Abstract
A comparative genomic approach was used to identify Helicobacter pylori 26695 open reading frames (ORFs) which are conserved in H. pylori J99 but highly diverged in other eubacteria. A survey of selected pathways of central intermediary metabolism was also carried out, and genes with a potentially selective role in H. pylori were identified. Forty-five ORFs identified in these two analyses were screened using a rapid vector-free allelic replacement mutagenesis technique, and 33 were shown to be essential in vitro. Notably, 13 ORFs gave essentiality results which are unexpected in view of their known or proposed functions, and phylogenetic analysis was used to investigate the annotation of 7 such ORFs which are highly diverged. We propose that the products of a number of these H. pylori-specific essential genes may be suitable targets for novel anti-H. pylori therapies.
The gastric pathogen Helicobacter pylori causes one of the most common infections of humans. In developed nations up to half of all individuals are infected with H. pylori, and this number rises to more than 90% in many developing nations (22). Although infected individuals are often asymptomatic, H. pylori is a causal agent of chronic gastritis in humans and is strongly associated with the development of both duodenal and gastric ulcers and the rare cancer gastric mucosa-associated lymphoid tissue lymphoma. It has also been established that there is a connection between H. pylori infection and gastric cancer, which is the second most common neoplastic cause of death worldwide. Various studies have even suggested that H. pylori infection has a role in nongastric diseases, including coronary heart disease and hypertension, although supporting evidence in these cases is often rather inconclusive. Eradication therapy is recommended for H. pylori-infected ulcer patients (39). Antibiotic monotherapies achieve only a low clearance rate, so the usual treatments are combinations of acid suppressors such as proton pump inhibitors with two antibiotics, often from metronidazole, tetracycline, amoxicillin, or clarithromycin. While they can achieve eradication rates of 85 to 90%, the efficacy of these treatments is being eroded by the rise in frequency of antibiotic-resistant isolates. Although resistance to amoxicillin and tetracycline is rare, substantial and increasing rates of resistance to metronidazole and clarithromycin are a significant cause of treatment failure (1). A further concern is that the use of broad-spectrum drug combinations to treat this chronic infection may promote resistance in other pathogenic bacteria, thereby compromising their utility in the treatment of more serious infections. As H. pylori produces an organ-specific infection which is not normally complicated by coinfection with other pathogens, it would seem ideally suited to treatment with a narrow-spectrum therapeutic agent which would not promote resistance in endogenous flora. Hence, there is considerable interest in developing novel therapies which are both more effective and specific for H. pylori.
The search for new ways to eradicate H. pylori has been greatly facilitated by the publication of the complete genome sequences of two H. pylori strains, 26695 (50) and J99 (2). We have used this information to drive two distinct approaches to identifying essential genes which are potential targets for H. pylori-specific antibiotics. Although the genome is only 1.65 Mb in size, it encodes many catabolic and anabolic capabilities which are equivalent to those of bacteria with much larger genomes, suggesting that H. pylori must have few functional redundancies and limited metabolic diversity. Hence, it is predicted that a relatively high percentage of genes involved in central metabolic pathways will be essential for its survival even if their direct equivalents in other organisms are not. Although the H. pylori 26695 and J99 genomes contain just 1,552 and 1,495 open reading frames (ORFs), respectively, the translational products of more than a third of these are only distantly related to their nearest eubacterial homologs or are of completely unknown function. Elucidation of the cellular roles of these unique genes may lead to the identification of completely novel, essential functions which are H. pylori specific. We therefore examined pathways of central intermediary metabolism for genes which are uniquely essential to H. pylori, and we also used a bioinformatic genome prioritization analysis to identify highly diverged H. pylori ORFs. We describe the allelic replacement mutagenesis of these genes and discuss the biological implications of our findings.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. pylori 26695 (ATCC 700392) was obtained from the American Type Culture Collection. H. pylori cultures were grown under microacrobic conditions (5% [vol/vol] O2, 5% [vol/vol] CO2, 90% [vol/vol] N2) at 37°C on Columbia agar (Oxoid, Basingstoke, United Kingdom) supplemented with 5% (vol/vol) defibrinated horse blood (Becton Dickinson, Sparks, Md.) and containing amphotericin B, vancomycin, and polymyxin B, each at 10 μg/ml. For selection of transformants, 20 μg of chloramphenicol per ml was also added. Supplements were used during the allelic replacement mutagenesis of particular ORFs as follows: 0.2 to 1 mM uracil with and without 0.3 mM orotidine monophosphate (OMP), 0.3 mM UMP, and/or 0.3 mM orotate (HP1257 [pyrE]); 0.01 to 1 mM p-aminobenzoic acid and 0.1 to 1 mM tyrosine, 0.02 to 1 mM tryptophan, and 1 to 4 mM phenylalanine (HP1038 [aroQ]); and 1 mM each adenine, guanine, xanthine, and hypoxanthine (HP0735 [gpt]). We found that increasing the concentrations of the medium supplements used during mutagenesis of HP0735 and HP1257 above these levels was toxic to wild-type cells.
Identification of highly diverged H. pylori ORFs.
A previously described relational bacterial genomic database was used to identify high-priority H. pylori gene targets in silico (11). The core of this database is an array of ORF-by-ORF sequence similarity scores (smallest sum probabilities) across multiple genomes as calculated by the Basic Local Alignment Search Tool (BLASTP) version 2.0 (3). Database queries in SQL language allow data mining of this database for various intergenomic protein evolutionary relationships. Predicted protein sequences of H. pylori 26695 were compared to those of H. pylori J99 (2), Campylobacter jejuni NCTC11168 (41), Escherichia coli K-12 (8), Bacillus subtilis (36), Haemophilus influenzae Rd (24), Enterococcus faecalis V583 (preliminary sequence data were obtained from The Institute for Genomic Research [TIGR] website at http://www.tigr.org), Yersinia pestis CO-92 biovar Orientalis (Y. pestis Sequencing Group, Sanger Centre, ftp://ftp.sanger.ac.uk/pub/pathogens/yp/), Klebsiella pneumoniae 573 (Genome Sequencing Center, Washington University, St. Louis, Mo., personal communication), Saccharomyces cerevisiae S288-C (28), Archeoglobus fulgidis VC-16 (35), Methanobacterium thermoautotrophicum ΔH (45), Methanococcus janaschii (12), Pyrococcus abyssi 29292 (R. Helig, unpublished data, http://www.genoscope.cns.fr.), and Pyrococcus horikoshii OT3 (34). H. pylori 26695 ORFs were selected for further target evaluation if they showed both highly significant homology to the top hit in H. pylori J99 (P[N] ≤ 1.0e−30) and low homology (P[N] ≤ 1.0e−15) to any ORF from most other bacterial genomes in the comparison profile.
Membrane protein predictions.
