Abstract
The antitumor efficacy of cancer immunotherapy can correlate with the presence of certain bacterial species within the gut microbiome. Yet, many of the molecular mechanisms that impact host response to immunotherapy remain elusive. Here, we show that members of the bacterial genus Enterococcus modulate response to checkpoint inhibitor immunotherapy in mouse tumor models. Active enterococci express and secrete orthologs of the NlpC/p60 peptidoglycan hydrolase SagA that generate immune-active muropeptides. Expression of SagA in non-protective E. faecalis was sufficient to promote anti–PD-L1 response, and its activity required the peptidoglycan sensor NOD2. Notably, SagA-engineered probiotics or synthetic muropeptides also augmented anti–PD-L1 antitumor efficacy. Together, our data suggest that microbiota species with unique peptidoglycan remodeling activity and muropeptide-based therapeutics may enhance cancer immunotherapy and could be leveraged as next-generation adjuvants.
One Sentence Summary
A conserved family of secreted NlpC/p60 peptidoglycan hydrolases from Enterococcus and muropeptides promote antitumor activity of checkpoint inhibitor immunotherapy.
Cancer immunotherapy harnesses the patient’s immune system to impede tumor growth and has demonstrated clinical success across a range of solid and hematological tumors (1-3). In particular, antibodies that target immune checkpoint inhibitor proteins such as CTLA-4 and PD-1/PD-L1 have been approved to treat a number of human cancers (3). However, patient response to immune checkpoint inhibitors is variable (3). The success of checkpoint blockade relies on numerous factors including mutational burden of the malignancy (4), successful tumor antigen presentation (5), recruitment and infiltration of lymphocytes (6), and signaling cues within the tumor microenvironment (7). Recently, the gut microbiota has emerged as a potent new factor associated with the efficacy of anti–CTLA4, anti–PD-1, and anti–PD-L1 treatment (8-14). In animal and human cohorts, the presence of specific microbial species was correlated with responsiveness to checkpoint blockade agents. The antitumor activity of these microbes was recapitulated in preclinical mouse models upon co-housing, fecal transplant, or direct inoculation, suggesting that the correlated microorganisms are direct causative agents of improved therapeutic response. Nevertheless, little is known about the molecular mechanisms by which immune modulatory microbes may act.
Multiple analyses of the commensal microbiome from human cohorts treated with immunotherapies targeting PD-1 have revealed that the bacterial genus Enterococcus is enriched in responding patients (12, 13). Although antibiotic-resistant strains of E. faecium and E. faecalis can be pathogenic (15), commensal strains of these bacteria have been used as probiotics in animals and humans (16). Recent studies also suggested that Enterococcus species can trigger immune signaling pathways and modulate infection (17-19), autoimmunity (20), and graft-versus-host-disease (21). These observations prompted our inquiry into whether specific Enterococcus species and strains can improve response to checkpoint inhibitor immunotherapy.
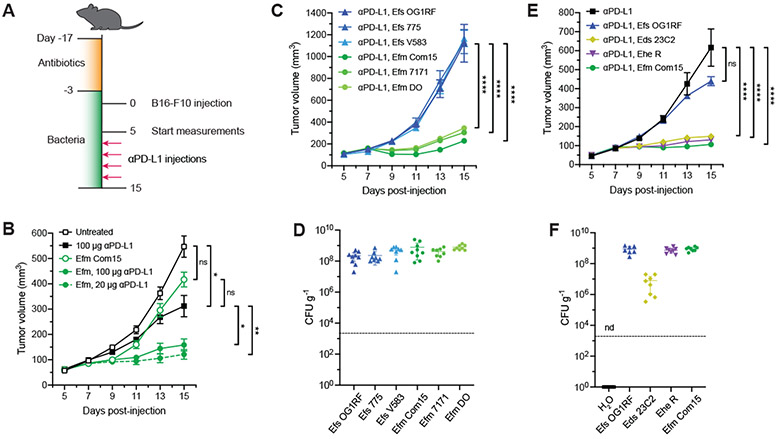
To evaluate specific enterococci and their mechanism of action, we utilized mouse tumor models and oral administration of bacteria. Here, specific pathogen-free (SPF) C57BL/6 mice from The Jackson Laboratory were pretreated for two weeks with a broad-spectrum antibiotic cocktail (ampicillin, colistin, and streptomycin) to clear resident microbial species that may confound effects caused by Enterococcus species and promote Enterococcus colonization. Animals were subsequently provided water supplemented with enterococci. Supplemented animals were then subcutaneously implanted with B16-F10 melanoma cells and treated with anti–PD-L1 (Fig. 1A). We first focused on the two most common Enterococcus species in the human gut microbiota: E. faecium (Efm) and E. faecalis (Efs). We found that supplementation with the human commensal E. faecium strain Com15 without therapeutic intervention did not alter tumor growth (Fig. 1B). However, treatment of supplemented animals with anti–PD-L1 immunotherapy at both high and low doses showed a significant decrease in tumor size compared to mice treated with anti–PD-L1 alone. We selected the low anti–PD-L1 dose protocol going forward and compared the activity of E. faecium with E. faecalis across multiple strains of each species—including human-isolated, reference type, and multi-drug resistant strains—to ascertain if the observed synergistic activity was unique to E. faecium Com15. Inhibition of tumor growth was observed across all three tested isolates of E. faecium and not for any strain of E. faecalis analyzed (Fig. 1C). To ensure that these effects were not due to differences in bacterial load, fecal samples were plated onto Enterococcus-selective medium and enumerated. All bacterial strains yielded similar amounts of colony forming units (CFU, Fig. 1D), indicating that the observed antitumor activity of E. faecium strains was not due to differences in bacterial amount in the gut. In addition to E. faecium, 16S rRNA analysis of gut microbiota from responsive human patients also uncovered the enrichment of other Enterococcus species (12). We found that antitumor activity was conserved across multiple species including E. durans (Eds), E. hirae (Ehe), and E. mundtii (Emi) (Figs. 1E and S1A). CFU analysis of the colonized animals again revealed that activity did not correlate with bacterial load (Figs. 1F and S1B).
