Abstract
Previous studies have shown that inhibition or depletion of N-acetyltransferase 1 (NAT1) in breast cancer cell lines leads to growth retardation both in vitro and in vivo, suggesting that NAT1 contributes to rapid growth of breast cancer cells. To understand molecular and cellular processes that NAT1 contributes to and generate novel hypotheses in regard to NAT1′s role in breast cancer, we performed an unbiased analysis of proteomes of parental MDA-MB-231 breast cancer cells and two separate NAT1 knockout (KO) cell lines. Among 4890 proteins identified, 737 proteins were found significantly (p < 0.01) upregulated, and 651 proteins were significantly (p < 0.01) downregulated in both NAT1 KO cell lines. We performed enrichment analyses to identify Gene Ontology biological processes, molecular functions, and cellular components that were enriched in each data set. Among the proteins upregulated in NAT1 KO cells, pathways associated with MHC (major histocompatibility complex) I-mediated antigen presentation were significantly enriched. This raises an interesting and new hypothesis that upregulation of NAT1 in breast cancer cells may aid them evade immune detection. Multiple pathways involved in mitochondrial functions were collectively downregulated in NAT1 KO cells, including multiple subunits of mitochondrial ATP synthase (Complex V of the electron transport chain). This was accompanied by a reduction in cell cycle-associated proteins and an increase in pro-apoptotic pathways in NAT1 KO cells, consistent with reported observations that NAT1 KO cells exhibit a slower growth rate both in vitro and in vivo. Thus, mitochondrial dysfunction in NAT1 KO cells likely contributes to growth retardation.
Abbreviations: NAT1, arylamine N-acetyltransferase 1; MHC-I, major histocompatibility complex I
Keywords: Proteomics, Mitochondria, ATP synthase, Cell cycle, Antigen presentation
Graphical Abstract
Highlights
-
•
NAT1 is frequently upregulated in breast cancer. Its inhibition leads to a decrease in breast cancer cell growth.
-
•
We performed an unbiased proteomics analysis of parental MDA-MB-231 breast cancer cells vs. NAT1 KO lines.
-
•
Proteins involved in antigen presentation were up in NAT1 KO cells, indicating they may become prone to immune detection.
-
•
Proteins involved in oxidative phosphorylation and mitochondrial biogenesis were collectively down in NAT1 KO.
-
•
Mitochondrial dysfunction, in part, can explain the growth retardation observed in NAT1 KO MDA-MB-231 breast cancer cells.
1. Introduction
Arylamine N-acetyltransferase 1 (NAT1) is a phase II metabolic enzyme that catalyzes acetyl coenzyme A-dependent biotransformation of aromatic amines and hydrazines [1], [2], [3]. In addition to its role in xenobiotic metabolism, recent studies have suggested that NAT1 has physiological roles. For instance, NAT1 is frequently upregulated in estrogen receptor-positive breast cancers as well as triple-negative breast cancers [4], [5], [6], [7]. These findings have led to further investigations of the role of NAT1 in cancer cell growth, morphology and metastasis [8], [9], [10], [11], [12], [13], [14], [15].
Small molecule-mediated inhibition and siRNA-induced silencing of NAT1, as well as CRISPR/Cas9 knockout (KO) technology, have been utilized to interrogate this novel link in various cancer cell lines, including those originating from breast cancers. Although discrepancies have been reported, studies from independent laboratories have shown that inhibition or depletion of NAT1 in cultured breast cancer cells results in a reduced cell growth and migration/invasion [12], [13], [14], [15], [16], [17]. In addition, inhibition or depletion of NAT1 leads to a marked decline in the ability of the cells grow in an anchorage-independent manner (i.e., in soft agar) [14], [17]. These results suggested that NAT1 promotes growth and metastasis of breast cancers. Our recent in vivo study [18], in which parental and NAT1 KO MDA-MB-231 cells were xenografted in immunocompromised mice, showed that both primary and secondary tumors of NAT1 KO cells grow significantly more slowly in vivo. However, their metastatic potential (indicated by lung metastasis) was not reduced in the absence of NAT1 [18]. These discrepancies highlight the fact that the role of NAT1 in breast cancer growth and development remains unclear, and novel hypotheses are needed to fill the gap in our knowledge.
In the current study, we conducted an unbiased analysis and comparison of proteomes of parental and NAT1 KO MDA-MB-231 cells in order to understand molecular and cellular processes that NAT1 contributes to and generate novel hypotheses in regard to its role in breast cancer.
2. Materials and methods
2.1. Cell culture
Generation and characterization of NAT1 KO MDA-MB-231 cell lines have been previously described [14]. NAT1 “KO2” and “KO5” cells represent two different KO cell lines generated using two unique guide RNAs and CRISPR/Cas9 technology. Parental and NAT1 KO MDA-MB-231 cells were cultured in DMEM media containing high glucose (4.5 g/L), fetal bovine serum (10 %), L-glutamine (4 mM), pyruvate (1 mM) and pen/strep (1 %). Cells were grown in a humidified incubator set at 37 °C with 5 % CO2.
