SUMMARY
Escherichia coli O157 is a human pathogen carried asymptomatically by cattle and shed in their faeces. Infection can occur from the consumption of contaminated beef or by direct contact. Large variations of E. coli O157 shedding in cattle exist and vary in the number of cattle positive for E. coli O157 and the amount of bacteria (c.f.u./g faeces) shed by positive animals. To investigate E. coli O157 shedding and super-shedding (>104 c.f.u./g) we used daily sampling over two 8-day periods; in January 2013 (n = 12) and February 2013 (n = 21). Samples were tested by direct faecal culture for enumeration and by immunomagnetic separation to detect lower levels of shedding. We identified three patterns of shedding, similar to previously observed descriptions: intermittent, transient and consistent. The most commonly observed pattern was intermittent shedding and variation in the level of shedding could be large. This extreme variation is demonstrated by a heifer from which E. coli O157 could be not detected one day, was super-shedding E. coli O157 the next and was detected as shedding >100 c.f.u./g the following day. Recto-anal mucosal swab testing did not predict super-shedding in this cohort of heifers. The variable individual patterns of shedding suggest that a common mechanism of infection may not operate within such a herd when considering previously described patterns and the inferred mechanisms. The sporadic and intermittent nature of shedding is a challenge to identifying risk factors and potential intervention strategies.
Key words: Food hygiene, Shiga-like toxin-producing E. coli
INTRODUCTION
Estimates of Escherichia coli O157 prevalence within cattle populations range widely as reviewed elsewhere [1–3]. The ‘super-shedder’ theory in which a few cattle shed high levels of E. coli O157, usually classified as >104 c.f.u./g [4], was introduced to explain these heterogeneous results [5]. Animals shedding at these high levels are attributed with increased environmental contamination, infection of cohorts, and increased public health risks at slaughter. Modelling has suggested that 20% of animals are responsible for 80% of transmission [5] and that ~4% of the population are super-shedders [6]. In prevalence studies super-shedding has been demonstrated at rates of 0·7–23% [7, 8]. Control methods targeted at these animals are therefore indicated to reduce potential exposure and the public health risk. Further research into the super-shedding phenomenon has led to the subsequent suggestion that animals do not persistently shed at such high levels [9]. Robinson et al. [10] used an intensive sampling strategy (~3 h over 5 days) and identified calves persistently shedding E. coli O157 at high levels (>103 c.f.u./g) and calves shedding intermittently. These findings raise more questions than they answer regarding the heterogeneous nature of E. coli O157 prevalence, the dynamics of infection and the significance of super-shedding in dissemination of E. coli O157 within a cohort.
Large differences in the overall prevalence within a cohort and the shedding levels of individual animals between weekly samplings has been previously noted [11, 12]. More information regarding the shedding events between two weekly sampling points could therefore provide much greater information on the dynamics of shedding. Previous studies have demonstrated the clearance of E. coli O157 within a short period following experimental infection. Following experimental inoculation (peroral gastric intubation) with 1 × 109 c.f.u. most calves were culture negative within 2 weeks of infection and this period was reduced following a second infection [13]. In another study calves were detected as shedding E. coli O157 for a mean of 30 days following experimental infection orally with 5 × 108 c.f.u.; following a second infection calves shed for 6–8 days after which it was not detected by the culture methods used [14]. This indicates that weekly sampling may not detect all shedding events. In an environment in which E. coli O157 is abundant, rapid changes in E. coli O157 shedding may be explained by environmental exposure leading to ingestion and clearance in a continuing cycle [15]. Other work has shown long-duration consistent shedding by some cattle, which has been associated with ‘colonization’. Lim et al. [16] described a naturally infected steer which was detected positive by recto-anal mucosal swab (RAMS) enrichment culture for every sample collected from weekly or bi-weekly sampling over 12 months. This study supported the earlier work of Naylor et al. [17] which identified the recto-anal junction (RAJ) as a primary location of E. coli O157 colonization in cattle. Associations between RAJ colonization, long-duration shedding, super-shedding and detection from RAMS (compared to faeces) have been proposed [16–19].
To further investigate the heterogeneity of E. coli O157 faecal shedding, two intensive sampling periods of 8 days each nested within a longitudinal study were conducted. Based on previous observations of variations from weekly sampling [11, 12], daily sampling was undertaken to provide more information on the time-frame over which shedding varies. Heifers included in the study were known to have shed E. coli O157 in faeces at frequent occasions or at high levels previously. The aims of this study were to clarify potential differences between colonization and transient shedding with re-infection, and to assess associations between consistently positive heifers, super-shedding and detection by RAMS.
