secondly, in some cases BAG3 recruits p62 to CHIP-HSC70/HSP70 complex. Due to this switching of degradation pathway from UPS to chaperone assisted selective autophagy (CASA) occur.85,90 Furthermore, BAG2 is a co-chaperone which inhibits the E3 ligase activity of CHIP that promotes the client protein HSP72 refolding by reducing its ubiquitination.91,92
HSP90 dependent mechanism
Abnormal proteins are also either refolded back into their native functional state through HSP90 mediated refolding mechanism or may get degraded with the involvement of CHIP-HSP90 complex. According to the literature review, both CHIP-HSC70/HSP70 and CHIP-HSP90 complexes competes with each other, to prove their importance in maintaining proteome health. But according to documented reports CHIP-HSC70 complex plays a crucial role in majority of the abnormal proteins poly-ubiquitination and degradation rather than CHIP-HSP90 complex. Thus, these studies concludes that 80%–90% of CHIP client proteins degradation were documented with the involvement of CHIP-HSC70 complex while 10%–20% of client proteins degradation were documented through CHIP-HSP90 complex.93,94 In a HSP90 dependent mechanism, HSP90 associates with major co-chaperones viz., p23 and HOP.28,94, 95, 96 Co-chaperone p23 associates with ATPase like activity domain of HSP90 and responsible for reducing its ATPase like activity, that results in stabilization of the ATP bound state of HSP90.8 Co-chaperone HSC70-HSP90 organizing protein (HOP) consists of two TPR domains 1 at N-terminus while other at its C-terminus. HSC70-HSP90 organizing protein (HOP) acts like a bridge between HSC70 and HSP90, both the chaperones work in collaboration to maintain cellular integrity of proteome. N-terminal TPR domain of HOP associates with the CTD of HSC70/HSP70 while C-terminal TPR domain of HOP associates with CTD of HSP90.28,78 Furthermore, in HSP90 dependent mechanism ‘early pre-folding complex’ is formed with the association of HSC70, HSP40, HIP and client protein. Both TPR domains of HOP unites the HSP90 chaperone with the early pre-folding complex that is known as ‘intermediate pre-folding complex’.28,29 This complex also contains co-chaperones associated with the HSP90 viz., p23, immunophilins etc. along with early pre-folding complex. With the utilization of ATP (through hydrolysis), dissociation of intermediate pre-folding complex occurs and transfer the client protein to HSP90-p23 complex known as ‘mature pre-folding complex’ or “HSP90-p23-client protein”. Mature pre-folding complex refolds the abnormal polypeptides into their functional native form.28,94,97, 98, 99 E3 ligase CHIP and p23 have their different binding sites at HSP90 but they are allosteric inhibitor to each other. Thus, the outcome of HSP90 dependent mechanism for folding/degradation is totally dependent on the competition between CHIP and p23. Furthermore, multiple reports states that if the chaperones are not able to restore unfolded/misfolded proteins back into their native form, then E3 ligase CHIP hijacks the HSP90 mediated folding mechanism by inhibiting the functionality of co-chaperones viz., p23 and HOP. Finally, CHIP-HSC70 complex containing client proteins gets ubiquitinated in the presence of Ubc4/Ubc5 E2's and degraded through the 26s proteasome.26,28,68
Direct mechanism
E3 ligase CHIP also degrades abnormal proteins in a chaperone independent manner. CHIP by consuming its internal binding ability to ubiquitinate, degrade the abnormal proteins through E2,s (Ubc5/Ubc4) and 26s proteasome dependent route without any involvement of molecular chaperones.48 Abnormal proteins those having slow refolding mechanism are directly ubiquitinated and disposed by E3 ligase CHIP viz., smad1,100 smad3101 and CLEC-2.102 Till date, there is no such evidence found related to the conditions under which cellular proteome will undergo any selected mechanisms for the degradation of client proteins. Thus, this is one of the major questions left to answer, we want to draw attention of readers to study the conditions under which CHIP is selecting the way of client protein degradation out of these three mechanisms.
Major functions of CHIP
CHIP is documented for its well-established roles in physiology as well as in various human diseases.103 Here, we enlightened the fate of E2s - E3s interactions; crucial physiological functions of CHIP as E4 ligase and in DNA repair, maintenance of cellular protein homeostasis and metabolism.
E2s and E3s interactions — rules the fortune of client proteins
E2s and E3s are mostly responsible for deciding the fortune of a client protein along with the involvement of other components of UPS. Ubiquitin binding domain of E2 family enzymes is highly conserved. These E2s work with E3s in a collaborative manner and decides the fortune of client proteins, by addition of ubiquitin moiety to specific lysine residue.104 Previously, E2 enzyme UbcH5 was known to be associated with E3 ligase CHIP without any lysine (K) residue specificity and it played a critical role in CHIP mediated ubiquitination, but the motifs for this interaction were not studied. Afterwards in many other reports, researchers found that ubiquitination of client proteins in the presence of UbcH5 and CHIP occurred via K-48 linkage.105 Furthermore, E3 ligase CHIP is also reported to associate with dimeric Ubc13-Uev1A E2 complex, and this interaction leads to poly-ubiquitination of client proteins via K-63 linkage. Additionally, researchers have successfully found a common motif (Ser-Pro-Ala) responsible for interaction between E2 enzymes (viz., UbcH5, UbcH4 & Ubc13-Uev1A) and Ubox domain of E3 ligase CHIP.104 A specific case of client protein ubiquitination via CHIP in association with both UbcH5 and UbcH6 E2 enzymes is also reported e.g. interferon regulatory factor 1 (IRF-1).106 Another group have found that eight E2 enzymes (viz., Ube2D1, Ube2D2, Ube2D3, Ube2E1, Ube2E2, Ube2E3, Ube2N and Ube2W) can specifically interact with Ubox domain of CHIP.107 Thus, CHIP can interact with a number of E2 enzymes and the outcome is too much complex and different for each interaction, hence much more research is needed in this area.
Can CHIP play like an E4 ligase?
Yes, CHIP can act like an E4 ligase and due to this it enhances the length of ubiquitin chain to the client proteins along with some other components of UPS.13 Most of the E3 ligases belongs to well-known families like HECT and RING, but in last two decades Ubox family of E3 ligases was documented as a third family represented as E4 ligases.50,51 UFD2 is the first Ubox containing protein having E4 ligase activity in yeast.50,108 A recent study revealed that CHIP is the first E3 ligase reported as an E4 ligase in mammalians e.g., CHIP mediates degradation of Parkin Associated Endothelin like Receptor (Pael receptor; Pael-R) along with Parkin in neurological disease Parkinson. Thus, this study clearly indicates that CHIP extends the ubiquitin length over Pael-R by acting as an E4 ligase and causes proteasomal degradation. Furthermore, E3 ligase CHIP also participates along with other E3 ligases in a mutual partnership.55,109
CHIP plays an important role in DNA repair
Base excision repair (BER) is a crucial pathway to re-structure smaller breaks of DNA such as single-strand breaks, base damage, and base loss. X-ray repair cross complementing group-1 (XRCC1), DNA polymerase β and DNA Ligase IIIα provides a platform to produce error less transcription and replication of damaged DNA.110, 111, 112, 113, 114 As reported in recent literature, CHIP plays an important role to control cellular levels of BER proteins. XRCC1 protein helps in the formation of DNA repair complex over the damaged DNA and stabilizes the BER proteins. CHIP in a chaperone independent manner ubiquitinates and degrades the excess BER proteins that are not taking part in DNA repair complex.115 Still the mechanism by which CHIP differentiates between excess and chromatin associated BER proteins is not documented yet. Thus, these findings suggests that CHIP plays a significant role in DNA repair system.
CHIP maintains the cell/tissue homeostasis
CHIP plays a crucial role in the maintenance of cellular homeostasis in normal as well as under various stress conditions. In the heat stress condition, apart from ubiquitination of mis-folded proteins, CHIP has been reported to interact and selectively degrade the misfolded/denatured proteins in a HSP70 independent manner. Surprisingly, CHIP stabilizes the HSP70-protein complexes to number folds in the presence of CHIP rather than its absence. Molecular mechanism underlying the contradicting dual role of CHIP in normal vs stress conditions is not fully understood yet.48 Additionally, CHIP also plays a protective role for maintaining the proteome health in induced oxidative stress conditions. Recently, it has been found that CHIP degrades the endonuclease G in normal condition but not in H2O2 induced oxidative stress. Still the question is remains to be answered that how CHIP is differentiating the endonuclease G in normal and oxidative stress conditions.116 However, in response to osmotic stress, it has been reported that activation of p38MAPK, JNK and ERKs occurs in mammalian cells.117 In sorbitol induced osmotic stress, MEKK is dephosphorylated and results in CHIP mediated ubiquitination and degradation.118 Thus, these findings suggests that CHIP plays a protective role in maintaining homeostasis against heat stress, oxidative stress, and osmotic stress. Furthermore, CHIP maintains cellular homeostasis in various stress conditions by regulating various stress induced disease factors classified as: enzymes (viz., ChAT,119 DLK,120 proto-Db,121 Endo G,122 Hsp70,123,124 Hsp72,123, 124 Hsp90,124,125 NOX4,126 SENP3,127 SirT6,128 SGK1,56 SGK3129 and SOD2130); signaling intermediates (viz., AIF,131, 132, 133 ASK1,134,135 ERK1,136 IRF-1 137, 138, 139 and MEKK2118); transcriptional factors (viz., GR67,140 and HIF-1 alpha43,141, 142, 143, 144); receptor proteins (viz., DAF-2/INSR145) and cytoskeleton protein Niemann-Pick C1146).
Additionally, CHIP also has its major role in maintaining the protein homeostasis and metabolism by regulating its various client proteins such as cytoskeleton proteins (viz., aquaporin-2 147, 148, 149, BER proteins,115,150 PrPC 151 and RHBDF2152); transcriptional factors (viz., β-catenin,153 eIF4E,154,155 hPXR T408D mutant,156 TEAD4157 and AhR125); receptor proteins (viz., GHR,158 nicotinic receptor,159 PPARγ160 and BMAL-1161); signaling intermediates (viz., MAPK3153 and phosphatase PPP3CA153); and enzymes (viz., AMPK proteins,162,163 CDK4,153 caspase 6,164 ClpP4,165 cytochrome P450A,166, 167, 168 cytochrome P450 3A4,169 NQO1,170 NIK171 and PRMT1153). Altogether, these documents conclude that CHIP maintains cellular protein metabolism and homeostasis in normal conditions as well as under stress situation.
CHIP in major human diseases
As discussed earlier, CHIP has several well established various physiological roles. In the recent past, research was focused mainly on its involvement in the initiation and progression of various human diseases such as immune disorders, neurological disorders, aging, inflammatory disorders, bone remodeling and cardiovascular disorders.
Immune system
CHIP is expressed in various immune cells such as macrophages, dendritic cells, natural killer (NK) cells, T cells, B cells etc. But still its exact role has not been studied yet in these immune cells.172,173 CHIP has its role in regulation of nuclear factor kappa-light-chain-enhancer of activated B cells.174,175 Previously, it was known that CHIP interacts with NF-kB-inducing kinase (NIK) through HSP70 and gets ubiquitinated and degraded. As the era proceeds, another group has also found that formation of the HSP70/NIK/CHIP dimer/TRAF3 complex increases the ubiquitination and degradation of NIK. They also found that degradation of NIK is not dependent on E3 ligase activity of CHIP. Thus, these studies suggests that CHIP forms a complex with other proteins including target proteins and degrades these clients without its E3 ligase activity.171,176,177 Toll like receptors play its crucial role in sensation and recognition of conserved structures of pathogens.178,179 It has been found that CHIP interacts with TLR4/9 and TLR2/7 complexes via HSP70 but does not interact with TLR3.180 CHIP binds with HSP70/protein kinase C (PKC)-ζ/SRC in TLR4/9 signaling pathway, resulting in K-63 dependent ubiquitination of both SRC and PKC-ζ, which leads to the activation of NF-κB signaling.180 Hence, these reports suggest that CHIP degrades TLR proteins and could affect the NF-κB signaling pathway.
CHIP is also known to activate the protein kinases (viz., Src/PKCζ,180 CARMA1181 and RIPK3182). K-63 linked ubiquitin chain and HSP70 are involved in the CHIP mediated recruitment and activation of Src/PKCζ protein kinase,180 while CHIP interacts with CARMA1 protein kinase through K-27 linked ubiquitin chain and promotes NF-kB activation.181 Additionally, CHIP mediates the RIPK3 ubiquitination and degradation through lysosomal pathway.182 Further, CHIP regulates various nitric oxide synthases such as iNOS,183,184 nNOS185 and eNOS.186 It interacts with iNOS and nNOS through both HSP70 and HSP90 chaperones dependent manner186 and promotes K-48 linked ubiquitination and proteasomal degradation,183, 184, 185 while eNOS interacts through HSP90 dependent manner and degraded by UPS.186 It also plays an essential role in the proteasome mediated ubiquitination and degradation of signaling intermediate FADD.187 Furthermore, it is found that CHIP mediates the regulation of various transcriptional factors (viz., HIF-1α,43,143,144 Foxp3,188, 189, 190 RFX-1,191 STAT4192 and Tat193) that are involved in immune system. It interacts with both Foxp3 and RFX-1 through K-48 linked ubiquitin chain in HSP70 dependent manner and leads to its degradation by UPS,188, 189, 190, 191 while transcriptional factor HIF-1α interacts via both K-48 and K-63 linked ubiquitin chain in HSP70 dependent manner and was found to be degraded through autophagy as well as UPS.43,143,144 CHIP also inhibits the HIV virus replication by proteasome mediated degradation of its viral component Tat.193 Thus, all these reports conclude that CHIP may play a significant role in the regulation of various immunological factors, transcriptional factors, protein kinases and various nitric oxide synthases in the immune system.
The body's defense system can acknowledge and govern the tumorigenic cells primarily through the production of cancer specific cytotoxic T lymphocytes.194,195 In a purpose to initiate anti-tumorigenic immune responses, various antigen presenting cells recognize and present the tumorigenic antigen to T cells. The activated cytotoxic T cells eliminate the tumorigenic cells and try to control the immune-suppressive tumor microenvironment.195, 196, 197 The tumor microenvironment is too much complex, mostly consisting of immune cells (viz., dendritic cells, neutrophils, T-reg cells, lymphocytes, monocytes and macrophages), adipose cells, fibroblasts, extracellular matrix and epithelial cells.198,199 Additionally, overwhelming of immune surveillance and extermination are hallmark of cancers.200,201 In the tumor micro-environment, these cells are functionally altered due to their interactions with each other, but the myeloid cells are plastic in nature in this context.198,202 Myeloid derived suppressor cells (MDSCs), neutrophils and tumor associated macrophages (TAMs) are the primary tumor associated myeloid cells and capable to suppress the immune system. The clear distinction between MDSCs and neutrophils in tumor is not well studied yet.203,204 However, monocytic-MDSCs are quite similar to TAMs, while polymorphonuclear-MDSCs are similar to tumor associated neutrophils.203 Furthermore, the release of granulocytic (G-CSF), macrophagic (M-CSF) and granulocyte-macrophage colony stimulating factor (GM-CSF) by tumor cells results in granulo-monocytopoiesis and extension of MDSCs in tumor microenvironment.205 The expansion of MDSCs in tumor microenvironment is also possible in a response to pro-inflammatory cytokines IFN-γ and IL-6.206,207 Additionally, various other cytokines (viz., IL-13 and IL-4) and TGF-β are also present in tumor microenvironment and promote the immunosuppressive role of MDSCs.208 On the other hand, TAMs are involved in immune linked cancer development as well as in tumor suppression. Presence of M1 pro-inflammatory phenotype of macrophages in tumor microenvironment encourages the tumorigenic transformation at the site of chronic inflammation, while the presence of M2 phenotype of macrophages promotes the immunosuppressive role.209, 210, 211, 212 Additionally, in most of the solid tumors, infiltration of macrophages is noticed199,209,212, 213, 214, 215 and responsible for poor prognosis in breast cancer, glioblastoma and lung cancer.216, 217, 218, 219, 220 Thus, this review concludes that immune system tightly resembles the cancer development and tumor microenvironment. However, the role of CHIP in immunity has been well explained in the previous section. CHIP is present in various immune cells of tumor microenvironment (viz., macrophages, T-cells, neutrophils, NK cells etc.), but its exact role in these cells is not studied yet.172,173 Also, the direct role of CHIP in immune response mediated tumor development and in tumor microenvironment is not studied well. CHIP mediates the indirect regulation of inflammatory mediators, tumorigenic signaling molecules and microenvironment factors, suggesting that CHIP might has its role in immune response mediated tumor biology. As discussed earlier, CHIP mediates the regulation of TLR proteins in immune cells and results in the activation of NF-κB signaling pathway in inflammation and tumor biology. This report does not mention the impact of NF-κB signaling activation in tumor development and tumor microenvironment. But the activation of NF-κB signaling suggests that CHIP might has its impact on the correlation between immune response and tumor development.171,176, 177, 178,180 Recently a research group has studied the Parkin mediated regulation of CHIP-IRF1 axis which controls the anti-tumorigenic immune response and found that removal of mitochondrial membrane protein Fam73b leads to the activation of IL-12 and causes the switching of mitochondrial morphology from fusion to fission. The mitochondrial dynamics is found to affect the parkin gene expression and its mitochondrial recruitment. E3 ligase Parkin inhibits the CHIP-IRF1axis and suggests the anti-tumorigenic immune response.138 Thus, all these reports suggests that CHIP has some indirect and unknown role in immune response mediated tumor development and tumor microenvironment. We would like to draw the kind attention of researchers to study the impact of CHIP upon the correlation between immune responses and tumor biology.
Aggresome formation and maturation
Cellular toxicity due to mis-folded proteins is operated by three different mechanisms. Firstly, mis-folded proteins try to get re-folded back to their nascent form with the help of chaperones. Secondly, if not possible to refold back, then it leads to its lysosome or proteasome mediated degradation. In the last mechanism, sequestration of mis-folded proteins/polypeptides occurs into juxtanuclear detergent-insoluble inclusion bodies called aggresome.221, 222, 223 Recently, it was found that CHIP plays another vital role in the aggresome formation and maturation in the context to iNOS and CFTRΔF508. Lacking of CHIP increases the formation of cellular protein aggregates and inhibits aggresome maturation.54,224 Thus, CHIP may play distinct role in aggresome formation and maturation.
Neurodegeneration
Neurodegenerative diseases such as Parkinson's disease (PD), Alzheimer's disease (AD), amyotrophic lateral sclerosis, and Huntington's disease occurs due to accumulation of non-functional, toxic and misfolded protein aggregates.225 In the removal of these mis-folded protein aggregates of neurodegenerative diseases various E3 ligases and proteasomes are involved.226,227 One of the E3 ligase CHIP plays an important role to maintain neural homeostasis with the involvement of HSP70, HSC70 and HSP90.62,225,228 It has been reported that CHIP plays an essential role in the ubiquitination of various important factors of these neurodegenerative diseases such as Tau and APP in AD,62,227,228 Ataxin-1 & Ataxin-3 in type-1 & type-3 spino-cerebellar ataxia229,230 and malin in Lafora disease.231,232 Mechanism of CHIP mediated protection of neural system from toxicity caused by misfolded protein aggregates is not yet fully understood. However, it has been reported that limited expression of CHIP causes neural toxicity due to protein aggregation, accumulation, and induction of stress.226 E3 ligase CHIP plays an important role to maintain neural homeostasis with the involvement of HSP70, HSC70 and HSP90. CHIP also has roles in various neurodegenerative diseases by ubiquitination and degradation of receptor proteins (AR)233, 234, 235, 236, 237; signaling intermediates (viz., Ataxin-1,229,238 LRRK2,61,239,240 MKKS,241 SCAR16227 and PINK1242); cytoskeleton proteins (viz., α-synuclein,65,228,243,244 FMR1,245 mHtt,246 Tau/Phosphorylated Tau225,231,247, 248, 249, 250, 251, 252 and PolyQ229,253); and various neuronal enzymes (viz., BACE1,254 Malin,232 nNOS,183, 184, 185,255,256 SOD1123,257, 258, 259 and HDAC6248). CHIP mediated protection of neural system from toxicity caused by misfolded protein aggregates is not yet completely understood. However, it has been reported that limited expression of CHIP causes neural toxicity due to protein aggregation and accumulation as well as induction of stress.62 Thus, multiple studies suggest that CHIP plays an essential role to maintain tissue homeostasis in nervous system.
Inflammation, bone remodeling and skin diseases
CHIP contributes in controlling inflammation through regulation of interleukins 4α receptor (IL-4Rα),260 TNFα261 and NF-κB.262,263 Both interleukins- 3 and 4 signaling pathways are regulated through a common IL-4Rα receptor. CHIP interacts with IL-4Rα and leads to its UPS dependent degradation that suggests CHIP regulates the inflammation by the degradation of IL-4Rα.264,265 In arthritis patients and inflammatory diseases, TNFα level is high which leads to the degeneration of bones and cartilages. Osterix is a novel zinc finger-containing transcription factor that is essential for osteoblast differentiation and bone formation. A recent study suggests that upon CHIP up-regulation, TNFα inhibits osteoblast differentiation due to degradation of osterix.261 CHIP has its magnificent role in bone remodeling by degradation of various signaling molecules such as multiple SMAD proteins,266,267 RUNX2128,268 and TRAF family members viz. TRAF2, TRAF5, TRAF6.263,269, 270, 271 CHIP mediates the degradation of multiple SMAD proteins and thus regulates TGFβ pathway and bone morphogenetic protein (BMP) signaling.266,267 Induction of RUNX2 degradation occurs by CHIP that results in inhibition of osteoblast differentiation. NF-κB signaling has its well-known role in regulation of bone remodeling.128,268 CHIP degrades various TRAF family proteins and leads to inhibition of NF-κB signaling. Authors also found that nuclear translocation of NF-κB subunit p65 is increased upon STUB1 gene knock out in mice, resulting in reduction of osteoblast formation.263,269, 270, 271 Thus, these findings suggest that CHIP may play a significant role in inflammation and bone remodeling. On the other hand, CHIP plays a tremendous role in skin diseases by regulating cytoskeleton protein (keratins272) and enzyme (NUCB1273). Thus, all these findings indicate that CHIP may play significant roles in inflammation, bone remodeling and skin diseases.
Aging and development
Aging is a phenomenon which is characterized by structural and functional changes in all tissues and organs, resulting in reduction of regenerative capacity of tissues. In the later stage of aging, increment in production and accumulation of toxic protein aggregates occurs due to reduction in chaperones and proteasomal activity.228,274 Recent studies reveal that HSP70 has a direct link with aging.275,276 CHIP plays its beneficiary role in cellular hemostasis as well as in the control of aging process. It has been reported that in STUB1 deficient mice, lagging related biomarkers are increased due to accumulation of toxic proteins.277 Expression of leucine rich repeat kinase-2 (LRRK2) is high in STUB1 KO mouse model of PD. Level of LRRK2 is down-regulated due to increased expression and activation of cellular CHIP.225,240 Additionally, CHIP controls aging induced hyper-activated biomarkers such as Sirtuin 6 (SIRT6)278,279 and insulin receptor.145,280 Thus, these findings concludes that CHIP plays an essential role in controlling the aging process. Further, CHIP regulates various developmental processes by regulating signaling intermediates (Ca2+/S100);281 transcriptional factors (myocardin282 and RUNX260); various enzymes (viz., NEK10,283 PABPN1,284 ribosomal Protein S3,285 TFEB286 and IRE1287); and cytoskeleton proteins (viz., KCNQ4,288 katanin-p60289 and plastid precursors290). All together these findings conclude that CHIP plays a necessary role in aging and developmental processes in the human body.
Aging is highly correlated with cancer where aging is considered as a pro-tumorigenic state.291,292 Number of cellular processes such as accumulation of senescent cells in tissue results into tissue degeneration. Changes in genes and chromatin structure, stem cell impairment and autophagy are probable factors responsible for aging mediated pro-tumorigenic environment.293, 294, 295, 296, 297, 298, 299 However, chaperone associated E3 ligase CHIP has its extensive role as an anti-aging agent as discussed in earlier section. But none of these reports has clearly discussed the role of CHIP on aging mediated tumor development and in tumor microenvironment, and hence requires immediate attention.
Cardiovascular diseases
CHIP is highly expressed in heart tissues and behaves like a cardio-protective agent in heart diseases.28 Recently in a couple of reports CHIP has been linked with cardiac crashes. Accumulation of p53 in cardiac myocytes is the major feature of ischemic heart diseases.298 This report suggests that CHIP is an important E3 ligase responsible for p53299,300 and HIF-1α300 degradation. In the coronary occlusion condition, hypoxia is generated in cardiac myocytes that leads to the up-regulation of HIF-1α and down-regulation of CHIP expression and eventually the ischemic heart diseases.300 Overexpression of CHIP in combination with HSP90 inhibition (using 17AAG) leads to the cardio-protective effects.299 Further, in various reports CHIP has been studied as a beneficiary factor in cardiac diseases by degrading its client proteins classified as transcription factors (viz., ICER301 and NFATc3302); cytoskeleton proteins (viz., HERG,303 Kv1.5304 and NaCl co-transporter305); and signaling intermediates (viz., Gαs306 and SHP-1307). Thus. CHIP is an eminent cardio-protective agent.
