Abstract
MiRNAs are short non-coding RNAs that regulate gene expression post-transcriptionally contributing to the development of different diseases including cancer. The miR-200 family consists of five members, miR-200a, miR-200b, miR-200c, miR-141, and miR-429. Their expression is dysregulated in cancer tissue and their level is altered in the body fluids of cancer patients. Moreover, the levels of miR-200 family members correlate with clinical parameters such as cancer patients’ survival which makes them potentially useful as diagnostic and prognostic biomarkers. MiRNAs can act as either oncomiRs or tumor suppressor miRNAs depending on the target genes and their role in the regulation of key oncogenic signaling pathways. In most types of cancer, the miR-200 family acts as tumor suppressor miRNA and regulates all features of cancer. In this review, we summarized the expression pattern of the miR-200 family in different types of cancer and their potential utility as biomarkers. Moreover, we comprehensively described the role of miR-200 family members in the regulation of all hallmarks of cancer proposed by Hanahan and Weinberg with the focus on the epithelial-mesenchymal transition, invasiveness, and metastasis of tumor cells.
Keywords: miR-200, miR-200 family, miRNA, hallmarks of cancer, tumor progression, invasiveness, metastasis
Introduction
MiRNAs (microRNAs, miRs) are small non-coding RNAs that regulate gene expression at the post-transcriptional level. They were discovered in 1993, when Lee et al. described an antisense RNA-RNA interaction between lin-4 transcripts complementary to the 3’ untranslated region (UTR) of lin-14 mRNA in Caenorhabditis elegans (1). Over the past 30 years, miRNAs have become the subject of intense research, including in the field of cancer research (2, 3). So far over 2000 human miRNAs have been described (4, 5). Numerous studies have focused on the role of miRNAs in cancer development in vitro and in vivo, as well as their potential use as diagnostic and prognostic biomarkers and therapeutic agents (6, 7).
MiRNA biogenesis is a multistep process resulting in the formation of mature single-stranded miRNA (8). It begins in the nucleus where pri-miRNA is transcribed with RNA polymerase II. MiRNAs may be monocistronic (encoded individually) or polycistronic (encoded in clusters). In the next step, pre-miRNA is generated with the participation of the Drosha complex (microprocessor) and transported to the cytoplasm where it is further processed by the Dicer complex to miRNA duplex. Only one strand of the miRNA duplex (mature miRNA) forms the miRNA-induced silencing complex (RISC) (9–11).
MiRNAs bind with their seed region to complementary sequences in the 3’ UTR of mRNAs (12). The seed region is a sequence of 2-7 nucleotides at the 5’ end of the miRNA that is responsible for mRNA binding (13). That leads to the degradation of mRNA or inhibition of protein translation (14). One miRNA may target several mRNAs, as well as an individual mRNA may be regulated by numerous miRNAs. The are several bioinformatical tools (e.g. TargetScan, Starbase) that enable the prediction of the targets of miRNAs in silico, but the result must be validated experimentally (15). Thus, miRNAs form a complex miRNA-mRNA regulatory network that influences key pathways in cells and controls all biological processes including carcinogenesis (2, 12).
The miR-200 family consists of five members, miR-200a, miR-200b, miR-200c, miR-141, and miR-429. They are divided into two clusters based on their seed sequence. MiR-200b, miR-200a, and miR-429 (cluster I) are located on chromosome 1 and have a common seed sequence (AAUACUG). While miR-200c and miR-141 belong to cluster II and are located on chromosome 11. Their seed sequence (AACACUG) differs only by one nucleotide from the seed sequence of cluster I ( Figure 1 ) (16). Hence the members of the miR-200 family can theoretically target similar mRNAs (13). This review aims to comprehensively describe the role of the miR-200 family members in regulating all hallmarks of cancer.
Figure 1.
The members of the miR-200 family. Genes encoding the miR-200 family form two clusters. One encoding miR-200b, miR-200a, and miR-429 is located in chromosome 1 while miR-200c and miR-141 are encoded by genes located in chromosome 11. Members of the miR-200 family are characterized by AA(U/C)ACUG seed sequence.
Expression of miR-200 family in cancer
Dysregulation of the expression of miRNAs in cancer
MiRNA expression is often dysregulated in cancer cells. New technologies such as high-throughput RNA sequencing enable to profile miRNAs expression (17). Several studies showed the general repression of miRNAs in cancer tissues since the majority of them are tumor suppressors (18–20). One of the miRNA families that suppress cancer development and progression is the miR-200 family which is known for targeting zinc-finger E-box binding homeobox (ZEB) and inhibiting epithelial-mesenchymal transition (EMT) (21). In contrast, miR-21 is a well-known oncomiR that targets mainly tumor suppressors (22). Notably, the individual miRNA expression and functions in cancer cells may be tissue-specific and differ between different tumor types (23).
Mechanisms of miRNAs dysregulation in cancer
The mechanisms of miRNAs expression regulation are complex and multistep (24). They may be divided into transcriptional mechanisms such as the influence of transcription factors, DNA methylation, and histone modifications (25). All of them control pri-miRNA transcription. Among numerous transcription factors that regulate miRNAs expression are p53, ZEB, Myc, and hormone receptors, including estrogen receptors (26). The next mechanism regulating miRNAs expression is DNA promoter methylation. Hypermethylation was associated with decreased expression of miRNAs, while hypomethylation caused the upregulation of miRNAs expression (27). Furthermore, there are posttranscriptional mechanisms that regulate miRNA expression by the regulation of pri-miRNA and pre-miRNA processing (26). The genes involved in miRNAs biogenesis, including DICER, are frequently dysregulated in cancer (28, 29). Downregulation of DICER is a prediction of poor prognosis for overall and progression-free survival (30).
Expression of miR-200 family
In our review, we collected articles that determined the expression of miR-200 family members in 25 cancer types in the tumor tissue compared to corresponding healthy tissue using the RT-qPCR (real-time quantitative PCR) method ( Table 1 ). MiR-200 family members were downregulated in 10 types of cancer and upregulated in 2 cancer types. In 12 types of cancer individual members of the miR-200 family had different levels of expression ( Table 1 ). MiRNAs from both miR-200b/miR-200a/miR-429 and miR200c/miR-141 clusters are expressed together as two different polycistronic pri-miRNA transcripts (133). Therefore, they have a similar pattern of expression (up- or downregulation) in most cancer types. The expression of the miR-200 family is regulated by different mechanisms, including DNA methylation. miR-200b, miR-200c, and miR-141 were described as epigenetically silenced by DNA methylation in breast cancer (134). Moreover, the miR-200b/miR-200a/miR-429 cluster was repressed in the mechanism of histone modifications in this model (135).
Table 1.
The expression of miR-200 family members in different types of cancer compared to the healthy tissue.
| Cancer | miR-200a expression | miR-200b expression | miR-200c expression | miR-141 expression | miR-429 expression | Ref. |
|---|---|---|---|---|---|---|
| Acute myeloid leukemia | ↓ | ↓ | n/d | n/d | ↓ | (31) |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | ↓ | ↑ | (32, 33) |
| Bladder cancer | ↓/↑ | ↑ | ↓/↑ | ↑ | ↑ | (34–41) |
| Breast cancer | ↓/↑ | ↓/↑ | ↓/↑ | ↓/↑ | n/d | (42, 43) |
| Cervical carcinoma | ↓ | ↓ | ↓/↑ | ↑ | ↓ | (44–50) |
| Cholangiocarcinoma | ↓ | ↓ | ↓ | n/d | n/d | (51, 52) |
| Colorectal cancer | ↑ | ↑ | ↑ | ↑ | ↑/↓ | (53–59) |
| Endometrial cancer | ↑ | ↑ | ↑ | ↑ | ↑ | (60, 61) |
| Esophageal carcinoma | ↓/↑ | ↓ | ↓ | ↓ | ↓ | (62–70) |
| Gastric cancer | ↓/↑ | ↓/↑ | ↓/↑ | ↓ | ↓/↑ | (71–76) |
| Glioma | ↓ | ↓ | ↓ | ↓ | ↑/↓ | (77–83) |
| Head and neck carcinoma | ↓ | n/d | n/d | ↓ | n/d | (84–86) |
| Liver cancer | ↓ | ↓ | ↓ | ↓ | ↓ | (87–91) |
| Melanoma | ↓ | ↓ | ↓ | ↓ | ↓ | (92–94) |
| Nephroblastoma | ↓ | ↓ | ↓ | ↓ | ↓ | (95–98) |
| Neuroblastoma | ↓ | n/d | n/d | n/d | n/d | (99) |
| Non-small cell lung cancer | ↓ | ↓ | ↓ | ↑/↓ | ↑ | (100) |
| Oral squamous cell carcinoma | ↓ | ↓ | ↓ | ↓ | ↓ | (101, 102) |
| Osteosarcoma | ↓ | ↓ | ↓ | ↓ | ↓ | (103–107) |
| Ovarian cancer | ↑ | ↑ | ↑ | ↑ | n/d | (108–113) |
| Pancreatic cancer | ↑ | ↑ | ↓ | ↑/↓ | ↓ | (114–118) |
| Prostate cancer | ↓/↑ | ↑ | ↓ | ↑ | n/d | (119–122) |
| Renal cell carcinoma | ↓ | ↓ | ↓ | ↓ | ↓ | (123–128) |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | n/d | n/d |
| Thyroid cancer | ↓/↑ | ↓/↑ | ↓/↑ | ↓/↑ | ↓/↑ | (129–132) |
↓ - downregulated expression; ↑ - upregulated expression; n/d – no data.
miRNAs in body fluids
Circulating miRNAs can be detected in various body fluids including serum, saliva, and urine. Thus, they are promising diagnostic and prognostic biomarkers. MiR-200 family members have been detected in different body fluids and were dysregulated in 18 cancer types ( Table 2 ). The level of miR-200 family members in serum was increased in 7 cancer types and decreased in serum in 3 cancer types. There are inconsistent data concerning serum miRNAs expression in bladder cancer, prostate cancer, and non-small lung carcinoma. Notably, the expression patterns of miR-200 family members in cancer tissue are consistent with their level in body fluids only in 5 cancer types. The observation is consistent with the studies that demonstrate that the miRNAs expression profiles of patients’ serum and cancer tissue are different (194, 195). Importantly, the levels of miR-200 family in different types of body fluids vary in cancer patients and may not correlate (196).
Table 2.
The level of miR-200 family members in the serum, urine or saliva of patients with different types of cancer.
| Cancer | miR-200a level | miR-200b level | miR-200c level | miR-141 level | miR-429 level | Ref. |
|---|---|---|---|---|---|---|
| Acute myeloid leukemia | n/d | n/d | n/d | n/d | n/d | n/d |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | n/d | n/d |
| Bladder cancer | ↓ (urine) | ↓ (urine) | ↓ (urine) | ↑/↓ (urine) | ↓ (urine) | (136–139) |
| Breast cancer | n/d | n/d | ↓ | n/d | n/d | (140, 141) |
| Cervical carcinoma | ↑ | n/d | ↑ | ↓ | n/d | (48, 142, 143) |
| Cholangiocarcinoma | ↑ | ↑ | ↑ | ↑ | n/d | (144) |
| Colorectal cancer | n/d | ↑ | ↑ | ↑ | n/d | (145–150) |
| Endometrial cancer | ↑ | n/d | n/d | ↑ | n/d | (61) |
| Esophageal carcinoma | n/d | n/d | ↑ | n/d | n/d | (151, 152) |
| Gastric cancer | ↑ | n/d | ↑ | ↓ | n/d | (153–158) |
| Glioma | ↓ | n/d | n/d | n/d | n/d | (81) |
| Head and neck carcinoma | n/d | n/d | n/d | n/d | n/d | n/d |
| Liver cancer | ↓ | n/d | n/d | n/d | ↑ | (159–162) |
| Melanoma | n/d | n/d | ↓ | n/d | n/d | (163) |
| Nephroblastoma | n/d | n/d | n/d | n/d | n/d | n/d |
| Neuroblastoma | n/d | n/d | n/d | n/d | n/d | n/d |
| Non-small cell lung cancer | n/d | ↑/↓ | n/d | ↑ | ↑/↓ | (164–169) |
| Oral squamous cell carcinoma | ↓ (saliva) | ↑ | n/d | n/d | n/d | (170, 171) |
| Osteosarcoma | n/d | n/d | n/d | n/d | n/d | n/d |
| Ovarian cancer | ↑ | ↑ | ↑ | ↑ | ↑ | (172–178) |
| Pancreatic cancer | ↑ | ↑ | ↑ | n/d | n/d | (179, 180) |
| Prostate cancer | ↑ | ↑/↓ | ↑/↓ | ↑ | n/d | (181–189) |
| Renal cell carcinoma | ↓ | n/d | n/d | ↓ | ↑ | (190–192) |
| Small cell lung carcinoma | n/d | n/d | n/d | ↑ | n/d | (193) |
| Thyroid cancer | n/d | n/d | n/d | n/d | n/d | n/d |
↓ - decreased level; ↑ - increased level; n/d – no data. Arrows refer to serum miRNA level unless otherwise indicated.
MiRNAs may be loaded into small vesicles, including exosomes. Exosomal miRNAs are distributed to body fluids where they play important role in the pathogenesis of cancer by regulating all hallmarks of cancer (197). The level of exosomal miR-200 family members may serve as prognostic or diagnostic markers in various tumor types. The level of exosomal miR-200 is upregulated in ovarian cancer patients’ serum (198) and in pleural effusion of patients diagnosed with lung adenocarcinoma (199). Moreover, the level of exosomal miR-200 correlates with the invasiveness of ovarian cancer (200). Exosomal miR-200b and miR-200c may serve as independent prognostic factors in pancreatic ductal adenocarcinoma as they correlate with overall survival of patients (180). Lower expression of exosomal miR-200c and miR-141 is associated with longer overall survival of colon cancer patients (201). Moreover, miR-200 family members may play role as biomarker of cholangiocarcinoma, in particular miR-200a/c correlates with tumor stage (144).
miRNAs as diagnostic and prognostic biomarkers
miRNAs may not only serve as diagnostic biomarkers that enable to distinguish between cancer and healthy tissue but also their levels correlate with clinical parameters and thus may have potential prognostic value (202). The combination of miRNAs may discriminate cancerous from healthy tissues with high accuracy (203, 204). The meta-analysis shows that miR-200c expression has a moderate diagnostic value in gastric cancer (205) and miR-141 may be a diagnostic marker of colorectal cancer (206). Notably, a meta-analysis of 58 articles with 8107 cancer patients revealed that in general higher expression of miR-200 family members is associated with worse survival (207, 208). Moreover, miR-200a expression was associated with unfavorable prognosis in breast cancer patients, and miR-429 correlated with shorter survival in liver cancer patients (207). Higher miR-200c expression was correlated with shorter overall survival in gastric cancer and non-small cell lung carcinoma (205, 209). However, in another meta-analysis, the expression of miR-200 family members was associated with a better prognosis in bladder cancer as it correlated with longer overall survival, recurrence-free survival, and cancer-specific survival (210). Moreover, miR-200c correlated with a better prognosis in ovarian cancer (211). Notably, it was found that serum miR-200c may be used as a marker of colorectal cancer tumor recurrence after surgery (147). Similarly, urine miR-200a was identified as marker the recurrence of non-muscle-invasive bladder cancer (137). In addition to miRNAs in the body fluids, it was found that the level of miR-141 in prostate cancer tissue may be associated with the tumor recurrence (212). Thus, more prospective clinical trials are required to determine the utility of miR-200 family members as biomarkers.
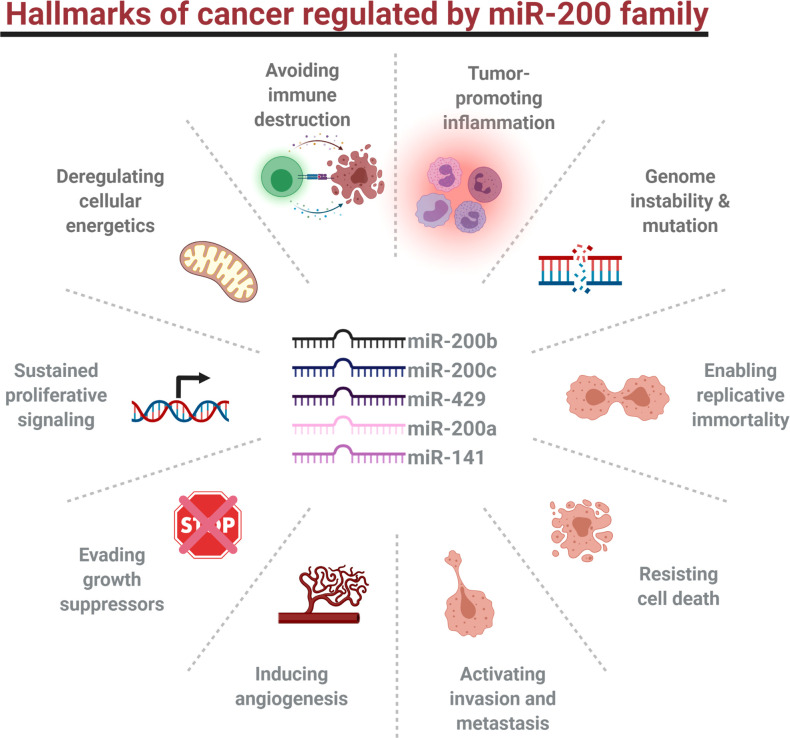
Hallmarks of cancer
Hallmarks of cancer have been first described in 2000 (213). In the updated version of their review, 14 features of human tumor development were proposed: sustaining proliferative signaling, evading tumor growth suppressors, enabling replicative immortality, activating invasion and metastasis, resisting cell death, inducing angiogenesis, avoiding immune destruction, tumor-promoting inflammation, genome instability and mutation, deregulating cellular energetics, unlocking phenotypic plasticity, polymorphic microbiomes, nonmutational epigenetic reprogramming, and senescence (214). All of them are regulated by miRNAs, including miR-200 family members ( Figure 2 ) (215, 216).
Figure 2.
Hallmarks of cancer regulated by miR-200 family members.
Regulation of cell proliferation by miR-200 family
The proliferation of cancer cells is one of the most important hallmarks of cancer. It is sustained by growth factors, constitutive activation of oncogenic signaling pathways, evading growth repressors like p53 or RB, and enabled replicative immortality (217). Proliferation is regulated by numerous pathways including PI3K/AKT, MEK/ERK, JAK/STAT, and β-Catenin/Wnt signaling, and by cell cycle regulators such as cyclin-dependent kinases (CDKs). The members of those pathways are regulated by miRNAs that may either promote or inhibit cancer cell proliferation. MiR-200 family has different effects on tumor cell proliferation in vitro depending on the cancer type ( Table 3 ). Based on our literature review, miR-200a increases the proliferation of 5 cancer types and inhibits the proliferation of 13 cancer types, and there are inconsistent results concerning colorectal cancer, non-small cell lung cancer, and renal cell carcinoma. In vivo, miR-200a inhibits tumor growth of gastric cancer, glioma, melanoma, neuroblastoma, and prostate cancer. Furthermore, miR-200b stimulates the proliferation of 4 cancer types, and inhibits 13 cancer types, and there are inconsistent results concerning colorectal and non-small cell lung, and ovarian cell carcinoma. In vivo studies show miR-200b increases tumor growth of cervical and ovarian cancer and decreases tumor growth of breast cancer, glioma, head and neck cancer, non-small lung cancer, prostate cancer, and thyroid cancer. MiR-200c acts as oncomiR in 3 cancer types and as tumor suppressor miR in 13 cancer types and there are inconsistent results concerning colorectal, endometrial, and renal cell carcinoma. It decreases tumor growth in vivo of 9 cancer types in vivo and increases tumor growth of head and neck carcinoma. Moreover, miR-141 increases the proliferation of 6 cancer types, and decreases in 12 cancer types, and there are discrepancies regarding the role of miR-141 in the regulation of colorectal, non-small cell lung, and ovarian carcinoma cell proliferation. In vivo, miR-141 acts mainly as tumor suppressor miR and inhibits the tumor growth of numerous cancer types including colorectal, gastric, and pancreatic cancer. miR-141 increases the tumor growth of cervical and non-small cell lung cancer. Furthermore, it increases the proliferation of 5 and decreases the proliferation of 14 cancer types. In vivo, miR-429 inhibits tumor growth of 7 cancer types and increases tumor growth of bladder cancer only. Based on that information, miR-200 family members are suppressive miRNAs in most cancer types, however, they might also stimulate the proliferation of some types of cancer cells.
Table 3.
