Abstract
The mechanical force between a virus and its host cell plays a critical role in viral infection. However, characterization of the virus-cell mechanical force at the whole-virus level remains a challenge. Herein, we develop a platform in which the virus is anchored with multivalence-controlled aptamers to achieve transfer of the virus-cell mechanical force to a DNA tension gauge tether (Virus-TGT). When the TGT is ruptured, the complex of binding module-virus-cell is detached from the substrate, accompanied by decreased host cell-substrate adhesion, thus revealing the mechanical force between whole-virus and cell. Using Virus-TGT, direct evidence about the biomechanical force between SARS-CoV-2 and the host cell is obtained. The relative mechanical force gap (<10 pN) at the cellular level between the wild-type virus to cell and a variant virus to cell is measured, suggesting a possible positive correlation between virus-cell mechanical force and infectivity. Overall, this strategy provides a new perspective to probe the SARS-CoV-2 mechanical force.
Keywords: SARS-CoV-2, host-pathogen interactions, DNA nano-assembly, aptamer, mechanical force, DNA tension gauge tether
Graphical abstract
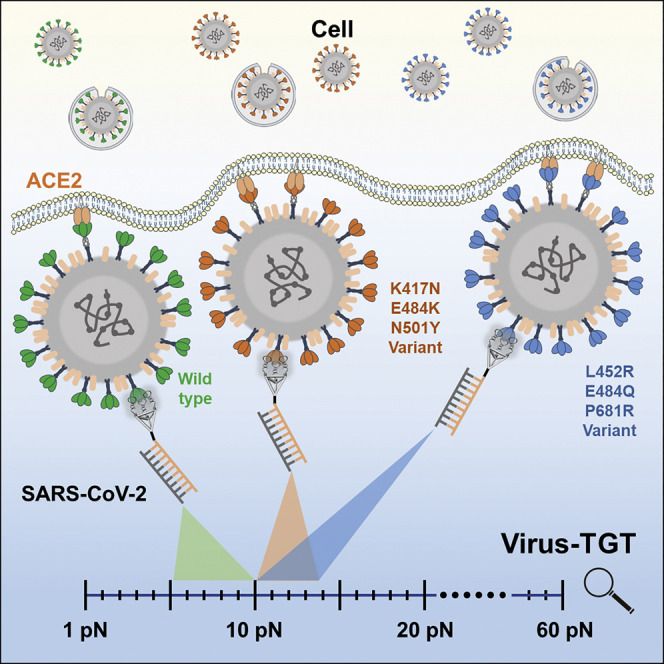
Leveraging multivalence-controlled-aptamers to anchor a virus, Zhang et al. develop a platform to achieve transfer of virus-cell mechanical force to a DNA tension gauge tether. This DNA nano-platform measures the mechanical force during SARS-CoV-2 infection at the whole-virus level, revealing a possible positive correlation between viral mechanical force and infectivity.
Introduction
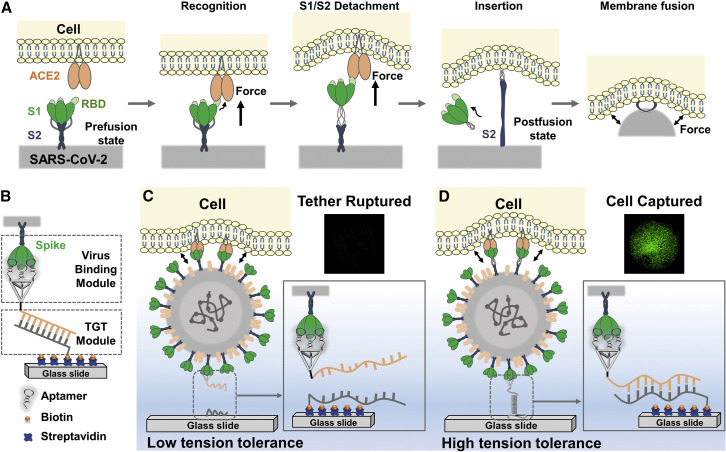
SARS-CoV-2 and its emerging mutant strains are spreading worldwide. The SARS-CoV-2 invasion into the host cell occurs in two vital processes: receptor (ACE2 [angiotensin-converting enzyme 2]) recognition by the spike protein (S protein) on SARS-CoV-2 and subsequent conformational changes of the S protein to mediate membrane fusion.1, 2, 3, 4, 5 Specifically, once a virion attaches to the host cell, the spike/ACE2 binding induces a pulling force to accelerate the detachment of the S1 subunit from the S2 subunit to rapidly form fusion machinery6 (Figure 1A). The accumulation of this process causes the bending of the host cell membrane, exerting a tensile force that inevitably impacts subsequent viral host recognition and invasion. Therefore, the mechanism of SARS-CoV-2 infection is associated with complex mechanical cues.
Figure 1.
Measurement of the mechanical force of SARS-CoV-2 and host cell using Virus-TGT
(A) Schematic diagram of SARS-CoV-2 spike S1/S2 detachment and subsequent membrane fusion.
(B) The composition of Virus-TGT mainly includes the binding module and the tension gauge tether (TGT) module.
(C and D) Working principle of Virus-TGT under (C) low tension tolerance or (D) high tension tolerance.
Although the dissociation constant (Kd) of the spike-ACE2 interaction has been widely studied,4 , 7 , 8 inconsistency between Kd value and infectivity has been found.6 To further understand the mechanism of SARS-CoV-2 infection, the interaction mechanical force of S protein and the ACE2/ACE2 expressed cell has been measured by single-molecule force spectroscopy (SMFS), such as with magnetic tweezers and atomic force microscopy.9, 10, 11, 12, 13 However, such specialized technical equipment-based strategies are often limited by the necessity of skilled technical staff to prepare functionalized probes.14, 15, 16 Moreover, holistic mechanical force characterization of SARS-CoV-2 virus and host cells is still missing. Lack of knowledge of the overall mechanical force between SARS-CoV-2 and the host cell raises questions about how tensile force affects viral infectivity, and whether the mechanical interactions of SARS-CoV-2 and its variants are different.
