Abstract
Background & Aims
The lipid oxidation is a key factor for damaging hepatocytes and causing cell death. However, the mechanisms underlying hepatocyte death and the role of the most popular lipid peroxidation product 4-hydroxy-2-nonenal (HNE) in nonalcoholic steatohepatitis (NASH) remains unclear.
Methods
We demonstrated using hepatoma cell lines, a NASH mouse model, HNE-treated monkeys, and biopsy specimens from patients with NASH that HNE induced hepatocyte death by disintegrating the lysosomal limiting membrane.
Results
The degree of HNE deposition in human NASH hepatocytes was more severe in cases with high lobular inflammation, ballooning, and fibrosis scores, and was associated with enlargement of the staining of lysosomes in hepatocytes. In in vitro experiments, HNE activated μ-calpain via G-protein coupled receptor (GPR) 120. The resultant rupture/permeabilization of the lysosomal limiting membrane induced the leakage of cathepsins from lysosomes and hepatocyte death. The blockade of G-protein coupled receptor 120 (GPR120) or μ-calpain expression suppressed lysosomal membrane damage and hepatocyte death by HNE. Alda-1, which activates aldehyde dehydrogenase 2 to degrade HNE, prevented HNE-induced hepatocyte death. Intravenous administration of HNE to monkeys for 6 months resulted in hepatocyte death by a mechanism similar to that of cultured cells. In addition, intraperitoneal administration of Alda-1 to choline-deficient, amino-acid defined treated mice for 8 weeks inhibited HNE deposition, decreased liver inflammation, and disrupted lysosomal membranes in hepatocytes, resulting in improvement of liver fibrosis.
Conclusions
These results provide novel insights into the mechanism of hepatocyte death in NASH and will contribute to the development of new therapeutic strategies for NASH.
Keywords: Alda-1, Cell Death, 4-hydroxy-2-nonenal, Lysosomal Membrane Rupture
Abbreviations used in this paper: ALDH2, aldehyde dehydrogenase 2; ALT, alanine aminotransferase; BSA, bovine serum albumin; CDAA, choline-deficient, amino-acid defined; CTSB, cathepsin B; EPI, Epirubicin; GPR, G-protein-coupled receptor; HNE, 4-Hydroxy-2-nonenal; Hsp70.1, heat shock protein 70.1; IgG, immunoglobulin G; IL, interleukin; LAMP2, lysosome-associated membrane protein 2; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; PBS, phosphate buffered saline; siRNA, small interfering ribonucleic acid
Graphical abstract
Summary.
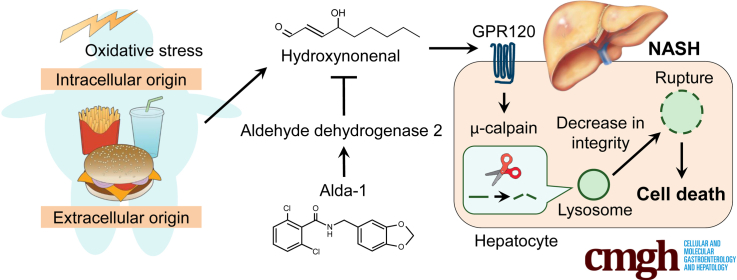
Hydroxynonenal is involved in the pathogenesis of nonalcoholic steatohepatitis by activating μ-calpain via G-protein coupled receptor 120 and disrupting the lysosome, leading to hepatocyte death. Administration of Alda-1 reduces liver fibrosis by inhibiting hydroxynonenal-induced hepatocyte death and liver inflammation.
Nonalcoholic steatohepatitis (NASH), one stage of nonalcoholic fatty liver disease (NAFLD), is characterized by excess adiposity, inflammation, and fibrosis of the liver and progresses to cirrhosis and hepatocellular carcinoma.1 The onset of NAFLD is associated with obesity, insulin resistance, and dyslipidemia, and its incidence is markedly increasing in developed countries.2 Because the incidence of NASH-related hepatocellular carcinoma is also increasing, it is emerging as an important public health issue.3 Multiple factors have been implicated in the pathophysiology of NAFLD. Oxidative stress is a crucial factor that damages the liver and contributes to the development of NASH. However, the mechanisms underlying hepatocyte degeneration/death that facilitate progression from simple fatty liver to NASH remain unclear. Therefore, the development of drugs that specifically target these pathways has been limited.
4-Hydroxy-2-nonenal (HNE) is one of the most cytotoxic aldehydes derived from ω-6 polyunsaturated fatty acids, such as linoleic and arachidonic acids. It directly induces functional damage to the proteins involved in cell death, the inhibition of enzymatic activities, and other biological processes.4 Protein modifications by HNE have been associated with many diseases, including atherosclerosis,5 ischemia-reperfusion injury,6,7 Parkinson’s disease,8 and Alzheimer’s disease.9,10 In addition, HNE inhibits deoxyribonucleic acid repair capacity through its direct interaction with repair proteins,11 causing genomic instability associated with carcinogenesis.12
HNE is also produced by overheating vegetable oil rich in linoleic acid,13 which is taken into our bodies orally. The recent increase in vegetable oil consumption14 suggests that HNE is deeply involved in our lives. In addition, HNE is pathologically detected in the hepatocytes of patients with NAFLD as an oxidative stress marker.15 Therefore, HNE may play an important role in pathogenesis of NAFLD. However, limited information is currently available on the effects of HNE on hepatocytes and the development of NASH.
We herein used an in vitro model with hepatoma cell lines and an in vivo HNE-treated Japanese macaque monkey model to demonstrate that HNE induces hepatocyte death by damaging cellular organelles, specifically the lysosomal membrane. We confirmed that the same mechanism led to HNE-induced cytotoxicity in the liver tissues of patients with NASH. Furthermore, we showed that intraperitoneal administration of Alda-1, which activates aldehyde dehydrogenase 2 (ALDH2) for HNE degradation,16,17 suppressed liver inflammation and fibrosis in the NASH mouse model. Collectively, the present results implicate HNE in hepatocyte degeneration/death and the resulting liver fibrosis; therefore, its inhibitors may block hepatocyte death and prevent progression from simple steatosis to NASH.
Results
Deposition of HNE in NAFLD Livers
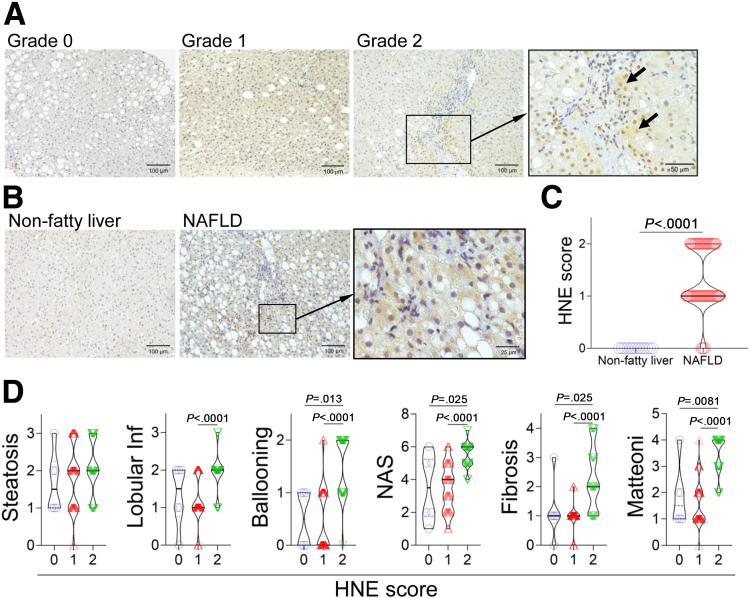
To investigate the relationship between HNE deposition in liver tissue and the NAFLD pathology, an immunohistochemical analysis was performed to detect HNE deposits in the livers of patients with nonfatty liver (n = 13) and NAFLD (n = 90) (Table 1). We semi-quantitatively classified the degree of HNE deposition into 3 levels (Figure 1, A). HNE deposits were not observed in nonfatty livers but were present in the cytoplasm of hepatocytes in most patients with NAFLD (Figure 1, B–C). The degree of HNE staining, scored using a 3-scale system (Figure 1, A), was significantly higher in the NAFLD group than in the nonfatty liver group (Figure 1, C). In the NAFLD group, lobular inflammation, the ballooning of hepatocytes, and fibrosis were noted in livers with extensive HNE staining. Livers with large HNE deposits had a high NAFLD activity score, reflecting disease activity in patients with NAFLD, and a high Matteoni’s classification score, indicating disease progression. However, no correlation was observed between the degree of HNE deposition and extent of fat accumulation (Figure 1, D).
Table 1.
Patient Profiles
| Variable | Nonfatty liver (n = 13) | NAFLD (n = 90) | P-value |
|---|---|---|---|
| Sex (male/female) | 7/6 | 53/37 | NS |
| Age, y | 53.0 ± 4.9 | 46.5 ± 1.7 | NS |
| Height, cm | 165.5 ± 1.7 | 163.2 ± 1.1 | NS |
| Weight, kg | 56.9 ± 3.3 | 73.6 ± 2.1 | < .0001 |
| BMI, kg/m2 | 20.1 ± 1.2 | 28.7 ± 0.6 | < .0001 |
| AST, IU/L | 17.0 ± 1.6 | 37.5 ± 5.6 | < .0001 |
| ALT, IU/L | 14.0 ± 2.4 | 49.0 ± 9.1 | < .0001 |
| Platelet count, ×104/mm3 | 22.5 ± 1.7 | 22.2 ± 0.7 | NS |
| Total protein, g/dL | 7.0 ± 0.2 | 7.1 ± 0.1 | NS |
| Albumin, g/dL | 4.3 ± 0.1 | 4.4 ± 0.0 | NS |
| PT, % | 102.0 ± 3.0 | 98.0 ± 1.6 | NS |
| HbA1c, % | 5.3 ± 0.1 | 6.7 ± 0.2 | .0002 |
| HOMA-IR | ND | 3.5 ± 0.6 | ND |
| Total cholesterol, mg/dL | 203.0 ± 9.7 | 200.0 ± 4.2 | NS |
| Triglyceride, mg/dL | 80.0 ± 13.6 | 136.0 ± 8.9 | .004 |
| HDL cholesterol, mg/dL | 57.0 ± 6.7 | 44.0 ± 1.3 | < .0001 |
| LDL cholesterol, mg/dL | 117.0 ± 8.1 | 122.0 ± 3.9 | NS |
| Histopathological findings | |||
| Fibrosis (0/1/2/3/4) | ND | 7/52/16/8/7 | ND |
| Steatosis (0/1/2/3) | ND | 2/28/37/23 | ND |
| Lobular inflammation (0/1/2/3) | ND | 6/37/42/5 | ND |
| Hepatocellular ballooning (0/1/2) | ND | 31/36/23 | ND |
| NAS score (0/1/2/3/4/5/6/7/8) | ND | 0/4/13/9/15/25/21/3/0 | ND |
Note: Data are expressed as number or median ± standard error of the median.
AST, Aspartate aminotransferase; ALT, alanine aminotransferase; HbA1c, hemoglobin A1c; HDL, high-density lipoprotein; HOMA-IR, homeostasis model assessment of insulin resistance [fasting serum insulin (μU/mL) × fasting plasma glucose (mg/dL)/405]; LDL, low-density lipoprotein; NAFLD, nonalcoholic fatty liver disease; NAS, NAFLD activity score; ND, not determined; NS, not significant; PT, prothrombin time.
Figure 1.
HNE is involved in the progression of disease in NASH.A, Semi-quantitative assessment of HNE immunoreactivity in the liver tissue of patients with NAFLD. The density of HNE immunoreactivity was scored into 3 grades as follows: no staining (Grade 0), weak and uniform staining of the entire tissue specimen (Grade 1), and intense spots (arrows) with uniform staining (Grade 2). The rectangle is magnified. B, HNE immunostaining in human liver tissue. The rectangle in the middle and its magnified image in the right, show focal HNE immunorecativity in the liver of a patient with NAFLD. C, The degree of HNE deposition was compared between patients with nonfatty liver (n = 13) and patients with NAFLD (n = 90). D, Relationships between pathological scores and HNE staining grades 0 (n = 6), 1 (n = 47), and 2 (n = 37) in patients with NAFLD. Lobular inf, lobular inflammation; Mattenoni, Matteoni’s classification; NAS, NAFLD activity score.