The role of the miR-200 family in proliferation and tumor growth.
| miRNAs | Cancer | Target | Cell proliferation in vitro | Tumor growth in vivo | Ref. |
|---|---|---|---|---|---|
| miR-200a | Acute myeloid leu-kemia | n/d | n/d | n/d | n/d |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | |
| Bladder cancer | STAT4 | ↓ | n/d | (218) | |
| Breast cancer | TFAM | ↓ | n/d | (219) | |
| Cervical cancer | EGLN1 | ↑ | n/d | (220) | |
| Cholangiocarcinoma | n/d | ↓ | n/d | (52) | |
| Colorectal cancer | FOXA1, RASSF2 | ↓/↑ | n/d | (56, 221, 222) | |
| Endometrial cancer | PTEN | ↑ | n/d | (60, 223) | |
| Esophageal carcinoma | CTNNB1, CDH1, APC, PTEN, CTNNA1, CRMP1 and SOD2 | ↑ | n/d | (70, 224) | |
| Gastric cancer | n/d | ↓ | ↓ | (225) | |
| Glioma | FOXA1, SIM2-s | ↓ | ↓ | (83, 226) | |
| Head and neck carcinoma | CD47 | ↓ | n/d | (86) | |
| Liver cancer | STAT4, FOXA2 | ↓ | n/d | (161, 162, 227) | |
| Melanoma | CDK6, GOLM1 | ↓ | ↓ | (93, 228) | |
| Nephroblastoma | n/d | n/d | n/d | n/d | |
| Neuroblastoma | AP-2γ | ↓ | ↓ | (99) | |
| Non-small cell lung cancer | RHPN2 | ↑/↓ | n/d | (229, 230) | |
| Oral squamous cell carcinoma | n/d | n/d | n/d | n/d | |
| Osteosarcoma | ZEB1 | ↓ | n/d | (104) | |
| Ovarian cancer | PDCH9 | ↑ | n/d | (111, 113) | |
| Pancreatic cancer | DEK | ↓ | n/d | (231, 232) | |
| Prostate cancer | BRD4 | ↓ | ↓ | (233, 234) | |
| Renal cell carcinoma | CBL, SIRT1, SPAG9, TGFβ2 | ↓/↑ | n/d | (124, 126, 127, 235, 236) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | FOXA1 | ↓ | n/d | (130) | |
| miR-200b | Acute myeloid leu-kemia | n/d | ↓ | n/d | (237) |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | |
| Bladder cancer | FSCN1 | ↓ | n/d | (238) | |
| Breast cancer | IKBKB, FUT4, radixin, SP1 | ↓ | ↓ | (239–242) | |
| Cervical cancer | FOXG1 | ↑ | ↑ | (243) | |
| Cholangiocarcinoma | n/d | ↑ | n/d | (244) | |
| Colorectal cancer | RECK, TUBB3, Wnt1 | ↑/↓ | n/d | (245–247) | |
| Endometrial cancer | PTEN | ↑ | n/d | (60, 223) | |
| Esophageal carcinoma | n/d | n/d | n/d | n/d | |
| Gastric cancer | ZEB2 | ↓ | n/d | (248) | |
| Glioma | CD133, CREB1, ERK5 | ↓ | ↓ | (249–251) | |
| Head and neck carcinoma | Notch1 | ↓ | ↓ | (252) | |
| Liver cancer | HMGB3 | ↓ | n/d | (90) | |
| Melanoma | n/d | ↓ | n/d | (253) | |
| Nephroblastoma | IKK-β | ↓ | n/d | (98) | |
| Neuroblastoma | n/d | n/d | n/d | n/d | |
| Non-small cell lung cancer | ABCA1, p70S6K1, RhoE | ↑/↓ | ↓ | (254–256) | |
| Oral squamous cell carcinoma | Kindlin-2, ZEB2 | ↓ | n/d | (257) | |
| Osteosarcoma | ZEB1 | ↓ | n/d | ||
| Ovarian cancer | ATAD2, ING5 | ↑/↓ | ↑ | (258, 259) | |
| Pancreatic cancer | n/d | n/d | n/d | n/d | |
| Prostate cancer | n/d | ↓ | ↓ | (233, 260–262) | |
| Renal cell carcinoma | n/d | ↑ | n/d | (124) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | RAP1B | ↓ | ↓ | (132) | |
| miR-200c | Acute myeloid leu-kemia | n/d | n/d | n/d | n/d |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | |
| Bladder cancer | BMI-1, E2F3, LDHA | ↓ | n/d | (36, 263) | |
| Breast cancer | BMI-1, KRAS, PDE7B, XIAP | ↓ | ↓ | (264–269) | |
| Cervical cancer | MAP4K4 | ↓ | n/d | (270) | |
| Cholangiocarcinoma | n/d | n/d | n/d | n/d | |
| Colorectal cancer | CDK2, RASSF2 | ↑/↓ | n/d | (56, 271) | |
| Endometrial cancer | PTEN, PTENP1, MALAT1 | ↑/↓ | ↓ | (60, 272; 273) | |
| Esophageal carcinoma | n/d | n/d | n/d | n/d | |
| Gastric cancer | EDNRA, FN1 | ↓ | n/d | (274, 275) | |
| Glioma | MSN | ↓ | ↓ | (79) | |
| Head and neck carcinoma | PTEN | ↑ | ↑ | (276) | |
| Liver cancer | MAD2L1 | ↓ | n/d | (88) | |
| Melanoma | n/d | ↓ | ↓ | (277) | |
| Nephroblastoma | FRS2, IKK-β | ↓ | n/d | (97, 98, 278) | |
| Neuroblastoma | n/d | n/d | n/d | n/d | |
| Non-small cell lung cancer | LDHA | ↓ | ↓ | (279, 280) | |
| Oral squamous cell carcinoma | n/d | ↓ | n/d | (102) | |
| Osteosarcoma | AKT2 | ↓ | ↓ | (105) | |
| Ovarian cancer | n/d | ↑ | ↓ | (281, 282) | |
| Pancreatic cancer | n/d | ↑ | n/d | (283) | |
| Prostate cancer | AMACR, ZEB2 | ↓ | ↓ | (122, 284, 285) | |
| Renal cell carcinoma | SLC6A1 | ↑/↓ | n/d | (124, 128) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | RAP1B | ↓ | ↓ | (132) | |
| miR-141 | Acute myeloid leu-kemia | RAB32 | ↓ | ↓ | (286) |
| Acute lymphoblastic leukemia | TRAF5 | ↓ | n/d | (32) | |
| Bladder cancer | n/d | n/d | n/d | n/d | |
| Breast cancer | ANP32E, HMGB1 | ↓ | ↓ | (287, 288) | |
| Cervical cancer | FOXA2 | ↑ | ↑ | (289) | |
| Cholangiocarcinoma | n/d | ↑ | n/d | (244) | |
| Colorectal cancer | PHLPP2, RASSF2 | ↑/↓ | ↓ | (56, 58, 290, 291) | |
| Endometrial cancer | n/d | ↑ | n/d | (60) | |
| Esophageal carcinoma | n/d | ↓ | n/d | (67) | |
| Gastric cancer | YAP1, TAZ | ↓ | ↓ | (75, 292) | |
| Glioma | SKA2 | ↓ | ↓ | (77) | |
| Head and neck carcinoma | EGFR | ↓ | ↓ | (85) | |
| Liver cancer | TGFβR1 | ↓ | n/d | (91) | |
| Melanoma | n/d | n/d | n/d | n/d | |
| Nephroblastoma | n/d | n/d | n/d | n/d | |
| Neuroblastoma | FUS | ↓ | ↓ | (293) | |
| Non-small cell lung cancer | HOXC13, KLF9 | ↑/↓ | ↑ | (193, 294, 295) | |
| Oral squamous cell carcinoma | n/d | n/d | n/d | n/d | |
| Osteosarcoma | GLI2 | ↓ | n/d | (106, 296) | |
| Ovarian cancer | n/d | ↑/↓ | n/d | (112, 297) | |
| Pancreatic cancer | MAP4K4 | ↓ | ↓ | (298) | |
| Prostate cancer | KLF9, RUNX1 | ↑ | n/d | (299, 300) | |
| Renal cell carcinoma | n/d | ↑ | n/d | (124) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | IRS2 | ↓ | ↓ | (129) | |
| miR-429 | Acute myeloid leukemia | n/d | n/d | n/d | n/d |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | |
| Bladder cancer | CDKN2B | ↑ | ↑ | (39) | |
| Breast cancer | FN1 | ↓ | n/d | (301) | |
| Cervical cancer | IKK-β, ZEB1 | ↓ | ↓ | (46, 302) | |
| Cholangiocarcinoma | n/d | n/d | n/d | n/d | |
| Colorectal cancer | HMGB3, Onecut2 | ↓ | ↓ | (55, 59) | |
| Endometrial cancer | n/d | ↑ | n/d | (60) | |
| Esophageal carcinoma | Bcl-2, SP1, Slug, RAB23 | ↓ | ↓ | (65, 68, 69) | |
| Gastric cancer | FSCN1, c-MYC | ↓ | ↓ | (71, 74, 303) | |
| Glioma | SOX2 | ↓ | n/d | (82) | |
| Head and neck carcinoma | n/d | ↓ | n/d | (304) | |
| Liver cancer | RAB23 | ↓ | n/d | (305) | |
| Melanoma | AKT1 | n/d | ↓ | (306) | |
| Nephroblastoma | c-MYC, IKK-β | ↓ | n/d | (96, 98) | |
| Neuroblastoma | IKK-β | ↓ | ↓ | (307) | |
| Non-small cell lung cancer | DLC−1, PTEN, RASSF8, TIMP2 | ↑ | n/d | (308, 309) | |
| Oral squamous cell carcinoma | ZEB1 | ↓ | n/d | (310) | |
| Osteosarcoma | ZEB1, HOXA9 | ↓ | n/d | (103, 107) | |
| Ovarian cancer | n/d | n/d | n/d | n/d | |
| Pancreatic cancer | NT-3 | ↓ | n/d | (117) | |
| Prostate cancer | p27Kip1 | ↑ | n/d | (311) | |
| Renal cell carcinoma | AKT1 | ↑ | ↓ | (124, 312) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | ZEB1 | ↓ | n/d | (131) |
↓ - suppression by miRNA; ↑ - promotion by miRNA; n/d – no data.
MiR-200 family members exert their effects by targeting several mRNAs associated with cancer cell proliferation. MiR-200a targets Cyclin-Dependent Kinase 6 (CDK6) in melanoma and thus causes cell-cycle arrest and decreases cancer cell proliferation (93). MiR-429 targets p27Kip1, an inhibitor of the cell cycle, and its overexpression promotes the proliferation of prostate cancer cells (311). Furthermore, the miR-200 family targets the members of key pathways regulating cancer cell proliferation. PI3K/AKT pathway promotes proliferation, migration, invasion, and EMT (epithelial-mesenchymal transition) (313). AKT1/2 is targeted by miR-200c in osteosarcoma (105), and by miR-429 in melanoma (306) and renal cell carcinoma (312). Downregulation of AKT1/2 by miRNAs inhibits cancer cell proliferation in vitro and tumor growth in vivo. Moreover, the miR-200 family targets PTEN, a key suppressor of the PI3K/AKT pathway (314). It is targeted by miR-200a in endometrial cancer (223) and esophageal carcinoma (70), miR-200b in endometrial cancer (223), miR-200c in endometrial cancer (273) and head and neck carcinoma (276), and miR-429 in non-small cell lung cancer (308) resulting in increased of cancer cells proliferation in vitro.
The second most important pathway regulating cell proliferation is MEK/ERK pathway. KRAS which activates MEK/ERK is targeted by miR-200c in breast cancer. MiR-200c thus has an inhibitory effect on breast cancer cell proliferation and tumor growth (265). Moreover, miR-200b targets p70S6K1 and inhibits lung cancer cell proliferation in vitro and tumor growth in vivo (256). Whereas, miR-429 targets RAB23 in esophageal and liver carcinomas resulting in the suppression of cell proliferation, migration, and invasion (69, 305). On the contrary, MEK/ERK inhibitor Ras Association Domain Family Member 2 (RASSF2) is targeted by miR-200a, miR-200c, miR-141 in colorectal cancer, and Ras Association Domain-Containing Protein 8 (RASSF8) by miR-429 in non-small lung carcinoma which results in increased proliferation (56, 308). Similarly, other signaling pathways regulating cell proliferation are targeted by miRNAs. MiR-200a binds STAT4 in bladder and liver cancer and decreases cell proliferation in vitro (162, 218). In esophageal carcinoma, however, miR-200a acts as oncomiR and targets APC, an inhibitor of β-Catenin/Wnt signaling (70).
Regulation of cell migration and invasiveness by miR-200 family
Cancer cell migration and invasion are multistep processes that involve cytoskeleton remodeling, interaction with extracellular matrix (ECM), and then its digestion. All these steps are regulated by miRNAs that suppress or promote cancer progression (7).
In this study, we summarized the knowledge about miR-200 family regulation of cancer cell migration and invasion in vitro ( Table 4 ). MiR-200 family members suppress cell migration and invasiveness in most types of cancer. MiR-200a increases the migration of 2 cancer types and the invasion of 3 cancer types and it decreases the migration of 12 and invasion of 13 cancer types. MiR-200b increases the migration of renal cell cancer and decreases the migration of 16 cancer types and invasion of 15 cancer types. Furthermore, miR-200c promotes the migration and invasion of head and neck carcinoma. On the other hand, miR-200c inhibits the migration of 15 and invasion of 13 types of cancers. MiR-141 decreases the migration of 13 types of cancer cells and invasion of 13 cancer types. However, it increases the migration of bladder and renal cell carcinoma, and it stimulates the invasion of bladder, cervical, and non-small lung cancer. Likewise, miR-429 decreases the migration and invasion of 15 cancer types, while it increases the migration of renal cell carcinoma and migration and invasion of non-small lung carcinoma.
Table 4.
Members of the miR-200 family regulating cancer cell migration and invasion.
| miRNAs | Cancer | Target | Cell migration in vitro | Cell invasion in vitro | Ref. |
|---|---|---|---|---|---|
| miR-200a | Acute myeloid leukemia | n/d | n/d | n/d | n/d |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | |
| Bladder cancer | DICER | n/d | ↑ | (41) | |
| Breast cancer | ELK3, EPHA2 | ↓ | ↓ | (315, 316) | |
| Cervical cancer | n/d | n/d | n/d | n/d | |
| Cholangiocarcinoma | n/d | n/d | ↓ | (52) | |
| Colorectal cancer | FOXA1 | ↓ | ↓ | (221, 222) | |
| Endometrial cancer | n/d | n/d | n/d | n/d | |
| Esophageal carcinoma | CTNNB1, CDH1, APC, PTEN, CTNNA1 and SOD2 | ↑ | ↑ | (70) | |
| Gastric cancer | n/d | ↓ | ↓ | (225) | |
| Glioma | FOXA1, SIM2-s | ↓ | ↓ | (83, 226) | |
| Head and neck carcinoma | CD47 | ↓ | ↓ | (86) | |
| Liver cancer | STAT4, Foxa2 | ↓ | ↓ | (161, 162, 227) | |
| Melanoma | GOLM1 | ↓ | ↓ | (228) | |
| Nephroblastoma | n/d | n/d | n/d | n/d | |
| Neuroblastoma | n/d | n/d | n/d | n/d | |
| Non-small cell lung cancer | HGF | ↓ | ↓ | (100) | |
| Oral squamous cell carcinoma | n/d | n/d | n/d | n/d | |
| Osteosarcoma | ZEB1 | ↓ | n/d | (104) | |
| Ovarian cancer | PDCH9 | ↑ | ↑ | (111, 113) | |
| Pancreatic cancer | DEK | ↓ | ↓ | (231, 232, 317) | |
| Prostate cancer | n/d | n/d | ↓ | (233) | |
| Renal cell carcinoma | TGFβ2 | ↓ | ↓ | (235) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | FOXA1 | ↓ | ↓ | (130) | |
| miR-200b | Acute myeloid leukemia | n/d | n/d | ↓ | (237) |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | |
| Bladder cancer | FSCN1 | ↓ | ↓ | (238) | |
| Breast cancer | IKBKB, FUT4, PKCα, radixin, | ↓ | ↓ | (240–242, 318, 319) | |
| Cervical cancer | FoxG1, RhoE | ↓/↑ | ↓/↑ | (45, 243, 320) | |
| Cholangiocarcinoma | SUZ12 and ROCK2 | ↓ | ↓ | (51) | |
| Colorectal cancer | TUBB3 | ↓ | ↓ | (247) | |
| Endometrial cancer | TIMP2 | ↓ | n/d | (321) | |
| Esophageal carcinoma | Kindlin-2 | ↓ | ↓ | (322) | |
| Gastric cancer | ZEB2 | ↓ | ↓ | (248) | |
| Glioma | CD133, ERK5 | ↓ | ↓ | (250, 251) | |
| Head and neck carcinoma | Notch1 | ↓ | ↓ | (252) | |
| Liver cancer | HMGB3 | ↓ | n/d | (90) | |
| Melanoma | n/d | ↓ | ↓ | (253) | |
| Nephroblastoma | IKK-β | ↓ | ↓ | (98) | |
| Neuroblastoma | n/d | n/d | n/d | n/d | |
| Non-small cell lung cancer | ABCA1, p70S6K1, RhoE, TIF1γ | ↑/↓ | ↑/↓ | (255, 256, 323, 324) | |
| Oral squamous cell carcinoma | Kindlin-2, ZEB2 | ↓ | ↓ | (257) | |
| Osteosarcoma | ZEB1 | ↓ | ↓ | ||
| Ovarian cancer | n/d | n/d | n/d | n/d | |
| Pancreatic cancer | n/d | n/d | n/d | n/d | |
| Prostate cancer | n/d | ↓ | ↓ | (233, 261, 262) | |
| Renal cell carcinoma | n/d | ↑ | n/d | (124) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | RAP1B | ↓ | ↓ | (132, 325) | |
| miR-200c | Acute myeloid leukemia | n/d | n/d | n/d | n/d |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | |
| Bladder cancer | BMI-1, E2F3, LDHA, RECK | ↑/↓ | ↑/↓ | (36, 38, 263) | |
| Breast cancer | n/d | ↓ | ↓ | (267, 268, 326) | |
| Cervical cancer | MAP4K4 | ↓ | n/d | (270) | |
| Cholangiocarcinoma | SUZ12 and ROCK2 | ↓ | ↓ | (51) | |
| Colorectal cancer | n/d | ↓ | ↓ | (327) | |
| Endometrial cancer | PTEN, PTENP1 | ↑/↓ | ↑/↓ | (272, 328; 273) | |
| Esophageal carcinoma | n/d | n/d | n/d | n/d | |
| Gastric cancer | EDNRA, FN1 | ↓ | ↓ | (274, 275) | |
| Glioma | MSN | ↓ | ↓ | (79) | |
| Head and neck carcinoma | PTEN | ↑ | ↑ | (276) | |
| Liver cancer | MAD2L1 | ↓ | ↓ | (88) | |
| Melanoma | n/d | ↓ | n/d | (277) | |
| Nephroblastoma | FRS2, IKK-β | ↓ | ↓ | (97, 98) | |
| Neuroblastoma | n/d | n/d | n/d | n/d | |
| Non-small cell lung cancer | LDHA | ↓ | n/d | (280) | |
| Oral squamous cell carcinoma | ZEB1 | ↓ | ↓ | (329) | |
| Osteosarcoma | AKT2 | ↓ | n/d | (105) | |
| Ovarian cancer | n/d | ↓ | ↓ | (281, 282) | |
| Pancreatic cancer | n/d | n/d | ↓ | (283, 330) | |
| Prostate cancer | AMACR, ZEB2 | ↓ | ↓ | (122, 284, 285) | |
| Renal cell carcinoma | SLC6A1 | ↑/↓ | ↓ | (124, 128) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | RAP1B | ↓ | ↓ | (132) | |
| miR-141 | Acute myeloid leukemia | n/d | n/d | n/d | n/d |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | |
| Bladder cancer | n/d | n/d | n/d | n/d | |
| Breast cancer | ANP32E, ZEB1/2 | ↓ | ↓ | (287, 331) | |
| Cervical cancer | FOXA2 | n/d | ↑ | (289) | |
| Cholangiocarcinoma | n/d | n/d | n/d | n/d | |
| Colorectal cancer | EGFR | ↓ | ↓ | (332) | |
| Endometrial cancer | n/d | n/d | n/d | n/d | |
| Esophageal carcinoma | n/d | ↓ | ↓ | (67) | |
| Gastric cancer | TAZ | ↓ | ↓ | (292) | |
| Glioma | SKA2 | ↓ | ↓ | (77) | |
| Head and neck carcinoma | EGFR | ↓ | ↓ | (85) | |
| Liver cancer | TGFβR1 | ↓ | ↓ | (91) | |
| Melanoma | n/d | n/d | n/d | n/d | |
| Nephroblastoma | n/d | n/d | n/d | n/d | |
| Neuroblastoma | FUS | ↓ | ↓ | (293) | |
| Non-small cell lung cancer | KLF9 | n/d | ↑ | (295) | |
| Oral squamous cell carcinoma | n/d | n/d | n/d | n/d | |
| Osteosarcoma | FUS | ↓ | ↓ | (296) | |
| Ovarian cancer | n/d | ↓ | ↓ | (297) | |
| Pancreatic cancer | MAP4K4, TM4SF1 | ↓ | ↓ | (298, 333) | |
| Prostate cancer | RUNX1 | ↓ | ↓ | (300) | |
| Renal cell carcinoma | n/d | ↑ | n/d | (124) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | IRS2 | ↓ | ↓ | (129) | |
| miR-429 | Acute myeloid leukemia | n/d | n/d | n/d | n/d |
| Acute lymphoblastic leukemia | n/d | n/d | n/d | n/d | |
| Bladder cancer | n/d | ↓ | ↓ | (334) | |
| Breast cancer | FN1 | ↓ | ↓ | (301) | |
| Cervical cancer | ZEB1 | ↓ | ↓ | (302) | |
| Cholangiocarcinoma | n/d | n/d | n/d | n/d | |
| Colorectal cancer | Onecut2, PAK6 | ↓ | ↓ | (55, 335) | |
| Endometrial cancer | n/d | n/d | n/d | n/d | |
| Esophageal carcinoma | Bcl-2, Slug, SP1 | ↓ | ↓ | (65, 68) | |
| Gastric cancer | Sp1, Notch1 | ↓ | ↓ | (336, 337) | |
| Glioma | BMK1, SOX2 | ↓ | ↓ | (82, 338) | |
| Head and neck carcinoma | n/d | ↓ | ↓ | (304) | |
| Liver cancer | CRKL, RAB23 | ↓ | ↓ | (89, 305) | |
| Melanoma | LIMK1 | ↓ | ↓ | (94) | |
| Nephroblastoma | IKK-β | ↓ | ↓ | (98) | |
| Neuroblastoma | IKKβ | ↓ | ↓ | (307) | |
| Non-small cell lung cancer | PTEN, RASSF8, TIF1γ and TIMP2, | ↑ | ↑ | (308, 323) | |
| Oral squamous cell carcinoma | n/d | n/d | n/d | n/d | |
| Osteosarcoma | ZEB1, HOXA9 | ↓ | ↓ | (103, 107) | |
| Ovarian cancer | p15PAF | ↓ | n/d | (339) | |
| Pancreatic cancer | NT-3 | n/d | ↓ | (117) | |
| Prostate cancer | n/d | n/d | n/d | n/d | |
| Renal cell carcinoma | n/d | ↑ | n/d | (124) | |
| Small cell lung carcinoma | n/d | n/d | n/d | n/d | |
| Thyroid cancer | ZEB1 | ↓ | ↓ | (131) |
↓ - suppression by miRNA; ↑ - promotion by miRNA; n/d – no data.