Herein, we describe a platform for studying virus-cell infection strengths by anchoring the virus with a multivalence-controlled aptamer. This allows mechanical force transfer between the virus-cell complex to a DNA tension gauge tether (Virus-TGT), enabling measurement of infection force between SARS-CoV-2 and its host cell (Figure 1). Previous TGT-based mechanical force analytical platforms simulate artificial biological membranes by conjugating the specific ligand to the tether-modified surface.17, 18, 19, 20, 21, 22 Thus, only the force between the specific ligand and receptor cell can be measured, rather than the overall mechanical force of virus to cell. For the first time, the Virus-TGT platform can measure the overall mechanical force between SARS-CoV-2 and host cell, rather than remaining at protein-level analysis as previously reported (Table S3).6 , 9, 10, 11, 12, 13 , 23 The protein-level analysis only considers the affinity between S protein and ACE2, but ignores the influence of viral membrane deformation, spike number, and spike distribution on viral infectivity and mechanical force. The overall mechanical force comprehensively involves the influence of many factors, so this developed mechanical force measurement enables more accurate comparison of the mutated strain/host cell interaction with that of the unmutated virus and cell.
Results and discussion
Assembly and characterization of the binding module
The Virus-TGT is composed of two modules: the binding module and the TGT module (Figure 1B). To transfer force from the virus-cell to the TGT, it is necessary to achieve a strong enough binding module to ensure that the virus and binding module do not separate during the force transfer, i.e., the binding force between virus and the binding module must be stronger than that of the virus and the cell. Therefore, we applied a ∼5.8 nm tetrahedral DNA framework (TDF) as a multivalent linker for an assembly of three SARS-CoV-2 aptamers.24 As a result of the TDF’s controllable multivalent topology,25 the synergetic effect of trivalent aptamers promotes tight binding of SARS-CoV-2 to the cell, enabling stable formation of the binding module-virus complex. Moreover, the valence-controlled binding module ensures a homogeneous tensile force between the binding module and the virus, which is essential for the final measurement.
The TGT module is formed by hybridizing the remaining pendant strand of TriApTDF with the substrate-immobilized strand. To determine the virus-cell force, we used a series of TGT modules with different defined pN-scale tensile forces26 , 27 (Figure S1). Once the mechanical force between the virus and cell is higher than the tensile force of the TGT module with low tension tolerance, the DNA tether will rupture, resulting in the detachment of the virus-cell complex from the solid surface (Figure 1C). In contrast, if the virus-cell mechanical force is smaller than that of the TGT module with high tension tolerance, the DNA tether will remain, holding the virus-cell complex on the surface (Figure 1D). Consequently, using a range of TGTs with tunable tension tolerances, we can determine the overall mechanical force between the virus and cell by counting the number of fluorescence-pre-stained cells retained on the surface.
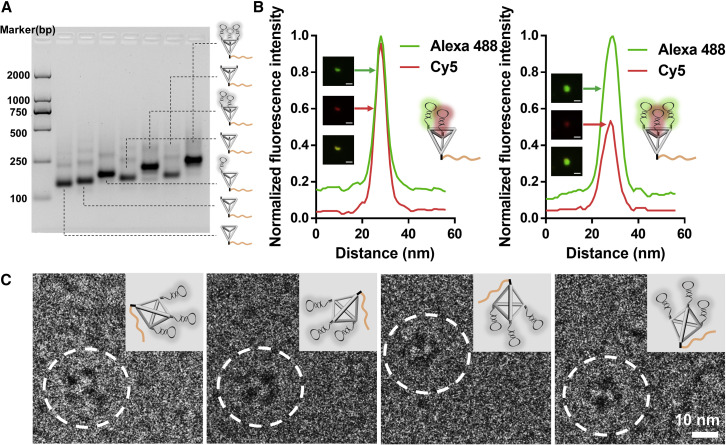
To construct a stable binding module that resists SARS-CoV-2 detachment during force transfer, we designed a multivalent binding module by anchoring three SARS-CoV-2 aptamers on the vertices of TDF (TriApTDF) with one-step self-assembly based on the quantitative ratio. The stepwise assembly of the binding module was characterized by agarose gel electrophoresis (AGE) analysis (Figures 2A and S2), showing a relatively high yield of over 85%. To probe the precise stoichiometry of aptamers on the binding module, we used a total internal reflection fluorescence microscope (TIRFM) to analyze the co-localization and fluorescence intensity of the aptamer labeled with different fluorophores. When the composition ratio of Alexa 488-labeled aptamer and Cy5-labeled aptamer was 2:1, the green dot was overlapped with the red dot, showing twice green fluorescence intensity enhancement (Figure 2B). The fluorescence intensity was comparable at the same site when Alexa 488-labeled aptamer and Cy5-labeled aptamer were assembled in equal proportions, indicating that aptamer assembly in TDF occurs with defined stoichiometry (Figure 2B). Moreover, the morphology of the binding module showed tetrahedral shape with four darker dots in cryoelectron microscopy images (Figure 2C), possibly due to the cantilever design of trivalent aptamers and the extending strands. Therefore, the successful formation of TriApTDF was consistent with controllable multivalent topology, thus guaranteeing both multivalency and homogeneity of mechanical forces between the binding module and the virus.
Figure 2.
Assembly and characterization of the binding module
(A) AGE analysis of the stepwise assembly of the binding module.
(B) TIRFM images and co-localization fluorescence curve of the binding module consisting of an Alexa 488-tagged aptamer and a Cy5-tagged aptamer. The light spots in the image enlarged to visualize the size of the original fluorophore, while their original sizes were at the nanometer level. Scale bar, 1 μm.
(C) Cryoelectron microscopy images and illustration of the binding module. Scale bar, 10 nm.
