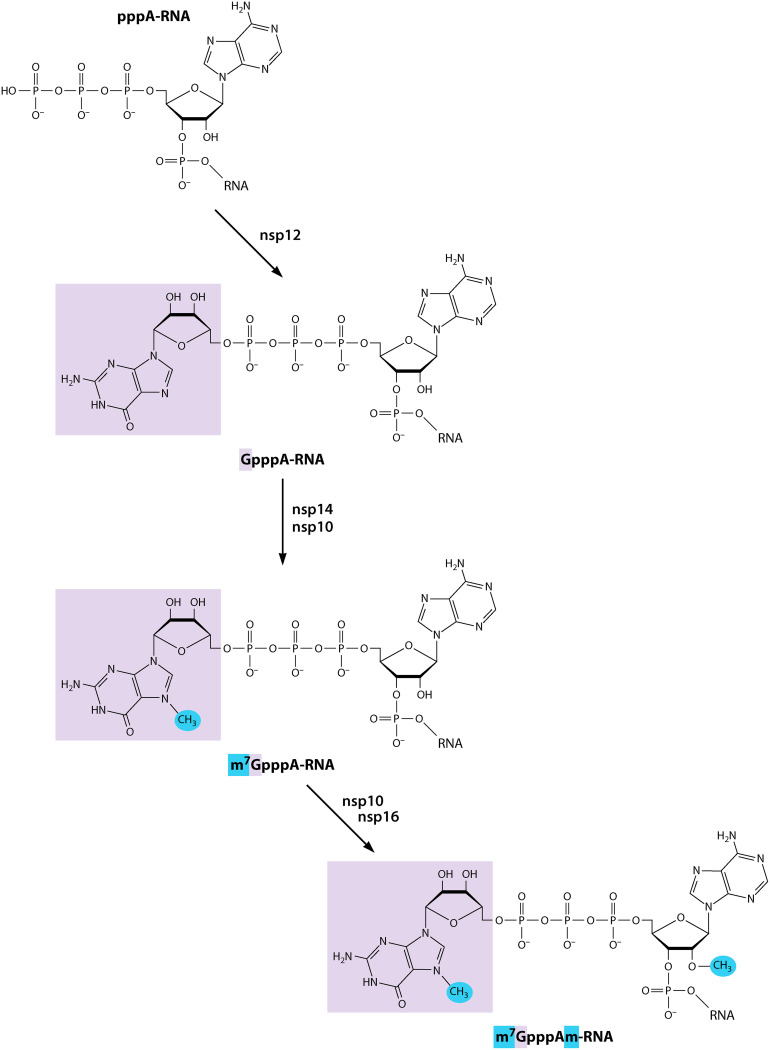
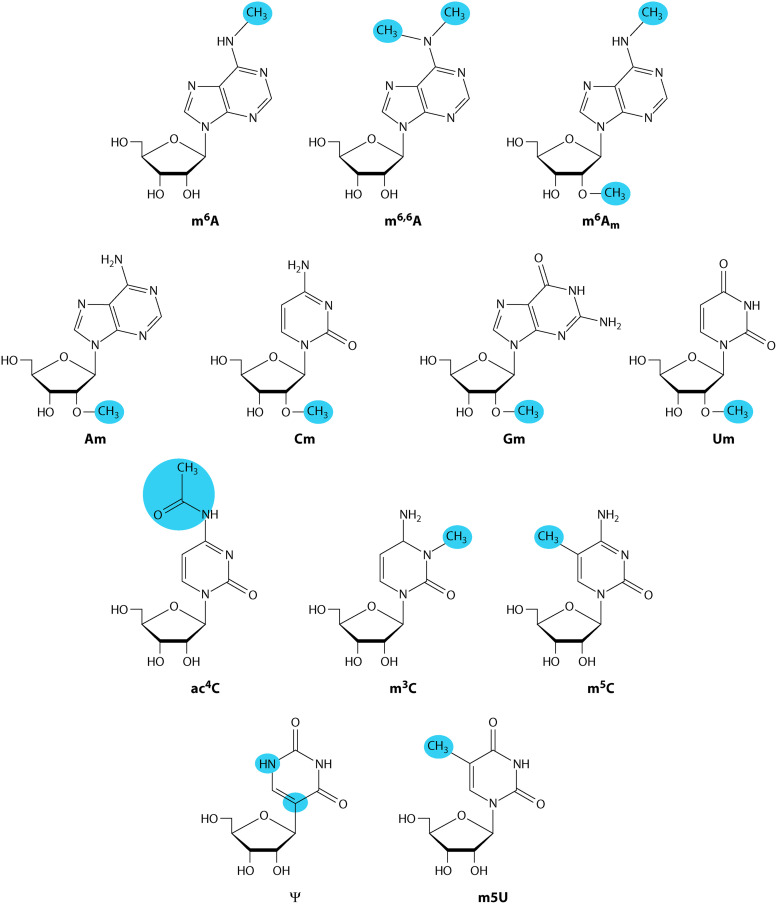
SUMMARY
SARS-CoV-2, the etiological agent responsible for the COVID-19 pandemic, is a member of the virus family Coronaviridae, known for relatively extensive (~30-kb) RNA genomes that not only encode for numerous proteins but are also capable of forming elaborate structures. As highlighted in this review, these structures perform critical functions in various steps of the viral life cycle, ultimately impacting pathogenesis and transmissibility. We examine these elements in the context of coronavirus evolutionary history and future directions for curbing the spread of SARS-CoV-2 and other potential human coronaviruses. While we focus on structures supported by a variety of biochemical, biophysical, and/or computational methods, we also touch here on recent evidence for novel structures in both protein-coding and noncoding regions of the genome, including an assessment of the potential role for RNA structure in the controversial finding of SARS-CoV-2 integration in “long COVID” patients. This review aims to serve as a consolidation of previous works on coronavirus and more recent investigation of SARS-CoV-2, emphasizing the need for improved understanding of the role of RNA structure in the evolution and adaptation of these human viruses.
KEYWORDS: coronavirus, RNA structure, SARS-CoV-2, epitranscriptomics, evolution
INTRODUCTION
The pandemic brought on by the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has resulted in over 270 million infections and 5 million deaths worldwide as of the end of the 2021 year (1). First identified as a string of novel pneumonia cases associated with a wet market in the city of Wuhan in the Hubei province of China late in 2019 (2), the disease now known as COVID-19 has crippled economies and disrupted societal equilibrium across the globe. Although the development of multiple vaccines has increased our capacity to curb the pandemic, the continual emergence of variants of concern (VoCs) during the last year likely presents a looming obstacle to eliminating completely the spread of COVID-19. This issue was more recently illuminated following the emergence of the Omicron variant (B.1.1.529) in South Africa in November of 2021, which exhibited numerous mutations associated with enhanced transmissibility, according to the Centers for Disease Control (3). The emergence of the Omicron variant, as well as the potential emergence of future variants, has accentuated the need for further surveillance and investigation to understand changes to the SARS-CoV-2 RNA genome in concert with alterations to disease pathology. Although proteins such as Spike have been continuously at the forefront of SARS-CoV-2 monitoring, the often unrecognized noncoding regions and structures within the RNA genome of the virus have been demonstrated to be critical to various viral functions and may play a role in the emergence and continued adaptation of the virus.
The ability of RNA to fold and form stable structures is critical to survival of RNA viruses in general, with elements of RNA structure playing roles ranging from transcription to packaging and interactions with the host cell. Whereas RNA can exist as a single-stranded molecule, it can also adopt secondary, tertiary, or even quaternary higher-order structures, similar to proteins, through a variety of base-pair interactions (4–8). Small molecule inhibitors have a history of successful use in targeting specific viral RNA structures in the prevention of infection in vitro and in vivo (9–13). RNA structures that make attractive therapeutic targets are conserved in nature and do not exhibit structural homology with the host that would pose a threat in the form of significant off-target effects (14). The use of RNA structure inhibitors is best exemplified in small-molecule studies inhibiting human immunodeficiency virus (HIV) infection (15, 16), the most extensively studied RNA virus to date (17). The HIV genome was also recently hypothesized to contain a significantly greater number of RNA structures than previously thought using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) mapping (18), revealing numerous additional potential targets within otherwise protein-coding regions of the genome. However, it remains to be seen whether these additional RNA structures could be targeted by inhibitors given evolutionary evidence (18) and bootstrap analysis of experimental SHAPE data (19), both of which posit a high level of uncertainty around the majority of these proposed RNA structures.
Chemical probing methods, such as SHAPE, rely on the treatment of RNA with a reagent that will selectively modify specific RNA nucleotides, typically either via cleavage of the RNA or via adduct formation; the reactivity of the reagents with each nucleotide, which is the measurement of interest, depends on accessibility as a consequence of the presence or absence of RNA interactions with other ligands. For example, dimethyl sulfide (DMS) modifies unpaired cytosines and adenines via the addition of a methyl group (20). Alternatively, SHAPE is capable of modifying all four RNA bases (21) through acylation of nucleotides whose ribose is exposed (22, 23). Although SHAPE modification is more robust than DMS modification in its ability to modify these additional bases, thus providing additional raw data DMS cannot, benchmarking of DMS and SHAPE modifications to determine RNA secondary structure demonstrated comparable or even improved error rates for DMS compared to SHAPE (24). This result is likely due to the fact that nucleotides engaged in noncanonical base pairing may be inaccessible to SHAPE modification but remain accessible to DMS modification (24). Thus, although SHAPE is considered the “gold standard” for experimental detection of RNA secondary structure (25), it may be beneficial to combine DMS and SHAPE data in order to increase the accuracy of certain RNA secondary structure algorithms (24). However, that is not to say that either method is without drawbacks: the false positive rate of these methods can be attributed to aberrations in environmental conditions and reagents relative to the conditions within the cell, to which RNA structures can be particularly sensitive (26). Transient structures can also form in certain conditions and so may not reflect legitimate in vivo structural formations or may form a range of alternating structures across an equilibrium of possible RNA structure conformations, which may confound RNA structure analysis searching for a single consensus structure (18). With respect to the former, more appropriate probes in SHAPE mapping have been designed for RNA structure determination in living cells (27); however, these agents vary in cellular permeability, potentially resulting in the opposite effect: a high rate of false negative detection (25). SHAPE-MaP and in vivo click SHAPE (icSHAPE), which utilize next generation sequencing for high-throughput analysis of heterogeneous populations (27, 28), result in a population average of probe reactivities, though they cannot distinguish distinct conformations and their frequencies within a population of homologous sequences (e.g., virus strains). In the case of icSHAPE, the addition of a biotin moiety to each RNA base that is SHAPE-modified permits enrichment for this modification, thus specifically isolating structured and/or modified RNA for downstream sequencing (27); however, icSHAPE reactivities in vivo can be affected by protein binding to target RNA (29). Additionally, given that any sufficiently long RNA will form some degree of secondary structure, even if transient (30), it is important to distinguish signal from noise to determine biologically relevant structural formations (28). To this end, SHAPE-MaP data can be analyzed in the context of Shannon entropy data whereby regions of both low reactivity and entropy indicate the presence of sustained RNA structure (as opposed to highly flexible), which can be interpreted as biologically relevant (28). While the presence of RNA structure can be inferred via molecular probing and Shannon entropy measurements for a single molecule, viruses can exhibit a high degree of sequence heterogeneity, calling into question the prevalence of a given structure within a sample or population. Combining these data with an additional layer of a priori evolutionary knowledge (18, 31, 32) can aid in the development of an enhanced understanding of“ druggable” RNA structures.
For RNA viruses, highly structured RNA regions are often associated with significantly less nucleotide variation than regions of RNA that lack structure, evident of the deleterious effects of nucleotide mutations and the selective pressure to maintain an RNA structure-function relationship. Because viruses are limited in molecular size, they are also limited in genomic size; RNA viruses have thus evolved to utilize the latter efficiently, with the same genomic subregion often accommodating multiple protein open reading frames (ORFs), as well as RNA structures. In order for mutations in these regions to survive, they must maintain all of these coding functions to a certain extent and can thus often be highly deleterious. However, strict conservation in highly functional RNA structures is not always observed (18, 31), as function can be maintained through evolution of the linear nucleotide sequence in the form of compensatory, or covarying, changes. In the instance that a single nucleotide change is either deleterious or reduces fitness of the virus, a second, compensatory change acting to restore amino acid interactions at the protein level, or nucleotide base-pair interactions at the level of the RNA structure, can restore function. Similar to HIV (33–36), several coronavirus RNA elements have been well characterized using the experimental approaches described above and exhibited a high degree of conservation; however, as we describe herein, the SARS-CoV-2 RNA genome has proven to be highly dynamic, whereby multiple conformations and functions, such as alternative higher-order RNA structures and long-range interactions, are able to coexist (37), some of which have been maintained through compensatory mutations. The conserved structural regions, whose roles in coronavirus replication are well characterized, as well as those less understood and perhaps unique to SARS-CoV-2, are described in this review in the context of the past, present, and future evolutionary history of the virus and importance in light of global viral dynamics and therapeutic strategies.
THE REGAL VIRUS FAMILY AND ITS RNA GENOME ARCHITECTURE
The Coronaviridae family of viruses is characterized by a positive-sense, single-stranded, nonsegmented RNA genome capable of infecting mammals and birds throughout the world (38). The most prominent Coronaviridae subfamily, Orthocoronavirinae, is further divided into four genera: Alpha-, Beta-, Delta-, and Gammacoronaviruses (38, 39). Clinically relevant CoVs with regard to human health are found in the Alphacoronavirus and Betacoronavirus genera, collectively encompassing seven CoVs known to infect humans (40). The appearance of SARS-CoV-1 (a Betacoronavirus in the subgenus Sarbecovirus) as the causative agent of the SARS outbreak in 2003 marked the establishment of known CoVs associated with severe respiratory complications that contributed to epidemics in human populations (41). SARS-CoV-1 and SARS-CoV-2 share <80% percent of their genomes (42), on the basis of which SARS-CoV-2 was deemed to be genetically distinct and thus a new human pathogen within the genus Betacoronavirus (43). The most closely related known CoV to SARS-CoV-2 is the bat coronavirus RaTG13, sharing 96.2% genetic similarity to SARS-CoV-2 (44), whereas the second most closely related sample lineage is that belonging to the pangolin (pangolin-CoV-2019) (45), both of which are highly suspect in the contribution to coinfection and multiple recombination events that resulted in the emergence of the unique SARS-CoV-2 genome (46).
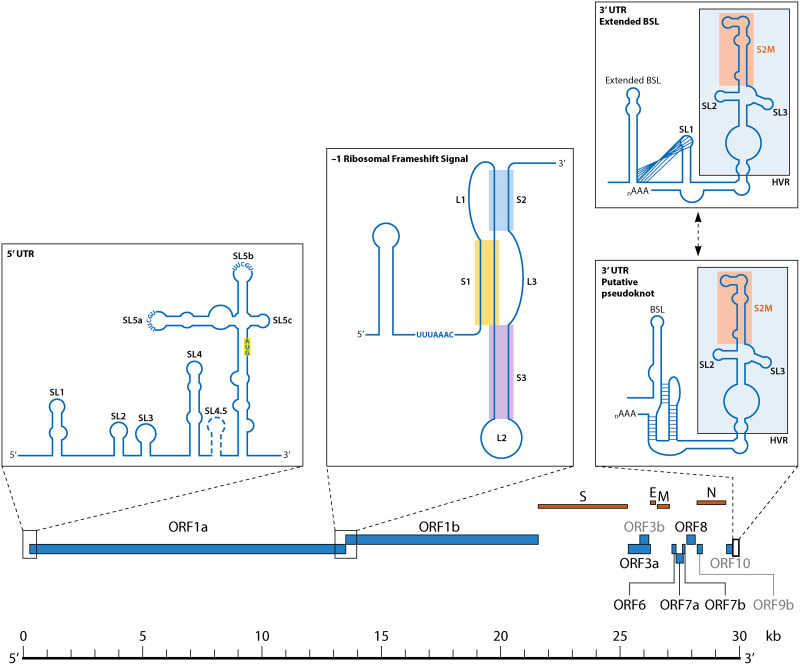
The SARS-CoV-2 RNA genome (reference sequence NC045512.2 used throughout this review) is approximately 29.9 kb in length, standard for CoVs, and is organized for the efficient production of proteins involved in replication following entry into the host cell (47, 48) (Fig. 1). The genome contains a 5′ cap, or N7-methylguanine (m7G) moiety, preceding the 5′ untranslated region (UTR), which is a critical structure for RNA stability and translation and common among RNA viruses. This moiety is followed by the open reading frames (ORF) ORF1a and ORF1b, which together encompass approximately two-thirds of the RNA genome (Fig. 1). The protein products of this first region are required for the assembly of the RNA-dependent RNA polymerase (RdRp), which is responsible for both replicating the genome and transcribing the subgenomic RNA (sgRNA). As with other CoVs, the genome contains a 3′ UTR followed by a poly-adenylated [poly(A)] tail (49) (Fig. 1). Aside from ORFs 1a and 1b, the SARS-CoV-2 genome contains 12 additional ORFs encoding for 29 proteins, including 4 structural proteins and a series of well-characterized accessory proteins, and putative proteins for which the function remains unknown (47, 50–52). RNA structure characterization has largely remained in the regions of the UTRs, as the majority of the SARS-CoV-2 genome is comprised of protein-coding RNA. However, the existence of conserved RNA structures in protein-coding regions in other viruses and availability of high-throughput methods for RNA secondary structure exploration have prompted the investigation of multifunctional RNA throughout the genome (53, 54), revealing potential additional RNA structure candidates that may be required for novel functions in distinct viral lineages (e.g., VoCs).
FIG 1.
Graphical representation of SARS-CoV-2 genome architecture emphasizing CoV RNA structures reviewed herein.
UNTRANSLATED, YET NOT UNEXPLORED
The dense population of RNA structures located within the 5′ and 3′ UTRs (Fig. 1) have been well characterized based on a combination of analyses using the linear sequence, secondary and tertiary predicted structures, and biochemical experimentation. These regions remain highly conserved among CoVs and are home to primarily cis-acting structures, defined as RNA elements that regulate the activity of the RNAs in which they reside (55), which are critical components of viral replication (56, 57). Many of these structures have been identified as a result of defective interfering (DI) RNA experiments. DI RNAs are erroneously generated during viral replication and are derived from the parental virus genome but lack the capacity for replication, thus relying on host cell or coinfecting viruses for replication machinery (58). Mutational studies using DI RNA have thus been useful in determining the cis-acting genomic components necessary for this function, particularly in CoV research using the Mouse Hepatitis Virus (MHV) model. MHV is a useful model, discussed throughout this review, because it belongs to the Betacoronavirus genus with SARS-CoV-1 and SARS-CoV-2 (although of the subgenus Embecovirus) and is a safer and more accessible virus (requiring biosafety level 2 rather than biosafety level 3, as with SARS-CoV-1 and SARS-CoV-2).
