Abstract
In Treponema denticola, a ribbon-like structure of cytoplasmic filaments spans the cytoplasm at all stages of the cell division process. Insertional inactivation was used as a first step to determine the function of the cytoplasmic filaments. A suicide plasmid was constructed that contained part of cfpA and a nonpolar erythromycin resistance cassette (ermF and ermAM) inserted near the beginning of the gene. The plasmid was electroporated into T. denticola, and double-crossover recombinants which had the chromosomal copy of cfpA insertionally inactivated were selected. Immunoblotting and electron microscopy confirmed the lack of cytoplasmic filaments. The mutant was further analyzed by dark-field microscopy to determine cell morphology and by the binding of two fluorescent dyes to DNA to assess the distribution of cellular nucleic acids. The cytoplasmic filament protein-deficient mutant exhibited pleiotropic defects, including highly condensed chromosomal DNA, compared to the homogeneous distribution of the DNA throughout the cytoplasm in a wild-type cell. Moreover, chains of cells are formed by the cytoplasmic filament-deficient mutant, and those cells show reduced spreading in agarose, which may be due to the abnormal cell length. The chains of cells and the highly condensed chromosomal DNA suggest that the cytoplasmic filaments may be involved in chromosome structure, segregation, or the cell division process in Treponema.
Treponema denticola is one of the major spirochete species associated with the periodontitis pandemic (45). The treponemes are invasive due to their unique motility in dense media and their ability to penetrate cell monolayers (47). This feature is associated with their helical or wave-shaped cell body and the periplasmic flagellar filament location (15, 24, 40).
The cytoplasmic filaments in treponemes are persistent and span the length of the cell (21). They are located underneath the periplasmic flagellar bundle in close apposition to the cytoplasmic membrane (8, 18). The filaments are composed of one major protein (33) that is well conserved among species (21), and the gene encoding them, cfpA, is preceded by a sigma 70 promoter (21, 53). In Treponema pallidum subsp. pallidum, the cytoplasmic filament protein, CfpA, is Tpn83, an antigen recognized by sera from humans with syphilis (37, 53). This cytoplasmic structure is found in cells at all growth stages and severs during cell division (21). Proposed to be involved in cell structure, cell motility, and cell division (8), the filamentous nature of the structure and the location of the cytoplasmic filaments also make them a candidate for involvement in chromosome structure or segregation.
In order to achieve a better understanding of the potential relationship between the cytoplasmic filaments and the chromosome, we insertionally inactivated T. denticola cfpA. The CfpA knockout was nonlethal and did not alter cell structure. The mutant predominantly formed chains of cells, and the chromosomal DNA was condensed in distinct areas. We propose that the cytoplasmic filament structure is actively involved in chromosome structure maintenance, segregation, or cell division.
MATERIALS AND METHODS
Strains, reagents, culture, and molecular methods.
T. denticola ATCC 33520 and the cytoplasmic filament-less mutant were grown in New Oral Spirochete medium (NOS) with 10% heat-inactivated rabbit serum and 10 μg of cocarboxylase per ml at 36°C in an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, Mich.) with an atmosphere of 85% nitrogen, 10% carbon dioxide, and 5% hydrogen. Oligonucleotides were synthesized by the Molecular Genetics Core Facility of the Wadsworth Center, using PerSeptive Biosystems 8909 (PE Biosystems, Foster City, Calif.). T. denticola chromosomal DNA and plasmid miniprep DNA were isolated by standard methods (32). PCR was performed using Taq polymerase, reagents, and thermal cyclers available from Perkin Elmer (Foster City, Calif.).
Construction of a plasmid containing T. denticola cfpA interrupted with a modified erythromycin resistance cassette.
The partial DNA sequence of T. denticola cfpA previously cloned in the pZero-2 vector (Invitrogen, Carlsbad, Calif.) (21) was used as the starting point for the construction of a suicide plasmid. The cfpA gene was interrupted at the beginning of the gene (DraI site). The erythromycin resistance cassette (ermF-ermAM cassette) used for insertional inactivation of T. denticola cfpA was described by Li and Kuramitsu (27) and was obtained by amplification of pJS97 using primers ERMBGLF and ERMBGLR (30). The ermF-ermAM cassette was then ligated to the DraI-digested pZero-2 plasmid that contained T. denticola cfpA (NTPHDE1N and DENTCF2R amplification product). Orientation of the cfpA gene and the ermF-ermAM cassette was determined by PCR using primers DENTCF4, located in the cfpA sequence (5′-AAATCGCTACCCTTCTTGATG-3′), and ERMFBGLR, located in the ermF sequence (5′-TATAAGATCTCAACCACCCGACTTTGAACTA-3′); only those clones containing the ermF-ermAM cassette in the same direction as cfpA were chosen for electroporation. Nonmethylated plasmid DNA for electroporation of T. denticola was prepared in Escherichia coli SCS110 (Stratagene) using Qiagen Midi columns (Qiagen Corp., Chatsworth, Calif.) and concentrated with Spin-X UF100 columns (Costar, Cambridge, Mass.).
Insertional inactivation of T. denticola cfpA using an ermF-ermAM cassette.
