SUMMARY
It is important to assess the suitability of sentinel sites for human disease; however, there have been few publications documenting the process of formal evaluation. We describe an approach to examining the representativeness of a single sentinel site employed for campylobacteriosis surveillance and source attribution, utilizing a selection of data sources and statistical comparisons of demographic, epidemiological and pathogen genotyping data across selected regions of New Zealand. Our findings showed that while this region captured the national variability in many variables, for example by containing sizable urban and rural populations, the relative frequency of these features did vary from other regions of New Zealand. We discuss the value of choosing a sentinel site that represents the national distribution of key variables, compared to a site that captures the broad features of the wider population, but provides greater power for the monitoring of sub-populations.
Key words: Campylobacter, infectious disease, molecular epidemiology, surveillance
INTRODUCTION
Data sourced from surveillance systems can help to determine priorities for effective disease control and prevention strategies [1]. Surveillance systems may have different goals and can be classified as either control-focused, aiming to identify outbreaks of disease and implement interventions, or strategy focused [2, 3]. Strategy-focused surveillance can provide data valuable for hypothesis-driven research, addressing questions related to: source attribution; risk factors for disease; spatial epidemiology; temporal patterns and trends; and the effect of interventions – thereby providing information for developing policy [3–5]. Thus, surveillance systems need to be fit for purpose with the importance of evaluating surveillance systems increasing [1].
The development of a sentinel site surveillance system for campylobacteriosis in New Zealand allowed strategy-focussed, targeted, intensive surveillance, in combination with molecular epidemiology, to answer a number of critical questions regarding source attribution and potential control measures for the disease [6–8]. The rationale for choosing the Manawatu region of New Zealand has been discussed previously [9]. Briefly, this region was selected based on the number of inhabitants, a population structure that captured features of a wider population, including both urban and rural areas, and the availability of a large number of cases with high-quality epidemiological data. This allowed hypothesis-driven research, providing evidence for interventions and policy development. Numerous studies conducted in the sentinel site region provided evidence that poultry meat was a significant source of human cases [6, 10–12]. These findings led to the implementation of a risk management strategy by the then New Zealand Food Safety Authority (NZFSA) in 2008 [13], which resulted in a drop in the number of notified campylobacteriosis cases in New Zealand [14, 15]. Assessments of the interventions provided an evaluation of the sentinel site in terms of usefulness (impact) and validity of data [14].
Given the outcome of the poultry interventions [13] and changes in population structure and livestock densities nationally, it was considered important to examine whether the Manawatu region remained sufficiently representative of the rest of New Zealand, and suitable as a sentinel site for campylobacteriosis surveillance in the post-intervention period.
The aim of this study was to evaluate the Manawatu region of New Zealand as a sentinel surveillance site for campylobacteriosis. The criteria for evaluation considered the ‘representativeness’ of the sentinel site, the completeness of data collected, and the purpose of the surveillance activities. The representativeness of a surveillance system is concerned with how well the features of population of interest are reflected in the surveillance data collected [1]. The system provides integrated surveillance across human, animal and environmental populations. To address this, we compared the demography of the human population, the distribution of factors that have been shown to be associated previously with the incidence and source attribution of campylobacteriosis, the epidemiology of campylobacteriosis, and the genotype distributions in human cases and other sources in the Manawatu region with other key regions in New Zealand.
METHODS
Regions
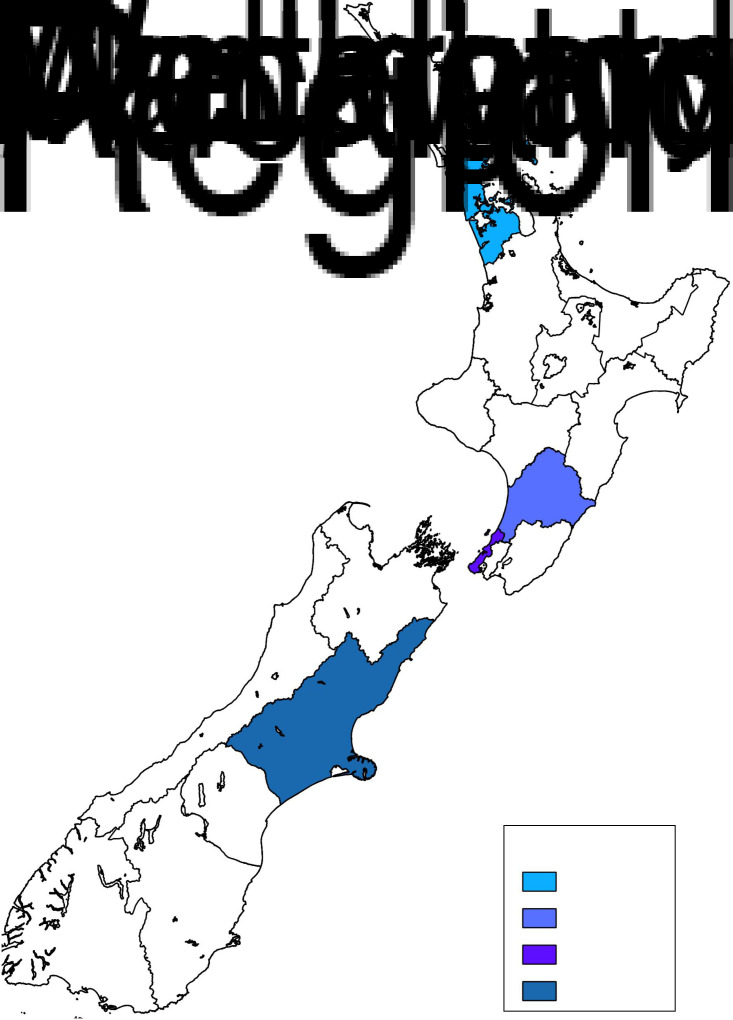
The representativeness of the Manawatu region [MidCentral District Health Board (DHB) including Otaki ward] was compared to other key regions in New Zealand that included: Auckland (Auckland, Waitemata and Counties Manukau DHBs combined) and Wellington (Capital and Coast DHB) in the North Island of New Zealand, and Canterbury (Canterbury DHB) in the South Island of New Zealand (Fig. 1).
