Abstract
Nitric oxide (NO⋅) is critical to numerous biological processes, including signal transduction and macrophage-mediated immunity. In this study, we have explored the biological effects of NO⋅-induced DNA damage on Escherichia coli. The relative importance of base excision repair, nucleotide excision repair (NER), and recombinational repair in preventing NO⋅-induced toxicity was determined. E. coli strains lacking either NER or DNA glycosylases (including those that repair alkylation damage [alkA tag strain], oxidative damage [fpg nei nth strain], and deaminated cytosine [ung strain]) showed essentially wild-type levels of NO⋅ resistance. However, apyrimidinic/apurinic (AP) endonuclease-deficient cells (xth nfo strain) were very sensitive to killing by NO⋅, which indicates that normal processing of abasic sites is critical for defense against NO⋅. In addition, recA mutant cells were exquisitely sensitive to NO⋅-induced killing. Both SOS-deficient (lexA3) and Holliday junction resolvase-deficient (ruvC) cells were very sensitive to NO⋅, indicating that both SOS and recombinational repair play important roles in defense against NO⋅. Furthermore, strains specifically lacking double-strand end repair (recBCD strains) were very sensitive to NO⋅, which suggests that NO⋅ exposure leads to the formation of double-strand ends. One consequence of these double-strand ends is that NO⋅ induces homologous recombination at a genetically engineered substrate. Taken together, it is now clear that, in addition to the known point mutagenic effects of NO⋅, it is also important to consider recombination events among the spectrum of genetic changes that NO⋅ can induce. Furthermore, the importance of recombinational repair for cellular survival of NO⋅ exposure reveals a potential susceptibility factor for invading microbes.
Nitric oxide (NO⋅) is an inorganic gas generated by the oxidation of L-arginine by a family of enzymes called NO⋅ synthases (32). NO⋅ is produced by many different mammalian cell types for a variety of biological functions, including its use as a signaling molecule in neurotransmission and vasodilation (3, 19). NO⋅, along with oxygen radicals, is also excreted at much higher concentrations by macrophages, as part of the mechanism for killing microbes and tumor cells. As the cytostatic effects of macrophages are greatly reduced by NO⋅ scavengers, it is thought that NO⋅ is a key mediator of macrophage-induced toxicity (reviewed in reference 30). NO⋅ is highly soluble in lipids and thus readily diffuses into microbes to cause lethal damage to proteins and DNA (24). To defend against the toxic effects of NO⋅ and reactive oxygen species, at least 15 genes are upregulated as part of the soxRS system in Escherichia coli (17, 49). In addition, NO⋅ induces the SOS response (29), which leads to increased expression of dozens of stress response genes (reviewed in reference 14). Some of the genes induced by the SOS and soxRS systems are involved in DNA repair (35) (reviewed in references 10 and 14). Nevertheless, little is known about the relative contributions of DNA repair pathways to the survival of microbes exposed to NO⋅.
NO⋅ does not react with DNA directly but becomes a potent DNA-damaging agent once it is oxidized or reacts with superoxide anion. Oxidation by molecular oxygen and auto-oxidation of NO⋅ result in the formation of nitrous anhydride, N2O3, a powerful nitrosating agent, while reaction of NO⋅ with superoxide anion generates peroxynitrite, a powerful oxidizing agent (reviewed in reference 4). In E. coli, N2O3 reacts with exocyclic amino groups to deaminate adenine, cytosine, and guanine to form hypoxanthine, uracil, and xanthine, respectively, all of which are potentially mutagenic if unrepaired (6); (reviewed in references 4 and 14). The Ung and AlkA DNA glycosylases excise uracil and hypoxanthine, respectively, to initiate the base excision repair (BER) pathway (it is not yet known how xanthine is repaired) (26, 40). After removal of deaminated bases by these DNA glycosylases or by spontaneous depurination, the phosphodiester backbone is cleaved by apyrimidinic/apurinic (AP) endonucleases at the abasic site. This results in a 3′-OH terminus, which can act as a primer for DNA synthesis, and a 5′-deoxyribose phosphate residue that is removed by a deoxyribose phosphodiesterase to facilitate repair DNA synthesis and ligation (reviewed in reference 51). The vast majority of lesions induced by N2O3 are thought to be deaminated bases (6, 18) (reviewed in reference 4), though it is noteworthy that nitrous acid (which is chemically similar to N2O3) can lead to intra- and interstrand DNA cross-links in vitro (42).
The second major pathway by which NO⋅ induces DNA damage is by reaction with superoxide to form peroxynitrite (ONOO−). Peroxynitrite causes oxidation and nitration of bases, most notably guanine (reviewed in reference 4), resulting in predominantly 8-oxoguanine (8-oxoG) and 8-nitroguanine (44, 53). 8-Nitroguanine is unstable and readily depurinates, with a half-life of 4 h, to form abasic sites that are substrates for the BER pathway (54). 8-oxoG is also repaired by BER, but in this case repair is initiated by the Fpg DNA glycosylase rather than by spontaneous depurination (reviewed in reference 51). However, it is possible that a significant portion of the 8-oxoG formed actually reacts again with peroxynitrite to form secondary oxidation products, the biological consequences of which are currently under investigation (5, 34, 48). In addition to base lesions, peroxynitrite can also lead to single-strand breaks, although the frequency of base damage is several orders of magnitude greater than the frequency of chemically induced strand breaks (21, 39, 44).
While base damage is generally removed via excision repair, damage that encompasses both strands of the DNA helix (i.e., double-strand breaks) is repaired via homologous recombination pathways. NO⋅ does not directly cause double-strand breaks. However, some of the minor lesions induced by NO⋅ are potentially recombinogenic (e.g., interstrand cross-links), and DNA excision repair intermediates might also become recombinogenic during DNA replication (e.g., abasic [AP] sites or nicks). Although the chemistry and biology of NO⋅ have been the subject of intensive investigation, to our knowledge, there are no reports of studies of the potential importance of various DNA repair systems in defending bacteria against NO⋅ toxicity. Hence, here we have compared the relative importance of BER, nucleotide excision repair (NER), and recombinational repair pathways in E. coli.
