Abstract
Microorganisms in natural environments are crucial in maintaining the material and energy cycle and the ecological balance of the environment. However, it is challenging to delineate environmental microbes' actual metabolic pathways and intraspecific heterogeneity because most microorganisms cannot be cultivated. Raman spectroscopy is a culture-independent technique that can collect molecular vibration profiles from cells. It can reveal the physiological and biochemical information at the single-cell level rapidly and non-destructively in situ. The first part of this review introduces the principles, advantages, progress, and analytical methods of Raman spectroscopy applied in environmental microbiology. The second part summarizes the applications of Raman spectroscopy combined with stable isotope probing (SIP), fluorescence in situ hybridization (FISH), Raman-activated cell sorting and genomic sequencing, and machine learning in microbiological studies. Finally, this review discusses expectations of Raman spectroscopy and future advances to be made in identifying microorganisms, especially for uncultured microorganisms.
Keywords: Raman spectroscopy, Environmental microorganisms, Single cells, Metabolic activities
Highlights
-
•
Raman spectroscopy can identify the phenotypes of environmental microbes in situ.
-
•
Raman bands corresponding to common substances in cells are integrated.
-
•
Raman spectroscopy combined with various technologies can achieve more functions.
-
•
Raman spectroscopy helps decipher microbial “dark matter” from the environment.
1. Introduction
Microorganisms are indispensable for maintaining the balance of the ecosystem [1]. Moreover, microorganisms are essential for human health [2], disease treatment [3], food processing [4], and treatment of environmental pollution [5,6]. While advances have been made in the species diversity and compositions of natural microbial populations, the physiological and biochemical properties of environmental microorganisms are still poorly known [7].
The studies of environmental microorganisms mainly include culture-dependent and culture-independent approaches. Traditional methods can reveal the physiological and biochemical characteristics of isolated microorganisms, which have significantly promoted the development of microbiology. However, more than 99% of microorganisms in the environment are difficult to isolate and grow in pure cultures under current technical conditions [8,9]. Some microorganisms, especially anaerobic microorganisms, grow slowly, increasing the time cost of microbial identification and the difficulty of isolation and cultivation [10]. In addition, ex situ culture changes the physiological status, metabolic characteristics, and interactions with other microbes in the community [11]. Therefore, it is necessary to apply more practical techniques to explore environmental microorganisms, especially uncultivated microorganisms.
The current omics technologies as culture-independent methods can explore the phylogenetic diversity, the metabolic pathways, and the potential physiological and biochemical functions of microorganisms in situ. Metagenomics can directly catalog and analyze all microorganisms in various environments, including unknown and uncultured ones [12]. But metagenomics can only infer the potential metabolic pathways of microorganisms because there are many factors affecting gene expression. The results of metagenomic sequencing cannot reveal dynamic properties of spatiotemporal activities and responses to environmental changes [13]. Transcriptomics can snapshot all transcripts present in a cell. Thus, this method can complement metagenomics in verifying the actual gene expression [14,15]. But currently, it is challenging to obtain full-length and low-abundance transcripts due to the high loss rate of mRNA [13]. Metabolomics is closely linked to phenotyping of an organism and intuitive in reflecting the response of living systems to the external environment [16,17]. However, metabolomics usually relies on mass spectrometry to analyze metabolites [18], so it inevitably faces the problems of large sample volume and poor reproducibility of test results. Qualitative analysis of large-scale products based on metabolome data has always been a key issue hindering the development of metabolomics [19].
In short, the current omics technologies brought many dazzling achievements to the field of environmental microbiology [12,13,20]. Metagenomics, transcriptomics, and metabolomics can complement each other in phenotypic and genotypic analyses of microbial populations. However, these technologies are generally biased towards features that represent the overall community. These technologies cannot analyze the proportion of active members and species attribution of metabolites in environmental samples. It is worth mentioning that single-cell multi-omics is on the rise. Although it still faces some challenges, such as low genome coverage in the current single-cell sequencing, the future is expected to witness profound new findings [21].
Raman spectroscopy is an effective method to complement the current omics technologies [22]. Raman spectroscopy is unsuitable for the non-targeted identification of microorganisms or for a comprehensive overview of metabolites, which are the advantages of the current omics technologies. But Raman spectroscopy can detect the metabolic activities and the physiological and biochemical characteristics of environmental microorganisms at the single-cell level [23]. Heterogeneous cells may contain special substances, or different cells may have different proportions of the same substances, which can be analyzed by Raman spectroscopy [24]. Moreover, omics technologies are unable to assist downstream experiments using the same batch of samples because they are destructive. Raman spectra can be interpreted by combining with other approaches or detecting standard samples of the presumed substance.
In addition to omics technologies, Raman spectroscopy can be combined with other biotechnologies to better understand microbial functions [25]. These complementary technologies include but are not limited to stable isotope probing (SIP), fluorescence in situ hybridization (FISH), Raman-activated cell sorting, and genomic sequencing. The combination of Raman spectroscopy and SIP can explore the metabolites and semi-quantify the metabolic activities of microorganisms [26]. Raman spectroscopy combined with FISH can quickly identify target microbes in complex samples by marking the DNA of a specific species [27]. Raman-activated cell sorting can collect target microbes in complex environmental samples according to the biomarker Raman bands [28]. The sorted cells still intact in cellular functions can be sequenced or used for cultivation. Thus, Raman spectroscopy is an effective targeted method to identify uncultivated microorganisms in situ. Combined with different techniques, Raman spectroscopy provides the possibility to obtain Raman fingerprints and whole-genome sequence of uncultured microorganisms, analyze the metabolic preferences of uncultivated species from the environment, and explore the heterogeneity of individuals in a community.
2. Raman spectroscopy
2.1. Principles of Raman scattering
Raman scattering was discovered by the Indian physicist Chandrasekhara Venkata Raman (C. V. Raman) in 1928 [29]. A sample is excited by a monochromatic laser in a Raman spectrometer. The molecules in the sample interact with the excited light to emit scattered light. Most scattered photons have the same frequency as the incident laser, called Elastic scattering or Rayleigh scattering. A few scattered photons (1/107) have different frequencies from the incident laser. This scattering is named Inelastic scattering or Raman scattering. The frequency variation of the scattered light depends on the vibrational characteristics of the chemical bonds of molecules in the sample.
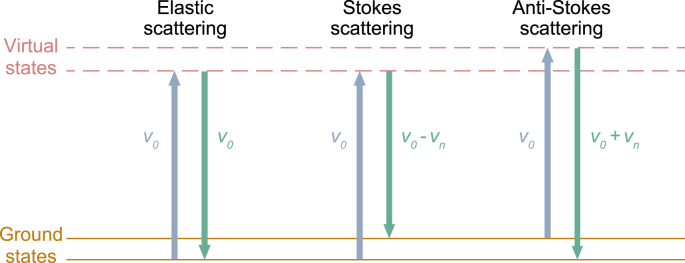
The Raman scattering can also be explained by the quantum mechanical model [30] (Fig. 1). The scattering phenomenon can be described as a two-photon process in a quantum mechanical model. First, an incident photon interacts with a molecule. The molecule absorbs the energy and undergoes a transition from the ground state to a higher energy state (excited virtual state). Second, the molecule in the excited virtual state releases a scattered photon after a short time and returns to the ground state. Rayleigh scattering means that the incident photon and the scattered photon have the same energy. In contrast, Raman scattering means the two photons have different energy.
Fig. 1.
The schematic diagram of Raman scattering. v0: The energy imparted to a molecule by excitation light; vn: Energy difference between ground and vibrational states.
Raman scattering can be divided into Stokes scattering and anti-Stokes scattering [31]. In Stokes scattering, the excited molecule does not completely return to the ground state. The energy of the scattered photon (v0 - vn) is less than incident photon (v0). In anti-Stokes scattering, the molecule has higher energy before being excited. When the excited molecule returns to the ground state, the energy of the scattered photon is v0 + vn. Compared with Stokes scattering, anti-Stokes scattering can only be caused by molecules with higher energy. The number of such molecules is limited, resulting in very low signal intensity of anti-Stokes scattering. Therefore, Stokes scattering is usually used as the signal source of Raman spectroscopy to reflect the material composition of the sample.
Raman spectroscopy can be regarded as a collection of molecular vibrations in the sample. Therefore, it can reflect ingredient and chemical information of the sample. Raman spectroscopy has high spatial resolution [32]. The laser diameter is around 1 μm with an objective of 100 × . It can obtain target microbes' physiological and biochemical information at the single-cell level [33,34]. The single-cell Raman spectra can illustrate a “fingerprint” and reveal the composition (such as proteins, nucleic acids, lipids) of each cell [35].
2.2. Common methods of Raman spectroscopy used in microbiology
Up to now, more than 25 different types of Raman spectroscopy have been developed [36]. Confocal Raman spectroscopy is commonly used for studying microorganisms because of its micron-level resolution and clear image quality. It has excellent potential for detecting single microbial cells in situ due to its non-destructive and water-free performance. Berg et al. [37] used confocal Raman spectroscopy to detect a Beggiatoa species under different growth conditions and plotted an image of the sulfide distribution in a living cell. They proved that the sulfur in Beggiatoa under culture conditions is stored in cyclooctasulfur (S8) and inorganic polysulfides (Sn2−). Serrano et al. [38] compared two strains of methanogenic archaea from permafrost and non-permafrost, respectively, by confocal Raman spectroscopy. Although the two strains have a high homology, their chemical compositions are significantly different.
Traditional Raman spectroscopy is relatively insensitive and requires long acquisition times due to the low Raman scattering cross section [39]. Molecular structures that are less abundant in cells are difficult to be detected. When the excitation wavelength is within the range of electron excitation of molecules, the resonant Raman scattering significantly improves the electric field and selectively enhances the Raman signal intensity of pigment-containing substances in microbial cells. Due to the high specificity and selectivity, the detection time and detection limit of intracellular substances are significantly reduced. It helps to detect molecular structures with suitable chromophores, including those less abundant in cells. Therefore, the vibration information of the target substance can be obtained by changing the wavelength of the exciter and combining it with spontaneous Raman spectroscopy. Mukherjee et al. [40] detected a new characteristic peak of Escherichia coli at 740 cm−1 under laser excitation at 633 nm using resonant Raman spectroscopy and verified that this peak may belong to the second subunit of cytochrome bd. This is the first study to use a red laser to achieve resonance enhancement in bacteria in situ, laying a foundation for real-time detection of intracellular cytochrome subunit changes and even intracellular respiration without labeling. Miyaoka et al. [41] detected in situ antibiotic Amphotericin B (AmB) produced by the marine bacteria Streptomyces nodosus. The Raman intensity of AmB correlates with the abundance of AmB in cells, which proves that Raman spectroscopy is an effective method for identifying antibiotic producers and (semi-) quantifying intracellular biological antibiotics [41].