Characterization of the stable formation of the binding module-virus complex
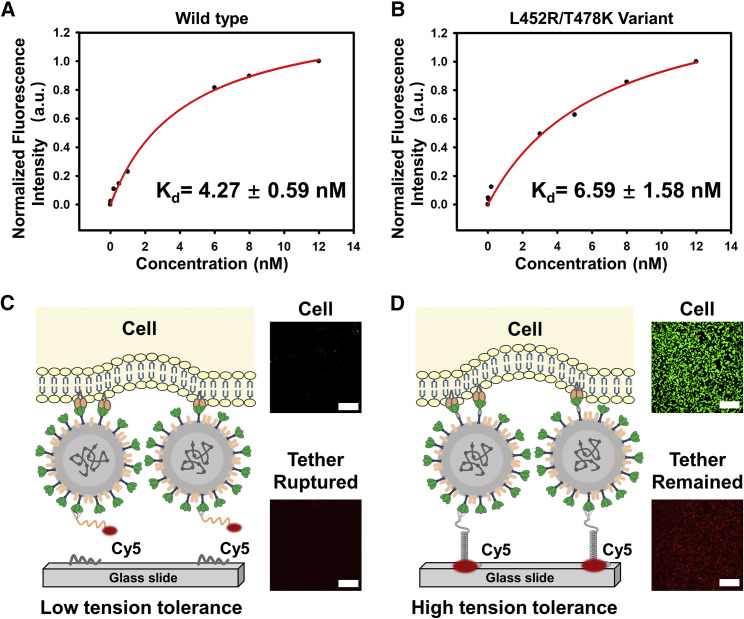
To evaluate the binding stability between the binding module and virus, we first analyzed the Kd values of TriApTDF to SARS-CoV-2 wild-type and two dominant mutant receptor binding domains (RBDs). The Kd value of TriApTDF against wild-type RBD (4.27 ± 0.59 nM, Figure 3A), is comparable with the Kd value against L452R/T478K mutant RBD of the Delta variant (6.59 ± 1.58 nM, Figure 3B). In addition, TriApTDF tightly binds to the K417N/E484K/N501Y mutant RBD (three critical mutations of the Beta variant), with a Kd value of 4.64 ± 0.75 nM (Figure S3). Benefitting from the multivalent synergy of aptamers, these nanomolar dissociation constant values indicate the outstanding binding adaptability of the binding module against SARS-CoV-2 variants. The reported Kd values of ACE2 and RBD/mutant RBDs ranged from 30 to 150 nM,7 , 9 , 28 , 29 suggesting that the firm binding of binding module-virus can be fulfilled theoretically due to the 10-fold increased binding affinity.
Figure 3.
Characterization of the stable formation of binding module-virus complex
(A and B) The binding curve and dissociation constant of TriApTDF against SARS-CoV-2 RBD-beads of (A) wild-type and (B) L452R/T478K mutant in binding buffer.
(C) Cy5-labeled binding module from substrate rupture induced by the interaction of ACE2-expressing HEK293T cells and SARS-CoV-2 at the low tensile force of TGT; almost invisible cell adherence on the substrate.
(D) Significant cell adhesion occurs as the TGT failed to rupture at high tensile force. Scale bar, 100 μm.
To further rule out the potential dissociation of the binding module and the virus, we employed a Cy5-labeled remaining pendant strand of TriApTDF to construct the TGT module. First, we separately constructed two Virus-TGT platforms with low and high tensile force tolerances (Figures 3C and 3D). Next, we added a constant particle concentration of SARS-CoV-2 pseudoviruses before mechanical force investigation with ACE2-expressing HEK293T cells (Figures S4–S6). After removing and rinsing the unbound cells (Figure S7), we analyzed the fluorescence images of Cy5-labeled binding module and pre-stained host cells. When the mechanical force between virus and cell was higher than the tensile force of TGT, the TGT was ruptured and there were almost invisible spotty fluorescence signals of both DNA and cells (Figure 3C), suggesting the complex of binding module-virus-cell was detached from the substrate. In contrast, when the mechanical force between virus and cell was lower than the tensile force of TGT, we observed that most Cy5-labeled binding modules remained on the surface accompanied by the virus-related marked cell adhesion (Figure 3D). As expected, formation of the virus-binding module was stable. A TGT other than the virus-binding module was ruptured when the mechanical force between virus and host cell was larger than the rupture force of TGT.
Mechanical force measurement between SARS-CoV-2 and host cell
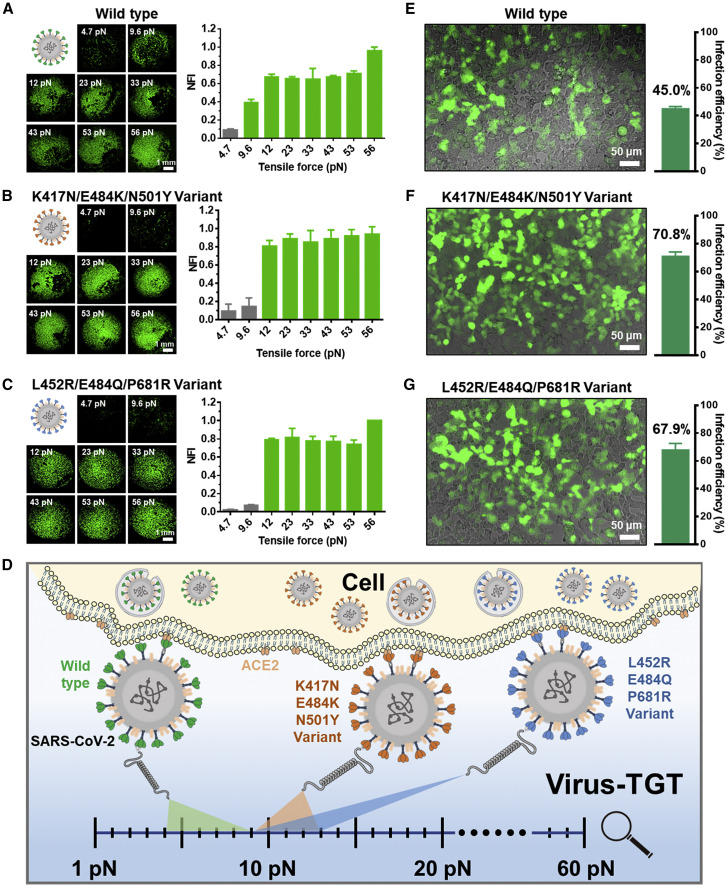
SARS-CoV-2 is a biosafety level 3 pathogen. To facilitate studies under routine laboratory conditions, we utilized a SARS-CoV-2 pseudovirus that had previously been shown to faithfully recapitulate features of virus infection.30 Using Virus-TGT, we investigated the mechanical forces between SARS-CoV-2 pseudoviruses and host cells. We obtained a set of force rulers consisting of a series of TGTs with defined tensile forces (4.7, 9.6, 12, 23, 33, 43, and 56 pN) by DNA hybridization (Figure S1; Tables S1 and S2). After pseudoviruses were incubated with binding module-functionalized TGT and subsequent wash, we added host cells for another incubation. The mechanical force-related cell binding events were evaluated after washing again (Figure S7). Since it is difficult to accurately control the ratio of virus to binding modules, and the number of binding modules bound to each virus is similar, tensile force of one TGT was used to represent the mechanical force in our work and in relevant literature.17 , 21 , 26 , 27 As shown in Figure 4A, rare cell adhesion was observed at a tensile force of 4.7 pN, while cells were obviously adhered with a force ≥9.6 pN, demonstrating that the mechanical force between wild-type SARS-CoV-2 pseudovirus and host cells ranged from 4.7 to 9.6 pN.