Fig. 1. Specific enterococci improve efficacy of anti–PD-L1 immunotherapy in B16-F10 melanoma model.
(A) Schematic of tumor growth model in SPF mice with antibiotic treatment and oral enterococci supplementation. Days are indexed based on the day of tumor injection. Mice were provided antibiotic-containing water ab libitum for two weeks followed by water supplemented with the indicated enterococci for the remainder of the experiment. Animals were then subcutaneously implanted with B16-F10 melanoma cells, and tumor volume measurements started when tumors reached ~50-100 mm3 (day 5). Mice were treated with anti–PD-L1 by intraperitoneal injection every other day starting two days after the start of the tumor measurement. For all data except for (B), 20 μg anti–PD-L1 was used for each injection. (B) B16-F10 tumor growth in antibiotic-treated animals that were supplemented with or without E. faecium (Efm) Com15 and treated with or without anti–PD-L1 starting on day 7 at doses indicated in the legend. n = 7-8 mice per group. (C) B16-F10 tumor growth in antibiotic-treated mice that were supplemented with the indicated E. faecalis (Efs) and E. faecium (Efm) strains and treated with anti–PD-L1 starting on day 7. n = 7-8 mice per group. (D) Colony forming unit (CFU) analysis of E. faecalis (Efs) and E. faecium (Efm) strains in fecal samples harvested from animals as treated in (C). (E) B16-F10 tumor growth in antibiotic-treated mice that were supplemented with the indicated enterococci strains and treated with anti–PD-L1 starting in day 7. n = 8-9 mice per group. (F) CFU analysis of enterococci in fecal samples harvested from animals as treated in (E). nd = not detected. For (B), (C), and (E), data represent mean ± s.e.m. and were analyzed by mixed-effects model with Tukey’s multiple comparisons post-hoc test. *P < 0.05, **P < 0.01, ****P < 0.0001, ns = not significant. For (D) and (F), each symbol represents one mouse. Dotted lines indicate the limit of detection (2,000 CFU g−1). Data represent means ± 95% confidence interval.
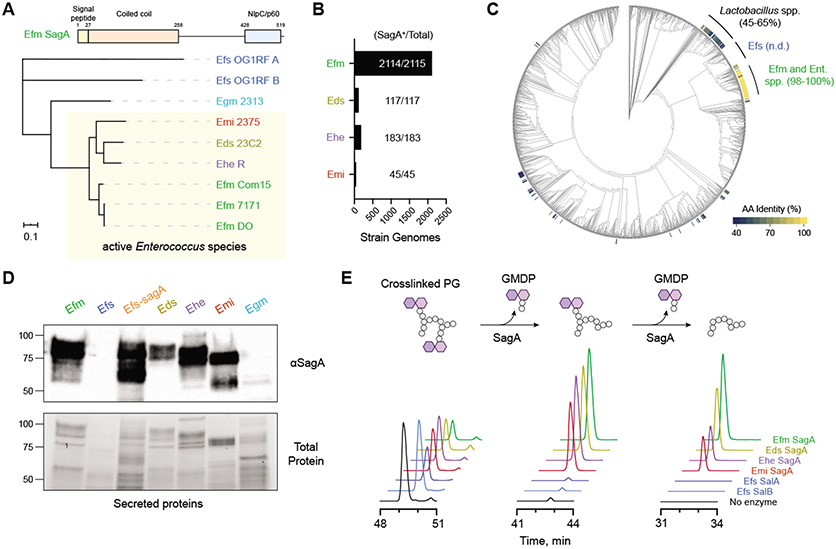
To explain the species-specific differences we observed across the Enterococcus genus, we examined possible sources of their immunomodulatory activity. Notably, our previous work indicated that E. faecium has unique peptidoglycan composition and remodeling capabilities to enhance host tolerance to enteric pathogens (17-19). To compare peptidoglycan composition across the enterococci, we isolated sacculus of each species and analyzed the digested peptidoglycan fragments by high performance liquid chromatography–mass spectrometry (HPLC-MS). All four of the immunotherapy-active enterococci (E. faecium, E. durans, E. hirae, and E. mundtii) showed similar peptidoglycan fragment patterns compared to the nonactive species E. faecalis and E. gallinarum (Egm) (Fig. S2A). Additionally, all four immunotherapy-active species showed an abundance of smaller, non-crosslinked peptidoglycan fragments, suggesting higher levels of peptidoglycan remodeling and turnover. The modification of peptidoglycan stem peptides is catalyzed by conserved families of amidases and peptidases (22), so we examined Enterococcus genomic assemblies for unique expression of peptidoglycan remodeling enzymes (Fig. S3, Tables S1 and S2). Based on this analysis, we identified a cluster of NlpC/p60 hydrolases that were highly conserved through all active enterococci (Fig. 2A). This group of related enzymes contained the peptidoglycan hydrolase secreted antigen A (SagA) from E. faecium, which we previously demonstrated improves host immunity against enteric infections (17-19). Using primary sequence alignment, we found that the putative SagA orthologs from other active enterococci were highly homologous, with greater than 90% sequence identity within the C-terminal NlpC/p60 hydrolase domain (Fig. S2B). Conversely, the closest related protein in E. gallinarum showed only 67% sequence identity in the predicted catalytic domain. No direct orthologs were found in E. faecalis, and the two most similar conserved enzymes, sagA-like proteins A and B (SalA and SalB) (23), possessed distinct C-terminal domains. Structural modeling revealed that the putative SagA orthologs also shared similar secondary structural organization compared to a crystal structure of the NlpC/p60 hydrolase domain from E. faecium SagA (Fig. S4).
Fig. 2. Protective enterococci express and secrete active orthologs of the peptidoglycan NlpC/p60 hydrolase SagA.