Surface-enhanced Raman spectroscopy can improve the electric field and molecular polarizability through the interaction between electromagnetic waves and metal, thus effectively enhancing the Raman signal [42]. This technique was first invented by Fleischmann et al. [43] in 1973. Metals (such as gold, silver, or copper) that are stable in the air are usually used as signal amplification materials. The strength of output signals is related to the type, particle size, and roughness of metals. Bodelon et al. [44] used surface-enhanced Raman spectroscopy to explore the metabolic changes of two pure culture strains, Chromobacterium violaceum (C. violaceum) and Pseudomonas aeruginosa (P. aeruginosa), in co-culture. Co-cultivation induces C. violaceum to produce violacein and inhibits the secretion of pyocyanin produced by P. aeruginosa due to the adverse effects of pyocyanin on the growth of C. violaceum. Liu et al. [45] designed a silver nanorod as a base of surface-enhanced Raman spectroscopy, detected 22 kinds of common pathogens, and obtained Raman spectra of these bacteria. Then the Raman spectra were effectively distinguished by various mathematical methods, which built a foundation for rapidly identifying pathogens from a complex environment. Other studies using surface-enhanced Raman spectroscopy are highlighted by Laucks et al. [46] who detected five polar psychrophilic marine bacteria and Xu et al. [47] who detected and identified seven kinds of marine pathogen Vibrio parahaemolyticus.
2.3. Advantages of Raman spectroscopy in studying environmental microorganisms
Raman spectroscopy can measure all molecular vibrations of a single microbial cell [48]. A single-cell Raman spectrum contains more than 1000 Raman bands reflecting the information of macromolecules, such as proteins, nucleic acids, lipids, and carbohydrates. The width of the Raman band is related to the structure and crystal form of the compound. Raman bands are much broader when detecting objects in solution. But the vast majority of Raman bands overlap each other on Raman spectra because the bands are concentrated in the fingerprint region. Only the most intense Raman peaks could be explained. Nonetheless, Raman spectra can still be used as effective microbial fingerprints, especially when ignoring individual Raman peaks and looking at a Raman spectrum as a whole.
The advantages of Raman spectroscopy in environmental microbiology are as follows:
-
1.
Detection at the single-cell level
The magnification of Raman microscopes can reach 100 times, the spot diameter of the laser can be less than 1 μm, and the resolution can be up to 0.4 μm. The diameter of most microbes is about 1 μm. Therefore, the Raman spectra of microorganisms can be obtained at the unicellular level and reflect physiological and biochemical information of microbes.
-
2.
Non-destructive to sample material
Raman spectroscopy can be a non-destructive method if the parameters are suitable. A laser with sufficient energy and adequate detection time is required to obtain obvious Raman signals. But excessive energy or detection time can easily lead to the destruction of chemical structures in cells, failing to collect Raman signals. During the first detection, the user needs to gradually increase the energy and detection time from low to high to obtain high-quality Raman signals without destroying the sample.
-
3.
Rapid detection with micro samples
In order to exceed the detection limit of compounds, most detection methods require the collection of sufficient target microbes in the environment. In contrast, Raman spectroscopy requires only a small volume of samples to detect compounds within a single cell.
-
4.
No interference by water
Microbial samples usually need to be processed in liquid. The strong infrared absorption peak of water will affect the spectral analysis for other substances in the microbe, so liquid samples are not suitable for infrared spectroscopy. Raman spectroscopy is an ideal technique for studying aqueous materials in that the Raman signal strength of water is very weak due to the asymmetry of chemical bonds of water molecules.
2.4. Analyses of single-cell Raman spectra (SCRS)
Based on the above advantages, Raman spectroscopy, combined with other technologies, can detect target single cells in environmental samples in situ, analyze the phenotypes [2], and perform subsequent genomic sequencing. Cells from pure cultures can be revived and reproduced after Raman detection and cell sorting [49]. The SCRS can be divided into three regions: fingerprint region (400–1800 cm−1), silent region (1800–2700 cm−1), and high wavenumber region (2700–3600 cm−1) [36]. The SCRS can reflect all the biological macromolecules in the cell (Fig. 2), including nucleic acids, proteins, lipids, carbohydrates, etc.
Fig. 2.
The Raman spectrum of uncultivated Marine Group II (MGII) archaea. Raman bands correspond to biological macromolecules, such as phenylalanine at 1004 cm−1, proteins and lipids at 1420–1470 cm−1. The silent region (1800–2700 cm−1) is significantly different from the fingerprint region (400–1800 cm−1) and the high wavenumber region (2700–3600 cm−1).
The Raman bands of most biological macromolecules are displayed in the fingerprint region, reflecting important physiological and biochemical information of a single cell (Table 1). For example, three types of amide bonds could reflect the secondary structure of the protein: Amide Ⅰ (1660 cm−1), Amide Ⅱ (1574 cm−1), and Amide Ⅲ (1235 cm−1) [50]. Five bases of deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) have strong Raman bands in the fingerprint region: adenine (735 cm−1), thymine (776 cm−1), cytosine (787 cm−1), guanine (651 cm−1), and uracil (771 cm−1). The characteristic Raman bands of exocyclic and endocyclic ring deformation of a monosaccharide are located at 540 cm−1 and 410 cm−1, and the characteristic Raman bands of the Cl–O–C4 glycosidic bond in glycogen are located at 480 cm−1. In addition, some biological macromolecules have special Raman bands, which can be used as biomarkers of the Raman spectrum, such as poly-β-hydroxybutyrate (PHB) (C O stretching of the ester functional group at 1735 cm−1) [51], carotenoids (C–CH3 deformation at 1003 cm−1, C–C stretching at 1155 cm−1, C C stretching at 1512 cm−1) [52], rhodopsin (the C–C stretching at 1187 cm−1, the C–C–H in-plane rocks at 1237 cm−1) [53], cytochrome C (pyrrole breathing at 750 cm−1) [54], and green fluorescent protein (GFP) (the –C C–C N– portion of the imidazolinone ring at 1552 cm−1) [55]. Thus, these notable Raman bands can be used to rapidly identify microorganisms from the community and for semi-quantitative enumeration of important intracellular components.
Table 1.
Substances corresponding to Raman bands in microorganisms.
| Wavenumber (cm−1) | Biological Assignment | Vibrational Assignment | Reference | |
|---|---|---|---|---|
| Fingerprint Region (400–1800 cm−1) | ||||
| 642 | Tyrosine | C–C twisting | [56] | |
| 650 | Amide IV | OCN bending | [50] | |
| 651 | Guanine | Ring stretching | [57] | |
| 700 | Amide V | Out-of-plane NH bending | [50] | |
| 728 | Tryptophan | Ring breathing | [58] | |
| 735 | Adenine | Ring breathing | [57] | |
| 748 | Tryptophan | Indole ring bending | [59] | |
| 750 | Cytochrome C | Pyrrole breathing | [54] | |
| 771 | Uracil | Ring breathing | [60] | |
| 776 | Thymine | Ring breathing | [57] | |
| 787 | Cytosine | Ring breathing | [57] | |
| 813 | A-type RNA of the viroid | C–O–P–O–C symmetric stretching | [61] | |
| 840–860 | Polysaccharide structure | 1,4-glycosidic link stretching | [62,63] | |
| 875 | Tryptophan | out-of-plane bending of indole ring and indole CH | [59] | |
| 951 | Proteins (α-helix) | CH3 symmetric stretching | [64] | |
| 1000 | Deoxyribose | C–O stretching | [65] | |
| 1003 | Carotenoid | C–CH3 deformation | [52] | |
| 1004 | Phenylalanine | Benzene ring breathing | [50] | |
| 1043 | HCO3− in sodium bicarbonate | symmetric stretching | [66] | |
| 1064 | Lipids | Skeletal C–C stretching | [67] | |
| 1078 | Glycogen | C–C stretching | [68] | |
| 1093 | DNA | PO2− symmetric stretching | [69] | |
| 1123 | Lipids and proteins | C–C stretching | [70] | |
| 1155 | Carotenoid | C–C stretching | [52] | |
| 1157 | Carotenoid | In-phase vibrations of the conjugated = C–C = | [71] | |
| 1172 | Tyrosine | C–H in-plane bending | [56] | |
| 1187 | Bacteriorhodopsin | C–C stretching | [72] | |
| 1199 | Tyrosine | Ring breathing | [58] | |
| 1228 | DNA | PO2− antisymmetric stretching | [56] | |
| 1235 | Amide Ⅲ | In-phase combination of the NH bending and CN stretching | [50] | |
| 1237 | Bacteriorhodopsin | C–C–H in-plane rocks | [72] | |
| 1260 | Lipids | CH2 in-plane deformation | [73] | |
| 1267 | Lipids | HC = in-plane deformation | [74] | |
| 1300 | Lipids | In-plane twisting | [62] | |
| 1304 | Lipids | CH2 deformation | [67] | |
| 1321 | Lipids | CH2 deformation | [75] | |
| 1358 | Tryptophan | Indole ring bending | [76] | |
| 1420–1450 | Lipids | CH2 scissoring | [77] | |
| 1420–1470 | Proteins and lipids | CH2 bending | [78] | |
| 1440 | Lipids | CH2 and CH3 deformation | [79] | |
| 1446 | Proteins and lipids | CH2 bending | [56] | |
| 1512 | Carotenoid | C=C stretching | [52] | |
| 1517 | β-Carotene | C–C stretching | [80] | |
| 1542 | Tryptophan | Indole ring stretching | [59] | |
| 1574 | Amide Ⅱ | Out-of-phase combination of the NH bending and the CN stretching | [50] | |
| 1625 | Chlorophyll | Stretching of conjugated vinyl groups | [81] | |
| 1660 | Amide Ⅰ | C=O stretching vibrations out-of-plane C–N stretching | [50] | |
| 1660 | Chlorophyll b | Stretching of formyl carbonyl group | [81] | |
| 1735 | PHB | C=O stretching of the ester functional group | [82] | |
| Silent Region (1800–2700 cm−1) | ||||
| 2040–2300 | C–D vibrations | [83] | ||
| High Wavenumber Region (2700–3600 cm−1) | ||||
| 2700–3100 | Lipids and amino acid side chains of proteins and carbohydrates | C–H stretching | [84] | |
| 2879 | Lipids and proteins | CH2 and CH stretching | [85] | |
| 2910–2965 | Proteins | CH3 stretching | [86] | |
| 3350–3550 | Water | OH stretching | [86] | |
Raman bands in the high wavenumber region usually represent –CH, –CH2, –CH3, –NH, and OH groups, which are very obvious and show up in Raman spectra of all microbes, such as the –CH2 symmetric stretch vibration near 2850 cm−1 and the Fermi resonance –CH2 stretch vibration near 2886 cm−1. Raman bands of these groups overlap strongly in the high wavenumber region, so there is generally little effective information that can identify biological macromolecules [87]. Proteins and lipids are mainly assigned at 2800–3000 cm−1. The relative contribution of DNA is small [88]. The Raman band of water overlaps with that of the OH group of carbohydrates at 3300–3500 cm−1 and with that of the N–H group of proteins at 3329 cm−1 [89]. In addition, the Raman band of the =CH vibration (3060 cm−1) from the unsaturated fatty acids is different to that of the –CH bond vibration (2880 cm−1) from saturated fatty acids. The ratio of the integral area of the two bands can represent the ratio of the unsaturated fatty acid to the total fatty acids in the cell, which is commonly used to characterize intracellular lipids.
