Abstract
A method was developed for protein localization in Mycoplasma pneumoniae by immunofluorescence microscopy. The P1 adhesin protein was revealed to be located at least at one cell pole in all adhesive cells, as has been observed by immunoelectron microscopy. Cell images were classified according to P1 localization and assigned by DNA content. Cells with a single P1 focus at one cell pole had a lower DNA content than cells with two foci, at least one of which was positioned at a cell pole. Those with one focus at each cell pole had the highest DNA content, suggesting that the nascent attachment organelle is formed next to the old one and migrates to the opposite cell pole before cell division. Double staining revealed that the accessory proteins for cytadherence—HMW1, HMW3, P30, P90, P40, and P65—colocalized with the P1 adhesin in all cells. The localization of cytadherence proteins was also examined in cytadherence-deficient mutant cells with a branched morphology. In M5 mutant cells, which lack the P90 and P40 proteins, HMW1, HMW3, P1, and P30 were focused at the cell poles of short branches, and P65 showed no signal. In M7 mutant cells, which produce a truncated P30 protein, HMW1, HMW3, P1, P90, and P40 were focused, and P65 showed no signal. In M6 mutant cells, which express no HMW1 and a truncated P30 protein, the P1 adhesin was distributed throughout the entire cell body, and no signal was detected for the other proteins. These results suggest that the cytadherence proteins are sequentially assembled to the attachment organelle with HMW1 first, HMW3, P1, P30, P90, and P40 next, and P65 last.
Mycoplasmas are parasitic bacteria with a small genome size and no peptidoglycan layer (36). Several mycoplasmas have terminal structures which enable them to adhere to the host cell surface for colonization and nutrient acquisition. The terminal structure of Mycoplasma pneumoniae, designated the attachment organelle, has been well described (19, 20). It is a membrane protrusion supported by a cytoskeleton-like structure and characterized by a dense cluster of the adhesin protein known as P1 (35).
Electron microscopic images have suggested that M. pneumoniae cells divide by binary fission and that the formation and migration of the attachment organelle are coordinated with the cell division process (6). However, the actual order of cell images relative to the cell cycle must be known, and information about the timing of DNA replication is required, in order to substantiate this model. In previous works we quantified and localized the chromosomal DNA through the observation of 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells of Mycoplasma capricolum by fluorescence microscopy (40, 41). This technique may also be useful for examining the cell division process of M. pneumoniae, although it does not provide the required information about the position of the attachment organelle. Recently, immunofluorescence microscopy was used to study the subcellular localization of bacterial proteins (27). This technique, combined with DAPI staining, may provide the crucial information for elucidating the cell reproduction scheme of mycoplasmas.
Several proteins including P1 adhesin, are thought to be essential for cytadherence, and some of them have been observed by immunoelectron microscopy to localize at the attachment organelle (19, 20, 35). However, we have little information about the localizing order and hierarchy of these proteins. The use of immunofluorescence microscopy in addition to electron microscopy might contribute to the study of this area, because fluorescence microscopy provides quick, sensitive, and quantitative analyses, including double staining.
We have developed a method for immunofluorescence microscopy of M. pneumoniae with staining of the cytadherence proteins and the chromosomal DNA. We demonstrated that the formation and migration of the attachment organelle were coordinated with the cell division process; furthermore, we describe the order of assembly of the cytadherence proteins into the attachment organelle.
MATERIALS AND METHODS
Cultivation.
To begin, 1-ml volumes of frozen stocks of M. pneumoniae M129 and its mutants were grown in 10 ml of Aluotto medium (2) for 2 or 3 days at 37°C, using plastic petri dishes and glass flasks, until about 107 to 108 CFU/ml was reached.
Preparation of antisera.
A mouse monoclonal antibody against P1 and rabbit polyclonal antibodies against other cytadherence proteins were kindly provided by P.-C. Hu and R. Herrmann, respectively (15, 22, 23, 32, 33). A mouse polyclonal antibody against the HU protein of M. pneumoniae was prepared by the following method. A fragment encoding the HU gene of M. pneumoniae (G12_orf109) was amplified by PCR from the chromosomal DNA with primers GGCCATGGAAAAAACAACAACATCG and CCAAGCTTAGTCTGCGTATTTCCAGCGT. This fragment codes for all 109 amino acid residues of the putative HU protein. The PCR product was digested with NcoI and HindIII and then inserted into the expression vector pET-30c(+) (Novagen, Madison, Wis.). The resulting plasmid, pET-HMp, was transformed into Escherichia coli BL21 (DE3) and induced with isopropyl-β-d-thiogalactopyranoside (IPTG). The histidine-tagged HU protein was purified with a Ni2+-nitrilotriacetic acid column under denaturing conditions according to the manufacturer's instructions. An antiserum against the HU protein was prepared in mice as described previously, (39). The specificity of serum was checked by immunoblot analysis (data not shown) (T. Kenri, T. Sasaki, and Y. Kano, Abstr. 12th Int. Cong. Int. Org. Mycoplasmol., abstr. D33, p. 137 [IOM Lett., vol. 5], 1998).
Immunofluorescence staining.