Currently, variety of chemotherapeutic agents has been developed and are used in clinical practice for cancer therapy viz., antimetabolites, platinum-based agents, alkylating agents, anti-microtubular agents, anthracyclines and antitumor antibiotics. Additionally, various targeting agents (viz., immune checkpoint inhibitors, small molecule inhibitors and HER2 inhibitors) are also developed for cancer therapy. However, the cardiotoxicity is noticed as a prominent side effect of these chemotherapeutics. Cardiotoxicity induced by chemotherapeutics may include cardiac cell damage, abnormalities in cardiac impulse conduction, reduction in left ventricle ejection rate and fraction, disturbance in blood pressure etc.308, 309, 310, 311, 312, 313 Sunitinib malate is a small molecule tyrosine kinase inhibitor having anti-tumorigenic activity against gastrointestinal, pancreatic, neuroendocrine tumors and advanced renal cell carcinoma. The use of sunitinib is noticed to produce cardiotoxic side effects such as vascular disturbances in thromboembolic events, hypertension, heart failure and occasionally may leads to death. In the management of these side effects, various other pharmacological agents are currently used. For thromboembolic events, low molecular weight heparins (LMWH), rivaroxaban and edoxaban are preferred; to manage hypertension, angiotensin inhibitors and calcium channel blockers are preferred; and in the management of heart failure clinicians are preferring angiotensin inhibitors and cardio-selective beta blockers.308 Recently, a research group has demonstrated the use of geldanamycin (HSP90 inhibitor) to attenuate the sunitinib induced cardiotoxicity and found significant reduction in sunitinib induced cytotoxicity in rat H9C2 cardiomyocytes. They also tried with the use of other HSP90 inhibitors such as tanespimycin, BIIB021 and ganetespib in sunitinib induced heart toxicity model. Sunitinib originated cardiotoxicity is due to the induction of autophagy in H9C2 cells, while in combination with geldanamycin sunitinib induced autophagy was inhibited and induced the degradation of various autophagy associated proteins viz., Atg7, ULK1 and Beclin-1. It also demonstrated the inhibitory effects of geldanamycin in sunitinib induced cardiotoxicity in pluripotent stem cell derived cardiomyocytes. Hence, these results conclude that HSP90 inhibitors in combination with sunitinib may be a potential strategy to attenuate the cardiotoxicity of sunitinib.314 Furthermore, doxorubicin (DOX, an anthracycline based antibiotic) is also preferred in chemotherapy against prostate, cervix, breast, brain cancers and in Hodgkin's lymphoma. Lower ventricle failure, arrhythmia and heart failure are primarily noticed cardio-toxic side effects in DOX treatment. Often clinicians prefer antiarrhythmic agents to manage DOX-induced arrhythmia, while beta blocker (carvedilol) and ACE inhibitor (enalapril) are much preferred to manage doxorubicin induced heart failure and ventricular dysfunction.308 Recently, a research group has studied the inhibitory action of 17-AAG in DOX-induced cardiotoxicity.315 And another group has demonstrated that doxorubicin induced cardiotoxicity may be attenuated upon the treatment of cardiomyocytes with 17-DMAG.316,317 Thus, these studies suggest that DOX-induced cardiotoxicity may be nullified, if the HSP90 inhibitors were used in combination. At present, HSP90 inhibitors in combination with DOX and sunitinib is only studied to prevent their cardiotoxicity.314,315,317 Thus, researchers and clinicians should explore more on this strategy with an objective to control cardiotoxicity of cancer chemotherapeutics.
We have enlightened above the responsibility of CHIP in normal physiology as well as in several diseases such neurodegenerative disorders, immunological disorders, cardiac disorders, aging, inflammatory disorders, bone remodeling and skin diseases to maintain the protein quality and control the cellular threshold of clients. But in the recent era of CHIP, it is studied for its fundamental role as a double agent in cancer and most of the substrates reported till date are involved in cancer initiation and progression. Mostly, CHIP is documented for its tumor suppressor role over its oncogenic actions. Further, we are interested to focus on clinical significance of CHIP in various cancers and its involvement as a double agent.
CHIP: a double agent in cancer
In the year of 1998, CHIP was screened and discovered in heart tissue by Ballinger and his group. It plays a critical role through maintenance of protein quality and control the cellular threshold of client proteins.28,67 Its role is totally un-predictable but depends upon the status of target proteins.28,67 After its discovery, several substrates of CHIP were documented, and its involvement was found in several human diseases as discussed in the earlier sections of this review. Here authors retrieved almost all the documented substrates till December 2020 and have represented all of them in a pictorial timeline, according to the year of discovery (Fig. 4).
Figure 4.
Timeline for client proteins of CHIP. The figure depict the timeline of all documented client proteins of CHIP identified till December 2020.
CHIP has its wide variety of functions in normal physiology as well as in several diseases including cancer.27 Also, the documented client proteins were also classified in context of various disease perspectives (Table S1). Surprisingly, it is found that in the last decade, most of its substrates were documented to be involved in cancer. It was found to have a double role (either as a tumor suppressor or as an oncogene) in many cancers. Hence, we classified the substrates further according to the hallmarks of cancer200,318,319 in the context of its role as an oncogene and a tumor suppressor144,172,320 (see Fig. 5). Here, we have tried to focus on both the opposing roles of CHIP in cancerous scenario.
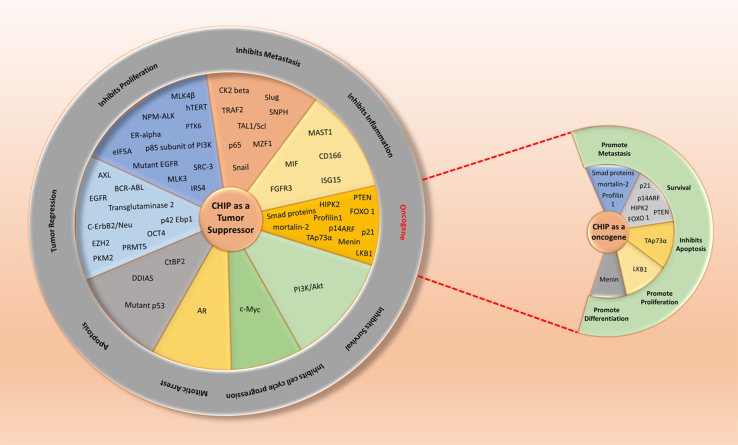
Figure 5.
Pie-in-pie chart of CHIP as a double agent in the context to cancer. CHIP client proteins classified according to the hallmarks of cancer and its dual role as a tumor suppressor (left hand sided pie chart) and as an oncogene (right hand sided pie chart) explained.
Acts as a tumor suppressor
In a pioneer study CHIP has been found as a key mediator that regulates the breast cancer progression. They found that CHIP level is negatively correlated with tumor growth and metastasis in a breast cancer model of nude mice.321 In support of this pioneer study, series of reports have been published showing the active involvement of CHIP in different cancers; this certainly creates an important debate. These reports suggest that CHIP may play as a tumor suppressor via interacting and proteasomal mediated degradation of onco-proteins. Here, in this study we made a survey of reported substrates of CHIP in cancer till date and classified them under various hallmarks of cancer (Fig. 5). In few reports, it has been observed that CHIP may acts as a tumor suppressor with the inhibition of inflammatory hallmark of cancer. In the aspect of tumor suppressive action of CHIP, it interacts with few inflammatory mediators, leading to ubiquitination and proteasome dependent degradation of target proteins viz., MIF,322 MAST1,323 CD166,324 FGFR3325 and ISG15.326 Often, MIF-HSP90 chaperone complex is reported as a highly stabilized complex in various human cancers and responsible for tumor aggressiveness. Inhibition of HSP90 chaperone with 17AAG or HSP90 specific siRNA, promotes the E3 ligase CHIP mediated MIF degradation and leads to the inhibition of inflammatory characteristics in breast cancer model.322 CHIP mediated regulation of FGFR3 also occurs through the same mechanism. Additionally, microtubule-associated serine/threonine kinase 1 (MAST1) is studied as a responsible factor for cisplatin resistance in multiple human cancers. Recently, through proteomic analysis a research group has shown HSP90B chaperone as an interacting partner and stabilizing factor of MAST1. Further, they have found E3 ligase CHIP mediates the degradation of MAST1, upon the inhibition of HSP90B chaperone and results in the improvement of cisplatin effectiveness. Thus, this report suggests that HSP90 inhibitors in combination with cisplatin may be an effective strategy to overcome the cisplatin resistance.323 Recently, a research group studied CD166 in head and neck (H&N) cancer to investigate the CHIP linked molecular mechanism and found that CHIP mediates the degradation of CD166 and leads to the inhibition of inflammatory characteristics of H&N cancer.324 Thus, all these mechanistic reports clearly indicate that how CHIP may govern the inflammatory hallmark of tumor biology.
Similarly, CHIP is negatively correlated with various substrates viz., AXL,327 C-ErbB2/Neu,173,320,328, 329, 330, 331 BCR-ABL,332,333 EGFR,334, 335, 336 EZH2,337 PKM2,338 p42 Ebp1,339 PRMT5,340 OCT4341 and trans-glutaminase 2342; results in the inhibition of tumor progression. Tyrosine kinase AXL receptor is highly expressed in multiple cancers and primarily responsible tumor progression. 17AAG dependent inhibition of HSP90 chaperone leads to destabilization of AXL through CHIP mediated degradation, resulting in inhibition of tumor progression.327 Furthermore, tyrosine kinase inhibitor imatinib is a primary drug in myeloid leukemia but resistant in chronic myeloid leukemia patients due to high expression of BCR-ABL protein. Chk1 inhibitors (AZD7762 and MK-8776) as well as oridonin promote CHIP mediated degradation of BCR-ABL protein in chronic myeloid leukemia cells. Thus, targeting Chk1 or treatment with oridonin may help to overcome the imatinib resistance in chronic myeloid leukemia patients.332,333 CHIP acts as a tumor suppressor through degradation of EGFR and its mutated forms those highly expressed in cancer.334, 335, 336 Next, CHIP can degrades oncogenic protein enhancer of zeste homolog 2 (EZH2) A novel EZH2 targeting molecule (derivative of gambogenic acid) is recently identified which covalently binds EZH2 at cys668 and promotes CHIP mediated degradation.337 Recently, through SILAC based proteomics study a research group has found pyruvate kinase iso-enzyme M2 (PKM2), an important regulatory factor for aerobic glycolysis (Warburg effect) and tumorigenesis, is a novel target protein of E3 ligase CHIP and maintain an inverse correlation in ovarian carcinoma. Thus, the Warburg effect is regulated through CHIP mediated degradation of PKM2.338 Furthermore, CHIP is expressed at low level in cancer stem cells that regulates the OCT4 stability through direct interaction and maintain an inverse correlation in breast cancer.341 Taken together, these mechanistic reports suggest that CHIP acts as a tumor suppressor that controls tumor regression hallmark in cancer.
It has been also found that CHIP inhibits cellular proliferation by ubiquitination and degradation of various substrates viz MLK4β,343 NPM-ALK,344 PTK6,345 ER-alpha,346, 347, 348, 349, 350, 351 mutant EGFR,334 eIF5A,352 hTERT,331 IRS4,353 MLK3,354 p85 subunit of PI3K355 and SRC-3.356,357 MLK3 and MLK4β are components of MAP kinase signaling pathway which regulates cellular responses like proliferation, invasion, and apoptosis. Expressions of MLK3 and MLK4β are inversely correlated with osmotic stress/thermo stress/HSP90 inhibition due to CHIP mediated degradation which results in decreased cellular proliferation of ovarian cancer cells.343,354 Similarly, inhibition of HSP90 chaperone with 17AAG promotes degradation of anaplastic lymphoma kinase (ALK)344 and PTK6,345 suggesting tumor suppressive action of CHIP.
In the screening of its substrates, it was also found that CHIP inhibits metastasis of tumor cells by targeting CK2β,358 slug,359 SNPH,360 TRAF2,361,362 TAL1/Scl,321 MZF1,363 p65 364 and snail.365 Activation of protein kinase CK2 promotes the TGFβ mediated epithelial mesenchymal transition. CHIP mediates the degradation of CK2β and results in inhibition of invasion and migration in tumor cells.358 Additionally, GSK3β mediates the slug phosphorylation that leads to its degradation by CHIP and prevent metastasis in lung cancer.359 CHIP negatively regulates the NF-κB signaling by downregulating TRAF2.362,363 A recent report indicated that CHIP and snail both are inversely correlated in ovarian carcinoma. CHIP inhibits EMT through downregulation of snail, a characteristic marker protein of EMT.365 Thus, CHIP acts as a tumor suppressor by inhibiting metastatic hallmark of cancer.
Additionally, CHIP promotes the apoptosis of tumor cells by ubiquitination and degradation of target proteins CtBP2,116 DDIAS366 and mutant p53.367 C-terminal binding protein 2 (CtBP2) and DNA damage induced apoptosis suppressor (DDIAS) are involved in cellular apoptosis, migration and tumorigenesis. These proteins are highly expressed in many cancers with low expression of CHIP. It was reported that overexpression of CHIP downregulates the CtBP2 and DDIAS proteins.116,366 CHIP mediated ubiquitination and degradation of Androgen Receptor (AR)368, 369, 370, 371, 372 results in mitotic arrest of prostate tumor cells. CHIP targets PI3K/Akt373 and inhibits the survival power of tumor cells. Onco-protein c-Myc plays its crucial role in oncogenesis. Our research group identified that CHIP targets and degrades c-Myc and results in the inhibition of cell cycle progression.374 Altogether, many reports clearly indicate that CHIP can act as a tumor suppressor in multiple cancer scenarios. CHIP mRNA and protein levels are differentially maintained in normal and breast cancer tissues. CHIP level is negatively correlated with the increment of breast cancer malignancy and overall survival of high CHIP expressing tumor bearing patients is much higher than the low CHIP expressing tumor bearing patients.375 Further, both mRNA and protein expression levels of CHIP are negatively correlated with its usual level in normal gastric tissues.58 Thus, all these reports suggest that CHIP can act as a tumor suppressor.
Acts as an oncogene
In adverse to above mentioned reports, CHIP is also seen to act like an oncogene in few of other reports. CHIP promotes proliferation and survival of tumor cells after the degradation of its substrates viz., PTEN,172,376 FOXO1,377 HIPK2,378 p14ARF379 and p21380 which are mainly engage in tumor suppression and apoptosis. Transcriptional factor FOXO1 is responsible to control the cellular proliferation and survival. Its cellular level and activity are controlled by UPS and PI3K/Akt. Overexpression of CHIP with TNF alpha treatment downregulates the FOXO1 and promotes the PI3K/Akt pathway mediated cellular proliferation and survival. Conversely, upon its knockdown, FOXO1 gets stabilized and results in the inhibition cellular proliferation and survival.377 Serine/threonine kinase HIPK2 plays crucial roles in DNA damage response and development. PARP1 regulates the HIPK2 protein stability. Furthermore, CHIP promotes the HIPK2 degradation in a PARP1 independent manner and suggests its oncogenic role.378 HSP90 in combination with CHIP promotes the lysosomal pathway mediated degradation of p14ARF. While the knockdown of CHIP in combination with HSP90 inhibition gives converse results in non-small cell lung cancer.379 Knockdown of CHIP inhibits degradation of p21 which targets the various stress related proteins and ultimately enhances the sensitivity of lung cancer cells towards radiotherapy. CHIP-HSP70-p21 axis may be potential target to improve the efficacy of radiotherapy.380
Similarly, CHIP is also reported to promote metastasis after the ubiquitination and degradation of SMAD proteins,100,267,381 Profilin1382 and Mortalin-2.383 A research group using surface plasmon resonance study established that the SMAD proteins physically interacts with CHIP. Further, another group observed that the overexpression of CHIP mediates downregulation of SMAD proteins and results in inhibition of BMP signaling.100,267,381 Profilin-1 is an important mediator of actin polymerization and regulates the cellular migration. Lower expression of profilin-1 is correlated with several malignancies. In recent study, poly-ubiquitination and degradation of profilin-1 is studied in the CHIP overexpressed breast cancer cells which results in the enhancement of cellular migration and metastasis. Converse results were obtained upon CHIP knockdown.382 Thus, all these reports indicate that CHIP may act like an oncogene by promoting metastatic hallmark of cancer. Subsequently, CHIP is found to increase cellular differentiation but not proliferation after the proteasome-mediated degradation of Menin.384, 385, 386 In another finding, CHIP mediates degradation of LKB1 and causes increment in the cellular proliferation,354,355,387 while TAp73α degradation causes anti-apoptotic action, and both finally leads to oncogenesis.388 Thus, all these reports clearly suggest that CHIP can play as an oncogene in several cancers.
Significantly high expression of CHIP was found in a metastatic lymph nodes of esophageal squamous cell carcinoma (ESCC) in comparison to primary tumors observed by using a tissue microarray approach. Also, low expression level of CHIP was found with the better survival of ESCC cancer.389 Similarly, CHIP expression level is higher in gall bladder tumor tissues with the overall decreased survival of gastric cancer patients.390 A similar phenomenon is also observed in prostate cancer.391 Hence, role of CHIP in different cancers is not that much straight forward to understand. Thus, much more study is needed to be done in this area for the deeper understanding of CHIP function in tumor biology.
Clinical significance and mechanistic details of CHIP in tumor biology
As reported in multiple cancerous tissues, CHIP expression level varies with type and grades of cancers. To inspect the variation of CHIP expression in different cancerous tissues, we screened almost every report published till December-2020 (Table 1). It was also found that tissues of breast cancer patients containing higher CHIP expression have higher overall survival rate than the patients having low expression level of CHIP.375,392,393 On the other hand, the expression level of CHIP is up-regulated in postmenopausal breast cancerous tissues.394 CHIP expression level in other cancerous tissues like gastric cancer,58,361,395,396 human renal cell carcinoma342,397 and non-small cell lung cancer398 were found to be lesser as compared to the respective normal tissues. Other cancerous tissues like glioma,399 gallbladder carcinoma,390 HBV-hepatocellular carcinoma,400 Head and neck cancer401 and colorectal cancer tissues364,402 were reported to have an increased CHIP expression level. So, we found that CHIP has wide diversity in its expression level and differential activity in different cancer scenario. Here, authors are interested to focus on the clinical significance and the mechanistic details of CHIP in tumor biology.
Table 1.
Expression of CHIP in various tumor tissues. Arrow headed (↑) and (↓) indicates up and down-regulation of CHIP.
| Year | Type of Cancer | Number of samples | In-vivo study (Yes/No) | CHIP Expression | Reference |
|---|---|---|---|---|---|
| 2010 | Breast cancer | 160 | No | ↓ | 375 |
| 2011 | Breast cancer | 183 | No | ↓ | 392 |
| Glioma | 40 | Yes | ↑ | 399 | |
| 2012 | Gastric cancer | 53 | No | ↓ | 395 |
| 2013 | Esophageal squamous cell carcinoma | 234 | No | No change | 389 |
| Gastric cancer | 640 | Yes | ↓ | 396 | |
| Gallbladder carcinoma | 78 | No | ↑ | 390 | |
| Colorectal cancer | 320 | No | ↓ | 364 | |
| 2015 | HBV-hepatocellular carcinoma | 79 | No | ↑ | 400 |
| Human renal cell carcinoma | 304 | Yes | ↓ | 397 | |
| Human renal cancer | 12 | Yes | ↓ | 342 | |
| Gastric cancer | 164 | No | ↓ | 58 | |
| 2016 | Non-small cell lung cancer | 106 | Yes | ↓ | 398 |
| Postmenopausal breast cancer | 272 | No | ↑ | 394 | |
| 2017 | Head and neck cancer | 101 | Yes | ↑ | 401 |
| 2018 | Colorectal cancer | 93 | Yes | ↑ | 402 |
| Prostate cancer | 90 | No | ↑ | 391 | |
| 2019 | Gastric cancer | 100 | Yes | ↓ | 361 |
| Breast cancer | 128 | Yes | ↓ | 393 |
Glioma
Malignant gliomas are studied as an invasive, aggressive, and most common primary brain tumors, having median survival time of 12–15 months for glioblastoma and 2–5 years for anaplastic glioma.403 In 2011, a research group has studied the CHIP expression level in 40 glioma patient samples. Among the various histological grades of glioma, authors found increase in CHIP expression level. The enhancement of colony formation and proliferation upon CHIP overexpression, while the knockdown of CHIP gives converse results in U87 and U251 glioma cell lines. Furthermore, intra-tumoral injection of CHIP shRNA containing lentiviral particles reduced the tumor size in tumor xenografts model, while opposite results were obtained after injecting CHIP overexpressed lentiviral particles. Thus, this pioneer report demonstrated that CHIP might have a tumorigenic role in human glioma.399 Furthermore, a report from our lab has demonstrated the tumor suppressive role of CHIP in glioma by degrading the proto-oncogenic transcription factor c-Myc through ubiquitin proteasome system. Here, we found that the N-terminal chaperone associated TPR domain of CHIP is responsible for interaction with c-Myc. Upon inhibition of HSP90 chaperone with 17-AAG, c-Myc protein level was decreased. Additionally, CHIP inhibits the transcriptional activity of c-Myc and results into reduction in malignant behavior of CHIP in C6 glioma cells, while knockdown of CHIP indicates opposite results. We demonstrated the molecular mechanism of its tumor suppressive role in glioma tumor biology.374 These opposing reports suggests that CHIP may acts a tumor suppressor or as an oncogene in glioma tumor biology.
Breast cancer
Breast cancer is one of the most common type of malignancy in women globally where CHIP through UPS indulged in the regulation of breast cancer prognosis.404 Till date, three clinical sample-based reports have been documented and found low expression of CHIP in breast tumor tissues than normal. Each research group concludes the overall variation in CHIP expression level. It is also noticed that breast cancer patients with increased expression of CHIP have higher overall survival rate.375,392,393 On the other hand, in another study CHIP expression level was studied in 272 postmenopausal breast cancerous tissues and up-regulation of CHIP expression level was observed in comparison to normal tissues.394 It is also reported that approximately 70% of primary breast cancer patients are ERα positive.405,406 CHIP was documented to degrade the ERα upon geldanamycin induced HSP90 inhibition.349,404,407,408 In addition to ERα, ErbB2 is also reported as a biomarker of breast cancer tissues. High expression of ErbB2 is also observed in 30% of primary breast cancer patients.409, 410, 411 CHIP is involved in ubiquitination and degradation of ErbB2. HSP90 inhibition leads to dissociation of ErbB2 from HSP90 chaperone complex and possible shifting to the HSP90 related HSP70 chaperone. Unfortunately, HSP70 associated E3 ligase CHIP hijacks ErbB2 and promotes its ubiquitination and degradation.173,320,392,412, 413, 414, 415 In addition to ERα and ErbB2, nuclear factor-κB (NF-κB) plays its crucial role in breast cancer.416,417 TNF receptor associated factor (TRAF) associated family proteins play important roles in the activation of NF-κB pathway. Upon CHIP knockdown in breast cancer cells, increment in TRAF2 protein level was observed along with enhanced invasiveness. But this enhanced invasiveness in CHIP knockdown breast cancer cells was significantly reduced upon NF-κB inhibition. Thus, these results suggest that CHIP is indulged in the regulation of TRAF2-NF-κB signaling pathway which leads cell invasion to metastasis in breast cancer.60,362,418, 419, 420
Prostate cancer
Prostate cancer is a second most cause of cancer associated deaths among the men in USA.421 A research group screened 90 prostate cancer patient samples for CHIP expression profiling and found that CHIP is highly expressed in prostate cancer tissues compared to the normal sample.391 Androgen receptor (AR) is highly expressed in prostate cancer and has crucial roles in prostate cancer progression. Thus, targeting androgen receptor may be attractive therapeutic strategy against prostate cancer. Recently a research group has documented the molecular mechanism of CHIP mediated ubiquitination and degradation of androgen receptor. They also observed that AR and CHIP interacts with each other through HSP70 chaperone.371,422,423 Furthermore, we established that CHIP degrades tumor suppressor protein PTEN in prostate cancer samples and cell lines. We also reported that TPR domain of CHIP is primarily responsible for interaction with PTEN. CHIP mediated down-regulation of PTEN activates the PI3K/AKT pathway that leads to enhanced proliferation and survival of prostate cancer cells.172 Thus, CHIP plays its dual role as a tumor suppressor as well as an oncogene in prostate tumor biology.
Gastric cancer
Most of the gastric cancer patients are diagnosed at advanced stages when tumor cells are resistant to radiotherapy and chemotherapy.424 Long time ago, a research group performed clinical sample-based CHIP expression profiling study and documented the association of reduced CHIP expression with aggressive gastric cancer phenotype and also observed the disappearance of CHIP expression in patients with high lymph node invasion. They also observed increased expression of CHIP in moderately differentiated gastric cancer samples compared to the poorly differentiated gastric cancer samples.395 Various other reports have been documented with CHIP expression profiling in variable gastric cancer samples58,364,395,402 (Table 1). Low CHIP expression in 100 gastric cancer patient samples was reported recently in 2019.361 Thus, from the above discussion it may be designated that CHIP may indulged in gastric cancer progression and can also serves as a significant diagnostic biomarker in gastric cancer patients.