3. Raman spectroscopy in combination with other technologies
3.1. Raman–SIP combination for analyzing the phenotypes of environmental microorganisms
Microorganisms gain energy by coupling electron donors and electron acceptors [90]. The principle of SIP is that microorganisms can incorporate the heavier isotope of an atom (13C vs. 12C, for example) into cell components when adding substrates labeled with the 13C isotope to environmental samples [91]. SIP is a powerful method to explore the physiological information and functional properties of individual microbes from complex environments [92,93]. As described above in the principle of Raman spectroscopy, the abscissa of Raman spectra is related to molecular vibration. The molecular mass increases when an atom is enriched by a heavier isotope, whereas the molecular vibration frequency decreases and the abscissa of the Raman peak shifts to the left. Raman–SIP can detect whether a given substrate is degraded and which microbes incorporate the heavier isotope. Redshifts caused by these changes can be found in Raman spectra of microorganisms labeled by the heavy isotope (Fig. 3). Therefore, this method can verify potential metabolic pathways and metabolic preferences of individuals in communities at the single-cell level, which contributes to uncovering the ecological role of individual microbes from natural habitats [[94], [95], [96], [97], [98], [99], [100], [101]].
Fig. 3.
The principle of Raman–SIP. a. Incubation of samples with stable isotope-labeled substrates. b. Acquisition and collection of single-cell Raman spectra. c. Raman spectra of stable isotope-labeled cells different from those of cells under natural conditions. Single-cell Raman spectra adapted from Ref. [102].
Heavy water (D2O) is an important substance to measure the metabolic activity of microbes. Moreover, compounds labeled by stable isotope deuterium (D) are commonly used to explore the ability of microbes to metabolize specific substances [103]. In general, the silent region of SCRS has no Raman bands. The C–D band (2040–2300 cm−1) in the “silent region” is easy to observe. Microbes with biological activity could incorporate D of D2O into the active cells through a H/D exchange reaction mediated by NADPH electron transfer chain. The C–D band can be observed in Raman spectra, which could semi-quantify the metabolic activity of microorganisms [94]. For fast-growing E. coli cells, the C–D band is detectable after 20 min of labeling [26]. D–labeled substrates have been used to analyze the products of several complex carbohydrates degraded by cecal microbes in mice [94]. D–glucose has been used as the sole carbon source to monitor lipid production in living cells [104]; D–amino acids have been used as substrates to detect the newly synthesized proteins and their proportion in total proteins [105,106]. In addition, if bacteria are still metabolically active in an environment with antibiotics, the bacteria can absorb heavy water, and the C–D band can be observed on their Raman spectra. Therefore, antibiotic-resistant bacteria in environmental samples can be detected by adding D2O and antibiotics at the lowest inhibitory concentration. This method has been applied to study antibiotic-resistant bacteria in the human gut and surface water samples [2,103].
15N– and 13C–labeled substrates are also commonly used to explore the metabolic characteristics of microorganisms. Studies have found that using 13C–glucose with different proportions as the sole carbon source of Pseudomonas fluorescens, redshifts of the Raman peaks are found in the fingerprint region of their Raman spectra. These redshifts are mainly manifested at the peak of phenylalanine and increase with the amount of 13C absorbed in the cell biomass [107]. 13C–naphthalene was used as the sole carbon source to cultivate Pseudomonas. It was found that Pseudomonas has a degrading effect on naphthalene in groundwater biofilms [27]. Cui et al. [102] used surface-enhanced Raman spectroscopy in combination with the stable isotope 15N to monitor different bacteria from multiple angles to assess their ability to assimilate various nitrogen sources. Raman signal intensity after redshift was linearly correlated with the content of 15N assimilated by single cells and was not affected by the type and concentration of nanoparticles used in this method. Cui et al. [95] cultivated soil samples by 15N2 and identified nitrogen-fixing bacteria in the soil. Milucka et al. [108] used confocal Raman spectroscopy combined with FISH, SIP, and other methods to verify that anaerobic oxidation of methane coupled with sulfate reduction in marine sediments was completed alone by methanotrophic archaea. Meanwhile, sulfate-reducing Deltaproteobacteria were first observed to dismutate zero-valent sulfur into sulfur dioxide. These results have important implications for understanding the global carbon and sulfur cycle.
Raman–SIP can identify photosynthetic bacteria in environmental samples and explore their carbon fixation ability and the heterogeneity of single cells in the population. Li et al. [109] detected single cells of 13C–labeled photosynthetic algae and photosynthetic bacteria by resonance Raman spectroscopy; the results showed that carotenoids are good biomarkers for Raman spectroscopy. The redshift of the Raman band can be detected when the 13C content of a single cell is only 10%. It is a non-invasive method to quickly and quantitatively detect the carbon fixation ability of single cells. On this basis, Jing et al. [110] isolated two typical bacteria, Synechococcus spp. and Pelagibacter spp. with redshift bands of 13C–carotenoid in seawater by Raman-activated cell ejection and linked phenotypes to genotypes of the carbon sequestrating capacity of uncultured marine microbes by reconstructing their genomes.
3.2. Raman–FISH combination for identifying the target microorganisms from natural environments
FISH can use fluorophore-bearing nucleotide fragments as probes to hybridize to 16S DNA or rRNA of target microorganisms, making it fluoresce under a fluorescence microscope, thus allowing us to identify a specific type of microorganisms from complex environmental samples [111,112]. Raman spectroscopy can be combined with FISH to obtain Raman spectra of target microbes in environmental samples and analyze their physiological and biochemical characteristics based on these Raman spectra. The traditional FISH-microautoradiography relies on radioisotopes such as 14C as substrates to label intracellular biological macromolecules, which has been used to detect the in situ absorption of the target microorganisms in complex environmental samples [113]. By contrast, as shown in Fig. 4, Raman–FISH does not reply on radioisotopes and can be a powerful method for non-destructive exploration of the physiological and biochemical characteristics of uncultivated microorganisms [27].
Fig. 4.
The principle of Raman–FISH. a. Nitrosopumilus maritimus SCM1 indistinguishable from other species under natural conditions. b. SCM1 cells distinguished under the fluorescent lens after being stained with a probe. c. Collection and integration of fifteen Raman spectra of fluorescent SCM1 cells.
Microorganisms have different metabolic pathways for the same substrate under different environmental conditions. Raman–FISH can quantify the metabolism of target microorganisms in situ. Studies have shown that Pseudomonas can degrade naphthalene in groundwater [27]. Gordon et al. [114] used this technique to study the differences in the growth rate of photosynthetic autotrophic bacteria within and between species under natural conditions and different culture conditions. Escudero et al. [115] verified the strong correlation between the abundance of Acidovorax and the distribution of pyrite by Raman–FISH to explore the interaction between deep underground microbes and minerals, indicating that ore will have an important impact on the distribution of underground microbes.
Some artificial environments, such as sewage treatment plants and bioreactors, use microorganisms to degrade or enrich pollutants, including enhanced biological phosphorus removal and microbial nitrogen fixation. According to the Raman signal intensity, Raman spectroscopy combined with quantitative FISH can quantify the proportion of species in the environment and detect the metabolic activity of species against specific elements or compounds. Fernando et al. [116] detected the phosphorus-accumulating microorganisms in the wastewater treatment systems of eight factories. In addition to Candidatus Accumulibacter, Tetrasphaera, which does not show the classic phenotypic characteristics of phosphorus-accumulating microorganisms, is also an important participant in enhanced biological phosphorus removal. In this study, Fernando and colleagues developed a method based on Raman–FISH to quantify intracellular polyphosphate (poly–P). Since the Raman signal intensity of poly–P at 1170 cm−1 is linearly related to the poly–P density after drying standard solutions of different concentrations, the intracellular poly–P content could be calculated by detecting the Raman signal intensity of target cells at 1170 cm−1. This method can explore the individual contribution of a variety of phosphorus-accumulating microorganisms during the comprehensive action and has reference significance for quantifying the material cycle of individual species in various ecosystems [116].
