ABSTRACT
Bacterial microcompartments (MCPs) are protein-based organelles that house the enzymatic machinery for metabolism of niche carbon sources, allowing enteric pathogens to outcompete native microbiota during host colonization. While much progress has been made toward understanding MCP biogenesis, questions still remain regarding the mechanism by which core MCP enzymes are enveloped within the MCP protein shell. Here, we explore the hypothesis that the shell protein PduB is responsible for linking the shell of the 1,2-propanediol utilization (Pdu) MCP from Salmonella enterica serovar Typhimurium LT2 to its enzymatic core. Using fluorescent reporters, we demonstrate that all members of the Pdu enzymatic core are encapsulated in Pdu MCPs. We also demonstrate that PduB is critical for linking the entire Pdu enzyme core to the MCP shell. Using MCP purifications, transmission electron microscopy, and fluorescence microscopy, we find that shell assembly can be decoupled from the enzymatic core, as apparently empty MCPs are formed in Salmonella strains lacking PduB. Mutagenesis studies reveal that PduB is incorporated into the Pdu MCP shell via a conserved, lysine-mediated hydrogen bonding mechanism. Finally, growth assays and system-level pathway modeling reveal that unencapsulated pathway performance is strongly impacted by enzyme concentration, highlighting the importance of minimizing polar effects when conducting these functional assays. Together, these results provide insight into the mechanism of enzyme encapsulation within Pdu MCPs and demonstrate that the process of enzyme encapsulation and shell assembly are separate processes in this system, a finding that will aid future efforts to understand MCP biogenesis.
IMPORTANCE MCPs are unique, genetically encoded organelles used by many bacteria to survive in resource-limited environments. There is significant interest in understanding the biogenesis and function of these organelles, both as potential antibiotic targets in enteric pathogens and also as useful tools for overcoming metabolic engineering bottlenecks. However, the mechanism by which these organelles are formed natively is still not completely understood. Here, we provide evidence of a potential mechanism in S. enterica by which a single protein, PduB, links the MCP shell and metabolic core. This finding is critical for those seeking to disrupt MCPs during pathogenic infections or for those seeking to harness MCPs as nanobioreactors in industrial settings.
KEYWORDS: 1, 2-propanediol utilization, Salmonella, bacterial microcompartment, enteric pathogens, metabolosome, organelle assembly, polar effect, shell protein
INTRODUCTION
Subcellular compartmentalization into organelles is an important feature of cellular life (1). Organelles allow cells to organize chemical transformations in space, enabling diverse metabolic processes to occur simultaneously within single cells. This functionality, sequestering diverse processes into membrane-delimited organelles, is most frequently associated with the eukaryotic domain (1). However, prokaryotes have diverse modes of subcellular compartmentalization as well, including both lipid (2)- and protein (3)-bound organelles.
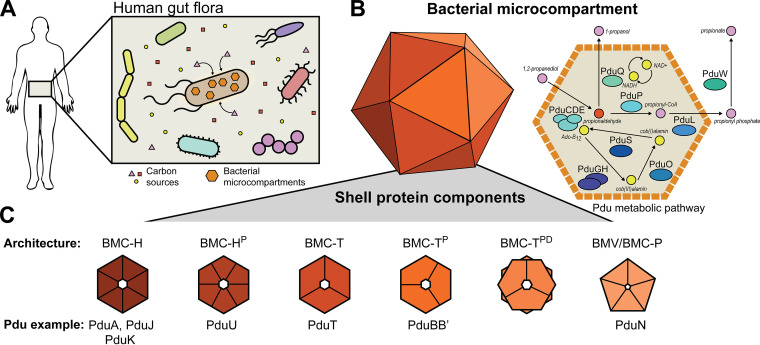
A prime example of a protein-delimited organelle is the bacterial microcompartment (MCP) (4). These protein-based organelles come in numerous varieties, including carbon-fixing carboxysomes found in cyanobacteria and carbon-harvesting metabolosomes often found in enteric pathogens (Fig. 1A) (5, 6). Metabolosomes consist of a proteinaceous shell surrounding an encapsulated enzymatic core (Fig. 1B) (6). Typically the enzymatic core consists of a specific metabolic pathway that utilizes a niche carbon source (Fig. 1B), often one that must move through a toxic aldehyde intermediate (Fig. 1B) (7). The MCP shell is thought to provide a diffusional barrier between the cytosol and enzymatic core, which both helps to increase flux through the encapsulated pathway and also protects the cell from the toxic intermediate (8, 9). In this way, MCPs are thought to be important for providing a growth advantage to pathogenic microbes during propagation within hosts (10).
FIG 1.
Schematic representation of cells containing Pdu MCPs. (A) Cells harboring MCPs can scavenge niche carbon sources from their environment. (B) The Pdu MCP encapsulates enzymes responsible for the metabolism of 1,2-propanediol to propionate and 1-propanol via the toxic intermediate propionaldehyde, which is sequestered by the MCP shell. The Pdu MCP also encapsulates various enzymes responsible for cofactor recycling. (C) MCPs are composed of a protein shell made of a variety of different shell protein architectures, including hexamers (BMC-H), trimers (BMC-T), and pentamers (BMC-P) and their various subtypes.
One of the most well-studied types of metabolosomes is the 1,2-propanediol utilization (Pdu) MCP found in Salmonella enterica serovar Typhimurium LT2 (here referred to as LT2). The Pdu MCP provides an excellent model for metabolosome study, as Pdu MCPs natively exist in a number of model bacterial hosts, such as the aforementioned LT2 as well as strains of Escherichia coli (11). Pdu MCPs are broadly distributed throughout many bacterial phyla, and numerous Pdu subtypes exist (12–14). The LT2 Pdu MCP is also a valuable model metabolosome because it contains representative shell proteins of all but one of the known shell protein subtypes (Fig. 1C), making Pdu-based studies broadly applicable to a variety of MCP systems (13).
Generally, Pdu MCP shells are irregular and polyhedral in geometry and roughly 140 nm in diameter (15). They are composed of small, assembled shell proteins that tile together around the enzymatic core into a perforated, mosaic surface containing small pores to allow metabolite diffusion (Fig. 1B and C) (16). The assembled shell proteins come in a number of basic archetypal categories (Fig. 1C) (13). These various architectures include six-sided hexameric or pseudohexameric proteins composed of pfam00936 domains as well as five-sided pentameric proteins (BMC-P or BMV) composed of pfam03319 domains (Fig. 1C) (17, 18). The hexagonal pfam00936 proteins can be further subdivided into simple hexamers (BMC-H), circularly permutated hexamers (BMC-HP), pseudohexameric trimers (BMC-T), circularly permutated pseudohexameric trimers (BMC-TP), and circularly permutated pseudohexameric trimer dimers (BMC-TDP) (Fig. 1C) (13). The LT2 Pdu operon contains eight total shell proteins, with representatives from all but the BMC-TDP subcategory (19).
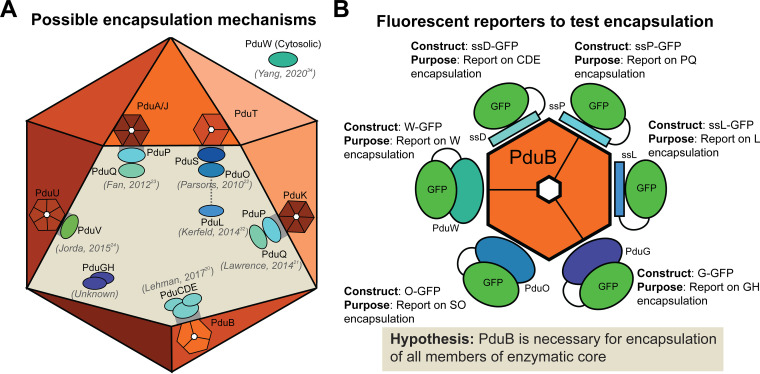
Although tremendous progress has been made toward understanding the biogenesis and assembly of the Pdu MCP, questions remain regarding the mechanism linking the shell and the enzymatic core. Numerous studies exist throughout the literature proposing nearly every single Pdu shell protein is responsible for encapsulating one or more of the enzymes that localize to the MCP core (Fig. 2A) (20–24). In this study, we expand on the hypothesis set forth by Lehman et al., which posited that the BMC-TP protein PduB is responsible for enzyme encapsulation in vitamin B12-dependent Pdu MCPs from LT2 (20). To build on this study, we created a suite of fluorescently tagged enzyme reporters that report on the encapsulation state of all enzyme interaction partners (i.e., at least one enzyme from sets of enzymes that are known to interact) (Fig. 2B), revealing that PduB plays a critical role in encapsulation. Through mutagenesis, we show that PduB is incorporated into the MCP shell via a conserved hydrogen bonding mechanism (25–27). We also show that signal sequences are sufficient for targeting to the enzymatic core, indicating that signal sequences interact with other members of the enzymatic core and not necessarily the MCP shell, as previously thought (21, 23). Interestingly, gene knockouts and MCP purifications demonstrate that the Pdu MCP shell still assembles in the absence of the enzymatic core, providing evidence that shell assembly and enzymatic core assembly are separate processes that can be decoupled. Finally, our work shows decoupling the enzymatic core from the MCP shell results in a surprising growth phenotype, differing from other strains expressing disrupted MCPs. Together, these results provide clarity to a field crowded with conflicting evidence regarding MCP enzyme encapsulation and organelle biogenesis.
