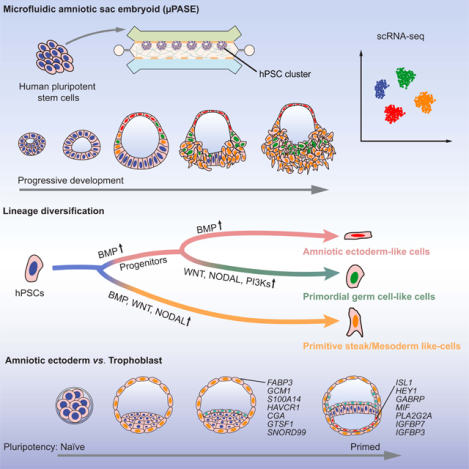
SUMMARY
Despite its clinical and fundamental importance, our understanding of early human development remains limited. Stem cell-derived, embryo-like structures (or embryoids) allowing studies of early development without using natural embryos can potentially help fill the knowledge gap of human development. Herein, transcriptome at the single-cell level of a human embryoid model was profiled at different time points. Molecular maps of lineage diversifications from the pluripotent human epiblast towards the amniotic ectoderm, primitive streak / mesoderm, and primordial germ cells were constructed and compared with in vivo primate data. The comparative transcriptome analyses reveal a critical role of NODAL signaling in human mesoderm and primordial germ cell specification, which is further functionally validated. Through comparative transcriptome analyses and validations with human blastocysts and in vitro cultured cynomolgus embryos, we further proposed stringent criteria for distinguishing between human blastocyst trophectoderm and early amniotic ectoderm cells.
Keywords: Human embryoid, single-cell transcriptome, NODAL signaling, primate development, amnion, primitive streak, primordial germ cell, mesoderm, trophoblast, microfluidics
Graphical Abstract
ETOC
Fu and colleagues profiled a stem cell-derived microfluidic amniotic sac embryoid (μPASE) using single-cell RNA Sequencing. Molecular maps of lineage diversifications from the pluripotent human epiblast towards the amniotic ectoderm, primitive streak / mesoderm, and primordial germ cells were constructed and compared with in vivo primate data.
INTRODUCTION
Development of multicellular organisms is one of nature’s greatest triumphs. Development is a tightly orchestrated process, following stereotypic lineage diversifications and morphogenetic tissue patterning events in a precise spatiotemporal order. Scientists commonly use animal models to study the key transcriptional and signaling activities that underlie pattern formation, morphogenesis, cell differentiation, and tissue growth (Gilbert, 2000; Schoenwolf, 2020; Solnica-Krezel, 2020). However, cross-species genetic and morphological divergence is evident between humans and commonly used animal models (Rossant, 2015; Rossant and Tam, 2017). To address this issue, there is a significant current interest in improving in vitro culture protocols of human and non-human primate (NHP) monkey embryos for experimental observations and mechanistic studies (Deglincerti et al., 2016; Ma et al., 2019; Niu et al., 2019; Shahbazi et al., 2016; Xiang et al., 2020; Yang et al., 2021). However, experimentations on human and NHP monkey embryos remain challenging due to limited access to and bioethical constraints on these natural specimens (Clark et al., 2021; Hyun et al., 2016; Lovell-Badge et al., 2021). As such, knowledge of human development remains limited; this is particularly true for early post-implantation human development, when the basic human body plan is laid down and when the human embryo in vivo is at its most inaccessible phase for experimentation.
Recently, stem cell-derived embryo-like structures (or embryoids) that could recapitulate certain aspects of mammalian early embryogenesis are emerging as tractable experimental tools for studying human development (Beccari et al., 2018; Haremaki et al., 2019; Harrison et al., 2017; Liu et al., 2021; Moris et al., 2020; Rivron et al., 2018; Shao et al., 2017a; Shao et al., 2017b; Simunovic et al., 2019; Warmflash et al., 2014; Xue et al., 2018; Yanagida et al., 2021; Yu et al., 2021; Zheng et al., 2019a; Zheng et al., 2019b). Particularly, we have recently developed a microfluidic post-implantation amniotic sac embryoid (μPASE) system (Zheng et al., 2019b), allowing the development of PASEs in a highly controllable, reproducible and scalable fashion.
RESULTS
Single-cell transcriptomic profiling of μPASE development
Development of μPASE involves successive cell morphogenetic and lineage specification events that recapitulate early post-implantation human development up to the onset of gastrulation (Figure 1A) (Zheng et al., 2021; Zheng et al., 2019b). Specifically, owing to their intrinsic lumenogenic property, each hPSC cluster undergoes lumenogenesis and epithelization to establish apical-basal polarity and form a single central apical lumen by t = 12 h (Figures 1B and S1A). hPSCs exposed directly to exogeneous BMP4 stimulation in each cluster initiate amniogenesis, evidenced by continuous flattening of cell morphology, resolving into a thin layer of squamous amniotic cells (Figure 1B). Inductive effects of AMLCs in the μPASEs lead to hPSCs at the opposite pole to undergo epithelial-mesenchymal transition (EMT) and gastrulation-like events (Zheng et al., 2021; Zheng et al., 2019b), with gastrulating cells disseminating away from μPASEs from t = 36 h onwards (Figure 1B), leading to disintegration of the μPASE structure. By t = 48 h, the μPASE contains only AMLCs, MeLCs and PGCLCs, without the presence of EpiLCs (Figures 1B and 1C).
Figure 1. Single-cell transcriptomic profiling of μPASE development.
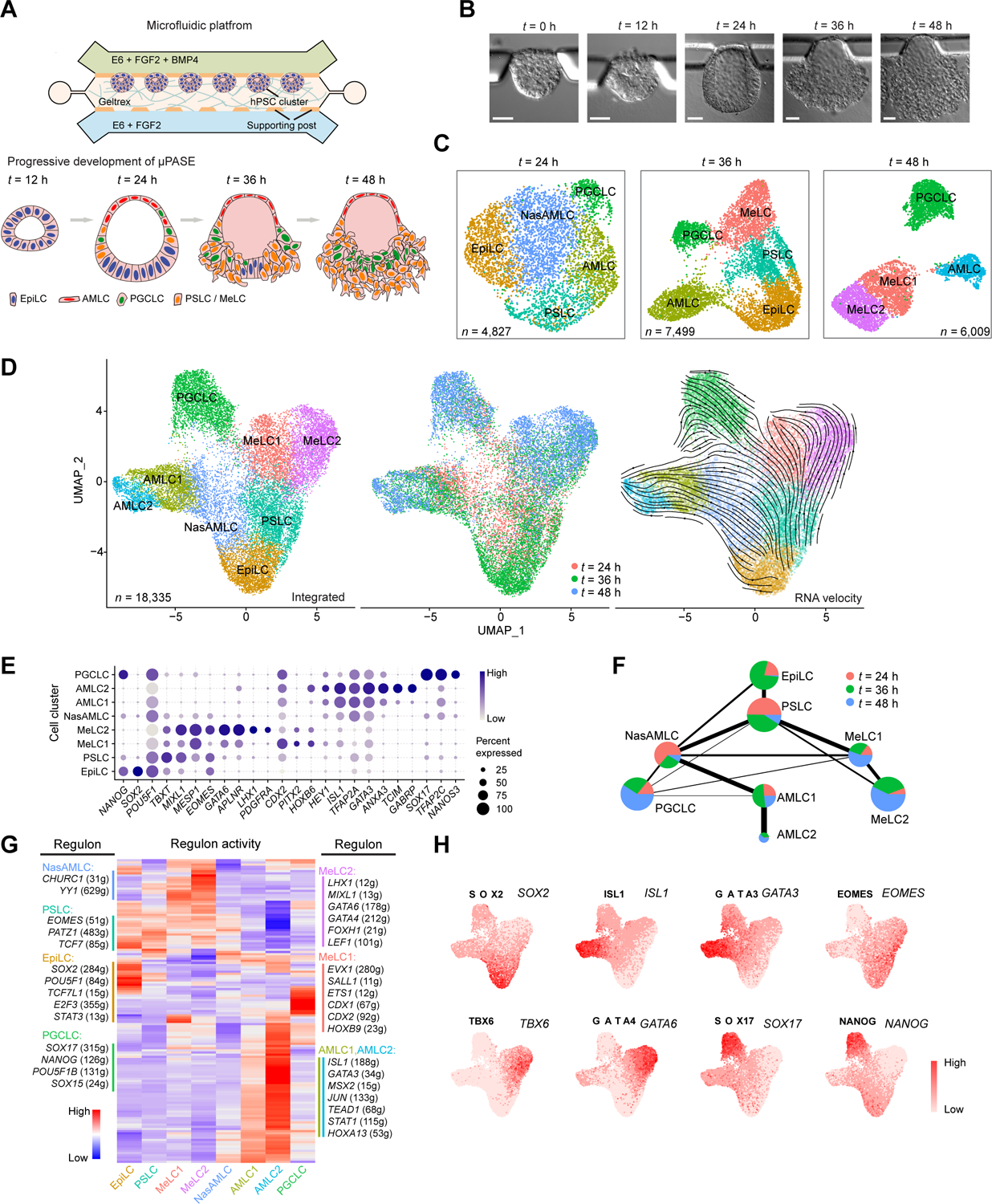
(A) Development of μPASEs. Single hPSCs were guided to form uniform-sized clusters at prescribed locations in the microfluidic device. Asymmetric stimulation with exogeneous BMP4 from t = 0 – 48 h led to progressive development of μPASEs. E6: Essential 6. See Methods for the μPASE protocol.
(B) Bright-field images showing progressive development of μPASEs over time, including thinning and flattening of the incipient amniotic ectoderm and thickening of the incipient mesoderm cells before their dissemination from the μPASE structure.
(C) Dimension reduction presentation via UMAP and cell identity annotations of single-cell transcriptome datasets obtained for μPASEs at indicated time points. n indicates cell numbers analyzed for each time point.
(D) UMAP of integrated single-cell transcriptome datasets of μPASEs from t = 24, 36 and 48 h (shown in (C)), color-coded according to cell identity annotations (left) or time points (middle). RNA velocity vectors were projected onto the UMAP-based embeddings (right). n indicates the total number of cells combined from the three time points.
(E) Dot plot showing expression of key marker genes across the cell clusters as indicated. The sizes and colors of dots indicate the proportion of cells expressing the corresponding genes and their averaged scaled values of log-transformed expression, respectively.
(F) Partition-based graph abstraction (PAGA) analysis of the integrated single-cell transcriptome dataset shown in (D). The thickness of lines connecting pairs of cell clusters indicates the degree of correlation between the cell cluster pairs. Lines with a correlation weight less than 0.05 are not shown. Pie charts for each cell cluster show percentages of indicated cell types from the three time points. Pie chart size is proportional to the total number of indicated cell types. See Mendeley Data Table1.
(G) Heatmap of regulon activities calculated from gene regulatory network interference. Selected master regulators of different cell clusters are depicted as indicated. See Mendeley Data Table2 and HTML Document 1.
(H) Gene set activity of selected regulons overlaid on the integrated UMAP plot from (D).
EpiLC: epiblast-like cell; PSLC: primitive streak-like cell; MeLC1/2: mesoderm-like cell 1/2; AMLC1/2: amniotic ectoderm-like cell 1/2; NasAMLC: nascent amniotic ectoderm-like cell; PGCLC: primordial germ cell-like cell. In (B), experiments were repeated more than twenty times with similar results. Scale bars, 50 μm.
To investigate dynamics of μPASE development at the transcriptome level, single-cell suspensions of μPASEs at t = 24 h, 36 h, and 48 h were prepared before single-cell RNA-sequencing (scRNA-seq) using 10× Genomics. We performed UMAP (uniform manifold approximation and projection) dimension reduction using the Seurat R package (Butler et al., 2018; Satija et al., 2015) for scRNA-seq datasets at each time point (Figure 1C), as well as for the integrated scRNA-seq dataset from all three time points (Figure 1D). These analyses reveal distinct cell clusters in the μPASE based on expression patterns of key lineage markers (Figures 1C–1E, S1B and S1C). Consistent with our previous findings (Zheng et al., 2021; Zheng et al., 2019b), in the μPASE, hPSCs develop progressively from a pluripotent epiblast-like cell (EpiLC) stage to three distinct cell populations by t = 48 h: amniotic ectoderm-like cells (AMLCs), mesoderm-like cells (MeLCs), and primordial germ cell-like cells (PGCLCs) (Figures 1C–1E, S1D and S1E).
To understand how hPSCs transit from the EpiLC stage to AMLCs, MeLCs and PGCLCs during μPASE development, RNA velocity analysis (Bergen et al., 2020; La Manno et al., 2018) was conducted for the integrated scRNA-seq dataset, with RNA velocity vectors overlaid on the integrated UMAP plot (Figure 1D). This RNA velocity analysis reveals developmental trajectories of the AMLC lineage (EpiLC → nascent AMLC or NasAMLC → AMLC1 → AMLC2) and MeLC lineage (EpiLC → primitive streak-like cell or PSLC → MeLC1 / MeLC2) (Figure 1D). However, developmental trajectory of PGCLCs is not as clear from the RNA velocity analysis (Figure 1D). To further examine lineage relations between different μPASE cell clusters, partition-based graph abstraction (PAGA) analysis (Wolf et al., 2018) was conducted, revealing that PGCLCs correlate best with the NasAMLC cluster (Figure 1F). To reveal gene regulatory network (GRN) underlying each cell cluster, we performed GRN analysis using SCENIC (Aibar et al., 2017) (single-cell regulatory network inference and clustering; Figures 1G and 1H).
Trajectory inference and gene expression dynamics analysis
To infer developmental trajectories of different μPASE cell lineages, we plotted a three-dimensional (3D) diffusion map based on PCA embeddings of the integrated scRNA-seq dataset (Figure 2A and Video S1) (Angerer et al., 2016). The AMLC, MeLC and PGCLC lineages display distinct and well separated trajectories in the 3D diffusion map (Figure 2A). To analyze transcriptome dynamics during AMLC lineage development, EpiLC, NasAMLC and AMLC1 / 2 clusters were isolated from the integrated scRNA-seq dataset and re-plotted using two-dimensional (2D) diffusion maps (Figure 2B). Similarly, to analyze MeLC lineage development, EpiLC, PSLC and MeLC1 / 2 clusters were isolated from the integrated scRNA-seq dataset before re-plotting using 2D diffusion maps (Figure 2C). Expression dynamics of selected genes related to AM and PS or Meso was plotted against diffusion pseudotime (dpt) (Figures 2D, 2E, S2A and S2B). Notably, in the AMLC lineage, expression of TFAP2A, MSX2 and ID2, which are commonly used AM markers (Ma et al., 2019; Sasaki et al., 2016; Yang et al., 2021), is quickly upregulated following exogeneous BMP4 stimulation (Figures 2D and S2A). Similarly, GATA3 and ISL1 become upregulated relatively early during AMLC lineage development (Figures 2D and S2A). In contrast, expression of GABRP, IGFBP3 and WNT6 shows delayed upregulation, whereas TBXT is only transiently expressed during early AMLC lineage development (Figures 2D and S2A).
Figure 2. Trajectory inference and gene expression dynamics during μPASE development.
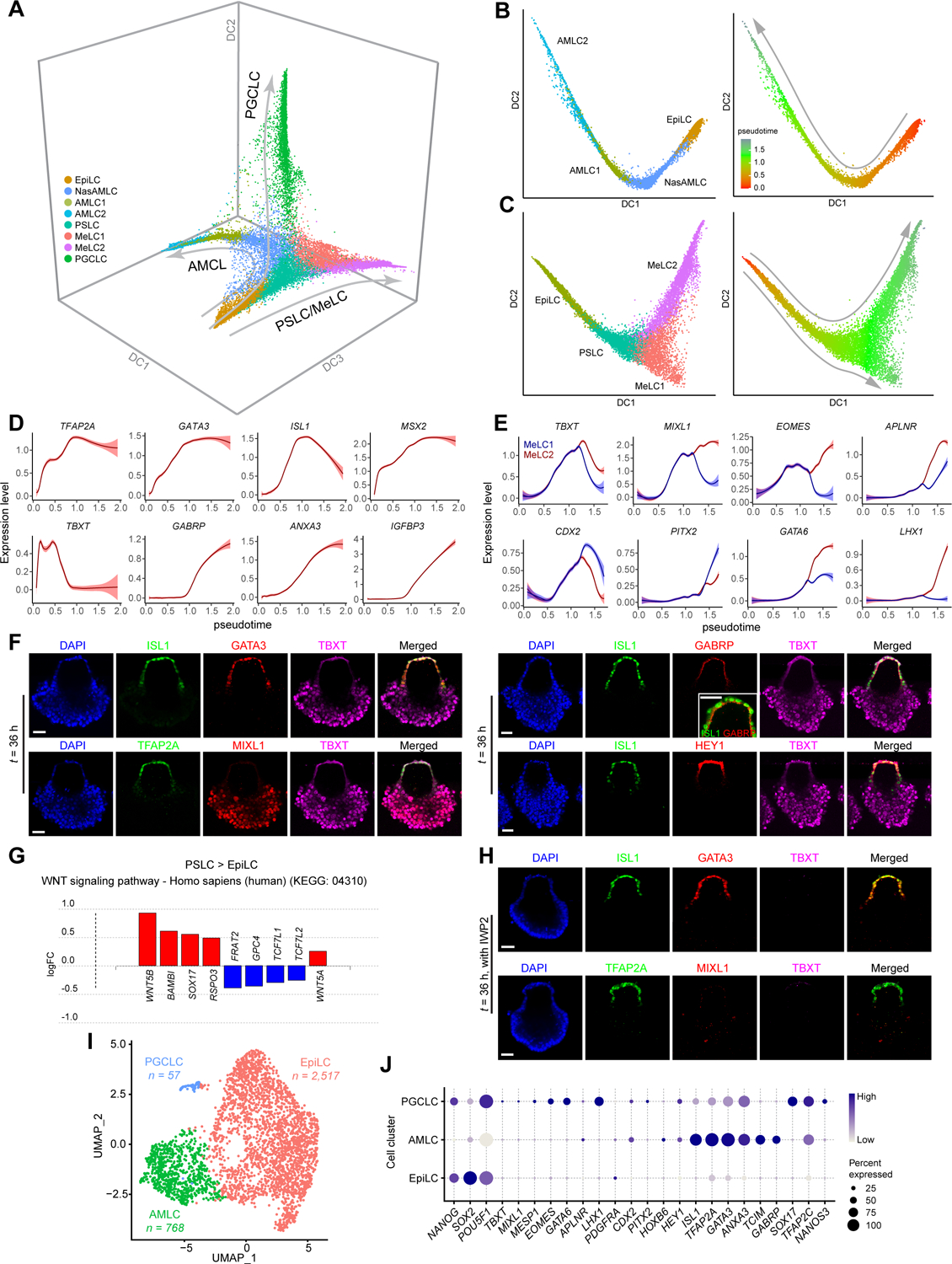
(A) Three-dimensional diffusion map based on embeddings of the UMAP plot in Figure 1D, showing developmental trajectories of AMLC, PSLC / MeLC and PGCLC lineages. The UMAP plot is color-coded according to cell identity annotations. See Video S1
(B) left: Trajectory inference (diffusion map) of AMLC lineage (EpiLC, NasAMLC, AMLC1 and AMLC2). right: Pseudotime analysis (color-coded) based on the AMLC lineage diffusion map.
