Abstract
COVID-19 (Coronavirus Disease 2019), a life-threatening viral infection, is caused by a highly pathogenic virus named SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2). Currently, no treatment is available for COVID-19; hence there is an urgent need to find effective therapeutic drugs to combat COVID-19 pandemic. Considering the fact that the world is facing a major issue of antimicrobial drug resistance, naturally occurring compounds have the potential to achieve this goal. Antimicrobial peptides (AMPs) are naturally occurring antimicrobial agents which are effective against a wide variety of microbial infections. Therefore, the use of AMPs is an attractive therapeutic strategy for the treatment of SARS-CoV-2 infection. This review sheds light on the potential of antimicrobial peptides as antiviral agents followed by a comprehensive description of effective antiviral peptides derived from various natural sources found to be effective against SARS-CoV and other respiratory viruses. It also highlights the mechanisms of action of antiviral peptides with special emphasis on their effectiveness against SARS-CoV-2 infection.
Keywords: Antimicrobial resistance, Antimicrobial peptide, Severe acute respiratory syndrome coronavirus, Antiviral peptides
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a highly pathogenic virus and has led to the emergence of Coronavirus Disease pandemic in 2019 (COVID-19). According to World Health Organization, there have been over 500 million reported cases of COVID-19 and 6 million deaths worldwide as of June 2022 (Anon, 2022). Vaccines have been developed to prevent the spread of COVID-19; however, no treatment for SARS-CoV-2 is available yet. A few already available antimicrobial drugs such as hydroxychloroquine, azithromycin, chloroquine, remdesivir (RDV), lopinavir, ritonavir, oseltamivir, umifenovir, dexamethasone and favipiravir are under investigation for the treatment of COVID-19. Even though drugs like dexamethasone and RDV have proven to be effective for the treatment of COVID-19, their use is still considered to be controversial (Ma et al., 2020). Moreover, antimicrobial drug resistance (ADR) is an emerging threat to the world due to a variety of technological, socio-economic, microbial, and environmental factors. Unavailability of affordable healthcare, microbial adaptation to drugs, growing populations of susceptible hosts, and unethical uses of antibiotics in cure as well as non-therapeutic regimens are few other factors responsible for ADR (Fletcher, 2015, Van Hoek et al., 2011). Microbes are developing resistance to existing armature of antimicrobial drugs through several mechanisms such as active efflux pump, decreased/blocked uptake of antibiotics, inactivation of enzymes and modification of targets such as DNA, RNA, and ribosomes (Kaur et al., 2022, Peterson and Kaur, 2018). It is estimated that approximately 10 million people will die worldwide by 2050 if antimicrobial drug resistance is not tackled (Dadgostar, 2019). Although ADR has been reported worldwide, developing countries are hit hard by this crisis due to the lack of availability of second and third-line treatments (Cox et al., 2017). This has led to considerable interest in the identification of natural compounds against multidrug-resistant microorganisms. The United Nations (UN) General Assembly pledged to tackle the issue of antimicrobial resistance in September 2016 by strengthening the regulation of antimicrobials and fostering new technologies and alternatives. In 2018, Singapore organized a meeting to discuss the challenges of antimicrobial resistance and the innovative solutions that could be employed (Yam et al., 2019). Naturally formulated compounds have become a popular choice as antimicrobial agents against multidrug-resistant microorganisms. While antibiotics, sometimes possess side effects such as immunosuppression, hypersensitivity and allergic reactions on the host, these antimicrobials of natural origin treat infections by overcoming the disadvantages of traditional antibiotics (Anand et al., 2019, Khameneh et al., 2019, Ncube et al., 2008). The use of antimicrobial peptides (AMPs) provides an attractive solution to combat the problem of antimicrobial resistance. These peptides are effective, broad-spectrum antimicrobials that establish themselves as new therapeutic agents, and hold potential to kill gram-negative and gram-positive bacteria, fungi, enclosed viruses, and even mutated or malignant cells (Boparai and Sharma, 2020, Di Somma et al., 2020). Mainly, this review will narrate the overview about AMPs, their mechanism of action, and their role as antiviral agents. Moreover, it will shed light on the possible role of AMPs exhibiting antiviral activity against SARS-CoV-2.
2. Antimicrobial peptides as antiviral agents
AMPs are small peptides consisting of 8–100 amino acids, amphiphilic in nature, and display broad-spectrum antimicrobial activity. Natural AMPs are found both in prokaryotes (e.g., bacteria) and eukaryotes (e.g., protozoans, fungi, plants, insects, and animals). The first AMP, gramicidin, was discovered in 1939 from a soil Bacillus strain, which provided protection against Pneumococcal infection in mice (Dubos, 1939). The first AMP, phagocytin, known to show bactericidal activity was isolated from animals (rabbit leukocytes) in 1956 (Hirsch, 1956). In the following years, AMPs from human leukocytes, lactoferrin from cow’s milk, and various others were also identified (Conesa et al., 2008, Weissmann et al., 1971). Recent evidence reveals that several AMPs of humans, insects and plants origin have antiviral activity against a broad range of viruses ( Table 1).
Table 1.
Antimicrobial peptides effective against viral infections.
| AMP | Target | Source | References |
|---|---|---|---|
| Gloverin | Autographa californica M nucleopolyhedrovirus (AcMNPV) | Trichoplusia ni larvae | Moreno-Habel et al. (2012) |
| α-defensin-5 (HD-5) | Human Immunodeficiency Virus-1 (HIV-1), Human papillomavirus 16 (HPV) | Humans | Furci et al. (2012); Wiens and Smith (2017) |
| Lactoferrin | Influenza virus, Hepatitis C virus (HCV), Dengue Virus (DENV) | Humans | Ammendolia et al. (2012);El-Fakharany et al. (2013);Chen et al. (2017) |
| Elafin and Trappin-2 | HIV-1 | Humans | Drannik et al. (2013) |
| Griffithsin (GRFT) | Hepatitis C Virus (HCV), Japanese encephalitis virus (JEV), Middle East respiratory syndrome coronavirus (MERS-CoV) | Griffithsia sp (red alga) | Takebe et al. (2013);Ishag et al. (2016);Millet et al. (2016);Derby et al. (2018) |
| Cycloviolacin Y5 and Cycloviolacin VY1 | Influenza A virus (IAV) | Viola yedoensis plant | Liu et al. (2014) |
| Hepcidin | IAV, HIV-1, HCV, and Hepatitis B Virus (HBV) | Humans | Armitage et al. (2014);Rodriguez et al. (2014) |
| Cathelicidin (LL-37) | IAV, HCV, Respiratory syncytial virus (RSV), DENV type 2, Venezuelan equine encephalitis virus (VEEV) | Humans | Tripathi et al. (2014);Harcourt et al. (2016);Matsumura et al. (2016);Alagarasu et al. (2017);Ahmed et al. (2019); |
| Lectin (NICTABA) | IAV, IBV, DENV type 2, Herpes Simplex Virus (HSV) types 1 and 2, HIV-1/2 | Nicotiana tabacum plant | Gordts et al. (2015) |
| Alstotides | Infectious bronchitis virus (IBV), DENV | Alstonia scholaris plant | Nguyen et al. (2015) |
| A. elatior lectin (AEL) | Vesicular Stomatitis Virus (VSV), Coxsackie Virus B4, and RSV | Aspidistra elatior plant | Xu et al. (2015) |
| Cathelicidins (Protegrin-1, and SMAP-29) | Human rhinoviruses (HRVs) | Humans | Sousa et al. (2017) |
| Cathelicidins (GF-17 and BMAP-18) | Zika virus (ZIKV) | Humans | He et al. (2018) |
| C-lysozyme | Nucleopolyhedrovirus (NPV) | Bombyx mori | Chen et al. (2018) |
| Lectin (TCLL) | Chikungunya virus (CHIKV) and Sindbis virus (SINV) | Tamarindus indica plant | Kaur et al. (2019) |
| Dermaseptins | HSV-1, HSV-2, HIV-1, and rabies virus | Agalychnis and Phyllomedusa family | Bartels et al. (2019) |
| Brevinin (1BYa and 1BYc) | Ebola virus, HIV-1 and HSV-1 | Rana boylii | Zohrab et al. (2019) |
| Temporin-SHa (SHa) and its synthetic analog [K3]SHa. | HSV-1 | North African ranid frog Pelophylax saharicus | Roy et al. (2019) |
| Brevinin-2GHk, | ZIKV | Fejervarya limnocharis | Xiong et al. (2021) |
| Fejerlectin | HIV-1 | Fejervarya limnocharis | Xiong et al. (2021) |
| AR-23 | HSV-1, Measles morbillivirus (MeV), Human parainfluenza virus (HPIV-2), HCoV-229E, and severe acute respiratory syndrome coronavirus (SARS-CoV-2) | Rana tagoi | Chianese et al. (2022) |
Certain evolutionary mechanisms in viruses lead to antiviral drug resistance and is a major cause of concern upon drug treatment. These mechanisms involve but not limited to high mutation rate due to frequent replication and proofreading errors caused by viral RNA polymerase as in case of Hepatitis C virus (HCV), error-prone reverse transcription leading to nucleotide substitution (e.g., Human Immunodeficiency Virus (HIV)), lack of proofreading during reverse transcription (e.g., Hepatitis B Virus (HBV)). Since the problem of antiviral drug resistance is a major concern in the development of therapeutic drugs, AMPs effective against viruses can be taken into consideration as antiviral agents (Kausar et al., 2021).