An immunofluorescence staining method was developed by modifying an approach designed for E. coli (1). At mid-log phase, liquid medium was replaced with fresh medium. The cells adhering to the bottom of the petri dishes were scraped into the fresh medium, recovered with the medium, passed through a 25-gauge needle several times, and filtered through a nitrocellulose membrane (pore size, 0.45 μm) to disperse cell aggregates (37). Cell suspensions were placed on coverslips for 1 to 4 h at 37°C. For cytadherence-deficient mutants, mid-log-phase cultures were suspended and filtered, and cell suspensions were placed on poly-l-lysine coated coverslips, because the mutant cells used in this study cannot bind to the glass surface and poly-l-lysine allows their attachment (14, 22, 24, 25). The medium was removed, and the cells bound to the coverslips were washed three times with phosphate-buffered saline (PBS). A fixation solution of 500 μl containing 3.0% paraformaldehyde (wt/vol) and 0.1% glutaraldehyde (vol/vol) in PBS was placed on the coverslip, and the cells were then incubated first for 10 min at room temperature and then for 50 min at 4°C. The cells were washed three times with PBS, overlaid with a permeabilizing solution containing 0.1% Triton X-100 (vol/vol) in PBS, and then incubated for 5 min at room temperature. The cells were again washed three times with PBS and were allowed to dry completely. Rehydration with PBS was carried out at room temperature for 5 min; then the PBS was replaced by a blocking solution containing 2% bovine serum albumin (BSA) (wt/vol) in PBS (PBS-BSA), and the cells were incubated for 10 min at room temperature. The PBS-BSA was removed, and the coverslips were incubated with antibodies and antisera diluted in PBS-BSA. A 2,000-fold dilution was used for the anti-P1 monoclonal antibody; a 200-fold dilution was used for the anti-HMW1, anti-HMW3, and anti-P30 antisera; a 100-fold dilution was used for the anti-P90, anti-P40, and anti-P65 antisera; and a 50-fold dilution was used for the anti-HU antiserum. After incubation for 60 min at room temperature, the cells were washed 10 times with PBS and then incubated with 1,000-fold-diluted goat anti-mouse or anti-rabbit antibodies labeled with Alexa 488 or Alexa 546 (Molecular Probes, Eugene, Oreg.) in PBS-BSA. After incubation for 60 min at room temperature in the dark, the cells were washed 10 times with PBS. For double staining, fixation of antibodies was carried out by incubation with 3.0% paraformaldehyde in PBS for 30 min at 4°C, and then the cells were washed five times with PBS before the second staining. The coverslips were mounted onto glass slides with 40% glycerol (vol/vol) containing 10 μg of DAPI/ml and were stored at −20°C if necessary. The cells were observed and photographed with an Olympus BX50 microscope using Fuji Super G400 (ISO 400) 35-mm film. Image files were produced by a personal computer equipped with a GT-9000 (Epson, Tokyo, Japan) flatbed scanner and a QuickScan 35 (Minolta, Osaka, Japan) film scanner.
Measurement of DNA content.
Cells stained with DAPI and an anti-P1 antibody were observed with the fluorescence microscope equipped with a charge-coupled device camera (WV-BP510; Panasonic, Osaka, Japan). The cell images were recorded with a digital videocassette recorder (WV-D9000; SONY, Tokyo, Japan), transferred to computer image files through an image capture card (DVRapter; Canopus, Kobe, Japan), and then analyzed by Scion Image PC beta 3 software. The fluorescence intensities of DAPI were measured by using the command “analyze particles” and taken as measurements of the DNA contents of individual cells. The fluorescence intensities of actual DAPI images were confirmed to be in the range of linearity of measurement by using Inspeck Microscope Image Intensity Calibration Kits (Molecular Probes).
RESULTS
Subcellular localization of the attachment organelle.
Immunofluorescence microscopy was used to localize the attachment organelle in M. pneumoniae cells. Since the attachment organelle is characterized by dense clusters of the P1 adhesin (35), we used P1 protein as a marker for the attachment organelle (Fig. 1A to E). Cells were fixed, permeabilized, stained with an anti-P1 antibody and DAPI, and then observed by fluorescence microscopy. A filamentous cell morphology was observed, as described previously (6, 19), while a small population of cells showed a flask shape. Immunofluorescence staining with an anti-P1 antibody revealed that at least one fluorescent focus was located at the end of a cell pole in all cells, with a slight distribution along the lateral cell extension, as has been reported in immunoelectron microscopic studies (3, 10, 15). DAPI staining occurred throughout the whole cell body. We tried phase-combined fluorescence microscopy to reduce the fluorescence intensity and localize the nucleoid as was done for M. capricolum (41), but the nucleoid was found to occupy almost the entire cell body (data not shown).
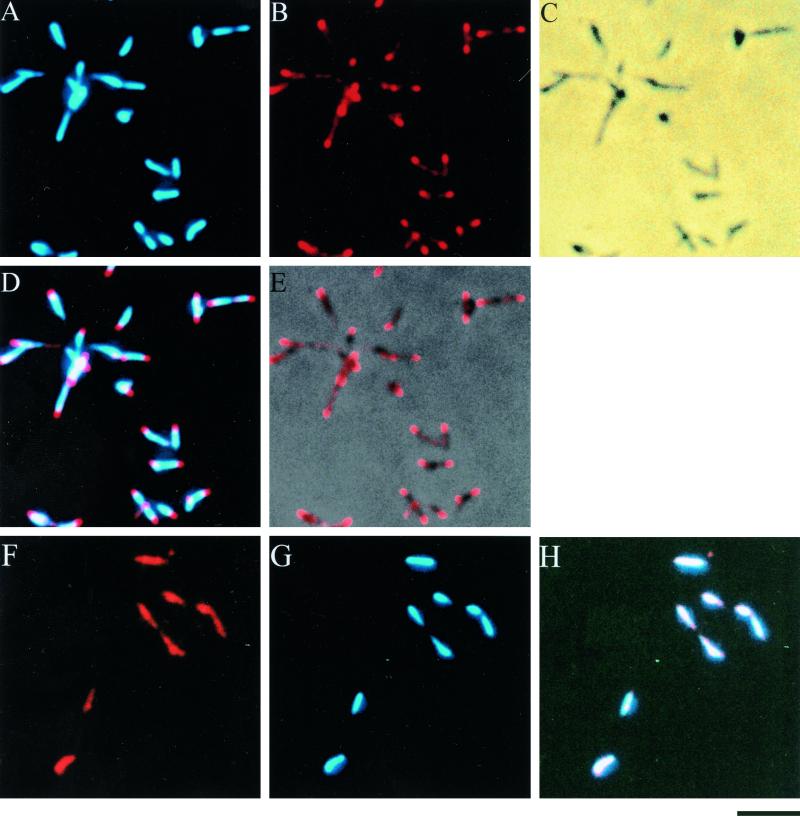
FIG. 1.
Subcellular localization of P1 adhesin (A to E) and HU protein (F to H) in M. pneumoniae. Cells were fixed, permeabilized, and stained with antibodies and DAPI. (A) DAPI-stained image; (B) anti-P1 antibody-stained image; (C) phase-contrast image; (D) merge of P1 and DAPI staining; (E) merge of P1 staining and phase-contrast image; (F) anti-HU antibody-staining image; (G) DAPI-staining image; (H) merge of HU and DAPI staining. Bar, 2 μm.