2.2. Proteome analysis
Proteomic studies were conducted using a TMT-labeling approach as previously described [19] to compare wild-type and two NAT1 KO MDA-MB-231 cell lines (n = 3 technical replicates per condition). Cells were lysed in 2% SDS dissolved in 100 mM Tris HCl, pH 8.5 and supplemented with 10 mM sodium butyrate and 1 × HALT™ (Thermo Fisher) protease inhibitors. Protein concentrations were estimated using a detergent compatible assay (BioRad, Cat. #500-0116) against a bovine serum albumin standard curve. Protein lysates (200 μg) were reduced with tris(2-carboxyethyl)phosphine (TCEP) (Thermo Fisher), denatured with 8 M urea and alkylated with iodoacetamide followed by centrifugation through a high molecular weight cutoff centrifugal filter (MilliporeSigma; 10k MWCO) [20]. After digestion with recombinant LysC and then sequencing grade Trypsin (Promega, Madison, WI), the digested proteins were collected and quantified by Nanodrop (Thermo Fisher) at A205 nm prior to concentration adjustment to 0.5 μg/µL using 100 mM triethylammonium bicarbonate. Protein digested samples (50 μg) were labeled with tandem mass tag (TMT) using TMT10plex™ Isobaric Label Reagent Set (Thermo Fisher); samples were then concentrated and desalted with Oasis HLB Extraction cartridges (Waters Corporation) using a modified protocol for extraction of the digested peptides [21], [22]. Samples were then subjected to high pH reversed phase separation with fraction concatenation on a Beckman System Gold LC system supplemented with 126 solvent module and 166 detector in tandem with a BioRad Model 2110 Fraction Collector. A total of 18 concatenated fractions were collected and analyzed by liquid chromatography-mass spectrometry (LC-MS) to detect and quantify TMT-labeled peptides. Briefly, every high pH reversed phase fraction was dissolved in 50 µL solution of 2 % v/v acetonitrile/0.1 % v/v formic acid and 2 µL of each fraction was analyzed on EASY-nLC 1000 UHPLC system (Thermo Fisher) and an Orbitrap Elite – ETD mass spectrometer (Thermo Fisher). Peptides were loaded onto an Acclaim PepMap 100 75 µm × 2 cm, nanoViper (C18, 3 µm, 100 Å) trap prior to separation at 250 nl/min on an Acclaim PepMap RSLC 50 µm × 15 cm, nanoViper (C18, 2 µm, 100 Å) separating column (Thermo Fisher) with a 2–40 % acetonitrile/0.1 % formic acid gradient using an EASY n-LC UHPLC system (Thermo Fisher). Eluted peptides were introduced into the Orbitrap ELITE mass spectrometer using a Nanospray Flex source (Thermo Fisher) with an ion transfer capillary temperature of 225 °C and 1.6 kV spray voltage. An Nth Order Double Play was created in Xcalibur v2.2. Scan event one of the method obtained an FTMS MS1 scan (normal mass range; 60,000 resolution, full scan type, positive polarity, centroid data type) for the range 300–2000 m/z. Scan event two obtained FTMS MS2 scans (HCD activation, 1.0 m/z isolation width, normalized collision energy of 40.0, default charge state of 2, 0.1 ms activation time, normal mass range, 60,000 resolution, centroid data type) on up to 10 peaks that had a minimum signal threshold of 5000 counts from scan event one. The lock mass option was enabled (0 % lock mass abundance) using the 371.101236 m/z polysiloxane peak as an internal calibrant. Proteome Discoverer v1.4.1.14 was used to direct the Mascot v2.5.1 (Matrix Science Ltd, London, UK) and SequestHT (Thermo Fisher) searches of the raw data using the 5/12/2017 version of the UniprotKB Homo sapiens reference proteome reviewed canonical and isoform sequences. The search criteria were the following: enzyme specified was trypsin (maximum two missed cleavages; inhibition by P) with Carbamidomethyl(C) and TMT10plex (K, N-term) set as a static modifications and Oxidation(M) as dynamic. Fragment tolerance was 0.05 Da (monoisotopic) and parent tolerance was 50 ppm (monoisotopic). A Target Decoy PSM Validator node was included in the Proteome Discoverer workflow. The result files from Proteome Discoverer were loaded into Scaffold Q + S v4.4.5. Scaffold was used to calculate the false discovery rate using the Scaffold Local FDR and Protein Prophet algorithms. Peptides were accepted if the identification had probability greater than 99.9 % and parent mass error within 2 ppm. Proteins were accepted if they had a probability greater than 99.9 % and at least one peptide. Proteins were grouped into clusters to satisfy the parsimony principle. TMT purity correction factors were obtained from Thermo. Intensity based normalization of reporter ions was done using the median calculation type, unique peptides blocking level, individual spectrum references, and normalization between samples. The reference value was required for use in the Scaffold Q module, and the minimum dynamic range of reporter ions was set to 1 %. The results were annotated with human gene ontology information from the Gene Ontology Annotations Database (ftp.ebi.ac.uk). This yielded 4890 proteins with a false discovery rate of 0.389 %.
Data Sharing- Acquired MS data (.RAW) files for 18 high pH reversed phase LC fractions, Uniprot Human (reviewed dates May 12, 2017) sequence database, metadata for proteomic sample handling and data acquisition protocols including sample key, Scaffold Q+S v4.5.sf3 file and comparative Scaffold search results exported as an excel file (.xlsx) files have been deposited as a study entitled as “Proteomic Analysis of Arylamine N-Acetyltransferase 1 Knockout Breast Cancer Cells” in the MassIVE (http://massive.ucsd.edu/) data repository (MassIVE ID: 000089446) with the Center for Computational Mass Spectrometry at the University of California, San Diego. Uploaded data maybe be accessed by reviewers using the following link (ftp://MSV000089446@massive.ucsd.edu). Shared data will be released from private embargo for public access upon acceptance for publication.
2.3. Data analysis
Levels of protein species in NAT1 KOs were transformed to log2 fold change relative to those in parental cells. MetaCore™ (https://clarivate.com/products/metacore/) software were used to analyze protein fold changes between cell lines and predict probable cellular pathways affected by NAT1 deletion. In addition to MetaCore™, we analyzed our data through PANTHER Classification System (www.pantherdb.org) [23], GOnet (tools.dice-database.org/GOnet/) [24], and reactome (ver 65) [25], [26]. These algorithms provide gene function, ontology, pathways and statistical analysis tools. Comparisons of select proteins were done using GraphPad Prism v6.0c (GraphPad Software). One-way ANOVA and a Bonferroni multiple comparison post-test were performed to calculate statistical significance in select proteins between parental (P) vs. KO2 or KO5 cell lines. The following symbols are used to indicate respective levels of significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
3. Results
In order to identify proteins that are differentially expressed in the absence of NAT1, we performed an unbiased proteomics analysis of parental vs. NAT1 KO MDA-MB-231 cells. The parental MDA-MB-231 cells (parental) were compared to two NAT1 KO cell lines (KO2 and KO5) which had been previously generated using two separate guide RNAs [9], [14]. A total of 4890 proteins were identified via quantitative proteomic analysis. Among these, protein species whose levels were concomitantly and significantly changed in both KO cell lines with the same direction of change were selected for further analysis. A total of 737 proteins were significantly upregulated (p < 0.01), and a total of 651 proteins were found significantly downregulated (p < 0.01) in both NAT1 KO2 and KO5 cell lines, compared to the parental cell line (Supplementary Tables S1 and S6). We analyzed the lists of differentially upregulated and downregulated proteins separately, using multiple algorithms, including MetaCore, PANTHER [23], Reactome [25], and GOnet [24], to identify biological processes or components that were significantly enriched in each set. These systems allowed combined analysis of gene function, ontology, and pathways, as well as statistical analysis.
3.1. Biological processes enriched among upregulated proteins in NAT1 KO cells
3.1.1. Immune response and antigen presentation
The enrichment analysis of proteins upregulated in NAT1 KO cells identified ‘antigen processing and presentation’ (GO:001988) and multiple, similar pathways involved in this category (Table 1; Supplementary Table S2). These included ‘antigen processing and presentation of exogenous antigen’ (GO:0019884), and ‘antigen processing and presentation of peptide antigen’ (GO:0048002). The enrichment of biological processes belonging to ‘antigen processing and presentation’ was largely contributed by collective upregulation of accessory proteins, such as TAP1, TAP2, tapasin (TAPBP), and calreticulin (CALR) (Fig. 1A; Supplementary Table S2). To confirm this finding, we performed Western blot analysis of parental and KO cell lines for tapasin and TAP1. The protein levels of both tapasin and TAP1 were relatively higher in both NAT1 KO cell lines, compared to the parental cells (Fig. 1B). Accordingly, GO Cellular Component terms, ‘TAP complex’ (GO:0042825) and ‘MHC class I peptide loading complex’ (GO:0042824) were the two of the most highly enriched terms (Supplementary Table S4). Consistent with these findings, GO Molecular Function terms, such as ‘ABC-type peptide transporter activity (GO:0015440)’ and ‘MHC class I protein binding (GO:0042288)’ (Supplementary Table S3) were also overrepresented. Similarly, Reactome (ver 65) pathway analysis identified ‘Antigen Presentation: folding, assembly and peptide loading of class I MHC (R-HSA-983170)’ and ‘MHC class II antigen presentation (R-HSA-2132295)’ (Supplementary Table S5). The latter was contributed by upregulation of HLA-DRA (major histocompatibility complex, class II, DR alpha) and HLA-DRB1 (major histocompatibility complex, class II, DR beta 1) in NAT1 KO cells.