(A) Domain structure and unrooted phylogenetic clustering of putative SagA ortholog protein sequences identified by global peptidoglycan peptidase analysis of enterococci species and strains along with the closest entries from E. gallinarum (Egm) and E. faecalis (Efs) based on IQ-Tree analysis. Numbers above each domain denote amino acid residue boundaries. Active strains are indicated by the yellow box. Scale bar represents sequence distance. (B) Bar plot and quantification of Enterococcus genomes containing SagA orthologs. (C) Cladogram of Human Microbiome Project isolates organized by 16S rRNA homology with heat map indicating amino acid (AA) sequence identity of putative SagA orthologs. n.d. = not detected; Ent. spp. = Enterococcus strains without an assigned species name. (D) Western blot detection of secreted SagA orthologs harvested from overnight cultures of the indicated enterococci using antiserum raised against E. faecium (Efm) Com15 SagA. Bottom panel shows total protein loading. Numbers indicate estimated molecular weight (kDa). (E) In vitro activity of purified SagA orthologs. Data are shown as extracted LC-MS ion chromatograms of a crosslinked peptidoglycan substrate and two iterative hydrolysis products after incubating a mixture of peptidoglycan fragments with purified SagA orthologs from the indicated species for 16 hours at 37 °C. Peak heights are shown as relative intensity of ion abundance, and all chromatograms are shown on the same scale.
To determine if these SagA-like enzymes are found in human patients treated with cancer immunotherapy, we performed sequence alignments using the NlpC/p60 domain of E. faecium Com15 SagA on clinical metagenomic data obtained from patients prior to checkpoint blockade treatment (10-14). However, we were unable to find SagA orthologs within the datasets, likely due to the low genome coverage of E. faecium across these samples (9.34%, Table S3). Therefore, we evaluated if SagA-like enzymes are conserved across all sequenced strains of active enterococci, which include both clinical and environmental isolates. Query of all RefSeq database genomes from E. faecium, E. durans, E. hirae, and E. mundtii showed SagA orthologs to be almost universally present and conserved (Fig. 2B, Table S4). We further confirmed that SagA orthologs are found in humans through alignment of the SagA NlpC/p60 domain with genomes within the Human Microbiome Project (24) (Fig. 2C, Table S5). As expected, we saw nearly 100% conservation of the sequence within E. faecium isolates as well as a complete lack of SagA orthologs within E. faecalis. Together, these data indicate that SagA orthologs are present and conserved across active Enterococcus species found in the human microbiome.
To directly detect the expression of SagA orthologs in enterococci, we performed Western blotting on both the secreted and cell-associated protein fractions from each species. Using antiserum raised against E. faecium SagA (19), strong signals were observed in the supernatant fractions of E. faecium, E. durans, E. hirae, and E. mundtii but not E. faecalis or E. gallinarum (Figs. 2D and S2C), suggesting that the similar proteins are both highly expressed and secreted by these species. The signal was confirmed to be SagA-dependent using a strain of E. faecalis OG1RF engineered with a chromosomal sagA insertion (Efs-sagA). As expected, all tested strains of E. faecium also showed similar expression and secretion patterns for SagA (Fig. S2D). To determine if these SagA orthologs were functional, we analyzed the hydrolytic activity of the purified recombinant proteins on peptidoglycan in vitro (Figs. 2E and S5). Proteins from E. durans, E. hirae, and E. mundtii showed d,l-endopeptidase activity against a model crosslinked peptidoglycan fragment similar to E. faecium SagA to produce the muropeptide GlcNAc-muramyl dipeptide (GMDP, Figs. 2E and S6). The hydrolytic activity of SagA was confirmed using a mutant construct lacking the cysteine active site residue, which was conserved in all orthologs (Fig. S7). E. faecalis SalB did not hydrolyze peptidoglycan at detectable levels in our assay, whereas SalA cleaved the crosslinked fragment in the cross-bridge region rather than the peptide stem (Figs. 2E and S8). These results show that immunotherapy-active enterococci possess similar peptidoglycan composition and remodeling activity.
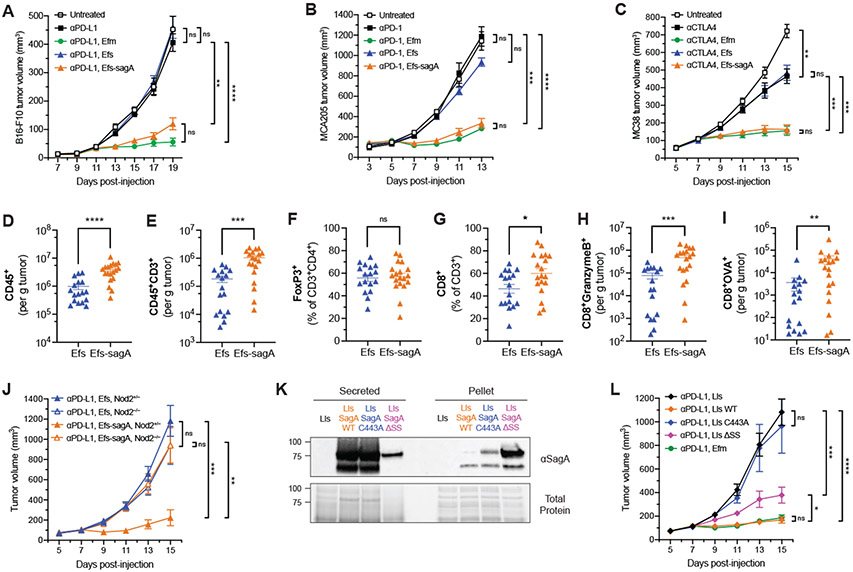
We then investigated whether SagA was sufficient to enhance the efficacy of anti-PD-L1 checkpoint inhibitor immunotherapy. Because sagA is an essential gene in E. faecium (25), we compared the inactive, parental E. faecalis OG1RF strain with the engineered, SagA-expressing strain E. faecalis–sagA (Fig. 2D). The peptidoglycan profile of E. faecalis–sagA showed changes consistent with increased d,l-endopeptidase activity (Fig. S9A), indicative of active SagA expression (Fig. 2D). Antibiotic-treated animals that were orally supplemented with E. faecalis–sagA showed a significant decrease in B16-F10 tumor growth upon anti–PD-L1 therapy compared to the parental E. faecalis strain (Fig. 3A). This antitumor activity was similar to the phenotype observed in E. faecium-supplemented