Abstract
Escherichia coli contains three biochemically distinct fumarases which catalyze the interconversion of fumarate to l-malate in the tricarboxylic acid cycle. Batch culture studies indicated that fumarase activities varied according to carbon substrate and cell doubling time. Growth rate control of fumarase activities in the wild type and mutants was demonstrated in continuous culture; FumA and FumC activities were induced four- to fivefold when the cell growth rate (k) was lowered from 1.2/h to 0.24/h at 1 and 21% O2, respectively. There was a twofold induction of FumA and FumC activities when acetate was utilized instead of glucose as the sole carbon source. However, these fumarase activities were still shown to be under growth rate control. Thus, the activity of the fumarases is regulated by the cell growth rate and carbon source utilization independently. Further examination of FumA and FumC activities in a cya mutant suggested that growth rate control of FumA and FumC activities is cyclic AMP dependent. Although the total fumarase activity increased under aerobic conditions, the individual fumarase activities varied under different oxygen levels. While FumB activity was maximal during anaerobic growth (k = 0.6/h), FumA was the major enzyme under anaerobic cell growth, and the maximum activity was achieved when oxygen was elevated to 1 to 2%. Further increase in the oxygen level caused inactivation of FumA and FumB activities by the high oxidized state, but FumC activity increased simultaneously when the oxygen level was higher than 4%. The same regulation of the activities of fumarases in response to different oxygen levels was also found in mutants. Therefore, synthesis of the three fumarase enzymes is controlled in a hierarchical fashion depending on the environmental oxygen that the cell encounters.
Fumarase, or fumarate hydratase (EC 4.2.1.2), is a component of the citric acid cycle. In facultative anaerobes such as Escherichia coli, fumarase also engages in the reductive pathway from oxaloacetate to succinate during anaerobic growth. Three fumarases, FumA, FumB, and FumC, have been reported in E. coli (7, 8, 17). Comparison of the fumA (FumA), fumB (FumB), and fumC (FumC) gene sequences and the biochemical characteristics of each gene product reveals that they comprise two biochemically distinct types of fumarase (34). The fumA and fumB genes are homologous and encode products of identical sizes which form thermolabile dimers of Mr 120,000. FumA and FumB are class I enzymes and are members of the iron-dependent hydrolases, which include aconitase and malate hydratase (17, 25, 34, 36). The active FumA contains a 4Fe-4S center, and it can be inactivated upon oxidation to give a 3Fe-4S center. However, FumA activity can be restored by anaerobic incubation with iron and thiol (4, 6, 36). FumC is a class II enzyme which does not require iron for its activity, and it forms thermostable tetramers containing identical subunits of Mr 50,000 (34). The fumC gene does not show any homology to either the fumA or fumB genes, but it exhibits extensive homology to the fumarase sequences of Bacillus subtilis, Saccharomyces cerevisiae, and mammals (13, 14, 18, 24, 26, 35). It also shows similarity with the aspartate gene, aspA, of E. coli (27, 33). These genes appear to belong to a family that encode structurally related enzymes.
The physiological function of each E. coli fumarase has been studied by using a triple mutant transformed with a plasmid containing one of the three fum genes (32). The FumA enzyme appeared to be a component of the tricarboxylic acid (TCA) cycle since it was synthesized predominantly under aerobic conditions (34). A decrease in FumA synthesis and catabolite control of the fumA promoter were observed when cells were grown in the presence of glucose (20, 32). Crp binding sites have been proposed within the fumA promoter region (32). The FumB enzyme was shown to be more abundant under anaerobic conditions, and expression of the fumB gene required Fnr as a transcriptional activator (32). However, less is known about the physiological significance of FumC, whose substrate affinity resembles that of FumA. It has been reported that synthesis of FumC increased with the addition of oxidizing agents, and this increase was assumed to be dependent upon the soxRS gene products (6, 15). The soxRS genes encode positive regulators for controlling synthesis of proteins in response to oxidative stress (5, 28). Recently, it has been demonstrated that both superoxide control and iron starvation control of fumC gene expression required the SoxR regulatory protein (20). FumC was thus proposed to substitute for FumA when environmental iron is limiting or when superoxide radicals accumulate.
In this study, we examined the three fumarase activities of E. coli in batch and continuous cultures at different oxygen levels, cell growth rates, and with various carbon substrates in the medium. The results of these studies demonstrate that the activity of the fumarases is regulated independently by the rate of cell growth and carbon source availability. The activity of FumA, FumB, and FumC also shows the hierarchical control which depends on the oxygen availability that the cell encounters. This study further elucidates the different roles of the three fumarases in E. coli.
MATERIALS AND METHODS
Bacterial strains and materials.