Here we show that none of the DNA glycosylases involved in the removal of damaged bases provides significant protection against NO⋅-induced toxicity, nor is there any significant sensitivity of cells deficient in NER. Although none of the DNA glycosylase mutants were sensitive to NO⋅, cells lacking AP endonuclease activity were very sensitive to NO⋅ toxicity, suggesting the formation of AP sites (generated by spontaneous base loss or by DNA glycosylases). Furthermore, we found that both the SOS response and recombinational repair are critical for defense against NO⋅ toxicity. By studying mutants deficient in specific aspects of homologous recombination, we provide evidence to support a model whereby recombinational repair serves to repair double-strand breaks that are likely to be formed when the replication machinery encounters nicked template DNA. A side effect of this repair pathway is that cells exposed to NO⋅ exhibit increased levels of recombination between misaligned sequences.
MATERIALS AND METHODS
Bacterial strains.
Strains used in this study are listed in Table 1. Most strains used for cytotoxic measurements are derived from AB1157, except for the fpg nei nth (parent BW35) and ung (parent MV1161) strains. The three parent strains were not significantly different from one another in their sensitivity to NO⋅ toxicity (data not shown). The phenotypic markers associated with the gene disruptions were confirmed by growth on the appropriate selection medium or treatment with agents such as UV light. Lactose minimal agar contained M9 salts, 1% lactose, thiamine (1 μg/ml), and 15 g of agar/liter.
TABLE 1.
E. coli strains used
| Strain | Relevant genotype | Source |
|---|---|---|
| AB1157 | thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 glnV44(AS) galK2(Oc) hisG4(Oc) rfbD1 mgl-51 rpoS396(Am) rpsL31 (Strr) kdgK51 xylA5 mtl-1 argE3(Oc) thi-1 | Lab stock |
| GC4803 | As AB1157 but tagA1::Tn5 his+ alkA1 | S. Boiteux |
| AB2500 | As AB1157 but uvrA6 deoB16 thyA12 | M. Marinus |
| GM5560 | As AB1157 but recA::Tn10 | M. Marinus |
| DM49 | As AB1157 but lexA3 | M. Marinus |
| JC9239 | As AB1157 but recF143 | M. Marinus |
| KM21 | As AB1157 but ΔrecBCD::Kan | M. Marinus |
| CS85 | As AB1157 but ruvC53 eda51::Tn10 | M. Marinus |
| BW9109 | As AB1157 but Δxth-pncA | B. Demple |
| BW527 | As AB1157 but nfo-1::Kan | B. Demple |
| BW528 | As AB1157 but Δxth-pncA nfo-1::Kan | B. Weiss |
| MV1161 | As AB1157 but rfa-550 | M. Volkert |
| MV3880 | As MV1161 but ung-152::Tn10 | M. Volkert |
| BW35 | Hfr KL16 PO-45 thi relA spoT1 | S. Wallace |
| KL16 nth nei fpg | As KL16 but nei::Ch1 nth::Kan fpg::Amp | S. Wallace |
| KL16 nth nei | As KL16 but nei::Ch1 nth::Kan | S. Wallace |
| KL16 fpg | As KL16 but fpg::Amp | S. Wallace |
| GM7330 | ΔlacMS286 φ80dIIlacBK1 ara thi(?) | M. Marinus |
Cytotoxicity assay.
Overnight cultures were diluted 100-fold and grown in Luria-Bertani (LB) medium until early log phase (optical density at 600 nm [OD600] = ∼0.26). Aliquots of 100 ml were transferred into delivery chambers and exposed to nitric oxide (Messer, Malvern, Pa.) at room temperature for 2 h. The details of the NO⋅ delivery system have been described earlier (46). Nitric oxide was delivered to the cell culture at a steady rate through silastic membrane tubing (Dow Corning Corp., Midland, Mich.). The delivery chamber has an aperture to facilitate diffusion of oxygen into the cell culture during treatment. Delivery rates were approximately 35 nmol/ml · min. NO⋅ delivery was confirmed by measuring the concentration of nitrate plus nitrite formed in the LB medium at various times during exposure (16) or by including a strain of known sensitivity to verify that exposure conditions were equitoxic with those of previous experiments wherein concentrations of nitrate and nitrite had been determined. After exposure of the cells to nitric oxide, appropriate dilutions of the culture in M9 salts were plated on LB plates and incubated. Surviving colonies were scored the following day.
NO⋅-induced recombination assay.
Strain GM7330 carries two different deletion alleles of the lac operon. The deletions are nonoverlapping, so that homologous recombination between the mutant alleles can restore the Lac+ phenotype (22). Overnight cultures of GM7330 were diluted 100-fold and grown in LB medium to early log phase (OD600 = ∼0.26) and exposed to NO⋅, as described above. After 2 h, 10 ml of the cell suspension was pelleted and resuspended in 10 ml of M9 salts solution. The cells were allowed to recover in M9 salts solution for 30 min, pelleted, and resuspended in M9 salts solution. Aliquots of 100 μl were plated on lactose agar, and appropriate dilutions were plated on LB agar to determine the number of survivors. The lactose plates were incubated at 37°C until recombinant colonies could be scored (∼4 days). Recombination events were calculated as the number of recombinants per survivor.
UV-induced recombination assay.
Overnight cultures of GM7330 were diluted 100-fold and grown in M9 minimal medium to early log phase (OD600 = ∼0.26). Aliquots of 20 ml were transferred onto glass plates and exposed to UV. After treatment, the cells were allowed to replicate for another 90 min, pelleted, and resuspended in 10 ml of M9 salts solution. The cells were allowed to recover in M9 salts for 30 min, pelleted, resuspended into M9 salts solution, and plated on LB medium and lactose agar, as described above.
RESULTS
NER does not protect E. coli from NO⋅-induced toxicity.