(C) left: Trajectory inferences (diffusion map) of PSLC / MeLC lineage (EpiLC, PSLC and MeLC1 or MeLC2). right: Pseudotime analysis (color-coded) based on the PSLC / MeLC lineage diffusion map.
(D) Expression dynamics (pseudotime) of selected genes during AMLC lineage development. Level of confidence (0.95) is indicated by band width.
(E) Expression dynamics (pseudotime) of selected genes during PSLC / MeLC lineage development. Level of confidence (0.95) is indicated by band width.
(F) Representative confocal micrographs showing μPASEs at t = 36 h stained for ISL1, GATA3 and TBXT (i); TFAP2A, MIXL1 and TBXT (ii); ISL1, GABRP and TBXT (iii, with zoom-in view showing apical expression of GABRP); ISL1, HEY1, and TBXT (iv).
(G) Differentially expressed genes (DEGs) related to WNT signaling pathway (KEGG: 04310) in PSLC compared to EpiLC.
(H) Representative confocal micrographs showing μPASEs at t = 36 h stained for ISL1, GATA3 and TBXT (top); TFAP2A, MIXL1 and TBXT (bottom) with IWP2 supplemented into the basal medium from t = 0 h.
(I) UMAP and cell identity annotations of single-cell transcriptome data obtained for μPASEs at t = 48 h with IWP2 supplemented into the basal medium from t = 0 h. n indicates cell numbers of indicated cell types.
(J) Dot plot showing expression of key marker genes across the cell clusters as indicated. The sizes and colors of dots indicate the proportion of cells expressing the corresponding genes and their averaged scaled values of log-transformed expression, respectively.
In F and H, experiments were repeated four times with similar results. Nuclei were counterstained with DAPI. Scale bars, 50 μm.
On the 2D diffusion map, the MeLC lineage branches into two separate paths corresponding to MeLC1 and MeLC2 clusters (Figure 2C). Both MeLC1 and MeLC2 lineages show upregulated expression of common Meso markers, yet with some key genes exhibiting distinct expression levels (Figures 2E and S2B). For example, compared with MeLC1, MeLC2 expresses relatively higher levels of MIXL1, EOMES and GATA6 (Figures 2E and S2B), presumably corresponding to a lateral plate / intermediate Meso fate (Daoud et al., 2014; Prummel et al., 2019). MeLC1 lineage, in contrast, expresses higher levels of CDX2 and HOXB6 (Figures 2E and S2B), presumably corresponding to a paraxial Meso fate (Casaca et al., 2016; Chawengsaksophak et al., 2004). We further conducted immunostaining for selected AM and Meso markers in μPASEs at t = 24 h, 36 h, and 48 h to validate lineage fate specification (Figures 2F and S2C–S2E). GATA6highTBXThighMIXL1highCDX2low MeLC2 appears as leading cells in the migratory gastrulating cell population, whereas GATA6lowTBXTlowMIXL1lowCDX2high MeLC1 appears as trailing cells in this population (Figure S2F).
Patterning of AMLCs and PSLCs was evident in the μPASE by t = 24 h, with positive immunostaining for ISL1, GATA3 and TFAP2A in incipient AMLCs and for TBXT and MIXL1 in incipient MeLCs (Figures S2C and S2D, Video S2). Notably, expression of AM marker GABRP (Yang et al., 2021) is restricted on the apical surface of AMLCs, whereas expression of HEY1, another AM marker (Yang et al., 2021), is evident in both the nucleus and cytoplasm of AMLCs (Figure 2F). AMLCs appear to actively proliferate during μPASE development (Figure S2G). Nonetheless, we could not exclude the possibility that EpiLCs continuously supply AMLCs during μPASE development. To reveal transcriptome changes during AMLC development, we conducted differentially expressed gene (DEG) and pathway enrichment analysis to compare NasAMLC, AMLC1 and AMLC2 (Figure S2H and Mendeley Data Table3).
To explore potential mechanisms underlying lineage choices between NasAMLCs vs. PSLCs for EpiLCs, we examined DEGs upregulated in NasAMLCs and PSLCs relative to EpiLCs and noticed significant overlaps (144 out of 342 genes for NasAMLCs, 144 out of 184 genes for PSLCs) (Figure S2I and Mendeley Data Table3). KEGG pathway analysis suggests that development of both NasAMLCs and PSLCs requires TGF-β signaling (KEGG: 04350; Figure S2J), which is not surprising given that exogenous BMP4 is supplemented in the μPASE protocol. However, WNT signaling pathway (KEGG: 04310) is evident only in PSLCs but not in NasAMLCs (Figure 2G), suggesting a critical role of WNT signaling in Meso induction but not in amniogenesis. When IWP2, a small molecule inhibitor blocking the transport, secretion or signaling activity of all WNT molecules (Chen et al., 2009; Kadowaki et al., 1996; Zhai et al., 2004), was supplemented into the microfluidic device, development of PSLCs and MeLCs in the μPASE was completely inhibited, as evidenced by negative immunostaining for TBXT or MIXL1 (Figure 2H). In contrast, AMLCs still emerge under IWP2 treatment, as evidenced by positive immunostaining for ISL1, TFAP2A and GATA3 in flattened, presumptive AMLCs directly exposed to exogeneous BMP4 stimulation (Figure 2H). Consistently, the scRNA-seq data obtained from IWP2-treated μPASEs at t = 48 h show absence of PSLCs or MeLCs; instead, the majority of cells remain pluripotent, retaining the EpiLC identity (Figures 2I and 2J), further supporting the requirement of WNT signaling in PSLC/MeLC development. It should be noted that under IWP2 treatment, AMLCs show lower expression of several AM marker genes, such as ISL1, GABRP and GATA3, and there are much less PGCLCs in IWP2-treated μPASEs (Figures S2K and 2I, and Mendeley Data Table3).
PGCLC specification
During μPASE development, incipient PGCLCs emerge together with NasAMLCs and PSLCs by t = 24 h (Figure 1C). To explore the origin and specification of PGCLCs in the μPASE, a 2D diffusion map with EpiLC, PSLC, MeLC1 and PGCLC clusters isolated from the integrated scRNA-seq dataset was plotted, which, however, did not show a continuous developmental trajectory connecting EpiLCs, PSLCs, MeLCs with PGCLCs (Figure S3A). In addition, in this 2D diffusion map, K-branch algorithm (Chlis et al., 2017) did not identify a branching point or branches with proper confidence (Figure S3A). Thus, PGCLCs in the μPASE are unlikely originated from PSLCs or MeLCs.
In contrast, 2D diffusion map analysis with EpiLC, NasAMLC, AMLC1 / 2 and PGCLC clusters clearly shows lineage progression and bifurcation of NasAMLCs into PGCLCs and AMLCs (Figure 3A). RNA velocity vectors were also computed and overlaid onto the diffusion map in Figure 3A, revealing that NasAMLCs have the closest lineage relation with PGCLCs, consistent with the PAGA analysis in Figure 1F. Our observation here is consistent with recent studies of cynomolgus monkey embryos reporting that primate PGCs could emerge in the nascent AM prior to the gastrulation (Sasaki et al., 2016). Expression of selected genes relevant to PGC development was also plotted against diffusion pseudotime, revealing their upregulated expression during PGCLC lineage development (Figures 3B and S3B). Immunofluorescence analyses of μPASEs at t = 24 h, 36 h, and 48 h further confirmed spatiotemporal expression of some key PGC markers, including TFAP2C, NANOG, SOX17 and BLIMP1, in incipient PGCLCs (Figures 3C and S3C). Correlation coefficient analysis based on PGC ontogenic genes identified from the cynomolgus embryo transcriptome data suggests that the transcriptome of PGCLCs in the μPASE is similar to that of Day 2 hPGCLCs derived from conventional protocols (Chen et al., 2019; Sasaki et al., 2015) (Figure S3D). It is worth noting that the μPASE essentially is a posteriorized embryonic-like structure, mimicking the posterior portion of the amnion and epiblast compartments. As such, the development of μPASE gives rise to a greater number of hPGCLCs but lack the development of ectoderm.
Figure 3.
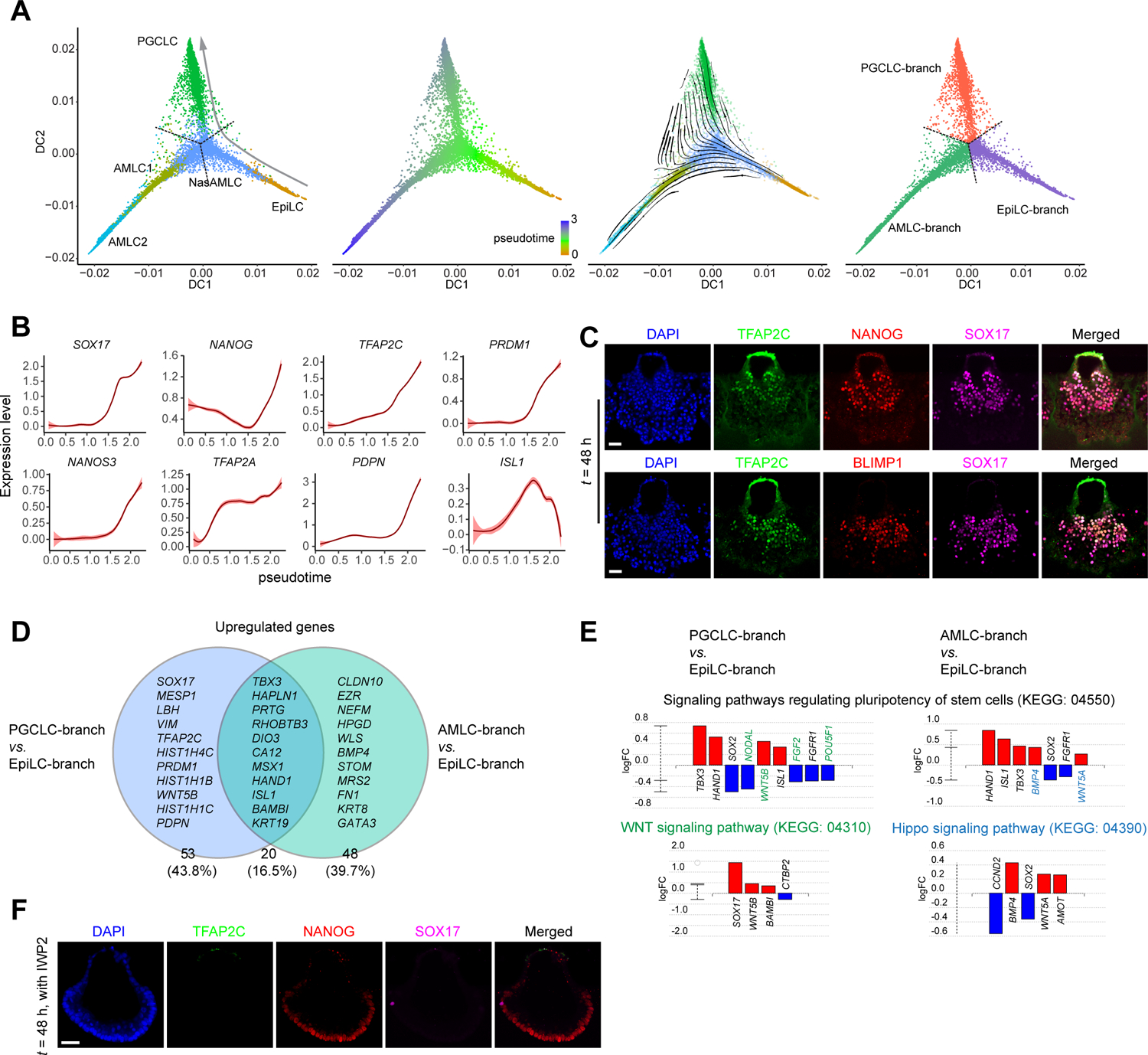
PGCLC specification during μPASE development. (A) Trajectory inference of PGCLC lineage. left: Diffusion map using EpiLC, NasAMLC, AMLC1, AMLC2, and PGCLC clusters from the UMAP plot in Figure 1D. Dotted lines show the branching point and branches identified by K-Branches algorithm. middle left: Pseudotime analysis based on the diffusion map. middle right: RNA velocity vectors overlaid on the diffusion map. right: Branches and the branching point identified by K-Branches algorithm. Note that NasAMLC cluster is separated into three branches, which after merging with EpiLC, AMLC1/2 and PGCLC, respectively, are annotated as EpiLC-branch NasAMLC, AMLC-branch NasAMLC and PGCLC-branch NasAMLC, respectively.
(B) Expression dynamics (pseudotime) of selected genes during PGCLC lineage development. Level of confidence (0.95) is indicated by band width.
(C) Representative confocal micrographs showing μPASEs at t = 48 h stained for TFAP2C, NANOG and SOX17 (top) and TFAP2C, BLIMP1 and SOX17 (bottom).
(D) Venn diagram showing upregulate genes of AMLC-branch and PGCLC-branch NasAMLCs when compared to EpiLC-branch NasAMLCs. Note that only a subset of these genes is specified in the diagram.
(E) Pathway analysis of DEGs in PGCLC-branch NasAMLCs and AMLC-branch NasAMLCs, as compared to EpiLC-branch NasAMLCs. Green and blue colors indicate genes or pathways identified only for PGCLC-branch NasAMLCs and AMLC-branch NasAMLCs, respectively.
(F) Representative confocal micrographs showing μPASEs at t = 48 h stained for TFAP2C, NANOG and SOX17, with IWP2 supplemented into the basal medium from t = 0 h.
In C and F, experiments were repeated four times with similar results. Nuclei were counterstained with DAPI. Scale bars, 50 μm.
Using K-branch algorithm, we identified developmental branches and the branching point for the 2D diffusion map with EpiLC, NasAMLC, AMLC1/2 and PGCLC clusters (Figure 3A). The K-branch analysis further allowed us to separate NasAMLCs into three sub-clusters, with each sub-cluster merged with EpiLCs, AMLCs and PGCLCs, respectively, and annotated as EpiLC-branch NasAMLC, AMLC-branch NasAMLC and PGCLC-branch NasAMLC, respectively (Figure 3A). To explore mechanisms underlying lineage choices between AMLCs vs. PGCLCs for NasAMLCs, DEGs upregulated in AMLC-branch NasAMLCs and PGCLC-branch NasAMLCs, as compared to EpiLC-branch NasAMLCs, were examined (Figure 3D; Mendeley Data Table4). KEGG pathway analysis of these DEGs reveals that PGCLC-branch NasAMLCs show upregulated WNT signaling, whereas AMLC-branch NasAMLCs exhibit upregulated Hippo activity (Figure 3E). We further performed pharmacological inhibition assays to explore the roles of different signaling pathways during μPASE development. When IWP2 was supplemented into the microfluidic device, development of PGCLCs was almost completely abolished in the μPASE (Figure 3F). PGCLCs show upregulated HIF-1 signaling activity compared to PGCLC-branch NasAMLCs (Figure S3E). When HIF-1 signaling was repressed by supplementing LY294002, which functions through binding to phosphoinositide 3-kinases (PI3Ks), in the microfluidic device, the percentage of PGCLCs was significantly reduced (Figures S3F and S3G). It should be noted that even though PI3K signaling functions downstream of HIF-1 signaling, it is also involved in other intracellular pathways. Thus, the involvements of HIF-1 and PI3K signaling in hPGCLC specification warrant further investigation.
Our scRNA-seq data analysis suggests that progenitors of PGCLCs likely pass through a transient transcriptome state similar to that of NasAMLCs before their full commitment to the PGCLC fate. Our data, however, do not exclude the possibility that cells at the junction between the AMLC and PSLC / MeLC compartments, or from the PSLC / MeLC pole of the μPASE, with a transient transcriptome state similar to that of NasAMLCs, can also give rise to PGCLCs. After all, NasAMLCs and PSLCs share a similar transcriptome, and cells in the early post-implantation human embryo might remain plastic (Chen et al., 2019; Irie et al., 2015; Kobayashi et al., 2017; Sasaki et al., 2015).
