Abstract
Approximately half of glioblastoma and more than two-thirds of grade II and III glioma tumors lack the DNA repair protein O6-methylguanine methyl transferase (MGMT). MGMT-deficient tumors respond initially to the DNA methylation agent temozolomide (TMZ) but frequently acquire resistance through loss of the mismatch repair (MMR) pathway. We report the development of agents that overcome this resistance mechanism by inducing MMR-independent cell killing selectively in MGMT-silenced tumors. These agents deposit a dynamic DNA lesion that can be reversed by MGMT but slowly evolves into an interstrand cross-link in MGMT-deficient settings, resulting in MMR-independent cell death with low toxicity in vitro and in vivo. This discovery may lead to new treatments for gliomas and may represent a new paradigm for designing chemotherapeutics that exploit specific DNA repair defects.
Genetic instability is a hallmark of cancer and typically arises from mutations in key DNA damage repair and/or reversal proteins [collectively referred to here as the DNA damage response (DDR)] (1). Intrinsic DDR defects can be exploited with DDR inhibitors through the concept of synthetic lethality, which is defined as a loss of viability resulting from the disruption of two genes or pathways that, if disrupted individually, are nonlethal (2, 3). Notable examples of synthetic lethal interactions between tumor-associated DDR defects and DDR inhibitors include (i) homologous recombination (HR)–defective tumors and inhibitors of polymerase θ (Pol θ)(4); (ii) ataxia-telangiectasia mutated (ATM)–mutant tumors and ataxia telangiectasia and Rad3-related (ATR) inhibitors (5); and (iii) mismatch repair (MMR)–deficient tumors and inactivation of Werner syndrome helicase (WRN) (6). HR-defective tumors also can be selectively targeted by inhibitors of poly(ADP-ribose) polymerase (PARP) (2, 3). However, the mechanistic basis for this toxicity appears to depend on trapping of PARP at sites of DNA damage, which likely induces more complex damage that requires HR for resolution (7). In each of these examples, selective tumor cell killing by means of the DDR protein inhibitor relies on either the induction or persistence of DNA damage or aberrant DNA structures.
These findings suggested to us that genotoxins could be tailored to exploit differential sensitivities that arise from specific tumor-associated DDR defects. This approach avoids the need to engage DDR proteins directly, circumventing resistance mechanisms that arise from mutations in the ligand binding site (8) while minimizing off-target effects in healthy, DDR-proficient cells. To achieve this, we conceived a mechanistic strategy in which a single agent modifies DNA through two successive chemical steps (Fig. 1A). The first chemical reaction is designed to generate a primary DNA lesion that is rapidly removed by healthy, DDR-proficient cells. The second chemical reaction is engineered to slowly transform the primary modification into a more toxic secondary lesion. We anticipated that if the rate of primary lesion repair were sufficiently rapid in healthy cells, the secondary lesion would accumulate only in the DNA of DDR-deficient tumor cells. This two-step pathway would overcome resistance mechanisms that mitigate the toxicity of the primary lesion, which have been implicated in resistance to various chemotherapies, including anthracyclines [impairment of nucleoside excision repair (NER)] (9), topoisomerase inhibitors (loss of nonhomologous end joining repair) (10), and antimetabolite (11) and platinum (12) agents (mutations in MMR). Furthermore, delivering these bespoke lesions by using established chemotherapy scaffolds could facilitate rapid translation into the clinic, owing to decades of use in cancer patients. We examine this strategy in the context of glioblastoma (GBM).
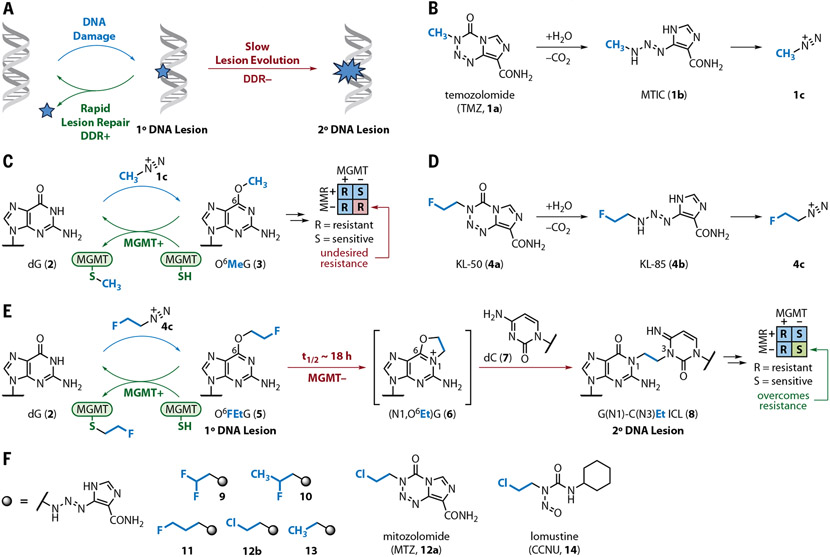
Fig. 1. Overview of mechanistic strategy and structures of agents used in this study.
(A) Underlying mechanistic design. Systemic administration of a bifunctional agent is envisioned to form a primary lesion that is rapidly resolved by healthy (DDR+) but not DDR-deficient (DDR−) cells. The persistence of the primary lesion allows it to evolve slowly to a more toxic secondary lesion. (B) Under physiological conditions, TMZ (1a) converts to MTIC (1b), which in turn decomposes to methyl diazonium (1c).(C) O6-Methylguanine (3) is the most clinical significant alkyl nucleobase generated by methyl diazonium (1c) and is rapidly reverted to dG (2) by MGMT. (D) Conversion of KL-50 (4a) to KL-85 (4b) in aqueous solution followed by decomposition to 2-fluoroethyl diazonium ion (4c).(E) 2-fluoroethyl diazonium ion (4c), derived from KL-50 (4a), is proposed to lead to the formation of G(N1)–C(N3)Et ICL 8 specifically in MGMT− cells. This mechanism is independent of MMR status. (F) Structures of the triazenes 9 to 13, mitozolomide 12a, and lomustine (CCNU; 14). Syntheses are available in the supplementary materials.
GBM is the most common and devastating form of brain cancer, with a 5-year survival rate of ~5%. Approximately half of GBMs lack the DDR protein O6-methylguanine methyltransferase (MGMT) through promoter hypermethylation. MGMT silencing occurs at an even greater frequency in grade II and III gliomas (more than 70% of cases), and these tumors are also largely incurable (13). Mechanistically, MGMT removes O6-alkylguanine adducts by transferring the alkyl adduct to an active site cysteine through an SN2 displacement. Patients with MGMT-deficient tumors (referred to here as MGMT− tumors) are treated with temozolomide (TMZ; 1a), a prodrug that converts under physiological conditions to the potent methylating agent methyl diazonium (1c) through the intermediacy of 3-methyl-(triazen-1- yl)-imidazole-4-carboxamide (MTIC; 1b) (Fig. 1B). N7-Methylguanine and N3-methyladenine are the major sites of methylation (70 and 9%, respectively) but are readily resolved by the base excision repair (BER) pathway. By contrast, although O6-methylguanine (O6MeG; 3) adducts derived from TMZ (1a) only compose ~5% of alkylation products, these lesions persist in the genome of MGMT− tumors [but are readily reversed by healthy (MGMT+) cells] (Fig. 1C). O6MeG (3) lesions are thought to induce formation of DNA double-strand breaks (DSBs) and tumor cell death through an MMR-dependent mechanism (14). MGMT status is a predictive biomarker for initial response to TMZ (1a) in GBM, with a significant overall survival benefit in the up-front setting for patients with these cancers (15). However, it is now well established (16) that acquired clinical resistance to TMZ (1a) through MMR mutations abrogates its efficacy, leading to recurrent GBM and death in nearly all patients. Frequently, TMZ (1a) is used as adjuvant therapy for grade III and high-risk grade II gliomas; however, it remains non-curative, with recurrences typically occurring over 2 to 10 years. In ~80% of patients, recurrences coincide with transformation to higher-grade tumors resistant to TMZ (1a), associated in ~50% of cases with acquisition of a distinct hypermutation signature secondary to MMR deficiency, resulting in reduced survival (17, 18).