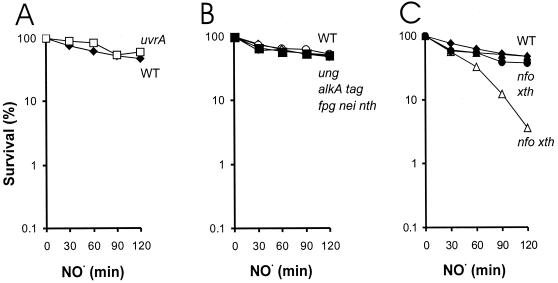
To investigate the potential importance of NER in protection against NO⋅ toxicity, log-phase NER-deficient uvrA E. coli and its parental strain were maintained for 2 h in an NO⋅ delivery chamber (NO⋅ gas is administered to the medium via a semipermeable membrane submerged in the culture [46]). There was no significant difference in the sensitivities of wild-type and uvrA E. coli strains to NO⋅ (Fig. 1A). Thus, NER does not appear to play a critical role in the removal of toxic NO⋅-induced lesions. Interestingly, using the same experimental system of NO⋅ delivery, NER-deficient uvrB Salmonella enterica serovar Typhimurium has been found to be more sensitive to NO⋅ than its wild-type counterparts (45). This suggests that, in serovar Typhimurium, toxic lesions can be removed from the genome via excision repair. Given that NER plays a more minor role in preventing NO⋅ toxicity in E. coli than in serovar Typhimurium, we suspected that other excision repair enzymes may be more relevant to E. coli (e.g., BER).
FIG. 1.
Relative NO⋅ sensitivity of E. coli strains carrying mutations in DNA excision repair pathways. Log-phase cells were exposed to 35 nmol of NO⋅/ml · min. (A) Survival of wild-type E. coli (black diamonds) and NER-deficient uvrA mutant cells (open squares) following NO⋅ exposure. (B) Survival following NO⋅ exposure of wild-type E. coli (black diamonds) and strains lacking expression of DNA glycosylases: alkA tag (open circles), fpg nei nth (open diamonds), and ung (black squares) strains. (C) Relative survival of wild-type E. coli (black diamonds), and strains lacking expression of AP endonucleases, xth (black circles), nfo (black triangles), and nfo xth (open triangles) strains, following NO⋅ exposure. For all panels, in cases where NO⋅ sensitivity is comparable to that of wild-type E. coli, the data presented are averages of at least two independent experiments. Data for wild-type AB1157 and the nfo xth strain are the averages of at least six independent experiments. There were no significant differences in sensitivity to NO⋅ among the wild-type strains that were used to create the mutant strains used in these studies (data not shown). WT, wild type.
DNA glycosylase mutants are not sensitive to NO⋅ toxicity.
Nitrous anhydride, formed after the reaction of NO⋅ with oxygen, can deaminate bases to form xanthine, hypoxanthine, and uracil (reviewed in reference 4). Uracil is removed by the Ung uracil DNA glycosylase (26), while hypoxanthine can be removed by the AlkA 3-methyladenine DNA glycosylase (40). It is not yet known how xanthine is repaired. To determine if the glycosylases involved in the removal of deaminated bases help prevent NO⋅ toxicity, we exposed strains deficient in uracil glycosylase (ung strain) or 3-methyladenine DNA glycosylases (alkA tag strain) to NO⋅. We observed that these glycosylase-deficient cells were not significantly sensitized to the effects of NO⋅ (Fig. 1B). This is consistent with the results of Tamir et al., who observed only a very slight increase in the sensitivity of ung E. coli to NO⋅ exposure (45). Likewise, alkA E. coli when exposed to nitrous acid displayed a sensitivity similar to that of wild-type cells (43). While Ung or AlkA may still remove potentially mutagenic NO⋅-induced base damage, neither Ung nor AlkA plays a critical role in the removal of potentially toxic NO⋅-induced base damage.
The peroxynitrite pathway leads primarily to oxidative DNA damage, rather than deaminated bases (reviewed in reference 4). Several DNA glycosylases are known to act on oxidized bases. The formamidopyrimidine-DNA glycosylase (fpg) repairs 8-oxoG (7), while the endonuclease III (EndoIII) (nth) (37) and EndoVIII (nei) (50) DNA glycosylases remove a broad spectrum of damage that primarily consists of oxidized pyrimidines, some of which are potentially cytotoxic (reviewed in reference 51). We exposed a strain deficient in all three of these oxidative damage glycosylases (fpg nei nth strain) (2) to NO⋅ and found no significant difference in survival between this triple mutant and its wild-type counterpart (Fig. 1B). Also, single-mutant fpg E. coli and double-mutant nei nth E. coli were no more sensitive to the toxic effects of NO⋅ than were their wild-type counterparts (data not shown). Despite the resistance of fpg E. coli to NO⋅, it is possible that the Fpg glycosylase indeed removes lesions created during exposure to NO⋅ but that these lesions are relatively nontoxic (or are removed by a redundant pathway in the absence of Fpg).
AP endonuclease activity is critical for preventing NO⋅ toxicity.
The removal of damaged bases by a DNA glycosylase or spontaneous base loss is the first step in the BER pathway. Base loss results in AP sites that are potentially toxic if they are not processed by downstream enzymes in the BER pathway, beginning with AP endonucleases that cleave the DNA backbone 5′ to the AP site. Glycosylases that repair oxidative damage, such as the Fpg DNA glycosylase, have an associated lyase activity that nicks the DNA backbone 3′ to the AP site (36). However, the nicked AP site cannot be extended by DNA polymerase unless there is a terminal 3′-OH group. AP endonucleases perform another critical function in the BER pathways by processing the 3′ end at nicked AP sites (created by the action of a DNA glycosylase with associated lyase activity, for example) to create a terminal 3′-OH group suitable for extension by DNA polymerase (12, 33).
E. coli has two AP endonucleases: exonuclease III (ExoIII, encoded by xth) and EndoIV (encoded by nfo). ExoIII accounts for 90% of the AP endonuclease activity in the cell (28). However, the residual EndoIV activity is sufficient to provide significant protection of xth mutant cells exposed to an agent that leads to the generation of AP sites (namely, methyl methanesulfonate) (9). In addition to its endonuclease and diesterase activities, ExoIII has an associated 3′-5′ exonuclease function of unknown significance.