Transcriptomic comparison between μPASEs and Carnegie Stage 7 human gastrula
The μPASE recapitulates certain aspects of human development from early implantation to the onset of gastrulation. Notably, single-cell transcriptome data of a CS7 human gastrula recently became available (Tyser et al., 2021). Thus, we conducted transcriptomic comparison between μPASEs and the CS7 human gastrula. We first downsampled the μPASE dataset by randomly selecting 100 cells from each cluster and integrated this downsampled dataset with the CS7 human gastrula dataset (Figures S4A and S4B). Based on the transcriptome proximity in the UMAP plot, the μPASE contains cells corresponding to “Epiblast”, “Primitive Streak”, “Nascent Mesoderm”, “Emergent Mesoderm”, “Amniotic/embryonic ectoderm”, and “PGC” cells in the human gastrula. We next selected only these cells from the CS7 human gastrula dataset to integrate with the whole μPASE dataset. A UMAP plot of the integrated scRNA-seq dataset shows cell clustering similar to that from the μPASE scRNA-seq data alone (Figure 4A). Cells from the CS7 human gastrula are evident in all cell clusters of the UMAP plot (Figure 4A). Furthermore, for each annotated cell cluster, expression patterns of key lineage markers are consistent between cells from the CS7 human gastrula and μPASEs (Figure 4B). Most of the cells from the CS7 human gastrula fall into different cell clusters consistent with their lineage annotations in the original publication (Tyser et al., 2021), except for some cells originally classified as “Epiblast” or “Primitive Streak” that fall into NasAM or AM1 clusters (Figure 4C). Notably, PGCs in the CS7 human gastrula can be easily distinguished in the PGC cluster of the integrated dataset in the UMAP plot (Figure 4A).
Figure 4. Transcriptomic comparison between μPASEs and Carnegie Stage 7 human gastrula.
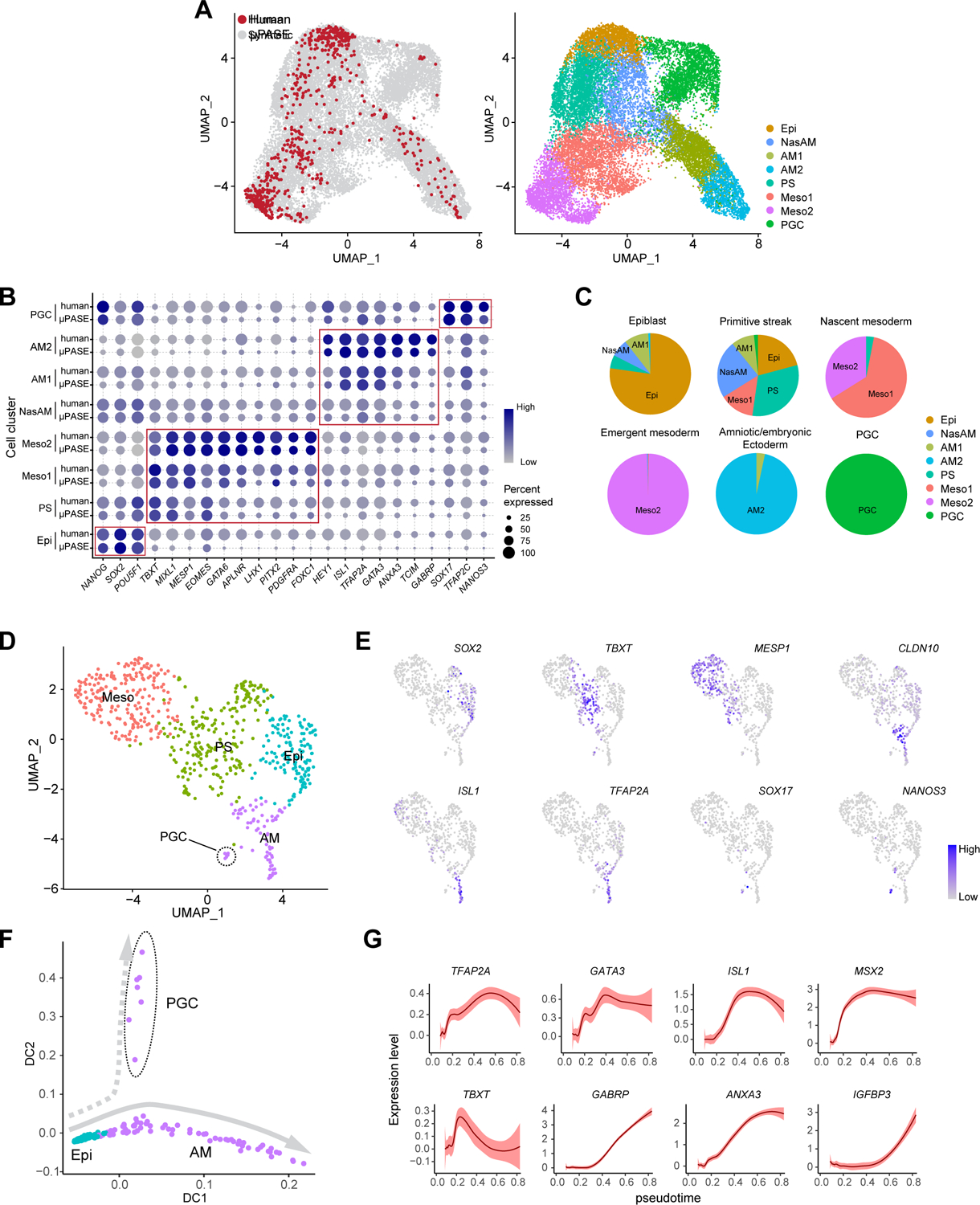
(A) left: UMAP of integrated dataset of μPASEs from t = 24, 36 and 48 h (18,335 cells; grey) and CS7 human gastrula (647 cells, excluding irrelevant cells; red). right: UMAP project of integrated dataset with cell identity annotations.
(B) Dot plot comparing expression of key marker genes across different cell clusters from μPASEs and CS7 human gastrula as indicated. The sizes and colors of dots indicate the proportion of cells expressing the corresponding genes and their averaged scaled values of log-transformed expression, respectively.
(C) Comparisons between human gastrula cell annotations in the original publication and annotations in the integrated dataset. The original annotations are indicated above the pie charts. See Mendeley Data Table5
(D) Re-analysis of related cells from CS7 human gastrula (647 cells). Cell identity annotations are color coded as indicated. Note that primordial germ cells (PGCs) appear in the amniotic/embryonic ectoderm (AM) cluster.
(E) Feature plots showing expression of selected lineage markers used for cell cluster annotations in (D).
(F) Trajectory inference (diffusion map) of the AM cluster (including PGCs) in (D).
(G) Expression dynamics (pseudotime) of selected genes during AM lineage development. Level of confidence (0.95) is indicated by band width.
Epi: Epiblast; PS: primitive steak; Meso: Mesoderm; AM: amniotic ectoderm; PGC: primordial germ cell.
In the original publication of the CS7 human gastrula (Tyser et al., 2021), a UMAP plot was generated with all the cells in the human gastrula, including those at relatively late developmental stages, such as “Hemogenic Endothelial Progenitors” and “Erythrocytes”, which could negatively affect the resolution of cell clustering analysis, especially for those closely related cell lineages. To address this issue, we re-generated a UMAP plot using only the “Epiblast”, “Primitive Streak”, “Nascent Mesoderm”, “Emergent Mesoderm”, “Amniotic/embryonic ectoderm” and “PGC” clusters from the human gastrula dataset (Figure 4D). Interestingly, this UMAP plot reveals distinct cell clusters corresponding to Meso and AM, supported by feature plots showing expression patterns of key Meso and AM markers (Figures 4D and 4E). Notably, human PGCs identified in the original publication is clustered together with AM cells in the UMAP plot (Figure 4D), supporting their close lineage relation. We further isolated the Epi, AM, and PGCs from the CS7 human gastrula to generate a diffusion map (Figure 4F). Surprisingly, AM and PGC seemingly display lineage bifurcation trajectories from the Epi (Figure 4F), similar to μPASEs in Figure 3A. In addition, as shown in Figure 4G, pseudotime gene expression dynamics of AM cells from the CS7 human gastrula is highly consistent with that of AMLCs in μPASEs in Figure 2D. However, due to the low number of PGCs, AM and PGC lineage bifurcation is not as evident as that shown in the μPASE diffusion map (Figure 3A); and we could not obtain pseudotime gene expression dynamics for PGCs.
To characterize pluripotency state transition in EpiLCs during the progressive development of the μPASE, we further performed scRNA-seq for cultured hPSCs and μPASEs at t = 0 h and t = 12 h. No distinguishable cell clusters or populations were observed when the scRNA-seq data were analyzed using Seurat R package (Mendeley Data Figure 1). Notably, although μPASEs at t = 12 h show upregulated TFAP2A expression owing to exogeneous BMP4 treatment, transcriptomes of cells in the μPASE have not yet changed sufficiently for the cells to emerge as distinct clusters in the UMAP. We then downsampled these datasets and compared them with transcriptome data of the human morula, pre-implantation epiblast from human blastocysts (Petropoulos et al., 2016), early post-implantation epiblast from days post-fertilization (d.p.f) 9 and d.p.f 11 in-vitro cultured human embryos (Molè et al., 2021), and late post-implantation epiblast from the CS7 human gastrula (Tyser et al., 2021). As suggested by the principal component analysis (PCA) plot (Figures S4C and S4D), cultured hPSCs and EpiLCs in the μPASE share comparable transcriptome states and are both at a developmental stage between d.p.f 11 and d.p.f 16 epiblast. Thus, there was no compelling evidence showing heterogeneous pluripotency states or transitions at early stages of μPASE development.
Transcriptomic coordination of early development across different primate species
NHP monkey embryos, including cynomolgus (Macaca fascicularis) embryos, have been used successfully as an in vivo model to study primate development (Ma et al., 2019; Nakamura et al., 2016; Sasaki et al., 2016; Yang et al., 2021). Compared with NHP monkeys, chimpanzees share much more of human DNA (99%), making them our closest living relatives (Gibbs et al., 2007; Mikkelsen et al., 2005). Thus, we sought to generate μPASEs from chimpanzee induced pluripotent stem cells (iPSCs) to examine whether chimpanzee μPASEs (or C_μPASEs) would develop in a fashion similar as μPASEs made from hPSCs (or H_μPASEs). To this end, the same microfluidic protocol for H_μPASE development was used for C_μPASE formation. Under exogeneous human BMP4 stimulation, C_μPASEs also display prominent molecular and morphogenetic asymmetry, with the pole exposed to BMP4 differentiating progressively into squamous, flattened AMLCs positive for AM markers ISL1, GATA3 and TFAP2A, and the opposite pole developing into germ layer lineages positive for TBXT and MIXL1 (Figure 5A). Similar to H_μPASEs, PGCLCs also emerge in C_μPASEs, as evidenced by positive immunostaining for SOX17, TFAP2C, NANOG and BLIMP1 (Figure 5A). However, distinct from H_μPASEs, FOXA2+BLIMP1+ endoderm-like cells (EndoLCs) also emerge in C_μPASEs, and these EndoLCs randomly intermix with MeLCs and PGCLCs (Figure 5A). scRNA-seq analysis of C_μPASEs obtained at t = 48 h further confirms the presence of EndoLCs in C_μPASEs (Figures 5B and 5C). It is worth noting that C_μPASEs do not develop a prominent central lumen (the pro-amniotic-like cavity) as in H_μPASEs (Figure 5A).
Figure 5. Transcriptomic coordination of primate early post-implantation development.
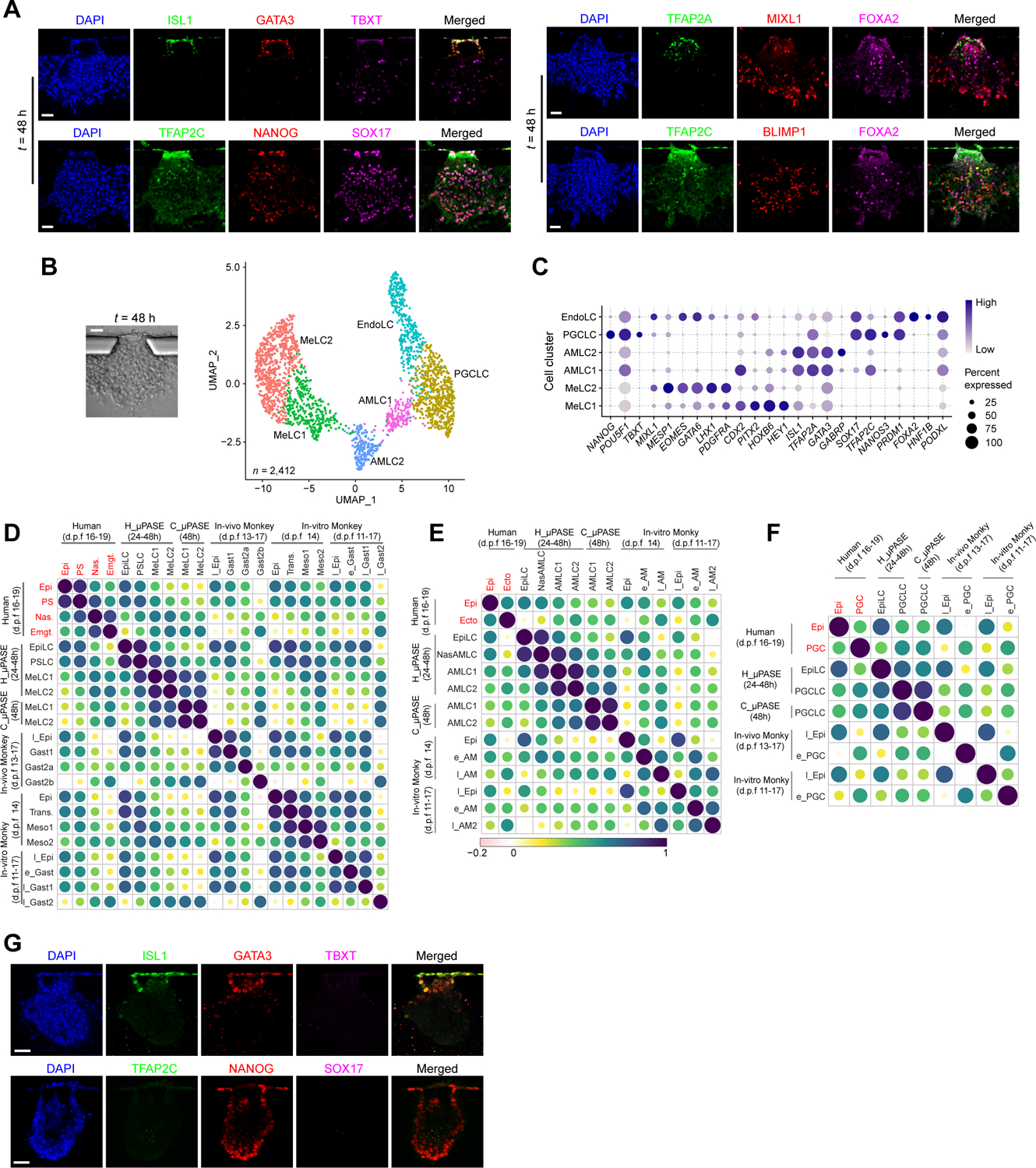
(A) Representative confocal micrographs showing chimpanzee μPASEs at t = 48 h stained for ISL1, GATA3 and TBXT (i); TFAP2A, MIXL1 and FOXA2 (ii); TFAP2C, NANOG, SOX17 (iii); TFAP2C, BLIMP1 and FOXA2 (iv).
(B) Left: Bright-field image showing a chimpanzee μPASE at t = 48 h. Right: UMAP plot of single-cell transcriptome dataset from chimpanzee μPASEs at t = 48 h. Cell identity annotations are color coded as indicated. n indicates the total cell number. EndoLC: Endoderm-like cell.
(C) Dot plot showing expression of key marker genes across the cell clusters as indicated. The sizes and colors of dots indicate the proportion of cells expressing the corresponding genes and their averaged scaled values of log-transformed expression, respectively.
(D) Heat map of correlation matrix for primitive streak / mesoderm-related lineages including those reported by others. Correlation coefficients between indicated cell types are calculated based on mesoderm ontogenic genes identified from cynomolgus embryo transcriptome data (131 in common). See Mendeley Data Table6.
(E) Heat map of correlation matrix for amnion-related lineages including those reported by others. Correlation coefficients between indicated cell types are calculated based on amnion ontogenic genes identified from cynomolgus embryo transcriptome data (142 in common). See Mendeley Data Table6.
(F) Heat map of correlation matrix for PGC-related lineages including those reported by others. Correlation coefficients between indicated cell types are calculated based on PGC ontogenic genes identified for cynomolgus embryo transcriptome data (194 in common). See Mendeley Data Table6.
(G) Representative confocal micrographs showing chimpanzee μPASEs at t = 48 h with IWP2 supplemented into the basal medium from t = 0 h, stained for ISL1, GATA3 and TBXT (top); TFAP2C, NANOG and SOX17 (bottom),
In D-F, correlation coefficients are calculated using average ontogenic gene expression of single cells. Original cell annotations in published datasets are used. Log2(reads per million + 1) and Log2 (transcripts per million + 1) are used for transcriptome datasets generated using 10× Chrome and Smart-seq2, respectively.
Human: human gastrula from Ref.(Tyser et al., 2021). Epi: epiblast; PS: primitive streak; Nas.: Nascent mesoderm; Emgt.: emergent mesoderm; Ecto.: amniotic/embryonic ectoderm; PGC: primordial germ cell. H_μPASE and C_μPASE: μPASEs generated from human and chimpanzee cells, respectively. In vivo monkey: in vivo cynomolgus embryo from Ref.(Nakamura et al., 2016; Sasaki et al., 2016). l_EPI: late epiblast; Gast: gastrulation; e_PGC: early primordial germ cell. In vitro monkey: in vitro cultured cynomolgus embryo from Ref.(Ma et al., 2019; Yang et al., 2021). Trans: Transition; Meso: mesoderm; l_EPI: late epiblast; e_Gast: early gastrulation; l_Gast: late gastrulation; e_AM: early amniotic ectoderm; l_AM: late amniotic ectoderm.
