Abstract
Different strains of Streptococcus suis serotypes 1 and 2 isolated from pigs either contained a restriction-modification (R-M) system or lacked it. The R-M system was an isoschizomer of Streptococcus pneumoniae DpnII, which recognizes nucleotide sequence 5′-GATC-3′. The nucleotide sequencing of the genes encoding the R-M system in S. suis DAT1, designated SsuDAT1I, showed that the SsuDAT1I gene region contained two methyltransferase genes, designated ssuMA and ssuMB, as does the DpnII system. The deduced amino acid sequences of M.SsuMA and M.SsuMB showed 70 and 90% identity to M.DpnII and M.DpnA, respectively. However, the SsuDAT1I system contained two isoschizomeric restriction endonuclease genes, designated ssuRA and ssuRB. The deduced amino acid sequence of R.SsuRA was 49% identical to that of R.DpnII, and R.SsuRB was 72% identical to R.LlaDCHI of Lactococcus lactis subsp. cremoris DCH-4. The four SsuDAT1I genes overlapped and were bounded by purine biosynthetic gene clusters in the following gene order: purF-purM-purN-purH-ssuMA-ssuMB-ssuRA-ssuRB-purD-purE. The G+C content of the SsuDAT1I gene region (34.1%) was lower than that of the pur region (48.9%), suggesting horizontal transfer of the SsuDAT1I system. No transposable element or long-repeat sequence was found in the flanking regions. The SsuDAT1I genes were functional by themselves, as they were individually expressed in Escherichia coli. Comparison of the sequences between strains with and without the R-M system showed that only the region from 53 bp upstream of ssuMA to 5 bp downstream of ssuRB was inserted in the intergenic sequence between purH and purD and that the insertion target site was not the recognition site of SsuDAT1I. No notable substitutions or insertions could be found, and the structures were conserved among all the strains. These results suggest that the SsuDAT1I system could have been integrated into the S. suis chromosome by an illegitimate recombination mechanism.
More than 3,000 restriction-modification (R-M) systems have been identified in a wide variety of microorganisms, where they are thought to protect the host from invasion by foreign DNA. Only a minority of R-M systems have been sequenced (6, 37, 56). Among them, some type II R-M systems, which recognize 4-bp palindromic sequence 5′-GATC-3′, involve a variety of isoschizomers. Three classes can be distinguished by their manners of DNA cleavage and susceptibilities to DNA methylation. The first class of isoschizomers, represented by Sau3AI, which was described for Staphylococcus aureus, is prevented from digesting host DNA by a cognate 5-methylcytosine methyltransferase and is not influenced by the modification of N6-methyladenine (48, 56). The second class, represented by DpnI, which was described for Streptococcus pneumoniae, is unique among restriction endonucleases in that it cleaves only at methylated DNA sequence 5′-GmeATC-3′, and thus the cells producing DpnI do not carry the corresponding methyltransferase gene (22, 23). The third class, including MboI, DpnII, and LlaDCHI, which were described for Moraxella bovis, S. pneumoniae, and Lactococcus lactis, respectively, contains restriction endonucleases complementary to the second class, in that they cleave at the same sequence, 5′-GATC-3′, but only when it is unmethylated (11, 22, 23, 30, 55). Bacterial cells expressing the third class of restriction endonucleases also contain an N6-methyladenine methyltransferase, which methylates adenine in the 5′-GATC-3′ sequences of the host DNA. The N6-methyladenine methyltransferase is functionally identical to the Dam methylase of Escherichia coli, and the evolutionary relatedness of these methylases has been discussed (29, 35).
Except for the second class, the type II R-M systems described above are generally composed of two structural genes, one for an endonuclease and a second for a methyltransferase, as is typical of other type II R-M systems (6, 56). However, two of these, DpnII and LlaDCHI, have genes that encode two methyltransferases (8, 24, 30). A study of well-characterized DpnII and a comparison of it with the LlaDCHI system indicated that in addition to the conventional double-stranded DNA methyltransferase, a single-stranded DNA methyltransferase was encoded, and this activity potentially facilitated the uptake of intact DNA via conjugation in the respective host bacteria. Another property of interest is that the two methyltransferases of the DpnII system, M.DpnII and M.DpnA, were highly homologous to those of the LlaDCHI system, M.LlaA and M.LlaB, respectively (75 and 88% identity). Thus the two M systems are thought to have the same origin, whereas the two restriction endonucleases showed relatively low homology (only 31% identity), suggesting the complexity of the evolutionary origin of restriction endonuclease genes (30).
Many genes for R-M systems have been found to be located on transferable elements such as plasmids and bacteriophages, or, in some cases, genes encoding proteins involved in DNA mobility, such as transposases, integrases, and invertases, are found in the vicinity of R-M systems (5, 7, 19, 26, 38, 46, 51, 54, 56). These genetic structures may facilitate the transfer of R-M systems and have led to the speculation that R-M genes could migrate among microorganisms of different genera. The acquisition or loss of an R-M system followed by a double-strand breakage may play a role in illegitimate recombination when the two genetic segments have a few base pairs of homology (21). The above findings, together with recent studies on the complete sequences of bacterial genomes, have led to a proposal that R-M systems are likely to be mobile genetic elements that mediate large-scale chromosomal conversions and that such genome plasticity may play a role in genome dynamics (20). However, these studies have thus far been conducted with only a few strains or with few different species (3, 17, 20, 47, 58). While the results of the above studies imply horizontal transfer of R-M systems, the precise locations of R-M systems in the chromosome and/or the genetic structures flanking the R-M systems have yet to be studied extensively. Therefore, further studies will be needed in order to make a more generalized statement relative to the mobility of R-M systems.
Streptococcus suis is a gram-positive, facultatively anaerobic coccus that has been implicated as the cause of a wide range of clinical disease syndromes in swine and other domestic animals. The disease syndromes caused by S. suis in swine include arthritis, meningitis, pneumonia, septicemia, endocarditis, polyserositis, abortions, and abscesses (9). S. suis has also been implicated in human disease (28). S. suis strains are currently classified into 35 serotypes on the basis of their capsular polysaccharide antigens (12, 13, 36). Among them, strains frequently isolated from diseased pigs and exhibiting a high degree of virulence mostly belong to serotype 2 (9, 15, 16). Although several genes of S. suis have been cloned and characterized (34, 41, 43–45, 49), the R-M system(s) of this bacterial species has not been described.
We found that some strains of serotypes 1 and 2 of S. suis possessed an R-M system that mimicked the DpnII and LlaDCHI systems, whereas the reference strain of serotype 2, NCTC10234, as well as other strains lacked this R-M system. This report describes the genetic organization of the R-M system and its flanking regions. The R-M system was found to contain two genes encoding methyltransferases similar to those of the DpnII and LlaDCHI systems and two genes encoding restriction endonucleases, one similar to R.DpnII and the other highly homologous to R.LlaDCHI. Comparison of the DNA sequences of the R-M system and its flanking regions among different strains containing the R-M system and those lacking the system indicated a horizontal transfer of the R-M system, which may have occurred by a unique mechanism of gene transfer.
MATERIALS AND METHODS
Bacterial strains, vectors, enzymes, media, and culture conditions.
S. suis strains of serotypes 1 and 2 used are listed in Table 1. Strain NCTC10234, a reference strain of serotype 2, was purchased from the National Collection of Type Cultures, Central Public Health Laboratory, London, England. Strain DAT1 was previously described (49). Other S. suis strains were independently isolated from diseases pigs in Japan and stocked in our laboratory. E. coli strains used were SCS110 (thr leu endA thi-1 lacY galK galT ara tonA tsx dam dcm supE44 rpsL Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15]) (Stratagene, La Jolla, Calif.), C600 (F− thr-1 thi-1 leuB6 lacY1 tonA21 supE44) (39), XL-1 blue MRF′ [Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10]) (Stratagene), XLOLR {Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10] Su−λ−} (Stratagene), and DH5α (F− endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA relA1 φ80lacZΔM15 Δ(lacZYA-argF) deoR+) (39). The plasmid vectors used for gene cloning were pUC19 (59), pCR2.1 (Invitrogen, Groningen, The Netherlands), and pHSG576 (50). Phagemid vector λZAP Express (Stratagene) was used for the construction of the genomic library. All restriction endonucleases and other enzymes were purchased from Takara Shuzo Co. Ltd. (Tokyo, Japan) except DpnI, which was purchased from Boehringer GmbH (Mannheim, Germany), and used according to the manufacturers' recommendations. S. suis strains were grown in Todd-Hewitt (TH) broth or agar medium (Difco Laboratories, Detroit, Mich.) at 37°C under 5% CO2 for 18 h. E. coli strains were cultured in Luria-Bertani broth or agar medium (Difco Laboratories) supplemented with, when necessary, ampicillin (50 μg/ml), kanamycin (25 μg/ml), and chloramphenicol (25 μg/ml) at 37°C for 18 h.