We hypothesized that we could deploy the strategy outlined in Fig. 1A to develop agents that overcome the resistance associated with MMR loss while maintaining TMZ’s (1a) selectivity for MGMT-silenced tumors. These agents would deposit a primary lesion susceptible to SN2-mediated removal by MGMT that could undergo a further chemical transformation to a secondary lesion capable of killing MGMT-deficient tumor cells in an MMR-independent manner. To maintain the therapeutic index (TI) between MGMT− tumor cells and MGMT+ healthy cells, the primary lesion must undergo MGMT-mediated repair faster than it undergoes transformation to the secondary lesion. With these considerations in mind, we developed an agent capable of depositing a 2-fluoroethyl lesion at O6-guanine. We expected that O6FEtG (5) would evolve slowly to the (N1,O6Et)G intermediate 6, which could then undergo ring-opening by N3 of the complementary cytidine base (7) to form the G(N1)–C(N3) interstrand cross-link (ICL) 8 (Fig. 1E). This hypothesis is based on the finding that the corresponding nucleoside, O6-(2-fluoroethyl)guanosine (S1), undergoes a hydrolytic rearrangement to N1-(2-hydroxyethyl)guanosine (S3) with a half-life of 18.5 hours (37°C, pH 7.4) (fig. S1A) (19) and the recent determination that oligonucleotides containing a single photocaged O6-(2-chloroethyl)guanosine residue can undergo G–C cross-linking through a parallel mechanistic pathway (20). Although the rates of reversal of O6FEtG (5) by MGMT are unknown, MGMT reacts rapidly with methylated calf thymus DNA (second-order rate constant = 1 × 109 M−1 min−1) (21); and oligonucleotides that contain O6-methyl-, O6-benzyl-, and O6-heteroarylmethylguanine residues possess nanomolar median inhibitory concentrations (IC50) against MGMT in vitro (22). Additionally, MGMT reverses a wide range of O6-alkylguanine substrates (23). These data suggested that MGMT-proficient cells should repair the O6FEtG lesion (5) before it transforms into ICL 8.
Results
Compound synthesis and in vitro evaluation
We synthesized the imidazotetrazine 4a (also known as KL-50) and the triazene 4b (also known as KL-85) as vehicles to deliver 2-fluoroethyl diazonium (4c) as well as a series of related agents to probe structure-activity relationships in tissue culture (Fig. 1, D and F). The 2-fluoropropyl- and 3-fluoropropyl-triazenes (10 and 11, respectively) were prepared through the diazotization of 4-aminoimidazole-5-carboxamide, followed by the addition of the respective amine (supplementary materials). All other triazenes were prepared according to literature procedures (supplementary materials). We evaluated the cytotoxicity of our compounds in short-term cell viability assays against four isogenic LN229 GBM cell lines engineered to be proficient or deficient in MGMT and/or MMR activity by using short hairpin RNAs (shRNAs) targeting MSH2 (referred to as MGMT+/−, MMR+/− cells here after) (fig. S2A). This approach allowed us to interrogate the relationship between MGMT and MMR status and compound activity.
The IC50 values of these agents are shown in Fig. 2A, and representative dose-response curves are shown in Fig. 2B (additional data are presented in fig. S2, B to G). KL-85 (4b) retained potency in MGMT−/MMR− cells (IC50 = 27.5 μM), whereas TMZ (1a) was essentially inactive (IC50 = 838 μM). Structure-activity studies were consistent with the mechanistic pathway shown in Fig. 1E. The 2,2-difluoroethyl triazene 9 and the 2-fluoropropyl triazene 10 possessed reduced potency in MGMT−/MMR− cells (fig. S2, B and C), which is in agreement with a reduced rate of displacement after introduction of an additional fluorine (24) or alkyl substituent. The 2-chloroethyl triazene 12b was modestly potent but not as selective for MGMT− cell lines (fig. S2E), which likely results from faster, nonselective ICL formation arising from chloride displacement. The 3-fluoropropyl triazene 11 was essentially inactive in all four cell lines, potentially because of elimination of the diazonium ion (fig. S2D). As expected, the ethyl triazene 13 was inactive (fig. S2F). This compound is known to rapidly undergo elimination to ethylene gas after conversion to ethyl diazonium (25).
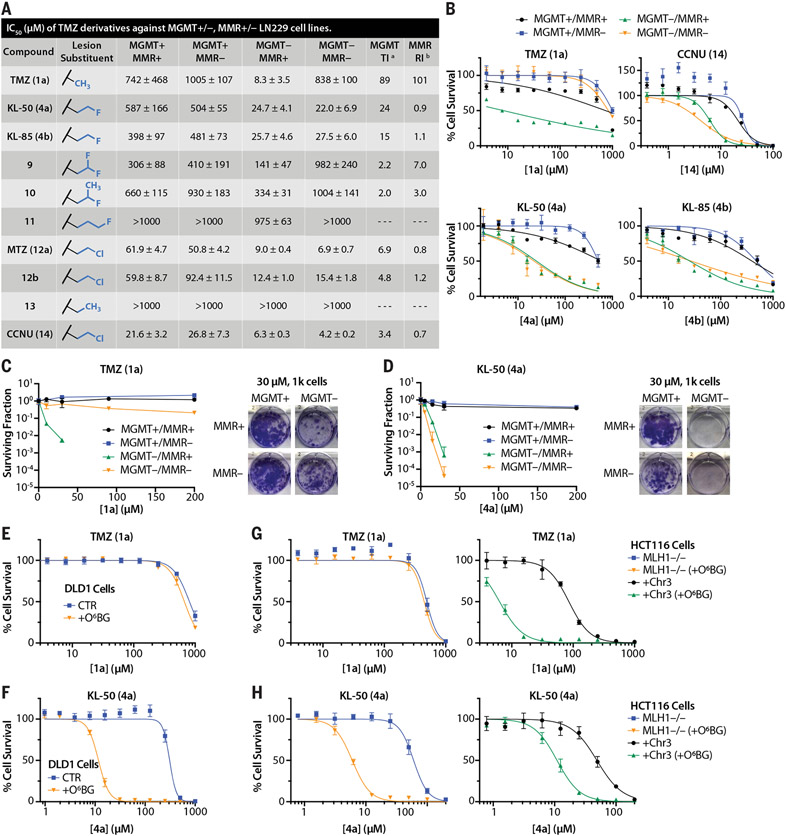
Fig. 2. KL-50 (4a) displays previously unseen MGMT-dependent, MMR-independent cytotoxicity in multiple isogenic cell models.