2.1. Human-derived antiviral peptides
During evolution, the tactics of mammals have slowly evolved for combating harmful microbes. AMPs are derived from various tissues in mammals, as exemplified by neutrophil granules, the intestinal tract, and secretions from mucosal membranes (Befus et al., 1999). In mammals, AMPs mainly belong to the class of cathelicidins and defensins. Cathelicidins are the AMPs derived from a variety of vertebrates, such as fish, birds, goats, pigs, rabbits, monkeys, horses, and human beings (Kościuczuk et al., 2012). They are generally found in epithelial cells, macrophages, and neutrophils. The most studied members of this class are Indolicidin and human cathelicidin LL-37 (Dürr et al., 2006, Ladokhin et al., 1999, Xhindoli et al., 2016). Cathelicidin LL-37 is a widely studied class of antimicrobial peptides commonly found in neutrophil granules (Nijnik and Hancock, 2009, Nizet et al., 2001). Cathelicidins are also known as myeloid AMPs because they were first identified in myeloid cells of the mammalian bone marrow (Gombart et al., 2005). Cathelicidin LL-37 derived from humans is found to be effective against a variety of viruses such as influenza A virus (IAV), HCV, Respiratory Syncytial Virus (RSV), dengue virus (DENV) and Venezuelan equine encephalitis virus (VEEV) (Ahmed et al., 2019, Alagarasu et al., 2017, Harcourt et al., 2016, Matsumura et al., 2016, Tripathi et al., 2014). Other human-derived cathelicidins known to exhibit antiviral activity are GF-17, BMAP-18, protegrin-1, and SMAP-29 (He et al., 2018, Sousa et al., 2017). Defensins are small cationic peptides, which usually consist of 18–45 amino acids. The first defensin was discovered in human neutrophils. These are generally derived from mast cells and tissues involved in host defense (Lehrer and Lu, 2012). Structurally, defensins are classified into α, β, and θ-defensins (Lehrer and Lu, 2012, Nguyen et al., 2003). Cysteine and arginine residues are abundantly found in all three classes of defensins. These three classes differ mainly in the position of cysteine residues linked to disulfide bridges (Silverstein et al., 2007). Human α-defensins are less cationic, more hydrophobic, and are shorter compared to human β-defensins (Hoover et al., 2000). Based on published data, human defensins possess effective antimicrobial activity against HIV-1 and Human Papillomavirus 16 (HPV) infection (Furci et al., 2012, Wiens and Smith, 2017).
2.2. Amphibian-derived antiviral peptides
AMPs derived from the skin of frogs acquire high antibacterial and antiviral properties. These peptides are produced by the granular glands and contain hydrophobic and positively charged amino acids. Most of these peptides belong to Pipidae family- Hymenochirus, Pseudhymenochirus, Pipa, Silurana, and Xenopus Belaid et al. (2002); Rinaldi (2002); Nicolas and Mor (1995)). Three amphibian-derived peptides, caerin 1.1, caerin1.9 and maculatin 1.1, showed significant inhibition of HIV infection in T cells even at low concentrations. These peptides were derived from the Pelodryadidae family and inhibited the transfer of HIV from dendritic cells to T-cells. Moreover, they showed antiviral activity against murine leukemia virus (MuLV) and Junín virus (JV) (VanCompernolle et al., 2005). Melittin is an amphipathic hexacosapeptide extracted from the bee venom and is identified as a therapeutic agent against a variety of virus such as IAV, HIV, HSV, herpes simplex virus (HSV), JV, RSV, vesicular stomatitis virus (VSV), and tobacco mosaic virus (TMV) (Memariani et al., 2020). Also, Magainins from Xenopus laevis and Cecropin A from Mythimna separata have been tested against HSV-1, HSV-2 and JV. The magainins inhibited HSV-1 and HSV-2 but remained inactive against JV, while Cecropin A has been found to be effective against JV (Albiol Matanic and Castilla, 2004). A 13-residue peptide, Temporin-SHA (Tb-SHA), known for its antimicrobial properties due to its alpha-helical structure, is found to interact with the microbial cytoplasmic membrane and induce pore formation in the membrane (Ladram and Nicolas, 2016). A study revealed the antiviral properties of Tb-SHA and its synthetic analog, [K3]SHa against HSV-1 in human keratinocyte cultures (Roy et al., 2019). Two peptides, Brevinin-2GHk (BR2GK) and Fejerlectin, derived from the skin secretion of Fejervarya limnocharis, were tested for antiviral properties by Xiong and his colleagues. Fejerlectin prevented HIV-1 entry into the host cells by inhibiting Env-mediated membrane fusion and BR2GK inhibited early and middle stages of Zika virus (ZIKV) (Xiong et al., 2021a, Xiong et al., 2021b). Recently, a study revealed that AR-23 could be a potential therapeutic drug against various DNA and RNA viruses due to its interaction with the viral particle leading to inhibition at early stages. It has been found to be effective against HSV-1, measles morbillivirus (MeV), human parainfluenza virus (HPIV-2), HCoV-229E, and SARS-CoV-2 (Chianese et al., 2022).
2.3. Plant-derived antiviral peptides
Plant-derived AMPs are categorized into distinct families based on differences in amino acid sequences. They are both positively and negatively charged (Pelegrini et al., 2011). They are classified into several classes, such as lipid transfer proteins, thionins, defensins, chitin-binding proteins, cyclotides, and others (Nawrot et al., 2014). Plant-derived AMPs are effective against a broad range of viral infections caused by HCV, Japanese encephalitis virus (JEV), (middle east respiratory syndrome coronavirus) MERS-CoV, HPV, IAV, infectious bronchitis virus (IBV), DENV, HSV types 1 and 2, HIV-1/2, VSV, coxsackie virus B4, RSV, chikungunya virus (CHIKV) Sindbis virus (SINV) (Derby et al., 2018, Gordts et al., 2015, Ishag et al., 2016, Kaur et al., 2019, Levendosky et al., 2015, Liu et al., 2014, Millet et al., 2016, Nguyen et al., 2015, Takebe et al., 2013, Xu et al., 2015). Cyclotides, a large family of plant-derived disulfide-rich peptides, are exceptionally stable and consist of cyclic cystine knot (CCK). Due to their extreme stability, they have been used in drug design applications and are known to be effective against HIV, DENV, and influenza virus (Craik and Du, 2017). Plant-derived lectins have predominantly been investigated against several viral infections. Some of the most promising plant-derived antiviral lectins include Griffithsin, A. elatior lectin, chitinase (chi)-like lectin from Tamarind (TCLL), Nicotiana tabacum (NICTABA) lectin, Cycloviolacin Y5 and VY1 (Derby et al., 2018, Gordts et al., 2015, Kaur et al., 2019, Liu et al., 2014, Xu et al., 2015).
