Proper reporting of metadata is essential to reproduce microscopy experiments, interpret results and share images1,2. The lack of methods reporting in microscopy is evident in that few research articles pass a test for the minimal information required to reproduce experiments1 (~17% of 240 articles with 1,500 figures with images). The problem is compounded by the number and variety of microscope modalities, options and associated components. Automation has distanced researchers from the technical parameters so it is difficult for them to know what information needs to be reported. MethodsJ2 is an ImageJ/Fiji based software tool that aims to improve reproducibility in microscopy.
To properly evaluate and reproduce microscopy images, information about sample preparation, experimental conditions, microscope hardware, image acquisition settings and image analysis parameters is required. This information is called “metadata” and is defined as “a set of data that describes and gives information about other data”. Researchers involved in the 4D Nucleome initiative3 and Bioimaging North America (BINA) (https://www.bioimagingna.org/) have developed extensive community driven Microscopy Metadata specifications4,5. These specifications build on a previous Open Microscopy Environment (OME) model6 and include an in-depth community driven Microscopy Metadata model for light microscopy termed “4DN-BINA-OME”4. The model scales with experimental design, instrument complexity and the degree to which image processing and quantitative image analysis is required for interpreting results. This ensures essential information is included while the burden on experimental scientists to collect and report metadata is minimized7.
Microscope Metadata guidelines8–10, examples of what can go wrong if metadata is not reported11 and the importance of measuring and reporting microscope quality control12 have been published. Increased awareness/education around Microscopy Metadata and straightforward accessible tools are vital for successful implementation. MethodsJ2 is an extensible open-source microscopy methods reporting software tool that runs in ImageJ/Fiji and builds on MethodsJ1,13,14. It captures Image Metadata from multiple sources, consolidates it and automatically generates methods text for publication. Integration with ImageJ/Fiji should make it broadly available to experimental scientists.
MethodsJ2 automatically gathers metadata from the image using OME BioFormats (e.g. pixel size, magnification) and captures Microscopy Metadata from a Microscope.JSON file generated using Micro-Meta App5,15. Micro-Meta App is a companion software tool that guides researchers step-by-step in the collection of community standardized Microscopy Metadata for a specific microscope4. MethodsJ2 also guides the user to enter specific Experimental/Sample Metadata (e.g. cell type, dyes). Finally, the software guides the user through a step-by-step validation of the metadata. To improve tracking of imaging facility impact, acknowledgement text, including a facility Research Resource ID (RRID, https://scicrunch.org/resources) can be added to the script. The methods text is then automatically generated but must be reviewed and edited.
