Nitrogen is a quantitatively important bioelement which is incorporated into the biosphere through assimilatory processes carried out by microorganisms and plants. Numerous nitrogen-containing compounds can be used by different organisms as sources of nitrogen. These include, for instance, inorganic ions like nitrate or ammonium and simple organic compounds like urea, amino acids, and some nitrogen-containing bases. Additionally, many bacteria are capable of fixing N2. Nitrogen control is a phenomenon that occurs widely among microorganisms and consists of repression of the pathways of assimilation of some nitrogen sources when some other, more easily assimilated source of nitrogen is available to the cells. Ammonium is the preferred nitrogen source for most bacteria, but glutamine is also a very good source of nitrogen for many microorganisms. Two thoroughly investigated nitrogen control systems are the NtrB-NtrC two-component regulatory system found in enterics and some other proteobacteria (80) and the GATA family global nitrogen control transcription factors of yeast and some fungi (75). Novel nitrogen control systems have, however, been identified in bacteria other than the proteobacteria, like Bacillus subtilis (26), Corynebacterium glutamicum (52), and the cyanobacteria. The cyanobacterial system is the subject of this review.
The cyanobacteria are prokaryotes that belong to the Bacteria domain and are characterized by the ability to perform oxygenic photosynthesis. Cyanobacteria have a wide ecological distribution, and they occupy a range of habitats, which includes vast oceanic areas, temperate soils, and freshwater lakes, and even extreme habitats like arid deserts, frigid lakes, or hot springs. Photoautotrophy, fixing CO2 through the Calvin cycle, is the dominant mode of growth of these organisms (109). A salient feature of the intermediary metabolism of cyanobacteria is their lack of 2-oxoglutarate dehydrogenase (109). As a consequence, they use 2-oxoglutarate mainly as a substrate for the incorporation of nitrogen, a metabolic arrangement that may have regulatory consequences. Notwithstanding their rather homogeneous metabolism, cyanobacteria exhibit remarkable morphological diversity, being found as either unicellular or filamentous forms and exhibiting a number of cell differentiation processes, some of which take place in response to defined environmental cues, as is the case for the differentiation of N2-fixing heterocysts (109).
Nitrogen control in cyanobacteria is mediated by NtcA, a transcriptional regulator which belongs to the CAP (the catabolite gene activator or cyclic AMP [cAMP] receptor protein) family and is therefore different from the well-characterized Ntr system. Interestingly, however, the signal transduction PII protein, which plays a key role in Ntr regulation, is found in cyanobacteria but with characteristics which differentiate it from proteobacterial PII. In the following paragraphs, we shall first briefly summarize our current knowledge of the cyanobacterial nitrogen assimilation pathways and of what is known about their regulation at the protein level. This description will introduce most of the known cyanobacterial nitrogen assimilation genes. We shall then describe the ntcA gene and the NtcA protein themselves to finally discuss NtcA function through a survey of the NtcA-regulated genes which participate in simple nitrogen assimilation pathways or in heterocyst differentiation and function.
GENES OF NITROGEN ASSIMILATION PATHWAYS RENDERING INTRACELLULAR AMMONIUM
The nitrogen sources most commonly used by cyanobacteria are nitrate, ammonium, urea, and dinitrogen (27). The unicellular cyanobacterium Synechococcus sp. strain PCC 7942 has also been shown to derive ammonium from cyanate (81). In this section, we shall describe the genetic structure of the systems involved in the assimilation of these nutrients, focusing on those cyanobacteria in which these systems have been studied in more detail.
Nitrate assimilation.
The assimilation of nitrate involves its incorporation into the cell through an active transport system and its intracellular two-step reduction to ammonium sequentially catalyzed by ferredoxin-nitrate reductase and ferredoxin-nitrite reductase. An ATP-binding cassette (ABC)-type transporter constituted by the products of the nrtA, nrtB, nrtC, and nrtD genes is involved in nitrate-nitrite uptake by the freshwater cyanobacteria Synechococcus sp. strain PCC 7942 and Anabaena sp. strain PCC 7120 (13, 35, 68, 72, 93), whereas a carrier belonging to the major facilitator superfamily mediates nitrate-nitrite uptake in the marine cyanobacteria Synechococcus sp. strain PCC 7002 (103) and Trichodesmium sp. strain WH9601 (122). The gene encoding this carrier has been named nrtP (103) or napA (122). Because a component of a well-known periplasmic nitrate reductase has previously been designated NapA (84), we suggest that these permeases be named NrtP. Whereas the ABC-type nitrate-nitrite transporter likely uses ATP as a source of energy (27), it is unknown whether the NrtP permeases use a gradient of H+ or Na+ (103). It has been suggested that nitrate utilization by cyanobacteria in saline environments may be facilitated by NapA (NrtP) rather than NrtABCD transporters (122), but NrtP homologues are also present in nonmarine cyanobacteria like Nostoc punctiforme (DOE Joint Genome Institute [http://www.jgi.doe.gov/]).
In Synechococcus sp. strain PCC 7942 and Anabaena sp. strain PCC 7120, the transporter-encoding genes are clustered together with the structural genes for nitrite reductase, nirA (67), and nitrate reductase, narB (102), constituting an operon, which we call the nir operon, with the structure nirA-nrtABCD-narB (13, 35, 70, 93, 112). This operon is expressed at high levels only when ammonium is not present in the growth medium, with nitrate or nitrite having some positive effect on the operon mRNA levels (35, 60). In both strains, upstream from the nir operon, in the opposite DNA strand, a number of genes whose mutation results in different degrees of impairment of expression of the nitrate and nitrite reductases have been found (36, 111). One of those genes, ntcB, encoding a protein which belongs to the LysR family of transcriptional regulators, is present in both strains and is ammonium repressible. NtcB has been suggested to mediate a positive effect of nitrate (via nitrite) on the expression of the nir operon in Synechococcus sp. strain PCC 7942 (1), but in Anabaena sp. strain PCC 7120, it is required for expression of the nir operon irrespective of the presence of nitrate (36). In the genome of Synechocystis sp. strain PCC 6803, the nrtABCD and narB genes are clustered together whereas the nirA gene is found in a different location (57).
In Synechococcus sp. strain PCC 7002, the nrtP gene is clustered with narB but they appear to be transcribed independently. The expression of both genes is, however, regulated in a similar way, being high in nitrate-containing medium and low in medium containing ammonium or urea (103). In Trichodesmium sp. strain WH9601, a gene cluster with the structure nirA-napA(nrtP)-narB is found but its expression has not yet been characterized (122).
A set of genes required for the biosynthesis of the molybdenum cofactor of nitrate reductase, bis-molybdopterin guanine dinucleotide, has been characterized in Synechococcus sp. strain PCC 7942. However, the expression of these genes appears not to be tightly regulated by the nitrogen source (100, 101).
Nitrogen fixation.
Many cyanobacteria fix atmospheric nitrogen, and a large number of them do so under aerobic conditions. Because nitrogenase, the enzymatic complex performing nitrogen fixation, is extremely oxygen sensitive, many cyanobacteria separate, either spatially or temporarily, the processes of oxygenic photosynthesis and nitrogen fixation. Whereas some filamentous cyanobacteria (e.g., those of the genera Anabaena and Nostoc) confine nitrogenase to heterocysts, differentiated cells specialized in nitrogen fixation, some other, unicellular as well as filamentous, strains express the nitrogenase activity in the dark periods of light-dark growth cycles. On the other hand, Trichodesmium sp., a nitrogen-fixing cyanobacterium of global ecological significance, fixes N2 aerobically and expresses nitrogenase in the light periods of light-dark growth cycles (15). Nitrogen fixation genes will be described here briefly, and the reader is invited to consult a number of recent reviews that cover different aspects of heterocyst formation and cyanobacterial nitrogen fixation (3, 5, 10, 27, 28, 42, 44, 127–129).
Nitrogen fixation genes have been identified in a number of cyanobacteria (reviewed in reference 28), but they have been characterized in more detail in two heterocyst formers, Anabaena sp. strain PCC 7120 and Anabaena variabilis strain ATCC 29413 (reviewed in reference 5), and in the unicellular organism Synechococcus sp. strain RF-1 (49). The nomenclature of the cyanobacterial nif genes is like that of other bacteria (5). Fifteen nitrogen fixation or nitrogen fixation-related genes, including the structural genes for nitrogenase, nifHDK, are clustered together as follows: nifB-fdxN-nifS-nifU-nifH-nifD- nifK-nifE-nifN-nifX-orf2-nifW-hesA-hesB-fdxH. These genes are organized in at least six transcriptional units: nifB-fdxN-nifS-nifU, nifHDK, nifEN, nifX-orf2, nifW-hesA-hesB, and fdxH (28, 44, 49). Upstream from nifB, in the opposite orientation, a nifP gene has been found in strain RF-1 (49), and downstream from fdxH, in the opposite orientation, a mop gene has been identified in the heterocyst formers (5). On the other hand, the nifJ gene and, clustered together, nifV, nifZ, and nifT are found in other locations in the chromosome of Anabaena sp. strain PCC 7120 (5). A gene designated glbN, which encodes a monomeric hemoglobin, has been found juxtaposed to nifU and nifH in Nostoc commune strain UTEX 584 (94) and is present in some other Nostoc strains as well (48).
The nif and nitrogen fixation-related genes are not expressed in cultures supplemented with combined nitrogen (28, 49). Regulation of expression of these genes, however, can be especially complex, since expression in some cyanobacteria appears to be subject to a circadian rhythm (17, 21, 49) and can be confined to the heterocysts in the heterocyst formers (24). Some heterocyst-forming cyanobacteria, like A. variabilis strain ATCC 29413, bear a second nif gene cluster that is expressed in the vegetative cells under microaerobic or anaerobic conditions (105, 117). This cyanobacterium also bears genes for a vanadium-dependent nitrogenase: vnfDG, vnfK, vnfE, and vnfN (115, 116).
Urea assimilation.
Urea likely represents an important nitrogen source for cyanobacteria in their natural environment (20, 27). Recent advances in our knowledge of cyanobacterial urea assimilation include the identification of the urease structural genes and of the genes encoding a urea transporter. The urease structural genes, ureA, ureB, and ureC, which would encode a typical bacterial urease, as well as all of the accessory genes required to produce an active urease, an enzyme which carries a bi-nickel center, have been identified in the marine cyanobacterium Synechococcus sp. strain WH7805 (20) and are also present in the genomes of Synechocystis sp. strain PCC 6803 (57) and Anabaena sp. strain PCC 7120 (Kazusa DNA Research Institute [http://www.kazusa.or.jp/cyano/anabaena/]). Both constitutive and ammonium-repressible ureases have been described in cyanobacteria (20, 27). An operon composed of five genes encoding an ABC-type transporter which exhibits high affinity for urea and is repressed by growth in the presence of ammonium has been characterized in Anabaena sp. strain PCC 7120 (A. Valladares, M. L. Montesinos, A. Herrero, and E. Flores, GenBank accession no. AJ271599).
Cyanate degradation.
Cyanase catalyzes the decomposition of cyanate into CO2 and ammonium. High levels of cyanase activity have been detected in Synechococcus sp. strain PCC 7942 and Synechocystis sp. strain PCC 6803 (43, 81), and the cynS gene encoding cyanase has been identified in both strains (43; F. Jalali and G. S. Espie, GenBank accession no. AF001333). In Synechococcus sp. strain PCC 7942, cynS is clustered together with three genes, cynABD, which might encode an ABC-type transporter. Cyanase activity and cynS and its accompanying genes are subject to repression by ammonium (43).
Ammonium uptake.