TABLE 1.
Strains of S. suis used and their R-M profiles
| Strainsa | Serotype |
|---|---|
| Strains expressing the R-M phenotype | |
| DAT1, 193, 195, 196, 197, 198, 199, 200, 202, 205, 220, 221, 222, 227, 228, 229, 230, 233, 234, 235, 236, 238, 239, 243, 244, 247 | 2 |
| 211, 212 | 1 |
| Strains with the null phenotype | |
| NCTC10234, DAT2, 194, 207, 209, 210, 213, 223, 226, 246 | 2 |
| 203, 204 | 1 |
Strains except NCTC10234 were isolated from diseased pigs in Japan from 1987 to 1996.
Analysis of DNA methylation.
Genomic DNA was isolated from S. suis and E. coli grown on agar medium by standard procedures as described previously (34, 42). Methylation of DNA was determined directly by testing the susceptibility of genomic DNA to the restriction endonucleases DpnI and MboI in vitro. Restriction digests were analyzed by agarose gel electrophoresis by standard procedures (39).
Restriction endonuclease assay in crude extracts.
The presence of restriction endonucleases could be detected in crude extracts of S. suis and E. coli. Extracts of S. suis were prepared according to the procedures described by Muckerman et al. (31). Briefly, 5-ml cultures of S. suis were grown in TH broth to an optical density at 600 nm of approximately 0.4. After centrifugation at 20,000 × g for 2 min at 4°C, the cell pellet was suspended in 0.1 ml of 10 mM Tris-HCl (pH 7.6)–50 mM NaCl. Then 2.5 μl of Triton X-100 was added, and the mixture was incubated at 30°C for 15 min for cell lysis. After incubation, the cells were removed by centrifugation at 20,000 × g for 5 min at 4°C, and the extracts were directly used for the assay. Extracts of E. coli were prepared as described by Schleif (40). Briefly, bacterial cells collected from 1 ml of an overnight shaking culture in Luria-Bertani broth supplemented with appropriate antibiotics were suspended in 0.1 ml of 10 mM Tris-HCl (pH 7.6)–10 mM 2-mercaptoethanol in Eppendorf tubes. The cell suspensions were then sonicated on ice with a Sonifier 250 (Branson Ultrasonics Corp., Danbury, Conn.) with a microtip. Sonication was carried out twice for 10 s with a 1-min interval and with a 50% duty cycle and an output of 7. After sonication, the cell debris was removed by centrifugation at 20,000 × g for 15 min at 4°C, and the supernatants were directly used for the assay. Enzyme reactions were performed by standard procedures for restriction endonuclease digests with a high-salt universal buffer (39).
PCR amplifications.
Synthetic oligonucleotide primers specifically designed for amplification of the R-M gene region in this study are listed in Table 2. Ex Taq polymerase (Takara) was used for the amplification according to the manufacturer's instructions except that the concentration of MgCl2 was 3 mM. DNA amplification was carried out in a Perkin-Elmer thermal cycler, model 2400 or 9600 (PE Biosystems Japan, Tokyo, Japan), and the program consisted of incubation for 1 min at 96°C, 30 cycles of 20 s at 96°C, 20 s at 60°C, and 3 min at 68°C, and final incubation for 5 min at 68°C.
TABLE 2.
Oligonucleotide primers
| Primer | Sequence (5′–3′)b | Location or description |
|---|---|---|
| dam-F | YRAARTGGRCWGGKGGHAARa | Degenerate primer |
| dam-R | CGDCCRWAYGGHACRTTRAAa | Degenerate primer |
| dam-in1 | TTTCCCCCTGCTCTGCAATT | purH region near BamHI site |
| dam-in2 | CAGTGGGTATGGCCTTAGAA | ssuMB region |
| purH3′ | CAGCTGCAGGTATCAAGGCTAT | 3′ end of purH gene |
| ssuMB3′ | CACATCTGGATACTTGTCAC | 5′ end of ssuRA gene |
| ssuRA5′Ec | GGAATTCGCTTCAACTCAAGAAGGAGA | 3′ end of ssuMB gene with EcoRI site |
| ssuRA3′Pst | AACTGCAGGTAATCTGGCGTTCGATTAG | 5′ end of ssuRB gene with PstI site |
| ssuRB5′Ec | GGAATTCTGGTGAAGGTTGGCAAAGGT | 3′ end of ssuRA gene with EcoRI site |
| ssuRB3′Pst | AACTGCAGCCTGCTCAGACTCTAACAAC | 5′ end of purD gene with PstI site |
| ssuRB3′ | CCTGCTCAGACTCTAACAAC | 5′ end of purD |
Y, C or T; R, A or G; W, A or T; K, G or T; H, A or C or T; D, A or G or T.
Unique restriction cleavage sites introduced in the oligonucleotides are underlined.
An LA PCR kit (Takara) was used for inverse PCR according to the manufacturer's instructions except that the concentration of MgCl2 was 3 mM. The rationale and the protocol for inverse PCR were essentially the same as those described elsewhere (32). Genomic DNA of S. suis strain DAT1 was digested with BamHI and self-ligated using a DNA ligation kit (Takara). The ligation mixture was used as a template for the inverse PCR.
Construction of genomic library and cloning procedures.
Genomic DNA of S. suis DAT1 was partially digested with Sau3AI and size fractionated by standard procedures (39). DNA fragments of approximately 5 to 7 kb were ligated with BamHI-digested dephosphorylated λZAP Express vector (Stratagene) and subjected to in vitro packaging with a GigaPack III kit (Stratagene) according to the instructions of the manufacturer. The library was screened via plaque hybridization using standard procedures (39). Phages from the hybridizing plaques were purified. The phagemids were rescued by helper phage EXASSIST (Stratagene) and used to infect E. coli XLOLR to create plasmid subclones by following the instructions of the manufacturer.
Construction of subclones was essentially accomplished by digestion and self-ligation procedures using a restriction cleavage site in a cloned fragment and the multicloning sites of the vector plasmid. The plasmids were digested with an appropriate restriction endonuclease and self-ligated to create subclones in which a part of the original insert fragment had been deleted. The combinations of restriction enzymes and the subclones constructed were as follows. For pDAM4, digestion with KpnI, SacI, PstI, ClaI, and SpeI generated pDAM4K, pDAM4Sa, pDAM4P, pDAM4C, and pDAM4Sp, respectively; for pDAM7, digestion with AvaI generated pDAM7Av; for pDAM9, digestion with KpnI, SacI, PstI, ClaI, and SpeI generated pDAM9K, pDAM9Sa, pDAM9P, pDAM9C, and pDAM9Sp, respectively; for pDAM10, digestion with PstI generated pDAM10P. Some of the subclones were modified further by the same procedures. pDAM4C and pDAM9C digested with PstI generated pDAM4CP and pDAM9CP, respectively. pDAM4Sa digested with EcoRI generated pDAM4SaE. A 1.5-kb HincII fragment of pDAM9K was subcloned in pUC19; the recombinant plasmid was designated pDAM9KHc. These subclones were used for sequencing the DNA of the cloned fragments using universal primers designed for the different vector plasmids.
Hybridization techniques.
Genomic Southern hybridization and plaque hybridization were performed as described previously (39) with a DIG DNA labeling and detection kit (Boehringer GmbH). Restriction endonuclease fragments were separated on agarose gels and transferred to positively charged nylon membranes (Nytran Plus; Schleicher & Schuell, Dassel, Germany) by using the vacuum transfer method with VacuGene (Pharmacia-LKB Biotechnology AB, Uppsala, Sweden) without depurination in accordance with the instructions of the manufacturer. Plaque hybridization was carried out on a positively charged nylon membrane (MAGNA LIFT; Micron Separations Inc., Westboro, Mass.). Prehybridization and hybridization were carried out at 63°C for 2 and 16 h, respectively. After hybridization, the sheets were washed twice at room temperature for 5 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7.2]) containing 0.1% sodium dodecyl sulfate (SDS), followed by two 15-min washes at 63°C in 0.1× SSC containing 0.1% SDS. DNA fragments cloned in plasmids were purified from agarose gels after electrophoresis and labeled with digoxigenin by essentially the same procedures described previously (42).
DNA sequencing and analysis.
The sequencing of cloned DNA fragments and various PCR products was carried out by dye terminator chemistry with specifically designed primers on an Applied Biosystems model 373A automated DNA sequencer (PE Biosystems). The sequences were assembled and analyzed with Sequencer, version 2 (Hitachi Software Engineering Co., Ltd., Yokohama, Japan), and Genetyx-Mac, version 10.1 (Software Developing Company, Tokyo, Japan). Sequences were searched against current DNA and protein databases by BLAST program network services available at the National Center for Biotechnology Information (Bethesda, Md.) (http://www.ncbi.nlm.nih.gov).