Table 1.
Selected GO biological processes enriched among upregulated proteins in NAT1 KO cells.a
| Category | GO term ID | GO term | P value | P FDR adj. |
|---|---|---|---|---|
| Antigen Presentation | GO:0019882 | antigen processing and presentation | 0.00E + 00 | 1.00E-09 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 2.79E-08 | 2.56E-06 | |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 1.27E-08 | 1.24E-06 | |
| GO:0048002 | antigen processing and presentation of peptide antigen | 0.00E + 00 | 1.00E-10 | |
| GO:0050851 | antigen receptor-mediated signaling pathway | 2.85E-04 | 7.08E-03 | |
| Innate Immune Response; Inflammation | GO:0006955 | immune response | 5.42E-05 | 1.79E-03 |
| GO:0002218 | activation of innate immune response | 8.24E-05 | 2.56E-03 | |
| GO:0045088 | regulation of innate immune response | 5.27E-06 | 2.56E-04 | |
| GO:0002758 | innate immune response-activating signal transduction | 1.57E-04 | 4.23E-03 | |
| GO:0070498 | interleukin-1-mediated signaling pathway | 4.96E-05 | 1.67E-03 | |
| GO:0002366 | leukocyte activation involved in immune response | 0.00E + 00 | 4.20E-09 | |
| GO:0002443 | leukocyte mediated immunity | 8.00E-10 | 9.86E-08 | |
| GO:0042098 | T cell proliferation | 4.43E-04 | 9.93E-03 | |
| Unfolded Protein Response; ERAD; Autophagy | GO:0034620 | cellular response to unfolded protein | 1.81E-05 | 7.32E-04 |
| GO:0006986 | response to unfolded protein | 9.89E-06 | 4.43E-04 | |
| GO:0036498 | IRE1-mediated unfolded protein response | 3.08E-05 | 1.14E-03 | |
| GO:0034975 | protein folding in endoplasmic reticulum | 3.00E-04 | 7.36E-03 | |
| GO:0030433 | ubiquitin-dependent ERAD pathway | 5.89E-05 | 1.93E-03 | |
| GO:0036503 | ERAD pathway | 1.11E-04 | 3.22E-03 | |
| GO:0006914 | autophagy | 4.55E-05 | 1.55E-03 | |
| GO:0010506 | regulation of autophagy | 1.80E-05 | 7.32E-04 | |
| Proteolysis | GO:0045862 | positive regulation of proteolysis | 1.43E-04 | 3.90E-03 |
| GO:0010498 | proteasomal protein catabolic process | 3.10E-09 | 3.43E-07 | |
| GO:0043248 | proteasome assembly | 6.06E-06 | 2.89E-04 | |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 4.00E-10 | 4.77E-08 | |
| GO:0051603 | proteolysis involved in cellular protein catabolic process | 0.00E + 00 | 1.00E-10 | |
| Vesicular Transport | GO:0016192 | vesicle-mediated transport | 9.78E-24 | 2.16E-20 |
| GO:0032527 | protein exit from endoplasmic reticulum | 2.18E-06 | 1.19E-04 | |
| GO:0051648 | vesicle localization | 1.68E-05 | 6.90E-04 | |
| GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 4.44E-07 | 2.93E-05 | |
| GO:0048199 | vesicle targeting, to, from or within Golgi | 2.42E-07 | 1.74E-05 | |
| GO:0048208 | COPII vesicle coating | 4.44E-07 | 2.93E-05 | |
| GO:0090114 | COPII-coated vesicle budding | 2.00E-07 | 1.49E-05 | |
| GO:1903513 | endoplasmic reticulum to cytosol transport | 3.29E-05 | 1.21E-03 | |
| GO:0048193 | Golgi vesicle transport | 0.00E + 00 | 0.00E + 00 | |
| GO:0030970 | retrograde protein transport, ER to cytosol | 3.29E-05 | 1.21E-03 |
Extended version of this table is available as Supplementary Table S2.
Fig. 1.
The antigen presentation by MHC class I pathway is upregulated inNAT1KO cells. A, proteomics analysis showed the levels of the indicated accessory proteins in the pathway (i.e., tapasin/TAPBP, TAP1, TAP2, and CALR) were significantly elevated in both KO cell lines (i.e., KO2 and KO5), compared to parental (P) MDA-MB-231 cells. The y-axis shows the relative fold change in log base 2 (Log2). One-way ANOVA and a Bonferroni multiple comparison post-test were performed to calculate statistical significance of the difference in the indicated proteins between parental (P) vs. KO2 or KO5 cell line. ***, p < 0.001. B, representative Western blots for tapasin and TAP1 in the indicated cell lines. GAPDH is shown as an internal control.
In addition, the proteins upregulated NAT1 KO cells were enriched in ‘immune response’ (GO:0006955) and related pathways, including ‘activation of innate immune response’ (GO:0002218), ‘regulation of innate immune response’ (GO:0045088), ‘leukocyte activation involved in immune response’ (GO:0002366), and ‘leukocyte mediated immunity’ (GO:0002443) (Table 1; Supplementary Table S2). This was, in part, due to upregulation of proteins, such as BCL10, MALT1 (mucosa-associated lymphoid tissue lymphoma translocation protein 1; paracaspase), NFKB1 (nuclear factor NF-kappa-b p105 subunit), PYCARD (a.k.a., apoptosis-associated speck-like protein containing a CARD), TOLLIP (toll interacting protein), and UBE2N. MALT1 is a caspase-like protease (‘paracaspase’) that binds to BCL10, and synergizes with BCL10 in promoting canonical NF-κB activation [27].
3.1.2. Membrane vesicle assembly and trafficking
Among the proteins upregulated in NAT1 KO cells, GO Biological Process terms pertaining to membrane vesicle assembly and trafficking were enriched. These included 'vesicle-mediated transport' (GO:0016192), 'vesicle localization' (GO:0051648), 'vesicle targeting, rough ER to cis-Golgi' (GO:0048207), 'Golgi vesicle transport' (GO:0048193), and 'COPII-coated vesicle budding' (GO:0090114) (Table 1; Supplementary Table S2). Accordingly, GO Cellular Component terms, including 'clathrin-coated vesicle' (GO:0030136), 'vesicle coat' (GO:0030120) and 'coated vesicle membrane' (GO:0030662), and GO Molecular Function terms, 'syntaxin binding' (GO:0019905), 'SNARE binding' (GO:0000149), and 'clathrin binding' (GO:0030276) were found enriched (Supplementary Tables S3 and S4).