Average hydropathy profiles 〈H〉 (51) were generated using GES hydropathy values (21) weighted using a trapezoid window. Using a process similar to the initial steps of the TopPred II algorithm (16), helical transmembrane segments (TMS) were predicted for each peptide sequence by selecting 19 amino acids centered on the highest 〈H〉 value (MaxH), masking these from further consideration, and repeating the process until no peaks with a 〈H〉 of >0.5 remained. Subcellular locations were assigned based on the peak MaxH value, number of segments with a 〈H〉 of >1.0, and distribution and peak 〈H〉 values of the putative TMS. A MaxH cutoff of 1.15 was chosen to maximize the discrimination between two SwissProtein release 34 test datasets containing transmembrane and cytoplasmic proteins, respectively (10). Proteins with a MaxH of <1.15 were classified as cytoplasmic, while those with a MaxH of >1.15 and at least three possible TMS were classified as membrane proteins. Anchored proteins were defined as having exactly two TMS, one starting before amino acid (aa) 35 and one having a 〈H〉 of >1.15 with the other having a 〈H〉 not lower than 0.5. Proteins containing a single N-terminal TMS with a 〈H〉 of >1.15 were classified as secreted, although no test for a signal sequence cleavage site was performed. Proteins not fitting into any other category were classified as unknown. This method does not detect gram-negative outer membrane proteins, which do not have multiple transmembrane helices, but outer membrane proteins identifiable by homology were eliminated on the grounds of gene prevalence or protein function at other stages of our analysis.
Phylogenetic analyses.
Publicly available databases including partial genomic sequences were searched for homologs of the highly divergent H. pylori ORFs HP1159 (fic), HP0785 (lolA), HP1553 (recB), HP0020 (nspC), HP1257 (pyrE), HP0735 (gpt), and HP1164 (trxB) using BLASTP, TBLASTN, and PSI-BLAST (3). Amino acid sequences were aligned using the program CLUSTALW (49) and then manually edited. Nonconserved indels (highly variable regions which constitute insertions or deletions with reference to the conserved blocks of the alignment) were removed from multiple-sequence alignments prior to phylogenetic analysis. Maximum-parsimony (MP) analysis was performed using PAUP∗ (47), while neighbor-joining (NJ) trees were based on pairwise distances between amino acid sequences calculated by the programs PROTDIST and NEIGHBOR of PHYLIP version 3.57c (23) (http://evolution.genetics.washington.edu/phylip.html). For both methods, confidence limits of branch points were estimated by 1,000 bootstrap replications. In PROTDIST the Dayhoff program option, which estimates the expected amino acid replacements per position using a model based on the Dayhoff 120 matrix, was invoked (18). Trees from all analyses were visualized and, where appropriate, converted into figures using TREEVIEW version 1.6.1 (40).
Annotation of target ORFs.
We utilized ORF annotations assigned by TIGR (further details on TIGR gene annotations can be obtained at http://www.tigr.org), with the following exceptions. The oor (HP0588-0591) and por (HP1108-1111) genes and HP0086 (mqo) are annotated as described by subsequent authors (31, 32). HP1138 is annotated as spo0J based on the homology of the translational product to B. subtilis Spo0J and its proximity to a soj homolog, HP1139. HP1563 is annotated as ahpC, which is the more usual nomenclature for bacterial alkyl hydroperoxide reductase enzymes. HP1553 and HP0785 are annotated as recB and lolA, respectively, based on phylogenetic analyses performed during this work (see Results and Discussion).
Generation of H. pylori allelic replacement mutants.
An invariant resistance cassette containing a Campylobacter coli chloramphenicol resistance gene (cat) (53) was PCR amplified using primers designated C1 (5′-GATATAGATTGAAAAGTGGAT-3′) and C2 (5′-TTATCAGTGCGACAAACTGGG-3′). H. pylori 26695 genomic DNA was isolated by washing cells scraped from five 48-h confluent plates, resuspending them in 3.5 ml of Qiagen (Valencia, Calif.) buffer B1, and then using Qiagen Genomic DNA kits as per the manufacturer's instructions. Two pairs of gene-specific primers designated P1-P2 and P3-P4 were used to PCR amplify the up- and downstream regions, respectively, of each target gene using Pwo DNA polymerase (Roche, Mannheim, Germany), in order to produce fragments on the order of 500 bp flanking the region to be deleted. P2 and P3 contained 5′ leaders complementary to C1 and C2, respectively, followed by 20 bp of gene-specific sequence. Each PCR product was purified using Qiaquick PCR purification cartridges. A template mixture containing 1 μg of each of the three purified products was then PCR amplified using primers P1 and P4 in a single reaction, to generate a linear construct in which the up- and downstream fragments are fused to the central cat gene. In order to maximize product recovery, the PCR conditions used, following a standard hot start, were 2 cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 5 min, followed by 40 cycles of 94°C for 45 s, 40°C for 1 min, and 72°C for 3 min, and finally 70°C for 10 min. The resulting allelic replacement construct was confirmed by sequencing and introduced into H. pylori 26695 by transformation (52), by scraping cells from 24-h confluent plates into 0.2 ml of brucella broth (Becton Dickinson) and spotting 100 μl of this high-density inoculum onto Columbia agar plates made as described above. After incubation with 2 μg of DNA for 12 h, the transformed cell mix was spread over the plates and incubated for a further 24 h. The cells were then scraped onto selective Columbia agar plates containing 20 μg of chloramphenicol per ml and incubated for 3 to 7 days.
For nonessential genes, typical frequencies of mutant recovery (representing the combined frequencies of transformation and double-crossover recombination) were 2 × 102 to 4 × 103 CFU/μg of transforming DNA. In these cases, the chloramphenicol-resistant (Cmr) transformant colonies were restreaked on selective plates. In order to confirm that the correct chromosomal rearrangement had occurred, transformants were initially screened by diagnostic PCR. Extracts were prepared by resuspending a loopful of cells in 20 μl of alkaline sodium dodecyl sulfate (0.05 M NaOH, 0.25% sodium dodecyl sulfate), incubating at 100°C for 10 min, and adding 200 μl of water (based on a method provided by J. Versalovic, personal communication). Two microliters of extract was used as the PCR template. The diagnostic PCRs comprised sets of DNA primers designed to hybridize within the cat cassette paired with primers hybridizing to distal chromosomal sequences to generate DNA products of characteristic size. In addition, primers designed to hybridize within the intact target gene were used to confirm that it had been deleted. Chromosomal DNA was isolated from selected clones which appeared to contain the correct mutation and further examined by diagnostic PCR and Southern blot analysis. Genes were described as nonessential if one or more mutants carrying a confirmed allelic replacement mutation of the target gene were isolated using these methods.
If no Cmr colonies were obtained in comparison with parallel control allelic replacement experiments targeting nonessential genes, the transformation was repeated at least twice more. Genes were described as essential if all three transformations were negative in comparison to the control.
If Cmr colonies were obtained but DNA analysis (see above) demonstrated that they contained an intact copy of the wild-type gene in addition to the correct mutation, the transformation was repeated at least twice more, and DNA analysis of 20 to 30 putative mutants was performed. In these cases mutant recovery frequency tended to be much lower than for nonessential genes. Genes were described as essential if no mutants with the wild-type gene deleted could be identified.
