Abstract
The Staphylococcus aureus genome encodes three ferric uptake repressor (Fur) homologues: Fur, PerR, and Zur. To determine the exact role of Fur in S. aureus, we inactivated the fur gene by allelic replacement using a tetracycline resistance cassette, creating strain MJH010 (fur). The mutant had a growth defect in rich medium, and this defect was exacerbated in metal-depleted CL medium. This growth defect was partially suppressed by manganous ion, a metal ion with known antioxidant properties. This suggests that the fur mutation leads to an oxidative stress condition. Indeed, MJH010 (fur) has reduced levels of catalase activity resulting from decreased katA transcription. Using a katA-lacZ fusion we have determined that Fur functions, either directly or indirectly, as an iron-dependent positive regulator of katA expression. Transcription of katA is coregulated by Fur and PerR, since in MJH010 (fur) transcription was still repressed by manganese while transcription in MJH201 (fur perR) was unresponsive to the presence of iron or manganese. Siderophore biosynthesis was repressed by iron in 8325-4 (wild-type) but in MJH010 (fur) was constitutive. A number of putative Fur-regulated genes were identified in the incomplete genome databases using known S. aureus Fur box sequences. Of those tested, the sstABCD and sirABC operons and the fhuD2 and orf4 genes were found to have Fur-regulated expression. MJH010 (fur) was attenuated (P < 0.04) in a murine skin abscess model of infection, as was double-mutant MJH201 (fur perR) (P < 0.03). This demonstrates the importance in vivo of iron homeostasis and oxidative stress resistance regulation in S. aureus.
The ability of a pathogen to successfully colonize tissues and proliferate is limited by iron availability in vivo. Iron, although abundant, is mostly bound to host carrier proteins, such as transferrin and lactoferrin (56). In addition, the limitation of metal ions, through nonspecific host responses to infection such as hypoferremia (6), reduces the ability of bacteria to replicate and increases their susceptibility to clearance by the immune system. Iron, together with manganese, is a cofactor for antioxidant defence enzymes of the pathogen, e.g., catalase, peroxidase, and superoxide dismutase (1). The reactivity of iron, however, means that it is potentially toxic to bacteria. For example, the Fenton reaction between intracellular iron and endogenously produced hydrogen peroxide produces the deleterious hydroxyl radical (29, 30, 43, 44).
Bacteria possess a number of iron-scavenging mechanisms to overcome iron limitation in vivo. The first of these is the secretion of high-affinity iron(III)-chelating ligands, called siderophores, that bind available iron and that are actively transported back into the cell via specific surface receptors (16, 24). A second mechanism, found in the non-siderophore-producing pathogens Neisseria meningitidis, Haemophilus influenzae, and Actinobacillus pleuropneumoniae (16, 56), involves direct contact between host transferrin and bacterial-cell-wall-located transferrin-binding proteins. A third mechanism involves importing iron(II) directly into the cell via membrane protein FeoB (9).
Staphylococcus aureus is capable of producing siderophores, removing iron from transferrin via cell wall transferrin-binding proteins (38, 52), and its genome encodes at least two FeoB homologues (http://www.genome.ou.edu; http://www.tigr.org). Siderophores aureochelin (17), staphyloferrin A (32, 35), and staphyloferrin B (19, 26) have been isolated from different strains of S. aureus, and purified staphyloferrin A can remove iron from transferrin (36). The 42-kDa cell wall transferrin-binding protein (Tpn) from S. aureus possesses glyceraldehyde-3-phosphate dehydrogenase activity and can sequester iron from transferrin in vitro (37). The removal of iron from transferrin was shown to be a receptor-mediated process involving primary receptor recognition of the N lobe of human transferrin (36). The importance of these iron acquisition processes in the virulence of S. aureus is not known.
Proteins that sense the levels of intracellular ions respond accordingly by modulating gene expression. These metalloregulatory proteins cluster into four distinct families represented by Fur (ferric uptake regulator), DtxR (diphtheria toxin repressor), MerR, and ArsR (42). The well-characterized DtxR and Fur proteins have similar roles with respect to iron homeostasis (49). S. aureus, like Bacillus subtilis (11, 12, 23, 42), encodes three Fur homologues (Fur, PerR, and Zur) and a DtxR homologue, called MntR. Zur functions as a zinc homeostasis regulator in S. aureus but is not important for pathogenicity (J. A. Lindsay and S. J. Foster, submitted for publication). We have shown that PerR functions as a peroxide stress regulator that controls iron storage proteins and that it is necessary for the virulence of S. aureus (M. J. Horsburgh, M. O. Clements, H. M. Crossley, E. Ingham, and S. J. Foster, submitted for publication). In addition, PerR was found to directly regulate the expression of Fur and to regulate its own expression. Purified S. aureus Fur protein binds in vitro to the promoter elements of the fhuC and sirA genes that encode homologues of iron-siderophore uptake genes (58). Expression of S. aureus Fur from a multicopy plasmid was shown to partially restore iron-responsive siderophore expression in a B. subtilis fur mutant (58). The exact role of Fur in S. aureus and its contribution to pathogenicity in vivo have not been determined.
Here we present our data that show that S. aureus Fur functions as the major regulator of iron supply and coordinately regulates catalase-mediated oxidative stress resistance with peroxide stress regulator PerR.
MATERIALS AND METHODS
Media and growth conditions.
