The formation of large von Willebrand factor (VWF) tubules is a critical step in formation and maturation of high-molecular-weight multimers that are required for proper hemostasis. Anderson et al describe the structure of the central tubulation, revealing novel findings about how tubule formation and concatemerization occur. These findings provide clues as to how several von Willebrand disease mutations result in abnormal bleeding.
Key Points
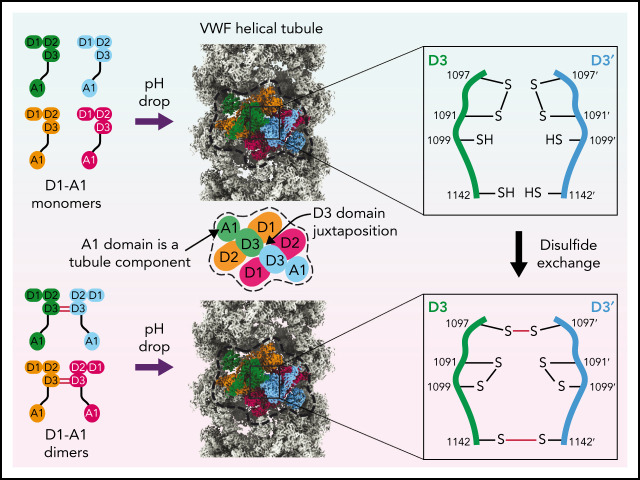
Structures of VWF tubules identify the cysteine residues responsible for concatemerization and reveal a disulfide bond exchange mechanism.
The A1 domain promotes tubule length by linking helical turns in the VWF tubule.
Visual Abstract
Abstract
von Willebrand factor (VWF) is an adhesive glycoprotein that circulates in the blood as disulfide-linked concatemers and functions in primary hemostasis. The loss of long VWF concatemers is associated with the excessive bleeding of type 2A von Willebrand disease (VWD). Formation of the disulfide bonds that concatemerize VWF requires VWF to self-associate into helical tubules, yet how the helical tubules template intermolecular disulfide bonds is not known. Here, we report electron cryomicroscopy (cryo-EM) structures of VWF tubules before and after intermolecular disulfide bond formation. The structures provide evidence that VWF tubulates through a charge–neutralization mechanism and that the A1 domain enhances tubule length by crosslinking successive helical turns. In addition, the structures reveal disulfide states before and after disulfide bond-mediated concatemerization. The structures and proposed assembly mechanism provide a foundation to rationalize VWD-causing mutations.
Introduction
von Willebrand disease (VWD) is the most common bleeding disorder worldwide and is characterized by excessive bleeding due to mutations in the gene encoding von Willebrand factor (VWF).1 VWF is an adhesive multidomain plasma glycoprotein (Figure 1A) that functions in hemostasis by acting as a stabilizer of coagulation factor VIII and a crosslinker of platelets and extracellular matrix components at sites of vascular injury. The ability of VWF to staunch bleeding is correlated with its ability to form long and polyvalent disulfide-linked concatemers.2 Short VWF concatemers are associated with the excessive bleeding of type 2A VWD.1 Maturation of VWF requires a remarkable intracellular biogenesis pathway that includes the formation of intra- and intermolecular disulfide bonds, glycosylation, and proteolytic removal of an N-terminal D1D2 prodomain3 (Figure 1A). Biogenesis starts in the endoplasmic reticulum (ER), where VWF dimerizes through disulfide bonds between C-terminal cysteine knot (CK) domains.4 Surprisingly, however, the disulfide bonds that covalently link VWF dimers into concatemers do not form in the oxidoreductase-rich ER3 but in the acidic environments of the trans-Golgi and Weibel-Palade bodies (WPBs).5,6 In these acidic environments, the C-terminal A2-CK domains in VWF zipper together to form a “dimeric bouquet,”7 and the dimers then assemble through their N-terminal portions into helical tubules.8 The helical tubules are proposed to template the formation of intermolecular disulfide bonds between adjacent D3 domains, leading to the formation of VWF concatemers.9 However, the mechanism by which tubules template concatemerization is incompletely understood, and there is uncertainty about which cysteine residues form the intermolecular disulfide linkages. Mass spectrometry showed that C1099 and C1142 were free in monomeric VWF D′D310 but not in mature VWF.11 Substitution of these 2 cysteine residues with alanine prevented disulfide-mediated dimerization of the D′D3 domain and concatemerization of full-length VWF.10 However, only C1142–C1142-linked peptides were observed in tryptic digests of plasma VWF.12 Further casting doubt on the presence of a C1099–C1099 linkage was a recent electron cryomicroscopy (cryo-EM) structure of human MUC2, a filament-forming mucin with an N-terminal head domain that resembles VWF, which indicated that C1142 and C1097 but not C1099 are responsible for intermolecular disulfide bonds.13 However, our previous crystal structure of monomeric VWF D′D3 showed that C1097 is engaged in an intradomain disulfide bond with C1091.14
Figure 1.
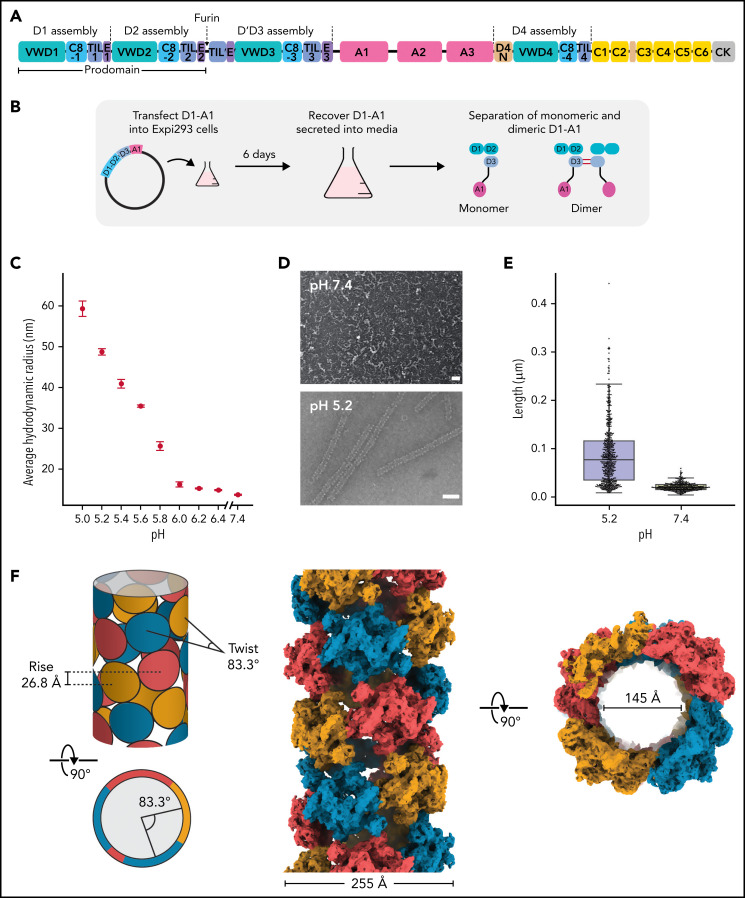
Structure of a VWF tubule. (A) Schematic of the domain organization of the VWF proprotein. Each D assembly except D′ is made of domains VWF type D domain, C8, TIL (trypsin inhibitor-like cysteine-rich domain), and E. D1 and D2 form a prodomain that is cleaved by furin protease before secretion. (B) Strategy to obtain VWF monomers and disulfide-linked dimers. VWF D1-A1 was expressed in Expi293 cells and purified from the media as monomers and disulfide-linked dimers. The furin cleavage site was mutated to ASAS to prevent prodomain dissociation. (C) DLS experiments of purified D1-A1 incubated at indicated pH showing an increase in average hydrodynamic radius with a decrease in pH. Error bars represent the difference between the means from 2 replicates. (D) Representative micrographs from negative-stain electron microscopy showing VWF dimers incubated at pH 7.4 and 5.2 (scale bars, 50 nm). (E) Quantification of (D) showing particle lengths measured from 3 micrographs from samples incubated together overnight. The bottom, middle, and top lines of the shaded boxes represent the first quartile, the median, and the third quartile of measured particle length, respectively. The length of the whiskers below and above the box plot represent the lesser of the range of data or 1.5 multiplied by the interquartile range. (F) Schematic and cryo-EM structure of the dimer-derived tubule showing the bead-like arrangement. The structure forms a right-handed helix with a helical rise of 26.8 Å and a helical twist of 83.3°. The cryo-EM map has been Gaussian filtered for visualization.