(A) IC50 values derived from short-term viability assays in LN229 MGMT+/−, MMR+/− cells treated with TMZ (1a) derivatives. aMGMT TI = IC50 (MGMT+/MMR+) divided by IC50 (MGMT−/MMR+). bMMR resistance index (RI) = IC50 (MGMT−/MMR−) divided by IC50 (MGMT−/MMR+). (B) Short-term viability assay curves for TMZ (1a), CCNU (14), KL-85 (4b), and KL-50 (4a) in LN229 MGMT+/−, MMR+/− cells. (C) Clonogenic survival curves for TMZ (1a) in LN229 MGMT+/−, MMR+/− cells, with representative images of wells containing 1000 plated cells treated with 30 μM TMZ (1a).(D) Clonogenic survival curves for KL-50 (4a) in LN229 MGMT+/−, MMR+/− cells, with representative images of wells containing 1000 plated cells treated with 30 μM KL-50 (4a). (E) Short-term viability assay curves for TMZ (1a) in DLD1 MSH6-deficient cells pretreated with 0.01% dimethyl sulfoxide (DMSO) control (CTR) or 10 μM O6BG (+O6BG) for 1 hour before TMZ (1a) addition to deplete MGMT. (F) Short-term viability assay curves for KL-50 (4a) in DLD1 MSH6-deficient cells pretreated with 0.01% DMSO control (CTR) or 10 μM O6BG (+O6BG) for 1 hour before KL-50 (4a) addition. (G) Short-term viability assay curves for TMZ (1a) in HCT116 MLH1−/− cells or HCT116 cells complemented with chromosome 3 carrying wild-type MLH1 (+Chr3) pretreated with 0.01% DMSO control or 10 μM O6BG (+O6BG) for 1 hour before TMZ (1a) addition. (H) Short-term viability assay curves for KL-50 (4a) in HCT116 MLH1−/− cells or HCT116 cells complemented with chromosome 3 carrying wild-type MLH1 (+Chr3) pretreated with 0.01% DMSO control or 10 μM O6BG (+O6BG) for 1 hour before KL-50 (4a) addition. For (B) to (H), points indicate the mean, and error bars indicate SD; n ≥ 3 technical replicates.
We prepared KL-50 (4a) through the diazotization of 4-aminoimidazole-5-carboxamide followed by the addition of (2-fluoroethyl) isocyanate (39% overall yield) (supplementary materials). The potency of 4a mirrored that of 4b in the four cell lines examined (Fig. 2B). To benchmark selectivity, we evaluated the experimental agent mitozolomide (MTZ; 12a) and the clinical nitrosourea lomustine (also known as CCNU; 14), which have been studied with hopes of addressing TMZ (1a) resistance (26, 27). However, these 2-chloroethylating agents were only approximately four- to sevenfold selective for MGMT-deficient cells (fig. S2G and Fig. 2B, respectively), as opposed to the ~25-fold selectivity seen with the 2-fluoroethylating agent KL-50 (4a).
We validated the antitumor activity of KL-50 (4a) in clonogenic survival assays (CSAs) and additional cell lines in vitro. TMZ (1a) possessed negligible activity in MGMT+ LN229 cells, irrespective of MMR status, and induced robust tumor cell killing in MGMT−, MMR+ cells that was abolished in isogenic cells lacking MMR (Fig. 2C). Lomustine (14) was effective in MMR− cells but was cytotoxic to MGMT+ cells (fig. S2H). By contrast, KL-50 (4a) demonstrated robust antitumor activity in MGMT− cells, independent of MMR status, with negligable toxicity in MGMT+ cells at doses of up to at least 200 μM (Fig. 2D). We observed a similar pattern of activities in several distinct cell lines across different tumor types with intrinsic or induced loss of MGMT and/or MMR activity. For example, TMZ (1a) was inactive in DLD1 cells, which possess MGMT but lack functional MMR (MSH6−), with or without induced depletion of MGMT by using O6-benzylguanine (O6BG) (Fig. 2E) (28). By contrast, KL-50 (4a) was efficacious in these cells, but only after O6BG-induced MGMT depletion (Fig. 2F). TMZ (1a) was inactive in HCT116 colorectal cancer cells, which lack the MMR protein MLH1, regardless of MGMT levels (Fig. 2G). Restoration of MMR activity by means of complementation with chromosome 3 containing MLH1 resulted in the expected enhanced sensitivity to TMZ (1a), which was further potentiated by MGMT depletion (Fig. 2G). By contrast, KL-50 (4a) induced selective tumor cell killing specifically in the setting of O6BG-induced MGMT suppression in both MLH1-deficient and MLH1-complemented cells (Fig. 2H). The MMR status and O6BG-induced loss of MGMT expression was confirmed with Western blot analysis for these cell lines (fig. S2, I and J). We also confirmed the activity of KL-50 (4a) in MGMT− LN229 cells engineered to lack expression of other key MMR proteins, including MSH6, MLH1, PMS2, and MSH3 (fig. S3). The inability of MSH3 depletion to induce TMZ (1a) resistance is an anticipated result, given that the primary role of the MSH2-MSH3 heterodimer is recognition of insertion-deletion loops, whereas single base pair mismatches induced by TMZ are processed by the MSH2-MSH6 heterodimer and subsequent downstream MMR proteins (29, 30). Last, we compared the cytotoxicity of KL-50 (4a) and TMZ (1a) in normal human fibroblast cells and observed no increase in toxicity with KL-50 (4a) (fig. S2K). These data suggest that KL-50 (4a) is a first-inclass molecule that overcomes MMR mutation–induced resistance while retaining selectivity for tumor cells that lack MGMT.