3.1.3. Proteolysis
Another group of biological processes that was overrepresented among upregulated proteins belonged to protein catabolic process, and it included 'proteolysis' (GO:0006508), 'cellular protein catabolic process' (GO:0044257), 'proteasome-mediated ubiquitin-dependent protein catabolic process' (GO:0043161), and 'proteasome assembly' (GO:0043248) (Table 1; Supplementary Table S2). This was largely due to upregulation of multiple subunits of 20S and 26S proteasome complexes, such as PSMA5, PSMD10, PSMD9, PSMD4, RPN2, PSMC1, PSMD2, PSME4, PSMB10, and PSMB8 (Supplementary Table S2). This suggested that NAT1 KO cells have increased protein catabolic process. Of note, these upregulated proteasome subunits contributed simultaneously to the enrichment of GO terms related to antigen processing and presentation (see above) as the proteasome mediates proteolytic cleavage of antigens [28], and thus, upregulation of proteasomal subunits may reflect the requirement for antigen processing.
3.1.4. Cell death and apoptosis
GO terms related to cell death and apoptotic pathways were also enriched in NAT1 KO cells. These included 'positive regulation of extrinsic apoptotic signaling pathway' (GO:2001238), 'neuron apoptotic process' (GO:0051402), 'neuron death' (GO:0070997), and 'regulation of oxidative stress-induced intrinsic apoptotic signaling pathway' (GO:1902175) (Table 1; Supplementary Table S2). Enrichment of apoptosis processes in NAT1 KO cells were contributed by upregulation of DR6(TNFRSF21), FAF1, DAP3, Apaf-1, Par-4 (PAWR), PEA15, CARD5 (PYCARD), DIABLO (SMAC), Caspase-7, Bcl-10, PARP-1, TAX1BP1, TIMP3, and TRAF3 (CAP1) (Supplementary Table S2). Reactome [26] pathway analysis also showed that 'DIABLO-mediated apoptotic process' (R-HSA-111469; R-HSA-111464; R-HSA-111463), as well as 'formation and regulation of apoptosome' (R-HSA-9627069; R-HSA-111458), were enriched more than 10-fold among proteins upregulated in NAT1 KO cells, and these apoptotic pathways belonged to top 10 Reactome pathways with respect to the fold enrichment (Supplementary Table S5). This indicates that NAT1 KO cells may exhibit a higher rate of apoptotic cell death, compared to the parental cells. The findings of our recently published study support this notion. When parental and NAT1 KO MDA-MB-231 cells were xenografted in immunocompromised mice, primary tumors of NAT1 KO expressed an increased level of activated/cleaved caspase 3 (pro-apoptotic) and a decreased level of an anti-apoptotic protein, BCL-B, suggesting that the decreased in vivo growth observed in NAT1 KO tumors is, in part, contributed by an increased rate of apoptotic cell death [18].
4. Biological processes enriched among downregulated proteins in NAT1 KO cells
4.1. Mitochondrial ATP synthase and mitochondrial biogenesis
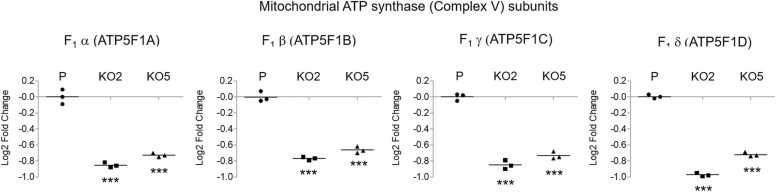
One of biological processes or pathways that were enriched among the downregulated proteins was subunits of mitochondrial ATP synthase, also known as F1/Fo ATPase or Complex V (EC 3.6.3.14) (Table 2; Supplementary Table S7). Multiple subunits of ATP synthase, including ATP5A1, ATP5B, ATP5C1, ATP5D, ATP5E, ATP5F1, ATP5H, ATP5J2, ATP5L, and ATP5O, were collectively downregulated in both NAT1 KO cell lines (Fig. 2; Supplementary Table S7). As a result, GO terms related to mitochondrial ATP synthesis were significantly enriched among the downregulated proteins. These included ‘ATP synthesis coupled proton transport’ (GO:0015986), ‘purine nucleoside triphosphate biosynthetic process’ (GO:0009145), and ‘oxidative phosphorylation’ (GO:0006119) (Table 2). Additionally, several subunits of Complex III of the electron transport chain were also downregulated in NAT1 KO cells. These included UQCRB, UQCRC1, UQCRC2, UQCRFS1, and UQCRQ, all of which belong to the ubiquinol-cytochrome-c reductase complex (cytochrome bc1 complex; Complex III), and accordingly, several GO terms falling under the biological process, ‘ubiquinol-cytochrome-c reductase complex,’ such as ‘mitochondrial electron transport, ubiquinol to cytochrome c' (GO:0006122), were enriched (Table 2; Supplementary Table S7). Collective downregulation of subunits of Complex III and V suggests that the electron transport chain and ATP synthesis via oxidative phosphorylation may be compromised in NAT1 KO cells.
Table 2.