Fig. 1.
A map of New Zealand with the Manawatu, Auckland, Wellington and Canterbury regions highlighted.
Data sources
To evaluate representativeness, a number of data sources were used to allow comparison of the human, livestock and genotype populations captured through the sentinel surveillance site.
Demographic
Demographic data were obtained from the 2006 Census for New Zealand (due to postponement of 2011 Census). The census data retrieved included: the usual resident population count and counts by age (in 5-year increments, from 0–4 to ⩾65 years), gender, and ethnicity. Ethnicity was provided in the Statistics New Zealand standard groups (Asian; European; Māori; Middle Eastern, Latin American and African; Pacific Peoples; Other), of which more than one group could be selected. Data on urban/rural profile and Social Deprivation index (SDI) score were obtained at the meshblock level, the smallest geographical unit of classification provided by Statistics New Zealand. SDI score is based on a combination of nine census variables to provide an ordinal scale that is coded from 1 to 10, with 1 representing the least and 10 representing the most deprived areas [16].
Livestock density and poultry supplies
Agricultural and land use data, including farm size and number of livestock, for the years 2005, 2008 and 2012 were obtained from AgriBase™ (AsureQuality, NZ) The farms' spatial location was linked to meshblock data using the geographical information system Quantum GIS v. 1.8.0 (http://qgis.org/downloads/). Poultry supply data were made available for the years 2005–2008, by the Poultry Industry Association of New Zealand. Data included information on the volume of supply from the two largest New Zealand companies to the Manawatu, Canterbury and Auckland regions.
Human campylobacteriosis cases
Anonymized data on human campylobacteriosis cases were provided from the national notifiable disease database (Episurv) by the Institute of Environmental Science and Research (ESR) Ltd, for the years 2005–2011 inclusive. Data included information on age, gender, occupation, ethnicity, contact with animals or sick animals, meshblock code, report date, urban/rural classification and water zone. Other data provided included exposure to untreated water and whether a case was overseas during the disease's incubation period.
Genotyping data
Genotyping data were collated from isolate databases held at ESR and Massey University. Data on human, ruminant and poultry Campylobacter isolates were available from 2000 to 2012 for the Manawatu and Canterbury regions. Primary human cases infected with Campylobacter jejuni that were resident in the Manawatu and Canterbury regions were typed using the seven-gene multilocus sequence typing scheme described by Dingle et al. [17].
Data analysis
Demography of the human population
The data on the three urban and four rural profile categories, were combined into a binary variable coded as urban (main urban area, independent urban area and satellite urban area) and rural (highly rural/remote area, rural area with high urban influence, rural area with moderate urban influence and rural area with low urban influence). Data classed as being outside of the urban/rural profile were excluded. Population counts by age, gender, ethnicity, urban/rural profile and SDI were summarized as counts and percentages for Auckland, Manawatu, Wellington and Canterbury regions. χ2 tests were used to compare the demographic variables by region; analyses were conducted in Stata v. 12 (Statacorp LP, USA) and the level of statistical significance was P < 0·01.
Livestock density and poultry supplies
The number of beef cattle, dairy cattle, sheep and total livestock were summarized at the meshblock level and densities created for each meshblock based on the number of livestock and the area (hectares) of the meshblock (using 2006 meshblock codes). Ruminant and poultry densities were created for 2005, 2008 and 2012. Quantum GIS (v. 1·8·0) was used to map the ruminant and poultry density, separately, for each year investigated.
Total livestock density was categorized into three groups based on the median density value of total livestock in 2012 (none, zero livestock; low, <median value; high, ⩾median). To compare the density of livestock across the regions being investigated these data were combined with the human case notification data stratified by region.
Human campylobacteriosis cases
Data were summarized as counts and percentages stratified by the Auckland, Manawatu, Wellington and Canterbury regions; a region coding for the rest of New Zealand was created. Cases that were recorded as being overseas during the incubation period were excluded from the analysis. Not all cases had complete data for each of the variables provided in the data extract; the total number of cases may differ for each variable investigated. Only regions with >50% completed data (excluding truly missing data) were included in the analysis. A previous report [12] had highlighted the difficulty of establishing whether answers marked as ‘unknown’ were truly unknown to the case or whether the question was not asked or the answer not recorded. Therefore, the ‘unknown’ data were included in the summaries and, where appropriate, some analyses were performed only on data known to be given by the case (cases having contact with farm animals, sick animals and drinking untreated water).
Age was categorized into 10-year age bands and ethnicity was collapsed into five groups due to low number of cases in the Middle Eastern, Latin American and African ethnic groups. χ2 tests were used to compare the demographic variables by region; the level of statistical significance was P < 0·01. Multiple correspondence analysis (MCA) was used to visualize the epidemiological variables of cases projected onto a two-dimensional plot. Multiple correspondence analysis is an exploratory technique used to describe how strongly and in which way variables are interrelated, based on a data matrix [18]. To identify if there were any changes in the demographics of cases post-intervention, a binary variable was created to code whether a case had occurred in the pre- (2005–2007) or post- (2008–2011) intervention period. To identify if the sentinel region was representative of cases in other regions in the post-intervention period, when the profile of cases may have changed, χ2 tests were used to compare the proportion of cases in the pre- and post-intervention periods, stratified by gender, ethnicity, and urban and rural profile. Denominator data for rates per 1000 population were obtained from the 2006 Census. Case rates per 1000 population and 95% confidence intervals (CIs) were determined for each region of interest, and for variables of interest stratified by region and time (pre- or post-intervention) period.
Genotyping data
The distribution of multilocus sequence types (STs) by region and source was summarized using counts and percentages. Diversity indices such as Shannon index and Simpson index were calculated using PAST v. 2.17 (http://folk.uio.no/ohammer/past/index_old.html) and rarefaction curves were used to compare the number of unique STs as a function of the number of individuals sampled, for C. jejuni human and poultry isolates, respectively, in the Manawatu and Canterbury regions. A Venn diagram was created using Venny [19] to identify the number of C. jejuni STs, for both human and poultry isolates, which were shared between the Manawatu and Canterbury regions.