Figure 4.
Mechanical force and infectivity comparison between SARS-CoV-2 and host cells
(A–C) Images of ACE2-expressing HEK293T cells captured on a circular area modified with (A) wild-type, (B) K417N/E484K/N501Y variant, and (C) L452R/E484Q/P681R variant SARS-CoV-2 and corresponding normalized fluorescence intensity (NFI) values versus a series of tensile forces. As the intensity of the tensile force of 56 pN for each virus was regarded as 1, normalized fluorescence intensity values were obtained. Scale bar, 1 mm.
(D) Illustration of evaluation of the mechanical forces between SARS-CoV-2 and host cells by Virus-TGT.
(E–G) Comparison of infection efficiency of (E) wild-type, (F) K417N/E484K/N501Y variant, and (G) L452R/E484Q/P681R variant under the same particle concentration. Scale bar, 50 μm. The error bars in each graph represent a standard deviation of three replicated experiments.
Since constantly emerging new SARS-CoV-2 variants circulate worldwide, study of the mechanical force difference between varieties of variants and host cells is critically important to combat new variants. Compared with the mechanical force between wild-type and host cells, both the K417N/E484K/N501Y variant (three critical mutations of Beta variant, Figure 4B) and the L452R/E484Q/P681R variant (three critical mutations of Kappa variant, Figure 4C) displayed higher mechanical forces with the range of 9.6–12 pN. Compared with the wild-type, the relatively higher mechanical force between the K417N/E484K/N501Y variant and the cell measured by Virus-TGT was similar to that between RBD and cell using SMFS-based strategy (<10 pN), revealing the promising application potential of Virus-TGT.11 The mechanical force gap between wild-type and variants may be mainly attributed to the alterations of amino acids and the number of S proteins per viral particle, indicating the potential differences of new variants in infectivity and transmissibility compared with the wild-type. Overall, we initially established a mechanical force analytical reference platform to study the overall viral infection mechanical forces (Figure 4D).
Infectivity comparison of SARS-CoV-2 and its variants
To explore the relationship between mechanical forces and viral infection, we determined the infectivity of wild-type, K417N/E484K/N501Y variant, and L452R/E484Q/P681R variant pseudovirus against ACE2-expressing HEK293T cells under a consistent particle concentration. Wild-type was determined as lower infectivity with an efficiency of 45.0% (Figure 4E), while the K417N/E484K/N501Y and L452R/E484Q/P681R variants displayed increased infection efficiency at 1.57-fold (70.8%, Figure 4F) and 1.51-fold (67.9%, Figure 4G), respectively. With Virus-TGT, we discovered that these two variants with significantly enhanced infectivity both displayed higher mechanical forces when interacted host cells, suggesting the possible positive correlation between mechanical force and infection efficiency. However, previous studies have found that some mutant strains with the increased infectivity have similar binding affinities to ACE2 as the wild-type.4 , 8 , 31 Taken together, the mechanical force of SARS-CoV-2 and host cell better correlates with viral infectivity, in contrast to the S protein-ACE2 binding affinity, suggesting the essential role of mechanical force in regulating SARS-CoV-2 infection and may be a better predictor for viral infectivity.
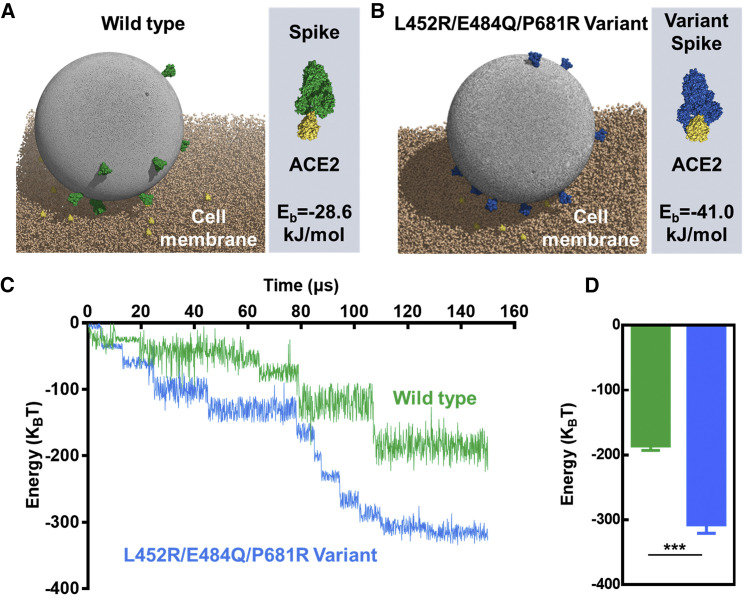
Interaction between SARS-CoV-2 and host cell in CG simulations
To further demonstrate the above-mentioned mechanical force difference, we conducted dissipative particle dynamics (DPD) simulations based on the coarse-grained (CG) model to probe the interaction energy between SARS-CoV-2 and an ACE2-expressed cell (Figure S8). Due to the specific interaction between S protein and ACE2, the wild-type (Figure 5A, Eb = −28.6 kJ/mol) and the L452R/E484Q/P681R variant (three critical mutations in S protein of Kappa variant, Figure 5B, Eb = −41.0 kJ/mol) could bind to the cell membrane in the final simulation. However, the enhancement of the interaction energy of wild-type and ACE2-embedded cell membrane occurred more slowly than that of the L452R/E484Q/P681R variant SARS-CoV-2 as time evolved (Figure 5C). Compared with the wild-type (−186 kBT), the interaction energy of the L452R/E484Q/P681R variant and the ACE2-embedded cell membrane showed 1.66-fold enhancement (−308 kBT) at the end of the simulation (Figure 5D). Aside from the conformational transition of S protein caused by mutated amino acids, this difference in interaction energy can likely be attributed to other factors, including membrane bending.32 These interaction energy gaps further verified the relative differences of mechanical forces of different typed SARS-CoV-2 and ACE2-expressed host cells.