Although ammonia appears to readily permeate biological membranes, ammonium transporters have been characterized in numerous organisms. They are monocomponent permeases which seem to be necessary for uptake of ammonium when it is present at low concentrations (i.e., below 1 μM) in the extracellular medium or when the organisms grow in a rather acidic medium (which is not the case for cyanobacteria). Three genes encoding ammonium permeases are found in the chromosome of Synechocystis sp. strain PCC 6803 (57, 82). The three genes are expressed at the highest levels when Synechocystis cells are incubated in a medium lacking a source of nitrogen, but one of them, amt1, is responsible for most of the [14C]methylammonium transport activity when this substrate is provided at micromolar concentrations (82). A homologue of Amt1 has been characterized as responsible for ammonium-methylammonium uptake in Synechococcus sp. strain PCC 7942 (M. F. Vázquez-Bermúdez, A. Herrero, and E. Flores, unpublished data), and amt homologues are present in heterocyst-forming cyanobacteria and unicellular marine strains whose genomes have been sequenced (Kazusa DNA Research Institute [http://www.kazusa.or.jp/cyano/anabaena/]; DOE Joint Genome Institute [http://www.jgi.doe.gov/]).
AMMONIUM INCORPORATION INTO CARBON SKELETONS
Glutamine synthetase-glutamate synthase pathway.
In cyanobacteria, ammonium, either taken up from the outer medium or produced intracellularly, is incorporated into carbon skeletons mainly through the glutamine synthetase-glutamate synthase cycle (130; reviewed in reference 27). The glnA gene encoding a typical eubacterial glutamine synthetase (type I) has been cloned from numerous cyanobacteria (reviewed in reference 27), but a second gene, glnN, encoding a type III glutamine synthetase is also present in some nondiazotrophic strains (97). Pseudanabaena sp. strain PCC 6903 has been found to carry glnN as its only glutamine synthetase gene (23). In general, the expression of glnA (reviewed in reference 28) and glnN (23, 98, 104) is lower in cells grown with ammonium than in those grown with nitrate or incubated in the absence of combined nitrogen.
The genes encoding a ferredoxin-dependent (glsF) and an NADH-dependent (gltB and gltD) glutamate synthase have been characterized in Plectonema boryanum (91). Synechocystis sp. strain PCC 6803 appears to bear similar genes (57). The contribution of ferredoxin-glutamate synthase to the photoassimilation of nitrogen is dominant over that of NADH-glutamate synthase when high levels of carbon are available (91). Only one glutamate synthase gene, glsF, encoding a ferredoxin-dependent enzyme has been found in Anabaena sp. strain PCC 7120 (74; Kazusa DNA Research Institute [http://www.kazusa.or.jp/cyano/anabaena/]). No regulation of glutamate synthase expression by the nitrogen source has been observed (27).
Carbon skeleton for nitrogen incorporation.
The carbon skeleton used for incorporation of ammonium through the glutamine synthetase-glutamate synthase cycle is 2-oxoglutarate, which is provided by NADP+-isocitrate dehydrogenase (86, 87). The expression of the icd gene encoding isocitrate dehydrogenase has been described to be highest under nitrogen stress in Synechocystis sp. strain PCC 6803 or under N2-fixing conditions in Anabaena sp. strain PCC 7120 (88). As mentioned above, cyanobacteria lack a 2-oxoglutarate dehydrogenase and use 2-oxoglutarate mainly for the biosynthesis of glutamate and glutamate-derived compounds (50, 108). Consistent with this, in short-term uptake assays carried out with a Synechococcus strain bearing an heterologous 2-oxoglutarate permease, most of the radioactivity from the 2-[14C]oxoglutarate taken up was found in glutamate and glutamine (119). Its predominant role as a carbon skeleton for nitrogen assimilation would make 2-oxoglutarate an appropriate sensor of the C-N balance of the cyanobacterial cell (30). Although 2-oxoglutarate cellular levels have not been extensively analyzed, available data indicate that they are indeed influenced by the nitrogen source (22, 76, 114).
POSTTRANSLATIONAL REGULATION
Most of the genes encoding nitrogen assimilation enzymes or permeases are repressed by ammonium in cyanobacteria, and this transcriptional control, which will be discussed below, is a major aspect of regulation of nitrogen assimilation in these organisms. Nonetheless, a few examples of posttranslational regulation are also well documented: modification of nitrogenase reductase; reversible inactivation, triggered by ammonium, of glutamine synthetase in Synechocystis sp. strain PCC 6803; and short-term inhibition by ammonium of nitrate uptake.
Nitrogenase reductase modification.
In cyanobacteria, the Fe protein of the nitrogenase complex is frequently resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis into two forms, the largest of which (modified form) appears to correspond, in some cases, to an inactive protein (reviewed in reference 3). Physiological conditions which lead to the appearance of the modified form include incubation in the dark, under high O2 tension, or in the presence of ammonium. Because of an apparent similarity to the inactivation of the Rhodospirillum rubrum nitrogenase Fe protein, ADP-ribosylation has been investigated as a possible inactivation mechanism but no proof for it has been obtained (3). Instead, it has recently been reported that modification of the nitrogenase Fe protein in the unicellular cyanobacterium Gloeothece sp. strain ATCC 27152 could operate through palmitoylation of the protein (38).
Inactivation of glutamine synthetase.
The presence of ammonium in the culture medium causes inactivation of the type I glutamine synthetase in Synechocystis sp. strain PCC 6803, with reactivation taking place in response to ammonium withdrawal (76). The in vivo-inactivated enzyme can be reactivated in vitro by several treatments, including incubation with a phosphatase, leading to the suggestion that inactivation involves the noncovalent binding of a phosphorylated compound (77). It has recently been shown that inactivation involves binding to the enzyme of one or two of the inactivating factors IF7 and IF17, the products of the gifA and gifB genes, respectively (40). These genes are quickly induced in response to the addition of ammonium to Synechocystis cells. Inactivation of the enzyme might simply be determined by the presence of adequate levels of IF7 and IF17 in the cells (40), whereas no role for a phosphorylation in this inactivation system has been described. IF7 and IF17 are homologous polypeptides, and a homologue is also found in Anabaena sp. strain PCC 7120. An ammonium-triggered decrease (other than repression) in glutamine synthetase has not been observed, however, in cyanobacteria, other than strain PCC 6803, in which the glutamine synthetase levels have been examined (78, 79; however, see reference 99).
Inhibition of nitrate uptake.
The nitrate-nitrite transporter of many cyanobacteria requires CO2 fixation by the cells for normal operation and is subject to quick and reversible inhibition by ammonium, which has to be metabolized through glutamine synthetase for the inhibitory effect to take place (reviewed in reference 27). Nitrate uptake appears, therefore, to be modulated by a regulatory system integrating the photosynthetic N and C assimilatory metabolisms of cyanobacteria (29). It has recently been shown that the signaling PII protein (glnB gene product) is required for the inhibition by ammonium of nitrate uptake in Synechococcus sp. strain PCC 7942 (63). PII is a trimeric protein which, in cyanobacteria, can be modified by phosphorylation of a serine residue in each subunit (32), a modification mechanism different from that previously known for PII, which, in some other bacteria, is uridylylated under low-nitrogen conditions (80). The phosphorylation degree of the trimer responds to the N and C supply of the cells so that PII is unphosphorylated in the presence of ammonium and phosphorylated to different degrees when the cells are incubated in the presence of nitrate or in the absence of any source of nitrogen and depending also on the CO2 supply (32). As is the case with Escherichia coli PII (56, 80), Synechococcus PII can bind ATP and 2-oxoglutarate in a mutually dependent manner (31). ATP- and 2-oxoglutarate-liganded Synechococcus PII appears to be the substrate of an ATP-dependent kinase, whereas phosphorylated PII in the absence of 2-oxoglutarate is the substrate of a phosphatase (33, 51). Thus, 2-oxoglutarate, a putative signal of the C-N balance of the cell, would determine the PII phosphorylation level (31). Unphosphorylated PII determines inhibition of nitrate uptake, whereas phosphorylated PII appears to be not inhibitory as long as the cells are incubated under conditions which likely determine high 2-oxoglutarate levels (64). The molecular mechanism through which PII inhibits nitrate uptake is unknown, but a direct interaction of PII with the nitrate-nitrite permease deserves investigation. One of the two ATPase components of the nitrate-nitrite transporter of strain PCC 7942, NrtC, is a polypeptide which bears a carboxyl extension that is homologous to the periplasmic substrate-binding protein of the system, NrtA. Deletion of this C-terminal domain of NrtC partially relieves the inhibition by ammonium of nitrate uptake (61). Interestingly, this study has also suggested an additional inhibitory effect of ammonium on nitrate reductase activity.
THE NTCA TRANSCRIPTIONAL REGULATOR
In the description of genes encoding elements of nitrogen assimilation pathways presented above, we have shown that repression by ammonium or combined nitrogen of the expression of those genes, so-called N control or N regulation, is a common phenomenon in cyanobacteria. Only glutamate synthase, the bis-molybdopterin guanine dinucleotide biosynthesis proteins, and, in some strains, urease appear not to be under N control. The repressive effect of ammonium or combined nitrogen on both the nitrate assimilation system and the nitrogen fixation machinery is abolished by inactivation of glutamine synthetase with methionine sulfoximine, indicating that repression requires the incorporation of ammonium into carbon skeletons via glutamine synthetase (47, 90, 110). This observation led to the notion that a common mechanism might operate the regulation of different nitrogen assimilation pathways in cyanobacteria.
Identification of ntcA.
Pleiotropic mutants of Synechococcus sp. strain PCC 7942 were isolated that, as a result of single mutations, were unable to induce the expression of genes involved in nitrogen assimilation that are subject to nutritional repression by ammonium (121). Complementation of one of those pleiotropic mutants with a gene library from the wild type led to the identification of a gene of 669 nucleotides that would encode a protein different from other regulators operating N control and was given the name ntcA (120, 121). Because spontaneous revertants, as well as complemented strains, derived from ntcA mutants always regained wild-type function and patterns of regulation of all of the proteins subject to N control, it was concluded that ntcA represents a positive-acting element required for the expression of the N-regulated genes (120, 121). The strain PCC 7942 ntcA gene would encode a 222-amino-acid protein homologous to members of the family of bacterial regulators, of which E. coli CAP is the best-characterized member. As is the case for other members of the family, NtcA appears to present, in its C-terminal part, a helix-turn-helix motif for interaction with DNA (120).
The ntcA gene was later identified through DNA-DNA hybridization and cloned from Synechocystis sp. strain PCC 6803 and Anabaena sp. strain PCC 7120 (37) and independently isolated from Anabaena sp. strain PCC 7120 through an in vivo transcriptional interference selection method for DNA-binding proteins (125). More recently, the ntcA gene has been isolated from, or identified in, A. variabilis strain PCC 7937 (ATCC 29413) (T. Thiel, GenBank accession no. U89516), N. punctiforme strain ATCC 29133 (J. C. Meeks, personal communication; DOE Joint Genome Institute [http://www.jgi.doe.gov/]), and the marine cyanobacteria Cyanothece sp. strain ATCC 51142-BH68K (a unicellular diazotrophic strain; 7), Synechococcus sp. strain WH7803 (66), Synechococcus sp. strain PCC 7002 (T. Sakamoto, S. Persson, K. Inoue-Sakamoto, K. Schone, and D. A. Bryant, GenBank accession no. AF195898), Trichodesmium sp. strain WH9601 (A. F. Post and H. Li, GenBank accession no. AF244902), and Prochlorococcus marinus strain MED4 (DOE Joint Genome Institute [http://www.jgi.doe.gov/]). DNA sequences homologous to ntcA have been detected in some other cyanobacteria as well (37), indicating wide distribution of this gene among cyanobacteria.
Transcription of ntcA.