The E. coli strains were kindly provided by the E. coli Genetic Stock Center at Yale University. W3110 is a fumA+B+C+ parental strain; EJ1535 is a fumA fumC mutant (34); DJ901 is a soxRS deletion strain (15); and CA8306 is a cya deletion strain (2). The cya mutant strains were constructed by introducing the cya mutation into W3110 by P1 transductions and then selecting for the appropriate phenotype (2).
To construct the fumA deletion strain, the 1.1-kbp SacI-KpnI fragment from upstream of the fumA gene was inserted into pMAK705. The 1.2-kbp KpnI-SphI fragment containing the sequence downstream of fumA was inserted into vector pUC19 which contained the 1.2-kbp fragment of the kanamycin resistance gene. This 2.4-kbp KpnI-SphI fragment was then inserted into pMAK705 to form plasmid pMA1KA2. To construct the fumB deletion strain, the 1.3-kbp SacI-KpnI fragment of the rep gene, which was upstream of the fumB gene, was inserted into pMAK705. The 1.6 kbp of fumB downstream gene was inserted into the pCRII-TOPO vector containing the kanamycin resistance gene to form the 2.8-kbp KpnI-SphI fragment and then inserted into pMAK705 to form plasmid pMB1KB2. To construct the fumC mutant, the 0.6-kbp XbaI-HpaI fragment of the sequence upstream of fumC was inserted into the XbaI and SmaI sites of pBluescript II KS(−) vector to form pBKS-C1. The 0.8-kbp KpnI-HpaI fragment of the sequence downstream of fumC was inserted into the EcoRV and KpnI sites of pBKS-C1 to form pBKS-C1C2. The 1.2-kbp PstI fragment of the kanamycin resistance gene was inserted into pBKS-C1C2 to form pBKS-C1KC2. The 2.7-kbp SacI-SphI fragment containing the fumC and kanamycin resistance genes was then inserted into pMAK705 to form pMC1KC2. To obtain the fumA, fumB, and fumC mutants, plasmids pMA1KA2, pMB1KB2, and pMC1KC2 were integrated into the chromosome as described by Hamilton et al. (8). Single colonies grown on L plates with kanamycin but not on plates with chloramphenicol were checked by PCR and Southern blotting of chromosomal DNA to confirm the mutations.
Cell growth.
For continuous culture experiments, a New Brunswick BiofloIII fermentor (New Brunswick Scientific Co., Inc.) was fitted with a 1.5-liter vessel and operated at a 1-liter liquid working volume as previously described (30). A modified Vogel-Bonner medium (pH 6.5) supplemented with Casamino Acids (0.25 mg/liter) and glucose (or acetate) (2.25 mM) was used to limit cell growth (carbon-limited medium). Aerobic continuous culture conditions were maintained by saturating the culture medium with sterile air at a rate of 300 ml/min. Anaerobic conditions were maintained by continuously sparging the vessel with oxygen-free nitrogen at a rate of 300 ml/min. To vary the cell growth rate, the medium addition rate was adjusted accordingly. The medium addition rates ranged from 4 to 20 ml/min (k = 0.24 to 1.2/h), which corresponded to cell generation times (g) of 173 and 35 min, respectively. To vary the degree of oxygen in the medium, the vessel was sparged with a stream of premixed gas in which the proportion of compressed air (21% O2) and compressed nitrogen (99.8%) was combined using a mixing manifold containing individually precalibrated flow meters for each gas (31). The percent oxygen in the medium was monitored with an Ingold oxygen probe (mode 1046), which was calibrated with 100% air (21% O2) and 99.8% nitrogen (0% O2) prior to inoculation of the vessel in each experiment. When cells were shifted to a new growth rate or oxygen level, steady state was generally achieved in six reactor residence times. This was confirmed by monitoring cell density at 600 nm and assaying the fumarase activity of harvested cells as an indicator that cells had reached equilibrium. The chemostat was maintained under the same conditions until the fumarase activity values varied no more than 5%. Casamino Acids was purchased from Difco Co., Detroit, Mich. All other chemicals used were of reagent grade.
Enzyme assay.
In batch culture, cells were grown aerobically with shaking in 20-ml culture volumes in 150-ml flasks which contained the different carbon substrates at 200 rpm. Flasks were inoculated with overnight cultures grown under the same conditions, and the cells were allowed to double four or five times under mid-log exponential phase prior to harvesting for the fumarase activity assay (11). In continuous culture, the fumarase activities obtained for each condition were independently determined at least twice, and there was less than 10% variation in activity. For cell sampling, 10-ml aliquots of culture were collected and placed on ice. After harvesting the cells by centrifugation, ultrasonicated extracts were prepared, and the supernatants were obtained after centrifuging at 15,000 × g for 20 min. Fumarase assays were performed at 25°C as described (11, 34). The protein concentration of cell extracts was determined by the method of Bradford (1) with Bio-Rad dye reagent using bovine serum albumin (Sigma) as a standard. One unit is defined as 1 μmol of fumarate formed per min at 25°C and pH 7.3. Since FumA and FumB are thermolabile enzymes, FumC activity was obtained by incubation of total enzymes at 18°C for 16 h; this treatment inactivates the fractions attributed to FumA and FumB, whereas the FumC activity is unaffected (15, 16, 34).
