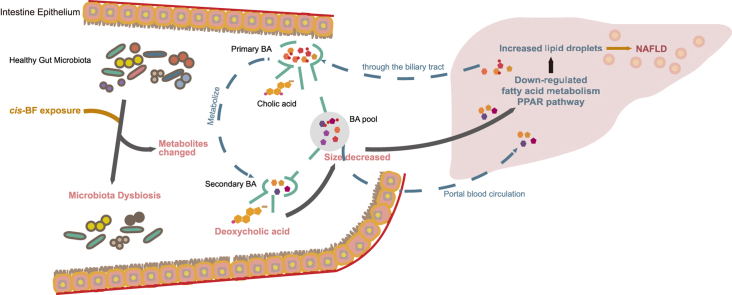
Abstract
A growing body of evidence has demonstrated the significance of the gut microbiota in host health, while the association between gut microbiota dysbiosis and multiple diseases is yet elusive in the scenario of exposure to widely used pesticides. Here, we show that gut microbiota dysbiosis involves in host's abnormal lipid metabolism and consequently the non-alcoholic fatty liver disease in Xenopus laevis upon exposure to cis-bifenthrin, one of the most prevalent pyrethroid insecticides in the world. With the guidance of gut microbiota analysis, we found that cis-bifenthrin exposure significantly perturbed the gut microbial community, and the specific taxa that served as biomarkers were identified. Metabolomics profiling and association analysis further showed that a significant change of intestinal metabolites involved in lipid metabolic pathways were induced along with the microbiota dysbiosis upon exposure to cis-bifenthrin. Detailed investigation showed an altered functional regulation of lipids in the liver after cis-bifenthrin exposure and the accumulation of lipid droplets in hepatocytes. Specifically, a change in deoxycholic acid alters bile acid hepatoenteral circulation, which affects lipid metabolism in the liver and ultimately causes the development of fatty liver disease. Collectively, these findings provide novel insight into the gut microbiota dysbiosis upon pesticide exposure and their potential implication in the development of chronic host diseases related to liver metabolic syndrome.
Keywords: Pyrethroid, Xenopus laevis, Metabolic disorders, Dysbiosis gut microbiota
Graphical abstract
Highlights
-
•
Cis-BF disturbed gut microbiota community and metabolite profile in frogs.
-
•
Cis-BF affected liver lipid metabolism and ultimately developed NAFLD in frogs.
-
•
Gut microbiota dysbiosis involves in host's NAFLD in frogs.
1. Introduction
A growing body of research has demonstrated the essential role of the gut microbiome in the normal development and overall health of the host organism [1,2]. The gut microbiota control physiology beyond the intestinal lumen as they can signal to cells throughout the host to regulate their activity through the production of metabolically active microbial metabolites [3,4]. Intestinal dysbiosis can contribute to hepatic metabolic diseases via various obesity-associated mechanisms, including abnormalities in lipid metabolism [5]. Current reports on the chemical disruption of host-associated microbiota have generally been concerned with persistent organic pollutants (POPs) or antibiotics [6,7], and fewer studies have well-characterized the microbiome-associated challenges to healthy hosts from pesticide exposure.
Pesticides are indispensable for modern agricultural production. In recent decades, synthetic pyrethroid (SP) use has increased continuously due to its insecticidal potency and shorter breakdown time than organophosphate pesticides [8]. Today, SPs have extensive indoor and outdoor applications, including in agriculture, public health, and residential usage [9]. SP contamination is extensively observed in soil, water, streambed sediments, and indoor dust. For instance, a maximum value of 901 ng L−1 of SP has been detected in the surface water samples of California coastal watersheds, with bifenthrin being the most commonly detected SP [10]. The maximum concentration of SP pesticides found in the surface waters of the Pearl River Basin (China) was 29.72 μg L−1 [11]. Eight SPs were also detected in the Liaohe River basin sediments, with a detected concentration of 2.2–102.5 μg kg−1 dry weight [12]. Additionally, SPs were detected in wild fish from Iberian River basins (Spain), with the highest concentration detected being 4938 ng g−1 lipid weight [13]. Moreover, it is worth noting that a considerable amount of research has shown that SPs ultimately pose a health threat to wildlife and humans. Mounting laboratory studies have reported the toxic effects of SPs, including developmental toxicity, neurotoxicity, and reproductive toxicity, in different nontarget organisms [[14], [15], [16], [17]]. In addition, in recent years, considering the indispensable and crucial role of the gut microbiota, specific attention has also been given to the influence of SPs on the abundance, composition, and metabolic function of the gut microbiota. A previous study indicated that low-concentration exposure to permethrin significantly changed the abundance of the gut microbiota in rats but also altered the metabolic profiles, including the acetic and propionic acid contents [18]. Besides, treatment with a mixture of pyrethroids increased the contents of triglycerides and high-density lipoprotein in the serum of rats [19]. However, the adverse influences of SPs on gut microbiota community structures and metabolite-mediated host metabolism are largely unknown. Mechanistic insight into the interplay of gut microbiota, host metabolism, and metabolic health has gained significant attention over the past decade [20]. The gut microbiota has been considered a “microbial organ” within the intestine that modulates the health phenotype of the host via a range of microbial metabolites. For instance, bile acids are classical examples of metabolites that are metabolized in the intestine by the gut microbiota into secondary bile acids and therefore regulate host metabolic pathways [20].
Since the gut microbiota has been shown to play indispensable roles in the maintenance of normal hepatic functions and the pathogenesis of the disease, greater insights into the mechanisms driving the interplay between SPs and the gut microbiota are needed to reveal SP-induced adverse effects and related susceptibilities. Therefore, the present study was conducted to address whether SP exposure would alter the gut microbiota community structure as well as the gut microbiota-related metabolic functions and whether SP-induced intestinal dysbiosis would further impact the metabolic process of the host liver. We applied 16S rRNA gene sequencing, liquid chromatography-mass spectrometry (LC-MS) metabolomics profiling, and transcriptomics to probe functional interactions between liver function and the gut microbiota after exposure to pyrethroids. Cis-bifenthrin (cis-BF) is among the most commonly used SPs [10,11]. Amphibians are suitable for studying the effects of stressors from environmental toxicants since they can provide more comprehensive toxicological endpoints [21]. The African clawed frog (Xenopus laevis, X. laevis) is a widely used amphibian model organism and is regarded as an invaluable tool for ecotoxicological assessment [22,23]. Therefore, cis-bifenthrin was chosen as the representative SP, and African clawed frogs were used as the bio model. Our study provides the first evidence of the functional interactions between liver function and the gut microbiota after pyrethroid exposure.
2. Methods
2.1. Animals and exposure
Adult African clawed frogs were obtained from Nasco (Fort Atkinson, WI, USA). To alleviate suffering, animals were maintained humanely following the guidelines of the American Society for Testing and Materials [24]. Adult frogs (2 years old) were exposed to the environmentally relevant concentrations of 0.06 and 0.3 μg L−1 cis-BF (Purity > 98%; Sigma, St. Louis, MO, USA). Stock solutions were prepared with dimethyl sulfoxide (Sigma, St. Louis, MO, USA), and the dimethyl sulfoxide (0.001%) was identical for all treatment groups. Each treatment consisted of 27 male frogs that were randomly assigned to three replicates. Experiments were carried out at the same photoperiod (12 h light: 12 h dark) and temperature (22 °C). During the three months of exposure, the exposure solutions were replaced daily to maintain the concentration of cis-BF. After exposure, the frogs were anesthetized in MS-222 and frozen in liquid nitrogen for subsequent analysis. All studies were approved by the Experimental Animal Ethics Committee of Zhejiang University.