Pulsed-field gel electrophoresis (PFGE) analysis.
Preparation of genomic DNAs and their restriction digests was performed as described previously (33) with some modifications. Briefly, S. suis strains grown on TH agar were harvested and suspended in TES (10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 0.15 M NaCl) to an optical density at 600 nm of 0.5. The cell suspensions were mixed with an equal volume of molten 3% low-melting-point preparative-grade agarose (Bio-Rad Laboratories, Richmond, Calif.) to obtain agarose plugs. Cells were lysed by submerging the plugs in lysis solution (N-acetylmuramidase [Seigagaku Kogyo, Tokyo, Japan] [30 μ/ml] and lysozyme [5 mg/ml] in TES) at 37°C for 1 h with gentle shaking. The lysis solution was replaced with ESP solution (0.5 M EDTA [pH 9.5], 1% [wt/vol] laurylsarcosine, 1 mg of proteinase K per ml), and the plugs were incubated at 50°C for 20 h. The plugs were washed several times with TE buffer (10 mM Tris-HCl, 2 mM EDTA [pH 8.0]) and stored at 4°C until use. The plugs were cut into small pieces, equilibrated with the recommended restriction buffer, placed in fresh restriction buffer supplemented with 0.02% [wt/vol] bovine serum albumin and 5 U of restriction enzyme SmaI, and then incubated at 37°C for 1 h. Contour-clamped homogeneous electric field (CHEF) electrophoresis was done in a Bio-Rad CHEF DRII system. Agarose gels (1.0% [wt/vol]) were electrophoresed in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA [pH 8.0]) for 20 h at 200 V and 10°C. The pulse times were ramped from 5 to 50 s.
Nucleotide sequence accession numbers.
The sequence of the entire R-M region and its flanking regions of S. suis strain DAT1 obtained in this study has been deposited in the DDBJ/EMBL/GenBank database under accession no. AB045609. The DDBJ/EMBL/GenBank accession numbers of the genetic regions flanking the R-M genes of the strains tested are as follows: 205, AB045610 and AB045611; 220, AB045612 and AB045613; 211, AB045614 and AB045615; 212, AB045616 and AB045617. The DNA sequences for the purH-purD loci of strains NCTC10234, 213, 246, 203, and 204 have been deposited in the DDBJ/EMBL/GenBank database with accession no. AB045618, AB045619, AB045620, AB045621, and AB045622, respectively.
RESULTS
Restriction enzyme phenotypes of S. suis.
The R-M phenotypes of S. suis strains were first examined by testing the susceptibility of their DNA to DpnI and MboI. Cleavage by DpnI and not by MboI indicates methylation of adenine at the 5′-GATC-3′ sequence and, hence, an MboI (or DpnII) methylase phenotype. The converse indicates the absence of methylation. Total DNA prepared from 40 strains was incubated with either DpnI or MboI. The results obtained are summarized in Table 1, and representative cleavage patterns of several strains are shown in Fig. 1A. Genomic DNA isolated from 12 of 40 strains was digested to small fragments by MboI; thus these bacteria had the unmethylated phenotype. On the other hand, the DNA from the remaining 28 strains could be digested only by DpnI, indicating that their DNAs were protected by methylation from MboI digestion.
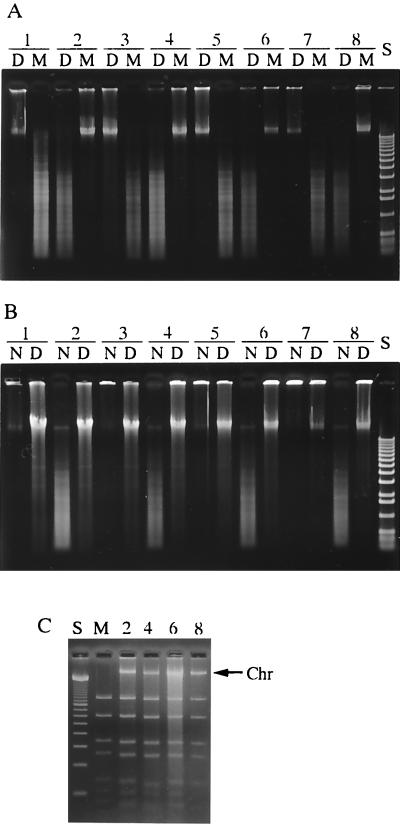
FIG. 1.
(A) Methylation of 5′-GATC-3′ sequence in the DNA of various strains of S. suis. Total DNAs from the following strains were treated with two complementary restriction endonucleases and analyzed by agarose gel electrophoresis: Lane 1, NCTC10234; lane 2, DAT1; lane 3, 204; lane 4, 205; lane 5, 207; lane 6, 211; lane 7, 213; lane 8, 220. D, digested with DpnI (specific for methylated sequence); M, digested with MboI (specific for unmethylated sequence). S, 1-kb ladder DNA size standards (GIBCO/BRL). (B) DNA cleavage activities of S. suis crude extracts using the total DNAs of S. suis NCTC10234 (N, unmethylated DNA) and DAT1 (D, methylated DNA) as substrates. Lane numbers for the strains from which the crude extracts were prepared are the same as in panel A. S, 1-kb ladder size standards (GIBCO/BRL). (C) Restriction cleavage activities of the crude extracts using unmethylated pUC19 as the substrate. The lane numbers of the strains from which the crude extracts were prepared match those in panel B. M, pUC19 digested with MboI; S, 100-bp ladder size standards (GIBCO/BRL); Chr, contaminating chromosomal DNA.
Crude extracts of the different strains were prepared, and their restriction endonuclease activities were assayed by digestion of the genomic DNA of strain NCTC10234, as an unmethylated DNA substrate, and that of DAT1, as a methylated DNA substrate. Restriction endonuclease activity was only detected in strains that exhibited the methylated DNA phenotype (Fig. 1B, lanes 2, 4, 6, and 8), whereas crude extracts of the strains that showed the unmethylated DNA phenotype digested neither methylated nor unmethylated DNA and were designated null phenotypes (Fig. 1B, lanes 1, 3, 5, and 7). These results were confirmed further by digestion of E. coli genomic DNA. E. coli K-12 strains are known to possess an orphan methylase called Dam, with a methylation property that is the same as that of M.MboI. Genomic DNA of E. coli C600, with a Dam+ phenotype, and of SCS110, a Dam-deficient mutant, was digested with crude extracts of S. suis; a restriction endonuclease activity that could digest only unmethylated SCS110 DNA was seen in extracts of the strains which showed the methylated phenotype, but no endonuclease activity was seen in extracts of the strains which showed the null phenotype (data not shown). To examine the enzymatic specificity of extracts of strains that showed restriction endonuclease activity, we compared the restriction endonuclease cleavage patterns of vector plasmid pUC19 prepared from Dam-deficient mutant SCS110. Representative cleavage patterns are shown in Fig. 1C. Although chromosomal contamination derived from S. suis strains appeared, the cleavage patterns of the crude extract were essentially identical to that of MboI digestion. Therefore, these strains were considered to possess an R-M system similar to the MboI (or DpnII) system. The R-M system of S. suis DAT1 was designated SsuDAT1I. The R-M phenotype did not correlate with the capsular serotype, nor did the virulence phenotype of the strains correlate with the R-M phenotype, since the strains were isolated from diseased pigs.
Presence and distribution of the R-M genes in S. suis strains.
Because the R-M phenotype found in 28 strains of S. suis was the same as that of the MboI system, degenerate primers, designated dam-F and dam-R, were designed on the basis of the nucleotide sequences of the related methylase genes that are found in the database (Table 2). PCR amplification was conducted with genomic DNA of strain DAT1 as a template, and the amplified fragment was cloned into pCR2.1 and sequenced. The nucleotide sequence of the cloned fragment was highly homologous to that of M.DpnII (71.8% identity) (data not shown). The fragment was then labeled with digoxigenin and used as a hybridization probe against genomic DNA from several S. suis strains that had been digested with either BamHI, EcoRI, or HindIII and separated by agarose gel electrophoresis. Strains expressing the R-M phenotype provided hybridizing DNA fragments of approximately 6.5, 25, and 10 kb, respectively, whereas no hybridizing fragments appeared in strains which exhibited the null phenotype (data not shown). These results suggested that strains expressing the R-M phenotype possessed a single copy of the gene and that strains with the null phenotype lacked the corresponding gene.
Cloning, sequencing, and genetic organization of the SsuDAT1I gene region.