Detailed MethodsJ2 workflow (Figure 1).
Figure 1: MethodsJ2 Workflow Overview.
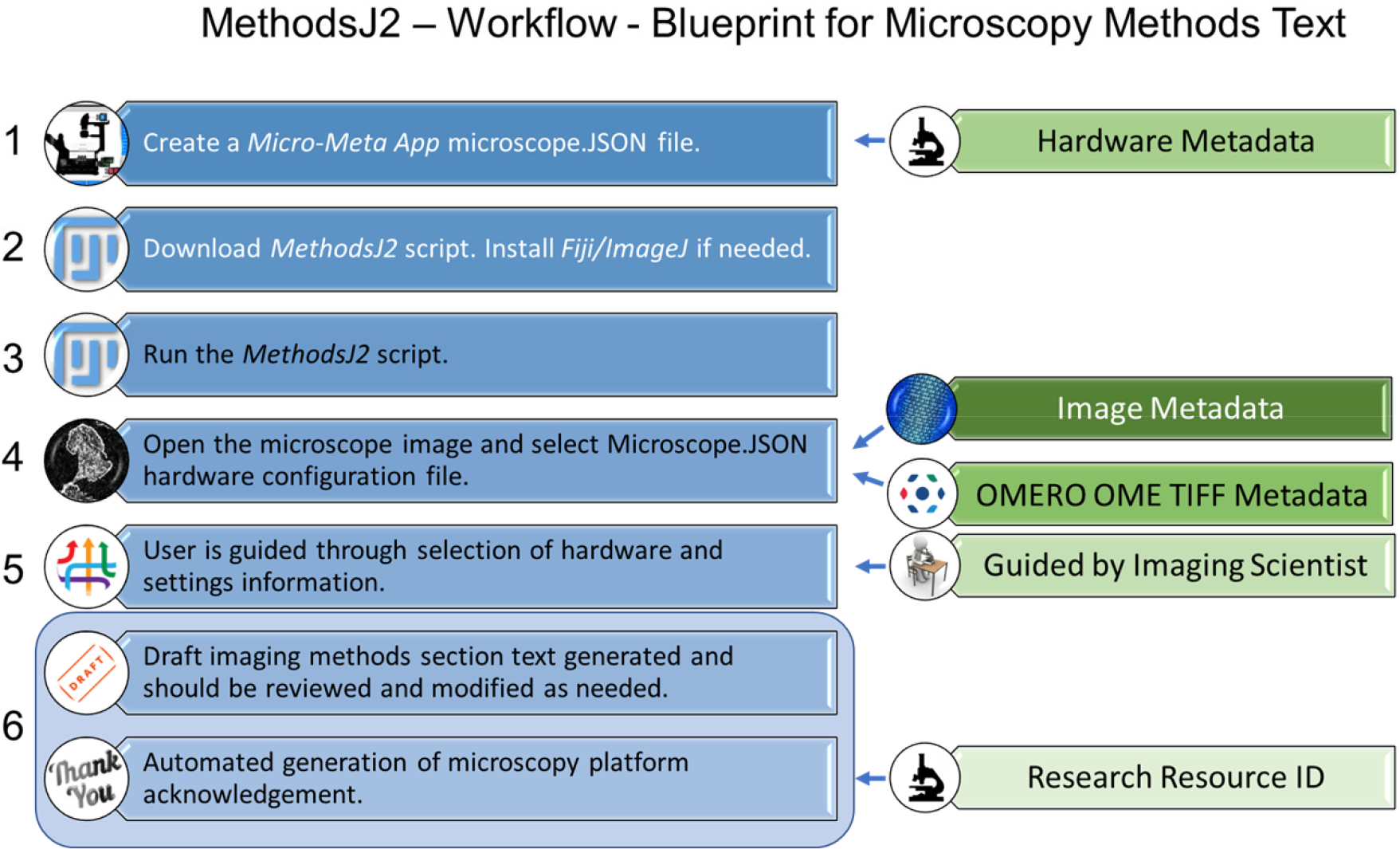
Steps required to automatically generate microscopy methods text. Image metadata is collected from the manufacturer metadata in the image file using the OME TIFF tools. Hardware metadata is collected from a Micro-Meta App Microscope.JSON file. It is recommended to have an experienced microscopist or imaging scientist guide researchers through the methods text generation and validation process. Acknowledgement text can be added to the script by imaging scientists in the microscopy platform including a RRID.
Supplemental materials provide a more detailed workflow and sample Microscope Metadata.
Use Micro-Meta App to create and save a Microscope.JSON file. Note: Give components detailed names as this text populates the methods text. Put “63x/1.4 NA Plan-Apochromatic oil immersion” not “63x”.
Download the MethodsJ2 script (file named: MethodsJ2_v1_2_.py), an example Microscope.JSON file and an example image file from GitHub (https://github.com/ABIF-McGill/MethodsJ2). Download and install ImageJ/Fiji (https://fiji.sc/).
Drag the MethodsJ2 script file and drop it on the ImageJ/Fiji toolbar. The Script Editor will open, then press “Run”.
Select an image file. The Image Metadata is automatically extracted. Sample information can be added manually. Select a Microscope.JSON file for the corresponding microscope.
The user is guided step-by-step to validate the metadata and input critical hardware and settings information. Note: Have an experienced microscope user or imaging scientist help with this step.
Click “OK”. Draft text is automatically generated and appears in a popup window, is copied to the clipboard and can be pasted into a manuscript. A .csv file of the Microscope Metadata is generated and saved. See the sample .csv file included as supplemental material and on the GitHub portal. Note: It is the responsibility of the experimental scientists to review the draft text to ensure it is accurate.
Comprehensive methods reporting is essential for reporting imaging data, sharing images and emerging new methods16–22. Progress along the path of rigor and reproducibility is essential for high quality microscope-based science and is a shared responsibility. Experimental scientists must use due diligence to understand the fundamentals of the technologies and required Microscope Metadata their research relies on. Imaging scientists need to educate experimental scientists, so they understand what metadata needs to be reported and why. Microscope manufacturers ought to integrate, automate and report Microscope Metadata. Scientific publishers and reviewers have a duty to promote community-based guidelines4,6,23 and ensure published microscope images meets a minimum standard. Funding agencies need to uphold high quality reproducible microscope images and ensure detailed Microscopy Metadata is available when images are publicly shared.