The size of the ntcA transcript(s) has been determined in Synechococcus sp. strain PCC 7942 (69), Anabaena sp. strain PCC 7120 (96, 124), and Cyanothece sp. strain ATCC 51142-BH68K (7). Whereas a single transcript of 1.3 kb has been observed in strain PCC 7942, transcripts of 1.4, 1.0, and 0.8 kb have been detected in strain PCC 7120 and transcripts of 1.3 and 1.0 kb have been observed in strain ATCC 51142. Because the size of ntcA is about 0.7 kb and, in strains PCC 7942 and PCC 7120, the transcription start point (tsp) for the gene is located less than 200 bp upstream from the translation start (see below), it follows that the ntcA transcript can extend 400 to 500 bp beyond the end of the gene. This seems to be not long enough, however, to accommodate a gene located downstream from ntcA in strains PCC 7942 (A. Herrero and E. Flores, unpublished data) and PCC 7120 (Kazusa DNA Research Institute [http://www.kazusa.or.jp/cyano/anabaena/]), as well as in Synechocystis sp. strain PCC 6803 (57), although the size of the ntcA transcript has not been reported for the latter organism.
Expression of ntcA is repressed by growth with ammonium in Synechococcus sp. strains PCC 7942 (69) and WH7803 (66). In the latter, the cellular levels of the ntcA mRNA found in nitrate-grown cells decrease rapidly (half-life, 1.28 ± 0.2 min) upon exposure to ammonium. In Cyanothece sp. strain ATCC 51142, levels of ntcA mRNA have been reported to be higher in cells grown with nitrate than in the absence of combined nitrogen, whereas in ammonium-grown cells the levels seem to depend on the concentration of ammonium used (7). A note of caution is necessary, however, on the interpretation of these data, since changes in the illumination of the cells were superimposed on those in the nitrogen supply. In Anabaena sp. strain PCC 7120, the 0.8-kb transcript is observed both in the presence and in the absence of combined nitrogen although hybridization signals in the range of 0.8 to 1.0 kb are stronger in the absence of combined nitrogen (96). On the other hand, transcripts up to 1.4 kb in length are observed in the presence of combined nitrogen (96). This complex distribution of ntcA transcripts in Anabaena sp. strain PCC 7120 appears to result from the use of different promoters (see below) and may also reflect differential stability of the transcripts. Nonetheless, the total amount of ntcA mRNA in this cyanobacterium does not notably change under different nitrogen regimens (96; J. E. Frías, E. Flores, and A. Herrero, unpublished data).
The NtcA protein.
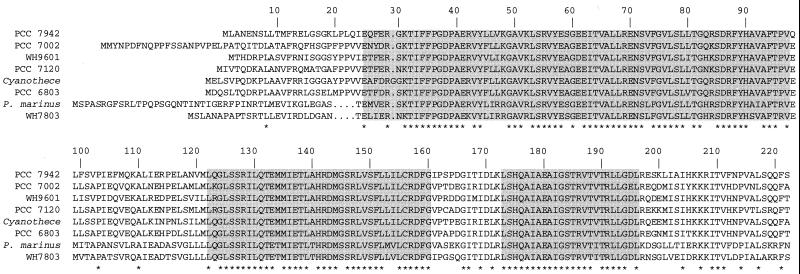
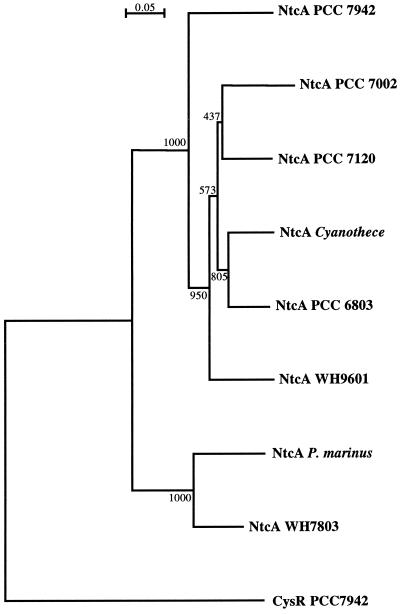
The predicted NtcA polypeptides (Fig. 1) are strongly similar proteins showing ≥61.4% identity in pairwise comparisons. Two groups of NtcA proteins are, however, evident (Fig. 2), with intergroup identities of ≤67%. One group includes the NtcA proteins from the unicellular marine organisms Synechococcus sp. strain WH7803 and P. marinus, which share 82.1% identity. The other group includes the NtcA proteins from the heterocyst formers and from Synechococcus sp. strains PCC 7942 and PCC 7002, Cyanothece sp. strain ATCC 51142, Synechocystis sp. strain PCC 6803, and Trichodesmium sp. strain WH9601, with pairwise identities of 76.9 to 88.4%. NtcA from strain PCC 7942, however, diverges from the NtcA proteins of the other strains in this group (Fig. 2).
FIG. 1.
Alignment of available amino acid sequences of NtcA proteins. The sequences shown are those of the NtcA proteins from the following cyanobacteria (see the text for references): Synechococcus sp. strain PCC 7942, Synechococcus sp. strain PCC 7002, Trichodesmium sp. strain WH9601, Anabaena sp. strain PCC 7120, Cyanothece sp. strain ATCC 51142-BH68K, Synechocystis sp. strain PCC 6803, Prochlorococcus marinus strain MED4, and Synechococcus sp. strain WH7803. The sequences of the NtcA proteins from A. variabilis strain PCC 7937 (ATCC 29413) and N. punctiforme strain ATCC 29133 are identical to that of the Anabaena sp. strain PCC 7120 NtcA protein. The alignment was obtained with the PileUp program. The numbering corresponds to that of strain PCC 7942 NtcA. Amino acids identical in all of the NtcA proteins are marked with asterisks. Shaded boxes denote conserved regions I, II, and III (see the text).
FIG. 2.
Phylogenetic tree of available NtcA proteins. The sequences used to derive the tree are shown in Fig. 1. A neighbor-joining tree derived from an alignment performed by the Clustal X (1.63b) program is presented, with bootstrap values (based on 1,000 bootstrap trials) shown at the nodes. The Synechococcus sp. strain PCC 7942 CysR protein, a homologue of NtcA, was used to root the tree. Scale bar, 0.05 amino acid substitution per position.
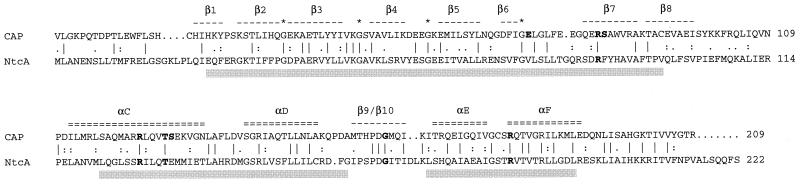
Three strongly conserved regions can be outlined in the NtcA protein (Fig. 1). Region I comprises residues 25 through 97 (numbering of Synechococcus sp. strain PCC 7942 NtcA), region II comprises residues 122 through 160, and region III comprises residues 173 through 196. Although no experimental data on the NtcA structure are available, an analysis of these conserved regions in the light of the known CAP structure (106, 123) is of interest. Conserved region I corresponds to the CAP region of β-roll structure (Fig. 3) and exhibits patterns of Gly and other amino acid residues characteristic of this type of structure, as found in cyclic-nucleotide-binding proteins (107). CAP residues Glu72 and Arg82 are essential, whereas Ser83 is important, for cAMP binding to this protein (83), but only the Arg residue is conserved in NtcA (Arg87 of strain PCC 7942 NtcA). It follows that NtcA may bear a β-roll structure and that this structural feature may represent the site of binding of a metabolite, but this can hardly be cAMP. Region II is predicted, with very high probability, to form two consecutive α-helices. The corresponding region in CAP bears α-helices C and D (Fig. 3), the former of which participates in protein dimerization (106) and contributes three residues (Arg123, Thr127, and Ser128) that contact cAMP (83, 123), the Arg and Thr residues being conserved in NtcA (Arg128 and Thr132, respectively, of strain PCC 7942 NtcA). Region III is predicted to contain the DNA-binding helix-turn-helix motif (120) and corresponds to the region carrying the helix-turn-helix motif (bearing α-helices E and F) of CAP. The amino acid sequences of the helix-turn-helix motifs of all available NtcA sequences are identical except for Val189 (numbering of strain PCC 7942 NtcA), which is replaced with Ile in the proteins from Synechococcus sp. strain WH7803 and P. marinus. It is anticipated that NtcA will recognize similar DNA sequences in the different cyanobacteria. Finally, it can be noted that CAP residue Gly162, located in transcription-activating region 1 (CAP residues 156 to 164; 12), is conserved in all of the available NtcA sequences (Gly166 of strain PCC 7942 NtcA).
FIG. 3.
Comparison of E. coli CAP and Synechococcus sp. strain PCC 7942 NtcA amino acid sequences. The sequences that conform to some of the secondary-structure features of CAP relevant for our discussion of NtcA (see the text) are indicated above the CAP sequence. CAP β strands 1 through 8 constitute a β-barrel structure which bears three amino acid residues (in boldface) involved in cAMP binding. Four Gly residues important for the β-barrel structure are indicated by asterisks. CAP α-helix C also contains three amino acid residues (highlighted in bold) that contact cAMP. CAP β strands 9 and 10 roughly correspond to CAP-activating region 1, which contains a Gly residue (in boldface) conserved in NtcA. CAP α-helices E and F constitute the DNA-binding helix-turn-helix motif, with an Arg residue (in boldface) in α-helix F that contacts a G nucleotide in the DNA-binding site and is conserved in NtcA. Shaded bars below the NtcA sequence correspond to boxes in Fig. 1.
NtcA binding to DNA.
The Synechococcus sp. strain PCC 7942 (69), Synechocystis sp. strain PCC 6803 (98), and Anabaena sp. strain PCC 7120 (85, 126) ntcA genes have been expressed in E. coli, and purified preparations of NtcA proteins have been obtained from E. coli strains bearing recombinant constructions carrying those genes. NtcA in E. coli cell extracts, as well as purified NtcA protein, is able to specifically bind to DNA fragments carrying sequences from the upstream region of a number of genes subject to nutritional repression by ammonium, as first demonstrated for Synechococcus sp. strain PCC 7942 NtcA (69). Some NtcA-binding sites on DNA have been determined by DNase footprinting and found to contain the palindromic sequence signature GTAN8TAC (69). These NtcA-binding sites were included in promoters of genes activated by NtcA. These promoters consist of a −10, E. coli ς70 consensus-like box in the form TAN3T and an NtcA-binding site located in place of the −35 box that is present in the E. coli ς70-type promoters (69). This structure of the NtcA-activated promoters is similar to that of class II, CAP-activated promoters in which the DNA-binding site for the activator is centered near position −42, overlapping and replacing the −35 DNA site for the binding of RNA polymerase (11). In this type of CAP-activated promoters, transcription activation operates through two different components, an increase in the binding constant of RNA polymerase and an increase in the rate constant for isomerization of the RNA polymerase-closed promoter complex to the transcriptionally active open complex, both effects operating through specific protein-protein contacts between CAP and RNA polymerase (reviewed in references 11, 12).
Binding to DNA of a protein factor, later identified as NtcA, present in partially purified preparations from cell extracts of Anabaena sp. strain PCC 7120 has been reported, and the sequence TGT(N9 or 10)ACA has been suggested as the consensus recognition sequence for this factor (95). This consensus partially overlaps the one described above, and the differences might arise from analysis of some Anabaena genes, like rbcL and xisA (see below), whose upstream regions bear binding sites for which NtcA would show weak binding (95).
In a recent study, the NtcA protein was used for in vitro selection of DNA fragments from a library of molecules which carried a sequence consisting of 13 random nucleotides followed by ACA cloned in a 480-bp fragment (55). Strong selection for fragments bearing the GTAN8TAC sequence and the GTN10AC subset of that sequence was observed. Although a few DNA fragments with a spacing of seven or nine nucleotides between the two triplets were also selected, comparison of the binding of NtcA to some of the selected DNA fragments led to the conclusion that the eight-nucleotide spacing is optimal (55).