Mechanism of action of KL-50 (4a)
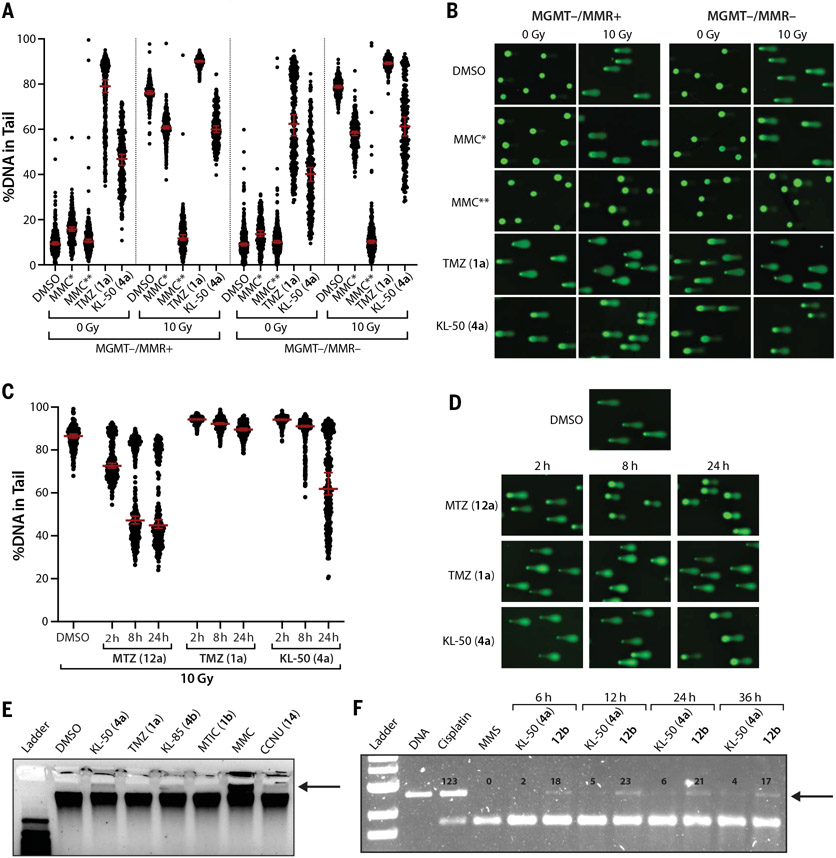
We used a well-established comet assay adapted for ICL detection to determine whether ICLs were formed in MGMT− cells treated with KL-50 (4a) (Fig. 3, A and B). In this assay, cells were sequentially exposed to genotoxins and ionizing radiation (IR) and then analyzed by means of single-cell alkaline gel electrophoresis. Attenuation of the IR–induced comet tail is indicative of ICL formation. As expected, in the absence of IR, TMZ (1a) (200 μM) and KL-50 (4a) (200 μM) both induced tailing in MGMT−/MMR+ cells, whereas mitomycin C (MMC) (0.1 or 50 μM) did not (31). Exposure to 50 μM MMC for 2 hours abolished the IR-induced comet tail, whereas exposure to 0.1 μM MMC [chosen to be ~10-fold greater than the IC50 for this drug, which is comparable with 200 μM KL-50 (4a) or TMZ (1a)] for 24 hours caused a partial reduction in the IR-induced comet tail. TMZ (1a) (200 μM) did not reduce DNA migration after IR, which is in agreement with its known function as a monoalkylation agent with no known cross-linking activity. By contrast, KL-50 (4a) (200 μM) reduced the percent of DNA in the tail after IR to levels similar to those seen for 0.1 μM MMC. We observed a similar pattern of comet tail migration for MMC and KL-50 (4a) in MGMT−/MMR− cells, which supports an MMR-independent cross-linking mechanism. Comparable results were observed in MGMT−/MMR+ cells treated with KL-85 (4b) (fig. S4, A and B).
Fig. 3. Unrepaired primary KL-50 (4a) lesions convert to DNA ICLs in the absence of MGMT.
(A) Scatter dot plots of the percent DNA in tail upon single-cell alkaline gel electrophoresis performed on LN229 MGMT−/MMR+ and MGMT−/MMR− cells treated with 0.2% DMSO control, 200 μM TMZ (1a), 200 μM KL-50 (4a), or 0.1 μM MMC (MMC*) for 24 hours or with 50 μM MMC (MMC**) for 2 hours. After cell lysis, comet slides were irradiated with 0 or 10 grays (Gy) before alkaline electrophoresis. Lines indicate median; error bars indicate 95% confidence interval (CI); n ≥ 160 comets per condition. (B) Representative comet images from (A). (C) Scatter dot plots of the percent DNA in tail upon single-cell alkaline gel electrophoresis performed on LN229 MGMT−/MMR− cells treated with 0.2% DMSO control, 200 μM MTZ (12a), 200 μM TMZ (1a), or 200 μM KL-50 (4a) for 2, 8, or 24 hours. After cell lysis, comet slides were irradiated with 10 Gy before alkaline electrophoresis. Lines indicate median; error bars indicate 95% CI; n ≥ 230 comets per condition. Data from samples treated with 0 Gy are shown in fig. S4, C and D. (D) Representative comet images from (C). (E) Denaturing gel electrophoresis of genomic DNA isolated from LN229 MGMT−/MMR+ cells treated with 0.2% DMSO control, 200 μM KL-50 (4a), 200 μM TMZ (1a), 200 μM KL-85 (4b), or 200 μM MTIC (1b) for 24 hours or with 50 μM MMC or 200 μM CCNU (14) for 2 hours. (F) Denaturing gel electrophoresis of linearized 100 ng pUC19 plasmid DNA treated in vitro with 100 μM cisplatin (36 hours), 100 μM MMS (36 hours), 100 μM of KL-50 (4a), or 12b for 6 to 36 hours. For (E) and (F), bands indicating cross-linked DNA are indicated with arrows. Quantification of bands in (F) is provided in fig. S4E.
We carried out this assay at varying time points (2 to 24 hours) to assess the rates of ICL formation in MGMT−/MMR− cells treated with KL-50 (4a), MTZ (12a), or TMZ (1a) (Fig. 3, C and D). The chloroethylating agent MTZ (12a) reduced DNA mobility within 2 hours, which is consistent with the cell line selectivities above and literature reports that this agent rapidly forms ICLs (32) through chloride displacement from sites of 2-chloroethylation other than O6G. TMZ (1a) did not induce a statistically significant decrease in DNA migration within 24 hours. However, we observed a time-dependent decrease in DNA mobility in cells treated with KL-50 (4a), with the largest difference observed between 8 and 24 hours; this is consistent with the reported half-life of O6-(2-fluoroethyl)guanosine (S1) (18.5 hours) (fig. S1A) (19) and that of O6FEtG (6) (18.5 hours). In the unirradiated samples, KL-50 (4a), MTZ (12a), and TMZ (1a) all induced maximal damage at 2 hours; this damage decreased over time, which is consistent with progressive DNA repair (fig. S4, C and D). Analysis of genomic DNA isolated from LN229 MGMT−/MMR+ cells treated with KL-50 (4a, 200 μM) or KL-85 (4b, 200 μM) by denaturing gel electrophoresis (33) demonstrated the presence of cross-linked DNA (Fig. 3E). TMZ (1a) and MTIC (1b) showed no evidence of ICL induction. Similarly, linearized pUC19 plasmid DNA treated with KL-50 (4a) (100 μM) also possessed ICLs, with delayed rates of formation relative to those of 12b (Fig. 3F). Collectively, these data support a mechanism of action for KL-50 (4a) that involves the slow generation of DNA ICLs in the absence of MGMT.
To probe for alternative mechanisms of action, we conducted experiments to assess for involvement of nucleotide excision repair (NER) or BER in lesion processing, generation of reactive oxygen species (ROS), and DNA duplex destabilization. Short-term cell viability assays in isogenic mouse embryonic fibroblasts (MEFs) proficient or deficient in XPA, a common shared NER factor, revealed no differential sensitivity, either with or without O6BG-induced MGMT depletion (fig. S5A). N7-MeG lesions induced by TMZ (1a) are prone to spontaneous depurination, apurinic (AP) site formation, and single-strand breaks (SSBs), which are all known BER substrates. To probe for potential differential induction of BER substrates by KL-50 (4a) compared with TMZ (1a), we performed in vitro supercoiled plasmid DNA assays to measure the formation of AP sites (34). We observed similar levels of spontaneous and enzyme-catalyzed SSBs from AP sites after treatment with KL-50 (4a) and TMZ (1a), suggesting comparable levels of depurination (fig. S5B). Cotreatment with increasing concentrations of the ROS scavenger N-acetylcysteine (NAC) did not rescue cell viability (fig. S5C). Melting point analysis did not reveal any notable differences in DNA stability resulting from fluoroethylation compared with methylation (fig. S5D). These data suggest that NER status, AP site induction, ROS, and altered DNA stability are peripheral or noncontributory to the effectiveness of KL-50 (4a).
