Abstract
Reversible ADP-ribosylation of dinitrogenase reductase, catalyzed by the dinitrogenase reductase ADP-ribosyl transferase–dinitrogenase reductase-activating glycohydrolase (DRAT-DRAG) regulatory system, has been characterized in Rhodospirillum rubrum and other nitrogen-fixing bacteria. To investigate the mechanisms for the regulation of DRAT and DRAG activities, we studied the heterologous expression of R. rubrum draTG in Klebsiella pneumoniae glnB and glnK mutants. In K. pneumoniae wild type, the regulation of both DRAT and DRAG activity appears to be comparable to that seen in R. rubrum. However, the regulation of both DRAT and DRAG activities is altered in a glnB background. Some DRAT escapes regulation and becomes active under N-limiting conditions. The regulation of DRAG activity is also altered in a glnB mutant, with DRAG being inactivated more slowly in response to NH4+ treatment than is seen in wild type, resulting in a high residual nitrogenase activity. In a glnK background, the regulation of DRAT activity is similar to that seen in wild type. However, the regulation of DRAG activity is completely abolished in the glnK mutant; DRAG remains active even after NH4+ addition, so there is no loss of nitrogenase activity. The results with this heterologous expression system have implications for DRAT-DRAG regulation in R. rubrum.
Biological nitrogen fixation, the conversion of atmospheric nitrogen to ammonium, is catalyzed by the nitrogenase complex, which consists of two proteins: dinitrogenase (or MoFe protein) and dinitrogenase reductase (or Fe protein) (7). It is a very energy-demanding process and is thus tightly regulated at both transcriptional and posttranslational levels.
Transcriptional regulation of the nif genes has been found in all studied nitrogen-fixing bacteria and is best characterized in Klebsiella pneumoniae, a free-living nitrogen-fixing bacterium, where it involves the general nitrogen regulation (ntr) system (36). Analysis of the ntr regulatory system in K. pneumoniae and Escherichia coli (36, 39) has shown that it controls the transcription of many genes involved in nitrogen fixation and assimilation, such as glnA (encoding glutamine synthetase [GS]) and nifA (encoding the transcriptional activator for the other nif genes). The ntr system involves a number of gene products, including those of glnD, ntrA, ntrB, ntrC, and glnB, glnD encodes a bifunctional, uridylyltransferase-uridylyl-removing enzyme (UTase-UR) that is believed to be the sensor of the intracellular concentration of glutamine in the cell. UTase-UR reversibly controls the activity of the PII protein (the gene product of glnB) by uridylylation or deuridylylation. PII is responsible for sensing α-ketoglutarate (α-KG) in E. coli (24), and it controls NtrB (NRII) activity. NtrB and NtrC (the gene products of ntrB and ntrC) belong to the family of two-component regulators. NtrB is a histidine kinase that phosphorylates NtrC (NRI) under nitrogen-limiting conditions and also can act as a phosphatase to dephosphorylate NtrC under nitrogen excess conditions. Both kinase and phosphatase activities of NtrB are regulated by PII in response to the level of α-KG in the cell (21). At low α-KG concentrations, the PII trimer (bound to only one molecule of α-KG) interacts with NtrB in vitro, thereby inhibiting its kinase activity and activating its phosphatase activity to dephosphorylate NtrC. However, at high α-KG concentrations, the PII trimer binds additional molecules of α-KG and thereby is unable to interact with NtrB, so that NtrB acts as a kinase to phosphorylate NtrC (21). The phosphorylated form of NtrC acts as a transcriptional activator of nifA, glnA, and other operons involved in nitrogen assimilation. The activation involves ς54, encoded by ntrA (also referred to as rpoN). PII, together with adenylytransferase (ATase), encoded by glnE, also controls GS activity by reversible adenylylation (22).
Besides the transcriptional regulation of nifA expression by the ntr system, NifA activity is also regulated. In K. pneumoniae and Azotobacter vinelandii, NifA activity is inhibited by a cotranscribed nif gene product, NifL, in response to NH4+ and oxygen, probably through a direct interaction (29, 37). Recently, a PII homolog (or paralog), GlnK, has been identified to be involved in the relief of NifL inhibition of NifA under N2-fixing conditions in K. pneumoniae (18, 20).
GlnK has been found in E. coli, K. pneumoniae, and many other Bacteria, as well as Archaea (47). GlnK and PII show very high sequence and structural similarity to one another (8, 34, 50). Each protein has been purified as a homotrimer (9, 34). Although these two proteins share some functions, such as the interaction with ATase to adenylylate GS (3, 48), they also have distinct functions in the cell. Only GlnK is involved in the relief of NifL inhibition in K. pneumoniae (18), although recent studies have shown that overexpressed PII can substitute for GlnK to relieve the NifL inhibition (2). In E. coli and K. pneumoniae, the expression of glnK is regulated in response to NH4+, but glnB is expressed constitutively (20, 48).
Nitrogen fixation is also regulated at the posttranslational level, which has been well characterized only in Rhodospirillum rubrum, Azospirillum brasilense, Azospirillum lipoferum, and Rhodobacter capsulatus, in which it involves reversible mono-ADP ribosylation of dinitrogenase reductase (33, 53). Dinitrogenase reductase ADP-ribosyl transferase (referred to here as DRAT, the gene product of draT) carries out the transfer of the ADP-ribose from NAD to the Arg-101 residue of one subunit of the dinitrogenase reductase homodimer, resulting in inactivation of that enzyme. Dinitrogenase reductase-activating glycohydrolase (referred to here as DRAG, the gene product of draG) removes the ADP-ribose group attached to dinitrogenase reductase, thus restoring nitrogenase activity. The DRAT-DRAG system negatively regulates nitrogenase activity in response to exogenous NH4+ or energy limitation in the form of a shift to darkness (in the cases of R. rubrum and R. capsulatus) or to anaerobic conditions (in A. brasilense) (13, 16, 17, 25, 30, 35, 40, 51, 54).
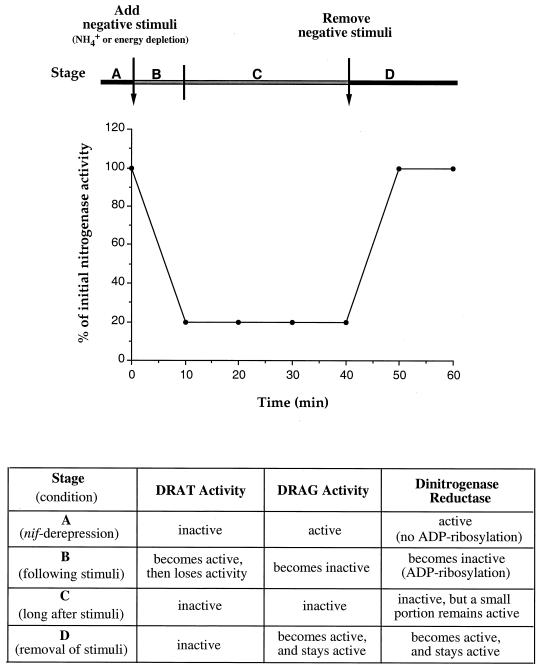
As illustrated in Fig. 1, the regulation of the ADP-ribosylation of dinitrogenase reductase is effected through the posttranslational regulation of both DRAT and DRAG activities. Under nitrogen-fixing conditions, DRAT is inactive and DRAG is active, so that dinitrogenase reductase is in its active form. Following a negative stimulus, such as exogenous NH4+ or energy depletion, DRAT is activated and DRAG becomes inactive, resulting in the loss of nitrogenase activity and the modification of dinitrogenase reductase. However, DRAT activation is only transient, and it becomes inactive again even in the continued presence of the negative stimulus. After removal of the negative stimulus, DRAG becomes active again, and it then reactivates dinitrogenase reductase by cleavage of the ADP-ribose group. Details of the mechanisms of regulation of DRAT and DRAG activities are still unknown. However, it has recently been found that the redox state of dinitrogenase reductase affects its ability to serve as substrate for DRAT and DRAG both in vitro and in vivo (14). DRAT can only modify oxidized dinitrogenase reductase and DRAG only removes ADP-ribosyl group from reduced dinitrogenase reductase. While the redox state of dinitrogenase reductase likely plays an important role in the regulation of DRAT and DRAG activities, it cannot be the only mode of regulation.