Permutational multivariate analysis of variance (PERMANOVA) was used to look at the variation in STs by region, based on a distance matrix of the allelic profiles, and was calculated using PERMANOVA+ (v. 1.0.4) in Primer 6, v. 6.1.14 (PRIMER-E Ltd). A second PERMANOVA was run with ST474 (the prevalent ST in Manawatu) excluded from the data to determine how much of the explained variation between the two regions was explained by the distribution of this dominant genotype.
A proportional similarity index (PSI) was used to calculate a measure of similarity of the frequency distribution of STs between source and regions. A PSI of 1 indicates the two populations are identical, whereas a PSI of 0 indicates that the two populations have no STs in common [6]. Based on a matrix of 1 – PSI distances a Neighbour-Net tree was produced using SplitsTree4 v. 4.11.3 (splitstree4.sharewarejunction.com/), based on region and source (ruminant, poultry, human). Multidimensional scaling bubble plots were generated (Primer 6, v. 6.1.14) to show the frequency of STs by human, ruminant and poultry sources, between the two regions. The size of the bubble indicates the frequency of the ST for that source and region.
RESULTS
Demography of human population
The total ‘usually resident population’ recorded in the 2006 Census for Manawatu, Auckland, Wellington and Canterbury regions was 159807, 1319349, 265695 and 466 404, respectively. The total population for each region stratified by the demographic variables of interest are shown in Table 1. There were a number of significant differences between regions for age, ethnicity, urban/rural profile and SDI. Manawatu and Canterbury regions had a higher percentage of people aged ⩾65 years, a higher percentage of people domiciled in rural areas and a higher percentage of people with SDI scores of 7–9 compared to the other regions.
Table 1.
Number and percentage of people in the Manawatu, Auckland, Wellington and Canterbury regions of New Zealand as reported in the 2006 census, stratified by gender, age, ethnicity, urban/rural profile and Social Deprivation index
| Manawatu | Auckland | Wellington | Canterbury | |||
|---|---|---|---|---|---|---|
| Variable | Level | n (%) | n (%) | n (%) | n (%) | P value |
| Gender | 0·23 | |||||
| Female | 82 314 (51·5) | 676 677 (51·3) | 238 953 (51·2) | 238 953 (51·2) | ||
| Male | 77 490 (48·5) | 642 669 (48·7) | 227 454 (48·7) | 227 454 (48·8) | ||
| Total | 159 804 | 1 319 346 | 466 407 | 466 407 | ||
| Age (years) | <0·001 | |||||
| 0–4 | 10 515 (6·6) | 95 205 (7·2) | 17 490 (6·5) | 29 403 (6·3) | ||
| 5–9 | 11 391 (7·1) | 96 582 (7·3) | 16 896 (6·3) | 30 078 (6·5) | ||
| 10–14 | 12 204 (7·6) | 100 968 (7·7) | 17 358 (6·5) | 32 208 (6·9) | ||
| 15–19 | 12 864 (8·1) | 100 389 (7·6) | 19 377 (7·2) | 33 918 (7·3) | ||
| 20–24 | 11 319 (7·1) | 99 693 (7·6) | 22 908 (8·6) | 32 595 (7·0) | ||
| 25–29 | 8847 (5·5) | 90 507 (6·9) | 20 196 (7·6) | 27 627 (5·9) | ||
| 30–34 | 9795 (6·1) | 99 681 (7·6) | 21 687 (8·1) | 32 250 (6·9) | ||
| 35–39 | 10 677 (6·7) | 106 446 (8·1) | 21 909 (8·2) | 35 640 (7·6) | ||
| 40–44 | 11 811 (7·4) | 106 725 (8·1) | 21 687 (8·1) | 36 144 (7·8) | ||
| 45–49 | 11 235 (7·0) | 94 143 (7·1) | 18 483 (6·9) | 34 347 (7·4) | ||
| 50–54 | 9834 (6·2) | 77 922 (5·9) | 15 564 (5·8) | 30 087 (6·5) | ||
| 55–59 | 9270 (5·8) | 69 252 (5·3) | 14 181 (5·3) | 28 305 (6·1) | ||
| 60–64 | 7581 (4·7) | 51 960 (3·9) | 10 407 (3·9) | 21 117 (4·5) | ||
| ⩾65 | 22 455 (14·1) | 129 876 (9·8) | 27 996 (10·5) | 62 688 (13·4) | ||
| Total | 159 798 | 1 319 349 | 265 698 | 466 407 | ||
| Ethnicityb | <0·001 | |||||
| Asian | 7014 (4·0) | 234 729 (17·0) | 25 746 (9·1) | 28 368 (5·8) | ||
| European | 114 201 (65·8) | 710 079 (51·3) | 177 357 (62·7) | 349 434 (71·9) | ||
| Māori | 26 748 (15·4) | 139 974 (10·1) | 26 457 (9·3) | 33 414 (6·9) | ||
| MELAAa | 963 (0·6) | 18 588 (1·3) | 3924 (1·3) | 3255 (0·7) | ||
| Pacific Peoples | 4608 (2·7) | 178 332 (12·9) | 21 942 (7·7) | 10 476 (2·2) | ||
| Other | 20 163 (11·6) | 101 667 (7·4) | 27 411 (9·6) | 61 104 (12·6) | ||
| Totalb | 173 697 | 1 383 369 | 282 837 | 486 051 | ||
| Urban/rural profile | <0·001 | |||||
| Rural | 29 067 (18·3) | 62 610 (4·7) | 2178 (0·8) | 60 330 (12·9) | ||
| Urban | 129 741 (81·7) | 1 256 658 (95·2) | 264 447 (99·2) | 406 014 (84·0) | ||
| SDI | <0·001 | |||||
| 1 | 11 904 (7·5) | 139 764 (10·6) | 51 861 (19·5) | 68 232 (14·6) | ||
| 2 | 13 560 (8·6) | 151 287 (11·5) | 30 366 (11·4) | 56 352 (12·1) | ||
| 3 | 13 116 (8·3) | 146 379 (11·1) | 28 950 (10·9) | 51 897(11·1) | ||
| 4 | 12 903 (8·1) | 129 219 (9·8) | 29 910 (11·2) | 52 533 (11·3) | ||
| 5 | 17 130 (10·8) | 121 368(9·2) | 25 683 (9·6) | 49 014 (10·5) | ||
| 6 | 18 252 (11·5) | 119 778 (9·1) | 23 706 (8·9) | 50 250 (10·8) | ||
| 7 | 18 081 (11·4) | 117 201(8·9) | 20 061 (7·5) | 40 107 (8·6) | ||
| 8 | 19 551(12·3) | 123 537 (9·4) | 17 151 (6·4) | 39 948 (8·6) | ||
| 9 | 19 404 (12·2) | 123 546 (9·4) | 14 367 (5·4) | 36 549 (7·8) | ||
| 10 | 14 667 (9·3) | 145 899 (11·1) | 23 574 (8·8) | 21 468 (4·6) |
SDI, Social Deprivation index.