2.2. Deoxycholic acid supplementation experiment
To verify, we also performed a dietary deoxycholic acid (DCA) supplementation study. Adult frogs were randomly divided into four groups: control group (normal diet); deoxycholic acid exposure group (diet supplemented with 0.2% deoxycholic acid; Purity ≥ 98.0%; ANPEL laboratory technologies Inc., Shanghai, China); 0.3 μg L−1 cis-BF exposure group (normal diet) and 0.3 μg L−1 cis-BF and deoxycholic acid co-exposure group (diet supplemented with 0.2% deoxycholic acid). After exposure, the blood was collected for the measurement of triglyceride and cholesterol. Liver tissues and gut intestinal contents were collected for qPCR and bile acid analysis, respectively.
2.3. Extraction and quantification of cis-BF
The cis-BF in the exposure solutions and intestines was extracted and detected according to a previous study with slight modifications [25]. For exposure solutions, 200 mL of exposure solution was mixed with sodium chloride (5.0 g) and dichloromethane (200 mL) in a 500 mL separatory funnel. The mixture was shaken for 5 min and left at room temperature for 5 min to separate the organic and aqueous phases. Then, the organic phase was collected, and the extraction procedure was repeated once again. The combined organic phase was evaporated to near dryness on a rotatory evaporator at 40 °C. Frog intestine contents (1 g) were homogenized and transferred into a 50 mL polypropylene tube preloaded with 0.25 g NaCl and 1 g MgSO4. Then, 5 mL acetonitrile was added to extract cis-BF, followed by sonication for 20 min. The mixture was centrifuged at 4500 rpm for 10 min, and the organic phase was transferred into a glass flask. The extraction procedure was repeated once again, and the organic phase was combined and subsequently evaporated almost to dryness on a rotatory evaporator at 40 °C. The residues from the exposure solution and tissues were reconstituted with 1 mL acetonitrile and transferred into a 1.5 mL polypropylene tube that contained PSA. After vortexing and centrifugation (12000 g, 10 min), the supernatant was filtered with a 0.22-μm polytetrafluoroethylene filter and transferred into vials for quantitative analysis. The limit of detection and quantitation for cis-BF were 0.0031 and 0.0094 ng g−1, respectively.
2.4. 16S rRNA gene sequencing
Total genomic DNA was extracted from the intestinal contents of frogs using the CTAB method. Extracted DNA was amplified to target the V3–V4 regions of the 16S rRNA gene of bacteria using the primer pair 341F (CCTAYGGGRBGCASCAG) and 806R (GGACTACNNGGGTATCTAAT). The amplicons were assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and sequenced on an Ion S5TM XL platform. To obtain high-quality clean reads, the raw reads were filtered on Cutadapt (Martin M., 2011). In addition, chimera sequences were removed with the UCHIME algorithm (UCHIME Algorithm). Uparse software (Uparse v7.0.1001) was used to choose operational taxonomic units (OTUs) with a threshold of 97% sequence similarity. A representative sequence from each OTU was selected for taxonomic annotation using the Silva Database (Version 132). The taxonomic information of the 16S rRNA gene sequences was obtained at different classification levels, including the phylum, class, order, family, and genus levels. Alpha diversity (observed species, Chao1, Shannon, Simpson, ACE, and good coverage indices) and beta diversity were calculated with QIIME (Version 1.7.0). Linear discriminant analysis (LDA) effect size (LEfSe) was performed using the LEfSe tool.
2.5. Metabolomics analysis
The intestinal contents (100 mg) were ground with liquid nitrogen, and the homogenate was resuspended in prechilled 80% methanol (−20 °C). After vortexing, the samples were incubated for 1 h at −20 °C and then centrifuged at 14000 g for 20 min at 4 °C. The supernatants were transferred to a new tube, dried in a vacuum concentrator, and then reconstituted in 60% methanol for LC-MS/MS analysis. LC-MS/MS analysis was performed on a Vanquish UHPLC system (Thermo Fisher, MA, USA) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher, MA, USA). The raw data were processed with Compound Discoverer 3.0 (CD 3.0, Thermo Fisher) for peak alignment, peak picking, and quantitation for each metabolite. Then, the molecular features were matched with the mzCloud (https://www.mzcloud.org/) and ChemSpider (http://www.chemspider.com/) databases.
2.6. Bile acid targeted metabolomics analysis
The intestinal content (100 mg) was ground into a powder with liquid nitrogen. After adding 900 μL water, the sample was vortexed for 30 s and diluted with water 10 times. Then, 100 μL of the sample was taken and transferred to EP tubes containing 300 μL of acetonitrile/methanol (8:2; v/v). The sample was vortexed and centrifuged at 12000 rpm for 10 min. Then, the supernatant passed through a 0.22-μm filter for subsequent analysis with HPLC-MS/MS.
2.7. Transcriptomics analysis
The mRNA-Seq experiment was completed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Total RNA was isolated from frog liver using TRIzol reagent (Takara Bio Inc., Kusatsu, Japan). The RNA purity and integrity were assessed by using the A260/A280 ratio and agarose-formaldehyde gel electrophoresis, and the RNA concentration was also measured. A total amount of 3 μg of RNA was used for RNA sample preparation, and the sequencing libraries were generated using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA). The quality of the sequencing library was assessed on the Agilent Bioanalyzer 2100 system. Then, the library was sequenced on an Illumina HiSeq platform to generate 125 bp/150 bp paired-end reads. Reads containing adapters and poly-N and low-quality reads were removed to obtain clean reads. FeatureCounts v1.5.0-p3 was employed to calculate the clean read numbers mapped to each gene, and the fragments per kilobase of exon model per million mapped fragments (FPKM) value of each gene was also calculated based on the length of the gene and the read counts mapped to this gene. The DESeq2 R package (1.16.1) was used to assess the differential expression analysis results, and genes with an adjusted p-value < 0.05 were considered differentially expressed. The statistical enrichment of differentially expressed genes (DEGs) in KEGG pathways was performed using the cluster profile R package.
2.8. Quantitative real-time PCR (qPCR) validation
Frog liver tissues (n = 3 replicates; each replicate contained three tissues) were homogenized with RNAiso plus (Takara Bio Inc, Kusatsu, Japan) to extract the total RNA. The total RNA contents were measured on an Epoch Microplate Spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA). The quality of the extracted total RNA was also examined via the 260/280 nm ratios and 1% agarose gel electrophoresis. The Prime Script TM RT reagent Kit (Takara Bio Inc, Kusatsu, Japan) was used to synthesize the cDNA. The real-time PCR was carried out on an ABI 7300 system (Applied Biosystems, CA, USA) using the SYBR Green kits (Takara Bio Inc, Kusatsu, Japan). Specific primers of the target genes were identified by the NCBI Primer-BLAST and showed in the SI (Table S1). The transcriptional stability of the reference gene was accessed using the GeNorm, and finally, the 18s was chosen as the housekeeping gene. The gene transcription levels were calculated using the 2−ΔΔCt method.
2.9. Statistical analysis
Data were presented as the mean ± standard error (SEM). SPSS 20.0 (SPSS, Chicago, IL, USA) was used to perform statistical analyses, and the comparison between the exposure groups and control group was performed using one-way analysis of variance (ANOVA) followed by Tukey's test. A p < 0.05 was considered to be significant.
3. Results
3.1. Exposure to cis-BF induced its accumulation in intestinal
The cis-BF concentrations were measured at 0 h and 24 h after renewing the exposure solutions, and the measured cis-BF concentrations were similar to the nominal exposure concentrations (Table S2). In the 0.06 and 0.3 μg L−1 exposure groups, the cis-BF concentrations in the intestinal contents were 170.35 ± 10.95 and 266.60 ± 30.15 μg kg−1 dry weight (DW), respectively. Cis-BF was not detected in the intestinal contents of the control group.