animals. Furthermore, fecal CFU analysis showed that E. faecalis–sagA exhibited similar bacterial load as the parental E. faecalis strain and E. faecium (Fig. S9B). To ensure that antitumor effects were not due to overall changes in microbiome composition, we performed 16S rRNA analysis of fecal samples collected from our antibiotics-treated model system. Antibiotic pre-treatment yielded a sharp decrease in read counts, with a maximal decreased observed at two weeks (Fig. S10A). Supplementation with the tested strains led to a high bacterial load by day 3 (Fig. S10A), with nearly 100% of the detected OTUs as Enterococcus (Figs. S10, B-E). Moreover, microbiomes maintained roughly 5% abundance of Enterococcus 14 days after the start of supplementation with an increase in spore-forming bacteria (Figs. S10, D and E), likely due to incomplete clearance by antibiotics. As expected, beta diversity analysis confirmed that the composition of the samples changed over time after antibiotic treatment and Enterococcus supplementation (Fig. S10F). Permutational multivariate ANOVA testing of the populations at day 14 showed no significant differences between treatment groups using both Bray-Curtis and weighted Unifrac principal coordinate analyses (PCoA) (Fig. S10G). Moreover, operational taxonomic unit (OTU) abundance quantification did not reveal other OTUs that consistently correlated with biological activity (Table S6), with most differentially prevalent OTUs found in only one of the two cages per condition.
Fig. 3. SagA improves checkpoint inhibitor immunotherapy and elicits an adaptive immune response.
(A) B16-F10 tumor growth in antibiotic-treated mice that were supplemented with the indicated enterococci and treated with anti–PD-L1 starting on day 9. n = 8 mice per group. (B) MCA205 tumor growth in antibiotic-treated mice that were supplemented with the indicated enterococci and treated with anti–PD-1 starting on day 5. n = 8 mice per group. (C) MC38 tumor growth in antibiotic-treated mice that were supplemented with the indicated enterococci and treated with anti–CTLA-4 starting on day 7. n = 8 mice per group. (D) to (I) Quantification of tumor infiltrating CD45+ cells (D), total CD3+ T cells (E), FoxP3+ regulatory T cells (F), CD8+ T cells (G), granzyme B+ CD8+ T cells (H), and OVA-specific CD8+ T cells (I) from B16-OVA tumors in mice supplemented with E. faecalis (Efs) or Efs-sagA harvested five days after the start of anti–PD-L1 treatment by flow cytometry. Data are pooled from two independent experiments of 7-10 mice per group per experiment; each symbol represents one mouse. (J) B16-F10 tumor growth in antibiotic-treated Nod2+/− or Nod2−/− mice that were supplemented with Efs or Efs-sagA and treated with anti–PD-L1 starting on day 7. n = 9-11 mice per group. (K) Western blot detection of ectopically expressed Efm Com15 SagA in secreted protein and cell pellet fractions harvested from overnight cultures of the indicated engineered Lactococcus lactis (Lls) strains using antiserum raised against Efm Com15 SagA. Bottom panels show total protein loading. Numbers indicate estimated molecular weight (kDa). WT = wild-type, ΔSS = signal sequence deletion. (L) B16-F10 tumor growth in antibiotic-treated mice that were supplemented with the indicated Lactococcus lactis (Lls) strains and treated with anti–PD-L1 starting on day 7. n = 9-11 mice per group. Data for (A)-(C) and (K)-(L) represent mean ± s.e.m. and analyzed by mixed-effects model with Tukey’s multiple comparisons post-hoc test. Data for (D)-(I) represent mean ± s.e.m. and analyzed by Mann-Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = not significant.
We then tested the outcome of SagA expression on checkpoint inhibitor therapy in an animal model with an intact, complex microbiota. SPF C57BL/6 mice from Taconic Biosciences were chosen as previous reports have found that these mice do not respond strongly to anti–PD-L1 therapy similar to germ-free animals (9, 13). Here, these mice were directly supplemented with enterococci-containing water without antibiotic pre-treatment prior to B16-F10 implantation (Fig. S11A). As seen in the antibiotic-pretreated model, animals supplemented with E. faecium or E. faecalis–sagA showed a significant decrease in tumor growth compared to treatment with antibody alone or antibody and parental E. faecalis (Fig. S11B). We found much lower overall Enterococcus CFU counts that did not significantly differ between the supplemented mice, suggesting that this effect did not require enteric domination by the newly administered, active enterococci (Fig. S11C). Moreover, we observed no major changes in observed OTU counts, alpha diversity, or taxonomic composition upon Enterococcus administration throughout the experiment (Fig. S11, D-F). No significant difference in microbiota composition was observed between samples at day 2 by beta diversity analysis (Fig. S11G). Permutational multivariant ANOVA testing of the weighted UniFrac PCoA at day 14 also showed no difference between groups (Fig. S11H). Finally, differential OTU abundance analysis revealed no OTUs that correlated with biological activity (Table S7), suggesting that the observed anti-tumor effects were Enterococcus-driven.