Here, we have determined cryo-EM structures of VWF helical tubules before and after intermolecular disulfide bond formation. The structures unambiguously identify the cysteine residues involved in concatemerization (C1142 and C1097) and reveal that tubulation templates concatemerization through a disulfide bond exchange mechanism. We propose a detailed assembly pathway that explains the pH-dependence of tubule formation, the role of the A1 domain in stabilizing tubules, and how mutations associated with type 2A VWD might impair VWF concatemerization by limiting tubule formation.
Methods
Expression and purification of VWF proteins
Sequences encoding VWF D1-D3 (residues 1-1264), D1-A1 (1-1464), and D1-A2 (1-1671) with a C-terminal hexahistidine tag and a furin cleavage site mutated to ASAS were cloned into PD2529 vectors. 1 L of Expi293 cells in Expi293 medium were transfected using FectoPRO (Polyplus, #101000014). Valproic acid and glucose were added to the culture 24 hours after transfection to final concentrations of 3 mM and 0.4%, respectively. Six days after transfection, secreted proteins were purified from the media by His-affinity in 20 mM Tris-HCl pH 8, 300 mM NaCl, and 10 mM CaCl2. After affinity chromatography, the purified protein was dialyzed into a low salt buffer (20 mM Tris-HCl pH 7.4, 50 mM NaCl, and 10 mM CaCl2). Next, anion-exchange chromatography was performed by elution with linearly increasing amounts of high salt buffer (20 mM Tris-HCl pH 7.4, 1 M NaCl, and 10 mM CaCl2). Final purification was performed by size-exclusion chromatography with a Sepharose 6 column (Cytiva) in 10 mM HEPES, pH 7.4, 100 mM NaCl, and 10 mM CaCl2. Protein purity was analyzed by SDS-PAGE (sodium dodecyl-sulfate polyacrylamide gel electrophoresis) (supplemental Figure 1A available on the Blood Web site).
Dynamic light scattering (DLS)
VWF proteins (22.5 μL at 0.11 mg/mL) were spiked with 3 μL 1 M sodium cacodylate buffer at various pHs and incubated at 37°C for 24 hours. Afterward, samples were placed in a 384 well black clear bottom plate (Aurora Microplates, #ABM2‐10100A). Measurements were taken on a DynaPro Plate Reader III (Wyatt Technology) at 0.1 µs intervals at 25°C. Average hydrodynamic radii were calculated using DYNAMICS (Wyatt Technology).
Negative-stain electron microscopy
VWF tubules (3 μL at 0.01 to 0.3 mg/mL) were deposited onto glow discharged (15 seconds at 30 mA voltage) grids (EMS, #CF200-Cu). After 1 minute, the grids were blotted with filter paper, washed twice with 3 μL of 1.5% uranyl formate (UF), incubated with 1.5% UF for 90 seconds, then blotted again. Grids were imaged on a Tecnai T12 microscope equipped with a Gatan UltraScan 895 camera. Particle lengths were measured using Fiji.15
Cryo-EM sample preparation and data collection
VWF proteins at 0.6 to 1 mg/mL were incubated in sodium cacodylate buffer at pH 5.2, 10 mM CaCl2, and 100 mM NaCl for 24 hours to allow tubule formation. After incubation, 3 μL of protein was deposited onto gold R2/1 grids (Quantifoil, #Q325AR1-2nm) precoated with continuous carbon that had been glow discharged for 15 seconds at 30 mA. The grids were blotted with a blot force of 2 to 5 and a blot time of 2 to 5 seconds using a Vitrobot Mark IV (Thermo Fisher Scientific) at 4°C and 100% humidity before being plunged into liquid ethane.
Cryo-EM data collection and image processing
Cryo-EM data for VWF D1-A1 tubules were collected on a Titan Krios microscope (Thermo Fischer Scientific) with a K3 detector with a BioQuantum Energy Filter (Gatan). Cryo-EM data for D1-A2 VWF tubules were collected on a Talos Arctica microscope with a K3 detector. Parameters used during data collection are provided in supplemental Table 1. All movies were motion-corrected using MotionCor2.16 Contrast transfer function (CTF) parameters were estimated with CTFFIND4.17 Particles were picked using crYOLO18 and imported into RELION-3.1.319 or RELION-4.0.20 Particles were extracted in 384- or 400-pixel boxes with an interbox distance of 123 or 197 pixels, respectively. Two- and 3-dimensional classification was used to remove false positives and contaminants. Three-dimensional refinement was initiated using a cylindrical reference with dimensions obtained from negative-stain electron microscopy (ns-EM) of in vitro assembled VWF tubules.21 Three-dimensional refinement for all datasets employed helical symmetry with a twist of 83.3° and a rise of 26.8 Å. CTF refinement and Bayesian polishing were used to improve map quality. Following helical processing, a binary mask applied to a central “bead” yielded maps with resolutions of 3.1, 3.2, and 4.7 Å for the D1A1 monomer, D1A1 dimer, and D1A2 dimer, respectively. Processing schemes are provided in supplemental Figures 1 and 2.
Model building and refinement
Maps were sharpened for model building and refinement using phenix.auto_sharpen.22 Model building was initiated by placing atomic models from the protein databank (PDB) of MUC2 (PDB: 6TM2)13 and the VWF A1 domain (PDB: 1AUQ)23 into the density using ChimeraX.24 Residues were mutated, inserted, or deleted to build the VWF polypeptide using Coot v.0.9.3.25 Atomic models were refined in ISOLDE26 and Phenix.real_space_refinement27 with symmetry constraints and restraints on Ramachandran angles and secondary structure.