Based on the protocol of Li and Kuramitsu as modified by Limberger et al. (30), 12.6 μg of nonmethylated plasmid DNA prepared in E. coli SCS110 was used to electroporate T. denticola ATCC 33520 competent cells. Briefly, 100 ml of T. denticola was grown to an optical density at 600 nm of 0.3. Cells were washed three times with cold 10% glycerol in water and resuspended in a final volume of 2 ml on ice. Electroporation was done using 1 μl of nonmethylated plasmid DNA, at a concentration of 12.6 μg/μl, in 100 μl of cells, using a Bio-Rad Gene Pulser at 1.8 kV, 200 Ω, and 25 μF and a 0.1-cm cuvette (Bio-Rad Laboratories, Hercules, Calif.). The time constant was 4.3; time constants of 4.1 to 4.6 are optimal. After overnight incubation in 10 ml of NOS broth without erythromycin, cells were cultured on NOS plates with 25 μg of erythromycin per ml. Colonies were visible after 7 days. Twenty colonies out of approximately 500 transformants were randomly selected, grown in liquid culture, and replated to ensure no cross-contamination by DNA present in the plated sample after electroporation. To check for the presence of the gene that confers resistance to erythromycin in T. denticola (ermF), a PCR was performed using primers ERMBGLF (5′-TATAAGATCTCCGATAGCTTCCGCTATTGC-3′) and ERMBGLR. To check the relative position of the antibiotic resistance cassette in the chromosome, four PCR assays were performed. The first one used primers DENTCF4 and ERMBGLF. One assay used primers ERMBGLR and DENTCF5 (5′-GCAGCCAAATCGTTAAAG-3′), located outside the cloned sequence in the pZero-2 vector in the 3′ end of the cfpA sequence. One assay used primers SP6 (5′-ATTTAGGTGACACTATAG-3′), located in the vector, and ERMBGLR. The last assay used primers T7 (5′-TAATACGACTCACTATAGGG-3′), located in the vector, and ERMAMF, located in ermAM sequence (5′-CAGCGGAATGCTTTCATC-3′).
Antibody production and immunoblotting.
Antiserum was raised against purified T. phagedenis CfpA purified filaments (21) in BALB/c mice, using RIBI adjuvant (RIBI Immunochemical Corp., Hamilton, Mont.) as described previously (31). Monoclonal antibody production was done using the Hybridoma Generation ClonaCell-HY kit (Vancouver, BC, Canada) and the spleen of a mouse immunized with purified T. phagedenis cytoplasmic filaments. A mid-log-phase culture of T. denticola was spun down for 1 min at 12,000 × g. The pellet was resuspended in 10 mM sodium phosphate (NaPi, pH 7.4) buffer and washed twice. The pellet was then resuspended in 1 mM EDTA–10 mM NaPi (pH 7.4) and sonicated for 1 min (Heat Systems Ultrasonics, Farmingdale, N.Y.). The quantity of protein was evaluated by the Bradford protein assay (Bio-Rad Laboratories). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was done using a Daiichi cassette electrophoresis unit and 10 to 20% Owl precast gels (Owl Separation System, Woburn, Mass.). Five micrograms of cell extract was loaded per lane, as well as 10-kDa protein ladders (Gibco-BRL, Grand Island, N.Y.). Gels were transferred onto 0.45-μm nitrocellulose membranes (Bio-Rad Laboratories) using the mini-transblot cell (Bio-Rad Laboratories). Immunoblots were performed using standard techniques with a monoclonal or polyclonal antibody directed against T. phagedenis CfpA and a goat anti-mouse immunoglobulin-alkaline phosphatase conjugate secondary antibody (Bio-Rad Laboratories).
Dark-field microscopy and motility test.
One milliliter of mid-log-phase culture was centrifuged for 1 min at 12,000 × g, and the pellet was suspended in 20 μl of reduced NOS or phosphate-buffered saline. Two hundred microliters of 0.5% methylcellulose (15 centipoise) was added (29). The cell mobility was observed by dark-field microscopy using a heated stage at 36°C (Fryer Co., Inc., Huntley, Ill.). Dark-field microscopy was performed using a Nikon Eclipse E600 microscope equipped with an oil dark-field condenser 1.43-1.20 and a Nikon Plan Fluor x100 0.5-1.3 oil objective. Pictures were taken with a Nikon FDX-35 camera mounted on a Nikon H-III unit (Nikon, Inc., Melville, N.Y.).
Visualization of chromosomal DNA.
Cells were grown in NOS broth, fixed with 1% glutaraldehyde, deposited on a poly(l-lysine)-treated slide, the DNA-binding dye Hoechst 33342 (Molecular Probes) (10 μg/ml) was added, and the slide was washed with water after 20 min. For fluorescence microscopy, a 100-W mercury lamp and a UV-2E/C filter were used (Nikon). An alternative protocol was also used. Cells were washed with 10 mM Tris-HCl (pH 7.4) buffer and suspended in 1 ml of 70% ethanol for fixation. The fixed cells were collected by centrifugation, washed with the same buffer, and deposited on a poly(l-lysine)- treated slide. Cells were stained with a DAPI (4′,6′-diamidino-2-phenylindole) solution (Molecular Probes) at 1 μg/ml in water.
Evaluation of the number of anucleated cytoplasmic cylinders.