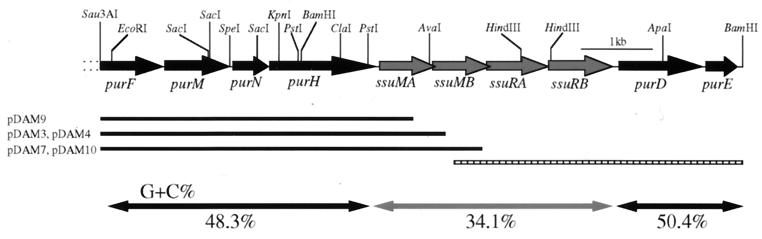
After plaque hybridization of the S. suis DAT1 genomic library with the labeled probe described above, 10 positive plaques were identified and the phagemids were rescued to create plasmid subclones as described in Materials and Methods. Five plasmid subclones, designated pDAM3, pDAM4, pDAM7, pDAM9, and pDAM10, were obtained (Fig. 2). Restriction digests of the five subclones showed that pDAM3 and pDAM4 were identical, whereas pDAM7 and pDAM10 contained the same insert in opposite orientations relative to the vector plasmid (Fig. 2). These subcloned fragments were sequenced, and the deduced translation products of several open reading frames (ORFs) were compared to amino acid sequences in databases using the BLAST program. The five subclones contained one or both of the methyltransferase genes, designated ssuMA and ssuMB. The region upstream of the methyltransferase genes contained four ORFs associated with purine biosynthesis (described in detail below). An inverse PCR was employed to amplify a fragment containing the downstream DNA region using synthetic oligonucleotides dam-in1 and dam-in2. An amplified fragment of approximately 4.5 kb was then directly sequenced. The entire DNA sequence of the SsuDAT1I genes and the flanking regions was 9,618 bp, and it included 10 putative ORFs (Fig. 2). These were identified on the basis of the adopted criterion that an ORF consists of at least 40 codons preceded by a potential Shine-Dalgarno sequence at an appropriate distance (6 to 15 bp) from one of the commonly used initiation codons (AUG, UUG, and GUG). The results of the database analysis of the deduced amino acid sequences are summarized in Table 3. As expected, a gene encoding a restriction endonuclease appeared just downstream of ssuMB. However, unexpectedly, downstream of this gene there was an additional restriction endonuclease gene. These two genes were designated ssuRA and ssuRB (Fig. 2). Other ORFs found in the sequenced region encoded products that were highly homologous to enzymes involved in purine biosynthetic pathways (Table 3). Genes identified in the entire sequenced region were in the order purF- purM-purN-purH-ssuMA-ssuMB-ssuRA-ssuRB-purD-purE, as shown in Fig. 2. All of these genes were encoded on the same DNA strand. No transposable element or long-repeat sequence was found in the 9,618-bp sequence. The average G+C content of the purine biosynthetic genes was 48.9%, which was higher than that of the total genome of S. suis (39 to 41%) (18), and the average G+C content of the SsuDAT1I region was much lower (34.1%) (Fig. 2). The codon usage pattern for the SsuDAT1I system genes was somewhat different from that of the purine biosynthetic gene clusters. For example, the most frequently used codon for leucine in the SsuDAT1I system was TTA (45% of the total leucine codons in SsuDAT1I genes), although TTA was the least-used codon for pur genes (8.4% of the total leucine codons in pur genes).
FIG. 2.
Physical and genetic map of the 9,618-bp sequence of S. suis DAT1 with the putative genes indicated. Shaded and solid arrows, ORFs of the SsuDAT1I genes and pur genes, respectively. Horizontal solid lines, positions of the cloned fragments of plasmids pDAM3, pDAM4, pDAM7, pDAM9, and pDAM10; striped bar, region amplified by an inverse PCR; shaded and solid double-headed arrows, average G+C content of the SsuDAT1I region and that of the pur region, respectively.
TABLE 3.
Predicted gene products and the homology exhibited by their potential protein products to amino acid sequences in the databasea
| Predicted gene | Coding position
|
Product size
|
Predicted pI | Homology
|
Description and function of closest relativeb | |||
|---|---|---|---|---|---|---|---|---|
| Start | Stop | aac | kDa | BLAST score | aa identityd | |||
| purF | <1 | 940 | >312 | >34.6 | 4.67 | e-124 | 73/300 | Phosphoribosylpyrophosphate amidotransferase precursor of L. lactis (EC 2.4.2.14) [U64311] |
| purM | 995 | 2017 | 340 | 36.4 | 4.54 | e-128 | 69/339 | Phosphoribosylformylglycineamide cycloligase of L. lactis subsp. cremoris (EC 6.3.3.1) [AF016634] |
| purN | 2014 | 2565 | 183 | 20.5 | 4.88 | 8e-58 | 71/150 | Phosphoribosylglycineamide formyltransferase homolog of S. pyogenes (EC 22.1.2.2) [U70775] |
| purH | 2575 | 4122 | 515 | 56.6 | 4.83 | e-169 | 58/513 | Phosphoribosylamidoimidazole carboxyamide formyltransferase of B. subtilis (EC 6.3.2.6) [J02732] |
| ssuMA | 4187 | 5017 | 276 | 31.8 | 4.93 | e-114 | 71/275 | M.LlaDCHI A of L. lactis [U16027] |
| e-110 | 70/274 | M.DpnII 1 of S. pneumoniae [M14339] | ||||||
| ssuMB | 5007 | 5822 | 271 | 31.2 | 9.50 | e-145 | 90/266 | M.DpnA of S. pneumoniae [M14339] |
| e-139 | 86/263 | M.LlaDCHI B of L. lactis [U16027] | ||||||
| ssuRA | 5800 | 6726 | 308 | 36.0 | 5.20 | 3e-78 | 49/299 | R.DpnII of S. pneumoniae [M14339] |
| ssuRB | 6726 | 7631 | 301 | 34.4 | 5.17 | e-126 | 72/300 | R.LlaDCHI of L. lactis [U16027] |
| purD | 7752 | 9014 | 420 | 45.3 | 4.48 | e-125 | 54/419 | Phosphoribosylamineglycine ligase of L. lactis (EC 6.3.4.13) [AJ000883] |
| purE | 9040 | 9528 | 162 | 16.8 | 6.36 | 4e-41 | 61/139 | Phosphoribosylamidoimidazole carboxylase catalytic subunit of B. subtilis (EC 4.1.1.21) [J02732] |
Proteins relating to purine biosynthesis have been named according to their closest relative, probable function, and E. coli gene designations (60). Coding region positions are numbered with respect to the entire 9,618-bp sequence determined (AB045609) and from the first base of the start codon to the last base of the stop codon. The Genetyx-Mac program (Software Developing Company) was used to make predictions of coding regions, molecular masses, and isoelectric points.
Numbers in blackets are accession numbers in the DDBJ/EMBL/GenBank database.
aa, amino acids.
Percent identity/number of amino acids evaluated.
Genetic structure and protein analysis of the SsuDAT1I system.
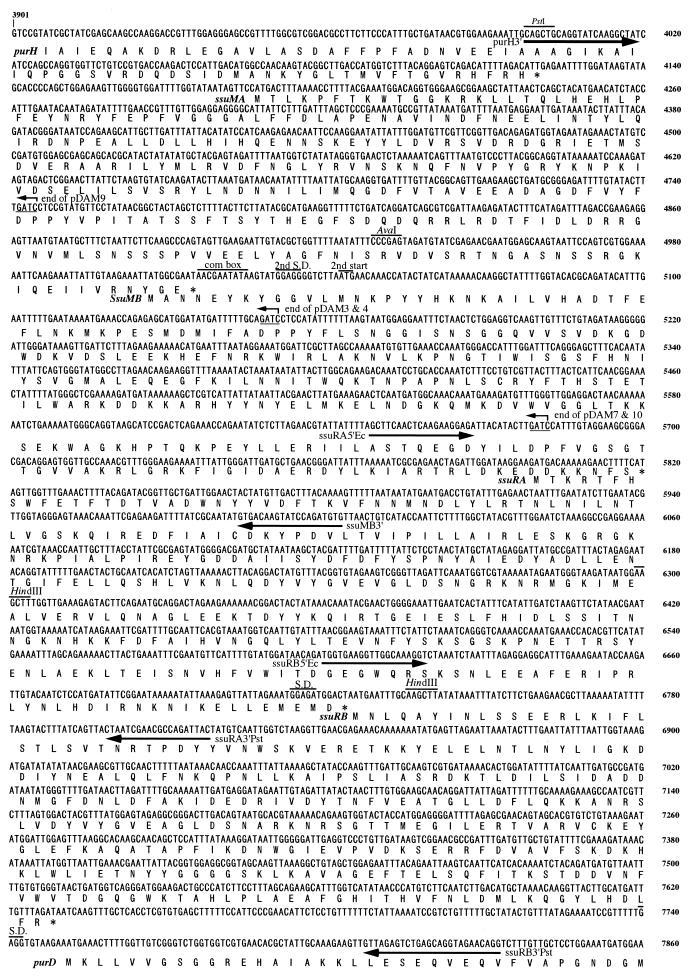
The complete DNA sequence and the deduced amino acid sequences of the SsuDAT1I region are shown in Fig. 3. The four ORFs in the SsuDAT1I gene region overlapped each other in a head-to-tail manner (Fig. 2 and 3). Only ssuRB had a typical Shine-Dalgarno sequence (Fig. 3). Interestingly, the competence-regulated sequence, the so-called combox, was present at the translation initiation site of the ssuMB gene. This sequence was originally found in several competence-regulated genes and was recently shown to be present in the methyltransferase gene of the DpnII system (25). The DNA sequence of the combox, including the first start codon, the second start codon, and a typical Shine-Dalgarno sequence prior to the second start codon, was completely identical to that described for the DpnII system (Fig. 3). A palindromic sequence, typical of an rho-independent terminator, could not be found at the end of the ssuRB gene.