S. aureus and Escherichia coli strains and plasmids are listed in Table 1. E. coli was grown in Luria-Bertani medium at 37°C. S. aureus was grown at 37°C with shaking at 250 rpm in brain heart infusion (BHI) broth (Oxoid), SSD medium (34), or chemically defined metal limitation medium (CL). The components of CL are (concentrations in milligrams per liter are in parentheses) Na2HPO4 (7,000), KH2PO4 (300), adenine sulfate (20), guanine-HCl (20), l-glutamic acid (2,220), l-aspartic acid (2,220), l-proline (2,220), glycine (2,220), l-threonine (2,220), l-serine (2,220), l-alanine (2,220), l-lysine-HCl (560), l-isoleucine (560), l-leucine (560), l-histidine (440), l-valine (440), l-arginine (330), l-cystine (220), l-phenylalanine (190), l-tyrosine (170), l-methionine (170), l-tryptophan (60), pyridoxal (0.8), pyridoxamine-2HCl (0.8), d-pantothenic acid (0.4), riboflavin (0.4), nicotinic acid (0.4), thiamine-2HCl (0.4), and biotin (0.02). CL was treated with 20 g of Chelex-100 (Sigma) liter−1 with stirring at room temperature for 4 h to deplete all divalent and trivalent metal ions. After removal of Chelex by filter sterilization, MgSO4 was aseptically added to the medium at a final concentration of 400 μM. CLR medium was prepared as described above but with the following metals added at 0.2 μM: calcium chloride, copper sulfate, ferrous sulfate, manganese sulfate, nickel sulfate, molybdenum sulfate, and zinc sulfate. Colonies from non-Chelex-treated CL agar plates were used to inoculate a CLR preculture. Experimental 25-ml cultures in acid-washed 250-ml flasks were inoculated to a starting optical density at 600 nm (OD600) of 0.002 prior to growth. When included, antibiotics were added at the following concentrations: ampicillin, 100 mg liter−1; kanamycin, 50 mg liter−1; neomycin, 50 mg liter−1; tetracycline, 5 mg liter−1; erythromycin and lincomycin, 5 and 25 mg liter−1, respectively.
TABLE 1.
Strains, plasmids, and primers used in this study
| Primer, strain, or plasmid | Genotype, description, or sequencea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | φ80 Δ(lacZ)M15 Δ(argF-lac)U169 endA1 recA1 | 46 |
| hsdR17(rK−mK+) deoR thi-1 supE44 gyrA96 relA1 | ||
| S. aureus | ||
| 8325-4 | Wild-type strain cured of prophages | Laboratory stock |
| RN4220 | Restriction-deficient transformation recipient | Laboratory stock |
| MJH001 | perR::kan | Horsburghb |
| MJH002 | ahpC::pAZ106 ahpC+ | Horsburgh |
| MJH005 | fur::pAZ106 fur+ | Horsburgh |
| MJH006 | katA::pAZ106 katA+ | Horsburgh |
| MJH007 | mrgA::pAZ106 mrgA+ | Horsburgh |
| MJH008 | perR::pAZ106 perR+ | Horsburgh |
| MJH010 | fur::tet | This study |
| MJH011 | fhuD2::pAZ106 fhuD2+ | This study |
| MJH012 | sirA::pAZ106 sirA+ | This study |
| MJH013 | orf4::pAZ106 orf4+ | This study |
| MJH014 | sstA::pAZ106 sstA+ | This study |
| MJH201 | fur::tet perR::kan | This study |
| MJH211 | fur::tet fhuD2::pAZ106 fhuD2+ | This study |
| MJH212 | fur::tet sirA::pAZ106 sirA+ | This study |
| MJH213 | fur::tet orf4::pAZ106 orf4+ | This study |
| MJH214 | fur::tet sstA::pAZ106 sstA+ | This study |
| MJH202 | fur::tet ahpC::pAZ106 ahpC+ | This study |
| MJH206 | fur::tet katA::pAZ106 katA+ | This study |
| MJH207 | fur::tet mrgA::pAZ106 mrgA+ | This study |
| MJH208 | fur::tet perR::pAZ106 perR+ | This study |
| MJH306 | fur::tet perR::kan katA::pAZ106 katA+ | |
| Plasmids | ||
| pAZ106 | Promoterless lacZ erm insertion vector | 59 |
| pAUL-A | Temperature-sensitive erm integrational shuttle vector | 13 |
| pMAL14 | OL 23-24 fhuD2 PCR fragment in pAZ106 (1.1 kb) | This study |
| pMAL15 | OL27-28 sirA PCR fragment in pAZ106 (1.01 kb) | This study |
| pMAL20 | OL38-39 orf4 PCR fragment in pAZ106 (0.75 kb) | This study |
| pMAL26 | OL51-26 sstA PCR fragment in pAZ106 (0.75 kb) | This study |
| pMAL17 | EcoRI-XhoI PCR fragment containing a tet cassette (2 kb) inserted in a KpnI site engineered into the fur gene by PCR | This study |
| Primers | ||
| OL22 | CCAGAATTCCGTAAGCACGTATAATTCCTTCTTG | This study |
| OL23 | CACAGGATCCAACGATTGCAACATTGCCAACTGTGC | This study |
| OL24 | CCAGAATTCGCTTGATATAATACTTCTCCACC | This study |
| OL26 | CCAGAATTCGAAGTTGCAATGGCAGCACCTAC | This study |
| OL27 | CCAGAATTCCCACTACATCCTGC | This study |
| OL28 | AATTGGATCCGGTACACGACTAGCACCGAT | This study |
| OL29 | AACCGCTCGAGTGATCGTTCAGAAGTGATTGCAGC | This study |
| OL30 | CCGGTACCTTCCAACGATGTCCACTCCCCTAC | This study |
| OL31 | CCGGTACCCATGGTGTGTGTGAAACGTGCCAA | This study |
| OL32 | CCGGTACCCGGATTTTATGACCGATGATGAAG | This study |
| OL33 | CCGGTACCTTAGAAATCCCTTTGAGAATGTTT | This study |
| OL38 | AATTGGATCCTATCTCTTCCTTGTAAAATCATCTC | This study |
| OL39 | CCAGAATTCGGCATGTATCTTGATGCATCTTCAG | This study |
| OL51 | CACAGGATCCATCTCATTGCGCACGAGTGCTG | This study |
Restriction enzyme sites used for cloning are underlined.