3.3. Raman activated cell sorting in combination with genomic sequencing for enhancing research in environmental microbiology
SCRS as microbial Raman fingerprints can reflect biomolecular vibrations inside specific single cells. By comparing with Raman spectra of putative substances, it is possible to explore the phenotypic characteristics of microorganisms to establish the association between phenotypes and genotypes. Meanwhile, based on the fingerprint region of SCRS, the targeted microbes can be sorted and collected by Raman-activated cell sorting. The collected cells can be genome amplified and sequenced for higher quality. High-quality genomes are constructed for analyzing potential metabolic pathways. The advantage of Raman-activated cell sorting is that the single cells of target uncultivated microbes can be isolated and collected from environmental samples without external labeling and then be sequenced or cultured [28]. Compared to traditional methods, single-cell genomic sequencing can get a complete genome of a single uncultivated species, and single-cell culture can avoid a complex screening process.
Up to now, different target microorganisms, such as antimicrobial-resistant bacteria and pathogenic bacteria, have been isolated from mixed flora, surface water, seawater, mouse intestinal tract, human intestinal tract, and human oral cavity. Their species identification and functional gene analysis have been carried out [103,117,118]. As shown in Fig. 5, there are three main types of Raman activated sorting technologies: Raman-activated cell ejection, Raman tweezers, and microfluidic cell sorting [28].
Fig. 5.
The principle of Raman-activated cell sorting. a. The principle of Raman-activated cell ejection (RACE): Observation and confirmation of target microorganisms by Raman spectroscopy followed by collection of target cells into a cell receptor. b. The principle of Raman tweezer: Capturing and moving of a cell from solution by optical tweezers to sort it out. c. The principle of microfluidic cell sorting: Allowing one cell to pass at a time through the capillary tube followed by sorting of the target cells into different regions based on Raman fingerprints.
Raman-activated cell ejection is a type of Raman-activated cell sorting coupled with a Raman spectroscopy to the cell isolation system (Fig. 5a). Wang et al. [117] used two independent approaches to achieve single-cell sorting: A 337 nm pulsed laser was applied to the thin water layer around a cell. The pulsed laser energy can evaporate the thin water layer and give the cell energy to move forward. The cell is then pushed from the slide onto the collection tube cover by a laser-induced forward transport device to achieve single-cell separation. Song et al. [118] used an all-in-one system to sort and collect single cells. They sequenced the obtained cells, and new functional genes were discovered in single-cell genomics. It is the first time that Raman-activated cell ejection coupled with single-cell genomics to explore uncultivated microorganisms.
Optical tweezers are optical traps integrated by laser focusing. Optical tweezers can capture micron-sized particles in fluids. When the microbial cells flow through the optical trap, they are fixed in the optical trap with non-contact. The cells can move with the position of the optical trap controlled by the laser source. Raman tweezer is the combination of optical tweezer and Raman spectroscopy (Fig. 5b). It can capture single target cells for Raman detection and monitor their dynamic changes in situ in real-time. Pilat et al. [119] used Raman tweezers to capture Staphylococcus aureus infected by bacteriophages and explored the interaction between bacteriophages and the host. Traditional phage detection methods require at least 45 min, while Pilat et al. [119] detected phage replicated in bacterial cells within 5 min after introducing the phage into the host cell. This provides a fast, non-destructive, and real-time detection method for understanding the mechanism of phage infection. Huang et al. [49] used Raman tweezers to identify and separate E. coli, Saccharomyces, and Pseudomonas in the mixture, then transferred the sorted target bacteria into the capillary tube. The accuracy of the Raman tweezer was verified by genome amplification, and the activity of separated cells was verified by isotope labeling and cell culturing.
The microfluidic cell sorting technique uses micron-sized pipes to create a microfluidic environment (Fig. 5c). It integrates microbial cell detection, separation, cultivation, and other independent operation processes into one chip. The chip can carry out many tasks in liquid, such as accurate cell counting, Raman detection, and single-cell sorting. The biomarker Raman bands of target microbes can be used as screening conditions to achieve rapid and high-throughput sorting of target species in environmental samples [120]. Mcilvenna et al. [120] developed a high-throughput sorting system that does not require fixed cells for Raman detection, simultaneously performing Raman signal acquisition, real-time identification, and cell sorting. They used photosynthetic bacteria as a type strain, and the sorting accuracy was 96.3%. Liao et al. [121] simplified the operation of the surface-enhanced Raman spectroscopy antimicrobial drug sensitivity testing by using a microfluidic system. They could separate the drug-sensitive and drug-resistant strains within 3.5 h and conduct the experiment in situ by changing the reagent on the chip to change the liquid environment and analyzing the bacterial response.
3.4. Single-cell Raman spectra of environmental microorganisms analyzed by machine learning
Machine learning refers to an algorithm model that completes certain functions through data training and is currently the mainstream artificial intelligence implementation method [122]. Machine learning has been successfully applied to the classification, clustering, and prediction of large data sets in many fields [[123], [124], [125]]. The composition and physiological and biochemical characteristics of environmental microorganisms are relatively complex, and traditional data processing cannot meet the needs of data analysis [126]. Many data visualization methods have been applied to the study of Raman spectroscopy. Due to the high sensitivity of Raman spectroscopy, when detecting two very similar cells, the Raman bands cannot be directly analyzed by simple data processing. To get accurate results, data visualization methods such as two-dimensional correlation analysis and linear discriminant analysis help us discover minor differences in Raman spectra. Raman spectroscopy can also connect with artificial neural networks [127], support vector machines [128], random forests [129], AdaBoost [130], and other algorithm models [131]. Herein, the Raman spectroscopy can classify and predict different microorganisms, extract their features, and analyze their “Raman fingerprints” in depth. These methods exhibit high efficiency and precision, possessing great potential in rapidly identifying uncultured pathogens and uncultured marine microorganisms [[132], [133], [134], [135], [136]].
Raman spectroscopy combined with machine learning has identified pathogens from the environment [[134], [135], [136], [137], [138]]. Ho et al. [134] used a convolutional neural network to identify 30 pathogens in low signal-to-noise ratio Raman spectra with an accuracy rate of over 82%, and identification accuracies of corresponding antibiotic treatment reached 97%. Tang et al. [135] compared the clustering and identification capabilities of various machine learning algorithms for nine species of Staphylococcus. Among these methods, density-based spatial clustering of applications with noise has the best clustering ability (Rand index 0.9733), and the convolutional neural network has the highest accuracy rate of identification (accuracy at 98.21%, area under the curve at 99.93%). Wang et al. [138] used confocal Raman spectroscopy (785 nm) combined with neural network technology to classify and identify Arcobacter. Arcobacter was selected from Campylobacter and Helicobacter by cluster analysis based on Raman spectra, and 18 species of Arcobacter were distinguished at the species level using the convolutional neural network, with an accuracy rate of 97.2%. They also constructed a fully connected artificial neural network based on SCRS to determine the actual proportion of specific Arcobacter in the bacterial mixture. These studies provided references for determining the proportion of target pathogens in environmental samples.
The combination of Raman spectroscopy and machine learning has also made progress in marine microbial research. Yu et al. [136] combined Raman spectroscopy with long and short-term memory neural networks. After optimizing the model parameters, they trained the model based on eight strains isolated from the marine organism Urechis unicinctus, and the accuracy of classification prediction exceeded 94%. Yu et al. [137] also fitted the Raman spectra of three marine strains (Staphylococcus hominis, Vibrio alginolyticus, and Bacillus licheniformis) to generative adversarial networks. The model only needed 100 SCRS of each strain. After being iteratively trained, the classification accuracy of these strains reached 100%. Liu et al. [139] collected ten species of marine actinomycetes and two species of non-marine actinomycetes, and detected their Raman spectra. They used one-dimensional convolutional neural networks and several traditional classification algorithms (support vector machines, principal component analysis, etc.) to classify the original data with an accuracy rate of 95%, and one-dimensional convolutional neural networks is preferred for processing the raw data. Heraud et al. [140] measured Dunaliella Tertiolecta cells with a 780 nm laser. The enhanced Raman bands of Chlorophyll a and beta-carotene in Raman spectra are indicative of whether nitrogen limitation exists. They successfully predicted more than 90% of the cellular nutritional status by partial least squares discriminant analysis and other multivariate classification methods. Our laboratory obtained the Raman spectra of nine types of archaea (seven halophilic archaea, one thermophilic archaeon, and one marine archaeon) by confocal Raman spectroscopy. We predicted the target microorganisms in the mixture of these organisms and verified the results by Raman-activated cell ejection and genomic sequencing. The accuracy rate of support vector machines was greater than 88%. It also was the first time that Raman–FISH is applied to obtain the Raman spectra of uncultured marine group II archaea [133].
4. Conclusion and outlook
Microorganisms are ubiquitous in the environment. They are the main driving force of the global biogeochemical cycles and are associated with almost all multicellular life forms [141]. Most microorganisms from nature cannot be studied in pure culture. These microorganisms are called microbial "dark matter". Although microbial "dark matter" has been brought to light by omics technologies, the physiological and biochemical research on microorganisms cannot be completely achieved only by metagenomic data analyses or cultivated strains. Moreover, it is essential to analyze the physiological and biochemical characteristics of individual microbial cells in situ because of the interaction among microorganisms in nature and the microbial host-specific metabolism. Detection at the single-cell level enables Raman spectroscopy as a method to complement the current omics technologies. Raman spectroscopy performs near-in-situ analysis of microbial phenotypes in the environment. Combined with downstream single-cell separation and sequencing, Raman spectroscopy provides new insights for the research of environmental microbiology. It enhances the possibility of deciphering environmental microbial "dark matter".
Raman spectroscopy in combination with other biotechnological methods is further expanding its range of application. Raman spectroscopy combined with SIP and FISH avoids the deficiency of simply detecting the material composition of cells. These integrated methods can accurately identify target microbes from the environment and quantify the metabolic activity of single cells on specific elements or nutrients according to the Raman bands of isotope-labeled substances.
Furthermore, with the increasing development of artificial intelligence, the combination of Raman spectroscopy and machine learning can improve the speed and accuracy of identifying target microbes. This combination promises rapid screening and classification of uncultivated bacteria and archaea and helps to visualize and fingerprint them from the natural environment.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. K19313901), the National Natural Science Foundation of China (91851210, 42141003), the State Key R&D project of China grant (No. 2018YFA0605800), the Stable Support Plan Program of Shenzhen Natural Science Fund (20200925173954005), the Guangdong-Shenzhen Joint Fund (2021B1515120080), the Shenzhen Key Laboratory of Marine Archaea Geo-Omics, Southern University of Science and Technology (ZDSYS201802081843490), and the financial support from China Postdoctoral Science Foundation (2020M682769).