THE MULTIPLE FACES OF SL1
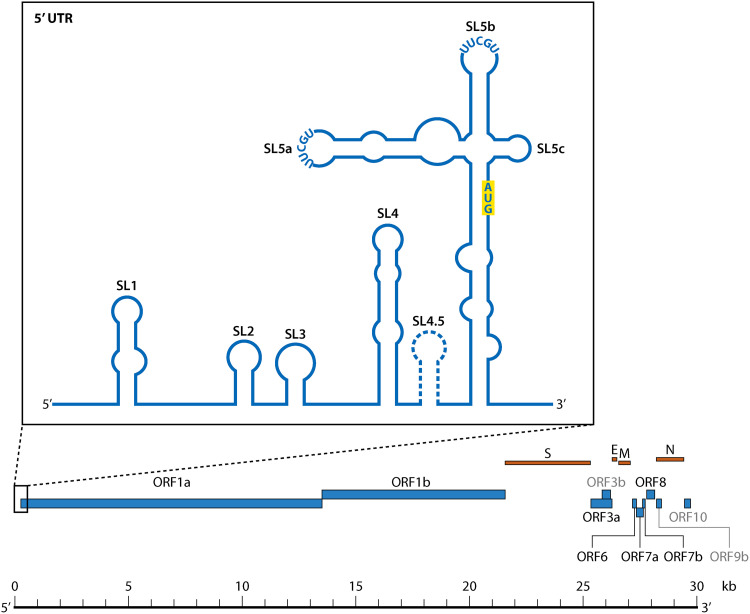
The 5′ UTR region is comprised of five distinct stem-loop structures (SL1 to SL5) that collectively play critical roles in RNA transcription, translation, and replication (Fig. 1 and 2). SL1 is approximately 27 nucleotides in length (positions 7 to 33 in the SARS-CoV-2 genome) and is universally present in the 5′ UTR of CoVs, though it is unique in that it demonstrates low thermodynamic stability (59). This diminished stability is attributed in part to the prevalence of adenine (A)- uracil (U) base-pair interactions within the stem region, which harbor fewer hydrogen bonds than guanine (G)-cytosine (C) pairs (59). The enrichment of adenine and uracil in the RNA genome is not isolated to SL1, nor to the Coronavirus family. APOBEC3 (apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3) proteins function to deaminate cytosines in mRNA (mRNA), resulting in an abundance of uracils that can disrupt viral RNA function. APOBEC3 has been found at high levels in the lung (60), where Betacoronaviruses preferentially replicate within the host (61). Moreover, temporal analysis of SARS-CoV-2 evolution has shown an increase in C-to-U genome mutations during the first 4 months following the first appearance of SARS-CoV-2, suggesting adaptation to APOBEC-mediated deamination and human infection (61, 62). The presence of a bulged adenine at the base of SL1 also contributes to the instability of the stem in MHV (position 35) (63). This destabilizing adenine was shown in MHV to be under relatively strong purifying selection; the emergence of mutations increasing stability in the stem region led to nonviable viral products owing to a significant decrease in synthesis of sgRNA, whereas mutations that rescued the instability in the stem maintained sgRNA production (64). Additionally, although the deletion of this site (similarly resulting in increased stability) was permitted, compensatory mutations were observed in virus rescuing SL1 stem instability as well as sgRNA synthesis (64). SARS-CoV-2 similarly contains a bulged adenine at position 27 (63), providing further evidence of a functionality ascribed to this small interference in the stem. The SL1 loop of SARS-CoV-2 was, however, originally described to contain two bulged nucleotides (A27 and C28) (63, 65) and an additional adenine bulge at residue 12, but this specific pattern is not consistent across differing variants of SARS-CoV-2 (65). Whether or not the heterogeneity in SL1 structure across variants contributes to differences in infectivity is unclear. As with other labile structures such as psi in HIV (66), the presence of these atypical interactions and relatively low stability has enabled SL1 in subgenuses Embecovirus and Buldecovirus to adopt multiple structural conformations with various functions, though this conformational change has been less investigated for SARS-CoV-2. One of these conformations is believed to permit long-range interactions between the 5′ and 3′ UTRs (64). Naturally arising compensatory mutations in MHV SL1 described above cooccurred with one of two mutations in the 3′ UTR, either A303G or A244G (counting from the 5′ end of the MHV 3′ UTR), providing additional evidence in support of an interaction between the two regions (64), though its role in SARS-CoV-2 replication remains unclear.
FIG 2.
Structural organization of the 5′-UTR stem loops SL1 through SL5abc in SARS-CoV-2. The start site (AUG) for the small upstream ORF described by Raman et al. (96) is highlighted. The controversial SL4.5 hairpin is represented by a dashed outline. TGEV, Transmissible gastroenteritis virus.
The SL1 region is critical for the translation of both gRNA and sgRNA across CoVs; specifically, the apical stem region of SL1 has been identified as being sufficient to protect SARS-CoV-2 viral mRNA against the suppression of mRNA translation by viral Nsp1 (67). Nsp1 is encoded by both Alphacoronaviruses and Betacoronaviruses and functions in a similar capacity to suppress host mRNA translation by binding to the host 40S ribosomal protein subunit (68) and inducing the endonucleolytic cleavage of the 5′ region of capped nonviral mRNA (69, 70). The ability of SL1 to promote distinct recognition of cellular and viral mRNA by Nsp1 was first characterized in SARS-CoV-1 (70) but has more recently been demonstrated in SARS-CoV-2 (71, 72) through observance of interactions between Nsp1 and SL1 (67, 73). Mutations in the apical stem region of SL1 that disrupt structural formation have also been observed to abrogate protection against Nsp1-associated inhibition (67). Analyses of binding kinetics and mutational profiling have further revealed that protection of viral mRNA is carried out by the interaction of SL1 with the carboxy-terminal domain of the Nsp1 bound into the mRNA channel of the 40S subunit (71), resulting in the rearrangement of the Nsp1 carboxy terminal and displacing it from the mRNA channel (67). The relative position of SL1 (between the 5′ cap and downstream SL2) has also been found to be important for the interaction with Nsp1, as swapping the positions of SL1 and SL2 within the 5′ UTR, or inserting five nucleotides between the 5′ cap and SL1, ablates protection against Nsp1 (73). The authors of this particular study thus propose a model in which the 5′ UTR is involved in allosterically modulating ribosomal binding, which is predicated on a specific distance between SL1 and the 5′ cap bound to the Nsp1-40S complex. Specifically targeting the 3′ region of SL1 with antisense oligonucleotides inhibited this binding and SARS-CoV-2 replication, revealing SL1 as a potential therapeutic target for SARS-CoV-2 infection (74). Indeed, intranasal administration of an antisense oligonucleotide (ASO) therapeutic targeting a region of the SARS-CoV-2 5′ UTR containing SL1 in a humanized COVID-19 mouse model resulted in the suppression of viral replication (75). The ASO specifically targets the stretch of stem following the two bulged nucleotides described above. A combined chemical probing/sequencing approach referred to as DMS-MaPseq was used to demonstrate successful ASO-mediated interruption of SL1 structure in a dose-dependent manner (75), presenting it as an ideal candidate for further human studies. Amiloride-based ligands have also been identified as small molecule inhibitors targeting the SL1 structure and consequently reducing SARS-CoV-2 replication in infected cells (76), potentially representing a promising therapeutic for not only SARS-CoV-2 but other coronaviruses given further testing in animal models.
SL2 AND SL3: THE LONG AND SHORT OF IT
Stem loop 2 (SL2), located 11 nucleotides downstream of SL1 in the SARS-CoV-2 genome, is conserved across all known CoVs and typically contains a 5 base-pair stretch [5′-(C/U)UUG(U/C)-3′ in/near the loop region of all Orthocoronaviruses and 5′-CUUGU-3′ in MHV] fixed atop a five base-pair helical stem (77, 78) (Fig. 1 and 2). Early nuclear magnetic resonance (NMR) studies demonstrated the formation of a uridine turn (U-turn)-like motif, characterized by a UNR sequence (N represents any nucleotide and R a purine [79]). UNR sequences form a sharp turn in the phosphate backbone (between the U and N) that is stabilized by interactions formed across the bend (77, 78). However, it was later demonstrated that the structure of MHV SL2 better fits a uYNMG(U)a-like or uCUYG(U)a-like tetraloop structure, in which G50 is stacked onto A52, and U51 is flipped out of the hairpin loop (80). The orientation of U51 was originally thought to be important in curating the stem-loop’s architecture by acting as a spacer element (80, 81), a region of RNA that does not have any specific sequence requirements but simply functions to spatially coordinate proximal RNA elements, permitting the stacking interaction between residues G50 and A52 (80). Indeed, mutations U51A, U51C, and U51G did not appear to impact viability of the virus (80). Alternatively, replacement of U48 with C48 was predicted to perturb the SL2 structure and ultimately proved lethal in MHV, resulting in the ablation of synthesis of viral sgRNA, whereas U48G was predicted to maintain SL2 structure and proved a viable mutation (77). Intriguingly, replacement of the MHV SL2 stem with the similar, yet somewhat more stable, SARS-CoV-1 SL2 restores the viability of the otherwise lethal U48C mutation (80), suggesting increased lability was not heavily selected for in MHV. Similar to MHV, in SARS-CoV-1 the SL2 apex is constituted by the 5′-CUUGU-3′ sequence, which also adopts a similar 5′-gCUYGc-3′ tetraloop fold and is located atop a 5 base-pair stem (78, 82). The SARS-CoV-1 structure also contains the flipped-out U51 (82), though deletion of U51 within the SARS-CoV-1 SL2 was viable, suggesting this residue is not necessary as a spacer. Although most evidence of conservation stems from analysis of MHV and SARS-CoV-1, the SL2 of SARS-CoV-2 is structurally and phylogenetically similar (tetraloop structure with 5′-CUUGU-3′ apex sequence atop a 5 base-pair stem) (65). The stem structure has been demonstrated to be critical to replication in MHV, though conservation of the linear sequence is not, as mutational analysis shows that maintained base-pairing along the stem, regardless of the interactions (i.e., AU versus GC), results in viable production of virus (77, 78). SL2 is thus highly permissible to plasticity in sequence but not structure. Despite this evidence, SL2 is the second most conserved region of the CoV 5′ UTR (83) (e.g., MHV and SARS-CoV-1 differ only by two nucleotides [80]). Given the experimental evidence suggesting a constraint on structure rather than sequence, the lack of frequent compensatory mutations in this region is unclear and may suggest an alternative function for this structure that relies on additional interactions requiring another layer of evolutionary constraints. The maintenance of SL2 structure in the absence of specificity for sequence has also been demonstrated in Alphacoronaviruses, wherein mutational analysis of the HCoV-229E SL2 stem region demonstrated the essential maintenance of overall base pairing, regardless of the specific bases involved, with the exception of a GC base pair located at the base (position 47) (84). For MHV, replacement of this particular residue with any nucleotide largely does not impact viability (80), nor do substitutions of U49C and U49G (the first U in the UNR motif); however, G50A and G50C do negatively affect viral replication (80), emphasizing the importance of the G50-A52 stacking interaction within the tetraloop. Owing to the conservation of these two nucleotides and the evolutionary conservation of an overall hairpin structure, as well as the imperative function, SL2 would seem to act as a suitable drug target. However, there currently exists no small molecule inhibitor targeting this particular tetraloop motif, and a predictive analysis of small molecule inhibitors of SARS-CoV-2 RNA secondary structures revealed no potential candidates for molecular inhibitors (85). The lack of inhibitors for SL2 is likely owing to its small size, as RNA molecules comprised primarily of secondary structure (in the absence of tertiary structure) contain small or shallow pockets. While it is still theoretically possible to target specific motifs in these structures, doing so presents challenges in much the same way as targeting the shallow grooves that characterize many protein-protein interactions and is thus a high-risk endeavor (86).
Similarly small, SL3 is comprised of a 15 nucleotide-length hairpin structure found in some Betacoronaviruses, notably SARS-CoV-1 and SARS-CoV-2, as well as bovine coronavirus (BCoV) and Gammacoronaviruses (83), and is located only one nucleotide downstream of SL2 in SARS-CoV-2 (Fig. 1 and 2). SL3 in these groups contains within its structure the leader transcriptional regulatory sequence (TRS-L), which is critical to the generation of sgRNA (78, 87). For SARS-CoV-2, this sequence is located in the 3′ region of the SL3 stem (59, 88). Distinct from SL1 and SL2, SL3 is involved in multiple long-range RNA interactions (37) and is actually not present in MHV (81, 89) or Middle East respiratory syndrome CoV (MERS-CoV) (90). The MHV TRS is found in a generally unstructured region; as such, unlike SL2, TRS for SARS-CoV-1 and MHV are noninterchangable (91). In other Betacoronaviruses, including SARS-CoV-2, the function of SL3 is the enhanced presentation of the TRS-L sequence for base pairing with the downstream TRS body (TRS-b) sequence regions found at the beginning of each gene and necessary for the generation of the sgRNA (92, 93). This process involves the discontinuous transcription of the negative strands of the sgRNA, whereby the RdRp will begin at the 3′ end of the genome and transcribe until reaching a TRS-b region, upon which the RdRp will, with some frequency, dissociate from the positive-sense genome template and reprime at the interacting TRS-L region in order to incorporate the 5′-leader (first ~70 nucleotides) sequence to the sgRNA (57, 94). Another long-range interaction involving SL3 occurs with a triple-helix junction found in the 3′ UTR (discussed in more depth below). A technique utilizing cross-linking of base-paired RNAs in tandem with deep sequencing (95) to uncover long-range RNA-RNA interactions applied to the SARS-CoV-2 genome demonstrated that the complete disruption of each of these two structures, required for the formation of this long-range interaction, results in the stable circularization of the SARS-CoV-2 genome (37). Additionally, mutational profiling with DMS has shown that residues within the SL3 stem display medium reactivity profiles, suggesting that SL3 readily alternates between folded and unfolded conformations in the context of the whole SARS-CoV-2 genome (88). The ability of SL3 to dissociate and reform this differing structure explains why SL3 itself is not stable at 37°C (77, 78), rendering it difficult to target therapeutically (59, 83, 90). SL3 is indeed not predicted to be efficiently targeted by small molecule inhibitors (76, 85), though this may also be due to its small stature.
SL4: STABILITY THROUGH EVOLUTIONARY INSTABILITY
As it has been alluded to, linear sequence conservation is not directly related to structural importance, as rapid evolving viruses can rely on compensatory mutations to maintain function, whether they occur simultaneously or are separated by a less stable, yet viable intermediate structure. SL4 is one such structure that falls into this category: composed of a relatively stable, long hairpin, which is commonly referred to in terms of the lower (SL4a) and upper stem (SL4b) regions. Combined, the SL4 stem has been recorded as variable lengths but is composed of approximately 44 nucleotides for SARS-CoV-2 (Fig. 1 and 2) and is conserved to some extent among Betacoronaviruses (83, 96) (particularly between MERS-CoV, SARS-CoV-1, and SARS-CoV-2). Unlike SL2, however, SL4 undergoes frequent coevolutionary changes, specifically among three base pairs within the stem region: R90-Y121, R97-U115, and G101-Y111 (65). DI RNA studies of SL4b in BCoV (though referred to previously as SL-III) have pointed to an important role for this structure in replication (78, 96), as disruption of base-pair interactions ablated replication, which could be rescued modestly through compensatory mutations (96). SL4b also contains a small (3 to 13 amino acid) upstream ORF (uORF) (96) (Fig. 1 and 2), the translation of which in MHV has been shown to attenuate ORF1 translation in vitro (97). Neither the SL4b portion of SL4 nor the uORF were believed to be critical to replication in MHV, however, as demonstrated through extensive mutational analysis altering both the uORF and SL4b structure (98). This finding was contrary to studies in BCoV using DI RNA replication assays, which reported that both the SL-III (SL4b) stem and the uORF were necessary to continually passage BCoV DI RNA (96). Later mutational analysis demonstrated that deletions in the uORF region also still produced viable virus, but the reemergence of the full uORF upon subsequent passages suggested that the uORF may not be an essential motif in MHV replication but is beneficial to viral fitness in some way (97). Deletion of the entire SL4 region was observed to be lethal in MHV (98), whereas replacement with a shorter and sequence-unrelated stem-loop was viable (98), suggesting the RNA structural element of SL4 may serve simply as a spacer element involved in the direction of sgRNA (78). Meanwhile, the uORF contained within SL4b has been predicted for all CoVs, although there is no direct evidence that it is actually translated (65, 96). DI RNA studies in which the uORF start codon was altered to be inefficient (via conversion of the uORF AUG to ACG) demonstrated that replication still occurred albeit with reduced efficiency (97). The impact of this specific mutation on the SL4 RNA structure was unclear.
SL4 is also hypothesized to be involved in the expression of sgRNA due to its location within the “5′-proximal hot spot,” a wide region canonically containing the TRS-L segment described above (99), which was identified in studies of BCoV as playing a role in discontinuous transcription (83, 99). This finding is also in contrast to Yang et al. (98), implicating SL4 as important to template switching during negative strand synthesis, particularly in CoVs that lack secondary structure in their TRS-L region (MHV, BCoV, and MERS-CoV) (83, 99), which is discussed in more depth later in this review.
SL5: LAST, BUT NOT LEAST
SL5 is the concluding structure within the 5′ UTR, encompassing 144 nucleotides in SARS-CoV-2, and is collectively comprised of three component substructural stem loop, SL5a, SL5b, and SL5c (Fig. 1 and 2), that have been observed in Alpha- and Betacoronaviruses (81, 83). The Embecovirus subgenus has previously been noted to lack these substructures (83, 100); however, the controversy surrounding this hypothesis is discussed below. In Betacoronaviruses, including SARS-CoV-2, SL5a and SL5b both contain a 5′-UUCGU-3′ pentaloop motif, whereas SL5c contains a GNRA tetraloop motif that in SARS-CoV-2 is composed of the sequence 5′-GAAA-3′ (59, 65, 101). The SL5 trifurcating a, b, and c stems form a four-way junction with the main SL5 stem, comprising a multitude of interactions that act to stabilize (78, 102) the complex SL5 structure (65). In silico analysis of the SL5 region indicated that the nucleotides comprising the basal stem are generally less structured than those that make up the individual substems (103). Consistent with this finding, substem SL5a was determined to harbor three base pairs that expressed significant covariance, suggesting structural importance (103). As discussed below, SL5 has been implicated as an important structure for the packaging of some CoVs (83, 100). Although the presence of a multistructured SL5 is conserved across CoVs, differences can be observed. Whereas SL5c in both SARS-CoV-1 and SARS-CoV-2 constitutes a GNRA tetraloop structure, MERS-CoV harbors a heptaloop structure (83, 104). In Alphacoronaviruses, each of the SL5 substructures, including SL5c, contains a conserved 5′-UUYCGU-3′ loop (104). A recent NMR-based screening of potential drug candidates for targeting known RNA structures across the SARS-CoV-2 RNA genome identified multiple small molecules that could potentially target substem regions of SL5 (85). Additionally, multiple potentially druggable pockets have been identified in the SL5 structure, implicating both the SL5 substems and the SL5 main stem as possible targets (105, 106). Amiloride-based ligands have also demonstrated successful targeting of SL5a in vitro (76), although neither studies have continued into animal models.
PACKAGING SIGNAL STRUCTURES: CARRIER CHAOS AMONG CoVs
The use of packaging signals, similar to other RNA viruses such as HIV (107), has been elucidated as important for the biological function of many CoVs (100). Packaging signals are cis-acting RNA elements involved in facilitating the selective encapsidation of the viral genomic RNA, though not always relying on the presence of a secondary or tertiary RNA structure for recognition and, as we discuss herein, not always functioning similarly, even among virus genera. SARS-CoV-1 and SARS-CoV-2 incorporate the nucleocapsid phosphoprotein (N) for recognition of the viral genome for selective packaging (108, 109), which for SARS-CoV-1 (and likely for SARS-CoV-2) associates with the genome to form a ribonucleoprotein complex that then interacts with the endodomain of the viral-encoded matrix (M) protein (108) to complete virion assembly (110, 111).
Investigation of the MHV packaging process, while not requiring the N protein (108, 112), has identified the presence of an RNA hairpin structure located 20.3 kb from the 5′ end of the genome within the region encoding protein Nsp15 (at the 3′ end of ORF1b) (113), which is conserved in all members of the subgenus Embecovirus (83, 114). This region in MHV, thought to function as the packaging signal, takes the form of a 95-nucleotide bulged stem-loop that contains four repeat AGC/GUAAU motifs, each separated at regular intervals by either a bulge or loop domain and displaying either an AA or GA bulge on the 3′ side (113). Akin to the linear sequence, these repeat structural components are conserved in all Embecoviruses (100). This structure is, however, thought to be absent in SARS-CoV-1 and SARS-CoV-2 (100).
For SARS-CoV-1, the packaging signal has been identified in upstream spanning nucleotides 19715 to 20294 (referred to as PS580, also in the coding region of Nsp15) (108, 115). Recent analysis has determined that the SARS-CoV-1 N protein binds a 151-nucleotide section of this packaging signal region in vivo (115), but further experimental analysis is needed to elucidate the full mechanism that dictates viral packaging for SARS-CoV-1, including resolution on whether or not an RNA structure is involved.
In contrast to the Embecoviruses and SARS-CoV-1, the 5′ most 598 nucleotides of Transmissible gastroenteritis virus (TGEV), an Alphacoronavirus, was demonstrated to be necessary for viral packaging (116). The exact structure within this region that dictates packaging was not determined (100), though all three SL5 substructures are predicted in TGEV, suggesting the SL5 structure plays a part in packaging for at least this particular virus.
