Abstract
Myxococcus xanthus responds to blue light by producing carotenoids. Several regulatory genes are known that participate in the light action mechanism, which leads to the transcriptional activation of the carotenoid genes. We had already reported the isolation of a carotenoid-less, Tn5-induced strain (MR508), whose mutant site was unlinked to the indicated regulatory genes. Here, we show that ΩMR508::Tn5 affects all known light-inducible promoters in different ways. It blocks the activation of two of them by light but makes the activity of a third one light independent. The ΩMR508 locus has been cloned and sequenced. The mutation had occurred at the promoter of a gene we propose is the M. xanthus ortholog of ihfA. This encodes the α subunit of the histone-like integration host factor protein. An in-frame deletion within ihfA causes the same effects as the ΩMR508::Tn5 insertion. Like other IhfA proteins, the deduced amino acid sequence of M. xanthus IhfA shows much similarity to HU, another histone-like protein. Sequence comparison data, however, and the finding that the M. xanthus gene is preceded by gene pheT, as happens in other gram-negative bacteria, strongly argue for the proposed orthology relationship. The M. xanthus ihfA gene shows some unusual features, both from structural and physiological points of view. In particular, the protein is predicted to have a unique, long acidic extension at the carboxyl terminus, and it appears to be necessary for normal cell growth and even vital for a certain wild-type strain of M. xanthus.
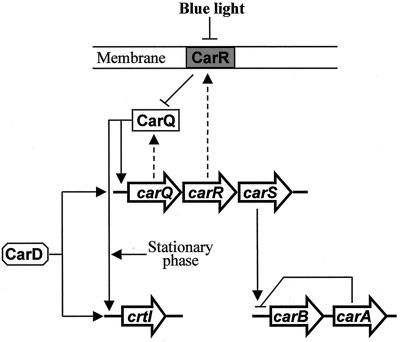
Cells of the gram-negative bacterium Myxococcus xanthus accumulate carotenoid pigments only when they are illuminated with blue light (13). This response is now understood as a case of differential gene expression. Light triggers a sequence of regulatory actions that activate transcription of the carotenoid genes. These actions are represented in the model shown in Fig. 1. The model is based mainly in the properties and interactions of mutations affecting the light response, expression studies using lacZ transcriptional fusions to different promoters, and sequence analysis of the involved genes (for reviews, see references 34 and 35).
FIG. 1.
Light induction of gene expression in M. xanthus. Three unlinked loci are represented (carQRS, crtI, and carB-carA). Genes are indicated by big arrows, which also indicate the direction of transcription. Discontinuous arrows connect some genes with their gene products. Continuous arrows, positive regulation; blunt-ended lines, negative regulation. Blue light inactivates the membrane-bound protein CarR; therefore, protein CarQ (an ECF-ς factor) can activate the promoters of crtI, which encodes an early enzyme for carotenoid synthesis, and the carQRS operon. This requires the action of protein CarD, which is produced in a light-independent manner. Light activation of crtI also requires entry into the stationary phase. The carB operon (represented by a single arrow) contains six genes coding for different carotenogenic enzymes. The carB promoter is repressed in the dark by the carA gene product. This negative action is somehow prevented by high expression of carS in the light.
All but one of the known carotenoid genes of M. xanthus are grouped together in a single, light-activated operon (the carB operon). Included in this operon are the genes required for the synthesis of phytoene (the first C40 carotene precursor) and for other steps downstream in the pathway (10, 70). The one exception is gene crtI (previously called carC) which is also controlled by a light-inducible promoter. The crtI gene product is required for phytoene dehydrogenation (19). The effect of light on the carB and crtI promoters is mediated by the action of several regulatory genes. The genes carQ, carR, and carS are grouped together in the carQRS operon, are unlinked to carB or crtI, and are also controlled by a light-inducible promoter (51). Protein CarQ is a member of the extracytoplasmic function (ECF) subfamily of ς factors (27, 48). Protein CarR is a negative regulator of the whole system (4, 19, 33). Evidence have been provided for CarR, a membrane-spanning protein, acting as an anti-ς factor which in the dark sequesters protein CarQ to the membrane (27). Illumination of the cells somehow results in loss of CarR, so CarQ is free to activate both the carQRS and the crtI promoters. These two promoters share two DNA segments, centered at the −31 and −10 positions, that most likely correspond to the binding sites of the ς-factor CarQ (49). These binding sites are absent from the carB promoter, which is activated by light through a different mechanism, only indirectly connected to the CarR-CarQ interplay. In the dark, the activity of the carB promoter is repressed by the product of carA, a gene which is located immediately downstream of the carB operon (4, 50). CarA is a DNA-binding protein related to the MerR family of transcriptional regulators (10). The negative action of CarA on the carB promoter is prevented by the product of gene carS, the third gene found at the carQRS operon. The action mechanism of protein CarS is unknown, but it seems to require the CarQ-dependent, high expression level of carS that takes place only in the light (51, 57).
The product of another gene, unlinked to the others and named carD, is required independently for light activation of the carQRS and the crtI promoters. Expression of gene carD itself is not regulated by light. Protein CarD, which contains a DNA-binding domain similar to that of eukaryotic HMGI(Y) proteins, has been shown to bind a particular cis-acting site of the carQRS promoter region (58). The gene carD was identified by screening a large collection of mutants for lack of carotenoid production. In the same screening, another mutation was found that was unlinked to carD or any other of the genes mentioned above (57). We report here data on the further characterization of this mutation, as well as on the cloning and identification of the mutated gene. The mutation alters the normal regulation of all known light-inducible promoters. DNA sequence analysis and other data on the cloned gene clearly indicate that it corresponds to the M. xanthus ortholog of ihfA. This codes for the α subunit of the integration host factor (IHF), a widespread, histone-like bacterial protein (55). Unlike other bacteria, M. xanthus requires an intact ihfA gene for normal vegetative growth. The predicted amino acid sequence of the IhfA protein of M. xanthus shows unique structural features, most notably a long, acidic extension of the carboxyl terminus.
MATERIALS AND METHODS
Bacterial strains, transducing phages, and growth conditions.
Most of the M. xanthus strains used in this study are listed in Table 1, together with their phenotype, genotype, and origin. Other strains are introduced in the text. The standard strains DK1050 and DK1622 show normal light-induced synthesis of carotenoids (Car+ phenotype). Car− stands for lack of carotenoid synthesis, and Carc stands for light-independent production of carotenoids. Some strains carried in vitro-constructed lacZ fusions that were integrated into the M. xanthus chromosome by homologous recombination (see below). The reporter gene retained the normal translation start signal preceded by stop codons in all three reading frames; therefore, it produced transcriptional but not translational fusions. Insertion of transposon Tn5 causes resistance both to kanamycin (Kmr) and phleomycin (Phlr), and insertion of Tn5-132 causes resistance to tetracycline (Tcr). For cloning purposes, Escherichia coli strains DH5α (30) and MC1061 (14) were used.
TABLE 1.
M. xanthus strains used in this study
| Group and straina | Phenotypeb | Genotype | Source, reference, or derivationc |
|---|---|---|---|
| Group I | |||
| DK1050 | Car+ | 69 | |
| DK1622 | Car+ | 37 | |
| MR151 | Carc | carR3 | 4 |
| MR358 | Car− | ihfA2 | This study |
| MR354 | Car− | carR3 ihfA2 | This study |
| MR508 | Car− Kmr Phlr | carR3 ΩMR508::Tn5 | 57 |
| MR521 | Car− Tcr | carR3 ΩMR508::Tn5-132 | P1::Tn5-132 × MR508→Tcr Kms |
| Group II | |||
| MR397 | Kmr LacZi | carQ::lacZ (pDAH217) | 33, 57 |
| MR533 | Kmr LacZi | carB::lacZ (pMAR113) | 70 |
| MR553 | Kmr LacZi | crtI::lacZ (pMAR206) | 19 |
| MR624 | Kmr LacZc | carD::lacZ (pMAR511) | 57 |
| MR395 | Kmr LacZc | carR3 carQ::lacZ | pDAH217 × MR151→Kmr LacZc |
| MR555 | Kmr LacZc | carR3 crtI::lacZ | 19 |
Strains are classified into two different groups for clarity. Group I includes standard strains, followed by strains carrying single and double Car mutations; group II includes strains carrying a lacZ transcriptional fusion (the integrated plasmid that carries the transcriptional fusion is indicated in parentheses in the genotype column). All mutant strains in group I and all of the strains in group II are derived from the standard strain DK1050.
Only the relevant phenotypes are indicated. LacZi, light-inducible synthesis of β-galactosidase; LacZc, high level of β-galactosidase in the dark and in the light.
A brief description is given for two of the strains constructed for this work (MR521 and MR395). In these cases, the plasmid or phage used as donor of the antibiotic marker is named in the first place.