We characterized the profile of DDR activation across our four isogenic cell lines after treatment with KL-50 (4a) or TMZ (1a). Our prior finding that the ATR-CHK1 signaling axis is activated in response to TMZ (1a)–induced replication stress in MGMT-deficient cells (35) prompted us to analyze the phosphorylation status of CHK1 and CHK2 in LN229 MGMT+/− and MMR+/− cells. KL-50 (4a) induced CHK1 and CHK2 phosphorylation in MGMT− cells regardless of MMR status, whereas TMZ (1a) only induced phospho-CHK1 and -CHK2 in MGMT−/MMR+ cells (fig. S6A). We analyzed foci formation of the DDR factors phospho-SER139-H2AX (γH2AX), p53 binding protein 1 (53BP1), and phospho-SER33-RPA2 (pRPA) over the period of 2 to 48 hours (Fig. 4, A to D, and fig. S6, B and C) (36). KL-50 (4a) induced a maximal foci response at 48 hours, specifically in MGMT− cells and irrespective of MMR status (4a). TMZ (1a) induced a comparable response in MGMT− cells, but this was abolished in the absence of functional MMR, which is consistent with known MMR-silencing–based resistance (16). We observed a reduced level of foci formation in MGMT+/MMR+ cells that was absent in MGMT+/MMR− cells, suggesting an MMR-dependent DDR in these cells. However, these foci dissipate at later time points (72 to 96 hours) (fig. S6D) and are not associated with appreciable cellular toxicity (Fig. 2, C and D).
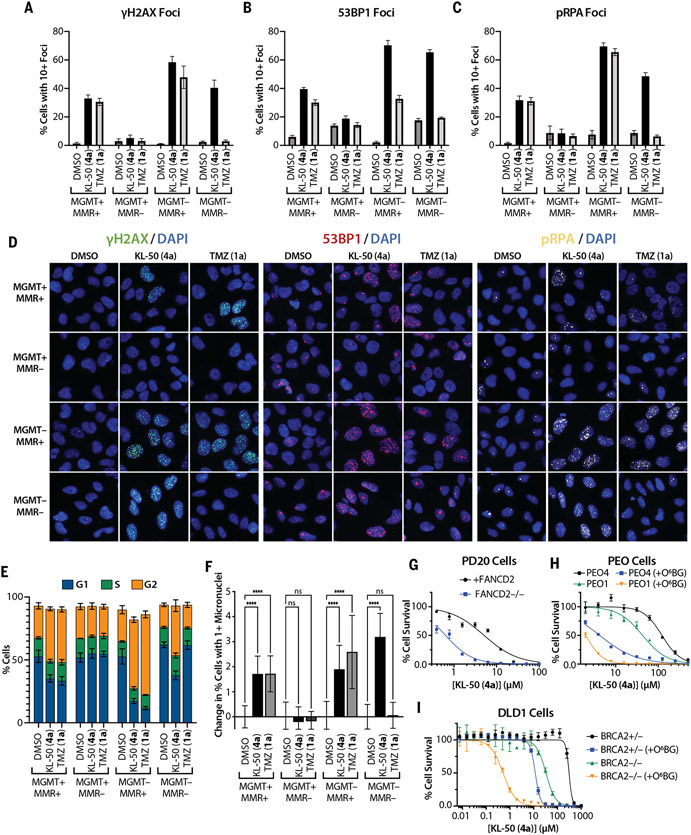
Fig. 4. KL-50 (4a) activates DDR pathways and cycle arrest in MGMT− cells, independent of MMR, and cells deficient in ICL or HR repair are sensitized to KL-50 (4a).
(A) γH2AX, (B) 53BP1, and (C) pRPA foci formation quantified by percent of cells with ≥10 foci in LN229 MGMT+/−, MMR+/− cells treated with 0.1% DMSO control, 20 μM KL-50 (4a), or 20 μM TMZ (1a) for 48 hours. Columns indicate the mean; error bars indicate SD; n > 5 technical replicates. Additional time course data are presented in fig. S6, B to D. (D) Representative foci images of data in (A) to (C). (E) Percentage of cells in G1, S, and G2 cell cycle phases after treatment as in (A) to (C), measured by using integrated nuclear (Hoechst) staining intensity. Columns indicate the mean; error bars indicate SD; n = 3 independent analyses. Additional time course data, cell cycle controls, and representative histograms are presented in fig. S7. (F) Change in percent cells with ≥1 micronuclei from baseline (DMSO control) after treatment as in (A) to (C). Columns indicate the mean; error bars indicate SD; n ≥ 15 technical replicates; ****P < 0.0001; ns, not significant. Additional validation is presented in fig. S9, A and B. (G) Short-term viability assay curves for KL-50 (4a) in PD20 cells, deficient in FANCD2 (FANCD2−/−) or complemented with FANCD2 (+FANCD2). (H) Short-term viability assay curves for KL-50 (4a) in PEO4 (BRCA2+) and PEO1 (BRCA2−/−) cells pretreated with 0.01% DMSO control or 10 mM O6BG (+O6BG) for 1 hour before KL-50 (4a) addition. (I) Short-term viability assay curves for KL-50 (4a) in DLD1 BRCA2+/− and BRCA2−/− cells pretreated with 0.01% DMSO control or 10 mM O6BG (+O6BG) for 1 hour before KL-50 (4a) addition. For (G) to (I), points indicate the mean, and error bars indicate SD; n = 3 technical replicates.
KL-50 (4a) induced increasing G2 arrest on progression from 24 to 48 hours in MGMT−/MMR+ cells as determined with simultaneous analysis of DNA content based on nuclear (Hoechst) staining in the foci studies above (Fig. 4E and fig.S7, A and B). As previously reported for other sources of DNA damage, KL-50 (4a) induced an attenuated G2 arrest in MGMT−/MMR− cells, which is consistent with a role of MMR in the G2-checkpoint (37). This effect in MGMT−/MMR− cells was absent after TMZ (1a) treatment. Both TMZ (1a) and KL-50 (4a) induced a moderate G2 arrest in MGMT+/MMR+ cells.
We quantified the levels of DDR foci across the individual cell cycle phases (fig. S8). KL-50 (4a) induced foci formation primarily in the S and G2 phases of the cell cycle, which is consistent with replication blocking by ICLs (38). Foci increased in MGMT− G1-phase cells at 48 hours, suggesting that a fraction of cells may progress through the S phase with unrepaired DNA damage. TMZ (1a) displayed a similar pattern of foci induction in the S and G2 phases, with smaller increases in G1-phase foci. We concurrently assessed micronuclei formation as an indication of replication stress-induced chromosomal instability (Fig. 4F). Consistent with previously reported data in MSH2−/− MEFs in which MMRloss suppressed micronuclei formation arising from replication-stress (39), MMR-deficient LN229 cells displayed suppression of micronuclei formation induced by KL-50 and TMZ in an MGMT+ background. However, KL-50 (4a), but not TMZ (1a), was able to induce micronuclei in MGMT−/MMR− cells, which suggests that the ICLs formed in these cells are able to induce chromosomal instability even in the absence of MMR. These findings are in agreement with the differential toxicity profiles of KL-50 (4a) and TMZ (1a): KL-50 (4a) induces multiple successive markers of DNA damage and engagement of the DDR in MGMT− cells, independent of MMR status, whereas the effects of TMZ (1a) are similar to those of KL-50 in MGMT−/MMR+ cells but absent in MMR− cells. Coupled with the ICL kinetics data presented above, these time-course data support a slow rate of ICL induction in situ by KL-50 (4a).