2.4. Insect-derived antiviral peptides
Insects use several defense mechanisms against microbial infections, including cellular and humoral immune responses. Localized melanization and coagulation, phagocytosis, and secretion of AMPs into the hemolymph are the cellular and humoral immune responses used by insects to counter infections (Eleftherianos and Revenis, 2011, Elrod-Erickson et al., 2000, Ishii et al., 2010). AMPs secreted by insects include cecropins, melittins, attacins, lysozymes, defensins, dipteracins, drosomycin, and metchikowins (Copley et al., 2007, Mojsoska et al., 2015, Tashmukhambetov, 2016). C-lysozyme is an example of an insect-derived antimicrobial peptide known to be effective against nucleopolyhedrovirus (NPV) infection. Lysozymes are enzymes that are important for the immune system of the organisms. While lysozymes break the peptidoglycan layer of the cell wall in bacteria, they are suggested to affect viruses through hydrolyzation of the viral structural proteins leading to the production of defective viruses (Chen et al., 2018). Moreover, gloverin proteins have predominantly been investigated against nucleopolyhedrovirus. They are usually small proteins consisting of 200 amino acids and are positively charged due to the presence of charged arginine and lysine residues (Moreno-Habel et al., 2012).
3. Antiviral peptides: mechanism of action
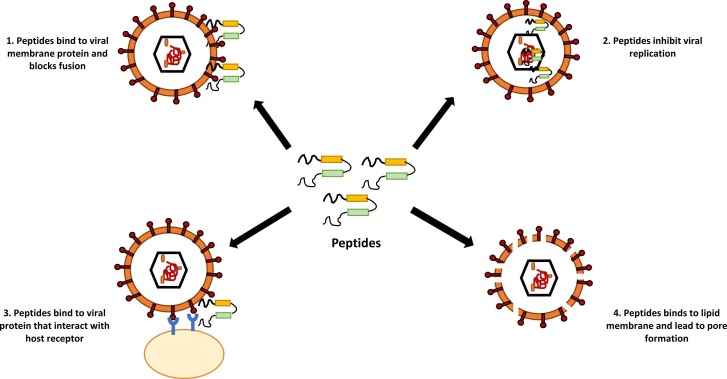
Despite having enormous diversity in their primary structure, antimicrobial peptides are characterized by a preponderance of positively charged amino acids lysine and arginine (Lei et al., 2019). They are also known as cationic host defense peptides (Brown and Hancock, 2006). Peptides tend to adopt different conformations depending upon environmental conditions. While AMPs adopt disordered conformations in solutions, they attain ordered structures with high α-helix content in membrane-mimicking environments (Vermeer et al., 2012, Teixeira et al., 2012). AMPs are considered effective antimicrobial agents due to their low propensity in the development of antimicrobial resistance. Unlike antibiotics, which target cellular activities (e.g., synthesis of proteins, DNA, or cell walls), these peptides bind to the lipopolysaccharide coating of the cell membrane. The entry of a virus into the host cells involves a series of steps including viral protein maturation, some non-specific interactions, endocytosis, conformational changes in the fusion protein, and viral membrane fusion [ Fig. 1].
Fig. 1.
Molecular mechanisms of action of antiviral peptides.
Antiviral AMPs deactivate enveloped RNA and DNA viruses through their integration with the viral envelope or the host cell membrane (Narayana and Chen, 2015, Taylor et al., 2005). They specifically target the lipids of the viral membrane and block their fusion and entry into the host cell. For instance, C5A and MP7-NH2 are the AMPs found to be effective against enveloped viruses such as HCV and RSV (Cheng et al., 2008, Sample et al., 2013). Various cationic amphiphilic AMPs bind to the viral membranes and lead to the formation of pores. In addition to disrupting viral envelopes and blocking viral receptors, certain antiviral AMPs can prevent viral particles from entering host cells by possessing unique mammalian cell receptors which can competitively interact with viral particles [ Fig.1] (Vigant et al., 2015). Such interactions lead to the stabilization or destabilization of the viral particles, thus preventing the interaction. A well-known example of such a receptor is heparan sulfate proteoglycans (HSPG), which are essential for attaching HSV viral particles to the host cell surface. The antiviral peptide interferes with the HSPG-virus interaction (Cagno et al., 2019). Since several AMPs have been used to inhibit viruses, they could potentially be used as a candidate against SARS-CoV-2 (Kurpe et al., 2020). The antiviral peptides restrict virus infection either through inhibition of virus attachment to the host cell, destruction of virus envelope, or inhibition of virus replication. Various research groups are trying to exploit the mechanism of inhibiting virus attachment to the host cell for the development of an effective treatment method against SARS-CoV-2.
4. AMPs against SARS-CoV-2 infection
The Coronaviridae Study Group (CSG) has classified SARS-CoV-2 as a member of Coronaviridae family (CSG, 2020). Seven known coronaviruses are known to infect humans and cause mild to severe symptoms. Four of these viruses (229E, NL63, OC43, and HKU1) cause common cold infection, while the other three coronaviruses (MERS-CoV, SARS-CoV and SARS-CoV-2) cause severe respiratory illness and even death. SARS-CoV-2 has a 30 kbp single stranded-RNA with 9860 amino acids encoding structural and non-structural proteins (Huang et al., 2020). ORF1a and ORF1b encode for 16 non-structural proteins (Nsp1–16) [ Fig. 2a]. These proteins are involved in viral replication and transcription. S, E, M, and N genes encode the structural proteins - spike, envelope, membrane, and nucleocapsid, respectively [Fig. 2b] (Mariano et al., 2020, Al-Qaaneh et al., 2021, Schütz et al., 2020). Spike protein (S-protein) contains 1273 amino acids and plays a vital role in the viral attachment and fusion to the host cell membrane. This protein has two subunits – S1 and S2. The S1 subunit is responsible for binding to angiotensin-converting enzyme 2 (ACE2) receptor of the host cell and the S2 helps in the fusion of the virus to the cell membrane. The S1 subunit consists of two domains – N-terminal domain (NTD) and C-terminal domain (CTD). C-terminal domain of S1 subunit has a receptor-binding domain (RBD) which binds to the ACE2 receptor (He et al., 2020, Huang et al., 2020, Walls et al., 2020). S2 subunit contains fusion protein (FP), heptapeptide repeat sequence 1(HR1), heptapeptide repeat sequence 1 (HR2), transmembrane domain (TM) and cytoplasmic domain (CT) [Fig. 2a].
Fig. 2.
SARS-CoV-2 (a) genomic organization and (b) structure.
During fusion, S-protein is activated through its cleavage into S1 and S2 subunits. This cleavage is mediated by TMPRSS2 and furin proteases of the host cell (Rani et al., 2022). Multiple furin cleavage sites have been found in highly pathogenic influenza viruses and SARS-CoV-2 (Hasan et al., 2020). Therefore, multiple furin cleavage sites could be a possible reason for the pathogenicity of SARS-CoV-2 (Kido et al., 2012, Heurich et al., 2014, Limburg et al., 2019). After cleavage, FP protein binds and fuses to the host cell membrane bringing HR1 closer to the host membrane. Next, HR1 and HR2 form a fusion core and trigger cell membrane fusion [ Fig. 3]. (Eckert and Kim, 2001, Harrison, 2015, Millet and Whittaker, 2018). Inside the host cell, viral RNA is translated to polyproteins using the host ribosomes. Viral 3 C-like protease (3CLPro) and papain like protease (PLpro) cleave polyproteins into Nsps that are required for transcription, replication, and packaging.
Fig. 3.
Mechanism of action of antiviral peptides developed against SARS-CoV-2.
RDV is the first drug approved by the FDA for COVID-19 treatment. It was discovered as an effective therapeutic drug for Ebola virus in 2017 (Siegel et al., 2017). It is a phosphonamidite prodrug which forms triphosphate to target viral RNA dependent RNA polymerase (RdRp) enzyme which is required for viral replication and delayed chain termination in SARS-CoV, MERS-CoV and SARS-CoV-2 (Gordon et al., 2020). Lopinavir-ritonavir (protease inhibitors) combination therapy was proposed for COVID-19 treatment as it inhibits 3CLPro of SARS-CoV-2. However, it did not show any significant effects on COVID-19 patients during clinical trials (Cao et al., 2020a, Cao et al., 2020b, Patel et al., 2021). Hence, there is a continuous need for the development of effective therapeutic solutions against SARS-CoV-2.