Factors associated with campylobacteriosis
The density of poultry and ruminants for the years 2005, 2008 and 2012 is shown in Figure 2. Overall, there were more areas with a high ruminant density in 2005 and 2008, and a reduction in the number of meshblocks with high density of ruminants in 2012. There was little change in the areas with high or low poultry density across the 3 years. Most of the poultry supplied to Auckland, Manawatu and Canterbury regions was from company A (82%, 70% and 95%, respectively), with Manawatu having the highest percentage (30%) from company B.
Fig. 2.
(a) Ruminant density (number per hectare) and (b) poultry density (number per hectare) in each meshblock in the years 2005, 2008 and 2012. Data sourced from AgriBase™.
The epidemiology of human campylobacteriosis cases
In total, 70 394 notifications of campylobacteriosis cases were recorded in New Zealand from 2005 to 2011. Overall, 1599 (2%) cases were reported to have been overseas during the incubation period and were excluded from the analysis; 45 917 (65%) cases were reported as ‘unknown’. There was ⩽10% of missing data for each region for the variables age, gender, and urban and rural profile, and <50% of missing data for the variable ‘source of untreated water’. The high percentage of ‘unknown’ reported in some regions for the variables ‘farm’, ‘sick’ and ‘water’ is shown in Table 2.
Table 2.
Notified campylobacteriosis cases in New Zealand from 2005 to 2011 by gender, age, contact with farm or sick animals, drinking untreated water and urban/rural profile, stratified by region
| Manawatu | Auckland | Wellington | Canterbury | Rest of NZ | |
|---|---|---|---|---|---|
| Variable | n (%) | n (%) | n (%) | n (%) | n (%) |
| Gender | |||||
| Female | 801 (44·2) | 10 016 (46) | 2676 (46·2) | 3632 (44·5) | 13 437 (44·5) |
| Male | 1011 (55·8) | 11 749 (54) | 3114 (53·8) | 4513 (55·4) | 16 735 (55·4) |
| Age, years | |||||
| 0–4 | 250 (13·7) | 2366 (10·8) | 826 (6·7) | 390 (10·1) | 3852 (12·7) |
| 5–9 | 100 (5·6) | 1135 (5·1) | 350 (4·3) | 253 (4·2) | 1419 (4·6) |
| 10–19 | 234 (12·8) | 2551 (11·6) | 973 (9·4) | 551 (11·9) | 3619 (11·9) |
| 20–29 | 257 (14·1) | 3942 (17·9) | 1498 (22) | 1286 (18·3) | 4797 (15·8) |
| 30–39 | 190 (10·4) | 3061 (13·9) | 1059 (15·5) | 902 (12·9) | 3565 (11·7) |
| 40–49 | 212 (11·6) | 2851 (12·9) | 1043 (14·2) | 833 (12·7) | 3823 (12·6) |
| 50–59 | 196 (10·8) | 2493 (11·3) | 952 (12·2) | 716 (11·6) | 3540 (11·6) |
| 60–69 | 177 (9·7) | 1879 (8·5) | 727 (8·6) | 5029 (8·8) | 2936 (9·6) |
| 70–79 | 132 (7·3) | 1146 (5·2) | 513 (5·1) | 296 (6·2) | 1896 (6·2) |
| ⩾80 | 73 (4·0) | 552 (2·5) | 233 (1·9) | 111 (2·8) | 884 (2·9) |
| Farm* | |||||
| No | 893 (49·0) | 255 (1·1) | 462 (7·9) | 3725 (45·4) | 8131 (26·7) |
| Yes | 620 (34·0) | 22 (0·1) | 73 (1·2) | 1276 (15·5) | 5084 (16·7) |
| Unknown | 308 (16·9) | 22 050 (98·7) | 5312 (90·8) | 3195 (38·9) | 17 192 (56·5) |
| Sick† | |||||
| No | 1315 (72·2) | 260 (1·16) | 427 (7·3) | 4146 (50·5) | 10 734 (35·3) |
| Yes | 152 (8·3) | 2 (0·01) | 11 (0·19) | 214 (2·6) | 651(2·14) |
| Unknown | 3836 (46·8) | 22 065 (98·8) | 5409 (92·5) | 3836 (46·8) | 19 022 (62·5) |
| Water‡ | |||||
| No | 870 (47·7) | 237 (1·06) | 460 (7·8) | 4016 (49) | 8702 (28·6) |
| Yes | 534 (29·3) | 30 (0·13) | 48 (0·82) | 494 (6·0) | 2434 (8) |
| Unknown | 417 (22·9) | 22 060 (98·8) | 5339 (91·3) | 3686 (44·9) | 19 271 (63·3) |
| Profile | |||||
| Rural | 325 (19·8) | 1,162 (5·5) | 46 (0·8) | 1150 (15·2) | 5,860 (21·6) |
| Urban | 1320 (80·2) | 20 097 (94·5) | 5569 (99·2) | 6405 (84·8) | 21 238 (78·4) |
Did case have contact with farm animals?