FIG 2.
Suite of fluorescent reporters is used to investigate hypotheses about enzyme encapsulation within Pdu MCPs. (A) A number of hypotheses have been proposed in the Pdu MCP literature regarding the mechanism of enzyme encapsulation. Six of the eight Pdu shell proteins have been suggested to have a role in enzyme encapsulation (20–24, 32, 34). (B) In order to test the hypothesis proposed in Lehman et al. (20), we created a suite of fluorescent reporters to inform on the encapsulation state on the entire Pdu MCP core.
RESULTS
Fluorescent reporters demonstrate PduB is critical for enzyme encapsulation.
In 2017, Lehman et al. (20) reported that deletion of the PduB N terminus reduced the apparent enzyme content of purified MCPs, as indicated by SDS-PAGE. While groundbreaking, this study did not directly demonstrate effects of PduB deletion on enzyme localization to MCPs, as malformed MCPs or mistargeted enzymes are copurified with MCPs in the differential centrifugation process (26). To build on this landmark study, we created a suite of green fluorescent protein (GFP) reporters to inform on which members of the enzymatic core are directly impacted by the loss of PduB. Numerous studies have indicated that many Pdu shell proteins directly bind and encapsulate various core enzymes. Thus, we reasoned that PduB may only be responsible for binding and encapsulating a select subset of Pdu enzymes. For example, studies have suggested that PduK (21), PduT (22), PduA (23), PduJ (23), and PduU (24) are responsible for binding and targeting one or more Pdu enzymes to the MCP core (Fig. 2A). However, the work by Lehman et al. (20) is the only study to date to directly show that a genetic alteration (deletion of the PduB N terminus in the pdu operon) leads to changes in enzyme localization. We therefore hypothesized that PduB is the most likely shell protein to be responsible for enzyme targeting (20).
To test the hypothesis that PduB is responsible for linking the Pdu MCP shell and enzymatic core, we created GFP fusions to report on each Pdu enzyme (Fig. 1B and 2B). Some enzymes or sets of enzymes contain known, short, N-terminal signal sequences that are sufficient for targeting enzymes to the MCP lumen. The signal sequence of PduD (ssD) is sufficient for targeting PduC, PduD, and PduE (28); the signal sequence for PduP (ssP) is sufficient for targeting PduP (29); and the signal sequence for PduL (ssL) is sufficient for targeting PduL (30). Since these are the minimal components necessary for targeting these proteins, they were fused to GFP and used to report on the enzyme’s change in encapsulation as a result of genetic manipulations in this study (Fig. 2B). Since PduD interacts with PduC and PduE, only ssD-GFP was necessary to report on these three proteins. In the same way, since PduP interacts with PduQ (31), ssP-GFP was used to report on this enzyme set (PduP and PduQ). PduL has been hypothesized to interact with PduS (32), but this has not been confirmed experimentally. However, PduS interacts with PduO (33), and since PduO is the more abundant member of this pair (34), it was fused to GFP and used as a reporter for PduS and PduO (O-GFP) (Fig. 2B). Similarly, PduG and PduH are subunits of a diol dehydratase reactivase enzyme and PduG is the more abundant member of this protein pair, so it was fused with GFP (G-GFP) and used for this study (Fig. 2B) (34–37). Finally, PduW, which is thought to be cytosolic (34), was fused C-terminally with GFP (W-GFP) to report on its encapsulation state (Fig. 2B).
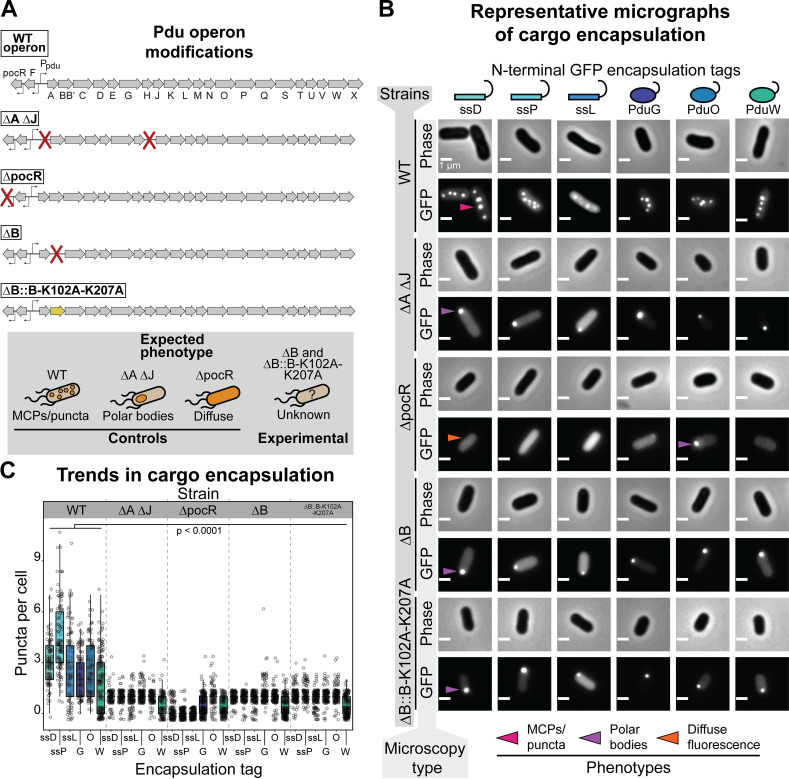
To test the hypothesis that PduB is necessary for the encapsulation of all members of the enzymatic core listed above, each fluorescent reporter was expressed in four strains of LT2 grown in rich medium with 1,2-propanediol (1,2-PD) added to simultaneously induce MCP expression: (i) wild-type (WT) LT2, (ii) a strain with the genes encoding the BMC-H proteins PduA and PduJ knocked out (ΔA ΔJ), (iii) a strain with the transcriptional activator PocR knocked out (ΔpocR), and (iv) an experimental strain with PduB knocked out (ΔB). This procedure informed on the effect of PduB loss on enzyme encapsulation (Fig. 3A). The WT strain serves as a positive control and enables visualization of which enzymes are normally encapsulated within MCPs (Fig. 3A). The ΔA ΔJ strain serves as a negative control, as this strain is incapable of forming assembled MCPs but still forms proto-MCP aggregates, which appear as polar bodies (Fig. 3A) (26). The ΔpocR strain serves as a negative control for MCP expression, as MCP proteins cannot be expressed from the genome in the presence of the native inducer 1,2-PD in this strain (Fig. 3A) (26, 38–41). This allows us to determine which GFP-tagged constructs formed aggregates when expressed in the absence of MCPs and to what degree. Note that these experiments are intended to test for MCP expression, enzyme encapsulation, and MCP assembly, not proper metabolic function of MCPs.
FIG 3.
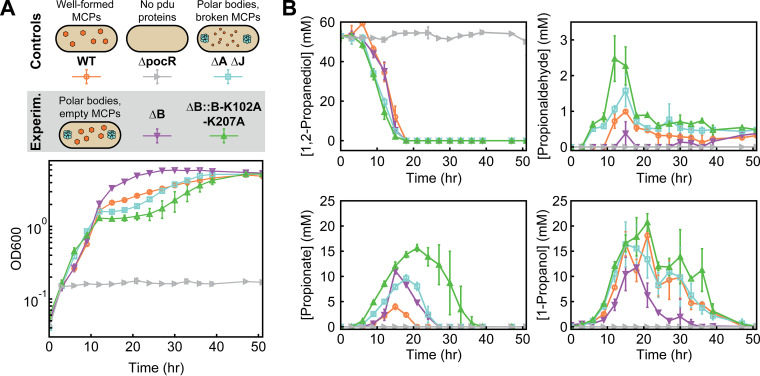
Loss of PduB disrupts enzyme encapsulation. (A) Schematic representation of the pdu operon and modifications to various LT2 strains used in this study, including the wild-type (WT) operon. ΔA ΔJ has the open reading frames encoding PduA and PduJ knocked out, disrupting MCP formation. The ΔpocR strain has the transcriptional activator for the pdu operon knocked out, disrupting Pdu MCP expression. The ΔB strain has PduB knocked out, and the ΔB::B-K102A-K207A strain has WT PduB replaced with a double mutant that disrupts PduB incorporation into the MCP shell. (B) Phase-contrast and GFP fluorescence micrographs of cells expressing different GFP reporter constructs (scale bar, 1 μm). Different LT2 strains are indicated by the far-left vertical axis label, and the micrograph type is indicated by the near-left vertical axis label. The reporter construct is indicated by the top horizontal labels. Representative bright, fluorescent puncta indicating MCPs are labeled with a pink arrow, polar bodies are labeled with a purple arrow, and diffuse fluorescence is indicated with a orange arrow. (C) Quantification of puncta per cell for different fluorescently tagged enzyme reporter constructs and various LT2 strains. The strain is indicated by the top, horizontal label, and the encapsulation tag is indicated by the bottom horizontal label. Note that each reporter construct is readily encapsulated to various degrees into the WT strain, whereas their encapsulation is significantly depleted in strains lacking PduB (P < 0.0001, one-tailed t test). Puncta were counted from three biological replicates, and >106 cells were counted per strain/construct.