Horsburgh et al., submitted.
Construction of strains.
The recombinant strains used in this study were constructed by PCR using Pwo polymerase (Roche) and standard cloning techniques (45). Derivatives of plasmid pAZ106, an integrating plasmid conferring resistance to erythromycin and containing a promoterless lacZ gene (59), and plasmid pAUL-A, a temperature-sensitive integrating plasmid conferring resistance to erythromycin, were constructed (13). A plasmid for disrupting fur was constructed by PCR amplification of two adjoining 1-kb fur partial fragments using primers OL29 with OL31 and OL30 with OL22 (Table 1), with incorporated XhoI, KpnI, KpnI, and EcoRI restriction sites, respectively, on the primers. A 1.5-kb tetracycline resistance cassette from pDG1513 (25) was amplified using primers OL32 and OL33, which contained KpnI restriction sites. Simultaneous ligation of the appropriately digested fur fragments and tetracycline cassette with SalI-EcoRI-digested pAUL-A was performed, and, following transformation of E. coli DH5α, tetracycline-resistant colonies were selected. Three identical clones, pMAL17, containing a tet cassette inserted into the fur gene were obtained. Transcriptional reporter fusions to the fhuD2, sirA, sstA, and orf4 genes were made by PCR amplification of suitable DNA fragments using the primers detailed in Table 1. Typically, between 0.5 and 1 kb of upstream DNA and 0.2 and 0.5 kb of the start of the gene were amplified using Pwo DNA polymerase (Roche). The purified DNA fragments were digested with BamHI and EcoRI and cloned into plasmid pAZ106 digested with the same enzymes. Transformation of S. aureus RN4220 was performed as described by Schenk and Ladagga (46), and phage transduction into recipient 8325-4 was performed as described by Novick (40) using φ11 as the transducing phage. MJH010 (fur) was isolated after transduction of an integrated S. aureus RN4220 transformant of pMAL17 into S. aureus 8325-4, selecting for Tetr Erys colonies. Southern blotting and PCR were used in each case to verify the location and correct integration of DNA at the chromosomal loci.
β-Galactosidase assays.
Levels of β-galactosidase activity were measured as described previously (14, 15) with the following modifications. Samples (0.1 ml) were harvested, and cell pellets were stored at −20°C. Thawed pellets were resuspended in 0.5 ml of ABT buffer (60 mM K2HPO4, 40 mM KH2PO4, 100 mM NaCl). The assay was started with the addition of 50 μl of freshly prepared 4-methylumbelliferyl-β-d-galactoside (10 mg ml−1), and the assay mixture was incubated at 25°C for 60 min. The assay was stopped with the addition of 0.5 ml of 0.4 M Na2CO3. The stopped assay mixture was then serially diluted in a 50:50 (vol/vol) mixture of ABT and Na2CO3 in 96-well microtiter plates (Nunc). Fluorescence was measured using a Victor plate reader (Wallac) with a 0.1-s count time and calibrated with standard concentrations of 4-methylumbelliferone (MU). One unit of β-galactosidase activity was defined as the amount of enzyme that catalyzed the production of 1 pmol of MU min−1 OD600 unit−1. The results presented here were representative of three independent experiments, which showed less than 20% variability.
Catalase assays, H2O2 resistance, and starvation survival.
Catalase activity was assayed spectrophotometrically at 240 nm as described by Beers and Sizer (5), and protein was measured by the method of Bradford (7) using bovine serum albumin (fraction V; Sigma) as the standard. Hydrogen peroxide resistance assays were carried out as described by Watson et al. (54) with the following modifications. Cells, grown to exponential phase (OD600 = 0.5) in CLR, were washed and diluted into phosphate-buffered saline (PBS) to an OD600 of 0.2. Following challenge with 7.5 mM H2O2, the cells were diluted in PBS containing catalase at 10 mg ml−1 and serially diluted in PBS and viability was assessed by overnight growth on BHI agar. Comparative starvation survival experiments were performed in glucose-limiting CDM medium with shaking at 250 rpm, at 37°C, as described by Watson et al. (54).
Detection of siderophore activity.
Siderophore activity in culture supernatants from cells grown in SSD medium was assayed by the liquid chrome azurol S (CAS) assay described by Schwyn and Neilands (47). Dilutions of supernatants were mixed with equal volumes of CAS shuttle solution. After 30 min of incubation at room temperature, the absorbance at 630 nm was determined using SSD medium as a blank and deferroxamine mesylate (Sigma) as a reference standard. Activity was measured as micromoles of deferroxamine equivalents per OD600 unit of the culture.
Virulence testing of strains in a murine skin abscess model.
S. aureus strains were grown to stationary phase in BHI (time, 15 h), harvested by centrifugation, and washed twice in PBS. The cell numbers were adjusted to 5 × 108 CFU ml−1, and then 200 μl of cell suspension was injected subcutaneously into female 6- to 8-week-old BALB/c mice. After 7 days, the mice were euthanized with CO2 and skin lesions were aseptically removed and stored frozen in liquid nitrogen. The lesions were weighed, chopped, and homogenized in a miniblender in 2.5 ml of cold PBS. After 1 h of incubation on ice the lesions were homogenized again before serial dilution of the suspension, and the total number of bacteria were determined by growth on BHI agar. The statistical significance of the percent recovery of strains was evaluated by using Student's t test and the Mann-Whitney U test, with a 5% confidence limit.
RESULTS
Isolation of an S. aureus fur mutant.
A homologue of the B. subtilis fur gene was identified in the S. aureus 8325 genome database (http://www.genome.ou.edu). To determine the role of fur in S. aureus, we introduced a tetracycline resistance cassette into the fur gene using allelic replacement to disrupt the chromosomal copy, creating strain MJH010.