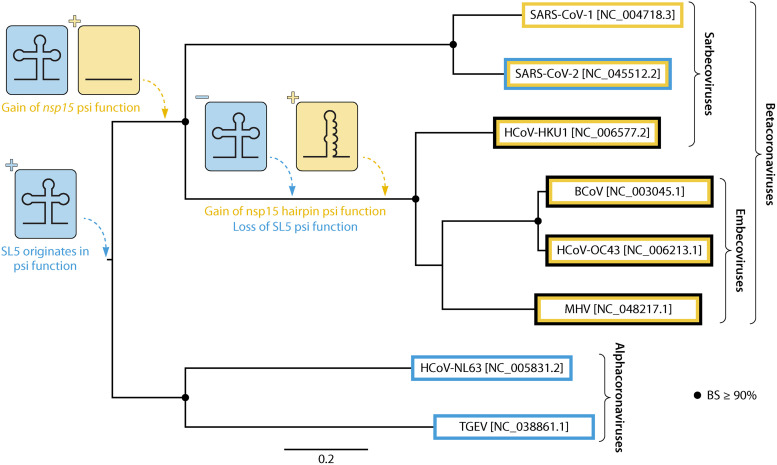
For SARS-CoV-2, the location of the packaging signal within the genome and role of RNA structure has not been elucidated, though RNA corresponding to both the 5′ UTR (as with TGEV) as well as nucleotides 19786 to 20361 (within nsp15, similar to SARS-CoV-1) have been demonstrated to associate with the N protein to form distinct condensate structures (117–119). The findings described thus far for the roles of RNA regions in packaging across coronavirus groups are thought to represent an inverse correlation between SL5 and ORF1ab presence (120). This relationship is consistent with a nonparsimonious pattern, or multiple independent gains and losses of structure/function, pertaining to packaging in the evolutionary history of CoVs (Fig. 3), a phenomenon that, if continued to persist, would render packaging a difficult viral process to target therapeutically going forward. Contrary to this hypothesis, more recent structure prediction for MHV using SHAPE analysis demonstrated the presence of a branched SL5 structure (81), confounding earlier analysis but lending to a less complicated evolutionary history than pictured in Fig. 3. If SL5 were to be observed to contribute to encapsidation across all Betacoronaviruses, the emergence of the hairpin structure in ORF1ab of MHV may be explained by a single, early evolutionary event that resulted in an altered encapsidation mechanism relative to Alphacoronaviruses, for which SL5 appears to act alone.
FIG 3.
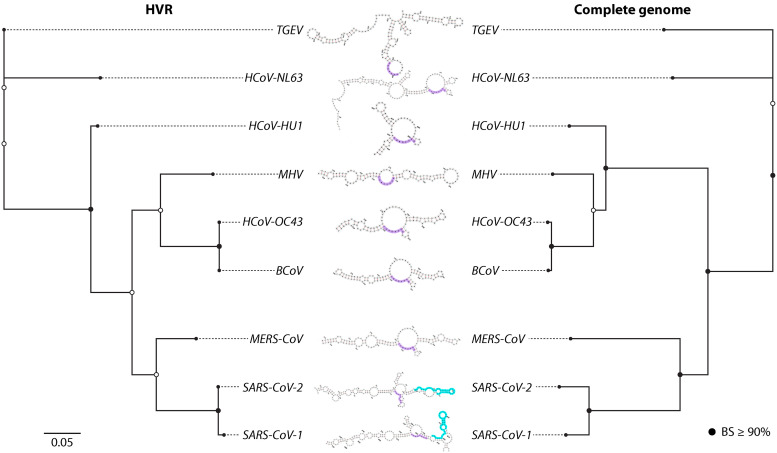
Parsimonious description of the gain and loss of packaging signal (psi) elements during the evolutionary history of select Alpha- and Betacoronaviruses. The maximum likelihood phylogeny was reconstructed (270) from available near-full-length genomes (accession numbers are given in square brackets) and bootstrapping (10,000 replicates [271]) used to provide support (BS). BS ≥ 90% are indicated by black circles. The most parsimonious pathway for the gain and loss of SL5 and nsp15 RNA structure and/or function in packaging is described. Branch lengths are scaled in substitutions/site (scale at bottom). It is important to note that role of the distinct SL5 structure in packaging has not been confirmed.
Analysis of the 5′ UTR RNA structure for SARS-CoV-2 has revealed contrasting evidence of an additional hairpin between SL4 and SL5, designated as SL4.5 (Fig. 1 and 2) located between positions 130 to 150, which comprises a highly conserved region across SARS-CoV-2 and closely related Betacoronaviruses (59, 103, 121). This motif has been presented using a variety of experimental methods; analyses of the 5′ UTR in viral RNA transcripts via in-line probing and RNase V1 enzymatic probing (65), as well as SHAPE-MaP (101), have upheld the formation of the SL4.5 as a short stem-loop, in stark contrast with the single-stranded predictions for this region determined using various SHAPE-based and DMS-based methods (88, 89, 105, 122). Similarly, a small stem-loop was also previously predicted to occur between SL4 and SL5 in MERS-CoV but was not predicted in SARS-CoV-1 (78). The position of SL4.5 has not been predicted to interfere with SL4 and SL5 formation; however, additional biochemical and evolutionary investigation is necessary to elucidate the true nature of this region and its implications.
3′ UTR: THREE PRIME’S A CHARM
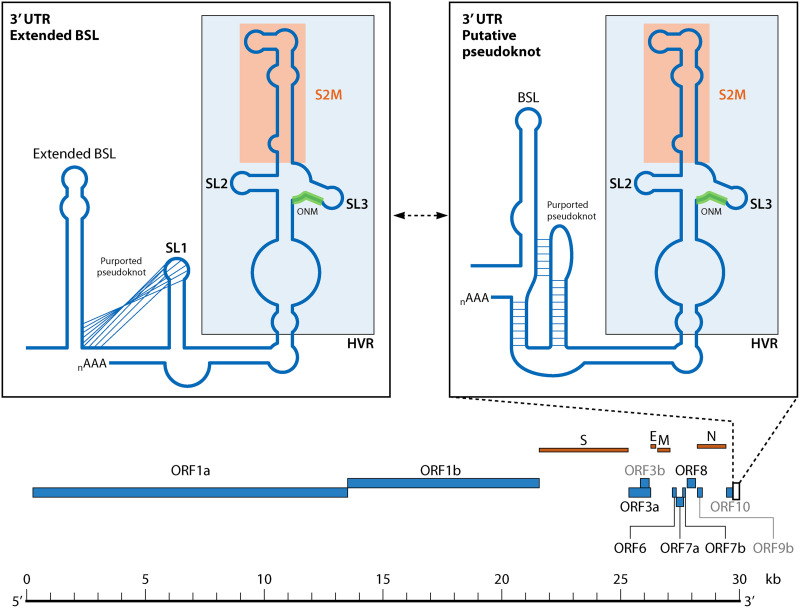
Because transcription initiates at the 3′ end of the genome for both the gRNA and the sgRNA, the 3′ UTR and its genomic structure is critical to the CoV life cycle. The 3′ UTR represents the stretch of genome between the 3′ terminus of the nucleocapsid (N) gene and the 5′ fragment of the poly(A) tail (123). The multitude of RNA structures in the 3′ UTR, believed to function as replication signals, are ubiquitous among Betacoronaviruses, specifically in MHV (124, 125) and SARS-CoV-1 (126). Three higher-order 3′-UTR structures have been specifically noted in SARS-CoV-1 and SARS-CoV-2 and consist of a bulged stem-loop (BSL) approximately 10 nucleotides downstream of the N protein stop codon; followed by a cis-acting pseudoknot or stem-loop (SL1); and ending with an elaborate structure containing the Hypervariable region (HVR), S2M domain (highy conserved across CoVs and other +ssRNA viruses) (127), and the octanucleotide motif (ONM) (123, 128) (Fig. 1 and 4). Five pairs of covarying nucleotides were also found between fractions of the 3′ UTR and the putative ORF10 coding regions (89), suggesting the potential presence of yet another RNA structure within this region. The 3′ UTR has been identified as containing multiple potential druggable binding pockets for which small molecule inhibitors could potentially be developed against (105).
FIG 4.
Structural organization of the 3′-UTR RNA structures within SARS-CoV-2. The two subpanel figures depict the two alternative structural forms that have been described: the extended bulged stem-loop (BSL) and the putative pseudoknot. The Hypervariable region (HVR) region, S2M region, and octanucleotide motif (ONM) motif are represented in light blue, orange, and green, respectively.
The cis-acting pseudoknot within the 3′ UTR is of particular interest, as initial studies of this region determined that it was phylogenetically conserved across CoVs and was essential for BCoV replication, likely functioning in the plus strand (129). A pseudoknot is formed when the single-stranded segments of RNA contained within a linearly isolated structure form complementary base pairing with an outside region of single-stranded RNA (Fig. 1 and 4). Variation in the lengths of the stem and loop regions in pseudoknots lend to the vast heterogeneity of pseudoknot types and stability (130). The BCoV cis-acting pseudoknot was found to be a Hairpin (H) type pseudoknot because the interacting, otherwise single-stranded regions occur as loops (referred to as L1, L2, and potentially L3) associated with separate hairpin stems (129, 131). These hairpin stems, referred to as S1 and S2, stack coaxially in a quasicontinuous helix (131, 132) (Fig. 1 and 4). In Alphacoronaviruses, the cis-acting pseudoknot was found to be highly conserved while the presence of the interacting BSL was not strictly conserved; contrastingly, in Gammacoronaviruses, a conserved stem-loop was predicted in the upstream region of the 3′ UTR (near the location of the BSL described above) and was deemed essential for replication with only a poor candidate for a predicted pseudoknot having been established (129, 132, 133). A somewhat extended, and also phylogenetically conserved, BSL (eBSL) structure (59) was proposed for Betacoronaviruses, encompassing the region predicted to comprise the pseudoknot in the other viruses. While the role of a downstream stem-loop (also referred to as SL1) in a pseudoknot interaction with eBSL was proposed, further analysis demonstrated that the pseudoknot formation was only marginally stable in comparison with an isolated SL1 (134). Though SHAPE and DMS-MaPseq analyses have also failed to verify the pseudoknot structure in the SARS-CoV-2 genome in infected cells (89, 101, 123), a switching mechanism between these two higher-order structures was hypothesized to be involved in the regulation of RNA synthesis (135). Specifically, modeling of the 3′ UTR pseudoknot produced a consistent conformation while modeling of the extended BSL did not reach a consensus structure (106). Previous experiments with MHV have demonstrated that the pseudoknot forms weakly at 25°C and not at all at 37°C (134), as well as only forming when the competing SL1 hairpin structure cannot form. The BSL, however, was deemed critical for the replication of DI RNA, and the structure of the BSL, but not necessarily the primary sequence, was critical for MHV replication (124). Principally, the conservation of base pairing through covariation among four base pairs in the stem structure of the BSL was critical for structure formation and maintenance (124).
While researchers seemed to navigate toward the formation of a pseudoknot in the 3′ UTR at least at some low rate, an additional conformation for this region was predicted with DMS data analyzed by the DREEM algorithm, originally developed to analyze seemingly stable HIV-1 RNA structures with alternative conformations (33), which predicted a new stem-loop formed by a stretch of nucleotides located between the established eBSL and the SL1, which would disrupt any pseudoknot formation (59). Previous analysis demonstrated that, while the SARS-CoV-1 3′ UTR could be substituted for the 3′ UTR in MHV resulting in chimeric virus with the capacity to synthesize viral RNA, the 3′ UTR of TGEV (an Alphacoronavirus) or the 3′ UTR of avian CoV (a Gammacoronavirus) was not tolerated (133), suggesting the presence of distinct, lineage-specific higher-order structures, rather than multiple conformations.
Despite its name, the otherwise considered “untranslated region” of the 3′ UTR putatively contains the open reading frame ORF10, found between the start of the BSL/eBSL and the start of the 3′ structure containing the HVR, overlapping with the pseudoknot/SL1 region. ORF10 encodes for 38 amino acids in SARS-CoV-2 but is interrupted by an early stop codon in SARS-CoV-1 and other SARS-CoV lineages (136). Examined at the protein level, ORF10 has demonstrated evidence of positive selection through excessive amino acid changes, although the function of this protein is currently unknown (136), begging the question of the relevance of these changes (and proposed selection) to the underlying RNA structure(s). It should also be noted that the expression of ORF10 is currently being investigated, as its expression has not been supported by both DNA nanoball sequencing and Nanopore Direct RNA sequencing (49).
Following the 3′-UTR SL1 is a complex secondary structure containing the HVR, the S2M domain, and the ONM (Fig. 1 and 4). Mutant strains of MHV in which this region was deleted (while maintaining all other structures of the 3′ UTR) were found to be viable, replicating at levels comparable to WT virus (132). The HVR region as a whole exhibits relatively poor sequence, as well as predicted secondary structure, conservation across all CoVs (Fig. 5), supporting this finding. The exceptions to this general rule are two particular motifs discussed herein, the ONM (104) and S2M (63, 137). The ONM can be found approximately 78 nucleotides upstream of the poly(A) tail in all CoVs, including SARS-CoV-2, and represents a highly conserved octameric sequence of 5′-GGAAGAGC-3′ (126). Mutational analysis of the ONM in MHV demonstrated its tolerance for single nucleotide mutations at every position, however, as well as random mutations at multiple positions (and even all eight bases), demonstrating yet again a seemingly unnecessary evolutionary constraint on the linear sequence (128). Despite the high degree of evolutionary conservation, deletion of this entire region does not deter replication competency (128), but reduced severity of pathogenesis in a mouse model (128) indicates at least a role for the ONM in infectivity.
FIG 5.
HVR structural evolution among select Alpha- and Betacoronaviruses. Maximum likelihood (Minh et al. [272]) phylogenies were reconstructed for the HVR region (left) and near-full-length genomes (right), with bootstrapping (10,000 replicates) used to provide support (BS). BS ≥90% are indicated by black circles. Within each predicted RNA structure, the ONM and S2M motifs have been highlighted in purple and teal, respectively, if present. Branch lengths are scaled in substitutions/site (scale at bottom left).
Phylogenetic analysis of the HVR region for different CoVs revealed a similar branch length distribution, representative of genetic diversity, to the full genome, consistent with lack of conservation of the region as a whole (Fig. 5). Differing branching patterns could also be observed, representing potentially differing evolutionary patterns, though reliable support could not be attributed to the alternative placement of HCoV-HKU1 within the HVR phylogeny (Fig. 5). The observed sequence diversity and phylogenetic relatedness of HVR sequences readily explain the variation in the predicted HVR secondary structures (using RNAfold) (138, 139), with the number of hairpin substructures ranging from two to as many as five in TGEV. The closely related SARS-CoV-1 and -2 exhibit similar placement of the ONM motifs in relatively small bulges and the presence of the S2M hairpin not represented in other lineages. The S2M motif is also observed among other RNA virus families outside of Coronaviridae, including Astroviridae, Caliciviridae, and Picornaviridae (127, 140), among which the sequence, secondary structure, and tertiary structure are all highly conserved (63, 106, 137). This motif has also demonstrated similarity, though unknown homology, to an rRNA loop, as well as a similar binding affinity to proteins involved in translation, suggesting that this structure functions to hijack the host translation machinery (59, 137). Despite the prevalence of S2M conservation among these viruses, a single nucleotide substitution (G29758U) can be found in the upper stem of SARS-CoV-2 S2M relative to SARS-CoV-1, predicted previously to alter the true structure (63, 137, 141), though a high level of structural conservation was contrastingly reported via modeling (106). This structural conservation partially explains why the S2M domain has been identified as a potentially promising pocket for targeting by small molecule inhibitors (105) and antisense oligonucleotides (ASOs; 142). Similar to the packaging signal, however, targeting of this region may prove problematic, as previous phylogenetic analysis of the S2M indicated that the appearance of S2M in the evolutionary history of these virus families is likely the result of multiple, independent acquisitions of the motif (140). The presence of the S2M domain in SARS-CoV-1, specifically, is thought to be the result of a horizontal transfer event with an Astrovirus, though the origins of the S2M motif in SARS-CoV-2 is unknown. On the other hand, analysis of the SARS-CoV-2 S2M region over the course of the COVID-19 pandemic revealed the instability of these acquisitions through several deletions, with lineages B.1.1.311 exhibiting complete S2M deletion (143). Moreover, two additional variations in the SARS-CoV-2 S2M have been reported, G29734C and G29742U, which are both predicted to contribute to destabilization of the S2M tertiary structure (144). Together, these results suggest the S2M motif may not provide an essential function for SARS-CoV-2 and, therefore, may not prove a successful drug target. The linear sequence in the RNA region upstream of the ONM motif for more distant relatives of SARS-CoV-1 and 2 was too dissimilar to be considered S2M motifs (140); however, stem loops were predicted herein for BCoV, HCoV-OC43, and MHV, suggesting similar function and warranting further investigation into the linear and structural constraints on S2M evolution in coronaviruses.
The presence of two additional stems within this 3′-UTR region immediately upstream and immediately downstream of SL1 was predicted by DMS-MaPSeq, but not by SHAPE-MaP (101, 105), though thought to be largely unstable (123). Additionally, mutations acting to partially destabilize either of the two helices were viable; however, disruption of both of the helices proved detrimental through the inability to direct the sgRNA synthesis (145). Specifically, disruption of base pair interactions in the first helix, formed via pairing of nucleotides 0 to 9 and 217 to 225 (counting from the poly(A) tail), was lethal. Disruption of the second helix, formed via pairing of nucleotides 18 to 29 and 171 to 183, produced either viable or lethal mutants, depending on the mutation (145), the 5′ end proving more necessary for replication. Secondary structure prediction revealed that the lethal mutations disrupting the 5′ end of the helical structure were potentially the result of a different folding pattern than that of the WT structure, but with similar thermodynamic stability. Contrary to what might be expected, the viable mutations that disrupted the 3′ end were predicted to unfold the helix entirely (145), suggesting the second helix is not necessary for replication but has the potential to alter replication capacity through enhanced or differential functionality with specific mutations in the 5′ region.
CoV POLY(A) TAIL: PERHAPS SIZE DOES MATTER
For SARS-CoV-2, the median length of the poly(A) tail has been reported to be approximately 47 residues in length, with gRNA having generally longer tail lengths than sgRNA and sgRNA representing two general populations of tail lengths of 30 and 45 nucleotides, respectively (49). Previous experiments identified a temporal dependence of different tail lengths in BCoV, recording average tail lengths of 45 nucleotides immediately after virus entry into cells, then peaking at approximately 65 nucleotides between 4 and 12 hours postinfection (hpi) and then decreasing in length linearly to approximately <30 nucleotides after 144 hpi (146). The authors suggested that the latter may be in part due to the degradation of the tail (49); however, Wu et al. (146) also demonstrated that BCoV poly(A) tail length was positively correlated with efficiency of translation, as they point out is also the case for eukaryotic mRNAs, and thus varying length of the poly(A) tail is similarly required for regulation of translation. The poly(A) tail has also been shown to be critical to RNA synthesis of the minus-strand RNA in MHV (147) and has been identified as an important cis-acting element in BCoV DI RNA replication, whereby as few as five adenines were sufficient for replication (78, 148). This finding may hold the explanation as to why the length of the SARS-CoV-2 poly(A) tail is relatively shorter than most CoVs. In other organisms such as bacteria, the poly(A) tail has been proposed to participate in the formation of stability-enhancing RNA structures, dictating translational efficiency and transcript stability (149). The impact of the shortened poly(A) tail in SARS-CoV-2 on the upstream RNA structures in the 3′ UTR, however, is unclear (49).
RNA structure in general plays an important role in viral recombination (e.g., 150), as the formation of certain structures can result in inefficient binding of the polymerase and movement of the polymerase between genomes in the neighboring area of synthesis, referred to as template switching. When the templates originate from different viruses, segments joined through recombination form entirely new viral strains. As with point mutations, these alterations may be beneficial to the virus. Additionally, insertions and deletions (indels) observed across the CoV genome have been associated with RNA structure formation in the respective region, indicating they might be artifacts of recombination (151). In MHV as a relevant example, analysis of spike deletion variants (SDVs) revealed a clustering of polymorphisms at the base of a stem-loop located in a hypervariable region of the coding region (152). Additional analysis revealed that the generation of SDVs ultimately resulted in the establishment of a stably heterogeneous population of MHV, or quasispecies, in persistently infected mice and that increasing diversity of SDV quasispecies was correlated with heightened disease severity (153). As the structured region of the 3′ UTR has previously been shown to be critical for the engagement of the RdRp (154), differences in the RNA architecture in this region are capable of influencing recombination and the ability of the virus to adapt (132, 155). Common 3′ replication-signaling RNA elements have been observed between MHV and BCoV (Embecoviruses), which are interchangeable in the production of viable virus despite a sequence divergence of 31% (124); MHV has already demonstrated its advantage as a helper virus for the replication of BCoV DI RNA, indicating recognition of these signals in BCoV by the MHV replication machinery (155). Also in this study, the BCoV DI RNA readily acquired the 5′ leader sequence of an HCoV-OC43 (itself also a member of the Embecovirus subgenus) helper virus, demonstrating not only in vivo recombination among CoVs, but recombination between a human and nonhuman CoV (155). “Leader switching” has been described as a high-frequency event in terms of recombination (156), so much so that it has been hypothesized to be a necessary part of the replicative process (157). Hence, though not a complex structure, additional investigation into the simple stretch of adenines at the end of the SARS-CoV-2 genome may reveal a significant role in viral evolution through its subtle impacts on neighboring RNA structures and the process of recombination that appears to play a major role in CoV replication.