A SURVEY OF NTCA-ACTIVATED GENES
Since the first identification of NtcA in Synechococcus sp. strain PCC 7942 and the description of its action, the number of genes recognized as positively and directly regulated by NtcA has been continually increasing. For that recognition, we ask that the following criteria need to be met: (i) that the gene be expressed from a tsp subject to repression by ammonium, (ii) that the NtcA protein be shown to specifically bind to that promoter, (iii) that the use of the tsp upon ammonium withdrawal not take place in an NtcA− background, and (iv) that the promoter structure upstream from the ammonium-regulated tsp conform to the structure of the NtcA-activated promoters described above. The latter represents a conclusion derived from experimentation that has become a prediction. Genes described to date that can be considered to be directly activated by NtcA are listed in Fig. 4. In this section, we shall discuss NtcA-activated genes which are involved in the assimilation of combined nitrogen, leaving genes specifically related to heterocysts for a later section.
FIG. 4.
Sequences of NtcA-activated promoters. Only promoters from genes whose putative tsps (lowercase, bold) have been experimentally determined are included (see the text for references). The location of the tsp with respect to the translation start of the corresponding gene is indicated. The derived consensus sequences for the NtcA-binding site (left) and the −10 promoter box (right) are presented at the bottom. Nucleotides matching those of the GTA and TAC triplets of the NtcA-binding site and the strongly conserved TA and T of the −10 box are in boldface.
Genes of Synechococcus sp. strain PCC 7942.
The nir operon of the cyanobacterium Synechococcus sp. strain PCC 7942 is expressed at higher levels in the absence than in the presence of ammonium from a tsp located at −32 (69; −29 in reference 112) with respect to the start of the nirA gene. Three NtcA-binding sites have been found centered at positions −40.5, −109.5, and −180.5 with respect to the tsp (69). The binding site centered at position −40.5 is included in a canonical NtcA-activated promoter structure (Fig. 4) and would direct N-regulated expression of the nir operon (69, 71). The site centered at position −109.5 has been implicated in NtcB-dependent enhancement by nitrite of the expression of the nir operon (71). The binding site centered at position −180.5 would be part of a different NtcA-activated promoter (Fig. 4) which would direct the expression of the divergent nirB and ntcB genes from a tsp located 30 bp upstream from nirB (71, 111).
The strain PCC 7942 ntcA transcript is synthesized from an N-regulated tsp located 109 bp upstream from the start of the ntcA gene. Another tsp, generating a weaker signal, is detected in the presence of ammonium at position −120 (69). Whereas the promoter for the −120 tsp resembles the canonical ς70-type promoters, the promoter for the −109 tsp is dependent on NtcA and exhibits the structure of the NtcA-type promoters (Fig. 4). NtcA is therefore positively autoregulated in strain PCC 7942. Also, the strain PCC 7942 glnB gene is transcribed from both a constitutive E. coli ς70-type promoter and an N-regulated, NtcA-dependent promoter (Fig. 4) with transcription start points located at positions −120 and −53, respectively, with respect to the start of the open reading frame (ORF) (65). The ς70-type promoters of these genes may be responsible for their expression in the presence of ammonium that is higher in the case of glnB than in that of ntcA.
The strain PCC 7942 glnA gene is transcribed mainly from an NtcA-activated promoter (Fig. 4) which generates a tsp located at nucleotide −147 with respect to the start of the glnA ORF (69; −146 in reference 18). The amt1 gene of this cyanobacterium is also expressed from an NtcA-activated promoter (Fig. 4) which binds NtcA in vitro (Vázquez-Bermúdez et al., unpublished data). The cynABDS gene cluster, possibly an operon involved in cyanate uptake and degradation, is preceded by a putative NtcA-type promoter (T. Omata, GenBank accession no. AB005890). Although the tsp for cynABDS has not been reported, the expression of these genes, tested with a cynS probe, has been shown to be NtcA dependent (43).
Genes of Anabaena sp. strain PCC 7120.
Anabaena sp. strain PCC 7120 is the second cyanobacterium from which an ntcA mutant was isolated (34, 124). Combined nitrogen assimilation genes or operons whose expression is NtcA dependent include the glnA gene and the nir (nitrate assimilation) and urt (urea transport) operons (Fig. 4). Transcription of glnA takes place from at least three transcription start points (reviewed in reference 28), the most proximal of which, located at nucleotide −92 with respect to the start of the gene and generating RNAI, corresponds to an NtcA-dependent promoter. NtcA-dependent promoters to which NtcA has been observed to bind in vitro have also been characterized for the nir and urt operons (35, 36; A. Valladares, A. Herrero, and E. Flores, unpublished data). Transcription of the Anabaena nir operon also requires NtcB, which can bind to the nir promoter just upstream from, and simultaneously to, NtcA (36). Expression of ntcB itself takes place from an NtcA-dependent promoter (Fig. 4) which has been shown to bind NtcA in vitro (36). It has been suggested that NtcB works as a booster of the NtcA-dependent expression of the nir operon (36).
The strain PCC 7120 ntcA gene has been reported to be transcribed from three N-regulated transcription start points (96). Additionally, a tsp located 136 nucleotides upstream from the start of the coding sequence has been detected with RNA isolated from cells grown with ammonium, nitrate, or dinitrogen (A. M. Muro-Pastor, A. Valladares, A. Herrero, and E. Flores, unpublished data). One of the N-regulated transcription start points is located at position −49, is preferentially used in the absence of combined nitrogen and early during heterocyst differentiation, and is active in mature heterocysts (96). Another is located at position −180 and seems to be transiently used early in heterocyst development but not in mature heterocysts (96). The third N-regulated tsp is located at position −190 and appears to be used increasingly during heterocyst differentiation and also in mature heterocysts (96). Activation of the −49 and −180 tsps in response to combined nitrogen deprivation is strictly dependent on NtcA, indicating that ntcA is also autoregulatory in strain PCC 7120 (Muro-Pastor et al., unpublished data). NtcA has been reported (96) to bind to a region covering nucleotides −131 to −155, which includes a sequence (GTAN8AAC) strongly similar to the consensus of NtcA-binding sites, but this sequence is located 88 bp upstream from the closer tsp (i.e., the −49 tsp). Thus, no canonical NtcA-activated promoter could be recognized upstream from any of the three ntcA transcription start points in strain PCC 7120. Therefore, the autoregulatory effect of NtcA in this strain might be exerted in an indirect manner or might involve a different type of NtcA-dependent promoter.
Figure 4 also includes the promoters of heterocyst development genes hetC and devBCA, which, as discussed below, appear to be directly regulated by NtcA.
Genes of Synechocystis sp. strain PCC 6803.
Several genes of the cyanobacterium Synechocystis sp. strain PCC 6803 have been shown to be transcribed from N-regulated tsps which are preceded by NtcA-type promoters (Fig. 4). They include amt1 (82), glnA (98), glnB (39), icd (88), and the rpoD2-V gene encoding a ς factor affecting survival under nitrogen stress (84a). Binding of NtcA to the promoter regions of these genes has been shown in every case. Although a fully segregated ntcA mutant of strain PCC 6803 has not been obtained, a partially segregated mutant has been reported to be impaired in the expression of glnA and glnB, supporting the occurrence of NtcA-dependent gene expression in this cyanobacterium (41).
Genes of other cyanobacterial strains.
N-regulated genes transcribed from tsps which are preceded by an NtcA-type promoter structure are also present in other cyanobacteria ntcA mutants of which have not been reported. The glnA gene of Calothrix sp. strain PCC 7601 bears a promoter structure very similar to that of the same gene in Anabaena sp. strain PCC 7120 (S. Liotenberg and N. Tandeau de Marsac, unpublished data). An N-regulated tsp named P1 is located at position −87 or −88 with respect to the start of the gene and is preceded by an NtcA-type promoter (Fig. 4). The Pseudanabaena sp. strain PCC 6903 glnN (23), the Synechococcus sp. strain PCC 7002 nrtP (103), and the Synechococcus sp. strain WH7803 ntcA (66) genes are transcribed from NtcA-type promoters (Fig. 4), but only in the case of strain PCC 6903 glnN has binding of NtcA to the promoter been demonstrated. Available data suggest that ntcA of Synechococcus sp. strain WH7803 would also be autoregulatory.
SEQUENCES OF NTCA-ACTIVATED PROMOTERS
Consensus sequence.
Scrutiny of the sequences presented in Fig. 4 shows that the palindromic sequence signature GTAN8TAC, defined for NtcA-binding sites on DNA (69), is strongly conserved. It should be noted that the spacing between the two strongly conserved GTA and TAC triplets is invariably eight nucleotides. The available data permit us to further define, by calculating the percentage of occurrence of each of the four bases at each position, the sequence of the NtcA-binding site (Fig. 4). In addition to the GTAN8TAC sequence signature, single bases that appear at a high frequency in some particular positions include C and A in the second and third positions, respectively, after the GTA triplet and A in the first and second positions after the TAC triplet. Despite some differences in the percentage of occurrence of the bases in some positions, it is evident that the two halves of the NtcA-binding site are rather complementary to each other, rendering a whole sequence that is palindromic in nature. It has been observed that purified NtcA protein behaves like a dimer in solution (126). This permits the assumption that, like other proteins in the CAP family, each subunit of NtcA interacts with one half of the binding site. Predictions made following the CAP model permit the suggestion that the strongly conserved G of the GTA triplet would be contacted by NtcA residue Arg186 (numbering of Synechococcus sp. strain PCC 7942 NtcA; Fig. 3), whereas Val187 would contact a T complementary to the A in the GTA triplet (69). These two amino acids are invariant in all of the available NtcA sequences (Fig. 1).
In NtcA-activated promoters, a −10 box can be identified that is usually separated from the tsp by five to seven nucleotides, although spacings of four to nine nucleotides have been observed (Fig. 4). The consensus that can be defined for these −10 boxes (Fig. 4) is very similar to that of the −10 box of the E. coli ς70-type promoters. The cyanobacterial major (vegetative) ς factor, SigA, is homologous to E. coli ς70 (8). Thus, it can be suggested that NtcA activates transcription mediated by RNA polymerase carrying a vegetative-type ς factor, although this has not been experimentally tested. On the other hand, the space between the −10 box and the GTAN8TAC sequence is usually 22 or 23 nucleotides, although spacings of 20, 21, and 24 nucleotides have also been observed (Fig. 4).
The essential role of the NtcA-binding site in NtcA-dependent activation of gene expression has been demonstrated. A deletion of nine nucleotides including the 5′ GTA triplet of the NtcA-binding site of the Synechococcus sp. strain PCC 7942 glnA promoter abolishes derepression of this gene (19), and replacement of the 5′ GTA triplet of the nir operon-proximal NtcA-binding site (−40.5 site), as well as of the nirB-proximal site (−180.5 with respect to the nir operon tsp), by CAT triplets also abolishes activation of the corresponding genes (71).
Promoters similar to NtcA-activated promoters.
Figure 5 shows a number of promoters of nitrogen assimilation genes that resemble, but do not match, the structure of the canonical NtcA-activated promoters. The tsp of the Synechocystis sp. strain PCC 6803 glnN gene has been localized to nucleotide −32 with respect to the start of the gene (98). The corresponding promoter is similar to NtcA-activated promoters, but the putative NtcA-binding site lacks a conserved TAC triplet, which is replaced with GTC (Fig. 5). Although a DNA fragment carrying this promoter has failed to bind NtcA in vitro (98), its use appears to be NtcA dependent (41). A similar NtcA-dependent promoter has been characterized for the glnN gene of Synechococcus sp. strain PCC 7942 (104). It is possible that a modified or activated (see below) NtcA protein is able to bind to these promoters in vivo. Alternatively, their activation by NtcA could require the participation of another factor. The same argument could be made for other genes, such as the N-regulated nifP gene of Synechococcus sp. strain RF-1, which is transcribed from a tsp (49) that is preceded by sequences bearing a putative −10 box and exhibiting some features of an NtcA-binding site (Fig. 5).