HU is a histone-like protein associated with the bacterial chromosome (9, 16, 18). As a control experiment, the localization of HU was examined (Fig. 1F to H). The fluorescent signal of the anti-HU protein antibody was located at the same position as the nucleoid stained with DAPI, which was different from that of the P1 adhesin. These results suggest that the subcellular localization of mycoplasma proteins can be visualized by immunofluorescence microscopy.
Cell typing based on the attachment organelle localization.
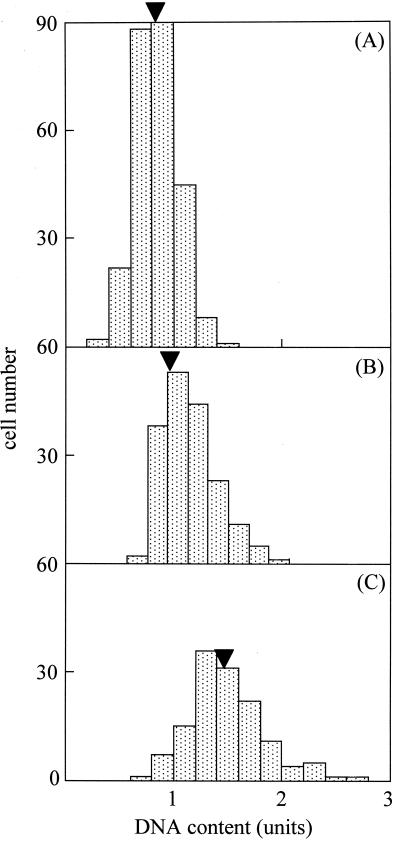
The images of cells stained for the P1 adhesin and DNA were classified into four types based on P1 localization (Fig. 2). The first type is a cell with a single P1 focus at one cell pole. The second has two P1 foci at one cell pole. The third has two P1 foci, only one of which is positioned at a cell pole. The fourth has one P1 focus at each cell pole. All cell images were classified as one of these four types; the proportions were 67.0, 5.5, 11.2, and 16.3%, respectively. The cells with two foci, only one of which was positioned at a cell pole, appeared to possess a bifurcated cell pole or a short branch along the lateral cell body, as described previously (6, 19). All cell images except the fourth type contained a single nucleoid, while three-quarters of the cells with one focus at each cell pole had two partitioned nucleoids. To address the question of subcellular positioning of the attachment organelle during the cell division process, the DNA contents of the individual cells were examined (Fig. 3). Assuming that the DNA content increases continuously during the cell division process (40, 41), cell images can be placed in the actual order of the process according to their DNA contents. The DNA contents of individual cell images showed a relationship with the cell types, i.e., those with one P1 focus at one cell pole, two P1 foci but not at both cell poles, and one P1 focus at each cell pole had 0.84, 1.04, and 1.48 times the average of total DNA content, respectively. The DNA content did not differ between cells with two foci at one cell pole and those with two foci, only one of which was positioned at a cell pole (data not shown). These results suggest that the nascent attachment organelle is formed next to the old one and that one organelle migrates to the opposite end before chromosome partitioning and cell division.
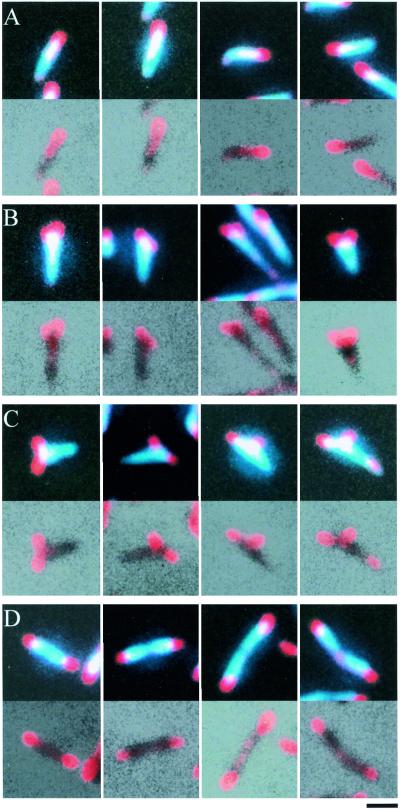
FIG. 2.
Cell image typing based on P1 localization. The upper and lower sections of each block show merges of P1 and DAPI staining and of P1 staining and phase-contrast images, respectively. Shown are cell images with a single focus at one cell pole (A), with two foci at one of the cell poles (B), with two foci, one of which is positioned at a distance from the cell poles (C), and with one focus at each cell pole (D). Bar, 1 μm.
FIG. 3.
DNA contents in individual cells of each type. (A) Cells with one P1 focus; (B) cells with two P1 foci, which are not positioned at both cell poles; (C) cells with one focus at each cell pole. Arrowhead indicates the average DNA content in the respective cell type. The average total DNA content of all cells was normalized to 1 U.
Subcellular localization of cytadherence accessory proteins.