HNE-induced Hepatocyte Death
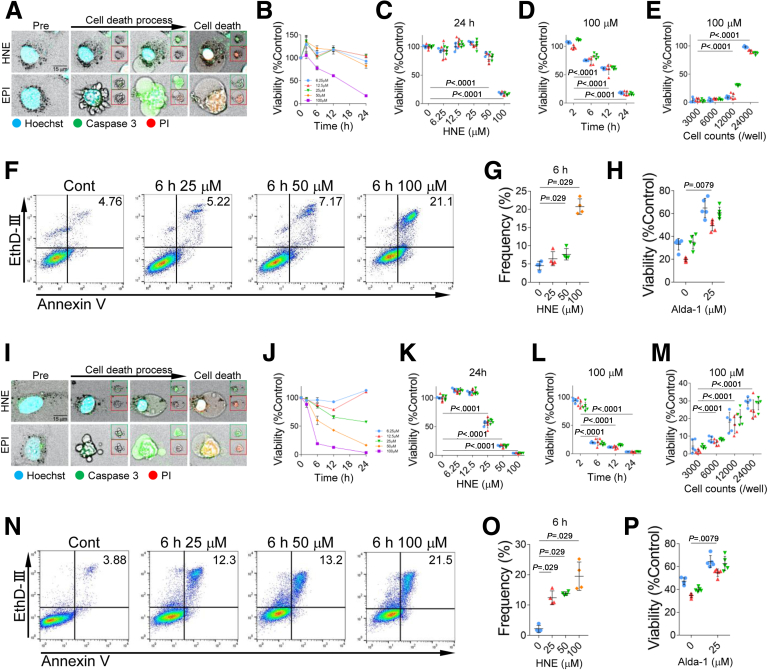
To assess the effects of HNE on hepatocytes, the HepG2 and Huh-7 hepatoma cell lines were exposed to HNE in vitro. Epirubicin (EPI) was used to induce apoptosis as a control.18 Time-lapse imaging revealed that the addition of EPI to the HepG2 culture induced cell death with the cell shrinkage and formation of blebbing. However, the addition of HNE induced “bursting” cell death without cell shrinkage or blebbing (Figure 2, A; Supplementary Movie 1). In analyses at the same cell density, cell viability decreased in time- and HNE concentration-dependent manners (Figure 2, B–D). However, as cell density increased, cell viability could be maintained at high levels even at seemingly high concentrations of HNE (100 μM). It should be noted that the HNE concentrations required to induce cell death were different (25–100 μM) because the cell densities varied with each assay, in the in vitro experiments in this study (Figure 2, E). The number of Annexin V- and ethidium homodimer III-positive dead cells in flow cytometry increased after the HNE treatment (Figure 2, F–G). Fluorescence time-lapse imaging showed that caspase-3 was activated by the addition of EPI or HNE (Figure 2, A; Supplementary Movie 2). Alda-1, which activates ALDH2 for HNE degradation,16,17 prevented HNE-induced reductions in cell viability of HepG2 (Figure 2, H). Similar results were obtained for Huh-7 cells after the addition of HNE (Figure 2, I–P).
Figure 2.
HNE induces hepatocyte death.A, The activation of caspase 3 was detected using NucView 488 Caspase-3 Substrate (green square) and cell death of HepG2 cells was visualized using propidium iodide (red square). Blue, nucleus; green, activated caspase 3, red, dead cell; yellow, merge. B, HepG2 cells viability was assayed 0, 2, 6, 12, and 24 hours after the addition of 6.25, 12.5, 25, 50, and 100 μM HNE. C, Statistical analysis of HepG2 cells viability 24 hours after the addition of 0, 6.25, 12.5, 25, 50, and 100 μM HNE is shown. D, Statistical analysis of HepG2 cells viability 2, 6, 12, and 24 hours after the addition of 100 μM HNE is shown. E, 3000, 6000, 12000, and 24000 HepG2 cells viability was assayed 24 hours after addition of 100 μM HNE. F, Cell death of HepG2 was assessed by staining cells with Annexin V and ethidium homodimer III (EthD-III), and analyzed by the flow cytometry. The analyses were done 6 hours after the addition of 25, 50, or 100 μM HNE. G, The difference in the frequency of double staining HepG2 cells with Annexin V and EthD-III 6 hours after the addition of 0, 25, 50, or 100 μM HNE is shown. H, Effect of Alda-1 on the viability of 40 μM HNE-treated HepG2 cells is shown. I, The activation of caspase 3 was detected using NucView 488 Caspase-3 Substrate (green square) and cell death of Huh-7 cells was visualized using propidium iodide (red square). Blue, nucleus; green, activated caspase 3; red, dead cell; yellow, merge. J, Huh-7 cells viability was assayed 0, 2, 6, 12, and 24 hours after the addition of 6.25, 12.5, 25, 50, and 100 μM HNE. K, Statistical analysis of Huh-7 cells viability 24 hours after the addition of 0, 6.25, 12.5, 25, 50, and 100 μM HNE is shown. L, Statistics analysis of Huh-7 cell viability 2, 6, 12, and 24 hours after the addition of 100 μM HNE is shown. M, 3000, 6000, 12000, and 24000 Huh-7 cells viability was assayed 24 hours after addition of 100 μM HNE. N, Cell death of Huh-7 was assessed by staining cells with Annexin V and EthD-III and analyzed by flow cytometry. The analyses were done 6 hours after the addition of 25, 50, or 100 μM HNE. O, The difference in the frequency of double staining Huh-7 cells with Annexin V and EthD-III 6 hours after the addition of 0, 25, 50, or 100 μM HNE is shown. P, Effect of Alda-1 on the viability of 40 μM HNE-treated Huh-7 cells is shown. C–E, H, K–M, P, The experiment was repeated 3 times, and blue, red, and green indicate the first, second, and third data, respectively.
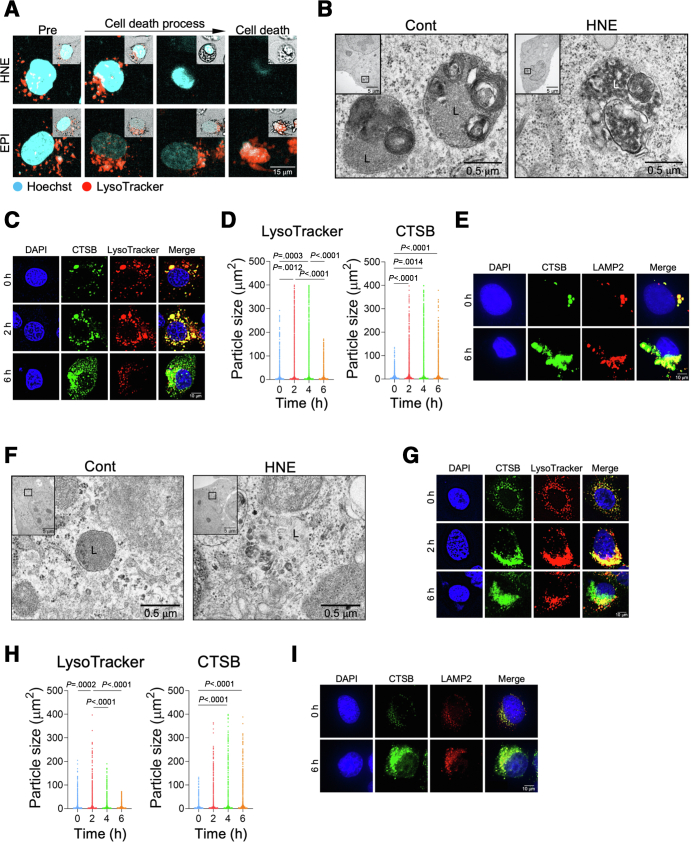
We then examined the mechanisms underlying hepatocyte death by HNE. HNE or EPI were added to the HepG2 culture, and the morphology of lysosomes was examined using time-lapse imaging by staining lysosomes with LysoTracker, which is a highly soluble small molecule that is retained in acidic intracellular compartments. The addition of HNE resulted in the gradual reduction and loss of lysosomes prior to cell death. However, lysosomes remained almost intact until the death of EPI-treated cells (Figure 3, A; Supplementary Movie 3). Electron microscopic observations revealed that HNE disrupted the lysosomal limiting membrane, resulting in the leakage of its content. In contrast, the ultrastructure of the lysosomal limiting membrane remained intact in HepG2 cells not exposed to HNE (Figure 3, B).
Figure 3.
Induction of lysosomal membrane permeabilization/rupture in hepatocytes by HNE.A, Changes in lysosomes of HepG2 cells after the addition of EPI and HNE were observed by time-lapse imaging using LysoTracker. Blue, Hoechst; red, LysoTracker. Square on the top right indicates morphological changes that were observed on bright field imaging. B, Electron microscopy images of HepG2 cells. Black squares show magnified images of membrane-bound lysosomes in the control (Cont) and disruption of the lysosomal limiting membrane in HNE-treated HepG2 cells (HNE), respectively. L, lysosome. C, HepG2 cells were treated with 25 μM HNE, and imaged 0, 2, and 6 hours later. Blue, DAPI; green, CTSB; red, LysoTracker; yellow, merge. D, Area occupied by LysoTracker and CTSB for each particle in HepG2 cells at a 20× image. The analysis was performed with 10 images in each group. E, Images show immunoreactivity of HepG2 cells for CTSB and LAMP2 before (0) and 6 hours after the treatment of 25 μM HNE. Blue, DAPI; green, CTSB; red, LAMP2; yellow, merge. F, Electron microscopy images of Huh-7. Black squares show magnified images of membrane-bound lysosomes in the control (Cont) and disruption of the lysosomal limiting membrane in HNE-treated Huh7 cells (HNE), respectively. L, lysosome. G, Huh-7 cells were treated with 25 μM HNE, and imaged 0, 2, and 6 hours later. Blue, DAPI; green, CTSB; red, LysoTracker; yellow, merge. H, Area occupied by LysoTracker and CTSB for each particle in Huh7 cells at a 20× image of Huh7. I, Images show immunoreactivity of Huh-7 cells for CTSB and LAMP2 before (0) and 6 hours after the treatment of 25 μM HNE. Blue, DAPI; green, CTSB; red, LAMP2; yellow, merge.
The morphology of lysosomes and translocalization of cathepsin, which is physiologically enclosed within lysosomes, were evaluated by immunofluorescence staining. Although the size of individual lysosomes transiently increased and then decreased, the granular area stained by cathepsin B (CTSB) increased after the HNE treatment (Figure 3, C– D). The CTSB-positive granular area was also co-stained by lysosome-associated membrane protein 2 (LAMP2), which is an abundant component of the lysosomal membrane, and the area of the merged color in the cytoplasm increased over time (Figure 3, E). A similar analysis was performed on Huh-7 cells (Figure 3, F–I) with the same results. Collectively, these results suggest that HNE disrupts the lysosomal limiting membrane to induce cell death by the leakage of cathepsins.
Mechanisms of Lysosomal Membrane Rupture
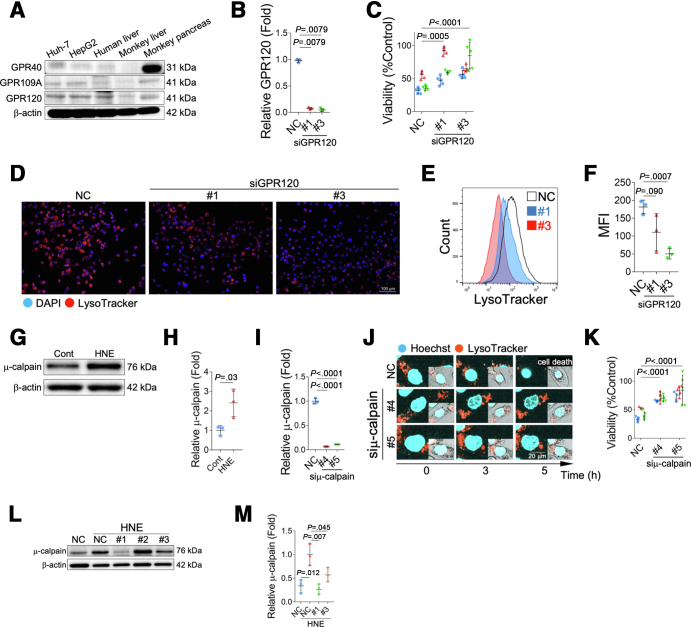
We next asked the mechanisms underlying HNE-induced lysosomal membrane rupture. HNE has been reported to activate the free fatty acid receptors, G-protein coupled receptor (GPR) 40 or GPR109A in neurons or pancreatic β-cells, and induces calpain activation, which is associated with disruptions in the stability of the lysosomal membrane, by increasing the intracellular Ca2+ concentration.13 We herein examined the expression of 3 GPRs; GPR40, GPR109A, and GPR120 (Figure 4, A). The expression of GPR120 was clearly expressed in HepG2, Huh-7 cells, and human and monkey liver tissues compared with the expression of GPR40 and GPR109A. Activation of GPR120 has been reported to rise the intracellular Ca2+ concentration,19,20 and may be involved in lysosomal membrane disruptions via activation of μ-calpain.
Figure 4.