These foci data suggest that KL-50 (4a) induces replication stress (as indicated by pRPA foci formation) and formation of DSBs (as indicated by γH2AX and 53BP1 foci), which are known to follow the formation of ICLs (38). Consistent with this, BRCA2- and FANCD2-deficient cells are hypersensitive to KL-50 (4a) (Fig. 4, G to I, and fig. S9, C to F). In two MGMT-proficient cell models, BRCA2 loss enhanced the toxicity of KL-50 (4a) after MGMT depletion through O6BG (Fig. 4, H and I). We observed FANCD2 ubiquitination by KL-50 (4a) specifically in MGMT− cells, suggesting activation of the Fanconi anemia (FA) ICL repair pathway (fig. S9G). As previously reported (40), TMZ (1a) also induced FANCD2 ubiquitination, but only in MGMT−/MMR+ cells. This is an expected result given the propensity of O6MeG (3) to induce stalled replication forks through the process of futile cycling (41).
KL-50 (4a) is active against TMZ (1a)-resistant tumors in vivo
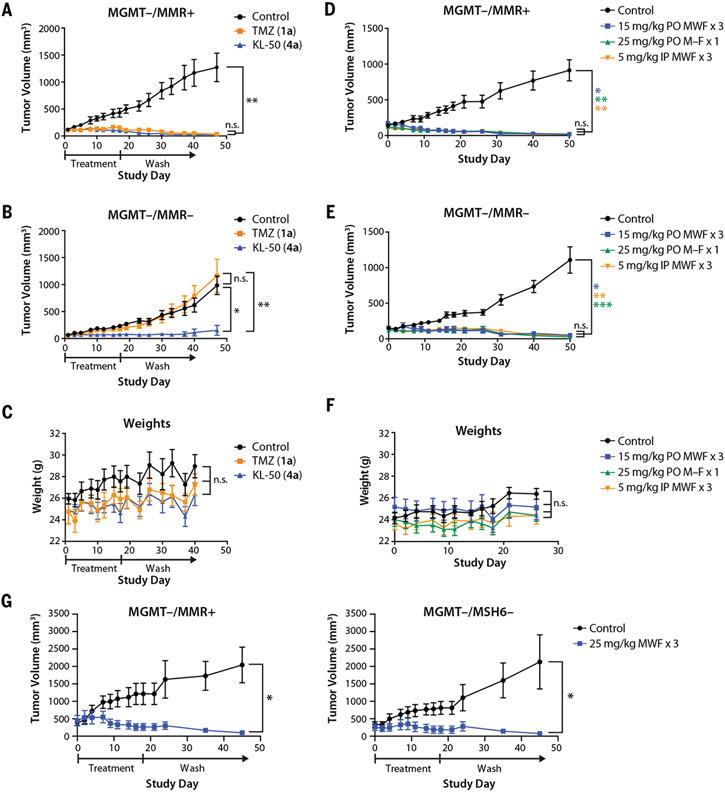
We evaluated the activity of KL-50 (4a) and TMZ (1a) in vivo using mouse flank tumor models derived from the isogenic LN229 MGMT− cell lines. We treated MGMT−/MMR+ and MGMT−/MMR− flank tumors with KL-50 (4a) or TMZ (1a) (5 mg/kg orally Monday, Wednesday, and Friday for 3 weeks) as previously described for TMZ (1a) (35). As expected, TMZ (1a) suppressed tumor growth in the MGMT−/MMR+ tumors (Fig. 5A). KL-50 (4a) was statistically noninferior to TMZ (1a), despite a 17% lower molar dosage owing to its higher molecular weight. In the MGMT−/MMR− tumors, TMZ (1a) demonstrated no efficacy, whereas KL-50 (4a) potently suppressed tumor growth (Fig. 5B). KL-50 (4a) treatment resulted in no statistically significant changes in body weight compared with that of TMZ (1a) or control (Fig. 5C). KL-50 (4a) was effective and nontoxic across different dosing regimens (5, 15, and 25 mg/kg), treatment schedules (Monday, Wednesday, and Friday for 3 weeks, and Monday through Friday for 1 week), and routes of drug administration (oral or intraperitoneal) in mice bearing MGMT−/MMR+ and MGMT−/MMR− flank tumors (Fig. 5, D to F). KL-50 (4a) (25 mg/kg orally Monday, Wednesday, and Friday for 3 weeks) potently suppressed the growth of large (~350 to 400 mm3) MGMT−/MMR+ and MGMT−/MSH6− tumors (Fig. 5G). KL-50 (4a) (25 mg/kg orally Monday through Friday for 1 week) was also effective in an orthotopic, intracranial LN229 MGMT−/MMR− model, whereas TMZ (1a) only transiently suppressed tumor growth (Fig. 6, A and B).
Fig. 5. KL-50 (4a) appears to be safe and efficacious in both MGMT−/MMR+ and MGMT−/MMR− flank tumors over a wide range of treatment regimens and conditions.
(A) Xenograft LN229 MGMT−/MMR+ flank tumors treated with three weekly cycles of oral administration of 10% cyclodextrin control (n = 7 mice), TMZ (1a) (n = 7 mice, 5 mg/kg) or KL-50 (4a) (n = 6 mice, 5 mg/kg) on Monday, Wednesday, and Friday (individual spider plots in fig. S10A). (B) Xenograft LN229 MGMT−/MMR− flank tumors treated with three weekly cycles of oral administration of 10% cyclodextrin control (n = 6 mice), TMZ (1a) (n = 5 mice, 5 mg/kg), or KL-50 (4a) (n = 5 mice, 5 mg/kg) on Monday, Wednesday, and Friday (individual spider plots in fig. S10B). (C) Mean body weight of mice during LN229 flank tumor experiments in (A) and (B). (D to F) Xenograft (D) LN229 MGMT−/MMR+ and (E) LN229 MGMT−/MMR− flank tumors treated with oral administration of 10% cyclodextrin control (n = 7 mice), KL-50 (4a) (n = 6 mice, three cycles of 15 mg/kg on Monday, Wednesday, and Friday), KL-50 (4a) (n = 6 mice, 1 cycle of 25 mg/kg Monday through Friday), or intraperitoneal administration of KL-50 (4a) (n = 7 mice, three cycles of 5 mg/kg on Monday, Wednesday, and Friday) revealed equal efficacy with no observable increases in toxicity, (F) as measured with mice systemic weights (individual spider plots are provided in fig. S10C). (G) Xenograft LN229 MGMT−/MMR+ and LN229 MGMT−/MSH6− flank tumors with a larger average starting tumor size of ~400 mm3 and ~350 mm3, respectively, treated with three weekly cycles of oral administration of 10% cyclodextrin (n = 4 mice) or KL-50 (4a) (n = 4 mice, 25 mg/kg on Monday, Wednesday, and Friday). The study period was limited by control groups that had to be euthanized for exceeding the ethical maximum allowed tumor size, thus ending the study. In (A) to (G), points indicate the mean, and error bars indicate SEM; *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant.