METHODS
Animals and samples
Heifers were selected from a cohort of 52 heifers enrolled in a 6-month longitudinal study of E. coli O157 shedding which were being raised as replacement dairy cows under standard operating practices within the commercial University herd. The management system under which these heifers were raised is representative of standard dairy practices in Australia. Sampling for the longitudinal study was undertaken weekly, and commenced 4 months prior to the first intensive sampling period. Heifers were maintained at pasture and received supplementary Lucerne hay and high-protein pellets ad libitum. Heifers were home-bred and aged between 7–11 months at the start of the first intensive sampling period. This age group has demonstrated a higher prevalence than usually observed in unweaned calves and adult cattle [20, 21]. Practical and economic constraints prohibited the inclusion of the entire cohort in these intensive studies, or more frequent sampling, hence heifers were selected on the basis of results from the larger longitudinal study (data not shown). Twelve heifers were selected for the first intensive study (IS1) in January 2013. Heifers selected had demonstrated at least one super-shedding event (defined as >104 c.f.u./g) or had tested positive on ⩾50% occasions to date during the longitudinal study. The second intensive study (IS2) was conducted in February 2013. Twenty-one heifers were selected including the 12 heifers from IS1 plus heifers which had demonstrated super-shedding or were identified as positive from ⩾50% of samples to date in the longitudinal study. Any heifers demonstrating shedding detectable by direct faecal culture (DFC) on the first sampling day, when the entire cohort of 52 heifers was sampled as part of the longitudinal study, were also included in IS2.
Each animal sampling point (ASP) represented a faecal and a RAMS sample from a heifer at a single sampling point. Faeces were tested by DFC for enumeration of shedding levels. Enriched faeces and RAMS were tested by immunomagnetic separation (IMS) for detection of shedding below the level of detection of DFC (100 c.f.u./g). Samples were obtained for eight consecutive days, between 07:00 and 08:00 hours each day, for both of the intensive sampling periods. RAMS were obtained prior to faecal sampling to minimize faecal contamination of the swab. The RAJ mucosa was swabbed with firm pressure using a sterile, cotton-tipped swab which was placed into 10 ml buffered peptone water (BPW). Faecal samples >10 g were obtained by digital rectal palpation and sealed into a zip-lock bag (generic). All samples were placed on ice for transportation to the laboratory and processed within 1 h of collection.
Ethical standards
The use of animals in this study was approved and monitored under University of Sydney Animals Ethics Committee Protocol number N00/4-2011/3/5487.
DFC
Faecal samples were diluted by weighing 10 g faeces into 90 ml BPW and shaken vigorously until faeces were observed to break up and disperse in the broth. A 100 μl aliquot of this dilution was spread-plated onto sorbitol MacConkey agar (Lab M, UK) supplemented with 0·05 mg/l cefixime and 2·5 mg/l tellurite (CT supplement, Lab M, UK) (CT-SMAC) and incubated at 37 °C overnight. Presumptive E. coli O157 colonies were identified, counted and confirmed using an E. coli O157 latex agglutination test (SSI, Denmark).
IMS
IMS was performed on enriched cultures following the manufacturer's instructions. Immediately following culture for enumeration, faecal dilutions in BPW were enriched at 37 °C for 6 h. RAMS in BPW were thoroughly mixed on a vortex mixer (~15 s) and enriched at 37 °C for 6 h. Enrichment broths were stored at 4 °C overnight. A 1·5 ml aliquot of the enrichment broth was taken and shaken well prior to IMS. Following a clarification spin at 500 g for 2 min, 1 ml of the supernatant was mixed with 20 μl of Dynabeads® (Invitrogen, USA) and separated on a plate magnet. The supernatant was removed and the beads resuspended in 1 ml of IMS wash buffer (phosphate buffered saline/0·05% Tween 20). This wash was repeated three times and the beads then resuspended in 100 μl wash buffer after the final wash. Of this final suspension, 50 μl was spread-plated onto CT-SMAC. After overnight incubation at 37 °C suspect colonies were identified and confirmed as E. coli O157 by latex agglutination. Heifers were classified as positive if either the faecal or RAMS sample (or both) was detected positive for E. coli O157.