When each fluorescent reporter is expressed in the WT strain it is localized to MCPs, as indicated by the presence of bright fluorescent puncta (Fig. 3B and C). This assay has been used as a readout of protein localization to MCPs and has been validated in numerous studies (26, 28, 29, 42). However, the relative encapsulation level of each enzyme set has not been quantitatively assessed using this assay before. Our results reveal that ssD-GFP and ssP-GFP are the most efficiently encapsulated, which is expected based on quantitative mass spectrometry analyses of purified MCPs (34). ssL-GFP is efficiently targeted to a relatively large number of MCPs, but puncta appear much dimmer qualitatively relative to ssD-GFP and ssP-GFP, as expected (Fig. 3B). G-GFP and O-GFP are also encapsulated into MCPs, albeit less efficiently than any of the signal sequence-tagged reporters. We hypothesize that this is due to their propensity to aggregate, as demonstrated by the formation of polar bodies in the ΔpocR strain (Fig. 3B and C). Surprisingly, W-GFP is targeted to MCPs as well despite it being reported to be primarily cytosolic (Fig. 3B and C) (34). It should be noted that it is only targeted to MCPs in a subset of cells, as indicated by the bimodal distribution in the MCP counts for this construct (Fig. 3C). Together these results demonstrate that all of the enzymatic members of the Pdu metabolic pathway are likely targeted to some degree to MCPs and that the GFP reporter constructs used here are adequate reporters of this targeting.
When the fluorescent reporter constructs are coexpressed with MCPs in the ΔB strain, only polar bodies are observed, similar to the ΔA ΔJ strain (Fig. 3B and C). Quantitatively, significantly fewer puncta are observed per cell for each fluorescent reporter in both the ΔB and ΔA ΔJ strains than the WT strain. Significantly more puncta are present in the WT strain than in the ΔpocR strain as well, as expected (Fig. 3C). This indicates that either MCP formation or enzyme encapsulation is significantly impacted by the loss of PduB. To confirm that PduB is responsible for the observed disruption to encapsulation, we performed a plasmid rescue experiment. In this experiment, PduB is overexpressed from a plasmid concomitantly with MCP expression and ssD-GFP is expressed from the pduD locus in the pdu operon to enable MCP visualization via fluorescence microscopy. PduB overexpression rescued compartment formation, evidenced by the presence of significantly more puncta in the ΔB strain with PduB overexpression (see Fig. S1 in the supplemental material). Interestingly, presence of the plasmid containing PduB alone partially rescued MCP formation, likely due to leaky PduB expression (Fig. S1).
The PduB knockout strain forms assembled but empty MCPs.
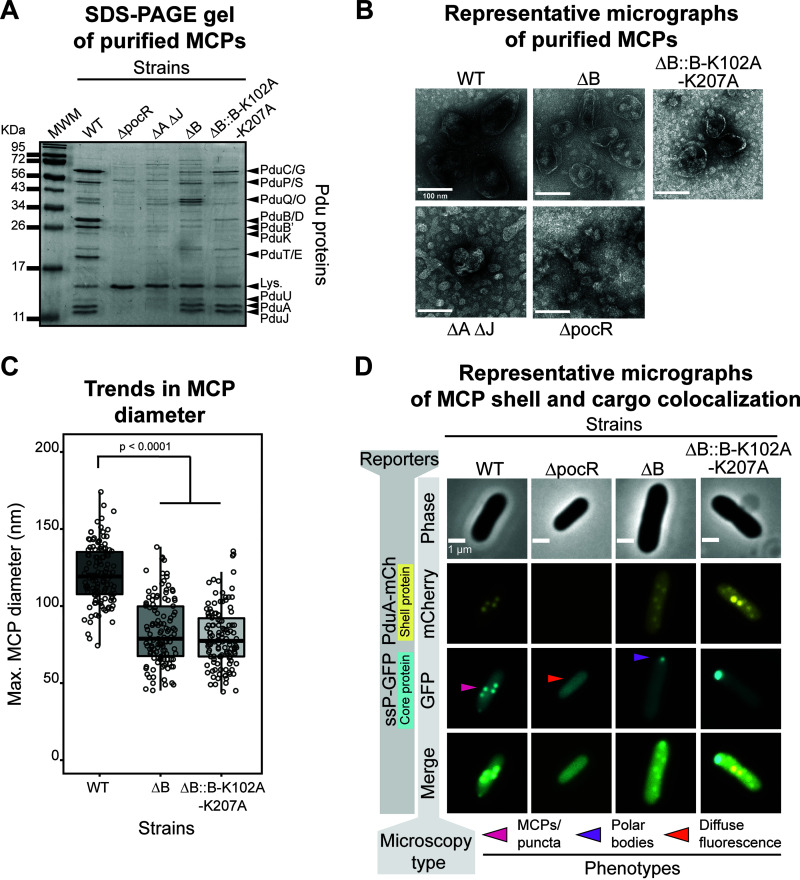
The encapsulation assay reveals that PduB loss dramatically alters the encapsulation state of all fluorescently tagged enzymes tested. However, this assay cannot distinguish between changes in encapsulation or changes in MCP formation. To test the hypothesis that PduB impacts encapsulation but not assembly, MCPs were purified using differential centrifugation from the ΔB strain grown in minimal medium supplemented with 1,2-PD to induce MCP expression (43, 44). When purified samples are analyzed by SDS-PAGE, the ΔB strain appears dramatically different from the standard MCP banding pattern apparent in the WT samples (Fig. 4A). The ΔB samples more closely resemble the ΔA ΔJ samples, which are known to contain malformed MCP aggregates (Fig. 4A and B) (26). An important distinction is that the ΔB sample has high apparent levels of PduA and PduJ (Fig. 4A). Note that weak bands corresponding to Pdu enzymes in the ΔB sample are likely due to copurified enzyme aggregates, which are also frequently present in the ΔA ΔJ assembly-deficient control samples (26).
FIG 4.
Loss of PduB decouples Pdu MCP shell assembly from the enzymatic core. (A) Coomassie-stained SDS-PAGE gel of MCP purifications from various LT2 strains (top horizontal label). Bands corresponding to various Pdu MCP components (right vertical labels) are present in the WT strain but absent from other strains. MWM, molecular weight marker. (B) Representative negative-stained transmission electron micrographs of Pdu MCP purifications from various LT2 strains. Well-formed MCP shells are irregular and polyhedral in shape, have a defined boundary, and are present in the WT, ΔB, and ΔB::B-K102A-K207A samples. (C) Pdu MCPs purified from WT are significantly larger in diameter than shells purified from the ΔB or ΔB::B-K102A-K207A strains (P < 0.0001, one-tailed t test). Measurements were collected on three biological replicates, and >120 MCPs were measured for each sample type. (D) Representative phase-contrast and fluorescence micrographs for various strains of LT2 expressing ssP-GFP and PduA-mCherry reporter constructs (scale bar, 1 μm). The strain is labeled on the top horizontal axis, and the microscopy type is labeled on the near left vertical axis. Bright, fluorescent puncta indicative of MCPs are labeled with a pink arrow, polar bodies are indicated with a purple arrow, and diffuse fluorescence is indicated with a orange arrow. mCherry fluorescence is pseudocolored in yellow and GFP fluorescence is pseudocolored in cyan. Overlapping fluorescence is indicated in the merged image.
Purified samples were then analyzed by transmission electron microscopy (TEM) to determine if MCPs or partially formed aggregates were produced in the ΔB strain (Fig. 4B). To our surprise, the ΔB sample shows many apparently well-formed MCPs. TEM micrographs from this sample contain structures with flat, distinguishable boundaries that appear roughly polyhedral in morphology (Fig. 4B). Interestingly, the ΔB shells appear smaller than the WT shells (83 ± 21 nm versus 121 ± 19 nm, respectively [mean ± standard deviation]) and have qualitatively less electron density in their center, indicating that they are empty (Fig. 4B and C). The structures in the ΔB sample are clearly distinct from the aggregates observed in the ΔA ΔJ sample and the background structures present in the ΔpocR sample. These results, coupled with the SDS-PAGE and encapsulation assay results, indicate that the ΔB strain forms empty MCP shells as well as separate aggregates of Pdu enzymes.
To confirm this surprising finding, colocalization assays were performed in which the shell protein PduA was fused with a fluorescent mCherry reporter protein and expressed from the PduA locus in the WT, ΔB, and ΔpocR strain backgrounds. As expected, PduA-mCherry is localized to MCPs, as indicated by the presence of bright fluorescent puncta (Fig. 4D) (45). PduA is known to be important for shell formation and is one of the most abundant members of the Pdu MCP shell (26, 34, 46). When expressed in the WT background with the tagged GFP reporter constructs, GFP and mCherry colocalize, indicating reporter targeting to MCPs, as expected (Fig. 4D). When expressed in the ΔpocR background, no mCherry puncta are present, as none of the proteins in the pdu operon, including PduA-mCherry, are expressed in this control (Fig. 4D). In the ΔB strain background, mCherry puncta are still present, verifying the finding that MCP shells were still formed in the absence of PduB. As expected based on the results described in Fig. 3, GFP reporter constructs are only found in polar bodies in the ΔB strain (Fig. 4D). These results conclusively demonstrate that PduB deletion impacts protein encapsulation but not MCP shell formation, and that these are two distinct processes that can be decoupled upon PduB deletion. Furthermore, this suggests that signal sequences are sufficient for targeting to the enzymatic core, challenging the idea that primary Pdu MCP signal sequence function is to bind to MCP shell proteins, enabling enzymatic core encapsulation (21, 23).