Mechanism of lysosomal disintegrity of cultured cells by HNE.A, The expression of GPR40, GPR109A, and GPR120 in Huh-7 cells, HepG2 cells, the human liver, monkey liver, and monkey pancreas was evaluated by a Western blotting analysis. B, The relative fold expressions by real-time PCR indicates that GPR120 in HepG2 cell is suppressed by siRNA (#1 and #3). C, Viability of HNE-treated HepG2 cells with (#1 and #3) and without (NC) the down-regulation of GPR120 by siRNA. D, Lysosomes of HepG2 cell, with 25 μM HNE treatment, in which GPR120 was suppressed by siRNA (#1 and #3) were stained with LysoTracker. Red, LysoTracker; blue, DAPI. E, GPR120 in HepG2 cells was down-regulated by siRNA (#1 and #3), and cells were treated with 100 μM HNE. Lysosomes were stained with LysoTracker 6 hours after the addition of HNE. Flow cytometry was used in the analysis, and results are shown in the histogram. NC means HNE-treated HepG2 cells without the down-regulation of GPR120. F, Flow cytometry was used in the analysis, and results are shown in the mean fluorescence intensity (MFI) of LysoTracker. G, A Western blotting analysis of activated μ-calpain in HepG2 cells is shown. H, Bands of panel G are quantified and shown as relative fold ratios. I, The relative fold expressions by real-time PCR indicates that μ-calpain in HepG2 cell is suppressed by siRNA (#4 and #5). J, μ-calpain in HepG2 cells was down-regulated by siRNA (#4 and #5) and cells were treated with 100 μM HNE. Lysosomes were stained with LysoTracker, and changes in the cell, and lysosomes were observed using time-lapse imaging. Blue, Hoechst; red, LysoTracker. Bright field images are shown in the square on the bottom right corner of each image. In the negative control without μ-calpain down-regulation (NC), lysosomes were no longer present, and HepG2 cells died 5 hours after the HNE treatment. In contrast, lysosomes were retained in cells in which μ-calpain was down-regulated by siRNA (#4 and #5), and many cells survived even after the HNE treatment. K, Viability of HNE-treated HepG2 cells with (#4 and #5) and without (NC) the down-regulation of μ-calpain by siRNA. The graph shows results of cell viability obtained 12 hours after the treatment with 40 μM HNE. L, A Western blotting analysis of activated μ-calpain in HepG2 cells with (#1-#3) and without (NC) the down-regulation of GPR120 by siRNA is shown. M, Bands of panel L are quantified and shown as relative fold ratios. NC, HNE-treated HepG2 cells without the down-regulation of μ-calpain.
GPR120 expression was suppressed by small interfering ribonucleic acid (siRNA) (Figure 4, B), and cell viability and changes in lysosomes were examined after the addition of HNE to HepG2 cells. When the expression of GPR120 was suppressed, cell viability improved after the addition of HNE (Figure 4, C). Based on LysoTracker immunofluorescence staining, the suppression of GPR120 did not significantly increase the size of lysosomes after the addition of HNE and resulted in protection of lysosomal disruptions (Figure 4, D–F). These results indicate that HNE primarily functions via GPR120 in HepG2 cells and is associated with lysosomal disruption and cell death.
Addition of HNE to HepG2 cells activates μ-calpain (Figure 4, G–H). Activated calpain was previously shown to disrupt the stability of the lysosomal membrane.13,21, 22, 23, 24, 25, 26 Therefore, the morphology of lysosomes and cell viability after the addition of HNE under the suppression of μ-calpain expression by siRNA (Figure 4, I) was examined. In the negative control with active μ-calpain expression, the number of lysosomes decreased after the HNE treatment (Figure 4, J). In contrast, it was maintained when the expression of μ-calpain was suppressed by siRNA (Figure 4, J), and cell viability improved (Figure 4, K). In addition, the addition of HNE to HepG2, in which GPR120 expression was suppressed (#1 and #3), did not activate μ-calpain (Figure 4, L–M). Three siRNAs (#1-#3) were used for GPR120 suppression, but #2 siRNA did not function well in suppressing GPR120 expression (data not shown). Inhibition of GPR120 with #2 siRNA did not result in lysosome disruption or suppression of cell viability (data not shown), which was considered to be attributed to insufficient calpain inhibition (Figure 4, L). These results suggest that the effects of HNE on lysosomal membrane rupture are regulated by activated μ-calpain via GPR120.
Lysosomal Membrane Rupture in Patients With NASH
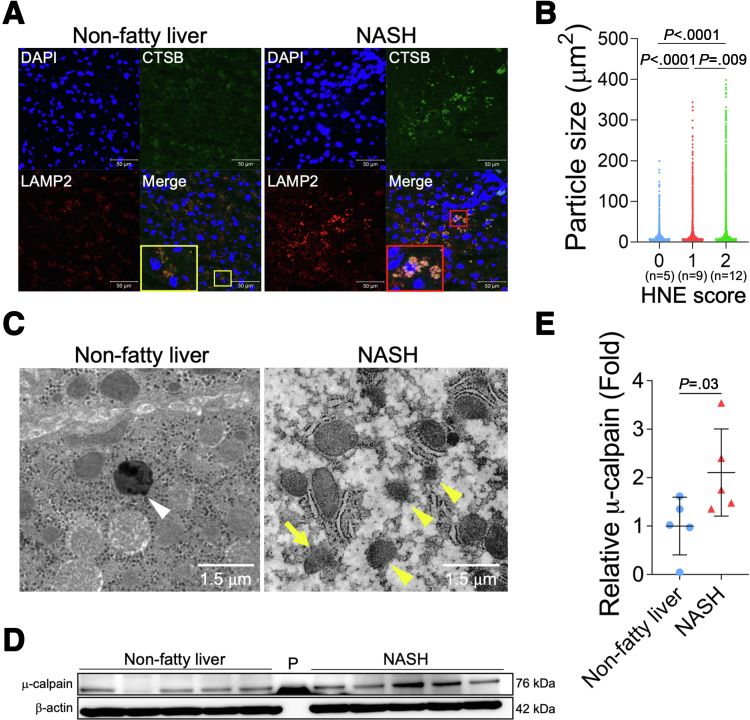
Liver tissues from patients with NASH were used to investigate the relationship between the presence of lysosomal disintegrity and HNE deposition. The double staining of CTSB and LAMP2 in control (nonfatty) livers showed that LAMP2 co-localized with small foci of CTSB in the cytoplasm of hepatocytes (Figure 5, A). On the other hand, in NASH livers, the merged color of CTSB and LAMP2 immunoreactivity expanded as coarse granules in the cytoplasm of hepatocytes. Increases in the stained area of LAMP2 indicated the collapse of the lysosomal limiting membrane, whereas those in the stained area of CTSB reflected its leakage from the lysosome into the cytoplasm; both were considered to indicate the permeabilization/disruption of the lysosomal limiting membrane. To investigate the relationship between HNE deposition and lysosomal disintegrity, the size of granules being stained by the LAMP2 antibody and the HNE staining score were both examined in 26 liver samples randomly selected from 90 patients with NAFLD (Table 2). In comparisons with liver tissue showing negligible HNE deposition, the areas stained by LAMP2 were larger in livers with significant HNE deposition (Figure 5, B).
Figure 5.
Lysosomal membrane permeabilization/rupture in hepatocytes of patients with NASH.A, Immunofluorescence staining of liver tissue from patients with nonfatty liver disease and NASH. Blue, DAPI; green, CTSB; red, LAMP2. The area highlighted in the yellow square is a magnified image of the nonfatty liver, whereas the red square is a magnified image of the NASH liver. B, Relationship between the HNE staining score (patient numbers; grade 0, n = 5; grade 1, n = 9; grade 2, n = 12) and granule sizes for LAMP2. The area of particles stained with LAMP2 in 20× image was calculated, respectively. C, Electron microscopy images of the non-fatty liver and NASH liver. Lysosomes with clear limiting membrane structures were observed in the nonfatty liver (white arrowheads). In contrast, lysosomes in the NASH liver showed disintegrity (yellow arrowheads). Furthermore, in some lysosomes, the lysosomal membrane was disrupted, and contents leaked out (yellow arrow). D, A Western blotting analysis of μ-calpain in the livers of 5 patients with nonfatty liver (normal) and 5 patients with NASH is shown. P, protein marker. E, Bands of panel D are quantified and shown as relative fold ratios.
Table 2.
Profiles of Patients Analyzed Showing a Relationship Between HNE Deposition and Lysosomal Disruption
| Variable | HNE0 (n = 5) | HNE1 (n = 9) | HNE2 (n = 12) |
|---|---|---|---|
| Sex (M/F) | 0/5 | 5/4 | 4/8 |
| Age, y | 53.0 ± 5.8 | 46.0 ± 5.5 | 46.0 ± 4.4 |
| Height, cm | 157.0 ± 2.6 | 162.4 ± 2.8 | 155.0 ± 3.3 |
| Weight, kg | 61.0 ± 5.5 | 69.9 ± 8.6 | 71.4 ± 7.3 |
| BMI, kg/m2 | 25.7 ± 1.8 | 26.9 ± 2.6 | 31.5 ± 2.2 |
| AST, IU/L | 21.0 ± 4.6 | 24.0 ± 2.7 | 49.0 ± 7.1a,b |
| ALT, IU/L | 23.0 ± 12.8 | 37.0 ± 8.5 | 77.0 ± 14.0 |
| Platelet count, ×104/mm3 | 18.0 ± 0.8 | 24.2 ± 2.2a | 22.3 ± 1.8 |
| Total protein, g/dL | 7.2 ± 0.2 | 6.8 ± 0.2 | 7.3 ± 0.1 |
| Albumin, g/dL | 4.4 ± 0.1 | 4.3 ± 0.2 | 4.4 ± 0.2 |
| PT, % | 96.0 ± 8.3 | 116.0 ± 4.9 | 92.0 ± 3.7 |
| HbA1c, % | 6.9 ± 0.7 | 5.9 ± 0.6 | 7.1 ± 0.7 |
| HOMA-IR | 2.1 ± 0.2 | 2.6 ± 1.4 | 6.9 ± 1.6 |
| Total cholesterol, mg/dL | 228.0 ± 7.9 | 192.0 ± 13.8 | 209.5 ± 9.6 |
| Triglyceride, mg/dL | 90.0 ± 14.2 | 156.0 ± 24.6 | 130.5 ± 36.3 |
| HDL cholesterol, mg/dL | 56.0 ± 1.2 | 41.0 ± 2.9 | 38.0 ± 5.5 |
| LDL cholesterol, mg/dL | 155.0 ± 7.9 | 111.2 ± 10.5a | 115.0 ± 7.7 |
| Histopathological findings | |||
| Fibrosis (0/1/2/3/4) | 1/3/0/1/0 | 3/5/1/0/0 | 0/3/4/3/2b |
| Steatosis (0/1/2/3) | 0/3/1/1 | 0/4/3/2 | 0/2/3/7 |
| Lobular inflammation (0/1/2/3) | 1/2/2/0 | 1/6/2/0 | 0/1/8/3b |
| Hepatocellular ballooning (0/1/2) | 3/2/0 | 9/0/0 | 0/5/7a,b |
| NAS score (0/1/2/3/4/5/6/7/8) | 0/1/2/0/0/1/1/0/0 | 0/1/2/3/3/0/0/0/0 | 0/0/0/0/0/1/8/3/0a,b |
Note: Data are expressed as number or median ± standard error of the median.
AST, Aspartate aminotransferase; ALT, alanine aminotransferase; PT, prothrombin time; HbA1c, hemoglobin A1c; HDL, high-density lipoprotein; HNE, 4-Hydroxy-2-nonenal; HOMA-IR, homeostasis model assessment of insulin resistance [fasting serum insulin (μU/mL) × fasting plasma glucose (mg/dL)/405]; LDL, low-density lipoprotein; NAFLD, nonalcoholic fatty liver disease; NAS, NAFLD activity score.
P < .05 vs the HNE0 group.
P < .05 vs the HNE1 group.
Lysosomes were observed in nonfatty and NASH livers by electron microscopy, and the disruption of the lysosomal limiting membrane was confirmed in the hepatocytes of NASH, but not nonfatty livers, which contained membrane-bound lysosomes (Figure 5, C). In addition, the expression of activated μ-calpain was significantly elevated in NASH livers compared with patients with nonfatty liver (Figure 5, D–E).