The rich medium CTT (12) and the exact culture conditions for growth of M. xanthus in the dark and in the light have been previously described (19). For generalized transduction between M. xanthus strains, phage Mx4-LA27 was used. The phage and the conditions used for transduction of Kmr and Tcr have been previously described (2). The transduction of the Tn5-encoded Phlr has not been previously reported in M. xanthus. About 2.108 phages grown on the donor strain were mixed with an equal number of recipient cells. Adsorption proceeded for 30 min at room temperature, and the transduction mixture was plated in CTT supplemented with 5 μg of phleomycin (the antibiotic was sterilized by filtration and added to the autoclaved bottom agar only after this cooled down to about 50°C) per ml. After 24 h, an additional 3 ml of CTT soft agar was poured, containing enough phleomycin to raise the average concentration in the plate to 15 μg/ml. Phage P1::Tn5-132 was used for the in situ replacement of a Tn5 inserted in the chromosome of M. xanthus with Tn5-132 (2). Coliphage P1clr100Cm (P1 in the text) was used to transfer plasmids from E. coli to M. xanthus (24).
Plasmids.
Cloning vector pDAH160 (33) carries a Kmr gene and the incompatibility region of P1 for transfer of the plasmid from E. coli to M. xanthus by P1-specialized transduction. Those two elements are also present in other plasmids used in this study. Plasmid pDAH274 (51) carries a lacZ transcriptional probe. Plasmid pDAH217 contains a lacZ transcriptional probe fused to the light-inducible promoter of the carQRS operon (see Fig. 1). Like all other plasmids used here, pDAH217 cannot replicate in M. xanthus cells but can integrate into the M. xanthus chromosome by homologous recombination. Integration of pDAH217 produces a tandem duplication of the cloned DNA; therefore, a normal copy of the carQRS operon is generated (33). Similarly, plasmids pMAR113, pMAR206, and pMAR511 contain a lacZ transcriptional fusion to the promoters of carB (70), crtI (previously called carC 19), and carD (57), respectively. For standard cloning procedures, plasmid pUC19 was used (59).
Nucleic acid manipulations.
The use of plasmid pDAH160 to clone wild-type DNA around a Tn5 insertion site has been described in detail previously (33). We used this procedure to obtain plasmid pMAR604, which carries a 9-kb DNA fragment from the M. xanthus wild-type strain DK1050 (Fig. 2). Different portions of the DNA cloned in pMAR604 were recloned by using the appropriated restriction enzymes or exonuclease III treatment. Southern analysis and all nucleic acid and enzymatic manipulations were done according to standard procedures (72). The primer extension reaction was carried out as described elsewhere (19). Exonuclease III came from Pharmacia. All other enzymes came from Roche.
FIG. 2.
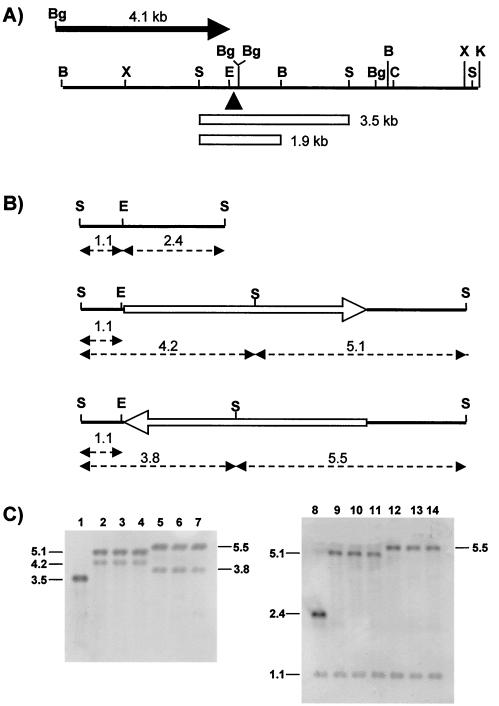
The phenotypic instability of mutant MR508 correlates with the in situ inversion of Tn5. (A) The black line is a physical map of the 9.0-kb DNA restriction fragment from wild-type M. xanthus that was cloned for this work (B, BamHI; Bg, BglII; C, ClaI; E, EcoRI; K, KpnI; S, SalI; X, XhoI). The black triangle indicates the insertion site of Tn5 in MR508. The two open rectangles correspond to restriction fragments mentioned in the text. The black arrow on top represents a 4.1-kb DNA fragment that extends from a BglII restriction site within the vector polylinker to a BglII site within the ihfA coding region. This fragment was cloned, in the orientation indicated by the arrow, in a lacZ promoter probe vector (see the text). (B) Numbers are distances in kilobases. At the top is shown the 3.5-kb, SalI restriction fragment in panel A. This was used as a probe in the Southern experiments shown in panel C. An EcoRI site located 35 bp upstream of the ΩMR508::Tn5 site is indicated. In the middle and at the bottom are representations of transposon Tn5 (open arrows) inserted, in the two possible orientations, at the ΩMR508 site. The single, asymmetrically located, SalI restriction site of Tn5 is indicated. Also indicated are the sizes of restriction fragments that should be detected with the 3.5-kb probe. (C) Southern analysis of the parental strain MR151 (lanes 1 and 8), three independent colonies of the MR508 mutant (lanes 2 to 4 and 9 to 11), and three independent MR508 revertants (lanes 5 to 7 and 12 to 14). DNA in lanes 1 to 7 was digested with SalI. DNA in lanes 8 to 14 was digested with both SalI and EcoRI.
Nucleotide sequencing and sequence analysis.
A DNA fragment of about 1.4 kb from plasmid pMAR604, which complements the Car− phenotype due to mutation ΩMR508::Tn5, was used as the starting material. The fragment was cloned into pUC19 in both orientations and progressively deleted with exonuclease III. The two strands were sequenced by using the dideoxy chain termination method (74). Each DNA sample was sequenced at least four times. To deal with compressions at G+C-rich DNA stretches, dGTP was replaced by dITP. The nucleotide sequence of the 1.4-kb DNA fragment appears in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession number AJ297483. In the cloning procedure mentioned above (33), a necessary intermediate product is a plasmid carrying part of the Tn5 inserted at the ΩMR508 site and the adjacent M. xanthus DNA. This plasmid was also sequenced, using a primer complementary to the appropriate DNA stretch from Tn5, to determine the exact location of insertion ΩMR508::Tn5. For comparison with databases, the BLAST programs provided by the BCM search launcher (http://www.hgsc.bcm.tmc.edu/ 1) were used. The helical propensity of the carboxyl end of M. xanthus IhfA (see Fig. 6) was analyzed by the nearest-neighbor prediction method (71) provided by the same launcher.
FIG. 6.
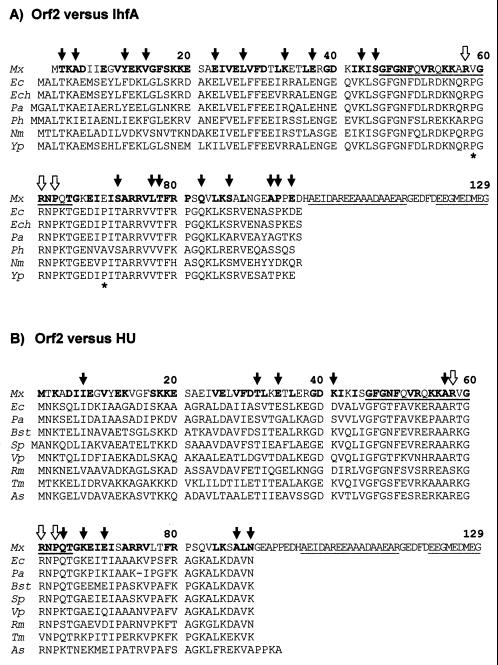
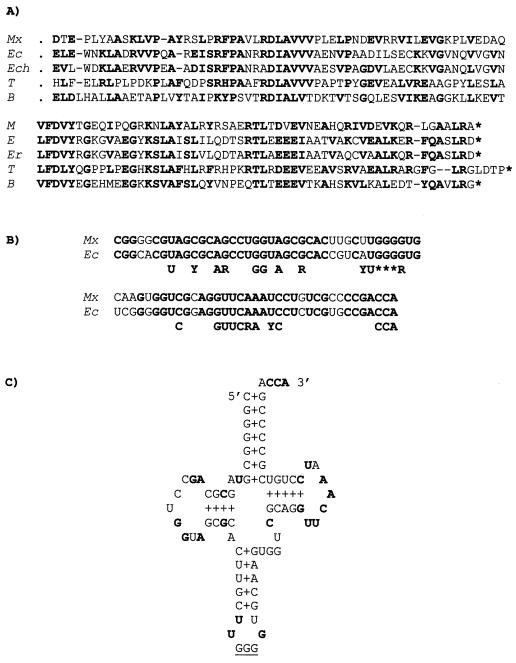
Alignment of the amino acid sequences of Orf2 (Fig. 4) and proteins IhfA (A) and HU (B) from different bacterial species. Numbers correspond to residues in Orf2. The amino acids of Orf2 which are similar or identical to those present at the same position in four or more members of the corresponding IhfA or HU families are in bold letters. The peptide stretch that should be missing in the ihfA2 deletion mutant (residues 46 to 65) and two peptide stretches with high helical propensity at the carboxyl end of Orf2 are underlined. The white arrows on top of the two groups of sequences (residues 58, 61, and 63) point to the amino acids known to be particularly important for DNA interaction, both for protein IhfA and HU (see the text). The black arrows in panel A mark Orf2 residues which are similar or identical to those found at the same position only in the IhfA protein family. The black arrows in panel B mark those Orf2 residues which are similar or identical to those found at the same position only in the HU protein family. The two asterisks below the IhfA sequences (panel A) mark two prolines (absent from Orf2) that are unique to the IhfA subfamily. Mx, M. xanthus (Orf2, Fig. 4); Ec, E. coli (IhfA 53; HU-1 38; Ech, E. chrysanthemi (SwissProt accession number P37982); Pa, P. aeruginosa (IhfA and HU 17); Ph, Pasteurella haemolytica (31); Nm, Neisseria meningitidis (63); Yp, Yersinia pseudotuberculosis (18); Bst, Bacillus stearothermophilus (61); Sp, Streptococcus pyogenes (78); Vp, Vibrio proteolyticus (22); Rm, Rhizobium meliloti (47); Tm, Thermotoga maritima (56); As, Anabaena sp. (54).