Researchers have been trying to exploit a variety of mechanisms for the development of antiviral peptides against SARS-CoV-2 such as targeting the viral envelope, S-glycoprotein, endosomal acidification inhibition and shielding the cell receptors for the host [Fig. 3] (Mahendran et al., 2020). Targeting S-protein, a potential strategy for drug development, has been studied by several research groups (He et al., 2020, Huang et al., 2020, Walls et al., 2020). Peptides binding to RBD of S1 subunit reduce the binding affinity of S1 to ACE2 receptor of the host cell, thereby preventing the spread of virus (Kurpe et al., 2020). A α5β1 integrin binding peptide ATN161 extracted from extracellular matrix fibronectin, disrupt spike protein and ACE2 interaction by binding to α5β1 attached to S-protein RBD and reduce viral infection (Beddingfield et al., 2021). A lipopeptide, EK1C4 is an example of an effective inhibitor against SARS-CoV-2. A molecule of cholesterol was covalently attached to EK1, an HR-targeting inhibitor, to induce solubility and activity. It targets HR1 viral protein and inhibits viral and host membrane fusion. Due to the identical nature of HR2 of SARS-CoV-1and SARS-CoV-2, 2019-nCov1-HR2P peptide derived from SAR-CoV-1 showed significant result in inhibiting viral infection against SARS-CoV-2 (Xia et al., 2020a, Xia et al., 2020b, Zhang et al., 2020). A study showed the cloaking of binding sites of ACE2 by human defensin-5 (HD-5), demonstrating innate defense mechanisms of intestinal epithelium against SARS-CoV-2 (Wang et al., 2020). Cathelicidin LL37 has also been shown to inhibit SARS-CoV-2 pseudo-virion infection by binding to the RBD of S1 subunits and ACE2 (Wang et al., 2021). Lactoferrin is an antimicrobial peptide known to be effective against SARS-CoV and is suggested to be a potential option due to its affordability, environmental safety, and efficiency (Campione et al., 2021).
TMPRSS2 and furin protease cleave surface spike protein into S1 and S2 subunits, which is a crucial step for the entry of SARS-CoV-2 to host cells. Aprotinin, a TMPRSS2 inhibitor isolated from bovine lung, and synthetic mimetic peptides MI-432 and MI-1900, showed significant suppression in S-protein activation and viral multiplication in Calu-3 human airway cells. Similar results were observed for the synthetic peptide MI-1851, a furin inhibitor, in Calu-3 cells. A combination therapy of MI-1851 and MI-432 reduced viral multiplication significantly at lower dose in Calu-3 cells in contrast to single inhibitor treatment (Bestle et al., 2020). Previously, mouse β-defensin-4 derived, P9 peptide, showed significant reduction in infection caused by SARS-CoV, MERS CoV, and influenza virus (Zhao et al., 2016). Recently, P9 and two potent peptides derived from mouse β-defensin-4 have been found to be effective therapeutic agents for COVID-19 treatment. P9R peptide has shown effective results in mice against lethal A (H1N1) pdm09 virus without inducing drug resistance in MDCK cells (Zhao et al., 2020). A synthetic peptide, brilacidin, inhibits SARS-CoV-2 infection through the inhibition of viral entry and disruption of viral integrity in Vero and Calu-3 cells. Moreover, its combination with RDV has a synergistic effect against SARS-CoV-2 (Bakovic et al., 2020). The AMP’s cathelicidin LL-37 and defensins reportedly take part in active viral defense. Additionally, two promising miniproteins-AHB1 and AHB2 were designed based on RBD binding motif using de-novo sequencing. Later, LCB1-LCB8 peptides were designed based on a similar strategy, and LCB1-LCB3 showed neutralization effect against SARS-CoV-2 (Cao et al., 2020a, Cao et al., 2020b). In 2020, the SARS BLOCK™ peptide scaffold was designed to mimic the RBD binding motif to inhibit virus entry to host cell and infection. This peptide inhibitor also displays an antibody neutralizing binding motif to induce immune response and acts as a therapeutic as well as immune stimulant peptide (Watson et al., 2020). Recently, researchers have discussed the potential of human-derived AMPs released by human mesenchymal stem cells (hMSC) in initiating pulmonary defense mechanisms against COVID-19. The hMSC-derived AMPs have possibly modulated cytokine storm in terminally ill COVID patients and led to effective therapeutic treatment (Ghosh and Weinberg, 2021). In a similar study antiviral traits of defensins have been highlighted in viral inhibition by binding to virion, modulating host cell receptor with intracellular signaling disruptions and alteration of immune responses by interacting with chemokine receptors and toll-like receptors (TLRs). The natural defensive mechanisms against invading viruses such as CoV-2 likely depend upon meticulous physical activity, which is similar to the immunity of athletes against pathogenic microorganisms offered by high concentrations of serum defensins (Laneri et al., 2021). Researchers have assessed one of the latest insights into the anti-corona virus activity of AMP DP7. This study revealed that drug-targeted sites of CoV-2 are divided into two categories, namely viral protease and others including CoV-2 S-protein, ACE-2 receptor, and a transmembrane protease. The researchers effectively demonstrated that DP7 prevented SARS-CoV/CoV-2 S-protein binding of ACE2–293 T cells. In addition, DP7 inhibited the protein-mediated host cell-cell fusion and inhibited SARS-CoV-2-Mpro (or 3CLpro). The 3CLPro plays an important role in CoV-2 viral replication and recently has been observed as a possible target of plant secondary metabolites and AMPs (Zhang et al., 2021, Mody et al., 2021, Jo et al., 2020). This could provide a revolutionary basis for AMP-based drug development against coronavirus infections. In yet another recent study bioengineered AMPs - glycocin F and lactococcine G derived from two probiotic bacterial strains Lactococcus lactis and Lactobacillus plantarum respectively were assessed as possible drugs for COVID-19. The study revealed that both glycocin F and Lactococcine G possess high binding affinities towards viral proteins and hence further experimental analyses against COVID-19 shall be performed (Balmeh et al., 2021). Recently, 8 alpha-helical peptides were tested to identify a potential therapeutic scaffold which can compete with S1 for the RBD binding site of the ACE2 receptor. This study revealed that S1-Dermaseptin-S9 (S1-DS9) complex could be a potential therapeutic peptide complex for SARS-CoV-2 treatment. It binds to S1 subunit of S-protein and prevent S1 binding to ACE2 receptor and inhibit virus entry to host cell (Sekar et al., 2022). Table 2 contains a list of antiviral peptides developed against SARS-CoV-2.
Table 2.
Antiviral peptides developed against SARS-CoV-2.
| Name | Target Protein | Source | Sequence derived from | Mechanism | References |
|---|---|---|---|---|---|
| DS9 | S1 subunit | Synthetic peptide | Dermaseptin | Impedes S1 binding to ACE2 receptor and regulates virus entry | Sekar et al. (2022) |
| AHB1 | S-protein | De-novo designed | ACE2 | Inhibits RBD binding to ACE2 | Cao et al. (2020) |
| LCB1 | S-protein RBD | De-novo designed | Based on ACE2 binding to RBD | Inhibits RBD binding to ACE2 | |
| LCB3 | S-protein RBD | Inhibits RBD binding to ACE2 | |||
| SARS-BLOCK | RBD binding site of ACE2 receptor | Synthetic peptide inhibitor that mimics S-protein RBD | S-protein RBD | Inhibits S-protein binding to ACE2 and stimulate immune response | Watson et al. (2020) |
| HD5 | ACE2 | Intestinal Paneth cells | Human defensin family peptide | Cloaking of binding of ACE2 and inhibits entry to the host cell | Wang et al. (2020) |
| LL37 | Carboxypeptidase domain of ACE2 and S protein RBD | Neutrophils and epithelial cells | Human cathelicidin family peptide | Inhibits the binding of RBD to ACE2 receptor | Wang et al. (2021) |
| Lactoferrin | Innate immunity and S-protein | Exocrine glands and neutrophils | Bovine lactoferrin (identical to human lectoferrin) | Prevents virus entry and anti-inflammatory and restore iron homeostasis | Campione et al. (2021) |
| Birlacidin | Not available (NA) | Synthetic nonpeptidic mimetic polymer | Host defense protein magainin | Inhibits virus entry and significantly effective in combination with remdesivir. | Bakovic et al. (2020) |
| EK1C4 | HR1 | Cholesterol molecule conjugated to the EK1 peptide (a modified OC43-HR2P peptide) | HR2 of OC43 | Inhibits S-protein mediated membrane fusion | Xia et al. (2020) |
| 2019-nCoV-HR2P | HR1 | Synthetic peptide derived from 2019-nCoV-HR2P | HR2 of SARS-CoV2/ 2019 nCoV (both sequences are identical) | Inhibits S-protein mediated membrane fusion | |
| ATN-161 | ACE2 receptor and S-protein RBD | Extracellular matrix component fibronectin | Fibronectin | Binds to integrin α5β1 and dirupt the interaction between ACE2 and S-protein RBD | Beddingfield et al. (2021) |
| Aprotinin | TMPRSS2 | Bovine lung | Aprotinin | Prevents proteolytic activation | Bestle et al. (2020) |
| MI-432 | Peptide mimetic inhibitors of TMPRSS2 | Protease inhibitor (peptidomimetic) | Prevents proteolytic activation | ||
| MI-1900 | Peptide mimetic inhibitors of TMPRSS2 | Prevents proteolytic activation | |||
| P9 | Cathepsin L | Synthetic peptide based on mouse β-defensin-4 | Mouse β-defensin-4 | Inhibition of endosomal acidification | Zhao et al. (2020) |
| P9R | |||||
| 8P9R | |||||
| MI-1851 | Furin | Peptide mimetic inhibitors of Furin | Protease inhibitor (peptidomimetic) | Inhibits proteolytic cleavage of S-protein into S1 and S2 subunits | Bestle et al. (2020) |
5. Conclusion
This review discusses the potential use of AMPs as antiviral peptides against SARS-CoV-2. Since AMPs are less susceptible to antimicrobial resistance, they have emerged as an effective strategy against microbes to tackle the problem of ADR. Successful use of AMPs against various pathogens has revealed its possible therapeutic interventions against antimicrobial resistance. Therefore, the potential application of antimicrobial peptides is required to be explored. AMPs have been successfully developed against several pathogenic viruses, including but not limited to HIV, DENV, IAV, IBV, HCV, HPV, HBV. The modes of action of antiviral peptides have been described in this review article and how they could be exploited to target SARS-CoV-2. Currently, a significant amount of effort is being devoted towards the development of antiviral peptides against SARS-CoV-2. Improvement of peptides is also achieved through de-novo designing and synthesizing them synthetically, considering their properties and target proteins (Sekar et al., 2022, Cao et al., 2020a, Cao et al., 2020b, Watson et al., 2020, Bakovic et al., 2020). In conclusion, AMPs are the future of therapeutics and an important area of research, especially during the emergence of the COVID-19 pandemic, which has affected millions of lives.