FIG. 3.
Coding sequence of the 3,960-bp region comprising a part of the purH gene, the complete SsuDAT1I genes, and a part of the purD gene. The deduced amino acid is indicated underneath the first nucleotide of each codon in the coding sequence. Arrows underneath the nucleotide sequence, positions of the primers used for the amplification and cloning of the restriction and modification genes; bent arrows, terminal ends of the cloned fragments of plasmids pDAM3, pDAM4, pDAM7, pDAM9, and pDAM10; lines above the sequence, sequences resembling a combox, the second Shine-Dalgarno sequence (S.D.), and the second start codon in the ssuMB gene. The coding sequence shown corresponds to nucleotides 3901 to 7860 of the sequence with accession no. AB045609.
Homology searches showed that the deduced protein encoded by ssuMA was 71 and 70% identical to M.LlaA and M.DpnII, respectively (Table 3). Alignments of the sequences of these three proteins revealed significant similarities among them (Fig. 4A). Thus, it was concluded that ssuMA encoded an N6-methyladenine methyltransferase, which was designated M.SsuMA. The deduced protein encoded by ssuMB was 90 and 86% identical to M.DpnA and M.LlaB, respectively (Table 3). Alignment of these three proteins highlighted striking similarities among them (Fig. 4B). Thus it was concluded that ssuMB encoded a second N6-methyladenine methyltransferase, which was designated M.SsuMB. The similarities among M.SsuMB, M.DpnA, and M.LlaB suggested that M.SsuMB methylates single-stranded DNA, as described for M.DpnA (8).
FIG. 4.
Sequence alignment of M.SsuMA, M.DpnII, and M.LlaA (A); M.SsuMB, M.DpnA, and M.LlaB (B); and R.SsuRA, R.DpnII, R.SsuRB, and R.LlaC (LlaDCHI) (C). Black background, amino acids identical among all the sequences aligned; dashes, gaps in the aligned sequences. DDBJ/EMBL/GenBank accession numbers of the sequences are given in Table 3.
The deduced amino acid sequence encoded by ssuRA was similar to that of R.DpnII, whereas the sequence encoded by ssuRB was highly homologous to that of R.LlaDCHI (Table 3). Both R.DpnII and R.LlaDCHI are endonucleases which specifically recognize sequence 5′-↓GATC-3′ and cleave prior to the 5′ guanine as indicated by the arrow. Thus, both ssuRA and ssuRB encode restriction endonucleases, which we named R.SsuRA and R.SsuRB, respectively. All currently characterized type II endonucleases displaying significant similarity are isoschizomers of each other, indicating that R.SsuRA and R.SsuRB likely recognize 5′-GATC-3′ sites and cleave them as R.DpnII and R.LlaDCHI do. Alignment of the amino acid sequences of these four endonucleases revealed similarities at their centers and near the C-terminal ends of the proteins (Fig. 4C). The conservation of these residues suggests that they may constitute the catalytically active sites of the enzymes. R.SsuRA and R.SsuRB have a limited homology (31%) with each other, and they do not have any significant homology with the GATC-specific type II endonucleases of other classes, such as Sau3AI and DpnI.
Organization of purine biosynthetic genes.
The genetic organization of the region flanking the SsuDAT1I genes revealed that the purine biosynthetic genes were clustered in this bacterium, as has been observed in other gram-positive bacteria such as Bacillus subtilis (4). The gene order, excluding the SsuDAT1I genes, is identical to that found in the genome sequence of Streptococcus pyogenes (B. A. Roe, S. P. Linn, L. Son, X. Yuan, S. Clifton, M. McShan, and J. Ferretti, Streptococcal Genome Sequencing Project, University of Oklahoma, Norman [http://www.genome.ou.edu/strep.html]) and similar to the gene order of the B. subtilis pur operon (4). All the pur genes found in S. suis except purF, of which the 5′ end was truncated, were preceded by typical Shine-Dalgarno sequences. These pur genes were separated by intergenic sequences of 13 to 25 bp, with the exception of purM and purN, which overlapped (Table 3). However, the intergenic sequences between purH and purD, which were interrupted by the SsuDAT1I genes, were longer than those found among pur genes, i.e., 64 and 120 bp (Table 3).
Expression of methylation and restriction genes in E. coli.
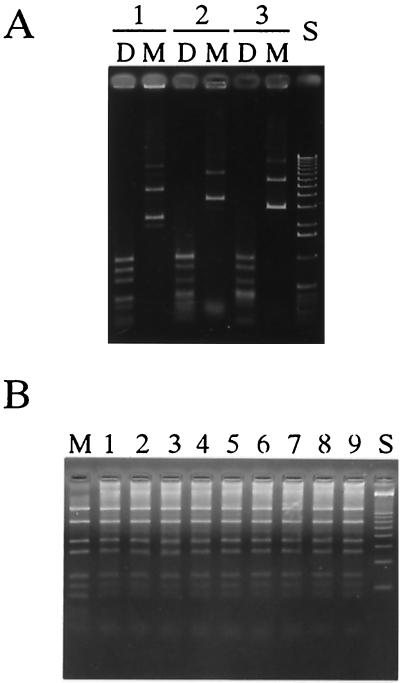
Plasmid pDAM4C, used for DNA sequencing, was digested with PstI (one PstI site is in the cloned insert and the other is in the multicloning sites of the vector), and the 1.2-kb fragment containing ssuMA was subcloned into pUC19 to create pSUMA1. The translational direction of ssuMA was opposite in orientation to that of the lacZ promoter on the vector. Introduction of pSUMA1 into E. coli SCS110 and restriction cleavage analysis of the pSUMA1 recovered from SCS110 using DpnI and MboI showed that SCS110 expressed the methyltransferase (Fig. 5A).
FIG. 5.
(A) Methylation of plasmid DNAs containing the cloned methylation gene(s) of the SsuDAT1I system. The plasmids recovered from E. coli SCS110 were digested with restriction endonucleases and analyzed by agarose gel electrophoresis. Lane 1, pSUMA1; lane 2, pSUMAB14; lane 3, pSUMB1. D, digested with DpnI; M, digested with MboI; S, 1-kb ladder size standards (GIBCO/BRL). (B) DNA cleavage activities of crude extracts prepared from E. coli strains carrying the following plasmids using unmethylated pUC19 as the substrate: lane 1, pSURA1; lane 2, pSURA2; lane 3, pSURA3; lane 4, pSURA4; lane 5, pSURB1; lane 6, pSURB2; lane 7, pSURB3; lane 8, pSURB4; lane 9, pSURB5. M, pUC19 digested with MboI; S, 100-bp ladder size standards (GIBCO/BRL).
The DNA fragment from approximately 100 bp upstream of ssuMA to approximately 100 bp downstream of ssuMB was amplified by PCR using primers purH3′ and ssuMB3′ (Fig. 3). The amplified 2.2-kb fragment was ligated to pCR2.1, and the ligation mixture was used to transform E. coli DH5α. Fifteen clones were picked. The translational direction of ssuMA and ssuMB was opposite to the orientation of the lacZ promoter in 14 clones. All 14 clones contained two or more nucleotide substitutions caused by misreading during the PCR amplification, resulting in undesired amino acid changes. Clone pSUMAB14 contained only one nucleotide substitution in the ORF of ssuMB (A at nucleotide 667 of ssuMB was replaced by G), which converted Ile223 to a similar amino acid, Val223, and this subclone was used for further study. pSUMAB14 was digested with AvaI, filled in with Klenow enzyme, self-ligated, and introduced into E. coli DH5α to create a plasmid carrying only ssuMB and designated pSUMB1. pSUMAB14 and pSUMB1 were then introduced into E. coli SCS110, and the plasmids recovered from SCS110 were digested with either DpnI or MboI. Both plasmids could be cleaved by DpnI but not by MboI (Fig. 5A), indicating that the methylation genes were functional in E. coli.