PduB is incorporated into MCPs by a conserved hydrogen bonding mechanism.
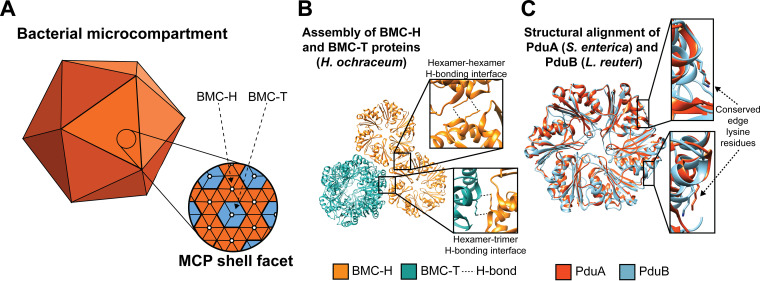
While PduB appears to be important for linking the MCP shell and enzymatic core, the mechanism by which PduB is incorporated into the shell (Fig. 5A) is not known. A groundbreaking structural study by Sutter et al. showed that BMC-TPD proteins similar to PduB appear to incorporate into the shell of a smaller, more regular, empty MCP from Haliangium ochraceum via a lysine-lysine hydrogen bonding motif (Fig. 5B) (25, 47). In this motif, lysine side chains on adjacent BMC proteins (in this case, a BMC-H protein and BMC-TPD protein) arrange antiparallel to each other and hydrogen bond with the backbone carbonyl oxygen of the adjacent lysine (Fig. 5B) (25, 47). Interestingly, analogous lysine residues (Fig. 5C) are also important for the self-assembly of BMC-H proteins PduA and PduJ and are necessary for their ability to drive shell assembly (26). For these reasons, we hypothesized that analogous lysine residues at these positions in the BMC-TP protein PduB (Fig. 5C) are important for PduB incorporation into the MCP shell, bridging the gap between shell and enzymatic core.
FIG 5.
PduB is incorporated in the Pdu MCP shell via a conserved hydrogen bonding motif. (A) Schematic representation of BMC-H (orange) and BMC-T (blue) proteins tiling together to form the facets of the MCP shell. (B) Lysine residues at the hexamer-hexamer and hexamer-trimer interfaces are predicted to play a role in facet tiling in an MCP from H. ochraceum (PDB entry 5V74) (25). BMC-H proteins are in orange and BMC-T proteins are in blue. (C) The edge lysine residue important for PduA (orange) self-assembly is also present in PduB (blue) (PduA PDB entry 3NGK [61]; PduB PDB entry 4FAY [62]). All structural visualization was done using Chimera (63).
To test this hypothesis, a double mutant version of PduB was cloned where the lysine residues at positions 102 and 207 were mutated to alanine residues. These residues appear to be well conserved among PduB homologs (25), so we predicted that mutations to alanine should be disruptive. We expect that this protein is well folded, as analogous mutations in other BMC proteins increase solubility and are used frequently in crystallographic studies (48, 49). The double mutant PduB (B-K102A-K207A) was then placed at the pduB locus in the pdu operon in place of WT pduB, generating the ΔB::B-K102A-K207A strain (Fig. 3A). The encapsulation assay, in which each fluorescent reporter is expressed simultaneously with MCPs, reveals that the ΔB::B-K102A-K207A strain has a phenotype similar to those of both the ΔB and ΔA ΔJ strains, where polar bodies are primarily observed rather than the scattered puncta indicative of MCPs (Fig. 3B and C). A plasmid rescue experiment also showed that PduB-K102A-K207A expressed from a plasmid was unable to recover MCP formation in the ΔB strain, confirming these results (Fig. S1). This indicates that the PduB double mutant is unable to incorporate into MCP shells, effectively recapitulating the ΔB phenotype.
To confirm the conclusion based on the encapsulation assay, MCPs were purified from the ΔB::B-K102A-K207A strain. Samples from this strain appear similar to the ΔB strain by SDS-PAGE, where bands corresponding to enzymes are greatly reduced, but bands for PduA and PduJ are still visible (Fig. 4A). Purified structures were then imaged by TEM, which reveals that apparently well-formed MCPs are present and similar in size and morphology to the MCPs purified from the ΔB strain (Fig. 4B and C). The PduB double mutant strain was also grown with PduA-mCherry expressed from the pduA locus, and this reveals bright puncta throughout the cytoplasm, similar to the ΔB strain, indicating the formation of MCP shells (Fig. 4D). As in the ΔB strain, GFP-tagged cargo appears only in polar bodies, again indicating well-formed shells decoupled from the enzymatic cargo (Fig. 4D). Together, these results strongly support the hypothesis that PduB is incorporated into the MCP shell by a mechanism similar to that of other BMC proteins and that disruption of the conserved lysine hydrogen bonding network disrupts this interaction.
Loss of PduB impacts LT2 growth on 1,2-propanediol as a carbon source.
Having established that PduB is essential for linking the core to the shell of the Pdu MCP, we next explored how decoupling of the core and shell impacts the function of the native Pdu pathway. To this end, we compared growth and metabolite profiles in culture conditions that promote buildup of the toxic propionaldehyde intermediate, thereby leading to a growth lag in strains with broken MCPs that cannot sequester this intermediate away from the cytosol (9, 16, 26). Specifically, cells were grown on 1,2-PD as a sole carbon source in minimal medium with excess adenosylcobalamin (Ado-B12), where the excess Ado-B12 drives the buildup of the toxic propionaldehyde intermediate by maximizing PduCDE activity (Fig. 1B). Under these growth conditions, we hypothesized that our strains with decoupled core and shell assembly would exhibit a growth phenotype similar to that of a strain expressing broken Pdu MCPs (ΔA ΔJ).
We tested this hypothesis by comparing the growth and metabolite profiles of three control strains (WT, ΔpocR, ΔA ΔJ) to two experimental strains (ΔB, ΔB::B-K102A-K207A). Our three control strains were the same as those used in the encapsulation assay: WT, which harbors functional MCPs, ΔpocR, which cannot express the pdu operon, and ΔA ΔJ, which forms proto-MCP aggregates but no shell and thus serves as our broken MCP control. Our two experimental strains (ΔB and ΔB::B-K102A-K207A), described above, both have decoupled MCP core and shell assembly. Our control strains behaved as expected. The ΔpocR strain, which is unable to express any Pdu pathway enzymes, does not grow or consume 1,2-PD (Fig. 6A and B). Our broken MCP control strain (ΔA ΔJ) initially grows and consumes 1,2-PD faster than the MCP-containing strain (WT), as expected; the absence of an MCP shell accelerates apparent pathway kinetics by providing more direct access to 1,2-PD (Fig. 6A and B). However, propionaldehyde buildup in the ΔA ΔJ strain eventually leads to a lag in growth from 12 to 30 h (Fig. 6A and B). During this time frame, the WT strain begins to outgrow the ΔA ΔJ strain, as the MCPs in the WT strain successfully sequester the toxic propionaldehyde intermediate away from the cytosol (Fig. 6A and B).
FIG 6.
PduB alterations change LT2 growth and metabolite profiles. (A) LT2 control and experimental strains containing different MCP morphologies (WT MCPs, polar bodies [ΔA ΔJ, ΔB, and ΔB::B-K102A-K207A], and no protein expression [ΔpocR]). These differing morphologies lead to various LT2 growth profiles, as measured by the optical density at 600 nm (OD600) at different time points. (B) Metabolite profiles of different LT2 strains during growth on 1,2-propanediol as a sole carbon source with excess adenosylcobalamin (150 nM). The concentration (mM) of four key metabolites (1,2-propanediol, propionaldehyde, propionate, and 1-propanol) was measured over the duration of the growth curve visualized in panel A.
Surprisingly, we find that our two experimental strains (ΔB and ΔB::B-K102A-K207A), both of which have decoupled MCP core and shell assembly, exhibit distinct growth and metabolite profiles. The growth and metabolite profiles of the PduB double mutant strain (ΔB::B-K102A-K207A) are similar to those of the broken MCP control (ΔA ΔJ), in agreement with our initial hypothesis. This strain (ΔB::B-K102A-K207A) grows rapidly at first and then suffers a growth lag after propionaldehyde accumulates after 9 h (Fig. 6A and B). Interestingly, the growth lag is longer (9 to 40 h) and the peak propionaldehyde level is higher (2.5 ± 0.6 mM) in the ΔB::B-K102A-K207A strain than in the ΔA ΔJ strain. More perplexing is the behavior of the ΔB strain: not only does this strain exhibit minimal propionaldehyde buildup but it also eventually outgrows the WT strain (Fig. 6A and B). Correspondingly, the downstream metabolites propionate and 1-propanol are consumed more rapidly in the ΔB strain than in other strains (Fig. 6A and B).
Kinetic modeling reconciles differences in assembly and growth phenotypes.
Given that ΔB and ΔB::B-K102A-K207A strains exhibit essentially identical behavior with respect to assembly phenotype (shown above), we turned to kinetic modeling to generate hypotheses that could explain this observed discrepancy in growth behavior and differences in propionaldehyde accumulation.