RESULTS
Effect of oxygen on FumA, FumB, and FumC activity.
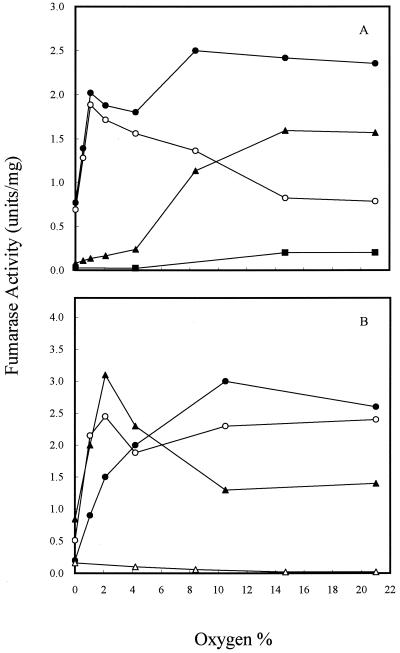
To determine how the three fumarase activities vary in the cell during growth at different oxygen levels, strains W3110, EJ1535, and DJ901 were grown in continuous culture at the same cell growth rate (k = 0.6/h) but the oxygen level was varied from 0 to 21% (Fig. 1). Strain W3110 is a parent strain which contains three fumarase genes. Strain EJ1535 was known to bear a mutation in the fumA gene that also abolishes expression of the fumC gene (34). It has been suggested that all fumarase activity in EJ1535 is due to FumB (16). In continuous culture, we found that the pattern of the three fumarase activities in strain W3110 varied significantly at different oxygen levels (Fig. 1A). Over the range from 0% (anaerobic) to 21% O2, the total fumarase activity was at a low basal level under anaerobic conditions and induced threefold at 21% O2. As the oxygen level was increased, FumA and FumB (FumA+B) activity increased until it achieved a maximum at 1% O2 (ca. 2.5-fold induction over the anaerobic level). However, when the oxygen level was increased to 15% O2, the activity was lowered to a level similar to the anaerobic level. Further increasing the oxygen to 21% did not lower FumA+B activity. The decrease in FumA+B activity may be due to oxidation by O2 under high oxygen levels (4). FumC activity was determined after inactivation of FumA+B activity at 18°C for 16 h as described by Woods et al. (34) and Liohevand and Fridovich (15, 16). The pattern of FumC activity at different oxygen levels differed from that observed for FumA+B. FumC activity remained at the lowest level when the oxygen level was below 4%, but it increased as the oxygen level was raised and achieved maximum activity when the oxygen level was 15%. Further increasing the oxygen to 21% did not change either FumA+B or FumC activities. FumC activity was also determined in a soxRS deletion strain, which showed that activity was maintained at a low level over the range of oxygen levels examined (Fig. 1A). The effect of oxygen tension on the activities of the three fumarases was also determined in fumA, fumB, and fumC deletion strains. Although the fumarase activities increased 30% in the fumA and fumC mutants, a reciprocal role for FumA and FumC was still seen in two mutants over the range of oxygen levels examined (Fig. 1B). In contrast to the pattern of varied fumarase activity in strain W3110, the maximum level of FumB activity in strain EJ1535, in which FumA and FumC were inactivated (16), was observed under completely anaerobic conditions (Fig. 1B). As the level of oxygen in the premixed gas stream was elevated above 4%, the FumB activity of strain EJ1535 was reduced by half. The basal level was about 10% of the maximum level of total fumerase activity seen under completely aerobic conditions. Even though FumB activity in strain EJ1535 was the highest under anaerobic conditions, it was relatively low compared to the FumA+B activity of strain W3110 and the fumC mutant under anaerobic growth conditions. A comparison of the fumarase activities of strains W3110, EJ1535, and the fumC mutant suggests that FumA is the major enzyme synthesized under anaerobic and microaerophilic conditions (ca. 0 to 4% O2), whereas FumC becomes more important under highly oxidized growth conditions.
FIG. 1.
Effect of oxygen on fumarase activity in continuous culture. Cells were grown in glucose (2.25 mM) minimum medium, and k was 0.6/h. (A) Total fumarase activity in W3110 (●); FumA+B activity in W3110 (○); FumC activity in W3110 (▴); FumC activity in DJ901 (■). (B) Total fumarase activity in EJ1535 (FumA− FumC−) (▵); total fumarase activity (FumB+C) in the fumA mutant (●); total fumarase activity (FumA+C) in the fumB mutant (○); total fumarase activity (FumA+B) in the fumC mutant (▴). Units of fumarase activity are expressed as micromoles of fumarate formed per minute at 25°C and pH 7.3.