Expression of ontogenic genes is included in Mendeley Data Table6. In A and G, experiments were repeated four times with similar results. Nuclei were counterstained with DAPI. Scale bars, 50 μm.
Transcriptomic comparisons between related corresponding cell clusters in H_μPASEs, C_μPASEs, and human and cynomolgus embryos (Ma et al., 2019; Nakamura et al., 2016; Sasaki et al., 2016; Tyser et al., 2021; Yang et al., 2021) reveal that such cell clusters from these in vitro and in vivo systems, regardless in the Meso (Figure 5D), AM (Figure 5E) or PGC (Figure 5F) lineages, show strong correlations based on ontogenic genes identified from cynomolgus embryo transcriptome data (Nakamura et al., 2016; Sasaki et al., 2016; Yang et al., 2021). Consistently, when IWP2 was supplemented into C_μPASEs, developments of PSLCs and PGCLCs were inhibited, whereas ISL1+GATA3+ AMLCs still emerged, similar with IWP2-treated H_μPASEs (Figure 5G).
NODAL is essential for mesoderm development
Our DEG and pathway analyses of μPASE scRNA-seq data in Figures 2G, 2H, 3D–3F, S2F and S2G suggest complex cell-cell interactions involved in early post-implantation human development. Thus, μPASE scRNA-seq data at t = 24 h were further analyzed using CellChat (Jin et al., 2021) for inference and analysis of ligand-receptor interactions (Figures S5A–S5C). Among cell clusters present in μPASEs at t = 24 h, AMLCs and PSLCs were identified as major sources of signaling ligands involved in key developmental pathways, such as BMP and WNT pathways (Ben-Haim et al., 2006; Bernardo et al., 2011; Clevers, 2006; Rivera-Perez and Magnuson, 2005; Wang et al., 2014; Zhao, 2003) (Figures S5A–S5C). This is consistent with a recent study of pre-gastrulation cynomolgus embryos (Yang et al., 2021), which shows that both AM and PS / Meso cells upregulate BMP4 and WNT5B and extraembryonic mesenchyme cells show high expression of BMP2 and BMP4 (Figures S5D–S5F). μPASEs lack extraembryonic mesenchyme cells, the effects of which might have been substituted by exogeneous supplementation of BMP4 in the μPASE protocol. For comparison, ligand-receptor interaction analysis was also conducted using CellChat on scRNA-seq data from E6.5 mouse embryos (Pijuan-Sala et al., 2019), revealing the extraembryonic ectoderm as the only source of BMP and WNT signals (Figures S5G–S5I). Interestingly, during the development of both μPASEs and pre-gastrulation cynomolgus embryos, non-canonical WNT pathways show a greater signaling strength than canonical WNT, whereas the opposite is observed for E6.5 mouse embryos (Figures S5A, S5D and S5G).
Our ligand-receptor interaction analysis for μPASEs using CellChat further reveals that, compared to BMP and WNT pathways identified as an “incoming signal” for multiple cell clusters, NODAL signals appear to be perceived only by PSLCs (Figures S5A and S5B), suggesting a critical role of NODAL signaling in PSLC development. To study the functional role of NODAL, NODAL-knockout (KO) hPSC lines were generated and used for μPASE development (Figures S6A–S6E). By t = 48 h, majority of cells in NODAL-KO μPASEs appear to have developed into flattened ISL1+GATA3+ AMLCs, and there are no cells disseminating away from the μPASE structure, in distinct contrast with wildtype control μPASEs (Figures 6A–6D and S6F). Consistently, scRNA-seq data obtained from NODAL-KO μPASEs at t = 48 h showed a dominant population of AMLCs at the expense of MeLCs (Figures 6B,6C,6E–6G), further supporting the critical role of NODAL in MeLC development. PGCLCs were still evident in NODAL-KO μPASEs at t = 48 h, albeit with a much less percentage compared with those in wildtype controls (8.1% vs. 28.2%; Figures 6D,6E–6G and S6G).
Figure 6. NODAL is essential for mesoderm development in μPASEs.
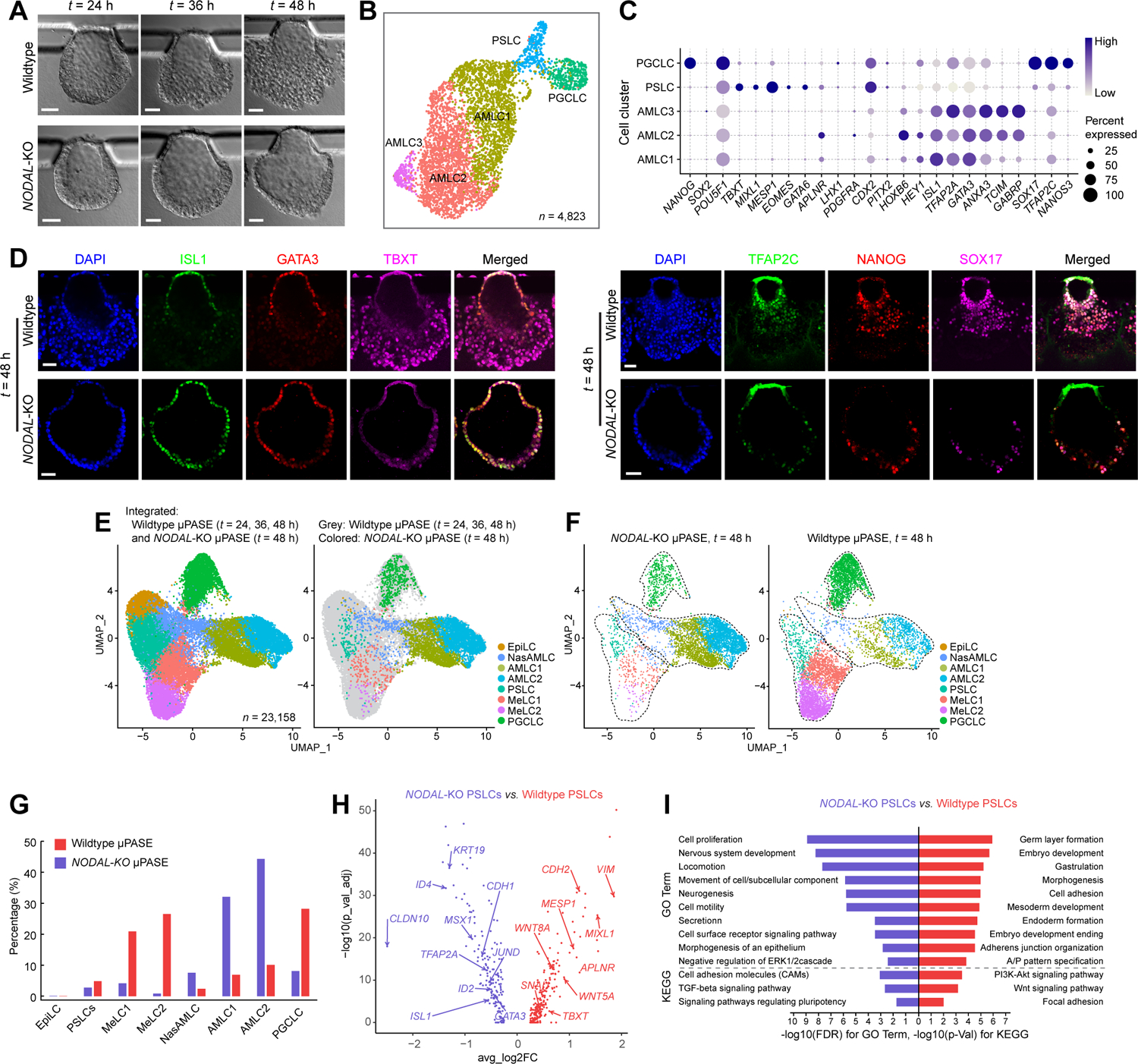
(A) Representative bight-field images showing progressive development of μPASEs generated from wildtype (top) and NODAL-KO (bottom) hPSCs.
(B) UMAP plot of single-cell transcriptome data from NODAL-KO μPASEs at t = 48 h, with cell identity annotations color coded. n indicates the total cell number.
(C) Dot plot showing expression of key marker genes across the cell clusters in NODAL-KO μPASEs as indicated. The sizes and colors of dots indicate the proportion of cells expressing the corresponding genes and their averaged scaled values of log-transformed expression, respectively.
(D) Representative confocal micrographs showing μPASEs generated from wildtype and NODAL-KO hPSCs at t = 48 h, stained for ISL1, GATA3 and TBXT (left) and TFAP2C, NANOG and SOX17 (right).
(E) left: Integrated UMAP plot of wildtype μPASEs at t = 24, 36, 48 h and NODAL-KO μPASEs at t = 48 h, color-coded according to cell identity annotations. right: data from wildtype and NODAL-KO μPASEs are shown in different colors as indicated.
(F) UMAP plots of NODAL-KO (left) and wildtype (right) μPASEs at t = 48 h, with data isolated from E. Doted lines contour cell clusters corresponding to AMLC, PSLC / MeLC and PGCLC lineages.
(G) Percentages of indicted cell types in wildtype (red) and NODAL-KO (blue) μPASEs.
(H) Volcano plots showing DEGs between PSLCs from wildtype and NODAL-KO μPASEs, with selected genes labelled.
(I) Enriched GO categories and KEGG pathways among DEGs between wildtype and NODAL-KO PSLCs.
In D, experiments were repeated three times with similar results. Nuclei were counterstained with DAPI. Scale bars, 50 μm.
We further conducted DEG and Gene Ontology (GO) enrichment analyses for PSLCs from NODAL-KO and wildtype control μPASEs (Figure 6H and 6I; Mendeley Data Table7). In NODAL-KO PSLCs, BMP target genes, such as ID2, TFAP2A and ISL1, are upregulated, whereas PS / Meso-related genes, such as MESP1 and MIXL1, and EMT related genes, including SNAI1 and VIM, are downregulated (Figure 6H). PI3K-Akt, WNT and focal adhesion signaling pathways appear to be downstream targets of NODAL signaling in PSLCs (Figure 6I). We speculate that lineage bifurcation between AMLCs and PSLCs from EpiLCs might be regulated by a competition between BMP and NODAL signaling. Absence or repression of NODAL signaling in the μPASE could lead to “hyper” BMP activities, which in turn causes excessive AMLC development (marked by an expanded ISL1 domain) and greater BMP activities in PSLCs. We also compared PGCLCs from wildtype and NODAL-KO μPASEs (Figure S6H and Mendeley Data Table7). Consistently, when SB431542, a pharmacological NODAL signaling inhibitor, was supplemented into the microfluidic device, development of PSLCs/MeLCs in μPASEs was completely inhibited (Figure S6I). Impaired development of MeLCs in NODAL-KO μPASEs can be efficiently reversed by supplementing ACTIVIN A, a NODAL pathway agonist, into the channel opposite to BMP4 stimulation. In addition to rescuing MeLC development, supplementing ACTIVIN A to NODAL-KO μPASEs also leads to the specification of BLIMP1+FOXA2+ EndoLCs (Figure S6J). Supplementing ACTIVIN A to wildtype μPASEs also results in specification of BLIMP1+FOXA2+ EndoLCs (Figure S6K). We also successfully generated μPASEs using a different hPSC line, including its corresponding NODAL-KO line, and a different chimpanzee iPSC line, and repeated IWP2 and SB431542 treatment assays with consistent results (Mendeley Data Figure 2). Consistently, we did not observe EndoLC development in μPASEs generated with these additional hPSC lines (Mendeley Data Figure 2).
Stringent criteria for distinguishing human trophoblast and amniotic ectoderm
In vivo, blastocyst trophoblast and pre-gastrulation AM both appear as flattened, squamous epithelium, and these two cell types share many lineage markers (Blakeley et al., 2015; Petropoulos et al., 2016; Tyser et al., 2021; Yang et al., 2021). Unique markers that can distinguish between these two cell types remain elusive, leading to confusions about true cell lineage identities in different human embryoids (Xu et al., 2002; Zhao et al., 2021). Through comparative transcriptome analysis of AM cells in the CS7 human gastrula (Tyser et al., 2021) and trophoblast cells in human blastocysts (Blakeley et al., 2015; Petropoulos et al., 2016), we identified a Trophoblast_Amnion ontogenic gene list (Figures S7A and S7B; Mendeley Data Table8) and applied this list to examine the properties of human trophoblast-like cells or AMLCs reported previously (Figure 7A). This ontogenic gene list contains several previously reported AM makers, such as ISL1, GABRP and IGFBP3 (Yang et al., 2021), and trophoblast makers, such as GCM1, HAVCR1 and CGA (Li et al., 2019; Okae et al., 2018; Pillai et al., 2019). Notably, several pan-preimplantation embryo markers, including DPPA3 and DNMT3L (Guo et al., 2021; Io et al., 2021; Yanagida et al., 2021) are found to be quite efficient markers for distinguishing between blastocyst trophoblast and pre-gastrulation AM. Based on quantified correlation coefficients between previously reported human trophoblast-like cells or AMLCs and human trophoblast or AM cells, we conclude that the transcriptome of BMP4-treated primed hPSCs, as in μPASEs, in 2D Transwell membrane-based AMLC differentiation assays (Zheng et al., 2019b) and in 2D patterned gastrulation models (Minn et al., 2020), is similar to that of human AM cells, whereas the transcriptome of trophoblast-like cells derived from naïve hPSCs (Dong et al., 2020; Guo et al., 2021; Io et al., 2021; Yanagida et al., 2021) is consistent with that of human blastocyst trophoblast. However, transcriptome of trophoblast-like cells derived from extended pluripotent stem (EPS) cells as well as that of trophoblast-like cells present in recently reported human blastocyst-like structures fall somewhat in-between (Liu et al., 2021; Yu et al., 2021).
Figure 7. Stringent criteria for identifying human trophoblast and amniotic ectoderm.
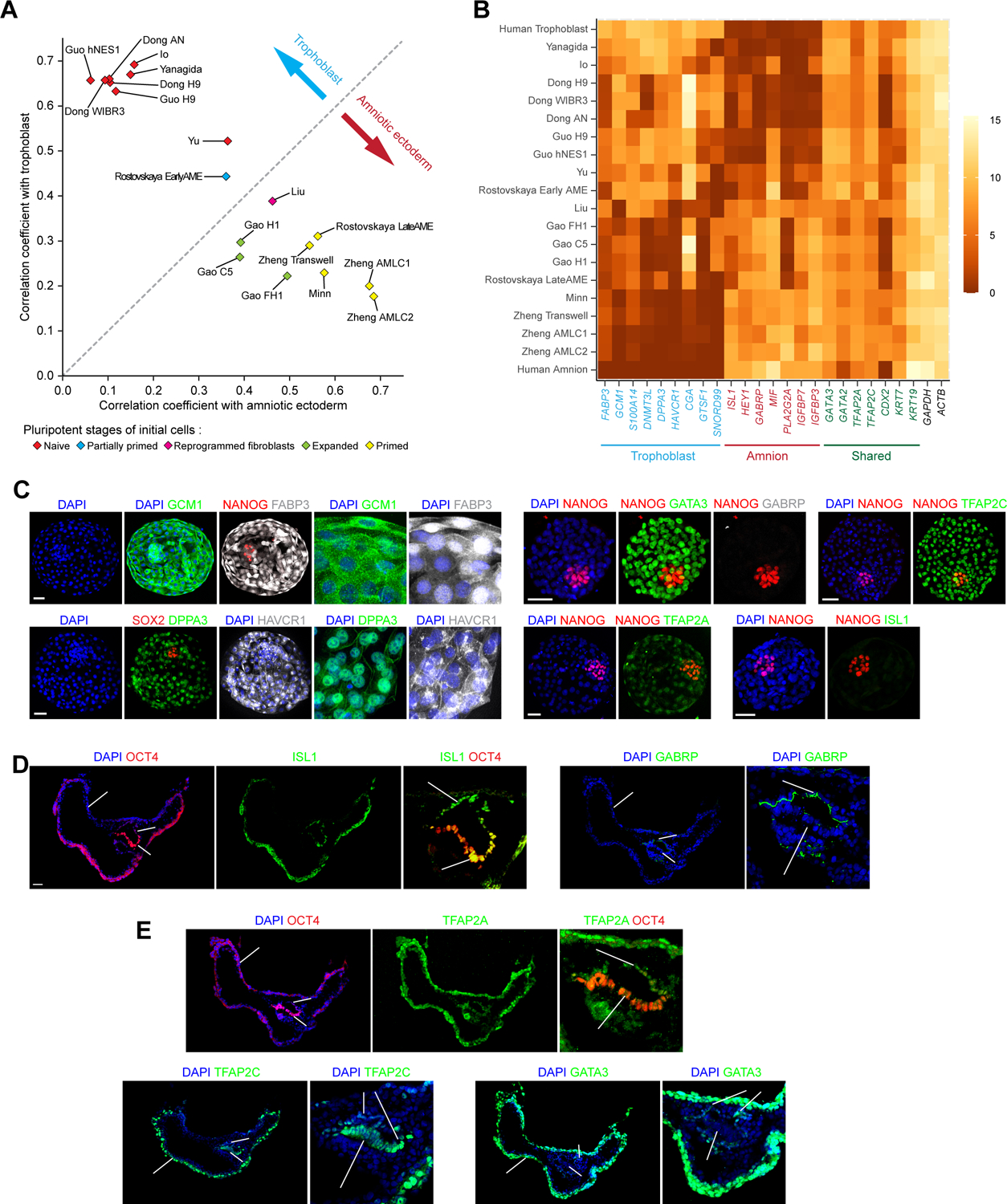
(A) Correlation coefficients of in vitro derived cells with human trophoblast and amniotic ectoderm, computed using Trophoblast_Amnion ontogenic genes. See Mendeley Data Table8.