Cells lacking EndoIV were not sensitive to NO⋅-induced killing, which is consistent with ExoIII being responsible for the majority of the AP endonuclease activity in E. coli (Fig. 1C). Similarly, cells lacking ExoIII had near-wild-type levels of resistance to NO⋅ toxicity. However, the double-mutant xth nfo cells were highly sensitive to NO⋅ toxicity (Fig. 1C). These results are consistent with previous studies showing that NO⋅ induces AP sites in mammalian cells and that ExoIII cleaves peroxynitrite-treated DNA (13, 45). Thus, it is clear that, in E. coli, exposure to NO⋅ results in the generation of AP sites (either spontaneously or via DNA glycosylase[s]) that are potentially toxic if they are not processed by ExoIII or EndoIV to generate a 3′-OH terminus that is suitable for extension by a DNA polymerase. Furthermore, the lack of sensitivity of the single xth mutant shows that the 3′-5′ exonuclease activity, which is specific to ExoIII, is not critical for defense against NO⋅ damage.
Recombinational repair is an important cellular defense against NO⋅ toxicity.
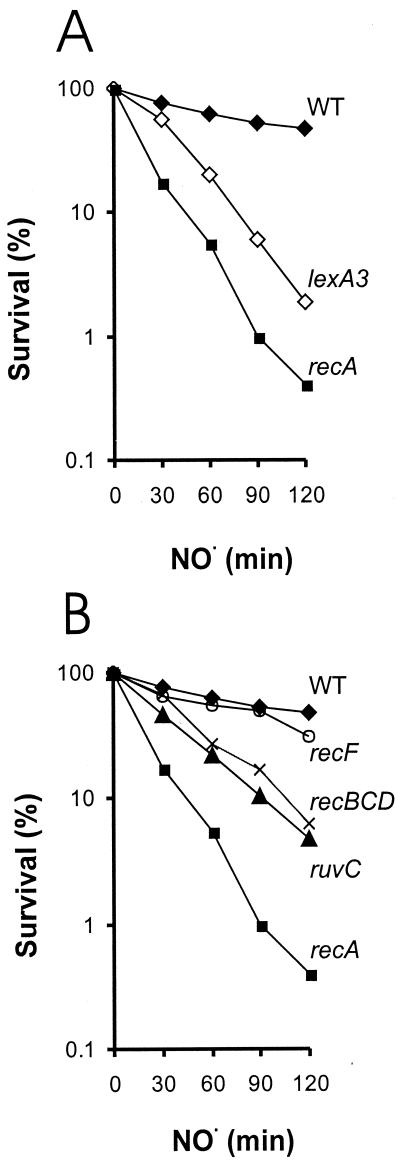
To investigate the potential importance of recombinational repair in preventing NO⋅ toxicity, RecA-deficient cells were exposed to NO⋅. RecA is essential to almost all homologous recombination processes, since it binds to single-stranded DNA to create a nucleoprotein filament that is essential for homology searching (reviewed in reference 23). Cells deficient in RecA were sensitive to NO⋅ (Fig. 2A). To our knowledge, this extreme sensitivity of recA to NO⋅ is far greater than that reported for any other DNA repair mutant E. coli strain. These data suggest that recombinational repair and/or the SOS response may help cells tolerate toxic DNA lesions induced by NO⋅.
FIG. 2.
Relative NO⋅ sensitivity of E. coli strains defective in SOS, recombination, and excision repair. (A) NO⋅ sensitivity of wild-type (WT) E. coli (black diamonds), recombination- and SOS-deficient recA mutant cells (black squares), and SOS-deficient lexA3 mutant cells (open diamonds). (B) NO⋅ sensitivity of wild-type E. coli (black diamonds) and ruvC (black triangles), recF (open circles), recBCD (crosses), and recA (black squares) mutant cells. Results are the averages of three or more independent experiments.
RecA initiates the SOS response by activating self-cleavage of the LexA repressor, which in turn leads to the upregulation of dozens of DNA repair and damage tolerance genes (reviewed in reference 14). To investigate the importance of the SOS response, we exploited lexA3 mutant cells, whose LexA protein is incapable of self-cleavage (making the cells deficient in the SOS response) (27). We found that the lexA3 mutant strain is sensitive to NO⋅ exposure, although not quite as sensitive as the recA strain (Fig. 2A). This implies that a major role of RecA in NO⋅ resistance is due to its ability to induce the SOS response. However, recombinational repair remains potentially quite important, particularly since the SOS-inducible recA, ruvA, and ruvB gene products play central roles in homologous recombination.
To test whether homologous recombination affords protection against NO⋅ toxicity, we monitored the survival of strains deficient in proteins involved in homologous recombination. In most homologous recombination processes, RecA initiates homology searching and strand invasion to form Holliday junctions that are resolved by the combined action of RuvA, RuvB, and RuvC or via RecG-mediated branch migration (reviewed in reference 23). Because some homologous recombination events may require Holliday junction resolution by a resolvase, such as RuvC, a ruvC mutant strain can potentially be used to monitor the involvement of homologous recombination in strain survival. The ruvC strain was sensitive to NO⋅ (Fig. 2B), demonstrating the importance of recombinational repair in preventing NO⋅ toxicity.
Almost all homologous recombination processes in E. coli involve RecA and RuvC, and they can be divided into two major processes: those initiated by single-strand gaps and those initiated by double-strand ends. Before RecA can be recruited to initiate homology searching, potential recombination substrates have to be processed to facilitate loading of RecA. The recBCD gene products process double-stranded ends (e.g., see references 1 and 47), while the recFOR gene products bind single-strand gaps (20, 31). In both cases, the action of these complexes facilitates RecA loading onto a single-stranded region (reviewed in reference 23). To determine if the majority of the recombination processes induced by NO⋅ are initiated by single-strand gaps or by double-strand ends, we monitored the relative survival of recBCD and recF mutant cells. The strain deficient in RecBCD was more sensitive to NO⋅ toxicity than was a strain deficient in RecF (indeed, recF mutant cells showed very little, if any, increased sensitivity to NO⋅). Thus, we conclude that unrepaired double-strand ends are primarily responsible for the toxicity observed in cells deficient in homologous recombinational repair.