Selected GO biological processes enriched among downregulated proteins in NAT1 KO cells.a
| Category | Sub-category | GO term ID | GO term | P value | P FDR adj. |
|---|---|---|---|---|---|
| Mitochondria |
Electron Transport Chain (Complex V and III) |
GO:0046034 | ATP metabolic process | 1.11E-06 | 6.78E-05 |
| GO:0006754 | ATP biosynthetic process | 6.00E-10 | 7.09E-08 | ||
| GO:0042776 | mitochondrial ATP synthesis coupled proton transport | 0.00E + 00 | 1.90E-09 | ||
| GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 8.69E-05 | 3.89E-03 | ||
| GO:0022900 | electron transport chain | 9.31E-04 | 2.64E-02 | ||
| Protein Synthesis (Mitochondrial Ribosome) | GO:0032543 | mitochondrial translation | 1.49E-07 | 1.07E-05 | |
| GO:0070126 | mitochondrial translational termination | 6.68E-06 | 3.64E-04 | ||
| GO:0070125 | mitochondrial translational elongation | 1.97E-07 | 1.36E-05 | ||
| GO:0070129 | regulation of mitochondrial translation | 1.17E-03 | 3.19E-02 | ||
| Membrane Organization | GO:0006839 | mitochondrial transport | 0.00E + 00 | 2.00E-10 | |
| GO:0007005 | mitochondrion organization | 2.00E-10 | 2.25E-08 | ||
| GO:0007006 | mitochondrial membrane organization | 0.00E + 00 | 4.20E-09 | ||
| GO:0070585 | protein localization to mitochondrion | 2.60E-04 | 9.48E-03 | ||
| GO:1990542 | mitochondrial transmembrane transport | 2.00E-10 | 2.32E-08 | ||
| GO:0070584 | mitochondrion morphogenesis | 5.00E-04 | 1.62E-02 | ||
| Ribosome & Ribonucleoprotein Complex | Ribosome Assembly | GO:0042254 | ribosome biogenesis | 3.32E-31 | 2.28E-28 |
| GO:0042273 | ribosomal large subunit biogenesis | 9.07E-16 | 0.00E + 00 | ||
| GO:0042255 | ribosome assembly | 9.36E-06 | 4.96E-04 | ||
| GO:0000054 | ribosomal subunit export from nucleus | 8.95E-04 | 2.56E-02 | ||
| rRNA Processing & Maturation | GO:0016072 | rRNA metabolic process | 7.78E-27 | 4.29E-24 | |
| GO:0006364 | rRNA processing | 6.13E-27 | 3.55E-24 | ||
| GO:0009303 | rRNA transcription | 1.84E-07 | 1.28E-05 | ||
| GO:0000469 | cleavage involved in rRNA processing | 5.91E-05 | 2.74E-03 | ||
| Ribonucleoprotein Complex | GO:0098781 | ncRNA transcription | 5.94E-05 | 2.74E-03 | |
| GO:0034470 | ncRNA processing | 1.24E-17 | 0.00E + 00 | ||
| GO:0071826 | ribonucleoprotein complex subunit organization | 0.00E + 00 | 3.60E-09 | ||
| GO:0022618 | ribonucleoprotein complex assembly | 0.00E + 00 | 3.60E-09 | ||
| GO:0022613 | ribonucleoprotein complex biogenesis | 5.08E-35 | 4.00E-32 | ||
| GO:0071426 | ribonucleoprotein complex export from nucleus | 0.00E + 00 | 1.00E-10 | ||
| Cell Cycle | GO:0007049 | cell cycle | 2.44E-08 | 2.05E-06 | |
| GO:0008283 | cell population proliferation | 1.50E-04 | 5.99E-03 | ||
| GO:0000075 | cell cycle checkpoint | 6.38E-04 | 1.93E-02 | ||
| GO:0044770 | cell cycle phase transition | 1.22E-04 | 5.25E-03 | ||
| GO:0044843 | cell cycle G1/S phase transition | 1.64E-04 | 6.43E-03 | ||
| GO:0006260 | DNA replication | 0.00E + 00 | 0.00E + 00 | ||
| DNA Damage Response | Checkpoints | GO:0031570 | DNA integrity checkpoint | 9.01E-04 | 2.57E-02 |
| GO:0044773 | mitotic DNA damage checkpoint | 2.62E-04 | 9.52E-03 | ||
| GO:0031571 | mitotic G1 DNA damage checkpoint | 1.04E-03 | 2.90E-02 | ||
| GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 2.60E-04 | 9.48E-03 | ||
| GO:0072422 | signal transduction involved in DNA damage checkpoint | 5.18E-04 | 1.66E-02 | ||
| DNA Repair | GO:0006281 | DNA repair | 0.00E + 00 | 0.00E + 00 | |
| GO:0006302 | double-strand break repair | 1.26E-07 | 9.10E-06 | ||
| GO:0000724 | double-strand break repair via homologous recombination | 7.16E-07 | 4.55E-05 | ||
| GO:0006974 | cellular response to DNA damage stimulus | 0.00E + 00 | 0.00E + 00 | ||
| GO:0006303 | double-strand break repair via nonhomologous end joining | 1.08E-04 | 4.73E-03 |
Extended version of this table is available as Supplementary Table S7.
Fig. 2.
The process involving mitochondrial oxidative phosphorylation is downregulated in NAT1 KO cells. Multiple subunits of the ATP synthase (Complex V) of the electron transport chain, including every subunit of its F1 domain (i.e., α, β, γ, δ, and ε subunits), were significantly downregulated in NAT1 KO cells. The graphs show the relative protein levels of α (ATP5F1A), β (ATP5F1B), γ (ATP5F1C), and δ (ATP5F1D) in parental (P) and NAT1 KO cell lines (KO2 and KO5). The y-axis shows relative fold change in log base 2 (Log2). ***, p < 0.001.
A number of mitochondrial ribosomal proteins (MRPL1, MRPL11, MRPL12, MRPL23, MRPL40, MRPL41, MRPL47, MRPL53, MRPL9, MRPS2, MRPS23, and MRPS27) were also downregulated collectively, and the biological processes such as, ‘mitochondrial translation’ (GO:0032543) and ‘mitochondrial translational elongation’ (GO:0070125) were enriched as a result (Table 2). Moreover, GO terms related to organization of mitochondrial membrane and mitochondrial protein localizations (e.g., GO:0007006 and GO:0070585) were also enriched among downregulated proteins in NAT1 KO cells (Table 2). For instance, TIMM50 (translocase of inner mitochondrial membrane 50; a.k.a., TIM50) and GRPEL1, which modulates mitochondrial heat shock protein 70, [29] were significantly downregulated in NAT1 KO cells.
4.2. Ribosome biogenesis
Another group of GO terms that stood out belonged to ribosome biogenesis. Downregulation of a set of proteins involved in ribosome and rRNA processing resulted in enrichment of the following GO terms: 'ribosome biogenesis' (GO:0042254 and GO:0042274), 'ncRNA and rRNA metabolic processing' (GO:0034660, GO:0034470, GO:0016072, and GO:0006364) (Table 2; Supplementary Table S7). In addition, GO terms related to both transcription and maturation of rRNA were also significantly enriched among downregulated proteins in NAT1 KO cells. These included as ‘rRNA transcription’ (GO:0009303), ‘maturation of LSU-rRNA’ (GO:0000470), and ‘maturation of 5.8S rRNA’ (GO:0000460) (Table 2; Supplementary Table S7). For example, the following factors involved in rRNA transcription were collectively downregulated in NAT1 KO cell lines: GTF3C4, GTF3C5, MARS, NIFK, POLR1E, SPIN1, and TCOF1 (Supplementary Table S7). Moreover, proteins involved in export of ribosome subunits and rRNA-containing ribonucleoprotein complexes, such as NPM, NUP88, RRS1, and SDAD1, were also found downregulated, and consequently, the following GO terms were enriched: ‘ribosomal subunit export from nucleus’ (GO:0000055 and GO:0000054) and ‘rRNA-containing ribonucleoprotein complex export from nucleus’ (GO:0071428) (Table 2; Supplementary Table S7). Taken together, collective downregulation of ribosomal subunits and rRNA, as well as factors involved in the export and assembly of ribosome suggests that NAT1 KO cells likely experience deficits in de novo synthesis of cellular (and mitochondrial) proteins necessary for their growth.
4.3. Cell cycle
Many proteins involved in cell cycle and DNA replication, such as CDK1, CCNH, KIF20A, NUMA1, PCNA, MKI67, MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7, were downregulated in NAT1 KO cells. As a result, a number of GO terms (e.g., GO:0000278, GO:0007049, GO:0022402, GO:0044786) related to the processes of cell cycle were enriched (Table 2; Supplementary Table S7).