MJH010 (fur) has a growth defect.
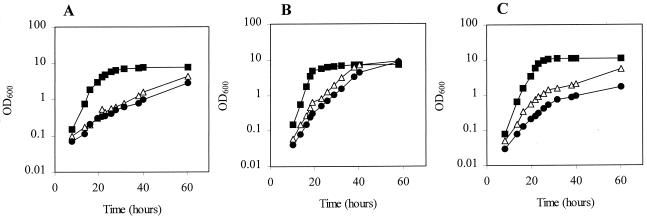
The growth yield of MJH010 (fur) was found to be much reduced compared to that of 8325-4 (wild type) after growth to stationary phase in complex media such as BHI broth (OD600 = 4.5 and 9.5, respectively) (data not shown). This phenotype was partially recovered in rich medium when a mutation in perR was introduced, producing strain MJH201 (fur perR) (OD600 = 7.8). The growth rates of MJH010 (fur) and MJH201 (fur perR) in chemically defined metal-depleted medium (CL) were greatly reduced compared to that of 8325-4 (wild type) (Fig. 1A). This reduced growth rate was improved in CLR, which contains 0.2 μM concentrations of a range of divalent metal ions (data not shown), or CL containing micromolar concentrations of manganese (Fig. 1B). In contrast, the addition of 20 μM iron sulfate did not improve the growth of MJH010 (fur) and further impaired the growth of MJH201 (fur perR) (Fig. 1C).
FIG. 1.
Growth of 8325-4 (wild type) (■), MJH010 (fur) (▵), and MJH201 (fur perR) (●) in CL medium (no added metals except magnesium) (A), CL with 20 μM manganese chloride (B), and CL with 20 μM iron sulfate (C).
MJH010 (fur) has reduced oxidative stress resistance.
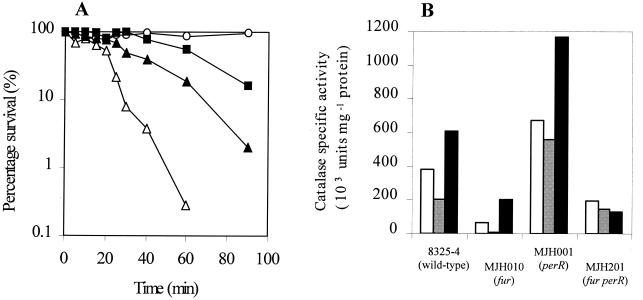
Since the impaired growth phenotype of MJH010 (fur) could be rescued by adding manganous ion, a metal ion with known antioxidant properties (3, 7, 22, 48), we hypothesized that the mutant had an oxidative stress defect. To test this further, MJH010 (fur), MJH201 (fur perR), MJH001 (perR), and 8325-4 (wild type) were assayed for resistance to 7.5 mM H2O2 and for levels of catalase activity after growth in CLR medium. Both MJH010 (fur) and MJH201 (fur perR) were significantly more sensitive to H2O2 than 8325-4 (wild type) (Fig. 2A), and the catalase-specific activity measured in MJH010 (fur) was found to be sixfold lower than that in 8325-4 (wild type). After growth in CLR medium, with either 20 μM manganese or 20 μM iron sulfate added, the levels of catalase in MJH010 (fur) were 30- and 3-fold less than that in 8325-4 (wild type), respectively (Fig. 2B). MJH001 (perR) showed the expected increase in catalase activity under all conditions. Catalase levels in MJH201 (fur perR) were found to be lower than those in 8325-4 (wild type) and MJH001 (perR) in each of the growth conditions tested, revealing a loss of response to the metal ions in the growth medium. These results suggest that Fur acts as a positive regulator of catalase expression.
FIG. 2.
(A) Effect of H2O2 (7.5 mM) on washed, exponential-phase cells of 8325-4 (wild type) (■), MJH010 (fur) (▵), MJH201 (fur perR) (▴), and MJH001 (perR) (○). (B) Total catalase activities of washed, lysed stationary-phase cells after growth in CLR medium (white bars), CL with 20 μM manganese chloride (grey bars), or CL medium with 20 μM iron sulfate (black bars).
The effect of Fur on katA expression.
Expression of katA in MJH206 (fur katA-lacZ) was reduced compared to that in MJH006 (katA-lacZ) during growth in CLR or CLR with 20 μM iron sulfate (Fig. 3A and B). Growth in CLR with 20 μM manganese added, a concentration that ensures PerR-mediated repression (M. J. Horsburgh, et al., submitted), virtually eliminated all transcription of katA in MJH206 (fur katA-lacZ) (Fig. 3B). Expression of katA in MJH306 (fur perR katA-lacZ) was found to be uniformly low in each of the media tested (Fig. 3C). These results support the function of Fur as a positive regulator of katA transcription.
FIG. 3.
Analysis of transcription from katA-lacZ fusions in different backgrounds during growth in CLR medium. Shown are growth (filled symbols) and expression (open symbols) of MJH006 (katA-lacZ) (A), MJH206 (fur katA-lacZ) (B), and MJH306 (fur perR katA-lacZ) (C) in CLR medium (triangles), CLR medium with 20 μM manganese chloride (diamonds), and CLR medium with 20 μM iron sulfate (circles).
The effect of Fur on control of the PerR regulon.