3.2. Lipid accumulation in the liver after cis-BF exposure
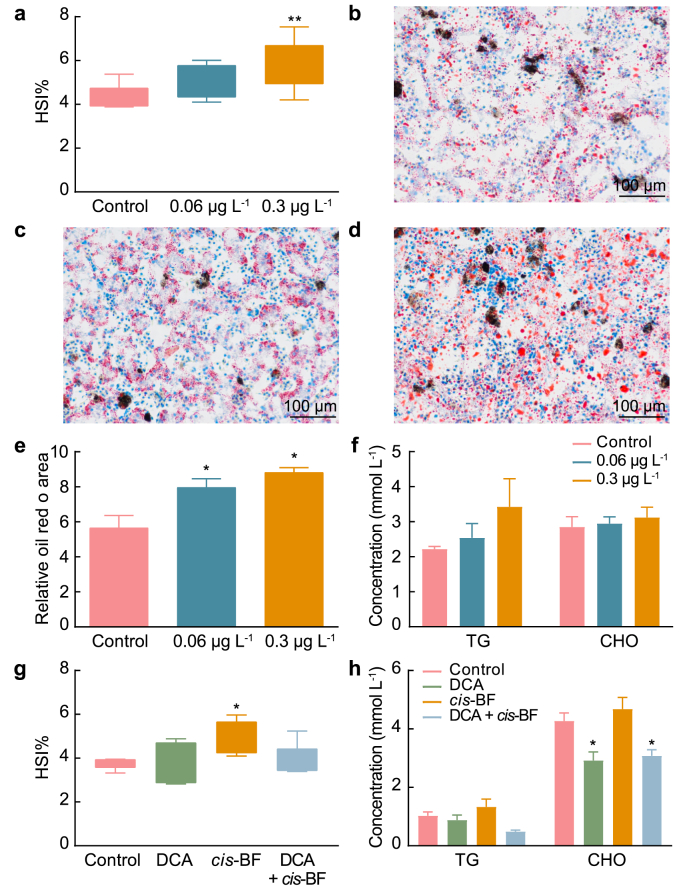
Exposure to 0.3 μg L−1 cis-BF increased the hepatosomatic index (HSI) of frogs (Fig. 1a). Compared with the control group (Fig. 1b), the hepatic lipid droplets in the liver of frogs in the 0.06 (Figs. 1c) and 0.3 μg L−1 (Fig. 1d) exposure groups were obviously increased (Fig. 1e). No significant alteration was observed for the contents of TG and CHO in the plasma of the frogs (Fig. 1f).
Fig. 1.
Hepatosomatic indices of frogs in the control and exposure groups (a, n = 9). Oil red O staining of livers of frogs treated with DMSO (b), 0.06 (c), and 0.3 μg L−1cis-BF (d). Magnification, × 200. e, Relative Oil Red O area in the livers (n = 3). f, The concentrations of TG and CHO in the plasma of frogs. g, Hepatosomatic indices of frogs in DCA supplemental study. h, The concentrations of TG and CHO in DCA supplemental study. Data are presented as mean ± SEM, ∗p < 0.05 and ∗∗p < 0.01 indicate significant differences compared with the control group.
In the validation study, no obvious changes were found for the HSI in DCA and DCA + cis-BF exposure groups compared with the control group (Fig. 1g). Compared with the cis-BF exposure group, lipid droplets in the liver of frogs decreased in DCA + cis-BF exposure groups (Fig. S1). Besides, the levels of CHO also decreased significantly in DCA and DCA + cis-BF exposure groups (Fig. 1h).
3.3. Gut microbial community was changed by cis-BF exposure
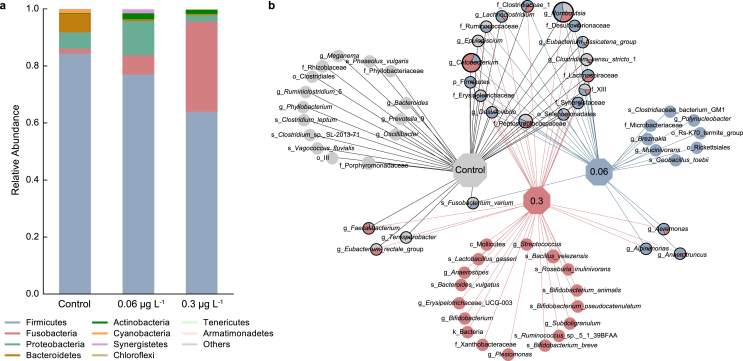
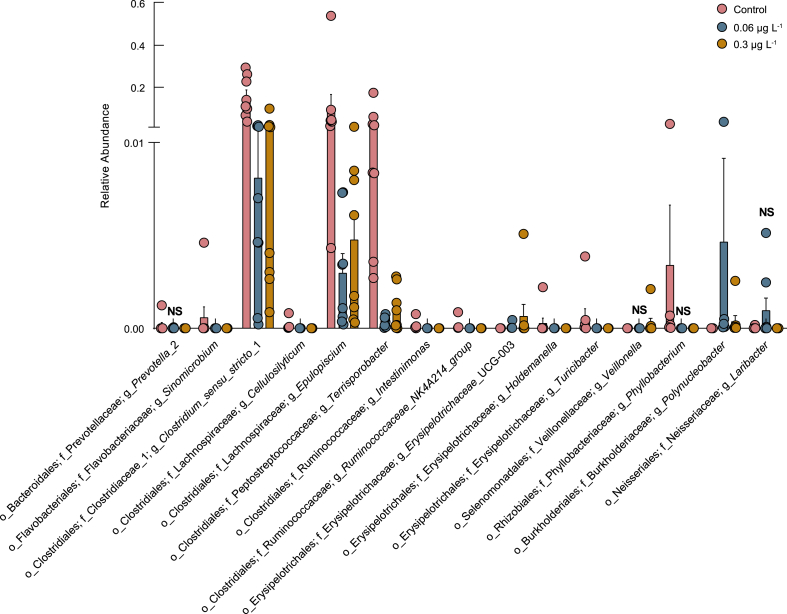
Cis-BF exposure significantly altered the composition, abundance, and diversity of the gut microbiota community. In terms of the gut microbial structure, obvious differences were observed between the control group and all cis-BF exposure groups. The 0.06 and 0.3 μg L−1 cis-BF exposure groups were more similar (ANOSIM; R = 0.07, p = 0.17), with all cis-BF exposure groups showing obvious differences from the control group (0.06: R = 0.18, p = 0.018; 0.3: R = 0.25, p = 0.013; Table S3). Compared with the control group, 0.3 μg L−1 cis-BF exposure significantly decreased the diversity of the microbiome (Simpson: p = 0.0379 and Shannon: p = 0.0312; Fig. S2 and Table S4). Fig. 2a illustrates the relative abundance of the top 10 microbiota at the phylum level in frogs. At the phylum level, Firmicutes and Fusobacteria were the most abundant phyla, followed by Proteobacteria, Bacteroidetes, Actinobacteria, Cyanobacteria, Synergistetes, Chloroflexi, Tenericutes, and Armatimonadetes. Obvious increases in the abundances of Chloroflexi and Armatimonadetes were observed in all cis-BF exposure groups. However, 0.3 μg L−1 cis-BF exposure significantly decreased the abundance of Bacteroidetes. A network based on the top 50 OTUs was also constructed to visualize the different bacteria in the intestinal contents of frogs in the control and cis-BF exposure groups. The result showed that 11 and 20 OTUs exclusively emerged in 0.06 and 0.3 μg L−1 exposure groups, respectively (Fig. 2b). At the genus level, several significantly changed genera are shown in Fig. 3. Exposure to cis-BF (0.06 and 0.3 μg L−1) also increased the abundances of the pathogenic bacteria Plesiomonas (Fig. S3a) and Aeromonas (Fig. S3b). LEfSe was performed to identify the specific bacteria that served as biomarkers in the control group and cis-BF treatment groups. The LEfSe results illustrated those 14 biomarkers with an LDA score > 4 were differentially abundant between the control group and the cis-BF treatment groups (Fig. S3c).