Effect of different carbon substrates on fumarase activity.
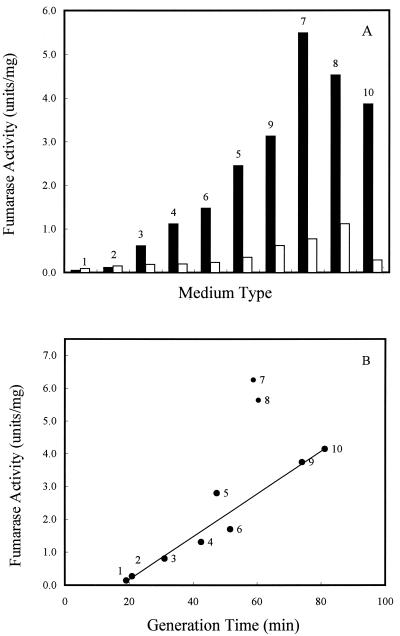
In aerobic batch cultures, FumA+B activities varied over a 50-fold range in strain W3110, but FumC activity did not vary more than 8-fold in all media tested (Fig. 2A). These observations are consistent with the fumA and fumC gene expression levels found during growth with different carbon substrates (20). The fumarase activity of strain EJ1535 was relatively low compared to the activity in strain W3110 (less than 5% in batch minimum medium with glucose as the carbon source) (Table 1). Therefore, the activity of FumA in W3110 is the major component of the FumA+B activity. Interestingly, when cells were grown in batch culture in different media, the total fumarase activity was higher in minimal medium containing two-, three-, or four-carbon compounds than in rich medium (Fig. 2B). The cell growth rates were determined for each type of medium, and when the generation times were graphed versus the level of the total fumarase activities, cell growth rate was inversely proportional to the total fumarase activity but generation time was directly proportional (Fig. 2B). Therefore, fumarase activity is proposed to be growth rate controlled.
FIG. 2.
Effect of cell growth rate and carbon substrate on total fumarase activity in batch culture. (A) W3110 cells were grown in minimal medium with the indicated carbon compounds (40 mM) or in buffered LB. Solid bars, FumA+B activities; open bars, FumC activities. (B) W3110 cells were grown in the indicated medium, and the cell generation time as well as the total fumarase activities were recorded. Medium and carbon compound (40 mM) used for cell growth: 1, LB plus glucose; 2, LB; 3, glucose plus Casamino Acids; 4, glucose; 5, glycerol; 6, xylose; 7, fumarate; 8, succinate; 9, galactose; 10, acetate. Units of fumarase activity are expressed as micromoles of fumarate formed per minute at 25°C and pH 7.3.
TABLE 1.
Total fumarase activity of strain EJ1535 in different media in batch culture
| Mediuma | Fumarase activityb (U/mg) | Generation timec (min) |
|---|---|---|
| LB + glucose | 0.02 | 21 |
| LB + pyruvate | 0.02 | 23 |
| LB | 0.03 | 23 |
| Glucose + Casamino Acids | 0.03 | 40 |
| Glucose | 0.03 | 47 |
| Xylose | 0.03 | 71 |
| Glycerol | 0.03 | 84 |
| Acetate | — | NG |
| Succinate | — | NG |
| Fumarate | — | NG |
Cells were grown aerobically in a minimal basal medium (pH 7.0) with the indicated carbon compounds added (40 mM).
Units are expressed as micromoles of fumarate formed per minute at 25°C and pH 7.3.
NG, no growth.
Effect of cell growth rate on fumarase activities.