Lysosomal Membrane Damage in vivo by HNE
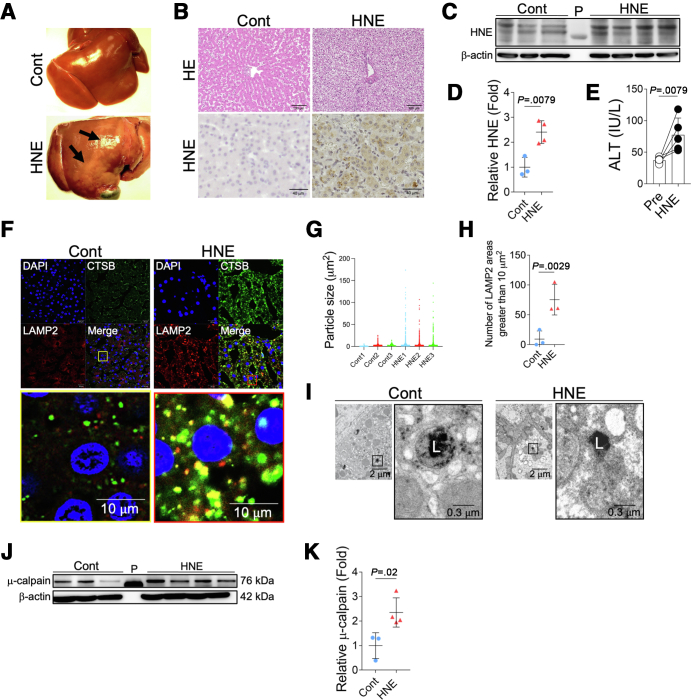
Based on similarities in GPR expression with humans, the non-human primate model (Japanese macaque monkey) was used to assess the degree of HNE-induced lysosomal disintegrity and resultant hepatocyte death in vivo. After intravenous injections of 5 mg of synthetic HNE once a week for 24 weeks, liver tissue was collected. In comparisons with control monkeys without HNE injections, macroscopic observations revealed a marked color change on the liver surface in monkeys treated with HNE (Figure 6, A). The marked degeneration of hepatocytes with the disruption of the cytoplasm was detected (Figure 6, B). HNE immunoreactivity also increased in HNE-injected monkeys (Figure 6, B). A Western blotting analysis showed that HNE adducts in liver tissue were significantly increased in HNE-treated monkeys over control monkeys (Figure 6, C–D). A blood analysis showed that the administration of HNE significantly increased alanine aminotransferase (ALT) levels (Figure 6, E). Immunofluorescence double staining of CTSB and LAMP2 revealed that the area of the merged color in lysosomes, which indicated the leakage of lysosomal contents, was larger in HNE monkeys than in the control (Figure 6, F). The staining area of LAMP2 was significantly increased in HNE-treated monkeys compared with control monkeys, suggesting lysosomal disruption (Figure 6, G–H). Electron microscopic observations confirmed that the lysosomal limiting membrane was disrupted with a loss of vivid lysosomes in the HNE group but remained intact in the control group (Figure 6, I). This was consistent with the results obtained from cultured cells (Figure 3, B) and patients with NASH (Figure 5, C). A Western blotting analysis showed that μ-calpain activation was significantly stronger in livers after HNE injections than in the control group (Figure 6, J–K). These results indicate that HNE leads to lysosome disruption in vivo, which may involve the activation of μ-calpain.
Figure 6.
HNE induces lysosomal disintegrity by activating μ-calpain in hepatocytes of Japanese macaque monkeys.A, Macroscopic findings of livers in the control (Cont) and in monkeys treated with HNE (HNE). Black arrows show regional discoloration. B, Hematoxylin and eosin staining and HNE immunostaining of liver tissue from the control group (Cont) and HNE-treated group (HNE). HNE immunoreactivity was observed in hepatocytes. C, The expression of liver HNE protein adducts in the control (Cont) and in monkeys treated with HNE (HNE) was evaluated by a Western blotting analysis. In the Western blotting analysis, HNE is depicted as HNE protein adducts of various molecular weights. P, protein marker. D, Bands of panel C are quantified and shown as relative fold ratios. E, Alterations in ALT levels before the HNE treatment and increases after the HNE treatment are shown. F, Comparison of immunofluorescence staining in the control group (Cont) and HNE-treated group (HNE). Blue, DAPI; green, CTSB; red, LAMP2; yellow, merge. The yellow square shows a magnified image of the liver in the control group, whereas the red square shows a magnified image of the liver in the HNE-treated group. G, The area of particles stained with LAMP2 is shown in 20× image for each monkey. H, The number of stained areas that were 10 μm2 or larger in panel G is shown. I, Electron microscopy images of livers in the control (Cont) and HNE-treated groups (HNE). Black squares show magnified images of membrane-bound lysosomes in the control (Cont) and disruption of the lysosomal limiting membrane in the HNE-treated group (HNE). L, lysosome. J, A Western blotting analysis showing the expression of activated μ-calpain in the livers of the control (Cont) and HNE-treated groups (HNE). P, protein marker. K, Bands of panel J are quantified and shown as relative fold ratios.
Alda-1 Improved Liver Inflammation and Fibrosis in Choline-deficient, Amino-acid Defined (CDAA) mice Model
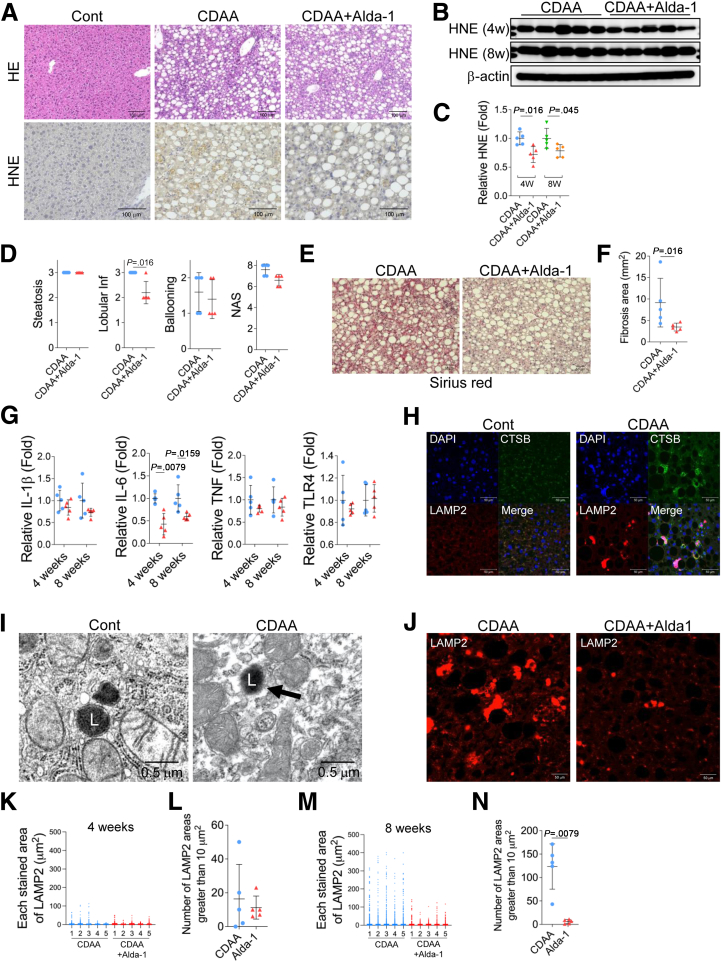
Finally, we assessed whether detoxification of HNE, the beginning of a series of cascades, would inhibit liver inflammation or fibrosis resulting from hepatocyte death. Here we used Alda-1, which resulted in inhibition of hepatocyte death by HNE in vitro (Figure 2, H and P). CDAA mice models were treated with 20 mg/kg Alda-1 intraperitoneally 3 times a week, and the histological images of the liver were compared after 8 weeks; HNE deposition was observed in CDAA mice but was reduced in Alda-1-treated CDAA mice (Figure 7, A). A Western blotting analysis showed that liver HNE protein adducts were significantly reduced in CDAA mice with Alda-1 treatment compared with CDAA mice (Figure 7, B–C). Lobular inflammation after 4 or 8 weeks of CDAA mice and liver fibrosis formed after 8 weeks of CDAA mice was ameliorated by intraperitoneal administration of Alda-1 (Figure 7, D–F). Using real-time polymerase chain reaction in mice liver tissue, the inflammation-related cytokines interleukin (IL)-6, tumor necrosis factor, and IL-1β were measured, and IL-6 was significantly lower in Alda-1-treated 4 or 8 weeks CDAA mice (Figure 7, G). There was no difference in toll-like receptor-4 expression (Figure 7, G).
Figure 7.
Administration of Alda-1 in CDAA mice suppresses liver fibrosis.A, Hematoxylin and eosin staining and HNE immunostaining of liver tissue from the control mice fed the standard diet (Cont), CDAA mice (CDAA) and Alda-1-treated CDAA mice (CDAA+Alda-1) for 8 weeks. B, The expressions of liver HNE protein adducts in CDAA mice (CDAA) and CDAA mice with Alda-1 treatment (CDAA+Alda-1) for 4 (4w) and 8 (8w) weeks were evaluated by a Western blotting analysis. C, Bands of panel B are quantified and shown as relative fold ratios. D, Comparison of histopathological scores of CDAA mice (CDAA) and Alda-1 treated CDAA mice (CDAA+Alda-1) for 8 weeks, such as steatosis score, lobular inflammation score (Lobular inf), ballooning score, and NAFLD activity score (NAS) is shown. E, Sirius red staining of liver tissue from CDAA mice (CDAA) and Alda-1-treated CDAA mice (CDAA+Alda-1) for 8 weeks. F, Area occupied by Sirius red staining in a 20× image were compared between the CDAA mice (CDAA) and Alda-1 treated CDAA mice (CDAA+Alda-1) for 8 weeks. The area was calculated for each of the 10 images and averaged. G, Real-time PCR analysis of IL-1β, IL-6, tumor necrosis factor (TNF), and toll-like receptor (TRL)-4 expression in CDAA mice (blue circle) and Alda-1 treated CDAA mice (red triangle) for 4 and 8 weeks. H, Immunofluorescence staining of livers in control mice (Cont) and the CDAA mice (CDAA). Blue, DAPI; green, CTSB; red, LAMP2. I, Electron microscopy images of liver tissue collected from control mice (Cont) and CDAA mice (CDAA) for 8 weeks. The black arrow shows the disruption of the lysosomal limiting membrane. L, lysosome. J, Immunofluorescence LAMP2 staining of liver tissue from CDAA mice (CDAA) and Alda-1-treated CDAA mice (CDAA+Alda-1) for 8 weeks. K, The area of particles stained with LAMP2 in CDAA mice (CDAA) and Alda-1 treated CDAA mice (CDAA+Alda-1) for 4 weeks is shown. L, The number of stained areas that were 10 μm2 or larger in panel K is shown. M, The area of particles stained with LAMP2 in CDAA mice (CDAA) and Alda-1 treated CDAA mice (CDAA+Alda-1) for 8 weeks is shown. N, The number of stained areas that were 10 μm2 or larger in panel M is shown.
In comparisons with the control group, hepatocytes in 8-week CDAA mice livers were characterized by enlarged lysosomes, and disruption of the lysosomal membrane resulted in the leakage of CTSB and scattering of LAMP2 into the cytoplasm (Figure 7, H). Disruption of the lysosomal limiting membrane in the hepatocytes of CDAA mice was also confirmed by electron microscopy (Figure 7, I). Immunofluorescence staining showed that the area of LAMP2 was decreased by Alda-1 treatment (Figure 7, J). The staining area of LAMP2 was calculated and the number whose area was greater than 10 μm2 was compared. At 4 weeks, there was no difference in the number of LAMP2 staining areas greater than 10 μm2 in CDAA mice and Alda-1-treated CDAA mice (Figure 7, K–L). However, at 8 weeks, the number of LAMP2 stained areas that were greater than 10 μm2, which increases in CDAA mice, was significantly suppressed by treatment with Alda-1 (Figure 7, M–N). These results suggested that Alda-1 treatment reduced permeabilization/disruption of the lysosomal limiting membrane.
Discussion
Lipid accumulation and oxidative stress due to dysregulated lipid metabolism have been implicated in the pathogenesis of NASH. The composition of fatty acids is altered in NASH livers, with an increase in the accumulation of fatty acids such as linoleic acid.27,28 HNE is an α, β-unsaturated aldehyde that is generated from highly unsaturated fatty acids, such as linoleic acid, and is one of the most cytotoxic of all aldehydes.4,9,29 HNE exerts dose-dependent effects on cellular homeostasis: low concentrations increase the susceptibility of proteins to proteolysis and removal by the proteasomal system. Under normal conditions, this system removes the majority of oxidatively damaged and modified proteins. However, under severe oxidative stress, the accumulation of damaged/modified proteins by high concentrations of HNE induces protein cross-linking, malfunctions in the proteolytic machinery, and failed autophagy.30, 31, 32, 33, 34, 35, 36
It is difficult to measure the reliable concentration of HNE in vivo, because of rapid metabolism, its efflux and its steady-state concentration in specific tissues.37,38 But its concentration is considered to increase substantially in the “local environment” under pathological conditions to over 100 μM,38 and hepatocytes are no exception, being markedly increased “locally” under oxidative stress.39 Cell death, whether apoptotic40 or non-apoptotic,41 is reported to occur sporadically in the liver of NAFLD. In the context of diseases associated with chronic hepatitis, such as NAFLD, we consider that various factors increase HNE “locally” in hepatocytes, leading to different cellular disorders including cell death. As shown in Table 3, the HNE concentrations used in previous articles ranged from 5 to 100 μM,42, 43, 44, 45, 46, 47, 48, 49 with varying cell concentration and incubation times, and did not clearly differ from the HNE concentrations we used (25–100 μM). These facts support that the HNE concentrations used in our in vitro experiments are within the appropriate concentration range.