Several attempts to clone the two restriction endonuclease genes into E. coli were unsuccessful. Thus, ssuRA and ssuRB were individually amplified by PCR using synthetic primers containing unique restriction cleavage sites, i.e., ssuRA5′Ec plus ssuRA3′Pst and ssuRB5′Ec plus ssuRB3′Pst, respectively. The positions of the primers are indicated in Fig. 3. The unique restriction sites introduced in the primers permitted their cloning into low-copy-number vector pHSG576 in the orientation opposite to that of the lacZ promoter. Four and five clones containing ssuRA and ssuRB, respectively, were obtained and were designated pSURA1∼pSURA4 and pSURB1∼pSURB5, respectively. These plasmids were then introduced into E. coli SCS110 carrying pSUMAB14. Crude extracts of these clones were prepared by sonication and examined for their DNA cleavage activities using unmethylated pUC19 DNA. All showed restriction endonuclease activity indistinguishable from that of MboI (Fig. 5B).
Comparison of the genetic region encoding the SsuDAT1I R-M system with the corresponding region of strains lacking the R-M system.
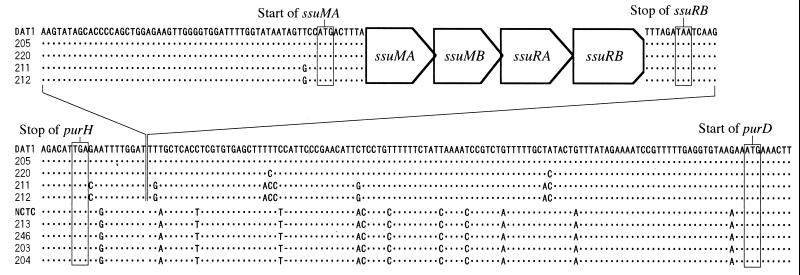
To confirm whether the genetic region encoding the SsuDAT1I system was conserved among other S. suis strains and to compare the corresponding regions of strains carrying the R-M system and strains with the null phenotype, we performed PCR using primers purH3′ and ssuRB3′ to amplify the entire R-M region with a part of the flanking regions. When genomic DNA isolated from strains carrying the R-M system was used, a 3.8-kb fragment was amplified from all the strains tested (data not shown). On the other hand, when genomic DNA from the strains with the null phenotype was used, a 323-bp fragment was amplified from all the strains tested (data not shown). These results indicated that the genetic organization of the R-M system was conserved among the strains tested and that the entire R-M system was missing in the strains with the null phenotype. Direct sequencing of the amplified fragments from four strains having the R-M genes and comparison among them showed that only a few nucleotide substitutions were present in this region (Fig. 6), indicating that the R-M region was conserved among the strains. Direct sequence determination of the 323-bp fragments amplified from five strains with the null phenotype and comparison with those of the R-M regions revealed the differences in DNA sequence between them. The 323-bp DNA sequences obtained from the five strains with the null phenotype were completely conserved. The unique DNA sequence of the R-M region comprising 3,503 bp, which extended from 53 bp upstream of the ssuMA gene to 5 bp downstream of the ssuRB gene, was inserted in an intergenic sequence between purH and purD. The insertion target site, 5′-GAT↓T(T/G)-3′, as indicated by the arrow, was highly conserved among the strains tested (Fig. 6). The sequence where the R-M genes were inserted was not the recognition site of SsuDAT1I. Comparison of the 323-bp DNA regions of both groups revealed several mismatches present in upstream and downstream sequences (Fig. 6). No other notable substitutions or insertions could be found in these regions. The only notable feature found in the junction region was that a 3-bp sequence at the 5′ end of the 3,503-bp sequence, AAG (Fig. 6, left end) was also present at the 3′ end of the sequence (Fig. 6, right end). The DNA sequences of the region flanking the R-M system showed no sequence homology to published sequences flanking the DpnII and LlaDCHI systems (24, 30).
FIG. 6.
Alignment of the sequences from the 3′ end of purH to the 5′ end of purD. Only the sequences that flank the SsuDAT1I genes are shown. The 3,503-bp sequences from 53 bp upstream of ssuMA to 5 bp downstream of ssuRB (top) are connected by the lines with the lower sequences flanking the SsuDAT1I region, which are aligned with the corresponding sequences of the strains lacking the R-M system. Strains are indicated to the left of the sequences (NCTC, NCTC10234). Dots, nucleotides identical to those of the aligned sequence of DAT1; dashes, gaps in the aligned sequences; boxes, stop and start codons of the genes appearing in these sequences.
PFGE analysis.
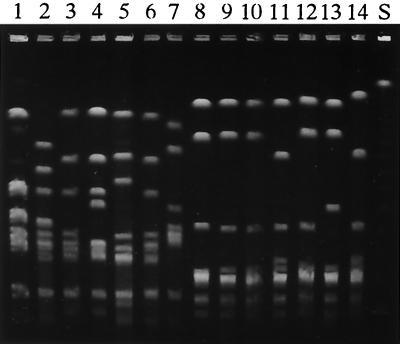
Genomic DNAs of S. suis strains were digested with SmaI and separated by PFGE. Representative cleavage patterns of several strains are shown in Fig. 7. The PFGE patterns of the strains having the R-M system were distinguishable from those obtained from the strains with the null phenotype. The PFGE patterns obtained from the strains with the null phenotype were similar to each other, and some of them were indistinguishable, suggesting that different isolates of the same strain were included. However, striking variations were seen among the PFGE patterns of the strains having the R-M system, demonstrating that the strains used were independent.
FIG. 7.
PFGE of the S. suis genomic DNA digested with SmaI. Electrophoresis was carried out with a CHEF system for 20 h at 200 V and 10°C with pulse time ramping from 5 to 50 s in a 1% agarose gel. Strains carrying the R-M system were as follows: DAT1 (lane 1), 205 (lane 2), 220 (lane 3), 227 (lane 4), 243 (lane 5), 222 (lane 6), and 236 (lane 7). Strains with the null phenotype were as follows: NCTC10234 (lane 8), 204 (lane 9), 207 (lane 10), 209 (lane 11), 210 (lane 12), 213 (lane 13), and 246 (lane 14). S, λ DNA concatemers as size standards.
DISCUSSION
We have demonstrated here the presence of an R-M system in S. suis. Cloning and DNA sequencing of the SsuDAT1I gene region revealed that the genetic organization was unique for all type II R-M systems described to date, i.e., the region contained two restriction endonuclease genes and two methyltransferase genes. Other R-M systems comprising two methyltransferase genes and one restriction endonuclease gene are known and are isoschizomers of SsuDAT1I, i.e., DpnII, LlaDCHI, and MboI. In DpnII and LlaDCHI, the order of the genes is similar to that of the SsuDAT1I system. The gene encoding a conventional methyltransferase for double-stranded DNA is upstream of the gene encoding a second methyltransferase for single-stranded DNA. The restriction endonuclease gene is downstream and the genes overlap, as seen in the SsuDAT1I genes (8, 24, 30). In MboI, the order of the genes is different. The gene encoding R.MboI is flanked by two methyltransferase genes, which are separated by 2-bp intergenic spaces (52). These differences suggest that the genetic rearrangement among the R-M genes could have occurred during the evolution of these R-M genes. R.SsuRA was similar to R.DpnII, and R.SsuRB showed extensive homology with R.LlaDCHI, whereas R.SsuRA (or R.DpnII) and R.SsuRB (or R.LlaDCHI) showed limited homology. The results suggest that an extra restriction endonuclease gene was acquired from some other bacterium and that this resulted in a composite genetic structure, i.e., the SsuDAT1I system. Alternatively, the SsuDAT1I system could be the prototype and a deletion of one restriction endonuclease gene resulted in the genetic structure represented by DpnII and LlaDCHI.
The amino acid sequences of the methyltransferases share many conserved structural motifs, and, on the basis of the presence of these motifs in an unknown protein, one can predict its function (6, 56). In this study, striking similarities among the methyltransferases indicated that M.SsuMA and M.SsuMB are highly related to the methyltransferases of the DpnII and LlaDCHI systems. On the other hand, different restriction endonucleases usually do not have extensive homology at the level of the amino acid sequence, even when they recognize the same DNA sequence (6, 56). Nevertheless, some exceptions are known; for example, the isospecific pairs EcoRI/RsrI (1), PstI/BsuBI (57), XmaI/Crf9I (56), NgoI/HaeII (47), and BanI/HgiCI (10) show some degree of homology. While R.DpnII and R.LlaDCHI have limited homology, they were found experimentally to have the same recognition sequence (30). R.SsuRA and R.SsuRB had significant similarities to one of these restriction endonucleases. This indicates that the specificity and other enzymatic properties are the same, although differences in enzymatic properties between DpnII and LlaDCHI have not been studied. To date, one example of an R-M system containing two restriction endonuclease genes has been reported. In that case, a gene encoding R.Eco47I, which recognizes 5′-GGWCC-3′ (W is A or T), was located in the vicinity of another R-M system, Eco47II, which recognizes similar but less specific sequence 5′-GGNCC-3′, where N is any nucleotide (46). In the SsuDAT1I system, R.SsuRA and R.SsuRB, whose genes were individually cloned and expressed in E. coli, could digest unmethylated pUC19 in the same way as MboI. Therefore, their sequence recognition specificities appeared identical.