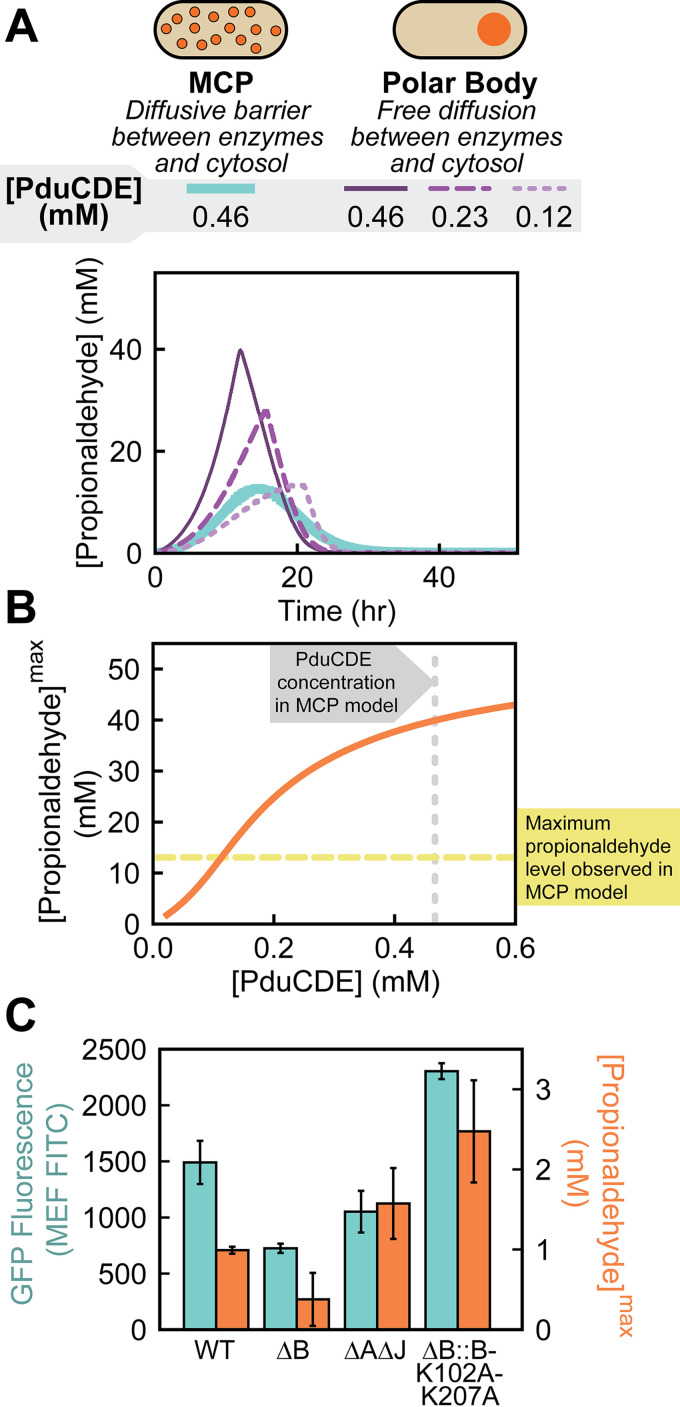
Using a system-level kinetic model of the Pdu pathway modified from previous work (8) (see Materials and Methods), we examined which features of the pathway or polar body geometry had the strongest impact on propionaldehyde buildup in cells containing polar bodies. We considered MCPs a control case, modeling these MCPs as spheres with a diffusive barrier at their surfaces that limits metabolite transport to the enzymatic core. Polar bodies, then, are modeled as one large sphere with free diffusion of metabolites at the surface (Fig. 7A). The volume of this polar body sphere was set to equal the total volume of MCPs in the cell (i.e., polar body volume = number of MCPs per cell × volume of a single MCP). We used this polar body model to assess the sensitivity of maximum propionaldehyde buildup to parameters affecting kinetics, geometry, and transport. Comparison of the MCP (Fig. S2) and polar body (Fig. S3) models reveals that the maximum propionaldehyde buildup in the polar body case is much more sensitive to the maximum velocity (Vmax) of the PduCDE enzyme than in the MCP case. As the concentration of PduCDE in the polar body is decreased (leading to an equivalent fractional decrease in Vmax), the predicted propionaldehyde profile for the polar body approaches that of the MCP (Fig. 7A). Indeed, decreasing the PduCDE concentration in the polar bodies decreases the maximum propionaldehyde concentration outside the cell (Fig. 7B). Specifically, we find that the maximum propionaldehyde concentration observed in the polar body model matches that of the MCP model if the PduCDE concentration is decreased by 76%, suggesting that variation in PduCDE concentration between strains can impact propionaldehyde buildup as strongly as Pdu pathway encapsulation in MCPs (Fig. 7B). We thus hypothesized that the difference between our strains containing polar bodies was the level of PduCDE expression. We hypothesized that this was the result of polar effects in which different alterations to the pduB locus differentially impact the expression of downstream pduCDE genes.
FIG 7.
System-level kinetic modeling reveals how a polar effect alters growth and metabolite profiles. (A) Two organizational strategies for Pdu enzymes (MCPs and polar bodies) were tested in a system-level kinetic model. Plotted here are propionaldehyde concentrations in the external media. PduCDE enzyme concentration was also varied in the polar body model case to account for the potential genetic polar effects that alter gene expression. (B) The kinetic model demonstrates that changes to PduCDE concentration (mM) impact the maximum propionaldehyde concentration (mM) observed in the external medium when pathway enzymes are contained in a polar body. (C) Comparison of GFP fluorescence (MEF FITC) measured by flow cytometry as a proxy for gene expression versus peak propionaldehyde concentration (mM) reveals that PduCDE expression is likely reduced due to a genetic polar effect in the ΔB strain.
We tested our hypothesis that PduCDE expression differed across strains containing polar bodies by measuring GFP expression at the pduD locus. Specifically, we replaced the pduD gene at the pduD locus with a gene encoding ssD-GFP in our five growth strains (WT, ΔpocR, ΔA ΔJ, ΔB, and ΔB::B-K102A-K207A) and used flow cytometry to quantify GFP expression. We find that the ΔB strain has the lowest expression at the pduD locus, with 49% ± 7% lower GFP fluorescence than the WT strain (P < 0.05). In contrast, the ΔB::B-K102A-K207A strain had higher apparent GFP expression at the pduD locus than the WT strain (P < 0.05). Finally, we find that our broken MCP control strain, ΔA ΔJ, has just slightly lower expression at the pduD locus than WT (P < 0.05) (Fig. 7C). Importantly, we note that increased GFP expression at the pduD locus correlates with peak propionaldehyde level in the strains with unencapsulated enzymes (Pearson’s correlation coefficient of 0.92), supporting our hypothesis that PduCDE expression causes differences in propionaldehyde buildup in these strains.
Together, these results highlight two important considerations for future work in the MCP field. First, modeling is a powerful tool for generating hypotheses and exploring parameter space, enabling rapid identification of the key governing parameters for a given subcellular organizational strategy. Second, polar effects can impact downstream enzyme expression, which, in turn, can dramatically alter observed pathway performance. Thus, quantifying these effects and minimizing them whenever possible is essential when conducting genetic studies of MCP function.
DISCUSSION
Metabolosomes are bacterial organelles confirmed to exist in a number of enteric bacteria, where their expression and activation has been linked to pathogenesis (10). Of the metabolosomes investigated to date, the Pdu MCP is the best studied and has served as a model system for understanding the basics of metabolosome assembly and metabolic function. However, even in this well-studied model system, a large body of conflicting evidence exists regarding the mechanism by which enzymatic cargo is loaded into the MCP shell (20–24).
Here, we used a comprehensive suite of fluorescent reporters to show that the shell protein PduB is responsible for linking the enzymatic core to the MCP shell. When PduB is absent or unable to incorporate into the shell, no Pdu enzymes are loaded into the MCP. This interaction between PduB and the enzymatic core likely happens via the N terminus of PduB, as previously suggested in Lehman et al., since deletion of this unique structural domain reduces enzyme loading into MCPs (20). Interestingly, the N terminus of PduB is not well conserved outside of Pdu and glycyl radical enzyme-associated (GRM) MCPs, based on in-depth bioinformatics studies done by Sutter et al. (25) and Axen et al. (for detailed information on the conservation of the PduB N terminus, see the supplemental material) (12–13). This implies that the mechanism by which the PduB N terminus mediates enzyme encapsulation is likely not conserved outside these systems.
In this study, we show that enzymes localize to proto-MCP aggregates, termed polar bodies, in the absence of PduB. Importantly, we find that N-terminal signal sequence tags such as those identified for PduD and PduP are sufficient for loading cargo to these proto-MCP aggregates (28, 29). This is significant, as these signal sequence tags were previously thought to exist primarily to link the shell and core of the MCP, although others have found that signal sequences lower the solubility of certain proteins, leading to aggregation (26, 50–52). We also report that although PduB is required for cargo loading, it is not necessary for MCP shell assembly, as empty MCP structures form in the absence of PduB. The fact that empty MCP shells and proto-MCP aggregates form separately in the absence of PduB suggests that MCP shell and core assembly are independent processes in the Pdu MCP system. Combined with previous work demonstrating that the self-assembling protein PduA or PduJ is required for Pdu MCP formation (26), this discovery brings us one step closer to realizing the components necessary to construct a minimal Pdu MCP. Specifically, two of the requisite components are (i) a hexameric protein with a strong propensity for self-assembly (PduA or PduJ) and (ii) a protein that links the core and shell by interacting with both the enzymatic core and the self-assembling hexameric proteins via a conserved hydrogen bonding mechanism. We expect that computational models of this assembly process will provide insight into the specific roles that these different proteins play in MCP assembly (46, 53).