Three samples representative of log-phase growth were evaluated for anucleated cytoplasmic cylinders in the wild type and the cytoplasmic filament-less mutant. To 100 μl of sample, 1 μl of Hoechst 33342 (10 mg/ml) was added (Molecular Probes, Inc.). After 15 min of incubation in the dark, the sample was observed by fluorescence and dark-field microscopy. One hundred twenty-six cells were evaluated for the presence of anucleated cytoplasmic cylinders in each of the three sample of the wild type and the cytoplasmic filament-less mutant.
Electron microscopy.
For electron microscopic visualization of T. denticola cells, 1 ml of logarithmio-phase culture was centrifuged for 1 min at 10,000 × g. The pellet was resuspended in the same amount of water with 0.5% deoxycholic acid sodium salt (Sigma-Aldrich, St. Louis, Mo.) and stored at 4°C overnight. The sample was centrifuged for 1 min at 10,000 × g, and the pellet was resuspended in 100 μl of sterile distilled water prior to use. Negative staining with sodium phosphotungstate was done as previously described (21). The samples were viewed in a Zeiss (LEO) 910 transmission electron microscope operating at 80 keV. The negatives were enlarged photographically.
RESULTS
Interruption of cfpA with an erythromycin resistance cassette.
To create a specific mutant of T. denticola that was deficient in CfpA, a suicide plasmid was constructed that contained an ermF-ermAM antibiotic resistance cassette inserted within T. denticola cfpA. The ermF-ermAM antibiotic resistance cassette does not terminate transcription or decrease the RNA level in T. denticola (30). Electroporation-mediated allelic exchange replaced the wild-type cfpA with the interrupted cfpA construct in the T. denticola chromosome. Several colonies out of approximately 500 were selected for further analysis by PCR and dark-field microscopy. Individual colonies were tested by PCR and were indistinguishable clones that had undergone a similar double-crossover recombination event to interrupt cfpA (data not shown). Therefore, one clone was used for all subsequent analyses.
Immunoblotting.
Immunoblots were done to assess whether CfpA was detectable in the cfpA-interrupted mutant. Using mouse polyclonal sera (data not shown) or a monoclonal antibody that reacts with T. denticola CfpA, the immunoblot revealed no trace of CfpA protein present in T. denticola cfpA-interrupted mutant cells (Fig. 1).
FIG. 1.
Immunoblot using monoclonal antibody M416 against T. phagedenis CfpA. (A) T. phagedenis cell extract. (B) Lane 1, T. denticola cell extract; lane 2, T. denticola cfpA mutant cell extract. The positions of size markers are shown (in kilodaltons).
Colony formation in agarose.
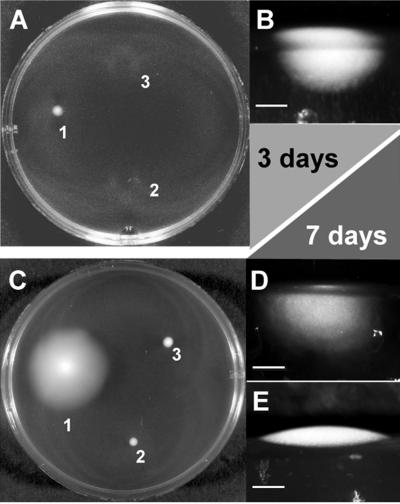
The pathogenicity of T. denticola is associated with its unique motility in dense medium, which is mediated by the periplasmic location of the flagellar filaments and the wave-shaped cell. After being plated on 0.5% agarose-NOS medium, T. denticola wild-type bacteria spread throughout the agarose (Fig. 2 and 3). No colony growth above the agarose was observed by macroscopic methods (Fig. 2B). In contrast, a previously described nonmotile mutant (30) formed colonies growing above the agarose over a limited surface (Fig. 2 and 3). T. denticola CfpA-deficient colonies spread within the agarose in a limited volume compared to the wild type (Fig. 2 and 3). Thus, T. denticola CfpA-deficient cells are characterized as having an altered motility in dense medium.
FIG. 2.
Macroscopic motility test in dense medium. (A) Plate after 3 days of incubation. A liquid culture (0.1 μl) of each sample was spotted on a 0.5% agarose–NOS plate and incubated at 36°C. 1, T. denticola wild type; 2, T. denticola CfpA-deficient mutant; 3, T. denticola fliK flagellar filament-less mutant (30). (B) T. denticola wild-type from a 3-day plated culture spread in agarose and propagated. Vertical section of the plate with side illumination. Bar, 1.5 mm. (C) Plate after 7 days of incubation, same strains as in panel A. (D) Vertical section of the plate with side illumination. Bar, 1 mm. T. denticola CfpA-deficient mutant from a 7-day plated culture does spread in the agarose but in a more limited volume. (E) Vertical section of the plate with side illumination. Bar, 1 mm. T. denticola fliK flagellar filament-less mutant from a 7-day plated culture was nonmotile and did not spread in the agarose. The vertical section photographs of the plate with side illumination were obtained using an SZ40 stereo microscope with a 35-mm camera mounted on a PM-20 exposure control unit (Olympus, Melville, N.Y.).
FIG. 3.