We could not clone the SsuDAT1I genes directly in E. coli from the phage library or from a PCR-derived fragment. However, the isoschizomeric LlaDCHI and MboI R-M genes have been cloned in E. coli on multicopy vectors (30, 52). E. coli possesses orphan methylase Dam, which has the same sequence specificity as the SsuDAT1I system and which protects the host DNA from attack by the corresponding restriction endonucleases. Recently, the Dam methylase was found to be involved in the regulation of gene expression, and hence not all the recognition sequences in the host chromosomal DNA were methylated (14). If the enzymatic activity of SsuDAT1I is stronger than those of the other isoschizomers, the cloning of the genes will inhibit the growth of the E. coli host cells. Indeed, the dual genes encoding the restriction endonucleases in the SsuDAT1I system might be expected to express twice as much of the restriction endonuclease activity due in essence to a gene dosage effect (53).
The SsuDAT1I system was not found in several S. suis strains, including NCTC10234. Moreover, the identity of the DNA sequences flanking this system, in conjunction with the differences in G+C content and codon usage between SsuDAT1I and the flanking regions, supports the notion that this system did not arise in this organism. The SsuDAT1I genes were not located on transferable elements such as plasmids and bacteriophages. Comparison of the corresponding DNA regions of strains with and without the R-M system revealed the precise location of the R-M genes. The SsuDAT1I genes have been inserted into a 125-bp intergenic region separating purH and purD. This may ensure constitutive expression of the R-M genes. In most bacteria, purine biosynthetic genes are part of an essential operon. The overall genetic organization of the pur operon, except for the SsuDAT1I genes, was identical to that found in S. pyogenes (Streptococcal Genome Sequencing Project, University of Oklahoma, Norman [http://www.genome.ou.edu/strep.html]) and was similar to that reported in B. subtilis (4). Because the pur regions of NCTC10234 and other S. suis strains lack the R-M genes and because the sequences flanking the R-M sequences in S. suis strains carrying the system are virtually identical, it appears that the latter S. suis strains have only recently acquired the sequence.
A natural competence for genetic transformation has not yet been demonstrated in S. suis. However, the combox (25), a competence-regulated sequence, was found in the initiation region of the ssuMB gene. This might support a hypothesis that the SsuDAT1I system is an exogenous element and that S. suis acquired the SsuDAT1I genes via a transformation event. However, the competence of some bacteria is coordinately expressed in response to the growth phase and culture conditions (2, 27). Further studies may be needed to determine whether S. suis has the potential to be competent for genetic transformation.
Recently, studies of a novel R-M system, Hpy188I, found in Helicobacter pylori strain J188 and comparison of its DNA sequence with those of other strains revealed that the R-M system has been horizontally transferred from other bacteria (58). The Hpy188I genes are flanked by 92-bp long direct repeats, suggesting that a transposition event involving the R-M system had taken place. In the region flanking SsuDAT1I, we could not find any long-repeat sequences or any notable nucleotide substitutions. The genetic structure was conserved among the S. suis strains tested. However, from the results of the PFGE analysis, we could not rule out the possibility that the strains harboring the R-M system were clonal. The structure of the genetic region flanking the SsuDAT1I system and stable maintenance of this genetic structure in the different strains of S. suis lead us to propose another hypothesis to explain the genetic conversion, i.e., illegitimate recombination. It is plausible that the SsuDAT1I genes were initially transferred on a transposon, followed by excision of the transposon, since no functions associated with DNA mobility, such as transposase, integrase, and invertase functions, were found in the vicinity of the SsuDAT1I genes. However, this hypothesis does not explain the facts that the conserved genetic structure of the R-M system was found in different strains and that no transposable element could be found in any of the strains tested. This indicated that excision of a transposable element did not occur following passage through the strains tested. It is also possible that the SsuDAT1I system was acquired by an ancestor of the S. suis DAT1 strain via illegitimate recombination from distant sources and that the R-M genes have been transferred among strains of S. suis en bloc along with the conserved flanking pur genes, perhaps by transformation, and incorporated into the recipient chromosome by homologous recombination.
ACKNOWLEDGMENTS
We are grateful to S. Yasuda and T. Hashimoto-Gotoh for providing us with the vector plasmid pHSG576. We thank T. Fujisawa for preparing photographs and M. Takahashi for technical assistance.
REFERENCES
- 1.Aiken C, Gumport R I. Restriction endonuclease RsrI from Rhodobacter sphaeroides, an isoschizomer of EcoRI: purification and properties. Nucleic Acids Res. 1988;16:7901–7916. doi: 10.1093/nar/16.16.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alloing G, Martin B, Granadel C, Claverys J-P. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol Microbiol. 1998;29:75–83. doi: 10.1046/j.1365-2958.1998.00904.x. [DOI] [PubMed] [Google Scholar]
- 3.Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 4.Anagnostopoulos C, Piggot P J, Hoch J A. The genetic map of Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 425–461. [Google Scholar]
- 5.Anton B P, Heiter D F, Benner J S, Hess E J, Greenough L, Moran L S, Slatko B E, Brooks J E. Cloning and characterization of the BglII restriction-modification system reveals a possible evolutionary footprint. Gene. 1997;187:19–27. doi: 10.1016/s0378-1119(96)00638-5. [DOI] [PubMed] [Google Scholar]
- 6.Bickle T A, Kruger D H. Biology of DNA restriction. Microbiol Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brassard S, Paquet H, Roy P H. A transposon-like sequence adjacent to the AccI restriction-modification operon. Gene. 1995;157:69–72. doi: 10.1016/0378-1119(94)00734-a. [DOI] [PubMed] [Google Scholar]
- 8.Cerritelli S, Springhorn S S, Lacks S A. DpnA, a methylase for single-strand DNA in the DpnII restriction system, and its biological function. Proc Natl Acad Sci USA. 1989;86:9223–9227. doi: 10.1073/pnas.86.23.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clifton-Hadley F A. Streptococcus suis type 2 infections. Br Vet J. 1983;139:1–5. doi: 10.1016/s0007-1935(17)30581-x. [DOI] [PubMed] [Google Scholar]
- 10.Erdmann D, Dusterhoft A, Kroger M. Cloning and molecular characterization of the HgiCI restriction/modification system from Herpetosiphon giganteus Hpg9 reveals high similarity to BanI. Eur J Biochem. 1991;202:1247–1256. doi: 10.1111/j.1432-1033.1991.tb16497.x. [DOI] [PubMed] [Google Scholar]
- 11.Gelinas R E, Myers P A, Roberts R J. Two sequence-specific endonucleases from Moraxella bovis. J Mol Biol. 1977;114:169–179. doi: 10.1016/0022-2836(77)90290-x. [DOI] [PubMed] [Google Scholar]
- 12.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J Clin Microbiol. 1991;29:2590–2594. doi: 10.1128/jcm.29.11.2590-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottschalk M, Higgins R, Jacques M, Mittal K R, Henrichsen J. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol. 1989;27:2633–2636. doi: 10.1128/jcm.27.12.2633-2636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 15.Higgins R, Gottschalk M. Distribution of Streptococcus suis capsular types in 1995. Can Vet J. 1996;37:242. [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins R, Gottschalk M, Beaudoin M, Rawluk S A. Distribution of Streptococcus suis capsular types in Quebec and western Canada. Can Vet J. 1992;33:27–30. [PMC free article] [PubMed] [Google Scholar]
- 17.Jeltsch A, Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J Mol Evol. 1996;42:91–96. doi: 10.1007/BF02198833. [DOI] [PubMed] [Google Scholar]
- 18.Kilpper-Balz R, Schleifer K H. Streptococcus suis sp. nov., nom. rev. Int J Sys Bacteriol. 1987;37:160–162. [Google Scholar]