The epithelial-mesenchymal process (EMT) is one of the crucial cellular processes in cancer cell invasion. During this process, epithelial cells gain mesenchymal capabilities including migration. The regulation of EMT is complex and is modulated by numerous signaling pathways including TGFβ and Wnt/β-catenin and transcription factors such as Snail, Slug, ZEB1/2, and TWIST1/2 (7, 340). MiR-200 family is a well-known regulator of EMT because of targeting ZEB1 and ZEB2 (341) which are key inducers of this process (342). ZEB1/2 is targeted by miR-200a [osteosarcoma (104)], 200b [gastric cancer, oral squamous cell carcinoma, osteosarcoma (248, 257)], miR-200c [oral squamous cell carcinoma, prostate cancer (285, 329)], miR-141 [breast cancer (331)], and miR-429 [cervical cancer, osteosarcoma, thyroid cancer (103, 131, 302)]. Moreover, the miR-200 family targets other regulators of EMT. MiR-200a targets TGFβ2 in renal cell carcinoma and inhibits the invasion and migration of cancer cells (235). MiR-141 suppresses liver cancer cell invasion and migration by targeting TGFβR1 (91). Moreover, miR-429 targets Slug, a transcription factor that belongs to the Snail family and activates EMT. Thus, miR-429 inhibits the migration and invasion of esophageal cancer cells (68). On the contrary, miR-200a targets Catenin Beta 1 (CTNNB1), the member of the Wnt/β-catenin pathway, and promotes the invasion and migration of esophageal cancer cells (70).
Cytoskeleton remodeling is a vital step in cells motility. It leads to the formation of cell membrane protrusions, then to cell contraction and retraction of rear-end (343). Moesin and radixin belong to ERM (ezrin-radixin-moesin) protein family that is responsible for cytoskeleton structure and cell migration (344). MiR-200c targets moesin in glioma (79) and miR-200b targets radixin in breast cancer cells (242). By targeting them, miR-200b and miR-200c suppress cancer progression (70). Fascin 1 (FSCN1) is an actin-bundling protein involved in the formation of protrusions. It is targeted by miR-200b leading to the decrease of cancer cell migration and invasion in bladder cancer (238). RhoE is a member of Ras superfamily and is responsible for actin reorganization. Both, miR-200b and miR-200c are described to target RhoE and inhibit the invasion of non−small cell lung cancer cells (344). Kindlin-2 is another protein involved in extracellular matrix regulation. It is reported to be directly targeted by miR-200b in esophageal carcinoma and in oral squamous cell carcinoma which causes the inhibition of cancer cell migration and invasion (79). TUBB3 (Tubulin Beta 3 Class III), a main cytoskeletal microtubule protein engaged in various processes including cancer invasion and migration, is targeted by miR-200b in colorectal cancer leading to the suppression of migration and invasion of cancer cells (247).
The ECM proteolysis and interactions with the cancer cell cytoskeleton play a crucial role in the migration and invasion of tumor cells (345). MiRNA-200c targets fibronectin (FN1), the component of the extracellular matrix, in gastric cancer cells and acts as a suppressor of invasion and migration (274). On the contrary, miR-200 family members also promote cancer progression by targeting the inhibitors of metalloproteinases that digest ECM. miR-200c targets RECK in bladder cancer (38), whereas TIMP2 is targeted by miR-200b in endometrial cancer cells (66, 257). Moreover, miR-200a regulates cell-to-cell adhesion by targeting CDH1 (cadherin 1) (70).
Regulation of apoptosis by miR-200 family
Apoptosis is so-called programmed cell death, essential for maintaining tissue homeostasis. The mechanism of apoptosis is complex and involves several steps. Each of these steps can be impaired, thus leading to carcinogenesis (346). The major role in apoptosis belongs to caspases, which may be both initiators and executioners of apoptosis (347). Numerous studies have reported the influence of miRNAs on the regulation of both the intrinsic and the extrinsic apoptosis pathways (348).
The miR-200 family members act mainly as apoptosis promoters ( Table 5 ). miR-200a promotes apoptosis in 6 cancer types and inhibits apoptosis only in cervical cancer. There are discordant results concerning the role of miR-200a in the regulation of non-small cell lung cancer. MiR-200b promotes apoptosis in 5 cancer types and miR-200c promotes apoptosis in 6 cancer types. miR-141 promotes apoptosis in 6 cancer types, and inhibits apoptosis in pancreatic cancer, but its role in prostate cancer is inconsistent. miR-429 promotes apoptosis in 9 cancer types and inhibits apoptosis only in colorectal cancer.
Table 5.
Members of the miR-200 family regulating apoptosis.
| miRNAs | Cancer | Target | Apoptosis in vitro | Ref. |
|---|---|---|---|---|
| miR-200a | Cervical cancer | EGLN1 | ↓ | (220) |
| Gastric cancer | A20 | ↑ | (349) | |
| Liver cancer | n/d | ↑ | (227) | |
| Nephroblastoma | CDC7 | ↑ | (350) | |
| Non-small cell lung cancer | HGF, RHPN2 | ↓/↑ | (100, 351) | |
| Prostate cancer | BRD4, SIRT1, SPAG9 | ↑ | (126, 234, 236) | |
| Renal cell carcinoma | CBL, SPAG9 | ↑ | (126, 127) | |
| Thyroid cancer | FOXA1 | ↑ | (130) | |
| miR-200b | Acute myeloid leukemia | n/d | ↑ | (237) |
| Breast cancer | Sp1 | ↑ | (239) | |
| Colorectal cancer | TUBB3, Wnt1 | ↑ | (246, 247) | |
| Oral squamous cell carcinoma | CDK2, PAF | ↑ | (352) | |
| Ovarian cancer | ATAD2 | ↑ | (259) | |
| miR-200c | Breast cancer | XIAP | ↑ | (264) |
| Colorectal cancer | CDK2 | ↑ | (271) | |
| Gastric cancer | EDNRA | ↑ | (275) | |
| Liver carcinoma | MAD2L1 | ↑ | (88) | |
| Nephroblastoma | n/d | ↑ | (278) | |
| Non-small cell lung cancer | n/d | ↑ | (279) | |
| miR-141 | Acute lymphoblastic leukemia | TRAF5 | ↑ | (32) |
| Colorectal cancer | EGFR | ↑ | (332) | |
| Gastric cancer | YAP1 | ↑ | (75) | |
| Head and neck carcinoma | EGFR | ↑ | (85) | |
| Osteosarcoma | GLI2 | ↑ | (106) | |
| Pancreatic cancer | MAP4K4 | ↓ | (298) | |
| Prostate cancer | KLF9 | ↓/↑ | (299, 300) | |
| Thyroid cancer | IRS2 | ↑ | (129) | |
| miR-429 | Gastric carcinoma | Bcl-2 | ↑ | (303, 353) |
| Glioblastoma | Bcl-2 | ↑ | ||
| Nephroblastoma | c-MYC | ↑ | (96) | |
| Colorectal cancer | HMGB3 | ↑ | (59) | |
| Osteosarcoma | ZEB1 | ↑ | (103) | |
| Glioblastoma | SOX2 | ↑ | (82) | |
| Esophageal carcinoma | Bcl-2, SP1 | ↑ | (65) | |
| Thyroid cancer | ZEB1 | ↑ | (131) | |
| Colorectal cancer | SOX2 | ↓ | (53) | |
| Cervical carcinoma | IKKb | ↑ | (46) |
↓ - suppression by miRNA; ↑ - promotion by miRNA; n/d – no data
MiR-200a enhances TRAIL-triggered apoptosis in gastric cancer cells by targeting TNFα-induced protein 3 (A20) (349). Moreover, miR-200a shows pro-apoptotic activity in the human hepatocellular carcinoma cell line (227). In Wilms tumor cells, miR-200a promotes apoptosis by targeting CDC7 (350). Similarly, miR-200a can promote apoptosis in prostate cancer cells through BRD4/AR signaling pathway (234), by directly targeting SIRT1 (236) or by targeting Sperm-associated antigen 9 (SPAG9) (100). The last-mentioned protein, SPAG9, is an oncogene protein that regulates renal cell carcinoma progression. Accordingly, its regulation by miR-200 also has a stimulatory effect on apoptosis in renal cell carcinoma (126). Moreover, miR-200a has been shown to promote apoptosis of renal cell carcinoma cells by targeting CBL (127). Furthermore, by targeting FOXA1, miR-200a also promotes cell apoptosis in anaplastic thyroid carcinoma (130). In contrast, miR-200a suppresses apoptosis and promotes the proliferation of cervical cancer cells by targeting EGLN1 (220). In the case of NSCLC, miR-200 demonstrates a two-fold action. It promotes apoptosis by downregulation of the hepatocyte growth factor (HGF) (100).
MiR-200b has been shown to have pro-apoptotic effects among all the studied types of cancer. In breast cancer, miR-200b induces apoptosis and inhibits cell proliferation by directly targeting Sp1 (239). Moreover, high expression of miR-200b appeared to be an independent prognostic factor for patients with breast cancer (239). Moreover, miR-200b-3p shows pro-apoptotic effects in colorectal cancer by inactivating the Wnt/β-catenin signaling pathway (246). Likewise, the same occurs in esophageal squamous cell carcinoma where miR-200b also modulates the Wnt/β-catenin signaling pathway by targeting CDK2 and PAF. Therefore, by inducing cell cycle arrest and apoptosis, miR-200b alleviates cancer cell growth (246). In ovarian cancer, miR-200b significantly increases the apoptosis rate of the cancer cells by targeting ATAD2 and regulating PI3K/AKT signaling pathway (259). miR-200b inhibited cancer growth and induced cell apoptosis in the oxaliplatin-resistant colorectal cancer cells through suppression of βIII-tubulin protein expression (247).
MiR-200c also has a pro-apoptotic effect in all studied types of cancer. In triple-negative breast cancer cells, microRNA-200c was found to downregulate XIAP expression and thus suppress proliferation and promote apoptosis of cancer (264). In gastric cancer, miR-200c promotes apoptosis of the tumor cells by downregulation of endothelin receptor A (EDNRA) expression (275). Similarly, in human hepatocellular carcinoma miR-200c induces apoptosis via suppressing MAD2L1 (88). Overexpression of miR-200c has been proven to promote apoptosis in Wilms tumors via downregulation of the Akt/GLUT1 signaling pathway (278). Furthermore, the high expression of miR-200c was also reported to enhance apoptosis in lung cancer tissues, where it caused activation of the JNK signaling pathway and upregulation of an ER stress-related protein, RECK.
MiR-141 demonstrates apoptosis-promoting activity in the majority of cancer types. However, miR-141 appears to modulate apoptosis in a bidirectional manner in prostate cancer. miR-141 induces prostate cancer cell apoptosis via targeting Runt-related transcription factor 1 (RUNX1) (300). However, another study reported that miR141 significantly reduced cell apoptosis, thus appearing to be a novel oncogene miRNA, which promotes prostate tumorigenesis via suppressing a key transcription factor kruppel-like factor-9 (KLF9) (299). Therefore, it remains to define the role that miR-141 plays in the apoptosis of prostate cancer cells. In T-cell acute lymphoblastic leukemia cells the upregulation of miR-141-3p significantly decreased cancer cell proliferation and promoted its apoptosis by direct targeting tumor necrosis factor receptor-associated factor 5 (TRAF5) (32). Furthermore, among colorectal cancer cells, miR-141- has been proven to reduce cell growth and induce apoptosis and differentiation of colorectal cancer cells by targeting EGFR (332). Similarly, in head and neck squamous cell carcinoma, where miR-141, also by suppressing EGFR signaling, inhibits tumor growth, and promotes apoptosis in cancer (85). Moreover, in pancreatic cancer cells, miR-141 acts as a tumor suppressor by targeting MAP4K4, which knockdown inhibits cell proliferation and induces G1 arrest and apoptosis (298). Moreover, since insulin receptor substrate 2 (IRS2), a known oncogene, was confirmed to be a direct target of miR-141, its overexpression blocks cell proliferation and induces cell apoptosis of thyroid cancer (129).
MiR-429 acts in a pro-apoptotic manner in the majority of cancer types. In colorectal cancer, miR-429 has been demonstrated to exert a two-fold effect. By mediating high mobility group box 3 (HMGB3) miR-429 promotes apoptosis in colorectal cancer cells (59). However, directly targeting SOX2 may prevent cell death (53). Therefore, it is unclear whether miR-429 plays an oncogenic or tumor-suppressive role in colorectal cancer. Remarkably, SOX2 is also directly targeted by miR-429 in glioblastoma multiforme, but in this cancer downregulation of SOX2 inhibits cell proliferation and induces apoptosis (82). Moreover, in glioblastoma miR-429 induces apoptosis also by targeting Bcl-2. Furthermore, downregulation of Bcl-2 by miR-429 induces apoptosis in gastric carcinoma (303, 353). In both types of cancers, overexpression of miR-429 inhibits Bcl-2-mediated cell survival (303, 353). In esophageal carcinoma upregulation of miR-429, by targeting both Bcl-2 and SP1, promotes apoptosis in cancer cells (65). Another direct target for miR-429 is ZEB1 which inhibition results in the induction of apoptosis in osteosarcoma (103) and thyroid cancer cells (131). Moreover, miR-429 enhances apoptosis in nephroblastoma, by targeting c-myc (96) and also in cervical carcinoma, through the NF-κB pathway by targeting IKKβ (46).
Regulation of angiogenesis by miR-200 family
Angiogenesis is the process of formation of new capillaries from a pre-existing vasculature, which is a crucial factor affecting tumor formation, progression, and metastasis (217). Malignant tumors create an environment that favors the predominance of proangiogenic factors over antiangiogenic factors, resulting in inappropriate vessel growth towards the neoplastic lesion. Many proangiogenic and antiangiogenic agents have been identified. They may be secreted by endothelial cells, tumor cells, or by the surrounding stroma. MiRNAs may target genes involved in angiogenesis, but on the other hand, their expression can be modulated via pro-angiogenic or anti-angiogenic factors (354).
miR-200 family members act both as angiogenesis inhibitors and promoters ( Table 6 ). miR-200a, miR-200b and miR-429 inhibit angiogenesis. miR-200c is described as an angiogenesis inhibitor, but it promotes angiogenesis in pancreatic cancer. Similarly, miR-141 is described to inhibit angiogenesis, but it also promotes angiogenesis in small-cell lung cancer and ovarian cancer.
Table 6.
Regulation of angiogenesis by miR-200 family members.
| miRNAs | Cancer | Target | Angiogenesis | Ref. |
|---|---|---|---|---|
| miR-200a | n/d | n/d | ↓ | (355) |
| miR-200b | n/d | n/d | ↓ | (355) |
| Prostate cancer | n/d | ↓ | (261) | |
| miR-200c | n/d | n/d | ↓ | (355) |
| Pancreatic cancer | n/d | ↑ | (356) | |
| miR-141 | Small cell lung cancer | KLF12 | ↑ | (193) |
| Ovarian cancer | n/d | ↑ | (357) | |
| n/d | CXCL12β, GAB1, GATA6, NRP1, TGFβ2 |
↓ | (355) | |
| miR-429 | n/d | n/d | ↓ | (355) |
↓ - suppression by miRNA; ↑ - promotion by miRNA; n/d – no data.
In 2011, Chan et al. first described the effect of miR-200b on the suppression of angiogenesis, thus identifying the first member of the miR-200 family to have an inhibitory effect on this process (358). Several years after this finding, it was confirmed that all members of the miR-200 family suppress angiogenesis (193). In prostate cancer cells, miR-200b reverses the angiogenic switch (261). Nevertheless, some members of the miR-200 family, more specifically miR-200c and miR-141, may demonstrate pro-angiogenic effects as well. For instance, increased expression of miR-200c in pancreatic cancer endothelial cells is observed and its inhibition significantly reduces cancer cell migration and angiogenesis, confirming the pro-angiogenic effects of miR-200c in pancreatic cancer (356). MiR-141 is another member of the miR-200 family that, in addition to its anti-angiogenic effects (355), also demonstrates angiogenesis-promoting activity in ovarian cancer. The ovarian cancer-secreted miR141-3p promotes endothelial cell angiogenesis by activating the JAK/STAT3 and NF-κB signaling pathways (357). Likewise, in small cell lung cancer (SCLC), miR-141 promotes angiogenesis via the KLF12 pathway (193).
Regulation of drug resistance by miR-200 family
Drug resistance in cancer cells, resulting in reduced or no response to the administered therapy and poorer overall survival of cancer patients is a limiting factor for this treatment approach. Furthermore, residual cancer cells surviving therapy gradually divide, thereby initiating recurrence of the disease, often having worse responses to treatment and a poorer prognosis. Chemoresistance can develop through various mechanisms, such as gene mutation, DNA methylation, and histone modification (359). Numerous studies focus on identifying and understanding the role that miRNAs play in the development of chemotherapy resistance. Members of the miR-200 family appear to be critical of this phenomenon. Depending on cancer and the miR-200 family member, the underlying mechanism and the effect they have on modulating the resistance are different.
miR-200 family members reduce drug resistance in most cases ( Table 7 ). miR-200a enhances drug resistance in breast and liver cancer, and it reduces drug resistance in glioma. miR-200b reduces drug resistance in 7 cancer types. miR-200c reduces drug resistance in 7 cancer types but it enhances chemoresistance in esophageal carcinoma. miR-141 increases drug resistance in breast cancer and glioma and it decreases drug resistance in neuroblastoma, colorectal and pancreatic cancer. miR-141 increases the resistance to cisplatin in non-small cell lung cancer and ovarian cancer. However, it reduces the resistance to other drugs in those cancer types. miR-429 reduces drug resistance in colorectal and ovarian cancers and increases drug resistance in endometrial cancer.
Table 7.
Regulation of drug resistance by miR-200 family members.
| miRNAs | Cancer | Target | Drug resistance | Ref. |
|---|---|---|---|---|
| miR-200a | Breast cancer | TP53INP1 | ↑ | (360) |
| Glioma | n/d | ↓ | (81) | |
| Liver cancer | DUSP6 | ↑ | (361) | |
| miR-200b | Bladder cancer | n/d | ↓ | (134) |
| Cholangiocarcinoma | SUZ12 and ROCK2 | ↓ | (51) | |
| Colorectal cancer | TUBB3 | ↓ | (247) | |
| Non-small cell lung cancer | ATG12, E2F3, p70S6K1, SUZ12 | ↓ | (254, 256, 362–365) | |
| Prostate cancer | n/d | ↓ | (262) | |
| Small cell lung cancer | ZEB2 | ↓ | (366) | |
| Ovarian cancer | DNMT3A/DNMT3B | ↓ | (367) | |
| miR-200c | Breast cancer | ↓ | (368) | |
| Gastric carcinoma | ZEB2 | ↓ | (369) | |
| Melanoma | n/d | ↓ | (277) | |
| Osteosarcoma | AKT2 | ↓ | (105) | |
| Ovarian cancer | DNMT3A/DNMT3B | ↓ | (367) | |
| Pancreatic cancer | n/d | ↓ | (330) | |
| Cholangiocarcinoma | SUZ12 and ROCK2 | ↓ | (51) | |
| Esophageal carcinoma | n/d | ↑ | (370) | |
| miR-141 | Colorectal cancer | EGFR | ↓ | (291) |
| Breast cancer | EIF4E | ↑ | (371) | |
| Glioma | TP53 | ↑ | (372) | |
| Neuroblastoma | FUS | ↓ | (293) | |
| Non-small cell lung cancer | PDCD4 | ↑/↓ | (365, 373) | |
| Ovarian cancer | KEAP1 | ↑/↓ | (374, 375) | |
| Pancreatic cancer | MAP4K4 | ↓ | (298) | |
| miR-429 | Colorectal cancer | DUSP4 | ↓ | (376) |
| Endometrial cancer | n/d | ↑ | (60) | |
| Ovarian cancer | ZEB1 | ↓ | (377) |
↓ - suppression by miRNA; ↑ - promotion by miRNA; n/d – no data.
MiR-200a demonstrates various effects on the modulation of drug resistance among different types of cancer. In breast cancer cells, Mir-200a, via antagonizing TP53INP1 and YAP1, contributes to increased resistance to chemotherapeutics (360). Moreover, inhibition of miR-200a enhances gemcitabine chemosensitivity in cancer cells (360). In human hepatocellular carcinoma, miR-200a-3p targets dual-specificity phosphatase 6 (DUSP6) to augment cancer cell resistance to 5-fluorouracil, doxorubicin, and cisplatin (361). In contrast, miR-200a has opposite effects on drug resistance in glioma. Moreover, downregulation of miR-200a is associated with decreased chemotherapeutic treatment efficacy (81).
MiR-200b appears to reduce the chemoresistance of all cancers. In cholangiocarcinoma, it was demonstrated that miR-200b, as well as miR-200c, reduces resistance to chemotherapeutics by directly targeting SUZ12 and ROCK2 (51). The same effect, but in a different mechanism, was reported with miR-200b-3p in colorectal cancer, where, via targeting TUBB3, it reduced resistance to oxaliplatin and promoted apoptosis and growth inhibition in resistant cancer cells (247). In bladder cancer cells, methylation of miR-200b was associated with resistance of this cancer to cisplatin (134). Moreover, it was suggested that epigenetic silencing of miR-200b might be a marker of cisplatin resistance in this tumor. In addition, miR-200b seems to play an essential role in the response of non-small lung cancer to treatment. It has been evidenced that induction of miR-200b, but also miR-141, increased sensitivity to nintedanib in nintedanib-resistant cells (365). Moreover, miR-200b increases the chemosensitivity of the docetaxel-resistant lung cancer cells by directly targeting autophagy-associated gene 12 (ATG12) (364). A further study revealed that expression of miR-200b, by direct targeting SUZ12 and through histone deacetylase 1/Sp1/miR-200b signaling pathway might lead to the formation of chemoresistant phenotype in docetaxel-resistant cancer cells. Moreover, histone deacetylase-mediated silencing of miR-200b increased chemoresistance in lung adenocarcinoma cells (254, 363). Furthermore, miR-200b in lung cancer cells inhibits chemoresistance and increased sensitivity to cisplatin via targeting p70S6K1 (256). This miRNA was also found to reverse the chemoresistance of docetaxel-resistant lung adenocarcinoma cells via targeting E2F3 (362). Moreover, also in small cell lung cancer, miR-200b reduces drug resistance namely by modulating ZEB2, which in small cell lung cancer leads to multidrug resistance of the tumor (366). By regulating B-cell-specific Moloney murine leukemia virus insertion site 1 (Bmi-1), miR-200b has been shown to enhance the chemosensitivity of prostate cancer cells to docetaxel (262). Besides, in ovarian cancer, miR-200b and miR-200c have been reported to be able to reverse cisplatin resistance by directly targeting DNMT3A/DNMT3B (367).