TRS-B: SELECTION FOR STRUCTURIZATION IN CIRCULARIZATION
For SARS-CoV-2, as well as other CoVs, all proteins following ORF1ab are translated from positive-sense sgRNA intermediates (51). The transcription of the sgRNAs relies on the presence of transcription regulation sequences of both the leader sequence (TRS-L, located in SL3) and of those that precede the 5′ edge of each gene (TRS-B) (94), as mentioned previously. The generated sgRNAs comprise a set of 3′ coterminal, nested transcripts, each with a leader RNA sequence derived from the 5′ UTR of the genome fused to the 3′ body sequence (57, 94). This fusion event is mediated in part by the cis-acting TRS elements, which contain a conserved core sequence that mediates the long-range template switch through sequence homology (57, 94). Upon reaching a TRS-B sequence during synthesis, the RdRp is thought to pause, subsequently “jumping” to the TRS-L sequence, mediating the recombination-like fusion event at the TRS sites (57, 158). It should be noted that noncanonical fusion events have been detected occurring outside of known TRS-B sites (e.g., references 159, 160); however, the biological relevance of these events, and whether or not they represent legitimate replicase jumps, remain to be elucidated (49). Previous analysis of the SARS-CoV-1 TRS regions demonstrated the presence of a minimal core sequence (CS) of 5′-ACGAAC-3′ (161, 162) or 5′-CUAAAC-3′ (163) mediating fusion, which are similar SARS-CoV-2 (may differ by up to one base pair [164]), indicating heavy selection. Seven of the nine TRS-B regions have been found to reside within stem-loop structures, with all but the TRS-B located at the N coding sequence containing their core sequence in the stem region of their respective loops facing the 5′ terminus of the genome (88). Abundance of sgRNA was determined to be positively correlated with the single-strandedness of its corresponding TRS-B (89), indicating a crucial role for TRS-B structure heterogeneity in sgRNA transcription (104). For this recombination-like process to occur effectively, the TRS-L and TRS-B sequences need to be in close proximity to each other; it is, therefore, hypothesized that the fusion is facilitated by the circularization of the RNA genome that results from RNA-RNA interactions at the 3′ and 5′ termini. Genome circularization is commonly observed for CoVs, including SARS-CoV-2 (37) as well as other RNA viruses, and is used to facilitate efficient viral replication and likely promotes recombination (57).
PRF STEM LOOPS: FRAME-SHIFTING INTO HIGH GEAR
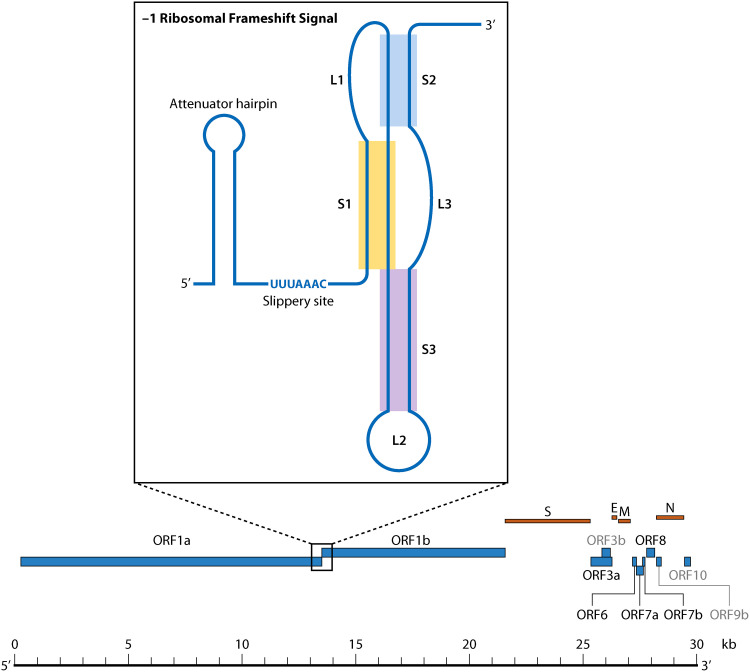
The utilization of overlapping reading frames to maximize coding potential is evolutionarily advantageous (165), and the switch between reading frames is often facilitated by RNA structure, typically involving RNA pseudoknots (166). As the individual proteins encoded by these ORFs are critical to viral replication, so are these frame-shifting elements (FSE). One of the most well-known FSE in RNA viruses belongs to HIV: read-through of the overlapping group antigen gene (gag) and polymerase gene (pol) reading frames to express the Gag-Pol fusion protein is mediated by an RNA pseudoknot (167). The function of pseudoknots as FSEs has been well characterized in SARS-CoV-1 and other CoVs, particularly involving the expression of ORF1b proteins (168, 169). Additionally, independent translation of the ORF1a and ORF1b regions requires a programmed −1 ribosomal frame-shifting (PRF) event mediated by the pseudoknot structure found between ORF1a and ORF1b (169) (Fig. 1 and 6). Downstream of the PRF element in SARS-CoV-1 and 2 is a heptanucleotide “slippery” sequence (170), which follows the general motif XXXYYYZ (171). For CoVs (including SARS-CoV-2) X represents any base, Y represents A or U, and Z represents either an A, C, or U (170, 172). This region is structured such that at some frequency the ribosome will pause upon encountering the pseudoknot and “slip” back, allowing the tRNA to repair in the −1 frame (166, 168, 170). The current model for this mechanism is the result of an equilibrium of tension across the system: as the ribosome begins to unwind the first stem of the pseudoknot, increased tension in the stretch of RNA between stem 2 and the RNA in the ribosomal active sites (169, 170, 173) leads to a “supercoiling” effect (described by Kelly et al. [169]) in the second stem. This phenomenon opposes forward motion of the ribosome, causing the ribosome to pause with its A- and P-sites bound to aminoacyl- and peptidyl tRNAs positioned above the slippery site. Slippage and consequent repairing of the tRNAs in the −1 frame relieve the tension, allowing for the read-through of the ORF1a stop codon and translation of ORF1b proteins, ultimately maintaining balance in expression of ORF1a to ORF1ab (169, 170).
FIG 6.
Ribosomal frameshift signal structure and genomic location within SARS-CoV-1 described by Kelly et al. (169).
The PRF element structure is highly conserved among known CoV members of the same genus, but not between genera; specifically, stem 1 length and GC composition are conserved among CoVs (notably between SARS-CoV-1, SARS-CoV-2, MHV, and BCoV), but these variables tend to vary more for stem 2 between CoV genera (174). The potential formation of a “kissing loop” structure between stem 2 with a sequence approximately 200 nucleotides downstream of the slippery site is considered to be prevalent among Betacoronaviruses (174) (Fig. 7A), although verification outside of HCoV-229E (175) and TGEV (176) is necessary given less conservation in the second stem. In SARS-CoV-2, a region 200 nucleotides downstream of the PRF element has been predicted to share a highly conserved RNA secondary structure with SARS-CoV-1 and closely related bat coronaviruses, which the authors speculate may be involved in a more elaborate structure akin to that described for HCoV-229E (59). The presence of a third stem-loop in the SARS-CoV-1 PRF element renders it more complex than other CoVs (168, 174, 177), although typical of PRF elements. Secondary structure predictions for this region indicate that all known CoVs have the capacity to form this third stem, despite the fact that the RNA nucleotide sequence of this region among the different individual viruses is not well conserved (174).
FIG 7.
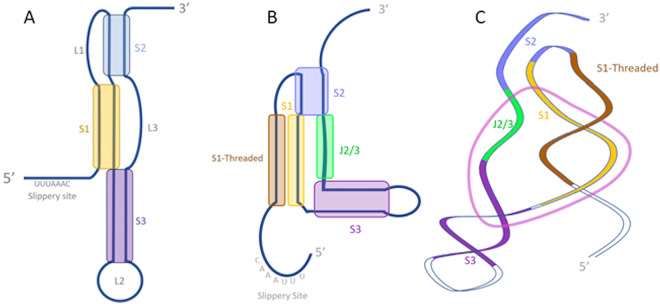
Differences in structure prediction for the SARS-CoV-2 frameshifting pseudoknot. (A) Secondary structure described by Plant et al. (174) using 2-dimensional nuclear magnetic resonance (NMR). (B) Secondary structure described by Zhang et al. (178) using SHAPE. (C) Tertiary structure described by Zhang et al. using cryo-EM map guided modeling. For panels B and C, the dark brown highlighted segment represents the stem strand described as “threaded” through the” ring” (displayed in light pink) formed by the other strand of stem 1 (light brown), stem 2 (blue), stem 3 (purple), and an unpaired region between stem 2 and stem 3 (J2/3) (green).
Single-particle cryo-electron microscopy (cryo-EM) examination of the frame-shifting element similarly demonstrated a three-stemmed pseudoknot; however, its tertiary structure was characterized by a distinct conformation for stem 3 that did not involve direct stacking with the stem 1-stem 2 pseudoknot (178) (Fig. 7B and C). In this description, the first strand of stem 1 is threaded through what the authors describe as a “ring” formed by the other strand of stem 1, the single-stranded region connecting stems 2 and 3, and the start of the double-stranded regions of stems 2 and stem 3. The in vitro results of Zhang et al. (178) were contradicted by SHAPE reactivities collected in vivo, which identified paired nucleotides in the cryo-EM model as unpaired, representing potentially unfolded regions of the frame-shifting element present in the context of the whole SARS-CoV-2 genome (105). Alternatively, DMS-MaPseq of an in vivo model of the PRF element in the context of the whole SARS-CoV-2 genome did not demonstrate the canonical three-stemmed pseudoknot, but rather the presence of an alternative stem 1 formed by the base pairing of the region immediately downstream of the slippery site with a complementary 10-bp region located 29 bases upstream of the slippery site (88). This model was generated in silico, based on predictions of the incorporation of upstream nucleotides in the FSE (103). Modeling of an extended FSE region in this way demonstrated putative heterogeneity in structural conformations (59). In an attempt to characterize these structural dynamics in SARS-CoV-2, recent experiments using optical tweezers have subjected the PRF element to tensions that simulate the −1 frame-shifting event. These experiments identified the formation of multiple structures that largely fall into two distinct fold topologies: a structure in which the 5′ end is threaded as described above, and another structure wherein the 5′ end remains unthreaded (179). The threaded conformation results in increased rigidity of the RNA in SARS-CoV-2 (179), though the biological implications of these dynamics are unclear.
Inhibition of the −1 ribosomal frameshift mechanism in SARS-CoV-1, as would be expected, effectively halts viral replication (180), and targeting this PRF with small molecule inhibitors has been shown to specifically ablate frame-shifting activity during transcription (181, 182). The primary sequences for the SARS-CoV-1 and SARS-CoV-2 pseudoknots only differ by a single nucleotide found within a loop region that is not predicted to affect the structure (170). The conservation of the −1 PRF structure across SARS-CoV-1 and SARS-CoV-2 and function of this pseudoknot across CoVs explains why small-molecule frameshift inhibitors previously demonstrated to inhibit −1 PRF activity in SARS-CoV-1 had a similar effect on −1 PRF activity in SARS-CoV-2 (170). Recent work has identified the compound merafloxacin as a potent inhibitor of the −1 PRF activity in SARS-CoV-2 as well as in others that use similar RNA structures to facilitate frame shifting (183). This drug was demonstrated to be effective against single-nucleotide substitutions in the coding region of the pseudoknot, which have been identified as recurrent in the SARS-CoV-2 viral population (183). Additionally, ASOs have been developed to target the RNA structure of the frameshifting element. Viral replication was diminished by generation of ASOs targeting regions of the pseudoknot intended to disrupt its RNA structure; specifically, an ASO targeting a region adjacent to stem 3 resulted in diminished viral replication (184) and ASOs generated to target stem 1 and the slippery site also diminished viral replication (178).
Upstream of the slippery sequence exists an attenuator hairpin, present in both SARS-CoV-1 and SARS-CoV-2 (101) (Fig. 6), which has been shown to help downregulate frame-shifting activity (170, 185). Efficiency of attenuation is closely tied to the stability of the hairpin as well as its proximity to the slippery sequence, with the current mechanistic model revolving around the unfolding and refolding of the hairpin as the RdRp moves through the region, the latter likely exerting tension in the 5′ direction (185). Increasing the thermodynamic stability of the hairpin decreases −1 PRF activity, and an analysis of small molecules targeting this region in SARS-CoV-2 by stabilizing the attenuator hairpin contributed to a decrease in frame-shifting efficiency, representing a therapeutic target that could disrupt SARS-CoV-2 translation (186). Significant differences in the primary nucleotide sequence have been observed in the region of this hairpin between SARS-CoV-1 and 2 that would potentially restrict the applicability of this drug to other CoVs - SHAPE-MaP analysis has shown that the SARS-CoV-2 hairpin even contains a distinct fold (101). However, the function of the attenuator hairpin is retained between SARS-CoV-1 and SARS-CoV-2 as a consequence of its conserved secondary structure (170). Alternatively, in silico analysis has suggested that both the slippery site and attenuator hairpin are found within a larger multibranched structure, which has previously been unreported (103).
SECONDARY STRUCTURE IN UNCHARACTERIZED REGIONS: BOLDLY GOING WHERE NO MAP HAS GONE BEFORE
As previously mentioned, early in the course of the pandemic, Rangan et al. (59) identified regions of sequence conservation within the 5′ and 3′ UTRs and frame-shifting element between SARS-CoV-1 and -2 and related bat coronavirus. These regions of conservation correspond to elements of RNA structure that are known to be shared among these closely related Betacoronaviruses, as enumerated above. In addition to these known regions, Rangan et al. identified multiple regions of sequence conservation that fall outside of well documented regions of RNA structure. Several of these regions were of sufficient length for the design of antisense oligonucleotides, showing promise as future drug targets. Specifically, 30 discrete regions were noted as sharing ≥99% sequence identity among all SARS-CoV-2 sequences (deposited as of March 18, 2020) and were also conserved across closely related bat coronaviruses and SARS-CoV-1.
More than 45% of the SARS-CoV-2 genome has been predicted to be highly structured according to RNAz (59) and ScanFold (103) analyses, which is generally a larger portion of the genome than other Riboviruses (103). Interspersed stretches of unpaired, yet conserved nucleotides were also posited by the authors to exhibit function in the form of recruitment of molecular machinery (59), contributing to the role of viral RNA in functions outside of mRNA-mediated protein production. Within this 45%, SHAPE reactivity analysis has identified 87 discrete, highly structured and thermodynamically favorable RNA regions across the genome, nine of which demonstrated significant covariation in at least two base pairs indicating natural selection (105). These 87 regions, representing approximately 19% of the SARS-CoV-2 genome, demonstrate a high degree of correlation between in vivo and in vitro SHAPE analysis reactivities, suggesting the sequence and thermodynamics alone account for folding (105). Additionally, ScanFold analysis largely agreed with these structured RNA regions, with 34 regions being identical between the two models (103). ScanFold analysis identified eight additional uncharacterized structures, which were predicted to possess more than one covarying base pair indicative of selection (103). One of these regions was previously targeted by an antisense oligonucleotide (ASO), which resulted in decreased viral infection in cells, indicating the value of investigating these regions of therapeutic interest (89, 103). Although, traditionally, ASO targeting of specific RNA sequences has been hindered by RNA structures, which may occlude the base pairing of the ASO to the target sequence, the RNA structure-based design of ASOs does not rely on canonical Watson Crick base (WC) pairing with the linear sequence (184). As mentioned previously, an ASO targeting stem 3 engages in major-groove base triples with bases along stem 1 and a Hoogsteen base-pair with the 3′ end of stem 1, resulting in greater inhibition of viral replication than an ASO strictly targeting the same sequence through strictly WC base pairing (184).
While 45% represents a larger-than-expected RNA structure presence given Ribovirus history, additional evidence has suggested that >84% of the SARS-CoV-2 genome is capable of forming structured, and even conformationally flexible, RNA as determined by the DREEM algorithm (35, 88). Naturally, a greater degree of variability in nucleotide reactivities is observed in the structures that comprise the difference between these two estimates, particularly in loop regions. This finding may be expected, however, if the majority of these additional structured regions do indeed exhibit multiple conformations. Experimental evidence supporting this hypothesis has not yet been demonstrated.
“LONG COVID”: THE LONG CON
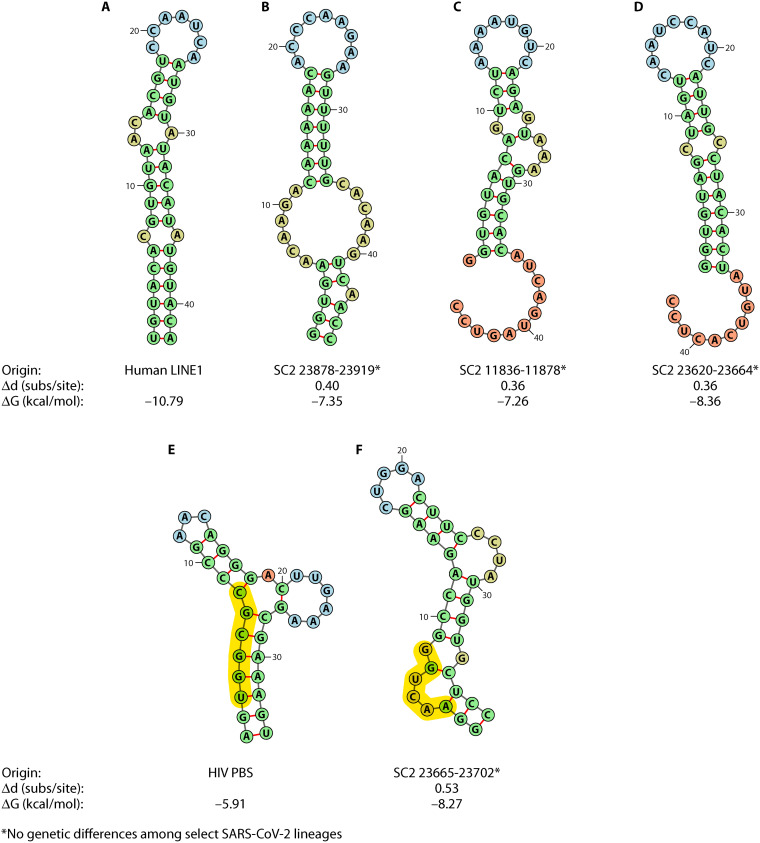
As illuminated by this review, the formation of RNA structures within the CoV genome, as well as other RNA viral genomes, is critical to their replication cycles and biological function. As mentioned previously, RNA structure is critical to the HIV replicative process known as reverse transcription and, ultimately, integration of the HIV genome into the host cell. Reverse transcription of HIV is initiated by the interaction of a host tRNA molecule with the HIV genome at a region referred to as the primer-binding site (PBS), located within the viral 5′ UTR. The PBS is a model of RNA functionality, as it has been shown to adopt multiple conformations with distinct functions: one in which an intramolecular helix and hairpin is formed following binding of the tRNA, and the other wherein an intermolecular primer activation signal helix forms between the viral RNA and tRNA (187, 188). This interaction is recognized by the HIV reverse transcriptase (RT), which carries out the formation of the double-stranded DNA molecule that is subsequently integrated into the host genome. The differing RNA conformations identified potentially serve as a regulation mechanism for the initiation of reverse transcription (187).
Non-retroviral RNA virus sequences have been detected in the genomes of many vertebrate species and/or have exhibited integration capability ex vivo (189–191), and recent analysis has put forth controversial evidence of potentially integrated SARS-CoV-2 (192). The transcription of this integrated provirus has been purported to explain the occurrence of prolonged viral RNA shedding seen in a fraction of infected individuals, referred to as individuals with “long COVID” (192–197). The prevalence of this phenomenon has not been fully characterized; however, a report examining large numbers of patients found extended viral RNA shedding after 21 days postonset of symptoms in 25.6% of patients (198). Integration of RNA viruses into human cells can occur in the absence of retroviruses such as HIV owing to the presence of endogenous retrovirus RTs. Alternatively, autonomous retrotransposition can occur across viruses and cellular RNA alike in elements referred to as retrotransposons, some of which can act as sources of reverse transcription for other nonautonomous elements. Long interspersed nuclear element 1 (LINE1) retrotransposons are prevalent in the human genome, accounting for approximately 17%, and are partially responsible for the “fossilization” of numerous species of nonretroviruses into the genome over course of human history, which now comprise as much as 8% of the human genome (199). Similar to PBS-recognition by HIV, Grechishnikova and Popstova (200) proposed LINE1 RT recognition of an RNA structure that was deemed conserved across human LINE1 3′ UTRs. The consensus stem-loop structure consists of a 5- to 7-base-pair loop (5′-CCAAUCA-3′ in humans) and average of 8- to 10-base-pair stem with an asymmetric bulge at a distance of 4 to 6 bp from the loop (Fig. 2), though the authors also found conserved stem-loop positions in other locations, such as the 5′ UTR, that may serve as potential candidates for reverse transcription. Grechishnikova and Popstova point out that the presence of the bulge was conserved in all reportedly active LINE1 transposons (not all elements are functional), comprising >50% of the analyzed 6,622 LINE1 elements from 27 families. Furthermore, previous experimental deletion of this bulge in eel transposons completely abolished transposition, leading the authors to believe the 3′-UTR stem-loop bulge to be critical in RT recognition.