Fig. 2.
Bacterial community compositions at the phylum level (top 10) of gut microbiota in frogs (a, n = 8), network-based analysis of top 50 OTUs of gut microbiota in frogs (b).
Fig. 3.
Significantly changed genera of gut microbiota in frogs after exposure to cis-BF (n = 8). NS means there is no significant difference compared with the control group.
3.4. The metabolic profile of the intestinal contents was altered by cis-BF exposure
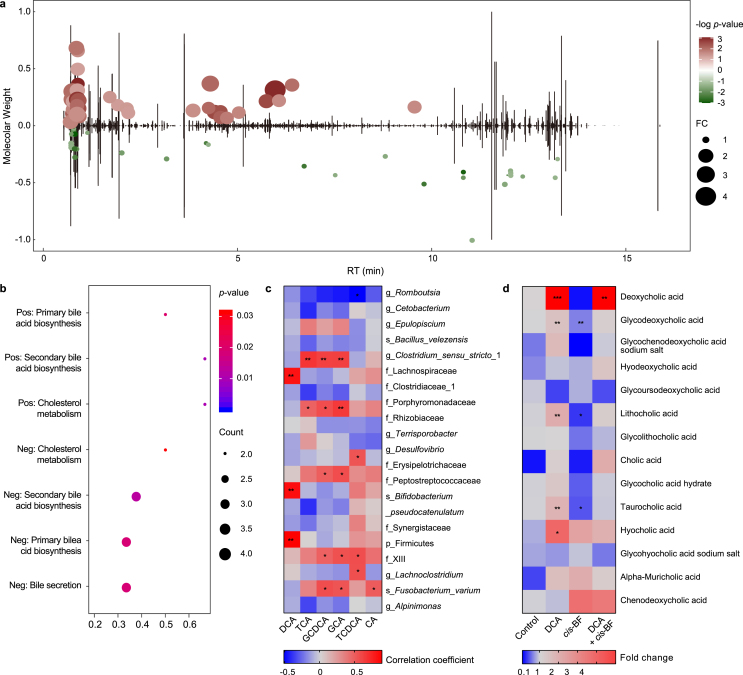
Three months of cis-BF exposure significantly disturbed the metabolic profiles of the intestinal contents. Specifically, 60 (pos: 36 and neg: 24) and 73 (pos: 36 and neg: 37) metabolites with a fold change > 1.5 and p < 0.05 were observed in frogs treated with 0.06 (Fig. S4a; Table S5) and 0.3 μg L−1 (Fig. 4a; Table S5) cis-BF, respectively.
Fig. 4.
Changes in the levels of metabolites (a, fold change > 1.5 and p < 0.05) in the intestinal contents of frogs in 0.3 μg L−1cis-BF exposure group (n = 6). b, KEGG pathway analysis in 0.3 μg L−1cis-BF exposure group. c, Correlations between bile acids concentrations and the relative abundance of top 20 genera. d, The bile acids concentrations in DCA supplemental study (n = 6). Data are presented as mean ± SEM, ∗p < 0.05 and ∗∗p < 0.01 indicate significant differences compared with the control group.
In the 0.3 μg L−1 cis-BF treatment group, the contents of taurocholic acid (TCA), glycochenodeoxycholic acid (GCDCA), and glycocholic acid (GCA) were lower than those in the control group. However, DCA, taurochenodeoxycholic acid (TCDCA), and cholic acid (CA) levels were unaffected by cis-BF (Fig. S4b). In the 0.06 μg L−1 cis-BF treatment group, there were no significant changes in terms of bile acids (Fig. S4b).
All metabolites that changed significantly upon cis-BF exposure were subjected to KEGG pathway enrichment analysis, and seven pathways were significantly enriched in the 0.3 μg L−1 cis-BF exposure group (Fig. 4b; Table S6). The significantly enriched metabolic pathways included primary bile acid biosynthesis, secondary bile acid biosynthesis, bile secretion, and cholesterol metabolism. In the 0.06 μg L−1 cis-BF treatment group, the pyrimidine metabolism pathway was significantly enriched (Fig. S4c; Table S6). Correlation analysis based on the top 20 OTUs and bile acids indicated that g_Clostridium_sensu_stricto_1, f_Lachnospiraceae, f_Porphyromonadaceae, g_Desulfovibrio, f_Erysipelotrichaceae, f_Peptostreptococcaceae, p_Firmicutes, f_XIII, g_Lachnoclostridium, and s_Fusobacterium_varium were positively correlated with bile acids. However, g_Romboutsia showed a negative correlation with taurochenodeoxycholic acid (Fig. 4c).
In the validation study, we measured the concentrations of several bile acids. As expected, the concentrations of DCA increased significantly in DCA and DCA + cis-BF exposure groups. Treatment with DCA also increased the levels of glycodeoxycholic acid, lithocholic acid, taurocholic acid, and hyocholic acid. However, exposure to cis-BF decreased the concentrations of glycodeoxycholic acid, lithocholic acid, and taurocholic acid (Fig. 4d).
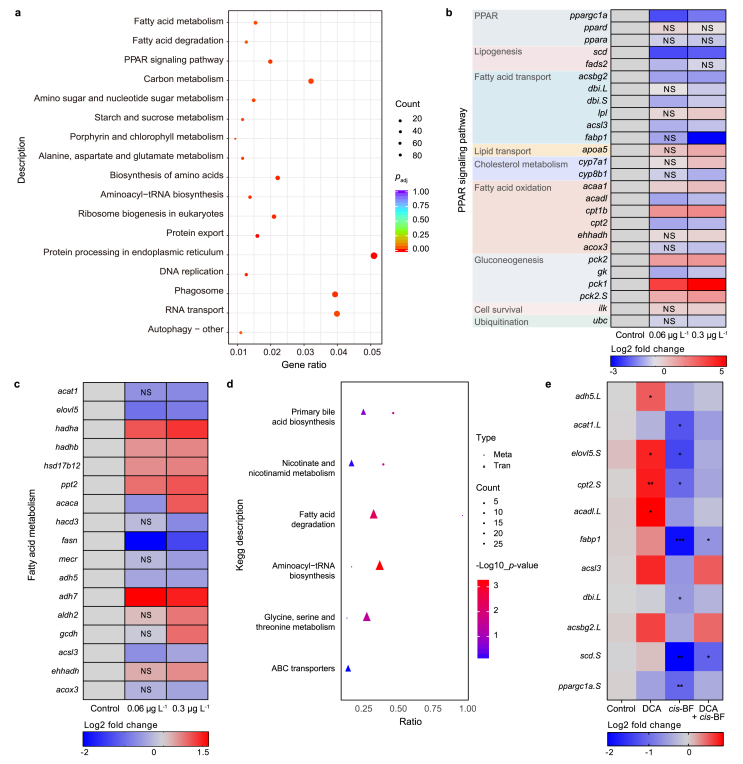
3.5. Hepatic transcriptome was disrupted after cis-BF exposure
We identified 5055 (2620 downregulated and 2435 upregulated genes; Figs. S5a) and 5188 (2570 downregulated and 2618 upregulated genes; Fig. S5b) DEGs in the 0.06 and 0.3 μg L−1 cis-BF treatment groups, respectively. All DEGs were assigned for KEGG pathway analysis, with the altered pathways shown in Table S7. The KEGG results showed that DEGs were mainly enriched in 34 and 17 pathways in the 0.06 (Table S7) and 0.3 μg L−1 (Fig. 5a) cis-BF treatment groups, respectively. Among these enriched pathways, the pathways associated with lipid metabolism were shown in Fig. S6a and Fig. S6b, including the fatty acid degradation pathway, fatty acid metabolism pathway, and peroxisome proliferator-activated receptor (PPAR) signaling pathway. For clarity, the DEGs involved in the PPAR signaling pathway and fatty acid metabolism pathway are shown in Heatmaps (Fig. 5b and c), respectively. In addition, DEGs and differential metabolites were annotated to KEGG pathways for conjoint analysis of transcriptome and metabolome data, and the pathways of primary bile acid biosynthesis and fatty acid degradation were significantly enriched (Fig. 5d and Fig. S6c). The gene expression levels obtained from the transcriptomics were validated by using qPCR, and the fold-changes of selected genes were in agreement with transcriptomic results (Table S8). In the DCA supplemental study, several genes involved in fatty acid metabolism were selected based on the DEGs of lipid and fatty acid metabolic-related pathways in cis-BF exposure study. Thereinto, ppargc1a, scd.S, acsbg2.L, dbi.L, fabp1, acadl.L, and cpt2.S belong to the PPAR pathway involved in lipogenesis, fatty acid transport, and fatty acid oxidation; acat1.L, adh5.L, acsl3, and elovl5.S belong to fatty acid metabolism pathway. Treatment with DCA increased the expression levels of acadl.L, cpt2.S, elovl5.S, and adh5.L. In the cis-BF exposure group, expressions of ppargc1a.S, scd.S, dbi.L, fabp1, cpt2.S, elovl5.S, and acat1.L were significantly down-regulated. However, in the DCA + cis-BF coexposure group, only expressions of scd.S and fabp1 were significantly down-regulated (Fig. 5e).