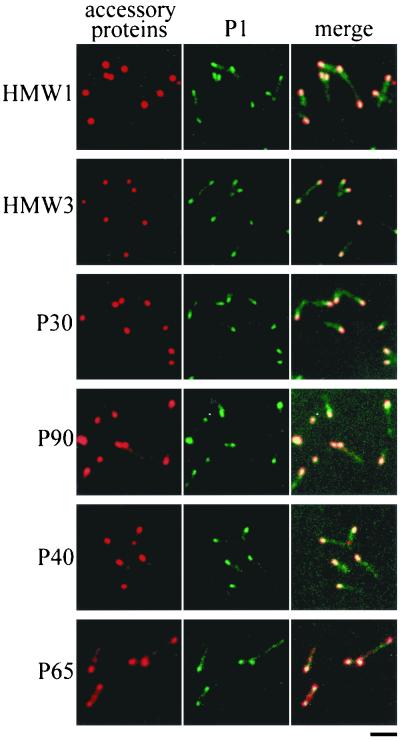
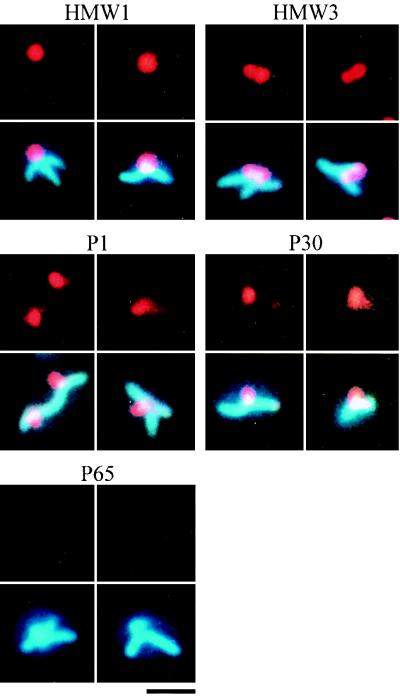
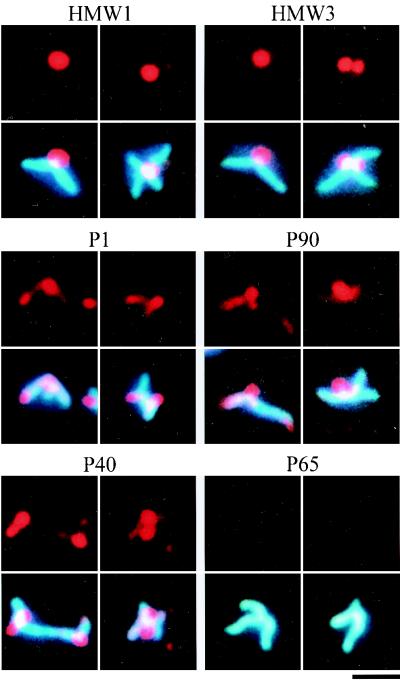
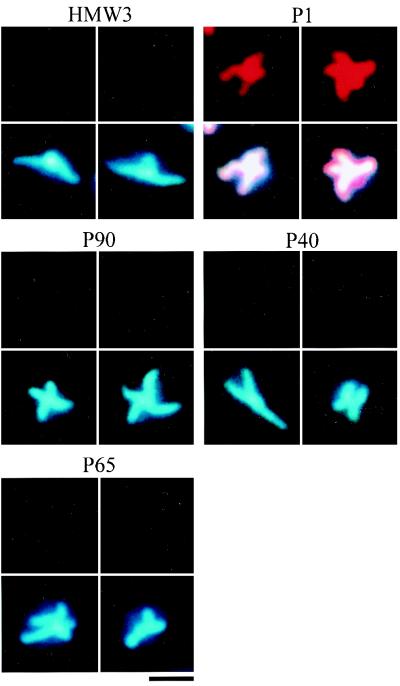
Several proteins, including the P1 adhesin, are thought to be essential for cytadherence, and HMW1, HMW3, P30, and P90 have been observed by immunoelectron microscopy to localize around the attachment organelle (4, 12, 44). However, we do not have adequate information on whether these proteins are always found at the attachment organelle, which is needed to address the order and hierarchy of assembly of cytadherence proteins. We used the immunofluorescence microscopic procedure described here to localize the cytadherence accessory proteins, i.e., the HMW1, HMW3, P30, P90, P40, and P65 proteins (Fig. 4). All the proteins involved were localized as one or two fluorescent foci at cell poles, a pattern similar to that seen with the P1 adhesin. The fluorescent foci of the HMW1, HMW3, and P30 proteins were obviously more condensed than those of P1. Those of P90, P40, and P65 were primarily located at cell poles, with some distribution along the cell extension. To examine the subcellular localization of these cytadherence accessory proteins, a double-staining procedure for the cytadherence accessory proteins and the P1 adhesin was carried out (Fig. 4). Most of the protein foci were found at the position where P1 adhesin was densely localized, suggesting that the localization of accessory proteins to the attachment organelle occurs in a short period in the cell reproduction cycle.
FIG. 4.
Subcellular localization of cytadherence accessory proteins. The left and middle columns in each panel show the same cells stained for accessory proteins and P1 adhesin, respectively. The right columns show these images merged. Bar, 2 μm.
Subcellular localization of cytadherence proteins in cytadherence-deficient mutants.
To examine the localization dependence of cytadherence proteins, their subcellular localization was analyzed in cytadherence-deficient mutants. One cytadherence-deficient mutant, M5, does not express either the P90 or the P40 protein (22). Phase-contrast microscopy showed that most mutant cells had branched shapes. DAPI staining of mutant cells revealed that the nucleoids were distributed throughout the entire cell body, including the branches (Fig. 5). Staining of the P90 and P40 proteins produced no signal, as expected (data not shown). Fluorescent foci of the HMW1, HMW3, P1, and P30 proteins were found at the poles of short branches. Staining of P65 resulted in faint fluorescence, while no change was detected in the protein expression level of P65 by immunoblot analysis (data not shown). These results suggest that P65 requires the function of P90 and/or P40 for subcellular localization but that the HMW1, HMW3, and P1 proteins do not.
FIG. 5.
Subcellular localization of cytadherence proteins in M5 mutant cells. Cells were stained with antibodies to the cytadherence proteins indicated (upper panels), and these images were then overlaid with DAPI staining images (lower panels). Bar, 1 μm.
In the M7 mutant, which expresses a truncated 22-kDa product of the p30 gene (25), most cells displayed a branched morphology. The nucleoid was distributed throughout the entire cell body, including the branches, as observed in the M5 mutant (Fig. 6). In the M7 mutant, the cytadherence proteins HMW1, HMW3, P1, P90, and P40 were recognized as fluorescent foci located at short branch poles. Staining of P65 produced no signal, while the level of the P65 protein observed by immunoblot analysis was similar to that of the wild-type strain (data not shown). The anti-P30 antibody we used has been reported to recognize the truncated 22-kDa protein of M7 mutant cells (25), and the truncated protein band was detected by immunoblotting, with an intensity similar to that of P30 in the wild-type strain, but no fluorescent signals were detected for the truncated P30 proteins (data not shown). These results suggest that the subcellular localization of the P65 and P30 proteins requires the proline-rich repeat sequences in the C-terminal part of the P30 protein missing in the M7 mutant but that the HMW1, HMW3, P1, P90, and P40 proteins can localize and assemble without them.