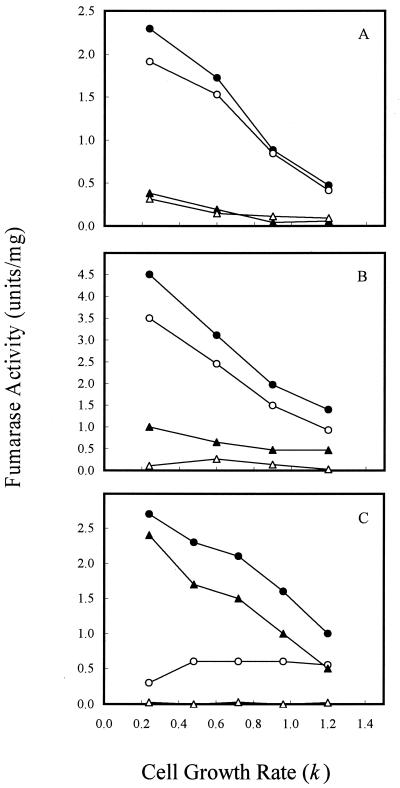
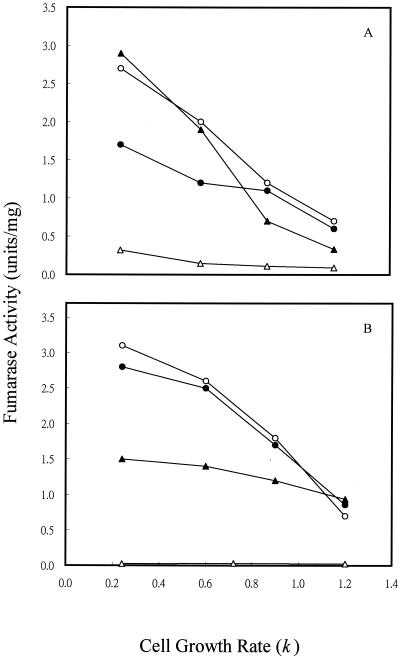
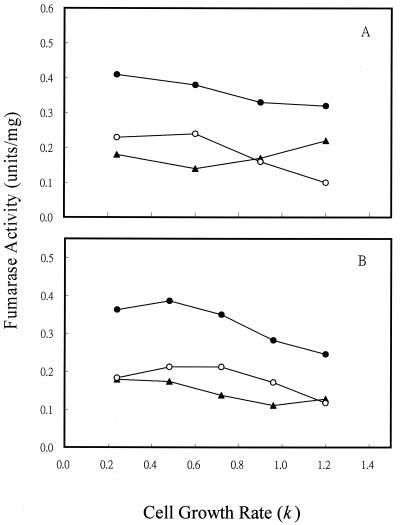
To determine if the variation in fumarase activity with different of carbon substrates was caused directly by the type of carbon compound used or indirectly by the change in cell growth rate, we examined the fumarase activity of cells grown in continuous culture (Fig. 3), where cell growth rate could be controlled by the specific medium flow rate when carbon is limited (i.e., by glucose or acetate). For cultures grown at 1% O2, which gave the maximum activity of FumA+B, both FumA+B and FumC activities were elevated by fourfold when the cell growth rate was shifted from 1.2/h to 0.24/h (Fig. 3A). In contrast, the growth rate control of FumB in strain EJ1535 remained constant: it did not vary by more than twofold at the different growth rates examined. Therefore, growth rate control of FumA+B activity in strain W3110 essentially reflects the growth rate regulation of FumA, since FumB activity was constant in strain EJ1535 under different growth rates. In addition, when strain W3110 was grown in acetate medium, the total fumarase and FumA+B activities still exhibited a fourfold growth rate control. The FumA+B activity was about twofold higher when cells were grown with acetate than with glucose at the same growth rates (Fig. 3B). This finding demonstrates that the growth rate control of FumA and FumC activities is independent of carbon substrate control. When cells were grown under higher agitation (21% O2), the total fumarase and FumC activities were still growth rate dependent. FumC activity was elevated about fourfold when the cell growth rate was decreased from 1.2/h to 0.24/h. However, FumA+B activity was growth rate independent, and its activity remained constant at the lowest level over the range of growth rates examined (Fig. 3C). The shift of FumA activity from growth rate dependent to independent at high oxygen levels may be caused by inactivation of FumA activity by the high oxidative state (4, 36). Growth rate-dependent regulation of fumarase activities was further demonstrated in the fumA, fumB, and fumC mutants. For cultures grown at 1% O2, the total fumarase activity of the fumA, fumB, and fumC mutants was elevated by two-, four-, and eightfold, respectively, when the cell growth rate was shifted from 1.2/h to 0.24/h (Fig. 4A). When cells were grown under higher agitation conditions (21% O2), the total fumarase activity in each of the three mutants remained growth rate dependent. Fumarase activities in the fumA and fumB mutants were each elevated about fourfold when the cell growth rate was decreased from 1.2/h to 0.24/h. However, the fumarase activity of the fumC mutant was only increased 50% over the range of growth rates examined (Fig. 4B). As the fumarase activities were repressed by glucose in batch and continuous cultures (Fig. 2A, 3A, and 3B), we tested whether the cya gene product contributes to this control. The results showed that fumarase activity decreased and became growth rate independent at various growth rates (Fig. 5). Therefore, the growth rate control of FumA and FumC activities was proposed to be cyclic AMP (cAMP) dependent.
FIG. 3.
Effect of cell growth rate on fumarase activity in continuous culture. (A) Cells were grown in glucose (2.25 mM) minimal medium at the indicated growth rates. The oxygen level was 1%. (B) Cells were grown in acetate (2.25 mM) minimal medium, and the oxygen level was 1%. (C) Cells were grown in glucose (2.25 mM) minimal medium, and the oxygen level was 21%. Total fumarase activity in W3110 (●), FumA+B activity in W3110 (○), FumC activity in W3110 (▴), and total fumarase activity in EJ1535 (FumA− FumC−) (▵) were determined.