Fig. 5.
a, Significantly enriched pathways. b, Heatmap of differently expressed genes of the PPAR signaling pathway. c, Heatmap of differently expressed genes of fatty acid metabolism pathway. NS means there is no significant difference compared with the control group. d, Significantly enriched pathways for both differentially expressed genes and differentially changed metabolites in 0.3 μg L−1cis-BF exposure group. e, The gene expressions in DCA supplemental study. Data are presented as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001 indicate significant differences compared with the control group.
4. Discussion
In this study, we showed that environmentally relevant levels of cis-BF exposure induced a significant change in the frog gut microbiota community and its metabolic profiles, leading to disorders of lipid metabolism in the liver. In addition, lipid metabolism disorders were strongly related to changes in bile acid production in the gut. Our results suggested that gut microbiota dysbiosis is involved in the abnormal lipid metabolism caused by SP.
Deposition of cis-BF in the gut contents of frogs indicates bioaccumulation and bioavailability of these compounds in the intestinal system, which suggests that cis-BF may interact with interstitial cells and the intestinal microbiota. Studies have suggested that a wide range of extrinsic factors, including drugs, probiotics, and xenobiotics, particularly environmental contaminants such as pesticides can induce the rapid change of microbiota community [26]. Our 16S rRNA gene sequencing results (ANOSIM) showed cis-BF exposure significantly altered the community structures of the frog gut microbiota. Exposure to environmental toxicants has been shown to alter the composition of gut bacteria. For instance, exposure to alphacypermethrin and permethrin significantly changed the bacterial composition of mosquitoes [27]. Fungicide propamocarb could alter gut microbial community structures both in cecal contents and feces of mice [28]. A reduction in bacterial diversity was observed in cis-BF-treated frogs, suggesting that cis-BF altered the patterns of the gut microbial ecosystems, which may alter the mutually beneficial interaction toward another stable but harmful balance [29]. We identified several bacterial genera perturbed in frogs after cis-BF exposure. Remarkably, several pathogenic bacteria of the Plesiomonas and Aeromonas genera were observed in treated frogs. Plesiomonas is a microorganism of aquatic origin and plays a role as a pathogen of both humans and animals causing occasional opportunistic infections and diarrhea [30]. Aeromonas species that are known to be pathogenic in humans are also pathogenic in fish [31]. Thus, our results showed that SP exposure induced the prevalence of several potentially pathogenic bacteria, which could increase the risk of disease development. In addition, several genera of the families Erysipelotrichaceae, Ruminococcaceae, and Lachnospiraceae, which belong to the Firmicutes phylum, decreased in the intestinal tract of frogs. Previous studies have confirmed that the members of the family Erysipelotrichaceae are associated with lipidemic profiles within the host [32,33]. The families Lachnospiraceae and Ruminococcaceae, which belong to Clostridiales within Firmicutes, have been associated with the maintenance of gut health [34]. A few members of the family Lachnospiraceae or Ruminococcaceae are known to convert conjugated primary bile acids to secondary bile acids. Specifically, the 7α-dehydroxylation reaction is the predominant procedure in the production of secondary bile acids by gut bacteria, and enzymes of the reaction are encoded by bile acid-inducible (bai) genes [35,36]. The previous study based on metagenome-assembled genomes (MAGs) has revealed that the majority of bai-containing genomes were associated with the Ruminococcaceae clade (more than 90%), and minority MAGs belonged to Lachnospiraceae [36]. The loss of Lachnospiraceae and Ruminococcaceae in acute and chronic intestinal diseases has been linked to bile acid metabolic changes [37]. For instance, a previous study reported that in pouch patients who experience ulcerative colitis, members of the family Ruminococcaceae were significantly depleted, and this condition was associated with secondary bile acid deficiency [38]. Mice fed with lard and bile acids showed changes in the dominant intestinal bacterial community, including a decline in the relative abundance of Lachnospiraceae linked to hepatic lipid rearrangements [39]. Therefore, our results suggested that microbiota dysbiosis in the intestinal tracts of frogs exposed to cis-BF likely involves abnormal metabolism in the host locally and systemically.
Metabolomics can capture the representative information of the functional state of biological systems’ responses to various contaminant-related stressors [40]. Consistent with the microbiota dysbiosis induced by cis-BF exposure, the metabolites profiles of intestinal contents were also significantly altered and enriched with several key lipid metabolism-related pathways. In particular, a perturbation was found in cholesterol metabolism, primary bile acid biosynthesis, secondary bile acid biosynthesis, and bile secretion in the intestinal of frogs treated at higher doses. Bile acids are metabolites of cholesterol that are synthesized in the liver and secreted into the intestine, and a portion of the total bile acids are dehydroxylated by intestinal bacteria to hydrophobic secondary bile acids [41]. Our results indicated that cis-BF possesses biological activities on the composition of the gut microbiota, which may cause changes in intestinal metabolites. Specifically, changes in several bile acids were strongly positively associated with the abundance of the Clostridium_sensu_stricto_1 bacterial genus (p < 0.01). Linear discriminant analysis effect size also suggested that Clostridium_sensu_stricto_1 was a typical biomarker found in the control group compared with the cis-BF exposure groups. A previous study reported that a small population of intestinal species in the genus Clostridium spp. can deconjugate conjugated BAs and metabolize bile acids [42]. In addition, the change in secondary bile acids DCA content was strongly correlated with the abundance of several bacterial families in Firmicutes phylum, including Lachnospiraceae and Peptostreptpcoccaceae. The form of secondary bile acid DCA by gut microbiota from cholic acid through a multi-step 7α-dehydroxylation reaction [35], and 7α-dehydroxylation enzymes are encoded by bile acid-inducible genes that were identified in bacterial populations from Lachnospiraceae and Peptostreptococcaceae [43]. Therefore, the perturbation in the gut microbiota caused by cis-BF exposure significantly affected its ability to metabolize bile acids and thus impacted the composition of secondary bile acids. And DCA is probably one of the most important metabolites that mediate other process changes induced by cis-BF. Environmental toxicants from various chemical families have been shown to affect the microbiota community and the metabolites of the feces [44]. For instance, diazinon was demonstrated to change the gut microbiota community and the associated metabolic profiles in the mice feces, which contribute to diazinon-induced neurotoxicity [45]. Aldicarb treatment induced the alteration of the gut microbiome along with the disrupted metabolome and lipidome in the feces of mice [46]. It is difficult to distinguish the metabolites of maternal and microbiota; however, many metabolites are exclusive microbial-derived, and abundant evidence still supports a relationship between the gut microbiota community and the metabolites profile of feces or gut content. Future studies are needed to address the role of the gut microbiota in the changes in its metabolites after exposure to multiple environmental toxicants. Gut microbiota-mediated secondary bile acids play an important role in the regulation of bile acid pool composition. We also verified whether DCA is a key factor in the change of bile acid pool induced by cis-BF. We found that diet supplemented with 0.2% DCA altered the composition and size of bile acid pool in the intestinal tract of frogs; in particular, DCA could ameliorate changes in the content of several bile acids induced by cis-BF, such as taurocholic acid, lithocholic acid, deoxycholic acid (itself) and its conjugates. The inhibition of the bacterial community that contained active DCA-inducible genes (e.g., 7α-dehydroxylation) might be the reason for the perturbation of bile acid pool composition and size.