Expression of β-galactosidase.
Rapid determination of β-galactosidase production was carried out by examining colony color on plates containing 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml. Quantitative analysis of β-galactosidase on dark- or light-grown liquid cultures was performed as previously described (4). The reported data of enzyme-specific activities, given in nanomoles of o-nitrophenol produced per minute per milligram of protein, are the average of three or more independent determinations. The standard deviations never exceed 15% of the average.
RESULTS
Instability of a new mutation affecting the M. xanthus response to blue light.
Colonies of the M. xanthus standard strain DK1050 turn red in the light due to the accumulation of carotenoids. Strain MR151 carries a mutation at gene carR (carR3), so it produces red colonies both in the dark and in the light (Fig. 1). Strain MR508 was obtained from MR151, by Tn5 mutagenesis and screening among the kanamycin-resistant (Kmr) colonies for a carotenoid-less (Car−) phenotype. MR508 was shown to carry a single insertion (ΩMR508::Tn5) that cosegregated with the Car− phenotype (57). MR508 is not completely blocked in carotenoid synthesis, as it produced slightly pink colonies. Chemical analysis of MR508 cells detected the same carotenoids as in MR151 but at a much lower level (about 3%; unpublished results). Insertion ΩMR508::Tn5 also blocked carotenoid synthesis in a wild-type genetic background. M. xanthus wild-type strain DK1050 was transduced for the Kmr marker of MR508. About 100 transductant colonies were checked, and they all failed to turn red upon illumination.
We frequently observed MR508-derived colonies that had spontaneously reverted to the parental, red phenotype. To quantify this phenomenon, six independent mutant colonies were grown in liquid and plated for single colonies, always in the presence of kanamycin. Red colonies were observed in all cases, at a frequency that varied between 3 × 10−4 and 1 × 10−3. Six of these red colonies, one from each culture, were used in experiments similar to the one just described, and they all gave back mutant colonies at a frequency that varied between 10−4 and 10−3. Chemical analysis of dark-grown cultures of some of the revertants (red colonies) detected the same carotenoids as in the parental strain MR151, at a level ca. 60% of that produced by MR151.
Strain MR508 was used for in situ replacement of Tn5 by Tn5-132 (resistance to tetracycline, Tcr). A total of 200 Tcr transductants were checked and, as expected, they were all sensitive to kanamycin. One of the transductants was purified and named MR521. MR521 was used as a recipient for the transduction of the Kmr marker from MR508 or several of the MR508-derived red revertants. For each transduction, ca. 100 Kmr transductants were checked, and they were all sensitive to tetracycline. These data argue against the phenotypic reversion being due to the mobilization of Tn5 to a new, distant site.
As explained later, the insertion site ΩMR508 is located at the promoter region of the M. xanthus gene ihfA. A possible explanation for the phenotypic instability of MR508 could be the in situ inversion of the inserted transposon, assuming that each Tn5 orientation had a different effect on the expression of the ihfA gene. That inversion might occur by intramolecular recombination between the 1.5-kb inverted repeats found at both ends of Tn5. This hypothesis was tested by Southern analysis, once the involved DNA region was cloned and characterized (Fig. 2A; see also below). Figure 2B depicts a SalI restriction fragment (3.5 kb) from wild-type M. xanthus, where insertion ΩMR508::Tn5 was located. Also shown in Fig. 2B are an EcoRI site in the M. xanthus DNA, very near the ΩMR508::Tn5 insertion site, and the single SalI site of Tn5, which is asymmetrically located with respect to the ends of the transposon. DNA was isolated from MR151, from five independent cultures of MR508, and from five independent, MR508-derived red revertants. DNA samples were treated with SalI or SalI-EcoRI and subjected to Southern analysis using as a probe the 3.5-kb SalI fragment. All of the MR508 samples produced the bands expected from Tn5 being inserted in a particular orientation, whereas the DNA samples from all of the revertants produced the bands expected from Tn5 being inserted at the same site but in the opposite orientation. Three representatives samples are shown in Fig. 2C.
Cloning of the M. xanthus gene ihfA allowed us to generate an in-frame deletion within the coding region of that gene (see below). The new mutation, which is not associated with any antibiotic resistance marker, was named ihfA2. It caused a Car− phenotype, and it behaved as a normal, stable mutation (reversion rate, <10−7). The wild-type and carR3 derivatives of M. xanthus carrying mutation ihfA2 were named MR358 and MR354, respectively.
The ihfA mutations alter normal expression from the light-inducible promoters.
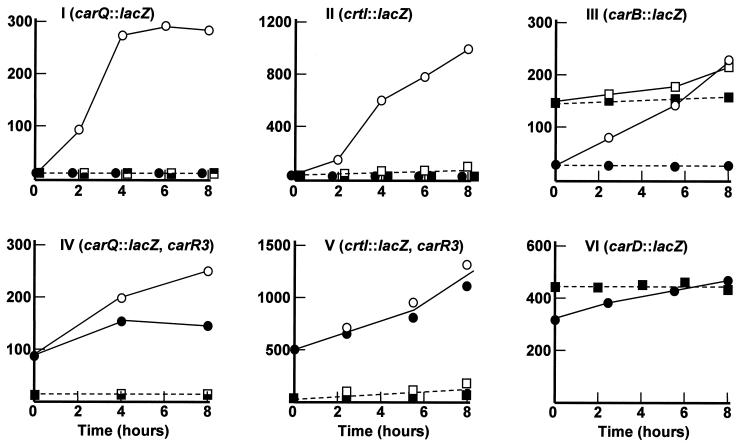
M. xanthus strains, which carry gene lacZ fused transcriptionally to the promoter of carQRS or crtI, have been obtained (Table 1). They were constructed by chromosomal integration (homologous recombination) of plasmid-based lacZ fusions to the corresponding light-inducible promoters. In both cases, plasmid integration was selected, taking advantage of a Kmr gene present in the plasmid vector. In addition to the Kmr gene, transposon Tn5 contains a gene that confers resistance to phleomycin (Phlr) (16). To test the effect of insertion ΩMR508::Tn5 on the two indicated promoters, the Phlr phenotype of MR508 was transduced into the corresponding Kmr LacZi strains. To test the effect of mutation ihfA2 on the same promoters, the original plasmids carrying the lacZ transcriptional fusions were separately transferred to strains MR358 and MR354 by P1-mediated transduction and selection for Kmr. For each transduction, 50 independent Phlr or Kmr colonies were tested on light- or dark-grown X-Gal plates. They all failed to develop the blue color indicative of a significant production of β-galactosidase, both in the dark and in the light. Quantitative analysis of lacZ expression confirmed that the ihfA mutations block the activation of the carQRS and crtI promoters by light (Fig. 3I and II), as well as the light-independent activity of those promoters which is normally caused by the carR3 mutation (Fig. 3IV and V).
FIG. 3.
Effect of ihfA mutations on the activity of different promoters. At time zero cell samples growing exponentially in the dark were divided in two, one for the dark (closed symbols) and the other for the light (open symbols). Samples in panels II and V were at the early stationary phase. Panel VI shows dark-grown samples only. The data are β-galactosidase specific activities from M. xanthus strains carrying a lacZ transcriptional fusion to the gene indicated in the corresponding panel. Also indicated is the carR3 mutation present in two of the strains (panels IV and V). Strains are as follows: I, MR397 (circles) and two derivatives carrying either insertion ΩMR508::Tn5 or the ihfA2 mutation (the same square symbols are used for both strains); II, MR553 (circles) and two derivatives carrying either insertion ΩMR508::Tn5 or the ihfA2 mutation (the same square symbols are used for both strains); III, MR533 (circles) and its ihfA2 derivative (squares); IV, MR395 (circles) and its ihfA2 derivative (squares); V, MR555 (circles) and its ihfA2 derivative (squares); and VI, MR624 (circles) and its ihfA2 derivative (squares). For clarity, some square and round symbols for low β-galactosidase activities in panels I and II have been displaced slightly to the left or to the right of the real time at which the samples were taken.