The reverse transcription and integration of SARS-CoV-2 described by Zhang et al. (192) was demonstrated in a HEK293T (human embryonic kidney) cell line that was transfected with a LINE1 expression vector. The majority of observed integration events involved sgRNA with LINE1 recognition sequences at a site near the junction, which the authors note would be in line with LINE1-mediated retrotransposition (192). Evidence of integration was also obtained in a calu3 (human lung epithelial cell) cell line (192). This report was the first to ascribe a retroviral property to a CoV (192). Although these results have many quizzical and interesting implications for both SARS-CoV-2 RNA structure and function, as well as COVID-19 pathology, the findings are still under investigation (201, 202), deemed as potential artifacts of the sequencing method (203). Further analysis investigating integration of SARS-CoV-2 in a HEK293T cell line found no evidence of integration, suggesting that if such integration events occur, they are likely incredibly rare (202). The importance of RNA secondary structure in LINE1 retrotransposition has previously been established whereby a 3′-UTR stem-loop has been demonstrated as essential (204, 205) and is proposed to serve as a common binding motif for LINE1 reverse transcriptases present in both LINE1 and Alu RNAs (200). If SARS-CoV-2 is indeed integrated via LINE1-mediated retrotransposition, it is likely that some homologous RNA secondary structure is involved. Analysis of RNA sequence and structural similarities (among the original strain and Alpha [MZ344997.1] and Delta [MZ359841.1] variants) revealed three potential positions within the SARS-CoV-2 genome that could act as a LINE1 element (Fig. 8A to D). The length of the hairpin, number of bulges, and bulge size differed across all three structures, and the minimum free energy estimates for each was greater than the human LINE1, suggesting less stable structures. Only the structure coinciding with position 11836 to 11878 within the SARS-CoV-2 genome (Fig. 8C) contained an asymmetric bulge. Similar to other LINE1 hairpins described in Grechishnikova and Popstova (200), this bulge contains an adenine and is located five base pairs from the loop. However, Zhang et al. (192) described integration of specifically the nucleocapsid protein (28274 to 29533). The downstream location of this region relative to the structure in Fig. 8 renders this structure unlikely to be utilized in the integration observed by Zhang et al. Similarly, both of the two remaining LINE1 possible structures are located upstream, suggesting LINE1 retrotransposition is not the likely mechanism. Not excluding RT recognition of a PBS-like element, the HIV-1 tRNA annealing arm of the RNA structure containing the PBS sequence was used in an alignment with SARS-CoV-2, revealing a somewhat similar sequence region (23665 to 23702) exhibiting greater stability than the original structure (Fig. 8E), though the six nucleotides determined to be the minimal sequence necessary for function by Wakefield et al. (206) (UGGCGC) were not similar to the aligned SARS-CoV-2 genome (AACUGG), nor did they form base pair interactions within the stem of the SARS-CoV-2 structure. As the PBS functions within the 5′ UTR of retroviruses, the upstream location of this structure relative to the nucleocapsid protein-coding region suggests this structure to be a better explanation than LINE1 for integration; however, further investigation is necessary to elucidate the biological reality of the capability of this structure in viral RNA reverse transcription and to contextualize the discrepancy of the conflicting results revolving around CoV integration.
FIG 8.
Predicted RNA secondary structures for regions within the SARS-CoV-2 (SC2) genome sharing similar sequence content with original human LINE1 hairpin (200) (A) and HIV-1 tRNA annealing arm containing the primer-binding site (PBS) (B). Genomic coordinates within the SARS-CoV-2 reference genomes, including the original Wuhan-Hu-1 reference sequence (MN908947.3), USA-WA12020 (MN985325.1) as used in Zhang et al., and variants Alpha (MZ344997.1) and Delta (MZ359841.1), are provided (B, C, D, and F), as well as the Hamming distance for the alignment of the original sequence (A and E) with the reference SARS-CoV-2 sequence (Δd) and the minimum free energy value for predicted RNA secondary structure (ΔG) in RNAfold (138, 139). PBS sequence is highlighted in yellow. No genetic differences were observed between MZ344997.1, MZ359841.1, MN985325.1, and NC045512.2. A double GC base pair was added to each SARS-CoV-2 sequence (flanking) to force stem formation and is included in the pictured structure.
SARS-CoV-2 EPITRANSCRIPTOMICS: THE NEW METHYLS ON THE BASE
Epigenetics, the field describing the modification of DNA nucleobases and histones, is a well characterized biological phenomenon that is essential to the regulation and expression of genes. Although comparatively still in its infancy, epitranscriptomics, a field exploring the plethora of modifications, which can be amended to RNA bases (the transcriptome) and their effect on gene expression (207), is rapidly gaining interest in line with increasing evidence of the numerous roles for RNA in essential biological processes (208). Currently, there are more than 150 known RNA base modifications (209), the majority of which are associated with rRNA and tRNA (210). For cellular mRNA, epitranscriptomic modification is primarily associated with the 5′-cap structure, although internal modifications can also occur in introns and coding regions (207). For eukaryotic and viral mRNA alike, the nucleotide that comprises the cap is typically a methylated guanosine (m7G) required for the efficient translation of the mRNA (207, 211), while the first and second residues adjacent to the cap can either be 2′-O-methylated (2′-O-Me) or, if the first of these residues is an adenosine, already having been modified to 2′-O-methyladenosine (Am), it can be additionally modified to N6,2′-O-dimethyladenosine (m6Am) (207). The latter, a reversible modification, is associated with an increase in transcript stability (212). In viruses, the 2′-O-methylation occurring on the penultimate nucleotide is implicated in discriminating self versus nonself mRNA, as the absence of this modification is recognized by the host cell as a pathogen-associated molecular pattern that triggers an antiviral response (213, 214).
For SARS-CoV-2, the gRNA and sgRNA both acquire a methylated 5′ cap (215, 216), which must be added by a virally encoded mechanism, as replication does not occur in the nucleus and so cannot utilize the host capping mechanism (217). This process is accomplished first by a guanylyltransferase, which has recently been identified as nsp12 in SARS-CoV-2 (218, 219). It should be noted that the results of Walker et al. (218) and Yan et al. (219) are the first indication that nsp12 possesses guanyltransferase activity in a coronavirus, as previous investigations attempting to elucidate the identity of CoV guanyltransferase, including experiments using SARS-CoV-1 nsp12 (220), did not indicate that it functioned as such (217). The novelty of this finding remains to be characterized in other CoVs. Following nsp12 function is the cooperative methyltransferase activity of nsp10 and nsp14 (221–223) and subsequent adenosine methylation by nsp10 and nsp16 (Fig. 9). The combined efforts of these viral enzymes, acting independently of host cellular capping enzymes, allow CoVs to effectively camouflage their genetic material as host mRNA, exhibiting the same nucleobase modifications found on cellular 5′ caps. Naturally, disruption of these modification processes has been found to disrupt the viral life cycle. Specifically, ablation of nsp16 function in SARS-CoV-1 was determined to attenuate viral replication, which was restored by deficient expression of viral mRNA-sensing enzymes both in vitro and in vivo (224). Inhibition of nsp16 2’-O-methyltransferase activity by nucleoside analogues such as sinefungin (221) similarly permits recognition of the viral RNA, leading to the induction of a type I interferon (IFN) antiviral response (225). Structural similarities and a high degree of conservation in the active site between nsp16 of SARS-CoV-2 and that of SARS-CoV-1, as well as similar binding patterns to sinefungin (226), suggest that inhibitors of nsp16 represent a promising direction for antivirals against SARS-CoV-2 and future CoVs.
FIG 9.
Current model for capping mechanism of SARS-CoV-2. Guanosine nucleobase added by guanylyltransferase nsp12 is highlighted in purple. Methyl groups added by the N7-methyltransferase nsp14 and 2’-O-methyltransferase nsp16 are highlighted in blue.
Outside of the 5′ cap, internal modifications can be made, playing a major role in the stability of RNA molecules such as mRNA (207, 227). N6-methyladenosine (m6A) and conversions from adenosine to inosine are the most common internal mRNA modifications. These, as well as other internal modifications, modulate a variety of RNA processes, including splicing and translation (207). Though m6A is potentially involved in multiple facets of mRNA activity (227), it is typically observed near stop codons and 3′ UTRs (228) and is associated with the decay of mRNA transcripts mediated by the cellular protein YTH-domain family member 2 (YTHDF2) (229, 230). Unlike capping, the addition of m6A to mRNA occurs primarily in the nucleus and is accomplished by the host m6A methyltransferase enzymes METTL3 and METTL14. The METTL3/METTL14 heterodimers, along with the WTAP cofactor (231, 232), are thus considered “writers” of m6A methylation, which target contextual adenine residues, discussed in more depth below. Like most cellular processes, the action of m6A writers is counteracted by the so called “erasers” of m6A-RNA demethylases. There are two primary demethylases: obesity-associated protein FTO, which typically targets m6A modifications adjacent to the 5′ cap, and ALKBH5, which targets more generalized m6A. ALKBH5 has been observed primarily in the nucleus (233) and has been shown to be upregulated upon viral infection, suggesting a role in cellular antiviral activity (234). It has been found that knockdown of ALKBH5 leads to a decrease of SARS-CoV-2 replication in vitro (235). m6A ‘readers’ have also been characterized, though, unlike their accompanied writers that exhibit preference for the nucleus, readers can be present in either the nucleus or cytoplasm, depending on their function (227). Notably, the YTH domain mentioned above is contained in the cytoplasmic proteins YTHDF1F2/F3, in the nucleic protein YTHDC1, but also in the protein YTHDC2, which can be observed in either location (227, 236). These reader proteins are able to bind m6A, primarily through a “tryptophan cage” surrounding the methyl group, and influence the ultimate fate of the modified RNA transcripts (227). The nature of the epitranscriptome as a potentially dynamic system comprised of a balance between writing, reading, and erasing modifications is still being investigated, and there is much work to be done to fully understand the laws governing RNA modification, particularly for viruses; however, for an authoritative review of cellular m6A modifications beyond the scope of this review, we defer to Zaccara et al. (227).
For viruses that replicate in the nucleus, m6A modification of viral transcripts is a somewhat well-characterized phenomenon. The 3′ end of the HIV-1 genome hosts multiple m6A modification sites, for which YTHDF protein interaction promotes viral expression (237). As a result of overexpression of YTHDF2, viral replication is enhanced in the HIV target cell population; CD4+ T lymphocytes (237). Additionally, influenza a virus (IAV) has been shown to express m6A-modified RNAs, and inhibition of m6A modification by depletion of METTL3 severely hampered IAV gene expression (238). Given that the primary location of m6A readers and writers is the nucleus, it is not a stretch to imagine how they may act on the genomes of viruses that replicate there. Moving out of this group of viruses, how then are RNA modifications deposited on both negative- and positive-sense RNA viruses such as Zika Virus (ZIKV) and respiratory syncytial virus (RSV), respectively (239), which operate solely in the cytoplasm? This intriguing question is being actively pursued. ZIKV replication can be inhibited by METTL3/METTL14 methylation as well as by recognition by YTHDF2, the silencing of these proteins acting to increase Zika replication (240). Despite this finding, in vitro infection of HEK293T cells with ZIKV did not lead to significant cytoplasmic localization of METTL3/14 and/or ALKBH5, although these proteins were detected in the cytoplasm of uninfected cells (240). Changes in the localization of these proteins were also not seen in in vitro infection of HeLa cells with RSV (241). For a further review of epitranscriptomic modifications in viral infection, please see Baquero-Perez et al. (239). In contrast to the results for ZIKV and RSV, in vitro infection of Huh7 and Vero E6 cells with SARS-CoV-2 resulted in the recruitment of METTL14 and ALKBH5 (235), as well as other m6A modifiers (242, 243), from the nucleus to the cytoplasm, although this finding could not be reproduced in A549+ACE2 and in Vero E6 cells (244). The former results readily explain the decreased replication observed for SARS-CoV-2 following depletion of writer METTL3 (244, 245) and readers YTHDF1 and YTHDF3 (244). This reduced activity is likely the result of decreased m6A levels, as was observed in Caco-2 cells by Li et al. (245) and Vero E6 cells by Zhang et al. (242, 243). In addition to changes in viral activity, Li et al. (245) found that in METTL3 knockdown cells, SARS-CoV-2 infection resulted in an increase in inflammatory cytokine and chemokine expression. The cytosolic RNA senser RIG-I was observed to bind SARS-CoV-2 RNA, which was enhanced upon METTL3 knockdown, suggesting m6A methylation acts to prevent recognition of SARS-CoV-2 RNA by RIG-I. Binding of RIG-I to SARS-CoV-2 RNA with mutations in m6A sites further led to increases in inflammatory gene induction (245). These results are consistent with previous studies indicating m6A-modified RNA is not bound efficiently by RIG-I, thus failing to initiate an immune response (246). COVID-19 infection has indeed been associated with upregulation of global mRNA m6A methylation, and more severe cases demonstrate higher methylation levels than mild cases (247). COVID-19 patients also exhibited an increase in the expression of certain m6A regulators, including METTL14 and, to a lesser extent, METTL3 (248). Unaltered, or even reduced, METTL3/14 expression, as was observed in lung biopsy samples and bronchoalveolar lavage fluid of severe COVID-19 patients (245), may then explain the inability to control SARS-CoV-2 infection through RIG-I-mediated viral sensing.
Sites identified in SARS-CoV-2 as m6A-modified or putative m6A-modified, largely through the efforts of direct RNA sequencing from infected human lung cell lines, can be observed in both the genome and subgenome (235, 242–244), though an increased frequency of this pattern observed for longer viral transcripts (corresponding to S, 3a, E, and M) relative to the shorter transcripts comprising the remaining 3′ regions, as well as for transcripts with shorter poly(A) tail lengths (49). Enrichment of m6A methylation sites in terms of the relative genomic location has also been observed in the 3′ end, similar to HIV (237). m6A modification has been detected in the SARS-CoV-2 negative sense RNA (235), also enriched at the 3′ genomic end, though generally at a later stage of infection (235). This finding has been contested by Baquero-Perez et al. (239), owing to the utilization by Liu et al. (245) of fragmented total RNA, the identification of m6A modification in SARS-CoV-2 could not be definitely ascribed to either gRNA or sgRNA. As hinted to above, m6A enzyme writers METTL3/METTL14 exhibit a preference for methylation of adenine residues in consensus stretches of nucleotides referred to as DRACH (D being an A, G, or U; R being an A or G; and H being an A, C, or U) (227), representative of A/G rich stretches of sequence, confirmed by additional studies by Kim et al. (49). Although the DRACH motif appears frequently across the cellular, and even SARS-CoV-2, transcriptome (as many as 30 in the N coding gene) (249), relatively few of these sites are actually methylated in the mRNA of either, and a complete view of the factors and mechanisms that govern the specificity of m6A methylation has yet to be elucidated (227, 250–252). Though RNA structure comes to mind, METTL3 and METTL14 do not show any preference for RNA secondary structure (231). Preference, however, does not imply lack of impact: the addition of m6A at the specific DRACH motif GGACU has been shown to affect RNA secondary structure via destabilization of A-U base pairing but also increasing stabilization of base stacking in unpaired regions of RNA adjacent to helical regions (253), warranting further investigation into RNA structural differences and immune recognition of viral RNA.
Although not as robustly characterized as m6A, other epitranscriptomic modifications have been detected for SARS-CoV-2 and other CoVs as well. The SARS-CoV-2 genome was found to contain multiple sites modified to 5-methylcytidine (m5C) (216). These modifications were found to consistently appear at regular positions across the different sgRNAs (216). This finding is in line with previous direct RNA sequencing of the HCoV-229E, which revealed the presence of consistent m5C methylation patterns across the sgRNAs (160). It should be noted that Viehweger et al. (160) represents an early (but nonetheless important) investigation into direct RNA sequencing of CoVs, as, at the time, detection of modified bases was limited to cytosines. Additional base modifications were detected by Li et al. (245) in the SARS-CoV-2 genome via mass spectrometry of viral RNA purified from Vero cells, including 2′-O-methylatation of all four possible nucleosides (Am, Um, Cm, and Gm); modified cytidine derivatives (ac4C, m3C, and m5C); modified uridine derivatives (pseudouridine [Ψ] and m5U); and alternatively modified adenosines (m6,6A) (245) (Fig. 10). These findings provide important insights into SARS-CoV-2 base modification; however, additional studies utilizing direct RNA sequencing (described in more detail below) to determine RNA base modifications will be required to fully elucidate these patterns of modification and potentially other modifications yet to be detected. Currently, there are multiple studies that have performed direct RNA-seq on SARS-CoV-2 (49, 254, 255); however, these investigations primarily focused on the transcriptome and largely omit details on epitranscriptomics and thus could be valuable sources of data on epitranscriptomics for subsequent analysis. The value in revisiting these data sets has already been demonstrated by Fleming et al. whereby in silico analysis of the direct RNA-seq data sets of SARS-CoV-2 provided by Kim et al. (49) and Miladi et al. (254) suggested the presence of multiple pseudouridine residues, which were not observed previously and were conserved across the sgRNA (256). The field of epitranscriptomics has grown immensely in the past few decades as new methods and technologies aiming to interrogate RNA modification have both emerged and progressed in rapid fashion. In recent years, the use of next-generation sequencing has allowed for the enhanced detection of RNA modifications based either on the recognition of modified bases using antibody immunoprecipitation (228, 257) or by using chemical labeling of modified bases (258–261). The primary drawback of these methods is their reliance on sequencing-by-synthesis (SBS), whereby information on base modification may be lost during the reverse transcription of RNA to cDNA (262). This obstacle has been overcome in recent years, however, by the use of direct RNA sequencing, primarily using Nanopore technology (160, 261), which allows for identification of modifications on native RNA while circumventing many of the biases of SBS by nature of its respectively minimal manipulation of input genomic material (262). This technology has shown the capacity for accurate detection of RNA modification at single-base resolution (263, 264), and the rapid improvement of this technology only enhances its future potential (262). In this manner, direct RNA sequencing can thus provide not only information on the identity of bases in an RNA sequence, but also any present base modifications. Although this represents a powerful new tool, it should be noted that there is still much progress to be made; however, the technology, and the methods and algorithms used to analyze the data it produces, are both progressing quickly (262).
FIG 10.
RNA base modifications discussed in the text. Individual modifications are highlighted in blue, and the abbreviation for each modification is found underneath.
As has already been discussed, RNA modification plays a prominent, yet underappreciated, role in the biology of SARS-CoV-2 and COVID-19 infections, as well as in many other RNA viruses. Despite the wealth of longitudinal sequencing that has been amassed globally, a consummate abundance of data on changes in RNA modification over the course of the pandemic is painfully lacking. Hence, longitudinal changes in the epitranscriptome of SARS-CoV-2, as well as any potential selective pressures acting on such, are not currently known. Accordingly, the complete view of SARS-CoV-2 evolutionary history is missing an important puzzle piece, without which our understanding of the virus’s evolution and our ability to holistically predict hot spots of future mutation in the SARS-CoV-2 genome are incomplete. In the future, the new data that can be generated from these technologies incorporating RNA base identity and base modification can be compounded with RNA secondary structure analysis, which will necessitate the development of new evolutionary models and will provide a more robust approach to understanding how these genetic elements interact to drive evolution.
CONCLUDING REMARKS
As illustrated in this review, the RNA structure of SARS-CoV-2 is critical to various steps of the viral replication cycle, ranging from transcription to translation, to adaptation through recombination. Historically, the extensive characterization of the untranslated regions of the genome provide a substantial foundation for investigating these regions in SARS-CoV-2 for their roles in transmission and infectivity, and their use in small-molecule drug-targeting. However, evolution of CoVs and resulting divergence of this virus from Alphaviruses, and even other Betacoronaviruses such as SARS-CoV-1, reveal the need for further study, as do the many additional RNA structures located within the remainder of the genome.
Several structures within SARS-CoV-2 present as opportunities for small-molecule inhibition of viral activity, though with specific and varying limitations. While the attenuator hairpin functioning in ribosomal frame-shifting for protein translation has shown drug-targeting capability (186), it is not well conserved and is thus potentially not ideal for broad application to other CoVs. SL3 within the 5′ UTR hypothetically would be a valuable target due to its ubiquitous involvement in the life cycle, including discontinuous transcription regulation and genome circularization required for transcription and genome replication (37); however, its small size severely limits drug options. Several small molecules have been designed to inhibit function of the universally present SL1 in the 5′ UTR, one of which has been tested in animal models, showing promising results (75). However, the impact of the diversity involving the bulged region within the stem of SL1 across SARS-CoV-2 variants (65) needs to be addressed. Alternatively, the S2M motif in the 3′ UTR is highly conserved across CoVs, and even virus families outside of CoV (e.g., Astroviruses) [127]), providing the ideal candidate for broad-use application, the drawbacks being the likely off-target effects owing to its similarity to the rRNA loop and frequent deletions observed throughout SARS-CoV-2 (and even CoV) evolutionary history. The remainder of the 3′ UTR is less explored than the 5′ UTR in terms of drug targeting, but also in terms of RNA structure and function in general, with multiple conformations and functions potentially existing in the 5′ region, the precise function of the two stem-loops flanking the S2M motif, and the impact of the shorter poly(A) tail for SARS-CoV-2 in 3′ UTR structure-mediated processes such as circularization and recombination.