Macroscopic measurement of cell spreading over time in dense medium. A liquid culture (0.1 μl) was spotted on the center of a 0.5% agarose–NOS plate and incubated at 36°C. The colony diameter was measured using a caliper. ●, T. denticola wild-type colony; ▴, T. denticola cytoplasmic filament-less mutant colony; ○, T. denticola fliK flagellar filament-less mutant colony (30). Error bars show the range of measured colony diameters.
Cell morphology, chaining and motility.
Direct observation of T. denticola wild-type and CfpA-deficient cells using dark-field microscopy showed no morphological difference at the single-cell level (Fig. 4A through C). Thus, the absence of the cytoplasmic filament ribbon did not alter the morphology of a single cell; in particular, the wave shape was retained. However, in the CfpA-deficient mutant, a single cell was rarely observed (less than 1%) at any stage of growth. A representative cell from a culture of the CfpA-deficient mutant is shown in Fig. 4D. The long cells are composed of several cells that have undergone cell division but are unable to separate. Using dark-field microscopy and a warm stage at 36°C, the motility phenotype of the CfpA-deficient mutant was observed. The cells in the chain were unable to move significantly compared to a single cell. The asynchronous crankshaft-like movement of the cell chain might by itself be the reason for the poor motility of these organisms in dense medium. However, a rarely observed cytoplasmic filament-less single cell, like the one shown in Fig. 4C, does not significantly differ in cell movement from the wild-type bacteria at 36°C.
FIG. 4.
T. denticola wild-type and CfpA-deficient cells from a mid-log-phase NOS broth culture as observed by dark-field microscopy. (A) Representative single cell of wild-type T. denticola. (B) Wild-type T. denticola undergoing cell division. (C) A rarely observed single cell of T. denticola CfpA-deficient mutant. (D) Representative cell of T. denticola CfpA-deficient mutant. Bars, 5 μm.
Electron microscopy.
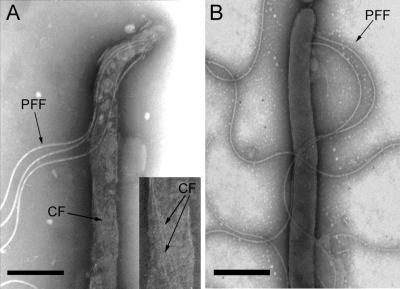
Electron microscopic analysis of the cytoplasmic filament-deficient mutant did not reveal any obvious structural or morphological differences compared to wild-type cells at the cell ends. The flagellar filament and flagellar basal body are indistinguishable from the wild type (Fig. 5), indicating that the cytoplasmic filaments are not needed for periplasmic flagellar synthesis and assembly despite their close association. No cytoplasmic filament was detectable, as expected (Fig. 5). The cells are longer than the wild-type cells, and usually several cytoplasmic cylinders can be observed under a unique outer membrane (data not shown).
FIG. 5.
Electron micrograph of wild-type T. denticola (A) and cytoplasmic filament-less mutant (B) cells. The outer membrane has been removed by detergent treatment of the cell using sodium deoxycholate 1% for 10 min, and the periplasmic flagellar filaments are freed. No cytoplasmic filaments were detected in the CfpA-deficient mutant. PFF, periplasmic flagellar filament; CF, cytoplasmic filament. Bar, 1 μM.
Visualization of chromosomal DNA.
To investigate the involvement of T. denticola cytoplasmic filaments in chromosome segregation or structure, T. denticola wild-type and CfpA-deficient cells were observed after incubation with Hoechst 33342, a fluorescent dye that binds to DNA. There was uniform distribution of the chromosomal DNA in T. denticola wild-type cells, which filled most of the cells, as shown in Fig. 6. However, the chromosomal DNA of the T. denticola CfpA-deficient cell was condensed in distinct areas (Fig. 6E and F). Similar results were observed using DAPI (data not shown).
FIG. 6.
Chromosomal DNA visualization and localization with Hoechst 33342 dye. (A) Dark-field microscopy of wild-type T. denticola and (B) the corresponding image by fluorescence. (C) Group of wild-type T. denticola cells showing a similar uniform DNA distribution. (D) Dark-field microscopy of T. denticola CfpA-deficient cells and (E) corresponding image by fluorescence. Arrows indicate condensed DNA areas. (F) T. denticola CfpA-deficient mutant cells, showing a typical pattern of distribution. Arrows indicate condensed DNA areas. Bars, 10 μm.
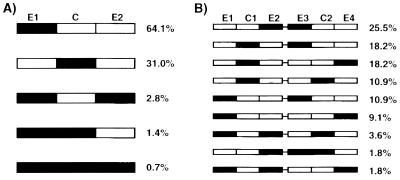
The location of the condensed chromosomal DNA in cells was investigated. As shown in Fig. 5E and F, a small portion of the cytoplasmic cylinder is occupied by the condensed chromosomal DNA labeled with the Hoescht 33342 dye. As mentioned above, under the same outer membrane there are multiple independent cytoplasmic cylinders. In a cell chain, each of the cytoplasmic cylinders is delineated by the pinch-like constrictions observed by dark-field microscopy along the cell (Fig. 6D). By comparing dark-field and Hoeschst 33342 fluorescence images, we were able to map the distribution of the condensed chromosomal DNA in the cytoplasmic cylinder of single or chained cells (Fig. 7). The cytoplasmic cylinder was divided into three sections, independently of the length of the cell. The cytoplasmic cylinder extremities were designated E1 and E2, and the center region was called C. Most of the cytoplasmic cylinders contained only one condensed DNA area (95%), with a random distribution: 31% found in the center and 64% found at one end of the cytoplasmic cylinder (Fig. 7A). When the distribution of the condensed DNA area was observed in two consecutive cytoplasmic cylinders (Fig. 7B), there was a preference for three patterns: the central region and one of the extremities in the other cytoplasmic cylinder (C1-E3 [18%] and C1-E4 [18%]) and two facing extremities (E2-E3 [25%]).