Transcription of both carQRS and crtI is driven by ς-factor CarQ (Fig. 1). Therefore, the effect of the ihfA mutations on crtI could be attributable to the reduced expression of the carQRS operon. A mutant condition is known in M. xanthus in which carQRS is transcribed in a light- and CarQ-independent manner. It corresponds to transposon insertion ΩDK1910::Tn5-132, which is close to, but does not interrupt, the carQRS operon. An outward promoter activity generated by the transposon results in carQRS being transcribed in a constitutive manner (33). This transposon insertion does not affect the light dependence of the crtI promoter, since CarQ remains inactive in the dark, due to the anti-ς factor CarR (57) (Fig. 1). In control experiments (results not shown) we first showed that the ihfA2 mutation did not affect the heterologous promoter activity generated by ΩDK1910::Tn5-132. This insertion was then transduced (Tcr selection) into the M. xanthus strain MR363 (crtI::lacZ ihfA2). When tested on X-Gal plates, all of the transductants showed low expression of β-galactosidase in the dark and high expression of β-galactosidase in the light. Quantitative analysis of randomly picked transductants confirmed that insertion ΩDK1910::Tn5-132 had counteracted the effect of ihfA2 on the crtI promoter (β-galactosidase specific activities of >900 U in cultures that have been illuminated for 8 h, which compared well with the activity of the crtI promoter in the wild-type, as shown in Fig. 3II).
We also tested the effect of the ihfA2 mutation on the activity of the carB promoter by using the previously reported carB::lacZ plasmid (Table 1). The behavior of all tested transductants on X-Gal plates and the results of β-galactosidase assays indicated that mutation ihfA2 causes high expression in the dark from the normally light-inducible carB promoter (Fig. 3III). Finally, the effect of the ihfA2 mutation on the expression of gene carD was tested, also using a previously described carD::lacZ plasmid (Table 1). No significant effect was detected in this case (Fig. 3VI).
Cloning and sequence analysis of the M. xanthus gene ihfA.
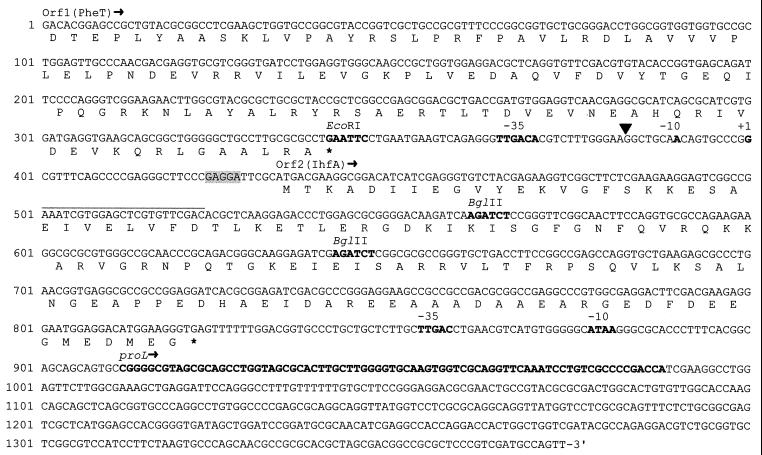
A 9.0-kb restriction fragment of wild-type DNA astride insertion site ΩMR508 was cloned in a plasmid vector (pDAH160, Kmr) that can be shuttled between E. coli and M. xanthus. The chimeric plasmid was named pMAR604. A restriction map of the cloned DNA is shown in Fig. 2A. The 9.0-kb fragment, as well as two shorter restriction fragments (3.5 and 1.9 kb, Fig. 2A) could complement the ΩMR508::Tn5 mutation. This mutation was also complemented by a 1.4-kb DNA fragment obtained by partial deletion (exonuclease III treatment) of the 1.9-kb fragment (results not shown). Both strands of the 1.4-kb DNA fragment were sequenced. A search for potential coding regions (open reading frames [ORFs]) within the sequenced DNA was carried out, taking into account the bias for G or C at the third codon positions which is predicted from the high G+C content of the M. xanthus DNA (9, 52). As shown in Fig. 4, the 3′ end of a truncated ORF (orf1) and a complete, nonoverlapping ORF (orf2) were detected. The two ORFs run in the same direction and are separated by 91 bp.
FIG. 4.
Sequence of the ihfA locus of M. xanthus. Nucleotide positions are shown on the left. The predicted amino acid sequence for the carboxyl end of Orf1 (PheT; see Fig. 5A) and for the complete Orf2 (IhfA; see Fig. 6A) are shown. Stop codons are indicated by asterisks. The putative ribosome-binding site for Orf2 is shadowed. The ihfA transcriptional start (nucleotide position 400) is marked +1. It was identified by extension of a primer complementary to DNA from positions 501 to 523 (upper line). The black triangle marks the insertion site of Tn5 in the mutant strain MR508. Other features, marked in bold letters, are as follows (from 5′ to 3′): the EcoRI site shown in Fig. 2A and B, the −35 and −10 sites of the ihfA promoter, the two BglII restriction sites used to generate the in-frame deletion mutation ihfA2 (the first one of these BglII sites was also used to insert a lacZ transcriptional probe, as mentioned in the text), the putative −35 and −10 sites of the proL promoter, and the DNA stretch (positions 912 to 988) that likely corresponds to the M. xanthus gene proL (see Fig. 5B and C).
The amino acid sequence predicted from the incomplete ORF showed strong similarity with the carboxyl end of the bacterial protein PheT (phenylalanyl-tRNA synthetase, β-chain) (Fig. 5A). The protein predicted from the complete ORF, which should be the one affected by insertion ΩMR508::Tn5, showed strong sequence similarity with the bacterial protein IhfA, the α subunit of the IHF (Fig. 6A). IhfA belongs to a family of histone-like bacterial proteins that includes protein HU. As shown in Fig. 6B, the Orf2 protein also showed strong sequence similarity with bacterial HU proteins. However, several lines of evidence strongly suggest that the M. xanthus gene is the ortholog of the bacterial gene for IhfA (see Discussion). Thus, it was named ihfA (85).
FIG. 5.
PheT and proL genes of M. xanthus. (A) Amino acid sequence similarity between the truncated Orf1 (Fig. 4) and the carboxyl end of protein PheT (phenyl-alanyl-tRNA synthetase, β-chain) from different bacterial species. Identical or conserved amino acids in at least three of the species are indicated in bold letters. Mx, M. xanthus; Ec, E. coli (53); Ech, Erwinia chrysanthemi (SwissProt, accession no. P37984); T, Thermus thermophilus (40); B, Bacillus subtilis (11). (B) Nucleotide sequence similarity between the tRNA from the proL gene (Pro-tRNA) of E. coli (45) and the RNA predicted from the alleged homologous gene of M. xanthus (Fig. 4). Residues which are particularly well conserved in all tRNA molecules are indicated below (77) (Y = C or T; R = G or A). The anticodon triplet is also indicated (asterisks). (C) Cloverleaf representation of the predicted Pro-tRNA of M. xanthus. The conserved residues indicated in panel B are in bold letters. The anticodon is underlined.
Eighty-eight nucleotides downstream of the ihfA stop codon a 77-residue-long DNA stretch was found that showed strong similarity (85% identity) with the proL gene of E. coli and other bacteria (not shown). Gene proL codes for one of the three Pro-tRNA molecules found in E. coli. Nucleotide positions conserved in tRNA molecules were all present in the M. xanthus sequence, as well as the expected triplet (GGG) in the anticodon loop (Fig. 5B). Moreover, in its RNA form, the indicated stretch of the M. xanthus DNA is predicted to generate the characteristic helix and loop domains of a tRNA molecule (Fig. 5C). When read in all possible frames, the rest of the sequenced DNA produced ORFs that were very short or that showed only weak similarities with known protein or DNA sequences.
The transcription start site of ihfA was identified by primer extension analysis. In this analysis, we used RNA isolated from the wild-type strain DK1050 and a synthetic primer complementary to a coding sequence downstream of the predicted ihfA start codon (Fig. 4). Three independent experiments produced the same, single extension product (not shown), which located the ihfA transcriptional start 32 nucleotides upstream of the start codon. At the −35 position, an hexanucleotide (TTGACA) that perfectly matches the consensus for the −35 binding site of the major bacterial ς factor was found (29). At the −10 position, an AT-rich sequence was found (considering the high G+C content of the M. xanthus DNA), but that sequence (AACAGT) does not conform to the consensus for the −10 binding site of the indicated ς factor (Fig. 4).