Since Fur apparently regulates katA, which was shown to be a member of the PerR regulon (M. J. Horsburgh et al., submitted), we investigated the effect of Fur on the expression of other known PerR-regulated genes. The fur mutation was transduced into lacZ fusion strains of some of the known PerR-regulated genes. MJH002 (ahpC-lacZ), MJH202 (fur ahpC-lacZ), MJH003 (bcp-lacZ), MJH203 (fur bcp-lacZ), MJH007 (mrgA-lacZ), MJH207 (fur mrgA-lacZ), MJH008 (perR-lacZ), and MJH208 (fur perR-lacZ) were grown in CLR medium to determine whether there was any Fur-regulated expression. In each case expression was at a higher level in the fur background than in the wild-type background (data not shown), in contrast to expression of katA, which was reduced in MJH206 (fur katA-lacZ). The increased expression of these PerR-regulated genes was at levels similar to that observed when katA was inactivated (M. J. Horsburgh et al., submitted). A similar observation was made for B. subtilis, where inactivation of katA or ahpC increases expression of the PerR regulon, possibly due to an intracellular accumulation of peroxide (2, 10).
Fur regulation of siderophore production.
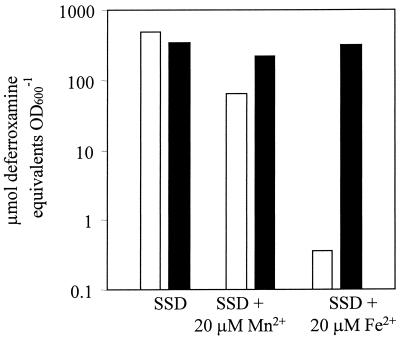
Since Fur is a known regulator of iron homeostasis in many bacteria, we examined the production of siderophores in MJH010 (fur). Siderophore biosynthesis was found to be constitutive in MJH010 (fur), whereas production was strongly repressed by iron and partially repressed by manganese in 8325-4 (wild type) (Fig. 4). Levels of siderophore production in MJH001 (perR) and 8325-4 (wild type) were the same in all conditions tested (data not shown).
FIG. 4.
Siderophore levels of 8325-4 (wild type) (open bars) and MJH010 (fur) (solid bars) strains after growth in metal-depleted SSD medium with or without manganese chloride or iron sulfate (20 μM each) supplementation.
Fur regulates iron uptake proteins in S. aureus.
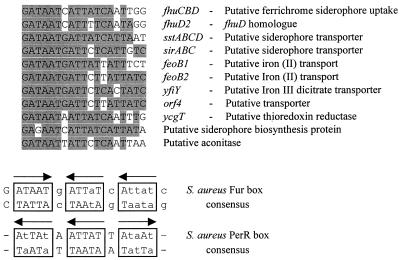
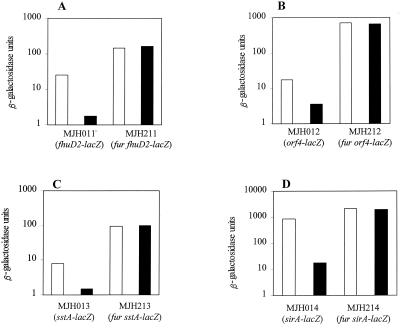
A search of the incomplete S. aureus 8325-4 genome (http://www.tigr.org and http://www.genome.ou.edu) revealed a large number of genes with homology to iron-regulated proteins from other bacteria. Many of these were preceded by sequences with homology to the putative Fur box previously identified for sirA (28) and to part of the fhuC promoter region protected by Fur (Fig. 5A) (58). To investigate whether these genes were regulated by Fur, lacZ fusions were constructed to monitor transcription from the promoter regions of sirA, fhuD2, sstA, and orf4, creating strains MJH011 (fhuD2-lacZ), MJH211 (fur fhuD2-lacZ), MJH012 (sirA-lacZ), MJH212 (fur sirA-lacZ), MJH013 (orf4-lacZ), MJH213 (fur orf4-lacZ), MJH014 (sstA-lacZ), and MJH214 (fur sstA-lacZ).
FIG. 5.
(A) Alignment of the putative Fur boxes identified in the incomplete S. aureus databases (http://www.tigr.org and http://www.genome.ou.edu). Sequences were identified using the Microsoft Word 2000 Find tool by inserting the “any character” function to enable mismatches. (B) The S. aureus consensus sequence was compiled from all of the sequences and is presented as described in reference 21. The sirA Fur box was taken from reference 28, the sstA gene name was taken from the sequence with GenBank accession no. AJ005352, the fhuC Fur box was from part of the protected region described in reference 58, and the PerR consensus sequence was from M. J. Horsburgh et al. (submitted).
Each of these genes was regulated in a Fur-dependent manner, and transcription was strongly repressed by the addition of micromolar levels of iron sulfate (Fig. 6). No further iron regulation of these genes was observed in the fur background, suggesting that Fur is likely to be the sole iron-dependent regulator of iron uptake transporters in S. aureus.
FIG. 6.
Effect of Fur on transcription of promoter-lacZ fusions to the fhuD2, orf4, sstA, and sirA genes during growth in CLR medium containing no iron (open bars) or 10 μM iron sulfate (solid bars). Values presented were taken during exponential growth (OD600 = 1) from growth curves sampled throughout growth. The data presented are representative of three independent experiments that showed less than 20% variability.
The importance of Fur in vivo.
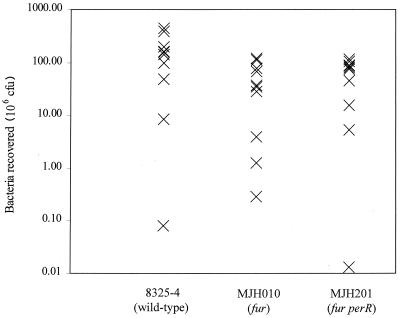
The pathogenicities of MJH010 (fur), MJH201 (fur perR), and 8325-4 (wild type) in a murine skin abscess model of infection were tested (Fig. 7). The mean percentages of recovery for the strains and Student's t test P values are as follows: 8325-4 (wild-type), 143%; MJH010 (fur), 45.7%, P < 0.04; MJH201 (fur perR), 38.9%, P < 0.03.
FIG. 7.