FIG. 4.
Effect of cell growth rate on fumarase activity in fumA, fumB, and fumC mutants. (A) Cells were grown in glucose (2.25 mM) minimal medium at the indicated growth rates. The oxygen level was 1%. (B) Cells were grown in glucose (2.25 mM) minimal medium, and the oxygen level was 21%. Total fumarase activity (FumB+C) in the fumA mutant (●), total fumarase activity (FumA+C) in the fumB mutant (○), total fumarase activity (FumA+B) in the fumC mutant (▴), and total fumarase activity in EJ1535 (FumA− FumC−) (▵) were determined.
FIG. 5.
Effect of cell growth rate on fumarase activity in a cya mutant strain. (A) Cells were grown in glucose (2.25 mM) minimal medium at the indicated growth rates. The oxygen level was 1%. (B) Cells were grown at 21% O2. Total fumarase activity (●), FumA+B activity (○), and FumC activity (▴) in the cya mutant strain were determined.
DISCUSSION
The existence of three genetically and biochemically distinct fumarases in E. coli poses questions concerning their physiological functions. Their differing affinities for fumarate and malate suggest that they function in reciprocal roles either in the overall oxidation of fumarate via the citric acid cycle (by FumA) or in the ultimate reduction of malate by providing fumarate as an anaerobic electron acceptor (by FumB) (32, 34). Also, it was previously shown that the fumA and fumC genes were expressed at higher levels under aerobic than anaerobic conditions, whereas fumB was expressed under anaerobic condition (20, 29, 32). In this study, we performed a more detailed analysis of three fumarase activities under different oxygen levels. The results indicate that FumA provides the major anaerobic fumarase activity (more than 70% of the total fumarase activity under anaerobic conditions), while the activity of FumB was also fully elevated under anaerobic conditions (Fig. 1). These physiological functions coincide with the results of studies using various fum-lacZ reporter fusions in which the level of fumA gene expression was seven- to eightfold higher than that of fumB even under anaerobic condition (20, 29). As the three fumarase activities in E. coli are regulated by oxygen differently, the cell utilizes a complex strategy for adaptation to diverse environmental oxygen levels. First, FumA activity was about fivefold higher than FumB activity under anaerobic growth conditions, while FumB activity was highest during anaerobiosis. FumA+B activities were elevated to the maximal level under microaerophilic conditions (1 to 2% O2) and then decreased back to anaerobic levels under aerobic conditions (>15% O2) (Fig. 1). These results suggest that FumA is the major fumarase enzyme under microaerophilic conditions (1 to 2% O2) and is constitutively synthesized under fermentation and aerobic growth conditions. FumB is an alternative enzyme during anaerobiosis, since it has a higher affinity for l-malate than for fumarate (34). Second, the observation that FumC activity remains quite low during anaerobic growth supports the model that the product of the fumC gene is not needed by the cell under these conditions because the cell can synthesize FumA and FumB to metabolize malate to fumarate (34). However, during aerobic growth (21% O2), FumC activity was elevated by 20-fold compared to the level under anaerobic conditions (Fig. 1). This elevated level of FumC activity reflects a strategy of the cell to produce an active fumarase, FumC, instead of iron-dependent FumA, which is inactivated under highly oxidative conditions (>4% O2) (4, 36). Higher fumarase activities in the fumA and fumC mutants than in the wild-type strain may be due to the products of the fumA and fumC genes involved in feedback regulation of the promoter of the fumA gene, which drives both fumA and fumC mRNA expression (20). Thus, the results of these studies imply that the three fumarase enzymes are controlled in a hierarchical manner depending on the oxidative conditions that the cell encounters. Therefore, E. coli appears to utilize highly responsive regulatory elements to adjust the levels of the fumarase enzymes during different cell growth conditions for energy generation.
Expression of the fumA and fumC genes has been previously found to be affected by the type of carbon used for cell growth (20). The same pattern of variation is also seen at the biochemical level for fumarase activity (Fig. 2A). Further examination showed that fumarase activity is controlled independently by cell growth rate and carbon substrate availability (Fig. 3 and 4). Prior studies reported that the β-galactosidase activity of fumA-lacZ varies over 20-fold in different media (20), which is similar to the variation of FumA activities in batch culture (Fig. 2A). This result suggests that the growth rate-dependent regulation of FumA activity occurred at the transcriptional level. However, in contrast to a more than 10-fold variation in fumAC-lacZ expression, the β-galactosidase activity of fumC-lacZ remained relatively constant in different media (20). These results suggest that the growth rate regulation of FumC activity is effected via readthrough from the transcription of the upstream fumA promoter instead of transcription from the internal fumC promoter.