More robust investigation is also required to fully characterize the SARS-CoV-2 packaging signal. The previously established inverse correlation between the presence of an ORF1ab packaging signal hairpin structure (in Embecoviruses [113]) and presence of SL5 substructures in the 5′ UTR (Sarbecoviruses [100]) attempts to de-mystify the circumstances of coronavirus packaging; however, the prediction of SL5 substructures in MHV by the recent SHAPE analysis conducted in Yang et al. (78) suggest these regions might work in concert across Betacoronaviruses. If the role of SL5 structure is truly universal across coronaviruses, SL5 remains a promising target for small molecule inhibitors (85).
Drugs targeting typical CoV replication would not prove beneficial in preventing the integration of the virus thought to be responsible for continued detection of viral RNA in patients considered to have “long COVID.” However, our analysis highlights that an RNA structure-mediated integration mechanism is unlikely owing to the relative positions and predicted structures of regions genetically similar to those used in retroviral integration and LINE1 retrotransposition (Fig. 8). Though additional DNA sequencing and experimental analysis are required to present evidence in opposition, results (202, 265) contradicting the findings of Zhang et al. are consistent with our findings, ultimately indicating that if SARS-CoV-2 is indeed being integrated into the genome of human cells, it is occurring with an unknown mechanism or via a less-stable LINE1 RNA structure, the latter of which may explain the hypothesized low rate of occurrence.
While this review represents a comprehensive collection of studies evaluating SARS-CoV-2 RNA structure (Table 1), it does not represent the full extent of structural investigations. We have focused on known structural features of the SARS-CoV-2 RNA genome most salient to its biological functions, but it is important to keep in mind that these represent only a fraction of the total structures predicted, yet uncharacterized, in the SARS-CoV-2 genome (266) and our knowledge of posttranscriptional modifications on these structures is lacking. Furthermore, given the rate of human spillover of CoVs in recent years (e.g., references 267, 268), tackling the structural, functional, and therapeutic characterization of the remaining RNA “structurome” would be infeasible in the amount of time needed before the next CoV outbreak. Integration of bioinformatic and machine learning approaches with experimental techniques would greatly benefit this realm of science and the current target of pan-coronavirus treatment and prevention strategies (269).
TABLE 1.
TABLE of current literature examining the structure of the SARS-CoV-2 genomea
| Region investigated | Author | Method/tool |
|---|---|---|
| Whole genome | Sanders et al. (90) | SHAPE-MaPseq |
| Whole genome | Huston et al. (101) | SHAPE-MaPseq |
| Whole genome | Rangan et al. (59) | MSA |
| Whole genome | Andrews et al. (103) | ScanFold |
| Whole genome | Manfredonia et al. (105) | SHAPE-MaPseq DMS-MaPseq |
| Whole genome | Alhatlani (273) | Mfold RNAfold |
| Whole genome | Sun et al. (89) | icSHAPE |
| Whole genome | Lan et al. (88) | DMS-MaPseq |
| Whole genome | Cao et al. (266) | vRIC-seq RNAstructure |
| Whole genome | Tavares et al. (274) | SuperFold |
| Whole genome | Ziv et al. (37) | COMRADES |
| Whole genome | Yang et al. (122) | SHAPE-MaP |
| SL1–8 in the extended 5′ UTR and the reverse complement of the 5′ UTR SL1–4, FSE, and 3′ UTR | Rangan et al. (106) | FARFAR2 |
| 5′ UTR | Miao et al. (65) | In-line probing RNAse V1 probing |
| 5′ Terminus and frame-shift element | Iserman et al. (119) | SHAPE-MaP |
| Nsp2 and 3′ UTR | Ali Hosseini Rad and McLellan (141) | RNAfold |
| Genome-scale ordered RNA structure (GORS) | Simmonds (275) | RNAfold |
| 5′ Terminus, frame-shift element, and 3′ UTR | Wacker et al. (63) | NMR DMS footprinting |
| 5′ UTR | Vankadari et al. (276) | RNAfold |
| 3′ UTR | Zhao et al. (123) | DMS-MaPseq |
| Frame-shift element | Zhang et al. (178) | SHAPE-MaPseq DMS-MaPseq |
aFSE, frame-shifting element; UTR, untranslated region.
ACKNOWLEDGMENTS
This work was supported by the Stephany W. Holloway University of Florida Chair and by funding from the University of Florida Office of Research and Health Science Center, National Science Foundation, Division of Environmental Biology (award no. 2028221), The AIDS Health Care Foundation (award no. OS00000633), and The Rockefeller Foundation (award 2021 HTH 012).
REFERENCES
- 1.Ritchie H, Mathieu E, Rodes-Guirao L, Appel C, Giattino C, Ortiz-Ospina E, Hasell J, Macdonald B, Beltekian D, Roser M. 2020. Coronavirus pandemic (COVID-19). OurWorldInData.org. https://ourworldindata.org/coronavirus. [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2021. Science brief: omicron (B.1.1.529) variant. CDC, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html. [PubMed] [Google Scholar]
- 4.Wang Q, Barr I, Feng G, Lee C. 2008. Evidence of a novel RNA secondary structure in the coding region of HIV-1 pol gene. RNA 14:2478–2488. 10.1261/rna.1252608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkhout B. 1992. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res 20:27–31. 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254–257. 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 7.Kuzembayeva M, Dilley K, Sardo L, Hu WS. 2014. Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454–455:362–370. 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witteveldt J, Blundell R, Maarleveld JJ, Mcfadden N, Evans DJ, Simmonds P. 2014. The influence of viral RNA secondary structure on interactions with innate host cell defences. Nucleic Acids Res 42:3314–3329. 10.1093/nar/gkt1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. 2011. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol 7:553–559. 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermann T. 2018. Viral RNA targets and their small molecule ligands. Top Med Chem 27:111–134. 10.1007/7355_2016_20. [DOI] [Google Scholar]
- 11.Wang L, Liang R, Gao Y, Li Y, Deng X, Xiang R, Zhang Y, Ying T, Jiang S, Yu F. 2019. Development of small-molecule inhibitors against zika virus infection. Front Microbiol 10:2725. 10.3389/fmicb.2019.02725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao Y, Zhang QC. 2020. Targeting RNA structures in diseases with small molecules. Essays Biochem 64:955–966. 10.1042/EBC20200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falese JP, Donlic A, Hargrove AE. 2021. Targeting RNA with small molecules: from fundamental principles towards the clinic. Chem Soc Rev 50:2224–2243. 10.1039/d0cs01261k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermann T. 2016. Small molecules targeting viral RNA. Wiley Interdiscip Rev RNA 7:726–743. 10.1002/wrna.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abulwerdi FA, Shortridge MD, Sztuba-Solinska J, Wilson R, Le Grice SFJ, Varani G, Schneekloth JS. 2016. Development of small molecules with a noncanonical binding mode to HIV-1 trans activation response (TAR) RNA. J Med Chem 59:11148–11160. 10.1021/acs.jmedchem.6b01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sztuba-Solinska J, Shenoy SR, Gareiss P, Krumpe LRH, Le Grice SFJ, O’Keefe BR, Schneekloth JS. 2014. Identification of biologically active, HIV TAR RNA-binding small molecules using small molecule microarrays. J Am Chem Soc 136:8402–8410. 10.1021/ja502754f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwetz TA, Fauci AS. 2019. The extended impact of human immunodeficiency virus/AIDS research. J Infect Dis 219:6–9. 10.1093/infdis/jiy441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rife Magalis B, Kosakovsky Pond SL, Summers MF, Salemi M. 2018. Evaluation of global HIV/SIV envelope gp120 RNA structure and evolution within and among infected hosts. Virus Evol 4:vey018. 10.1093/ve/vey018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kladwang W, Vanlang CC, Cordero P, Das R. 2011. Understanding the errors of SHAPE-directed RNA structure modeling. Biochemistry 50:8049–8056. 10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tijerina P, Mohr S, Russell R. 2007. DMS footprinting of structured RNAs and RNA-protein complexes. Nat Protoc 2:2608–2623. 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. 2009. Influence of nucleotide identity on ribose 2’-hydroxyl reactivity in RNA. RNA 15:1314–1321. 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by Selective 2’-Hydroxyl Acylation and Primer Extension (SHAPE). J Am Chem Soc 127:4223–4231. 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 23.McGinnis JL, Dunkle JA, Cate JHD, Weeks KM. 2012. The mechanisms of RNA SHAPE chemistry. J Am Chem Soc 134:6617–6624. 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordero P, Kladwang W, Vanlang CC, Das R. 2012. Quantitative dimethyl sulfate mapping for automated RNA secondary structure inference. Biochemistry 51:7037–7039. 10.1021/bi3008802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee B, Flynn RA, Kadina A, Guo JK, Kool ET, Chang HY. 2017. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA 23:169–174. 10.1261/rna.058784.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casiano-Negroni A, Sun X, Al-Hashimi HM. 2007. Probing Na+-induced changes in the HIV-1 TAR conformational dynamics using NMR residual dipolar couplings: new insights into the role of counterions and electrostatic interactions in adaptive recognition. Biochemistry 46:6525–6535. 10.1021/bi700335n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flynn RA, Zhang QC, Spitale RC, Lee B, Mumbach MR, Chang HY. 2016. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat Protoc 11:273–290. 10.1038/nprot.2016.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegfried NA, Busan S, Rice GM, Nelson JAE, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11:959–965. 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. 2015. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519:486–490. 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doty P, Boedtker H, Fresco JR, Haselkorn R, Litt M. 1959. Secondary structure in ribonucleic acids. Proc Natl Acad Sci USA 45:482–499. 10.1073/pnas.45.4.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rollins C, Levengood JD, Rife BD, Salemi M, Tolbert BS. 2014. Thermodynamic and phylogenetic insights into hnRNP A1 recognition of the HIV-1 exon splicing silencer 3 element. Biochemistry 53:2172–2184. 10.1021/bi500180p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedersen JS, Meyer IM, Forsberg R, Simmonds P, Hein J. 2004. A comparative method for finding and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res 32:4925–4936. 10.1093/nar/gkh839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomezsko PJ, Corbin VDA, Gupta P, Swaminathan H, Glasgow M, Persad S, Edwards MD, Mcintosh L, Papenfuss AT, Emery A, Swanstrom R, Zang T, Lan TCT, Bieniasz P, Kuritzkes DR, Tsibris A, Rouskin S. 2020. Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature 582:438–442. 10.1038/s41586-020-2253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716. 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomezsko P, Swaminathan H, Rouskin S. 2020. Viral RNA structure analysis using DMS-MaPseq. Methods 183:68–75. 10.1016/j.ymeth.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huthoff H, Berkhout B. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7:143–157. 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziv O, Price J, Shalamova L, Kamenova T, Goodfellow I, Weber F, Miska EA. 2020. The short- and long-range RNA-RNA interactome of SARS-CoV-2. Mol Cell 80:1067–1077.e5. 10.1016/j.molcel.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burrell CJ, Howard CR, Murphy FA. 2017. Chapter 31–coronaviruses, p 437–446. In Burrell CJ, Howard CR, Murphy FA (ed), Fenner and White’s medical virology (5th ed). Academic Press, New York. 10.1016/B978-0-12-375156-0.00031-X. [DOI] [Google Scholar]
- 39.International Committee on Taxonomy of Viruses (ICTV). 2019. Virus taxonomy: 2019 release, EC 51. ICTV. https://talk.ictvonline.org/taxonomy. [Google Scholar]
- 40.King A. 2020. An uncommon cold. New Sci 246:32–35. 10.1016/S0262-4079(20)30862-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 42.Gralinski LE, Menachery VD. 2020. Return of the coronavirus: 2019-nCoV. Viruses 12:135. 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam TTY, Jia N, Zhang YW, Shum MHH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ, Jiang BG, Wei W, Yuan TT, Zheng K, Cui XM, Li J, Pei GQ, Qiang X, Cheung WYM, Li LF, Sun FF, Qin S, Huang JC, Leung GM, Holmes EC, Hu YL, Guan Y, Cao WC. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583:282–285. 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 46.Tagliamonte MS, Abid N, Borocci S, Sangiovanni E, Ostrov DA, Kosakovsky Pond SL, Salemi M, Chillemi G, Mavian C. 2021. Multiple recombination events and strong purifying selection at the origin of SARS-CoV-2 spike glycoprotein increased correlated dynamic movements. Int J Mol Sci 22:80. 10.3390/ijms22010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, Tamir H, Achdout H, Stein D, Israeli O, Beth-Din A, Melamed S, Weiss S, Israely T, Paran N, Schwartz M, Stern-Ginossar N. 2021. The coding capacity of SARS-CoV-2. Nature 589:125–130. 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 48.Jungreis I, Nelson CW, Ardern Z, Finkel Y, Krogan NJ, Sato K, Ziebuhr J, Stern-Ginossar N, Pavesi A, Firth AE, Gorbalenya AE, Kellis M. 2021. Conflicting and ambiguous names of overlapping ORFs in the SARS-CoV-2 genome: a homology-based resolution. Virology 558:145–151. 10.1016/j.virol.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. 2020. The architecture of SARS-CoV-2 transcriptome. Cell 181:914–921.e10. 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, Sauter D, Gifford RJ, Nakagawa S, Sato K. 2020. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep 32:108185. 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. 2021. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 19:155–170. 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, Cinatl J, Münch C. 2020. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 583:469–472. 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dinger ME, Gascoigne DK, Mattick JS. 2011. The evolution of RNAs with multiple functions. Biochimie 93:2013–2018. 10.1016/j.biochi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 54.Smyth RP, Negroni M, Lever AM, Mak J, Kenyon JC. 2018. RNA structure-a neglected puppet master for the evolution of virus and host immunity. Front Immunol 9:2097. 10.3389/fimmu.2018.02097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gebauer F, Preiss T, Hentze MW. 2012. From cis-regulatory elements to complex RNPs and back. Cold Spring Harb Perspect Biol 4:a012245. 10.1101/cshperspect.a012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed F, Sharma M, Al-Ghamdi AA, Al-Yami SM, Al-Salami AM, Refai MY, Warsi MK, Howladar SM, Baeshen MN. 2020. A comprehensive analysis of cis-acting RNA elements in the SARS-CoV-2 genome by a bioinformatics approach. Front Genet 11:572702. 10.3389/fgene.2020.572702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicholson BL, White KA. 2014. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol 12:493–504. 10.1038/nrmicro3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pathak KB, Nagy PD. 2009. Defective interfering RNAs: foes of viruses and friends of virologists. Viruses 1:895–919. 10.3390/v1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rangan R, Zheludev IN, Hagey RJ, Pham EA, Wayment-Steele HK, Glenn JS, Das R. 2020. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA 26:937–959. 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res 38:4274–4284. 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei Y, Silke JR, Aris P, Xia X. 2020. Coronavirus genomes carry the signatures of their habitats. PLoS One 15:e0244025. 10.1371/journal.pone.0244025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forni D, Cagliani R, Pontremoli C, Clerici M, Sironi M. 2022. The substitution spectra of coronavirus genomes. Briefings Bioinform 23:bbab382. 10.1093/bib/bbab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wacker A, Weigand JE, Akabayov SR, Altincekic N, Bains JK, Banijamali E, Binas O, Castillo-Martinez J, Cetiner E, Ceylan B, Chiu LY, Davila-Calderon J, Dhamotharan K, Duchardt-Ferner E, Ferner J, Frydman L, Fürtig B, Gallego J, Tassilo Grün J, Hacker C, Haddad C, Hähnke M, Hengesbach M, Hiller F, Hohmann KF, Hymon D, de Jesus V, Jonker H, Keller H, Knezic B, Landgraf T, Löhr F, Luo L, Mertinkus KR, Muhs C, Novakovic M, Oxenfarth A, Palomino-Schätzlein M, Petzold K, Peter SA, Pyper DJ, Qureshi NS, Riad M, Richter C, Saxena K, Schamber T, Scherf T, Schlagnitweit J, Schlundt A, Schnieders R, et al. 2020. Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy. Nucleic Acids Res 48:12415–12435. 10.1093/nar/gkaa1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L, Kang H, Liu P, Makkinje N, Williamson ST, Leibowitz JL, Giedroc DP. 2008. Structural lability in stem-loop 1 drives a 5’ UTR-3’ UTR interaction in coronavirus replication. J Mol Biol 377:790–803. 10.1016/j.jmb.2008.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miao Z, Tidu A, Eriani G, Martin F. 2021. Secondary structure of the SARS-CoV-2 5’-UTR. RNA Biol 18:447–456. 10.1080/15476286.2020.1814556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding P, Kharytonchyk S, Waller A, Mbaekwe U, Basappa S, Kuo N, Frank HM, Quasney C, Kidane A, Swanson C, Van V, Sarkar M, Cannistraci E, Chaudhary R, Flores H, Telesnitsky A, Summers MF. 2020. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc Natl Acad Sci USA 117:17737–17746. 10.1073/pnas.2008519117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tidu A, Janvier A, Schaeffer L, Sosnowski P, Kuhn L, Hammann P, Westhof E, Eriani G, Martin F. 2021. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA 27:253–264. 10.1261/rna.078121.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16:1134–1140. 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakagawa K, Makino S. 2021. Mechanisms of coronavirus nsp1-mediated control of host and viral gene expression. Cells 10:300–318. 10.3390/cells10020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. 2011. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog 7:e1002433. 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Mühlemann O, Ban N. 2020. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27:959–966. 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 72.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, MacKens-Kiani T, Cheng J, Straub JH, Stürzel CM, Fröhlich T, Berninghausen O, Becker T, Kirchhoff F, Sparrer KMJ, Beckmann R. 2020. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369:1249–1256. 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, Bhat P, Ollikainen N, Quinodoz SA, Loney C, Thai J, Miller ZD, Lin AE, Schmidt MM, Stewart DG, Goldfarb D, De Lorenzo G, Rihn SJ, Voorhees RM, Botten JW, Majumdar D, Guttman M. 2020. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183:1325–1339.e21. 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vora SM, Fontana P, Mao T, Leger V, Zhang Y, Fu TM, Lieberman J, Gehrke L, Shi M, Wang L, Iwasaki A, Wu H. 2022. Targeting stem-loop 1 of the SARS-CoV-2 50 UTR to suppress viral translation and Nsp1 evasion. Proc Natl Acad Sci USA 119:e2117198119. 10.1073/pnas.2117198119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu C, Lee JY, Woo JZ, Xu L, Nguyenla X, Yamashiro LH, Biering SB, Van Dis E, Gonzalez F, Fox D, Rustagi A, Pinsky BA, Blish CA, Chiu C, Harris E, Sadreyev RI, Kauppinen S, Rouskin S, Näär AM. 2021. An intranasal ASO therapeutic targeting SARS-CoV-2. bioRxiv. 10.1101/2021.05.17.444397. [DOI] [PMC free article] [PubMed]
- 76.Zafferani M, Haddad C, Luo L, Davila-Calderon J, Chiu LY, Mugisha CS, Monaghan AG, Kennedy AA, Yesselman JD, Gifford RJ, Tai AW, Kutluay SB, Li ML, Brewer G, Tolbert BS, Hargrove AE. 2021. Amilorides inhibit SARS-CoV-2 replication in vitro by targeting RNA structures. Sci Adv 7:eabl6096. 10.1126/sciadv.abl6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu P, Li L, Millership JJ, Kang H, Leibowitz JL, Giedroc DP. 2007. A U-turn motif-containing stem-loop in the coronavirus 5’ untranslated region plays a functional role in replication. RNA 13:763–780. 10.1261/rna.261807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang D, Leibowitz JL. 2015. The structure and functions of coronavirus genomic 3’ and 5’ ends. Virus Res 206:120–133. 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gutell RR, Cannone JJ, Konings D, Gautheret D. 2000. Predicting U-turns ribosomal RNA with comparative sequence analysis. J Mol Biol 300:791–803. 10.1006/jmbi.2000.3900. [DOI] [PubMed] [Google Scholar]