RESULTS AND DISCUSSION
Identification of ORFs unique to H. pylori.
In order to identify H. pylori-specific ORFs for essentiality testing, we performed a global bioinformatics analysis of the H. pylori 26695 genome using a series of prioritization steps based on various biological criteria. H. pylori strains are genetically diverse, and a previous comparison of the H. pylori 26695 and J99 genomes showed that approximately 7% of the genes are strain specific (2), so a requirement for the chosen ORFs was that they be present in both available genomes at a BLASTP confidence level of 1.0e−30 or less (3). For the same reason, genes associated with the variable cag pathogenicity island (14) were excluded. The translational products of ORFs common to both strains were screened against 13 other microbial genomes to identify unique and highly diverged ORFs. An ORF was defined as having a homolog in another genome if the top hit had a BLASTP score equal to or less than 1.0e−15. Homology to archeabacterial or yeast ORFs was recorded for information but was not used as an exclusion criterion. H. pylori 26695 ORFs were retained if they were either unique or had homologs only in the related organism C. jejuni or in only one or two additional eubacterial genomes. These first two steps reduced the number of target ORFs from 1,552 to 606. TMS predictions using hydropathy profiles showed that H. pylori 26695 encodes 1,221 cytoplasmic proteins, while the remaining 21% of ORF translational products are either noncytoplasmic or of unknown subcellular localization (E. coli contains 27% noncytoplasmic proteins). Since membrane proteins tend to be poorly soluble outside the membrane and therefore less amenable to in vitro biochemical study, ORFs encoding probable integral membrane proteins were excluded from the gene set (with the exception of HP0740 [murF]), further reducing the number of target ORFs to 505. ORFs whose only detectable homologs were genes of unknown or hypothetical function were also excluded, since the absence of clues to function would make biochemical exploitation difficult, leaving 172 ORFs. Finally, ORFs known to encode proteins involved in nonessential processes (such as flagellar biosynthesis, hemolysis, urease production, capsular biosynthesis, and restriction-modification), or whose highest homologs in the public databases were genes encoding such proteins, were excluded. From the remaining 73 highly diverged potential target ORFs, 29 were chosen at random for essentiality testing.
Identification of key genes in central intermediary metabolism.
Biochemical and in silico pathway reconstruction studies have indicated that there are gaps in some H. pylori central intermediary metabolic pathways. We selected 13 ORFs encoding key enzymes in these pathways for further study. These included HP1099 (eda) and HP1100 (edd), which are thought to encode the Entner-Doudoroff (ED) pathway enzymes 2-keto-3-deoxy-6-phosphogluconate aldolase (EC 4.1.2.14) and 6-phosphogluconate dehydratase (EC 4.2.1.12), respectively; HP1385 (fbp), which is thought to encode the Embden-Meyerhof pathway enzyme fructose-1,6-bisphosphatase (EC 3.1.3.11); HP0086 (mqo), encoding malate quinone oxidoreductase (EC 1.1.99.16); and HP0509 (glcD), which is thought to encode glycolate oxidase subunit (EC 1.1.3.15). We also selected genes which encode the Krebs cycle enzymes pyruvate:flavodoxin oxidoreductase (EC 1.2.7.1) (HP1108-1111 [porCDAB]) and 2-oxoglutarate oxidoreductase (EC 1.2.7.3) (HP0588-0591 [oorDABC]). The porB (HP1111) and oorA (HP0590) genes have previously been shown to be essential (31). Some of these ORFs (oorDABC [HP0588-0591], porCDAB [HP1108-1111], and mqo [HP0086]) are highly diverged and also ranked highly in our global genome bioinformatic analysis, but HP0509 (glcD), HP1099 (eda), HP1100 (edd), and HP1385 (fbp) are moderately or highly conserved in other bacterial species.
Design of allelic replacement mutagenesis cassettes.
As a result of these analyses, 29 highly diverged ORFs and 13 ORFs encoding key products in central intermediary metabolism were chosen for further study. Three more genes whose essentiality or nonessentiality in H. pylori has already been reported in the literature (HP1027 [fur], HP1021 [response regulator], and HP1364 [histidine kinase]) were also included as controls. Allelic replacement constructs targeting each of these 45 ORFs were made by PCR amplification. In order to minimize potential polar effects, oligonucleotide primers were chosen so that flanking genes and intergenic regions including potential promoters would remain intact in the deletion mutant. The translation start sites of each ORF assigned by TIGR were checked for optimal spacing from potential ribosome binding sites, and where possible potential promoter sequences conforming to the H. pylori −10 region consensus TAtaaT were identified (25). Flanking genes were identified and in most cases were consistent with the gene order given by TIGR. However for certain ORFs with several possible start sites and those adjacent to short unannotated ORFs, the most conservative option was used when designing the primers.
Six of the target ORFs (HP0616, HP0372, HP0785, HP1164, HP1563, and HP1027) are monocistronic, making it extremely unlikely that replacement of these genes would have a polar effect on expression of neighboring genes. However, the rest appear to be transcriptionally or, in 16 cases, translationally coupled with one or both of their neighbors. Hence, in addition to the precautions we have described, transcriptional termination signals were removed from the chloramphenicol resistance gene marker (cat), and the cassettes were designed to integrate in the same orientation as the target genes in order to ensure transcription of the downstream region. The success of our approach is demonstrated by the fact that we detected essential and nonessential genes of both the monocistronic and polycistronic types. Furthermore, in at least one case we detected both an essential (HP1140 [birA]) and a nonessential (HP1138 [spo0J]) gene in the same operon (fmt birA soj spo0J).
Allelic replacement mutagenesis of potential target genes.
In order to determine gene essentiality, the allelic replacement constructs were directly transformed into H. pylori 26695. Typically, essential genes were so annotated following three independent, controlled transformations in which no mutants were recovered. However, in the case of 14 genes which we have designated essential (HP0020, HP0509, HP0588, HP0589, HP0590, HP0808, HP0975, HP1108, HP1109, HP1418, HP1563, HP1553, HP0740, and HP1159), a small number of chloramphenicol-resistant colonies was recovered following transformation with the allelic replacement cassette. In each case PCR and Southern blot analysis of 20 to 30 putative mutants demonstrated that they contained an apparently intact wild-type copy of the target gene in addition to the correct mutation, suggesting that only mutants which contain a functional second gene copy are viable. During allelic replacement mutagenesis studies for a further three ORFs (HP1257 [pyrE], HP1038 [aroQ], and HP0735 [gpt]), the transformed cells were plated onto media with and without nutritional supplements appropriate to the proposed gene function. A few chloramphenicol-resistant aroQ and gpt putative mutant colonies were obtained on supplemented media, but further subculture demonstrated that they did not require the presence of supplements for growth, and they too were found to contain a copy of the wild-type target gene. This type of gene duplication has been observed during similar allelic replacement mutagenesis studies with Streptococcus pneumoniae (15), and it was postulated that this might be explained by the stabilization under selective pressure of spontaneous tandem unstable chromosomal duplications, similar to those which have been reported to occur between repetitive sequences at moderately high frequency in E. coli and Salmonella enterica serovar Typhimurium (29). The essentiality results for all 45 ORFs tested are shown in Table 1.