In most cases, miR-200c decreases drug resistance. In breast cancer, miR-200c increases sensitivity to chemotherapy as well as sensitizes HER2+ cancer cells to trastuzumab (368). Moreover, miR-200c, by directly targeting and thus downregulating ZEB2, increases the sensitivity of gastric cancer tissues to cisplatin (369). In melanoma cells, overexpression of miR-200c significantly reduces resistance to chemotherapy via downregulation of Bmi-1 (277). This miRNA was also revealed to enhance osteosarcoma chemosensitivity to cisplatin by targeting AKT2 (105). In pancreatic cells, miR-200c sensitizes cancer cells to chemotherapy (330). In contrast, overexpression of miR-200c in esophageal cancer induces chemoresistance to cisplatin by activation of the Akt signaling pathway (370).
Similarly, miR-141 has been reported to have significant effects on the modulation of drug resistance. In neuroblastoma cells, miR-141 increases cancer cell sensitivity to cisplatin (293). Also in pancreatic cancer, miR-141, by directly targeting MAP4K4, increases the chemosensitivity of the cancer cells (298). However, in breast cancer upregulation of miR-141 has been found to exacerbate docetaxel resistance of cancer cells (371). In non-small cell lung carcinoma, the function of miR-141 is more complex as, on the one hand, it increases sensitivity to nintedanib in nintedanib-resistant cancer cells (365), but on the other hand, via upregulation of PDCD4, it reverses cisplatin resistance (373). Moreover, in ovarian cancer, the action of miR-141 is also bidirectional. Transfection with inhibitors of miR141, as well as of inhibitors of miR-200c, in ovarian cancer cell lines induced cell resistance to paclitaxel and carboplatin (375); however, it has been described that expression of miR-141, via regulating KEAP1, can increase resistance to cisplatin chemotherapy in ovarian cancer cells (374). In glioma cells, miR-141 renders resistance to temozolomide therapy by targeting TP53 (372).
The last member of the miR-200 family is miR-429, whose effect on the modulation of drug resistance has been observed in colorectal, endometrial, and ovarian cancers. In colorectal cancer, overexpression of miR-429 has been found to target DUSP4, block the JNK pathway, and thereby increase cancer cell sensitivity to nintedanib (376). Also in ovarian cancer, overexpression of miR-429 appears to sensitize cancer cells to cisplatin by targeting ZEB1 (377). However, in endometrial cancer, the effect of miR-429 on drug resistance of this tumor is the opposite, as transfection with anti-miR-429 increased the cytotoxic effect of cisplatin in cancer cells, thus improving treatment efficacy (60).
Regulation of immune response by miR-200 family
Tumor cells evade immune response via multiple mechanisms. Programmed Cell Death Protein 1 (PD-1) and its ligand, PD-L1, are key immune checkpoint molecules suppressing anti-tumor immune response (378). Several miRNAs were identified to regulate the PD-1/PD-L1 axis, including miR-200 (379). By targeting PD-L1, miR-200 enhances CD8+ cytotoxic T-cells activity in the tumor microenvironment and regulates the metastatic potential of tumor cells (379). Moreover, PD-L1 is targeted by miR-429 in gastric cancer (380).
MiR-200 family regulates also myeloid cells in the tumor microenvironment. MiR-200c by targeting PTEN and FOG2 induces the expansion and enhances the immunoregulatory properties of myeloid-derived suppressor cells (MDSCs) (381). It induces, among others, the expression of arginase 1, a key immunoregulatory enzyme of MDSCs (382), potentiating suppressive effects on T-cells (381). Moreover, miR-200c suppresses the expression of multiple cytokines by tumor-associated macrophages (383). Restoration of miR-200c upregulates cytokines and promotes M1 polarization of macrophages (383). Notably, cytokines suppressed by miR-200c predict favorable survival of TNBC patients (383). Similarly, miR-200a promotes phagocytosis of tumor cells by macrophages by targeting CD47, a “do not eat me” signal protein overexpressed in tumor cells (86). Nonetheless, more research is required to dissect the role of the miR-200 family in the antitumor immune response as well as patients’ response to immunotherapy.
Genome instability and mutations
Various miRNAs regulate the expression of DNA damage proteins leading to genomic instability (384). However, little is known regarding the role of the miR-200 family in the regulation of genomic instability and mutations. It was demonstrated that miR-200a regulates DNA repair in aging keratinocytes (385), nonetheless, its role in genome instability in cancer requires further studies.
Novel hallmarks of cancer
Recently, five emerging hallmarks of cancer have been suggested, including dysregulation of cellular energetic, unlocking phenotypic plasticity, dysregulation of the microbiome, nonmutational epigenetic reprogramming, and senescence (214). Despite little being known about the role of the miR-200 family in novel hallmarks of cancer, some studies suggested that they may be important regulators of these features.
MiRNAs, including the miR-200 family, are involved in the reprogramming of cancer cell metabolism. For instance, miR-200b suppresses lactate dehydrogenase A which suppresses glycolysis leading to the inhibited proliferation and invasion of glioma cells (386). Moreover, miR-200b/miR-200c regulates EMT differentiation and proliferation by modulation of metabolic properties of colorectal cancer cells (387). Notably, the miR-200 family also regulates the cellular senescence of cancer cells. It was demonstrated that suppression or loss of the miR-200 family in cancer cells induces morphological changes, cell cycle arrest, and induces cell senescence (388).
Recently, more and more studies have stated that the multifactorial impact of polymorphic microbiomes on cancer regulation is linked to their bidirectional interference with miRNAs in which those two factors interact resulting in apoptosis or proliferation of tumor cells (389, 390). A prominent example is an ovarian cancer where tissue-specific bacteria, L. lactis, seem to be in control of miR-200b and TLR-4 downregulation, which is connected to the progression of ovarian cancer (389). Another example of miRNA-microbiome interaction is exosomal miR-200c which was identified as a tumor suppressor of colorectal cancer cells, but only after it came in contact with lipopolysaccharide (LPS) that is a component of the bacterial outer membrane (391). Nonetheless, more research is required to determine the role of the miR-200 family in the regulation of these hallmarks of cancer.
Conclusions
Members of the miR-200 family are crucial regulators of hallmarks of cancer. Several studies described alterations in their expression in human tumors and determined their utility as biomarkers in cancer. Our literature review summarized known functions and biological targets of the miR-200 family in different types of cancer. Furthermore, it identified gaps and limitations in current knowledge indicating the directions of further research. More studies are necessary to determine the role of members of the miR-200 family in the regulation of recently added hallmarks of cancer. Additionally, preclinical studies are required to determine the therapeutic potential of the miR-200 family.
Author contributions
KK and TG are co-first authors. All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This research was funded by the Medical University of Warsaw, grant number 1MN/1/M/MBM/N/21. The APC was funded by the Medical University of Warsaw. TG is supported by the PRELUDIUM grant funded by the National Center of Science (UMO-2021/41/N/NZ6/02774) and by the Foundation for Polish Science (FNP).
Acknowledgments
Figures were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Lee RC, Feinbaum RL, Ambros V. The c. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell (1993) 75:843–54. doi: 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 2. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer (2015) 15:321–33. doi: 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shi Y, Liu Z, Lin Q, Luo Q, Cen Y, Li J, et al. MiRNAs and cancer: Key link in diagnosis and therapy. Genes (Basel) (2021) 12. doi: 10.3390/genes12081289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev (2015) 87:3–14. doi: 10.1016/j.addr.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alles J, Fehlmann T, Fischer U, Backes C, Galata V, Minet M, et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res (2019) 47:3353–64. doi: 10.1093/nar/gkz097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov (2017) 16:203–22. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 7. Grzywa TM, Klicka K, Włodarski PK. Regulators at every step-how microRNAs drive tumor cell invasiveness and metastasis. Cancers (Basel) (2020) 12. doi: 10.3390/cancers12123709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol (2005) 6:376–85. doi: 10.1038/nrm1644 [DOI] [PubMed] [Google Scholar]
- 9. Michlewski G, Cáceres JF. Post-transcriptional control of miRNA biogenesis. Rna (2019) 25:1–16. doi: 10.1261/rna.068692.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen PS, Lin SC, Tsai SJ. Complexity in regulating microRNA biogenesis in cancer. Exp Biol Med (Maywood) (2020) 245:395–401. doi: 10.1177/1535370220907314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matsuyama H, Suzuki HI. Systems and synthetic microRNA biology: From biogenesis to disease pathogenesis. Int J Mol Sci (2020) 21:132. doi: 10.3390/ijms21010132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis (2015) 35:3–11. doi: 10.1055/s-0034-1397344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol (2008) 5:115–9. doi: 10.4161/rna.5.3.6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pager CT, Wehner KA, Fuchs G, Sarnow P. MicroRNA-mediated gene silencing. Prog Mol Biol Transl Sci (2009) 90:187–210. doi: 10.1016/S1877-1173(09)90005-9 [DOI] [PubMed] [Google Scholar]
- 15. Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. a comprehensive review. EMBO Mol Med (2012) 4:143–59. doi: 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Senfter D, Madlener S, Krupitza G, Mader RM. The microRNA-200 family: still much to discover. Biomol Concepts (2016) 7:311–9. doi: 10.1515/bmc-2016-0020 [DOI] [PubMed] [Google Scholar]
- 17. Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev (2013) 23:3–11. doi: 10.1016/j.gde.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature (2005) 435:834–8. doi: 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 19. Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene (2008) 27:1788–93. doi: 10.1038/sj.onc.1210809 [DOI] [PubMed] [Google Scholar]
- 20. Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell (2010) 141:1195–207. doi: 10.1016/j.cell.2010.05.017 [DOI] [PubMed] [Google Scholar]
- 21. Toda H, Seki N, Kurozumi S, Shinden Y, Yamada Y, Nohata N, et al. RNA-Sequence-based microRNA expression signature in breast cancer: tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol Oncol (2020) 14:426–46. doi: 10.1002/1878-0261.12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pfeffer SR, Yang CH, Pfeffer LM. The role of miR-21 in cancer. Drug Dev Res (2015) 76:270–7. doi: 10.1002/ddr.21257 [DOI] [PubMed] [Google Scholar]
- 23. Rosignolo F, Memeo L, Monzani F, Colarossi C, Pecce V, Verrienti A, et al. MicroRNA-based molecular classification of papillary thyroid carcinoma. Int J Oncol (2017) 50:1767–77. doi: 10.3892/ijo.2017.3960 [DOI] [PubMed] [Google Scholar]
- 24. Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Trans Med (2016) 14:143. doi: 10.1186/s12967-016-0893-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer (2009) 125:2737–43. doi: 10.1002/ijc.24638 [DOI] [PubMed] [Google Scholar]
- 26. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci (2020) 21 1723. doi: 10.3390/ijms21051723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He XX, Kuang SZ, Liao JZ, Xu CR, Chang Y, Wu YL, et al. The regulation of microRNA expression by DNA methylation in hepatocellular carcinoma. Mol Biosyst (2015) 11:532–9. doi: 10.1039/C4MB00563E [DOI] [PubMed] [Google Scholar]
- 28. Lai HH, Chen PS. Dual mechanism of DICER downregulation facilitates cancer metastasis. Mol Cell Oncol (2018) 5:e1472056. doi: 10.1080/23723556.2018.1472056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramírez-Moya J, Wert-Lamas L, Riesco-Eizaguirre G, Santisteban P. Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene (2019) 38:5486–99. doi: 10.1038/s41388-019-0804-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shan W, Sun C, Zhou B, Guo E, Lu H, Xia M, et al. Role of dicer as a prognostic predictor for survival in cancer patients: a systematic review with a meta-analysis. Oncotarget (2016) 7:72672–84. doi: 10.18632/oncotarget.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou JD, Zhang LC, Zhang TJ, Gu Y, Wu DH, Zhang W, et al. Dysregulation of miR-200s clusters as potential prognostic biomarkers in acute myeloid leukemia. J Transl Med (2018) 16:135. doi: 10.1186/s12967-018-1494-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou R, Mo W, Wang S, Zhou W, Chen X, Pan S. miR-141-3p and TRAF5 network contributes to the progression of T-cell acute lymphoblastic leukemia. Cell Transplant (2019) 28:59s–65s. doi: 10.1177/0963689719887370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang L-C, Zhong C, Ma Y-J, Liu M, Nong W-X. [Expression of MiR-429 in patients with acute lymphoblastic leukemia and its prognostic value]. Zhongguo shi yan xue ye xue za zhi (2020) 28:119–24. doi: 10.19746/j.cnki.issn.1009-2137.2020.01.020 [DOI] [PubMed] [Google Scholar]
- 34. Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer (2013) 132:2479–91. doi: 10.1002/ijc.27949 [DOI] [PubMed] [Google Scholar]
- 35. Ratert N, Meyer HA, Jung M, Lioudmer P, Mollenkopf HJ, Wagner I, et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J Mol Diagn (2013) 15:695–705. doi: 10.1016/j.jmoldx.2013.05.008 [DOI] [PubMed] [Google Scholar]
- 36. Liu L, Qiu M, Tan G, Liang Z, Qin Y, Chen L, et al. miR-200c inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3. J Transl Med (2014) 12:305. doi: 10.1186/s12967-014-0305-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou H, Tang K, Xiao H, Zeng J, Guan W, Guo X, et al. A panel of eight-miRNA signature as a potential biomarker for predicting survival in bladder cancer. J Exp Clin Cancer Res (2015) 34:53. doi: 10.1186/s13046-015-0167-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheng Y, Zhang X, Li P, Yang C, Tang J, Deng X, et al. MiR-200c promotes bladder cancer cell migration and invasion by directly targeting RECK. Onco Targets Ther (2016) 9:5091–9. doi: 10.2147/OTT.S101067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang J, Liu Y, He A, Liu Y, Wu J, Liao X, et al. Hsa-miR-429 promotes bladder cancer cell proliferation via inhibiting CDKN2B. Oncotarget (2017) 8:68721–9. doi: 10.18632/oncotarget.19878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen W, Jiang T, Mao H, Gao R, Zhang H, He Y, et al. SNHG16 regulates invasion and migration of bladder cancer through induction of epithelial-to-mesenchymal transition. Hum Cell (2020) 33:737–49. doi: 10.1007/s13577-020-00343-9 [DOI] [PubMed] [Google Scholar]
- 41. Yang R, Xu J, Hua X, Tian Z, Xie Q, Li J, et al. Overexpressed miR-200a promotes bladder cancer invasion through direct regulating Dicer/miR-16/JNK2/MMP-2 axis. Oncogene (2020) 39:1983–96. doi: 10.1038/s41388-019-1120-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen L, Li Y, Fu Y, Peng J, Mo MH, Stamatakos M, et al. Role of deregulated microRNAs in breast cancer progression using FFPE tissue. PloS One (2013) 8:e54213. doi: 10.1371/journal.pone.0054213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu F, He H, Huang W, Lin Y, Luo S, Du Q, et al. Decreased expression of MicroRNA-200 family in human breast cancer is associated with lymph node metastasis. Clin Transl Oncol (2016) 18:283–8. doi: 10.1007/s12094-015-1364-1 [DOI] [PubMed] [Google Scholar]
- 44. Li MY, Hu XX. Meta-analysis of microRNA expression profiling studies in human cervical cancer. Med Oncol (2015) 32:510. doi: 10.1007/s12032-015-0510-5 [DOI] [PubMed] [Google Scholar]
- 45. Cheng YX, Zhang QF, Hong L, Pan F, Huang JL, Li BS, et al. MicroRNA-200b suppresses cell invasion and metastasis by inhibiting the epithelial-mesenchymal transition in cervical carcinoma. Mol Med Rep (2016) 13:3155–60. doi: 10.3892/mmr.2016.4911 [DOI] [PubMed] [Google Scholar]
- 46. Fan JY, Fan YJ, Wang XL, Xie H, Gao HJ, Zhang Y, et al. miR-429 is involved in regulation of NF-κBactivity by targeting IKKβ and suppresses oncogenic activity in cervical cancer cells. FEBS Lett (2017) 591:118–28. doi: 10.1002/1873-3468.12502 [DOI] [PubMed] [Google Scholar]
- 47. Liu SS, Chan KKL, Chu DKH, Wei TN, Lau LSK, Ngu SF, et al. Oncogenic microRNA signature for early diagnosis of cervical intraepithelial neoplasia and cancer. Mol Oncol (2018) 12:2009–22. doi: 10.1002/1878-0261.12383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Okoye JO, Ngokere AA, Onyenekwe CC, Erinle CA. Comparable expression of miR-let-7b, miR-21, miR-182, miR-145, and p53 in serum and cervical cells: Diagnostic implications for early detection of cervical lesions. Int J Health Sci (Qassim) (2019) 13:29–38. [PMC free article] [PubMed] [Google Scholar]
- 49. Sommerova L, Anton M, Bouchalova P, Jasickova H, Rak V, Jandakova E, et al. The role of miR-409-3p in regulation of HPV16/18-E6 mRNA in human cervical high-grade squamous intraepithelial lesions. Antiviral Res (2019) 163:185–92. doi: 10.1016/j.antiviral.2019.01.019 [DOI] [PubMed] [Google Scholar]
- 50. Bozgeyik E, Tepe NB, Bozdag Z. Identification of microRNA expression signature for the diagnosis and prognosis of cervical squamous cell carcinoma. Pathol Res Pract (2020) 216:153159. doi: 10.1016/j.prp.2020.153159 [DOI] [PubMed] [Google Scholar]
- 51. Peng F, Jiang J, Yu Y, Tian R, Guo X, Li X, et al. Direct targeting of SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma tumourigenesis and metastasis. Br J Cancer (2013) 109:3092–104. doi: 10.1038/bjc.2013.655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen C, Yang D, Wang Q, Wang X. Expression and clinical pathological significance of miR-200a in concurrent cholangiocarcinoma associated with hepatolithiasis. Med Sci Monit (2015) 21:3585–90. doi: 10.12659/MSM.895013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li J, Du L, Yang Y, Wang C, Liu H, Wang L, et al. MiR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2. Cancer Lett (2013) 329:84–90. doi: 10.1016/j.canlet.2012.10.019 [DOI] [PubMed] [Google Scholar]
- 54. Diaz T, Tejero R, Moreno I, Ferrer G, Cordeiro A, Artells R, et al. Role of miR-200 family members in survival of colorectal cancer patients treated with fluoropyrimidines. J Surg Oncol (2014) 109:676–83. doi: 10.1002/jso.23572 [DOI] [PubMed] [Google Scholar]
- 55. Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu Z, et al. MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol Cell Biochem (2014) 390:19–30. doi: 10.1007/s11010-013-1950-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Carter JV, O'brien SJ, Burton JF, Oxford BG, Stephen V, Hallion J, et al. The microRNA-200 family acts as an oncogene in colorectal cancer by inhibiting the tumor suppressor RASSF2. Oncol Lett (2019) 18:3994–4007. doi: 10.3892/ol.2019.10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ranković B, Zidar N, Žlajpah M, Boštjančič E. Epithelial-mesenchymal transition-related MicroRNAs and their target genes in colorectal cancerogenesis. J Clin Med (2019) 8. doi: 10.3390/jcm8101603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fang F, Cheng L, Wu X, Ye M, Zhang H. miR-141 promotes colon cancer cell proliferation by targeted PHLPP2 expression inhibitionn. Cancer Manag Res (2020) 12:11341–50. doi: 10.2147/CMAR.S256670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tian X, Chang J, Zhang N, Wu S, Liu H, Yu J. MicroRNA-429 acts as a tumor suppressor in colorectal cancer by targeting high mobility group box 3. Oncol Lett (2021) 21:250. doi: 10.3892/ol.2021.12511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee JW, Park YA, Choi JJ, Lee YY, Kim CJ, Choi C, et al. The expression of the miRNA-200 family in endometrial endometrioid carcinoma. Gynecol Oncol (2011) 120:56–62. doi: 10.1016/j.ygyno.2010.09.022 [DOI] [PubMed] [Google Scholar]