As a class of host-derived metabolites, BAs are signaling molecules through a variety of receptors to maintain metabolic homeostasis, such as hepatic lipid and energy homeostasis [47]. Disorders in bile acid metabolism could cause cholestatic liver diseases, dyslipidemia, fatty liver diseases, diabetes, etc. [35]. In addition, a defect in the synthesis of the bile acids (such as cholic acid or chenodeoxycholic acid) in the liver would cause cholestasis and the malabsorption of fat and fat-soluble vitamin [48]. It was also observed that bacterial dysbiosis observed in cirrhosis is linked to low bile acid levels entering the intestine [42]. Microbe-derived metabolites and the signaling pathways they affect might play important roles in the development of nonalcoholic fatty liver disease [49]. Therefore, the modification of BA metabolites after cis-BF exposure may perturb the normal physiological process of hepatic metabolism in frogs. Transcriptomic analysis of the liver was performed in parallel, and differentially expressed genes were identified. KEGG analysis revealed that fatty acid metabolism-related terms were significantly enriched among these differentially expressed genes. For example, cis-BF treatment in frogs altered the activation of fatty acid metabolism, fatty acid degradation, and PPAR signaling pathway. PPARs are nuclear hormone receptors that modulate the expression of genes and are involved in adipogenesis and maintenance of metabolic homeostasis [50]. In particular, the mRNA of genes associated with fatty acid transport processes and the PPARG coactivator 1 alpha (ppargc1a) gene were downregulated significantly. Thereinto, the enzymes encoded by acsbg2.L, acsl3, and dbi.L possess long-chain acyl-CoA synthetase and/or binding activity. The fabp1 gene functions as an intracellular carrier to facilitate long-chain fatty acid transport [51]. The protein encoded by ppargc1a interacts with PPARγ, acts as a transcriptional coactivator that regulates the genes downstream of PPAR [52]. The inhibition of the expression levels of these signaling pathway-related genes may decrease disorders of fatty acid oxidation and lipid catabolism, resulting in increased lipid deposition in the liver. The association between the hepatic gene enriched pathways and the gut metabolic profile was evaluated, and significant enrichment was found in the primary bile acid biosynthesis pathway. Bile acids circulate between the liver and the intestine and back to the liver, which is called the enterohepatic circulation of the bile, play an essential role in the liver, biliary, and intestinal function and disease [35]. Clinical studies indicate that dysregulation of bile acid homeostasis and its related signaling pathways is associated with the occurrence of liver disease [49]. Our results suggest the important role of bile acids in liver metabolism, especially when challenged by environmental pollutants, and bile acid disorders have a part in inducing liver metabolism disorder. Although the regulatory mechanism remains unclear, previous reports have suggested that bile acids can induce the expression of the human PPAR alpha gene and contribute to liver disease by altering nuclear bile acid receptor farnesoid X receptor (FXR) signaling [53,54], indicating that the suppressed BA-FXR signaling pathway might participate in the regulation of fatty acid metabolism in the liver. In our study, a diet supplemented with 0.2% DCA changed the genes related to fatty acid transport/metabolism processes in the liver, and after co-exposure with cis-BF, DCA regulates the changes of fatty acid transport/metabolism induced by cis-BF. Our results suggest that DCA changes play a key role in the cis-BF-induced metabolic capacity disorder. In other words, the changes in the metabolic capacities of intestinal microbes could reduce the lipid metabolism function of the frog liver.
Lipids are stored within lipid droplets in cells [55]. To further investigate whether the hepatic metabolic pathway changes that were induced by cis-BF exposure led to alterations in lipid storage, we measured the formation of lipid droplets in hepatocytes as well as the plasma lipid levels in the plasma. We observed an increased tendency in the amount of triglyceride in the plasma but no statistically significant difference. However, liver sections from exposed frogs showed increased lipid droplets by using oil red O staining. A previous study reported that lipid droplet accumulation is the hallmark of nonalcoholic fatty liver disease [55]. Our results indicated that the inhibition of metabolism-related signaling pathways in the liver induces the accumulation of lipids, which leads to cellular dysfunction and liver disease, such as hepatic steatosis. Interestingly, in the DCA supplement study, decreased lipid droplets accumulation was observed in the group exposed to both DCA and cis-BF. Consistently, DCA was applied for aesthetic appearance use in submental fat reduction since it could disrupt adipocytes membranes and reduce fat cells in the tissue [56]. Therefore, we suggest that BAs produced by gut microbiota can serve as a signal to regulate the adverse effects in the gut-liver axis caused by cis-BF. The functional role of intestinal function in the biological response to xenobiotics is certainly worthy of further discussion.
5. Conclusions
In summary, the present study demonstrates that environmentally relevant levels of cis-BF exposure significantly altered the structure and composition of intestinal microorganisms, resulting in changes in the intestinal metabolites involved in lipid metabolic pathways, especially bile acid metabolism, and ultimately interfered with lipid metabolism in the frog liver. Our results indicated the crucial role of gut microbiota in the host response to environmental toxicants. In particular, the interplay between gut microbes and xenobiotics may promote the unexpected development of toxicity, which is generally overlooked due to the lack of information on the direct-action sites of xenobiotic compounds in organisms. Collectively, our results highlight the potential implication of intestinal microbiota dysbiosis on pesticide exposure-induced chronic diseases related to liver metabolic syndrome, which unveils the unexpected ecological risks from the widely applied pesticides.