Conflict of interest
Author declares no conflicts of interest in submitting this article in Microbiology Research. There are also no financial interests.
References
- Ahmed A., Siman-Tov G., Keck F., Kortchak S., Bakovic A., Risner K., Lu T.K., Bhalla N., de la Fuente-Nunez C., Narayanan A. Human cathelicidin peptide LL-37 as a therapeutic antiviral targeting Venezuelan equine encephalitis virus infections. Antivir. Res. 2019;164:61–69. doi: 10.1016/j.antiviral.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Alagarasu K., Patil P.S., Shil P., Seervi M., Kakade M.B., Tillu H., Salunke A. In-vitro effect of human cathelicidin antimicrobial peptide LL-37 on dengue virus type 2. Peptides. 2017;92:23–30. doi: 10.1016/j.peptides.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Albiol Matanic V.C., Castilla V. Antiviral activity of antimicrobial cationic peptides against Junin virus and herpes simplex virus. Int. J. Antimicrob. Agents. 2004;23:382–389. doi: 10.1016/j.ijantimicag.2003.07.022. [DOI] [PubMed] [Google Scholar]
- Al-Qaaneh A.M., Alshammari T., Aldahhan R., Aldossary H., Alkhalifah Z.A., Borgio J.F. Genome composition and genetic characterization of SARS-CoV-2. Saudi J. Biol. Sci. 2021;28:1978–1989. doi: 10.1016/j.sjbs.2020.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammendolia M.G., Agamennone M., Pietrantoni A., Lannutti F., Siciliano R.A., De Giulio B., Amici C., Superti F. Bovine lactoferrin-derived peptides as novel broad-spectrum inhibitors of influenza virus. Pathog. Glob. Health. 2012;106:12–19. doi: 10.1179/2047773212Y.0000000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand U., Jacobo-Herrera N., Altemimi A., Lakhssassi N. A comprehensive review on medicinal plants as antimicrobial therapeutics: potential avenues of biocompatible drug discovery. Metabolites. 2019;9:258. doi: 10.3390/metabo9110258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AnonWorld Health Organization, June 2022. WHO Coronavirus (COVID-19) Dashboard. 〈https://covid19.who.int〉.
- Armitage A.E., Stacey A.R., Giannoulatou E., Marshall E., Sturges P., Chatha K., Smith N.M., Huang X., Xu X., Pasricha S.R. Distinct patterns of hepcidin and iron regulation during HIV-1, HBV, and HCV infections. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12187–12192. doi: 10.1073/pnas.1402351111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakovic A., Risner K., Bhalla N., Alem F., Chang T.L., Weston W., Narayanan A. Brilacidin, a COVID-19 drug candidate, exhibits potent in vitro antiviral activity against SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.10.29.352450. [DOI] [Google Scholar]
- Balmeh N., Mahmoudi S., Fard N.A. Manipulated bio antimicrobial peptides from probiotic bacteria as proposed drugs for COVID-19 disease. Inf. Med. Unlocked. 2021;23 doi: 10.1016/j.imu.2021.100515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels E., Dekker D., Amiche M. Dermaseptins, multifunctional antimicrobial peptides: a review of their pharmacology, effectivity, mechanism of action, and possible future directions. Front. Pharmacol. 2019;10:1421. doi: 10.3389/fphar.2019.01421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddingfield B.J., Iwanaga N., Chapagain P.P., Zheng W., Roy C.J., Hu T.Y., Kolls J.K., Bix G.J. The integrin binding peptide, ATN-161, as a novel therapy for SARS-CoV-2 infection. J. Am. Coll. Cardiol. Basic Trans. Sci. 2021;6:1–8. doi: 10.1016/j.jacbts.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Befus A.D., Mowat C., Gilchrist M., Hu J., Solomon S., Bateman A. Neutrophil defensins induce histamine secretion from mast cells: mechanisms of action. J. Immunol. 1999;163:947–953. [PubMed] [Google Scholar]
- Belaid A., Aouni M., Khelifa R., Trabelsi A., Jemmali M., Hani K. In vitro antiviral activity of dermaseptins against herpes simplex virus type 1. J. Med. Virol. 2002;66:229–234. doi: 10.1002/jmv.23450. [DOI] [PubMed] [Google Scholar]
- Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Klenk H.D., Garten W., Steinmetzer T., Böttcher-Friebertshäuser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boparai J.K., Sharma P.K. Mini review on antimicrobial peptides, sources, mechanism and recent applications. Protein Pept. Lett. 2020;27:4–16. doi: 10.2174/0929866526666190822165812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.L., Hancock R.E. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 2006;18:24–30. doi: 10.1016/j.coi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Cagno V., Tseligka E.D., Jones S.T., Tapparel C. Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias? Viruses. 2019;11:596. doi: 10.3390/v11070596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campione E., Lanna C., Cosio T., Rosa L., Conte M.P., Iacovelli F., Romeo A., Falconi M., Del Vecchio C., Franchin E., Lia M.S., Minieri M., Chiaramonte C., Ciotti M., Nuccetelli M., Terrinoni A., Iannuzzi I., Coppeda L., Magrini A., Bernardini S., Bianchi L. Lactoferrin against SARS-CoV-2: in vitro and in silico evidences. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.666600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Goreshnik I., Coventry B., Case J.B., Miller L., Kozodoy L., Chen R.E., Carter L., Walls A.C., Park Y.J., Strauch E.M., Stewart L., Diamond M.S., Veesler D., Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Sci. (N. Y., N. Y. ) 2020;370:426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.M., Fan Y.C., Lin J.W., Chen Y.Y., Hsu W.L., Chiou S.S. Bovine lactoferrin inhibits dengue virus infectivity by interacting with heparan sulfate, low-density lipoprotein receptor, and DC-SIGN. Int. J. Mol. Sci. 2017;18:1957. doi: 10.3390/ijms18091957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.T., Tan L.R., Hu N., Dong Z.Q., Hu Z.G., Jiang Y.M., Chen P., Pan M.H., Lu C. C-lysozyme contributes to antiviral immunity in Bombyx mori against nucleopolyhedrovirus infection. J. Insect Physiol. 2018;108:54–60. doi: 10.1016/j.jinsphys.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Cheng G., Montero A., Gastaminza P. A virocidal amphipathic α-helical peptide that inhibits hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3088–3093. doi: 10.1073/pnas.0712380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chianese A., Zannella C., Monti A., De Filippis A., Doti N., Franci G., Galdiero M. The Broad-Spectrum Antiviral Potential of the Amphibian Peptide AR-23. Int. J. Mol. Sci. 2022;23:883. doi: 10.3390/ijms23020883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa C., Sánchez L., Rota C., Pérez M.D., Calvo M., Farnaud S., Evans R.W. Isolation of lactoferrin from milk of different species: calorimetric and antimicrobial studies. Biochem. Mol. Biol. 2008;150:131–139. doi: 10.1016/j.cbpb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Copley R.R., Totrov M., Linnell J., Field S., Ragoussis J., Udalova I.A. Functional conservation of Rel binding sites in drosophilid genomes. Genome Res. 2007;17:1327–1335. doi: 10.1101/gr.6490707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.A., Vlieghe E., Mendelson M., Wertheim H., Ndegwa L., Villegas M.V., Hara G.L. Antibiotic stewardship in low-and middle-income countries: the same but different? Clin. Microbiol. Infect. 2017;23:812–818. doi: 10.1016/j.cmi.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Craik D.J., Du J. Cyclotides as drug design scaffolds. Curr. Opin. Chem. Biol. 2017;38:8–16. doi: 10.1016/j.cbpa.2017.01.018. [DOI] [PubMed] [Google Scholar]