Growth rate regulation of succinate dehydrogenase, 2-ketoglutarate dehydrogenase, and NADH dehydrogenase activities has been reported in E. coli (12). Growth rate control of several TCA cycle genes has also recently been reported (21, 22, 30), but the regulation is still unclear. All of them show the same regulatory pattern as the FumA and FumC activities in continuous cultures. It is well known that during rapid aerobic growth, E. coli produces acetate as a by-product. The metabolism of E. coli switches from respiration to fermentation when cells grow at a high specific growth rate with a low glucose concentration even under aerobic conditions (3, 10). It has been suggested that an increase in cell growth rate presumably increases glucose uptake rate and intracellular glucose concentration (23). Recently, the intracellular cAMP concentration has been found to decrease as the dilution rate increases in glucose-limited chemostat (19). Expression of the fumA gene is catabolite controlled (20); in this study, the substrate glucose also caused a twofold suppression of the fumarase activities in batch (Fig. 2A) and continuous (Fig. 3) cultures. In addition, the fumarase activities decreased sixfold at a low growth rate (k = 0.24/h) and did not appreciably alter in a cya mutant at all cell growth rates examined (Fig. 5). Since Crp binding sites have been proposed to reside within the fumA promoter region (32), it is suggested that growth rate-dependent regulation of fumarase activity and catabolite inhibition at high growth rates are controlled by changing intracellular cAMP levels. In addition, the growth rate control of the fumarase activities parallels the previous report that E. coli decreases the respiratory quotient at high growth rates (12). Thus, the decrease in fumarase activity at a high cell growth rate could also account for the decrease in the respiratory quotient values.
ACKNOWLEDGMENTS
This work was supported by grants NSC86-2321-B-009-001 and NSC87-2321-B-009-001 from the National Science Council of the Republic of China.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:28–32. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Brickman E, Soll L, Beckwith J. Genetic characterization of mutations which affect catabolite-sensitive operons in Escherichia coli, including deletions of the gene for adenyl cyclase. J Bacteriol. 1973;116:582–587. doi: 10.1128/jb.116.2.582-587.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Mansi E M T, Holms W H. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J Gen Microbiol. 1989;135:2875–2883. doi: 10.1099/00221287-135-11-2875. [DOI] [PubMed] [Google Scholar]
- 4.Flint D H, Emptage M H, Guest J R. Fumarase A from Escherichia coli: purification and characterization as an iron-sulfur cluster containing enzyme. Biochemistry. 1992;31:10331–10337. doi: 10.1021/bi00157a022. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg J T, Monach P, Chou J H, Josephy P D, Demple B. Positive control of a global anti-oxidant defense regulon activated by superoxide-generated agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruer M J, Guest J R. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology. 1994;140:2531–2541. doi: 10.1099/00221287-140-10-2531. [DOI] [PubMed] [Google Scholar]
- 7.Guest J R, Miles J S, Roberts R E, Woods S A. The fumarase genes of Escherichia coli: location of the fumB gene and discovery of a new gene (fumC) J Gen Microbiol. 1985;131:2971–2984. doi: 10.1099/00221287-131-11-2971. [DOI] [PubMed] [Google Scholar]
- 8.Guest J R, Roberts R E. Cloning and mapping and expression of the fumarase gene of Escherichia coli K-12. J Bacteriol. 1983;153:588–596. doi: 10.1128/jb.153.2.588-596.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han K, Lim H C, Hong J. Acetic acid formation in Escherichia coli fermentation. Biotechnol Bioeng. 1992;39:663–671. doi: 10.1002/bit.260390611. [DOI] [PubMed] [Google Scholar]
- 11.Hill R L, Bradshaw R H. Fumarase. Methods Enzymol. 1969;13:91–99. [Google Scholar]
- 12.Hollywood N, Doelle H W. Effect of specific growth rate and glucose concentration on growth and glucose metabolism of Escherichia coli. K-12. Microbios. 1976;17:23–33. [PubMed] [Google Scholar]
- 13.Kinsella B T, Doonan S. Nucleotide sequence of a cDNA coding for mitochondrial fumarase from human liver. Biosci Rep. 1986;6:921–929. doi: 10.1007/BF01116247. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi K, Yamanishi T. Physicochemical, catalytic, and immunochemical properties of fumarases crystallized separately from mitochondrial and cytosolic fractions of rat liver. J Biochem. 1981;89:1923–1931. doi: 10.1093/oxfordjournals.jbchem.a133394. [DOI] [PubMed] [Google Scholar]