Figure 5.
Interaction between SARS-CoV-2 and host cell in coarse-grained (CG) simulations
(A and B) Representative final snapshots of (A) wild-type and (B) L452R/E484Q/P681R variant SARS-CoV-2 interacting with an ACE2-embedded cell membrane. Illustration of the interaction between different S proteins and ACE2 on the right side of each figure. Eb, binding energy.
(C) Energy of wild-type or L452R/E484Q/P681R variant SARS-CoV-2 interacting with ACE2-embedded cell membrane as function of time.
(D) Final energy of receptor-ligand interaction of wild-type (green bar) or L452R/E484Q/P681R variant (blue bar) SARS-CoV-2 and ACE2-embedded cell membrane (n = 10). ∗∗∗p < 0.001 (Student’s t test).
In conclusion, leveraging a multivalence-controlled aptamer and a range of tethers with tunable tension tolerances, we have developed a platform named Virus-TGT to study the mechanical force between SARS-CoV-2 and its host cell. With multivalent synergy and controllable assembly of superior aptamers, the virus can be firmly grasped by the binding module, thus realizing the force transfer from cell-virus to the TGT module, enabling study of viral infection mechanical force at the whole-virus level. Using Virus-TGT, we measured the holistic mechanical force between SARS-CoV-2 and host cell, rather than remaining at protein-level analysis as reported previously6 , 9, 10, 11, 12, 13 , 23 (Table S3). This overall mechanical force measurement avoids the limitation of only considering the S protein-ACE2 affinity, and takes into account the influence of other factors comprehensively. Accordingly, the developed strategy provides direct evidence that the mutated strain has a stronger biomechanical strength on the host cell rather than that of wild-type SARS-CoV-2. Using the Virus-TGT and infectivity characterization, the positive correlation between viral mechanical force and infection efficiency was shown for the first time. Therefore, the developed strategy holds great potential for deeper understanding of biomechanical force during viral infection, which is conducive to in-depth exploration of the infection mechanism and the impact of viral mutants. In addition, the developed strategy can be used to evaluate the efficacy of neutralizing antibodies and other neutralizing agents. Moreover, leveraging delicate DNA nanostructures and superior aptamers,33, 34, 35, 36, 37, 38, 39, 40, 41, 42 multifaceted efforts are expected to unravel the infection mechanisms of more disease-causing pathogens.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yanling Song (ylsong@xmu.edu.cn).
Materials availability
All materials generated in this study are available from the lead contact without restriction.
Materials and reagents
His-tagged RBD-conjugated Ni beads were obtained from GE Healthcare (cat. 17-5268-01). SARS-CoV-2 spike RBD-His recombinant protein (cat. 40592-V08B), SARS-CoV-2 spike RBD (K417N/E484K/N501Y)-His recombinant protein (cat. 40592-V08H85), SARS-CoV-2 spike Delta RBD (L452R/T478K)-His recombinant protein (cat. 40592-V08H90), and biotinylated ACE2 (cat. 10108-H08H-B) were obtained from Sino Biological (China). ACE2-expressing HEK293T cells, pseudovirus-SARS-CoV-2 (cat. FNV215), pseudovirus-SARS-CoV-2 (K417N/E484K/N501Y) (cat. FNV3327), and pseudovirus-SARS-CoV-2 (L452R/E484Q/P681R) (cat. FNV3609) were obtained from Fubio Biological Technology, Shanghai, China. Streptavidin (SA) and (3-mercaptopropyl) methyldimethoxysilane (MPTS) were obtained from Sigma-Aldrich. N-γ-Maleimidobutyryl-oxysuccinimide ester (GMBS) was purchased from Thermo Fisher Scientific (USA). DNA marker (20–500 bp, cat. 3420A) was purchased from Takara. DNA marker (100–2,000 bp) and other common chemical reagents were purchased from Sangon Biotechnology, Shanghai, China. Binding buffer was 0.55 mM MgCl2 in PBS (136.8 mM NaCl, 10.1 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4 [pH 7.4]).
Preparation and characterization of the binding module
All DNA sequences shown in Table S1 were synthesized and purified by HPLC from Sangon Biotechnology. Each DNA complex was obtained by mixing all component strands with corresponding ratios in TM buffer (20 mM Tris, 50 mM MgCl2 [pH 8.0]) (Table S2). In a thermal cycler, the mixture solution was annealed by incubating at 95°C for 10 min, then quickly cooled to 4°C for 20 min. Formation of all DNA nanostructures was characterized by 2% AGE in 1× TBE (10 mM magnesium acetate) at 100 V for 1 h. The yield of DNA nanostructures was analyzed by Image J software. To obtain a series of the complexes of binding module-TGT module, the binding module was separately incubated with the substrate-immobilized strands (named 4.7, 9.6, 12, 23, 33, 43, 53, and 56 pN; Tables S1 and S2) at a final concentration of 1 μM at 37°C for 1 h.
Cryoelectron microscopy characterization
Four microliters of the binding module (400 nM) was placed on the glow-discharged copper grid (cat. 01895-F, Lacey carbon, 300 mesh, copper, Ted Pella, USA) and blotted for 3 s (blot force: −2; wait time: 1 s; blot total: 1; drain time: 0 s; temperature: 22°C; humidity: 100%). Subsequently, sample-loaded grids were rapidly frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). All grids were observed on a cryoelectron microscope (Talos F200C G2, Thermo Fisher Scientific) equipped with 4k × 4k Ceta CCD (charge-coupled device) camera. The samples were imaged at 200 kV with an absolute magnification of 57,000×.
TIRFM characterization
For fluorescence co-localization characterization of DNA nanostructures, 10 μL of 40 nM samples were applied onto the pre-cleaned cover glass slide and imaged by TIRFM at 647 and 488 nm excitation. Molecular fluorescence co-localization and fluorescence intensity were analyzed by Image J software.