(B) Heatmap showing expression levels of selected genes related to human trophoblast and amniotic ectoderm. Identified human trophoblast and amniotic ectoderm markers are highlighted in blue and red, respectively. Markers shared between human trophoblast and amniotic ectoderm are highlighted in green.
(C) Representative confocal micrographs showing E6 human blastocysts, stained for GCM1, NANOG and FABP3; SOX2, DPPA3 and HAVCR1 (Images on the right show magnified views of the trophectoderm); NANOG, GATA3 and GABRP; NANOG and TFAP2C; NANOG and TFAP2A; and NANOG and ISL1.
(D) Representative confocal micrographs showing in vitro cultured D14 cynomolgus embryos, stained for OCT4 and ISL1; and GABRP. Images on the right show magnified views of the epiblast and amniotic ectoderm.
(E) Representative confocal micrographs showing in vitro cultured D14 cynomolgus embryos, stained for OCT4 and TFAP2A, TFAP2C and GATA3. Images on the right show magnified views of the epiblast and amniotic ectoderm.
Human trophoblast (Ref.(Blakeley et al., 2015; Petropoulos et al., 2016)), Human amnion (Ref.(Tyser et al., 2021)), Zheng Transwell (Ref.(Zheng et al., 2019b)), Gao C5, Gao H1, Gao FH1(Ref.(Gao et al., 2019)), Minn (Ref.(Minn et al., 2020)), Liu (Ref.(Liu et al., 2021)), Yu (Ref.(Yu et al., 2021)), Guo H9, Guo hNES1(Ref.(Guo et al., 2021)), Io (Ref.(Io et al., 2021)), Yanagida (Ref.(Yanagida et al., 2021)), Dong H9, Dong WIBR3, Dong AN (Ref.(Dong et al., 2020)), Rostovskaya EarlyAME, Rostovskaya LateAME (Ref.(Rostovskaya et al., 2022))
Correlation coefficients and gene expression heatmap were calculated based on averages of experimental repeats, if any. When the published work contains transcriptome datasets from multiple time points, datasets from the experimental endpoint are utilized.
In C-E, experiments were repeated twice with similar results. Nuclei were counterstained with DAPI. Scale bars, 50 μm.
Using stringent criteria, we further identified a subset of DEGs within the Trophoblast_Amnion ontogenic gene list useful for distinguishing between human blastocyst trophoblast and pre-gastrulation AM and thus between human trophoblast-like cells and AMLCs (see Methods; Figures 7B, S7A and S7B). We should note that although AM transcriptome is obtained from the CS7 human gastrula (Tyser et al., 2021), which remains as the only bona fide human pre-gastrulation AM data currently available, most of pre-gastrulation AM markers identified here, including ISL1, HEY1, GABRP and IGFBP3, are upregulated in the pre-gastrulation AM of cynomolgus embryos (Yang et al., 2021).
We further conducted immunofluorescence analyses of IVF human blastocysts (D6), in vitro cultured cynomolgus embryos (D14), trophoblast stem cells (TSCs) (Okae et al., 2018), and AMLCs derived from BMP4-treated primed hPSCs (Zheng et al., 2019b), to ascertain these newly identified human blastocyst trophoblast and pre-gastrulation AM markers (Figures 7C–7E and S7C). Consistently, trophoblast cells of D6 human blastocysts show positive immunostaining for GCM1, FABP3, DPPA3 and HAVCR1, but are negative for ISL1 or GABRP (Figure 7C). In contrast, AM cells in D14 cynomolgus embryos show clear immunostaining for ISL1 and GABRP (Figure 7D). GATA3, TFAP2A and TFAP2C show positive immunostaining in both trophoblast cells of D6 human blastocysts and AM cells of D14 cynomolgus embryos (Figures 7C and 7E). Immunofluorescence analyses of TSCs (Okae et al., 2018), AMLCs (Zheng et al., 2019b) and μPASEs at t = 36 h further support that ISL1, GCM1 and HAVCR1 can be utilized for distinguishing between trophoblast-like cells and AMLCs, whereas GATA3, TFAP2A or TFAP2C could not (Figure S7C and S7D). We should note that ISL1 is a particular useful nuclear marker for distinguishing between human blastocyst trophoblast and pre-gastrulation AM and thus between human trophoblast-like cells and AMLCs (Figures 7C–7E and S7C). The function of ISL1 in the AM in inducing Meso development in cynomolgus embryos has recently been documented (Yang et al., 2021).
DISCUSSION
Experimentation on human embryoids can lead to a better understanding of the mechanisms of human development and offers opportunities for functional genomic studies of disease-causing mechanisms, identification of therapeutic targets, and preclinical modeling of advanced therapeutics for precision medicine (Fu et al., 2021; Rossant and Tam, 2021). When developing new embryoids or improving existing embryoids, rigorous scientific scrutiny must be implemented to avoid incorrect interpretations or overstatements (Posfai et al., 2021; Zhao et al., 2021). Currently, there are few lineage markers accepted for distinguishing between human trophectoderm and AM cells, and most existing embryoids are still limited in imitating sequential cell lineage diversifications and 3D tissue organizations exhibited stereotypically in natural embryos. Besides using cell lineage markers, validation and authentication of embryoids are currently commonly conducted through comparative transcriptome studies (e.g., through integration of scRNA-seq datasets from embryoids and in vivo models) (Posfai et al., 2021; Zhao et al., 2021). However, caution should be taken when interpreting such integrated datasets, since, although seemingly “unbiased”, existing scRNA-seq data analysis tools still have notable artifacts depending on the cell types present, cell numbers and percentages of each cell type, and specific computational algorithms used in the analysis tools. For instance, AM cells only account for a very small cell population in post-implantation primate embryos; such a small AM cell number can cause significant issues when scRNA-seq data of AM cells are integrated with human blastocyst datasets wherein trophoblast cells are abundant. Thus, we propose that in order to validate cell identities in peri-implantation human embryoids, expression of key cell fate markers need to be clearly demonstrated, in addition to transcriptome comparison based on scRNA-seq data. The cell identity markers established in this work for distinguishing between human trophoblast and AM cells represent one step towards this important direction.
LIMITATIONS OF STUDY
As demonstrated in this study, μPASEs provide a promising tractable experimental model for exploring previously inaccessible phases of early post-implantation human development. However, μPASEs lack a few key embryonic and extraembryonic lineages that present in the post-implantation human embryo, including the hypoblast (or extraembryonic endoderm), extraembryonic mesoderm, or trophoblast cells. Therefore, μPASEs only recapitulate certain aspects of the lineage diversification and development of the posterior end of the embryonic sac during the early post-implantation human development. Additionally, μPASEs disintegrate and lose their embryonic-like structure soon after 48 h in culture, as differentiating MeLCs that are undergoing EMT emigrate from the μPASE structure, which limits the potential of μPASEs for prolonged culture to investigate cell lineage development and embryonic tissue formation at later developmental stages. It should also be noted that cell type annotations and lineage inference analyses of the μPASE in this work are solely based on transcriptomic studies. Thus, caution should be taken when interpreting the data included in this Resource.
STAR★Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jianping Fu (jpfu@umich.edu).
Materials availability
Requests for NODAL-knockout H9 hPSCs should be addressed to Yi Zheng and Jianping Fu (yzheng88@syr.edu, jpfu@umich.edu).
Data and code availability
scRNA-seq data have been deposited at the Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Interactive visualization of processed datasets is available at https://umichibbl.shinyapps.io/shinyapp/.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-ISL1 antibody | DSHB | Cat# 39.4D5, RRID:AB_2314683 |
| Rabbit anti-GATA3 antibody | Cell Signaling Technology | Cat# 5852, RRID:AB_10835690 |
| Mouse anti-GATA3 antibody | Thermo Fisher Scientific | Cat# MA1–028, RRID:AB_2536713 |
| Rabbit anti-GABRP antibody | Thermo Fisher Scientific | Cat# PA5–46830, RRID:AB_2610501 |
| Rabbit anti-MIXL1 antibody | Atlas Antibodies | Cat# HPA005662, RRID:AB_2667382 |
| Rabbit anti-HEY1 antibody | Abcam | Cat# ab154077, RRID:AB_2893447 |
| Mouse anti-TFAP2A antibody | Santa Cruz Biotechnology | Cat# sc-12726, RRID:AB_667767 |
| Goat anti-TBXT antibody | Thermo Fisher Scientific | Cat# PA5–46984, RRID:AB_2610378 |
| Rabbit anti-NANOG antibody | Cell Signaling Technology | Cat# 4903, RRID:AB_10559205 |
| Goat anti-NANOG antibody | R and D Systems | Cat# AF1997, RRID:AB_355097 |
| Mouse anti-TFAP2C antibody | Santa Cruz Biotechnology | Cat# sc-12762, RRID:AB_667770 |
| Goat anti-SOX17 antibody | R&D Systems | Cat# AF1924, RRID:AB_355060 |
| Rabbit anti-BLIMP1 antibody | Cell Signaling Technology | Cat# 9115, RRID:AB_2169699 |
| Rabbit anti-FOXA2 antibody | Cell Signaling Technology | Cat# 8186, RRID:AB_10891055 |
| Mouse anti-OCT4 antibody | Santa Cruz Biotechnology | Cat# sc-5279, RRID:AB_628051 |
| Rabbit anti-OCT4 antibody | Santa Cruz Biotechnology | Cat# sc-9081, RRID:AB_2167703 |
| Rabbit anti-SOX2 antibody | Millipore | Cat# AB5603, RRID:AB_2286686 |
| Mouse anti-DPPA3 antibody | Millipore | Cat# MAB4388, RRID:AB_2094156 |
| Mouse anti-GCM1 antibody | Santa Cruz Biotechnology | Cat# sc-101173, RRID:AB_2108121 |
| Goat anti-HAVCR antibody | R and D Systems | Cat# AF1750, RRID:AB_2116561 |
| Rabbit anti-FABP3 antibody | Proteintech | Cat# 10676-1-AP, RRID:AB_2102309 |
| Goat anti-GATA6 antibody | R and D Systems | Cat# AF1700, RRID:AB_2108901 |
| Mouse anti-NODAL antibody | Abcam | Cat# ab55676, RRID:AB_2151660 |
| Mouse anti- GAPDH antibody | Sigma-Aldrich | Cat# G8795, RRID:AB_1078991 |
| HRP-conjugated anti-mouse IgG antibody | Santa Cruz Biotechnology | Cat# sc-516102, RRID:AB_2687626 |
| Donkey anti-Rabbit 546 | Thermo Fisher Scientific | Cat# A10040, RRID:AB_2534016 |
| Donkey anti-Mouse 488 | Thermo Fisher Scientific | Cat# A-21202, RRID:AB_141607 |
| Donkey anti-Goat 647 | Thermo Fisher Scientific | Cat# A-21447, RRID:AB_2535864 |
| Donkey anti-Rabbit 568 | Thermo Fisher Scientific | Cat# A10042, RRID:AB_2534017 |
| Donkey anti-Mouse 647 | Thermo Fisher Scientific | Cat# A-31571, RRID:AB_162542 |
| Goat anti-Mouse IgG1 488 | Thermo Fisher Scientific | Cat# A-21121, RRID:AB_2535764 |
| Goat anti-Mouse IgG2b 647 | Thermo Fisher Scientific | Cat# A-21242, RRID:AB_2535811 |
| Biological samples | ||
| Human embryo | First People’s Hospital of Yunnan Province | N/A |
| cynomolgus monkeys (Macaca fascicularis) | Yunnan Key Laboratory of Primate Biomedical Research (LPBR) in China. Astrid Fagræus laboratory of the Karolinska Institutet in Sweden. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Epidermal growth factor (EGF) | R&D System | Cat# 236-EG-200 |
| CHIR99021 | Cayman Chemical | Cat# 13122 |
| A83-01 | Tocris Bioscience | Cat# 2939 |
| SB431542 | STEMCELL Technologies | Cat# 72234 |
| Y27632 | Tocris | Cat# 1254 |
| FGF2 | Thermo Fisher Scientific | Cat# PHG0261 |
| BMP4 | R&D Systems | Cat# 314-BP-050 |
| IWP2 | Tocris | Cat# 3533 |
| LY294002 | Selleckchem | Cat# S1105 |
| ACTVIN A | R&D Systems | Cat# 338-AC-050 |
| Critical commercial assays | ||
| LookOut Mycoplasma PCR Detection Kit | Sigma-Aldrich | Cat# MP0035-1KT |
| Click-iT™ Plus EdU Cell Proliferation Kit | Thermo Fisher Scientific | Cat# C10637 |
| Deposited data | ||
| Data files for RNA sequencing | Gene Expression Omnibus | GSE185643 |
| Processed dataset | Gene Expression Omnibus | GSE185643, https://umichibbl.shinyapps.io/shinyapp/ |
| Raw data for plots | Mendeley Data | http://dx.doi.org/10.17632/cvv5rzymhd.1 |
| Experimental models: Cell lines | ||
| Human: H9 (WA09) (female) | WiCell | NIH registration number: 0062 |
| Human: ESI-017 (male) | BioTime, Inc. | NIH registration number: 0093 |
| Chimpanzee: Chimpanzee iPSC lines (C3651, male and C4955, female) | (Gallego Romero et al., 2015; Pavlovic et al., 2018) | N/A |
| Human: Human trophoblast stem cells (hTSCs) | (Okae et al., 2018) | N/A |
| Human: ESI-017 NODAL-KO hPSC line | (Chhabra et al., 2019) | N/A |
| Oligonucleotides | ||
| NODAL_crRNA_1: 5’-AGGCUCAGCAUGUACGCCAG-3’ | Thermo Fisher Scientific | N/A |
| NODAL_crRNA_2: 5’-AGACAUCAUCCGCAGCCUAC-3’ | Thermo Fisher Scientific | N/A |
| NODAL exon 1 Forward Primer: 5’-CTTCCTTCTGCACGCCTGGTGG-3’ | Thermo Fisher Scientific | N/A |
| NODAL exon 1 Reverse Primer: 5’-CCAACCCACAGCACTTCCCGAG-3’ | Thermo Fisher Scientific | N/A |
| Recombinant DNA | ||
| pCXLE-EGFP expression plasmid | Addgene | plasmid # 27082; RRID: Addgene_27082 |
| Software and algorithms | ||
| R | open source | https://www.r-project.org |
| Python | open source | https://www.python.org/ |
| ImageJ | open source | https://imagej.net/software/fiji/ |
| Other | ||
| mTeSR™1 | STEMCELL Technologies | Cat# 85850 |
| mTeSR™ Plus | STEMCELL Technologies | Cat# 100-0276 |
| Gibco™ Geltrex™ LDEV-Free, hESC-Qualified, Reduced Growth Factor Basement Membrane Matrix | Thermo Fisher Scientific | Cat# A1413302 |
| Corning™ Matrigel™ hESC-Qualified Matrix | Thermo Fisher Scientific | Cat# 08-774-552 |
| KnockOut™ Serum Replacement | Thermo Fisher Scientific | Cat# 10828028 |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS -X) | Thermo Fisher Scientific | Cat# 51500056 |
| Essential 6 medium (E6) | Thermo Fisher Scientific | Cat# A1516401 |
This paper does not report original codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Raw data for the plots were deposited on Mendeley at http://dx.doi.org/10.17632/cvv5rzymhd.1
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
Cell lines used in this study are listed in the Key Resources Table. Culture/growth conditions are described in “METHOD DETAILS”.
Human embryos
IVF human blastocysts (D6) were obtained from couples who had at least one healthy baby after in vitro fertilization with signed consents. Human embryo experiments were approved by the Medicine Ethics Committee of the First People’s Hospital of Yunnan Province (KHLL2020-KY064).
Monkey embryos
IVF cynomolgus macaque embryos were obtained from healthy female cynomolgus monkeys aged 5 to 8 years old. All animals were housed either at the facility of the Yunnan Key Laboratory of Primate Biomedical Research (LPBR) in China, or at the Astrid Fagræus laboratory of the Karolinska Institutet in Sweden. Both facilities are accredited by AAALAC international. Experimental protocols for using cynomolgus macaque embryos were approved by the Institutional Animal Care and Use Committee of LPBR in China (KBI K001115033/01,01) and by the Jordbruksverket in Sweden (Ethical Permit Number N277/14).
METHOD DETAILS
Ethics statement
μPASE lacks both the primitive endoderm and trophoblast and thus cannot form the yolk sac or placenta. Therefore, μPASE does not have human organismal form or potential. Furthermore, μPASE disassembles and lose structural integrity after t = 48 h, and all experiments were terminated by no later than t = 72 h. All protocols with hPSCs were approved by the Human Pluripotent Stem Cell Research Oversight Committee at the University of Michigan, Ann Arbor. Protocols of human embryo experiments were approved by the Medicine Ethics Committee of the First People’s Hospital of Yunnan Province (KHLL2020-KY064). This Medicine Ethics Committee has 13 members, including lawyers, scientists and clinicians with relevant expertise. This committee evaluated the scientific merits and ethical justification of human embryo experiments and conducted a full review of the donation and use of human embryo samples. All human embryos donated to this study were surplus frozen embryos from couples who had at least one healthy baby after in vitro fertilization. All donor couples signed informed consents for voluntary donations at the Department of Reproductive Medicine in the First People’s Hospital of Yunnan Province. No economic benefits were offered during the process. Couples were informed that their embryos would be used for experimental studies of human development and that their donation would not affect their in vitro fertilization processes.