Results
Cryo-EM reconstruction of VWF tubules
Prior work had shown that the VWF prodomain (D1D2) is essential for concatemerization5,28,29 and that D1-D3 constructs are sufficient for VWF tubulation in vitro, although the inclusion of the A1 domain led to longer tubules.21 We, therefore, recombinantly expressed and purified human D1-A1 (the VWF “head”) with the furin cleavage site mutated to ASAS to prevent prodomain cleavage in human Expi293 cells (Figure 1A-B). As seen previously,30 the C-terminally truncated VWF purified as size-separable monomers and disulfide-linked dimers (supplemental Figure 1A). Using DLS (Figure 1C) and ns-EM (Figure 1D-E), we demonstrated that the D1-A1 dimer could form tubules in low pH buffers containing calcium. We thus used pH 5.2 and 10 mM CaCl2 to generate VWF tubules for cryo-EM analysis (supplemental Figure 1C). Using this sample, we were able to resolve the structure of the VWF tubule to 3.3-Å resolution (Figure 1F; supplemental Table 1; supplemental Movie 1). The structure forms a 255-Å wide right-handed helix with an internal diameter of 145 Å. The overall dimensions, architecture, and helical parameters (83.3° twist and 26.8 Å rise) closely match cryo-ET reconstructions of VWF tubules inside WPBs,31 demonstrating that our in vitro assembled tubules recapitulate native intracellular tubules.
Structure of a bead
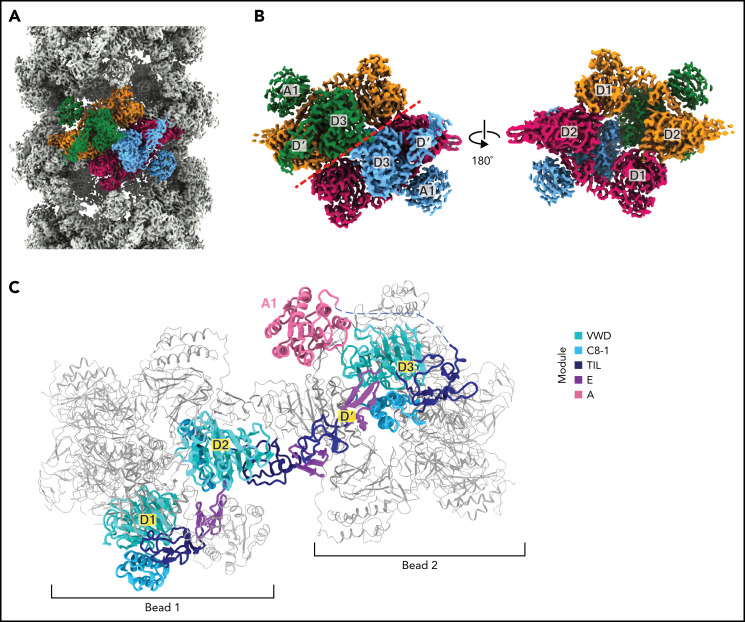
Like reconstructions of VWF tubules at low resolution,21,31 we observed a repeating bead-like density (Figure 2A; supplemental Movie 1). Each bead has C2 symmetry and is formed by parts of 4 different VWF monomers (Figure 2B), while a single VWF monomer spans 2 beads (Figure 2C). Two antiparallel D1D2 prodomains form the layer of the bead on the inner face of the tubule, while 2 D′D3 domains from neighboring molecules cradle between the D1 and D2 domains and form the outer layer of the bead (Figure 2B). A similar organization of D assemblies is found in MUC2.13 The positioning of the 2 D3 domains atop the D1D2 cradle juxtaposes them for disulfide linkage to one another and explains the critical role of the prodomain in concatemerization.29
Figure 2.
Organization of VWF within a tubule. (A) Cryo-EM map of the dimer-derived VWF tubule in gray with a single bead colored by each of the 4 monomeric VWF subunits from which it is formed. Beads have twofold symmetry. (B) A single bead. The left view has the same orientation as in (A) and has a red dashed line separating symmetrical halves. The right view shows the luminal facing portion of the bead, which is formed by 2 antiparallel D1D2 prodomains that form a cradle for the binding of 2 juxtaposed D′D3 domains. (C) A VWF monomer, colored by domain, adopts an extended conformation that spans 2 beads. The D1D2 prodomain is in 1 bead, and the D′D3 and A1 domains are in a neighboring bead. Domains of neighboring VWF molecules are colored gray and are shown with thinner loops, helices, and strands.
The A1 domain is a VWF tubule constituent
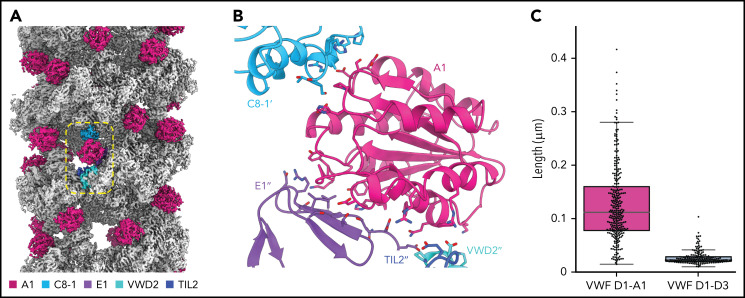
Our structure revealed that the A1 domain, a globular domain of 6 amphipathic α-helices surrounding a hydrophobic 6-stranded β-sheet core,23 locates to the outer tubule wall and participates in tubule packing by linking successive helical turns (Figure 3A). Specifically, each A1 domain makes trans interactions with E1, TIL2, and VWD2 domains of a VWF molecule in its own bead, as well as with a C8-1 domain 4 beads away in the next helical turn (consistent with the tubule having 4.2 beads per helical turn) (Figure 3B; supplemental Movie 1). Indeed, without the A1 domain, there would be no contacts between neighboring turns of the tubule. To test the contribution of the A1 domain to tubule formation, we assessed the ability of a construct truncated at the end of the D3 domain to form tubules. As expected from prior work,21 loss of the A1 domain did not prevent tubule formation but led to significantly shorter tubules (Figure 3C; supplemental Figure 2B). The involvement of the A1 domain in tubule packing and the promotion of long tubules may explain why D1-A1 constructs form longer WPB-like granules than D1-D3 constructs in cells.6
Figure 3.
The A1 domain is a component of the tubule and links helical repeats. (A) Cryo-EM map of the dimer-derived VWF tubule in gray with A1 domains in pink. A single A1 domain is boxed together with the domains of the neighboring molecules with which it interacts. (B) Details of the trans interactions an A1 domain makes with 2 neighboring VWF molecules. Distinct molecules are denoted by no apostrophe, 1 apostrophe (’), or 2 apostrophes (’’). (C) Quantification of tubule length observed by negative-stain electron microscopy with and without the A1 domain. Particle lengths were measured from 3 micrographs from samples incubated together overnight. Whisker plots are as described in the Figure 1E legend.