Cell lines and culture
ACE2-expressing HEK293T cells were cultured in high glucose DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin solution (Gibco). All cell lines used in this study were cultured at 37°C, 5% CO2.
Flow cytometry analysis
RBD-beads were obtained by the co-incubation of His-tagged RBD and Ni-beads in 1× PBS (0.1 mol/L K2HPO4, 0.1 mol/L KH2PO4 [pH 8.0]) for 2 h. To obtain the binding affinities of SARS-CoV-2 spike RBD (wild-type, K417N/E484K/N501Y, and L452R/T478K) toward ligand (TriApTDF), RBD-beads were incubated with different concentrations of Alexa 488-labeled ligands in 200 μL binding buffer at room temperature for 30 min. After washing twice and suspension in 200 μL binding buffer, the mean fluorescence intensity of beads was analyzed using flow cytometry (FACSVerse, BD). To evaluate the Kd values, the mean fluorescence intensity (Y) and the concentration of ligands (X) were fitted with the following equation Y = Bmax ×/(Kd + X) using SigmaPlot software.
SA slide chip modification
As shown in Figure S4, glass slides were ultrasonically cleaned by 10% glass detergent. Subsequently, glass slides were ultrasonically cleaned with ultrapure water before drying. As previously reported,43 glass slides were incubated with MPTS (5% in ethanol) for 2 h after plasma treatment. Then, glass slides were rinsed with ethanol before drying at 90°C for 0.5 h. Subsequently, fresh GMBS (0.5 mg/mL in ethanol) was applied for 30 min. To prepare the SA slide chip, all glass slides were incubated with SA (25 μg/mL in PBS) immediately for 2 h or overnight at 4°C after washing once with ultrapure water and PBS. All modification processes were carried out at room temperature except as noted in additional instructions.
Virus-TGT measurement
As shown in Figure S7, first, the SA slide chip was incubated with 3% BSA for 30 min. Second, the SA-Chip was washed with ultrapure water before blowing dry with N2. Next, 1.5 μL/spot of 1 μM solutions of a series of the complexes of binding module-TGT module was added to the surface of the chip for 45 min, respectively. After washing once with PBS, approximately 106 particles/mL SARS-CoV-2 pseudovirus was plated for 1 h. Calcein-dyed ACE2-transfected 293T cells were added to the pre-prepared functional interface for 3 h after washing again with PBS. Next, the images of spots were acquired with a fluorescence microscope (Nikon Ts2R-FL, Japan) after washing again. Finally, the fluorescence intensities of all spots were evaluated by Image J software.
For the optimization of SARS-CoV-2 pseudovirus particle concentration (Figure S5), 1.5 μL/spot 100 ng/mL biotinylated ACE2 was added to the surface of the chip for 1 h. Next, diluted groups (105, 106, 107, and 108 particles/mL) were spotted for 1 h. Subsequent procedures were carried out as mentioned above with one wash using PBS between each step. All fabrication processes were conducted at room temperature.
For inducing DNA capture or rupture by the interaction of pseudovirus and ACE2-expressing HEK293T cells, Cy5-labeled binding module was chosen to verify the stable formation of binding module and virus. Next, fluorescence intensities of spots were evaluated using a fluorescence microscope (Nikon Ts2R-FL, Japan).
SARS-CoV-2 pseudovirus infection assay
About 104 ACE2-expressing HEK293T cells were seeded into a 384-well plate for culturing for at least 12 h. Then, different types of pseudoviruses with the same particle concentration were separately added to infect ACE2-expressing HEK293T cells. After 8 h co-incubation with pseudovirus, the culture medium was replaced with fresh medium, and cells were cultured for an additional 48 h. Finally, after imaging with a fluorescence microscope (Nikon Ts2R-FL), the infection efficiencies were evaluated by counting the infectious cells.
Construction of CG models in the simulations
To probe the interaction between SARS-CoV-2 and the host cell, we first constructed several CG models based on different components. All components of 12 spike proteins and 20 ACE2s are built base on CG Martini force field (v.2.1). For modeling the cell membrane, two different lipids, DLPG and DOPC, were used in our simulation, the CG lipid models were described in other articles.44, 45, 46 The head group of DPPC was coarse grained to Qo, Qa, and Na beads, while the DPPG head group was CG to P4, Qa, and Na beads, with one negatively charged. Considering the specific interaction between membrane protein ACE2 and SARS-CoV-2 spike protein, we used structure-based elastic network models for subsequent analysis. Then, we introduced a patterned cylinder representing ACE2 based on the protein crystal structure (PDB: 1R4L). The cylinder consisted of a hydrophobic middle part and two specific hydrophilic beads at the ends. In the simulation, the surface density of ACE2 on the cell membrane was approximately 0.01 nm−2.
For modeling different typed SARS-CoV-2 viruses, we constructed a sphere of lipid molecules with particle diameter of 90 nm and randomly assigned 12 spike proteins on the sphere.47 For comparison of the differences of wild-type and variant SARS-CoV-2 virus interaction with host cells, we further constructed the CG model of the spike protein based on the protein crystal structure of the wild-type (PDB: 7DDN) and the L452R/E484Q/P681R variant (PDB: 7VXE).
Dissipative particle dynamics simulations
In this study, we carried out a CG simulation technique, named dissipative particle dynamics (DPD). Utilizing hydrodynamic interaction, the elementary units depended on Newton’s equations of motion in the DPD simulations. To distinguish the hydrophilic and hydrophobic property of each bead, the repulsive parameter aii in conservative force as 25 kBT/rc was assigned for any two beads of the same type, while aij = 100 kBT/rc was assigned for any two beads of different types.
As previous work reported,48 we introduced coulomb force into our DPD simulations to include electrostatic interaction between charged beads. To simulate the receptor-ligand-specific interaction, the Lennard-Jones potential was applied with a strength of 15.0 kBT. Furthermore, to ensure the integration of proteins on each membrane, the harmonic spring interaction s = −ks(1 − ri, i+1/l0) i, i+1 (ks is the spring constant and l0 is the equilibrium length and the parameters ks = 128 kBT, l0 = 0.5rc) was used between neighboring beads in a single molecule. In addition, a three-body bond angle potential Ua = ka (1-cos (−)) was applied to depict the rigidity of lipid tails (ka = 10.0 kBT and = 180°) and spike trimer protein (ka = 2.0 kBT, = 180°), where is the angle formed by three adjacent beads in the same molecule and is the equilibrium value of the angle.