Did case have contact with sick animals?
Did case drink untreated water?
There was a significant difference in the percentage of male and female cases by region (P = 0·004) (Table 2). The age distribution of cases differed significantly by region (P < 0·001), with the highest proportion of cases in Manawatu being aged 0–4 or 20–29 years, while Wellington and Canterbury regions had high proportions of cases aged 20–29 years (Table 2).
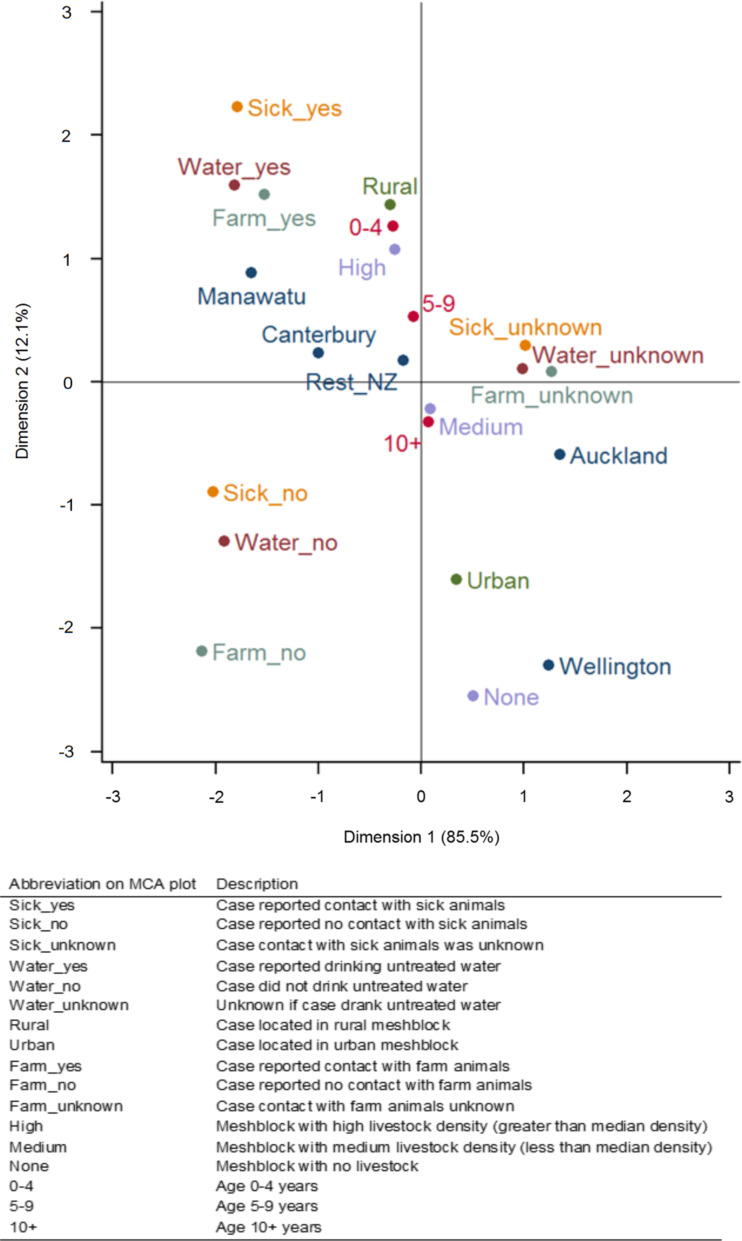
The percentage of cases reporting contact with farm animals, sick animals or exposure to untreated water differed significantly by region (P < 0·001) (Table 2). Analysis of these variables with the ‘unknown’ data excluded showed significant differences (P < 0·001) between regions. Of the cases that reported drinking untreated water, most cases in Manawatu (42%, 223), Wellington (45%, 17) and Rest of New Zealand (40%, 900) reported the source as ‘roof’, while 46% (186) of cases in Canterbury reported ‘ground’ as the origin. In Wellington and Auckland the proportion of cases residing in rural areas was lower than Manawatu, Canterbury and the rest of New Zealand (Table 2). Results of the multiple correspondence analysis are shown in Figure 3. Cases that had contact with sick animals, farm animals and drank untreated water cluster together and also cluster close to the Manawatu region, a rural location, aged 0–4 years and a meshblock with high livestock density.
Fig. 3.
Multiple correspondence analysis of epidemiological variables of notified campylobacteriosis cases occurring between 2005 and 2011.
In the post-intervention period a higher percentage of cases were male (56%) compared to female, compared to the pre-intervention period (54%, P < 0·001). Rural cases increased from 11% in the pre-intervention period to 18% in the post-intervention period (P < 0·001) and non-European cases rose post-intervention (18%) compared to pre-intervention (12%, P < 0·001).
The campylobacteriosis notification rates for both males and females in the post-intervention period were lower in Manawatu compared to Auckland and Wellington. The notification rates for males were similar in Auckland and Canterbury, and highest in Wellington, in the post-intervention period (Supplementary Fig. S1). In the post-intervention period the campylobacteriosis notification rates in rural (6·5/1000 people, 95% CI 5·6–7·5) and urban (4·6/1000 people, 95% CI 4·2–5·0) areas were lower in Manawatu compared to Auckland and Wellington (Supplementary Fig. S2), while urban notification rates in Manawatu were similar to those in Canterbury (4·9/1000 people, 95% CI 4·7–5·1).
Comparison of genotype distributions
There were 3290 fully multilocus sequence typed C. jejuni isolates available from 2000 to 2012 for analysis, of which 1635, 725 and 930 were from human, ruminant and poultry sources, respectively. Overall, 268 different C. jejuni STs were identified, of which 117 were from human isolates. Within the regions, 70 and 76 different C. jejuni STs were identified in Manawatu and Canterbury, respectively. The frequency of C. jejuni STs from human isolates differed between the Canterbury and Manawatu regions (Supplementary Fig. S3). Overall, the most common ST identified in the Manawatu region was ST474 (26%, 226/860), compared to ST45 (14%, 111/775) and ST50 (12%, 96/775) in the Canterbury region. In the post-intervention period (2008–2012), ST474 was the most common (18%, 63/348) ST identified in the Manawatu region followed by ST50 (13%, 39/348), while in Canterbury ST45 (15%, 85/580) and ST474 (11%, 64/580) were the most common STs identified.