We next interrogated whether SagA-expressing bacteria would also improve the efficacy of other checkpoint antibodies against different cancer cell types beyond anti–PD-L1 treatment of B16-F10 melanoma. Subcutaneous tumors were established with MCA205 fibrosarcoma or MC38 colorectal carcinoma cells in antibiotic-pretreated animals colonized by enterococci and then treated with anti–PD-1 or anti–CTLA-4 immunotherapy, respectively. In both cases, we also observed a significant decrease in tumor growth when animals were cotreated with SagA-expressing enterococci and checkpoint inhibitor (Fig. 3, B and C). Given the broad efficacy of these enterococci with different targeted therapies, we then asked whether this effect was mediated by an adaptive immune response. Using our antibiotic-pretreated model, we subcutaneously implanted B16-OVA tumor cells into animals colonized with either parental E. faecalis or E. faecalis–sagA and then treated with anti–PD-L1. Tumors were harvested the day following the third antibody treatment (five days after the start of treatment), and tumor-infiltrating lymphocytes were quantified by flow cytometric analysis (Fig. S12). Animals colonized with E. faecalis–sagA showed an overall increase in the absolute amount of intratumoral CD45+ leukocytes as well as CD3+ lymphocytes (Fig. 3, D and E). The composition of tumor-infiltrating CD3+ lymphocytes showed an increase in the proportion of CD8+ T cells but no change in CD4+FoxP3+ regulatory T cells (Fig. 3, F and G). Tumors contained higher amounts of CD8+ T cells that expressed granzyme B (Fig. 3H), a marker for activated cytotoxic T lymphocytes (26). Tetramer staining also revealed a significant increase in the number of OVA-specific CD8+ T cells (Fig. 3I), consistent with enhanced priming of a tumor antigen-specific immune response (9, 13).
To characterize the microbial mechanism of immune activation, we first examined whether SagA contributed to bacterial dissemination, as Enterococcus translocation from the gut has been implicated during autoimmunity (20), chemotherapy treatment (27), and alcoholic hepatitis (28). CFU analysis of mesenteric lymph nodes and whole spleens of supplemented animals only showed low levels of live bacteria in proximal tissues (Fig. S9C). The bacterial load in mesenteric lymph nodes was independent of SagA expression, suggesting that SagA did not improve barrier transit for live bacteria in our studies. We then turned our attention to peptidoglycan remodeling by SagA and the generation of muropeptides such as GMDP as a potential mechanism of action (19). Indeed, we found that NOD2, a key pattern recognition receptor for muropeptides (29, 30), was required for the anti–PD-L1 antitumor activity in animals supplemented with E. faecalis–sagA (Fig. 3J). To evaluate the enzymatic activity and utility of SagA to improve checkpoint blockade, we investigated heterologous expression in probiotic bacteria. Lactococcus lactis (Lls) has been explored extensively as a live, oral probiotic to deliver bioactive proteins and enzymes (31). Therefore, we produced L. lactis strains that chromosomally expressed wild-type, catalytically inactive (C443A), or secretion-deficient (ΔSS) SagA. All three constructs expressed well, and the wild-type and C443A mutant SagA constructs showed higher signal in the secreted fraction as expected (Fig. 3K). Antibiotic-pretreated animals were orally supplemented with these strains as well as parental L. lactis or E. faecium and used to monitor B16-F10 responsiveness to anti–PD-L1. Animals supplemented with L. lactis expressing wild-type SagA showed similar tumor growth inhibition as E. faecium-treated animals (Fig. 3L). L. lactis expressing catalytically inactive SagA (C443A) were not able to recapitulate the antitumor phenotype, indicating that the enzymatic activity of SagA is required to impede tumor growth. We found that the SagA secretion-deficient strain (ΔSS) did slow tumor growth, which suggests that the low amount of SagA secreted by this strain may have been sufficient to partially inhibit tumor growth. Alternatively, active, non-secreted SagA may escape from L. lactis due to cell lysis in the gut.
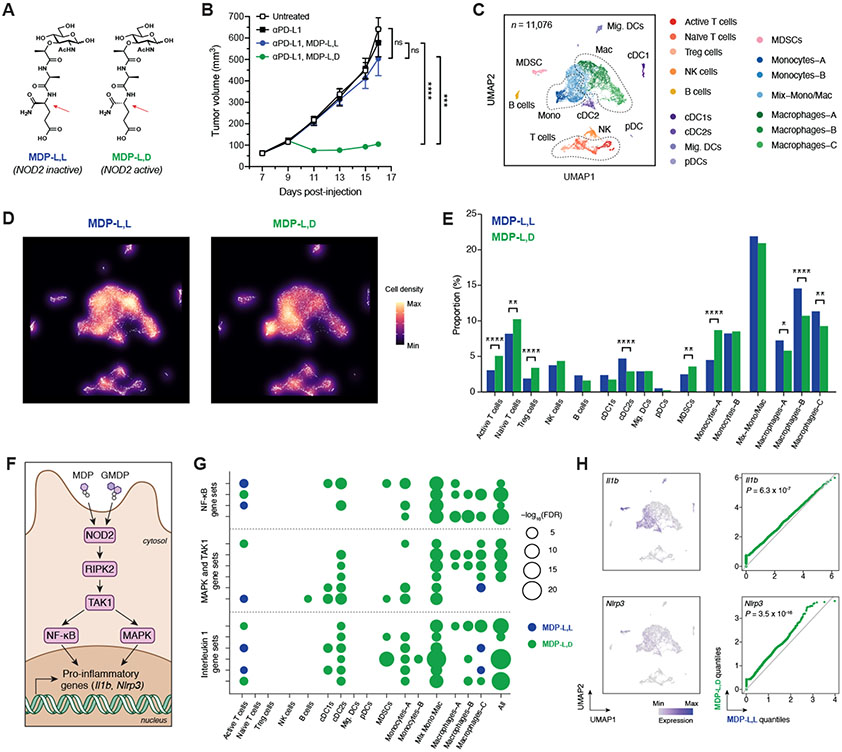
Because SagA NlpC/p60 hydrolase activity was required, we asked whether synthetic muropeptide analogs of SagA enzymatic products such as muramyl dipeptide (MDP) could also elicit an improved response to checkpoint therapy. For these experiments, the NOD2-active MDP-l,d isomer or the inactive MDP-l,l diastereomer (Fig. 4A) were co-administered by intraperitoneal injection with anti–PD-L1. Animals that received active MDP-l,d along with anti–PD-L1 showed a significant antitumor effect that was not observed upon co-administration of the MDP-l,l negative control, suggesting the NOD2-active muropeptide agonist was sufficient to improve checkpoint blockade (Fig. 4B). To better understand the mechanism(s) by which muropeptides augment checkpoint blockade, we profiled tumor-infiltrating leukocytes in the B16 melanoma model using single cell-based RNA sequencing (scRNA-seq). After three treatments of either MDP stereoisomer with anti–PD-L1, CD45+ cells from dissociated tumors were sorted, pooled, and sequenced (Figs. 4C and S13), Tumors treated with the active MDP-l,d isomer showed a significant increase in the proportion of intratumoral T lymphocytes (Fig. 4, D and E), similar to our results using flow cytometry (Fig. 3F). The MDP-l,d sample also showed higher levels of checkpoint genes Ctla4 and Pdcd1, consistent with increased TCR-mediated signaling (Fig. S14A). Moreover, marker genes of cytotoxic T cell activity including Ifng, Gzmb, Ltb, and Prf1 were enriched in MDP-l,d–treated tumors (Fig. S14B). We also observed