The insertion site of Tn5 in the mutant strain MR508 was located by cloning and sequencing the appropriate DNA fragment from that strain. The results (not shown) indicated that Tn5 had inserted at the promoter region, 18 residues upstream of the transcription start site.
The ihfA2 mutation.
To confirm the involvement of the ihfA gene in the Car− phenotype of the MR508 strain, an in-frame deletion of that gene was generated. For this, we took advantage of two BglII restriction sites that were located within the ihfA coding region. They were separated by 81 bp, and they were the only BglII restriction sites present in the 3.5-kb fragment shown in Fig. 2A. Deleting the BglII fragment would result in a protein lacking a 27-residue internal peptide. This peptide includes amino acid residues which are critical for the activity of the IhfA protein (see Discussion), so the mutated product was expected to be largely inactive. The indicated fragment was digested with BglII, religated, cloned in the appropriate plasmid vector, and separately transduced (Kmr selection) into the wild-type strain DK1050 and the carR3 mutant strain MR151. The plasmid cannot replicate in M. xanthus, so the Kmr transductants should have integrated the plasmid by homologous recombination. In these merodiploid transductants, intramolecular recombination events would result in the removal of both the plasmid vector and either the wild-type or the deleted version of the ihfA gene. In the first case, a Car− and kanamycin-sensitive (Kms) cell should arise, given that the ihfA gene were required for normal carotenoid synthesis. Independent merodiploid transductants were grown separately for about 50 generations in the absence of kanamycin and plated for a Car− phenotype. Colonies showing that phenotype were obtained from all cultures of the DK1050- and MR151-derived merodiploids, the average frequency being about 5%. When checked for sensitivity to kanamycin, most of the Car− colonies proved to be Kms. Southern analysis (not shown) confirmed the presence of the expected deletion in several independent Car− Kms colonies. The deletion mutation, named ihfA2, was the one used in the experiments described above.
We noticed that the ihfA2 mutant colonies, both from DK1050 and from MR151, showed an abnormal appearance. They were much smaller than those produced by the parental strains, and they also showed the characteristic phenotype of a nonmotile colony. Normal M. xanthus colonies are surrounded by a thin layer of gliding cells, moving outward, individually or as a group. This produces a characteristic, irregularly edged “halo” around the colony (32). To the naked eye, the ihfA2 colonies were sharply round. Under the microscope, no single cells or group of cells could be seen outside the sharp edge of the mutant colonies. The small size of the ihfA2 colonies was not simply due to lack of cell motility. When cultured in rich liquid medium (CTT), the generation time of the ihfA2 mutants varied between 9.5 and 10 h, whereas that of the parental strains DK1050 and MR151 varied at between 3.5 and 4 h (not shown).
As in other myxobacteria, M. xanthus responds to starvation conditions by building complex multicellular structures (fruiting bodies) where the vegetative cells differentiate into myxospores (76). Most studies on multicellular development in M. xanthus have used DK1622 as the standard strain. This is different from, although related to, our standard strain DK1050 (69, 82). To test for a role of ihfA in the developmental process, we tried to introduce the ihfA2 deletion in DK1622, using the same procedure described above. Five independent merodiploid transductants were grown for about 50 generations in the absence of kanamycin and plated for a Car− phenotype. More than 100,000 colonies were screened, and none was found to be Car−. About 1,000 of these colonies were tested for a Kms phenotype, and six of them (from three independent merodiploids) were found. Southern analysis (not shown) confirmed that the six Kms colonies had lost the integrated plasmid and the ihfA2 deletion, having retained the ihfA normal allele.
Negative autoregulation of the ihfA gene.
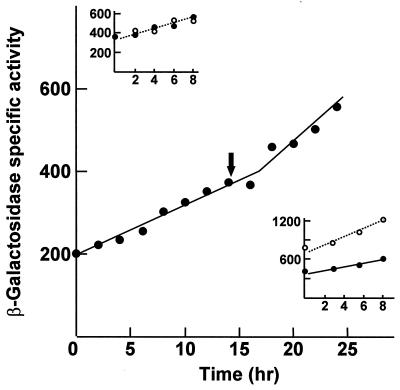
The expression of ihfA was analyzed by β-galactosidase quantitative assays of M. xanthus strains carrying an ihfA::lacZ transcriptional fusion. A BglII DNA fragment that extends from 4 kb upstream to 130 bp downstream of the ihfA start codon (Fig. 2A and 4) was cloned into the unique EcoRI site (blunt ends) of the plasmid vector pDAH274. The EcoRI site is located just upstream of the promoter-less lacZ gene of the vector. The correct orientation for expression of ihfA was assigned by restriction analysis. The chimeric plasmid, named pMAR609, was transferred to the M. xanthus wild-type strain by transduction and selection for Kmr. The transductants should carry a normal copy of the ihfA gene and the ihfA::lacZ fusion. About 100 independent transductants were plated on X-Gal plates, and they all seemed to express the reporter gene at a similar level. Quantitative assays of β-galactosidase were carried out on dark- and light-grown cultures of randomly picked transductants. The assays all produced similar results. A representative example is shown in Fig. 7. The β-galactosidase specific activity increased steadily with cell growth. Entry into the stationary phase produced a moderate stimulation of ihfA expression. Illumination with blue light did not affect that expression (Fig. 7, top inset).
FIG. 7.
Time course expression of the M. xanthus gene ihfA. The β-galactosidase specific activity of a dark-grown culture of a wild-type derivative having lacZ inserted at the ihfA gene is shown. Stationary phase was reached at the time indicated by the arrow. (Top inset) β-Galactosidase expression in the same strain growing exponentially in the dark (closed symbols) or the light (open symbols). (Bottom inset) β-Galactosidase expression in exponential cultures of the same strain (closed symbols) and of a second strain carrying also the ihfA2 mutation (open symbols).
Plasmid pMAR609 was also transduced into different M. xanthus strains carrying mutations at genes that play a regulatory role in the response to blue light. None of the mutations (carQ, carR, carS, carA, or carD) produced a significant effect on the expression of the ihfA gene (results not shown). The same plasmid was also transduced into the ihfA2 mutant strain MR358. As shown in Fig. 7 (bottom inset), the ihfA2 mutation resulted in a twofold increase in β-galactosidase expression, clearly suggesting a negative regulatory action of the ihfA gene on its own expression.
DISCUSSION
The M. xanthus mutant strain MR508 carries a Tn5-induced mutation that blocks blue light-induced carotenoid synthesis. The mutation had been mapped at a site unlinked to all genes previously implicated in the light response (57) (Fig. 1). We show here that the insertion has occurred at the promoter region of the ihfA gene, 18 bp upstream of the transcription start site of that gene. Thus, the phenotype of MR508 could be explained by assuming that the Tn5 insertion reduces the expression of ihfA and that the ihfA gene product is positively required for the normal response to blue light. This explanation is strongly supported by the fact that mutation ihfA2, an in-frame deletion within ihfA, also results in a carotenoid-less phenotype.
Strain MR508 is strikingly unstable, reverting to the parental phenotype at high frequency. This is not a true reversion phenomenon, since the transposon remains at the same site in the pseudorevertants. There is a correlation between the phenotypic reversion and the in situ inversion of the Tn5 DNA. This inversion could be caused by recombination between the two long inverted repeats, IS50L and IS50R, of Tn5 (84). A simple explanation for the observed correlation is that Tn5 generates an outward promoter activity, facing ihfA, when inserted in one orientation, but not in the other. Transcription from within Tn5 has been reported both in E. coli (83) and M. xanthus (33). The function of a mutated version of Tn5 as a recombinational switch for expression of a nearby gene (7) and the preferential action of IS50L versus IS50R to activate downstream genes (41) have also been reported. Judging from the frequency of phenotypic reversion, the in situ inversion of Tn5 is about 50 times more frequent in the M. xanthus strain MR508 than in Rec+ E. coli (84). This may be a context-dependent phenomenon, but even so it is one to be aware of when dealing with other Tn5-induced mutations in M. xanthus.
As deduced from expression analysis of a lacZ transcriptional fusion, both insertion ΩMR508::Tn5 and mutation ihfA2 block almost completely the activation of the carQRS promoter by light. This indicates that in the wild type, the ihfA gene product functions as a positive element at an early stage of the regulatory cascade initiated by the light stimulus (Fig. 1). At present, little is known about how the anti-ς-factor CarR is inactivated by blue light or how CarR blocks ς-factor CarQ in the dark. A role for ihfA in either of those two steps is dismissed, as the ihfA mutations are epistatic over a carR mutation, both for the effect on the Car phenotype and for the effect on the carQRS promoter (Fig. 3IV). Instead, our data suggest that the ihfA gene product participates more directly in the activation of the carQRS promoter, once the action of CarR has been blocked by blue light.