Pathogenicity of S. aureus strains in a murine skin abscess model of infection. Approximately 108 CFU of each strain was inoculated subcutaneously into 6- to 8-week-old BALB/c mice (10 for each strain). Seven days after infection mice were euthanized, lesions were removed and homogenized, and viable bacteria were counted after dilution and growth on BHI agar plates.
DISCUSSION
The S. aureus genome contains three fur homologues and one dtxR homologue, which encode Fur, PerR, and Zur and MntR, respectively. To date Zur has been shown to regulate zinc homeostasis (J. A. Lindsay and S. J. Foster, submitted), and PerR functions as a manganese-dependent repressor of a regulon of proteins required for oxidative stress resistance and iron storage (M. J. Horsburgh et al., submitted). The role of Fur in S. aureus as a regulator of iron-siderophore gene transcription was proposed by Xiong et al. (58) on the basis of purified Fur protein binding to Fur box sequences located in the promoters of the fhuC and sirA genes.
Our results demonstrate that S. aureus Fur functions as an iron-dependent transcriptional repressor of genes encoding iron uptake proteins. Fur mediates iron-dependent repression of the putative hydroxamate siderophore uptake gene, fhuD2, and the putative iron-siderophore transport operons sirABC and sstABCD. In addition orf4, which encodes a putative metal transporter, was also Fur regulated. As with many other bacteria, iron represses siderophore biosynthesis in S. aureus; significantly, this iron-dependent repression was abolished in MJH010 (fur). We propose that Fur is the primary regulator of iron uptake in S. aureus. A search of the incomplete S. aureus genomes with the two known S. aureus Fur box sequences identified many genes likely to be members of the Fur regulon (Fig. 5), and of those tested all were confirmed to be Fur regulated. Heinrichs et al. (28) demonstrated that the sirABC operon Fur box was sufficient for Fur-dependent regulation in E. coli.
In S. aureus, expression of the oxidative stress resistance enzymes, catalase, alkyl hydroperoxide reductase, thiol-dependent peroxidase (Bcp), and thioredoxin reductase (TrxB), is controlled through manganese-dependent PerR-mediated transcriptional repression (M. J. Horsburgh et al., submitted; 31). The S. aureus fur mutant was found to have low levels of catalase activity that were repressed by manganese, but not iron, in a PerR-dependent manner. In a perR mutant catalase levels are increased during growth in high iron in a Fur-dependent manner but are no longer repressed by manganese. This demonstrates that both Fur and PerR regulate the transcription of katA, with Fur acting, either directly or indirectly, as an iron-responsive activator of transcription. PerR acts as a manganese-dependent transcriptional repressor, and the PerR regulon is induced during growth in elevated levels of iron (M. J. Horsburgh et al., submitted). While the induction of katA in response to high iron levels was mediated by Fur, no such induction of other PerR genes was observed since in the fur mutant background the expression of other PerR-regulated genes was increased.
An explanation for this regulation in S. aureus is that elevated iron produces significant oxidative stress through formation of deleterious hydroxyl radicals via the Fenton reaction. An increased level of catalase through peroxide-induced, PerR-mediated derepression of katA (M. J. Horsburgh, et al., submitted) coupled with iron-Fur-mediated induction of katA will effectively reduce the Fenton reaction by lowering the intracellular level of hydrogen peroxide. In addition to this, the increased levels of iron produce derepression of PerR-regulated iron storage protein ferritin and ferritin-like Dps protein MrgA (M. J. Horsburgh et al., submitted), allowing this excess iron to be more safely stored and further limiting hydroxyl radical formation. Elevated concentrations of manganese repress katA transcription in a PerR-dependent manner. The antioxidant properties of manganese complexes have been demonstrated clearly (3, 7, 22, 48). Lactobacillus plantarum has been shown to accumulate high intracellular levels (30 μM) of manganese and does not require iron (3). Treponema pallidum has also been suggested to utilize manganese but not iron for growth (41). The sensitivity of MJH010 (fur) to hydrogen peroxide is likely to be exacerbated by the unregulated uptake of iron into the cell, which has been shown to confer sensitivity in an E. coli fur mutant (51).
The Fur protein of S. aureus, like that of E. coli, has been shown to bind a number of metals in vitro (4, 58). The role of Fur in vivo has been demonstrated to be predominantly iron homeostasis; however, some manganese regulation has been observed in E. coli for a limited number of Fur-dependent loci (4, 21). The Fur-dependent loci tested here were not found to display significant manganese regulation (data not shown), further suggesting that the major role of Fur in S. aureus is iron uptake. The consensus putative S. aureus PerR box element (M. J. Horsburgh et al., submitted) and the consensus putative S. aureus Fur box element bear a striking similarity in terms of modular composition (Fig. 5B) when interpreted as arrays of three repeats of 6 bp using the method of Escolar et al. (21).
The dual regulation of catalase synthesis by PerR and Fur has been shown in Campylobacter jejuni, where both of these proteins function as iron-responsive repressors of catalase expression (53). However, it is not clear whether in C. jejuni this repression of activity is due to the direct or indirect regulation of katA transcription by both PerR and Fur. Similarly, in S. aureus it is not yet clear if the positive regulation of katA transcription by Fur is direct or indirect.
Until recently, Fur was believed to function solely as a repressor of transcription. Indeed Escolar et al. (21) discuss the fact that there have been reports of Fur-positive regulation (4, 39), but as yet there is no confirmation of this activity at the DNA level. A recent report by Dubrac and Touati (20) has confirmed that in E. coli the sodB gene, encoding the iron-containing superoxide dismutase, is positively regulated, in part or in whole, posttranscriptionally by Fur in an iron-dependent manner; no obvious Fur box is located in the sodB promoter region. A deletion analysis demonstrated that an AT-rich region was required for positive regulation by Fur; however, no Fur-DNA binding analysis at this promoter was undertaken, and the authors do not exclude an indirect effect. We note that the S. aureus katA gene does not have an obvious Fur box similar to the E. coli sodB promoter region.