FIG. 1.
Scheme for the regulation of DRAT, DRAG, and dinitrogenase reductase activities in R. rubrum. The top panel shows the idealized time course of the regulation of nitrogenase activity following the treatment of negative stimuli, such as exogenous NH4+ and energy depletion. The bottom panel shows the regulation of DRAT, DRAG, and dinitrogenase reductase activities at different stages. Under nitrogen-fixing conditions (stage A), dinitrogenase reductase is in an active form without ADP-ribosylation, because DRAG is active and DRAT is inactive under these conditions. Following treatment with a negative stimulus (stage B), DRAT is activated and DRAG is inactivated, resulting in the loss of nitrogenase activity and the modification of dinitrogenase reductase. DRAT activity is only transient. In the period after the treatment with negative stimuli (stage C), both DRAT and DRAG have become inactive, and a steady residual nitrogenase activity remains. After cultures were returned to nitrogen-fixing conditions (stage D), DRAG becomes activated, and it removes the ADP-ribose from dinitrogenase reductase, thus restoring its activity.
draTG mutants have been constructed and physiologically characterized in R. rubrum, R. capsulatus, and A. brasilense (30, 35, 51, 54), confirming their functions in vivo. When draTG genes from A. lipoferum and R. rubrum were transferred into K. pneumoniae, which lacks this regulatory system, the nitrogenase activity was reversibly regulated in response to NH4+ (12). These results suggest that the regulatory signals that control DRAT-DRAG are conserved among these organisms and that analysis of the DRAT-DRAG system in the well-studied K. pneumoniae would provide valuable insights into that regulation.
There are, however, significant differences in the ntr systems of the two organisms with respect to nitrogen fixation. In R. rubrum and A. brasilense, PII is required for the activation of NifA activity (31, 55, 56); in R. rubrum, a strain with an alteration in glnB (PII-Y51F, where the tyrosine that serves as the site of uridylylation was altered) had low nitrogenase activity, and only slight effects on the regulation of DRAT activity are seen (54). In R. rubrum and A. brasilense, ntrBC mutations have no effect on nif expression, whereas in K. pneumoniae, NtrBC proteins are required for nif expression (31, 36, 55). However, ntrBC mutations in A. brasilense do alter the regulation of DRAG activity and cause a slower inactivation of DRAG activity by NH4+ (52), which indicates that some parts of the ntr system are involved in DRAG regulation.
Because of their key roles in the ntr system in response to an NH4+ signal, PII and GlnK are very reasonable candidates for serving as the factor in R. rubrum that actually causes the observed effects on DRAT-DRAG regulation. glnB and glnK of K. pneumoniae have therefore been examined for their effects on the heterologously expressed DRAT-DRAG system. The results demonstrate a striking effect of glnK mutations, strongly implying a role of PII (or GlnK) homologs of R. rubrum in this regulation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. Antibiotics were used at the following concentrations (in milligrams/liter): streptomycin, 25; kanamycin, 12.5; and tetracycline, 10 (for K. pneumoniae); and streptomycin, 25; kanamycin, 25; and tetracycline, 12.5 (for E. coli).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotype and/or genotype and descriptiona | Reference |
|---|---|---|
| K. pneumoniae | ||
| UNF122 | hisD2 Δlac-2002; referred to as wild type | 19 |
| UNF1537 | hisD2 Δlac-2002 glnB502 glnB21::Tn5; lacks PII; Kmr | 19 |
| UNF3432 | hisD2 Δlac-2002 glnK1::KIXX; lacks GlnK; Kmr | 20 |
| UN5507 | UNF122 (wild type) with pCH1 (R. rubrum draTGB); Smr | This report |
| UN5508 | UNF1537 (glnB) with pCH1 (R. rubrum draTGB); Kmr Smr | This report |
| UN5509 | UNF3432 (glnK) with pCH1 (R. rubrum draTGB); Kmr Smr | This report |
| UN5510 | UNF122 (wild type) with pCH3 (R. rubrum draT); Smr | This report |
| UN5511 | UNF1537 (glnB) with pCH3 (R. rubrum draT); Kmr Smr | This report |
| UN5512 | UNF3432 (glnK) with pCH3 (R. rubrum draT); Kmr Smr | This report |
| UN5513 | UNF122 (wild type) with pCK3 (K. pneumoniae nifA), Tcr | This report |
| UN5514 | UNF1537 (glnB) with pCK3 (K. pneumoniae nifA); Kmr Tcr | This report |
| UN5515 | UNF3432 (glnK) with pCK3 (K. pneumoniae nifA); Kmr Tcr | This report |
| UN5516 | UNF122 (wild type) with pCH1 (R. rubrum draTGB) and pCK3 (K. pneumoniae nifA); Smr Tcr | This report |
| UN5517 | UNF1537 (glnB) with pCH1 (R. rubrum draTGB) and pCK3 (K. pneumoniae nifA); Kmr Smr Tcr | This report |
| UN5518 | UNF3432 (glnK) with pCH1 (R. rubrum draTGB) and pCK3 (K. pneumoniae nifA); Kmr Smr Tcr | This report |
| UN5519 | UNF122 (wild type) with pCH3 (R. rubrum draT) and pCK3 (K. pneumoniae nifA); Smr Tcr | This report |
| UN5520 | UNF1537 (glnB) with pCH3 (R. rubrum draT) and pCK3 (K. pneumoniae nifA); Kmr Smr Tcr | This report |
| UN5521 | UNF3432 (glnK) with pCH3 (R. rubrum draT) and pCK3 (K. pneumoniae nifA); Kmr Smr Tcr | This report |
| UN5522 | UNF122 (wild type) with pCH7 (R. rubrum draTG); Smr | This report |
| UN5523 | UN1537 (glnB) with pCH7 (R. rubrum draTG); Kmr Smr | This report |
| UN5524 | UN3432 (glnK) with pCH7 (R. rubrum draTG); Kmr Smr | This report |
| UN5525 | UNF122 (wild type) with pCH7 (R. rubrum draTG) and pCK3 (K. pneumoniae nifA); Smr Tcr | This report |
| UN5526 | UN1537 (glnB) with pCH7 (R. rubrum draTG) and pCK3 (K. pneumoniae nifA); Kmr Smr Tcr | This report |
| UN5527 | UN3432 (glnK) with pCH7 (R. rubrum draTG) and pCK3 (K. pneumoniae nifA); Kmr Smr Tcr | This report |
| Plasmids | ||
| pCH1 | pEX21 derivative containing R. rubrum draTGB expressed under Ptac promoter; Smr | 15 |
| pCH3 | pEX21 derivative containing R. rubrum draT expressed under Ptac promoter; Smr | 15 |
| pCH7 | pEX21 derivative containing R. rubrum draTG expressed under Ptac promoter; Smr | This report |
| pCK3 | pRK290 derivative containing K. pneumoniae nifA; Tcr | 26 |
Kmr, kanamycin resistant; Smr, streptomycin resistant; Tcr, tetracycline resistant.
Growth conditions and whole-cell nitrogenase activity assay.
K. pneumoniae was first grown in rich LC solid medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl, and 1.5% Bacto agar) at 30°C and then inoculated into 3 ml of minimal medium containing 0.2% ammonium acetate (KBSa) (38) and grown overnight at 30°C. A 0.5-ml portion of culture was inoculated into 25 ml of KBSa medium in a 125-ml flask and grown on a shaker at 250 rpm overnight. The cells were collected by centrifugation and resuspended in 125 ml of minimal medium containing 0.015% of l-serine to replace ammonium acetate (KBSser). To induce the expression of draTG, a low level (10 or 50 μM as indicated) of isopropyl-β-d-thiogalactopyranoside (IPTG) was added in KBSser medium. A 15 to 25-ml culture was transferred to a 60-ml serum vial with a rubber stopper, degassed and flushed with argon, and then derepressed for nitrogenase for 4 to 5 h under anaerobic conditions at 30°C. Assay of whole-cell nitrogenase activity has been described previously (15). To maintain plasmids, appropriate antibiotics were added in all media. Because all K. pneumoniae strains used in this study have the hisD2 mutation (19), l-histidine was added in all minimal media at a final concentration of 25 μg/ml.