The simulations employed the velocity-Verlet integration algorithm and the integration time step Δt = 0.02τ. Moreover, the cutoff radius rc, bead mass m, and energy kBT were chosen as the simulation units. All simulations were conducted in the NVT ensembles. We also adopted the periodic boundary conditions in three directions. All simulations in this study were performed by the Martini model in gromacs.
Acknowledgments
We give thanks for the National Natural Science Foundation of China grants (22022409, 21735004, and 21874089) and the Program for Changjiang Scholars and Innovative Research Team in University (grant IRT13036) for their financial support.
Author contributions
J.Z. designed and performed the main experiments and wrote the manuscript. Y.H. performed partial cell corresponding assays and data analysis. M.S., S.W., and T.S. contributed to pseudovirus neutralization analysis and data analysis. C.Y. provided important insights and revised the manuscript. Y.S. designed the study, performed the analysis, and co-wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: September 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrp.2022.101048.
Supplemental information
Data and code availability
This study did not generate any datasets. Any requests for data will be fulfilled by the lead contact upon reasonable request.
References
- 1.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Xu C., Wang Y., Hong Q., Zhang C., Li Z., Xu S., Zuo Q., Liu C., Huang Z., Cong Y. Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM. Nat. Commun. 2021;12:7345. doi: 10.1038/s41467-021-27350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu W., Zhang Y., Fei P., Zhang T., Yao D., Gao Y., Liu J., Chen H., Lu Q., Mudianto T., et al. Mechanical activation of spike fosters SARS-CoV-2 viral infection. Cell Res. 2021;31:1047–1060. doi: 10.1038/s41422-021-00558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J., Petitjean S.J.L., Koehler M., Zhang Q., Dumitru A.C., Chen W., Derclaye S., Vincent S.P., Soumillion P., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao W., Dong C., Kim S., Hou D., Tai W., Du L., Im W., Zhang X.F. Biomechanical characterization of SARS-CoV-2 spike RBD and human ACE2 protein-protein interaction. Biophys. J. 2021;120:1011–1019. doi: 10.1016/j.bpj.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian F., Tong B., Sun L., Shi S., Zheng B., Wang Z., Dong X., Zheng P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife. 2021;10:e69091. doi: 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koehler M., Ray A., Moreira R.A., Juniku B., Poma A.B., Alsteens D. Molecular insights into receptor binding energetics and neutralization of SARS-CoV-2 variants. Nat. Commun. 2021;12:6977. doi: 10.1038/s41467-021-27325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer M.S., Gruber S., Hausch A., Gomes P.S.F.C., Milles L.F., Nicolaus T., Schendel L.C., Navajas P.L., Procko E., Lietha D., et al. A tethered ligand assay to probe SARS-CoV-2:ACE2 interactions. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2114397119. e2114397119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B., O'Brien C., Mudiyanselage A.P.K.K.K., Li N., Bagheri Y., Wu R., Sun Y., You M. Visualizing intercellular tensile forces by DNA-based membrane molecular probes. J. Am. Chem. Soc. 2017;139:18182–18185. doi: 10.1021/jacs.7b11176. [DOI] [PubMed] [Google Scholar]
- 15.Joonaki E., Hassanpouryouzband A., Heldt C.L., Areo O. Surface chemistry can unlock drivers of surface stability of SARS-CoV-2 in a variety of environmental conditions. Chem. 2020;6:2135–2146. doi: 10.1016/j.chempr.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alsteens D., Gaub H.E., Newton R., Pfreundschuh M., Gerber C., Müller D.J. Atomic force microscopy-based characterization and design of biointerfaces. Nat. Rev. Mater. 2017;2:17008. doi: 10.1038/natrevmats.2017.8. [DOI] [Google Scholar]
- 17.Liu Y., Blanchfield L., Ma V.P.Y., Andargachew R., Galior K., Liu Z., Evavold B., Salaita K. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc. Natl. Acad. Sci. USA. 2016;113:5610–5615. doi: 10.1073/pnas.1600163113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Qiu Y., Blanchard A.T., Chang Y., Brockman J.M., Ma V.P.Y., Lam W.A., Salaita K. Platelet integrins exhibit anisotropic mechanosensing and harness piconewton forces to mediate platelet aggregation. Proc. Natl. Acad. Sci. USA. 2018;115:325–330. doi: 10.1073/pnas.1710828115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glazier R., Brockman J.M., Bartle E., Mattheyses A.L., Destaing O., Salaita K. DNA mechanotechnology reveals that integrin receptors apply pN forces in podosomes on fluid substrates. Nat. Commun. 2019;10:4507. doi: 10.1038/s41467-019-12304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma V.P.Y., Salaita K. DNA nanotechnology as an emerging tool to study mechanotransduction in living systems. Small. 2019;15:e1900961. doi: 10.1002/smll.201900961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Zhang C., Hu Y., Liu P., Sun F., Chen W., Zhang X., Ma J., Wang W., Wang L., et al. A reversible shearing DNA probe for visualizing mechanically strong receptors in living cells. Nat. Cell Biol. 2021;23:642–651. doi: 10.1038/s41556-021-00691-0. [DOI] [PubMed] [Google Scholar]