The diversity indices indicated the C. jejuni human isolates to be slightly more diverse with greater species richness in Canterbury (Simpson index: 0·93, 95% CI 0·92–0·94; Shannon index: 3·17, 95% CI 3·07–3·25) compared to Manawatu (Simpson index: 0·90, 95% CI 0·88–0·91; Shannon index: 2·94, 95% CI 2·85–3·03). The diversity indices indicated the C. jejuni poultry isolates to be slightly more diverse with greater species richness in Manawatu (Simpson index: 0·90, 95% CI 0·89–0·91; Shannon index: 2·72, 95% CI 2·62–2·82) compared to Canterbury (Simpson index: 0·78, 95% CI 0·75–0·81; Shannon index: 2·25, 95% CI 2·11–2·38). A Venn diagram indicated 34 STs from human isolates and 17 STs from poultry isolates were shared by both Manawatu and Canterbury. PERMANOVA analysis showed that region explained 10% (P = 0·001) of the molecular variation in the C. jejuni human isolates. After removing ST474 from the analysis, 5% (P = 0·014) of the variation was accounted for by region.
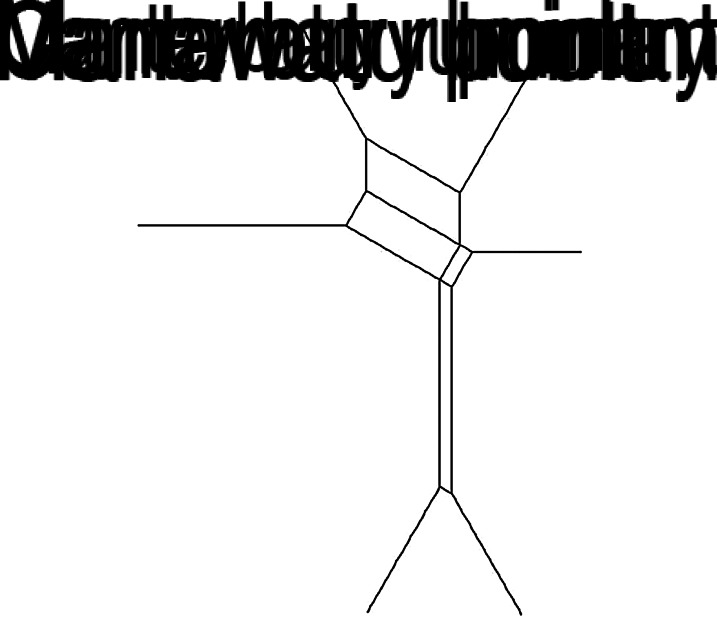
When comparing the genotype distributions using the PSI, high similarities were observed for ruminant sources (PSI 0·70, 95% CI 0·61–0·72) in Manawatu and Canterbury, while poultry sources in both regions had lower similarities (PSI 0·57, 95% CI 0·56–0·65). Both human and poultry sources in Manawatu showed high similarities (PSI 0·61, 95% CI 0·56–0·65), as did poultry sources in Manawatu and human sources in Canterbury (PSI 0·61, 95% CI 0·55–0·64). The least similarity was observed between ruminant and poultry sources in both regions (PSI 0·28 and 0·31). The Neighbour-Net tree showed evidence of variation in poultry isolates and ruminant isolates in both regions (Fig. 4). There was a split separating Manawatu human, poultry isolates and Canterbury human from Canterbury poultry isolates and the two ruminant sources.
Fig. 4.
Neighbour-Net of 1 – PSI for Campylobacter sequence types grouped by region and source.
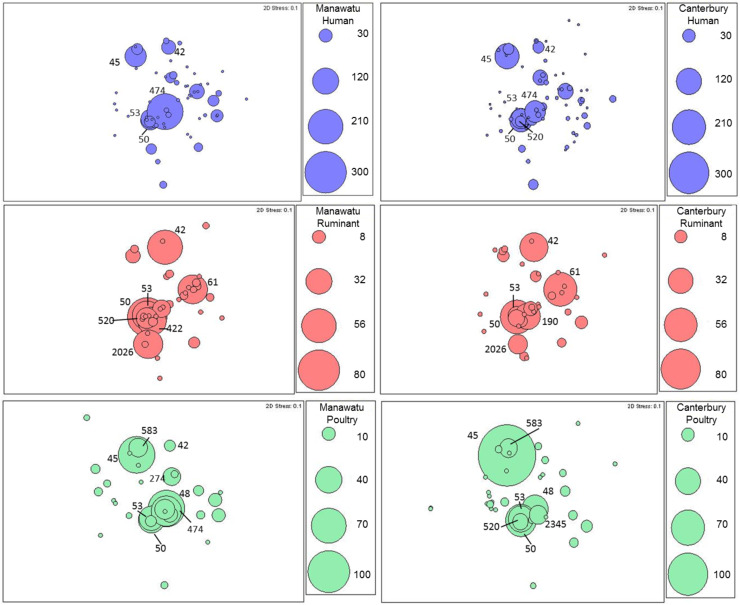
Multidimensional scaling bubble plots showed different patterns of STs by source and within source by region (Fig. 5). For human isolates, the dominant ST in the Manawatu region was ST474, followed by ST45 and ST53, while ST45 was more common in human isolates from Canterbury (Fig. 5). Different patterns were seen for poultry isolates between regions, with ST45 being more common in Canterbury compared to Manawatu, while ST45 and ST48 showed a similar frequency in Manawatu. The pattern of the common STs (ST42, ST61, ST50) from ruminant isolates was similar for both regions.
Fig. 5.
Multidimensional scaling bubble plots of sequence type frequency by region (Manawatu and Canterbury) and source (human, ruminant and poultry).