Gallbladder carcinoma
Gallbladder carcinoma is typically diagnosed at later stages and is signified as a fatal disease.425 Despite of surgical resection, 10–30% of gallbladder carcinoma patients have survival rate of 5 years.426,427 Cyclooxygenase-2 and HIF are two molecular biomarkers of gallbladder carcinoma.428 CHIP expression profiling was done using 78 gallbladder carcinoma patient samples, found increased CHIP expression in carcinoma samples in comparison to normal gallbladder tissues and nicely demonstrated the relationship of CHIP expression level with clinico-pathological characteristics and patient survival rate.390 Retrospective investigations with large number of sample size are needed to be done, to understand the exact role of CHIP in gallbladder tumor biology.
Esophageal squamous cell carcinoma (ESCC)
ESCC is responsible for most of the esophageal malignant tumors, designated as most common cancer worldwide.429 To establish a molecular biomarker of ESCC, a research group has demonstrated the relationship between CHIP expression level and ESCC. They observed no significant change in CHIP expression level in ESCC primary tumors and normal epithelial tissue but increased level in ESCC metastatic lymph nodes. The higher CHIP expression in metastatic lymph nodes was found to be a prognostic factor which is not favorable in stage-III ESCC patients. CHIP expression level may represent a significant prognostic molecular biomarker in metastatic lymph nodes in ESCC.389
No definite mechanism is established yet for its dichotomous role in any type of cancer. As discussed above in different cancers, CHIP may act as a novel therapeutic target or a diagnostic molecular biomarker. Role of CHIP varies from one type to another type of cancer. Literature review suggests that it may act as tumor suppressor or as an oncogene or even both in same cancer type with individual perspectives. Its role is totally unpredicted and solely depends upon downstream targets.
Therapeutic perspectives
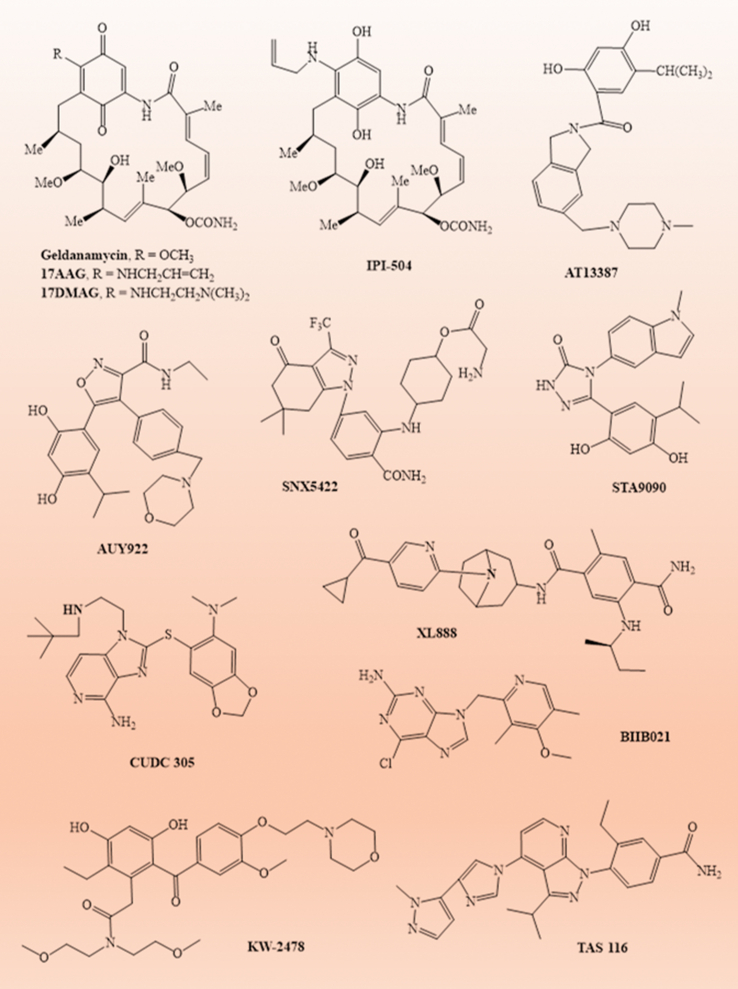
Significance of CHIP has been studied in several pathological conditions such as neurological disorders (viz., Parkinson's, Alzheimer's, Huntington's, Lafora, Atherosclerosis, Spino-cerebellar ataxia type-1 and hypo-gonadism), cardiac disorders, oxidative stress, muscular dystrophies, hyperthermia, bone related disorders and in cancer.27 Now a days, cancer is one of the prominent, non-curable and life-threatening disease.200,319,430 Till date, there is no definite, curable and life-saving strategy in cancer therapy.431,432 Furthermore, as reported till date, CHIP has its anti-tumorigenic effects by ubiquitination and degradation of several target onco-proteins in different cancers. Subsequently, it has been found that CHIP overexpression is negatively correlated with tumor growth in multiple cancers such as breast cancer and gastric cancer.58,278, 279, 280 Thus, CHIP overexpression leads to the inhibition of cancerous growth. Small molecule inhibitor of HSP90 also has anti-tumorigenic effect via stimulating the CHIP mediated degradation of oncoproteins. The reported HSP90 inhibitors in clinical trials are geldanamycin,433 17AAG,434, 435, 436, 437, 438, 439, 440, 441, 442, 443, 444, 445, 446, 447, 448, 449, 450, 451, 452, 453, 454, 455, 456, 457, 458, 459, 460, 461, 462 17DMAG,442,463, 464, 465, 466, 467 AT13387,468, 469, 470, 471, 472, 473, 474 AUY922,475, 476, 477, 478, 479, 480, 481, 482, 483, 484, 485 BIIB021,486,487 BIIB028,488 Debio0932,489 HSP990,490 IPI-504,491, 492, 493 KW2478,494,495 SNX5422,496, 497, 498, 499, 500, STA9090,501, 502, 503, 504, 505, 506, 507, 508, 509, 510, 511, 512, 513, 514, 515, 516 PU-H71,517,518 TAS-116,519, NVP-BEP800,520 CUDC 305,521, TQB3474,522 PEN8667,523 RTA-901 524 and XL888 525,526 (Fig. 6). All these compounds have tumor growth inhibitory property when used alone or in combination with standard anticancer drugs for clinical advancement (Table 2). These compounds act by blocking the ATP binding site of HSP90 which leads to CHIP mediated degradation of oncogenic client proteins results in an anti-tumorigenic effect.527 The details of clinical advancement of these inhibitors during the period 2005–2021 are presented in Table 2. Thus, inhibition of HSP90 in combination with over-expression of CHIP may be an alternate novel and ideal anti-cancer strategy for cancer therapy.
Figure 6.
Chemical structures of HSP90 inhibitors. Figure depicts the chemical structures of various HSP90 inhibitors viz., geldanamycin, 17AAG, 17DMAG, IPI-504, AT13387, AUY922, SNX5422, STA9090, XL888, CUDC 305, BIIB021, KW-2478 and TAS-116, act as anti-cancer agents. All the structures were drawn by using chem draw software.
Table 2.
Clinical advancement of HSP90 inhibitors for cancer therapy in a combinatorial approach.
| HSP90 inhibitor | Year | Combination | Clinical status (Phases) | NCT no. | Status | Outcome/Indication | References |
|---|---|---|---|---|---|---|---|
| 17AAG | 2005 | I | NCT00003969 | C | Advanced cancer | 434 | |
| I | NCT00003969 | C | Advanced cancer | 435 | |||
| I | NCT00003969 | C | Advanced malignancy | 436 | |||
| I | NCT00003969 | C | Solid tumors | 437 | |||
| 2006 | I | NCT00003969 | C | Advanced cancer | 438 | ||
| II | N/A | – | Renal cell carcinoma | 439 | |||
| 2007 | I | NCT00003969 | C | Advanced cancer | 440 | ||
| Trastuzumab | I | N/A | – | HER2-positive BC | 441 | ||
| I | NCT00003969 | C | Advanced cancers | 442 | |||
| I | NCT00079404 | C | Pediatric solid tumors | 443 | |||
| I | NCT00079404 | C | Pediatric solid tumors | 444 | |||
| 2008 | Irinotecan | I | NCT00119236 | C | Solid tumors | 445 | |
| II | NCT00118092 | C | Prostate cancer | 446 | |||
| II | NCT00104897 | C | Metastatic melanoma | 447 | |||
| Paclitaxel | I | NCT00087217 | C | Solid tumors | 448 | ||
| 2010 | Bortezomib | II | N/A | – | Multiple melanoma | 449 | |
| Sorafenib | I | NCT00121264 | C | Solid tumors | 450 | ||
| I | NCT0051437/NCT00546780 | C | Multiple myeloma | 451 | |||
| Gemcitabine or cisplatin | I | NCT00047047 | C | Solid tumors | 452 | ||
| 2011 | Trastuzumab | II | NCT00773344 | C | HER2-positive BC | 453 | |
| Cytarabine | I | NCT00098423 | C | Acute leukemia | 454 | ||
| Docetaxel | I | NCT00058253 | C | Solid tumors | 455 | ||
| Gemcitabine | II | NCT00093496 | C | EPC and PPC | 456 | ||
| Bortezomib | I/II | NCT00514371 | C | Multiple myeloma | 457 | ||
| 2012 | II | N/A | – | Breast cancer | 458 | ||
| II | NCT00104897 | C | Metastatic melanoma | 459 | |||
| 2013 | Bortezomib | I | NCT00096005 | T | Solid malignancies | 460 | |
| Bortezomib | I | N/A | – | AML | 461 | ||
| 2015 | Gemcitabine | II | NCT00577889 | C | Pancreatic cancer | 462 | |
| 17-DMAG | 2010 | I | NCT00089362 | C | Solid tumors | 442 | |
| I | NCT00088868 | C | Advanced malignancy | 463 | |||
| I | NCT0008927/NCT00088868 | C | AML | 464 | |||
| 2011 | I | NCT00248521 | U | Solid tumors | 465 | ||
| 2012 | Trastuzumab | I | N/A | – | Solid tumors | 466 | |
| 2016 | I | NCT01126502 | W | CLL/ALL | 467 | ||
| AT13387 | 2015 | I | NCT01246102 | C | Solid tumors | 468 | |
| I | NCT00878423 | C | Solid tumors | 469 | |||
| 2016 | Talazoparib | I | NCT02627430 | W | Ovarian cancer, FTC, PPC, TNBC | 470 | |
| 2021 | AT7519 | I | NCT02503709 | A | Solid tumors | 471 | |
| Olaparib | I | NCT02898207 | A | Ovarian cancer, FTC, PPC and TNBC | 472 | ||
| Cisplatin and radiation | I | NCT02381535 | A | SCC of the head and neck | 473 | ||
| Paclitaxel | I | NCT02474173 | A | TNBC | 474 | ||
| AUY922 | 2013 | I | N/A | Solid tumors | 475 | ||
| 2014 | 89Zr-trastuzumab/bevacizumab | I | NCT01081613/NCT01081600 | C | Breast cancer | 476 | |
| 2015 | Bortezomib | I/Ib | NCT00708292 | C | Multiple myeloma | 477 | |
| Capecitabine | I | NCT01226732 | C | Solid tumors | 478 | ||
| 2016 | II | NCT01404650 | C | GIST | 479 | ||
| II | NCT01124864 | C | NSCLC | 480 | |||
| II | NCT01668173 | T | MPNs | 481 | |||
| 2017 | II | NCT01485536 | T | Lymphoma | 482 | ||
| 2019 | II | NCT01646125 | T | NSCLC | 483 | ||
| Erlotinib | I/II | NCT01259089 | C | Lung ADC, NSCLC | 484 | ||
| 2020 | Pemetrexed | I | NCT01784640 | C | NSCLC, lung cancer | 485 | |
| BIIB021 | 2013 | II | NCT00618319 | C | GIST | 486 | |
| 2014 | I | NCT00618735 | C | Solid tumors | 487 | ||
| BIIB028 | 2013 | I | NCT00725933 | C | Solid tumors | 488 | |
| Debio0932 | 2015 | I | NCT01168752 | C | Advanced cancer | 489 | |
| HSP990 | 2015 | I | NCT00879905 | C | Solid tumors | 490 | |
| IPI-504 | 2011 | I | NCT00113204 | C | Multiple myeloma | 491 | |
| 2013 | I | NCT00276302 | C | GIST and sarcomas | 492 | ||
| 2017 | II | NCT01228435 | T | Lung cancer | 493 | ||
| KW-2478 | 2014 | Bortezomib | I/II | NCT01063907 | C | Multiple melanoma | 494 |
| 2016 | I | NCT00457782 | C | B-cell malignancy | 495 | ||
| SNX-5422 | 2011 | I | NCT00647764 | C | ST's and lymphomas | 496 | |
| 2013 | I | NCT00595686 | C | Malignant hematology | 497 | ||
| 2017 | I/II | NCT01848756 | T | HER2 positive cancers | 498 | ||
| 2018 | II | NCT02612285 | T | TP53 null cancers | 499 | ||
| 2019 | Lbrutinib | I | NCT02973399 | T | CLL | 500 | |
| STA-9090 | 2013 | I | NCT00687934 | C | Solid tumors | 501 | |
| II | NCT01562015 | C | Advanced NSCLC | 502 | |||
| 2014 | II | NCT01111838 | C | Metastatic CRC | 503 | ||
| II | NCT01227018 | T | Pancreas cancer | 507 | |||
| 2015 | Docetaxel | II | NCT01348126 | T | Advanced NSCLC | 504 | |
| 2016 | II | NCT01270880 | C | Prostate cancer | 505 | ||
| I | NCT02334319 | T | Head and neck cancer | 508 | |||
| Paclitaxel | II | NCT02637375 | W | Breast cancer | 509 | ||
| 2017 | II | NCT01551693 | T | Melanoma | 510 | ||
| 2018 | II | NCT01200238 | C | Ocular melanoma | 511 | ||
| Paclitaxel/Trastuzumab | I | NCT02060253 | C | HER2-positive BC | 512 | ||
| Ziv-aflibercept | I | NCT02192541 | T | UC, GIST & NSCLC | 513 | ||
| 2019 | II | NCT01173523 | C | SCLC | 514 | ||
| I/II | NCT02012192 | T | EPC, FTC and PPC | 515 | |||
| Sirolimus | I/II | NCT02008877 | C | MPNST, sarcoma | 516 | ||
| 2020 | Docetaxel | III | NCT01798485 | T | NSCLC | 506 | |
| PU-H71 | 2017 | I | NCT01581541 | T | ST's, lymphoma | 517 | |
| 2020 | Ruxolitinib | I | NCT03935555 | R | PMF, Post-PV MF, Post-ET MF | 518 | |
| TAS-116 | 2019 | I | NCT02965885 | C | Solid tumors | 519 | |
| NVP-BEP800 | 2020 | – | NCT04437420 | R | T-ALL, B-ALL | 520 | |
| CUDC 305 | 2019 | I | NCT03675542 | R | Psoriasis vulgaris | 521 | |
| TQB3474 | 2019 | I | NCT04144855 | R | Solid tumors | 522 | |
| PEN-866 | 2021 | I/II | NCT03221400 | R | Solid tumors | 523 | |
| RTA-901 | 2017 | I | NCT02666963 | C | Healthy adults | 524 | |
| XL888 | 2020 | Pembrolizumab | I | NCT03095781 | R | GIST | 525 |
| 2021 | Vemurafenib /Cobimetinib |
I | NCT02721459 | A | Melanoma | 526 |
Abbreviations C: Completed; T: Terminated; R: Recruiting; U: Unknown; A: Active; W: Withdrawn; N/A: Not available; CLL: Chronic lymphocytic leukemia; SLL: Small lymphocytic lymphoma; NSCLC: Non-small cell lung cancer; GIST: Gastrointestinal stromal tumors; T-ALL: T-cell acute lymphocytic leukemia; B-ALL: B-cell acute lymphocytic leukemia; PMF: Primary myelofibrosis; Post-PV MF: Post-polycythemia vera myelofibrosis; Post-ET MF: Post-essential thrombocythemia myelofibrosis; MPNST: Malignant peripheral nerve sheath tumor; SCLC: Small cell lung cancer; TP53: Tumor protein P53; TNBC: Triple negative breast cancer; SCC: Squamous cell carcinoma; CRC: Colorectal cancer; MPNs: Myeloproliferative neoplasms; EPC: Epithelial ovarian cancer; FTC: Fallopian tube cancer; PPC: Primary peritoneal cancer; BC: Breast cancer; AML: Acute myloid leukemia; ST's: Solid tumors; UC: Urothelial carcinoma; Lung ADC: Lung adenocarcinoma.
Basic disputes and unresolved mysteries of CHIP
In last two decades, CHIP has been established as a distinct protein enzyme which has huge number of substrates. CHIP interacts with its substrates through direct or indirect manner and performs a key role in various cellular processes such as apoptosis, proliferation, metabolism and in cellular signaling. In various reports, it is observed that CHIP regulates the substrate proteins in a chaperone dependent as well as in independent manner. It is still a key question to study how CHIP interacts with proteins and the selectivity in the complex cellular proteome. Further, it is found that chaperones are involved in recruitment of several proteins for folding or CHIP mediated ubiquitination and degradation. Still, a doubt remains to be resolved whether other auxiliary factors like co-chaperones are involved in the substrate selection for recruitment under diseased context. Both TPR and Ubox are structural domains of CHIP, responsible for determining its functional abilities. As reported, CHIP helps in binding the ubiquitin molecule to the substrate proteins. Although, it seems to be some specific known sequence elements for CHIP mediated ubiquitination of substrates, this question is not yet answered. Additionally, CHIP is responsible for its dual actions as a co-chaperone as well as for E3/E4 ligase activity. It is a subject of study, how CHIP decides to perform its dual role inside the cells. How CHIP along with other chaperones acknowledge mis-folded proteins to be degraded, is still a crucial question to be answered. Thus, several studies needed to be conducted in this direction, for better understanding of the probable ways to pick up cellular proteins upon the background of organizational and physiological situations. Number of studies have been performed to distinguish the normal proteins from the mis-folded ones, whether they are to be altered or degraded. Significant answers have not yet been found according to our current knowledge. CHIP also has its decisive roles in maintenance of cellular homeostasis and serves as a safety guard from the stress induced various diseases. Till date, major research on CHIP was focused upon its role but critical investigation about its disease specific differential regulation and gene expression is yet to be confirmed. It is urgent now to study about how and in which conditions CHIP is acting like a dual agent viz. tumor suppressor and oncogene. Thus, much work is needed to be done to resolve all these unresolved mysteries about CHIP for future perspectives.
Conclusion
CHIP acts as a central player in the multiple cellular processes including apoptosis, immunity, stress-response, inflammation, protein trafficking and morphogenesis. Its role has been convincingly studied in numerous morbid condition such as neurological disorders, cardiac diseases, bone remodeling and in cancer. Several studies indicate that CHIP can act as a dual agent like tumor suppressor as well as an oncogene in tumor biology. Furthermore, in multiple reports CHIP also manifests its defensive role by helping cells to remove deleterious forms of several proteins, especially under stress situation. Also, recent studies suggest that CHIP may be a novel therapeutic target to remove the numerous onco-proteins in multiple cancers. Thus, CHIP over-expression in combination with nano-encapsulated HSP90 inhibitors seems to be a valuable combinatorial strategy for next generation cancer therapy.
Author contributions
Conceived the idea and review structure: SK, MB and MKG; Wrote the manuscript: SK and MKG; Revised the manuscript: MB and MKG. All authors have read and agreed to the final draft of the manuscript.
Conflict of interests
There is no competing interest.
Funding
This work is jointly supported by the Department of Science and Technology (NanoMission: DST/NM/NT/2018/105(G); SERB: EMR/2017/000992) and HCT-Focused Basic Research (FBR), (Sanction No. 31-2(274)2020-21/Bud-II, dated- 03.03.2021), CSIR, Govt. of India.
Acknowledgements
Authors are happy to acknowledge Ms. Pratyasha Ghosh, Department of Economics, Bethune College, University of Calcutta for carefully editing the manuscript.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2021.08.003.