FIG. 7.
Schematic representation of the distribution of condensed chromosomal DNA in the T. denticola CfpA-minus mutant. (A) Distribution in a single cytoplasmic cylinder (142 cytoplasmic cylinders). E1 and E2, extremities of the cytoplasmic cylinder; C, center region. For example, in Fig. 5F, the upper right corner cell has one cytoplasmic cylinder and bears the pattern E1-C-E2. (B) Distribution in two adjacent cytoplasmic cylinders (55 cytoplasmic cylinder pairs). The bar between the two schematic cytoplasmic cylinders indicates that they are under the same outer membrane. E1 to E4, extremities of the cytoplasmic cylinder; C1 and C2, center regions. For example, in Fig. 5F, the lower left corner cell bears two patterns, E4-C1 and C1-C2.
When single cells with a single cytoplasmic cylinder, as defined above by comparison of dark-field and fluorescence microscopy, was observed, a pattern distribution similar to that shown in Fig. 6A was found (18 cells, E1 [58%], C [39%], and E1-C-E2 [5%]).
When single cells with two cytoplasmic cylinders, as described above, were observed, only the E1-E3 pattern was not represented (24 cells, E2-E3 [30%], C1-E3 [30%], C1-E4 [12%], C1-C2 [4%], E1-E4 [8%], E1-E2-C2 [8%], E2-E3-C2 [4%], and E1-E2-E4 [4%]).
At mid-log phase, the percentage of anucleated cytoplasmic cylinders was 4.74% ± 0.74% for the cytoplasmic filament-less mutant, versus 2.74% ± 0.47% for wild-type T. denticola.
DISCUSSION
Discovered by electron microscopy, cytoplasmic filaments have been observed in treponemes (16, 18) and certain other spirochetes (3, 17). However, no function has been associated with these distinctive structures. Their proposed hypothetical functions were involvement in cell structure, cell motility, and cell division (8). Other cytoplasmic filament structures such as microtubules, tubules, fibers, fibrils, and filaments have been described in prokaryotes, but their function is unknown. The shape, length, striation, and number of these structures vary (for a review, see reference 4). In T. denticola, the cytoplasmic filaments are permanent and are found at all stages of cell division and growth; together with their organization in a ribbon-like structure, their apparent lack of involvement in cell shape differentiates them from the filament structures observed in E. coli and Bacillus subtilis, such as the ones formed by FtsZ. The CfpA-deficient mutant phenotype includes the presence of chained cells, altered motility, and chromosome condensation. Thus, we hypothesize that the cytoplasmic filaments are involved in cell division, chromosome segregation, or structure maintenance.
Due to chromosomal DNA condensation in the cytoplasmic filament-deficient mutant, we were able to analyze the distribution of the nucleoid in the cytoplasm of a population undergoing exponential growth. Chromosome segregation, arrangement, structure, and duplication are critical elements for the cell. For duplication, the two replication forks initiate from oriC and meet at the opposite side in the terminus region (12, 34). The oriC and terminus region have a defined location in the cell (36, 52), and between them, the chromosome regions are laid out according to the order of replication (36, 46). The factory model of replication proposed by Lemon and Grossman (26) in B. subtilis implies that the replisome should be located at the cell center. Electron microscopy autoradiography experiments lead to the same conclusion for E. coli (25). In Mycoplasma capricolum, the nucleoid is centrally located (43). The presence of a replisome cannot yet be generalized to all bacteria. However, if a replisome does exist, it should migrate after cell septation from the previous central region (designated C in Fig. 7A) that became two end regions of the two newly formed cytoplasmic cylinders (E2 and E3 in Fig. 7B), to the new central regions in each newly formed cytoplasmic cylinders (C1 and C2 in Fig. 7B). In the cytoplasmic filament-less mutant, the distribution of the condensed DNA in two consecutive cytoplasmic cylinders follows the rules of this concept, as shown in Fig. 7. We propose that the condensed chromosomal DNA progresses from pattern C to pattern E2-E3 or C1-C2. However, not all the cells possess a pattern directly described by the model. These can be explained as follows. In a cell with multiple cytoplasmic cylinders, two adjacent cytoplasmic cylinders at different stages of cell division would result in a C1-E3 pattern. The patterns C1-E4 and E1-E4 would be the expected result of two adjacent cytoplasmic cylinders (with pattern E2-E3) which have undergone a non-synchronized cell division process. The pattern E1-E3 could be the result of two adjacent cytoplasmic cylinders (with pattern E2-E3) in which only one has undergone the cell division process. Other patterns could be the result of unsuccessful chromosome transfer events resulting in cells without a nucleoid. In the absence of cytoplasmic filaments, chromosomal DNA replication continues, and the location of the condensed DNA after replication is in accordance with the presence of a replisome in T. denticola.