We also investigated how LT2 growth on 1,2-PD as a sole carbon source in minimal medium supplemented with Ado-B12 was impacted by the decoupling of MCP core and shell assembly. As expected, when enzymes are localized to polar bodies, buildup of the toxic aldehyde intermediate is not as well controlled as it is in MCPs. However, using a combination of system-level kinetic modeling and experiments, we also discovered that strains that contain polar bodies rather than MCPs exhibit dramatically different growth profiles depending on PduCDE expression level. In our strains, we believe the differences in PduCDE expression we observed are a consequence of polar effects on gene expression, as the genes encoding pduCDE are immediately downstream of the pduB gene. Specifically, we find that while our pduB knockout strain (ΔB) and our pduB double mutant strain (ΔB::B-K102A-K207A) both result in a similar decoupling of MCP core and shell assembly, the strain with the more dramatic genetic alteration at the pduB locus (ΔB, the knockout) decreases expression at the pduD locus substantially, whereas the strain with minimal genetic alteration at the pduB locus (ΔB::B-K102A-K207A) does not. This result emphasizes an important point of concern for studies in this field, namely, that polar effects on downstream gene expression can confound interpretation of experimental results and, thus, must be considered. Indeed, it has been shown that alterations to the pduL locus, which encodes enzymatic cargo, can disrupt Pdu MCP shell assembly, likely by modifying expression levels of downstream shell proteins like PduN (42). Conversely, we show here that modifications to the pduB locus, which encodes a shell protein, can alter enzyme concentrations in the core, altering Pdu pathway performance. Thus, the identification of point mutants that minimally disrupt operon sequence, but prevent specific protein functions, will be essential in future MCP studies.
MATERIALS AND METHODS
Plasmid and strain creation.
All plasmids used for this study were created using the Golden Gate cloning (54) method into a Golden Gate-compatible pBAD33t parent vector (chloramphenicol resistance, p15A origin of replication). All strains, plasmids, and primers are listed and described in Tables S1 to S3, respectively, in the supplemental material. For constructs in which a signal sequence was appended to the N terminus of GFPmut2, a BsaI cut site and the DNA sequence encoding the signal sequence were added to GFP using PCR (see Table S3 for primer sequence information). For strains in which GFPmut2 was appended to the C terminus of a full enzyme, a two-piece Golden Gate reaction was carried out. This is true for all GFP-enzyme fusions with the exception of PduG-GFP, which was cloned using SacI and XbaI restriction sites. First, forward and reverse primers were used to amplify the enzyme and GFPmut2 open reading frames. Each primer encoded compatible sticky ends to enable ligation into the pBAD33t parent vector in the proper orientation. A glycine-serine (GS) linker was also encoded between the enzyme and GFPmut2 open reading frames. Golden Gate reactions were carried out using FastDigest Eco31I (Thermo Fisher Scientific) and T4 DNA ligase (New England Biolabs), transformed into E. coli DH10b cells, purified using Zyppy plasmid miniprep kit (Zymo Research), and sequence verified by Sanger sequencing (Genewiz).
All modified LT2 strains used in this study are listed in Table S1 and were generated using λ red recombineering as described in previous reports (26, 55). Briefly, a marker encoding dual selection (cat-sacB) was PCR amplified containing flanking homologous overhangs matching the target gene locus and inserted into the pdu operon. The cat-sacB selectable marker was then either replaced with a gene of interest or knocked out, leaving only the C terminus of the target gene to reduce polar effects on downstream gene expression. For the PduB-K102A-K207A double mutant strain, QuikChange was used (with KOD hot start DNA polymerase [Sigma-Aldrich]) to perform site-directed mutagenesis on the two lysine residues on a plasmid backbone first before it was amplified and inserted into the pdu operon. Modified strains were sequence confirmed using Sanger sequencing (Genewiz).
Fluorescence and phase-contrast microscopy.
Fluorescence and phase-contrast micrographs were collected on a Nikon Eclipse Ni-U upright microscope, 100× oil immersion objective, and an Andor Clara digital camera with NIS Elements software (Nikon). GFP fluorescence micrographs were collected using a C-FL Endow GFP HYQ bandpass filter, and mCherry fluorescence micrographs were collected using a C-FL Y-2E/C filter. Exposure times of 100 ms were used for ssD-GFP and ssP-GFP, 300 ms for ssL-GFP, 400 ms for G-GFP, O-GFP, and W-GFP, and 500 ms for mCherry. For experiments where ssD-GFP was expressed from the pduD locus, an exposure time of 1 to 2 s was used. Cell culture samples were placed onto Fisherbrand frosted microscope slides and covered using 22-mm-by-22-mm, number 1.5 thickness coverslips. All images were equally adjusted within experiments and sample type for brightness and contrast using ImageJ (56). Puncta counts were collected on brightness- and contrast-adjusted images.
Microcompartment expression and purification.
MCP expression and purification was done as described in detail previously (43). Briefly, overnight starter cultures were started from single colonies streaked out onto lysogeny broth, Miller (LB-M; Thermo Fisher Scientific) agar plates. These 5-mL LB-M cultures (with 34 μg/mL chloramphenicol added when appropriate) were grown in 24-well blocks for 15 to 18 h at 37°C and shaking at 225 rpm. Saturated starter cultures were used to subculture expression cultures. If cultures were used for microscopy experiments, cultures were subcultured 1:500 in 5 mL LB-M supplemented with 0.02% (wt/vol) final concentration of l-(+)-arabinose to induce fluorescent reporter expression, 0.4% (vol/vol) final concentration 1,2-PD to induce MCP expression, and 34 μg/mL chloramphenicol. l-(+)-arabinose was left out of cultures where GFP was expressed from the genome. Cultures were then grown for a minimum of 6 h at 37°C and 225 rpm before imaging. For cultures used for MCP purification, saturated overnight cultures were diluted 1:1,000 into 200 mL No Carbon Essential (NCE) medium supplemented with 50 μM ferric citrate, 42 mM succinate as a carbon source, 1 mM magnesium sulfate, and 55 mM 1,2-PD to induce MCP expression. The cultures were grown in 1-liter glass Erlenmeyer flasks at 37°C and 225 rpm until cultures reached an optical density at 600 nm (OD600) of at least 1. MCPs were purified using an established differential centrifugation method described previously (43). Briefly, cells were pelleted at 5,000 × g and lysed chemically using an octylthioglucoside (OTG) solution. Cell lysate was clarified at 12,000 × g (4°C, 5 min), and MCPs were pelleted at 21,000 × g (4°C, 20 min). Purified MCPs were stored in a buffered saline solution containing 50 mM Tris (pH 8.0), 50 mM potassium chloride, 5 mM magnesium chloride, and 1% (vol/vol) 1,2-PD at 4°C. Cultures used for flow cytometry were subcultured 1:1,000 overnight into 5 mL of the same NCE medium used for compartment purifications and grown for 16 h at 37°C at 225 rpm prior to analysis.
TEM.
Transmission electron microscopy (TEM) on purified MCPs was done as described in detail in a prior study (15). Samples containing purified MCPs were applied and fixed onto 400-mesh Formvar-coated copper grids (no. FF400-Cu; EMS). Fixation was done using 2% (vol/vol) glutaraldehyde, and staining was done using 1% (wt/vol) uranyl acetate. Samples were then imaged using a JEOL 1230 transmission electron microscope with a Gatan 831 bottom-mounted charge-coupled device (CCD) camera. MCP diameter measurements were collected using the Ferret diameter tool in ImageJ (56).
SDS-PAGE.
For analysis of purified MCPs using SDS-PAGE, samples were first normalized by protein concentration (measured by bicinchoninic acid assay) before loading onto the 15 wt% polyacrylamide Tris-glycine minigels. SDS-PAGE was then run a second time with sample loading normalized by densitometry on the PduA band to correct for potential contaminants within the samples. Samples were boiled at 95°C for 5 min in Laemmli buffer and then immediately run at 120 V for 90 min. Gels were stained with Coomassie brilliant blue R-250 and imaged using the Bio-Rad ChemiDoc XRS+ system.
Growth assays and metabolite analysis.