Table 3.
Concentrations of HNE Used in Previous Articles for HepG2 Cells
| Plate | Cell concentration | HNE concentration | Incubation time | Reference |
|---|---|---|---|---|
| 25-cm2 plate | 2×105 cells/cm2 | 2–50 μM | 24 hours | Muzio et al.42 |
| 96-well | – | 5–20 μM | 1, 2, 4, 8, 24 hours | Gallagher et al.43 |
| 6-well | 5 × 106 cells/well | 20–100 μM | 15, 30, 60 minutes | Stewart et al.44 |
| – | 1 × 107 cells/well | 100 μM | 1 hour | Shearn et al.45 |
| – | 1 × 106–1 × 107 cells/well | 100 μM | 1 hour | Shearn et al.46 |
| 12-well | 5 × 105 cells/well | 12.5–100 μM | 16 hours | Shearn et al.47 |
| – | – | 40 μM | 24 hours | Chaudhary et al.48 |
| 96-well | 2 × 103 cells/well | 70 μM | 24 hours | Zhang et al.49 |
HNE, 4-Hydroxy-2-nonenal.
Dash indicates that it was not described.
Lysosomes are membrane-bound organelles that mediate the degradation and recycling of damaged/aged/misfolded proteins. Maintaining lysosomal membrane integrity is crucial for the cell homeostasis and survival.50,51 The present study using in vitro and in vivo experimental paradigms and the liver tissue of patients with NASH demonstrated that HNE induced calpain activation via GPR120, resulting in lysosomal membrane permeabilization/rupture and cell death.
It is well-known that GPR120 generally reacts with fatty acids with 14−16 carbons. However, this receptor protein is also known to react with ligands of various chemical structures,52 especially conjugated linoleic acid, even if the number of carbons is small (C9 or C10).53 Therefore, it is quite possible that HNE with 9 carbons may affect hepatocytes via GPR120. In this study, we showed that suppression of GPR120 by siRNA suppressed HNE-induced μ-calpain activation, lysosomal damage, and cell death. Furthermore, calpain activation has been reported to lead to the cleavage of heat shock protein 70.1 (Hsp70.1),13,21, 22, 23 LAMP2,24,25 and v-ATPase subunit b2,26 which are associated with lysosomal limiting membrane integrity under physiological conditions. Thus, we hypothesized that the GPR120-mediated activation of calpain may contribute to the generation of lysosomal membrane rupture/permeabilization by cleaving proteins involved in stabilizing the lysosomal membrane.
We previously reported that Hsp70.1, particularly after HNE-induced oxidation (carbonylation), was susceptible to calpain-mediated cleavage, which disrupted the stability of the lysosomal limiting membrane of neurons. This induced the leakage of cathepsins, which damage cell constitutive proteins, membranes, and organelles, resulting in neuronal death.13,21, 22, 23 This is reasonable based on the similar interaction between this cascade and lysosomal rupture was confirmed using Caenorhabditis elegans neurodegeneration models, in which the loss of function of the proteases CLP-1 and TRA-3 (equivalent to calpains in C. elegans) as well as ASP-3 and ASP-4 (equivalent to cathepsins) was neuroprotective.54 The calpain-mediated cleavage of Hsp70.1 from low-species animals to primates is physiologically indispensable for the turnover of a chaperone protein itself, but is detrimental to cell survival when excessive.13,21, 22, 23
Several mechanisms of lysosomal membrane destabilization in NAFLD hepatocytes are known. Free fatty acids transfer cytoplasmic Bax, which induces channel formation,55 to lysosomes, which may increase the permeability of lysosomal membranes.56 In addition, the activity of acidic sphingomyelinase, which is involved in lysosomal metabolism, regulation of membrane structure, and signal transduction,57 leads to lysosomal membrane instability.58 Lysosomal destabilization based on the “calpain-cathepsin hypothesis”13, 22 involving calpain activation by HNE demonstrated in this study adds new insights into the understanding of the pathophysiology of NAFLD.
The toxicity of HNE suggests that its detoxification may be a new target for therapeutic intervention in NAFLD. Enzymes involved in the degradation of HNE are glutathione-S-transferase,59 alcohol dehydrogenase, and ALDH.60 ALDH2 has been reported to be the most effective in detoxifying HNE compared with other ALDH isoforms.61 In addition, inactive ALDH2∗2 allele may potentially be a risk factor for NAFLD.62 Therefore, we focused on Alda-1, which activates wild-type ALDH2 and recovers mutant ALDH2⁄2 to near-wild-type activity.17 In vitro experiments showed that Alda-1 suppressed HNE-induced cell death, and in vivo experiments showed that intraperitoneal administration of Alda-1 to CDAA-mice suppressed liver inflammation and fibrosis. These results are further supported by the other liver injury models shown below. Increasing the activity of ALDH-2 by Alda-1 in hepatic ischemia/reperfusion injury, which increases the production of HNE in hepatocytes, eliminated the accumulation of HNE and suppressed cell death.63 In addition, in alcohol intoxicated mice where HNE is deposited in hepatocytes, treatment of Alda-1 reduced hepatic HNE levels, restored steatosis, and suppressed cell death.64
In obesity and NAFLD, autophagy in hepatocyte is reported to be suppressed65 due to excess triglycerides66 and free fatty acids.67 One of the mechanism of the dysfunction of autophagy, which plays an important role in liver metabolism,68 may be the reduction of autophagy clearance.69 Since lysosomes are important factors in autophagy flux, the disruption of lysosomes in hepatocytes by HNE shown in this study may be a phenomenon that explains not only cell death due to CTSB leakage, but also an unknown mechanism of autophagic impairment in NAFLD.
There are several limitations to this study. First, we were unable to clarify whether the main source of HNE production involved in the pathogenesis of NASH is intracellular (eg, oxidative stress) or extracellular (eg, diet). Second, the mechanisms underlining hepatocyte death in NASH by HNE remain unclear. A variety of cell deaths have been reported to be involved in the pathogenesis of NASH, including apoptosis,40 necrosis,70 necroptosis,71 pyroptosis,72 and ferroptosis.41 In this study, we found that HNE leads to cell death that is morphologically distinct from apoptosis, although it results in activation of caspase-3. We surmise that both the rupture of lysosomes resulting from HNE-induced calpain activation and the subsequent leakage of cathepsin result in cell death. Lysosomal rupture and cathepsin release are involved in the activation of effectors, such as ROS, Bax, and iron, causing various types of cell death, including apoptosis, pyroptosis, ferroptosis, and necrosis.73 Elucidating the detailed mechanisms of HNE-induced hepatocyte death is an important topic for further investigation. Third, we were unable to clarify what the actual concentration of HNE in cells is sufficient to cause cell death. This is because there is no technology to measure the concentration of HNE in individual cells.
We herein demonstrated for the first time that HNE-induced hepatocyte death due to lysosomal membrane permeabilization/rupture via the calpain activation. The present results provide novel insights into the mechanisms responsible for hepatocyte death in NASH and will contribute to the development of new therapeutic strategies for NASH.
Materials and Methods
Human Samples
Liver tissue samples were collected by ultrasound-guided subcutaneous biopsy from 103 patients, including 13 patients with nonfatty liver and 90 patients with NAFLD (Table 1). Forty-nine of the 90 patients with NAFLD met the criteria for NASH defined by a NAFLD activity score of 5 or more. None of the study participants tested positive for the hepatitis B surface antigen or hepatitis C virus antibody or had any other chronic liver diseases. All participants had a daily alcohol intake of less than 20 g and no long-term history of steatogenic medication. Pathological examinations were performed independently by 2 pathologists who scored tissue samples based on Matteoni’s classification,74 the NAFLD activity score (steatosis, lobular inflammation, and hepatocellular ballooning),75 and fibrosis score. Written informed consent was obtained from all participants according to the Declaration of Helsinki, and the study was approved by the regional Ethics Committee (Medical Ethics Committee of Kanazawa University, No. 2418).
Fasting blood tests were performed prior to liver biopsy. Tissues collected by biopsy were submerged in OCT compound and stored at −80 °C. Some of the tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, and stored at room temperature.
The reasons for performing liver biopsy in 13 patients with nonfatty liver were a liver transplant donor preoperative examination (n = 6), an examination for elevated gamma-glutamyl transferase (n = 2), a background liver evaluation at liver tumor biopsy (n = 4), and an examination for slightly elevated ALT (n = 1).
Mouse Model
Male wild-type C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME). CDAA-diet mice were prepared according to a previously published protocol.76 After weaning at 8 weeks, mice were randomly assigned to be in the control or CDAA-diet group and were kept 8 weeks. Control mice were fed a commercial standard diet for 8 weeks, whereas CDAA-diet mice were fed a choline-deficient, L-amino acid-defined, high-fat diet with 0.1% methionine (CDAHFD; A0671302, Research Diets, New Brunswick, NJ) for 8 weeks. For Alda-1 administration, another random assignment was made. For the administration of Alda-1, the mice were randomly divided into 3 groups and kept 8 weeks: control, CDAA-diet, or CDAA-diet with Alda-1, for 8 weeks. The control group and the CDAA-diet group were injected intraperitoneally with a solvent solution of Alda-1, and the CDAA-diet with Alda-1 group was injected intraperitoneally with Alda-1 3 times a week.
Monkey Model
After the referee of animal experimentation about the ethical or animal welfare, nine young (4–5 years: comparable with human teenagers) female Japanese macaque monkeys (Macaca fuscata) were supplied by the National Bio-Resource Project “Japanese monkey.” Because females were previously reported to be more resistant to NASH than males because estrogen is protective in mice,77 we intentionally only used female monkeys. Japanese macaque monkeys are characterized by an average life span of 25 to 35 years and gene sequence homology of approximately 94% with humans.
Monkeys were reared in a wide cage with autofeeding and autodrainage machines as well as appropriate toys to play with for at least for 1 year to facilitate acclimation. Room temperature was maintained at 22 °C to 24 °C with a humidity of 40% to 50%. They were fed approximately 100 g × 2/day of a non-purified diet (CLEA Old World Monkey Diet CMK-2 containing 344.7 kcal/100 g, but only 4.05% crude fat, CLEA Japan, Inc, Tokyo, Japan) for 1 year to facilitate acclimation. Apples, pumpkins, or sweet potatoes were given twice every week. In the morning and afternoon, animal care staff and the first 2 authors monitored the health and well-being of the animals to check the consumption of foods, the pupilar reflex to the light, and the conditions of standing and jumping.
At 5 to 6 years of age, monkeys were randomly divided into sham-operated controls (n = 4) and those undergoing HNE injections (n = 5). In 5 monkeys, 5 mg/week of synthetic hydroxynonenal (Cayman Chemical, Ann Arbor, MI) was intravenously injected every week for 24 weeks. In our prior experiments, monkeys treated with intravenous injections of 1 or 2 mg of HNE once a week for 6 months showed no significant differences from controls in bloods tests and histological evaluations of multiple organs. By contrast, monkeys treated with intravenous injections of 5 mg of HNE once a week for 6 months showed similar pathology to cultured cells and mouse and human samples. When 5 mg of HNE is intravenously administered to a monkey, the serum HNE concentration is 60 μM, as calculated from the blood volume converted from the monkey’s body weight. On the basis of a report that the human serum concentration of HNE in a certain disease is 20 μM13,78 and the fact that HNE is rapidly metabolized, we can assume that the concentration of HNE in monkey serum in our experimental system was not too far from the pathological state in humans. In each monkey, venous blood was collected every month for 6 months from the lower leg vein and ALT levels were measured. All experimental procedures were strictly in accordance with the guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health, which met the ‘International Guiding Principles for Biomedical Research Involving Animals,’ as issued by the council for the International Organizations of Medical Sciences. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Kanazawa University Graduate School of Medical Sciences (Protocol Number: AP-153613). The samples available for this experiment were 5 of HNE-treated monkeys for blood analysis, 3 of controls, and 4 of HNE-treated monkeys for Western blotting analysis, and 3 of controls and HNE-treated monkeys for fluorescent staining.