Statistical analysis
The prevalence of E. coli O157 shedding across both intensive sampling periods demonstrated normal distributions. Pearson correlation statistics were used to investigate the correlation between herd prevalence of E. coli O157 at sampling points and prevalence of E. coli O157 at previous sampling points within the same sampling period, and the prevalence of high shedding (>100 c.f.u./g). Results at the individual sample level were tested for associations with the individual's previous results using logistic regression. In this model all results were considered as a binomial outcome, positive by any test and heifer was included as a random variable. All analysis was undertaken in GenStat 14th edition (VSN International, UK).
RESULTS
Prevalence
The results of E. coli O157 detection from IS1 and IS2 are shown in Table 1. For IS1 daily prevalence of E. coli O157 ranged from 16·7% to 100% of heifers with a combined average of 54·2%. During IS1, all heifers were detected positive at some point during sampling but no heifers were detected to be super-shedding. For IS2, daily prevalence ranged from 40·9% to 76·2% with a combined average of 54·8%. The daily prevalence of super-shedding ranged from 0% to 9·5% with a combined average of 4·2%. Seven super-shedding events were identified from four heifers. Two heifers were detected super-shedding on one occasion during IS2, one heifer was detected super-shedding twice, and one three times.
Table 1.
Results of E. coli O157 detection during two intensive sampling periods by direct faecal culture (DFC) and immunomagnetic separation (IMS)
| Trial | Day | No. of samples | Not detected n (%) | Low shedding n (%) | High shedding n (%) | Super-shedding n (%) | Total positive n (%) |
|---|---|---|---|---|---|---|---|
| IS1 | 1 | 12 | 9 (75) | 2 (16·7) | 1 (8·3) | 0 (0) | 3 (25) |
| IS1 | 2 | 12 | 10 (83·3) | 2 (16·7) | 0 (0) | 0 (0) | 2 (16·7) |
| IS1 | 3 | 12 | 6 (50) | 5 (41·7) | 1 (8·3) | 0 (0) | 6 (50) |
| IS1 | 4 | 12 | 1 (8·3) | 9 (75) | 2 (16·7) | 0 (0) | 11 (91·7) |
| IS1 | 5 | 12 | 0 (0) | 11 (91·7) | 1 (8·3) | 0 (0) | 12 (100) |
| IS1 | 6 | 12 | 4 (33·3) | 8 (66·7) | 0 (0) | 0 (0) | 8 (66·7) |
| IS1 | 7 | 12 | 6 (50) | 6 (50) | 0 (0) | 0 (0) | 6 (50) |
| IS1 | 8 | 12 | 8 (66·7) | 3 (25) | 1 (8·3) | 0 (0) | 4 (33·3) |
| IS1 | Total | 96 | 44 (45·8) | 46 (47·9) | 6 (6·3) | 0 (0) | 52 (54·2) |
| IS2 | 1 | 21 | 9 (42·9) | 6 (28·6) | 5 (23·8) | 1 (4·8) | 12 (57·1) |
| IS2 | 2 | 21 | 5 (23·8) | 7 (33·3) | 7 (33·3) | 2 (9·5) | 16 (76·2) |
| IS2 | 3 | 21 | 10 (47·6) | 8 (38·1) | 2 (9·5) | 1 (4·8) | 11 (52·4) |
| IS2 | 4 | 21 | 7 (33·3) | 10 (47·6) | 4 (19) | 0 (0) | 14 (66·7) |
| IS2 | 5 | 21 | 13 (61·9) | 4 (19) | 3 (14·3) | 1 (4·8) | 8 (38·1) |
| IS2 | 6 | 21 | 10 (47·6) | 7 (33·3) | 3 (14·3) | 1 (4·8) | 11 (52·4) |
| IS2 | 7 | 21 | 11 (52·4) | 5 (23·8) | 5 (23·8) | 0 (0) | 10 (47·6) |
| IS2 | 8 | 21 | 11 (52·4) | 7 (33·3) | 2 (9·5) | 1 (4·8) | 10 (47·6) |
| IS2 | Total | 168 | 76 (45·2) | 54 (32·1) | 31 (18·5) | 7 (4·2) | 92 (54·8) |
From enumeration by DFC, samples were classed as high shedding (⩾100 c.f.u./g) or super-shedding (⩾104 c.f.u./g). Low shedding (<100 c.f.u./g) was detected by IMS only.