We next aimed to exclude the possibility that the A2 domain might also contribute to tubule formation. To do this, we purified the dimeric form of the D1-A2 construct (supplemental Figures 1A and 2A) and showed using DLS and cryo-EM that it could form tubules but with a mean length less than the D1-A1 construct (supplemental Figure 2B). Next, we determined a 4.8-Å resolution structure of tubules formed by the purified D1-A2 dimer (supplemental Figure 2C-D). The structure was nearly identical to the tubule formed with D1-A1 dimers (supplemental Figure 2E) and showed no additional density near the A1 domain that could correspond to the A2 domain (supplemental Figure 2F). This observation demonstrated that the A1 but not A2 domain is a structural component of the VWF helical tubule. In addition, the inability to resolve the A2 domain suggested that the 31 residues between the A1 and A2 domains form a flexible linker, in agreement with ns-EM of C-terminal VWF dimeric bouquets containing the A1 and A2 domains7 and explaining the absence of density for the C-terminal portion of VWF in reconstructions of native VWF tubules in WPBs.31
Structural basis for the pH dependence of tubule formation
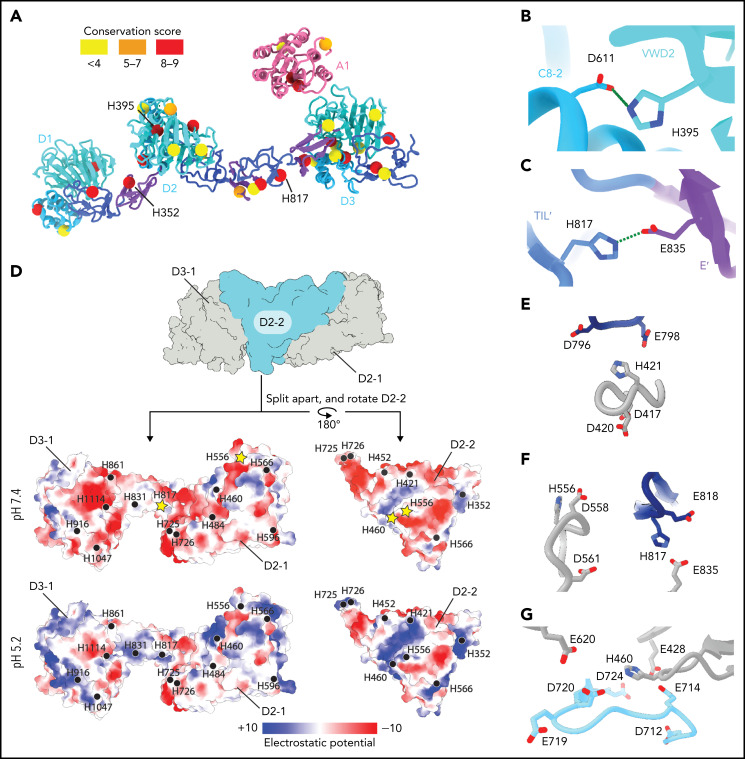
VWF tubulation is pH-dependent.21,32 Histidine protonation has been implicated in this phenomenon based on histidine having a pKa (6.0) between the neutral pH of the ER and the acidic pH of the trans-Golgi and WPB.33 One possible mechanism for pH-dependent tubulation is that protonated histidine sidechains form intermolecular salt bridges at acidic pH that stabilize the helical tubule. Therefore, we analyzed the 33 histidine residues that are resolved in our structure (Figure 4A; supplemental Table 2). First, we determined their conservation across VWF sequences.34 Second, we calculated electrostatic potential and structure-based pKas for each histidine to determine which histidine residues are specifically protonated by a decrease in pH from 7.4 to 5.2. Third, we analyzed their location in the VWF tubule. This analysis revealed that only 1 histidine residue (H352) is in a potential interfacial salt bridge. Together with prior work showing substitution of H352 with alanine did not impair VWF concatemerization,33 we conclude that interfacial salt bridges involving histidine residues are not required for VWF tubulation.
Figure 4.
Structural basis for pH-dependent VWF tubule formation. (A) The Cα positions of the 33 histidine residues resolved in the structure are depicted as spheres color-coded based on sequence conservation obtained from 140 homologs using the ConSurf server.47 (B) Details of the intramonomer salt bridge made by H395. (C) Details of the intramonomer salt bridge made by H817. (D) Interface between D2-D3 of one molecule (denoted D2-1 and D3-1; gray) and the D2 domain of another molecule (D2-2; teal) (top). The interfaces are separated and colored by electrostatic potential calculated at pH 7.4 (middle) and 5.2 (bottom). Electrostatic potentials were calculated using default coulombic parameters in ChimeraX 1.3.24 The positions of surface-exposed histidine residues are marked with a black circle or a yellow star if highlighted in panels (E-G). (E-G) Examples of histidine residues—(E) H421, (F) H556 and H817, and (G) H460—that change protonation state upon pH change from 7.4 to 5.2 in electronegative local environments.
An alternative mechanism is that histidine protonation forms intramolecular salt bridges that stabilize a VWF conformation capable of self-associating into helical tubules. Evidence for this mechanism comes from our observation that H395, a highly conserved residue in the VWD2 domain, forms a potential salt bridge with D611 in the C8-2 domain that might lead to a tighter association of these domains at acidic pH than at a neutral pH (Figure 4B). Consistent with this model, a H395A substitution completely abolished VWF concatemerization.33 H817 in the TIL′ domain also forms an apparent intramolecular salt bridge with E835 of the E′ domain (Figure 4C), which could stabilize the bridge between neighboring beads.
Our structure also reveals a third possible mechanism for how acidic pH permits tubulation. When we analyzed the interfaces between VWF molecules, we found that most interfaces shared an electronegative charge at pH 7.4 (Figure 4D). Repulsion between these interfaces at neutral pH might prevent tubulation. We observed that several histidine residues occur within these electronegative environments (H421, H460, H556, and H817) (Figure 4E-G). This leads us to propose a charge–neutralization model; as the pH drops in the trans-Golgi, the electronegative charge is neutralized (Figure 4D) by histidine, aspartic acid, and glutamic acid protonation, allowing tubules to form. The charge–neutralization model emphasizes that other residues besides histidine may regulate tubulation. Although we expected to see histidine residues in key interactions in tubules, we instead found that only a small number are found in such contacts, and these are restricted to intramolecular interfaces (Figure 4B-C). As pH is lowered in the trans and cis Golgi, it is not only histidine residues but also aspartic and glutamic acid residues that are protonated. The ratio of (Asp + Glu):His residues in the D1-A1 domains is 5:1, and thus protonation of acidic residues may also have an important effect on tubulation.