FIG. 6.
Subcellular localization of cytadherence proteins in M7 mutant cells. Cells were stained with antibodies to the cytadherence proteins indicated (upper panels), and these images were then overlaid with DAPI-staining images (lower panels). Bar, 1 μm.
The M6 mutant cannot synthesize HMW1 protein because of a frameshift mutation, and it produces a 25-kDa protein product of the truncated p30 gene (24). Phase-contrast microscopy showed branched cells, as has been reported previously (14), and DAPI staining showed that the nucleoids occupied the entire cell body, including the branches (Fig. 7). Immunofluorescence staining for the HMW3, P90, P40, and P65 proteins did not produce signals while the P1 adhesin was distributed throughout the entire cell body. Staining of P30 yielded no signal, although the antibody can detect the truncated protein by immunoblot analysis (24), and the truncated protein band was detected by immunoblotting, with an intensity similar to that of P30 in the wild-type strain (data not shown). The steady-state levels of cytadherence proteins were not affected in the M6 mutant in comparison to the wild-type strain (data not shown), indicating that the loss of function of HMW1 and P30 has no effect on the stability of these proteins. Considering the results for the M7 mutant, where the truncation of P30 had no effect on the localization of cytadherence-associated proteins (Fig. 6 and Table 1) the results for the M6 mutant suggest that the HMW1 protein is essential for the subcellular localization of HMW3, P1, P90, P40, and P65.
FIG. 7.
Subcellular localization of cytadherence proteins in M6 mutant cells. Cells were stained with antibodies to the cytadherence proteins indicated (upper panels), and these images were then overlaid with DAPI-staining images (lower panels). Bar, 1 μm.
TABLE 1.
Localization of cytadherence proteins in cytadherence-deficient mutant cellsa
| Cytadherence protein | Localizationb in cells of the following typec:
|
|||
|---|---|---|---|---|
| WT | M5 mutant (p90 and p40)c | M6 mutant (hmw1 and p30) | M7 mutant (p30) | |
| HMW1 | + | + | − | + |
| HMW3 | + | + | − | + |
| P1 | + | + | (−)d | + |
| P30 | + | + | − | − |
| P90 | + | − | − | + |
| P40 | + | − | − | + |
| P65 | + | − | − | − |
Plus signs, fluorescent foci were recognized in the cell images; minus signs, no fluorescent foci were recognized in the cell images.
WT, wild type. For mutants, the mutated genes are given in parentheses.
A signal was detected but was distributed throughout the entire cell body.
To address the possibility that the missing of some protein signals in mutants was caused by loss of proteins in the staining process, we examined whether the proteins were removed in the procedure. The wild-type and mutant cells were collected, suspended in PBS, and subjected to the same procedure as that for fluorescence staining except that the cells were treated in suspensions and were not incubated with antibodies. The fixed cells, Triton extracts, and final cell suspensions were analyzed by indirect enzyme-linked immunosorbent assay (ELISA) for the detection of all the cytadherence proteins that we studied except P90 and P40 in the M5 mutant and HMW1 in the M6 mutant. The results did not depend on the protein or the strains. The final cell suspensions showed ELISA signals with levels equivalent to those for the fixed cells, and the Triton extracts showed negligible signals (data not shown). We also analyzed the content of P65 by immunoblotting. The cells fixed with 3% paraformaldehyde were treated in suspension by the same procedure as that for immunofluorescence staining but without the addition of antibodies. The cross-linking was removed as previously reported (26), and immunoblotting was performed. The final cell suspensions showed bands with intensities more than 90% of those for the fixed cell suspensions in all strains (data not shown).
DISCUSSION
In this study, we developed an immunofluorescence microscopy technique for the visualization of the M. pneumoniae attachment organelle during cell division and for the subcellular localization of individual cytadherence-associated proteins. Fluorescent foci of the P1 adhesin were recognized by an immunofluorescence staining method described by Feldner et al. (10). We applied this procedure to the other cytadherence-associated proteins by using specific antibodies but failed to stain the cells. Presumably, the topology of the P1 adhesin, which projects from the cell membrane, confers an advantage for this type of assay. Therefore, we had to develop a method applicable to all kinds of mycoplasma proteins. Basically, we used the staining procedure for walled bacteria and modified it so as to avoid breakage of the cell structure, which might be caused by centrifugation. Mycoplasma cells were allowed to adhere to coverslips and fixed. Permeabilization was done with Triton X-100 without pretreatment by lysozyme because of the complete lack of the peptidoglycan layer in mycoplasmas. The permeabilization step is indispensable for efficient staining, because some of the cytadherence-associated proteins were not detected reproducibly without it (data not shown).
Previously, we demonstrated that M. capricolum cells divide into two daughter cells by binary fission with accurate partition of the replicated chromosomes (29, 40, 41). In this study, we observed that M. pneumoniae nucleoids could be stained with DAPI (Fig. 1). The nucleoids occupied the whole cell interior, and no condensation was observed, even when phase-combined fluorescence microscopy, which can reduce the fluorescence intensity and localize the nucleoid, was used. However, the nucleoid images suggest that the daughter cells receive an equal amount of DNA at binary fission in M. pneumoniae, because the standard deviation of DNA content was 0.35 of the average, indicating that the highest content was close to twice that of the lowest.