Abbreviations
- ADP
Adenosine di-phosphate
- AhR
Aryl hydrocarbon receptor
- AIF
Apoptosis inducing factor
- AMPK
AMP-activated protein kinase
- AR
Androgen receptor
- ASK1
Apoptosis signal-regulating kinase 1
- ATP
Adenosine tri-phosphate
- BACE1
Beta-secretase 1
- BAG 1, 2 and 3
BCL2 associated athano-Gene 1, 2 and 3
- BER
Base excision repair
- BMAL-1
Brain and muscle ARNT-Like 1
- CARMA1
Caspase recruitment domain-containing membrane-associated guanylate kinase protein-1
- CASA
Chaperone assisted selective autophagy
- CDK4
Cyclin-dependent kinase 4
- CFTR
Cystic fibrosis transmembrane conductance regulator
- ChAT
Choline acetyltransferase
- CHIP
Carboxy-terminus of Hsp70-interacting protein
- CK2β
Casein kinase protein subunit 2β
- CLEC-II
C-type lectin-like type II transmembrane receptor
- ClpP4
Chloroplast protease subunit P4
- CMA
Chaperone-mediated autophagy
- CtBP2
C-terminal-binding protein 2
- C-ErbB2/Neu
Receptor tyrosine-protein kinase erbB-2
- DDIAS
DNA Damage Induced Apoptosis Suppressor
- DLK
Dual leucine zipper kinase
- DNA
Deoxy-ribose nucleic acid
- E1's
Ubiquitin activating enzymes
- E2's
Ubiquitin conjugating enzymes
- E3's
Ubiquitin ligase enzymes
- EGFR
Epidermal growth factor receptor
- Endo G
Endonuclease G
- ER
Endoplasmic reticulum
- ERα
Estrogen receptor alpha
- ERKs
Extracellular-signal-regulated kinase
- EZH2
Enhancer of zeste homolog 2
- eIF4E
Eukaryotic translation initiation factor 4E
- eIF5A
Eukaryotic translation initiation factor 5A
- FADD
FAS-associated via death domain
- FGFR3
Fibroblast growth factor receptor 3
- FMR1
Fragile X mental retardation 1
- FOXO1
Forkhead box protein O1
- Foxp3
Forkhead box P3
- GHR
Growth hormone receptor
- GHS
Gorden holmes syndrome
- GR
Glucocorticoid receptor
- Gαs
Gs alpha subunit
- HIV
Human immunodeficiency virus
- HECT
Homologous to the E6-AP carboxyl terminus
- HDAC6
Histone deacetylase 6
- HERG
human Ether-à-go-go-Related Gene
- HIF1α
Hypoxia inducible factor-1 alpha
- HIP
Hsp70-interacting protein
- HIPK2
Homeodomain-interacting protein kinase 2
- HOP
Hsp70-Hsp90 organizing protein
- HSP70, 40, 72, 40 and 90
Heat shock protein-70, 40, 72, 40 and 90
- H2O2
Hydrogen peroxide
- hPXR T408D mutant
Phosphomimetic mutation at threonine-408
- hTERT
human Telomerase reverse transcriptase
- IA
Intracranial aneurysm
- ICER
Inducible cAMP Early Repressor
- IL-4α
Interleukin 4
- IRE1
Inositol-requiring enzyme 1
- IRF-1
Interferon regulatory factor 1
- IRS4
Insulin receptor substrate 4
- ISG15
Interferon-stimulated gene 15
- JNK
C-jun N-terminal kinase
- KCNQ4
potassium voltage-gated channel subfamily KQT member 4
- LKB1
liver kinase B1
- LRRK2
Leucine rich repeat kinase 2
- MAPK3
Mitogen-activated protein kinase 3
- MAST1
Microtubule-associated serine/threonine-protein kinase 1
- MEKK
Mitogen-activated protein kinase kinase
- MIF
Migration inhibitory factor
- MJD
Machado-joseph disease
- MLK3
Mixed-lineage kinase-3
- MLK4β
Mixed lineage kinase 4β
- MZF1
Myeloid Zinc Finger 1
- mHtt
Mutant Huntington
- Niemann-Pick C1
Niemann-pick disease, type C1
- NIK
NF-κB-inducing kinase
- NEK10
NIMA related kinase 10
- NFATc3
Nuclear factor of activated T-cells, cytoplasmic 3
- NOX4
NADPH oxidase 4
- NPM-ALK
Nucleophosmin-Anaplastic lymphoma kinase
- NQO1
NAD(P)H dehydrogenase [quinone] 1
- NUCB1
Nucleobindin 1
- OCT4
Octamer-binding transcription factor 4
- OTUD1
OTU Deubiquitinase 1
- PABPN1
Poly(A) binding protein nuclear 1
- Pae1R
Parkin associated endothelin like receptor
- PINK1
PTEN-induced kinase 1
- PI3K
Phosphoinositide 3-kinase
- PKC-ζ/SRC
Protein kinase C, zeta type
- PKM2
Pyruvate kinase M2
- PolyQ
Polyglutamine tract
- PRMT1,15
Protein arginine methyl-transferase 1, 15
- PrPC
Cellular Prion Protein
- PPARγ
Peroxisome proliferator-activated receptor γ
- PTK6
Protein Tyrosine Kinase 6
- PTEN
Phosphatase and tensin homolog
- p65
Transcription factor p65 (RELA)
- p38MAPK
p38 mitogen-activated protein kinases
- p42 Ebp1
Tumor suppressor p42, ErbB3-binding protein 1
- phosphatase PPP3CA
Serine/threonine-protein phosphatase 2B catalytic subunit alpha
- RHBDF2
Rhomboid 5 Homolog 2
- RFX-1
Regulatory factor X-1
- RIG-1
Retinoic acid-inducible gene I
- RING
Really interesting new gene
- RIPK3
Receptor-interacting serine/threonine-protein kinase 3
- RNF216
Ring finger protein 216
- RUNX 1 and 2
Runt-related transcription factor 1 and 2
- SCAR16
Spinocerebellar ataxia autosomal recessive type 16
- SENP3
Sentrin/SUMO-specific protease
- SOD2
Superoxide dismutase 2
- SGK1 and 3
Serum/glucocorticoid regulated kinase 1 and 3
- SirT6
Sirtuin 6
- SHP-1
Src homology region 2 domain-containing phosphatase-1
- SMAD 1 and 3
Small mothers against dpp 1 and 3
- SNPH
Syntaphilin
- SRC-3
Steroid receptor coactivator-3
- STAT4
Signal transducer and activator of transcription 4
- STUB1
STIP1 homology and Ubox containing protein 1
- Tat
Trans-activator of transcription
- TAL1/Scl
stem cell leukemia/T-cell acute lymphoblastic leukemia 1
- TAp73α
p73α antibody specifically recognizes human and monkey full-length p73α (TAp73α) protein
- TEAD4
TEA Domain Transcription Factor 4
- TFEB
Transcription factor EB
- TLR
Toll-like receptor
- TNFα
Tumor necrosis factor alpha
- TPR
Tetratricopeptide
- TRAF3
TNF receptor-associated factor 3
- UPS
Ubiquitin proteasome system
- XRCC1
X-ray repair cross complementing group-1
- 17AAG
17-N-allylamino-17-demethoxygeldanamycin
- 17DMAG
17-Dimethylaminoethylamino-17-demethoxygeldanamycin
- β-APP
β-amyloid precursor protein
- (i, n, e)NOS
(inducible, neuronal, endothelial) Nitric oxide synthase
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Multimedia component 1
References
- 1.Broadley S.A., Hartl F.U. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583(16):2647–2653. doi: 10.1016/j.febslet.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 2.Brodsky J.L., Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6(11):1215–1225. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 3.Ellis R.J., Hemmingsen S.M. Molecular chaperones: proteins essential for the biogenesis of some macromolecular structures. Trends Biochem Sci. 1989;14(8):339–342. doi: 10.1016/0968-0004(89)90168-0. [DOI] [PubMed] [Google Scholar]
- 4.Martin J., Hartl F.U. Molecular chaperones in cellular protein folding. Bioessays. 1994;16(9):689–692. doi: 10.1002/bies.950160916. [DOI] [PubMed] [Google Scholar]
- 5.Loureiro J., Ploegh H.L. Antigen presentation and the ubiquitin-proteasome system in host–pathogen interactions. Adv Immunol. 2006;92:225–305. doi: 10.1016/S0065-2776(06)92006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amm I., Sommer T., Wolf D.H. Protein quality control and elimination of protein waste: the role of the ubiquitin–proteasome system. Biochim Biophys Acta BBA-Mol Cell Res. 2014;1843(1):182–196. doi: 10.1016/j.bbamcr.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 7.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17(24):7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lecker S.H., Goldberg A.L., Mitch W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17(7):1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 9.Roos-Mattjus P., Sistonen L. The ubiquitin-proteasome pathway. Ann Med. 2004;36(4):285–295. doi: 10.1080/07853890310016324. [DOI] [PubMed] [Google Scholar]
- 10.Wolf D.H., Sommer T., Hilt W. Death gives birth to life: the essential role of the ubiquitin-proteasome system in biology. Biochim Biophys Acta. 2004;1695(1–3):1–2. doi: 10.1016/j.bbamcr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Maupin-Furlow J.A., Wilson H.L., Kaczowka S.J., Ou M.S. Proteasomes in the archaea: from structure to function. Front Biosci. 2000;5:D837–D865. doi: 10.2741/furlow. [DOI] [PubMed] [Google Scholar]
- 12.Nandi D., Tahiliani P., Kumar A., Chandu D. The ubiquitin-proteasome system. J Biosci. 2006;31(1):137–155. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- 13.Koegl M., Hoppe T., Schlenker S., Ulrich H.D., Mayer T.U., Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96(5):635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 14.Hershko A., Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 15.Johnson E.S., Bartel B., Seufert W., Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11(2):497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson E.S., Ma P.C., Ota I.M., Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270(29):17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 17.Lee I., Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134(2):268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 18.Olsen S.K., Lima C.D. Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer. Mol Cell. 2013;49(5):884–896. doi: 10.1016/j.molcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheffner M., Nuber U., Huibregtse J.M. Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature. 1995;373(6509):81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson K.D. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11(3):141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 21.Xu P., Duong D.M., Seyfried N.T., et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaushik S., Cuervo A.M. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22(8):407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin M., Liu X., Klionsky D.J. SnapShot: selective autophagy. Cell. 2013;152(1-2):368-368.e2. doi: 10.1016/j.cell.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaushik S., Cuervo A.M. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19(6):365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng L., Meng T., Chen L., Wei W., Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5(1):11. doi: 10.1038/s41392-020-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edkins A.L. CHIP: a co-chaperone for degradation by the proteasome. Subcell Biochem. 2015;78:219–242. doi: 10.1007/978-3-319-11731-7_11. [DOI] [PubMed] [Google Scholar]
- 27.Paul I., Ghosh M.K. The E3 ligase CHIP: insights into its structure and regulation. BioMed Res Int. 2014;2014:918183. doi: 10.1155/2014/918183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballinger C.A., Connell P., Wu Y., et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheufler C., Brinker A., Bourenkov G., et al. Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell. 2000;101(2):199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 30.Allan R.K., Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones. 2011;16(4):353–367. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamb J.R., Tugendreich S., Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20(7):257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 32.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capriotti E., Calabrese R., Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22(22):2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- 34.Ronnebaum S.M., Patterson C., Schisler J.C. Emerging evidence of coding mutations in the ubiquitin–proteasome system associated with cerebellar ataxias. Hum Genome Var. 2014;1:14018. doi: 10.1038/hgv.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y., Wang J., Li J.D., et al. Identification of CHIP as a novel causative gene for autosomal recessive cerebellar ataxia. PLoS One. 2013;8(12):e81884. doi: 10.1371/journal.pone.0081884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durcan T.M., Fon E.A. Ataxin-3 and its E3 partners: implications for Machado-Joseph disease. Front Neurol. 2013;4:46. doi: 10.3389/fneur.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.França M.C., Jr., Emmel V.E., D'Abreu A., et al. Normal ATXN3 allele but not CHIP polymorphisms modulates age at onset in Machado–Joseph disease. Front Neurol. 2012;3:164. doi: 10.3389/fneur.2012.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su L., Zhang Y., Zhang C.Y., et al. Genetic screening for mutations in the chip gene in intracranial aneurysm patients of Chinese Han nationality. Asian Pac J Cancer Prev APJCP. 2013;14(3):1687–1689. doi: 10.7314/apjcp.2013.14.3.1687. [DOI] [PubMed] [Google Scholar]
- 39.Shi C.H., Schisler J.C., Rubel C.E., et al. Ataxia and hypogonadism caused by the loss of ubiquitin ligase activity of the U box protein CHIP. Hum Mol Genet. 2014;23(4):1013–1024. doi: 10.1093/hmg/ddt497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudarsky L., Coutinho P. Machado-Joseph disease. Clin Neurosci N Y NY. 1995;3(1):17–22. [PubMed] [Google Scholar]
- 41.Dürr A., Stevanin G., Cancel G., et al. Spinocerebellar ataxia 3 and Machado-Joseph disease: clinical, molecular, and neuropathological features. Ann Neurol. 1996;39(4):490–499. doi: 10.1002/ana.410390411. [DOI] [PubMed] [Google Scholar]
- 42.de Roux N., Carel J.C., Léger J. Congenital hypogonadotropic hypogonadism: a trait shared by several complex neurodevelopmental disorders. Endocr Dev. 2016;29:72–86. doi: 10.1159/000438875. [DOI] [PubMed] [Google Scholar]
- 43.Ferreira J.V., Fôfo H., Bejarano E., et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9(9):1349–1366. doi: 10.4161/auto.25190. [DOI] [PubMed] [Google Scholar]
- 44.Sikorski R.S., Boguski M.S., Goebl M., Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60(2):307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 45.Das A.K., Cohen P.W., Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein–protein interactions. EMBO J. 1998;17(5):1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonough H., Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8(4):303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H., Amick J., Chakravarti R., et al. A bipartite interaction between Hsp70 and CHIP regulates ubiquitination of chaperoned client proteins. Structure. 2015;23(3):472–482. doi: 10.1016/j.str.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosser M.F., Washburn E., Muchowski P.J., Patterson C., Cyr D.M. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem. 2007;282(31):22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- 49.Nikolay R., Wiederkehr T., Rist W., Kramer G., Mayer M.P., Bukau B. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J Biol Chem. 2004;279(4):2673–2678. doi: 10.1074/jbc.M311112200. [DOI] [PubMed] [Google Scholar]
- 50.Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K.I. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276(35):33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 51.Hatakeyama S., Nakayama K.I. U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun. 2003;302(4):635–645. doi: 10.1016/s0006-291x(03)00245-6. [DOI] [PubMed] [Google Scholar]
- 52.Passmore L.A., Barford D. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem J. 2004;379(Pt 3):513–525. doi: 10.1042/BJ20040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aravind L., Koonin E.V. The U box is a modified RING finger—a common domain in ubiquitination. Curr Biol. 2000;10(4):R132–R134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 54.Meacham G.C., Patterson C., Zhang W., Younger J.M., Cyr D.M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3(1):100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 55.Imai Y., Soda M., Hatakeyama S., et al. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10(1):55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- 56.Belova L., Sharma S., Brickley D.R., Nicolarsen J.R., Patterson C., Conzen S.D. Ubiquitin-proteasome degradation of serum-and glucocorticoid-regulated kinase-1 (SGK-1) is mediated by the chaperone-dependent E3 ligase CHIP. Biochem J. 2006;400(2):235–244. doi: 10.1042/BJ20060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai Q., Zhang C., Wu Y., et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22(20):5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu F., Zhou J., Zhou P., Chen W., Guo F. The ubiquitin ligase CHIP inactivates NF-κB signaling and impairs the ability of migration and invasion in gastric cancer cells. Int J Oncol. 2015;46(5):2096–2106. doi: 10.3892/ijo.2015.2893. [DOI] [PubMed] [Google Scholar]
- 59.Shang Y., Zhao X., Xu X., et al. CHIP functions an E3 ubiquitin ligase of Runx1. Biochem Biophys Res Commun. 2009;386(1):242–246. doi: 10.1016/j.bbrc.2009.06.043. [DOI] [PubMed] [Google Scholar]
- 60.Li X., Huang M., Zheng H., et al. CHIP promotes Runx2 degradation and negatively regulates osteoblast differentiation. J Cell Biol. 2008;181(6):959–972. doi: 10.1083/jcb.200711044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding X., Goldberg M.S. Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS One. 2009;4(6):e5949. doi: 10.1371/journal.pone.0005949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar P., Ambasta R.K., Veereshwarayya V., et al. CHIP and HSPs interact with β-APP in a proteasome-dependent manner and influence Aβ metabolism. Hum Mol Genet. 2007;16(7):848–864. doi: 10.1093/hmg/ddm030. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y., Mandelkow E. Degradation of tau protein by autophagy and proteasomal pathways. Biochem Soc Trans. 2012;40(4):644–652. doi: 10.1042/BST20120071. [DOI] [PubMed] [Google Scholar]
- 64.Qian S.B., McDonough H., Boellmann F., Cyr D.M., Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440(7083):551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shin Y., Klucken J., Patterson C., Hyman B.T., McLean P.J. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates α-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280(25):23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 66.Mayer M.P., Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Connell P., Ballinger C.A., Jiang J., et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 68.Kundrat L., Regan L. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry. 2010;49(35):7428–7438. doi: 10.1021/bi100386w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marques C., Guo W., Pereira P., et al. The triage of damaged proteins: degradation by the ubiquitin-proteasome pathway or repair by molecular chaperones. FASEB J. 2006;20(6):741–743. doi: 10.1096/fj.05-5080fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wickner S., Maurizi M.R., Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286(5446):1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 71.Strickland E., Qu B.H., Millen L., Thomas P.J. The molecular chaperone Hsc70 assists the in VitroFolding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1997;272(41):25421–25424. doi: 10.1074/jbc.272.41.25421. [DOI] [PubMed] [Google Scholar]
- 72.Landry S.J., Gierasch L.M. Polypeptide interactions with molecular chaperones and their relationship to in vivo protein folding. Annu Rev Biophys Biomol Struct. 1994;23(1):645–669. doi: 10.1146/annurev.bb.23.060194.003241. [DOI] [PubMed] [Google Scholar]
- 73.Cheetham M.E., Jackson A.P., Anderton B.H. Regulation of 70-kDa heat-shock-protein ATPase activity and substrate binding by human DnaJ-like proteins, HSJ1a and HSJ1b. Eur J Biochem. 1994;226(1):99–107. doi: 10.1111/j.1432-1033.1994.tb20030.x. [DOI] [PubMed] [Google Scholar]
- 74.Hiromura M., Yano M., Mori H., Inoue M., Kido H. Intrinsic ADP-ATP exchange activity is a novel function of the molecular chaperone, Hsp70. J Biol Chem. 1998;273(10):5435–5438. doi: 10.1074/jbc.273.10.5435. [DOI] [PubMed] [Google Scholar]
- 75.Kettern N., Dreiseidler M., Tawo R., Höhfeld J. Chaperone-assisted degradation: multiple paths to destruction. Biol Chem. 2010;391(5):481–489. doi: 10.1515/BC.2010.058. [DOI] [PubMed] [Google Scholar]
- 76.Balchin D., Hayer-Hartl M., Hartl F.U. In vivo aspects of protein folding and quality control. Science. 2016;353(6294):aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 77.Mayer M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38(10):507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 78.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70(1):603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 79.Kriegenburg F., Ellgaard L., Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 2012;279(4):532–542. doi: 10.1111/j.1742-4658.2011.08456.x. [DOI] [PubMed] [Google Scholar]
- 80.Esser C., Alberti S., Höhfeld J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim Biophys Acta. 2004;1695(1–3):171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 81.Höhfeld J., Cyr D.M., Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2(10):885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meimaridou E., Gooljar S.B., Chapple J.P. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J Mol Endocrinol. 2009;42(1):1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- 83.Demand J., Alberti S., Patterson C., Höhfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol. 2001;11(20):1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 84.Lüders J., Demand J., Höhfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem. 2000;275(7):4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- 85.Arndt V., Dick N., Tawo R., et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20(2):143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 86.Alberti S., Esser C., Höhfeld J. BAG-1-a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones. 2003;8(3):225–231. doi: 10.1379/1466-1268(2003)008<0225:bnefoh>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doong H., Rizzo K., Fang S., Kulpa V., Weissman A.M., Kohn E.C. CAIR-1/BAG-3 abrogates heat shock protein-70 chaperone complex-mediated protein degradation: accumulation of poly-ubiquitinated Hsp90 client proteins. J Biol Chem. 2003;278(31):28490–28500. doi: 10.1074/jbc.M209682200. [DOI] [PubMed] [Google Scholar]
- 88.Gässler C.S., Wiederkehr T., Brehmer D., Bukau B., Mayer M.P. Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J Biol Chem. 2001;276(35):32538–32544. doi: 10.1074/jbc.M105328200. [DOI] [PubMed] [Google Scholar]
- 89.Wiederkehr T., Bukau B., Buchberger A. Protein turnover: a CHIP programmed for proteolysis. Curr Biol. 2002;12(1):R26–R28. doi: 10.1016/s0960-9822(01)00644-3. [DOI] [PubMed] [Google Scholar]
- 90.Gamerdinger M., Hajieva P., Kaya A.M., Wolfrum U., Hartl F.U., Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28(7):889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arndt V., Daniel C., Nastainczyk W., Alberti S., Höhfeld J. BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol Biol Cell. 2005;16(12):5891–5900. doi: 10.1091/mbc.E05-07-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schönbühler B., Schmitt V., Huesmann H., Kern A., Gamerdinger M., Behl C. BAG2 interferes with CHIP-mediated ubiquitination of HSP72. Int J Mol Sci. 2016;18(1):69. doi: 10.3390/ijms18010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stankiewicz M., Nikolay R., Rybin V., Mayer M.P. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J. 2010;277(16):3353–3367. doi: 10.1111/j.1742-4658.2010.07737.x. [DOI] [PubMed] [Google Scholar]
- 94.Garcia-Carbonero R., Carnero A., Paz-Ares L. Inhibition of HSP90 molecular chaperones: moving into the clinic. Lancet Oncol. 2013;14(9):e358–e369. doi: 10.1016/S1470-2045(13)70169-4. [DOI] [PubMed] [Google Scholar]
- 95.Smith D.F., Whitesell L., Nair S.C., Chen S., Prapapanich V., Rimerman R.A. Progesterone receptor structure and function altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol. 1995;15(12):6804–6812. doi: 10.1128/mcb.15.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pratt W.B., Toft D.O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228(2):111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 97.Young J.C., Moarefi I., Hartl F.U. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154(2):267–274. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhan S., Wang T., Ge W. Multiple functions of the E3 ubiquitin ligase CHIP in immunity. Int Rev Immunol. 2017;36(5):300–312. doi: 10.1080/08830185.2017.1309528. [DOI] [PubMed] [Google Scholar]
- 99.Murata S., Chiba T., Tanaka K. CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell Biol. 2003;35(5):572–578. doi: 10.1016/s1357-2725(02)00394-1. [DOI] [PubMed] [Google Scholar]
- 100.Li R.F., Zhang F., Lu Y.J., Sui S.F. Specific interaction between Smad1 and CHIP: a surface plasmon resonance study. Colloids Surf B Biointerfaces. 2005;40(3–4):133–136. doi: 10.1016/j.colsurfb.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 101.Xin H., Xu X., Li L., et al. CHIP controls the sensitivity of transforming growth factor-β signaling by modulating the basal level of Smad3 through ubiquitin-mediated degradation. J Biol Chem. 2005;280(21):20842–20850. doi: 10.1074/jbc.M412275200. [DOI] [PubMed] [Google Scholar]