The ihfA gene encodes the α subunit of the IHF. This heterodimeric protein functions as an architectural factor in many processes that involve higher-order protein-DNA complexes, including site-specific recombination, transcriptional regulation, and replication (55). The architectural function of IHF depends on its ability to induce a sharp bend in the DNA, which facilitates the interaction of other components assembled in the nucleoprotein complexes (67). Particularly well known is the role of that bending in the activator-mediated stimulation of ς54-dependent promoters (26, 46, 64). CarQ, the ς factor for the carQRS promoter, is a member of the ECF subfamily of ς factors (27, 49). Molecular details on the functioning of ECF-ς-dependent promoters have not been worked out, but the participation of IHF has been reported at least in one case, the promoter of the Pseudomonas aeruginosa algD gene (86). The activation of this promoter depends on the formation of a high-order looped structure that allows multivalent contacts between RNA polymerase and activator proteins (5, 6). The activation of the carQRS promoter might also depend on multivalent contacts, since it has been shown to require the binding of at least another protein, CarD (58). Thus, the IHF may be an essential architectural element of the appropriated macromolecular complex at the carQRS promoter. We cannot discard, however, the possibility that IHF exerts its effect on carQRS indirectly, for example, by controlling the expression of a yet-unknown gene (we have found that the expression of gene carD itself is not affected by the ihfA mutations).
The ihfA mutations also block the activation of the crtI promoter by blue light or by a carR mutation (Fig. 3). This explains the Car− phenotype of the ihfA mutants, since the crtI gene product is required for an early step in the carotenoid pathway, before the first colored carotene precursor is formed. The crtI promoter is also regulated by ς factor CarQ (19, 49). The effect of the ihfA mutations on crtI is overcome when carQ is expressed from a heterologous, ihfA-independent promoter. Therefore, that effect is due to the lack of CarQ caused by the ihfA mutations and not to the direct involvement of IHF in the activation of the crtI promoter. Unlike carQRS, the crtI gene is transcriptionally activated by light only when the cells have reached the stationary phase or when they have been starved for a carbon source (19). All of this points to a certain versatility of the CarQ-dependent transcription machinery, insofar as the molecular partners it can accommodate.
Mutating the ihfA gene also causes the constitutive expression at a high level of the normally light-inducible carB promoter. The precise molecular mechanism for the light dependence of this promoter is not known. However, it has been established that the carB promoter is repressed in the dark by the product of the nearby gene carA and that this repression is somehow canceled out in the light by the high expression of CarS (4, 51) (Fig. 1). CarA is predicted to contain a helix-turn-helix DNA-binding domain (10), but direct evidence for binding of CarA to the carB promoter is lacking. CarS shows little similarity with previously known proteins (51). The effect of the ihfA mutations on the carQRS operon, which should result in a great reduction in the expression of CarS, appears to be contradictory with the effect of the same mutations on the carB promoter. This raises the possibility that IHF plays a role as a corepressor of the carB promoter that is independent from its role on carQRS. Other cases in which IHF helps to downregulate a promoter have been reported (15, 36, 65). As commented upon for the carQRS operon, an indirect effect, for example, the requirement of IHF for the correct expression of carA, cannot be discarded. Strains and DNA clones obtained in this work should be instrumental in exploring those two alternatives.
As shown in Fig. 6, the predicted amino acid sequence of the protein we have named IhfA shows strong similarity with both the IHF α subunit and the HU proteins from other bacteria. Protein HU is a nonspecific DNA-binding protein that also bends DNA. It is generally found as a homodimeric protein, although in some bacterial species it is formed by two, very similar subunits (60). Data from X-ray crystallography and nuclear magnetic resonance studies have revealed a similar structure for IHF and the homodimeric HU protein (67, 80, 81). The two subunits form a compact body from which two long β-ribbon arms extend. The crystal structure of an IHF-DNA complex has been solved (67). The DNA is wrapped around the body of the protein, thus executing a U-turn, and the two β-ribbon arms curl around the DNA and interact with the minor groove. Most of the bending occurs at two large kinks, where a proline at the tip of the arm of each subunit is intercalated between base pairs. This proline (P65 in E. coli IhfA) is conserved in all members of the IHF/HU family, as well as two arginines (R60 and R63 in E. coli IhfA) that make direct hydrogen bonds to DNA. The three indicated residues are present, at the expected positions, in the M. xanthus protein (open arrows in Fig. 6).
Two lines of evidence strongly support that the gene identified here is the M. xanthus ortholog of ihfA. First, the M. xanthus protein shows higher overall similarity to IhfA than to HU (71 versus 61 identical or conserved residues). In particular, the M. xanthus protein shows the same residue as IhfA, or a chemically related one, at 18 conserved positions at which the HU proteins usually contain a chemically unrelated amino acid (three of the positions, located at the carboxyl terminus of IhfA, are missing in HU; Fig. 6A). Second, and more reassuring from a genetic point of view, the M. xanthus gene is located directly downstream of (and runs in the same direction as) gene pheT (Fig. 4 and 5A). This is exactly the situation found in other gram-negative bacteria, such as E. coli and Salmonella spp. The evolutionary conservation of this chromosomal region does not extend to the short DNA stretch we have sequenced downstream of ihfA. Here, a tRNA gene showing strong similarity to proL is found in M. xanthus, but not in E. coli or Salmonella (8, 73) (Fig. 4 and 5). Other properties shared by the ihfA genes of M. xanthus and E. coli are the steady increase in expression along the growth cycle, the moderate stimulation of that rate when the cells reach the stationary phase, and the negative autoregulation (3) (Fig. 7).
Some unusual features are observed in the predicted amino acid sequence of the M. xanthus protein. The most noticeable is the unique, long extension at the carboxyl end. The extension is very acidic (aspartic or glutamic acid in 15 out of 33 positions) and includes two stretches with high helical propensity (Fig. 6). These features are typical of the protein-protein interaction domain that forms the activating region of certain transcription factors, mainly from eukaryotes (21, 66, 79). A direct interaction of IHF with RNA polymerase has been proposed in a few cases in which IHF stimulates transcription without the involvement of other factors (23, 26). This is a controversial issue, however, since evidence has been provided for alternative explanations based in IHF-mediated DNA transactions (62, 75). The acidic extension of the IhfA of M. xanthus might indicate that interacting with other proteins is important for some of the molecular actions of IHF in this organism.
Also noticeable in the M. xanthus IhfA is an unusually pronounced HU-like character. Several amino acids of the M. xanthus protein are identical or similar to those found at conserved positions of the protein HU, at which the IhfA proteins usually contain a chemically unrelated amino acid (Fig. 6B). In fact, many of these positions are particularly well conserved in the IhfA proteins (residues T31, Q64, K67, A88, and N90 in M. xanthus IhfA; Fig. 6). To explain why the β-ribbon arm of IhfA but not that of HU recognizes a specific DNA sequence, Rice et al. (67) pointed out two prolines (asterisks in Fig. 6A) which are present only in the subfamily of IhfA proteins. The two prolines, that “may rigidify the arm, making it more effective in filtering DNA sequences,” are missing in the M. xanthus protein. From all of these observations, one may predict that M. xanthus IHF will be less specific in binding to DNA than its counterparts from other bacteria. It should be noted that, after exploring a long stretch of the carQRS promoter region (51), we were unable to find a sequence that may reasonably match the consensus for the IHF binding site (ATCAANNNNTTR 25). We did find, however, a matching sequence (gTCAgAGGGTTG) at the promoter region of the autoregulated ihfA gene (nucleotide positions 355 to 366 in Fig. 4). This sequence partially overlaps the putative −35 region of the ihfA promoter, what may be related to the negative effect of IhfA on its own expression.
In addition to hindering the light response, the lack of IhfA produces other physiological effects on M. xanthus. These effects were observed when the cells carry mutation ihfA2, but not when they carry insertion ΩMR508::Tn5. The latter does not affect the coding region of ihfA, so the cells might produce a normal IhfA protein at a low level. The peptide stretch covered by the ihfA2 deletion (Fig. 6) is coincident with the arm of IhfA where the intercalating proline, and the arginines that make direct contact to DNA, are normally found (67; also see above). So, the mutant protein should be quite inactive. Cells carrying the ihfA2 mutation are grossly impaired in cell motility. As mentioned above, IHF plays a critical role in the activity of ς54-dependent promoters, so the motility defect could be due to the effect of ihfA2 on the expression of ς54-dependent motility genes. Several ς54-dependent genes, as well as various ς54 activator proteins, have been identified in M. xanthus (28, 39, 42, 68). At least one of those genes is required for gliding motility (87). Gliding is controlled in M. xanthus by two distinct genetic systems called adventurous (A) motility and social (S) motility. Thus, two independent mutations are normally required to completely abolish cell motility. Our standard strain DK1050 was derived from strain FB, which carries a mutation at the S-motility gene pilQ (82). Thus, the ihfA gene product might be required only for A motility.