NO⋅ induces recombination in E. coli.
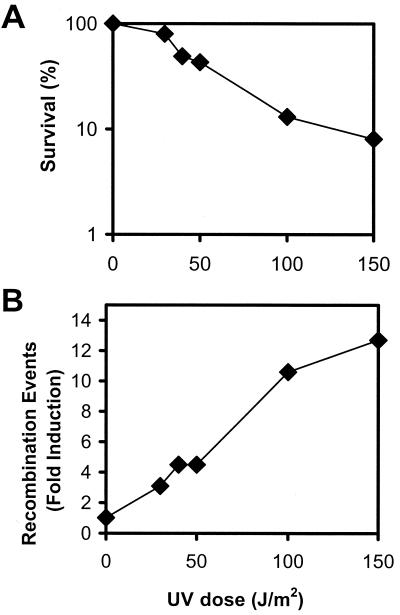
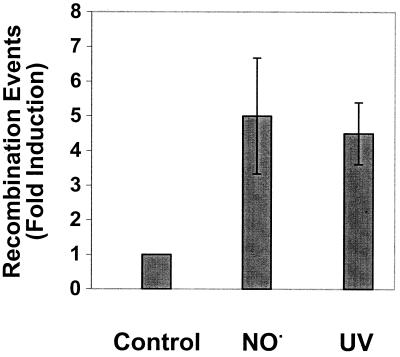
The GM7330 strain of E. coli carries two copies of the lac operon, each with a nonoverlapping deletion so that these cells are Lac− (22). At one locus, the deletion inactivates the lacA and lacY genes, while the other locus carries a deletion in the lacZ region. A Lac+ phenotype can be reconstituted by a recombination event between the two incomplete lac loci. To our knowledge, only one study using this strain for measurements of DNA damage-induced recombination events has been reported previously (55). To be able to compare our results with NO⋅ to those for a known recombinogen, we exposed GM7330 to UV and plated bacteria on both lactose plates (to monitor recombinants) and LB plates (to monitor percent survival). Recombination events can then be scored as the number of recombinants per survivor. Figure 3 shows that the frequency of recombinants rose with increasing UV dose (and decreasing survival), which is consistent with studies of another microbe, Saccharomyces cerevisiae, where UV has been shown elsewhere to induce recombination in a dose-dependent manner (41). Subsequently, we determined the frequency of recombination events induced by NO⋅. At a dose of NO⋅ at which ∼50% of the cells survive, the frequency of recombination was approximately fivefold greater than that in untreated cells, showing that NO⋅ was comparable to UV in its ability to induce recombination (Fig. 4).
FIG. 3.
UV-induced recombination at the lac operon. (A) UV-induced toxicity in E. coli GM7330. (B) Fold induction of recombination events relative to untreated cells. Recombination was monitored by reconstitution of a Lac+ phenotype in E. coli GM7330. Data are the averages of two or more independent experiments. Fold induction is calculated by dividing the number of DNA damage-induced recombination events by the number of recombination events in the untreated control sample.
FIG. 4.
UV- and NO⋅-induced recombination in E. coli monitored by lacZ reconstitution. Shown is relative recombinogenicity of UV and NO⋅ in GM7330 at doses that cause a ∼50% reduction in survival (43% survival for NO⋅- and 45% survival for UV-exposed cells). The data are the averages of six or more independent experiments. Fold induction is calculated as for Fig. 3. Error bars show 1 standard deviation.
DISCUSSION
Extensive investigation has shown that NO⋅ exerts its biological effects via damage to proteins and direct damage to DNA. NO⋅ reacts with iron-sulfur centers and sulfhydryl groups to inactivate proteins that contribute to DNA stability, such as ribonucleotide reductase (necessary for normal DNA replication) and several enzymes involved in DNA repair (e.g., Fpg DNA glycosylase, DNA repair methyltransferase, and DNA ligase) (15, 25, 38, 52). In addition to the potentially deleterious effects of loss of function of these important DNA-metabolizing proteins, NO⋅ can induce covalent modifications to the DNA such as nicks, cross-links, and base damage (reviewed in reference 4). Although strand breaks and cross-links are potentially toxic to cells, the majority of lesions induced by NO⋅ are base modifications, such as deamination or oxidation products (21, 39, 44). Base damage is generally repaired by the NER and BER pathways. We therefore initially investigated the potential of excision repair pathways to defend against NO⋅-induced toxicity.
Despite the broad substrate range of NER, E. coli cells deficient in NER did not show increased sensitivity to NO⋅, raising the possibility that the BER process may be relevant to repair of NO⋅-induced base damage. However, none of the glycosylase-deficient strains examined in this study showed increased sensitivity to NO⋅ toxicity. Thus, it appears that BER of NO⋅-induced base damage may be important for removal of potentially mutagenic lesions but not for removal of toxic lesions. Alternatively, there is sufficient redundancy among DNA glycosylases (or between excision repair pathways) to prevent base damage-induced toxicity in the absence of certain DNA glycosylases.
We also examined the importance of enzymes that act downstream of the DNA glycosylases in the BER pathway. Although E. coli strains lacking either the EndoIV or the ExoIII AP endonucleases were resistant to NO⋅-induced toxicity, cells carrying mutations in both of these AP endonucleases (xth nfo cells) were very sensitive to NO⋅-induced cell killing. In contrast, previous studies by Nunoshiba et al. showed that E. coli strains deficient in nfo alone are quite sensitive to macrophage-mediated toxicity (35). Perhaps the very long-term nature of these experiments, involving activated macrophages, accounts for the sensitivity of the nfo mutant E. coli in these experiments.