significant shifts in myeloid cell populations upon MDP-l,d administration (Fig. 4, D and E), with decreases in all macrophage clusters and an increase in a specific monocyte population characterized by Cx3cr1 and Nr4a1 expression (Figs. S14C). NOD2 activation has previously been linked to a similar increase of Cx3cr1+ monocytes with patrolling-like activities in circulation (32), and these results provide direct evidence that this monocyte subclass can accumulate in inflammatory microenvironments. Infrequent but detectable Nod2 expression was observed across multiple myeloid populations as expected (29, 30) (Fig. S14D). Significant enrichment of hallmark gene sets (33) for inflammatory response and NF-κB signaling were found in the MDP-l,d dataset (Table S8). Further uncurated gene set enrichment analysis revealed widespread changes in inflammatory, metabolic, and other innate immune pathways within the myeloid cell clusters (Tables S9-S24). NOD2 signal transduction occurs through the activation of transcription factor NF-κB and mitogen-activated protein kinase (MAPK) phosphorylation cascades via RIPK2 and TAK1 (Fig. 4F) (29, 30). Accordingly, we found enrichment of multiple canonical gene sets involving these proteins, particularly within the monocyte and macrophage clusters that shifted upon muropeptide treatment (Fig. 4G). Further confirming increased NF-κB activation, we found that NF-κB target transcripts Il1b and Nlrp3 (34) were increased in the MDP-l,d sample along with IL-1β pro-inflammatory gene sets throughout the intratumoral myeloid compartment (Fig. 4, G and H).
Fig. 4. Peptidoglycan fragment MDP enhances checkpoint blockade efficacy and generates a pro-inflammatory tumor microenvironment.
(A) Chemical structures of the active (l,d) and inactive (l,l) diastereomers of muramyl dipeptide (MDP). Arrows indicate the single altered stereocenter. (B) B16-F10 tumor growth in antibiotic-treated, non-supplemented mice treated with anti–PD-L1 and either MDP-l,d or MDP-l,l starting on day 9. n = 7-8 mice per group. (C) Uniform manifold approximation and projection (UMAP) plots of scRNA-seq data from CD45+ tumor-infiltrating cells after treatment with anti–PD-L1 and MDP diastereomers. n = 11,076 total cells pooled from six animals per condition. (D) Density plot and (E) quantification of immune cell clusters. (F) Schematic of NOD2-dependent signaling of pro-inflammatory genes. GMDP = GlcNAc-muramyl dipeptide. (G) Bubble plot for enrichment of curated canonical pathway gene sets involving NF-κB, MAPK/TAK1, and interleukin 1 across cell clusters. (H) UMAP plots and paired quantile-quantile plots of NF-κB target genes Il1b and Nlrp3. For (B), data represent mean ± s.e.m. and were analyzed by mixed-effects model with Tukey’s multiple comparisons post-hoc test. For (E), data represent absolute cell counts and were analyzed by Pearson’s chi-squared test for count data with Holm’s correction for multiple comparisons. For (G), false discovery rates (FDR) were obtained by model-based analysis of single transcriptomics (MAST). For (H), data were analyzed with two-sided Wilcoxon rank-sum test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = not significant.
Together, our data indicate that enterococci with unique NlpC/p60 peptidoglycan hydrolase activity can generate NOD2-active muropeptides and modulate the efficacy of checkpoint blockade immunotherapy in vivo (Fig. S15). Although bacteria enriched in responding patients do not correlate well by phylogeny (35), specific enterococci and other microbiota species with privileged cell wall composition and remodeling activity could provide functional indicators of therapeutic efficacy. Our results demonstrating the prevalence of conserved SagA-like enzymes throughout human-associated Enterococcus species indicate that the production of immune active muropeptides may be prevalent across human microbiomes. Beyond enterococci, the detection of potential NlpC/p60 orthologs in other genera such as Lactobacillus (Fig. 2C, Table S5), which has also been identified in immunotherapy-responsive patients (13), may provide additional specific microbiota correlations to predict and improve patient outcomes. In addition, recent genetic analyses of Bifidobacterium bifidium strains that synergized with PD-1 blockade showed an enrichment of peptidoglycan biosynthetic genes (36). These observations suggests that peptidoglycan remodeling may be a broad mechanism to augment immunotherapy efficacy, which requires additional functional studies of specific microbiota peptidoglycan remodeling factors. As peptidoglycan fragments can disseminate into circulation and prime systemic immune responses (37, 38), the presence of NlpC/p60 hydrolases and NOD2-active muropeptides may be clinically relevant biomarkers for predicting personalized therapeutic responses. In addition, our results suggest that peptidoglycan remodeling enzymes may be utilized to reprogram probiotic bacteria as a novel therapeutic approach for enhancing the efficacy of checkpoint blockade inhibitors. Lastly, our findings corroborate a wealth of emerging evidence that NOD2 stimulation by MDP and its analogs can alter host immunity through multiple pathways including the direct activation of macrophages for tumor cell clearance (39), epigenetic reprogramming of monocytes (40, 41), generation of conventional type 1 dendritic cells (42), and priming of dendritic cells for cross-presentation to CD8+ T cells (43), which together have been implicated in trained immunity (44). Therefore, our work underscores how microbially-produced or synthetic small molecules that can activate peptidoglycan pattern recognition receptors could be employed as next-generation adjuvants for immunotherapy.