DISCUSSION
This study was designed to evaluate the suitability of a sentinel surveillance site for campylobacteriosis (Manawatu region), through the utilization of multiple data sources and statistical analysis of demographic, epidemiological and genotyping data. For ‘suitability’ we considered both the ‘representativeness’ of the sentinel site and the purpose of the surveillance activities. If the rationale of a surveillance system is strategy-focused (as with the Manawatu sentinel site) and aims to detect changes in the epidemiology of a disease in defined sub-populations it may be better to choose a site that captures a broad diversity of sub-populations with differing risk factors, than a site that has an identical distribution of risk factors to the rest of the country.
The demographic data for the regions studied indicated that there were differences in the underlying population structure; age, ethnicity and urban and rural profile varying between regions. Specifically, while most of the regions were predominantly urban areas, Manawatu and Canterbury had a similar, and higher, percentage of rural population compared to the other regions. Notification patterns and the rate of disease have been shown to vary between urban and rural areas [8, 10, 14, 20]. Therefore, if one of the aims of a sentinel site is to capture the diversity of risk factors, as with the Manawatu region, then ‘oversampling’ these sub-populations is of benefit for understanding the determinants of trends in, for example, both urban and rural populations. As such, the Manawatu region may be considered suitable for purpose. The disadvantage of this over-representation of some sub-populations is that the overall picture may be biased and less representative of the wider population as a whole. SDI is associated with the notification rates of campylobacteriosis in New Zealand, with lower rates in deprived urban areas [8]. The broad representation of sub-populations with different levels of deprivation, and similarity with other regions, demonstrated the Manawatu region was suitable for monitoring trends across the demographic range of SDI. Thus, aside from gender, and to a lesser extent SDI, the Manawatu region may not be representative of the demographic structure of populations in all the regions studied, and New Zealand as a whole. However, the Manawatu region does capture the diversity of the population in the area, through the distribution of age, ethnicity, SDI score and urban and rural profile.
While there was no marked difference in the poultry supply to the Manawatu region compared to other regions, as determined by the relative contribution of different companies (although the farms and processing plants sourced by the companies that supply each region will inevitably differ), an increase in the amount supplied by company B to the region was apparent in 2007 and 2008. The data available precluded a detailed examination of the poultry industry in each region. It is possible therefore that any dissimilarity in poultry supply pre- and post-intervention may explain differences in the Campylobacter genotypes in both poultry and human cases.
The results of the livestock data analysis showed an apparent change in the ruminant densities across New Zealand in recent years. However, while some reductions in sheep and dairy numbers were evident, care should be taken in the interpretation of these data due to voluntary reporting of livestock numbers by farmers. The MCA indicated that the density of livestock clustered with notified cases by region. Previous reports have shown the density of ruminants in rural areas to be associated with notifications and the risk of campylobacteriosis [8, 10]. Therefore, some areas within the Manawatu region may have different risks of exposure to potential environmental pathways, compared to that of the other regions.
The MCA showed the Manawatu region clustered with rural dwelling, high livestock density, 0–4 years age group, exposure to farm animals, untreated water and sick animals, highlighting the association between the region's cases and variables linked with rural residence and the risk of campylobacteriosis [8]; making it suitable for examining the epidemiology of campylobacteriosis in rural sub-populations. Furthermore, the Manawatu region had a similar age distribution as notified cases in Auckland and the rest of New Zealand, and a similar gender distribution to cases in Canterbury
A comparison of the notified cases in the pre- and post-intervention period indicated a significant difference in the percentage of cases by age, ethnicity and urban and rural profile. Previous studies [14, 15] have reported the success of the intervention in the poultry industry in New Zealand at reducing cases of campylobacteriosis by 50%. More people may have sought medical care thereby creating increased reporting of cases associated with a public awareness campaign [4, 13]. Furthermore, the intervention was successful at reducing poultry-associated cases in urban areas, with less impact on rural areas [14]. In support of this, the Manawatu region appeared to be representative of campylobacteriosis notification rates in urban areas in Auckland and Canterbury post-intervention.
Notified case data should be interpreted with caution due to the variability in reporting across different DHBs [9, 21], which may have resulted in reporting bias. Furthermore, surveillance of campylobacteriosis was enhanced in the Manawatu region from 2007, resulting in a 97% case contact rate and 96–100% completeness of notification data fields [22]. Missing data or a lack of follow-up in some regions may have resulted in under-reporting of risk factors for campylobacteriosis compared to the Manawatu region. Although the percentage of truly missing data was low (<10%), the main issue was the use of the category ‘unknown’, specifically for reporting exposure to farm or sick animals or untreated water. A previous report highlighted that ‘unknown’ could be supplied by the case or reported by the health protection officer if the question was not asked, [12, 22], which could contribute to the amount of missing data.
The results indicated the Manawatu region was likely to be representative of isolates from ruminant sources in Canterbury, while there were differences in the distribution and frequency of STs from human and poultry isolates. The Canterbury region had a slightly more diverse population in human cases with greater species richness compared to Manawatu. The PERMANOVA analysis indicated that half the variation in STs between the two regions was due to the high prevalence of ST474 in the Manawatu region [2]. While this may indicate some bias in Manawatu due to ST474, there were still differences between regions when this ST was excluded from the analysis. Therefore, it is possible that the genotype distribution of campylobacteriosis in Manawatu may differ from other regions in New Zealand. The strong association between poultry supplier and ST could explain both spatial and temporal variation in genotypes in poultry and humans [23], and this should be taken into consideration when assessing the suitability of the Manawatu region as a sentinel site.