Cell culture
H9 (WA09, WiCell; NIH registration number: 0062, female) and ESI-017 (BioTime, Inc.; NIH registration number: 0093, male) hPSCs and chimpanzee iPSC lines (C3651, male and C4955, female) (Gallego Romero et al., 2015; Pavlovic et al., 2018) were maintained in a feeder-free culture system using mTeSR medium (STEMCELL Technologies). For hPSCs, culture plate was coated with 1% lactate dehydrogenase-elevating virus (LDEV)-free, hESC cell-qualified reduced growth factor basement membrane matrix Geltrex (Thermo Fisher Scientific; derived from Engelbreth-Holm-Swarm mouse tumors) before cell seeding. For chimpanzee iPSCs, culture plate was coated with 1% Matrigel (Thermo Fisher Scientific; extracted from Engelbreth-Holm-Swarm mouse sarcoma). Cells were visually examined during each passage to ensure absence of spontaneously differentiated, mesenchymal-like cells in culture. Cells were used before P70. Both human and chimpanzee cells have been authenticated by original sources as well as in-house by immunostaining for pluripotency markers and successful differentiation into the three germ layers. Cells were maintained for at least ten passages and authenticated as karyotypically normal. Karyotype analysis was performed by Cell Line Genetics. Both human and chimpanzee cell lines were tested negative for mycoplasma contamination (LookOut Mycoplasma PCR Detection Kit, Sigma-Aldrich).
Generation of NODAL-knockout hPSCs
To generate NODAL-knockout (KO) hPSCs, a 58-bp portion of genomic DNA within NODAL exon 1 was deleted by CRISPR/Cas9 using two crRNA purchased from Thermo Fisher Scientific [NODAL_crRNA_1: 5’-AGGCUCAGCAUGUACGCCAG-3’; NODAL_crRNA_2: 5’-AGACAUCAUCCGCAGCCUAC-3’] (Figures S7A–S7C). Deplexes of crRNA:tracrRNA were prepared using a standard protocol and introduced into H9 hPSCs with the Cas9 enzyme and the pCXLE-EGFP expression plasmid (a gift from Shinya Yamanaka; Addgene plasmid # 27082; RRID: Addgene_27082) for constitutional expression of EGFP using the NEON electroporation system (Thermo Fisher Scientific). EGFP-expressing single cells were collected and seeded onto Matrigel-coated 96-well plates by fluorescence-activated cell sorting (FACSAria Fusion, BD Biosciences) with CloneR single-cell culture supplement diluted with mTeSR Plus medium (STEMCELL Technologies). To detect the anticipated deletion, genomic DNA was isolated from single-cell derived clones and subjected to PCR using the following primers designed for amplification of NODAL exon 1 [Forward Primer: 5’-CTTCCTTCTGCACGCCTGGTGG-3’; Reverse Primer: 5’-CCAACCCACAGCACTTCCCGAG-3’]. Resulting amplicons were subjected to Sanger sequencing using a primer 5’-CTTCCTTCTGCACGCCTGGTGG-3’. ESI-017 NODAL-KO hPSC line is a generous gift from Aryeh Warmflash at Rice University (Chhabra et al., 2019).
Western blotting
Wildtype and NODAL-KO hPSCs were exposed to GSK3 inhibitor CHIR99021 (CHIR, 10 μM; Cayman Chemical) for 24 h to augment expression of NODAL protein before cells were lysed with RIPA buffer containing the cOmplete protease inhibitor cocktail (Roche). Protein concentration was determined by the Bradford assay using Protein Assay Dye Reagent Concentrate (Bio-Rad). An equal amount of protein (80 μg) from cell lysates of widetype and NODAL-KO hPSCs was resolved on 10% SDS-PAGE and transferred onto PVDF membranes (Thermo Fisher Scientific). Immunostaining was performed by blocking PVDF membranes with 5% skim milk for 1 h at room temperature followed by incubation overnight at 4 °C with mouse monoclonal antibodies to human NODAL (Abcam ab55676; 1:500 dilution) or human GAPDH (Sigma-Aldrich G8795; 1:20000 dilution) diluted in 5% skim milk. Membranes were washed with phosphate buffered saline (PBS) containing 0.1% Tween-20 (PBS-T) and stained with HRP-conjugated anti-mouse IgG antibody (Santa Cruz sc-516102; 1:3000 dilution) diluted in 5% skim milk for 1 h at room temperature. Protein bands were detected by a chemiluminescence assay using SuperSignal West Pico Plus and SuperSignal West Femto reagents (Thermo Fisher Scientific).
Cynomolgus macaque
Healthy cynomolgus monkeys (Macaca fascicularis), aged from 5 to 8 years old, were used in this study. All animals were housed either at the facility of the Yunnan Key Laboratory of Primate Biomedical Research (LPBR) in China, or at the Astrid Fagræus laboratory of the Karolinska Institutet in Sweden. Both facilities are accredited by AAALAC international. Experimental protocols for using cynomolgus macaque embryos were approved by the Institutional Animal Care and Use Committee of LPBR in China (KBI K001115033/01,01) and by the Jordbruksverket in Sweden (Ethical Permit Number N277/14). Animals involved in this study were never used for other treatments.
In vitro fertilization and culture of cynomolgus macaque embryo
In vitro fertilized cynomolgus macaque embryos were generated as described previously (Niu et al., 2014). Briefly, healthy female cynomolgus monkeys aged 5 to 8 years old with regular menstrual cycles were selected as oocyte donors. Cynomolgus monkeys were treated with recombinant human follicle stimulation hormone (Merck, Gonal-f) for 8 days, followed by administration of recombinant human chorionic gonadotropin (Merck, Ovidrel) on day 9. After 32–35 h, oocytes were collected by laparoscopic follicular aspiration. Metaphase II (MII) oocytes were used for intracytoplasmic sperm injection to generate zygotes, and fertilization was confirmed by the presence of two pronuclei. Zygotes were cultured in embryo culture medium-9 (polyvinyl alcohol (0.1 mg/mL), calcium chloride (1.9 mM), magnesium chloride (0.46 mM), potassium chloride (3.0 mM), sodium chloride (113.8 mM), sodium bicarbonate (25.0 mM), sodium lactate (4.5 mM), Minimum Essential Medium (MEM) amino acid, MEM nonessential amino acid, and gentamicin (10 mg/mL)) containing 10% fetal calf serum in 37 °C incubator supplied with 5% CO2 until the blastocyst stage. In vitro culture of NHP monkey blastocysts beyond the implantation stage has been described previously (Yang et al., 2021). In brief, frozen NHP monkey blastocysts were thawed using Thawing Media (Kizatato) and cultured in blastocyst culture medium (Origio) for at least 4 h to recover. Blastocysts were then treated with Acidic Tyrode’s solution (Sigma) to remove zona pellucida before being transferred onto an ibiTreat 8-well μ-plate (Ibidi) containing 300 μL of pre-equilibrated in vitro culture medium 1 (advanced DMEM/F12, 20% FBS, l-Glutamine, l-cysteine, 1x Penicillin/Streptomycin, 1x ITS-X, supplemented with beta-estradiol, progesterone). On the second day, 150 μL of culture medium was aspirated, before 200 μL of pre-equilibrated in vitro culture medium 2 (advanced DMEM/F12, 30% KSR, l-Glutamine, l-cysteine, 1x Penicillin/Streptomycin, 1x ITS-X, supplemented with beta-estradiol, progesterone) was added into the ibiTreat 8-well μ-plate. Embryo growth was recorded daily, and culture medium was replenished every two days till Day 14.
NHP monkey embryo cryosection and immunocytochemistry
Day 14 NHP monkey embryos were fixed using 2% paraformaldehyde (PFA; buffered in PBS) overnight at 4 °C before being washed with PBS. Fixed embryos were dehydrated by 30% sucrose overnight at 4 °C before embedded in Tissue-Tek O.C.T. compound (Sakura) and frozen in liquid nitrogen. Frozen blocks were used for cryosection using CryoStar NX70 Cryostat (Thermo Fisher Scientific) according to manufacturer’s protocol. Immunofluorescence staining was then performed according to a standard procedure. Briefly, sections were thawed and air-dried at room temperature. After washing with PBS, sections were incubated in blocking buffer (3% FBS) diluted in PBS with 0.1% Triton X-100) for 1 h at room temperature and then incubated overnight at 4 °C with primary antibodies diluted in blocking buffer. The sections were washed with PBS-T and incubated with secondary antibodies diluted in blocking buffer for 2 h at room temperature. After washing thoroughly with PBS-T, sections were mounted and imaged. Secondary antibodies were used in dilution of 1:500. Images were acquired by a Zeiss 700 LSM Confocal Microscope and analyzed by iMaris.
Immunocytochemistry for D6 human embryos
D6 human embryos were fixed with 4% PFA for 20 min at room temperature in a 96-well plate, washed with PBS, and then permeabilized and blocked by 0.2% Triton X-100 supplemented with 3% bovine serum albumin (BSA) overnight at 4 °C. Embryos were then transferred onto a new well with primary antibodies for 16 – 18 h at 4 °C. Embryos were washed 3 times with PBS containing 0.05% Tween-20, 15 min every time, before being transferred to secondary antibody solutions for 4 h at room temperature. Embryos were then washed 3 times in PBS containing 0.05% Tween-20, 15 min every time, before being transferred onto an ibiTreat 8-well μ-plate. All antibodies were diluted by 1% BSA solutions.
Generation of μPASEs
The microfluidic device was fabricated by bonding a polydimethylsiloxane (PDMS) structure layer to a glass coverslip. Singly dissociated hPSCs or chimpanzee iPSCs were loaded into the cell loading channel and guided to settle into preformed, concave Geltrex pockets by tilting the device by 90° for 10 min. hPSCs were then maintained in mTeSR containing 10 μM Y27632 (Tocris) for 18 h to allow for cell clustering. At t = 0 h, culture medium in all medium reservoirs of the device was switched to a fresh basal medium comprising Essential 6 medium (E6; Thermo Fisher Scientific) and FGF2 (20 ng mL−1; Thermo Fisher Scientific), with additional 50 ng mL−1 BMP4 (R&D Systems) supplemented only in the cell loading channel. A detailed protocol for microfluidic device fabrication and generation of μPASEs can be found elsewhere(Zheng et al., 2021; Zheng et al., 2019b). To examine possible involvements of different signaling pathways involved in the development of μPASEs, IWP2 (10 μM; Tocris), SB431542 (10 μM; STEMCELL Technologies) or LY294002 (20 μM; Selleckchem) was supplemented into basal medium from t = 0 h. For assays with ACTVIN A, 100 ng mL−1 ACTIVIN A (R&D Systems) was supplemented into the channel opposite to BMP4 stimulation from t = 0 h. For in situ proliferation measurements, Click-iT EdU Kit (Invitrogen) was used as per manufacturer’s protocol. Diluted EdU solution (10 μM) was introduced into all reservoirs of the device at t = 24 h. After 3 h of incubation, the μPASEs was fixed and imaged.
Immunocytochemistry for μPASEs
μPASEs were fixed with 4% PFA for 12 h before being permeabilized in 0.1% SDS solution (sodium dodecyl sulphate, dissolved in PBS) for 3 h. μPASEs were then blocked in 4% donkey serum (Sigma-Aldrich) at 4 °C for 24 h, followed by incubation with primary antibody solutions at 4 °C for another 24 h. Samples were then labelled with donkey-raised secondary antibodies (1:500 dilution) at 4 °C for 24 h. 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) was used for counterstaining cell nuclei. Both primary and secondary antibodies were prepared in 4% donkey serum supplemented with 0.1% NaN3. 70 μL antibody solutions were added to each medium reservoir of the microfluidic device for immunostaining. All primary and secondary antibodies used in this study are listed in KEY RESOURCES TABLE. All confocal micrographs of μPASEs were acquired by a NIKON A1SI Confocal Microscope equipped with a photomultiplier tube (PMT) detector and processed using ImageJ 1.53c.
Quantification of SOX17+ cells
μPASEs at t = 48 h with or without LY294002 were stained for SOX17 using the protocol described above. All confocal micrographs captured at the central focal plane of structure (50 μm above the microfluidic device bottom surface) were used for quantification. The percentage of SOX17+ cells was calculated as the ratio between the area of SOX17 channel and DAPI channel for each μPASE. The binarization and measurement of the images was conducted using ImageJ 1.53c. Quantitative results were analyzed using independent, two-tailed Student’s t-test in Excel (Microsoft). P < 0.05 was considered statistically significant.
Trophoblast stem cells and derivation of AMLCs using BMP4
Human trophoblast stem cells (hTSCs) derived from blastocysts were generously provided by Dr. H. Okae and Dr. T. Arima (Okae et al., 2018). hTSCs were maintained in 1% Geltrex coated 6-well plates in DMEM/F12 supplemented with 0.05 mM 2-mercaptoethanol, 0.2% fetal bovine serum (FBS), 0.5% knockout serum replacement (KSR), 0.5% penicillin-streptomycin, 0.3% BSA, 1% ITS-X supplement, 1.5 μg Ml−1 L-ascorbic acid, 50 ng mL−1 epidermal growth factor (EGF), 2 μM CHIR99021, 1 μM A83-01, 1 μM SB431542, 0.8 mM valproic acid (VPA) and 5 μM Y27632, and passaged using TrypLE.
To obtain AMLCs by treating hPSCs with BMP4, singly dissociated hPSCs were suspended in mTesR1 containing 10 μM Y27632 and seeded in a 1% Geltrex coated 6-well plate at a density of 2.5 × 103 cells cm−2. Note that this cell seeding density is optimized to avoid extensive cell death (when cell density is too low) or emergence of PSLCs in culture through an unspecified inductive effect of incipient AMLCs (Zheng et al., 2019b). 18 h after cell seeding, culture medium was switched to Essential 6 medium containing FGF2 (20 ng mL−1) and BMP4 (50 ng mL−1). Resulting AMLCs were fixed and stained after 48 h of BMP4 treatment.
Single cell dissociation and RNA-sequencing
μPASEs (H9) at different time points were washed twice with DMEM/F12 for 10 min and incubated with Accutase for 1 h. After incubation, μPASEs in the microfluidic device were dissociated into single cells by gentle agitating. Single cells from six microfluidic devices were collected and pooled into PBS containing 0.5% BSA before being centrifuged at 300 g for 5 min. The resultant cell pellet was re-suspended in PBS containing 0.5% BSA. Within 1 h after cell dissociation, cells were loaded into the 10X Genomics Chromium system (10X Genomics). 10X Genomics v.3 libraries were prepared according to the manufacturer’s instructions. Libraries were then sequenced using paired-end sequencing with a minimum coverage of 20,000 raw reads per cell using an Illumina NovaSeq-6000. scRNA-seq data were aligned and quantified using Cell Ranger Single-Cell Software Suite (v.3.1.0, 10X Genomics) against the Homo sapiens (human) genome assembly GRCh38.p13 from ENSEMBL. Chimpanzee μPASEs (C3651) were dissociated and sequenced following the same protocol. scRNA-seq data from chimpanzee μPASEs were aligned against Pan_tro_3.0 from ENSEMBL.
Data integration, dimensionality reduction and clustering
Analysis of scRNA-seq data and integration of scRNA-seq datasets were performed using Seurat R package (v.4.0.0.0, https://satijalab.org/seurat/) (Butler et al., 2018; Satija et al., 2015). Default setups were used unless noted otherwise. Briefly, a single batch of scRNA-seq dataset was filtered based on total number of genes detected and percentage of total mitochondrial genes. Gene expression was then calculated by normalizing the raw count with the total count before multiplying by 10,000 and log transformed. Cell cycle was regressed out based on cell cycle scores (CellCycleScoring) during data scaling process (ScaleData). PCA analysis (RunPCA) was then performed on filtered data followed by embedding into low dimensional space with Uniform Manifold Approximation and Projection (UMAP; RunUMAP). Identification of cell clusters by a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm was achieved using the FindClusters function in the Seurat R package. For integration of different scRNA-seq datasets, count matrices of different datasets were filtered and normalized separately before being integrated using the IntegrateData function based on 2,000 anchor features. After integration, the integrated scRNA-seq dataset was analyzed following the standard Seurat pipeline. Annotation of cell clusters was based on expression of canonical lineage marker genes.
RNA velocity analysis and partition-based graph abstraction analysis (PAGA)
Bam files generated by the Cell Ranger pipeline was used for RNA velocity analysis. Genome annotations GRCh38.p13 were used for counting spliced and unspliced mRNA of individual cells. Python package scVelo (v.0.2.2, https://scvelo.readthedocs.io) was employed to perform RNA velocity analysis using dynamical modeling (scv.tl.velocity) (Bergen et al., 2020; La Manno et al., 2018). Function ‘scv.pl.velocity_embedding_stream’ was used to project RNA velocities onto UMAP plots or diffusion maps. All default parameters were used unless noted otherwise. Python package Scanpy (v.1.8.0, https://scanpy.readthedocs.io/en/stable/) was used for evaluating the relationship between different cell clusters by the partition-based graph abstraction (PAGA) analysis (Wolf et al., 2018). Briefly, Seurat object was converted to “Loom” file and passed to Scanpy. Neighborhood graph of observations was then computed using 20 PCs (sc.pp.neighbors). Finally, PAGA graph was plotted with a “eq_tree” layout.
Gene regulatory network analysis
Regulatory activity of transcription factors associated with specific cell types was assessed using the R-package SCENIC (Single Cell rEgulatory Network Inference and Clustering, v.1.1.2–2, https://github.com/aertslab/SCENIC) (Aibar et al., 2017). Briefly, regulatory modules were first identified by inferring co-expression with transcription factors using GENIE3. Each co-expression module was then analyzed using cis-regulatory motif analyses (RcisTarget). Only modules with significant motif enrichment of the correct upstream regulator were retained. The human motif collection v9 and the cisTarget databases for hg38 were used in the pipeline (https://resources.aertslab.org/cistarget/). Filtered counts of the integrated Seurat object were used as input of SCENIC. All default parameters were used in SCENIC unless noted otherwise.