Because of the unique structure of the cytoplasmic filaments, their location, and their participation in the cell division process, it is unclear how the filaments interact with the chromosome. Even if the model of duplication and separation of oriC and ter differs between E. coli and B. subtilis, the existence of an active chromosome segregation mechanism seems unambiguous (10, 23, 36, 44, 51). Many genes are involved in chromosomal DNA segregation. The mechanism that spatially and temporally controls and directs the oriC region is unknown; however, it seems likely that this mechanism involves the action of a hypothetical spindle or mitotic-like apparatus and “motor” proteins (44). The genetic analysis of mutants shows a potential nucleoid segregation role for numerous genes. Some of these genes are involved in DNA replication, cell division, structural maintenance of chromosome, or SOS system, or they encode histone-like proteins (1, 5, 6, 9, 11, 20, 22, 23, 54). Others were associated with the formation of anucleate cells (13, 14) or polyploidy (48). There is no clear understanding of how the genes interact, their degree of involvement, and when they intervene. However, none of them are candidates to form a filamenteous structure involved in a mitotic apparatus.
Filamentous structures that may be involved in nucleoid segregation are CafA, CfpA, and some noncharacterized structures (4, 8, 38, 53). There is no phenotypic change in a cafA deletion mutant (49, 50). However, the cytoplasmic filament-deficient mutant phenotype described in this report includes chained cells and condensed DNA areas along the cytoplasmic cylinder, indicating a cell division defect as well as a chromosome structure alteration. The cytoplasmic filaments may be part of a mitosis-like apparatus involved in chromosome segregation in T. denticola, allowing proper organization of the chromosomal DNA in the cytoplasm. We found a nearly twofold increase in anucleated cells, which could suggest a segregation defect. However, the observed increase in the number of anucleated cells in the cytoplasmic filament-less mutant compared to the wild type in log phase suggests that the filaments do not have a critical role in the chromosome segregation process.
The structure of the nucleoid is the result of DNA packaging, which is the result of short- and long-range structure of the DNA and RNA, macromolecular crowding and the effect of proteins that directly bind to the DNA or actively modify chromosomal DNA topology (39). We were not able to determine the number of chromosomal DNA copies in the wild type compared to the cytoplasmic filament-deficient mutant. However, it seems that in the CfpA-deficient cells, the DNA is highly condensed. The cytoplasmic filament protein consequently could be involved in “spreading” the DNA in the cytoplasm. The structural consequences of this topological arrangement would be a distribution of the chromosome subdomains on a greater distance on the long axis of the cell than in E. coli or B. subtilis.
The helical or flat wave-shaped treponemal cell, in conjunction with the movement created by the periplasmic flagellar filaments, is proposed to be the determinant of the unique ability of spirochetes to penetrate dense media and cell layers (15, 47). In Borrelia burgdorferi, a spirochete lacking cytoplasmic filaments, a flagellar filament-deficient mutant has rod-shaped cells (35, 42). In T. phagedenis and T. denticola, the absence of periplasmic flagellar filaments does not significantly alter the cell shape other than the distal cell ends; there is release of the stress applied to the outer membrane, and the cell body maintains the waved or helical shape (7, 30, 41). One proposal (8) was that the cytoplasmic filaments were involved in maintaining the helical cell shape. The results presented here show that the absence of cytoplasmic filaments does not significantly alter T. denticola cell shape.
The cell motility generated by the rotation of periplasmic flagellar filaments is unique to spirochetes (2, 28). Since the absence of the periplasmic flagellar filaments eliminates motility in dense media, the close apposition of the cytoplasmic filament to the inner membrane beneath the periplasmic flagellar filaments (8, 18, 19) raised the question of a relationship between the two structures during cell propulsion. The results presented here show that under dark-field microscopy at 36°C at the single-cell level in T. denticola, there is no obvious difference in motility between the wild-type bacteria and the cytoplasmic filament-less mutant.
In summary, the evidence presented here suggests a role for the cytoplasmic filaments in the cell division process or in chromosome structure or segregation. However, the precise role of the cytoplasmic filaments is not yet clearly understood. There is no similarity between the cytoplasmic filament protein sequence and any other open reading frame currently known. Evolution may have allowed different molecules to maintain the same function or use different strategies. The study of this persistent structure and its protein partners will bring new critical data on cell division and chromosome structure maintenance and an understanding of the mechanisms involved in transient or permanent structures to be discovered in other bacterial genera.
ACKNOWLEDGMENTS
We thank Linda L. Slivienski and the Wadsworth Center Molecular Genetics, Electron Microscopy, Molecular Immunology, and Photography core facilities.
This work was supported by Public Health Service grant AI34354 from the National Institutes of Health.