The mean number of times a heifer included in both studies (n = 12, 16 sampling points) was detected positive was 8·2 [95% confidence interval (CI) 6·1–10·3] from 16 sampling points. The mean number of times a heifer included in IS2 only (n = 9) was detected positive was 5·1 (95% CI 2·9–7·3) from eight sampling points.
Prevalence correlations and patterns
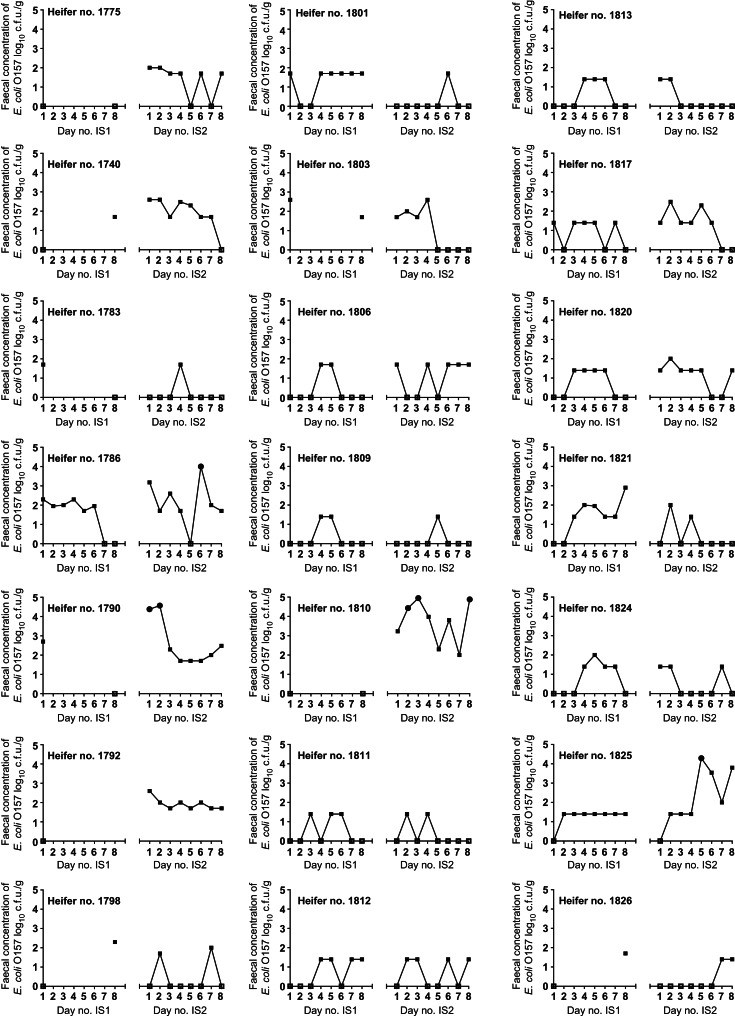
The total number of samples obtained from the two studies was 264. Data from the early points of the studies could not be analysed with respect to previous shedding as this data was not available. The numbers included in individual modelling were therefore 231 samples to 1 day prior, 198 samples to 2 days prior, 165 samples to 3 days prior, and 132 samples to 4 days prior. The shedding levels of E. coli O157 detected from individual heifers over the studies are shown in Figure 1. Correlations between the number of heifers positive (from which E. coli O157 was detected by any test) at a sampling point and the numbers positive in the preceding days were not consistent (Table 2). Correlations between the number of positives and the number of high-shedding events prior to sampling, and vice versa, are shown in Tables 3 and 4, respectively. These results were variable. Generally, moderate to strong positive correlations (0·6–0·9) were observed between the number of heifers shedding ⩾100 c.f.u./g the preceding one or two days and the number detected positive at the sampling point. No associations or patterns were apparent between the number of heifers detected shedding ⩾100 c.f.u./g and the number positive in the preceding days. A strong negative association (–0·84) was observed between DFC positive animals (shedding >100 c.f.u./g) and the number of heifers detected positive for E. coli O157 3 days prior during IS1, but a strong positive correlation (0·95) was observed for the same analysis during IS2 and for 5 days prior (0·94). A negative correlation was observed 4 days prior (−0·67).
Fig. 1.