In contrast to NO⋅ exposure, where xth strains showed near-wild-type levels of resistance, xth mutant cells are very sensitive to H2O2 (11). Exposure of cells to H2O2 causes radical-induced breakdown of deoxyribose, which requires 3′-end processing by ExoIII (ExoIII is responsible for ∼99% of the end-processing activity in E. coli [33]). While the generation of similar direct single-strand breaks through peroxynitrite radicals cannot be excluded, chemically induced strand breaks are rare in comparison to the number of strand breaks generated through the action of repair enzymes (21, 39, 44). Furthermore, the observation that ExoIII and EndoIV can compensate for one another in the case of exposure to NO⋅ is consistent with their roles as AP endonucleases and makes it less likely that ExoIII is playing a critical role in processing damaged 3′ ends. In any case, the finding that nfo xth E. coli strains are sensitive to NO⋅ toxicity indicates that AP sites are induced by NO⋅ exposure and that repair of these sites is critical to cellular survival. Further studies are necessary to determine the relative contributions of spontaneous base loss and specific DNA glycosylases to the generation of these AP sites.
Excision repair is effective for lesions that occur on one strand, but recombinational repair is critical for the repair of lesions that encompass both strands of the DNA duplex, such as interstrand cross-links, double-strand breaks, daughter-strand gaps, and collapsed replication forks (reviewed in reference 23). Almost all homologous recombination events require the function of RecA. Cells deficient in RecA were very sensitive to NO⋅ toxicity; however, RecA-deficient cells are sensitive to other DNA-damaging agents as well. Nevertheless, for many other DNA-damaging agents, excision repair pathways play a much more substantial role in suppressing toxicity (e.g., NER for UV damage and DNA glycosylases for methylation damage). For RecA to start filament formation and initiate homologous recombination, it needs to be assisted by RecBCD or RecF. While RecBCD mediates repair of double-strand ends, RecFOR mediates repair of daughter-strand gaps. The results of the studies presented here show that cells deficient in RecF were, at most, only slightly sensitive to the toxic effects of NO⋅, whereas cells lacking RecBCD were as sensitive to NO⋅ as were cells deficient in RuvC. The function of a resolvase is required to process Holliday junctions formed during homologous recombination, and RuvC is the major E. coli Holliday junction resolvase. These results suggest that the vast majority of NO⋅-induced Holliday junctions are formed during RecBCD-mediated repair of double-strand ends. It is noteworthy that RecF is critical both for the repair of daughter-strand gaps that form when replication is inhibited in the lagging strand and for tolerance of lesions that inhibit DNA replication in the leading strand (8, 20, 31). Consequently, cells lacking RecF are very sensitive to agents that create lesions that inhibit DNA replication, such as UV (8). The apparent resistance of the recF mutant cells to NO⋅ exposure suggests that inhibition of DNA replication is not the major cause of NO⋅-induced recombination.
What type of NO⋅ damage might lead to double-strand ends and recombination events? Two of the most potent inducers of recombination events are double-strand breaks and interstrand cross-links. As mentioned before, NO⋅ does not directly form double-strand breaks (reviewed in reference 4). Although there is evidence that NO⋅ can create interstrand cross-links, it is unlikely that such cross-links are pivotal in this process, since NER-deficient cells are not sensitive to NO⋅ (Fig. 1A) and the major pathway for repair of interstrand cross-links involves NER (reviewed in reference 14). A remaining candidate for recombination induction is damage that inhibits DNA replication. Given the resistance of recF mutant cells to NO⋅-induced killing, which is required to restart replication (8), it seems unlikely that lesions that inhibit DNA replication are primarily responsible for NO⋅-induced recombination.
We propose the following model for the mechanism of NO⋅-induced recombination. NO⋅ creates base damage, such as deamination products, that are processed by enzymes in the BER pathway, creating nicks and gaps in the process. If such BER intermediates are encountered by the replication machinery, this may result in the collapse of the replication fork, which would be repaired by the RecBCD pathway or lead to cell death. If the replication fork finds homology in a homologous chromosome or at another locus in the genome, this would result in a rearrangement of DNA sequences. Indeed, we observed that NO⋅ does induce such rearrangements between two mutant lac loci to restore a Lac+ phenotype. It is also possible that single-strand breaks formed directly by peroxynitrite might become recombinogenic during DNA replication.
The implications of the work presented here can be summarized as follows. We have shown that recombinational repair is pivotal for preventing NO⋅-induced killing of microbes. To our knowledge, the recA mutation in cells has a far greater effect on cellular survival at physiological levels of NO⋅ exposure than that of any other gene mutation previously reported. These results suggest that the recA gene product, which is key both to the SOS response and to recombinational repair, may be an effective target as an adjuvant to antibiotic therapies. In addition, we have shown that NO⋅ induces recombination, at least in E. coli, which indicates that DNA rearrangements must be included when considering the mutational spectrum of NO⋅ and, in terms of mammalian biology, introduces a novel potential mechanism for the underlying link between inflammation and cancer.
ACKNOWLEDGMENTS
This research was partially supported by NIH grant CA26371-21. T.L.W. is supported by T32-ES07020. E.J.S. is supported by NIH grant R01CA79827-0. M.G.M. is supported by a Howard Hughes Medical Institute Research Resources Program for Medical Schools Award to the University of Massachusetts Medical School. B.P.E. is partially supported by the Samuel A. Goldblith Career Development Professorship.
We thank Carrie A. Hendricks for helpful discussions and Karie Ng for her technical help in the preparation of the manuscript. We thank S. Wallace, M. Volkert, S. Boiteux, B. Demple, and B. Weiss for helpful discussions and the strains listed in Table 1.