DNA techniques.
A freeze-thaw protocol was used for the transformation of plasmids into K. pneumoniae (46). pCH7 (Ptac-draTG) was constructed by digestion of pCH1 (Ptac-draTGB) (15) with NcoI and religated, resulting in the deletion of draB.
Immunoblotting of dinitrogenase reductase.
A trichloroacetic acid precipitation method was used to extract protein quickly as described previously (51). Low-cross-linker sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (the ratio of acrylamide to bisacrylamide was 172 to 1) was used for protein separation to obtain better resolution of the modified and unmodified subunits of dinitrogenase reductase, since the modified subunit migrates more slowly. Proteins from SDS-PAGE were electrophoretically transferred onto a nitrocellulose membrane, immunoblotted with polyclonal antibody against dinitrogenase reductase, and visualized with horseradish peroxidase color detection reagents (Bio-Rad, Richmond, Calif.).
RESULTS
Effect of a glnB mutation on the DRAT-DRAG regulatory system.
Plasmid pCH1, bearing R. rubrum draTGB, was transferred into UNF122 (wild type), UNF1537 (glnB insertion mutant which lacks PII), and UNF3432 (glnK insertion mutant which lacks GlnK), yielding UN5507, UN5508, and UN5509, respectively. Similar to their parental strains lacking draTGB, UN5507 and UN5508 showed high initial nitrogenase activity, but glnK mutants (UNF1537 and UN5509) showed little nitrogenase activity (Table 2). The loss of nitrogenase activity in glnK mutants is caused by the failure to relieve NifL inhibition to NifA under N-limiting conditions (18, 20).
TABLE 2.
Nitrogenase activity in K. pneumoniae strains and its response to NH4Cl in the presence or in the absence of R. rubrum draT or draTGB with 50 μM IPTG in the derepression medium
| Strain | Chromosomal genotype | Gene(s) on plasmids | Nitrogenase activitya
|
|
|---|---|---|---|---|
| Initial | 30 min after NH4Cl was added (%)b | |||
| UNF122 | Wild type | None | 900 | 1,280 (142) |
| UNF1537 | glnB | None | 890 | 1,250 (140) |
| UNF3432 | glnK | None | <10 | <10 |
| UN5507 | Wild type | draTGB | 800 | 30 (4) |
| UN5508 | glnB | draTGB | 610 | 350 (57) |
| UN5509 | glnK | draTGB | <10 | <10 |
| UN5510 | Wild type | draT | 780 | 15 (2) |
| UN5511 | glnB | draT | 65 | <10 |
| UN5512 | glnK | draT | <10 | <10 |
Each unit of nitrogenase activity is expressed as nanomoles of ethylene produced per hour per milliliter of cells at an optical density at 600 nm of 1. Each activity value is from at least five replicate assays from different individually grown cultures. The deviation is <15%.
NH4Cl was added at a final concentration of 10 mM. The percentage of initial nitrogenase activity that remained after the NH4Cl treatments is indicated in parentheses.
The regulation of nitrogenase activity by NH4+ was studied in these mutants. As we expected, no loss of nitrogenase activity was seen in UNF122 and UNF1537 (Table 2), since there is no posttranslational regulation of nitrogenase activity (such as the DRAT-DRAG system) in K. pneumoniae. Depending on the time frame of the derepression, we often see a higher nitrogenase activity (100 to 150% of the initial activity) after NH4+ treatment in strains such as UNF122 and UNF5137 (Table 2), which probably reflects de novo synthesis of nitrogenase. With heterologous expression of R. rubrum draTGB, K. pneumoniae wild type (UN5507) showed essentially complete loss of nitrogenase activity after 10 mM NH4+ treatment. However, NH4+ caused only partial loss of nitrogenase activity in the glnB mutant background (UN5508 in Table 2), suggesting that regulation of DRAT and/or DRAG is altered in this background.
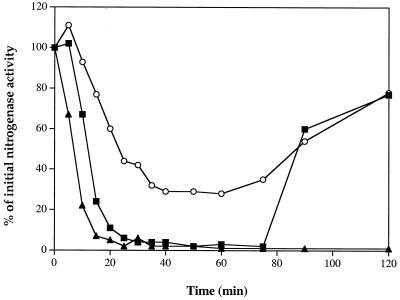
While it is easiest to compare strains at fixed time points after NH4+ addition, it can be deceptive if the kinetics of loss and recovery of nitrogenase activity are different in these strains. Therefore, a time course of loss and recovery of nitrogenase activity following NH4+ treatment was also monitored in UN5507 and UN5508 (Fig. 2). Addition of 1 mM NH4Cl caused complete loss of nitrogenase activity in UN5507, but a partial NH4+ response was seen in UN5508, with about 30 to 40% of residual nitrogenase activity. Nitrogenase activity was recovered after NH4+ exhaustion (ca. 80 min). A similar pattern of NH4+ response was seen in these strains when a low concentration of NH4Cl (0.2 mM) was used (data not shown). This low level of NH4+ still can cause almost complete loss of nitrogenase activity in UN5507, but a partial loss of nitrogenase activity was seen in UN5508, with a high residual nitrogenase activity (60%). It look a short time (ca. 40 min) to completely recover nitrogenase activity in both UN5507 and UN5508. These results again show a significant difference in the regulation of nitrogenase activity between wild-type and glnB backgrounds.
FIG. 2.
Regulation of nitrogenase activity by NH4Cl in K. pneumoniae UN5507 (wild type with draTGB) (■), UN5508 (glnB mutant with draTGB) (○), and UN5510 (wild type with draT) (▴). At t = 0, NH4Cl was added to a final concentration of 1 mM. Samples (1 ml) of the cells were withdrawn and assayed for nitrogenase activity for 2 min at the times indicated. The initial activities of UN5507, UN5508, and UN5510 were about 900, 700, and 700 nmol, respectively, of ethylene produced per h per ml of cells at an optical density at 600 nm of 1.0. IPTG was added to final concentration of 50 μM in the derepression medium.
Previous studies have shown that both DRAG and DRAT activities are subject to posttranslational regulation (see model in Fig. 1). Specifically, prior to a negative stimulus (such as NH4+), DRAG is active and DRAT is inactive. After treatment with a negative stimulus, DRAT is activated and DRAG become inactive. Differences in residual nitrogenase activity in the wild type and a glnB mutant after NH4+ treatment therefore could reflect altered regulation of DRAT and/or DRAG activities. In order to characterize the effect of a glnB mutation on DRAT, pCH3, bearing R. rubrum draT alone, was transferred into these K. pneumoniae backgrounds. Because these strains lack DRAG, any response differences would indicate an effect of PII on DRAT activity.
pCH3 was transferred into K. pneumoniae UNF122 (wild type), UNF1537 (glnB mutant), and UNF3432 (glnK mutant), yielding UN5510, UN5511, and UN5512, respectively. The nitrogenase activity results in these strains and its response to NH4+ addition are presented in Table 2. UN5510 (wild type with draT) showed a high initial nitrogenase activity, indicating that DRAT is “tightly” regulated in the wild-type background, as reported previously for R. rubrum, A. brasilense, and other K. pneumoniae strains (15, 30, 51, 54). However, a low initial nitrogenase activity was seen in UN5511 (glnB mutant with draT), indicating that at least some DRAT escapes its normal regulation and becomes active under N-limiting conditions in this glnB background. A similar level of dinitrogenase reductase was accumulated in UN5510 and UN5511, as estimated by Western blotting of dinitrogenase reductase (data not shown), confirming that low initial nitrogenase activity in UN5511 reflects regulation of activity rather than gene expression. Both UN5510 and UN5511 showed the loss of nitrogenase activity after NH4+ addition, indicating the activation of DRAT activity.
To compare the kinetics of DRAT activation, a time course of loss of nitrogenase activity following NH4+ treatment was also monitored in UN5510 (Fig. 2). UN5510 showed almost a complete loss of nitrogenase activity after treatment with 1 mM NH4Cl. Because of the absence of DRAG, UN5510 has a slightly faster rate of loss of nitrogenase activity than UN5507 does, and it showed no recovery of nitrogenase activity after NH4+ exhaustion. Recovery of nitrogenase activity was also absent in UN5510 when a low concentration of NH4Cl (0.2 mM) was used (data not shown). These results showed that DRAT is regulated normally in response to NH4+ in the wild-type background, but not in the glnB background.