TABLE 1.
H. pylori target ORFs and essentiality testing resultsa
| Category | HP no.b | Genec | Functiond | Product length (aa) | Spectrume | Localizationf | Essentialityg |
|---|---|---|---|---|---|---|---|
| Allelic replacement controls | 1021 | RR | Response regulator | 298 | C. jejuni | C | NE |
| 1027 | fur | Ferric uptake regulator | 150 | C. jejuni, E. coli, H. influenzae | C | NE | |
| 1364 | HK | Histidine kinase | 397 | C. jejuni | A | NE | |
| 1111 | porB | Pyruvate oxidoreductase subunit B | 314 | Archaebacteria | C | E | |
| 589 | oorA | 2-Oxoglutarate oxidoreductase subunit A | 375 | C. jejuni, archaebacteria | C | E | |
| Nucleic acid metabolism | 1553 | recB∗ | Helicase | 945 | C. jejuni | C | E |
| 1231 | holB | DNA polymerase III δ′ subunit | 218 | — | C | E | |
| 1138 | spo0J∗ | Partition-related protein | 290 | C. jejuni, B. subtilis | C | NE | |
| 1448 | rnpA | RNase P protein component | 161 | — | C | E | |
| 440 | topA | Topoisomerase I | 677 | C. jejuni, B. subtilis, H. influenzae, E. coli, archaebacteria | C | NE | |
| Protein synthesis and modification | 785 | lolA∗ | Lipoprotein chaperone | 184 | — | C | NE |
| 830 | gatA | Glu-tRNAGln amidotransferase subunit A | 453 | C. jejuni, B. subtilis, S. cerevisiae, archaebacteria | C | E | |
| 658 | gatB | Glu-tRNAGln amidotransferase subunit B | 475 | C. jejuni, B. subtilis, S. cerevisiae, archaebacteria | C | E | |
| 975 | gatC | Glu-tRNAGln amidotransferase subunit C | 93 | — | C | E | |
| 657 | ymxG | Putative processing protease | 432 | C. jejuni, B. subtilis | S | E | |
| 470 | pepF | Oligoendopeptidase F | 578 | C. jejuni, B. subtilis | C | NE | |
| 1140 | birA | Biotin acetyl coenzyme A carboxylase | 212 | C. jejuni | C | E | |
| 74 | lspA | Lipoprotein signal peptidase | 157 | C. jejuni | S | E | |
| Cell wall biosynthesis and cell division | 1159 | fic | Cell filamentation protein | 177 | H. influenzae | C | E |
| 740 | murF | UDP-MurNAc-pentapeptide presynthetase | 493 | C. jejuni, B. subtilis | M | E | |
| 1372 | mreC | Rod-shape-determining protein | 248 | C. jejuni | S | E | |
| 1418 | murB | UDP-N-acetylenolpyruvoyl glucosamine reductase | 259 | C. jejuni, B. subtilis | C | E | |
| Energy metabolism | 1099 | eda | 2-Keto-3-deoxy-6-phosphogluconate aldolase | 208 | B. subtilis, H. influenzae, E. coli | C | E |
| 1100 | edd | 6-Phosphogluconate dehydratase | 608 | C. jejuni, B. subtilis, H. influenzae, E. coli, archaebacteria, S. cerevisiae | C | E | |
| 1385 | fbp | Fructose-1,6-bisphosphatase | 290 | C. jejuni, H. influenzae, E. coli, S. cerevisiae | C | E | |
| 86 | mqo | Malate:quinone oxidoreductase | 450 | C. jejuni | C | E | |
| 588 | oorD | 2-Oxoglutarate oxidoreductase subunit D | 113 | C. jejuni | C | E | |
| 590 | oorB | 2-Oxoglutarate oxidoreductase subunit B | 273 | C. jejuni, archaebacteria | C | E | |
| 591 | oorC | 2-Oxoglutarate oxidoreductase subunit C | 186 | C. jejuni, archaebacteria | C | E | |
| 1108 | porC | Pyruvate oxidoreductase subunit C | 186 | Archaebacteria | C | E | |
| 1109 | porD | Pyruvate oxidoreductase subunit D | 130 | — | C | E | |
| 1110 | porA | Pyruvate oxidoreductase subunit A | 407 | C. jejuni, E. coli, archaebacteria | C | E | |
| 509 | glcD | Glycolate oxidase | 459 | C. jejuni, B. subtilis, H. influenzae, E. coli, archaebacteria, S. cerevisiae | U | E | |
| 1495 | tal | Transaldolase | 316 | C. jejuni | C | NE | |
| Intermediary metabolism | 735 | gpt | Xanthine-guanine PRTase | 153 | C. jejuni | C | E |
| 1257 | pyrE | Orotate PRTase | 201 | C. jejuni | C | E | |
| 1038 | aroQ | 3-Dehydroquinase | 167 | C. jejuni, B. subtilis, H. influenzae | U | E | |
| 20 | nspC | Carboxynorspermidine decarboxylase | 405 | C. jejuni | C | E | |
| 832 | speE | Spermidine synthase | 262 | — | C | NE | |
| 801 | moaD | Molybdopterin-converting factor subunit 1 | 74 | C. jejuni | C | NE | |
| 372 | dcd | Deoxycytidine triphosphate deaminase | 190 | C. jejuni | C | NE | |
| 808 | acpS | Holo-acyl carrier protein synthase | 119 | C. jejuni | C | E | |
| Response to exogenous stimuli | 1164 | trxB | Thioredoxin reductase | 324 | C. jejuni | C | E |
| 1563 | ahpC∗ | Alkyl hydroperoxide reductase subunit | 198 | C. jejuni, E. coli, B. subtilis, S. cerevisiae | C | E | |
| 616 | cheV | Chemotaxis protein | 313 | C. jejuni, B. subtilis | C | NE |
ORFs in boldface are discussed in more detail in the text.
H. pylori 26695 locus number assigned by TIGR.
An asterisk indicates that the gene name used in this study differs from previous annotations (see text for details).
Function of predicted gene product according to gene annotation.
Genomes screened which contain homologs of the gene with a confidence score of better than 1.0e−15. Archaebacteria are listed in the text. —, no hits at this confidence level.
C, cytoplasmic; A, anchored; S, secreted; U, unknown; M, membrane bound.
E, essential in vitro; NE, not essential in vitro.
Control allelic replacement studies.