REFERENCES
- 1.Arnold D A, Kowalczykowski S C. Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J Biol Chem. 2000;275:12261–12265. doi: 10.1074/jbc.275.16.12261. [DOI] [PubMed] [Google Scholar]
- 2.Blaisdell J O, Hatahet Z, Wallace S S. A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G→T transversions. J Bacteriol. 1999;181:6396–6402. doi: 10.1128/jb.181.20.6396-6402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenman J E, Bredt D S. Synaptic signaling by nitric oxide. Curr Opin Neurobiol. 1997;7:374–378. doi: 10.1016/s0959-4388(97)80065-7. [DOI] [PubMed] [Google Scholar]
- 4.Burney S, Caulfield J L, Niles J C, Wishnok J S, Tannenbaum S R. The chemistry of DNA damage from nitric oxide and peroxynitrite. Mutat Res. 1999;424:37–49. doi: 10.1016/s0027-5107(99)00006-8. [DOI] [PubMed] [Google Scholar]
- 5.Burney S, Niles J C, Dedon P C, Tannenbaum S R. DNA damage in deoxynucleosides and oligonucleotides treated with peroxynitrite. Chem Res Toxicol. 1999;12:513–520. doi: 10.1021/tx980254m. [DOI] [PubMed] [Google Scholar]
- 6.Caulfield J L, Wishnok J S, Tannenbaum S R. Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J Biol Chem. 1998;273:12689–12695. doi: 10.1074/jbc.273.21.12689. [DOI] [PubMed] [Google Scholar]
- 7.Chung M H, Kasai H, Jones D S, Inoue H, Ishikawa H, Ohtsuka E, Nishimura S. An endonuclease activity of Escherichia coli that specifically removes 8-hydroxyguanine residues from DNA. Mutat Res. 1991;254:1–12. doi: 10.1016/0921-8777(91)90035-n. [DOI] [PubMed] [Google Scholar]
- 8.Courcelle J, Carswell-Crumpton C, Hanawalt P C. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham R P, Saporito S M, Spitzer S G, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demple B. Genetic responses against nitric oxide toxicity. Braz J Med Biol Res. 1999;32:1417–1427. doi: 10.1590/s0100-879x1999001100013. [DOI] [PubMed] [Google Scholar]
- 11.Demple B, Halbrook J, Linn S. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J Bacteriol. 1983;153:1079–1082. doi: 10.1128/jb.153.2.1079-1082.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demple B, Johnson A, Fung D. Exonuclease III and endonuclease IV remove 3′ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc Natl Acad Sci USA. 1986;83:7731–7735. doi: 10.1073/pnas.83.20.7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epe B, Ballmaier D, Roussyn I, Briviba K, Sies H. DNA damage by peroxynitrite characterized with DNA repair enzymes. Nucleic Acids Res. 1996;24:4105–4110. doi: 10.1093/nar/24.21.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 15.Graziewicz M, Wink D A, Laval F. Nitric oxide inhibits DNA ligase activity: potential mechanisms for NO⋅-mediated DNA damage. Carcinogenesis. 1996;17:2501–2505. doi: 10.1093/carcin/17.11.2501. [DOI] [PubMed] [Google Scholar]
- 16.Green L C, Wagner D A, Glowgowski J, Skipper P L, Wishnok J S, Tannenbaum S R. Analysis of nitrate, nitrite, and [15N]-nitrate in biological fluids. Anal Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg J T, Monach P, Chou J H, Josephy P D, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grishko V I, Druzhyna N, LeDoux S P, Wilson G L. Nitric oxide-induced damage to mtDNA and its subsequent repair. Nucleic Acids Res. 1999;27:4510–4516. doi: 10.1093/nar/27.22.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobbs A J, Ignarro L J. Nitric oxide-cyclic GMP signal transduction system. Methods Enzymol. 1996;269:134–148. doi: 10.1016/s0076-6879(96)69016-8. [DOI] [PubMed] [Google Scholar]
- 20.Horii Z, Clark A J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy L J, Moore K, Jr, Caulfield J L, Tannenbaum S R, Dedon P C. Quantitation of 8-oxoguanine and strand breaks produced by four oxidizing agents. Chem Res Toxicol. 1997;10:386–392. doi: 10.1021/tx960102w. [DOI] [PubMed] [Google Scholar]
- 22.Konrad E B. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J Bacteriol. 1977;130:167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lancaster J R., Jr A tutorial on the diffusibility and reactivity of free nitric oxide. Nitric Oxide. 1997;1:18–30. doi: 10.1006/niox.1996.0112. [DOI] [PubMed] [Google Scholar]
- 25.Laval F, Wink D A. Inhibition by nitric oxide of the repair protein, O6-methylguanine-DNA-methyltransferase. Carcinogenesis. 1994;15:443–447. doi: 10.1093/carcin/15.3.443. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci USA. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Little J W, Harper J E. Identification of the lexA gene product of Escherichia coli K-12. Proc Natl Acad Sci USA. 1979;76:6147–6151. doi: 10.1073/pnas.76.12.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ljungquist S, Lindahl T, Howard-Flanders P. Methyl methane sulfonate-sensitive mutant of Escherichia coli deficient in an endonuclease specific for apurinic sites in deoxyribonucleic acid. J Bacteriol. 1976;126:646–653. doi: 10.1128/jb.126.2.646-653.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobysheva I I, Stupakova M V, Mikoyan V D, Vasilieva S V, Vanin A F. Induction of the SOS DNA repair response in Escherichia coli by nitric oxide donating agents: dinitrosyl iron complexes with thiol-containing ligands and S-nitrosothiols. FEBS Lett. 1999;454:177–180. doi: 10.1016/s0014-5793(99)00777-2. [DOI] [PubMed] [Google Scholar]
- 30.MacMicking J, Xie Q W, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 31.Mahdi A A, Lloyd R G. Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol Gen Genet. 1989;216:503–510. doi: 10.1007/BF00334397. [DOI] [PubMed] [Google Scholar]