Notably, the pI of the A1 domain (8.8) is substantially more basic than D1-D3 (pI 5.1). Charge neutralization by the A1 domain may therefore provide another mechanism by which the A1 domain promotes VWF tubulation. However, this effect is not essential to the core mechanism of tubulation because tubules can form in the absence of the A1 domain.21
Identification of the disulfide bonds required for concatemerization
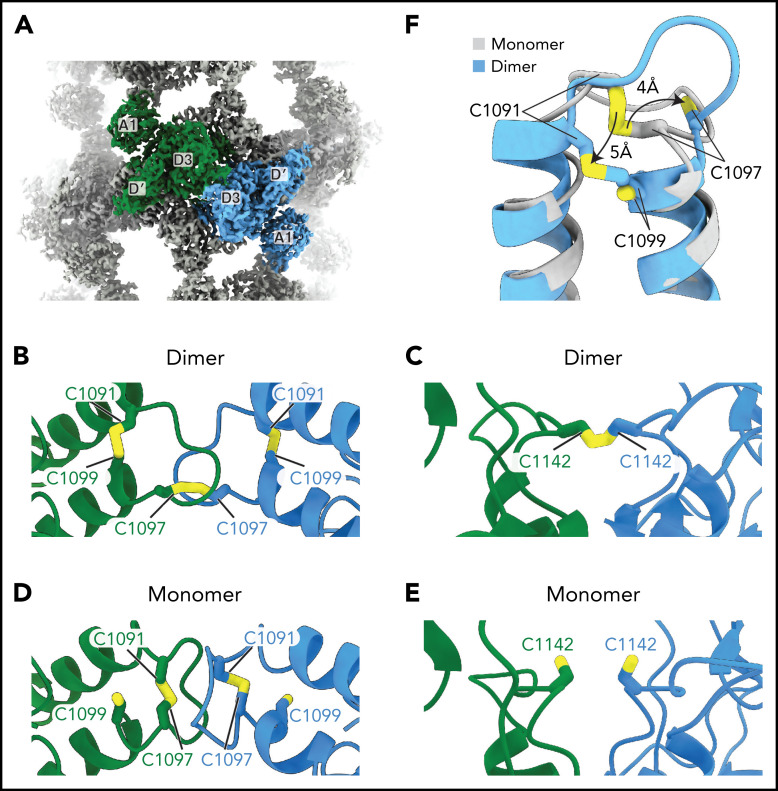
The structure conclusively demonstrates that C1097 and C1142 in the D3 domain are responsible for the disulfide-mediated concatemerization of VWF (Figure 5A-C). Density shows that both residues form intermolecular disulfide bonds with their equivalent residues in the neighboring D3 domain (supplemental Figure 3A-B). Thus, VWF and MUC2 multimers are disulfide-bonded through equivalent cysteine residues.13 We further observed that C1099, which was previously speculated to form an intermolecular disulfide bond,10 instead forms an intramolecular disulfide bond with C1091 (Figure 5B; supplemental Figure 3C).
Figure 5.
VWF concatemerization proceeds through a disulfide exchange mechanism. (A) Overview showing the juxtaposition of domains D′-A1 from separate molecules in the center of a single bead. (B-C) Arrangement of intra- and intermolecular disulfide bonds in VWF tubules generated with dimeric D1-A1. (D-E) Arrangement of interfacial cysteines and intramolecular disulfide in VWF tubules generated with monomeric D1-A1. (F) Superposition of D3 domains showing the rearrangement of the 1091-1097 loop between monomer and dimer states. Model superposition was performed using the matchmake function of ChimeraX.
To observe the conformation of VWF before the disulfide linkage of D3 domains, we reconstituted VWF tubules with our monomeric D1-A1 construct for cryo-EM analysis (supplemental Figure 1D). VWF retained its monomeric form within these tubules (supplemental Figure 1B). This observation confirms that prior disulfide-mediated concatemerization is not a prerequisite for VWF tubulation.8 From the monomer-derived VWF tubules, we obtained a 3.2-Å resolution structure that revealed no gross morphological changes and similar helical parameters to VWF tubules reconstituted with disulfide-linked dimers (supplemental Figure 2C-D). The only changes are at the interface between neighboring D3 domains (Figure 5A-F). In general, the D3 interface between monomers is less ordered than in the dimer-derived tubule, presumably because of increased flexibility caused by an absence of covalent linkages. Notable changes occur in the 910-923 and 1092-1098 loops (Figure 5F; supplemental Figure 4A-C). Compared with before intersubunit disulfide bonding, the 910-923 loop shifts into a hydrophobic pocket of the VWD3 domain (supplemental Figure 4A-B), and the 1092-1098 loop switches conformation (supplemental Figure 4C). The position of the 1092-1098 loop in the monomer-derived tubule matches the D′D3 monomer crystal structure.14 However, the C1142-containing loop in the crystal structure adopts a different conformation from that observed in either tubule structure (supplemental Figure 4D). In the monomeric crystal structure, determined at mildly alkaline pH, the loop conformation buries C1142 in a hydrophobic pocket. This conformation would protect against premature disulfide bond formation in the ER.
The structure of the monomer-derived tubule confirms that C1099 and C1142 are unpaired (supplemental Figure 3D-E), as expected from their ability to be alkylated in purified monomers.10 In contrast to their disulfide-bonded state in the dimer-derived tubules, the sulfhydryl groups from neighboring C1142 residues are clearly unbonded (Figure 5E; supplemental Figure 3E). C1097, the other residue responsible for VWF concatemerization, is confirmed to be in an intramolecular disulfide bond with C1091 (Figure 5D; supplemental Figure 3F), as observed in the D′D3 monomer crystal structure.14
VWF concatemerizes through a disulfide exchange mechanism
The observation that C1097 forms an intramolecular disulfide bond before concatemerization but forms an intermolecular disulfide bond in the concatemer confirmed our hypothesis that VWF concatemerization proceeds through a disulfide bond exchange mechanism.14 The details of this mechanism remain to be defined; however, we can rule out a suggestion that the D1 or D2 assemblies of VWF are oxidoreductases.35 This proposal was based on the presence of CXXC sequences in both the prodomain and in the active site sequence of oxidoreductase family members and that insertion of a single glycine into CXXC motifs in either the D1 or D2 assemblies inhibited VWF concatemerization.35 However, there is no overall structural homology between VWF D assemblies and oxidoreductases. Furthermore, the cysteine residues in these motifs are disulfide-bonded in the VWF monomer and >35 Å from the disulfide bonds at the D3:D3 interface (supplemental Figure 5A-C).
Interestingly, we observed additional density extending from the sulfhydryl group of unpaired C1099 in the monomer-derived tubule (supplemental Figure 3D) that could correspond to S-glutathionylation or formation of a disulfide with a component of the culture medium such as cysteine during D1-A1 protein production.