FIG. 5.
Sequences of some promoters similar to NtcA-activated promoters. Tsps are indicated by boldface lowercase letters (see the text for references). The location of the tsp with respect to the translation start of the corresponding gene is indicated. Shaded boxes denote the putative NtcA-binding sites (left boxes) and −10 promoter hexamers (right box), respectively. The consensus sequence for the NtcA-activated promoters derived from data in Fig. 4 is presented at the bottom for comparison. W, A or T; Y, C or T; R, A or G.
NTCA-ACTIVATED GENES IN HETEROCYST DIFFERENTIATION AND FUNCTION
As previously mentioned, some filamentous cyanobacteria that are able to fix N2 develop specialized cells called heterocysts, which differentiate at semiregular intervals in response to combined nitrogen deprivation. Heterocysts exhibit substantial structural changes, as well as new metabolic capabilities with respect to vegetative cells, the process of heterocyst differentiation involving changes in the transcriptional program of the vegetative cells (44, 129).
Heterocyst development requires NtcA.
NtcA has been shown to be required for differentiation of heterocysts in Anabaena sp. strain PCC 7120 (34, 124), A. variabilis strain ATCC 29413 (T. Thiel, personal communication), and N. punctiforme strain ATCC 29133 (J. C. Meeks, personal communication). It can be suggested that NtcA represents a regulatory link between nitrogen nutrition and heterocyst development. Induction of the heterocyst development positive regulatory gene hetR, which takes place shortly after nitrogen deprivation (4, 9), is impaired in an ntcA mutant (34), but the basis for such dependence on NtcA is not known. Expression of hetR in Anabaena sp. strain PCC 7120 takes place from several promoters, none of which shows the typical structure of an NtcA-activated promoter (W. J. Buikema and R. Haselkorn, unpublished data; A. M. Muro-Pastor, A. Valladares, A. Herrero, and E. Flores, unpublished data). Thus, the requirement for NtcA of hetR induction might be indirect.
NtcA-regulated genes involved in heterocyst differentiation and function.
Induction of a number of genes whose expression is required for heterocyst development but takes place later than that of hetR is dependent on an intact hetR gene (14). The nature of this dependence is unknown, although HetR has been shown to exhibit protease activity in vitro (131). Because NtcA is required for hetR induction and heterocyst development, impairment in an ntcA mutant of the expression of genes normally expressed in the heterocyst could be a consequence of the lack of heterocyst differentiation in such a mutant. However, as shown below, the NtcA protein can be directly involved in the transcriptional activation of some genes that are expressed at different stages of heterocyst development or in the mature heterocyst.
The Anabaena sp. strain PCC 7120 hetC gene would encode a product which acts early in heterocyst development and is homologous to ABC exporters, although its putative substrate is unknown (59). Expression of hetC is not depressed in a hetR mutant (59) but is impaired in an ntcA mutant and takes place from a typical NtcA-activated promoter (Fig. 4) which generates a transcript whose 5′ end corresponds to nucleotide −571 with respect to the putative gene start (85). Anabaena sp. strain PCC 7120 genes whose expression in the mature heterocyst appears to be activated by NtcA include petH (encoding ferredoxin:NADP+ reductase) and glnA. Ferredoxin:NADP+ reductase, which can contribute to the provision of the reduced ferredoxin required for the nitrogenase reaction, and glutamine synthetase, responsible for the incorporation of fixed nitrogen into carbon skeletons, are critical for the assimilation of nitrogen in heterocysts. RNAI of glnA (discussed above) and tsp2 of petH are detected with heterocyst RNA, but their use is independent of HetR (118; A. Valladares, A. M. Muro-Pastor, A. Herrero, and E. Flores, unpublished data). The promoters originating these transcripts are, however, NtcA dependent, bind the NtcA protein in vitro (although binding is weaker for petH than for glnA), and match, in the case of glnA RNAI (Fig. 4), or show some similarity to, in the case of petH tsp2 (Fig. 5), the structure of the NtcA-activated promoters.
Some genes or operons whose expression is dependent on both NtcA and HetR might also be directly regulated by NtcA. The devBCA operon encodes an ABC transporter which is involved in the maturation of the heterocyst envelope (25, 73), and expression of devA has been shown to be blocked in a hetR mutant of Anabaena sp. strain PCC 7120 (14). This operon is transcribed in strain PCC 7120 from a tsp located at −704 which is not used in an ntcA mutant and is preceded by sequences that bind NtcA in vitro and match those of the NtcA-activated promoters (Fig. 4; G. Fiedler, A. M. Muro-Pastor, E. Flores, and I. Maldener, unpublished data). The nifHDK operon is expressed in Anabaena sp. strain PCC 7120 exclusively in the heterocysts (24) and consistently is not expressed in ntcA (34) or hetR (Valladares et al., unpublished data) mutants. Weak binding of NtcA to DNA from the region upstream of nifH has been observed (95, 118). As is the case with the petH promoter discussed above, the nifHDK promoter shows some similarity to, but does not match, the structure of the NtcA-activated promoters (Fig. 5). Additionally, as shown in Fig. 5, sequences with some similarity to the NtcA-binding site can be recognized in the promoters of nif gene cluster ORF1 from strain PCC 7120 (6) and of the vnfDG gene involved in the expression of the alternative vanadium-containing nitrogenase of A. variabilis strain ATCC 29413 (115).
Binding of NtcA to three sites in the upstream region of xisA, encoding the Anabaena sp. strain PCC 7120 site-specific recombinase responsible for excision of the nifD 11-kb intervening element (44), has been described (16, 95). In these sites, the spacing between the palindromic outer sequences is shorter than that found in the typical NtcA-activated promoters (e.g., TGTN9ACA instead of GTN10AC). NtcA has also been shown to bind to a region upstream from the glbN gene of N. commune that bears an NtcA-binding site-like sequence also with a shorter intervening sequence, GTAN7TAC (48). In any case, the function of the interaction of NtcA with these sites is unknown since no information is available on the transcription start point(s) of xisA or glbN.
Consistent with its putative role in gene expression in the heterocysts, ntcA appears to be expressed in fully developed, mature heterocysts (2, 95, 96). An additional interesting observation is that although overexpression of hetR results in heterocyst development independent of the nitrogen source, only in a medium without combined nitrogen are the heterocysts active in nitrogen fixation (45). This observation implies operation of N regulation beyond induction of hetR, consistent with direct regulation by NtcA of some genes involved in heterocyst function. On the other hand, a number of promoters putatively regulated by NtcA that are used in the heterocyst do not match the consensus sequence defined for NtcA-activated promoters (e.g., petH tsp2, and nifHDK; Fig. 5). Interestingly, most of the promoter sequences included in Fig. 5 contain a good half site for NtcA binding. As discussed above, it is possible that a modified or activated NtcA protein could bind to these promoters. Alternatively, NtcA assisted by another factor, or even a heterodimer of NtcA and another transcription factor, could constitute the actual activator. Of interest in this context is the recent identification of devH, a gene whose expression is NtcA and HetR dependent and whose disruption leads to the development of nonfunctional heterocysts (46). DevH is homologous to NtcA and represents a putative DNA-binding protein (46). Because the sequence of the second helix of the helix-turn-helix motif of DevH is strongly similar to that of NtcA, DevH might recognize a binding site on DNA similar to that of NtcA.
NTCA REPRESSOR SITES
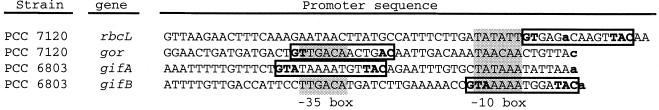
NtcA-binding sites that appear to have a repressor role have been identified in Anabaena sp. strain PCC 7120 and Synechocystis sp. strain PCC 6803. In Anabaena sp. strain PCC 7120, NtcA has been shown to bind to two sites in the promoter of the rbcL gene, which encodes a subunit of ribulose bisphosphate carboxylase/oxygenase (16, 95). At least one of these sites could be a repressor site, since it maps to the region extending from −12 to +12 with respect to the tsp (Fig. 6; see reference 95). Also, a sequence, TGTAN8AACA, that could represent an NtcA-binding site is located 60 nucleotides downstream from a tsp, which has been detected with RNA from ammonium-grown cells, for the Anabaena hanA gene (58, 89). The hanA gene encodes the histone-like HU protein, and its mutation results in a highly pleiotropic phenotype that includes lack of heterocyst development (58). Because ribulose bisphosphate carboxylase/oxygenase (129) and the HU protein (89) are absent from heterocysts, repression of rbcL and hanA can represent another role for NtcA in the mature heterocyst. Finally, a sequence, TGTN9ACA, located at a position that would be compatible with an NtcA repressor site (Fig. 6) has been reported for a promoter of the Anabaena gor gene, encoding glutathione reductase, that is preferentially used in ammonium-grown cells (53). This sequence is similar to those discussed above for the NtcA-binding sites of xisA and glbN and could represent a weak binding site for NtcA (55).
FIG. 6.
Sequences of some promoters reported to carry NtcA repressor sites. Shaded boxes denote −10 and, when they can be discerned, −35 promoter boxes. Putative NtcA-binding sites are indicated by open boxes, and nucleotides matching those of the GTA and TAC triplets are in boldface. See the text for references. Tsps are indicated by boldface lowercase letters.
Expression of the gifA and gifB genes from Synechocystis sp. strain PCC 6803, encoding glutamine synthetase-inactivating factors IF7 and IF17, respectively (see above), has been shown to be repressed in the absence of combined nitrogen, with full expression taking place in the presence of ammonium (41). Repression of these genes is not observed in a partially segregated ntcA mutant (41). Binding of NtcA to the promoters of gifA and gifB has been shown, and in both cases, the position of the binding site is closer to the tsp than in the case of NtcA-activated promoters (Fig. 6). In fact, the NtcA-binding site found in the gifB promoter overlaps the −10 box (41). These results demonstrate a repressor effect of NtcA, which can therefore act as both an activator and a repressor, as is also the case for other regulators of the CAP family (62).
REGULATION OF NTCA
As described above, the ntcA gene has been found to be autoregulatory in those cyanobacteria in which ntcA expression has been investigated. Constitutive expression of NtcA in Synechococcus sp. strain PCC 7942 derivatives bearing a Ptrc-ntcA construct is not able to promote the activation of NtcA-dependent genes in the presence of ammonium (M. F. Vázquez-Bermúdez, I. Luque, A. Herrero, and E. Flores, unpublished data). The NtcA protein could be activated upon nitrogen stepdown, becoming competent in promoting the use of NtcA-dependent promoters and, therefore, its own expression. It might be suggested that NtcA responds to a signal which senses the presence of ammonium in the medium. However, as was previously shown for nitrate reductase (47), inactivation of glutamine synthetase with MSX induces expression of the nir operon transcript (112), indicating that the repressor effect of ammonium requires incorporation of this ion into carbon skeletons. Furthermore, the glnB NtcA-dependent promoter of Synechococcus sp. strain PCC 7942 has been shown to be used differentially, depending not only on the nitrogen source but also on the carbon supply (65). Whereas this promoter is used during nitrogen starvation but not in the presence of ammonium, its use in the presence of nitrate depends on an extra supply of CO2 (air plus 3% CO2). Therefore, NtcA appears to respond, directly or indirectly, to a signal of the C-N balance of the cell rather than to ammonium. As discussed above, 2-oxoglutarate may represent a C-N balance signal in cyanobacteria. Recent results from our laboratory have shown that the affinity of NtcA for the Synechococcus glnA promoter is increased by 2-oxoglutarate in the presence of Mg2+ ions (Vázquez-Bermúdez et al., unpublished data), but the participation of other regulatory elements in the modulation of NtcA activity or in NtcA-dependent transcriptional control is also possible. In this context, it is worth noting that the PII protein appears not to be an element required for nitrogen-dependent transcriptional control in cyanobacteria, since this control is not altered in a PII-null mutant (63).