- 32.Marletta M A, Yoon P S, Iyengar R, Leaf C D, Wishnok J S. Macrophage oxidation of L-arginine to nitrite and nitrate: nitric oxide is an intermediate. Biochemistry. 1988;27:8706–8711. doi: 10.1021/bi00424a003. [DOI] [PubMed] [Google Scholar]
- 33.Milcarek C, Weiss B. Mutants of Escherichia coli with altered deoxyribonucleases. I. Isolation and characterization of mutants for exonuclease 3. J Mol Biol. 1972;68:303–318. doi: 10.1016/0022-2836(72)90215-x. [DOI] [PubMed] [Google Scholar]
- 34.Niles J C, Burney S, Singh S P, Wishnok J S, Tannenbaum S R. Peroxynitrite reaction products of 3′,5′-di-O-acetyl-8-oxo-7,8-dihydro-2′-deoxyguanosine. Proc Natl Acad Sci USA. 1999;96:11729–11734. doi: 10.1073/pnas.96.21.11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nunoshiba T, deRojas-Walker T, Wishnok J S, Tannenbaum S R, Demple B. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc Natl Acad Sci USA. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connor T, Laval J. Physical association of the 2,6-diamino-4-hydroxy-5N-formamidopyrimidine-DNA glycosylase of Escherichia coli and an activity nicking DNA at apurinic/apyrimidinic sites. Proc Natl Acad Sci USA. 1989;86:5222–5226. doi: 10.1073/pnas.86.14.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radman M. An endonuclease from Escherichia coli that introduces single polynucleotide chain scissions in ultraviolet-irradiated DNA. J Biol Chem. 1976;251:1438–1445. [PubMed] [Google Scholar]
- 38.Roy B, Lepoivre M, Henry Y, Fontecave M. Inhibition of ribonucleotide reductase by nitric oxide derived from thionitrites: reversible modifications of both subunits. Biochemistry. 1995;34:5411–5418. doi: 10.1021/bi00016a012. [DOI] [PubMed] [Google Scholar]
- 39.Salgo M G, Stone K, Squadrito G L, Battista J R, Pryor W A. Peroxynitrite causes DNA nicks in plasmid pBR322. Biochem Biophys Res Commun. 1995;210:1025–1030. doi: 10.1006/bbrc.1995.1759. [DOI] [PubMed] [Google Scholar]
- 40.Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci USA. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiestl R H, Gietz R D, Mehta R D, Hastings P J. Carcinogens induce intrachromosomal recombination in yeast. Carcinogenesis. 1989;10:1445–1455. doi: 10.1093/carcin/10.8.1445. [DOI] [PubMed] [Google Scholar]
- 42.Shapiro R, Dubelman S, Feinberg A M, Crain P F, McCloskey J A. Isolation and identification of cross-linked nucleosides from nitrous acid treated deoxyribonucleic acid. J Am Chem Soc. 1977;99:302–303. doi: 10.1021/ja00443a080. [DOI] [PubMed] [Google Scholar]
- 43.Sidorkina O, Saparbaev M, Laval J. Effects of nitrous acid treatment on the survival and mutagenesis of Escherichia coli cells lacking base excision repair (hypoxanthine-DNA glycosylase-ALK A protein) and/or nucleotide excision repair. Mutagenesis. 1997;12:23–28. doi: 10.1093/mutage/12.1.23. [DOI] [PubMed] [Google Scholar]
- 44.Spencer J P, Wong J, Jenner A, Aruoma O I, Cross C E, Halliwell B. Base modification and strand breakage in isolated calf thymus DNA and in DNA from human skin epidermal keratinocytes exposed to peroxynitrite or 3-morpholinosydnonimine. Chem Res Toxicol. 1996;9:1152–1158. doi: 10.1021/tx960084i. [DOI] [PubMed] [Google Scholar]
- 45.Tamir S, Burney S, Tannenbaum S R. DNA damage by nitric oxide. Chem Res Toxicol. 1996;9:821–827. doi: 10.1021/tx9600311. [DOI] [PubMed] [Google Scholar]
- 46.Tamir S, Lewis R S, deRojas Walker T, Deen W M, Wishnok J S, Tannenbaum S R. The influence of delivery rate on the chemistry and biological effects of nitric oxide. Chem Res Toxicol. 1993;6:895–899. doi: 10.1021/tx00036a021. [DOI] [PubMed] [Google Scholar]
- 47.Taylor A F, Schultz D W, Ponticelli A S, Smith G R. RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation-dependence of the cutting. Cell. 1985;41:153–163. doi: 10.1016/0092-8674(85)90070-4. [DOI] [PubMed] [Google Scholar]
- 48.Tretyakova N Y, Niles J C, Burney S, Wishnok J S, Tannenbaum S R. Peroxynitrite-induced reactions of synthetic oligonucleotides containing 8-oxoguanine. Chem Res Toxicol. 1999;12:459–466. doi: 10.1021/tx980235c. [DOI] [PubMed] [Google Scholar]
- 49.Tsaneva I R, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallace S S. AP endonucleases and DNA glycosylases that recognize oxidative DNA damage. Environ Mol Mutagen. 1988;12:431–477. doi: 10.1002/em.2860120411. [DOI] [PubMed] [Google Scholar]
- 51.Wilson D M, III, Engelward B P, Samson L. Prokaryotic base excision repair. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair: biochemistry, genetics, and cell biology. I. Totowa, N.J: Humana Press, Inc.; 1998. pp. 29–64. [Google Scholar]
- 52.Wink D A, Laval J. The Fpg protein, a DNA repair enzyme, is inhibited by the biomediator nitric oxide in vitro and in vivo. Carcinogenesis. 1994;15:2125–2129. doi: 10.1093/carcin/15.10.2125. [DOI] [PubMed] [Google Scholar]
- 53.Yermilov V, Rubio J, Becchi M, Friesen M D, Pignatelli B, Ohshima H. Formation of 8-nitroguanine by the reaction of guanine with peroxynitrite in vitro. Carcinogenesis. 1995;16:2045–2050. doi: 10.1093/carcin/16.9.2045. [DOI] [PubMed] [Google Scholar]
- 54.Yermilov V, Rubio J, Ohshima H. Formation of 8-nitroguanine in DNA treated with peroxynitrite in vitro and its rapid removal from DNA by depurination. FEBS Lett. 1995;376:207–210. doi: 10.1016/0014-5793(95)01281-6. [DOI] [PubMed] [Google Scholar]
- 55.Zdraveski Z Z, Mello J A, Marinus M G, Essigmann J M. Multiple pathways of recombination define cellular responses to cisplatin. Chem Biol. 2000;7:39–50. doi: 10.1016/s1074-5521(00)00064-8. [DOI] [PubMed] [Google Scholar]