The effect of a glnB mutation on DRAT regulation is dependent on the amount of DRAT in the cell.
We routinely use 50 μM IPTG to induce the expression of draT or draTGB, which results in a low level of DRAT (and DRAG) protein accumulation, and the pattern of regulation of DRAT and DRAG activities in K. pneumoniae background is very similar to that seen in R. rubrum (15). In the glnB mutant, some DRAT escapes the regulation to become active under derepression conditions, resulting in a low initial nitrogenase activity in UN5511 (Table 2). It is possible that the loss of the regulation of DRAT activity was due to a decreased level of negative effector for DRAT activity in this mutant. We therefore tested different levels of IPTG to see if different levels of DRAT had any effect on the regulation of its activity.
As shown in Table 3, different levels of IPTG (from 0 to 50 μM) have little effect on the initial nitrogenase activity (before NH4+ addition) in UN5510 (wild type with draT); 10 μM IPTG caused the accumulation of enough DRAT to inhibit nitrogenase activity after NH4+ treatment, although it showed a higher residual activity than that seen with 50 μM IPTG. However, with UN5511 (glnB mutant with draT), increasing the IPTG level significantly decreases the initial nitrogenase activity, indicating that the regulation of DRAT is altered in this mutant when excess DRAT is present in the cell. At 10 μM IPTG, DRAT in UN5511 is regulated normally under N-limiting conditions, and it could be activated after NH4+ treatment. As more DRAT is induced at higher levels of IPTG, some DRAT escapes the regulation and becomes active even under N-limiting conditions. It has been our hypothesis that both DRAT and DRAG activities are regulated by loosely binding inhibitors, since both DRAT and DRAG always are active in in vitro assays either in extracts or when purified (32, 45). One possibility for the results in Table 3 is that the level of this negative inhibitor for DRAT is significantly lower in a glnB mutant than in the wild type, resulting in the altered regulation of DRAT activity when more DRAT is induced in the cell. We are unable to precisely quantitate the level of DRAT, since DRAT levels are too low to be detected by immunoblotting even when the enhanced chemiluminescence detection method was used (Amersham, Arlington Heights, III.). The fact that very little DRAT (even at 50 μM IPTG) could titrate out such a signal suggests that this signal is either a rare small molecule or a protein effector in this glnB mutant background.
TABLE 3.
Effect of IPTG level on the nitrogenase activity in K. pneumoniae strains and its response to NH4Cl treatments
| Strain and genotype/ gene on plasmidsa | IPTG level (μM) | Nitrogenase activityb
|
|
|---|---|---|---|
| Initial | 30 min after NH4Cl was added (%)c | ||
| UN5510 | |||
| wt/draT | 0 | 980 | 1,170 (120) |
| wt/draT | 10 | 990 | 240 (24) |
| wt/draT | 20 | 940 | 40 (4) |
| wt/draT | 30 | 980 | 20 (2) |
| wt/draT | 50 | 780 | 15 (2) |
| UN5511 | |||
| glnB/draT | 0 | 820 | 800 (98) |
| glnB/draT | 10 | 600 | 75 (12) |
| glnB/draT | 20 | 540 | 20 (4) |
| glnB/draT | 30 | 400 | 15 (4) |
| glnB/draT | 50 | 60 | 6 (10) |
draT is from R. rubrum. wt, wild type.
Each unit of nitrogenase activity is expressed as nanomoles of ethylene produced per hour per milliliter of cells at an optical density at 600 nm of 1. Each activity value is from at least three replicate assays from different individually grown cultures. The deviation is <15%.
NH4Cl was added at a final concentration of 10 mM. The percentage of initial nitrogenase activity that remained after the NH4Cl treatments is indicated in parentheses.
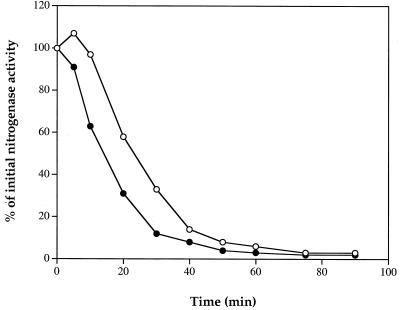
To compare the kinetics of DRAT activation in response to NH4+ in the wild type and a glnB mutant, a time course of loss of nitrogenase activity following NH4+ treatment was monitored in UN5510 and UN5511 at 10 μM IPTG (Fig. 3). At this low IPTG level most DRAT is properly regulated in both strains under N-limiting conditions, resulting in substantial initial nitrogenase activity. Because there is less DRAT protein at 10 μM IPTG, the rate of loss of nitrogenase activity is slower (UN5510 in Fig. 3) than that at 50 μM IPTG (UN5510 in Fig. 2). UN5511 showed a slightly faster rate of loss of nitrogenase activity than did UN5510 (Fig. 3), which suggests that DRAT is activated faster in UN5511 than in UN5510. These results indicate that DRAT is functioning normally in response to NH4+ in this glnB mutant, so that the partial NH4+ response in UN5508 must be caused by altered regulation of DRAG activity. Apparently, the regulation of both DRAT and DRAG activities is altered in a K. pneumoniae glnB background.
FIG. 3.
Regulation of nitrogenase activity by NH4Cl in K. pneumoniae UN5510 (wild type with draT) (○) and UN5511 (glnB mutant with draT) (●). At t = 0, NH4Cl was added to a final concentration of 10 mM. The initial nitrogenase activities of UN5510 and UN5511 were ca. 1,000 and 600 nmol, respectively, of ethylene produced per h per ml of cells at an optical density at 600 nm of 1.0. IPTG was added to final concentration of 10 μM in the derepression medium.
Effect of a glnK mutation on the regulation of nitrogenase activity.
Because a very low nitrogenase activity was detected in K. pneumoniae glnK mutants (UNF3432 in Table 2), we are unable to study the modification of dinitrogenase reductase by the DRAT-DRAG regulatory system in these backgrounds. As reported previously, GlnK is necessary for the dissociation of NifL from NifA, which allows NifA activity under N-limiting conditions (18, 20). In a glnK mutant, NifL always inhibits NifA activity, probably forming a tight complex that cannot be dissociated under N-limiting conditions. We therefore introduced a multicopy plasmid with nifA (without nifL) into a glnK mutant background, so that excess NifA (relative to NifL) would allow NifA to be constitutively active. A plasmid, pCK3, which carries K. pneumoniae nifA expressed from a constitutive promoter (26), was transferred into these K. pneumoniae backgrounds, and the nitrogenase activities from these strains are shown in Table 4. As expected, in the presence of pCK3, glnK mutants (UN5515) showed a high nitrogenase activity, similar to that in wild-type and glnB backgrounds (UN5513 and UN5514). UN5515 showed no loss of nitrogenase activity after NH4+ treatment, since it lacks the DRAT-DRAG regulatory system. The expression of K. pneumoniae nifA from pCK3 showed no significant effect on nitrogenase activity in wild-type and glnB backgrounds, and the nitrogenase activities in these strains (UN5513 and UN5514 in Table 4) are very similar to those in their parental strains (UNF122 and UNF1537 in Table 2). As seen before (Table 2), a higher nitrogenase activity (100 to 150% of the initial activity) was seen after NH4+ treatment in these K. pneumoniae strains and that probably reflects de novo synthesis of nitrogenase enzymes.
TABLE 4.