- 102.Shao M., Li L., Song S., et al. E3 ubiquitin ligase CHIP interacts with C-type lectin-like receptor CLEC-2 and promotes its ubiquitin-proteasome degradation. Cell Signal. 2016;28(10):1530–1536. doi: 10.1016/j.cellsig.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 103.Wang T., Wang W., Wang Q., Xie R., Landay A., Chen D. The E3 ubiquitin ligase CHIP in normal cell function and in disease conditions. Ann N Y Acad Sci. 2020;1460(1):3–10. doi: 10.1111/nyas.14206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu Z., Kohli E., Devlin K.I., Bold M., Nix J.C., Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Page R.C., Pruneda J.N., Amick J., Klevit R.E., Misra S. Structural insights into the conformation and oligomerization of E2 ubiquitin conjugates. Biochemistry. 2012;51(20):4175–4187. doi: 10.1021/bi300058m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Narayan V., Pion E., Landré V., Müller P., Ball K.L. Docking-dependent ubiquitination of the interferon regulatory factor-1 tumor suppressor protein by the ubiquitin ligase CHIP. J Biol Chem. 2011;286(1):607–619. doi: 10.1074/jbc.M110.153122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Soss S.E., Yue Y., Dhe-Paganon S., Chazin W.J. E2 conjugating enzyme selectivity and requirements for function of the E3 ubiquitin ligase CHIP. J Biol Chem. 2011;286(24):21277–21286. doi: 10.1074/jbc.M111.224006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120(1):73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 109.Hoppe T. Multiubiquitylation by E4 enzymes:‘one size’doesn't fit all. Trends Biochem Sci. 2005;30(4):183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 110.Robertson A.B., Klungland A., Rognes T., Leiros I. DNA repair in mammalian cells. Cell Mol Life Sci. 2009;66(6):981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Caldecott K.W., McKeown C.K., Tucker J.D., Ljungquist S., Thompson L.H. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14(1):68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sancar A., Sancar G.B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- 113.Lindahl T., Wood R.D. Quality control by DNA repair. Science. 1999;286(5446):1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 114.Wood R.D., Mitchell M., Sgouros J., Lindahl T. Human DNA repair genes. Science. 2001;291(5507):1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 115.Parsons J.L., Tait P.S., Finch D., Dianova, Allinson S.L., Dianov G.L. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol Cell. 2008;29(4):477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 116.Lee J.S., Yoo S.J. C-terminus of Hsc70-interacting protein regulates C-terminal binding protein 2 and the expression of its target genes. Biochem Biophys Res Commun. 2013;432(3):418–424. doi: 10.1016/j.bbrc.2013.01.124. [DOI] [PubMed] [Google Scholar]
- 117.Burg M.B., Ferraris J.D., Dmitrieva N.I. Cellular response to hyperosmotic stresses. Physiol Rev. 2007;87(4):1441–1474. doi: 10.1152/physrev.00056.2006. [DOI] [PubMed] [Google Scholar]
- 118.Maruyama T., Kadowaki H., Okamoto N., et al. CHIP-dependent termination of MEKK2 regulates temporal ERK activation required for proper hyperosmotic response. EMBO J. 2010;29(15):2501–2514. doi: 10.1038/emboj.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Morey T.M., Winick-Ng W., Seah C., Rylett R.J. Chaperone-mediated regulation of choline acetyltransferase protein stability and activity by HSC/HSP70, HSP90, and p97/VCP. Front Mol Neurosci. 2017;10:415. doi: 10.3389/fnmol.2017.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Daviau A., Proulx R., Robitaille K., et al. Down-regulation of the mixed-lineage dual leucine zipper-bearing kinase by heat shock protein 70 and its co-chaperone CHIP. J Biol Chem. 2006;281(42):31467–31477. doi: 10.1074/jbc.M607612200. [DOI] [PubMed] [Google Scholar]
- 121.Niu X., Su L., Qi S., Gao Z., Zhang Q., Zhang S. GRP75 modulates oncogenic Dbl-driven endocytosis derailed via the CHIP-mediated ubiquitin degradation pathway. Cell Death Dis. 2018;9(10):971. doi: 10.1038/s41419-018-1039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee J.S., Seo T.W., Yi J.H., Shin K.S., Yoo S.J. CHIP has a protective role against oxidative stress-induced cell death through specific regulation of Endonuclease G. Cell Death Dis. 2013;4(6):e666. doi: 10.1038/cddis.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ishigaki S., Niwa J., Yamada S., et al. Dorfin-CHIP chimeric proteins potently ubiquitylate and degrade familial ALS-related mutant SOD1 proteins and reduce their cellular toxicity. Neurobiol Dis. 2007;25(2):331–341. doi: 10.1016/j.nbd.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 124.Kundrat L., Regan L. Identification of residues on Hsp70 and Hsp90 ubiquitinated by the cochaperone CHIP. J Mol Biol. 2010;395(3):587–594. doi: 10.1016/j.jmb.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morales J.L., Perdew G.H. Carboxyl terminus of hsc70-interacting protein (CHIP) can remodel mature aryl hydrocarbon receptor (AhR) complexes and mediate ubiquitination of both the AhR and the 90 kDa heat-shock protein (hsp90) in vitro. Biochemistry. 2007;46(2):610–621. doi: 10.1021/bi062165b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gil Lorenzo A.F., Costantino V.V., Appiolaza M.L., et al. Heat shock protein 70 and CHIP promote Nox4 ubiquitination and degradation within the losartan antioxidative effect in proximal tubule cells. Cell Physiol Biochem. 2015;36(6):2183–2197. doi: 10.1159/000430184. [DOI] [PubMed] [Google Scholar]
- 127.Yan S., Sun X., Xiang B., et al. Redox regulation of the stability of the SUMO protease SENP3 via interactions with CHIP and Hsp90. EMBO J. 2010;29(22):3773–3786. doi: 10.1038/emboj.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ronnebaum S.M., Wu Y., McDonough H., Patterson C. The ubiquitin ligase CHIP prevents SirT6 degradation through noncanonical ubiquitination. Mol Cell Biol. 2013;33(22):4461–4472. doi: 10.1128/MCB.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Y., Xu W., Zhou D., Neckers L., Chen S. Coordinated regulation of serum- and glucocorticoid-inducible kinase 3 by a C-terminal hydrophobic motif and hsp90-cdc37 chaperone complex. J Biol Chem. 2014;289(8):4815–4826. doi: 10.1074/jbc.M113.518480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zemanovic S., Ivanov M.V., Ivanova L.V., et al. Dynamic phosphorylation of the C terminus of Hsp70 regulates the mitochondrial import of SOD2 and redox balance. Cell Rep. 2018;25(9):2605–2616. doi: 10.1016/j.celrep.2018.11.015. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oh K.H., Yang S.W., Park J.M., et al. Control of AIF-mediated cell death by antagonistic functions of CHIP ubiquitin E3 ligase and USP2 deubiquitinating enzyme. Cell Death Differ. 2011;18(8):1326–1336. doi: 10.1038/cdd.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim C., Lee J., Ko Y.U., Oh Y.J. Cyclin-dependent kinase 5-mediated phosphorylation of CHIP promotes the tAIF-dependent death pathway in rotenone-treated cortical neurons. Neurosci Lett. 2018;662:295–301. doi: 10.1016/j.neulet.2017.10.053. [DOI] [PubMed] [Google Scholar]
- 133.Kim C., Yun N., Lee J., et al. Phosphorylation of CHIP at Ser20 by Cdk5 promotes tAIF-mediated neuronal death. Cell Death Differ. 2016;23(2):333–346. doi: 10.1038/cdd.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gao Y., Han C., Huang H., et al. Heat shock protein 70 together with its co-chaperone CHIP inhibits TNF-α induced apoptosis by promoting proteasomal degradation of apoptosis signal-regulating kinase1. Apoptosis. 2010;15(7):822–833. doi: 10.1007/s10495-010-0495-7. [DOI] [PubMed] [Google Scholar]
- 135.Hwang J.R., Zhang C., Patterson C. C-terminus of heat shock protein 70-interacting protein facilitates degradation of apoptosis signal-regulating kinase 1 and inhibits apoptosis signal-regulating kinase 1-dependent apoptosis. Cell Stress Chaperones. 2005;10(2):147–156. doi: 10.1379/CSC-90R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang Y., Huang X., Wang J., et al. Nitration-induced ubiquitination and degradation control quality of ERK1. Biochem J. 2019;476(13):1911–1926. doi: 10.1042/BCJ20190240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao B., Wang Y., Xu W., Li S., Li Q., Xiong S. Inhibition of histone deacetylase activity suppresses IFN-γ induction of tripartite motif 22 via CHIP-mediated proteasomal degradation of IRF-1. J Immunol. 2013;191(1):464–471. doi: 10.4049/jimmunol.1203533. [DOI] [PubMed] [Google Scholar]
- 138.Gao Z., Li Y., Wang F., et al. Mitochondrial dynamics controls anti-tumour innate immunity by regulating CHIP-IRF1 axis stability. Nat Commun. 2017;8(1):1805. doi: 10.1038/s41467-017-01919-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Landré V., Pion E., Narayan V., Xirodimas D.P., Ball K.L. DNA-binding regulates site-specific ubiquitination of IRF-1. Biochem J. 2013;449(3):707–717. doi: 10.1042/BJ20121076. [DOI] [PubMed] [Google Scholar]
- 140.Wang X., DeFranco D.B. Alternative effects of the ubiquitin-proteasome pathway on glucocorticoid receptor down-regulation and transactivation are mediated by CHIP, an E3 ligase. Mol Endocrinol. 2005;19(6):1474–1482. doi: 10.1210/me.2004-0383. [DOI] [PubMed] [Google Scholar]
- 141.Biswas K., Sarkar S., Said N., Brautigan D.L., Larner J.M. Aurora B kinase promotes CHIP-dependent degradation of HIF1α in prostate cancer cells. Mol Canc Ther. 2020;19(4):1008–1017. doi: 10.1158/1535-7163.MCT-19-0777. [DOI] [PubMed] [Google Scholar]
- 142.Im S., Kim D.W. Nkx3.2 induces oxygen concentration-independent and lysosome-dependent degradation of HIF-1α to modulate hypoxic responses in chondrocytes. Cell Signal. 2017;36:127–138. doi: 10.1016/j.cellsig.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 143.Bento C.F., Fernandes R., Ramalho J., et al. The chaperone-dependent ubiquitin ligase CHIP targets HIF-1α for degradation in the presence of methylglyoxal. PLoS One. 2010;5(11):e15062. doi: 10.1371/journal.pone.0015062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Luo W., Zhong J., Chang R., Hu H., Pandey A., Semenza G.L. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1α but not HIF-2α. J Biol Chem. 2010;285(6):3651–3663. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tawo R., Pokrzywa W., É Kevei, et al. The ubiquitin ligase CHIP integrates proteostasis and aging by regulation of insulin receptor turnover. Cell. 2017;169(3):470–482. doi: 10.1016/j.cell.2017.04.003. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nakasone N., Nakamura Y.S., Higaki K., Oumi N., Ohno K., Ninomiya H. Endoplasmic reticulum-associated degradation of Niemann-Pick C1: evidence for the role of heat shock proteins and identification of lysine residues that accept ubiquitin. J Biol Chem. 2014;289(28):19714–19725. doi: 10.1074/jbc.M114.549915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dema A., Faust D., Lazarow K., et al. Cyclin-dependent kinase 18 controls trafficking of aquaporin-2 and its abundance through ubiquitin ligase STUB1, which functions as an AKAP. Cells. 2020;9(3):673. doi: 10.3390/cells9030673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Centrone M., Ranieri M., Di Mise A.D., et al. AQP2 abundance is regulated by the E3-ligase CHIP via HSP70. Cell Physiol Biochem. 2017;44(2):515–531. doi: 10.1159/000485088. [DOI] [PubMed] [Google Scholar]
- 149.Wu Q., Moeller H.B., Stevens D.A., et al. CHIP regulates aquaporin-2 quality control and body water homeostasis. J Am Soc Nephrol. 2018;29(3):936–948. doi: 10.1681/ASN.2017050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sobol R.W. CHIPping away at base excision repair. Mol Cell. 2008;29(4):413–415. doi: 10.1016/j.molcel.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 151.Gimenez A.P., Richter L.M., Atherino M.C., et al. Identification of novel putative-binding proteins for cellular prion protein and a specific interaction with the STIP1 homology and U-Box-containing protein 1. Prion. 2015;9(5):355–366. doi: 10.1080/19336896.2015.1075347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang Y., Li Y., Yang X., et al. Uev1A-Ubc13 catalyzes K63-linked ubiquitination of RHBDF2 to promote TACE maturation. Cell Signal. 2018;42:155–164. doi: 10.1016/j.cellsig.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 153.Bhuripanyo K., Wang Y., Liu X., et al. Identifying the substrate proteins of U-box E3s E4B and CHIP by orthogonal ubiquitin transfer. Sci Adv. 2018;4(1):e1701393. doi: 10.1126/sciadv.1701393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Murata T., Shimotohno K. Ubiquitination and proteasome-dependent degradation of human eukaryotic translation initiation factor 4E. J Biol Chem. 2006;281(30):20788–20800. doi: 10.1074/jbc.M600563200. [DOI] [PubMed] [Google Scholar]
- 155.Seo T.W., Lee J.S., Choi Y.N., Jeong D.H., Lee S.K., Yoo S.J. A novel function of cIAP1 as a mediator of CHIP-driven eIF4E regulation. Sci Rep. 2017;7(1):9816. doi: 10.1038/s41598-017-10358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sugatani J., Noguchi Y., Hattori Y., Yamaguchi M., Yamazaki Y., Ikari A. Threonine-408 regulates the stability of human pregnane X receptor through its phosphorylation and the CHIP/chaperone-autophagy pathway. Drug Metab Dispos. 2016;44(1):137–150. doi: 10.1124/dmd.115.066308. [DOI] [PubMed] [Google Scholar]
- 157.Kim N.-G., Gumbiner B.M. Cell contact and Nf2/Merlin-dependent regulation of TEAD palmitoylation and activity. Proc Natl Acad Sci Unit States Am. 2019;116(20):9877–9882. doi: 10.1073/pnas.1819400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Slotman J.A., da Silva Almeida A.C., Hassink G.C., et al. Ubc13 and COOH terminus of Hsp70-interacting protein (CHIP) are required for growth hormone receptor endocytosis. J Biol Chem. 2012;287(19):15533–15543. doi: 10.1074/jbc.M111.302521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Teng Y., Rezvani K., De Biasi M. UBXN2A regulates nicotinic receptor degradation by modulating the E3 ligase activity of CHIP. Biochem Pharmacol. 2015;97(4):518–530. doi: 10.1016/j.bcp.2015.08.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kim J.H., Shin S., Seo J., et al. C-terminus of HSC70-interacting protein (CHIP) inhibits adipocyte differentiation via ubiquitin-and proteasome-mediated degradation of PPARγ. Sci Rep. 2017;7:40023. doi: 10.1038/srep40023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ullah K., Chen S., Lu J., et al. The E3 ubiquitin ligase STUB1 attenuates cell senescence by promoting the ubiquitination and degradation of the core circadian regulator BMAL1. J Biol Chem. 2020;295(14):4696–4708. doi: 10.1074/jbc.RA119.011280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kim S.M., Grenert J.P., Patterson C., Correia M.A. CHIP(-/-)- mouse liver: adiponectin-AMPK-FOXO-activation overrides CYP2E1-elicited JNK1-activation, delaying onset of NASH: therapeutic implications. Sci Rep. 2016;6:29423. doi: 10.1038/srep29423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schisler J.C., Rubel C.E., Zhang C., Lockyer P., Cyr D.M., Patterson C. CHIP protects against cardiac pressure overload through regulation of AMPK. J Clin Invest. 2013;123(8):3588–3599. doi: 10.1172/JCI69080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ravalin M., Theofilas P., Basu K., et al. Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases. Nat Chem Biol. 2019;15(8):786–794. doi: 10.1038/s41589-019-0322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shen G., Yan J., Pasapula V., et al. The chloroplast protease subunit ClpP4 is a substrate of the E3 ligase AtCHIP and plays an important role in chloroplast function. Plant J. 2007;49(2):228–237. doi: 10.1111/j.1365-313X.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- 166.Kim S.-M., Acharya P., Engel J.C., Correia M.A. Liver cytochrome P450 3A ubiquitination in vivo by gp78/autocrine motility factor receptor and C terminus of Hsp70-interacting protein (CHIP) E3 ubiquitin ligases: physiological and pharmacological relevance. J Biol Chem. 2010;285(46):35866–35877. doi: 10.1074/jbc.M110.167189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Y., Guan S., Acharya P., et al. Ubiquitin-dependent proteasomal degradation of human liver cytochrome P450 2E1: identification of sites targeted for phosphorylation and ubiquitination. J Biol Chem. 2011;286(11):9443–9456. doi: 10.1074/jbc.M110.176685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang Y., Guan S., Acharya P., et al. Multisite phosphorylation of human liver cytochrome P450 3A4 enhances its gp78-and CHIP-mediated ubiquitination: a pivotal role of its Ser-478 residue in the gp78-catalyzed reaction. Mol Cell Proteomics. 2012;11(2) doi: 10.1074/mcp.M111.010132. M111.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang Y., Kim S.-M., Trnka M.J., Liu Y., Burlingame A.L., Correia M.A. Human liver cytochrome P450 3A4 ubiquitination: molecular recognition by UBC7-gp78 autocrine motility factor receptor and UbcH5a-CHIP-Hsc70-Hsp40 E2-E3 ubiquitin ligase complexes. J Biol Chem. 2015;290(6):3308–3332. doi: 10.1074/jbc.M114.611525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tsvetkov P., Adamovich Y., Elliott E., Shaul Y. E3 ligase STUB1/CHIP regulates NAD (P) H: quinone oxidoreductase 1 (NQO1) accumulation in aged brain, a process impaired in certain Alzheimer disease patients. J Biol Chem. 2011;286(11):8839–8845. doi: 10.1074/jbc.M110.193276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jiang B., Shen H., Chen Z., Yin L., Zan L., Rui L. Carboxyl terminus of HSC70-interacting protein (CHIP) down-regulates NF-κB-inducing kinase (NIK) and suppresses NIK-induced liver injury. J Biol Chem. 2015;290(18):11704–11714. doi: 10.1074/jbc.M114.635086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ahmed S.F., Deb S., Paul I., et al. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J Biol Chem. 2012;287(19):15996–16006. doi: 10.1074/jbc.M111.321083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu W., Marcu M., Yuan X., Mimnaugh E., Patterson C., Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci U S A. 2002;99(20):12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sun S.-C. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21(1):71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yin L., Wu L., Wesche H., et al. Defective lymphotoxin-β receptor-induced NF-κB transcriptional activity in NIK-deficient mice. Science. 2001;291(5511):2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- 176.Willmann K.L., Klaver S., Doğu F., et al. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat Commun. 2014;5:5360. doi: 10.1038/ncomms6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zarnegar B.J., Wang Y., Mahoney D.J., et al. Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9(12):1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 179.Mukherjee S., Karmakar S., Babu S.P.S. TLR2 and TLR4 mediated host immune responses in major infectious diseases: a review. Braz J Infect Dis. 2016;20(2):193–204. doi: 10.1016/j.bjid.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang M., Wang C., Zhu X., et al. E3 ubiquitin ligase CHIP facilitates Toll-like receptor signaling by recruiting and polyubiquitinating Src and atypical PKCζ. J Exp Med. 2011;208(10):2099–2112. doi: 10.1084/jem.20102667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang S., Li Y., Hu Y.H., et al. STUB1 is essential for T-cell activation by ubiquitinating CARMA 1. Eur J Immunol. 2013;43(4):1034–1041. doi: 10.1002/eji.201242554. [DOI] [PubMed] [Google Scholar]
- 182.Seo J., Lee E.W., Sung H., et al. CHIP controls necroptosis through ubiquitylation- and lysosome-dependent degradation of RIPK3. Nat Cell Biol. 2016;18(3):291–302. doi: 10.1038/ncb3314. [DOI] [PubMed] [Google Scholar]
- 183.Chen L., Kong X., Fu J., et al. CHIP facilitates ubiquitination of inducible nitric oxide synthase and promotes its proteasomal degradation. Cell Immunol. 2009;258(1):38–43. doi: 10.1016/j.cellimm.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 184.Yao X., Li G., Lü C., Xu H., Yin Z. Arctigenin promotes degradation of inducible nitric oxide synthase through CHIP-associated proteasome pathway and suppresses its enzyme activity. Int Immunopharm. 2012;14(2):138–144. doi: 10.1016/j.intimp.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 185.Peng H.-M., Morishima Y., Jenkins G.J., et al. Ubiquitylation of neuronal nitric-oxide synthase by CHIP, a chaperone-dependent E3 ligase. J Biol Chem. 2004;279(51):52970–52977. doi: 10.1074/jbc.M406926200. [DOI] [PubMed] [Google Scholar]
- 186.Jiang J., Cyr D., Babbitt R.W., Sessa W.C., Patterson C. Chaperone-dependent regulation of endothelial nitric-oxide synthase intracellular trafficking by the co-chaperone/ubiquitin ligase CHIP. J Biol Chem. 2003;278(49):49332–49341. doi: 10.1074/jbc.M304738200. [DOI] [PubMed] [Google Scholar]
- 187.Seo J., Lee E.-W., Shin J., et al. K6 linked polyubiquitylation of FADD by CHIP prevents death inducing signaling complex formation suppressing cell death. Oncogene. 2018;37(36):4994–5006. doi: 10.1038/s41388-018-0323-z. [DOI] [PubMed] [Google Scholar]
- 188.Zhang Y., Chen Z., Luo X., Wu B., Li B., Wang B. Cimetidine down-regulates stability of Foxp3 protein via Stub1 in Treg cells. Hum Vaccin Immunother. 2016;12(10):2512–2518. doi: 10.1080/21645515.2016.1191719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chen Z., Barbi J., Bu S., et al. The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity. 2013;39(2):272–285. doi: 10.1016/j.immuni.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhao Y., Guo H., Qiao G., Zucker M., Langdon W.Y., Zhang J. E3 ubiquitin ligase Cbl-b regulates thymic-derived CD4+ CD25+ regulatory T cell development by targeting Foxp3 for ubiquitination. J Immunol. 2015;194(4):1639–1645. doi: 10.4049/jimmunol.1402434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Guo Y., Zhao M., Lu Q. Transcription factor RFX1 is ubiquitinated by E3 ligase STUB1 in systemic lupus erythematosus. Clin Immunol. 2016;169:1–7. doi: 10.1016/j.clim.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 192.Zhao K., Zhang Q., Li X., et al. Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP-mediated degradation of RIG-I. J Immunol. 2016;196(3):1209–1217. doi: 10.4049/jimmunol.1501224. [DOI] [PubMed] [Google Scholar]
- 193.Ali A., Farooqui S.R., Banerjea A.C. The host cell ubiquitin ligase protein CHIP is a potent suppressor of HIV-1 replication. J Biol Chem. 2019;294(18):7283–7295. doi: 10.1074/jbc.RA118.007257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kim R., Emi M., Tanabe K. Cancer immunoediting: from immune surveillance to immune escape. Immunology. 2007;121(1):1–14. doi: 10.1111/j.1365-2567.2007.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jackaman C., Bundell C.S., Kinnear B.F., et al. IL-2 intratumoral immunotherapy enhances CD8+ T cells that mediate destruction of tumor cells and tumor-associated vasculature: a novel mechanism for IL-2. J Immunol. 2003;171(10):5051–5063. doi: 10.4049/jimmunol.171.10.5051. [DOI] [PubMed] [Google Scholar]
- 196.Jackaman C., Lew A.M., Zhan Y., et al. Deliberately provoking local inflammation drives tumors to become their own protective vaccine site. Int Immunol. 2008;20(11):1467–1479. doi: 10.1093/intimm/dxn104. [DOI] [PubMed] [Google Scholar]
- 197.Steer H.J., Lake R.A., Nowak A.K., Robinson B.W.S. Harnessing the immune response to treat cancer. Oncogene. 2010;29(48):6301–6313. doi: 10.1038/onc.2010.437. [DOI] [PubMed] [Google Scholar]
- 198.Li H., Fan X., Houghton J. Tumor microenvironment: the role of the tumor stroma in cancer. J Cell Biochem. 2007;101(4):805–815. doi: 10.1002/jcb.21159. [DOI] [PubMed] [Google Scholar]
- 199.Jackaman C., Cornwall S., Lew A.M., Zhan Y., Robinson B.W., Nelson D.J. Local effector failure in mesothelioma is not mediated by CD4+ CD25+ T-regulator cells. Eur Respir J. 2009;34(1):162–175. doi: 10.1183/09031936.00101008. [DOI] [PubMed] [Google Scholar]
- 200.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 201.Teng M.W., Galon J., Fridman W.H., Smyth M.J. From mice to humans: developments in cancer immunoediting. J Clin Invest. 2015;125(9):3338–3346. doi: 10.1172/JCI80004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mantovani A., Sica A., Allavena P., Garlanda C., Locati M. Tumor-associated macrophages and the related myeloid-derived suppressor cells as a paradigm of the diversity of macrophage activation. Hum Immunol. 2009;70(5):325–330. doi: 10.1016/j.humimm.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 203.Bronte V., Brandau S., Chen S.H., et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Pillay J., Tak T., Kamp V.M., Koenderman L. Immune suppression by neutrophils and granulocytic myeloid-derived suppressor cells: similarities and differences. Cell Mol Life Sci. 2013;70(20):3813–3827. doi: 10.1007/s00018-013-1286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Strauss L., Sangaletti S., Consonni F.M., et al. RORC1 regulates tumor-promoting “emergency” granulo-monocytopoiesis. Cancer Cell. 2015;28(2):253–269. doi: 10.1016/j.ccell.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 206.Grizzle W.E., Xu X., Zhang S., et al. Age-related increase of tumor susceptibility is associated with myeloid-derived suppressor cell mediated suppression of T cell cytotoxicity in recombinant inbred BXD12 mice. Mech Ageing Dev. 2007;128(11–12):672–680. doi: 10.1016/j.mad.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 207.Voronov E., Dotan S., Krelin Y., et al. Unique versus redundant functions of IL-1α and IL-1β in the tumor microenvironment. Front Immunol. 2013;4:177. doi: 10.3389/fimmu.2013.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Biswas S.K., Sica A., Lewis C.E. Plasticity of macrophage function during tumor progression: regulation by distinct molecular mechanisms. J Immunol. 2008;180(4):2011–2017. doi: 10.4049/jimmunol.180.4.2011. [DOI] [PubMed] [Google Scholar]