- 61. Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, Cassandrini P, et al. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int J Cancer (2013) 132:1633–45. doi: 10.1002/ijc.27840 [DOI] [PubMed] [Google Scholar]
- 62. Smith CM, Watson DI, Leong MP, Mayne GC, Michael MZ, Wijnhoven BP, et al. miR-200 family expression is downregulated upon neoplastic progression of barrett's esophagus. World J Gastroenterol (2011) 17:1036–44. doi: 10.3748/wjg.v17.i8.1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ, Wang TY, et al. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett (2013) 5:1639–42. doi: 10.3892/ol.2013.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saad R, Chen Z, Zhu S, Jia P, Zhao Z, Washington MK, et al. Deciphering the unique microRNA signature in human esophageal adenocarcinoma. PloS One (2013) 8:e64463. doi: 10.1371/journal.pone.0064463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang Y, Li M, Zang W, Ma Y, Wang N, Li P, et al. MiR-429 up-regulation induces apoptosis and suppresses invasion by targeting bcl-2 and SP-1 in esophageal carcinoma. Cell Oncol (Dordr) (2013) 36:385–94. doi: 10.1007/s13402-013-0144-6 [DOI] [PubMed] [Google Scholar]
- 66. Zhang HF, Zhang K, Liao LD, Li LY, Du ZP, Wu BL, et al. miR-200b suppresses invasiveness and modulates the cytoskeletal and adhesive machinery in esophageal squamous cell carcinoma cells via targeting kindlin-2. Carcinogenesis (2014) 35:292–301. doi: 10.1093/carcin/bgt320 [DOI] [PubMed] [Google Scholar]
- 67. Tan H, Zhu Y, Zhang J, Peng L, Ji T. miR141 expression is downregulated and negatively correlated with STAT5 expression in esophageal squamous cell carcinoma. Exp Ther Med (2016) 11:1803–8. doi: 10.3892/etm.2016.3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zong M, Liu Y, Zhang K, J Y, Chen L. The effects of miR-429 on cell migration and invasion by targeting slug in esophageal squamous cell carcinoma. Pathol Res Pract (2019) 215:152526. doi: 10.1016/j.prp.2019.152526 [DOI] [PubMed] [Google Scholar]
- 69. Wang Y, Yu XJ, Zhou W, Chu YX. MicroRNA-429 inhibits the proliferation and migration of esophageal squamous cell carcinoma cells by targeting RAB23 through the NF-κB pathway. Eur Rev Med Pharmacol Sci (2020) 24:1202–10. doi: 10.26355/eurrev_202002_20172 [DOI] [PubMed] [Google Scholar]
- 70. Yang B, Liu Y, Li L, Deng H, Xian L. MicroRNA−200a promotes esophageal squamous cell carcinoma cell proliferation, migration and invasion through extensive target genes. Mol Med Rep (2020) 21:2073–84. doi: 10.3892/mmr.2020.11002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sun T, Wang C, Xing J, Wu D. miR-429 modulates the expression of c-myc in human gastric carcinoma cells. Eur J Cancer (2011) 47:2552–9. doi: 10.1016/j.ejca.2011.05.021 [DOI] [PubMed] [Google Scholar]
- 72. Chang L, Guo F, Huo B, Lv Y, Wang Y, Liu W. Expression and clinical significance of the microRNA-200 family in gastric cancer. Oncol Lett (2015) 9:2317–24. doi: 10.3892/ol.2015.3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen Z, Liu X, Hu Z, Wang Y, Liu M, Liu X, et al. Identification and characterization of tumor suppressor and oncogenic miRNAs in gastric cancer. Oncol Lett (2015) 10:329–36. doi: 10.3892/ol.2015.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang M, Dong BB, Lu M, Zheng MJ, Chen H, Ding JZ, et al. miR-429 functions as a tumor suppressor by targeting FSCN1 in gastric cancer cells. Onco Targets Ther (2016) 9:1123–33. doi: 10.2147/OTT.S91879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Du F, Yu C, Li R, Ding D, He L, Wen G. Expression of miR-141 and YAP1 in gastric carcinoma and modulation of cancer cell proliferation and apoptosis. Int J Clin Exp Pathol (2019) 12:559–67. [PMC free article] [PubMed] [Google Scholar]
- 76. Li D, Wang J, Zhang M, Hu X, She J, Qiu X, et al. LncRNA MAGI2-AS3 is regulated by BRD4 and promotes gastric cancer progression via maintaining ZEB1 overexpression by sponging miR-141/200a. Mol Ther Nucleic Acids (2020) 19:109–23. doi: 10.1016/j.omtn.2019.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bian EB, Ma CC, He XJ, Wang C, Zong G, Wang HL, et al. Epigenetic modification of miR-141 regulates SKA2 by an endogenous 'sponge' HOTAIR in glioma. Oncotarget (2016) 7:30610–25. doi: 10.18632/oncotarget.8895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sun X, Li Z, Chen Y. The potential prognostic value of MicroRNA-429 for human gliomas. Ann Clin Lab Sci (2016) 46:44–8. [PubMed] [Google Scholar]
- 79. Qin Y, Chen W, Liu B, Zhou L, Deng L, Niu W, et al. MiR-200c inhibits the tumor progression of glioma via targeting moesin. Theranostics (2017) 7:1663–73. doi: 10.7150/thno.17886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kong X, Gong S, Yan T, Yang Y. MicroRNA-200b expression level is negatively associated with pathological grading in human gliomas. Cancer Manag Res (2018) 10:2825–34. doi: 10.2147/CMAR.S171137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang C, Kang L, Wang X, Liu Y, Zhao X. Expression of miR-200a and chemotherapeutic treatment efficacy of glioma. Oncol Lett (2018) 15:5767–71. doi: 10.3892/ol.2018.8063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dong H, Hao X, Cui B, Guo M. MiR-429 suppresses glioblastoma multiforme by targeting SOX2. Cell Biochem Funct (2017) 35:260–8. doi: 10.1002/cbf.3271 [DOI] [PubMed] [Google Scholar]
- 83. Chen X, Liu K, Yang P, Kuang W, Huang H, Tu E, et al. microRNA-200a functions as a tumor suppressor by targeting FOXA1 in glioma. Exp Ther Med (2019) 17:221–9. doi: 10.3892/etm.2018.6895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nurul-Syakima AM, Yoke-Kqueen C, Sabariah AR, Shiran MS, Singh A, Learn-Han L. Differential microRNA expression and identification of putative miRNA targets and pathways in head and neck cancers. Int J Mol Med (2011) 28:327–36. doi: 10.3892/ijmm.2011.714 [DOI] [PubMed] [Google Scholar]
- 85. Zhao Z, Gao D, Ma T, Zhang L. MicroRNA-141 suppresses growth and metastatic potential of head and neck squamous cell carcinoma. Aging (Albany NY) (2019) 11:921–32. doi: 10.18632/aging.101791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhao Y, Yu X, Tang H, Han R, Wang X, Wang J, et al. MicroRNA-200a promotes phagocytosis of macrophages and suppresses cell proliferation, migration, and invasion in nasopharyngeal carcinoma by targeting CD47. BioMed Res Int (2020) 2020:3723781. doi: 10.1155/2020/3723781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Feng J, Wang J, Chen M, Chen G, Wu Z, Ying L, et al. miR-200a suppresses cell growth and migration by targeting MACC1 and predicts prognosis in hepatocellular carcinoma. Oncol Rep (2015) 33:713–20. doi: 10.3892/or.2014.3642 [DOI] [PubMed] [Google Scholar]
- 88. Li Y, Bai W, Zhang J. MiR-200c-5p suppresses proliferation and metastasis of human hepatocellular carcinoma (HCC) via suppressing MAD2L1. BioMed Pharmacother (2017) 92:1038–44. doi: 10.1016/j.biopha.2017.05.092 [DOI] [PubMed] [Google Scholar]
- 89. Guo C, Zhao D, Zhang Q, Liu S, Sun MZ. miR-429 suppresses tumor migration and invasion by targeting CRKL in hepatocellular carcinoma via inhibiting Raf/MEK/ERK pathway and epithelial-mesenchymal transition. Sci Rep (2018) 8:2375. doi: 10.1038/s41598-018-20258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang LK, Xie XN, Song XH, Su T, Chang XL, Xu M, et al. Upregulation of miR-200b inhibits hepatocellular carcinoma cell proliferation and migration by targeting HMGB3 protein. Technol Cancer Res Treat (2018) 17:1533033818806475. doi: 10.1177/1533033818806475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhao Y, Xu Z, Zhou J, Yang H. miR−141 inhibits proliferation, migration and invasion in human hepatocellular carcinoma cells by directly downregulating TGFβR1. Oncol Rep (2019) 42:1656–66. doi: 10.3892/or.2019.7325 [DOI] [PubMed] [Google Scholar]
- 92. Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer (2012) 106:553–61. doi: 10.1038/bjc.2011.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bustos MA, Ono S, Marzese DM, Oyama T, Iida Y, Cheung G, et al. MiR-200a regulates CDK4/6 inhibitor effect by targeting CDK6 in metastatic melanoma. J Invest Dermatol (2017) 137:1955–64. doi: 10.1016/j.jid.2017.03.039 [DOI] [PubMed] [Google Scholar]
- 94. Sheng H, Guo YH, Cao DS, Li XJ, Zhao Y, Ding H, et al. MiR-429-5p attenuates the migration and invasion of malignant melanoma by targeting LIMK1. Eur Rev Med Pharmacol Sci (2020) 24:2625–31. doi: 10.26355/eurrev_202003_20531 [DOI] [PubMed] [Google Scholar]
- 95. Senanayake U, Das S, Vesely P, Alzoughbi W, Fröhlich LF, Chowdhury P, et al. miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated and their common target ACVR2B is strongly expressed in renal childhood neoplasms. Carcinogenesis (2012) 33:1014–21. doi: 10.1093/carcin/bgs126 [DOI] [PubMed] [Google Scholar]
- 96. Wang HF, Wang WH, Zhuang HW, Xu M. MiR-429 regulates the proliferation and apoptosis of nephroblastoma cells through targeting c-myc. Eur Rev Med Pharmacol Sci (2018) 22:5172–9. doi: 10.26355/eurrev_201808_15713 [DOI] [PubMed] [Google Scholar]
- 97. Li T, Zhao P, Li Z, Wang CC, Wang YL, Gu Q. miR-200c-3p suppresses the proliferative, migratory, and invasive capacities of nephroblastoma cells via targeting FRS2. Biopreserv Biobank (2019) 17:444–51. doi: 10.1089/bio.2019.0009 [DOI] [PubMed] [Google Scholar]
- 98. Liu P, Chen S, Huang Y, Xu S, Song H, Zhang W, et al. LINC00667 promotes wilms' tumor metastasis and stemness by sponging miR-200b/c/429 family to regulate IKK-β. Cell Biol Int (2020) 44:1382–93. doi: 10.1002/cbin.11334 [DOI] [PubMed] [Google Scholar]
- 99. Gao SL, Wang LZ, Liu HY, Liu DL, Xie LM, Zhang ZW. miR-200a inhibits tumor proliferation by targeting AP-2γ in neuroblastoma cells. Asian Pac J Cancer Prev (2014) 15:4671–6. doi: 10.7314/APJCP.2014.15.11.4671 [DOI] [PubMed] [Google Scholar]
- 100. Du M, Wang J, Chen H, Wang S, Chen L, Xu Y, et al. MicroRNA−200a suppresses migration and invasion and enhances the radiosensitivity of NSCLC cells by inhibiting the HGF/c−Met signaling pathway. Oncol Rep (2019) 41:1497–508. doi: 10.3892/or.2018.6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Arunkumar G, Deva Magendhra Rao AK, Manikandan M, Prasanna H, Srinivasa Rao, Subbiah S, Ilangovan R, et al. Dysregulation of miR-200 family microRNAs and epithelial-mesenchymal transition markers in oral squamous cell carcinoma. Oncol Lett (2018) 15:649–57. doi: 10.3892/ol.2017.7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yan Y, Yan F, Huang Q. MiR-200c inhibited the proliferation of oral squamous cell carcinoma cells by targeting akt pathway and its downstream Glut1. Arch Oral Biol (2018) 96:52–7. doi: 10.1016/j.archoralbio.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 103. Deng Y, Luan F, Zeng L, Zhang Y, Ma K. MiR-429 suppresses the progression and metastasis of osteosarcoma by targeting ZEB1. Excli J (2017) 16:618–27. doi: 10.17179/excli2017-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Liu C, Pan C, Cai Y, Wang H. Interplay between long noncoding RNA ZEB1-AS1 and miR-200s regulates osteosarcoma cell proliferation and migration. J Cell Biochem (2017) 118:2250–60. doi: 10.1002/jcb.25879 [DOI] [PubMed] [Google Scholar]
- 105. Liu Y, Zhu ST, Wang X, Deng J, Li WH, Zhang P, et al. MiR-200c regulates tumor growth and chemosensitivity to cisplatin in osteosarcoma by targeting AKT2. Sci Rep (2017) 7:13598. doi: 10.1038/s41598-017-14088-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang N, Li P, Liu W, Wang N, Lu Z, Feng J, et al. miR-141-3p suppresses proliferation and promotes apoptosis by targeting GLI2 in osteosarcoma cells. Oncol Rep (2018) 39:747–54. doi: 10.3892/or.2017.6150 [DOI] [PubMed] [Google Scholar]
- 107. Sun L, Wang L, Luan S, Jiang Y, Wang Q. miR-429 inhibits osteosarcoma progression by targeting HOXA9 through suppressing wnt/β-catenin signaling pathway. Oncol Lett (2020) 20:2447–55. doi: 10.3892/ol.2020.11766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res (2007) 67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936 [DOI] [PubMed] [Google Scholar]
- 109. Chen Y, Zhang L, Hao Q. Candidate microRNA biomarkers in human epithelial ovarian cancer: systematic review profiling studies and experimental validation. Cancer Cell Int (2013) 13:86. doi: 10.1186/1475-2867-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cao Q, Lu K, Dai S, Hu Y, Fan W. Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer. Int J Clin Exp Pathol (2014) 7:2392–401. [PMC free article] [PubMed] [Google Scholar]
- 111. Zhu CL, Gao GS. miR-200a overexpression in advanced ovarian carcinomas as a prognostic indicator. Asian Pac J Cancer Prev (2014) 15:8595–601. doi: 10.7314/APJCP.2014.15.20.8595 [DOI] [PubMed] [Google Scholar]
- 112. Chen JL, Chen F, Zhang TT, Liu NF. Suppression of SIK1 by miR-141 in human ovarian cancer cell lines and tissues. Int J Mol Med (2016) 37:1601–10. doi: 10.3892/ijmm.2016.2553 [DOI] [PubMed] [Google Scholar]
- 113. Shi C, Yang Y, Zhang L, Yu J, Qin S, Xu H, et al. MiR-200a-3p promoted the malignant behaviors of ovarian cancer cells through regulating PCDH9. Onco Targets Ther (2019) 12:8329–38. doi: 10.2147/OTT.S220339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ali S, Saleh H, Sethi S, Sarkar FH, Philip PA. MicroRNA profiling of diagnostic needle aspirates from patients with pancreatic cancer. Br J Cancer (2012) 107:1354–60. doi: 10.1038/bjc.2012.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhu ZM, Xu YF, Su QJ, Du JD, Tan XL, Tu YL, et al. Prognostic significance of microRNA-141 expression and its tumor suppressor function in human pancreatic ductal adenocarcinoma. Mol Cell Biochem (2014) 388:39–49. doi: 10.1007/s11010-013-1897-y [DOI] [PubMed] [Google Scholar]
- 116. Paik WH, Song BJ, Kim HW, Kim HR, Hwang JH. MicroRNA-200c as a prognostic biomarker for pancreatic cancer. Korean J Gastroenterol (2015) 66:215–20. doi: 10.4166/kjg.2015.66.4.215 [DOI] [PubMed] [Google Scholar]
- 117. Liu D, Song L, Dai Z, Guan H, Kang H, Zhang Y, et al. MiR-429 suppresses neurotrophin-3 to alleviate perineural invasion of pancreatic cancer. Biochem Biophys Res Commun (2018) 505:1077–83. doi: 10.1016/j.bbrc.2018.09.147 [DOI] [PubMed] [Google Scholar]
- 118. Diaz-Riascos ZV, Ginesta MM, Fabregat J, Serrano T, Busquets J, Buscail L, et al. Expression and role of MicroRNAs from the miR-200 family in the tumor formation and metastatic propensity of pancreatic cancer. Mol Ther Nucleic Acids (2019) 17:491–503. doi: 10.1016/j.omtn.2019.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Basu S, Majumder S, Bhowal A, Ghosh A, Naskar S, Nandy S, et al. A study of molecular signals deregulating mismatch repair genes in prostate cancer compared to benign prostatic hyperplasia. PloS One (2015) 10:e0125560. doi: 10.1371/journal.pone.0125560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Bian X, Shen Y, Zhang G, Gu C, Cai Y, Wang C, et al. Expression of dicer and its related miRNAs in the progression of prostate cancer. PloS One (2015) 10:e0120159. doi: 10.1371/journal.pone.0120159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Song CJ, Chen H, Chen LZ, Ru GM, Guo JJ, Ding QN. The potential of microRNAs as human prostate cancer biomarkers: A meta-analysis of related studies. J Cell Biochem (2018) 119:2763–86. doi: 10.1002/jcb.26445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Xie H, Nie L, Zhang M, Su Z, Chen X, Xu M, et al. Suppression of α-methylacyl-coenzyme a racemase by miR200c inhibits prostate adenocarcinoma cell proliferation and migration. Exp Ther Med (2020) 19:1806–16. doi: 10.3892/etm.2019.8406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Nakada C, Matsuura K, Tsukamoto Y, Tanigawa M, Yoshimoto T, Narimatsu T, et al. Genome-wide microRNA expression profiling in renal cell carcinoma: significant down-regulation of miR-141 and miR-200c. J Pathol (2008) 216:418–27. doi: 10.1002/path.2437 [DOI] [PubMed] [Google Scholar]
- 124. Yoshino H, Enokida H, Itesako T, Tatarano S, Kinoshita T, Fuse M, et al. Epithelial-mesenchymal transition-related microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. J Hum Genet (2013) 58:508–16. doi: 10.1038/jhg.2013.31 [DOI] [PubMed] [Google Scholar]
- 125. Butz H, Szabó PM, Khella HW, Nofech-Mozes R, Patocs A, Yousef GM. miRNA-target network reveals miR-124as a key miRNA contributing to clear cell renal cell carcinoma aggressive behaviour by targeting CAV1 and FLOT1. Oncotarget (2015) 6:12543–57. doi: 10.18632/oncotarget.3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang X, Jiang F, Song H, Li X, Xian J, Gu X. MicroRNA-200a-3p suppresses tumor proliferation and induces apoptosis by targeting SPAG9 in renal cell carcinoma. Biochem Biophys Res Commun (2016) 470:620–6. doi: 10.1016/j.bbrc.2016.01.095 [DOI] [PubMed] [Google Scholar]
- 127. Ding M, Sun X, Zhong J, Zhang C, Tian Y, Ge J, et al. Decreased miR-200a-3p is a key regulator of renal carcinoma growth and migration by directly targeting CBL. J Cell Biochem (2018) 119:9974–85. doi: 10.1002/jcb.27326 [DOI] [PubMed] [Google Scholar]
- 128. Maolakuerban N, Azhati B, Tusong H, Abula A, Yasheng A, Xireyazidan A. MiR-200c-3p inhibits cell migration and invasion of clear cell renal cell carcinoma via regulating SLC6A1. Cancer Biol Ther (2018) 19:282–91. doi: 10.1080/15384047.2017.1394551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Dong S, Meng X, Xue S, Yan Z, Ren P, Liu J. microRNA-141 inhibits thyroid cancer cell growth and metastasis by targeting insulin receptor substrate 2. Am J Transl Res (2016) 8:1471–81. [PMC free article] [PubMed] [Google Scholar]
- 130. Gou L, Zou H, Li B. Long noncoding RNA MALAT1 knockdown inhibits progression of anaplastic thyroid carcinoma by regulating miR-200a-3p/FOXA1. Cancer Biol Ther (2019) 20:1355–65. doi: 10.1080/15384047.2019.1617567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wu G, Zheng H, Xu J, Guo Y, Zheng G, Ma C, et al. miR-429 suppresses cell growth and induces apoptosis of human thyroid cancer cell by targeting ZEB1. Artif Cells Nanomed Biotechnol (2019) 47:548–54. doi: 10.1080/21691401.2018.1564320 [DOI] [PubMed] [Google Scholar]
- 132. Zhou B, Xu J, Chen Y, Gao S, Feng X, Lu X. miR-200b/c-RAP1B axis represses tumorigenesis and malignant progression of papillary thyroid carcinoma through inhibiting the NF-κB/Twist1 pathway. Exp Cell Res (2020) 387:111785. doi: 10.1016/j.yexcr.2019.111785 [DOI] [PubMed] [Google Scholar]
- 133. Gregorova J, Vychytilova-Faltejskova P, Sevcikova S. Epigenetic regulation of MicroRNA clusters and families during tumor development. Cancers (Basel) (2021) 13. doi: 10.3390/cancers13061333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Shindo T, Niinuma T, Nishiyama N, Shinkai N, Kitajima H, Kai M, et al. Epigenetic silencing of miR-200b is associated with cisplatin resistance in bladder cancer. Oncotarget (2018) 9:24457–69. doi: 10.18632/oncotarget.25326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lim YY, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E, et al. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J Cell Sci (2013) 126:2256–66. doi: 10.1242/jcs.122275 [DOI] [PubMed] [Google Scholar]
- 136. Wang G, Chan ES, Kwan BC, Li PK, Yip SK, Szeto CC, et al. Expression of microRNAs in the urine of patients with bladder cancer. Clin Genitourin Cancer (2012) 10:106–13. doi: 10.1016/j.clgc.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 137. Yun SJ, Jeong P, Kim WT, Kim TH, Lee YS, Song PH, et al. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol (2012) 41:1871–8. doi: 10.3892/ijo.2012.1622 [DOI] [PubMed] [Google Scholar]
- 138. Du L, Jiang X, Duan W, Wang R, Wang L, Zheng G, et al. Cell-free microRNA expression signatures in urine serve as novel noninvasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Oncotarget (2017) 8:40832–42. doi: 10.18632/oncotarget.16586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ghorbanmehr N, Gharbi S, Korsching E, Tavallaei M, Einollahi B, Mowla SJ. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate (2019) 79:88–95. doi: 10.1002/pros.23714 [DOI] [PubMed] [Google Scholar]