Cell Culture
Human hepatoma cell lines (HepG2 and Huh-7) were maintained in Dulbecco’s Modified Eagle Medium (Nacalai Tesque, Kyoto, Japan) containing 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% L-glutamine at 37 °C in a humidified 5% CO2 incubator. We seeded 2 × 105 cells/2 mL of medium in each well of a 6-well plate, 2 × 103 cells/100 μL of medium in each well of a 96-well plate, and 1 × 104 cells/500 μL of medium in each well of a 4- or 8-chamber slide. Cells were seeded and incubated overnight before performing each assay.
Cell Viability Assay
Cell viability was assayed using the Cell Counting Kit-8 kit (CK04; Dojindo Co, Ltd, Kumamoto, Kumamoto, Japan). Briefly, cells (2 × 104/mL) were seeded on 96-well plates at 100 μL/well. After the HNE treatment, 10 μL of Cell Counting Kit-8 reagent was added to each well and a blank control well was established. After an incubation at 37 °C in a humidified 5% CO2 incubator for 2 hours, absorbance at 450 nm was measured and that at 620 nm was used as the reference. The absorbance of each well was defined as the difference in absorbance between 450 and 650 nm. Regarding data normalization, the absorbance of control wells without HNE was set to ‘100’ and other wells were normalized to control wells. Measurements were taken from 5 wells for the control and test groups, and the experiment was repeated at least 3 times.
HNE Treatment
HNE was stored in 99% ethanol. Prior to use, the required amount of HNE was prepared by evaporating ethanol under a gradual nitrogen flow and subsequently dissolved in phosphate buffered saline (PBS) before its addition to the cell culture medium.
Alda-1 Treatment
Alda-1 (SML0462-5MG, Sigma-Aldrich, St Louis, MO) was dissolved in dimethyl sulfoxide. Assays on culture cells were performed such that the final concentration of Alda-1 did not exceed 25 μM. Cells were incubated with Alda-1 for 1 hour prior to experiments. For CDAA-diet mice, Alda-1 was dissolved in olive oil and administered intraperitoneally at 20 mg/g 3 times a week for 8 weeks.
Flow Cytometry Analysis
Dead cells were stained with Annexin V and ethidium homodimer III contained in the Apoptotic/Necrotic/Healthy Cells Detection Kit (PK-CA707-30018, PromoCell GmbH, Heidelberg, Germany). Cells were seeded on 6-well plates and treated with HNE. After cells were collected by trypsinization, they were centrifuged at 1000 rpm for 10 minutes and then dissolved in 100 μL of the buffer and 5 μL of each antibody. Cells were incubated for 15 minutes in the dark, and flow cytometry was performed with BD FACSCalibur (BD Biosciences, Franklin Lakes, NJ) using the FL1 channel for Annexin V and FL4 channel for ethidium homodimer III to measure mean fluorescence intensity.
The dynamics of lysosomes were analyzed by staining lysosomes with LysoTracker Red DND-99 (L7528, Thermo Fisher Scientific, Waltham, MA). Cells were treated with HNE and incubated with LysoTracker at 37°C for 1 hour in the dark. Cells were subsequently trypsinized in the dark and analyzed using the FL2 channel on BD FACSCalibur (BD Biosciences, Franklin Lakes, NJ) to measure mean fluorescence intensity.
Time-lapse Imaging
Cells were seeded on 8-chamber slides, stained, and subsequently treated with either 3 μM EPI or 100 μM HNE. Time-lapse imaging was performed using a confocal quantitative image cytometer and a live cell imaging microscope. Dead cells were stained with propidium iodide (PI) (P4864-10ML, Sigma-Aldrich, St. Louis, MO), lysosomes were stained with LysoTracker, cell nuclei were stained with Hoechst 33342 (H3570, Thermo Fisher Scientific, Waltham, MA), and activated caspase 3 was stained with Nuc ViewTM 488 Caspase-3 Substrate (10403; Biotium, Inc, Fremont, CA). Cells stained with propidium iodide and LysoTracker were imaged every 5 minutes, and cells stained with Nuc ViewTM 488 Caspase-3 Substrate were imaged every 7 minutes. Cells were incubated with LysoTracker at 37°C for 1 hour and with Hoechst and Nuc ViewTM 488 Caspase-3 Substrate for 30 minutes for staining.
SiRNA Transfection
siRNA was used to knockdown the following genes: G-protein-coupled receptor (GPR)120 (#1; FFAR4HSS139006, sequence UCACAUUUGCUAAUUCAGCCCUAAA, #2; FFAR4HSS139007, sequence UGACUUGUCGAUUAUUUCUGGCUAA, #3; FFAR4HSS139008, sequence GGAAAUUUCGAUUUGCACACUGAUU) and μ-calpain (#4; CAPN1HSS101345, sequence CCGUACACUUGAAGCGUGACUUCUU, #5; CAPN1HSS188701, sequence CAGAGUGGAACAACGUGGACCCAUA, #6; CAPN1HSS188702, sequence GCGGUCGACUUUGAUAAUUUCGUUU). Stealth RNAiTM siRNA Negative Control Lo GC (12935200; Thermo Fisher Scientific, Waltham, MA) was used as a control.
All materials were purchased from Thermo Fisher Scientific. Cells were seeded on 6-well plates, and siRNA was transfected using LipofectamineTM RNAiMAX Transfection Reagent (2167741; Thermo Fisher Scientific, Waltham, MA) and GlicoTMOpti-MEMTM I Reduced Serum Medium (31985070; Thermo Fisher Scientific, Waltham, MA) such that the final concentration of siRNA reached 100 nM. Cell viability assays were performed by seeding cells onto 96-well plates 24 hours after the transfection of siRNA, and immunostaining was performed after seeding cells on 4-chamber slides. Each assay was performed 48 hours after the transfection of siRNA.
Real-time Quantitative Reverse-Transcription PCR Analysis
The expression of GPR120, μ-calpain, IL-1β, IL-6, tumor necrosis factor, and too-like receptor-4 was analyzed by using quantitative real-time PCR, which was performed on a CFX384 machine (Bio-Rad, Hercules, CA) using SYBR Green Master Mix (Applied Biosystems). Total RNA was extracted from cultured cells using High Pure RNA Isolation Kit (Roche Diagnostics K.K., Tokyo, Japan) according to the manufacturer's protocol. cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). Each sample was determined in triplicate and normalized relative to β-actin expression. The following probes were used: GPR120 (Hs00699184_m1), μ-calpain (Hs00559804_m1), IL-1β (Mm01336189_ml), IL-6 (Mm00446190_ml), tumor necrosis factor (Mm00443258_ml), and toll-like receptor-4 (Mm00445273).
Immunohistochemistry
Liver tissues that were fixed in formalin and embedded in paraffin were sliced into 2 μm-thick sections. Antigen activation was performed in an autoclave (121 °C, 10 minutes) using 0.01 M citric acid buffer at pH 6.0. Endogenous peroxidase was blocked using Dako REAL Peroxidase-Blocking Solution. Tissue sections were incubated with Dako Protein Block Serum-Free for 10 minutes to prevent non-specific staining. Tissue sections were subsequently stained with the primary antibody for 90 minutes and with the secondary antibody for 30 minutes. The mouse monoclonal antibody against human HNE (JaICA) was used as the primary antibody at a dilution of 250. Tissue sections were incubated with the secondary antibody against mouse immunoglobulin G (IgG) conjugated to a peroxidase-labeled polymer (Histofine Simple Stain MAX PO(M)) for 30 minutes. Color was developed with the DAB substrate kit. Tissues were then counterstained with hematoxylin.
Sirius red staining was performed according to the following procedure; liver tissue sections after initial deparaffinization and hydration were incubated in picro-sirius red for 60 minutes and subsequently washed with acetic acid. Tissue sections were then counterstained with hematoxylin for 5 minutes.
Assessment of HNE Deposition
Stained slides were observed under the All-in-One Fluorescence Microscope BZ-X800 (KEYENCE, Osaka, Japan). The extent of HNE staining was evaluated using a scoring system (0: no staining, 1: mild, and 2: strong) (Figure 1, A)
Immunofluorescence Staining
Frozen sections (3 μm thick) were incubated in 4% paraformaldehyde for 15 minutes, followed by 1% bovine serum albumin (BSA)/PBS for 1 hour to prevent nonspecific staining. Tissues were stained with primary antibodies at 4 °C overnight. After washing with PBS, tissue sections were incubated with secondary antibodies at room temperature for 30 minutes. Tissue sections were then stained with DAPI and incubated with the Autofluorescence Quenching Kit (Vector Laboratories, Burlingame, CA) at room temperature for 5 minutes to suppress autofluorescence. The following primary antibodies were used: a mouse monoclonal antibody against African green monkey LAMP2 (ab25631, Abcam, Cambridge, UK) at a dilution of 250, a rabbit monoclonal antibody against human cathepsin B (CTSB) (D1C7Y, Cell Signaling, Danvers, MA) at a dilution of 800, a rat monoclonal antibody against mouse LAMP2 (6A430, Santa Cruz Biotechnology, Dallas, TX) at a dilution of 500, and a rabbit monoclonal antibody against mouse CTSB (D1C7Y, Cell Signaling, Danvers, MA) at a dilution of 1000. The following secondary antibodies were used: mouse IgG conjugated to Alexa Fluor®594 and rabbit IgG conjugated to Alexa Fluor 488 diluted 500× in 1% BSA/PBS.
Regarding the immunofluorescence staining of cultured cells, HepG2 and Huh-7 cells were seeded on 4-chamber slides and treated with 25 μM HNE. Cells were subsequently washed with PBS and incubated with 4% paraformaldehyde for 15 minutes. Cells were washed with 0.05% PBST (PBS containing 0.05% Tween 20) and incubated with a protein block for 5 minutes. Cells were then incubated with the primary antibody at 37 °C for 1 hour, washed 3 times with 0.05% PBST, and incubated with the secondary antibody at 37 °C for 30 minutes. Chambers were removed from slides, cells were stained with DAPI, and then observed after the placement of a cover glass.
The following primary antibodies for LAMP2, CTSB, μ-calpain, and were used: a mouse monoclonal antibody against African green monkey LAMP2 (ab25631, Abcam, Cambridge, UK), a rabbit monoclonal antibody against human CTSB (D1C7Y, Cell Signaling, Danvers, MA), a rabbit antiserum anti-human activated μ-calpain antibody (order made by PEPTIDE Institute, Ibaraki, Osaka, Japan) that binds with the activated (76 kDa), not inactivated (80 kDa) form of μ-calpain. Mouse IgG conjugated to Alexa Fluor 594 (Thermo Fisher Scientific, Waltham, MA) and rabbit IgG conjugated to Alexa Fluor 488 (Thermo Fisher Scientific, Waltham, MA) were used as secondary antibodies. In staining with LysoTracker, LysoTracker was added 1 hour prior to staining with primary antibodies, and cells were stained at 37 °C for 1 hour and fixed with 4% paraformaldehyde. For the lysosomal membrane destabilization by immunofluorescence staining, stained tissue sections were observed using a confocal microscope at 400× magnification. Three regions of interest were randomly selected to image LAMP2 staining and images were processed on ImageJ to measure each red area of staining for LAMP2. The lower and upper limits of the area of staining were set to 4 and 400 μm2, respectively.
Western Blotting
Total protein was extracted using a protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO) and PhosSTOP phosphatase inhibitor cocktail tablets (Roche, Germany). Twenty micrograms of liver proteins were separated by SDS-PAGE on a SuperSep (TM) Ace 5-20% gel (Wako, Japan) at 40 mA for 1 hour. Proteins were transferred to a PVDF membrane (Millipore, Burlington, MA). Transferred proteins were detected using Ponceau S solution (Sigma-Aldrich, St Louis, MO). Membranes were blocked with 1% BSA for 1 hour and then incubated with a rabbit anti-human activated μ-calpain antibody (order made by PEPTIDE Institute, Japan) diluted 1:250, or a mouse monoclonal antibody human HNE antibody (JaICA) diluted 1:500 overnight. β-actin was utilized as an internal control (Sigma-Aldrich, St Louis, MO). Membranes were subsequently incubated for 1 hour with anti-mouse (Santa Cruz, Dallas, TX) or anti-rabbit IgG (Sigma-Aldrich, St Louis, MO) diluted 1:10000. An enhanced chemiluminescence HRP substrate detection kit (Millipore, Burlington, MA) was used to visualize reactive protein bands. The bands were quantified using ImageJ and compared by relative fold ratio.
Electron Microscopy
Samples of cultured cells and hepatocytes collected from mice, monkeys, and humans were fixed in 2.5% glutaraldehyde for 24 hours, washed with 0.1 M PBS (pH 7.4), and fixed with 1% osmium tetroxide. Samples were dehydrated with graded alcohol, immersed in propylene oxide, and embedded in epoxy resin. Thin sections were stained with toluidine blue for trimming to make ultrathin sections. Ultrathin sections were then stained with 2% uranyl acetate followed by 1% lead citrate. Stained sections were observed under a transmission electron microscope (H-7650, Hitachi, Tokyo, Japan).