Disease mechanisms
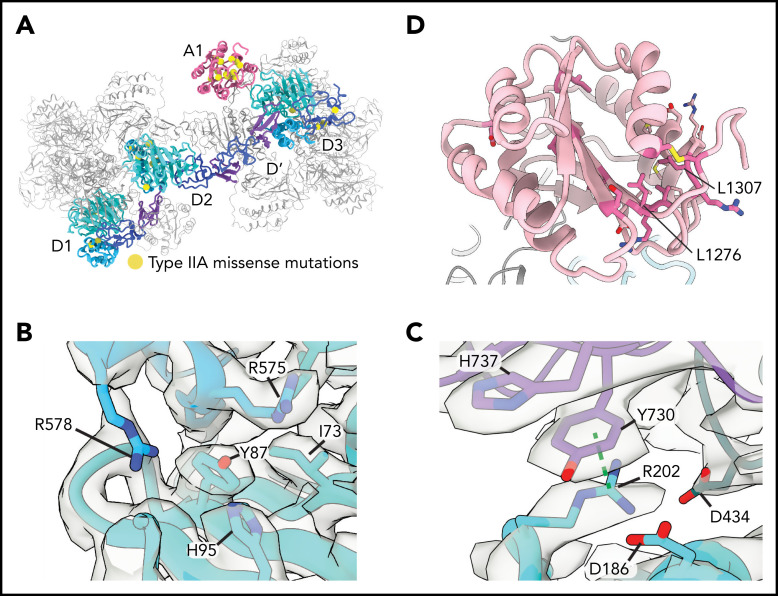
The loss of ultralong VWF concatemers in type 2A VWD results from impaired intracellular multimerization of VWF or increased susceptibility of secreted VWF to proteolysis by ADAMTS13. Our structures provided an opportunity to interpret, at the atomic level, a well-annotated list of clinical mutations36 (Figure 6A; supplemental Table 3). We focused on type 2A VWD mutations that did not substitute cysteine residues, which likely affect VWF folding by disrupting normal disulfide bonding. We note that several noncysteine mutations map to interfaces between neighboring molecules, including Y87S and R202W. The Y87S mutation has been shown to disrupt tubular packing and block concatemerization.6,37 Y87 lines a crevice in the VWD1 domain into which an α-helix protrudes from the C8-2 domain of a neighboring molecule (Figure 6B). The Y87S substitution would admit solvent and destabilize this packing interaction, potentially causing decreased tubule formation. R202 sits at the nexus between 3 VWF molecules at the interface between beads and forms both cation-π interactions (with Y730 of the neighboring E2 domain) and electrostatic interactions with D186 and D434 (Figure 6C). Substitution to tryptophan38 (R202W) would disrupt these interactions, consistent with our model that charge neutralization is required for effective tubulation.
Figure 6.
Type 2A mutations. (A) Positions of VWD type 2A missense mutations mapped onto the structure of VWF D1-A1 as it occurs in a tubule. (B) Y87 lines a crevice in the VWD1 domain. The type 2A VWD mutation Y87S would allow solvent into this crevice, potentially disrupting tubule formation. (C) R202 forms intra- and intermolecular interactions with 2 additional VWF molecules denoted in purple and dark teal. R202W, a mutation causing type 2A VWD, would disrupt these interactions. A dashed green line shows a π–cation bond. (D) A1 domain in light pink with residues altered in type 2A VWD colored in dark pink. Leucine residues described in the text are labeled.
A number of type 2A VWD mutations with unknown pathomechanisms map to the A1 domain (Figure 6D; supplemental Table 3). Our observation that the A1 domain is a constituent of the tubule wall and enhances tubule length in vitro (supplemental Figure 3C) suggests that mutations that impair the stability of the A1 domain might limit VWF concatemerization by generating short tubules. Consistent with this model, the VWD-associated L1307P substitution, which inserts a proline into the hydrophobic core of the A1 domain, has short and round WPB-like granules in cells that are indicative of reduced VWF tubule length.39 L1276P likely functions through a similar mechanism by disrupting the β1 strand of A140 (Figure 6D).
Discussion
Here, we have leveraged the propensity of C-terminally truncated VWF constructs to form tubules in vitro to determine structures of VWF before and after disulfide bond formation between D3 domains. These structures, together with prior information, allow us to generate a multistep model for the assembly of full-length VWF into concatemers (Figure 7).
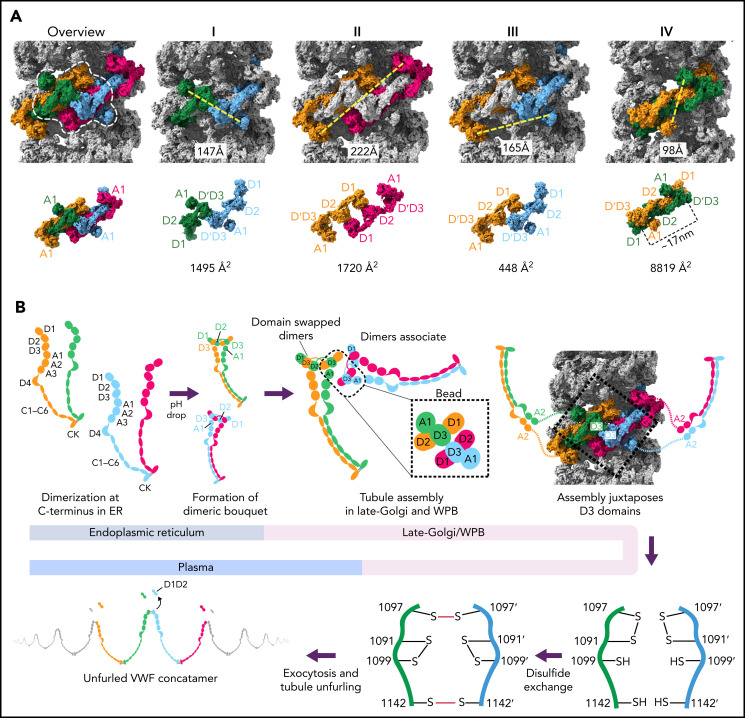
Figure 7.
Model for VWF tubule assembly. (A) Options I-IV show the possible molecules that could be disulfide bonded at their C-termini in native tubules generated with full-length VWF. Distances between A1 domains (calculated between A1464 Cα positions) are shown below, together with surface representations of potential C-terminally–linked dimers and their quantified interfacial surface area. An overview is provided with a single bead denoted with a dashed white outline. Interfacial surface areas were calculated using PDBePISA.41 (B) Schematic showing a hypothetical maturation pathway for full-length VWF based on our cryo-EM structures and prior work. In this model, the acidic environment of the Golgi induces 2 changes in the VWF dimer: the A2-CK domains zipper together, and the N-terminal heads form a “dimer swapped” conformation with D3 of one monomer nestling in a D1-D2 cradle of the other monomer. N-terminal dimer association leads to tubule assembly. Within the tubule, D3 domains from different dimers are juxtaposed. Once positioned, a disulfide exchange occurs where C1097 is liberated from an intramolecular disulfide bond to form 1 of 2 disulfides that concatemerize VWF dimers at their D3 domain. The second disulfide, between adjacent C1142 residues, occurs without disulfide exchange. With dimers linked at their D3 domains, the VWF tubule can unfurl into a high molecular weight VWF concatemer upon exocytosis into the blood plasma.
Although our structures were generated from N-terminal domains only, a process of elimination can be used to determine which VWF molecules belong to the same C-terminally linked dimer in vivo. We can exclude the subunits that contribute D3 domains to a single bead (Figure 7AI), as their disulfide bonding would generate a closed dimer. We can also exclude the 2 subunits that contribute prodomains to a single bead (Figure 7AII) because their extended and antiparallel configuration places their A1 domains 22 nm apart, which is more than twice the distance between A2 domains in the dimeric A1-CK bouquet.7 It is unlikely that this distance can be bridged by the 31-residue flexible linker between the A1 and A2 domains. Similarly, the distance between A1 domains makes Figure 7AIII unlikely. Instead, we propose that the dimer corresponds to symmetry-related subunits with prodomains in adjacent beads (Figure 7AIV), where the 10-nm distance between A1 domains matches the distance between A2 domains in the dimeric A1-CK bouquet.7
The interface between VWF monomers in the most feasible model for linking dimeric bouquets (Figure 7AIV) is ∼8830 Å2, more than 5× larger than in alternative models (Figure 7A). This large size makes it highly preferred over the alternatives.41 Notably, this dimer is also domain-swapped (ie, D3 of one monomer in the dimeric bouquet is nestled in the D1-D2 cradle of the other monomer) (Figure 7A-B). Domain swapping was also seen in MUC2 filaments13 and occurred because the connection between the D2 domain and the D′ domain is shorter in the extended conformation of the D1-D3 unit shown in Figure 7A than it would be if the monomer doubled back to allow its own D3 to interact with D1-D2.