Contributor Information
Yi Wang, Email: wangy67@sustech.edu.cn.
Chuanlun Zhang, Email: zhangcl@sustech.edu.cn.
References
- 1.Cavicchioli R., Ripple W.J., Timmis K.N., Azam F., Bakken L.R., Baylis M., Behrenfeld M.J., Boetius A., Boyd P.W., Classen A.T., Crowther T.W., Danovaro R., Foreman C.M., Huisman J., Hutchins D.A., Jansson J.K., Karl D.M., Koskella B., Welch D.B.M., Martiny J.B.H., Moran M.A., Orphan V.J., Reay D.S., Remais J.V., Rich V.I., Singh B.K., Stein L.Y., Stewart F.J., Sullivan M.B., van Oppen M.J.H., Weaver S.C., Webb E.A., Webster N.S. Scientists' warning to humanity: microorganisms and climate change. Nat. Rev. Microbiol. 2019;17(9):569–586. doi: 10.1038/s41579-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Xu J.B., Kong L.C., Li B., Li H., Huang W.E., Zheng C.M. Raman-activated sorting of antibiotic-resistant bacteria in human gut microbiota. Environ. Microbiol. 2020;22(7):2613–2624. doi: 10.1111/1462-2920.14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marmann A., Aly A.H., Lin W.H., Wang B.G., Proksch P. Co-Cultivation-A powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs. 2014;12(2):1043–1065. doi: 10.3390/md12021043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamang J.P., Watanabe K., Holzapfel W.H. Review: diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emadian S.M., Onay T.T., Demirel B. Biodegradation of bioplastics in natural environments. Waste Manag. 2017;59:526–536. doi: 10.1016/j.wasman.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Yin K., Wang Q.N., Lv M., Chen L.X. Microorganism remediation strategies towards heavy metals. Chem. Eng. J. 2019;360:1553–1563. doi: 10.1016/j.cej.2018.10.226. [DOI] [Google Scholar]
- 7.Naylor D., Sadler N., Bhattacharjee A., Graham E.B., Anderton C.R., McClure R., Lipton M., Hofmockel K.S., Jansson J.K. Soil microbiomes under climate change and implications for carbon cycling. Annu. Rev. Environ. Resour. 2020;45:29–59. doi: 10.1146/annurev-environ-012320-082720. [DOI] [Google Scholar]
- 8.Solden L., Lloyd K., Wrighton K. The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 2016;31:217–226. doi: 10.1016/j.mib.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 9.Huang W.E., Ferguson A., Singer A.C., Lawson K., Thompson I.P., Kalin R.M., Larkin M.J., Bailey M.J., Whiteley A.S. Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell Raman-fluorescence in situ hybridization. Appl. Environ. Microbiol. 2009;75(1):234–241. doi: 10.1128/Aem.01861-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leadbetter J.R. Cultivation of recalcitrant microbes: cells are alive, well and revealing their secrets in the 21st century laboratory. Curr. Opin. Microbiol. 2003;6(3):274–281. doi: 10.1016/S1369-5274(03)00041-9. [DOI] [PubMed] [Google Scholar]
- 11.Pinto D., Santos M.A., Chambel L. Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit. Rev. Microbiol. 2015;41(1):61–76. doi: 10.3109/1040841x.2013.794127. [DOI] [PubMed] [Google Scholar]
- 12.Bharti R., Grimm D.G. Current challenges and best-practice protocols for microbiome analysis. Briefings Bioinf. 2021;22(1):178–193. doi: 10.1093/bib/bbz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguiar-Pulido V., Huang W.R., Suarez-Ulloa V., Cickovski T., Mathee K., Narasimhan G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol. Bioinf. Online. 2016;12:5–16. doi: 10.4137/Ebo.S36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poretsky R.S., Bano N., Buchan A., LeCleir G., Kleikemper J., Pickering M., Pate W.M., Moran M.A., Hollibaugh J.T. Analysis of microbial gene transcripts in environmental samples. Appl. Environ. Microbiol. 2005;71(7):4121–4126. doi: 10.1128/Aem.71.7.4121-4126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe R., Shirley N., Bleackley M., Dolan S., Shafee T. Transcriptomics technologies. PLoS Comput. Biol. 2017;13(5) doi: 10.1371/journal.pcbi.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiehn O. Metabolomics - the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48(1–2):155–171. doi: 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- 17.Bernini P., Bertini I., Luchinat C., Nepi S., Saccenti E., Schafer H., Schutz B., Spraul M., Tenori L. Individual human phenotypes in metabolic space and time. J. Proteome Res. 2009;8(9):4264–4271. doi: 10.1021/pr900344m. [DOI] [PubMed] [Google Scholar]
- 18.Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016;15(7):473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 19.Shahaf N., Rogachev I., Heinig U., Meir S., Malitsky S., Battat M., Wyner H., Zheng S., Wehrens R., Aharoni A. The WEIZMASS spectral library for high-confidence metabolite identification. Nat. Commun. 2016;7:12423. doi: 10.1038/ncomms12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansson J.K., Hofmockel K.S. The soil microbiome - from metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018;43:162–168. doi: 10.1016/j.mib.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Hu Y.J., An Q., Sheu K., Trejo B., Fan S.X., Guo Y. Single cell multi-omics technology: methodology and application. Front. Cell Dev. Biol. 2018;6 doi: 10.3389/fcell.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theophilou G., Paraskevaidi M., Lima K.M.G., Kyrgiou M., Martin-Hirsch P.L., Martin F.L. Extracting biomarkers of commitment to cancer development: potential role of vibrational spectroscopy in systems biology. Expert Rev. Mol. Diagn. 2015;15(5):693–713. doi: 10.1586/14737159.2015.1028372. [DOI] [PubMed] [Google Scholar]
- 23.Lee K.S., Pereira F.C., Palatinszky M., Behrendt L., Alcolombri U., Berry D., Wagner M., Stocker R. Optofluidic Raman-activated cell sorting for targeted genome retrieval or cultivation of microbial cells with specific functions. Nat. Protoc. 2021;16(2) doi: 10.1038/s41596-020-00427-8. [DOI] [PubMed] [Google Scholar]
- 24.Yan S.S., Qiu J.X., Guo L., Li D.Z., Xu D.P., Liu Q. Development overview of Raman-activated cell sorting devoted to bacterial detection at single-cell level. Appl. Microbiol. Biotechnol. 2021;105(4):1315–1331. doi: 10.1007/s00253-020-11081-1. [DOI] [PubMed] [Google Scholar]
- 25.Harrison J.P., Berry D. Vibrational spectroscopy for imaging single microbial cells in complex biological samples. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.David Berry E.M., Lee Tae Kwon, Woebken Dagmar, Wang Yun, Zhu Di. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. U.S.A. 2015:E194–E203. doi: 10.1073/pnas.1420406112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W.E., Stoecker K., Griffiths R., Newbold L., Daims H., Whiteley A.S., Wagner M. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ. Microbiol. 2007;9(8):1878–1889. doi: 10.1111/j.1462-2920.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- 28.Song Y.Z., Yin H.B., Huang W.E. Raman activated cell sorting. Curr. Opin. Chem. Biol. 2016;33:1–8. doi: 10.1016/j.cbpa.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Raman C.V., Krishnan K.S. A new type of secondary radiation. Nature. 1928;121:501–502. doi: 10.1038/121501c0. [DOI] [Google Scholar]
- 30.Frezzotti M.L., Tecce F., Casagli A. Raman spectroscopy for fluid inclusion analysis. J. Geochem. Explor. 2012;112:1–20. doi: 10.1016/j.gexplo.2011.09.009. [DOI] [Google Scholar]
- 31.Zrimsek A.B., Chiang N.H., Mattei M., Zaleski S., McAnally M.O., Chapman C.T., Henry A.I., Schatz G.C., Van Duyne R.P. Single-molecule chemistry with surface- and tip-enhanced Raman spectroscopy. Chem. Rev. 2017;117(11):7583–7613. doi: 10.1021/acs.chemrev.6b00552. [DOI] [PubMed] [Google Scholar]
- 32.Das R.S., Agrawal Y.K. Raman spectroscopy: recent advancements, techniques and applications. Vib. Spectrosc. 2011;57(2):163–176. doi: 10.1016/j.vibspec.2011.08.003. [DOI] [Google Scholar]
- 33.Schuster K.C., Reese I., Urlaub E., Gapes J.R., Lendl B. Multidimensional information on the chemical composition of single bacterial cells by confocal Raman microspectroscopy. Anal. Chem. 2000;72(22):5529–5534. doi: 10.1021/ac000718x. [DOI] [PubMed] [Google Scholar]
- 34.Jones R.R., Hooper D.C., Zhang L.W., Wolverson D., Valev V.K. Raman techniques: fundamentals and frontiers. Nanoscale Res. Lett. 2019;14 doi: 10.1186/s11671-019-3039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camp C.H., Cicerone M.T. Chemically sensitive bioimaging with coherent Raman scattering. Nat. Photonics. 2015;9(5):295–305. doi: 10.1038/Nphoton.2015.60. [DOI] [Google Scholar]
- 36.Saletnik A., Saletnik B., Puchalski C. Overview of popular techniques of Raman spectroscopy and their potential in the study of plant tissues. Molecules. 2021;26(6) doi: 10.3390/molecules26061537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berg J.S., Schwedt A., Kreutzmann A.C., Kuypers M.M.M., Milucka J. Polysulfides as intermediates in the oxidation of sulfide to sulfate by Beggiatoa spp. Appl. Environ. Microbiol. 2014;80(2):629–636. doi: 10.1128/Aem.02852-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serrano P., Hermelink A., Lasch P., de Vera J.P., Konig N., Burckhardt O., Wagner D. Confocal Raman microspectroscopy reveals a convergence of the chemical composition in methanogenic archaea from a Siberian permafrost-affected soil. FEMS Microbiol. Ecol. 2015;91(12) doi: 10.1093/femsec/fiv126. [DOI] [PubMed] [Google Scholar]