- 210.Ojalvo L.S., Whittaker C.A., Condeelis J.S., Pollard J.W. Gene expression analysis of macrophages that facilitate tumor invasion supports a role for Wnt-signaling in mediating their activity in primary mammary tumors. J Immunol. 2010;184(2):702–712. doi: 10.4049/jimmunol.0902360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Qian B.Z., Pollard J.W. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Szebeni G.J., Vizler C., Kitajka K., Puskas L.G. Inflammation and cancer: extra- and intracellular determinants of tumor-associated macrophages as tumor promoters. Mediators Inflamm. 2017;2017:e9294018. doi: 10.1155/2017/9294018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jackaman C., Yeoh T.L., Acuil M.L., Gardner J.K., Nelson D.J. Murine mesothelioma induces locally-proliferating IL-10(+) TNF-α(+) CD206(-) CX3CR1(+) M3 macrophages that can be selectively depleted by chemotherapy or immunotherapy. OncoImmunology. 2016;5(6):e1173299. doi: 10.1080/2162402X.2016.1173299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mantovani A., Sozzani S., Locati M., Allavena P., Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 215.Sica A., Schioppa T., Mantovani A., Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer. 2006;42(6):717–727. doi: 10.1016/j.ejca.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 216.Cortez-Retamozo V., Etzrodt M., Newton A., et al. Origins of tumor-associated macrophages and neutrophils. Proc Natl Acad Sci U S A. 2012;109(7):2491–2496. doi: 10.1073/pnas.1113744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.De Palma M. Origins of brain tumor macrophages. Cancer Cell. 2016;30(6):832–833. doi: 10.1016/j.ccell.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 218.Movahedi K., Laoui D., Gysemans C., et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C (high) monocytes. Cancer Res. 2010;70(14):5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 219.Shand F.H., Ueha S., Otsuji M., et al. Tracking of intertissue migration reveals the origins of tumor-infiltrating monocytes. Proc Natl Acad Sci U S A. 2014;111(21):7771–7776. doi: 10.1073/pnas.1402914111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhou W., Ke S.Q., Huang Z., et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat Cell Biol. 2015;17(2):170–182. doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Beaudoin S., Goggin K., Bissonnette C., Grenier C., Roucou X. Aggresomes do not represent a general cellular response to protein misfolding in mammalian cells. BMC Cell Biol. 2008;9:59. doi: 10.1186/1471-2121-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Johnston J.A., Ward C.L., Kopito R.R. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143(7):1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sha Y., Pandit L., Zeng S., Eissa N.T. A critical role for CHIP in the aggresome pathway. Mol Cell Biol. 2009;29(1):116–128. doi: 10.1128/MCB.00829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Luciani A., Villella V.R., Esposito S., et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12(9):863–875. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- 225.Dickey C.A., Patterson C., Dickson D., Petrucelli L. Brain CHIP: removing the culprits in neurodegenerative disease. Trends Mol Med. 2007;13(1):32–38. doi: 10.1016/j.molmed.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 226.Petrucelli L., Dickson D., Kehoe K., et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13(7):703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 227.Kanack A.J., Newsom O.J., Scaglione K.M. Most mutations that cause spinocerebellar ataxia autosomal recessive type 16 (SCAR16) destabilize the protein quality-control E3 ligase CHIP. J Biol Chem. 2018;293(8):2735–2743. doi: 10.1074/jbc.RA117.000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kalia L.V., Kalia S.K., Chau H., Lozano A.M., Hyman B.T., McLean P.J. Ubiquitinylation of α-synuclein by carboxyl terminus Hsp70-interacting protein (CHIP) is regulated by Bcl-2-associated athanogene 5 (BAG5) PLoS One. 2011;6(2):e14695. doi: 10.1371/journal.pone.0014695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Choi J.Y., Ryu J.H., Kim H.S., et al. Co-chaperone CHIP promotes aggregation of ataxin-1. Mol Cell Neurosci. 2007;34(1):69–79. doi: 10.1016/j.mcn.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 230.Williams A.J., Knutson T.M., Colomer Gould V.F., Paulson H.L. In vivo suppression of polyglutamine neurotoxicity by C-terminus of Hsp70-interacting protein (CHIP) supports an aggregation model of pathogenesis. Neurobiol Dis. 2009;33(3):342–353. doi: 10.1016/j.nbd.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Shimura H., Schwartz D., Gygi S.P., Kosik K.S. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J Biol Chem. 2004;279(6):4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 232.Rao S.N., Sharma J., Maity R., Jana N.R. Co-chaperone CHIP stabilizes aggregate-prone malin, a ubiquitin ligase mutated in Lafora disease. J Biol Chem. 2010;285(2):1404–1413. doi: 10.1074/jbc.M109.006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Adachi H., Waza M., Tokui K., et al. CHIP overexpression reduces mutant androgen receptor protein and ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model. J Neurosci. 2007;27(19):5115–5126. doi: 10.1523/JNEUROSCI.1242-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cardozo C.P., Michaud C., Ost M.C., et al. C-terminal Hsp-interacting protein slows androgen receptor synthesis and reduces its rate of degradation. Arch Biochem Biophys. 2003;410(1):134–140. doi: 10.1016/s0003-9861(02)00680-x. [DOI] [PubMed] [Google Scholar]
- 235.DaSilva J., Gioeli D., Weber M.J., Parsons S.J. The neuroendocrine-derived peptide parathyroid hormone-related protein promotes prostate cancer cell growth by stabilizing the androgen receptor. Cancer Res. 2009;69(18):7402–7411. doi: 10.1158/0008-5472.CAN-08-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.He B., Bai S., Hnat A.T., et al. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP) J Biol Chem. 2004;279(29):30643–30653. doi: 10.1074/jbc.M403117200. [DOI] [PubMed] [Google Scholar]
- 237.Rees I., Lee S., Kim H., Tsai F.T. The E3 ubiquitin ligase CHIP binds the androgen receptor in a phosphorylation-dependent manner. Biochim Biophys Acta BBA- 2006;1764(6):1073–1079. doi: 10.1016/j.bbapap.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 238.Al-Ramahi I., Lam Y.C., Chen H.K., et al. CHIP protects from the neurotoxicity of expanded and wild-type ataxin-1 and promotes their ubiquitination and degradation. J Biol Chem. 2006;281(36):26714–26724. doi: 10.1074/jbc.M601603200. [DOI] [PubMed] [Google Scholar]
- 239.Rudenko I.N., Kaganovich A., Langston R.G., et al. The G2385R risk factor for Parkinson's disease enhances CHIP-dependent intracellular degradation of LRRK2. Biochem J. 2017;474(9):1547–1558. doi: 10.1042/BCJ20160909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ko H.S., Bailey R., Smith W.W., et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc Natl Acad Sci U S A. 2009;106(8):2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hirayama S., Yamazaki Y., Kitamura A., et al. MKKS is a centrosome-shuttling protein degraded by disease-causing mutations via CHIP-mediated ubiquitination. Mol Biol Cell. 2008;19(3):899–911. doi: 10.1091/mbc.E07-07-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yoo L., Chung K.C. The ubiquitin E3 ligase CHIP promotes proteasomal degradation of the serine/threonine protein kinase PINK1 during staurosporine-induced cell death. J Biol Chem. 2018;293(4):1286–1297. doi: 10.1074/jbc.M117.803890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Dimant H., Zhu L., Kibuuka L.N., Fan Z., Hyman B.T., McLean P.J. Direct visualization of CHIP-mediated degradation of alpha-synuclein in vivo: implications for PD therapeutics. PLoS One. 2014;9(3):e92098. doi: 10.1371/journal.pone.0092098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Tetzlaff J.E., Putcha P., Outeiro T.F., et al. CHIP targets toxic α-synuclein oligomers for degradation. J Biol Chem. 2008;283(26):17962–17968. doi: 10.1074/jbc.M802283200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Choi Y.N., Jeong D.H., Lee J.S., Yoo S.J. Regulation of fragile X mental retardation 1 protein by C-terminus of Hsc70-interacting protein depends on its phosphorylation status. Biochem Biophys Res Commun. 2014;453(1):192–197. doi: 10.1016/j.bbrc.2014.09.099. [DOI] [PubMed] [Google Scholar]
- 246.Zhao T., Hong Y., Yin P., Li S., Li X.J. Differential HspBP1 expression accounts for the greater vulnerability of neurons than astrocytes to misfolded proteins. Proc Natl Acad Sci U S A. 2017;114(37):E7803–E7811. doi: 10.1073/pnas.1710549114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Saidi L.-J., Polydoro M., Kay K.R., et al. Carboxy terminus heat shock protein 70 interacting protein reduces tau-associated degenerative changes. J Alzheimers Dis. 2015;44(3):937–947. doi: 10.3233/JAD-142094. [DOI] [PubMed] [Google Scholar]
- 248.Cook C., Gendron T.F., Scheffel K., et al. Loss of HDAC6, a novel CHIP substrate, alleviates abnormal tau accumulation. Hum Mol Genet. 2012;21(13):2936–2945. doi: 10.1093/hmg/dds125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Dickey C.A., Koren J., Zhang Y.J., et al. Akt and CHIP coregulate tau degradation through coordinated interactions. Proc Natl Acad Sci U S A. 2008;105(9):3622–3627. doi: 10.1073/pnas.0709180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Grune T., Botzen D., Engels M., et al. Tau protein degradation is catalyzed by the ATP/ubiquitin-independent 20S proteasome under normal cell conditions. Arch Biochem Biophys. 2010;500(2):181–188. doi: 10.1016/j.abb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Sahara N., Murayama M., Mizoroki T., et al. In vivo evidence of CHIP up-regulation attenuating tau aggregation. J Neurochem. 2005;94(5):1254–1263. doi: 10.1111/j.1471-4159.2005.03272.x. [DOI] [PubMed] [Google Scholar]
- 252.Sakagami Y., Kudo T., Tanimukai H., et al. Involvement of endoplasmic reticulum stress in tauopathy. Biochem Biophys Res Commun. 2013;430(2):500–504. doi: 10.1016/j.bbrc.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 253.Miller L.M., D’esposito M. Perceptual fusion and stimulus coincidence in the cross-modal integration of speech. J Neurosci. 2005;25(25):5884–5893. doi: 10.1523/JNEUROSCI.0896-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Singh A.K., Pati U. CHIP stabilizes amyloid precursor protein via proteasomal degradation and p53-mediated trans-repression of β-secretase. Aging Cell. 2015;14(4):595–604. doi: 10.1111/acel.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Clapp K.M., Peng H.M., Morishima Y., et al. C331A mutant of neuronal nitric-oxide synthase is labilized for Hsp70/CHIP (C terminus of HSC70-interacting protein)-dependent ubiquitination. J Biol Chem. 2010;285(44):33642–33651. doi: 10.1074/jbc.M110.159178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Matsumoto K., Nishiya T., Maekawa S., et al. The ECS (SPSB) E3 ubiquitin ligase is the master regulator of the lifetime of inducible nitric-oxide synthase. Biochem Biophys Res Commun. 2011;409(1):46–51. doi: 10.1016/j.bbrc.2011.04.103. [DOI] [PubMed] [Google Scholar]
- 257.Choi J.S., Cho S., Park S.G., Park B.C., Lee D.H. Co-chaperone CHIP associates with mutant Cu/Zn-superoxide dismutase proteins linked to familial amyotrophic lateral sclerosis and promotes their degradation by proteasomes. Biochem Biophys Res Commun. 2004;321(3):574–583. doi: 10.1016/j.bbrc.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 258.Urushitani M., Kurisu J., Tateno M., et al. CHIP promotes proteasomal degradation of familial ALS-linked mutant SOD1 by ubiquitinating Hsp/Hsc70. J Neurochem. 2004;90(1):231–244. doi: 10.1111/j.1471-4159.2004.02486.x. [DOI] [PubMed] [Google Scholar]
- 259.Jana N.R., Dikshit P., Goswami A., et al. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem. 2005;280(12):11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- 260.Burgos P.I., Causey Z.L., Tamhane A., et al. Association of IL4R single-nucleotide polymorphisms with rheumatoid nodules in African Americans with rheumatoid arthritis. Arthritis Res Ther. 2010;12(3):R75. doi: 10.1186/ar2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Xie J., Gu J. Identification of C-terminal Hsp70-interacting protein as a mediator of tumour necrosis factor action in osteoblast differentiation by targeting osterix for degradation. J Cell Mol Med. 2015;19(8):1814–1824. doi: 10.1111/jcmm.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Li S., Shu B., Zhang Y., et al. CHIP regulates osteoclast formation through promoting TRAF6 protein degradation. Arthritis Rheumatol Hoboken NJ. 2014;66(7):1854–1863. doi: 10.1002/art.38521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wang T., Li S., Yi D., et al. CHIP regulates bone mass by targeting multiple TRAF family members in bone marrow stromal cells. Bone Res. 2018;6:10. doi: 10.1038/s41413-018-0010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yadava K., Marsland B.J. IL-4Rα, a STUB-strat for proteasomal degradation: understanding the termination of cytokine signaling in asthma. Am JRespirCrit CareMed. 2014;189(1):4–6. doi: 10.1164/rccm.201311-2083ED. [DOI] [PubMed] [Google Scholar]
- 265.Oh C.K., Geba G.P., Molfino N. Investigational therapeutics targeting the IL-4/IL-13/STAT-6 pathway for the treatment of asthma. Eur Respir Rev. 2010;19(115):46–54. doi: 10.1183/09059180.00007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wang L., Liu Y.T., Hao R., et al. Molecular mechanism of the negative regulation of Smad1/5 protein by carboxyl terminus of Hsc70-interacting protein (CHIP) J Biol Chem. 2011;286(18):15883–15894. doi: 10.1074/jbc.M110.201814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Li L., Xin H., Xu X., et al. CHIP mediates degradation of Smad proteins and potentially regulates Smad-induced transcription. Mol Cell Biol. 2004;24(2):856–864. doi: 10.1128/MCB.24.2.856-864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Schroeder T.M., Jensen E.D., Westendorf J.J. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75(3):213–225. doi: 10.1002/bdrc.20043. [DOI] [PubMed] [Google Scholar]
- 269.Liu T., Zhang L., Joo D., Sun S.C. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ye H., Arron J.R., Lamothe B., et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418(6896):443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 271.Kadono Y., Okada F., Perchonock C., et al. Strength of TRAF6 signalling determines osteoclastogenesis. EMBO Rep. 2005;6(2):171–176. doi: 10.1038/sj.embor.7400345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Löffek S., Wöll S., Höhfeld J., et al. The ubiquitin ligase CHIP/STUB1 targets mutant keratins for degradation. Hum Mutat. 2010;31(4):466–476. doi: 10.1002/humu.21222. [DOI] [PubMed] [Google Scholar]
- 273.Xue F., Wu Y., Zhao X., et al. CHIP mediates down-regulation of nucleobindin-1 in preosteoblast cell line models. Cell Signal. 2016;28(8):1058–1065. doi: 10.1016/j.cellsig.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 274.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Esser C., Scheffner M., Höhfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem. 2005;280(29):27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- 276.Strahler JR Kuick R., Hanash S.M. Diminished phosphorylation of a heat shock protein (HSP 27) in infant acute lymphoblastic leukemia. Biochem Biophys Res Commun. 1991;175(1):134–142. doi: 10.1016/s0006-291x(05)81211-2. [DOI] [PubMed] [Google Scholar]
- 277.Min J.N., Whaley R.A., Sharpless N.E., Lockyer P., Portbury A.L., Patterson C. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol Cell Biol. 2008;28(12):4018–4025. doi: 10.1128/MCB.00296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kopp Y., Lang W.H., Schuster T.B., et al. CHIP as a membrane-shuttling proteostasis sensor. Elife. 2017;6 doi: 10.7554/eLife.29388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Frye R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273(2):793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 280.Branicky R., Hekimi S. Proteostasis or aging: let the CHIPs fall where they may. Dev Cell. 2017;41(2):126–128. doi: 10.1016/j.devcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 281.Shimamoto S., Kubota Y., Yamaguchi F., Tokumitsu H., Kobayashi R. Ca2+/S100 proteins act as upstream regulators of the chaperone-associated ubiquitin ligase CHIP (C terminus of hsc70-interacting protein) J Biol Chem. 2013;288(10):7158–7168. doi: 10.1074/jbc.M112.436758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Xie P., Fan Y., Zhang H., et al. CHIP represses myocardin-induced smooth muscle cell differentiation via ubiquitin-mediated proteasomal degradation. Mol Cell Biol. 2009;29(9):2398–2408. doi: 10.1128/MCB.01737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Porpora M., Sauchella S., Rinaldi L., et al. Counterregulation of cAMP-directed kinase activities controls ciliogenesis. Nat Commun. 2018;9(1):1224. doi: 10.1038/s41467-018-03643-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Shi C., Huang X., Zhang B., et al. The Inhibition of Heat Shock Protein 90 Facilitates the Degradation of Poly-Alanine Expanded Poly (A) Binding Protein Nuclear 1 via the Carboxyl Terminus of Heat Shock Protein 70-Interacting Protein. PLoS One. 2015;10(9):e0138936. doi: 10.1371/journal.pone.0138936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Hwang I., Cho S.W., Ahn J.Y. Chaperone-E3 ligase complex HSP70-CHIP mediates ubiquitination of ribosomal protein S3. Int J Mol Sci. 2018;19(9):2723. doi: 10.3390/ijms19092723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Sha Y., Rao L., Settembre C., Ballabio A., Eissa N.T. STUB 1 regulates TFEB-induced autophagy–lysosome pathway. EMBO J. 2017;36(17):2544–2552. doi: 10.15252/embj.201796699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zhu X., Zhang J., Sun H., et al. Ubiquitination of inositol-requiring enzyme 1 (IRE1) by the E3 ligase CHIP mediates the IRE1/TRAF2/JNK pathway. J Biol Chem. 2014;289(44):30567–30577. doi: 10.1074/jbc.M114.562868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Gao Y., Yechikov S., Vazquez A.E., Chen D., Nie L. Distinct roles of molecular chaperones HSP90α and HSP90β in the biogenesis of KCNQ4 channels. PLoS One. 2013;8(2):e57282. doi: 10.1371/journal.pone.0057282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Yang S.W., Oh K.H., Park E., et al. USP47 and C terminus of Hsp70-interacting protein (CHIP) antagonistically regulate katanin-p60-mediated axonal growth. J Neurosci. 2013;33(31):12728–12738. doi: 10.1523/JNEUROSCI.0698-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lee S., Lee D.W., Lee Y., et al. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell. 2009;21(12):3984–4001. doi: 10.1105/tpc.109.071548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Hoeijmakers J.H. DNA damage, aging, and cancer. N Engl J Med. 2009;361(15):1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 292.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zinger A., Cho W.C., Ben-Yehuda A. Cancer and aging-the inflammatory connection. Aging Dis. 2017;8(5):611–627. doi: 10.14336/AD.2016.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Soria-Valles C., López-Soto A., Osorio F.G., López-Otin C. Immune and inflammatory responses to DNA damage in cancer and aging. Mech Ageing Dev. 2017;165 (Pt A):10–16. doi: 10.1016/j.mad.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 295.Lasry A., Ben-Neriah Y. Senescence-associated inflammatory responses: aging and cancer perspectives. Trends Immunol. 2015;36(4):217–228. doi: 10.1016/j.it.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 296.Chung H.Y., Kim H.J., Jung K.J., et al. The inflammatory process in aging. Rev Clin Gerontol. 2000;10(3):207–222. [Google Scholar]
- 297.Rao C.V., Asch A.S., Yamada H.Y. Emerging links among Chromosome Instability (CIN), cancer, and aging. Mol Carcinog. 2017;56(3):791–803. doi: 10.1002/mc.22539. [DOI] [PubMed] [Google Scholar]
- 298.Crow M.T. Revisiting P53 and its Effectors in Ischemic Heart Injury. Cardiovasc Res. 2006;70(3):401–403. doi: 10.1016/j.cardiores.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 299.Naito A.T., Okada S., Minamino T., et al. Promotion of CHIP-mediated p53 degradation protects the heart from ischemic injury. Circ Res. 2010;106(11):1692. doi: 10.1161/CIRCRESAHA.109.214346. [DOI] [PubMed] [Google Scholar]
- 300.Long X., Boluyt M.O., Hipolito M.L., et al. p53 and the hypoxia-induced apoptosis of cultured neonatal rat cardiac myocytes. J Clin Invest. 1997;99(11):2635–2643. doi: 10.1172/JCI119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Woo C.H., Le N.T., Shishido T., et al. Novel role of C terminus of Hsc70-interacting protein (CHIP) ubiquitin ligase on inhibiting cardiac apoptosis and dysfunction via regulating ERK5-mediated degradation of inducible cAMP early repressor. Faseb J. 2010;24(12):4917–4928. doi: 10.1096/fj.10-162636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Chao C.N., Lai C.H., Badrealam K.F., et al. CHIP attenuates lipopolysaccharide-induced cardiac hypertrophy and apoptosis by promoting NFATc3 proteasomal degradation. J Cell Physiol. 2019;234(11):20128–20138. doi: 10.1002/jcp.28614. [DOI] [PubMed] [Google Scholar]
- 303.Iwai C., Li P., Kurata Y., et al. Hsp90 prevents interaction between CHIP and HERG proteins to facilitate maturation of wild-type and mutant HERG proteins. Cardiovasc Res. 2013;100(3):520–528. doi: 10.1093/cvr/cvt200. [DOI] [PubMed] [Google Scholar]
- 304.Li P., Kurata Y., Maharani N., et al. E3 ligase CHIP and Hsc70 regulate Kv1.5 protein expression and function in mammalian cells. J Mol Cell Cardiol. 2015;86:138–146. doi: 10.1016/j.yjmcc.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 305.Donnelly B.F., Needham P.G., Snyder A.C., et al. Hsp70 and Hsp90 multichaperone complexes sequentially regulate thiazide-sensitive cotransporter endoplasmic reticulum-associated degradation and biogenesis. J Biol Chem. 2013;288(18):13124–13135. doi: 10.1074/jbc.M113.455394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Feng C.C., Liao P.H., Tsai H.I., et al. Tumorous imaginal disc 1 (TID1) inhibits isoproterenol-induced cardiac hypertrophy and apoptosis by regulating c-terminus of hsc70-interacting protein (CHIP) mediated degradation of Gαs. Int J Med Sci. 2018;15(13):1537–1546. doi: 10.7150/ijms.24296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wang L., Zhang T.-P., Zhang Y., et al. Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein. Sci Rep. 2016;6:28399. doi: 10.1038/srep28399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Mudd T.W., Khalid M., Guddati A.K. Cardiotoxicity of chemotherapy and targeted agents. Am J Cancer Res. 2021;11(7):3461–3474. [PMC free article] [PubMed] [Google Scholar]
- 309.Santoni M., Guerra F., Conti A., et al. Incidence and risk of cardiotoxicity in cancer patients treated with targeted therapies. Cancer Treat Rev. 2017;59:123–131. doi: 10.1016/j.ctrv.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 310.Shah C.P., Moreb J.S. Cardiotoxicity due to targeted anticancer agents: a growing challenge. Ther Adv Cardiovasc Dis. 2019;13 doi: 10.1177/1753944719843435. 1753944719843435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Verma S., Ewer M.S. Is cardiotoxicity being adequately assessed in current trials of cytotoxic and targeted agents in breast cancer? Ann Oncol. 2011;22(5):1011–1018. doi: 10.1093/annonc/mdq607. [DOI] [PubMed] [Google Scholar]
- 312.Berardi R., Caramanti M., Savini A., et al. State of the art for cardiotoxicity due to chemotherapy and to targeted therapies: a literature review. Crit Rev Oncol Hematol. 2013;88(1):75–86. doi: 10.1016/j.critrevonc.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 313.Chen Z.I., Ai D.I. Cardiotoxicity associated with targeted cancer therapies. Mol Clin Oncol. 2016;4(5):675–681. doi: 10.3892/mco.2016.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kimura T., Uesugi M., Takase K., Miyamoto N., Sawada K. Hsp90 inhibitor geldanamycin attenuates the cytotoxicity of sunitinib in cardiomyocytes via inhibition of the autophagy pathway. Toxicol Appl Pharmacol. 2017;329:282–292. doi: 10.1016/j.taap.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 315.Gabrielson K., Bedja D., Pin S., et al. Heat shock protein 90 and ErbB2 in the cardiac response to doxorubicin injury. Cancer Res. 2007;67(4):1436–1441. doi: 10.1158/0008-5472.CAN-06-3721. [DOI] [PubMed] [Google Scholar]
- 316.Pecoraro M., Sorrentino R., Franceschelli S., Del Pizzo M., Pinto A., Popolo A. Doxorubicin-mediated cardiotoxicity: role of mitochondrial connexin 43. Cardiovasc Toxicol. 2015;15(4):366–376. doi: 10.1007/s12012-014-9305-8. [DOI] [PubMed] [Google Scholar]
- 317.Robles A.I., Wright M.H., Gandhi B., et al. Schedule-dependent synergy between the heat shock protein 90 inhibitor 17-(dimethylaminoethylamino)-17-demethoxygeldanamycin and doxorubicin restores apoptosis to p53-mutant lymphoma cell lines. Clin Cancer Res. 2006;12(21):6547–6556. doi: 10.1158/1078-0432.CCR-06-1178. [DOI] [PubMed] [Google Scholar]
- 318.Fouad Y.A., Aanei C. Revisiting the hallmarks of cancer. Am J Cancer Res. 2017;7(5):1016–1036. [PMC free article] [PubMed] [Google Scholar]
- 319.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 320.Zhou P., Fernandes N., Dodge I.L., et al. ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol Chem. 2003;278(16):13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 321.Kajiro M., Hirota R., Nakajima Y., et al. The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat Cell Biol. 2009;11(3):312–319. doi: 10.1038/ncb1839. [DOI] [PubMed] [Google Scholar]