The E. coli cells deficient in any two of three histone-like proteins, IHF, HU, and H-NS, are viable, and only the simultaneous deficiency of all three of the proteins is lethal (88). The situation in M. xanthus may be different, since we have failed in numerous attempts to introduce the ihfA2 deletion into the standard strain DK1622. This is an interesting result, considering the HU-like character of the M. xanthus IhfA and considering also that a bacterial species is known that contains a single sequence homologue to the HU/IHF family (20, 44). The rpoN gene encoding ς54 in DK1622 has been identified. In contrast to the situation in other bacterial species, rpoN was found to be a vital gene for the indicated M. xanthus strain (43). It is currently not known for which essential genes ς54 may be required, but the apparent lethal effect of ihfA2 could be easily explained if IHF participates in the correct expression of those genes. The same procedure used to introduce ihfA2 into DK1622 worked successfully when DK1050-derived strains were used. However, the growth rate of the ihfA2 mutants in rich medium was much lower than the growth rate of the parental strains. The M. xanthus standard strains DK1050 and DK1622 are not isogenic (82). Whatever may be the explanation for the lethal effect of ihfA2, DK1050 seems to carry an extra gene function, or a suppressor allele, that partially counteracts that effect. The viability of some ihfA2 mutants opens the way to uncover the IHF-dependent genes related to cell growth and to investigate their relationships to the essential, ς54-dependent genes of M. xanthus.
ACKNOWLEDGMENTS
We thank David A. Hodgson for providing some plasmids. We also thank José A. Madrid and Ana C. García for technical assistance.
This work was supported by the Spanish Dirección General de Enseñanza Superior (grant PB96-1096 and fellowship to A.J.M.).
REFERENCES
- 1.Altschul S F, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- 2.Avery L, Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol Gen Genet. 1983;191:99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- 3.Aviv M, Giladi H, Schreiber G, Oppenheim A B, Glaser G. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol Microbiol. 1994;14:1021–1031. doi: 10.1111/j.1365-2958.1994.tb01336.x. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre J M, Ruiz-Vázquez R M, Murillo F J. Light induction of gene expression in Myxococcus xanthus. Proc Natl Acad Sci USA. 1987;84:2359–2362. doi: 10.1073/pnas.84.8.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baynham P J, Wozniak D J. Identification and characterisation of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol. 1996;22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 6.Baynham P J, Brown A L, Hall L L, Wozniak D J. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol Microbiol. 1999;33:1069–1080. doi: 10.1046/j.1365-2958.1999.01550.x. [DOI] [PubMed] [Google Scholar]
- 7.Berg D E. Control of gene expression by a mobile recombinational switch. Proc Natl Acad Sci USA. 1980;77:4880–4884. doi: 10.1073/pnas.77.8.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, 9th ed. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1715–1902. [Google Scholar]
- 9.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein coding regions. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 10.Botella J A, Ruiz-Vázquez R M, Murillo F J. A cluster of structural and regulatory genes for light-induced carotenogenesis in Myxococcus xanthus. Eur J Biochem. 1995;223:238–248. doi: 10.1111/j.1432-1033.1995.238_1.x. [DOI] [PubMed] [Google Scholar]
- 11.Brakhage A A, Wozny M, Putzer H. Structure and nucleotide sequence of the Bacillus subtilis phenylalanyl-tRNA synthase genes. Biochimie. 1991;73:127. doi: 10.1016/0300-9084(91)90085-f. [DOI] [PubMed] [Google Scholar]
- 12.Bretscher A P, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978;133:763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burchard R P, Dworkin M. Light-induced lysis and carotenogenesis in Myxococcus xanthus. J Bacteriol. 1966;91:535–545. doi: 10.1128/jb.91.2.535-545.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in E. coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 15.Colland F, Barth M, Hengge-Aronis R, Kolb A. ς-Factor selectivity of Escherichia coli RNA polymerase: role for CRP, IHF and Lrp transcription factors. EMBO J. 2000;19:3028–3037. doi: 10.1093/emboj/19.12.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collis C M, Hall R M. Identification of a Tn5 determinant conferring resistance to phleomycins, bleomycins, and tallysomycins. Plasmid. 1985;14:143–151. doi: 10.1016/0147-619x(85)90074-5. [DOI] [PubMed] [Google Scholar]
- 17.Delic-Attree I, Toussaint B, Vignais P M. Cloning and sequence analyses of the genes coding for the integration host factor (IHF) and HU proteins of Pseudomonas aeruginosa. Gene. 1995;154:61–64. doi: 10.1016/0378-1119(94)00875-s. [DOI] [PubMed] [Google Scholar]
- 18.Devalckenaere A, Odaert M, Trieu-Cuot P, Simonet M. Characterization of IS1541-like elements in Yersinia enterocolitica and Yersinia pseudotuberculosis. FEMS Microbiol Lett. 1999;176:229–233. doi: 10.1111/j.1574-6968.1999.tb13666.x. [DOI] [PubMed] [Google Scholar]
- 19.Fontes M, Ruiz-Vázquez R M, Murillo F J. Growth phase dependence of the activation of a bacterial gene for carotenoid synthesis by blue light. EMBO J. 1993;12:1265–1275. doi: 10.1002/j.1460-2075.1993.tb05771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 21.Gann A A F, Himmelfarb H J, Ptashne M. GAL11, GAL11P, and the action of GAL4. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 931–948. [Google Scholar]
- 22.Giladi H, Wang W, Oppenheim A B. Isolation and characterization of the hupA gene coding for the HU of Aeromonas proteolytica. Nucleic Acid Res. 1992;20:4092–4092. doi: 10.1093/nar/20.15.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giladi H, Koby S, Prag G, Engelhorn M, Geiselmann J, Oppenheim A B. Participation of IHF and a distant UP element in the stimulation of the phage lambda PL promoter. Mol Microbiol. 1998;30:443–451. doi: 10.1046/j.1365-2958.1998.01079.x. [DOI] [PubMed] [Google Scholar]
- 24.Gill R E, Cull M G, Fly S. Genetic identification and cloning of a gene required for developmental cell interactions in Myxococcus xanthus. J Bacteriol. 1988;170:5279–5288. doi: 10.1128/jb.170.11.5279-5288.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodrich J A, Chwartz M L, McClure W R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF) Nucleic Acid Res. 1990;18:4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goosen N, van de Putte P. The regulation of transcription initiation by integration host factor. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 27.Gorham H C, McGowan S J, Robson P R H, Hodgson D A. Light-induced carotenogenesis in Myxococcus xanthus: light-dependent membrane sequestration of ECF sigma factor CarQ by antisigma factor CarR. Mol Microbiol. 1996;19:171–186. doi: 10.1046/j.1365-2958.1996.360888.x. [DOI] [PubMed] [Google Scholar]
- 28.Gorski L, Kaiser D. Targeted mutagenesis of ς54 activator proteins in Myxococcus xanthus. J Bacteriol. 1998;180:5896–5905. doi: 10.1128/jb.180.22.5896-5905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross C A, Lonetto M, Losick R. Bacterial sigma factors. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 129–176. [Google Scholar]
- 30.Hanaham D. Studies of transformation of E. coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 31.Highlander S K, Garza O, Brown B J, Koby S, Oppenheim A B. Isolation and characterization of the integration host factor genes of Pasteurella haemolytica. FEMS Microbiol Lett. 1997;146:181–188. doi: 10.1111/j.1574-6968.1997.tb10190.x. [DOI] [PubMed] [Google Scholar]
- 32.Hodgkin J, Kaiser D. Cell-cell stimulation of movement in non motile mutants of Myxococcus. Proc Natl Acad Sci USA. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodgson D A. Light-induced carotenogenesis in Myxococcus xanthus: genetic analysis of the carR region. Mol Microbiol. 1993;7:471–488. doi: 10.1111/j.1365-2958.1993.tb01138.x. [DOI] [PubMed] [Google Scholar]
- 34.Hodgson D A, Berry A E. Light regulation of carotenoid synthesis in Myxococcus xanthus. In: Caddick M X, Baumber S, Hodgson D A, Phillips-Jones M K, editors. Microbial responses to light and time. Cambridge, United Kingdom: Cambridge University Press; 1998. pp. 186–211. [Google Scholar]
- 35.Hodgson D A, Murillo F J. Genetics of regulation and pathway of synthesis of carotenoids. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C.: American Society for Microbiology; 1993. pp. 157–181. [Google Scholar]
- 36.Huang L, Tsui P, Freundlich M. Integration host factor is a negative effector of in vivo and in vitro expression of ompC in E. coli. J Bacteriol. 1990;172:5293–5298. doi: 10.1128/jb.172.9.5293-5298.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kano Y, Yoshino S, Wada M, Yokoyama K, Nobuhara M, Imamoto F. Molecular cloning and nucleotide sequence of the HU-1 gene of Escherichia coli. Mol Gen Genet. 1985;201:360–362. doi: 10.1007/BF00425687. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan H B, Plamann L. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol Lett. 1996;139:89–95. doi: 10.1111/j.1574-6968.1996.tb08185.x. [DOI] [PubMed] [Google Scholar]