Notes
The authors declare no competing financial interest. The raw datasets supporting the results of this article have been deposited in the NCBI and the project numbers are PRJNA725147 and PRJNA725067.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (grant no. 22176173) and the Natural Science Foundation of Zhejiang Province (grant no. LY22B070008).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ese.2022.100185.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ba Q., Li M., Chen P., Huang C., Duan X., Lu L., Li J., Chu R., Xie D., Song H., Wu Y., Ying H., Jia X., Wang H. Sex-Dependent effects of cadmium exposure in early life on gut microbiota and fat accumulation in mice. Environ. Health Perspect. 2017;125(3):437–446. doi: 10.1289/EHP360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao B., Bian X., Mahbub R., Lu K. Sex-Specific effects of organophosphate diazinon on the gut microbiome and its metabolic functions. Environ. Health Perspect. 2017;125(2):198–206. doi: 10.1289/EHP202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramírez-Pérez O., Cruz-Ramón V., Chinchilla-López P., Méndez-Sánchez N. The role of the gut microbiota in bile acid metabolism. Ann. Hepatol. 2017;16:S21–S26. doi: 10.5604/01.3001.0010.5672. [DOI] [PubMed] [Google Scholar]
- 4.Wang M., Cerna Va T. Overhauling the assessment of agrochemical-driven interferences with microbial communities for improved global ecosystem integrity. Environ. Sci. Ecotechnol. 2020;4:100061. doi: 10.1016/j.ese.2020.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amanda C.S., Omar R.L., Riezu-Boj J.I., Milagro F.I., Alfredo M.J. Diet, gut microbiota, and obesity: links with host genetics and epigenetics and potential applications. Adv. Nutr. 2019;10:S17–S30. doi: 10.1093/advances/nmy078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Nichols R.G., Correll J., Murray I.A., Tanaka N., Smith P.B., Hubbard T.D., Sebastian A., Albert I., Hatzakis E., Gonzalez F.J., Perdew G.H., Patterson A.D. Persistent organic pollutants modify gut microbiota–host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Environ. Health Perspect. 2015;123(7):679–688. doi: 10.1289/ehp.1409055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou L., Limbu S.M., Shen M., Zhai W., Qiao F., He A., Du Z., Zhang M. Environmental concentrations of antibiotics impair zebrafish gut health. Environ. Pollut. 2018;235:245–254. doi: 10.1016/j.envpol.2017.12.073. [DOI] [PubMed] [Google Scholar]
- 8.Bertotto L.B., Richards J., Gan J., Volz D.C., Schlenk D. Effects of bifenthrin exposure on the estrogenic and dopaminergic pathways in zebrafish embryos and juveniles. Environ. Toxicol. Chem. 2017;37(1):236–246. doi: 10.1002/etc.3951. [DOI] [PubMed] [Google Scholar]
- 9.Saillenfait A.M., Ndiaye D., Sabaté J.P. Pyrethroids: exposure and health effects – an update. Int. J. Hyg Environ. Health. 2015;218(3):281–292. doi: 10.1016/j.ijheh.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Delgado-Moreno L., Lin K., Veiga-Nascimento R., Gan J. Occurrence and toxicity of three classes of insecticides in water and sediment in two Southern California coastal watersheds. J. Agric. Food Chem. 2011;59(17):9448–9456. doi: 10.1021/jf202049s. [DOI] [PubMed] [Google Scholar]
- 11.Tang W., Wang D., Wang J., Wu Z., Li L., Huang M., Xu S., Yan D. Pyrethroid pesticide residues in the global environment: an overview. Chemosphere. 2018;191:990–1007. doi: 10.1016/j.chemosphere.2017.10.115. [DOI] [PubMed] [Google Scholar]
- 12.He Y., Guo C., Lv J., Deng Y., Xu J. Occurrence, sources, and ecological risks of three classes of insecticides in sediments of the Liaohe River basin, China. Environ. Sci. Pollut. Res. 2021;28(44):62726–62735. doi: 10.1007/s11356-021-15060-5. [DOI] [PubMed] [Google Scholar]
- 13.Corcellas C., Eljarrat E., Barceló D. First report of pyrethroid bioaccumulation in wild river fish: a case study in Iberian river basins (Spain) Environ. Int. 2015;75:110–116. doi: 10.1016/j.envint.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Xiang D., Zhong L., Shen S., Song Z., Zhu G., Wang M., Wang Q., Zhou B. Chronic exposure to environmental levels of cis-bifenthrin: enantioselectivity and reproductive effects on zebrafish (Danio rerio) Environ. Pollut. 2019;251:175–184. doi: 10.1016/j.envpol.2019.04.089. [DOI] [PubMed] [Google Scholar]
- 15.Li M., Wu Q., Wang Q., Xiang D., Zhu G. Effect of titanium dioxide nanoparticles on the bioavailability and neurotoxicity of cypermethrin in zebrafish larvae. Aquat. Toxicol. 2018;199:212–219. doi: 10.1016/j.aquatox.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y., Zhang Y., Vinturache A., Wang Y., Shi R., Chen L., Qin K., Tian Y., Gao Y. Effects of environmental pyrethroids exposure on semen quality in reproductive-age men in Shanghai, China. Chemosphere. 2020;245:125580. doi: 10.1016/j.chemosphere.2019.125580. [DOI] [PubMed] [Google Scholar]
- 17.Magnuson J.T., Cryder Z., Andrzejczyk N.E., Harraka G., Schlenk D. Metabolomic profiles in brains of juvenile steelhead (Oncorhynchus mykiss) following bifenthrin treatment. Environ. Sci. Technol. 2020;54(19):12245–12253. doi: 10.1016/j.aquaculture.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 18.Nasuti C., Coman M.M., Olek R.A., Fiorini D., Verdenelli M.C., Cecchini C., Silvi S., Fedeli D., Gabbianelli R. Changes on fecal microbiota in rats exposed to permethrin during postnatal development. Environ. Sci. Pollut. Res. 2016;23:10930–10937. doi: 10.1007/s11356-016-6297-x. [DOI] [PubMed] [Google Scholar]
- 19.Ravula A.R., Yenugu S. Effect of oral administration of a mixture of pyrethroids at doses relevant to human exposure on the general and male reproductive physiology in the rat. Ecotoxicol. Environ. Saf. 2021;208:111714. doi: 10.1016/j.ecoenv.2020.111714. [DOI] [PubMed] [Google Scholar]
- 20.Dabke K., Hendrick G., Devkota S. The gut microbiome and metabolic syndrome. J. Clin. Invest. 2019;129(10):4050–4057. doi: 10.1172/JCI129194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M., Zhu J., Fang H., Wang M., Wang Q., Zhou B. Coexposure to environmental concentrations of cis-bifenthrin and graphene oxide: adverse effects on the nervous system during metamorphic development of Xenopus laevis. J. Hazard Mater. 2020;381:120995. doi: 10.1016/j.jhazmat.2019.120995. [DOI] [PubMed] [Google Scholar]
- 22.Evariste L., Lagier L., Gonzalez P., Mottier A., Mouchet F., Cadarsi S., Lonchambon P., Daffe G., Chimowa G., Sarrieu C., Ompraret E., Galibert A., Ghimbeu C.M., Pinelli E., Flahaut E., Gauthier L. Thermal reduction of graphene oxide mitigates its in vivo genotoxicity toward Xenopus laevis tadpoles. Nanomaterials. 2019;9(4):584. doi: 10.3390/nano9040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Alvarez I., Islas-Flores H., Manuel Gomez-Olivan L., Barcelo D., Miren L.D.A., Perez Solsona S., Sanchez-Aceves L., Sanjuan-Reyes N., Galar-Martinez M. Determination of metals and pharmaceutical compounds released in hospital wastewater from Toluca, Mexico, and evaluation of their toxic impact. Environ. Pollut. 2018;240:330–341. doi: 10.1016/j.envpol.2018.04.116. [DOI] [PubMed] [Google Scholar]
- 24.Bantle J., Sabourin T. FETAX); 1991. Standard Guide for Conducting the Frog Embryo Teratogenesis Assay-Xenopus. [Google Scholar]
- 25.Yang Y., Ye X., He B., Liu J. Cadmium potentiates toxicity of cypermethrin in zebrafish. Environ. Toxicol. Chem. 2016;35:435–445. doi: 10.1002/etc.3200. [DOI] [PubMed] [Google Scholar]