- 39.Min W., Freudiger C.W., Lu S.J., Xie X.S. Coherent nonlinear optical imaging: beyond fluorescence microscopy. Annu. Rev. Phys. Chem. 2011;62:507–530. doi: 10.1146/annurev.physchem.012809.103512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee R., Verma T., Nandi D., Umapathy S. Identification of a resonance Raman marker for cytochrome to monitor stress responses in Escherichia coli. Anal. Bioanal. Chem. 2020;412(22):5379–5388. doi: 10.1007/s00216-020-02753-y. [DOI] [PubMed] [Google Scholar]
- 41.Miyaoka R., Hosokawa M., Ando M., Mori T., Hamaguchi H.O., Takeyama H. In situ detection of antibiotic Amphotericin B produced in Streptomyces nodosus using Raman microspectroscopy. Mar. Drugs. 2014;12(5):2827–2839. doi: 10.3390/md12052827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albrecht M.G., Creighton J.A. Anomalously intense Raman-spectra of pyridine at a silver electrode. J. Am. Chem. Soc. 1977;99(15):5215–5217. doi: 10.1021/ja00457a071. [DOI] [Google Scholar]
- 43.Fleischmann M., Hendra P.J., Mcquillan A.J. Raman-spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974;26(2):163–166. doi: 10.1016/0009-2614(74)85388-1. [DOI] [Google Scholar]
- 44.Bodelon G., Montes-Garcia V., Costas C., Perez-Juste I., Perez-Juste J., Pastoriza-Santos I., Liz-Marzan L.M. Imaging bacterial interspecies chemical interactions by surface-enhanced Raman scattering. ACS Nano. 2017;11(5):4631–4640. doi: 10.1021/acsnano.7b00258. [DOI] [PubMed] [Google Scholar]
- 45.Liu S., Hu Q., Li C., Zhang F., Gu H., Wang X., Li S., Xue L., Madl T., Zhang Y., Zhou L. Wide-range, rapid, and specific identification of pathogenic bacteria by surface-enhanced Raman spectroscopy. ACS Sens. 2021;6(8):2911–2919. doi: 10.1021/acssensors.1c00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laucks M.L., Sengupta A., Junge K., Davis E.J., Swanson B.D. Comparison of psychro-active arctic marine bacteria and common mesophillic bacteria using surface-enhanced Raman spectroscopy. Appl. Spectrosc. 2005;59(10):1222–1228. doi: 10.1366/000370205774430891. [DOI] [PubMed] [Google Scholar]
- 47.Xu J.J., Turner J.W., Idso M., Biryukov S.V., Rognstad L., Gong H., Trainer V.L., Wells M.L., Strom M.S., Yu Q.M. In situ strain-level detection and identification of Vibrio parahaemolyticus using surface-enhanced Raman spectroscopy. Anal. Chem. 2013;85(5):2630–2637. doi: 10.1021/ac3021888. [DOI] [PubMed] [Google Scholar]
- 48.Cheng J.X., Xie X.S. Vibrational spectroscopic imaging of living systems: an emerging platform for biology and medicine. Science. 2015;350(6264) doi: 10.1126/science.aaa8870. [DOI] [PubMed] [Google Scholar]
- 49.Huang W.E., Ward A.D., Whiteley A.S. Raman tweezers sorting of single microbial cells. Env. Microbiol. Rep. 2009;1(1):44–49. doi: 10.1111/j.1758-2229.2008.00002.x. [DOI] [PubMed] [Google Scholar]
- 50.Kuhar N., Sil S., Umapathy S. Potential of Raman spectroscopic techniques to study proteins. Spectroc. Acta Pt. A-Molec. Biomolec. Spectr. 2021;258 doi: 10.1016/j.saa.2021.119712. [DOI] [PubMed] [Google Scholar]
- 51.Xu J.B., Yu T., Zois C.E., Cheng J.X., Tang Y.G., Harris A.L., Huang W.E. Unveiling cancer metabolism through spontaneous and coherent Raman spectroscopy and stable isotope probing. Cancers. 2021;13(7) doi: 10.3390/cancers13071718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Oliveira V.E., Castro H.V., Edwards H.G.M., de Oliveira L.F.C. Carotenes and carotenoids in natural biological samples: a Raman spectroscopic analysis. J. Raman Spectrosc. 2010;41(6):642–650. doi: 10.1002/jrs.2493. [DOI] [Google Scholar]
- 53.Alsafadi D., Khalili F.I., Juwhari H., Lahlouh B. Purification and biochemical characterization of photo-active membrane protein bacteriorhodopsin from Haloarcula marismortui, an extreme halophile from the Dead Sea. Int. J. Biol. Macromol. 2018;118:1942–1947. doi: 10.1016/j.ijbiomac.2018.07.045. [DOI] [PubMed] [Google Scholar]
- 54.Okada M., Smith N.I., Palonpon A.F., Endo H., Kawata S., Sodeoka M., Fujita K. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proc. Natl. Acad. Sci. U.S.A. 2012;109(1):28–32. doi: 10.1073/pnas.1107524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandair G.S., Han A.L., Keller E.T., Morris M.D. Raman microscopy of bladder cancer cells expressing green fluorescent protein. J. Biomed. Opt. 2016;21(11) doi: 10.1117/1.Jbo.21.11.115001. Artn 115001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stone N., Stavroulaki P., Kendall C., Birchall M., Barr H. Raman spectroscopy for early detection of laryngeal malignancy: preliminary results. Laryngoscope. 2000;110(10 Pt 1):1756–1763. doi: 10.1097/00005537-200010000-00037. [DOI] [PubMed] [Google Scholar]
- 57.Mussi V., Ledda M., Convertino A., Lisi A. Raman mapping of biological systems interacting with a disordered nanostructured surface: a simple and powerful approach to the label-free analysis of single DNA bases. Micromachines. 2021;12(3) doi: 10.3390/mi12030264. ARTN 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nawaz H., Bonnier F., Meade A.D., Lyng F.M., Byrne H.J. Comparison of subcellular responses for the evaluation and prediction of the chemotherapeutic response to cisplatin in lung adenocarcinoma using Raman spectroscopy. Analyst. 2011;136(12):2450–2463. doi: 10.1039/c1an15104e. [DOI] [PubMed] [Google Scholar]
- 59.Koch H., Polepil S., Eisen K., Will S. Raman microspectroscopy and multivariate data analysis: optical differentiation of aqueous D- and L-tryptophan solutions. Phys. Chem. Chem. Phys. 2017;19(45):30533–30539. doi: 10.1039/c7cp02321a. [DOI] [PubMed] [Google Scholar]
- 60.Perez-Estebanez M., Cheuquepan W., Cuevas-Vicario J.V., Hernandez S., Heras A., Colina A. Double fingerprint characterization of uracil and 5-fluorouracil. Electrochim. Acta. 2021;388 doi: 10.1016/j.electacta.2021.138615. [DOI] [Google Scholar]
- 61.Gaston H.B.H. Application of NIR Raman spectroscopy to probe the flexibility of RNA structure. Methods Mol. Biol. 2020;2113:149–164. doi: 10.1007/978-1-0716-0278-2_12. [DOI] [PubMed] [Google Scholar]
- 62.Gniadecka M., Wulf H.C., Mortensen N.N., Nielsen O.F., Christensen D.H. Diagnosis of basal cell carcinoma by Raman spectroscopy. J. Raman Spectrosc. 1997;28(2–3):125–129. doi: 10.1002/(SICI)1097-4555(199702)28. [DOI] [Google Scholar]
- 63.Maquelin K., Choo-Smith L.P., van Vreeswijk T., Endtz H.P., Smith B., Bennett R., Bruining H.A., Puppels G.J. Raman spectroscopic method for identification of clinically relevant microorganisms growing on solid culture medium. Anal. Chem. 2000;72(1):12–19. doi: 10.1021/ac991011h. [DOI] [PubMed] [Google Scholar]
- 64.Lakshmi R.J., Kartha V.B., Krishna C.M., Solomon J.G.R., Ullas G., Devi P.U. Tissue Raman spectroscopy for the study of radiation damage: brain irradiation of mice. Radiat. Res. 2002;157(2):175–182. doi: 10.1667/0033-7587(2002)157. [DOI] [PubMed] [Google Scholar]
- 65.Rao S., Raj S., Balint S., Fons C.B., Campoy S., Llagostera M., Petrov D. Single DNA molecule detection in an optical trap using surface-enhanced Raman scattering. Appl. Phys. Lett. 2010;96(21) doi: 10.1063/1.3431628. [DOI] [Google Scholar]
- 66.de Veij M., Vandenabeele P., De Beer T., Remonc J.P., Moens L. Reference database of Raman spectra of pharmaceutical excipients. J. Raman Spectrosc. 2009;40(3):297–307. doi: 10.1002/jrs.2125. [DOI] [Google Scholar]
- 67.Stone N., Kendall C., Smith J., Crow P., Barr H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004;126:141–157. doi: 10.1039/b304992b. [DOI] [PubMed] [Google Scholar]
- 68.Chiriboga L., Xie P., Yee H., Vigorita V., Zarou D., Zakim D., Diem M. Infrared spectroscopy of human tissue. I. Differentiation and maturation of epithelial cells in the human cervix. Biospectroscopy. 1998;4(1):47–53. doi: 10.1002/(Sici)1520-6343(1998)4:1<47::Aid-Bspy5>3.3.Co;2-1. [DOI] [PubMed] [Google Scholar]
- 69.Chan J.W., Taylor D.S., Zwerdling T., Lane S.M., Ihara K., Huser T. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys. J. 2006;90(2):648–656. doi: 10.1529/biophysj.105.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stone N., Kendall C., Shepherd N., Crow P., Barr H. Near-infrared Raman spectroscopy for the classification of epithelial pre-cancers and cancers. J. Raman Spectrosc. 2002;33(7):564–573. doi: 10.1002/jrs.882. [DOI] [Google Scholar]
- 71.Puppels G.J., Garritsen H.S.P., Kummer J.A., Greve J. Carotenoids located in human lymphocyte subpopulations and natural-killer-cells by Raman microspectroscopy. Cytometry. 1993;14(3):251–256. doi: 10.1002/cyto.990140303. [DOI] [PubMed] [Google Scholar]