Growth assays and metabolite analyses were carried out as previously described (9, 16, 26), with some modifications. Overnight starter cultures of LT2 strains of interest were started from a single colony and grown in test tubes in 5 mL of Terrific broth (Dot Scientific, Inc.) without glycerol. Overnight cultures were grown 15 to 16 h at 37°C at 225 rpm with orbital shaking. These cultures were subcultured into 50 mL NCE medium supplemented with 50 μM ferric citrate, 1 mM magnesium sulfate, 150 nM adenosylcobalamin (Santa Cruz Biotechnology), and 55 mM 1,2-PD to an OD600 of 0.05. 1,2-PD was added to serve as the sole carbon source, and Ado-B12 was added to support PduCDE activity. This enables interrogation of whether Pdu MCPs can successfully mediate propionaldehyde toxicity. Cultures were grown in 250-mL unbaffled Erlenmeyer flasks at 37°C, 225 rpm. At each time point, 500 μL of cell culture was taken for OD600 measurement and high-performance liquid chromatography (HPLC) analysis. OD600 measurements were taken using a BioTek Synergy HTX multimode plate reader. After 9 h, cultures from all strains except the ΔpocR strain were diluted 1:5 in fresh NCE to ensure measurements were within the linear range of the instrument. Error bars on growth curves represent standard deviations over three biological replicates. The remainder of the cell culture sample (not used for OD600 measurement) was centrifuged at 13,000 × g for 5 min to pellet cells. The supernatant was collected and frozen at −20°C. After completion of the growth experiment, supernatant samples were thawed and filtered (Corning Costar spin-X LC filters) in preparation for HPLC analysis. Samples were run on an Agilent 1260 HPLC system. Metabolites were separated using a Rezex ROA-organic acid H+ (8%) LC column (Phenomenex) at 35°C in 5 mM sulfuric acid flowing at 0.4 mL/min. Metabolites were detected with a refractive index detector (RID) as previously described (9). Peak areas were calculated using the Agilent ChemLab software and converted to metabolite concentrations using standards of the metabolites of interest (1,2-propanediol, propionaldehyde, propionate, and 1-propanol) at 200 mM, 100 mM, 50 mM, 20 mM, and 5 mM. Error bars on metabolite concentrations represent standard deviations over three biological replicates.
Modeling.
The kinetic model used here is modified from previous work, which described MCP function in the cell using a reaction-diffusion framework (8). Modifications include an assumption that the cytosol is well mixed to produce a compartmental model and the incorporation of the effects of cell growth. We analyze how metabolite profiles evolve over time, while previous studies focused on steady state. A detailed description of the model used in this work is provided here and in the supplemental material.
We model cells as capsule-shaped (cylinders with a hemisphere at either end) and allow passive transport of 1,2-propanediol, propionaldehyde, propionyl-coenzyme A (CoA), 1-propanol, and propionate at the cell surface, specified by the permeabilities included in Table S4. MCPs are modeled as spheres 140 nm in diameter (15 per cell) that have a specified permeability at the surface; this permeability is assumed to be the same for all metabolites. Polar bodies are modeled as a single sphere per cell 345 nm in diameter (selected such that the volume of the polar body is equal to that of 15 MCPs) and allow free diffusion of all metabolites at their surfaces. Recognizing that diffusion in the MCP, cytosol, and medium are much faster than the transport processes and enzyme reaction rates, we assume that the concentration is uniform in each of these locations.
We assume that conversion of 1,2-propanediol to propionaldehyde by PduCDE is irreversible. Similarly, we assume that conversion of propionyl-CoA to propionate by PduL and PduW occurs in a single step and that this reaction is also irreversible. Reverse reactions are included for conversion of propionaldehyde to 1-propanol and propionyl-CoA by PduQ and PduP, respectively. Reactions catalyzed by PduCDE, PduP, and PduQ are assumed to occur only in the MCP or polar body volume, whereas the reaction catalyzed by PduL/PduW is assumed to occur only in the cytosol. All enzymes are assumed to have Michaelis-Menten kinetics.
Cell concentration and growth are calculated using the data from the growth curves. The model was built and implemented in Python (57). All codes used are available on GitHub at https://github.com/cemills/MCP-vs-PolarBody. A detailed description of the equations used in the model are available in the supplemental material. The parameters used for calculations presented here are available in Table S4. Metabolite profiles over time of 1,2-propanediol, propionaldehyde, propionate, 1-propanol for the PduCDE concentrations shown in Fig. 7A can be found in Fig. S5.
Flow cytometry.
Prior to measurement, cells were diluted to an OD600 of 0.03 into 200 μL phosphate-buffered saline (PBS) supplemented with 2 mg/mL kanamycin to stop translation. Samples were prepared in a U-bottomed 96-well plate and kept away from light until measurement. A total of 10,000 events were collected per sample on an Attune NxT acoustic focusing cytometer paired with an Attune NxT auto sampler. Cell populations were gated using forward and side scatter channels, and average reported fluorescence was calculated using the geometric mean of the population. Values were converted to molecules of equivalent fluorescence (MEF) using BD Sphero rainbow calibration particles (catalog number BDB559123; Fisher Scientific) according to the manufacturer’s instructions. Analysis was performed using FlowJo software (www.FlowJo.com).
Bioinformatic analysis of PduB N terminus.
A list of 45 diverse PduB homologs was curated based on the comprehensive list available in Sutter et al. (13). In this study, MCP shell proteins were divided into subclades and given a color designator based on the color palette available online at https://xkcd.com/color/rgb/. This study also expanded on the work published in Axen et al. (12) by assigning MCP shell protein homologs to diverse MCP subtypes based on operon structure. For the analysis done here, a single representative from each subclade was chosen. The full set of sequences, including their subclade/color designation, MCP subtype, and UniProt accession number, is available in Table S5. This set of PduB homologs was then aligned using Clustal Omega, available at the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) webpage (https://www.ebi.ac.uk/Tools/msa/clustalo/) (58). The multiple-sequence alignment was analyzed and viewed using JalView software (59) and used to make a phylogenetic tree using the neighbor-joining method.
Data availability.
All scripts used to generate modeling data for this paper are available on GitHub at https://github.com/cemills/MCP-vs-PolarBody (60). All strains, plasmids, and underlying data files are available upon request.
ACKNOWLEDGMENTS
We thank and acknowledge members of the Tullman-Ercek and Mangan groups for insightful advice and discussion related to designing experiments associated with this study as well as preparing the manuscript. We specifically acknowledge Taylor M. Nichols, Eddy Kim, Chris Jakobson, and Mike Vincent for strains, plasmids, and primers used in this study.
This work was funded in part by the Army Research Office (grant W911NF-19-1-0298 to D.T.E.) and the Department of Energy (grant DE-SC0019337 to D.T.E. and N.M.M.). N.W.K. was funded by the National Science Foundation Graduate Research Fellowship Program (grant DGE-1842165) and by a National Institutes of Health training grant (T32GM008449) via the Northwestern University Biotechnology Training Program.
CRediT contributor roles: N.W.K., conceptualization, formal analysis, funding acquisition, investigation, methodology, project administration, validation, visualization, writing–original draft, writing–review, and editing; C.E.M., conceptualization, formal analysis, investigation, methodology, project administration, software, validation, visualization, writing–original draft, writing–review and editing; C.H.A., conceptualization, investigation, methodology, writing–review and editing; A.A., conceptualization, formal analysis, software, writing–review and editing; S.S., conceptualization, formal analysis, software, writing–review and editing; M.C.J., funding acquisition, resources, supervision, writing–review and editing; N.M.M., conceptualization, funding acquisition, resources, supervision, writing–review and editing; D.T.E., conceptualization, funding acquisition, resources, supervision, writing–review and editing.
Footnotes
Supplemental material is available online only.
Contributor Information
Danielle Tullman-Ercek, Email: ercek@northwestern.edu.