- 22.Duan Y., Glazier R., Bazrafshan A., Hu Y., Rashid S.A., Petrich B.G., Ke Y., Salaita K. Mechanically triggered hybridization chain reaction. Angew Chem. Int. Ed. Engl. 2021;60:19974–19981. doi: 10.1002/anie.202107660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petitjean S.J.L., Chen W., Koehler M., Jimmidi R., Yang J., Mohammed D., Juniku B., Stanifer M.L., Boulant S., Vincent S.P., Alsteens D. Multivalent 9-O-Acetylated-sialic acid glycoclusters as potent inhibitors for SARS-CoV-2 infection. Nat. Commun. 2022;13:2564. doi: 10.1038/s41467-022-30313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun M., Liu S., Wei X., Wan S., Huang M., Song T., Lu Y., Weng X., Lin Z., Chen H., et al. Aptamer blocking strategy inhibits SARS-CoV-2 virus infection. Angew Chem. Int. Ed. Engl. 2021;60:10266–10272. doi: 10.1002/anie.202100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M., Ding H., Lin M., Yin F., Song L., Mao X., Li F., Ge Z., Wang L., Zuo X., et al. DNA framework-programmed cell capture via topology-engineered receptor-ligand interactions. J. Am. Chem. Soc. 2019;141:18910–18915. doi: 10.1021/jacs.9b11015. [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin F., Li M., Mao X., Li F., Xiang X., Li Q., Wang L., Zuo X., Fan C., Zhu Y. DNA framework-based topological cell sorters. Angew Chem. Int. Ed. Engl. 2020;59:10406–10410. doi: 10.1002/anie.202002020. [DOI] [PubMed] [Google Scholar]
- 28.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Xiao T., Cai Y., Lavine C.L., Peng H., Zhu H., Anand K., Tong P., Gautam A., Mayer M.L., et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021;374:1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leong T., Voleti C., Peng Z. Coarse-grained modeling of coronavirus spike proteins and ACE2 receptors. Front. Physiol. 2021;9:680983. doi: 10.3389/fphy.2021.680983. [DOI] [Google Scholar]
- 33.Dey S., Fan C., Gothelf K.V., Li J., Lin C., Liu L., Liu N., Nijenhuis M.A.D., Saccà B., Simmel F.C., et al. DNA origami. Nat. Rev. Methods Primers. 2021;1:13. doi: 10.1038/s43586-020-00009-8. [DOI] [Google Scholar]
- 34.Kwon P.S., Ren S., Kwon S.J., Kizer M.E., Kuo L., Xie M., Zhu D., Zhou F., Zhang F., Kim D., et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat. Chem. 2020;12:26–35. doi: 10.1038/s41557-019-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw A., Hoffecker I.T., Smyrlaki I., Rosa J., Grevys A., Bratlie D., Sandlie I., Michaelsen T.E., Andersen J.T., Högberg B. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat. Nanotechnol. 2019;14:184–190. doi: 10.1038/s41565-018-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonchhe S., Pandey S., Beneze C., Emura T., Sugiyama H., Endo M., Mao H. Dissection of nanoconfinement and proximity effects on the binding events in DNA origami nanocavity. Nucleic Acids Res. 2022;50:697–703. doi: 10.1093/nar/gkab1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kacherovsky N., Yang L.F., Dang H.V., Cheng E.L., Cardle I.I., Walls A.C., McCallum M., Sellers D.L., DiMaio F., Salipante S.J., et al. Discovery and characterization of spike N-terminal domain-binding aptamers for rapid SARS-CoV-2 detection. Angew Chem. Int. Ed. Engl. 2021;60:21211–21215. doi: 10.1002/anie.202107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., Zhang Z., Gu J., Stacey H.D., Ang J.C., Capretta A., Filipe C.D.M., Mossman K.L., Balion C., Salena B.J., et al. Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res. 2021;49:7267–7279. doi: 10.1093/nar/gkab574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung Y.W., Röthlisberger P., Mechaly A.E., Weber P., Levi-Acobas F., Lo Y., Wong A.W.C., Kinghorn A.B., Haouz A., Savage G.P., et al. Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker. Proc. Natl. Acad. Sci. USA. 2020;117:16790–16798. doi: 10.1073/pnas.2003267117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peinetti A.S., Lake R.J., Cong W., Cooper L., Wu Y., Ma Y., Pawel G.T., Toimil-Molares M.E., Trautmann C., Rong L., et al. Direct detection of human adenovirus or SARS-CoV-2 with ability to inform infectivity using DNA aptamer-nanopore sensors. Sci. Adv. 2021;7:eabh2848. doi: 10.1126/sciadv.abh2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bujold K.E., Lacroix A., Sleiman H.F. DNA nanostructures at the interface with biology. Chem. 2018;4:495–521. doi: 10.1016/j.chempr.2018.02.005. [DOI] [Google Scholar]
- 42.Dutta P.K., Zhang Y., Blanchard A.T., Ge C., Rushdi M., Weiss K., Zhu C., Ke Y., Salaita K. Programmable multivalent DNA-origami tension probes for reporting cellular traction forces. Nano Lett. 2018;18:4803–4811. doi: 10.1021/acs.nanolett.8b01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J., Lin B., Wu L., Huang M., Li X., Zhang H., Song J., Wang W., Zhao G., Song Y., Yang C. DNA nanolithography enables a highly ordered recognition interface in a microfluidic chip for the efficient capture and release of circulating tumor cells. Angew Chem. Int. Ed. Engl. 2020;59:14115–14119. doi: 10.1002/anie.202005974. [DOI] [PubMed] [Google Scholar]
- 44.He X., Li L., Yang Y., Dong Z., Wang L., Qu Z., Xu F. Tailoring patchy nanoparticle design to modulate serum albumin adsorption and membrane interaction. Soft Matter. 2021;17:2071–2080. doi: 10.1039/d0sm01889a. [DOI] [PubMed] [Google Scholar]
- 45.He X., Gu Z., Wang L., Qu Z., Xu F. Coarse-grained molecular dynamics simulation of dendrimer transmembrane transport with temperature-dependent membrane phase states. Int. J. Heat Mass Tran. 2020;155:119797. doi: 10.1016/j.ijheatmasstransfer.2020.119797. [DOI] [Google Scholar]
- 46.Zhang Q., Tian F., Wang F., Guo Z., Cai M., Xu H., Wang H., Yang G., Shi X., Shan Y., Cui Z. Entry dynamics of single ebola virus revealed by force tracing. ACS Nano. 2020;14:7046–7054. doi: 10.1021/acsnano.0c01739. [DOI] [PubMed] [Google Scholar]
- 47.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu L., Ding H., Qu X., Shi X., Yang J., Huang M., Zhang J., Zhang H., Song J., Zhu L., et al. Fluidic multivalent membrane nanointerface enables synergetic enrichment of circulating tumor cells with high efficiency and viability. J. Am. Chem. Soc. 2020;142:4800–4806. doi: 10.1021/jacs.9b13782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any datasets. Any requests for data will be fulfilled by the lead contact upon reasonable request.