- Dadgostar P. Antimicrobial resistance: implications and costs. Infect. Drug Resist. 2019;12:3903–3910. doi: 10.2147/IDR.S234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby N., Lal M., Aravantinou M., Kizima L., Barnable P., Rodriguez A., Lai M., Wesenberg A., Ugaonkar S., Levendosky K., Mizenina O., Kleinbeck K., Lifson J.D., Peet M.M., Lloyd Z., Benson M., Heneine W., O'Keefe B.R., Robbiani M., Martinelli E., Grasperge B., Blanchard J., Gettie A., Teleshova N., Fernández-Romero J.A., Zydowsky T.M. Griffithsin carrageenan fast dissolving inserts prevent SHIV HSV-2 and HPV infections in vivo. Nat. Commun. 2018;9:3881. doi: 10.1038/s41467-018-06349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Somma A., Moretta A., Canè C., Cirillo A., Duilio A. Antimicrobial and antibiofilm peptides. Biomolecules. 2020;10:652. doi: 10.3390/biom10040652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drannik A.G., Nag K., Sallenave J.M., Rosenthal K.L. Antiviral activity of trappin-2 and elafin in vitro and in vivo against genital herpes. J. Virol. 2013;87:7526–7538. doi: 10.1128/JVI.02243-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos R.J. Studies on a bactericidal agent extracted from a soil bacillus: II. Protective effect of the bactericidal agent against experimental Pneumococcus infections in mice. J. Exp. Med. 1939;70:11–17. doi: 10.1084/jem.70.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr U.H., Sudheendra U.S., Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta. 2006;1758:1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Eleftherianos I., Revenis C. Role and importance of phenoloxidase in insect hemostasis. J. Innate Immun. 2011;3:28–33. doi: 10.1159/000321931. [DOI] [PubMed] [Google Scholar]
- El-Fakharany E.M., Sanchez L., Al-Mehdar H.A., Redwan E.M. Effectiveness of human camel bovine and sheep lactoferrin on the hepatitis C virus cellular infectivity: comparison study. Virol. J. 2013;10:199. doi: 10.1186/1743-422X-10-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod-Erickson M., Mishra S., Schneider D. Interactions between the cellular and humoral immune responses in Drosophila. Curr. Biol. 2000;10:781–784. doi: 10.1016/s0960-9822(00)00569-8. [DOI] [PubMed] [Google Scholar]
- Fletcher S. Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ. Health Prev. Med. 2015;20:243–252. doi: 10.1007/s12199-015-0468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furci L., Tolazzi M., Sironi F., Vassena L., Lusso P. Inhibition of HIV-1 infection by human alpha-defensin-5, a natural antimicrobial peptide expressed in the genital and intestinal mucosae. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.K., Weinberg A. Ramping up antimicrobial peptides against severe acute respiratory syndrome coronavirus-2. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.620806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombart A.F., Borregaard N., Koeffler H.P. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up‐regulated in myeloid cells by 1, 25–dihydroxyvitamin D3. FASEB J. 2005;19:1067–1077. doi: 10.1096/fj.04-3284com. [DOI] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordts S.C., Renders M., Férir G., Huskens D., Van Damme E.J.M., Peumans W., Balzarini J., Schols D. NICTABA and UDA, two GlcNAc-binding lectins with unique antiviral activity profiles. J. Antimicrob. Chemother. 2015;70:1674–1685. doi: 10.1093/jac/dkv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J.L., McDonald M., Svoboda P., Pohl J., Tatti K., Haynes L.M. Human cathelicidin, LL-37, inhibits respiratory syncytial virus infection in polarized airway epithelial cells. BMC Res. Notes. 2016;9:11. doi: 10.1186/s13104-015-1836-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B.A., Hussain A., Qadir F.A., Attar F., Aziz F.M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A.A., Falahati M. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020;22:1–9. doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Tao H., Yan Y., Huang S.Y., Xiao Y. Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses. 2020;12:428. doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Zhang H., Li Y., Wang G., Tang B., Zhao J., Huang Y., Zheng J. Cathelicidin-derived antimicrobial peptides inhibit zika virus through direct inactivation and interferon pathway. Front. Immunol. 2018;9:722. doi: 10.3389/fimmu.2018.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J.G. Phagocytin: a bactericidal substance from polymorphonuclear leucocytes. J. Exp. Med. 1956;103:589–611. doi: 10.1084/jem.103.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover D.M., Rajashankar K.R., Blumenthal R., Puri A., Oppenheim J.J., Chertov O., Lubkowski J. The structure of human β-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 2000;275:32911–32918. doi: 10.1074/jbc.M006098200. [DOI] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishag H.Z., Li C., Wang F., Mao X. Griffithsin binds to the glycosylated proteins (E and prM) of Japanese encephalitis virus and inhibit its infection. Virus Res. 2016;215:50–54. doi: 10.1016/j.virusres.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Hamamoto H., Kamimura M., Nakamura Y., Noda H., Imamura K., Sekimizu K. Insect cytokine paralytic peptide (PP) induces cellular and humoral immune responses in the silkworm Bombyx mori. J. Biol. Chem. 2010;285:28635–28642. doi: 10.1074/jbc.M110.138446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. PMID: 31724441; PMCID: PMC6882434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R., Neetu, Mudgal R., Jose J., Kumar P., Tomar S. Glycan-dependent chikungunya viral infection divulged by antiviral activity of NAG specific chi-like lectin. Virology. 2019;526:91–98. doi: 10.1016/j.virol.2018.10.009. [DOI] [PubMed] [Google Scholar]
- Kaur R., Rani P., Atanasov A.G., Alzahrani Q., Gupta R., Kapoor B., Gulati M., Chawla P. Discovery and development of antibacterial agents: fortuitous and designed. Mini Rev. Med. Chem. 2022;22(7):984–1029. doi: 10.2174/1570193X19666211221150119. [DOI] [PubMed] [Google Scholar]
- Kausar S., Said Khan F., Ishaq Mujeeb Ur Rehman M., Akram M., Riaz M., Rasool G., Hamid Khan A., Saleem I., Shamim S., Malik A. A review: mechanism of action of antiviral drugs. Int. J. Immunopathol. Pharmacol. 2021;35 doi: 10.1177/20587384211002621. 20587384211002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khameneh B., Iranshahy M., Soheili V., Fazly Bazzaz B.S. Review on plant antimicrobials: a mechanistic viewpoint. Antimicrob. Resist. Infect. Control. 2019;8:118. doi: 10.1186/s13756-019-0559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido H., Okumura Y., Takahashi E., Pan H.Y., Wang S., Yao D., Yao M., Chida J., Yano M. Role of host cellular proteases in the pathogenesis of influenza and influenza-induced multiple organ failure. Biochim Biophys. Acta. 2012;1824:186–194. doi: 10.1016/j.bbapap.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Kościuczuk E.M., Lisowski P., Jarczak J., Strzałkowska N., Jóźwik A., Horbańczuk J., Krzyżewski J., Zwierzchowski L., Bagnicka E. Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012;39:10957–10970. doi: 10.1007/s11033-012-1997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpe S.R., Grishin S.Y., Surin A.K., Panfilov A.V., Slizen M.V., Chowdhury S.D., Galzitskaya O.V. Antimicrobial and amyloidogenic activity of peptides. Can antimicrobial peptides be used against sars-cov-2? Int. J. Mol. Sci. 2020;21:9552. doi: 10.3390/ijms21249552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladokhin A.S., Selsted M.E., White S.H. CD spectra of indolicidin antimicrobial peptides suggest turns, not polyproline helix. Biochemistry. 1999;38:12313–12319. doi: 10.1021/bi9907936. [DOI] [PubMed] [Google Scholar]