- 40.Keller B, Kast P, Hennecke H. Cloning and sequence analysis of the phenylalanyl-tRNA synthetase genes (pheST) from Thermus thermophilus. FEBS Lett. 1992;301:83–88. doi: 10.1016/0014-5793(92)80215-3. [DOI] [PubMed] [Google Scholar]
- 41.Kendrick K E, Reznikoff W S. Transposition of IS50L activates downstream genes. J Bacteriol. 1988;170:1965–1968. doi: 10.1128/jb.170.4.1965-1968.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keseler I M, Kaiser D. An early A-signal dependent gene in Myxococcus xanthus has a ς54-like promoter. J Bacteriol. 1995;177:4638–4644. doi: 10.1128/jb.177.16.4638-4644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keseler I M, Kaiser D. ς54, a vital protein for Myxococcus xanthus. Proc Natl Acad Sci USA. 1997;94:1979–1984. doi: 10.1073/pnas.94.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobryn K, Naigamwalla D Z, Chaconas G. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol Microbiol. 2000;37:145–155. doi: 10.1046/j.1365-2958.2000.01981.x. [DOI] [PubMed] [Google Scholar]
- 45.Komine Y, Adachi T, Inokuchi H, Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K-12. J Mol Biol. 1990;212:579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- 46.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of ς54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laine B, Belaiche D, Khanaka H, Sautiere P. Primary structure of the DNA-binding protein HRm from Rhizobium meliloti. Eur J Biochem. 1983;131:325–331. doi: 10.1111/j.1432-1033.1983.tb07265.x. [DOI] [PubMed] [Google Scholar]
- 48.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martínez-Argudo I, Ruiz-Vázquez R M, Murillo F J. The structure of an ECF-ς-dependent, light-inducible promoter from the bacterium Myxococcus xanthus. Mol Microbiol. 1998;30:883–893. doi: 10.1046/j.1365-2958.1998.01129.x. [DOI] [PubMed] [Google Scholar]
- 50.Martínez-Laborda A, Murillo F J. Genic and allelic interactions in the carotenogenic response of Myxococcus xanthus to blue light. Genetics. 1989;122:801–806. doi: 10.1093/genetics/122.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGowan S J, Gorham H C, Hodgson D A. Light-induced carotenogenesis in Myxococcus xanthus: DNA sequence analysis of the carR region. Mol Microbiol. 1993;10:713–735. doi: 10.1111/j.1365-2958.1993.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 52.Mesbah M, Premachambran U, Whitman W B. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol. 1989;39:159–167. [Google Scholar]
- 53.Miller H I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenyl-alanyl-tRNA synthetase operon. Cold Spring Harb Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- 54.Nagaraja R, Haselkorn R. Protein HU from the cyanobacterium Anabaena. Biochimie. 1994;6:1082–1089. doi: 10.1016/0300-9084(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 55.Nash H A. The HU and IHF proteins: accessory factors for complex protein-DNA assemblies. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Company; 1996. pp. 149–179. [Google Scholar]
- 56.Nelson K E, Clayton R A, Gill S R, et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 57.Nicolás F J, Ruiz-Vázquez R M, Murillo F J. A genetic link between light response and multicellular development in the bacterium Myxococcus xanthus. Genes Dev. 1994;8:2375–2387. doi: 10.1101/gad.8.19.2375. [DOI] [PubMed] [Google Scholar]
- 58.Nicolás F J, Cayuela M L, Martínez-Argudo I M, Ruiz-Vázquez R M, Murillo F J. High mobility group I (Y)-like DNA-binding domain on a bacterial transcription factor. Proc Natl Acad Sci USA. 1996;93:6881–6885. doi: 10.1073/pnas.93.14.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 60.Oberto J, Rouviere-Yaniv J. Serratia marcescens contains a heterodimeric HU protein like Escherichia coli and Salmonella typhimurium. J Bacteriol. 1996;178:293–297. doi: 10.1128/jb.178.1.293-297.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Padas P M, Wilson K S, Vorgias C E. The DNA-binding protein HU from mesophilic and thermophilic bacilli: gene cloning, overproduction and purification. Gene. 1992;117:39–44. doi: 10.1016/0378-1119(92)90487-a. [DOI] [PubMed] [Google Scholar]
- 62.Parekh B S, Hatfield G W. Transcriptional activation by protein-induced DNA bending: evidence for a DNA structural transmission model. Proc Natl Acad Sci USA. 1996;93:1173–1177. doi: 10.1073/pnas.93.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parkhill J, Achtman M, James K D, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 64.Pérez-Martín J, de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu Rev Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- 65.Pratt T S, Steiner T, Feldman L S, Walker K A, Osuna R. Deletion analysis of the fis promoter region in Escherichia coli: antagonistic effects of integration host factor and Fis. J Bacteriol. 1997;179:6367–6377. doi: 10.1128/jb.179.20.6367-6377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ptashne M. How eukaryotic transcriptional activation works. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 67.Rice P A, Yang S Y, Mizuuchi K, Nash H. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 68.Romeo J M, Zusman D R. Transcription of the myxobacterial hemagglutinin gene is mediated by a ς54-like promoter and a cis-acting upstream regulatory region of DNA. J Bacteriol. 1991;173:2969–2976. doi: 10.1128/jb.173.9.2969-2976.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruiz-Vázquez R M, Murillo F J. Abnormal motility and fruiting behavior of Myxococcus xanthus bacteriophage-resistant strains induced by a clear plaque mutant of bacteriophage Mx8. J Bacteriol. 1984;160:818–821. doi: 10.1128/jb.160.2.818-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruiz-Vázquez R M, Fontes M, Murillo F J. Clustering and co-ordinated activation of carotenoid genes in Myxococcus xanthus by blue light. Mol Microbiol. 1993;10:25–34. doi: 10.1111/j.1365-2958.1993.tb00900.x. [DOI] [PubMed] [Google Scholar]
- 71.Salamov A A, Solovyev V V. Prediction of protein secondary structure by combining nearest-neighbour algorithms and multiply sequence alignments. J Mol Biol. 1995;247:11–15. doi: 10.1006/jmbi.1994.0116. [DOI] [PubMed] [Google Scholar]
- 72.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 73.Sanderson K E, Hessel A, Liu S-L, Rudd K E. The genetic map of Salmonella typhimurium, 8th ed. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular Biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1903–1999. [Google Scholar]
- 74.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheridan S D, Benham C J, Hatfield G W. Activation of gene expression by a novel structural transmission mechanism that requires supercoiling-induced DNA duplex destabilization in an upstream activating sequence. J Biol Chem. 1998;273:21298–21308. doi: 10.1074/jbc.273.33.21298. [DOI] [PubMed] [Google Scholar]
- 76.Shimkets L J. Social and developmental biology of the myxobacteria. Microbiol Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stinson M W, McLaughlin R, Choi S H, Juarez Z E, Barnard J. Streptococcal histone-like protein: primary structure of hlpA and protein binding to lipoteichoic acid and epithelial cells. Infect Immun. 1998;66:259–265. doi: 10.1128/iai.66.1.259-265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Struhl K. Yeast GCN4 transcriptional activator protein. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. p. 833859. [Google Scholar]
- 80.Tanaka I, Appelt K, Dijk J, White S W, Wilson K S. 3-Å resolution structure of a protein with histone-like properties in prokaryotes. Nature. 1984;310:376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- 81.Vis H, Mariani M, Vorgias C E, Wilson K S, Kaptein R, Boelen R. Solution structure of the HU protein from Bacillus stearothermophilus. J Mol Biol. 1995;254:692–703. doi: 10.1006/jmbi.1995.0648. [DOI] [PubMed] [Google Scholar]
- 82.Wall D, Kolenbrander P E, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang A, Roth J R. Activation of silent genes by transposons Tn5 and Tn10. Genetics. 1988;120:875–885. doi: 10.1093/genetics/120.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weber P C. Analysis of Tn5 inversion events in Escherichia coli plasmids. Mol Gen Genet. 1995;248:459–470. doi: 10.1007/BF02191646. [DOI] [PubMed] [Google Scholar]
- 85.Weisberg R A, Freundlich M, Friedman D, Gardner J, Goosen N, Nash H. Nomenclature of the genes encoding IHF. Mol Microbiol. 1996;19:642. doi: 10.1046/j.1365-2958.1996.t01-2-442924.x. [DOI] [PubMed] [Google Scholar]
- 86.Wozniak D J. Integration host factor and sequences downstream of the Pseudomonas aeruginosa algD transcription start site are required for expression. J Bacteriol. 1994;174:5068–5076. doi: 10.1128/jb.176.16.5068-5076.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu S S, Kaiser D. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yasuzawa K, Hayashi N, Goshima N, Kohno K, Imamoto F, Kano Y. Histone-like proteins are required for cell growth and constraint of supercoils in DNA. Gene. 1992;122:9–15. doi: 10.1016/0378-1119(92)90026-l. [DOI] [PubMed] [Google Scholar]