- 80.Liu P, Li L, Keane SC, Yang D, Leibowitz JL, Giedroc DP. 2009. Mouse hepatitis virus stem-loop 2 adopts a uYNMG(U)a-like tetraloop structure that is highly functionally tolerant of base substitutions. J Virol 83:12084–12093. 10.1128/JVI.00915-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang D, Liu P, Wudeck EV, Giedroc DP, Leibowitz JL. 2015. Shape analysis of the rna secondary structure of the mouse hepatitis virus 5’ untranslated region and n-terminal nsp1 coding sequences. Virology 475:15–27. 10.1016/j.virol.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee CW, Li L, Giedroc DP. 2011. The solution structure of coronaviral stem-loop 2 (SL2) reveals a canonical CUYG tetraloop fold. FEBS Lett 585:1049–1053. 10.1016/j.febslet.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen SC, Olsthoorn RCL. 2010. Group-specific structural features of the 5’-proximal sequences of coronavirus genomic RNAs. Virology 401:29–41. 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Madhugiri R, Karl N, Petersen D, Lamkiewicz K, Fricke M, Wend U, Scheuer R, Marz M, Ziebuhr J. 2018. Structural and functional conservation of cis-acting RNA elements in coronavirus 5’-terminal genome regions. Virology 517:44–55. 10.1016/j.virol.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sreeramulu S, Richter C, Berg H, Wirtz Martin MA, Ceylan B, Matzel T, Adam J, Altincekic N, Azzaoui K, Bains JK, Blommers MJJ, Ferner J, Fürtig B, Göbel M, Grün JT, Hengesbach M, Hohmann KF, Hymon D, Knezic B, Martins JN, Mertinkus KR, Niesteruk A, Peter SA, Pyper DJ, Qureshi NS, Scheffer U, Schlundt A, Schnieders R, Stirnal E, Sudakov A, Tröster A, Vögele J, Wacker A, Weigand JE, Wirmer-Bartoschek J, Wöhnert J, Schwalbe H. 2021. Exploring the druggability of conserved RNA regulatory elements in the SARS-CoV-2 genome. Angew Chem Int Ed Engl 60:19191–19200. 10.1002/anie.202103693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Warner KD, Hajdin CE, Weeks KM. 2018. Principles for targeting RNA with drug-like small molecules. Nat Rev Drug Discov 17:547–558. 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Liao CL, Lai MM. 1994. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J Virol 68:4738–4746. 10.1128/JVI.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lan TCT, Allan MF, Malsick LE, Woo JZ, Zhu C, Zhang F, Khandwala S, Nyeo SSY, Sun Y, Guo JU, Bathe M, Näär A, Griffiths A, Rouskin S. 2022. Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells. Nat Commun 13:1128. 10.1038/s41467-022-28603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun L, Li P, Ju X, Rao J, Huang W, Ren L, Zhang S, Xiong T, Xu K, Zhou X, Gong M, Miska E, Ding Q, Wang J, Zhang QC. 2021. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell 184:1865–1883.e20. 10.1016/j.cell.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanders W, Fritch EJ, Madden EA, Graham RL, Vincent HA, Heise MT, Baric RS, Moorman NJ. 2020. Comparative analysis of coronavirus genomic RNA structure reveals conservation in SARS-like coronaviruses. bioRxiv. 10.1101/2020.06.15.153197. [DOI]
- 91.Kang H, Feng M, Schroeder ME, Giedroc DP, Leibowitz JL. 2006. Putative cis-acting stem-loops in the 5’ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J Virol 80:10600–10614. 10.1128/JVI.00455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sola I, Almazán F, Zúñiga S, Enjuanes L. 2015. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2:265–288. 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gallaher WR. 2020. A palindromic RNA sequence as a common breakpoint contributor to copy-choice recombination in SARS-COV-2. Arch Virol 165:2341–2348. 10.1007/s00705-020-04750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. J Virol 81:20–29. 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ziv O, Gabryelska MM, Lun ATL, Gebert LFR, Sheu-Gruttadauria J, Meredith LW, Liu ZY, Kwok CK, Qin CF, MacRae IJ, Goodfellow I, Marioni JC, Kudla G, Miska EA. 2018. COMRADES determines in vivo RNA structures and interactions. Nat Methods 15:785–788. 10.1038/s41592-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raman S, Bouma P, Williams GD, Brian DA. 2003. Stem-loop III in the 5’ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA eeplication. J Virol 77:6720–6730. 10.1128/jvi.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu HY, Guan BJ, Su YP, Fan YH, Brian DA. 2014. Reselection of a genomic upstream open reading frame in mouse hepatitis coronavirus 5’-untranslated-region mutants. J Virol 88:846–858. 10.1128/JVI.02831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang D, Liu P, Giedroc DP, Leibowitz J. 2011. Mouse hepatitis virus stem-loop 4 functions as a spacer element required to drive subgenomic RNA synthesis. J Virol 85:9199–9209. 10.1128/JVI.05092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu HY, Ozdarendeli A, Brian DA. 2006. Bovine coronavirus 5’-proximal genomic acceptor hotspot for discontinuous transcription is 65 nucleotides wide. J Virol 80:2183–2193. 10.1128/JVI.80.5.2183-2193.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Masters PS. 2019. Coronavirus genomic RNA packaging. Virology 537:198–207. 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huston NC, Wan H, Strine MS, de Cesaris Araujo Tavares R, Wilen CB, Pyle AM. 2021. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol Cell 81:584–598.e5. 10.1016/j.molcel.2020.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Laing C, Schlick T. 2009. Analysis of four-way junctions in RNA structures. J Mol Biol 390:547–559. 10.1016/j.jmb.2009.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Andrews RJ, O’Leary CA, Tompkins VS, Peterson JM, Haniff HS, Williams C, Disney MD, Moss WN. 2021. A map of the SARS-CoV-2 RNA structurome. NAR Genom Bioinform 3:lqab043. 10.1093/nargab/lqab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Manfredonia I, Incarnato D. 2021. Structure and regulation of coronavirus genomes: state-of-the-art and novel insights from SARS-CoV-2 studies. Biochem Soc Trans 49:341–352. 10.1042/BST20200670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Manfredonia I, Nithin C, Ponce-Salvatierra A, Ghosh P, Wirecki TK, Marinus T, Ogando NS, Snijder EJ, van Hemert MJ, Bujnicki JM, Incarnato D. 2020. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res 48:12436–12452. 10.1093/nar/gkaa1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rangan R, Watkins AM, Chacon J, Kretsch R, Kladwang W, Zheludev IN, Townley J, Rynge M, Thain G, Das R. 2021. De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures. Nucleic Acids Res 49:3092–3108. 10.1093/nar/gkab119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, Summers MF. 2015. Structure of the HIV-1 RNA packaging signal. Science 348:917–921. 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hsieh PK, Chang SC, Huang CC, Lee TT, Hsiao CW, Kou YH, Chen IY, Chang CK, Huang TH, Chang MF. 2005. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol 79:13848–13855. 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ye Q, West AMV, Silletti S, Corbett KD. 2020. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci 29:1890–1901. 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N, Tsao SW, Nicholls JM, Altmeyer R, Peiris JSM, Bruzzone R, Nal B. 2008. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol 82:11318–11330. 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fehr AR, Perlman S. 2015. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 1282:1–23. 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Narayanan K, Chen CJ, Maeda J, Makino S. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J Virol 77:2922–2927. 10.1128/jvi.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuo L, Masters PS. 2013. Functional analysis of the murine coronavirus genomic RNA packaging signal. J Virol 87:5182–5192. 10.1128/JVI.00100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Woo K, Joo M, Narayanan K, Kim KH, Makino S. 1997. Murine coronavirus packaging signal confers packaging to nonviral RNA. J Virol 71:824–827. 10.1128/JVI.71.1.824-827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Woo J, Lee EY, Lee M, Kim T, Cho YE. 2019. An in vivo cell-based assay for investigating the specific interaction between the SARS-CoV N-protein and its viral RNA packaging sequence. Biochem Biophys Res Commun 520:499–506. 10.1016/j.bbrc.2019.09.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Morales L, Mateos-Gomez PA, Capiscol C, del Palacio L, Enjuanes L, Sola I. 2013. Transmissible gastroenteritis coronavirus genome packaging signal is located at the 5’ end of the genome and promotes viral RNA incorporation into virions in a replication-independent process. J Virol 87:11579–11590. 10.1128/JVI.01836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lu S, Ye Q, Singh D, Cao Y, Diedrich JK, Yates JR, Villa E, Cleveland DW, Corbett KD. 2021. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat Commun 12:502. 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jack A, Ferro LS, Trnka MJ, Wehri E, Nadgir A, Nguyenla X, Fox D, Costa K, Stanley S, Schaletzky J, Yildiz A. 2021. SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA. PLoS Biol 19:e3001425. 10.1371/journal.pbio.3001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iserman C, Roden CA, Boerneke MA, Sealfon RSG, McLaughlin GA, Jungreis I, Fritch EJ, Hou YJ, Ekena J, Weidmann CA, Theesfeld CL, Kellis M, Troyanskaya OG, Baric RS, Sheahan TP, Weeks KM, Gladfelter AS. 2020. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol Cell 80:1078–1091.e6. 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rehman SU, Shafique L, Ihsan A, Liu Q. 2020. Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. Pathogens 9:240. 10.3390/pathogens9030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chan AP, Choi Y, Schork NJ. 2020. Conserved genomic terminals of SARS-CoV-2 as coevolving functional elements and potential therapeutic targets. mSphere 5:e00754-20. 10.1128/mSphere.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang SL, DeFalco L, Anderson DE, Zhang Y, Aw JGA, Lim SY, Lim XN, Tan KY, Zhang T, Chawla T, Su Y, Lezhava A, Merits A, Wang LF, Huber RG, Wan Y. 2021. Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. Nat Commun 12:5113. 10.1038/s41467-021-25357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhao J, Qiu J, Aryal S, Hackett JL, Wang J. 2020. The RNA architecture of the SARS-CoV-2 3’-untranslated region. Viruses 12:1473. 10.3390/v12121473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hsue B, Masters PS. 1997. A bulged stem-loop structure in the 3’ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J Virol 71:7567–7578. 10.1128/JVI.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hsue B, Hartshorne T, Masters PS. 2000. Characterization of an Essential RNA Secondary Structure in the 3’ Untranslated Region of the Murine Coronavirus Genome. J Virol 74:6911–6921. 10.1128/jvi.74.15.6911-6921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Enjuanes L. 2005. Coronavirus replication and reverse genetics: preface. Springer, New York. [Google Scholar]
- 127.Jonassen CM, Jonassen T, Grinde B. 1998. A common RNA motif in the 3’ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J Gen Virol 79:715–718. 10.1099/0022-1317-79-4-715. [DOI] [PubMed] [Google Scholar]
- 128.Goebel SJ, Miller TB, Bennett CJ, Bernard KA, Masters PS. 2007. A hypervariable region within the 3’ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J Virol 81:1274–1287. 10.1128/JVI.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Williams GD, Chang RY, Brian DA. 1999. A phylogenetically conserved hairpin-type 3’ untranslated region pseudoknot functions in coronavirus RNA replication. J Virol 73:8349–8355. 10.1128/JVI.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Staple DW, Butcher SE. 2005. Pseudoknots: RNA structures with diverse functions. PLoS Biol 3:e213. 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brierley I, Pennell S, Gilbert RJC. 2007. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol 5:598–610. 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Züst R, Miller TB, Goebel SJ, Thiel V, Masters PS. 2008. Genetic interactions between an essential 3’ cis -acting RNA pseudoknot, replicase gene products, and the extreme 3’ end of the mouse coronavirus genome. J Virol 82:1214–1228. 10.1128/JVI.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Goebel SJ, Taylor J, Masters PS. 2004. The 3’ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J Virol 78:7846–7851. 10.1128/JVI.78.14.7846-7851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stammler SN, Cao S, Chen SJ, Giedroc DP. 2011. A conserved RNA pseudoknot in a putative molecular switch domain of the 3’-untranslated region of coronaviruses is only marginally stable. RNA 17:1747–1759. 10.1261/rna.2816711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Goebel SJ, Hsue B, Dombrowski TF, Masters PS. 2004. Characterization of the RNA components of a putative molecular switch in the 3’ untranslated region of the murine coronavirus genome. J Virol 78:669–682. 10.1128/jvi.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cagliani R, Forni D, Clerici M, Sironi M. 2020. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect Genet Evol 83:104353. 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Robertson MP, Igel H, Baertsch R, Haussler D, Ares M, Scott WG. 2005. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol 3:e5. 10.1371/journal.pbio.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6:26. 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res 36:W70–74. 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tengs T, Kristoffersen AB, Bachvaroff TR, Jonassen CM. 2013. A mobile genetic element with unknown function found in distantly related viruses. Virol J 10:132. 10.1186/1743-422X-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ali Hosseini Rad SM, McLellan AD. 2020. Implications of SARS-CoV-2 mutations for genomic RNA structure and host microRNA targeting. Int J Mol Sci 21:4807. 10.3390/ijms21134807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lulla V, Wandel MP, Bandyra KJ, Ulferts R, Wu M, Dendooven T, Yang X, Doyle N, Oerum S, Beale R, O’Rourke SM, Randow F, Maier HJ, Scott W, Ding Y, Firth AE, Bloznelyte K, Luisi BF. 2021. Targeting the conserved stem loop 2 motif in the SARS-CoV-2 genome. J Virol 95:e00663-21. 10.1128/JVI.00663-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gilbert C, Tengs T. 2021. No species-level losses of s2m suggests critical role in replication of SARS-related coronaviruses. Sci Rep 11:16145. 10.1038/s41598-021-95496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ryder SP, Morgan BR, Coskun P, Antkowiak K, Massi F. 2021. Analysis of emerging variants in structured regions of the SARS-CoV-2 genome. Evol Bioinform Online 17:11769343211014167. 10.1177/11769343211014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu P, Yang D, Carter K, Masud F, Leibowitz JL. 2013. Functional analysis of the stem loop S3 and S4 structures in the coronavirus 3’UTR. Virology 443:40–47. 10.1016/j.virol.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wu HY, Ke TY, Liao WY, Chang NY. 2013. Regulation of coronaviral poly(A) tail length during infection. PLoS One 8:e70548. 10.1371/journal.pone.0070548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lin YJ, Liao CL, Lai MM. 1994. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol 68:8131–8140. 10.1128/JVI.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Spagnolo JF, Hogue BG. 2000. Host Protein Interactions with the 3’ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J Virol 74:5053–5065. 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Moqtaderi Z, Geisberg JV, Struhl K. 2014. Secondary structures involving the poly(A) tail and other 3’ sequences are major determinants of mRNA isoform stability in yeast. Microb Cell 1:137–139. 10.15698/mic2014.04.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Simon-Loriere E, Holmes EC. 2011. Why do RNA viruses recombine? Nat Rev Microbiol 9:617–626. 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chrisman BS, Paskov K, Stockham N, Tabatabaei K, Jung JY, Washington P, Varma M, Sun MW, Maleki S, Wall DP. 2021. Indels in SARS-CoV-2 occur at template-switching hotspots. BioData Min 14:20. 10.1186/s13040-021-00251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rowe CL, Fleming JO, Nathan MJ, Sgro JY, Palmenberg AC, Baker SC. 1997. Generation of coronavirus spike deletion variants by high-frequency recombination at regions of predicted RNA secondary structure. J Virol 71:6183–6190. 10.1128/JVI.71.8.6183-6190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rowe CL, Baker SC, Nathan MJ, Fleming JO. 1997. Evolution of mouse hepatitis virus: detection and characterization of spike deletion variants during persistent infection. J Virol 71:2959–2969. 10.1128/JVI.71.4.2959-2969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Masters PS. 2006. The molecular biology of coronaviruses. Adv Virus Res 65:193–292. 10.1016/s0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu HY, Guy JS, Yoo D, Vlasak R, Urbach E, Brian DA. 2003. Common RNA replication signals exist among group 2 coronaviruses: Evidence for in vivo recombination between animal and human coronavius molecules. Virology 315:174–183. 10.1016/S0042-6822(03)00511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brian DA, Spaan WJM. 1997. Recombination and coronavirus defective interfering RNAs. Semin Virol 8:101–111. 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stirrups K, Shaw K, Evans S, Dalton K, Cavanagh D, Britton P. 2000. Leader switching occurs during the rescue of defective RNAs by heterologous strains of the coronavirus infectious bronchitis virus. J Gen Virol 81:791–801. 10.1099/0022-1317-81-3-791. [DOI] [PubMed] [Google Scholar]
- 158.Spaan W, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, van der Zeijst BA, Siddell SG. 1983. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J 2:1839–1844. 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bentley K, Keep SM, Armesto M, Britton P. 2013. Identification of a Noncanonically Transcribed subgenomic mRNA of infectious bronchitis virus and other gammacoronaviruses. J Virol 87:2128–2136. 10.1128/JVI.02967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Viehweger A, Krautwurst S, Lamkiewicz K, Madhugiri R, Ziebuhr J, Hölzer M, Marz M. 2019. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res 29:1545–1554. 10.1101/gr.247064.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thiel V, Ivanov KA, Putics Á, Hertzig T, Schelle B, Bayer S, Weißbrich B, Snijder EJ, Rabenau H, Doerr HW, Gorbalenya AE, Ziebuhr J. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol 84:2305–2315. 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 162.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, hsin Chen M, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TCT, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Günther S, Osterhaus ADHE, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399. 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 163.Marra MA, Jones SJM, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YSN, Khattra J, Asano JK, Barber SA, Chan SY, Cloutier A, Coughlin SM, Freeman D, Girn N, Griffith OL, Leach SR, Mayo M, McDonald H, Montgomery SB, Pandoh PK, Petrescu AS, Robertson AG, Schein JE, Siddiqui A, Smailus DE, Stott JM, Yang GS, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth TF, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples GA, Tyler S, et al. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399–1404. 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 164.Yang Y, Yan W, Hall AB, Jiang X. 2021. Characterizing transcriptional regulatory sequences in coronaviruses and their role in recombination. Mol Biol Evol 38:1241–1248. 10.1093/molbev/msaa281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Krakauer DC. 2000. Stability and evolution of overlapping genes. Evolution 54:731–739. 10.1111/j.0014-3820.2000.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 166.Namy O, Moran SJ, Stuart DI, Gilbert RJC, Brierley I. 2006. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441:244–247. 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wills NM, Gesteland RF, Atkins JF. 1994. Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J 13:4137–4144. 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Baranov PV, Henderson CM, Anderson CB, Gesteland RF, Atkins JF, Howard MT. 2005. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology 332:498–510. 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kelly JA, Woodside MT, Dinman JD. 2021. Programmed −1 ribosomal frameshifting in coronaviruses: a therapeutic target. Virology 554:75–82. 10.1016/j.virol.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kelly JA, Olson AN, Neupane K, Munshi S, Emeterio JS, Pollack L, Woodside MT, Dinman JD. 2020. Structural and functional conservation of the programmed −1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J Biol Chem 295:10741–10748. 10.1074/jbc.AC120.013449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yu CH, Noteborn MHM, Olsthoorn RCL. 2010. Stimulation of ribosomal frameshifting by antisense LNA. Nucleic Acids Res 38:8277–8283. 10.1093/nar/gkq650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Brierley I, Jenner AJ, Inglis SC. 1992. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol 227:463–479. 10.1016/0022-2836(92)90901-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Plant EP, Jacobs KLM, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, Petrov AN, Dinman JD. 2003. The 9-Å solution: How mRNA pseudoknots promote efficient programmed -1 ribosomal frameshifting. RNA 9:168–174. 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Plant EP, Pérez-Alvarado GC, Jacobs JL, Mukhopadhyay B, Hennig M, Dinman JD. 2005. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol 3:e172. 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Herold J, Siddell SG. 1993. An “elaborated” pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res 21:5838–5842. 10.1093/nar/21.25.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Eleouet JF, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. 1995. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology 206:817–822. 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Su MC, Chang CT, Chu CH, Tsai CH, Chang KY. 2005. An atypical RNA pseudoknot stimulator and an upstream attenuation signal for -1 ribosomal frameshifting of SARS coronavirus. Nucleic Acids Res 33:4265–4275. 10.1093/nar/gki731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang K, Zheludev IN, Hagey RJ, Haslecker R, Hou YJ, Kretsch R, Pintilie GD, Rangan R, Kladwang W, Li S, Wu MT-P, Pham EA, Bernardin-Souibgui C, Baric RS, Sheahan TP, D'Souza V, Glenn JS, Chiu W, Das R. 2021. Cryo-EM and antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. Nat Struct Mol Biol 28:747–754. 10.1038/s41594-021-00653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Neupane K, Zhao M, Lyons A, Munshi S, Ileperuma SM, Ritchie DB, Hoffer NQ, Narayan A, Woodside MT. 2021. Structural dynamics of single SARS-CoV-2 pseudoknot molecules reveal topologically distinct conformers. Nat Commun 12:4749. 10.1038/s41467-021-25085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Plant EP, Sims AC, Baric RS, Dinman JD, Taylor DR. 2013. Altering SARS coronavirus frameshift efficiency affects genomic and subgenomic RNA production. Viruses 5:279–294. 10.3390/v5010279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Park SJ, Kim YG, Park HJ. 2011. Identification of RNA pseudoknot-binding ligand that inhibits the -1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J Am Chem Soc 133:10094–10100. 10.1021/ja1098325. [DOI] [PubMed] [Google Scholar]