Nitrogenase activity in K. pneumoniae strains with pCK3 (K. pneumoniae nifA) and its response to NH4Cl in the presence of either 10 or 50 μM IPTG in the derepression medium
| IPTG concn (μM) and strain | Chromosomal genotypea | Gene(s) on plasmidsb | Nitrogenase activityc
|
|
|---|---|---|---|---|
| Initial | 40 min after NH4Cl was added (%)d | |||
| 50 | ||||
| UN5513 | wt | nifA | 950 | 980 (104) |
| UN5514 | glnB | nifA | 870 | 980 (113) |
| UN5515 | glnK | nifA | 800 | 1,200 (150) |
| UN5516 | wt | draTGB, nifA | 1,000 | 8 (1) |
| UN5517 | glnB | draTGB, nifA | 700 | 320 (46) |
| UN5518 | glnK | draTGB, nifA | 990 | 1,180 (119) |
| UN5519 | wt | draT, nifA | 580 | 6 (1) |
| UN5520 | glnB | draT, nifA | 80 | 6 (8) |
| UN5521 | glnK | draT, nifA | 630 | 7 (1) |
| 10 | ||||
| UN5516 | wt | draTGB, nifA | 810 | 80 (10) |
| UN5517 | glnB | draTGB, nifA | 780 | 500 (64) |
| UN5518 | glnK | draTGB, nifA | 930 | 1,400 (150) |
| UN5519 | wt | draT, nifA | 850 | 9 (1) |
| UN5520 | glnB | draT, nifA | 480 | 5 (1) |
| UN5521 | glnK | draT, nifA | 1,010 | 7 (1) |
wt, wild type.
nifA was from K. pneumoniae; draT, draG, and draB were from R. rubrum.
Each unit of nitrogenase activity is expressed as nanomoles of ethylene produced per hour per milliliter of cells at an optical density at 600 nm of 1. Each activity value is from at least three replicate assays from different individually grown cultures. The deviation is <15%.
NH4Cl was added at a final concentration of 10 mM. The percentage of initial nitrogenase activity that remained after the NH4Cl treatments is indicated in parentheses.
When both R. rubrum draTGB and K. pneumoniae nifA (on compatible plasmids) were introduced into wild-type, glnB, and glnK backgrounds, all strains showed a high initial nitrogenase activity (UN5516, UN5517, and UN5518 in Table 4). In response to NH4+ addition, UN5516 (wild type) showed complete loss of nitrogenase activity and UN5517 (glnB) showed a partial loss of nitrogenase activity, a result similar to that seen in UN5507 and UN5508 (without pCK3; Table 2). However, UN5518 (glnK) showed a complete lack of NH4+ response (Table 4), indicating that the regulation of DRAT and/or DRAG is dramatically altered in this mutant.
A time course of loss of nitrogenase activity following NH4+ treatment was also monitored in UN5516, UN5517, and UN5518. Similar to the results seen in UN5507 and UN5508 (Fig. 2), UN5516 and UN5517 showed complete or partial loss of nitrogenase activity after NH4+ treatment, but no NH4+ response was seen in UN5518 (data not shown). These results clearly indicate that the regulation of nitrogenase activity by NH4+ is completely abolished in a K. pneumoniae glnK background.
The effect of a glnK mutation on the regulation of nitrogenase activity is due to altered regulation of DRAG activity but not of DRAT activity.
pCH3, bearing R. rubrum draT alone, was transferred into a K. pneumoniae glnK background, which also contains K. pneumoniae nifA. This allowed us to study the effect of a glnK mutation on DRAT activity alone. Unlike the glnB mutants (UN5520 in Table 4 and UN5511 in Table 2), UN5521 (glnK with draT and nifA) showed a substantial initial nitrogenase activity at 50 μM IPTG, a level similar to that seen in UN5519 (wild type with draT and nifA). This suggested that most DRAT is inactive under N-limiting conditions in this glnK mutant. However, the nitrogenase activity in both UN5519 and UN5521 (both containing draT) is lower than that seen in UN5516 and UN5518 (both containing draTGB). As will be shown below, some ADP-ribosylation of dinitrogenase reductase exists in UN5519 and UN5521 but not in UN5516 and UN5518. We believe that this small amount of modification of dinitrogenase reductase is caused by the l-histidine used in the derepression medium. l-Histidine is a nitrogen source and could cause activation of some DRAT to modify dinitrogenase reductase even under derepression conditions, especially at 50 μM IPTG. Because of the lack of DRAG in UN5519 and UN5521, a small portion of active DRAT caused some modification of dinitrogenase reductase in these strains. In contrast, the presence of active DRAG in UN5516 and UN5518 can compensate for this low DRAT activity, so that no modification of dinitrogenase reductase was seen in these strains under derepression conditions (see below). Both UN5519 and UN5521 showed a normal NH4+ response and a complete loss of nitrogenase activity after NH4+ treatment (Table 4). To further analyze the DRAT regulation and its response to NH4+, a time course of the loss of nitrogenase activity following NH4+ treatment was also monitored in UN5519 and UN5521. Both UN5519 (wild type) and UN5521 (glnK) showed a fast rate of NH4+ response similar to that in UN5510 (Fig. 2) (data not shown). These results indicate that the absence of glnK has no significant effect on DRAT regulation, so that a lack of NH4+ response in UN5518 must be caused by altered DRAG regulation. While DRAG in the wild type becomes inactive after NH4+ treatment, DRAG in this K. pneumoniae glnK background appears to stay active even in the presence of NH4+. All of these results indicate that mutation of glnK showed no significant effect on the regulation of DRAT activity but has a profound effect on the regulation of DRAG activity.
Different levels of IPTG have no effect on DRAG regulation in the glnK mutant background.
We also examined the regulation of DRAT and DRAG activities in these glnK mutants in the presence of pCK3 at a low IPTG level (10 μM) and to see if it was also dependent on the amount of DRAT and DRAG proteins in the cell. As shown in Table 4, in the presence of draTGB, a very similar pattern of NH4+ response was seen in UN5516, UN5517, and UN5518 at both 10 and 50 μM IPTG levels, with only a small difference in residual activity in UN5516 and UN5517. No NH4+ response was seen in UN5518 in either condition. In the presence of draT alone, all strains have a higher initial nitrogenase activity at 10 μM IPTG than that seen at 50 μM IPTG, especially UN5520. This is consistent with the results seen in UN5511 (without pCK3) in Tables 2 and 3 and might be due to less DRAT expression at 10 μM IPTG. The NH4+ response in the wild type and the glnK mutant was exactly the same at these two different levels of IPTG. Unlike the altered regulation of DRAT activity in glnB mutant, the effect of a glnK mutation on the regulation of DRAG is independent of the level of DRAG protein in the cell. Together these results suggest that the regulation of DRAT and DRAG have different mechanisms.
Correlation of ADP-ribosylation of dinitrogenase reductase with the regulation of nitrogenase activity in K. pneumoniae strains containing K. pneumoniae nifA and R. rubrum draTGB or draT.
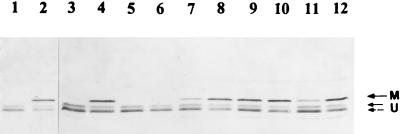
In addition to the assay of nitrogenase activity to monitor DRAT and DRAG activities indirectly, a direct assay for DRAT and DRAG activities is the ADP-ribosylation of dinitrogenase reductase, which can be monitored directly by immunoblotting. In R. rubrum, the ADP-ribosylated (inactive) form of dinitrogenase reductase runs as two bands on SDS-PAGE, because the ADP-ribosylated subunit migrates slower, while active dinitrogenase reductase has two identical subunits that migrate as a single band (55). In contrast, active dinitrogenase reductase in K. pneumoniae migrated as two bands (labeled “U” in lanes 1, 3, and 5 in Fig. 4). These two bands were also found in UNF122 (wild type without draT) (data not shown) and were reported previously (43); the reason for this doublet is unknown. As we expected, UN5516, UN5517, and UR5518 showed no modification of dinitrogenase reductase before NH4+ addition (Fig. 4). After NH4+ treatment, both UN5516 and UN5517 showed a modified (ADP-ribosylated) upper band (labeled “M” in Fig. 4). However, very little modification of dinitrogenase reductase was found in UN5518 after NH4+ treatment, a finding consistent with the lack of NH4+ effect on nitrogenase activity in this glnK mutant (Table 4).
FIG. 4.
Western immunoblot of dinitrogenase reductase in K. pneumoniae UN5516 (wild type with R. rubrum draTGB and K. pneumoniae nifA) (lanes 1 and 2), UN5517 (glnB mutant with R. rubrum draTGB and K. pneumoniae nifA) (lanes 3 and 4), UN5518 (glnK mutant with R. rubrum draTGB and K. pneumoniae nifA) (lanes 5 and 6), UN5519 (wild type with R. rubrum draT and K. pneumoniae nifA) (lanes 7 and 8), UN5520 (glnB mutant with R. rubrum draT and K. pneumoniae nifA) (lanes 9 and 10), and UN5521 (glnK mutant with R. rubrum draT and K. pneumoniae nifA) (lanes 11 and 12). Samples of dinitrogenase reductase were collected by trichloroacetic acid precipitation from cultures grown in derepression medium in the presence of 50 μM IPTG in derepression medium before (lanes 1, 3, 5, 7, 9, and 11) and after (lanes 2, 4, 6, 8, 10, and 12) treatment of 10 mM NH4Cl for 40 min. Arrow “M” indicates the position of the modified (ADP-ribosylated) subunit, and arrow “U” indicates that of the unmodified subunits, as explained in the text.