Four of the five genes whose essentiality in H. pylori has previously been reported behaved as expected in our allelic mutagenesis system. We confirmed the essentiality of porB (HP1111) and oorA (HP0589) (31) and additionally demonstrated that each of the six remaining por and oor subunit genes is individually required for cell viability in vitro. We also recovered and confirmed mutants of the nonessential genes HP1027 (fur) (7) and HP1364 (histidine kinase) (6). However, we found that the orphan response regulator gene HP1021 is not essential for viability, in contrast to a previous study in which putative HP1021 mutant colonies were recovered but could not be further passaged (6). It is possible that the discrepancy is due to some difference in our respective protocols, and indeed we note that those authors used clinical isolate strains as mutation hosts and a more oxygen-rich atmosphere (5% CO2, 95% air) for cell culture. However we were unable to test the hypothesis that HP1021 is essential only under conditions of higher oxygen tension, as in our hands neither the HP1021 mutants nor wild-type H. pylori 26695 grew under these conditions.
Overview of target gene essentiality data.
Of the remaining 40 ORFs, 31 were essential and 9 were nonessential in vitro. In five cases (HP1140 [birA], HP0657 [ymxG], HP0830 [gatA], HP0658 [gatB], and HP0975 [gatC]) this analysis represents to our knowledge the first description of essentiality of the gene or its homologs in any bacterial species. In most other cases the essentiality data are consistent with the proposed function of the ORF. The results for these 27 ORFs are listed in Table 1 together with their putative function but are not further discussed. However, the behavior of 13 ORFs in our allelic mutagenesis system (shown in boldface in Table 1) was unexpected based on existing knowledge of the gene products or their orthologs. Six of these ORFs either have been biochemically characterized in H. pylori (HP0086 [mqo] and HP1038 [aroQ]), or are well conserved among eubacteria (HP1099 [eda], HP1100 [edd], HP1385 [fbp], and HP1563 [ahpC]). The remaining seven (HP1257 [pyrE], HP0735 [gpt], HP1553 [recB], HP1159 [fic], HP1164 [trxB], HP0020 [nspC], and HP0785 [lolA]) are highly diverged, so their primary structural relationships were investigated using phylogenetic analysis.
Essentiality of H. pylori genes of known function.
HP0086 has been shown in complementation studies to encode malate:quinone oxidoreductase (Mqo) (32). Although malate dehydrogenase activity has been reported in H. pylori extracts (43), no mdh gene has been detected in the H. pylori genomes, so our finding that HP0086 is essential is consistent with the argument that H. pylori is solely dependent on Mqo for malate oxidation.
The product of HP1038 (aroQ) has been purified and confirmed to be a type II dehydroquinase from the shikimate pathway (9). Disruption of shikimate pathway genes is well documented in other bacteria and has been shown to cause auxotrophy for aromatic amino acids and p-aminobenzoic acid, but we were unable to obtain HP1038 mutants on rich media even when these supplements were supplied exogenously. It therefore appears either that H. pylori is impaired in its ability to take up these supplements or that elimination of the shikimate pathway is lethal. Chorismate is a precursor for synthesis of the quinone components of the electron transport chain, and our results could be explained if H. pylori cannot obtain these molecules from the horse blood-supplemented Columbia plates used here.
Essentiality of conserved genes.
The ED pathway is responsible for catabolism of glucose, the only carbohydrate utilized by H. pylori (37). The pathway is constitutively expressed in H. pylori, and genes for key enzymes in the alternative glycolytic and pentose phosphate pathways (phosphofructokinase, pyruvate kinase, and 6-phosphogluconate dehydrogenase) cannot be detected in the genome, suggesting that it has a particular importance in this organism. The ED enzymes 6-phosphogluconate dehydratase and 2-keto-3-deoxy-6-phosphogluconate aldolase have been detected in H. pylori extracts (38) and are thought to be encoded by HP1099 (edd) and HP1100 (eda), respectively. Neither gene is essential in E. coli, although the accumulation of 2-keto-3-deoxy-6-phosphogluconate in eda mutants has a bacteriostatic effect on growth (26). The fact that we were unable to recover mutants of HP1099 or HP1100 is consistent with the hypothesis that catabolism of glucose in H. pylori occurs exclusively via the ED pathway.
The conserved ORF HP1385 is thought to encode fructose-1,6-bisphosphatase (Fbp). The relative activities in H. pylori extracts of this enzyme and the phosphofructokinase enzyme, which catalyzes the reverse reaction, suggest that gluconeogenesis is favored over glycolysis (30), and indeed no phosphofructokinase gene has been detected in the genome (37). The fbp gene is not essential in E. coli or B. subtilis (27), but in Pseudomonas aeruginosa, where Fbp is additionally required for catabolism of glyceraldehyde-3-phosphate, fbp null mutants could not be recovered (5). We found that HP1385 is essential, suggesting that Fbp may have a similarly enhanced cell role in H. pylori.
The conserved ORF HP1563 is thought to encode the AhpC subunit of alkylhydroperoxide reductase (AhpR), which catalyzes the reduction of organic hydroperoxides and hydrogen peroxide. ahpC is not essential in C. jejuni (4), but as C. jejuni ahpC mutants are hypersusceptible to oxidative stress, it is possible that HP1563 mutants were generated but were not able to proliferate in the 5% O2 microaerobic atmosphere used. It therefore appears either that the mechanisms for controlling oxidative stress in H. pylori are exquisitely sensitive to oxygen concentration or that they play an unusually critical role in cell viability.
Essentiality of highly diverged ORFs.