Mohamed Y. El-Naggar, University of Southern California
REFERENCES
- 1.Gabaldón T, Pittis AA. 2015. Origin and evolution of metabolic sub-cellular compartmentalization in eukaryotes. Biochimie 119:262–268. 10.1016/j.biochi.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uebe R, Schüler D. 2016. Magnetosome biogenesis in magnetotactic bacteria. Nat Rev Microbiol 14:621–637. 10.1038/nrmicro.2016.99. [DOI] [PubMed] [Google Scholar]
- 3.Greening C, Lithgow T. 2020. Formation and function of bacterial organelles. Nat Rev Microbiol 18:677–689. 10.1038/s41579-020-0413-0. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy NW, Mills CE, Nichols TM, Abrahamson CH, Tullman-Ercek D. 2021. Bacterial microcompartments: tiny organelles with big potential. Curr Opin Microbiol 63:36–42. 10.1016/j.mib.2021.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Kerfeld CA, Melnicki MR. 2016. Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol 31:66–75. 10.1016/j.pbi.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, Sutter M. 2018. Bacterial microcompartments. Nat Rev Microbiol 16:277–290. 10.1038/nrmicro.2018.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asija K, Sutter M, Kerfeld CA. 2021. A survey of bacterial microcompartment distribution in the human microbiome. Front Microbiol 12:669024. 10.3389/fmicb.2021.669024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakobson CM, Tullman-Ercek D, Slininger MF, Mangan NM. 2017. A systems-level model reveals that 1,2-propanediol utilization microcompartments enhance pathway flux through intermediate sequestration. PLoS Comput Biol 13:e1005525. 10.1371/journal.pcbi.1005525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampson EM, Bobik TA. 2008. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol 190:2966–2971. 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prentice MB. 2021. Bacterial microcompartments and their role in pathogenicity. Curr Opin Microbiol 63:19–28. 10.1016/j.mib.2021.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Stewart KL, Stewart AM, Bobik TA. 2020. Prokaryotic organelles: bacterial microcompartments in E. coli and Salmonella. EcoSal Plus 9:ecosalplus.ESP-0025-2019. 10.1128/ecosalplus.ESP-0025-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axen SD, Erbilgin O, Kerfeld CA. 2014. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol 10:e1003898. 10.1371/journal.pcbi.1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutter M, Melnicki MR, Schulz F, Woyke T, Kerfeld CA. 2021. A catalog of the diversity and ubiquity of bacterial microcompartments. Nat Commun 12:3809. 10.1038/s41467-021-24126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFarland AG, Kennedy NW, Mills CE, Tullman-Ercek D, Huttenhower C, Hartmann EM. 2022. Density-based binning of gene clusters to infer function or evolutionary history using GeneGrouper. Bioinformatics 38:612–620. 10.1093/bioinformatics/btab752. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy NW, Hershewe JM, Nichols TM, Roth EW, Wilke CD, Mills CE, Jewett MC, Tullman-Ercek D. 2020. Apparent size and morphology of bacterial microcompartments varies with technique. PLoS One 15:e0226395. 10.1371/journal.pone.0226395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slininger Lee MF, Jakobson CM, Tullman-Ercek D. 2017. Evidence for improved encapsulated pathway behavior in a bacterial microcompartment through shell protein engineering. ACS Synth Biol 6:1880–1891. 10.1021/acssynbio.7b00042. [DOI] [PubMed] [Google Scholar]
- 17.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936–938. 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 18.Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO. 2013. Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci 22:660–665. 10.1002/pro.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1, 2-propanediol degradation. J Bacteriol 181:5967–5975. 10.1128/JB.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehman BP, Chowdhury C, Bobik TA. 2017. The N terminus of the PduB protein binds the protein shell of the Pdu microcompartment to its enzymatic core. J Bacteriol 199:e00785-16. 10.1128/JB.00785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue W-F, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, Warren MJ. 2014. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol 3:454–465. 10.1021/sb4001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons JB, Lawrence AD, McLean KJ, Munro AW, Rigby SEJ, Warren MJ. 2010. Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment. PLoS One 5:e14009. 10.1371/journal.pone.0014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan C, Cheng S, Sinha S, Bobik TA. 2012. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci USA 109:14995–15000. 10.1073/pnas.1207516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorda J, Liu Y, Bobik TA, Yeates TO. 2015. Exploring bacterial organelle interactomes: a model of the protein-protein interaction network in the Pdu microcompartment. PLoS Comput Biol 11:e1004067. 10.1371/journal.pcbi.1004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutter M, Greber B, Aussignargues C, Kerfeld CA. 2017. Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356:1293–1297. 10.1126/science.aan3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy NW, Ikonomova SP, Slininger Lee M, Raeder HW, Tullman-Ercek D. 2021. Self-assembling shell proteins PduA and PduJ have essential and redundant roles in bacterial microcompartment assembly. J Mol Biol 433:166721. 10.1016/j.jmb.2020.11.020. [DOI] [PubMed] [Google Scholar]
- 27.Pang A, Frank S, Brown I, Warren MJ, Pickersgill RW. 2014. Structural insights into higher order assembly and function of the bacterial microcompartment protein PduA. J Biol Chem 289:22377–22384. 10.1074/jbc.M114.569285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan C, Bobik TA. 2011. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol 193:5623–5628. 10.1128/JB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. 2010. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci USA 107:7509–7514. 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Jorda J, Yeates TO, Bobik TA. 2015. The PduL phosphotransacylase is used to recycle coenzyme A within the Pdu microcompartment. J Bacteriol 197:2392–2399. 10.1128/JB.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng S, Fan C, Sinha S, Bobik TA. 2012. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One 7:e47144. 10.1371/journal.pone.0047144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerfeld CA, Erbilgin O. 2015. Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol 23:22–34. 10.1016/j.tim.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Sampson EM, Johnson CLV, Bobik TAY. 2005. Biochemical evidence that the pduS gene encodes a bifunctional cobalamin reductase. Microbiology (Reading) 151:1169–1177. 10.1099/mic.0.27755-0. [DOI] [PubMed] [Google Scholar]
- 34.Yang M, Simpson DM, Wenner N, Brownridge P, Harman VM, Hinton JCD, Beynon RJ, Liu L-N. 2020. Decoding the stoichiometric composition and organisation of bacterial metabolosomes. Nat Commun 11:1976. 10.1038/s41467-020-15888-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda S, Toraya T, Fukui S. 1980. In situ reactivation of glycerol-inactivated coenzyme B12-dependent enzymes, glycerol dehydratase and diol dehydratase. J Bacteriol 143:1458–1465. 10.1128/jb.143.3.1458-1465.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toraya T, Mori K. 1999. A reactivating factor for coenzyme B12-dependent diol dehydratase. J Biol Chem 274:3372–3377. 10.1074/jbc.274.6.3372. [DOI] [PubMed] [Google Scholar]
- 37.Walter D, Ailion M, Roth J. 1997. Genetic characterization of the pdu operon: use of 1, 2-propanediol in Salmonella typhimurium. J Bacteriol 179:1013–1022. 10.1128/jb.179.4.1013-1022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen P, Andersson DI, Roth JR. 1994. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol 176:5474–5482. 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bobik TA, Ailion M, Roth JR. 1992. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol 174:2253–2266. 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rondon MR, Escalante-Semerena JC. 1992. The poc locus is required for 1,2-propanediol-dependent transcription of the cobalamin biosynthetic (cob) and propanediol utilization (pdu) genes of Salmonella typhimurium. J Bacteriol 174:2267–2272. 10.1128/jb.174.7.2267-2272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim EY, Jakobson CM, Tullman-Ercek D. 2014. Engineering transcriptional regulation to control Pdu microcompartment formation. PLoS One 9:e113814. 10.1371/journal.pone.0113814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nichols TM, Kennedy NW, Tullman-Ercek D. 2020. A genomic integration platform for heterologous cargo encapsulation in 1,2-propanediol utilization bacterial microcompartments. Biochem Eng J 156:107496. 10.1016/j.bej.2020.107496. [DOI] [Google Scholar]
- 43.Nichols TM, Kennedy NW, Tullman-Ercek D. 2019. Cargo encapsulation in bacterial microcompartments: methods and analysis. Methods Enzymol 617:155–186. 10.1016/bs.mie.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha S, Cheng S, Fan C, Bobik TA. 2012. The PduM protein is a structural component of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella enterica. J Bacteriol 194:1912–1918. 10.1128/JB.06529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ. 2010. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell 38:305–315. 10.1016/j.molcel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Kennedy NW, Li S, Mills CE, Tullman-Ercek D, Olvera de la Cruz M. 2021. Computational and experimental approaches to controlling bacterial microcompartment assembly. ACS Cent Sci 7:658–670. 10.1021/acscentsci.0c01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greber BJ, Sutter M, Kerfeld CA. 2019. The plasticity of molecular interactions governs bacterial microcompartment shell assembly. Structure 27:749–763. 10.1016/j.str.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowdhury C, Chun S, Pang A, Sawaya MR, Sinha S, Yeates TO, Bobik TA. 2015. Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc Natl Acad Sci USA 112:2990–2995. 10.1073/pnas.1423672112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chowdhury C, Chun S, Sawaya MR, Yeates TO, Bobik TA. 2016. The function of the PduJ microcompartment shell protein is determined by the genomic position of its encoding gene. Mol Microbiol 101:770–783. 10.1111/mmi.13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tobimatsu T, Kawata M, Toraya T. 2005. The N-terminal regions of β and γ subunits lower the solubility of adenosylcobalamin-dependent diol dehydratase. Biosci Biotechnol Biochem 69:455–462. 10.1271/bbb.69.455. [DOI] [PubMed] [Google Scholar]
- 51.Lee MJ, Brown IR, Juodeikis R, Frank S, Warren MJ. 2016. Employing bacterial microcompartment technology to engineer a shell-free enzyme-aggregate for enhanced 1,2-propanediol production in Escherichia coli. Metab Eng 36:48–56. 10.1016/j.ymben.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Juodeikis R, Lee MJ, Mayer M, Mantell J, Brown IR, Verkade P, Woolfson DN, Prentice MB, Frank S, Warren MJ. 2020. Effect of metabolosome encapsulation peptides on enzyme activity, coaggregation, incorporation, and bacterial microcompartment formation. MicrobiologyOpen 9:e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohajerani F, Sayer E, Neil C, Inlow K, Hagan MF. 2021. Mechanisms of scaffold-mediated microcompartment assembly and size control. ACS Nano 15:4197–4212. 10.1021/acsnano.0c05715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engler C, Gruetzner R, Kandzia R, Marillonnet S. 2009. Golden Gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4:e5553. 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datta S, Costantino N, Court DL. 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379:109–115. 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 56.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0 Contributors. 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17:261–272. 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madeira F, Mi Park Y, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kennedy NW, Mills CE, Abrahamson CH, Archer A, Jewett MC, Mangan NM, Tullman-Ercek D. 2021. Cemills/MCP-vs-polarbody. Linking the Salmonella enterica 1,2-propanediol utilization bacterial microcompartment shell to the enzymatic core via the shell protein PduB. https://github.com/cemills/MCP-vs-PolarBody. Accessed 9 November 2021. [DOI] [PMC free article] [PubMed]
- 61.Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO. 2010. Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J Biol Chem 285:37838–37846. 10.1074/jbc.M110.160580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pang A, Liang M, Prentice MB, Pickersgill RW. 2012. Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallogr D Biol Crystallogr 68:1642–1652. 10.1107/S0907444912039315. [DOI] [PubMed] [Google Scholar]
- 63.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplemental material. Download jb.00576-21-s001.pdf, PDF file, 0.9 MB (1,015.4KB, pdf)
Data Availability Statement
All scripts used to generate modeling data for this paper are available on GitHub at https://github.com/cemills/MCP-vs-PolarBody (60). All strains, plasmids, and underlying data files are available upon request.