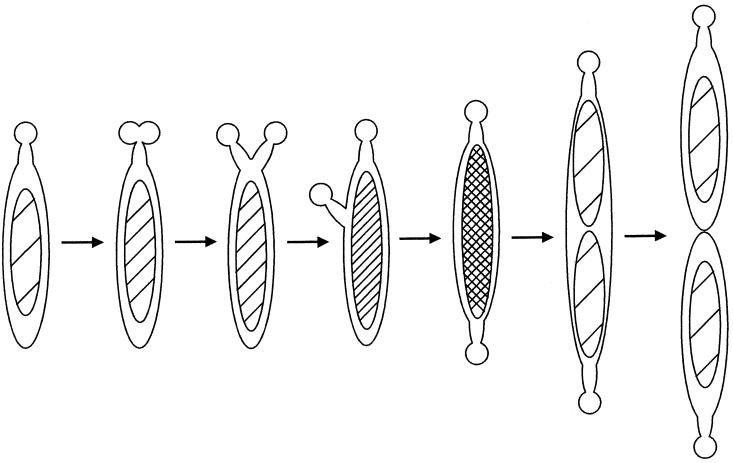
The electron microscopic images of M. pneumoniae suggest that the formation and migration of the attachment organelle are coordinated with cell division (6). However, more information is needed for the models to be substantiated adequately. We classified the cell images by the position of the attachment organelle and assigned them by their DNA contents (Fig. 2 and 3). This ordering can provide a model for the coupling of the attachment organelle formation and cell division (Fig. 8). At the first stage, the nascent attachment organelle is formed next to the old one. Next, one of the attachment organelles migrates to the opposite end along the lateral cell body, and then nucleoid partitioning and cell division occur. Boatman observed cells possessing two attachment organelles adjacent to one another at one cell pole and cells possessing one attachment organelle at each cell pole by electron microscopy of M. pneumoniae (6). Bredt observed that bifurcation of one cell pole occurred at the first step of cell division in living cells (7). Our present model of the formation and migration of the attachment organelle in M. pneumoniae is consistent with Bredt's observations.
FIG. 8.
Model for cell division scheme in M. pneumoniae in relation to the formation and migration of attachment organelles.
What motive force propels the attachment organelle? One possibility is gliding motility, by which some mycoplasmas, including M. pneumoniae, slide on solid surfaces towards the attachment organelle (17, 38). Considering that the attachment organelle is the leading end for gliding motility, gliding motility might be involved in the migration of the attachment organelle. Actually, Bredt observed that gliding cells of M. pneumoniae showed binary fission and that the bifurcation of the cell pole occurred during gliding motility (7). Another possibility might be the involvement of filamentous structures in the division process. These structures, which are composed of protein, can be observed in detergent-treated M. pneumoniae cells (13, 28, 30). They may maintain the extension of the attachment organelle and promote its migration by polymerization and depolymerization of the protein monomers.
A phenomenon analogous to the behavior of the attachment organelle (Fig. 8) is reported for the origin of replication of the chromosome in walled bacteria (27). The origin is replicated near a cell pole, after which one copy migrates to the opposite pole in a stage of chromosome partitioning. Movement from one pole to the other during cell reproduction may be a common phenomenon in a wide variety of bacteria. The attachment organelle of Mycoplasma gallisepticum has been suggested to bind chromosomal DNA (34). It is possible that the attachment organelle also works for chromosome partitioning in mycoplasmas.
In addition to the P1 adhesin, several other proteins are thought to be essential for cytadherence activity, and for most of them, subcellular localization has been examined by immunoelectron microscopy (19, 20). In this study, we examined the subcellular localization of cytadherence proteins HMW1, HMW3, P30, P90, P40, and P65 by immunofluorescence microscopy (Fig. 4). Although the intensity and condensation of fluorescence were not uniform, all proteins involved were localized to regions overlapping the P1 adhesin. The results suggest that these proteins are components of the attachment organelle and that localization is characteristic for each individual protein. Our results concerning the location of the HMW3, P30, and P90 proteins are consistent with those described previously (4, 12, 44). Stevens and Krause reported that the HMW1 protein is sometimes found at the cell extension of the other pole as well as at the cell extension of the attachment organelle (20, 43). However, our observation by immunofluorescence microscopy revealed that all fluorescent foci of the HMW1 protein were found at the attachment organelle (Fig. 4). The subcellular localizations of P40 and P65 have not yet been precisely elucidated, although chemical cross-linking studies suggest that they are associated with the attachment organelle (23, 26). Our observations support the results obtained by cross-linking studies.
Most cells of the M6 mutant demonstrated branched structures, as observed previously (14, 37). A similar morphology was observed for most M5 and M7 mutant cells (Fig. 5 and 6). The branched structure of the M7 mutant is consistent with a previous report that disruption of the p30 gene induces branch formation (14, 37). Assuming that the branch is a form of incorrectly located cell extension of the attachment organelle, these observations suggest that P30, P90, and P40 are not involved in the cell extension formation. In the M6 mutant, the clustering of all cytadherence-associated proteins in a certain region of the cell was completely lost, but branches were formed (Fig. 6), suggesting that the other cytadherence proteins, including HMW1, HMW3, P1, or P65, are not necessary for cell extension formation, either.
Aberrant cell morphology coupled with cell adhesion deficiency is also reported for Mycoplasma mobile (30). Four of 10 mutants isolated based on gliding motility were revealed to have deficiencies in both adhesion and normal formation of a “head-like structure” which is believed to have a function similar to that of the attachment organelle of M. pneumoniae. Abnormal formation of the cell shape may cause incorrect assignment of cytadherence proteins, resulting in adherence deficiency in these types of mutants.
What mechanism is involved in multibranching? Previously, we demonstrated that multibranched cell morphology is induced by nucleoside starvation in M.capricolum (40). We proposed a branching scheme whereby nucleoids which remain at the division site inhibit constriction and division potential: cytoskeleton and lipid synthesis, for instance, induce the development of new branches (29, 40, 41). It is possible that the abnormal formation of the attachment organelle affects the process of cell division in the cytadherence mutants and causes branching. Another possibility is that P30, P90, and P40 participate directly in the inhibition of branch formation. Detergent-treated and sectioned images of M. pneumoniae suggest that the extended structure of the attachment organelle is supported by an electron-dense core anchoring to the end of the attachment organelle, which has been designated the terminal button (5, 13, 28, 45) and suggested to include HMW3 as a component (44). The electron-dense core may be anchored to the terminal button by the functions of the P30, P90, and P40 proteins in wild-type cells. According to this model, the loss of these proteins induces the release and abnormal arrangement of the electron-dense cores in the mutants.