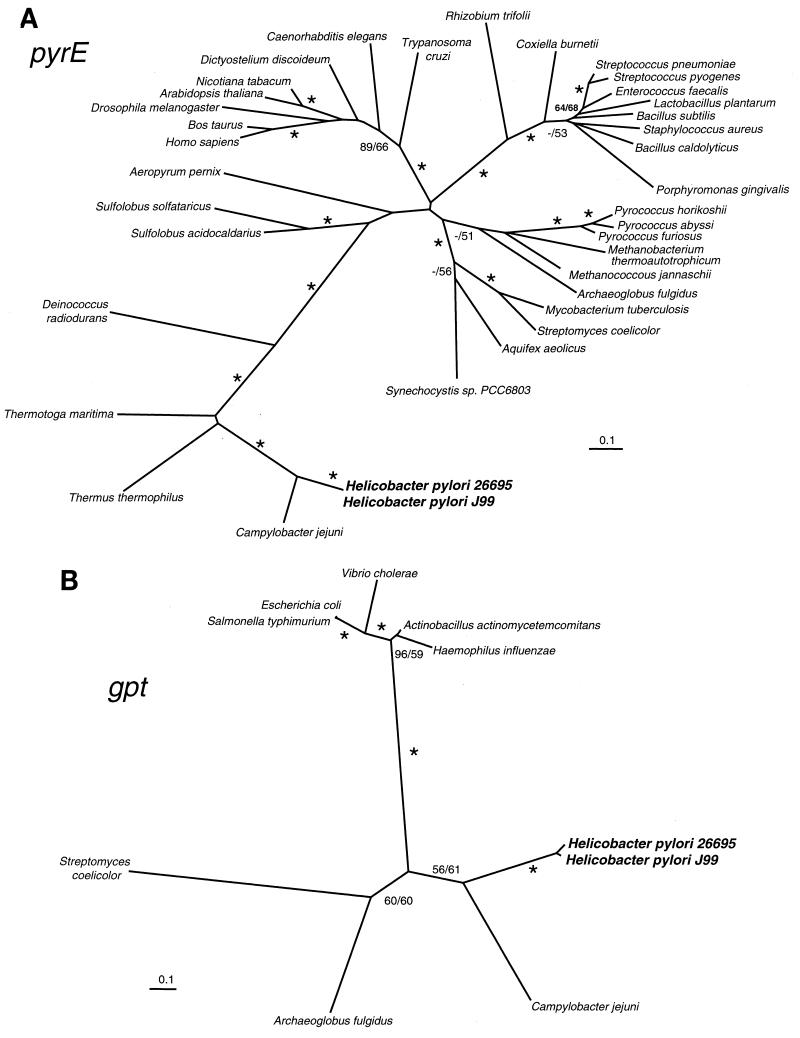
Phylogenetic analysis (Fig. 1A) supports the annotation of HP1257 as pyrE, encoding orotate phosphoribosyl transferase (PRTase), although it is very distantly related to other pyrE genes. This enzyme catalyzes the production of OMP, which is a precursor for UMP, a central intermediate in pyrimidine nucleotide metabolism. Bacterial pyrE mutants are usually uracil auxotrophs (54), but we were unable to obtain HP1257 mutants on rich media in the presence of exogenous uracil, even with supplemental OMP, UMP, and orotate. H. pylori has no homologs of the pyrimidine salvage enzymes uracil PRTase (Upp) or uridine kinase (Udk), which convert uracil to UMP (37). Although our results could be explained by poor uptake of medium supplements, they are consistent with recent reports that the conserved gene HP1084 (pyrB), encoding aspartate carbamoyltransferase, is also essential (13) and that inhibitors of dihydroorotate dehydrogenase (PyrD) are lethal to H. pylori (17). Taken together these data appear to demonstrate that de novo pyrimidine biosynthesis is required for survival of H. pylori.
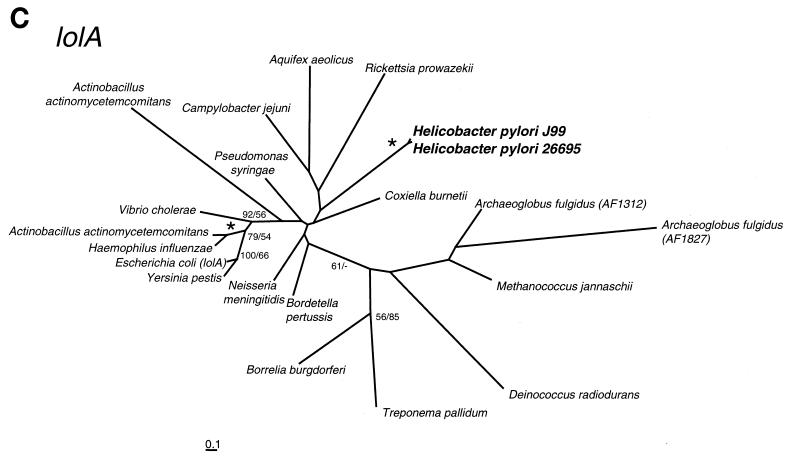
FIG. 1.
NJ phylogenetic trees, which show the divergence of H. pylori genes HP1257 (pyrE) (A), HP0735 (gpt) (B), and HP0785 (lolA) (C). The numbers at nodes represent the percent occurrence of nodes in 1,000 bootstrap resamplings of the MP and NJ trees. The NJ trees shown here were largely congruent with MP trees constructed using the program PAUP (47). Nodes occurring in more than 70% of resamplings by both methods are marked with an asterisk; values of <50% are marked with a dash or not shown. The scale bar represents 0.1 expected amino acid replacement per site as estimated by PROTDIST using the Dayhoff option. The trees were drawn using the program NEIGHBOR from PHYLIP version 3.57c (22).
(47). Nodes occurring in more than 70% of resamplings by both methods are marked with an asterisk; values of <50% are marked with a dash or not shown. The scale bar represents 0.1 expected amino acid replacement per site as estimated by PROTDIST using the Dayhoff option. The trees were drawn using the program NEIGHBOR from PHYLIP version 3.57c (22).
HP0735 (gpt) is thought to encode xanthine/guanine PRTase, which is involved in the salvage of nucleotides and nucleosides. Phylogenetic analysis (Fig. 1B) shows that the product of HP0735 is highly diverged from other proteobacterial Gpt enzymes. H. pylori does not appear to encode hypoxanthine PRTase (hpt), so it is possible that in this organism Gpt has a relaxed substrate specificity (37). E. coli gpt and gpt hpt mutants are viable (20), but no HP0735 mutants were recovered on rich media even in the presence of exogenous adenine, guanine, xanthine, and hypoxanthine. It therefore appears either that the medium supplements were not taken up or that nucleotide salvage has a particular importance for the survival of H. pylori.
Phylogenetic analysis (not shown) demonstrates that the helicase encoded by the essential gene HP1553 is a homolog of RecB, a subunit of the ExoV recombinase. The protein is clearly identifiable by sequence motifs specific to members of the RecB helicase subfamily, including the conserved C-terminal region at aa 876 to 901. This is surprising both because H. pylori does not appear to encode the other ExoV subunits RecC and RecD and also because ExoV is not an essential enzyme. Essential helicases have been described for other bacteria, including DnaB in E. coli and DnaB and PcrA in B. subtilis (42). Our finding suggests that despite its RecB-like structure and the fact that H. pylori encodes several other helicases, including the products of HP1478 (uvrD) and HP0911, the HP1553 helicase plays a critical role in cell function.
The Fic protein regulates cell division, although its exact function is unknown. Phylogenetic analysis (not shown) demonstrates that the products of HP1159 (fic) and E. coli fic are orthologous but highly divergent. E. coli fic mutants are viable, although they form shorter rods than wild-type cells (33), so our finding that HP1159 is essential suggests that there may be less functional redundancy among cell division proteins in H. pylori than in E. coli.
Reduced thioredoxin is required for cellular processes such as hydrogen peroxide reduction and reduction of catalytic disulfide bonds in cytoplasmic proteins. H. pylori 26695 contains two genes annotated as trxB (thioredoxin reductase), HP0825 and HP1164. The two proteins are only 16% identical, but HP0825 is more closely related to known thioredoxin reductases and forms an operon with trxA (HP0824). In E. coli trxB mutants, disulfide bond formation is controlled by TrxA and TrxC (46), but we found that HP1164 is essential. Since some thioredoxin reductases show limited homology to AhpF, the electron transfer subunit of AhpR (44), and as H. pylori does not encode an AhpF homolog, it is possible that HP1164 fulfils this function. As discussed above, the mechanisms for controlling oxidative stress in H. pylori appear to be unusually critical for cell viability.