A consequence of the intertwined, domain-swapped arrangement is that the 2 D3 domains from a single dimer are kept apart (with ∼17 nm between equivalent C1142 residues), thereby preventing closed dimer formation. Noncovalent association of the VWF heads in this manner would also reduce the risk of heads from the same dimer becoming incorporated into different tubules, which would lead to tubule bundling inconsistent with the regularly aligned arrangement of VWF tubules in WPBs.42 It is for these reasons that we hypothesize that noncovalent head pairing occurs as a discrete step before tubule formation. Intradimer head pairing could be the major pH-regulated step that, once formed, is sufficient for tubule formation. However, histidine residues experimentally important for tubule formation33 (eg, H95 and H737) are found outside the putative intradimer interface as well as within it (H395, H460, and H817) (supplemental Table 2), implying that head pairing alone may not fully explain the pH dependence of tubule formation.
Having rationalized the C-terminally linked VWF dimer in the VWF tubule, we propose a model (Figure 7B) wherein the first step of VWF maturation after leaving the ER is zippering together of the C-terminal A2-CK domains into a dimeric bouquet.7 We propose that 2 dimeric bouquets next associate through their N-terminal heads to form helical tubules. In our model, the pairing of C-terminally disulfide-linked dimers through their N-terminal heads recruits additional dimers through D1-A1 interactions to form a tubule that can polymerize at either tubule end. The dimerized A2-CK tails would spiral outwards from the central tubule but with sufficient flexibility conferred by the A1-A2 linker to not be seen in tomographic reconstructions.31 The A1 domain crosslinks successive helical turns, a property that reinforces the tubule, explaining how the A1 domain promotes increased tubule length21 (Figure 3C) and why some mutations within the A1 domain cause type 2A VWD through the loss of ultralong concatemers.
A consequence of tubule formation is that D3 domains from neighboring dimers are brought sufficiently close to allow intersubunit disulfide bonding. We show that disulfide bonding must occur through a disulfide bond exchange mechanism as one of the paired cysteines, C1097, is initially in an intramolecular disulfide bond with C1091. C1099 is likely critical to this reshuffling as it eventually becomes paired with C1091 after intersubunit disulfide bonding. We propose that nucleophilic attack by C1099 on the C1091-C1097 disulfide liberates C1097 for the formation of the C1097–C1097' disulfide in concatemers as previously proposed43 (Figure 7B). The disulfide linkage between C1142 residues is promoted by their close juxtaposition in the tubule arrangement and can occur without disulfide exchange. Further work will be required to clarify a more detailed mechanism, including identifying which enzymes or small molecules act as electron acceptors for the cysteine sulfhydryl groups. With its D3 disulfide bonds formed, VWF is stored in WPBs. During secretion, the pH in WPBs rises to that in the bloodstream, allowing VWF tubules to uncoil, during which the mature VWF concatemer and the cleaved prodomain dissociate.
While preparing this manuscript, 2 other groups reported cryo-EM structures of short VWF tubules reconstituted in vitro from D1D2 prodomains and dimeric D′D3.44-46 These reports agree with our assignment of the cysteines responsible for VWF concatemerization. However, without the stabilization provided by the A1 domain, their tubules contained only a few helical turns. Furthermore, helical parameters of tubules assembled without the A1 domain less closely match helical paramaters of VWF tubules in vivo compared to our reconstructions including the A1 domain. A key finding from our study is that the A1 domain plays a structural role in VWF concatemerization, suggesting that type 2A mutations within the A1 domain may cause defective concatemer genesis.
Conclusions
We elucidate key steps in VWF biogenesis through cryo-EM structures of VWF tubules before and after intermolecular disulfide bond formation. From these structures, we present a model for how charge neutralization in the acidic environment of the trans-Golgi allows tubule formation in which the D3 domains are juxtaposed for disulfide bonding through a disulfide bond exchange mechanism.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
Cryo-EM data were collected at The Harvard Cryo-EM Center for Structural Biology. The authors thank Sarah Sterling, Richard Walsh, and Shaun Rawson for microscopy and computing assistance.
J.R.A., T.A.S., and A.B. are supported by grants from the National Institutes of Health (F30-HL162128, R01-HL148755, and R01-GM141109, respectively).
Footnotes
Atomic models of VWF tubules formed with monomeric D1-A1 and dimeric D1-A1 have been deposited to the PDB with accession codes 8D3C and 8D3D, respectively. Composite maps for the VWF tubules formed with monomeric D1-A1 or dimeric D1-A1 have been deposited to the Electron Microscopy Data Bank (EMDB) with accession codes EMD-27156 and EMD-27157, respectively. These density maps are composites derived from stitching together a central masked bead to create a tubule with a length similar to the helical reconstruction. The unstitched map for the VWF tubule formed from dimeric D1-A2 has been deposited to the EMDB with accession code EMD-27158.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.R.A. collected and processed all electron microscopy data and built atomic models; J.L. purified proteins, performed light scattering experiments, and assembled tubules; T.A.S. and A.B. supervised the research; and J.R.A., T.A.S., and A.B. wrote the paper with contributions from J.L.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy A. Springer, Boston Children’s Hospital, 3 Blackfan Circle, Room 3103, Boston, MA 02115; e-mail: springer@crystal.harvard.edu; and Alan Brown, Harvard Medical School, 240 Longwood Ave, Room C249A, Boston, MA 02115; e-mail: alan_brown@hms.harvard.edu.