With draT alone, both UN5519 and UN5521 showed some modified dinitrogenase reductase before NH4+ addition (Fig. 4), and this is the reason that a lower initial nitrogenase activity was seen in these strains than that seen in UN5516 and UN5518 (Table 4). As mentioned before, most DRAT is inactive in wild-type and glnK backgrounds under N-limiting conditions even at the 50 μM IPTG level, but a small portion of DRAT was activated with the l-histidine used in the derepression medium. In the absence of DRAG, this small amount of active DRAT can cause some modification of dinitrogenase reductase in UN5519 and UN5521. In contrast, the presence of active DRAG in UN5516 and UN5518 can compete with this low DRAT activity, so that no modification of dinitrogenase reductase was apparent in these strains under derepression conditions. However, UN5520 (glnB with draT) showed almost complete modification of dinitrogenase reductase even without NH4+ addition, a result consistent with a very low initial nitrogenase activity in this mutant (Table 4).
Overall, UN5519 (wild type with draT) and UN5520 (glnB with draT) accumulated very similar amounts of dinitrogenase reductase (lanes 7 and 9 in Fig. 4) in the presence of 50 μM IPTG, so that the different initial nitrogenase activities in these strains (Table 4) are mainly due to the modification status of dinitrogenase reductase rather than to the level of nitrogenase proteins themselves.
DRAB plays little role in the effect of GlnK and PII on the regulation of DRAG activity.
draB is an open reading frame located in downstream of draTG in R. rubrum, and it is apparently cotranscribed with draTG (D. P. Lies and G. P. Roberts, unpublished data). draB shows high sequence similarity to nifO of A. vinelandii (44) and some similarity to arsC of E. coli (4, 10). While the function of nifO of A. vinelandii is unknown, it might be involved in molybdenum metabolism (44). arsC encodes an arsenate reductase, which catalyzes the reduction of arsenate to arsenite (10). In R. rubrum the mutation of draB caused a decrease in DRAG activity and protein level, and the total DRAG activity in draB mutant is approximately 35% of that of the wild type (30). Recently, DRAG has been shown to have a binuclear manganese center when it is treated with Mn2+ (1). Based on its sequence similarity to other metal processing proteins, therefore, DRAB might be involved in the processing of a metal center in DRAG. To investigate if DRAB is involved in the regulation of DRAG activity by PII and GlnK, plasmid pCH7, carrying R. rubrum draTG (no draB), was transferred into different K. pneumoniae backgrounds. Nitrogenase activity in these strains and its response to NH4+ was monitored, and the results are shown in Table 5. Overall, there was no significant difference in nitrogenase activity and its response to NH4+ between K. pneumoniae strains with draTGB and those with draTG alone. A partial NH4+ response was also seen in glnB mutants with draTG alone (UN5523 and UN5526), but the residual activity in these strains is lower than that seen in glnB mutants with R. rubrum draTGB (UN5508 in Table 2 and UN5517 in Table 4). This might due to the effect of draB on DRAG activity itself, as reported previously (30). The glnK mutant with R. rubrum draTG (UN5527) showed no NH4+ response, a result similar to that seen in the glnK mutant with R. rubrum draTGB (UN5518 in Table 4). These results indicate that draB has no significant effect on the regulation of DRAG activity by PII and GlnK under the conditions employed for these experiments.
TABLE 5.
Nitrogenase activity in K. pneumoniae strains with pCH7 (R. rubrum draTG) and its response to NH4Cl in the presence of 50 μM IPTG in the derepression medium
| Strains | Chromosomal genotypea | Gene(s) on plasmidsb | Nitrogenase activityc
|
|
|---|---|---|---|---|
| Initial | 40 min after NH4Cl was added (%)d | |||
| UN5522 | wt | draTG | 810 | 34 (4) |
| UN5523 | glnB | draTG | 760 | 250 (33) |
| UN5524 | glnK | draTG | <10 | <10 |
| UN5525 | wt | draTG, nifA | 1,000 | 10 (1) |
| UN5526 | glnB | draTG, nifA | 750 | 190 (25) |
| UN5527 | glnK | draTG, nifA | 800 | 1,100 (138) |
wt, wild type.
draTG was from R. rubrum; nifA was from K. pneumoniae.
Each unit of nitrogenase activity is expressed as nanomoles of ethylene produced per hour per milliliter of cells at an optical density at 600 nm of 1. Each activity value is from at least five replicate assays from different individually grown cultures. The deviation is <15%.
NH4Cl was added at a final concentration of 10 mM. The percentage of initial nitrogenase activity that remained after NH4Cl treatments is given in parentheses.
DISCUSSION
Although the general function of DRAT and DRAG in the regulation of nitrogenase activity has been characterized in R. rubrum, A. brasilense, and R. capsulatus, the mechanisms for the regulation of DRAT and DRAG activities are still unknown. When draTG genes from A. lipoferum or R. rubrum were transferred into K. pneumoniae, the nitrogenase activity was reversibly regulated in response to NH4+ (12, 15), indicating that the regulatory effectors in the signal transduction pathways for the regulation of DRAT and DRAG should be present in K. pneumoniae. Because PII and GlnK play very important roles in sensing NH4+ status to regulate nif expression in K. pneumoniae, it seemed reasonable to consider that these proteins may also be involved in the regulation of the DRAT-DRAG system in response to NH4+ as well. Indeed, the results presented here show significant effects on the DRAT-DRAG system by mutations in both glnB and glnK. These results provide a testable hypothesis that PII and its homolog(s) probably have a similar function in the regulation of DRAT and DRAG activity in organisms that normally contain the DRAT-DRAG regulatory system.
The mutation of glnB in K. pneumoniae showed effects on the regulation of both DRAT and DRAG activities. DRAT escapes normal regulation and becomes active under N-limiting conditions in this glnB mutant, and this altered regulation of DRAT activity is probably due to the decreased level of some inhibitor for DRAT in this mutant, because elevated DRAT levels exacerbate this effect. It has been our hypothesis that both DRAT and DRAG activities are regulated by loosely binding negative inhibitors, since both DRAT and DRAG always are active in in vitro assays either in extracts or when purified. It is possible that the expression of the negative effector for DRAT is affected by PII. The level of this negative effector might be significantly lower in a glnB mutant than in the wild type, resulting in the altered regulation of DRAT activity when more DRAT is induced in the cell. Using this glnB mutant as a control, it is possible to screen for potential candidates of the negative effector for DRAT in vitro.
The regulation of DRAG activity is also altered in a glnB mutant, with DRAG being inactivated more slowly by NH4+ in this mutant than in the wild type, resulting in a high residual nitrogenase activity. A K. pneumoniae glnK mutation showed an even more profound effect on the regulation of DRAG activity but no significant effect on the regulation of DRAT activity. It has been previously shown that glnK is normally expressed in a glnB mutant under N-limiting conditions (20), and Western blots also showed that the level of GlnK in this glnB mutant is very similar to that seen in the wild type (data not shown), so that the effects of the glnB mutation are not due to direct or indirect effects on GlnK. These data indicate that both PII and GlnK are required for the maximum inactivation of DRAG activity in response to NH4+. Though it is unknown if the effects of PII and GlnK on DRAG are direct, we have added purified PII and GlnK to an in vitro DRAG activity assay (which measures the conversion of modified dinitrogenase reductase into active form), but no inhibition of DRAG activity was seen (data not shown). The effect of PII and GlnK on DRAG might be indirect and may involve the regulation of an unknown negative effector that regulates DRAG activity. The lack of regulation of DRAG activity in a K. pneumoniae glnK mutant could be caused by either the absence of the negative effector for DRAG (transcriptional regulation) or the accumulation of the inactive form of this effector (posttranslational regulation). Regardless of which regulatory mechanism is involved, the lack of the active form of the negative effector in a glnK mutant should allow us to screen for such small molecules or protein effectors in vitro.