The importance of S. aureus Fur as a central regulator of iron homeostasis was confirmed by the reduced virulence of MJH010 (fur) in a murine skin abscess model of infection. The reduced virulence is unlikely to be merely a consequence of the reduced levels of catalase since a katA mutant is not attenuated, at least in this model of infection (M. J. Horsburgh et al., submitted). Instead, the reduced growth rate and the unregulated uptake of iron into the cell in MJH010 (fur) coupled with a diminished ability to prevent toxic hydroxyl radical formation by catalase-mediated dismutation of hydrogen peroxide may be more significant.
This study has begun to reveal the complex interplay between metal ion homeostasis and stress resistance in S. aureus. Both of these mechanisms are important for pathogenesis, and their regulation will be crucial as part of the host-pathogen interaction. It is the adaptive ability of S. aureus that enables it to be such a versatile and successful pathogen.
ACKNOWLEDGMENTS
We thank the BBSRC (M.J.H.) and the Royal Society (S.J.F.) for funding this research.
We thank the S. aureus Genome Sequencing Project (8325) and B. A. Roe, Y. Qian, A. Dorman, F. Z. Najar, S. Clifton, and J. Iandolo. Preliminary sequence data of S. aureus (COL) were obtained from The Institute For Genomic Research website at http://www.tigr.org.
REFERENCES
- 1.Agranoff D D, Krishna S. Metal ion homeostasis and intracellular parasitism. Mol Microbiol. 1998;8:403–412. doi: 10.1046/j.1365-2958.1998.00790.x. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann H, Engelmann S, Schmid R, Hecker M. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol. 1996;178:6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archibald F S, Fridovich I. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J Bacteriol. 1981;145:442–451. doi: 10.1128/jb.145.1.442-451.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagg A, Neilands J B. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 5.Beers R F, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 6.Beisel W R. Magnitude of the host nutritional responses to infection. Am J Clin Nutr. 1977;30:1236–1247. doi: 10.1093/ajcn/30.8.1236. [DOI] [PubMed] [Google Scholar]
- 7.Berlett B S, Chock P B, Yim M B, Stadtman E R. Manganese (II) catalyses the bicarbonate-dependent oxidation of amino acids by hydrogen peroxide and the amino acid-facilitated dismutation of hydrogen peroxide. Proc Natl Acad Sci USA. 1990;87:389–393. doi: 10.1073/pnas.87.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and quantitative method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Braun V, Hantke K, Koster W. Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst. 1998;35:67–145. [PubMed] [Google Scholar]
- 10.Bsat N, Chen L, Helmann J D. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J Bacteriol. 1996;178:6579–6586. doi: 10.1128/jb.178.22.6579-6586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 12.Bsat N, Helmann J D. Interaction of Bacillus subtilis Fur (ferric uptake repressor) with the dhb operator in vitro and in vivo. J Bacteriol. 1999;181:4299–4307. doi: 10.1128/jb.181.14.4299-4307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraborty T, Leimeister-Wachter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan P F, Foster S J. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus 8325-4. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clements M O, Watson S P, Foster S J. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol. 1999;181:3898–3903. doi: 10.1128/jb.181.13.3898-3903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelissen C N, Sparling P F. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol Microbiol. 1994;14:843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 17.Courcol R J, Trivier D, Bissinger M C, Martin G R, Brown M R W. Siderophore production by Staphylococcus aureus and the identification of iron-regulated proteins. Infect Immun. 1997;65:1944–1948. doi: 10.1128/iai.65.5.1944-1948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalet K, Gouin E, Cenatiempo Y, Cossart P, Hechard Y. Characterization of a new operon encoding a Zur-like protein and an associated ABC zinc permease in Listeria monocytogenes. FEMS Microbiol Lett. 1999;170:111–116. doi: 10.1111/j.1574-6968.1999.tb13556.x. [DOI] [PubMed] [Google Scholar]
- 19.Dreschel H, Freund S, Nicholson G, Haag H, Jung O, Zahner H, Jung G. Purification and characterisation of staphyloferrin B, a hydrophilic siderophore from staphylococci. Biometals. 1993;6:185–192. doi: 10.1007/BF00205858. [DOI] [PubMed] [Google Scholar]
- 20.Dubrac S, Touati D. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J Bacteriol. 2000;182:3802–3808. doi: 10.1128/jb.182.13.3802-3808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escolar L, Perez-Martin J, De Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faulkner K M, Liochev S I, Fridovich I. Stable Mn (III) porphyrins mimic superoxide dismutase in vitro and substitute for it in vivo. J Biol Chem. 1994;269:23471–23476. [PubMed] [Google Scholar]
- 23.Gaballa A, Helmann J D. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths E. The iron-uptake systems of pathogenic bacteria. In: Bullen J J, Griffiths E, editors. Iron and infection: molecular, physiological and clinical aspects. Chichester, United Kingdom: John Wiley & Sons; 1987. pp. 69–138. [Google Scholar]
- 25.Guerot-Fleury A-M, Shazand K, Frandsen N, Stragier P. Antibiotic resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 26.Haag H, Fiedler H P, Meiwes J, Zahner H. Isolation and characterisation of staphyloferrin B, a compound with siderophore activity from staphylococci. FEMS Microbiol Lett. 1994;115:125–130. doi: 10.1111/j.1574-6968.1994.tb06626.x. [DOI] [PubMed] [Google Scholar]