Carboxynorspermidine decarboxylase (NspC) catalyzes the last step in synthesis of norspermidine. Our finding that HP0020 (nspC) is essential suggests that H. pylori, like members of the genus Vibrio, may utilize norspermidine as its primary polyamine (55). Alternatively, as H. pylori does not encode homologs of the SpeB, −C, or −D enzymes, which provide precursors for spermidine synthase SpeE, and as we have shown that HP0832 (speE) is not essential, it is possible that NspC is solely responsible for spermidine biosynthesis (19).
All of the ORFs described above are essential in H. pylori even though their orthologs in other organisms are nonessential. HP0785 is the only example of a nonessential ORF which might be expected to be essential. The product of HP0785 is homologous to the periplasmic lipoprotein chaperone LolA, although phylogenetic analysis (Fig. 1C) shows that the H. pylori protein is highly divergent from its proteobacterial equivalents. LolA localizes outer membrane-specific lipoproteins following their translocation to the periplasmic side of the inner membrane, in conjunction with the LolB receptor protein. The lolA gene is essential in E. coli (48). Our result that the lipoprotein signal peptidase gene HP0074 is essential demonstrates that lipoproteins are important for H. pylori survival, so our finding that HP0785 is not essential suggests that H. pylori may encode more than one such chaperone.
Perspectives.
The data presented here demonstrate that stepwise prioritization of genome ORFs using simple biological criteria, particularly in the case of small genomes such as that of H. pylori, can be an effective way of quickly reducing the number of genes of interest to an experimentally manageable number. The essentiality testing of a relatively large number of gene targets was accomplished by the application in H. pylori of a rapid and robust vector-free allelic replacement mutagenesis method. When combined with a set of initial selection criteria which are directly relevant to antibacterial drug discovery, this process is an efficient way to enrich for potential target genes and identify those which are critical for normal cell function.
The essentiality results reported for most of the ORFs are consistent with their proposed functions. However, we have identified 12 essential genes which might be expected to be nonessential by analogy with orthologous genes in other bacteria, including two genes whose products have previously been directly characterized in H. pylori (aroQ and mqo) and one nonessential ORF (lolA) whose closest homolog is essential in E. coli. It is possible that the most highly diverged of these ORFs have been misannotated, but in general our phylogenetic analyses support the ORF annotation given by TIGR. The exceptions are HP0785 and HP1553, which we have designated lolA and recB, respectively, but this refinement of the functional annotation has not clarified the reason for their essentiality. These results reflect the fact that H. pylori has a relatively small genome with little room for functional redundancy and must therefore contain a higher proportion of essential genes with core functions.
The high degree of sequence divergence of many of the essential gene products described here suggests that it may be possible to design specific chemical inhibitors which do not affect the growth of other organisms. This is recognized as a desirable attribute for any new anti-H. pylori therapeutic agent, as it would reduce the severity of gastrointestinal sequelae caused by destruction of the normal flora, and also minimize the spread of drug resistance to other pathogens. Importantly, a further degree of selectivity may be conferred by the fact that the nearest neighbors of some of these genes are not required for viability in other bacteria. The potential of this type of approach has been graphically illustrated by the recent identification of pyrazole-based anti-H. pylori compounds which specifically inhibit the pyrimidine biosynthesis enzyme PyrD in H. pylori but are inactive against orthologous enzymes in other bacteria and humans (17). We therefore suggest that the products of these highly diverged essential genes merit further investigation as potential targets for novel, highly specific anti-H. pylori agents.
ACKNOWLEDGMENTS
We are grateful to John Throup for useful discussions, Peter Morrison for assistance with phylogenetic analysis, Thomas Mathie for synthesis of oligonucleotide primers, and Stephanie Van Horn for sequence confirmation. We thank the Genome Sequencing Center, Washington University, St. Louis, Mo., for communication of K. pneumoniae DNA sequence data prior to publication.
Sequencing of H. pylori 26695 was accomplished with support from TIGR. Sequencing of E. faecalis V583 was accomplished with support from NIAID.
REFERENCES
- 1.Alarcon T, Domingo D, Lopez-Brea M. Antibiotic resistance problems with Helicobacter pylori. Int J Antimicrob Agents. 1999;12:19–26. doi: 10.1016/s0924-8579(99)00051-5. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L-S L, Moir D T, King N L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelson M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baillon M-L A, van Vliet A H M, Ketley J M, Constantinidou C, Penn C W. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J Bacteriol. 1999;181:4798–4804. doi: 10.1128/jb.181.16.4798-4804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee P C. Fructose-bisphosphatase-deficient mutants of mucoid Pseudomonas aeruginosa. Folia Microbiol. 1989;34:81–86. doi: 10.1007/BF02823683. [DOI] [PubMed] [Google Scholar]
- 6.Beier D, Frank R. Molecular characterization of two-component systems of Helicobacter pylori. J Bacteriol. 2000;182:2068–2076. doi: 10.1128/jb.182.8.2068-2076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bereswill S, Lichte F, Greiner S, Waidner B, Fassbinder F, Kist M. The ferric uptake regulator (Fur) homologue of Helicobacter pylori: functional analysis of the coding gene and controlled production of the recombinant protein in Escherichia coli. Med Microbiol Immunol. 1999;188:31–40. doi: 10.1007/s004300050102. [DOI] [PubMed] [Google Scholar]
- 8.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Bottomley J R, Clayton C L, Chalk P A, Kleanthous C. Cloning, sequencing, expression, purification and preliminary characterization of a type II dehydroquinase from Helicobacter pylori. Biochem J. 1996;319:559–565. doi: 10.1042/bj3190559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd D, Schierle C, Beckwith J. How many membrane proteins are there? Protein Sci. 1998;7:201–205. doi: 10.1002/pro.5560070121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown J R, Warren P V. Antibiotic discovery: is it in the genes? Drug Disc Today. 1998;3:564–566. [Google Scholar]
- 12.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 13.Burns B P, Hazell S L, Mendz G L, Kolesnikow T, Tillet D, Neilan B A. The Helicobacter pylori pyrB gene encoding aspartate carbamoyltransferase is essential for bacterial survival. Arch Biochem Biophys. 2000;380:78–84. doi: 10.1006/abbi.2000.1920. [DOI] [PubMed] [Google Scholar]
- 14.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalker A F, Lupas A, Ingraham K, So C Y, Lunsford R D, Li T, Bryant A, Holmes D J, Marra A, Pearson S C, Ray J, Burnham M K R, Palmer L M, Biswas S, Zalacain M. Genetic characterization of Gram-positive homologs of the XerCD site-specific recombinases. J Mol Microbiol Biotechnol. 2000;2:225–233. [PubMed] [Google Scholar]