While the results presented above provide compelling evidence that PII and GlnK might be central to DRAT-DRAG regulation in organisms that normally contain this regulatory system, the specific roles of these PII homologs are probably not identical to that seen in K. pneumoniae. In R. rubrum, creation of PII-Y51F (in which the tyrosine site for uridylylation of PII is changed to phenylalanine) allows some DRAT to escape regulation under N-limiting conditions (56). Unlike the case in K. pneumoniae, however, a glnB mutation in R. rubrum showed only a small effect on the regulation of nitrogenase activity (Y. Zhang and G. P. Roberts, unpublished data). Some other aspect of the ntr system of R. rubrum and A. brasilense must be involved in the regulation of DRAT-DRAG system, since the mutation of ntrBC had a significant effect on the regulation of DRAG activity, causing a slower inactivation of DRAG activity by NH4+ (52). One candidate for this regulator is the PII homolog, GlnK. Consistent with this hypothesis, R. rubrum and R. sphaeroides cbbM mutants (defective in the Calvin-Benson-Bassham pathway) showed high nitrogenase activity even in the presence of NH4+ (23), indicating an alteration in nif expression and DRAT-DRAG regulation in this R. rubrum cbbM mutant. The expression of glnB and glnK were also affected in the cbbM mutant in R. sphaeroides (41). Therefore, the GlnB and GlnK families, although falling into evolutionarily distinct groupings, might not have specific functions that are as consistently grouped. The eventual analysis of the effects on the DRAT-DRAG system of different PII and GlnK homologs from different species should begin to clarify the sequence features that define the functional differences in these proteins.
The effect of PII and its homologs on the posttranslational regulation of nitrogenase activity has been reported in the Archaea Methanococcus maripaludis (27). In M. maripaludis, a deletion mutant of two glnB homologs has no effect on nif expression but a significant effect on the posttranslational regulation of nitrogenase activity in response to NH4+. Unfortunately, the mechanism for the posttranslational regulation of nitrogenase activity has not been characterized in M. maripaludis, but it may be also involved in the ADP-ribosylation of dinitrogenase reductase. DRAG homologs have also been found in other Bacteria and some Archaea, such as Archaeoglobus fulgidus, Methanococcus jannaschii, E. coli, Aquifex aeolicus, Deinococcus radiodurans, Listeria monocytogenes, and Streptomyces coelicolor (4–6, 11, 28, 42, 49).
In conclusion, the results presented here are that (i) PII of K. pneumoniae affects DRAT regulation and this effect is dependent on levels of DRAT protein, suggesting that regulation is through a molecule (an inhibitor for DRAT) that is nonabundant; (ii) PII also affects the regulation of DRAG activity, but less dramatically than does GlnK; (iii) the absence of GlnK alters DRAG, but not DRAT, and this effect is independent of the levels of DRAG; (iv) overexpression of nifA of K. pneumoniae overcomes the effect of glnK mutations on expression of other nif genes; (v) ADP-ribosylation of dinitrogenase reductase correlates with the loss of nitrogenase activity in K. pneumoniae, a result consistent with the view that there is no other posttranslational regulation of nitrogenase activity in this organism in response to NH4+; (vi) DRAB, whose absence has modest effects on DRAG activity in R. rubrum, does not appear to be involved in the PII or GlnK effects on DRAT-DRAG seen in K. pneumoniae; and (vii) the precise functions of PII and GlnK appear to be different in different organisms.
ACKNOWLEDGMENTS
This work was supported by the College of Agricultural and Life Sciences, University of Wisconsin-Madison; Department of Agriculture grant 99-35305-8010 to G.P.R.; and NIGMS grant 54910 to P.W.L.
We thank B. S. Antharavally for technical help and advice, M. Merrick for generously providing K. pneumoniae strains, A. J. Ninfa for kindly providing GlnK and PII proteins, W. C. van Heeswijk for kindly providing PII antibody, and C. Kennedy for kindly providing pCK3 plasmid.
REFERENCES
- 1.Antharavally B S, Poyner R R, Ludden P W. EPR spectral evidence for a binuclear Mn(II) center in dinitrogenase reductase-activating glycohydrolase from Rhodospirillum rubrum. J Am Chem Soc. 1998;120:8897–8898. [Google Scholar]
- 2.Arcondeguy T, van Heeswijk W C, Merrick M. Studies on the roles of GlnK and GlnB in regulating Klebsiella pneumoniae NifL-dependent nitrogen control. FEMS Microbiol Lett. 1999;180:263–270. doi: 10.1111/j.1574-6968.1999.tb08805.x. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson M R, Ninfa A J. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol Microbiol. 1998;29:431–447. doi: 10.1046/j.1365-2958.1998.00932.x. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1473. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Brehm K, Ripio M T, Kreft J, Vazquez-Boland J A. The bvr locus of Listeria monocytogenes mediates virulence gene repression by β-glucosides. J Bacteriol. 1999;181:5024–5032. doi: 10.1128/jb.181.16.5024-5032.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Burris R H. Nitrogenases. J Biol Chem. 1991;266:9339–9342. [PubMed] [Google Scholar]
- 8.Carr P D, Cheah E, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. X-ray structure of the signal transduction protein PII from Escherichia coli at 1.9 Å. Acta Crystallogr D Biol Crystallogr. 1998;52:93–104. doi: 10.1107/S0907444995007293. [DOI] [PubMed] [Google Scholar]
- 9.Cheah E, Carr P D, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. Structure of the Escherichia coli signal transducing protein PII. Structure. 1994;2:981–990. doi: 10.1016/s0969-2126(94)00100-6. [DOI] [PubMed] [Google Scholar]
- 10.Chen C M, Misra T K, Silver S, Rosen B P. Nucleotide sequence of the structural genes for an anion pump. The plasmid-encoded arsenical resistance operon. J Biol Chem. 1986;261:15030–15038. [PubMed] [Google Scholar]
- 11.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 12.Fu H, Burris R H, Roberts G P. Reversible ADP-ribosylation is demonstrated to be a regulatory mechanism in prokaryotes by heterologous expression. Proc Natl Acad Sci USA. 1990;87:1720–1724. doi: 10.1073/pnas.87.5.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu H A, Hartmann A, Lowery R G, Fitzmaurice W P, Roberts G P, Burris R H. Posttranslational regulatory system for nitrogenase activity in Azospirillum spp. J Bacteriol. 1989;171:4679–4685. doi: 10.1128/jb.171.9.4679-4685.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbleib C M, Zhang Y, Ludden P W. Regulation of dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase-activating glycohydrolase by a redox-dependent conformational change of nitrogenase Fe protein. J Biol Chem. 2000;275:3493–3500. doi: 10.1074/jbc.275.5.3493. [DOI] [PubMed] [Google Scholar]
- 15.Halbleib C M, Zhang Y, Roberts G P, Ludden P W. Effects of perturbations of the nitrogenase electron transfer chain on reversible ADP-ribosylation of nitrogenase Fe protein in Klebsiella pneumoniae strains bearing the Rhodospirillum rubrum dra operon. J Bacteriol. 2000;182:3681–3687. doi: 10.1128/jb.182.13.3681-3687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartmann A, Burris R H. Regulation of nitrogenase activity by oxygen in Azospirillum brasilense and Azospirillum lipoferum. J Bacteriol. 1987;169:944–948. doi: 10.1128/jb.169.3.944-948.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartmann A, Fu H, Burris R H. Regulation of nitrogenase activity by ammonium chloride in Azospirillum spp. J Bacteriol. 1986;165:864–870. doi: 10.1128/jb.165.3.864-870.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He L, Soupene E, Ninfa A, Kustu S. Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J Bacteriol. 1998;180:6661–6667. doi: 10.1128/jb.180.24.6661-6667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtel A, Merrick M. Identification of the Klebsiella pneumoniae glnB gene: nucleotide sequence of wild-type and mutant alleles. Mol Gen Genet. 1988;215:134–138. doi: 10.1007/BF00331314. [DOI] [PubMed] [Google Scholar]