Daily shedding (log10 c.f.u./g) of E. coli O157 by each heifer over the intensive study periods (IS1 and IS2). Samples from which E. coli O157 was not detected are represented by open squares (□). Super-shedding events are represented by circles (●). Detection at between <1 c.f.u./g and >104 c.f.u./g is represented by solid squares (■).
Table 2.
Pearson's correlations of numbers positive for E. coli O157 by any test at a given sampling point to numbers detected positive in previous days
| Number of days prior to sampling point | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Sampling period 1 | 0·589 | −0·390 | −0·981 | −0·796 | −0·720 |
| Sampling period 2 | −0·094 | 0·849 | −0·317 | 0·736 | −0·327 |
Correlation coefficients >0·5 appear in bold font.
Table 3.
Pearson's correlations between the number of positives detected and the number positive by direct faecal culture (DFC) and super-shedding (SS) in the days prior to sampling
| Time-frame | Test level | Number of days prior to sampling point | |||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | ||
| Sampling period 1 | DFC | 0·646 | 0·602 | −0·206 | −0·872 | −0·683 | * |
| Sampling period 2 | DFC | 0·646 | 0·048 | 0·932 | −0·203 | 0·462 | 0·000 |
| Sampling period 2 | SS | 0·347 | 0·344 | 0·550 | −0·183 | 0·500 | −0·500 |
Correlation coefficients >0·5 are shown in bold font.
Insufficient data.
Table 4.
Direct faecal culture (DFC) positive or super-shedding (SS) detected correlated to the number of positives in the days prior to sampling (Pearson's correlation)
| Time-frame | Test level | Number of days prior to sampling point | |||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | ||
| Sampling period 1 | DFC | 0·646 | 0·138 | −0·713 | −0·841 | −0·303 | 0·693 |
| Sampling period 2 | DFC | 0·646 | −0·010 | −0·270 | 0·951 | −0·667 | 0·945 |
| Sampling period 2 | SS | 0·347 | 0·186 | −0·104 | −0·203 | 0·676 | −0·982 |
Correlation coefficients >0·5 appear in bold font.
From the combined dataset, logistic regression demonstrated a significant association between shedding 1 day prior and current shedding (Table 5). Models (general linear mixed model) could not be fitted to the combined trial data adjusting for trial, probably due to substantial differences in results between the trials.
Table 5.
Generalized linear mixed model of the association between shedding (detected positive by any test on a given sampling day) and shedding in the days prior from the same heifer. Heifer was included in the model as a random effect
| Time-frame | Parameter* | Cases† (n) | OR | 95% CI | P | Intra-class correlation |
|---|---|---|---|---|---|---|
| Day -1 | n.d. | 35 | 1 | |||
| Positive | 94 | 3·308 | 1·851–5·911 | <0·001 | 0·169 | |
| Day -2 | n.d. | 39 | 1 | |||
| Positive | 72 | 1·252 | 0·674–2·326 | 0·474 | 0·255 | |
| Day -3 | n.d. | 40 | 1 | |||
| Positive | 54 | 1·317 | 0·674–2·577 | 0·418 | 0·214 | |
| Day -4 | n.d. | 33 | 1 | |||
| Positive | 36 | 2·108 | 0·971–4·579 | 0·059 | 0·233 |
OR, Odds ratio; CI, confidence interval, n.d., not detected as shedding in prior days (reference level).
Parameter is based on detection by any test in the days prior to detection.
Cases were defined as positive on the sampling day by any test (<1 c.f.u./g).
Detection by RAMS IMS
Throughout the duration of the studies, a total of 144 (54·5%) samples were detected positive by any test. Of these, 56 (38·9%) were detected positive from RAMS IMS including 40 (27·8%) detected positive from both RAMS and faecal IMS. Therefore a total of 16 samples were identified positive by RAMS IMS alone.
DISCUSSION
In both study periods, every heifer was detected positive for E. coli O157 on at least one occasion. Over the course of IS1 the number of positive heifers increased with each consecutive sampling day, peaking at day 5 with all (n = 12) heifers positive, then reducing again over the following 3 days. No obvious pattern was observed during IS2.
Regression analysis indicated a heifer was 3·3 times more likely to be detected as positive (by any test) if it had been positive the previous day, compared to if it was not detected positive the previous day. No significant association between shedding of E. coli O157 and the shedding status 2 days prior to sampling was found. The lack of associations identified 2 days and beyond may be attributable to a lack of power in the analysis due to decreasing numbers in the datasets. This supports the hypothesis that shedding can be a short-term phenomenon and very dynamic. Correlation between the number of heifers detected positive and those shedding at high levels in the preceding days, and vice versa, showed variable results between the two sampling periods. No particular patterns or trends were found in this analysis. This may be due to the small number of samples analysed or may be indicative of the random nature of shedding.