- 27.Hantke K. Selection procedure for downregulated iron transport mutants (fur) in Escherichia coli K-12: fur not only affects iron metabolism. Mol Gen Genet. 1987;210:135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- 28.Heinrichs J H, Gatlin L E, Kunsch C, Choi G H, Hanson M S. Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J Bacteriol. 1999;181:1436–1443. doi: 10.1128/jb.181.5.1436-1443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoepelman I M, Bezemer W A, Vandenbroucke-Grauls C M, Marx J J, Verhoef J. Bacterial iron enhances oxygen-radical mediated killing of Staphylococcus aureus by phagocytes. Infect Immun. 1990;58:26–31. doi: 10.1128/iai.58.1.26-31.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imlay J, Linn S. Toxic DNA damage through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 31.Jeong W, Cha M, Kim I. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem. 2000;275:2924–2930. doi: 10.1074/jbc.275.4.2924. [DOI] [PubMed] [Google Scholar]
- 32.Konnetschy-Rapp S, Jung G, Meiwes J, Zahner H. Staphyloferrin A: a structurally new siderophore from staphylococci. Eur J Biochem. 1991;191:65–74. doi: 10.1111/j.1432-1033.1990.tb19094.x. [DOI] [PubMed] [Google Scholar]
- 33.Kornblum J, Kreisworth B N, Projan S J, Ross H, Novick R P. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 373–402. [Google Scholar]
- 34.Lindsay J A, Riley T V. Staphylococcal iron requirements, siderophore production, and iron-regulated protein expression. Infect Immun. 1994;62:2309–2314. doi: 10.1128/iai.62.6.2309-2314.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meiwes J, Fiedler H P, Zahner H, Konnetschy-Rapp S, Jung G. Isolation and characterisation of staphyloferrin A, a compound with siderophore activity from Staphylococcus hyicus DSM 20459. FEMS Microbiol Lett. 1990;67:201–206. doi: 10.1111/j.1574-6968.1990.tb13863.x. [DOI] [PubMed] [Google Scholar]
- 36.Modun B, Evans R W, Joannou C L, Williams P. Receptor-mediated recognition and uptake of iron from human transferrin by Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 1998;66:3591–3596. doi: 10.1128/iai.66.8.3591-3596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modun B, Williams P. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect Immun. 1999;67:1086–1092. doi: 10.1128/iai.67.3.1086-1092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Modun B, Morrissey J, Williams P. The staphylococcal transferrin receptor: a glycolytic enzyme with novel functions. Trends Microbiol. 2000;8:231–237. doi: 10.1016/s0966-842x(00)01728-5. [DOI] [PubMed] [Google Scholar]
- 39.Niederhoffer E C, Naranjo C M, Bradley K L, Fee J A. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol. 1990;172:1930–1938. doi: 10.1128/jb.172.4.1930-1938.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 41.Posey J E, Hardham J M, Norris S J, Gherardini F C. Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc Natl Acad Sci USA. 1999;96:10887–10892. doi: 10.1073/pnas.96.19.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Que Q, Helmann J D. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol. 2000;35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 43.Repine J E, Fox R B, Berger E M. Hydrogen peroxide kills Staphylococcus aureus by reacting with staphylococcal iron to form hydroxyl radical. J Biol Chem. 1981;256:7094–7096. [PubMed] [Google Scholar]
- 44.Repine J E, Fox R B, Berger E M, Harada R N. Effect of staphylococcal iron content on the killing of Staphylococcus aureus by polymorphonuclear leukocytes. Infect Immun. 1981;32:407–410. doi: 10.1128/iai.32.1.407-410.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Schenk S, Ladagga R A. Improved methods for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;94:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 47.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 48.Stadtman E R, Berlett B S, Chock P B. Manganese-dependent disproportionation of hydrogen peroxide in bicarbonate buffer. Proc Natl Acad Sci USA. 1990;87:384–388. doi: 10.1073/pnas.87.1.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao X, Schiering N, Zeng H Y, Ringe D, Murphy J R. Iron, DtxR, and regulation of diphtheria toxin expression. Mol Microbiol. 1999;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 50.Telenti A, Philipp W J, Sreevatsan S, Bernasconi C, Stockbauer K E, Wieles B, Musser J M, Jacobs W R. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med. 1997;3:567–570. doi: 10.1038/nm0597-567. [DOI] [PubMed] [Google Scholar]
- 51.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trivier D, Courcol R J. Iron depletion and virulence in Staphylococcus aureus. FEMS Microbiol Lett. 1996;141:117–127. doi: 10.1111/j.1574-6968.1996.tb08373.x. [DOI] [PubMed] [Google Scholar]
- 53.van Vliet A H M, Baillon M A, Penn C W, Ketley J. Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress genes by the PerR repressor. J Bacteriol. 1999;181:6371–6376. doi: 10.1128/jb.181.20.6371-6376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watson S P, Clements M O, Foster S J. Characterization of the starvation survival response of Staphylococcus aureus. J Bacteriol. 1998;180:1750–1758. doi: 10.1128/jb.180.7.1750-1758.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watson S P, Antonio M A, Foster S J. Isolation and characterisation of Staphylococcus aureus starvation-induced, stationary phase mutants defective in survival or recovery. Microbiology. 1998;144:3159–3169. doi: 10.1099/00221287-144-11-3159. [DOI] [PubMed] [Google Scholar]
- 56.Williams P, Griffiths E. Bacterial transferrin receptor—structure, function and contribution to virulence. Med Microbiol Immunol. 1992;181:301–322. doi: 10.1007/BF00191543. [DOI] [PubMed] [Google Scholar]
- 57.Wooldridge K G, Williams P H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 58.Xiong A, Singh V K, Cabrera G, Jayaswal R K. Molecular characterisation of the ferric uptake regulator, Fur, from Staphylococcus aureus. Microbiology. 2000;146:659–668. doi: 10.1099/00221287-146-3-659. [DOI] [PubMed] [Google Scholar]
- 59.Zuberi A R, Moir A, Feavers I M. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene. 1987;51:1–11. doi: 10.1016/0378-1119(87)90468-9. [DOI] [PubMed] [Google Scholar]