Supplementary Material
Acknowledgments:
We thank S. Mazel, A. Keprova, M. Jaimes, D. Tran, and the Rockefeller Flow Cytometry Resource Center for assistance with flow cytometric experiments, G. Putzel and the Weill Cornell Microbiome Core for assistance with 16S rRNA sequencing experiments and analysis, C. Steckler and H. Molina of the Rockefeller Proteomics Resource Center for assistance with peptidoglycan analysis, K. Eckartt for assistance in homology modeling, B. Ostendorf for assistance with scRNA-seq data analysis, A. Griffin for assistance with bacterial genomic data preparation and analysis, and M. Huse, T. Merghoub, K. Cadwell, and D. Mucida for helpful discussions.
Funding:
This work was supported by the National Institutes of Health (1R01CA245292-01, H.C.H.) and in part by the Melanoma Research Foundation (Career Development Award, M.E.G.). M.E.G. is a Hope Funds for Cancer Research Fellow supported by the Hope Funds for Cancer Research (HCFR-19-03-02). This work is also partially funded by an NIH research service award training grant (A1070084, J.E.).
Footnotes
Competing interests: M.E.G. and H.C.H. have filed a patent application (PCT/US2020/019038) for the commercial use of SagA-bacteria to improve checkpoint blockade immunotherapy. Rise Therapeutics (J.J. and G.R.F.) has licensed the patent to develop immunological-based biologics.
Data and materials availability: All 16S rRNA and scRNA-seq sequencing data are available via BioProject PRJNA749064. All other data are available in the main text and supplementary materials.
References and Notes:
- 1.Sahin U, Türeci Ö, Personalized vaccines for cancer immunotherapy. Science. 359, 1355–1360 (2018). [DOI] [PubMed] [Google Scholar]
- 2.June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC, CAR T cell immunotherapy for human cancer. Science. 359, 1361–1365 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Ribas A, Wolchok JD, Cancer immunotherapy using checkpoint blockade. Science. 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA, Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, Rizvi NA, Merghoub T, Levine AJ, Chan TA, Wolchok JD, Greenbaum BD, A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 551, 517–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spranger S, Bao R, Gajewski TF, Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 523, 231–235 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF, Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med 24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CPM, Poirier-Colame V, Roux A, Becharef S, Formenti S, Golden E, Cording S, Eberl G, Schlitzer A, Ginhoux F, Mani S, Yamazaki T, Jacquelot N, Enot DP, Bérard M, Nigou J, Opolon P, Eggermont A, Woerther P-L, Chachaty E, Chaput N, Robert C, Mateus C, Kroemer G, Raoult D, Boneca IG, Carbonnel F, Chamaillard M, Zitvogel L, Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre M-L, Chang EB, Gajewski TF, Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, Koh AY, Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 19, 848–855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA, Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria J-C, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L, Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science. 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, Luke JJ, Gajewski TF, The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science. 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters BA, Wilson M, Moran U, Pavlick A, Izsak A, Wechter T, Weber JS, Osman I, Ahn J, Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Med. 11, 61 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebreton F, Manson AL, Saavedra JT, Straub TJ, Earl AM, Gilmore MS, Tracing the enterococci from paleozoic origins to the hospital. Cell. 169, 849–861.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanchi H, Mottawea W, Sebei K, Hammami R, The genus Enterococcus: between probiotic potential and safety concerns-an update. Front. Microbiol 9, 1791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rangan KJ, Pedicord VA, Wang Y-C, Kim B, Lu Y, Shaham S, Mucida D, Hang HC, A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science. 353, 1434–1437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedicord VA, Lockhart AAK, Rangan KJ, Craig JW, Loschko J, Rogoz A, Hang HC, Mucida D, Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci. Immunol 1, eaai7732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim B, Wang Y-C, Hespen CW, Espinosa J, Salje J, Rangan KJ, Oren DA, Kang JY, Pedicord VA, Hang HC, Enterococcus faecium secreted antigen A generates muropeptides to enhance host immunity and limit bacterial pathogenesis. eLife. 8, e45343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, Costa FRC, Tiniakou E, Greiling T, Ruff W, Barbieri A, Kriegel C, Mehta SS, Knight JR, Jain D, Goodman AL, Kriegel MA, Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 359, 1156–1161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein-Thoeringer CK, Nichols KB, Lazrak A, Docampo MD, Slingerland AE, Slingerland JB, Clurman AG, Armijo G, Gomes ALC, Shono Y, Staffas A, Burgos da Silva M, Devlin SM, Markey KA, Bajic D, Pinedo R, Tsakmaklis A, Littmann ER, Pastore A, Taur Y, Monette S, Arcila ME, Pickard AJ, Maloy M, Wright RJ, Amoretti LA, Fontana E, Pham D, Jamal MA, Weber D, Sung AD, Hashimoto D, Scheid C, Xavier JB, Messina JA, Romero K, Lew M, Bush A, Bohannon L, Hayasaka K, Hasegawa Y, Vehreschild MJGT, Cross JR, Ponce DM, Perales MA, Giralt SA, Jenq RR, Teshima T, Holler E, Chao NJ, Pamer EG, Peled JU, van den Brink MRM, Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science. 