Cyanate has been proposed as the metabolic signal for the ammonium-promoted repression of the nir operon in Synechococcus sp. strain PCC 7942 (113), but the observation that the negative effect of cyanate is not manifest in a cyanase mutant has made this suggestion questionable (92). On the other hand, based on a requirement for a reducing agent in the buffer for protein-DNA-binding assays, Anabaena sp. strain PCC 7120 NtcA has been suggested to be subject to redox regulation (54), a proposal that requires further investigation.
CONCLUDING REMARKS AND PROSPECTS
Cyanobacteria assimilate ammonium in preference to other nitrogen sources, like nitrate or N2, and utilize combined nitrogen in preference to N2. Some strains that can assimilate urea also seem to use this simple organic compound as a preferred nitrogen source. The CAP family transcriptional regulator NtcA has been found to constitute a key element that operates N control in cyanobacteria. The ntcA mutants generally require ammonium as a nitrogen source, NtcA being necessary for the expression of genes encoding proteins involved in the assimilation of alternative nitrogen sources. NtcA is required for heterocyst differentiation to take place, and recent results have indicated that NtcA can activate the expression not only of early development genes but also of other genes involved in later steps of the differentiation process and in the function of mature heterocysts. NtcA is also required for the expression of ammonium permease and for full expression of the ammonium-assimilating enzyme glutamine synthetase when the cells are incubated without ammonium. Additionally, ntcA is a positively autoregulatory gene. On the other hand, NtcA can act as a repressor of some genes which are expressed only in the presence of ammonium or that are not expressed in the heterocysts.
A substantial amount of work has been devoted to the characterization of NtcA-activated promoters permitting the definition of a clear structure for this type of promoters (Fig. 4). Most of the NtcA-activated promoters identified to date generate transcripts whose start points are located 27 to 264 nucleotides upstream from the putative translation start of the corresponding genes. However, for some genes of Anabaena sp. strain PCC 7120, the NtcA-dependent transcription start points have been localized farther upstream (−460 to −704). The role, if any, of the corresponding long, presumably untranslated transcript fragments remains to be investigated.
How NtcA perceives the nitrogen status (or the C-N balance) of the cell, so that it activates transcription only when ammonium is not available, is unknown. The observed effect of 2-oxoglutarate on NtcA binding deserves further investigation, since this metabolite represents a putative indicator of the C-N balance of the cyanobacterial cell. In fact, 2-oxoglutarate plays a key role in determining the phosphorylation degree of the signal transduction PII protein in cyanobacteria. PII also senses 2-oxoglutarate in enterobacteria, but in these organisms N control additionally involves sensing of glutamine by the PII uridylyltransferase/uridylyl-removing enzyme (56), a protein which is not present in cyanobacteria. On the other hand, PII appears not to be involved in NtcA function, in contrast to the key role of PII in the enterobacterial Ntr system.
Under certain conditions, NtcA is able to activate transcription from some promoters while being inactive on others. A clear example of this is repression by nitrate of heterocyst differentiation, an NtcA-dependent process, while the NtcA-dependent nitrate assimilation system is expressed. We are currently investigating whether different affinities of NtcA for different promoters could be responsible for a hierarchy of expression of nitrogen assimilation genes. Additionally, cooperation of other factors, like the LysR family NtcB protein, in NtcA-dependent activation of gene expression may lead to the preferential use of some promoters under some defined physiological conditions.
ACKNOWLEDGMENTS
We thank D. Bryant, W. J. Buikema, J. C. Meeks, A. Post, J. C. Reyes, N. Tandeau de Marsac, and T. Thiel for communicating results previous to publication; E. Martínez-Force and A. Serrano for help with data bank protein analysis; and J. E. Frías, I. Luque, A. Valladares, and M. F. Vázquez-Bermúdez for their numerous unpublished contributions to this review.
Work in our laboratory is currently supported by grants PB97-1137 and PB98-0481 from the Ministerio de Ciencia y Tecnología, Spain.
REFERENCES
- 1.Aichi M, Omata T. Involvement of NtcB, a LysR family transcription factor, in nitrite activation of nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1997;179:4671–4675. doi: 10.1128/jb.179.15.4671-4675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer C C, Haselkorn R. Vectors for determining the differential expression of genes in heterocysts and vegetative cells of Anabaena sp. strain PCC 7120. J Bacteriol. 1995;177:3332–3336. doi: 10.1128/jb.177.11.3332-3336.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergman B, Gallon J R, Rai A N, Stal L J. N2 fixation by non-heterocystous cyanobacteria. FEMS Microbiol Rev. 1997;19:139–185. [Google Scholar]
- 4.Black T A, Cai Y, Wolk C P. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol. 1993;9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 5.Böhme H. Regulation of nitrogen fixation in heterocyst-forming cyanobacteria. Trends Plant Sci. 1998;3:346–351. [Google Scholar]
- 6.Borthakur D, Basche M, Buikema W J, Borthakur P B, Haselkorn R. Expression, nucleotide sequence and mutational analysis of two open reading frames in the nif gene region of Anabaena sp. strain PCC7120. Mol Gen Genet. 1990;221:227–234. doi: 10.1007/BF00261725. [DOI] [PubMed] [Google Scholar]
- 7.Bradley R L, Reddy K J. Cloning, sequencing, and regulation of the global nitrogen regulator gene ntcA in the unicellular diazotrophic cyanobacterium Cyanothece sp. strain BH68K. J Bacteriol. 1997;179:4407–4410. doi: 10.1128/jb.179.13.4407-4410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahamsha B, Haselkorn R. Isolation and characterization of the gene encoding the principal sigma factor of the vegetative cell RNA polymerase from the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991;173:2442–2450. doi: 10.1128/jb.173.8.2442-2450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buikema W J, Haselkorn R. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 1991;5:321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 10.Buikema W J, Haselkorn R. Molecular genetics of cyanobacterial development. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:33–52. [Google Scholar]
- 11.Busby S, Ebright R H. Transcription activation at class II CAP-dependent promoters. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 12.Busby S, Ebright R H. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 13.Cai Y, Wolk C P. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J Bacteriol. 1997;179:258–266. doi: 10.1128/jb.179.1.258-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Y, Wolk C P. Anabaena sp. strain PCC 7120 responds to nitrogen deprivation with a cascade-like sequence of transcriptional activations. J Bacteriol. 1997;179:267–271. doi: 10.1128/jb.179.1.267-271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capone D G, Zehr J P, Paerl H W, Bergman B, Carpenter E J. Trichodesmium, a globally significant marine cyanobacterium. Science. 1997;276:1221–1229. [Google Scholar]
- 16.Chastain C J, Brusca J S, Ramasubramanian T S, Wei T-F, Golden J W. A sequence-specific DNA-binding factor (VF1) from Anabaena sp. strain PCC 7120 vegetative cells binds to three adjacent sites in the xisA upstream region. J Bacteriol. 1990;172:5044–5051. doi: 10.1128/jb.172.9.5044-5051.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y-B, Dominic B, Mellon M T, Zehr J P. Circadian rhythm of nitrogenase gene expression in the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. strain IMS 101. J Bacteriol. 1998;180:3598–3605. doi: 10.1128/jb.180.14.3598-3605.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen-Kupiec R, Gurevitz M, Zilberstein A. Expression of glnA in the cyanobacterium Synechococcus sp. strain PCC 7942 is initiated from a single nif-like promoter under various nitrogen conditions. J Bacteriol. 1993;175:7727–7731. doi: 10.1128/jb.175.23.7727-7731.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen-Kupiec R, Zilberstein A, Gurevitz M. Characterization of cis elements that regulate the expression of glnA in Synechococcus sp. strain PCC 7942. J Bacteriol. 1995;177:2222–2226. doi: 10.1128/jb.177.8.2222-2226.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collier J L, Brahamsha B, Palenik B. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology. 1999;145:447–459. doi: 10.1099/13500872-145-2-447. [DOI] [PubMed] [Google Scholar]
- 21.Colón-López M S, Sherman D M, Sherman L A. Transcriptional and translational regulation of nitrogenase in light-dark- and continuous-light-grown cultures of the unicellular cyanobacterium Cyanothece sp. strain ATCC 51142. J Bacteriol. 1997;179:4319–4327. doi: 10.1128/jb.179.13.4319-4327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coronil T, Lara C, Guerrero M G. Shift in carbon flow and stimulation of amino-acid turnover induced by nitrate and ammonium assimilation in Anacystis nidulans. Planta. 1993;189:461–467. doi: 10.1007/BF00194446. [DOI] [PubMed] [Google Scholar]
- 23.Crespo J L, García-Domínguez M, Florencio F J. Nitrogen control of the glnN gene that codes for GS type III, the only glutamine synthetase in the cyanobacterium Pseudanabaena sp. PCC 6903. Mol Microbiol. 1998;30:1101–1112. doi: 10.1046/j.1365-2958.1998.01143.x. [DOI] [PubMed] [Google Scholar]
- 24.Elhai J, Wolk C P. Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J. 1990;9:3379–3388. doi: 10.1002/j.1460-2075.1990.tb07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiedler G, Arnold M, Hannus S, Maldener I. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol. 1998;27:1193–1202. doi: 10.1046/j.1365-2958.1998.00762.x. [DOI] [PubMed] [Google Scholar]
- 26.Fisher S H. Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence! Mol Microbiol. 1999;32:223–232. doi: 10.1046/j.1365-2958.1999.01333.x. [DOI] [PubMed] [Google Scholar]
- 27.Flores E, Herrero A. Assimilatory nitrogen metabolism and its regulation. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 487–517. [Google Scholar]
- 28.Flores E, Muro-Pastor A M, Herrero A. Cyanobacterial nitrogen assimilation genes and NtcA-dependent control of gene expression. In: Peschek G A, Löffelhardt W, Schmetterer G, editors. The phototrophic prokaryotes. New York, N.Y: Plenum Publishing Corporation; 1999. pp. 463–477. [Google Scholar]
- 29.Flores E, Romero J M, Guerrero M G, Losada M. Regulatory interaction of photosynthetic nitrate utilization and carbon dioxide fixation in the cyanobacterium Anacystis nidulans. Biochim Biophys Acta. 1983;725:529–532. [Google Scholar]
- 30.Forchhammer K. The PII protein in Synechococcus PCC 7942 senses and signals 2-oxoglutarate under ATP-replete conditions. In: Peschek G A, Löffelhardt W, Schmetterer G, editors. The phototrophic prokaryotes. New York, N.Y: Plenum Publishing Corporation; 1999. pp. 549–553. [Google Scholar]
- 31.Forchhammer K, Hedler A. Phosphoprotein PII from cyanobacteria. Analysis of functional conservation with PII signal-transduction protein from Escherichia coli. Eur J Biochem. 1997;244:869–875. doi: 10.1111/j.1432-1033.1997.00869.x. [DOI] [PubMed] [Google Scholar]