- 322.Schulz R., Marchenko N.D., Holembowski L., et al. Inhibiting the HSP90 chaperone destabilizes macrophage migration inhibitory factor and thereby inhibits breast tumor progression. J Exp Med. 2012;209(2):275–289. doi: 10.1084/jem.20111117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Pan C., Chun J., Li D., et al. Hsp90B enhances MAST1-mediated cisplatin resistance by protecting MAST1 from proteosomal degradation. J Clin Invest. 2019;129(10):4110–4123. doi: 10.1172/JCI125963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Xiao M., Yan M., Zhang J., et al. Cancer stem-like cell related protein CD166 degrades through E3 ubiquitin ligase CHIP in head and neck cancer. Exp Cell Res. 2017;353(1):46–53. doi: 10.1016/j.yexcr.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Laederich M.B., Degnin C.R., Lunstrum G.P., Holden P., Horton W.A. Fibroblast growth factor receptor 3 (FGFR3) is a strong heat shock protein 90 (Hsp90) client: implications for therapeutic manipulation. J Biol Chem. 2011;286(22):19597–19604. doi: 10.1074/jbc.M110.206151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Yoo L., Yoon A.R., Yun C.O., Chung K.C. Covalent ISG15 conjugation to CHIP promotes its ubiquitin E3 ligase activity and inhibits lung cancer cell growth in response to type I interferon. Cell Death Dis. 2018;9:97. doi: 10.1038/s41419-017-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Krishnamoorthy G.P., Guida T., Alfano L., et al. Molecular mechanism of 17-allylamino-17-demethoxygeldanamycin (17-AAG)-induced AXL receptor tyrosine kinase degradation. J Biol Chem. 2013;288(24):17481–17494. doi: 10.1074/jbc.M112.439422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Carraway K.L., III E3 ubiquitin ligases in ErbB receptor quantity control. Semin Cell Dev Biol. 2010;21(9):936–943. doi: 10.1016/j.semcdb.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Jeong J.-H., An J.Y., Kwon Y.T., Li L.-Y., Lee Y.J. Quercetin-induced ubiquitination and down-regulation of Her-2/neu. J Cell Biochem. 2008;105(2):585–595. doi: 10.1002/jcb.21859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Jung Y., Xu W., Kim H., Ha N., Neckers L. Curcumin-induced degradation of ErbB2: a role for the E3 ubiquitin ligase CHIP and the Michael reaction acceptor activity of curcumin. Biochim Biophys Acta BBA-Mol Cell Res. 2007;1773(3):383–390. doi: 10.1016/j.bbamcr.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 331.Lee J.H., Khadka P., Baek S.H., Chung I.K. CHIP promotes human telomerase reverse transcriptase degradation and negatively regulates telomerase activity. J Biol Chem. 2010;285(53):42033–42045. doi: 10.1074/jbc.M110.149831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Lei H., Jin J., Liu M., et al. Chk1 inhibitors overcome imatinib resistance in chronic myeloid leukemia cells. Leuk Res. 2018;64:17–23. doi: 10.1016/j.leukres.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 333.Huang H., Weng H., Dong B., Zhao P., Zhou H., Qu L. Oridonin triggers chaperon-mediated proteasomal degradation of BCR-ABL in leukemia. Sci Rep. 2017;7:41525. doi: 10.1038/srep41525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Chung C., Yoo G., Kim T., et al. The E3 ubiquitin ligase CHIP selectively regulates mutant epidermal growth factor receptor by ubiquitination and degradation. Biochem Biophys Res Commun. 2016;479(2):152–158. doi: 10.1016/j.bbrc.2016.07.111. [DOI] [PubMed] [Google Scholar]
- 335.Wang T., Yang J., Xu J., et al. CHIP is a novel tumor suppressor in pancreatic cancer through targeting EGFR. Oncotarget. 2014;5(7):1969–1986. doi: 10.18632/oncotarget.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Hou J., Deng Q., Zhou J., et al. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene. 2017;36(8):1134–1144. doi: 10.1038/onc.2016.280. [DOI] [PubMed] [Google Scholar]
- 337.Wang X., Cao W., Zhang J., et al. A covalently bound inhibitor triggers EZH 2 degradation through CHIP-mediated ubiquitination. EMBO J. 2017;36(9):1243–1260. doi: 10.15252/embj.201694058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Shang Y., He J., Wang Y., et al. CHIP/Stub1 regulates the Warburg effect by promoting degradation of PKM2 in ovarian carcinoma. Oncogene. 2017;36(29):4191–4200. doi: 10.1038/onc.2017.31. [DOI] [PubMed] [Google Scholar]
- 339.Hwang I., Kim C.K., Ko H.R., Park K.W., Cho S.W., Ahn J.Y. C-terminal domain of p42 Ebp1 is essential for down regulation of p85 subunit of PI3K, inhibiting tumor growth. Sci Rep. 2016;6:30626. doi: 10.1038/srep30626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Zhang H.T., Zeng L.F., He Q.Y., Tao W.A., Zha Z.G., Hu C.D. The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochim Biophys Acta. 2016;1863(2):335–346. doi: 10.1016/j.bbamcr.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Cho Y., Kang H.G., Kim S.J., et al. Post-translational modification of OCT4 in breast cancer tumorigenesis. Cell Death Differ. 2018;25(10):1781–1795. doi: 10.1038/s41418-018-0079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Min B., Park H., Lee S., et al. CHIP-mediated degradation of transglutaminase 2 negatively regulates tumor growth and angiogenesis in renal cancer. Oncogene. 2016;35(28):3718–3728. doi: 10.1038/onc.2015.439. [DOI] [PubMed] [Google Scholar]
- 343.Blessing N.A., Kasturirangan S., Zink E.M., Schroyer A.L., Chadee D.N. Osmotic and heat stress-dependent regulation of MLK4β and MLK3 by the CHIP E3 ligase in ovarian cancer cells. Cell Signal. 2017;39:66–73. doi: 10.1016/j.cellsig.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Bonvini P., Dalla Rosa H., Vignes N., Rosolen A. Ubiquitination and proteasomal degradation of nucleophosmin-anaplastic lymphoma kinase induced by 17-allylamino-demethoxygeldanamycin: role of the co-chaperone carboxyl heat shock protein 70-interacting protein. Cancer Res. 2004;64(9):3256–3264. doi: 10.1158/0008-5472.can-03-3531. [DOI] [PubMed] [Google Scholar]
- 345.Kang S.A., Cho H.S., Yoon J.B., Chung I.K., Lee S.T. Hsp90 rescues PTK6 from proteasomal degradation in breast cancer cells. Biochem J. 2012;447(2):313–320. doi: 10.1042/BJ20120803. [DOI] [PubMed] [Google Scholar]
- 346.Dhamad A.E., Zhou Z., Zhou J., Du Y. Systematic proteomic identification of the heat shock proteins (Hsp) that interact with estrogen receptor alpha (ERα) and biochemical characterization of the ERα-Hsp70 interaction. PLoS One. 2016;11(8):e0160312. doi: 10.1371/journal.pone.0160312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wu H., Yao S., Zhang S., et al. Elevated expression of Erbin destabilizes ERα protein and promotes tumorigenesis in hepatocellular carcinoma. J Hepatol. 2017;66(6):1193–1204. doi: 10.1016/j.jhep.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 348.Black K.L., Witty C.F., Daniel J.M. Previous midlife oestradiol treatment results in long-term maintenance of hippocampal oestrogen receptor α levels in ovariectomised rats: mechanisms and implications for memory. J Neuroendocrinol. 2016;28(10) doi: 10.1111/jne.12429. 10.1111/jne.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Fan M., Park A., Nephew K.P. CHIP (carboxyl terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-α. Mol Endocrinol. 2005;19(12):2901–2914. doi: 10.1210/me.2005-0111. [DOI] [PubMed] [Google Scholar]
- 350.Tateishi Y., Sonoo R., Sekiya Y.I., et al. Turning off estrogen receptor β-mediated transcription requires estrogen-dependent receptor proteolysis. Mol Cell Biol. 2006;26(21):7966–7976. doi: 10.1128/MCB.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Zhang X., Qian S.B. Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol Biol Cell. 2011;22(18):3277–3288. doi: 10.1091/mbc.E11-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Shang Y., Zhao X., Tian B., et al. CHIP/Stub1 interacts with eIF5A and mediates its degradation. Cell Signal. 2014;26(5):1098–1104. doi: 10.1016/j.cellsig.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 353.Li X., Zhong L., Wang Z., et al. Phosphorylation of IRS4 by CK1γ2 promotes its degradation by CHIP through the ubiquitin/lysosome pathway. Theranostics. 2018;8(13):3643–3653. doi: 10.7150/thno.26021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Blessing N.A., Brockman A.L., Chadee D.N. The E3 ligase CHIP mediates ubiquitination and degradation of mixed-lineage kinase 3. Mol Cell Biol. 2014;34(16):3132–3143. doi: 10.1128/MCB.00296-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ko H.R., Kim C.K., Lee S.B., et al. P42 Ebp1 regulates the proteasomal degradation of the p85 regulatory subunit of PI3K by recruiting a chaperone-E3 ligase complex HSP70/CHIP. Cell Death Dis. 2014;5(3):e1131. doi: 10.1038/cddis.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Nie L., Wu H., Sun X.H. Ubiquitination and degradation of Tal1/SCL are induced by notch signaling and depend on Skp2 and CHIP. J Biol Chem. 2008;283(2):684–692. doi: 10.1074/jbc.M704981200. [DOI] [PubMed] [Google Scholar]
- 357.Terme J.M., Lhermitte L., Asnafi V., Jalinot P. TGF-β induces degradation of TAL1/SCL by the ubiquitin-proteasome pathway through AKT-mediated phosphorylation. Blood J Am Soc Hematol. 2009;113(26):6695–6698. doi: 10.1182/blood-2008-07-166835. [DOI] [PubMed] [Google Scholar]
- 358.Kim S., Ham S., Yang K., Kim K. Protein kinase CK2 activation is required for transforming growth factor β-induced epithelial–mesenchymal transition. Mol Oncol. 2018;12(10):1811–1826. doi: 10.1002/1878-0261.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Kao S.H., Wang W.L., Chen C.Y., et al. GSK3β controls epithelial–mesenchymal transition and tumor metastasis by CHIP-mediated degradation of Slug. Oncogene. 2014;33(24):3172–3182. doi: 10.1038/onc.2013.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Seo J.H., Agarwal E., Bryant K.G., et al. Syntaphilin ubiquitination regulates mitochondrial dynamics and tumor cell movements. Cancer Res. 2018;78(15):4215–4228. doi: 10.1158/0008-5472.CAN-18-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Dai H., Chen H., Xu J., et al. The ubiquitin ligase CHIP modulates cellular behaviors of gastric cancer cells by regulating TRAF2. Cancer Cell Int. 2019;19:132. doi: 10.1186/s12935-019-0832-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Jang K.W., Lee K.H., Kim S.H., et al. Ubiquitin ligase CHIP induces TRAF2 proteasomal degradation and NF-κB inactivation to regulate breast cancer cell invasion. J Cell Biochem. 2011;112(12):3612–3620. doi: 10.1002/jcb.23292. [DOI] [PubMed] [Google Scholar]
- 363.Luan H., Mohapatra B., Bielecki T.A., et al. Loss of the nuclear pool of ubiquitin ligase CHIP/STUB1 in breast cancer unleashes the MZF1-cathepsin pro-oncogenic program. Cancer Res. 2018;78(10):2524–2535. doi: 10.1158/0008-5472.CAN-16-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Wang Y., Ren F., Wang Y., et al. CHIP/Stub1 functions as a tumor suppressor and represses NF- B-mediated signaling in colorectal cancer. Carcinogenesis. 2014;35(5):983–991. doi: 10.1093/carcin/bgt393. [DOI] [PubMed] [Google Scholar]
- 365.Park S.M., Park S.H., Ryu K.J., et al. Downregulation of CHIP promotes ovarian cancer metastasis by inducing Snail-mediated epithelial–mesenchymal transition. Mol Oncol. 2019;13(5):1280–1295. doi: 10.1002/1878-0261.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Won K.J., Im J.Y., Kim B.K., et al. Stability of the cancer target DDIAS is regulated by the CHIP/HSP70 pathway in lung cancer cells. Cell Death Dis. 2018;8(1):e2554. doi: 10.1038/cddis.2016.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Li D., Marchenko N.D., Schulz R., et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9(5):577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Kwegyir-Afful A.K., Ramalingam S., Purushottamachar P., Ramamurthy V.P., Njar V.C. Galeterone and VNPT55 induce proteasomal degradation of AR/AR-V7, induce significant apoptosis via cytochrome c release and suppress growth of castration resistant prostate cancer xenografts in vivo. Oncotarget. 2015;6(29):27440–27460. doi: 10.18632/oncotarget.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Wang J., Zhang H., Zhang X., et al. PC-1 works in conjunction with E3 ligase CHIP to regulate androgen receptor stability and activity. Oncotarget. 2016;7(49):81377–81388. doi: 10.18632/oncotarget.13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Moon S.J., Jeong B.C., Kim H.J., Lim J.E., Kwon G.Y., Kim J.H. DBC1 promotes castration-resistant prostate cancer by positively regulating DNA binding and stability of AR-V7. Oncogene. 2018;37(10):1326–1339. doi: 10.1038/s41388-017-0047-5. [DOI] [PubMed] [Google Scholar]
- 371.Sarkar S., Brautigan D.L., Parsons S.J., Larner J.M. Androgen receptor degradation by the E3 ligase CHIP modulates mitotic arrest in prostate cancer cells. Oncogene. 2014;33(1):26–33. doi: 10.1038/onc.2012.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Sarkar S., Brautigan D.L., Larner J.M. Aurora kinase A promotes AR degradation via the E3 ligase CHIP. Mol Cancer Res. 2017;15(8):1063–1072. doi: 10.1158/1541-7786.MCR-17-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Su C.H., Lan K.H., Li C.P., et al. Phosphorylation accelerates geldanamycin-induced Akt degradation. Arch Biochem Biophys. 2013;536(1):6–11. doi: 10.1016/j.abb.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 374.Paul I., Ahmed S.F., Bhowmik A., Deb S., Ghosh M.K. The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity. Oncogene. 2013;32(10):1284–1295. doi: 10.1038/onc.2012.144. [DOI] [PubMed] [Google Scholar]
- 375.Patani N., Jiang W., Newbold R., Mokbel K. Prognostic implications of carboxyl-terminus of Hsc70 interacting protein and lysyl-oxidase expression in human breast cancer. J Carcinog. 2010;9:9. doi: 10.4103/1477-3163.72505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Ahmed S.F., Das N., Sarkar M., Chatterjee U., Chatterjee S., Ghosh M.K. Exosome-mediated delivery of the intrinsic C-terminus domain of PTEN protects it from proteasomal degradation and ablates tumorigenesis. Mol Ther. 2015;23(2):255–269. doi: 10.1038/mt.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Li F., Xie P., Fan Y., et al. C terminus of Hsc70-interacting protein promotes smooth muscle cell proliferation and survival through ubiquitin-mediated degradation of FoxO1. J Biol Chem. 2009;284(30):20090–20098. doi: 10.1074/jbc.M109.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Choi J.R., Shin K.S., Choi C.Y., Kang S.J. PARP1 regulates the protein stability and proapoptotic function of HIPK2. Cell Death Dis. 2016;7(10):e2438. doi: 10.1038/cddis.2016.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Han S.Y., Ko A., Kitano H., et al. Molecular chaperone HSP90 is necessary to prevent cellular senescence via lysosomal degradation of p14ARF. Cancer Res. 2017;77(2):343–354. doi: 10.1158/0008-5472.CAN-16-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Biswas K., Sarkar S., Du K., Brautigan D.L., Abbas T., Larner J.M. The E3 ligase CHIP mediates p21 degradation to maintain radioresistance. Mol Cancer Res. 2017;15(6):651–659. doi: 10.1158/1541-7786.MCR-16-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Shang Y., Xu X., Duan X., et al. Hsp70 and Hsp90 oppositely regulate TGF-β signaling through CHIP/Stub1. Biochem Biophys Res Commun. 2014;446(1):387–392. doi: 10.1016/j.bbrc.2014.02.124. [DOI] [PubMed] [Google Scholar]
- 382.Choi Y.N., Lee S.K., Seo T.W., Lee J.S., Yoo S.J. C-terminus of Hsc70-interacting protein regulates profilin1 and breast cancer cell migration. Biochem Biophys Res Commun. 2014;446(4):1060–1066. doi: 10.1016/j.bbrc.2014.03.061. [DOI] [PubMed] [Google Scholar]
- 383.Sane S., Hafner A., Srinivasan R., et al. UBXN2A enhances CHIP-mediated proteasomal degradation of oncoprotein mortalin-2 in cancer cells. Mol Oncol. 2018;12(10):1753–1777. doi: 10.1002/1878-0261.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Canaff L., Vanbellinghen J.F., Kanazawa I., et al. Menin missense mutants encoded by the MEN1 gene that are targeted to the proteasome: restoration of expression and activity by CHIP siRNA. J Clin Endocrinol Metab. 2012;97(2):E282–E291. doi: 10.1210/jc.2011-0241. [DOI] [PubMed] [Google Scholar]
- 385.Matkar S., Thiel A., Hua X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem Sci. 2013;38(8):394–402. doi: 10.1016/j.tibs.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Niki T., Tsutsui S., Hirose S., et al. Galectin-9 is a high affinity IgE-binding lectin with anti-allergic effect by blocking IgE-antigen complex formation. J Biol Chem. 2009;284(47):32344–32352. doi: 10.1074/jbc.M109.035196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Gaude H., Aznar N., Delay A., et al. Molecular chaperone complexes with antagonizing activities regulate stability and activity of the tumor suppressor LKB1. Oncogene. 2012;31(12):1582–1591. doi: 10.1038/onc.2011.342. [DOI] [PubMed] [Google Scholar]
- 388.Tanwar K., Pati U. Inhibition of apoptosis via CHIP-mediated proteasomal degradation of TAp73α. J Cell Biochem. 2019;120(7):11091–11103. doi: 10.1002/jcb.28386. [DOI] [PubMed] [Google Scholar]
- 389.Wen J., Luo K.J., Hu Y., Yang H., Fu J.H. Metastatic lymph node CHIP expression is a potential prognostic marker for resected esophageal squamous cell carcinoma patients. Ann Surg Oncol. 2013;20(5):1668–1675. doi: 10.1245/s10434-012-2733-4. [DOI] [PubMed] [Google Scholar]
- 390.Liang Z.L., Kim M., Huang S.M., Lee H.J., Kim J.M. Expression of carboxyl terminus of Hsp70-interacting protein (CHIP) indicates poor prognosis in human gallbladder carcinoma. Oncol Lett. 2013;5(3):813–818. doi: 10.3892/ol.2013.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Cheng L., Zang J., Dai H.J., Li F., Guo F. Ubiquitin ligase CHIP functions as an oncogene and activates the AKT signaling pathway in prostate cancer. Int J Oncol. 2018;53(1):203–214. doi: 10.3892/ijo.2018.4377. [DOI] [PubMed] [Google Scholar]
- 392.Jan C.I., Yu C.C., Hung M.C., et al. Tid1, CHIP and ErbB2 interactions and their prognostic implications for breast cancer patients. J Pathol. 2011;225(3):424–437. doi: 10.1002/path.2921. [DOI] [PubMed] [Google Scholar]
- 393.Xu J., Wang H., Li W., et al. E3 ubiquitin ligase CHIP attenuates cellular proliferation and invasion abilities in triple-negative breast cancer cells. Clin Exp Med. 2020;20(1):109–119. doi: 10.1007/s10238-019-00594-3. [DOI] [PubMed] [Google Scholar]
- 394.Kurozumi S., Yamaguchi Y., Hayashi S.I., et al. Prognostic value of the ubiquitin ligase carboxyl terminus of the Hsc70-interacting protein in postmenopausal breast cancer. Cancer Med. 2016;5(8):1873–1882. doi: 10.1002/cam4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Gan L., Liu D.B., Lu H.F., et al. Decreased expression of the carboxyl terminus of heat shock cognate 70 interacting protein in human gastric cancer and its clinical significance. Oncol Rep. 2012;28(4):1392–1398. doi: 10.3892/or.2012.1957. [DOI] [PubMed] [Google Scholar]
- 396.Wang S., Wu X., Zhang J., et al. CHIP functions as a novel suppressor of tumour angiogenesis with prognostic significance in human gastric cancer. Gut. 2013;62(4):496–508. doi: 10.1136/gutjnl-2011-301522. [DOI] [PubMed] [Google Scholar]
- 397.Sun C., Li H., Chen H., et al. Decreased expression of CHIP leads to increased angiogenesis via VEGF-VEGFR2 pathway and poor prognosis in human renal cell carcinoma. Sci Rep. 2015;5:9774. doi: 10.1038/srep09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Qian T., Wang J., Wang Q., et al. CHIP involves in non-small cell lung cancer prognosis through VEGF pathway. Biomed Pharmacother. 2016;83:271–276. doi: 10.1016/j.biopha.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 399.Xu T., Zhou Q., Zhou J., et al. Carboxyl terminus of Hsp70-interacting protein (CHIP) contributes to human glioma oncogenesis. Cancer Sci. 2011;102(5):959–966. doi: 10.1111/j.1349-7006.2011.01888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Jin Y., Zhou L., Liang Z.Y., Jin K.M., Zhou W.X., Xing B.C. Clinicopathologic and prognostic significance of carboxyl terminus of Hsp70-interacting protein in HBV-related hepatocellular carcinoma. Asian Pac J Cancer Prev APJCP. 2015;16(9):3709–3713. doi: 10.7314/apjcp.2015.16.9.3709. [DOI] [PubMed] [Google Scholar]
- 401.Xiao M., Yan M., Zhang J., Xu Q., Chen W. Carboxy-terminus Hsc70 interacting protein exerts a tumor inhibition function in head and neck cancer. Oncol Rep. 2017;38(3):1629–1636. doi: 10.3892/or.2017.5827. [DOI] [PubMed] [Google Scholar]
- 402.Xu J., Zhou J., Dai H., et al. CHIP functions as an oncogene by promoting colorectal cancer metastasis via activation of MAPK and AKT signaling and suppression of E-cadherin. J Transl Med. 2018;16(1):169. doi: 10.1186/s12967-018-1540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Wen P.Y., Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 404.Yi X., Wei W., Wang S.Y., Du Z.Y., Xu Y.J., Yu X.D. Histone deacetylase inhibitor SAHA induces ERα degradation in breast cancer MCF-7 cells by CHIP-mediated ubiquitin pathway and inhibits survival signaling. Biochem Pharmacol. 2008;75(9):1697–1705. doi: 10.1016/j.bcp.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 405.Allred D.C., Mohsin S.K. Biological features of premalignant disease in the human breast. J Mammary Gland Biol Neoplasia. 2000;5(4):351–364. doi: 10.1023/a:1009573710675. [DOI] [PubMed] [Google Scholar]
- 406.Harvey J.M., Clark G.M., Osborne C.K., Allred D.C. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17(5):1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 407.Pratt W.B., Toft D.O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18(3):306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 408.Beliakoff J., Bagatell R., Paine-Murrieta G., Taylor C.W., Lykkesfeldt A.E., Whitesell L. Hormone-refractory breast cancer remains sensitive to the antitumor activity of heat shock protein 90 inhibitors. Clin Cancer Res. 2003;9(13):4961–4971. [PubMed] [Google Scholar]
- 409.Paik S., Kim C., Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358(13):1409–1411. doi: 10.1056/NEJMc0801440. [DOI] [PubMed] [Google Scholar]
- 410.Hayes D.F., Thor A.D. c-erbB-2 in breast cancer: development of a clinically useful marker. Semin Oncol. 2002;29(3):231–245. doi: 10.1053/sonc.2002.32899. [DOI] [PubMed] [Google Scholar]
- 411.Camp R.L., Dolled-Filhart M., King B.L., Rimm D.L. Quantitative analysis of breast cancer tissue microarrays shows that both high and normal levels of HER2 expression are associated with poor outcome. Cancer Res. 2003;63(7):1445–1448. [PubMed] [Google Scholar]
- 412.Xu W., Mimnaugh E., Rosser M.F., et al. Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J Biol Chem. 2001;276(5):3702–3708. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- 413.Mimnaugh E.G., Chavany C., Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem. 1996;271(37):22796–22801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 414.Miller P., DiOrio C., Moyer M., et al. Depletion of the erbB-2 gene product p185 by benzoquinoid ansamycins. Cancer Res. 1994;54(10):2724–2730. [PubMed] [Google Scholar]
- 415.Zheng F.F., Kuduk S.D., Chiosis G., et al. Identification of a geldanamycin dimer that induces the selective degradation of HER-family tyrosine kinases. Cancer Res. 2000;60(8):2090–2094. [PubMed] [Google Scholar]
- 416.Huber M.A., Azoitei N., Baumann B., et al. NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114(4):569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Park B.K., Zhang H., Zeng Q., et al. NF-κB in breast cancer cells promotes osteolytic bone metastasis by inducing osteoclastogenesis via GM-CSF. Nat Med. 2007;13(1):62–69. doi: 10.1038/nm1519. [DOI] [PubMed] [Google Scholar]
- 418.Chen S., Guttridge D.C., Tang E., Shi S., Guan K., Wang C.Y. Suppression of tumor necrosis factor-mediated apoptosis by nuclear factor κB-independent bone morphogenetic protein/Smad signaling. J Biol Chem. 2001;276(42):39259–39263. doi: 10.1074/jbc.M105335200. [DOI] [PubMed] [Google Scholar]
- 419.Wu C.J., Conze D.B., Li X., Ying S.X., Hanover J.A., Ashwell J.D. TNF-α induced c-IAP1/TRAF2 complex translocation to an Ubc6-containing compartment and TRAF2 ubiquitination. EMBO J. 2005;24(10):1886–1898. doi: 10.1038/sj.emboj.7600649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Habelhah H., Frew I.J., Laine A., et al. Stress-induced decrease in TRAF2 stability is mediated by Siah2. EMBO J. 2002;21(21):5756–5765. doi: 10.1093/emboj/cdf576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M.J. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 422.Prins G.S., Sklarew R.J., Pertschuk L.P. Image analysis of androgen receptor immunostaining in prostate cancer accurately predicts response to hormonal therapy. J Urol. 1998;159(3):641–649. [PubMed] [Google Scholar]
- 423.Gaughan L., Logan I.R., Neal D.E., Robson C.N. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005;33(1):13–26. doi: 10.1093/nar/gki141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Paoletti X., Oba K., Burzykowski T., et al. Benefit of adjuvant chemotherapy for resectable gastric cancer: a meta-analysis. JAMA. 2010;303(17):1729–1737. doi: 10.1001/jama.2010.534. [DOI] [PubMed] [Google Scholar]
- 425.Gourgiotis S., Kocher H.M., Solaini L., Yarollahi A., Tsiambas E., Salemis N.S. Gallbladder cancer. Am J Surg. 2008;196(2):252–264. doi: 10.1016/j.amjsurg.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 426.Balachandran P., Agarwal S., Krishnani N., et al. Predictors of long-term survival in patients with gallbladder cancer. J Gastrointest Surg. 2006;10(6):848–854. doi: 10.1016/j.gassur.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 427.Park J.S., Yoon D.S., Kim K.S., et al. Analysis of prognostic factors after curative resection for gallbladder carcinoma. Korean J Gastroenterol. 2006;48(1):32–36. [PubMed] [Google Scholar]