Longitudinal studies have previously estimated that cattle shed E. coli O157 for up to a month. Short-term variations as observed in data from the current intensive studies indicate that sampling interval will affect such results, and provides little indication of events between monthly or weekly sampling. This is supported by the variations in point (daily) prevalence observed over each week (16·7–100% and 38·1–76·2% for IS1 and IS2, respectively). Weekly sampling intervals may provide a guide to shedding levels, but since this can change on a daily basis more frequent sampling is required to provide information on shedding dynamics. This highlights the issues associated with cross-sectional studies in which only a snapshot of a very dynamic process is obtained.
Various patterns of E. coli O157 shedding were observed from individuals during the sampling periods. Previous studies have used various terms to describe results and relate them to mechanisms, and the majority of studies have observed variable results. Some distinctions in the definition of patterns can be observed; for example the focus may be the duration of shedding [22], or the repeatability of results [23]. We have clarified these terms to describe all potential shedding patterns in terms of observed results.
‘Persistent’ shedders consistently excreting E. coli O157 at low levels have been associated with the potential for high environmental contamination [23]. We defined ‘consistent’, as heifers which began or ceased shedding during the week or remained positive throughout the week. We identified three heifers (ID nos. 1790, 1792, 1810) which were consistently positive during IS2 and had tested positive for 3 weeks prior to the start of IS2. These heifers had not been included in IS1 having been detected positive only by IMS on limited occasions prior to the start of IS1. Each of these heifers was detected as super-shedding in the 2 weeks of sampling (during IS2 or in the weekly sampling preceding it). This may indicate colonization but without ongoing sampling to identify persistent shedding this is purely speculation. Longer term sampling, such as daily sampling over at least a month, would be required to clarify the distinction between consistent and persistent shedding. The duration of persistent shedding has been investigated [22]; including following experimental infection (~400 c.f.u. administered orally) in which calves consistently shed for ~70 days, and shed intermittently after that [24]. Persistence has been associated with colonization [17, 25, 26]. Cobbold et al. [18] described an association between RAJ colonization and super-shedding, including at least four consecutive RAMS samples, in the definition of a super-shedder.
Transient shedding has been used to describe the scenario in which cattle are exposed to E. coli O157 and passively shed until it is cleared from the intestinal system, generally estimated to take <1 month [15, 21, 27, 28]. We defined transient shedding as consecutive positive samples identified between samples from which E coli O157 was not detected. This specifies a short-term but consistent pattern of shedding, as demonstrated by heifer ID nos. 1820 and 1824. Smith et al. [29] identified 92 cattle (aged <24 months) positive on more than one occasion during a longitudinal study sampling monthly. Of these, 58 showed a transient pattern with consecutive samples positive. Many studies describing transient shedding do also acknowledge that shedding is often intermittent [21, 27, 28].
Sporadic, variable or intermittent shedding has been noted previously following natural and experimental infection [10, 14, 21, 28, 30]. We defined intermittent shedding as one or more samples not detected positive between positive samples. Cray & Moon [28] attributed the intermittent ‘negative’ samples to failure of their enrichment culture detection method to identify low levels of shedding. Wray et al. [30] infer that actual shedding is intermittent given the sensitivity of their detection method using IMS. Given the number of studies which have identified intermittent shedding [9, 10, 12, 14, 15, 21, 28, 30] and the increased sensitivity of current detection methods, it is likely that the intermittent shedding patterns observed are related to variations in shedding levels, but are potentially exacerbated by failures in the diagnostic methods used.
Patterns of intermittent shedding have been described in detail from monthly sampling [29], including a pattern of alternating status (positive/negative) over 7 months. We identified intermittent shedding over a much shorter time-frame, consistent with the previous results of Robinson et al. [10]. We identified 14 samples as positive where the heifer was not detected as shedding the day before or the day following the positive sample, further demonstrating the sporadic, and sometimes very short-lived (e.g. heifer ID nos. 1783 and 1809) nature of shedding.