- Ladram A., Nicolas P. Antimicrobial peptides from frog skin: Biodiversity and therapeutic promises. Front. Biosci. 2016;21:1341–1371. doi: 10.2741/446. [DOI] [PubMed] [Google Scholar]
- Laneri S., Brancaccio M., Mennitti C., De Biasi M.G., Pero M.E., Pisanelli G., Scudiero O., Pero R. Antimicrobial peptides and physical activity: a great hope against COVID 19. Microorganisms. 2021;9(7):1415. doi: 10.3390/microorganisms9071415. Jun 30. PMID: 34209064; PMCID: PMC8304224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer R.I., Lu W. α‐Defensins in human innate immunity. Immunol. Rev. 2012;245:84–112. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- Lei J., Sun L., Huang S., Zhu C., Li P., He J., Mackey V., Coy D.H., He Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019;11:3919–3931. [PMC free article] [PubMed] [Google Scholar]
- Levendosky K., Mizenina O., Martinelli E., Jean-Pierre N., Kizima L., Rodriguez A., Kleinbeck K., Bonnaire T., Robbiani M., Zydowsky T.M., O'Keefe B.R., Fernández-Romero J.A. Griffithsin and carrageenan combination to target herpes simplex virus 2 and human papillomavirus. Antimicrob. Agents Chemother. 2015;59:7290–7298. doi: 10.1128/AAC.01816-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limburg H., Harbig A., Bestle D., Stein D.A., Moulton H.M., Jaeger J., Janga H., Hardes K., Koepke J., Schulte L., Koczulla A.R., Schmeck B., Klenk H.D., Böttcher-Friebertshäuser E. TMPRSS2 is the major activating protease of influenza a virus in primary human airway cells and influenza B virus in human type II pneumocytes. J. Virol. 2019;93 doi: 10.1128/JVI.00649-19. –19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.Z., Yang Y., Zhang S.X., Tang L., Wang H.M., Chen C.J., Shen Z.F., Cheng K.D., Kong J.Q., Wang W. A cyclotide against influenza A H1N1 virus from Viola yedoensis. Acta Pharm. Sin. 2014;49:905–912. [PubMed] [Google Scholar]
- Ma L.L., Yin X., Li B.H., Yang J.Y., Jin Y.H., Huang D., Deng T., Wang Y.Y., Ren X.Q., Ji J., Zeng X.T. Coronavirus disease 2019 related clinical studies: a cross-sectional analysis. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.540187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendran A., Lim Y.S., Fang C.M., Loh H.S., Le C.F. The potential of antiviral peptides as COVID-19 therapeutics. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.575444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariano G., Farthing R.J., Lale-Farjat S., Bergeron J. Structural characterization of SARS-CoV-2: where we are, and where we need to be. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T., Sugiyama N., Murayama A., Yamada N., Shiina M., Asabe S., Wakita T., Imawari M., Kato T. Antimicrobial peptide LL-37 attenuates infection of hepatitis C virus. Hepatol. Res. 2016;46:924–932. doi: 10.1111/hepr.12627. [DOI] [PubMed] [Google Scholar]
- Memariani H., Memariani M., Moravvej H., Shahidi-Dadras M. Melittin: a venom-derived peptide with promising anti-viral properties. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:5–17. doi: 10.1007/s10096-019-03674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/j.virol.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Séron K., Labitt R.N., Danneels A., Palmer K.E., Whittaker G.R., Dubuisson J., Belouzard S. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antivir. Res. 2016;133:1–8. doi: 10.1016/j.antiviral.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody V., Ho J., Wills S., Mawri A., Lawson L., Ebert M.C.C.J.C., Fortin G.M., Rayalam S., Taval S. Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun. Biol. 2021;4(1):93. doi: 10.1038/s42003-020-01577-x. Jan 20. PMID: 33473151; PMCID: PMC7817688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojsoska B., Zuckermann R.N., Jenssen H. Structure-activity relationship study of novel peptoids that mimic the structure of antimicrobial peptides. Antimicrob. Agents Chemother. 2015;59:4112–4120. doi: 10.1128/AAC.00237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Habel D.A., Biglang-awa I.M., Dulce A., Luu D.D., Garcia P., Weers P.M., Haas-Stapleton E.J. Inactivation of the budded virus of Autographa californica M nucleopolyhedrovirus by gloverin. J. Invertebr. Pathol. 2012;110:92–101. doi: 10.1016/j.jip.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana J.L., Chen J.Y. Antimicrobial peptides: possible anti-infective agents. Peptides. 2015;72:88–94. doi: 10.1016/j.peptides.2015.05.012. [DOI] [PubMed] [Google Scholar]
- Nawrot R., Barylski J., Nowicki G., Broniarczyk J., Buchwald W., Goździcka-Józefiak A. Plant antimicrobial peptides. Folia Microbiol. 2014;59:181–196. doi: 10.1007/s12223-013-0280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ncube N.S., Afolayan A.J., Okoh A.I. Assessment techniques of antimicrobial properties of natural compounds of plant origin: current methods and future trends. Afr. J. Biotechnol. 2008;7:1797–1806. [Google Scholar]
- Nguyen P.Q., Ooi J.S., Nguyen N.T., Wang S., Huang M., Liu D.X., Tam J.P. Antiviral cystine knot α-amylase inhibitors from alstonia scholaris. J. Biol. Chem. 2015;290:31138–31150. doi: 10.1074/jbc.M115.654855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.X., Cole A.M., Lehrer R.I. Evolution of primate theta-defensins: a serpentine path to a sweet tooth. Peptides. 2003;24:1647–1654. doi: 10.1016/j.peptides.2003.07.023. [DOI] [PubMed] [Google Scholar]
- Nicolas P., Mor A. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu. Rev. Microbiol. 1995;49:277–304. doi: 10.1146/annurev.mi.49.100195.001425. [DOI] [PubMed] [Google Scholar]
- Nijnik A., Hancock R.E. The roles of cathelicidin LL-37 in immune defences and novel clinical applications. Curr. Opin. Hematol. 2009;16:41–47. doi: 10.1097/moh.0b013e32831ac517. [DOI] [PubMed] [Google Scholar]
- Nizet V., Ohtake T., Lauth X., Trowbridge J., Rudisill J., Dorschner R.A., Pestonjamasp V., Piraino J., Huttner K., Gallo R.L. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Patel T.K., Patel P.B., Barvaliya M., Saurabh M.K., Bhalla H.L., Khosla P.P. Efficacy and safety of lopinavir-ritonavir in COVID-19: a systematic review of randomized controlled trials. J. Infect. Public Health. 2021;14:740–748. doi: 10.1016/j.jiph.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrini P.B., Del Sarto R.P., Silva O.N., Franco O.L., Grossi-De-Sa M.F. Antibacterial peptides from plants: what they are and how they probably work. Biochem. Res. Int. 2011;2011 doi: 10.1155/2011/250349. 250349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E., Kaur P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 2018;9:2928. doi: 10.3389/fmicb.2018.02928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani P., Kapoor B., Gulati M., Atanasov A.G., Alzahrani Q., Gupta R. Antimicrobial peptides: a plausible approach for COVID-19 treatment. Expert Opin. Drug Discov. 2022;17:473–487. doi: 10.1080/17460441.2022.2050693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi A.C. Antimicrobial peptides from amphibian skin: an expanding scenario. Curr. Opin. Chem. Biol. 2002;6:799–804. doi: 10.1016/s1367-5931(02)00401-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez R., Jung C.L., Gabayan V., Deng J.C., Ganz T., Nemeth E., Bulut Y. Hepcidin induction by pathogens and pathogen-derived molecules is strongly dependent on interleukin-6. Infect. Immun. 2014;82:745–752. doi: 10.1128/IAI.00983-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M., Lebeau L., Chessa C., Damour A., Ladram A., Oury B., Boutolleau D., Bodet C., Lévêque N. Comparison of anti-viral activity of frog skin anti-microbial peptides temporin-sha and [K³]SHa to LL-37 and temporin-Tb against herpes simplex virus type 1. Viruses. 