- 182.Ritchie DB, Soong J, Sikkema WKA, Woodside MT. 2014. Anti-frameshifting ligand reduces the conformational plasticity of the SARS virus pseudoknot. J Am Chem Soc 136:2196–2199. 10.1021/ja410344b. [DOI] [PubMed] [Google Scholar]
- 183.Sun Y, Abriola L, Niederer RO, Pedersen SF, Alfajaro MM, Monteiro VS, Wilen CB, Ho YC, Gilbert WV, Surovtseva YV, Lindenbach BD, Guo JU. 2021. Restriction of SARS-CoV-2 replication by targeting programmed -1 ribosomal frameshifting. Proc National Acad Sci USA 118:e2023051118. 10.1073/pnas.2023051118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Y, Garcia G, Arumugaswami V, Guo F. 2021. Structure-based design of antisense oligonucleotides that inhibit SARS-CoV-2 replication. bioRxiv. 10.1101/2021.08.23.457434. [DOI]
- 185.Cho CP, Lin SC, Chou MY, Hsu HT, Chang KY. 2013. Regulation of programmed ribosomal frameshifting by co-translational refolding RNA hairpins. PLoS One 8:e62283. 10.1371/journal.pone.0062283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Haniff HS, Tong Y, Liu X, Chen JL, Suresh BM, Andrews RJ, Peterson JM, O'Leary CA, Benhamou RI, Moss WN, Disney MD. 2020. Targeting the SARS-COV-2 RNA genome with small molecule binders and ribonuclease targeting chimera (RiboTAC) degraders. ACS Cent Sci 6:1713–1721. 10.1021/acscentsci.0c00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Larsen KP, Choi J, Prabhakar A, Puglisi EV, Puglisi JD. 2019. Relating structure and dynamics in RNA biology. Cold Spring Harb Perspect Biol 11:a032474. 10.1101/cshperspect.a032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Coey A, Larsen K, Puglisi JD, Puglisi EV. 2016. Heterogeneous structures formed by conserved RNA sequences within the HIV reverse transcription initiation site. RNA 22:1689–1698. 10.1261/rna.056804.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shimizu A, Nakatani Y, Nakamura T, Jinno-Oue A, Ishikawa O, Boeke JD, Takeuchi Y, Hoshino H. 2014. Characterisation of cytoplasmic DNA complementary to non-retroviral RNA viruses in human cells. Sci Rep 4:5074. 10.1038/srep05074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Geuking MB, Weber J, Dewannieux M, Gorelik E, Heidmann T, Hengartner H, Zinkernagel RM, Hangartner L. 2009. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science 323:393–396. 10.1126/science.1167375. [DOI] [PubMed] [Google Scholar]
- 191.Klenerman P, Hengartner H, Zinkernagel RM. 1997. A non-retroviral RNA virus persists in DNA form. Nature 390:298–301. 10.1038/36876. [DOI] [PubMed] [Google Scholar]
- 192.Zhang L, Richards A, Inmaculada Barrasa M, Hughes SH, Young RA, Jaenisch R. 2021. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc Natl Acad Sci USA 118:e2105968118. 10.1073/pnas.2105968118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, Yin H, Xiao Q, Tang Y, Qu X, Kuang L, Fang X, Mishra N, Lu J, Shan H, Jiang G, Huang X. 2020. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 5:434–435. 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhou B, She J, Wang Y, Ma X. 2020. Duration of viral shedding of discharged patients with severe COVID-19. Clin Infect Dis 71:2240–2242. 10.1093/cid/ciaa451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nakajima Y, Ogai A, Furukawa K, Arai R, Anan R, Nakano Y, Kurihara Y, Shimizu H, Misaki T, Okabe N. 2021. Prolonged viral shedding of SARS-CoV-2 in an immunocompromised patient. J Infect Chemother 27:387–389. 10.1016/j.jiac.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McKie AM, Jones TPW, Sykes C. 2020. Prolonged viral shedding in an immunocompetent patient with COVID-19. BMJ Case Rep 13:e237357. 10.1136/bcr-2020-237357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Widders A, Broom A, Broom J. 2020. SARS-CoV-2: the viral shedding vs infectivity dilemma. Infect Dis Health 25:210–215. 10.1016/j.idh.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Xu K, Chen Y, Yuan J, Yi P, Ding C, Wu W, Li Y, Ni Q, Zou R, Li X, Xu M, Zhang Y, Zhao H, Zhang X, Yu L, Su J, Lang G, Liu J, Wu X, Guo Y, Tao J, Shi D, Yu L, Cao Q, Ruan B, Liu L, Wang Z, Xu Y, Liu Y, Sheng J, Li L. 2020. Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19). Clin Infect Dis 71:799–806. 10.1093/cid/ciaa351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Horie M, Tomonaga K. 2011. Non-retroviral fossils in vertebrate genomes. Viruses 3:1836–1848. 10.3390/v3101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Grechishnikova D, Poptsova M. 2016. Conserved 3’ UTR stem-loop structure in L1 and Alu transposons in human genome: possible role in retrotransposition. BMC Genom 17:992. 10.1186/s12864-016-3344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Parry R, Gifford RJ, Lytras S, Ray SC, Coin LJM. 2021. No evidence of SARS-CoV-2 reverse transcription and integration as the origin of chimeric transcripts in patient tissues. Proc Natl Acad Sci USA 118:e2109066118. 10.1073/pnas.2109066118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Smits N, Rasmussen J, Bodea GO, Amarilla AA, Gerdes P, Sanchez-Luque FJ, Ajjikuttira P, Modhiran N, Liang B, Faivre J, Deveson IW, Khromykh AA, Watterson D, Ewing AD, Faulkner GJ. 2021. No evidence of human genome integration of SARS-CoV-2 found by long-read DNA sequencing. Cell Rep 36:109530. 10.1016/j.celrep.2021.109530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yan B, Chakravorty S, Mirabelli C, Wang L, Trujillo-Ochoa JL, Chauss D, Kumar D, Lionakis MS, Olson MR, Wobus CE, Afzali B, Kazemian M. 2021. Host-virus chimeric events in SARS-CoV-2-infected cells are infrequent and artifactual. J Virol 95:e0029421. 10.1128/JVI.00294-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Osanai M, Takahashi H, Kojima KK, Hamada M, Fujiwara H. 2004. Essential motifs in the 3’ untranslated region required for retrotransposition and the precise start of reverse transcription in non-long-terminal-repeat retrotransposon SART1. Mol Cell Biol 24:7902–7913. 10.1128/MCB.24.18.7902-7913.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hayashi Y, Kajikawa M, Matsumoto T, Okada N. 2014. Mechanism by which a LINE protein recognizes its 3’ tail RNA. Nucleic Acids Res 42:10605–10617. 10.1093/nar/gku753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wakefield JK, Rhim H, Morrow CD. 1994. Minimal sequence requirements of a functional human immunodeficiency virus type 1 primer binding site. J Virol 68:1605–1614. 10.1128/JVI.68.3.1605-1614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Anreiter I, Mir Q, Simpson JT, Janga SC, Soller M. 2021. New twists in detecting mRNA modification dynamics. Trends Biotechnol 39:72–89. 10.1016/j.tibtech.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Boo SH, Kim YK. 2020. The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med 52:400–408. 10.1038/s12276-020-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, Kurkowska M, Shirvanizadeh N, Destefanis E, Groza P, Avşar G, Romitelli A, Pir P, Dassi E, Conticello SG, Aguilo F, Bujnicki JM. 2022. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res 50:D231–D235. 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. 2012. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol 13:175. 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Cowling VH. 2010. Regulation of mRNA cap methylation. Biochemistry J 425:295–302. 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, Gross SS, Elemento O, Debart F, Kiledjian M, Jaffrey SR. 2017. Reversible methylation of m6 Am in the 5’ cap controls mRNA stability. Nature 541:371–375. 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hyde JL, Diamond MS. 2015. Innate immune restriction and antagonism of viral RNA lacking 2’-O methylation. Virology 479–480:66–74. 10.1016/j.virol.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr, Shi PY, Diamond MS. 2010. 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Walker AP, Fan H, Keown JR, Knight ML, Grimes JM, Fodor E. 2021. The SARS-CoV-2 RNA polymerase is a viral RNA capping enzyme. Nucleic Acids Res 49:13019–13030. 10.1093/nar/gkab1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Taiaroa G, Rawlinson D, Featherstone L, Pitt M, Caly L, Druce J, Purcell D, Harty L, Tran T, Roberts J, Catton M, Williamson D, Coin L, Duchene S. 2020. Direct RNA sequencing and early evolution of SARS-CoV-2. bioRxiv. 10.1101/2020.03.05.976167. [DOI]
- 217.Chen Y, Guo D. 2016. Molecular mechanisms of coronavirus RNA capping and methylation. Virol Sin 31:3–11. 10.1007/s12250-016-3726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Walker AP, Fan H, Keown JR, Grimes JM, Fodor E. 2021. Identification of guanylyltransferase activity in the SARS-CoV-2 RNA polymerase. bioRxiv. 10.1101/2021.03.17.435913v1. [DOI]
- 219.Yan L, Ge J, Zheng L, Zhang Y, Gao Y, Wang T, Huang Y, Yang Y, Gao S, Li M, Liu Z, Wang H, Li Y, Chen Y, Guddat LW, Wang Q, Rao Z, Lou Z. 2021. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell 184:184–193.e10. 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jin X, Chen Y, Sun Y, Zeng C, Wang Y, Tao J, Wu A, Yu X, Zhang Z, Tian J, Guo D. 2013. Characterization of the guanine-N7 methyltransferase activity of coronavirus nsp14 on nucleotide GTP. Virus Res 176:45–52. 10.1016/j.virusres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, Decroly E. 2010. In vitro reconstitution of sars-coronavirus mRNA cap methylation. PLoS Pathog 6:e1000863. 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ogando NS, Kazzi PE, Zevenhoven-Dobbe JC, Bontes BW, Decombe A, Posthuma CC, Thiel V, Canard B, Ferron F, Decroly E, Snijder EJ. 2021. Structure–function analysis of the nsp14 N7–guanine methyltransferase reveals an essential role in betacoronavirus replication. Proc Natl Acad Sci USA 118:e2108709118. 10.1073/pnas.2108709118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bouvet M, Lugari A, Posthuma CC, Zevenhoven JC, Bernard S, Betzi S, Imbert I, Canard B, Guillemot JC, Lécine P, Pfefferle S, Drosten C, Snijder EJ, Decroly E, Morelli X. 2014. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J Biol Chem 289:25783–25796. 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Menachery VD, Yount BL, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-O-methyltransferase activity. J Virol 88:4251–4264. 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Krafcikova P, Silhan J, Nencka R, Boura E. 2020. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun 11:3717. 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zaccara S, Ries RJ, Jaffrey SR. 2019. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20:608–624. 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 228.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149:1635–1646. 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. 2014. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505:117–120. 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. 2016. YTHDF2 destabilizes m 6 A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun 7:12626. 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. 2014. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10:93–95. 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE, Darnell RB. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31:990–1006. 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RPG, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49:18–29. 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rubio RM, Depledge DP, Bianco C, Thompson L, Mohr I. 2018. RNA m 6 A modification enzymes shape innate responses to DNA by regulating interferon β. Genes Dev 32:1472–1484. 10.1101/gad.319475.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Liu J, Xu YP, Li K, Ye Q, Zhou HY, Sun H, Li X, Yu L, Deng YQ, Li RT, Cheng ML, He B, Zhou J, Li XF, Wu A, Yi C, Qin CF. 2021. The m6A methylome of SARS-CoV-2 in host cells. Cell Res 31:404–414. 10.1038/s41422-020-00465-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Patil DP, Pickering BF, Jaffrey SR. 2018. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol 28:113–127. 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kennedy EM, Bogerd HP, Kornepati AVR, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, Cullen BR. 2016. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19:675–685. 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, Cullen BR. 2017. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe 22:377–386.e5. 10.1016/j.chom.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Baquero-Perez B, Geers D, Díez J. 2021. From a to m6a: the emerging viral epitranscriptome. Viruses 13:1049. 10.3390/v13061049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM. 2016. Dynamics of human and viral RNA methylation during zika virus infection. Cell Host Microbe 20:666–673. 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Xue M, Zhao BS, Zhang Z, Lu M, Harder O, Chen P, Lu Z, Li A, Ma Y, Xu Y, Liang X, Zhou J, Niewiesk S, Peeples ME, He C, Li J. 2019. Viral N 6-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat Commun 10:4595. 10.1038/s41467-019-12504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zhang T, Yang Y, Xiang Z, Gao CC, Wang W, Wang C, Xiao X, Wang X, Qiu WN, Li WJ, Ren L, Li M, Zhao YL, Chen YS, Wang J, Yang YG. 2021. N 6-methyladenosine regulates RNA abundance of SARS-CoV-2. Cell Discov 7:7–4. 10.1038/s41421-020-00241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang X, Hao H, Ma L, Zhang Y, Hu X, Chen Z, Liu D, Yuan J, Hu Z, Guan W. 2021. Methyltransferase-like 3 modulates severe acute respiratory syndrome coronavirus-2 RNA N6-methyladenosine modification and replication. mBio 2:e0106721. 10.1128/mbio.01067-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Burgess HM, Depledge DP, Thompson L, Srinivas KP, Grande RC, Vink EI, Abebe JS, Blackaby WP, Hendrick A, Albertella MR, Kouzarides T, Stapleford KA, Wilson AC, Mohr I. 2021. Targeting the m6A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes Dev 35:1005–1019. 10.1101/gad.348320.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Li N, Hui H, Bray B, Gonzalez GM, Zeller M, Anderson KG, Knight R, Smith D, Wang Y, Carlin AF, Rana TM. 2021. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep 35:109091. 10.1016/j.celrep.2021.109091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Durbin AF, Wang C, Marcotrigiano J, Gehrke L. 2016. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 7:e00833-16. 10.1128/mBio.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Meng Y, Zhang Q, Wang K, Zhang X, Yang R, Bi K, Chen W, Diao H. 2021. RBM15-mediated N6-methyladenosine modification affects COVID-19 severity by regulating the expression of multitarget genes. Cell Death Dis 12:1–10. 10.1038/s41419-020-03229-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.An S, Xie Z, Liao Y, Jiang J, Dong W, Yin F, Li WX, Ye L, Lin J, Liang H. 2022. Systematic analysis of clinical relevance and molecular characterization of m6A in COVID-19 patients. Genes Dis. 10.1016/j.gendis.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Campos JHC, Maricato JT, Braconi CT, Antoneli F, Janini LMR, Briones MRS. 2021. Direct RNA sequencing reveals SARS-CoV-2 m6A sites and possible differential DRACH motif methylation among variants. Viruses 13:2108. 10.3390/v13112108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bayoumi M, Munir M. 2021. Evolutionary conservation of the DRACH signatures of potential N6-methyladenosine (m6A) sites among influenza A viruses. Sci Rep 11:4548. 10.1038/s41598-021-84007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Shulman Z, Stern-Ginossar N. 2020. The RNA modification N 6-methyladenosine as a novel regulator of the immune system. Nat Immunol 21:501–512. 10.1038/s41590-020-0650-4. [DOI] [PubMed] [Google Scholar]
- 252.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12:767–772. 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. 2015. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc 137:2107–2115. 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Miladi M, Fuchs J, Maier W, Weigang S, Díaz i Pedrosa N, Weiss L, Lother A, Nekrutenko A, Ruzsics Z, Panning M, Kochs G, Gilsbach R, Grüning B. 2020. The landscape of SARS-CoV-2 RNA modifications. bioRxiv. 10.1101/2020.07.18.204362v1. [DOI]
- 255.Vacca D, Fiannaca A, Tramuto F, Cancila V, La Paglia L, Mazzucco W, Gulino A, La Rosa M, Maida CM, Morello G, Belmonte B, Casuccio A, Maugeri R, Iacopino G, Balistreri CR, Vitale F, Tripodo C, Urso A. 2022. Direct RNA nanopore sequencing of SARS-CoV-2 extracted from critical material from swabs. Life 12:69. 10.3390/life12010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Fleming AM, Mathewson NJ, Howpay Manage SA, Burrows CJ. 2021. Nanopore dwell time analysis permits sequencing and conformational assignment of pseudouridine in sARS-CoV-2. ACS Cent Sci 7:1707–1717. 10.1021/acscentsci.1c00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485:201–206. 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 258.Marchand V, Ayadi L, Ernst FGM, Hertler J, Bourguignon-Igel V, Galvanin A, Kotter A, Helm M, Lafontaine DLJ, Motorin Y. 2018. AlkAniline-Seq: profiling of m7G and m3C RNA modifications at single nucleotide resolution. Angew Chem Int Ed 57:16785–16790. 10.1002/anie.201810946. [DOI] [PubMed] [Google Scholar]
- 259.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, Fink G, Regev A. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159:148–162. 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kumar S, Mohapatra T. 2021. Deciphering epitranscriptome: modification of mRNA bases provides a new perspective for post-transcriptional regulation of gene expression. Front Cell Dev Biol 9:628415. 10.3389/fcell.2021.628415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, Novoa EM. 2019. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 10:4079. 10.1038/s41467-019-11713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. 2017. The RNA modification landscape in human disease. RNA 23:1754–1769. 10.1261/rna.063503.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Pratanwanich PN, Yao F, Chen Y, Koh CWQ, Wan YK, Hendra C, Poon P, Goh YT, Yap PML, Chooi JY, Chng WJ, Ng SB, Thiery A, Goh WSS, Göke J. 2021. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat Biotechnol 39:1394–1402. 10.1038/s41587-021-00949-w. [DOI] [PubMed] [Google Scholar]
- 264.Leger A, Amaral PP, Pandolfini L, Capitanchik C, Capraro F, Miano V, Migliori V, Toolan-Kerr P, Sideri T, Enright AJ, Tzelepis K, van Werven FJ, Luscombe NM, Barbieri I, Ule J, Fitzgerald T, Birney E, Leonardi T, Kouzarides T. 2021. RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat Commun 12:7198. 10.1038/s41467-021-27393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Briggs E, Ward W, Rey S, Law D, Nelson K, Bois M, Ostrov N, Lee HH, Laurent JM, Mita P. 2021. Assessment of potential SARS-CoV-2 virus integration into human genome reveals no significant impact on RT-qPCR COVID-19 testing. Proc Natl Acad Sci USA 118:e2113065118. 10.1073/pnas.2113065118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Cao C, Cai Z, Xiao X, Rao J, Chen J, Hu N, Yang M, Xing X, Wang Y, Li M, Zhou B, Wang X, Wang J, Xue Y. 2021. The architecture of the SARS-CoV-2 RNA genome inside virion. Nat Commun 12:3917. 10.1038/s41467-021-22785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lednicky JA, Tagliamonte MS, White SK, Elbadry MA, Alam MM, Stephenson CJ, Bonny TS, Loeb JC, Telisma T, Chavannes S, Ostrov DA, Mavian C, Beau De Rochars VM, Salemi M, Morris JG. 2021. Independent infections of porcine deltacoronavirus among Haitian children. Nature 600:133–137. 10.1038/s41586-021-04111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lednicky JA, Tagliamonte MS, White SK, Blohm GM, Alam MM, Iovine NM, Salemi M, Mavian C, Morris JG. 2021. Isolation of a novel recombinant canine coronavirus from a visitor to haiti: further evidence of transmission of coronaviruses of zoonotic origin to humans. Clin Infect Dis ciab924. 10.1093/cid/ciab924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Li X, Zhang L, Chen S, Ouyang H, Ren L. 2021. Possible targets of pan-coronavirus antiviral strategies for emerging or re-emerging coronaviruses. Microorganisms 9:1479. 10.3390/microorganisms9071479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R, Teeling E. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Minh BQ, Nguyen MAT, Von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Alhatlani BY. 2020. In silicoidentification of conserved cis-acting RNA elements in the SARS-CoV-2 genome. Future Virol 15:409–417. 10.2217/fvl-2020-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Tavares R, Mahadeshwar G, Wan H, Huston NC, Pyle AM. 2021. The global and local distribution of RNA structure throughout the SARS-CoV-2 genome. J Virol 95:e02190-20. 10.1128/JVI.02190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Simmonds P. 2020. Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses. mBio 11:1–15. 10.1128/mBio.01661-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Vankadari N, Jeyasankar NN, Lopes WJ. 2020. Structure of the SARS-CoV-2 Nsp1/5’-untranslated region complex and implications for potential therapeutic targets, a vaccine, and virulence. J Phys Chem Lett 11:9659–9668. 10.1021/acs.jpclett.0c02818. [DOI] [PubMed] [Google Scholar]