- 15.Liohevand S I, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liohevand S I, Fridovich I. Modulation of the fumarases of Escherichia coli in response to oxidative stress. Arch Biochem Biophys. 1993;301:379–384. doi: 10.1006/abbi.1993.1159. [DOI] [PubMed] [Google Scholar]
- 17.Miles J S, Guest J R. Complete nucleotide sequence of the fumarase gene fumA of Escherichia coli. Nucleic Acids Res. 1984;12:3631–3642. doi: 10.1093/nar/12.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miles J S, Guest J R. Complete nucleotide sequence of the fumarase gene (citG) of Bacillus subtilis 168. Nucleic Acids Res. 1985;13:131–140. doi: 10.1093/nar/13.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notley-McRobb L, Death A, Ferenci T. The relationship between external glucose concentration and cAMP levels inside Escherichia coli: implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology. 1997;143:1909–1918. doi: 10.1099/00221287-143-6-1909. [DOI] [PubMed] [Google Scholar]
- 20.Park S-J, Gunsalus R P. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: role of the arcA, fnr, and soxR gene products. J Bacteriol. 1995;177:6255–6262. doi: 10.1128/jb.177.21.6255-6262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S-J, Cotter P A, Gunsalus R P. Regulation of malate dehydrogenase (mdh) gene expression in Escherichia coli in response to oxygen, carbon, and heme availability. J Bacteriol. 1995;177:6652–6656. doi: 10.1128/jb.177.22.6652-6656.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park S-J, Tseng C P, Gunsalus R P. Regulation of the Escherichia coli succinate dehydrogenase (sdhCDAB) operon in response to anaerobiosis and medium richness: role of the ArcA and Fnr proteins. Mol Microbiol. 1995;15:473–482. doi: 10.1111/j.1365-2958.1995.tb02261.x. [DOI] [PubMed] [Google Scholar]
- 23.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacchettini J C, Frazier M W, Chiara D C, Banazak L J, Grant P A. Amino acid sequence of porcine heart fumarases. Biochem Biophys Res Commun. 1988;153:435–440. doi: 10.1016/s0006-291x(88)81243-9. [DOI] [PubMed] [Google Scholar]
- 25.Shibata H, Gardiner W E, Schwartzbach S D. Purification, characterization, and immunochemical properties of fumarase from Euglena gracilis var. bacillaris. J Bacteriol. 1985;164:762–768. doi: 10.1128/jb.164.2.762-768.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki T, Sato M, Yoshida T, Tudoi S. Rat liver mitochondrial and cytosolic fumarases with identical amino acid sequences are encoded by a single gene. J Biol Chem. 1989;264:2581–2586. [PubMed] [Google Scholar]
- 27.Takagi J S, Ida N, Tokushige M, Sakamoto A, Shimura Y. Cloning and nucleotide sequence of the aspartase gene of Escherichia coli W. Nucleic Acids Res. 1985;13:2063–2074. doi: 10.1093/nar/13.6.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsaneva I R, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng C P. Regulation of fumarase (fumB) gene expression in Escherichia coli in response to oxygen, iron and heme availability: role of the arcA, fur, and hemA gene products. FEMS Microbiol Lett. 1997;157:67–72. doi: 10.1111/j.1574-6968.1997.tb12754.x. [DOI] [PubMed] [Google Scholar]
- 30.Tseng C-P, Hansen A K, Cotter P G, Gunsalus R P. Effect of cell growth rate on expression of the anaerobic respiratory pathway operons frdABCD, dmsABC, and narGHJI of Escherichia coli. J Bacteriol. 1994;176:6599–6605. doi: 10.1128/jb.176.21.6599-6605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tseng C-P, Albrecht J, Gunsalus R P. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE, cydAB) and anaerobic (narGHJI, frdABCD, dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol. 1996;178:1094–1098. doi: 10.1128/jb.178.4.1094-1098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woods S A, Guest J R. Differential roles of the Escherichia coli fumarases and fnr-dependent expression of fumarase B and aspartase. FEMS Microbiol Lett. 1988;48:219–224. [Google Scholar]
- 33.Woods S A, Miles J S, Roberts R E, Guest J R. Structural and functional relationships between fumarase and aspartase: nucleotide sequence of the fumarase (fumC) and aspartase (aspA) genes of Escherichia coli K-12. Biochem J. 1986;237:547–557. doi: 10.1042/bj2370547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woods S A, Schwartzbach S D, Guest J R. Two biochemically distinct classes of fumarases in Escherichia coli. Biochim Biophys Acta. 1988;954:14–26. doi: 10.1016/0167-4838(88)90050-7. [DOI] [PubMed] [Google Scholar]
- 35.Wu M, Tzagoloff A. Mitochondrial and cytoplasmic fumarases in Saccharomyces cerevisiae are encoded by a single nuclear gene, FUM1. J Biol Chem. 1987;262:12275–12282. [PubMed] [Google Scholar]
- 36.Yuji U, Yumoto N, Tokushige M, Fukui K, Ohya-Nishiguchi H. Purification and characterization of two types of fumarases from Escherichia coli. J Biochem. 1991;109:728–733. doi: 10.1093/oxfordjournals.jbchem.a123448. [DOI] [PubMed] [Google Scholar]