2019;11:77. doi: 10.3390/v11010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample C.J., Hudak K.E., Barefoot B.E. A mastoparan-derived peptide has broad-spectrum antiviral activity against enveloped viruses. Peptides. 2013;48:96–105. doi: 10.1016/j.peptides.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz D., Ruiz-Blanco Y.B., Münch J., Kirchhoff F., Sanchez-Garcia E., Müller J.A. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv. Drug Deliv. Rev. 2020;167:47–65. doi: 10.1016/j.addr.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar P.C., Srinivasan E., Chandrasekhar G., Paul D.M., Sanjay G., Surya S., Kumar N., Rajasekaran R. Probing the competitive inhibitor efficacy of frog-skin alpha helical AMPs identified against ACE2 binding to SARS-CoV-2 S1 spike protein as therapeutic scaffold to prevent COVID-19. J. Mol. Model. 2022;28:128. doi: 10.3389/fchem.2021.753146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- Silverstein K.A., Moskal W.A., Jr, Wu H.C., Underwood B.A., Graham M.A., Town C.D., VandenBosch K.A. Small cysteine‐rich peptides resembling antimicrobial peptides have been under‐predicted in plants. Plant J. 2007;51:262–280. doi: 10.1111/j.1365-313X.2007.03136.x. [DOI] [PubMed] [Google Scholar]
- Sousa F.H., Casanova V., Findlay F., Stevens C., Svoboda P., Pohl J., Proudfoot L., Barlow P.G. Cathelicidins display conserved direct antiviral activity towards rhinovirus. Peptides. 2017;95:76–83. doi: 10.1016/j.peptides.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe Y., Saucedo C.J., Lund G., Uenishi R., Hase S., Tsuchiura T., Kneteman N., Ramessar K., Tyrrell D.L., Shirakura M., Wakita T., McMahon J.B., O'Keefe B.R. Antiviral lectins from red and blue-green algae show potent in vitro and in vivo activity against hepatitis C virus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashmukhambetov, B., 2016. An investigation of the effects of an antimicrobial peptide on the survival of Acanthamoeba and intracellular bacteria associated with Cystic Fibrosis. Doctoral dissertation, University of Essex, December.
- Taylor D.R., Puig M., Darnell M.E., Mihalik K., Feinstone S.M. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J. Virol. 2005;79:6291–6298. doi: 10.1128/JVI.79.10.6291-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira V., Feio M.J., Bastos M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012;51:149–177. doi: 10.1016/j.plipres.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Tripathi S., Verma A., Kim E.J., White M.R., Hartshorn K.L. LL-37 modulates human neutrophil responses to influenza A virus. J. Leukoc. Biol. 2014;96:931–938. doi: 10.1189/jlb.4A1113-604RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoek A.H., Mevius D., Guerra B., Mullany P., Roberts A.P., Aarts H.J. Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2011;2:203. doi: 10.3389/fmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanCompernolle S.E., Taylor R.J., Oswald-Richter K., Jiang J., Youree B.E., Bowie J.H., Tyler M.J., Conlon J.M., Wade D., Aiken C., Dermody T.S., KewalRamani V.N., Rollins-Smith L.A., Unutmaz D. Antimicrobial peptides from amphibian skin potently inhibit human immunodeficiency virus infection and transfer of virus from dendritic cells to T cells. J. Virol. 2005;79:11598–11606. doi: 10.1128/JVI.79.18.11598-11606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer L.S., Lan Y., Abbate V., Ruh E., Bui T.T., Wilkinson L.J. Conformational flexibility determines selectivity and antibacterial, antiplasmodial, and anticancer potency of cationic α-helical peptides. J. Biol. Chem. 2012;287:34120–34133. doi: 10.1074/jbc.M112.359067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigant F., Santos N.C., Lee B. Broad-spectrum antivirals against viral fusion. Nat. Rev. Microbiol. 2015;13:426–437. doi: 10.1038/nrmicro3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang S., Li D., Wei D.Q., Zhao J., Wang J. Human intestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2. Gastroenterology. 2020;159:1145. doi: 10.1053/j.gastro.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang S., Li D., Chen P., Han S., Zhao G., Wang J. Human cathelicidin inhibits SARS-CoV-2 infection: killing two birds with one stone. ACS Infect. Dis. 2021;7:1545–1554. doi: 10.1021/acsinfecdis.1c00096. [DOI] [PubMed] [Google Scholar]
- Watson, A., Ferreira, L., Hwang, P., Xu, J., Stroud, R., 2020. Peptide Antidotes to SARS-CoV-2 (COVID-19). bioRxiv 08.06.238915.
- Weissmann G., Zurier R.B., Spieler P.J., Goldstein I.M. Mechanisms of lysosomal enzyme release from leukocytes exposed to immune complexes and other particles. J. Exp. Med. 1971;134:149–165. [PMC free article] [PubMed] [Google Scholar]
- Wiens M.E., Smith J.G. Alpha-defensin HD5 inhibits human papillomavirus 16 infection via capsid stabilization and redirection to the lysosome. MBio. 2017:8. doi: 10.1128/mBio.02304-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhindoli D., Pacor S., Benincasa M., Scocchi M., Gennaro R., Tossi A. The human cathelicidin LL-37—A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta. 2016;1858:546–566. doi: 10.1016/j.bbamem.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Li J., Feng Y., Chai J., Wu J., Hu Y., Tian M., Lu W., Xu X., Zou M. Brevinin-2GHk, a peptide derived from the skin of Fejervarya limnocharis, inhibits zika virus infection by disrupting viral integrity. Viruses. 2021;13:2382. doi: 10.3390/v13122382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Zhou C., Yin S., Chai J., Zeng B., Wu J., Li Y., Li L., Xu X. Fejerlectin, a Lectin-like Peptide from the Skin of Fejervarya limnocharis, Inhibits HIV-1 Entry by Targeting Gp41. ACS Omega. 2021;6:6414–6423. doi: 10.1021/acsomega.1c00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.C., Zhang Z.W., Chen Y.E., Yuan M., Yuan S., Bao J.K. Antiviral and antitumor activities of the lectin extracted from Aspidistra elatior. Z. Nat. C. J. Biosci. 2015;70:7–13. doi: 10.1515/znc-2014-4108. [DOI] [PubMed] [Google Scholar]
- Yam E.L.Y., Hsu L.Y., Yap E.P.H., Yeo T.W., Lee V., Schlundt J., Wilder-Smith A. Antimicrobial Resistance in the Asia Pacific region: a meeting report. Antimicrob. Resist. Infect. Control. 2019;8:202. doi: 10.1186/s13756-019-0654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G., Pomplun, S., Loftis, A.R., Tan, X., Loas, A., Pentelute, B.L., 2020. Investigation of ACE2 N-terminal fragments binding to SARS-CoV-2 Spike RBD. BioRxiv.
- Zhang R., Jiang X., Qiao J., et al. Antimicrobial peptide DP7 with potential activity against SARS coronavirus infections. Signal Transduct. Target. Ther. 2021;6:140. doi: 10.1038/s41392-021-00551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhou J., Zhang K., Chu H., Liu D., Poon V.K., Chan C.C., Leung H.C., Fai N., Lin Y.P., Zhang A.J., Jin D.Y., Yuen K.Y., Zheng B.J. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016;6:22008. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., To K., Sze K.H., Yung T.T., Bian M., Lam H., Yeung M.L., Li C., Chu H., Yuen K.Y. A broad-spectrum virus- and host-targeting peptide against respiratory viruses including influenza virus and SARS-CoV-2. Nat. Commun. 2020;11:4252. doi: 10.1038/s41467-020-17986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohrab F., Askarian S., Jalili A., Kazemi Oskuee R. Biological properties, current applications and potential therapeautic applications of brevinin peptide superfamily. Int. J. Pept. Res. Ther. 2019;25(1):39–48. doi: 10.1007/s10989-018-9723-8. [DOI] [PMC free article] [PubMed] [Google Scholar]