REFERENCES
- 1.Leebeek FWG, Eikenboom JCJ. von Willebrand’s disease. N Engl J Med. 2016;375(21):2067-2080. [DOI] [PubMed] [Google Scholar]
- 2.Federici AB, Bader R, Pagani S, Colibretti ML, De Marco L, Mannucci PM. Binding of von Willebrand factor to glycoproteins Ib and IIb/IIIa complex: affinity is related to multimeric size. Br J Haematol. 1989;73(1):93-99. [DOI] [PubMed] [Google Scholar]
- 3.Wagner DD, Marder VJ. Biosynthesis of von Willebrand protein by human endothelial cells. Identification of a large precursor polypeptide chain. J Biol Chem. 1983;258(4):2065-2067. [PubMed] [Google Scholar]
- 4.Marti T, Rösselet SJ, Titani K, Walsh KA. Identification of disulfide-bridged substructures within human von Willebrand factor. Biochemistry. 1987;26(25):8099-8109. [DOI] [PubMed] [Google Scholar]
- 5.Mayadas TN, Wagner DD. In vitro multimerization of von Willebrand factor is triggered by low pH. Importance of the propolypeptide and free sulfhydryls. J Biol Chem. 1989;264(23):13497-13503. [PubMed] [Google Scholar]
- 6.Michaux G, Abbitt KB, Collinson LM, Haberichter SL, Norman KE, Cutler DF. The physiological function of von Willebrand’s factor depends on its tubular storage in endothelial Weibel-Palade bodies. Dev Cell. 2006;10(2):223-232. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y-F, Eng ET, Nishida N, Lu C, Walz T, Springer TA. A pH-regulated dimeric bouquet in the structure of von Willebrand factor. EMBO J. 2011;30(19):4098-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner DD, Saffaripour S, Bonfanti R, et al. Induction of specific storage organelles by von Willebrand factor propolypeptide. Cell. 1991;64(2):403-413. [DOI] [PubMed] [Google Scholar]
- 9.Springer TA. von Willebrand factor, jedi knight of the bloodstream. Blood. 2014;124(9):1412-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purvis AR, Gross J, Dang LT, et al. Two Cys residues essential for von Willebrand factor multimer assembly in the Golgi. Proc Natl Acad Sci USA. 2007;104(40):15647-15652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legaz ME, Schmer G, Counts RB, Davie EW. Isolation and characterization of human factor VIII (antihemophilic factor). J Biol Chem. 1973;248(11):3946-3955. [PubMed] [Google Scholar]
- 12.Dong Z, Thoma RS, Crimmins DL, McCourt DW, Tuley EA, Sadler JE. Disulfide bonds required to assemble functional von Willebrand factor multimers. J Biol Chem. 1994;269(9):6753-6758. [PubMed] [Google Scholar]
- 13.Javitt G, Khmelnitsky L, Albert L, et al. Assembly mechanism of mucin and von Willebrand factor polymers. Cell. 2020;183(3):717-729.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong X, Leksa NC, Chhabra ES, et al. The von Willebrand factor D’D3 assembly and structural principles for factor VIII binding and concatemer biogenesis. Blood. 2019;133(14):1523-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14(4):331-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192(2):216-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner T, Lusnig L, Pospich S, Stabrin M, Schönfeld F, Raunser S. Two particle-picking procedures for filamentous proteins: SPHIRE-crYOLO filament mode and SPHIRE-STRIPER. Acta Crystallogr D Struct Biol. 2020;76(Pt 7):613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zivanov J, Nakane T, Forsberg BO, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimanius D, Dong L, Sharov G, Nakane T, Scheres SHW. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem J. 2021;478(24):4169-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang R-H, Wang Y, Roth R, et al. Assembly of Weibel-Palade body-like tubules from N-terminal domains of von Willebrand factor. Proc Natl Acad Sci USA. 2008;105(2):482-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terwilliger TC, Sobolev OV, Afonine PV, Adams PD. Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr D Struct Biol. 2018;74(Pt 6):545-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley J, Cruz M, Handin R, Liddington R. Crystal structure of the von Willebrand factor A1 domain and implications for the binding of platelet glycoprotein Ib. J Biol Chem. 1998;273(17):10396-10401. [DOI] [PubMed] [Google Scholar]
- 24.Pettersen EF, Goddard TD, Huang CC, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown A, Long F, Nicholls RA, Toots J, Emsley P, Murshudov G. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 1):136-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croll TI. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol. 2018;74(Pt 6):519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afonine PV, Poon BK, Read RJ, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74(Pt 6):531-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verweij CL, Hart M, Pannekoek H. Expression of variant von Willebrand factor (vWF) cDNA in heterologous cells: requirement of the pro-polypeptide in vWF multimer formation. EMBO J. 1987;6(10):2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise RJ, Pittman DD, Handin RI, Kaufman RJ, Orkin SH. The propeptide of von Willebrand factor independently mediates the assembly of von Willebrand multimers. Cell. 1988;52(2):229-236. [DOI] [PubMed] [Google Scholar]
- 30.Purvis AR, Sadler JE. A covalent oxidoreductase intermediate in propeptide-dependent von Willebrand factor multimerization. J Biol Chem. 2004;279(48):49982-49988. [DOI] [PubMed] [Google Scholar]
- 31.Berriman JA, Li S, Hewlett LJ, et al. Structural organization of Weibel-Palade bodies revealed by cryo-EM of vitrified endothelial cells. Proc Natl Acad Sci USA. 2009;106(41):17407-17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner DD, Mayadas T, Marder VJ. Initial glycosylation and acidic pH in the Golgi apparatus are required for multimerization of von Willebrand factor. J Cell Biol. 1986;102(4):1320-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang LT, Purvis AR, Huang R-H, Westfield LA, Sadler JE. Phylogenetic and functional analysis of histidine residues essential for pH-dependent multimerization of von Willebrand factor. J Biol Chem. 2011;286(29):25763-25769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glaser F, Pupko T, Paz I, et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19(1):163-164. [DOI] [PubMed] [Google Scholar]
- 35.Mayadas TN, Wagner DD. Vicinal cysteines in the prosequence play a role in von Willebrand factor multimer assembly. Proc Natl Acad Sci USA. 1992;89(8):3531-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hampshire DJ, Goodeve AC. The international society on thrombosis and haematosis von Willebrand disease database: an update. Semin Thromb Hemost. 2011;37(5):470-479. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg JB, Haberichter SL, Jozwiak MA, et al. The role of the D1 domain of the von Willebrand factor propeptide in multimerization of VWF. Blood. 2002;100(5):1699-1706. [DOI] [PubMed] [Google Scholar]
- 38.Baronciani L, Federici AB, Punzo M, et al. Type 2A (IIH) von Willebrand disease is due to mutations that affect von Willebrand factor multimerization. J Thromb Haemost. 2009;7(7):1114-1122. [DOI] [PubMed] [Google Scholar]
- 39.Wang J-W, Bouwens EAM, Pintao MC, et al. Analysis of the storage and secretion of von Willebrand factor in blood outgrowth endothelial cells derived from patients with von Willebrand disease. Blood. 2013;121(14):2762-2772. [DOI] [PubMed] [Google Scholar]
- 40.Meyer D, Fressinaud E, Gaucher C, et al. ; INSERM Network on Molecular Abnormalities in von Willebrand Disease . Gene defects in 150 unrelated French cases with type 2 von Willebrand disease: from the patient to the gene. Thromb Haemost. 1997;78(1):451-456. [PubMed] [Google Scholar]
- 41.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774-797. [DOI] [PubMed] [Google Scholar]
- 42.Streetley J, Fonseca A-V, Turner J, et al. Stimulated release of intraluminal vesicles from Weibel-Palade bodies. Blood. 2019;133(25):2707-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong X, Springer TA. Disulfide exchange in multimerization of von Willebrand factor and gel-forming mucins. Blood. 2021;137(9):1263-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shu Z, Zeng J, Xia L, Cai H, Zhou A. Structural mechanism of VWF D’D3 dimer formation. Cell Discov. 2022;8(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng J, Shu Z, Liang Q, et al. Structural basis of von Willebrand factor multimerization and tubular storage. Blood. 2022;139(22):3314-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Javitt G, Fass D. Helical self-assembly of a mucin segment suggests an evolutionary origin for von Willebrand factor tubules. Proc Natl Acad Sci USA. 2022;119(15):e2116790119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashkenazy H, Abadi S, Martz E, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44(W1):W344-W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.