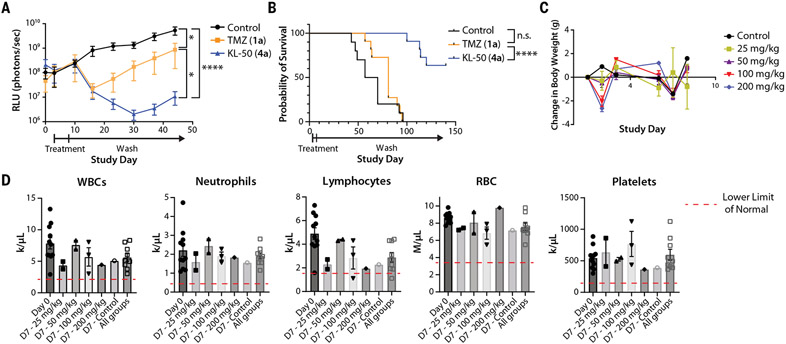
Fig. 6. KL-50 (4a) is efficacious in an LN229 MGMT−/MMR− intracranial model and is well tolerated with limited myelosuppression at supratherapeutic doses.
(A) Mean tumor size as measured with bioluminescent imaging as relative light units (RLU; photons/s) with SEM of xenograft LN229 MGMT−/MMR− intracranial tumors treated with 5 consecutive days of oral administration of 10% cyclodextrin control (n = 10 mice), TMZ (1a) (n = 11 mice, 25 mg/kg), or KL-50 (4a) (n = 11 mice, 25 mg/kg) (individual spider plots are provided in fig. S10D). (B) Kaplan-Meier analysis of intracranial xenograft tumor-bearing mice in (A), treated with 5 consecutive days of oral administration of 10% cyclodextrin control (n = 10 mice), TMZ (1a) (n = 11 mice, 25 mg/kg), or KL-50 (4a) (n = 11 mice, 25 mg/kg) demonstrating a significant survival benefit for KL-50 (4a) compared with both control and TMZ (1a) groups. (C) Mean body weight change with SEM of mice during maximum tolerated dose experiment in non-tumor-bearing mice. (D) Complete blood counts for mice before treatment and 7 days after treatment with escalations of single-dose KL-50 (4a) delivered orally. White blood cells (WBC) lower limit of normal (LLN), 2.2 k/μL; neutrophil LLN, 0.42 k/μl; lymphocyte LLN, 1.7 k/μl; red blood cells (RBC) LLN, 3.47 M/μl; platelet LLN, 155 k/μl. *P < 0.05; ****P < 0.0001; n.s.; not significant.
A focused maximum tolerated dose study revealed that KL-50 (4a) is well tolerated. Healthy mice were treated with escalating doses of KL-50 (4a) (0, 25, 50, 100, and 200 mg/kg orally for 1 dose) and monitored over time for changes in both weights and hematologic profiles. Mice in the higher dosage groups (100 or 200 mg/kg) experienced a greater than 10% weight loss after treatment, which regressed to baseline at the end of 1 week (Fig. 6C). Two of three mice in the 200 mg/kg treatment group became observably ill, warranting euthanasia, but no evidence of toxicity was observed in the remaining cohorts. Because the main dose-limiting systemic toxicity of TMZ (1a) is myelosuppression, we measured complete blood counts for all mice on day 0 before treatment and subsequently on day 7 after drug administration. Overall, there was a trend toward reduced neutrophil and lymphocyte counts with KL-50 (4a) treatment, although average blood counts were within normal physiological ranges (defined as values falling within 2 SD of the average for healthy mice) for all cohorts (Fig. 6D). Taken together, these data demonstrate in vivo efficacy, acceptable systemic tolerability, and central nervous system penetrance of KL-50 (4a).
Discussion
MMR mutation–induced resistance to alkylating agents has been a major barrier to treatment efficacy since the introduction of TMZ (1a) in the early 1990s. Chloroethyl alkylating agents, such as lomustine (14) and MTZ (12a), lack a TI owing to indiscriminate cross-linking activity through chloride displacement (32, 42). Fluoride is not normally susceptible to bimolecular displacement [the homolytic bond dissociation energies of aliphatic C–F and C–Cl bonds are ~115 kcal/mol (481 kJ/mol) and 83.7 kcal/mol (350 kJ/mol), respectively]. However, the appropriate positioning of hydrogen bond donors or covalently attached nucleophiles can promote substitution (43). Thus, the intramolecular displacement pathway (5→6) appears to provide an essential acceleration in the formation of ICLs by KL-50 (4a) while maintaining sufficient lifetime for MGMT-mediated repair of the O6FEtG (5) adduct. This hypothesis is consistent with the reported half-lives of O6-(2-fluoroethyl)guanosine (S1) and O6-(2-chloroethyl)guanosine (S4) (t1/2 = ~18.5 hours and ~18 min) (fig. S1, A and B) (19, 44) and the reported resistance of N7-(2-fluoroethyl)guanosine (S5) to bimolecular fluoride displacement (fig. S1C) (45). We hypothesize that the rate of MGMT reversal of O6FEtG (5) is comparable with the corresponding ethyl and n-propyl adducts, which are processed at 25 to 50% the rate of O6MeG (3) in vitro (46). It has been reported that the mean lethal dose of ICLs in HeLa cells is 230 (47), and TMZ (1a) has been demonstrated to yield 20,600 O6MeG (3) adducts per cell at a dose of 20 μM (48). Assuming that a similar level of O6FEtG (5) lesions are induced by KL-50 (4a) at the IC50 (~20 μM) in MGMT−/MMR− LN229 cells, the number of adducts required to convert to ICLs to generate the mean lethal dose is ~1 in 90, or ~1.1% crosslinking efficiency.
Beyond MGMT-silenced recurrent glioma, it is possible that there may be additional beneficial indications for selective targeting of cancer cells with KL-50 (4a). MGMT silencing has been reported in 40% of colorectal cancers and 25% of non-small cell lung cancer, lymphoma, and head and neck cancers (49). MGMT mRNA expression is also reduced in subsets of additional cancer types, including breast carcinoma, bladder cancer, and leukemia (50). MMR loss, as reported by microsatellite instability, is a well-established phenomenon in multiple cancer types and leads to resistance to various standard-of-care agents.
Our data also suggest that KL-50 (4a) will display a higher TI in tumors with MGMT deficiency and impaired ICL repair, including HR deficiency. The TI of KL-50 (4a) in the DLD1 isogenic model, as measured by the ratio of IC50 values in MGMT+/BRCA2+ cells compared with MGMT−/BRCA2− cells, was ~600-fold, which is larger than canonical cross-linking agents cisplatin (42-fold) or MMC (26-fold). A similar amplification of the TI was seen in the PEO1/4 model with KL-50 (4a) (62-fold) versus cisplatin (13-fold) or MMC (7-fold). HR-related gene mutations have been detected in a substantial number of tumors across multiple cancer types (17.4% in 21 cancer lineages), and methods have been developed to assess for tumor-associated HR deficiency (51).