REFERENCES
- 1.Akerlund T, Bernander R, Nordstrom K. Cell division in Escherichia coli minB mutants. Mol Microbiol. 1992;6:2073–2083. doi: 10.1111/j.1365-2958.1992.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 2.Berg H C. How spirochetes may swim. J Theor Biol. 1976;56:269–273. doi: 10.1016/s0022-5193(76)80074-4. [DOI] [PubMed] [Google Scholar]
- 3.Bermudes D, Chase D, Margulis L. Morphology as a basis for taxonomy of large spirochetes symbiotic in wood-eating cockroaches and termites: Pillotina gen. nov., nom. rev.; Pillotina calotermitidis sp. nov., nom. rev.; Diplocalyx gen. nov., nom. rev.; Diplocalyx calotermitidis sp. nov., nom. rev.; Hollandina gen. nov., nom. rev.; Hollandina pterotermitidis sp. nov., nom. rev.; and Clevelandina reticulitermitidis gen. nov., sp. nov. Int J Syst Bacteriol. 1988;38:291–302. doi: 10.1099/00207713-38-3-291. [DOI] [PubMed] [Google Scholar]
- 4.Bermudes D, Hinkle G, Margulis L. Do prokaryotes contain microtubules? Microbiol Rev. 1994;58:387–400. doi: 10.1128/mr.58.3.387-400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi E F, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 6.Britton R A, Lin D C, Grossman A D. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charon N C, Goldstein S F, Curci K, Limberger R J. The bent-end morphology of Treponema phagedenis is associated with short, left-handed, periplasmic flagella. J Bacteriol. 1991;173:4820–4826. doi: 10.1128/jb.173.15.4820-4826.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eipert S R, Black S H. Characterization of the cytoplasmic fibrils of Treponema refringens (Nichols) Arch Microbiol. 1979;120:205–214. doi: 10.1007/BF00423067. [DOI] [PubMed] [Google Scholar]
- 9.Gayda R C, Henk M C, Leong D. C-shaped cells caused by expression of an ftsA mutation in Escherichia coli. J Bacteriol. 1992;174:5362–5370. doi: 10.1128/jb.174.16.5362-5370.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaser P, Sharpe M E, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- 11.Grompe M, Versalovic J, Koeuth T, Lupski J R. Mutations in the Escherichia coli dnaG gene suggest coupling between DNA replication and chromosome partitioning. J Bacteriol. 1991;173:1268–1278. doi: 10.1128/jb.173.3.1268-1278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill T. Features of the chromosomeal terminus region. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 1602–1614. [Google Scholar]
- 13.Hiraga S, Niki H, Imamura R, Ogura T, Yamanaka K, Feng J, Ezaki B, Jaffe A. Mutants defective in chromosome partitioning in E. coli. Res Microbiol. 1991;142:189–194. doi: 10.1016/0923-2508(91)90029-a. [DOI] [PubMed] [Google Scholar]
- 14.Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holt S C. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;42:114–160. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hovind-Hougen K. Further observations on the ultrastructure of Treponema pallidum nichols. Acta Pathol Microbiol Scand Sect B Microbiol. 1972;80:297–304. doi: 10.1111/j.1699-0463.1972.tb00163.x. [DOI] [PubMed] [Google Scholar]
- 17.Hovind-Hougen K. Leptospiraceae, a new family to include Leptospira Noguchi 1917 and Leptonema gen. nov. Int J Syst Bacteriol. 1979;29:245–251. [Google Scholar]
- 18.Hovind-Hougen K. The ultrastructure of cultivable treponemes. I. Treponema phagedenis, Treponema vincentii, and Treponema refringens. Acta Pathol Scand Microbiol Sect B Microbiol. 1974;82:329–344. [PubMed] [Google Scholar]
- 19.Hovind-Hougen K, Birch-Andersen A. Electron microscopy of the endoflagella and microtubules in Treponema Reiter. Acta Pathol Microbiol Scand Sect B Microbiol. 1971;79:37–50. doi: 10.1111/j.1699-0463.1971.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 20.Huisman O, Faelen M, Girard D, Jaffe A, Toussaint A, Rouviere-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J Bacteriol. 1989;171:3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izard J, Samsonoff W A, Kinoshita M B, Limberger R J. Genetic and structural analysis of the cytoplasmic filaments of wild-type and flagellar fillament mutant of Treponema phagedenis. J Bacteriol. 1999;181:6739–6746. doi: 10.1128/jb.181.21.6739-6746.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaffe A, D'Ari R, Hiraga S. Minicell-forming mutants of Escherichia coli: production of minicells and anucleate rods. J Bacteriol. 1988;170:3094–3101. doi: 10.1128/jb.170.7.3094-3101.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen R B, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci USA. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klitorinos A, Noble P, Siboo R, Chan E C. Viscosity-dependent locomotion of oral spirochetes. Oral Microbiol Immunol. 1993;8:242–244. doi: 10.1111/j.1399-302x.1993.tb00567.x. [DOI] [PubMed] [Google Scholar]
- 25.Koppes L J, Woldringh C L, Nanninga N. Escherichia coli contains a DNA replication compartment in the cell center. Biochimie. 1999;81:803–810. doi: 10.1016/s0300-9084(99)00217-5. [DOI] [PubMed] [Google Scholar]
- 26.Lemon K P, Grossman A D. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Kuramitsu H K. Development of a gene transfer system in Treponema denticola by electroporation. Oral Microbiol Immunol. 1996;11:161–165. doi: 10.1111/j.1399-302x.1996.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 28.Lighthill J. Helical distribution of stokeslets. J Eng Maths. 1996;30:35–78. [Google Scholar]