Statistical Analysis
Data are expressed as the mean ± standard standard error of the median. Statistical analyses were performed with GraphPad Prism 8 (GraphPad Software, San Diego, CA). The Mann-Whitney U test was used to compare the HNE staining score between the NAFLD and control groups and to compare in vitro cell culture assay results between the 2 groups. The Kruskal-Wallis test was used to assess whether the HNE staining score was associated with liver pathology and lysosomal disintegrity. P < .05 was considered to be significant.
Acknowledgment
The authors thank Mr. Jun Uchimoto for the daily care of monkeys and autopsy assistance, Mrs. Katsumi Hara, Mrs. Nami Nishiyama, and Mrs. Masayo Baba for tissue preparation, and Mrs. Rie Nishioka and Mrs. Mai Nakayama for their secretarial work.
CRediT Authorship Contributions
Takuya Seike, MD, PhD (Conceptualization: Equal; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead).
Piyakarn Boontem, MMSc (Investigation: Lead; Methodology: Lead; Validation: Equal; Visualization: Equal; Writing – original draft: Supporting).
Hidenori Kido, MD (Investigation: Equal; Resources: Equal; Validation: Equal; Visualization: Equal; Writing – original draft: Supporting).
Masahiro Yanagi, MD (Formal analysis: Equal; Investigation: Equal; Resources: Equal; Validation: Equal; Visualization: Equal; Writing – original draft: Supporting).
Daisuke Yamamiya, MD, PhD (Investigation: Supporting; Resources: Supporting; Validation: Supporting; Visualization: Supporting).
Hidetoshi Nakagawa, MD, PhD (Visualization: Equal; Writing – original draft: Supporting; Writing – review & editing: Supporting).
Tatsuya Yamashita, MD, PhD, (Project administration: Equal; Resources: Equal; Supervision: Lead; Writing – review & editing: Supporting).
Shihui Li, MD (Investigation: Supporting; Methodology: Supporting; Validation: Supporting).
Hikari Okada, PhD (Investigation: Supporting; Methodology: Supporting; Resources: Supporting; Validation: Supporting).
Kenichi Harada, MD, PhD (Investigation: Supporting; Validation: Equal).
Mitsuru Kikuchi, MD, PhD (Investigation: Supporting). Yoshitake Shiraishi, MMSc (Investigation: Supporting; Methodology: Supporting).
Noriyuki Ozaki, MD, PhD (Investigation: Supporting).
Shuichi Kaneko MD, PhD (Conceptualization: Lead; Funding acquisition: Lead; Project administration: Lead; Supervision: Lead; Writing – review & editing: Lead).
Tetsumori Yamashima, MD, PhD (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Supporting; Methodology: Equal; Project administration: Lead; Supervision: Lead; Validation: Supporting; Visualization: Supporting; Writing – original draft: Supporting; Writing – review & editing: Lead).
Eishiro Mizukoshi MD, PhD (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Supporting; Methodology: Equal; Project administration: Lead; Supervision: Lead; Validation: Supporting; Visualization: Supporting; Writing – original draft: Supporting; Writing – review & editing: Lead).
Conflicts of interest
The authors disclose no conflicts.
Funding
This study was supported by a grant from the Mitani Foundation for Research and Development, and the Japanese Ministry of Education, Culture, Sports, Science and Technology (19H04029).
Contributor Information
Tetsumori Yamashima, Email: yamashima215@gmail.com.
Eishiro Mizukoshi, Email: eishirom@m-kanazawa.jp.
Supplementary Material
References
- 1.Chalasani N., Younossi Z., Lavine J.E., Charlton M., Cusi K., Rinella M., Harrison S.A., Brunt E.M., Sanyal A.J. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease: meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 3.Estes C., Anstee Q.M., Arias-Loste M.T., Bantel H., Bellentani S., Caballeria J., Colombo M., Craxi A., Crespo J., Day C.P., Eguchi Y., Geier A., Kondili L.A., Kroy D.C., Lazarus J.V., Loomba R., Manns M.P., Marchesini G., Nakajima A., Negro F., Petta S., Ratziu V., Romero-Gomez M., Sanyal A., Schattenberg J.M., Tacke F., Tanaka J., Trautwein C., Wei L., Zeuzem S., Razavi H. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016-2030. J Hepatol. 2018;69:896–904. doi: 10.1016/j.jhep.2018.05.036. [DOI] [PubMed] [Google Scholar]
- 4.Czerwińska J., Poznański J., Dębski J., Bukowy Z., Bohr V.A., Tudek B., Speina E. Catalytic activities of Werner protein are affected by adduction with 4-hydroxy-2-nonenal. Nucleic Acids Res. 2014;42:11119–11135. doi: 10.1093/nar/gku783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gargiulo S., Gamba P., Testa G., Rossin D., Biasi F., Poli G., Leonarduzzi G. Relation between TLR4/NF-κB signaling pathway activation by 27-hydroxycholesterol and 4-hydroxynonenal, and atherosclerotic plaque instability. Aging Cell. 2015;14:569–581. doi: 10.1111/acel.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas D.T., Szweda L.I. Cardiac reperfusion injury: aging, lipid peroxidation, and mitochondrial dysfunction. Proc Natl Acad Sci U S A. 1998;95:510–514. doi: 10.1073/pnas.95.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma H., Guo R., Yu L., Zhang Y., Ren J. Aldehyde dehydrogenase 2 (ALDH2) rescues myocardial ischaemia/reperfusion injury: role of autophagy paradox and toxic aldehyde. Eur Heart J. 2011;32:1025–1038. doi: 10.1093/eurheartj/ehq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoritaka A., Hattori N., Uchida K., Tanaka M., Stadtman E.R., Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc Natl Acad Sci U S A. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro J.P., Jung T., Grune T., Siems W. 4-Hydroxynonenal (HNE) modified proteins in metabolic diseases. Free Radic Biol Med. 2017;111:309–315. doi: 10.1016/j.freeradbiomed.2016.10.497. [DOI] [PubMed] [Google Scholar]
- 10.Di Domenico F., Tramutola A., Butterfield D.A. Role of 4-hydroxy-2-nonenal (HNE) in the pathogenesis of alzheimer disease and other selected age-related neurodegenerative disorders. Free Radic Biol Med. 2017;111:253–261. doi: 10.1016/j.freeradbiomed.2016.10.490. [DOI] [PubMed] [Google Scholar]
- 11.Feng Z., Hu W., Tang M.S. Trans-4-hydroxy-2-nonenal inhibits nucleotide excision repair in human cells: a possible mechanism for lipid peroxidation-induced carcinogenesis. Proc Natl Acad Sci U S A. 2004;101:8598–8602. doi: 10.1073/pnas.0402794101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lord C.J., Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 13.Yamashima T., Ota T., Mizukoshi E., Nakamura H., Yamamoto Y., Kikuchi M., Yamashita T., Kaneko S. Intake of ω-6 polyunsaturated fatty acid-rich vegetable oils and risk of lifestyle diseases. Adv Nutr. 2020;11:1489–1509. doi: 10.1093/advances/nmaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blasbalg T.L., Hibbeln J.R., Ramsden C.E., Majchrzak S.F., Rawlings R.R. Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century. Am J Clin Nutr. 2011;93:950–962. doi: 10.3945/ajcn.110.006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seki S., Kitada T., Yamada T., Sakaguchi H., Nakatani K., Wakasa K. In situ detection of lipid peroxidation and oxidative DNA damage in non-alcoholic fatty liver diseases. J Hepatol. 2002;37:56–62. doi: 10.1016/s0168-8278(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 16.Chen C.H., Budas G.R., Churchill E.N., Disatnik M.H., Hurley T.D., Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321:1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Miller S., Younus H., Vanam R., Chen C.H., Mochly-Rosen D., Hurley T.D. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat Struct Mol Biol. 2010;17:159–164. doi: 10.1038/nsmb.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang T.C., Chiu P.R., Chang W.T., Hsieh B.S., Huang Y.C., Cheng H.L., Huang L.W., Hu Y.C., Chang K.L. Epirubicin induces apoptosis in osteoblasts through death-receptor and mitochondrial pathways. Apoptosis. 2018;23:226–236. doi: 10.1007/s10495-018-1450-2. [DOI] [PubMed] [Google Scholar]
- 19.Hirasawa A., Tsumaya K., Awaji T., Katsuma S., Adachi T., Yamada M., Sugimoto Y., Miyazaki S., Tsujimoto G. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat Med. 2005;11:90–94. doi: 10.1038/nm1168. [DOI] [PubMed] [Google Scholar]
- 20.Kang S., Huang J., Lee B.K., Jung Y.S., Im E., Koh J.M., Im D.S. Omega-3 polyunsaturated fatty acids protect human hepatoma cells from developing steatosis through FFA4 (GPR120) Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:105–116. doi: 10.1016/j.bbalip.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Oikawa S., Yamada T., Minohata T., Kobayashi H., Furukawa A., Tada-Oikawa S., Hiraku Y., Murata M., Kikuchi M., Yamashima T. Proteomic identification of carbonylated proteins in the monkey hippocampus after ischemia-reperfusion. Free Radic Biol Med. 2009;46:1472–1477. doi: 10.1016/j.freeradbiomed.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 22.Yamashima T., Oikawa S. The role of lysosomal rupture in neuronal death. Prog Neurobiol. 2009;89:343–358. doi: 10.1016/j.pneurobio.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Zhu H., Yoshimoto T., Yamashima T. Heat shock protein 70.1 (Hsp70.1) affects neuronal cell fate by regulating lysosomal acid sphingomyelinase. J Biol Chem. 2014;289:27432–27443. doi: 10.1074/jbc.M114.560334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan M., Zhu W., Zheng X., Li Y., Tang L., Lu B., Chen W., Qiu P., Leng T., Lin S., Yan G., Yin W. Effect of glutamate on lysosomal membrane permeabilization in primary cultured cortical neurons. Mol Med Rep. 2016;13:2499–2505. doi: 10.3892/mmr.2016.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerónimo-Olvera C., Montiel T., Rincon-Heredia R., Castro-Obregón S., Massieu L. Autophagy fails to prevent glucose deprivation/glucose reintroduction-induced neuronal death due to calpain-mediated lysosomal dysfunction in cortical neurons. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnandis T., Ferrer-Vicens I., García-Trevijano E.R., Miralles V.J., García C., Torres L., Viña J.R., Zaragozá R. Calpains mediate epithelial-cell death during mammary gland involution: mitochondria and lysosomal destabilization. Cell Death Differ. 2012;19:1536–1548. doi: 10.1038/cdd.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puri P., Baillie R.A., Wiest M.M., Mirshahi F., Choudhury J., Cheung O., Sargeant C., Contos M.J., Sanyal A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46:1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 28.Yamada K., Mizukoshi E., Sunagozaka H., Arai K., Yamashita T., Takeshita Y., Misu H., Takamura T., Kitamura S., Zen Y., Nakanuma Y., Honda M., Kaneko S. Characteristics of hepatic fatty acid compositions in patients with nonalcoholic steatohepatitis. Liver Int. 2015;35:582–590. doi: 10.1111/liv.12685. [DOI] [PubMed] [Google Scholar]
- 29.Mattson M.P. Roles of the lipid peroxidation product 4-hydroxynonenal in obesity, the metabolic syndrome, and associated vascular and neurodegenerative disorders. Exp Gerontol. 2009;44:625–633. doi: 10.1016/j.exger.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L.L., Chen H., Wang J., Xia T., Sun H., Yuan C.H., Liu S.L., Chen J.B. 4-HNE induces apoptosis of human retinal pigment epithelial cells by modifying HSP70. Curr Med Sci. 2019;39:442–448. doi: 10.1007/s11596-019-2057-8. [DOI] [PubMed] [Google Scholar]