Sanderson et al. [14] noted that following multiple experimental infections, calves would shed E. coli O157 for much shorter periods after the initial infection. This suggests a pattern of re-infection and clearance, or passive shedding for these heifers whereby ingested E. coli O157 pass through the gastrointestinal tract without adhering to or colonizing intestinal or RAJ mucosa. In a pasture-based system with lower stocking densities than in housed systems, direct animal contact and direct ingestion of faecal matter is less likely. Therefore E. coli O157 survival within the environment would be necessary to maintain the cycle and the degree of environmental contamination has been shown to be associated with transmission [31]. That our results showed shorter duration than previous studies of housed animals may also indicate lower dose rates associated with environmental contamination in an extensive grazing system.
The first intensive sampling period did not identify a super-shedding heifer. Given the low number of heifers sampled and the low occurrence of super-shedding, this is to be expected. Super-shedding has been observed previously at 0·6–0·7% at slaughter in Scotland [8, 32, 33] The heifers were selected based on previous super-shedding and high-shedding occurrences, therefore this result may suggest that super-shedding is not associated with specific animals, and previous high-shedding incidences are not an indicator that an animal will shed at high levels again.
Seven super-shedding events were detected during the second intensive sampling period and two heifers were detected super-shedding on multiple occasions. The two heifers detected super-shedding on multiple occasions were included in the study due to super-shedding prior to (ID no. 1810) or at the start of (ID no. 1790) IS2. Neither heifer had demonstrated high-level or recurrent shedding prior to this and had not been included in IS1. These observations support the observation that long-term colonization and high-level or super-shedding may not necessarily be related.
This also suggests that colonization of the RAJ and detection by RAMS IMS is not necessarily a pre-cursor to super-shedding as has been previously proposed [18, 19]. This is further supported by the low numbers of samples detected positive by RAMS IMS only and that only three heifers were detected by RAMS IMS only on more than one occasion (data not shown).
The two heifers detected as super-shedding on one occasion each during IS2 (ID nos. 1786 and 1825) had both been sporadically positive throughout the longitudinal study, although both were detected positive at the majority of sampling points during the intensive sampling periods. This does not support a hypothesis of colonization and persistent shedding being associated with super-shedding, or may be indicative of colonization also being associated with intermittent shedding. A recent study by Munns et al. [9] also identified super-shedding to be sporadic and short-lived. This study did not have background shedding information on the steers tested beyond the identification of super-shedding 4 days prior to enrolment in the study but supports our observations that super-shedding is sporadic and not necessarily associated with specific animals.
CONCLUSIONS
Three patterns of E. coli O157 shedding were identified in this study; transient, intermittent and consistent. The most frequently observed was intermittent shedding and it is likely that an extended sampling period may have demonstrated that transient shedders would have recurred as intermittent shedders. Intermittent shedding is likely to be associated with environmental exposure and passive excretion; variations in intermittent exposure may be due to variations in environmental contamination. The highly variable nature of shedding and the sporadic manner in which super-shedding occurs provide little potential for predicting prevalence and super-shedding. Shedding by a heifer was observed to be associated with it shedding on the previous day. This supports the conclusion that shedding may be highly variable and dynamic within a cohort over a short time-frame. Super-shedding was also observed to be highly dynamic and short-lived, and therefore may not be as important in on-farm dynamics as previously suspected. Factors affecting shedding may therefore be difficult to identify due to this variability.