Approximately half of patients with recurrent MGMT-methylated glioma acquire TMZ (1a) resistance through loss of MMR (18, 52). KL-50 (4a) is designed to fill this therapeutic void. We expect KL-50 (4a) to be amenable to derivatization for improved drug pharmacokinetic properties on the basis of prior work with the imidazotetrazine scaffold (53), facilitating translation to clinical trials. Broadly, incorporating a consideration of the relative rates of DNA modification and DNA repair pathways into the design of therapeutics may lead to the development of additional selective chemotherapies.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Ganesa for her help in generating MMR-deficient isogenic cell lines used in this work. We thank the Yale Center for Molecular Discovery for their assistance with high-throughput immunofluorescence microscopy and image analysis.
Funding:
We gratefully acknowledge financial support from the National Institutes of Health (R35-GM131913 to S.B.H, T32-GM007205 and F30-CA254158 to K.L, and 5RO1CA215453-02 and U19CA264362 to R.S.B.), the Yale Cancer Center (Pilot grant to S.B.H. and R.S.B.), and the CureSearch for Children’s Cancer (Catapult Award to R.S.B.). The Yale Center for Molecular Discovery is supported in part by an NCI Cancer Center Support Grant NIH P30 CA016359. Equipment and libraries were supported in part by the Program in Innovative Therapeutics for Connecticut’s Health.
Footnotes
Competing interest: S.B.H., R.S.B., and K.L. are cofounders and hold equity in Aztek Biosciences, which has exclusive rights to the findings herein. K.L., S.E.G., R.K.S., E.D.H., R.S.B., and S.B.H. are inventors on US Provisional Application 63/290,572 submitted by Yale University, which covers the mechanism of action and use of compound 4a for the treatment of MGMT−/MMR− GBM. K.L., R.S.B., and S.B.H are inventors on US Provisional Applications 63/209,763, 63/212,410, 63/247,645, 63/290,622, 63/290,627, and 63/290,630, which cover compounds 4a, 4b, and related derivatives for the treatment of MGMT− tumors.
Data and materials availability: Crystallographic data for KL-50 (4a) has been deposited in the Cambridge Crystallographic Data Centre as CCDC 2122008/ 2122009. All other data are included in the manuscript and supplementary materials.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Curtin NJ, Nat. Rev. Cancer 12, 801–817 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Bryant HE et al. , Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Farmer H et al. , Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Zatreanu D et al. , Nat. Commun 12, 3636 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reaper PM et al. , Nat. Chem. Biol 7, 428–430 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Lieb S et al. , eLife 8, e43333 (2019).30910006 [Google Scholar]
- 7.Murai J et al. , Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dias MP, Moser SC, Ganesan S, Jonkers J, Nat. Rev. Clin. Oncol 18, 773–791 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Sabatino MA et al. , Mol. Cancer 9, 259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adachi N, So S, Koyama H, J. Biol. Chem 279, 37343–37348 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Fordham SE, Matheson EC, Scott K, Irving JAE, Allan JM, Leukemia 25, 1046–1049 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Sawant A, Kothandapani A, Zhitkovich A, Sobol RW, Patrick SM, DNA Repair 35, 126–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell EH et al. , JAMA Oncol. 4, 1405–1409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G-M, Cell Res. 18, 85–98 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Hegi ME et al. , N. Engl. J. Med 352, 997–1003 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Yoshimoto K et al. , Front. Oncol 2, 186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Touat M et al. , Nature 580, 517–523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Y et al. , Neuro-oncol. 23, 1872–1884 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong WP, Kirk MC, Ludlum DB, Biochem. Pharmacol 32, 2011–2015 (1983). [DOI] [PubMed] [Google Scholar]
- 20.Hentschel S, Alzeer J, Angelov T, Schärer OD, Luedtke NW, Angew. Chem. Int. Ed 51, 3466–3469 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Chan CL, Wu Z, Ciardelli T, Eastman A, Bresnick E, Arch. Biochem. Biophys 300, 193–200 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Shibata T et al. , Nucleic Acids Res. 34, 1884–1891 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pegg AE, Chem. Res. Toxicol 24, 618–639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez H, Rebeyrol A, Nelms TB, Dolbier WR Jr., J. Fluor. Chem 135, 167–175 (2012). [Google Scholar]
- 25.Wang Y, Wheelhouse RT, Zhao L, Langnel DAF, Stevens MFG, J. Chem. Soc., Perkin Trans. 1 10, 1669–1676 (1998). [Google Scholar]
- 26.Double JA, Bibby MC, J. Natl. Cancer Inst 81, 988–994 (1989). [DOI] [PubMed] [Google Scholar]
- 27.Fairbairn LJ, Chinnasamy N, Lashford LS, Chinnasamy D, Rafferty JA, Cancer Gene Ther. 7, 233–239 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Liu L, Markowitz S, Gerson SL, Cancer Res. 56, 5375–5379 (1996). [PubMed] [Google Scholar]
- 29.Jiricny J, Cold Spring Harb. Perspect. Biol 5, a012633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesa S, Sule A, Sundaram RK, Bindra RS, Sci. Rep 12, 5827 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna DJ, Gallus M, McKeown SR, Downes CS, McKelvey-Martin VJ, DNA Repair 2, 879–890 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Brent TP, Gonzaga PE, Smith DG, in DNA Repair Mechanisms and Their Biological Implications in Mammalian Cells, Lambert MW, Laval J, Eds. (Springer, 1989), pp. 619–638. [Google Scholar]
- 33.Bossuet-Greif N et al. , mBio 9, e02393–17 (2018).29559578 [Google Scholar]
- 34.Xue M, Wernke KM, Herzon SB, Biochemistry 59, 892–900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson CB et al. , Cancer Res. 79, 4331–4338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothkamm K et al. , Environ. Mol. Mutagen 56, 491–504 (2015). [DOI] [PubMed] [Google Scholar]
- 37.O’Brien V, Brown R, Carcinogenesis 27, 682–692 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Vare D et al. , DNA Repair 11, 976–985 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Matsuno Y et al. , Nat. Commun 10, 3925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CC, Taniguchi T, D’Andrea A, J. Mol. Med 85, 497–509 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Mojas N, Lopes M, Jiricny J, Genes Dev 21, 3342–3355 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leelatian N, Hong CS, Bindra RS, Neurosurg. Clin. N. Am 32, 171–180 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Pan Y, ACS Med. Chem. Lett 10, 1016–1019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker S, Kirk MC, Ludlum DB, Biochem. Biophys. Res. Commun 148, 1124–1128 (1987). [DOI] [PubMed] [Google Scholar]
- 45.Ludlum DB, Tong WP, Biochem. Pharmacol 27, 2391–2394 (1978). [DOI] [PubMed] [Google Scholar]
- 46.Morimoto K, Dolan ME, Scicchitano D, Pegg AE, Carcinogenesis 6, 1027–1031 (1985). [DOI] [PubMed] [Google Scholar]
- 47.Lawley PD, Phillips DH, Mutation Res. 355, 13 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Stratenwerth B et al. , Mol. Cancer Ther 20,1789–1799 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG, Cancer Res. 59, 793–797 (1999). [PubMed] [Google Scholar]
- 50.Thomas A et al. , Cancer Res. 77, 823–826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies H et al. , Nat. Med 23, 517–525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cahill DP, Codd PJ, Batchelor TT, Curry WT, Louis DN, Clin. Neurosurg 55, 165–171 (2008). [PubMed] [Google Scholar]
- 53.Svec RL et al. , ACS Chem. Biol 13, 3206–3216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.