- 140. Antolín S, Calvo L, Blanco-Calvo M, Santiago MP, Lorenzo-Patiño MJ, Haz-Conde M, et al. Circulating miR-200c and miR-141 and outcomes in patients with breast cancer. BMC Cancer (2015) 15:297. doi: 10.1186/s12885-015-1238-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Markou A, Zavridou M, Sourvinou I, Yousef G, Kounelis S, Malamos N, et al. Direct comparison of metastasis-related miRNAs expression levels in circulating tumor cells, corresponding plasma, and primary tumors of breast cancer patients. Clin Chem (2016) 62:1002–11. doi: 10.1373/clinchem.2015.253716 [DOI] [PubMed] [Google Scholar]
- 142. Wang WT, Zhao YN, Yan JX, Weng MY, Wang Y, Chen YQ, et al. Differentially expressed microRNAs in the serum of cervical squamous cell carcinoma patients before and after surgery. J Hematol Oncol (2014) 7:6. doi: 10.1186/1756-8722-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Jia W, Wu Y, Zhang Q, Gao GE, Zhang C, Xiang Y. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol Clin Oncol (2015) 3:851–8. doi: 10.3892/mco.2015.560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Shen L, Chen G, Xia Q, Shao S, Fang H. Exosomal miR-200 family as serum biomarkers for early detection and prognostic prediction of cholangiocarcinoma. Int J Clin Exp Pathol (2019) 12:3870–6. [PMC free article] [PubMed] [Google Scholar]
- 145. Brunet Vega A, Pericay C, Moya I, Ferrer A, Dotor E, Pisa A, et al. microRNA expression profile in stage III colorectal cancer: circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep (2013) 30:320–6. doi: 10.3892/or.2013.2475 [DOI] [PubMed] [Google Scholar]
- 146. Zhang GJ, Zhou T, Liu ZL, Tian HP, Xia SS. Plasma miR-200c and miR-18a as potential biomarkers for the detection of colorectal carcinoma. Mol Clin Oncol (2013) 1:379–84. doi: 10.3892/mco.2013.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, et al. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg (2014) 259:735–43. doi: 10.1097/SLA.0b013e3182a6909d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Sun Y, Liu Y, Cogdell D, Calin GA, Sun B, Kopetz S, et al. Examining plasma microRNA markers for colorectal cancer at different stages. Oncotarget (2016) 7:11434–49. doi: 10.18632/oncotarget.7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wang YN, Chen ZH, Chen WC. Novel circulating microRNAs expression profile in colon cancer: a pilot study. Eur J Med Res (2017) 22:51. doi: 10.1186/s40001-017-0294-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Ardila HJ, Sanabria-Salas MC, Meneses X, Rios R, Huertas-Salgado A, Serrano ML. Circulating miR-141-3p, miR-143-3p and miR-200c-3p are differentially expressed in colorectal cancer and advanced adenomas. Mol Clin Oncol (2019) 11:201–7. doi: 10.3892/mco.2019.1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Tanaka K, Miyata H, Yamasaki M, Sugimura K, Takahashi T, Kurokawa Y, et al. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann Surg Oncol (2013) 20 Suppl 3:S607–15. doi: 10.1245/s10434-013-3093-4 [DOI] [PubMed] [Google Scholar]
- 152. Yu H, Duan B, Jiang L, Lin M, Sheng H, Huang J, et al. Serum miR-200c and clinical outcome of patients with advanced esophageal squamous cancer receiving platinum-based chemotherapy. Am J Transl Res (2013) 6:71–7. [PMC free article] [PubMed] [Google Scholar]
- 153. Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Haz M, et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med (2012) 10:186. doi: 10.1186/1479-5876-10-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Zhang HP, Sun FB, Li SJ. Serum miR-200c expression level as a prognostic biomarker for gastric cancer. Genet Mol Res (2015) 14:15913–20. doi: 10.4238/2015.December.7.2 [DOI] [PubMed] [Google Scholar]
- 155. Zhang ZZ, Wang CJ, Niu L, Xu J, Wang M, Cao H, et al. Analysis of plasma MicroRNAs to identifying early diagnostic molecule for gastric cancer. Int J Clin Exp Med (2015) 8:3700–6. [PMC free article] [PubMed] [Google Scholar]
- 156. Wang J, Zhang H, Zhou X, Wang T, Zhang J, Zhu W, et al. Five serum-based miRNAs were identified as potential diagnostic biomarkers in gastric cardia adenocarcinoma. Cancer biomark (2018) 23:193–203. doi: 10.3233/CBM-181258 [DOI] [PubMed] [Google Scholar]
- 157. Ghaedi H, Mozaffari MAN, Salehi Z, Ghasemi H, Zadian SS, Alipoor S, et al. Co-Expression profiling of plasma miRNAs and long noncoding RNAs in gastric cancer patients. Gene (2019) 687:135–42. doi: 10.1016/j.gene.2018.11.034 [DOI] [PubMed] [Google Scholar]
- 158. Wang T, Zhang J, Tian J, Hu S, Wei R, Cui L. Low expression levels of plasma miR-141 are associated with susceptibility to gastric cancer. Oncol Lett (2019) 18:629–36. doi: 10.3892/ol.2019.10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wang Y, Gao Y, Shi W, Zhai D, Rao Q, Jia X, et al. Profiles of differential expression of circulating microRNAs in hepatitis b virus-positive small hepatocellular carcinoma. Cancer biomark (2015) 15:171–80. doi: 10.3233/CBM-140451 [DOI] [PubMed] [Google Scholar]
- 160. Li L, Chen J, Chen X, Tang J, Guo H, Wang X, et al. Serum miRNAs as predictive and preventive biomarker for pre-clinical hepatocellular carcinoma. Cancer Lett (2016) 373:234–40. doi: 10.1016/j.canlet.2016.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Chen SY, Ma DN, Chen QD, Zhang JJ, Tian YR, Wang ZC, et al. MicroRNA-200a inhibits cell growth and metastasis by targeting Foxa2 in hepatocellular carcinoma. J Cancer (2017) 8:617–25. doi: 10.7150/jca.17394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Chen S, Zhang J, Chen Q, Cheng J, Chen X, Mao Y, et al. MicroRNA-200a and microRNA-141 have a synergetic effect on the suppression of epithelial-mesenchymal transition in liver cancer by targeting STAT4. Oncol Lett (2021) 21:137. doi: 10.3892/ol.2020.12398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Fogli S, Polini B, Carpi S, Pardini B, Naccarati A, Dubbini N, et al. Identification of plasma microRNAs as new potential biomarkers with high diagnostic power in human cutaneous melanoma. Tumour Biol (2017) 39:1010428317701646. doi: 10.1177/1010428317701646 [DOI] [PubMed] [Google Scholar]
- 164. Zhu W, He J, Chen D, Zhang B, Xu L, Ma H, et al. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PloS One (2014) 9:e87780. doi: 10.1371/journal.pone.0087780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Nadal E, Truini A, Nakata A, Lin J, Reddy RM, Chang AC, et al. A novel serum 4-microRNA signature for lung cancer detection. Sci Rep (2015) 5:12464. doi: 10.1038/srep12464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Halvorsen AR, Bjaanæs M, Leblanc M, Holm AM, Bolstad N, Rubio L, et al. A unique set of 6 circulating microRNAs for early detection of non-small cell lung cancer. Oncotarget (2016) 7:37250–9. doi: 10.18632/oncotarget.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Arab A, Karimipoor M, Irani S, Kiani A, Zeinali S, Tafsiri E, et al. Potential circulating miRNA signature for early detection of NSCLC. Cancer Genet (2017) 216-217:150–8. doi: 10.1016/j.cancergen.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 168. Zhao Y. The diagnostic and prognostic role of circulating miR-141 expression in non-small-cell lung cancer patients. Int J Clin Exp Pathol (2018) 11:2597–604. [PMC free article] [PubMed] [Google Scholar]
- 169. Zou JG, Ma LF, Li X, Xu FL, Fei XZ, Liu Q, et al. Circulating microRNA array (miR-182, 200b and 205) for the early diagnosis and poor prognosis predictor of non-small cell lung cancer. Eur Rev Med Pharmacol Sci (2019) 23:1108–15. doi: 10.26355/eurrev_201902_17001 [DOI] [PubMed] [Google Scholar]
- 170. Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res (2009) 15:5473–7. doi: 10.1158/1078-0432.CCR-09-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Sun G, Cao Y, Wang P, Song H, Bie T, Li M, et al. miR-200b-3p in plasma is a potential diagnostic biomarker in oral squamous cell carcinoma. Biomarkers (2018) 23:137–41. doi: 10.1080/1354750X.2017.1289241 [DOI] [PubMed] [Google Scholar]
- 172. Meng X, Joosse SA, Müller V, Trillsch F, Milde-Langosch K, Mahner S, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer (2015) 113:1358–66. doi: 10.1038/bjc.2015.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zuberi M, Mir R, Das J, Ahmad I, Javid J, Yadav P, et al. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol (2015) 17:779–87. doi: 10.1007/s12094-015-1303-1 [DOI] [PubMed] [Google Scholar]
- 174. Meng X, Müller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget (2016) 7:16923–35. doi: 10.18632/oncotarget.7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Meng X, Müller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Circulating cell-free miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Adv Exp Med Biol (2016) 924:3–8. doi: 10.1007/978-3-319-42044-8_1 [DOI] [PubMed] [Google Scholar]
- 176. Pendlebury A, Hannan NJ, Binder N, Beard S, Mcgauran M, Grant P, et al. The circulating microRNA-200 family in whole blood are potential biomarkers for high-grade serous epithelial ovarian cancer. BioMed Rep (2017) 6:319–22. doi: 10.3892/br.2017.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Fitriawan AS, Kartika AI, Chasanah SN, Aryandono T, Haryana SM. Expression of circulating MicroRNA-141 in epithelial ovarian cancer. Malays J Med Sci (2020) 27:27–38. doi: 10.21315/mjms2020.27.6.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Xiong J, He X, Xu Y, Zhang W, Fu F. MiR-200b is upregulated in plasma-derived exosomes and functions as an oncogene by promoting macrophage M2 polarization in ovarian cancer. J Ovarian Res (2021) 14:74. doi: 10.1186/s13048-021-00826-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Li A, Omura N, Hong SM, Vincent A, Walter K, Griffith M, et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res (2010) 70:5226–37. doi: 10.1158/0008-5472.CAN-09-4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Reese M, Flammang I, Yang Z, Dhayat SA. Potential of exosomal microRNA-200b as liquid biopsy marker in pancreatic ductal adenocarcinoma. Cancers (Basel) (2020) 12. doi: 10.3390/cancers12010197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer (2012) 106:768–74. doi: 10.1038/bjc.2011.595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Selth LA, Townley S, Gillis JL, Ochnik AM, Murti K, Macfarlane RJ, et al. Discovery of circulating microRNAs associated with human prostate cancer using a mouse model of disease. Int J Cancer (2012) 131:652–61. doi: 10.1002/ijc.26405 [DOI] [PubMed] [Google Scholar]
- 183. Cheng HH, Mitchell PS, Kroh EM, Dowell AE, Chéry L, Siddiqui J, et al. Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PloS One (2013) 8:e69239. doi: 10.1371/journal.pone.0069239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Kelly BD, Miller N, Sweeney KJ, Durkan GC, Rogers E, Walsh K, et al. A circulating MicroRNA signature as a biomarker for prostate cancer in a high risk group. J Clin Med (2015) 4:1369–79. doi: 10.3390/jcm4071369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Li Z, Ma YY, Wang J, Zeng XF, Li R, Kang W, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther (2016) 9:139–48. doi: 10.2147/OTT.S95565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Souza MF, Kuasne H, Barros-Filho MC, Cilião HL, Marchi FA, Fuganti PE, et al. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PloS One (2017) 12:e0184094. doi: 10.1371/journal.pone.0184094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Danarto R, Astuti I, Umbas R, Haryana SM. Urine miR-21-5p and miR-200c-3p as potential non-invasive biomarkers in patients with prostate cancer. Turk J Urol (2020) 46:26–30. doi: 10.5152/tud.2019.19163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Jin W, Fei X, Wang X, Chen F, Song Y. Circulating miRNAs as biomarkers for prostate cancer diagnosis in subjects with benign prostatic hyperplasia. J Immunol Res (2020) 2020:5873056. doi: 10.1155/2020/5873056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Li W, Dong Y, Wang KJ, Deng Z, Zhang W, Shen HF. Plasma exosomal miR-125a-5p and miR-141-5p as non-invasive biomarkers for prostate cancer. Neoplasma (2020) 67:1314–8. doi: 10.4149/neo_2020_191130N1234 [DOI] [PubMed] [Google Scholar]
- 190. Wang ZK, Luo L, Du ZJ, Zhang GM, Sun LJ. MiR429 expression level in renal cell cancer and its correlation with the prognosis of patients. J buon (2017) 22:1428–33. [PubMed] [Google Scholar]
- 191. Yadav S, Khandelwal M, Seth A, Saini AK, Dogra PN, Sharma A. Serum microRNA expression profiling: Potential diagnostic implications of a panel of serum microRNAs for clear cell renal cell cancer. Urology (2017) 104:64–9. doi: 10.1016/j.urology.2017.03.013 [DOI] [PubMed] [Google Scholar]
- 192. Wang C, Ding M, Zhu Y-Y, Hu J, Zhang C, Lu X, et al. Circulating miR-200a is a novel molecular biomarker for early-stage renal cell carcinoma. ExRNA (2019) 1:25. doi: 10.1186/s41544-019-0023-z [DOI] [Google Scholar]
- 193. Mao S, Lu Z, Zheng S, Zhang H, Zhang G, Wang F, et al. Exosomal miR-141 promotes tumor angiogenesis via KLF12 in small cell lung cancer. J Exp Clin Cancer Res (2020) 39:193. doi: 10.1186/s13046-020-01680-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Cookson VJ, Bentley MA, Hogan BV, Horgan K, Hayward BE, Hazelwood LD, et al. Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. Cell Oncol (2012) 35:301–8. doi: 10.1007/s13402-012-0089-1 [DOI] [PubMed] [Google Scholar]
- 195. Zhu J, Zheng Z, Wang J, Sun J, Wang P, Cheng X, et al. Different miRNA expression profiles between human breast cancer tumors and serum. Front Genet (2014) 5:149. doi: 10.3389/fgene.2014.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Savolainen K, Scaravilli M, Ilvesmäki A, Staff S, Tolonen T, Mäenpää JU, et al. Expression of the miR-200 family in tumor tissue, plasma and urine of epithelial ovarian cancer patients in comparison to benign counterparts. BMC Res Notes (2020) 13:311. doi: 10.1186/s13104-020-05155-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinf (2015) 13:17–24. doi: 10.1016/j.gpb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Choi PW, Ng SW. The functions of MicroRNA-200 family in ovarian cancer: Beyond epithelial-mesenchymal transition. Int J Mol Sci (2017) 18. doi: 10.3390/ijms18061207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Hydbring P, De Petris L, Zhang Y, Brandén E, Koyi H, Novak M, et al. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer (2018) 124:45–52. doi: 10.1016/j.lungcan.2018.07.018 [DOI] [PubMed] [Google Scholar]
- 200. Kobayashi M, Salomon C, Tapia J, Illanes SE, Mitchell MD, Rice GE. Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of let-7 miRNA and miR-200. J Transl Med (2014) 12:4. doi: 10.1186/1479-5876-12-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Santasusagna S, Moreno I, Navarro A, Martinez Rodenas F, Hernández R, Castellano JJ, et al. Prognostic impact of miR-200 family members in plasma and exosomes from tumor-draining versus peripheral veins of colon cancer patients. Oncology (2018) 95:309–18. doi: 10.1159/000490726 [DOI] [PubMed] [Google Scholar]
- 202. Macha MA, Seshacharyulu P, Krishn SR, Pai P, Rachagani S, Jain M, et al. MicroRNAs (miRNAs) as biomarker(s) for prognosis and diagnosis of gastrointestinal (GI) cancers. Curr Pharm Des (2014) 20:5287–97. doi: 10.2174/1381612820666140128213117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Usuba W, Urabe F, Yamamoto Y, Matsuzaki J, Sasaki H, Ichikawa M, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci (2019) 110:408–19. doi: 10.1111/cas.13856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Yang X, Zhang Q, Zhang M, Su W, Wang Z, Li Y, et al. Serum microRNA signature is capable of early diagnosis for non-small cell lung cancer. Int J Biol Sci (2019) 15:1712–22. doi: 10.7150/ijbs.33986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Huang ZS, Guo XW, Zhang G, Liang LX, Nong B. The diagnostic and prognostic value of miR-200c in gastric cancer: A meta-analysis. Dis Markers (2019) 2019:8949618. doi: 10.1155/2019/8949618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Peng Z, Zhu W, Dai J, Ju F. MicroRNA-200 as potential diagnostic markers for colorectal cancer: meta-analysis and experimental validation. Cell Mol Biol (Noisy-le-grand) (2018) 64:77–85. doi: 10.14715/cmb/2018.64.6.14 [DOI] [PubMed] [Google Scholar]
- 207. Huang GL, Sun J, Lu Y, Liu Y, Cao H, Zhang H, et al. MiR-200 family and cancer: From a meta-analysis view. Mol Aspects Med (2019) 70:57–71. doi: 10.1016/j.mam.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 208. Yu C, Wan H, Shan R, Wen W, Li J, Luo D, et al. The prognostic value of the MiR-200 family in colorectal cancer: A meta-analysis with 1882 patients. J Cancer (2019) 10:4009–16. doi: 10.7150/jca.27529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Tejero R, Navarro A, Campayo M, Viñolas N, Marrades RM, Cordeiro A, et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PloS One (2014) 9:e101899. doi: 10.1371/journal.pone.0101899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Mei Y, Zheng J, Xiang P, Liu C, Fan Y. Prognostic value of the miR-200 family in bladder cancer: A systematic review and meta-analysis. Med (Baltimore) (2020) 99:e22891. doi: 10.1097/MD.0000000000022891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Shi C, Zhang Z. The prognostic value of the miR-200 family in ovarian cancer: a meta-analysis. Acta Obstet Gynecol Scand (2016) 95:505–12. doi: 10.1111/aogs.12883 [DOI] [PubMed] [Google Scholar]
- 212. Richardsen E, Andersen S, Melbø-Jørgensen C, Rakaee M, Ness N, Al-Saad S, et al. MicroRNA 141 is associated to outcome and aggressive tumor characteristics in prostate cancer. Sci Rep (2019) 9:386. doi: 10.1038/s41598-018-36854-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell (2000) 100:57–70. doi: 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 214. Hanahan D. Hallmarks of cancer: New dimensions. Cancer Discovery (2022) 12:31–46. doi: 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- 215. Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett (2009) 285:116–26. doi: 10.1016/j.canlet.2009.04.031 [DOI] [PubMed] [Google Scholar]
- 216. Van Roosbroeck K, Calin GA. Cancer hallmarks and MicroRNAs: The therapeutic connection. Adv Cancer Res (2017) 135:119–49. doi: 10.1016/bs.acr.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 217. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 218. Li M, Li J, Ye C, Wu W, Cheng Y. miR-200a-3p predicts prognosis and inhibits bladder cancer cell proliferation by targeting STAT4. Arch Med Sci (2019). doi: 10.5114/aoms.2019.89969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Yao J, Zhou E, Wang Y, Xu F, Zhang D, Zhong D. microRNA-200a inhibits cell proliferation by targeting mitochondrial transcription factor a in breast cancer. DNA Cell Biol (2014) 33:291–300. doi: 10.1089/dna.2013.2132 [DOI] [PubMed] [Google Scholar]
- 220. Su X, Lang C, Luan A, Zhao P. MiR-200a promotes proliferation of cervical cancer cells by regulating HIF-1α/VEGF signaling pathway. J buon (2020) 25:1935–40. [PubMed] [Google Scholar]
- 221. Wu G, Zhao H, He N, Han H. Expression of miR-200a in colorectal carcinoma cell lines and its effect on LoVo cells. Nan Fang Yi Ke Da Xue Xue Bao (2015) 35:450–4. [PubMed] [Google Scholar]
- 222. Di Z, Di M, Fu W, Tang Q, Liu Y, Lei P, et al. Integrated analysis identifies a nine-microRNA signature biomarker for diagnosis and prognosis in colorectal cancer. Front Genet (2020) 11:192. doi: 10.3389/fgene.2020.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Wu Q, Lu RL, Li JX, Rong LJ. MiR-200a and miR-200b target PTEN to regulate the endometrial cancer cell growth in vitro. Asian Pac J Trop Med (2017) 10:498–502. doi: 10.1016/j.apjtm.2017.05.007 [DOI] [PubMed] [Google Scholar]
- 224. Zang Y, Tai Y, Wan B, Jia X. miR-200a-3p promotes the proliferation of human esophageal cancer cells by post-transcriptionally regulating cytoplasmic collapsin response mediator protein-1. Int J Mol Med (2016) 38:1558–64. doi: 10.3892/ijmm.2016.2758 [DOI] [PubMed] [Google Scholar]