ACKNOWLEDGEMENTS
The authors acknowledge support from The University of Sydney, in particular technical and farm staff. Funding for this study was provided by Meat & Livestock Australia Ltd, project number A.MFS.0257.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Ferens WA, Hovde CJ. Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathogens and Disease; 8: 465–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renter DG, Sargeant JM. Enterohemorrhagic Escherichia coli O157: epidemiology and ecology in bovine production environments. Animal Health Research Reviews 2002; 3: 83–94. [PubMed] [Google Scholar]
- 3.Meyer-Broseta S, et al. Review of epidemiological surveys on the prevalence of contamination of healthy cattle with Escherichia coli serogroup O157:H7. International Journal of Hygiene and Environmental Health 2001; 203: 347–361. [DOI] [PubMed] [Google Scholar]
- 4.Chase-Topping M, et al. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nature Reviews Microbiology 2008; 6: 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews L, et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proceedings of the National Academy of Sciences USA 2006; 103: 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthews L, et al. Super-shedding cattle and the transmission dynamics of Escherichia coli O157. Epidemiology and Infection 2006; 134: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cernicchiaro N, et al. Assessment of diagnostic tools for identifying cattle shedding and super-shedding Escherichia coli O157:H7 in a longitudinal study of naturally infected feedlot steers in Ohio. Foodborne Pathogens and Disease 2011; 8: 239–248. [DOI] [PubMed] [Google Scholar]
- 8.Omisakin F, et al. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Applied and Environmental Microbiology 2003; 69: 2444–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munns KD, et al. Are super-shedder feedlot cattle really super? Foodborne Pathogens and Disease 2014; 11. [DOI] [PubMed] [Google Scholar]
- 10.Robinson SE, et al. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. Journal of Applied Microbiology 2004; 97: 1045–1053. [DOI] [PubMed] [Google Scholar]
- 11.Widiasih DA, et al. Duration and magnitude of faecal shedding of Shiga toxin-producing Escherichia coli from naturally infected cattle. Epidemiology and Infection 2003; 132: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shere JA, Bartlett KJ, Kaspar CW. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Applied and Environmental Microbiology 1998; 64: 1390–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonsson ME, et al. Experimental infection in calves with a specific subtype of verocytotoxin-producing Escherichia coli O157:H7 of bovine origin. Acta Veterinaria Scandinavica 2009; 51: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanderson MW, et al. Fecal Escherichia coli O157:H7 shedding patterns of orally inoculated calves. Veterinary Microbiology 1999; 69: 199–205. [DOI] [PubMed] [Google Scholar]
- 15.Besser TE, et al. Duration of detection of fecal excretion of Escherichia coli O157:H7 in cattle. Journal of Infectious Diseases 1997; 175: 726–729. [DOI] [PubMed] [Google Scholar]
- 16.Lim JY, et al. Escherichia coli O157:H7 colonization at the rectoanal junction of long-duration culture-positive cattle. Applied and Environmental Microbiology 2007; 73: 1380–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naylor SW, et al. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infection and Immunity 2003; 71: 1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cobbold RN, et al. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Applied and Environmental Microbiology 2007; 73: 1563–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice DH, et al. Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. Journal of Clinical Microbiology 2003; 41: 4924–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock DD, et al. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiology and Infection 1997; 118: 193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mechie SC, Chapman PA, Siddons CA. A fifteen month study of Escherichia coli O157:H7 in a dairy herd. Epidemiology and Infection 1997; 118: 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grauke LJ, et al. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Applied and Environmental Microbiology 2002; 68: 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson BA, et al. Escherichia coli O157:H7 strains that persist in feedlot cattle are genetically related and demonstrate an enhanced ability to adhere to intestinal epithelial cells. Applied and Environmental Microbiology 2009; 75: 5927–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Besser TE, et al. Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiology and Infection 2001; 127: 555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baines D, Lee B, McAllister T. Heterogeneity in enterohemorrhagic Escherichia coli O157:H7 fecal shedding in cattle is related to Escherichia coli O157:H7 colonization of the small and large intestine. Canadian Journal of Microbiology 2008; 54: 984–995. [DOI] [PubMed] [Google Scholar]
- 26.Cornick NA, et al. Persistent colonization of sheep by Escherichia coli O157:H7 and other E. coli pathotypes. Applied and Environmental Microbiology 2000; 66: 4926–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown CA, et al. Experimental Escherichia coli O157:H7 carriage in calves. Applied and Environmental Microbiology 1997; 63: 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cray WC Jr., Moon HW. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Applied and Environmental Microbiology 1995; 61: 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith RP, Paiba GA, Ellis-Iversen J. Longitudinal study to investigate VTEC O157 shedding patterns in young cattle. Research in Veterinary Science 2010; 88: 411–414. [DOI] [PubMed] [Google Scholar]
- 30.Wray C, et al. Natural and experimental infection of normal cattle with Escherichia coli O157. Veterinary Record 2000; 147: 65–68. [DOI] [PubMed] [Google Scholar]
- 31.Gautam R, et al. Transmission of Escherichia coli O157 in cattle is influenced by the level of environmental contamination. Epidemiology and Infection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogden ID, MacRae M, Strachan NJC. Is the prevalence and shedding concentrations of E. coli O157 in beef cattle in Scotland seasonal? FEMS Microbiology Letters 2004; 233: 297–300. [DOI] [PubMed] [Google Scholar]
- 33.Low JC, et al. Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. Applied and Environmental Microbiology 2005; 71: 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]