5. Discussion
One of the interesting findings of the present study was the upregulation of the components of the major histocompatibility complex (MHC) class I (MHC-I) synthesis and antigen processing in NAT1 KO cells, including TAP1, TAP2, tapasin (TAPBP), and calreticulin (CALR) (see Fig. 1). These accessory proteins play essential roles in synthesis of and antigen presentation by MHC-I. For instance, tapasin and calreticulin function as chaperones and are required to stabilize the MHC-I complex in endoplasmic reticulum (ER) [30]. TAP (transporter associated with antigen processing) proteins (e.g., TAP1 and TAP2) transport antigenic peptides into ER, and the interaction between tapasin and TAP proteins allows loading of peptides into the MHC-I binding groove [30]. One way by which cancer cells avoid immune detection is downregulation of tumor-associated antigen presentation via MHC-I [31]. MHC-I plays an essential role in the initiation of the adaptive immune response, and cancer cells with downregulated MHC-I potentially evade immune system, for they are less likely to be recognized and targeted by CD8+ T-cells [31]. Accordingly, downregulation of MHC-I has been described in multiple types of human cancer, including breast cancer [32], [33], [34], and often correlates with poor prognosis [35], [36], [37]. Although the mechanism remains unknown, our results show that MDA-MB-231 breast cancer cells that are devoid of NAT1 exhibit increased expression of MHC-I and its accessory proteins, which would facilitate presentation of tumor-associated antigens and thereby become subject to cytotoxic T cell-mediated ablation. The present finding raises an interesting possibility that the breast cancer cells may become more prone to immune detection and ablation upon inhibition or silencing of NAT1.
According to this study’s proteomic data, NAT1 KO MDA-MB-231 cells appear to have mitochondrial dysfunction in multiple aspects. Proteins that play essential roles in 1) mitochondrial biogenesis, 2) mitochondrial membrane organization, 3) mitochondrial protein transport/localization, 4) electron transport chain complexes, and 5) mitochondrial ribosome biogenesis were collectively downregulated in both NAT1 KO cell lines. One of the cellular components that are heavily affected by the absence of NAT1 in MDA-MB-231 breast cancer cells was the mitochondrial ATP synthase and the electron transport chain. The human mitochondrial ATP synthase, also known as F1/Fo ATPase or complex V (EC 3.6.3.14), is composed of 29 subunits with a combined molecular weight of approximately 592 kDa [38]. The human mitochondrial ATP synthase complex is the fifth (and last) component of oxidative phosphorylation chain that harnesses the electrochemical energy to power ATP synthesis [12]. ATP synthase is comprised of two main functional domains, F1 and Fo. The F1 domain, which is located in the mitochondrial matrix, serves as the catalytic core, while the Fo domain contains the membrane proton channel, linked together by central and peripheral stalks [38]. According to our results, every subunit of F1 domain (i.e., α, β, γ, δ, and ε subunits) of the ATP synthase was significantly downregulated in both NAT1 KO cell lines at the protein level (see Fig. 2 and Table 2). Also, multiple subunits that comprise Fo domain, including subunits b, f, d, g, F6 and O (OSCP), were concomitantly downregulated in both NAT1 KO cell lines (see Supplementary Tables S6 and S7). Quantitatively, there was approximately 40–45 % reduction in the ATPase subunits in NAT1 KO cells, compared to the parental cells. According to our previous transcriptomics study on the same cell lines, the aforementioned subunits of the ATP synthases are also downregulated at the mRNA level in both NAT1 KO cell lines [39]. This further corroborates our current finding and suggests that the decreased protein levels of ATP synthase subunits are contributed by a decrease in transcription of the corresponding genes. NAT1 KO cell lines also showed reduced levels of some of the subunits of Complex III of the electron transport chain, also known as coenzyme Q:cytochrome c–oxidoreductase or cytochrome bc1 complex (EC 1.10.2.2). Specifically, cytochrome b-c1 complex subunit 1 (UQCRC1), 2 (UQCRC2), 7 (UQCRB), and Rieske (UQCRFS1) levels were significantly decreased in both NAT1 KO cell lines. Although it could not be confirmed by this current proteomic study, our previous transcriptomic study also found that some subunits of Complex I (e.g., NDUFA6, NDUFAB1, NDUFB1, NDUFB4, and NDUFS4) and Complex IV (e.g., COX7C) are also downregulated at the mRNA level in NAT1 KO cells [39].
GO terms related to organization of mitochondrial membrane and mitochondrial protein localizations were also enriched among downregulated proteins in NAT1 KO cells (see Table 2 and Supplementary Table S7). For instance, TIMM50 (translocase of inner mitochondrial membrane 50; a.k.a., TIM50) was significantly downregulated in NAT1 KO cells. TIMM50 functions as a subunit of the TIM23 inner mitochondrial membrane translocase complex which recognizes the mitochondrial targeting signal on protein cargos and translocates them to the mitochondrial inner membrane and matrix [40]. A recent study showed that TIMM50 deficiency results in a severe mitochondrial dysfunction and is essential for several aspects of mitochondrial physiology, including mitochondrial morphology, electron transport chain assembly, and mitochondrial respiratory capacity [41]. Similarly, a nucleotide exchange factor, GRPEL1, which plays a crucial role in modulating mitochondrial heat shock protein 70 (mtHsp70; HSPA9) [29], was also downregulated in both NAT1 KO cell lines. Notably, mtHsp70 facilitates the translocation of nuclear-encoded mitochondrial proteins across the mitochondrial membrane system into the matrix [42].
Collective downregulation of the electron transport chain complexes and mitochondrial proteome suggests that the oxidative phosphorylation process and ATP production in mitochondria is likely perturbed in NAT1 KO cells. In support of this notion, our previous metabolomics study on the same cell lines [10] suggested that the process of mitochondrial β‐oxidation of fatty acids is compromised in NAT1 KO cells. The NAT1 KO cells exhibit significant accumulation of β‐oxidation intermediates and a decrease in acyl carnitine species [10], which can be attributed to mitochondrial dysfunction. The mitochondrial dysfunction in NAT1 KO cancer cells have been also reported by others. A group examined the effects of NAT1 deletion on mitochondrial function in two human cell lines, MDA-MB-231 and HT-29 (colorectal adenocarcinoma) [15], [43], [44]. They found that NAT1 deletion resulted in decreased oxidative phosphorylation with a significant loss in respiratory reserve capacity in both cell lines [15]. There also was a decrease in glycolysis without a change in glucose uptake. These changes in mitochondrial function were attributed to a decrease in pyruvate dehydrogenase activity [15]. Taken together, these previous reports and our current findings suggest that the mitochondrial function is compromised at multiple levels in NAT1 KO cells. Although discrepancies exist in the NAT1-breast cancer literature, one of the most reproducible findings is that silencing or inhibition of NAT1 via multiple approaches results in a reduced growth of cancer cells, both in vitro and in vivo [13], [14], [16], [17], [18], [45]. We speculate that, in combination with increased pro-apoptotic mediators, the apparent mitochondrial dysfunction observed in NAT1 KO cells, at least in part, can contribute to the growth retardation.