Trajectory interference using diffusion map and pseudotime
Diffusion maps were obtained by the R-package Density (https://bioconductor.org/packages/release/bioc/html/destiny.html), which computes kernel density estimates with parametric starts and asymmetric kernels (Angerer et al., 2016). To generate three-dimensional diffusion maps, PCA embeddings of the integrated Seurat object were used as input of ‘diffmap’ function. Cells in the three-dimensional diffusion map were color-coded consistently with the UMAP plot of the integrated Seurat object. Three-dimensional visualization was realized using R-package rgl (3D Visualization Using OpenGL). For trajectory inference and pseudotime analysis of specific cell lineages, relevant cell clusters of the integrated Seurat object were extracted using Subset function (Seurat). PCA embeddings of selected cell clusters were then used as the input of ‘diffmap’ function. To visualize gene expression dynamics, diffusion pseudotime (dpt) was utilized. The roots of the diffusion map were automatically chosen by the algorithm; and the EpiLC cluster was arbitrarily made as the initial cell type. Expression levels of selected genes were fitted as a function of the pseudotime with “loess” method by using ‘geom_smooth’ function in ggplot2 R package (v.3.3.3). Cells with extreme dpt values were counted as outliner and excluded from gene expression dynamic analysis. RNA velocity vectors were computed as described above and superimposed onto the diffusion maps. In the case of lineage bifurcation, branching point and branches of different lineages were identified by locally fitting half-lines to single-cell data in the diffusion map using K-Branches R package (https://github.com/theislab/kbranches) (Chlis et al., 2017).
Differential gene expression, gene ontology enrichment and pathway analyses
Gene expression data depicted in feature plots and dot plots in this paper were calculated from raw counts after NormalizeData function followed by ScaleData function of the Seurat package, unless noted otherwise. Differentially expressed genes (DEGs) between different cell types were identified using FindMarkers function (Seurat), with a minimal fold difference of 0.25 in the logarithmic scale and > 10% detection rate in either of the two cell types under comparison. Gene ontology (GO) enrichment and pathway analyses were performed using online tool iPathwayGuide (Advaita Bioinformatics) referencing AmiGO Gene Ontology database and KEGG PATHWAY Database, respectively.
Integration, re-analysis and PCA of published human embryo data
The scRNA-seq dataset of the CS7 human gastrula was integrated with μPASE scRNA-seq datasets. Since μPASEs contain AMLCs, PGCLCs and PSLCs / MeLCs, only relevant cell types from the human gastrula (Amniotic/embryonic ectoderm, Epiblast, Primitive Streak, Nascent Mesoderm, Emergent Mesoderm) were extracted according to the annotations in the original publication. Human gastrula scRNA-seq dataset was generated using Smart-seq2. To compare with μPASE scRNA-seq datasets generated using the 10X Genomics Chromium system, raw counts of each cell in the human gastrula were normalized to exon sizes before being utilized to create the Seurat object (input count = raw count / exon size × 1,000). Exon size information was obtained from GRCh38.p13, ENSEMBL. To integrate scRNA-seq data from the human gastrula with those of μPASEs, μPASE scRNA-seq datasets from the three time points (t = 24 h, 36 h and 48 h) were first filtered. Normalization, Scaling (including cell cycle regression) and PCA were then performed separately on each of the datasets after which they were combined using the reciprocal PCA approach (IntegrateData function, Seurat) based on 30 dimensions and 2,000 anchor features. Dimensionality reduction and clustering were then performed as described above. Downsampling of large scRNA-seq datasets was performed using Seurat R package (Subset), which randomly selects 100 cells from every cluster in the original datasets to form a new Seurat object. This step helps prevent larger datasets from dominating the downstream analysis.
For re-analysis of human gastrula scRNA-seq dataset, relevant cell types were processed with the default Seurat pipeline. Trajectory inference using diffusion map and pseudotime analysis was achieved using the same pipeline as for μPASEs, as descried earlier. For PCA, large datasets were first downsampled; then the normalized counts of epiblast ontogenetic genes (Mendeley Data Table5) from different datasets were merged into a matrix. The matrix was scaled to ensure each gene has zero average and unit variance. PCAs was calculated and visualized using Seurat R package.
Cross-species comparison
Cross-species comparisons utilize scRNA-seq datasets from the CS7 human gastrula, human μPASEs (t = 24 h, 36 h and 48 h), chimpanzee μPASEs (t = 48 h), in vivo cynomolgus embryos (D13 – 17), and in vitro cultured cynomolgus embryos (D11 – 17) (Ma et al., 2019; Nakamura et al., 2016; Sasaki et al., 2016; Tyser et al., 2021; Yang et al., 2021). To mitigate artifacts resulted from different sequencing platforms, for datasets generated using Smart-seq2, raw counts were first normalized to exon length, as described above. Exon size information of cynomolgus monkey was obtained from Macaca_fascicularis_6.0, ENSEMBLE. Average raw counts normalized by exon length for specific cell types in different datasets were then transformed into log2(counts per million reads + 1) and used as gene expression levels for calculating the correlation coefficient matrix and heat map plotting.
Ontogenic genes of mesoderm and PGC lineages were identified through DEG analysis of in vivo cynomolgus embryo datasets. Briefly, for mesoderm lineage ontogenic genes, DEGs of “Gast1”, “Gast2a” and “Gast2b” as compared to “PostL-EPI” were identified using FindMarkers function (Seurat)(Nakamura et al., 2016). All DEGs, with a minimal fold difference of 2 in the logarithmic scale (logfc), > 10% detection rate in either of the two cell types under comparison, and adjusted P-value < 0.05, were used as mesoderm lineage ontogenic genes (Mendeley Data Table6). To identify PGC lineage ontogenic genes, DEGs of “ePGC” as compared to “PostL-EPI”(Sasaki et al., 2016), with a minimal fold difference of 2 in the logarithmic scale (logfc), > 10% detection rate in either of the two cell types under comparison, and adjusted P-value < 0.05, were used as PGC lineage ontogenic genes (Mendeley Data Table6). Amniotic ectoderm ontogenic genes were obtained through DEG analysis of Day 14 in vitro cultured cynomolgus embryos (Yang et al., 2021). Specifically, DEGs of “early-Amnion” and “late-Amnion” as compared to “EPI” were identified using FindMarkers function (Seurat). All DEGs, with a minimal fold difference of 0.75 in the logarithmic scale (logfc), > 10% detection rate in either of the two cell types under comparison, and adjusted P-value < 0.01, were used as amniotic lineage ontogenic genes (Mendeley Data Table6).
Analysis of cell-cell interactions
R package CellChat was used to perform cell-cell communication analysis (http://www.cellchat.org/) (Jin et al., 2021). Briefly, based on manually curated databases that consider known structural compositions of ligand-receptor interactions, CellChat infers and analyzes intercellular communication networks from scRNA-seq data using network analysis and pattern recognition. Seurat object including count matrix and clustering results from each dataset is imported to CellChat. The default human database was used for μPASE and cynomolgus embryo dataset analyses, whereas the default mouse database was used for mouse embryo dataset analysis. For both databases, only secreted signaling pathways from Kyoto Encyclopedia of Genes and Genomes (KEGG) were used. Default values were used for all parameters, except that the truncated mean was lowered to 5% to increase algorithm sensitivity.
Comparisons between human trophoblast and amniotic ectoderm
Several published RNA-seq datasets related to human blastocyst trophoblast and amniotic ectoderm were surveyed in this work. Datasets generated from bulk RNA-sequencing or Smart-seq2 were first normalized by exon sizes as described earlier. Specifically, blastocyst trophoblast cells annotated in (Blakeley et al., 2015; Petropoulos et al., 2016) were utilized as the reference of human blastocyst trophoblast. Both two datasets were analyzed in this work to mitigate batch effects and systematic variations. Amniotic/embryonic ectoderm cells from the CS7 human gastrula were utilized as the reference of human amniotic ectoderm (Tyser et al., 2021). DEGs between trophoblast and amniotic ectoderm were first identified using trophoblast cells from (Blakeley et al., 2015; Petropoulos et al., 2016), respectively, as compared to amniotic/embryonic ectoderm cells from the CS7 human gastrula, with a minimal fold difference of 0.75 in the logarithmic scale (logfc) and adjusted P-value < 0.05. Overlapped DEGs identified from both comparative transcriptome analyses were used as Trophoblast_Amnion ontogenic genes (n = 299) to calculate correlation coefficients of published datasets as well as AMLCs from this work with human blastocyst trophoblast and amniotic/embryonic ectoderm (Mendeley Data Table8). Correlation coefficients and gene expression heatmap were calculated based on averages of experimental repeats, if any. The average gene expression level of the two trophoblast datasets was used to calculate correlation coefficients of surveyed cells with the human blastocyst trophoblast. To identify the most reliable markers for distinguishing trophoblast cells from amniotic ectoderm cells, more stringent criteria were applied than those for identifying Trophoblast_Amnion ontogenic genes. Among Trophoblast_Amnion ontogenic genes with logfc > 2, only the genes that are negative in the other cell type (normalized count < 0.05) were identified as reliable markers. Overlapped markers revealed for both trophoblast datasets were proposed as markers for distinguishing human trophoblast cells from amniotic ectoderm cells.
Quantification and statistical analysis
Quantitative results were analyzed using independent, two-tailed Student’s t-test in Excel (Microsoft). P < 0.05 was considered statistically significant.
Supplementary Material
Supplemental Information Video 2_3D staining: Three-dimensional reconstruction of μPASE at t = 36 h, stained for ISL1 (green), TBXT (red) and SOX17 (magenta). Related to Figure 2
Supplemental Information Video 1_diffusion_map: Three-dimensional visualization showing developmental trajectories of AMLC, PSLC / MeLC and PGCLC lineages. Related to Figure 2
Highlights.
Single-cell transcriptional atlas of microfluidic amniotic sac embryoid (μPASE)
Single-cell transcriptional characterization of chimpanzee μPASE
Critical role of NODAL in human mesoderm and primordial germ cell specification
Stringent criteria for distinguishing between trophectoderm and amniotic ectoderm
ACKNOWLEDGEMENTS
Studies of μPASE are supported by the Michigan-Cambridge Research Initiative (J.F.), the National Institutes of Health (R21 NS113518 and R21 HD100931, J.F.), the National Science Foundation (CMMI 1917304 and CBET 1901718, J.F.), and the 21st Century Jobs Trust Fund received through the Michigan Strategic Fund from the State of Michigan (Grant CASE-315037, J.F.). Studies of human blastocysts are supported by the Open Project of Yunnan Provincial Reproductive and Obstetrics and Gynecology Clinical Medicine Center (zx2019-01-01 and 2020LCZXKF-SZ04, L.X.). Non-human primate monkey studies are supported by the National Key Research and Development Program of China (2016YFA0101401), the Major Basic Research of Yunnan from China (2019FY002), and a Grant for Swedish-Chinese Collaboration from the Swedish Research Council (Dnr 539-2013-7002). M.K. and T.S. are partially supported by the RICBAC Foundation. K.C. is partially supported by a Grant for Distinguished Professors from the Swedish Research Council (Dnr 541-2013-8351). A.M.R.I. is partially supported by the National Science Foundation Graduate Research Fellowship under grant no. DGE 1256260. A.M.R.I. and S.N.E. are partially supported by the University of Michigan Rackham Predoctoral Fellowship. The Lurie Nanofabrication Facility at the University of Michigan is acknowledged for support with microfabrication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS
Two patents related to this work have been filed (US20190321415 / WO2018106997 by J. Fu, Y. Zheng and S.N. Esfahani; US2020049721 / PCT / US20/49721 by J. Fu and Y. Zheng).
REFERENCES
- Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer P, Haghverdi L, Buttner M, Theis FJ, Marr C, and Buettner F (2016). destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243. [DOI] [PubMed] [Google Scholar]
- Beccari L, Moris N, Girgin M, Turner DA, Baillie-Johnson P, Cossy AC, Lutolf MP, Duboule D, and Arias AM (2018). Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562, 272–276. [DOI] [PubMed] [Google Scholar]
- Ben-Haim N, Lu C, Guzman-Ayala M, Pescatore L, Mesnard D, Bischofberger M, Naef F, Robertson EJ, and Constam DB (2006). The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell 11, 313–323. [DOI] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 38. [DOI] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, et al. (2011). BRACHYURY and CDX2 Mediate BMP-Induced Differentiation of Human and Mouse Pluripotent Stem Cells into Embryonic and Extraembryonic Lineages. Cell Stem Cell 9, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeley P, Fogarty NME, del Valle I, Wamaitha SE, Hu TX, Elder K, Snell P, Christie L, Robson P, and Niakan KK (2015). Defining the three cell lineages of the human blastocyst by single-cell RNA-seq (vol 142, pg 3151, 2015). Development 142, 3613–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaca A, Nóvoa A, and Mallo M (2016). Hoxb6 can interfere with somitogenesis in the posterior embryo through a mechanism independent of its rib-promoting activity. Development 143, 437–448. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, and Beck F (2004). Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci U S A 101, 7641–7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan C-W, Wei S, Hao W, Kilgore J, Williams NS, et al. (2009). Small molecule–mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nature Chemical Biology 5, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Sun N, Hou L, Kim R, Faith J, Aslanyan M, Tao Y, Zheng Y, Fu J, Liu W, et al. (2019). Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell Rep 29, 4568–4582 e4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra S, Liu L, Goh R, Kong X, and Warmflash A (2019). Dissecting the dynamics of signaling events in the BMP, WNT, and NODAL cascade during self-organized fate patterning in human gastruloids. PLOS Biology 17, e3000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlis NK, Wolf FA, and Theis FJ (2017). Model-based branching point detection in single-cell data by K-branches clustering. Bioinformatics 33, 3211–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AT, Brivanlou A, Fu JP, Kato K, Mathews D, Niakan KK, Rivron N, Saitou M, Surani A, Tang FC, et al. (2021). Human embryo research, stem cell-derived embryo models and in vitro gametogenesis: Considerations leading to the revised ISSCR guidelines. Stem Cell Rep 16, 1416–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H (2006). Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480. [DOI] [PubMed] [Google Scholar]
- Daoud G, Kempf H, Kumar D, Kozhemyakina E, Holowacz T, Kim DW, Ionescu A, and Lassar AB (2014). BMP-mediated induction of GATA4/5/6 blocks somitic responsiveness to SHH. Development 141, 3978–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED, and Brivanlou AH (2016). Self-organization of the in vitro attached human embryo. Nature 533, 251–254. [DOI] [PubMed] [Google Scholar]
- Dong C, Beltcheva M, Gontarz P, Zhang B, Popli P, Fischer LA, Khan SA, Park K. m., Yoon E-J, Xing X, et al. (2020). Derivation of trophoblast stem cells from naïve human pluripotent stem cells. Elife 9, e52504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Warmflash A, and Lutolf MP (2021). Stem-cell-based embryo models for fundamental research and translation. Nat Mater 20, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego Romero I, Pavlovic BJ, Hernando-Herraez I, Zhou X, Ward MC, Banovich NE, Kagan CL, Burnett JE, Huang CH, Mitrano A, et al. (2015). A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. Elife 4, e07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XF, Nowak-Imialek M, Chen X, Chen DS, Herrmann D, Ruan DG, Chen ACH, Eckersley-Maslin MA, Ahmad S, Lee YL, et al. (2019). Establishment of porcine and human expanded potential stem cells. Nat Cell Biol 21, 687–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. (2007). Evolutionary and biomedical insights from the rhesus macaque genome. Science 316, 222–234. [DOI] [PubMed] [Google Scholar]
- Gilbert S (2000). Developmental Biology, 6th edition (Sinauer Associates; ). [Google Scholar]
- Guo G, Stirparo GG, Strawbridge SE, Spindlow D, Yang J, Clarke J, Dattani A, Yanagida A, Li MA, Myers S, et al. (2021). Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 28, 1040–1056 e1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haremaki T, Metzger JJ, Rito T, Ozair MZ, Etoc F, and Brivanlou AH (2019). Self-organizing neuruloids model developmental aspects of Huntington’s disease in the ectodermal compartment. Nat Biotechnol 37, 1198–1208. [DOI] [PubMed] [Google Scholar]