- 29.Limberger R J. Periplasmic flagella of Treponema phagedenis. Ph.D. thesis. Morgantown, W.Va: West Virginia University; 1984. [Google Scholar]
- 30.Limberger R J, Slivienski L L, Izard J, Samsonoff W A. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J Bacteriol. 1999;181:3743–3750. doi: 10.1128/jb.181.12.3743-3750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limberger R J, Slivienski L L, Samsonoff W A. Genetic and biochemical analysis of the flagellar hook of Treponema phagedenis. J Bacteriol. 1994;176:3631–3637. doi: 10.1128/jb.176.12.3631-3637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 33.Masuda K, Kawata T. Isolation and characterization of cytoplasmic fibrils from treponemes. Microbiol Immunol. 1989;33:619–630. doi: 10.1111/j.1348-0421.1989.tb02012.x. [DOI] [PubMed] [Google Scholar]
- 34.Messer W, Weigel C. Intiation of chromosome replication. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 1579–1601. [Google Scholar]
- 35.Motaleb M A, Corum L, Bono J L, Elias A F, Rosa P, Samuels D S, Charon N W. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci USA. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niki H, Hiraga S. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 1998;12:1036–1045. doi: 10.1101/gad.12.7.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norris S J. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Microbiol Rev. 1993;57:750–779. doi: 10.1128/mr.57.3.750-779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada Y, Wachi M, Hirata A, Suzuki K, Nagai K, Matsuhashi M. Cytoplasmic axial filaments in Escherichia coli cells: possible function in the mechanism of chromosome segregation and cell division. J Bacteriol. 1994;176:917–922. doi: 10.1128/jb.176.3.917-922.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettijohn D E. The nucleoid. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 158–166. [Google Scholar]
- 40.Riviere G R, Weisz K S, Adams D F, Thomas D D. Pathogen-related oral spirochetes from dental plaque are invasive. Infect Immun. 1991;59:3377–3380. doi: 10.1128/iai.59.10.3377-3380.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruby J D, Li H, Kuramitsu H, Norris S J, Goldstein S F, Buttle K F, Charon N W. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol. 1997;179:1628–1635. doi: 10.1128/jb.179.5.1628-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadziene A, Thomas D D, Bundoc V G, Holt S C, Barbour A G. A flagella-less mutant of Borrelia burgdorferi: structural, molecular and in vitro functional characterization. J Clin Investig. 1991;88:82–92. doi: 10.1172/JCI115308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seto S, Miyata M. Partitioning, movement, and positioning of nucleoids in Mycoplasma capricolum. J Bacteriol. 1999;181:6073–6080. doi: 10.1128/jb.181.19.6073-6080.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharpe M E, Errington J. Upheaval in the bacterial nucleoid: an active chromosome segregation mechanism. Trends Genet. 1999;15:70–74. doi: 10.1016/s0168-9525(98)01660-6. [DOI] [PubMed] [Google Scholar]
- 45.Simonson L G, Goodman C H, Bial J J, Morton H E. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect Immun. 1988;56:726–728. doi: 10.1128/iai.56.4.726-728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teleman A A, Graumann P L, Lin D C-h, Grossman A D, Losick R. Chromosome arrangement within a bacterium. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 47.Thomas D D, Navab M, Haake D A, Fogelman A M, Miller J N, Lovett M A. Treponema pallidum invades intracellular junctions of endothelial cell monolayers. Proc Natl Acad Sci USA. 1988;8:3608–3612. doi: 10.1073/pnas.85.10.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trun N J, S. G. Characterization of Escherichia coli mutants with altered ploidy. Res Microbiol. 1991;142:195–200. doi: 10.1016/0923-2508(91)90030-e. [DOI] [PubMed] [Google Scholar]
- 49.Wachi M, Doi M, Okada Y, Matsuhashi M. New mre genes mreC and mreD, responsible for formation of the rod shape of Echerichia coli cells. J Bacteriol. 1989;171:6511–6516. doi: 10.1128/jb.171.12.6511-6516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wachi M, Umitsuki G, Nagai K. Functional relationship between Escherichia coli RNase E and the CafA protein. Mol Gen Genet. 1997;253:515–519. doi: 10.1007/s004380050352. [DOI] [PubMed] [Google Scholar]
- 51.Webb C D, Graumann P L, Kahana J A, Teleman A A, Silver P A, Losick R. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol Microbiol. 1998;28:883–892. doi: 10.1046/j.1365-2958.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 52.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C, Grossman A D, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 53.You Y, Elmore S, Colton L L, Mackenzie C, Stoops J K, Weinstock G M, Norris S J. Characterization of the cytoplasmic filament protein gene (cfpA) of Treponema pallidum subsp. pallidum. J Bacteriol. 1996;178:3177–3187. doi: 10.1128/jb.178.11.3177-3187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zyskind J W, Svitil A L, Stine W B, Biery M C, Smith D W. RecA protein of Escherichia coli and chromosome partitioning. Mol Microbiol. 1992;6:2525–2537. doi: 10.1111/j.1365-2958.1992.tb01429.x. [DOI] [PubMed] [Google Scholar]