- 19.Kita K, Tsuda J, Kato T, Okamoto K, Yanase H, Tanaka M. Evidence of horizontal transfer of the EcoO1091 restriction-modification gene to Escherichia coli chromosomal DNA. J Bacteriol. 1999;181:6822–6827. doi: 10.1128/jb.181.21.6822-6827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome—restriction-modification systems as mobile genetic elements. Curr Opin Genet Dev. 1999;9:649–656. doi: 10.1016/s0959-437x(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 21.Kusano K, Sakagami K, Yokochi T, Naito T, Tokunaga Y, Ueda E, Kobayashi I. A new type of illegitimate recombination is dependent on restriction and homologous interaction. J Bacteriol. 1997;179:5380–5390. doi: 10.1128/jb.179.17.5380-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacks S, Greenberg B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J Biol Chem. 1975;250:4060–4066. [PubMed] [Google Scholar]
- 23.Lacks S, Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977;114:153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- 24.Lacks S A, Mannarelli B M, Springhorn S S, Greenberg B. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell. 1986;46:993–1000. doi: 10.1016/0092-8674(86)90698-7. [DOI] [PubMed] [Google Scholar]
- 25.Lacks S A, Ayalew S, de la Campa A G, Greenberg B. Regulation of competence for genetic transformation in Streptococcus pneumoniae: expression of dpnA, a late competence gene encoding a DNA methyltransferase of the DpnII restriction system. Mol Microbiol. 2000;35:1089–1098. doi: 10.1046/j.1365-2958.2000.01777.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee K F, Shaw P C, Picone S J, Wilson G G, Lunnen K D. Sequence comparison of the EcoHK31I and EaeI restriction-modification systems suggests an intergenic transfer of genetic material. Biol Chem. 1998;379:437–441. doi: 10.1515/bchm.1998.379.4-5.437. [DOI] [PubMed] [Google Scholar]
- 27.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutticken R, Temme N, Hahn G, Bartelheimer E W. Meningitis caused by Streptococcus suis: case report and review of the literature. Infection. 1986;14:181–185. doi: 10.1007/BF01645260. [DOI] [PubMed] [Google Scholar]
- 29.Mannarelli B M, Balganesh T S, Greenberg B, Springhorn S S, Lacks S A. Nucleotide sequence of the DpnII DNA methylase gene of Streptococcus pneumoniae and its relationship to the dam gene of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:4468–4472. doi: 10.1073/pnas.82.13.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moineau S, Walker S A, Vedamuthu E R, Vandenbergh P A. Cloning and sequencing of LlaDCHI restriction/modification genes from Lactococcus lactis and relatedness of this system to the Streptococcus pneumoniae DpnII system. Appl Environ Microbiol. 1995;61:2193–2202. doi: 10.1128/aem.61.6.2193-2202.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muckerman C C, Springhorn S S, Greenberg B, Lacks S A. Transformation of restriction endonuclease phenotype in Streptococcus pneumoniae. J Bacteriol. 1982;152:183–190. doi: 10.1128/jb.152.1.183-190.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochman H, Gerberi A S, Hartl L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogata K, Aminov R I, Nagamine T, Sugiura M, Tajima K, Mitsumori M, Sekizaki T, Kudo H, Minato H, Benno Y. Construction of a Fibrobacter succinogenes genome map and demonstration of diversity at the genomic level. Curr Microbiol. 1997;35:22–27. doi: 10.1007/s002849900205. [DOI] [PubMed] [Google Scholar]
- 34.Osaki M, Takamatsu D, Tsuji N, Sekizaki T. Cloning and characterization of the gene encoding O-acetylserine lyase from Streptococcus suis. Curr Microbiol. 2000;40:67–71. doi: 10.1007/s002849910013. [DOI] [PubMed] [Google Scholar]
- 35.Palmer B R, Marinus M G. The dam and dcm strains of Escherichia coli—a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 36.Perch B, Pedersen K B, Henrichsen J. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J Clin Microbiol. 1983;17:993–996. doi: 10.1128/jcm.17.6.993-996.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts R J, Macelis D. REBASE-restriction enzymes and methylases. Nucleic Acids Res. 2000;28:306–307. doi: 10.1093/nar/28.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rochepeau P, Selinger L B, Hynes M F. Transposon-like structure of a new plasmid-encoded restriction-modification system in Rhizobium leguminosarum VF39SM. Mol Gen Genet. 1997;256:387–396. doi: 10.1007/s004380050582. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schleif R. Assaying of organisms for the presence of restriction endonucleases. Methods Enzymol. 1980;65:19–23. doi: 10.1016/s0076-6879(80)65004-6. [DOI] [PubMed] [Google Scholar]
- 41.Segers R P, Kenter T, de Haan L A, Jacobs A A. Characterisation of the gene encoding suilysin from Streptococcus suis and expression in field strains. FEMS Microbiol Lett. 1998;167:255–261. doi: 10.1111/j.1574-6968.1998.tb13236.x. [DOI] [PubMed] [Google Scholar]
- 42.Sekizaki T, Ito H, Asawa T, Nonomura I. DNA sequence of type 1 fimbrin, Fpul1, gene from a chicken pathogenic Escherichia coli serotype O78. J Vet Med Sci. 1993;55:395–400. doi: 10.1292/jvms.55.395. [DOI] [PubMed] [Google Scholar]
- 43.Serhir B, Dugourd D, Jacques M, Higgins R, Harel J. Cloning and characterization of a dextranase gene (dexS) from Streptococcus suis. Gene. 1997;190:257–261. doi: 10.1016/s0378-1119(97)00004-8. [DOI] [PubMed] [Google Scholar]
- 44.Smith H E, Vecht U, Gielkens A L J, Smits M A. Cloning and nucleotide sequence of the gene encoding the 136-kilodalton surface protein (muramidase-released protein) of Streptococcus suis type 2. Infect Immun. 1992;60:2361–2367. doi: 10.1128/iai.60.6.2361-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith H E, Damman M, van der Velde J, Wagenaar F, Wisselink H J, Stockhofe-Zurwieden N, Smits M A. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67:1750–1756. doi: 10.1128/iai.67.4.1750-1756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stankevicius K, Povilionis P, Lubys A, Menkevicius S, Janulaitis A. Cloning and characterization of the unusual restriction-modification system comprising two restriction endonucleases and one methyltransferase. Gene. 1995;157:49–53. doi: 10.1016/0378-1119(94)00796-u. [DOI] [PubMed] [Google Scholar]
- 47.Stein D C, Gunn J S, Piekarowicz A. Sequence similarities between the genes encoding the S.NgoI and HaeII restriction/modification systems. Biol Chem. 1998;379:575–578. [PubMed] [Google Scholar]
- 48.Sussenbach J S, Monfoort C H, Schiphof R, Stobberingh E E. A restiction endonuclease from Staphylococcus aureus. Nucleic Acids Res. 1976;3:3193–3202. doi: 10.1093/nar/3.11.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takamatsu D, Osaki M, Sekizaki T. Sequence analysis of a small cryptic plasmid isolated from Streptococcus suis serotype 2. Curr Microbiol. 2000;40:61–66. doi: 10.1007/s002849910012. [DOI] [PubMed] [Google Scholar]
- 50.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ α-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 51.Twomey D P, McKay L L, O'Sullivan D J. Molecular characterization of the Lactococcus lactis LlaKR2I restriction-modification system and effect of an IS982 element positioned between the restriction and modification genes. J Bacteriol. 1998;180:5844–5854. doi: 10.1128/jb.180.22.5844-5854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueno T, Ito H, Kimizuka F, Kotani H, Nakajima K. Gene structure and expression of the MboI restriction-modification system. Nucleic Acids Res. 1993;21:2309–2313. doi: 10.1093/nar/21.10.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uhlin B E, Nordstrom K. R plasmid gene dosage effects in Escherichia coli K12: copy mutants of the R plasmid R1drd-19. Plasmid. 1977;1:1–7. doi: 10.1016/0147-619x(77)90003-8. [DOI] [PubMed] [Google Scholar]
- 54.Vaisvila R, Vilkaitis G, Janulaitis A. Identification of a gene encoding a DNA invertase-like enzyme adjacent to the PaeR7I restriction-modification system. Gene. 1995;157:81–84. doi: 10.1016/0378-1119(94)00793-r. [DOI] [PubMed] [Google Scholar]
- 55.Vovis G F, Lacks S. Complementary action of restriction enzymes Endo R.DpnI and Endo R.DpnII on bacteriophage f1 DNA. J Mol Biol. 1977;115:525–538. doi: 10.1016/0022-2836(77)90169-3. [DOI] [PubMed] [Google Scholar]
- 56.Wilson G G, Murray N E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 57.Xu G-L, Kapfer W, Walter J, Trautner T A. BsuBI—an isospecific restriction and modification system of PstI: characterization of the BsuBI genes and enzymes. Nucleic Acids Res. 1992;20:6517–6523. doi: 10.1093/nar/20.24.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Q, Stickel S, Roberts R J, Blaser M J, Morgan R D. Purification of the novel endonuclease, Hpy188I, and cloning of its restriction-modification genes reveal evidence of its horizontal transfer to the Helicobacter pylori genome. J Biol Chem. 2000;275:17086–17093. doi: 10.1074/jbc.M910303199. [DOI] [PubMed] [Google Scholar]
- 59.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 60.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 561–579. [Google Scholar]