366, 1143–1149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermassen A, Leroy S, Talon R, Provot C, Popowska M, Desvaux M, Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol 10, 331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohamed JA, Teng F, Nallapareddy SR, Murray BE, Pleiotrophic effects of 2 Enterococcus faecalis sagA-like genes, salA and salB, which encode proteins that are antigenic during human infection, on biofilm formation and binding to collagen type i and fibronectin. J. Infect. Dis 193, 231–240 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Consortium THMJRS, A catalog of reference genomes from the human microbiome. Science. 328, 994–999 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng F, Kawalec M, Weinstock GM, Hryniewicz W, Murray BE, An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect. Immun 71, 5033–5041 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman J, The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat. Rev. Immunol 3, 361–370 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CPM, Flament C, Lepage P, Roberti MP, Routy B, Jacquelot N, Apetoh L, Becharef S, Rusakiewicz S, Langella P, Sokol H, Kroemer G, Enot D, Roux A, Eggermont A, Tartour E, Johannes L, Woerther P-L, Chachaty E, Soria J-C, Golden E, Formenti S, Plebanski M, Madondo M, Rosenstiel P, Raoult D, Cattoir V, Boneca IG, Chamaillard M, Zitvogel L, Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 45, 931–943 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, White RC, Clarke TH, Nguyen K, Torralba M, Shao Y, Liu J, Hernandez-Morales A, Lessor L, Rahman IR, Miyamoto Y, Ly M, Gao B, Sun W, Kiesel R, Hutmacher F, Lee S, Ventura-Cots M, Bosques-Padilla F, Verna EC, Abraldes JG, Brown RS, Vargas V, Altamirano J, Caballería J, Shawcross DL, Ho SB, Louvet A, Lucey MR, Mathurin P, Garcia-Tsao G, Bataller R, Tu XM, Eckmann L, van der Donk WA, Young R, Lawley TD, Stärkel P, Pride D, Fouts DE, Schnabl B, Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 575, 505–511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE, NOD proteins: regulators of inflammation in health and disease. Nat. Rev. Immunol 14, 9–23 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Caruso R, Warner N, Inohara N, Núñez G, NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 41, 898–908 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bron PA, Kleerebezem M, Lactic acid bacteria for delivery of endogenous or engineered therapeutic molecules. Front. Microbiol 9, 1821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lessard A-J, LeBel M, Egarnes B, Préfontaine P, Thériault P, Droit A, Brunet A, Rivest S, Gosselin J, Triggering of NOD2 receptor converts inflammatory Ly6Chigh into Ly6Clow monocytes with patrolling properties. Cell Reports. 20, 1830–1843 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P, The molecular signatures database hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taniguchi K, Karin M, NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol 18, 309–324 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA, The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 33, 570–580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S-H, Cho S-Y, Yoon Y, Park C, Sohn J, Jeong J-J, Jeon B-N, Jang M, An C, Lee S, Kim YY, Kim G, Kim S, Kim Y, Lee GB, Lee EJ, Kim SG, Kim HS, Kim Y, Kim H, Yang H-S, Kim S, Kim S, Chung H, Moon MH, Nam MH, Kwon JY, Won S, Park J-S, Weinstock GM, Lee C, Yoon KW, Park H, Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol 6, 277–288 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN, Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med 16, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Z, Wang J, Xu X, Wang H, Qiao Y, Chu WC, Xu S, Chai L, Cottier F, Pavelka N, Oosting M, Joosten LAB, Netea M, Ng CYL, Leong KP, Kundu P, Lam K-P, Pettersson S, Wang Y, Antibody neutralization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat. Microbiol 4, 766–773 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Fidler IJ, Sone S, Fogler WE, Barnes ZL, Eradication of spontaneous metastases and activation of alveolar macrophages by intravenous injection of liposomes containing muramyl dipeptide. Proc. Natl. Acad. Sci. U.S.A 78, 1680–1684 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinnijenhuis J, Quintin J, Preijers F, Joosten LAB, Ifrim DC, Saeed S, Jacobs C, van Loenhout J, de Jong D, Stunnenberg HG, Xavier RJ, van der Meer JWM, van Crevel R, Netea MG, Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U.S.A 109, 17537–17542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Priem B, van Leent MMT, Teunissen AJP, Sofias AM, Mourits VP, Willemsen L, Klein ED, Oosterwijk RS, Meerwaldt AE, Munitz J, Prévot G, Vera Verschuur A, Nauta SA, van Leeuwen EM, Fisher EL, de Jong KAM, Zhao Y, Toner YC, Soultanidis G, Calcagno C, Bomans PHH, Friedrich H, Sommerdijk N, Reiner T, Duivenvoorden R, Zupančič E, Di Martino JS, Kluza E, Rashidian M, Ploegh HL, Dijkhuizen RM, Hak S, Pérez-Medina C, Bravo-Cordero JJ, de Winther MPJ, Joosten LAB, van Elsas A, Fayad ZA, Rialdi A, Torre D, Guccione E, Ochando J, Netea MG, Griffioen AW, Mulder WJM, Trained immunity-promoting nanobiologic therapy suppresses tumor growth and potentiates checkpoint inhibition. Cell. 183, 786–801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prescott D, Maisonneuve C, Yadav J, Rubino SJ, Girardin SE, Philpott DJ, NOD2 modulates immune tolerance via the GM-CSF–dependent generation of CD103 + dendritic cells. Proc. Natl. Acad. Sci. U.S.A, 201912866 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asano J, Tada H, Onai N, Sato T, Horie Y, Fujimoto Y, Fukase K, Suzuki A, Mak TW, Ohteki T, Nucleotide oligomerization binding domain-like receptor signaling enhances dendritic cell-mediated cross-priming in vivo. J. Immunol 184, 736–745 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Netea MG, Domínguez-Andrés J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, Joosten LAB, van der Meer JWM, Mhlanga MM, Mulder WJM, Riksen NP, Schlitzer A, Schultze JL, Stabell Benn C, Sun JC, Xavier RJ, Latz E, Defining trained immunity and its role in health and disease. Nat. Rev. Immunol 20, 375–388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.