Factors involved in various phases of the cell cycle were collectively downregulated in NAT1 KO MDA-MB-231 cells (see Table 2). This is also supported by our previous transcriptomics study of the same cell lines [39]. Reactome (ver 65) analysis of the transcripts that are significantly downregulated (p < 0.01) in both NAT1 KO cell lines shows overrepresentation of several reactome pathways for the mitotic cell cycle, especially the G1/S transition (i.e., R-HSA-69205, R-HSA-453279, and R-HSA-1640170; p < 0.001, FDR < 0.01) (see Supplementary Table S10). This suggests that the cell cycle entry (i.e., cell proliferation) is decreased or delayed in the absence of NAT1 in MDA-MB-231 cells. Our recently published study showed that when the cells are grown in vivo as a xenograft, both NAT1 KO cell lines exhibit slower tumor growth, and the relative level of PCNA protein (a commonly used histological marker of cell proliferation) is decreased, compared to the parental cells [18]. This is consistent with our current proteomics data which shows downregulation of numerous cell cycle proteins, including PCNA (see Supplementary Table S7). However, the levels of other cell cycle markers we examined in our previous study (i.e., cyclin D1, cyclin B1, and Ki67) were not consistently downregulated, which led us to conclude that the rate of cell proliferation in vivo is not significantly altered in NAT1 KO primary tumors [18]. Our current proteomics data suggest that the cell cycle entry is delayed in the absence of NAT1 in MDA-MB-231 cells in culture. Such discrepancy may reflect the difference in growth conditions between in vitro culture and in vivo. Regardless, the retarded growth of NAT1 KO MDA-MB-231 cells observed in both in vitro [14] and in vivo [18] appears to be contributed by reduced cell proliferation and/or increased cell death (apoptosis) (see under Section 3.1.4 above). These findings highlight that NAT1 is required for rapid growth of MDA-MB-231 breast cancer cells.
Additional studies are required to further corroborate and test the findings and hypotheses generated from the present study. It would be of interest to co-culture parental and NAT1 KO MDA-MB-231 cells with cytotoxic T cells or NK cells to test the role of NAT1 in antigen presentation and tumoral immunity. We and others have previously reported on changes in the mitochondrial respiration in NAT1 KO cells [9], [15]. However, the findings from these studies contrasted with each other, and thus, additional endpoints of mitochondrial function and structure would need to be investigated further to corroborate downregulation of multiple mitochondrial components observed in NAT1 KO cells.
CRediT authorship contribution statement
Kyung Hong: Writing – original draft, Formal analysis, Validation, Visualization. Jonathan Q. Gardner, Marcus W. Stepp and Mark A. Doll: Investigation, Data curation, Formal analysis, Validation, Visualization, Project administration, Writing – review & editing. Danny Wilkey, Frederick W. Benz, and Jian Cai: Investigation, Data curation, Formal analysis. Michael L. Merchant: Conceptualization, Supervision, Project administration, Writing – review & editing. David Hein: Conceptualization, Supervision, Project administration, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Institutes of Health, USA [NIEHS T32-ES011564, NCI R25-CA134283, NIEHS P30-ES030283 and NIGMS P20-GM113226 to D.W.H., and NIAAA P50-AA024337 and NIAAA R01-AA028436 to M.L.M.].
Handling Editor: Dr. Lawrence Lash
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.toxrep.2022.07.010.
Appendix A. Supplementary material
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
References
- 1.Hein D.W. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat. Res. 2002;506–507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 2.Hein D.W., Doll M.A., Fretland A.J., et al. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol. Prev. Biomark. 2000;9(1):29–42. [PubMed] [Google Scholar]
- 3.Sim E., Abuhammad A., Ryan A. Arylamine N-acetyltransferases: from drug metabolism and pharmacogenetics to drug discovery. Br. J. Pharm. 2014;171(11):2705–2725. doi: 10.1111/bph.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fei H., Chen S., Xu C. RNA-sequencing and microarray data mining revealing: the aberrantly expressed mRNAs were related with a poor outcome in the triple negative breast cancer patients. Ann. Transl. Med. 2020;8(6):363. doi: 10.21037/atm.2020.02.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perou C.M., Jeffrey S.S., van de Rijn M., et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl. Acad. Sci. USA. 1999;96(16):9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakefield L., Robinson J., Long H., et al. Arylamine N-acetyltransferase 1 expression in breast cancer cell lines: a potential marker in estrogen receptor-positive tumors. Genes Chromosomes Cancer. 2008;47(2):118–126. doi: 10.1002/gcc.20512. [DOI] [PubMed] [Google Scholar]
- 7.Zhao H., Langerød A., Ji Y., et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol. Biol. Cell. 2004;15(6):2523–2536. doi: 10.1091/mbc.e03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlisle S.M., Trainor P.J., Yin X., et al. Untargeted polar metabolomics of transformed MDA-MB-231 breast cancer cells expressing varying levels of human arylamine N-acetyltransferase 1. Metabolomics. 2016;12(7) doi: 10.1007/s11306-016-1056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlisle S.M., Trainor P.J., Doll M.A., Stepp M.W., Klinge C.M., Hein D.W. Knockout of human arylamine N-acetyltransferase 1 (NAT1) in MDA-MB-231 breast cancer cells leads to increased reserve capacity, maximum mitochondrial capacity, and glycolytic reserve capacity. Mol. Carcinog. 2018;57(11):1458–1466. doi: 10.1002/mc.22869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlisle S.M., Trainor T., Hong K.U., Doll M.A., Hein D.W. CRISPR/Cas9 knockout of human arylamine N-acetyltransferase 1 in MDAMB-231 breast cancer cells suggests a role in cellular metabolism. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-66863-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li P., Butcher N.J., Minchin R.F. Arylamine N-acetyltransferase 1 regulates expression of matrix metalloproteinase 9 in breast cancer cells: role of hypoxia-inducible factor 1-α. Mol. Pharm. 2019;96(5):573–579. doi: 10.1124/mol.119.117432. [DOI] [PubMed] [Google Scholar]