- 32.Forchhammer K, Tandeau de Marsac N. The PII protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J Bacteriol. 1994;176:84–91. doi: 10.1128/jb.176.1.84-91.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forchhammer K, Tandeau de Marsac N. Phosphorylation of the PII protein (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942: analysis of in vitro kinase activity. J Bacteriol. 1995;177:5812–5817. doi: 10.1128/jb.177.20.5812-5817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frías J E, Flores E, Herrero A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 35.Frías J E, Flores E, Herrero A. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:477–486. doi: 10.1128/jb.179.2.477-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frías J E, Flores E, Herrero A. Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors. Mol Microbiol. 2000;38:613–625. doi: 10.1046/j.1365-2958.2000.02156.x. [DOI] [PubMed] [Google Scholar]
- 37.Frías J E, Mérida A, Herrero A, Martín-Nieto J, Flores E. General distribution of the nitrogen control gene ntcA in cyanobacteria. J Bacteriol. 1993;175:5710–5713. doi: 10.1128/jb.175.17.5710-5713.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallon J R, Cheng J, Dougherty L J, Gallon V A, Hilz H, Pederson D M, Richards H M, Rüggerberg S, Smith C J. A novel covalent modification of nitrogenase in a cyanobacterium. FEBS Lett. 2000;468:231–233. doi: 10.1016/s0014-5793(00)01229-1. [DOI] [PubMed] [Google Scholar]
- 39.García-Domínguez M, Florencio F J. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol. 1997;35:723–734. doi: 10.1023/a:1005846626187. [DOI] [PubMed] [Google Scholar]
- 40.García-Domínguez M, Reyes J C, Florencio F J. Glutamine synthetase inactivation by protein-protein interaction. Proc Natl Acad Sci USA. 1999;96:7161–7166. doi: 10.1073/pnas.96.13.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García-Domínguez M, Reyes J C, Florencio F J. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol Microbiol. 2000;35:1192–1201. doi: 10.1046/j.1365-2958.2000.01789.x. [DOI] [PubMed] [Google Scholar]
- 42.Golden J W, Yoon H-S. Heterocyst formation in Anabaena. Curr Opin Microbiol. 1998;1:623–629. doi: 10.1016/s1369-5274(98)80106-9. [DOI] [PubMed] [Google Scholar]
- 43.Harano Y, Suzuki I, Maeda S-I, Kaneko T, Tabata S, Omata T. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J Bacteriol. 1997;179:5744–5750. doi: 10.1128/jb.179.18.5744-5750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haselkorn R, Buikema W J. Nitrogen fixation in cyanobacteria. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. pp. 166–190. [Google Scholar]
- 45.Haselkorn R, Schlictman D, Jones K, Buikema W J. Heterocyst differentiation and nitrogen fixation in cyanobacteria. In: Elmerich C, Kondorosi A, Newton W E, editors. Biological nitrogen fixation for the 21st century. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 93–96. [Google Scholar]
- 46.Hebbar P B, Curtis S E. Characterization of devH, a gene encoding a putative DNA binding protein required for heterocyst function in Anabaena sp. strain PCC 7120. J Bacteriol. 2000;182:3572–3581. doi: 10.1128/jb.182.12.3572-3581.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrero A, Flores E, Guerrero M G. Regulation of nitrate reductase levels in the cyanobacteria Anacystis nidulans, Anabaena sp. strain 7119, and Nostoc sp. strain 6719. J Bacteriol. 1981;145:175–180. doi: 10.1128/jb.145.1.175-180.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hill D R, Belbin T J, Thorsteinsson M V, Bassam D, Brass S, Ernst A, Böger P, Paerl H, Mulligan M E, Potts M. GlbN (cyanoglobin) is a peripheral membrane protein that is restricted to certain Nostoc spp. J Bacteriol. 1996;178:6587–6598. doi: 10.1128/jb.178.22.6587-6598.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang T-C, Lin R-F, Chu M-K, Chen H-M. Organization and expression of nitrogen-fixation genes in the aerobic nitrogen-fixing unicellular cyanobacterium Synechococcus sp. strain RF-1. Microbiology. 1999;145:743–753. doi: 10.1099/13500872-145-3-743. [DOI] [PubMed] [Google Scholar]
- 50.Ihlenfeldt M J A, Gibson J. Acetate uptake by the unicellular cyanobacteria Synechococcus and Aphanocapsa. Arch Microbiol. 1977;113:231–241. doi: 10.1007/BF00492030. [DOI] [PubMed] [Google Scholar]
- 51.Irmler A, Sanner S, Dierks H, Forchhammer K. Dephosphorylation of the phosphoprotein PII in Synechococcus PCC 7942: identification of an ATP and 2-oxoglutarate-regulated phosphatase activity. Mol Microbiol. 1997;26:81–90. doi: 10.1046/j.1365-2958.1997.5521918.x. [DOI] [PubMed] [Google Scholar]
- 52.Jakoby M, Nolden L, Meier-Wagner J, Krämer R, Burkovski A. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Mol Microbiol. 2000;37:964–977. doi: 10.1046/j.1365-2958.2000.02073.x. [DOI] [PubMed] [Google Scholar]
- 53.Jiang F, Hellman U, Sroga G E, Bergman B, Mannervik B. Cloning, sequencing, and regulation of the glutathione reductase gene from the cyanobacterium Anabaena PCC 7120. J Biol Chem. 1995;270:22882–22889. doi: 10.1074/jbc.270.39.22882. [DOI] [PubMed] [Google Scholar]
- 54.Jiang F, Mannervik B, Bergman B. Evidence for redox regulation of the transcription factor NtcA, acting both as an activator and a repressor, in the cyanobacterium Anabaena PCC 7120. Biochem J. 1997;327:513–517. doi: 10.1042/bj3270513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang F, Wisén S, Widersten M, Bergman B, Mannervik B. Examination of the transcription factor NtcA-binding motif by in vitro selection of DNA sequences from a random library. J Mol Biol. 2000;301:783–793. doi: 10.1006/jmbi.2000.4000. [DOI] [PubMed] [Google Scholar]
- 56.Jiang P, Peliska J A, Ninfa A J. Reconstitution of the signal-transduction bicyclic cascade responsible for the regulation of Ntr gene transcription in Escherichia coli. Biochemistry. 1998;37:12795–12801. doi: 10.1021/bi9802420. [DOI] [PubMed] [Google Scholar]
- 57.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 58.Khudyakov I, Wolk C P. Evidence that the hanA gene coding for HU protein is essential for heterocyst differentiation in, and cyanophage A-4(L) sensitivity of, Anabaena sp. strain PCC 7120. J Bacteriol. 1996;178:3572–3577. doi: 10.1128/jb.178.12.3572-3577.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khudyakov I, Wolk C P. hetC, a gene coding for a protein similar to bacterial ABC protein exporters, is involved in early regulation of heterocyst differentiation in Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:6971–6978. doi: 10.1128/jb.179.22.6971-6978.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kikuchi H, Aichi M, Suzuki I, Omata T. Positive regulation by nitrite of the nitrate assimilation operon in the cyanobacteria Synechococcus sp. strain PCC 7942 and Plectonema boryanum. J Bacteriol. 1996;178:5822–5825. doi: 10.1128/jb.178.19.5822-5825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobayashi M, Rodríguez R, Lara C, Omata T. Involvement of the C-terminal domain of an ATP-binding subunit in the regulation of the ABC-type nitrate/nitrite transporter of the cyanobacterium Synechococcus sp. strain PCC 7942. J Biol Chem. 1997;272:27197–27201. doi: 10.1074/jbc.272.43.27197. [DOI] [PubMed] [Google Scholar]
- 62.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 63.Lee H-M, Flores E, Herrero A, Houmard J, Tandeau de Marsac N. A role for the signal transduction protein PII in the control of nitrate/nitrite uptake in a cyanobacterium. FEBS Lett. 1998;427:291–295. doi: 10.1016/s0014-5793(98)00451-7. [DOI] [PubMed] [Google Scholar]
- 64.Lee H-M, Flores E, Forchhammer K, Herrero A, Tandeau de Marsac N. Phosphorylation of the signal transducer PII protein and an additional effector are required for the PII-mediated regulation of nitrate and nitrite uptake in the cyanobacterium Synechococcus sp. PCC 7942. Eur J Biochem. 2000;267:591–600. doi: 10.1046/j.1432-1327.2000.01043.x. [DOI] [PubMed] [Google Scholar]
- 65.Lee H-M, Vázquez-Bermúdez M F, Tandeau de Marsac N. The global nitrogen regulator NtcA regulates transcription of the signal transducer PII (GlnB) and influences its phosphorylation level in response to nitrogen and carbon supplies in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1999;181:2697–1702. doi: 10.1128/jb.181.9.2697-2702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindell D, Padan E, Post A F. Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J Bacteriol. 1998;180:1878–1886. doi: 10.1128/jb.180.7.1878-1886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luque I, Flores E, Herrero A. Nitrite reductase gene from Synechococcus sp. PCC 7942: homology between cyanobacterial and higher-plant nitrite reductases. Plant Mol Biol. 1993;21:1201–1205. doi: 10.1007/BF00023618. [DOI] [PubMed] [Google Scholar]
- 68.Luque I, Flores E, Herrero A. Nitrate and nitrite transport in the cyanobacterium Synechococcus sp. PCC 7942 are mediated by the same permease. Biochim Biophys Acta. 1994;1184:296–298. [Google Scholar]
- 69.Luque I, Flores E, Herrero A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luque I, Herrero A, Flores E, Madueño F. Clustering of genes involved in nitrate assimilation in the cyanobacterium Synechococcus. Mol Gen Genet. 1992;232:7–11. doi: 10.1007/BF00299130. [DOI] [PubMed] [Google Scholar]
- 71.Maeda S-I, Kawaguchi Y, Ohe T-A, Omata T. cis-acting sequences required for NtcB-dependent, nitrite-responsive positive regulation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1998;180:4080–4088. doi: 10.1128/jb.180.16.4080-4088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maeda S-I, Omata T. Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in the transport of nitrate and nitrite. J Biol Chem. 1997;272:3036–3041. doi: 10.1074/jbc.272.5.3036. [DOI] [PubMed] [Google Scholar]
- 73.Maldener I, Fiedler G, Ernst A, Fernández-Piñas F, Wolk C P. Characterization of devA, a gene required for the maturation of proheterocysts in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1994;176:7543–7549. doi: 10.1128/jb.176.24.7543-7549.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martín-Figueroa E, Navarro F, Florencio F J. The GS-GOGAT pathway is not operative in the heterocysts. Cloning and expression of glsF gene from the cyanobacterium Anabaena sp. PCC 7120. FEBS Lett. 2000;476:282–286. doi: 10.1016/s0014-5793(00)01722-1. [DOI] [PubMed] [Google Scholar]
- 75.Marzluf G A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mérida A, Candau P, Florencio F J. Regulation of glutamine sythetase activity in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 by the nitrogen source: effect of ammonium. J Bacteriol. 1991;173:4095–4100. doi: 10.1128/jb.173.13.4095-4100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mérida A, Candau P, Florencio F J. In vitro reactivation of in vivo ammonium-inactivated glutamine synthetase from Synechocystis sp. PCC 6803. Biochem Biophys Res Commun. 1991;181:780–786. doi: 10.1016/0006-291x(91)91258-e. [DOI] [PubMed] [Google Scholar]
- 78.Mérida A, Flores E, Florencio F J. Regulation of Anabaena sp. strain PCC 7120 glutamine synthetase activity in a Synechocystis sp. strain PCC 6803 derivative strain bearing the Anabaena glnA gene and a mutated host glnA gene. J Bacteriol. 1992;174:650–654. doi: 10.1128/jb.174.2.650-654.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mérida A, Leurentop L, Candau P, Florencio F J. Purification and properties of glutamine synthetases from the cyanobacteria Synechocystis sp. strain PCC 6803 and Calothrix sp. strain PCC 7601. J Bacteriol. 1990;172:4732–4735. doi: 10.1128/jb.172.8.4732-4735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller A G, Espie G S. Photosynthetic metabolism of cyanate by the cyanobacterium Synechococcus UTEX 625. Arch Microbiol. 1994;162:151–157. [Google Scholar]
- 82.Montesinos M L, Muro-Pastor A M, Herrero A, Flores E. Ammonium/methylammonium permeases of a cyanobacterium. Identification and analysis of three nitrogen-regulated amt genes in Synechocystis sp. PCC 6803. J Biol Chem. 1998;273:31463–31470. doi: 10.1074/jbc.273.47.31463. [DOI] [PubMed] [Google Scholar]