MethodsJ2 and two companion software tools, Micro-Meta App15 and OMERO.mde23, advance rigor and reproducibility in microscopy (Supplemental Figure), but there are still challenges. Microscope Metadata is often limited, not in standard formats, not accessible due to proprietary microscope manufacturer software and/or lost when images are saved and opened with third-party software4. Microscope manufacturers need to work with the global community through organizations like Quality Assessment and Reproducibility for Instruments & Images in Light Microscopy (QUAREP-LiMi)24,25 to automate metadata collection, ensure it conforms to community standards4,6,23 and make it readily available. Implementation and evolution of MethodsJ2, Micro-Meta App15 and OMERO.mde23, will advance rigor and reproducibility in microscopy, promote transparency and reproducibility and help stakeholders ensure Microscopy Metadata is documented and reported.
Supplementary Material
ACKNOWLEDGEMENTS:
We thank our microscopy core facility staff and users of McGill University Advanced BioImaging Facility (ABIF) (RRID:SCR_017697), University Imaging Centers of the University of Minnesota (RRID:SCR_020997), MicRoN (Microscopy Resources on the North Quad) Core at Harvard Medical School, UNC Neuroscience Microscopy Core (RRID:SCR_019060) (supported, in part, NIH-NINDS Neuroscience Center Support Grant P30 NS045892, NIH-NICHD Intellectual and Developmental Disabilities Research Center Support Grant P50 HD103573) and Duke University Light Microscopy Core Facility. Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation supports C.M.B. (Grant# 2020–225398), C.S.-D.-C. (Grant# 2019–198155 (5022)) and M.S.I. (Grant# 2019– 198107). NIH Grants #2U01CA200059–06, #1U01EB021238 to C.S.-D.-C.
Footnotes
AUTHOR CONTRIBUTIONS
(based on CRediT Contributor Roles Taxonomy https://casrai.org/credit/):
J.R. Conceptualization, Software Development, Validation, Data Curation, Writing Review & Editing, T.P. Conceptualization, Software Development, Validation, A.R. Conceptualization, SoftwareDevelopment, Validation, P.M.L. Conceptualization, Validation, Writing Review & Editing, M.S.I. Conceptualization, Validation, L.A.C. Conceptualization, Validation, Writing Review & Editing, G.M. Conceptualization, Validation, C.S.-D.-C. Conceptualization, Software, Validation, Supervision, Funding Acquisition, M.A.S. Conceptualization, Writing Review & Editing, C.M.B. Conceptualization, Validation, Writing Original Draft, Writing Review & Editing, Supervision, Project Administration, Funding Acquisition
DATA AND CODE AVAILABILITY STATEMENT:
Data in the form of a sample image and MICROSCOPE.JSON file are available at https://github.com/ABIF-McGill/MethodsJ2. Full source code and step-by-step instructions are available at https://github.com/ABIF-McGill/MethodsJ2 and 10.5281/zenodo.5172827.
REFERENCES
- 1.Marques G, Pengo T & Sanders MA Imaging methods are vastly underreported in biomedical research. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JY & Kitaoka M A beginner’s guide to rigor and reproducibility in fluorescence imaging experiments. Mol Biol Cell 29, 1519–1525, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’Shea CC, Park PJ, Ren B et al. The 4D nucleome project. Nature 549, 219–226, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammer M, Huisman M, Rigano A, Boehm U, Chambers JJ, Gaudreault N, North AJ, Pimentel JA, Sudar D, Bajcsy P et al. Towards community-driven metadata standards for light microscopy: tiered specifications extending the OME model. bioRxiv, 2021.2004.2025.441198, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigano A, Boehm U, Chambers J, Gaudreault N, North A, Pimentel J, Sudar D, Bajcsy P, Brown C, Corbett A et al. 4DN-BINA-OME (NBO) Tiered Microscopy Metadata Specifications - v2.01, <https://github.com/WU-BIMAC> (2021).