- Harrison SE, Sozen B, Christodoulou N, Kyprianou C, and Zernicka-Goetz M (2017). Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356, eaal1810. [DOI] [PubMed] [Google Scholar]
- Hyun I, Wilkerson A, and Johnston J (2016). Revisit the 14-day rule. Nature 533, 169–171. [DOI] [PubMed] [Google Scholar]
- Io S, Kabata M, Iemura Y, Semi K, Morone N, Minagawa A, Wang B, Okamoto I, Nakamura T, Kojima Y, et al. (2021). Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell 28, 1023–1039 e1013. [DOI] [PubMed] [Google Scholar]
- Irie N, Weinberger L, Tang WWC, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, and Surani MA (2015). SOX17 Is a Critical Specifier of Human Primordial Germ Cell Fate. Cell 160, 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nat Commun 12, 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Wilder E, Klingensmith J, Zachary K, and Perrimon N (1996). The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes & Development 10, 3116–3128. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Zhang H, Tang WWC, Irie N, Withey S, Klisch D, Sybirna A, Dietmann S, Contreras DA, Webb R, et al. (2017). Principles of early human development and germ cell program from conserved model systems. Nature 546, 416–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kurosawa O, and Iwata H (2019). Establishment of human trophoblast stem cells from human induced pluripotent stem cell-derived cystic cells under micromesh culture. Stem Cell Research & Therapy 10, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Tan JP, Schroder J, Aberkane A, Ouyang JF, Mohenska M, Lim SM, Sun YBY, Chen J, Sun G, et al. (2021). Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 591, 627–632. [DOI] [PubMed] [Google Scholar]
- Lovell-Badge R, Anthony E, Barker RA, Bubela T, Brivanlou AH, Carpenter M, Charo RA, Clark A, Clayton E, Cong YL, et al. (2021). ISSCR Guidelines for Stem Cell Research and Clinical Translation: The 2021 update. Stem Cell Rep 16, 1398–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zhai J, Wan H, Jiang X, Wang X, Wang L, Xiang Y, He X, Zhao ZA, Zhao B, et al. (2019). In vitro culture of cynomolgus monkey embryos beyond early gastrulation. Science 366, eaax7890. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hillier LW, Eichler EE, Zody MC, Jaffe DB, Yang SP, Enard W, Hellmann I, Lindblad-Toh K, Altheide TK, et al. (2005). Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87. [DOI] [PubMed] [Google Scholar]
- Minn KT, Fu YC, He S, Dietmann S, George SC, Anastasio MA, Morris SA, and Solnica-Krezel L (2020). High-resolution transcriptional and morphogenetic profiling of cells from micropatterned human ESC gastruloid cultures. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molè MA, Coorens THH, Shahbazi MN, Weberling A, Weatherbee BAT, Gantner CW, Sancho-Serra C, Richardson L, Drinkwater A, Syed N, et al. (2021). A single cell characterisation of human embryogenesis identifies pluripotency transitions and putative anterior hypoblast centre. Nat Commun 12, 3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris N, Anlas K, van den Brink SC, Alemany A, Schroder J, Ghimire S, Balayo T, van Oudenaarden A, and Martinez Arias A (2020). An in vitro model of early anteroposterior organization during human development. Nature 582, 410–415. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, and Saitou M (2016). A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537, 57–62. [DOI] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843. [DOI] [PubMed] [Google Scholar]
- Niu Y, Sun N, Li C, Lei Y, Huang Z, Wu J, Si C, Dai X, Liu C, Wei J, et al. (2019). Dissecting primate early post-implantation development using long-term in vitro embryo culture. Science 366, eaaw5754. [DOI] [PubMed] [Google Scholar]
- Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, and Arima T (2018). Derivation of Human Trophoblast Stem Cells. Cell Stem Cell 22, 50–63.e56. [DOI] [PubMed] [Google Scholar]
- Pavlovic BJ, Blake LE, Roux J, Chavarria C, and Gilad Y (2018). A Comparative Assessment of Human and Chimpanzee iPSC-derived Cardiomyocytes with Primary Heart Tissues. Sci Rep 8, 15312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S, Edsgard D, Reinius B, Deng QL, Panula SP, Codeluppi S, Reyes AP, Linnarsson S, Sandberg R, and Lanner F (2016). Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 165, 1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, Mulas C, Ibarra-Soria X, Tyser RCV, Ho DLL, et al. (2019). A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai VV, Siqueira LGB, Das M, Kei TG, Tu LN, Herren AW, Phinney BS, Cheong SH, Hansen PJ, and Selvaraj V (2019). Physiological profile of undifferentiated bovine blastocyst-derived trophoblasts. Biology Open 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai E, Schell JP, Janiszewski A, Rovic I, Murray A, Bradshaw B, Yamakawa T, Pardon T, El Bakkali M, Talon I, et al. (2021). Evaluating totipotency using criteria of increasing stringency. Nat Cell Biol 23. [DOI] [PubMed] [Google Scholar]
- Prummel KD, Hess C, Nieuwenhuize S, Parker HJ, Rogers KW, Kozmikova I, Racioppi C, Brombacher EC, Czarkwiani A, Knapp D, et al. (2019). A conserved regulatory program initiates lateral plate mesoderm emergence across chordates. Nat Commun 10, 3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Perez JA, and Magnuson T (2005). Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev Biol 288, 363–371. [DOI] [PubMed] [Google Scholar]
- Rivron NC, Frias-Aldeguer J, Vrij EJ, Boisset JC, Korving J, Vivie J, Truckenmuller RK, van Oudenaarden A, van Blitterswijk CA, and Geijsen N (2018). Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111. [DOI] [PubMed] [Google Scholar]
- Rossant J (2015). Mouse and human blastocyst-derived stem cells: vive les differences. Development 142, 9–12. [DOI] [PubMed] [Google Scholar]
- Rossant J, and Tam PPL (2017). New Insights into Early Human Development: Lessons for Stem Cell Derivation and Differentiation. Cell Stem Cell 20, 18–28. [DOI] [PubMed] [Google Scholar]
- Rossant J, and Tam PPL (2021). Opportunities and challenges with stem cell-based embryo models. Stem Cell Rep 16, 1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovskaya M, Andrews S, Reik W, and Rugg-Gunn PJ (2022). Amniogenesis occurs in two independent waves in primates. Cell Stem Cell 29, 744–759.e746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Nakamura T, Okamoto K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T, et al. (2016). The Germ Cell Fate of Cynomolgus Monkeys Is Specified in the Nascent Amnion. Dev Cell 39, 169–185. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, et al. (2015). Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell 17, 178–194. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenwolf G, Bleyl S, Brauer P, Francis-West P (2020). Larsen’s Human Embryology 6th Edition (Elsevier; ). [Google Scholar]
- Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty NME, Campbell A, Devito LG, Ilic D, et al. (2016). Self-organization of the human embryo in the absence of maternal tissues (vol 18, pg 700, 2016). Nat Cell Biol 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Taniguchi K, Gurdziel K, Townshend RF, Xue X, Yong KMA, Sang J, Spence JR, Gumucio DL, and Fu J (2017a). Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat Mater 16, 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Taniguchi K, Townshend RF, Miki T, Gumucio DL, and Fu J (2017b). A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat Commun 8, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M, Metzger JJ, Etoc F, Yoney A, Ruzo A, Martyn I, Croft G, You DS, Brivanlou AH, and Siggia ED (2019). A 3D model of a human epiblast reveals BMP4-driven symmetry breaking. Nat Cell Biol 21, 900–910. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L (2020). Gastrulation: From Embryonic Pattern to Form (Academic Press; ). [Google Scholar]
- Tyser RCV, Mahammadov E, Nakanoh S, Vallier L, Scialdone A, and Srinivas S (2021). Single-cell transcriptomic characterization of a gastrulating human embryo. Nature 600, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RN, Green J, Wang Z, Deng Y, Qiao M, Peabody M, Zhang Q, Ye J, Yan Z, Denduluri S, et al. (2014). Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis 1, 87–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmflash A, Sorre B, Etoc F, Siggia ED, and Brivanlou AH (2014). A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods 11, 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Yin Y, Zheng Y, Ma Y, Li Y, Zhao Z, Guo J, Ai Z, Niu Y, Duan K, et al. (2020). A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 577, 537–542. [DOI] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, and Thomson JA (2002). BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20, 1261–1264. [DOI] [PubMed] [Google Scholar]
- Xue X, Sun Y, Resto-Irizarry AM, Yuan Y, Aw Yong KM, Zheng Y, Weng S, Shao Y, Chai Y, Studer L, et al. (2018). Mechanics-guided embryonic patterning of neuroectoderm tissue from human pluripotent stem cells. Nat Mater 17, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida A, Spindlow D, Nichols J, Dattani A, Smith A, and Guo G (2021). Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell 28, 1016–1022 e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Goedel A, Kang Y, Si C, Chu C, Zheng Y, Chen Z, Gruber PJ, Xiao Y, Zhou C, et al. (2021). Amnion signals are essential for mesoderm formation in primates. Nat Commun 12, 5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Wei Y, Duan J, Schmitz DA, Sakurai M, Wang L, Wang K, Zhao S, Hon GC, and Wu J (2021). Blastocyst-like structures generated from human pluripotent stem cells. Nature 591, 620–626. [DOI] [PubMed] [Google Scholar]
- Zhai L, Chaturvedi D, and Cumberledge S (2004). Drosophila Wnt-1 Undergoes a Hydrophobic Modification and Is Targeted to Lipid Rafts, a Process That Requires Porcupine. Journal of Biological Chemistry 279, 33220–33227. [DOI] [PubMed] [Google Scholar]
- Zhao C, Reyes AP, Schell JP, Weltner J, Ortega N, Zheng Y, Björklund ÅK, Rossant J, Fu J, Petropoulos S, et al. (2021). Reprogrammed iBlastoids contain amnion-like cells but not trophectoderm. bioRxiv, 2021.2005.2007.442980. [Google Scholar]
- Zhao GQ (2003). Consequences of knocking out BMP signaling in the mouse. Genesis 35, 43–56. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Shao Y, and Fu J (2021). A microfluidics-based stem cell model of early post-implantation human development. Nat Protoc 16, 309–326. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Xue X, Resto-Irizarry AM, Li Z, Shao Y, Zheng Y, Zhao G, and Fu J (2019a). Dorsal-ventral patterned neural cyst from human pluripotent stem cells in a neurogenic niche. Sci Adv 5, eaax5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Xue X, Shao Y, Wang S, Esfahani SN, Li Z, Muncie JM, Lakins JN, Weaver VM, Gumucio DL, et al. (2019b). Controlled modelling of human epiblast and amnion development using stem cells. Nature 573, 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information Video 2_3D staining: Three-dimensional reconstruction of μPASE at t = 36 h, stained for ISL1 (green), TBXT (red) and SOX17 (magenta). Related to Figure 2
Supplemental Information Video 1_diffusion_map: Three-dimensional visualization showing developmental trajectories of AMLC, PSLC / MeLC and PGCLC lineages. Related to Figure 2
Data Availability Statement
scRNA-seq data have been deposited at the Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Interactive visualization of processed datasets is available at https://umichibbl.shinyapps.io/shinyapp/.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-ISL1 antibody | DSHB | Cat# 39.4D5, RRID:AB_2314683 |
| Rabbit anti-GATA3 antibody | Cell Signaling Technology | Cat# 5852, RRID:AB_10835690 |
| Mouse anti-GATA3 antibody | Thermo Fisher Scientific | Cat# MA1–028, RRID:AB_2536713 |
| Rabbit anti-GABRP antibody | Thermo Fisher Scientific | Cat# PA5–46830, RRID:AB_2610501 |
| Rabbit anti-MIXL1 antibody | Atlas Antibodies | Cat# HPA005662, RRID:AB_2667382 |
| Rabbit anti-HEY1 antibody | Abcam | Cat# ab154077, RRID:AB_2893447 |
| Mouse anti-TFAP2A antibody | Santa Cruz Biotechnology | Cat# sc-12726, RRID:AB_667767 |
| Goat anti-TBXT antibody | Thermo Fisher Scientific | Cat# PA5–46984, RRID:AB_2610378 |
| Rabbit anti-NANOG antibody | Cell Signaling Technology | Cat# 4903, RRID:AB_10559205 |
| Goat anti-NANOG antibody | R and D Systems | Cat# AF1997, RRID:AB_355097 |
| Mouse anti-TFAP2C antibody | Santa Cruz Biotechnology | Cat# sc-12762, RRID:AB_667770 |
| Goat anti-SOX17 antibody | R&D Systems | Cat# AF1924, RRID:AB_355060 |
| Rabbit anti-BLIMP1 antibody | Cell Signaling Technology | Cat# 9115, RRID:AB_2169699 |
| Rabbit anti-FOXA2 antibody | Cell Signaling Technology | Cat# 8186, RRID:AB_10891055 |
| Mouse anti-OCT4 antibody | Santa Cruz Biotechnology | Cat# sc-5279, RRID:AB_628051 |
| Rabbit anti-OCT4 antibody | Santa Cruz Biotechnology | Cat# sc-9081, RRID:AB_2167703 |
| Rabbit anti-SOX2 antibody | Millipore | Cat# AB5603, RRID:AB_2286686 |
| Mouse anti-DPPA3 antibody | Millipore | Cat# MAB4388, RRID:AB_2094156 |
| Mouse anti-GCM1 antibody | Santa Cruz Biotechnology | Cat# sc-101173, RRID:AB_2108121 |
| Goat anti-HAVCR antibody | R and D Systems | Cat# AF1750, RRID:AB_2116561 |
| Rabbit anti-FABP3 antibody | Proteintech | Cat# 10676-1-AP, RRID:AB_2102309 |
| Goat anti-GATA6 antibody | R and D Systems | Cat# AF1700, RRID:AB_2108901 |
| Mouse anti-NODAL antibody | Abcam | Cat# ab55676, RRID:AB_2151660 |
| Mouse anti- GAPDH antibody | Sigma-Aldrich | Cat# G8795, RRID:AB_1078991 |
| HRP-conjugated anti-mouse IgG antibody | Santa Cruz Biotechnology | Cat# sc-516102, RRID:AB_2687626 |
| Donkey anti-Rabbit 546 | Thermo Fisher Scientific | Cat# A10040, RRID:AB_2534016 |
| Donkey anti-Mouse 488 | Thermo Fisher Scientific | Cat# A-21202, RRID:AB_141607 |
| Donkey anti-Goat 647 | Thermo Fisher Scientific | Cat# A-21447, RRID:AB_2535864 |
| Donkey anti-Rabbit 568 | Thermo Fisher Scientific | Cat# A10042, RRID:AB_2534017 |
| Donkey anti-Mouse 647 | Thermo Fisher Scientific | Cat# A-31571, RRID:AB_162542 |
| Goat anti-Mouse IgG1 488 | Thermo Fisher Scientific | Cat# A-21121, RRID:AB_2535764 |
| Goat anti-Mouse IgG2b 647 | Thermo Fisher Scientific | Cat# A-21242, RRID:AB_2535811 |
| Biological samples | ||
| Human embryo | First People’s Hospital of Yunnan Province | N/A |
| cynomolgus monkeys (Macaca fascicularis) | Yunnan Key Laboratory of Primate Biomedical Research (LPBR) in China. Astrid Fagræus laboratory of the Karolinska Institutet in Sweden. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Epidermal growth factor (EGF) | R&D System | Cat# 236-EG-200 |
| CHIR99021 | Cayman Chemical | Cat# 13122 |
| A83-01 | Tocris Bioscience | Cat# 2939 |
| SB431542 | STEMCELL Technologies | Cat# 72234 |
| Y27632 | Tocris | Cat# 1254 |
| FGF2 | Thermo Fisher Scientific | Cat# PHG0261 |
| BMP4 | R&D Systems | Cat# 314-BP-050 |
| IWP2 | Tocris | Cat# 3533 |
| LY294002 | Selleckchem | Cat# S1105 |
| ACTVIN A | R&D Systems | Cat# 338-AC-050 |
| Critical commercial assays | ||
| LookOut Mycoplasma PCR Detection Kit | Sigma-Aldrich | Cat# MP0035-1KT |
| Click-iT™ Plus EdU Cell Proliferation Kit | Thermo Fisher Scientific | Cat# C10637 |
| Deposited data | ||
| Data files for RNA sequencing | Gene Expression Omnibus | GSE185643 |
| Processed dataset | Gene Expression Omnibus | GSE185643, https://umichibbl.shinyapps.io/shinyapp/ |
| Raw data for plots | Mendeley Data | http://dx.doi.org/10.17632/cvv5rzymhd.1 |
| Experimental models: Cell lines | ||
| Human: H9 (WA09) (female) | WiCell | NIH registration number: 0062 |
| Human: ESI-017 (male) | BioTime, Inc. | NIH registration number: 0093 |
| Chimpanzee: Chimpanzee iPSC lines (C3651, male and C4955, female) | (Gallego Romero et al., 2015; Pavlovic et al., 2018) | N/A |
| Human: Human trophoblast stem cells (hTSCs) | (Okae et al., 2018) | N/A |
| Human: ESI-017 NODAL-KO hPSC line | (Chhabra et al., 2019) | N/A |
| Oligonucleotides | ||
| NODAL_crRNA_1: 5’-AGGCUCAGCAUGUACGCCAG-3’ | Thermo Fisher Scientific | N/A |
| NODAL_crRNA_2: 5’-AGACAUCAUCCGCAGCCUAC-3’ | Thermo Fisher Scientific | N/A |
| NODAL exon 1 Forward Primer: 5’-CTTCCTTCTGCACGCCTGGTGG-3’ | Thermo Fisher Scientific | N/A |
| NODAL exon 1 Reverse Primer: 5’-CCAACCCACAGCACTTCCCGAG-3’ | Thermo Fisher Scientific | N/A |
| Recombinant DNA | ||
| pCXLE-EGFP expression plasmid | Addgene | plasmid # 27082; RRID: Addgene_27082 |
| Software and algorithms | ||
| R | open source | https://www.r-project.org |
| Python | open source | https://www.python.org/ |
| ImageJ | open source | https://imagej.net/software/fiji/ |
| Other | ||
| mTeSR™1 | STEMCELL Technologies | Cat# 85850 |
| mTeSR™ Plus | STEMCELL Technologies | Cat# 100-0276 |
| Gibco™ Geltrex™ LDEV-Free, hESC-Qualified, Reduced Growth Factor Basement Membrane Matrix | Thermo Fisher Scientific | Cat# A1413302 |
| Corning™ Matrigel™ hESC-Qualified Matrix | Thermo Fisher Scientific | Cat# 08-774-552 |
| KnockOut™ Serum Replacement | Thermo Fisher Scientific | Cat# 10828028 |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS -X) | Thermo Fisher Scientific | Cat# 51500056 |
| Essential 6 medium (E6) | Thermo Fisher Scientific | Cat# A1516401 |
This paper does not report original codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Raw data for the plots were deposited on Mendeley at http://dx.doi.org/10.17632/cvv5rzymhd.1