To investigate the effect of the loss of particular cytadherence proteins on the location of other proteins involved in the attachment process, their localization was examined in cytadherence-deficient mutants by the same staining procedure that was used for wild-type cells, and it was found that the signals of some proteins could not be detected in the mutants (Fig. 5, 6, and 7). Titration of these proteins using an indirect ELISA and immunoblotting showed that the disappearance of fluorescent signals for cytadherence-associated proteins was not caused by loss of proteins in the staining process. Presumably, it was caused by the dispersion of protein molecules, i.e., the signals were detectable only when they were concentrated at small spots over the detection threshold. P1 adhesin signals were detected in the entire cell bodies of the M6 mutant and did not depend on the source of the antibody, i.e., mouse monoclonal or rabbit polyclonal antibodies (data not shown), suggesting that these signals are far more intense than those of other proteins, a fact which is related to the antigenic character of the P1 adhesin.
As summarized in Table 1, HMW1 protein is essential for the localization of the other cytadherence proteins, while P65 requires all other proteins. This is consistent with a previous report showing a requirement of HMW1 for P1 localization (14). Baseman et al. examined P1 adhesin localization in a class III mutant possessing a genetic background similar to that of the M5 mutant, but they could not find a P1 adhesin cluster (3), unlike our result (Fig. 5). Possibly, this discrepancy is due to the increased sensitivity of immunofluorescence microscopy compared to that of immunoelectron microscopy. An analogous phenomenon has been reported for the localization of FtsZ protein, a bacterial cytoskeletal protein (1). Alternatively, genetic difference may exist between the M5 mutant and the strain used by Baseman et al. which apparently have very similar backgrounds. P40 and P90 have been reported to be essential for the association of P1 with the Triton shell (22). Considering this observation, our results may suggest that the P1 protein can assemble independently from the association of P1 with the Triton shell.
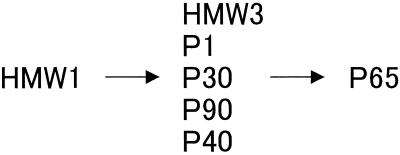
Our results show that proteins HMW3, P1, P30, P90, and P40 can assemble independently of each other. Focusing on protein assembly, our results suggest the sequential assembly of cytadherence proteins to form the attachment organelle (Fig. 9). During the formation of the attachment organelle, the HMW1 protein may be translocated first, followed by assembly of the HMW3 protein, P1, P30, P90, and P40. P65 might be the last component which localizes to the attachment organelle. This assembly sequence may be related to the observations made by electron microscopy that the cell poles of hmw1 mutants are round and different from those of wild-type and p30 mutant strains (14, 37). The control mechanism of the HMW1 protein is not known, but phosphorylation (8, 21) and degradation dependent on HMW2 (11, 31) have been reported. Caulobacter crescentus, which presents a distinct cell cycle, controls the function of CtrA, the key protein for cell differentiation, by protein phosphorylation and degradation (42). It is possible that M. pneumoniae has a similar mechanism to control HMW1 protein.
FIG. 9.
Scheme of assembly of the cytadherence proteins to the attachment organelle.
ACKNOWLEDGMENTS
We are grateful to P.-C.Hu of the University of North Carolina and to R. Herrmann of the Universität Heidelberg for the antibodies to mycoplasma proteins.
This work was partly supported by a Sasakawa Scientific Research Grant to S.S., a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Science, Sports, and Culture to M.M., and a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science to M.M.
REFERENCES
- 1.Addinall S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aluotto B B, Wittler R G, Williams C O, Faber J E. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int J Syst Bacteriol. 1970;20:35–58. [Google Scholar]
- 3.Baseman J B, Cole R M, Krause D C, Leith D K. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J Bacteriol. 1982;151:1514–1522. doi: 10.1128/jb.151.3.1514-1522.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baseman J B, Morrison-Plummer J, Drouillard D, Puleo-Scheppke B, Tryon V V, Holt S C. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr J Med Sci. 1987;23:474–479. [PubMed] [Google Scholar]
- 5.Biberfeld G, Biberfeld P. Ultrastructural features of Mycoplasma pneumoniae. J Bacteriol. 1970;102:855–861. doi: 10.1128/jb.102.3.855-861.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boatman E S. Morphology and ultrastructure of the mycoplasmatales. In: Barile M F, Razin S, editors. The mycoplasmas. I. New York, N.Y: Academic Press; 1979. pp. 63–102. [Google Scholar]
- 7.Bredt W. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol Microbiol. 1968;32:321–326. doi: 10.1159/000162074. [DOI] [PubMed] [Google Scholar]
- 8.Dirksen L B, Krebes K A, Krause D C. Phosphorylation of cytadherence-accessory proteins in Mycoplasma pneumoniae. J Bacteriol. 1994;176:7499–7505. doi: 10.1128/jb.176.24.7499-7505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drlica K, Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldner J, Göbel U, Bredt W. Mycoplasma pneumoniae adhesin localized to tip structure by monoclonal antibody. Nature. 1982;298:765–767. doi: 10.1038/298765a0. [DOI] [PubMed] [Google Scholar]
- 11.Fisseha M, Göhlmann H W, Herrmann R, Krause D C. Identification and complementation of frameshift mutations associated with loss of cytadherence in Mycoplasma pneumoniae. J Bacteriol. 1999;181:4404–4410. doi: 10.1128/jb.181.14.4404-4410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzoso G, Hu P C, Meloni G A, Barile M F. The immunodominant 90-kilodalton protein is localized on the terminal tip structure of Mycoplasma pneumoniae. Infect Immun. 1993;61:1523–1530. doi: 10.1128/iai.61.4.1523-1530.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Göbel U, Speth V, Bredt W. Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J Cell Biol. 1981;91:537–543. doi: 10.1083/jcb.91.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn T W, Willby M J, Krause D C. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J Bacteriol. 1998;180:1270–1276. doi: 10.1128/jb.180.5.1270-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu P C, Cole R M, Huang Y S, Graham J A, Gardner D E, Collier A M, Clyde W A., Jr Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science. 1982;216:313–315. doi: 10.1126/science.6801766. [DOI] [PubMed] [Google Scholar]