- 428.Choi S.Y., Jo Y.S., Huang S.M., et al. L1 cell adhesion molecule as a novel independent poor prognostic factor in gallbladder carcinoma. Hum Pathol. 2011;42(10):1476–1483. doi: 10.1016/j.humpath.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 429.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 430.Folkman J., Kalluri R. Cancer without disease. Nature. 2004;427(6977):787. doi: 10.1038/427787a. [DOI] [PubMed] [Google Scholar]
- 431.Samet J., Hunt W.C., Key C., Humble C.G., Goodwin J.S. Choice of cancer therapy varies with age of patient. JAMA. 1986;255(24):3385–3390. [PubMed] [Google Scholar]
- 432.Wang M.D., Shin D.M., Simons J.W., Nie S. Nanotechnology for targeted cancer therapy. Expert Rev Anticancer Ther. 2007;7(6):833–837. doi: 10.1586/14737140.7.6.833. [DOI] [PubMed] [Google Scholar]
- 433.Fukuyo Y., Hunt C.R., Horikoshi N. Geldanamycin and its anti-cancer activities. Cancer Lett. 2010;290(1):24–35. doi: 10.1016/j.canlet.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 434.Ramanathan R.K., Trump D.L., Eiseman J.L., et al. Phase I pharmacokinetic-pharmacodynamic study of 17-(allylamino)-17-demethoxygeldanamycin (17AAG, NSC 330507), a novel inhibitor of heat shock protein 90, in patients with refractory advanced cancers. Clin Cancer Res. 2005;11(9):3385–3391. doi: 10.1158/1078-0432.CCR-04-2322. [DOI] [PubMed] [Google Scholar]
- 435.Goetz M.P., Toft D., Reid J., et al. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. J Clin Oncol. 2005;23(6):1078–1087. doi: 10.1200/JCO.2005.09.119. [DOI] [PubMed] [Google Scholar]
- 436.Banerji U., O'Donnell A., Scurr M., et al. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J Clin Oncol. 2005;23(18):4152–4161. doi: 10.1200/JCO.2005.00.612. [DOI] [PubMed] [Google Scholar]
- 437.Grem J.L., Morrison G., Guo X.D., et al. Phase I and pharmacologic study of 17-(allylamino)-17-demethoxygeldanamycin in adult patients with solid tumors. J Clin Oncol. 2005;23(9):1885–1893. doi: 10.1200/JCO.2005.12.085. [DOI] [PubMed] [Google Scholar]
- 438.Nowakowski G.S., McCollum A.K., Ames M.M., et al. A phase I trial of twice-weekly 17-allylamino-demethoxy-geldanamycin in patients with advanced cancer. Clin Cancer Res. 2006;12(20):6087–6093. doi: 10.1158/1078-0432.CCR-06-1015. [DOI] [PubMed] [Google Scholar]
- 439.Ronnen E.A., Kondagunta G.V., Ishill N., et al. A phase II trial of 17-(Allylamino)-17-demethoxygeldanamycin in patients with papillary and clear cell renal cell carcinoma. Invest N Drugs. 2006;24(6):543–546. doi: 10.1007/s10637-006-9208-z. [DOI] [PubMed] [Google Scholar]
- 440.Solit D.B., Ivy S.P., Kopil C., et al. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. Clin Cancer Res. 2007;13(6):1775–1782. doi: 10.1158/1078-0432.CCR-06-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Modi S., Stopeck A.T., Gordon M.S., et al. Combination of trastuzumab and tanespimycin (17-AAG, KOS-953) is safe and active in trastuzumab-refractory HER-2–overexpressing breast cancer: a phase I dose-escalation study. J Clin Oncol. 2007;25(34):5410–5417. doi: 10.1200/JCO.2007.11.7960. [DOI] [PubMed] [Google Scholar]
- 442.Ramanathan R.K., Egorin M.J., Erlichman C., et al. Phase I pharmacokinetic and pharmacodynamic study of 17-dimethylaminoethylamino-17-demethoxygeldanamycin, an inhibitor of heat-shock protein 90, in patients with advanced solid tumors. J Clin Oncol. 2010;28(9):1520–1526. doi: 10.1200/JCO.2009.25.0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Weigel B.J., Blaney S.M., Reid J.M., et al. A phase I study of 17-allylaminogeldanamycin in relapsed/refractory pediatric patients with solid tumors: a Children's Oncology Group study. Clin Cancer Res. 2007;13(6):1789–1793. doi: 10.1158/1078-0432.CCR-06-2270. [DOI] [PubMed] [Google Scholar]
- 444.Bagatell R., Gore L., Egorin M.J., et al. Phase I pharmacokinetic and pharmacodynamic study of 17-N-allylamino-17-demethoxygeldanamycin in pediatric patients with recurrent or refractory solid tumors: a pediatric oncology experimental therapeutics investigators consortium study. Clin Cancer Res. 2007;13(6):1783–1788. doi: 10.1158/1078-0432.CCR-06-1892. [DOI] [PubMed] [Google Scholar]
- 445.Tse A.N., Klimstra D.S., Gonen M., et al. A phase 1 dose-escalation study of irinotecan in combination with 17-allylamino-17-demethoxygeldanamycin in patients with solid tumors. Clin Cancer Res. 2008;14(20):6704–6711. doi: 10.1158/1078-0432.CCR-08-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Heath E.I., Hillman D.W., Vaishampayan U., et al. A phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with hormone-refractory metastatic prostate cancer. Clin Cancer Res. 2008;14(23):7940–7946. doi: 10.1158/1078-0432.CCR-08-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Solit D.B., Osman I., Polsky D., et al. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res. 2008;14(24):8302–8307. doi: 10.1158/1078-0432.CCR-08-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Ramalingam S.S., Egorin M.J., Ramanathan R.K., et al. A phase I study of 17-allylamino-17-demethoxygeldanamycin combined with paclitaxel in patients with advanced solid malignancies. Clin Cancer Res. 2008;14(11):3456–3461. doi: 10.1158/1078-0432.CCR-07-5088. [DOI] [PubMed] [Google Scholar]
- 449.Richardson P.G., Badros A.Z., Jagannath S., et al. Tanespimycin with bortezomib: activity in relapsed/refractory patients with multiple myeloma. Br J Haematol. 2010;150(4):428–437. doi: 10.1111/j.1365-2141.2010.08264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Vaishampayan U.N., Burger A.M., Sausville E.A., et al. Safety, efficacy, pharmacokinetics, and pharmacodynamics of the combination of sorafenib and tanespimycin. Clin Cancer Res. 2010;16(14):3795–3804. doi: 10.1158/1078-0432.CCR-10-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Richardson P.G., Chanan-Khan A.A., Alsina M., et al. Tanespimycin monotherapy in relapsed multiple myeloma: results of a phase 1 dose-escalation study. Br J Haematol. 2010;150(4):438–445. doi: 10.1111/j.1365-2141.2010.08265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Hubbard J., Erlichman C., Toft D.O., et al. Phase I study of 17-allylamino-17 demethoxygeldanamycin, gemcitabine and/or cisplatin in patients with refractory solid tumors. Invest N Drugs. 2011;29(3):473–480. doi: 10.1007/s10637-009-9381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Modi S., Stopeck A., Linden H., et al. HSP90 inhibition is effective in breast cancer: a phase II trial of tanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumab. Clin Cancer Res. 2011;17(15):5132–5139. doi: 10.1158/1078-0432.CCR-11-0072. [DOI] [PubMed] [Google Scholar]
- 454.Kaufmann S.H., Karp J.E., Litzow M.R., et al. Phase I and pharmacological study of cytarabine and tanespimycin in relapsed and refractory acute leukemia. Haematologica. 2011;96(11):1619–1626. doi: 10.3324/haematol.2011.049551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Iyer G., Morris M.J., Rathkopf D., et al. A phase I trial of docetaxel and pulse-dose 17-allylamino-17-demethoxygeldanamycin in adult patients with solid tumors. Cancer Chemother Pharmacol. 2012;69(4):1089–1097. doi: 10.1007/s00280-011-1789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Hendrickson A.E., Oberg A.L., Glaser G., et al. A phase II study of gemcitabine in combination with tanespimycin in advanced epithelial ovarian and primary peritoneal carcinoma. Gynecol Oncol. 2012;124(2):210–215. doi: 10.1016/j.ygyno.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Richardson P.G., Chanan-Khan A.A., Lonial S., et al. Tanespimycin and bortezomib combination treatment in patients with relapsed or relapsed and refractory multiple myeloma: results of a phase 1/2 study. Br J Haematol. 2011;153(6):729–740. doi: 10.1111/j.1365-2141.2011.08664.x. [DOI] [PubMed] [Google Scholar]
- 458.Gartner E.M., Silverman P., Simon M., et al. A phase II study of 17-allylamino-17-demethoxygeldanamycin in metastatic or locally advanced, unresectable breast cancer. Breast Cancer Res Treat. 2012;131(3):933–937. doi: 10.1007/s10549-011-1866-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Pacey S., Gore M., Chao D., et al. A Phase II trial of 17-allylamino, 17-demethoxygeldanamycin (17-AAG, tanespimycin) in patients with metastatic melanoma. Invest N Drugs. 2012;30(1):341–349. doi: 10.1007/s10637-010-9493-4. [DOI] [PubMed] [Google Scholar]
- 460.Schenk E., Hendrickson A.E., Northfelt D., et al. Phase I study of tanespimycin in combination with bortezomib in patients with advanced solid malignancies. Invest N Drugs. 2013;31(5):1251–1256. doi: 10.1007/s10637-013-9946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Walker A.R., Klisovic R., Johnston J.S., et al. Pharmacokinetics and dose escalation of the heat shock protein inhibitor 17-allyamino-17-demethoxygeldanamycin in combination with bortezomib in relapsed or refractory acute myeloid leukemia. Leuk Lymphoma. 2013;54(9):1996–2002. doi: 10.3109/10428194.2012.760733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Pedersen K.S., Kim G.P., Foster N.R., Wang-Gillam A., Erlichman C., McWilliams R.R. Phase II trial of gemcitabine and tanespimycin (17AAG) in metastatic pancreatic cancer: a Mayo Clinic Phase II Consortium study. Invest N Drugs. 2015;33(4):963–968. doi: 10.1007/s10637-015-0246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Kummar S., Gutierrez M.E., Gardner E.R., et al. Phase I trial of 17-dimethylaminoethylamino-17-demethoxygeldanamycin (17-DMAG), a heat shock protein inhibitor, administered twice weekly in patients with advanced malignancies. Eur J Cancer. 2010;46(2):340–347. doi: 10.1016/j.ejca.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Lancet J.E., Gojo I., Burton M., et al. Phase I study of the heat shock protein 90 inhibitor alvespimycin (KOS-1022, 17-DMAG) administered intravenously twice weekly to patients with acute myeloid leukemia. Leukemia. 2010;24(4):699–705. doi: 10.1038/leu.2009.292. [DOI] [PubMed] [Google Scholar]
- 465.Pacey S., Wilson R.H., Walton M., et al. A phase I study of the heat shock protein 90 inhibitor alvespimycin (17-DMAG) given intravenously to patients with advanced solid tumors. Clin Cancer Res. 2011;17(6):1561–1570. doi: 10.1158/1078-0432.CCR-10-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Jhaveri K., Miller K., Rosen L., et al. A phase I dose-escalation trial of trastuzumab and alvespimycin hydrochloride (KOS-1022; 17 DMAG) in the treatment of advanced solid tumors. Clin Cancer Res. 2012;18(18):5090–5098. doi: 10.1158/1078-0432.CCR-11-3200. [DOI] [PubMed] [Google Scholar]
- 467.Maddocks K., Hertlein E., Chen T.L., et al. A phase I trial of the intravenous Hsp90 inhibitor alvespimycin (17-DMAG) in patients with relapsed chronic lymphocytic leukemia/small lymphocytic lymphoma. Leuk Lymphoma. 2016;57(9):2212–2215. doi: 10.3109/10428194.2015.1129536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Do K., Speranza G., Chang L.C., et al. Phase I study of the heat shock protein 90 (Hsp90) inhibitor onalespib (AT13387) administered on a daily for 2 consecutive days per week dosing schedule in patients with advanced solid tumors. Invest N Drugs. 2015;33(4):921–930. doi: 10.1007/s10637-015-0255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Shapiro G.I., Kwak E., Dezube B.J., et al. First-in-human phase I dose escalation study of a second-generation non-ansamycin HSP90 inhibitor, AT13387, in patients with advanced solid tumors. Clin Cancer Res. 2015;21(1):87–97. doi: 10.1158/1078-0432.CCR-14-0979. [DOI] [PubMed] [Google Scholar]
- 470.National Cancer Institute (NCI) 2016. A Phase 1 Study of PARP Inhibitor BMN 673 and HSP90 Inhibitor AT13387 for Treatment of Advanced Solid Tumors with Expansion in Patients with Recurrent Epithelial Ovarian, Fallopian Tube, Peritoneal Cancer or Recurrent Triple-Negative Breast Cancer. [Google Scholar]
- 471.National Cancer Institute (NCI). A Phase 1 Trial of the Combination of the Heat Shock Protein-90 (HSP90) Inhibitor Onalespib (AT13387) and the Cyclin-dependent Kinase (CDK) Inhibitor AT7519M in Patients with Advanced Solid Tumors; 2021.
- 472.National Cancer Institute (NCI). A Phase 1 Study of PARP Inhibitor Olaparib and HSP90 Inhibitor AT13387 for Treatment of Advanced Solid Tumors with Expansion in Patients with Recurrent Epithelial Ovarian, Fallopian Tube, Peritoneal Cancer or Recurrent Triple-Negative Breast Cancer; 2021.
- 473.National Cancer Institute (NCI). A Phase I Trial of AT13387 in Patients with Locoregionally Advanced Squamous Cell Carcinoma of the Head and Neck (LA-SCCHN) Receiving Concurrent Radiation and Cisplatin; 2021.
- 474.National Cancer Institute (NCI). Phase 1b Study of HSP90 Inhibitor, AT13387 (Onalespib) in Combination with Paclitaxel in Patients with Advanced, Triple Negative Breast Cancer; 2021.
- 475.Sessa C., Shapiro G.I., Bhalla K.N., et al. First-in-human phase I dose-escalation study of the HSP90 inhibitor AUY922 in patients with advanced solid tumors. Clin Canc Res. 2013;19(13):3671–3680. doi: 10.1158/1078-0432.CCR-12-3404. [DOI] [PubMed] [Google Scholar]
- 476.Gaykema S.B., Schröder C.P., Vitfell-Rasmussen J., et al. 89Zr-trastuzumab and 89Zr-bevacizumab PET to evaluate the effect of the HSP90 inhibitor NVP-AUY922 in metastatic breast cancer patients. Clin Cancer Res. 2014;20(15):3945–3954. doi: 10.1158/1078-0432.CCR-14-0491. [DOI] [PubMed] [Google Scholar]
- 477.Seggewiss-Bernhardt R., Bargou R.C., Goh Y.T., et al. Phase 1/1 B trial of the heat shock protein 90 inhibitor NVP-AUY 922 as monotherapy or in combination with bortezomib in patients with relapsed or refractory multiple myeloma. Cancer. 2015;121(13):2185–2192. doi: 10.1002/cncr.29339. [DOI] [PubMed] [Google Scholar]
- 478.SCRI Development Innovations, LLC. A Phase I Study of the Hsp90 Inhibitor AUY922 Plus Capecitabine for the Treatment of Patients with Advanced Solid Tumors; 2015. [DOI] [PubMed]
- 479.SCRI Development Innovations, LLC. A Phase II Study of Hsp90 Inhibitor AUY922 for the Treatment of Patients Eith Refractory Gastrointestinal Stromal Tumor; 2016.
- 480.Novartis Pharmaceuticals. A Phase II, Multi-Center, Open-Label Study of AUY922 Administered IV on a Once-Weekly Schedule in Patients with Advanced Non-small-cell Lung Cancer Who Have Received at Least Two Lines of Prior Chemotherapy; 2016.
- 481.Memorial Sloan Kettering Cancer Center. A phase II study of the HSP90 inhibitor, AUY922, in patients with primary myelofibrosis (PMF), post-polycythemia vera myelofibrosis (post-PV MF), post-essential thrombocythemia myelofibrosis (post-ET MF), and refractory PV/ET; 2016.
- 482.Anderson Cancer Center MD. A Phase II Study of the HSP90 Inhibitor AUY922 in Patients with Relapsed and Refractory Lymphoma; 2016.
- 483.Novartis Pharmaceuticals. A Multicenter, Open-Label, Randomized Phase II Study to Evaluate the Efficacy of AUY922 vs Pemetrexed or Docetaxel in NSCLC Patients with EGFR Mutations Who Have Progressed on Prior EGFR TKI Treatment; 2019.
- 484.Northwestern University. A Phase I/II Trial of HSP 90 Inhibitor AUY-922 in Patients with Lung Adenocarcinoma with “Acquired Resistance” to EGFR Tyrosine Kinase Inhibitors; 2019.
- 485.Jonsson Comprehensive Cancer Center. A Phase IB Dose-Escalationstudy of Pemetrexed and AUY922 in Previously-Treated Patients with Metastatic Non-squamous, Non-small Cell Lung Cancer; 2020.
- 486.Dickson M.A., Okuno S.H., Keohan M.L., et al. Phase II study of the HSP90-inhibitor BIIB021 in gastrointestinal stromal tumors. Ann Oncol. 2013;24(1):252–257. doi: 10.1093/annonc/mds275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Saif M.W., Takimoto C., Mita M., et al. A phase 1, dose-escalation, pharmacokinetic and pharmacodynamic study of BIIB021 administered orally in patients with advanced solid tumors. Clin Cancer Res. 2014;20(2):445–455. doi: 10.1158/1078-0432.CCR-13-1257. [DOI] [PubMed] [Google Scholar]
- 488.Hong D., Said R., Falchook G., et al. Phase I study of BIIB028, a selective heat shock protein 90 inhibitor, in patients with refractory metastatic or locally advanced solid tumors. Clin Cancer Res. 2013;19(17):4824–4831. doi: 10.1158/1078-0432.CCR-13-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Isambert N., Delord J.P., Soria J.C., et al. Debio0932, a second-generation oral heat shock protein (HSP) inhibitor, in patients with advanced cancer results of a first-in-man dose-escalation study with a fixed-dose extension phase. Ann Oncol. 2015;26(5):1005–1011. doi: 10.1093/annonc/mdv031. [DOI] [PubMed] [Google Scholar]
- 490.Spreafico A., Delord J.P., De Mattos-Arruda L., et al. A first-in-human phase I, dose-escalation, multicentre study of HSP990 administered orally in adult patients with advanced solid malignancies. Br J Cancer. 2015;112(4):650–659. doi: 10.1038/bjc.2014.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Siegel D., Jagannath S., Vesole D.H., et al. A phase 1 study of IPI-504 (retaspimycin hydrochloride) in patients with relapsed or relapsed and refractory multiple myeloma. Leuk Lymphoma. 2011;52(12):2308–2315. doi: 10.3109/10428194.2011.600481. [DOI] [PubMed] [Google Scholar]
- 492.Wagner A.J., Chugh R., Rosen L.S., et al. A phase I study of the HSP90 inhibitor retaspimycin hydrochloride (IPI-504) in patients with gastrointestinal stromal tumors or soft-tissue sarcomas. Clin Cancer Res. 2013;19(21):6020–6029. doi: 10.1158/1078-0432.CCR-13-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Sequist LV. A Phase II Study of IPI-504, A Novel Hsp90 Inhibitor in NSCLC Patients with ALK Translocations; 2017.
- 494.Kyowa Hakko Kirin Pharma, Inc. An Open Label, Dose Escalation, Multicenter Phase 1/2 Study of KW-2478 in Combination with Bortezomib in Subjects with Relapsed And/or Refractory Multiple Myeloma; 2014.
- 495.Yong K., Cavet J., Johnson P., et al. Phase I study of KW-2478, a novel Hsp90 inhibitor, in patients with B-cell malignancies. Br J Cancer. 2016;114(1):7–13. doi: 10.1038/bjc.2015.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Rajan A., Kelly R.J., Trepel J.B., et al. A phase I study of PF-04929113 (SNX-5422), an orally bioavailable heat shock protein 90 inhibitor, in patients with refractory solid tumor malignancies and lymphomas. Clin Cancer Res. 2011;17(21):6831–6839. doi: 10.1158/1078-0432.CCR-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Reddy N., Voorhees P.M., Houk B.E., Brega N., Hinson J.M., Jr., Jillela A. Phase I trial of the HSP90 inhibitor PF-04929113 (SNX5422) in adult patients with recurrent, refractory hematologic malignancies. Clin Lymphoma, Myeloma & Leukemia. 2013;13(4):385–391. doi: 10.1016/j.clml.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 498.Esanex Inc. A Single Arm, Phase 1/2 Study of SNX-5422 in Subjects with Selected HER2 Positive Cancers; 2016.
- 499.Esanex Inc. A Single Arm Study of SNX-5422 in Subjects with TP53 Null Cancers; 2018.
- 500.Esanex Inc. A Phase 1, Open-Label Study of SNX-5422 Added to Ibrutinib in Chronic Lymphocytic Leukemia Subjects with Residual Disease; 2019.
- 501.Goldman J.W., Raju R.N., Gordon G.A., et al. A first in human, safety, pharmacokinetics, and clinical activity phase I study of once weekly administration of the Hsp90 inhibitor ganetespib (STA-9090) in patients with solid malignancies. BMC Cancer. 2013;13:152. doi: 10.1186/1471-2407-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Socinski M.A., Goldman J., El-Hariry I., et al. A multicenter phase II study of ganetespib monotherapy in patients with genotypically defined advanced non–small cell lung cancer. Clin Cancer Res. 2013;19(11):3068–3077. doi: 10.1158/1078-0432.CCR-12-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Cercek A., Shia J., Gollub M., et al. Ganetespib, a novel Hsp90 inhibitor in patients with KRAS mutated and wild type, refractory metastatic colorectal cancer. Clin Colorectal Cancer. 2014;13(4):207–212. doi: 10.1016/j.clcc.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Ramalingam S., Goss G., Rosell R., et al. A randomized phase II study of ganetespib, a heat shock protein 90 inhibitor, in combination with docetaxel in second-line therapy of advanced non-small cell lung cancer (GALAXY-1) Ann Oncol. 2015;26(8):1741–1748. doi: 10.1093/annonc/mdv220. [DOI] [PubMed] [Google Scholar]
- 505.Thakur M.K., Heilbrun L.K., Sheng S., et al. A phase II trial of ganetespib, a heat shock protein 90 Hsp90) inhibitor, in patients with docetaxel-pretreated metastatic castrate-resistant prostate cancer (CRPC)-a prostate cancer clinical trials consortium (PCCTC) study. Invest N Drugs. 2016;34(1):112–118. doi: 10.1007/s10637-015-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Pillai R.N., Fennell D.A., Kovcin V., et al. Randomized phase III study of ganetespib, a heat shock protein 90 inhibitor, with docetaxel versus docetaxel in advanced non–small-cell lung cancer (GALAXY-2) J Clin Oncol. 2020;38(6):613–622. doi: 10.1200/JCO.19.00816. [DOI] [PubMed] [Google Scholar]
- 507.MD DC. Phase II Study of STA-9090 as Second or Third-Line Therapy for Metastatic Pancreas Cancer; 2014.
- 508.Saba NF. A Molecularly Driven Pilot Study of Preoperative Ganetespib in Resectable Squamous Cell Carcinoma of the Head and Neck (SCCHN); 2016.
- 509.University of Chicago. A Pilot Preoperative Trial of Ganetespib with Paclitaxel for Triple-Negative Breast Cancer; 2016.
- 510.MD FSH. This trial is an open label, parallel cohort, phase ii study evaluating the efficacy of the heat shock protein 90 (HSP90) inhibitor STA-9090 in patients with 1546 S. Kumar et al. unresectable stage III or stage IV melanoma who were intolerant of, or progressed on, prior tyrosine kinase inhibitor treatment. Two cohorts will enroll concurrently. One cohort will be composed of patients with melanoma expressing a mutation in the potein BRAF and the other cohort will be composed of patients with melanoma expressing wild-type BRAF; 2017.
- 511.MD FSH. A Phase II Study of the HSP Inhibitor STA-9090 in Metastatic Ocular Melanoma; 2018.
- 512.Memorial Sloan Kettering Cancer Center. A Phase I Clinical Trial of Ganetespib (Heat Shock Protein 90 Inhibitor) in Combination with Paclitaxel, Trastuzumab and Pertuzumab in Human Epidermal Growth Factor Receptor-2 Positive (HER2þ) Metastatic Breast Cancer; 2018.
- 513.MD AC. Phase I Study of Ganetespib and Ziv-Aflibercept in Patients with Advanced Gastrointestinal Carcinomas, Nonsquamous Non-small Cell Lung Carcinomas, Urothelial Carcinomas, and Sarcomas; 2018.
- 514.MD AC. Phase I Study of Ganetespib and Ziv-Aflibercept in Patients with Advanced Gastrointestinal Carcinomas, Nonsquamous Non-small Cell Lung Carcinomas, Urothelial Carcinomas, and Sarcomas. 2018.
- 515.Concin N. A Two-Part, Multicentre, International Phase I and II Trial Assessing the Safety and Efficacy of the Hsp90 Inhibitor Ganetespib in Combination with Paclitaxel Weekly in Women with High-Grade Serous, High-Grade Endometrioid, or Undifferentiated, Platinum-Resistant Epithelial Ovarian, Fallopian Tube or Primary Peritoneal Cancer; 2019.
- 516.Sarcoma Alliance for Research through Collaboration. A Phase I/II Trial of Ganetespib in Combination with the MTOR Inhibitor Sirolimus for Patients with Recurrent or Refractory Sarcomas Including Unresectable or Metastatic Malignant Peripheral Nerve Sheath Tumors; 2019.
- 517.M.D AC. Phase I Study of the Hsp90 Inhibitor, PU-H71, in Patients with Refractory Solid Tumors and Low-Grade Non-hodgkin’s Lymphoma; 2017.
- 518.Samus Therapeutics, Inc. Phase 1b Study of PU-H71 for the Treatment of Subjects with Primary Myelofibrosis (PMF), Post-polycythemia Vera Myelofibrosis (Post-PV MF), Post-essential Thrombocythemia Myelofibrosis Post-ET MF), Treated with Ruxolitinib; 2020.
- 519.Taiho Oncology, Inc. A Phase IA/IB Study Evaluating TAS-116 in Patients with Advanced Solid Tumors; 2019.
- 520.Centre Hospitalier Universitaire Dijon. Preclinical Project on the Traitment of Acute Lymphoblastique Leukemia with NVP-Bep800, an Inhibitor of the Heat Shock Protein HSP90; 2020.
- 521.Bregnhoej A. A Phase 1b, Open-Label, Single-Arm, Dose-Selection, Proof-Of-Concept Study to Assess the Safety and Efficacy of a Novel HSP90 Inhibitor (CUDC-305) in the Treatment of Moderate to Severe Plaque Psoriasis; 2019.
- 522.Chia Tai Tianqing Pharmaceutical Group Co., Ltd. A Phase I, Open-Label, Multicenter, Dose Escalation and Expansion Study to Evaluate the Tolerance and Pharmacokinetics of TQB3474 Injection; 2019.
- 523.Tarveda Therapeutics. A Phase 1/2a, Open-Label, Multicenter Study to Assess the Safety, Tolerability, Pharmacokinetics, Pharmacodynamics, and Preliminary Anti-tumor Activity of PEN-866 in Patients with Advanced Solid Malignancies; 2021.
- 524.Reata Pharmaceuticals, Inc. A Study of the Safety, Tolerability, and Pharmacokinetics of Single and Multiple Doses of RTA 901 in Healthy Adults; 2017.
- 525.El-Rayes B. Phase Ib Trial of Pembrolizumab and XL888 in Patients with Advanced Gastrointestinal Malignancies; 2021.
- 526.H. Lee Moffitt Cancer Center and Research Institute. Phase I Study of Escalating Doses of XL888 with Vemurafenib Plus Cobimetinib for Patients with Unresectable BRAF Mutated Stage III/IV Melanoma; 2021.
- 527.Li Y., Zhang T., Schwartz S.J., Sun D. New developments in Hsp90 inhibitors as anti-cancer therapeutics: mechanisms, clinical perspective and more potential. Drug Resist Updates. 2009;12(1–2):17–27. doi: 10.1016/j.drup.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1