The work presented in this study provides an evaluation of the Manawatu region as a sentinel site for campylobacteriosis in New Zealand, in terms of its representativeness and the extent to which it captured features of the population of New Zealand. Potential limitations of the data should be viewed in the context of the purpose of the Manawatu sentinel site, which was primarily for informing policy and source attribution rather than local disease control. Although the region has a mixed population of urban and rural dwellers, across all age groups, and captures variation in many demographic variables, the relative frequency of these features did vary, often markedly, from other regions and the population of New Zealand as a whole. This suggests that, if the aim is to monitor a sentinel population that is representative of the wider population, other surveillance sites may be required in order to adequately inform future control policies. If, however, the aim is to monitor a site that captures the broad features of the wider population, and enables monitoring of both large and small sub-populations in an efficient way, then the single (Manawatu) sentinel site used in this study, may be considered fit for purpose. This study has demonstrated the use of multiple statistical techniques and the integration of epidemiological and genotyping data to evaluate the representativeness of a sentinel surveillance site. For similar evaluations of other surveillance systems, it is recommended that representativeness is evaluated using data sources that capture multiple features of the populations at risk, as well as the pathogens they are exposed to, and take full advantage of an array of multivariate methods, including molecular epidemiological tools, to allow a formal assessment of any apparent differences.
ACKNOWLEDGEMENTS
The authors thank the team at ESR Ltd for providing the genotyping data for Campylobacter isolates from Christchurch and Esther Lim (ESR) for preparation and provision of EpiSurv data. We acknowledge AsureQuality and the Poultry Industry Association of New Zealand (PIANZ) for provision of data. Thanks are also due to the laboratory staff in mEpiLab in the Hopkirk Institute, and Jonathan Marshall for his statistical advice and assistance with the poultry data. Thanks goes to Tui Shadbolt and colleagues at MidCentral DHB for consultation regarding the collection of human epidemiological data in the Manawatu region.
This work was supported by the Ministry for Primary Industries.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268814003173.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Drewe JA, et al. Evaluation of animal and public health surveillance systems: a systematic review. Epidemiology & Infection 2012; 140: 575–590. [DOI] [PubMed] [Google Scholar]
- 2.Muellner P, et al. Molecular-based surveillance of campylobacteriosis in New Zealand – from source attribution to genomic epidemiology. Eurosurveillance 2013; 18: pii = 20365. [PubMed] [Google Scholar]
- 3.Baker M, Easther S, Wilson N. A surveillance sector review applied to infectious diseases at a country level. BMC Public Health 2010; 10: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. The global view of campylobacteriosis: report of expert consultation, Utrecht, Netherlands, 9 to 11 July 2012. World Health Organization, Geneva, Switzerland (http://apps.who.int/iris/handle/10665/80751).
- 5.Campbell DM, van der Logt P, Hathaway S. Surveillance for action – managing foodborne Campylobacter in New Zealand. Western Pacific Surveillance and Response Journal 2012; 3: 7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muellner P, et al. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infection, Genetics and Evolution 2009; 9: 1311–1319. [DOI] [PubMed] [Google Scholar]
- 7.Muellner P, et al. Molecular and spatial epidemiology of human campylobacteriosis: source association and genotype-related risk factors. Epidemiology & Infection 2010; 138: 1372–1383. [DOI] [PubMed] [Google Scholar]
- 8.Spencer S, et al. The spatial and temporal determinants of campylobacteriosis notifications in New Zealand, 2001–2007. Epidemiology and Infection 2012; 140: 1663–1677. [DOI] [PubMed] [Google Scholar]
- 9.French NP, Marshall J. A review of work carried out to determine the source of human campylobacteriosis. Technical report, Ministry of Primary Industries: MAF Agreement 11777, Schedule 1A 2012.
- 10.Muellner P, et al. Molecular and spatial epidemiology of human campylobacteriosis: source association and genotype-related risk factors. Epidemiology and Infection 2010; 138: 1372–1383. [DOI] [PubMed] [Google Scholar]
- 11.Muellner P, et al. Source attribution of food borne zoonosis in New Zealand: a modified Hald model. Risk Analysis 2009; 29: 970–984. [DOI] [PubMed] [Google Scholar]
- 12.French N. The Molecular Epidemiology and Public Health Group. Enhancing surveillance of potentially foodborne enteric diseases in New Zealand: human campylobacteriosis in the Manawatu. Final report for the New Zealand Food safety Authority for project: FDI/236/2005. p 1–56. Technical report, New Zealand Food Safety Authority (http://www.foodsafety.govt.nz/elibrary/industry/enhancing-surveillance-potentially-research-projects-2/Campy_Attribution_Manawatu.pdf). 2008.
- 13.Anon. New Zealand Food Safety Authority Campylobacter Risk Management Strategy 2008–2011. 2008. (http://www.foodsafety.govt.nz/elibrary/industry/Campylobacter_Risk-Aims_Acheive.pdf).
- 14.Muellner P, et al. Utilizing a combination of molecular and spatial tools to assess the effect of a public health intervention. Preventive Veterinary Medicine 2011; 102: 242–253. [DOI] [PubMed] [Google Scholar]
- 15.Sears A, et al. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. Emerging Infectious Diseases 2011; 17: 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmond C, Crampton P, Atkinson J. NZDep2006 Index of Deprivation. Department of Public Health, University of Otago, Wellington, 2007.
- 17.Dingle KE, et al. Multilocus sequence typing system for Campylobacter jejuni. Journal of Clinical Microbiology 2001; 39: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenacre M. Correspondence Analysis in Practice, 2nd edn. Boca Raton: Taylor and Francis Group, 2007, pp. 137–144. [Google Scholar]
- 19.Oliveros JC. VENNY. An interactive tool for comparing lists with Venn diagrams (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Accessed 14 February 2013.
- 20.Strachan NJC, et al. Attribution of Campylobacter infections in northeast Scotland to Specific sources by use of multilocus sequence typing. Journal of Infectious Diseases 2009; 199: 1205–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McBride G, et al. Campylobacter in Food and the Environment: Examining the Link with Public Health MAF Technical Paper No: 2011/61 Technical report, (http://wwwfoodsafetygovtnz/elibrary/industry/examining-link-with-public-health/campylobacter-in-food-and-the-environmentpdf), 2011.
- 22.Shadbolt T, Enhanced surveillance of potentially food borne enteric disease within a New Zealand public health service. Palmerston North: MSc thesis, Massey University; 2009. [Google Scholar]
- 23.Mullner P, et al. Molecular epidemiology of Campylobacter jejuni in a geographically isolated country with a uniquely structured poultry industry. Applied and Environmental Microbiology 2010; 76: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268814003173.
click here to view supplementary material