- 225. Liu X, Du P, Han L, Zhang A, Jiang K, Zhang Q. Effects of miR-200a and FH535 combined with taxol on proliferation and invasion of gastric cancer. Pathol Res Pract (2018) 214:442–9. doi: 10.1016/j.prp.2017.12.004 [DOI] [PubMed] [Google Scholar]
- 226. Su Y, He Q, Deng L, Wang J, Liu Q, Wang D, et al. MiR-200a impairs glioma cell growth, migration, and invasion by targeting SIM2-s. Neuroreport (2014) 25:12–17. doi: 10.1097/WNR.0000000000000032 [DOI] [PubMed] [Google Scholar]
- 227. Zhong C, Li MY, Chen ZY, Cheng HK, Hu ML, Ruan YL, et al. MicroRNA-200a inhibits epithelial-mesenchymal transition in human hepatocellular carcinoma cell line. Int J Clin Exp Pathol (2015) 8:9922–31. [PMC free article] [PubMed] [Google Scholar]
- 228. Chen WY, Xu YY, Zhang XY. Targeting GOLM1 by microRNA-200a in melanoma suppresses cell proliferation, invasion and migration via regulating PI3K/Akt signaling pathway and epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci (2019) 23:6997–7007. doi: 10.26355/eurrev_201908_18740 [DOI] [PubMed] [Google Scholar]
- 229. Chen Y, Peng W, Lu Y, Chen J, Zhu YY, Xi T. MiR-200a enhances the migrations of A549 and SK-MES-1 cells by regulating the expression of TSPAN1. J Biosci (2013) 38:523–32. doi: 10.1007/s12038-013-9351-6 [DOI] [PubMed] [Google Scholar]
- 230. Huang Y, Bao T, Li Z, Ji G, Zhang L. Function of miR-200a in proliferation and apoptosis of non-small cell lung cancer cells. Oncol Lett (2020) 20:1256–62. doi: 10.3892/ol.2020.11649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Wu X, Wu G, Wu Z, Yao X, Li G. MiR-200a suppresses the proliferation and metastasis in pancreatic ductal adenocarcinoma through downregulation of DEK gene. Transl Oncol (2016) 9:25–31. doi: 10.1016/j.tranon.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Hu B, Qiu-Lan H, Lei RE, Shi C, Jiang HX, Qin SY. Interleukin-9 promotes pancreatic cancer cells proliferation and migration via the miR-200a/Beta-Catenin axis. BioMed Res Int (2017) 2017:2831056. doi: 10.1155/2017/2831056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Zhang Z, Lanz RB, Xiao L, Wang L, Hartig SM, Ittmann MM, et al. The tumor suppressive miR-200b subfamily is an ERG target gene in human prostate tumors. Oncotarget (2016) 7:37993–8003. doi: 10.18632/oncotarget.9366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Guan H, You Z, Wang C, Fang F, Peng R, Mao L, et al. MicroRNA-200a suppresses prostate cancer progression through BRD4/AR signaling pathway. Cancer Med (2019) 8:1474–85. doi: 10.1002/cam4.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Lu R, Ji Z, Li X, Qin J, Cui G, Chen J, et al. Tumor suppressive microRNA-200a inhibits renal cell carcinoma development by directly targeting TGFB2. Tumour Biol (2015) 36:6691–700. doi: 10.1007/s13277-015-3355-9 [DOI] [PubMed] [Google Scholar]
- 236. Fu H, Song W, Chen X, Guo T, Duan B, Wang X, et al. MiRNA-200a induce cell apoptosis in renal cell carcinoma by directly targeting SIRT1. Mol Cell Biochem (2018) 437:143–52. doi: 10.1007/s11010-017-3102-1 [DOI] [PubMed] [Google Scholar]
- 237. Ning F, Zhou Q, Chen X. miR-200b promotes cell proliferation and invasion in t-cell acute lymphoblastic leukemia through NOTCH1. J Biol Regul Homeostatic Agents (2018) 32:1467–71. [PubMed] [Google Scholar]
- 238. Gao R, Zhang N, Yang J, Zhu Y, Zhang Z, Wang J, et al. Long non-coding RNA ZEB1-AS1 regulates miR-200b/FSCN1 signaling and enhances migration and invasion induced by TGF-β1 in bladder cancer cells. J Exp Clin Cancer Res (2019) 38:111. doi: 10.1186/s13046-019-1102-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Yao Y, Hu J, Shen Z, Yao R, Liu S, Li Y, et al. MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med (2015) 19:760–9. doi: 10.1111/jcmm.12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Wu H, Wang G, Wang Z, An S, Ye P, Luo S. A negative feedback loop between miR-200b and the nuclear factor-κB pathway via IKBKB/IKK-β in breast cancer cells. FEBS J (2016) 283:2259–71. doi: 10.1111/febs.13543 [DOI] [PubMed] [Google Scholar]
- 241. Zheng Q, Cui X, Zhang D, Yang Y, Yan X, Liu M, et al. miR-200b inhibits proliferation and metastasis of breast cancer by targeting fucosyltransferase IV and α1,3-fucosylated glycans. Oncogenesis (2017) 6:e358. doi: 10.1038/oncsis.2017.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Yuan J, Xiao C, Lu H, Yu H, Hong H, Guo C, et al. miR-200b regulates breast cancer cell proliferation and invasion by targeting radixin. Exp Ther Med (2020) 19:2741–50. doi: 10.3892/etm.2020.8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Zeng F, Xue M, Xiao T, Li Y, Xiao S, Jiang B, et al. MiR-200b promotes the cell proliferation and metastasis of cervical cancer by inhibiting FOXG1. BioMed Pharmacother (2016) 79:294–301. doi: 10.1016/j.biopha.2016.02.033 [DOI] [PubMed] [Google Scholar]
- 244. Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology (2006) 130:2113–29. doi: 10.1053/j.gastro.2006.02.057 [DOI] [PubMed] [Google Scholar]
- 245. Pan Y, Liang H, Chen W, Zhang H, Wang N, Wang F, et al. microRNA-200b and microRNA-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with kazal motifs. RNA Biol (2015) 12:276–89. doi: 10.1080/15476286.2015.1017208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Chen L, Wang X, Zhu Y, Zhu J, Lai Q. miR−200b−3p inhibits proliferation and induces apoptosis in colorectal cancer by targeting Wnt1. Mol Med Rep (2018) 18:2571–80. doi: 10.3892/mmr.2018.9287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Wu YZ, Lin HY, Zhang Y, Chen WF. miR-200b-3p mitigates oxaliplatin resistance via targeting TUBB3 in colorectal cancer. J Gene Med (2020) 22:e3178. doi: 10.1002/jgm.3178 [DOI] [PubMed] [Google Scholar]
- 248. Kurashige J, Kamohara H, Watanabe M, Hiyoshi Y, Iwatsuki M, Tanaka Y, et al. MicroRNA-200b regulates cell proliferation, invasion, and migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg Oncol (2012) 19 Suppl 3:S656–664. doi: 10.1245/s10434-012-2217-6 [DOI] [PubMed] [Google Scholar]
- 249. Peng B, Hu S, Jun Q, Luo D, Zhang X, Zhao H, et al. MicroRNA-200b targets CREB1 and suppresses cell growth in human malignant glioma. Mol Cell Biochem (2013) 379:51–8. doi: 10.1007/s11010-013-1626-6 [DOI] [PubMed] [Google Scholar]
- 250. Wu J, Cui H, Zhu Z, Wang L. MicroRNA-200b-3p suppresses epithelial-mesenchymal transition and inhibits tumor growth of glioma through down-regulation of ERK5. Biochem Biophys Res Commun (2016) 478:1158–64. doi: 10.1016/j.bbrc.2016.08.085 [DOI] [PubMed] [Google Scholar]
- 251. Zhao C, Ma ZG, Mou SL, Yang YX, Zhang YH, Yao WC. Targeting effect of microRNA on CD133 and its impact analysis on proliferation and invasion of glioma cells. Genet Mol Res (2017) 16. doi: 10.4238/gmr16019281 [DOI] [PubMed] [Google Scholar]
- 252. Yang X, Ni W, Lei K. miR-200b suppresses cell growth, migration and invasion by targeting Notch1 in nasopharyngeal carcinoma. Cell Physiol Biochem (2013) 32:1288–98. doi: 10.1159/000354527 [DOI] [PubMed] [Google Scholar]
- 253. Zhao H, Xing G, Wang Y, Luo Z, Liu G, Meng H. Long noncoding RNA HEIH promotes melanoma cell proliferation, migration and invasion via inhibition of miR-200b/a/429. Biosci Rep (2017) 37. doi: 10.1042/BSR20170682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Chen DQ, Huang JY, Feng B, Pan BZ, De W, Wang R, et al. Histone deacetylase 1/Sp1/microRNA-200b signaling accounts for maintenance of cancer stem-like cells in human lung adenocarcinoma. PloS One (2014) 9:e109578. doi: 10.1371/journal.pone.0109578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Tang Q, Li M, Chen L, Bi F, Xia H. miR-200b/c targets the expression of RhoE and inhibits the proliferation and invasion of non-small cell lung cancer cells. Int J Oncol (2018) 53:1732–42. doi: 10.3892/ijo.2018.4493 [DOI] [PubMed] [Google Scholar]
- 256. Jin HF, Wang JF, Song TT, Zhang J, Wang L. MiR-200b inhibits tumor growth and chemoresistance via targeting p70S6K1 in lung cancer. Front Oncol (2020) 10:643. doi: 10.3389/fonc.2020.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Ren W, Gao L, Qiang C, Li S, Zheng J, Wang Q, et al. Kindlin-2-mediated upregulation of ZEB2 facilitates migration and invasion of oral squamous cell carcinoma in a miR-200b-dependent manner. Am J Transl Res (2018) 10:2529–41. [PMC free article] [PubMed] [Google Scholar]
- 258. Guan W, Cui H, Huang P, Chun WJ, Lee JW, Kim H, et al. miR-200b/200a/429 cluster stimulates ovarian cancer development by targeting ING5. J Oncol (2020) 2020:3404059. doi: 10.1155/2020/3404059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Wang AQ, Lv M, Xu YH, Xie PM, Dong YY. MiR-200b-5p inhibits proliferation of ovarian cancer cells by targeting ATAD2 and regulating PI3K/AKT signaling pathway. Eur Rev Med Pharmacol Sci (2020) 24:9860–8. doi: 10.26355/eurrev_202010_23196 [DOI] [PubMed] [Google Scholar]
- 260. He M, Liu Y, Deng X, Qi S, Sun X, Liu G, et al. Down-regulation of miR-200b-3p by low p73 contributes to the androgen-independence of prostate cancer cells. Prostate (2013) 73:1048–56. doi: 10.1002/pros.22652 [DOI] [PubMed] [Google Scholar]
- 261. Williams LV, Veliceasa D, Vinokour E, Volpert OV. miR-200b inhibits prostate cancer EMT, growth and metastasis. PloS One (2013) 8:e83991. doi: 10.1371/journal.pone.0083991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Yu J, Lu Y, Cui D, Li E, Zhu Y, Zhao Y, et al. miR-200b suppresses cell proliferation, migration and enhances chemosensitivity in prostate cancer by regulating bmi-1. Oncol Rep (2014) 31:910–8. doi: 10.3892/or.2013.2897 [DOI] [PubMed] [Google Scholar]
- 263. Yuan D, Zheng S, Wang L, Li J, Yang J, Wang B, et al. MiR-200c inhibits bladder cancer progression by targeting lactate dehydrogenase a. Oncotarget (2017) 8:67663–9. doi: 10.18632/oncotarget.18801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Ren Y, Han X, Yu K, Sun S, Zhen L, Li Z, et al. microRNA-200c downregulates XIAP expression to suppress proliferation and promote apoptosis of triple-negative breast cancer cells. Mol Med Rep (2014) 10:315–21. doi: 10.3892/mmr.2014.2222 [DOI] [PubMed] [Google Scholar]
- 265. Song C, Liu LZ, Pei XQ, Liu X, Yang L, Ye F, et al. miR-200c inhibits breast cancer proliferation by. Oncotarget (2015) 6:34968–78. doi: 10.18632/oncotarget.5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Wu D, Ji N, Zhang L, Zhang L. Differential expression of miR-200c in breast cancer stem cells. Int J Clin Exp Pathol (2017) 10:10085–91. [PMC free article] [PubMed] [Google Scholar]
- 267. Zhang J, Li G, Chen Y, Fang L, Guan C, Bai F, et al. Metformin inhibits tumorigenesis and tumor growth of breast cancer cells by upregulating miR-200c but downregulating AKT2 expression. J Cancer (2017) 8:1849–64. doi: 10.7150/jca.19858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Yuan X, Liu J, Ye X. Effect of miR-200c on the proliferation, migration and invasion of breast cancer cells and relevant mechanisms. J buon (2019) 24:61–7. [PubMed] [Google Scholar]
- 269. Zhang DD, Li Y, Xu Y, Kim J, Huang S. Phosphodiesterase 7B/microRNA-200c relationship regulates triple-negative breast cancer cell growth. Oncogene (2019) 38:1106–20. doi: 10.1038/s41388-018-0499-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Mei J, Wang DH, Wang LL, Chen Q, Pan LL, Xia L. MicroRNA-200c suppressed cervical cancer cell metastasis and growth via targeting MAP4K4. Eur Rev Med Pharmacol Sci (2018) 22:623–31. doi: 10.26355/eurrev_201802_14286 [DOI] [PubMed] [Google Scholar]
- 271. Gao Z, Zhou H, Wang Y, Chen J, Ou Y. Regulatory effects of lncRNA ATB targeting miR-200c on proliferation and apoptosis of colorectal cancer cells. J Cell Biochem (2020) 121:332–43. doi: 10.1002/jcb.29180 [DOI] [PubMed] [Google Scholar]
- 272. Li Q, Zhang C, Chen R, Xiong H, Qiu F, Liu S, et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett (2016) 383:28–40. doi: 10.1016/j.canlet.2016.09.019 [DOI] [PubMed] [Google Scholar]
- 273. Chen R, Zhang M, Liu W, Chen H, Cai T, Xiong H, et al. Estrogen affects the negative feedback loop of PTENP1-miR200c to inhibit PTEN expression in the development of endometrioid endometrial carcinoma. Cell Death Dis (2018) 10:4. doi: 10.1038/s41419-018-1207-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Zhang H, Sun Z, Li Y, Fan D, Jiang H. MicroRNA-200c binding to FN1 suppresses the proliferation, migration and invasion of gastric cancer cells. Biomed Pharmacother (2017) 88:285–92. doi: 10.1016/j.biopha.2017.01.023 [DOI] [PubMed] [Google Scholar]
- 275. Wei W, Shi L, Chen W, Hu L, Chen D, Shi X, et al. miR-200c regulates the proliferation, apoptosis and invasion of gastric carcinoma cells through the downregulation of EDNRA expression. Int J Mol Med (2018) 41:1619–26. doi: 10.3892/ijmm.2017.3317 [DOI] [PubMed] [Google Scholar]
- 276. Chen P, Guo X, Zhang L, Zhang W, Zhou Q, Tian Z, et al. MiR-200c is a cMyc-activated miRNA that promotes nasopharyngeal carcinoma by downregulating PTEN. Oncotarget (2017) 8:5206–18. doi: 10.18632/oncotarget.14123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Liu S, Tetzlaff MT, Cui R, Xu X. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am J Pathol (2012) 181:1823–35. doi: 10.1016/j.ajpath.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Zhao GZ, Niu YQ, Li ZM, Kou D, Zhang L. MiR-200c inhibits proliferation and promotes apoptosis of wilms tumor cells by regulating akt signaling pathway. Eur Rev Med Pharmacol Sci (2020) 24:6623–31. doi: 10.26355/eurrev_202006_21648 [DOI] [PubMed] [Google Scholar]
- 279. Bai T, Dong DS, Pei L. Synergistic antitumor activity of resveratrol and miR-200c in human lung cancer. Oncol Rep (2014) 31:2293–7. doi: 10.3892/or.2014.3090 [DOI] [PubMed] [Google Scholar]
- 280. Lei W, Kang W, Nan Y, Lei Z, Zhongdong L, Demin L, et al. The downregulation of miR-200c promotes lactate dehydrogenase a expression and non-small cell lung cancer progression. Oncol Res (2018) 26:1015–22. doi: 10.3727/096504018X15151486241153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Ibrahim FF, Jamal R, Syafruddin SE, Ab Mutalib NS, Saidin S, Mdzin RR, et al. MicroRNA-200c and microRNA-31 regulate proliferation, colony formation, migration and invasion in serous ovarian cancer. J Ovarian Res (2015) 8:56. doi: 10.1186/s13048-015-0186-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Yang C, Li H, Zhang T, Chu Y, Chen D, Zuo J. miR-200c overexpression inhibits the invasion and tumorigenicity of epithelial ovarian cancer cells by suppressing lncRNA HOTAIR in mice. J Cell Biochem (2020) 121:1514–23. doi: 10.1002/jcb.29387 [DOI] [PubMed] [Google Scholar]
- 283. Yu J, Ohuchida K, Mizumoto K, Sato N, Kayashima T, Fujita H, et al. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol Cancer (2010) 9:169. doi: 10.1186/1476-4598-9-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Shi R, Xiao H, Yang T, Chang L, Tian Y, Wu B, et al. Effects of miR-200c on the migration and invasion abilities of human prostate cancer Du145 cells and the corresponding mechanism. Front Med (2014) 8:456–63. doi: 10.1007/s11684-014-0353-z [DOI] [PubMed] [Google Scholar]
- 285. Zhang J, Zhang H, Qin Y, Chen C, Yang J, Song N, et al. MicroRNA-200c-3p/ZEB2 loop plays a crucial role in the tumor progression of prostate carcinoma. Ann Transl Med (2019) 7:141. doi: 10.21037/atm.2019.02.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Bao J, Li X, Li Y, Huang C, Meng X, Li J. MicroRNA-141-5p acts as a tumor suppressor via targeting RAB32 in chronic myeloid leukemia. Front Pharmacol (2020) 10. doi: 10.3389/fphar.2019.01545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Li P, Xu T, Zhou X, Liao L, Pang G, Luo W, et al. Downregulation of miRNA-141 in breast cancer cells is associated with cell migration and invasion: involvement of ANP32E targeting. Cancer Med (2017) 6:662–72. doi: 10.1002/cam4.1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Sun S, Ma J, Xie P, Wu Z, Tian X. Hypoxia-responsive miR-141-3p is involved in the progression of breast cancer via mediating the HMGB1/HIF-1α signaling pathway. J Gene Med (2020) 22:e3230. doi: 10.1002/jgm.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Li JH, Zhang Z, Du MZ, Guan YC, Yao JN, Yu HY, et al. microRNA-141-3p fosters the growth, invasion, and tumorigenesis of cervical cancer cells by targeting FOXA2. Arch Biochem Biophys (2018) 657:23–30. doi: 10.1016/j.abb.2018.09.008 [DOI] [PubMed] [Google Scholar]
- 290. Long ZH, Bai ZG, Song JN, Zheng Z, Li J, Zhang J, et al. miR-141 inhibits proliferation and migration of colorectal cancer SW480 cells. Anticancer Res (2017) 37:4345–52. doi: 10.21873/anticanres.11828 [DOI] [PubMed] [Google Scholar]
- 291. Ye J, Wang Z, Zhao J, Chen W, Wu D, Wu P, et al. MicroRNA-141 inhibits tumor growth and minimizes therapy resistance in colorectal cancer. Mol Med Rep (2017) 15:1037–42. doi: 10.3892/mmr.2017.6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Zuo QF, Zhang R, Li BS, Zhao YL, Zhuang Y, Yu T, et al. MicroRNA-141 inhibits tumor growth and metastasis in gastric cancer by directly targeting transcriptional co-activator with PDZ-binding motif, TAZ. Cell Death Dis (2015) 6:e1623. doi: 10.1038/cddis.2014.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Wang Z, Lei H, Sun Q. MicroRNA-141 and its associated gene FUS modulate proliferation, migration and cisplatin chemosensitivity in neuroblastoma cell lines. Oncol Rep (2016) 35:2943–51. doi: 10.3892/or.2016.4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Yao Y, Luo J, Sun Q, Xu T, Sun S, Chen M, et al. HOXC13 promotes proliferation of lung adenocarcinoma via modulation of CCND1 and CCNE1. Am J Cancer Res (2017) 7:1820–34. [PMC free article] [PubMed] [Google Scholar]
- 295. Kong YJ, Tan XX, Zhang Y, He QJ, Zhao L, Meng Q. MiR-141 promotes cell proliferation and invasion in non-small cell lung cancer by targeting KLF9. Eur Rev Med Pharmacol Sci (2019) 23:10370–8. doi: 10.26355/eurrev_201912_19676 [DOI] [PubMed] [Google Scholar]
- 296. Wang L. MiR-141-3p overexpression suppresses the malignancy of osteosarcoma by targeting FUS to degrade LDHB. Biosci Rep (2020) 40. doi: 10.1042/BSR20193404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Ye Q, Lei L, Shao L, Shi J, Jia J, Tong X. MicroRNA−141 inhibits epithelial−mesenchymal transition, and ovarian cancer cell migration and invasion. Mol Med Rep (2017) 16:6743–9. doi: 10.3892/mmr.2017.7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Zhao G, Wang B, Liu Y, Zhang JG, Deng SC, Qin Q, et al. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Mol Cancer Ther (2013) 12:2569–80. doi: 10.1158/1535-7163.MCT-13-0296 [DOI] [PubMed] [Google Scholar]
- 299. Li JZ, Li J, Wang HQ, Li X, Wen B, Wang YJ. MiR-141-3p promotes prostate cancer cell proliferation through inhibiting kruppel-like factor-9 expression. Biochem Biophys Res Commun (2017) 482:1381–6. doi: 10.1016/j.bbrc.2016.12.045 [DOI] [PubMed] [Google Scholar]
- 300. Xu S, Ge J, Zhang Z, Zhou W. miR-141 inhibits prostatic cancer cell proliferation and migration, and induces cell apoptosis via targeting of RUNX1. Oncol Rep (2018) 39:1454–60. doi: 10.3892/or.2018.6209 [DOI] [PubMed] [Google Scholar]
- 301. Zhang L, Liu Q, Mu Q, Zhou D, Li H, Zhang B, et al. MiR-429 suppresses proliferation and invasion of breast cancer via inhibiting the wnt/β-catenin signaling pathway. Thorac Cancer (2020) 11:3126–38. doi: 10.1111/1759-7714.13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Wang Y, Dong X, Hu B, Wang X-J, Wang Q, Wang W-L. The effects of micro-429 on inhibition of cervical cancer cells through targeting ZEB1 and CRKL. Biomed Pharmacother (2016) 80:311–21. doi: 10.1016/j.biopha.2016.03.035 [DOI] [PubMed] [Google Scholar]
- 303. Liu D, Xia P, Diao D, Cheng Y, Zhang H, Yuan D, et al. MiRNA-429 suppresses the growth of gastric cancer cells in vitro . J BioMed Res (2012) 26:389–93. doi: 10.7555/JBR.26.20120029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Wang Z, Zhu Z, Lin Z, Luo Y, Liang Z, Zhang C, et al. miR-429 suppresses cell proliferation, migration and invasion in nasopharyngeal carcinoma by downregulation of TLN1. Cancer Cell Int (2019) 19:115. doi: 10.1186/s12935-019-0831-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Xue H, Tian GY. MiR-429 regulates the metastasis and EMT of HCC cells through targeting RAB23. Arch Biochem Biophys (2018) 637:48–55. doi: 10.1016/j.abb.2017.11.011 [DOI] [PubMed] [Google Scholar]