- 20.Jack R, De Zamaroczy M, Merrick M. The signal transduction protein GlnK is required for NifL-dependent nitrogen control of nif gene expression in Klebsiella pneumoniae. J Bacteriol. 1999;181:1156–1162. doi: 10.1128/jb.181.4.1156-1162.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang P, Ninfa A J. Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J Bacteriol. 1999;181:1906–1911. doi: 10.1128/jb.181.6.1906-1911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang P, Peliska J A, Ninfa A J. The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry. 1998;37:12802–12810. doi: 10.1021/bi980666u. [DOI] [PubMed] [Google Scholar]
- 23.Joshi H M, Tabita F R. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc Natl Acad Sci USA. 1996;93:14515–14520. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamberov E S, Atkinson M R, Ninfa A J. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J Biol Chem. 1995;270:17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 25.Kanemoto R H, Ludden P W. Effect of ammonia, darkness, and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J Bacteriol. 1984;158:713–720. doi: 10.1128/jb.158.2.713-720.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy C, Drummond M H. The use of cloned nif regulatory elements from Klebsiella pneumoniae to examine nif regulation in Azotobacter vinelandii. J Gen Microbiol. 1985;131:1787–1795. [Google Scholar]
- 27.Kessler P S, Leigh J A. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics. 1999;152:1343–1351. doi: 10.1093/genetics/152.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 29.Lei S, Pulakat L, Gavini N. Genetic analysis of nif regulatory genes by utilizing the yeast two-hybrid system detected formation of a NifL-NifA complex that is implicated in regulated expression of nif genes. J Bacteriol. 1999;181:6535–6539. doi: 10.1128/jb.181.20.6535-6539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang J H, Nielsen G M, Lies D P, Burris R H, Roberts G P, Ludden P W. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J Bacteriol. 1991;173:6903–6909. doi: 10.1128/jb.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang Y Y, Arsène F, Elmerich C. Characterization of the ntrBC genes of Azospirillum brasilense Sp7: their involvement in the regulation of nitrogenase synthesis and activity. Mol Gen Genet. 1993;240:188–196. doi: 10.1007/BF00277056. [DOI] [PubMed] [Google Scholar]
- 32.Lowery R G, Ludden P W. Purification and properties of dinitrogenase reductase ADP-ribosyltransferase from the photosynthetic bacterium Rhodospirillum rubrum. J Biol Chem. 1988;263:16714–16719. [PubMed] [Google Scholar]
- 33.Ludden P W, Roberts G P. Regulation of nitrogenase activity by reversible ADP ribosylation. Curr Top Cell Regul. 1989;30:23–56. doi: 10.1016/b978-0-12-152830-0.50004-9. [DOI] [PubMed] [Google Scholar]
- 34.MacPherson K H, Xu Y, Cheah E, Carr P D, van Heeswijk W C, Westerhoff H V, Luque E, Vasudevan S G, Ollis D L. Crystallization and preliminary X-ray analysis of Escherichia coli GlnK. Acta Crystallogr D Biol Crystallogr. 1998;54:996–998. doi: 10.1107/s0907444998001887. [DOI] [PubMed] [Google Scholar]
- 35.Masepohl B, Krey R, Klipp W. The draTG gene region of Rhodobacter capsulatus is required for post-translational regulation of both the molybdenum and the alternative nitrogenase. J Gen Microbiol. 1993;139:2667–2675. doi: 10.1099/00221287-139-11-2667. [DOI] [PubMed] [Google Scholar]
- 36.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Money T, Jones T, Dixon R, Austin S. Isolation and properties of the complex between the enhancer binding protein NIFA and the sensor NIFL. J Bacteriol. 1999;181:4461–4468. doi: 10.1128/jb.181.15.4461-4468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nieva-Gómez D, Roberts G P, Klevickis S, Brill W J. Electron transport to nitrogenase in Klebsiella pneumoniae. Proc Natl Acad Sci USA. 1980;77:2555–2558. doi: 10.1073/pnas.77.5.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ninfa A J, Atkinson M R, Kamberov E S, Feng J, Ninfa E G. Control of nitrogen assimilation by the NRI-NRII two-component system of enteric bacteria. In: Hoch J A, Silhavy T J, editors. Two component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 67–88. [Google Scholar]
- 40.Pierrard J, Ludden P W, Roberts G P. Posttranslational regulation of nitrogenase in Rhodobacter capsulatus: existence of two independent regulatory effects of ammonium. J Bacteriol. 1993;175:1358–1366. doi: 10.1128/jb.175.5.1358-1366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian Y, Tabita F R. Expression of glnB and a glnB-like gene (glnK) in a ribulose bisphosphate carboxylase/oxygenase-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1998;180:4644–4649. doi: 10.1128/jb.180.17.4644-4649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 43.Roberts G P, MacNeil T, MacNeil D, Brill W J. Regulation and characterization of protein products coded by the nif (nitrogen fixation) genes of Klebsiella pneumoniae. J Bacteriol. 1978;136:267–279. doi: 10.1128/jb.136.1.267-279.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Quinones F, Bosch R, Imperial J. Expression of the nifBfdxNnifOQ region of Azotobacter vinelandii and its role in nitrogenase activity. J Bacteriol. 1993;175:2926–2935. doi: 10.1128/jb.175.10.2926-2935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saari L L, Triplett E W, Ludden P W. Purification and properties of the activating enzyme for iron protein of nitrogenase from the photosynthetic bacterium Rhodospirillum rubrum. J Biol Chem. 1984;259:15502–15508. [PubMed] [Google Scholar]
- 46.Shokolenko I N, Alexeyev M F. Transformation of Escherichia coli TG1 and Klebsiella oxytoca VN13 by freezing-thawing procedure. BioTechniques. 1995;18:596–598. [PubMed] [Google Scholar]
- 47.Thomas G, Coutts G, Merrick M. The glnKamtB operon. A conserved gene pair in prokaryotes. Trends Genet. 2000;16:11–14. doi: 10.1016/s0168-9525(99)01887-9. [DOI] [PubMed] [Google Scholar]
- 48.van Heeswijk W C, Stegeman B, Hoving S, Molenaar D, Kahn D, Westerhoff H V. An additional PII in Escherichia coli: a new regulatory protein in the glutamine synthetase cascade. FEMS Microbiol Lett. 1995;132:153–157. doi: 10.1111/j.1574-6968.1995.tb07825.x. [DOI] [PubMed] [Google Scholar]
- 49.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, Cheah E, Carr P D, van Heeswijk W C, Westerhoff H V, Vasudevan S G, Ollis D L. GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J Mol Biol. 1998;282:149–165. doi: 10.1006/jmbi.1998.1979. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Burris R H, Ludden P W, Roberts G P. Posttranslational regulation of nitrogenase activity by anaerobiosis and ammonium in Azospirillum brasilense. J Bacteriol. 1993;175:6781–6788. doi: 10.1128/jb.175.21.6781-6788.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Burris R H, Ludden P W, Roberts G P. Posttranslational regulation of nitrogenase activity in Azospirillum brasilense ntrBC mutants: ammonium and anaerobic switch-off occurs through independent signal transduction pathways. J Bacteriol. 1994;176:5780–5787. doi: 10.1128/jb.176.18.5780-5787.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Burris R H, Ludden P W, Roberts G P. Regulation of nitrogen fixation in Azospirillum brasilense. FEMS Microbiol Lett. 1997;152:195–204. doi: 10.1111/j.1574-6968.1997.tb10428.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Burris R H, Roberts G P. Cloning, sequencing, mutagenesis, and functional characterization of draT and draG genes from Azospirillum brasilense. J Bacteriol. 1992;174:3364–3369. doi: 10.1128/jb.174.10.3364-3369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Cummings A D, Burris R H, Ludden P W, Roberts G P. Effect of an ntrBC mutation on the posttranslational regulation of nitrogenase activity in Rhodospirillum rubrum. J Bacteriol. 1995;177:5322–5326. doi: 10.1128/jb.177.18.5322-5326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Pohlmann E L, Ludden P W, Roberts G P. Mutagenesis and functional characterization of the glnB, glnA, and nifA genes from the photosynthetic bacterium Rhodospirillum rubrum. J Bacteriol. 2000;182:983–992. doi: 10.1128/jb.182.4.983-992.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]