- 83.Moore J, Kantorow M, Vanderzwaag D, McKenney K. Escherichia coli cyclic AMP receptor protein mutants provide evidence for ligand contacts important in activation. J Bacteriol. 1992;174:8030–8035. doi: 10.1128/jb.174.24.8030-8035.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moreno-Vivián C, Cabello P, Martínez-Luque M, Blasco R, Castillo F. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol. 1999;181:6573–6584. doi: 10.1128/jb.181.21.6573-6584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84a.Muro-Pastor, A. M., A. Herrero, and E. Flores. A nitrogen-regulated group 2 sigma factor from Synechocystis sp. strain PCC 6803 involved in survival under nitrogen stress. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 85.Muro-Pastor A M, Valladares A, Flores E, Herrero A. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J Bacteriol. 1999;181:6664–6669. doi: 10.1128/jb.181.21.6664-6669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muro-Pastor M I, Florencio F J. Purification and properties of NADP+-isocitrate dehydrogenase from the unicellular cyanobacterium Synechocystis sp. PCC 6803. Eur J Biochem. 1992;203:99–105. doi: 10.1111/j.1432-1033.1992.tb19833.x. [DOI] [PubMed] [Google Scholar]
- 87.Muro-Pastor M I, Florencio F J. NADP+-isocitrate dehydrogenase from the cyanobacterium Anabaena sp. strain PCC 7120: purification and characterization of the enzyme and cloning, sequencing, and disruption of the icd gene. J Bacteriol. 1994;176:2718–2726. doi: 10.1128/jb.176.9.2718-2726.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muro-Pastor M I, Reyes J C, Florencio F J. The NADP+-isocitrate dehydrogenase gene (icd) is nitrogen regulated in cyanobacteria. J Bacteriol. 1996;178:4070–4076. doi: 10.1128/jb.178.14.4070-4076.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nagaraja R, Haselkorn R. Protein HU from the cyanobacterium Anabaena. Biochimie. 1994;76:1082–1089. doi: 10.1016/0300-9084(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 90.Nagatani H H, Haselkorn R. Molybdenum independence of nitrogenase component synthesis in the non-heterocystous cyanobacterium Plectonema. J Bacteriol. 1978;134:597–605. doi: 10.1128/jb.134.2.597-605.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okuhara H, Matsumura T, Fujita Y, Hase T. Cloning and inactivation of genes encoding ferredoxin- and NADH-dependent glutamate synthases in the cyanobacterium Plectonema boryanum. Imbalances in nitrogen and carbon assimilations caused by deficiency of the ferredoxin-dependent enzyme. Plant Physiol. 1999;120:33–41. doi: 10.1104/pp.120.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Omata T. Transcriptional and post-translational regulation of nitrate utilization in the cyanobacterium Synechococcus sp. strain PCC 7942. In: Kimiyuki S, Murata N, editors. Stress responses of photosynthetic organisms. Amsterdam, The Netherlands: Elsevier; 1998. pp. 197–214. [Google Scholar]
- 93.Omata T, Andriesse X, Hirano A. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. PCC 7942. Mol Gen Genet. 1993;236:193–202. doi: 10.1007/BF00277112. [DOI] [PubMed] [Google Scholar]
- 94.Potts M, Angeloni S V, Ebel R E, Bassam D. Myoglobin in a cyanobacterium. Science. 1992;256:1690–1692. doi: 10.1126/science.256.5064.1690. [DOI] [PubMed] [Google Scholar]
- 95.Ramasubramanian T S, Wei T-F, Golden J W. Two Anabaena sp. strain PCC 7120 DNA-binding factors interact with vegetative cell- and heterocyst-specific genes. J Bacteriol. 1994;176:1214–1223. doi: 10.1128/jb.176.5.1214-1223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ramasubramanian T S, Wei T-F, Oldham A K, Golden J W. Transcription of the Anabaena sp. strain PCC 7120 ntcA gene: multiple transcripts and NtcA binding. J Bacteriol. 1996;178:922–926. doi: 10.1128/jb.178.3.922-926.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reyes J C, Florencio F J. A new type of glutamine synthetase in cyanobacteria: the protein encoded by the glnN gene supports nitrogen assimilation in Synechocystis sp. strain PCC 6803. J Bacteriol. 1994;176:1260–1267. doi: 10.1128/jb.176.5.1260-1267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reyes J C, Muro-Pastor M I, Florencio F J. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J Bacteriol. 1997;179:2678–2689. doi: 10.1128/jb.179.8.2678-2689.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rowell P, Sampaio M J A M, Ladha J K, Stewart W D P. Alteration of cyanobacterial glutamine synthetase activity in vivo in response to light and NH4+ Arch Microbiol. 1979;120:195–200. [Google Scholar]
- 100.Rubio L M, Flores E, Herrero A. The narA locus of Synechococcus sp. strain PCC 7942 consists of a cluster of molybdopterin biosynthesis genes. J Bacteriol. 1998;180:1200–1206. doi: 10.1128/jb.180.5.1200-1206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rubio L M, Flores E, Herrero A. Molybdopterin guanine dinucleotide cofactor in Synechococcus sp. nitrate reductase: identification of mobA and isolation of a putative moeB gene. FEBS Lett. 1999;462:358–362. doi: 10.1016/s0014-5793(99)01556-2. [DOI] [PubMed] [Google Scholar]
- 102.Rubio L M, Herrero A, Flores E. A cyanobacterial narB gene encodes a ferredoxin-dependent nitrate reductase. Plant Mol Biol. 1996;30:845–850. doi: 10.1007/BF00019017. [DOI] [PubMed] [Google Scholar]
- 103.Sakamoto T, Inoue-Sakamoto K, Bryant D A. A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol. 1999;181:7363–7372. doi: 10.1128/jb.181.23.7363-7372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sauer J, Dirmeier U, Forchhammer K. The Synechococcus strain PCC 7942 glnN product (glutamine synthetase III) helps recovery from prolonged nitrogen chlorosis. J Bacteriol. 2000;182:5615–5619. doi: 10.1128/jb.182.19.5615-5619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schrautemeier B, Neveling U, Schmitz S. Distinct and differently regulated Mo-dependent nitrogen-fixing systems evolved for heterocysts and vegetative cells of Anabaena variabilis ATCC 29413: characterization of the fdxH1/2 gene regions as part of the nif1/2 gene clusters. Mol Microbiol. 1995;18:357–369. doi: 10.1111/j.1365-2958.1995.mmi_18020357.x. [DOI] [PubMed] [Google Scholar]
- 106.Schultz S C, Shields G C, Steitz T A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90°. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 107.Shabb J B, Corbin J D. Cyclic nucleotide-binding domains in proteins having diverse functions. J Biol Chem. 1992;267:5723–5726. [PubMed] [Google Scholar]
- 108.Smith A J, London J, Stainer R Y. Biochemical basis of obligate autotrophy in blue-green algae and thiobacilli. J Bacteriol. 1967;94:972–983. doi: 10.1128/jb.94.4.972-983.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stanier R Y, Cohen-Bazire G. Phototrophic prokaryotes: the cyanobacteria. Annu Rev Microbiol. 1977;31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- 110.Stewart W D P, Rowell P. Effects of l-methionine-dl-sulphoximine on the assimilation of newly fixed NH3, acetylene reduction and heterocyst production in Anabaena cylindrica. Biochem Biophys Res Commun. 1975;65:846–856. doi: 10.1016/s0006-291x(75)80463-3. [DOI] [PubMed] [Google Scholar]
- 111.Suzuki I, Horie N, Sugiyama T, Omata T. Identification and characterization of two nitrogen-regulated genes of the cyanobacterium Synechococcus sp. strain PCC7942 required for maximum efficiency of nitrogen assimilation. J Bacteriol. 1995;177:290–296. doi: 10.1128/jb.177.2.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suzuki I, Sugiyama T, Omata T. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 1993;34:1311–1320. [Google Scholar]
- 113.Suzuki I, Sugiyama T, Omata T. Regulation by cyanate of the genes involved in carbon and nitrogen assimilation in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1996;178:2688–2694. doi: 10.1128/jb.178.9.2688-2694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tapia M I, Ochoa de Alda J A G, Llama M J, Serra J L. Changes in intracellular amino acids and organic acids induced by nitrogen starvation and nitrate or ammonium resupply in the cyanobacterium Phormidium laminosum. Planta. 1996;198:526–531. doi: 10.1007/BF00262638. [DOI] [PubMed] [Google Scholar]
- 115.Thiel T. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J Bacteriol. 1993;175:6276–6286. doi: 10.1128/jb.175.19.6276-6286.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thiel T. Isolation and characterization of the vnfEN genes of the cyanobacterium Anabaena variabilis. J Bacteriol. 1996;178:4493–4499. doi: 10.1128/jb.178.15.4493-4499.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thiel T, Lyons E M, Erker J C, Ernst A. A second nitrogenase in vegetative cells of a heterocyst-forming cyanobacterium. Proc Natl Acad Sci USA. 1995;92:9358–9362. doi: 10.1073/pnas.92.20.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Valladares A, Muro-Pastor A M, Fillat M F, Herrero A, Flores E. Constitutive and nitrogen-regulated promoters of the petH gene encoding ferredoxin:NADP+ reductase in the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 1999;449:159–164. doi: 10.1016/s0014-5793(99)00404-4. [DOI] [PubMed] [Google Scholar]
- 119.Vázquez-Bermúdez M F, Herrero A, Flores E. Uptake of 2-oxoglutarate in Synechococcus strains transformed with the Escherichia coli kgtP gene. J Bacteriol. 2000;182:211–215. doi: 10.1128/jb.182.1.211-215.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vega-Palas M A, Flores E, Herrero A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of transcriptional regulators. Mol Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 121.Vega-Palas M A, Madueño F, Herrero A, Flores E. Identification and cloning of a regulatory gene for nitrogen assimilation in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1990;172:643–647. doi: 10.1128/jb.172.2.643-647.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang Q, Li H, Post A F. Nitrate assimilation genes of the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp. strain WH9601. J Bacteriol. 2000;182:1764–1767. doi: 10.1128/jb.182.6.1764-1767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weber I T, Steitz T A. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 Å resolution. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- 124.Wei T-F, Ramasubramanian T S, Golden J W. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol. 1994;176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wei T-F, Ramasubramanian T S, Pu F, Golden J W. Anabaena sp. strain PCC 7120 bifA gene encoding a sequence-specific DNA-binding protein cloned by in vivo transcriptional interference selection. J Bacteriol. 1993;175:4025–4035. doi: 10.1128/jb.175.13.4025-4035.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wisén S, Jiang F, Bergman B, Mannervik B. Expression and purification of the transcription factor NtcA from the cyanobacterium Anabaena PCC 7120. Protein Expr Purif. 1999;17:351–357. doi: 10.1006/prep.1999.1131. [DOI] [PubMed] [Google Scholar]
- 127.Wolk C P. Heterocyst formation. Annu Rev Genet. 1996;30:59–78. doi: 10.1146/annurev.genet.30.1.59. [DOI] [PubMed] [Google Scholar]
- 128.Wolk C P. Heterocyst formation in Anabaena. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 2000. pp. 83–104. [Google Scholar]
- 129.Wolk C P, Ernst A, Elhai J. Heterocyst metabolism and development. In: Bryant D A, editor. The Molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 769–823. [Google Scholar]
- 130.Wolk C P, Thomas J, Shaffer P W, Austin S M, Galonsky A. Pathway of nitrogen metabolism after fixation of 13N-labeled nitrogen gas by the cyanobacterium, Anabaena cylindrica. J Biol Chem. 1976;251:5027–5034. [PubMed] [Google Scholar]
- 131.Zhou R, Wei X, Jiang N, Li H, Dong Y, Hsi K-L, Zhao J. Evidence that HetR protein is an unusual serine-type protease. Proc Natl Acad Sci USA. 1998;95:4959–4963. doi: 10.1073/pnas.95.9.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]