- 12.Li P., Butcher N.J., Minchin R.F. Effect arylamine N-acetyltransferase 1 on morphology, adhesion, migration, and invasion of MDA-MB-231 cells: role of matrix metalloproteinases and integrin αV. Cell Adhes. Migr. 2020;14(1):1–11. doi: 10.1080/19336918.2019.1710015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stepp M.W., Doll M.A., Carlisle S.M., States J.C., Hein D.W. Genetic and small molecule inhibition of arylamine N-acetyltransferase 1 reduces anchorage-independent growth in human breast cancer cell line MDA-MB-231. Mol. Carcinog. 2018;57(4):549–558. doi: 10.1002/mc.22779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stepp M.W., Salazar-González R.A., Hong K.U., Doll M.A., Hein D.W. N-Acetyltransferase 1 knockout elevates acetyl coenzyme A levels and reduces anchorage-independent growth in human breast cancer cell lines. J. Oncol. 2019;2019:3860426. doi: 10.1155/2019/3860426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Minchin R.F., Essebier P.J., Butcher N.J. Loss of human arylamine N-acetyltransferase I regulates mitochondrial function by inhibition of the pyruvate dehydrogenase complex. Int. J. Biochem. Cell Biol. 2019;110:84–90. doi: 10.1016/j.biocel.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Tiang J.M., Butcher N.J., Minchin R.F. Effects of human arylamine N-acetyltransferase I knockdown in triple-negative breast cancer cell lines. Cancer Med. 2015;4(4):565–574. doi: 10.1002/cam4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiang J.M., Butcher N.J., Cullinane C., Humbert P.O., Minchin R.F. RNAi-mediated knock-down of arylamine N-acetyltransferase-1 expression induces E-cadherin up-regulation and cell-cell contact growth inhibition. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0017031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doll M.A., Ray A.R., Salazar-González R.A., et al. Deletion of arylamine N-acetyltransferase 1 in MDA-MB-231 human breast cancer cells reduces primary and secondary tumor growth in vivo with no significant effects on metastasis. Mol. Carcinog. 2022;61(5):481–493. doi: 10.1002/mc.23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin J., Wahlang B., Shi H., et al. Dioxin-like and non-dioxin-like PCBs differentially regulate the hepatic proteome and modify diet-induced nonalcoholic fatty liver disease severity. Med. Chem. Res. Int. J. Rapid Commun. Des. Mech. Action Biol. Act. Agents. 2020;29:1247–1263. doi: 10.1007/s00044-020-02581-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 21.Keshishian H., Burgess M.W., Gillette M.A., et al. Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol. Cell. Proteom. MCP. 2015;14(9):2375–2393. doi: 10.1074/mcp.M114.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDowell G.S., Gaun A., Steen H. iFASP: combining isobaric mass tagging with filter-aided sample preparation. J. Proteome Res. 2013;12(8):3809–3812. doi: 10.1021/pr400032m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with PANTHER classification system. Nat. Protoc. 2013;8(8):1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pomaznoy M., Ha B., Peters B. GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinform. 2018;19(1):470. doi: 10.1186/s12859-018-2533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griss J., Viteri G., Sidiropoulos K., Nguyen V., Fabregat A., Hermjakob H. ReactomeGSA – efficient multi-omics comparative pathway analysis. Mol. Cell. Proteom. MCP. 2020;19(12):2115–2125. doi: 10.1074/mcp.TIR120.002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabregat A., Sidiropoulos K., Viteri G., et al. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinform. 2017;18(1):142. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucas P.C., Yonezumi M., Inohara N., et al. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J. Biol. Chem. 2001;276(22):19012–19019. doi: 10.1074/jbc.M009984200. [DOI] [PubMed] [Google Scholar]
- 28.Kloetzel P.M. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2001;2(3):179–188. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- 29.Srivastava S., Savanur M.A., Sinha D., et al. Regulation of mitochondrial protein import by the nucleotide exchange factors GrpEL1 and GrpEL2 in human cells. J. Biol. Chem. 2017;292(44):18075–18090. doi: 10.1074/jbc.M117.788463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cresswell P., Bangia N., Dick T., Diedrich G. The nature of the MHC class I peptide loading complex. Immunol. Rev. 1999;172(1):21–28. doi: 10.1111/j.1600-065X.1999.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 31.Cornel A.M., Mimpen I.L., Nierkens S. MHC class I downregulation in cancer: underlying mechanisms and potential targets for cancer immunotherapy. Cancers. 2020;12(7):1760. doi: 10.3390/cancers12071760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrido M.A., Rodriguez T., Zinchenko S., et al. HLA class I alterations in breast carcinoma are associated with a high frequency of the loss of heterozygosity at chromosomes 6 and 15. Immunogenetics. 2018;70(10):647–659. doi: 10.1007/s00251-018-1074-2. [DOI] [PubMed] [Google Scholar]
- 33.Maleno I., Cabrera C.M., Cabrera T., et al. Distribution of HLA class I altered phenotypes in colorectal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Immunogenetics. 2004;56(4):244–253. doi: 10.1007/s00251-004-0692-z. [DOI] [PubMed] [Google Scholar]
- 34.Romero J.M., Jiménez P., Cabrera T., et al. Coordinated downregulation of the antigen presentation machinery and HLA class I/beta2-microglobulin complex is responsible for HLA-ABC loss in bladder cancer. Int. J. Cancer. 2005;113(4):605–610. doi: 10.1002/ijc.20499. [DOI] [PubMed] [Google Scholar]
- 35.Andersson E., Villabona L., Bergfeldt K., et al. Correlation of HLA-A02* genotype and HLA class I antigen down-regulation with the prognosis of epithelial ovarian cancer. Cancer Immunol. Immunother. CII. 2012;61(8):1243–1253. doi: 10.1007/s00262-012-1201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson N.F.S., Ramage J.M., Madjd Z., et al. Immunosurveillance is active in colorectal cancer as downregulation but not complete loss of MHC class I expression correlates with a poor prognosis. Int. J. Cancer. 2006;118(1):6–10. doi: 10.1002/ijc.21303. [DOI] [PubMed] [Google Scholar]
- 37.Turcotte S., Katz S.C., Shia J., et al. Tumor MHC class I expression improves the prognostic value of T-cell density in resected colorectal liver metastases. Cancer Immunol. Res. 2014;2(6):530–537. doi: 10.1158/2326-6066.CIR-13-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He J., Ford H.C., Carroll J., et al. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc. Natl. Acad. Sci. USA. 2018;115(12):2988–2993. doi: 10.1073/pnas.1722086115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlisle S.M., Trainor P.J., Doll M.A., Hein D.W. Human arylamine N-acetyltransferase 1 (NAT1) knockout in MDA-MB-231 breast cancer cell lines leads to transcription of NAT2. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.803254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaudhuri M., Tripathi A., Gonzalez F.S. Diverse functions of Tim50, a component of the mitochondrial inner membrane protein translocase. Int. J. Mol. Sci. 2021;22(15):7779. doi: 10.3390/ijms22157779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tort F., Ugarteburu O., Texidó L., et al. Mutations in TIMM50 cause severe mitochondrial dysfunction by targeting key aspects of mitochondrial physiology. Hum. Mutat. 2019;40(10):1700–1712. doi: 10.1002/humu.23779. [DOI] [PubMed] [Google Scholar]
- 42.Kang P.J., Ostermann J., Shilling J., Neupert W., Craig E.A., Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348(6297):137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 43.Lichter J., Golka K., Sim E., Blömeke B. Recent progress in N-acetyltransferase research: 7th international workshop on N-acetyltransferases (NAT): workshop report. Arch. Toxicol. 2017;91(7):2715–2718. doi: 10.1007/s00204-017-1957-2. [DOI] [PubMed] [Google Scholar]
- 44.Wang L., Minchin R.F., Butcher N.J. Arylamine N-acetyltransferase 1 protects against reactive oxygen species during glucose starvation: role in the regulation of p53 stability. PLoS One. 2018;13(3) doi: 10.1371/journal.pone.0193560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiang J.M., Butcher N.J., Minchin R.F. Small molecule inhibition of arylamine N-acetyltransferase type I inhibits proliferation and invasiveness of MDA-MB-231 breast cancer cells. Biochem. Biophys. Res. Commun. 2010;393(1):95–100. doi: 10.1016/j.bbrc.2010.01.087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.