- 16.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 17.Copeland R A, Marcinkeviciene J, Haque T S, Kopcho L M, Jiang W, Wang K, Ecret L D, Sizemore C, Amsler K A, Foster L, Tadesse S, Combs A P, Stern A M, Trainor G L, Slee A, Rogers M J, Hobbs F. Helicobacter pylori-selective antibacterials based on inhibition of pyrimidine biosynthesis. J Biol Chem. 2000;275:33373–33378. doi: 10.1074/jbc.M004451200. [DOI] [PubMed] [Google Scholar]
- 18.Dayhoff M O, Eck R V, Park C M. A model of evolutionary change in proteins. In: Dayhoff M O, editor. Atlas of protein sequence and structure. Vol. 5. Washington, D.C.: National Biomedical Research Foundation; 1972. pp. 89–99. [Google Scholar]
- 19.Doig P, de Jonge B L, Alm R A, Brown E D, Uria-Nickelsen M, Noonan B, Mills S D, Tummino P, Carmel G, Guild B C, Moir D T, Vovis G F, Trust T J. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol Mol Biol Rev. 1999;63:675–707. doi: 10.1128/mmbr.63.3.675-707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn I S. Pseudomonas aeruginosa plasmids as suicide vectors in Escherichia coli: resolution of genomic cointegrates through short regions of homology. Gene. 1991;108:109–114. doi: 10.1016/0378-1119(91)90494-v. [DOI] [PubMed] [Google Scholar]
- 21.Engelman D M, Steitz T A, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 22.Feldman R A, Eccersley A J P, Hardie J M. Transmission of Helicobacter pylori. Curr Opin Gastroenterol. 1997;13S1:8–12. [Google Scholar]
- 23.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.57c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 24.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 25.Forsyth M H, Cover T L. Mutational analysis of the vacA promoter provides insight into gene transcription in Helicobacter pylori. J Bacteriol. 1999;181:2261–2266. doi: 10.1128/jb.181.7.2261-2266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuhrman L K, Wanken A, Nickerson K W, Conway T. Rapid accumulation of intracellular 2-keto-3-deoxy-6-phosphogluconate in an Entner-Doudoroff aldolase mutant results in bacteriostasis. FEMS Microbiol Lett. 1998;159:261–266. doi: 10.1111/j.1574-6968.1998.tb12870.x. [DOI] [PubMed] [Google Scholar]
- 27.Fujita Y, Yoshida K, Miwa Y, Yanai N, Nagakawa E, Kasahara Y. Identification and expression of the Bacillus subtilis fructose-1,6-bisphosphatase gene (fbp) J Bacteriol. 1998;180:4309–4313. doi: 10.1128/jb.180.16.4309-4313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goffeau A, et al. The yeast genome directory. Nature. Vol. 387 1997. , suppl. 5. [PubMed] [Google Scholar]
- 29.Haack K R, Roth J R. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics. 1995;141:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman P S, Goodwin A, Johnsen J, Magee K, Veldhuyzen van Zanten S J O. Metabolic activities of metronidazole-sensitive and -resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J Bacteriol. 1996;178:4822–4829. doi: 10.1128/jb.178.16.4822-4829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes N J, Clayton C L, Chalk P A, Kelly D J. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J Bacteriol. 1998;180:1119–1128. doi: 10.1128/jb.180.5.1119-1128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kather B, Stingl K, van der Rest M E, Altendorf K, Molenaar D. Another unusual type of citric acid cycle enzyme in Helicobacter pylori: the malate:quinone oxidoreductase. J Bacteriol. 2000;182:3204–3209. doi: 10.1128/jb.182.11.3204-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamukai M, Matsuda H, Fujii W, Nishida T, Izumoto Y, Himeno M, Utsumi R, Komano T. Cloning of the fic-1 gene involved in cell filamentation induced by cyclic AMP and construction of a Δfic Escherichia coli strain. J Bacteriol. 1988;170:3864–3869. doi: 10.1128/jb.170.9.3864-3869.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 35.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger H H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D'Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 36.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S-K, Codani J-J, Connerton I F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 37.Marais A, Mendz G L, Hazell S L, Megraud F. Metabolism and genetics of Helicobacter pylori: the genomic era. Microbiol Mol Biol Rev. 1999;63:642–674. doi: 10.1128/mmbr.63.3.642-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendz G L, Hazell S L, Burns B P. The Entner-Doudoroff pathway in Helicobacter pylori. Arch Biochem Biophys. 1994;312:349–356. doi: 10.1006/abbi.1994.1319. [DOI] [PubMed] [Google Scholar]
- 39.NIH Consensus Conference. Helicobacter pylori in peptic ulcer disease. NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. JAMA. 1994;272:65–69. [PubMed] [Google Scholar]
- 40.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 41.Parkhill J, Wren B W, Mungall K, Ketley J M, Churcher C, Basham D, Chillingworth T, Davies R M, Feltwell T, Holroyd S, Jagels K, Karlyshev A V, Moule S, Pallen M J, Penn C W, Quail M A, Rajandream M A, Rutherford K M, van Vliet A H, Whitehead S, Barrell B G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 42.Petit M A, Dervyn E, Rose M, Entian K D, McGovern S, Ehrlich S D, Bruand C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 43.Pitson S M, Mendz G L, Srinivasan S, Hazell S L. The tricarboxylic acid cycle of Helicobacter pylori. Eur J Biochem. 1999;260:258–267. doi: 10.1046/j.1432-1327.1999.00153.x. [DOI] [PubMed] [Google Scholar]
- 44.Poole L B, Godzik A, Nayeem A, Schmitt J D. AhpF can be dissected into two functional units: tandem repeats of two thioredoxin-like folds in the N-terminus mediate electron transfer from the thioredoxin reductase-like C-terminus to AhpC. Biochemistry. 2000;39:6602–6615. doi: 10.1021/bi000405w. [DOI] [PubMed] [Google Scholar]
- 45.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart E J, Aslund F, Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swofford D L. PAUP∗. Phylogenetic Analysis Using Parsimony (∗and Other Methods), version 4. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 48.Tajima T, Yokota N, Matsuyama S, Tokuda H. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett. 1998;439:51–54. doi: 10.1016/s0014-5793(98)01334-9. [DOI] [PubMed] [Google Scholar]
- 49.Thompson J D, Higgins D G, Gibson T J. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 51.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Roos P, Taylor D E. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993;139:2485–2493. doi: 10.1099/00221287-139-10-2485. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Taylor D E. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- 54.Watrin L, Lucas S, Purcarea C, Legrain C, Prieur D. Isolation and characterization of pyrimidine auxotrophs, and molecular cloning of the pyrE gene from the hyperthermophilic archeon Pyrococcus abyssi. Mol Gen Genet. 1999;262:378–381. doi: 10.1007/s004380051096. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto S, Sugahara T, Tougou K, Shinoda S. Cloning and nucleotide sequence of the carboxynorspermidine decarboxylase gene from Vibrio alginolyticus. Microbiology. 1994;140:3117–3124. doi: 10.1099/13500872-140-11-3117. [DOI] [PubMed] [Google Scholar]