- 72.Marcus M.A., Lewis A. Resonance Raman spectroscopy of the retinylidene chromophore in bacteriorhodopsin (bR570), bR560, M421, and other intermediates: structural conclusions based on kinetics, analogues, models, and isotopically labeled membranes. Biochemistry. 1978;17(22):4722–4735. doi: 10.1021/bi00615a019. [DOI] [PubMed] [Google Scholar]
- 73.Koljenovic S., Schut T.B., Vincent A., Kros J.M., Puppels G.J. Detection of meningioma in dura mater by Raman spectroscopy. Anal. Chem. 2005;77(24):7958–7965. doi: 10.1021/ac0512599. [DOI] [PubMed] [Google Scholar]
- 74.van Manen H.J., Kraan Y.M., Roos D., Otto C. Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes. Proc. Natl. Acad. Sci. U.S.A. 2005;102(29):10159–10164. doi: 10.1073/pnas.0502746102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kamemoto L.E., Misra A.K., Sharma S.K., Goodman M.T., Luk H., Dykes A.C., Acosta T. Near-infrared micro-Raman spectroscopy for in vitro detection of cervical cancer. Appl. Spectrosc. 2010;64(3):255–261. doi: 10.1366/000370210790918364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maiti N., Thomas S., Jacob J.A., Chadha R., Mukherjee T., Kapoor S. DFT and surface-enhanced Raman scattering study of tryptophan-silver complex. J. Colloid Interface Sci. 2012;380(1):141–149. doi: 10.1016/j.jcis.2012.04.071. [DOI] [PubMed] [Google Scholar]
- 77.Uzunbajakava N., Lenferink A., Kraan Y., Willekens B., Vrensen G., Greve J., Otto C. Nonresonant Raman imaging of protein distribution in single human cells. Biopolymers. 2003;72(1):1–9. doi: 10.1002/bip.10246. [DOI] [PubMed] [Google Scholar]
- 78.Lau D.P., Huang Z.W., Lui H., Man C.S., Berean K., Morrison M.D., Zeng H.S. Raman spectroscopy for optical diagnosis in normal and cancerous tissue of the nasopharynx - preliminary findings. Laser Surg. Med. 2003;32(3):210–214. doi: 10.1002/lsm.10084. [DOI] [PubMed] [Google Scholar]
- 79.Hanlon E.B., Manoharan R., Koo T.W., Shafer K.E., Motz J.T., Fitzmaurice M., Kramer J.R., Itzkan I., Dasari R.R., Feld M.S. Prospects for in vivo Raman spectroscopy. Phys. Med. Biol. 2000;45(2):R1–R59. doi: 10.1088/0031-9155/45/2/201. [DOI] [PubMed] [Google Scholar]
- 80.Silveira L., Sathaiah S., Zangaro R.A., Pacheco M.T.T., Chavantes M.C., Pasqualucci C.A.G. Correlation between near-infrared Raman spectroscopy and the histopathological analysis of atherosclerosis in human coronary arteries. Laser Surg. Med. 2002;30(4):290–297. doi: 10.1002/lsm.10053. [DOI] [PubMed] [Google Scholar]
- 81.Schulz H., Baranska M. Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib. Spectrosc. 2007;43(1):13–25. doi: 10.1016/j.vibspec.2006.06.001. [DOI] [Google Scholar]
- 82.Ciobota V., Burkhardt E.M., Schumacher W., Rosch P., Kusel K., Popp J. The influence of intracellular storage material on bacterial identification by means of Raman spectroscopy. Anal. Bioanal. Chem. 2010;397(7):2929–2937. doi: 10.1007/s00216-010-3895-1. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y., Song Y.Z., Tao Y.F., Muhamadali H., Goodacre R., Zhou N.Y., Preston G.M., Xu J., Huang W.E. Reverse and multiple stable isotope probing to study bacterial metabolism and interactions at the single cell level. Anal. Chem. 2016;88(19):9443–9450. doi: 10.1021/acs.analchem.6b01602. [DOI] [PubMed] [Google Scholar]
- 84.Maquelin K., Kirschner C., Choo-Smith L.P., van den Braak N., Endtz H.P., Naumann D., Puppels G.J. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods. 2002;51(3):255–271. doi: 10.1016/s0167-7012(02)00127-6. [DOI] [PubMed] [Google Scholar]
- 85.Huang N.Y., Short M., Zhao J.H., Wang H.Q., Lui H., Korbelik M., Zeng H.S. Full range characterization of the Raman spectra of organs in a murine model. Opt Express. 2011;19(23):22892–22909. doi: 10.1364/Oe.19.022892. [DOI] [PubMed] [Google Scholar]
- 86.Caspers P.J., Lucassen G.W., Carter E.A., Bruining H.A., Puppels G.J. In vivo confocal Raman microspectroscopy of the skin: noninvasive determination of molecular concentration profiles. J. Invest. Dermatol. 2001;116(3):434–442. doi: 10.1046/j.1523-1747.2001.01258.x. [DOI] [PubMed] [Google Scholar]
- 87.Krafft C., Neudert L., Simat T., Salzer R. Near infrared Raman spectra of human brain lipids. Spectroc. Acta Pt. A-Molec. Biomolec. Spectr. 2005;61(7):1529–1535. doi: 10.1016/j.saa.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 88.Demul F.F.M., Vanwelie A.G.M., Otto C. J. Mud and J. Greve. Micro-Raman spectroscopy of chromosomes. J. Raman Spectrosc. 1984;15(4):268–272. doi: 10.1002/jrs.1250150412. [DOI] [Google Scholar]
- 89.Leikin S., Parsegian V.A., Yang W.H., Walrafen G.E. Raman spectral evidence for hydration forces between collagen triple helices. Proc. Natl. Acad. Sci. U.S.A. 1997;94(21):11312–11317. doi: 10.1073/pnas.94.21.11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmidt T.M. The maturing of microbial ecology. Int. Microbiol. 2006;9(3):217–223. [PubMed] [Google Scholar]
- 91.Neufeld J.D., Wagner M., Murrell J.C. Who eats what, where and when? Isotope-labelling experiments are coming of age. ISME J. 2007;1(2):103–110. doi: 10.1038/ismej.2007.30. [DOI] [PubMed] [Google Scholar]
- 92.Berry D., Loy A. Stable-isotope probing of human and animal microbiome function. Trends Microbiol. 2018;26(12):999–1007. doi: 10.1016/j.tim.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y., Huang W.E., Cui L., Wagner M. Single cell stable isotope probing in microbiology using Raman microspectroscopy. Curr. Opin. Biotechnol. 2016;41:34–42. doi: 10.1016/j.copbio.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 94.Berry D., Mader E., Lee T.K., Woebken D., Wang Y., Zhu D., Palatinszky M., Schintlmeister A., Schmid M.C., Hanson B.T., Shterzer N., Mizrahi I., Rauch I., Decker T., Bocklitz T., Popp J., Gibson C.M., Fowler P.W., Huang W.E., Wagner M. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. U.S.A. 2015;112(2):E194–E203. doi: 10.1073/pnas.1420406112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cui L., Yang K., Li H.Z., Zhang H., Su J.Q., Paraskevaidi M., Martin F.L., Ren B., Zhu Y.G. Functional single-cell approach to probing nitrogen-fixing bacteria in soil communities by resonance Raman spectroscopy with 15N2 labeling. Anal. Chem. 2018;90(8):5082–5089. doi: 10.1021/acs.analchem.7b05080. [DOI] [PubMed] [Google Scholar]
- 96.Wang Y., Xu J., Kong L., Liu T., Yi L., Wang H., Huang W.E., Zheng C. Raman-deuterium isotope probing to study metabolic activities of single bacterial cells in human intestinal microbiota. Microb. Biotechnol. 2020;13(2):572–583. doi: 10.1111/1751-7915.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Notingher I., Hench L.L. Raman microspectroscopy: a noninvasive tool for studies of individual living cells in vitro. Expet Rev. Med. Dev. 2006;3(2):215–234. doi: 10.1586/17434440.3.2.215. [DOI] [PubMed] [Google Scholar]
- 98.Radajewski S., Ineson P., Parekh N.R., Murrell J.C. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403(6770):646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- 99.Manefield M., Whiteley A.S., Griffiths R.I., Bailey M.J. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 2002;68(11):5367–5373. doi: 10.1128/aem.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wilmes P., Heintz-Buschart A., Bond P.L. A decade of metaproteomics: where we stand and what the future holds. Proteomics. 2015;15(20):3409–3417. doi: 10.1002/pmic.201500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wegener G., Bausch M., Holler T., Thang N.M., Prieto Mollar X., Kellermann M.Y., Hinrichs K.-U., Boetius A. Assessing sub-seafloor microbial activity by combined stable isotope probing with deuterated water and 13C-bicarbonate. Environ. Microbiol. 2012;14(6):1517–1527. doi: 10.1111/j.1462-2920.2012.02739.x. [DOI] [PubMed] [Google Scholar]
- 102.Cui L., Yang K., Zhou G.W., Huang W.E., Zhu Y.G. Surface-enhanced Raman spectroscopy combined with stable isotope probing to monitor nitrogen assimilation at both bulk and single-cell level. Anal. Chem. 2017;89(11):5794–5801. doi: 10.1021/acs.analchem.6b04913. [DOI] [PubMed] [Google Scholar]
- 103.Song Y.Z., Cui L., Lopez J.A.S., Xu J.B., Zhu Y.G., Thompson I.P., Huang W.E. Raman-Deuterium Isotope Probing for in-situ identification of antimicrobial resistant bacteria in Thames River. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-16898-x. ARTN 16648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jehlička J., Edwards H.G.M., Oren A. Raman spectroscopy of microbial pigments. Appl. Environ. Microbiol. 2014;80(11):3286–3295. doi: 10.1128/AEM.00699-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Manen H.-J., Lenferink A., Otto C. Noninvasive imaging of protein metabolic labeling in single human cells using stable isotopes and Raman microscopy. Anal. Chem. 2008;80(24):9576–9582. doi: 10.1021/ac801841y. [DOI] [PubMed] [Google Scholar]