- 6.Goldberg IG, Allan C, Burel JM, Creager D, Falconi A, Hochheiser H, Johnston J, Mellen J, Sorger PK & Swedlow JR The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol 6, R47, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huisman M, Hammer M, Rigano A, Boehm U, Chambers JJ, Gaudreault N, North AJ, Pimentel JA, Sudar D, Bajcsy P et al. A perspective on Microscopy Metadata: data provenance adn quality control. ArXiv abs/1910.11370, (2021). [Google Scholar]
- 8.Aaron JS & Chew T-L A guide to accurate reporting in digital image acquisition: can anyone replicate your microscopy data? J Cell Sci 134, (2021). [DOI] [PubMed] [Google Scholar]
- 9.Linkert M, Rueden CT, Allan C, Burel JM, Moore W, Patterson A, Loranger B, Moore J, Neves C, Macdonald D et al. Metadata matters: access to image data in the real world. J Cell Biol 189, 777–782, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heddleston JM, Aaron JS, Khuon S & Chew T-L A guide to accurate reporting in digital image processing: can anyone reproduce your quantitative analysis? J Cell Sci 134, (2021). [DOI] [PubMed] [Google Scholar]
- 11.Montero Llopis P, Senft RA, Ross-Elliott TJ, Stephansky R, Keeley DP, Koshar P, Marques G, Gao YS, Carlson BR, Pengo T et al. Best practices and tools for reporting reproducible fluorescence microscopy methods. Nat Methods, (2021). [DOI] [PubMed] [Google Scholar]
- 12.Nelson G, Gelman L, Faklaris O, Nitschke R & Laude A Interpretation of Confocal ISO 21073: 2019 confocal microscopes: Optical data of fluorescence confocal microscopes for biological imaging- Recommended Methodology for Quality Control. arXiv, (2020). [Google Scholar]
- 13.Ryan J, Pengo T, Rigano A, Llopis PM, Itano MS, Cameron LA, Marqués G, Strambio-De-Castillia C, Sanders MA & Brown CM MethodsJ2: A Software Tool to Improve Microscopy Methods Reporting. bioRxiv 1, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan J, Pengo T, Rigano A, Llopis PM, Itano MS, Cameron LA, Marqués G, Strambio-De-Castillia C, Sanders MA & Brown CM MethodsJ2: v1.2. Zenodo 1, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rigano A, Ehmsen S, Ozturk SU, Ryan J, Balashov A, Hammer M, Kirli K, Bellve K, Boehm U, Brown CM et al. Micro-Meta App: an interactive software tool to facilitate the collection of microscopy metadata based on community-driven specifications. bioRxiv, 2021.2005.2031.446382, (2021). [Google Scholar]
- 16.Ellenberg J, Swedlow JR, Barlow M, Cook CE, Sarkans U, Patwardhan A, Brazma A & Birney E A call for public archives for biological image data. Nat Methods 15, 849–854, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyakawa T No raw data, no science: another possible source of the reproducibility crisis. Mol Brain 13, 24, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sansone SA, McQuilton P, Rocca-Serra P, Gonzalez-Beltran A, Izzo M, Lister AL, Thurston M & Community FA FAIRsharing as a community approach to standards, repositories and policies. Nat Biotechnol 37, 358–367, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Botvinik-Nezer R, Holzmeister F, Camerer CF, Dreber A, Huber J, Johannesson M, Kirchler M, Iwanir R, Mumford JA, Adcock RA et al. Variability in the analysis of a single neuroimaging dataset by many teams. Nature 582, 84–88, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheen MR, Fields JL, Northan B, Lacoste J, Ang LH, Fiering S & Reproducibility Project: Cancer, B. Replication Study: Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosselin RD Statistical Analysis Must Improve to Address the Reproducibility Crisis: The ACcess to Transparent Statistics (ACTS) Call to Action. Bioessays 42, e1900189, (2020). [DOI] [PubMed] [Google Scholar]
- 22.Gibney E This AI researcher is trying to ward off a reproducibility crisis. Nature 577, 14, (2020). [DOI] [PubMed] [Google Scholar]
- 23.Kunis S, Hänsch S, Schmidt C, Wong F & Weidtkamp-Peters S OMERO.mde in a use case for microscopy metadata harmonization: Facilitating FAIR principles in practical application with metadata annotation tools. arXiv, (2021). [Google Scholar]
- 24.Boehm U, Nelson G, Brown CM, Bagley S, Bajcsy P, Bischof J, Dauphin A, Dobbie IM, Eriksson JE, Faklaris O et al. QUAREP-LiMi: a community endeavor to advance quality assessment and reproducibility in light microscopy. Nat Methods, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson G, Boehm U, Bagley S, Bajcsy P, Bischof J, Brown CM, Dauphin A, Dobbie IM, Eriksson JE, Faklaris O et al. QUAREP-LiMi: A community-driven initiative to establish guidelines for quality assessment and reproducibility for instruments and images in light microscopy. J Microsc, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data in the form of a sample image and MICROSCOPE.JSON file are available at https://github.com/ABIF-McGill/MethodsJ2. Full source code and step-by-step instructions are available at https://github.com/ABIF-McGill/MethodsJ2 and 10.5281/zenodo.5172827.