- 31.Gautam J., Banskota S., Shah S., Jee J.G., Kwon E., Wang Y., Kim D.Y., Chang H.W., Kim J.A. 4-Hydroxynonenal-induced GPR109A (HCA(2) receptor) activation elicits bipolar responses, G(αi) -mediated anti-inflammatory effects and G(βγ) -mediated cell death. Br J Pharmacol. 2018;175:2581–2598. doi: 10.1111/bph.14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji Y., Dai Z., Wu G., Wu Z. 4-Hydroxy-2-nonenal induces apoptosis by activating ERK1/2 signaling and depleting intracellular glutathione in intestinal epithelial cells. Sci Rep. 2016;6 doi: 10.1038/srep32929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu P.S., Yen J.H., Kou M.C., Wu M.J. Luteolin and apigenin attenuate 4-hydroxy-2-nonenal-mediated cell death through modulation of UPR, Nrf2-ARE and MAPK pathways in PC12 Cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0130599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kashyap M.P., Singh A.K., Yadav D.K., Siddiqui M.A., Srivastava R.K., Chaturvedi V., Rai N. 4-Hydroxy-trans-2-nonenal (4-HNE) induces neuronal SH-SY5Y cell death via hampering ATP binding at kinase domain of Akt1. Arch Toxicol. 2015;89:243–258. doi: 10.1007/s00204-014-1260-4. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhary P., Sharma R., Sharma A., Vatsyayan R., Yadav S., Singhal S.S., Rauniyar N., Prokai L., Awasthi S., Awasthi Y.C. Mechanisms of 4-hydroxy-2-nonenal induced pro- and anti-apoptotic signaling. Biochemistry. 2010;49:6263–6275. doi: 10.1021/bi100517x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubinina E.E., Dadali V.A. Role of 4-hydroxy-trans-2-nonenal in cell functions. Biochemistry (Mosc) 2010;75:1069–1087. doi: 10.1134/s0006297910090014. [DOI] [PubMed] [Google Scholar]
- 37.Siems W., Grune T. Intracellular metabolism of 4-hydroxynonenal. Mol Aspects Med. 2003;24:167–175. doi: 10.1016/s0098-2997(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 38.Dalleau S., Baradat M., Guéraud F., Huc L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013;20:1615–1630. doi: 10.1038/cdd.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siems W.G., Zollner H., Grune T., Esterbauer H. Metabolic fate of 4-hydroxynonenal in hepatocytes: 1,4-dihydroxynonene is not the main product. J Lipid Res. 1997;38:612–622. [PubMed] [Google Scholar]
- 40.Feldstein A.E., Canbay A., Angulo P., Taniai M., Burgart L.J., Lindor K.D., Gores G.J. Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology. 2003;125:437–443. doi: 10.1016/s0016-5085(03)00907-7. [DOI] [PubMed] [Google Scholar]
- 41.Tsurusaki S., Tsuchiya Y., Koumura T., Nakasone M., Sakamoto T., Matsuoka M., Imai H., Yuet-Yin Kok C., Okochi H., Nakano H., Miyajima A., Tanaka M. Hepatic ferroptosis plays an important role as the trigger for initiating inflammation in nonalcoholic steatohepatitis. Cell Death Dis. 2019;10:449. doi: 10.1038/s41419-019-1678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muzio G., Trombetta A., Maggiora M., Martinasso G., Vasiliou V., Lassen N., Canuto R.A. Arachidonic acid suppresses growth of human lung tumor A549 cells through down-regulation of ALDH3A1 expression. Free Radic Biol Med. 2006;40:1929–1938. doi: 10.1016/j.freeradbiomed.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 43.Gallagher E.P., Huisden C.M., Gardner J.L. Transfection of HepG2 cells with hGSTA4 provides protection against 4-hydroxynonenal-mediated oxidative injury. Toxicol In Vitro. 2007;21:1365–1372. doi: 10.1016/j.tiv.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart B.J., Roede J.R., Doorn J.A., Petersen D.R. Lipid aldehyde-mediated cross-linking of apolipoprotein B-100 inhibits secretion from HepG2 cells. Biochim Biophys Acta. 2009;1791:772–780. doi: 10.1016/j.bbalip.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shearn C.T., Fritz K.S., Reigan P., Petersen D.R. Modification of Akt2 by 4-hydroxynonenal inhibits insulin-dependent Akt signaling in HepG2 cells. Biochemistry. 2011;50:3984–3996. doi: 10.1021/bi200029w. [DOI] [PubMed] [Google Scholar]
- 46.Shearn C.T., Smathers R.L., Stewart B.J., Fritz K.S., Galligan J.J., Hail N., Jr., Petersen D.R. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) inhibition by 4-hydroxynonenal leads to increased Akt activation in hepatocytes. Mol Pharmacol. 2011;79:941–952. doi: 10.1124/mol.110.069534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shearn C.T., Reigan P., Petersen D.R. Inhibition of hydrogen peroxide signaling by 4-hydroxynonenal due to differential regulation of Akt1 and Akt2 contributes to decreases in cell survival and proliferation in hepatocellular carcinoma cells. Free Radic Biol Med. 2012;53:1–11. doi: 10.1016/j.freeradbiomed.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaudhary P., Sharma R., Sahu M., Vishwanatha J.K., Awasthi S., Awasthi Y.C. 4-Hydroxynonenal induces G2/M phase cell cycle arrest by activation of the ataxia telangiectasia mutated and Rad3-related protein (ATR)/checkpoint kinase 1 (Chk1) signaling pathway. J Biol Chem. 2013;288:20532–20546. doi: 10.1074/jbc.M113.467662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang B., Dong J.L., Chen Y.L., Liu Y., Huang S.S., Zhong X.L., Cheng Y.H., Wang Z.G. Nrf2 mediates the protective effects of homocysteine by increasing the levels of GSH content in HepG2 cells. Mol Med Rep. 2017;16:597–602. doi: 10.3892/mmr.2017.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence R.E., Zoncu R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat Cell Biol. 2019;21:133–142. doi: 10.1038/s41556-018-0244-7. [DOI] [PubMed] [Google Scholar]
- 51.Ballabio A., Bonifacino J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol. 2020;21:101–118. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 52.Carullo G., Mazzotta S., Vega-Holm M., Iglesias-Guerra F., Vega-Pérez J.M., Aiello F., Brizzi A. GPR120/FFAR4 pharmacology: focus on agonists in type 2 diabetes mellitus drug discovery. J Med Chem. 2021;64:4312–4332. doi: 10.1021/acs.jmedchem.0c01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su H., Zhao W., Zhang F., Song M., Liu F., Zheng J., Ling M., Yang X., Yang Q., He H., Chen L., Lai X., Zhu X., Wang L., Gao P., Shu G., Jiang Q., Wang S. cis 9, trans 11, but not trans 10, cis 12 CLA isomer, impairs intestinal epithelial barrier function in IPEC-J2 cells and mice through activation of GPR120-[Ca2+]i and the MLCK signaling pathway. Food Funct. 2020;11:3657–3667. doi: 10.1039/d0fo00376j. [DOI] [PubMed] [Google Scholar]
- 54.Syntichaki P., Xu K., Driscoll M., Tavernarakis N. Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature. 2002;419:939–944. doi: 10.1038/nature01108. [DOI] [PubMed] [Google Scholar]
- 55.Narita M., Shimizu S., Ito T., Chittenden T., Lutz R.J., Matsuda H., Tsujimoto Y. Bax interacts with the permeability transition pore to induce permeability transition and cytochrome c release in isolated mitochondria. Proc Natl Acad Sci U S A. 1998;95:14681–14686. doi: 10.1073/pnas.95.25.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feldstein A.E., Werneburg N.W., Canbay A., Guicciardi M.E., Bronk S.F., Rydzewski R., Burgart L.J., Gores G.J. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha expression via a lysosomal pathway. Hepatology. 2004;40:185–194. doi: 10.1002/hep.20283. [DOI] [PubMed] [Google Scholar]
- 57.Truman J.P., Al Gadban M.M., Smith K.J., Hammad S.M. Acid sphingomyelinase in macrophage biology. Cell Mol Life Sci. 2011;68:3293–3305. doi: 10.1007/s00018-011-0686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fucho R., Martínez L., Baulies A., Torres S., Tarrats N., Fernandez A., Ribas V., Astudillo A.M., Balsinde J., Garcia-Rovés P., Elena M., Bergheim I., Lotersztajn S., Trautwein C., Appelqvist H., Paton A.W., Paton J.C., Czaja M.J., Kaplowitz N., Fernandez-Checa J.C., García-Ruiz C. ASMase regulates autophagy and lysosomal membrane permeabilization and its inhibition prevents early-stage non-alcoholic steatohepatitis. J Hepatol. 2014;61:1126–1134. doi: 10.1016/j.jhep.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayes J.D., Flanagan J.U., Jowsey I.R. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 60.Hartley D.P., Ruth J.A., Petersen D.R. The hepatocellular metabolism of 4-hydroxynonenal by alcohol dehydrogenase, aldehyde dehydrogenase, and glutathione S-transferase. Arch Biochem Biophys. 1995;316:197–205. doi: 10.1006/abbi.1995.1028. [DOI] [PubMed] [Google Scholar]
- 61.Breitzig M., Bhimineni C., Lockey R., Kolliputi N. 4-Hydroxy-2-nonenal: a critical target in oxidative stress? Am J Physiol Cell Physiol. 2016;311:C537–C543. doi: 10.1152/ajpcell.00101.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oniki K., Morita K., Watanabe T., Kajiwara A., Otake K., Nakagawa K., Sasaki Y., Ogata Y., Saruwatari J. The longitudinal effect of the aldehyde dehydrogenase 2∗2 allele on the risk for nonalcoholic fatty liver disease. Nutr Diabetes. 2016;6 doi: 10.1038/nutd.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Z., Ye S., Zhong X., Wang W., Lai C.H., Yang W., Yue P., Luo J., Huang X., Zhong Z., Xiong Y., Fan X., Li L., Wang Y., Ye Q. Pretreatment with the ALDH2 activator Alda-1 protects rat livers from ischemia/reperfusion injury by inducing autophagy. Mol Med Rep. 2020;22:2373–2385. doi: 10.3892/mmr.2020.11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong W., Zhang W., Li Q., Xie G., Sun Q., Sun X., Tan X., Sun X., Jia W., Zhou Z. Pharmacological activation of aldehyde dehydrogenase 2 by Alda-1 reverses alcohol-induced hepatic steatosis and cell death in mice. J Hepatol. 2015;62:1375–1381. doi: 10.1016/j.jhep.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L., Li P., Fu S., Calay E.S., Hotamisligil G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park H.W., Park H., Semple I.A., Jang I., Ro S.H., Kim M., Cazares V.A., Stuenkel E.L., Kim J.J., Kim J.S., Lee J.H. Pharmacological correction of obesity-induced autophagy arrest using calcium channel blockers. Nat Commun. 2014;5:4834. doi: 10.1038/ncomms5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ueno T., Komatsu M. Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14:170–184. doi: 10.1038/nrgastro.2016.185. [DOI] [PubMed] [Google Scholar]
- 69.Inami Y., Yamashina S., Izumi K., Ueno T., Tanida I., Ikejima K., Watanabe S. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem Biophys Res Commun. 2011;412:618–625. doi: 10.1016/j.bbrc.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 70.Yetti H., Naito H., Jia X., Shindo M., Taki H., Tamada H., Kitamori K., Hayashi Y., Ikeda K., Yamori Y., Nakajima T. High-fat-cholesterol diet mainly induced necrosis in fibrotic steatohepatitis rat by suppressing caspase activity. Life Sci. 2013;93:673–680. doi: 10.1016/j.lfs.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 71.Afonso M.B., Rodrigues P.M., Carvalho T., Caridade M., Borralho P., Cortez-Pinto H., Castro R.E., Rodrigues C.M. Necroptosis is a key pathogenic event in human and experimental murine models of non-alcoholic steatohepatitis. Clin Sci (Lond) 2015;129:721–739. doi: 10.1042/CS20140732. [DOI] [PubMed] [Google Scholar]
- 72.Xu B., Jiang M., Chu Y., Wang W., Chen D., Li X., Zhang Z., Zhang D., Fan D., Nie Y., Shao F., Wu K., Liang J. Gasdermin D plays a key role as a pyroptosis executor of non-alcoholic steatohepatitis in humans and mice. J Hepatol. 2018;68:773–782. doi: 10.1016/j.jhep.2017.11.040. [DOI] [PubMed] [Google Scholar]
- 73.Wang F., Gómez-Sintes R., Boya P. Lysosomal membrane permeabilization and cell death. Traffic. 2018;19:918–931. doi: 10.1111/tra.12613. [DOI] [PubMed] [Google Scholar]
- 74.Matteoni C.A., Younossi Z.M., Gramlich T., Boparai N., Liu Y.C., McCullough A.J. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116:1413–1419. doi: 10.1016/s0016-5085(99)70506-8. [DOI] [PubMed] [Google Scholar]
- 75.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 76.Koteish A., Diehl A.M. Animal models of steatosis. Semin Liver Dis. 2001;21:89–104. doi: 10.1055/s-2001-12932. [DOI] [PubMed] [Google Scholar]
- 77.Hewitt K.N., Pratis K., Jones M.E., Simpson E.R. Estrogen replacement reverses the hepatic steatosis phenotype in the male aromatase knockout mouse. Endocrinology. 2004;145:1842–1848. doi: 10.1210/en.2003-1369. [DOI] [PubMed] [Google Scholar]
- 78.McGrath L.T., McGleenon B.M., Brennan S., McColl D., Mc I.S., Passmore A.P. Increased oxidative stress in Alzheimer’s disease as assessed with 4-hydroxynonenal but not malondialdehyde. QJM. 2001;94:485–490. doi: 10.1093/qjmed/94.9.485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.