- 26.Ursell L.K., Haiser H.J., Treuren W.V., Garg N., Reddivari L., Vanamala J., Dorrestein P.C., Turnbaugh P.J., Knight R. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology. 2014;146(6):1470–1476. doi: 10.1053/j.gastro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dada N., Lol J.C., Benedict A.C., López F., Sheth M., Dzuris N., Padilla N., Lenhart A. Pyrethroid exposure alters internal and cuticle surface bacterial communities in Anopheles albimanus. ISME J. 2019;13(10):2447–2464. doi: 10.1038/s41396-019-0445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S., Luo T., Wang S., Zhou J., Ni Y., Fu Z., Jin Y. Chronic exposure to fungicide propamocarb induces bile acid metabolic disorder and increases trimethylamine in C57BL/6J mice. Sci. Total Environ. 2018;642:341–348. doi: 10.1016/j.scitotenv.2018.06. [DOI] [PubMed] [Google Scholar]
- 29.Alexis M., Marion L., Hugot J.P. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front. Microbiol. 2016;7:455. doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Y.W., Sussman M., Liu D., Poxton I., Merritt A. second ed. Molecular Medical Microbiology; 2014. Molecular Medical Microbiology. second ed. [Google Scholar]
- 31.Fernández-Bravo A., Figueras M.J. An update on the genus aeromonas: taxonomy, epidemiology, and pathogenicity. Microorganisms. 2020;8(1):129. doi: 10.3390/microorganisms8010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaakoush N.O. Insights into the role of erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 2015;5:84. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez I., Perdicaro D.J., Brown A.W., Hammons S., Carden T.J., Carr T.P., Eskridge K.M., Walter J. Diet-Induced alterations of host cholesterol metabolism are likely to affect the gut microbiota composition in hamsters. Appl. Environ. Microbiol. 2013;79(2):516–524. doi: 10.1128/AEM.03046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biddle A., Stewart L., Blanchard J., Leschine S. Untangling the genetic basis of fibrolytic specialization by lachnospiraceae and ruminococcaceae in diverse gut communities. Diversity. 2013;5(3):627–640. doi: 10.3390/d5030627. [DOI] [Google Scholar]
- 35.Doden H.L., Wolf P.G., Gaskins H.R., Anantharaman K., Alves J., Ridlon J.M. Completion of the gut microbial epi-bile acid pathway. Gut Microb. 2021;13(1):1–20. doi: 10.1080/19490976.2021.1907271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vital M., Rud T., Rath S., Pieper D.H., Schlüter D. Diversity of bacteria exhibiting bile acid-inducible 7α-dehydroxylation genes in the human gut. Comput. Struct. Biotec. 2019;17:1016–1019. doi: 10.1016/j.csbj.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suchodolski J.S. Diagnosis and interpretation of intestinal dysbiosis in dogs and cats. Vet. J. 2016;215:30–37. doi: 10.1016/j.tvjl.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Sinha S.R., Haileselassie Y., Nguyen L.P., Tropini C., Wang M., Becker L.S., Sim D., Jarr K., Spear E.T., Singh G., Namkoong H., Bittinger K., Fischbach M.A., Sonnenburg J.L., Habtezion A. Dysbiosis-Induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27(4):659–670. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Just S., Mondot S., Ecker J., Wegner K., Rath E., Gau L., Streidl T., Hery-Arnaud G., Schmidt S., Lesker T.R., Bieth V., Dunkel A., Strowig T., Hofmann T., Haller D., Liebisch G., Gérard P., Rohn S., Lepage P., Clavel T. The gut microbiota drives the impact of bile acids and fat source in diet on mouse metabolism. Microbiome. 2018;6(1):134. doi: 10.1186/s40168-018-0510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., Qian L., Ding L., Wang L., Wong M.H., Tao H. Ecological and toxicological assessments of anthropogenic contaminants based on environmental metabolomics. Environ. Sci. Ecotechnol. 2021;5:100081. doi: 10.1016/j.ese.2021.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Gao X., Zhang X., Xiao Y., Huang J., Yu D., Li X., Hu H., Ge T., Li D., Zhang T. Gut microbiota dysbiosis is associated with altered bile acid metabolism in infantile cholestasis. mSystems. 2019;4(6) doi: 10.1128/mSystems.00463-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ridlon J.M., Kang D.J., Hylemon P.B., Bajaj J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014;30(3):332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ridlon J.M., Harris S.C., Bhowmik S., Kang D.J., Hylemon P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microb. 2016;7(1):22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Claus S.P., Guillou H., Ellero-Simatos S. The gut microbiota: a major player in the toxicity of environmental pollutants? Npj Biofilms Microbi. 2016;2(1):1–11. doi: 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao B., Bian X., Mahbub R., Lu K. Sex-Specific effects of organophosphate diazinon on the gut microbiome and its metabolic functions. Environ. Health Perspect. 2017;125(2):198–206. doi: 10.1289/EHP202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao B., Chi L., Tu P., Gao N., Lu K. The carbamate aldicarb altered the gut microbiome, metabolome, and lipidome of C57BL/6J mice. Chem. Res. Toxicol. 2019;32(1):67–79. doi: 10.1021/acs.chemrestox.8b00179. [DOI] [PubMed] [Google Scholar]
- 47.Chiang J. Bile acid metabolism and signaling. Compr. Physiol. 2013;3(3):1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelking L.R. third ed. 2015. Textbook of Veterinary Physiological Chemistry; pp. 584–589. [Google Scholar]
- 49.Chu H., Duan Y., Yang L., Schnabl B. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut. 2019;68(2):359–370. doi: 10.1136/gutjnl-2018-316307. [DOI] [PubMed] [Google Scholar]
- 50.Daniele F., Valeria A., Stefano C. The interplay between metabolism, PPAR signaling pathway, and cancer. PPAR Res. 2017;2017:1–2. doi: 10.1155/2017/1830626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L., Li L., Jiang J., Wang Y., Zhong T., Chen Y., Wang Y., Zhang H. Molecular characterization and different expression patterns of the FABP gene family during goat skeletal muscle development. Mol. Biol. Rep. 2015;42(1):201–207. doi: 10.1007/s11033-014-3759-4. [DOI] [PubMed] [Google Scholar]
- 52.Lvarez-Nava F., Salinas M., Bastidas D., Vicua Y., Racines-Orbe M. PPARGC1A promoter DNA-methylation level and glucose metabolism in Ecuadorian women with Turner syndrome. Horm. Mol. Biol. Clin. Invest. 2020;42(2):159–165. doi: 10.1515/hmbci-2020-0076. [DOI] [PubMed] [Google Scholar]
- 53.Torra I.P., Claudel T., Duval C., Kosykh V., Fruchart J.C., Staels B. Bile acids induce the expression of the human peroxisome Proliferator-Activated receptor gene via activation of the farnesoid x receptor. Mol. Endocrinol. 2003;(2):259–272. doi: 10.1210/me.2002-0120. [DOI] [PubMed] [Google Scholar]
- 54.Chiang J.Y.L. Bile acid metabolism and signaling in liver disease and therapy. Liver Res. 2017;1(1):3–9. doi: 10.1016/j.livres.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seebacher F., Zeigerer A., Kory N., Krahmer N. Hepatic lipid droplet homeostasis and fatty liver disease. Semin. Cell Dev. Biol. 2020;108:72–81. doi: 10.1016/j.semcdb.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 56.Farina G.A., Cherubini K., de Figueiredo M.A.Z., Salum F.G. Deoxycholic acid in the submental fat reduction: a review of properties, adverse effects, and complications. J. Cosmet. Dermatol. 2020;19(10):2497–2504. doi: 10.1111/jocd.13619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.