- 306. Huang D, Wang F, Wu W, Lian C, Liu E. MicroRNA-429 inhibits cancer cell proliferation and migration by targeting the AKT1 in melanoma. Cancer biomark (2019) 26:63–8. doi: 10.3233/CBM-190289 [DOI] [PubMed] [Google Scholar]
- 307. Zhou X, Lu H, Li F, Hao X, Han L, Dong Q, et al. MicroRNA-429 inhibits neuroblastoma cell proliferation, migration and invasion via the NF-κB pathway. Cell Mol Biol Lett (2020) 25:5. doi: 10.1186/s11658-020-0202-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Lang Y, Xu S, Ma J, Wu J, Jin S, Cao S, et al. MicroRNA-429 induces tumorigenesis of human non-small cell lung cancer cells and targets multiple tumor suppressor genes. Biochem Biophys Res Commun (2014) 450:154–9. doi: 10.1016/j.bbrc.2014.05.084 [DOI] [PubMed] [Google Scholar]
- 309. Xiao P, Liu W, Zhou H. miR-429 promotes the proliferation of non−small cell lung cancer cells via targeting DLC-1 retraction in /10.3892/ol.2021.12806. Oncol Lett (2016) 12:2163–8. doi: 10.3892/ol.2016.4904 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 310. Lei W, Liu YE, Zheng Y, Qu L. MiR-429 inhibits oral squamous cell carcinoma growth by targeting ZEB1. Med Sci Monit (2015) 21:383–9. doi: 10.12659/MSM.893412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Ouyang Y, Gao P, Zhu B, Chen X, Lin F, Wang X, et al. Downregulation of microRNA-429 inhibits cell proliferation by targeting p27Kip1 in human prostate cancer cells. Mol Med Rep (2015) 11:1435–41. doi: 10.3892/mmr.2014.2782 [DOI] [PubMed] [Google Scholar]
- 312. Su Z, Jiang G, Chen J, Liu X, Zhao H, Fang Z, et al. MicroRNA-429 inhibits cancer cell proliferation and migration by targeting AKT1 in renal cell carcinoma. Mol Clin Oncol (2020) 12:75–80. doi: 10.3892/mco.2020.2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh Migr (2015) 9:317–24. doi: 10.1080/19336918.2015.1016686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets (2008) 8:187–98. doi: 10.2174/156800908784293659 [DOI] [PubMed] [Google Scholar]
- 315. Tsouko E, Wang J, Frigo DE, Aydoğdu E, Williams C. miR-200a inhibits migration of triple-negative breast cancer cells through direct repression of the EPHA2 oncogene. Carcinogenesis (2015) 36:1051–60. doi: 10.1093/carcin/bgv087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Kim HK, Park JD, Choi SH, Shin DJ, Hwang S, Jung HY, et al. Functional link between miR-200a and ELK3 regulates the metastatic nature of breast cancer. Cancers (Basel) (2020) 12. doi: 10.3390/cancers12051225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Lu Y, Lu J, Li X, Zhu H, Fan X, Zhu S, et al. MiR-200a inhibits epithelial-mesenchymal transition of pancreatic cancer stem cell. BMC Cancer (2014) 14:85. doi: 10.1186/1471-2407-14-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Humphries B, Wang Z, Oom AL, Fisher T, Tan D, Cui Y, et al. MicroRNA-200b targets protein kinase cα and suppresses triple-negative breast cancer metastasis. Carcinogenesis (2014) 35:2254–63. doi: 10.1093/carcin/bgu133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Hong H, Yu H, Yuan J, Guo C, Cao H, Li W, et al. MicroRNA-200b impacts breast cancer cell migration and invasion by regulating ezrin-Radixin-Moesin. Med Sci Monit (2016) 22:1946–52. doi: 10.12659/MSM.896551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Cheng YX, Chen GT, Chen C, Zhang QF, Pan F, Hu M, et al. MicroRNA-200b inhibits epithelial-mesenchymal transition and migration of cervical cancer cells by directly targeting RhoE. Mol Med Rep (2016) 13:3139–46. doi: 10.3892/mmr.2016.4933 [DOI] [PubMed] [Google Scholar]
- 321. Dai Y, Xia W, Song T, Su X, Li J, Li S, et al. MicroRNA-200b is overexpressed in endometrial adenocarcinomas and enhances MMP2 activity by downregulating TIMP2 in human endometrial cancer cell line HEC-1A cells. Nucleic Acid Ther (2013) 23:29–34. doi: 10.1089/nat.2012.0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Zhang H-F, Zhang K, Liao L-D, Li L-Y, Du Z-P, Wu B-L, et al. miR-200b suppresses invasiveness and modulates the cytoskeletal and adhesive machinery in esophageal squamous cell carcinoma cells via targeting kindlin-2. Carcinogenesis (2013) 35:292–301. doi: 10.1093/carcin/bgt320 [DOI] [PubMed] [Google Scholar]
- 323. Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer (2018) 17:140. doi: 10.1186/s12943-018-0889-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Liu K, Zhang W, Tan J, Ma J, Zhao J. MiR-200b-3p functions as an oncogene by targeting ABCA1 in lung adenocarcinoma. Technol Cancer Res Treat (2019) 18:1533033819892590. doi: 10.1177/1533033819892590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Tamagawa S, Enomoto K, Gunduz E, Gunduz M, Sato F, Uchino S, et al. MicroRNA 200b promotes mesenchymal-to-epithelial transition in anaplastic thyroid carcinoma. Oncol Lett (2020) 20:3. doi: 10.3892/ol.2020.11864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Zhang T, Wan JG, Liu JB, Deng M. MiR-200c inhibits metastasis of breast tumor via the downregulation of Foxf2. Genet Mol Res (2017) 16. doi: 10.4238/gmr16038971 [DOI] [PubMed] [Google Scholar]
- 327. Chen ML, Liang LS, Wang XK. miR-200c inhibits invasion and migration in human colon cancer cells SW480/620 by targeting ZEB1. Clin Exp Metastasis (2012) 29:457–69. doi: 10.1007/s10585-012-9463-7 [DOI] [PubMed] [Google Scholar]
- 328. Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther (2009) 8:1055–66. doi: 10.1158/1535-7163.MCT-08-1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Xie NN, Liu ZX, Wu C, Wang PL, Song GT, Chen Z. MicroRNA-200c suppresses tumor metastasis in oral squamous carcinoma by inhibiting epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci (2018) 22:3415–22. doi: 10.26355/eurrev_201806_15164 [DOI] [PubMed] [Google Scholar]
- 330. Ma C, Huang T, Ding YC, Yu W, Wang Q, Meng B, et al. MicroRNA-200c overexpression inhibits chemoresistance, invasion and colony formation of human pancreatic cancer stem cells. Int J Clin Exp Pathol (2015) 8:6533–9. [PMC free article] [PubMed] [Google Scholar]
- 331. Zhang Y, Li J, Jia S, Wang Y, Kang Y, Zhang W. Down-regulation of lncRNA-ATB inhibits epithelial-mesenchymal transition of breast cancer cells by increasing miR-141-3p expression. Biochem Cell Biol (2019) 97:193–200. doi: 10.1139/bcb-2018-0168 [DOI] [PubMed] [Google Scholar]
- 332. Xing Y, Jing H, Zhang Y, Suo J, Qian M. MicroRNA-141-3p affected proliferation, chemosensitivity, migration and invasion of colorectal cancer cells by targeting EGFR. Int J Biochem Cell Biol (2020) 118:105643. doi: 10.1016/j.biocel.2019.105643 [DOI] [PubMed] [Google Scholar]
- 333. Xu L, Li Q, Xu D, Wang Q, An Y, Du Q, et al. Hsa-miR-141 downregulates TM4SF1 to inhibit pancreatic cancer cell invasion and migration. Int J Oncol (2014) 44:459–66. doi: 10.3892/ijo.2013.2189 [DOI] [PubMed] [Google Scholar]
- 334. Wu CL, Ho JY, Chou SC, Yu DS. MiR-429 reverses epithelial-mesenchymal transition by restoring e-cadherin expression in bladder cancer. Oncotarget (2016) 7:26593–603. doi: 10.18632/oncotarget.8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Tian X, Wei Z, Wang J, Liu P, Qin Y, Zhong M. MicroRNA-429 inhibits the migration and invasion of colon cancer cells by targeting PAK6/cofilin signaling. Oncol Rep (2015) 34:707–14. doi: 10.3892/or.2015.4039 [DOI] [PubMed] [Google Scholar]
- 336. Ni J, Yang Y, Liu D, Sun H, Jin S, Li J. MicroRNA-429 inhibits gastric cancer migration and invasion through the downregulation of specificity protein 1. Oncol Lett (2017) 13:3845–9. doi: 10.3892/ol.2017.5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Hang Q, Lu J, Zuo L, Liu M. Linc00641 promotes the progression of gastric carcinoma by modulating the miR-429/Notch-1 axis. Aging (Albany NY) (2021) 13:8497–509. doi: 10.18632/aging.202661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Chen W, Zhang B, Guo W, Gao L, Shi L, Li H, et al. miR-429 inhibits glioma invasion through BMK1 suppression. J Neurooncol (2015) 125:43–54. doi: 10.1007/s11060-015-1887-x [DOI] [PubMed] [Google Scholar]
- 339. Chen H, Xia B, Liu T, Lin M, Lou G. KIAA0101, a target gene of miR-429, enhances migration and chemoresistance of epithelial ovarian cancer cells. Cancer Cell Int (2016) 16:74. doi: 10.1186/s12935-016-0353-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev (2009) 28:15–33. doi: 10.1007/s10555-008-9169-0 [DOI] [PubMed] [Google Scholar]
- 341. Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther (2010) 10:219–22. doi: 10.4161/cbt.10.3.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep (2008) 9:582–9. doi: 10.1038/embor.2008.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Caswell PT, Zech T. Actin-based cell protrusion in a 3D matrix. Trends Cell Biol (2018) 28:823–34. doi: 10.1016/j.tcb.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Adada M, Canals D, Hannun YA, Obeid LM. Sphingolipid regulation of ezrin, radixin, and moesin proteins family: implications for cell dynamics. Biochim Biophys Acta (2014) 1841:727–37. doi: 10.1016/j.bbalip.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Chambers AF, Groom AC, Macdonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer (2002) 2:563–72. doi: 10.1038/nrc865 [DOI] [PubMed] [Google Scholar]
- 346. Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis (2000) 21:485–95. doi: 10.1093/carcin/21.3.485 [DOI] [PubMed] [Google Scholar]
- 347. Wong RSY. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res (2011) 30:87. doi: 10.1186/1756-9966-30-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc (2009) 84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x [DOI] [PubMed] [Google Scholar]
- 349. Guo T, Zhang Y, Qu X, Che X, Li C, Fan Y, et al. miR-200a enhances TRAIL-induced apoptosis in gastric cancer cells by targeting A20. Cell Biol Int (2018) 42:506–14. doi: 10.1002/cbin.10924 [DOI] [PubMed] [Google Scholar]
- 350. Liang X-L, Wang Y-L, Wang P-R. MiR-200a with CDC7 as a direct target declines cell viability and promotes cell apoptosis in wilm’s tumor via wnt/β-catenin signaling pathway. Mol Cell Biochem (2021) 476:2409–20. doi: 10.1007/s11010-021-04090-9 [DOI] [PubMed] [Google Scholar]
- 351. Wang T, Tang X, Liu Y. LncRNA-ATB promotes apoptosis of non-small cell lung cancer cells through MiR-200a/β-Catenin. J buon (2019) 24:2280–6. [PubMed] [Google Scholar]
- 352. Zhang HF, Alshareef A, Wu C, Jiao JW, Sorensen PH, Lai R, et al. miR-200b induces cell cycle arrest and represses cell growth in esophageal squamous cell carcinoma. Carcinogenesis (2016) 37:858–69. doi: 10.1093/carcin/bgw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Zhu P, Zhang J, Zhu J, Shi J, Zhu Q, Gao Y. MiR-429 induces gastric carcinoma cell apoptosis through bcl-2. Cell Physiol Biochem (2015) 37:1572–80. doi: 10.1159/000438524 [DOI] [PubMed] [Google Scholar]
- 354. Caporali A, Emanueli C. MicroRNA regulation in angiogenesis. Vascul Pharmacol (2011) 55:79–86. doi: 10.1016/j.vph.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 355. Dong H, Weng C, Bai R, Sheng J, Gao X, Li L, et al. The regulatory network of miR-141 in the inhibition of angiogenesis. Angiogenesis (2019) 22:251–62. doi: 10.1007/s10456-018-9654-1 [DOI] [PubMed] [Google Scholar]
- 356. Li L, Li B, Chen D, Liu L, Huang C, Lu Z, et al. miR-139 and miR-200c regulate pancreatic cancer endothelial cell migration and angiogenesis. Oncol Rep (2015) 34:51–8. doi: 10.3892/or.2015.3945 [DOI] [PubMed] [Google Scholar]
- 357. Masoumi-Dehghi S, Babashah S, Sadeghizadeh M. microRNA-141-3p-containing small extracellular vesicles derived from epithelial ovarian cancer cells promote endothelial cell angiogenesis through activating the JAK/STAT3 and NF-κB signaling pathways. J Cell Commun Signal (2020) 14:233–44. doi: 10.1007/s12079-020-00548-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem (2011) 286:2047–56. doi: 10.1074/jbc.M110.158790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther (2010) 17:523–31. doi: 10.1038/cgt.2010.18 [DOI] [PubMed] [Google Scholar]
- 360. Yu SJ, Yang L, Hong Q, Kuang XY, Di GH, Shao ZM. MicroRNA-200a confers chemoresistance by antagonizing TP53INP1 and YAP1 in human breast cancer. BMC Cancer (2018) 18:74. doi: 10.1186/s12885-017-3930-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Lee H, Kim C, Kang H, Tak H, Ahn S, Yoon SK, et al. microRNA-200a-3p increases 5-fluorouracil resistance by regulating dual specificity phosphatase 6 expression. Exp Mol Med (2017) 49:e327. doi: 10.1038/emm.2017.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Feng B, Wang R, Song HZ, Chen LB. MicroRNA-200b reverses chemoresistance of docetaxel-resistant human lung adenocarcinoma cells by targeting E2F3. Cancer (2012) 118:3365–76. doi: 10.1002/cncr.26560 [DOI] [PubMed] [Google Scholar]
- 363. Chen DQ, Pan BZ, Huang JY, Zhang K, Cui SY, De W, et al. HDAC 1/4-mediated silencing of microRNA-200b promotes chemoresistance in human lung adenocarcinoma cells. Oncotarget (2014) 5:3333–49. doi: 10.18632/oncotarget.1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Pan B, Feng B, Chen Y, Huang G, Wang R, Chen L, et al. MiR-200b regulates autophagy associated with chemoresistance in human lung adenocarcinoma. Oncotarget (2015) 6:32805–20. doi: 10.18632/oncotarget.5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Nishijima N, Seike M, Soeno C, Chiba M, Miyanaga A, Noro R, et al. miR-200/ZEB axis regulates sensitivity to nintedanib in non-small cell lung cancer cells. Int J Oncol (2016) 48:937–44. doi: 10.3892/ijo.2016.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Fang S, Zeng X, Zhu W, Tang R, Chao Y, Guo L. Zinc finger e-box-binding homeobox 2 (ZEB2) regulated by miR-200b contributes to multi-drug resistance of small cell lung cancer. Exp Mol Pathol (2014) 96:438–44. doi: 10.1016/j.yexmp.2014.04.008 [DOI] [PubMed] [Google Scholar]
- 367. Liu J, Zhang X, Huang Y, Zhang Q, Zhou J, Zhang X, et al. miR-200b and miR-200c co-contribute to the cisplatin sensitivity of ovarian cancer cells by targeting DNA methyltransferases. Oncol Lett (2019) 17:1453–60. doi: 10.3892/ol.2018.9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Tang H, Song C, Ye F, Gao G, Ou X, Zhang L, et al. miR-200c suppresses stemness and increases cellular sensitivity to trastuzumab in HER2+ breast cancer. J Cell Mol Med (2019) 23:8114–27. doi: 10.1111/jcmm.14681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Jiang T, Dong P, Li L, Ma X, Xu P, Zhu H, et al. MicroRNA-200c regulates cisplatin resistance by targeting ZEB2 in human gastric cancer cells. Oncol Rep (2017) 38:151–8. doi: 10.3892/or.2017.5659 [DOI] [PubMed] [Google Scholar]
- 370. Hamano R, Miyata H, Yamasaki M, Kurokawa Y, Hara J, Moon JH, et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the akt signaling pathway. Clin Cancer Res (2011) 17:3029–38. doi: 10.1158/1078-0432.CCR-10-2532 [DOI] [PubMed] [Google Scholar]
- 371. Yao YS, Qiu WS, Yao RY, Zhang Q, Zhuang LK, Zhou F, et al. miR-141 confers docetaxel chemoresistance of breast cancer cells via regulation of EIF4E expression. Oncol Rep (2015) 33:2504–12. doi: 10.3892/or.2015.3866 [DOI] [PubMed] [Google Scholar]
- 372. Zhou X, Wu W, Zeng A, Nie E, Jin X, Yu T, et al. MicroRNA-141-3p promotes glioma cell growth and temozolomide resistance by directly targeting p53. Oncotarget (2017) 8:71080–94. doi: 10.18632/oncotarget.20528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Fu WF, Chen WB, Dai L, Yang GP, Jiang ZY, Pan L, et al. Inhibition of miR-141 reverses cisplatin resistance in non-small cell lung cancer cells via upregulation of programmed cell death protein 4. Eur Rev Med Pharmacol Sci (2016) 20:2565–72. [PubMed] [Google Scholar]
- 374. Van Jaarsveld MT, Helleman J, Boersma AW, Van Kuijk PF, Van Ijcken WF, Despierre E, et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene (2013) 32:4284–93. doi: 10.1038/onc.2012.433 [DOI] [PubMed] [Google Scholar]
- 375. Brozovic A, Duran GE, Wang YC, Francisco EB, Sikic BI. The miR-200 family differentially regulates sensitivity to paclitaxel and carboplatin in human ovarian carcinoma OVCAR-3 and MES-OV cells. Mol Oncol (2015) 9:1678–93. doi: 10.1016/j.molonc.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Cheng G, Li Y, Liu Z, Song X. The microRNA-429/DUSP4 axis regulates the sensitivity of colorectal cancer cells to nintedanib. Mol Med Rep (2021) 23. doi: 10.3892/mmr.2021.11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Zou J, Liu L, Wang Q, Yin F, Yang Z, Zhang W, et al. Downregulation of miR-429 contributes to the development of drug resistance in epithelial ovarian cancer by targeting ZEB1. Am J Transl Res (2017) 9:1357–68. [PMC free article] [PubMed] [Google Scholar]
- 378. Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res (2020) 10:727–42. [PMC free article] [PubMed] [Google Scholar]
- 379. Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn Y-H, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun (2014) 5:5241. doi: 10.1038/ncomms6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380. Lv J, Guo T, Qu X, Che X, Li C, Wang S, et al. PD-L1 under regulation of miR-429 influences the sensitivity of gastric cancer cells to TRAIL by binding of EGFR. Front Oncol (2020) 10. doi: 10.3389/fonc.2020.01067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Mei S, Xin J, Liu Y, Zhang Y, Liang X, Su X, et al. MicroRNA-200c promotes suppressive potential of myeloid-derived suppressor cells by modulating PTEN and FOG2 expression. PloS One (2015) 10:e0135867. doi: 10.1371/journal.pone.0135867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. Grzywa TM, Sosnowska A, Matryba P, Rydzynska Z, Jasinski M, Nowis D, et al. Myeloid cell-derived arginase in cancer immune response. Front Immunol (2020) 11:938. doi: 10.3389/fimmu.2020.00938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383. Williams MM, Christenson JL, O’neill KI, Hafeez SA, Ihle CL, Spoelstra NS, et al. MicroRNA-200c restoration reveals a cytokine profile to enhance M1 macrophage polarization in breast cancer. NPJ Breast Cancer (2021) 7:64. doi: 10.1038/s41523-021-00273-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384. Li Y, Tong Y, Liu J, Lou J. The role of MicroRNA in DNA damage response. Front Genet (2022) 13. doi: 10.3389/fgene.2022.850038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385. Tinaburri L, D’errico M, Sileno S, Maurelli R, Degan P, Magenta A, et al. miR-200a modulates the expression of the DNA repair protein OGG1 playing a role in aging of primary human keratinocytes. Oxid Med Cell Longevity (2018) 2018:9147326. doi: 10.1155/2018/9147326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Hu S, Jiang Q, Luo D, Zhao L, Fu X, Chen Y, et al. miR-200b is a key regulator of tumor progression and metabolism targeting lactate dehydrogenase a in human malignant glioma. Oncotarget (2016) 7:48423–31. doi: 10.18632/oncotarget.10301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Gollavilli PN, Parma B, Siddiqui A, Yang H, Ramesh V, Napoli F, et al. The role of miR-200b/c in balancing EMT and proliferation revealed by an activity reporter. Oncogene (2021) 40:2309–22. doi: 10.1038/s41388-021-01708-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388. Yu L, Cao C, Li X, Zhang M, Gu Q, Gao H, et al. Complete loss of miR-200 family induces EMT associated cellular senescence in gastric cancer. Oncogene (2022) 41:26–36. doi: 10.1038/s41388-021-02067-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Allegra A, Musolino C, Tonacci A, Pioggia G, Gangemi S. Interactions between the MicroRNAs and microbiota in cancer development: Roles and therapeutic opportunities. Cancers (Basel) (2020) 12. doi: 10.3390/cancers12040805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Xing J, Liao Y, Zhang H, Zhang W, Zhang Z, Zhang J, et al. Impacts of MicroRNAs induced by the gut microbiome on regulating the development of colorectal cancer. Front Cell Infect Microbiol (2022) 12. doi: 10.3389/fcimb.2022.804689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391. Jiang Y, Ji X, Liu K, Shi Y, Wang C, Li Y, et al. Exosomal miR-200c-3p negatively regulates the migration and invasion of lipopolysaccharide (LPS)-stimulated colorectal cancer (CRC). BMC Mol Cell Biol (2020) 21:48. doi: 10.1186/s12860-020-00291-0 [DOI] [PMC free article] [PubMed] [Google Scholar]