- 16.Kenri T, Sasaki T, Kano Y. Identification and characterization of HU protein from Mycoplasma gallisepticum. Biochem Biophys Res Commun. 1998;249:48–52. doi: 10.1006/bbrc.1998.9090. [DOI] [PubMed] [Google Scholar]
- 17.Kirchhoff H. Motility. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas—molecular biology and pathogenesis. Washington, D.C.: American Society for Microbiology; 1992. pp. 289–306. [Google Scholar]
- 18.Köhler P, Marahiel M A. Association of the histone-like protein HBsu with the nucleoid of Bacillus subtilis. J Bacteriol. 1997;179:2060–2064. doi: 10.1128/jb.179.6.2060-2064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause D C. Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle. Trends Microbiol. 1998;6:15–18. doi: 10.1016/S0966-842X(97)01168-2. [DOI] [PubMed] [Google Scholar]
- 20.Krause D C. Mycoplasma pneumoniae cytadherence: unraveling the tie that binds. Mol Microbiol. 1996;20:247–253. doi: 10.1111/j.1365-2958.1996.tb02613.x. [DOI] [PubMed] [Google Scholar]
- 21.Krebes K A, Dirksen L B, Krause D C. Phosphorylation of Mycoplasma pneumoniae cytadherence-accessory proteins in cell extracts. J Bacteriol. 1995;177:4571–4574. doi: 10.1128/jb.177.15.4571-4574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Layh-Schmitt G, Harkenthal M. The 40- and 90-kDa membrane proteins (ORF6 gene product) of Mycoplasma pneumoniae are responsible for the tip structure formation and P1 (adhesin) association with the Triton shell. FEMS Microbiol Lett. 1999;174:143–149. doi: 10.1111/j.1574-6968.1999.tb13561.x. [DOI] [PubMed] [Google Scholar]
- 23.Layh-Schmitt G, Herrmann R. Spatial arrangement of gene products of the P1 operon in the membrane of Mycoplasma pneumoniae. Infect Immun. 1994;62:974–979. doi: 10.1128/iai.62.3.974-979.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Layh-Schmitt G, Hilbert H, Pirkl E. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence-accessory protein HMW1. J Bacteriol. 1995;177:843–846. doi: 10.1128/jb.177.3.843-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Layh-Schmitt G, Himmelreich R, Leibfried U. The adhesin related 30-kDa protein of Mycoplasma pneumoniae exhibits size and antigen variability. FEMS Microbiol Lett. 1997;152:101–108. doi: 10.1111/j.1574-6968.1997.tb10415.x. [DOI] [PubMed] [Google Scholar]
- 26.Layh-Schmitt G, Podtelejnikov A, Mann M. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiology. 2000;146:741–747. doi: 10.1099/00221287-146-3-741. [DOI] [PubMed] [Google Scholar]
- 27.Margolin W. A green light for the bacterial cytoskeleton. Trends Microbiol. 1998;6:233–238. doi: 10.1016/s0966-842x(98)01280-3. [DOI] [PubMed] [Google Scholar]
- 28.Meng K E, Pfister R M. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J Bacteriol. 1980;144:390–399. doi: 10.1128/jb.144.1.390-399.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyata M, Seto S. Cell reproduction cycle of mycoplasma. Biochimie. 1999;81:873–878. doi: 10.1016/s0300-9084(99)00209-6. [DOI] [PubMed] [Google Scholar]
- 30.Miyata M, Yamamoto H, Shimizu T, Uenoyama A, Citti C, Rosengarten R. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology. 2000;146:1311–1320. doi: 10.1099/00221287-146-6-1311. [DOI] [PubMed] [Google Scholar]
- 31.Popham P L, Hahn T W, Krebes K A, Krause D C. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc Natl Acad Sci USA. 1997;94:13979–13984. doi: 10.1073/pnas.94.25.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Proft T, Herrmann R. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol Microbiol. 1994;13:337–348. doi: 10.1111/j.1365-2958.1994.tb00427.x. [DOI] [PubMed] [Google Scholar]
- 33.Proft T, Hilbert H, Layh-Schmitt G, Herrmann R. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J Bacteriol. 1995;177:3370–3378. doi: 10.1128/jb.177.12.3370-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinlan D C, Maniloff J. Membrane association of the deoxyribonucleic acid growing-point region in Mycoplasma gallisepticum. J Bacteriol. 1972;112:1375–1379. doi: 10.1128/jb.112.3.1375-1379.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Razin S, Jacobs E. Mycoplasma adhesion. J Gen Microbiol. 1992;138:407–422. doi: 10.1099/00221287-138-3-407. [DOI] [PubMed] [Google Scholar]
- 36.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero-Arroyo C E, Jordan J, Peacock S J, Willby M J, Farmer M A, Krause D C. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J Bacteriol. 1999;181:1079–1087. doi: 10.1128/jb.181.4.1079-1087.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosengarten R, Kirchhoff H. Gliding motility of Mycoplasma sp. nov. strain 163K. J Bacteriol. 1987;169:1891–1898. doi: 10.1128/jb.169.5.1891-1898.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Seto S, Miyata M. Cell reproduction and morphological changes in Mycoplasma capricolum. J Bacteriol. 1998;180:256–264. doi: 10.1128/jb.180.2.256-264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seto S, Miyata M. Partitioning, movement, and positioning of nucleoids in Mycoplasma capricolum. J Bacteriol. 1999;181:6073–6080. doi: 10.1128/jb.181.19.6073-6080.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro L, Losick R. Protein localization and cell fate in bacteria. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- 43.Stevens M K, Krause D C. Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1 and HMW4 in the cytoskeletonlike Triton shell. J Bacteriol. 1991;173:1041–1050. doi: 10.1128/jb.173.3.1041-1050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens M K, Krause D C. Mycoplasma pneumoniae cytadherence phase-variable protein HMW3 is a component of the attachment organelle. J Bacteriol. 1992;174:4265–4274. doi: 10.1128/jb.174.13.4265-4274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson M H, Collier A M. Ultrastructural study of Mycoplasma pneumoniae in organ culture. J Bacteriol. 1976;125:332–339. doi: 10.1128/jb.125.1.332-339.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]