- 106.Wei L., Yu Y., Shen Y., Wang M.C., Min W. Vibrational imaging of newly synthesized proteins in live cells by stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. U.S.A. 2013;110(28):11226–11231. doi: 10.1073/pnas.1303768110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang W.E., Griffiths Robert I., Thompson Ian P., Bailey a. Mark J., Whiteley A.S. Raman microscopic analysis of single microbial cells. Anal. Chem. 2004;76(15):4452–4458. doi: 10.1021/ac049753k. [DOI] [PubMed] [Google Scholar]
- 108.Milucka J., Ferdelman T.G., Polerecky L., Franzke D., Wegener G., Schmid M., Lieberwirth I., Wagner M., Widdel F., Kuypers M.M.M. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature. 2012;491(7425):541. doi: 10.1038/nature11656. + [DOI] [PubMed] [Google Scholar]
- 109.Li M.Q., Canniffe D.P., Jackson P.J., Davison P.A., FitzGerald S., Dickman M.J., Burgess J.G., Hunter C.N., Huang W.E. Rapid resonance Raman microspectroscopy to probe carbon dioxide fixation by single cells in microbial communities. ISME J. 2012;6(4):875–885. doi: 10.1038/ismej.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jing X.Y., Gou H.L., Gong Y.H., Su X.L., Xu L., Ji Y.T., Song Y.Z., Thompson I.P., Xu J., Huang W.E. Raman-activated cell sorting and metagenomic sequencing revealing carbon-fixing bacteria in the ocean. Environ. Microbiol. 2018;20(6):2241–2255. doi: 10.1111/1462-2920.14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moter A., Gobel U.B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods. 2000;41(2):85–112. doi: 10.1016/S0167-7012(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 112.Frickmann H., Zautner A.E., Moter A., Kikhney J., Hagen R.M., Stender H., Poppert S. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: a review. Crit. Rev. Microbiol. 2017;43(3):263–293. doi: 10.3109/1040841x.2016.1169990. [DOI] [PubMed] [Google Scholar]
- 113.Lee N., Nielsen P.H., Andreasen K.H., Juretschko S., Nielsen J.L., Schleifer K.H., Wagner M. Combination of fluorescent in situ hybridization and microautoradiography - a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 1999;65(3):1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taylor G.T., Suter E.A., Li Z.Q., Chow S., Stinton D., Zaliznyak T., Beaupré S.R. Single-cell growth rates in photoautotrophic populations measured by stable isotope probing and resonance Raman microspectrometry. Front. Microbiol. 2017;8:1449. doi: 10.3389/fmicb.2017.01449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Escudero C., Del Campo A., Ares J.R., Sanchez C., Martinez J.M., Gomez F., Amils R. Visualizing microorganism-mineral interaction in the iberian pyrite belt subsurface: the Acidovorax case. Front. Microbiol. 2020;11:572104. doi: 10.3389/fmicb.2020.572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fernando E.Y., McIlroy S.J., Nierychlo M., Herbst F.A., Petriglieri F., Schmid M.C., Wagner M., Nielsen J.L., Nielsen P.H. Resolving the individual contribution of key microbial populations to enhanced biological phosphorus removal with Raman-FISH. ISME J. 2019;13(8):1933–1946. doi: 10.1038/s41396-019-0399-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Y., Ji Y.T., Wharfe E.S., Meadows R.S., March P., Goodacre R., Xu J., Huang W.E. Raman activated cell ejection for isolation of single cells. Anal. Chem. 2013;85(22):10697–10701. doi: 10.1021/ac403107p. [DOI] [PubMed] [Google Scholar]
- 118.Song Y.Z., Kaster A.K., Vollmers J., Song Y.Q., Davison P.A., Frentrup M., Preston G.M., Thompson I.P., Murrell J.C., Yin H.B., Hunter C.N., Huang W.E. Single-cell genomics based on Raman sorting reveals novel carotenoid-containing bacteria in the Red Sea. Microb. Biotechnol. 2017;10(1):125–137. doi: 10.1111/1751-7915.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pilat Z., Jonas A., Pilatova J., Klementova T., Bernatova S., Siler M., Manka T., Kizovsky M., Ruzicka F., Pantucek R., Neugebauer U., Samek O., Zemanek P. Analysis of bacteriophage-host interaction by Raman tweezers. Anal. Chem. 2020;92(18):12304–12311. doi: 10.1021/acs.analchem.0c01963. [DOI] [PubMed] [Google Scholar]
- 120.McIlvenna D., Huang W.E., Davison P., Glidle A., Cooper J., Yin H.B. Continuous cell sorting in a flow based on single cell resonance Raman spectra. Lab Chip. 2016;16(8):1420–1429. doi: 10.1039/c6lc00251j. [DOI] [PubMed] [Google Scholar]
- 121.Liao C.C., Chen Y.Z., Lin S.J., Cheng H.W., Wang J.K., Wang Y.L., Han Y.Y., Huang N.T. A microfluidic microwell device operated by the automated microfluidic control system for surface-enhanced Raman scattering-based antimicrobial susceptibility testing. Biosens. Bioelectron. 2021;191:113483. doi: 10.1016/j.bios.2021.113483. [DOI] [PubMed] [Google Scholar]
- 122.Camacho D.M., Collins K.M., Powers R.K., Costello J.C., Collins J.J. Next-generation machine learning for biological networks. Cell. 2018;173(7):1581–1592. doi: 10.1016/j.cell.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 123.Butler K.T., Davies D.W., Cartwright H., Isayev O., Walsh A. Machine learning for molecular and materials science. Nature. 2018;559(7715):547–555. doi: 10.1038/s41586-018-0337-2. [DOI] [PubMed] [Google Scholar]
- 124.Min S., Lee B., Yoon S. Deep learning in bioinformatics. Briefings Bioinf. 2017;18(5):851–869. doi: 10.1093/bib/bbw068. [DOI] [PubMed] [Google Scholar]
- 125.Libbrecht M.W., Noble W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015;16(6):321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lussier F., Thibault V., Charron B., Wallace G.Q., Masson J.F. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. Trac. Trends Anal. Chem. 2020;124 doi: 10.1016/j.trac.2019.115796. [DOI] [Google Scholar]
- 127.Schmidhuber J. Deep learning in neural networks: an overview. Neural Network. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 128.Cortes C., Vapnik V. Support-vector networks. Mach. Learn. 1995;20(3):273–297. doi: 10.1007/Bf00994018. [DOI] [Google Scholar]
- 129.Breiman L. Random forests. Mach. Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 130.Freund Y., Schapire R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997;55(1):119–139. doi: 10.1006/jcss.1997.1504. [DOI] [Google Scholar]
- 131.Yang K.K., Wu Z., Arnold F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods. 2019;16(8):687–694. doi: 10.1038/s41592-019-0496-6. [DOI] [PubMed] [Google Scholar]
- 132.Xu J.B., Yi X.F., Jin G.L., Peng D., Fan G.Y., Xu X.G., Chen X., Yin H.B., Cooper J.M., Huang W.E. High-speed diagnosis of bacterial pathogens at the single cell level by Raman microspectroscopy with machine learning filters and denoising autoencoders. ACS Chem. Biol. 2022;17(2):376–385. doi: 10.1021/acschembio.1c00834. [DOI] [PubMed] [Google Scholar]
- 133.Wang Y., Xu J., Cui D., Kong L., Chen S., Xie W., Zhang C. Classification and identification of archaea using single-cell Raman ejection and artificial intelligence: implications for investigating uncultivated microorganisms. Anal. Chem. 2021;93(51):17012–17019. doi: 10.1021/acs.analchem.1c03495. [DOI] [PubMed] [Google Scholar]
- 134.Ho C.S., Jean N., Hogan C.A., Blackmon L., Jeffrey S.S., Holodniy M., Banaei N., Saleh A.A.E., Ermon S., Dionne J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-12898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tang J.W., Liu Q.H., Yin X.C., Pan Y.C., Wen P.B., Liu X., Kang X.X., Gu B., Zhu Z.B., Wang L. Comparative analysis of machine learning algorithms on surface enhanced Raman spectra of clinical Staphylococcus species. Front. Microbiol. 2021;12:696921. doi: 10.3389/fmicb.2021.696921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yu S.X., Li X., Lu W.L., Li H.F., Fu Y.V., Liu F.H. Analysis of Raman spectra by using deep learning methods in the identification of marine pathogens. Anal. Chem. 2021;93(32):11089–11098. doi: 10.1021/acs.analchem.1c00431. [DOI] [PubMed] [Google Scholar]
- 137.Yu S.X., Li H.F., Li X., Fu Y.V., Liu F.H. Classification of pathogens by Raman spectroscopy combined with generative adversarial networks. Sci. Total Environ. 2020:726. doi: 10.1016/j.scitotenv.2020.138477. [DOI] [PubMed] [Google Scholar]
- 138.Wang K.D., Chen L., Ma X.Y., Ma L.N., Chou K.C., Cao Y.K., Khan I.U.H., Golz G., Lu X.N. Arcobacter identification and species determination using Raman spectroscopy combined with neural networks. Appl. Environ. Microbiol. 2020;86(20) doi: 10.1128/AEM.00924-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu Y.Y., Xu J.J., Tao Y., Fang T., Du W.B., Ye A.P. Rapid and accurate identification of marine microbes with single-cell Raman spectroscopy. Analyst. 2020;145(9):3297–3305. doi: 10.1039/c9an02069a. [DOI] [PubMed] [Google Scholar]
- 140.Heraud P., Beardall J., McNaughton D., Wood B.R. In vivo prediction of the nutrient status of individual microalgal cells using Raman microspectroscopy. FEMS Microbiol. Lett. 2007;275(1):24–30. doi: 10.1111/j.1574-6968.2007.00861.x. [DOI] [PubMed] [Google Scholar]
- 141.Zhang C.L.L., Xie W., Martin-Cuadrado A.B., Rodriguez-Valera F. Marine Group II Archaea, potentially important players in the global ocean carbon cycle. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.01108. [DOI] [PMC free article] [PubMed] [Google Scholar]